- 1Key Laboratory of Forest Protection of National Forestry and Grassland Administration, Ecology and Nature Conservation Institute, Chinese Academy of Forestry, Beijing, China
- 2Panzhihua City Academy of Agricultural and Forest Sciences, Panzhihua, China
A novel Gram-stain-negative, aerobic, motile bacterial strain, D13-10-4-6T, was isolated from the bark sample of Populus × euramericana. The strain could grow at 15–35°C, at pH 6–10 and in 0–4% (w/v) NaCl, and the strain tested positive for oxidase and catalase activities. The main polar lipids were phosphatidylmonomethylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, and phosphatidylethanolamine. The main respiratory quinone was Q-10, and the predominant fatty acid was C18:1 ω7c. The phylogenetic analyses showed that the strain belonged to the genus Pseudogemmobacter of the family Rhodobacteraceae. The family Rhodobacteraceae is an ecologically diverse group that includes bacteria from aquatic to terrestrial ecosystems. As a consequence, the classification of the family Rhodobacteraceae is difficult, not least when the early taxonomy work relied heavily on 16S rRNA gene analysis. Recently, the taxonomic status of many members of the family has been revised based on the genome analysis; however, there are still some classification conflicts due to the lack of genome sequences and parallel publication time. In this study, phylogenetic trees based on 16S rRNA gene, gyrB gene, and 120 concatenated proteins, the average amino acid identity (AAI) and percentage of conserved proteins (POCP) have been used for the analysis of strain D13-10-4-6T and other members of 15 genera within the family to further clarify their taxonomic relationships. For the data of phylogeny, AAI, and POCP, the taxonomic proposals are (1) reclassification of Rhodobacter tardus as the type species of a novel genus, Stagnihabitans gen. nov., as Stagnihabitans tardus comb. nov.; (2) reclassification of Tabrizicola alkalilacus, Tabrizicola sediminis, Tabrizicola algicola into a novel genus, Pseudotabrizicola gen. nov., as Pseudotabrizicola alkalilacus comb. nov., Pseudotabrizicola sediminis comb. nov., Pseudotabrizicola algicola comb. nov.; (3) reclassification of Rhodobacter sediminicola into the genus Cereibacter as Cereibacter sediminicola comb. nov.; (4) reclassification of Rhodobacter flagellatus, Rhodobacter thermarum, and Xinfangfangia soli into the genus Tabrizicola as Tabrizicola flagellatus comb. nov., Tabrizicola thermarum comb. Nov., and Tabrizicola soli comb. nov.; (5) reclassification of Xinfangfangia humi into the genus Pseudogemmobacter as Pseudogemmobacter humicola comb. nov.; (6) classification of strain D13-10-4-6T as a novel species of the genus Pseudogemmobacter, for which the name P. hezensis sp. nov. is proposed, the type strain is D13-10-4-6T (= CFCC 12033T = KCTC 82215T).
Introduction
Populus × euramericana cakers on poplar trees were found in China for many years, and the stem or branch bark of the diseased tree was cracked and exuded frothy fluid. During our investigation of the bacterial diversity in Populus × euramericana caker, strain D13-10-4-6T was isolated from the symptomatic bark of Populus × euramericana caker. The phylogenetic analyses showed that the strain belonged to the genus Pseudogemmobacter of the family Rhodobacteraceae. The family Rhodobacteraceae, described by Garrity et al. (2005), the so-called purple non-sulfur bacteria (Imhoff et al., 1998), is one of the major subdivisions of the class Alphaproteobacteria. It is ecologically and phenotypically diverse, and most of the members of the family have been found in various marine environments, including seawater, sea sediments, sea ice, coastal biofilms, marine animal tissues, and seaweeds (Selje et al., 2004; Buchan et al., 2005; Brinkhoff et al., 2008). At the time of writing, the family included more than 180 genera with validated names.1
The early classification of the genera within the family Rhodobacteraceae relied heavily on the analysis of 16S rRNA gene sequence and resulted in several non-monophyletic genera, for instance, the genus Rhodobacter (Imhoff et al., 1984). The genus Rhodobacter was reclassified by Suresh et al. (2019) and Hördt et al. (2020) based on the genome analysis. The members of the genus were divided into five distinct clades in the 16S rRNA gene-based phylogenetic tree constructed by Suresh et al. (2019). The Cereibacter sphaeroides (formerly Rhodobacter sphaeroides) clade was reclassified into the genus Luteovulum (Suresh et al., 2019) and then transferred into the genus Cereibacter by Hördt et al. (2020). However, Rhodobacter alkalitolerans, which belongs to the C. sphaeroides clade, was not reclassified due to the lack of genomic sequence. At present, the genus Rhodobacter contains 13 species with validated names according to the List of Prokaryotic names with Standing in Nomenclature (LSPN).2 The taxonomic status of the recently described Rhodobacter species, R. thermarum (Khan et al., 2019), R. flagellatus (Xian et al., 2020), R. sediminicola (Suresh et al., 2020), and R. tardus (Sheu et al., 2020), was also not included in the early reclassification of Rhodobacter due to the nearly parallel time of description, causing confusion in the classification of the genus Rhodobacter.
The genus Xinfangfangia, described by Hu et al. (2018), is closely related to the genera of Rhodobacter and Tabrizicola within the family Rhodobacteraceae (Hu et al., 2018). It contains only two species with validated names, namely, Xinfangfangia soli and Xinfangfangia humi (Kämpfer et al., 2019). The genus Pseudogemmobacter described by Suman et al. (2019) contains only one species with validly published names, namely, Pseudogemmobacter bohemicus. While X. humi and P. bohemicus, which were proposed almost simultaneously, share a high 16S rRNA gene sequence similarity of 99.2%. Therefore, the relationship of P. bohemicus and X. Humi needs to be studied.
Along with the advances in whole-genome sequencing technologies, several methods for taxonomic classification at the species and genus levels have been developed. The new standards for species recognition are developed using digital whole-genome comparisons, such as average nucleotide identities (ANIs) (Konstantinidis and Tiedje, 2005) and genome-to-genome-distance calculations (GGDCs) (Richter and Rosselló-Móra, 2009; Meier-Kolthoff et al., 2013). The average amino acid identity (AAI) (Luo et al., 2014; Rodriguez-R and Konstantinidis, 2014) and percentage of conserved proteins (POCPs) (Qin et al., 2014), which are methods of measuring amino acid-level genomic similarity between protein-coding regions, have been used in the delineation of prokaryotic organisms at the genus level. Furthermore, the phylogenetic analysis based on the whole-genome sequence has been recently encouraged for the taxonomy of prokaryotes owing to its robustness and repeatability (Chun et al., 2018). Nowadays, along with those methods, the reclassification of prokaryotes at class (Hördt et al., 2020), order (Orata et al., 2018), family (Liang et al., 2021), and genus (Suresh et al., 2019) levels has been done.
In this study, phylogenetic analysis based on the 16S rRNA gene, gyrB gene, and genomes sequence, as well as ANI, AAI, and POCP, was used to confirm the taxonomic relationship of the novel strain and its closely related members (e.g., members of the genus Xinfangfangia and Rhodobacter) in the family Rhodobacteraceae.
Materials and Methods
Strain and Culture Conditions
Strain D13-10-4-6T was isolated from the bark sample of Populus × euramericana collected from Heze, Shandong Province, China (34° 82′N, 115° 46′E) as previously described (Li et al., 2015). In brief, the bark samples were sterilized for 30 s with 70% ethanol, and then exposed to 4% (v/v) sodium hypochlorite for 3 min. After rinsing in sterile water three times, the samples with 2 ml sterile water were transferred to sterile mortar and ground with a pestle, respectively. The obtained solution was then shaken for 30 min at 30°C. The suspension with a dilution series was spread on tryptic soy agar (TSA, Difco). After 2 days of incubation on TSA plates at 30°C, single colonies were selected and cultured on a new plate, and were then preserved at -80°C with a supplement of 20% (v/v) glycerol.
Genome Sequencing
The genomes of the strains D13-10-4-6T and X. soli ZQBWT were sequenced by Illumina NovaSeq PE150 (Novogene, Co., Ltd., Beijing, China). Low-quality reads in the raw data were filtered by readfq (version 10), then the genome assembly with high-quality reads was performed using SOAPdenovo (version 2.04) (Li et al., 2008; Li et al., 2010), SPAdes (Bankevich et al., 2012), ABySS (Simpson et al., 2009), and then the results were integrated with CISA (Lin and Liao, 2013). The gap of the genome assembly was filled using gapclose (version 1.12).
Phylogenetic Analysis
The 16S rRNA gene of strain D13-10-4-6T was amplified by the primers 27F/1492R (Lane, 1991). The similarity of the 16S rRNA gene sequence between the strain D13-10-4-6T and the validly published bacterial species was determined using EzBio-Cloud’s identify service3 (Yoon et al., 2017). The 16S rRNA gene sequence of the related strains was obtained from GenBank for the phylogenetic analysis. After multiple sequence alignment with Clustal W, the phylogenetic analysis was carried out using MEGA X by the neighbor-joining, maximum-likelihood, and maximum-parsimony methods (Kumar et al., 2018). Aquidulcibacter paucihalophilus TH1-2T was used as an outgroup. The phylogenetic trees were evaluated by 1,000 bootstrap resamplings.
The gyrB gene sequences of the strain D13-10-4-6T were obtained from its genomic sequences according to Altschul et al. (1990), and a 1,050 bp sequence was obtained. The gyrB gene sequences of the related strains were obtained from GenBank or their genome sequences. The phylogenetic trees based on the gyrB gene sequence were constructed using the maximum-likelihood, neighbor-joining, and maximum-parsimony methods as a description of 16S rRNA gene phylogenetic analysis.
Concatenated protein tree has a higher recognition than single phylogenetic marker gene tree (e.g., 16S rRNA and gyrB) for bacterial taxonomy (Ciccarelli et al., 2006; Thiergart et al., 2014), and has been widely used in solving bacterial taxonomy (Hördt et al., 2020; Xu et al., 2021). The genome sequences of the strain D13-10-4-6T and its related strains retrieved from GenBank (Supplementary Table 1) were used to construct the phylogenetic tree. A concatenated alignment of 120 ubiquitous single-copy proteins of the related strains was performed by GTDB-Tk v1.5.14 using the classify_wf command (Chaumeil et al., 2020). The alignment file was used to construct the maximum-likelihood tree with IQ-TREE 2.1.4-beta (Minh et al., 2020), and the best model was automatically selected by ModerFinder for the ML tree. The tree was visualized and edited with iTOL (Letunic and Bork, 2021).
Phylogenomic Metric Calculations
Average nucleotide identity (ANI is a measure of similarity between two genomic sequences, which is a useful tool to differentiate bacterial species in common with DNA-DNA hybridization (DDH) (Goris et al., 2007; Richter and Rosselló-Móra, 2009). The ANI values among the novel strain D13-10-4-6T and its closely related reference strains (P. bohemicus Cd-10T, X. humi IMT-291T) were determined using OrthoANI (Lee et al., 2016). The GGDC5 was used to calculate the dDDH values among the novel and its closely related reference strains (Meier-Kolthoff et al., 2013). The analysis of the average AAI and POCP among the strains in this work was carried out with CompareM6 and a Python script (POCP)7 (Xu et al., 2020), respectively. The pan-genome analysis was carried out with BPGA (Chaudhari et al., 2016) with default parameters.
Chemotaxonomic Characterization
The strain D13-10-4-6T was shaken for 48 h in a tryptic soy broth (TSB; Difco) at 30°C, then collected by centrifuging at 10,000 rpm for 4 min. The harvested cells were freeze-dried and used to analyze the polar lipid and respiratory quinone. Polar lipids were analyzed by two-dimensional thin-layer chromatography as described by Minnikin et al. (1984). Isoprenoid quinones were extracted from the strain D13-10-4-6T as reported by Collins et al. (1977), analyzed by high-performance liquid chromatography (Groth et al., 1997; Du et al., 2013), and confirmed by liquid chromatography/mass spectrometry. After culturing for 2 days in TSB at 30°C, the cells were harvested at exponential phase and used for cellular fatty acids. Cellular fatty acids were extracted as reported by Kuykendall et al. (1988), analyzed using the Sherlock Microbial Identification System (Sasser, 1990).
Phenotypic Characterization
Growth conditions of the strain D13-10-4-6T were determined at different temperature, pH, and salinity levels according to the method described by Li et al. (2016). The growth temperature was set at 4, 10, 15, 20, 25, 28, 30, 37, 41, and 45°C. The pH values for growth were adjusted to various pH values (pH 4.0–11.0, at intervals of 1.0 pH unit) by the buffers (Delory and King, 1945; Gomori, 1955) citrate and Na2HPO4 buffer (pH 4.0–5.0), phosphate buffer (pH 6.0–7.0), Tris buffer (pH 8.0–9.0), and Na2HPO4/NaOH (pH 10.0–11.0). The salinity was determined in the range of 0–9% (w/v, intervals of 1%). Gram staining was performed according to the method described by Jenkins et al. (2003). To examine the anaerobic growth, the strain was incubated on TSA plates at 30°C for 1 week in an anaerobic jar (Li et al., 2016). The activities of catalase and oxidase were determined by the methods described by Smibert and Krieg (1994). Enzymatic activity, carbon source utilization, and acid production were performed by API ZYM, API 20 NE, and API 50 CH (bioMérieux) according to the manufacturer’s instructions, respectively.
Results and Discussion
Genome Information
The genome of strains D13-10-4-6T and X. soli ZQBWT were sequenced and analyzed. In total, 63 contigs with a total sequence length of 4,683,906 bp for the strain X. soli ZQBWT were obtained, which was predicted to have 4,455 protein-coding genes, 47 tRNA genes, 3 rRNA genes, and 3 other RNA genes. The DNA G + C contents was 67.6%. The strain D13-10-4-6T genome produced 66 contigs with a total sequence length of 4,605,234 bp, which was predicted to have 4,206 protein-coding genes, 45 tRNA genes, 3 rRNA genes, and 3 other RNA genes. The DNA G + C content of the strain D13-10-4-6T was 62.9%, which was similar to P. bohemicus Cd-10T (63.2%).
Phylogenetic Analyses
In this study, we have constructed phylogenetic trees based on the 16S rRNA gene, gyrB gene, and concatenated proteins (Figures 1–3) for representative members of the family Rhodobacteraceae encompassing 15 genera. The main groups clustering with the members of Rhodobacteraceae in 16S rRNA gene-based tree, gyrB gene-based tree, and concatenated proteins-based tree are almost consistent. The strains D13-10-4-6T, P. bohemicus Cd-10T, and X. humi IMT-291T form one monophyletic group to in turn form Pseudogemmobacter clade with strong bootstrap support in all three phylogenetic trees (Figures 1–3), which is far removed from the branch of X. soli (the type species of the genus Xinfangfangia). X. humi IMT-291T forms a distinct branch from the strains D13-10-4-6T and P. bohemicus Cd-10T in the Pseudogemmobacter clade. The results suggested that X. humi IMT-291T should be a species belonging to the genus Pseudogemmobacter, although P. bohemicus Cd-10T and X. humi IMT-291T were published almost simultaneously and shared 99.26% 16S rRNA gene sequence similarity with each other. The strain D13-10-4-6T forms a distinct branch from P. bohemicus Cd-10T and X. humi IMT-291T in all phylogenetic trees, and it has the highest 16S rRNA gene sequence similarity to P. bohemicus Cd-10T (97.6%) and X. humi IMT-291T (97.4%), and shares a less than 97% sequence similarity with all other validly published species. The results indicate that the strain D13-10-4-6T should belong to a novel species of the genus Pseudogemmobacter.
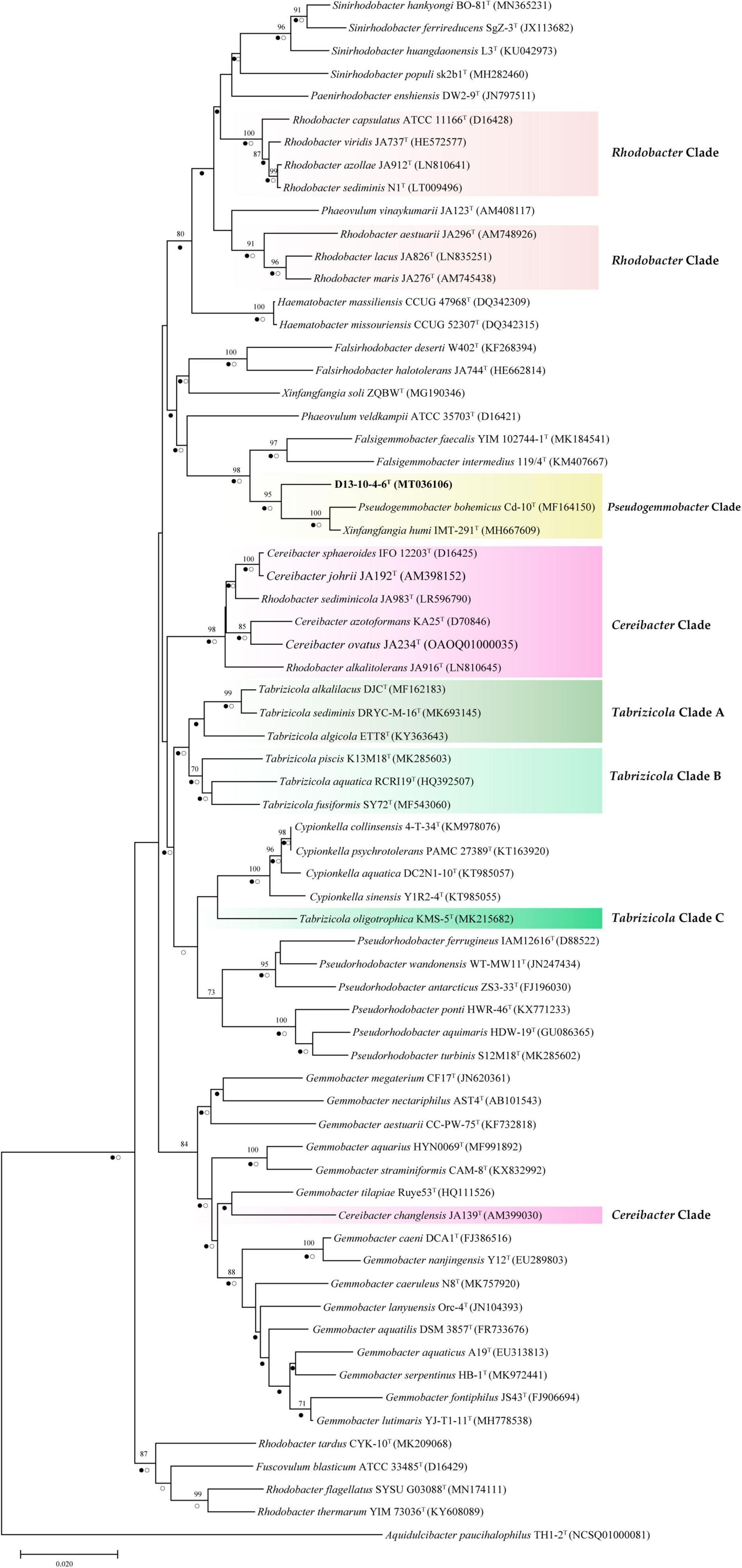
Figure 1. Neighbor-joining tree showing phylogenetic relationships among strain D13-10-4-6T and reference strains based on 16S rRNA gene sequences. Aquidulcibacter paucihalophilus TH1-2T was used as an outgroup. Only bootstrap values over 70% (based on 1,000 resamplings) are shown. The scale bar corresponds to 0.01 substitutions per nucleotide site. Filled circles indicate branches recovered by maximum-likelihood method and open circles at branches recovered by the maximum-parsimony method.
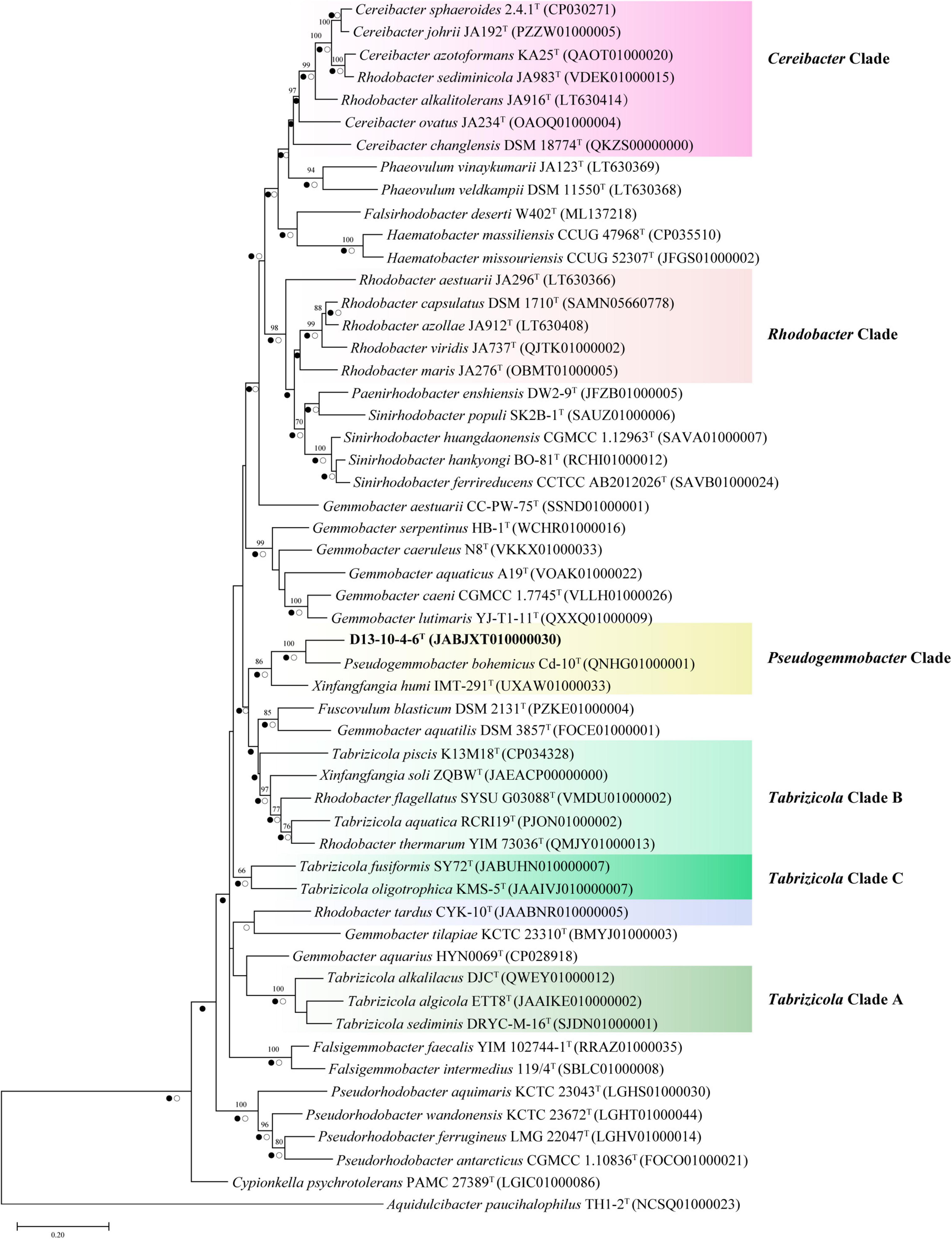
Figure 2. Neighbor-joining phylogenetic trees based on partial gyrB gene sequences showing the position of strain D13-10-4-6T and reference strains. Bootstrap values over 70% (expressed as percentages of 1,000 replications) are shown. The scale bar corresponds to 0.05 substitutions per nucleotide site. Filled circles indicate branches recovered by maximum-likelihood method and open circles at branches recovered by the maximum-parsimony method.
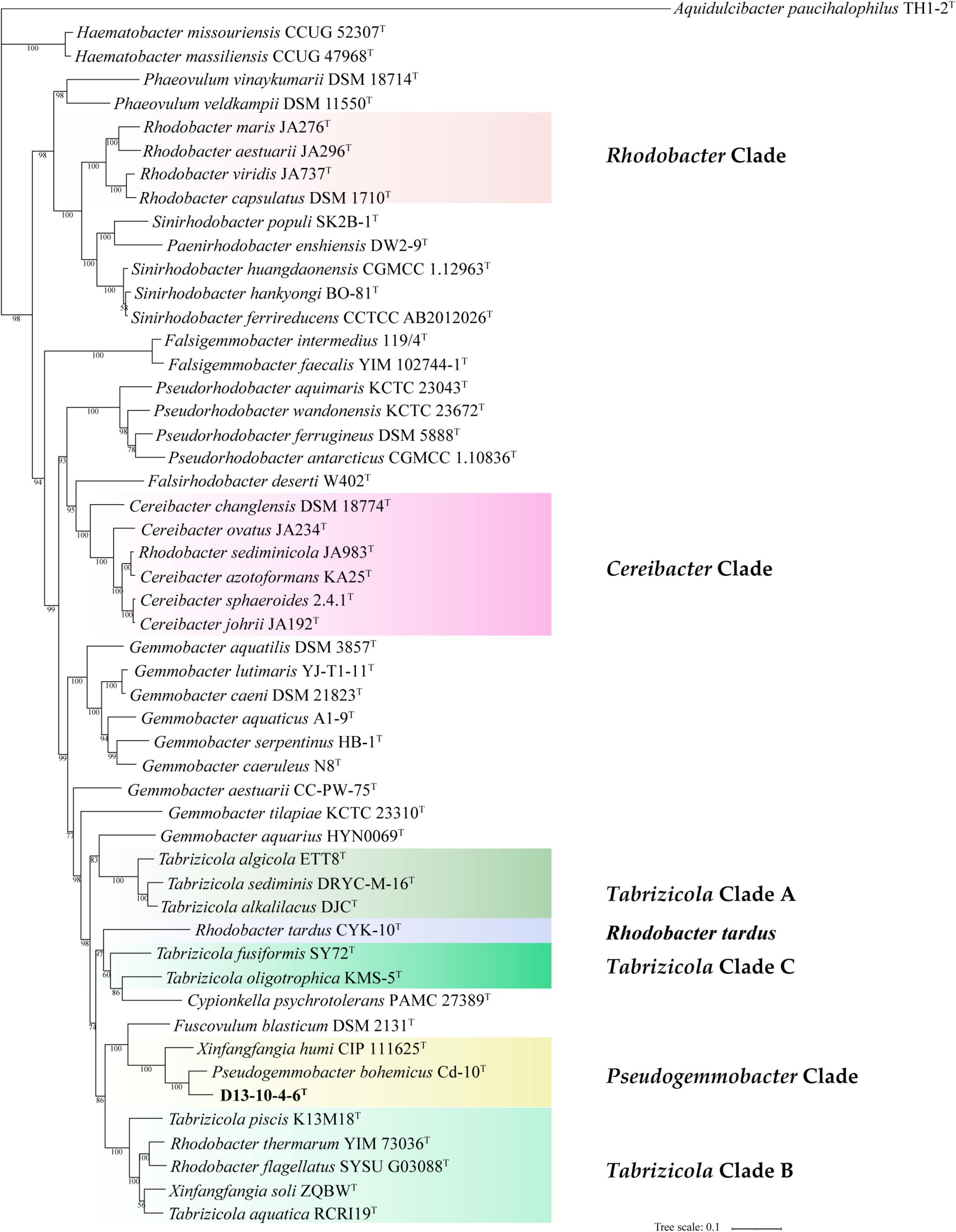
Figure 3. Phylogenetic tree among strain D13-10-4-6T and reference strains based on a concatenated alignment of 120 ubiquitous single-copy proteins. Aquidulcibacter paucihalophilus TH1-2T was used as an outgroup. The scale bar corresponds to 0.1 substitutions per amino acid position.
Several genera are non-monophyletic, such as Rhodobacter, Tabrizicola, and Xinfangfangia. In most of the cases, the 16S rRNA gene-based tree shows its low discriminatory power. For instance, the species of the genus Tabrizicola are divided into two branches in the 16S rRNA gene-based tree, but they are clustered into three distinct branches, not least T. aquatica RCRI19T (type species of the genus) and T. piscis K13M18T are grouped together with R. thermarum YIM 73036T, R. flagellatus SYSU G03088T, and X. soli ZQBWT in trees based on gyrB gene and concatenated proteins tree with strong support.
It can be seen from the trees based on the 16S rRNA gene, gyrB gene, and concatenated proteins that Rhodobacter and Tabrizicolaa are two closely related non-monophyletic genera. The members of the genus Tabrizicola are observed in three clades, clades A, B, and C, which are labeled in Figures 1–3. Clade A, formed by Tabrizicola alkalilacus DJCT, Tabrizicola sediminis DRYC-M-16T, and Tabrizicola algicola ETT8T, is next to the Gemmobacter clade and is far removed from two other Tabrizicola clades with a strong support in 16S rRNA gene-based, gyrB gene-based, and concatenated proteins-based trees. These results suggest that clade A should belong to a novel genus of the family Rhodobacteraceae. Tabrizicola clade B, grouped by T. aquatica RCRI19T (type species of the genus), T. piscis K13M18T, R. thermarum YIM 73036T, R. flagellatus SYSU G03088T, and X. soli ZQBWT, is a monophyletic cluster found in trees based on the gyrB gene and concatenated proteins with a strong support, indicating that R. thermarum YIM 73036T, R. flagellatus SYSU G03088T, and X. soli ZQBWT should be transferred to the genus Tabrizicola. Tabrizicola fusiformis SY72T, located in clade B in the 16S rRNA gene-based tree, is clustered together with Tabrizicola oligotrophica KMS-5T to form clade C in both trees based on the gyrB gene and concatenated proteins, demonstrating that T. fusiformis SY72T and T. oligotrophica KMS-5T may be a novel genus of the family Rhodobacteraceae.
In the 16S rRNA gene-based tree, Cereibacter clade include R. alkalitolerans JA916T, R. sediminicola JA983T), and members of the genus Cereibacter, except for Cereibacter changlensis JA139T (the type species). The species C. changlensis JA139T is observed in the Gemmobacter clade, which is similar to the results reported by Suresh et al. (2015). While C. changlensis JA139T is grouped in the Cereibacter clade and located at the edge of the clade in trees inferred from the gyrB gene and concatenated proteins, indicating that it should belong to the genus Cereibacter, which is consistent to the results described by Suresh et al. (2015). R. sediminicola JA983T is clustered in the Cereibacter clade in all the trees based on the 16S rRNA gene, gyrB gene, and concatenated proteins, indicating that they should be transferred to the genus Cereibacter.
The genus Rhodobacter proposed by Imhoff et al. (1984) contains 13 species with validated names according to the LSPN. In trees based on the 16S rRNA gene, gyrB gene, and concatenated protein, R. tardus CYK-10T forms one distinct branch from other clades, suggesting that it should belong to a novel genus of the family Rhodobacteraceae. Three members (R. sediminicola JA983T, R. thermarum YIM73036T, R. flagellatus SYSU G03088T) are clustered into the Tabrizicola clade and Cereibacter clade, respectively. The other eight members of the genus Rhodobacter are temporarily classified into the genus Rhodobacter because of the absence of genome sequence of R. azollae, R. lacus, R. alkalitolerans, and R. sediminis, although they are grouped into two clusters in the 16S rRNA gene tree.
Genomic, Chemotaxonomic, and Physiological Analysis of the Novel Strain
The ANI values between the strain D13-10-4-6T and its three closely related strains range from 74.4 to 81.2%, which are lower than the recommended ANI species boundary cutoff value (95–96%). The dDDH values between the strain D13-10-4-6T and its closely related strains are 19.7–24.3%, lower than the threshold for species (70%). Those data indicate that the strain D13-10-4-6T should belong to a novel species of the genus Pseudogemmobacter. Besides, P. bohemicus Cd-10T and X. humi IMT-291T share a 99.26% 16S rRNA gene sequence similarity, while their ANI and dDDH values are 79.1 and 22.1%, respectively (Table 1), which are lower than the species boundary cutoff values. Therefore, P. bohemicus Cd-10T and X. humi IMT-291T should belong to a different species of the genus Pseudogemmobacter.
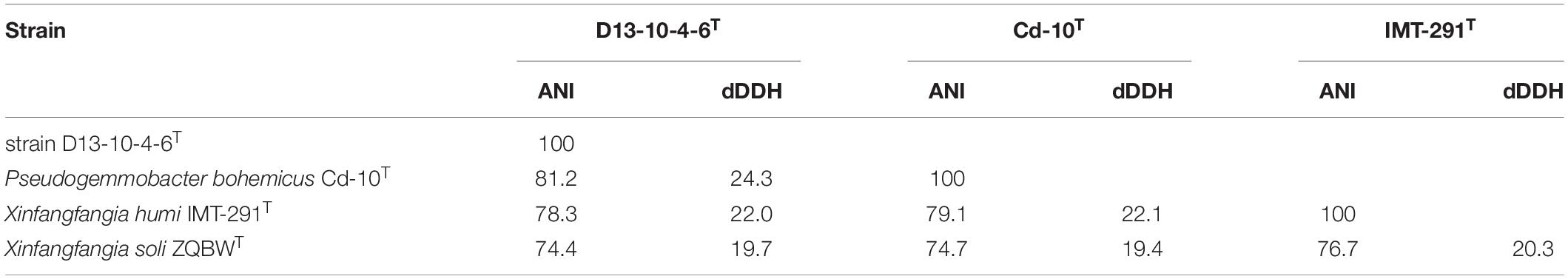
Table 1. Average nucleotide identity (ANI), digital DNA–DNA hybridization (dDDH) values among D13-10-4-6T, P. bohemicus Cd-10T, X. humi IMT-291T, and Xinfangfangia soli ZQBWT.
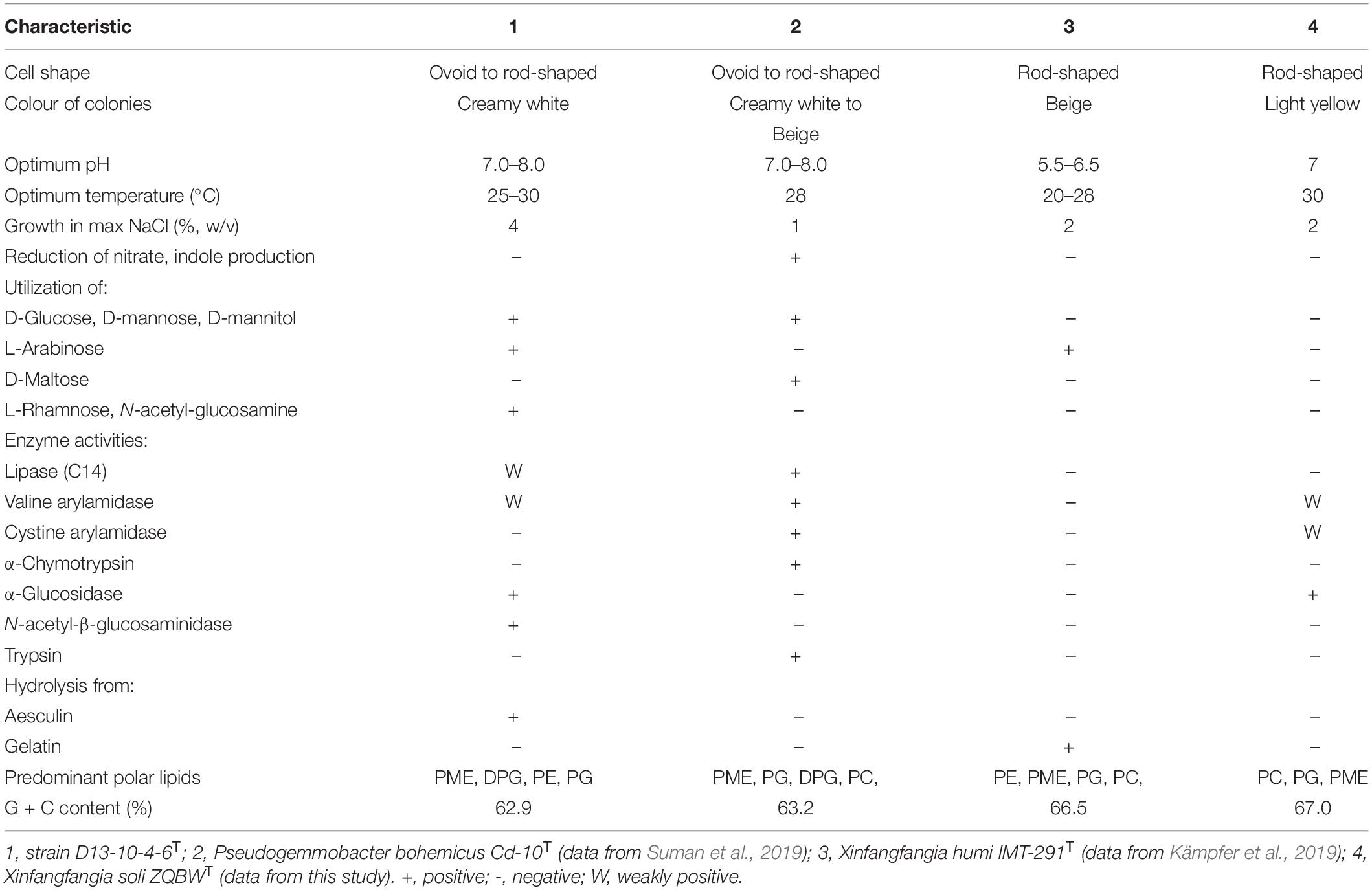
The polar lipids of the strain D13-10-4-6T are phosphatidy- lmonomethylethanolamine (PME), diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylcholine (PC), an unidentified phospholipid (PL), and six unidentified lipids (L) (Supplementary Figure 4). The presence of PE in the strain D13-10-4-6T is a useful characteristic to distinguish it from P. bohemicus and X. soli. The presence of DPG and absence of PC in the strain D13-10-4-6T are important characteristics to differentiate it from X. humi. The respiratory quinones detected in the strain D13-10-4-6T are Q-10 (91.3%) and Q-9 (8.7%), which are similar to P. bohemicus Cd-10T and X. humi IMT-291T. X. soli contains the only respiratory quinone of Q-10, which is different from the strains D13-10-4-6T, P. bohemicus Cd-10T, and X. humi IMT-291T. The phenotypic characterization of the strain D13-10-4-6T is listed in Table 2 and in the species description.
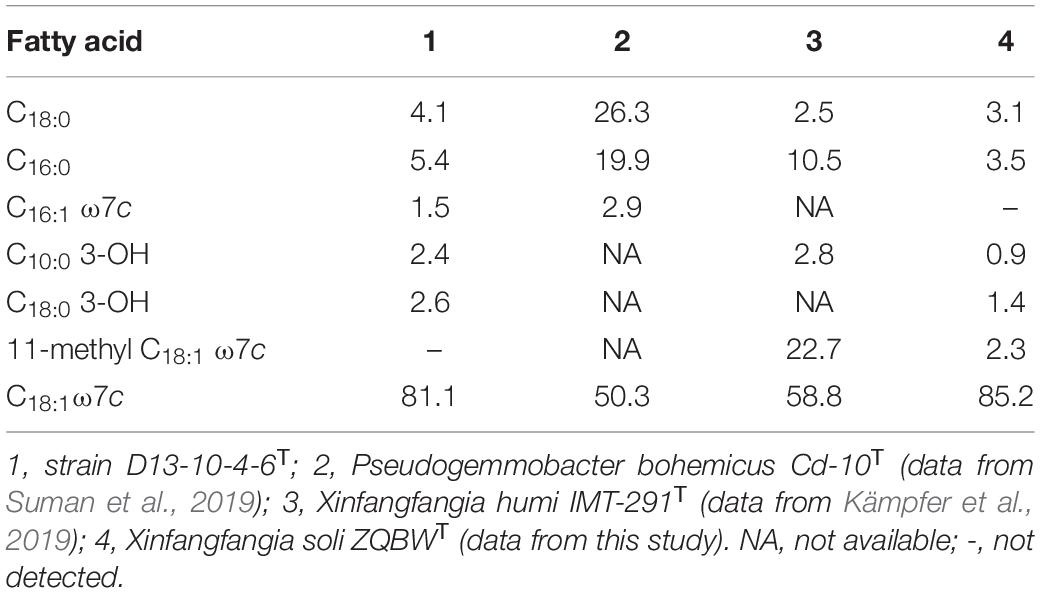
The predominant fatty acids of the strain D13-10-4-6T are C18:1 ω7c (81.1%), C16:0 (5.4%), and C18:0 (4.1%). The detailed and differential fatty acids data of strain D13-10-4-6T and its related species are listed in Table 3. The percentage of C18:1ω7c in the novel strain can be used to distinguish it from P. bohemicus Cd-10T and X. humi IMT-291T. The absence of 11-methyl C18:1 ω7c in the strain D13-10-4-6T is a useful characteristic to differentiate it from X. soli ZQBWT.
Phylogenomic Metric Analysis
Average AAI is one of the well-established methods to separate prokaryotic genera (Luo et al., 2014; Rodriguez-R and Konstantinidis, 2014). It is proposed to be 65% AAI value for genera delineation of Bacteria and Archaea (Konstantinidis et al., 2017). However, the category thresholds of AAI for genus delineation are variable in many genera. For example, the value of 70% AAI is used to separate the genus Geomonas from the other genera of the family Geobacteraceae (Xu et al., 2020), and a range of 64.6–77.0% AAI is used to delineate different genera of the family Geobacteraceae (Xu et al., 2019). In this study, the values of AAI among 52 type strains from 15 related genera of the family Rhodobacteraceae have been determined and are listed in Table 4 and Supplementary Table 2. It can be seen from Table 4 that a gradient of 63.5–75.3% and 74.2–98.7% AAI values is found among the different clade (genera) and in the same clade of the family Rhodobacteraceae. Those data are consistent to the results of phylogeny based on concatenated proteins.
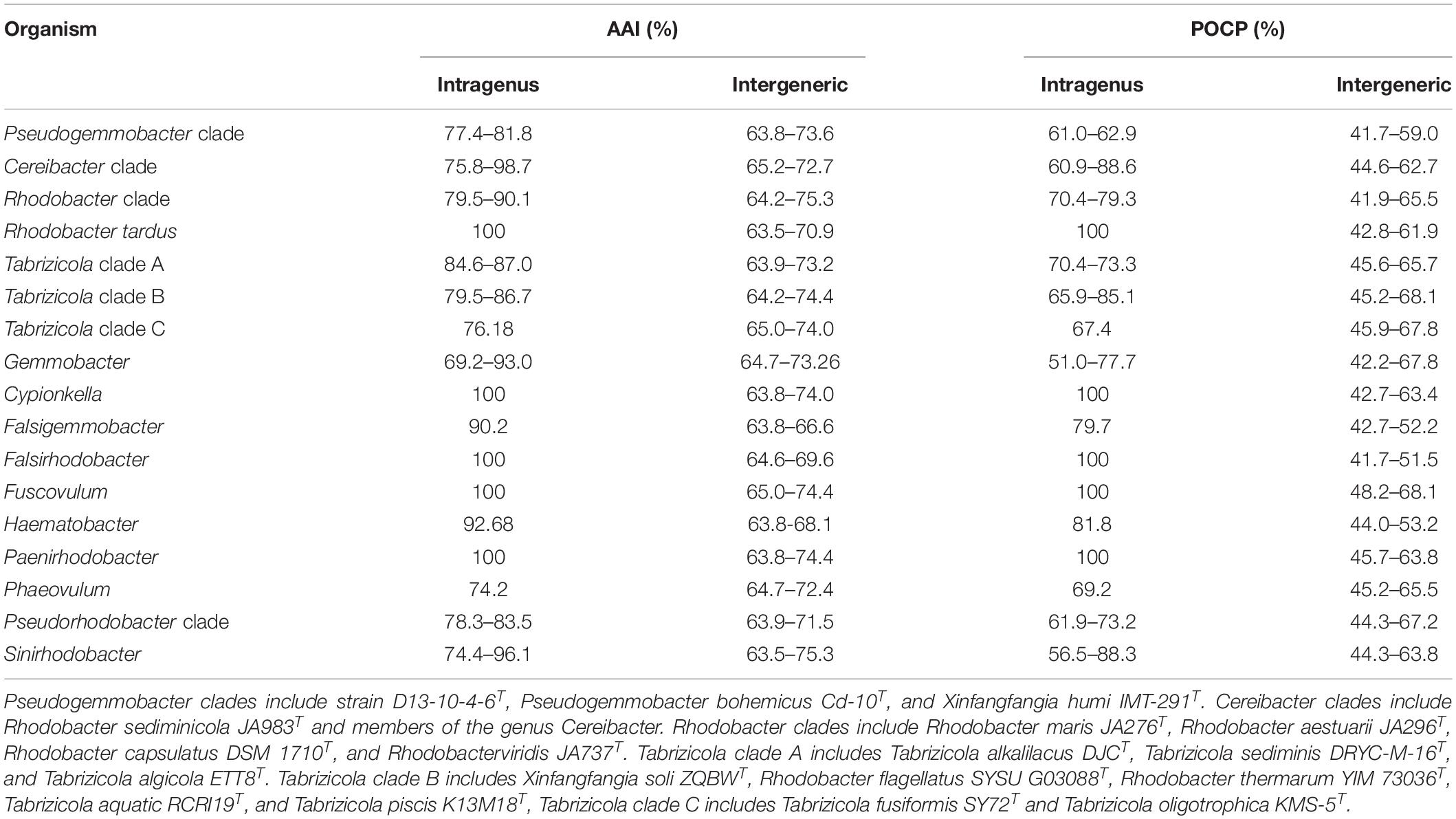
Table 4. Average amino acid identity (AAI) values and percentage of conserved protein (POCP) values of all strains for the intragenus and intergeneric comparisons.
The Pseudogemmobacter clade, including strains D13-10-4-6T, P. bohemicus Cd-10T and X. humi IMT-291T, has 77.4–81.8% AAI values among each other and shows 63.8–73.6% AAI values among the members of other clades in this study (Table 4 and Figure 4), which is consistent to the results of phylogeny based on the 16S rRNA gene, gyrB gene, and concatenated proteins (Figures 1–3). The Cereibacter clade, including R. sediminicola JA983T and members of the genus Cereibacter, has 75.8–98.7% AAI values among the members of the clade and 65.2–72.7% AAI values among the members from the other clades in this work, indicating that R. sediminicola should be transferred to the genus Cereibacter (Table 4 and Supplementary Figure 1). Similarly, the AAI values within the Tabrizicola clade A and Tabrizicola clade B can also distinguish them from the other strains (Table 4 and Supplementary Figures 2, 3).
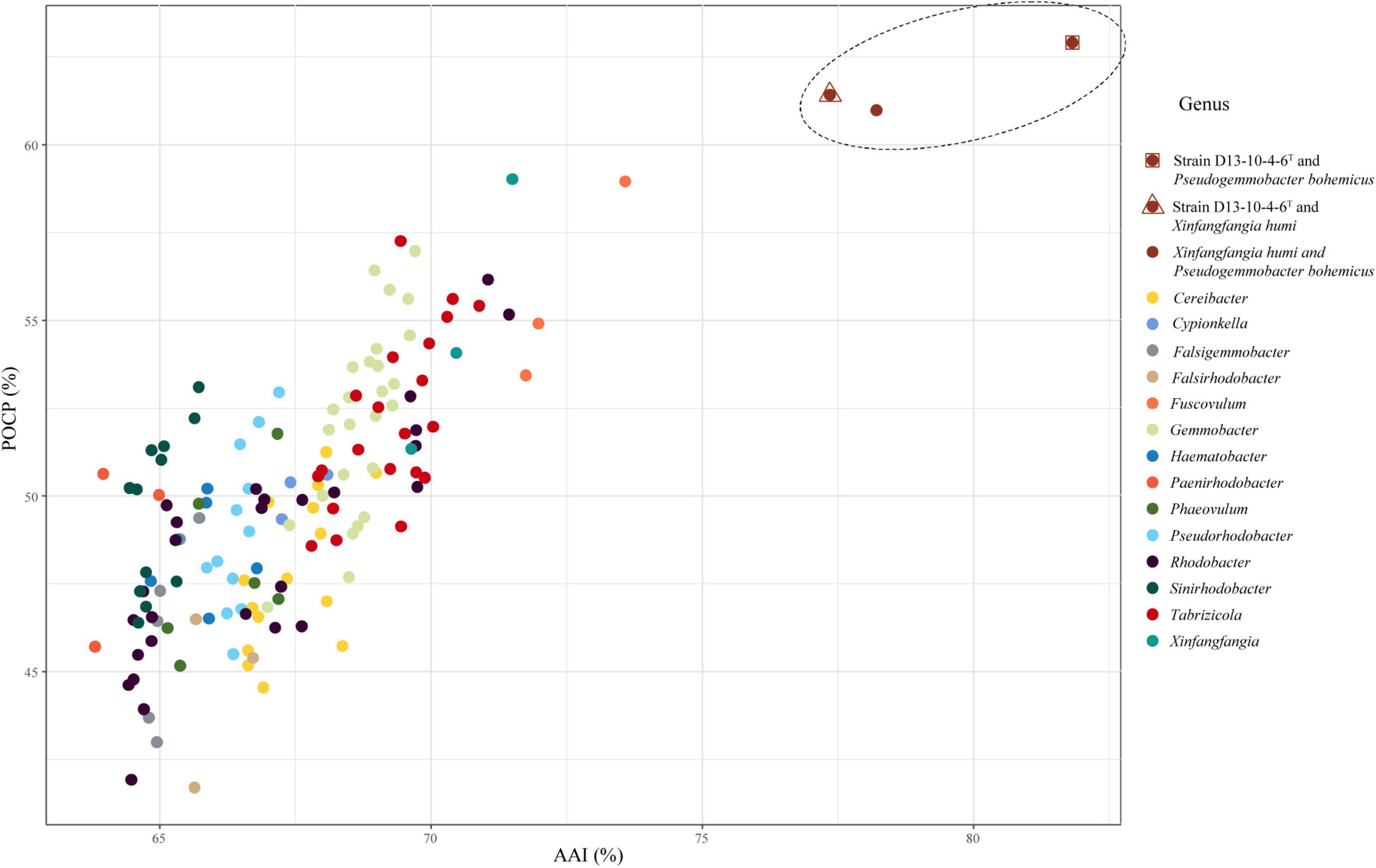
Figure 4. The relationship of AAI and POCP between strain D13-10-4-6T, P. bohemicus Cd-10T, X. humi IMT-291T and the related strains in the family Rhodobacteraceae. The dots inside the dashed line represent the values between strain D13-10-4-6T, P. bohemicus Cd-10T and X. humi IMT-291T, and those outside represent the values between the three strains and strains in the related genera of the family Rhodobacteraceae. A total of 52 genomes were included in this analysis.
Percentage of conserved protein is another method for genus delineation of prokaryote, and the value for genera delineation of POCP is proposed to be 50% (Qin et al., 2014). While the thresholds of POCP for genus delineation are also variable in many genera. The value of 65% POCP was used to separate the genus Geomonas from the other genus of the family Geobacteraceae (Xu et al., 2020), and most of the POCP values within the Roseobacter group comparisons were greater than 50% of the family Rhodobacteraceae (Wirth and Whitman, 2018). In this work, we also calculated the POCP values among the 52 type strains of 15 related genera of the family Rhodobacteraceae (Supplementary Table 2). The results show that a gradient of 41.7–68.1% POCP, except the values between, was found among the different clade (genera) of the family Rhodobacteraceae, and 56.5–88.6% among the species of the same clade. Therefore, it is hard to use the same thresholds for genus delineation because they show a broad range of values from both intragenus and intergeneric. But for several clades, it is useful to distinguish one group from the others, for instance, members of the Pseudogemmobacter clade show the values of POCP from 61.0 to 62.9% among each other and have 41.7–59% POCP values among members from the other clades (Table 4 and Figure 4). The same goes for the Cereibacter clade and Tabrizicola clade A (Table 4 and Supplementary Figures 1, 2).
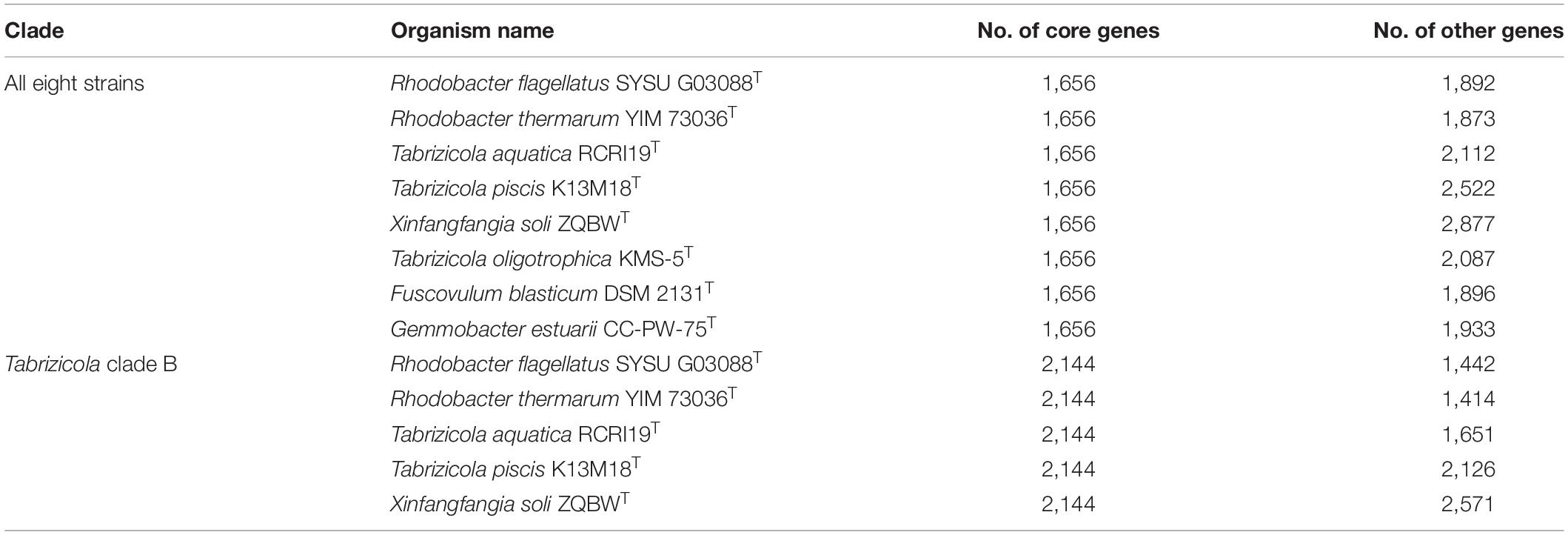
The POCP values in this study are not always consistent to the phylogenetic analysis, as exemplified by the Tabrizicola clade B. Members of the Tabrizicola clade B show 45.2–68.1% POCP values among each other, and 65.9–85.1% POCP values among the members of other clades, respectively (Table 4). The POCP values can distinguish Tabrizicola clade B from other related members except for T. oligotrophica KMS-5T, Fuscovulum blasticum DSM 2131T, and Gemmobacter aestuarii CC-PW-75T (Supplementary Figure 3). The POCP values between X. soli ZQBWT (belonging to Tabrizicola clade B) and T. oligotrophica KMS-5T, F. blasticum DSM 2131T, and G. aestuarii CC-PW-75T were slightly higher than those between X. soli ZQBWT and other members in Tabrizicola clade B (Supplementary Figure 3). Due to X. soli ZQBWT forming a stable clade within Tabrizicola clade B and forming a distinct clade with T. oligotrophica KMS-5T, F. blasticum DSM 2131T, and G. aestuarii CC-PW-75T, we analyzed the pan-genome of Tabrizicola clade B in a supplementary analysis.
The pan-genome analysis was used in the classification of bacteria (Awan et al., 2018; Suresh et al., 2019). The amount of core genes was sensitive to heterogeneous in the core- and pan-genome analysis (Inglin et al., 2018; Suresh et al., 2019). The core gene numbers within were Tabrizicola clade B were considerably higher than those within the Tabrizicola clade B and T. oligotrophica KMS-5T, F. blasticum DSM 2131T, and G. aestuarii CC-PW-75T (Table 5), indicating that the relationships of Tabrizicola clade B and the three species were heterogeneous and the Tabrizicola clade B should belong to a genus different from the three species. The pan-genome analysis reinforces the results of gyrB and concatenated protein phylogenetic trees. In conclusion, Tabrizicola clade B should belong to the same genus.
Conclusion
Phylogenetic trees based on the 16S rRNA gene, gyrB gene, and concatenated alignment of 120 ubiquitous single-copy proteins were constructed to clarify the relationship of the members from the 15 closely related genera within the family Rhodobacteraceae. The AAI, POCP, and ANI analyses as well as chemotaxonomic and physiological tests were also performed and used as supplementary evidence. On the basis of the data obtained, the taxonomic proposals were (1) reclassification of R. tardus as the type species of a novel genus, Stagnihabitans gen. nov., as Stagnihabitans tardus comb. nov.; (2) reclassification of T. alkalilacus, Tabrizicola sediminis, and Tabrizicola algicola into a novel genus, Pseudotabrizicola gen. nov., as Pseudotabrizicola alkalilacus comb. nov., Pseudotabrizicola sediminis comb. nov., Pseudotabrizicola algicola comb. nov.; (3) reclassification of Rhodobacter sediminicola into the genus Cereibacter as Cereibacter sediminicola comb. nov.; (4) reclassification of Rhodobacter flagellatus, Rhodobacter thermarum, and X. soli into the genus Tabrizicola as Tabrizicola flagellatus comb. nov., Tabrizicola thermarum comb. nov., and Tabrizicola soli comb. nov.; (5) reclassification of X. humi into the genus Pseudogemmobacter as Pseudogemmobacter humicola comb. nov.; and (6) classification of strain D13-10-4-6T as a novel species of the genus Pseudogemmobacter, for which the name Pseudogemmobacter hezensis sp. nov. is proposed; the type strain is D13-10-4-6T (= CFCC 12033T = KCTC 82215T).
Description of Pseudogemmobacter hezensis Sp. nov.
Pseudogemmobacter hezensis (he.zen’sis. N.L. masc./fem. adj. hezensis, of Heze, a city in Shandong Province, China, where the organism was isolated).
Cells are Gram-stain-negative, aerobic, non-motile, catalase- and oxidase-positive, ovoid to rod-shaped, 1.6–2.0 μm in length and 0.8–1.0 μm in width. Colonies are creamy white, circular, smooth, with entire margins after incubation for 2 days at 28°C on TSA. The strain can grow at 15–37°C (optimum, 25–30°C), at pH 6–10 (optimum, pH 7–8). Growth occurs at a concentration of 0–4% (w/v) NaCl. It is positive for the activity of alkaline phosphatase, esterase lipase (C8), esterase (C4), leucine arylamidase, valine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-glucosidase, N-acetyl-β-glucosaminidase; weakly positive for lipase (C14), β-galactosidase; negative for cystine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-glucuronidase, β-glucosidase, α-mannosidase, and α-fucosidase (API ZYM). It is negative for reduction of nitrate to nitrogen, reduction of nitrate to nitrite, indole production, gelatin hydrolysis, and the activity of urease, arginine dihydrolase; positive for the utilization of glucose, D-mannose, L-arabinose, D-mannitol, N-acetyl-glucosamine, malic acid (API 20 NE). Acid is produced from L-arabinose, D-xylose, D-galactose, D-fructose, L-rhamnose, D-lyxose, D-fucose, L-fucose; weakly positive for erythritol, D-arabinose, D-ribose, and L-sorbose (API 50 CH). The polar lipids are PME, DPG, PE, PG, PC, PL1, and six unidentified lipids (L). The respiratory quinones are Q-10 and Q-9. The predominant fatty acids are C18:1ω7c. The type strain is D13-10-4-6T (= CFCC 12033T = KCTC82215T), isolated from the bark samples of Populus × euramericana in Shandong Province, China. The DNA G + C content is 62.9%.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene and genome sequences for strain D13-10-4-6T is MT036106 and JABJXT000000000, respectively.
Description of Pseudogemmobacter humi Comb. nov.
Pseudogemmobacter humi (hu’mi. L. gen. fem. n. humi, of/from soil, the isolation source of the type strain).
Basonym: Xinfangfangia humi (Kämpfer et al., 2019).
The description of Pseudogemmobacter humi is the same as that given for X. humi by Kämpfer et al. (2019). The type strain is IMT-291T (= LMG 30636T = CIP 111625T = CCM 8858T).
Emended Description of the Genus Pseudogemmobacter
The description as given by Suman et al. (2019) remains correct except that the species are positive or negative for catalase and nitrate reductase.
Description of C. sediminicola Comb. nov.
Cereibacter sediminicola (se.di.mi.ni’co.la. L. neut. n. sedimen, sediment; L. masc./fem. suff. -cola, inhabitant, dweller; from L. masc./fem. n. incola, dweller; N.L. masc./fem. n. sediminicola, dweller of sediments).
Basonym: Rhodobacter sediminicola (Suresh et al., 2020).
The description of C. sediminicola is the same as that given for R. sediminicola by Suresh et al. (2020). The type strain is JA983T (= KCTC 15782T = NBRC 113843T).
Description of Stagnihabitans Gen. nov.
Stagnihabitans (Sta.gni.ha’bi.tans. L. neut. n. stagnum, a small area of water, pond; L. pres. part. Habitans, an inhabitant; N.L. masc. n. Stagnihabitans, an inhabitant of pond water).
Cells are Gram-strain-negative, aerobic, non-motile, oxidase-positive, catalase-negative, ovoid to rod-shaped and divide by binary fission, sometimes forming chains. The predominant respiratory quinone is Q-10. The major cellular fatty acid is C18:1ω7c. PE, PG, and PC are the major polar lipids. The DNA G + C content is 66%. The member of the genus is separated from Rhodobacter based on the 16S rRNA, gyrB and concatenated protein phylogenetic trees, genome comparison. The type species is S. tardus comb. nov.
Description of S. tardus Comb. nov.
Stagnihabitans tardus (tar’dus. L. masc.adj. tardus, slow, referring to the slow growth of the organism).
Basonym: Rhodobacter tardus (Sheu et al., 2020).
The description of S. tardus is the same as that given for R. tardus by Sheu et al. (2020). The type strain is CYK-10T (= BCRC 81191T = LMG 31336T).
Description of Pseudotabrizicola Gen. nov.
Pseudotabrizicola (Pseu.do.ta.bri.zi.co.la. Gr. masc./fem. adj. pseudês, false; N.L. fem. n. Tabrizicola, a bacterial generic name; N.L. fem. n. Pseudotabrizicola, false Tabrizicola).
Cells are Gram-strain-negative, aerobic, non-motile, catalase- and oxidase-positive, rod-shaped. PG, DPG, PE, and PC are the major polar lipids. The predominant respiratory quinone is Q-10. The major cellular fatty acids are usually iso-C18:0, C18:1 ω7c, and/or C18:1 ω6c. The DNA G + C content is 62.9–64.4%. Members of the genus are separated from Tabrizicola based on the 16S rRNA, gyrB, and concatenated proteins phylogenetic trees, genome comparison. The type species is P. sediminis comb. nov.
Description of P. sediminis Comb. nov.
Pseudotabrizicola sediminis (se.di’mi.nis. L. gen. net. n. sediminis, of a sediment).
Basonym: Tabrizicola sediminis (Liu et al., 2019).
The description of P. sediminis is the same as that given for Tabrizicola sediminis by Liu et al. (2019). The type strain is DRYC-M-16T (= CGMCC 1.13881T = KCTC 72105T).
Description of P. alkalilacus Comb. nov.
Pseudotabrizicola alkalilacus (al.ka.li.la’cus. N.L. neut. n. alkali from Arabic article al, the; Arabic n. qaliy, ashes of saltwort, alkali; L. masc. n. lacus, a lake; N.L. gen. masc. n. alkalilacus of analkaline lake).
Basonym: Tabrizicola alkalilacus (Phurbu et al., 2019).
The description of Pseudotabrizicola alkalilacus is the same as that given for T. alkalilacus by Phurbu et al. (2019). The type strain is DJCT (= CICC 24242T = KCTC 62173T).
Description of P. algicola Comb. nov.
Pseudotabrizicola algicola (al.gi’co.la. L. fem. n. algae, an alga; L. masc./fem. suff.-cola, dweller; from L. masc./fem. n. incola an inhabitant; N.L. masc./fem. n. algicola an inhabitant of algae).
Basonym: Tabrizicola algicola (Park et al., 2020).
The description of P. algicola is the same as that given for Tabrizicola algicola by Park et al. (2020). The type strain is ETT8T (= KCTC 72206T = JCM 31893T = MCC 4339T).
Description of T. flagellatus Comb. nov.
Tabrizicola flagellatus (fla.gel.la’tus. L. masc. part. adj. flagellatus, flagellated).
Basonym: Rhodobacter flagellatus (Xian et al., 2020).
The description of T. flagellatus is the same as that given for Rhodobacter flagellatus by Xian et al. (2020). The type strain is SYSU G03088T (= CGMCC 1.16876T = KCTC 72354T).
Description of T. thermarum Comb. nov.
Tabrizicola thermarum (ther.ma’rum. L. gen. fem. pl. n. thermarum, of hot springs).
Basonym: Rhodobacter thermarum (Khan et al., 2019).
The description of T. thermarum is the same as that given for Rhodobacter thermarum by Khan et al. (2019). The type strain is YIM 73036T (= KCTC 52712T = CCTCC AB 2016298T).
Description of T. soli Comb. nov.
Tabrizicola soli (so’li. L. neut. n. soli of soil, the source of the type strain).
Basonym: Xinfangfangia soli (Hu et al., 2018).
The description of T. soli is the same as that given for X. soli by Hu et al. (2018). The type strain is ZQBWT (= KCTC 62102T = CCTCC AB 2017177T).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories andaccession number(s) can be found below: NCBI—MT036106, JABJXT000000000, JAEACP000000000.
Author Contributions
CP and YL designed the experiments, provided the methods, and revised the manuscript. TM finished the manuscript and completed most of the experiments. HX helped to reconstructed and analyzed the gene trees. DB finished the fatty acid profiles. CL and MY collected the samples. All authors read and approved the final version of the manuscript.
Funding
This research was supported by the National Infrastructure of Microbial Resources (NMRC-2021-7) from the Ministry of Science and Technology of the People’s Republic of China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Yan Liu, Fengjuan, Wang, and Wenjieng Li (China Forestry Culture Collection Center) for the material preparation and strain preservation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.849695/full#supplementary-material
Footnotes
- ^ https://lpsn.dsmz.de/family/rhodobacteraceae
- ^ https://lpsn.dsmz.de/genus/rhodobacter
- ^ https://www.ezbiocloud.net/identify
- ^ https://github.com/Ecogenomics/GtdbTk
- ^ http://ggdc.dsmz.de
- ^ https://github.com/dparks1134/CompareM
- ^ https://github.com/2015qyliang/POCP
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Awan, F., Dong, Y., Liu, J., Wang, N., Mushtaq, M. H., Lu, C., et al. (2018). Comparative genome analysis provides deep insights into Aeromonas hydrophila taxonomy and virulence-related factors. BMC genomics 19:712. doi: 10.1186/s12864-018-5100-4
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Brinkhoff, T., Giebel, H.-A., and Simon, M. (2008). Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch. Microbiol. 189, 531–539. doi: 10.1007/s00203-008-0353y
Buchan, A., González, J. M., and Moran, M. A. (2005). Overview of the marine Roseobacter lineage. Appl. Environ. Microb. 71, 5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6, 1–10. doi: 10.1038/srep24373
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., and Parks, D. (2020). GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Ciccarelli, F. D., Doerks, T., Von Mering, C., Creevey, C. J., Snel, B., and Bork, P. (2006). Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283–1287. doi: 10.1126/science.1123061
Collins, M. D., Pirouz, T., Goodfellow, M., and Minnikin, D. E. (1977). Distribution of menaquinones in Actinomycetes and corynebacteria. Microbiology 100, 221–230. doi: 10.1099/00221287-100-2-221
Delory, G. E., and King, E. J. (1945). A sodium carbonate-bicarbonate buffer for alkaline phosphatases. Biochem. J. 39, 245. doi: 10.1146/annurev.bi.14.070145.003533
Du, H. J., Zhang, Y. Q., Liu, H. Y., Su, J., Wei, Y. Z., Ma, B. P., et al. (2013). Allonocardiopsis opalescens gen. nov., sp. nov., a new member of the suborder Streptosporangineae, from the surface-sterilized fruit of a medicinal plant. Int. J. Syst. Evol. Microbiol. 63, 900–904. doi: 10.1099/ijs.0.041491-0
Garrity, G., Bell, J., and Lilburn, T. (2005). “Family I. rhodobacteraceae fam. nov,” in Bergey’s Manual of Systematic Bacteriology, 2nd Edn, Vol. 2 (The Proteobacteria), part C (The Alpha-, Beta-, Delta-, and Epsilonproteobacteria), eds N. R. Krieg, J. T. Staley, and G. M. Garrity (New York:NY: Springer), 161.
Gomori, G. (1955). Preparation of buffers for use in enzyme studies. Method. Enzymol. 1, 138–146. doi: 10.1016/0076-6879(55)01020-3
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Groth, I., Schumann, P., Rainey, F., Martin, K., Schuetze, B., and Augsten, K. (1997). Demetria terragena gen. nov., sp. nov., a new genus of actinomycetes isolated from compost soil. Int. J. Syst. Evol. Microbiol. 47, 1129–1133. doi: 10.1099/00207713-47-4-1129
Hördt, A., López, M. G., Meier-Kolthoff, J. P., Schleuning, M., Weinhold, L.-M., Tindall, B. J., et al. (2020). Analysis of 1,000+ type-strain genomes substantially improves taxonomic classification of alphaproteobacteria. Front. Microbiol. 11:468. doi: 10.3389/fmicb.2020.00468
Hu, Q., Zhang, L., Hang, P., Zhou, X.-Y., Jia, W.-B., Li, S.-P., et al. (2018). Xinfangfangia soli gen. nov., sp. nov., isolated from a diuron-polluted soil. Int. J. Syst. Evol. Microbiol. 68, 2622–2626. doi: 10.1099/ijsem.0.002887
Imhoff, J. F., Petri, R., and Süling, J. (1998). Reclassification of species of the spiral-shaped phototrophic purple non-sulfur bacteria of the α-Proteobacteria: description of the new genera Phaeospirillum gen. nov. nov. Rhodospirillum mediosalinum to Roseospira mediosalina comb. nov. Int. J. Syst. Evol. Microbiol. 48, 793–798. doi: 10.1099/00207713-48-3-793
Imhoff, J. F., Trüper, H. G., and Pfennig, N. (1984). Rearrangement of the species and genera of the phototrophic “purple nonsulfur bacteria”. Int. J. Syst. Evol. Mcrobiol. 34, 340–343. doi: 10.1099/00207713-34-3-340
Inglin, R. C., Meile, L., and Stevens, M. J. (2018). Clustering of pan-and core-genome of Lactobacillus provides novel evolutionary insights for differentiation. BMC genomics 19:284. doi: 10.1186/s12864-018-4601-5
Jenkins, D., Richard, M. G., and Daigger, G. T. (2003). “Methods,” in Manual on the Causes and Control Of Activated Sludge Bulking, Foaming, and Other Solids Separation Problems, Chap. 2, eds D. Jenkins, M. G. Richard, and G. T. Daigger (Boca Raton, FL: CRC Press).
Kämpfer, P., Busse, H.-J., McInroy, J. A., Criscuolo, A., Clermont, D., and Glaeser, S. P. (2019). Xinfangfangia humi sp. nov., isolated from soil amended with humic acid. Int. J. Syst. Evol. Microbiol. 69, 2070–2075. doi: 10.1099/ijsem.0.003434
Khan, I. U., Habib, N., Xiao, M., Li, M.-M., Xian, W.-D., Hejazi, M. S., et al. (2019). Rhodobacter thermarum sp. nov., a novel phototrophic bacterium isolated from sediment of a hot spring. Antonie van Leeuwenhoek 112, 867–875. doi: 10.1007/s10482-018-01219-7
Konstantinidis, K. T., Rosselló-Móra, R., and Amann, R. (2017). Uncultivated microbes in need of their own taxonomy. ISME J. 11, 2399–2406. doi: 10.1038/ismej.2017.113
Konstantinidis, K. T., and Tiedje, J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 2567–2572. doi: 10.1073/pnas.0409727102
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kuykendall, L. D., Roy, M. A., O’neill, J. J., and Devine, T. E. (1988). Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int. J. Syst. Evol. Microbiol. 38, 358–361. doi: 10.1099/00207713-38-4-358
Lane, D. J. (1991). “16S/23S RRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (Chichester, UK: Wiley), 115–175.
Lee, I., Kim, Y. O., Park, S.-C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, R., Li, Y., Kristiansen, K., and Wang, J. (2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714. doi: 10.1016/S0953-5438(03)00036-5
Li, R., Zhu, H., Ruan, J., Qian, W., Fang, X., Shi, Z., et al. (2010). De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272. doi: 10.1101/gr.097261.109
Li, Y., Fang, W., Xue, H., Liang, W.-X., Wang, L.-F., Tian, G.-Z., et al. (2015). Brenneria populi sp. nov., isolated from symptomatic bark of Populus× euramericana canker. Int. J. Syst. Evol. Microbiol. 65, 432–437. doi: 10.1099/ijs.0.066068-0
Li, Y., Song, L. M., Guo, M. W., Wang, L. F., and Liang, W. X. (2016). Sphingobacterium populi sp. nov., isolated from bark of Populus× euramericana. Int. J. Syst. Evol. Microbiol. 66, 3456–3462. doi: 10.1099/ijsem.0.001217
Liang, K. Y., Orata, F. D., Boucher, Y. F., and Case, R. J. (2021). Roseobacters in a sea of poly-and paraphyly: whole genome-based taxonomy of the family Rhodobacteraceae and the proposal for the split of the “Roseobacter clade” into a novel family. Roseobacteraceae fam. nov. Front. Microbiol. 12:683109. doi: 10.3389/fmicb.2021.683109
Lin, S.-H., and Liao, Y.-C. (2013). CISA: contig integrator for sequence assembly of bacterial genomes. PLoS One 8:e60843. doi: 10.1371/journal.pone.0060843
Liu, Z.-X., Dorji, P., Liu, H.-C., Li, A.-H., and Zhou, Y.-G. (2019). Tabrizicola sediminis sp. nov., one aerobic anoxygenic photoheterotrophic bacteria from sediment of saline lake. Int. J. Syst. Evol. Microbiol. 69, 2565–2570. doi: 10.1099/ijsem.0.003542
Luo, C., Rodriguez-r, L. M., and Konstantinidis, K. T. (2014). MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 42:73. doi: 10.1093/nar/gku169
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Minnikin, D. E., O’donnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Meth. 2, 233–241. doi: 10.1016/0167-7012(84)90018-6
Orata, F. D., Meier-Kolthoff, J. P., Sauvageau, D., and Stein, L. Y. (2018). Phylogenomic analysis of the gammaproteobacterial methanotrophs (order Methylococcales) calls for the reclassification of members at the genus and species levels. Front. Microbiol. 9:3162. doi: 10.3389/fmicb.2018.03162
Park, C.-Y., Chun, S.-J., Jin, C., Le, V. V., Cui, Y., Kim, S.-Y., et al. (2020). Tabrizicola algicola sp. nov. isolated from culture of microalga ettlia sp. Int. J. Syst. Evol. Microbiol. 70, 6133–6141. doi: 10.1099/ijsem.0.004508
Phurbu, D., Wang, H., Tang, Q., Lu, H., Zhu, H., Jiang, S., et al. (2019). Tabrizicola alkalilacus sp. nov., isolated from alkaline Lake Dajiaco on the Tibetan Plateau. Int. J. Syst. Evol. Microbiol. 69, 3420–3425. doi: 10.1099/ijsem.0.003635
Qin, Q.-L., Xie, B.-B., Zhang, X.-Y., Chen, X.-L., Zhou, B.-C., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. bacteriol. 196, 2210–2215. doi: 10.1128/JB.01688-14
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Rodriguez-R, L. M., and Konstantinidis, K. T. (2014). Bypassing cultivation to identify bacterial species. Microbe 9, 111–118. doi: 10.1128/microbe.9.111.1
Sasser, M. (1990). Identification of Bacteria By Gas Chromatography Of Cellular Fatty Acids. Newark:DE: MIDI Inc.
Selje, N., Simon, M., and Brinkhoff, T. (2004). A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427, 445–448. doi: 10.1038/nature02272
Sheu, C., Li, Z.-H., Sheu, S.-Y., Yang, C.-C., and Chen, W.-M. (2020). Tabrizicola oligotrophica sp. nov. and Rhodobacter tardus sp. nov., two new species of bacteria belonging to the family rhodobacteraceae. Int. J. Syst. Evol. Microbiol. 70, 6266–6283. doi: 10.1099/ijsem.0.004526
Simpson, J. T., Wong, K., Jackman, S. D., Schein, J. E., Jones, S. J., and Birol, I. (2009). ABySS: a parallel assembler for short read sequence data. Genome Res. 19, 1117–1123. doi: 10.1101/gr.089532.108
Smibert, R.M and Krieg NR (1994). “Phenotypic characterization,” in Methods for General and Molecular Bacteriology, eds P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (Washington:US: American Society for Microbiology).
Suman, J., Zubrova, A., Rojikova, K., Pechar, R., Svec, P., Cajthaml, T., et al. (2019). Pseudogemmobacter bohemicus gen. nov., sp. nov., a novel taxon from the Rhodobacteraceae family isolated from heavy-metal-contaminated sludge. Int. J. Syst. Evol. Microbiol. 69, 2401–2407. doi: 10.1099/ijsem.0.003493
Suresh, G., Kumar, D., Krishnaiah, A., and Ch, S. (2020). Rhodobacter sediminicola sp. nov., isolated from a fresh water pond. Int. J. Syst. Evol. Microbiol. 70, 1294–1299. doi: 10.1099/ijsem.0.003913
Suresh, G., Lodha, T. D., Indu, B., Sasikala, C., and Ramana, C. V. (2019). Taxogenomics resolves conflict in the genus Rhodobacter: a two and half decades pending thought to reclassify the genus Rhodobacter. Front. Microbiol. 10:2480. doi: 10.3389/fmicb.2019.02480
Suresh, G., Sasikala, C., and Ramana, C. V. (2015). Reclassification of Gemmobacter changlensis to a new genus as Cereibacter changlensis gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 65, 794–798. doi: 10.1099/ijs.0.000016
Thiergart, T., Landan, G., and Martin, W. F. (2014). Concatenated alignments and the case of the disappearing tree. BMC Evol. Biol. 14:266. doi: 10.1186/s12862-014-0266-0
Wirth, J. S., and Whitman, W. B. (2018). Phylogenomic analyses of a clade within the roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter, and the proposal of six novel genera. Int. J. Syst. Evol. Microbiol. 68, 2393–2411. doi: 10.1099/ijsem.0.002833
Xian, W.-D., Liu, Z.-T., Li, M.-M., Liu, L., Ming, Y.-Z., Xiao, M., et al. (2020). Rhodobacter flagellatus sp. nov., a thermophilic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 70, 1541–1546. doi: 10.1099/ijsem.0.003929
Xu, Z., Masuda, Y., Hayakawa, C., Ushijima, N., Kawano, K., Shiratori, Y., et al. (2020). Description of Three novel members in the family Geobacteraceae. Microorganisms 8, 634. doi: 10.3390/microorganisms8050634
Xu, Z., Masuda, Y., Itoh, H., Ushijima, N., Shiratori, Y., and Senoo, K. (2019). Geomonas oryzae gen. nov., sp. nov. Front. Microbiol. 10:2201. doi: 10.3389/fmicb.2019.02201
Xu, Z., Masuda, Y., Wang, X., Ushijima, N., Shiratori, Y., Senoo, K., et al. (2021). Genome-based taxonomic rearrangement of the rrder Geobacterales including the description of Geomonas azotofigens sp. nov. and Geomonas diazotrophica sp. nov. Front. Microbiol. 12:737531. doi: 10.3389/fmicb.2021.737531
Keywords: Xinfangfangia, cereibacter, Rhodobacter, Tabrizicola, Pseudotabrizicola, GTDB-Tk, average amino acid identity
Citation: Ma T, Xue H, Piao C, Liu C, Yang M, Bian D and Li Y (2022) Reclassification of 11 Members of the Family Rhodobacteraceae at Genus and Species Levels and Proposal of Pseudogemmobacter hezensis sp. nov. Front. Microbiol. 13:849695. doi: 10.3389/fmicb.2022.849695
Received: 06 January 2022; Accepted: 04 March 2022;
Published: 13 April 2022.
Edited by:
Baokai Cui, Beijing Forestry University, ChinaReviewed by:
Stefan Spring, German Collection of Microorganisms and Cell Cultures GmbH (DSMZ), GermanyQiliang Lai, Third Institute of Oceanography, State Oceanic Administration, China
Copyright © 2022 Ma, Xue, Piao, Liu, Yang, Bian and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Li, bHlseDc4QGhvdG1haWwuY29t
 Tengfei Ma
Tengfei Ma Han Xue1
Han Xue1 Danran Bian
Danran Bian Yong Li
Yong Li