- 1Department of Infectious Diseases, Istituto Superiore di Sanità, Rome, Italy
- 2Department of Science, Roma Tre University, Rome, Italy
- 3Department of Veterinary and Animal Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Frederiksberg, Denmark
- 4Unit of Microbiology, Policlinico S. Orsola, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Bologna, Italy
- 5Unit of Clinical Microbiology, S. Agostino-Estense Hospital Baggiovara, AUSL Modena, Modena, Italy
- 6Department of Experimental and Clinical Medicine, University of Florence, Florence, Italy
- 7Santa Lucia Foundation (IRCCS), Rome, Italy
Staphylococcus aureus is the most common pathogen causing osteomyelitis (OM). The aim of this study was to explore the clonal complex (CC) distribution and the pattern of virulence determinants of S. aureus isolates from OM in Italy. Whole-genome sequencing was performed on 83 S. aureus isolates from OM cases in six hospitals. Antibiotic susceptibility tests showed that 30.1% of the isolates were methicillin-resistant S. aureus (MRSA). The most frequent CCs detected were CC22, CC5, CC8, CC30, and CC15, which represent the most common lineages circulating in Italian hospitals. MRSA were limited in the number of lineages (CC22, CC5, CC8, and CC1). Phylogenetic analysis followed the sequence type-CC groupings and revealed a non-uniform distribution of the isolates from the different hospitals. No significant difference in the mean number of virulence genes carried by MRSA or MSSA isolates was observed. Some virulence genes, namely cna, fib, fnbA, coa, lukD, lukE, sak, and tst, were correlated with the CC. However, different categories of virulence factors, such as adhesins, exoenzymes, and toxins, were frequently detected and unevenly distributed among all lineages. Indeed, each lineage carried a variable combination of virulence genes, likely reflecting functional redundancy, and arguing for the importance of those traits for the pathogenicity in OM. In conclusion, no specific genetic trait in the most frequent lineages could explain their high prevalence among OM isolates. Our findings highlight that CCs detected in OM isolates follow the epidemiology of S. aureus infections in the country. It is conceivable that any of the most common S. aureus CC can cause a variety of infections, including OM.
Introduction
Osteomyelitis (OM) is an infective and inflammatory process of the bone, which can progress to osteonecrosis and bone destruction. Treatment of OM is challenging due to a variety of factors including the scarce penetration of antibiotics into the bone tissue, the biofilm-like presentation of the infection, and the increasing antibiotic resistance of bacterial pathogens. Despite the improvement in the diagnosing of OM, the incidence of OM has been increasing over the last few decades. This could be ascribed to the increase of certain risk factors such as diabetes and surgery procedures such as arthroplasty (Kurtz et al., 2012; Geraghty and LaPorta, 2019). The number of orthopaedic surgeries is constantly increasing. In the United States (US), the average orthopaedic surgical volume increased of 38% in the last 3 years (Shukla et al., 2021) and is estimated that by 2030, 572,000 hip revisions and 3.48 million knee revisions could be performed with an increase of 174 and 673%, respectively (Kurtz et al., 2012). In line with the increase of arthroplasty, the incidence of OM increased from 11.4 to 24.2 cases per 100,000 person-years in four decades in United States (Kremers et al., 2015). If a prosthetic-joint infection develops, revision surgery for removal of the implant is often required, which leads to prolonged hospitalization and a high risk of re-infection (Trampuz and Zimmerli, 2008).
S. aureus is the principal cause of OM, being responsible for 30–60% of cases (Karwowska et al., 1998; Grammatico et al., 2008; Byren et al., 2009; Howard-Jones and Isaacs, 2013). OM caused by S. aureus is associated with severe prognosis and persistent infections in approximately 40% of patients (Kremers et al., 2015). During bone infection, S. aureus attachment and colonization are facilitated by adhesins. The colonization of bone occurs through direct interaction of S. aureus with the bone cells or the extracellular matrix (ECM). After bone colonization, S. aureus can grow as biofilm and produce toxins, which facilitate persistence and dissemination of the infection in the host, respectively (Patti et al., 1994; Tuchscherr et al., 2019). The success of S. aureus in bone infections is related to a multitude of virulence factors involved in critical steps of the pathogenesis, including adhesion to surfaces, invasion of host tissues, evasion of the immune system, and biofilm formation (Ricciardi et al., 2018).
S. aureus strains from different sources carry various combinations of virulence genes (Montanaro et al., 1999; Peacock et al., 2002; Campoccia et al., 2008), and some studies were conducted to identify and characterize the most frequent S. aureus clones cause of OM (reviewed by Pimentel de Araujo et al., 2021). Pandemic lineages, including CC5, CC8, CC22, CC30, and CC45, were the most common in OM, and their distribution greatly differed among the countries reflecting the local epidemiology of S. aureus and the MSSA heterogeneity (Pimentel de Araujo et al., 2021). However, the characterization of S. aureus clones causing bone and joint infections in Italy is limited to only few studies (Campoccia et al., 2008; Montanaro et al., 2016).
It is unknown to date if the ability to cause OM of some lineages is correlated with the presence of certain combination of virulence factors, or it is only a reflection of the local epidemiology of clones. The purpose of this study was to identify the pattern of virulence determinants and the clonal distribution of S. aureus isolates from OM in Italy, by using whole-genome sequencing (WGS) for a detailed genetic characterization.
Materials and Methods
Staphylococcus aureus Collection
From February 2019 to February 2020, a total of 83 non-duplicate S. aureus isolates were collected from patients with OM, in six hospitals in two Italian Regions (Tuscany and Emilia-Romagna). A positive case of S. aureus OM was ascertained upon identification of S. aureus from bone biopsy, aspirate of intraosseous abscess, intraoperative swabs, prosthetic implants, or any osteosynthesis material. S. aureus isolates were isolated and identified by the hospital laboratories according to the local procedures, including both MALDI-TOF and the Vitek2® system (BioMérieux, Marcy l’Etoile, France). Isolates were shipped to Istituto Superiore di Sanità (ISS, Rome, Italy) and Roma Tre University for the phenotypic and genotypic characterization, respectively. Strains have been screened for oxacillin resistance according to EUCAST protocol (version 11.0, 2021).1 The study was approved by the Ethics Committee of ISS (n° 0013802 18/03/2019) and the participating hospitals.
Antibiotic Susceptibility
Antibiotic susceptibility testing was preliminary performed using the Vitek2® system (BioMérieux, Marcy l’Etoile, France) or MicroScan Walkaway (Beckman, United States) at the participating hospital laboratories, and subsequently by the broth microdilution method using commercially available microplates (MERLIN Diagnostika GmbH, Germany). Discrepant results were resolved by testing individual isolates with the disk diffusion method. Results were interpreted according to the EUCAST breakpoints (version 11.0, 2021; see footnote 1).
DNA Isolation, Whole-Genome Sequencing, and Genotyping
Genomic DNA of the 83 S. aureus isolates was extracted using the QIAamp DNA Mini Kit (QIAGEN srl, Milan, Italy) according to the manufacturer’s protocol, with the only modification of the addition of 50 μg/ml of lysostaphin (Sigma Aldrich, Milan, Italy) for the lysis step. Sequencing was performed by using Illumina MiSeq (Illumina, San Diego, CA, United States). Processed FASTQ reads were de novo assembled using SPAdes pipeline (Bankevich et al., 2012) through the ARIES public Galaxy server.2 Sequence Types (ST)s, spa types, and Staphylococcal Cassette Chromosome mec (SCCmec) types were identified by MLST v2.0, spaTyper v1.0 and SCCmec Finder v1.2 pipelines, respectively, available at the CGE website.3 Clonal complex (CC) grouping was performed using pubMLST website.4 Antimicrobial resistance genes were searched in the assembled genomes of all isolates using ABRicate v1.0.1 and both ResFinder and CARD databases (Zankari et al., 2012; Jia et al., 2017).5 Positive hits were selected based on >95% nucleotide sequence identity. Point mutations in gyrA and grlA genes conferring resistance to quinolones, and in rpoB conferring rifampicin resistance, were searched in the assembled genomes using reference gene sequences from S. aureus NCTC 8325 (GenBank accession no. CP000253.1). Translated protein sequences were aligned using ClustalW in MEGA X v.10.2.1 with default parameters (Kumar et al., 2018) and inspected for known substitutions conferring resistance to quinolones and rifampicin (Aubry-Damon et al., 1998; Tanaka et al., 2000).
To screen for virulence factors, a custom database based on the allele library previously described by Strauß et al. (2016) was constructed. The database comprised 1,205 allelic variants of 122 known virulence genes (Supplementary Table S1), including those encoding exoenzymes, toxins, adhesins superantigens, capsule, regulators, and biofilm formation. The presence of virulence determinants was ascertained using ABRicate v1.0.1 and alignment results with identity scores greater than 95% were selected as positive matches.
Core genome MLST (cgMLST)-based genotyping was performed on the 1,861 target genes of the S. aureus cgMLST scheme (Leopold et al., 2014). Isolates showing less than 11 allelic differences in the core genome were considered as genetically indistinguishable (Leopold et al., 2014). A neighbor-joining (NJ) tree was created based on the allelic profiles of the cgMLST target genes. All analyses were conducted in SeqSphere+ v8.0.1 (Ridom GmbH, Germany).
Statistical Analysis
Data analysis was performed in R v4.1.1. Normality distribution of virulence factors among CCs was evaluated using the Shapiro–Wilk’s method. Normally and non-normally distributed data were compared with Student’s t-test and Mann–Whitney-Wilcoxon’s test, respectively, with Benjamini-Hochberg’s correction for false discovery rate. Adjusted p ≤ 0.05 were considered as significant.
Data Availability
WGS data for the 83 S. aureus isolates have been submitted to the NCBI SRA under BioProject PRJNA784720.
Results
Demographic Characteristics of the Patients
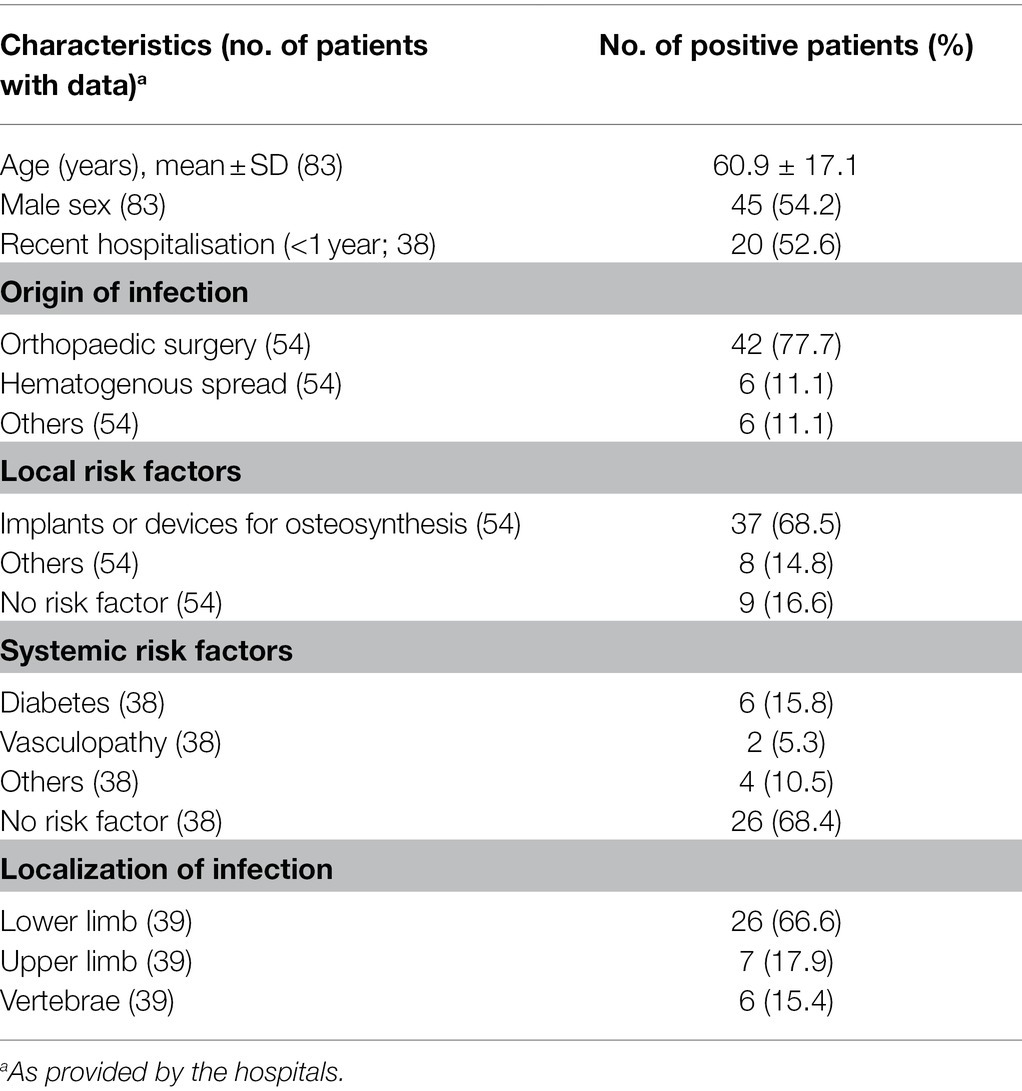
The main characteristics of OM patients are summarized in Table 1. Most patients were adults (mean age 60.9 ± 17.1 years, range 22–94), and 54.2% were male. Additional information was available only for a portion of the patients/isolates. The most common source of infection was orthopaedic surgery (42/54 patients, 77.7%) in the presence of a prosthetic implant (37/54, 68.5%); hematogenous OM was observed only in 11.1% (6/54) of the patients. The systemic risk factor most reported was diabetes (6/38, 15.8%). OM affected primarily lower limbs (26/39, 66.6%), followed by upper limbs and vertebrae (17.9 and 15.4% of the patients, respectively).
Antibiotic Susceptibility
The results of antimicrobial susceptibility testing of the 83 S. aureus isolates are shown in Table 2. All isolates were susceptible to vancomycin, and 30.1% were resistant to methicillin (MRSA). MRSA isolates showed elevated frequencies of resistance to levofloxacin (84.0%), erythromycin (64.0%), clindamycin (64.0%) and gentamicin (32.0%; Table 2). The majority of MSSA isolates were resistant to penicillin (62.1%) and, less frequently, to levofloxacin (17.2%), erythromycin (13.8%) and clindamycin (13.8%; Table 2).
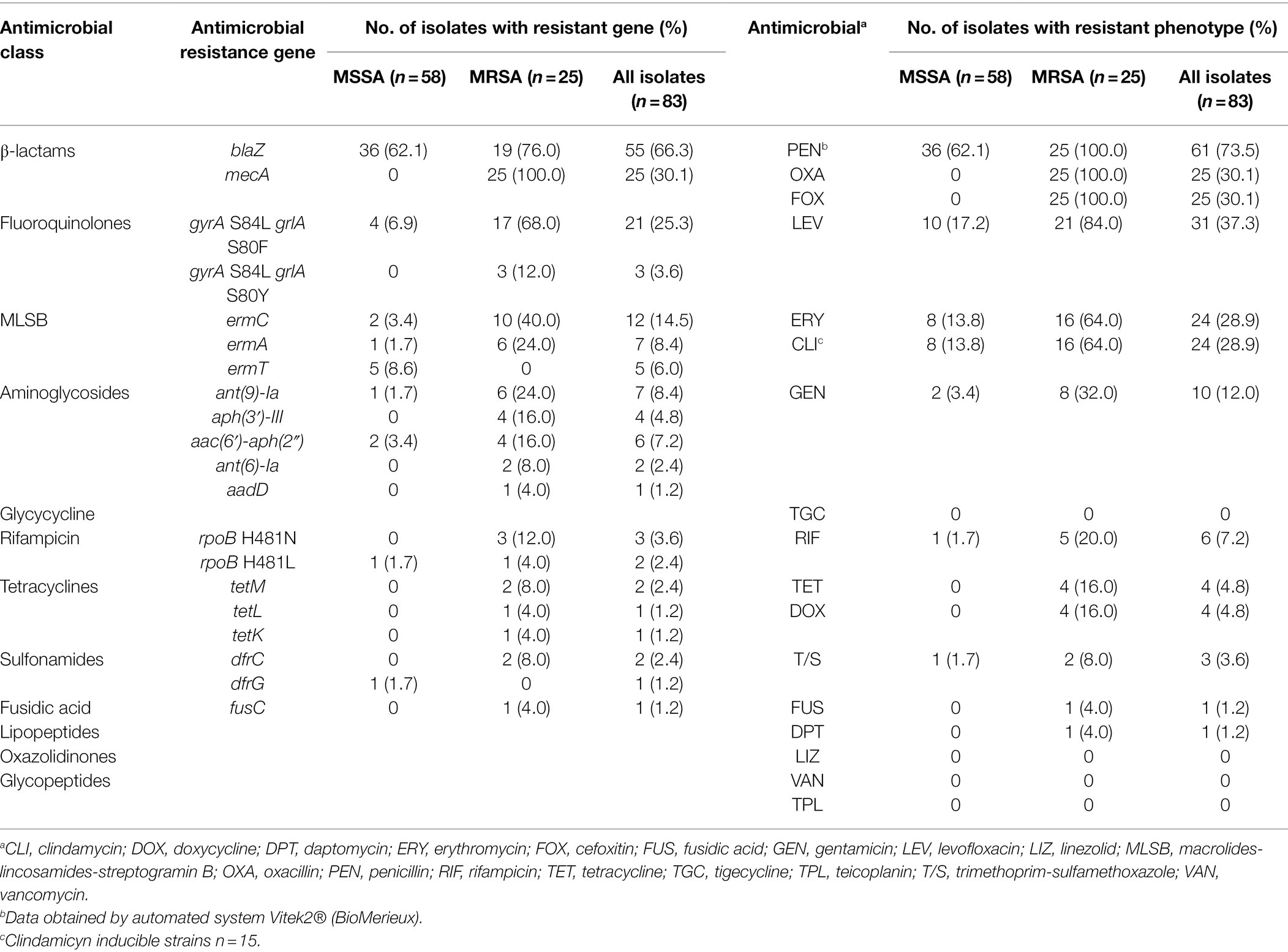
Table 2. Resistance genes and antimicrobial-susceptibility profile in 83 Staphylococcus aureus isolates from OM.
Staphylococcus aureus Genotyping
The CC distribution for MRSA and MSSA is shown in Figure 1. Overall, 53 different spa types were detected (Supplementary Table S3), and isolates clustered into 26 STs and nine CCs (Figure 1). CC22 was the most frequent (25.3% of the isolates), followed by CC5 (15.7%), CC8 (12.0%), CC30 (12.0%), and CC15 (9.6%). The largest variability was observed among MSSA isolates, which were distributed into 20 STs and eight CCs. CC30 and CC22 were the most frequent CCs among MSSA and MRSA, respectively, whereas CC5, CC8, and CC22 were detected among both MSSA and MRSA isolates.
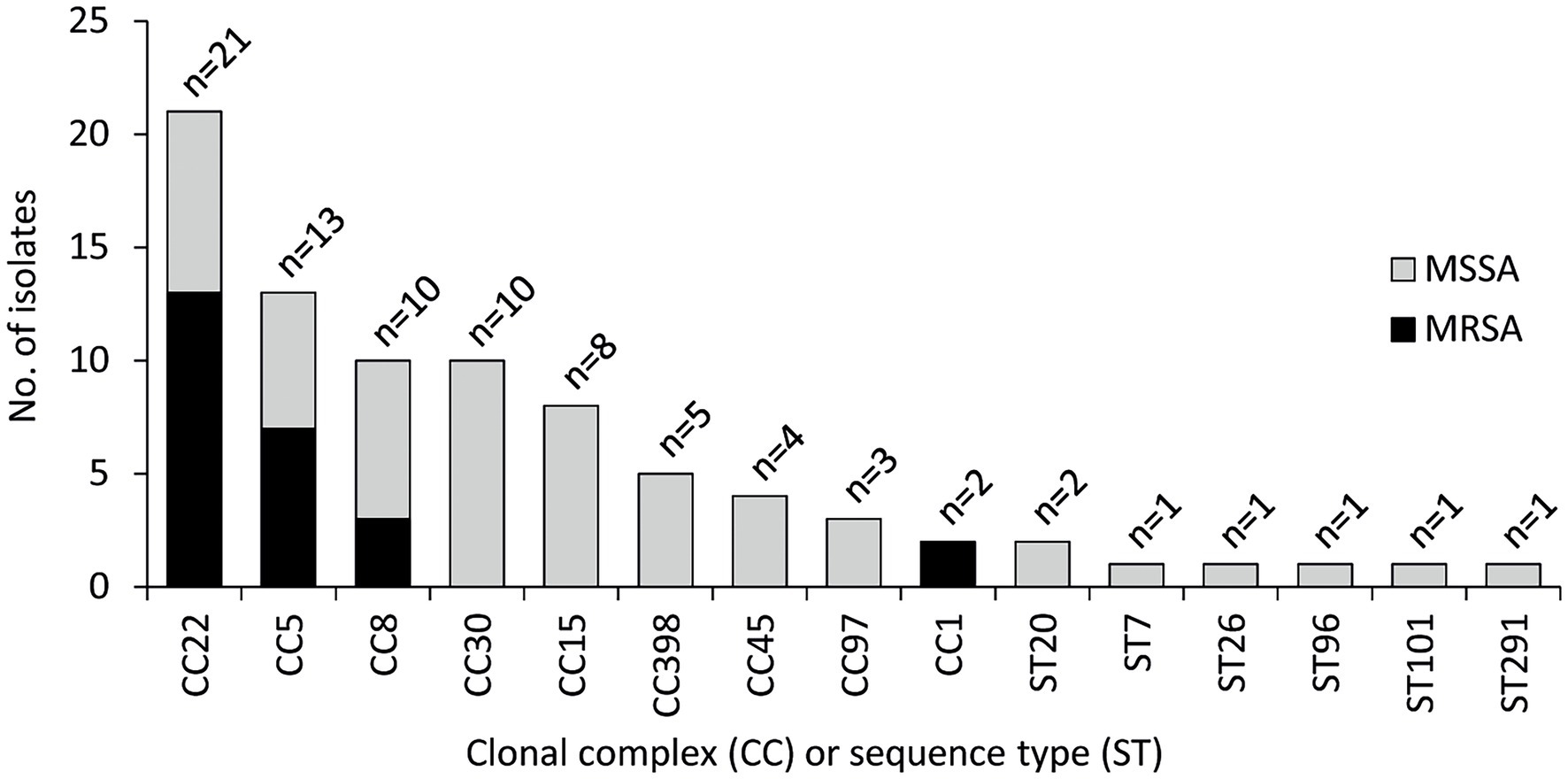
Figure 1. Clonal complex (CC) and sequence type (ST) distribution of 83 Staphylococcus aureus isolates from OM. CC22 (ST22, n = 20; ST3863, n = 1), CC5 (ST5, n = 9; ST105, n = 2; ST228, n = 2), CC8 (ST8, n = 7; ST72, n = 1; ST368, n = 1; ST789, n = 1), CC30 (ST30, n = 7; ST34, n = 1; ST4391, n = 1; ST7297, n = 1), CC15 (ST15, n = 5; ST582, n = 3), CC398 (ST398, n = 5), CC45 (ST45, n = 4), CC97 (ST97, n = 3), CC1 (ST1, n = 1; ST6927, n = 1), ST20 (n = 2), ST7 (n = 1), ST26 (n = 1), ST96 (n = 1), and ST101 (n = 1).
The phylogenetic relationship based on cgMLST of the isolates is shown in Figure 2. Overall, phylogeny follows the ST-CC groupings and reveals a non-uniform distribution of the isolates from the six hospitals, which appeared intermingled throughout the phylogeny and did not cluster according to the geographic origin (see hospital ID in Figure 2). Interestingly, five cgMLST clusters of genetically indistinguishable isolates were observed (no. of allelic differences ≤3), containing a total of 10 isolates (two isolates per cluster), both MSSA and MRSA (Supplementary Table S2). In three cases, isolates were collected from the same hospitals (SAO42/SAO45, SAO74/SAO75, and SAO82/SAO84), whereas two clusters contained isolates originating from different hospitals (SAO22/SAO23 and SAO24/SAO26; Supplementary Table S2) in the same Italian region (H2, H5, and H6; see Supplementary Table S3).
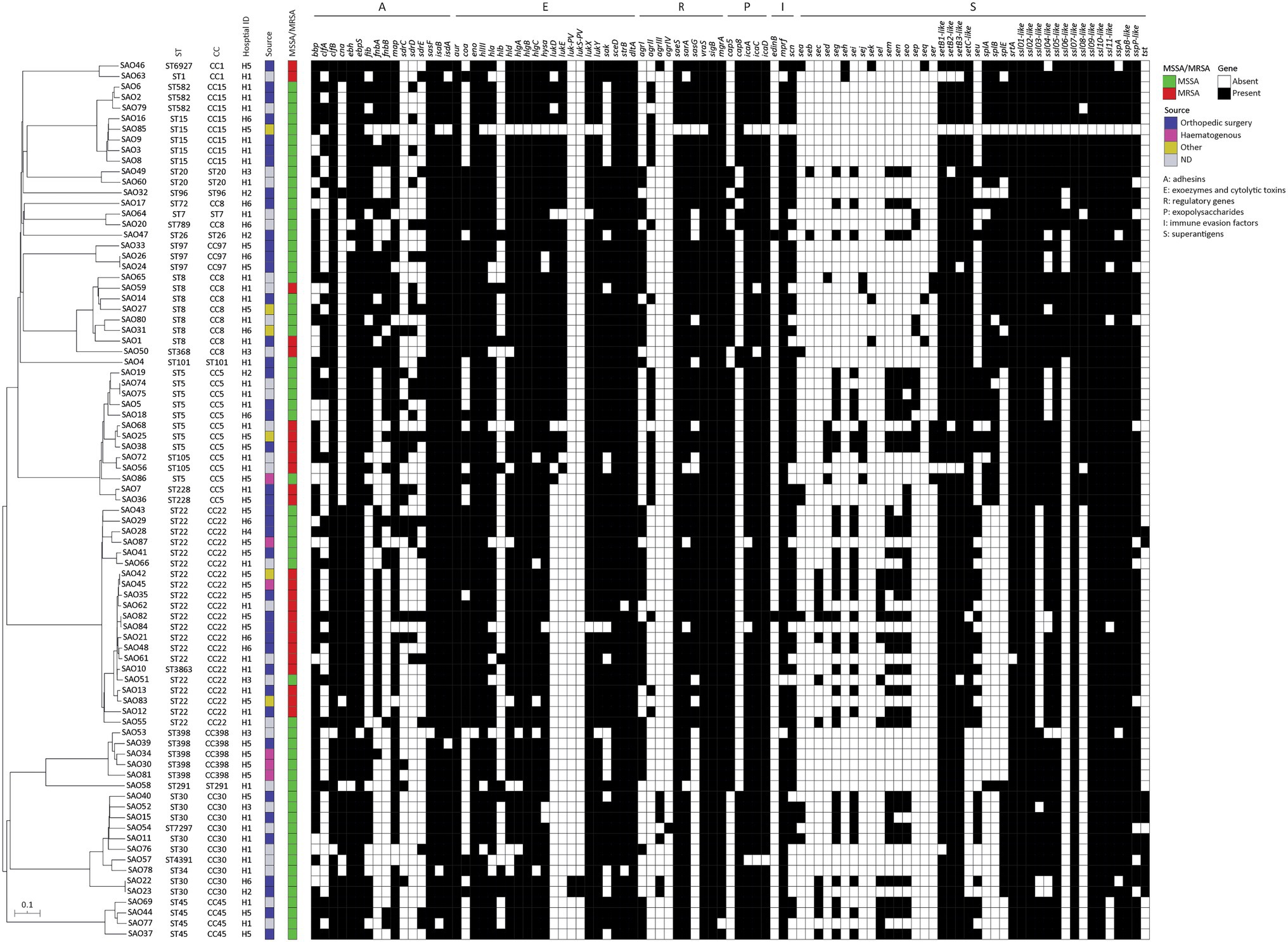
Figure 2. Neighbor joining (NJ) tree based on the allelic profiles of the cgMLST target genes (n = 1,861) of 83 Staphylococcus aureus isolates from osteomyelitis in Italy, and associated heat-map of in silico detected virulence genes (indicated on top).
Detection of Virulence-Related Genes
A dataset of the 122 virulence-related genes (Figure 2; Supplementary Table S3) was used to screen 83 S. aureus strains from OM. An average of 53.7 ± 7 virulence-related genes was detected in the whole collection (coefficient of Variation = 14.5%); of these, 20 genes were detected in ≥95% of the isolates and eight in ≤10%. The most frequent genes encoded different categories of virulence factors, namely haemolysins (hlIII, hlgA, and hlgBIII), proteases (lukX and lukY), several staphylococcal superantigen-like proteins (ssl01-like, ssl02-like, ssl05-like, ssl09-like, and ssl10-like), exoenzymes (srtA), cell wall anchored proteins (CWA) endowed with adhesive properties (ebh, ebps, sasF, isaB, and isdA), and regulatory genes (saeS, vraS, sigB, and mgrA; Figure 2). Genes encoding exotoxins such as Panton–Valentine leukocidin (lukS-PV and lukF-PV), epidermal cell differentiation inhibitor (edinB), and superantigens (seb, seh, sej, sek, seq, and ser) were less frequently detected.
No significant difference in the mean number of virulence genes carried by MRSA or MSSA isolates was observed. Overall, CC8 carried a significantly higher number of virulence genes than CC15, CC22, and CC30 isolates (Figure 3A).
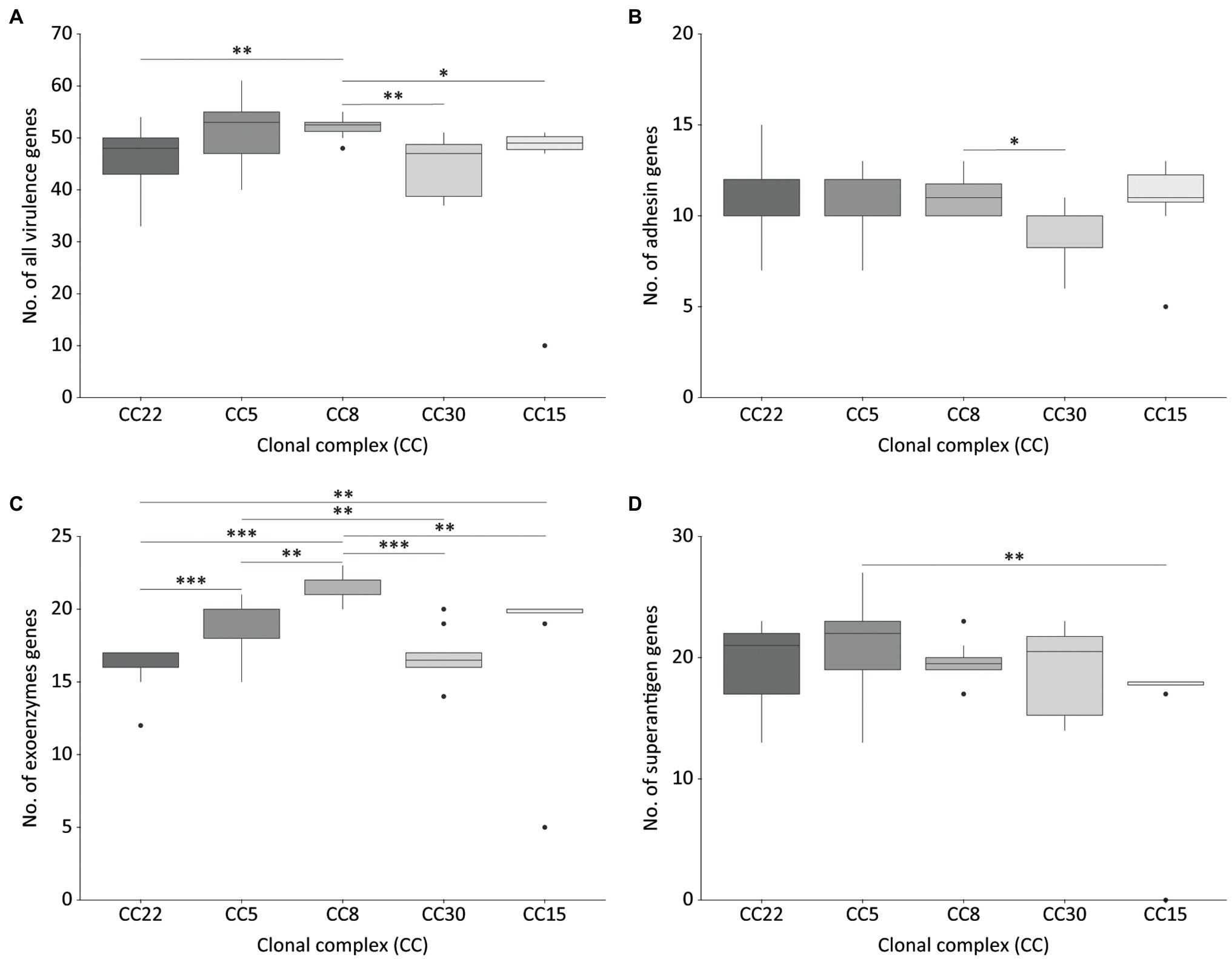
Figure 3. Virulence-related genes detected in isolates belonging to the most frequent Staphylococcus aureus clonal complexes (CC) causing OM. All virulence genes (A), adhesin genes (B), exoenzymes genes (C), and superantigen genes (D). CC22, n = 21; CC5, n = 13; CC8, n = 10; CC30, n = 10; CC15, n = 8. The significance of the differences in the number of virulence factors between CCs was assessed using the Mann–Whitney-Wilcoxon’s test (non-normally distributed data) for all pairwise comparisons with the exception of CC15 vs. CC30, for which Student’s t-test was employed (normally distributed data). Boxes denote the second and third quartiles, vertical lines (whisker) the smallest and largest values of the first and fourth quartiles, with outliers marked by dots. *p < 0.05, **p < 0.01, and ***p < 0.001.
Adhesins
Genes coding for microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) and other CWA proteins were similarly distributed among all major S. aureus CCs, except for CC30 which showed significantly lower adhesin genes than CC8 (Figure 3B; Supplementary Table S3). The adhesin genes clfB, ebh, ebpS, sasF, isaB, and isdA were widespread in the whole collection, being present in ≥95% of the isolates, while the fibronectin-binding protein genes (fnbA, fnbB) and collagen adhesin gene (cna) were detected in nearly half of the strains (range 43.7–47.1%), and in different combinations. Some adhesins genes such as cna and fib were differently associated with CC (p < 0.001). The cna gene was detected in CC1, CC22, CC30, CC45, and CC398, fib in CC1, CC5, CC8, CC15, CC30, CC45, CC97, and CC398.
Exoenzymes and Toxins
Virulence factors with enzymatic or lytic properties responsible for nutrient acquisition, bacterial survival and dissemination were detected in all CCs. Haemolysins and lytic toxins (aur, eno, hlIII, hlgA, hlgB, hlgC, hysa, sceD, strB, and dltA) were present in most of the strains (≥90%). The presence of some exoenzymes genes as coa, lukD, lukE, and sak was correlated with the CC (p < 0.001). Isolates belonging to CC8, CC22, CC30, CC45, and CC97 were associated with coa. The lukD and lukE genes were associated with CC8 and CC97. The sak gene was detected in more than 80% of CC5, CC8, CC22, and CC97. CC8 harbored a significantly higher number of exoenzyme and toxin genes compared to the other CCs (Figure 3C; Supplementary Table S3). Panton-Valentin leukocidin (PVL) was detected in only two isolates (2.4%) from different hospitals and belonging to CC30, which clustered together within the cgMLST phylogeny and showed a similar virulence pattern (Figure 2).
Superantigens
The enterotoxin gene cluster egc (seg, sei, sem, sen, seo, and seu) was detected in 31.3% of the isolates, mostly belonging to CC22, CC30, and CC5. The cluster was absent in all CC398 isolates and all but one CC8 isolate. Among the predominant CCs, a significant difference in the number of superantigen genes was observed between CC5 and CC15 (Figures 2, 3D; Supplementary Table S3). The staphylococcal superantigen-like genes were present in all CCs with minor differences. The toxic shock syndrome toxin gene (tst) was associated with CC30 and was detected in 13.3% of the isolates, all belonging to CC30 and CC22.
Biofilm and Capsular Genes
Capsular genes were detected in all the isolates and the most frequent capsular type was type 5 (cap5; 65.0%), associated with CC5, CC8, CC22, CC97, and CC398. Capsular type 8 (cap8) was found in CC1, CC15, CC30, and CC45. The complete icaACD locus encoding genes involved in biofilm production was detected in 77 out of 83 strains (92.7%). Two strains belonging to CC30 and CC15 did not carry any of the ica genes, while four strains belonging to CC8 (two isolates), CC22, and CC97 (one isolate each) were negative for at least one gene of the locus.
Regulatory Genes
The accessory gene regulator system (agr) responsible for the regulation and expression of toxins and exoenzymes and biofilm was detected in 86.7% of the isolates (72/83). The most frequent agr type was agrI (48.2% of the isolates), associated with CC8, CC22, CC45, CC97, CC398, and CC30 (only one strain); agrII was most frequently detected among CC5, CC15 and less in CC8 and CC22; agrIII was detected in CC1 and CC30, and agrIV in a single isolate belonging to CC30.
Resistance Genes
The mecA gene was detected in 30.1% of the isolates belonging to CC22, CC5, CC8, and CC1. The majority of MRSA isolates carried the SCCmec type IV (72.0%, 18/25), while few isolates carried type I (12.0%, 3/25), type II (8.0%, 2/25), type III (4.0%, 1/25), and type V (4.0%, 1/25; Supplementary Table S3). MRSA strains belonging to CC22 harbored a lower number of resistance genes compared to the other CCs (Supplementary Figure S1). Besides resistance to β-lactams, the most frequent resistance genes detected in MRSA were aminoglycoside resistance genes (68.0%, 17/25 isolates) followed by macrolide resistance genes (64.0%, 16/25 isolates) and tetracycline resistance genes (16.0%, 4/25 isolates; Table 2). Point mutations conferring resistance to quinolones (gyrA and grlA genes) and rifampicin (rpoB gene) were detected in 80.0% (20/25) and 16% (4/25) of the MRSA isolates, respectively. Discrepancies between the susceptibility phenotype and resistance gene carriage were noticed in few cases for levofloxacin (n = 7 isolates), gentamicin (n = 2 isolates), rifampicin (n = 1 isolate), and daptomycin (n = 1 isolate), presumably due to limitations associated with the in silico prediction of resistant genes.
Discussion
S. aureus is the most frequent pathogen causing OM, and a wide range of virulence factors involved in adhesion, host cells damage, and evasion of the immune system are likely to account for its success in OM. Given the multifactorial nature of S. aureus pathogenesis, this study was aimed at understanding whether S. aureus isolates from OM were characterized by (i) distinctive epidemiological traits and (ii) definite repertoire(s) of virulence-related genes. To this purpose, the clonal characteristics and the virulence determinants inferred from WGS data of 83 S. aureus isolates responsible for OM in Italy were investigated. The most frequent CCs were CC22, followed by CC5, CC8 and CC30, which are among the prevalent lineages in Italian nosocomial settings (Giufrè et al., 2017). The phylogenetic relationship based on cgMLST revealed a non-uniform distribution of the isolates that clustered together according to the ST-CC grouping, regardless of the geographic origin or methicillin resistance status. Close genetic relatedness between isolates from either the same or different hospitals was rarely observed, suggesting limited transmission of S. aureus strains causing OM within the same hospital and between hospitals in the same geographical area.
The S. aureus lineages identified in the present study are largely consistent with those responsible for bone and joint infections worldwide (Pimentel de Araujo et al., 2021), and essentially match the most frequent lineages causing both invasive and non-invasive infections in Italian hospitals (Gagliotti et al., 2012; Grundmann et al., 2014; Giufrè et al., 2017). Although ST8/CC8 is the most common clone involved in OM in several countries (Luedicke et al., 2010; Senneville et al., 2014; Gaviria-Agudelo et al., 2015), the most common lineages in Italy were CC22 and CC5. MSSA isolates showed polyclonality with CC30 being the most represented CC, as previously reported in OM isolates (Post et al., 2014; Valour et al., 2014; Montanaro et al., 2016). MRSA clones belonging to the CC1 and CC8 lineages carried a high number of resistance determinants. Conversely, isolates belonging to CC22, which is one of the most common MRSA lineages in hospital settings in Italy, carried a limited number of resistant genes.
Studies attempting to characterize the virulence of OM strains identified some genetic traits that could be associated with a greater propensity to cause bone infections, such as the cna and bbp adhesins genes (Post et al., 2014; Montanaro et al., 2016). In our collection cna and bbp gene were present in 48.2 and 77.1% of the isolates, respectively. In particular, isolates belonging to CC22, the prevalent clone in our collection, showed a wide pattern of adhesin genes, including those coding for collagen adhesin (Cna), bone sialoprotein-binding protein (BbP), and fibronectin binding protein A (FnbA). However, it is worth noticing that each lineage expressed a different combination of adhesin genes, likely reflecting functional redundancy. Previous studies showed how these adhesin genes were frequently associated with invasive infections including bone infections (Palmqvist et al., 2005; Rieg et al., 2013; Foster et al., 2014; Post et al., 2014). Cna has previously been identified in few S. aureus clones (Peacock et al., 2002) and has been shown to play an important role in bone tropism in a murine model of haematogenous OM and in the development of septic arthritis (Patti et al., 1994; Elasri et al., 2002). Similarly, BbP is involved in bone tropism and in the initial adhesion to osteoblasts due to its ability to bind bone sialoprotein and fibrogen (Tung et al., 2000; Testoni et al., 2011). FnbA and FnbB have frequently been detected in clinical S. aureus isolates (Peacock et al., 2002). FnbA mediates the internalization of S. aureus into osteoblasts and adhesion to implant materials enhancing the severity of bone infection and its chronicity (Ahmed et al., 2001; Palmqvist et al., 2005; Testoni et al., 2011; Gries et al., 2020).
After the initial adhesion of the bacteria to bone tissue, the subsequent infection step is biofilm formation. The locus icaACD involved in the production of the polysaccharide intercellular adhesin (PIA), which promotes the aggregation of the bacterial cells in biofilm (Heilmann et al., 1996), was detected in over 95% of the S. aureus isolates examined in our study. Indeed, the presence of the ica locus is very frequent among strains from biofilm-related infection (90%) and less frequent in isolates from carriers (43%; VictoriaMartín-López et al., 2002). Capsular genes were detected in all the isolates, and cap5 was the most frequent irrespective of the CC. Other important virulence factors, such as the PVL toxin genes, which are a hallmark of community-acquired MRSA (Otto, 2013), were rarely detected, being present in only two CC30 strains. This is probably because OM patients in our study were adults, while PVL genes were more frequently detected in S. aureus from paediatric OM (Kechrid et al., 2011; Bouras et al., 2018), and only occasionally in isolates from adults (Senneville et al., 2014; Valour et al., 2014). Other leucocidin genes, such as lukX and lukY, and haemolysin genes aur, hla, hlII and hlgA/B/C, were ubiquitous among the strains. The enterotoxins cluster seg-sei-sem-sen-seo, which has been documented to increase the commensal fitness of S. aureus (Nowrouzian et al., 2015), was detected in the most frequent lineages CC5, CC22, and CC30. The expression of several virulence genes such as alpha-toxin (hla), gamma-hemolysin (hlg), leucocidins (lukX and lukY), and adhesins is under the control of the agr system (Cheung et al., 2011; Jenul and Horswill, 2019), which has been directly associated with the pathogenesis of OM (Gillaspy et al., 1995). The agr system was detected in nearly 90% of the isolates. In this study, the higher frequency of isolates carrying agr I and agrII, compared with agrIII and agrIV, is in accordance with previous reports with other collections of isolates from bone infections (Montanaro et al., 2010; Kawamura et al., 2011).
Studies on the association between S. aureus genetic traits and clinical manifestations are challenging and often generated contradictory results. Some studies support that certain S. aureus lineages harboring specific sets of virulence genes are more successful than others in causing invasive disease (Rasmussen et al., 2013; Tasse et al., 2018). However, the limit of these studies is the consensus repertoire of virulence genes that are shared within the lineage. Thus, the association of virulence factors and disease can be biased by an uneven distribution of CC between the group of isolates investigated (hitchhiker effect; Lindsay et al., 2006). Strains causing OM show substantial heterogeneity of virulence factors as it is for isolates causing colonization. A recent study showed that the commensal nasal isolates shared the same CC and genetic determinants with isolates from joint infection, suggesting that commensal S. aureus clones can cause bone and joint infections (Wildeman et al., 2020). Similarly, there is no evidence that a particular lineage or a single virulence factor or a combination of factors were distinctive of isolates from bone and implant infections (Luedicke et al., 2010) or from invasive infections (van Belkum et al., 2009). Although belonging to several lineages and being characterized by heterogeneous virulence profiles, strains isolated from carriers can become invasive under certain circumstances (Mehraj et al., 2016; Deinhardt-Emmer et al., 2018). Given that S. aureus clones causing OM originate from commensal clones which are characterized by an uneven distribution of virulence genes, it is not surprising that S. aureus strains from our collection show a broadly diversified pattern of virulence-related traits.
Conclusion
In summary, this is the first Italian study providing a genome-level characterization of a large collection of S. aureus isolates from bone infections. The prevalence of the different CCs in OM follows the epidemiology of S. aureus infections in Italy. It is conceivable that the prevalence of certain S. aureus lineages in OM is due to their more frequent circulation among patients rather than to a particular pattern of associated virulence factors. Understanding the virulence of S. aureus and the consequent infection tropism(s) continues to be a challenging topic for the scientific community. A better understanding of this aspect could be useful for the development of prevention and treatment strategies for OM.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA784720.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics committee of Istituto Superiore di Sanità no. 0013802 of 18 April 2019. Patients were required to sign an informed consent that included the acceptance of the collection and analysis of clinical and microbiological data for epidemiologic and scientific purposes. Patients’ data were anonymized.
Author Contributions
FPA, AP, and PV conceived the study. AP, PV, MM, and MDG contributed to the design of the study. SA, DL, TC, RG, and ER provided the isolates and patients’ data. FPA performed the experiments and drafted the manuscript. FPA, MP, and MDG performed the WGS. FPA and MP analysed the data. FPA, MP, AP, PV, MDG, and MM revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported in part by the Excellence Departments grant (art. 1, commi 314–337 Legge 232/2016) to the Department of Science, Roma Tre University, and grant PRIN 2017 (Prot. 20177J5Y3P) to PV, both from the Italian Ministry of Education, University and Research (MIUR), and in part by the Italian Ministry of Health, Centro Controllo Malattie (CCM), 2019 project “Sostegno alla Sorveglianza delle Infezioni correlate all’assistenza anche a supporto del PNCAR.”
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors wish to thank Pierluigi Viale (Infectious Diseases Unit, Department for Integrated Infectious Risk Management, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Italy), Nicolò Rossi (Infectious Diseases Unit, Department for Integrated Infectious Risk Management, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Italy), Domenico Tigani (Department of Orthopaedic Surgery, Ospedale Maggiore, Bologna, Italy), Calogero Alfonso (Orthopedics Unit, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Italy), Mauro Girolami (Bentivoglio Orthopaedic Ward, IRCCS Istituto Ortopedico Rizzoli, Bologna, Italy), Carlo Impallomeni (Orthopedics Unit, Imola Hospital, Bologna, Italy), Gian Maria Rossolini (Department of Experimental and Clinical Medicine, University of Florence, Italy), Domenico Andrea Campanacci (Department of Orthopaedic Oncology and Reconstructive Surgery, Careggi University Hospital, Firenze, Italy), Alberto Farese (Infectious and Tropical Diseases Unit, Careggi University Hospital, Florence, Italy) for their contribution in data collection. The valuable support of Fabio D’Ambrosio (Department of Infectious Diseases, Istituto Superiore di Sanità, Rome, Italy) and the technical staff of the Unit of Clinical Microbiology, S. Agostino-Estense, Hospital Baggiovara, Modena, Italy is also acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.846167/full#supplementary-material
Footnotes
1. ^https://www.eucast.org/clinical_breakpoints/
2. ^https://w3.iss.it/site/aries/
References
Ahmed, S., Meghji, S., Williams, R. J., Henderson, B., Brock, J. H., and Nair, S. P. (2001). Staphylococcus aureus fibronectin binding proteins are essential for internalization by osteoblasts but do not account for differences in intracellular levels of bacteria. Infect. Immun. 69, 2872–2877. doi: 10.1128/IAI.69.5.2872-2877.2001
Aubry-Damon, H., Soussy, C.-J., and Courvalin, P. (1998). Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 42, 2590–2594. doi: 10.1128/AAC.42.10.2590
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bouras, D., Doudoulakakis, A., Tsolia, M., Vaki, I., Giormezis, N., Petropoulou, N., et al. (2018). Staphylococcus aureus osteoarticular infections in children: an 8-year review of molecular microbiology, antibiotic resistance and clinical characteristics. J. Med. Microbiol. 67, 1753–1760. doi: 10.1099/jmm.0.000859
Byren, I., Bejon, P., Atkins, B. L., Angus, B., Masters, S., McLardy-Smith, P., et al. (2009). One hundred and twelve infected arthroplasties treated with ‘DAIR’ (debridement, antibiotics and implant retention): antibiotic duration and outcome. J. Antimicrob. Chemother. 63, 1264–1271. doi: 10.1093/jac/dkp107
Campoccia, D., Baldassarri, L., Pirini, V., Ravaioli, S., Montanaro, L., and Arciola, C. R. (2008). Molecular epidemiology of Staphylococcus aureus from implant orthopaedic infections: Ribotypes, agr polymorphism, leukocidal toxins and antibiotic resistance. Biomaterials 29, 4108–4116. doi: 10.1016/j.biomaterials.2008.07.006
Cheung, G. Y. C., Wang, R., Khan, B. A., Sturdevant, D. E., and Otto, M. (2011). Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 79, 1927–1935. doi: 10.1128/IAI.00046-11
Deinhardt-Emmer, S., Sachse, S., Geraci, J., Fischer, C., Kwetkat, A., Dawczynski, K., et al. (2018). Virulence patterns of Staphylococcus aureus strains from nasopharyngeal colonization. J. Hosp. Infect. 100, 309–315. doi: 10.1016/j.jhin.2017.12.011
Elasri, M. O., Thomas, J. R., Skinner, R. A., Blevins, J. S., Beenken, K. E., Nelson, C. L., et al. (2002). Staphylococcus aureus collagen adhesin contributes to the pathogenesis of osteomyelitis. Bone 30, 275–280. doi: 10.1016/S8756-3282(01)00632-9
Foster, T. J., Geoghegan, J. A., Ganesh, V. K., and Höök, M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62. doi: 10.1038/nrmicro3161
Gagliotti, C., Monaco, M., Sabia, C., Gargiulo, R., Sarti, M., Sanchini, A., et al. (2012). Staphylococcus aureus in a northern Italian region: phenotypic and molecular characterization. Scand. J. Infect. Dis. 44, 24–28. doi: 10.3109/00365548.2011.603744
Gaviria-Agudelo, C., Aroh, C., Tareen, N., Wakeland, E. K., Kim, M., and Copley, L. A. (2015). Genomic heterogeneity of methicillin resistant Staphylococcus aureus associated with variation in severity of illness among children with acute hematogenous osteomyelitis. PLoS One 10:e0130415. doi: 10.1371/journal.pone.0130415
Geraghty, T., and LaPorta, G. (2019). Current health and economic burden of chronic diabetic osteomyelitis. Expert Rev. Pharmacoecon. Outcomes Res. 19, 279–286. doi: 10.1080/14737167.2019.1567337
Gillaspy, A. F., Hickmon, S. G., Skinner, R. A., Thomas, J. R., Nelson, C. L., and Smeltzer, M. S. (1995). Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63, 3373–3380. doi: 10.1128/iai.63.9.3373-3380.1995
Giufrè, M., Ricchizzi, E., Accogli, M., Barbanti, F., Monaco, M., Pimentel de Araujo, F., et al. (2017). Colonization by multidrug-resistant organisms in long-term care facilities in Italy: a point-prevalence study. Clin. Microbiol. Infect. 23, 961–967. doi: 10.1016/j.cmi.2017.04.006
Grammatico, L., Baron, S., Rusch, E., Lepage, B., Surer, N., Desenclos, J. C., et al. (2008). Epidemiology of vertebral osteomyelitis (VO) in France: analysis of hospital-discharge data 2002-2003. Epidemiol. Infect. 136, 653–660. doi: 10.1017/S0950268807008850
Gries, C. M., Biddle, T., Bose, J. L., Kielian, T., and Lo, D. D. (2020). Staphylococcus aureus fibronectin binding protein A mediates biofilm development and infection. Infect. Immun. 88, e00859–e00819. doi: 10.1128/IAI.00859-19
Grundmann, H., Schouls, L. M., Aanensen, D. M., Pluister, G. N., Tami, A., Chlebowicz, M., et al. (2014). The dynamic changes of dominant clones of Staphylococcus aureus causing bloodstream infections in the European region: results of a second structured survey. Eur. Secur. 19:20987. doi: 10.2807/1560-7917.ES2014.19.49.20987
Heilmann, C., Schweitzer, O., Gerke, C., Vanittanakom, N., Mack, D., and Götz, F. (1996). Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x
Howard-Jones, A. R., and Isaacs, D. (2013). Systematic review of duration and choice of systemic antibiotic therapy for acute haematogenous bacterial osteomyelitis in children. J. Paediatr. Child Health 49, 760–768. doi: 10.1111/jpc.12251
Jenul, C., and Horswill, A. R. (2019). Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 7, 1–21. doi: 10.1128/microbiolspec.GPP3-0031-2018
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Karwowska, A., Davies, H. D., and Jadavji, T. (1998). Epidemiology and outcome of osteomyelitis in the era of sequential intravenous-oral therapy. Pediatr. Infect. Dis. J. 17, 1021–1026. doi: 10.1097/00006454-199811000-00012
Kawamura, H., Nishi, J., Imuta, N., Tokuda, K., Miyanohara, H., Hashiguchi, T., et al. (2011). Quantitative analysis of biofilm formation of methicillin-resistant Staphylococcus aureus (MRSA) strains from patients with orthopaedic device-related infections. FEMS Immunol. Med. Microbiol. 63, 10–15. doi: 10.1111/j.1574-695X.2011.00821.x
Kechrid, A., Pérez-Vázquez, M., Smaoui, H., Hariga, D., Rodríguez-Baños, M., Vindel, A., et al. (2011). Molecular analysis of community-acquired methicillin-susceptible and resistant Staphylococcus aureus isolates recovered from bacteraemic and osteomyelitis infections in children from Tunisia. Clin. Microbiol. Infect. 17, 1020–1026. doi: 10.1111/j.1469-0691.2010.03367.x
Kremers, H. M., Nwojo, M. E., Ransom, J. E., Wood-Wentz, C. M., Melton, L. J., and Huddleston, P. M. (2015). Trends in the epidemiology of osteomyelitis: a population-based study, 1969 to 2009. J. Bone Joint Surg. Am. 97, 837–845. doi: 10.2106/JBJS.N.01350
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kurtz, S. M., Lau, E., Watson, H., Schmier, J. K., and Parvizi, J. (2012). Economic burden of periprosthetic joint infection in the United States. J. Arthroplast. 27, 61–65.e1. doi: 10.1016/j.arth.2012.02.022
Leopold, S. R., Goering, R. V., Witten, A., Harmsen, D., and Mellmann, A. (2014). Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J. Clin. Microbiol. 52, 2365–2370. doi: 10.1128/JCM.00262-14
Lindsay, J. A., Moore, C. E., Day, N. P., Peacock, S. J., Witney, A. A., Stabler, R. A., et al. (2006). Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J. Bacteriol. 188, 669–676. doi: 10.1128/JB.188.2.669-676.2006
Luedicke, C., Slickers, P., Ehricht, R., and Monecke, S. (2010). Molecular fingerprinting of Staphylococcus aureus from bone and joint infections. Eur. J. Clin. Microbiol. Infect. Dis. 29, 457–463. doi: 10.1007/s10096-010-0884-4
Mehraj, J., Witte, W., Akmatov, M. K., Layer, F., Werner, G., and Krause, G. (2016). Epidemiology of Staphylococcus aureus nasal carriage patterns in the community. Curr. Top. Microbiol. Immunol. 398, 55–87. doi: 10.1007/82_2016_497
Montanaro, L., Arciola, C. R., Baldassarri, L., and Borsetti, E. (1999). Presence and expression of collagen adhesin gene (cna) and slime production in Staphylococcus aureus strains from orthopaedic prosthesis infections. Biomaterials 20, 1945–1949. doi: 10.1016/s0142-9612(99)00099-x
Montanaro, L., Ravaioli, S., Ruppitsch, W., Campoccia, D., Pietrocola, G., Visai, L., et al. (2016). Molecular characterization of a prevalent ribocluster of methicillin-sensitive Staphylococcus aureus from orthopedic implant infections. Correspondence with MLST CC30. Front. Cell. Infect. Microbiol. 6:8. doi: 10.3389/fcimb.2016.00008
Montanaro, L., Speziale, P., Campoccia, D., Pirini, V., Ravaioli, S., Cangini, I., et al. (2010). Polymorphisms of agr locus correspond to distinct genetic patterns of virulence in Staphylococcus aureus clinical isolates from orthopedic implant infections. J. Biomed. Mater. Res. A 94, 825–832. doi: 10.1002/jbm.a.32764
Nowrouzian, F. L., Ali, A., Badiou, C., Dauwalder, O., Lina, G., and Josefsson, E. (2015). Impacts of enterotoxin gene cluster-encoded superantigens on local and systemic experimental Staphylococcus aureus infections. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1443–1449. doi: 10.1007/s10096-015-2371-4
Otto, M. (2013). Community-associated MRSA: what makes them special? Int. J. Med. Microbiol. 303, 324–330. doi: 10.1016/j.ijmm.2013.02.007
Palmqvist, N., Foster, T., Fitzgerald, J. R., Josefsson, E., and Tarkowski, A. (2005). Fibronectin-binding proteins and fibrinogen-binding clumping factors play distinct roles in staphylococcal arthritis and systemic inflammation. J. Infect. Dis. 191, 791–798. doi: 10.1086/427663
Patti, J. M., Bremell, T., Krajewska-Pietrasik, D., Abdelnour, A., Tarkowski, A., Rydén, C., et al. (1994). The Staphylococcus aureus collagen adhesin is a virulence determinant in experimental septic arthritis. Infect. Immun. 62, 152–161. doi: 10.1128/iai.62.1.152-161.1994
Peacock, S. J., Moore, C. E., Justice, A., Kantzanou, M., Story, L., Mackie, K., et al. (2002). Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 70, 4987–4996. doi: 10.1128/IAI.70.9.4987-4996.2002
Pimentel de Araujo, F., Monaco, M., Del Grosso, M., Pirolo, M., Visca, P., and Pantosti, A. (2021). Staphylococcus aureus clones causing osteomyelitis: a literature review (2000–2020). J. Glob. Antimicrob. Resist. 26, 29–36. doi: 10.1016/j.jgar.2021.03.030
Post, V., Wahl, P., Uçkay, I., Ochsner, P., Zimmerli, W., Corvec, S., et al. (2014). Phenotypic and genotypic characterisation of Staphylococcus aureus causing musculoskeletal infections. Int. J. Med. Microbiol. 304, 565–576. doi: 10.1016/j.ijmm.2014.03.003
Rasmussen, G., Monecke, S., Ehricht, R., and Söderquist, B. (2013). Prevalence of clonal complexes and virulence genes among commensal and invasive Staphylococcus aureus isolates in Sweden. PLoS One 8:e77477. doi: 10.1371/journal.pone.0077477
Ricciardi, B. F., Muthukrishnan, G., Masters, E., Ninomiya, M., Lee, C. C., and Schwarz, E. M. (2018). Staphylococcus aureus evasion of host immunity in the setting of prosthetic joint infection: biofilm and beyond. Curr. Rev. Musculoskelet. Med. 11, 389–400. doi: 10.1007/s12178-018-9501-4
Rieg, S., Jonas, D., Kaasch, A. J., Porzelius, C., Peyerl-Hoffmann, G., Theilacker, C., et al. (2013). Microarray-based genotyping and clinical outcomes of Staphylococcus aureus bloodstream infection: an exploratory study. PLoS One 8:e71259. doi: 10.1371/journal.pone.0071259
Senneville, E., Brière, M., Neut, C., Messad, N., Lina, G., Richard, J.-L., et al. (2014). First report of the predominance of clonal complex 398 Staphylococcus aureus strains in osteomyelitis complicating diabetic foot ulcers: a national French study. Clin. Microbiol. Infect. 20, O274–O277. doi: 10.1111/1469-0691.12375
Shukla, D., Patel, S., Clack, L., Smith, T. B., and Shuler, M. S. (2021). Retrospective analysis of trends in surgery volumes between 2016 and 2019 and impact of the insurance deductible: cross-sectional study. Ann. Med. Surg. 63:102176. doi: 10.1016/j.amsu.2021.02.022
Strauß, L., Ruffing, U., Abdulla, S., Alabi, A., Akulenko, R., Garrine, M., et al. (2016). Detecting Staphylococcus aureus virulence and resistance genes: a comparison of whole-genome sequencing and DNA microarray technology. J. Clin. Microbiol. 54, 1008–1016. doi: 10.1128/JCM.03022-15
Tanaka, M., Wang, T., Onodera, Y., Uchida, Y., and Sato, K. (2000). Mechanism of quinolone resistance in Staphylococcus aureus. J. Infect. Chemother. 6, 131–139. doi: 10.1007/s101560070010
Tasse, J., Trouillet-Assant, S., Josse, J., Martins-Simões, P., Valour, F., Langlois-Jacques, C., et al. (2018). Association between biofilm formation phenotype and clonal lineage in Staphylococcus aureus strains from bone and joint infections. PLoS One 13:e0200064. doi: 10.1371/journal.pone.0200064
Testoni, F., Montanaro, L., Poggi, A., Visai, L., Campoccia, D., and Arciola, C. R. (2011). Internalization by osteoblasts of two Staphylococcus aureus clinical isolates differing in their adhesin gene pattern. Int. J. Artif. Organs 34, 789–798. doi: 10.5301/ijao.5000058
Trampuz, A., and Zimmerli, W. (2008). Diagnosis and treatment of implant-associated septic arthritis and osteomyelitis. Curr. Infect. Dis. Rep. 10, 394–403. doi: 10.1007/s11908-008-0064-1
Tuchscherr, L., Pöllath, C., Siegmund, A., Deinhardt-Emmer, S., Hoerr, V., Svensson, C.-M., et al. (2019). Clinical S. aureus isolates vary in their virulence to promote adaptation to the host. Toxins 11:135. doi: 10.3390/toxins11030135
Tung, H. S., Guss, B., Hellman, U., Persson, L., Rubin, K., and Rydén, C. (2000). A bone sialoprotein-binding protein from Staphylococcus aureus: a member of the staphylococcal Sdr family. Biochem. J. 345, 611–619. doi: 10.1042/bj3450611
Valour, F., Tasse, J., Trouillet-Assant, S., Rasigade, J.-P., Lamy, B., Chanard, E., et al. (2014). Methicillin-susceptible Staphylococcus aureus clonal complex 398: high prevalence and geographical heterogeneity in bone and joint infection and nasal carriage. Clin. Microbiol. Infect. 20, O772–O775. doi: 10.1111/1469-0691.12567
van Belkum, A., Melles, D., Nouwen, J., van Leeuwen, W., van Wamel, W., Vos, M., et al. (2009). Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect. Genet. Evol. 9, 32–47. doi: 10.1016/j.meegid.2008.09.012
VictoriaMartín-López, J., Pérez-Roth, E., Claverie-Martín, F., Díez Gil, O., Batista, N., Morales, M., et al. (2002). Detection of Staphylococcus aureus clinical isolates harboring the Ica gene cluster needed for biofilm establishment. J. Clin. Microbiol. 40, 1569–1570. doi: 10.1128/JCM.40.4.1569-1570.2002
Wildeman, P., Tevell, S., Eriksson, C., Lagos, A. C., Söderquist, B., and Stenmark, B. (2020). Genomic characterization and outcome of prosthetic joint infections caused by Staphylococcus aureus. Sci. Rep. 10:5938. doi: 10.1038/s41598-020-62751-z
Keywords: Staphylococcus aureus, virulence genes, osteomyelitis, clones, antibiotic resistance, whole genome sequencing
Citation: Pimentel de Araujo F, Pirolo M, Monaco M, Del Grosso M, Ambretti S, Lombardo D, Cassetti T, Gargiulo R, Riccobono E, Visca P and Pantosti A (2022) Virulence Determinants in Staphylococcus aureus Clones Causing Osteomyelitis in Italy. Front. Microbiol. 13:846167. doi: 10.3389/fmicb.2022.846167
Edited by:
Giovanni Gherardi, Campus Bio-Medico University, ItalyReviewed by:
Luca A. Vitali, University of Camerino, ItalyFrancesco Iannelli, University of Siena, Italy
Copyright © 2022 Pimentel de Araujo, Pirolo, Monaco, Del Grosso, Ambretti, Lombardo, Cassetti, Gargiulo, Riccobono, Visca and Pantosti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paolo Visca, cGFvbG8udmlzY2FAdW5pcm9tYTMuaXQ=
†These authors have contributed equally to this work
 Fernanda Pimentel de Araujo
Fernanda Pimentel de Araujo Mattia Pirolo
Mattia Pirolo Monica Monaco
Monica Monaco Maria Del Grosso
Maria Del Grosso Simone Ambretti
Simone Ambretti Donatella Lombardo4
Donatella Lombardo4 Paolo Visca
Paolo Visca Annalisa Pantosti
Annalisa Pantosti