- 1Division of Molecular Genetics and Biochemistry, Molecular Biology Group, ICMR-National Institute of Cancer Prevention and Research, Noida, India
- 2ICMR-AIIMS Computational Genomics Centre, Division of Biomedical Informatics, Indian Council of Medical Research (ICMR), New Delhi, India
- 3Molecular Biology Group, ICMR-National Institute of Cancer Prevention and Research, Noida, India
Smokeless tobacco products (STPs) carry assorted microbial population that contributes to carcinogens synthesis like tobacco-specific nitrosamines (TSNAs). Extensive exploration of microbiota-harboring STPs is required to understand their full carcinogenic potential. Here, we applied 16S rRNA gene sequencing to investigate bacteriome present in moist STPs immensely consumed in India (Khaini, Moist-snuff, Qiwam, and Snus). Further, the functional metagenome was speculated by PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) to assign the abundance of genes related to nitrogen metabolism, bacterial toxins, antibiotic drug resistance and other pro-inflammatory molecules. Highly diverse bacterial communities were observed in all moist STPs. Taxonomic analysis revealed a total of 549 genera belonging to four major phyla Proteobacteria, Firmicutes, Bacteroidetes and Actinobacteria. Overall, the core bacterial genera Acinetobacter, Bacillus, Prevotella, Acetobacter, Lactobacillus, Paracoccus, Flavobacterium, and Bacteroides were significantly abundant in moist STPs. Elevated moisture-holding products like Moist-snuff and Qiwam harbor rich bacterial species diversity and showed similar bacteriome composition. Furthermore, Qiwam products showed the highest level of genes associated with nitrogen metabolism, antibiotic resistance, toxins, and pro-inflammation (predicted by PICRUSt) which can contribute to the synthesis of TSNAs and induction of oral cancer. The present broad investigation of moist STPs-associated bacteriome prevalence and their detailed metabolic potential will provide novel insight into the oral carcinogenesis induced by STPs.
Introduction
Non-combustible form of tobacco (smokeless tobacco) is used by >300 million people worldwide and majorly (>85%) in South Asian countries (Siddiqi et al., 2020). A report on global tobacco consumption documented that there are 199.4 million smokeless tobacco (SLT) users in India (GATS-2, 2017). SLT consumption is associated with cancer, cardiovascular diseases, nicotine addiction, diabetes mellitus, oral diseases, neuronal and reproductive defects (Carlsson et al., 2017; Centers for Disease Control and Prevention, 2020). These pathological effects are generally assigned to carcinogens present in STPs essentially tobacco-specific nitrosamine (TSNAs), benzo(a)pyrene and heavy metals (Critchley and Unal, 2003; Stepanov et al., 2008). Among 233 unique chemical compounds reported in STPs, TSNAs are the most abundant and potent carcinogens due to their high toxicity and ability to reprogram normal cells into neoplastic cells (Kaur et al., 2019; Sarlak et al., 2020). The STPs consumed in the South Asian region have higher TSNAs content in contrast to STPs available in Western countries (Nasrin et al., 2020).
The moist STPs are the most commonly used non-combustible variants of tobacco in India (GATS-2, 2017). The various categories of moist SLT products available in India include Khaini, Moist-snuff, Qiwam (Kiwam), and Snus (Gupta and Ray, 2003). The SLTChemDB database provides details about the physicochemical properties, biological information, toxicological effects and information of chemicals found in moist STPs (Kaur et al., 2019). The moist STPs are not only different in their composition and manufacturing process but also possess variable levels of TSNAs (Stepanov et al., 2017; Kaur et al., 2019; Shaik and Maddu, 2019). Moisture content and storage temperature are significantly responsible for the synthesis of high levels of TSNAs in tobacco products (Shi et al., 2013; Law et al., 2016). Storage conditions and aging also influence the TSNA levels in STPs that may be due to changing microbial diversity within these products (Chong et al., 2020).
The microbiota has been previously reported in cured tobacco leaves and STPs using both culture-dependent and culture-independent approaches (Di Giacomo et al., 2007; Han et al., 2016; Tyx et al., 2016; Smyth et al., 2017; Mehra et al., 2020). The microbiome of tobacco products participates in the synthesis of TSNAs. These microbes produce nitrite from nitrate and these nitrite molecules further react with various alkaloids available in the tobacco products and generate TSNAs (Shi et al., 2013; Chopyk et al., 2017). Studies on the SLT associated bacteriome showed the presence of species belonging to clinically relevant bacterial classes like Actinobacteria (Corynebacterium, Mycobacterium, and Propionibacterium), Bacteroidia (Prevotella and Porphyromonas), Bacilli (Bacillus, Listeria, Staphylococcus, and Streptococcus), Chlamydiae, Clostridia (Clostridium), Fusobacteria (Fusobacterium and Streptobacillus), α-Proteobacteria (Brucella and Rickettsia), β-Proteobacteria (Neisseria and Spririllum), γ-Proteobacteria (Haemophilus and Pseudomonas), Spirochete (Borrelia and Leptospira) and Mollicutes (Mycoplasma) (Gholizadeh et al., 2016; Al-Hebshi et al., 2017; Monika et al., 2020). Furthermore, SLT-associated microbes can influence systemic inflammation and supplementary signaling pathways associated with oral cancer progression because they produce toxins and other pro-inflammatory molecules (Rivera et al., 2020; Sajid et al., 2021). Additionally, SLT-associated microbiota can be contaminated with pathogenic microbes and possibly a source of antibiotic resistance threat (Rivera et al., 2020).
Despite the significant potential of STP associated microbes to form TSNAs, their presence in various STPs is still not well explored and significant metagenome analysis has not been conducted on STPs, especially those available in India. Therefore in this study, we have selected 11 moist SLT products based on high consumption and elevated moisture content which can facilitate microbial growth inside these STPs. The bacterial community structure was investigated by 16S rDNA and prediction of critical functional genes was performed related to nitrogen metabolism, antibiotic drug resistance, production of toxins and pro-inflammatory molecules. Hence, comprehensive metagenome analysis was performed using the MicrobiomeAnalyst platform to unveil the structural and functional bacteriome present in moist STPs especially consume in India.
Materials and Methods
Tobacco Samples and DNA Extraction
The domestic moist STPs were purchased from vendors in the Delhi and Uttar Pradesh state of India. The 16S rDNA sequencing techniques were used to analyze the bacterial diversity of 11 moist STPs from 4 different categories including Khaini (K1, K2, and K3), Moist-snuff (MS1, MS2, and MS3), Qiwam (Q1, Q2, and Q3), and Snus (S1 and S2) samples. The STPs were stored at −20°C to inhibit the further growth of microorganisms. The STPs were open in the sterilized conditions and microbial metagenomic DNA was isolated with a Power-soil DNA isolation kit as per the protocol provided by the manufacturer (Qiagen, Bangalore, India). The purity and quantity of isolated metagenomic DNA from the different STPs was checked by NanoDrop (Thermo, Bangalore, India). Further, the quality of metagenomic DNA was confirmed agarose gel (1%) before amplification by PCR. The metagenomic DNA concentration of all tested SLT products was found to be >30 ng/μl.
Bacterial 16S rDNA Amplification and Library Preparation
The PCR was performed to amplify the 16S rDNA V3–V4 region of metagenomic DNA isolated from the different moist STPs. The extracted metagenomic DNA (40 ng) was amplified with a pair of universal primers (10 pM of each) (FP: 5′-AGAGTTTGATGMTGGCTCAG-3′ and RP: 5′-TTACCGCGGCMGCSGGCAC-3′) as described earlier (Kroes et al., 1999). Along with primers and metagenomic DNA, a master mix was added containing dNTPs (0.5 mM), MgCl2 (3.2 mM), high-fidelity DNA polymerase and PCR enzyme buffer. The PCR amplification was performed with the following condition: 95°C for 3 min chase by 25 cycles at 95°C/15 s, 60°C/15 s and 72°C for 120 s and with a final elongation at 72°C for 10 min. The amplified 16S PCR amplicons were purified and subjected to agarose gel (2%) and NanoDrop for quality check.
Sequencing of Prepared Library
The Ampure beads (Beckman Coulter Inc., Indianapolis, IN, United States) were used to purify the amplicons of each sample by eliminating the unused primers and to prepare the sequencing libraries an additional 8 cycles of PCR was executed via Illumina barcoded adapters (Supplementary Table 1). Further, prepared libraries were purified by Ampure beads and quantified with the help of a QuDye-dsDNA HS assay kit (Thermo Fisher, Bangalore, India). The Illumina Miseq with a 2 × 300PE v3 sequencing kit was used for sequencing at Biokart India Pvt. Ltd., Bengaluru, India. The raw data was submitted to NCBI Short Read Archive (SRA) under BioProject accession number PRJNA767533.
Bioinformatics Analysis
The raw data binary base call (BCL) file received from the sequencer was de-multiplexed into FASTQ format. Quality control checks of raw sequenced data were performed by FastQC (Version 0.11.9) and MultiQC (Version 1.10.1) tools. Next, removal of contaminant adapters and trimming of low-quality reads were done by TrimGalore (Version 0.6.6)1. The QC passed samples were again analyzed by Quantitative Insights Into Microbial Ecology (QIIME version 1.9.0) workflow including fusion of paired-end reads, chimera elimination, OTU clustering and taxonomy assignment (Caporaso et al., 2010). The QIIME workflow facilitates precise exploration at the genus level. The Kraken2 with database NCBI was used as a reference for OTU picking (Wood et al., 2019). Further, data was filtered on a web-based platform MicrobiomeAnalyst to eliminate the low quality or uninformative features using minimum count = 4, 20% prevalence and low variance filter (Inter-Quartile range – 10%) (Chong et al., 2020). A total of 148 low abundance features were separated based on prevalence and a total of 28 low variance features were removed based on inter-quartile range. The 247 number features remain after the data filtering step. All analyses like rarefaction curve, α-diversity, principal coordinate analysis (PCoA), core microbiome, cluster study, random forest, Linear discriminant analysis Effect Size (LEfSe) and Sparse Correlations for Compositional data (SparCC) were performed by MicrobiomeAnalyst2. Subsequently, the metabolic pathway analysis was executed using the PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) algorithm (Langille et al., 2013). The functional genes associated with bacteriome of moist STPs were derived from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa and Goto, 2000). Next, Taxon Set Enrichment Analysis (TSEA) module of MicrobiomeAnalyst was performed to identify the biologically or ecologically meaningful patterns of STP associated bacteriome by analyzing them with context to pre-defined taxon set. To check the reproducibility of sequencing results, duplicates of Snus samples (S1_dup and S2_dup) were taken.
Statistical Analysis
The relative abundance percentage of each OTUs of taxonomic classification was calculated and plotted using Origin software. For data set phyla, we removed OTUs < 10 reads and for the data set genus, we removed OTUs < 100 reads. Analysis of Variance (ANOVA) was used to determine differences between products. A p-value < 0.05 was considered statistically considerable.
Results
Bacterial Community Composition and Diversity in Moist Smokeless Tobacco Products
After all pre-processing steps, an OTU table was generated and the number of amplicons obtained for each moist STPs varies between 10,601 and 131,082 (Supplementary Table 2 and Supplementary Figure 1). Most of the moist STPs showed an increase in observed bacterial species richness and diversity (Supplementary Figure 2). The numbers of OTUs were found to increase as the sequencing depth increases and saturation of species richness was observed in the rarefaction curves for all the tested moist STPs (Supplementary Figure 2). However, the numbers of observed bacterial species were less in K3 and S2 as compared to other moist STPs. Interestingly, the STPs having the highest moisture content such as Moist-snuff and Qiwam exhibited higher overall species diversity than Khaini and Snus which have comparatively low moisture content (Supplementary Figure 2 and Supplementary Table 3).
Further, OTUs richness was estimated by determining α-diversity (within-sample diversity) indices (Chao1, Fisher, Shannon, and Simpson) of STP-associated bacteriome. The Moist-snuff and Qiwan showed increased diversity indices as compared to Khaini and Snus suggesting that the Moist-snuff and Qiwam products have higher expected species richness of the bacteriome (Supplementary Figure 3 and Table 1). However, there was no statistically significant change observed among the four groups of moist STPs (Supplementary Figure 3). All the moist STPs displayed Good’s estimator values > 99% suggesting that the majority of bacterial species in the sample have been detected (Table 1).
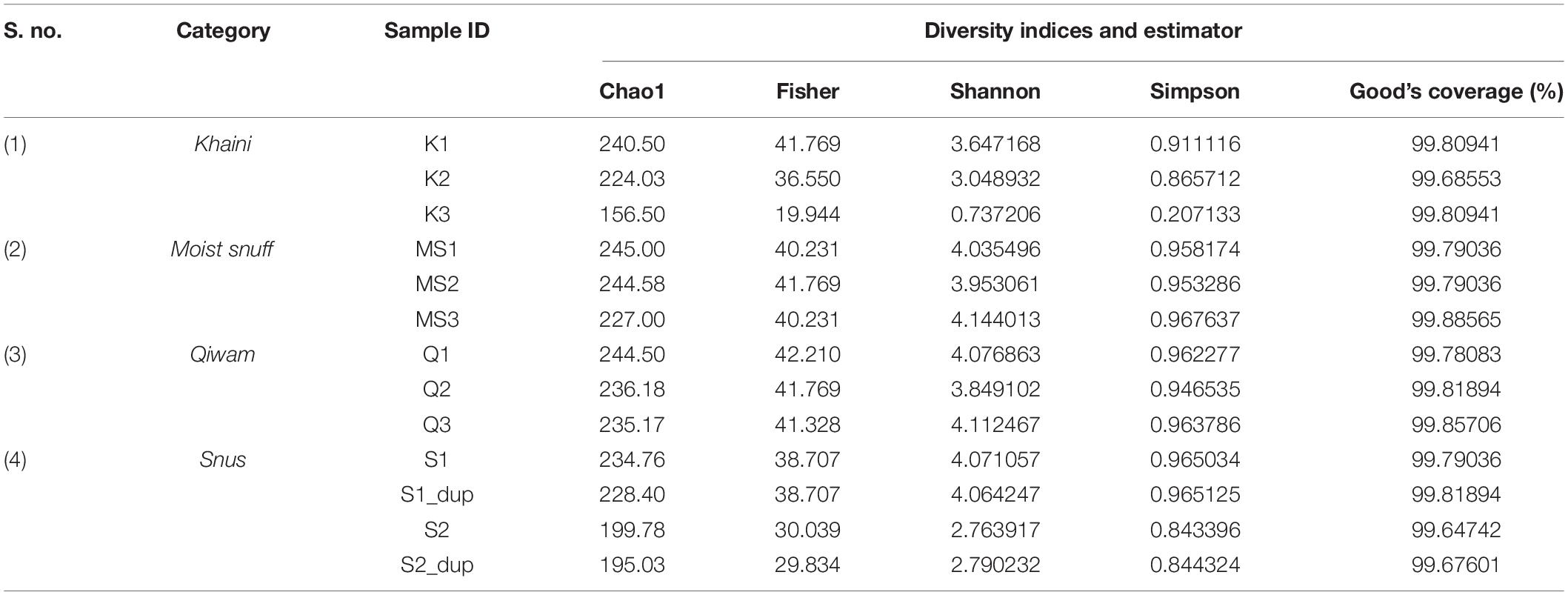
The β–diversity indicates differences in the bacterial community profile between the samples. To calculate β–diversity among the moist STPs, the Bray–Curtis dissimilarity metric was determined from the OTU abundance and exploited in Principal Component Analysis (Goodrich et al., 2014). The Permutational Multivariate Analysis of Variance (PERMANOVA) algorithm on Bray–Curtis dissimilarity was applied to construct Principal Coordinate Analysis (PCoA) plots (Kelly et al., 2015). PERMANOVA analysis of Bray–Curtis dissimilarities revealed that the bacteriome of each group was highly dissimilar (PERMANOVA; F = 2.9354, R2 = 0.49456, p < 0.004) (Supplementary Figure 4). The 3D-PCoA plot displayed that the Moist-snuff and Qiwam samples showed close association to each other and therefore, restrain more related bacteriome profiles (Figure 1A). Next, one sample from Khaini (K1) and one sample from Snus (S1) was also found to be close to each other and clustered together with Moist-snuff and Qiwam products and the sample K3 and S2 clustered separately from the other samples (Figure 1A). The interactive PCoA 3D plots at the level of the genus were constructed and dissimilar samples K3 and S2 showed a higher abundance of genera Actinobacteria and Prevotella, respectively (Figures 1B,C).
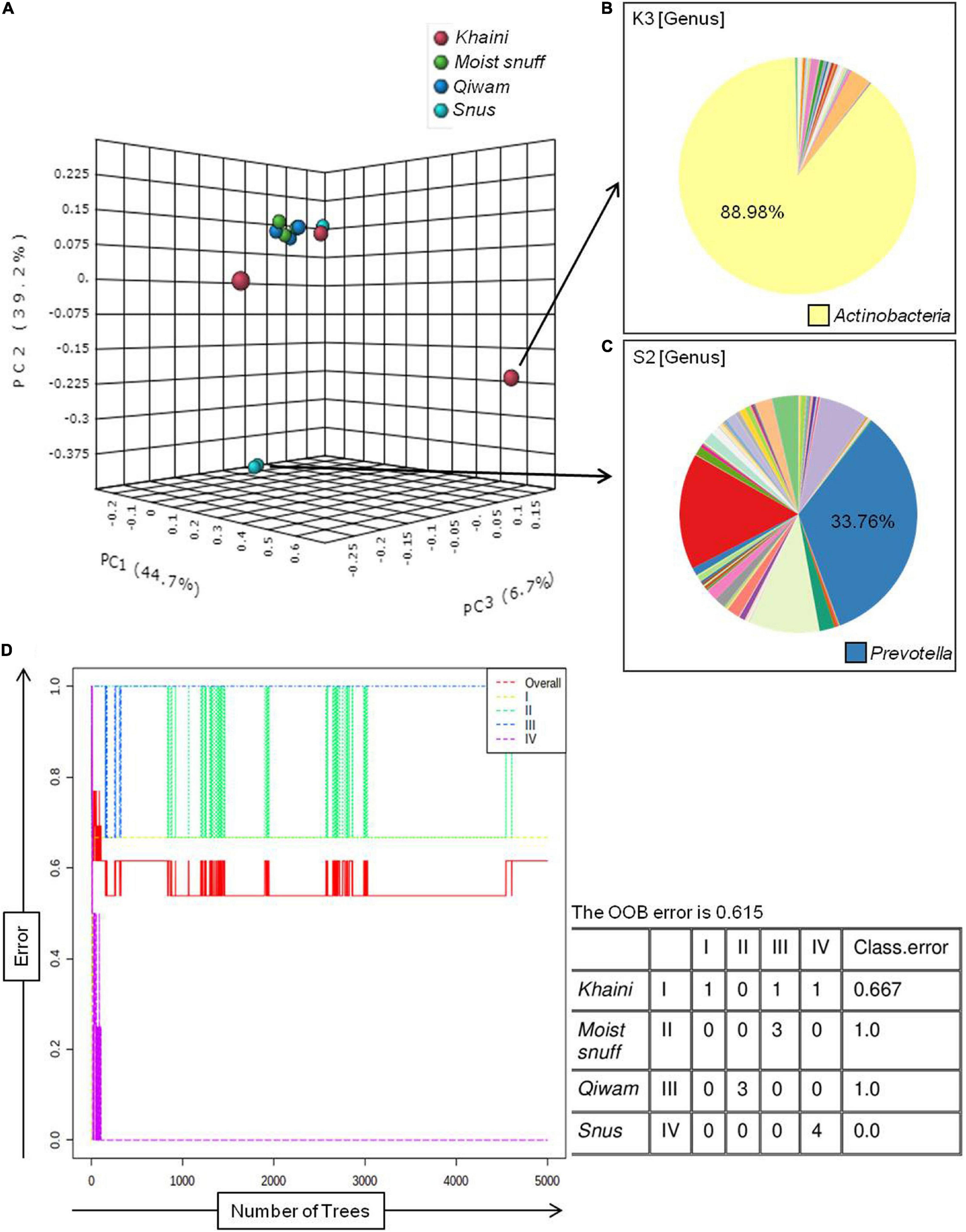
Figure 1. β–diversity among moist smokeless tobacco products. Interactive 3D-Principal Coordinate Analysis (PCoA) plot for bacterial β-diversity in moist STPs and pie chart generated by the MicrobiomeAnalyst. (A) PCoA plot of 11 moist STPs derived from Bray–Curtis index showing the distance of bacterial communities present in Khaini, Moist-snuff, Snus, and Qiwam samples. The samples of each group are represented by different color as indicated on the above of the figure. The pie charts, (B) Khaini sample, K3 and (C) Snus sample, S2 are shown at the level of genera. (D) The error plot originated from random forest analysis. Overall genera present in moist STPs was represented by a red line, yellow-line indicate the distinct genera present in Khaini, green-line showed specific genera of Moist-snuff, blue-line represent unique genera of Qiwam and magenta line specify exclusive genera of Snus.
Further, a ‘Random Forest’ algorithm was applied to validate the similarity and dissimilarity in bacteriome among the four groups of moist STPs (Figure 1D). The decision tree constructed from the random forest classification recognized distinctive bacteriome in moist STPs. In the error plots identified from random forest analysis, the red line represents overall genera present in moist STPs while distinct genera present in Khaini, Moist-snuff, Qiwam, and Snus were indicated by yellow, green, blue, and magenta-line, respectively (Figure 1D). Among all three samples of Khaini, one sample contained unique genera and two samples demonstrated overlapping genera with Qiwam and Snus; whereas Moist-snuff and Qiwam products showed resemblance with each other. The Snus products exhibited a unique genera profile (Figure 1D).
Taxonomic Distribution of Dominant Bacterial Communities in Moist Smokeless Tobacco Products
Bacterial populations recognized in moist STPs were first analyzed at the phyla level (Figure 2A). There were 4 major phyla Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes (range 89–98%) were observed in all moist STPs. The other notable phyla were Acidobacteria, Chloroflexi, Cyanobacteria, Fusobacteria, Gemmatimonadetes, Spirochaetes, and Verrucomicrobia. The relative abundance of phylum Proteobacteria was found to be majorly present in all STPs except S2 (Figure 2A). The highest proportion of Proteobacteria was observed in the Khaini group (K1–91%, K2–57%, and K3–50%). Moist-snuff (MS1–41%, MS2–49%, and MS3–43%), Qiwam (Q-41% and Q2–50%), and Snus (S1–39%) also showed an increased level of Proteobacteria phyla compared to other phyla. In addition, phyla Firmicutes was the predominant phyla in S2 (60%) with the lowest presence in K3 (4%) (Figure 2A). The third most prevalent phylum identified in all STPs was Bacteroidetes (Figure 2A). However, Khaini had a lower abundance (range 2–14%) of Bacteroidetes as compared to Moist-snuff, Qiwam, and Snus (range 15–23%). The phyla Actinobacteria was noticeably present in all moist STPs ranging from 1 to 10% (Figure 2A).
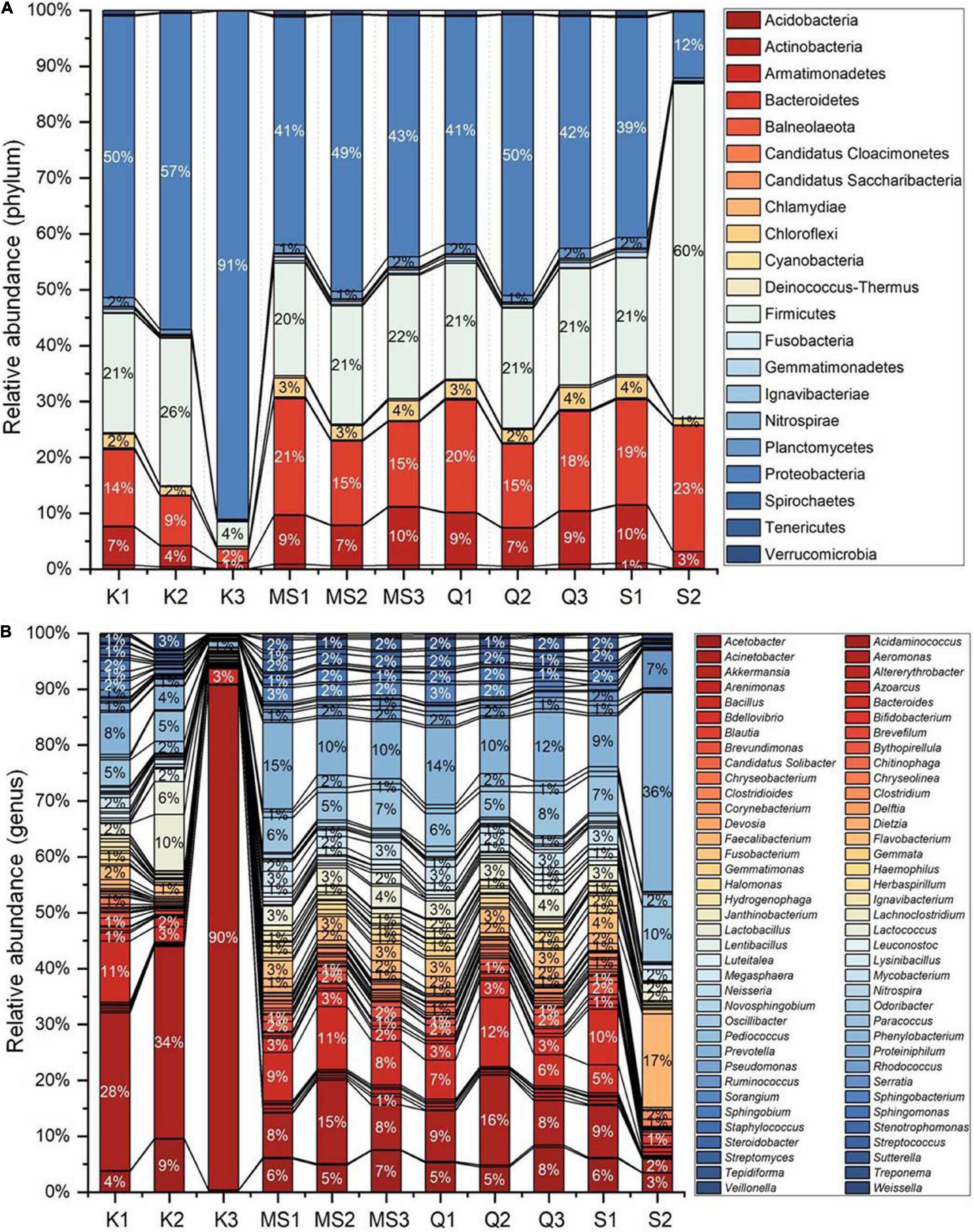
Figure 2. Bacterial phyla of moist smokeless tobacco products. (A) The stacked bar showed the relative abundance of bacterial phyla identified in each STPs. The OTUs > 10 reads were represented in their relative abundance. (B) The stacked bar showed the relative abundance of bacterial genera identified in each STPs. The OTUs > 100 reads were represented in their relative abundance. Each bacterial phylum is symbolized as a sequential color (red to blue) in the stacked bar graphs with the connecting lines. The entire relative abundance was calculated as 100% for each product.
A total of 549 genera were identified in all moist STPs and OTUs > 100 reads showed 61 genera were in relative abundance (Figure 2B). The genus Acinetobacter was abundantly observed in Khaini (90% of K3, 34% of K2, and 28% of K1). Another genus Prevotella was found to be significantly high in one of the Snus (36% of S2). However, a moderate level of Prevotella was observed in Moist-snuff (MS1-15%, 10% of MS1 and MS2), Qiwam (14% of Q1, 12% of Q3, and 10% of Q2) and Snus (9% of S1). The other important genera observed in STPs were Faecalibacterium (17% of S2), Bacillus (12% of Q2, 11% of K1 and MS2, and 9% of MS1), Lactobacillus (10% of K2), and Ruminococcus (7% of S2) (Figure 2B).
Core Bacteriome of Moist Smokeless Tobacco Products
Despite inter-product variability, there was a core bacteriome identified in the moist STPs that remain unchanged in their composition across different groups of moist STPs. Core bacteriome investigation was executed at the genus level according to sample prevalence ≥ 20% and relative abundance ≥ of 0.2% (Figure 3 and Supplementary Table 4). The 22 core bacterial genera were identified and the prevalence of Acinetobacter, Bacillus, and Prevotella genera was observed in moist STPs (Figure 3). Additionally, Acetobacter, Lactobacillus, Paracoccus, Flavobacterium, and Bacteroides were found to be the dominant core bacteria in moist STPs (Figure 3).
Figure 3. Core bacteriome of moist smokeless tobacco products. The core bacterial genera in moist STPs determined by applying the parameters sample dominance (≥20%) and relative abundance (≥0.2%). Heatmap illustrating the detection threshold and relative abundances of the most dominant bacterial genera in tested moist STPs. The color key shows the range of threshold relative abundance of the individual values.
Clustering Analysis of Moist Smokeless Tobacco Products and Their Associated Bacterial Genera
The best correlation among samples of moist STPs at the OTU level was determined using the Bray–Curtis index (Supplementary Figure 5). The dendrogram showed similarities between the products and 11 moist STPs clustered into three groups. Group-I consists of two Khaini products K1 and K2 whereas K3 was clustered with Q2 and MS2 into a subgroup of Group-II. The high moisture containing products such as Moist-snuff and Qiwam clustered together into subgroups of Group-II (Supplementary Table 3 and Supplementary Figure 5). Therefore, the high moisture content may lead to similar bacterial diversity in moist STPs. Group-III comprises Snus samples S2 and S_2 dup whereas S1 clustered with their respective sample in Group-II which confirmed that the sequencing method produced reproducible data (Supplementary Figure 5).
Additionally, a hierarchical clustering heat map was generated for improved visualization of the distinct bacterial genera abundance across different moist STPs (Supplementary Figure 6). For Khaini samples the dominant genera included Acinetobacter, Staphylococcus, Panacibacter, Citrobacter, Pediococcus, Lactococcus, Weissella, Lactobacillus, and Leuconostoc. The Moist-snuff samples showed the abundance of genera such as Mycoplasma, Pantoea, Filifactor, Trueperella, Lysinibacillus, Marinobacter, Frankia, Campylobacter, Olsenella, Aeromonas, Dolosigranulum, Agromyces, Collinsella, and Fuerstia (Supplementary Figure 6). The Qiwam products demonstrated the dominance of Chelativorans, Proteus, Capnocytophaga, Microvirga, Methylobacterium, Turicibacter, Parvibaculum, and Simkania genera. The Snus products illustrated the occurrence of genera Tabrizicola, Sulfuritortus, Fimbrimonas, Anaeromyxobacter, Caloramator, Immudisolibacter, Mannheimia, Dermabacter, Ruminococcus, and Lachnoclostridium (Supplementary Figure 6). The genera having clinical relevance like Pseudomonas, Haemophilus, Actinomyces, Neisseria, Streptococcus, Campylobacter, Corynebacterium, Porphyromonas, and Fusobacterium were abundant in Moist-snuff products while Prevotella, Faecalibacterium and Clostridium were high in Snus product S2. The genus Capnocytophaga and Bacillus was elevated in Q1 and Q2 product, respectively (Supplementary Figure 6).
Co-occurrence Network of Bacterial Genera Associated With Moist Smokeless Tobacco Products
To identify potential interactions within the moist STPs bacterial communities, a co-occurrence network at the level of genus was constructed using a compositional robust method SparCC correlation coefficient that formulate a strong assumption of a sparse correlation network (Friedman and Alm, 2012). Overall, the prevalence of 247 genera were found to be considerably different between the groups of moist STPs and to aid interpretation, nodes were colored according to their phylum (Supplementary Figure 7A). Altogether, 245 positive and 238 negative considerable correlations (coefficient correlation > 0.3 and p-value < 0.05) were observed between 247 genera (Supplementary Table 5). The co-occurrence pattern of relevant genera, significantly associated with pre-cancer lesion or oral cancer, in moist STPs were examined in detail (Halboub et al., 2020; Sarkar et al., 2021; Srivastava et al., 2021). The genus Prevotella was found to be correlated positively with Clostridium (SparCC = 0.9719, p = 0.0099), Bifidobacterium (SparCC = 0.9332, p = 0.0099), Treponema (SparCC = 0.9332, p = 0.0099), Mycobacterium (SparCC = 0.8094, p = 0.0099), Rhodococcus (SparCC = 0.7948, p = 0.0099), and Lactobacillus (SparCC = 0.7294, p = 0.0396) while it was negatively correlated with Corynebacterium (SparCC = −0.9622, p = 0.0099) and Capnocytophaga (SparCC = −0.6792, p = 0.0297) (Figure 4 and Supplementary Table 5). Streptococcus was positively correlated with Veillonella (SparCC = 0.9237, p = 0.0099) and Haemophilus (SparCC = 0.8739, p = 0.0099), whereas Fusobacterium showed positive association with Capnocytophaga (SparCC = 0.7415, p = 0.0297) and Lautropia (SparCC = 0.9473, p = 0.0099). Another important genera Pseudomonas displayed positive concurrence with Treponema (SparCC = 0.9149, p = 0.0198), Mycobacterium (SparCC = 0.9027, p = 0.0099), Haemophilus (SparCC = 0.8982, p = 0.0297), Prevotella (SparCC = 0.8000, p = 0.0099), Acinetobacter (SparCC = 0.8247, p = 0.0099), Bifidobacterium (SparCC = 0.8186, p = 0.0099), Streptomyces (SparCC = 0.8592, p = 0.0099), and Clostridium (SparCC = 0.8345, p = 0.0099) (Figure 4 and Supplementary Table 5). The correlation network plot is interactive and the genera Prevotella showed the highest abundance in Snus products compared to Khaini, Moist-snuff, and Qiwam (Supplementary Figure 7B).
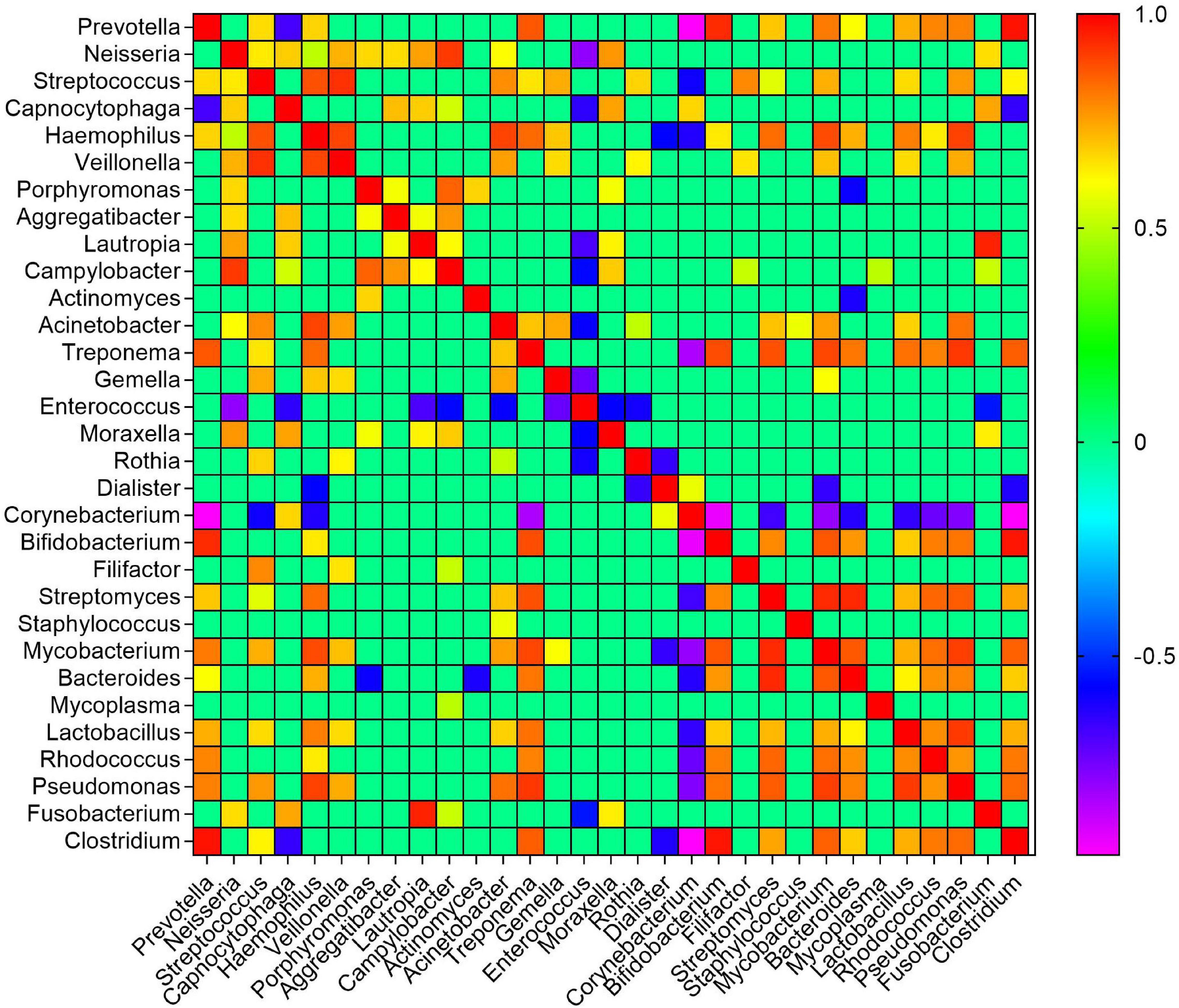
Figure 4. Co-occurrence of bacterial genera in moist smokeless tobacco products. The SparCC correlation of clinically relevant genera was generated and plotted in a heatmap. The scale bar on the right of the plot showed calculated positive and negative correlation values to generate the heatmap. The correlation threshold | >0.3 and p-value < 0.05.
Biomarker Detection by Linear Discriminant Analysis Effect Size
The robust biomarker of moist STPs was identified using a non-parametric statistical method LEfSe (Segata et al., 2011). LEfSe method discovers features with considerable differential abundance across the moist STPs and established the biomarker bacteriome at the genus level. Fifteen significant taxa were recognized as per the cutoffs values: FDR-adjusted p-value < 0.1 and linear discriminant analysis (LDA) > 2.0 (Figure 5A). The LDA score for Acinetobacter and Lactococcus were highest in Khaini products whereas Streptococcus and Neisseria had high LDA scores in Moist-snuff samples (Figure 5A). Further, the LDA score of Sphingobacterium, Janthinobacterium, and Pseudomonas was dominant in Qiwam, whilst that of Prevotella, Faecalibacterium, Oscillibacter, Ruminococcus, Blautia, Clostridium, Megasphaera, and Clostridioides was highest in Snus products (Figure 5A).
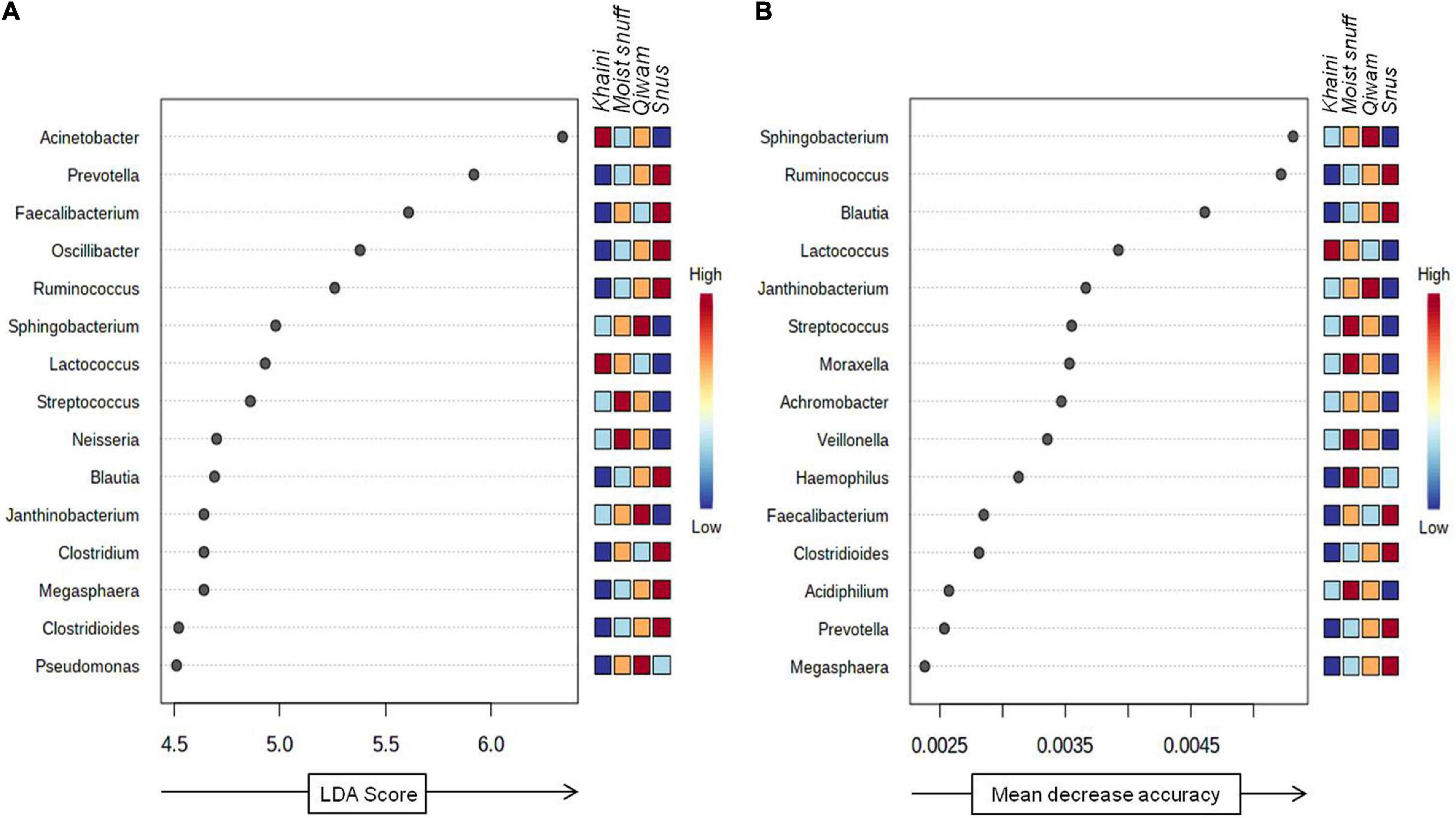
Figure 5. Biomarker analysis of moist smokeless tobacco products-linked bacteriome. (A) Linear discriminant analysis Effect Size (LEfSe) of bacteriome present in moist STPs. The significant 15 genera were ranked in declining order as per their LDA scores (x-axis). (LEfSe parameters; Taxonomic level-genus, FDR-adjusted p-value cut off <0.1, log LDA score > 2.0). (B) The significant feature was identified by random forest analysis. The variable importance calculated by mean decrease in accuracy of predictor genera in the Random Forest model and top 15 genera were ranked in increasing order as per their mean decrease accuracy value (x-axis). The right heatmap plot designates whether the genera abundance were high (red) or low (blue) in each group of moist smokeless tobacco products.
Next, to identify bacterial genera that differentiate between phenotypes, a random forest algorithm was applied to bacteriome data (Knights et al., 2011). The significant genera identified with random forest showed a pattern of changes across different groups of moist STPs (Figure 5B). At the genus level, the random forest model brought up Sphingobacterium, Ruminococcus, Blautia, Lactococcus, Janthinobacterium, Streptococcus, Moraxella, Achromobacter, Veillonella, Haemophilus, Faecalibacterium, Clostridioides, Acidiphilium, Prevotella, and Megasphaera (Figure 5B). The genera Sphingobacterium, Ruminococcus, and Blautia were the most decisive discriminated genera because of their higher predictive values.
Functional Capacity of Moist Smokeless Tobacco Products-Linked Bacteriome
The 16s rDNA sequencing data were used to infer the metabolic potential of bacteriome with PICRUSt which is derived from a phylogenetic distance or sequence similarity of identified microbes with the related microorganism whose whole genome has been sequenced and based on Greengenes annotated OTUs (Langille et al., 2013). The MicrobiomeAnalyst utilizes the PICRUSt algorithm and metagenome contributions were computed for all moist STPs based on Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology (KO term) (Kanehisa et al., 2013; Dhariwal et al., 2017). The result containing KO abundance level was generated and in sum 3695 KO terms were observed in the imputed metagenome of moist STPs (Supplementary Table 6). Several KEGG metabolic pathways were identified in moist STPs and their relative abundance in different moist STPs was monitored by MicrobiomeAnalyst (Supplementary Figures 8A,B).
Nitrogen Metabolism Potential of Moist Smokeless Tobacco-Linked Bacteriome
Microbial reduction of nitrates to nitrite that leads to formation of TSNAs involves the nitrogen metabolism pathway.
The two important pathways involved in the extracellular accumulation of nitrite are (i) dissimilatory nitrate reduction nar operon that includes regulators (narXL), transporters (narK) and nitrate reductases (narZHJI); (ii) periplasmic nitrate reductase nap operon (González et al., 2006). The dissimilatory nitrate reduction pathway genes (narK, narZ, narJ, narI, and narH) were abundant in the Q3 product and Q1, Q2, and MS2 products showed a significant number of imputed genes of the nitrate reduction pathway (Figure 6 and Supplementary Table 6). The periplasmic nitrate reductases gene napA was prevalent in one Snus product (S2) and noticeably present in Q3, Q1, and MS2 products. The assimilatory nitrate reductases genes nasA and nasB were also predicted by PICRUSt and abundantly monitored in Qiwam (Q1, Q2, and Q3), Moist-snuff (MS2) and Snus (S2) products. The predicted genes of nitrite reductases including nirA, nirB, nirD, nirK, and nrfA were found to be abundant in all three Qiwam products and one Moist-snuff (MS2) product, whereas nirB was also prevalent in one Snus product (S2) (Figure 6 and Supplementary Table 6). Another important step in nitrogen metabolism is denitrification in which nitrogenous compounds (nitrate, nitrite, and ammonia) were converted to nitrogen gas (N2). All moist STPs contained imputed genes related to denitrification nosZ (K00376), norB (K04561), and norC (K02305) but their abundance was very low (Supplementary Table 4). Further, nitrogen fixation related genes nifD1 (K02586), nifH (K02588), and nifK (K02591) were present in all moist STPs and the abundance of nifD1 and nifK were high in Q3, Q1, MS2, Q2, and K1 whereas nifH was prevalent in Q3, K2, K3, Q1, MS2, Q2, and K1 (Figure 6 and Supplementary Table 6).
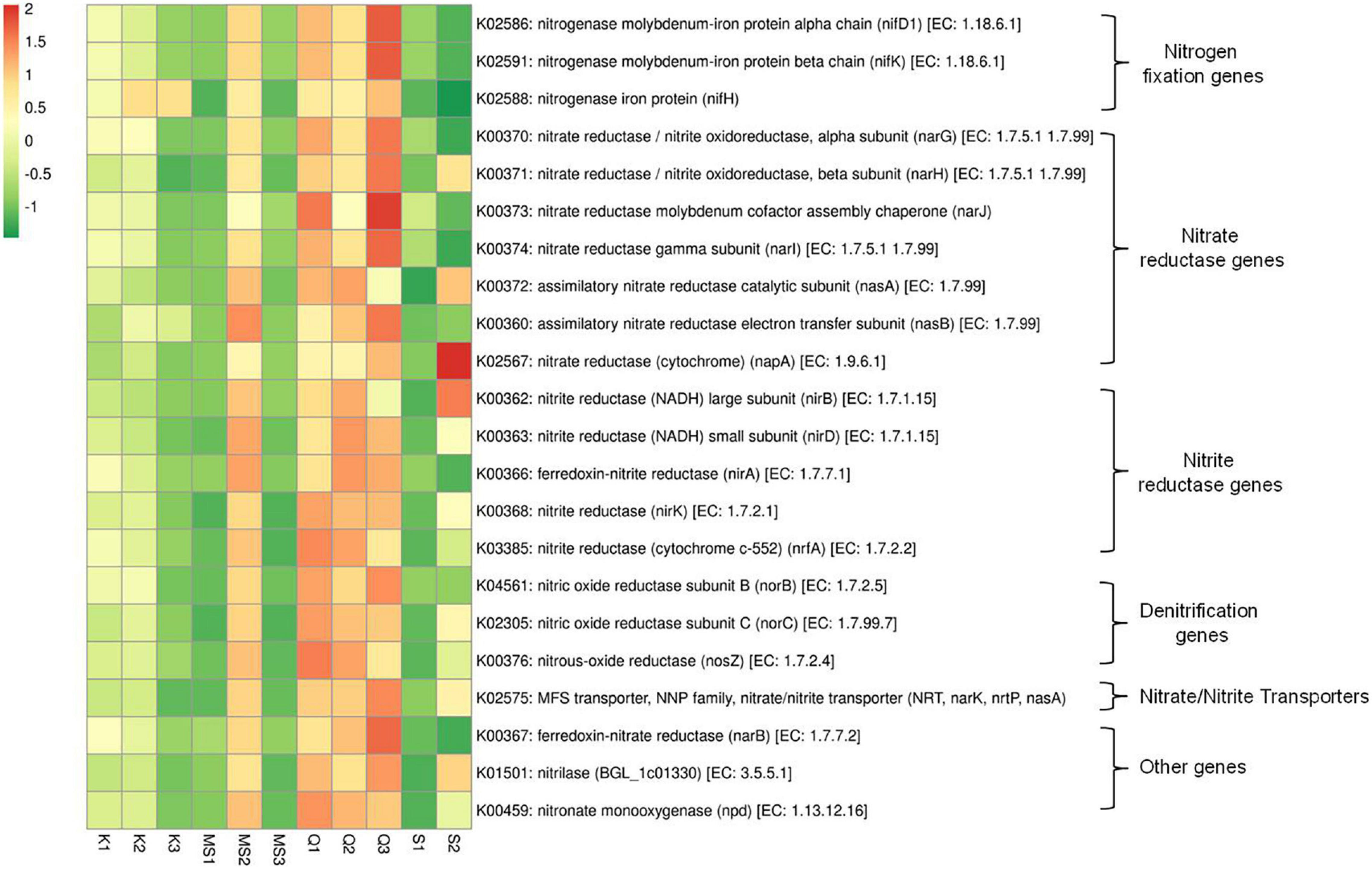
Figure 6. Relative abundance of nitrogen metabolism pathway genes in moist smokeless tobacco products. Heatmap displaying the predicted genes identified using PICRUSt (y-axis) based on KEGG database in each sample of moist smokeless tobacco products (x-axis). Each column represents a SLT sample and each row nitrogen metabolism gene with relative abundance indicated by color bar.
Antibiotic Drug Resistance Abundance in Bacteriome of Moist Smokeless Tobacco Products
The imputed metagenome of moist STPs through PICRUSt assists in the prediction of genes involved in antibiotic drug resistance. The highest number of antibiotic resistance genes was observed in all three products of Qiwam, one Moist-snuff product (MS2) and one Snus product (S2) as compared to Khaini products (K1, K2, and K3), MS1, MS3, and S1 products (Figure 7 and Supplementary Table 6). The multiple antibiotic resistance protein marc (K05595), penicillin-binding protein 1A mrcA (K05366) and penicillin-binding protein-4 (serine-type D-Ala-D-Ala carboxypeptidase/endopeptidase) dacB (K07259) was the two most abundant antibiotic resistance genes (Supplementary Table 6). The Q3 product showed genes having higher predicted prevalence such as macrolide-specific efflux system (macA, K13888), macrolide phosphotransferase (mph, K06979), macrolide efflux protein (ykuC, K08217), zinc D-Ala-D-Ala carboxypeptidase (vanY, K07260) and multidrug resistance protein (bcr/tcaB, K07552) (Figure 7 and Supplementary Table 6). The products Q2 and MS2 showed the prevalence of antibiotic resistance genes like multidrug-resistance protein (mdtG, K08161), aminoglycoside 3-N-acetyltransferase (aacC, K00662), penicillin-binding protein 1B (mrcB, K05365), and penicillin-binding protein 5/6 (dacC/dacA, K07258) (Figure 7 and Supplementary Table 6). The sample Q1 displayed the presence of zinc D-Ala-D-Ala dipeptidase (K08641), beta-lactamase induction signal transducer (K082184), and multidrug resistance proteins (emrB, K03446 and norB, K08170). The S2 products displayed antibiotic resistance genes including vancomycin resistance associated response regulator (vraR, K07694), small multi-drug resistance pump (emrE, K03297), macrolide transport system ATP binding/permease protein (macA, K05685), fosmidomycin resistance protein (tsr, K08223), and penicillin-binding protein activator (lpoB, K07337) (Figure 7 and Supplementary Table 6).
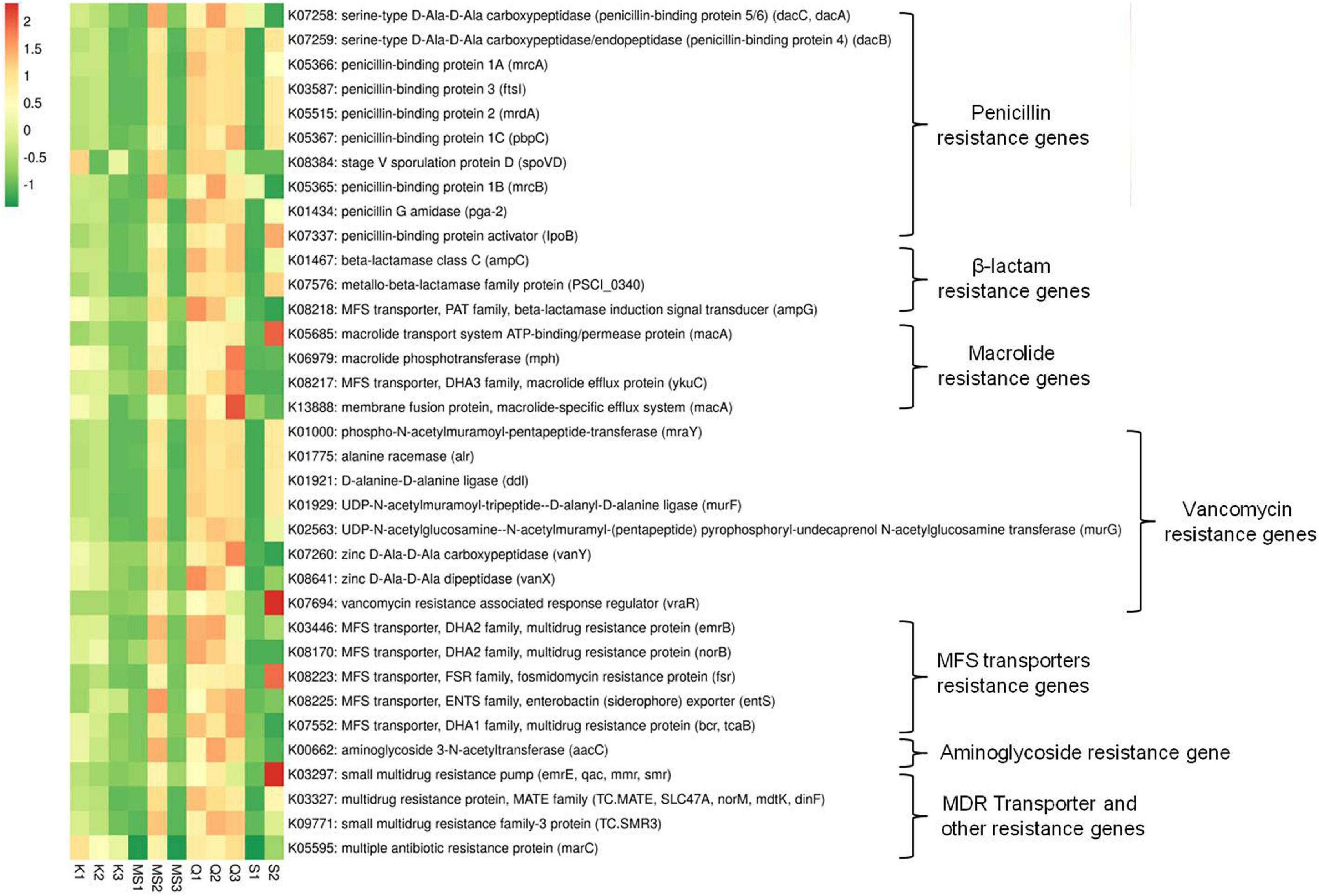
Figure 7. Relative abundance of antibiotics resistance genes in moist smokeless tobacco products. Heatmap displaying the predicted genes identified using PICRUSt (y-axis) based on KEGG database in each sample of moist smokeless tobacco products (x-axis). Each column represents a SLT sample and each row antibiotic resistance gene with relative abundance indicated by color bar.
Pro-inflammatory and Toxic Effect of Moist Smokeless Tobacco Products-Associated Bacteriome
Gram-negative bacteria lipopolysaccharide (LPS) is involved in the progression and migration of oral squamous cell carcinoma (OSCC) (He et al., 2015). The LPS inflammatory activity is due to the Lipid-A component that is known to activate pro-inflammatory cytokines (Zhang and Ghosh, 2000; Karpiński, 2019). The ABC transporter complex (lptBFG) responsible for LPS transport from the inner to the outer membrane was found in moist STPs and its abundance was high in Qiwam and MS2 products. In moist STPs several genes related to LPS biosynthesis were observed (Figure 8 and Supplementary Table 6). The abundance of genes Lipid-A-disaccharide synthase (lpxB) and Kdo2-lipid IVA lauroyltransferase/acyltransferase (lpxL) which were involved in LPS synthesis significantly increased in Qiwam (Q1, Q2, and Q3), MS2 and S2 products (Figure 8 and Supplementary Table 6).
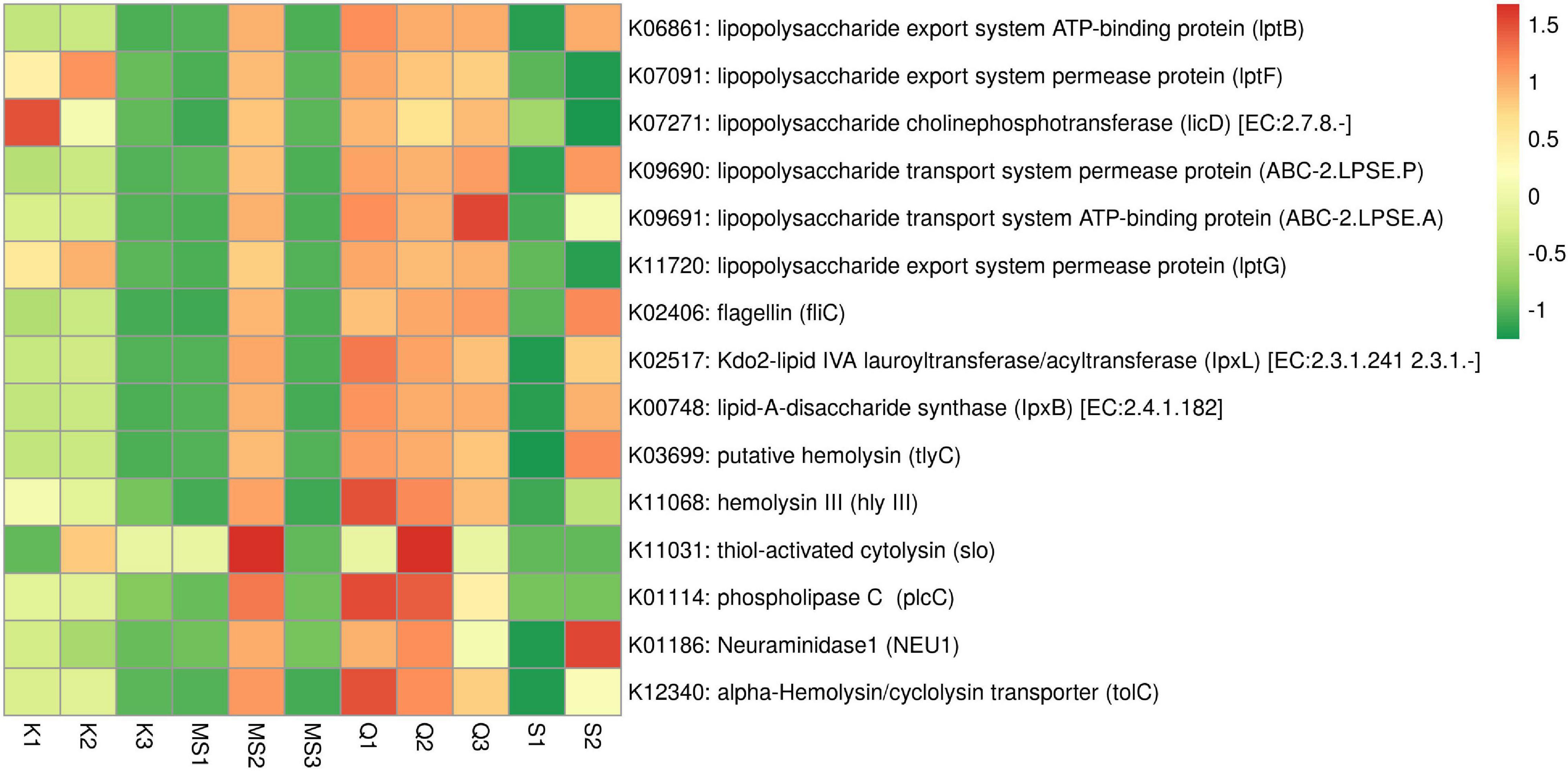
Figure 8. Relative abundance of imputed genes encoding toxins and pro-inflammatory molecules in moist smokeless tobacco products. Heatmap displaying the predicted genes identified using PICRUSt (y-axis) based on KEGG database in each sample of moist smokeless tobacco products (x-axis). Each column represents a moist STP sample and each row toxin/pro-inflammatory gene with relative abundance indicated by color bar.
Another potent pro-inflammatory molecule is Flagellin which participates in the motility of bacteria. Flagellin coding gene fliC was found in all moist STPs and their abundance was high in S2, Q3, Q2, MS2, and Q1 as compared to K2, K1, S1, MS3, K3, and MS1 (Figure 8 and Supplementary Table 6). However, the genes involved in the synthesis of other pro-inflammatory molecules like peptidoglycan, teichoic acid and lipoteichoic acid (K03739 and K03740) were not observed in any moist STPs (Supplementary Table 6).
Further, an important category of genes is related to bacterial toxins because these toxins can participate in the pathogenesis of inflammation leading to several diseases (toxinoses) and can induce genomic damage that can lead to neoplastic transformation of epithelial cells (La Rosa et al., 2020). The STP-associated bacterial toxins identification based on KEGG orthology displayed that bacterial species having toxins genes were present in moist STPs. The highest sequence hits were observed for alpha-Hemolysin/cyclolysin transporter gene tolC (K12340), followed by putative hemolysin gene (tlyC, K03699), phospholipase-C gene (plcC, K01114), Neuraminidase1 gene (NEU1, K01186), hemolysin III gene (hly III, K11068) and thiol-activated cytolysin gene (slo, K11031) (Figure 8 and Supplementary Table 6). The moist STPs Q1, Q2, MS2, and S2 showed a high abundance of toxin genes whereas Q3, K1, and K2 also contain a significant prevalence of these genes compared to K3, MS1, MS3, and S1 products (Figure 8 and Supplementary Table 6).
Taxon Set Enrichment Analysis of Moist Smokeless Tobacco Product Bacteriome
Next, we performed a Taxon Set Enrichment Analysis (TSEA) of all significant genera using the MicrobiomeAnalyst tool and examined them across 239 known taxon set linked with host-intrinsic factors such as age and diseases (Chong et al., 2020). The bacteriome of moist STPs showed strong correlations with colorectal cancer, Crohn’s disease and Rheumatoid arthritis (Table 2). Further, we parsed the bacteriome of moist STPs with 53 taxon sets associated with microbiome-intrinsic factors such as microbe motility and shape. The bacteriome of moist STPs displayed a significant correlation with indole producers and mucin degraders (Table 3).
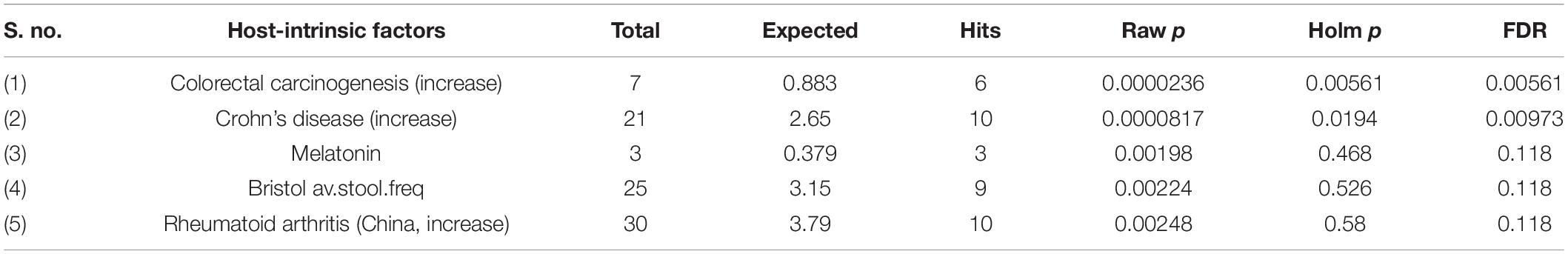
Table 2. Correlation of host-intrinsic factors (diseases) with bacteriome of moist smokeless tobacco products.
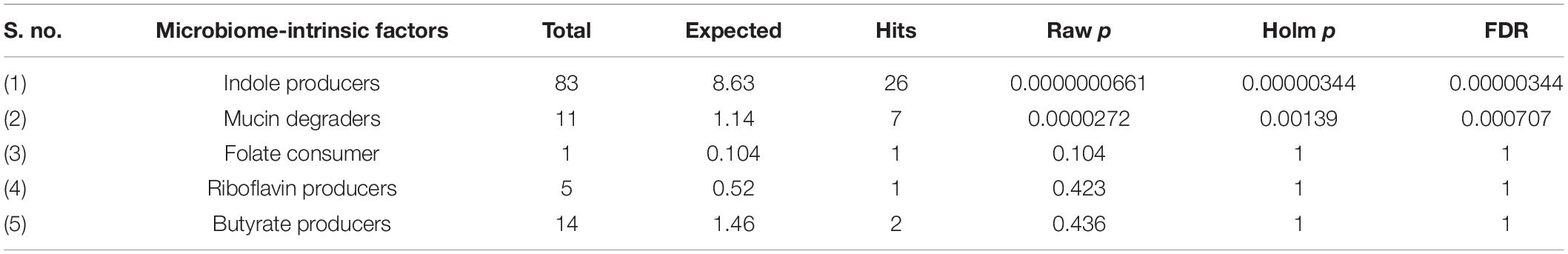
Table 3. Correlation of microbiome-intrinsic factor with bacteriome of moist smokeless tobacco products.
Discussion
The bacteriome of moist STPs was established by high throughput sequencing of 16S rDNA (V3-V4 region) sequence present in genomic DNA isolated from the STPs and first time expansively analyzed (community profiling, comparative analysis, and functional prediction) by MicrobiomeAnalyst tool (Dhariwal et al., 2017). Several previous studies have identified culturable and non-culturable microorganisms in STPs (Rubinstein and Pedersen, 2002; Han et al., 2016; Onuorah Samuel, 2016; Tyx et al., 2016, 2020). The genera such as Bacillus, Lactobacillus, Staphylococcus, Corynebacterium, Streptococcus, Prevotella, Rothia, Pantoea, Veillonella, Propionibacterium, Fusobacterium, Actinomyces, Lactobacillus, Sphingomonas, Marinilactibacillus, Oceanobacillus, and Porphyromonas were recognized in different STPs (Rivera and Tyx, 2021; Sajid et al., 2021).
In our study, we focused on the moist STPs because high moisture content plays a significant role in the increased level of TSNAs by facilitating the growth of the microorganisms (Gholizadeh et al., 2016; Han et al., 2016). An in vitro assay using epithelial cells (AMOL-III) from the oral leukoplakia of a Khaini user and treatment with aqueous extract of Khaini showed alteration in the expression of proteins involved in cell cycle regulation and DNA methylation, suggesting the role of Khaini in oral carcinogenesis (Rohatgi et al., 2005). Other moist STPs category consumed as Snus contains high levels of carcinogenic TSNAs (23.1–61.2 μg/g) (Stepanov et al., 2015). Qiwam products also retain a significant level of TSNAs (5.43–22.2 mg/kg) (Tricker and Preussmann, 1989).
The α–diversity of bacteriome present in different moist STPs was determined and we have observed that the species richness (α-diversity) was more in STPs having high moisture content such as Moist-snuff and Qiwam compared to products having low moisture levels like Khaini and Snus. In contrast, Tyx et al. (2016) observed that American Dry snuff products exhibited elevated overall species diversity compared to Moist-snuff samples. Al-Hebshi et al. (2017) found the highest species richness in Swedish Snus products and lowest for the Yemeni Shammah product. Likewise, Monika et al. (2020) observed that Indian Snus products have increased α–diversity as compared to less moist STPs like Chewable tobacco and Snuff. The prevalence of bacteria in moist STPs may be due to the hot and high humid ambiance of India which can facilitate the growth of microorganisms involved in increasing the fermentation rate of alkaloids present in STPs to carcinogenic TSNAs. Further, β–diversity analysis of moist STPs, based on Bray–Curtis dissimilarity metric, revealed that the products of Moist-snuff (MS1, MS2, and MS3) and Qiwam (Q1, Q2, and Q3) clustered together and a clear bacterial community similarity was localized between Moist-snuff and Qiwam products whereas distinct separation of Khaini and Snus products was observed. Following our study, American Moist-snuff products also clustered mutually after PCoA analysis based on Weighted UniFrac distances (Tyx et al., 2016; Al-Hebshi et al., 2017).
The moist STPs contain complex communities of bacterial species. The dominant phylum in all moist STPs was Proteobacteria followed by Firmicutes, the findings being similar to a previous study (Zhou et al., 2020). However, studies conducted on American moist STPs reported Firmicutes as the most abundant phylum (Han et al., 2016; Tyx et al., 2016; Al-Hebshi et al., 2017). In our study, we targeted the V3-V4 section of the 16S rRNA gene and observed the several abundant bacterial genera Acetobacter, Acinetobacter, Bacillus, Bacteroides, Faecalibacterium, Lactobacillus, Oscillibacter, Paracoccus, Prevotella, Pseudomonas, and Ruminococcus. However, a study on American Moist-snuff products metagenomic analysis (based on the V4 region of the 16S gene alone) observed several predominant genera Tetragenococcus, Aerococcus, Alloiococcus, and Staphylococcus (Tyx et al., 2016). While, another metagenomic study on American Moist-snuff products using the V1–V3 segment of the 16S rRNA gene identified genera Paenibacillus, Oceanobacillus, and Bacillus (Al-Hebshi et al., 2017). Further, a study on Indian STPs (Chewable tobacco, Snus, and Snuff) using entire 16S gene sequencing and analysis established the abundance of genera Staphylococcus, Bacillus, Corynebacterium, Virgibacillus, Brevibacterium, Rothia, Veillonella, and Fusobacterium (Monika et al., 2020).
The genera co-occurrence network analysis identified several significant relationships within the bacteriome of moist STPs. As revealed in our correlation analysis, the genera involved in the development of oral diseases showed a positive correlation independent of their phyla. Compared with previous studies on bacteriome of STPs, our study, for the first time identified numbers of distinguishing genera using the LEfSe method and random forest analysis. We have identified 15 genera associated directly with the moist STPs.
The most important pathway involved in TSNAs formation is the nitrogen metabolism pathway. Microbial fermentation forms nitrite from nitrate which reacts with numerous tobacco alkaloids to form different carcinogenic TSNAs molecules (Wang et al., 2017). During microbial respiration under anoxic conditions, nitrate reduction is an alternative respiratory pathway, nitrate acts as a terminal electron acceptor in place of oxygen, contributing to the oxidation of NADH (Igamberdiev and Hill, 2004). The nitrate is converted to nitrite by cytosolic nitrate reductase (nas, nap, and nar) and by membrane-bound nitrate reductase (Igamberdiev and Hill, 2004). Expression of nitrate reductases was found during the hypoxic condition that may result in extracellular nitrite accumulation during aging or storage of tobacco/tobacco products (Nishimura et al., 2007). Further, several bacteria contain nitrite exporting enzymes to regulate the level of nitrite as it can be toxic to the microbial cell. The nitrate and nitrite transport process are carried out by nitrate/nitrite anti-porters or nitrite extrusion transporters and determine the extracellular nitrite levels (Alvarez et al., 2019). Excreted nitrite can be metabolize by microbes having assimilatory or dissimilatory (denitrifying) pathways (Averill, 1996; Luque-Almagro et al., 2011). Furthermore, under optimal conditions N-nitrosation reaction of alkaloids with nitrite results in the formation of TSNAs (Wang et al., 2017). A few studies identified nitrogen metabolism genes in American STPs and Sudanese Toombak by whole metagenome and 16S rRNA gene metagenomics (Tyx et al., 2016; Rivera et al., 2020). Tyx et al. (2016) observed that the nitrate reductase genes (narGHJI), nitrite reductase genes (nirABC) and nitrate/nitrite transporters genes were significantly abundant in American dry snuff products. In our study, we have observed that narGHJI and nirABDK were abundant in moist STPs like Qiwam and Moist-snuff. The enhanced level of TSNAs in Qiwam may be due to a high level of the nitrogen metabolizing enzymes being able to contribute to the synthesis of TSNAs. Therefore, the identification of bacterial species performing nitrogen metabolism is crucial to decipher the carcinogenic potential of bacteria present in moist STPs.
A serious global threat of antimicrobial resistance spread lead to the emergence of multidrug-resistance bacteria or “Superbug” (Aslam et al., 2018). The spread of antibiotic resistance genes to oral microbiota of SLT users can be attributed to the practice of smokeless tobacco consumption (Lacoma et al., 2019). The whole metagenome sequencing and analysis of American-STPs showed the presence of several antibiotic resistance genes associated with resistance to β–lactam, penicillin, vancomycin, macrolides, aminoglycosides antibiotics, and other genes encode for multidrug transporters and efflux pumps (Rivera et al., 2020). The imputed metagenome of Indian-moist STPs like Q1, Q2, Q3, MS2, and S2 displayed a prevalence of antibiotic resistance genes having the potential to deactivate antibiotics. The multidrug efflux pumps of the major facilitator superfamily (MFS) can uniport small molecules and provide a noteworthy mechanism of bacterial resistance to antimicrobial compounds (Kumar et al., 2016). Several MFS transporters resistance genes were identified in moist STPs. Hence, moist STPs used in India can be a source of antibiotic resistance genes and are capable of spreading these genes to human microbiota and make them difficult to treat.
Bacterial association with mucosal lining can deliver bacterial products, like LPS (Gram-negative bacteria) that stimulate many cell types and can contribute to OSCC progression (Kurago et al., 2008). LPS can induce cytokines discharge (IL-6, IL-1β, IL-8, and TNF-α) upon attachment with the toll-like receptor (TLR receptor) causing the LPS induced inflammation (Zhang and Ghosh, 2000; Karpiński, 2019). Additionally, LPS activate TLR-4 of cancer cells and assist tumor cells immune-escape by preventing the action of cytotoxic T cells or natural killer (NK) cells (Huang et al., 2005). Gram-negative bacterium Shigella flexneri can inhibit apoptosis by suppressing the effector caspase activity by direct attachment of lipopolysaccharide (LPS) with caspases (Günther et al., 2020). The predicted metagenome of moist STPs include several genes related to LPS transport (ABC transporter complex, lptBFG) and their dominance was elevated in Qiwam and Moist-snuff. The abundance of LPS in moist STPs can provide cancer supporting environment and help in the progression of oral cancer in SLT users.
Studies are suggesting that the microbial-derived toxins (endotoxins, exotoxins and mycotoxins) may contribute to the health risks of STPs (Pauly and Paszkiewicz, 2011; Han et al., 2016). Gram-positive bacterial genera were found to produce hemolysins (pore-forming toxins). An extensively studied Pneumolysin (thio-activated cytolysin, K11031) secreted by Streptococcus pneumonia can induce cell death and inflammation by pore-forming cytolytic activity or by provoking the necrosis and program cell death pathways (Nishimoto et al., 2020). We observed that the abundance of Pneumolysin was high in Q2 and MS2 products. Therefore, the presence of Pneumolysin in these products can be attributed to generating host tissue injury especially oral layer damage during chewing of STPs.
In this study bacteriome of moist STPs had a strong correlation with increased colorectal cancer in humans. This observation is corroborated with previous observation where Fusobacterium nucleatum (a Gram-negative bacterium) was found to be associated with colorectal cancer and oral cancer (Shang and Liu, 2018; Fujiwara et al., 2020). As we observe that the Fusarium genus is abundant in the Moist-snuff products, therefore, it can be postulated that the presence of Fusarium sp. in moist STPs products can contribute to the development of colorectal and oral cancer. Further, the mucous layer provides protection to oral cavity against pathogens. Initiation of pathogenesis is linked with mucin degradation by the bacteria because it would damage the protective host mucosal surfaces (Derrien et al., 2010). We have observed that TSEA analysis reveal the presence of mucin-degrading bacteria in moist STPs. Hence, mucin degradation in the oral cavity of SLT users by mucin-degrading bacteria can contribute to oral carcinogenesis.
Further, in this study, we collected the STPs from a restricted geographical location due to Covid-19 pandemic travel constraints. Since microbial content of STPs is dependent upon the climatic and storage conditions which may vary across the country. Therefore, a broad spectrum of STPs from different regions of India needs to be inspected for a more definitive outcome.
Conclusion
All moist STPs harbor diverse bacteriome with the prevalence of harmful bacteria genera Acinetobacter, Bacillus, Prevotella, Faecalibacterium, and Pseudomonas. Moist-snuff, and Qiwam products were more significantly diverse and showed similar bacterial diversity than Khaini and Snus products irrespective of brand, type and manufacturer of the products. The core bacteriome was present in most or all moist STPs tested and showed an abundance of genera Acinetobacter, Bacillus, Prevotella, Acetobacter, Lactobacillus, Paracoccus, Flavobacterium, and Bacteroides. The STP-associated bacteriome has significant metabolic potential to contribute to TSNAs synthesis by nitrogen metabolism. The presence of antibiotic drug resistance microbes in the STPs can passively transfer resistance to the oral microbiota of SLT users and may contribute to oral cancer. Moreover, delivery of several bacterial pro-inflammatory components and toxins molecules to the host during STP intake can contribute to the development of oral cancer. Hereafter, identification of carcinogenic potential of bacterial population and their products will provide a detailed insight into oral cancer induction in SLT users and provide a basis to regulate the use of STPs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra; PRJNA767533.
Author Contributions
MS: experiments, data curation, data analysis, literature review, and writing original draft. SS: literature review and manuscript preparation. AmK: data validation and review and editing. AnK and HS: review and editing. MB: conceptualization, supervision, investigation, project administration, funding acquisition, data interpretation, validation, review, and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Indian Council of Medical Research (ICMR), India covered by the Task-Force project (ISRM/14(04)/TF/2018) funding acquisition to MB.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to the Director of the National Institute of Cancer Prevention and Research for providing the amenities to carry out this study. The Biokart India Pvt. Ltd., Bengaluru, India services of metagenomics sequencing and data processing are thankfully acknowledged. We thank Ravi Yadav and Lata Joshi for necessary assistance in the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.784841/full#supplementary-material
Supplementary Table 1 | Barcode sequences.
Supplementary Table 2 | OTUs table.
Supplementary Table 3 | Characteristics of moist smokeless tobacco products.
Supplementary Table 4 | Core microbiome values.
Supplementary Table 5 | Correlation values and correlation matrix.
Supplementary Table 6 | KEGG orthology (KO) terms.
Footnotes
- ^ https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- ^ https://www.microbiomeanalyst.ca/
References
Al-Hebshi, N. N., Alharbi, F. A., Mahri, M., and Chen, T. (2017). Differences in the bacteriome of smokeless tobacco products with different oral carcinogenicity: compositional and predicted functional analysis. Genes 8:106. doi: 10.3390/genes8040106
Alvarez, L., Sanchez-Hevia, D., Sánchez, M., and Berenguer, J. (2019). A new family of nitrate/nitrite transporters involved in denitrification. Int. Microbiol. 22, 19–28. doi: 10.1007/s10123-018-0023-0
Aslam, B., Wang, W., Arshad, M. I., Khurshid, M., Muzammil, S., Rasool, M. H., et al. (2018). Antibiotic resistance: a rundown of a global crisis. Infect. Drug Resist. 11, 1645–1658. doi: 10.2147/IDR.S173867
Averill, B. A. (1996). Dissimilatory nitrite and nitric oxide reductases. Chem. Rev. 96, 2951–2964. doi: 10.1021/cr950056p
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carlsson, S., Andersson, T., Araghi, M., Galanti, R., Lager, A., Lundberg, M., et al. (2017). Smokeless tobacco (snus) is associated with an increased risk of type 2 diabetes: results from five pooled cohorts. J. Intern. Med. 281, 398–406. doi: 10.1111/joim.12592
Centers for Disease Control and Prevention (2020). Smokeless Tobacco: Health Effects. Available online at: https://www.cdc.gov/tobacco/data_statistics/fact_sheets/smokeless/health_effects/index.htm (accessed March 15, 2021).
Chong, J., Liu, P., Zhou, G., and Xia, J. (2020). Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 15, 799–821. doi: 10.1038/s41596-019-0264-1
Chopyk, J., Chattopadhyay, S., Kulkarni, P., Smyth, E. M., Hittle, L. E., Paulson, J. N., et al. (2017). Temporal variations in cigarette tobacco bacterial community composition and tobacco-specific nitrosamine content are influenced by brand and storage conditions. Front. Microbiol. 8:358. doi: 10.3389/fmicb.2017.00358
Critchley, J. A., and Unal, B. (2003). Health effects associated with smokeless tobacco: a systematic review. Thorax 58, 435–443. doi: 10.1136/thorax.58.5.435
Derrien, M., van Passel, M. W., van de Bovenkamp, J. H., Schipper, R. G., de Vos, W. M., and Dekker, J. (2010). Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 1, 254–268. doi: 10.4161/gmic.1.4.12778
Dhariwal, A., Chong, J., Habib, S., King, I. L., Agellon, L. B., and Xia, J. (2017). MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 45, W180–W188. doi: 10.1093/nar/gkx295
Di Giacomo, M., Paolino, M., Silvestro, D., Vigliotta, G., Imperi, F., Visca, P., et al. (2007). Microbial community structure and dynamics of dark fire-cured tobacco fermentation. Appl. Environ. Microbiol. 73, 825–837. doi: 10.1128/aem.02378-06
Friedman, J., and Alm, E. J. (2012). Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8:e1002687. doi: 10.1371/journal.pcbi.1002687
Fujiwara, N., Kitamura, N., Yoshida, K., Yamamoto, T., Ozaki, K., and Kudo, Y. (2020). Involvement of fusobacterium species in oral cancer progression: a literature review including other types of cancer. Int. J. Mol. Sci. 21:6207. doi: 10.3390/ijms21176207
GATS-2 (2017). Global Adult Tobacco Survey. Available online at: https://ntcp.nhp.gov.in/assets/document/surveys-reports-publications/Global-Adult-Tobacco-Survey-Second-Round-India-2016-2017.pdf (accessed September, 2020).
Gholizadeh, P., Eslami, H., Yousefi, M., Asgharzadeh, M., Aghazadeh, M., and Kafil, H. S. (2016). Role of oral microbiome on oral cancers, a review. Biomed. Pharmacother. 84, 552–558. doi: 10.1016/j.biopha.2016.09.082
González, P. J., Correia, C., Moura, I., Brondino, C. D., and Moura, J. J. (2006). Bacterial nitrate reductases: molecular and biological aspects of nitrate reduction. J. Inorg. Biochem. 100, 1015–1023. doi: 10.1016/j.jinorgbio.2005.11.024
Goodrich, J. K., Di Rienzi, S. C., Poole, A. C., Koren, O., Walters, W. A., Caporaso, J. G., et al. (2014). Conducting a microbiome study. Cell 158, 250–262. doi: 10.1016/j.cell.2014.06.037
Günther, S. D., Fritsch, M., Seeger, J. M., Schiffmann, L. M., Snipas, S. J., Coutelle, M., et al. (2020). Cytosolic Gram-negative bacteria prevent apoptosis by inhibition of effector caspases through lipopolysaccharide. Nat. Microbiol. 5, 354–367. doi: 10.1038/s41564-019-0620-5
Gupta, P. C., and Ray, C. S. (2003). Smokeless tobacco and health in India and South Asia. Respirology 8, 419–431. doi: 10.1046/j.1440-1843.2003.00507.x
Halboub, E., Al-Ak’hali, M. S., Alamir, A. H., Homeida, H. E., Baraniya, D., Chen, T., et al. (2020). Tongue microbiome of smokeless tobacco users. BMC Microbiol. 20:201. doi: 10.1186/s12866-020-01883-8
Han, J., Sanad, Y. M., Deck, J., Sutherland, J. B., Li, Z., Walters, M. J., et al. (2016). Bacterial populations associated with smokeless tobacco products. Appl. Environ. Microbiol. 82, 6273–6283. doi: 10.1128/aem.01612-16
He, Z., Deng, R., Huang, X., Ni, Y., Yang, X., Wang, Z., et al. (2015). Lipopolysaccharide enhances OSCC migration by promoting epithelial-mesenchymal transition. J. Oral Pathol. Med. 44, 685–692. doi: 10.1111/jop.12285
Huang, B., Zhao, J., Li, H., He, K. L., Chen, Y., Chen, S. H., et al. (2005). Toll-like receptors on tumor cells facilitate evasion of immune surveillance. Cancer Res. 65, 5009–5014. doi: 10.1158/0008-5472.can-05-0784
Igamberdiev, A. U., and Hill, R. D. (2004). Nitrate, NO and haemoglobin in plant adaptation to hypoxia: an alternative to classic fermentation pathways. J. Exp. Bot. 55, 2473–2482. doi: 10.1093/jxb/erh272
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kanehisa, M., Goto, S., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2013). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205. doi: 10.1093/nar/gkt1076
Karpiński, T. M. (2019). Role of oral microbiota in cancer development. Microorganisms 7:20. doi: 10.3390/microorganisms7010020
Kaur, J., Sharma, A., Kumar, A., Bhartiya, D., Sinha, D. N., Kumari, S., et al. (2019). SLTChemDB: a database of chemical compounds present in Smokeless tobacco products. Sci. Rep. 9:7142. doi: 10.1038/s41598-019-43559-y
Kelly, B. J., Gross, R., Bittinger, K., Sherrill-Mix, S., Lewis, J. D., Collman, R. G., et al. (2015). Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics 31, 2461–2468. doi: 10.1093/bioinformatics/btv183
Knights, D., Costello, E. K., and Knight, R. (2011). Supervised classification of human microbiota. FEMS Microbiol. Rev. 35, 343–359. doi: 10.1111/j.1574-6976.2010.00251.x
Kroes, I., Lepp, P. W., and Relman, D. A. (1999). Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. U.S.A. 96, 14547–14552. doi: 10.1073/pnas.96.25.14547
Kumar, S., He, G., Kakarla, P., Shrestha, U., Ranjana, K. C., Ranaweera, I., et al. (2016). Bacterial multidrug efflux pumps of the major facilitator superfamily as targets for modulation. Infect. Disord. Drug Targets 16, 28–43. doi: 10.2174/1871526516666160407113848
Kurago, Z. B., Lam-Ubol, A., Stetsenko, A., De La Mater, C., Chen, Y., and Dawson, D. V. (2008). Lipopolysaccharide-squamous cell carcinoma-monocyte interactions induce cancer-supporting factors leading to rapid STAT3 activation. Head Neck Pathol. 2, 1–12. doi: 10.1007/s12105-007-0038-x
La Rosa, G. R. M., Gattuso, G., Pedullà, E., Rapisarda, E., Nicolosi, D., and Salmeri, M. (2020). Association of oral dysbiosis with oral cancer development. Oncol. Lett. 19, 3045–3058. doi: 10.3892/ol.2020.11441
Lacoma, A., Edwards, A. M., Young, B. C., Domínguez, J., Prat, C., and Laabei, M. (2019). Cigarette smoke exposure redirects Staphylococcus aureus to a virulence profile associated with persistent infection. Sci. Rep. 9:10798. doi: 10.1038/s41598-019-47258-6
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., Mcdonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Law, A. D., Fisher, C., Jack, A., and Moe, L. A. (2016). Tobacco, microbes, and carcinogens: correlation between tobacco cure conditions, tobacco-specific nitrosamine content, and cured leaf microbial community. Microb. Ecol. 72, 120–129. doi: 10.1007/s00248-016-0754-4
Luque-Almagro, V. M., Gates, A. J., Moreno-Vivián, C., Ferguson, S. J., Richardson, D. J., and Roldán, M. D. (2011). Bacterial nitrate assimilation: gene distribution and regulation. Biochem. Soc. Trans. 39, 1838–1843. doi: 10.1042/bst20110688
Mehra, R., Mohanty, V., Balappanavar, A. Y., and Kapoor, S. (2020). Bacterial contamination of packaged smokeless tobacco sold in India. Tob. Prev. Cessat. 6:11. doi: 10.18332/tpc/115064
Monika, S., Dineshkumar, T., Priyadharini, S., Niveditha, T., Sk, P., and Rajkumar, K. (2020). Smokeless Tobacco Products (STPs) harbour bacterial populations with potential for oral carcinogenicity. Asian Pac. J. Cancer Prev. 21, 815–824. doi: 10.31557/apjcp.2020.21.3.815
Nasrin, S., Chen, G., Watson, C. J. W., and Lazarus, P. (2020). Comparison of tobacco-specific nitrosamine levels in smokeless tobacco products: high levels in products from Bangladesh. PLoS One 15:e0233111. doi: 10.1371/journal.pone.0233111
Nishimoto, A. T., Rosch, J. W., and Tuomanen, E. I. (2020). Pneumolysin: pathogenesis and therapeutic target. Front. Microbiol. 11:1543. doi: 10.3389/fmicb.2020.01543
Nishimura, T., Vertès, A. A., Shinoda, Y., Inui, M., and Yukawa, H. (2007). Anaerobic growth of Corynebacterium glutamicum using nitrate as a terminal electron acceptor. Appl. Microbiol. Biotechnol. 75, 889–897. doi: 10.1007/s00253-007-0879-y
Onuorah Samuel, O. M. (2016). Microbial contamination of locally-prepared snuff sold at Eke-Awka Market, Anmbra State, Nigeria. Am. J. Life Sci. Res. 4, 74–77. doi: 10.21859/ajlsr-040301
Pauly, J. L., and Paszkiewicz, G. (2011). Cigarette smoke, bacteria, mold, microbial toxins, and chronic lung inflammation. J. Oncol. 2011:819129. doi: 10.1155/2011/819129
Rivera, A. J., and Tyx, R. E. (2021). Microbiology of the american smokeless tobacco. Appl. Microbiol. Biotechnol. 105, 4843–4853. doi: 10.1007/s00253-021-11382-z
Rivera, A. J., Tyx, R. E., Keong, L. M., Stanfill, S. B., and Watson, C. H. (2020). Microbial communities and gene contributions in smokeless tobacco products. Appl. Microbiol. Biotechnol. 104, 10613–10629. doi: 10.1007/s00253-020-10999-w
Rohatgi, N., Kaur, J., Srivastava, A., and Ralhan, R. (2005). Smokeless tobacco (khaini) extracts modulate gene expression in epithelial cell culture from an oral hyperplasia. Oral. Oncol. 41, 806–820. doi: 10.1016/j.oraloncology.2005.04.010
Rubinstein, I., and Pedersen, G. W. (2002). Bacillus species are present in chewing tobacco sold in the United States and evoke plasma exudation from the oral mucosa. Clin. Diagn. Lab. Immunol. 9, 1057–1060. doi: 10.1128/cdli.9.5.1057-1060.2002
Sajid, M., Srivastava, S., Joshi, L., and Bharadwaj, M. (2021). Impact of smokeless tobacco-associated bacteriome in oral carcinogenesis. Anaerobe 70, 1–9. doi: 10.1016/j.anaerobe.2021.102400
Sarkar, P., Malik, S., Laha, S., Das, S., Bunk, S., Ray, J. G., et al. (2021). Dysbiosis of oral microbiota during oral squamous cell carcinoma development. Front. Oncol. 11:614448. doi: 10.3389/fonc.2021.614448
Sarlak, S., Lalou, C., Amoedo, N. D., and Rossignol, R. (2020). Metabolic reprogramming by tobacco-specific nitrosamines (TSNAs) in cancer. Semin. Cell Dev. Biol. 98, 154–166. doi: 10.1016/j.semcdb.2019.09.001
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shaik, F. B., and Maddu, N. (2019). Smokeless tobacco products profile and pictorial warning labels in India: a review. Popul. Med. 1, 1–6. doi: 10.18332/popmed/114940
Shang, F. M., and Liu, H. L. (2018). Fusobacterium nucleatum and colorectal cancer: a review. World J. Gastrointest. Oncol. 10, 71–81. doi: 10.4251/wjgo.v10.i3.71
Shi, H., Wang, R., Bush, L. P., Zhou, J., Yang, H., Fannin, N., et al. (2013). Changes in TSNA contents during tobacco storage and the effect of temperature and nitrate level on TSNA formation. J. Agric. Food Chem. 61, 11588–11594. doi: 10.1021/jf404813m
Siddiqi, K., Husain, S., Vidyasagaran, A., Readshaw, A., Mishu, M. P., and Sheikh, A. (2020). Global burden of disease due to smokeless tobacco consumption in adults: an updated analysis of data from 127 countries. BMC Med. 18:222. doi: 10.1186/s12916-020-01677-9
Smyth, E. M., Kulkarni, P., Claye, E., Stanfill, S., Tyx, R., Maddox, C., et al. (2017). Smokeless tobacco products harbor diverse bacterial microbiota that differ across products and brands. Appl. Microbiol. Biotechnol. 101, 5391–5403. doi: 10.1007/s00253-017-8282-9
Srivastava, A., Mishra, S., and Verma, D. (2021). Characterization of oral bacterial composition of adult smokeless tobacco users from healthy indians using 16S rDNA analysis. Microb. Ecol. 82, 1061–1073. doi: 10.1007/s00248-021-01711-0
Stepanov, I., Gupta, P. C., Dhumal, G., Yershova, K., Toscano, W., Hatsukami, D., et al. (2015). High levels of tobacco-specific N-nitrosamines and nicotine in Chaini Khaini, a product marketed as snus. Tob. Control. 24, e271–e274. doi: 10.1136/tobaccocontrol-2014-051744
Stepanov, I., Gupta, P. C., Parascandola, M., Yershova, K., Jain, V., Dhumal, G., et al. (2017). Constituent variations in smokeless tobacco purchased in Mumbai, India. Tob. Regul. Sci. 3, 305–314. doi: 10.18001/TRS.3.3.6
Stepanov, I., Jensen, J., Hatsukami, D., and Hecht, S. S. (2008). New and traditional smokeless tobacco: comparison of toxicant and carcinogen levels. Nicotine Tob. Res. 10, 1773–1782. doi: 10.1080/14622200802443544
Tricker, A. R., and Preussmann, R. (1989). The occurrence of N-nitroso compounds in kiwam tobacco. Cancer Lett. 46, 221–224. doi: 10.1016/0304-3835(89)90134-1
Tyx, R. E., Rivera, A. J., Keong, L. M., and Stanfill, S. B. (2020). An exploration of smokeless tobacco product nucleic acids: a combined metagenome and metatranscriptome analysis. Appl. Microbiol. Biotechnol. 104, 751–763. doi: 10.1007/s00253-019-10232-3
Tyx, R. E., Stanfill, S. B., Keong, L. M., Rivera, A. J., Satten, G. A., and Watson, C. H. (2016). Characterization of bacterial communities in selected smokeless tobacco products using 16S rDNA analysis. PLoS One 11:e0146939. doi: 10.1371/journal.pone.0146939
Wang, J., Yang, H., Shi, H., Zhou, J., Bai, R., Zhang, M., et al. (2017). Nitrate and nitrite promote formation of tobacco-specific nitrosamines via nitrogen oxides intermediates during postcured storage under warm temperature. J. Chem. 2017:6135215. doi: 10.1155/2017/6135215
Wood, D. E., Lu, J., and Langmead, B. (2019). Improved metagenomic analysis with Kraken 2. Genome Biol. 20:257. doi: 10.1186/s13059-019-1891-0
Zhang, G., and Ghosh, S. (2000). Molecular mechanisms of NF-kappaB activation induced by bacterial lipopolysaccharide through Toll-like receptors. J. Endotoxin Res. 6, 453–457. doi: 10.1179/096805100101532414
Keywords: smokeless tobacco products (STPs), tobacco-specific nitrosamines (TSNAs), nitrogen metabolism genes, smokeless tobacco-associated bacteriome, antibiotic-resistance genes, toxins, oral cancer
Citation: Sajid M, Srivastava S, Kumar A, Kumar A, Singh H and Bharadwaj M (2021) Bacteriome of Moist Smokeless Tobacco Products Consumed in India With Emphasis on the Predictive Functional Potential. Front. Microbiol. 12:784841. doi: 10.3389/fmicb.2021.784841
Received: 28 September 2021; Accepted: 24 November 2021;
Published: 24 December 2021.
Edited by:
Spyridon Ntougias, Democritus University of Thrace, GreeceReviewed by:
Digvijay Verma, Babasaheb Bhimrao Ambedkar University, IndiaChristopher L. Hemme, University of Rhode Island, United States
Lateef Salam, Elizade University, Nigeria
Copyright © 2021 Sajid, Srivastava, Kumar, Kumar, Singh and Bharadwaj. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mausumi Bharadwaj, bWF1c3VtaS5iaGFyYWR3YWpAZ21haWwuY29t
 Mohammad Sajid
Mohammad Sajid Sonal Srivastava
Sonal Srivastava Amit Kumar2
Amit Kumar2 Anuj Kumar
Anuj Kumar Harpreet Singh
Harpreet Singh Mausumi Bharadwaj
Mausumi Bharadwaj