- State Key Laboratory of Microbial Technology, Shandong University, Qingdao, China
The filamentous fungus Trichoderma reesei secretes large quantities of cellulases and hemicellulases that have found wide applications in industry. Compared with extensive studies on the mechanism controlling cellulase gene expression, less is known about the regulatory mechanism behind xylanase gene expression. Herein, several putative sugar transporter encoding genes that showed significant upregulation on xylan were identified in T. reesei. Deletion of one such gene, gat1, resulted in increased xylanase production but hardly affected cellulase induction. Further analyses demonstrated that deletion of gat1 markedly increased XYNI production at the transcriptional level and only exerted a minor effect on XYNII synthesis. In contrast, overexpressing gat1 caused a continuous decrease in xyn1 expression. Deletion of gat1 also affected the expression of xyn1 and pectinase genes when T. reesei was cultivated with galacturonic acid as the sole carbon source. Transcriptome analyses of Δgat1 and its parental strain identified 255 differentially expressed genes that are enriched in categories of glycoside hydrolases, lipid metabolism, transporters, and transcriptional factors. The results thus implicate a repressive role of the sugar transporter GAT1 in xyn1 expression and reveal that distinct regulatory mechanisms may exist in controlling the expression of different xylanase genes in T. reesei.
Introduction
Lignocellulose including cellulose, hemicellulose, and pectin is the most abundant renewable carbon resource in nature (Rubin, 2008). Bioconversion of lignocellulose into fermentable sugars is of great significance regarding biofuel and bio-based chemical production for various industrial and synthetic applications (Himmel, 2007). In nature, a number of microorganisms are capable of efficiently degrading lignocellulose by secreting a large quantity of lignocellulolytic enzymes. Among others, the filamentous fungus Trichoderma reesei is one of the most prolific producers of cellulases and hemicellulases (mainly xylanases), which have found applications in many industrial fields for a long history (Bischof et al., 2016; Liu and Qu, 2021). Whereas T. reesei produces a much lower amount of xylanases compared to cellulases, these enzyme components play an indispensable role in increasing the accessibility of cellulases to cellulose that is embedded in hemicellulose matrix mainly composed of xylan, and therefore enhancing the overall efficiency of lignocellulose hydrolysis (Gupta et al., 2016). Trichoderma reesei secrets five endo-β-1,4-xylanases including XYNI–XYNV (Tenkanen et al., 1992, 2013; Torronen et al., 1992; Xu et al., 1998; Ogasawara et al., 2006; Herold et al., 2013) and two ß-xylosidases BXLI (Herrmann et al., 1997) and BXLII (Derntl et al., 2015) for efficient xylan degradation. XYNI and XYNII represent the major xylanases that are responsible for more than 90% of the total extracellular xylanolytic activity (Tenkanen et al., 1992; Torronen et al., 1992). Whereas the expression of XYNI and XYNII encoding genes (xyn1 and xyn2) has been known to be readily triggered by xylan or Avicel, how the induction cascade is initiated is not clear yet. Understanding the underlying regulatory mechanism involved in xylanolytic gene expression would contribute to the knowledge-based strain design for increasing xylanase production.
During the past decades, extensive studies have been performed to investigate the molecular mechanism underlying the induced expression of cellulolytic and xylanolytic genes. Several transcription factors including XYR1 (Stricker et al., 2006; Furukawa et al., 2009; Derntl et al., 2013), ACE1 (Saloheimo et al., 2000; Rauscher et al., 2006), ACE2 (Aro et al., 2001; Würleitner et al., 2003; Mach-Aigner et al., 2010), ACE3 (Häkkinen et al., 2014; Zhang et al., 2019; Xue et al., 2020), and CRE1 (Strauss et al., 1995; Mach et al., 1996; Rauscher et al., 2006) have been shown to be involved in the regulation of most cellulase and xylanase genes (for details please refer to reviews Bischof et al., 2016; Shida et al., 2016). Whereas most transcription factors target the expression of both cellulase and xylanase genes, there are indications that distinct mechanisms involving specific regulatory factors are acting in the regulation of xylanase gene expression (Derntl et al., 2013). In this respect, two repressors, Xpp1 and SxlR, have been found to exert specific regulatory effects on the expression of xylanase genes but not cellulase genes. While Xpp1 fished out by the xyn2 promoter in a pull-down assay acts as a repressor for both xyn1 and xyn2 (Derntl et al., 2015), SxlR plays a critical role in inhibiting the expression of GH11 xylanase genes including xyn1, xyn2, and xyn5 (Liu et al., 2017).
The soluble mono- or oligo-saccharides released from lignocellulose enter the cytoplasm via membrane sugar transporters, acting as either nutrient molecules to provide energy and building blocks or as signaling molecules to regulate the biosynthesis of hydrolytic enzymes (Huberman et al., 2016). Therefore, sugar transporters have been considered to play important roles in coordinating nutrient acquisition and stringent control of enzyme production in the process of lignocellulose utilization. Whereas a number of T. reesei sugar transporters have been characterized regarding their in vitro activities in transporting sugars released from lignocellulose hydrolysis (Saloheimo et al., 2007; Ivanova et al., 2013; Zhang et al., 2013; Huang et al., 2015; Sloothaak et al., 2016; Nogueira et al., 2018; Havukainen et al., 2020, 2021a,b), relatively few of them have been found to be involved in the regulation of lignocellulolytic enzyme production. In particular, the cellobiose transporter Crt1 plays an essential role in activation of cellulase genes in T. reesei, regardless of the inducing carbon sources used (Ivanova et al., 2013; Zhang et al., 2013; Havukainen et al., 2020). Deletion of crt1, however, does not compromise but improves xylanase production (Zhang et al., 2013), implicating a difference in sugar transporter-involved induction mechanisms between cellulase and xylanase genes. On the other hand, the xylose transporter TrSTR1 has been demonstrated to play an important role in xylanase synthesis (Huang et al., 2015). Moreover, the sugar transporter Tr_69957 is not only involved in uptake of cellobiose, xylose, and mannose, but also participates in the expression regulation of both cellulase and xylanase genes in the presence of sugarcane bagasse (Nogueira et al., 2018). Given that T. reesei genome contains numerous genes encoding putative sugar transporters (Martinez et al., 2008), it is most likely that specific sugar transporters exist to be involved in regulating xylanase production. Identification of such transporters and unraveling the underlying regulatory mechanism would help to optimize the ratio of xylanase-to-cellulase in the T. reesei enzyme cocktail to improve the efficiency of lignocellulose saccharification.
In this study, several putative sugar transporter encoding genes that showed significant upregulated expression on xylan were identified in T. reesei. Deletion of gat1, which was previously identified to encode a galacturonic acid transporter, increased xylanase production on xylan with a predominant effect on xyn1 expression. We also found that GAT1 is involved in the regulation of both pectinase and xylanase gene expression when T. reesei was cultivated with galacturonic acid as carbon source. Finally, transcriptome analyses of Δgat1 was performed to identify a gene set regulated by GAT1.
Materials and Methods
Strains, Media, and Culture Conditions
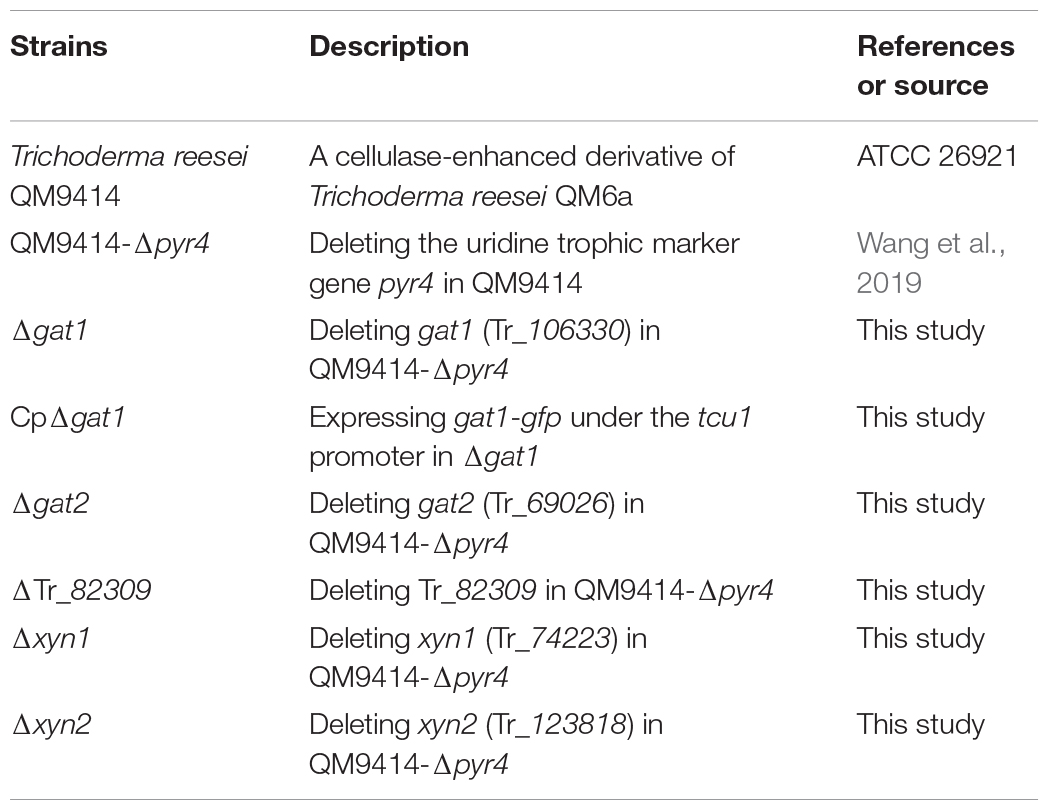
Trichoderma reesei QM9414 (ATCC 26921), a cellulase-enhanced derivative of the original strain QM6a, was used as the control strain throughout the study. QM9414–Δpyr4, a strain created by deleting the uridine trophic marker gene pyr4 in QM9414 (Wang et al., 2019), was used as the parental strain in this study. All T. reesei strains were maintained on malt extract agar. For transcription and enzyme production analyses, T. reesei cells were pre-cultured at 30°C for 36 h on a rotary shaker (200 rpm) in 1 L Erlenmeyer flasks containing 250 mL of Mandels-Andreotti (MA) medium (Mandels et al., 1962) and supplemented with 1% (v/v) glycerol. The mycelia were then harvested through filtration and washed twice with medium without any carbon source. Equal amounts of mycelia were transferred to fresh MA media containing 0.5% (w/v) beechwood xylan (Biosynth Carbosynth, United Kingdom), 1% (w/v) Avicel, 5 mM xylose, 0.5% (w/v) galacturonic acid, or 0.5% (w/v) beechwood xylan plus glucuronic acid (1 or 5 mM) as the sole carbon source, and were cultivated for the indicated time periods.
Escherichia coli DH5a was used for routine plasmid construction and amplification. Cells were cultured in lysogeny broth in a rotary shaker (200 rpm) at 37°C.
Construction of Plasmids and Trichoderma reesei Mutant Strains
To delete gat1 (Tr_106330), two DNA fragments corresponding to approximately 2.4 and 2.1 kb of gat1 up- and downstream non-coding regions were amplified from QM9414 genomic DNA with the primer pairs F106330-up/R106330-up and F106330-down/R106330-down, respectively, and successively ligated into pDonorpyr4 (Zhang et al., 2013) via BP-cloning (Invitrogen, United States). The resultant plasmid pDonor106330pyr4 was used to transform T. reesei QM9414-Δpyr4 after linearization with I-SceI. Deletion plasmids for gat2 (Tr_69026), Tr_82309, and xyn1 were similarly constructed. To construct the deletion plasmid for gat2, two DNA fragments corresponding to approximately 2.2 and 2.4 kb of gat2 up- and downstream non-coding regions were amplified with the primer pairs F69026up/R69026-up and F69026-down/R69026-down, respectively, and successively ligated into pDonorpyr4 via BP-cloning to yield the disruption vector pDonor69026pyr4. For Tr_82309, approximately two 2.3 kb of up- and downstream fragments corresponding to Tr_82309 non-coding regions were amplified with the primer pairs F82309-up/R82309-up and F82309-down/R82309-down, respectively, and successively ligated into pDonorpyr4 via BP-cloning to create pDonor82309pyr4. For xyn1, approximately two 2.4 kb of up- and downstream of xyn1 non-coding regions were amplified with the primer pairs Fxyn1-up/Rxyn1-up and Fxyn1-down/Rxyn1-down, respectively, and successively ligated into pDonorpyr4 via BP-cloning to construct the pDonorxyn1pyr4. To delete xyn2, two DNA fragments corresponding to approximately 2.4 kb of xyn2 up- and downstream non-coding regions were amplified with the primer pairs Fxyn2-up/Rxyn2-up and Fxyn2-down/Rxyn2-down, respectively. The upstream fragment was inserted into the HindIII/PmeI site of pUC19-pyr4 (Lv et al., 2015) to generate pUC19-pyr4-xyn2up. The downstream fragment was then inserted into EcoRI-linearized pUC19-pyr4-xyn2up to obtain pUC19-pyr4-xyn2. The deletion plasmids including pDonor69026pyr4, pDonor82309pyr4, and pDonorxyn1pyr4 were linearized with I-SceI and the plasmid pUC19-pyr4-xyn2 was linearized with SspI, followed by T. reesei transformation.
To complement the Δgat1 strain and detect the subcellular localization of GAT1, a 2.4 kb fragment of full-length gat1 without stop codon was amplified from QM9414 genomic DNA with the primer pair Fre106330/Rre106330, and then inserted into the AscI/PmeI site of pMDPtcu-gfp-TtrpC (Lv et al., 2015) to yield the complementation plasmid wherein gat1-gfp was under the control of the tcu1 promoter. All the primers used were listed in Supplementary Table 1.
Transformation of T. reesei was performed as previously described (Zhou et al., 2012). The transformants were selected on minimal medium for either uridine prototroph or for resistance to hygromycin (120 mg/mL). Anchored PCR was used to verify the correct integration events. Strains constructed in this study were listed in Table 1.
Growth Assays
To analyze the growth in liquid culture, T. reesei strains were pre-cultured in MA medium supplemented with 1% (v/v) glycerol at 30°C for 36 h. Mycelia were collected through filtration and washed twice with medium without any carbon source. Equal amounts of mycelia were then transferred to fresh MA medium containing 0.5% (w/v) beechwood xylan (Biosynth Carbosynth, United Kingdom) or 0.5% (w/v) galacturonic acid as the sole carbon source. After cultivation for the indicated periods, mycelia were filtered, dried at 80°C for 48 h, and finally weighed.
Fluorescence Microscopy
To visualize GAT1-GFP, spores of CpΔgat1 were inoculated into minimal medium containing 1% (w/v) glucose. After 16 h-cultivation, the mycelia were subjected to fluorescence microscopy analyses. The fluorescence of GAT1-GFP was detected with a Nikon Eclipse 80i fluorescence microscope (Nikon, Melville, NY, United States), and the images were captured and processed by NIS-ELEMENTSAR software.
Enzyme Activity and Protein Analyses
Cellobiohydrolase and β-glucosidase activities were determined by measuring the amount of released p-nitrophenol with p-nitrophenyl-D-cellobioside (pNPC; Sigma-Aldrich, United States) and p-nitrophenyl-β-D-glucopyranoside (pNPG; Sigma-Aldrich, United States) as substrates, respectively. Reaction was carried out in a 160 μL-reaction system containing 40 μL of diluted culture supernatant and 40 μL of substrate plus 40 μL of 50 mM sodium acetate buffer (pH 4.8). The mixture was incubated at 45°C for 30 min. For measurement of cellobiohydrolase activity, D-glucono-1,5-δ-lactone (1 mg/mL) was added to inhibit the activity of β-glucosidase (Deshpande et al., 1984). One unit (U) of pNPCase or pNPGase activity was defined as the amount of enzyme releasing 1 μmol of pNP per minute. For endoglucanase activity, measurement was carried out in a 120 μL-reaction mixture containing 60 μL of culture supernatant and 60 μL of 0.5% carboxymethylcellulose sodium salt (CMC, Sigma Aldrich, United States) dissolved in 50 mM sodium acetate buffer (pH 4.8) and was incubated at 50°C for 30 min. With glucose as standard, the release of reducing sugar in the mixture was determined using DNS method (Bailey et al., 1992). One unit of enzyme activity was defined as the amount of enzyme capable of releasing 1 μmol of glucose per minute. Xylanase activities were determined using beechwood xylan (Biosynth Carbosynth, United Kingdom) as substrate by measuring the amount of xylose released. Briefly, the assay was carried out in a 120 μL-reaction mixture containing 60 μL of diluted culture supernatant and 60 μL of 0.5% (w/v) xylan dissolved in 50 mM sodium acetate buffer (pH 4.8). The reaction mixture were incubated at 50°C for 10 min and the release of reducing sugar was determined using DNS method with xylose as standard. One unit of xylanase activity was defined as the amount of enzyme capable of releasing 1 μmol of xylose per minute.
SDS-PAGE and western blot were performed according to standard protocols. XYNI and XYNII were immunoblotted using polyclonal antibodies raised against peptides of XYNI (amino acids of 52–65) and XYNII (amino acids of 79–92), respectively.
Quantitative Reverse Transcription PCR
Total RNA was extracted using TRIzol reagent (Sangong Biotech, China) and purified using the TURBO DNA-free kit (Ambion, United States) to remove gDNA according to the manufacturer’s instructions. Reverse transcription was performed using the PrimeScript RT reagent Kit (Takara Bio, Japan) according to the instructions. Quantitative PCR was performed on a Bio-Rad myIQ2 thermocycler (Bio-Rad, United States). Data was analyzed using the relative quantitation/comparative CT (ΔΔCT) method and was normalized to an endogenous control (actin). Three biological replicates were performed for each analysis and the results. Statistical analysis was performed using the student’s t-test analysis.
Transcriptome Analysis
To analyze and compare the gene expression profiles of T. reesei on glucose and xylan, QM9414 was pre-cultivated in MA medium supplemented with 1% (v/v) glycerol for 36 h. An aliquot of mycelia was transferred to a fresh MA medium containing 1% (w/v) glucose or 0.5% (w/v) xylan, respectively, followed by continuous incubation at 30°C for 15 h. Similarly, to compare the gene expression profiles of Δgat1 and QM9414 on xylan, strains were pre-cultivated in MA medium supplemented with 1% (v/v) glycerol for 36 h and then transferred to a fresh MA medium containing 0.5% (w/v) xylan for further 15 h-cultivation. Filtered mycelia were quickly frozen in liquid nitrogen and stored at –80°C. RNA was extracted and RNA sequencing was performed at the Beijing Genome Institute (BGI; Shenzhen, China). Briefly, poly-(A) mRNA was purified and enriched from total RNA using Oligo-(dT) magnetic beads. Then random hexamer-primers were used to synthesize first-strand cDNA and second-strand cDNA by reverse transcriptase and DNA polymerase I. Double-stranded cDNA was then repaired to blunt ends, phosphorylated at the 5′ end and added the sticky end of “A” at the 3′ end. Suitable fragments were amplified by PCR using specific primers. The PCR products were thermally denatured into single strand and were circularized to obtain a single-stranded circular DNA library. Sequencing was performed by the BGISEQ-500 platform. Gene expression levels were calculated using FPKM (Fragments Per Kilobase of exon model per Million mapped fragments). Log2Ratio was the ratio of FPKM values for the treatment and control samples and indicates the degree of differential expression between two samples. The false discovery rate (FDR) was used to determine the P-value threshold in multiple testing. Differentially expressed genes were defined by default as those with FDR ≤ 0.001 and differences of more than two fold. Clean data was deposited in the NCBI Sequence Read Archive under the accession number of PRJNA771532 (QM9414 data on glucose/xylan) and PRJNA766839 (Δgat1/QM9414 data on xylan).
Sequence Analysis
Amino acid sequences from T. reesei were obtained from the NCBI1 or JGI2 databases.
Results
Identification of Putative Sugar Transporter Encoding Genes With Significant Induction on Xylan in Trichoderma reesei
In order to search for putative sugar transporters involved in the induced xylanase production, gene expression profiles of T. reesei QM9414 cells cultivated on glucose and xylan, respectively, were determined via transcriptome analyses. As expected, xylanolytic genes including xyn1, xyn2, xyn4, xyn5, and bxl1 were all significantly induced on xylan compared to that on glucose (Supplementary Table 2). In addition, among the differentially expressed genes between glucose and xylan cultivation, eleven genes encoding putative sugar transporters displayed a significantly upregulated expression (Table 2). Given that the two top-ranked genes, Tr_50894 and Tr_104072, have been reported to play roles in xylanase production or xylose transport (Saloheimo et al., 2007; Huang et al., 2015), we therefore chose the following three genes, Tr_82309, Tr_106330, and Tr_69206, to analyze their potential involvement in regulation of xylanase expression. While Tr_106330 and Tr_69206 have been recently demonstrated to encode sugar transporters (GAT1 and GAT2, respectively) capable of transporting galacturonic and glucuronic acids (Havukainen et al., 2021b), their roles in xylanase production is unclear.
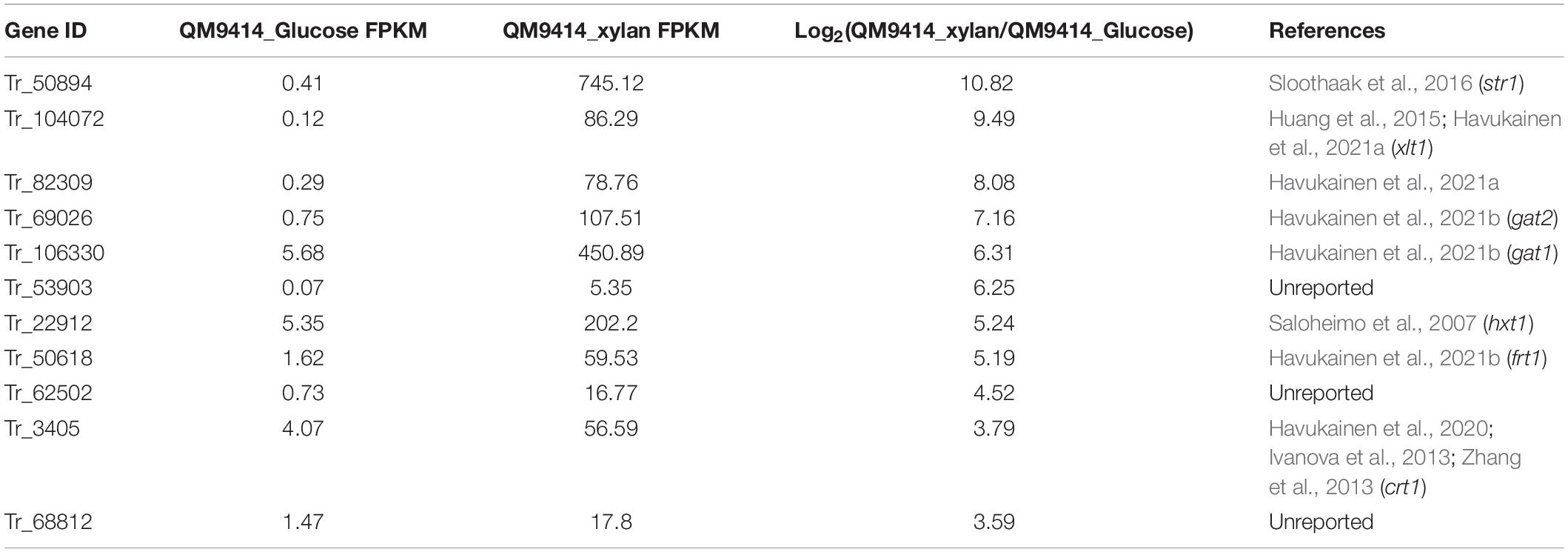
Table 2. Putative sugar transporter encoding genes with significant induction on xylan in Trichoderma reesei QM9414.
Disruption of gat1 Improved Xylanase Production of Trichoderma reesei on Xylan
To test the roles of Tr_82309, gat1, and gat2 in xylanase production in T. reesei, they were individually deleted to result in the mutant strains ΔTr_82309, Δgat1, and Δgat2, respectively. Determination of extracellular xylanase activities demonstrated that, whereas disruption of gat2 or Tr_82309 had hardly any effect on xylanase production (Figures 1A,B), disruption of gat1 resulted in a 38% increase in xylan hydrolytic activities compared to that of QM9414 strain (Figure 1C). Biomass accumulation analysis revealed little effect of gat1 deletion on mycelial growth during the whole cultivation period (Figure 1D), indicating that the increment in xylanase production caused by gat1 deletion did not result from differences in mycelial growth. Complementation of Δgat1 with an expression cassette of gat1 fused to gfp revealed that the fluorescence signal was mainly dispersed at the periphery of the complemented mycelia (Figures 2A,B), indicating that GAT1 is appropriately located on the plasma membrane. Moreover, the restored expression of GAT1 fully reverted the elevated xylanase activity to the level of QM9414 (Figure 2C), supporting that the observed enhanced xylanase production was indeed caused by gat1 deletion.
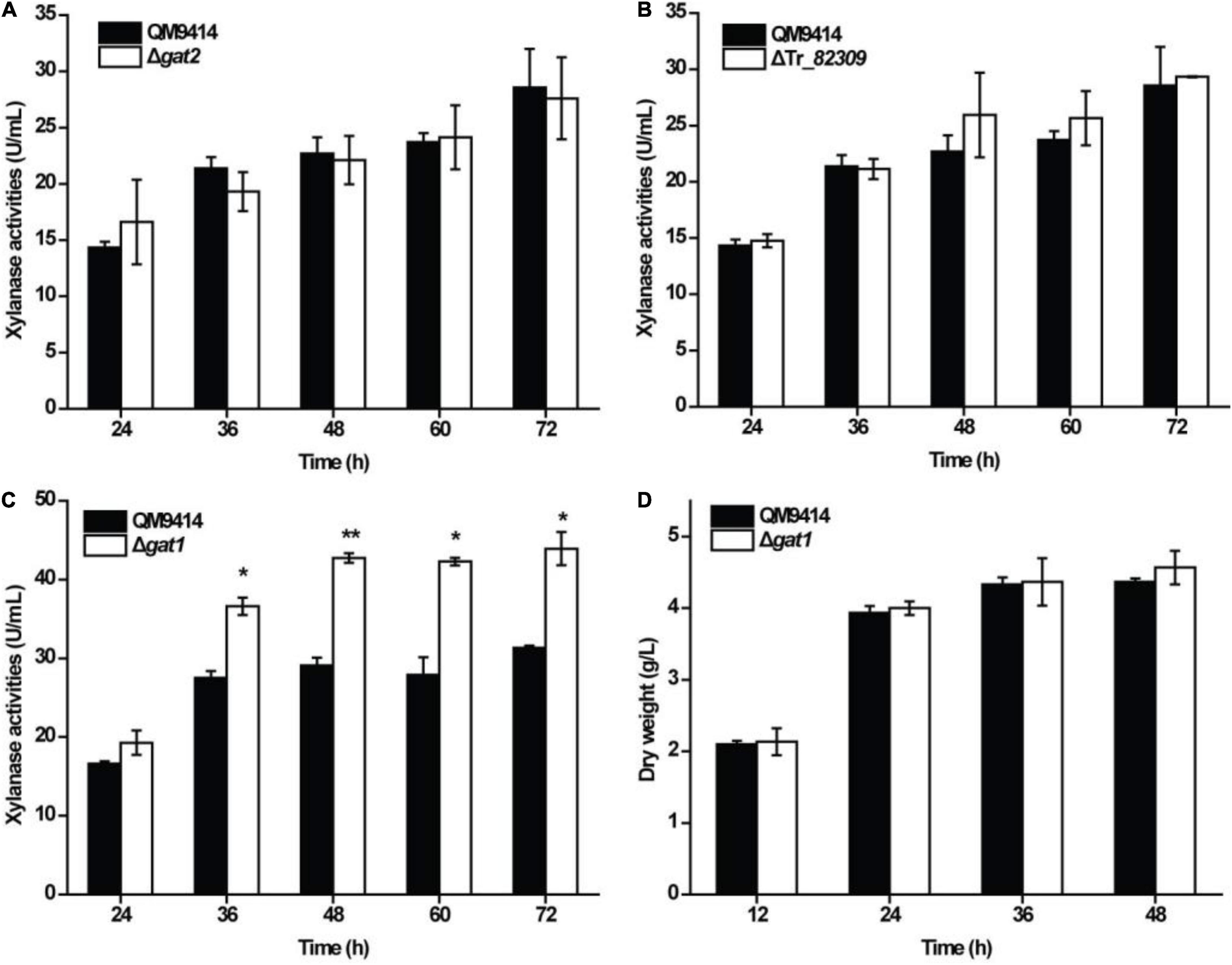
Figure 1. Deletion of gat1 improved xylanase production of Trichoderma reesei on xylan. (A–C) Xylanase activity analysis of the culture supernatant from the QM9414 and mutant strains including Δgat2 (A), ΔTr_82309 (B), and Δgat1 (C) cultivated on 0.5% (w/v) xylan for the indicated time periods. (D) Growth analyses of QM9414 and Δgat1 cultured on 0.5% (w/v) xylan. Values in this figure are the mean of three biological replicates. Error bars are the SD from these replicates. Significant differences (t-test *P < 0.05, **P < 0.01) were detected in the extracellular xylanase activities between QM9414 and Δgat1. No significant differences were detected in the extracellular xylanase activities of Δgat2 or ΔTr_82309 compared with that of QM9414. No significant differences were observed in biomass accumulation between Δgat1 and QM9414 when grown on xylan.
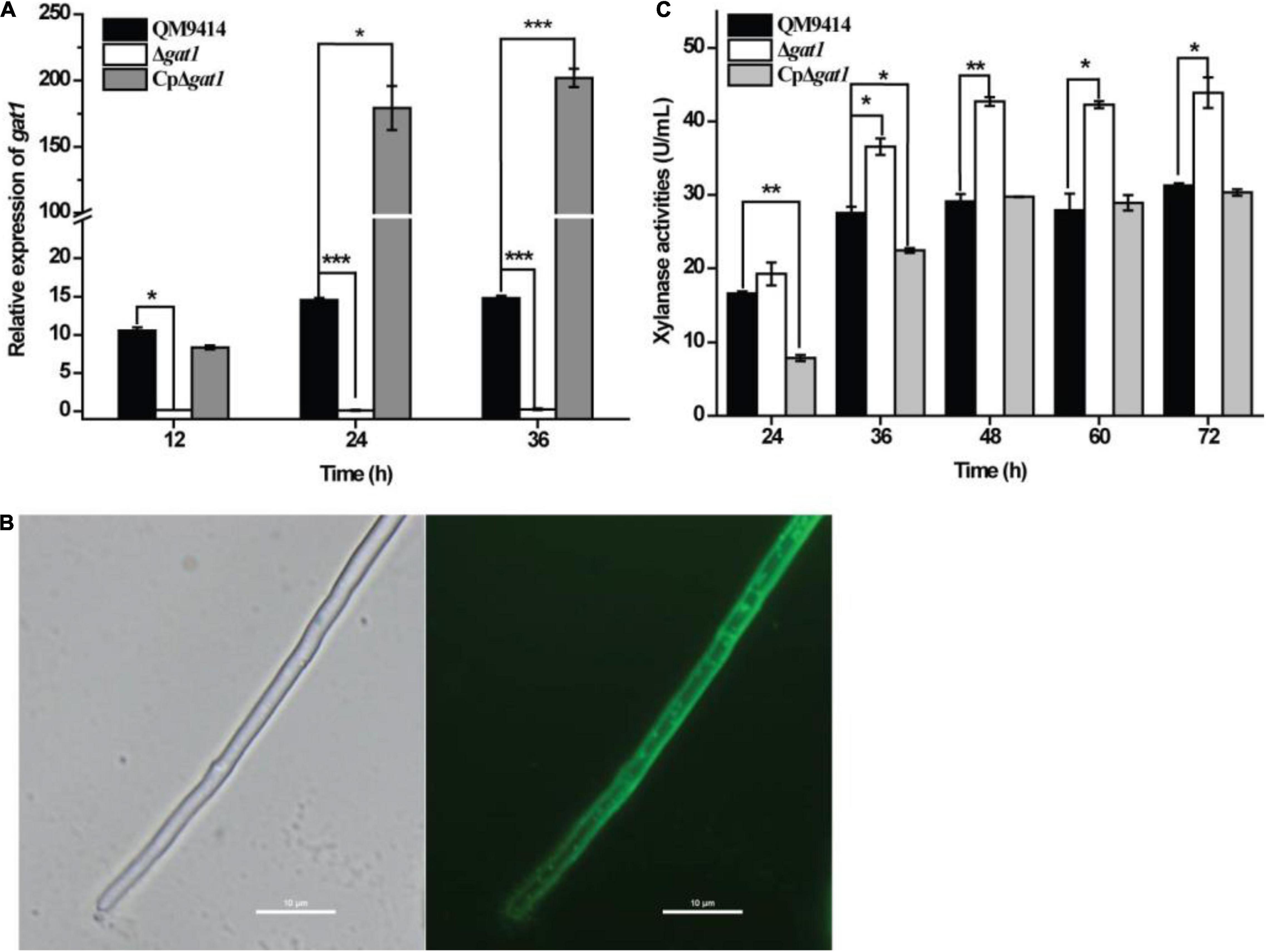
Figure 2. Complementation of Δgat1 with gat1-gfp overexpression reverted the elevated xylanase activity to the level of QM9414. (A) Quantitative RT-PCR analyses of the relative transcriptional levels of gat1 in QM9414, Δgat1, and CpΔgat1 cultivated on 0.5% xylan. (B) Fluorescence microscopic analyses of CpΔgat1 expressing gat1-gfp after cultivation on minimal medium containing 1% glucose for 16 h. (C) Xylanase activity analyses of the culture supernatant from QM9414, Δgat1 and CpΔgat1 strains cultivated on 0.5% xylan. Values in this figure are the mean of three biological replicates. Error bars are the SD from these replicates. Significant differences (t-test, *P < 0.05, **P < 0.01, and ***P < 0.0001) were detected in the transcriptional level of gat1 and xylanase activities between QM9414 and Δgat1 or between QM9414 and CpΔgat1.
To evaluate the role of GAT1 in cellulase production, Δgat1 was cultivated on Avicel cellulose and its performance was compared with that of QM9414. In contrast with the increased production of xylanase, Δgat1 showed quite similar cellulase production to that of QM9414 as determined by either extracellular cellulolytic activities or the relative transcriptional levels of the main cellulase genes (Figure 3).
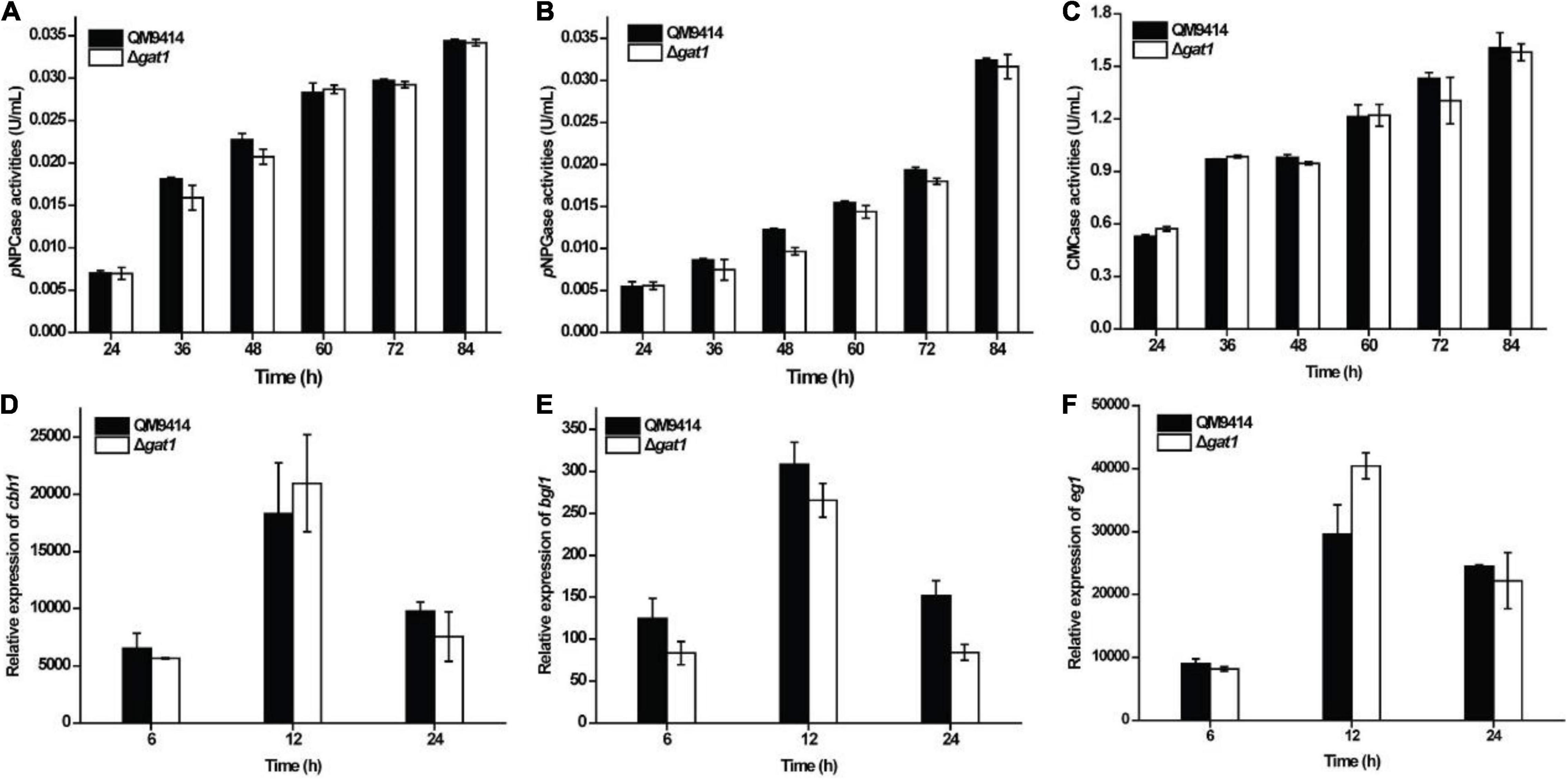
Figure 3. Deletion of gat1 hardly affected cellulase production on Avicel cellulose. (A–C) Extracellular cellobiohydrolase (A), ß-glucosidase (B), and endoglucanase (C) activities of the culture supernatant from QM9414 and Δgat1. (D–F) Transcriptional analyses of cellulase encoding genes including cbh1 (D), bgl1 (E) and eg1 (F) using quantitative RT-PCR. Strains were cultivated on 1% (w/v) Avicel cellulose for the indicated time periods. Values in this figure are the mean of three biological replicates. Error bars are the SD from these replicates. No significant differences were observed in cellulase activities or gene transcription between Δgat1 and QM9414.
Deletion of gat1 Predominantly Enhanced XYNI Production
In order to further verify the promoting effect on xylanase production caused by gat1 deletion, antibodies were raised against XYNI and XYNII, respectively, and western blot analyses were performed to detect the extracellular levels of these two main xylanases in T. reesei. To verify the specificity of antibodies, Δxyn1 and Δxyn2 were constructed in the background of QM9414–Δpyr4, respectively. Western blot analyses verified that each antibody specifically picked out the corresponding xylanase and no cross-reactivity existed (Supplementary Figure 1). Further western blot analyses demonstrated that while the production level of XYNII in Δgat1 was quite similar to that of QM9414, XYNI production was significantly elevated in the absence of GAT1 (Figures 4A,B). Consistently, quantitative reverse transcription PCR (RT-PCR) analyses showed that disruption of gat1 in QM9414 led to an up to 4-fold-enhancement in the expression of xyn1 (Figure 4C). In contrast with xyn1, gat1 deletion caused a moderate increase in xyn2 expression only at the early growth phase (Figure 4D). Since xylose has been demonstrated to act as an inducer to trigger xylanase gene expression when present in lower concentrations (Mach-Aigner et al., 2010), the expression levels of xyn1 and xyn2 were analyzed in QM9414 and Δgat1 cultured with 0.5 mM xylose, respectively, and the results showed that their expression levels in Δgat1 were increased by 4.5- and 2.6-fold, respectively (Figures 4E,F), suggesting that deletion of gat1 could also promote the expression of xylanase genes, especially xyn1, when in response to xylose.
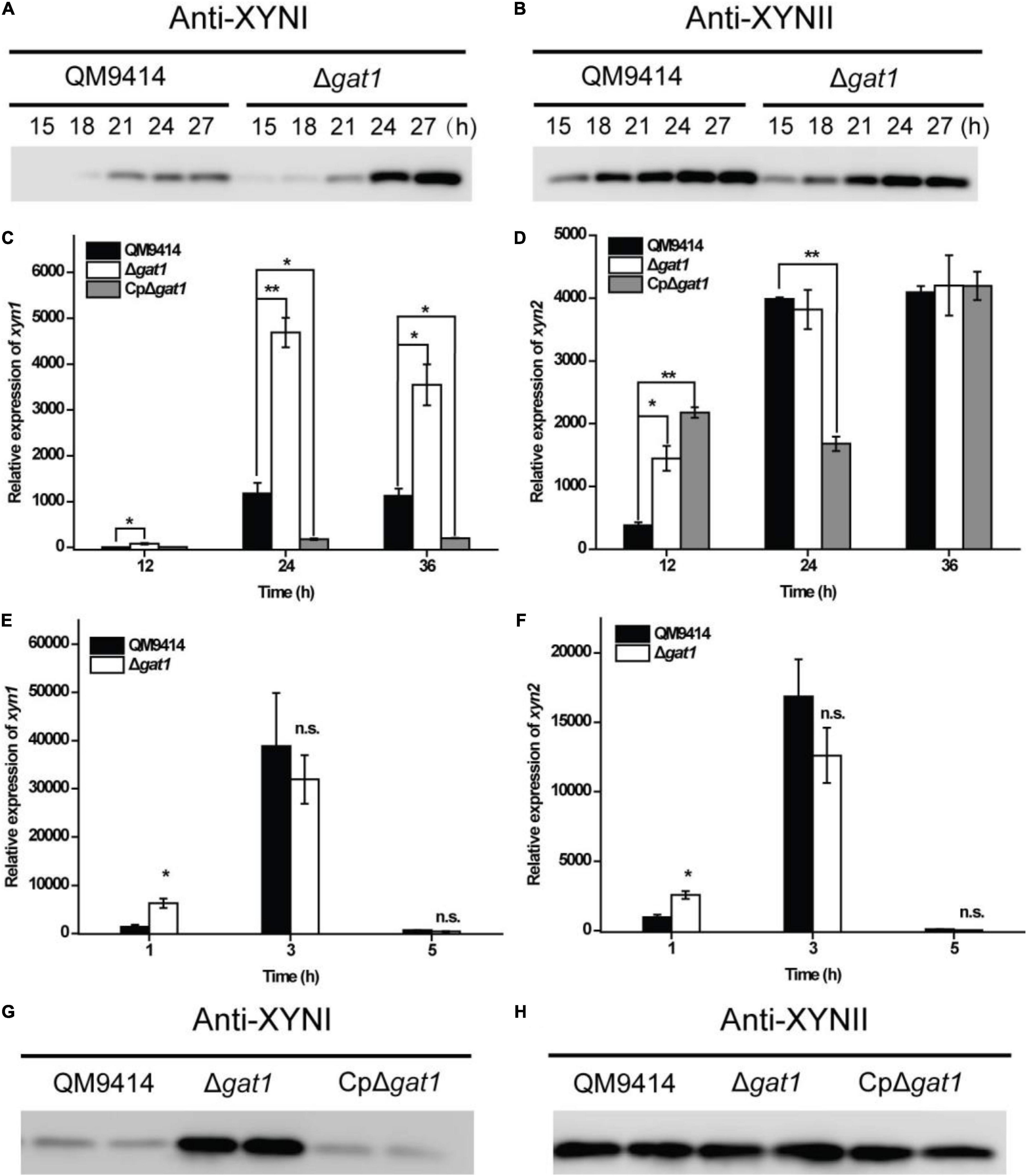
Figure 4. Deletion of gat1 predominantly increased XYNI production but exerted a minor effect on XYNII synthesis. (A,B) Western blot analyses of XYNI (A) and XYNII (B) production in the culture supernatant of QM9414 and Δgat1 that were cultivated on 0.5% xylan for the indicated time periods. (C,D) Quantitative RT-PCR analyses of the relative transcriptional levels of xyn1 (C) and xyn2 (D) of QM9414, Δgat1 and CpΔgat1 cultivated on xylan. (E,F) Quantitative RT-PCR analyses of the relative transcriptional levels of xyn1 (E) and xyn2 (F) of QM9414 and Δgat1 cultivated on 0.5 mM xylose. (G,H) Western blot analyses of XYNI (G) and XYNII (H) production in the culture supernatant of QM9414, Δgat1, and CpΔgat1 cultivated on xylan for 60 h. Values in this figure are the mean of three biological replicates. Error bars are the SD from these replicates. Significant differences (t-test, *P < 0.05, and **P < 0.01) were detected in the transcription of xyn1 or xyn2 between QM9414 and Δgat1, or between QM9414 and CpΔgat1. No significant difference (n.s.) was observed in the transcription level of xyn1 or xyn2 between QM9414 and Δgat1 after cultivation on 5 mM xylose for 3–5 h. For western blot analyses, equal amounts of culture supernatant were loaded for all strains. Three independent assays with quite similar results were performed and one representative figure is shown.
It was also observed that overexpression of gat1–gfp in Δgat1, to create the complemented strain, reversed the enhanced production of XYNI (Figure 4G). In consistence with reduced expression of xyn1 (Figure 4C), the extracellular xylanase activity during the earlier growth stage was even markedly reduced in the complemented strain (Figure 2B). Again, the GAT1-GFP overexpression exerted only a minor effect on XYNII production (Figure 4H). Taken together, the above results demonstrated that T. reesei GAT1 is involved in the regulation of xylanase genes expression with xyn1 as the major regulatory target.
Deletion of gat1 Affected Expression of Both Pectinase Genes and xyn1 on Galacturonic Acid
Given that GAT1 has been characterized as a galacturonic acid transporter (Havukainen et al., 2021b), we analyzed and compared the growth of Δgat1 and QM9414 using galacturonic acid as the sole carbon source. In consistence with the reported transporting activity of GAT1 toward galacturonic acid, Δgat1 displayed compromised biomass accumulation compared with QM9414 (Figure 5A), which was most probably caused by the impaired galacturonic acid transportation. Considering that galacturonic acid is a main hydrolysis product from pectin, the transcriptional levels of all four putative pectinase encoding genes, pec1, pec2, pec3, and pec4 (Beier et al., 2020), were analyzed. Results showed that while all the four pectin hydrolytic genes can be readily induced by galacturonic acid in QM9414, disruption of gat1 exerted differential effects on their transcriptional levels. Specifically, gat1 deletion significantly increased the expression of pec1 and pec2, but resulted in a reduced induction in the expression of pec3 and pec4 (Figures 5B–E). An induction of xyn1 by galacturonic acid was also observed in QM9414 (Figure 5F). Interestingly, the absence of GAT1 caused a significantly enhanced transcription of xyn1 compared to that of QM9414 (Figure 5F). By contrast, no increment in xyn2 transcription was observed, but rather a moderate decrease was observed at 36 h of cultivation (Figure 5G). Together these data indicate that as a galacturonic acid transporter, GAT1 participates in the regulation of xyn1 and pectinase gene expression when confronted with the hydrolytic products of pectin.
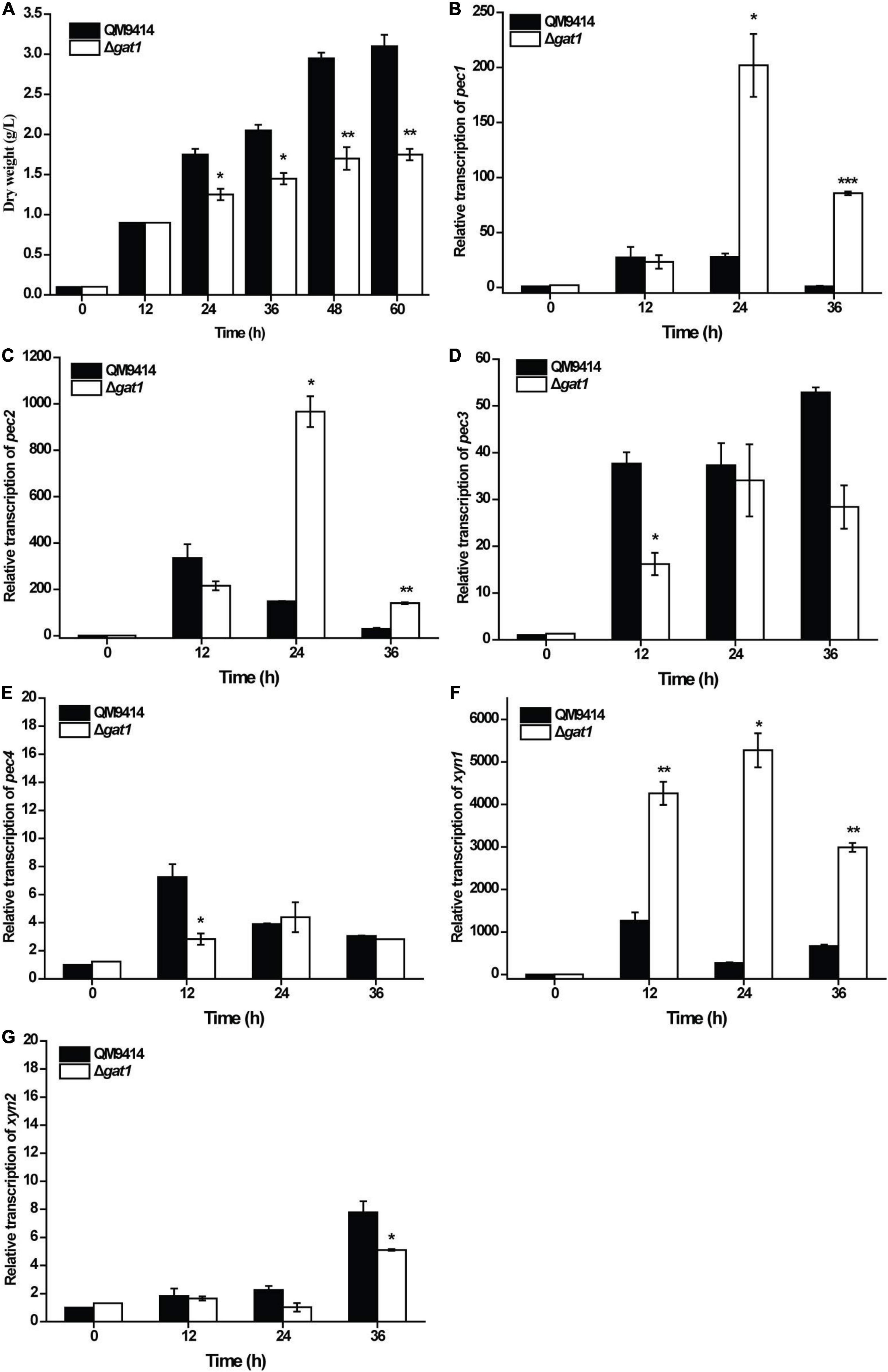
Figure 5. Deletion of gat1 affected the expression of both pectinase genes xylanase genes upon galacturonic acid. (A) Growth analyses of QM9414 and Δgat1 with 0.5% (w/v) galacturonic acid as the sole carbon source. (B–F) Quantitative RT-PCR analyses of the relative transcriptional levels of pec1 (B), pec2 (C), pec3 (D), pec4 (E), xyn1 (F) and xyn2 (G) in QM9414 and Δgat1 cultivated with 0.5% (w/v) galacturonic acid as the sole carbon source. Values in this figure are the mean of three biological replicates. Error bars are the SD from these replicates. Significant differences (t-test, *P < 0.05, **P < 0.01, and ***P < 0.001) were detected in the biomass accumulation and relative transcription of the above genes between QM9414 and Δgat1.
Transcriptome Analysis of the Δgat1 Strain
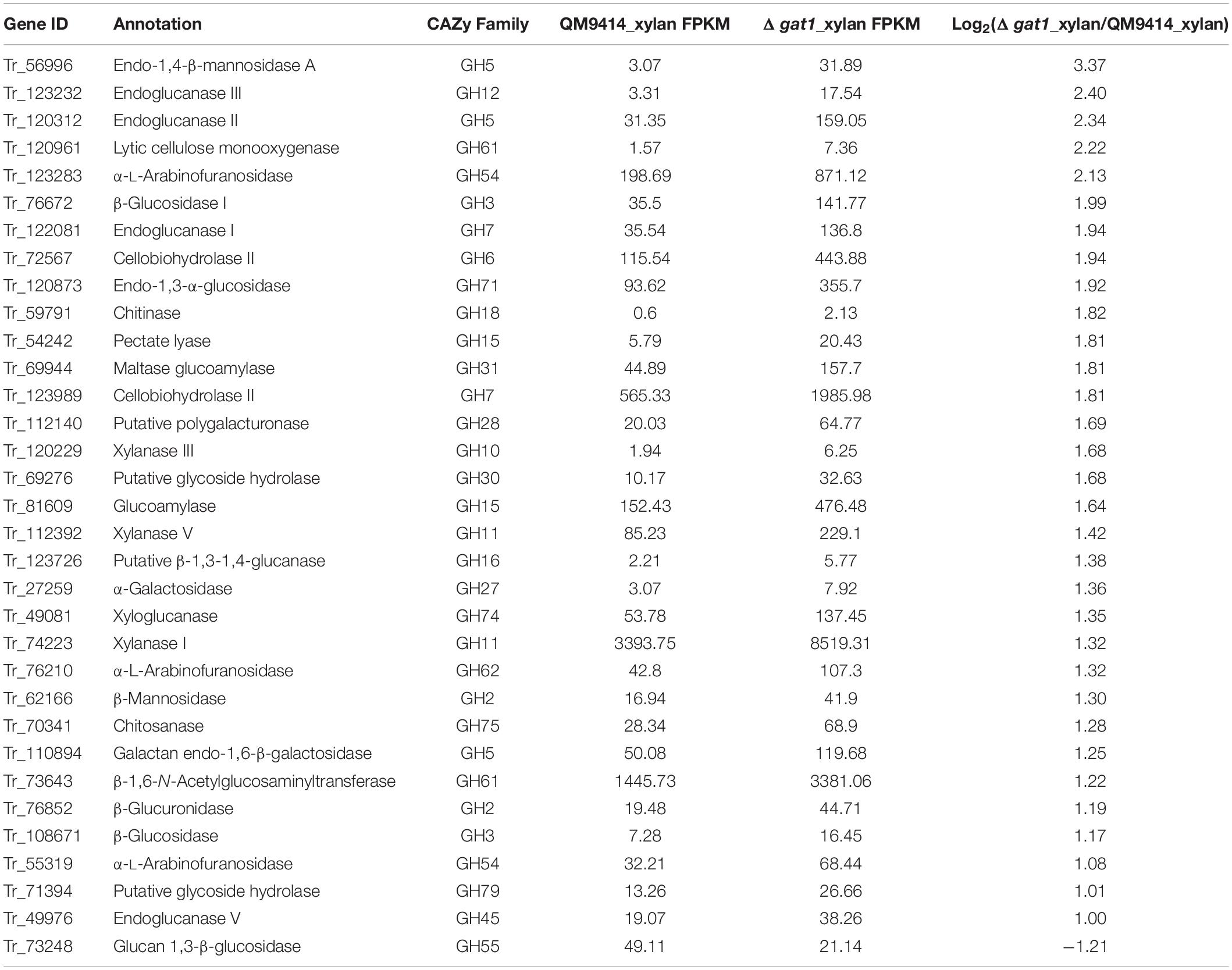
Given that the absence of GAT1 enhanced xylanase production on xylan, we further undertook RNA sequencing (RNA-seq) to identify gene sets that may be regulated by GAT1 via profiling mRNA abundances in both QM9414 and Δgat1 strains after cultivation on xylan for 15 h. A Total of 255 genes were differentially expressed in Δgat1, including 154 upregulated genes and 101 downregulated genes (Supplementary Database 1). Functional cluster analyses indicated that these differentially expressed genes are mainly enriched in the categories of glycoside hydrolases, lipid metabolism, transporters, amino acid metabolism, and transcription factors (Figure 6). In agreement with the above quantitative RT-PCR analyses, xyn1 expression in the profile of Δgat1 was increased by ∼2.5-fold while no significant increase in xyn2 expression was observed. The expression of xyn5 was also increased by ∼2.5-fold. In addition, a number of genes encoding glycoside hydrolases involved in cellulose and hemicellulose degradation also showed significant upregulation in Δgat1 (Table 3), suggesting that GAT1 has a relatively extensive effect on expression of cellulase and hemicellulase genes in T. reesei under the cultivation condition of xylan.
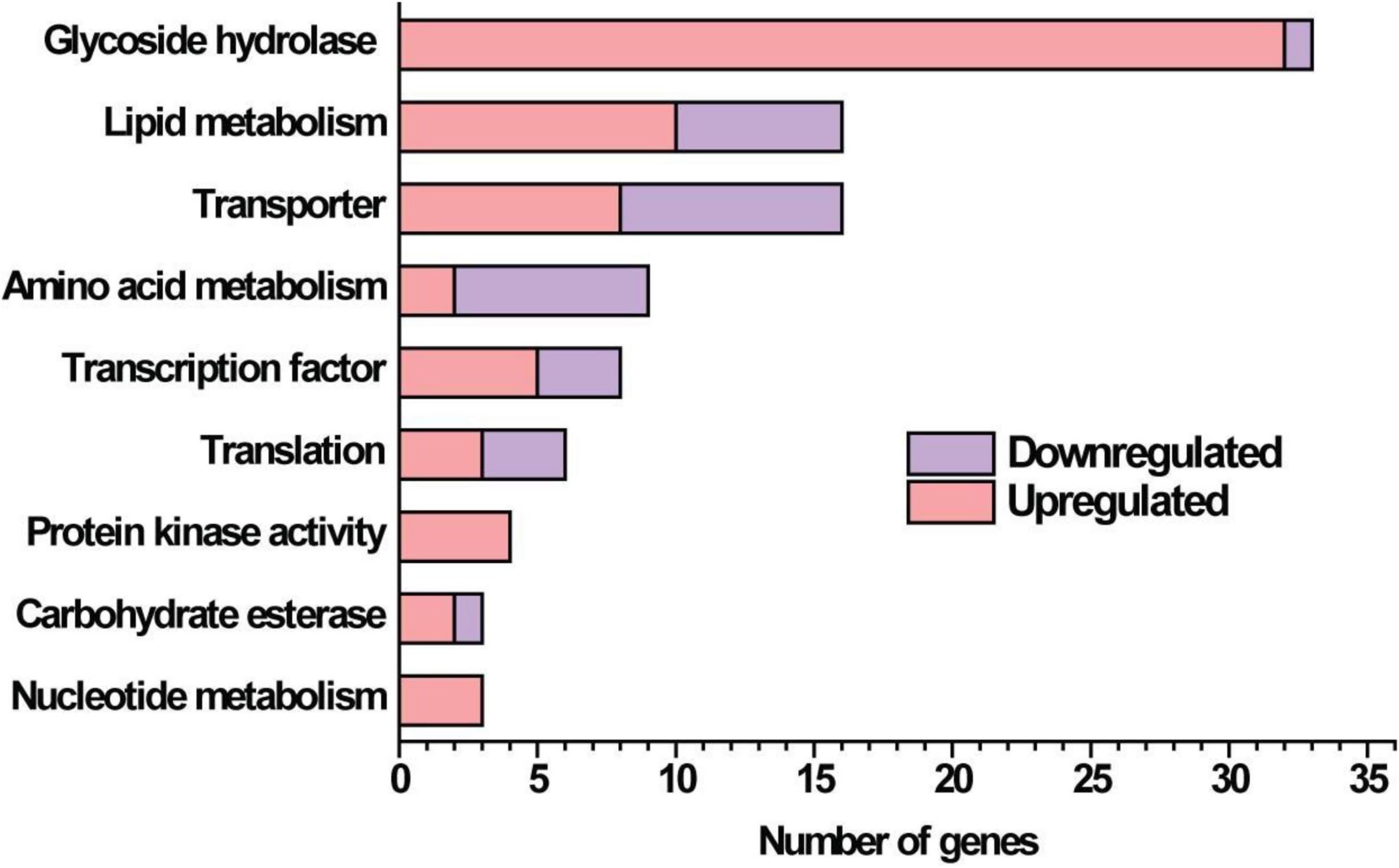
Figure 6. Functional category of differentially expressed genes in Δgat1 compared with QM9414. Transcriptome analysis identified a total of 255 differentially expressed genes between Δgat1 and QM9414. The 130 genes that encode hypothetical proteins and 27 unclassified proteins are not included in this figure.
Of the 16 putative transporter encoding genes with changed expression between Δgat1 and QM9414, four genes correspond to putative sugar transporters encoding genes (1 upregulated and 3 downregulated, Table 4). The gene encoding the sugar transporter Tr_69957 that is involved in cellobiose, mannose, and xylose absorption and cellulase and hemicellulase gene expression (Nogueira et al., 2018), showed an approximate 4.7-fold-upregulation. On the other hand, Tr_62380 encoding the well-characterized glucose/xylose transporter STR3 (Sloothaak et al., 2016; Havukainen et al., 2021b) displayed a significant downregulation (∼11 fold) in Δgat1, and the other two genes Tr_76800 and Tr_80875 encoding two putative sugar transporters with unknown functions were also downregulated. Moreover, eight transcriptional factor encoding genes were also differentially expressed in Δgat1 compared with QM9414 (Table 5). Tr_122879, the gene encoding the repressor Xpp1 (Derntl et al., 2015), was upregulated by ∼3-fold. Except for xpp1, none of the remaining seven genes has been described and their involvement in xylanase gene expression is unclear.
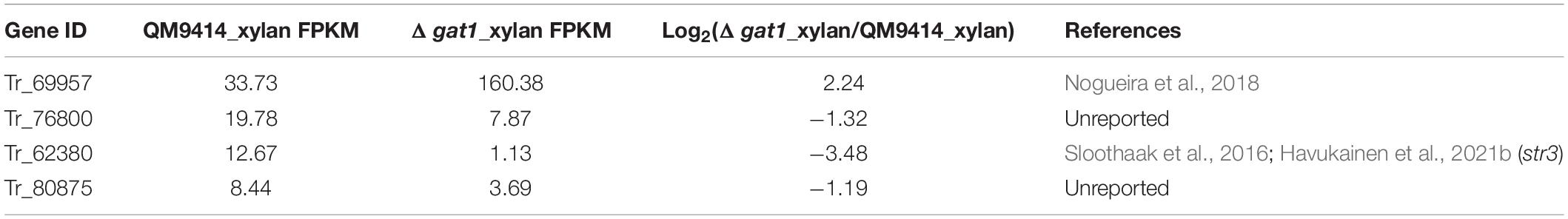
Table 4. Sugar transporter encoding genes with differential expression between Δgat1 and QM9414 on xylan.
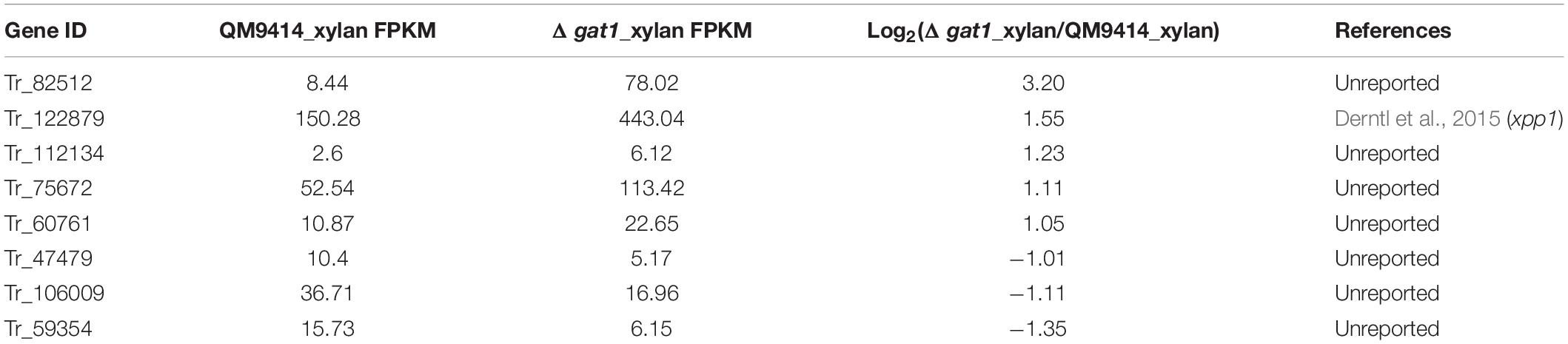
Table 5. Transcription factor encoding genes with differential expression between Δgat1 and QM9414 on xylan.
Discussion
XYNI and XYNII, the two main extracellular xylanases of T. reesei, share quite a few structural and biochemical characteristics (Tenkanen et al., 1992; Torronen et al., 1992; Torronen and Rouvinen, 1995). Although expression of both xyn1 and xyn2 is highly induced on xylan and has been known to be co-regulated by several transcriptional factors including XYR1 (Stricker et al., 2006; Furukawa et al., 2009; Derntl et al., 2013; Xue et al., 2020), ACE3 (Häkkinen et al., 2014; Zhang et al., 2019; Xue et al., 2020), Xpp1 (Derntl et al., 2015), and SxlR (Liu et al., 2017), subtle differences have been noticed regarding their induction patterns and regulatory mechanisms. In particular, xyn1 is virtually silenced in the presence of glucose while a partially constitutive expression of xyn2 is allowed under the non-inducing carbon sources (Zeilinger et al., 1996). This phenomenon is consistent with the fact that xyn1 is under direct control of the well-known carbon catabolite repressor CRE1 whereas xyn2 is not (Mach et al., 1996). Moreover, the transcriptional repressor ACE1 only binds to the promoter of xyn1 but not that of xyn2 (Rauscher et al., 2006). To the contrary, the activator ACE2 does not exert apparent regulatory effect on xyn1 expression while it contributes to the basal and induced expression of xyn2 via binding to the xylanase activating element (XAE) in the xyn2 promoter (Aro et al., 2001; Würleitner et al., 2003; Rauscher et al., 2006; Stricker et al., 2008). These published data implicate that different regulatory mechanisms work to fine tune the expression of xyn1 and xyn2. In this respect, we found that a sugar transporter GAT1 exerts differential effects on the expression of xyn1 and xyn2 in response to either xylan or galacturonic acid. The absence of GAT1 caused a significant change in the expression of xyn1 while xyn2 expression was hardly affected. Given that the absence of another similarly functional galacturonic/glucuronic acid transporter GAT2 (Tr_69026) has no effect on xylanase production in T. reesei (Figure 1A), it is speculated that the role of GAT1 in regulation of xylanase expression is probably independent of its galacturonic/glucuronic acid-transporting activity. We also tested the effect of glucuronic acid, which might be released from beechwood xylan side chains into medium, on the production of xylanases upon xylan. Addition of glucuronic acid (1 or 5 mM) did not compromise but slightly promoted xylanase production in QM9414 (Supplementary Figure 2). These data excluded the possibility that the increment of XYNI production in Δgat1 resulted from the alleviated intracellular uptake of glucuronic acid which might repress xylanase gene expression.
Whereas previous study (Havukainen et al., 2021b) and our own assay (data not shown) showed that GAT1 does not have detectable xylose-transporting activity, its absence resulted in transcriptional changes of four putative sugar transporters encoding genes. Among others, the corresponding proteins of Tr_69957 and Tr_62380 have been previously demonstrated to play a role in xylose uptake (Sloothaak et al., 2016; Nogueira et al., 2018), which might consequently exert an effect on xylanase gene expression. Indeed, it has been reported that deletion of Tr_69957 affects the expression of cellulase and xylanase genes in the presence of complex carbon source or simple sugars (Nogueira et al., 2018). Particularly, when cultivated with xylose as the sole carbon source, quite opposite changes in the transcription of xyn1 and xyn2 were observed in the Tr_69957-null mutant (Nogueira et al., 2018), implicating a different response pattern of xylanase gene expression to xylose. Although the upregulated expression of Tr_69957 in Δgat1 may not cause a significant change in xylose uptake as shown by the similar growth of Δgat1 to QM9414 (Figure 1D), possibility exists that a subtle alteration in intracellular or extracellular xylose concentration together with the changed expression of the other sugar transporters, e.g., downregulated expression of Tr_62380, would consequently affected the expression of xylanase genes. On the other hand, it is also possible that deletion of gat1 caused some variation in membrane sugar sensors yet to be known, which might not participate in direct sugar transportation, but act in the process of signal sensing and transduction from xylose or xylooligosaccharides molecules or their derivatives, and consequently affected xylanase gene expression.
Considering that transcriptional factors are well known to play direct regulatory roles in gene expression, we also identified changes in the expression of putative transcription factors encoding genes. While a total of eight relevant genes are differentially expressed between Δgat1 and QM9414, no significant changes in the transcriptional levels of genes encoding XYR1, ACE1, ACE2, or CRE1 were observed. Instead, the gene encoding the transcriptional repressor Xpp1 for both xyn1 and xyn2 was upregulated. Of note, previous reports showed that Xpp1 regulates the transcription of xylanase genes only at later cultivation stages (Derntl et al., 2015). In our results, deletion of gat1 increased the transcription level of xyn1 over the entire cultivation period (Figure 4C). Therefore, it was deemed that potential repressive effects brought by the upregulated expression of Xpp1 is probably overridden by other unknown factors. Specific transcriptional factors and the precise mechanism involved in this differential regulation of xylanase genes warrant further investigation.
GAT1 exhibits significant transporting activity toward galacturonic acid (Havukainen et al., 2021b), which is a main product from pectin hydrolysis. Given that xylan and pectin are both widely present in plant biomass, it is speculated that upregulated expression of GAT1 and also GAT2 on xylan may facilitate the simultaneous pectin utilization via galacturonic acid absorption. We found that galacturonic acid not only triggered expression of pectinase genes, but also that of xyn1. Deletion of gat1 not only remarkably upregulated the expression of two major pectin hydrolytic genes, but also significantly increased the transcriptional level of xyn1 on galacturonic acid. The precise mechanism remains unclear although it may involve as yet unknown mechanisms similar to that upon induction by xylan. One can surmise that a coordinated regulation of pectinase and xylanase genes were present in T. reesei to ensure efficient degradation of hemicellulosic biomass.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
WL and WZ designed the project. WX, YF, MD, and YR performed the experiments. WZ and WX wrote the manuscript. WL revised the manuscript. All authors analyzed the data, read, and approved the final manuscript.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2018YFA0900500), National Natural Science Foundation of China (31970029, 31770047, and 31970071), Major Basic Research Projects of Natural Science Foundation of Shandong Province (ZR2019ZD19), and Open Projects Fund from State Key Laboratory of Microbial Technology of Shandong University (Project No. M2021-05).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.810066/full#supplementary-material
Footnotes
References
Aro, N., Saloheimo, A., Ilmén, M., and Penttilä, M. (2001). ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276, 24309–24314. doi: 10.1074/jbc.M003624200
Bailey, M. J., Biely, P., and Poutanen, K. (1992). Interlaboratory testing of methods for assay of xylanase activity. J. Biotechnol. 23, 257–270. doi: 10.1016/0168-1656(92)90074-J
Beier, S., Hinterdobler, W., Bazafkan, H., Schillinger, L., and Schmoll, M. (2020). CLR1 and CLR2 are light dependent regulators of xylanase and pectinase genes in Trichoderma reesei. Fungal Genet. Biol. 136:103315. doi: 10.1016/j.fgb.2019.103315
Bischof, R. H., Ramoni, J., and Seiboth, B. (2016). Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Fact. 15:106. doi: 10.1186/s12934-016-0507-6
Derntl, C., Gudynaite-Savitch, L., Calixte, S., White, T., Mach, R. L., and Mach-Aigner, A. R. (2013). Mutation of the Xylanase regulator 1 causes a glucose blind hydrolase expressing phenotype in industrially used Trichoderma strains. Biotechnol. Biofuels 6:62. doi: 10.1186/1754-6834-6-62
Derntl, C., Rassinger, A., Srebotnik, E., Mach, R. L., and Mach-Aigner, A. R. (2015). Xpp1 regulates the expression of xylanases, but not of cellulases in Trichoderma reesei. Biotechnol. Biofuels 8:112. doi: 10.1186/s13068-015-0298-8
Deshpande, M. V., Eriksson, K. E., and Pettersson, L. G. (1984). An assay for selective determination of exo-1,4,-beta-glucanases in a mixture of cellulolytic enzymes. Anal. Biochem. 138, 481–487. doi: 10.1016/0003-2697(84)90843-1
Furukawa, T., Shida, Y., Kitagami, N., Mori, K., Kato, M., Kobayashi, T., et al. (2009). Identification of specific binding sites for XYR1, a transcriptional activator of cellulolytic and xylanolytic genes in Trichoderma reesei. Fungal Genet. Biol. 46, 564–574. doi: 10.1016/j.fgb.2009.04.001
Gupta, V. K., Kubicek, C. P., Berrin, J. G., Wilson, D. W., Couturier, M., Berlin, A., et al. (2016). Fungal enzymes for bio-products from sustainable and waste biomass. Trends Biochem. Sci. 41, 633–645. doi: 10.1016/j.tibs.2016.04.006
Häkkinen, M., Valkonen, M. J., Westerholm-Parvinen, A., Aro, N., Arvas, M., Vitikainen, M., et al. (2014). Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol. Biofuels 7:14. doi: 10.1186/1754-6834-7-14
Havukainen, S., Pujol-Giménez, J., Valkonen, M., Hediger, M. A., and Landowski, C. P. (2021a). Functional characterization of a highly specific L-arabinose transporter from Trichoderma reesei. Microb. Cell Fact. 20:177. doi: 10.1186/s12934-021-01666-4
Havukainen, S., Pujol-Giménez, J., Valkonen, M., Westerholm-Parvinen, A., Hediger, M. A., and Landowski, C. P. (2021b). Electrophysiological characterization of a diverse group of sugar transporters from Trichoderma reesei. Sci. Rep. 11:14678. doi: 10.1038/s41598-021-93552-7
Havukainen, S., Valkonen, M., Koivuranta, K., and Landowski, C. P. (2020). Studies on sugar transporter CRT1 reveal new characteristics that are critical for cellulase induction in Trichoderma reesei. Biotechnol. Biofuels 13:158. doi: 10.1186/s13068-020-01797-7
Herold, S., Bischof, R., Metz, B., Seiboth, B., and Kubicek, C. P. (2013). Xylanase gene transcription in Trichoderma reesei is triggered by different inducers representing different hemicellulosic pentose polymers. Eukaryotic Cell 12, 390–398. doi: 10.1128/Ec.00182-12
Herrmann, M. C., Vrsanska, M., Jurickova, M., Hirsch, J., Biely, P., and Kubicek, C. P. (1997). The beta-D-xylosidase of Trichoderma reesei is a multifunctional beta-D-xylan xylohydrolase. Biochem. J. 321, 375–381. doi: 10.1042/bj3210375
Himmel, M. E. (2007). Biomass recalcitrance: engineering plants and enzymes for biofuels production (vol 315, pg 804, 2007). Science 316:982. doi: 10.1126/science.1137016
Huang, Z. B., Chen, X. Z., Qin, L. N., Wu, H. Q., Su, X. Y., and Dong, Z. Y. (2015). A novel major facilitator transporter TrSTR1 is essential for pentose utilization and involved in xylanase induction in Trichoderma reesei. Biochem. Biophys. Res. Commun. 460, 663–669. doi: 10.1016/j.bbrc.2015.03.087
Huberman, L. B., Liu, J., Qin, L. N., and Glass, N. L. (2016). Regulation of the lignocellulolytic response in filamentous fungi. Fungal Biol. Rev. 30, 101–111. doi: 10.1016/j.fbr.2016.06.001
Ivanova, C., Bååth, J. A., Seiboth, B., and Kubicek, C. P. (2013). Systems analysis of lactose metabolism in Trichoderma reesei Identifies a Lactose Permease that is essential for cellulase induction. PLoS One 8:e62631. doi: 10.1371/journal.pone.0062631
Liu, G., and Qu, Y. (2021). Integrated engineering of enzymes and microorganisms for improving the efficiency of industrial lignocellulose deconstruction. Eng. Microbiol. 1:100005 doi: 10.1016/j.engmic.2021.100005
Liu, R., Chen, L., Jiang, Y. P., Zou, G., and Zhou, Z. H. (2017). A novel transcription factor specifically regulates GH11 xylanase genes in Trichoderma reesei. Biotechnol. Biofuels 10:19. doi: 10.1186/s13068-017-0878-x
Lv, X. X., Zheng, F. L., Li, C. Y., Zhang, W. X., Chen, G. J., and Liu, W. F. (2015). Characterization of a copper responsive promoter and its mediated overexpression of the xylanase regulator 1 results in an induction-independent production of cellulases in Trichoderma reesei. Biotechnol. Biofuels 8:67. doi: 10.1186/s13068-015-0249-4
Mach, R. L., Strauss, J., Zeilinger, S., Schindler, M., and Kubicek, C. P. (1996). Carbon catabolite repression of xylanase I (xyn1) gene expression in Trichoderma reesei. Mol. Microbiol. 21, 1273–1281. doi: 10.1046/j.1365-2958.1996.00094.x
Mach-Aigner, A. R., Pucher, M. E., and Mach, R. L. (2010). D-Xylose as a repressor or inducer of xylanase expression in Hypocrea jecorina (Trichoderma reesei). Appl. Environ. Microbiol. 76, 1770–1776. doi: 10.1128/AEM.02746-09
Mandels, M. L., Parrish, F. W., and Reese, E. T. (1962). Sophorose as an inducer of cellulase in Trichoderma viride. J. Bacteriol. 83:400. doi: 10.1128/jb.83.2.400-408.1962
Martinez, D., Berka, R. M., Henrissat, B., Saloheimo, M., Arvas, M., Baker, S. E., et al. (2008). Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26:1193.
Nogueira, K. M. V., de Paula, R. G., Antoniêto, A. C. C., dos Reis, T. F., Carraro, C. B., Silva, A. C., et al. (2018). Characterization of a novel sugar transporter involved in sugarcane bagasse degradation in Trichoderma reesei. Biotechnol. Biofuels 11:84 doi: 10.1186/s13068-018-1084-1
Ogasawara, W., Shida, Y., Furukawa, T., Shimada, R., Nakagawa, S., Kawamura, M., et al. (2006). Cloning, functional expression and promoter analysis of xylanase III gene from Trichoderma reesei. Appl. Microbiol. Biotechnol. 72, 995–1003. doi: 10.1007/s00253-006-0365-y
Rauscher, R., Würleitner, E., Wacenovsky, C., Aro, N., Stricker, A. R., Zeilinger, S., et al. (2006). Transcriptional regulation of xyn1, encoding xylanase I, in Hypocrea jecorina. Eukaryotic Cell 5, 447–456. doi: 10.1128/EC.5.3.447-456.2006
Saloheimo, A., Aro, N., Ilmén, M., and Penttilä, M. (2000). Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J. Biol. Chem. 275, 5817–5825. doi: 10.1074/jbc.275.8.5817
Saloheimo, A., Rauta, J., Stasyk, O. V., Sibirny, A. A., Penttilä, M., and Ruohonen, L. (2007). Xylose transport studies with xylose-utilizing Saccharomyces cerevisiae strains expressing heterologous and homologous permeases. Appl. Microbiol. Biotechnol. 74, 1041–1052. doi: 10.1007/s00253-006-0747-1
Shida, Y., Furukawa, T., and Ogasawara, W. (2016). Deciphering the molecular mechanisms behind cellulase production in Trichoderma reesei, the hyper-cellulolytic filamentous fungus. Biosci. Biotechnol. Biochem. 80, 1712–1729. doi: 10.1080/09168451.2016.1171701
Sloothaak, J., Tamayo-Ramos, J. A., Odoni, D. I., Laothanachareon, T., Derntl, C., Mach-Aigner, A. R., et al. (2016). Identification and functional characterization of novel xylose transporters from the cell factories Aspergillus niger and Trichoderma reesei. Biotechnol. Biofuels 9:148. doi: 10.1186/s13068-016-0564-4
Strauss, J., Mach, R. L., Zeilinger, S., Hartler, G., Stöffler, G., Wolschek, M., et al. (1995). Crel, the carbon catabolite repressor protein from Trichoderma-Reesei. Febs Lett. 376, 103–107. doi: 10.1016/0014-5793(95)01255-5
Stricker, A. R., Grosstessner-Hain, K., Würleitner, E., and Mach, R. L. (2006). Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and D-xylose metabolism in Hypocrea jecorina. Eukaryotic Cell 5, 2128–2137. doi: 10.1128/Ec.00211-06
Stricker, A. R., Trefflinger, P., Aro, N., Penttilä, M., and Mach, R. L. (2008). Role of Ace2 (Activator of Cellulases 2) within the xyn2 transcriptosome of Hypocrea jecorina. Fungal Genet. Biol. 45, 436–445. doi: 10.1016/j.fgb.2007.08.005
Tenkanen, M., Puls, J., and Poutanen, K. (1992). Two major Xylanases of Trichoderma-Reesei. Enzyme Microb. Technol. 14, 566–574. doi: 10.1016/0141-0229(92)90128-B
Tenkanen, M., Vršanská, M., Siika-aho, M., Wong, D. W., Puchart, V., Penttilä, M., et al. (2013). Xylanase XYN IV from Trichoderma reesei showing exo- and endo-xylanase activity. FEBS J. 280, 285–301. doi: 10.1111/febs.12069
Torronen, A., and Rouvinen, J. (1995). Structural comparison of two major endo-1,4-xylanases from Trichoderma reesei. Biochemistry 34, 847–856. doi: 10.1021/bi00003a019
Torronen, A., Mach, R. L., Messner, R., Gonzalez, R., Kalkkinen, N., Harkki, A., et al. (1992). The two major xylanases from Trichoderma reesei: characterization of both enzymes and genes. Biotechnology (N Y) 10, 1461–1465. doi: 10.1038/nbt1192-1461
Wang, L., Lv, X. X., Cao, Y. L., Zheng, F. L., Meng, X. F., Shen, Y., et al. (2019). A novel transcriptional regulator RXE1 modulates the essential transactivator XYR1 and cellulase gene expression in Trichoderma reesei. Appl. Microbiol. Biotechnol. 103, 4511–4523. doi: 10.1007/s00253-019-09739-6
Würleitner, E., Pera, L., Wacenovsky, C., Cziferszky, A., Zeilinger, S., Kubicek, C. P., et al. (2003). Transcriptional regulation of xyn2 in Hypocrea jecorina. Eukaryotic Cell 2, 150–158. doi: 10.1128/EC.2.1.150-158.2003
Xu, J., Takakuwa, N., Nogawa, M., Okada, H., and Morikawa, Y. (1998). A third xylanase from Trichoderma reesei PC-3-7. Appl. Microbiol. Biotechnol. 49, 718–724. doi: 10.1007/s002530051237
Xue, Y., Han, J., Li, Y. Y., Liu, J., Gan, L. H., and Long, M. N. (2020). Promoting cellulase and hemicellulase production from Trichoderma orientalis EU7-22 by overexpression of transcription factors Xyr1 and Ace3. Bioresour. Technol. 296:122355. doi: 10.1016/j.biortech.2019.122355
Zeilinger, S., Mach, R. L., Schindler, M., Herzog, P., and Kubicek, C. P. (1996). Different inducibility of expression of the two xylanase genes xyn1 and xyn2 in Trichoderma reesei. J. Biol. Chem. 271, 25624–25629. doi: 10.1074/jbc.271.41.25624
Zhang, J. J., Chen, Y. M., Wu, C., Liu, P., Wang, W., and Wei, D. Z. (2019). The transcription factor ACE3 controls cellulase activities and lactose metabolism via two additional regulators in the fungus Trichoderma reesei. J. Biol. Chem. 294, 18435–18450. doi: 10.1074/jbc.RA119.008497
Zhang, W. X., Kou, Y. B., Xu, J. T., Cao, Y. L., Zhao, G. L., Shao, J., et al. (2013). Two major facilitator superfamily sugar transporters from Trichoderma reesei and their roles in induction of cellulase biosynthesis. J. Biol. Chem. 288, 32861–32872. doi: 10.1074/jbc.M113.505826
Keywords: Trichoderma reesei, xylanase, sugar transporter, GAT1, XYNI
Citation: Xu W, Fang Y, Ding M, Ren Y, Meng X, Chen G, Zhang W and Liu W (2022) Elimination of the Sugar Transporter GAT1 Increased Xylanase I Production in Trichoderma reesei. Front. Microbiol. 13:810066. doi: 10.3389/fmicb.2022.810066
Received: 05 November 2021; Accepted: 04 January 2022;
Published: 26 January 2022.
Edited by:
Birgitta Elisabeth Ebert, University of Queensland, AustraliaReviewed by:
Chaoguang Tian, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences (CAS), ChinaShuji Tani, Osaka Prefecture University, Japan
Copyright © 2022 Xu, Fang, Ding, Ren, Meng, Chen, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weixin Zhang, emhhbmd3eEBzZHUuZWR1LmNu
 Wenqiang Xu
Wenqiang Xu Yu Fang
Yu Fang Guanjun Chen
Guanjun Chen Weixin Zhang
Weixin Zhang Weifeng Liu
Weifeng Liu