Abstract
Translation initiation factors and, in particular, the eIF4E family are the primary source of recessive resistance to potyviruses in many plant species. However, no eIF4E-mediated resistance to this virus genus has been identified in potato (Solanum tuberosum L.) germplasm. As in tomato, the potato eIF4E gene family consists of eIF4E1, its paralog eIF4E2, eIF(iso)4E, and nCBP. In tomato, eIF4E1 knockout (KO) confers resistance to a subset of potyviruses, while the eIF4E1/2 double KO, although conferring a broader spectrum of resistance, leads to plant developmental defects. Here, the tetraploid potato cv. Desirée owning the dominant Ny gene conferring resistance to potato virus Y (PVY) strain O but not NTN was used to evaluate the possibility to expand its PVY resistance spectrum by CRISPR-Cas9-mediated KO of the eIF4E1 susceptibility gene. After a double process of plant protoplast transfection-regeneration, eIF4E1 KO potatoes were obtained. The knockout was specific for the eIF4E1, and no mutations were identified in its eIF4E2 paralog. Expression analysis of the eIF4E family shows that the disruption of the eIF4E1 does not alter the RNA steady-state level of the other family members. The eIF4E1 KO lines challenged with a PVYNTN isolate showed a reduced viral accumulation and amelioration of virus-induced symptoms suggesting that the eIF4E1 gene was required but not essential for its multiplication. Our data show that eIF4E1 editing can be usefully exploited to broaden the PVY resistance spectrum of elite potato cultivars, such as Desirée, by pyramiding eIF4E-mediated recessive resistance.
Introduction
Potato is a member of the Solanaceae family and, on a global scale, is the fourth most cultivated species as a food source right after the three main cereal crops: rice, wheat, and corn.1 Like other vegetatively propagated plants, potato is prone to diseases, and potato virus Y (PVY), the type member of the Potyvirus genus, is the most harmful pathogen affecting potato yield and quality (Valkonen, 1997, 2007; Kreuze et al., 2020).
The PVY genome consists of a 9.7 kb monopartite positive-sense single-stranded RNA (ssRNA) encoding 11 functional proteins, one of which the VPg is covalently linked to the 5′ end of the genome and is a virulence factor. PVY isolates can be grouped into at least seven strains (PVYO, PVYC, PVYZ, PVYE, PVYN, PVYN–Wi, and PVYNTN) based on their biological properties (Lacomme and Jacquot, 2017).
PVY is non-persistently transmitted by more than sixty aphid species (Lacomme and Jacquot, 2017). Since insecticides are ineffective to manage non-persistently transmitted viruses (Kirchner et al., 2014) the spread of PVY in the field is difficult to control unless resistant plants are used.
Two types of dominant PVY resistance have been described in potatoes, the hypersensitive response (HR) and the extreme resistance (ER) conferred by N (Ny, Nc, and Nz) and Ry (Ryadg, Rysto, and Rycbc) genes, respectively (Karasev and Gray, 2013). N genes were the first used in potato-breeding programs for PVY resistance; hence, most elite potato cultivars possess one or more N genes (Valkonen et al., 2017). Ny, Nc, and Nz genes confer strain-group-specific HR to virus isolates belonging to the PVYO, PVYC, and PVYZ strains, respectively (Singh et al., 2008). Notably, recombinant PVY strains, such as the PVYNTN, inducing the potato tuber necrosis ringspot disease (PTRND) and overcoming all three N genes, have emerged and become prevalent in fields exposing well-accepted consumer potato cultivars to PVY disease (Funke et al., 2017).
In addition to dominant resistance genes, potential sources of virus resistance are plant genes encoding proviral factors required by the virus to complete its entire infection cycle. This class of host genes is known as susceptibility (S) genes. Mutations in S genes that preclude the virus’s ability to use them for its infection purpose act as virus-resistance genes. In nature, the virus-resistant trait of a mutated susceptibility gene usually occurs in homozygosity, thus being genetically recessive.
The eukaryotic translation initiation proteins from the eIF4E family are major resistance factors for RNA viruses, particularly to Potyviridae (Truniger and Aranda, 2009). In flowering plants, the eIF4E family comprises three forms, eIF4E, its isoform eIF(iso)4E, and nCBP (new cap-binding protein), and all are 7-methylguanosine triphosphate (m7GTP) cap-binding proteins, although nCBP does not appear to function in canonical translation (Patrick and Browning, 2012; Browning and Bailey-Serres, 2015). Significantly, eIF4E and eIF(iso)4E functions partially overlap, as shown by gene knockout (KO) and RNA silencing experiments (Duprat et al., 2002; Combe et al., 2005; Atarashi et al., 2020; Miroshnichenko et al., 2020).
The eIF4E family of potato, tomato, and pepper consists of four genes, eIF4E1 and its paralog eIF4E2, eIF(iso)4E, and nCBP. Notably, natural eIF4E alleles conferring PVY resistance have been discovered in pepper and tomato (Ruffel et al., 2002, 2005) but not in S. tuberosum L. germplasm (Baebler et al., 2020). The differences between an eIF4E susceptible and resistant allele are often few (1–5) non-synonymous amino acid substitutions located mainly near the domain involved in m7GTP cap recognition (Poulicard et al., 2016). The interaction between an eIF4E and the VPg of several potyviruses is closely linked to infection success (Charron et al., 2008; Poulicard et al., 2016), although the precise role(s) of eIF4E in potyvirus infection is still under debate (Bastet et al., 2017).
Based on the evidence of eIF4E requirement for PVY infectivity and the partial gene redundancy between eIF4E and eIF(iso)4E for their translational function, mutagenesis experiments were performed to introduce resistance traits in solanaceous crops by knocking out eIF4E genes. A tomato eIF4E1 EMS splicing mutant was immune to the isolate PVY-LE90 but not to PVY-LE84 (Piron et al., 2010), while the double eIF4E1/eIF4E2 mutant, although showing a broader PVY resistance, has developmental defects (Gauffier et al., 2016). Similarly, tobacco eIF4E1 EMS-KO lines were PVY resistant (Julio et al., 2015; Zhao et al., 2020). Conversely, no eIF4E KO potatoes were produced, and our knowledge on eIF4E involvement in PVY resistance comes from two opposite transgenic approaches based on eIF4E1: (a) silencing of the native susceptible eIF4E1/2 (Miroshnichenko et al., 2020); and (b) overexpression of a resistant eIF4E1 allele from other Solanaceae species (Cavatorta et al., 2011; Duan et al., 2012; Zhang C. et al., 2021) or a potato eIF4E1 mimicking pepper and tomato resistant alleles (Cavatorta et al., 2011; Arcibal et al., 2016; Gutierrez Sanchez et al., 2020). While silencing of eIF4E expression partially mimics the effect of a KO mutant, a more complex and not yet well-understood mechanism underlines the resistance gained through overexpression (Cavatorta et al., 2011; Duan et al., 2012; Gutierrez Sanchez et al., 2020; Zhang C. et al., 2021).
The advent of CRISPR-Cas editing tools has revolutionized our ability to specifically modify plant genomes, boosting our knowledge of genes function and resulting in crops with improved characters (Zhu et al., 2020; Zhang D. et al., 2021). In particular, editing the eIF4E gene family has recently been exploited to confer virus resistance in crops. In cucumber, eIF4E KO confers resistance to the potyviruses zucchini yellow mosaic virus and papaya ringspot mosaic virus-W (Chandrasekaran et al., 2016), whereas in tomato it induces a reduced accumulation of PVYN but not PVYO (Atarashi et al., 2020). Similarly, eIF4E1 editing in the tomato cv. MicroTom confers resistance to pepper mottle virus but not to tobacco etch virus (Yoon et al., 2020). In addition to the use of eIF4E1, eIF4E2 KO has been shown to reduce the accumulation of some isolates of pepper veinal mottle virus in cherry tomato (Kuroiwa et al., 2022), while editing cassava nCBP1 and nCBP2 was shown to reduce the severity and incidence of cassava brown streak disease symptoms (Gomez et al., 2019). Thus, depending on the crop and the selected eIF4E gene, resistance to some potyviruses can be introduced.
In this paper, we investigated the impact of eIF4E1 gene disruption on the ability to broaden the PVY resistance spectrum of the elite tetraploid potato cv. Desirée.2 The cv. Desirée is resistant to PVYO but not to PVYNTN, among others (Singh et al., 2008; Jones and Vincent, 2018). Protoplasts were transiently transfected with a CRISPR-Cas9 construct targeting eIF4E1. After a double plant-transfection regeneration process, eIF4E1 KO potato plants were obtained. Expression data show that knockout of the eIF4E1 gene does not alter the mRNA steady-state levels of the other members of the potato eIF4E gene family. According to the recessive resistance character of eIF4E1, only plants mutated in all four eIF4E1 copies were partially resistant to PVYNTN. The data demonstrate that eIF4E1 editing can be profitably used to extend the virus resistance spectrum of elite potato cultivars.
Materials and Methods
Plant Material and Potato Virus Y Strain
Nodal stem explants with axillary buds were collected from sprouted certified potato tubers of cv. Desirée (NAK; plantenpaspoort zp-d2/a6/a13 model 2 643.454.408).3 Explants sterilization and in vitro culture were performed as described in Tavazza and Ancora (1986).
PVY Pa36 was kindly provided by Dr. Massimo Turina and Dr. Marina Ciuffo (Istituto per la Protezione Sostenibile delle Piante IPSP, CNR Torino Italy). PVY Pa36 is an NTN recombinant type. In addition, the 5′ genomic region of PVY Pa36 was PCR amplified using the appropriate primers (Supplementary Table 1) and sequenced to identify which NTN recombinant type it belonged to Green et al. (2017). The sequence shows that the P1 genomic region derives from a PVYN isolate; thus, Pa36 is an NTNa recombinant type (Green et al., 2017).
sgRNA Design and Plasmid Construction
Potato eIF4E1 sequences were retrieved from the NCBI database and aligned to identify conserved Cas9 targets. RNA-seq data of the potato cv. Desirée4 (Ali et al., 2014) were analyzed with the online free software package Galaxy5 (Afgan et al., 2016) to evaluate the presence of SNPs in the selected target sequences. Bowtie2 was run using as reference gene the eIF4E1 sequence FN666436 and the output (BAM file) used to visualize the distribution of the RNA-seq data on the eIF4E1 gene using the Integrative Genomic Viewer (IGV) software6. Routine molecular cloning procedures were followed for plasmid construction. The HBT-pcoCas9-SteIF4E target6 (HBT-pcoCas9-SteIF4E_6) construct was created from the HBT-pcoCas9 plasmid vector (Addgene_52254; Li et al., 2013).
First, the sgRNA expressing cassette was generated by overlapping PCR on the pUC119-gRNA plasmid (Addgene_52255; Li et al., 2013) using appropriate primers (Supplementary Table 1). The first two PCR reactions were assembled using PrimerF1-XhoI + PrR1-SteIF4E-2 and PrF2-SteIF4E-2 + PrimerR2, respectively. In the final PCR reaction, products of the above PCRs were mixed and used as templates with PrimerF1-XhoI + PrimerR2 primers. The final PCR product was digested with XhoI and AatII and cloned into the HBT-pcoCas9 plasmid.
Protoplasts Isolation, Transfection, and Regeneration of Potato Plants
Leaf mesophyll protoplasts of S. tuberosum cv. Desirée were isolated and cultured following the method and media reported by Tavazza and Ancora (1986) with slight modification. Briefly, before enzyme digestion, leaves were preplasmolysed in the culture medium for 0.5–1 h, and then chopped and incubated overnight in the enzyme solution in the dark at 28°C. After filtration through 297 and 88 μm nylon sieves, the filtrate was diluted with an equal volume of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl and 2 mM MES, pH 5.8) and centrifuged at 70 × g for 5 min. The pellet was washed twice with W5 solution and centrifuged at 70 × g for 5 min. The harvested protoplasts were resuspended in W5 solution, stored at room temperature (RT), flat, for 2–3 h in the dark. After centrifugation at 70 × g for 5 min, protoplasts were resuspended in MMG solution (4 mM MES pH 5.7, 0.4 M mannitol, 15 mM MgCl2) to bring the final concentration to 1.6 × 105 protoplasts ml–1 as calculated by microscope on a hemocytometer. PEG-mediated transfection was performed according to Yoo et al. (2007) with modifications. Briefly, 1.6 × 105 protoplasts (200 μl) were mixed with 10 μg of HBT-pcoCas9-SteIF4E_6 (20 μl) and an equal volume (220 μl) of 40% w/v PEG 4000 (Duchefa, Haarlem, The Netherlands), mixed gently and incubated at RT for 10 min. Transfection reaction was stopped by adding 7.5 ml of W5 solution to protoplasts solution followed by centrifugation 5 min at 70 × g. Protoplasts were resuspended carefully on 1 ml of culture medium (Tavazza and Ancora, 1986) containing 50 μg ml–1 of cefotaxime (Claforan), placed in 12-well cell culture plates, and incubated for 72 h at 24°C in the dark. Protoplasts culture and regeneration were carried out as reported by Tavazza and Ancora (1986).
Detection of CRISPR-Cas9 Potato Mutants
Multiplex PCR-based high-resolution fragment analysis (MHRFA) (Nicolia et al., 2015) was used to screen the potato regenerants. DNA extracted as described in Edwards et al. (1991) was used to assemble four PCR reactions using a 5′ 6-FAM- or VIC-labeled forward primer (eIF4E_LEI_Fw20_Fam or eIF4E_LEI_Fw20_Vic) together with a 5′G stabilized reverse primer (eIF4E_LEI_Rev201G or eIF4E_LEI_Rev246G) (Supplementary Table 1). The PCR conditions were: 94°C 2’, 35 cycles (94°C 30″, 60°C 30″, 72°C 30″) followed by a 30′ final extension at 72°C. The amount and correctness of PCR products was evaluated by agarose gel electrophoresis and pools of four reactions were equally mixed, run on the Applied Biosystems™ 3130xl Genetic Analyzer (Genechron)7 and analyzed using the Peak Scanner Software v1.0 (Applied Biosystems®). Selected mutants were further investigated by ICE (Hsiau et al., 2019) analyzing the PCR fragments generated by the primers eIF4E_LEI_Fw20 × eIF4E_LEI_Rev246 or eIF4E_LEI_Fw20 × eIF4e_LEI_Rev297 (Supplementary Table 1).
Gene Expression Analysis of the eIF4E Gene Family
For gene expression analysis, in vitro multiplicated wild-type (wt) and eIF4E1 mutated potato plants were adapted on soil and grown in a growth chamber set with a photoperiod of 16/8 h (light/dark), and 22/25°C (night/day) temperature regime. The third leaf from the apical meristem was sampled from three plants per genotype and after carefully removing the midvein, all leaf lamina samples were immediately frozen in liquid nitrogen. Total RNA was extracted with the RNeasy Plant Mini Kit (QIAGEN) according to manufacturer’s specifications, and 1 μg was used for cDNA synthesis with QuantiTect Reverse Transcription Kit (QIAGEN). RT-qPCR was performed with SYBR Green PCR Master Mix (Applied Biosystems) on ABI Prism 7900HT (Applied Biosystems) according to the manufacturer’s instructions. Assays were performed in triplicate using 0.3 μl of diluted cDNA (1:5) and 300 nM of both primers in a reaction volume of 10 μl in 384 multiwell plates. PCR primers were designed using the Primer-Blast software.8 All primers used for RT-qPCR are listed in Supplementary Table 1. Relative quantification values were calculated with the 2–ΔΔCt method (Livak and Schmittgen, 2001) using eEF1-a (AB061263) for normalization (Nicot et al., 2005; Joseph et al., 2018). The relative expression levels of target genes in mutant lines were normalized respect to wt. All the RT-qPCR values shown are from six independent biological replicates from two independent experiments. Kruskal–Wallis statistical test was applied to determine significant differences compared to the wt. P < 0.05 was considered statistically significant.
Virus-Resistance Analysis
In vitro multiplicated potatoes were adapted and grown in greenhouse with a photoperiod of 16/8 h (light/dark) and 17/21–25°C (night/day) temperature regime. PVY Pa36 was initially multiplicated in Nicotiana benthamiana plants and frozen infected leaf stocks were used as inoculum source. Two leaves of each potato plant, 10–15 cm in height, were inoculated with PVY-infected leaf stock grounded in 50 mM phosphate buffer pH 7.0, 5 mM Na2SO3 and 1% celite. Four independent infection experiments were performed. The upper leaves of plants were sampled at 16–18 and 28–31 days post inoculation (dpi). Infections were assessed by Northern blot analysis or by RT-qPCR. For Northern blot, total nucleic acids (TNA) were extracted from sampled leaves as described in Brunetti et al. (2001); the probe was a DIG labeled PCR fragment obtained with primers PVY-Fw9054 and PVY-Rev9444 (Supplementary Table 1) on a cDNA derived from reverse transcription of TNA extracted from a PVY Pa36 infected Nicotiana tabacum plant. RT-qPCR analysis was performed as described above for eIF4E gene family using PVY Univ Fw and PVY Univ Rev primers (Dai et al., 2013) and eEF1-a (AB061263) for normalization (Nicot et al., 2005; Joseph et al., 2018); primers are listed in Supplementary Table 1. Kruskal–Wallis statistical test was applied to determine significant differences compared to the wild type. P < 0.05 was considered statistically significant.
VPg Sequencing
Two micrograms of TNA extracted from PVY Pa36-infected leaves were treated with DNaseI Amplification Grade (Invitrogen) and reverse transcribed with SuperScript III RT (Invitrogen) using random hexamers as recommended by supplier. First strand cDNA was used as template in PCR reactions to amplify the VPg region with Phusion High-Fidelity DNA Polymerase (Thermo Scientific) and primers PVY_NTNab_VPg_Fw and PVY_NTNab_VPg_Rev (Supplementary Table 1). PCR conditions were: 98°C 30″, 35 cycles (98°C 10″, 62°C 20″, 72°C 20″) followed by a 5′ final extension at 72°C. The amplified fragments were sequenced on both strands with the same primers used for PCR.
Results and Discussion
Identification of the Target Sequence for Cas9-Mediated eIF4E1 Knockout
In order to verify whether the eIF4E1 protein is required or essential for PVY infection and to which extent eIF4E1 Cas9 inactivation can broaden the PVY resistance spectrum, it was crucial to exclude: (a) Cas9 targeting of other genes of the potato eIF4E family, in particular, its paralog eIF4E2; (b) possible effects from unwanted truncated proteins acting as dominant-negative mutants.
The BLAST homology search shows that the potato eIF4E1, eIF4E2, eIF(iso)4E and nCBP genes are located on chromosome 3, 2, 9, and 10, respectively. Notably, the 5′ end of the eIF4E1 gene is divergent from the other members of the eIF4 gene family. In addition, previous works showed that N-terminal truncated (aa 38–51) eIF4E proteins retain the ability to bind the m7GTP cap analog and VPg (Monzingo et al., 2007; Ashby et al., 2011; Walter et al., 2019). These data supported the idea that a frameshift mutation or the introduction of a stop codon in the first 114 nt (38 aa) of the gene should not generate a dominant-negative mutant. Thus, we decided to identify Cas9 targets in the 5′ of the eIF4E1 gene for both aspects mentioned above.
As common tetraploid potato cultivars display a highly heterozygous genome, RNAseq data from cv. Desirée (Ali et al., 2014) were used to assess inter-allelic polymorphism. Several single nucleotide polymorphisms (SNPs) were identified along the eIF4E1 gene. In particular, four SNPs were present in the first 114 nt, indicating that the Desirée genome possesses at least two eIF4E1 alleles (Supplementary Figure 1). The first 114 nt of eIF4E1 contain eight potential (20 nt) Cas9 targets, six in the plus and two in the minus strand. All potential Cas9 targets except target-6 (GTAGACGATGAACTTGAAGAAGG) contain at least one SNP (Supplementary Figure 2A) in the so-called crRNA “seed” region (Semenova et al., 2011; Wiedenheft et al., 2011; Jiang et al., 2015).
The target-6 sequence, which has 100% homology among eIF4E1 alleles, the best score as assessed by CRISPR-P analysis9 (Supplementary Figure 2B) and no significant homology with eIF4E2 sequences (Supplementary Figure 3) was chosen to design the Cas9-guide RNA (gRNA_6).
Potato eIF4E1 Knockout Is Compatible With Life
High heterozygosity of potatoes coupled with the tetrasomic inheritance (Muthoni et al., 2015) makes outcrossing of the transgene challenging to achieve without affecting the agronomic characteristics of the cultivar. For this reason, to generate eIF4E1 mutated Desirée plants, Cas9 and gRNA-6 were transiently expressed in protoplasts using the HBT-pcoCas9-SteIF4E_6 construct (Supplementary Figure 4). Four hundred seventy-nine and three hundred fifty-nine shoots were regenerated from two independent transfection experiments. Regenerants were screened for eIF4E1 mutations by multiplex PCR-based high-resolution fragment analysis (MHRFA) (Figure 1A). In total, 18 mutated clones were identified with an overall mutagenesis frequency of 2.1% (Supplementary Figure 5). However, a marked difference in mutagenesis frequency was observed between the two experiments, 3.3% in the first and 0.6% in the second transfection experiment. The mutation frequency found in the first experiment is in the range with that reported previously (Andersson et al., 2017; Veillet et al., 2019). Thus, the low number of mutated plants obtained in the second experiment seems unrelated to target-6 efficiency. Collectively, eleven clones possessed one mutated eIF4E1 copy and seven two mutated ones; no plants were mutated in all the four copies. Lines 47, 122, 1,554, and 1,612 were characterized by frameshift mutations in two of the four eIF4E1 copies (Supplementary Figure 5). Several physical parameters can influence transfection-mediated Cas9 targeting. Andersson et al. (2017) showed that PEG, DNA, protoplasts concentration and DNA incubation time could affect the chance of targeting all four potato gene copies. In particular, they showed, using three different Cas9 targets, that fully knocked out plants could not be recovered with 40% PEG (Andersson et al., 2017). Thus, it could be that our inability to obtain fully knocked out plants in a single round of transfection was derived by the PEG concentration used.
FIGURE 1

Molecular screening of potato plants regenerated from protoplasts and characterization of line 122 possessing two copies of the eIF4E1 gene knocked out. (A) Schematic representation of high-resolution fragment analysis (MHRFA) to detect Cas9-generated indels. The scheme shows the results expected from wild-type plants. Relative positions of oligonucleotides and CRISPR-Cas9 target site (in red) on Desirée genome. Blue and green represent labeling of forward primers Fw1 and Fw2 with FAM or VIC, respectively. Each peak corresponds to a PCR fragment the size of which is indicated in the upper bar. (B) MHRFA of potato line 122. Identification of Δ2 mutations. Each peak corresponds to a PCR fragment the size of which is indicated in the upper bar. The shape of the wild-type peak identifies the presence of two alleles differing in nucleotide polymorphism. (C) Potential translation of eIF4E1 Δ2 alleles. Open reading frames are highlighted in red. Stop, premature stop codons.
Clone 122, carrying two eIF4E1 copies characterized by a deletion of two nucleotides (Δ2, Δ2, wt, wt) as bolstered by MHRFA analysis (Figure 1B) and showing a phenotype similar to wt plants was analyzed in more detail. The eIF4E1 gene region spanning the introduced Cas9 mutation was amplified from genomic DNA and evaluated by ICE analysis. The amplified genomic DNA fragment confirmed that two out of the four copies of the eIF4E1 gene had a Δ2 mutation (Supplementary Figure 6A). Interestingly, when the same fragment was amplified from the cDNA 99% of the fragments were derived from wt transcripts (Supplementary Figure 6B). Two non-mutually exclusive hypotheses can explain the ICE results: (a) the two copies of the mutated gene were transcribed at a very low level; (b) the eIF4E1 mutated transcripts were potentially targeted to degradation, possibly by the Nonsense-mediated mRNA Decay (NMD) by the presence of premature termination codons (PTCs) generated by the Δ2 frame-shift (Figure 1C). In plants, similarly to mammals, PTCs, long 3′UTR, and introns > 50–55 nt downstream of TCs can trigger NMD (Shaul, 2015) and these three features are all generated in the Δ2 alleles.
Line 122 which does not contain plasmid derived sequences integrated in its genome was further evaluated for the absence of Cas9-induced mutations in the eIF4E2 gene. The eIF4E2 sequence encompassing the potential site (target-6) for Cas9 cutting was amplified and sequenced. No indels or mutations were identified in the eIF4E2 sequence overlapping the potential Cas9 target-6 site (see below).
Based on the overall evidence, we selected clone 122 for the second round of protoplasts transfection-plant regeneration process. Two hundred sixty-nine shoots were regenerated. MHRFA screening identified 18 plants (C series) with additional mutated eIF4E1 copies (Supplementary Figure 7). The overall mutagenesis efficiency was 6.7%. Half of the mutated plantlets were mutated in all eIF4E1 alleles, three of which C14, C29, and C249 were out-of-frame (OF) mutants. Line C249, which showed premature senescence and leaf abscission, was not used in further experiments. The altered phenotype of line C249 appears unrelated to the eIF4E1 mutation as lines C14 and C29 did not show this phenomenon, thus suggesting somaclonal variations occurring during the regeneration process the cause. Fossi et al. (2019) reported that all potato (Desirée) plants regenerated from protoplasts regardless of showing an altered phenotype have their genomes affected by aneuploidy or structural chromosomal changes.
Lines C14 and C29 (Figure 2A) as bolstered by MHRFA analysis (Figure 2B) were further investigated by ICE analysis (Figure 2C) confirming that the eIF4E1 genotype of C14 and C29 was (Δ2, Δ2, Δ2, Δ2) and (Δ2, Δ2, Δ4, + 1), respectively, and that, as observed in the progenitor line 122, no mutations were present in the eIF4E2 sequence overlapping the potential Cas9 target-6 site (Supplementary Figure 8). Thus, only eIF4E1 and not its paralog eIF4E2 was mutated in the C14 and C29 lines. Our data show that the knockout of the potato eIF4E1 gene is compatible with life. This result is in agreement with the data obtained on another Solanaceae species, the Solanum lycopersicon (Piron et al., 2010).
FIGURE 2

Characterization of the tetraploid potato cv. Desirée plants knockout in the eIF4E1 gene. (A) C14 and C29 potato plant lines knockout in the eIF4E1 gene together with their progenitor line 122, and wild-type plant. (B) High-resolution fragment analysis of the four eIF4E1 gene copies present in the eIF4E1 knockout plant lines C14 and C29. (C) ICE (https://ice.synthego.com/#/) analysis of PCR fragments amplified from genomic DNA of the C14 and C29 lines.
A Non-negligible Fraction of the Potato Edited Plants Have Integrated Vector Sequences in Their Genome
Potato genome editing experiments based on the transfection of protoplasts with plasmid DNA can led to stable integration of vector sequences into the plant genome (Clasen et al., 2016; Andersson et al., 2017). The number, size, and area of each eIF4E1 fragment/peak inferred from MHRFA analysis of mutated clones (Supplementary Figure 5 and data not shown) suggest that plasmid DNA integration, if present, was not at the eIF4E1 target site. To verify if plasmid DNA was integrated into the genome of eIF4E1 mutated plants, two different regions of the HBT-pcoCas9-SteIF4E_6 construct encompassing Cas9 and gRNA-6 sequences were investigated by PCR analysis. Potato clones possessing two (C13, C28, and C172) and four (C14, C29, and C173) mutated eIF4E1 copies were initially selected. Only C14 and C29 were transgenic (Supplementary Figure 9). To know to which extent plasmid sequences integration occurred, all 18 mutated potato C clones were analyzed for the presence of HBT-pcoCas9-SteIF4E_6 sequences. PCR analysis showed that in 36% of the potato clones, the vector sequences were integrated into the genome (data not shown). The same percentage of transgenics was observed in plants with three or four eIF4E1 copies mutated. Thus, the percentage of vector integration does not seem to correlate with the number of mutated eIF4E1 copies.
Plasmid DNA integration could originate from DNA breaks generated during the transfection-regeneration process (Fossi et al., 2019), Cas9 target activity (Andersson et al., 2017), and micro-homology between plasmid and potato sequences. Our data suggest that DNA integration events were independent of Cas9 activity on the eIF4E1 target. Differently from our data, Andersson et al. (2017) reported that about 10% (3 out 29) of regenerated potato plants mutated in the granule-bound starch synthase gene had insertion of plasmid DNA sequences into the predicted Cas9 target site; however, no data were reported for random integration of plasmid sequences into the potato genome (Andersson et al., 2017). Similarly, about 10% of potato plants mutated in the amylose-producing StGBSSI gene have been shown to have plasmid-derived sequences integrated into their genome (Veillet et al., 2019). In another potato gene editing work, Clasen et al. (2016) reported that approximately 60% of the TALEN-edited potato plants for the vacuolar invertase gene (VInv) had vector sequences integrated; these transgenic plants were similarly distributed among those that had one to four edited VInv gene copies. Thus, the 36% frequency of vector DNA integration observed here is within the average of those previously reported (Clasen et al., 2016; Andersson et al., 2017; Veillet et al., 2019). Recently, Andersson et al. (2018) showed that transgene-free edited potato plants could be obtained using the Cas9 ribonucleoprotein complex, thus overcoming the bottleneck of vector DNA integration. In our case, the frequency of vector DNA integration was limiting in obtaining transgene-free eIF4E1 KO plants. Our results together with previous data on potato-edited plants through plasmid-mediated protoplasts transfection suggest that, whenever possible, the use of DNA vectors should be avoided, thus maximizing the output of transgene-free edited potato plants.
The Disruption of eIF4E1 Does Not Affect the mRNA Steady-State Levels of Other eIF4E Family Members
The knockout or the knockdown of an eIF4 family member has been shown in some cases to modulate the expression of another gene family member, suggesting the existence of possible regulatory feedback mechanisms (Duprat et al., 2002; Combe et al., 2005; Lellis et al., 2019). An early indication (Duprat et al., 2002) pointed out a possible regulation at the post-transcriptional level. However, an mRNA steady-state level analysis on the consequence of KO of an eIF4 member to the other family members is scarce.
Here, RT-qPCR analysis was conducted to evaluate the possible impact of eIF4E1 disruption on the mRNA steady-state level of eIF4E2, eIF(iso)4E, and nCBP. As shown in Figure 3, wt, 122, C14, and C29 plants accumulate similar amounts of transcripts from each tested gene. Thus, at least under the conditions tested, knockout of the eIF4E1 gene does not impact the mRNA steady-state levels of the other three potato cap-binding proteins. It remains to be established if a post-transcriptional compensatory regulation mechanism such as that described by Duprat et al. (2002) in the eIF(iso)4E Arabidopsis null mutant also applies to the eIF4E1 potato mutants. The compensatory effect among eIF4s is not only associated with an increased expression of another family member. A double eIF(iso)4G1 and eIF(iso)4G2 Arabidopsis KO mutant was shown to accumulate less eIF(iso)4E (Lellis et al., 2019). In tobacco, Combe et al. (2005) showed that the reduction of about 60% of the eIF(iso)4E-a and -b proteins, through an antisense approach, led to a compensatory increase in eIF4E levels of approximately 200%. However, the impact of the reduction of eIF(iso)4E-a and -b proteins on the eIF4E mRNA level was not investigated. Conversely, the down-regulation of the eIF4E does not trigger a reciprocal increase in eIFiso4E levels (Combe et al., 2005). Thus, our data are in line, at least for the lack of variation in eIF(iso)4E, with those observed by Combe et al. (2005). To our knowledge, this is the first time that the consequence of eIF4E1 KO on the mRNA steady-state levels of IF4 family members was investigated.
FIGURE 3
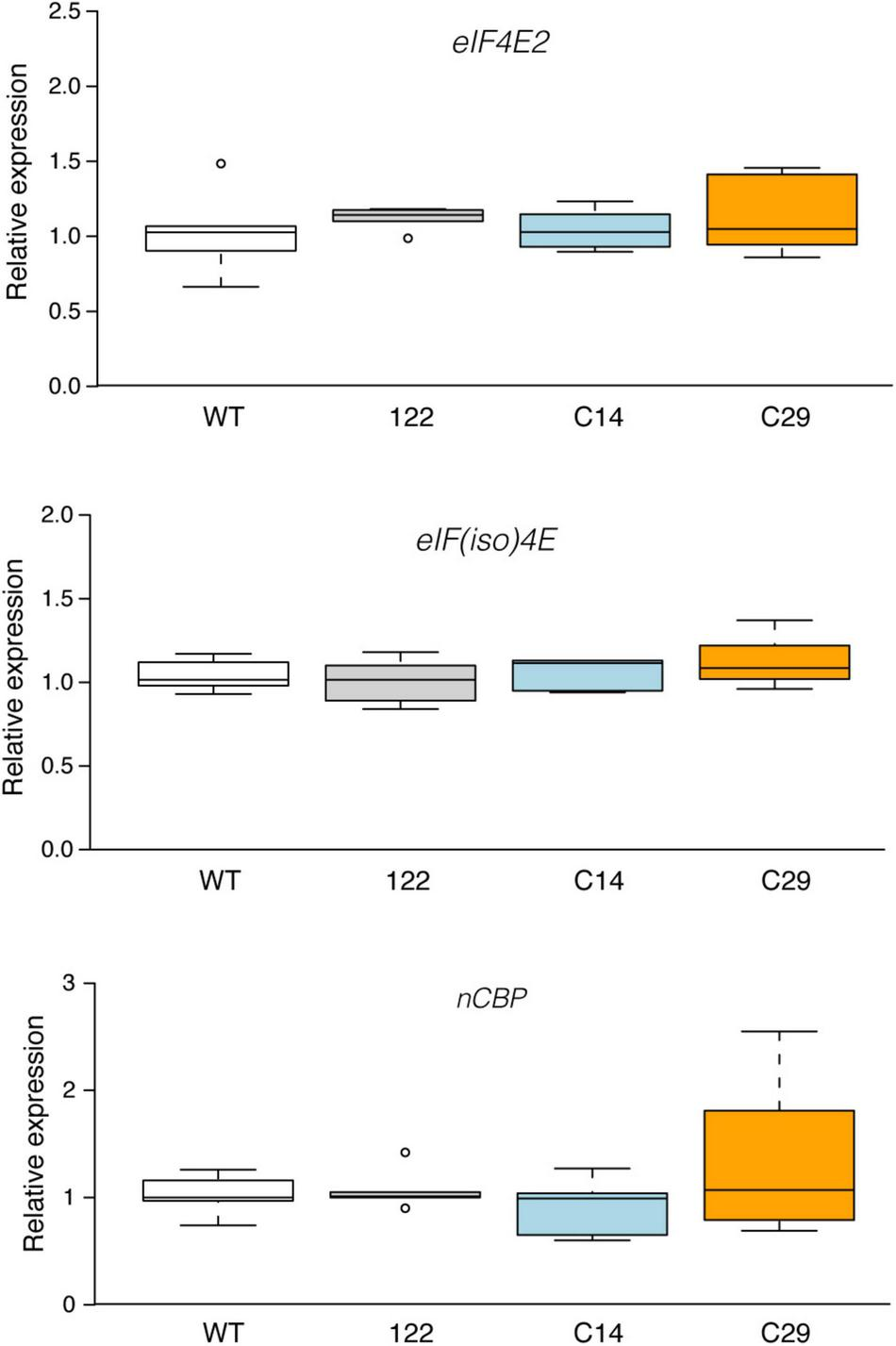
Expression analysis of eIF4E2, eIF(iso)4E, and nCBP genes in eIF4E1 mutated potato plants. Relative expression levels of the target genes were normalized to the wild-type. Six independent biological replicates from two independent experiments were tested for each genotype. Box plot was generated using the online resource (http://shiny.chemgrid.org/boxplotr/; Spitzer et al., 2014). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; dots represent outliers. Statistical analyses according to Kruskal-Wallis (P < 0.05) reveal no significant differences between wild-type and knockout lines.
eIF4E1 Knockout Reduces Potato Virus Y Pa36 Multiplication and Ameliorates Virus-Induced Symptoms
Virus resistance, in plants mutated for a gene of the eIF4E family, can range from immunity to a reduction in viral accumulation, and can be restricted to a few viral isolates or show a broad spectrum depending on: (a) the gene(s) of the eIF4E family mutated; (b) the type of mutation introduced; (c) the viral isolates used to challenge the plants (Bastet et al., 2017).
The recessive nature of eIF4E mediated resistance predicts that only potato plants mutated in all the eIF4E1 gene copies can show resistance to PVY. Thus, we expected line 122 to behave like wt plants regard to PVY virus resistance.
To initially test the degree of virus resistance of eIF4E1 mutated potatoes, three wt, 122, C14, and C29 plants were mechanically inoculated with an isolate of the PVYNTNa recombinant type, the PVY Pa36. At 28 days post-inoculation (dpi), wt and 122 plants showed mosaic on the upper leaves, and a slight distortion of leaves margin, C14 plants a pale yellowing, and C29 had very mild symptoms (Figure 4A). Total nucleic acids were extracted from each plant and analyzed by Northern blot for the presence of viral genomic RNA (Figure 4B). In agreement with the recessive nature of eIF4E resistance, line 122, the progenitor of C14 and C29, possessing two unmutated copies of the eIF4E1 gene, was susceptible to PVY Pa36 like the wt plants. Conversely, in both fully mutated eIF4E1 lines, PVY Pa36 accumulation was reduced, albeit with some plant-to-plant difference in the C14 line (Figure 4B).
FIGURE 4

PVY Pa36 multiplication and symptom development are reduced in eIF4E1 knockout plants. (A) Symptoms in PVY Pa36 inoculated plants at 28 dpi. (B) Northern blot of TNA extracted from PVY Pa36 inoculated plants at 28 dpi. C, uninoculated wild-type plant.
A further experiment was conducted by challenging seven wt, C14, and C29 plants with PVY Pa36 and analyzing virus accumulation by RT-qPCR at 16 and 29 dpi. At 16 dpi, the wt but not C14 and C29 plants showed initial distortion of leaves margin of the non-inoculated upper leaves. RT-qPCR analysis showed a reduction of virus accumulation (difference in the medians) in both eIF4E1 mutated lines; in C29 plants, the difference was statistically supported (Figure 5). At 29 dpi, a more pronounced distortion of the leaves margin and some localized yellowing of the leaves were present on the wt plants. Conversely, no apparent symptoms were observed in C14 and C29 plants. Coherently with the previous experiment, C14 and C29 plants accumulated less virus than wt plants (Figure 5), thus supporting the notion that the eIF4E1 KO plants interfere with virus multiplication and-or spread.
FIGURE 5
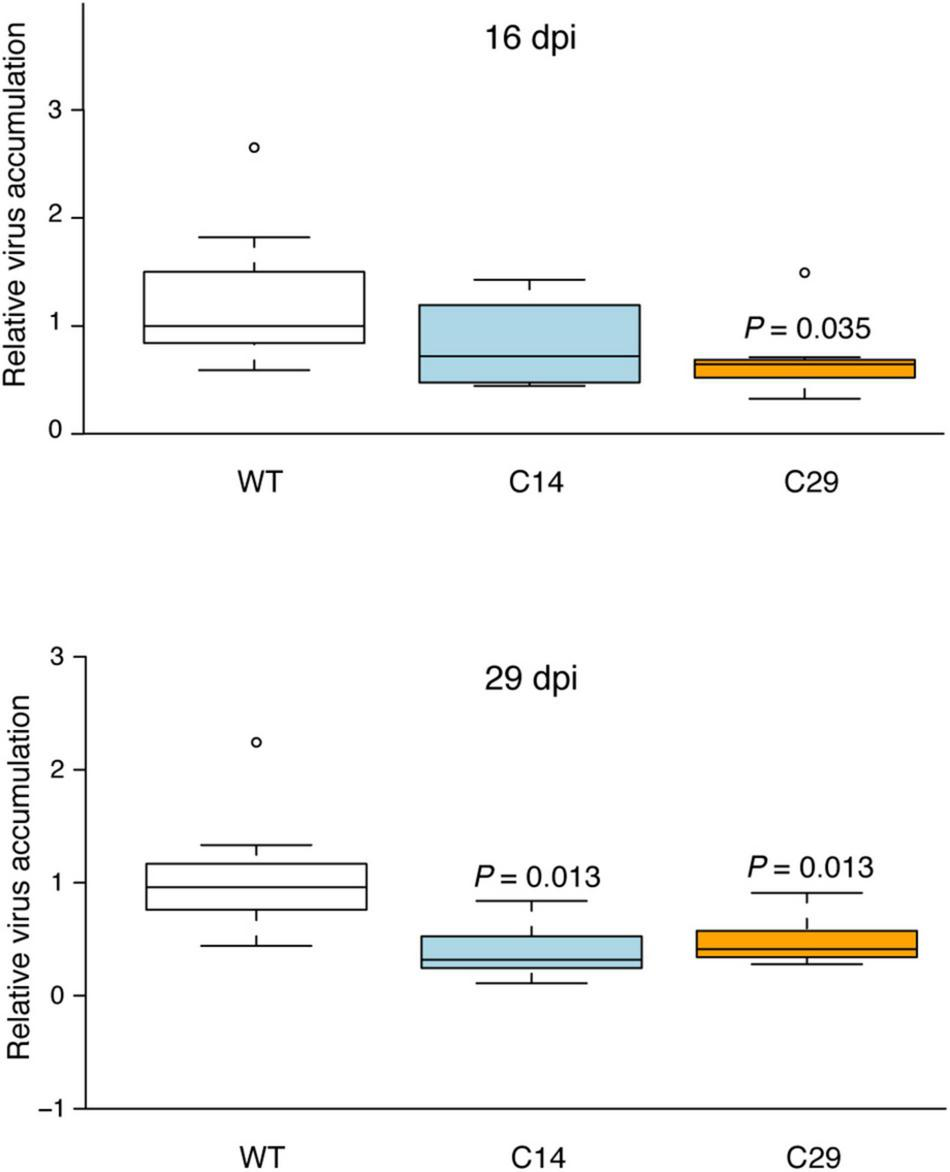
Viral accumulation of PVY Pa36 in eIF4E1 mutated plants at 16- and 29-days post-inoculation (dpi). Viral accumulation was quantified by RT-qPCR. Seven plants were tested for each genotype. Box plot was generated using the online resource (http://shiny.chemgrid.org/boxplotr/; Spitzer et al., 2014). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; dots represent outliers. P-value of samples with significant difference from wild-type according to Kruskal-Wallis statistical analyses (P < 0.05) is shown above the box.
The reduction of virus accumulation in eIF4E1 mutated plants was further confirmed in two additional experiments (Supplementary Figures 10, 11). Again, only mild symptoms were observed in eIF4E1 mutated potato plants at the end of the experiments (30–31 dpi).
The adaptation of PVY to eIF4E-mediated resistance in tobacco, pepper, and tomato has been reported to involve mutations in the VPg, most often in its central domain (Moury et al., 2004; Charron et al., 2008; Janzac et al., 2014). We then investigated whether the partial resistance observed was due to a rapid virus evolution to replicate and move in the absence of eIF4E1. For this purpose, TNA extracted from two C14 and C29 plants at 28–30 dpi were used to amplify and sequence the viral genome region encoding the VPg. All four virus sequences were identical to the original PVY Pa36 (Supplementary Figure 12). In addition, leaves sap of PVY Pa36-infected C14 plants at 28 dpi was used to back-inoculate four wt, C14, and C29 plants. Again, only mild symptoms were visible on C14 and C29 plants at the end of the experiment (31 dpi) (Supplementary Figure 13). The absence of mutations in the VPg sequence of viruses isolated from C14 and C29 plants, coupled with the inability to induce resistance-breaking symptoms in the back-inoculation experiment, suggests that the reduced inhibition of PVY Pa36 accumulation is not due to the emergence of viral resistance-breaking genotypes. The overall data show that potato plants KO for the eIF4E1 are partially resistant to PVY Pa36, an isolate belonging to the PVYNTNa recombinant type (Green et al., 2017). The eIF4E1 KO lines showed a marked reduction of virus-induced symptoms although the resistance was only partial. It remains to be established whether eIF4E1 KO can prevent potato tuber necrosis ringspot disease (Beczner et al., 1984).
Our data are in line with those obtained on tomato mutants for the eIF4E1 gene. A tomato eIF4E1 frameshift mutant generated by CRISPR-Cas9 editing and encoding the first 144 aa of the protein showed a reduced virus accumulation (Atarashi et al., 2020). Similarly, a tomato eIF4E1 EMS splicing mutant coding for a truncated eIF4E1 protein impaired in cap-binding activity confers resistance to the PVY-LE90 isolate (Piron et al., 2010). Further experiments with different potyviruses will be needed to understand the breadth of resistance of C14 and C29 lines. Preliminary infection data show that C14 and C29 are susceptible as wt and 122 plants to the potato virus V (Lucioli and Tavazza unpublished results). We did not expect Desirée eIF4E1 KO plants to have a broad-spectrum of resistance to potyviruses. Indeed, in tomatoes, broad-spectrum resistance to potyviruses has been shown to be hampered by eIF4E gene redundancy (Piron et al., 2010; Mazier et al., 2011; Gauffier et al., 2016; Atarashi et al., 2020). As suggested previously (Gauffier et al., 2016; Bastet et al., 2017), the elective strategy to achieve broad-spectrum resistance to potyviruses should mimic the functional eIF4E1 resistant allele found in natural diversity such as that found in pepper germplasm. Non-synonymous mutations characterize these eIF4E1 resistant alleles, most if not all the mutated proteins being impaired in their interaction with potyviral VPg (Wang and Krishnaswamy, 2012). It should be noted that this genetic variability is unknown in potato germplasm and that the obtainment of edited potato plants to mimic resistance alleles found in other Solanaceae species could be challenging (Gallois personal communication).
Here we show that the knockout of the potato eIF4E1 confers a limited resistance to PVY Pa36, suggesting that the eIF4E1 is required but not essential for virus infectivity. It remains to be established whether the reduced accumulation of PVY Pa36 in eIF4E1 KO potato plants results from a partial impairment of viral replication or spread. Our data support the idea that PVY Pa36 might use another eIF4E protein in addition to eIF4E1 for its infection cycle. Examples of potyviruses whose VPg can interact with both eIF4E1 and eIF4E2 proteins have been described (Mazier et al., 2011). In particular, PVY-LYE84 VPg, unlike that of PVY-LE90, has been shown to interact with both tomato eIF4E1 and eIF4E2 protein (Mazier et al., 2011). Similar results were observed with a PVYNTN isolate in potatoes (Lebedeva et al., 2021). In addition, the requirement of simultaneous mutations of the pepper eIF4E and eIF(iso)4E genes to confer Chili veinal mottle virus and Pepper veinal mottle virus resistance suggests that these viruses may utilize both proteins (Ruffel et al., 2006; Hwang et al., 2009; Rubio et al., 2009).
Conclusion
Our data show that the knockout of the eIF4E1 can broaden the resistance spectrum of the elite cv. Desirée conferred by the Ny gene to PVYO and that eIF4E1 KO does not alter the mRNA steady-state level of other members of the potato eIF4E gene family. The limited resistance observed to PVY Pa36 suggests that this virus could profitably use other proteins of the eIF4E family beyond eIF4E1. A more refined editing strategy based on mimicking natural eIF4E resistance alleles will be required to extend the spectrum of resistance to most potyviruses (Bastet et al., 2017, 2019); in addition, this approach should avoid the potential exposure of eIF4E knocked out plants to threats of potyviruses capable of recruiting alternative eIF4E copies, as recently shown to occur in Arabidopsis (Zafirov et al., 2021). New gene-editing approaches (Bastet et al., 2019; Veillet et al., 2020) and a better understanding of the fine interplay between the eIF4E proteins, VPgs (Lebedeva et al., 2021) and potyviruses infection should help to achieve this goal. At the same time, to preserve the integrity of the elite potato cultivars’ traits, improved transformation-regeneration protocols will be required to minimize somaclonal variations incurring during potato plants regeneration (Fossi et al., 2019).
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
JB, KF, and MT conceived the idea and designed the strategy. RT performed the potato transfection and regeneration of Cas9 mutants. AL carried out the screening and the molecular analyses of the regenerated plants and performed the PVY infections and northern analyses. SB carried out the RT-qPCR analysis of eIF4E expression and PVY resistance. MT, AL, RT, and SB analyzed the data. MT supervised the work and wrote the manuscript. RT, AL, SB, KF, and JB helped to improve the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially funded by the Italian Ministry of Agricultural, Food, and Forestry Policies, BIOTECH Programme D.M. 15924.
Acknowledgments
We thank M. Ludman for the construction of the HBT-pcoCas9-SteIF4E_6 plasmid, D. Giorgi and S. Lucretti for the flow cytometric analysis. We thank Prof. K. S. Browning for helpful suggestions and material support, and V. Ilardi, C. Lico, G. Aprea, and L. Nardi for helpful suggestions and technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.873930/full#supplementary-material
Footnotes
1.^http://www.fao.org/faostat/en/#data/QC
2.^http://varieties.ahdb.org.uk/varieties/view/Desiree
4.^http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-1712/samples/?full=truet
6.^http://software.broadinstitute.org/software/igv/
References
1
AfganE.BakerD.Van den BeekM.BlankenbergD.BouvierD.ČechM.et al (2016). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update.Nucl. Acids Res.44W3–W10. 10.1093/nar/gkw343
2
AliA.AlexanderssonE.SandinM.ResjöS.LenmanM.HedleyP.et al (2014). Quantitative proteomics and transcriptomics of potato in response to Phytophthora infestans in compatible and incompatible interactions.BMC Genomics15:497. 10.1186/1471-2164-15-497
3
AnderssonM.TuressonH.NicoliaA.FältA. S.SamuelssonM.HofvanderP. (2017). Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts.Plant Cell Rep.36117–128. 10.1007/s00299-016-2062-3
4
AnderssonM.TuressonH.OlssonN.FältA. S.OhlssonP.GonzalezM. N.et al (2018). Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery.Physiol. Plant.164378–384. 10.1111/ppl.12731
5
ArcibalE.Morey GoldK.FlahertyS.JiangJ.JahnM.RakotondrafaraA. M. (2016). A mutant eIF4E confers resistance to potato virus y strains and is inherited in a dominant manner in the potato varieties atlantic and russet norkotah.Am. J. Potato Res.9364–71. 10.1007/s12230-015-9489-x
6
AshbyJ. A.StevensonC. E.JarvisG. E.LawsonD. M.MauleA. J. (2011). Structure-based mutational analysis of eIF4E in relation to sbm1 resistance to pea seed-borne mosaic virus in pea.PLos One6:e15873. 10.1371/journal.pone.0015873
7
AtarashiH.JayasingheW. H.KwonJ.KimH.TaninakaY.IgarashiM.et al (2020). Artificially edited alleles of the eukaryotic translation initiation factor 4E1 gene differentially reduce susceptibility to cucumber mosaic virus and potato virus Y in tomato.Front. Microbiol.11:3075. 10.3389/fmicb.2020.564310
8
BaeblerŠCollA.GrudenK. (2020). Plant molecular responses to Potato Virus Y: A continuum of outcomes from sensitivity and tolerance to resistance.Viruses12:217. 10.3390/v12020217
9
BastetA.RobagliaC.GalloisJ. L. (2017). eIF4E resistance: natural variation should guide gene editing.Trends Plant Sci.22411–419. 10.1016/j.tplants.2017.01.008
10
BastetA.ZafirovD.GiovinazzoN.Guyon-DebastA.NoguéF.RobagliaC.et al (2019). Mimicking natural polymorphism in eIF 4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses.Plant Biotechnol. J.171736–1750. 10.1111/pbi.13096
11
BecznerL.HorváthJ.RomhanyiI.FörsterH. (1984). Studies on the etiology of tuber necrotic ringspot disease in potato.Potato Res.27339–352.
12
BrowningK. S.Bailey-SerresJ. (2015). Mechanism of cytoplasmic mRNA translation.Arabidopsis Book13:e0176. 10.1199/tab.0176
13
BrunettiA.TavazzaR.NorisE.LucioliA.AccottoG. P.TavazzaM. (2001). Transgenically expressed T-Rep of tomato yellow leaf curl Sardinia virus acts as a trans-dominant-negative mutant, inhibiting viral transcription and replication.J. Virol.7510573–10581. 10.1128/JVI.75.22.10573-10581.2001
14
CavatortaJ.PerezK. W.GrayS. M.Van EckJ.YeamI.JahnM. (2011). Engineering virus resistance using a modified potato gene.Plant Biotechnol. J.91014–1021. 10.1111/j.1467-7652.2011.00622.x
15
ChandrasekaranJ.BruminM.WolfD.LeibmanD.KlapC.PearlsmanM.et al (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology.Mol. Plant Pathol.171140–1153. 10.1111/mpp.12375
16
CharronC.NicolaïM.GalloisJ. L.RobagliaC.MouryB.PalloixA.et al (2008). Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg.Plant J.5456–68. 10.1111/j.1365-313X.2008.03407.x
17
ClasenB. M.StoddardT. J.LuoS.DemorestZ. L.LiJ.CedroneF.et al (2016). Improving cold storage and processing traits in potato through targeted gene knockout.Plant Biotechnol. J.14169–176. 10.1111/pbi.12370
18
CombeJ. P.PetracekM. E.van EldikG.MeulewaeterF.TwellD. (2005). Translation initiation factors eIF4E and eIFiso4E are required for polysome formation and regulate plant growth in tobacco.Plant Mol. Biol.57749–760. 10.1007/s11103-005-3098-x
19
DaiJ.PengH.ChenW.ChengJ.WuY. (2013). Development of multiplex real-time PCR for simultaneous detection of three Potyviruses in tobacco plants.J. Appl. Microbiol.114502–508. 10.1016/j.jviromet.2008.01.025
20
DuanH.RichaelC.RommensC. M. (2012). Overexpression of the wild potato eIF4E1 variant Eva1 elicits Potato virus Y resistance in plants silenced for native eIF4E1.Transgenic Res.21929–938. 10.1007/s11248-011-9576-9
21
DupratA.CarantaC.ReversF.MenandB.BrowningK. S.RobagliaC. (2002). The Arabidopsis eukaryotic initiation factor (iso) 4E is dispensable for plant growth but required for susceptibility to potyviruses.Plant J.32927–934. 10.1046/j.1365-313X.2002.01481.x
22
EdwardsK.JohnstoneC.ThompsonC. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis.Nucleic Acids Res.19:1349. 10.1093/nar/19.6.1349
23
FossiM.AmundsonK.KuppuS.BrittA.ComaiL. (2019). Regeneration of Solanum tuberosum plants from protoplasts induces widespread genome instability.Plant Physiol.18078–86. 10.1104/pp.18.00906
24
FunkeC. N.NikolaevaO. V.GreenK. J.TranL. T.Chikh-AliM.Quintero-FerrerA.et al (2017). Strain-specific resistance to Potato virus Y (PVY) in potato and its effect on the relative abundance of PVY strains in commercial potato fields.Plant Dis.10120–28. 10.1094/PDIS-06-16-0901-RE
25
GauffierC.LebaronC.MorettiA.ConstantC.MoquetF.BonnetG.et al (2016). A tilling approach to generate broad-spectrum resistance to potyviruses in tomato is hampered by eIF4E gene redundancy.Plant J.85717–729. 10.1111/tpj.13136
26
GomezM. A.LinZ. D.MollT.ChauhanR. D.HaydenL.RenningerK.et al (2019). Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence.Plant Biotechnol. J.17421–434. 10.1111/pbi.12987
27
GreenK. J.BrownC. J.GrayS. M.KarasevA. V. (2017). Phylogenetic study of recombinant strains of Potato virus y.Virology50740–52. 10.1016/j.virol.2017.03.018
28
Gutierrez SanchezP. A.BabujeeL.Jaramillo MesaH.ArcibalE.GannonM.HaltermanD.et al (2020). Overexpression of a modified eIF4E regulates potato virus Y resistance at the transcriptional level in potato.BMC Genomics21:18. 10.1186/s12864-019-6423-5
29
HsiauT.ConantD.RossiN.MauresT.WaiteK.YangJ.et al (2019). Inference of CRISPR edits from Sanger trace data.BioRxiv5251082. 10.1101/251082
30
HwangJ.LiJ.LiuW. Y.AnS. J.ChoH.HerN. H.et al (2009). Double mutations in eIF4E and eIFiso4E confer recessive resistance to Chilli veinal mottle virus in pepper.Mol. Cells27329–336. 10.1007/s10059-009-0042-y
31
JanzacB.TribodetM.LacroixC.MouryB.VerrierJ. L.JacquotE. (2014). Evolutionary pathways to break down the resistance of allelic versions of the PVY resistance gene va.Plant Dis.981521–1529. 10.1094/PDIS-11-13-1126-RE
32
JiangF.ZhouK.MaL.GresselS.DoudnaJ. A. (2015). A Cas9–guide RNA complex preorganized for target DNA recognition.Science3481477–1481. 10.1126/science.aab1452
33
JonesR. A.VincentS. J. (2018). Strain-specific hypersensitive and extreme resistance phenotypes elicited by Potato virus Y among 39 potato cultivars released in three world regions over a 117-year period.Plant Dis.102185–196. 10.1094/PDIS-06-17-0901-RE
34
JosephJ. T.PoolakkalodyN. J.ShahJ. M. (2018). Plant reference genes for development and stress response studies.J. Biosci.43173–187. 10.1007/s12038-017-9728-z
35
JulioE.CotucheauJ.DecorpsC.VolpattiR.SentenacC.CandresseT.et al (2015). A eukaryotic translation initiation factor 4E (eIF4E) is responsible for the “va” tobacco recessive resistance to potyviruses.Plant Mol. Biol. Rep.33609–623. 10.1007/s11105-014-0775-4
36
KarasevA. V.GrayS. M. (2013). Continuous and emerging challenges of Potato virus y in potato.Ann. Rev. Phytopathol.51571–586. 10.1146/annurev-phyto-082712-102332
37
KirchnerS. M.HiltunenL. H.SantalaJ.DöringT. F.KetolaJ.KankaalaA.et al (2014). Comparison of straw mulch, insecticides, mineral oil, and birch extract for control of transmission of potato virus y in seed potato crops.Potato Res.5759–75. 10.1371/journal.pone.0071030
38
KreuzeJ. F.Souza-DiasJ. A. C.JeevalathaA.FigueiraA. R.ValkonenJ. P. T.JonesR. A. C. (2020). “Viral diseases in potato,” in The Potato Crop, eds Edn, edsCamposH.OrtizO. (Berlin: Springer), 389–430. 10.1016/j.virusres.2017.01.015
39
KuroiwaK.ThenaultC.NoguéF.PerrotL.MazierM.GalloisJ. L. (2022). CRISPR-based knock-out of eIF4E2 in a cherry tomato background successfully recapitulates resistance to pepper veinal mottle virus.Plant Sci.316:111160. 10.1016/j.plantsci.2021.111160
40
LacommeC.JacquotE. (2017). “General characteristics of Potato virus Y (PVY) and its impact on potato production: an overview,” in Potato Virus y: Biodiversity, Pathogenicity, Epidemiology and Management, eds Edn, edsLacommeC.GlaisL.BellstedtD. U.DupuisB.KarasevA. V.JacquotE. (Basel: Springer), 1–19.
41
LebedevaM. V.NikonovaE. Y.TerentievA. A.TaranovV. V.BabakovA. V.NikonovO. S. (2021). VPg of potato virus y and potato cap-binding eif4e factors: selective interaction and its supposed mechanism.Biochemistry (Moscow)861128–1138. 10.1134/S000629792109008X
42
LellisA. D.PatrickR. M.MayberryL. K.LorenceA.CampbellZ. C.RooseJ. L.et al (2019). Eifiso4g augments the synthesis of specific plant proteins involved in normal chloroplast function.Plant Physiol.18185–96. 10.1104/pp.19.00557
43
LiJ. F.NorvilleJ. E.AachJ.McCormackM.ZhangD.BushJ.et al (2013). Multiplex and homologous recombination–mediated genome editing in arabidopsis and nicotiana benthamiana using guide RNA and Cas9.Nat. Biotechnol.31688–691. 10.1038/nbt.2654
44
LivakK. J.SchmittgenT. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method.Methods25402–408. 10.1006/meth.2001.1262
45
MazierM.FlamainF.NicolaïM.SarnetteV.CarantaC. (2011). Knock-down of both eIF4E1 and eIF4E2 genes confers broad-spectrum resistance against potyviruses in tomato.PLos One6:e29595. 10.1371/journal.pone.0029595
46
MiroshnichenkoD.TimerbaevV.OkunevaA.KlementyevaA.SidorovaT.PushinA.et al (2020). Enhancement of resistance to PVY in intragenic marker-free potato plants by RNAi-mediated silencing of eIF4E translation initiation factors.Plant Cell Tissue Organ Cult.140691–705. 10.1007/s11240-019-01746-9
47
MonzingoA. F.DhaliwalS.Dutt-ChaudhuriA.LyonA.SadowJ. H.HoffmanD. W.et al (2007). The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond.Plant Physiol.1431504–1518. 10.1104/pp.106.093146
48
MouryB.MorelC.JohansenE.GuilbaudL.SoucheS.AymeV.et al (2004). Mutations in Potato virus Y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum.Mol. Plant Mic. Int.17322–329. 10.1094/MPMI.2004.17.3.322
49
MuthoniJ.KabiraJ.ShimelisH.MelisR. (2015). Tetrasomic inheritance in cultivated potato and implications in conventional breeding.Austr. J. Crop Sci.9185–190. 10.3316/informit.07520680241382
50
NicoliaA.Proux-WéraE.ÅhmanI.OnkokesungN.AnderssonM.AndreassonE.et al (2015). Targeted gene mutation in tetraploid potato through transient talen expression in protoplasts.J. Biotechnol.20417–24. 10.1016/j.jbiotec.2015.03.021
51
NicotN.HausmanJ. F.HoffmannL.EversD. (2005). Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress.J. Experiment. Bot.562907–2914. 10.1093/jxb/eri285
52
PatrickR. M.BrowningK. S. (2012). The eIF4F and eIFiso4F complexes of plants: an evolutionary perspective.Int J. Genomics2012:287814. 10.1155/2012/287814
53
PironF.NicolaïM.MinoïaS.PiednoirE.MorettiA.SalguesA.et al (2010). An induced mutation in tomato eIF4E leads to immunity to two potyviruses.PLos One5:e11313. 10.1371/journal.pone.0011313
54
PoulicardN.PaciosL. F.GalloisJ. L.PiñeroD.Garcia-ArenalF. (2016). Human management of a wild plant modulates the evolutionary dynamics of a gene determining recessive resistance to virus infection.PLoS Genet.12:e1006214. 10.1371/journal.pgen.1006214
55
RubioM.NicolaïM.CarantaC.PalloixA. (2009). Allele mining in the pepper gene pool provided new complementation effects between pvr2-eIF4E alleles and pvr6-eIF (iso) 4E for resistance to the Pepper veinal mottle virus.J. Gen. Virol.902808–2814. 10.1099/vir.0.013151-0
56
RuffelS.DussaultM. H.PalloixA.MouryB.BendahmaneA.RobagliaC.et al (2002). A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E).Plant J.321067–1075. 10.1046/j.1365-313X.2002.01499.x
57
RuffelS.GalloisJ. L.LesageM. L.CarantaC. (2005). The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene.Mol. Genet. Genom.274346–353. 10.1007/s00438-005-0003-x
58
RuffelS.GalloisJ. L.MouryB.RobagliaC.PalloixA.CarantaC. (2006). Simultaneous mutations in translation initiation factors eIF4E and eIF (iso) 4E are required to prevent pepper veinal mottle virus infection of pepper.J. Gen. Virol.872089–2098. 10.1099/vir.0.81817-0
59
SemenovaE.JoreM. M.DatsenkoK. A.SemenovaA.WestraE. R.WannerB.et al (2011). Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence.Proc. Natl. Acad. Sci. U.S.A.10810098–10103. 10.1073/pnas.1104144108
60
ShaulO. (2015). Unique aspects of plant nonsense-mediated mRNA decay.Trend Plant Sci.20767–779. 10.1016/j.tplants.2015.08.011
61
SinghR. P.ValkonenJ. P.GrayS. M.BoonhamN.JonesR. A. C.KerlanC.et al (2008). Discussion paper: The naming of Potato virus Y strains infecting potato.Arch. Virol.1531–13. 10.1007/s00705-007-1059-1
62
SpitzerM.WildenhainJ.RappsilberJ.TyersM. (2014). BoxPlotR: a web tool for generation of box plots.Nat. Methods11121–122. 10.1038/nmeth.2811
63
TavazzaR.AncoraG. (1986). Plant regeneration from mesophyll protoplasts in commercial potato cultivars (primura. kennebec, spunta, desirée).Plant Cell Rep.5243–246. 10.1007/BF00269812
64
TrunigerV.ArandaM. A. (2009). Recessive resistance to plant viruses.Adv. virus Res.75119–231. 10.1016/S0065-3527(09)07504-6
65
ValkonenJ. P. T. (1997). Novel resistances to four potyviruses in tuber-bearing potato species, and temperature-sensitive expression of hypersensitive resistance to potato virus Y.Ann. Appl. Biol.13091–104. 10.1111/j.1744-7348.1997.tb05785.x
66
ValkonenJ. P. T. (2007). “Viruses: economical losses and biotechnological potential,” in Potato Biology and Biotechnology, eds Edn, edsVreugdenhilD.BradshawJ.GebhardtC.GoversF.MackerronD. K. L.TaylorM. A.et al (Amsterdam: Elsevier), 619–641.
67
ValkonenJ. P. T.GebhardtC.Zimnoch-GuzowskaE.WatanabeK. N. (2017). “Resistance to Potato virus Y in potato,” in Potato virus Y: Biodiversity, Pathogenicity, Epidemiology and Management, eds Edn, edsLacommeC.GlaisL.BellstedtD. U.DupuisB.KarasevA. V.JacquotE. (Basel: Springer), 207–241.
68
VeilletF.PerrotL.ChauvinL.KermarrecM. P.Guyon-DebastA.ChauvinJ. E.et al (2019). Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor.Int. J. Mol. Sci.20:402. 10.3390/ijms20020402
69
VeilletF.PerrotL.Guyon-DebastA.KermarrecM. P.ChauvinL.ChauvinJ. E.et al (2020). Expanding the CRISPR toolbox in P. patens using SpCas9-NG variant and application for gene and base editing in solanaceae crops.Int. J. Mol. Sci.21:1024. 10.3390/ijms21031024
70
WalterJ.CharonJ.HuY.LachatJ.LegerT.LafforgueG.et al (2019). Comparative analysis of mutational robustness of the intrinsically disordered viral protein VPg and of its interactor eIF4E.PLos One14:e0211725. 10.1371/journal.pone.0211725
71
WangA.KrishnaswamyS. (2012). Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement.Mol. Plant Pathol.13795–803. 10.1111/j.1364-3703.2012.00791.x
72
WiedenheftB.van DuijnE.BultemaJ. B.WaghmareS. P.ZhouK.BarendregtA.et al (2011). RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions.Proc. Natl. Acad. Sci. U.S.A.10810092–10097. 10.1073/pnas.1102716108
73
YooS. D.ChoY. H.SheenJ. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis.Nat. Protoc.21565–1572. 10.1038/nprot.2007.199
74
YoonY. J.VenkateshJ.LeeJ. H.KimJ.LeeH. E.KimD. S.et al (2020). Genome editing of eIF4E1 in tomato confers resistance to pepper mottle virus.Front. Plant Sci.11:1098. 10.3389/fpls.2020.01098
75
ZafirovD.GiovinazzoN.BastetA.GalloisJ. L. (2021). When a knockout is an achilles’ heel: resistance to one potyvirus species triggers hypersusceptibility to another one in arabidopsis thaliana.Mol. Plant Pathol.22334–347. 10.1111/mpp.13031
76
ZhangC.ZarkaK. A.ZarkaD. G.WhitworthJ. L.DouchesD. S. (2021). Expression of the tomato pot-1 gene confers potato virus Y (PVY) resistance in susceptible potato varieties.Am. J. Potato Res.9842–50. 10.1007/s12230-020-09815-y
77
ZhangD.ZhangZ.UnverT.ZhangB. (2021). CRISPR/Cas: A powerful tool for gene function study and crop improvement.J. Adv. Res.29207–221. 10.1016/j.jare.2020.10.003
78
ZhaoL.LiW.WangB.GaoY.SuiX.LiuY.et al (2020). Development of a PVY resistant flue-cured tobacco line via EMS mutagenesis of eIF4E.Agronomy10:36. 10.3390/agronomy10010036
79
ZhuH.LiC.GaoC. (2020). Applications of Crispr–Cas in agriculture and plant biotechnology.Nat. Rev. Mol. Cell Biol.21661–677. 10.1038/s41580-020-00288-9
Summary
Keywords
potato, translation initiation factors, eIF4E, genome editing, potyvirus resistance, PVY
Citation
Lucioli A, Tavazza R, Baima S, Fatyol K, Burgyan J and Tavazza M (2022) CRISPR-Cas9 Targeting of the eIF4E1 Gene Extends the Potato Virus Y Resistance Spectrum of the Solanum tuberosum L. cv. Desirée. Front. Microbiol. 13:873930. doi: 10.3389/fmicb.2022.873930
Received
11 February 2022
Accepted
10 May 2022
Published
01 June 2022
Volume
13 - 2022
Edited by
Jesús Navas-Castillo, La Mayora Experimental Station (CSIC), Spain
Reviewed by
Miguel A. Aranda, Spanish National Research Council (CSIC), Spain; David B. Collinge, University of Copenhagen, Denmark
Updates
Copyright
© 2022 Lucioli, Tavazza, Baima, Fatyol, Burgyan and Tavazza.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mario Tavazza, mario.tavazza@enea.it
This article was submitted to Microbe and Virus Interactions with Plants, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.