- 1Department of Pulmonary and Critical Care Medicine, The Third Medical Center of Chinese PLA General Hospital, Beijing, China
- 2College of Pulmonary and Critical Care Medicine, Chinese PLA General Hospital, Beijing, China
- 3Section of Pulmonary and Critical Care and Sleep Medicine, Department of Medicine, Yale University School of Medicine, New Haven, CT, United States
- 4Department of Oncology, The Second Medical Center of Chinese PLA General Hospital, Beijing, China
- 5College of Tuberculosis Medicine, Chinese PLA General Hospital, Beijing, China
Klebsiella species cause infections at multiple sites, including lung, urinary tract, bloodstream, wound or surgical site, and brain. These infections are more likely to occur in people with preexisting health conditions. Klebsiella pneumoniae (K. pneumoniae) has emerged as a major pathogen of international concern due to the increasing incidences of hypervirulent and carbapenem-resistant strains. It is imperative to understand risk factors, prevention strategies, and therapeutic avenues to treat multidrug-resistant Klebsiella infections. Here, we highlight the epidemiology, risk factors, and control strategies against K. pneumoniae infections to highlight the grave risk posed by this pathogen and currently available options to treat Klebsiella-associated diseases.
Introduction
Klebsiella pneumoniae is a Gram-negative, encapsulated, non-motile, facultatively anaerobic bacteria (Ray and Ryan, 2004). It was first isolated from the airways of a patient dying of pneumonia by Edwin Klebs in 1875 and later described by Carl Friedländer in 1882, leading it to be called Friedlander’s bacillus for some time (Köhler and Mochmann, 1987). Klebsiella species include Klebsiella ozaenae, Klebsiella rhinoscleroma, and K. pneumoniae, the last of which is an important opportunistic and iatrogenic infectious pathogen with major clinical implications (Bagley, 1985). In humans, Klebsiella often colonizes the nasal and digestive tract without causing any symptomatic disease. However, the colonization can turn into an infection when the host immunity fails to control the pathogen growth, examples of which include patients with diabetes, on glucocorticoid therapy, and those who have received organ transplantation. This mini-review will discuss critical aspects of K. pneumoniae biology related to its pathogenesis and control strategies.
Clinical Epidemiology
Colonization and Infection
Klebsiella species are present abundantly in nature and commonly found in soil, water, and other surfaces (Martin and Bachman, 2018). In humans, K. pneumoniae often colonizes at various mucosal surfaces, including the upper respiratory tract and the gut, where colonization rates vary widely among individuals based on their habitat and exposures (Bagley, 1985; Podschun et al., 2001; Martin and Bachman, 2018). Recent studies showed that the prevalence of Klebsiella colonization ranges from 18.8 to 87.7% in Asia and 5 to 35% in Western countries (Lin et al., 2012; Russo and Marr, 2019). In the non-hospital settings, the carrier rate of Klebsiella in fecal samples ranges from 5 to 38%, while the carrier rates in the nasopharynx range from 1 to 6% (Fuxench-López and Ramírez-Ronda, 1978; Podschun and Ullmann, 1998). The presentation of Klebsiella involving the skin is rare, which is considered transient rather than persistent (Kloos and Musselwhite, 1975). In hospitalized patients, colonization rates in the nasopharynx rise to 19%, while it can be as high as 77% in the gastrointestinal tract (Pollack et al., 1972; Podschun and Ullmann, 1998). Due to its extensive presence in humans, gastrointestinal colonization serves as a major reservoir for transmission and infection to other sites (Martin et al., 2016; Dorman and Short, 2017). The experimental evidence of gastrointestinal sources of infection came from a study by Selden et al. (1971), which demonstrated that K. pneumoniae infections often show similar serotypes as colonizing bacteria in the intestinal tract. More recent studies have confirmed the relationship between colonizing strains of K. pneumoniae and strains obtained from the infection sites (Martin et al., 2016; Dorman and Short, 2017). A longitudinal study confirmed these studies where investigators determined the colonization of Klebsiella in the gastrointestinal tract of a cohort of 1765 patients and followed this cohort for 3 months to assess the respiratory tract, urinary tract, or blood infections. The results demonstrated that 21 of 406 patients (5.2%) had colonization developed a subsequent infection compared to only 1.3% of the subjects without colonization. Genetic sequencing revealed that most of the Klebsiella infections originated from the colonization sites in the same patient.
Similarly, Gorrie et al. (2017) analyzed the relationship between colonization and susceptibility to infection by K. pneumoniae in 498 ICU patients. They found that 16% of the patients colonized with K. pneumoniae were found to be infected, compared to only 3% in the non-carriers (Gorrie et al., 2017). Whole-genome sequencing revealed that the patients were infected with the same strain they carried in the form of colonization. From a genomic perspective, these studies demonstrated that gastrointestinal microbiota is a prominent source of nosocomial K. pneumoniae infections, 80% of which are caused by self-colonizing strains. The transition from colonization to infection is primarily due to the impairment of host defense contributed by underlying diseases or immunomodulatory therapies (Tsay et al., 2002; Meatherall et al., 2009). To support this notion, a study by Lee et al. (1994) observed 101 patients diagnosed with Klebsiella bacteremia and found that 36% of patients had diabetes, and 26% had malignant tumors (Tsai et al., 2010).
Discriminating between colonization and infection has always been a puzzle for clinicians and researchers, making it difficult for subsequent intervention strategies. However, several factors can be considered to distinguish colonization from infection. These factors include:
1. Detection of Klebsiella in the bloodstream indicates an active infection. Circulating blood remains sterile, unlike the respiratory, urinary, and digestive tracts, which harbor colonizing K. pneumoniae.
2. The patient’s symptoms, clinical signs, laboratory examination, and imaging data should be employed to differentiate colonization from infection. For example, respiratory infection with Klebsiella is diagnosed if the patient has a fever, cough, sputum production, high WBC, and imaging evidence of pneumonia in the lung.
3. When patients have underlying diseases such as COPD, diabetes, heart disease, organ transplantation, or a recent history of steroid use or antimicrobial drugs, the infection of Klebsiella should be considered when positive cultures are obtained. In summary, infections by K. pneumoniae often derive from the colonizing bacteria within the host, and both clinical and bacteriological factors should be considered to distinguish active infection from colonization.
Hypervirulent K. pneumoniae
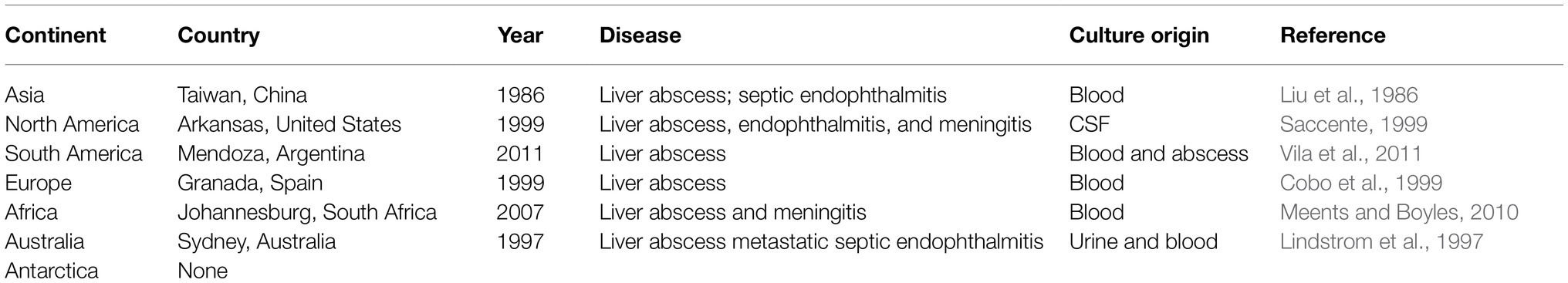
Hypervirulent K. pneumoniae (hvKp) indicates a K. pneumoniae strain that can cause infections in relatively healthy subjects, often in community settings. The infection is often manifested in multiple organs (Struve et al., 2015). HvKp was first reported in 1986 in Taiwan, which caused pneumonia complicated with liver abscess, meningitis, and endophthalmitis (Liu et al., 1986). This strain causing liver abscess was also known as hypermucoviscous K. pneumoniae due to its hypermucoviscous phenotype (string test >5 mm), as described by Fang et al. (2004). Unique sequences on the plasmid can distinguish hvKp from classic K. pneumoniae strains (Russo et al., 2018). The high virulence of these strains is recapitulated in animal models as demonstrated by a 50% lethal dose (LD50) being as low as 103 colony forming units (Nassif and Sansonetti, 1986). The hypermucoviscous phenotype is contributed by a unique plasmid in some of these strains (Nassif et al., 1989; Nassrf et al., 1989). In this regard, the first strain isolated by Friedlander in 1882 was considered as hvKp because it was highly pathogenic and could infect multiple sites in the body (Russo and Marr, 2019). However, the subsequent studies demonstrated that not all hvKp have hypermucoviscous phenotype, and some common K. pneumoniae strains were hypermucoviscous, indicating that hypermucoviscous phenotype can be present in non-hypervirulent strains of Klebsiella (Catalán-Nájera et al., 2017; Russo et al., 2018). At present, cases of hvKp have already been reported from Europe, Asia, and the United States (Russo and Marr, 2019).
The first case of hvKp infection in North America was a 38-year-old African American man with headache and fever diagnosed with a liver abscess caused by hvKp and complicated by endophthalmitis and meningitis (Saccente, 1999; Table 1). In a retrospective study of 56 cases of liver abscess at Elmhurst Hospital in New York City, 36 percent of the patients were caused by hvKp using imaging and culture of blood or liver aspiration (Pastagia and Arumugam, 2008). Subsequently, more and more hvKp infections were reported (Pastagia and Arumugam, 2008; Mccabe et al., 2010; Fierer et al., 2011; Vila et al., 2011). Increasing reports of the multiorgan clinical manifestation associated with K. pneumoniae such as liver abscess, bacteremia, meningitis, endophthalmitis, and necrotizing fasciitis indicate a high prevalence hvKp, which has become a global disease that requires immediate attention. The earliest clinical clues for hvKp include the presence of liver abscess and bacteremia in patients with a positive culture. Due to high lethality, early diagnosis and intervention play a key role in limiting the disease severity and death due to hvKp.
Carbapenem-Resistant K. pneumoniae
Pathogens employ multiple mechanisms to develop antibiotic resistance, including the production of beta-lactamase, loss of susceptible outer membrane proteins, change of target, biofilm formation, efflux pump, and integron (Thomson and Bonomo, 2005). Exposure to antibiotics at sublethal concentrations may lead to the development of resistance among exposed pathogens. In addition to the clinical misuse of antimicrobial agents, the human population is often exposed to a wide range of non-iatrogenic antibacterial drugs in daily life, including antibiotic exposures to the livestock in the meat industry, which leads to increased drug resistance in pathogens. Increased use of antibiotics in both clinical and non-clinical settings is associated with an increased number of clinically isolated drug-resistant strains, including carbapenem-resistant K. pneumoniae (CRKP; Fair and Tor, 2014; Li and Webster, 2018).
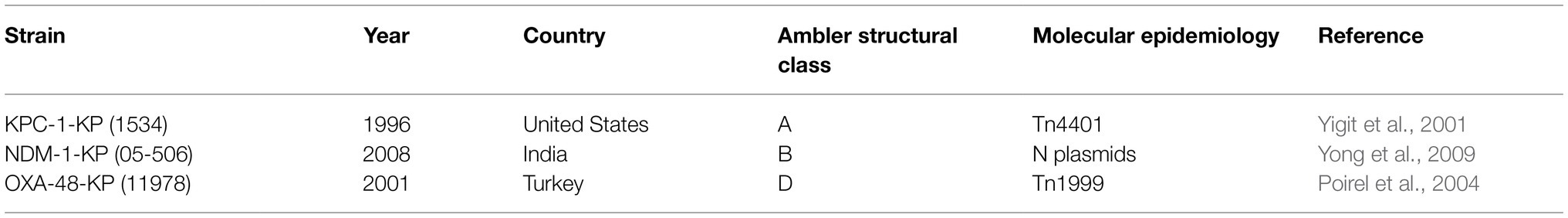
In recent years, the extensive use of carbapenems in clinical practice increased carbapenem resistance in K. pneumoniae. Since its emergence in the 1990s, it has gradually become prevalent worldwide with high mortality (Table 2). The presence of carbapenem-resistance in the infecting pathogens is an independent risk factor of mortality among patients with nosocomial infection. There are several mechanisms responsible for carbapenem resistance among K. pneumoniae. The primary mechanism is the production of a carbapenemase classified into the Ambler class A (K. pneumoniae carbapenemase, KPC), B (the metalloenzymes NDM, VIM, and IMP), and D (oxacillin enzyme, OXA; Spagnolo et al., 2014; Logan and Weinstein, 2017).
The coding gene of KPC is blaKPC, which can be transferred by the plasmid among K. pneumoniae strains through Tn3-based transposon Tn4401 (Spagnolo et al., 2014; Logan and Weinstein, 2017). The blaKPC gene can also be transmitted to other bacteria like Enterobacter and Pseudomonas aeruginosa (Virgincar et al., 2011). The blaKPC-K. pneumoniae strains have been isolated across the globe in recent years, indicating their global presence. The first reported blaKPC-K. pneumoniae strain was isolated in a hospital located in North Carolina in 1996 and submitted to the Centers for Disease Control and Prevention (CDC) through the intensive care antimicrobial resistance epidemiology program (ICARE; Yigit et al., 2001). Naas et al. (2005) reported the first French case of CRKP in 2005, an 80-year-old man with prostate cancer and metastasis. These cases demonstrated the transmission of blaKPC-K. pneumoniae among different continents.
New Delhi Metallo-beta-lactamase 1 (NDM-1) K. pneumoniae is a newly emerged highly resistant bacteria that can produce NDM-1 capable of breaking down beta-lactam antibiotics, belonging to Ambler class B carbapenemase (Kumarasamy et al., 2010). The bacteria with NDM-1 genes were considered “superbugs” due to their lack of susceptibility to almost all available antibiotics (Moellering, 2010). The resistance gene was located not only in the bacterial genome but also in plasmids. Thus, the strains that are initially susceptible to antibiotics may quickly become resistant through horizontal gene transfer. The first NDM-1 K. pneumoniae was reported in a 59-year-old Indian male with type II diabetes and multiple strokes. The patient was a resident of Sweden with multiple visits to India (Yong et al., 2009). The strain was isolated from the urine sample on January 9, 2008, when the patient had no apparent urinary tract infection symptoms. To date, NDM-1 K. pneumoniae has also been reported across the world including in China, Australia, the United States, Canada, Europe, and Africa (Dhainaut et al., 2005; Burgner et al., 2006; Vered et al., 2014; Hammer et al., 2018).
OXA-48 is an Ambler class D carbapenemase encoded by the blaOXA-48 gene, which has been reported increasingly among the Enterobacteriaceae family of bacteria in recent years. Although this enzyme has relatively weak β-lactamase activity, it can hydrolyze penicillin and cannot be inhibited by β-lactamase inhibitors. It can spread widely among bacteria through the Tn199-based plasmid containing the blaOXA-48 gene flanked with the IS1999 sequence (Pfeifer et al., 2012). OXA-48 was first identified in Turkey in a K. pneumoniae in 2001, which was resistant to almost all beta-lactams, including penicillin, cephalosporins, monocyclic lactams, and carbapenems (Poirel et al., 2004). Further analysis revealed that SHV-2a, TEM-1, and OXA-47 were expressed, and antibiotic susceptible extracellular membrane proteins were absent, leading to their resistance to various antibiotics. In subsequent years, outbreaks of the OXA-48 strains occurred in various regions of Turkey. In addition to Turkey, OXA-48 has been reported in other European countries, America, Asia, Oceania, and Africa (Nordmann et al., 2011; Espedido et al., 2013; Van Duin and Doi, 2017). Notably, the incidence of the OXA-48-expressing strains may be underestimated because it is difficult to recognize the presence of OXA-48-like enzymes due to their low levels of carbapenem resistance.
It has been reported that overexpression of efflux pumps, decreased permeability of outer membrane proteins, and production of the β-lactamase enzymes are also important reasons for CRKP (Zhang et al., 2014; Hamzaoui et al., 2018). Carbapenems are described as our last line of defense against drug-resistant bacteria. Most frightening of all, CRKP can pass on its resistance to other bacteria through horizontal gene transfer by various methods, including bacterial conjugation, leading to drug resistance. In particular, the emergence of highly virulent CRKP strains named ST11 CR-HvKp, which are highly virulent and resistant to carbapenems, poses a major challenge to our ability to control Klebsiella infections (Yao et al., 2018). The ST11 CR-HvKp strain infects the lungs and causes pneumonia, and invades the blood and other organs, potentially resulting in incurable and fatal infections in relatively healthy individuals.
Risk Factors
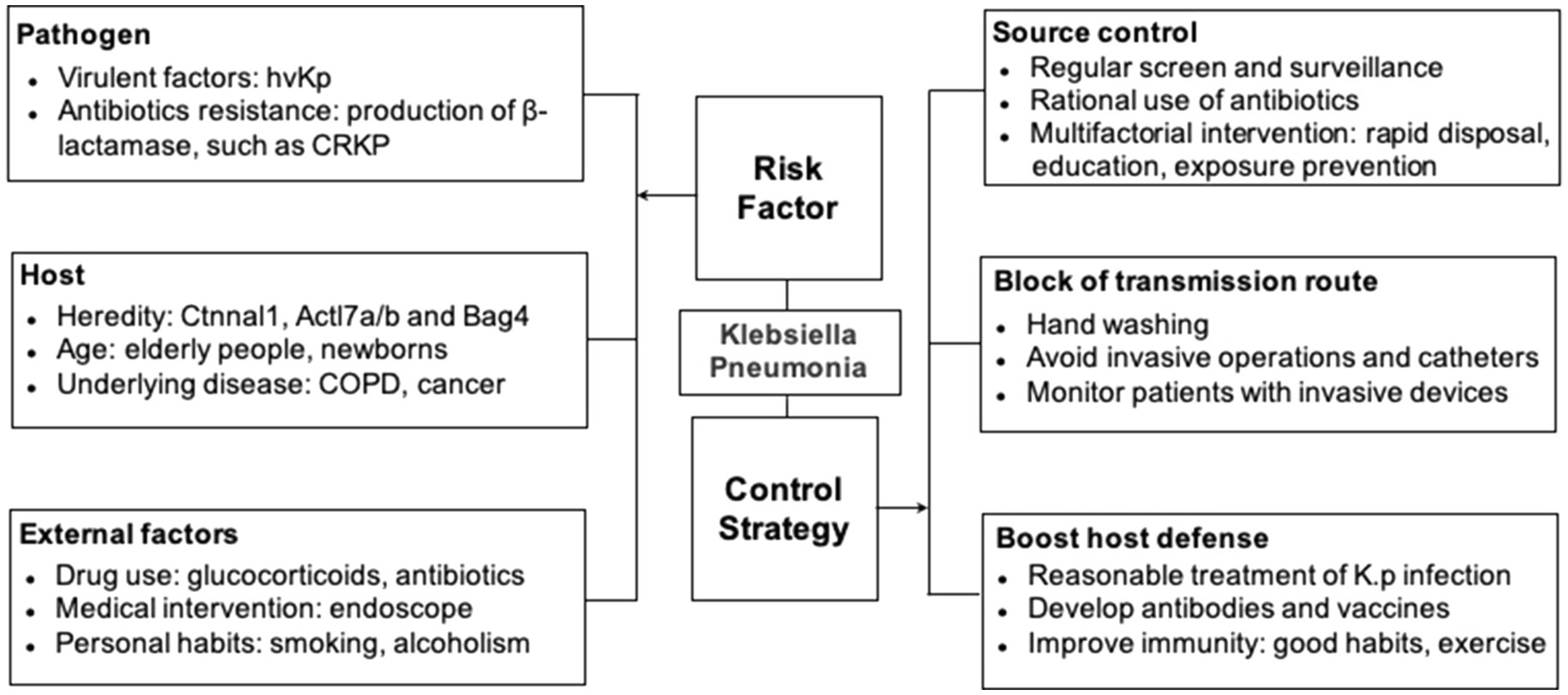
Susceptibility to K. pneumoniae infection is determined by pathogen variables (such as virulence factors and antibiotics resistance), host intrinsic (such as genetics, age, and immune status), and extrinsic factors (such as antibiotic use, environmental exposure, nutrition, and alcoholism), among others (Jong et al., 1995; Paczosa and Mecsas, 2016; Martin and Bachman, 2018; Liu and Guo, 2019).
Various virulence factors have been demonstrated to aid in the infectivity of K. pneumoniae. These virulence factors include capsule, lipopolysaccharide, adhesin, and siderophores, which are more frequent in CRKP/hvKp, leading to various immune responses and related phenotypes observed in the hvKp strains (Paczosa and Mecsas, 2016; Gomez-Simmonds and Uhlemann, 2017; Martin and Bachman, 2018).
The host provides nutrition and shelter to the bacteria. At the same time, the effective immune system controls bacterial replication to prevent infection. Host factors associated with increased host susceptibility include heredity, age, and underlying diseases (Dhainaut et al., 2005; Burgner et al., 2006; Hammer et al., 2018). Vered et al. (2014) studied the host’s susceptibility to K. pneumoniae using quantitative trait locus (QTL) mapping and collaborative cross (CC) mice. They identified host candidate genes for K. pneumoniae infection, including Ctnnal1, Actl7a, Actl7b, and Bag4 (Vered et al., 2014). Newborns, especially those born prematurely or in intensive care units, are at increased risk due to underdeveloped immune systems and immature mucosal barriers of the gastrointestinal tract. On the other end of the spectrum, the elderly subjects have the highest risk of death from K. pneumoniae. It is estimated that a mortality rate of 30% in older subjects following hospitalization due to K. pneumoniae infection is mainly caused by aspiration of oropharyngeal flora (Afroza, 2006; Collado et al., 2015). Studies on patients whose mean ages were more than 60 years showed that 17.2% of all CAPs and 6.5–11.6% of all HAPs were caused by K. pneumoniae (Teramoto et al., 2015). Moreover, diabetes, malignancy, liver and gallbladder disease, chronic obstructive pulmonary disease, renal failure, and nutritional status are additional risk factors, often associated with aging, toward increased susceptibility (Paczosa and Mecsas, 2016).
External factors include antibiotics and glucocorticoids, chemotherapy, transplantation, dialysis, hospital and ICU stays, personal habits, invasive medical procedures such as an endoscope, hypodermic injection, percutaneous surgery, and implantation (Tietjen et al., 2003; Li et al., 2019). Many of these procedures can either contribute to the disruption of the mucosal barrier at the colonization site and allow the pathogen to escape the colonizing site to establish infections, or they directly give the pathogens access to body sites such as intubation.
Control Strategies
Source Control
Identifying and eliminating the source of K. pneumoniae can effectively avoid the infection by these bacteria. However, major challenges remain in the identification and elimination of the source of infection. Specimen culture is still the primary method for most hospitals to screen for presence of K. pneumoniae (Podschun and Ullmann, 1998; Lolans et al., 2010). Some molecular approaches have been reported to identify chromosomal genes such as blaSHV, blaLEN, blaOKP, and their side-chain genes (deoR) by multiplex polymerase chain reaction (Fonseca et al., 2017). It is critical to control the sources of hvKp and CRKP. The Carb NP test and molecular identification have been used to screen Enterobacteriaceae producing carbohydrase, especially for CRKP in asymptomatic carriers (Dortet et al., 2014).
Extensive screening, identification, education, and multifactorial intervention are essential to control the spread of K. pneumoniae from the source (Spagnolo et al., 2014; Figure 1). Exposure prevention should be conducted by timely identifying the infected individuals and following standard precautions including the use of personal protective equipment such as gowns, gloves, and masks. Contact tracing should be employed whenever possible to minimize further exposure to uninfected subjects, both in hospital and community settings. Education on hand hygiene is also vital for medical workers (Akova et al., 2012; Shobowale et al., 2016; Kang et al., 2019).
The use of antibiotics should be regulated strictly under the guidelines and principles, especially for initial empirical treatment (Guven and Uzun, 2003; Kollef and Micek, 2005). The treatment of hvKp and CRKP should follow their specific treatment guidelines (Calfee, 2017). Rational and standardized use of antibiotics, which includes particular indications, adequate dose, sufficient duration of treatment, cautious change of antibiotics, and other interventions such as surgical drainage and removal of implants, whenever possible, should be followed (Weichman et al., 2013). Moreover, abuse of antibiotics should be minimized, which occurs in the medical setting and during livestock breeding (Levy and Marshall, 2004). Controlling antibiotic use in non-human environments can contribute to limiting the source of CRKP.
Prevention of Transmission/Block of the Transmission Route
As mentioned above, hand washing is considered the most critical and practical measure to prevent the spread of pathogens (Pittet, 2001; WHO, 2009). Microbes found on healthcare workers include Klebsiella, Staphylococcus aureus, Clostridium difficile, and other Gram-negative bacteria (Haque et al., 2018). Healthcare workers’ hands can be contaminated by direct contact with patients or touching contaminated surfaces in the hospital setting.
The use of invasive surgeries and indwelling devices, including central venous catheters, endotracheal catheters, should be avoided and limited whenever possible or used for a minimum possible duration (O’Grady et al., 2002; Trautner and Darouiche, 2004).
For patients with indwelling devices, specimens should be screened for the presence of bacterial pathogens from relevant sites, such as skin, urine, sputum, and wound secretions (Baron et al., 2013). The transmission among patients can be blocked by monitoring the compliance of contact precautions and specimen culture results, which should be communicated with the healthcare workers to make appropriate and timely decisions in situations, where unexpected transmission is observed. Contact precautions should be taken for the CRKP/HvKp, especially in a population with a high risk of transmission or those who come in contact with infected individuals (Magiorakos et al., 2017). High-risk patients should be screened at admission and periodically afterward for CRKP/HvKp during their hospital stay. Standard contact precautions should be taken for patients with a low risk of transmission and people who have an epidemiological connection with unrecognized or infected or colonized CRKP/HvKp patients. Pre-emptive contact precautions can be taken, while admission monitoring test results are pending, especially for patients admitted from other hospitals known to have HvKp/CRE (Spagnolo et al., 2014).
Host Defense and Protection of the Susceptible Population
Klebsiella pneumoniae is covered with polysaccharides, including the capsule and lipopolysaccharides, which are ideal candidate antigens that can be targeted by vaccine or host immunity (Podschun and Ullmann, 1998). The antibodies and vaccines against capsular polysaccharides are being developed but facing difficulties due to a total of 77 different capsules and nine different kinds of LPS serotypes present in the K. pneumoniae (Ahmad et al., 2012; Diago-Navarro et al., 2017, 2018; Hegerle et al., 2018). It is crucial to identify highly conserved antigens of K. pneumoniae strains to develop antibodies or vaccines with comprehensive coverage and extensive protection across strains (Lundberg et al., 2013).
Other methods to improve immunity include maintaining a healthy lifestyle, including regular exercise, ensuring enough sleep, smoking cessation, healthy diets, including fruits and vegetables (Monye and Adelowo, 2020). Due to high resistance against a wide range of antibiotics, alternative therapies should be explored. Bacteriophage therapy is a new therapeutic method that can replace or supplement traditional antibiotics (Matsuzaki et al., 2005; Hung et al., 2011; Gu et al., 2012). Phase therapy has demonstrated many advantages, including its specificity, high efficiency, fewer side effects, and lower cost.
The antibiotic treatment regimen against K. pneumoniae is usually determined by bacterial culture and subsequent antibiotic sensitivity tests (Giuliano et al., 2019). For patients with gangrene, abscess, and empyema, surgery or interventional treatment should be performed. For community-acquired pneumonia, the empirical antimicrobial therapy should provide adequate coverage for potential Gram-negative pathogens. Third-generation cephalosporin or quinolones should be used for at least 2 weeks, either alone or with aminoglycosides (Metlay et al., 2019). For patients with hospital-acquired K. pneumonia, appropriate antibiotic regimens should be used either alone or in combination for at least 14 days, including imipenem, third-generation cephalosporin, quinolones, or aminoglycosides (Qureshi, 2015). Quinolones could be administered intravenously if patients respond rapidly. Carbapenems should be considered the treatment of choice against Extended-Spectrum Beta-Lactamase (ESBL)-producer stains, colistin, tigecycline, and intravenous fosfomycin should be chosen for the strains producing carbapenems (Yu and Chuang, 2015).
In summary, K. pneumoniae is an important pathogen for respiratory tract infections, often leading to severe pneumonia and multiorgan infections. It can also cause urinary tract infection, meningitis, sepsis, and biliary tract infection in hospitalized patients, entering the human body through the contaminated respirator, atomizer, or catheters in addition to the self-contamination from the colonized bacteria. In recent years, the emergence of HvKp and CRKP strains has become a major challenge in clinical practice. Further studies on virulence and resistance determinants, genetic lineage information, dissemination mechanisms, effective diagnosis methods, potential antibacterial targets, and prevention measures are needed to help reduce the occurrence and spread of the K. pneumoniae infection and associated morbidity and mortality.
Author Contributions
DC wrote the first draft of the manuscript. LS, CD, and DZ wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by funding from The National Key Research and Development Program of China (SQ2021YFC2300197), Beijing Nova Program Interdisciplinary Cooperation Project (DC; No. Z191100001119021), and the Chinese PLA General Hospital Youth Project (DC; No. QNF19074).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Afroza, S. (2006). Neonatal sepsis--a global problem: an overview. Mymensingh Med. J. 15, 108–114. doi: 10.3329/mmj.v15i1.2
Ahmad, T. A., El-Sayed, L. H., Haroun, M., Hussein, A. A., and El Sayed, H. (2012). Development of immunization trials against Klebsiella pneumoniae. Vaccine 30, 2411–2420. doi: 10.1016/j.vaccine.2011.11.027
Akova, M., Daikos, G., Tzouvelekis, L., and Carmeli, Y. (2012). Interventional strategies and current clinical experience with carbapenemase-producing gram-negative bacteria. Clin. Microbiol. Infect. 18, 439–448. doi: 10.1111/j.1469-0691.2012.03823.x
Bagley, S. T. (1985). Habitat association of Klebsiella species. Infect. Control. 6, 52–58. doi: 10.1017/S0195941700062603
Baron, E. J., Miller, J. M., Weinstein, M. P., Richter, S. S., Gilligan, P. H., Thomson, R. B., et al. (2013). A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the infectious diseases society of America (IDSA) and the American society for microbiology (ASM) a. Clin. Infect. Dis. 57, e22–e121. doi: 10.1093/cid/cit278
Burgner, D., Jamieson, S. E., and Blackwell, J. M. (2006). Genetic susceptibility to infectious diseases: big is beautiful, but will bigger be even better? Lancet Infect. Dis. 6, 653–663. doi: 10.1016/S1473-3099(06)70601-6
Calfee, D. P. (2017). Recent advances in the understanding and management of Klebsiella pneumoniae. F1000Res 6:1760. doi: 10.12688/f1000research.11532.1
Catalán-Nájera, J. C., Garza-Ramos, U., and Barrios-Camacho, H. (2017). Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence 8, 1111–1123. doi: 10.1080/21505594.2017.1317412
Cobo, F. M., Aliaga, L. M., Díaz, F. M., Mediavilla, J. G., and Arrebola, J. N. (1999). Liver abscess caused by Klebsiella pneumoniae in diabetic patients. Rev. Clin. Esp. 199, 517–519.
Collado, M. C., Cernada, M., Neu, J., Pérez-Martínez, G., Gormaz, M., and Vento, M. (2015). Factors influencing gastrointestinal tract and microbiota immune interaction in preterm infants. Pediatr. Res. 77, 726–731. doi: 10.1038/pr.2015.54
Dhainaut, J.-F., Claessens, Y.-E., Janes, J., and Nelson, D. R. (2005). Underlying disorders and their impact on the host response to infection. Clin. Infect. Dis. 41, S481–S489. doi: 10.1086/432001
Diago-Navarro, E., Calatayud-Baselga, I., Sun, D., Khairallah, C., Mann, I., Ulacia-Hernando, A., et al. (2017). Antibody-based immunotherapy to treat and prevent infection with hypervirulent Klebsiella pneumoniae. Clin. Vaccine Immunol. 24, e00456–e00516. doi: 10.1128/CVI.00456-16
Diago-Navarro, E., Motley, M. P., Ruiz-Peréz, G., Yu, W., Austin, J., Seco, B. M., et al. (2018). Novel, broadly reactive anticapsular antibodies against carbapenem-resistant Klebsiella pneumoniae protect from infection. MBio 9, e00091–e00118. doi: 10.1128/mBio.00091-18
Dorman, M. J., and Short, F. L. (2017). Genome watch: Klebsiella pneumoniae: when a colonizer turns bad. Nat. Rev. Microbiol. 15:384. doi: 10.1038/nrmicro.2017.64
Dortet, L., Bréchard, L., Cuzon, G., Poirel, L., and Nordmann, P. (2014). Strategy for rapid detection of carbapenemase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 58, 2441–2445. doi: 10.1128/AAC.01239-13
Espedido, B. A., Steen, J. A., Ziochos, H., Grimmond, S. M., Cooper, M. A., Gosbell, I. B., et al. (2013). Whole genome sequence analysis of the first Australian OXA-48-producing outbreak-associated Klebsiella pneumoniae isolates: the resistome and in vivo evolution. PLoS One 8:e59920. doi: 10.1371/journal.pone.0059920
Fair, R. J., and Tor, Y. (2014). Antibiotics and bacterial resistance in the 21st century. Perspect. Medicin. Chem. 6, 25–64. doi: 10.4137/PMC.S14459
Fang, C.-T., Chuang, Y.-P., Shun, C.-T., Chang, S.-C., and Wang, J.-T. (2004). A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199, 697–705. doi: 10.1084/jem.20030857
Fierer, J., Walls, L., and Chu, P. (2011). Recurring Klebsiella pneumoniae pyogenic liver abscesses in a resident of San Diego, California, due to a K1 strain carrying the virulence plasmid. J. Clin. Microbiol. 49, 4371–4373. doi: 10.1128/JCM.05658-11
Fonseca, E. L., Da Veiga Ramos, N., Andrade, B. G. N., Morais, L. L., Marin, M. F. A., and Vicente, A. C. P. (2017). A one-step multiplex PCR to identify Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae in the clinical routine. Diagn. Microbiol. Infect. Dis. 87, 315–317. doi: 10.1016/j.diagmicrobio.2017.01.005
Fuxench-López, Z., and Ramírez-Ronda, C. H. (1978). Pharyngeal flora in ambulatory alcoholic patients: prevalence of gram-negative bacilli. Arch. Intern. Med. 138, 1815–1816. doi: 10.1001/archinte.1978.03630370033017
Giuliano, C., Patel, C. R., and Kale-Pradhan, P. B. (2019). A guide to bacterial culture identification and results interpretation. Pharmacy Ther. 44, 192–200.
Gomez-Simmonds, A., and Uhlemann, A.-C. (2017). Clinical implications of genomic adaptation and evolution of carbapenem-resistant Klebsiella pneumoniae. J. Infect. Dis. 215, S18–S27. doi: 10.1093/infdis/jiw378
Gorrie, C. L., Mirčeta, M., Wick, R. R., Edwards, D. J., Thomson, N. R., Strugnell, R. A., et al. (2017). Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 65, 208–215. doi: 10.1093/cid/cix270
Gu, J., Liu, X., Li, Y., Han, W., Lei, L., Yang, Y., et al. (2012). A method for generation phage cocktail with great therapeutic potential. PLoS One 7:e31698. doi: 10.1371/journal.pone.0031698
Guven, G., and Uzun, O. (2003). Principles of good use of antibiotics in hospitals. J. Hosp. Infect. 53, 91–96. doi: 10.1053/jhin.2002.1353
Hammer, C. C., Brainard, J., and Hunter, P. R. (2018). Risk factors and risk factor cascades for communicable disease outbreaks in complex humanitarian emergencies: a qualitative systematic review. BMJ Glob. Health 3:e000647. doi: 10.1136/bmjgh-2017-000647
Hamzaoui, Z., Ocampo-Sosa, A., Martinez, M. F., Landolsi, S., Ferjani, S., Maamar, E., et al. (2018). Role of association of OmpK35 and OmpK36 alteration and blaESBL and/or blaAmpC genes in conferring carbapenem resistance among non-carbapenemase-producing Klebsiella pneumoniae. Int. J. Antimicrob. Agents 52, 898–905. doi: 10.1016/j.ijantimicag.2018.03.020
Haque, M., Sartelli, M., Mckimm, J., and Bakar, M. A. (2018). Health care-associated infections–an overview. Infect. Drug Resist. 11, 2321–2333. doi: 10.2147/IDR.S177247
Hegerle, N., Choi, M., Sinclair, J., Amin, M. N., Ollivault-Shiflett, M., Curtis, B., et al. (2018). Development of a broad spectrum glycoconjugate vaccine to prevent wound and disseminated infections with Klebsiella pneumoniae and Pseudomonas aeruginosa. PLoS One 13:e0203143. doi: 10.1371/journal.pone.0203143
Hung, C.-H., Kuo, C.-F., Wang, C.-H., Wu, C.-M., and Tsao, N. (2011). Experimental phage therapy in treating Klebsiella pneumoniae-mediated liver abscesses and bacteremia in mice. Antimicrob. Agents Chemother. 55, 1358–1365. doi: 10.1128/AAC.01123-10
Jong, G.-M., Hsiue, T.-R., Chen, C.-R., Chang, H.-Y., and Chen, C.-W. (1995). Rapidly fatal outcome of bacteremic Klebsiella pneumoniae pneumonia in alcoholics. Chest 107, 214–217. doi: 10.1378/chest.107.1.214
Kang, J. S., Yi, J., Ko, M. K., Lee, S. O., Lee, J. E., and Kim, K.-H. (2019). Prevalence and risk factors of carbapenem-resistant Enterobacteriaceae acquisition in an emergency intensive care unit in a tertiary hospital in Korea: a case-control study. J. Korean Med. Sci. 34:e140. doi: 10.3346/jkms.2019.34.e140
Kloos, W. E., and Musselwhite, M. S. (1975). Distribution and persistence of staphylococcus and micrococcus species and other aerobic bacteria on human skin. Appl. Microbiol. 30, 381–395. doi: 10.1128/am.30.3.381-395.1975
Köhler, W., and Mochmann, H. (1987). Carl Friedländer (1847–1887) and the discovery of the Pneumococcus–in memory of the centenary of his death. Z. Arztl. Fortbild. 81, 615–618.
Kollef, M. H., and Micek, S. T. (2005). Strategies to prevent antimicrobial resistance in the intensive care unit. Crit. Care Med. 33, 1845–1853. doi: 10.1097/01.CCM.0000171849.04952.79
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. doi: 10.1016/S1473-3099(10)70143-2
Lee, K. H., Hui, K. P., Tan, W. C., and Lim, T. K. (1994). Klebsiella bacteraemia: a report of 101 cases from national university hospital, Singapore. J. Hosp. Infect. 27, 299–305. doi: 10.1016/0195-6701(94)90117-1
Levy, S. B., and Marshall, B. (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10, S122–S129. doi: 10.1038/nm1145
Li, S., Guo, F.-Z., Zhao, X.-J., Wang, Q., Wang, H., An, Y.-Z., et al. (2019). Impact of individualized active surveillance of carbapenem-resistant Enterobacteriaceae on the infection rate in intensive care units: a 3-year retrospective study in a teaching hospital of people’s republic of China. Infect. Drug Resist. 12, 1407–1414. doi: 10.2147/IDR.S201644
Li, B., and Webster, T. J. (2018). Bacteria antibiotic resistance: new challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 36, 22–32. doi: 10.1002/jor.23656
Lin, Y. T., Siu, L. K., Lin, J. C., Chen, T. L., Tseng, C. P., Yeh, K. M., et al. (2012). Seroepidemiology of Klebsiella pneumoniae colonizing the intestinal tract of healthy Chinese and overseas Chinese adults in Asian countries. BMC Microbiol. 12:13. doi: 10.1186/1471-2180-12-13
Lindstrom, S., Healey, P., and Chen, S. (1997). Metastatic septic endophthalmitis complicating pyogenic liver abscess caused by Klebsiella pneumoniae. Aust. NZ J. Med. 27, 77–78. doi: 10.1111/j.1445-5994.1997.tb00920.x
Liu, Y.-C., Cheng, D.-L., and Lin, C.-L. (1986). Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 146, 1913–1916. doi: 10.1001/archinte.1986.00360220057011
Liu, C., and Guo, J. (2019). Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann. Clin. Microbiol. Antimicrob. 18:4. doi: 10.1186/s12941-018-0302-9
Logan, L. K., and Weinstein, R. A. (2017). The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215, S28–S36. doi: 10.1093/infdis/jiw282
Lolans, K., Calvert, K., Won, S., Clark, J., and Hayden, M. K. (2010). Direct ertapenem disk screening method for identification of KPC-producing Klebsiella pneumoniae and Escherichia coli in surveillance swab specimens. J. Clin. Microbiol. 48, 836–841. doi: 10.1128/JCM.01988-09
Lundberg, U., Senn, B. M., Schüler, W., Meinke, A., and Hanner, M. (2013). Identification and characterization of antigens as vaccine candidates against Klebsiella pneumoniae. Hum. Vaccin. Immunother. 9, 497–505. doi: 10.4161/hv.23225
Magiorakos, A., Burns, K., Baño, J. R., Borg, M., Daikos, G., Dumpis, U., et al. (2017). Infection prevention and control measures and tools for the prevention of entry of carbapenem-resistant Enterobacteriaceae into healthcare settings: guidance from the European centre for disease prevention and control. Antimicrob. Resist. Infect. Control 6:113. doi: 10.1186/s13756-017-0259-z
Martin, R. M., and Bachman, M. A. (2018). Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 8:4. doi: 10.3389/fcimb.2018.00004
Martin, R. M., Cao, J., Brisse, S., Passet, V., Wu, W., Zhao, L., et al. (2016). Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere 1, e00261–e00316. doi: 10.1128/mSphere.00261-16
Matsuzaki, S., Rashel, M., Uchiyama, J., Sakurai, S., Ujihara, T., Kuroda, M., et al. (2005). Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J. Infect. Chemother. 11, 211–219. doi: 10.1007/s10156-005-0408-9
Mccabe, R., Lambert, L., and Frazee, B. (2010). Invasive Klebsiella pneumoniae infections, California, USA. Emerg. Infect. Dis. 16, 1490–1491. doi: 10.3201/eid1609.100386
Meatherall, B. L., Gregson, D., Ross, T., Pitout, J. D., and Laupland, K. B. (2009). Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am. J. Med. 122, 866–873. doi: 10.1016/j.amjmed.2009.03.034
Meents, E. F., and Boyles, T. (2010). Schistosoma haematobium prevalence in school children in the rural eastern cape Province, South Africa. South. Afr. J. Epidemiol. Infect. 25, 28–29. doi: 10.1080/10158782.2010.11441409
Metlay, J. P., Waterer, G. W., Long, A. C., Anzueto, A., Brozek, J., Crothers, K., et al. (2019). Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American thoracic society and infectious diseases society of America. Am. J. Respir. Crit. Care Med. 200, e45–e67. doi: 10.1164/rccm.201908-1581ST
Moellering, R. C. (2010). NDM-1—a cause for worldwide concern. N. Engl. J. Med. 363, 2377–2379. doi: 10.1056/NEJMp1011715
Monye, I., and Adelowo, A. B. (2020). Strengthening immunity through healthy lifestyle practices: recommendations for lifestyle interventions in the management of COVID-19. Lifesty. Med. 1:e7. doi: 10.1002/lim2.7
Naas, T., Nordmann, P., Vedel, G., and Poyart, C. (2005). Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 49, 4423–4424. doi: 10.1128/AAC.49.10.4423-4424.2005
Nassif, X., Fournier, J., Arondel, J., and Sansonetti, P. (1989). Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect. Immun. 57, 546–552. doi: 10.1128/iai.57.2.546-552.1989
Nassif, X., and Sansonetti, P. J. (1986). Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 54, 603–608. doi: 10.1128/iai.54.3.603-608.1986
Nassrf, X., Honore, N., Vasselon, T., Cole, S., and Sansonetti, P. (1989). Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Kiebsiella pneumoniae. Mol. Microbiol. 3, 1349–1359. doi: 10.1111/j.1365-2958.1989.tb00116.x
Nordmann, P., Naas, T., and Poirel, L. (2011). Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17, 1791–1798. doi: 10.3201/eid1710.110655
O'Grady, N. P., Alexander, M., Dellinger, E. P., Gerberding, J. L., Heard, S. O., Maki, D. G., et al. (2002). Guidelines for the prevention of intravascular catheter-related infections. Centers for disease control and prevention. MMWR Recomm. Rep. 51, 1–29.
Paczosa, M. K., and Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/MMBR.00078-15
Pastagia, M., and Arumugam, V. (2008). Klebsiella pneumoniae liver abscesses in a public hospital in Queens, New York. Travel Med. Infect. Dis. 6, 228–233. doi: 10.1016/j.tmaid.2008.02.005
Pfeifer, Y., Schlatterer, K., Engelmann, E., Schiller, R. A., Frangenberg, H. R., Stiewe, D., et al. (2012). Emergence of OXA-48-type carbapenemase-producing Enterobacteriaceae in German hospitals. Antimicrob. Agents Chemother. 56, 2125–2128. doi: 10.1128/AAC.05315-11
Pittet, D. (2001). Improving adherence to hand hygiene practice: a multidisciplinary approach. Emerg. Infect. Dis. 7, 234–240. doi: 10.3201/eid0702.010217
Podschun, R., Pietsch, S., Höller, C., and Ullmann, U. (2001). Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl. Environ. Microbiol. 67, 3325–3327. doi: 10.1128/AEM.67.7.3325-3327.2001
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/CMR.11.4.589
Poirel, L., Héritier, C., Tolün, V., and Nordmann, P. (2004). Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48, 15–22. doi: 10.1128/AAC.48.1.15-22.2004
Pollack, M., Charache, P., Nieman, R. E., Jett, M. P., Reimhardt, J. A., and Hardy, P. H. (1972). Factors influencing colonisation and antibiotic-resistance patterns of gram-negative bacteria in hospital patients. Lancet 2, 668–671. doi: 10.1016/S0140-6736(72)92084-3
Qureshi, S. (2015). Klebsiella Infections Treatment & Management. Retrieved November 29, 2015.
Ray, C. G., and Ryan, K. J. (2004). Sherris Medical Microbiology: An Introduction to Infectious Diseases. NY: McGraw-Hill.
Russo, T. A., and Marr, C. M. (2019). Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32, e00001–e00119. doi: 10.1128/CMR.00001-19
Russo, T. A., Olson, R., Fang, C.-T., Stoesser, N., Miller, M., Macdonald, U., et al. (2018). Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J. Clin. Microbiol. 56, e00776–e00818. doi: 10.1128/JCM.00776-18
Saccente, M. (1999). Klebsiella pneumoniae liver abscess, endophthalmitis, and meningitis in a man with newly recognized diabetes mellitus. Clin. Infect. Dis. 29, 1570–1571. doi: 10.1086/313539
Selden, R., Lee, S., Wang, W. L., Bennett, J. V., and Eickhoff, T. C. (1971). Nosocomial Klebsiella infections: intestinal colonization as a reservoir. Ann. Intern. Med. 74, 657–664. doi: 10.7326/0003-4819-74-5-657
Shobowale, E. O., Adegunle, B., and Onyedibe, K. (2016). An assessment of hand hygiene practices of healthcare workers of a semi-urban teaching hospital using the five moments of hand hygiene. Niger. Med. J. 57, 150–154. doi: 10.4103/0300-1652.184058
Spagnolo, A. M., Orlando, P., Panatto, D., Perdelli, F., and Cristina, M. L. (2014). An overview of carbapenem-resistant Klebsiella pneumoniae: epidemiology and control measures. Rev. Med. Microbiol. 25, 7–14. doi: 10.1097/MRM.0b013e328365c51e
Struve, C., Roe, C. C., Stegger, M., Stahlhut, S. G., Hansen, D. S., Engelthaler, D. M., et al. (2015). Mapping the evolution of hypervirulent Klebsiella pneumoniae. MBio 6:e00630. doi: 10.1128/mBio.00630-15
Teramoto, S., Yoshida, K., and Hizawa, N. (2015). Update on the pathogenesis and management of pneumonia in the elderly-roles of aspiration pneumonia. Respir. Investig. 53, 178–184. doi: 10.1016/j.resinv.2015.01.003
Thomson, J. M., and Bonomo, R. A. (2005). The threat of antibiotic resistance in gram-negative pathogenic bacteria: β-lactams in peril! Curr. Opin. Microbiol. 8, 518–524. doi: 10.1016/j.mib.2005.08.014
Tietjen, L., Bossemeyer, D., and Mcintosh, N. (2003). Infection Prevention: Guidelines for Healthcare Facilities with Limited Resources. Jhpiego Corporation.
Trautner, B. W., and Darouiche, R. O. (2004). Catheter-associated infections: pathogenesis affects prevention. Arch. Intern. Med. 164, 842–850. doi: 10.1001/archinte.164.8.842
Tsai, S. S., Huang, J. C., Chen, S. T., Sun, J. H., Wang, C. C., Lin, S. F., et al. (2010). Characteristics of Klebsiella pneumoniae bacteremia in community-acquired and nosocomial infections in diabetic patients. Chang Gung Med. J. 33, 532–539.
Tsay, R. W., Siu, L. K., Fung, C. P., and Chang, F. Y. (2002). Characteristics of bacteremia between community-acquired and nosocomial Klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. Arch. Intern. Med. 162, 1021–1027. doi: 10.1001/archinte.162.9.1021
Van Duin, D., and Doi, Y. (2017). The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8, 460–469. doi: 10.1080/21505594.2016.1222343
Vered, K., Durrant, C., Mott, R., and Iraqi, F. A. (2014). Susceptibility to Klebsiella pneumonaie infection in collaborative cross mice is a complex trait controlled by at least three loci acting at different time points. BMC Genomics 15:865. doi: 10.1186/1471-2164-15-865
Vila, A., Cassata, A., Pagella, H., Amadio, C., Yeh, K. M., Chang, F. Y., et al. (2011). Appearance of Klebsiella pneumoniae liver abscess syndrome in Argentina: case report and review of molecular mechanisms of pathogenesis. Open Microbiol. J. 5, 107–113. doi: 10.2174/1874285801105010107
Virgincar, N., Iyer, S., Stacey, A., Maharjan, S., Pike, R., Perry, C., et al. (2011). Klebsiella pneumoniae producing KPC carbapenemase in a district general hospital in the UK. J. Hosp. Infect. 78, 293–296. doi: 10.1016/j.jhin.2011.03.016
Weichman, K. E., Levine, S. M., Wilson, S. C., Choi, M., and Karp, N. S. (2013). Antibiotic selection for the treatment of infectious complications of implant-based breast reconstruction. Ann. Plast. Surg. 71, 140–143. doi: 10.1097/SAP.0b013e3182590924
WHO (2009). “WHO Guidelines on Hand Hygiene in Health Care: World Alliance for Patient Safety ”. Geneva: WHO Press.
Yao, H., Qin, S., Chen, S., Shen, J., and Du, X.-D. (2018). Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis. 18:25. doi: 10.1016/S1473-3099(17)30628-X
Yigit, H., Queenan, A. M., Anderson, G. J., Domenech-Sanchez, A., Biddle, J. W., Steward, C. D., et al. (2001). Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45, 1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Yu, W.-L., and Chuang, Y.-C. (2015). Clinical features, diagnosis, and treatment of Klebsiella pneumoniae infection. UpToDate.
Keywords: Klebsiella pneumoniae, clinical epidemiology, risk factors, control strategies, hvKp, CRKP
Citation: Chang D, Sharma L, Dela Cruz CS and Zhang D (2021) Clinical Epidemiology, Risk Factors, and Control Strategies of Klebsiella pneumoniae Infection. Front. Microbiol. 12:750662. doi: 10.3389/fmicb.2021.750662
Edited by:
David Andes, University of Wisconsin-Madison, United StatesReviewed by:
Aleksandr G. Bulaev, Federal Center Research Fundamentals of Biotechnology, Russian Academy of Sciences (RAS), RussiaVictor H. Bustamante, National Autonomous University of Mexico, Mexico
Copyright © 2021 Chang, Sharma, Dela Cruz and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: De Chang, Y2hhbmdkZTU1MDFAZm94bWFpbC5jb20=; Lokesh Sharma, bG9rZXNoa3VtYXIuc2hhcm1hQHlhbGUuZWR1; Charles S. Dela Cruz, Y2hhcmxlcy5kZWxhY3J1ekB5YWxlLmVkdQ==; Dong Zhang, emhhbmdkb25nNjc2N0BvdXRsb29rLmNvbQ==
 De Chang
De Chang Lokesh Sharma
Lokesh Sharma Charles S. Dela Cruz
Charles S. Dela Cruz Dong Zhang4,5*
Dong Zhang4,5*