- 1Key Lab of Food Quality and Safety of Jiangsu Province—State Key Laboratory Breeding Base, Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, Nanjing, China
- 2Fungal Genomics Laboratory (FungiG), Jiangsu Provincial Key Lab of Organic Solid Waste Utilization, Nanjing Agricultural University, Nanjing, China
- 3College of Food and Bioengineering, Henan University of Science and Technology, Luoyang, China
- 4Jinan Academy of Agricultural Sciences, Jinan, China
- 5Genome Evolution Laboratory, Maynooth University, Maynooth, Ireland
- 6School of Environment and Safety Engineering, Jiangsu University, Zhenjiang, China
Biological control is a promising approach to suppress diseases caused by Pythium spp. such as Pythium soft rot of ginger caused by P. myriotylum. Unusually for a single genus, it also includes species that can antagonize Pythium plant pathogens, such as Pythium oligandrum. We investigated if a new isolate of P. oligandrum could antagonize P. myriotylum, what changes occurred in gene expression when P. oligandrum (antagonist) and P. myriotylum (host) interacted, and whether P. oligandrum could control soft-rot of ginger caused by P. myriotylum. An isolate of P. oligandrum, GAQ1, recovered from soil could antagonize P. myriotylum in a plate-based confrontation assay whereby P. myriotylum became non-viable. The loss of viability of P. myriotylum coupled with how P. oligandrum hyphae could coil around and penetrate the hyphae of P. myriotylum, indicated a predatory interaction. We investigated the transcriptional responses of P. myriotylum and P. oligandrum using dual-RNAseq at a stage in the confrontation where similar levels of total transcripts were measured from each species. As part of the transcriptional response of P. myriotylum to the presence of P. oligandrum, genes including a subset of putative Kazal-type protease inhibitors were strongly upregulated along with cellulases, elicitin-like proteins and genes involved in the repair of DNA double-strand breaks. In P. oligandrum, proteases, cellulases, and peroxidases featured prominently in the upregulated genes. The upregulation along with constitutive expression of P. oligandrum proteases appeared to be responded to by the upregulation of putative protease inhibitors from P. myriotylum, suggesting a P. myriotylum defensive strategy. Notwithstanding this P. myriotylum defensive strategy, P. oligandrum had a strong disease control effect on soft-rot of ginger caused by P. myriotylum. The newly isolated strain of P. oligandrum is a promising biocontrol agent for suppressing the soft-rot of ginger. The dual-RNAseq approach highlights responses of P. myriotylum that suggests features of a defensive strategy, and are perhaps another factor that may contribute to the variable success and durability of biological attempts to control diseases caused by Pythium spp.
Introduction
Biological control is a promising strategy to suppress diseases in an environmentally friendly manner whereby microorganisms, the biocontrol agents (BCAs), can antagonize a plant pathogen and prime plant defenses (Köhl et al., 2019). The application of biocontrol strategies in the field is often hampered by a lack of robustness where the previously observed disease protection effect is not maintained. One area needing further investigation is molecular aspects of the interaction between a plant pathogen and a biocontrol agent or in terms appropriate to their ecological interactions—the host or prey and the microbial antagonist.
One prominent biocontrol species is the oomycete Pythium oligandrum, which has several known mechanisms of antagonism contributing to its ability to control plant diseases as well as plant-mediated mechanisms. Recently, the review of Thambugala et al. (2020) summarized plant diseases caused by fungi or oomycetes that could be controlled by P. oligandrum. In particular, with regard to diseases caused by oomycetes, most of the diseases where P. oligandrum has a control effect are caused by P. ultimum, which causes diseases on a broad range of crop species (Thambugala et al., 2020). E.g., P. oligandrum has been reported to control pre- and post-emergence damping-off of wheat caused by P. ultimum (Abdelzaher et al., 1997). To our knowledge, there are no reports in the literature of antagonism of P. oligandrum toward P. myriotylum or to the control of diseases caused by P. myriotylum. An unpublished report showed that an isolate of P. oligandrum was antagonistic toward P. myriotylum in plate confrontation assays, but there was no control effect on Pythium soft-rot (PSR) of ginger caused by Pythium myriotylum (Le, 2016).
P. myriotylum is a broad host range pathogen that can infect a range of monocot and dicot plant species (van der Plaats-Niterink, 1981). One of these monocot hosts is ginger (Zingiber officinale) and recently, it was shown that P. myriotylum can be the most frequently found oomycete species from infected ginger rhizomes showing symptoms of PSR in the major ginger growing regions in China (Daly et al., 2021). Ginger is a spice and an important crop (De-Guzman and Siemonsma, 1999), with global production in 2019 estimated at approximately four million metric tons (FAOSTAT, 2019). Control strategies for PSR of ginger were comprehensively reviewed by Le et al. (2014) and are primarily based on chemical pesticide treatments. There is also potential for biological control methods such as a report of using a hypocrealean fungus Trichoderma harzianum to control PSR of ginger (Singh, 2011), but as reviewed by Le et al. (2014), the efficacies of the biological treatments appear lower in field conditions. In light of this, there is a need to identify novel biological control agents to control PSR of ginger.
Recently, the genome of the P. oligandrum ATCC 38472 strain was sequenced by long-read sequencing technology (Faure et al., 2020), and previously the P. oligandrum strains Po37 (Berger et al., 2016) and CBS 530.74 (Kushwaha et al., 2017) were sequenced by short-read sequencing technology. Note that the ATCC 38472 strain of P. oligandrum is used in the commercial oospore-containing product Polyversum®. In a comparative genomics analysis of P. oligandrum with other oomycetes, there was an expansion of carbohydrate-active enzyme (CAZy) families (Lombard et al., 2014) involved in the degradation of cellulose (Liang et al., 2020), perhaps related to the ability of P. oligandrum to degrade the cellulose in the cell wall of an oomycete host or prey. P. oligandrum is often referred to as a mycoparasite as its antagonism includes the coiling of P. oligandrum hyphae around hyphae of its host (Berry et al., 1993). P. oligandrum possesses an arsenal of hydrolytic enzymes such as cellulases and proteases that are important for antagonism toward particular hosts. The CAZyme content of P. oligandrum was described recently by Liang et al. (2020). The production of antimicrobial compounds by P. oligandrum also contributes to the antagonism (Benhamou et al., 1999). Interestingly, P. oligandrum can enter and colonize the root tissue before it rapidly degrades or degenerates itself (Benhamou et al., 2012). Presumably, this rapid degeneration of P. oligandrum can limit the damage that its cellulases can cause to the plant cell wall, of which cellulose is a major component. P. oligandrum appears to have a priming role in stimulating plant defense responses by microbe-associated molecular pattern (MAMP) production such as the elicitin-like protein oligandrin (Benhamou et al., 2001). Recently, type II NLP toxin proteins from P. oligandrum were found to suppress Phytophthora capsici infection (Yang et al., 2021). Expression of putative effectors was found when P. oligandrum interacted with heat-killed Phytophthora infestans mycelia (Horner et al., 2012), and these could potentially antagonize the host or mediate plant defense responses.
Dual transcriptomics simultaneously analyses the transcriptomes of two or more interacting organisms and is frequently used in host-pathogen interaction (Westermann et al., 2017). This can uncover expression differences in interacting organisms such as the upregulation of genes related to antagonism when three saprobic ascomycete fungi were cultured together (Daly et al., 2017). Dual-RNAseq was used to uncover the transcriptional responses of the biocontrol rhizobacterium Lysobacter capsici and the pathogen Phytophthora infestans (Tomada et al., 2017). In this interaction, L. capsici genes involved in host colonization and degradation, and detoxification were upregulated while in contrast the host P. infestans was not considered to have mounted a major defensive or counter-attacking response but a transcriptomic response perhaps related to cell death (Tomada et al., 2017).
Here we investigated if a new isolate of P. oligandrum could antagonize P. myriotylum and used dual-RNAseq to measure the changes in gene expression that occurred when the two species confronted each other, alongside investigating if P. oligandrum had a control effect on the disease caused by P. myriotylum.
Materials and Methods
Pythium Strains and Routine Culturing
P. oligandrum GAQ1 CGMCC No. 17470 (Wei et al., 2019) was isolated from soil from a field where infected ginger was growing in Laiwu district, Jinan City, Shandong Province, China. The molecular marker sequences of the GAQ1 isolate (MK774755.1, MZ891585.1, MZ891586.1 and MZ869812.1) have been deposited in GenBank. The P. myriotylum SWQ7 CGMCC No. 21459 isolate was described previously by Daly et al. (2021). The Pythium species were routinely cultured on 10% V8 juice agar at 25°C and maintained on V8 juice agar slants at 12°C.
Identification of P. oligandrum GAQ1
The CTAB protocol of Doyle (1990) was used to extract DNA from mycelial mats growing on V8 juice agar. The ITS region including the 5.8S rRNA subunit was amplified using the two universal primers ITS1 and ITS4 (White et al., 1990). The CoxI gene was amplified using OomCoxILevup and OomCoxI-Levlo primers (Robideau et al., 2011), the CoxII gene using FM66 and F58 primers (Martin and Tooley, 2003), and the β-tubulin gene using TUBUF2 and TUBUR1 primers (Kroon et al., 2004). PCR reactions were performed using a proof-reading polymerase with standard reaction and cycling conditions. All PCR products were run on an agarose gel to confirm the presence of an amplified product. The sequencing of the PCR products, BLAST searches and phylogenetic analysis were performed as described previously (Daly et al., 2021).
Visual Analysis of P. oligandrum and P. myriotylum Interaction
From fresh pre-culture plates on V8 juice media, an agar plug from the hyphal front was used to inoculate onto opposite ends of V8 agar on 9 cm plates and incubated at 25°C. For the macroscopic observation of the colonies, 20 mL of V8 agar was added to the plate, and for the microscopic visualization, 4.5 ml of V8 agar was added to the plates to provide a thinner layer. 5 mL of a 0.01% w/v trypan blue solution was used to cover the mycelium and agar surface. Trypan blue is a type of vitality stain, and it stains non-viable tissues with a blue color. The plates were gently rotated to ensure that the dye and mycelium were in full contact, and then incubated without shaking at room temperature for 3 min. Then the staining solution was decanted, and any remaining dye was pipetted off. The stained mycelial cultures were washed twice with 5 ml of water by incubating for 5 min without shaking. The staining was done after contact of the hyphal fronts and at various stages afterward. Microscopic visualization was performed using a brightfield microscope (NE900, Nexcope). At least three replicate cultures were used for the visualization work. A cryo-scanning electron microscope (cryo-SEM) (Quorum PP3010T system integrated onto a Hitachi SU8010 FE-SEM) was also used to visualize the interaction between P. oligandrum and P. myriotylum.
Dual-RNAseq Transcriptomics Experiment
Pre-cultures were grown on 10% V8 juice agar at 25°C. From the edge of the colony of P. myriotylum SWQ7 or P. oligandrum GAQ1, 0.5 cm mycelial plugs were transferred to 10% V8 juice agar covered with a 14.2 cm diameter polycarbonate membrane with a 0.1 μm pore size (GVS filter technology, Cat. no. 1215304) in a 14 cm Petri dish. The mycelial plugs were placed equidistant from each other and the edge of the Petri dish. The Petri dishes were inoculated with either two plugs of the same species (self-confrontation) or one plug from each species (mixed species confrontation). The cultures were incubated at 25°C in the dark, and mycelia from the confrontation zone was sampled as shown by the dotted-lines (Supplementary Figure 1). The mycelium was flash-frozen in liquid nitrogen before being ground using a Tissue Lyser. The sample was mixed with 1 mL of Trizol before adding 200 μl chloroform: isoamyl alcohol (24:1). The RNA was precipitated by mixing 0.8 volume of isopropanol with the aqueous phase from the previous step and the resulting pellet was washed once with 70% EtOH before resuspension in DEPC-treated water. The RNA was then cleaned up using a Tiangen Plant RNA extraction kit which included an on-column DNase treatment. RNA integrity was visualized using an agarose gel and a Bioanalyzer, and RNA quantity and purity using a NanoDrop spectrophotometer.
RNA Sequencing and Data Analysis
A paired library 2 × 150 bp was prepared using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) and sequenced using the MGISEQ-2000 platform (MGI Tech Co. Ltd). The raw reads were processed using Trimmomatic (Bolger et al., 2014). The reads containing poly-N and the low-quality reads were removed to obtain the clean reads. Downstream analysis of the cleaned reads was performed using the following software tools in UseGalaxy.eu (Afgan et al., 2018). For all samples, reads that passed the filters were mapped using HISAT2 Galaxy Version 2.1.0 (Kim et al., 2019) onto a composite reference genome that consisted of the genomes of both species. The composite genome was made by concatenating into a single file those Fasta files made of the sequences for the scaffolds from P. oligandrum ATCC 38472 GCA_005966545.1_ASM596654v1_genomic.fasta (Faure et al., 2020) and P. myriotylum SWQ7 JAAS_PmSWQ7_1_0_assembly.fsa (PRJNA692555) (manuscript in preparation). The use of this composite genome facilitated the exclusion from subsequent analysis of reads that were not unique to a particular species as these reads would not be uniquely mapped in the composite genome. Uniquely mapped read counts (at the MAPQ10 threshold) for each gene were calculated using HTSeq-count Galaxy version 0.9.11 (Anders et al., 2015). Read counts were calculated for each species separately using a genome annotation file for each species that contained the known gene exon coordinates for the genes for that species. The genome annotation file used for P. oligandrum was GCA_005966545.1_ASM596654v1_genomic.gff (Faure et al., 2020). Principal component analysis (PCA) indicated if the biological replicates were sufficiently similar for subsequent statistical analysis. The read counts of genes values for each species were subsequently used for statistical analysis using DESeq2 Galaxy Version 2.11.40.6 (Love et al., 2014). Note that the Cook’s distance cut-off was not used for outlier filtering. The criteria for a differentially expressed (DE) gene were an FPKM > 10 in one condition, DESeq padj value < 0.05 and a DESeq log2 FC > 1 or < −1. FPKM normalized read counts were defined originally by Mortazavi et al. (2008). In our study, an FPKM value for a gene is the number of uniquely mapped (to the composite genome) fragments per kilobase of gene model per million uniquely mapped (to the composite genome) fragments onto gene models from a species. The RNA-seq reads from this project were submitted to the GEO database (GEO accession GSE179387).
Gene Annotations of P. myriotylum and P. oligandrum Genes
For the P. oligandrum genes, the annotations of Faure et al. (2020) for the ATCC 38472 strain were used. The P. myriotylum genes were functionally annotated primarily using InterProScan (Jones et al., 2014), and the CAZymes were annotated using dbCAN2 (Zhang et al., 2018; manuscript in preparation). The annotations for the P. oligandrum and P. myriotylum genes are listed in Supplementary Table 1.
qPCR Validation of Expression Differences in Dual-RNAseq
The qPCR validation was performed using RNA samples extracted from triplicate repeat cultures using the same experimental conditions and extraction method as described for the RNAseq experiment. cDNA was synthesized using the EasyScript one step gDNA removal and cDNA synthesis kit (TransGen Biotech) using random primers and 2.5 μg of total RNA according to the manufacturer’s instructions. The qPCR was performed with a LightCycler 96 Instrument (Roche Life Sciences) using the ChamQ SYBR qPCR Master Mix (VAZyme). The cycling conditions were 95°C for 3 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. A melt curve analysis step was included to confirm the specificity of the primer pairs used. A standard curve of fivefold serial dilutions of the gDNA from P. myriotylum SWQ7 and P. oligandrum GAQ1 was used to determine the primer efficiency. The data was analyzed using the LightCycler 96 software. To quantify the abundance of the target in the cDNA samples, the relative standard curve method was used, and the expression levels of the gene of interest were normalized to two reference genes. A tubulin gene (Pm_g6466.t1) and a gapdh gene (Pm_17033.t1) were used for P. myriotylum, and a succinate dehydrogenase gene (Po_g2472.t1) and a g6pdh gene (Po_g6912.t1) were used for P. oligandrum. A pair of primers that amplified from gDNA was used to confirm the efficiency of the gDNA removal treatments for the gDNA from each species. Primers were designed using Primer-BLAST software (Ye et al., 2012) at NCBI and the primers used are listed in Supplementary Table 2. The primer efficiency for the primer pairs used was at least 86% and generally > 90%, as detailed in Supplementary Table 2. Melt curve and agarose gel analysis were used to confirm the specificity of the primer pairs along with sequencing of the PCR products. A confirmation that the primers designed for P. myriotylum were unable to amplify from P. oligandrum and vice-versa was also performed.
Ginger Pot-Trial With P. myriotylum and P. oligandrum to Determine Biocontrol Effect
P. myriotylum was cultured on 10% V8 juice agar for 1–2 day, and then twice-autoclaved wheat seeds (which had previously been soaked overnight in water) were plated onto the mycelial mat of P. myriotylum and incubated for a further 7 d or until the seeds were fully covered with mycelia. The wheat seeds from the P. myriotylum cultures (after scraping away the excess mycelium on the outside of the seed) were used to inoculate the vermiculite near the roots at a depth of 2–3 cm. P. oligandrum was inoculated using four mycelia-covered balls from liquid culture. The use of the wheat seed inoculum method for P. myriotylum was based on the sorghum seed inoculum method of Le et al. (2016). To produce the P. oligandrum inoculum, P. oligandrum was incubated on 10% V8 juice agar for 1 day at 25°C, and then 20 agar plugs of a diameter of 0.5 cm were used to inoculate 50 mL of 10% V8 juice medium in a 150 mL flask, then incubated for 3 d at 25°C and 100 rpm. A mycelium-based inoculum was used for P. oligandrum because oospore production has not been optimized to achieve a yield that can practically be used as an inoculum. Ginger plants (“Laiwu” variety) were derived from tissue culture and transplanted to autoclaved vermiculite in 100 mL pots with an inverted Petri dish lid placed underneath each pot (to hold the water and prevent cross-contamination of the pathogen), and grown in 16 h light and 8 h dark cycles at 25°C in a growth chamber. Periodically, the ginger plants were given a water-soluble fertilizer (NPK 20-20-20 + TE). Eighteen ginger plants (grown in individual pots) were used for each of the six test or control conditions (Table 1). After 7 days from deflasking of the ginger seedlings, the experiment was initiated by inoculating three conditions with P. oligandrum, and two of these P. oligandrum-inoculated conditions were later inoculated with P. myriotylum at either 7 or 14 days while the third P. oligandrum-inoculated condition was not inoculated with P. myriotylum. Two of the other conditions were inoculated only with P. myriotylum at either 7 or 14 days, and the last condition was not inoculated with either of the two species.
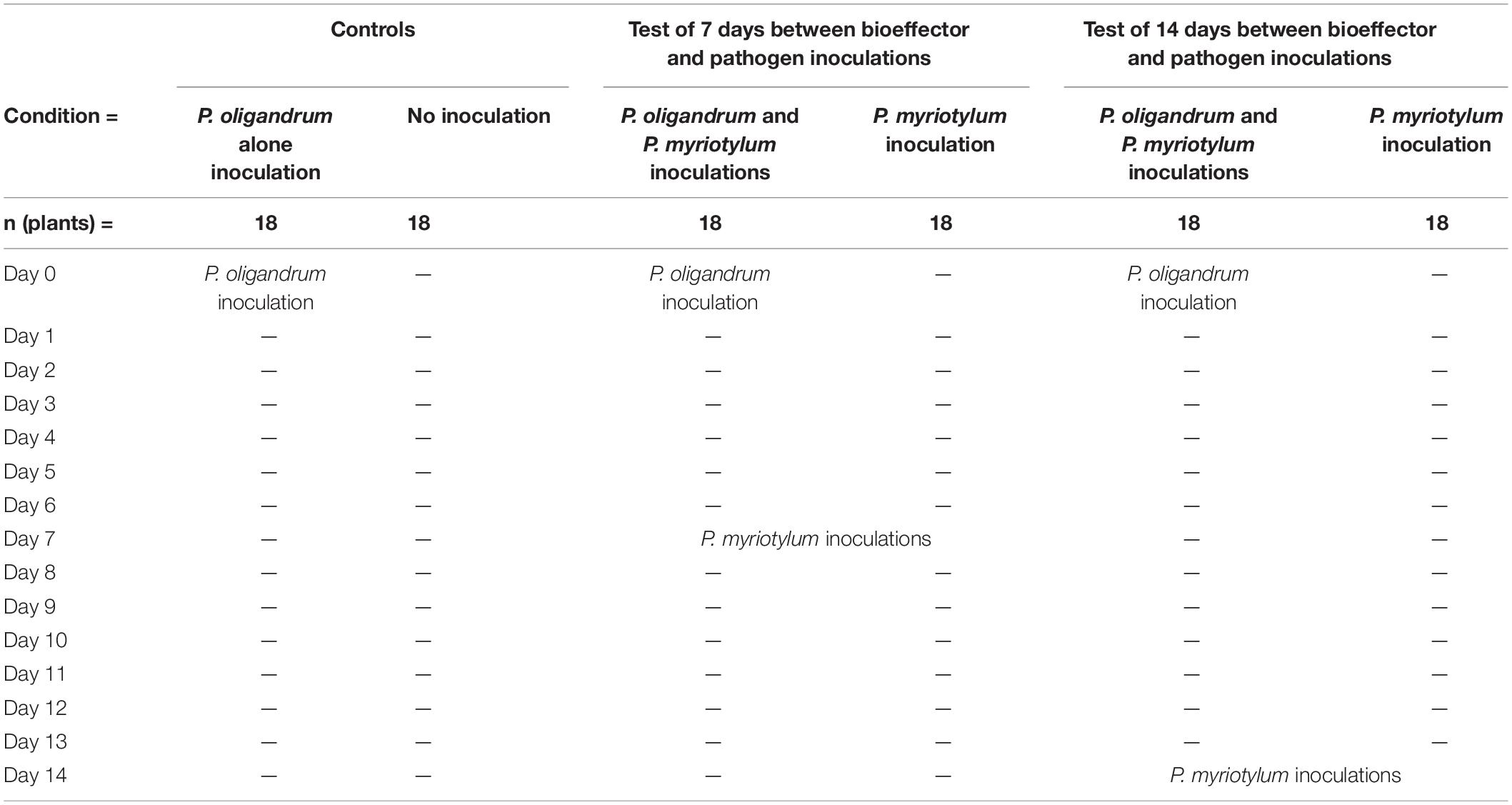
Table 1. Experimental design for ginger pot-trial with P. myriotylum and P. oligandrum to determine biocontrol effect.
Two different P. myriotylum inoculation time-points were used to show the biocontrol effect after different time periods between pathogen and bioeffector inoculation, and from ginger plants of different ages. Seven days after inoculation with P. myriotylum, the plants were scored for 5 days with the following scale: 0 = plants remain green and healthy; 1 = leaf sheath collar discolored and lower leaves turned yellow; 2 = plants alive, but shoots either totally yellow or dead; and 3 = all shoots dead (Stirling et al., 2009). The disease index (DI) was calculated using the equation DI = Σ 18i=1 Xi/54 where DI is the disease index rating from 0 (healthy) to 1 (dead), Xi is the disease rating of the ith replicate (from 1 to 18), and 54 is equal to the number of replicates multiplied by the highest rating scale of 3. The disease control rate was calculated by the following equation: (“P. myriotylum inoculated plants” - “P. oligandrum and P. myriotylum inoculated plants”)diseaseindex/P. myriotylumdiseaseindex × 100%.
To demonstrate that P. oligandrum could be recovered from around the roots of plants where there was a biocontrol effect on the PSR, the vermiculite from around the roots was collected into water containing the following five antimicrobial compounds: 100 μg/mL ampicillin, 50 μg/mL nystatin, 10 μg/mL pentachloronitrobenzene, 50 μg/mL rifampicin and 100 μg/mL carbenicillin. Serial dilutions of the suspension were plated onto 10% V8 agar containing the same antibiotics as above, and isolated colonies were sub-cultured onto V8 agar for DNA extraction using a CTAB-based extraction method. For PCR from the gDNA samples, ITS1 and ITS4 primers (White et al., 1990) were used to identify the species present. The 25 μL PCR reactions consisted of a 2X Taq Master Mix (VAZyme), 0.4 μM of each primer and 2 μL of gDNA template or water. The PCR cycling conditions were 95°C for 3 min, followed by 35 cycles of 95°C for 15 s, 56°C for 15 s, 72°C for 1 min, and then 72°C for 5 min. After agarose gel analysis, PCR products from the reactions using the ITS1 and ITS4 primers were sequenced using Sanger sequencing (Sangon Biotech).
Results and Discussion
The GAQ1 Isolate Belongs to P. oligandrum
The P. oligandrum isolate GAQ1 was recovered from soil from a field where infected ginger with symptoms of PSR disease was growing. The ITS region sequence of GAQ1 (MK774755.1) is 99.87% similar (788/789) to the strain of P. oligandrum CBS 382.34 (AY598618.2) used in the taxonomic monograph of the Pythium genus of van der Plaats-Niterink (1981). For three other molecular marker sequences compared to the CBS 382.34 strain, the GAQ1 isolate was 99.69% similar to the CoxI sequence, 99.81% similar to the CoxII, and 99.87% similar to the β-tubulin sequence. In a multi-locus phylogenetic analysis (using the ITS region, CoxI, CoxII and β-tubulin sequences), GAQ1 formed a clade with the P. oligandrum CBS 382.34 strain and with genome-sequenced P. oligandrum strains (Supplementary Figure 2). Previous reports of the ability of P. oligandrum to control other diseases caused by Pythium spp. such as P. ultimum, e.g., Abdelzaher et al. (1997), prompted an investigation into whether the newly recovered isolate of P. oligandrum could antagonize P. myriotylum, and what transcriptional changes occurred during their interaction, and whether P. oligandrum could control PSR of ginger caused by P. myriotylum.
Visualization of P. oligandrum and P. myriotylum Interaction
We were interested in understanding if the P. oligandrum GAQ1 isolated from soil where infected ginger was growing could control diseases of ginger such as Pythium soft rot. Previously, we recovered a virulent isolate of P. myriotylum named SWQ7 that could infect and kill ginger plants (Daly et al., 2021). To understand whether the GAQ1 isolate could antagonize P. myriotylum, we used plate-based confrontation assays and microscopy coupled with vitality staining using trypan blue. Both P. oligandrum and P. myriotylum had similar growth rates on V8 medium. Before contact of the mycelial fronts of both species, blue staining with trypan blue (indicating the presence of non-viable mycelium) was not observed from either species. Several hours after contact, the confrontation zone showed staining with the trypan blue (Figure 1), indicating a loss of viability in some of the hyphae. As the incubation of other cultures were continued for longer periods, the blue-stained area was seen to expand to cover the entire side of the plate where P. myriotylum had previously been growing on. The pattern of the trypan blue staining suggested successful antagonism by P. oligandrum of P. myriotylum as the loss of viability of the hyphae only appeared to occur in the part of the cultures where P. myriotylum was growing before the confrontation with P. oligandrum. Of note, when P. oligandrum or P. myriotylum was confronted with itself, the occurrence of trypan blue staining was not observed (Figure 1).
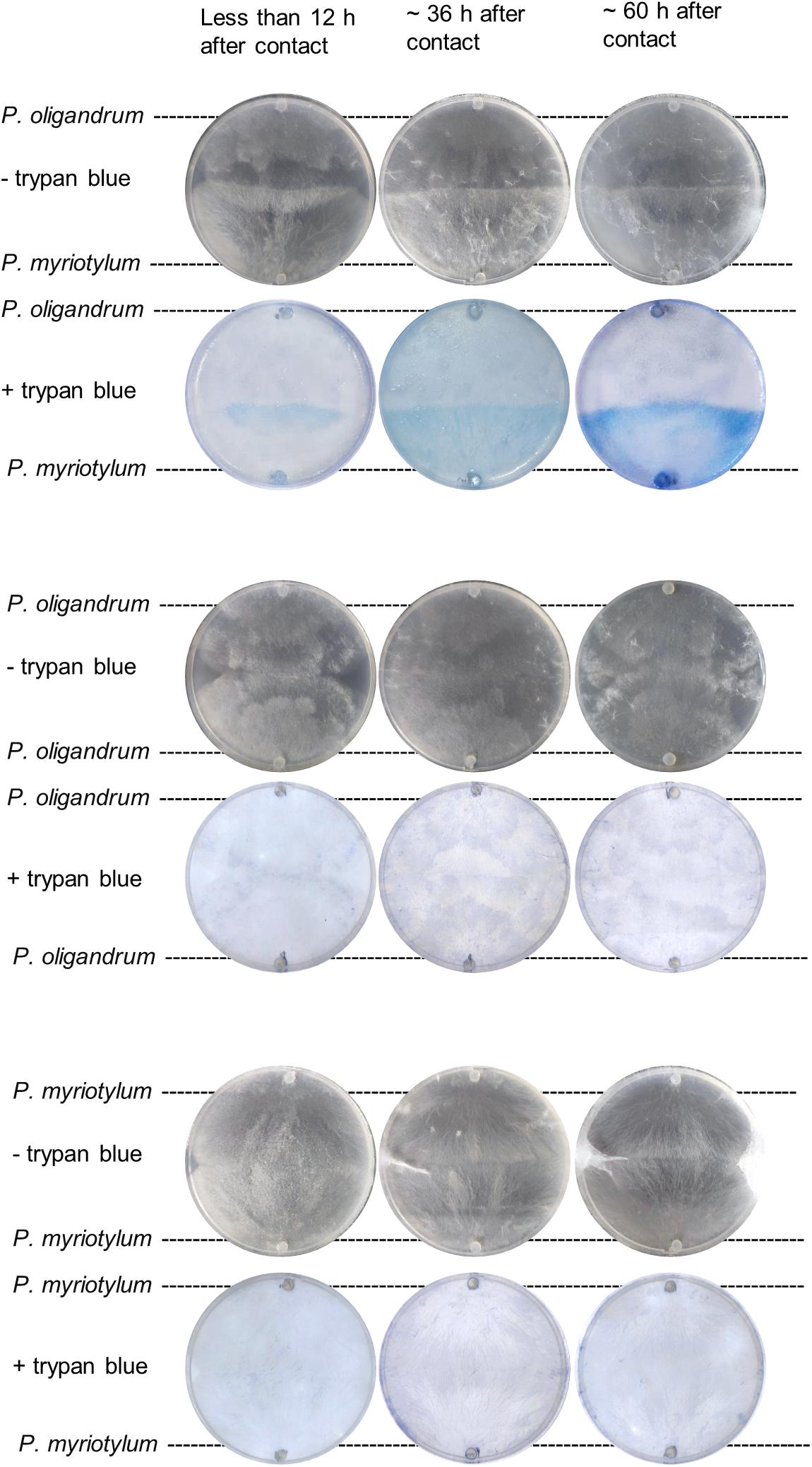
Figure 1. Representative images of trypan blue staining of mycelia from P. myriotylum and P. oligandrum self- or mixed-species confrontation demonstrating that P. myriotylum mycelium becomes non-viable in the presence of P. oligandrum. Images before and after trypan blue staining of the cultures are shown from less than 12 h, from ∼ 36 h and from ∼60 h after contact of the hyphal fronts of the colonies. The brightness and contrast of the images was adjusted consistently. The images from the control self-confrontation cultures demonstrate that the blue-colored staining does not appear in the self-confrontation.
The brightfield microscopy revealed that the trypan blue staining occurred on the thicker hyphae which are generally found from P. myriotylum while the thinner hyphae of P. oligandrum lack the trypan blue stain. Figures 2A,B demonstrate that the size of P. myriotylum hyphae is larger than those of P. oligandrum. Clear occurrences of hyphal coiling were observed where the thinner hyphae of P. oligandrum coiled around the thicker trypan blue-stained hyphae of P. myriotylum, with also occurrences of penetration of the P. myriotylum hyphae by P. oligandrum (Figure 2C). Cryo-SEM was used to obtain higher magnification and resolution images of the hyphal coiling (Figures 2D–F). In the brightfield images, there were also occurrences of the spiny oogonia on the part of the plate where P. myriotylum had previously been growing. Spiny oogonia are one of the characteristic features of P. oligandrum and related species (van der Plaats-Niterink, 1981) and are not formed by P. myriotylum. The observation here of the spiny oogonia further demonstrates the colonization by P. oligandrum of the part of the plate where P. myriotylum had previously been growing.
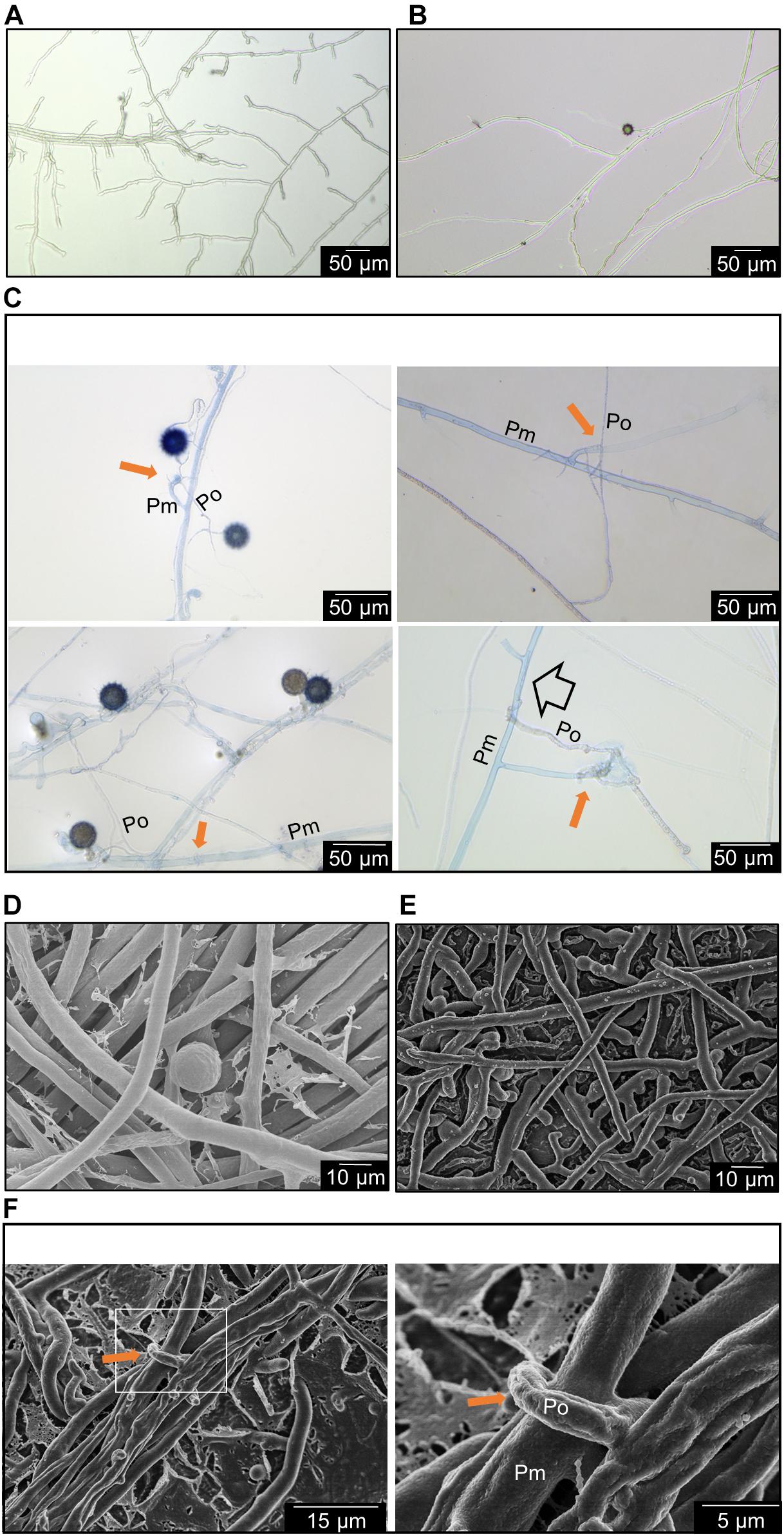
Figure 2. Representative features of the interaction between P. myriotylum (Pm) and P. oligandrum (Po). Here hyphae are imaged in the region where the two colonies have recently made contact using either brightfield microscopy coupled with trypan blue staining (A–C) or cryo-scanning electron microscopy (D–F). (A) P. myriotylum from self-confrontation, (B) P. oligandrum from self-confrontation with spiny oogonia visible, (C) coiling of P. oligandrum hypha around the hypha of P. myriotylum and apparent penetration by P. oligandrum of P. myriotylum hypha. The brightfield images of the cultures from where P. oligandrum and P. myriotylum confront each other were stained with trypan blue. (D) P. myriotylum from self-confrontation, (E) P. oligandrum from self-confrontation, (F) coiling of a P. oligandrum hypha around a hypha of P. myriotylum at two different magnifications. The cryo-SEM images were taken from cultures growing on polycarbonate membranes using the same experimental set-up as was used for the Dual-RNAseq (see Supplementary Figure 1 for images of these cultures). The solid amber arrow indicates examples of hyphal coiling and the empty arrow indicates an example of penetration.
The analysis here suggests a clear parasitic interaction of the P. oligandrum GAQ1 isolate on P. myriotylum. The macroscopic observations of the loss of vitality of the P. myriotylum hyphae after contact with P. oligandrum coupled with the microscopic observations of hyphal coiling support this parasitic mechanism. Previously, there are reports of the P. oligandrum isolate CGH1 antagonizing multiple Pythium spp. with observances of hyphal coiling and hyphal penetration (Berry et al., 1993). Interestingly, in the study of Berry et al. (1993), a lack of antagonism from the P. oligandrum isolate toward the plant pathogen P. aphanidermatum was found, and instead P. aphanidermatum coiled around the hyphae of P. oligandrum. The two species of P. aphanidermatum and P. myriotylum are relatively closely related, being from the Pythium clades A and B, respectively (Lévesque and De Cock, 2004), but in our study, we did not find occurrences of P. myriotylum hyphae coiling around the hyphae of the P. oligandrum GAQ1 isolate.
Overview of Transcriptome Analysis of the Interaction Between P. oligandrum and P. myriotylum
The interaction between P. oligandrum and P. myriotylum as observed macro- and microscopically was used as the basis to sample mycelia for transcriptomic analysis when P. myriotylum and P. oligandrum confronted each other or themselves. The part of the mycelia indicated by the black dotted-line in Supplementary Figure 1. were sampled for RNAseq which corresponds to an early time-point in the interaction between P. oligandrum and P. myriotylum. The P. oligandrum GAQ1 isolate had a flatter colony morphology with a notable chrysanthemal pattern to the mycelia, compared to the P. myriotylum SWQ7 isolate, which had a fluffier colony appearance with considerable numbers of aerial hyphae (Supplementary Figure 1). Approximately 20 million reads were obtained from each sample with ∼ 90% of the reads aligning to one location in a concatenation of the genome assemblies of the two species (Supplementary Table 3). When the reads that aligned uniquely to gene models from each species from the mixed species confrontation samples were counted, similar numbers of reads were counted on gene models for each species, with the biggest difference being only twofold in replicate two (Supplementary Table 3). From the single-species cultures of P. oligandrum or P. myriotylum, very few reads were counted on the gene models from the species not found in the culture, i.e., at between 0.005 and 0.06% of the reads that were counted on the gene models of the species present in the single culture (Supplementary Table 3). The above analysis indicated that the genes from the two species are sufficiently different in sequence to be distinguished from each other. The clustering pattern from a principal component analysis of the expression levels of the genes from each species showed separate clustering of the replicates from the self-confrontation compared to when the two species confronted each other (Figure 3A). In terms of the number of differentially expressed genes, there were 707 genes upregulated and 315 genes downregulated in P. myriotylum in the interaction with P. oligandrum. In P. oligandrum, there were 433 genes upregulated and 122 genes downregulated in the interaction with P. myriotylum (Figure 3B and Supplementary Table 1).
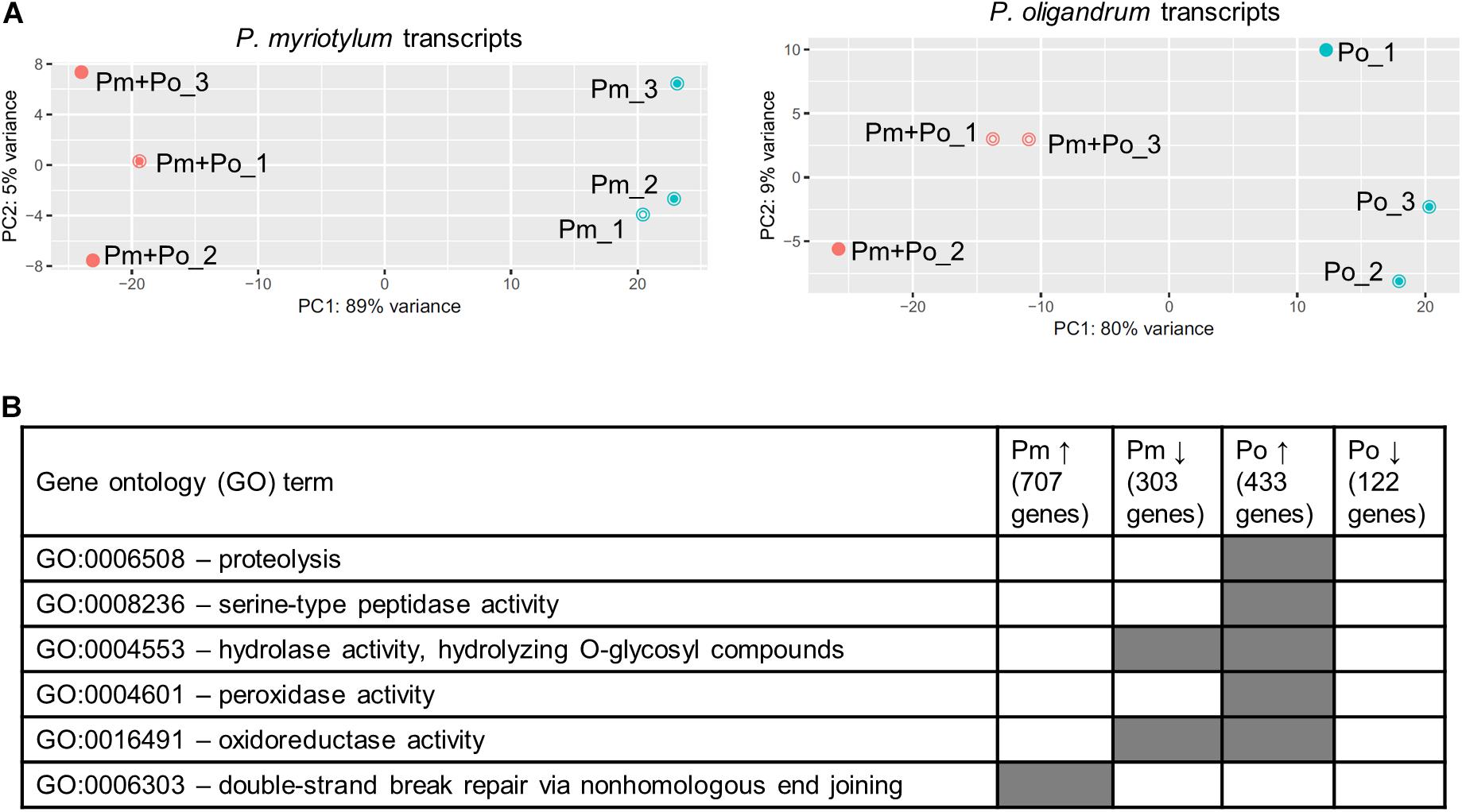
Figure 3. (A) Overview of the clustering pattern from principal component (PC) analysis of the transcripts from the replicate cultures from P. oligandrum (Po) and P. myriotylum (Pm) self- or mixed-species confrontations, (B) summary of interesting gene ontology (GO) terms enriched in the differentially expressed genes in P. myriotylum and P. oligandrum when they confront each other compared to a self-confrontation. The full list of enriched GO terms can be found in Supplementary Table 4.
Transcriptomic Aspects of Parasitic Mechanisms From P. oligandrum
Cellulose is a key component of the oomycete cell wall (Mélida et al., 2013), and a potential major target of any antagonist of an oomycete like P. myriotylum. Based on CAZy, signal peptide and transmembrane domain annotations, P. oligandrum has 50 putative host/prey targeted cellulases. Host/prey targeted cellulases were considered those cellulases which contained a signal peptide and lacked transmembrane domains indicating that they are likely secreted into the extracellular environment (where they could degrade cellulose found in the cell walls of both a plant and oomycete host) and are unlikely to be involved in remodeling of the P. oligandrum cell wall. These host/prey targeted cellulases were from CAZy families GH1, GH3, GH5, GH6, GH7, GH30_1, GH131 and AA9. Of the putative host/prey targeted cellulases, 11/50 were significantly upregulated in the interaction with P. myriotylum, and notably, none of these 50 was significantly downregulated (Supplementary Table 1C). In general, the P. oligandrum putative host/prey targeted cellulases were expressed but were not upregulated to a high level in the presence of P. myriotylum. Only two of the upregulated host/prey targeted cellulases had an FPKM > 100 (two GH5_14 members that likely encode for β-glucosidase activity). Two members of the GH30_1 sub-family that likely also encode for β-glucosidase were constitutively expressed with FPKM > 100 but there were no good examples of upregulated host/prey targeted cellulases that likely have activity on the cellulose polymer (e.g., cellobiohydrolases or endoglucanases). P. oligandrum protease or peptidase encoding genes were a major category of genes that were upregulated in the interaction with P. myriotylum e.g., the GO term for—proteolysis (GO:0006508) was enriched in the P. oligandrum genes upregulated in the interaction with P. myriotylum (Figure 3B). The putative role of P. oligandrum proteases will be described in more detail in a later section.
The limited upregulation of the host/prey targeted cellulases in P. oligandrum in the confrontation with P. myriotylum contrasts somewhat with their expression in P. oligandrum in the interaction with P. infestans from the study of Liang et al. (2020). Here the majority of the AA9 cellulases were upregulated (with a subset expressed at a high level) at 12 and 24 h after contact between P. oligandrum and P. infestans compared to control samples before contact was made (Liang et al., 2020). Differences in the timing of the sampling for expression analysis, using a different P. oligandrum strain (CBS530.74), or other hosts (e.g., differences in the cell wall composition between the hosts) could contribute to the differences in cellulase gene expression when P. oligandrum confronted P. myriotylum compared to when P. infestans was confronted. The cellulolytic and protease gene expression measured from the GAQ1 isolate likely contributes to the effects seen in the microscopy images whereby there are clear signs of a loss of viability of the P. myriotylum mycelium from the trypan blue staining (Figures 1, 2) and examples of hyphal penetration by P. oligandrum of the P. myriotylum hyphae (Figure 2C).
Apart from genes involved in a parasitic mechanism, several known genes from P. oligandrum with a role in mediating the biocontrol of diseases via stimulation of plant defenses, were highly expressed at a constitutive level. e.g., oligandrin Po_g9166.t1 (Oli−D1) and Po_g9168.t1 (POD-2) (there is no evidence that the presence of P. myriotylum downregulated these genes). Oli-D1 and POD-2 are elicitin/elicitin-like proteins, and seven other P. oligandrum elicitin-like genes were upregulated in the confrontation with P. myriotylum, but their expression level was 10–100-fold less than Oli-D1 and POD-2 (Supplementary Table 1).
Features of the Transcriptomic Response of P. myriotylum When Parasitized by P. oligandrum
A subset of P. myriotylum putative cellulase genes were upregulated in the confrontation with P. oligandrum. In contrast, to the P. oligandrum upregulated cellulases described in the previous section, a smaller number of P. myriotylum cellulases were upregulated, and at least half seemed likely to have a cellulose remodeling or intracellular role rather than as part of a counter-attacking defensive strategy. Six P. myriotylum genes annotated as putative cellulases were upregulated: Pm_g1192.t1 (GH5_14), Pm_g1197.t1 (GH5_14), Pm_g7072.t1 (GH5_20), Pm_g7136.t1 (GH5_20), Pm_g7449.t1 (AA9), and Pm_g14120.t1 (GH5_20). Two of the putative cellulases were annotated with a signal peptide (Pm_g1192.t1 and Pm_g7449.t1) and three of those without a signal peptide were annotated with transmembrane domains (Pm_g7072.t1, Pm_g7136.t1, and Pm_g14120.t1), suggesting these three are membrane-bound proteins. The six upregulated P. myriotylum putative cellulases were annotated as either GH5_14, GH5_20, or AA9 family members. The characterized activities within the GH5_14 family include β-glucosidase and exo-β-1,3-glucosidase activities.2 The β-glucosidase activity could act on the cellobiose released from cellulose but cellulose does not contain β-1,3 linked glucans. The GH5_20 members appear to only be found in Stramenopiles and appear most closely related to GH5_1 members, of which many have been characterized as endo-β-1,4-glucanases (Aspeborg et al., 2012) but no GH5_20 members appear to have been characterized to date. The expression levels of the three putative cellulases from the GH5_20 family encoded by Pm_g7072.t1, Pm_g7136.t1, and Pm_g14120.t1 were validated by qPCR (Figure 4A). For the other CAZy family found amongst the upregulated cellulases; generally, proteins containing an AA9 domain have oxidative activity toward crystalline cellulose (Quinlan et al., 2011). The putative transmembrane domain-containing cellulases upregulated in P. myriotylum when confronted with P. oligandrum may remodel the cellulose in the P. myriotylum cell wall as part of a defensive response to the P. oligandrum antagonism. In mycoparasitic interactions involving Trichoderma atroviride, the remodeling of prominent fungal cell wall polysaccharides chitin and chitosan was important for the mycoparasitic ability of the fungus by contributing to self-defense (Kappel et al., 2020).
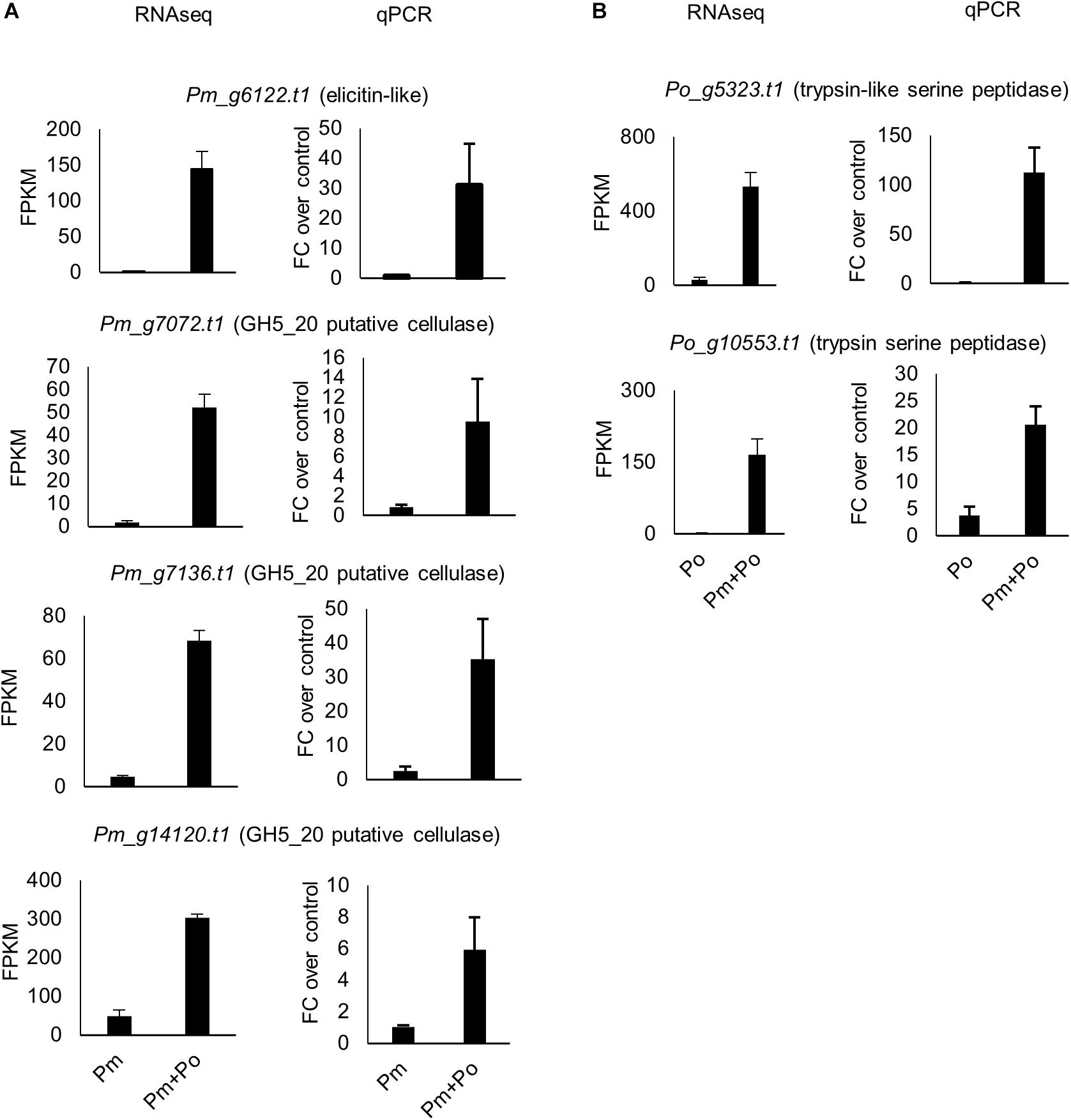
Figure 4. Expression levels from RNAseq and qPCR for selected (A) P. myriotylum genes and (B) P. oligandrum genes upregulated in the mixed-species confrontation compared to the self-confrontation. The error bars represent standard errors (n = 3). The expression levels of P. myriotylum were normalized using the two reference genes Pm_g6466.t1 (tubulin) and Pm_17033.t1 (gapdh) and the expression levels of P. oligandrum were normalized using Po_g2472.t1 (succinate dehydrogenase) and Po_g6912.t1 (g6pdh). FC, fold change; Pm, P. myriotylum; Po, P. oligandrum.
A subset of nine P. myriotylum elicitin-like putative pathogen-associated molecular pathogen (PAMP) molecules were upregulated in the confrontation with P. oligandrum. The upregulation of the elicitin-like Pm_g6122.t1 was validated by qPCR (Figure 4A). Of note, other elicitin-like proteins are highly constitutively expressed, such as Pm_g10365.t1 with an FPKM ∼ 20,000 that are more likely to function in P. myriotylum in the major primary role of the elicitin-like proteins in scavenging for sterols (Janků et al., 2020). If the upregulation of a subset of the P. myriotylum elicitin-likes found in the in vitro confrontation was replicated in the tri-partite interaction of the two Pythium spp. and a plant host of P. myriotylum (e.g., ginger), there is the potential to modulate plant immunity and the virulence of P. myriotylum, as elicitin-like proteins from other oomycete species can function as PAMPs (Derevnina et al., 2016). This, of course, would be an unintentional consequence of the interaction of P. oligandrum on P. myriotylum as a bioeffector does not intentionally alter the virulence of the plant pathogen it antagonizes as instead, the plant pathogen is a nutrient source or a competitor for ecological resources.
The GO term for double-strand break repair via non-homologous end-joining (GO:0006303) was enriched in the genes upregulated in P. myriotylum in the interaction with P. oligandrum (Supplementary Table 4). The two genes responsible for this enrichment, Pm_g12183.t1 and Pm_g10950.t1, were annotated with domains found in Ku70/Ku80 proteins (PF02735, PF03730 and PF03731). Ku proteins are evolutionarily conserved proteins that mediate repair of double-stranded DNA by non-homologous end-joining of the broken ends of the DNA (Doherty and Jackson, 2001). Although these two genes were also expressed in P. myriotylum when it confronts itself, their upregulation in the confrontation with P. oligandrum suggests there could be a greater requirement to repair double-strand breaks.
In P. myriotylum, there were relatively few protease-encoding genes upregulated in the confrontation with P. oligandrum. Instead, one stand-out part of the interaction between P. myriotylum and P. oligandrum was the high upregulation of P. myriotylum putative protease inhibitors along with the upregulation or constitutive expression of P. oligandrum proteases, which will be described in detail in the next section.
High Upregulation of a Sub-Set of P. myriotylum Kazal-Type Protease Inhibitors and P. oligandrum Peptidases
In the P. myriotylum genome, there are 23 genes annotated with a Kazal-type protease inhibitor domain (Supplementary Table 1 and Figure 5A). One of the most striking features of the genes induced in the interaction between P. myriotylum and P. oligandrum was the upregulation of six of the P. myriotylum genes annotated with a Kazal-type protease inhibitor domain in the interaction with P. oligandrum (Pm_g8255.t1, Pm_g8256.t1, Pm_g8257.t1, Pm_g8263.t1, Pm_g8265.t1, and Pm_g8266.t1). Three of these genes (Pm_g8256.t1, Pm_g8257.t1, and Pm_g8265.t1) were induced greater than 1,000-fold to a high FPKM value of greater than 1000 FPKM. All six of these Kazal-type protease inhibitor genes were annotated with a signal peptide, and only Pm_g8257.t1 is predicted to possesses a transmembrane domain, indicating that the other five are likely secreted into the environment around P. myriotylum. The induction of the genes annotated with a Kazal-type protease inhibitor domain was validated by qPCR (Figure 5B). Of the other Kazal-type protease inhibitor genes that were not upregulated, one of the genes (Pm_g16340.t1) appeared to be constitutively expressed when P. myriotylum was confronted by itself or by P. oligandrum, and two other Kazal-type protease inhibitor genes (Pm_g8268.t1 and Pm_g1759.t1) were downregulated when P. myriotylum was confronted by P. oligandrum. Overall, for the P. myriotylum genes annotated with a Kazal-type protease inhibitor domain, the dominant feature was high upregulation of a subset of these when P. myriotylum was confronted by P. oligandrum. Of the genes upregulated in P. myriotylum in the interaction with P. oligandrum, the largest fold-change upregulated and highest expression was seen for these genes annotated as Kazal-type protease inhibitors.
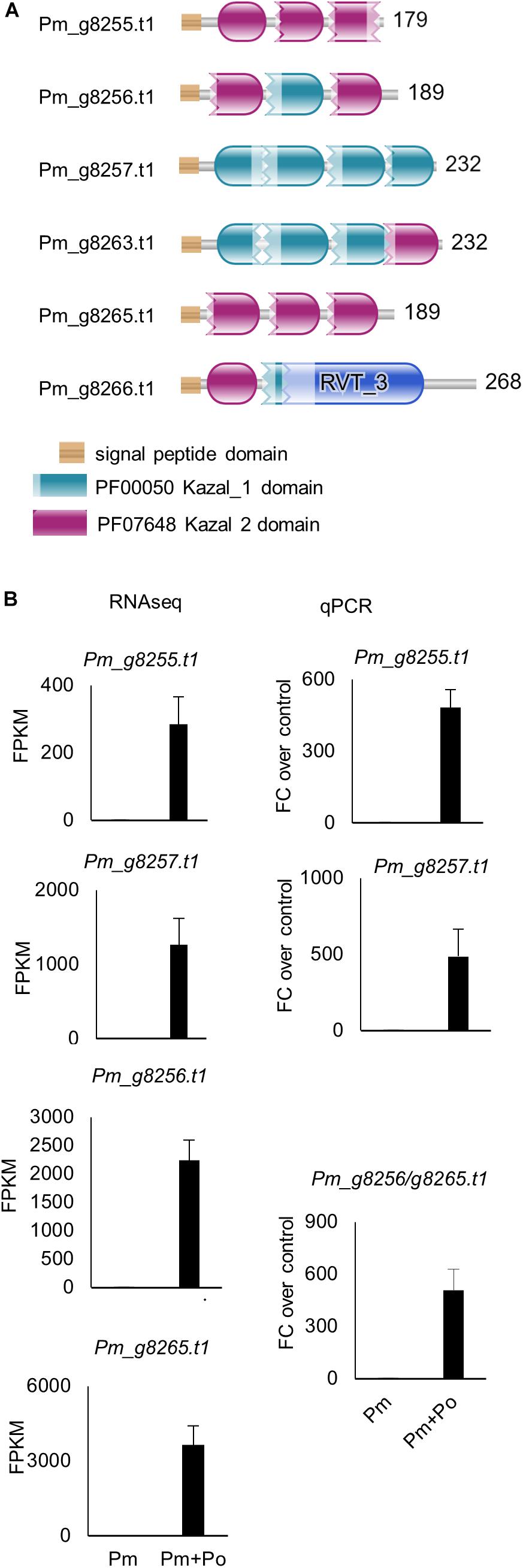
Figure 5. (A) Protein sequences of the confrontation-upregulated P. myriotylum Kazal-type protease inhibitors highlighting the locations of the Kazal domains. (B) Expression levels from RNAseq and qPCR of a sub-set of the P. myriotylum Kazal-type protease inhibitors upregulated in the confrontation with P. oligandrum compared to the self-confrontation. The error bars represent standard errors (n = 3). The expression levels of P. myriotylum were normalized using two reference genes Pm_g6466.t1 (tubulin) and Pm_17033.t1 (gapdh). Pm, P. myriotylum; Po, P. oligandrum.
In contrast to the P. myriotylum Kazal-type protease inhibitor genes, in P. oligandrum, none of the 13 genes annotated with a Kazal domain were differentially expressed, while one gene annotated with a Kazal domain (Po_g7532.t1) appeared to be constitutively expressed to a relatively high level of ∼500 FPKM. This gene, Po_g7532.t1, is a match to two EST sequences (EV245133.1 and EV243682.1) from an EST library sequenced from an in vitro interaction between P. oligandrum with heat-killed P. infestans mycelia (Horner et al., 2012). Another EST from the same library EV245779.1, annotated with a Kazal domain, matched Po_g10361.t1, which was expressed constitutively in the conditions at ∼70 FPKM.
Kazal-type protease inhibitors can inhibit serine proteases (Tian and Kamoun, 2005) and whether there was any upregulation of the class of proteases in P. oligandrum that the P. myriotylum Kazal-type protease inhibitors could inhibit was of interest. Interestingly, the GO enrichment analysis showed that the GO term for serine-type peptidase activity (GO:0008236) was enriched in the P. oligandrum genes upregulated in the confrontation with P. myriotylum (Figure 3B). The expression level of P. oligandrum putative serine proteases was assessed using the following Pfam annotations: Pfam00089 (Trypsin), Pfam13365 (Trypsin-like peptidase domain), Pfam00082 (Subtilase family), Pfam00450 (Serine carboxypeptidase), and Pfam05577 (Serine carboxypeptidase S28). Of these 142 P. oligandrum serine proteases, there were 23 upregulated when P. oligandrum confronted P. myriotylum compared to when P. oligandrum confronted itself. All of these 23 putative serine proteases were annotated with a signal peptide sequence (Supplementary Table 1). The upregulation of the peptidase with one of the largest fold changes in upregulation (Po_g10553.t1, a trypsin family serine protease) and the peptidase that had the highest expression level in the confrontation with P. myriotylum (Po_g5323.t1, a trypsin-like serine protease) was validated by qPCR (Figure 4B).
There is potential for these proteins with Kazal-type protease inhibitor domains to be involved in inhibition of the serine peptidases upregulated or constitutively expressed in P. oligandrum. Previously in the interaction between the biocontrol rhizobacterium L. capsici and the pathogen P. infestans (Tomada et al., 2017) there was an upregulation of a subset of Kazal-type protease inhibitors, and it was hypothesized that these could be involved in inhibiting lytic activities from L. capsici. Although, notably, Tomada et al. (2017) did not consider overall an active response from P. infestans to the antagonism from L. capsici. Consistent with a prominent role for P. oligandrum proteases in the antagonism of P. myriotylum was a lack of upregulation of the P. oligandrum Kazal-type protease inhibitors, as their upregulation could inhibit the P. oligandrum protease activity. Horner et al. (2012) considered the ESTs annotated with a Kazal domain as part of a defense or counter-defensive response of P. oligandrum to heat-killed P. infestans mycelia. However, in the confrontation between P. oligandrum and the living P. myriotylum mycelia, the P. oligandrum putative Kazal-type inhibitors appear to play a minor role.
While the transcriptomic response of P. myriotylum in the confrontation with P. oligandrum suggested aspects of a defensive or counter-attacking strategy, this appears insufficient to defend against P. oligandrum as clearly in the plate confrontation assays, P. oligandrum caused progressive loss of viability in the hyphae of P. myriotylum (Figure 1). As the ability to antagonize on plate cultures suggested it could have a control effect on diseases caused by P. myriotylum, the ability to control the Pythium soft-rot of ginger caused by P. myriotylum was investigated using a pot-trial experiment.
P. oligandrum GAQ1 Has Control Effect on Pythium Soft-Rot of P. myriotylum Infected Ginger
As P. oligandrum demonstrated an ability to antagonize P. myriotylum in plate confrontation assays, its ability to control PSR of ginger was investigated. P. oligandrum was inoculated first followed by inoculations of P. myriotylum at two time points after the inoculation of P. oligandrum, and this also led to inoculations of P. myriotylum onto plants of two different ages and sizes. Clearly from the data on the above-ground disease symptoms in Figure 6, the plants inoculated with P. myriotylum developed the disease symptoms. The disease was more severe in the younger and smaller ginger plants as a higher proportion of the plants developed the more severe symptoms whereby all the aerial parts of the plant appeared dead (Figure 6A). In both of the time-points analyzed, the presence of P. oligandrum resulted in a clear and significant (P < 0.001) reduction in the disease symptoms whereby > 80% of the plants had no disease symptoms (Figure 6A). The disease control rate was ∼80% in both inoculation time points (Figure 6B). To show that P. oligandrum could be recovered from plants where there was a biocontrol effect on the PSR, P. oligandrum was re-isolated from around the roots that had been infected with P. myriotylum (Supplementary Figure 3).
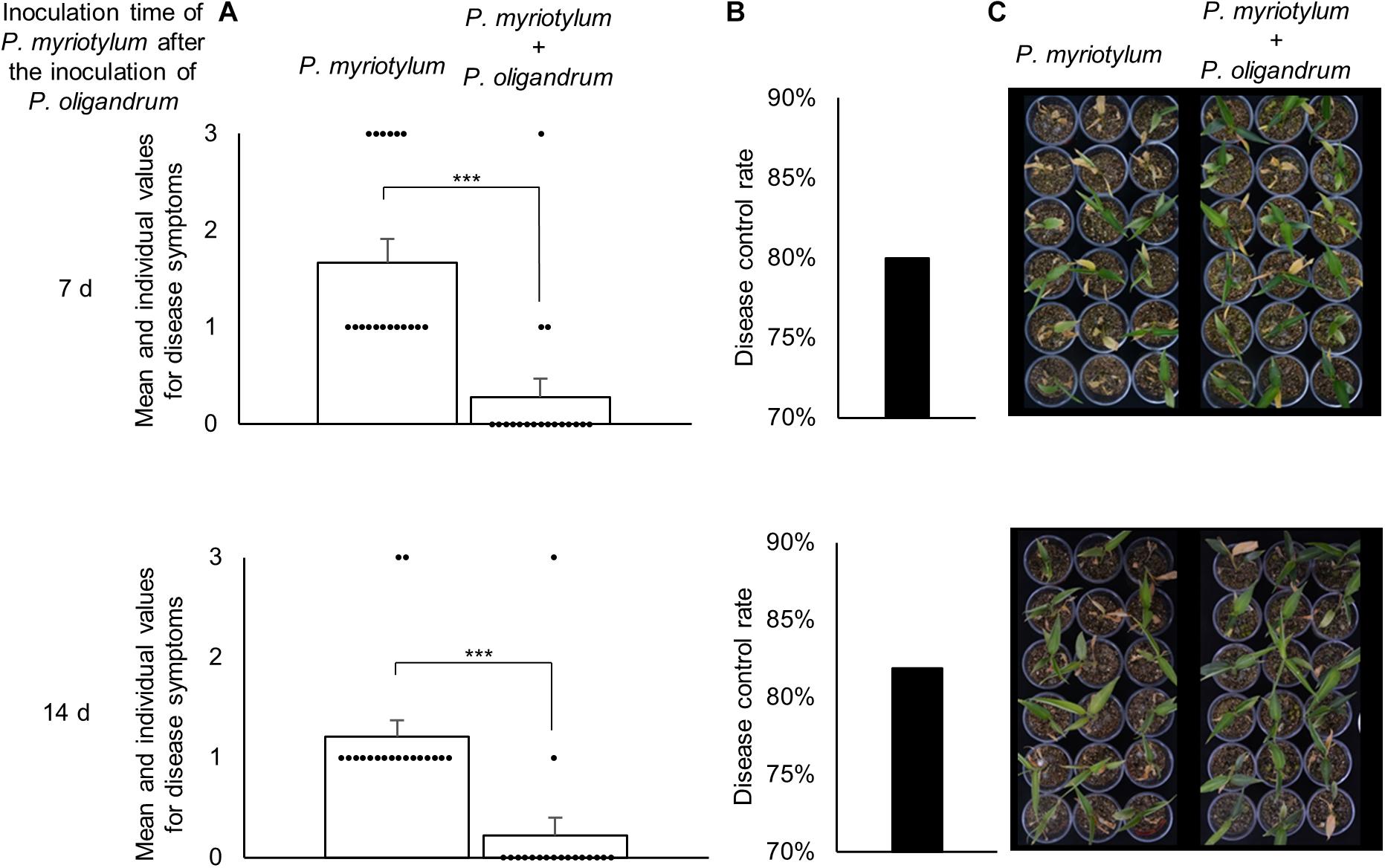
Figure 6. Biocontrol effect of P. oligandrum on Pythium soft-rot (PSR) of ginger disease caused by P. myriotylum from two different inoculation time-points of P. myriotylum after the inoculation of P. oligandrum. (A) The mean values and raw data from the scoring of the disease symptoms (scale from 0 to 3) in the 18 replicate plants for each condition. ***corresponds to a P ≤ 0.001 from a Mann-Whitney Rank Sum Test (n = 18). (B) The disease control rate of P. oligandrum on PSR of ginger. (C) Images of the 18 replicate plants inoculated with P. myriotylum or P. myriotylum and P. oligandrum. The data and images were recorded 11 d after the inoculation of the plants with P. myriotylum. The scoring for the disease symptoms was always zero for the plants that were inoculated with only P. oligandrum, and for the plants that were not inoculated with either of the Pythium spp.
A similar experiment using a different P. oligandrum isolate to control PSR soft-rot of ginger caused by a P. myriotylum isolated from infected ginger in Australia did not reduce the disease symptoms on ginger plants (Le, 2016). It was speculated by Le (2016) that the lack of a control effect may have been in part due to poor colonization of the soil by the P. oligandrum isolate (vermiculite was used to grow the ginger plants inoculated with GAQ1) or the ineffective application of the oospore suspension by soil drenching (mycelia of the GAQ1 isolate was added directly into the vermiculite). However, there is support for P. oligandrum to control other diseases caused by P. myriotylum where previously it was shown that P. oligandrum was able to reduce soybean damping-off caused by P. myriotylum (You et al., 2019).
Part of the rationale for the suite of analyses undertaken in this study involving microscopy, dual-transcriptomics, and biocontrol was to look for further insights into why biocontrol strategies in the field are often hampered by a lack of robustness or durability. Of course, dominant reasons for this include interactions between the bioeffector and a complex microbiome, varying abilities of the bioeffector to propagate, and changes in host strain or isolate (i.e., the plant pathogen) from year to year. The biocontrol effect from the controlled laboratory conditions is promising, with a disease control rate of ∼80% for the application of P. oligandrum to control PSR of ginger, although the ability of P. oligandrum to suppress PSR of ginger in field conditions has yet to be tested. Suppose one were to extrapolate the microscopic and transcriptomic observations from the in vitro confrontation assays to the tri-partite interaction in the pot trial experiment. In that case, there is likely P. oligandrum parasitism or predation of P. myriotylum contributing to the suppression of the disease. The defensive or counter-attacking strategies suggested from the dual-transcriptomics, such as the Kazal-type protease inhibitor upregulation, do not appear to manifest themselves into a successful defense. In the in vitro plate confrontation assays, P. oligandrum progressively parasitized P. myriotylum mycelia, and a relatively high control rate of ∼80% was maintained in the pot-trial experiment. However, these defensive or counter-attacking strategies suggested by the dual-transcriptomics are another factor that is worthwhile in considering for the robustness and durability of biocontrol strategies in the field.
Conclusion
The P. oligandrum isolate GAQ1 is a promising bioeffector of PSR disease of ginger caused by P. myriotylum and has parasitic abilities that likely contribute to the control of PSR disease of ginger. Likely side-by-side comparisons of different P. oligandrum strains would be required to determine whether the GAQ1 isolate is superior for control of particular diseases (e.g., whether P. oligandrum GAQ1 which was isolated from a field with infested ginger is superior to other strains in controlling the PSR of ginger). For field-based application, an oospore-based preparation of P. oligandrum GAQ1 would likely ensure better shelf-life of the bioeffector compared to the use of a mycelial-based inoculum. The use of dual-transcriptomics is a powerful approach to uncover differentially expressed genes in the interaction between an antagonist and its prey or host. The differentially expressed genes such as Kazal-type protease inhibitors are promising candidates to investigate further molecular aspects of the antagonism and defensive responses between host and prey.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
PD, SC, QZ, JZ, JL, and TX performed the experiments. PD, DF, and JM performed bioinformatic analyses. PD and SC wrote the manuscript and analyzed the results. XW and XS sampled the soil for microbe isolation. LW obtained funding for the research. TS, DZ, SD, MJ, ID, and LW contributed to the discussion and manuscript revision. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Natural Science Foundation of China (32050410305), the China Agriculture Research System of MOF and MARA (CARS-24-C-01), the Jiangsu Agricultural Science and Technology Innovation Fund [CX(18)2005], and the China Postdoctoral Science Foundation (2020M671388).
Conflict of Interest
LW, DZ, and QZ were co-authors on a patent application for P. oligandrum (Patent application number CN201910757035.2) relating to the biocontrol of plant diseases using the P. oligandrum GAQ1 isolate.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Assistance from OE Biotech company for next-generation sequencing and the electron microscopy facility staff at Nanjing Agricultural University for the microscopy work is acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.765872/full#supplementary-material
Footnotes
- ^ http://www-huber.embl.de/users/anders/HTSeq/doc/count.html
- ^ http://www.cazy.org/GH5_14_characterized.html
References
Abdelzaher, H. M. A., Elnaghy, M. A., and Fadl-Allah, E. M. (1997). Isolation of Pythium oligandrum from Egyptian soil and its mycoparasitic effect on Pythium ultimum var. ultimum the damping-off organism of wheat. Mycopathologia 139:97. doi: 10.1023/A:1006836703594
Afgan, E., Baker, D., Batut, B., Van den beek, M., Bouvier, D., Čech, M., et al. (2018). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. doi: 10.1093/nar/gky379
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Aspeborg, H., Coutinho, P., Wang, Y., Brumer, H., and Henrissat, B. (2012). Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 12:186. doi: 10.1186/1471-2148-12-186
Benhamou, N., Bélanger, R. R., Rey, P., and Tirilly, Y. (2001). Oligandrin, the elicitin-like protein produced by the mycoparasite Pythium oligandrum, induces systemic resistance to Fusarium crown and root rot in tomato plants. Plant Physiol. Biochem. 39, 681–696. doi: 10.1016/s0981-9428(01)01283-9
Benhamou, N., Le Floch, G., Vallance, J., Gerbore, J., Grizard, D., and Rey, P. (2012). Pythium oligandrum: an example of opportunistic success. Microbiology 158, 2679–2694. doi: 10.1099/mic.0.061457-0
Benhamou, N., Rey, P., Picard, K., and Tirilly, Y. (1999). Ultrastructural and cytochemical aspects of the interaction between the mycoparasite Pythium oligandrum and soilborne plant pathogens. Phytopathology 89, 506–517. doi: 10.1094/PHYTO.1999.89.6.506
Berger, H., Yacoub, A., Gerbore, J., Grizard, D., Rey, P., Sessitsch, A., et al. (2016). Draft genome sequence of biocontrol agent Pythium oligandrum strain Po37, an oomycota. Genome Announcements 4, e00215–e00216. doi: 10.1128/genomeA.00215-16
Berry, L. A., Jones, E. E., and Deacon, J. W. (1993). Interaction of the mycoparasite Pythium oligandrum with other Pythium species. Biocontrol Sci. Technol. 3, 247–260.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Daly, P., Chen, Y., Zhang, Q., Zhu, H., Li, J., Zhang, J., et al. (2021). Pythium myriotylum is recovered most frequently from Pythium soft-rot infected ginger rhizomes in China. Plant Disease. Online ahead of print. doi: 10.1094/PDIS-05-21-0924-RE
Daly, P., Van Munster, J. M., Kokolski, M., Sang, F., Blythe, M. J., Malla, S., et al. (2017). Transcriptomic responses of mixed cultures of ascomycete fungi to lignocellulose using dual RNA-seq reveal inter-species antagonism and limited beneficial effects on CAZyme expression. Fungal Genet. Biol. 102, 4–21. doi: 10.1016/j.fgb.2016.04.005
De-Guzman, C. C., and Siemonsma, J. S. (1999). Plant Resources of South East Asia: Spices. Leiden: Backhuys Publishers.
Derevnina, L., Dagdas, Y. F., De La Concepcion, J. C., Bialas, A., Kellner, R., Petre, B., et al. (2016). Nine things to know about elicitins. New Phytol. 212, 888–895. doi: 10.1111/nph.14137
Doherty, A. J., and Jackson, S. P. (2001). DNA repair: how Ku makes ends meet. Curr. Biol. 11, R920–R924. doi: 10.1016/s0960-9822(01)00555-3
FAOSTAT (2019). Food and Agricultural Organisation of the United Nations Statistical Database. Available online at: http://faostat.fao.org/ (accessed 24 July, 2021).
Faure, C., Veyssiere, M., Boelle, B., San Clemente, H., Bouchez, O., Lopez-Roques, C., et al. (2020). Long-read genome sequence of the sugar beet rhizosphere mycoparasite Pythium oligandrum. G3 (Bethesda) 10, 431–436. doi: 10.1534/g3.119.400746
Horner, N., Grenville-Briggs, L., and Van West, P. (2012). The oomycete Pythium oligandrum expresses putative effectors during mycoparasitism of Phytophthora infestans and is amenable to transformation. Fungal Biol. 116, 24–41. doi: 10.1016/j.funbio.2011.09.004
Janků, M., Činčalová, L., Luhová, L., Jan, L., and Petřivalský, M. (2020). Biological effects of oomycetes elicitins. Plant Protect. Sci. 1, 1–8. doi: 10.17221/21/2019-pps
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., Mcanulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kappel, L., Münsterkötter, M., Sipos, G., Escobar Rodriguez, C., and Gruber, S. (2020). Chitin and chitosan remodeling defines vegetative development and Trichoderma biocontrol. PLoS Pathogens 16:e1008320. doi: 10.1371/journal.ppat.1008320
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Köhl, J., Kolnaar, R., and Ravensberg, W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci. 10:845. doi: 10.3389/fpls.2019.00845
Kroon, L. P., Bakker, F. T., Van Den Bosch, G. B., Bonants, P. J., and Flier, W. G. (2004). Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genet. Biol. 41, 766–782. doi: 10.1016/j.fgb.2004.03.007
Kushwaha, S. K., Vetukuri, R. R., and Grenville-Briggs, L. J. (2017). Draft genome sequence of the mycoparasitic oomycete Pythium oligandrum strain CBS 530.74. Genome Announcements 5, e346–e317. doi: 10.1128/genomeA.00346-17
Le, D. P., Smith, M., Hudler, G. W., and Aitken, E. (2014). Pythium soft rot of ginger: detection and identification of the causal pathogens, and their control. Crop Protection 65, 153–167. doi: 10.1016/j.cropro.2014.07.021
Le, D. P., Smith, M. K., and Aitken, E. A. B. (2016). An assessment of Pythium spp. associated with soft rot disease of ginger (Zingiber officinale) in Queensland, Australia. Australasian Plant Pathol. 45, 377–387. doi: 10.1007/s13313-016-0424-5
Le, D. P. (2016). Characterization of Species Associated With Pythium Soft Rot of Ginger and Evaluation of Pythium oligandrum as a Biocontrol. Ph.D thesis, Brisbane, QLD: University of Queensland
Lévesque, A. C., and De Cock, A. W. A. M. (2004). Molecular phylogeny and taxonomy of the genus Pythium. Mycol. Res. 108, 1363–1383. doi: 10.1017/s0953756204001431
Liang, D., Andersen, C. B., Vetukuri, R. R., Dou, D., and Grenville-Briggs, L. J. (2020). Horizontal gene transfer and tandem duplication shape the unique cazyme complement of the mycoparasitic oomycetes Pythium oligandrum and Pythium periplocum. Front. Microbiol. 11:581698. doi: 10.3389/fmicb.2020.581698
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495.
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Martin, F. N., and Tooley, P. W. (2003). Phylogenetic relationships among Phytophthora species inferred from sequence analysis of mitochondrially encoded cytochrome oxidase I and II genes. Mycologia 95, 269–284. doi: 10.1080/15572536.2004.11833112
Mélida, H., Sandoval-Sierra, J. V., Diéguez-Uribeondo, J., and Bulone, V. (2013). Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryot. Cell 12, 194–203. doi: 10.1128/EC.00288-12
Mortazavi, A., Williams, B. A., Mccue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Meth 5, 621–628. doi: 10.1038/nmeth.1226
Quinlan, R., Sweeney, M., Lo Leggio, L., Otten, H., Poulsen, J.-C., Johansen, K., et al. (2011). Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. U S A. 108, 15079–15084. doi: 10.1073/pnas.1105776108
Robideau, G. P., De Cock, A. W. A. M., Coffey, M. D., Voglmayr, H., Brouwer, H., Bala, K., et al. (2011). DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol. Ecol. Resources 11, 1002–1011. doi: 10.1111/j.1755-0998.2011.03041.x
Singh, A. (2011). Management of rhizome rot caused by Pythium, Fusarium and Ralstonia spp in ginger (Zingiber officinale) under natural field conditions. Ind. J. Agricultural Sci. 81, 268–270.
Stirling, G. R., Turaganivalu, U., Stirling, A. M., Lomavatu, M. F., and Smith, M. K. (2009). Rhizome rot of ginger (Zingiber officinale) caused by Pythium myriotylum in Fiji and Australia. Australasian Plant Pathol. 38, 453–460. doi: 10.1071/ap09023
Thambugala, K. M., Daranagama, D. A., Phillips, A. J. L., Kannangara, S. D., and Promputtha, I. (2020). Fungi vs. fungi in biocontrol: an overview of fungal antagonists applied against fungal plant pathogens. Front. Cell. Infect. Microbiol. 10:604923. doi: 10.3389/fcimb.2020.604923
Tian, M., and Kamoun, S. (2005). A two disulfide bridge Kazal domain from Phytophthora exhibits stable inhibitory activity against serine proteases of the subtilisin family. BMC Biochem. 6:15. doi: 10.1186/1471-2091-6-15
Tomada, S., Sonego, P., Moretto, M., Engelen, K., Pertot, I., Perazzolli, M., et al. (2017). Dual RNA-Seq of Lysobacter capsici AZ78 - Phytophthora infestans interaction shows the implementation of attack strategies by the bacterium and unsuccessful oomycete defense responses. Environ. Microbiol. 19, 4113–4125. doi: 10.1111/1462-2920.13861
Wei, L., Zhang, Q., and Zhou, D. (2019). Bio-Controlling Pythium oligandrum and its Application (translated). Patent Application Number CN201910757035.2. Beijing: China National Intellectual Property Administration.
Westermann, A. J., Barquist, L., and Vogel, J. (2017). Resolving host-pathogen interactions by dual RNA-seq. PLoS Pathog 13:e1006033. doi: 10.1371/journal.ppat.1006033
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York, NY: Academic Press), 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Yang, K., Dong, X., Li, J., Wang, Y., Cheng, Y., Zhai, Y., et al. (2021). Type 2 Nep1-like proteins from the biocontrol oomycete Pythium oligandrum suppress Phytophthora capsici infection in solanaceous plants. J. Fungi 7:496. doi: 10.3390/jof7070496
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., and Madden, T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi: 10.1186/1471-2105-13-134
You, X., Barraud, J., and Tojo, M. (2019). Suppressive effects of Pythium oligandrum on soybean damping off caused by P. aphanidermatum and P. myriotylum. Ann. Report Kansai Plant Protection Soc. 61, 9–13. doi: 10.4165/kapps.61.9
Keywords: Pythium oligandrum, bioeffector, dual RNAseq, antagonism, ginger, Pythium myriotylum
Citation: Daly P, Chen S, Xue T, Li J, Sheikh TMM, Zhang Q, Wang X, Zhang J, Fitzpatrick DA, McGowan J, Shi X, Deng S, Jiu M, Zhou D, Druzhinina IS and Wei L (2021) Dual-Transcriptomic, Microscopic, and Biocontrol Analyses of the Interaction Between the Bioeffector Pythium oligandrum and the Pythium Soft-Rot of Ginger Pathogen Pythium myriotylum. Front. Microbiol. 12:765872. doi: 10.3389/fmicb.2021.765872
Received: 27 August 2021; Accepted: 20 October 2021;
Published: 16 November 2021.
Edited by:
Vaibhav Srivastava, Royal Institute of Technology, SwedenReviewed by:
Filippo De Curtis, Università degli Studi del Molise, ItalyLauren Sara McKee, Royal Institute of Technology, Sweden
Copyright © 2021 Daly, Chen, Xue, Li, Sheikh, Zhang, Wang, Zhang, Fitzpatrick, McGowan, Shi, Deng, Jiu, Zhou, Druzhinina and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihui Wei, d2VpbGlodWlAamFhcy5hYy5jbg==
†These authors have contributed equally to this work
 Paul Daly
Paul Daly Siqiao Chen
Siqiao Chen Taiqiang Xue
Taiqiang Xue Jingjing Li3
Jingjing Li3 Taha Majid Mahmood Sheikh
Taha Majid Mahmood Sheikh David A. Fitzpatrick
David A. Fitzpatrick Jamie McGowan
Jamie McGowan Irina S. Druzhinina
Irina S. Druzhinina Lihui Wei
Lihui Wei