- 1College of Bioscience and Biotechnology, Hunan Agricultural University, Changsha, China
- 2Institute of Agricultural Product Quality Standard and Testing Research, Tibet Academy of Agricultural and Animal Husbandry Sciences, Lhasa, China
- 3College of Food Science and Technology, Hunan Agricultural University, Changsha, China
- 4State Key Laboratory of Utilization of Woody Oil Resource, Hunan Academy of Forestry, Changsha, China
The enhancement of nitrogen fixation activity of diazotrophs is essential for safe crop production. Lysine succinylation (KSuc) is widely present in eukaryotes and prokaryotes and regulates various biological process. However, knowledge of the extent of KSuc in nitrogen fixation of Azotobacter chroococcum is scarce. In this study, we found that 250 mg/l of rhamnolipid (RL) significantly increased the nitrogen fixation activity of A. chroococcum by 39%, as compared with the control. Real-time quantitative reverse transcription PCR (qRT-PCR) confirmed that RL could remarkably increase the transcript levels of nifA and nifHDK genes. In addition, a global KSuc of A. chroococcum was profiled using a 4D label-free quantitative proteomic approach. In total, 5,008 KSuc sites were identified on 1,376 succinylated proteins. Bioinformatics analysis showed that the addition of RL influence on the KSuc level, and the succinylated proteins were involved in various metabolic processes, particularly enriched in oxidative phosphorylation, tricarboxylic acid cycle (TCA) cycle, and nitrogen metabolism. Meanwhile, multiple succinylation sites on MoFe protein (NifDK) may influence nitrogenase activity. These results would provide an experimental basis for the regulation of biological nitrogen fixation with KSuc and shed new light on the mechanistic study of nitrogen fixation.
Introduction
Nitrogen has been considered as a critical factor to restrict agricultural production. Biological nitrogen fixation (BNF) is a process in which diazotrophs catalyze the reduction of atmospheric N2 to NH3 through nitrogenase enzyme systems (Bloch et al., 2020), which is the main source of nitrogen uptake for crops. Azotobacters are the previously discovered and studied free-living, obligately aerobic diazotrophs, which have been used as nitrogen fixation fertilizer for more than a century (Das, 2019). Azotobacter chroococcum was isolated from soils and the rhizosphere of various plants (Lenart, 2012; Rizvi and Khan, 2018). The species of A. chroococcum promote crop growth and fruit development through secretion of growth hormones or nitrogen fixation (Kumar et al., 2019; Song et al., 2020). Because of the relatively low nitrogen fixation efficiency of Azotobacters compared with Rhizobia, which hardly satisfies the nitrogen demand of crops, the improvement of its efficiency is crucial. The use of molecular biotechnology to construct efficient nitrogen fixation strains is an effective way to improve the efficiency of nitrogen fixation (Peralta et al., 2004; Orikasa et al., 2010; Deng et al., 2018; Wang T. et al., 2018). Besides, the use of chemical materials or reagents can also improve the nitrogen fixation efficiency of A. chroococcum. For instance, the addition of 5% perfluorodecalin and 4 μg/ml of carbon dots can effectively improve the nitrogen fixation activity of A. chroococcum (Bakulin et al., 2007; Wang H. et al., 2018). The non-ionic surfactant Alk 3 could enhance the nitrogen fixation activity of Rhizobium trtfolii in soils (Fisher et al., 1978), but there have been no studies of surfactant on the nitrogen fixation activity of A. chroococcum.
Posttranslational modifications (PTMs) are the process of reversible covalent modification on the main and side chains of amino acid residues of proteins (Wagner and Payne, 2013; Gil et al., 2017). PTMs are a key feature of bacteria that have the ability to regulate the functions of proteins. PTMs mainly include acetylation, methylation, phosphorylation, ubiquitination, succinylation, crotonylation, 2-hydroxyisobutyrylation, and benzoylation. Lysine is one of the most common targets for covalent modification among the 20 amino acids of proteins (Macek et al., 2019). Lysine succinylation (KSuc) is a novel acyl modification and has been found widely in eukaryotes and prokaryotes (Zhang et al., 2011; Xie et al., 2012; Weinert et al., 2013), which is involved in many important metabolic processes, such as the ribosomal complex, glycolysis, gluconeogenesis, tricarboxylic acid cycle (TCA) cycle, and carbon metabolism (Xu et al., 2017; Ren et al., 2018; Zhang et al., 2019). Furthermore, BNF is a relatively complex process, and apart from the direct involvement of nitrogenase enzyme systems (Mus et al., 2018), it also requires the synergistic action of several biological processes, such as respiration, hydrogenases, electron transport, carbon metabolism, transport systems, and secretion systems. Therefore, we hypothesized that KSuc may be present in diazotrophs.
In this study, we evaluated the effects of anionic surfactant sodium dodecyl sulfate (SDS), non-ionic surfactant Triton X-100 (Tr), and non-ionic biosurfactant rhamnolipid (RL) on A. chroococcum and found that RL could effectively improve the nitrogen fixation activity. Meanwhile, a 4D label-free quantitative proteomic analysis was conducted to explore the mechanism of RL enhancing nitrogen fixation activity. This study provides not only a new technical approach to improve the nitrogen fixation activity of diazotrophs but also a reference for KSuc involved in the nitrogen fixation process.
Materials and Methods
Strains, Media, and Cultivation Conditions
Azotobacter chroococcum strain HR1 was isolated by our laboratory. The strain HR1 was cultured in nitrogen-free medium (20 g sucrose, 0.2 g MgSO4⋅7H2O, 0.1 g K2HPO4, 0.4 g KH2PO4, 0.1 g NaCl, 0.01 g FeCl3, 0.002 g Na2MoO4⋅H2O, and 1,000 ml ddH2O) (Hong et al., 2009) at 30°C with shaking at 100 rpm for 36 h.
Cell Growth and Viability
The different types of surfactants (SDS, Tr, and RL) were accurately weighed and prepared in dH2O to form mother liquors of 10 g/l SDS, 10% Tr (v/v), and 10 g/l RL, respectively. Prior to the experiments, SDS and Tr were sterilized by heat steam and RL was filtered by 0.22 μm pore size filters. The different surfactants were added to the nitrogen-free medium, and the low and high concentrations of SDS (10 and 100 mg/l), Tr (0.025 and 0.1%), and RL (25 and 250 mg/l) could be adjusted according to experimental requirements.
The effect of different surfactants (SDS, Tr, and RL) on the growth of A. chroococcum was investigated using a Bioscreen C Microbiological Growth Analyser (Labsystems, Helsinki, Finland) in 100-well honeycomb plates. A. chroococcum cells were grown to log phase in nitrogen-free medium, and the cells were collected by centrifugation (4°C, 3,500 rpm, and 5 min), washed twice with ddH2O, and adjusted to a concentration of ∼109 cells/ml. Each well of the honeycomb plate contained 25 μl cell suspension and 175 μl of the nitrogen-free medium with or without different surfactants (SDS, Tr, and RL). The plates were incubated at 30°C for 72 h with shaking at medium amplitude, and the OD600 values were recorded every 4 h. All of the experiments were conducted triplicate.
The antibacterial activity of A. chroococcum was evaluated by colony-forming unit (CFU) counting. The cells were treated with different surfactants (SDS, Tr, and RL). A. chroococcum cells were grown to log phase in nitrogen-free medium or nitrogen-free medium supplemented with different concentrations of surfactants (SDS, Tr, and RL). After that, the cells were collected and adjusted to a concentration of ∼109 cells/ml. The cell suspensions were applied with a 108-fold serial dilution of ddH2O. Onto the nitrogen-free medium plates, 200 μl cell suspensions were spread and incubated at 30°C for 36 h. The number of colonies per treatment was counted. All the experiments were tested in triplicate.
Nitrogen Fixation Activity by Acetylene Reduction Assay
The nitrogen fixation activity was determined by acetylene reduction assay (Hardy et al., 1973). The cell suspensions were diluted with nitrogen-free liquid medium to a final OD600 of ∼0.3. A 5-ml aspirate of the nitrogen-free medium with or without surfactants was placed inside a serum bottle with a rubber cap, and 10% of the air in the tubes was removed and replaced with acetylene. After incubation at 30°C for 20 h with shaking (150 r/min), 100 μl of gas in these bottles was extracted, and the amount of ethylene and acetylene was determined by gas chromatography. The nitrogen fixation activity is expressed as nanomole C2H4/h/OD600. Data presented were representative of the means from three replicates.
The nitrogen fixation activity analysis was carried out by a SCION 456-GC (Techcomp, Beijing, China) gas chromatograph with an FID detector. An HP-PLOT capillary column (30 m × 0.32 mm × 20.00 μm) was used for the chromatographic separation, and the carrier gas was ultra high purity nitrogen. Gas flow rate and split ratios were 1.5 ml/min and 10:1, respectively. The temperatures of the inlet, oven, and detector were 200, 60, and 250°C, respectively.
Real-Time PCR Analysis
To compare the transcript levels of nif genes, A. chroococcum cells were grown in nitrogen-free medium (with or without different surfactants) for 22 h and harvested for total RNA isolation and RT-PCR analysis. Total RNA was extracted using the TRIzol reagent, and cDNAs were synthesized using Quantscript RT Kit (Tiangen, Beijing, China). Real-time quantitative reverse transcription PCR (qRT-PCR) was performed with three independent RNA preparations using Applied Biosystems 7300 Real-Time System and detected using GoTaq® qPCR Master Mix (Promega, Madison, CT, United States). The 16S RNA gene was used as a stable reference gene for the normalization of samples. The relative transcript levels were calculated by the 2–△ △ CT method and were normalized. PCR primers for nif genes and 16S rRNA are shown in Supplementary Table 1.
Protein Extraction
The A. chroococcum cells treated with RL (250 mg/l) and without RL were grinded by liquid nitrogen and resolved in lysis buffer [8 M urea, 50 mM nicotinamide (NAM), 3 μM trichostatin A (TSA), and 1% Protease Inhibitor Cocktail]. Cells were sonicated three times on ice using a high-intensity ultrasonic processor (Scientz, Ningbo, China). The supernatant was collected by centrifugation at 12,000 × g for 10 min at 4°C. Protein concentration was determined using a BCA kit (Tiangen, Beijing, China).
Western Blotting Analysis
The protein extracts (20 μg) were separated by 12% SDS-PAGE gel and then transferred to PVDF membranes. The membrane was incubated with pan anti-succinyl lysine antibody (1:1,000, PTM Biolabs, Hangzhou, Zhejiang, China) at 4°C overnight. The membrane was washed five times with Tris-buffered saline with Tween 20 (TBST) (25 mM Tris–HCl, pH 8.0; 125 mM NaCl; and 0.1% Tween 20) containing 5% (w/v) BSA for 10 min and subsequently incubated with goat anti-mouse IgG (1:1,000, Thermo Fisher Scientific, Waltham, MA, United States) at an ambient temperature for 2 h. After the membrane was washed with TBST buffer, the SuperSignal West Pico kit (Pierce, Dallas, TX, United States) was used for visualizing.
Trypsin Digestion and HPLC Fractionation
The collected proteins were added with dithiothreitol (DTT, 5 mM), reduced at 56°C for 30 min, and then alkylated with iodoacetamide (11 mM) for 15 min at an ambient temperature in the dark. The proteins were then diluted by adding 100 mM tetraethylammonium bromide (TEAB) to a urea concentration less than 2 M and digested with trypsin (Promega, Madison, WI, United States) to a trypsin/protein ratio of 1:50 (w/w) at 37°C overnight. After digestion, peptides were fractionated into fractions using a Thermo BetaSil C18 column (250 mm × 10 mm × 5 μm) with a gradient of 8% to 32% acetonitrile (pH 9.0) over 60 min into 60 fractions. Then, the peptides were combined into six fractions and dried by vacuum centrifugation.
Affinity Enrichment and Liquid Chromatography With Tandem Mass Spectrometry Analysis
The peptides were dissolved in NETN buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris–HCl, and 0.5% NP-40, pH 8.0) and mixed with conjugated pan anti-succinyl lysine agarose antibody beads (PTM Biolabs, Hangzhou, Zhejiang, China) at 4°C overnight with gentle shaking. The beads were washed four times with NETN buffer and twice with dH2O subsequently. The peptides bound were eluted from the beads with 0.1% trifluoroacetic acid (TFA) and desalted with C18 Zip-Tips (Millipore, Billerica, MA, United States).
Liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis was performed at PTM Biolabs, Hangzhou, Zhejiang, China. Briefly, the enriched peptides were dissolved in buffer A [0.1% (v/v) formic acid in dH2O] and loaded directly onto a home-made reversed-phase analytical column (15-cm length and 75-mm diameter) using an EASY-nLC 1000 UPLC system (Thermo Fisher Scientific, Waltham, MA, United States) for elution. The gradient was 6–22% buffer B (0.1% formic acid in 98% acetonitrile) for 44 min, 22–30% for 12 min, 30–80% for 2 min, and finally held at 80% for 2 min. The flow rate of the column was maintained at 300 nl/min.
The resultant peptides were then analyzed by MS/MS in timsTOF Pro (Bruker, Billerica, MA, United States) coupled online to the UHPLC. The electrospray voltage was set at 1.4 kV. Precursors and fragments were analyzed at the TOF detector, with an MS/MS scan range set as 100–1,700 m/z. The Bruker timsTOF Pro was operated in parallel accumulation serial fragmentation (PASEF) mode. Precursors with charge states 0–5 were selected for fragmentation, and 10 PASEF-MS/MS scans were acquired per cycle. The dynamic exclusion was set to 24 s. Three biological replicates were performed.
Proteomics Database Search
The resulting MS/MS data were processed using the MaxQuant search engine (v.1.6.6.0). Tandem mass spectra were searched against the UniProt A. chroococcum (strain NCIMB 8003) database downloaded from NCBI (4,584 protein sequences, released 2015) and concatenated with a reverse decoy database. Trypsin/P was specified as a cleavage enzyme and up to four missing cleavages. The mass tolerance of precursor ions for the first search and main search was set as 40 ppm, and the mass tolerance for fragment ions was 0.04 Da. Carbamidomethyl on Cys was specified as a fixed modification, and acetylation on protein N-terminal, succinylation, deamidation, and oxidation on methionine were specified as variable modifications. Peptide and protein false discovery rate (FDR) was adjusted to <1%, and the minimum score of modified peptides was more than 40 (Cox and Mann, 2008).
Bioinformatics Analysis
KSuc sites (10 amino acids upstream and downstream of the motif site) were analyzed using MoMo (Chou and Schwartz, 2011). Gene Ontology (GO) annotations of the succinylated proteins were compared against the UniProt-GOA database1 using InterProScan (v.5.14–53.0). The subcellular localization prediction analysis of succinylated proteins was using CELLO. The GO, domains, and Kyoto Encyclopedia of Genes and Genomes (KEGG) were used for functional enrichment analyses. Two-tailed Fisher’s exact tests were applied to each category to obtain p values for the analysis of functional enrichment, the terms (p < 0.05) were considered significant. According to the succinylation quantification analysis, a 1.5-fold change was set as the differential expression multiple.
The further hierarchical clustering was based on differentially succinylated protein functional classification (GO, domains, and KEGG). Those categories, which were at least enriched in one of the clusters with p value <0.05, were filtered. This filtered p value matrix was transformed by the function x = −log10 (p value). These x values were z-transformed for each functional category. These z scores were then clustered by one-way hierarchical clustering (Euclidean distance and average linkage clustering) in Genesis. Protein–protein interaction (PPI) networks were performed by STRING database (Szklarczyk et al., 2011) and visualized by Cytoscape v. 3.7.2 (Shannon et al., 2003). The interactions that had confidence scores of 0.7 (high confidence) were fetched.
Results
The Effects of Surfactants on A. chroococcum
To study the effects of surfactants (RL, SDS, and Tr) on A. chroococcum, the growth curves and survival rate of A. chroococcum under low and high concentrations of surfactants were determined. As shown in Figures 1A,B, the growth of A. chroococcum was significantly inhibited by both low and high concentrations of SDS and Tr. The effects of low concentrations of RL were comparable to the control. However, high concentrations of RL promoted the growth of A. chroococcum with the extension of the incubation time. At the same time, the CFU were used to validate the survival rate of A. chroococcum cells with different surfactants, and the result indicated that SDS and Tr had obvious antibacterial effects on A. chroococcum cells (Figure 1C).
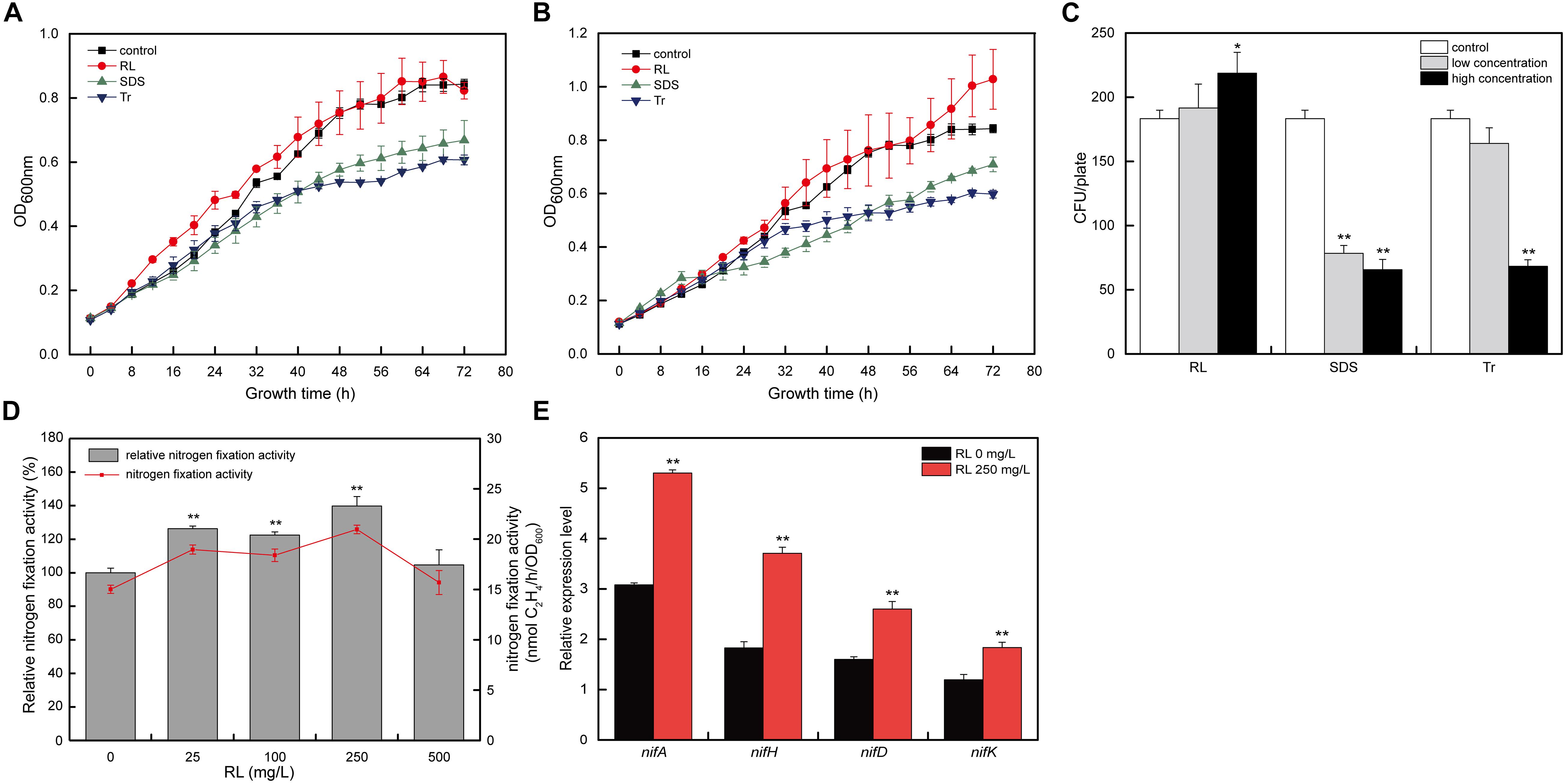
Figure 1. The effects of different surfactants on A. chroococcum. The growth curves of A. chroococcum cells were treated with (A) low concentrations of surfactant (10 mg/l SDS, 0.025% Tr, and 25 mg/l RL) and (B) high concentrations of surfactant (100 mg/l SDS, 0.1% Tr, and 250 mg/l RL), respectively. (C) The CFU analyses of A. chroococcum. The cells were incubated in nitrogen-free medium (control) or nitrogen-free medium added with both low and high concentration of SDS (10 and 100 mg/l), Tr (0.025 and 0.1%), and RL (25 and 250 mg/l), respectively. (D) The nitrogen fixation activity of A. chroococcum with different concentrations of RL (0, 25, 100, 250, and 500 mg/l). Relative nitrogen fixation activity of each sample represents 100% that of A. chroococcum cells treated without RL. (E) Relative transcript levels of nif genes in A. chroococcum with or without RL (250 mg/l). (*p < 0.05; **p < 0.01). SDS, sodium dodecyl sulfate; Tr, Triton X-100; RL, rhamnolipid; CFU, colony-forming unit.
Because RL showed a positive effect on the growth and activity of A. chroococcum, and the survival of A. chroococcum is an important prerequisite for nitrogen fixation, RL should be applied in nitrogen fixation. To further investigate the effect of RL on A. chroococcum, different concentrations of RL (0, 25, 100, 250, and 500 mg/l) were selected to determine its effects on nitrogen fixation activity. As compared with the control, different concentrations of RL could improve the nitrogen fixation activity of A. chroococcum, and the effect of 250 mg/l of RL (increased by 39.7%) was the most significant (Figure 1D).
The expression and synthesis of nitrogenase rely on the coordinated regulation of a series of genes (Curatti et al., 2005), which mainly include the positive regulation gene (nifA) and the structure gene (nifHDK) (Ribbe et al., 2014). To investigate the mechanisms of the enhancement of nitrogen fixation activity of A. chroococcum by RL, the effects of RL on the transcript levels of nitrogen-fixing enzyme genes were studied. The transcript levels of nitrogen fixation genes (nifA, nifH, nifD, and nifK) of A. chroococcum upon 250 mg/l of RL were significantly higher than those of the control (Figure 1E). The results indicated that RL could increase the transcript levels of these nitrogen fixation genes.
Identification of KSuc in A. chroococcum
BNF was involved in multiple metabolic pathways (e.g., respiration, electron transport, carbon metabolism, nitrogen metabolism, etc.), and succinylated proteins play regulatory roles in various metabolic processes as previously described. Thus, the A. chroococcum cells were treated with RL (250 mg/l) and without RL, and the protein extracts were investigated using anti-succinyl lysine antibody by the western blotting assay. The results showed stronger reactions to the anti-succinyl lysine in all treatments, indicating the presence of KSuc in A. chroococcum (Figure 2A). To identify the KSuc sites in A. chroococcum, we chose 0 h (RL0) and 22 h (RL22) of 250 mg/l of RL treatments and without RL treatment as the control. Figure 2B shows the workflow of a mass spectrometry-based high-throughput proteomics approach to the identification of KSuc. In order to ensure the reliability of the data, the samples were tested for reproducibility using principal component analysis (PCA). The degree of aggregation between each set of replicates was good, which indicated a good reproducibility between each set of replicates samples (Figure 2C). A total of 5,008 KSuc sites on 1,376 proteins were yielded (Supplementary Table 2). There were 1,376 KSuc sites on 645 proteins and 1,372 KSuc sites on 639 proteins found in CK22/CK0 and RL22/RL0, respectively (Figure 2D and Supplementary Table 3). The results indicated that a large number of succinylated proteins were presented in A. chroococcum and RL may alter the KSuc level. These results imply that KSuc is likely linked to nitrogen fixation in A. chroococcum, although the mechanism for this is unclear.
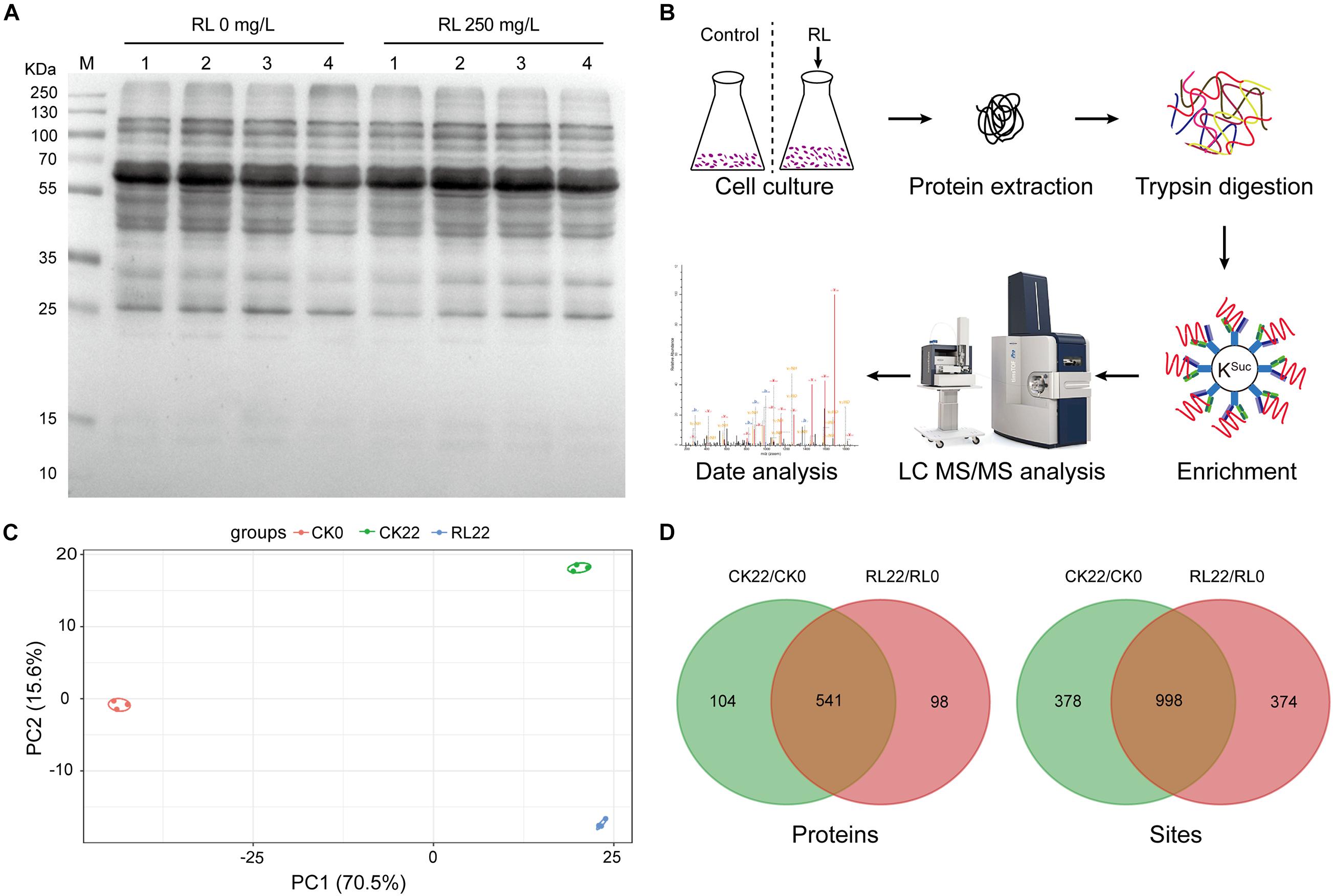
Figure 2. Identification of succinylation in A. chroococcum. (A) Detection of succinylated proteins using the pan anti-succinyl lysine antibody in A. chroococcum treated with or without RL (250 mg/l) for 0 h (lane 1), 4 h (lane 2), 12 h (lane 3), and 22 h (lane 4), respectively. (B) The workflow of an integrated strategy for global mapping of KSuc in A. chroococcum. (C) PCA of KSuc. (D) Survey of succinylated proteins and KSuc sites. CK0, A. chroococcum cells were treated without RL for 0 h; CK22, A. chroococcum cells were treated without RL for 22 h; RL0, A. chroococcum cells were treated without RL for 0 h; RL22, A. chroococcum cells were treated with RL for 22 h. KSuc, lysine succinylation; PCA, principal component analysis.
Analyses of the Sites of KSuc in A. chroococcum
To identify the motifs of KSuc, lysine (K) was frequently found both upstream and downstream of KSuc, while arginine (R) was frequently found downstream of KSuc (Figure 3A and Supplementary Table 4). A total of nine conserved motifs were ∗K∗KSuc*, ∗KSuc*R∗, and ∗KSuc*K∗ [asterisk (∗) indicates multiple random amino acid residues]. To further assess the enrichment or depletion of specific amino acids neighboring the KSuc sites, the basic amino acids [lysine (K) and arginine (R)] were significantly enriched surrounding KSuc sites, but R was greatly depleted at the −1 and +1 positions, respectively. Moreover, the acidic amino acid aspartic acid (D) was enriched in positions +3 of the KSuc sites, and aliphatic amino acid valine (V) was significantly enriched at the −1 and +2 positions (Figure 3B). Conserved amino acid sequences surrounding the KSuc sites suggested the preference of enzymes that catalyze the succinylation modifications.
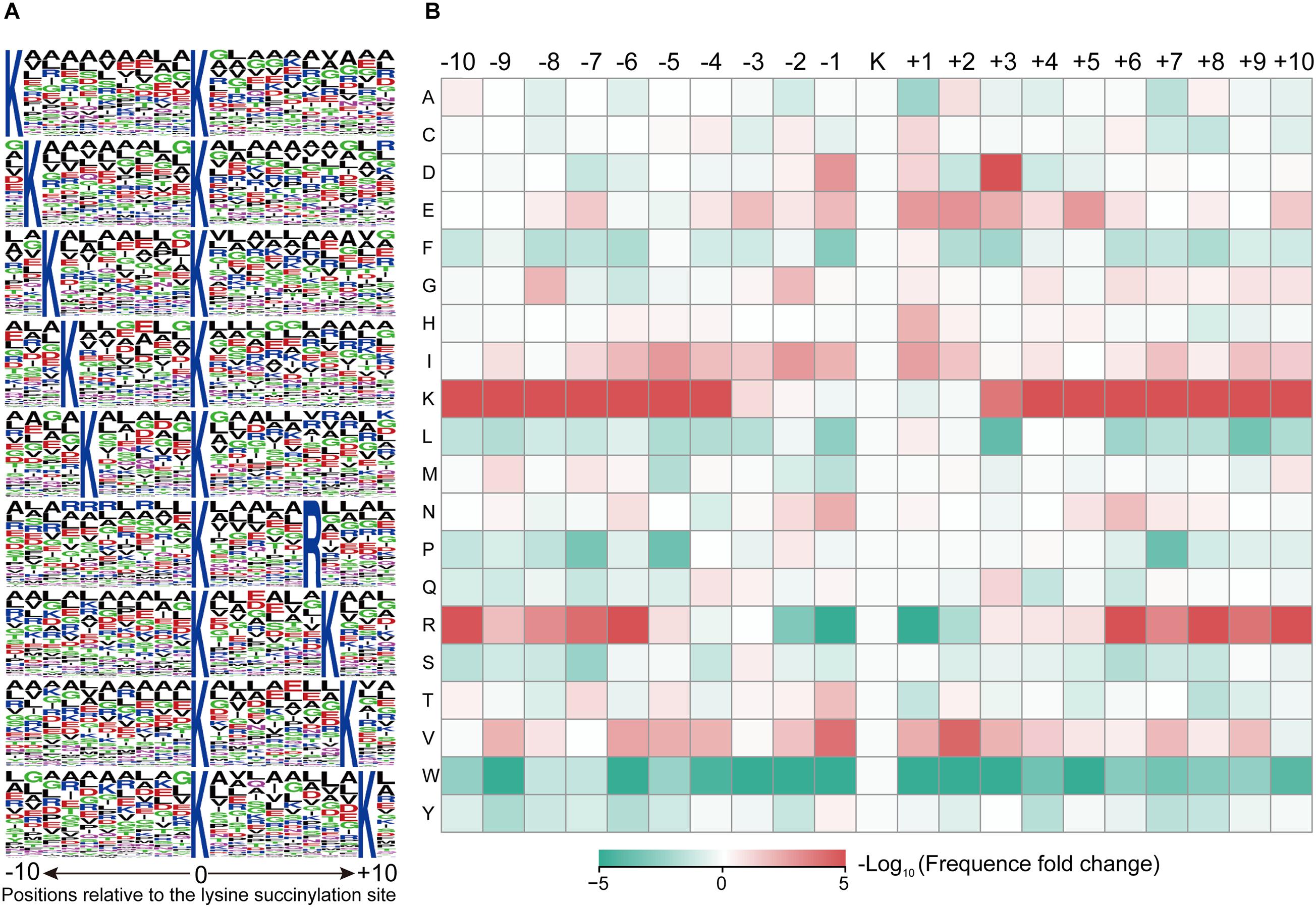
Figure 3. MoMo analysis of the lysine-succinylated sites. (A) Sequence probability of significantly enriched KSuc motifs for ± 10 amino acids around the KSuc sites. (B) Heat map of the sequence motif of KSuc sites. The colors represent the enrichment (red) or depletion (green) of amino acids at specific positions.
GO and Subcellular Localization of Succinylated Proteins
For the purpose of understanding the biological functions of succinylated proteins in A. chroococcum, GO classification of all succinylated proteins was performed (Figure 4 and Supplementary Table 5). In cellular composition, intracellular (34%), cell periphery (14%), and plasma membrane (12%) were accounted for the major part of succinylated proteins (Figure 4A). In terms of molecular function, 60 and 26% of succinylated proteins were associated with binding function and catalytic activity, respectively (Figure 4B). For biological process, 65% of succinylated proteins were associated with metabolism, among which 15% of succinylated proteins were involved in the nitrogen compound metabolic process (Figure 4C). Further analysis showed that the major classes were similarly to RL treatments, but the number of succinylated proteins was different from each individual component. In addition, 21 succinylated proteins associated with intrinsic components of the membrane were present in CK22/CK0, while 35 succinylated proteins associated with response to abiotic stimulus were present in RL22/RL0. The results showed that the level of KSuc in A. chroococcum was altered in response to RL (Supplementary Figure 1 and Supplementary Tables 6, 7).
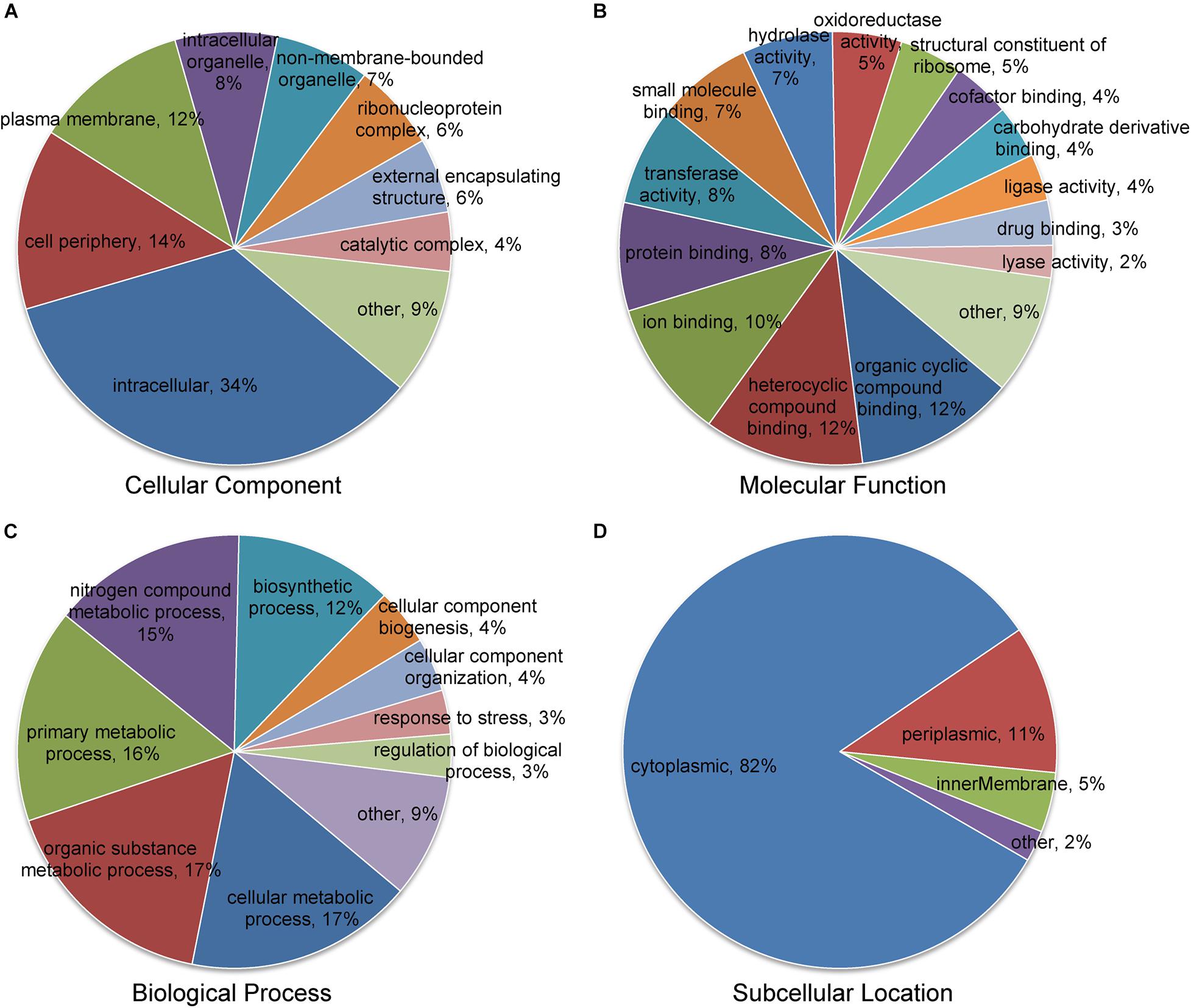
Figure 4. Classification of the succinylated proteins based on Gene Ontology (GO) and subcellular location. (A) Classification of the succinylated proteins based on the cellular component. (B) Classification of the succinylated proteins based on the molecular function. (C) Classification of succinylated proteins based on the biological process. (D) Subcellular location classification of the succinylated proteins.
The subcellular localization analysis indicated that the succinylated proteins in A. chroococcum were predominantly located in cytoplasm (82%), followed by periplasm (11%), inner membrane (5%), and other subcellular components (2%) (Figure 4D). The majority of succinylated proteins were involved in intracellular material transport, metabolism, and cellular activity, which suggested that the succinylated proteins played an important role in the growth and development of A. chroococcum.
Enrichment-Based Clustering Analysis of Succinylated Proteins
To reveal the mechanisms of the enhancement of nitrogen fixation activity in A. chroococcum by RL, GO, protein domain, and KEGG pathway enrichment-based clustering analyses were performed (Figure 5 and Supplementary Table 8).
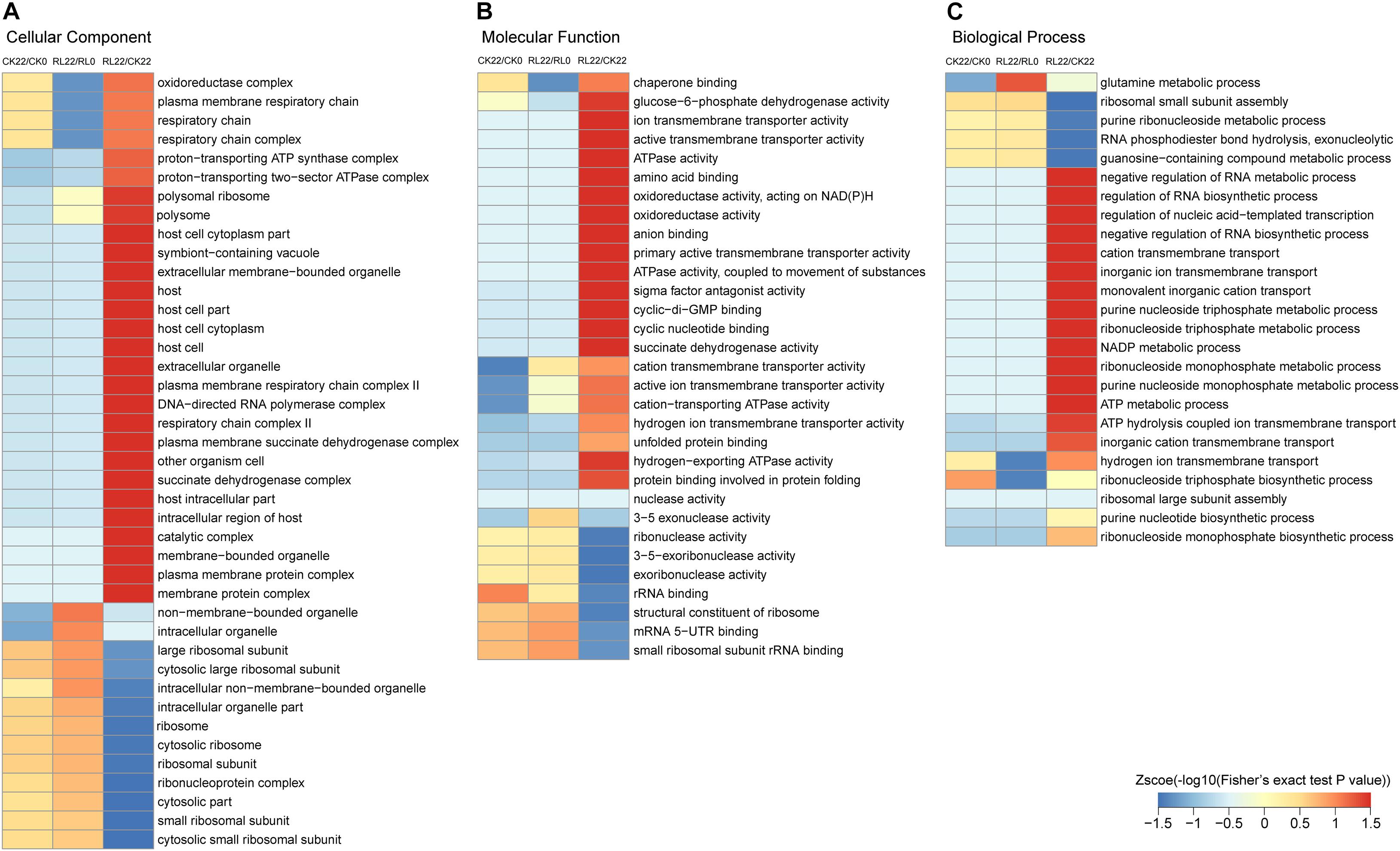
Figure 5. Gene Ontology enrichment-based clustering analysis of the succinylated proteins. (A) Cellular component analysis. (B) Molecular function analysis. (C) Biological process analysis.
Gene Ontology enrichment-based clustering analysis showed various degrees of enrichment of succinylated proteins in a different category. In the cellular component category, the enrichment of succinylated proteins in CK22/CK0 and RL22/RL0 was similar, and only a few succinylated proteins were significantly different, such as oxidoreductase complexes, intracellular organelles, and those associated with the respiratory chain. However, the succinylated proteins were mainly enriched in components that were related to cellular respiration and membranes in RL22/CK22 (Figure 5A). The enrichment analysis of the molecular function category revealed significant differences in succinylated proteins, which were associated with the transmembrane activity and 3–5 exonuclease activity in CK22/CK0 and RL22/RL0; the succinylated proteins were significantly enriched in terms that were related to energy metabolisms in RL22/CK22, such as glucose-6-phosphate dehydrogenase activity, ATPase activity, oxidoreductase activity, and succinate dehydrogenase activity (Figure 5B). In the biological process category, succinylated proteins were significantly different in glutamine metabolism, hydrogen ion transport across membranes, and ribonucleoside triphosphate biosynthesis in CK22/CK0 and RL22/RL0; the enriched succinylated proteins were mainly involved in the cellular nucleic acid metabolic process in RL22/CK22, such as ion transmembrane transport and energy metabolic process (Figure 5C).
The domain enrichment analysis of succinylated proteins indicated that the majority of protein domains were similarly enriched in CK22/CK0 and RL22/RL0. However, the enriched protein domains mainly relate to nitrogenase (nitrogenase component type 1 oxidoreductase and 4Fe-4S dicluster domain) and energy metabolism (citrate synthase, glucose-6-phosphate dehydrogenase, NAD-binding domain; glucose-6-phosphate dehydrogenase, C-terminal domain; and ATP synthase alpha/beta family, nucleotide-binding domain) in RL22/CK22, followed by 2-oxoacid dehydrogenases acyltransferase, catalytic domain; chalcone and stilbene synthases, C-terminal domain; and glutamine synthetase, catalytic domain (Figure 6A). In the KEGG enrichment analysis, the succinylated proteins were mainly enriched in the ribosome, thiamine metabolism, and starch and sucrose metabolism in CK22/CK0. The ribosome, oxidative phosphorylation, and citrate cycle (TCA cycle) were the most dominant pathways in RL22/RL0. Conversely, the nitrogen metabolism, oxidative phosphorylation, and TCA cycle were significantly enriched in RL22/CK22 (Figure 6B).
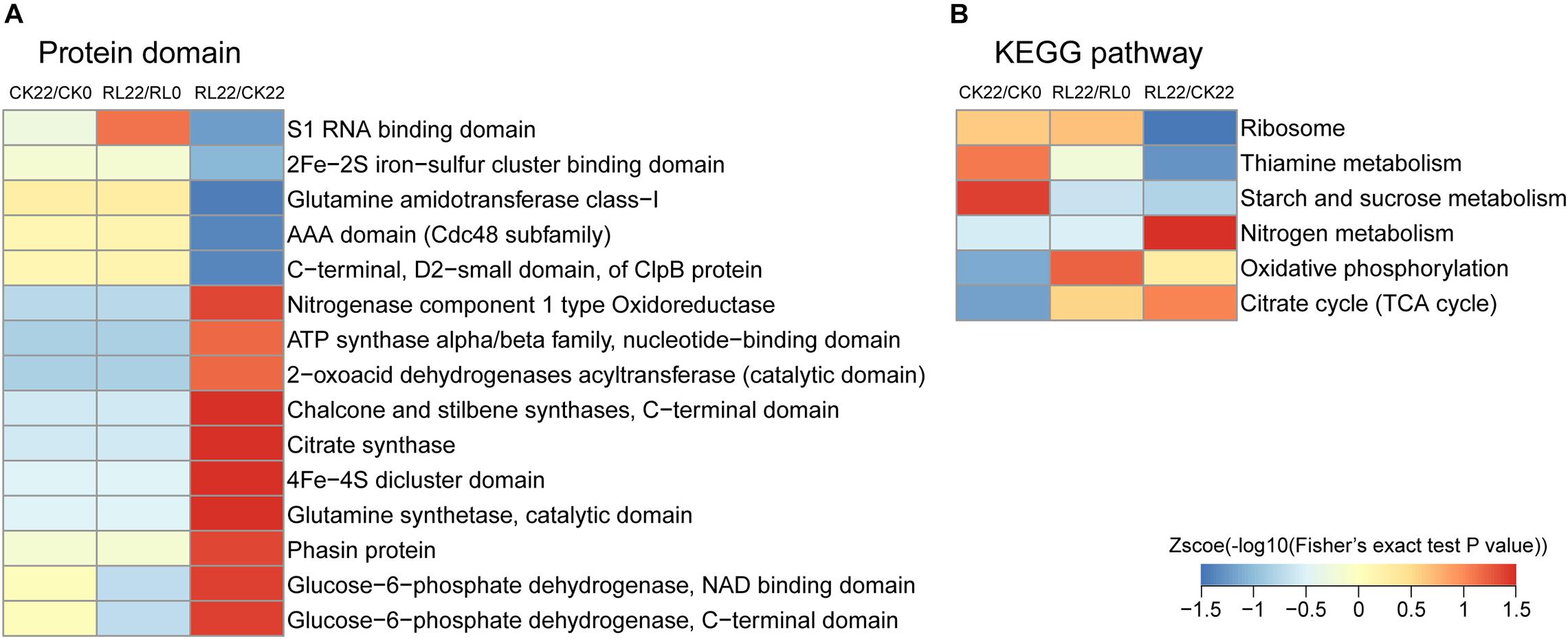
Figure 6. Functional enrichment-based clustering analysis of the quantified proteins. (A) Protein domain. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway.
Protein Interaction Networks of Succinylated Proteins in A. chroococcum
To reveal the relationships of succinylated proteins and the molecular mechanisms of the enhancement of nitrogen fixation activity in A. chroococcum with RL treatment, the PPI networks were constructed (Figure 7 and Supplementary Table 9).
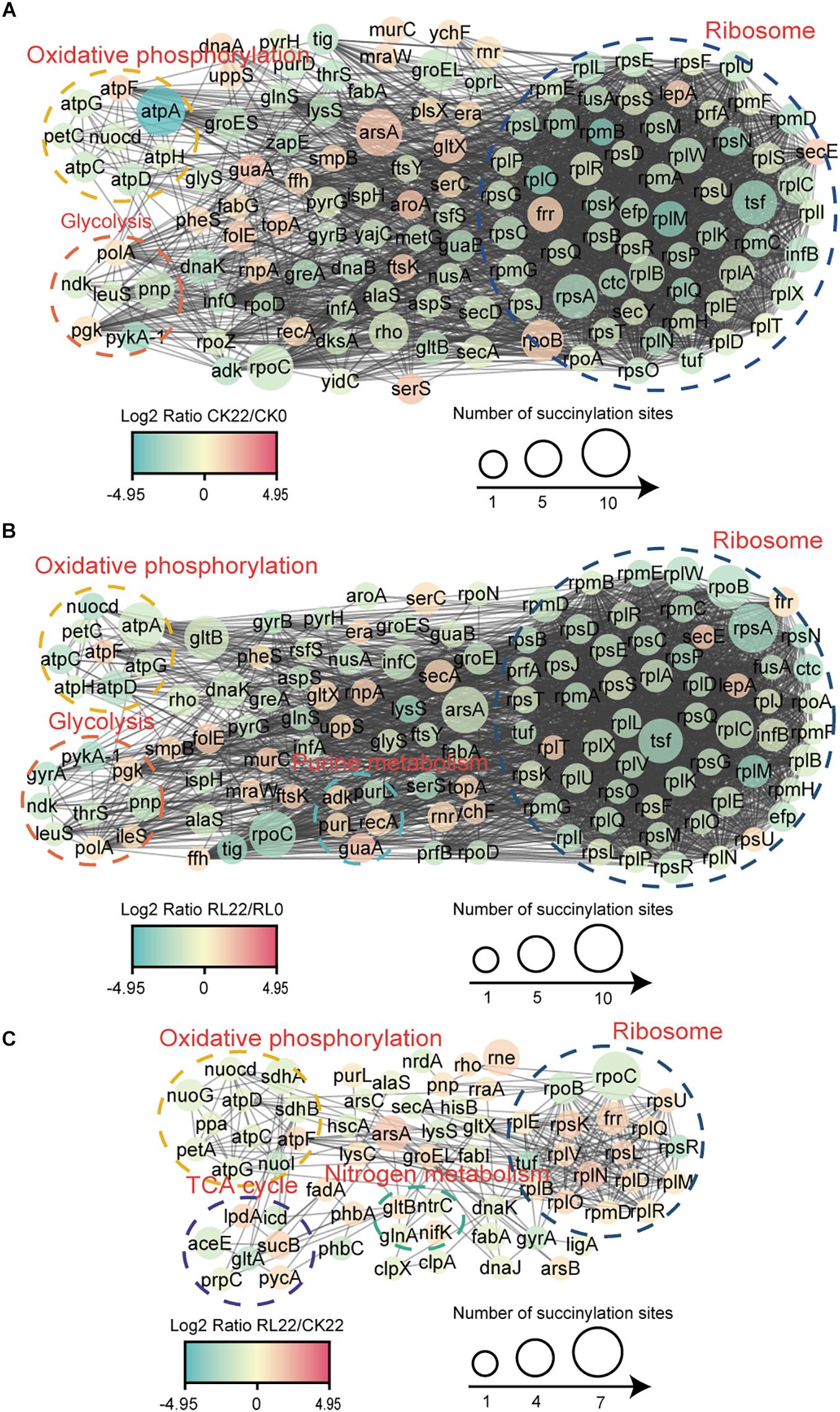
Figure 7. Protein–protein interaction (PPI) network of the succinylated proteins identified in A. chroococcum. (A) The PPI network of CK22/CK0. (B) The PPI network of RL22/RL0. (C) The PPI network of RL22/CK22.
A total of 135 succinylated proteins interacting with each other in CK22/CK0 were profiled, and the predominant interrelated clusters were oxidative phosphorylation, glycolysis, and ribosomes (Figure 7A), while five succinylated proteins, which were enriched in purine metabolisms, were also present in RL22/RL0 (Figure 7B). In RL22/CK22, succinylated proteins were significantly clustered in TCA cycle and nitrogen metabolism, except in the oxidative phosphorylation and ribosomal complexes (Figure 7C).
To further reveal the relationship between succinylated proteins and nitrogen fixation activity, the network involved in the nitrogen fixation process in A. chroococcum with RL treatment was mapped, which was based on the enrichment of succinylated proteins in KEGG pathway (Supplementary Figure 2) and the interactions between succinylated proteins. As shown in Figure 8, succinylated proteins were involved in almost all parts of oxidative phosphorylation and TCA cycle. It is noteworthy that there were multiple modification sites of KSuc on the MoFe protein (NifDK) (succinylated at K330, K341, and K433 on NifD and K300 on NifK) and flavodoxin (NifF) (succinylated at K3 and K160) (Supplementary Table 10). Therefore, the level of KSuc on NifDK may have a great correlation with the nitrogen fixation activity of A. chroococcum.
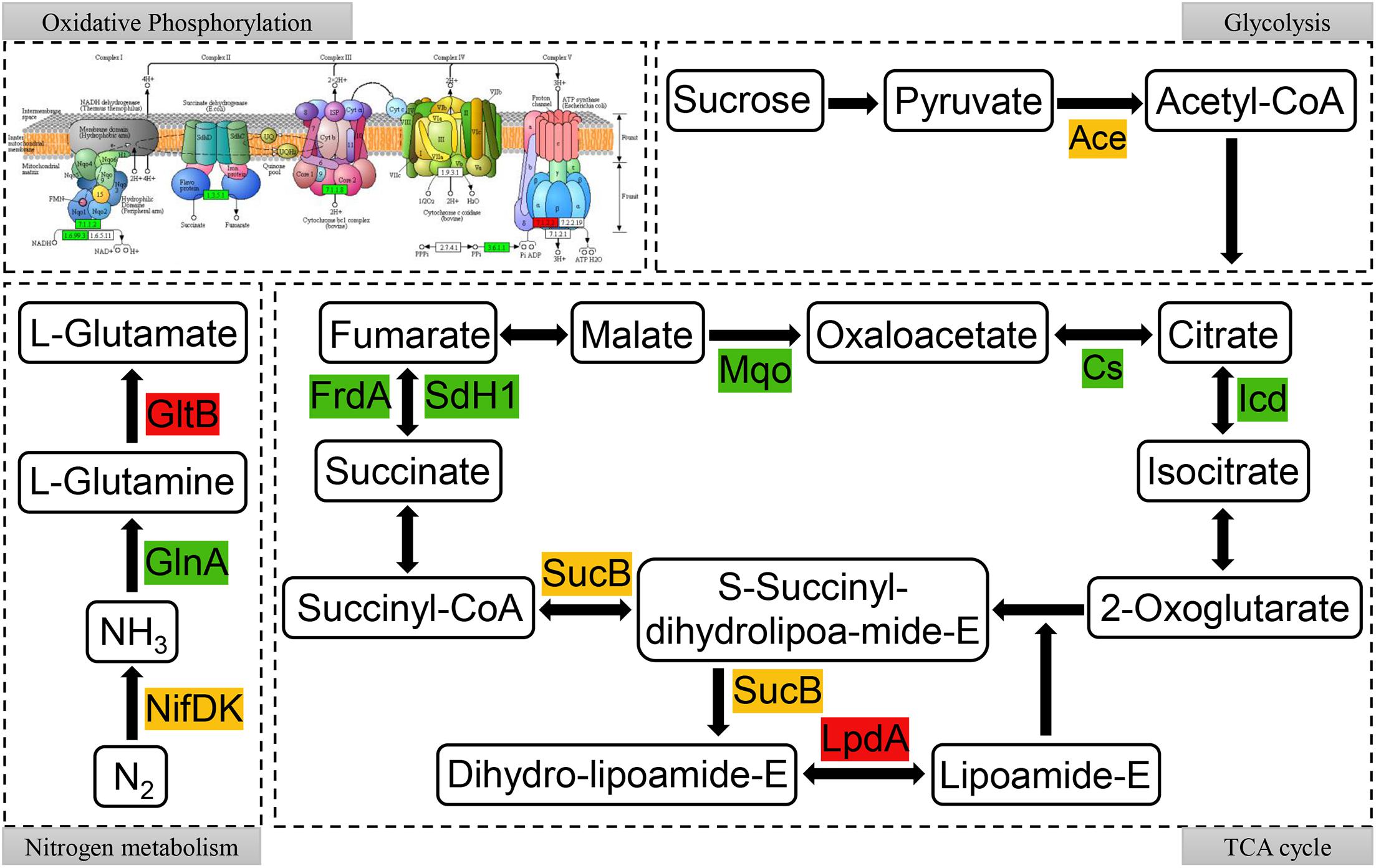
Figure 8. Succinylated proteins in oxidative phosphorylation, glycolysis, nitrogen metabolism, and tricarboxylic acid (TCA) cycle metabolic pathways in RL22/CK22. Different colors represent the upregulated (red), downregulated (green), or up/downregulated (yellow) succinylated proteins.
Discussion
Azotobacter species were model organisms to study nitrogenase and the mechanisms of nitrogen fixation (Baars et al., 2016; Danyal et al., 2016; Rebelein et al., 2018). Meanwhile, Azotobacter species is important as free-living, nitrogen-fixing bacteria and potential bacterial biofertilizer (Sumbul et al., 2020; Aasfar et al., 2021). In addition, A. chroococcum has other characteristics, such as phosphorus hydrolysis, synthetic indoles, and alginate (Romero-Perdomo et al., 2017; Gawin et al., 2020). RL was mainly produced by Pseudomonas aeruginosa (Abbasi et al., 2012) and belonged to biosurfactant, which is capable of inhibiting plant pathogen and improving composting efficiency (Sachdev and Cameotra, 2013; Inès and Dhouha, 2016; Thakur et al., 2021). In this study, we found that RL could significantly enhance the nitrogen-fixing activity of A. chroococcum, in which the nitrogen fixation activity was increased by 39.7% with RL (250 mg/l) treatment. Therefore, RL has the potential to improve the nitrogen fixation activity and agricultural production of diazotrophs.
The 4D label-free quantitative proteomic approach is highly sensitive and capable of accurate quantification (Meier et al., 2018; Lesur et al., 2021; Loginov et al., 2021). This approach can detect more low-abundance proteins, but many low-abundance proteins were not identified with western blotting (Kurien and Scofield, 2015). Western blotting qualitatively or semiquantitatively identifies proteins, that is, the corresponding size of protein band has many target proteins (Hnasko and Hnasko, 2015). The size of the differentially succinylated proteins may be higher or lower, which meant that they may counteract each other. Although in this paper the results of western blotting exhibited similar succinylation levels between CK22 and RL22, the detailed proteomic analysis showed differences between CK22 and RL22. KSuc is an important protein PTM that is closely associated with multiple biological processes. LC-MS/MS was used for quantitative proteomics to perform a global succinylation analysis of A. chroococcum, and 5,008 succinylation sites in 1,376 proteins were identified. Several reports had shown that PTMs could be changed in response to environmental factors. For example, acetylation of Escherichia coli was the most significant process in PTMs cultured in ethanol-enriched media (Soufi et al., 2015), and the level of KSuc in Bacillus subtilis was varied in response to different carbon sources (Kosono et al., 2015). We did not find studies of other modifications in diazotrophs, but found quantitative analysis of the modified proteins in other organisms with a fold change of 1.3 (Jiang et al., 2018; Zhang et al., 2019; Li et al., 2020). Therefore, the 1.5-fold change in succinylation is relevant enough in our study. We found that RL could change the enrichment of KSuc in A. chromococcus, which indicated that environmental factors played a key role in PTMs. Succinylated proteins were involved in various biological process in A. chroococcum and significantly enriched in metabolism. However, most succinylated proteins were associated with the function and activity of enzyme. KSuc played an important role in regulating the activities of enzymes (Ren et al., 2018; Zhou et al., 2019), and we judged that KSuc might be involved in the nitrogen fixation process in A. chroococcum by regulating the structure and function of proteins in this study.
Under the optimal conditions, the overall reaction catalyzed by nitrogenase was described as follows: N2 + 8H+ + 16MgATP + 8e−→2NH3 + H2 + 16MgADP + 16Pi (Ribbe et al., 2014). Therefore, BNF requires large amounts of ATP and e–. As nitrogen fixation proceeds, the expression of glnA (encoding glutamine synthetase I) and genes in respiration was increased while the TCA cycle was decreased (Sarkar and Reinhold-Hurek, 2014; Shi et al., 2016). In RL22/CK22, the oxidative phosphorylation and TCA cycles contained succinylated proteins, which could provide the ATP and e– in nitrogen fixation process. In addition, there was KSuc enrichment of glutamine synthetase (GlnA) and glutamate synthase (GltB) in glutamate metabolism. The fixed nitrogen by A. chroococcum could enter into other metabolic pathways after its assimilation by GlnA and GltB, so we speculate that KSuc was involved in the nitrogen fixation process, which was altered by RL.
The nifA is the positively regulated gene for nif operon expression in A. vinelandii (Mitra et al., 2005). The nitrogenase is composed of Fe protein encoded by nifH and MoFe protein encoded by nifDK (Einsle et al., 2002). In this study, RL was able to significantly increase the transcript levels of nifA and nifHDK, which could promote the synthesis of nitrogenase and then enhance the efficiency of nitrogen fixation. Nitrogenase type 1 oxidoreductase is the domain of nitrogenase, and the 4Fe-4S cluster is essential for nitrogenase cofactor assembly (Hu and Ribbe, 2013; Rettberg et al., 2020). Protein domains were significantly enriched in nitrogenase component type 1 oxidoreductase and 4Fe-4S dicluster domain in RL22/CK22. Proteins with these enriched domains play a key role in the synthesis of nitrogenase (Ribbe et al., 2014; Sickerman et al., 2017), so we speculate that KSuc regulates the nitrogen fixation activity of A. chroococcum by affecting the activity of nitrogenase. NifDK is the catalytic center of nitrogenase (Spatzal et al., 2011), and the reduced flavodoxin (NifF) is the direct electron donor to Fe protein (Neuer and Bothe, 1985), all of them are the key enzymes that directly affect the nitrogen fixation. Multiple succinylation sites were identified on NifDK and NifF, which indicated that KSuc may play a direct role in the regulation of nitrogen fixation in A. chroococcum.
Based on previous studies, we hypothesized a model of KSuc involved in the regulation process of nitrogen fixation in A. chroococcum (Figure 9). When A. chroococcum cells are treated with RL, RL penetrate the cytoplasm and lead to altered KSuc level. The KSuc could increase the amount of transcript levels of nifA gene by modifying nucleic acid transcriptional regulatory proteins, which in turn upregulated the nifHDK genes and promoted the nitrogenase synthesis in A. chroococcum. On the other hand, succinylated proteins participated in the nitrogen fixation process through oxidative phosphorylation and TCA cycle, which could provide ATP and e– and ensure the smooth progress of nitrogen fixation. At the same time, KSuc modifies the NifDK of nitrogenase and NifF, which may affect the activity of these two enzymes and alter the catalytic process. Finally, nitrogenase converts nitrogen into ammonia, which then enters into other metabolic pathways.
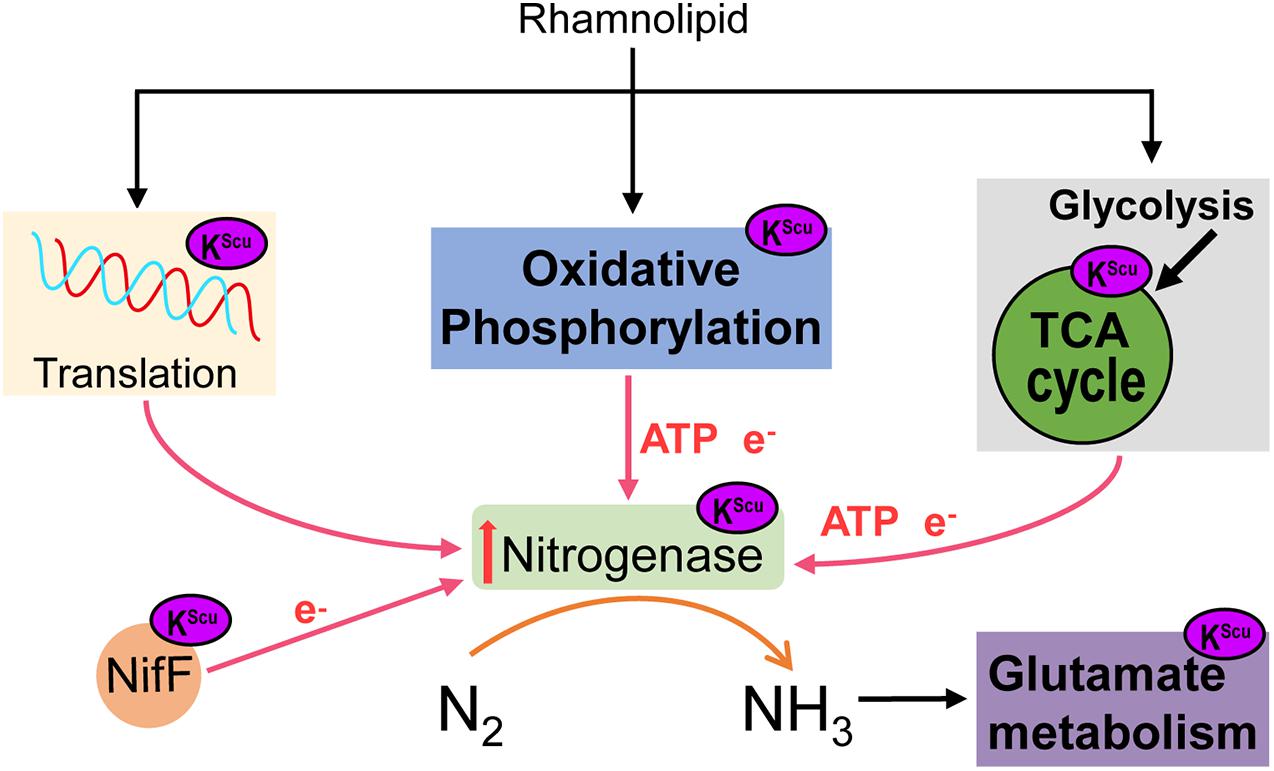
Figure 9. A model of the role of RL in mediating KSuc involved in the regulation process of nitrogen fixation in A. chroococcum.
In summary, we hypothesized that KSuc may play a key role in regulating the process of nitrogen fixation. KSuc has a greater impact on protein function than lysine methylation or acetylation (Yang and Gibson, 2019). We have shown that there is NifDK of nitrogenase by KSuc. As for the modification sites of KSuc on NifDK influencing nitrogenase activity needs further verification. Therefore, we will analyze the succinylation sites on NifDK to investigate the potential role of KSuc in nitrogen fixation.
Conclusion
In this study, we found that RL has a positive impact on the nitrogen-fixing activity in A. chroococcum. Further studies indicate that RL altered the KSuc level and succinylated proteins were involved in important biological pathways. The study shows that succinylation sites on NifDK may influence nitrogenase activity. To our knowledge, this is the first reported data on KSuc in A. chroococcum. Our study provides a foundation for further research on the role of KSuc in regulating biological nitrogen fixation. However, more studies are needed to reveal the effects of succinylated proteins in A. chroococcum and to better understand the underlying mechanisms of nitrogen fixation in control of protein succinylation’s ability.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
XL and YT initiated and designed the research. JL and HP performed the experiments and drafted the manuscript. JL, HY, CW, HL, HZ, PL, and CL analyzed the data. JL, HP, and HY prepared materials. All of the authors read and approved the final manuscript.
Funding
This work was supported by the Science and Technology Department of Hunan Province (Nos. 2018NK1030 and 2019GK4018), the National Key R&D Program of China (Nos. 2019YFC1604903 and 2019YFC1604905), the National Natural Science Foundation of China (Nos. 31971042 and 21808052), the State Key Laboratory of Utilization of Woody Oil Resource (Nos. 2019XK2002), the China Postdoctoral Science Foundation (No. 2019TQ0088), the Hunan Provincial Natural Science Foundation of China (No. 2019JJ50227), the Scientific Research Foundation of Hunan Provincial Education Department (Nos. 18B090 and 18K039), the Training Program for Excellent Young Innovators of Changsha (No. kq2009019), and the Double First-Class Construction Project of Hunan Agricultural University (No. SYL201802002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.697963/full#supplementary-material
Footnotes
References
Aasfar, A., Bargaz, A., Yaakoubi, K., Hilali, A., Bennis, I., Zeroual, Y., et al. (2021). Nitrogen fixing Azotobacter species as potential soil biological enhancers for crop nutrition and yield stability. Front. Microbiol. 12:628379. doi: 10.3389/fmicb.2021.628379
Abbasi, H., Hamedi, M. M., Lotfabad, T. B., Zahiri, H. S., Sharafi, H., Masoomi, F., et al. (2012). Biosurfactant-producing bacterium, Pseudomonas aeruginosa MA01 isolated from spoiled apples: Physicochemical and structural characteristics of isolated biosurfactant. J. Biosci. Bioeng. 113, 211–219. doi: 10.1016/j.jbiosc.2011.10.002
Baars, O., Zhang, X., Morel, F. M., and Seyedsayamdost, M. R. (2016). The siderophore metabolome of Azotobacter vinelandii. Appl. Environ. Microbiol. 82, 27–39. doi: 10.1128/aem.03160-15
Bakulin, M. K., Grudtsyna, A. S., and Pletneva, A. (2007). Biological fixation of nitrogen and growth of bacteria of the genus Azotobacter in liquid media in the presence of perfluorocarbons. Prikl. Biokhim. Mikrobiol. 43, 443–449.
Bloch, S. E., Clark, R., Gottlieb, S. S., Wood, L. K., Shah, N., Mak, S. M., et al. (2020). Biological nitrogen fixation in maize: optimizing nitrogenase expression in a root-associated diazotroph. J. Exp. Bot. 71, 4591–4603. doi: 10.1093/jxb/eraa176
Chou, M. F., and Schwartz, D. (2011). Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinformatics Chapter 13, Unit 13.15–24.
Cox, J., and Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. doi: 10.1038/nbt.1511
Curatti, L., Brown, C. S., Ludden, P. W., and Rubio, L. M. (2005). Genes required for rapid expression of nitrogenase activity in Azotobacter vinelandii. Proc. Nat. Acad. Sci. U.S.A. 102, 6291–6296. doi: 10.1073/pnas.0501216102
Danyal, K., Shaw, S., Page, T. R., Duval, S., Horitani, M., Marts, A. R., et al. (2016). Negative cooperativity in the nitrogenase Fe protein electron delivery cycle. Proc. Nat. Acad. Sci. U.S.A. 113, E5783–E5791.
Das, H. K. (2019). Azotobacters as biofertilizer. Adv. Appl. Microbiol. 108, 1–43. doi: 10.1016/bs.aambs.2019.07.001
Deng, L., Michelle, L., Jingjie, Y., Pakrasi, H. B., Maitrayee, B. P., and Papoutsakis, E. T. (2018). Engineering nitrogen fixation activity in an oxygenic phototroph. mBio 9:e01029-18.
Einsle, O., Tezcan, F. A., and Andrade, S. L. A. (2002). Nitrogenase MoFe-protein at 1.16 a resolution: a central ligand in the FeMo-cofactor. Science 297, 1696–1700. doi: 10.1126/science.1073877
Fisher, D. J., Hayes, A. L., and Jones, C. A. (1978). Effects of some surfactant fungicides on Rhizobium trtfolii and its symbiotic relationship with white clover. Ann. Appl. Biol. 90, 73–84. doi: 10.1111/j.1744-7348.1978.tb02612.x
Gawin, A., Tietze, L., Aarstad, O. A., Aachmann, F. L., Brautaset, T., and Ertesvag, H. (2020). Functional characterization of three Azotobacter chroococcum alginate-modifying enzymes related to the Azotobacter vinelandii AlgE mannuronan C-5-epimerase family. Sci. Rep. 10:12470.
Gil, J., Ramírez-Torres, A., and Encarnación-Guevara, S. (2017). Lysine acetylation and cancer: a proteomics perspective. J. Proteomics 150, 297–309. doi: 10.1016/j.jprot.2016.10.003
Hardy, R. W. F., Burns, R. C., and Holsten, R. D. (1973). Applications of the acetylene-ethylene assay for measurement of nitrogen fixation. Soil Biol. Biochem. 5, 47–81. doi: 10.1016/0038-0717(73)90093-x
Hong, Y. Y., Ma, Y. C., Zhou, Y. G., Gao, F., Liu, H. C., and Chen, S. F. (2009). Paenibacillus sonchi sp. nov., a nitrogen-fixing species isolated from the rhizosphere of Sonchus oleraceus. Int. J. Syst. Evol. Microbiol. 59, 2656–2661. doi: 10.1099/ijs.0.009308-0
Hu, Y., and Ribbe, M. W. (2013). Biosynthesis of the iron-molybdenum cofactor of nitrogenase. J. Biol. Chem. 288, 13173–13177.
Inès, M., and Dhouha, G. (2016). Glycolipid biosurfactants: main properties and potential applications in agriculture and food industry. J. Sci. Food Agric. 96, 4310–4320. doi: 10.1002/jsfa.7759
Jiang, J., Gai, Z., Wang, Y., Fan, K., Sun, L., Wang, H., et al. (2018). Comprehensive proteome analyses of lysine acetylation in tea leaves by sensing nitrogen nutrition. BMC Genomics 19:840. doi: 10.1186/s12864-018-5250-4
Kosono, S., Tamura, M., Suzuki, S., Kawamura, Y., Yoshida, A., Nishiyama, M., et al. (2015). Changes in the acetylome and succinylome of bacillus subtilis in response to carbon source. PLoS One 10:e0131169. doi: 10.1371/journal.pone.0131169
Kumar, U., Kaviraj, M., Panneerselvam, P., Priya, H., Chakraborty, K., Swain, P., et al. (2019). Ascorbic acid formulation for survivability and diazotrophic efficacy of Azotobacter chroococcum Avi2 (MCC 3432) under hydrogen peroxide stress and its role in plant-growth promotion in rice (Oryza sativa L.). Plant Physiol. Biochem. 139, 419–427. doi: 10.1016/j.plaphy.2019.04.003
Kurien, B. T., and Scofield, R. H. (2015). Western blotting: an introduction. Methods Mol. Biol. 1312, 17–30. doi: 10.1007/978-1-4939-2694-7_5
Lenart, A. M. (2012). In Vitro effects of various xenobiotics on Azotobacter chroococcum strains isolated from soils of southern Poland. J. Environ. Sci. Health B. 47, 7–12. doi: 10.1080/03601234.2012.601942
Lesur, A., Schmit, P. O., Bernardin, F., Letellier, E., Brehmer, S., Decker, J., et al. (2021). Highly multiplexed targeted proteomics acquisition on a TIMS-QTOF. Anal. Chem. 93, 1383–1392. doi: 10.1021/acs.analchem.0c03180
Li, H., Li, Y., Sun, T., Du, W., Zhang, Z., Li, D., et al. (2020). Integrative proteome and acetylome analyses of murine responses to Cryptococcus neoformans infection. Front. Microbiol. 11:575. doi: 10.3389/fmicb.2020.00575
Loginov, D. S., Fiala, J., Chmelik, J., Brechlin, P., Kruppa, G., and Novak, P. (2021). Benefits of ion mobility separation and parallel accumulation-serial fragmentation technology on timsTOF Pro for the needs of fast photochemical oxidation of protein analysis. ACS Omega 6, 10352–10361. doi: 10.1021/acsomega.1c00732
Macek, B., Forchhammer, K., Hardouin, J., Weber-Ban, E., Grangeasse, C., and Mijakovic, I. (2019). Protein post-translational modifications in bacteria. Nat. Rev. Microbiol. 17, 651–664.
Meier, F., Brunner, A. D., Koch, S., Koch, H., Lubeck, M., Krause, M., et al. (2018). Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteomics 17, 2534–2545. doi: 10.1074/mcp.tir118.000900
Mitra, R., Das, H. K., and Dixit, A. (2005). Identification of a positive transcription regulatory element within the coding region of the nifLA operon in Azotobacter vinelandii. Appl. Environ. Microbiol. 71, 3716–3724. doi: 10.1128/aem.71.7.3716-3724.2005
Mus, F., Alleman, A. B., Pence, N., Seefeldt, L. C., and Peters, J. W. (2018). Exploring the alternatives of biological nitrogen fixation. Metallomics 10, 523–538. doi: 10.1039/c8mt00038g
Neuer, G., and Bothe, H. (1985). Electron donation to nitrogenase in heterocysts of cyanobacteria. Arch. Microbiol. 143, 185–191. doi: 10.1007/bf00411045
Orikasa, Y., Nodasaka, Y., Ohyama, T., Okuyama, H., Ichise, N., Yumoto, I., et al. (2010). Enhancement of the nitrogen fixation efficiency of genetically-engineered Rhizobium with high catalase activity. J. Biosci. Bioeng. 110, 397–402. doi: 10.1016/j.jbiosc.2010.04.007
Peralta, H., Mora, Y., Salazar, E., Encarnacion, S., Palacios, R., and Mora, J. (2004). Engineering the nifH promoter region and abolishing poly-β-hydroxybutyrate accumulation in Rhizobium etli enhance nitrogen fixation in symbiosis with Phaseolus vulgaris. Appl. Environ. Microbiol. 70, 3272–3281. doi: 10.1128/aem.70.6.3272-3281.2004
Rebelein, J. G., Lee, C. C., Newcomb, M., Hu, Y., and Ribbe, M. W. (2018). Characterization of an M-cluster-substituted nitrogenase VFe protein. mBio 9:e00310-18.
Ren, S., Yang, M., Yue, Y., Ge, F., Li, Y., Guo, X., et al. (2018). Lysine succinylation contributes to aflatoxin production and pathogenicity in Aspergillus flavus. Mol. Cell. Proteomics 17, 457–471. doi: 10.1074/mcp.ra117.000393
Rettberg, L. A., Wilcoxen, J., Jasniewski, A. J., Lee, C. C., Tanifuji, K., Hu, Y., et al. (2020). Identity and function of an essential nitrogen ligand of the nitrogenase cofactor biosynthesis protein NifB. Nat. Commun. 11:1757.
Ribbe, M. W., Hu, Y., Hodgson, K. O., and Hedman, B. (2014). Biosynthesis of nitrogenase metalloclusters. Chem. Rev. 114, 4063–4080. doi: 10.1021/cr400463x
Rizvi, A., and Khan, M. S. (2018). Heavy metal induced oxidative damage and root morphology alterations of maize (Zea mays L.) plants and stress mitigation by metal tolerant nitrogen fixing Azotobacter chroococcum. Ecotoxicol. Environ. Saf. 157, 9–20. doi: 10.1016/j.ecoenv.2018.03.063
Romero-Perdomo, F., Abril, J., Camelo, M., Moreno-Galvan, A., Pastrana, I., Rojas-Tapias, D., et al. (2017). Azotobacter chroococcum as a potentially useful bacterial biofertilizer for cotton (Gossypium hirsutum): effect in reducing N fertilization. Rev. Argent. Microbiol. 49, 377–383. doi: 10.1016/j.ram.2017.04.006
Sachdev, D. P., and Cameotra, S. S. (2013). Biosurfactants in agriculture. Appl. Microbiol. Biotechnol. 97, 1005–1016. doi: 10.1007/s00253-012-4641-8
Sarkar, A., and Reinhold-Hurek, B. (2014). Transcriptional profiling of nitrogen fixation and the role of NifA in the diazotrophic endophyte Azoarcus sp. strain BH72. PLoS One 9:e86527. doi: 10.1371/journal.pone.0086527
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shi, H. W., Wang, L. Y., Li, X. X., Liu, X. M., Hao, T. Y., He, X. J., et al. (2016). Genome-wide transcriptome profiling of nitrogen fixation in Paenibacillus sp. WLY78. BMC Microbiol. 16:25. doi: 10.1186/s12866-016-0642-6
Sickerman, N. S., Rettberg, L. A., Lee, C. C., Hu, Y., and Ribbe, M. W. (2017). Cluster assembly in nitrogenase. Essays Biochem. 61, 271–279. doi: 10.1042/ebc20160071
Song, Y., Liu, J., and Chen, F. (2020). Azotobacter chroococcum inoculation can improve plant growth and resistance of maize to armyworm, Mythimna separata even under reduced nitrogen fertilizer application. Pest Manag. Sci. 76, 4131–4140. doi: 10.1002/ps.5969
Soufi, B., Krug, K., Harst, A., and Macek, B. (2015). Characterization of the E. coli proteome and its modifications during growth and ethanol stress. Front. Microbiol. 6:103. doi: 10.3389/fmicb.2015.00103
Spatzal, T., Aksoyoglu, M., Zhang, L., Andrade, S. L. A., Schleicher, E., Weber, S., et al. (2011). Evidence for interstitial carbon in nitrogenase FeMo cofactor. Science 334, 940–940. doi: 10.1126/science.1214025
Sumbul, A., Ansari, R. A., Rizvi, R., and Mahmood, I. (2020). Azotobacter: a potential bio-fertilizer for soil and plant health management. Saudi J. Biol. Sci. 27, 3634–3640. doi: 10.1016/j.sjbs.2020.08.004
Szklarczyk, D., Franceschini, A., Kuhn, M., Simonovic, M., Roth, A., Minguez, P., et al. (2011). The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568.
Thakur, P., Saini, N. K., Thakur, V. K., Gupta, V. K., Saini, R. V., and Saini, A. K. (2021). Rhamnolipid the glycolipid biosurfactant: emerging trends and promising strategies in the field of biotechnology and biomedicine. Microb. Cell Fact. 20:1. doi: 10.1007/978-981-13-1289-2_1
Wagner, G. R., and Payne, R. M. (2013). Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 288, 29036–29045. doi: 10.1074/jbc.m113.486753
Wang, H., Li, H., Zhang, M., Song, Y., Huang, J., Huang, H., et al. (2018). Carbon dots enhance the nitrogen fixation activity of Azotobacter chroococcum. ACS Appl. Mater. Interfaces 10, 16308–16314. doi: 10.1021/acsami.8b03758
Wang, T., Zhao, X., Shi, H., Sun, L., Li, Y., Li, Q., et al. (2018). Positive and negative regulation of transferred nif genes mediated by indigenous GlnR in Gram-positive Paenibacillus polymyxa. PLoS Genet. 14:e1007629. doi: 10.1371/journal.pgen.1007629
Weinert, B. T., Scholz, C., Wagner, S. A., Iesmantavicius, V., Su, D., Daniel, J. A., et al. (2013). Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4, 842–851. doi: 10.1016/j.celrep.2013.07.024
Xie, Z., Dai, J., Dai, L., Tan, M., Cheng, Z., Wu, Y., et al. (2012). Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics 11, 100–107. doi: 10.1074/mcp.m111.015875
Xu, X., Liu, T., Yang, J., Chen, L., Liu, B., Wei, C., et al. (2017). The first succinylome profile of Trichophyton rubrum reveals lysine succinylation on proteins involved in various key cellular processes. BMC Genomics 18:577. doi: 10.1186/s12864-017-3977-y
Yang, Y., and Gibson, G. E. (2019). Succinylation links metabolism to protein functions. Neurochem. Res. 44, 2346–2359. doi: 10.1007/s11064-019-02780-x
Zhang, H., Li, P., Ren, S., Cheng, Z., and Zhao, W. (2019). ScCobB2-mediated lysine desuccinylation regulates protein biosynthesis and carbon metabolism in Streptomyces coelicolor. Mol. Cell. Proteomics 18, 2003–2017. doi: 10.1074/mcp.ra118.001298
Zhang, Z., Tan, M., Xie, Z., Dai, L., Chen, Y., and Zhao, Y. (2011). Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63. doi: 10.1038/nchembio.495
Keywords: nitrogen fixation, Azotobacter chroococcum, posttranslational modifications, lysine succinylation (KSuc), rhamnolipid
Citation: Li J, Pan H, Yang H, Wang C, Liu H, Zhou H, Li P, Li C, Lu X and Tian Y (2021) Rhamnolipid Enhances the Nitrogen Fixation Activity of Azotobacter chroococcum by Influencing Lysine Succinylation. Front. Microbiol. 12:697963. doi: 10.3389/fmicb.2021.697963
Received: 20 April 2021; Accepted: 28 June 2021;
Published: 30 July 2021.
Edited by:
Harold J. Schreier, University of Maryland, Baltimore County, United StatesReviewed by:
Xiangmin Lin, Fujian Agriculture and Forestry University, ChinaLuis M. Rubio, Center for Plant Biotechnology and Genomics, National Institute of Agricultural and Food Research and Technology, Spain
Copyright © 2021 Li, Pan, Yang, Wang, Liu, Zhou, Li, Li, Lu and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangyang Lu, eGlhbmd5YW5nY25AMTYzLmNvbQ==; Yun Tian, dGlhbnl1bkBodW5hdS5lZHUuY24=
 Jin Li1
Jin Li1 Yun Tian
Yun Tian