- 1Department of Otolaryngology-Head and Neck Surgery, Eye Ear Nose and Throat Hospital, Fudan University, Shanghai, China
- 2Key Laboratory of Medical Molecular Virology (MOE/NHC/CAMS), Department of Medical Microbiology and Parasitology, School of Basic Medical Sciences, Shanghai Medical College of Fudan University, Shanghai, China
- 3Department of Infectious Diseases, Huashan Hospital, Fudan University, Shanghai, China
- 4National Clinical Research Center for Aging and Medicine, Huashan Hospital, Fudan University, Shanghai, China
- 5State Key Laboratory of Genetic Engineering, School of Life Science, Fudan University, Shanghai, China
- 6Key Laboratory of Medical Molecular Virology (MOE/MOH/CAMS) and Institutes of Biomedical Sciences, Shanghai Medical College, Fudan University, Shanghai, China
- 7Department of Gynecology, Obstetrics and Gynecology Hospital of Fudan University, Shanghai, China
- 8Department of Otolaryngology, The Third Affiliated Hospital of Wenzhou Medical University, Ruian, Zhejiang, China
Methicillin-resistant Staphylococcus aureus (MRSA) is a common cause of both community- and hospital-associated infections. The antibiotic resistance and virulence characteristics of MRSA are largely regulated by two-component signal transduction systems (TCS) including the graRS TCS. To make a relatively comprehensive insight into graRS TCS in MRSA, the bioinformatics analysis of dataset GSE26016 (a S. aureus HG001 WT strain vs. the ΔgraRS mutant) from Gene Expression Omnibus (GEO) database was performed, and a total of 563 differentially expressed genes (DEGs) were identified. GO analysis revealed that the DEGs were mainly enriched in the “de novo” IMP biosynthetic process, lysine biosynthetic process via diaminopimelate, and pathogenesis; and they were mainly enriched in purine metabolism, lysine biosynthesis, and monobactam biosynthesis in KEGG analysis. WGCNA suggested that the turquoise module was related to the blue module, and the genes in these two modules were associated with S. aureus virulence and infection. To investigate the role of graRS in bacterial virulence, a graRS knockout mutant (ΔgraRS) was constructed using MRSA USA500 2,395 strain as a parent strain. Compared to the wild-type strain, the USA500ΔgraRS showed reduced staphyloxanthin production, retarded coagulation, weaker hemolysis on blood agar plates, and a decreased biofilm formation. These altered phenotypes were restored by the complementation of a plasmid-expressed graRS. Meanwhile, an expression of the virulence-associated genes (coa, hla, hlb, agrA, and mgrA) was downregulated in the ΔgraRS mutant. Consistently, the A549 epithelial cells invasion of the ΔgraRS mutant was 4-fold lower than that of the USA500 wild-type strain. Moreover, on the Galleria mellonella infection model, the survival rate at day 5 post infection in the USA500ΔgraRS group (55%) was obviously higher than that in the USA500 group (20%), indicating graRS knockout leads to a decreased virulence in vivo. In addition, the deletion of the graRS in the MRSA USA500 strain resulted in its increased susceptibilities to ampicillin, oxacillin, vancomycin, and gentamicin. Our work suggests that the graRS TCS plays an important role in regulating S. aureus virulence in vitro and in vivo and modulate bacterial resistance to various antibiotics.
Introduction
Staphylococcus aureus (S. aureus) is a major Gram-positive pathogen causing both community-acquired and hospital-acquired infections (Tong et al., 2015). The prevalence of methicillin-resistant Staphylococcus aureus (MRSA) has aroused more concerns over the past decades because of their antibiotic resistance and virulence (Otto, 2013). Staphylococcus aureus has 16 two-component systems (TCSs), which are significant for bacteria and commonly used to sense and respond to environmental changes (Capra and Laub, 2012; Burgui et al., 2018; Villanueva et al., 2018; Yan et al., 2019). The TCSs have been implicated in stress conditions, pathogenesis, and essential cellular pathways (Groisman, 2016). The graRS (Glycopeptide Resistance Associated) TCS has been shown to play an important role in response to CAMPs (cationic antimicrobial peptides) and promoting resistance to CAMPS by controlling the expression of mprF and dlt operons (Howden et al., 2008; Falord et al., 2011; Jousselin et al., 2013; Muzamal et al., 2014; Keinhorster et al., 2019). The graRS is also involved in regulating the susceptibility to vancomycin and daptomycin in MRSA (Doddangoudar et al., 2011; Cafiso et al., 2012; Mensa et al., 2014; Muller et al., 2018). However, the role of graRS in the regulation of bacterial virulence remains to be determined.
With the development of high throughput microarray and RNA-seq technologies, transcription profiles data provided by researchers worldwide are valuable resources for data mining, which can provide clues for investigations of gene functions. In this study, bioinformatics analysis including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), protein–protein interaction (PPI) networks, and weighted gene co-expression network analysis (WGCNA) were applied to identify the potential genes and pathways associated with graRS in the dataset GSE26016 (Falord et al., 2011). To further investigate the role of graRS in S. aureus, the graRS knockout mutant was constructed using the MRSA USA500 strain as the parent strain. The virulence-related phenotypes measurements in vitro, cell invasion assay, and Galleria mellonella model were carried out to investigate the role of graRS in regulating S. aureus virulence.
Materials and Methods
Microarray Materials
Dataset GSE26016 downloaded from Gene Expression Omnibus (GEO) database was performed to screen the hub genes related to graRS.1 GSE26016 is based on GPL11308 (BaSysBio Staphylococcus aureus T1 385K array) platform and includes three S. aureus HG001 wild type (WT) strains and three ΔgraRS mutant strains, which cultured to mid-exponential phase in Tryptone soya broth (TSB) with 50 μg/ml colistin.
Differentially Expressed Genes Identification
The raw microarray data of GSE26016 was processed and normalized in R software (version 3.5.2, USA). Then, the “limma” package was utilized to select differentially expressed genes (DEGs) between ΔgraRS mutant strains and WT strains. |Log2 FC| (fold change) > 1 and adj. p < 0.05 were considered as statistically significant.
GO and KEGG Enrichment Analyses of DEGs
To further understand the function of the DEGs in GSE26016, the Database for Annotation, Visualization, and Integrated Discovery (DAVID) online tool were used to perform the GO and KEGG pathway analyses (Zeng et al., 2019). GO analysis consists of three items, cellular component (CC), biological processes (BP), and molecular function (MF). R software was applied to visualize the results.
Protein–Protein Interaction Network Construction and Hub Genes Identification
The STRING (Search Tool for the Retrieval of Interacting Genes) was used to analyze the interactions between the DEGs.2 The cut-off criterion of interaction with a combined score was set as >0.4. Cytoscape software (version 3.7.1, UAS) was applied to construct a PPI network and visualize the results. Moreover, the top four significant modules were identified by MCODE (molecular complex detection), a plug-in of Cytoscape software (Bandettini et al., 2012).
Co-Expression Network Analysis
To identify the potential function and clusters of highly correlated genes in GSE26016, the co-expression network was constructed by the “WGCNA” package in R software. Topological Overlap Matrix (TOM) was applied to detect gene modules. Modules were regarded as branches and hierarchical clustering results, which were cut by the Dynamic Tree-Cut algorithm.
Bacterial Strains and Culture Media
To make a relatively comprehensive insight into the effect of graRS in S. aureus, USA500 2395 strain was selected for further experiments. Staphylococcus aureus USA500 2395 strain was provided by Prof. Ying Zhang at Johns Hopkins University, and S. aureus ATCC 29213 was provided by Dr. Jinxin Zheng at Fudan University. TSB (Oxoid, United States) was used for bacterial cultivation. Mueller-Hinton Broth (MHB) and cation adjusted Mueller-Hinton Broth (CAMHB) were applied for antibiotic susceptibility tests.
Construction of the Gene Knockout Mutant and Complementation Strains
To further explore the role of graRS in S. aureus USA500 2395 strain, the graRS knockout mutant and complementation strains were constructed according to the method we previously reported (Bai et al., 2019). Briefly, the graRS genes in methicillin-resistant S. aureus USA500 2395 strain were deleted by homologous recombination using the pKOR-1, which is a temperature-sensitive vector. The regions flanking graRS gene were amplified by PCR and then inserted into the pKOR-1. The recombinant plasmid pKOR-graRS was transformed to the Escherichia coli DC10B strain and then into the S. aureus USA500 strain by electroporation. The USA500ΔgraRS was verified by PCR and RT-PCR. The vector pRB475 was used for graRS complementation. The DNA fragments of the graRS genes were amplified by PCR and then inserted into the vector. To form the complementary strain USA500ΔgraRS:: pRB475-graRS (PgraRS), the plasmid was transformed by electroporation into the USA500ΔgraRS.
RNA Extraction and Quantitative Real-Time-PCR
Total RNA was extracted from the S. aureus USA500 strain, the graRS knock-out mutant USA500ΔgraRS, and the graRS complementation strain USA500ΔgraRS:: pRB475-graRS. According to the results of bioinformatics analysis and the direction of our laboratory research, genes related to graRS, β-lactam resistance, and virulence were verified by quantitative real-time (qRT)-PCR. RNA extraction and qRT-PCR were carried out according to the previously established protocol (Bai et al., 2019). Briefly, overnight cultured USA500, USA500ΔgraRS, and USA500ΔgraRS:: pRB475-graRS were diluted 1:200 with TSB and incubated at 37°C with shaking (180 rpm) to an OD600 of 0.6 (4 h). Bacterial cells were collected and washed three times with ice-cold normal saline and homogenized using a Mini-BeadBeater-16 (Biospec, United States). The bacterial RNA was purified by using a RNeasy kit (Qiagen, Germany). DNase I-treated RNA was reversed to cDNA by using PrimeScript RT Master Mix (Takara, Japan). All samples were quantified by qRT-PCR with SYBR green reagents (Takara, Japan) and ABI 7500 real-time PCR system (Applied Biosystems, United States). The primers used in this study are shown in Table 1. The housekeeping gene gyrB was used as an internal control. All experiments were performed in triplicate and the relative expression level of mRNA was calculated by the 2−ΔΔCT method.
Carotenoid Pigment Measurement
Colonies of S. aureus USA500, USA500ΔgraRS, and USA500ΔgraRS:: pRB475-graRS were cultured in TSB at 37°C for 12 h with shaking (180 rpm). The bacterial cells were collected by centrifugation (10,000 × g, 2 min), and washed three times with double distilled water. To extract carotenoids, 100% methanol was added to the cell pellets and then heating in a water bath at 55°C for 5 min. The supernatant containing extracted carotenoid pigment was quantified by measuring the OD462 values with a microplate reader (BioRAD, United States).
Coagulase Test
The coagulase test was carried out using the freeze-dried rabbit plasma (Haibo, China). The overnight cultured S. aureus USA500, USA500ΔgraRS, and USA500ΔgraRS::pRB475-graRS were adjusted to the same OD600 value, and 0.8 ml suspension of each strain was added to the tubes, respectively. The tubes were shaken slightly to make sure the freeze-dried rabbit plasma dissolved completely. Afterward, they were incubated at 37°C and checked for visual clot formation every half hour until up to 6 h.
Hemolytic Phenotype Analysis
The S. aureus USA500, USA500ΔgraRS, and USA500ΔgraRS::pRB475-graRS strains were inoculated on blood agar plates (BioMérieux, France) and cultured at 37°C for 48 h. Then the results of the hemolytic phenotype of these three strains were observed and photographed.
Detection of Bacterial Biofilm Formation With the Microplate Assay
The overnight culture of bacterial strains was diluted 1:200 with TSB which supplemented with 1% glucose, and then added to a 96-well polyethylene microplate (200 μl bacterial suspension in each well). After incubation at 37°C for 24 h and 48 h, the biofilms were washed three times with phosphate-buffered saline (PBS) after removal of non-adhered bacteria. To fix the adhered cells, each well was added with 200 μl of 100% methanol. After the methanol was removed and the biofilms were stained with 2% crystal violet, the wells were washed with running tap water. The OD570 values of stained biofilms were measured by a microtiter-plate reader (Beckman Coulter, United States).
Observation of Staphylococcus aureus Biofilms by Confocal Laser-Scanning Microscopy
The effect of graRS knockout on the S. aureus biofilms was evaluated by LIVE/DEAD staining. The bacterial strains were cultivated in TSB supplemented with 1% glucose in glass-bottomed fluorodishes for 48 h. The biofilms were washed with PBS 3 times, stained with 1 μM of SYTO9 and 1 μM of propidium iodide (PI) for 20 min, and then visualized by confocal laser-scanning microscopy (CLSM) with a 63× 1.4-NA oil immersion objective (Leica TCS SP8 Confocal Laser Scanning Platform, Leica Microsystems, Germany). The three-dimensional biofilm images were generated with IMARIS 7.0 software (Bitplane, United States). The thicknesses of the biofilms and the fluorescence intensities were determined using Leica Application Suite 1.0 software (Leica Microsystem).
Antimicrobial Susceptibility Testing
The susceptibilities of S. aureus USA500, USA500ΔgraRS, and USA500ΔgraRS:: pRB475-graRS strains to various antibiotics were measured by the broth microdilution method based on the protocols of the American Clinical and Laboratory Standards Institute (CLSI). A 2-fold microdilution assay was performed to measure the minimal inhibitory concentrations (MICs) of the antibiotics. The range of the antibiotic’s concentrations was from 8 to 0.125 mg/L in the 96-well microplates containing CAMHB. Overnight cultured bacteria were diluted 1:200 with MHB and incubated at 37°C for 4 h. Then the 0.5 McFarland bacterial suspension was inoculated 1:200 into the CAMHB (for oxacillin, CAMHB+2% NaCl) and incubated at 35°C for 16 h or 24 h. S. aureus ATCC 29213 was included as controls.
Epithelial Cell Invasion Assay
Human alveolar epithelial cells A549 were cultured in DMEM media (Hyclone, United States), which was supplemented with 10% FBS (fetal bovine serum; Gibco, United States). One day before infection, cells were seeded in 24 wells plates (Costar, United States) and cultured overnight at 37°C with 5% CO2. The cells infected at a MOI (multiplicity of infection) of 10:1 were centrifuged at 500 × g for 3 min and then incubated at 37°C with 5% CO2 for 2 h. To kill the extracellular bacteria, the medium containing 40 μg/ml lysostaphin and 200 μg/ml gentamicin was added to each well, which was previously washed three times with 500 μl PBS. The plates were incubated at 37°C for 30 min, and then washed twice with 500 μl PBS. The 0.1% Triton X-100 was applied to lyse the monolayer cells. After 20 min incubation, the lysates were diluted with PBS and plated on TSA plates to count the number of the intracellular bacteria.
Virulence Assessment in Galleria mellonella Model
Galleria mellonella (G. mellonella) larvae were used as an invertebrate infection model to investigate the virulence of USA500, USA500ΔgraRS, and USA500ΔgraRS:: pRB475-graRS strains referring to previous reports (Insua et al., 2013; Mannala et al., 2018). Briefly, a total of 100 healthy G. mellonella larvae weighing between 250 and 300 mg were selected and randomly divided into five groups (blank control, PBS control, USA500, USA500ΔgraRS, and USA500ΔgraRS:: pRB475-graRS; 20 G. mellonella larvae/group). Each bacterial suspension (105 CFU/ml) and the PBS control were injected into the four groups of larvae, respectively. The liquid was injected to the last left proleg of each larva with a 10 μl Hamilton syringe. The G. mellonella larvae were incubated at 37°C and monitored daily for survival for 5 days.
Statistical Analysis
The analyses of GSE26016 were performed by R software (version 3.5.2, United States). GraphPad Prism version 7.0 software (La Jolla, United States) was used for statistical analysis. Kaplan–Meier analysis was performed for the survival plots of in vivo infection. A log-rank test was carried out for comparison of survival curves.
Results
DEGs Between Staphylococcus aureus HG001 Wild-Type Strains and ΔgraRS Mutant Strains
GSE26016 includes three S. aureus HG001 WT strains and three ΔgraRS mutant strains, which were grown with shaking (180 rpm) at 37°C in TSB containing 50 μg/ml colistin until OD600 of 1.0 was obtained. A total of 563 DEGs were identified by R software (adj. p < 0.05, |log2 FC| > 1), which were shown in the heatmap (Figure 1A). Among them, 269 genes were upregulated, and 294 genes were downregulated (Figure 1B).
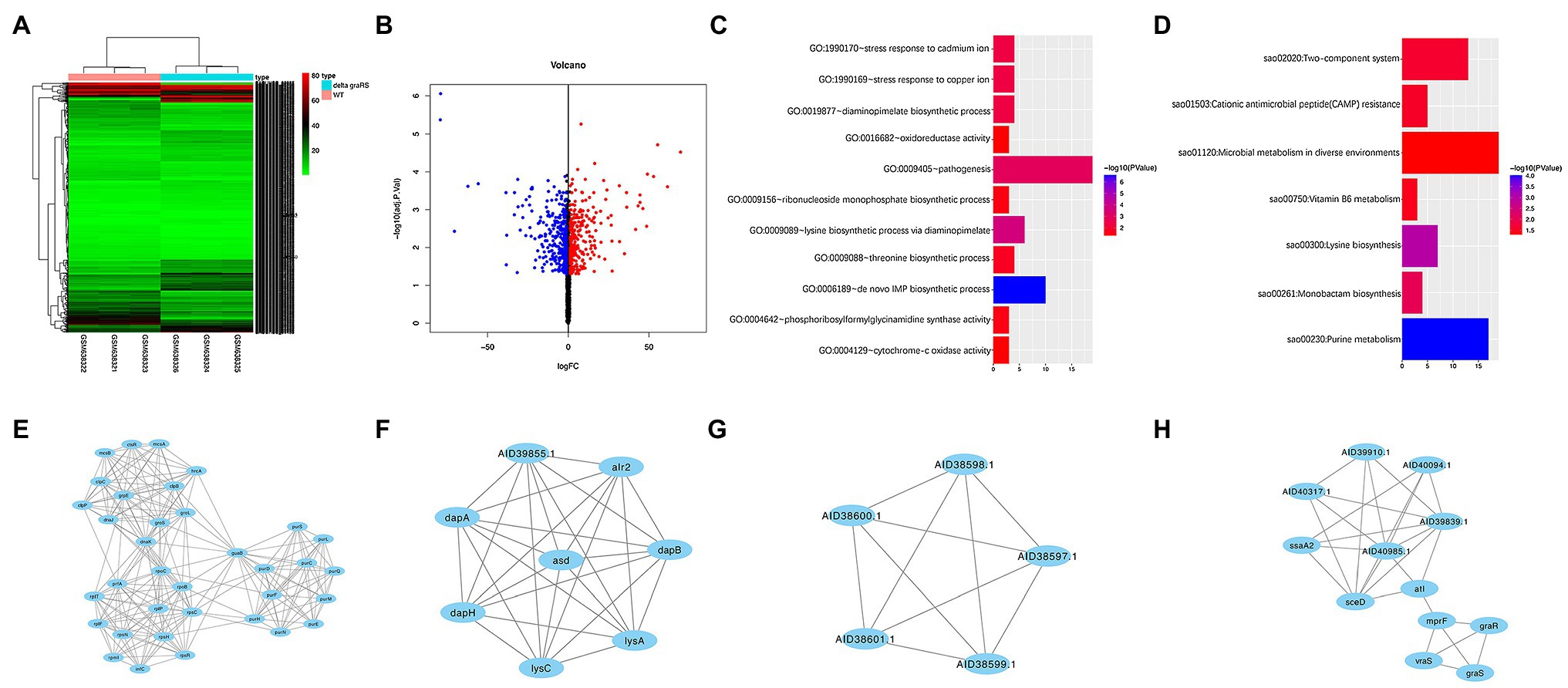
Figure 1. Bioinformatic analysis of GSE26016 dataset. (A) Heatmap representation of differentially expressed genes in Staphylococcus aureus HG001 wild-type strain and the ΔgraRS mutant. The highly expressed genes of HG001 ΔgraRS mutant strain compared with HG001 WT were represented by the red areas and the lowly expressed genes were represented by the green areas. (B) Volcano plot of the differentially expressed genes (DEGs) in GSE26016 identified by “limma” package in R software. Blue dots represent the down-regulated genes and red dots represent the up-regulated genes of HG001 ΔgraRS mutant strain compared with HG001 WT. (C) Gene Ontology (GO) functional enrichment result of DEGs in GSE26016. (D) Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment result of DEGs in GSE26016. (E–H) Four significant modules in GSE26016 obtained by protein–protein interaction (PPI) of 563 DEGs: (E) The most significant module detected by MCODE (score = 13.176). (F) The second significant module detected by MCODE (score = 8). (G) The third significant module detected by MCODE (score = 5). (H) The fourth significant module detected by MCODE (score = 4.909).
GO and KEGG Pathway Enrichment of DEGs Between Staphylococcus aureus HG001 Wild-Type Strains and ΔgraRS Mutant Strains
GO and KEGG analyses were carried out by DAVID online tools to determine the biological functions of the 563 DEGs. GO analysis revealed that the DEGs were mainly enriched in “de novo” biosynthetic process of inosine monophosphate (IMP), lysine biosynthetic process via diaminopimelate, and pathogenesis; while in KEGG analysis they were mainly enriched in purine metabolism, lysine biosynthesis, and monobactam biosynthesis (Figures 1C,D).
PPI Network of DEGs Between Staphylococcus aureus HG001 Wild-Type Strains and ΔgraRS Mutant Strains
PPI network of DEGs was constructed based on the STRING database and the four most significant modules were obtained by MCODE. The IMP dehydrogenase guaB is relevant to metabolic and redox enzymes. The aspartate semialdehyde dehydrogenase (asd) is essential for the biosynthesis of lysine (Oogai et al., 2016). lysC is related to vancomycin resistance in S. aureus (Shoji et al., 2011). The graR and graS are relevant to mprF and vraS, and autolysin (atl) is involved in biofilm formation (Figures 1E–H; Herbert et al., 2007).
Co-expression Network Analysis of GSE26016
There were 2,892 original mRNAs in GSE26016. After deletion and outlier values were eliminated, 2,825 original mRNAs remained. To group the coherent expression genes into modules, a hierarchical clustering algorithm was performed (Pu et al., 2020). Finally, 26 modules were obtained (Supplementary Figure S1A), and the number of mRNAs contained in the top six modules is shown in Table 2. Meanwhile, the blue module was related to the turquoise one (Supplementary Figures S1B,C). GO and KEGG analyses were performed in these two modules to further understand the correlation of these genes (Table 3).
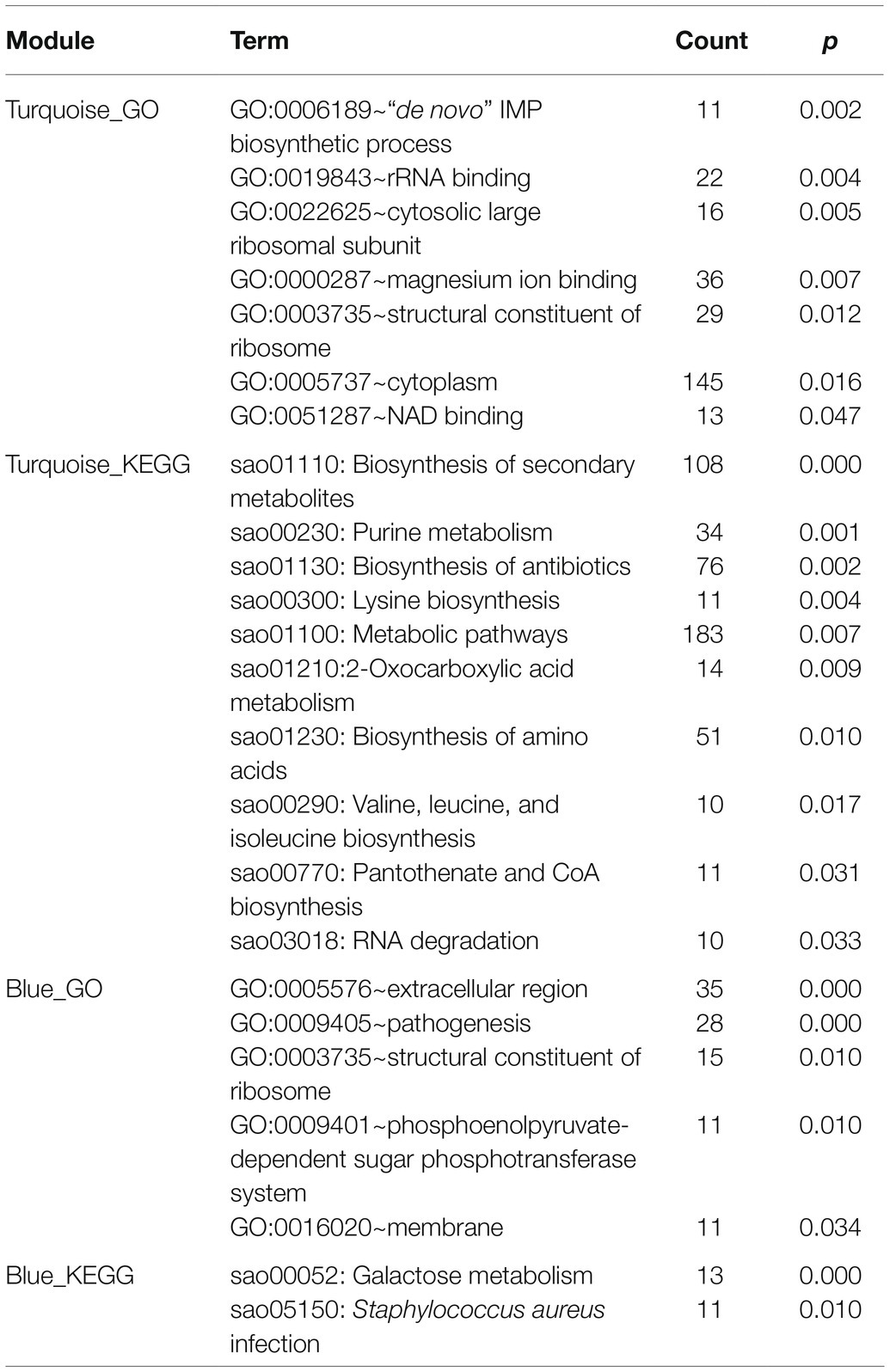
Table 3. GO and KEGG enrichment analyses of module blue and turquoise constructed by WGCNA (p < 0.05, count ≥ 10).
Effect of graRS Mutation in the MRSA USA500 Strain on Bacterial Virulence in vitro
According to the bioinformatics analysis, the DEGs in GSE26016 were enriched in pathogenesis category in GO analysis (Figure 1C). Meanwhile, the blue module and the turquoise module in WGCNA were associated with S. aureus infection and virulence (Table 3). As we know, S. aureus produces numerous virulence factors, including coagulase, alpha and beta hemolysin, etc. (Lan et al., 2010). To investigate the effects of graRS on the expression levels of associated genes in the graRS knockout mutant of the USA500 2395 strain, qRT-PCR was applied. As shown Figure 2A, the expression of coa, hla, and hlb were significantly downregulated in the ΔgraRS mutation strain (greater than 6-fold). In addition, the virulence-related phenotypes, such as the formation of staphyloxanthin, coagulase, and hemolysis were examined. Compared with USA500 wild-type strain, the USA500ΔgraRS mutant showed a reduced staphyloxanthin production (p < 0.05), which was restored in the graRS complementation strain (Figure 2B). In the coagulase test, when rabbit plasma was incubated with each diluted bacterial suspension, respectively, for 0.5 h, there was obvious flocculation precipitation in the tubes with USA500 and the graRS complementation strains, while no precipitation was seen in the tube with USA500ΔgraRS. Then the tube with USA500ΔgraRS showed a little visible white precipitate at 1 h, which as much less than those in the tubes with USA500 and the graRS complementation strains. All three tubes had immobile clots formed at 6 h (Figure 2C). In the hemolysis test, a clear hemolytic ring with a diameter of larger than 7 mm was observed around the USA500 colonies on blood agar plates, whereas a smaller ring (a diameter of 5 mm) was formed around USA500ΔgraRS colonies. The phenotype was restored in the graRS complementation strain (Figure 2D).
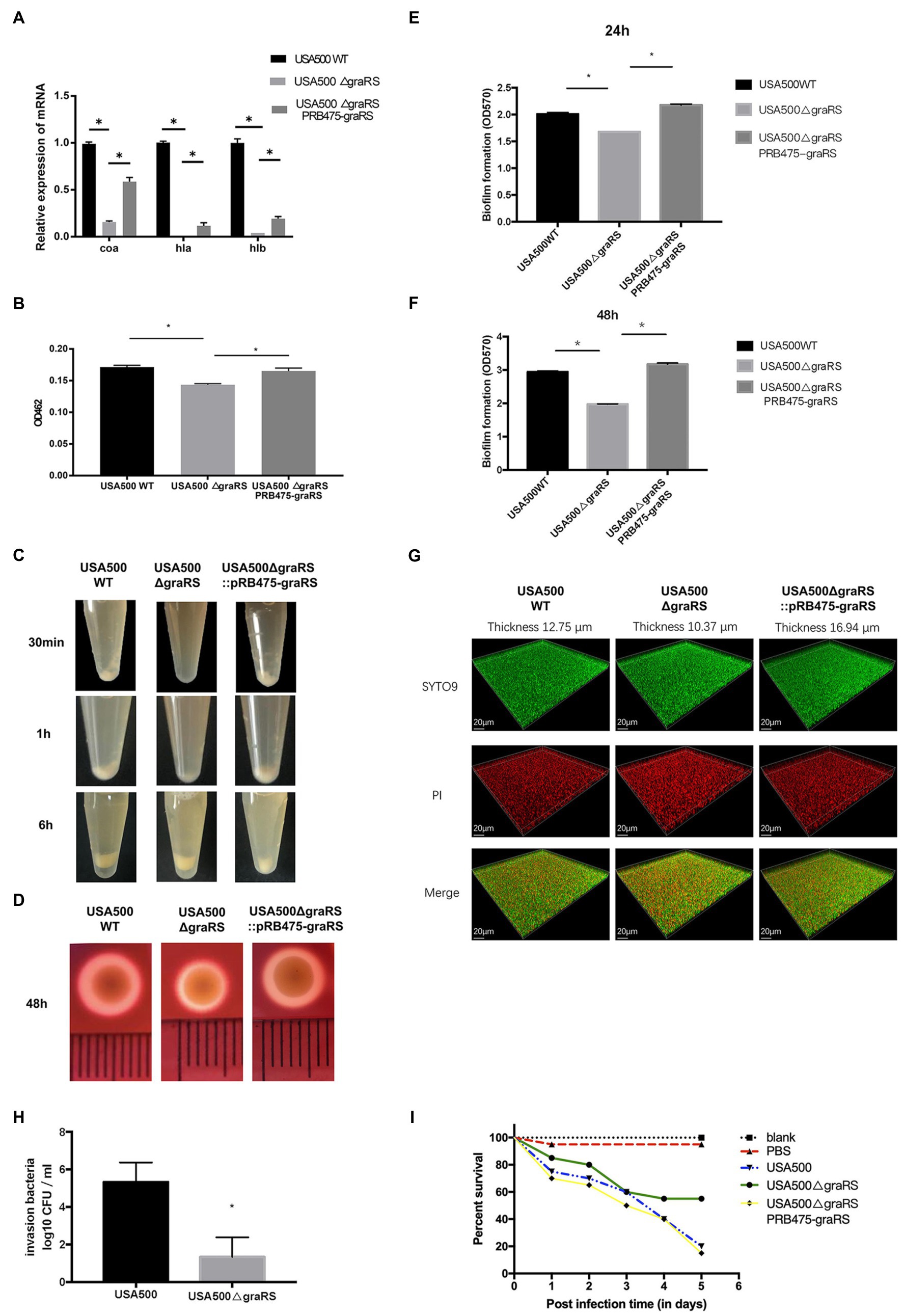
Figure 2. The effect of graRS knockout on the expression of the virulence-associated genes, virulence phenotypes and biofilm formation. (A) Transcriptional levels of the virulence genes in USA500WT, USA500ΔgraRS, and USA500ΔgraRS::pRB475-graRS strains were detected by qRT-PCR. (B) The effect of graRS deletion in the USA500 on the carotenoid pigment formation. (C) The effect of graRS knockout in the USA500 on the coagulase activities at 30 min, 1, and 6 h. (D) The effect of graRS FIGURE 2deletion in the USA500 on β-hemolysis on the blood agar at 48 h. (E,F) The microtiter plate assay was carried out to detect the ability to form biofilms of USA500WT, USA500ΔgraRS, and USA500ΔgraRS::pRB475-graRS strains in TSB supplemented with 1% glucose at 24 h (E) and 48 h (F). (G) Confocal laser-scanning microscopy (CLSM) was performed to visualize the three-dimensional structure of biofilms formed by the three strains. The bacterial strains were cultivated in TSB supplemented with 1% glucose in glass-bottomed fluorodishes for 48 h. The biofilms were stained with 1 μM of SYTO9 and 1 μM of propidium iodide (PI) for 20 min, and then observed by CLSM with a 63× 1.4-NA oil immersion objective. The three-dimensional biofilm images were generated with IMARIS 7.0 software. (H) Epithelial cell invasion assay: Human alveolar epithelial cells A549 infected by USA500 or USA500ΔgraRS at a MOI (multiplicity of infection) of 10:1 were centrifuged at 500 × g for 3 min and then incubated at 37°C with 5% CO2 for 2 h. Then the medium containing 40 μg/ml lysostaphin and 200 μg/ml gentamicin was used to kill the extracellular bacteria. The cells were lysed by 0.1% Triton X-100, and the intracellular bacteria were diluted with PBS and plated on TSA plates for counting. (I) Virulence assessment in the Galleria mellonella infection model: A total of 100 healthy G. mellonella larvae were randomly divided into five groups (20 Galleria mellonella larvae/group). Among them, three groups were injected with the bacterial suspension of USA500, USA500ΔgraRS, or USA500ΔgraRS::pRB475-graRS (105 CFU/ml), respectively. Another group was injected with sterile PBS (PBS control). The last group did not receive any treatment (untreated control). The five groups of G. mellonella larvae were incubated at 37°C and monitored daily for survival for 5 days (*p < 0.05).
Effect of graRS Mutation on Bacterial Biofilm Formation
To investigate the effects of graRS on S. aureus biofilm formation, a microtiter plate assay was performed to detect the biofilms of the USA500, USA500ΔgraRS, and USA500ΔgraRS::pRB475-graRS strains, using TSB supplemented with 1% glucose as the culture medium. After 24 h incubation, the biofilm of USA500ΔgraRS (OD570 = 1.678 ± 0.00) was decreased compared to wild-type strain (OD570 = 2.009 ± 0.03), and graRS complementation restored the biofilm formation to the wild-type level (OD570 = 2.178 ± 0.02; Figure 2E). After 48 h incubation, biofilm produced by USA500ΔgraRS (OD570 = 1.975 ± 0.01) was significantly less than its wild-type counterpart (OD570 = 2.944 ± 0.03) and was also restored by graRS complementation (OD570 = 3.171 ± 0.04; Figure 2F). Consistently, when biofilms of the USA500, USA500ΔgraRS, and USA500ΔgraRS::pRB475-graRS strains were cultured in TSB supplemented with 1% glucose in fluorodishes and were observed under CLSM with live/dead staining (Figure 2G), the thickness of the graRS mutant biofilm (10.37 μm) was less than that of the parent strain (12.75 μm), and the thickness was restored by complementation with pRB475-graRS (16.94 μm).
Effect of graRS Mutation on the Bacterial Invasion of the Epithelial Cells
The ability of bacteria to invade epithelial cells partly reflects the virulence of these strains. To further identify the effect of graRS knockout on S. aureus virulence, the invasion capacity of the USA500 and USA500ΔgraRS strains was evaluated by the gentamicin protection assay on A549 cells. Bacterial strains were separately inoculated into A549 cells at a MOI of 10, incubated for 2 h, and then gentamicin and lysostaphin were added to kill extracellular bacteria. It showed that the intracellular bacterial cells of the USA500 and the ΔgraRS mutant were log10 CFUs of 5.333 ± 0.60 and 1.333 ± 0.60 (n = 3, p < 0.01), respectively, indicating that graRS knockout led to a 4-fold reduction in the cell invasion rate (Figure 2H).
Effect of graRS Mutation on MRSA Infections in the Galleria mellonella Model
To further investigate the influence of graRS knockout on in vivo infection, the G. mellonella larvae infection model that has been widely applied in studies of microbial pathogenesis was employed (Jonsson et al., 2017). The G. mellonella larvae were divided into three infection groups (USA500, USA500ΔgraRS, USA500ΔgraRS:: pRB475-graRS) and two control groups (blank control and PBS control), with 20 G. mellonella larvae per group. Galleria mellonella larvae were incubated at 37°C and monitored daily for survival for 5 days. On day 1, 2, 3, 4, and 5, the survival rate in the USA500 infection group was 75, 70, 60, 40, and 20%, respectively, while that in the USA500ΔgraRS infection group was 85, 80, 60, 55, and 55%, respectively. The reduction in survival of G. mellonella after 5 days in the USA500ΔgraRS group was obviously lower than that in the USA500 group (p < 0.05). The survival rate in the graRS complementation group was similar to that in the wild-type group. The blank control (untreated group) and the PBS treated group (uninfected group) showed 100 and 95% survival rates at day 5, respectively (Figure 2I).
Effect of graRS Mutation in USA500 on the Antimicrobial Susceptibilities
According to the bioinformatics analysis, the DEGs in GSE26016 were related to monobactam biosynthesis. Monobactams are beta-lactam antibiotics that contain a monocyclic beta-lactam nucleus and could cause cell wall impairment in MRSA (Liu et al., 2018; Wang et al., 2019). To investigate the effect of graRS two-component system on antibiotics resistance, antimicrobial susceptibility test using the broth microdilution method was carried out. The minimum inhibitory concentration (MICs) of ampicillin, oxacillin, vancomycin, and gentamicin for the USA500∆graRS mutant were 2, 2, 0.25, and 0.5 μg ml, respectively, while were >8, >8, 1, and 4 μg/ml for the parent strain USA500. The gene complementation by plasmid expressing graRS in USA500∆graRS restored the MIC values to the wild-type levels (Table 4). In addition, the mRNA levels of β-lactam resistance-related genes were detected by qRT-PCR. The results indicated that the transcriptional levels of the genes encoding penicillin-binding proteins PBP1, PBP2, and PBP4, the PBP2a (or PBP2’) encoding gene mecA, peptidoglycan synthesis genes femA, autolytic activity related gene llm, global regulator genes mgrA, spx, agrA, and sigB, the cationic antimicrobial peptide (CAMP) resistance-related genes dltX, dltA, mprF, vraF, and vraG, were significantly down-regulated in USA500ΔgraRS mutant strain (Figures 3A,B; Supplementary Figure S2).
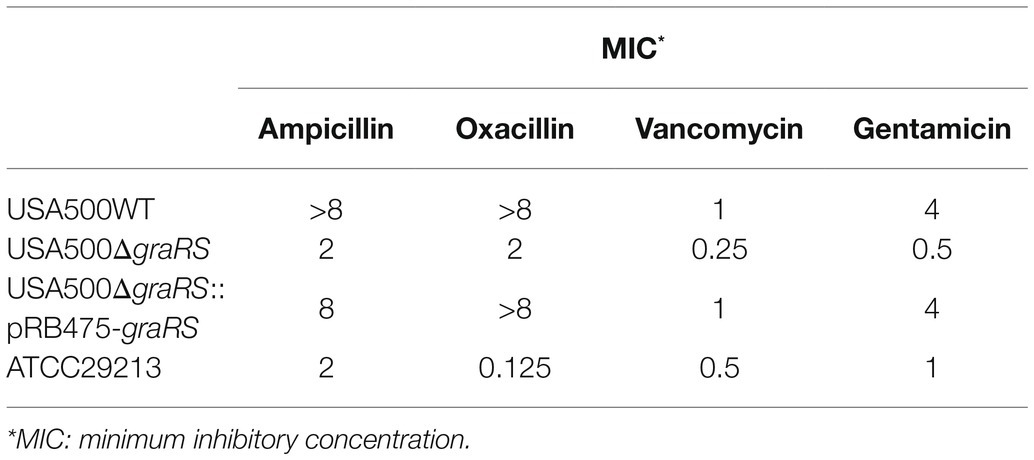
Table 4. The MIC of ampicillin, gentamicin, oxacillin, and vancomycin for different Staphylococcus aureus strains (the broth microdilution method).
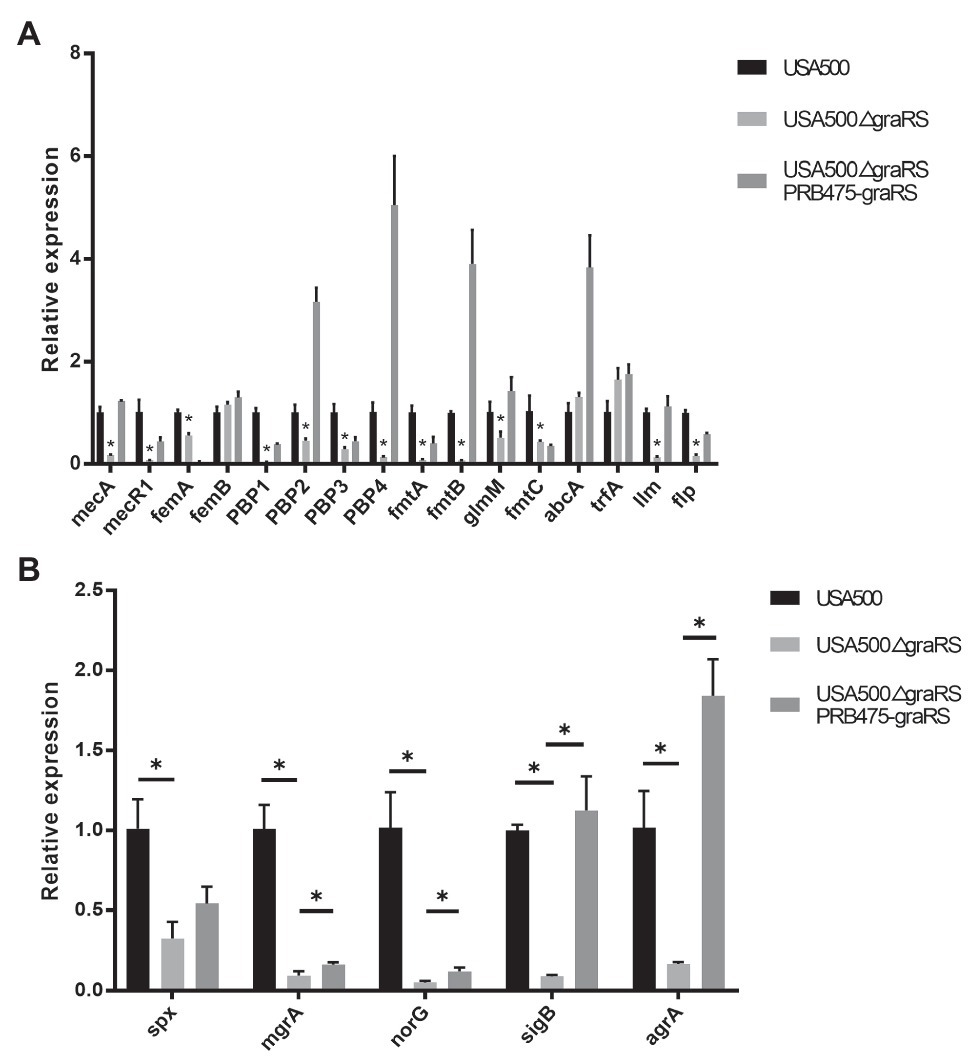
Figure 3. The effect of graRS knockout on the mRNA levels of the antibiotic resistance-related genes. The mRNA of the USA500WT, USA500ΔgraRS, and USA500ΔgraRS::pRB475-graRS strains cultured in TSB at 4 h was extracted, then qRT-PCR was performed to detected the mRNA levels of β-lactam resistance-related genes (A) and the antibiotic resistance-related global regulatory genes (B).
Discussion
The two-component system graRS of S. aureus belongs to the IM-HK (intramembrane-sensing histidine kinase) family and is conserved within the firmicutes. Previous studies of the graRS TCS mainly focused on its regulatory role in the resistance to host defense CAMPs and vancomycin (Kawada-Matsuo et al., 2011; Kolar et al., 2011; Cheung et al., 2014). In recent decades, with the development of high-throughput microarray technology, transcriptome and bioinformatics analysis have been widely used in disease biology areas. Falord, M et al. reported the differentially expressed genes between the S. aureus parental HG001 strain and the ΔgraRS mutant (mid-exponential phase in TSB with 50 μg/ml colistin; Falord et al., 2011), while the detailed description of the interaction between DEGs needs further exploration. In our present study, to make a better understanding of the function of graRS in S. aureus, the dataset GSE26016, which including three Staphylococcus aureus HG001 WT strains and three ΔgraRS mutant strains, was analyzed in detail. As a result, 563 DEGs were identified between WT group and ΔgraRS mutant group, including 269 upregulated DEGs and 294 downregulated DEGs (adj. p < 0.05, |log2FC| > 1). According to GO and KEGG analyses, the DEGs were mostly enriched in pathogenesis and microbial metabolism in diverse environments, respectively (Figures 1C,D). The PPI results showed that GraR and GraS proteins were relevant to MprF and VraS (Figures 1E,H). Meanwhile, WGCNA suggested that the turquoise module was related to the blue module, and the genes in these two modules were associated with S. aureus virulence and infection (Supplementary Figures S1A–C; Table 3). The above analyses in GSE26016 showed that the expression levels of some virulence-associated genes were significantly downregulated in ΔgraRS mutant strains in the condition of 50 μg/ml colistin. In addition, DEGs function enrichment analysis results suggest that graRS plays a role in pathogenesis and antibiotic resistance. To make a relatively comprehensive insight into the effect of graRS on bacterial virulence phenotypes and antibiotic resistance, in the present study, S. aureus USA500 2395 strain was selected and cultured without antibiotic interference. USA500ΔgraRS showed a weaker β-hemolysis on blood agar plates, and the same trend was shown in its ability of coagulase and pigment production (Figures 2B–D). β-hemolysis is related with Staphylococcal α-hemolysin, a pore-forming toxin, which is encoded by hla and expressed in an agr-dependent manner. According to the results of qRT-PCR, the transcription of hla and agrA was significantly downregulated in ΔgraRS mutant, which is consistent with the weaker β-hemolysis phenotype. Coagulase (encoded by coa), another important virulence factor produced by S. aureus, is a polypeptide that binds to and activates prothrombin, thereby converting fibrinogen to fibrin and promoting clotting of plasma or blood. We found that coa transcription was significantly downregulated in ΔgraRS mutant, which is correlated with its coagulase production. It has been reported that another toxin, β-toxin, encoded by hlb gene, is required for the lethal effect of S. aureus culture on silkworm larvae. Our work showed that the transcription level of hlb was significantly lower in the ΔgraRS mutant, compared to that in the USA500 WT strain. We further established the A549 epithelial cell invasion model and the G. mellonella infection model to assess the effects of graRS on bacterial cell invasion and in vivo infection, and the results showed the graRS knockout was associated with a decreased number of intracellular bacteria and a higher survival rate of G. mellonella (Figures 2H,I). Our data indicate that GraRS plays a vital role in regulating various aspects of S. aureus virulence.
Boles et al. (2010) reported that transposon insertion mutations in the graS gene of the S. aureus strains SH1000 and LAC resulted a significant decrease in biofilm formation in a flow cell apparatus. Consistently, we found that the graRS knockout mutation led to reduced biofilm formation, as the microtiter plate assay showed a lower OD570 value in the USA500ΔgraRS than that of the USA500 WT strain and the graRS complementation strain (Figures 2E,F). The low expression of dlt operons, ica, and atlA was related to decreased biofilm formation (Herbert et al., 2007; Boles and Horswill, 2008), while agr mutation enhanced biofilm formation (Yarwood et al., 2004; Boles and Horswill, 2008; Jenul and Horswill, 2019). In our study, qRT-PCR results showed that the expression of dltX, dltA, and agrA was significantly decreased in USA500ΔgraRS mutation strain (Supplementary Figure S2; Figure 3B). It could make a combined influence on biofilm formation which needs further exploration.
According to the results of the antimicrobial susceptibility test in this study, the TCS graRS knockout in MRSA strain USA500 provides increased sensitivity to ampicillin, gentamicin, oxacillin, and vancomycin, which suggest an important role of graRS in regulating antibiotic resistance in MRSA (Table 4). The mRNA levels of the relevant genes including mprF, mgrA, spx, and vraF were significantly downregulated in USA500ΔgraRS mutant strain. Previous studies have shown that vraFG (encoding a peptide efflux pump), mprF (encoding the MprF enzyme and modulating the charge of cell membrane outer surface), and the dlt operon (encoding components modulating the cell wall charge) are regulated by the graRS sensory system (Li et al., 2007; Pietiainen et al., 2009; Bayer et al., 2016). The graRS system plays an important role in cell wall stress responses (Meehl et al., 2007; Yang et al., 2012). Furthermore, the graRS system seems to be important to neutrophil killing, as the dltX and mprF play an important role in response to cationic antimicrobial peptides (CAMPs; Kraus et al., 2008; Ernst et al., 2009; Rajagopal et al., 2016; Kamar et al., 2017). According to the modulation of anionic charge of cell surface components, S. aureus can obtain protection from the harmful effects of the CAMPs activity (Peschel and Sahl, 2006). Meanwhile, mprF is a hub gene that confers resistance to daptomycin (Mehta et al., 2012). In some reports, as a global transcriptional regulator, mgrA can affect the expression of virulence genes and has a negative regulation in autolysis genes (Trotonda et al., 2009; Sun et al., 2011; Crosby et al., 2016). Bai et al. (2019) have reported that silence spx results in a significant increase in oxacillin susceptibility of USA300 and USA500 strains.
In conclusion, the graRS knockout in the MRSA USA500 strain repressed bacterial virulence in vitro, in the cell model and in the G. mellonella infection model. Meanwhile, the ΔgraRS mutant was more susceptible to various antibiotics. The transcriptional profile analysis data of the ΔgraRS mutant was consistent with the altered phenotypes. These findings provide novel insights into the role of the graRS system in regulating S. aureus virulence and antibiotic resistance.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding author/s.
Author Contributions
YW, KZ, DQ, and CZ designed the work and revised the manuscript. LC performed bioinformatic analysis. LC, ZW, TX, HG, FZ, XZ, and XL completed all the experiments. LC, ZW, KZ, and YW performed the statistical analysis, made the figures, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Natural Science Foundation of Shanghai (no. 20ZR1410000) and National Natural Science Foundation of China (No. 82072249, 81991532, 81671982, and 81970855).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.727104/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | WGCNA analysis and the mRNA levels of interested genes in GSE26016. (A) Clustering dendrogram of genes in GSE26016. (B) Clustering tree based on the module eigengenes of modules. (C) Network heatmap plot in the co-expression modules. (D) The mRNA levels of hla, hlb, and coa in GSE26016. (E) The mRNA levels of mprF, dltX, dltA, and agrA in GSE26016. WGCNA: weighted gene co-expression network analysis.
SUPPLEMENTARY FIGURE S2 | The effect of graRS knockout on the mRNA levels of the CAMP resistance-associated genes. The mRNA of the USA500WT and USA500ΔgraRS cultured in TSB at 4 h was extracted, then qRT-PCR was performed to detected the mRNA levels of the graR, graS, vraF, vraG, dltX, and dltA.
Footnotes
References
Bai, J., Zhu, X., Zhao, K., Yan, Y., Xu, T., Wang, J., et al. (2019). The role of ArlRS in regulating oxacillin susceptibility in methicillin-resistant Staphylococcus aureus indicates it is a potential target for antimicrobial resistance breakers. Emerg. Microbes Infect. 8, 503–515. doi: 10.1080/22221751.2019.1595984
Bandettini, W. P., Kellman, P., Mancini, C., Booker, O. J., Vasu, S., Leung, S. W., et al. (2012). MultiContrast delayed enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J. Cardiovasc. Magn. Reson. 14:83. doi: 10.1186/1532-429X-14-83
Bayer, A. S., Mishra, N. N., Cheung, A. L., Rubio, A., and Yang, S. J. (2016). Dysregulation of mprF and dltABCD expression among daptomycin-non-susceptible MRSA clinical isolates. J. Antimicrob. Chemother. 71, 2100–2104. doi: 10.1093/jac/dkw142
Boles, B. R., and Horswill, A. R. (2008). Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. doi: 10.1371/journal.ppat.1000052
Boles, B. R., Thoendel, M., Roth, A. J., and Horswill, A. R. (2010). Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. doi: 10.1371/journal.pone.0010146
Burgui, S., Gil, C., Solano, C., Lasa, I., and Valle, J. (2018). A systematic evaluation of the two-component systems network reveals That ArlRS is a key regulator of catheter colonization by Staphylococcus aureus. Front. Microbiol. 9:342. doi: 10.3389/fmicb.2018.00342
Cafiso, V., Bertuccio, T., Spina, D., Purrello, S., Campanile, F., Di Pietro, C., et al. (2012). Modulating activity of vancomycin and daptomycin on the expression of autolysis cell-wall turnover and membrane charge genes in hVISA and VISA strains. PLoS One 7:e29573. doi: 10.1371/journal.pone.0029573
Capra, E. J., and Laub, M. T. (2012). Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66, 325–347. doi: 10.1146/annurev-micro-092611-150039
Cheung, A. L., Bayer, A. S., Yeaman, M. R., Xiong, Y. Q., Waring, A. J., Memmi, G., et al. (2014). Site-specific mutation of the sensor kinase GraS in Staphylococcus aureus alters the adaptive response to distinct cationic antimicrobial peptides. Infect. Immun. 82, 5336–5345. doi: 10.1128/IAI.02480-14
Crosby, H. A., Schlievert, P. M., Merriman, J. A., King, J. M., Salgado-Pabon, W., and Horswill, A. R. (2016). The Staphylococcus aureus global regulator MgrA modulates clumping and virulence by controlling surface protein expression. PLoS Pathog. 12:e1005604. doi: 10.1371/journal.ppat.1005604
Doddangoudar, V. C., Boost, M. V., Tsang, D. N. C., and O'Donoghue, M. M. (2011). Tracking changes in the vraSR and graSR two component regulatory systems during the development and loss of vancomycin non-susceptibility in a clinical isolate. Clin. Microbiol. Infect. 17, 1268–1272. doi: 10.1111/j.1469-0691.2011.03463.x
Ernst, C. M., Staubitz, P., Mishra, N. N., Yang, S. J., Hornig, G., Kalbacher, H., et al. (2009). The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 5:e1000660. doi: 10.1371/journal.ppat.1000660
Falord, M., Mader, U., Hiron, A., Debarbouille, M., and Msadek, T. (2011). Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS One 6:e21323. doi: 10.1371/journal.pone.0021323
Groisman, E. A. (2016). Feedback control of two-component regulatory systems. Annu. Rev. Microbiol. 70, 103–124. doi: 10.1146/annurev-micro-102215-095331
Herbert, S., Bera, A., Nerz, C., Kraus, D., Peschel, A., Goerke, C., et al. (2007). Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog. 3:e102. doi: 10.1371/journal.ppat.0030102
Howden, B. P., Stinear, T. P., Allen, D. L., Johnson, P. D., Ward, P. B., and Davies, J. K. (2008). Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob. Agents Chemother. 52, 3755–3762. doi: 10.1128/AAC.01613-07
Insua, J. L., Llobet, E., Moranta, D., Perez-Gutierrez, C., Tomas, A., Garmendia, J., et al. (2013). Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth galleria mellonella. Infect. Immun. 81, 3552–3565. doi: 10.1128/IAI.00391-13
Jenul, C., and Horswill, A. R. (2019). Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.GPP3-0031-2018
Jonsson, R., Struve, C., Jenssen, H., and Krogfelt, K. A. (2017). The wax moth galleria mellonella as a novel model system to study Enteroaggregative Escherichia coli pathogenesis. Virulence 8, 1894–1899. doi: 10.1080/21505594.2016.1256537
Jousselin, A., Kelley, W. L., Barras, C., Lew, D. P., and Renzoni, A. (2013). The Staphylococcus aureus thiol/oxidative stress global regulator Spx controls trfA, a gene implicated in cell wall antibiotic resistance. Antimicrob. Agents Chemother. 57, 3283–3292. doi: 10.1128/AAC.00220-13
Kamar, R., Rejasse, A., Jehanno, I., Attieh, Z., Courtin, P., Chapot-Chartier, M. P., et al. (2017). DltX of Bacillus thuringiensis is essential for D-Alanylation of teichoic acids and resistance to antimicrobial response in insects. Front. Microbiol. 8:1437. doi: 10.3389/fmicb.2017.01437
Kawada-Matsuo, M., Yoshida, Y., Nakamura, N., and Komatsuzawa, H. (2011). Role of two-component systems in the resistance of Staphylococcus aureus to antibacterial agents. Virulence 2, 427–430. doi: 10.4161/viru.2.5.17711
Keinhorster, D., George, S. E., Weidenmaier, C., and Wolz, C. (2019). Function and regulation of Staphylococcus aureus wall teichoic acids and capsular polysaccharides. Int. J. Med. Microbiol. 309, 151333. doi: 10.1016/j.ijmm.2019.151333
Kolar, S. L., Nagarajan, V., Oszmiana, A., Rivera, F. E., Miller, H. K., Davenport, J. E., et al. (2011). NsaRS is a cell-envelope-stress-sensing two-component system of Staphylococcus aureus. Microbiology 157, 2206–2219. doi: 10.1099/mic.0.049692-0
Kraus, D., Herbert, S., Kristian, S. A., Khosravi, A., Nizet, V., Gotz, F., et al. (2008). The GraRS regulatory system controls Staphylococcus aureus susceptibility to antimicrobial host defenses. BMC Microbiol. 8:85. doi: 10.1186/1471-2180-8-85
Lan, L., Cheng, A., Dunman, P. M., Missiakas, D., and He, C. (2010). Golden pigment production and virulence gene expression are affected by metabolisms in Staphylococcus aureus. J. Bacteriol. 192, 3068–3077. doi: 10.1128/JB.00928-09
Li, M., Cha, D. J., Lai, Y., Villaruz, A. E., Sturdevant, D. E., and Otto, M. (2007). The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 66, 1136–1147. doi: 10.1111/j.1365-2958.2007.05986.x
Liu, J., Yang, L., Hou, Y., Soteyome, T., Zeng, B., Su, J., et al. (2018). Transcriptomics study on Staphylococcus aureus biofilm Under low concentration of ampicillin. Front. Microbiol. 9:2413. doi: 10.3389/fmicb.2018.02413
Mannala, G. K., Koettnitz, J., Mohamed, W., Sommer, U., Lips, K. S., Sproer, C., et al. (2018). Whole-genome comparison of high and low virulent Staphylococcus aureus isolates inducing implant-associated bone infections. Int. J. Med. Microbiol. 308, 505–513. doi: 10.1016/j.ijmm.2018.04.005
Meehl, M., Herbert, S., Gotz, F., and Cheung, A. (2007). Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51, 2679–2689. doi: 10.1128/AAC.00209-07
Mehta, S., Cuirolo, A. X., Plata, K. B., Riosa, S., Silverman, J. A., Rubio, A., et al. (2012). VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 92–102. doi: 10.1128/AAC.00432-10
Mensa, B., Howell, G. L., Scott, R., and DeGrado, W. F. (2014). Comparative mechanistic studies of Brilacidin, Daptomycin, and the antimicrobial peptide LL16. Antimicrob. Agents Chemother. 58, 5136–5145. doi: 10.1128/AAC.02955-14
Muller, A., Grein, F., Otto, A., Gries, K., Orlov, D., Zarubaev, V., et al. (2018). Differential daptomycin resistance development in Staphylococcus aureus strains with active and mutated gra regulatory systems. Int. J. Med. Microbiol. 308, 335–348. doi: 10.1016/j.ijmm.2017.12.002
Muzamal, U., Gomez, D., Kapadia, F., and Golemi-Kotra, D. (2014). Diversity of two-component systems: insights into the signal transduction mechanism by the Staphylococcus aureus two-component system GraSR. F1000Res 3:252. doi: 10.12688/f1000research.5512.2
Oogai, Y., Yamaguchi, M., Kawada-Matsuo, M., Sumitomo, T., Kawabata, S., and Komatsuzawa, H. (2016). Lysine and threonine biosynthesis from aspartate contributes to Staphylococcus aureus growth in calf serum. Appl. Environ. Microbiol. 82, 6150–6157. doi: 10.1128/AEM.01399-16
Otto, M. (2013). Community-associated MRSA: what makes them special? Int. J. Med. Microbiol. 303, 324–330. doi: 10.1016/j.ijmm.2013.02.007
Peschel, A., and Sahl, H. G. (2006). The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4, 529–536. doi: 10.1038/nrmicro1441
Pietiainen, M., Francois, P., Hyyrylainen, H. L., Tangomo, M., Sass, V., Sahl, H. G., et al. (2009). Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics 10:429. doi: 10.1186/1471-2164-10-429
Pu, L., Wang, M., Li, K., Feng, T., Zheng, P., Li, S., et al. (2020). Identification micro-RNAs functional modules and genes of ischemic stroke based on weighted gene co-expression network analysis (WGCNA). Genomics 112, 2748–2754. doi: 10.1016/j.ygeno.2020.03.011
Rajagopal, M., Martin, M. J., Santiago, M., Lee, W., Kos, V. N., Meredith, T., et al. (2016). Multidrug intrinsic resistance factors in Staphylococcus aureus identified by profiling fitness within high-diversity transposon libraries. MBio 7, e00950–e00916. doi: 10.1128/mBio.00950-16
Shoji, M., Cui, L., Iizuka, R., Komoto, A., Neoh, H. M., Watanabe, Y., et al. (2011). walK and clpP mutations confer reduced vancomycin susceptibility in Staphylococcus aureus. Antimicrob. Agents Chemother. 55, 3870–3881. doi: 10.1128/AAC.01563-10
Sun, F., Zhou, L., Zhao, B. C., Deng, X., Cho, H., Yi, C., et al. (2011). Targeting MgrA-mediated virulence regulation in Staphylococcus aureus. Chem. Biol. 18, 1032–1041. doi: 10.1016/j.chembiol.2011.05.014
Tong, S. Y., Davis, J. S., Eichenberger, E., Holland, T. L., and Fowler, V. G. Jr. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661. doi: 10.1128/CMR.00134-14
Trotonda, M. P., Xiong, Y. Q., Memmi, G., Bayer, A. S., and Cheung, A. L. (2009). Role of mgrA and sarA in methicillin-resistant Staphylococcus aureus autolysis and resistance to cell wall-active antibiotics. J. Infect. Dis. 199, 209–218. doi: 10.1086/595740
Villanueva, M., García, B., Valle, J., Rapún, B., Ruiz de los Mozos, I., Solano, C., et al. (2018). Sensory deprivation in Staphylococcus aureus. Nat. Commun. 9:523. doi: 10.1038/s41467-018-02949-y
Wang, J., Wang, J., Wang, Y., Sun, P., Zou, X., Ren, L., et al. (2019). Protein expression profiles in methicillin-resistant Staphylococcus aureus (MRSA) under effects of subminimal inhibitory concentrations of imipenem. FEMS Microbiol. Lett. 366:fnz195. doi: 10.1093/femsle/fnz195
Yan, H., Wang, Q., Teng, M., and Li, X. (2019). The DNA-binding mechanism of the TCS response regulator ArlR from Staphylococcus aureus. J. Struct. Biol. 208:107388. doi: 10.1016/j.jsb.2019.09.005
Yang, S. J., Bayer, A. S., Mishra, N. N., Meehl, M., Ledala, N., Yeaman, M. R., et al. (2012). The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect. Immun. 80, 74–81. doi: 10.1128/IAI.05669-11
Yarwood, J. M., Bartels, D. J., Volper, E. M., and Greenberg, E. P. (2004). Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186, 1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004
Keywords:Staphylococcus aureus, graRS, virulence, biofilm, two-component signal transduction system
Citation: Chen L, Wang Z, Xu T, Ge H, Zhou F, Zhu X, Li X, Qu D, Zheng C, Wu Y and Zhao K (2021) The Role of graRS in Regulating Virulence and Antimicrobial Resistance in Methicillin-Resistant Staphylococcus aureus. Front. Microbiol. 12:727104. doi: 10.3389/fmicb.2021.727104
Edited by:
Santi M. Mandal, Indian Institute of Technology Kharagpur, IndiaReviewed by:
Hongyi Zhu, Shanghai Jiao Tong University, ChinaHarshad Harshad Lade, Hallym University, South Korea
Alaguvel Valliammai, Alagappa University, India
Copyright © 2021 Chen, Wang, Xu, Ge, Zhou, Zhu, Li, Qu, Zheng, Wu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Wu, eWFuZ3d1QGZ1ZGFuLmVkdS5jbg==; Keqing Zhao, cmhpbm9yZXNlYXJjaEAxNjMuY29t; Chunquan Zheng, emhlbmdfZW50OTZAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Le Chen
Le Chen Zihui Wang
Zihui Wang Tao Xu
Tao Xu Hongfei Ge
Hongfei Ge Fangyue Zhou
Fangyue Zhou Xiaoyi Zhu
Xiaoyi Zhu Xianhui Li
Xianhui Li Di Qu
Di Qu Chunquan Zheng
Chunquan Zheng Yang Wu
Yang Wu Keqing Zhao
Keqing Zhao