- 1Jiangsu Key Laboratory for Eco-Agricultural Biotechnology Around Hongze Lake, School of Life Science, Huaiyin Normal University, Huai’an, China
- 2Jiangsu Collaborative Innovation Center of Regional Modern Agriculture & Environmental Protection, Huai’an, China
- 3Agro-Tech Extension and Service Center, Huai’an, China
- 4College of Plant Protection, Nanjing Agricultural University, Nanjing, China
- 5Huaiyin Institute of Agricultural Sciences of Xuhuai Region in Jiangsu, Huaian Academy of Agricultural Sciences, Huai’an, China
Biofilm plays important roles in the life cycle of Bacillus species, such as promoting host and object surface colonization and resisting heavy metal stress. This study utilized transcriptomics to evaluate the impacts of cadmium on the components, morphology, and function of biofilms of Bacillus subtilis strain 1JN2. Under cadmium ion stress, the morphology of the B. subtilis 1JN2 biofilm was flattened, and its mobility increased. Moreover, differential gene expression analysis showed that the main regulator of biofilm formation, Spo0A, decreased in expression under cadmium ion stress, thereby inhibiting extracellular polysaccharide synthesis through the SinI/SinR two-component regulatory system and the AbrB pathway. Cadmium ion treatment also increased the SigD content significantly, thereby increasing the expression of the flagella encoding and assembly genes in the strain. This promoted poly-γ-glutamic acid production via the DegS/DegU two-component regulatory system and the conversion of biofilm extracellular polysaccharide to poly-γ-glutamic acid. This conferred cadmium stress tolerance in the strain. Additionally, the cadmium ion-mediated changes in the biofilm composition affected the colonization of the strain on the host plant root surface. Cadmium ions also induced surfactin synthesis. These findings illustrate the potential of Bacillus species as biocontrol strains that can mitigate plant pathogenic infections and heavy metal stress. The results also provide a basis for the screening of multifunctional biocontrol strains.
Background
Bacillus is an important genus of plant growth-promoting rhizobacteria (PGPR) owing to its production of a variety of active metabolites and its strong resistance to environmental factors. Over the past years, more and more PGPRs have been used to assist host plants in resisting abiotic stress such as drought, cold, and heavy metal pollution (Yang et al., 2009).In this respect, biofilm plays a crucial role along with the occurrence of Bacillus in nature (Yang et al., 2018). A biofilm can protect an individual bacterium from environmental stress, such as antibiotic compounds (Høiby et al., 2010), and aid bacterial colonization of host plant roots (Bais et al., 2004; Beauregard et al., 2013). The latter is considered the first and most important defense technique that biocontrol agents use against soil-borne pathogens. Biofilm-enhanced mutants have been reported to exhibit enhanced biocontrol activities against Ralstonia, while biofilm-deletion mutants have reduced disease prevention capacity (Yang et al., 2018). Zhang et al. (2021) found that Bacillus subtilis B12 can secrete more surfactin under Cd stress to regulate biofilm formation to absorb more Cd. Kaçar et al. (2002) found that the enrichment of Cd2+ by Bacillus can reach 21.4% of its dry weight. However, there is no clear conclusion on which component of biofilm is involved in bacterial colonization and heavy metal adsorption.
Bacterial biofilms exist on the surface of bacteria, as aggregates formed by the extracellular polymers secreted by the bacteria themselves (Koo et al., 2017), including polysaccharides, proteins, nucleic acids, and lipids, which account for more than 90% of bacterial biofilms (Sutherland, 2001). The regulatory pathway for biofilm formation is well-defined in B. subtilis (Vlamakis et al., 2013). Briefly, the environmental signals (like plant root secretions) are perceived by sensory histidine kinases (such as KinC/KinD), which activate the global master regulator, Spo0A, through protein phosphorylation (López et al., 2009; Mcloon et al., 2011; Chen et al., 2012; Beauregard et al., 2013; Shemesh and Chai, 2013). The phosphorylated Spo0A (Spo0A-P) induces biofilm formation via two independent mechanisms that antagonize the two main repressors (SinR and AbrB) that inhibit biofilm formation (Hamon and Lazazzera, 2001; Chai et al., 2011). While in other species, several regulators were involved in biofilm formation as well as flagella assembly. For example, Zur is required for Zinc uptake and ensure the expression of flagella and biofilm formation in Salmonella enterica (Ammendola et al., 2016). Fur, the cytoplasmic ferric uptake regulator protein, ensure the mutant to form more mature biofilm than the parent in Pseudomonas aeruginosa (Banin et al., 2005). Like Fur, IdeR binds iron and then interacts with a specific sequence in the operator regions of iron-regulated genes to control their transcription coordinating iron homeostasis and morphological differentiation (Cheng et al., 2018). Another iron binding regulator, XibR in Xanthomonas campestris, not only can promote biofilm formation but also regulate the virulence of the pathogen (Pandey et al., 2016).
Abbreviations: EPS, extracellular polysaccharides; γ-PGA, poly-γ-glutamic acids; FPKM, fragments per kilobase of exon model per million mapped fragments.
Poly-γ-glutamic acid (γ-PGA), ranging from about 10 to 1,000 kDA in size, is an important component of B. subtilis biofilm matrix (Morikawa et al., 2006; Poo et al., 2010; Ogunleye et al., 2015). Its biosynthesis in B. subtilis is regulated by the conserved operon pgsB-pgsC-pgsA-pgsE (previously named ywsC-ywtA-ywtB-ywtC) (Ashiuchi and Misono, 2002). These genes are highly conserved across different Bacillus species, including B. cereus and B. anthracis (Makino et al., 1989; Stanley and Lazazzera, 2005). In B. subtilis, the pgs operon is regulated by two cascades (ComA-ComP and DegS-DegU) of the two-component system (Stanley and Lazazzera, 2005). It has been shown that regulatory mutants of B. subtilis 168 degU and B. subtilis 168 ccpA are more resistant to the addition of divalent metal ions (Ca2+, Mg2+, Mn2+, and Zn2+) than wild-type B. subtilis (Dinh et al., 2019).
Furthermore, γ-PGA biosynthesis may interact with the regulatory pathway of extracellular polysaccharide (EPS) production. Yu et al. (2016) reported that down-regulated expression of epsD and yqxM, which are responsible for biofilm formation, have minimal effects on the expression of the γ-PGA synthesis gene ywtB. In contrast, overproduction of γ-PGA in B. amyloliquefaciens C06 and its epsA and tasA mutants resulted in reduced EPS and TasA levels (Liu et al., 2010). Moreover, KinC and KinD negatively regulate γ-PGA production in B. subtilis by activating Spo0A, which down-regulates AbrB activities. Thus, this pathway positively regulates biofilm formation but negatively controls γ-PGA production, suggesting the existence of a noteworthy switch-like mechanism of EPS and γ-PGA production. In contrast, the reverse regulatory pathway mediated by the DegS-DegU two-component system positively regulates γ-PGA production but strongly inhibits biofilm matrix genes (Yu et al., 2016).
Many environmental factors affect the formation of bacterial biofilms. Ma and Sun (2021) found that the expression levels of many biofilm formation-related genes, including those that encode proteins involved in flagellar assembly, signal transduction, bacterial secretion, and the TonB-dependent transfer system, were significantly upregulated when facing Cd stress, indicating their critical roles in determining bacterial biofilm formation and enhancing Cd resistance. Li et al. (2020) also found that the upregulation of genes related to biofilm formation in Pseudomonas chenduensis strain MBR (i.e., algK, algX, and alg44) enabled Cd absorption and contributed to Cd bioremediation. However, Alviz-Gazitua et al. (2019) found that inhibition of the initiation of biofilm formation and EPS production by Cupriavidus metallidurans CH34 is positively correlated with cadmium concentration.
Poly-γ-glutamic acid plays different roles in various B. subtilis isolates. Deleting ywsAB and ywsC, homologs of pgsBCA, resulted in weak biofilm formation in B. subtilis JH642; however, there were no differences in biofilm formation between pgsBCA mutant and wild-type B. subtilis 3,610 (Stanley and Lazazzera, 2005; Branda et al., 2006). Moreover, γ-PGA has been proven essential for B. amyloliquefaciens C06 and B. subtilis isolate colonization of apple (Liu et al., 2010) and tomato root surfaces (Yu et al., 2016), respectively. Similarly, EPS, another important component of biofilms, has also been shown to help biocontrol strains achieve effective colonization (Zhu et al., 2020).
Although the regulatory pathways of EPS production and γ-PGA biosynthesis have been well studied, the cause of their production shift is still unclear. It is also unknown whether changes between these two biofilm components might affect other activities, such as colonization and biocontrol ability of these strains. B. subtilis strain 1JN2 was isolated in our previous work and found to be an efficient biocontrol agent against Ralstonia wilt (Yang et al., 2012). In addition, with the acceleration of industrialization and the continuous growth of population, heavy metal pollution has become a global problem (Chen et al., 2015). Among them, cadmium is the main culprit threatening agricultural production. According to the latest “National Soil Pollution Survey Bulletin” (2014), cadmium ranks first among inorganic pollutants and the area of cadmium contaminated farmland has exceeded 2.0 × 104 hm2. Owing to the seriousness of cadmium pollution in China, we tested the resistance of this strain to cadmium ion contamination as well as its ability to protect host plants; although the biofilm morphology of this strain changed after cadmium treatment, it did not affect its control effect on Ralstonia wilt (Yang et al., 2015). Comparative transcriptomics was used in the present study to evaluate the impacts of cadmium on the components, morphology, and function of biofilms of a B. subtilis strain 1JN2. It was demonstrated that high concentrations of Cd2+ induced γ-PGA biosynthesis and inhibited EPS production in B. subtilis 1JN2. The shift between EPS and γ-PGA production enabled the strains to adapt to heavy metal contamination and persistently control host plant diseases.
Materials and methods
Strains, reagents, and media conditions
We used a wild B. subtilis strain 1JN2, previously isolated by Yang et al. (2018) and deposited in the China General Microbiological Culture Collection Center (CGMCC, NO 9759), for the present experiments. Competent 1JN2 strains containing chloramphenicol resistance were prepared and then electrotransformed with a green fluorescent plasmid (pGFP 4,412) for fluorescent microscopy using the method described by Xue (2008). All strains were routinely grown in Luria-Bertani (LB) broth (10 g/l of tryptone, 5 g/l of yeast extract, and 5/L g of NaCl) or LB medium with 1.5% agar. Conversely, biofilm formation was conducted using LBGM medium (LB + 1% (v/v) glycerol +0.1 mM MnSO4) (Shemesh and Chai, 2013).
Biofilm formation assay
A stock solution containing 1 M Cd2+ was prepared using cadmium sulfate (3CdSO4-8H2O) and sterilized by filtration through a 0.22-μm bacterial filter followed by storage at 4°C.
To analyze the impacts of Cd2+ on biofilm formation, five μL of 1JN2 suspension (106 CFU/ml) was distributed as drops on each LBGM agar plate containing different Cd2+ concentrations, including 0, 1, 2, 3, 4, and 5 mM. Each concentration was repeated three times. The plates were incubated at 30°C for 72 h, and then the diameter and the height of each biofilm were measured by Vernier caliper.
Tomato root colonization by Bacillus subtilis 1JN2
Three treatment categories, (A) water, (B) B. subtilis 1JN2, and (C) B. subtilis 1JN2 + Cd2+, were used to detect the colonization ability of tomato plants (obtained from the Shanghai Cooperation 903 seed market, Shanghai, China) by B. subtilis 1JN2 after cadmium ion treatment. Each treatment was conducted in three replicates with 20 plants per replicate. Tomato plants with three to four true leaves were transferred from seedling trays into experimental pots (10 cm height × 10 cm diameter) filled with 0.5 kg of nutrient soil. Each pot was supplied with 20 ml of B. subtilis 1JN2 culture (107 CFU/ml) 7 days after transplantation. Thereafter, pots in the treatment C category were supplemented with 20 ml of Cd2+ solution (6 mM) 14 days after transplantation. Greenhouse growth conditions included a temperature of 30°C with 16/8 h of light/dark photoperiod.
After treatment, the plant roots were collected 21 days post-transplantation and divided into two parts. The first part of the root sample was immersed in sterile water, and then, its population of 1JN2 was measured by dilution spreading on chloramphenicol-containing plates. The second part of the root sample was analyzed using a ZEISS LSM 700 confocal microscope (Carl Zeiss AG, Oberkochen, Germany) after fixation. The colonization ability of 1JN2 was evaluated under treatments B and C by using a fluorescent analysis described by Yang et al. (2018). GFP signal was selected via a Smart Setup, and the excitation and emission wavelengths were 488 nm and 493–550 nm, respectively.
Collection of Cd2+-treated bacterial cells
We inoculated 0.5 ml of 1JN2 suspension (106 CFU/ml) into 50 ml of LB broth supplemented with 3 mM Cd2+ and incubated the cells at 30°C for 24 h. The inoculation was conducted in twelve replicates, among which three replicates each were used for bacterial cells collection at 6, 12, 18, and 24 h after inoculation. Bacterial cells were also collected from a blank LB broth (control) inoculated in triplicate without Cd2+ at 24 h post-inoculation. The cell collection was conducted by centrifugation at 5000 rpm for 5 min and then stored in liquid nitrogen prior to RNA extraction. The obtained supernatants were used for EPS and γ-PGA content detection.
EPS and γ-PGA content detection following 1JN2 treatment with Cd2+
The same treatment described above was conducted for the purpose of EPS and γ-PGA content detection, and each treatment consisted of three replicates. For EPS detection, 2 ml of the supernatant was transferred into test tubes and 6 ml of anthrone reagent was added to each test tube. The solutions were then properly mixed and heated in boiling water for 15 min, followed by cooling in ice water for 15 min. The absorbance of each sample was measured at a wavelength of 625 nm, and their sugar contents were determined using a glucose standard curve.
Meanwhile, for γ-PGA detection, 3 ml of methylene blue (color developing solution; 10 mg/l) was added into test tubes containing 3 ml of the supernatants and vortexed for 5 min at 25°C. A wavelength of 664 nm was used for measuring the absorbance of each sample, and the γ-PGA content of the samples was determined based on a γ-PGA standard curve.
RNA extraction and preparation
Total RNA was isolated from the collected cells using Tiangen reagent (Tiangen, Beijing, China) and subsequently purified using a QIAGEN RNeasy MINI kit (QIAGEN, Hilden, Germany) according to the manufacturers’ instructions. The RNA quality and quantity were determined at a 260 nm wavelength via UV spectroscopy (NanoPhotometer® spectrophotometer; IMPLEN, Westlake Village, CA, United States) and analyzed on an RNA 6000 Nano Labchip using 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, United States).
Library preparation for strand-specific transcriptome sequencing
Library preparation was conducted at Genepioneer Biotechnologies Co., Ltd., Nanjing, China, using 3 μg of RNA from each sample based on a protocol described by Yang et al. (2019). Illumina sequencing libraries were generated using a NEBNext® Ultra™ Directional RNA Library Prep Kit (NEB, Ipswich, MA, United States). Briefly, a random hexamer primer and M-MuLV Reverse Transcriptase (RNase H–) were used for first-strand cDNA synthesis, while the second cDNA strand was synthesized using DNA polymerase I and RNase H. NEBNext Adaptors (NEB) with hairpin loop structure were ligated to the cDNA fragments after adenylation to prepare for hybridization. The library fragments were then purified using the AMPure XP system (Beckman Coulter, Beverly, MA, United States), and their quality was assessed on the Agilent Bioanalyzer 2100 system (Lei et al., 2018).
Clustering and sequencing
The samples were index-coded and clustered on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina, San Diego, CA, United States), according to the manufacturer’s instructions. An Illumina HiSeq platform (Cloud Health, Nanjing, China)1 was then used for library sequencing and paired-end read generation.
Data analysis
The initial raw data obtained in fastq format were processed using in-house Perl scripts. Subsequently, HISAT2 2.0.5 (Zhang et al., 2016) was used for reference genome indexing and aligning clean reads to the reference genome. The reads mapped to each gene were counted using String Tie (Pertea et al., 2016). The fragments per kilobase of exon model per million mapped fragments (FPKM) value of each gene was then calculated based on their length and the read counts mapped to the gene (Lei et al., 2018). Differential expression analysis of the groups was performed using the DESeq R package (1.18.0), while the GOseq R package (with corrected gene length bias) was utilized for Gene Ontology (GO) enrichment analysis of the differentially expressed genes. BLAST software was used to test the statistical significance of enrichment of the differentially expressed genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Li et al., 2016).
Quantitative real-time PCR analysis
The expression levels of five selected genes of B. subtilis 1JN2 cultured under similar conditions were validated by real-time RT-PCR using an iCyclerMyiQ Real-Time PCR System (Bio-Rad, Hercules, CA, United States). The PCR conditions were set as follows: initial denaturation at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 65°C for 15 s and extension at 72°C for 20 s. Melting curve analysis of the amplicons was performed at the end of each PCR run to ensure that unique products had been amplified.
Results
Cd2+ treatment changed biofilm formation by Bacillus subtilis 1JN2
We found that biofilm formation by B. subtilis 1JN2 varied with the Cd2+ concentration (Figure 1). In general, as cadmium ion concentration increased, the biofilm gradually became smaller and the surface gradually became smoother. Increasing Cd2+ concentration from 0 to 2 mM moderately inhibited biofilm formation by 1JN2, but the biofilm morphology did not change markedly; however, Cd2+ concentrations of 3 mM or higher significantly inhibited biofilm formation by 1JN2 (Supplementary Table 1), and not only the diameter but also the height decreased significantly. In contrast to biofilm formation, there was no significant difference in the growth of 1JN2 under a Cd2+ concentration of 0 to 2 mM Cd2+ (Yang et al., 2018).
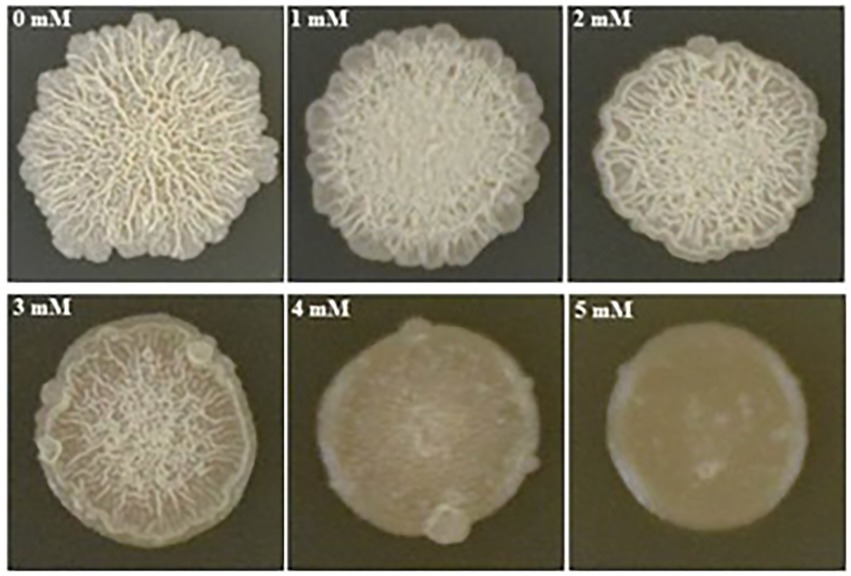
Figure 1. Impacts of Cd2+ on the biofilm colonies of B. subtilis 1JN2. 5 ul of 1JN2 suspension (106 CFU/ml) was spotted on the surface of LBGM agar plate with a Cd2+ gradient of 0, 1, 2, 3, 4 and 5 mM, the picture was taken at 72 h post incubation at 30°C.
The biofilm forms were also affected by Cd2+ treatment. Biofilm surfaces of the Cd2+-treated cells were complanate compared to the convex surfaces of the group without Cd2+. Moreover, the viscosity of the biofilms also increased with the Cd2+ concentration.
Cd2+ treatment influenced tomato root colonization by Bacillus subtilis 1JN2
Because biofilm formation is an important step in root colonization, it was also imperative to evaluate the effect of Cd2+ on the root colonization ability of B. subtilis 1JN2. A GFP-tagged 1JN2 was constructed for the root colonization experiment in the greenhouse. The population of B. subtilis 1JN2, which colonized the tomato roots, decreased with Cd2+ treatment (Supplementary Figure 1), indicating that biofilm inhibition caused by Cd2+ affected the colonization ability of 1JN2. However, as shown in Figure 2, the Cd2+ treatment only affected the colonization ability on the surface of tomato roots. A large number of bacterial cells entered the inside of tomato roots, and the difference was not significant compared with the treatment group without cadmium ions (Figure 2). In addition, Compared with the control group without cadmium ions, we can see that the strain could colonized on the surface of tomato roots in the control group, so the effect of chelating cadmium ions by other components in the rhizosphere can be excluded.
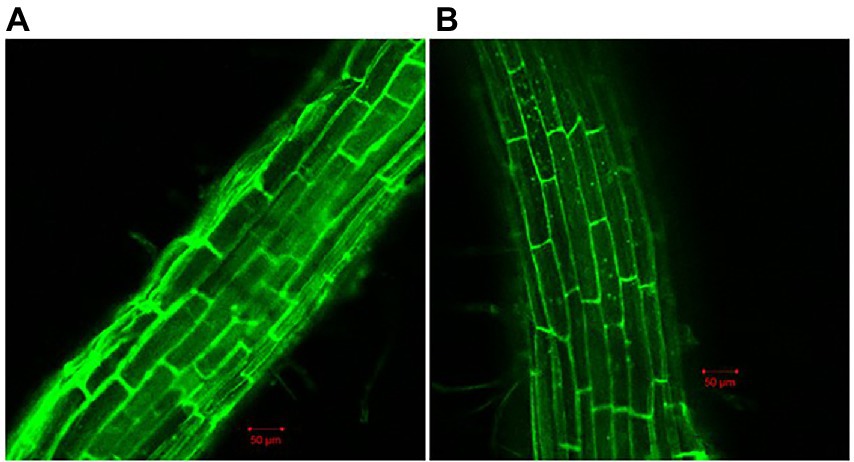
Figure 2. Confocal microscope detection of B. subtilis 1JN2 on the root of tomato with (B) or without (A) Cd2+ treatment. Fluorescent analysis was carried out using ZEISS LSM 700. GFP was selected in Smart Setup, the excitation wavelength is 488 nm, and the emission wavelength is 493–550 nm.
RNA sequencing and identification of differentially expressed genes
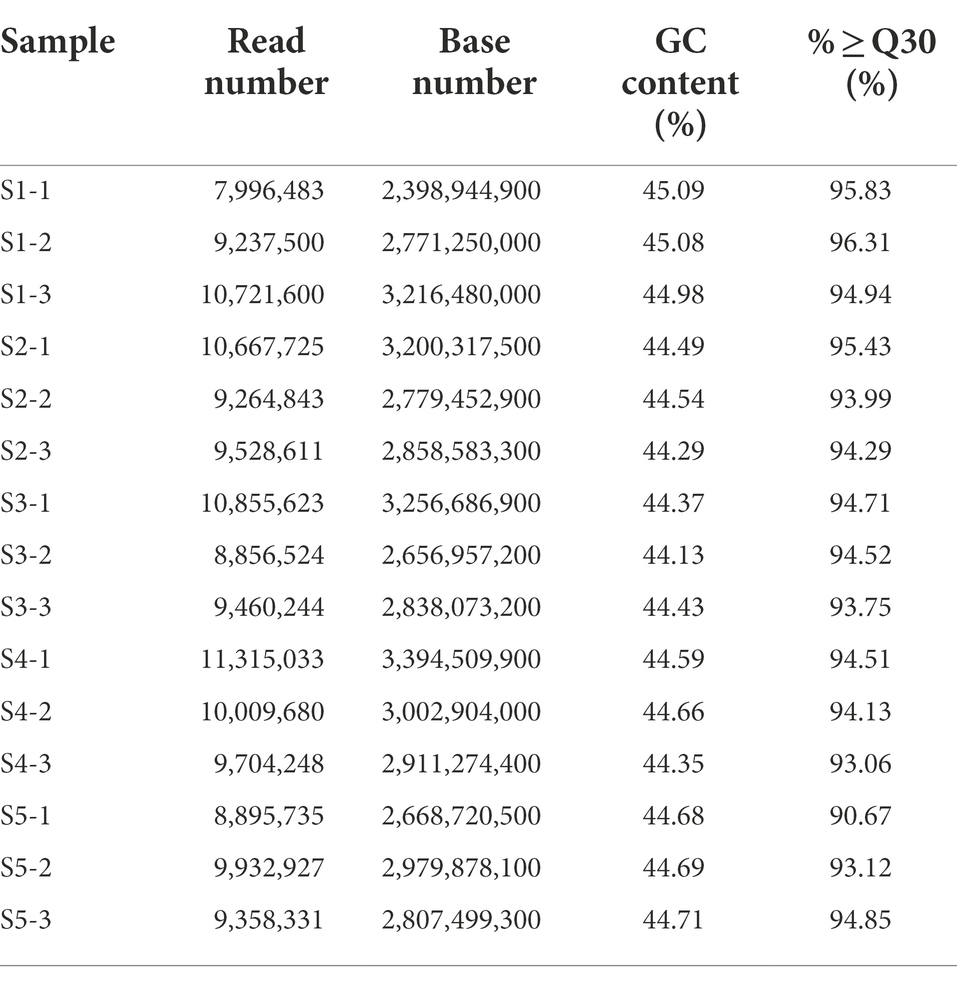
Cell samples were collected at four time points after Cd2+ treatment; the samples collected at 6, 12, 18, and 24 h were denoted S2, S3, S4, and S5, respectively, while the blank control without Cd2+ was designated S1. The sequence data are summarized in Table 1. More than 93% of the clean reads from each sample mapped to the reference genome, indicating that the obtained transcriptome data were suitable for further analysis. The raw data were deposited to the NCBI SRA database under accession number PRJNA646606.2
The numbers of differentially expressed genes (DEGs) between the various time points after Cd2+ treatment of B. subtilis 1JN2 were visualized as a Venn diagram (Figure 3A). The number of DEGs between S1 and S2 decreased with the prolongation of treatment time. There were 659 DEGs between S1 and S2, among which 308 were up-regulated, and 351 were down-regulated. However, the number of DEGs between S2 and S3, S3 and S4, and S4 and S5 were 178, 36 and 6, respectively (Figure 3B). This implies that the cells undergo an important adjustment phase during the initial period of exposure to Cd2+ and later acquire resistance to Cd2+ stress. The same trend was observed in the principal component analysis results (Supplementary Figure 2), which showed greater similarity among S3, S4, and S5, consistent with a lack of significant differences in their gene expression.
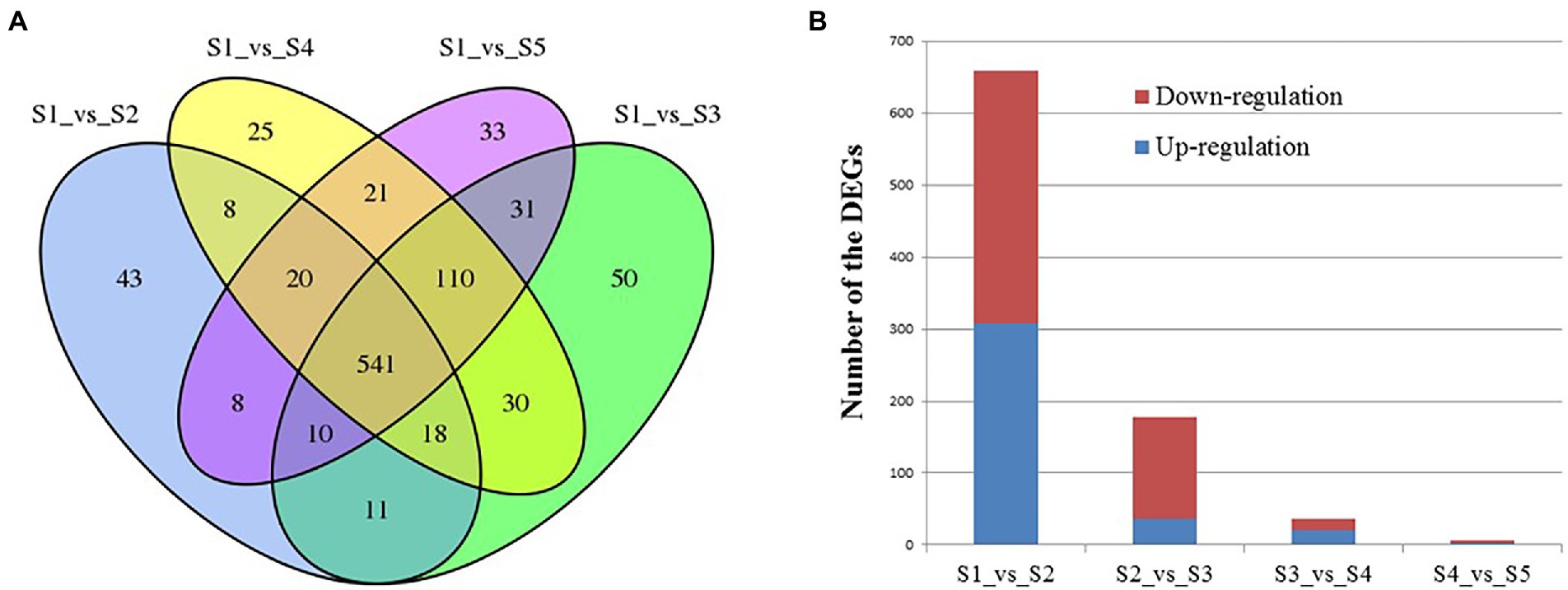
Figure 3. Analysis of the differentially expressed genes of B. subtilis 1JN2 after Cd2+ treatment. (A) Venn diagram analysis, (B) numbers of DEGs between time points. Samples collected at 6 h, 12 h, 18 h, and 24 h were named as S2, S3, S4, S5 and the blank control that without Cd2+ was named as S1. Comparison between S1 and S2 means the DEGs between the two samples, and so on.
Cell mobility-related genes changed significantly after Cd2+ treatment
Because Cd2+ treatment modified the morphological features of strain 1JN2 biofilm, we checked whether it could also affect the mobility of the strain. The Cluster of Orthologous Groups of Proteins (COG) database is constructed based on the phylogenetic relationship between bacteria, archaea and eukaryotes. Applying BLAST to the database allows for orthologous classification of gene products. The COG database BLAST results showed that the most significant changes in gene products of the strain after exposure to Cd2+ were related to cell mobility, followed by chromatin structure and dynamics, amino acid and nucleotide transport, and metabolism (Figure 4). Moreover, a comparison between S1 and S2 indicated that the strain could perceive and respond to Cd2+ stress after some period of exposure via regulatory metabolism. These results are consistent with the observed biofilm morphological changes of the strain.
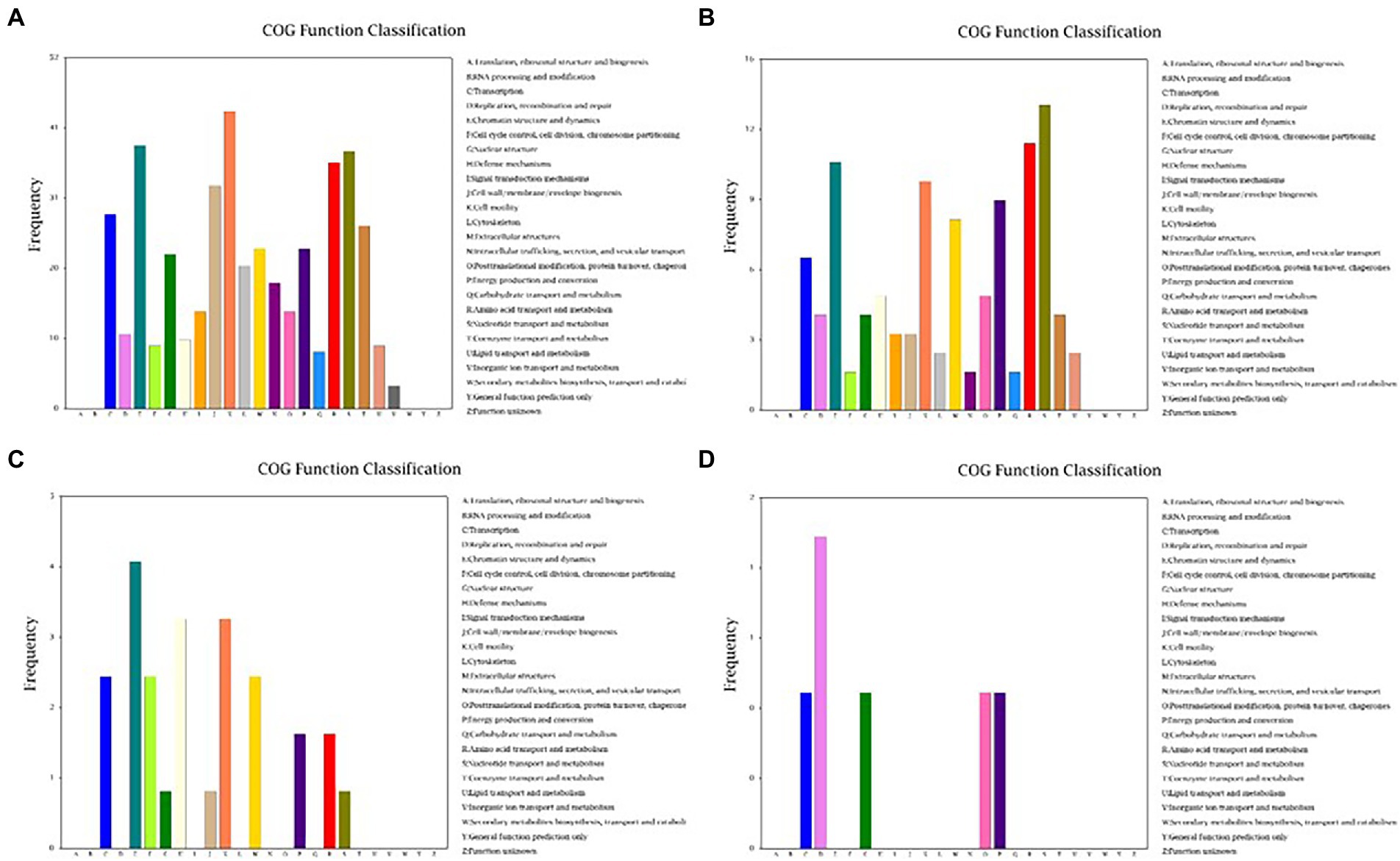
Figure 4. Cluster of Orthologous Groups of Proteins (COG) function classification of the DEGs of B. subtilis 1JN2 after Cd2+ treatment. (A–D) Means the comparison of DEGs according to COG function classification between S1 and S2, S2 and S3, S3 and S4, S4 and S5 separately.
Expression of flagella synthesis-and assembly-related genes increased in strain 1JN2 after Cd2+ treatment
The flagellum is the most important motor organ, which plays an important role in the mobility and chemotaxis of bacterial cells. Owing to the observed significant effects of Cd2+ on the strain’s mobility, we assessed its effects on flagella synthesis and assembly genes. KEGG is a database for systematic gene function analysis and genomic information. Significant pathway enrichment can indicate the main biochemical, metabolic, or signal transduction pathways associated with differentially expressed genes. According to KEGG analysis, flagella synthesis and assembly genes were significantly enhanced relative to the control group 12 h after Cd2+ treatment (Figure 5). The differential genes at the later sampling time points were mainly involved in metabolism-related reactions, such as glycolysis/gluconeogenesis and biosynthesis of valine, leucine, isoleucine, pantothenate, and CoA. Figure 6 also illustrates similar results.
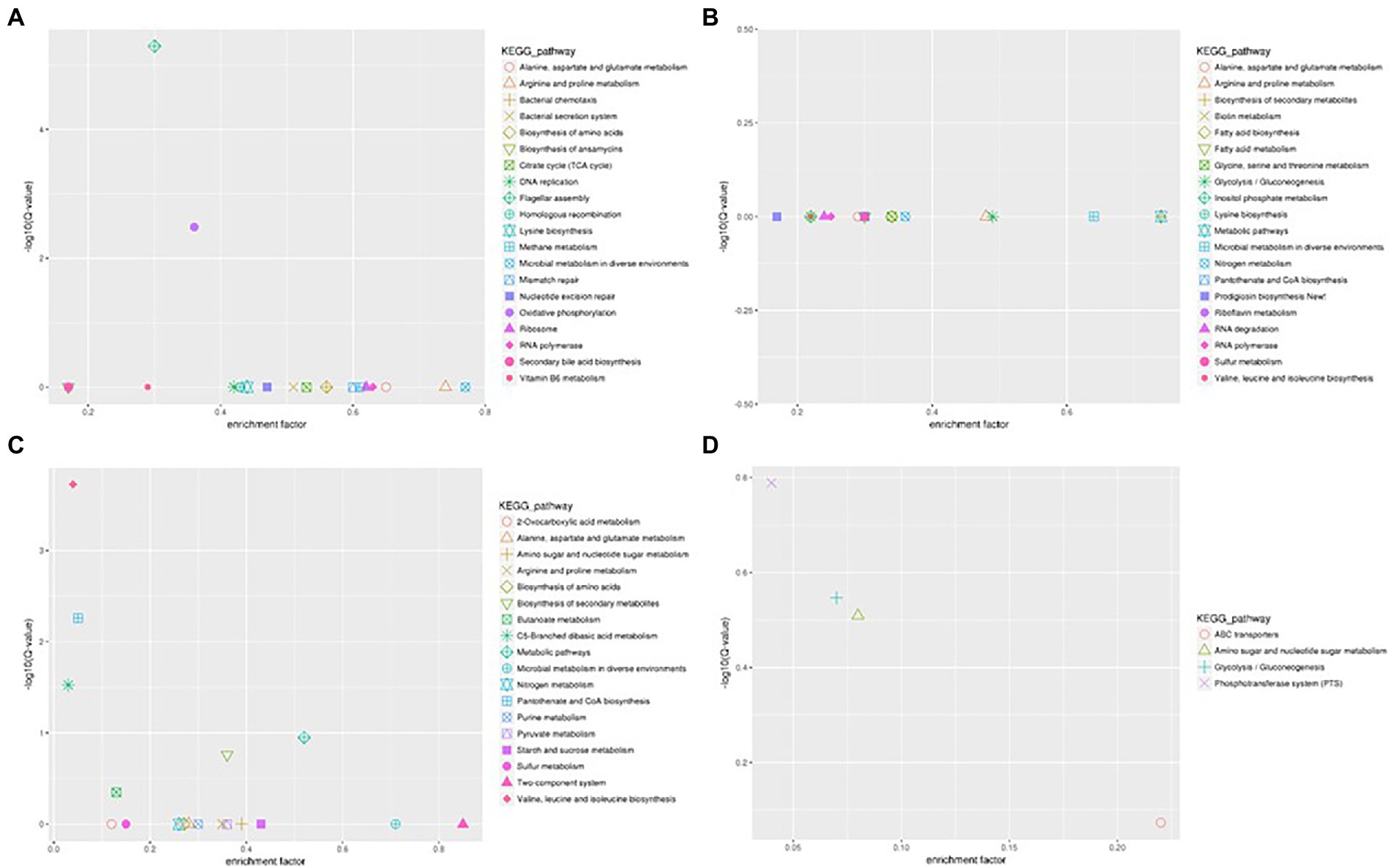
Figure 5. Scatter plot of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of the DEGs of B. subtilis 1JN2 after Cd2+ treatment. (A–D) Means the comparison of DEGs according to KEGG pathway enrichment between S1 and S2, S2 and S3, S3 and S4, S4 and S5 separately.
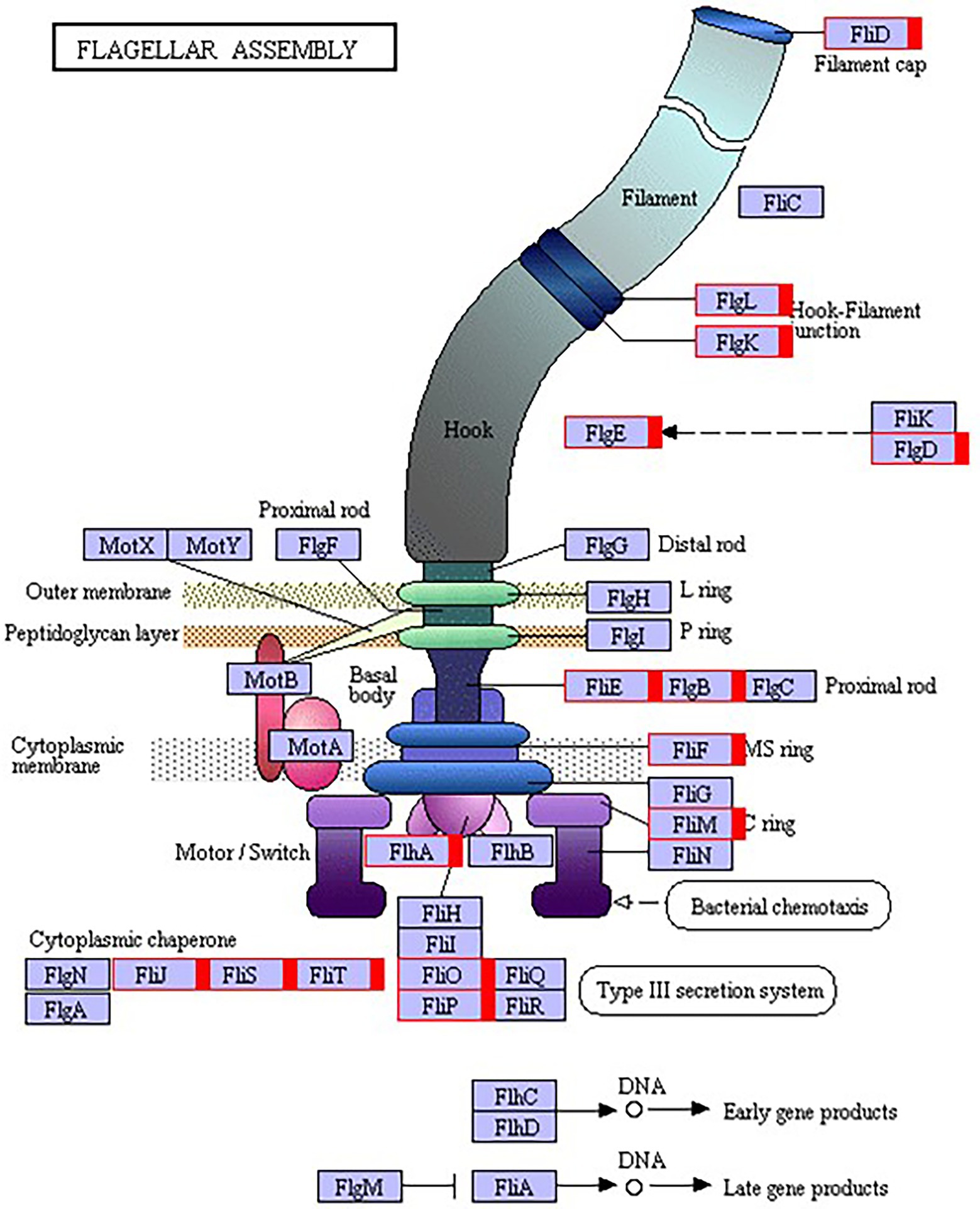
Figure 6. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation of the genes involved in flagella encoding and assembly of B. subtilis 1JN2 after Cd2+ treatment. Genes markered with red indicate up-regulation.
Effect of Cd2+ on the extracellular secretions of strain 1JN2
We evaluated the effects of Cd2+ stress on the extracellular secretions (polysaccharides and proteins) of strain 1JN2 and found that the extracellular protein-related genes (tapA-sipW-tasA) were not expressed in the strain. However, the expression of EPS-encoding genes (epsA-O) decreased significantly after Cd2+ treatment, indicating that Cd2+ reduces polysaccharides levels in B. subtilis 1JN2 biofilm (Figure 7). According to Yu et al. (2016), γ-PGA plays an important role in polymorphism of B. subtilis biofilm; therefore, we assessed the expression of the γ-PGA-encoding genes in this study. Moreover, the genes encoding γ-PGA (pgsB-pgsC-pgsA) were highly expressed after Cd2+ treatment, but there was no significant difference in the expression across the last four sampling time points. This indicated that changes in the extracellular secretions mainly occurred within a certain period after 1JN2 exposure to Cd2+ treatment. Similar results were obtained by comparing the changes in EPS and γ-PGA levels at different time points after Cd2+ treatment. The EPS levels remained unchanged at very low concentrations, while γ-PGA content increased significantly compared to the control group Figure 8).
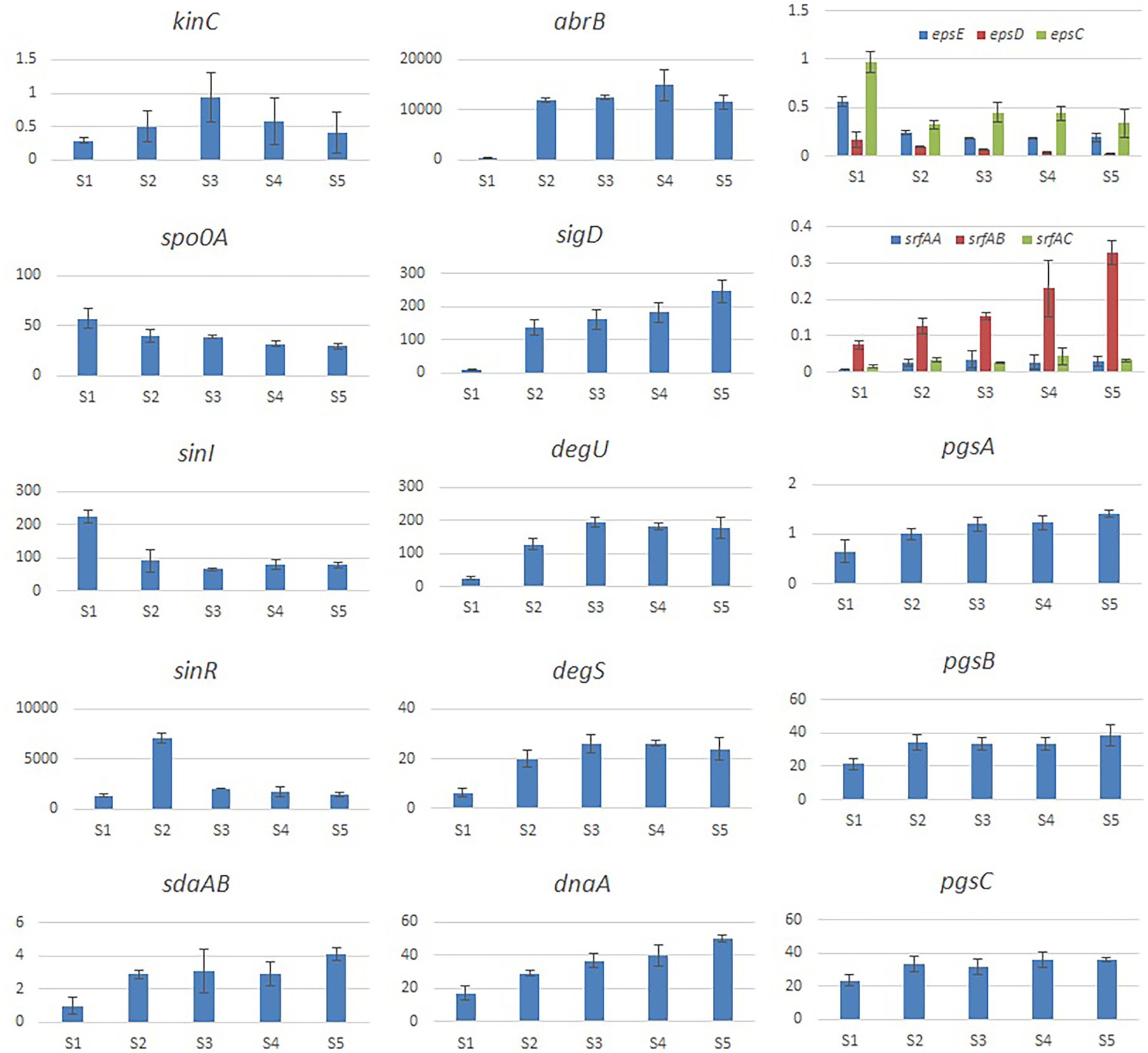
Figure 7. Relative expression level of the genes involved in extracellular secretion and regulation of B. subtilis 1JN2 after Cd2+ treatment. The histogram was based on the expression level in FPKM (Fragments per Kilobase of Transcript per Million Fragments Mapped) value.
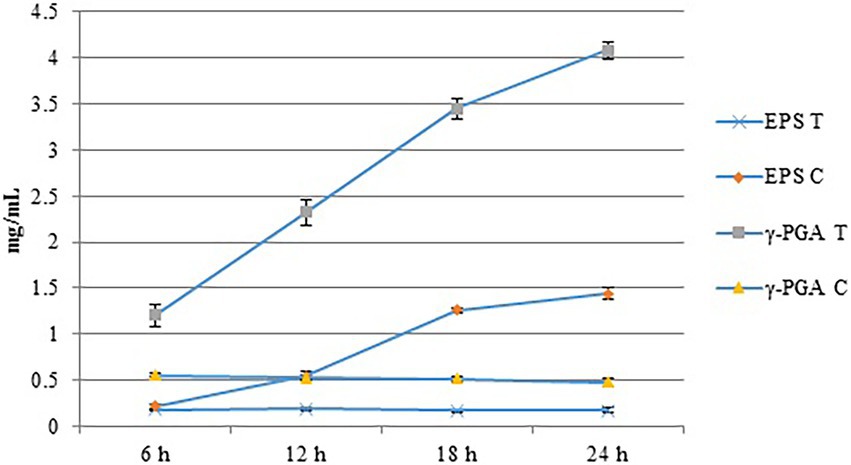
Figure 8. The content of EPS and γ-PGA of the strain 1JN2 after Cd2+ treatment. 3 mM Cd2+ was added in the broth and the supernatants at 6, 12, 18 and 24 h after inoculation were used to detect the content of EPS and γ-PGA. T means treated with Cd2+ and C means control without Cd2+.
Additionally, studies have reported that surfactant secretion by Bacillus increases after exposure to cations; thus, we evaluated the expression of surfactin-encoding genes. The expression levels of the genes related to the surfactin-related genes (srfAA-srfAB-srfAC) were also significantly increased following Cd2+ treatment (Figure 7).
Effect of Cd2+ on the regulation pathway of 1JN2 biofilm formation
According to related reports, multiple histidine kinases (KinA, KinB, KinC, KinD, and KinE) mainly sense exogenous environmental signals and collectively act on Spo0A either directly through protein phosphorylation or indirectly via a phospho-relay (Mcloon et al., 2011). We examined the expression of the regulatory factors of the biofilm formation pathway of 1JN2 and found that the expression of kinC significantly increased after exposure to Cd2+, reaching its highest level at 24 h (Figure 7). This shows that KinC plays an important role in detecting the change in exogenous ion signals. It has been reported that KinC induces low levels of Spo0A-P in response to potassium cation evacuation caused by surfactin-generated pores in the B. subtilis membrane (López et al., 2009). However, the main regulatory factor showed a downward trend. Therefore, increased kinC levels reduced the expression of Spo0A after exposure to Cd2+ (Figure 7). To explain this, we also examined the trends in Sda-and DnaA-encoding genes following Cd2+ treatment. Sda is a critical protein that controls the sporulation or biofilm states of bacterial cells. It can prevent the transfer of phosphate groups from histidine kinase to Spo0F, thereby blocking or delaying Spo0A activity (Whitten et al., 2007; Yan et al., 2016). The replication initiation protein, DnaA, activates sda, which effectively prevents phosphate group accumulation and activates Spo0A, thereby preventing cells from premature sporulation (Burkholder et al., 2001). Sda expression and activity are reduced when the cell enters the stationary phase, thus reversing the Spo0A and proteolytic activities of the existing Sda protein (Ruvolo et al., 2006). Accordingly, the increased expression levels of dnaA and sdaAB after exposure to Cd2+may have led to the changes in Spo0A. Meanwhile, the two-component regulatory system SinI/SinR limited the production of extracellular polysaccharides, similar to the effects of increased expression of AbrB-encoding genes after Cd2+ treatment.
We also evaluated the changes in the γ-PGA regulatory factors and found that SigD significantly increased after Cd2+ treatment. This activated the expression of flagella encoding and assembly genes by regulating γ-PGA production through another two-component regulatory system called DegU/S. Expression of the components of this regulatory pathway (sigD, degS, and degU) increased consistently, resulting in (n of flagella encwhich in turn likely enhanced the mobility of the strain. The changes in the expression levels of γ-PGA and EPS are consistent with the previously reported switch-like mechanism (Yu et al., 2016).
Real-time PCR validation of the selected DEGs
Five genes were selected for qPCR assay to confirm the reproducibility and accuracy of the transcriptome data. We found only minor differences between the two data sets from RNA-seq and RT-PCR analyses (Figure 9).
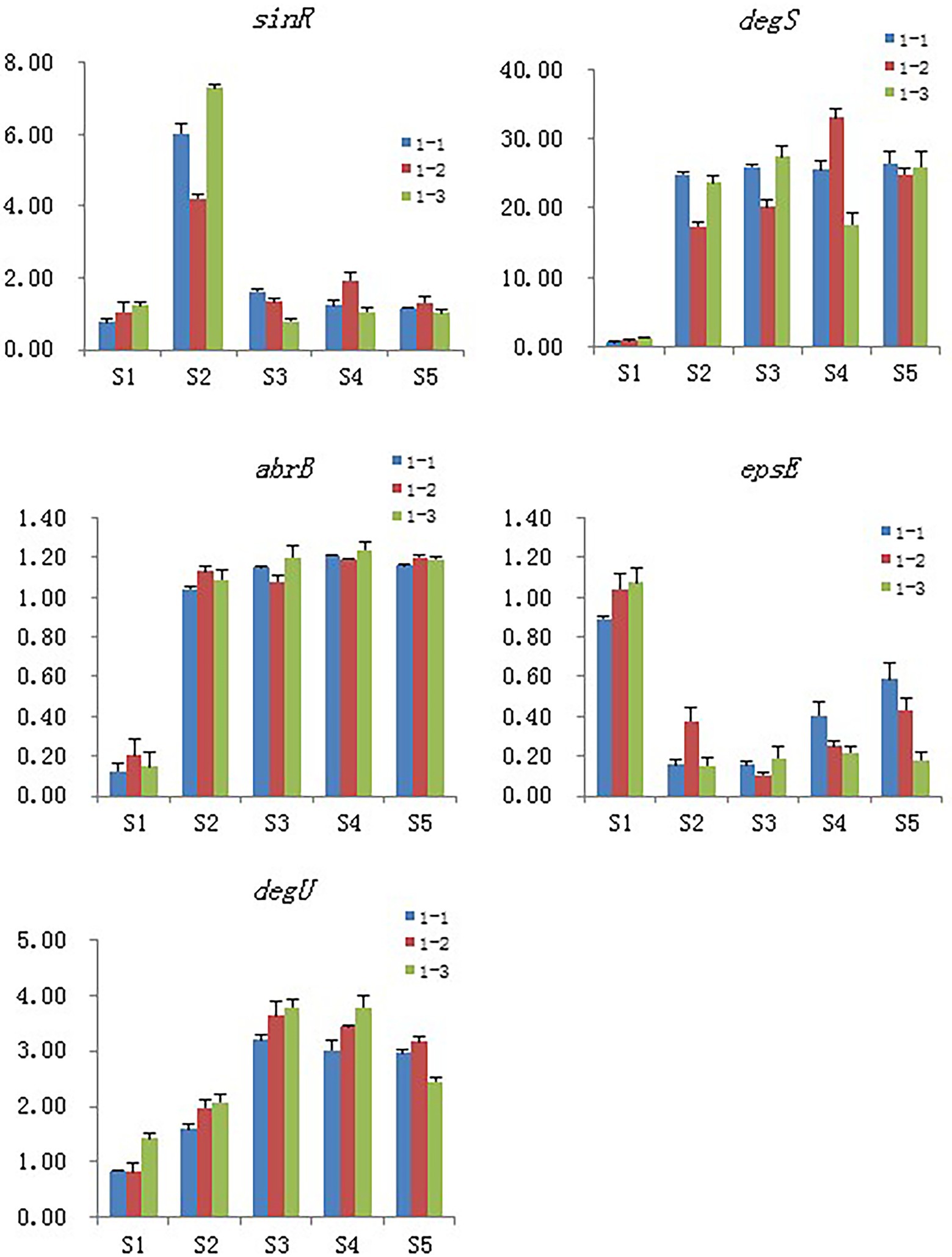
Figure 9. Validation of the selected DEGs by real-time PCR. Samples collected at 6 h, 12 h, 18 h, and 24 h were named as S2, S3, S4, S5 and the blank control that without Cd2+ was named as S1, 1–1, 1–2 and 1–3 means the three replicates of each sample.
Discussion
Biofilms play important roles in the life cycle of Bacillus (Branda et al., 2001; Vlamakis et al., 2013), such as protection from antibiotics (Høiby et al., 2010) and adherence to the surface of objects. Bacillus is a preferable biocontrol agent of plant pathogens because of its strong survivability; therefore, various Bacillus species have been screened from the laboratory and applied in the field (Chen et al., 2012). Colonization is a very important factor in biocontrol efficacy since the level of colonization directly affects the inhibition capacity against plant pathogens of biocontrol agents (Chen et al., 2013). Various factors such as light, pH, and metal ions affect the colonization ability of biocontrol strains in the field. These factors also represent bottlenecks limiting the application of laboratory-generated biocontrol strains in the field.
Among the main components of Bacillus biofilm, both EPS and γ-PGA were reported to help the strain to colonize the host roots, thus inhibiting plant pathogens effectively (Yu et al., 2016). Wild-type Bacillus species exhibit different biofilm morphologies owing to differences in their extracellular secretions. Moreover, the role of these components in biofilm formation and morphological changes has been demonstrated using deletion mutants of different extracellular secretory components (Yan et al., 2016; Yu et al., 2016). Our previous work demonstrated that Cd2+ can change the biofilm morphology of B. subtilis 1JN2 (Yang et al., 2015). High Cd2+ concentrations increased the viscosity and flattened the surface of the 1JN2 biofilm, showing that exogenous heavy metal ions can affect the composition of the extracellular secretion of the strain. Accordingly, it is necessary to evaluate the mechanisms involved in the extracellular component change and determine whether the biological functions of the strain are affected by the change.
B. subtilis 1JN2 has proven to be a significant biocontrol agent against Ralstonia via host colonization (Yang et al., 2012). In a follow-up study, we found that the strain can effectively adsorb cadmium ions in the environment to relieve the stress to host plants (Yang et al., 2015). After cadmium ion treatment, the biofilm morphology of the strain changed significantly, but its biocontrol effect on bacterial wilt did not change significantly. Thus,we assessed whether the biofilm morphological changes caused by Cd2+ impact the colonization ability of this strain. The results show that the change in extracellular components caused by Cd2+ treatment reduced tomato root surface colonization by the strain. However, as mentioned above, the effect of Cd2+ on the biocontrol efficiency of B. subtilis 1JN2 against Ralstonia was not significant. Therefore, we further explored the mechanism by which this strain can simultaneously control bacterial wilt and assist the host to alleviate cadmium stress.
The transcriptome sequencing results show that the expression levels of the EPS-encoding genes were significantly reduced, but that of γ-PGA genes increased after Cd2+ treatment. Further analysis revealed that KinC is the receptor of exogenous Cd2+ in B. subtilis 1JN2, which activates the main regulatory factor Spo0A through phosphorylation. This activation process is regulated by the critical protein Sda, which also controls sporulation or biofilm formation in bacterial cells. The decline in Spo0A levels inhibited the EPS synthesis through the SinI/SinR two-component regulatory system and increased AbrB levels. According to previous reports, metal ions generally positively regulate biofilm formation. For example, in Salmonella enterica and Erwinia amylovora, the regulator Zur specifically regulates the uptake of zinc ions and promotes the formation of bacterial biofilms (Ammendola et al., 2016; Kharadi and Sundin, 2020). For some pathogenic microorganisms, ion acquisition is critical for survival within the host. Therefore, specialized ion uptake regulators such as Fur and XibR can help these pathogens obtain enough metal ions and can regulate their adhesion and pathogenicity (Pandey et al., 2016; Teper et al., 2021).
Cadmium ion treatment significantly increased the expression of SigD, thus enhancing the expression of the flagella encoding and assembly genes of the strain. This further activated the DegS/DegU two-component regulatory system for γ-PGA production, enhancing the mobility of the strain and conversion of EPS to γ-PGA. Both EPS and γ-PGA have been reported to improve the colonization ability of Bacillus strains. However, in our results, we found that the bacterial population that colonized the tomato root surface was decreased. Accordingly, we speculate that the roles of γ-PGA and EPS in colonization are interchangeable in some Bacillus strains but not for strain 1JN2 on tomato root surfaces. As such, there may be other reasons why this strain can simultaneously control bacterial wilt and assist the host in alleviating cadmium stress.
The expression of surfactin synthesis-related genes was also significantly increased after Cd2+ treatment. Previous studies also reported that metal ions could induce surfactant secretion by Bacillus (López et al., 2009). As an important active protein product, surfactin plays a key role in alleviating heavy metal stress and controlling plant diseases (Almoneafy et al., 2014).According to our results, increased surfactin levels may improve the biocontrol ability of the strain by resisting metal ion stress and exerting inhibitory activity against Ralstonia solanacearum.
Generally, the main changes caused by Cd2+ treatment in B. subtilis 1JN2 biofilm were decreased EPS content, increased γ-PGA and surfactin levels, and enhanced cell mobility. Based on this, we deduce that the adaptive changes by Bacillus do not affect its disease control activity within a certain range of cadmium ion concentrations. Through surfactin secretion, B. subtilis can alleviate pathogen infections and heavy metal stress for the host plant. Exploring additional mechanisms involved in B. subtilis biofilm changes and screening of multifunctional biocontrol strains will be the focus of our future work.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
WY and YL designed the experiments. WY, HY, GD, ZL, and DZ performed the experiments. WY, CJ, DG, and DN analyzed the results and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Natural Science Foundation for Higher Education Institutions of Jiangsu Province (21KJA210005), Natural Science Foundation of Jiangsu Province (BK20170467), and Jiangsu University Student Innovation Training Program (201910323013Z). These funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Acknowledgments
We appreciated Genepioneer Biotechnologies Co., Ltd. Nanjing, China, for the help in data analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can befound online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1002482/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Colonization of B. subtilis 1JN2 on the root of tomato with (C) or without (B) Cd2+ treatment.
SUPPLEMENTARY FIGURE S2 | PCA analysis of the differentially expressed genes of B. subtilis 1JN2 after Cd2+ treatment. Samples collected at 6h, 12h, 18h, and 24h were named as S2, S3, S4, S5 and the blank control that without Cd2+ was named as S1.
Footnotes
References
Almoneafy, A. A., Kakar, K. U., Nawaz, Z., Li, B., saand, M. A., Chun-lan, Y., et al. (2014). Tomato plant growth promotion and antibacterial related-mechanisms of four rhizobacterial Bacillus strains against Ralstonia solanacearum. Symbiosis 63, 59–70. doi: 10.1007/s13199-014-0288-9
Alviz-Gazitua, P., Fuentes-Alburquenque, S., Rojas, L. A., Turner, R. J., Guiliani, N., and Seeger, M. (2019). The response of Cupriavidus metallidurans CH34 to cadmium involves inhibition of the initiation of biofilm formation, decrease in intracellular c-di-GMP levels, and a novel metal regulated phosphodiesterase. Front. Microbiol. 10:1499. doi: 10.3389/fmicb.2019.01499
Ammendola, S., D’Amico, Y., Chirullo, B., Drumo, R., Civardelli, D., Pasquali, P., et al. (2016). Zinc is required to ensure the expression of flagella and the ability to form biofilms in salmonella enterica sv typhimurium. Metallomics 8, 1131–1140. doi: 10.1039/C6MT00108D
Ashiuchi, M., and Misono, H. (2002). Biochemistry and molecular genetics of poly-γ-glutamate synthesis. Appl. Microbiol. Biotechnol. 59, 9–14. doi: 10.1007/s00253-002-0984-x
Bais, H. P., Fall, R., and Vivanco, J. M. (2004). Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 134, 307–319. doi: 10.1104/pp.103.028712
Banin, E., Vasil, M. L., and Greenberg, E. P. (2005). Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. 102, 11076–11081. doi: 10.1073/pnas.0504266102
Beauregard, P. B., Chai, Y., Vlamakis, H., Losick, R., and Kolter, R. (2013). Bacillus subtilis biofilm induction by plant polysaccharides. Proc. Natl. Acad. Sci. U. S. A. 110, 1621–1630. doi: 10.1073/pnas.1218984110
Branda, S. S., Chu, F., Kearns, D. B., Losick, R., and Kolter, R. (2006). A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59, 1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x
Branda, S. S., Gonzálezpastor, J. E., Benyehuda, S., Losick, R., and Kolter, R. (2001). Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 98, 11621–11626. doi: 10.1073/pnas.191384198
Burkholder, W. F., Kurtser, I., and Grossman, A. D. (2001). Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cells 104, 269–279. doi: 10.1016/S0092-8674(01)00211-2
Chai, Y., Norman, T., Kolter, R., and Losick, R. (2011). Evidence that metabolism and chromosome copy number control mutually exclusive cell fates in Bacillus subtilis. EMBO J. 30, 1402–1413. doi: 10.1038/emboj.2011.36
Chen, Y., Cao, S., Chai, Y., Clardy, J., Kolter, R., Guo, J. H., et al. (2012). A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol. Microbiol. 85, 418–430. doi: 10.1111/j.1365-2958.2012.08109.x
Chen, H., Teng, Y., Lu, S., Wang, Y., and Wang, J. (2015). Contamination features and health risk of soil heavy metals in China. Sci. Total Environ. 512, 143–153. doi: 10.1016/j.scitotenv.2015.01.025
Chen, Y., Yan, F., Chai, Y., Liu, H., Kolter, R., Losick, R., et al. (2013). Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol. 15, 848–864. doi: 10.1111/j.1462-2920.2012.02860.x
Cheng, Y., Yang, R., Lyu, M., Wang, S., Liu, X., Wen, Y., et al. (2018). IdeR, a DtxR family iron response regulator, controls iron homeostasis, morphological differentiation, secondary metabolism, and the oxidative stress response in Streptomyces avermitilis. Appl. Environ. Microbiol. 84:e01503-18. doi: 10.1128/AEM.01503-18
Dinh, T. L., Akhmetova, G. R., Martykanova, D. S., Rudakova, N. L., and Sharipova, M. R. (2019). Influence of divalent metal ions on biofilm formation by Bacillus subtilis. Bionanoscience 9, 521–527. doi: 10.1007/s12668-019-00621-3
Hamon, M. A., and Lazazzera, B. A. (2001). The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 42, 1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x
Høiby, N., Bjarnsholt, T., Givskov, M., Molin, S., and Ciofu, O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332. doi: 10.1016/j.ijantimicag.2009.12.011
Kaçar, Y., Arpa, Ç., Tan, S. M., Denizli, A., Genç, Ö., and Arıca, M. Y. (2002). Biosorption of hg(II) and cd(II) from aqueous solutions: comparison of biosorptive capacity of alginate and immobilized live and heat inactivated Phanerochaete chrysosporium. Process Biochem. 37, 601–610. doi: 10.1016/S0032-9592(01)00248-5
Kharadi, R. R., and Sundin, G. W. (2020). Cyclic-di-GMP regulates autoaggregation through the putative peptidoglycan hydrolase, EagA, and regulates transcription of the znuABC zinc uptake gene cluster in Erwinia amylovora. Front. Microbiol. 11:605265. doi: 10.3389/fmicb.2020.605265
Koo, H., Allan, R. N., Howlin, R. P., Stoodley, P., and Hall-Stoodley, L. (2017). Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 15, 740–755. doi: 10.1038/nrmicro.2017.99
Lei, Z., Wu, Y., Nie, W., Yin, D., Yin, X., Guo, Y., et al. (2018). Transcriptomic analysis of xylan oligosaccharide utilization systems in Pediococcus acidilactici strain BCC-1. J. Agric. Food Chem. 66, 4725–4733. doi: 10.1021/acs.jafc.8b00210
Li, L., Lin, Q., Li, T., He, X., Peng, S., and Tao, Y. (2020). Transcriptional response of pseudomonas chenduensis strain MBR to cadmium toxicity. Appl. Microbiol. Biotechnol. 104, 9749–9757. doi: 10.1007/s00253-020-10928-x
Li, Y., Zhang, W., Chang, L., Han, Y., Sun, L., Gong, X., et al. (2016). Vitamin C alleviates aging defects in a stem cell model for Werner syndrome. Protein Cell 7, 478–488. doi: 10.1007/s13238-016-0278-1
Liu, J., He, D., Li, X. Z., Gao, S., Wu, H., Liu, W., et al. (2010). Gamma-polyglutamic acid (gamma-PGA) produced by Bacillus amyloliquefaciens C06 promoting its colonization on fruit surface. Int. J. Food Microbiol. 142, 190–197. doi: 10.1016/j.ijfoodmicro.2010.06.023
López, D., Fischbach, M. A., Chu, F., Losick, R., and Kolter, R. (2009). Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 106, 280–285. doi: 10.1073/pnas.0810940106
Ma, N., and Sun, C. (2021). Cadmium sulfide nanoparticle biomineralization and biofilm formation mediate cadmium resistance of the deep-sea bacterium Pseudoalteromonas sp. MT33b. Environ. Microbiol. Rep. 13, 325–336. doi: 10.1111/1758-2229.12933
Makino, S., Uchida, I., Terakado, N., Sasakawa, C., and Yoshikawa, M. (1989). Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J. Bacteriol. 171, 722–730. doi: 10.1128/jb.171.2.722-730.1989
Mcloon, A. L., Kolodkin-Gal, I., Rubinstein, S. M., Kolter, R., and Losick, R. (2011). Spatial regulation of histidine kinases governing biofilm formation in Bacillus subtilis. J. Bcteriol. 193, 679–685. doi: 10.1128/JB.01186-10
Morikawa, M., Kagihiro, S., Haruki, M., Takano, K., Branda, S., Kolter, R., et al. (2006). Biofilm formation by a Bacillus subtilis strain that produces gamma-polyglutamate. Microbiology 152, 2801–2807. doi: 10.1099/mic.0.29060-0
Ogunleye, A., Bhat, A., Irorere, V. U., Hill, D., Williams, C., and Radecka, I. (2015). Poly-γ-glutamic acid: production, properties and applications. Microbiology 161, 1–17. doi: 10.1099/mic.0.081448-0
Pandey, S. S., Patnana, P. K., Lomada, S. K., Tomar, A., and Chatterjee, S. (2016). Co-regulation of iron metabolism and virulence associated functions by iron and XibR, a novel iron binding transcription factor, in the plant pathogen Xanthomonas. PLoS Pathog. 12:e1006019. doi: 10.1371/journal.ppat.1006019
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Poo, H., Park, C., Kwak, M. S., Choi, D. Y., Hong, S. P., Lee, I. H., et al. (2010). New biological functions and applications of high-molecular-mass poly-gamma-glutamic acid. Chem. Biodivers. 7, 1555–1562. doi: 10.1002/cbdv.200900283
Ruvolo, M. V., Mach, K. E., and Burkholder, W. F. (2006). Proteolysis of the replication checkpoint protein Sda is necessary for the efficient initiation of sporulation after transient replication stress in Bacillus subtilis. Mol. Microbiol. 60, 1490–1508. doi: 10.1111/j.1365-2958.2006.05167.x
Shemesh, M., and Chai, Y. (2013). A combination of glycerol and manganese promotes biofilm formation in Bacillus subtilis via histidine kinase KinD signaling. J. Bacteriol. 195, 2747–2754. doi: 10.1128/JB.00028-13
Stanley, N. R., and Lazazzera, B. A. (2005). Defining the genetic differences between wild and domestic strains of Bacillus subtilis that affect poly-gamma-dl-glutamic acid production and biofilm formation. Mol. Microbiol. 57, 1143–1158. doi: 10.1111/j.1365-2958.2005.04746.x
Sutherland, I. W. (2001). The biofilm matrix-an immobilized but dynamic microbial environment. Trends Microbiol. 9, 222–227. doi: 10.1016/S0966-842X(01)02012-1
Teper, D., Pandey, S. S., and Wang, N. (2021). The HrpG/HrpX regulon of Xanthomonads – an insight to the complexity of regulation of virulence traits in phytopathogenic bacteria. Microorganisms 9:187. doi: 10.3390/microorganisms9010187
Vlamakis, H., Chai, Y., Beauregard, P., Losick, R., and Kolter, R. (2013). Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168. doi: 10.1038/nrmicro2960
Whitten, A. E., Jacques, D. A., Hammouda, B., Hanley, T., King, G. F., Guss, J. M., et al. (2007). The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition. J. Mol. Biol. 368, 407–420. doi: 10.1016/j.jmb.2007.01.064
Xue, Q. Y. (2008). Studies on Screening Strategy of Antagonists against Ralstonia solanacearum and Biocontrol Mechanism of Antagonistic Strain xy21. Nanjing: Nanjing Agricultural University.
Yan, F., Yu, Y. Y., Wang, L. Y., Luo, Y., Guo, J. H., and Chai, Y. (2016). The comER gene plays an important role in biofilm formation and sporulation in both Bacillus subtilis and Bacillus cereus. Front. Microbiol. 7:1025. doi: 10.3389/fmicb.2016.01025
Yang, J., Kloepper, J. W., and Ryu, C. M. (2009). Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 14, 1–4. doi: 10.1016/j.tplants.2008.10.004
Yang, Y. W., Mi, J. D., Liang, J. D., Liao, X. D., Ma, B. H., Zou, Y. D., et al. (2019). Changes in the carbon metabolism of Escherichia coli during the evolution of doxycycline resistance. Front. Microbiol. 10:2506. doi: 10.3389/fmicb.2019.02506
Yang, W., Xu, Q., Liu, H. X., Wang, Y. P., Wang, Y. M., Yang, H. T., et al. (2012). Evaluation of biological control agents against Ralstonia wilt on ginger. Biol. Control 62, 144–151. doi: 10.1016/j.biocontrol.2012.05.001
Yang, W., Yan, H. X., Wang, J. L., Ji, L. L., and Wang, X. F. (2015). Selection of a Bacillus subtilis strain alleviating Cd2+ stress in host plants and the mechanism. Jiangsu J. Agric. Sci. 31, 806–810. doi: 10.3969/j.issn.1000-4440.2015.04.015
Yang, W., Yan, H. X., Zhang, J., Gao, Y., Xu, W., Shang, J., et al. (2018). Inhibition of biofilm formation by Cd2+ on Bacillus subtilis 1JN2 depressed its biocontrol efficiency against Ralstonia wilt on tomato. Microbiol. Res. 215, 1–6. doi: 10.1016/j.micres.2018.06.002
Yu, Y. Y., Yan, F., Chen, Y., Jin, C., Guo, J. H., and Chai, Y. (2016). Poly-γ-glutamic acids contribute to biofilm formation and plant root colonization in selected environmental isolates of Bacillus subtilis. Front. Microbiol. 7:1811. doi: 10.3389/fmicb.2016.01811
Zhang, Y., Chen, S., Hao, X., Su, J. Q., Xue, X., Yan, Y., et al. (2016). Transcriptomic analysis reveals adaptive responses of an Enterobacteriaceae strain LSJC7 to arsenic exposure. Front. Microbiol. 7:636. doi: 10.3389/fmicb.2016.00636
Zhang, Y. J., He, L. Y., Zeng, Q., and Sheng, X. F. (2021). Surfactin production by Bacillus subtilis B12 under cd stress and its effect on biofilm formation and cd removal. Microbiol. China 48, 1883–1894. doi: 10.13344/j.microbiol.china.200966
Keywords: Bacillus subtilis, biofilm, cadmium, stress response, adaptation, transcriptomics analysis
Citation: Yang W, Yan H, Dong G, Li Z, Jiang C, Gu D, Niu D, Zhou D and Luo Y (2022) Comparative transcriptomics reveal different genetic adaptations of biofilm formation in Bacillus subtilis isolate 1JN2 in response to Cd2+ treatment. Front. Microbiol. 13:1002482. doi: 10.3389/fmicb.2022.1002482
Edited by:
Jorge Barriuso, Spanish National Research Council (CSIC), SpainReviewed by:
Mohini Prabha Singh, Punjab Agricultural University, IndiaSheo Shankar Pandey, University of Florida, United States
Copyright © 2022 Yang, Yan, Dong, Li, Jiang, Gu, Niu, Zhou and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuming Luo, eXVtaW5nbHVvQDE2My5jb20=
 Wei Yang1,2
Wei Yang1,2 Chunhao Jiang
Chunhao Jiang Yuming Luo
Yuming Luo