- 1Institute for Physiology and Cell Biology, University of Veterinary Medicine, Hannover, Germany
- 2Department of Molecular Systems Biology, Helmholtz Centre for Environmental Research (UFZ), Leipzig, Germany
- 3Institute for Biometry, Epidemiology and Information Processing, University of Veterinary Medicine, Hannover, Germany
- 4Institute of Animal Nutrition, Friedrich-Loeffler-Institut (FLI), Federal Research Institute for Animal Health, Brunswick, Germany
- 5German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, Leipzig, Germany
- 6Faculty of Life Sciences, Institute of Biochemistry, University of Leipzig, Leipzig, Germany
The use of the herbicide glyphosate and its formulations on protein-rich feedstuff for cattle leads to a considerable intake of glyphosate into the rumen of the animals, where glyphosate may potentially impair the 5-enolpyruvylshikimate-3-phosphate pathway of the commensal microbiota, which could cause dysbiosis or proliferation of pathogenic microorganisms. Here, we evaluated the effects of pure glyphosate and the formulations Durano TF and Roundup® LB plus in different concentrations on the fermentation pattern, community composition and metabolic activity of the rumen microbiota using the Rumen Simulation Technique (RUSITEC). Application of the compounds in three concentrations (0.1 mg/l, 1.0 mg/l or 10 mg/l, n = 4 each) for 9 days did not affect fermentation parameters such as pH, redox potential, NH3-N concentration and production of short-chain fatty acids compared to a control group. Microbial protein synthesis and the degradation of different feed fractions did not vary among the treatments. None of the used compounds or concentrations did affect the microbial diversity or abundance of microbial taxa. Metaproteomics revealed that the present metabolic pathways including the shikimate pathway were not affected by addition of glyphosate, Durano TF or Roundup® LB plus. In conclusion, neither pure glyphosate, nor its formulations Durano TF and Roundup® LB plus did affect the bacterial communities of the rumen.
Introduction
Glyphosate is an herbicide acting by inhibition of the 5-enolpyruvylshikimate-3-phosphate pathway (Amrhein et al., 1980a; Boocock and Coggins, 1983). Glyphosate-containing herbicides are among the most-used herbicides worldwide. Glyphosate use in the United States has increased 300-fold in 30 years since its market introduction. Due to the development of glyphosate-resistant crops (Benbrook, 2016), the use of this herbicide is especially popular in soybean and maize production. In Europe, the application of glyphosate is being intensely discussed in regard to potential detrimental environmental or health effects, though it still accounts for one-third of the herbicide volume in 2017 (Antier et al., 2020). The mode of action of Glyphosate is that it directly inhibits 5-enolpyruvylshikimate-3-phosphate synthase and therefore hinders aromatic amino acid synthesis (Amrhein et al., 1980b). Although the 5-enolpyruvylshikimate-3-phosphate pathway is not present in human and animal cells, the microbial community of the gastrointestinal tract of animals and humans also contains glyphosate-sensitive members (Motta et al., 2018; Leino et al., 2021; Mesnage et al., 2021). In honeybees, for example glyphosate decreased the abundance of Snodgrasella alvi favouring infection with Serratia marescens but not with Nosema ceranae (Motta et al., 2018; Blot et al., 2019).
Ruminants livestock are fed protein-rich feed components such as rapes and soybeans which are highly exposed to glyphosate (Benbrook, 2016; Antier et al., 2020). In ruminants, the microbial community of the forestomach is vital in the degradation of plant-derived carbohydrates to short-chain fatty acids (SCFA), a major energy source for the host, as well as providing the host with microbially synthesised protein and water soluble vitamins. Glyphosate could potentially affect and disrupt the microbial community in the forestomach, and therefore might lead to an adverse health outcome for the host. Earlier, several reports on an association between glyphosate and so-called ‘visceral botulism’ in cows (Krüger et al., 2013; Ackermann et al., 2015) have been published.
In vivo, von Soosten et al. (2016) measured a glyphosate intake between 0.08 mg/d and 6.67 mg/d in dairy cows under conventional feeding conditions, of which 61% were excreted by faeces, but 6–36% disappeared during rumen passage indicating a potential interaction or degradation by rumen microorganisms. In sheep, Hüther et al. (2005) did not observe effects of feeding 0.77 g/kg of dry matter glyphosate on rumen fermentation parameters, such as pH or SCFA concentration, or in sacco degradation of feedstuff. Using the Rumen simulation technique (RUSITEC) as in vitro model for rumen fermentation, Riede et al. (2016) reported a decrease in NH3-N concentrations and a trend towards increasing acetate production; however, glyphosate application (3.31 mg/l) did not affect the composition of the Clostridia population. High glyphosate levels (10 mg/l) did also not affect establishment of Escherichia coli or Salmonella Ser. Typhimurium in the RUSITEC system (Bote et al., 2019). Schnabel et al. (2017) did not observe adverse effects of a glyphosate-contaminated diet (intake up to 84.5 mg/d) on performance or energy balance of dairy cows, neither did this affect leucocytes or erythrocytes (Schnabel et al., 2020) or the rumen microbiome (Billenkamp et al., 2021).
Commercial herbicides contain a mixture of glyphosate and adjuvants, e.g., polyethoxylene amine (POEA), which are assumed to enhance toxicity of glyphosate or exhibit additional effects themselves (Defarge et al., 2018; Mesnage et al., 2019). Different formulations vary regarding their effects on the gut community and their metabolome profile in rats (Mesnage et al., 2021). To our knowledge, no data comparative on the effects of different glyphosate formulations on the rumen microbiome are available and this might account for differences in the results of previous studies. In this study, we compare the effects of pure glyphosate and two widely used commercial formulations on the rumen biochemical functions as well as rumen microbiome taxonomic composition and functionality at concentrations comparable to and above the observed in vivo levels reported by von Soosten et al. (2016).
Materials and Methods
Animals
Rumen contents were obtained from four ruminal fistulated German Holstein cattle housed at the Institute for Animal Nutrition, Federal Research Institute for Animal Nutrition, Brunswick, Germany. Fistulation of the animals was approved by the Lower Saxony State Office for Costumer Protection and Food Safety, Oldenburg, Germany (33.9-42502-04-12/0876). All procedures involving animals were carried out in accordance with the German Animal Welfare Act.
RUSITEC Experiments
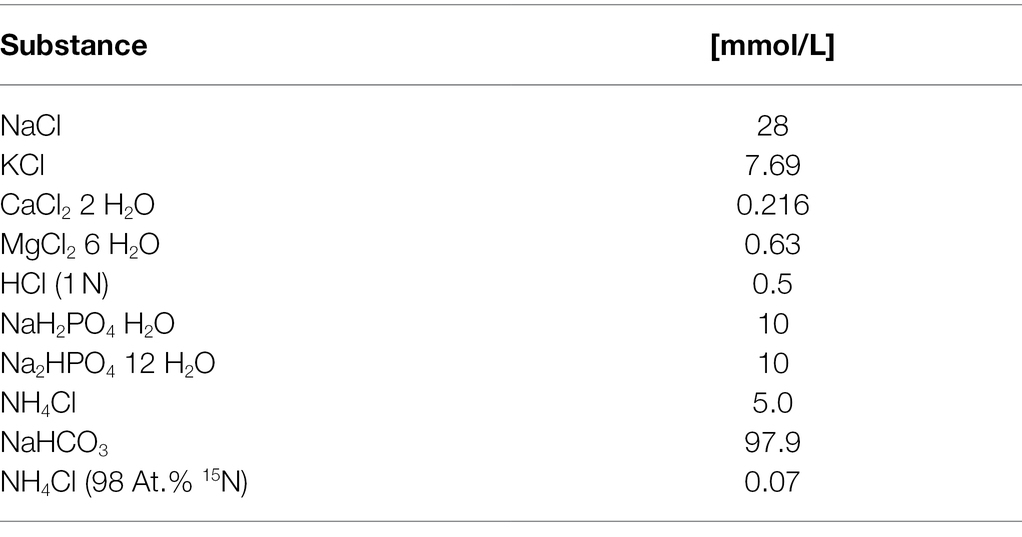
Four RUSITEC experiments were carried out using two RUSITEC systems with in total 12 fermenters. Inoculation of the fermentation vessels was carried out as described previously (Eger et al., 2018), with the modification that for each experiment rumen contents of one of the four cow were used. Briefly, rumen ingesta were squeezed through two layers of medical gauze (Gazin®, 80 cm × 5 cm, Lohmann & Rauscher International GmbH Co.KG, Rengsdorf, Germany) to separate the liquid and solid phase. Rumen contents were transported in preheated insulated containers to avoid heat losses. For each fermenter, a nylon bag (R712 Forage Bags in situ, ANKOM Technology, Gesellschaft für Analysetechnik HLS, Salzwedel, Germany) was filled with 70 g of solid rumen ingesta and placed in the inner fermentation vessel. A second nylon bag containing the daily feed was added. Feed bags contained 15 g of substrate. Components and chemical contents of the daily substrate are presented in Table 1. All components were shown to be free from glyphosate and aminomethylphosphonic acid (AMPA) before use (< 0.01 mg/kg, Wessling GmbH, Hannover). In each experiment, three fermentation vessels served as control, and one vessel each was assigned to one of the following nine treatments: 0.1 mg/l, 1.0 mg/l or 10 mg/l of pure glyphosate (40% N-(Phosphonomethyl) glycine, monoisopropylamine salt solution, Sigma-Aldrich, Darmstadt, Germany), 0.1 mg/l, 1.0 mg/l or 10 mg/l of glyphosate as Durano TF (052389-83/MOT, Bayer Crop Science, Langenfeld, Germany), or 0.1 mg/l, 1.0 mg/l or 10 mg/l of glyphosate as Roundup® LB plus (024142-60/MOT, Nordland Agrar, Süderlügüm, Germany). The first two concentrations were chosen to meet the concentrations of 0.08 mg/d to 6.78 mg/d reported for in vivo uptake of glyphosate by von Soosten et al. (2016), the highest concentration exceeds in vivo uptake levels by 10-fold to provide a safety margin. The compounds were dissolved in methanol. The experiments consisted of an equilibration period of 6 days, a control day (day 7) and the experimental period from day 8 to day 16, during which, on each day either the respective glyphosate treatment was added to the fermentation vessels or the same amount of methanol (control vessels). A physiological buffer solution was infused into the fermenters to achieve a liquid turnover of once per day (Table 2). In order to measure microbial protein synthesis, 15N was included in the buffer solution (Table 2).
Measurements
The pH and redox potential were measured daily throughout the experiment (Digital-pH-Meter 646, Knick GmbH & Co. KG, Berlin, Germany, electrodes: InLab® Routine, InLab® Redox Pro, Mettler-Toledo GmbH, Gießen, Germany). At control day and during the experimental period, fluid flow-through of the fermenters was recorded daily and samples for the analysis of NH3-N and short-chain fatty acids (SCFA) were collected daily from the overflow. Short-chain fatty acid concentrations were measured by gas chromatography as described by Koch et al. (2006). The concentration of NH3-N was determined by photometry as described by Riede et al. (2013). From day 10 to day 13, feed bags were collected for the analysis of nutrient degradation. To determine the degradation of organic matter (OM), crude fat (XL), crude protein (XP), crude fibre (XF), neutral detergent fibre (NDF) and acid detergent fibre (ADF) feed residues from the 4 days were pooled, analysed by Weender analysis (Chair of Animal Nutrition, Technical University of Munich, Munich, Germany) and compared to undigested substrate.
Microbial Protein Synthesis
From day 10 to day 16, 20 ml of fermenter fluid was collected daily for the separation of liquid-associated microorganism (LAM) and frozen at −20°C. Solid-associated microorganism (SAM) was collected from the feed bags at days 14 and 15 by methyl cellulose incubation (Boguhn et al., 2013) and also frozen until further treatment. Liquid samples and the methyl cellulose samples were then thawed and subjected to differential centrifugation (Brandt and Rohr, 1981). First samples were centrifuged at 2,000 × g for 5 min. The supernatant was again frozen at −20°C and the pellet was discarded. The supernatant was thawed in a fridge, centrifuged at 2,000 × g and 5 min again and the supernatant was collected in a new tube. The samples were then centrifuged at 27,500 × g for 15 min. The supernatants of LAM samples, which represented the fermenter liquid, were stored for later 15N-analysis, while the pellets representing LAMs or SAMs were washed with 0.9% NaCl three times. The remaining pellets were weighted and frozen at −20°C. For the analysis, LAM and SAM pellets and fermenter liquid were freeze-dried and grinded with a glass stick. The amount of 15N was analysed using a mass spectrometer (TracerMAT, Thermo Fisher Scientific GmbH, Dreieich Germany) after incineration in an elementary analyser (EA1108, Fisons Instruments GmbH, Mainz-Kastel, Germany).
N assimilation in LAMs was calculated using the following formula:
Where 15Ninput is input by the buffer solution; 15Noutput is the content in the fermenter liquid; and 15Nplateau is the 15N percentage in the reference microbes.
The N assimilation in SAM was calculated using the following formula:
Where 15NFR is the measured 15N content in the feed residues; 15Nnat is the natural content of 0.3663%; NFR is the N content of the feed residues; and 15NSAM is the measured 15N content in the reference microbes.
Microbial protein synthesis was calculated by multiplying N content by 6.25 (Boguhn et al., 2006). The efficiency of microbial protein synthesis was calculated by dividing totally synthesised protein by fermented organic matter.
Statistical Analysis of Fermentation Data
Statistical analysis was performed using the Statistical Analysis System for Windows SAS, version 9.4 by using the SAS Enterprise Guide version 7.1 (SAS Institute Inc., Cary, NC, United States). A three-factorial analysis of variance was applied by using the procedure mixed to determine effects of ‘treatment’ (=glyphosate formulation), ‘concentration’ and ‘day’ for parameters measured daily. Treatment and concentration were set as independent factors. Statistical analysis applies for days 7 to 16; however, only days 7 and 16 (before application and end of application) are displayed in graphs and tables to improve readability. For parameters obtained once per treatment or from pooled samples, only effects of ‘treatment’ and ‘concentration’ were analysed by two-factorial analysis of variance. The levels of significance were set at *p < 0.05, **p < 0.01 and ***p < 0.001. Data are presented as means ± SD.
Microbiome DNA and Protein Extraction
For microbiome and metaproteome analysis, 10 ml of fermenter liquid was collected at days 10–16 and SAM was collected at days 14 and 15. Bacteria pellets were thawed and lysed, followed by protein and DNA extraction as previously described (Haange et al., 2020). Briefly, samples were resuspended in 1 ml lysis-buffer (50 mm Tris, 5 mm EDTA, 0.4% SDS, 50 mm NaCl and 1 mm PMSF, pH = 8). This was followed by lysis using a Fastprep 24 (MP Biomedicals, Santa Ana, CA, United States) set to three cycles, with each cycle consisting of 1 min disruption at a speed of 5.5 m/s and 1 min at rest. The resulting extract was heated in a thermomixer (Thermomixer comfort 5,355, Eppendorf, Hamburg, Germany) at 60°C with shaking at 1400 rpm for 15 min and spun at 10,000 × g at 4°C for 10 min. Supernatants, containing the DNA and protein content, were stored at −20°C.
For DNA purification, half of the supernatant was thawed and 260 μl 10 M ammonium acetate was added, mixed and incubated on ice for 5 min. Samples were then spun at 20,000 × g for 10 min at 4°C and the supernatants were kept. Isopropanol in equal volume was added to the supernatants, mixed thoroughly and incubated on ice for 30 min. The samples were then centrifuged at 20,000 × g for 15 min at 4°C to pellet the DNA. The pelleted DNA was washed with 10 μl pure ethanol, dried in a speed vac and finally dissolved overnight at 4°C in TE-Buffer (1 mm EDTA and 10 mm Tris, pH = 8). The dissolved DNA was purified and proteins removed using the QIAmp DNA Mini Kit (QIAGEN, United States) and following the manufacturer’s instructions. Finally, the DNA content recovered from each sample was quantified using NanoDrop (NanoDrop2000, Thermo Scientific, Rockford, IL, United States).
16S rRNA Gene Sequencing
The 16S gene region V3 to V4 of Bacteria and Archaea was amplified with the universal primers 341F and 806R (5′-3′, forward: CCTACGGGAGGCAGCAG; reverse: GGACTACHVGGGTWTCTAAT; Takahashi et al., 2014) from the purified DNA samples. Amplicons were sequenced by an Illumina MiSeq DNA sequencer (Illumina, San Diego, United States). Amplification and sequencing were done by StarSeq GmbH (Mainz, Germany). Raw data analysis was done by StarSeq GmbH (Mainz, Germany) following their standard data analysis pipeline. Briefly, raw sequencing data were de-multiplexed, quality checked by FastQC and primers trimmed. Paired-end reads were joined, low-quality reads were removed, reads were corrected, chimaeras removed and Amplicon Sequence Variants (ASVs) were obtained by the deblur workflow. Taxonomy was annotated to the ASVs using the SILVA database (Quast et al., 2013). Read counts of ASVs were normalised and relative abundances determined, followed by taxonomic binning of ASVs using the R scripts Rhea (Lagkouvardos et al., 2017).
Metaproteomics
The concentration of protein in the supernatants from the protein and DNA extraction was determined with bicinchoninic acid assay according to the manufacturer’s instructions (Pierce™ BCA Protein Assay Kit, Thermo Fisher Scientific, Waltham, United States). Then, 100 μg protein was precipitated in acetone 1:5 (V/V) at −20°C overnight and then centrifuged (10 min, 14,000 × g). The precipitate was dissolved in Laemmli buffer and used for SDS-PAGE analysis, in-gel digestion and protein purification with ZipTip® treatment (Haange et al., 2020). The peptide lysate concentration was determined by NanoDrop (NanoDrop2000, Thermo Fisher Scientific, Rockford, IL, United States) and 2 μg peptide lysate was injected into nanoHPLC (UltiMate 3,000 RSLCnano, Dionex, Thermo Fisher Scientific, Waltham, United States). Peptides were separated on a C18-reverse phase trapping column (C18 PepMap100, 300 μm × 5 mm, particle size 5 μm, nano viper, Thermo Fischer Scientific, Waltham, United States), followed by a C18-reverse phase analytical column (Acclaim PepMap® 100, 75 μm × 25 cm, particle size 3 μm, nanoViper, Thermo Fisher Scientific, Waltham, United States; Haange et al., 2019). Mass spectrometric analysis of peptides was performed on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, Waltham, United States) coupled to a TriVersa NanoMate (Advion, Ltd., Harlow, United Kingdom) source in LC chip coupling mode. LC gradient, ionisation mode and mass spectrometry mode are described in Haange et al. (2019). Raw data were processed with Proteome Discoverer (v 2.2, Thermo Fischer Scientific, Waltham, United States). Parameters for Sequest HT search engine were set to: Trypsin (Full), Max. Missed Cleavage: 2, precursor mass tolerance: 10 ppm and fragment mass tolerance: 0.02 Da. Protein identification was done as previously described using a protein sequence data base constructed from all protein coding sequences of microbial genera identified in the 16S rRNA gene profiling as well as from plants commonly found in animal feed available on the UniProt (23.10.2017)1 data repository (Haange et al., 2019). Protein grouping was enabled, with protein group requiring at least one unique peptide. Relative abundance of protein groups was calculated as normalised intensity divided by the sum of all protein group intensities in the sample. Protein function and metabolic pathway relative abundances were calculated by summing all relative abundances of the associated protein groups.
Quantification of Glyphosate in Reactor Medium
Quantification of glyphosate in the fermenter medium was done as previously described (Krause et al., 2020). Briefly, for the extraction of glyphosate, 1,000 μl of methanol:acetonitrile:water (2:3:1) was added to 100 μl of specimen. Samples were vortexed, sonicated and centrifuged at 14,000 × g. The supernatant was dried in a vacuum centrifuge (Concentrator Plus, Eppendorf AG Hamburg, Germany) and re-dissolved in 100 μl of Milli-Q water afterwards. A 10 μl of each sample of the resuspended extract was injected into a BEH Amide column (2.1 × 100 mm, 1.7 μm) supplied by Waters (Milford, United States). Chromatographic separation was performed with a gradient of solvent A (66% H2O, 33% acetonitrile, 10 mm ammonium acetate and 0.04% ammonium hydroxide; pH 9) and solvent B (10% H2O, 90% acetonitrile, 10 mm ammonium acetate and 0.04% ammonium hydroxide; pH 9). The LC was run at a constant flow rate of 0.4 ml/min. Initial equilibration for 2 min with 0% B, within 2.5 min gradient from 0 to 100% solvent B, hold 100% solvent B for 2 min, back to 0% solvent B for 3.4 min. Targeted multi-reaction monitoring (MRM) measurement of glyphosate was done on a QTRAP® 5,500 (AB Sciex, Framingham, United States) in negative ionisation mode. Analyst® Software (AB Sciex, Framingham, United States, version 1.6.2) was used for data acquisition and analysis.
Statistical Analysis of Omics Data
Alpha-diversity and principal component analysis (PCA) were done in R (Ihaka and Gentleman, 1996) using the basic functions and the vegan package (Dixon, 2003). Statistics were done in R using in-house written scripts as previously described (Haange et al., 2020). Briefly, the statistical tests used were for complete sample data analysis Permutational Multivariate Analysis of Variance (PERMANOVA) using the adonis function in the vegan R package, and for single variables, Kruskal-Wallis group test followed by a post-hoc pairwise Dunn test. Where appropriate (number of tests >20), p-values were corrected for multi-testing by the Benjamini-Hochberg method (Benjamini and Hochberg, 1995). Heat maps were constructed with pheatmap R package (version 1.10)2 and all other figures were constructed using the R package ggplots2 (Wickham, 2011).
Results
Fermentation Parameters
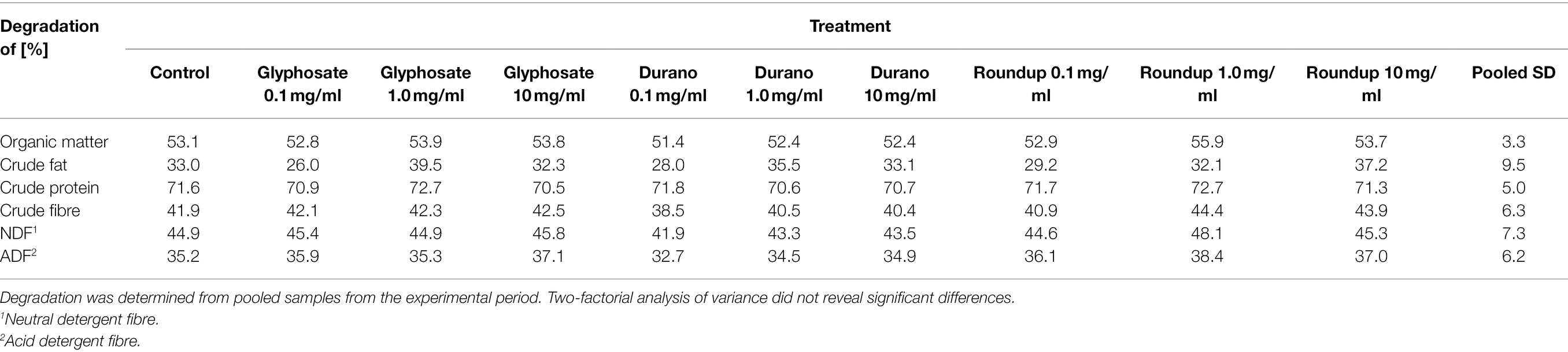
The effects of different formulations and concentrations of glyphosate on rumen fermentation were assessed by measuring pH (Figure 1A), redox potential (Figure 1B), NH3-N concentration (Figure 1C) and production of SCFA (Figure 1D). None of these parameters were significantly affected by the factors ‘glyphosate formulation’ or ‘concentration’. Slight variations over time were observed for all four parameters. The pH value slightly increased over time (p < 0.001) with a value of 6.58 ± 0.09 (mean ± SD) at day 7 and 6.66 ± 0.07 at day 16 (Figure 1A). Although this difference was significant, the change in pH value was very small and all values were within the physiological range. The redox potential also varied significantly over time (p < 0.001); however, all values were within physiological levels (Figure 1B). The concentration range of NH3-N was between 11.4 ± 2.3 mmol/l at day 7 and 12.4 ± 1.9 mmol/l at day 16 (p < 0.001). The production of SCFA varied between 44.0 ± 4.5 mmol/d and 41.4 ± 4.3 mmol/d (effect of time p < 0.01). For all four mentioned parameters, no significant interactions were present between the factors. The molar proportions of the individual SCFAs were influenced neither by glyphosate formulation, nor by concentration, nor were interactions revealed. While the proportion of acetate remained unchanged at about 45–50% throughout the whole experiment, the proportions of propionate, butyrate, isobutyrate, valerate and isovalerate varied slightly over time (at least p < 0.05, means are presented in Supplementary Figure 1). The proportions of propionate ranged between 23.3 ± 3.1% and 23.6 ± 3.0%, butyrate accounted for 16.7 ± 1.1%–16.4 ± 1.5% of the SCFA mixture, while isobutyrate was below 1%. The proportion of valerate varied between 6.3 ± 0.9% and 6.6 ± 0.7% and the proportion of isovalerate between 4.5 ± 0.7% and 4.6 ± 0.8%. Although significant time-dependent alterations were observed for nearly all biochemical parameters, these were relatively small and most probable of low biological impact.
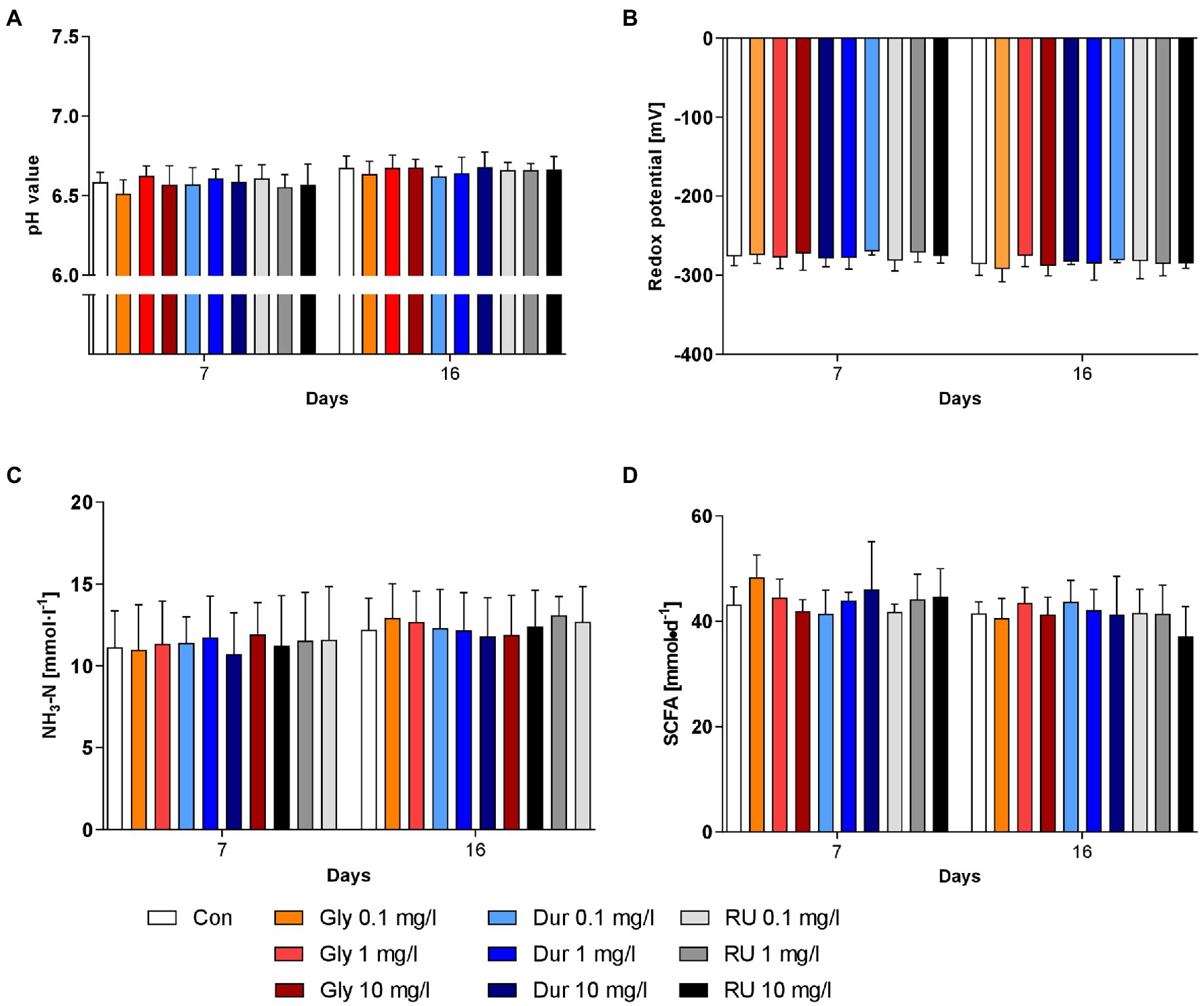
Figure 1. Effects of glyphosate on biochemical parameters. Effects of pure glyphosate (Gly), Durano TF (Dur) and Roundup® LB plus (RU) at different concentrations on pH (A), redox potential (B), NH3-N concentration (C) and SCFA production (D) were compared to control group (Con) before addition (day 7) and after 9 days of addition at day 16. Data are presented as means ± SD.
Feed Degradation
The degradation of organic matter did not differ based on treatment, glyphosate concentration or interaction of both (Table 3). A degradation of 51.4–55.9% was observed during the 48 h incubation period. The degradation of crude fat was more variable with 26.0–39.5% (Table 3). The degradation of crude protein reached between 70.5 and 72.0% and between 38.5 and 44.4% of crude fibre were degraded (Table 3). Within the fibre fraction, 41.9–48.1% of NDF and 32.7–38.4% of ADF were degraded (Table 3). None of these parameters was significantly altered by treatment, glyphosate concentration or an interaction of both (Table 3).
Microbial Protein Synthesis
Rumen microorganisms produced about 1 g of protein per day in each fermenter. Most of this protein (781.8 ± 66.5 mg/d) was produced by the LAM (Figure 2A), while SAM produced only about 225.0 ± 41.7 mg/d (Figure 2B). For both, LAM and SAM, more than 80% of N for microbial protein synthesis was derived from the NH3-pool (Figures 2C,D). The efficiency of microbial protein synthesis (Figure 2E) reached about 158.0 ± 35.5 g/kg degraded dry matter. None of the parameters for microbial protein synthesis was affected by treatment, glyphosate concentration or interaction of both.
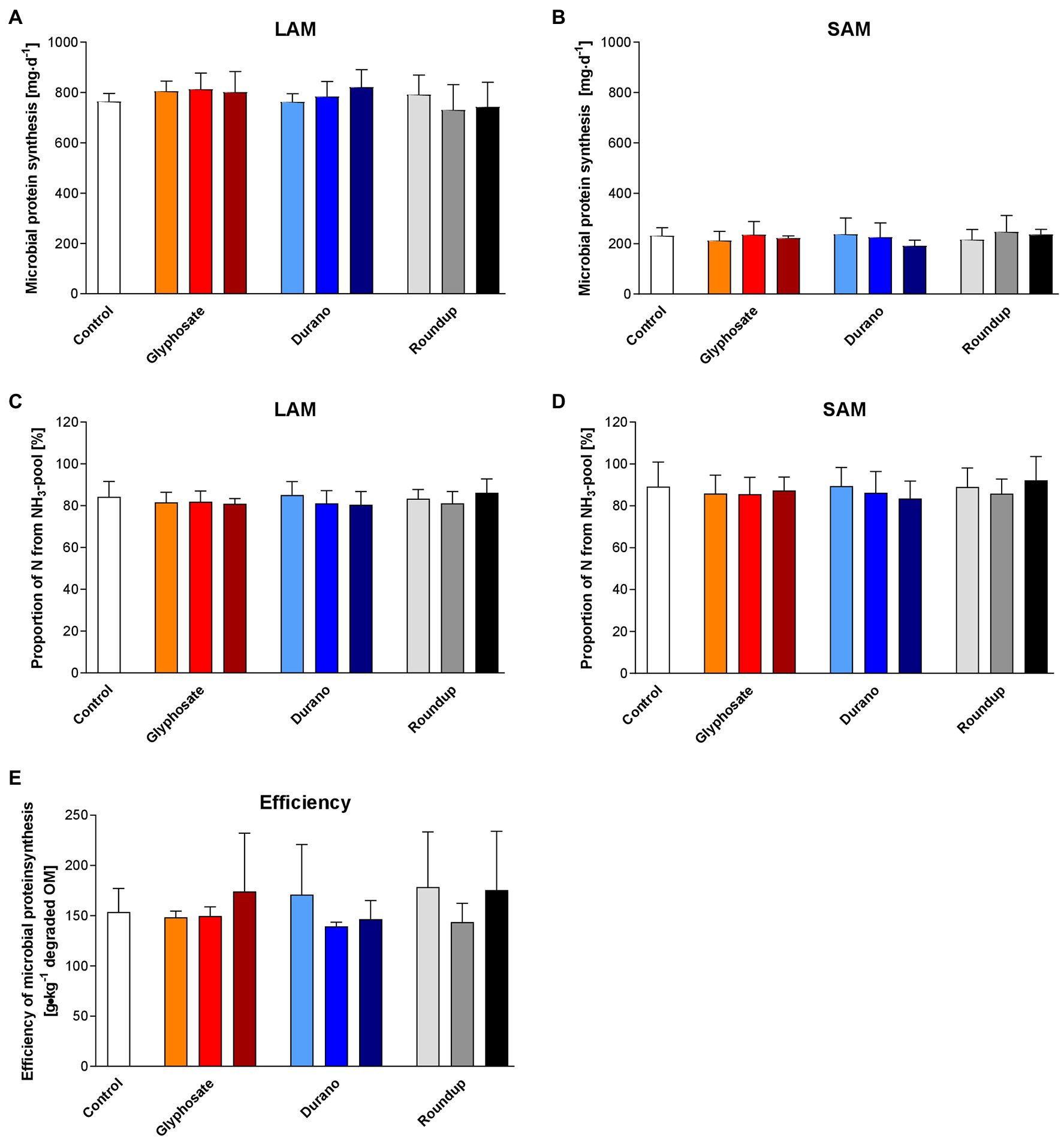
Figure 2. Effects of glyphosate on microbial protein synthesis. The effects of pure glyphosate, Durano TF and Roundup® LB plus on microbial protein synthesis of liquid-associated microorganism (LAM; A), solid-associated microorganisms (SAM; B), the proportion of N derived from the NH3-N pool for LAM (C) and SAM (D) and the efficiency of microbial protein synthesis (E) were evaluated. For each formulation, three different concentrations were used as: 0.1 mg/l (orange, light blue and light grey), 1 mg/l (red, blue and grey) and 10 mg/l (dark red, dark blue and black). Data are presented as means ± SD.
Microbial Community Structure
To investigate any shifts in taxonomic structure of the microbiome associated with treatment with glyphosate or its formulations Durano and Roundup, 16S rRNA gene profiling was performed. The mean number of taxa based on amplicon sequencing variants (ASVs) was 508 ± 105 for LAM (Figure 3A) and 468 ± 70 for SAM (Figure 3B). There was no significant difference (LAM: p = 0.887, SAM: p = 0.874, Kruskal-Wallis test) in the number of taxa present for either the LAM or SAM when treated with glyphosate or a glyphosate formulation. A decrease in alpha-diversity, a combination of the number of taxa present in a sample and their distribution is often a sign of dysbiosis. Therefore, alpha-diversity based on Shannon-Effective was calculated (Figures 3C,D), but no significant change in Shannon-Effective between the control samples and those cultivated with different concentrations of glyphosate or a glyphosate formulation (LAM: p = 0.767, SAM: p = 0.651, Kruskal-Wallis test) was observed. To investigate if there was a global shift in taxonomic distribution between the control samples and the glyphosate and glyphosate formulation exposed microbial community’s, beta-diversity analysis based on principal component analysis was done. No clustering was observed between the control communities and those exposed to glyphosate at different concentrations of glyphosate. Neither for glyphosate, Durano nor Roundup exposure at the concentrations of 0.1 mg/l, 1 mg/l or 10 mg/l were significant differences in global community structure observed in the LAM (Figures 3E–G) or SAM (Figures 3H–J) communities.
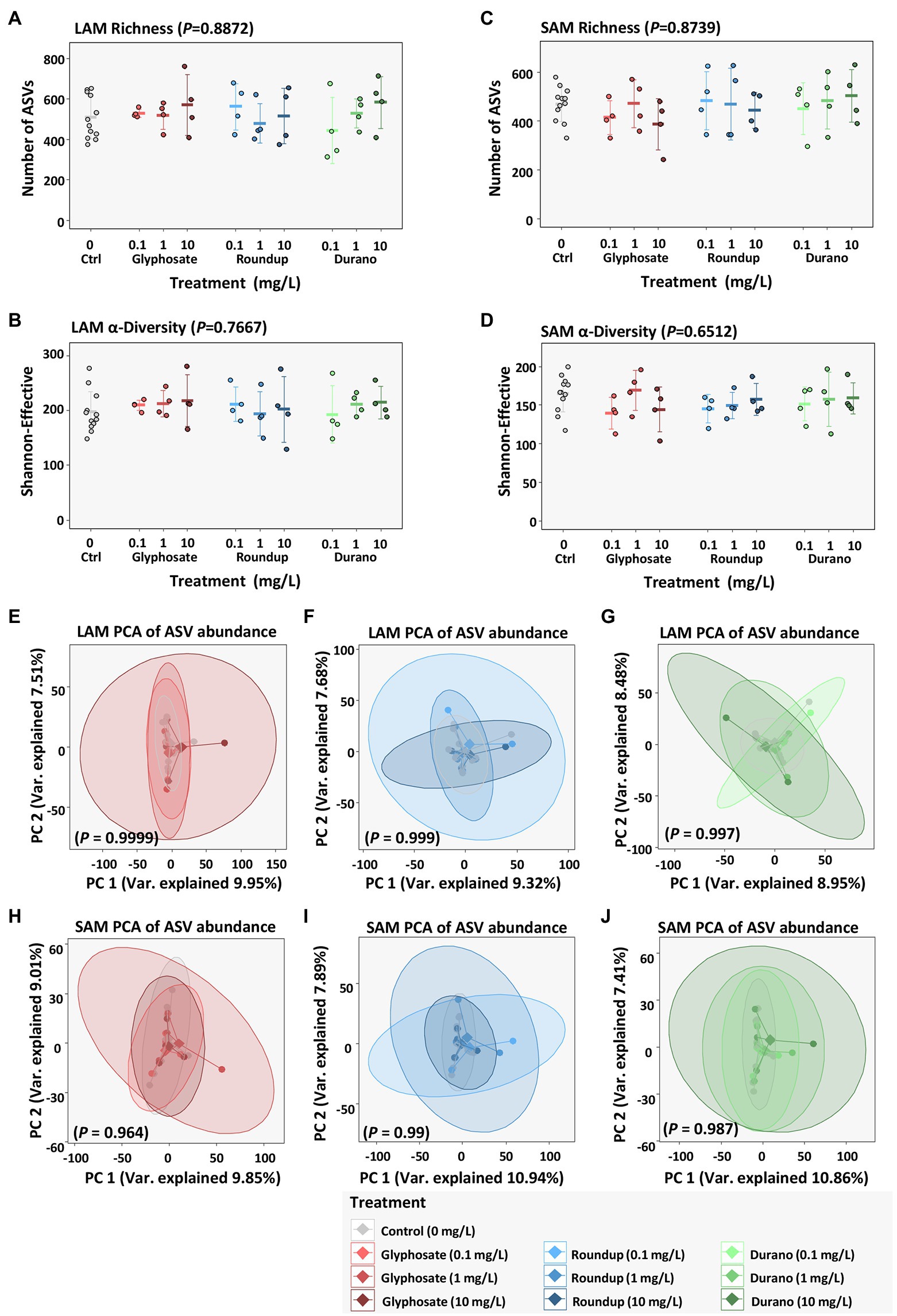
Figure 3. Influence of glyphosate and glyphosate formulations on diversity of bacterial communities based on abundances of amplicon sequencing variants (ASVs). LAM richness (A) and alpha-diversity (B) as well as SAM richness (C) and alpha-diversity (D), beta-diversity based on principal component analysis (PCA) of LAM treated with pure glyphosate (E), Roundup® LB plus (F) and Durano TF (G) as well as SAM communities treated with pure glyphosate (H), Roundup® LB plus (I) and Durano TF (J). For each formulation, three different concentrations were used as: 0.1 mg/l, 1 mg/l and 10 mg/l and compared to untreated controls.
On the taxonomic level of family, the most abundant families in the non-treated LAM communities were from the Firmicutes Ruminococcaceae (15.1 ± 4.3%, mean ± SD), Lachnospiraceae (12.9 ± 3.6%) and Veillonellaceae (6.8 ± 2.1%), and from the Bacteroidetes the families Prevotellaceae (15.2 ± 3.6%), Rikenellaceae (4.9 ± 1.1%) and the F082 (2.6 ± 0.8%). Other abundant families included Succinivibrionaceae (5.4 ± 6.6%), Spirochaetaceae (4.7 ± 2.2%), Bifidobacteriaceae (2 ± 2.2%) and the archaea family Methanobacteriaceae (2.9 ± 1.6%; Figure 4A). The SAM non-treated community exhibited a different distribution in bacterial families than the LAM. Here, the most abundant families were Lachnospiraceae (32.6 ± 3.2%), Prevotellaceae (15.6 ± 2.8%) and Veillonellaceae (10.8 ± 1.2%), Ruminococcaceae (7.5 ± 2.5%), Lactobacillaceae (6.6 ± 2.7%), Spirochaetaceae (6.5 ± 1.6%), Bifidobacteriaceae (4.8 ± 1.8%) and the archaeal family Methanobacteriaceae (2 ± 0.6%; Figure 4B). No significant changes in relative abundance of any bacterial family were observed in either LAM or SAM communities with treatment with glyphosate, Durano or Roundup.
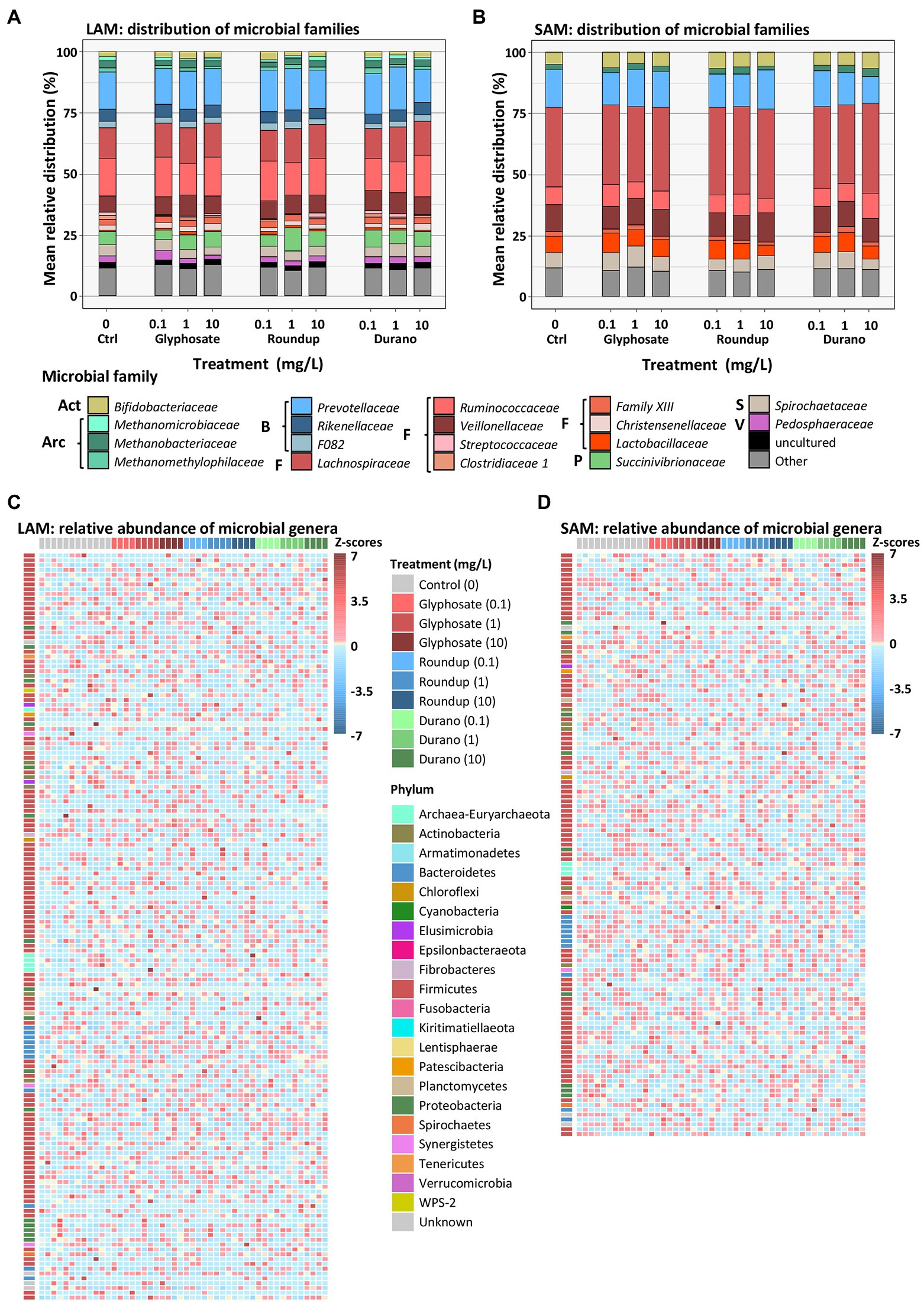
Figure 4. Relative distribution of microbial families in LAM (A) and SAM (B) communities as well as relative abundance of microbial genera in the LAM (C) and SAM (D) communities treated with pure glyphosate, Roundup® LB plus and Durano TF. For each formulation, three different concentrations were used as: 0.1 mg/l, 1 mg/l and 10 mg/l and compared to untreated controls. (A,B): Act = Actinobacteria, Arc = Archaea, B = Bacteroidetes, F = Firmicutes, S = Spirochaetota, V = Verrucomicrobia. The (C,D) depict relative abundance based on z-scores.
To investigate if changes in taxa distribution could only be observed at higher taxonomic resolution, an analysis on genus level was conducted. In total, we identified 155 genera in LAM (Figure 4C) and 121 genera in SAM (Figure 4D) with a relative abundance of at least 0.01% in one sample. No significant change in abundance was observed for any single genus in either LAM or SAM treated with glyphosate or a glyphosate formulation.
Microbial Functional Analysis
Metaproteomics was performed to analyse if glyphosate or glyphosate formulation exposure affected the functions exhibited by the LAM or SAM communities. In the LAM communities, a total of 8,052 microbial protein groups were detected, whereas in the SAM communities, 5,992 microbial protein groups were detected. To determine global metaproteome wide significant changes associated with glyphosate exposure or its formulations, principle component analysis was performed with the relative abundance of protein groups. PERMANOVA was used to determined significance. No global significant differences between the untreated control LAM (Figures 5A–C) or the untreated SAM communities (Figures 5D–F) with those treated with glyphosate, Durano or Roundup were observed. To investigate possible specific functional changes, protein groups were annotated to Kyoto Encyclopedia of Genes and Genomes (KEGG)3 (Ogata et al., 1999) metabolic pathways and the relative abundances summed and compared between the treatment groups. In total, 134 KEGG metabolic pathways with a functional coverage of at least 10% were identified in LAM (Figure 5G), while 106 were seen in SAM (Figure 5H). Glyphosate, Durano or Roundup treatment at any of the three concentrations (0.1 mg/l, 1 mg/l and 10 mg/l) did not significantly enrich any KEGG functional pathway when compared to the untreated controls. The KEGG pathway map 00400 was investigated more closely since it included the shikimate pathways and the 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase, which is the specific target for glyphosate. We identified 16 KEGG protein functions including the EPSP synthase in the metaproteomes of LAM and SAM (Figure 6A), but none of them were significantly altered depending on the abundance by glyphosate, Durano or Roundup treatment.
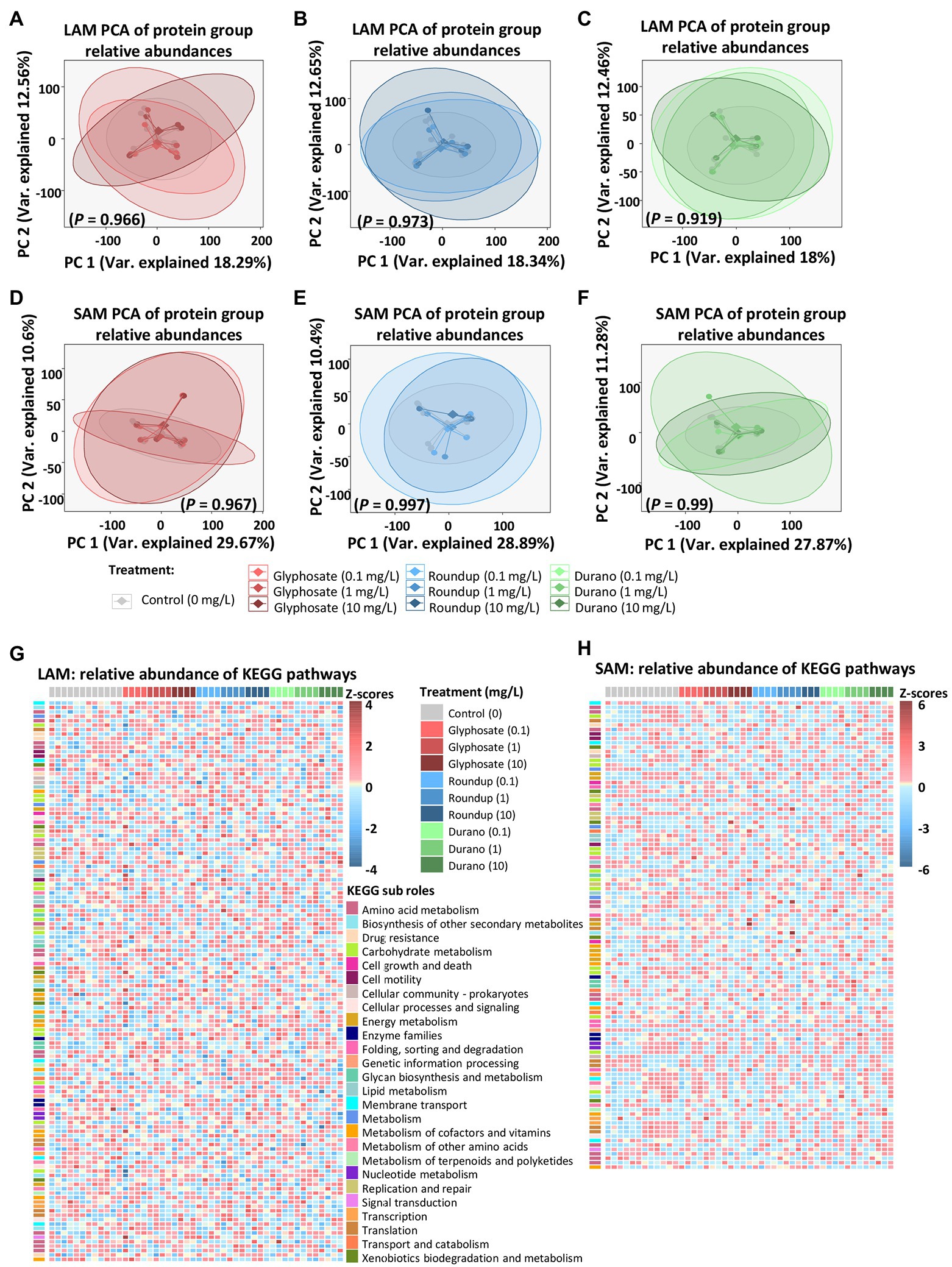
Figure 5. Influence of glyphosate and formulations on functions of microbial communities based on metaproteomic data. Principal component analysis (PCA) of protein group relative abundances of LAM treated with pure glyphosate (A), Roundup® LB plus (B) and Durano TF (C) as well as SAM communities treated with pure glyphosate (D), Roundup® LB plus (E) and Durano TF (F). Relative abundances based on z-scores of KEGG metabolic pathways present in LAM (G) and SAM (H) communities. For each formulation, three different concentrations were used as: 0.1 mg/l, 1 mg/l and 10 mg/l and compared to untreated controls.
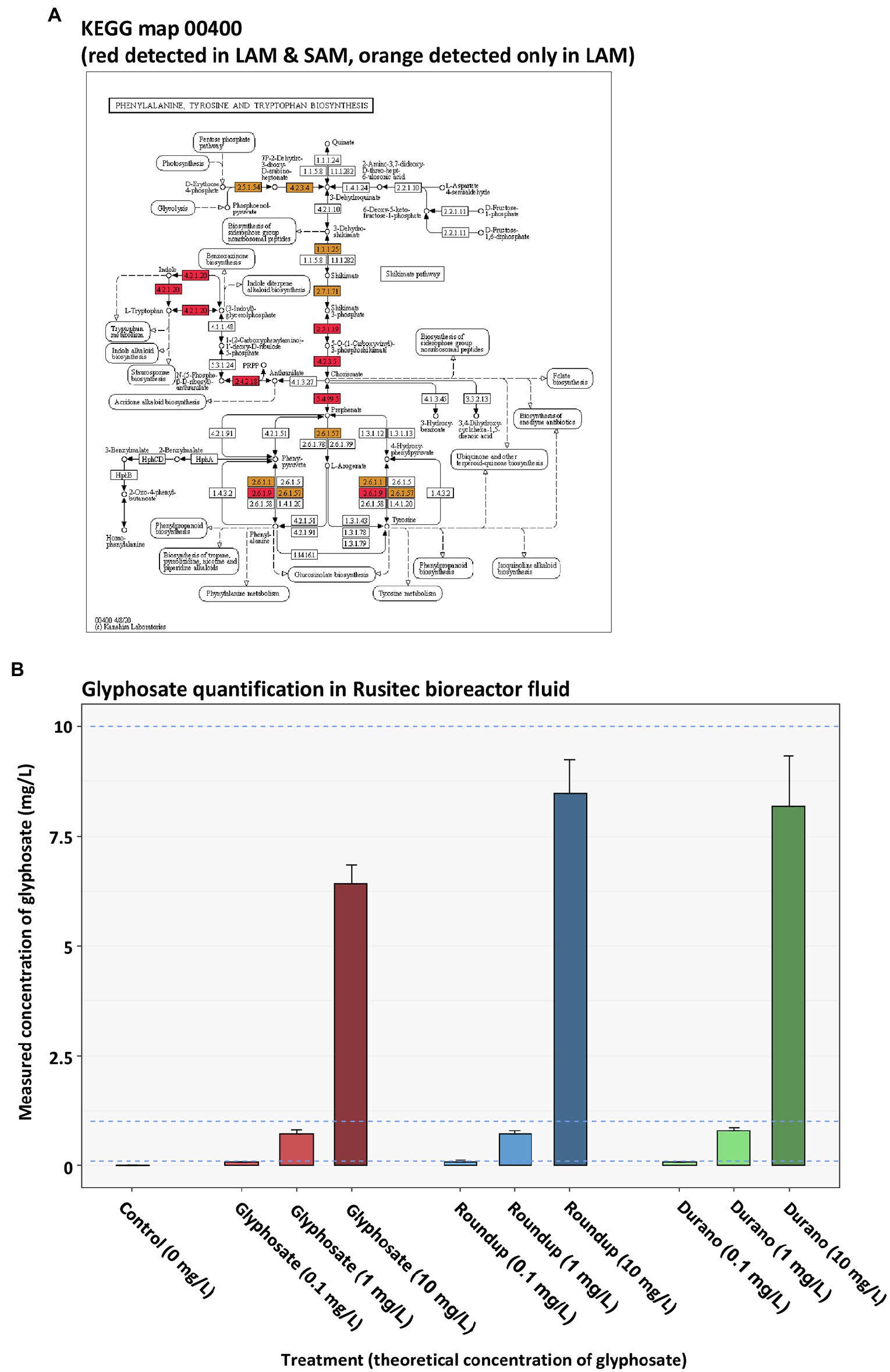
Figure 6. KEGG map 00400 of the phenylalanine, tyrosine and tryptophan biosynthesis (A) depicting all the protein functions identified in LAM and SAM (red) as well as only in LAM (orange). Quantification of glyphosate present in reactor medium at sampling time point (B).
Glyphosate Quantification and Correlation Analysis
One potential reason for the lack of changes in the taxonomic or functional community structure of LAM or SAM observed after glyphosate, or glyphosate formulation treatment, could be a potential degradation of glyphosate in the microbial community. To control for this, we quantified glyphosate in the bioreactor supernatant by targeted multi-reaction monitoring mass spectrometry. As expected, in the untreated control samples, no glyphosate was detected (Figure 6B). The spiked in concentration in the bioreactor medium was 10 mg/l, 1 mg/l and 0.1 mg/l. In the glyphosate or glyphosate formulation treated bioreactors, we detected somewhat lower levels of glyphosate than expected. The concentrations in the bioreactors with pure glyphosate were 6.414 mg/l ± 0.438 mg/l, 0.717 mg/l ± 0.105 mg/l and 0.068 mg/l ± 0.009 mg/l. In the bioreactors with Roundup, the measured concentrations were 8.478 mg/l ± 0.753 mg/l, 0.791 mg/l ± 0.088 mg/l and 0.074 mg/l ± 0.011 mg/l. In the bioreactors containing Durano, glyphosate concentrations after the run were measured at 8.179 mg/l ± 1.152 mg/l, 0.791 mg/l ± 0.088 mg/l and 0.074 mg/l ± 0.011 mg/l. These determined concentrations values were consistent with no or little degradation of the glyphosate when taking the accuracy tolerance (78% relative recovery) for the glyphosate quantification method into account. This suggested that the majority of glyphosate was still present in the medium. To reveal potential correlations among glyphosate concentration in the fermenter, microbial abundances and functions, the measured glyphosate values were correlated with the relative abundance of the microbial genera detected by 16S rRNA gene profiling and the relative abundance of KEGG metabolic pathways in the metaproteomics study. No significant positive or negative correlations between the concentration of glyphosate and the abundance of genera or KEGG pathways in the LAM or SAM were identified (data not shown). In addition, the relative abundances of protein functions of the KEGG 00400 metabolic pathway map (aromatic amino acid biosynthesis) identified in LAM and SAM were not significantly positively or negatively associated with the glyphosate concentration (data not shown).
Discussion
The rumen microbiota consists of bacteria, fungi, protozoa and archaea, containing members, which are potentially vulnerable to inhibition of the 5-enolpyruvylshikimate-3-phosphate pathway by glyphosate and/or adverse effects of adjuvants in commercial formulations. Therefore, the aim of this study was to investigate the effects of pure glyphosate and two commercially available formulations on rumen fermentation and rumen microbiota.
Neither the type of glyphosate applied, nor the concentration of the three formulations affected biochemical parameters such as pH, redox potential and concentration of NH3-N or SCFA production. Previous studies on the impact of glyphosate on these parameters are variable. The mentioned parameters were not altered by application of Roundup Ultra® (0.77 mg/d) in sheep (Hüther et al., 2005). Administration of Roundup® LB plus to a bioreactor model for the pig colon microbiota did also not change SCFA production (Krause et al., 2020). In contrast, Riede et al. (2016) applied Plantaclean® 360 in two doses (about 0.46 mg/l and 3.9 mg/l) in a RUSITEC-trial and detected no effects on pH, redox potential and most SCFAs, but reported a decrease in NH3-N concentration and in propionate proportion, a tendency for increased total SCFA and acetate production, and an increase in isovalerate proportion. In vivo, Billenkamp et al. (2021) observed no effects of Roundup Record® [about 80 mg/d (Schnabel et al., 2017)] on pH, NH3-N and the proportions of the main SCFA; however, total SCFA production and the proportion of isovalerate decreased. Summarising these studies, there were no consistent effects of different glyphosate formulations on rumen fermentation parameters. Glyphosate dosages in these studies were close to our medium dosage. Although no previous study comparing several formulations is available, the results of the mentioned studies are based on different formulations and agree with our result that there is no difference among glyphosate formulations.
Small variations in biochemical parameters were observed over time, e.g., pH varied about 0.08 units during the experimental period. Comparable variations have been observed in other Rusitec experiments without glyphosate additions (Orton et al., 2020) and are far below diurnal variations observed in vivo (Leedle et al., 1982). Therefore, these observations are probably of negligible biological relevance.
Microbial protein synthesis was to our knowledge not measured previously under glyphosate treatment. However, the lack of effect by glyphosate on microbial protein synthesis agrees with the stable NH3-N concentration. The degradation of organic matter, crude nutrients and fibre fractions was also unaffected by glyphosate in the present study. This agrees with the results on in sacco degradation of dry matter and NDF in sheep (Hüther et al., 2005).
Previous studies indicated an effect of glyphosate on bacterial communities, e.g., in soil (Lancaster et al., 2010), in the honeybee gut (Motta et al., 2018; Blot et al., 2019) or in the rat cecum (Mesnage et al., 2021). Therefore, we also investigated the rumen bacterial community, its metabolic activity and the potential inhibition of the EPSP pathway. We did not observe any effect of glyphosate, Durano or Roundup on the abundance of bacterial families and genera, nor on alpha or beta-diversity of the prokaryotic community.
Previous reports on the effects of glyphosate on rumen microorganisms were mainly based on short-time studies using isolated cultures of single strains or batch cultures (Krüger et al., 2013; Ackermann et al., 2015). The RUSITEC system, which was used here, is a more complex system, which is able to maintain the core bacterial community of the rumen over a longer time (Wetzels et al., 2018). Our results agree with previous RUSITEC studies, which observed only minor effects of glyphosate on the rumen bacterial population and no effects on the growth of pathogenic bacteria (Riede et al., 2016; Bote et al., 2019). In a recent in vivo study with dairy cattle, Billenkamp et al. (2021) observed differences for only four taxa, which, however, appeared to be linked closer to experimental conditions than to glyphosate application. One of the formulations used in the present study, Roundup® LB plus was also tested in a pig colon microbiota in vitro model, where also no effect on the microbial community composition was detected (Krause et al., 2020). Metaproteome analysis confirmed the presence of the enzymes of the shikimate pathway and the EPSP synthase in the rumen microbiota; however, expression of these enzymes was not influenced by addition of pure glyphosate or its formulations at any concentration, which also agrees with the study by Krause et al. (2020) for the pig gut. Until now, there is no study available using untargeted metabolomics to focus on the end products of microbial fermentation pathways, this might be a future approach to support the current observations. Comparing the results of the mentioned studies, it appears that the effects of glyphosate on microbiota may be more pronounced, when single strains or simple microbial communities are investigated, while for more diverse communities, such as in the RUSITEC system or in the rumen in vivo, glyphosate effects are rarely observed. In addition, microbiomes in amino acid poor environments, such as soil or the honeybee gut, may rely more heavily on synthesis of aromatic amino acids, and therefore could be more susceptible to glyphosate inhibition. In contrast, microbiomes in amino acid rich environments such as the stomach and gut of livestock, especially if the host livestock is provided with protein-rich feed, may be resilient towards glyphosate inhibition.
In conclusion, glyphosate treatment, independent whether pure glyphosate, or the formulation Durano TF or Roundup® LB plus was applied, did neither influence the rumen procaryotic community, nor its fermentation, nor expression of the EPSP synthase pathway in the RUSITEC system even in concentrations far above common intake levels. This underlines the high stability of the complex rumen microbial community.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD032166. 16S rRNA gene sequencing data was deposited in the SRA repository and can be found under the identifier PRJNA802980.
Ethics Statement
The animal study was reviewed and approved by Lower Saxony State Office for Costumer Protection and Food Safety (approval number: 33.9-42502-04-12/0876).
Author Contributions
GB and MvB planned the study and acquired the funding. GB and SR planned the experiments. DS and SR were involved in performance of the Rusitec experiments. S-BH did 16S rRNA gene analysis. S-BH and NJ performed proteomics analysis. BE and UR-K did glyphosate measurements. SR, KR, MB and S-BH analysed the data. MB and S-BH wrote the paper. All authors approved the final version of the manuscript.
Funding
This project was funded by the BMEL (BLE grant numbers 314-06.01-2815HS016 and 314-06.01-2815HS018). MvB is grateful for the support by Novo Nordisk Foundation grant NNF21OC0066551.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Markus Pez and Marion Burmester for their help in performing the Rusitec trials.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.873101/full#supplementary-material
Footnotes
References
Ackermann, W., Coenen, M., Schrödl, W., Shehata, A. A., and Krüger, M. (2015). The influence of glyphosate on the microbiota and production of botulinum neurotoxin during ruminal fermentation. Curr. Microbiol. 70, 374–382. doi: 10.1007/s00284-014-0732-3
Amrhein, N., Deus, B., Gehrke, P., and Steinrücken, H. C. (1980a). The site of the inhibition of the shikimate pathway by glyphosate: II. Interference of glyphosate with chorismate formation in vivo and in vitro. Plant Physiol. 66, 830–834. doi: 10.1104/pp.66.5.830
Amrhein, N., Schabe, J., and Steinrücken, H. C. (1980b). The mode of action of the herbicide glyphosate. Naturwissenschaften 67, 356–357. doi: 10.1007/BF01106593
Antier, C., Kudsk, P., Reboud, X., Ulber, L., Baret, P. V., and Messéan, A. (2020). Glyphosate use in the European agricultural sector and a framework for its further monitoring. Sustainability 12:5682. doi: 10.3390/su12145682
Benbrook, C. (2016). Trends in glyphosate herbicide use in the United States and globally. Environ. Sci. Eur. 28:3. doi: 10.1186/s12302-016-0070-0
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Billenkamp, F., Schnabel, K., Hüther, L., Frahm, J., von Soosten, D., Meyer, U., et al. (2021). No hints at glyphosate-induced ruminal dysbiosis in cows. NPJ Biofilms Micro. 7:30. doi: 10.1038/s41522-021-00198-4
Blot, N., Veillat, L., Rouzé, R., and Delatte, H. (2019). Glyphosate, but not its metabolite AMPA, alters the honeybee gut microbiota. PLoS One 14:e0215466. doi: 10.1371/journal.pone.0215466
Boguhn, J., Kluth, H., and Rodehutscord, M. (2006). Effect of Total mixed ration composition on fermentation and efficiency of Ruminal microbial crude protein synthesis In vitro. J. Dairy Sci. 89, 1580–1591. doi: 10.3168/jds.S0022-0302(06)72225-1
Boguhn, J., Zuber, T., and Rodehutscord, M. (2013). Effect of donor animals and their diet on in vitro nutrient degradation and microbial protein synthesis using grass and corn silages. J. Anim. Physiol. Anim. Nutr. 97, 547–557. doi: 10.1111/j.1439-0396.2012.01295.x
Boocock, M. R., and Coggins, J. R. (1983). Kinetics of 5-enolpyruvylshikimate-3-phosphate synthase inhibition by glyphosate. FEBS Lett. 154, 127–133. doi: 10.1016/0014-5793(83)80888-6
Bote, K., Pöppe, J., Riede, S., Breves, G., and Roesler, U. (2019). Effect of a glyphosate-containing herbicide on Escherichia coli and salmonella Ser. Typhimurium in an in vitro rumen simulation system. Eur. J. Microbiol. Immunol. 9, 94–99. doi: 10.1556/1886.2019.00010
Brandt, M., and Rohr, K. (1981). Belträge zur Quantifizierung der N-Umsetzungen in den Vormägen von Milchkühen: bestimmung des Mikrobenstickstoffs im Duodenalchymus mit Hilfe von 15N. Z. Tierphysiol. Tierernahr. Futtermittelkd. 46, 39–48. doi: 10.1111/j.1439-0396.1981.tb01335.x
Defarge, N., Spiroux de Vendômois, J., and Séralini, G. E. (2018). Toxicity of formulants and heavy metals in glyphosate-based herbicides and other pesticides. Toxicol. Rep. 5, 156–163. doi: 10.1016/j.toxrep.2017.12.025
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Eger, M., Riede, S., and Breves, G. (2018). Induction of a transient acidosis in the rumen simulation technique. J. Anim. Physiol. Anim. Nutr. (Berl.) 102, 94–102. doi: 10.1111/jpn.12662
Haange, S. B., Jehmlich, N., Hoffmann, M., Weber, K., Lehmann, J., von Bergen, M., et al. (2019). Disease development is accompanied by changes in bacterial protein abundance and functions in a refined model of dextran sulfate sodium (DSS)-induced colitis. J. Proteome Res. 18, 1774–1786. doi: 10.1021/acs.jproteome.8b00974
Haange, S. B., Jehmlich, N., Krügel, U., Hintschich, C., Wehrmann, D., Hankir, M., et al. (2020). Gastric bypass surgery in a rat model alters the community structure and functional composition of the intestinal microbiota independently of weight loss. Microbiome 8:13. doi: 10.1186/s40168-020-0788-1
Hüther, L., Drebes, S., and Lebzien, P. (2005). Effect of glyphosate contaminated feed on rumen fermentation parameters and in sacco degradation of grass hay and corn grain. Arch. Anim. Nutr. 59, 73–79. doi: 10.1080/17450390512331342403
Ihaka, R., and Gentleman, R. (1996). R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314. doi: 10.1080/10618600.1996.10474713
Koch, M., Strobel, E., Tebbe, C. C., Heritage, J., Breves, G., and Huber, K. (2006). Transgenic maize in the presence of ampicillin modifies the metabolic profile and microbial population structure of bovine rumen fluid in vitro. Br. J. Nutr. 96, 820–829. doi: 10.1017/bjn20061889
Krause, J. L., Haange, S.-B., Schäpe, S. S., Engelmann, B., Rolle-Kampczyk, U., Fritz-Wallace, K., et al. (2020). The glyphosate formulation roundup® LB plus influences the global metabolome of pig gut microbiota in vitro. Sci. Total Environ. 745:140932. doi: 10.1016/j.scitotenv.2020.140932
Krüger, M., Shehata, A. A., Schrödl, W., and Rodloff, A. (2013). Glyphosate suppresses the antagonistic effect of enterococcus spp. on clostridium botulinum. Anaerobe 20, 74–78. doi: 10.1016/j.anaerobe.2013.01.005
Lagkouvardos, I., Fischer, S., Kumar, N., and Clavel, T. (2017). Rhea: a transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ 5:e2836. doi: 10.7717/peerj.2836
Lancaster, S. H., Hollister, E. B., Senseman, S. A., and Gentry, T. J. (2010). Effects of repeated glyphosate applications on soil microbial community composition and the mineralization of glyphosate. Pest Manag. Sci. 66, 59–64. doi: 10.1002/ps.1831
Leedle, J. A., Bryant, M. P., and Hespell, R. B. (1982). Diurnal variations in bacterial numbers and fluid parameters in ruminal contents of animals fed low- or high-forage diets. Appl. Environ. Microbiol. 44, 402–412. doi: 10.1128/aem.44.2.402-412.1982
Leino, L., Tall, T., Helander, M., Saloniemi, I., Saikkonen, K., Ruuskanen, S., et al. (2021). Classification of the glyphosate target enzyme (5-enolpyruvylshikimate-3-phosphate synthase) for assessing sensitivity of organisms to the herbicide. J. Hazard. Mater. 408:124556. doi: 10.1016/j.jhazmat.2020.124556
Mesnage, R., Benbrook, C., and Antoniou, M. N. (2019). Insight into the confusion over surfactant co-formulants in glyphosate-based herbicides. Food Chem. Toxicol. 128, 137–145. doi: 10.1016/j.fct.2019.03.053
Mesnage, R., Teixeira, M., Mandrioli, D., Falcioni, L., Ducarmon, Q. R., Zwittink, R. D., et al. (2021). Use of shotgun Metagenomics and metabolomics to evaluate the impact of glyphosate or roundup MON 52276 on the gut microbiota and serum Metabolome of Sprague-Dawley rats. Environ. Health Perspect. 129:017005. doi: 10.1289/ehp6990
Motta, E. V. S., Raymann, K., and Moran, N. A. (2018). Glyphosate perturbs the gut microbiota of honey bees. Proc. Natl. Acad. Sci. U. S. A. 115, 10305–10310. doi: 10.1073/pnas.1803880115
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., and Kanehisa, M. (1999). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34. doi: 10.1093/nar/27.1.29
Orton, T., Rohn, K., Breves, G., and Brede, M. (2020). Alterations in fermentation parameters during and after induction of a subacute rumen acidosis in the rumen simulation technique. J. Anim. Physiol. Anim. Nutr. 104, 1678–1689. doi: 10.1111/jpn.13412
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Riede, S., Boguhn, J., and Breves, G. (2013). Studies on potential effects of fumaric acid on rumen microbial fermentation, methane production and microbial community. Arch. Anim. Nutr. 67, 368–380. doi: 10.1080/1745039x.2013.830518
Riede, S., Toboldt, A., Breves, G., Metzner, M., Köhler, B., Bräunig, J., et al. (2016). Investigations on the possible impact of a glyphosate-containing herbicide on ruminal metabolism and bacteria in vitro by means of the 'Rumen simulation Technique'. J. Appl. Microbiol. 121, 644–656. doi: 10.1111/jam.13190
Schnabel, K., Schmitz, R., Frahm, J., Meyer, U., Breves, G., and Dänicke, S. (2020). Functionality and DNA-damage properties of blood cells in lactating cows exposed to glyphosate contaminated feed at different feed energy levels. Arch. Anim. Nutr. 74, 87–106. doi: 10.1080/1745039x.2020.1718474
Schnabel, K., Schmitz, R., von Soosten, D., Frahm, J., Kersten, S., Meyer, U., et al. (2017). Effects of glyphosate residues and different concentrate feed proportions on performance, energy metabolism and health characteristics in lactating dairy cows. Arch. Anim. Nutr. 71, 413–427. doi: 10.1080/1745039X.2017.1391487
Takahashi, S., Tomita, J., Nishioka, K., Hisada, T., and Nishijima, M. (2014). Development of a prokaryotic universal primer for simultaneous analysis of bacteria and Archaea using next-generation sequencing. PLoS One 9:e105592. doi: 10.1371/journal.pone.0105592
von Soosten, D., Meyer, U., Hüther, L., Dänicke, S., Lahrssen-Wiederholt, M., Schafft, H., et al. (2016). Excretion pathways and ruminal disappearance of glyphosate and its degradation product aminomethylphosphonic acid in dairy cows. J. Dairy Sci. 99, 5318–5324. doi: 10.3168/jds.2015-10585
Wetzels, S. U., Eger, M., Burmester, M., Kreienbrock, L., Abdulmawjood, A., Pinior, B., et al. (2018). The application of rumen simulation technique (RUSITEC) for studying dynamics of the bacterial community and metabolome in rumen fluid and the effects of a challenge with Clostridium perfringens. PLoS One 13:e0192256. doi: 10.1371/journal.pone.0192256
Keywords: rumen, glyphosate, fermentation, metaproteome, bacterial community, 16S rRNA gene sequencing
Citation: Brede M, Haange S-B, Riede S, Engelmann B, Jehmlich N, Rolle-Kampzczyk U, Rohn K, von Soosten D, von Bergen M and Breves G (2022) Effects of Different Formulations of Glyphosate on Rumen Microbial Metabolism and Bacterial Community Composition in the Rumen Simulation Technique System. Front. Microbiol. 13:873101. doi: 10.3389/fmicb.2022.873101
Edited by:
Leonardo Erijman, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Robin Mesnage, King’s College London, United KingdomChristophe Calvayrac, Université de Perpignan – Domitia, France
Copyright © 2022 Brede, Haange, Riede, Engelmann, Jehmlich, Rolle-Kampzczyk, Rohn, von Soosten, von Bergen and Breves. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melanie Brede, bWVsYW5pZS5icmVkZUB0aWhvLWhhbm5vdmVyLmRl; Sven-Bastiaan Haange, c3Zlbi5oYWFuZ2VAdWZ6LmRl
†These authors have contributed equally to this work and share first authorship
 Melanie Brede
Melanie Brede Sven-Bastiaan Haange
Sven-Bastiaan Haange Susanne Riede1
Susanne Riede1 Beatrice Engelmann
Beatrice Engelmann Nico Jehmlich
Nico Jehmlich Ulrike Rolle-Kampzczyk
Ulrike Rolle-Kampzczyk Karl Rohn
Karl Rohn Martin von Bergen
Martin von Bergen