- Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL, United States
Clostridioides difficile flagellin FliC is associated with toxin gene expression, bacterial colonization, and virulence, and is also involved in pleiotropic gene regulation during in vivo infection. However, how fliC expression is regulated in C. difficile remains unclear. In Bacillus subtilis, flagellin homeostasis and motility are coregulated by flagellar assembly factor (FliW), flagellin Hag (FliC homolog), and Carbon storage regulator A (CsrA), which is referred to as partner-switching mechanism “FliW-CsrA-Hag.” In this study, we characterized FliW and CsrA functions by deleting or overexpressing fliW, csrA, and fliW-csrA in C. difficile R20291. We showed that fliW deletion, csrA overexpression in R20291, and csrA complementation in R20291ΔWA (fliW-csrA codeletion mutant) dramatically decreased FliC production, but not fliC gene transcription. Suppression of fliC translation by csrA overexpression can be relieved mostly when fliW was coexpressed, and no significant difference in FliC production was detected when only fliW was complemented in R20291ΔWA. Further, loss of fliW led to increased biofilm formation, cell adhesion, toxin production, and pathogenicity in a mouse model of C. difficile infection (CDI), while fliW-csrA codeletion decreased toxin production and mortality in vivo. Our data suggest that CsrA negatively modulates fliC expression and FliW indirectly affects fliC expression through inhibition of CsrA post-transcriptional regulation. In light of “FliW-CsrA-Hag” switch coregulation mechanism reported in B. subtilis, our data also suggest that “FliW-CsrA-fliC/FliC” can regulate many facets of C. difficile R20291 pathogenicity. These findings further aid us in understanding the virulence regulation in C. difficile.
Introduction
Clostridioides difficile (Lawson et al., 2016; Oren and Garrity, 2018) is a Gram-positive, spore-forming, toxin-producing, anaerobic bacterium that is a leading cause of nosocomial antibiotic-associated diarrhea in the developed countries (Sebaihia et al., 2006). Clostridioides difficile infection (CDI) can result in a spectrum of symptoms, ranging from mild diarrhea to pseudomembranous colitis and potential death (Lessa et al., 2012). Clostridioides difficile has many virulence factors, among which toxin A (TcdA) and toxin B (TcdB) are the major ones (Lyras et al., 2009; Kuehne et al., 2010). These toxins can disrupt the actin cytoskeleton of intestinal cells through glucosylation of the Rho family of GTPases, and induce mucosal inflammation and symptoms associated with CDI (Peniche et al., 2013).
CsrA, the carbon storage regulator A, has been reported to control various physiological processes, such as flagella synthesis, virulence, central carbon metabolism, quorum sensing, motility, and biofilm formation in pathogens including Pseudomonas aeruginosa, Pseudomonas syringae, Borrelia burgdorferi, Salmonella typhimurium, and Proteus mirabilis (Sabnis et al., 1995; Pessi et al., 2001; Lawhon et al., 2003; Lucchetti-Miganeh et al., 2008; Timmermans and Van Melderen, 2010; Karna et al., 2011; Morris et al., 2013; Ferreiro et al., 2018). Recently, the role of CsrA on carbon metabolism and virulence-associated processes in C. difficile 630Δerm was analyzed by overexpressing the csrA gene (Gu et al., 2018). Authors showed that the csrA overexpression resulted in flagella defect, poor motility, and induced carbon metabolism change. Oppositely, toxin production and cell adherence increased in the csrA overexpression strain. CsrA is a widely distributed RNA binding protein that post-transcriptionally modulates gene expression through regulating mRNA stability and/or translation initiation of target mRNA (Romeo et al., 1993; Liu et al., 1995; Timmermans and Van Melderen, 2010). It typically binds to multiple specific sites that are located nearby or overlapping the cognate Shine–Dalgarno (SD) sequence in the target transcripts (Sorger-Domenigg et al., 2007; Yakhnin et al., 2007). The roles of CsrA in Bacillus subtilis have been well-studied (Yakhnin et al., 2007; Mukherjee et al., 2011; Oshiro et al., 2019). Flagellin Hag (FliC homolog), a main structure flagellar component, has been reported to be regulated by CsrA in B. subtilis. Yakhnin et al. (2007) first reported that CsrA in B. subtilis can regulate translation initiation of Hag by preventing ribosome binding to the hag transcript. Mukherjee et al. (2011) elucidated that the interaction between CsrA and FliW could govern flagellin homeostasis and checkpoint on flagellar morphogenesis in B. subtilis. FliW, the first protein antagonist of CsrA activity, was also identified and characterized in B. subtilis. They elegantly demonstrated a novel regulation system “a partner-switching mechanism” (Hag-FliW-CsrA) on flagellin synthesis in B. subtilis. Briefly, following the flagellar assembly checkpoint of hook completion, FliW was released from a FliW-Hag complex. Afterward, FliW binds to CsrA which will relieve CsrA-mediated hag translation repression for flagellin synthesis concurrent with filament assembly. Thus, flagellin homeostasis restricts its own expression on the translational level. Results also suggested that CsrA has an ancestral role in flagella assembly and has evolved to coregulate multiple cellular processes with motility. Oshiro et al. (2019) further quantitated the interactions in the Hag-FliW-CsrA system. They found that Hag-FliW-CsrAdimer functions at nearly 1:1:1 stoichiometry in B. subtilis. The Hag-FliW-CsrAdimer system is hypersensitive to the cytoplasmic Hag concentration and is robust to perturbation.
Clostridioides difficile flagellin gene fliC is associated with toxin gene expression, bacterial colonization, and virulence, and is responsible for pleiotropic gene regulation during in vivo infection (Tasteyre et al., 2001; Aubry et al., 2012; Baban et al., 2013; Barketi-Klai et al., 2014; Stevenson et al., 2015). The delicate regulations among fliC gene expression, toxin production, bacterial motility, colonization, and pathogenicity in C. difficile are indicated. Though the important roles of CsrA in flagellin synthesis and flagellin homeostasis have been studied in other bacteria (Yakhnin et al., 2007; Mukherjee et al., 2011; Oshiro et al., 2019), the regulation of FliW, CsrA, and FliC and the function of fliW in C. difficile remain unclear.
In this communication, we aimed to study the involvement of FliW and CsrA in fliC expression and C. difficile virulence and physiology by constructing and analyzing fliW and fliW-csrA deletion mutants of C. difficile R20291. We evaluated these mutants in the expression of fliC, motility, adhesion, biofilm formation, toxin production, sporulation, germination, and pathogenicity in a mouse model of CDI.
Materials and Methods
Bacteria, Plasmids, and Culture Conditions
Table 1 lists the strains and plasmids used in this study. Clostridioides difficile strains were cultured in BHIS media (brain heart infusion broth supplemented with 0.5% yeast extract and 0.1% L-cysteine, and 1.5% agar for agar plates) at 37°C in an anaerobic chamber (90% N2, 5% H2, and 5% CO2). For spores preparation, C. difficile strains were cultured in Clospore media and purified as described earlier (Perez et al., 2011). Escherichia coli DH5α and E. coli HB101/pRK24 were grown aerobically at 37°C in LB media (1% tryptone, 0.5% yeast extract, and 1% NaCl). Escherichia coli DH5α was used as a cloning host, and E. coli HB101/pRK24 was used as a conjugation donor host. Antibiotics were added when needed for E. coli, 15μg/ml chloramphenicol; for C. difficile, 15μg/ml thiamphenicol, 250μg/ml D-cycloserine, 50μg/ml kanamycin, 8μg/ml cefoxitin, and 500ng/ml anhydrotetracycline.
DNA Manipulations and Chemicals
DNA manipulations were carried out according to standard techniques (Chong, 2001). Plasmids were conjugated into C. difficile as described earlier (Heap et al., 2010). The DNA markers, protein markers, PCR product purification kit, DNA gel extraction kit, restriction enzymes, cDNA synthesis kit, and SYBR Green RT-qPCR kit were purchased from ThermoFisher Scientific (Waltham, United States). PCRs were performed with the high-fidelity DNA polymerase NEB Q5 Master Mix, and PCR products were assembled into target plasmids with NEBuilder HIFI DNA Assembly Master Mix (New England, United Kingdom). Primers (Supplementary Table 1) were purchased from IDT (Coralville, United States). All chemicals were purchased from Sigma-Aldrich (St. Louis, United States) unless those stated otherwise.
Gene Deletion, Complementation, and Overexpression in R20291
Gene edit plasmid pDL-1 containing Cas12a (AsCpfI) under the control of tetracycline-inducing promoter was constructed and used for C. difficile gene deletion according to a previous report (Hong et al., 2018). The target sgRNA was designed with an available website tool,1 and the off-target prediction was analyzed on the Cas-OFFinder website.2 The sgRNA, up- and down-homologous arms, were assembled into pDL-1. Two target sgRNAs for one gene deletion were selected and used for gene deletion plasmid construction in C. difficile, respectively. Briefly, the gene deletion plasmid was constructed in the cloning host E. coli DH5α and was transformed into the donor host E. coli HB101/pRK24, and subsequently was conjugated into R20291. Potential successful transconjugants were selected with selective antibiotic BHIS-TKC plates (15μg/ml thiamphenicol, 50μg/ml kanamycin, and 8μg/ml cefoxitin). The transconjugants were cultured in BHIS-Tm broth (15μg/ml thiamphenicol) to log phase, then the subsequent cultures were diluted with PBS serially and plated on the inducing plates (BHIS-Tm-ATc: 15μg/ml thiamphenicol and 500ng/ml anhydrotetracycline). The plates were incubated at 37°C in the anaerobic chamber for 24–48h, then 20–40 colonies were used as templates for colony PCR test with check primers for correct gene deletion colony isolation. The correct gene deletion colony was sub-cultured into BHIS broth without antibiotics and was passaged several times to cure the deletion plasmid, and then the cultures were plated on BHIS plates and subsequent colonies were replica plated on BHIS-Tm plates to isolate pure gene deletion mutants. The genome of R20291ΔfliW (referred hereafter as R20291ΔW) and R20291ΔfliW-csrA (referred hereafter as R20291ΔWA) were isolated and used as templates for the PCR test with check primers, and the PCR products were sequenced to confirm the correct gene deletion.
The fliW (396bp; primers 3-F/R), csrA (213bp; primers 4-F/R), and fliW-csrA (599bp; primers 5-F/R) genes were amplified and assembled into SacI-BamHI digested pMTL84153 plasmid, yielding the complementation plasmid pMTL84153-fliW, pMTL84153-csrA, and pMTL84153-fliW-csrA, and were subsequently conjugated into R20291ΔWA, R20291ΔW, and R20291 yielding complementation strain R20291ΔWA/pMTL84153-fliW (referred as R20291ΔWA-W), R20291ΔWA/pMTL84153-csrA (R20291ΔWA-A), R20291ΔWA/pMTL84153-fliW-csrA (R20291ΔWA-WA), and R20291ΔW/pMTL84153-fliW (R20291ΔW-W), and overexpression strain R20291/pMTL84153-fliW (R20291-W), R20291/pMTL84153-csrA (R20291-A), and R20291/pMTL84153- fliW-csrA (R20291-WA).
Growth Profile, Motility, and Biofilm Assay
Clostridioides difficile strains were incubated to an optical density of OD600 of 0.8 in BHIS media and were diluted to an OD600 of 0.2. Then, 1% of the culture was inoculated into fresh BHIS, followed by measuring OD600 for 32h.
To examine the effect of fliW and fliW-csrA deletion on C. difficile motility, R20291, R20291ΔWA, and R20291ΔW were cultured to an OD600 of 0.8. For swimming analysis, 2μl of C. difficile culture was penetrated into soft BHIS agar (0.175%) plates, meanwhile, 2μl of culture was dropped onto 0.3% BHIS agar plates for swarming analysis. The swimming assay plates were incubated for 24h, and the swarming plates were incubated for 48h, respectively.
For biofilm formation analysis, wild-type and mutant strains were cultured to an OD600 of 0.8, and 1% of C. difficile cultures were inoculated into reinforced clostridial medium (RCM) with eight-well repeats in a 96-well plate and incubated in the anaerobic chamber at 37°C for 48h. Biofilm formation was analyzed by crystal violet dye. Briefly, C. difficile cultures were removed by pipette carefully. Then, 100μl of 2.5% glutaraldehyde was added into the well to fix the bottom biofilm, and the plate was kept at room temperature for 30min. Next, the wells were washed with PBS three times and dyed with 0.25% (w/v) crystal violet for 10min. The crystal violet solution was removed, and the wells were washed five times with PBS, followed by the addition of acetone into wells to dissolve the crystal violet of the cells. The dissolved solution was further diluted with ethanol 2–4 times, and biomass was determined at OD570.
Adherence of C. difficile Vegetative Cells to HCT-8 Cells
Clostridioides difficile adhesion ability was evaluated with HCT-8 cells (ATCC CCL-244; Janvilisri et al., 2010). Briefly, HCT-8 cells were grown to 95% confluence (2×105/well) in a 24-well plate and then moved into the anaerobic chamber, followed by infecting with 6×106 of log phase of C. difficile vegetative cells at a multiplicity of infection (MOI) of 30:1. The plate was cultured at 37°C for 30min. After incubation, the infected cells were washed with 300μl of PBS three times, and then suspended in RPMI media with trypsin and plated on BHIS agar plates to enumerate the adhered C. difficile cells. The adhesion ability of C. difficile to HCT-8 cells was calculated as follows: CFU of adhered bacteria/total cell numbers.
To visualize the adherence of C. difficile to HCT-8 cells, C. difficile vegetative cells were labeled with the chemical 5(6)-CFDA (5- and−6)-Carboxyfluorescein diacetate (Fuller et al., 2000). Briefly, C. difficile strains were cultured to an OD600 of 0.8, then washed with PBS 3 times and resuspended in fresh BHIS supplemented with 50-mM 5(6)-CFDA, followed by incubation at 37°C for 30min in the anaerobic chamber. After post-incubation, the labeled C. difficile cells were collected and washed with PBS three times, and then resuspended in RPMI medium. Afterward, the labeled C. difficile cells were used for the infection experiment as described above. After 30-min post-infection, the fluorescence of each well was scanned by the multi-mode reader (excitation, 485nm; emission, 528nm), the relative fluorescence unit (RFU) was recorded as F0. Following, the plates were washed with PBS three times to remove unbound C. difficile cells, then the plates were scanned, and the RFU was recorded as F1. The adhesion ratio was calculated as follows: F1/F0. After scanning, the infected cell plates were further detected by the fluorescence microscope.
fliC Expression Assay
For fliC transcription analysis, 2ml of 24-h post-inoculated C. difficile cultures were centrifuged at 4°C, 12,000×g for 5min, respectively. Then, the total RNA of different strains was extracted with TRIzol reagent. The transcription of fliC was measured by RT-qPCR with primers Q-fliC-F/R. All RT-qPCRs were repeated in triplicate, independently. Data were analyzed by the comparative CT (2-∆∆CT) method with 16s rRNA as a control.
To analyze the FliC protein level, C. difficile cell lysates from overnight cultures were used for Western blot analysis. Briefly, overnight C. difficile cultures were collected and washed three times with PBS and then resuspended in 5ml of distilled water. The suspensions were lysed with TissueLyser LT (Qiagen), followed centrifuged at 4°C, 25,000×g for 1h. The final pellets were resuspended in 30μl of PBS, and the total protein concentration was measured by using a BCA protein assay (Thermo Scientific, Suwanee, GA, United States). Protein extracts were subjected to 10% SDS-PAGE. Sigma A protein (SigA) was used as a loading control protein in SDS-PAGE (Mukherjee et al., 2013). FliC and SigA proteins on the gel were detected with anti-FliC and anti-SigA primary antibody (1:1,000, a generous gift from Dr. Daniel Kearns at Indiana University) and horseradish peroxidase-conjugated secondary antibody goat anti-mouse (Cat: ab97023, IgG, 1:3,000, Abcam, Cambridge, MA, United States) by Western blot, respectively. Anti-FliC antibody used in the Western blot analysis is an anti-FliCD serum, generated in the laboratory. FliCD is a fusion protein containing C. difficile FliC and FliD (Wang et al., 2018). The relative intensity of blot bands was analyzed by ImageJ software, and FliC relative intensity was normalized to SigA control.
Toxin Expression Assay
To evaluate toxin expression in C. difficile strains, one single colony from each strain was inoculated into 25ml of BHIS and incubated in an anaerobic chamber at 37°C, and 10ml of C. difficile cultures from different strains were collected at 24- and 48-h post-incubation. The cultures were adjusted to the same OD600 value with fresh BHIS. Then, the collected C. difficile cultures were centrifuged at 4°C, 8,000×g for 15min, filtered with 0.22μm filters, and used for ELISA. Anti-TcdA (PCG4.1, Novus Biologicals, United States) and anti-TcdB (AI, Gene Tex, United States) were used as coating antibodies for ELISA, and HRP-Chicken anti-TcdA and HRP-Chicken anti-TcdB (Gallus Immunotech, United States) were used as detection antibodies.
For toxin transcription analysis, 2ml of 24- and 48-h post-inoculated C. difficile cultures were centrifuged at 4°C, 12,000×g for 5min, respectively. Next, the total RNA of different strains was extracted with TRIzol reagent. The transcription of tcdA and tcdB was measured by RT-qPCR with primers Q-tcdA-F/R and Q-tcdB-F/R, respectively. All RT-qPCRs were repeated in triplicate, independently. Data were analyzed by using the comparative CT (2-∆∆CT) method with 16s rRNA as a control.
Germination and Sporulation Assay
Clostridioides difficile germination and sporulation analysis were conducted as reported earlier (Zhu et al., 2019). Briefly, for C. difficile sporulation analysis, C. difficile strains were cultured in Clospore media for 4days. Afterward, the CFU of cultures from 48 and 96h were counted on BHIS plates with 0.1% TA to detect sporulation ratio, respectively. The sporulation ratio was calculated as CFU (65°C heated, 20min)/CFU (no heated). For C. difficile germination analysis, C. difficile spores were collected from 2-week Clospore media-cultured bacteria and purified with sucrose gradient layer (50, 45, 35, 25, and 10%). The heated purified spores were diluted to an OD600 of 1.0 in the germination buffer [10mM Tris (pH 7.5), 150mM NaCl, 100mM glycine, and 10mM taurocholic acid (TA)] to detect the germination ratio. The value of OD600 was monitored immediately (0min, t0), and was detected once every 2min (tx) for 20min at 37°C. The germination ratio was calculated as OD600 (tx)/OD600 (T0). Spores in germination buffer without TA were used as the negative control.
R20291, R20291ΔWA, and R20291ΔW Virulence in the Mouse Model of C. difficile Infection
C57BL/6 female mice (6weeks old) were ordered from Charles River Laboratories, Cambridge, MA. All studies were approved by the Institutional Animal Care and Use Committee of University of South Florida. The experimental design and antibiotic administration were conducted as described earlier (Sun et al., 2011). Briefly, 30 mice were divided into three groups in six cages. Group 1 mice were challenged with R20291 spores, group 2 mice with R20291ΔWA spores, and group 3 mice with R20291ΔW spores, respectively. Mice were given an orally administered antibiotic cocktail (kanamycin 0.4mg/ml, gentamicin 0.035mg/ml, colistin 0.042mg/ml, metronidazole 0.215mg/ml, and vancomycin 0.045mg/ml) in drinking water for 4days. After 4days of antibiotic treatment, all mice were given autoclaved water for 2days, followed by one dose of clindamycin (10mg/kg, intraperitoneal route) 24h before spores challenge (Day 0). After that mice were orally gavaged with 106 spores and monitored daily for a week for changes in weight, diarrhea, and mortality. If body weight loss was equal to or greater than 20%, the mouse was euthanized and counted as a dead one. Mortality also included mice that were succumbed to disease. Diarrhea was defined as soft or watery feces. All survived mice were humanely euthanized on day 7 of post-C. difficile challenge.
Enumeration of C. difficile Spores and Determination of Toxin Level in Feces
Fecal pellets from post-infection day 0 to day 7 were collected from each mouse and stored at −80°C. To enumerate C. difficile spores, feces were diluted with PBS at a final concentration of 0.1g/ml, followed by adding 900μl of absolute ethanol into 100μl of the fecal solution, and kept at room temperature for 1h to inactivate vegetative cells. Afterward, 200μl of vegetative cells inactivated fecal solution from the same group and the same day was mixed. Then, fecal samples were serially diluted and plated on BHIS-CCT plates (250μg/ml D-cycloserine, 8μg/ml cefoxitin, and 0.1% TA). After 48-h incubation, colonies were counted and expressed as CFU/g feces. To evaluate toxin tilter in feces, 0.1g/ml of the fecal solution was diluted two times with PBS, followed by examining TcdA and TcdB ELISA.
Statistical Analysis
The reported experiments were conducted in independent biological triplicates, and each sample was additionally taken in technical triplicates. Animal survivals were analyzed by Kaplan–Meier survival analysis and compared by the log-rank test. One-way ANOVA with post hoc Tukey test was used for more than two groups’ comparison. Results were expressed as mean±SEM. Differences were considered statistically significant if p<0.05 (*).
Results
Highly Conserved fliW and csrA Genes in C. difficile
DNA and protein sequences of fliW and csrA from 10 C. difficile strains belonging to different ribotypes (RTs), including RT106, RT027, RT001, RT078, RT009, RT012, RT046, and RT017 were selected and aligned to those of R20291 (Table 2). We found that fliW and csrA genes are broadly found in C. difficile genomes, and both DNA and protein sequences of fliW and csrA are conserved across different C. difficile strains. These results motivated us to investigate the functions of fliW and csrA in C. difficile.
Construction of fliW and fliW-csrA Deletion Mutants and Complementation Strains
The C. difficile R20291 flagellar gene operon was analyzed through the IMG/M website,3 and the late-stage flagellar genes (F1) are drawn as Figure 1A (Stevenson et al., 2015). Among them, fliW and csrA genes have a 10bp overlap and were demonstrated as cotranscription by RT-PCR (Supplementary Figure 1).
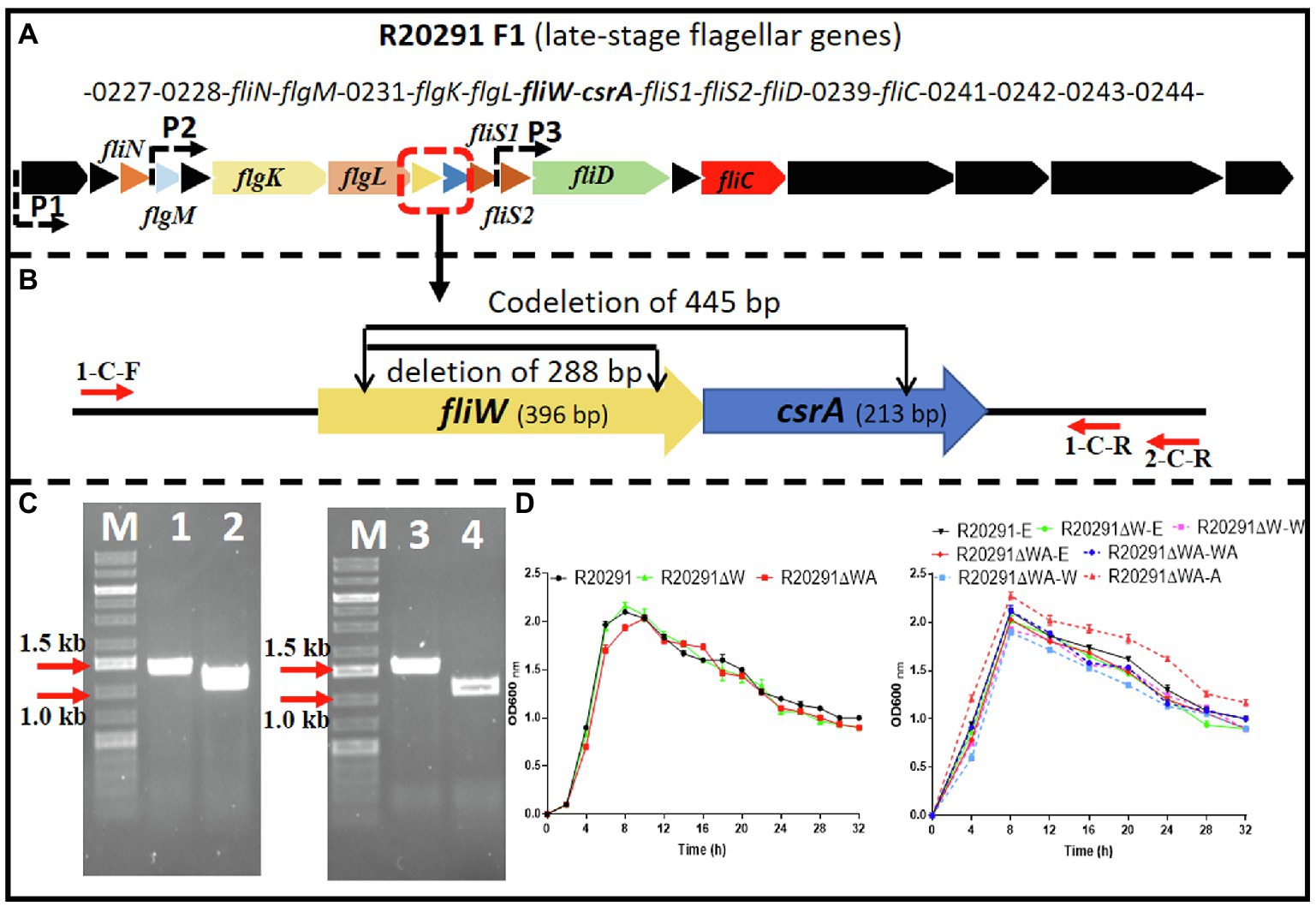
Figure 1. R20291 late-stage flagellar genes (F1) and fliW and fliW-csrA deletion. (A) Schematic representation of late-stage flagellar genes (F1). Dotted arrows (P1, P2, and P3) indicate the potential promoters in F1. (B) Deletion of fliW and fliW-csrA genes. 1-C-F/R were used to verify fliW deletion, and 1-C-F and 2-C-R were used to test fliW-csrA codeletion. (C) Verification of fliW and fliW-csrA deletions by PCR. M, DNA ladder; 1, R20291 genome as PCR template; 2, R20291∆W genome as PCR template; 3, R20291 genome as PCR template; and 4, R20291∆WA genome as PCR template. (D) Growth profile of parent strain and gene deletion mutants. Experiments were independently repeated thrice. Bars stand for mean±SEM. One-way ANOVA with post hoc Tukey test was used for statistical significance.
To analyze the role of fliW and csrA in R20291 (NC_013316.1), CRISPR-AsCpfI-based plasmid pDL1 (pMTL82151-Ptet-AscpfI) was constructed for gene deletion in C. difficile (Zhu et al., 2021). pDL1-fliW and pDL1-csrA gene deletion plasmids were constructed, and the fliW gene (288bp deletion; R20291ΔW) was deleted successfully. However, after several trials, we could not get the csrA gene deletion mutant possibly due to its small size (213bp) or particularly unknown roles for R20291. We also tried to use Clostron and pyrE gene edit system to delete csrA gene, but failed to get the correct mutant. Therefore, we constructed fliW-csrA codeletion plasmid pDL1-fliW-csrA. Part of fliW-csrA (445bp deletion) gene was codeleted, and the plasmid curing mutant R20291ΔWA was obtained (Figure 1B,C). To study the role of csrA in R20291, the single gene complementation strain R20291ΔWA-W and R20291ΔWA-A were constructed. R20291, R20291-pMTL84153 (R20291-E), R20291ΔW-pMTL84153 (R20291ΔW-E), and R20291ΔWA-pMTL84153 (R20291ΔWA-E) were used as control strains when needed.
The effects of fliW and fliW-csrA deletion on R20291 growth were evaluated. Figure 1D shows that there was no significant difference in bacterial growth between parent strain and mutants in BHIS media.
Effects of fliW and fliW-csrA Deletions on C. difficile Motility and Biofilm Formation
To characterize the effects of fliW and fliW-csrA deletions on C. difficile motility, swimming, and swarming motilities of R20291, R20291ΔWA, and R20291ΔW were first analyzed at 24 and 48-h post-inoculation (Figure 2A; Supplementary Figure 2), respectively. The diameter of the swimming halo of R20291ΔWA increased by 27.2% (p<0.05), while that of R20291ΔW decreased by 58.4% (p<0.05) compared to that of R20291. Next, we examined the motility of the complementation strains (Figure 2B; Supplementary Figure 2), and similar results were obtained among R20291-E, R20291ΔWA-E (with the swimming halo increased by 74.8%, p<0.05), and R20291ΔW-E (with the swimming halo decreased by 59.2%, p<0.05; Figure 2B). No significant difference was detected between complementation strain R20291ΔWA-WA, R20291ΔWA-W, R20291ΔW-W, and the parent strain R20291-E except R20291ΔWA-A which decreased by 52.0% (p<0.05) in swimming halo (Figure 2B). The swarming (48h) and swimming (24h) motilities analyzed on agar plates are shown in Supplementary Figure 2.
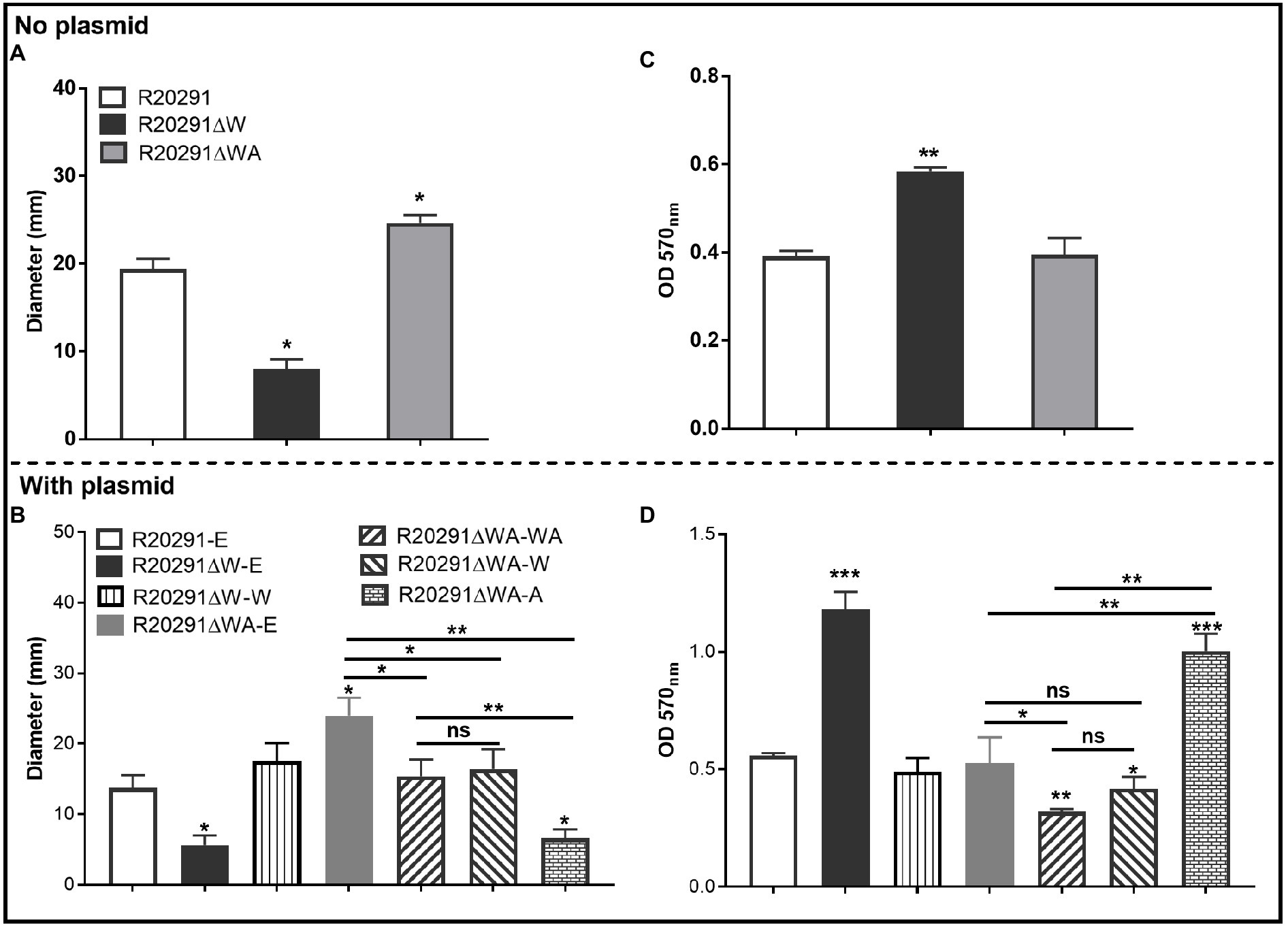
Figure 2. Motility and biofilm analysis. (A,B): Halo diameter of motility (swimming analysis on 0.175% agar plate). (C,D): Biofilm formation analysis. Bars stand for mean±SEM (*p<0.05, **p<0.01, and ***p<0.001). One-way ANOVA with post hoc Tukey test was used for statistical significance. ***directly upon the column means the significant difference of the experimental strain compared to R20291 or R20291-E.
The effects of fliW and fliW-csrA deletions on C. difficile biofilm formation were also analyzed. In comparison with R20291, the biofilm formation of R20291ΔW increased by 49.5% (p<0.01), and no significant difference in biofilm formation was detected in R20291ΔWA (Figure 2C). The biofilm formation of R20291ΔW-E increased 112.3% (p<0.001) and R20291ΔWA-A increased by 79.9% (p<0.001) compared to R20291-E (Figure 2D). Meanwhile, the biofilm formation of R20291ΔWA-WA and R20291ΔWA-W decreased by 42.8% (p<0.01) and 25.2% (p<0.05), respectively.
Together, these data indicate that loss of FliW impairs C. difficile motility, and increases biofilm production. The decrease of motility and increase in biofilm production were also detected in R20291ΔWA-A, which was largely restored by coexpressing fliW with csrA in R20291ΔWA (Figures 2B,D), indicating that FliW could antagonize CsrA to regulate bacterial motility and biofilm production.
Effects of fliW and fliW-csrA Deletions on Bacterial Adherence in vitro
The ability of C. difficile vegetative cells to adhere to HCT-8 cells in vitro was analyzed. Figure 3A shows that the mean adhesion number of R20291 was 2.40±0.70 bacteria/cell, while that of R20291ΔW was 7.17±0.61, which was 3.0-fold (p < 0.0001) of R20291. No significant difference was detected between R20291ΔWA and R20291. In the complementation strains, we detected a similar result which showed that the mean adhesion number of R20291ΔW-E (6.17±0.64) was 3.20-fold (p < 0.0001) of R20291-E (1.93±0.25; Figure 3B). The adhesion ability of complementation strains nearly recovered to that of wild-type strain except for R20291ΔWA-A (7.13±0.66, p < 0.0001) which was 3.69-fold of R20291-E in the mean adhesion number (Figure 3B).
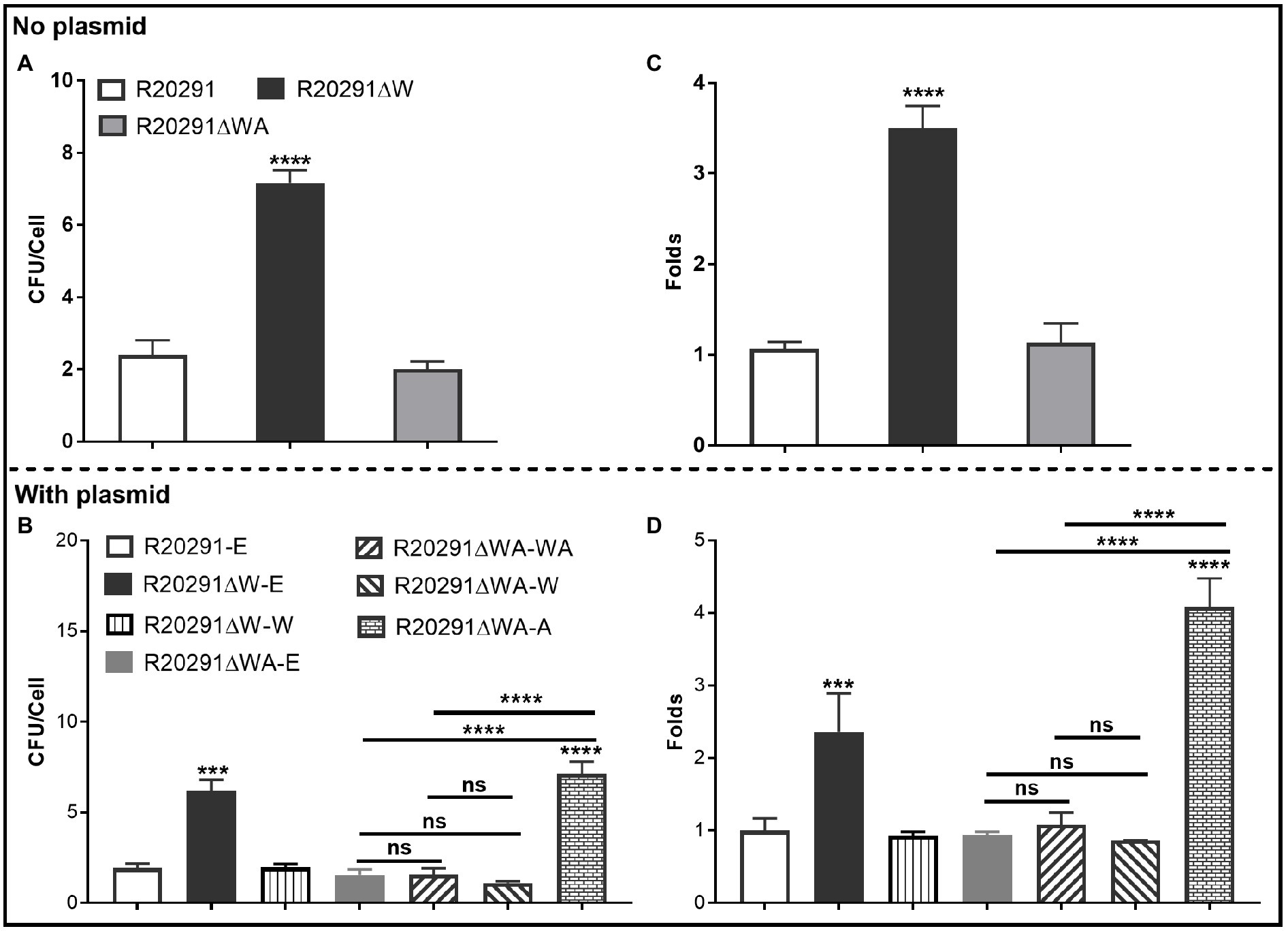
Figure 3. Adhesion analysis (A,B): Adherence of C. difficile vegetative cells to HCT-8 cells in vitro. (C,D): Adhesion analysis with 5(6)-CFDA dye. The fluorescence intensity was scanned by the multi-mode reader (excitation, 485nm; emission, 528nm). The original relative fluorescence unit (RFU) was recorded as F0, after PBS wash, the RFU was recorded as F1. The adhesion ratio was calculated as follows: F1/F0. Experiments were independently repeated thrice. Bars stand for mean±SEM (*p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001). One-way ANOVA with post hoc Tukey test was used for statistical significance. * directly upon the column means the significant difference of the experimental strain compared to R20291 or R20291-E.
To visualize the adhesion of C. difficile to HCT-8 cells, the C. difficile vegetative cells were labeled with the chemical 5(6)-CFDA. Figures 3C,D shows that the fluorescence intensity of R20291ΔW was 3.50-fold (p < 0.0001) of that in R20291, and the fluorescence intensity of R20291ΔW-E was 2.36-fold (p < 0.001), and R20291ΔWA-A was 4.08-fold (p < 0.0001) of that in R20291-E, respectively, which is consistent with the results shown in Figures 3A,B. Meanwhile, the adherence of C. difficile to HCT-8 cells was also visualized by fluorescence microscopy (Supplementary Figure 3).
Our data showed that FliW negatively affects bacterial adherence. CsrA complementation in R20291ΔWA increased adherence, while the phenotype change can be recovered partially when fliW was coexpressed with csrA in R20291ΔWA, suggesting that FliW could antagonize CsrA to regulate bacterial adherence. The results from bacterial adherence analysis were consistent with biofilm production analysis indicating the close relation between biofilm production and adherence in C. difficile.
Effects of Deletion and Overexpression of fliW and fliW-csrA on fliC Expression
In B. subtilis, FliW interacts with CsrA to regulate hag (a homolog of fliC) translation. We reasoned that FliW and CsrA would also regulate fliC expression in C. difficile. As shown in Figure 4A, the transcription of fliC in R20291ΔWA increased 1.12-fold (p<0.05), while the fliW deletion impaired the fliC transcription slightly while no significant difference. Figure 4B shows the production of FliC in R20291ΔW dramatically decreased (10.4-fold reduction, p<0.001), while that of R20291ΔWA increased significantly (increased by 27.5%, p<0.05). To further determine the role of the single-gene csrA on FliC synthesis, csrA and fliW were complemented into R20291ΔWA or overexpressed in R20291, respectively. Results showed that the significant difference of fliC transcription could only be detected in R20291ΔWA-E (increased by 32.3%, p<0.05; Figure 4C) and R20291-W (increased by 69.8%) compared to R20291-E (Figure 4E). Interestingly, the FliC production of R20291ΔWA-A decreased 3.2-fold (p<0.001) compared to that of R20291-E, while that of R20291ΔWA-WA only decreased by 14.3% (p<0.05), and no significant difference of FliC production in R20291ΔWA-W was detected (Figure 4D). As shown in Figures 4E,F, the fliC transcription of R20291-A was not affected compared to R20291-E, but the FliC production in R20291-A decreased 5.3-fold (p<0.0001). The decrease in FliC production in R20291-A can be partially recovered when fliW was coexpressed with csrA (R20291-WA decreased by 16.2%, p<0.05).
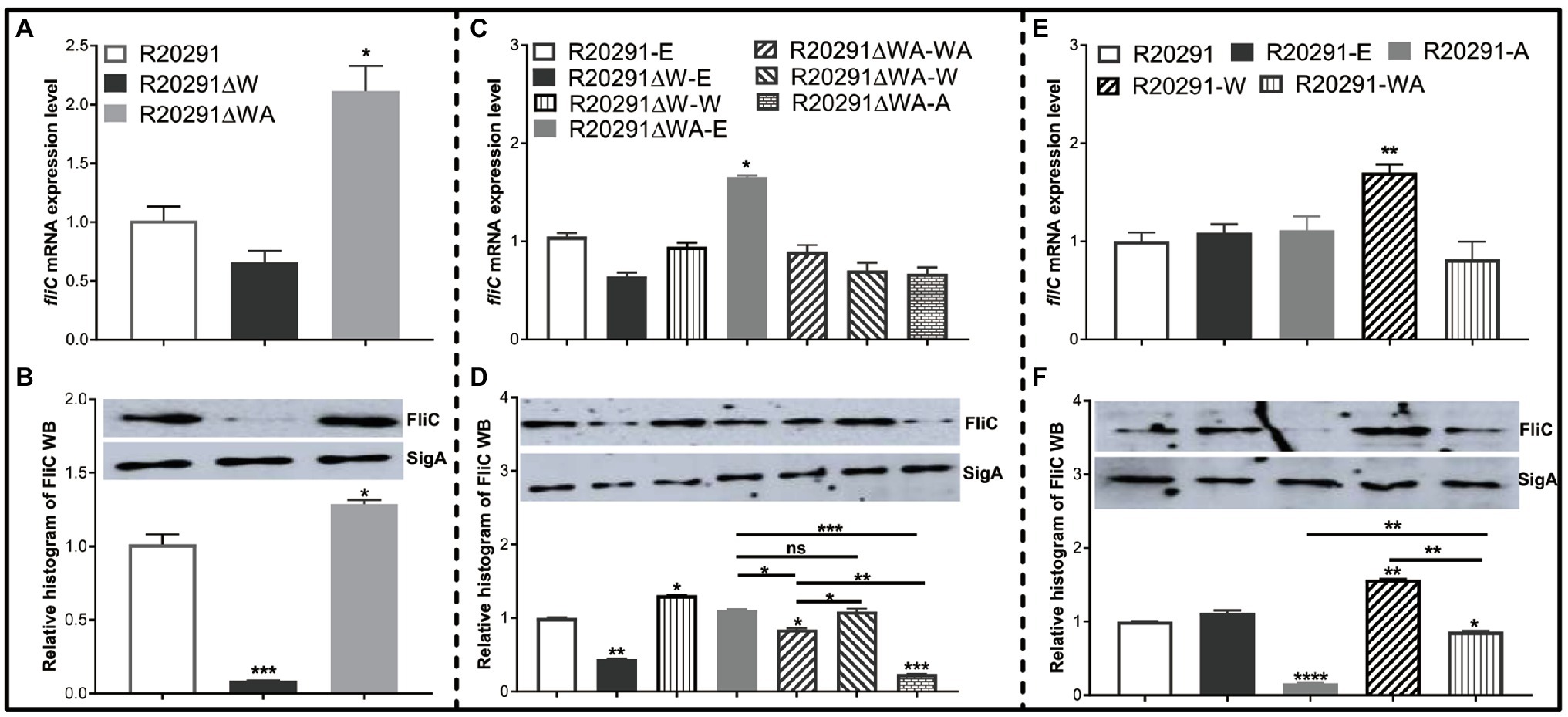
Figure 4. fliC expression analysis. (A,C,E) Analysis of fliC expression on transcription level. (B,D,F) Analysis of fliC expression on translation level by Western blot. SigA protein was used as a loading control. Experiments were independently repeated thrice. Bars stand for mean±SEM (*p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001). One-way ANOVA with post hoc Tukey test was used for statistical significance. **** upon the column directly means the significant difference of experimental strain compared to R20291 or R20291-E.
Collectively, our data indicate that CsrA negatively modulates fliC expression post-transcriptionally and FliW antagonizes CsrA to regulate fliC expression possibly through inhibiting CsrA-mediated negative post-transcriptional regulation.
Effects of fliW and fliW-csrA Deletions on Toxin Expression
It has been reported that the expression of csrA could affect toxin expression in C. difficile (Gu et al., 2018). To evaluate the effects of fliW and fliW-csrA deletions on toxin production, the supernatants of C. difficile cultures were collected at 24- and 48-h post-inoculation, and the toxin concentration was determined by ELISA. Figure 5A shows that the TcdA concentration of R20291ΔWA decreased by 28.6% (p < 0.05), while R20291ΔW increased by 65.1% (p < 0.01) compared to R20291 at 24-h post-inoculation. However, after 48-h incubation, no significant difference was detected. In Figure 5B, TcdB concentration of R20291ΔWA decreased by 26.4% (p < 0.05) at 24-h post-inoculation, while that of R20291ΔW increased by 93.6% (p < 0.01) at 24h and 33.0% (p < 0.05) at 48h. Similar results were also detected in the complementation strains group (Figures 5C,D). As shown in Figures 5C,D, after 24-h post-inoculation, TcdA (Figure 5C) concentration of R20291ΔWA-E and R20291ΔWA-W decreased by 33.0% (*p < 0.05) and 47.7% (p < 0.01), and TcdB (Figure 5D) concentration of R20291ΔWA-E and R20291ΔWA-W decreased by 37.9% (p < 0.05) and 31.3% (p < 0.05), respectively, while TcdA concentration of R20291ΔW-E, R20291ΔWA-A, and R20291ΔW-W increased by 83.1% (p < 0.01), 64.7% (p < 0.05), and 56.5% (p < 0.05), respectively. Meanwhile, TcdB concentration of R20291ΔW-E increased by 100.2% (p < 0.01). At 48-h post-inoculation, though no significant difference in TcdA production was detected among different C. difficile strains, TcdB concentration of R20291ΔWA-A increased by 28.5% (p < 0.05) compared to R20291-E.
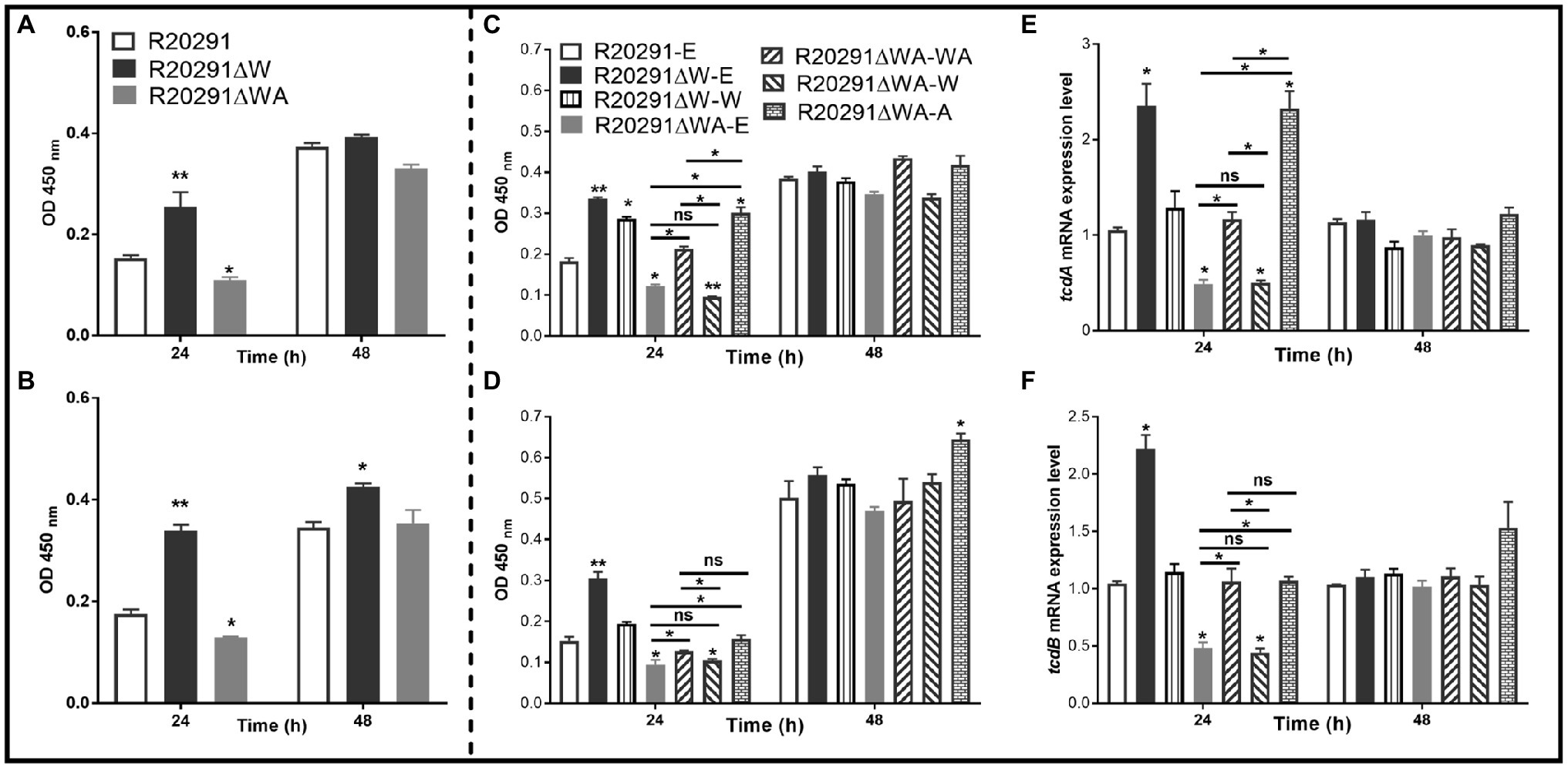
Figure 5. Toxin expression analysis. (A) TcdA concentration in the supernatants of R20291, R20291∆WA, and R20291∆W. (B) TcdB concentration in the supernatants of R20291, R20291∆WA, and R20291∆W. (C) TcdA concentration in the supernatants of parental and gene complementation strains. (D) TcdB concentration in the supernatants of parental and gene complementation strains. (E) Transcription of tcdA in the supernatants of parental and gene complementation strains. (F) Transcription of tcdB in the supernatants of parental and gene complementation strains. Experiments were independently repeated thrice. Bars stand for mean±SEM (*p<0.05, **p<0.01). One-way ANOVA with post hoc Tukey test was used for statistical significance. ** upon the column directly means the significant difference of experimental strain compared to R20291 or R20291-E.
To analyze the transcription of tcdA and tcdB in the complementation strains, RT-qPCR was performed. As shown in Figures 5E,F, the transcription of tcdA and tcdB of R20291ΔWA-E and R20291ΔWA-W decreased significantly (p < 0.05), while that of R20291ΔW-E increased significantly (p < 0.05). Interestingly, the tcdA transcription of R20291ΔWA-A also showed a significant increase (p < 0.05) compared to the wild-type strain. Our data indicate that FliW negatively regulates toxin expression, while CsrA plays a positive regulation role in toxin expression.
Effects of fliW and fliW-csrA Deletions on Sporulation and Germination
To assay the sporulation ratio of C. difficile strains, R20291, R20291ΔWA, and R20291ΔW were cultured in Clospore media for 48 and 96h, respectively. Results (Supplementary Figure 4A) showed that no significant difference in the sporulation ratio was detected between the wild-type strain and the mutants. The germination ratio of C. difficile spores was evaluated as well. Purified spores of R20291, R20291ΔWA, and R20291ΔW were incubated in the germination buffer supplemented with taurocholic acid (TA). As shown in Supplementary Figure 4B, there was no significant difference in the germination ratio between the wild-type strain and the mutants.
Evaluation of fliW and fliW-csrA Deletions on Bacterial Virulence in the Mouse Model of CDI
To evaluate the effects of fliW and fliW-csrA deletions on C. difficile virulence in vivo, the mouse model of CDI was used. Thirty mice (n=10 per group) were orally challenged with R20291, R20291ΔWA, or R20291ΔW spores (1×106 spores/mouse) after antibiotic treatment. As shown in Figure 6A, the R20291ΔW infection group lost more weight at post-challenge days 1 (p < 0.05), and the R20291ΔWA infection group lost less weight at post-challenge days 3 (p < 0.05) compared to the R20291 infection group. Figure 6B shows that 60% of mice succumbed to severe disease within 4days in the R20291ΔW infection group and 20% in the R20291ΔWA infection group compared to 50% mortality in the R20291 infection group (no significant difference with log-rank analysis, p=0.1629). Meanwhile, 100% of mice developed diarrhea in both the R20291ΔW and R20291 infection groups vs. 80% in the R20291ΔWA infection group at post-challenge days 2 (Figure 6C). As shown in Figure 6D, the spores CFU of the R20291ΔW infection group increased in the fecal shedding samples at post-challenge days 1 and 2 (p < 0.05), while the spores CFU of the R20291ΔWA infection group decreased at post-challenge days 1, 5, and 6 (p < 0.05) compared to the R20291 infection group. Interestingly, while we did not detect significant differences in bacterial growth, germination, and sporulation between the wild-type strain and mutants, the spore numbers from different infection groups were different (Figure 6D). This kind of difference implied that the culture media we used in vitro cannot simulate the complicated intestine environment well, which can lead to the different outcomes in bacterial physiology between in vitro and in vivo analysis. CsrA, as a carbon storage regulator, its regulation on carbon metabolization, and other potential roles in the complicated gut environment in vivo remain to be further studied.
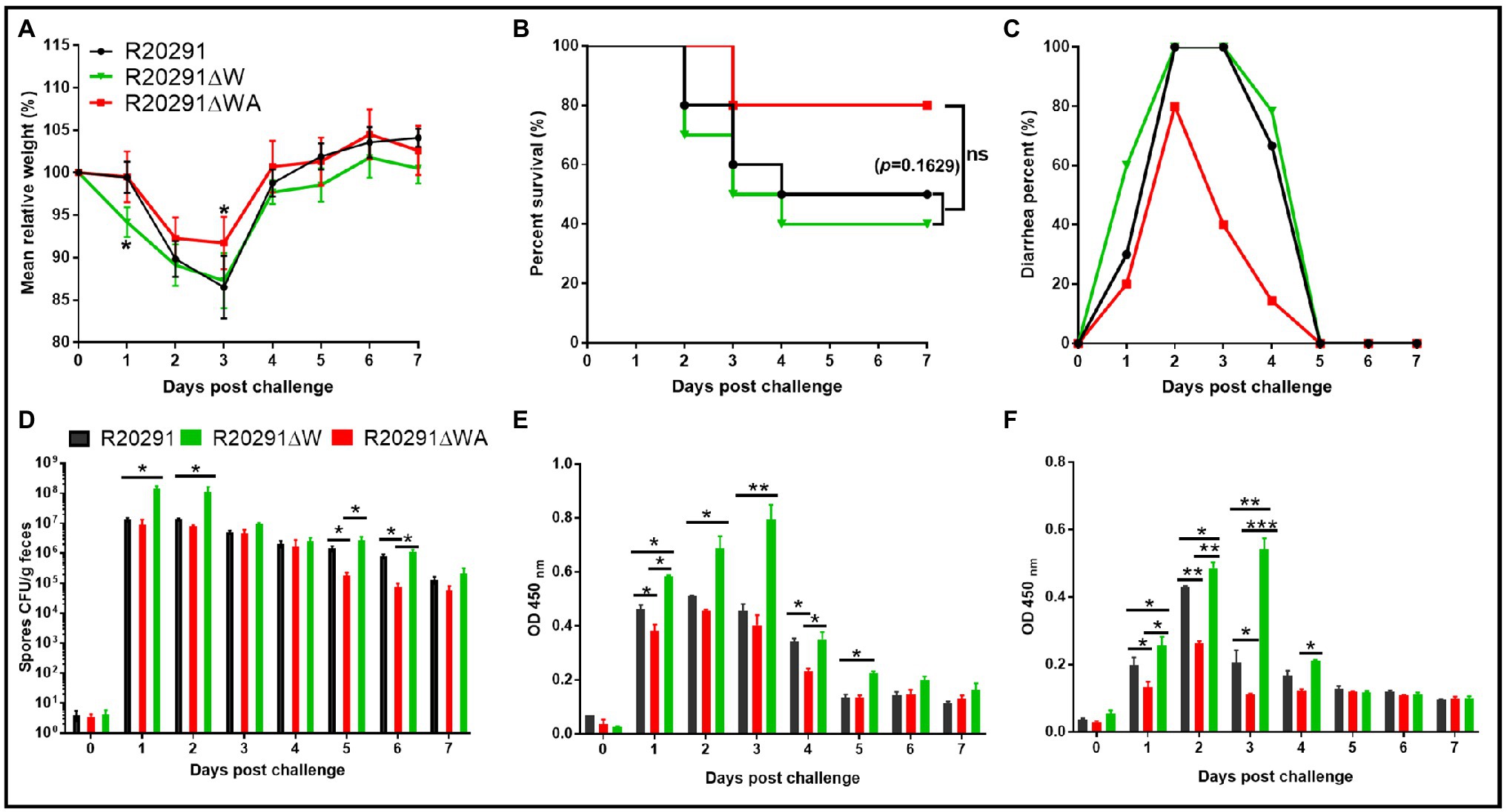
Figure 6. Effects of fliW and fliW-csrA deletion on C. difficile virulence in mice. (A) Mean relative weight changes. (B) Survival curve. (C) Diarrhea percentage. (D) Clostridioides difficile in feces. (E) TcdA titer of fecal sample. (F) TcdB titer of fecal sample. Bars stand for mean±SEM (*p<0.05, **p<0.01, ***p<0.001). One-way ANOVA with post hoc Tukey test was used for statistical significance. Animal survivals were analyzed by Kaplan–Meier survival analysis with a log-rank test of significance.
To evaluate the toxin level in the gut, the concentrations of TcdA and TcdB in the feces were measured by ELISA. In comparison with the R20291 infection group, the TcdA of the R20291ΔW infection group increased significantly at post-challenge days 1 (p < 0.05), 2 (p < 0.05), 3 (p < 0.01), and 5 (p < 0.05; Figure 6E), while the TcdA of the R20291ΔWA infection group decreased significantly at post-challenge days 1 (p < 0.05) and 4 (p < 0.05; Figure 6E). As shown in Figure 6F, the TcdB concentration of the R20291ΔWA infection group decreased significantly at post-challenge days 1 (p < 0.05), 2 (p < 0.05), and 3 (p < 0.05), and that of the R20291ΔW increased significantly at post-challenge days 1 (p < 0.05), 2 (p < 0.01), and 3 (p < 0.01). Taken together, our results indicate that the FliW defect increases R20291 pathogenicity in vivo, while the fliW-csrA codeletion impairs R20291 pathogenicity.
Discussion
In this study, we sought to characterize the impacts of FliW, CsrA, and FliC on C. difficile pathogenicity. Our data suggest that CsrA negatively modulates fliC expression post-transcriptionally, and FliW affects fliC expression possibly through inhibiting CsrA-mediated negative post-transcriptional regulation. Our data also indicate that FliW negatively affects C. difficile pathogenicity possibly by antagonizing CsrA in vivo. Based on our current pleiotropic phenotype analysis, a similar partner-switching mechanism “FliW-CsrA-fliC/FliC” (FliC binds FliW, FliW binds CsrA, and CsrA regulates fliC translation by binding to 5′ untranslated region of fliC transcripts) is predicted in C. difficile, though more direct experimental data are needed to uncover the molecular interactions of CsrA, FliW, and fliC/FliC in C. difficile (Supplementary Figure 5).
It has been reported that overexpression of the csrA gene could result in flagella defects, poor motility, and increased toxin production and adhesion in C. difficile 630Δerm (Gu et al., 2018). In our study, we found that CsrA and FliW widely exist in C. difficile (Table 2), even in the C. difficile strains without flagella like C. difficile M120 (Stabler et al., 2009), indicating a potentially important role of FliW-CsrA in C. difficile. Interestingly, while there are no flagella in C. difficile M120, six flagellar structure genes (fliS, fliN, flgK, flgL, fliC, and fliD) are still found in the genome, which inspired us to explore the potential roles of fliW, csrA, and fliC in C. difficile by deleting or overexpressing fliW, csrA, and fliW-csrA genes. However, after several trials with different gene edit methods in C. difficile, we could not get the csrA gene deletion mutant possibly due to its small size. This result motivated us to construct fliW-csrA double deletion mutant. While we did not get the single csrA gene deletion, we complemented the single fliW gene in the fliW-csrA double deletion mutant for simulation of the csrA deletion effects. The important roles of CsrA in flagellin synthesis and flagellin homeostasis have been reported (Yakhnin et al., 2007; Mukherjee et al., 2011; Gu et al., 2018; Oshiro et al., 2019). A previous study had shown that the overexpression of the csrA gene can cause a dramatic motility reduction and a significant Hag decrease in B. subtilis (Yakhnin et al., 2007). FliW (the first protein regulator of CsrA activity) deletion abolished the B. subtilis swarming and swimming motility and decreased the number of flagella and flagellar length (Mukherjee et al., 2011, 2016). In this study, we obtained similar results that FliW defect impaired R20291 motility significantly (Figure 2A) and increased biofilm formation (Figures 2C,D). Interestingly, the csrA gene complementation in R20291ΔWA dramatically suppressed bacterial motility and showed a similar result to R20291ΔW, indicating that CsrA can suppress C. difficile motility and increase biofilm production, while FliW antagonizes csrA to regulate bacteria motility and biofilm formation indirectly.
The partner-switching mechanism “Hag-FliW-CsrA” on flagellin synthesis was elucidated in B. subtilis, and the intracellular concentration of the flagellar filament protein Hag is restricted tightly by the Hag-FliW-CsrA system (Mukherjee et al., 2011). To investigate whether FliW and CrsA coregulate the fliC expression in C. difficile, we evaluated both the transcriptional and translational expression level of fliC gene. Our data (Figure 4) showed that the fliW deletion resulted in a 10.4-fold decrease in FliC accumulation, while the fliW-csrA codeletion increased FliC production, indicating that CsrA could suppress the fliC translation and FliW antagonizes CsrA to regulate FliC production. In csrA, fliW, and fliW-csrA overexpression experimental groups, we found that the csrA overexpression dramatically decreased FliC production (5.3-fold reduction) and the reduction in FliC production in R20291-A can be partially recovered when fliW-csrA was coexpressed. The FliW complementation in R20291ΔWA did not affect FliC production, but the fliW overexpression in R20291 increased FliC production. Taken together, our data suggest that CsrA negatively modulates fliC expression post-transcriptionally and FliW antagonizes CsrA to regulate fliC expression through inhibiting CsrA-mediated negative post-transcriptional regulation, indicating a similar partner-switching mechanism “FliW-CsrA-FliC” in C. difficile. In B. subtilis, two CsrA binding sites (BS1: A51 to A55; BS2: C75 to G82) were identified in the hag leader of the mRNA (Yakhnin et al., 2007). Based on the hag 5’-UTR sequence and CsrA conserved binding sequence, a 91bp 5’-UTR structure with two potential CsrA binding sites (BS1: 5’-TGACAAGGATGT-3′, BS2: 5’-CTAAGGAGGG-3′) of fliC gene was predicted (Supplementary Figure 6; Dubey et al., 2005). Recently, it was also reported that cytoplasmic Hag levels play a central role in maintaining proper intracellular architecture, and the Hag-FliW-CsrAdimer system works at nearly 1:1:1 stoichiometry in B. subtilis (Oshiro et al., 2019). Further studies on the exquisite interactions of CsrA, FliW, and fliC/FliC in C. difficile are still needed.
Flagella play multiple roles in bacterial motility, colonization, growth, toxin production, and survival optimization (Harshey, 2003; Duan et al., 2013; Stevenson et al., 2015). Recently, several papers have reported that the flagellar genes can affect toxin expression in C. difficile, but results from different research groups were controversial (Aubry et al., 2012; Baban et al., 2013; Stevenson et al., 2015). Aubry et al. (2012) reported that disruption of some early-stage flagellar genes (F3), such as fliF, fliG, and fliM, could lead to a significant reduction in tcdR, tcdE, tcdA, and tcdB expression in C. difficile 630Δerm, but no significant difference of tcdC expression was detected. Inversely, disruption of late-stage flagellar genes (F1) such as fliC increased toxin expression in C. difficile 630Δerm. In 2013, Baban et al. (2013) reported that the mutation of flgE (one of the F3 genes) resulted in a tenfold reduction in tcdA expression and corroborated that the expression of tcdA in a fliC mutant increased 44.4-fold compared to the wild-type strain C. difficile 630Δerm. Surprisingly, Aubry et al. (2012) found that a glycosylation gene (CD0240, one of F2 region genes) mutation, which can totally abolish C. difficile 630 motility, but did not change toxin expression. Meanwhile, cyclic diguanylate (C-di-GMP), a cellular second messenger, was also reported to be involved in bacterial motility, biofilm formation, and toxin production by repressing the expression of flagellar genes in C. difficile (Purcell et al., 2012; McKee et al., 2013). While we did not detect the C-di-GMP concentration in C. difficile, it could be perturbed by fliW and csrA deletion affecting C. difficile physiology. It was hypothesized that the regulation of the flagellar genes on toxin expression could be caused by the direct change or loss of flagellar genes (such as fliC gene deletion) rather than loss of the functional flagella (Stevenson et al., 2015). Future study about fliC deletion in M120 will be very interesting and will further address the fliC gene function in C. difficile as there are no flagella in RT078 strains. In our study, data indicate that CsrA negatively modulates fliC translation and also plays a positive regulation in toxin expression. Inversely, FliW works against CsrA to regulate fliC expression, which can negatively regulate toxin production. While studies of flagellar effects on motility and toxin production in C. difficile from different groups were controversial, the role of the flagella in C. difficile pathogenicity cannot be overlooked. Dingle et al. (2011) and Baban et al. (2013) both showed higher mortality of the fliC mutant in the animal model of CDI compared to the wild-type strains. Our study showed results similar to the published data suggesting that R20291ΔW whose FliC production was dramatically suppressed exhibited higher fatality, while R20291ΔWA showed a decreased pathogenicity compared to R20291 (Figure 6). In 2014, Barketi-Klai et al. (2014) examined the pleiotropic roles of the fliC gene in R20291 during colonization in mice. Interestingly, the transcription of fliW and csrA in the fliC mutant was 2.03- and 4.36-fold, respectively, of that in R20291 in vivo experiment (Barketi-Klai et al., 2014), which further corroborated that there is a coregulation among fliC, fliW, and csrA. Surprisingly, transcription of treA, a trehalose-6-phosphate hydrolase, increased 177.63-fold in the fliC mutant compared to that of R20291 during in vivo infection (Barketi-Klai et al., 2014). Recently, Collins et al. (2018) hypothesized that dietary trehalose can contribute to the virulence of epidemic C. difficile. The relationship of FliW, CsrA, FliC, and trehalose metabolization is another interesting question in C. difficile, and some other carbon metabolism affected by the fliC mutation could also facilitate C. difficile pathogenesis in vivo. Previous studies have also highlighted that the flagella of C. difficile play an important role in toxin production, biofilm formation, and bacterial adherence to the host (Tasteyre et al., 2001; Dingle et al., 2011; Aubry et al., 2012; Baban et al., 2013; Ethapa et al., 2013). In this study, we showed that the FliW defect led to a significant motility decrease, while the biofilm, adhesion, and toxin production increased significantly. Inversely, R20291ΔWA-W, which can imitate the csrA gene deletion, showed an increase in motility and a decrease in biofilm formation, toxin production, and adhesion.
In conclusion, we characterized the function of FliW and CsrA and showed the pleiotropic functions of FliW and CsrA in R20291. Our data suggest that fliW and csrA play important roles in flagellin (FliC) synthesis, which could contribute to C. difficile pathogenicity. Currently, in vitro study of the interactions of CsrA, FliW, and fliC/FliC in C. difficile is underway in our group.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of University of South Florida.
Author Contributions
DZ and XS designed the experiments. DZ wrote the manuscript. DZ and SW performed the experiments. DZ and XS revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the National Institutes of Health grants (R01-AI132711 and R01-AI149852).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Dr. Abhraham L. Sonnenshein at Tufts University, Dr. Joseph Sorg at Texas A&M, and Dr. Daniel Kearns at Indiana University for the gifts C. difficile R20291, E.coli HB101/pRK24, and anti-SigA primary antibody, respectively. We thank Dr. Nigel Minton at the University of Nottingham for the gift plasmids pMTL84151 and pMTL83353. We also thank Jessica Bullock and Dr. Heather Danhof for their mindful revision and comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.735616/full#supplementary-material
Footnotes
References
Aubry, A., Hussack, G., Chen, W., KuoLee, R., Twine, S. M., Fulton, K. M., et al. (2012). Modulation of toxin production by the flagellar regulon in Clostridium difficile. Infect. Immun. 80, 3521–3532. doi: 10.1128/IAI.00224-12
Baban, S. T., Kuehne, S. A., Barketi-Klai, A., Cartman, S. T., Kelly, M. L., Hardie, K. R., et al. (2013). The role of flagella in Clostridium difficile pathogenesis: comparison between a non-epidemic and an epidemic strain. PLoS One 8:e73026. doi: 10.1371/journal.pone.0073026
Barketi-Klai, A., Monot, M., Hoys, S., Lambert-Bordes, S., Kuehne, S. A., Minton, N., et al. (2014). The flagellin FliC of Clostridium difficile is responsible for pleiotropic gene regulation during in vivo infection. PLoS One 9:e96876. doi: 10.1371/journal.pone.0096876
Chong, L. (2001). Molecular cloning—A laboratory manual, 3rd edition. Science 292:446. doi: 10.1126/science.1060677
Collins, J., Robinson, C., Danhof, H., Knetsch, C. W., van Leeuwen, H. C., Lawley, T. D., et al. (2018). Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature 553, 291–294. doi: 10.1038/nature25178
Dingle, T. C., Mulvey, G. L., and Armstrong, G. D. (2011). Mutagenic analysis of the Clostridium difficile flagellar proteins, FliC and FliD, and their contribution to virulence in hamsters. Infect. Immun. 79, 4061–4067. doi: 10.1128/IAI.05305-11
Duan, Q., Zhou, M., Zhu, L., and Zhu, G. (2013). Flagella and bacterial pathogenicity. J. Basic Microbiol. 53, 1–8. doi: 10.1002/jobm.201100335
Dubey, A. K., Baker, C. S., Romeo, T., and Babitzke, P. (2005). RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11, 1579–1587. doi: 10.1261/rna.2990205
Ethapa, T., Leuzzi, R., Ng, Y. K., Baban, S. T., Adamo, R., Kuehne, S. A., et al. (2013). Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J. Bacteriol. 195, 545–555. doi: 10.1128/JB.01980-12
Ferreiro, M. D., Nogales, J., Farias, G. A., Olmedilla, A., Sanjuan, J., and Gallegos, M. T. (2018). Multiple CsrA proteins control key virulence traits in pseudomonas syringae pv. Tomato DC3000. Mol. Plant-Microbe Interact. 31, 525–536. doi: 10.1094/MPMI-09-17-0232-R
Fuller, M. E., Streger, S. H., Rothmel, R. K., Mailloux, B. J., Hall, J. A., Onstott, T. C., et al. (2000). Development of a vital fluorescent staining method for monitoring bacterial transport in subsurface environments. Appl. Environ. Microbiol. 66, 4486–4496. doi: 10.1128/AEM.66.10.4486-4496.2000
Gu, H., Qi, H., Chen, S., Shi, K., Wang, H., and Wang, J. (2018). Carbon storage regulator CsrA plays important roles in multiple virulence-associated processes of Clostridium difficile. Microb. Pathog. 121, 303–309. doi: 10.1016/j.micpath.2018.05.052
Harshey, R. M. (2003). Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57, 249–273. doi: 10.1146/annurev.micro.57.030502.091014
Heap, J. T., Kuehne, S. A., Ehsaan, M., Cartman, S. T., Cooksley, C. M., Scott, J. C., et al. (2010). The ClosTron: mutagenesis in clostridium refined and streamlined. J. Microbiol. Methods 80, 49–55. doi: 10.1016/j.mimet.2009.10.018
Heap, J. T., Pennington, O. J., Cartman, S. T., and Minton, N. P. (2009). A modular system for clostridium shuttle plasmids. J. Microbiol. Methods 78, 79–85. doi: 10.1016/j.mimet.2009.05.004
Hong, W., Zhang, J., Cui, G., Wang, L., and Wang, Y. (2018). Multiplexed CRISPR-Cpf1-mediated genome editing in Clostridium difficile toward the understanding of pathogenesis of C. difficile infection. ACS Synth. Biol. 7, 1588–1600. doi: 10.1021/acssynbio.8b00087
Janvilisri, T., Scaria, J., and Chang, Y. F. (2010). Transcriptional profiling of Clostridium difficile and Caco-2 cells during infection. J. Infect. Dis. 202, 282–290. doi: 10.1086/653484
Karna, S. L., Sanjuan, E., Esteve-Gassent, M. D., Miller, C. L., Maruskova, M., and Seshu, J. (2011). CsrA modulates levels of lipoproteins and key regulators of gene expression critical for pathogenic mechanisms of Borrelia burgdorferi. Infect. Immun. 79, 732–744. doi: 10.1128/IAI.00882-10
Kuehne, S. A., Cartman, S. T., Heap, J. T., Kelly, M. L., Cockayne, A., and Minton, N. P. (2010). The role of toxin A and toxin B in Clostridium difficile infection. Nature 467, 711–713. doi: 10.1038/nature09397
Lawhon, S. D., Frye, J. G., Suyemoto, M., Porwollik, S., McClelland, M., and Altier, C. (2003). Global regulation by CsrA in salmonella typhimurium. Mol. Microbiol. 48, 1633–1645. doi: 10.1046/j.1365-2958.2003.03535.x
Lawson, P. A., Citron, D. M., Tyrrell, K. L., and Finegold, S. M. (2016). Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prevot 1938. Anaerobe 40, 95–99. doi: 10.1016/j.anaerobe.2016.06.008
Lessa, F. C., Gould, C. V., and McDonald, L. C. (2012). Current status of Clostridium difficile infection epidemiology. Clin. Infect. Dis. 55, S65–S70. doi: 10.1093/cid/cis319
Liu, M. Y., Yang, H., and Romeo, T. (1995). The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J. Bacteriol. 177, 2663–2672. doi: 10.1128/jb.177.10.2663-2672.1995
Lucchetti-Miganeh, C., Burrowes, E., Baysse, C., and Ermel, G. (2008). The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology 154, 16–29. doi: 10.1099/mic.0.2007/012286-0
Lyras, D., O'Connor, J. R., Howarth, P. M., Sambol, S. P., Carter, G. P., Phumoonna, T., et al. (2009). Toxin B is essential for virulence of Clostridium difficile. Nature 458, 1176–1179. doi: 10.1038/nature07822
McKee, R. W., Mangalea, M. R., Purcell, E. B., Borchardt, E. K., and Tamayo, R. (2013). The second messenger cyclic Di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD. J. Bacteriol. 195, 5174–5185. doi: 10.1128/JB.00501-13
Morris, E. R., Hall, G., Li, C., Heeb, S., Kulkarni, R. V., Lovelock, L., et al. (2013). Structural rearrangement in an RsmA/CsrA ortholog of Pseudomonas aeruginosa creates a dimeric RNA-binding protein, RsmN. Structure 21, 1659–1671. doi: 10.1016/j.str.2013.07.007
Mukherjee, S., Babitzke, P., and Kearns, D. B. (2013). FliW and FliS function independently to control cytoplasmic flagellin levels in Bacillus subtilis. J. Bacteriol. 195, 297–306. doi: 10.1128/JB.01654-12
Mukherjee, S., Oshiro, R. T., Yakhnin, H., Babitzke, P., and Kearns, D. B. (2016). FliW antagonizes CsrA RNA binding by a noncompetitive allosteric mechanism. Proc. Natl. Acad. Sci. U. S. A. 113, 9870–9875. doi: 10.1073/pnas.1602455113
Mukherjee, S., Yakhnin, H., Kysela, D., Sokoloski, J., Babitzke, P., and Kearns, D. B. (2011). CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol. Microbiol. 82, 447–461. doi: 10.1111/j.1365-2958.2011.07822.x
Oren, A., and Garrity, G. M. (2018). Notification of changes in taxonomic opinion previously published outside the IJSEM. Int. J. Syst. Evol. Microbiol. 68, 2137–2138. doi: 10.1099/ijsem.0.002830
Oshiro, R. T., Rajendren, S., Hundley, H. A., and Kearns, D. B. (2019). Robust stoichiometry of FliW-CsrA governs flagellin homeostasis and cytoplasmic organization in Bacillus subtilis. MBio 10, e00533–e00519. doi: 10.1128/mBio.00533-19
Peniche, A. G., Savidge, T. C., and Dann, S. M. (2013). Recent insights into Clostridium difficile pathogenesis. Curr. Opin. Infect. Dis. 26, 447–453. doi: 10.1097/01.qco.0000433318.82618.c6
Perez, J., Springthorpe, V. S., and Sattar, S. A. (2011). Clospore: a liquid medium for producing high titers of semi-purified spores of Clostridium difficile. J. AOAC Int. 94, 618–626. doi: 10.1093/jaoac/94.2.618
Pessi, G., Williams, F., Hindle, Z., Heurlier, K., Holden, M. T., Camara, M., et al. (2001). The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183, 6676–6683. doi: 10.1128/JB.183.22.6676-6683.2001
Purcell, E. B., McKee, R. W., McBride, S. M., Waters, C. M., and Tamayo, R. (2012). Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J. Bacteriol. 194, 3307–3316. doi: 10.1128/JB.00100-12
Romeo, T., Gong, M., Liu, M. Y., and Brun-Zinkernagel, A. M. (1993). Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 175, 4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993
Sabnis, N. A., Yang, H., and Romeo, T. (1995). Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J. Biol. Chem. 270, 29096–29104. doi: 10.1074/jbc.270.49.29096
Sebaihia, M., Wren, B. W., Mullany, P., Fairweather, N. F., Minton, N., Stabler, R., et al. (2006). The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38, 779–786. doi: 10.1038/ng1830
Sorger-Domenigg, T., Sonnleitner, E., Kaberdin, V. R., and Blasi, U. (2007). Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem. Biophys. Res. Commun. 352, 769–773. doi: 10.1016/j.bbrc.2006.11.084
Stabler, R. A., He, M., Dawson, L., Martin, M., Valiente, E., Corton, C., et al. (2009). Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10:R102. doi: 10.1186/gb-2009-10-9-r102
Stevenson, E., Minton, N. P., and Kuehne, S. A. (2015). The role of flagella in Clostridium difficile pathogenicity. Trends Microbiol. 23, 275–282. doi: 10.1016/j.tim.2015.01.004
Sun, X., Wang, H., Zhang, Y., Chen, K., Davis, B., and Feng, H. (2011). Mouse relapse model of Clostridium difficile infection. Infect. Immun. 79, 2856–2864. doi: 10.1128/IAI.01336-10
Tasteyre, A., Barc, M. C., Collignon, A., Boureau, H., and Karjalainen, T. (2001). Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 69, 7937–7940. doi: 10.1128/IAI.69.12.7937-7940.2001
Timmermans, J., and Van Melderen, L. (2010). Post-transcriptional global regulation by CsrA in bacteria. Cell. Mol. Life Sci. 67, 2897–2908. doi: 10.1007/s00018-010-0381-z
Wang, Y., Wang, S., Bouillaut, L., Li, C., Duan, Z., Zhang, K., et al. (2018). Oral immunization with nontoxigenic Clostridium difficile strains expressing chimeric fragments of TcdA and TcdB elicits protective immunity against C. difficile infection in both mice and hamsters. Infect. Immun. 86, e00489–e00418. doi: 10.1128/IAI.00489-18
Williams, D. R., Young, D. I., and Young, M. (1990). Conjugative plasmid transfer from Escherichia coli to clostridium acetobutylicum. J. Gen. Microbiol. 136, 819–826. doi: 10.1099/00221287-136-5-819
Yakhnin, H., Pandit, P., Petty, T. J., Baker, C. S., Romeo, T., and Babitzke, P. (2007). CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol. Microbiol. 64, 1605–1620. doi: 10.1111/j.1365-2958.2007.05765.x
Zhu, D., Bullock, J., He, Y., and Sun, X. (2019). Cwp22, a novel peptidoglycan cross-linking enzyme, plays pleiotropic roles in Clostridioides difficile. Environ. Microbiol. 21, 3076–3090. doi: 10.1111/1462-2920.14706
Keywords: Clostridioides difficile, FliW, FliC, CsrA, R20291, virulence
Citation: Zhu D, Wang S and Sun X (2021) FliW and CsrA Govern Flagellin (FliC) Synthesis and Play Pleiotropic Roles in Virulence and Physiology of Clostridioides difficile R20291. Front. Microbiol. 12:735616. doi: 10.3389/fmicb.2021.735616
Edited by:
Meina Neumann-Schaal, German Collection of Microorganisms and Cell Cultures GmbH (DSMZ), GermanyReviewed by:
Susanne Sievers, University of Greifswald, GermanySarah Anna Kuehne, University of Birmingham, United Kingdom
Joshua Robert Fletcher, University of Minnesota Twin Cities, United States
Copyright © 2021 Zhu, Wang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingmin Sun, c3VuNUB1c2YuZWR1
 Duolong Zhu
Duolong Zhu Shaohui Wang
Shaohui Wang Xingmin Sun
Xingmin Sun