- 1Fisheries College, Jimei University, Xiamen, China
- 2College of Environment and Public Health, Xiamen Huaxia University, Xiamen, China
- 3Key Laboratory of Special Aquatic Feed for Fujian, Fujian Tianma Technology Company Limited, Fuzhou, China
Pseudomonas plecoglossicida is the causative agent of “visceral white spot disease” in cultured fish and has resulted in serious economic losses. tonB gene plays a crucial role in the uptake of nutrients from the outer membranes in Gram-negative bacteria. The previous results of our lab showed that the expression of tonB gene of P. plecoglossicida was significantly upregulated in the spleens of infected Epinephelus coioides. To explore the effect of tonB gene on the virulence of P. plecoglossicida and the immune response of E. coioides, tonB gene of P. plecoglossicida was knocked down by RNAi; and the differences between the wild-type strain and the tonB-RNAi strain of P. plecoglossicida were investigated. The results showed that all of the four mutants of P. plecoglossicida exhibited significant decreases in mRNA of tonB gene, and the best knockdown efficiency was 94.0%; the survival rate of E. coioides infected with the tonB-RNAi strain was 20% higher than of the counterpart infected with the wild strain of P. plecoglossicida. Meanwhile, the E. coioides infected with the tonB-RNAi strain of P. plecoglossicida carried less pathogens in the spleen and less white spots on the surface of the spleen; compared with the wild-type strain, the motility, chemotaxis, adhesion, and biofilm formation of the tonB-RNAi strain were significantly attenuated; the transcriptome data of E. coioides infected with the tonB-RNAi strain were different from the counterpart infected with the wild strain of P. plecoglossicida; the antigen processing and presentation pathway and the complement and coagulation cascade pathway were the most enriched immune pathways. The results indicated that tonB was a virulence gene of P. plecoglossicida; tonB gene was involved in the regulation of motility, chemotaxis, adhesion, and biofilm formation; tonB gene affected the immune response of E. coioides to P. plecoglossicida infection.
Introduction
Pseudomonas plecoglossicida is the causative agent of “visceral white spot disease” in Epinephelus coioides and Larimichthys crocea under the water temperatures of 15–20°C and has resulted in high mortality and heavy economic loss (Zhang et al., 2018; Huang et al., 2020). To alleviate the harm caused by P. plecoglossicida, its pathogenic mechanism has attracted much attention. The pathogenicity of pathogens was reported to be controlled by different genes. So far, many genes have been shown to have a strong relationship with the virulence regulation of aquatic pathogens, such as sodA and sodB to Aeromonas hydrophila (Zhang et al., 2019); secA and cheB to Vibrio alginolyticus (Guo et al., 2018); ssaV to Edwardsiella piscicida (Edrees et al., 2018); toxA and toxB to Vibrio parahaemolyticus (Singhapol and Tinrat, 2020); and secY and tssD-1 to P. plecoglossicida (Luo et al., 2020; Ye et al., 2021). The previous transcriptome data (NCBI, SRP115064) of our lab showed that tonB gene of P. plecoglossicida was highly expressed in the spleen of E. coioides, which suggested that it might play a role in the virulence of P. plecoglossicida.
tonB gene encodes TonB, which is an element of the TonB system (TonB-ExbB-ExbD). The TonB system occupies a crucial position in the transport of nutrients, including iron, carbohydrates, hemin, transition metal elements, and vitamin B12 (Schauer et al., 2008; Huang and Wilks, 2017; Gomez-Santos et al., 2019). In Gram-negative bacteria, the TonB system and tonB-dependent transporter (TBDT) accomplish jointly transport of nutrients. TBDT can acquire energy to transport nutrients from the outer membrane with the help of the TonB system (Oeemig et al., 2018; Kopp and Postle, 2020; Samantarrai et al., 2020). Dong et al. (2019) found that the pathogenicity of A. hydrophila was attenuated due to the deletion of tonB gene. However, there have been no reports about P. plecoglossicida tonB gene.
Considering the great harms of P. plecoglossicida to aquaculture and the potentially important role of tonB in the pathogenicity of P. plecoglossicida, this article is devoted to exploring the contribution of tonB in the virulence of P. plecoglossicida and the immune response of E. coioides to P. plecoglossicida infection. tonB gene of P. plecoglossicida was stably knocked down by RNAi; the spleens at 3 and 5 day post injection (dpi) infected with the wild-type strain or the tonB-RNAi strain were sampled and subjected to RNA-seq to monitor the transcriptomes of E. coioides, and the transcriptome data were compared and analyzed.
Materials and Methods
Bacterial Strains and Culture Conditions
The virulent wild-type strain of P. plecoglossicida (NZBD9) was isolated from the spleen of large yellow croaker suffered from “visceral white spot disease” and was stored at −80°C (Huang et al., 2019). The tonB-RNAi strain of P. plecoglossicida was constructed from the wild-type strain of P. plecoglossicida. P. plecoglossicida was routinely grown in Luria Bertani (LB) broth under 18 or 28°C with shaking at 220 rpm. Escherichia coli DH5α was obtained from TransGen Biotech (Beijing, China) and grown in LB broth at 37°C with shaking at 220 rpm.
RNAi-Induced Knockdown of Pseudomonas plecoglossicida tonB Gene
Pseudomonas plecoglossicida tonB gene was knocked down according to the methods described by Choi and Schweizer (2006) and Darsigny et al. (2010), with minor modifications. Four oligonucleotides complementary to short hairpin RNA (shRNA) sequences targeting tonB gene (Supplementary Table 1) were designed by BLOCK-iTTMRNAi Designer1 and synthesized by Shanghai Generay Biotech Co., Ltd. (Shanghai, China). The oligonucleotides were ligated to pCM130/tac vector using T4 DNA ligase (Takara Biomedical Technology, Beijing, China), according to the manufacturer’s recommendations. The recombinant plasmids were transformed into competent E. coli DH5α cells by heat shock and electroporated into P. plecoglossicida NZBD9 as described by Luo et al. (2019). The mRNA level of tonB gene in four mutants was determined by quantitative real-time polymerase chain reaction (qRT-PCR).
Growth Rate Assay
The bacterial suspension were adjusted to optical density at a wavelength of 600 nm (OD600) = 0.3 and diluted 1,000-fold with LB broth. Aliquot of 200 μl of the bacterial diluent was added into per well of 96-well plate and incubated at 28°C. The OD600 values of bacterial culture were measured hourly for 48 h (Zuo et al., 2019). Ten replicates were carried out for each group.
Bacterial Chemotaxis Assay
The bacterial chemotaxis assay for P. plecoglossicida was performed with fine-tuning as described by Zhang et al. (2020). The overnight culture of P. plecoglossicida was adjusted to OD600 nm ≈ 1.0 with sterile phosphate-buffered saline (PBS), and 0.25 ml of bacterial suspension was aspirated into a 1-ml syringe. Then, a capillary tube (inner diameter of 0.1 mm, one end sealed) filled with mucus was dipped into the bacterial suspension and incubated at 28°C for 1 h. Finally, the mucus in the capillary was blown out, and the colony-forming unit (cfu) number of P. plecoglossicida in the mucus was determined by dilution method of plate counting. Three replicates were performed.
Biofilm Formation Assay
Biofilm formation assay of P. plecoglossicida was performed according to the method described by Mao et al. (2020), with some modifications. P. plecoglossicida at exponential growth period was adjusted to OD600 = 0.2 by fresh LB broth. Then, 100 μl of diluted bacterial suspension was added into per well of microtiter plate and incubated at 28°C for 24 h. After that, each well was washed twice with sterile PBS, dyed with 175 μl of crystal violet (0.1%) for 15 min, washed twice with sterile PBS, and air-dried. Finally, the stained biofilm was solubilized into 200 μl of 33% acetic acid, and the OD values of each well were measured at 590 nm. Eight independent replicates were performed.
Motility Assay
The soft agar plate motility assay for P. plecoglossicida was performed as described by Zhang et al. (2019), with minor modifications. The overnight culture in LB was diluted with PBS and adjusted to OD600 = 0.3. Bacterial suspension of 1 μl was inoculated on LB semisolid agar plates supplemented with 0.4% agar and incubated at 28°C for 16–20 h. The diameters of bacterial colonies were measured. Biological replicates were carried out in triplicate for each group.
Adhesion Assay
The bacterial adhesion assay was conducted according to the method depicted by Guo et al. (2018), with some modifications. Aliquot of 20 μl of sterile mucus was spread uniformly over the 22 × 22 mm glass slide area. After the mucus was air-dried, 4% methanol was used to fix mucus for 30 min. Bacterial suspension of 200 μl (OD600 = 0.3) was spread equably on the region of mucus on glass slides. Sterile PBS of 200 μl instead of bacterial suspension was used as the negative control. After incubation at 28°C for 2 h in a damp chamber, the slides were washed three times with PBS to remove the un-adhered bacterial cells. The adhering bacterial cells on the slide were fixed by 200 μl of 4% methanol for 30 min and dyed with 0.1% crystal violet for 3 min. After the unstained crystal violet were washed with PBS, the adhering bacterial cells in 10 randomly selected fields were counted under a microscope (×1,000). Five replicates were performed.
Epinephelus coioides Infection and Sampling
All E. coioides infection experiments were executed completely following the proposals in the “Guide for the Care and Use of Laboratory Animals” set by the National Research Council (Copyright 1996 by the National Academy of Sciences). The animal protocols were officially ratified by the Animal Ethics Committee of Jimei University (Acceptance No. JMULAC201159).
Healthy weight-matched E. coioides were obtained from Zhangzhou (Fujian, China) and were acclimatized at 18 ± 1°C for 10 days in recirculating aquaculture systems.
For survival assay, size-matched E. coioides (60 fish per group) were intraperitoneally injected with the wild-type strain or tonB-RNAi strain of P. plecoglossicida at a dose of 5 × 104 cfu/fish. In addition, 60 E. coioides intraperitoneally injected with PBS were used as the negative control. The daily mortality of experimental fish was observed and recorded until 10 dpi.
For RNA-seq, six spleens of three different groups (wild-type strain group, tonB-RNAi strain group, and PBS group) were sampled at 3 and 5 dpi, and two spleens were mixed into one sample. All of the samples were sent to Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China) for sequencing.
For the pathogen load assay and tonB expression assay, six spleens of E. coioides intraperitoneally infected with the wild-type strain or tonB-RNAi strain of P. plecoglossicida were randomly sampled at 1, 2, 3, 4, 5, and 6 dpi. P. plecoglossicida cultured at 18°C in vitro was considered as the control.
Quantitative Real-Time Polymerase Chain Reaction
Quantitative real-time polymerase chain reaction was performed by a QuantStudio 6 Flex real-time PCR system (Life Technologies, Carlsbad, CA, United States). Primers are synthesized by Xiamen Borui Biotechnology, and primer sequences are provided in Supplementary Table 2. The 16S rDNA (Sun et al., 2019) was applied to normalize tonB gene expression levels of P. plecoglossicida. The pathogen load of P. plecoglossicida in the infected spleens was assessed by the copy number of housekeeping gene gyrB (Xin et al., 2020). The relative expression of gene in different groups was calculated using 2–Δ Δ CT method (Luo et al., 2020).
Transcriptomic Analysis
A TruSeqTM RNA sample preparation Kit (Illumina, San Diego, CA, United States) was used to prepare the RNA-seq libraries under protocols of the Kit. The RNA quality was detected and quantified by the Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA, United States) and the ND-2000 instrument (NanoDrop Technologies, Thermo Fisher Scientific, Waltham, MA, United States) separately. The high-quality RNA samples [OD260/280 = 2.03–2.09, RNA integrity number (RIN) ≥ 9.0, 28S:18S ≥ 1.0, ≥1.6 μg) met the construction of individual sequencing libraries. The rRNA-depleted RNA samples were fragmented in fragmentation buffer, and cDNA synthesis was carried out with protocols supplied with the SuperScript double-stranded cDNA synthesis kit (Invitrogen, Carlsbad, CA, United States). The cDNA libraries were amplified by Phusion DNA polymerase (NEB) after end repair, phosphorylation, and poly(A) addition. Sequencing was performed on the Illumina HiSeq4000 sequencing platform at Majorbio Biotech Co., Ltd. (Shanghai, China). The trimming and quality control of the raw Illumina reads were performed using SeqPrep2 and Sickle3 with the default settings. The mapped reads were used for de novo assembly as the unigenes of E. coioides.
To investigate the biological processes, the BLAST2GO software4 was used for the Gene Ontology (GO) annotation (Conesa et al., 2005), which accomplished the molecular annotation of differentially expressed transcripts of E. coioides. The differentially expressed mRNAs (DEMs) met the standards [| log2FC| ≥ 1 and false discovery rate (FDR) < 0.05] was deemed as significant. The GO enrichment analysis visualization of E. coioides transcriptome data was performed by the clusterProfiler R package (Yu et al., 2012). Finally, metabolic pathways were analyzed with Kyoto Encyclopedia of Genes and Genomes (KEGG) (Okuda et al., 2008). Furthermore, 10 genes were randomly selected from E. coioides to verify the reliability of RNA-seq by qRT-PCR (Supplementary Figure 1).
Statistical Analyses
The experimental data are showed as means ± SD and dissected with one-way ANOVA followed by Dunnett’s test using IBM SPSS Statistics 26.0 (Armonk, NY, United States). p < 0.05 was considered as statistically significant.
Data Access
The RNA sequencing results were put aside in the GenBank SRA database under accession number SRP315640.
Results
Effect of RNAi on tonB mRNA Level of Pseudomonas plecoglossicida
After shRNA sequence design and synthesis, recombinant plasmid construction, and electrical transfer, four RNAi mutant strains (tonB-RNAi-419, tonB-RNAi-424, tonB-RNAi-663, and tonB-RNAi-675) were successfully constructed. The results of qRT-PCR showed that tonB gene mRNA levels of four mutants were lower than those of the wild-type strain of P. plecoglossicida (Figure 1A). The mutant named tonB-RNAi-663 (hereafter called the tonB-RNAi strain) exhibited the best silencing efficiency (94.0%) and was chosen for further research.
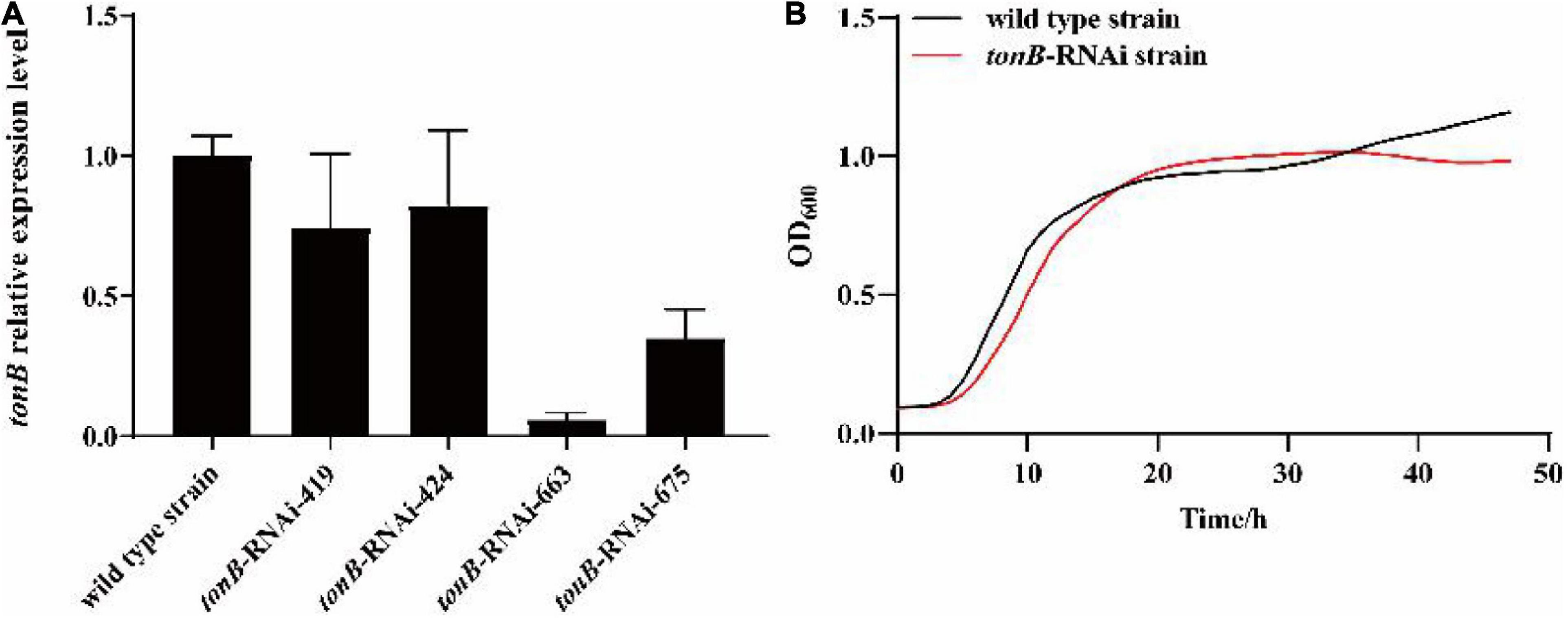
Figure 1. Construction and growth curve of the tonB-RNAi strain of Pseudomonas plecoglossicida. (A) The tonB mRNA levels of four mutant strains. (B) Growth curve of the wild-type strain and tonB-RNAi strain.
Effects of tonB Gene Silencing on the Growth of Pseudomonas plecoglossicida
The growth curve of the tonB-RNAi strain and wild-type strain of P. plecoglossicida under the same culture conditions was determined. The results illustrated that there was no significant difference between the growth rate of the tonB-RNAi strain and wild-type strain of P. plecoglossicida (Figure 1B).
Effects of tonB Gene Silencing on the Characteristics of Pseudomonas plecoglossicida
The motility, chemotaxis, and adhesion of the tonB-RNAi strain of P. plecoglossicida were enervated compared with the wild-type strain; and the motility, chemotaxis, and adhesion of tonB have weakened by 18.17% (Figures 2A,E), 39.99% (Figure 2B), and 59.34% (Figures 2C,F), respectively, at corresponding periods. Meanwhile, the tonB-RNAi strain of P. plecoglossicida showed an extremely significant difference (p < 0.001) in biofilm formation as compared with the wild-type strain. When the expression of tonB was inhibited, the biofilm formation ability of P. plecoglossicida decreased at 38.35% (Figure 2D).
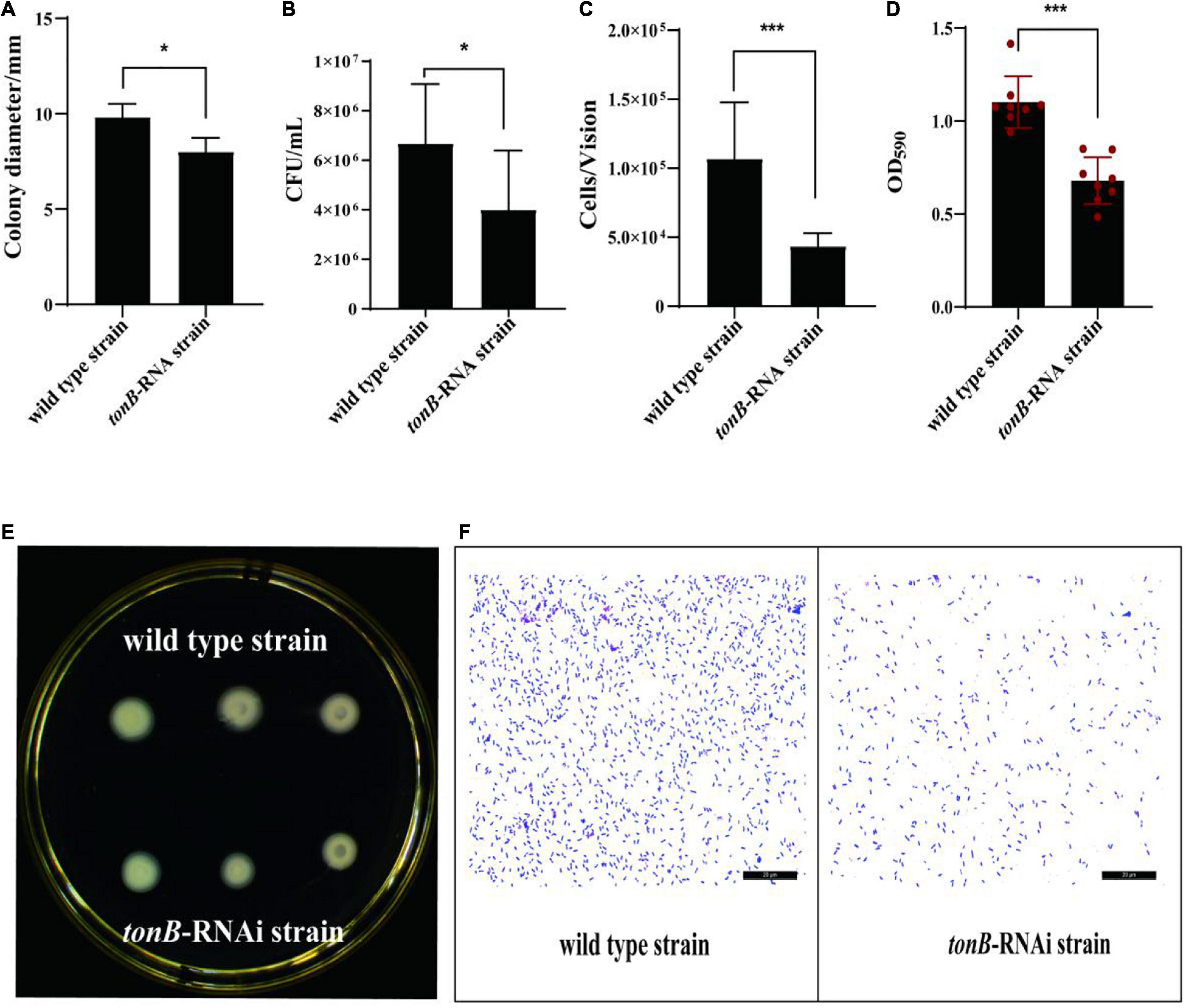
Figure 2. The characteristics of two strains of Pseudomonas plecoglossicida. (A) Colony diameters of wild-type strain and tonB-RNAi strain. (B) Chemotaxis capacity of wild-type strain and tonB-RNAi strain. (C) Adhesion ability of wild-type strain and tonB-RNAi strain. (D) Biofilm formation ability of wild-type strain and tonB-RNAi strain. (E) Colonies of wild-type strain and tonB-RNAi strain on a plate. (F) Adhering bacteria under the microscope. Data are presented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
Effects of tonB Gene Silencing on the Virulence of Pseudomonas plecoglossicida
Infection of the wild-type strain of P. plecoglossicida caused the death of E. coioides. The first death was recorded at 2 dpi, and the mortality reached 100% at 7.5 dpi. Infection of same dose of the tonB-RNAi strain of P. plecoglossicida resulted in 1 day delay in the time of first death and 20% decrease in cumulative mortality. No death of E. coioides injected with PBS was recorded (Figure 3A).
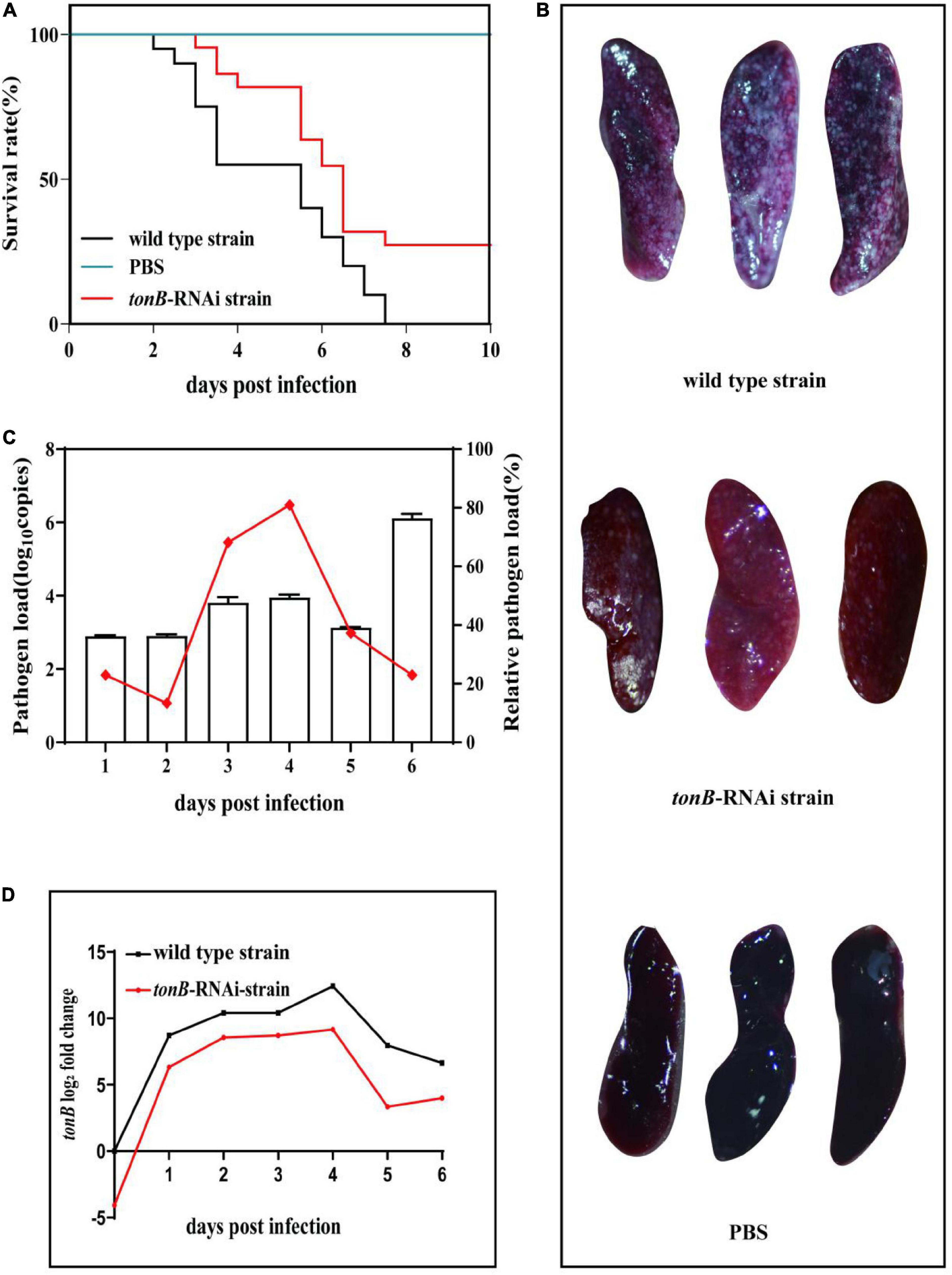
Figure 3. Pathogenicity of two strains of Pseudomonas plecoglossicida to Epinephelus coioides. (A) Survival rate of E. coioides infected with wild-type strain or tonB-RNAi strain. (B) Symptoms of E. coioides spleen after infection of wild-type strain or tonB-RNAi strain. (C) Pathogen load of tonB-RNAi strain of P. plecoglossicida in the spleens of E. coioides during infection. The bar graph represents the pathogen load and is represented by the copy number of gyrB gene; the line graph represents the relative pathogen load, which is represented by (copy number of gyrB gene of tonB-RNAi strain/copy number of gyrB gene of wild-type strain). (D) mRNA level of tonB gene of P. plecoglossicida in E. coioides spleen during infection.
The surface of E. coioides spleens injected with the wild-type strain of P. plecoglossicida was covered with numerous white nodules, while much fewer white nodules were found on the surface of counterpart spleens infected with the tonB-RNAi strain, and no white nodule was found on the surface of counterpart spleens injected with PBS (Figure 3B).
The pathogen loads of the tonB-RNAi strain of P. plecoglossicida were always lower than those of the wild-type strain during the whole infection. The relative pathogen load (pathogen load of the tonB-RNAi strain/pathogen load of the wild-type strain) of P. plecoglossicida peaked at 4 dpi. Although the tonB-RNAi strain had a higher pathogen load at 6 dpi than on other times, the relative pathogen load at 6 dpi was lower than that on other times except that at 2 dpi (Figure 3C). The mRNA level of tonB gene of P. plecoglossicida in the spleen of E. coioides was always higher than that in vitro, and the highest values were recorded at 4 dpi. Simultaneously, the expression levels of tonB in the tonB-RNAi strain were always lower than those in the wild-type strain (Figure 3D).
The Effects of tonB Gene on the Immune Response of Epinephelus coioides to Pseudomonas plecoglossicida Infection
Quality Control of RNA-Seq Data
The E. coioides’s spleens were subjected to RNA-seq after being infected with the tonB-RNAi strain or wild-type strain of P. plecoglossicida. The A/T/G/C base content distribution was balanced, and N% met the normative range (Supplementary Figure 2). The main criterion of evaluating the quality of reads was Q20, which fulfilled the requirement of sequencing data of each sample (Q20 > 98%) (Supplementary Figure 3). The base error rate of the sequencing data was <0.1%. Pearson’s correlation coefficients (r) showed that three biological replicate samples were closely correlated (r > 0.9) (Supplementary Figure 4). The quality of sequence data satisfied the requirements for the subsequent data process and analysis steps.
Analysis of Differentially Expressed mRNAs
DESeq2 was used for the analysis of significantly differentially expressed transcripts of E. coioides. The criteria of the statistically significant of mRNA expression level changes in the transcriptome data were FDR < 0.05 and | log2FC| ≥ 1. GO and KEGG pathway enrichment analyses were performed for DEMs.
The first analyzed transcriptome data from the E. coioides’s spleens were sampled at 3 dpi. There exist 375 DEMs between the spleen infected with tonB-RNAi strains and wild-type strains of P. plecoglossicida, which included 291 upregulated mRNAs and 84 downregulated mRNAs (Figure 4). Further KEGG analysis showed that 20 KEGG pathways involved in immune response were enriched (Figure 5). Of the total DEMs, 26.67% were enriched in immune system pathways, including Toll and Imd signaling pathway, Intestinal immune network for IgA production, B-cell receptor signaling pathway, and antigen processing and presentation. The antigen processing and presentation pathway (KO ID: ko04612) was significantly enriched based on the p-value of Fisher’s exact test.
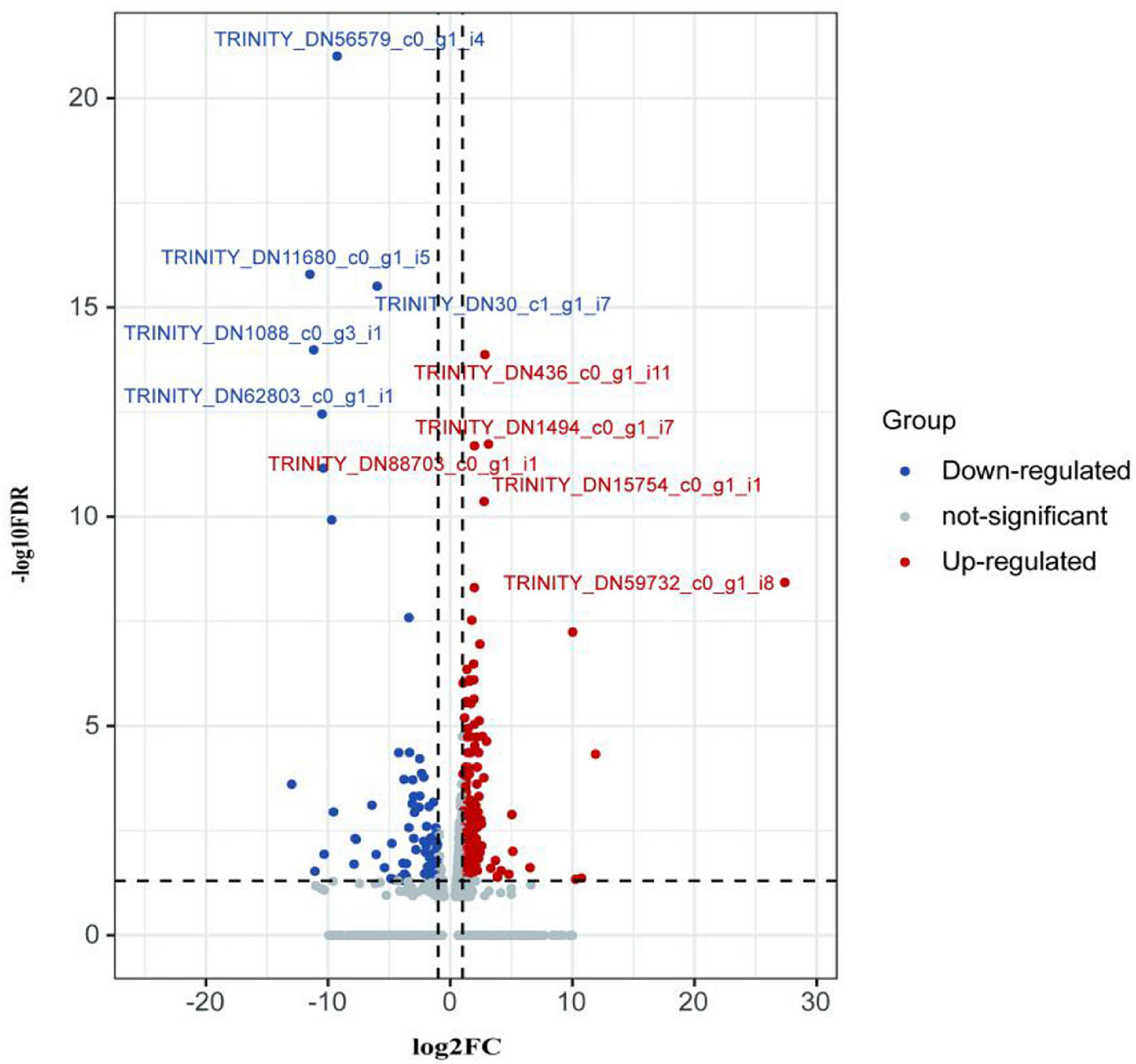
Figure 4. Volcano plot obtained from the DESeq2 analysis of Epinephelus coioides’s spleen RNA pools (3 dpi).
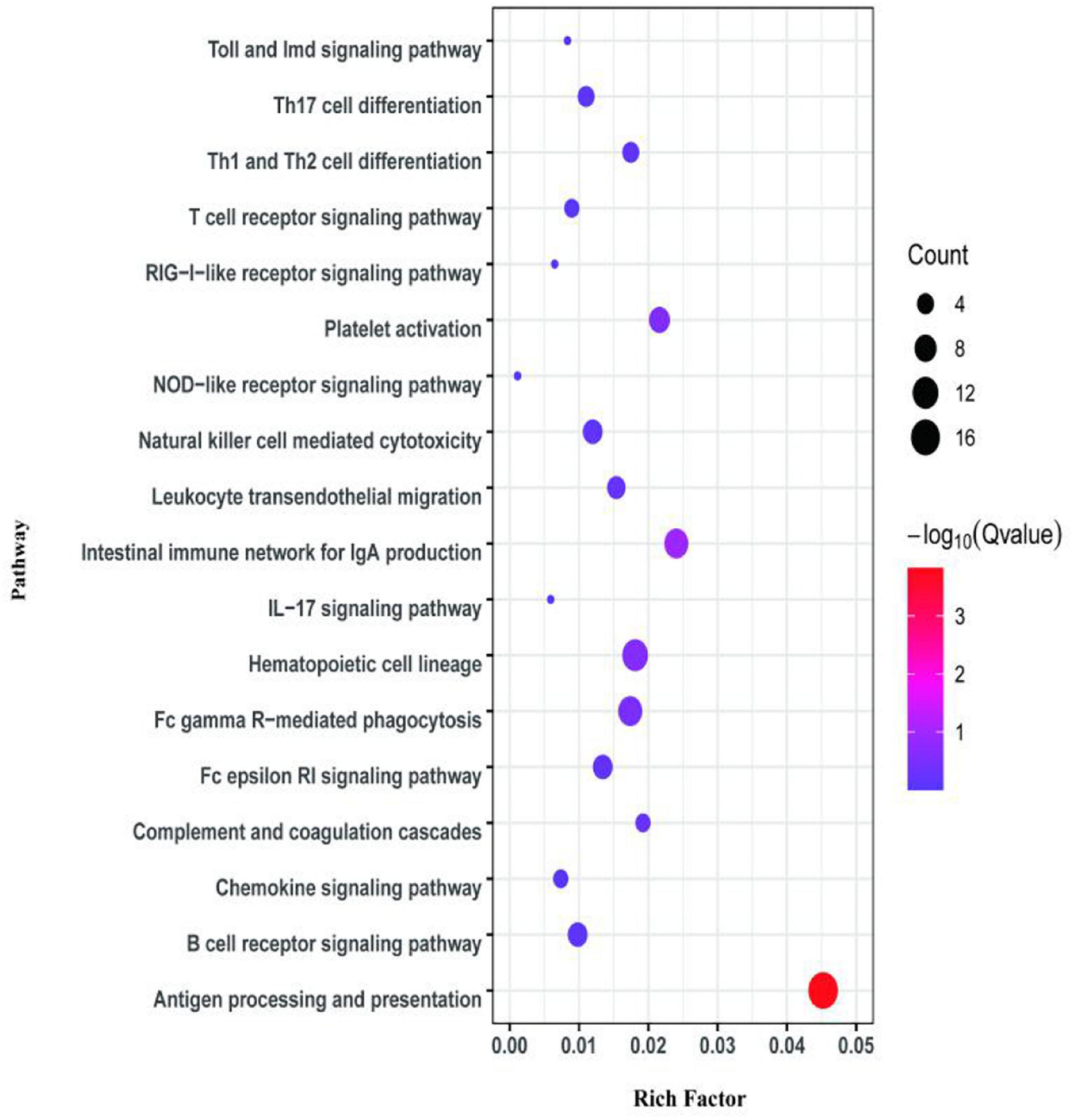
Figure 5. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis for differentially expressed mRNAs (DEMs) of transcriptome at 3 dpi.
The second analyzed transcriptome data from the E. coioides’s spleens were sampled at 5 dpi. A total of 218 mRNAs collected from the spleen infected with the tonB-RNAi strain of P. plecoglossicida were identified as significant differences in expression compared with the spleen infected with the wild-type strain, which included 43 upregulated mRNAs and 175 downregulated mRNAs (Figure 6). According to the GO annotation conventions, DEMs fall into three categories: biological processes, cellular components, and molecular functions. A total of 117 GO terms were enriched, including 29 significantly enriched GO terms. The 29 notably enriched GO terms of the three categories were selected for statistical analysis. The GO analysis results showed that tonB gene had a great influence on the immune system, because more than half of the immune GO terms were enriched in biological processes (Figure 7). Of the DEMs, 19.3% were enriched in immune GO terms. According to the KEGG database, 145 KEGG pathways were enriched, including 14 immune-related KEGG pathways. Of the total DEMs, 35.78% were enriched in immune system pathways (Figure 8). Complement and coagulation cascade pathway (ko04610), which included the greatest number of DEMs, was significantly enriched according to the p-value of Fisher’s exact test.
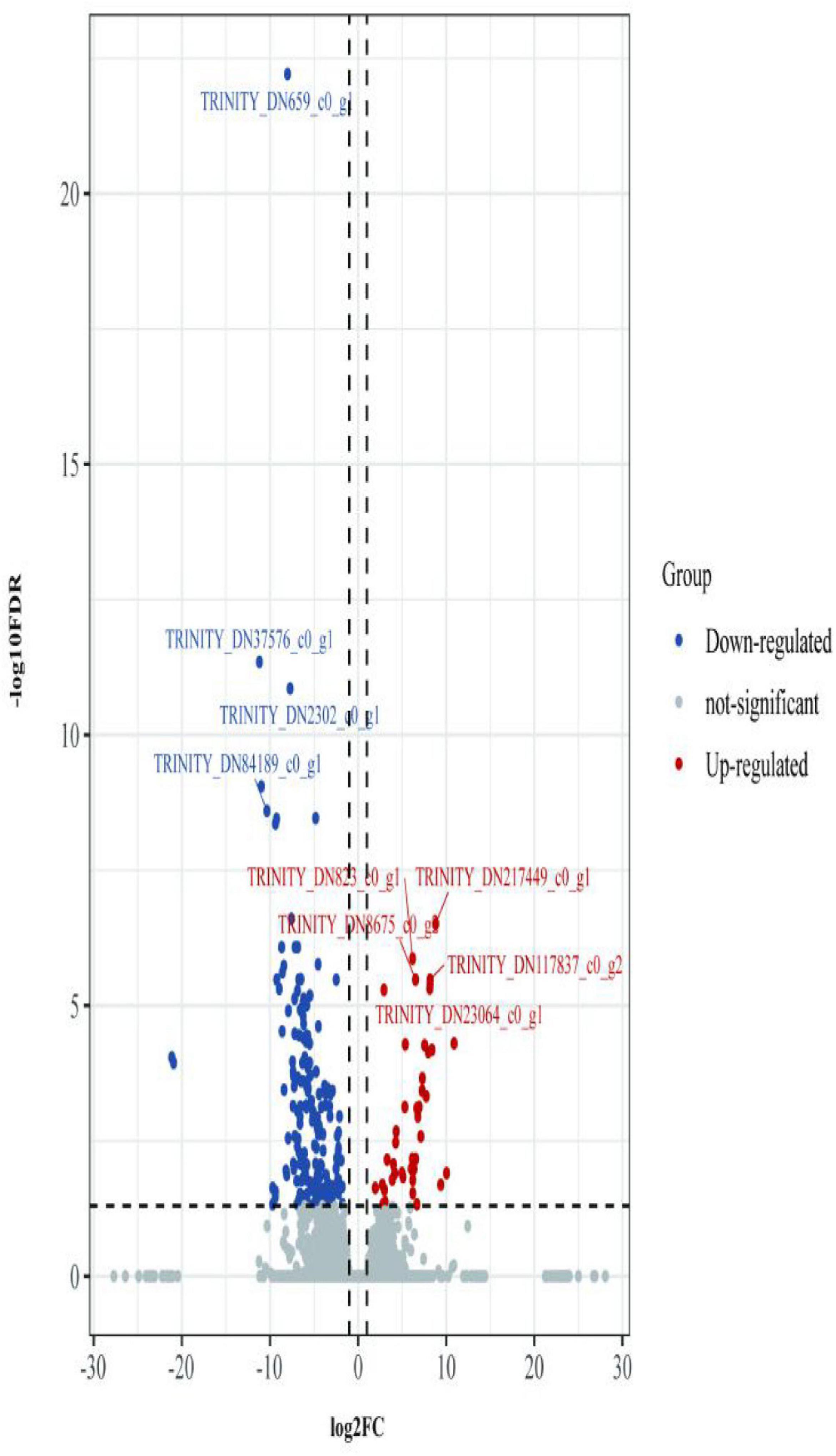
Figure 6. Volcano plot obtained from the DESeq2 analysis of Epinephelus coioides’s spleen RNA pools (5 dpi).
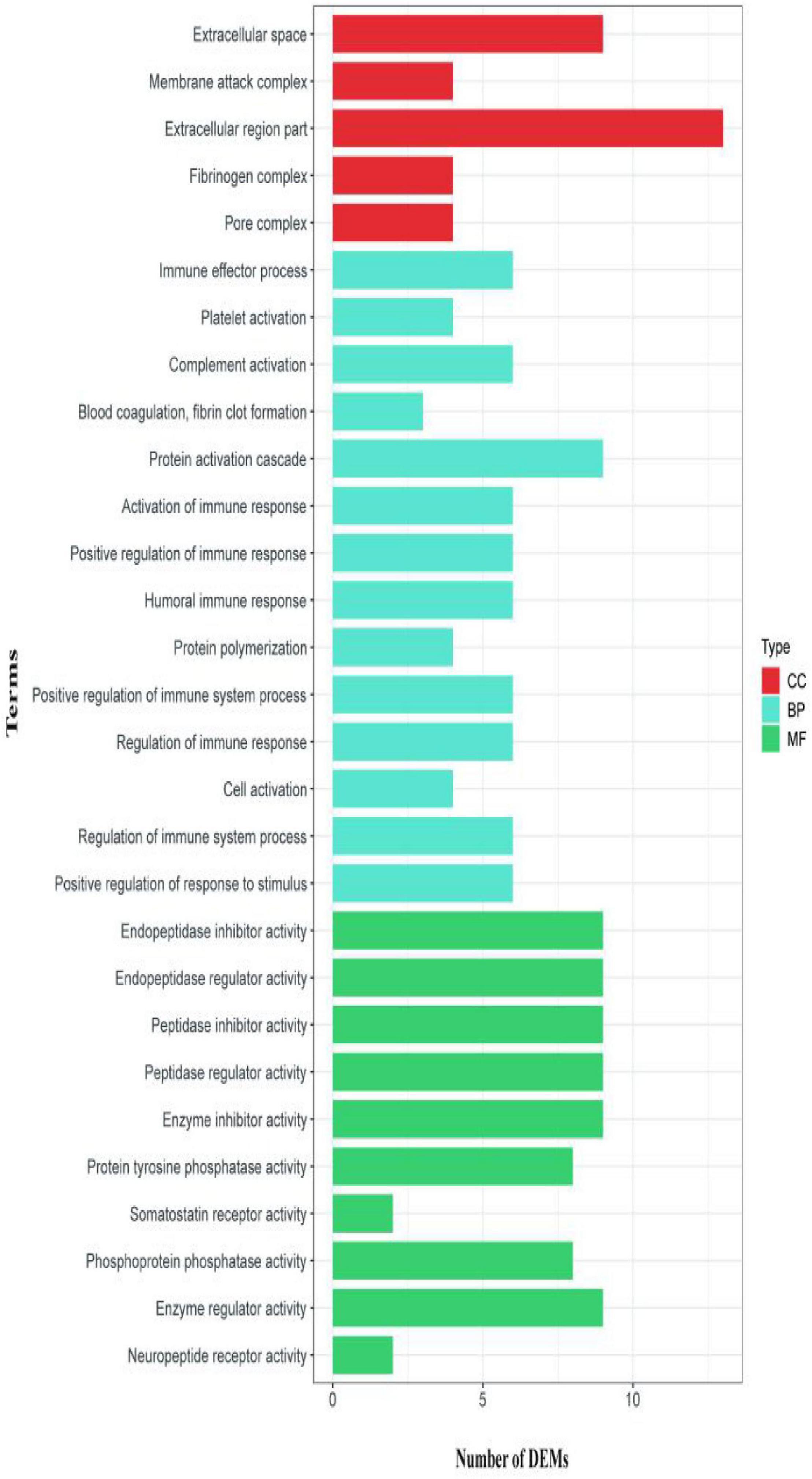
Figure 7. Gene Ontology (GO) enrichment analysis for differentially expressed mRNAs (DEMs) of transcriptome at 5 dpi.
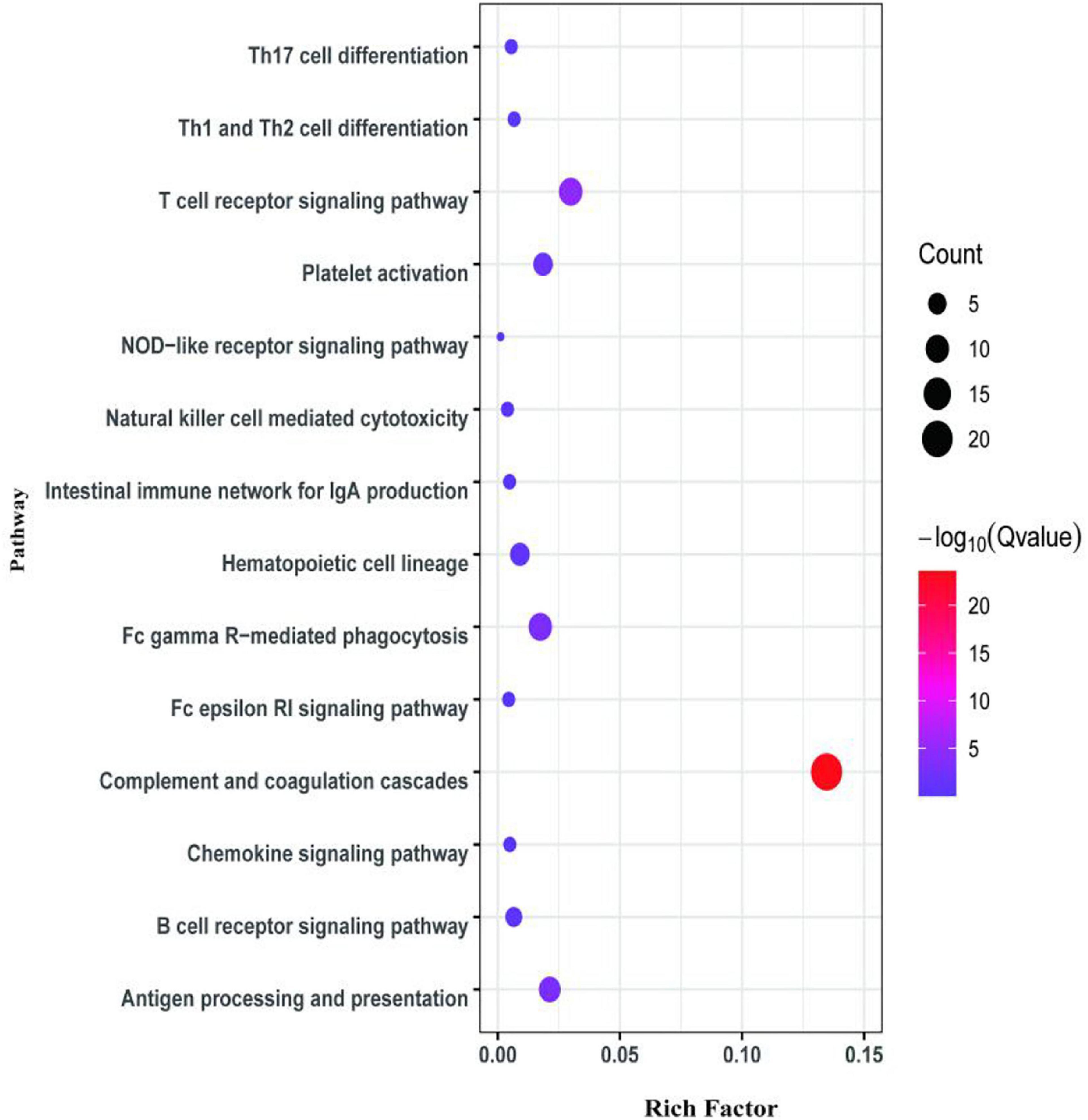
Figure 8. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis for differentially expressed mRNAs (DEMs) of the second transcriptome (5 dpi).
The most significant enrichment pathways were complement and coagulation cascades pathway and antigen processing and presentation pathway according to the KEGG enrichment analysis of the transcriptome data that from the E. coioides’s spleens were sampled at 3 and 5 dpi, respectively. Antigen processing and presentation pathway enriched 17 DEMs significantly, which including six downregulated DEMs (such as MHCII and TAP1/2) and 11 upregulated DEMs (such as MHCI, AEP, CTSB/L/S, TCR, TAPBP, and HSP70) (Figure 9A). Complement and coagulation cascades pathway enriched 21 downregulated DEMs significantly (such as FI, C3, C5, C6, C7, C8, and C9) (Figure 9B).
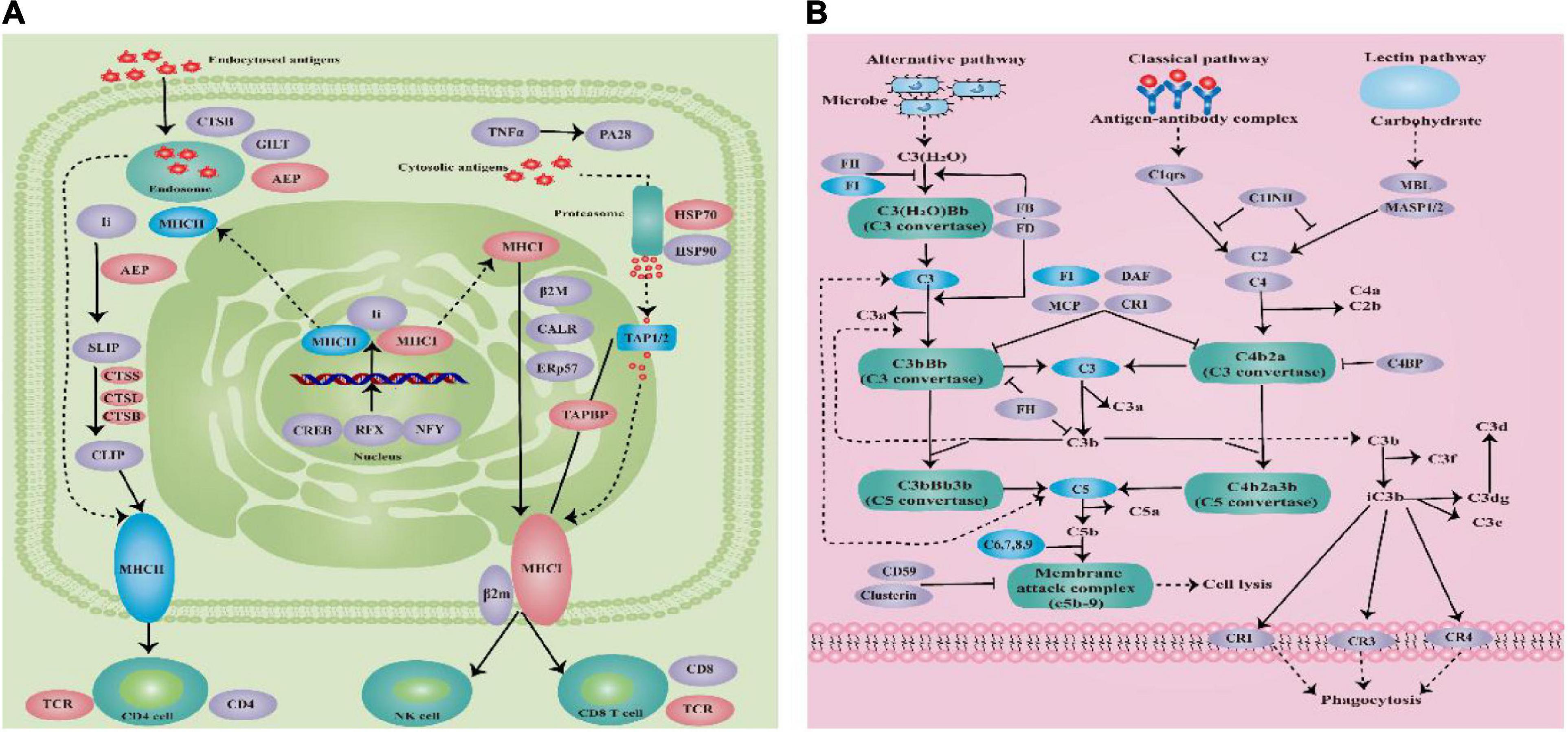
Figure 9. (A) Schematic diagram of antigen processing and presentation pathway. (B) Schematic diagram of complement and coagulation cascades pathway (blue indicates downregulation, red indicates upregulation, and purple indicates no significant change).
Discussion
RNA interference has been used for many fields to explore the functions of genes (Belles, 2010; Cooper et al., 2021). In this study, four shRNAs exhibited different silence efficiencies to tonB gene, which were consistent with the previous RNAi results (Liu et al., 2020; Xin et al., 2020). The best silence efficiency of four shRNAs to tonB gene was 94%, which was higher than that of pvdE gene (Xin et al., 2020) but lower than that of impB gene (Liu et al., 2020). The stability of gene silencing is crucial to the study of gene function. In the present study, tonB gene in the tonB-RNAi strain of P. plecoglossicida was persistently silenced during the infection process, and the relative expressions of tonB gene in the tonB-RNAi strain were always lower than those of the wild-type strain. These results indicated that RNAi of tonB gene was reliable and laid the foundation for subsequent research.
The TonB protein, encoded by tonB gene, has amalgamated to form the TonB system with ExbB and ExbD proteins (Liao et al., 2015). Abdelhamed et al. (2017) investigated the virulence of tonB gene of Edwardsiella ictaluri, and their findings show that the E. ictaluri mutant defective in tonB has a 2.16-fold reduction in virulence as compared with the wild-type of E. ictaluri. Wang et al. (2008) found that the LD50 value of three tonB mutant strains of V. alginolyticus was increased by 11-fold, 14-fold, and 25-fold, respectively. RNAi of tonB gene caused a 20% increase in the survival rate of E. coioides to P. plecoglossicida infection and had no significant effect on the growth rate of P. plecoglossicida, which suggested that the decline in mortality of E. coioides was due to the decrease of P. plecoglossicida virulence, not due to the decrease of bacterial growth rate. These results suggested that tonB gene contributed to the pathogenicity of P. plecoglossicida. These results agreed with previous studies of tonB gene contribution to the virulence in other bacterial strains. The different influence degrees of tonB gene on the pathogenic of different bacteria might be due to the bacterial species, the host species, and the conditions of infection. The results of pathogen load and symptoms agreed with the mortality result. Several genes have been verified to associate with the virulence of P. plecoglossicida (Luo et al., 2019). The silencing of some of these genes in P. plecoglossicida resulted in lower mortality in experimental fish infected with the mutants (Tang et al., 2019; Wang et al., 2019), and silencing of other virulence genes did not result in the death of E. coioides infected with the mutants of P. plecoglossicida (Liu et al., 2020; Xin et al., 2020).
RNAi of tonB gene attenuated bacterial motility, chemotaxis, adhesion, and biofilm formation of P. plecoglossicida. Bacterial motility (Luo et al., 2016), chemotaxis (Zhang et al., 2020), adhesion (Guo et al., 2018), and biofilm formation (Zhang et al., 2019) have been demonstrated to be related to the pathogenic of pathogen. Some studies showed that tonB mutant had great influences on motility, adhesion, and in vivo virulence due to its ability of helping the formation of type IV pili (Huang et al., 2004; Duong-Nu et al., 2016).
Nowadays, transcriptome analysis is an important way to reveal the mechanisms of host immune response to infection (Wu and Chen, 2016; Song et al., 2017; Li et al., 2021). In this study, compared with the transcriptome data from E. coioides infected with the wild strain of P. plecoglossicida, significant changes of transcriptome were observed in the spleen of E. coioides infected with the tonB-RNAi strain. The GO analysis results showed that the tonB-RNAi strain had a great influence on the immune system, and more than half of the immune pathways were enriched in biological processes. The KEGG pathway analysis results showed that the most significant pathways for enrichment were complement and coagulation cascades pathway and antigen processing and presentation pathway.
Complement and coagulation cascades pathway is an immune defense mechanism of the host, and the complement cascades are closely associated with coagulation cascades to jointly achieve effective protection of the host (Lupu et al., 2014; Wiegner et al., 2016). It is widely known that the activation and generation of C3a and C5a have serious proinflammatory effects (Pontrelli et al., 2020). Compared with the transcriptome data from E. coioides infected with the wild-type strain of P. plecoglossicida, all of the DEMs in transcriptome data from E. coioides infected with the tonB-RNAi strain were significantly downregulated in the complement and coagulation cascades pathway, which indicated that the inflammatory reaction significantly declined in E. coioides infected with the tonB-RNAi strain of P. plecoglossicida.
The antigen processing and presentation pathway plays a crucial role in immunological process, which is presented by major histocompatibility complexes (MHCs) (Park and Jae-Hwan, 2011; Wieczorek et al., 2017). MHCI relies on proteasomal proteolysis to present foreign peptides that come from degradation of endocellular microbial pathogens; MHCII depends upon lysosomal degradation to accomplish the presentation and processing of extracellular antigens (Wilson, 2017). Therefore, the determining factor of the successful pathogen elimination obviously depends on the recognition of MHCs in the immune system (Unanue, 2002). Compared with the transcriptome data from E. coioides infected with the wild-type strain of P. plecoglossicida, most of the DEMs in transcriptome data from E. coioides infected with the tonB-RNAi strain related with MHCI and MHCII were significantly upregulated in the antigen processing and presentation pathway. These results indicated that E. coioides’s immune system could more efficiently identify the tonB-RNAi strain of P. plecoglossicida, which might cause the immune system to remove the mutant strain more efficiently.
Conclusion
In conclusion, tonB is a virulence gene of P. plecoglossicida; tonB gene is involved in the regulation of bacterial motility, chemotaxis, adhesion, and biofilm formation of P. plecoglossicida; RNAi of tonB gene significantly affected the immune response of E. coioides to P. plecoglossicida infection; complement and coagulation cascades pathway and antigen processing and presentation pathway are the most affected immune pathways.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of Jimei University (Acceptance No. JMULAC201159).
Author Contributions
All authors contributed to the article. QY, LL, ZZ, and XW conceived the experiments. LFH, LL, and LZ conducted the experiments. All authors assisted in the collection and interpretation of data. LFH, ZZ, and QY wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China under contract No. 31972836, open fund of Fujian Province Key Laboratory of Special Aquatic Formula Feed under contract No. TMKJZ2002, and the Fund of Fujian Key Laboratory of Functional Aquafeed and Culture Environment Control (FACE20200007).
Conflict of Interest
JZ and QY were employed by company Fujian Tianma Technology Company Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.720967/full#supplementary-material
Supplementary Figure 1 | Verification of transcriptome data with qRT-PCR.
Supplementary Figure 2 | Bases content along raw reads.
Supplementary Figure 3 | Mean errors distribution along raw reads.
Supplementary Figure 4 | Correlation of transcriptional data.
Supplementary Table 1 | The sequences of four shRNAs for tonB gene.
Supplementary Table 2 | The sequence of Primers for PCR and qRT-PCR.
Footnotes
- ^ http://rnaidesigner.thermofisher.com/rnaiexpress/setOption.do?designOption=shrna&pid=708587103220684543
- ^ https://github.com/jstjohn/SeqPrep
- ^ https://github.com/najoshi/sickle
- ^ https://www.blast2go.com/
References
Abdelhamed, H., Lawrence, M. L., and Karsi, A. (2017). The role of Edwardsiella ictaluri TonB mutant in Edwardsiella ictaluri virulence. Front. Physiol. 8:1066. doi: 10.3389/fphys.2017.01066
Belles, X. (2010). Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu. Rev. Entomol. 55, 111–128. doi: 10.1093/bfgp/elp052
Choi, K.-H., and Schweizer, H. P. (2006). mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1, 153–161. doi: 10.1038/nprot.2006.24
Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Cooper, A. M. W., Song, H., Shi, X., Yu, Z., Lorenzen, M., Silver, K., et al. (2021). Characterization, expression patterns, and transcriptional responses of three core RNA interference pathway genes from Ostrinia nubilalis. J. Insect Physiol. 129, 104181–104181. doi: 10.1016/j.jinsphys.2020.104181
Darsigny, M., Babeu, J.-P., Seidman, E. G., Gendron, F.-P., Levy, E., Carrier, J., et al. (2010). Hepatocyte nuclear factor-4α promotes gut neoplasia in mice and protects against the production of reactive oxygen species. Cancer Res. 70, 9423–9433. doi: 10.1158/0008-5472.CAN-10-1697
Dong, Y., Geng, J., Liu, J., Pang, M., Awan, F., Lu, C., et al. (2019). Roles of three TonB systems in the iron utilization and virulence of the Aeromonas hydrophila Chinese epidemic strain NJ-35. Appl. Microbiol. Biotechnol. 103, 4203–4215. doi: 10.1007/s00253-019-09757-4
Duong-Nu, T.-M., Jeong, K., Hong, S. H., Hong-Vu, N., Van-Hoan, N., Min, J.-J., et al. (2016). All three TonB systems are required for Vibrio vulnificus CMCP6 tissue invasiveness by controlling flagellum expression. Infect. Immun. 84, 254–265. doi: 10.1128/iai.00821-15
Edrees, A., Abdelhamed, H., Nho, S. W., Park, S. B., Karsi, A., Austin, F. W., et al. (2018). Construction and evaluation of type III secretion system mutants of the catfish pathogen Edwardsiella piscicida. J. Fish Dis. 41, 805–816. doi: 10.1111/jfd.12784
Gomez-Santos, N., Glatter, T., Koebnik, R., Swiatek-Polatynska, M. A., and Sogaard-Andersen, L. (2019). A TonB-dependent transporter is required for secretion of protease PopC across the bacterial outer membrane. Nat. Commun. 10:1360.
Guo, L., Huang, L., Su, Y., Qin, Y., Zhao, L., and Yan, Q. (2018). secA, secD, secF, yajC, and yidC contribute to the adhesion regulation of Vibrio alginolyticus. Microbiologyopen 7:e00551. doi: 10.1002/mbo3.551
Huang, B. X., Ru, K., Yuan, Z., Whitchurch, C. B., and Mattick, J. S. (2004). tonB3 is required for normal twitching motility and extracellular assembly of type IV pili. J. Bacteriol. 186, 4387–4389. doi: 10.1128/jb.186.13.4387-4389.2004
Huang, L., Zhao, L., Qi, W., Xu, X., Zhang, J., Zhang, J., et al. (2020). Temperature-specific expression of cspA1 contributes to activation of sigX during pathogenesis and intracellular survival in Pseudomonas plecoglossicida. Aquaculture 518:734861. doi: 10.1016/j.aquaculture.2019.734861
Huang, L., Zuo, Y., Jiang, Q., Su, Y., Qin, Y., Xu, X., et al. (2019). A metabolomic investigation into the temperature-dependent virulence of Pseudomonas plecoglossicida from large yellow croaker (Pseudosciaena crocea). J. Fish Dis. 42, 431–446. doi: 10.1111/jfd.12957
Huang, W., and Wilks, A. (2017). Extracellular heme uptake and the challenge of bacterial cell membranes. Annu. Rev. Biochem. 86, 799–823. doi: 10.1146/annurev-biochem-060815-014214
Kopp, D. R., and Postle, K. (2020). The intrinsically disordered region of ExbD is required for signal transduction. J. Bacteriol. 202:e00687–19.
Li, C., Jiang, J., Xie, J., Yang, W., and Wang, Y. (2021). Transcriptome profiling and differential expression analysis of the immune-related genes during the acute phase of infection with Mycobacterium marinum in the goldfish (Carassius auratus L.). Aquaculture 533:736198. doi: 10.1016/j.aquaculture.2020.736198
Liao, H., Liu, M., and Cheng, A. (2015). Structural features and functional mechanism of TonB in some Gram-negative bacteria-A review. Wei Sheng Wu Xue Bao 55, 529–536.
Liu, Z., Zhao, L., Huang, L., Qin, Y., Zhang, J., Zhang, J., et al. (2020). Integration of RNA-seq and RNAi provides a novel insight into the immune responses of Epinephelus coioides to the impB gene of Pseudomonas plecoglossicida. Fish Shellfish Immunol. 105, 135–143. doi: 10.1016/j.fsi.2020.06.023
Luo, G., Huang, L., Su, Y., Qin, Y., Xu, X., Zhao, L., et al. (2016). flrA, flrB, and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerg. Microbes and Infect. 5, e85. doi: 10.1038/emi.2016.82
Luo, G., Sun, Y., Huang, L., Su, Y., Zhao, L., Qin, Y., et al. (2020). Time-resolved dual RNA-seq of tissue uncovers Pseudomonas plecoglossicida key virulence genes in host-pathogen interaction with Epinephelus coioides. Environ. Microbiol. 22, 677–693. doi: 10.1111/1462-2920.14884
Luo, G., Zhao, L., Xu, X., Qin, Y., Huang, L., Su, Y., et al. (2019). Integrated dual RNA-seq and dual iTRAQ of infected tissue reveals the functions of a diguanylate cyclase gene of Pseudomonas plecoglossicida in host-pathogen interactions with Epinephelus coioides. Fish Shellf. Immunol. 95, 481–490. doi: 10.1016/j.fsi.2019.11.008
Lupu, F., Keshari, R. S., Lambris, J. D., and Coggeshall, K. M. (2014). Crosstalk between the coagulation and complement systems in sepsis. Thromb. Res. 133, S28–S31.
Mao, L., Qin, Y., Kang, J., Wu, B., Huang, L., Wang, S., et al. (2020). Role of LuxR-type regulators in fish pathogenic Aeromonas hydrophila. J. Fish Dis. 43, 215–225. doi: 10.1111/jfd.13114
Oeemig, J. S., Ollila, O. H. S., and Iwai, H. (2018). NMR structure of the C-terminal domain of TonB protein from Pseudomonas aeruginosa. PeerJ 6:e5412. doi: 10.7717/peerj.5412
Okuda, S., Yamada, T., Hamajima, M., Itoh, M., Katayama, T., Bork, P., et al. (2008). KEGG atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 36, W423–W426.
Park, H.-L., and Jae-Hwan, N. (2011). Regulation of innate immunity via MHC class II-mediated signaling; non-classical role of MHC class II in innate immunity. J. Bacteriol. Virol. 41, 205–207. doi: 10.4167/jbv.2011.41.3.205
Pontrelli, P., Grandaliano, G., and Van Kooten, C. (2020). Editorial: kidney transplantation and innate immunity. Front. Immunol. 11:603982. doi: 10.3389/fimmu.2020.603982
Samantarrai, D., Sagar, A. L., Gudla, R., and Siddavattam, D. (2020). TonB-dependent transporters in sphingomonads: unraveling their distribution and function in environmental adaptation. Microorganisms 8:359. doi: 10.3390/microorganisms8030359
Schauer, K., Rodionov, D. A., and de Reuse, H. (2008). New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 33, 330–338. doi: 10.1016/j.tibs.2008.04.012
Singhapol, C., and Tinrat, S. (2020). Virulence genes analysis of Vibrio parahaemolyticus and anti-vibrio activity of the citrus extracts. Curr. Microbiol. 77, 1390–1398. doi: 10.1007/s00284-020-01941-4
Song, X., Hu, X., Sun, B., Bo, Y., Wu, K., Xiao, L., et al. (2017). A transcriptome analysis focusing on inflammation-related genes of grass carp intestines following infection with Aeromonas hydrophila. Sci. Rep. 7:40777.
Sun, Y., Zhuang, Z., Wang, X., Huang, H., Fu, Q., and Yan, Q. (2019). Dual RNA-seq reveals the effect of the flgM gene of Pseudomonas plecoglossicida on the immune response of Epinephelus coioides. Fish Shellfish Immunol. 87, 515–523. doi: 10.1016/j.fsi.2019.01.041
Tang, Y., Sun, Y., Zhao, L., Xu, X., Huang, L., Qin, Y., et al. (2019). Mechanistic insight into the roles of Pseudomonas plecoglossicida clpV gene in host-pathogen interactions with Larimichthys crocea by dual RNA-seq. Fish Shellf. Immunol. 93, 344–353. doi: 10.1016/j.fsi.2019.07.066
Unanue, E. R. (2002). Perspective on antigen processing and presentation. Immunol. Rev. 185, 86–102. doi: 10.1034/j.1600-065x.2002.18510.x
Wang, L., Sun, Y., Zhao, L., Xu, X., Huang, L., Qin, Y., et al. (2019). Dual RNA-seq uncovers the immune response of Larimichthys crocea to the secY gene of Pseudomonas plecoglossicida from the perspective of host-pathogen interactions. Fish Shellf. Immunol. 93, 949–957. doi: 10.1016/j.fsi.2019.08.040
Wang, Q., Liu, Q., Cao, X., Yang, M., and Zhang, Y. (2008). Characterization of two TonB systems in marine fish pathogen Vibrio alginolyticus: their roles in iron utilization and virulence. Arch. Microbiol. 190, 595–603. doi: 10.1007/s00203-008-0407-1
Wieczorek, M., Abualrous, E. T., Sticht, J., Alvaro-Benito, M., Stolzenberg, S., Noe, F., et al. (2017). Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front. Immunol. 8:292. doi: 10.3389/fimmu.2017.00292
Wiegner, R., Chakraborty, S., and Huber-Lang, M. (2016). Complement-coagulation crosstalk on cellular and artificial surfaces. Immunobiology 221, 1073–1079. doi: 10.1016/j.imbio.2016.06.005
Wilson, A. B. (2017). MHC and adaptive immunity in teleost fishes. Immunogenetics 69, 521–528. doi: 10.1007/s00251-017-1009-3
Wu, C.-C., and Chen, B.-S. (2016). A systems biology approach to the coordination of defensive and offensive molecular mechanisms in the innate and adaptive host-pathogen interaction networks. PLoS One 11:e0149303. doi: 10.1371/journal.pone.0149303
Xin, G., Wang, F., Zhao, L., Qin, Y., Huang, L., and Yan, Q. (2020). Integration of RNA-seq and RNAi provides a novel insight into the effect of pvdE gene to the pathogenic of Pseudomonas plecoglossicida and on the immune responses of orange-spotted grouper (Epinephelus coioides). Aquaculture 529:735695. doi: 10.1016/j.aquaculture.2020.735695
Ye, H., Xu, Z., Tao, Z., Li, W., Li, Y., Yang, A., et al. (2021). Efficacy and safety of Pseudomonas plecoglossicida mutant Delta tssD-1 as a live attenuated vaccine for the large yellow croaker (Larimichthys crocea). Aquaculture 531, 1–7.
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. doi: 10.1089/omi.2011.0118
Zhang, B., Luo, G., Zhao, L., Huang, L., Qin, Y., Su, Y., et al. (2018). Integration of RNAi and RNA-seq uncovers the immune responses of Epinephelus coioides to L321_RS19110 gene of Pseudomonas plecoglossicida. Fish Shellfish Immunol. 81, 121–129. doi: 10.1016/j.fsi.2018.06.051
Zhang, M., Kang, J., Wu, B., Qin, Y., Huang, L., Zhao, L., et al. (2020). Comparative transcriptome and phenotype analysis revealed the role and mechanism of ompR in the virulence of fish pathogenic Aeromonas hydrophila. Microbiologyopen 9:e1041.
Zhang, M., Qin, Y., Huang, L., Yan, Q., Mao, L., Xu, X., et al. (2019). The role of sodA and sodB in Aeromonas hydrophila resisting oxidative damage to survive in fish macrophages and escape for further infection. Fish Shellfish Immunol. 88, 489–495. doi: 10.1016/j.fsi.2019.03.021
Keywords: Pseudomonas plecoglossicida, tonB, pathogenicity, Epinephelus coioides, immune response
Citation: Hu L, Zhao L, Zhuang Z, Wang X, Fu Q, Huang H, Lin L, Huang L, Qin Y, Zhang J and Yan Q (2021) The Effect of tonB Gene on the Virulence of Pseudomonas plecoglossicida and the Immune Response of Epinephelus coioides. Front. Microbiol. 12:720967. doi: 10.3389/fmicb.2021.720967
Received: 05 June 2021; Accepted: 21 July 2021;
Published: 16 August 2021.
Edited by:
Xinhua Chen, Fujian Agriculture and Forestry University, ChinaReviewed by:
Xiangmin Lin, Fujian Agriculture and Forestry University, ChinaMinh-Thu Nguyen, Institute of Medical Microbiology, University Hospital Münster, Germany
Copyright © 2021 Hu, Zhao, Zhuang, Wang, Fu, Huang, Lin, Huang, Qin, Zhang and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingpi Yan, eWFucXBAam11LmVkdS5jbg==; Lili Lin, bGlubGxAaHh4eS5lZHUuY24=
 Lingfei Hu
Lingfei Hu Lingmin Zhao1
Lingmin Zhao1 Lixing Huang
Lixing Huang Yingxue Qin
Yingxue Qin Qingpi Yan
Qingpi Yan