- State Key Laboratory of Crop Stress Biology for Arid Areas and College of Plant Protection, Northwest A&F University, Yangling, China
Dicer proteins are mainly responsible for generating small RNAs (sRNAs), which are involved in gene silencing in most eukaryotes. In previous research, two DCL proteins in Valsa mali, the pathogenic fungus causing apple tree Valsa canker, were found associated with both the pathogenicity and generation of sRNAs. In this study, the differential expression of small interfering RNAs (siRNAs) and miRNA-like RNAs (milRNAs) was analyzed based on the deep sequencing of the wild type and Vm-DCL2 mutant, respectively. Overall, the generation of 40 siRNAs and 18 milRNAs was evidently associated with Vm-DCL2. The target genes of milRNAs were then identified using degradome sequencing; according to the prediction results, most candidate targets are related to pathogenicity. Further, expression of Vm-PC-3p-92107_6 was confirmed in the wild type but not in the Vm-DCL2 mutant. Moreover, the pathogenicity of Vm-PC-3p-92107_6 deletion mutants (ΔVm-PC-3p-92107_6) and the over-expression transformants (Vm-PC-3p-92107_6-OE) was significantly increased and decreased, respectively. Based on those degradome results, vacuolar protein sorting 10 (Vm-VPS10) was identified as the target of Vm-PC-3p-92107_6. Co-expression analysis in tobacco leaves further confirmed that Vm-PC-3p-92107_6 could suppress the expression of Vm-VPS10. Meanwhile, the expression levels of Vm-PC-3p-92107_6 and Vm-VPS10 displayed divergent trends in ΔVm-PC-3p-92107_6 and Vm-PC-3p-92107_6-OE, respectively. Perhaps most importantly, ΔVm-VPS10 featured a significant reduction in pathogenicity. Taken together, our results indicate that a DCL2-dependent milRNA Vm-PC-3p-92107_6 plays roles in pathogenicity by regulating the expression of Vm-VPS10. This study lays a foundation for the comprehensive analysis of pathogenic mechanisms of V. mali and deepens our understanding of the generation and function of fungal sRNA.
Introduction
RNA interference (RNAi) is a conserved gene silencing mechanism in most eukaryotes (Dahlmann and Kueck, 2015). In the RNAi pathway, Dicer or Dicer-like (DCL) proteins are responsible for cutting long double-stranded RNAs, or those RNAs with a typical stem-loop structure, into small RNAs (sRNAs; Shabalina and Koonin, 2008). These sRNAs are then loaded into AGO proteins, after which the guide strand sRNA directs the RNA-induced silencing complex (RISC) to match and cleave the complementary mRNAs or suppress their translation (Fire et al., 1998; Chang et al., 2012). There are two major regulatory classes of sRNAs: short interfering RNAs (siRNAs) and microRNAs (miRNAs; Gent et al., 2010). The siRNAs generally guide gene silencing by binding perfectly to the complementary mRNAs. In addition to degrading the target gene’s mRNA, miRNAs can also suppress the translation of that target gene, but miRNAs do not need to be perfect complementarity to the mRNA (Nakayashiki et al., 2006). Both siRNAs and miRNAs play important roles in most eukaryotes with respect to the growth, development, and response to biotic or abiotic stresses (Doench et al., 2003; Ghildiyal and Zamore, 2009).
Fungi are fundamental evolved branch of eukaryotic organisms. Given that core RNAi components have been found in a wide range of fungal species, corresponding functional RNAi pathways may also exist in fungi (Mochizuki and Gorovsky, 2005). Previous studies have shown that Dicer proteins might be involved in various biological processes in fungi. The association of Dicer proteins with siRNA production and vegetative growth of mycelia were confirmed in both Neurospora crassa and Mucor circinelloides (Catalanotto et al., 2004; Nicolas et al., 2007, 2010). Yet the functioning of DCL proteins was redundant in N. crassa (Catalanotto et al., 2004) while their functions in M. circinelloides were divergent. For example, DCL1 is mainly responsible for regulating vegetative development and other biological functions of mycelia, while DCL2 is mainly responsible for generating sRNAs (Nicolas et al., 2007; de Haro et al., 2009).
The generation of sRNAs is essential because they act as the source switch of RNAi. It is now widely accepted that generation mechanisms of sRNAs are very complex in fungi (Lee et al., 2010; Jin et al., 2019). In N. crassa, the siRNAs could be generated in both Dicer-dependent and Dicer-independent pathways (Lee et al., 2010). Further, the production of milR-1 was completely dependent on Dicer, QDE-2, QIP, and MRPL3, while that of milR-2 did not depend on Dicers but did require QDE-2; conversely, the production of milR-3 was completely dependent on Dicers but the biogenesis of milR-4 depended only partly on Dicers (Lee et al., 2010). Moreover, in Verticillium dahlia, the biogenesis of VdmilR1 requires an RNase III domain-containing protein VdR3, but not Dicer-like or Argonaute proteins (Jin et al., 2019).
Since the discovery of sRNAs in fungi, some of their various functions have been revealed. In N. crassa, sRNAs play key roles in genome defense and gene regulation via post-transcriptional gene silencing activity (Fulci and Macino, 2007). Nonetheless, sRNAs could also affect the vegetative growth, pathogenicity, and toxin synthesis of certain fungi. For example, the milRNAs in Penicillium marneffei are capable of regulating the growth and development of mycelia, in addition to their participation in fungal pathogenicity and hormone secretion (Lau et al., 2013). Another role of milRNAs is to regulate mycotoxin biosynthesis and mycelium growth, as demonstrated in Aspergillus flavus (Bai et al., 2015), while a study of Curvularia lunata revealed that its milRNAs might contribute to pathogen infection and mycelial growth (Liu et al., 2016). In Fusarium oxysporum f. sp. niveum, the Fon-miR7696a-3p and Fon-miR6108a were found associated with trichothecene and NEP1 biosynthesis (Jiang et al., 2017). Recently, the milR236 of Magnaporthe oryzae was found able to influence both appressorium formation and pathogenicity by regulating the expression of the histone acetyltransferase gene MoHat1 (Li et al., 2020). Other works have shown that PstmilR1 of Puccinia striiformis sp. tritici could inhibit the plant immune response by suppressing the expression of PR2 in a cross-kingdom RNAi pathway (Wang et al., 2017). More importantly, several DCL2-dependent milRNAs in Fusarium graminearum are known to be relevant to sexual reproduction (Zeng et al., 2018). In the fungus V. dahliae, a Dicer-independent milRNA, VdmilR1, was shown to regulate its pathogenicity by promoting the histone H3K9 methylation of VdHy1 and transcriptional inhibition of the 3′ UTR of the protein-coding gene VdHy1 (Jin et al., 2019). Nevertheless, the generation and functional mechanism of sRNAs in fungi are still largely unknown.
Valsa mali (Cytospora spp.) is a critical species of ascomycete as it causes the severest of apple tree trunk disease (Wang et al., 2014). Previous studies revealed that key RNAi components DCL and AGO proteins are involved in the stress responses and pathogenicity of this fungal species (Feng et al., 2017a,b). In particular, the Vm-DCL2 deletion mutants showed a significant reduction in pathogenicity and sRNA abundance (Feng et al., 2017a). In the present study, the sRNAs in both the wild type and Vm-DCL2 deletion mutant were analyzed to identify the Vm-DCL2-dependent sRNAs. We prove that a Vm-DCL2-dependent milRNA, Vm-PC-3p-92107_6, is involved in pathogenicity by regulating the expression of the vesicle pathway-related gene Vm-VPS10.
Materials and Methods
Strains and Growth Conditions
The V. mali wild-type strain 03–8 and Vm-DCL2 deletion mutant strains were kept in storage by the Research Team of Pathogen Biology and Integrated Control of Fruit Trees, at the College of Plant Protection, Northwest A&F University, China. All strains were cultured in a PDA medium at 25°C in darkness. Escherichia coli DH5α was cultured in LB medium at 37°C.
sRNAs Sequence Data Analysis
The sRNAs libraries of wild-type 03–8 (MVm) and Vm-DCL2 deletion mutant (MD2) strains were constructed in previous study (Feng et al., 2017a). Raw data were first processed through custom Perl and python scripts. During this step, clean data were obtained by removing reads containing ploy-N, reads with 5′ adapter contaminants, reads without 3′ adapters or insert tags, reads containing ploy A, T, G, or C, and low-quality reads from the raw data. The clean reads were mapped into the reference sequence using Bowtie (Langmead et al., 2009) without a mismatch to confirm the sequence accuracy. The number of total unique sRNAs from the different samples was calculated to compare the difference between the MVm and MD2. To preliminarily estimate the sRNAs varieties, the length distribution of sRNAs in MVm and MD2 was also analyzed.
Expression Profiles of Vm-milRNAs
Raw data were processed using the Illumina pipeline filter (Solexa 0.3). The ensuing data were subjected to the ACGT101-miR (LC Sciences, Houston, TX, United States) to remove any adapter dimers, junk, common RNA families, low complexity, and repeats. The specific screening process applied to milRNA in V. mali was consistent with that already described in a previous study (Xu et al., 2020).
The differential expression of milRNAs was determined according to the relative expression abundance of each miRNA in the MVm and MD2. When the |log2 (fold change)| of miRNA was ≥1 and value of p≤0.01, the expression was judged significantly different. Normalized expression level was calculated as mapped read count/total reads × 106, with the value of p calculated this way:
where N1 denotes the expression level of miRNAs in the wild-type strain (MVm), while N2 denotes the expression level of miRNAs in the Vm-DCL2 deletion mutant (MD2); x represents all the miRNAs sequenced in the sample of the wild-type strain, and y represents all the miRNAs sequenced in the sample of the DCL2 mutant.
Target Gene Identification of Vm-milRNAs by Degradome Sequencing
To verify the target genes of Vm-milRNAs, degradome sequencing was used. Samples of total RNA (each 20μg)—from the RNAs used for sRNA libraries construction—were used to construct the degradome sequencing library by following the protocols described previously (German et al., 2009). The specific degradome sequencing and data analysis methods were consistent with those used in a previous study (Xu et al., 2020).
Sequence Alignment and Phylogenetic Analysis
Homologous protein sequences were searched by using the Blast function in NCBI Web site.1 For those proteins, their conserved domains were predicted by SMART and using the conserved domain database at NCBI.2 The alignment of multiple protein sequences was done with DNAMAN software, and the phylogenetic tree was built using the neighbor-joining method in MEGA 6 (bootstrap values were set as 1,000).
Co-expression of Vm-PC-3p-92107_6 and Vm-VPS10 in Nicotiana Benthamiana Leaves
Vm-PC-3p-92107_6 and the predicted target region of Vm-VPS10 were separately inserted into the empty pCAMBIA1302 vector with GFP as the reporter gene. Then, these two recombinant vectors were co-transformed into the same site of N. benthamiana leaves via the Agrobacterium-mediated transfection system (GV3101), as described by Weiberg et al. (2013). Vm-milR9 with no sequence similarity to Vm-PC-3p-92107_6 and mutated Vm-PC-3p-92107_6 were used as controls, respectively. Confocal images were taken at 48h post-Agrobacterium infiltration. The quantitative GFP intensity is proportional to the expression level of the candidae target gene. To further verify the expression of GFP, Western blot analysis was done using Anti-GFP (Sungene Biotech, Tianjin, China), with horseradish peroxidase-conjugated goat anti-mouse IgG (Cwbiotech, Beijing, China) used as the secondary antibody. The co-expression experiment was repeated twice, independently, for which all the primers can be found in Supplementary Table S1.
Relative Expression of Pre-Vm-PC-3p-92107_6, Vm-PC-3p-92107_6 and Vm-VPS10
Total RNA was extracted using the miRcute Plant miRNA Isolation Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. For the expression of Vm-PC-3p-92107_6, it was detected with stem-loop qRT-PCR, as described by Xu et al. (2020). The first strand cDNA was synthesized by implementing the first strand cDNA synthesis of miRNA (Stem-Loop Method; Sangon Biotech, Shanghai, China) with the stem-loop RT primer, according to the manufacturer’s instructions. PCR amplification was performed using the Vm-PC-3p-92107_6-specific forward primers and universal reverse primers; small nuclear RNA U6 served as a control. To determine the transcript levels of precursor of Vm-PC-3p-92107_6 (pre-Vm-PC-3p-92107_6) and Vm-VPS10, a sequence-specific primer and oligo(dT) primer were used to carry out the reverse-transcription using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, United States) according to the manufacturer’s instructions. The transcriptional level of pre-Vm-PC-3p-92107_6 and Vm-VPS10 was analyzed by qRT-PCR, for which the glucose-6-phosphate dehydrogenase gene (G6PDH) served as the reference gene (Yin et al., 2013). The quantitative PCR was run on LightCycler 96 real-time PCR instrument (Roche, Basel, Switzerland). Relative expression levels were calculated by applying the 2-ΔΔCt method (Shabalina and Koonin, 2008). All primers used in this study are in Supplementary Table S1.
Generation of Vm-PC-3p-92107_6 Over-Expression Transformants, Vm-PC-3p-92107_6, Vm-VPS10 Deletion Mutants, and the Complement Transformants
To over-express Vm-milRNA, the pre-Vm-PC-3p-92107_6 and its forward and reverse sequences of 200bp (31,247bp to 31,766bp in the V. mali wild-type strain genome contig 451) were amplified using Phusion High-fidelity DNA polymerase. The methodology used to derive the over-expression constructs in plasmid PDL2 is consistent with that described in Xu et al. (2020). The constructs were verified by sequencing and transformed into the wild-type strain. For the generation of mutated Vm-PC-3p-92107_6 (Mut-Vm-PC-3p-92107_6) constructs, the methods described by Xu et al. (2020) were also referred to and applied. All primers used for over-expression analyses are listed in Supplementary Table S1.
Double-joint PCR was used to build the deletion construction, and the specific construction process is consistent with already described by Yu et al. (2004). Vm-VPS10 deletion mutants were generated using the strain ΔVm-Ku80, which provided for highly enhanced target gene deletion efficiency but did not affect either vegetative growth or virulence (Xu et al., 2016). Four types of PCR detections were conducted to confirm that both Vm-PC-3p-92107_6 and Vm-VPS10 were indeed deleted. To construct the complement vector, the gene was amplified, and then connected to the PDL2 vector that had been digested by the homologous recombination method. The positive vectors extracted from the competent state DH5α of E. coli were confirmed by sequencing, and then transformed into the corresponding deletion mutants. PCR was used detect the complement transformants. All primers used for genes deletion are given in Supplementary Table S1.
Vegetative Growth and Pathogenicity Tests of Mutants
The vegetative growth and pathogenicity of gene deletion mutants and over-expression transformants were analyzed as previous described (Feng et al., 2017a). Briefly, fungal colony diameters were measured at 48h post-cultivation. The assay was independently performed three times, and each experiment had three replicates. Pathogenicity was tested using “Fuji” apple twigs, as described by Feng et al. (2017a). Lesion length was measured at 5days post-inoculation (dpi). The pathogenicity test was repeated three times, and each experiment had three replicates. Next, the significant difference in means was analyzed by a t-test (for two independent sample groups) and ANOVA (for three or more independent sample groups; at p≤0.05) in GraphPad Prism 6.0 software.
Results
Analysis of Small RNAs’ Abundance in MVm and MD2
The sequence data of MVm were deposited in NCBI (GEO accession no. GSM3757989) with the publication of a previous study (Xu et al., 2020). Referring to the previous study, 12,818,591 raw read sequences and 1,023,889 valid sequences were obtained in MVm, while in MD2 library, 11,565,746 raw read sequences were obtained and then after removing the repetitive sequence and junk sequences, valid was for only 593,982, which indicated the deletion of DCL2 could affect the generation of sRNAs in V. mali (Supplementary Table S2; Supplementary Figure S1A). Analyzing the length distribution of sRNAs revealed they had a consistent length of 19–24nt in both libraries (Supplementary Figure S1B); however, the abundance of sRNAs of a given same length differed. The sRNAs in MVm were mostly 20nt, 21nt, and 22nt, while those in MD2 were mainly composed of those 22nt and 23nt in length (Supplementary Figure S1C). Hence, we speculated that deletion of Vm-DCL2 could somehow affect the generation of sRNA, but there may be an as of yet unknown complementary pathway that could compensate for part function of sRNAs generation.
Isolation and Identification of DCL2-Associated siRNAs and milRNAs
The differential expression of siRNAs and milRNAs between the two libraries was first analyzed. Based on the sequencing results, 3,243 siRNAs were isolated and identified, of which 3,186 were not detected in MD2 (data not shown). Among these siRNAs, 25 were significantly upregulated in MD2 compared with MVm, including four siRNAs specifically expressed in MD2, and 15 were downregulated, with one siRNA not detected in MD2 (Supplementary Figure S2, Supplementary Table S6). Thus, the generation of siRNAs was greatly changed when Vm-DCL2 was deleted. By comparing the difference in expression of milRNAs between MVm and MD2, 33 milRNAs were not detected in MD2 while 28 milRNAs were detected in both MVm and MD2 (Figure 1A, Supplementary Table S3). The statistical analysis revealed that 1 and 17 milRNAs, respectively, exhibited upregulated and downregulated expression in MD2 compared with MVm (p<0.05; Figure 1B). Importantly, 10 milRNAs were not detected in MD2, which were identified as being DCL2-dependent milRNAs (Table 1).
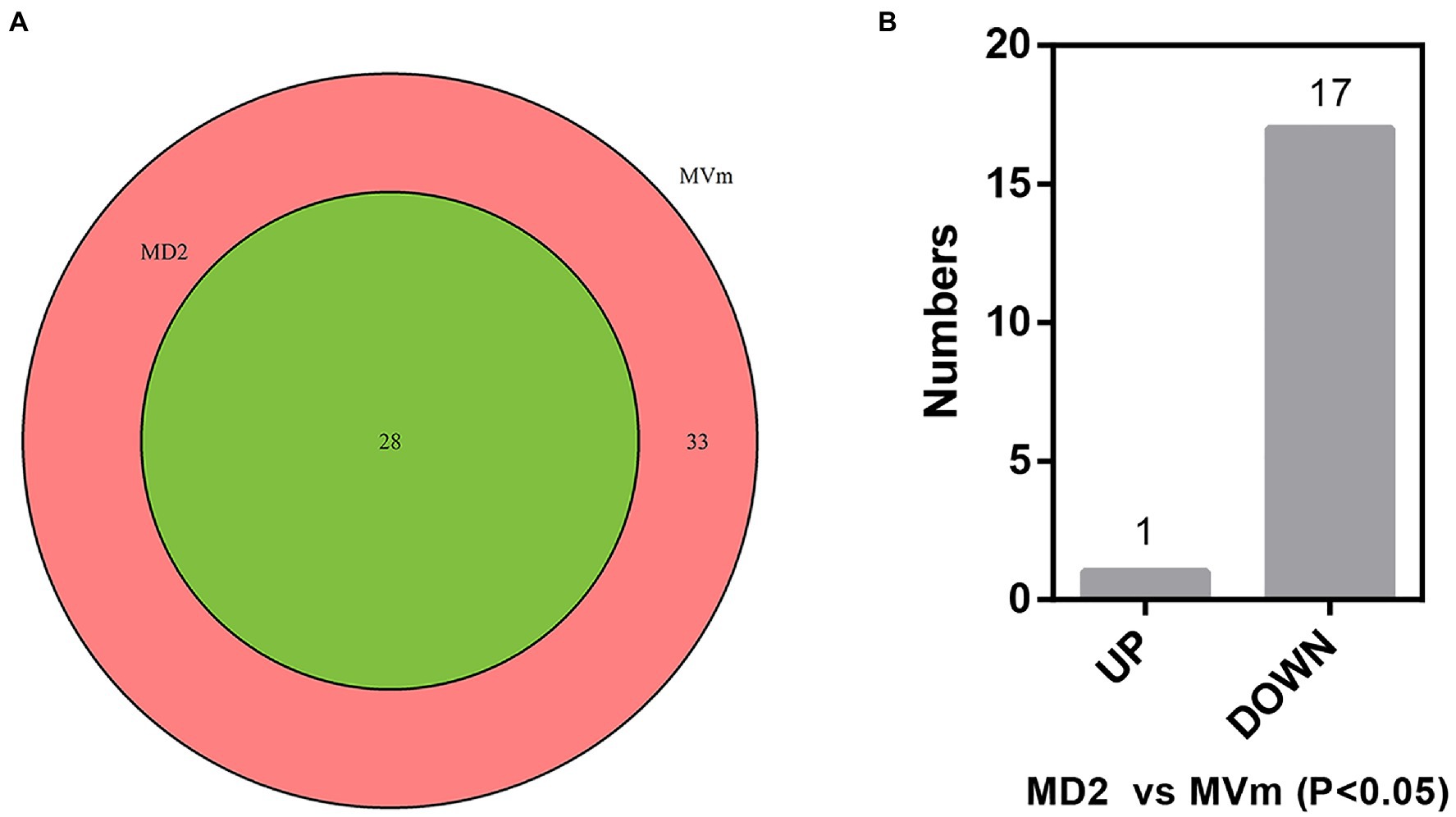
Figure 1. (A) Venn diagrams of detected miRNA-like RNAs (milRNAs). (B) The bar plot of differentially expressed milRNAs in the MD2 and MVm (P<0.05). MD2: ΔVm-DCL2; MVm: WT.
Target Prediction of DCL2-Associated milRNAs
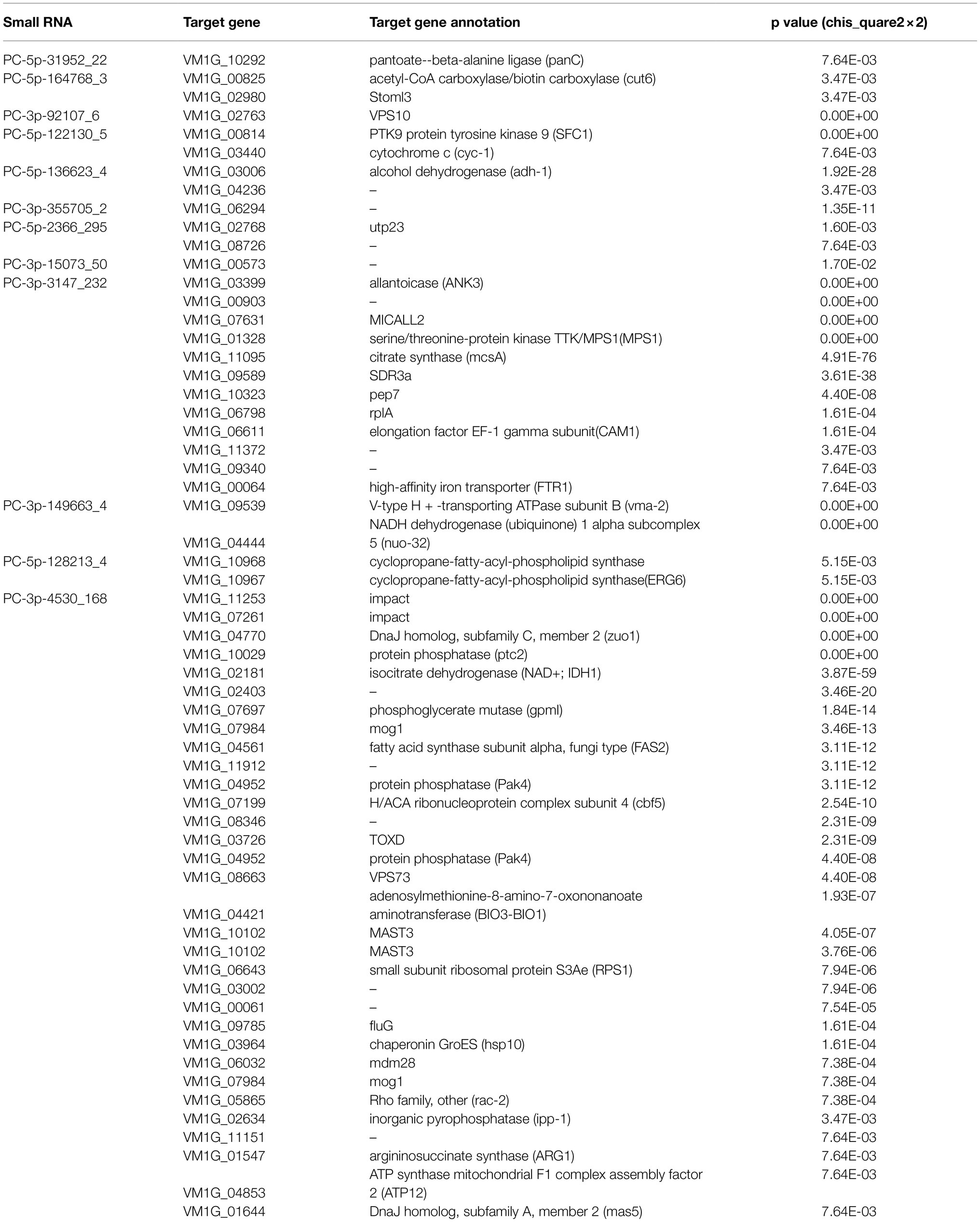
The target genes of DCL2-associated milRNAs were identified by high-throughput degradome sequencing technology. In total, the target genes of 12 milRNAs were distinguishable. These target genes were annotated as follows: histidine kinase, serine/threonine-protein kinase, pectin esterase, glucose transport regulator, ATP-dependent RNA helicase, myosin, and AP-2 complex subunit, etc. They mainly participate in 35 different groups at the three categories of biological process, molecular function, and cell component (Supplementary Table S5). We found that PC-5p-164768_3, PC-5p-122130_5, PC-5p-136623_4, PC-5p-2366_295, PC-3p-3147_232, PC-3p-149663_4, PC-5p-128213_4, and PC-3p-4530_168 could target more than one gene. For example, PC-3p-4530_168 could target 31 genes, and these main target genes were associated with protein phosphatase, isocitrate dehydrogenase, phosphoglycerate mutase, etc. In stark contrast, PC-5p-31952_22, PC-3p-92107_6, PC-3p-355705_2, and PC-3p-15073_50 could only target one gene, such as Vm-PC-3p-92107_6 targeting VM1G_02763, the latter predicted to be VPS10 (Table 2). Evidently, the regulatory network of milRNAs is very complex.
Vm-PC-3p-92107_6 Is a DCL2-Dependent milRNA
Based on the sequencing results, we found that Vm-PC-3p-92107_6 could not be detected in MD2. Accordingly, we speculated that Vm-PC-3p-92107_6 is a DCL2-dependent milRNA. To test this hypothesis, the relative expression levels of pre-Vm-PC-3p-92107_6 (Vm-PC-3p-92107_6-P) and Vm-PC-3p-92107_6 were detected by qRT-PCR in the wild type and DCL2 mutant (ΔVm-DCL2), respectively. The expression of pre-Vm-PC-3p-92107_6 showed no significant difference between the wild type and ΔVm-DCL2, whereas that of Vm-PC-3p-92107_6 was barely detected in ΔVm-DCL2 (Figures 2A,B). Meanwhile, the expression of Vm-PC-3p-92107_6 was detectable in the Vm-DCL1 mutant (ΔVm-DCL1) but at a similar level as in the wild type; likewise, no significant differences were found in the expression level of Vm-PC-3p-92107_6 in ΔVm-DCL1 relative to the wild type (Figure 2C). These results further proved that Vm-PC-3p-92107_6 is specifically dependent on Vm-DCL2 in V. mali.
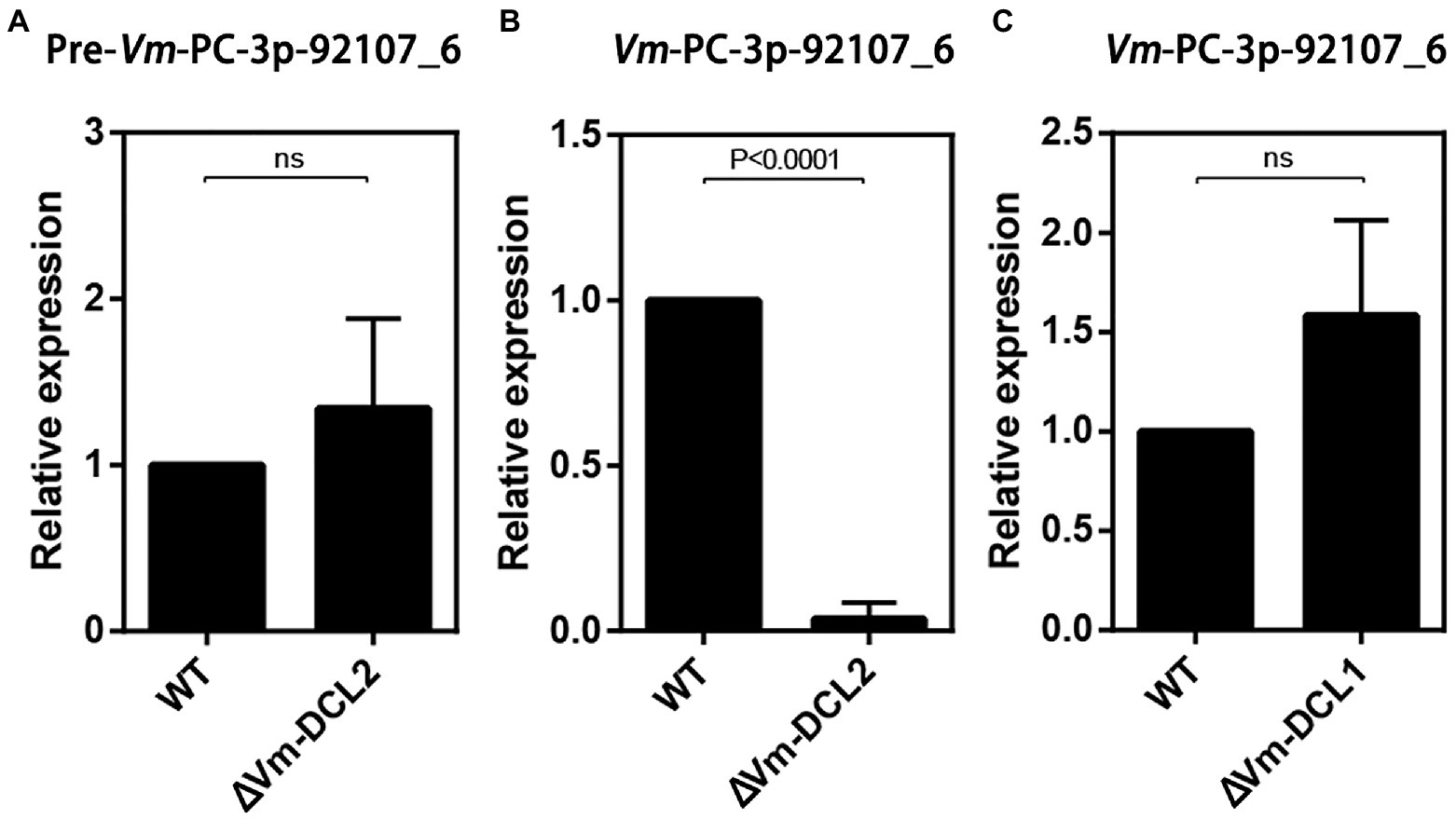
Figure 2. Relative expressions of pre-Vm-PC-3p-92107_6 and Vm-PC-3p-92107_6 by quantitative real-time PCR. (A) Relative expression of pre-Vm-PC-3p-92107_6 in Vm-DCL2 deleted mutant (ΔVm-DCL2). Vm-G6PDH was selected to reference gene and the relative expression level of pre-Vm-PC-3p-92107_6 in WT was set to 1 by the 2−ΔΔCt method. (B) Relative expression of Vm-PC-3p-92107_6 in Vm-DCL2 deleted mutant (ΔVm-DCL2). Vm-U6 was selected to reference gene and the relative expression level of Vm-PC-3p-92107_6 in WT was set to 1. (C) Relative expression of Vm-PC-3p-92107_6 in Vm-DCL1 deleted mutant (ΔVm-DCL1). Vm-U6 was selected to reference gene and the relative expression level of Vm-PC-3p-92107_6 in WT was set to 1.
Vm-PC-3p-92107_6 Played Important Roles in Vegetative Growth and Pathogenicity
To clarify the function of Vm-PC-3p-92107_6, the Vm-PC-3p-92107_6 deletion mutants (ΔVm-PC-3p-92107_6) and complement transformants (ΔVm-PC-3p-92107_6-C) were constructed (Supplementary Figure S3, S4). Compared with the wild type, the colony diameter of ΔVm-PC-3p-92107_6 was significantly reduced, yet there was little difference in either their colony morphology or density of aerial hyphae (Figures 3A,B). Moreover, compared with the wild type, the pathogenicity of ΔVm-PC-3p-92107_6 was significantly augmented, but after the Vm-PC-3p-92107_6 complementation, it was similar to the wild type (Figures 3C,E).
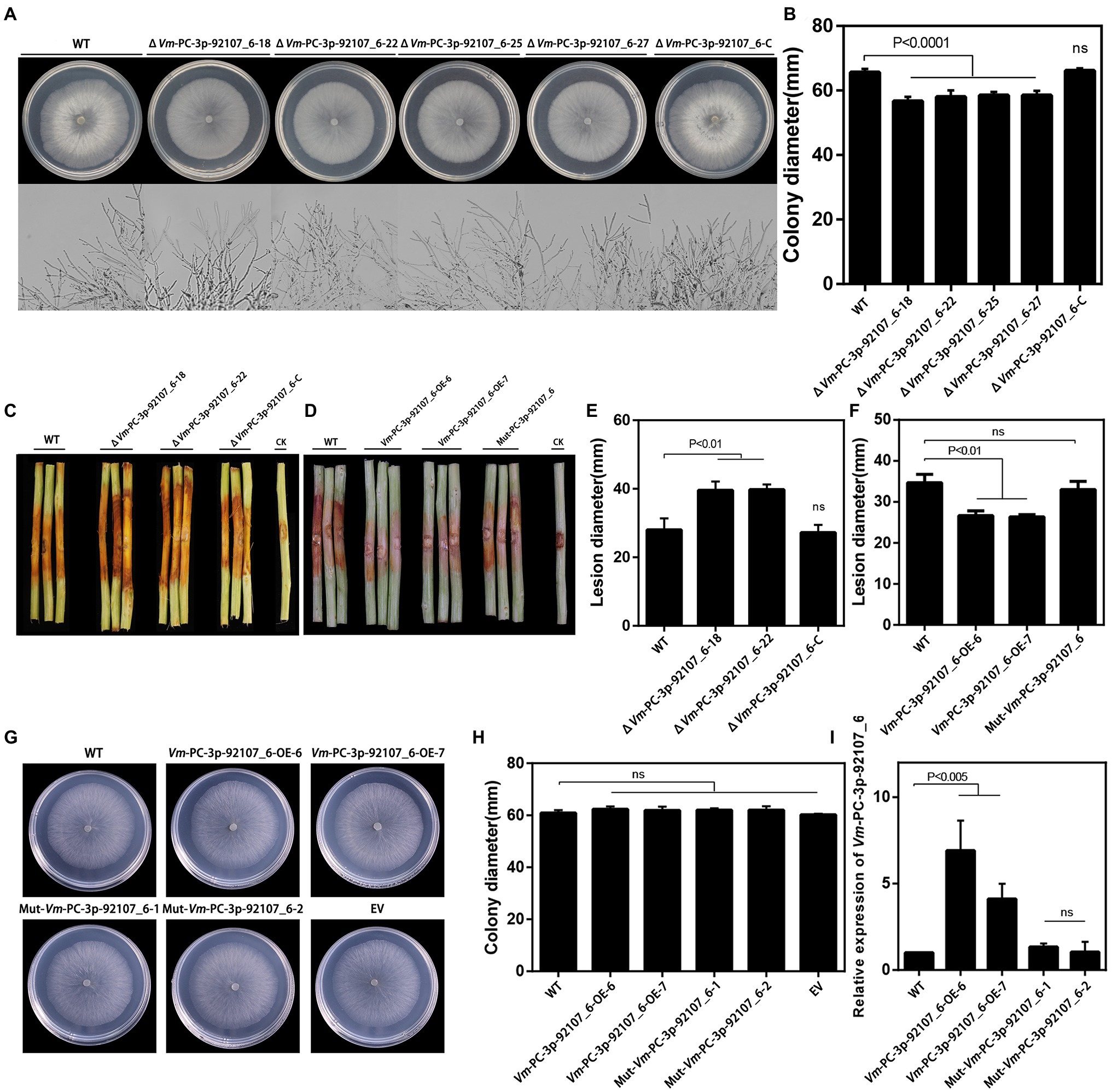
Figure 3. Phenotypical analysis of Vm-PC-3p-92107_6 deleted mutants (ΔVm-PC-3p-92107_6), complement transformants (ΔVm-PC-3p-92107_6-C), over-expression transformants (Vm-PC-3p-92107_6-OE), and the wild type (WT). (A) and (B) Colony morphology of WT, Vm-PC-3p-92107_6 deleted mutants (ΔVm-PC-3p-92107_6–18, ΔVm-PC-3p-92107_6–22, ΔVm-PC-3p-92107_6–25, and ΔVm-PC-3p-92107_6–27), and complement transformants (ΔVm-PC-3p-92107_6-C) after 48h (hr) incubation in PDA at 25°C. Related colony diameters were measured after 48h incubation. Data represent mean±SD. The experiment was repeated three times, each time with three plates. (C) and (E) Pathogenicity test of WT, ΔVm-PC-3p-92107_6–18, ΔVm-PC-3p-92107_6–22, and ΔVm-PC-3p-92107_6-C after 5days post-inoculation. Three representative diseased twigs are shown. The pathogenicity test was independently repeated three times, each time with six replicates. CK represents a negative control. Data represent mean±SD. (D) and (F) Pathogenicity test of WT, Vm-PC-3p-92107_6-OE-6, Vm-PC-3p-92107_6-OE-7, and Mut-Vm-PC-3P-92107_6 after 5dpi. Three representative diseased twigs are shown. The pathogenicity test was independently repeated three times, each time with six replicates. CK represents a negative control. Data represent mean±SD. (G) and (H) Colony morphology of WT, Vm-PC-3p-92107_6 over-expression transformants (Vm-PC-3p-92107_6-OE-6 and Vm-PC-3p-92107_6-OE-7), mutated Vm-PC-3p-92107_6 (Mut-Vm-PC-3P-92107_6–1 and Mut-Vm-PC-3P-92107_6–2), and empty vector transformant (EV) after 48h incubation in PDA at 25°C. Related colony diameters were measured after 48h incubation. Data represent mean±SD. The experiment was repeated three times, each time with three plates. (I) The relative expression level of Vm-PC-3p-92107_6 in WT, Vm-PC-3p-92107_6 over-expression transformants (Vm-PC-3p-92107_6-OE-6 and Vm-PC-3p-92107_6-OE-7), and mutated Vm-PC-3p-92107_6 (Mut-Vm-PC-3P-92107_6-1 and Mut-Vm-PC-3P-92107_6-2). Vm-U6 was selected to reference, the relative expression level of WT is set to 1 as control group using the 2-ΔΔCt method. Data represent mean ± SD.
To further confirm the function of Vm-PC-3p-92107_6, the Vm-PC-3p-92107_6 over-expression transformants (Vm-PC-3p-92107_6-OE) and the Mut-Vm-PC-3p-92107_6 mutants (Mut-Vm-PC-3p-92107_6) were also constructed (Supplementary Figure S5). In comparison with the wild type, the colony diameter of Vm-PC-3p-92107_6-OE, Mut-Vm-PC-3p-92107_6, and EV showed no significant differences (Figures 3G,H). More importantly, the pathogenicity of Vm-PC-3p-92107_6-OE was significantly reduced, whereas it was not significantly different between Mut-Vm-PC-3p-92107_6 and EV (Figures 3D,F). This confirmed that Vm-PC-3p-92107_6 could regulate the vegetative growth and pathogenicity of V. mali.
Vm-VPS10 Was Identified as the Target of Vm-PC-3p-92107_6
Based on the degradome sequencing results, only the transcript VM1G_02763 was identified to be the target of Vm-PC-3p-92107_6. The bioinformatics analysis indicated that VM1G_02763 encoded a protein with three low complexity regions, two VPS10 regions, and two transmembrane regions, which we designated here as Vm-VPS10 (Figure 4A). The phylogeny of vacuolar protein sorting protein (VPS) between V. mali and other 23 filamentous fungi was investigated by constructing a neighbor-joining phylogenetic tree. This revealed that Vm-VPS10 (KUI67952) is highly homologous to VPS10 of Valsa pyri (KUI60044; Figure 4B).
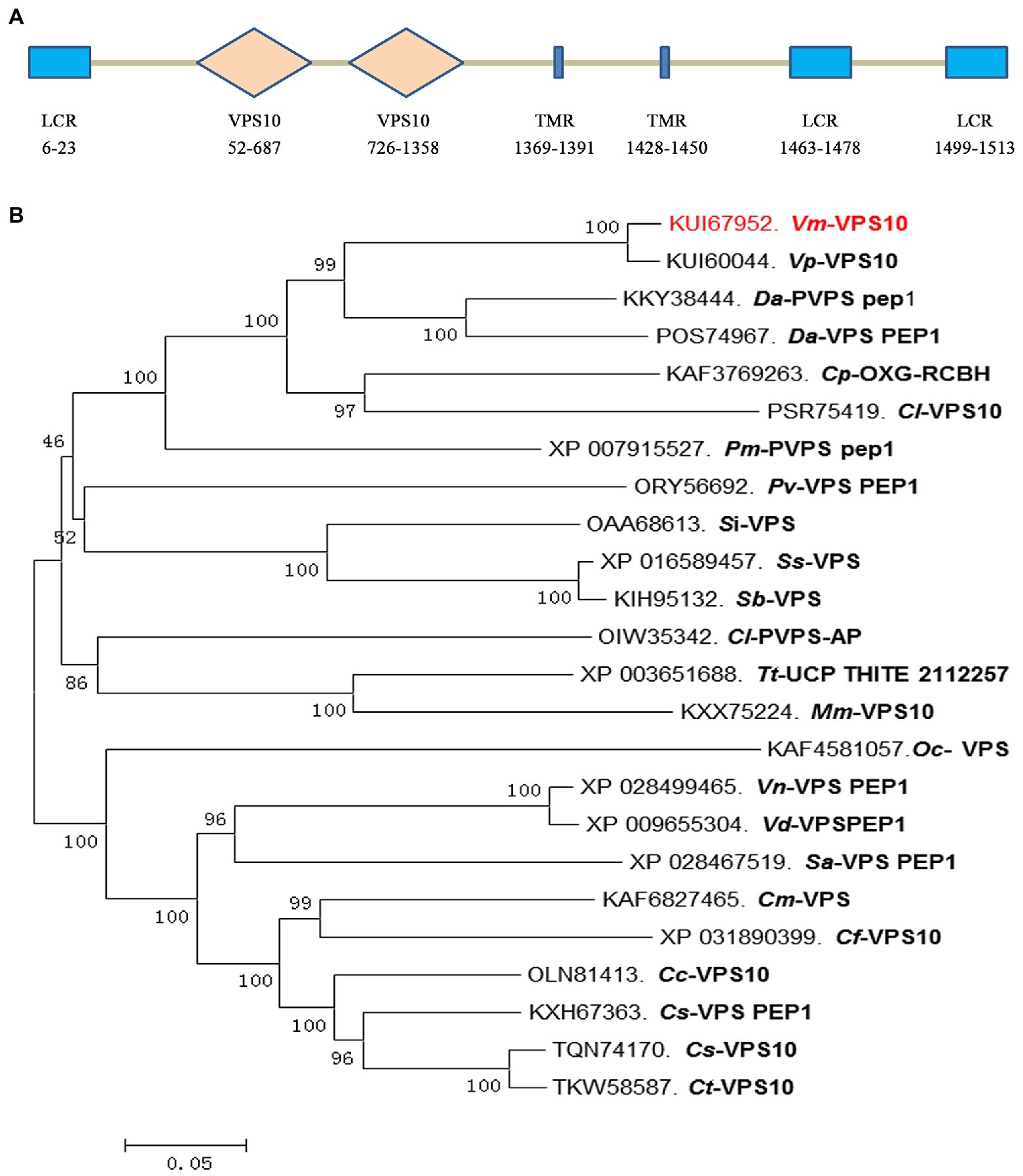
Figure 4. Protein sequence characterization and phylogeny analysis of the Vm-VPS10. (A) The typical domains of Vm-VPS10. Vm-VPS10 contains three conserved domains, including low complexity region (LCR), VPS10, and transmembrane region (TMR). VPS10 is the core domain, which is a receptor domain, and TMR is an important transmembrane helix region. The numbers below the structure indicate the amino acid sites of each domain. (B) The phylogenetic tree was constructed with neighbor-joining method using MEGA 6. Bootstrap values were set as 1,000. Vm-VPS10 is highlighted in red. Vm, Valsa mali; Vp, Valsa pyri; Da, Diaporthe ampelina; Dh, Diaporthe helianthi; Cp, Cryphonectria parasitica; Cl, Coniella lustricola; Pm, Phaeoacremonium minimum; Pv, Pseudomassariella vexata; Si, Sporothrix insectorum; Sc, Sporothrix schenckii; Sb, Sporothrix brasiliensis; Tt, Thermothielavioides terrestris; Mm, Madurella mycetomatis; Oc, Ophiocordyceps camponoti-floridani; Vn, Verticillium nonalfalfae; Sa, Sodiomyces alkalinus; Cm, Colletotrichum musicola; Cf, Colletotrichum fructicola; Cc, Colletotrichum chlorophyte; Cs, Colletotrichum salicis; and Ct, Colletotrichum tanaceti.
To further verify the regulatory relationship between Vm-PC-3p-92107_6 and Vm-VPS10, the expression vectors were successfully constructed and co-transformed into N. benthamiana leaves (Supplementary Figure S6). These results clearly showed green fluorescence visible on the leaves injected with Vm-VPS10 alone. However, when Vm-PC-3p-92107_6 and Vm-VPS10 were co-expressed, the intensity of green fluorescence was significantly diminished and the controls indistinguishable from Vm-VPS10 alone. Meanwhile, when Vm-milR9, which has no sequence similarity with Vm-PC-3P-92107_6, was co-expressed with Vm-VPS10, the intensity of green fluorescence was similar with Vm-VPS10 alone. The result of Mut-Vm-PC-3p-92107_6 also showed similar result (Figure 5A; Supplementary Figure S7). Further, the expression of GFP in the tissues co-expressing Vm-PC-3p-92107_6 and Vm-VPS10 was significantly reduced in the Western blot analysis (Figure 5C). Collectively, these results indicated that Vm-PC-3p-92107_6 could suppress the expression of Vm-VPS10.
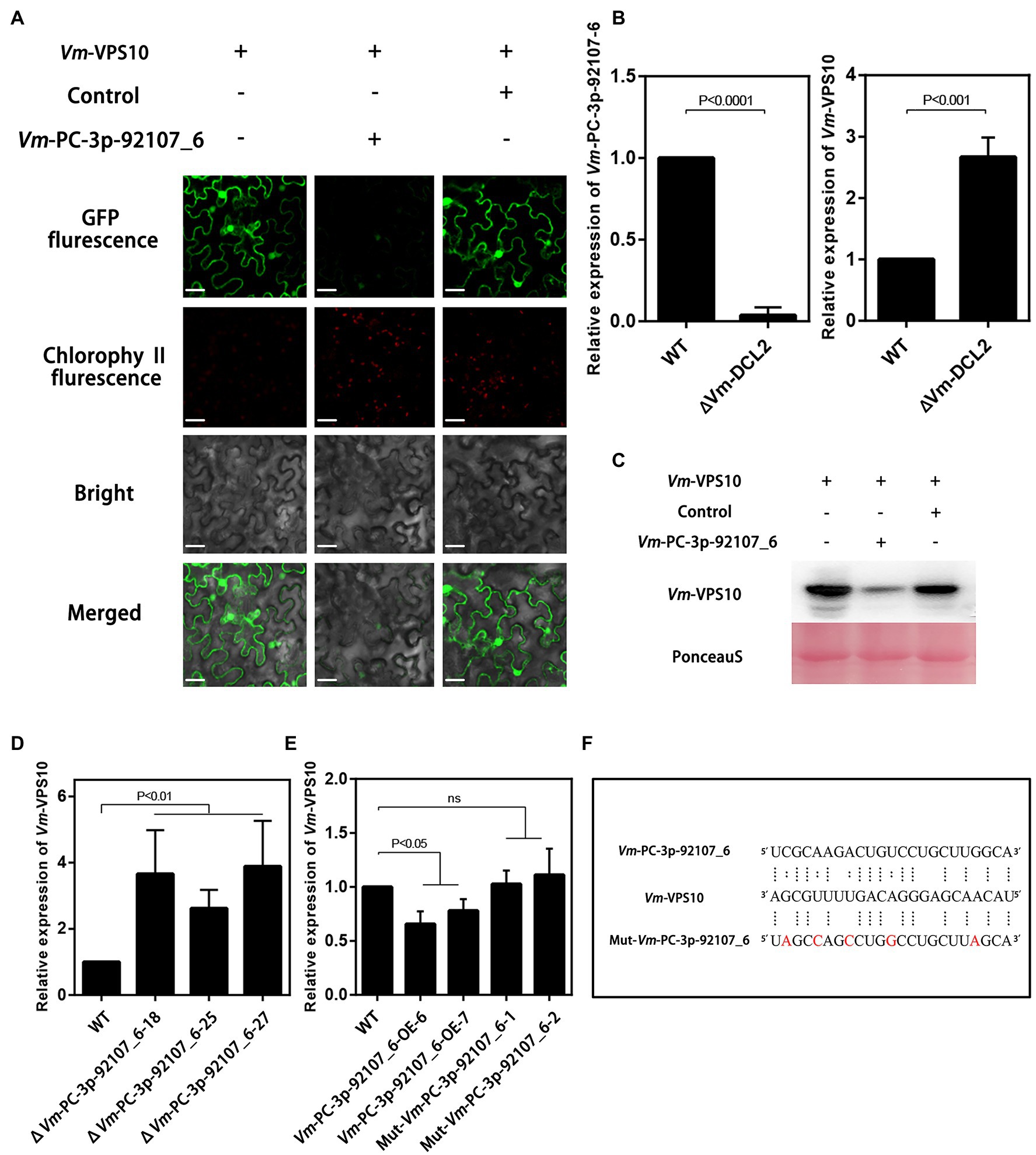
Figure 5. The regulatory relationship analysis between Vm-PC-3p-92107_6 and Vm-VPS10. (A) Vm-VPS10 expression vectors and Vm-PC-3p-92107_6-OE expression vectors were constructed, respectively. Vm-milR9 with no sequence similarity was used as the control. Confocal imaging was performed 48h after Agrobacterium infiltration. (B) The relative expression of Vm-PC-3p-92107_6 and Vm-VPS10 in WT and Vm-DCL2 deleted mutants (ΔVm-DCL2). Vm-U6 and Vm-G6PDH were selected to reference gene using 2-ΔΔCt method. And the relative expressions of Vm-PC-3p-92107_6 and Vm-VPS10 in WT were set to 1 which is taken as control. (C) Western blot analysis of eGFP-Vm-VPS10. Anti-GFP antibodies were used for analysis. The co-expression experiment was repeated twice and similar results were obtained. (D) The relative expression of Vm-VPS10 in WT and Vm-PC-3p-92107_6 deleted mutants (ΔVm-PC-3p-92107_6–18, ΔVm-PC-3p-92107_6–25, and ΔVm-PC-3p-92107_6–27). (E) The relative expression of Vm-VPS10 in WT and over-expression transformants of Vm-PC-3p-92107_6 (Vm-PC-3p-92107_6-OE-6 and Vm-PC-3p-92107_6-OE-7), and mutated Vm-PC-3p-92107_6 (Mut-Vm-PC-3p-92107_6-1 and Mut-Vm-PC-3p-92107_6-2). (F) Sequence alignment of Vm-VPS10 with Vm-PC-3p-92107_6 and mutated Vm-PC-3p-92107_6 (Mut-Vm-PC-3p-92107_6).
Next, the relative expression levels of Vm-VPS10 and Vm-PC-3p-92107_6 were determined by qRT-PCR in the wild type and ΔVm-DCL2, respectively. When compared with the wild type, the expression level of Vm-VPS10 in ΔVm-DCL2 was significantly upregulated (Figure 5B). Meanwhile, the relative expression of Vm-VPS10 was also detected in both ΔVm-PC-3p-92107_6 and Vm-PC-3p-92107_6-OE mutants. Compared with the wild type, the relative expression of Vm-VPS10 was significantly increased in ΔVm-PC-3p-92107_6, whereas it was significantly decreased in Vm-PC-3p-92107_6-OE (Figures 5D,E). Moreover, during the infection progress of V. mali, Vm-PC-3p-92107_6 was significantly downregulated at 12, 24, and 36 hpi (Figure 6A), yet Vm-VPS10 had enhanced transcript levels during infection (Figure 6B). These results provided compelling evidence that the expression of Vm-VPS10 could be regulated by Vm-PC-3p-92107_6.
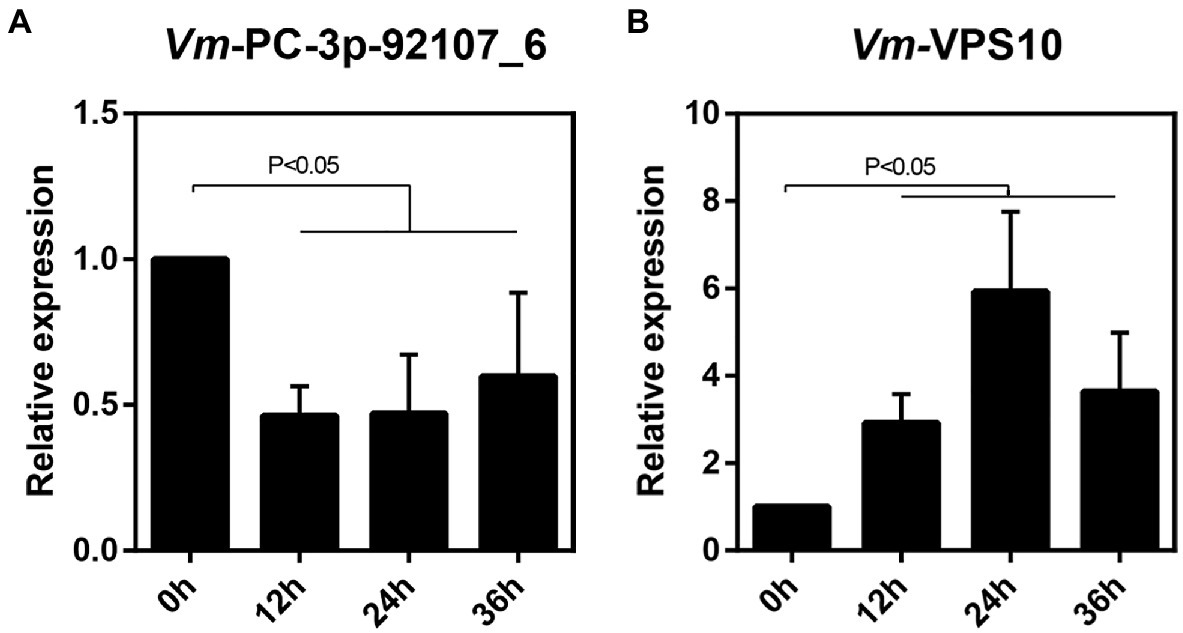
Figure 6. The relative expression pattern of Vm-PC-3p-92107_6 (A) and Vm-VPS10 (B) during V. mali infection.
Vm-VPS10 Contributed to Vegetative Growth and Pathogenicity
Finally, to explore the function of Vm-VPS10, a gene deletion mutant (ΔVm-VPS10) was constructed using ΔVmKu80 (Supplementary Figure S8). Compared with ΔVmKu80, the colony diameter of ΔVm-VPS10 was significantly reduced, but both the density and morphology of airborne mycelia were not affected (Figures 7A,B). Notably, the pathogenicity was significantly lower for ΔVm-VPS10 than ΔVmKu80 (Figures 7C,D).
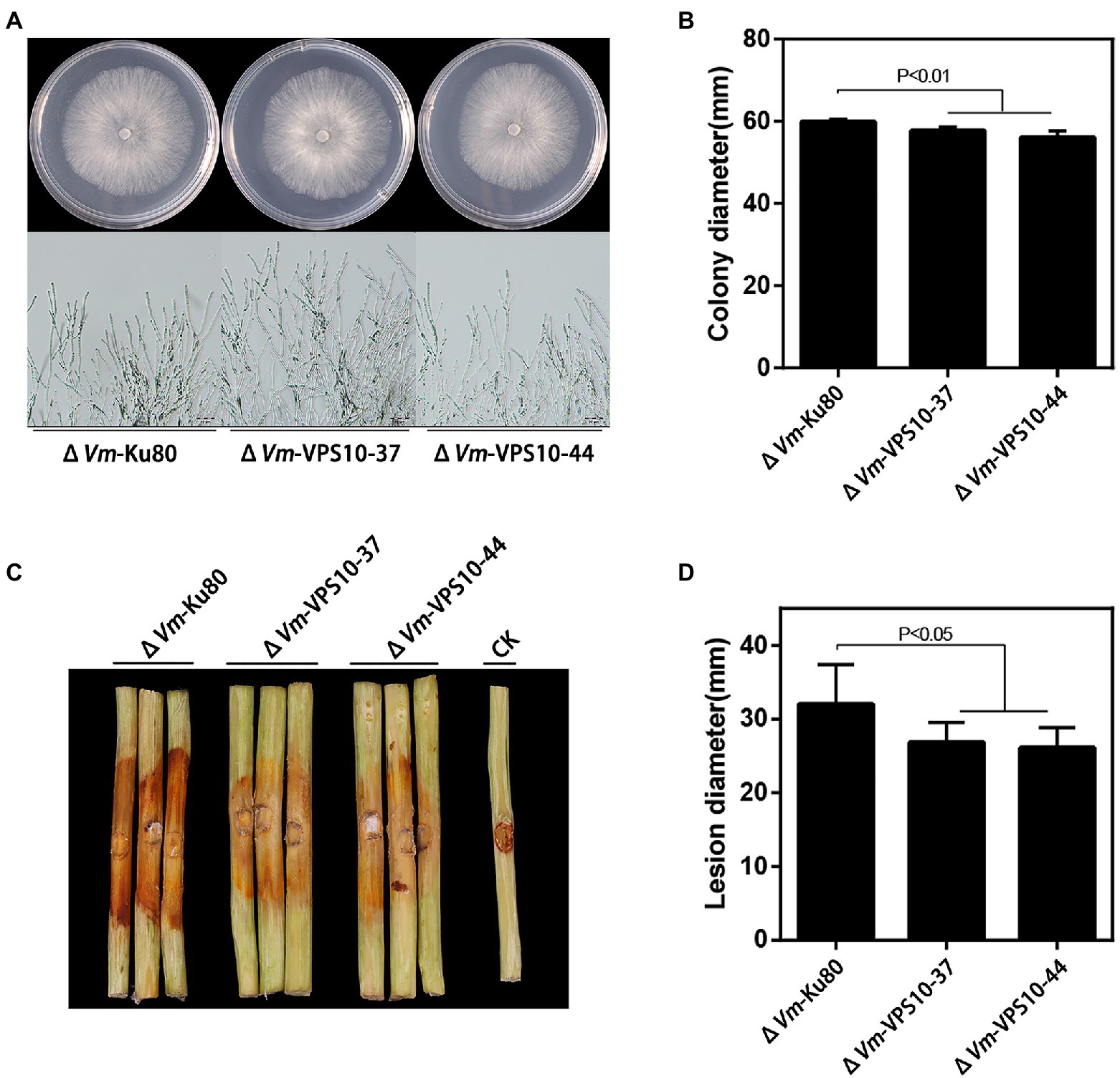
Figure 7. Phenotypical analysis of Vm-VPS10 deleted mutants (ΔVm-VPS10) and ΔVm-Ku80. (A) Colony morphology of ΔVm-Ku80 and Vm-VPS10 deleted mutants (ΔVm-VPS10-37 and ΔVm-VPS10-44) after 48h incubation in PDA at 25°C. (B) Colony diameters of ΔVm-Ku80 and ΔVm-VPS10-37, ΔVm-VPS10-44 after 48h incubation. Data represent mean ± SD. (C) and (D) Pathogenicity test of ΔVm-Ku80 andΔVm-VPS10-37, ΔVm-VPS10-44 after 5dpi. Three representative diseased twigs are shown. The pathogenicity test was independently repeated three times, each time with six replicates. CK represents a negative control. Data represent mean ± SD.
Discussion
As important non-coding RNA regulators, sRNAs play key roles in many biological processes, such as development regulation, transposon inhibition, environmental response, and host–pathogen interactions (Mallory and Vaucheret, 2006; Inui et al., 2010). As their key switch, Dicer proteins play crucial roles in the generation process of sRNAs (Nicolas et al., 2010). Still, the generation of sRNA is a more complex phenomenon in fungi. Since the first discovery of siRNA in N. crassa (Cogoni and Macino, 1999), research on the isolation of fungi sRNAs has expanded immensely, especially concerning the generation mechanism and functioning of sRNAs. In N. crassa, there are at least four pathways by which milRNAs are generated, including those that are DCL-dependent and DCL-independent (Lee et al., 2010). In F. graminearum, the generation of milRNAs was confirmed to be associated with FgAGO1 and FgDicer2 (Chen et al., 2015), and the Dicer-independent pathway to generate milRNA also exists in M. circinelloides (Nicolas and Ruiz-Vazquez, 2013). In V. mali, it has been shown that Vm-DCL1 and Vm-DCL2 deletion mutants could significantly reduce the production of sRNAs (Feng et al., 2017a). In our study, the differences in sRNAs between the wild type and Vm-DCL2 deletion mutant were investigated. The results supported the view that the deletion of Vm-DCL2 could affect the generation of sRNAs. Going further, we speculated another complementary pathway might exist that compensates for part function of sRNA generation when Vm-DCL2 was deleted.
The miRNAs in plants and animals are crucially involved in their growth, development, reproduction, and responses to biotic and abiotic stresses, by inhibiting their corresponding target genes via transcriptional inhibition, mRNA cutting, or translation inhibition (Carrington and Ambros, 2003; Ghildiyal and Zamore, 2009; D’Ario et al., 2017; Wang et al., 2017; Song et al., 2019; Wang and Galili, 2019). However, the functions and mechanisms of milRNAs in fungi are still largely unknown. Some studies have shown that gene expression in fungi could be regulated at the post-transcriptional level to enable their adaption to various environments (Tan and Oliver, 2017). Similarly, the expression of many virulence genes could also be regulated by sRNAs in pathogenic fungi. For example, the expression of pathogenicity-related genes in Magnaporthe grisea can be regulated by sRNAs to affect its growth, development, and pathogenicity of this fungus (Nunes et al., 2011; Raman et al., 2013), with similar results reportedly found in Trichoderma reesei and V. dahliae (Kang et al., 2013; Jin et al., 2019). In V. mali, Vm-milR16 was identified to adaptively regulate the expression of its virulence genes VmSNF1, VmDODA, and VmHy1, thereby contributing to the infection ability of V. mali (Xu et al., 2020). In the present study, a DCL2-dependent milRNA, Vm-PC-3p-92107_6, was clearly associated with fungal vegetative growth and pathogenicity by regulating the expression of a vacuolar protein sorting protein (Vm-VPS10). VPS was first isolated in Saccharomyces cerevisiae; it encodes a type I transmembrane receptor protein, which is sequentially aggregated by binding to soluble proteins in cells (Cooper and Stevens, 1996). As such, it can figure prominently in protein transport, in which most proteins entering the vacuole are transported from the endoplasmic reticulum to the Golgi complex along with secreted proteins and are then sorted by sorting apparatus from other secretory traffic in the late Golgi lumen for specific transport to the vacuole. In filamentous fungi, the involvement of VPS74 of F. gramineae in mycelia growth, conidia production, sexual reproduction, toxin production, and pathogenic process has been confirmed (Kim et al., 2015). In our study, functional analysis of Vm-VPS10 suggested it could influence the growth and pathogenicity of V. mali that are regulated by milRNAs at the post-transcriptional level, which further broadens our understanding of the regulatory mechanism underpinning VPS10 activity. Although the exact regulation mechanism is still unknown, we do know that Vm-VPS10’s expression could be suppressed by Vm-PC-3p-92107_6 to some extent. As mentioned above, the generation mechanism of sRNAs in fungi is more complex than that in plants and animals. Thus, we speculate the action mechanism of fungal sRNAs may also be more intricate, that is, to say, there may be more than three inhibition pathways by which fungal sRNAs impact their target genes.
Overall then, this study proved that DCL2-dependent milRNAs exist in V. mali. Among them, as a representative milRNA, Vm-PC-3p-92107_6 was confirmed to be involved in both vegetative growth and pathogenicity by regulating the expression of Vm-VPS10. This study enhances our understanding of the pathogenic mechanism of V. mali and helps to pave the way for fully revealing the generation and regulation mechanisms of fungal sRNA. Nonetheless, the functions of other DCL2-dependent sRNAs, especially their relationships, are still unclear and need further study.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at NCBI GEO, accession no: GSM3757989.
Author Contributions
FG and HF conceived and designed the work. FG, JL, MX, and GZ performed the experiments. FG and JL analyzed and interpreted the data. JL drafted the manuscript. HF and LH critically revised the manuscript for intellectual content. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Natural Science Basis Research Plan in Shaanxi Province of China (no. 2019JM-418), the New Star of Youth Science and Technology of Shaanxi Province (no. 2021KJXX-10), and the China Postdoctoral Science Foundation funded project (no. 2015M580883).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Jin-Rong Xu at Purdue University for providing plasmids pDL2.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.721399/full#supplementary-material
Footnotes
1. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome
References
Bai, Y. H., Lan, F. X., Yang, W. Q., Zhang, F., Yang, K. L., Li, Z. G., et al. (2015). sRNA profiling in aspergillus flavus reveals differentially expressed miRNA-like RNAs response to water activity and temperature. Fungal Genet. Biol. 81, 113–119. doi: 10.1016/j.fgb.2015.03.004
Carrington, J. C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301, 336–338. doi: 10.1126/science.1085242
Catalanotto, C., Pallotta, M., ReFalo, P., Sachs, M. S., Vayssie, L., Macino, G., et al. (2004). Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 24, 2536–2545. doi: 10.1128/MCB.24.6.2536-2545.2004
Chang, S. S., Zhang, Z., and Liu, Y. (2012). RNA interference pathways in fungi: mechanisms and functions. Annu. Rev. Microbiol. 66, 305–323. doi: 10.1146/annurev-micro-092611-150138
Chen, Y., Gao, Q., Huang, M., Liu, Y., Liu, Z., Liu, X., et al. (2015). Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum. Sci. Rep. 5, 413–420. doi: 10.1038/srep12500
Cogoni, C., and Macino, G. (1999). Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399, 166–169. doi: 10.1038/20215
Cooper, A. A., and Stevens, T. H. (1996). VPS10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J. Cell Biol. 133, 529–541.
D’Ario, M., Griffiths-Jones, S., and Kim, M. J. T. I. P. S. (2017). Small RNAs: big impact on plant development. Trends Plant Sci. 22, 1056–1068. doi: 10.1016/j.tplants.2017.09.009
Dahlmann, T. A., and Kueck, U. (2015). Dicer-dependent biogenesis of small RNAs and evidence for microRNA-like RNAs in the penicillin producing fungus Penicillium chrysogenum. PLoS One 10:e0125989. doi: 10.1371/journal.pone.0125989
de Haro, J. P., Calo, S., Cervantes, M., Nicolas, F. E., Torres-Martinez, S., and Ruiz-Vazquez, R. M. (2009). A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in Mucor circinelloides. Eukaryot. Cell 8, 1486–1497. doi: 10.1128/EC.00191-09
Doench, J. G., Petersen, C. P., and Sharp, P. A. (2003). siRNAs can function as miRNAs. Genes Dev. 17, 438–442. doi: 10.1101/gad.1064703
Feng, H., Xu, M., Liu, Y., Dong, R., Gao, X., and Huang, L. (2017a). Dicer-Like genes are required for H2O2 and KCl stress responses, pathogenicity and small RNA generation in Valsa mali. Front. Microbiol. 8:1166. doi: 10.3389/fmicb.2017.01166
Feng, H., Xu, M., Liu, Y., Gao, X., Yin, Z., Voegele, R. T., et al. (2017b). The distinct roles of Argonaute protein 2 in the growth, stress responses and pathogenicity of the apple tree canker pathogen. For. Pathol. 47:e12354. doi: 10.1111/efp.12354
Fire, A., Xu, S. Q., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
Fulci, V., and Macino, G. (2007). Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa. Curr. Opin. Microbiol. 10, 199–203. doi: 10.1016/j.mib.2007.03.016
Gent, J. I., Lamm, A. T., Pavelec, D. M., Maniar, J. M., Parameswaran, P., Tao, L., et al. (2010). Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans Soma. Mol. Cell 37, 679–689. doi: 10.1016/j.molcel.2010.01.012
German, M. A., Luo, S., Schroth, G., Meyers, B. C., and Green, P. J. (2009). Construction of parallel analysis of RNA ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 4, 356–362. doi: 10.1038/nprot.2009.8
Ghildiyal, M., and Zamore, P. D. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108. doi: 10.1038/nrg2504
Inui, M., Martello, G., and Piccolo, S. (2010). MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 11, 252–263. doi: 10.1038/nrm2868
Jiang, X., Qiao, F., Long, Y., Cong, H., and Sun, H. (2017). MicroRNA-like RNAs in plant pathogenic fungus Fusarium oxysporum f. sp niveum are involved in toxin gene expression fine tuning. 3 Biotech 7, 1–12. doi: 10.1007/s13205-017-0951-y
Jin, Y., Zhao, J.-H., Zhao, P., Zhang, T., Wang, S., and Guo, H. S. (2019). A fungal milRNA mediates epigenetic repression of a virulence gene in Verticillium dahliae. Philos. Trans. Royal Soc. B-Biol. Sci. 374:20180309. doi: 10.1098/rstb.2018.0309
Kang, K., Zhong, J., Jiang, L., Liu, G., Gou, C. Y., Wu, Q., et al. (2013). Identification of microRNA-like RNAs in the filamentous fungus Trichoderma reesei by solexa sequencing. PLoS One 8:e76288. doi: 10.1371/journal.pone.0076288
Kim, H. K., Kim, K. W., and Yun, S.H.J.J.o. M. (2015). Multiple roles of a putative vacuolar protein sorting associated protein 74, FgVPS74, in the cereal pathogen Fusarium graminearum. J. Microbiol. 53, 243–249. doi: 10.1007/s12275-015-5067-7
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, 1–10. doi: 10.1186/gb-2009-10-3-r25
Lau, S. K. P., Chow, W. N., Wong, A. Y. P., Yeung, J. M. Y., Bao, J., Zhang, N., et al. (2013). Identification of microRNA-like RNAs in mycelial and yeast phases of the thermal dimorphic fungus Penicillium marneffei. PLoS Negl. Trop. Dis. 7:e2398. doi: 10.1371/journal.pntd.0002398
Lee, H. C., Li, L., Gu, W., Xue, Z., Crosthwaite, S. K., Pertsemlidis, A., et al. (2010). Diverse pathways generate microRNA-like RNAs and dicer-independent small interfering RNAs in fungi. Mol. Cell 38, 803–814. doi: 10.1016/j.molcel.2010.04.005
Li, Y., Liu, X., Yin, Z., You, Y., Zou, Y., Liu, M., et al. (2020). MicroRNA-like milR236, regulated by transcription factor MoMsn2, targets histone acetyltransferase MoHat1 to play a role in appressorium formation and virulence of the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 137:103349. doi: 10.1016/j.fgb.2020.103349
Liu, T., Hu, J., Zuo, Y., Jin, Y., and Hou, J. (2016). Identification of microRNA-like RNAs from Curvularia lunata associated with maize leaf spot by bioinformation analysis and deep sequencing. Mol. Gen. Genomics. 291, 587–596. doi: 10.1007/s00438-015-1128-1
Mallory, A. C., and Vaucheret, H. (2006). Functions of microRNAs and related small RNAs in plants. Nat. Genet. 38:850. doi: 10.1038/ng0706-850b
Mochizuki, K., and Gorovsky, M. A. (2005). A dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 19, 77–89. doi: 10.1101/gad.1265105
Nakayashiki, H., Kadotani, N., and Mayama, S. (2006). Evolution and diversification of RNA silencing proteins in fungi. J. Mol. Evol. 63, 127–135. doi: 10.1007/s00239-005-0257-2
Nicolas, F. E., de Haro, J. P., Torres-Martinez, S., and Ruiz-Vazquez, R. M. (2007). Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet. Biol. 44, 504–516. doi: 10.1016/j.fgb.2006.09.003
Nicolas, F. E., Moxon, S., de Haro, J. P., Calo, S., Grigoriev, I. V., Torres-Martinez, S., et al. (2010). Endogenous short RNAs generated by dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic Acids Res. 38, 5535–5541. doi: 10.1093/nar/gkq301
Nicolas, F. E., and Ruiz-Vazquez, R. M. (2013). Functional diversity of RNAi-associated sRNAs in fungi. Int. J. Mol. Sci. 14, 15348–15360. doi: 10.3390/ijms140815348
Nunes, C. C., Gowda, M., Sailsbery, J., Xue, M., Chen, F., Brown, D. E., et al. (2011). Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae. BMC Genomics 12, 1–20. doi: 10.1186/1471-2164-12-288
Raman, V., Simon, S. A., Romag, A., Demirci, F., Mathioni, S. M., Zhai, J., et al. (2013). Physiological stressors and invasive plant infections alter the small RNA transcriptome of the rice blast fungus Magnaporthe oryzae. BMC Genomics 14, 1–18. doi: 10.1186/1471-2164-14-326
Shabalina, S. A., and Koonin, E. V. (2008). Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 23, 578–587. doi: 10.1016/j.tree.2008.06.005
Song, X., Li, Y., Cao, X., and Qi, Y. (2019). MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 70, 489–525. doi: 10.1146/annurev-arplant-050718-100334
Tan, K. C., and Oliver, R. P. (2017). Regulation of proteinaceous effector expression in phytopathogenic fungi. PLoS Pathog. 13:e1006241. doi: 10.1371/journal.ppat.1006241
Wang, B., Sun, Y., Song, N., Zhao, M., Liu, R., Feng, H., et al. (2017). Puccinia striiformis f. sp tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 215, 338–350. doi: 10.1111/nph.14577
Wang, X., Zang, R., Yin, Z., Kang, Z., and Huang, L. (2014). Delimiting cryptic pathogen species causing apple Valsa canker with multilocus data. Ecol. Evol. 4, 1369–1380. doi: 10.1002/ece3.1030
Wang, W., and Galili, G. (2019). Tuning the orchestra: miRNAs in plant immunity. Trends Plant Sci. 24, 189–191. doi: 10.1016/j.tplants.2019.01.009
Weiberg, A., Wang, M., Lin, F. M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Xu, M., Yin, Z., Gao, M., Lu, W., Gao, X., and Huang, L. (2016). Construction of enhanced gene deletion frequency recipient strain ΔVmKu80 in Valsa mali. Acta. Agric. Boreali-occidentalis. Sinica. 25, 298–305. doi: 10.7606/j.issn.1004-1389.2016.02.021
Xu, M., Guo, Y., Tian, R., Gao, C., Guo, F., Voegele, R. T., et al. (2020). Adaptive regulation of virulence genes by microRNA-like RNAs in Valsa mali. New Phytol. 227, 899–913. doi: 10.1111/nph.16561
Yin, Z., Ke, X., Huang, D., Gao, X., Voegele, R. T., Kang, Z., et al. (2013). Validation of reference genes for gene expression analysis in Valsa mali var. mali using real-time quantitative PCR. World. J. Microbiol. Biotechnol. 29, 1563–1571. doi: 10.1007/s11274-013-1320-6
Yu, J. H., Hamari, Z., Han, K. H., Seo, J. A., Reyes-Domínguez, Y., and Scazzocchio, C. J. F. G. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. doi: 10.1016/j.fgb.2004.08.001
Keywords: Dicer, micro-like RNA, pathogenicity, RNA silencing, Valsa mali
Citation: Guo F, Liang J, Xu M, Zhang G, Huang L and Feng H (2021) A Novel DCL2-Dependent Micro-Like RNA Vm-PC-3p-92107_6 Affects Pathogenicity by Regulating the Expression of Vm-VPS10 in Valsa mali. Front. Microbiol. 12:721399. doi: 10.3389/fmicb.2021.721399
Edited by:
Eva Varallyay, National Agricultural Research and Innovation Centre, HungaryReviewed by:
Caixia Wang, Qingdao Agricultural University, ChinaMing Wang, University of California, United States
Copyright © 2021 Guo, Liang, Xu, Zhang, Huang and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Feng, eGlhb3NvbmcwNDAwNUAxNjMuY29t; Lili Huang, aHVhbmdsaWxpQG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work
 Feiran Guo†
Feiran Guo† Jiahao Liang
Jiahao Liang Ming Xu
Ming Xu Lili Huang
Lili Huang Hao Feng
Hao Feng