- 1Henan International Joint Laboratory of Crop Protection, Henan Biopesticide Engineering Research Center, Institute of Plant Protection Research, Graduate T&R Base of Zhengzhou University, Henan Academy of Agricultural Sciences, Zhengzhou, China
- 2School of Agricultural Sciences, Zhengzhou University, Zhengzhou, China
- 3Institute of Horticulture, Henan Academy of Agricultural Sciences, Zhengzhou, China
- 4School of Environmental Sciences, University of Guelph, Guelph, ON, Canada
Cucumber wilt caused by Fusarium oxysporum f.sp. cucumerinum (Foc) is a highly destructive disease that leads to reduced yield in cucumbers. In this study, strain YB-04 was isolated from wheat straw and identified as Bacillus subtilis. It displayed strong antagonistic activity against F. oxysporum f.sp. cucumerinum in dual culture and exhibited significant biocontrol of cucumber Fusarium wilt with a higher control effect than those of previously reported Bacillus strains and displayed pronounced growth promotion of cucumber seedlings. B. subtilis YB-04 could secrete extracellular protease, amylase, cellulose, and β-1,3-glucanase and be able to produce siderophores and indole acetic acid. Inoculation with B. subtilis YB-04 or Foc increased cucumber defense-related enzyme activities for PPO, SOD, CAT, PAL, and LOX. However, the greatest increase was with the combination of B. subtilis YB-04 and Foc. Sequencing the genome of B. subtilis YB-04 showed that it had genes for the biosynthesis of various secondary metabolites, carbohydrate-active enzymes, and assimilation of nitrogen, phosphorous, and potassium. B. subtilis YB-04 appears to be a promising biological control agent against the Fusarium wilt of cucumber and promotes cucumber growth by genomic, physiological, and phenotypic analysis.
Introduction
Cucumber (Cucumis sativus L.) is an important vegetable crop worldwide. Cucumber wilt caused by Fusarium oxysporum f.sp. cucumerinum (Foc) is one of the most destructive diseases of cucumber that can lead to severe losses in yield and quality (Zhou et al., 2017). Foc enters root tissues by direct penetration or wounds causing visible symptoms, including necrotic lesions, vascular and root wilt, and ultimately death (Ahn et al., 1998). It can survive up to 20 years in soil (Zhao et al., 2017). Furthermore, there are no commercially available cucumber cultivars with resistance against Fusarium wilt. Therefore, biological control of cucumber Fusarium wilt using antagonistic microorganisms has been considered to be a promising alternative.
Beneficial microorganisms can be used as biological control agents (BCAs) against different plant diseases. Many of these are species of Bacillus, which are also plant growth-promoting bacteria (PGPB) that can improve plant growth by producing secondary metabolites, such as fengycin, surfactin, bacillaene, and macrolactin, siderophores, and indole acetic acid (IAA) and secreting hydrolases to suppress plant pathogens and promote plant growth (Blake et al., 2021). More importantly, Bacillus species have several advantages, such as rapid growth, spore production, safe, non-pathogenic nature, and adaptation to broader environmental conditions (Nayak, 2021). However, there are several problems in the field application of microbial agents, including lack of high-efficiency biocontrol, relatively short shelf life, and variable control effectiveness. Therefore, understanding the growth-promotion and biocontrol mechanisms of beneficial microorganisms can significantly contribute to improved application efficacy of BCAs.
One way to better understand these organisms is through whole genome sequencing allowing for the discovery of genes for the production of bioactive compounds responsible for biocontrol and growth promotion (Bauman et al., 2021). For instance, sequencing the complete genome of the BCA Bacillus velezensis 9912D revealed gene clusters for secondary metabolite synthesis, including several potentially new lantibiotics (Pan et al., 2017). Similarly, the complete genome of Bacillus subtilis 7PJ-16 revealed genes for biosynthesis of antimicrobial metabolites and promoting plant growth traits, indicating its ability to act as a BCA and PGPB (Xu et al., 2019). The genome of B. subtilis 9407 showed that it had genes for the biocontrol mechanism against bacterial fruit blotch, including genes for a newly identified subtilosin A, bacilysin, and bacillaene (Gu et al., 2021).
In this study, strain YB-04 was isolated from wheat straw. A number of BCA and PGPB traits were screened for YB-04 in culture. The genome of strain YB-04 was sequenced to identify a number of genes associated with BCA and PGPB traits. Plant growth promotion of cucumber seedlings by soil inoculation of YB-04 was assessed based on chlorophyll content and growth of shoots, roots, stems, and leaves. Biocontrol activity by YB-04 against Fusarium wilt of cucumber was assessed based on disease severity and disease index. Furthermore, the activities of cucumber defense-related enzymes activities, both by YB-04 alone and in combination with Foc inoculation, were examined. The discovery and characterization of B. subtilis YB-04 indicate that it is a promising BCA and PGPB of cucumber.
Materials and Methods
Isolation of Strain YB-04 and in vitro Antagonism Test
Strain YB-04 was isolated from wheat straw and cultured in LB broth by a dilution plate method at 37°C, collected from a field (E113°97’, N35°05’) at the Henan Academy of Agricultural Sciences in Xinxiang, Henan, China in June. Foc was obtained from the College of Plant Protection, Henan Agricultural University. Antagonistic activity against Foc was performed by a dual culture where Foc was grown on PDA at 28°C for 5 days, and then 5 mm agar plugs were excised and transferred to the center of another PDA plate. Strain YB-04 was placed 3 cm away from the edge of the Petri dish, and the growth rate of Foc was measured relative to the control, which was Foc without strain on a plate (Xu et al., 2020).
Biocontrol Efficiency of Strain YB-04 Against Cucumber Fusarium Wilt and Growth Promotion on Cucumber Seedlings
Strain YB-04 was cultured in LB broth for 24 h at 37°C with shaking at 180 rpm and harvested by centrifugation (4,000 × g for 5 min), washed once with LB broth, and adjusted to 108 CFU/ml based on OD at 595 nm. Foc was grown on PDA at 28°C for 5 days, and then ten 5 mm agar plugs were excised and transferred to 100 ml PDB. The broths were incubated at 28°C in a shaker at 180 rpm for 3 days. The Foc cultures were filtered through 4 layers of sterile gauze, and the filtered spores were adjusted to 105 spores/ml using a hemacytometer (XB-K-25, Qiujing, Shanghai, China).
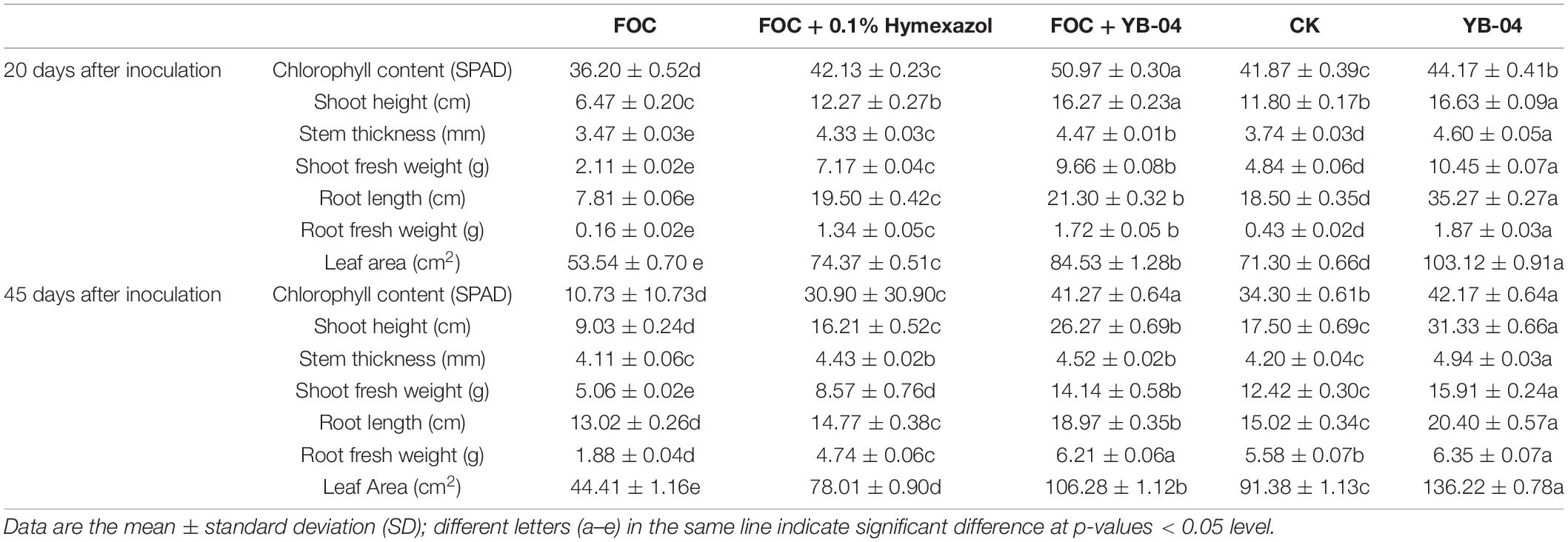
Cucumber seeds of cultivar Chuancui No. 3 were surface-sterilized in 75% ethanol (v/v) for 30 s and then rinsed with sterile water three times. The seeds were air-dried and each seed was planted in a separate pot (10 cm high, 10 cm diameter) filled with a 400 g sterilized mixture of soil. The plants were grown in the greenhouse at 25°C with a 16 h light/8 h dark photoperiod. After 10 days, each pot of cucumber seedlings was treated as follows: (1) drenching with 15 ml of YB-04 suspension; (2) first drenching with 15 ml l of YB-04 suspension and 24 h of later drenching with 15 ml Foc spore suspension; (3) first drenching with 15 ml of 0.1% hymexazol and 24 h later drenching with 15 ml Foc spore suspension; (4) drenching with 15 ml of sterile distilled water; or (5) drenching with 15 ml of sterile distilled water and 24 h later drenching with 15 ml of Foc spore suspension. Each treatment was performed using 12 plants with 3 replicates. At 20 and 45 days post inoculation (dpi) with YB-04, chlorophyll content, shoot height and fresh weight, root length and fresh weight, stem thickness, and leaf area were measured, and disease severity and disease index were recorded for plants inoculated with Foc at 45 days post YB-04 inoculation (Chen et al., 2010). In brief, disease severity was assessed using a 0–4 disease scale; 0 = leaf asymptomatic; 1 = leaf wilting below 1/4 of cucumber seedling; 2 = leaf wilting in 1/4 to 1/2 of cucumber seedling; 3 = leaf wilting above 1/2 of cucumber seedling; 4 = the whole plant was wilted and died. The disease index was calculated using DI = [[(0 × N0) + (1 × N1) + (2 × N2) + (3 × N3) + (4 × N4)]/T × 4] × 100, where N is the number of cucumber seedlings for each disease score and T is the total number of cucumber seedlings. Disease incidence = [N1 + N2 + N3 + N4]/T × 100%. Control efficacy = (DI of control - DI of treatment)/DI of control × 100%. The chlorophyll content of leaves was measured by using a SPAD-502 Plus chlorophyll content meter (Konica Minolta, Tokyo, JP). Root length and shoot height were measured with a ruler. The fresh weight of root and shoot was recorded with an analytical balance (ME203E, Mettler Toledo, Shanghai, China). Stem thickness was measured at 2 cm from the crown with a vernier caliper (MNT-200, Shanghai Meinaite Metals Instruments Co., Shanghai, China).
Determination of Defense Enzyme Activities in the Cucumber Leaves
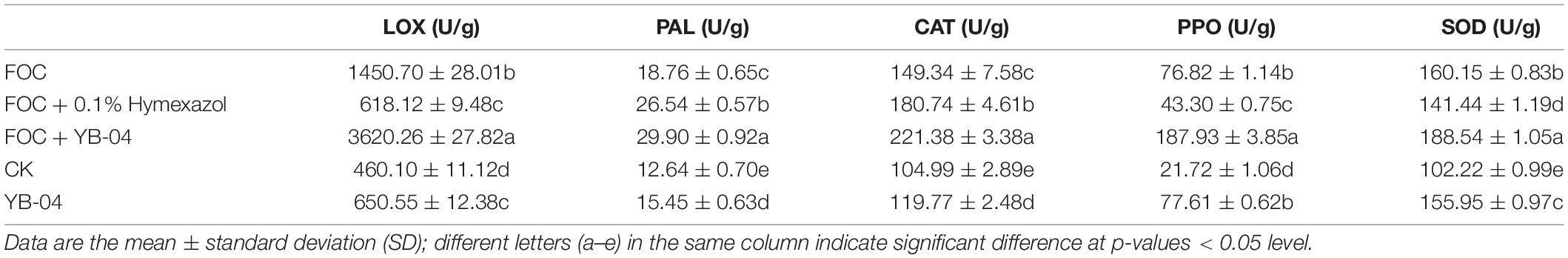
After 20 days post YB-04 inoculation, leaves were harvested and stored at –80°C. In brief, 0.5 g leaves were ground in liquid nitrogen, and 1 ml of extraction buffer was added. After centrifugation at 8,000 × g for 10 min, the supernatant was removed for enzyme assays. Enzyme activities were measured using assay kits for PPO (Cat. No. BC0195), SOD (Cat. No. BC0175), CAT (Cat. No. BC0205), PAL (Cat. No. BC0215), and LOX (Cat. No. BC0325) following the procedures of the manufacturer (Solarbio, Beijing, China). Absorbance was determined by using a plate reader (Tecan Spark, Tecan, Switzerland).
Detection of Plant Growth-Promoting Bacteria and Biological Control Agents Traits
Protease activity was detected with single colonies of YB-04 grown at 30°C for 5 days on skim milk agar (0.1 g CaCl2, 5.0 g NaCl, 10.0 g skim milk, 10.0 g peptone, and 18.0 g of agar per liter, pH 7.2). Protease activity was observed as clear zones around the colonies (Kazanas, 1968). Amylase activity was detected with single colonies grown at 30°C for 48 h on starch agar (10.0 g soluble starch, 10.0 g tryptone, 5.0 g glucose, 5.0 g NaCl, 5.0 g beef extract, and 18.0 g of agar per liter, pH 7.2). Lugol’s iodine solution (1% iodine in 2% potassium iodide w/v) was added to the starch agar plate, and amylase activity was observed as a colorless halo (Al-Naamani et al., 2015). Cellulose activity was assayed with single colonies grown for 7 days at 30°C on carboxymethylcellulose agar (5.0 g CMC-Na, 0.1 g MgSO4⋅7H2O, 0.25 g (NH4)2SO4, 0.25 g K2HPO4, and 18.0 g of agar per liter, pH 5.5). The plates were flooded with 1% (m/v) Congo Red, and then washed with sterilized distilled water, and cellulose activity was detected as a clear zone (Teather and Wood, 1982). The β-Glucanase activity was assayed with single colonies grown at 30°C for 2 days on β-glucan agar (0.05 g glucose, 0.5 g yeast extract, 1 g peptone, 0.5 g NaCl, 0.01 g Congo Red, and 18.0 g of agar per liter, pH 7.0). The β-Glucanase activity was indicated by a clear zone around the colonies (Teather and Wood, 1982). Siderophore production was determined with single colonies grown at 30°C for 2 days in the dark on Chrome Azurol S blue agar (10 ml 20% sucrose solution, 30 ml 10% acid hydrolyzed casein, 1 ml 1 mmol/L CaCl2, 5 ml 0.1 mol/L phosphate-buffered saline (pH 6.8), 50 ml CAS dyeing solution, and 18 g of agar per liter, pH 7.2). Siderophore production was indicated by a change from blue to orange around the colonies (Schwyn and Neilands, 1987). IAA production was measured with single colonies grown at 30°C for 2 days on L-tryptophan nutrient broth (3 g beef extract, 10 g peptone, 5 g NaCl,0.5 g L-tryptophan per liter, pH 7.2). After centrifugation at 14,000 × g for 10 min,1 ml of supernatant was mixed with 2 ml of Salkowski stain, and then kept at room temperature in the dark for 30 min (Glickmann and Dessaux, 1995). All of the above reagents were of analytical grade and produced by China National Pharmaceutical Group Corp., Shanghai, China.
Genome Sequencing and Assembly of Strain YB-04
Strain YB-04 was grown in LB broth for 16 h at 37°C by shaking at 180 rpm. Genomic DNA was extracted with a Mini-BEST Bacterial Genomic DNA Extraction Kit Ver. 3.0 following the manufacturer’s instructions (Takara, Beijing, China). An approximately 10 kb insert sequencing library was constructed and sequencing was performed using the PacBio Sequel II system (Pacific Biosciences, Menlo Park, CA, United States) by Frasergen (Wuhan, Hubei, China). Sequencing reads were de novo assembled by using HGAP4 (Chin et al., 2013) and the Canu (v.1.6) (Koren et al., 2017) software. The depth of genome coverage was analyzed by using the align tool (BLASR, v0.4.1) (Chaisson and Tesler, 2012). The assembled complete genome sequence was deposited in NCBI GenBank (Accession number CP072525). A circular map of the genome was constructed by using Circos (v0.64) (Krzywinski et al., 2009).
Genome Annotation of Strain YB-04
The genome of strain YB-04 was annotated using Glimmer (v3.02) (Delcher et al., 2007). The tRNA and rRNA genes were identified by tRNAscan-SE (v2.0) (Lowe and Eddy, 1997) and RNAmmer (v1.2) (Lagesen et al., 2007), respectively. Functional descriptions of putative protein encoding genes were done by BLASTx with an E-value threshold of 1e-5 using the NCBI Non-Redundant protein database (NR), Swiss-Prot, Clusters of Orthologous Groups (COG), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ontology (GO).
Phylogenetic Relationship of Strain YB-04
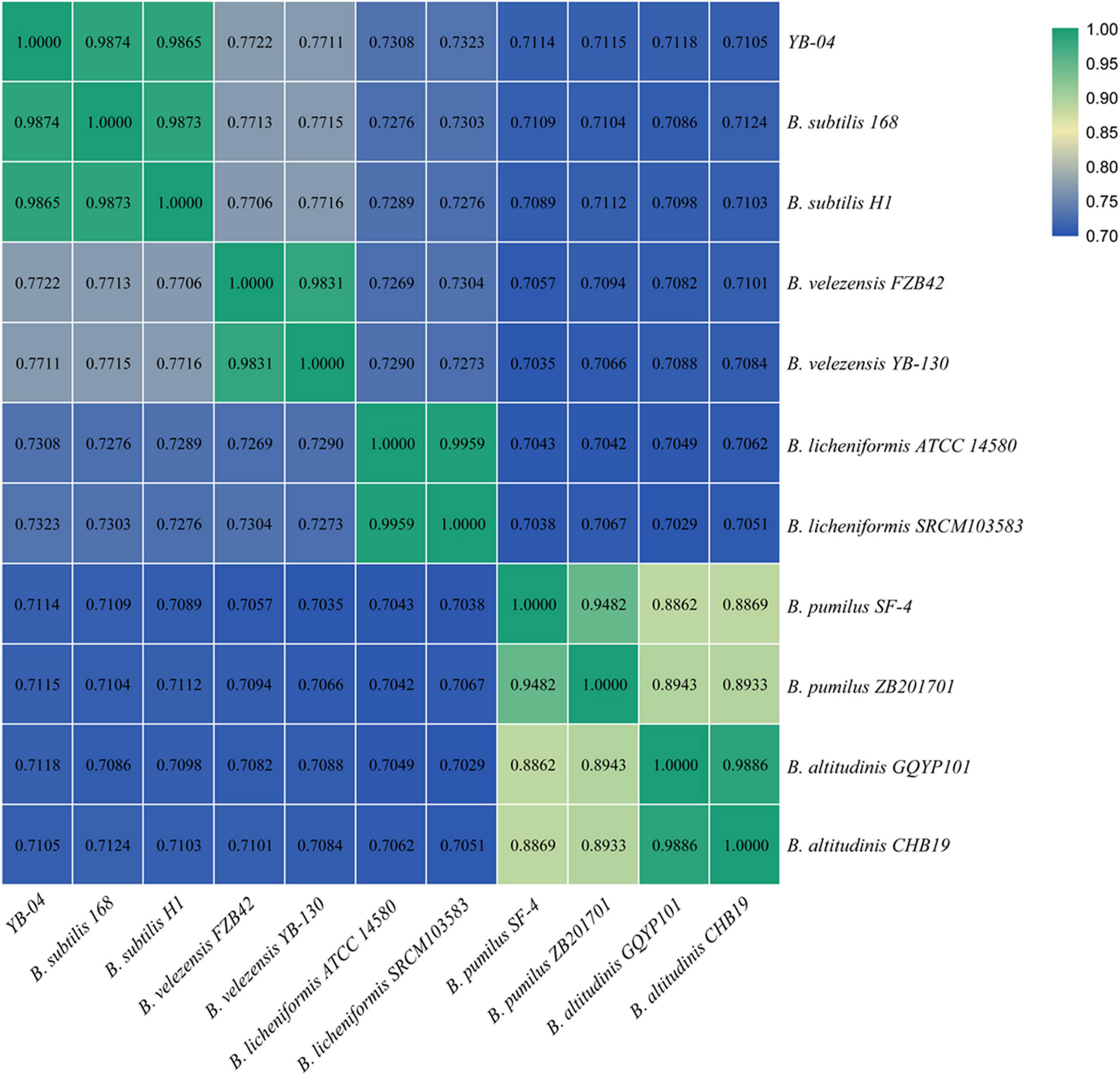
The 16S rRNA gene sequences of strain YB-04 and B. velezensis FZB42, B. velezensis YB-130, B. subtilis H1, B. subtilis 168, B. licheniformis SRCM103583, B. licheniformis ATCC 14580, B. altitudinis CHB19, B. altitudinis GQYP101, B. pumilus SF-4, and B. pumilus ZB201701 were obtained from the genomes (GenBank IDs: CP000560.2, CP054562.1, CP026662.1, NC_000964.3, CP035404.1, CP034569.1, CP043559.1, CP040514.1, CP047089.1, and CP029464.1, respectively). A tree of the 16S rRNA gene sequences was constructed with MEGA 7.0 using the Neighbor Joining method (Kumar et al., 2016). Average Nucleotide Identity (ANI) was calculated using an ANI calculator (Yoon et al., 2017).
Analysis of Genes Encoding CAZymes and Gene Clusters Responsible for the Biosynthesis of Secondary Metabolites
Protein-coding genes in the genome of strain YB-04 were aligned with the carbohydrate active enZYme (CAZy) database (Lombard et al., 2014) using dbCAN2 (Zhang et al., 2018) and HMMER (v3.1b2) (Finn et al., 2011) with an E-value threshold of 1e-15. Identification of gene clusters for the synthesis of secondary metabolites was analyzed by using antiSMASH5.0 (Blin et al., 2019).
Statistical Analysis
Statistical analysis was performed using SPSS v21.0 by one-way analysis of variance (ANOVA). Means were compared with Duncan’s multiple range tests at a probability of p ≤ 0.05.
Results
Isolation of YB-04 and Wilt Disease Biocontrol Activity in vitro and in vivo
Dilution plating from surface-sterilized wheat straw yielded numerous colonies with different colony appearances. Twenty strains were purified and screened for antagonistic activity against Foc in dual culture and reduced wilt severity of cucumber inoculated with Foc in the greenhouse (data not shown). Strain YB-04 was selected based on having the greatest antagonistic activity against Foc in culture (Figure 1) and reduced wilt severity of cucumber seedlings at 20 days after Foc inoculation (Figure 2). Disease incidence, disease index, and control efficacy at 45 dpi revealed that YB-04 significantly reduced wilt symptoms caused by Foc to levels slightly less than the chemical fungicide hymexazol (Table 1).
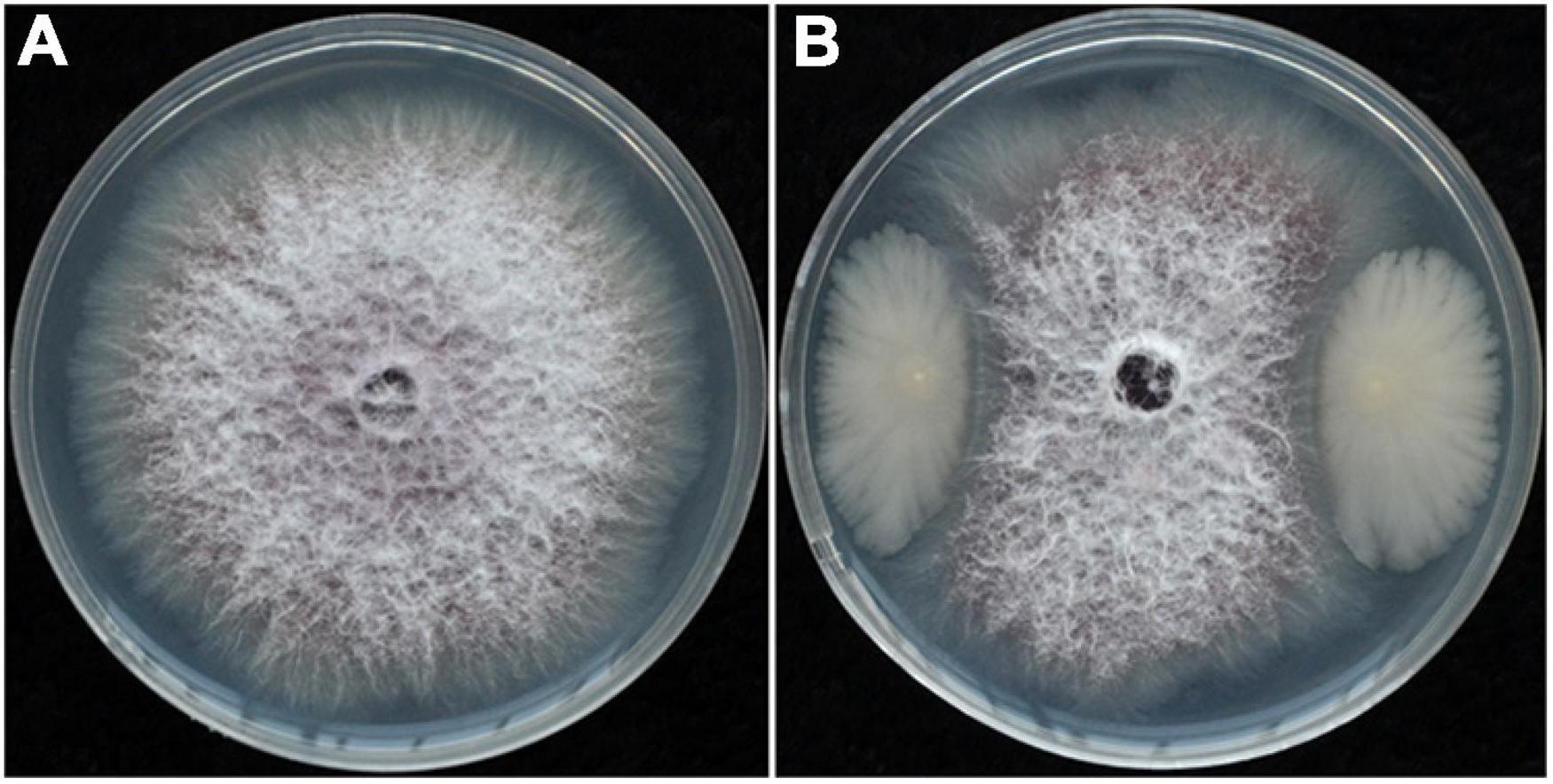
Figure 1. Colony morphology of Foc co-cultivated with or without strain YB-04. (A) Colony morphology of Foc in PDA; (B) inhibition of strain YB-04 on Foc growth.

Figure 2. Effect of strain YB-04 against Fusarium wilt and on growth-promotion of Cucumber Seedlings. (A) Only Foc inoculation; (B) Foc inoculation and hymexazol treatment; (C) inoculation of strain YB-04 and Foc; (D) sterile distilled water; (E) only strain YB-04 inoculation.
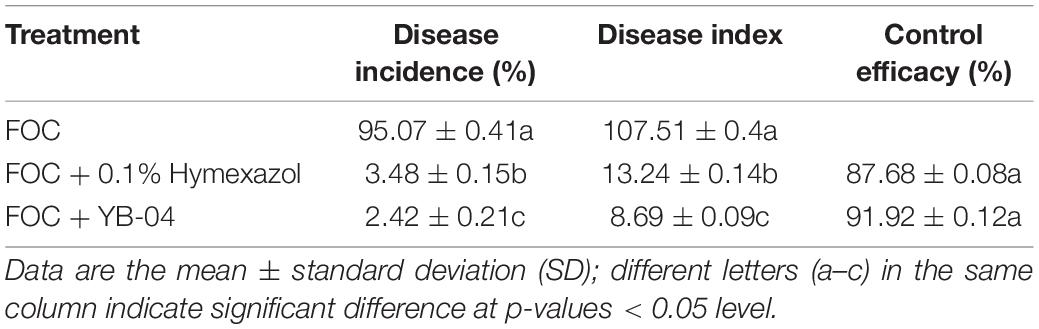
Table 1. Disease incidence, disease index, and control efficacy of B. subtilis YB-04 against cucumber Fusarium wilt.
Growth-Promotion Activity of Strain YB-04
At 20 and 45 dpi with strain YB-04, there was a significant increase in chlorophyll content, height and fresh weight of shoot, root length and fresh weight, stem thickness, and leaf area compared to non-treated cucumber seedlings (Figure 2 and Table 2). At 20 dpi, the greatest increases were observed for the fresh weight of shoots and roots at 115.91 and 334.88%, respectively. At 45 dpi, the greatest increases were observed for shoot height and leaf area at 79.03 and 49.07%, respectively.
At 20 and 45 dpi with strain YB-04 and Foc inoculation, there was also a significant increase in chlorophyll content, height, and fresh weight of shoot, root length and fresh weight, stem thickness, and leaf area compared to that of the Foc inoculated cucumber seedlings (Figure 2 and Table 2). This was also observed with the Foc inoculated seedlings treated with hymexazol. However, strain YB-04 treatment of the Foc-inoculated seedlings resulted in significantly higher chlorophyll content, shoot height and fresh weight, root length and fresh weight, and leaf area than Foc-inoculated seedlings with hymexazol. However, there was no significant difference in the stem thickness of Foc-inoculated seedlings with strain YB-04 or hymexazol at 45 dpi.
Effect of Strain YB-04 on Activities of Defense-Related Enzymes in Cucumber Seedlings
At 20 dpi with strain YB-04, cucumber seedlings showed significantly higher activities of SOD, CAT, PAL, and LOX, but not PPO, compared to non-treated seedlings (Table 3). Inoculation with Foc also significantly increased those enzyme activities, except PPO, compared to seedlings without Foc inoculation. The highest activities were observed with Foc inoculation and strain YB-04 treatment, which was significantly higher for all the enzymes compared to Foc inoculation. However, the activities of PAL and CAT significantly increased but LOX, PPO, and SOD significantly decreased with Foc inoculation and hymexazol treatment compared to Foc inoculation.
Detection of in vitro Antifungal and Growth-Promoting Traits
Strain YB-04 could secrete protease (Figure 3A), amylase (Figure 3B), cellulose (Figure 3C), and β-1, 3-glucanase (Figure 3D). In addition, it could produce siderophores (Figure 3E) and indole acetic acid (Figure 3F).
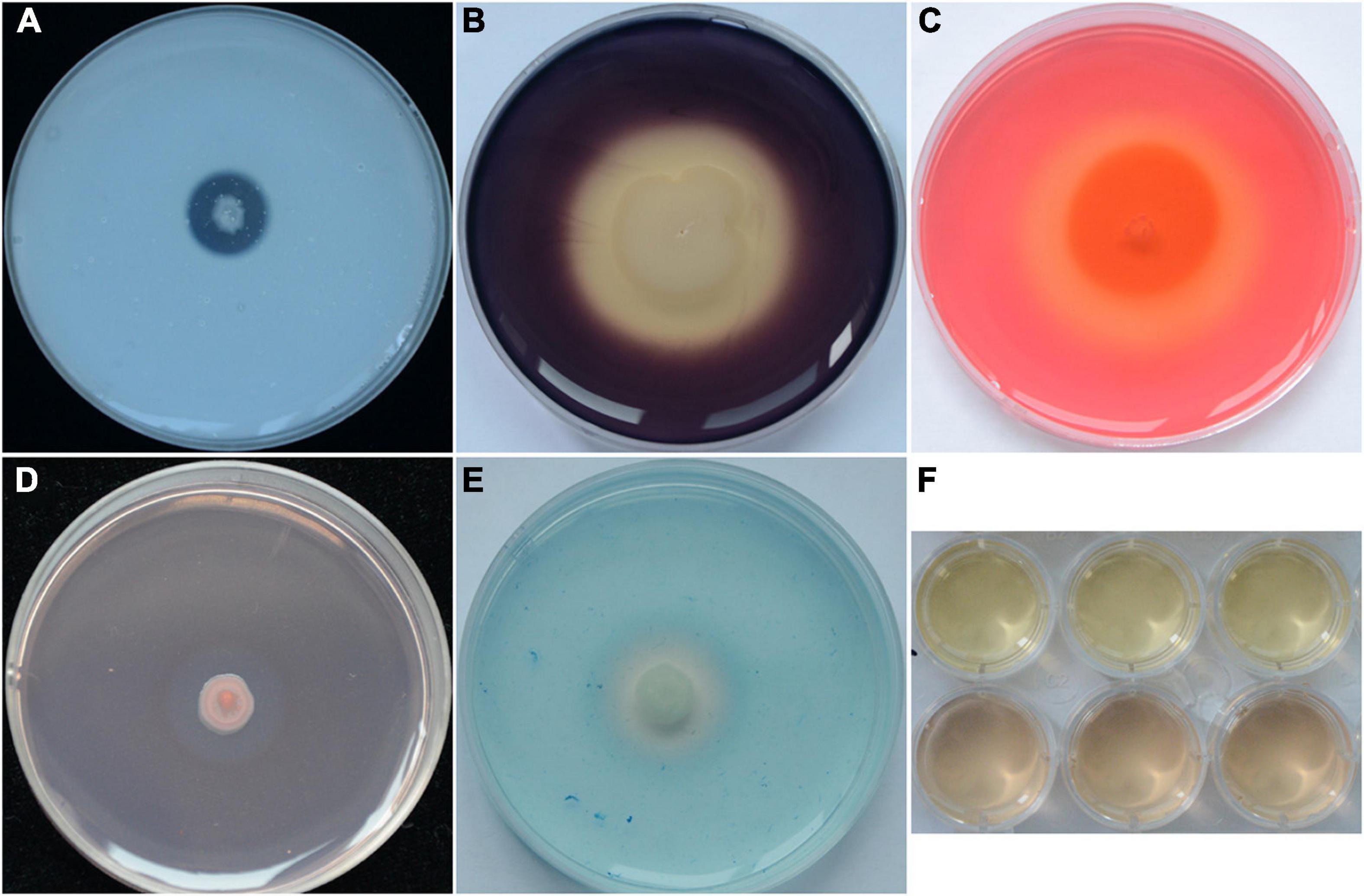
Figure 3. Antifungal and PGP traits of strain YB-04. (A) Protease production; (B) amylase production; (C) cellulose production; (D) β-1,3-glucanase production; (E) siderophore production; (F) IAA production.
Genome Sequencing, Assembly, and Identification of Strain YB-04
A total of 387,797 high-quality sequencing long reads with a mean length of 10,962 bp and an N50 of 13,374 bp were generated from the genomic DNA of strain YB-04 by the Pacbio sequencing platform. Total base pairs were 4,251,215,058 bp with an 882.47X genome coverage. The YB-04 genome consisted of a single circular chromosome of 4,156,177 bp with a GC content of 43.83% (Figure 4). There were 4,325 protein-coding genes covering 88.62% of the genome with an average gene length of 851.6 bp, which included 87 tRNAs, 30 rRNAs (5S, 16S, 23S), and 22 sRNAs. Four gene islands, three CRISPRs, and four prophages were detected (Supplementary Tables 1–3). For the predicted protein encoding genes, 99.70, 90.73, 75.45, 64.30, and 53.87% could be annotated with the NR, Swiss-Prot, COG, GO, and KEGG databases, respectively (Supplementary Table 4).
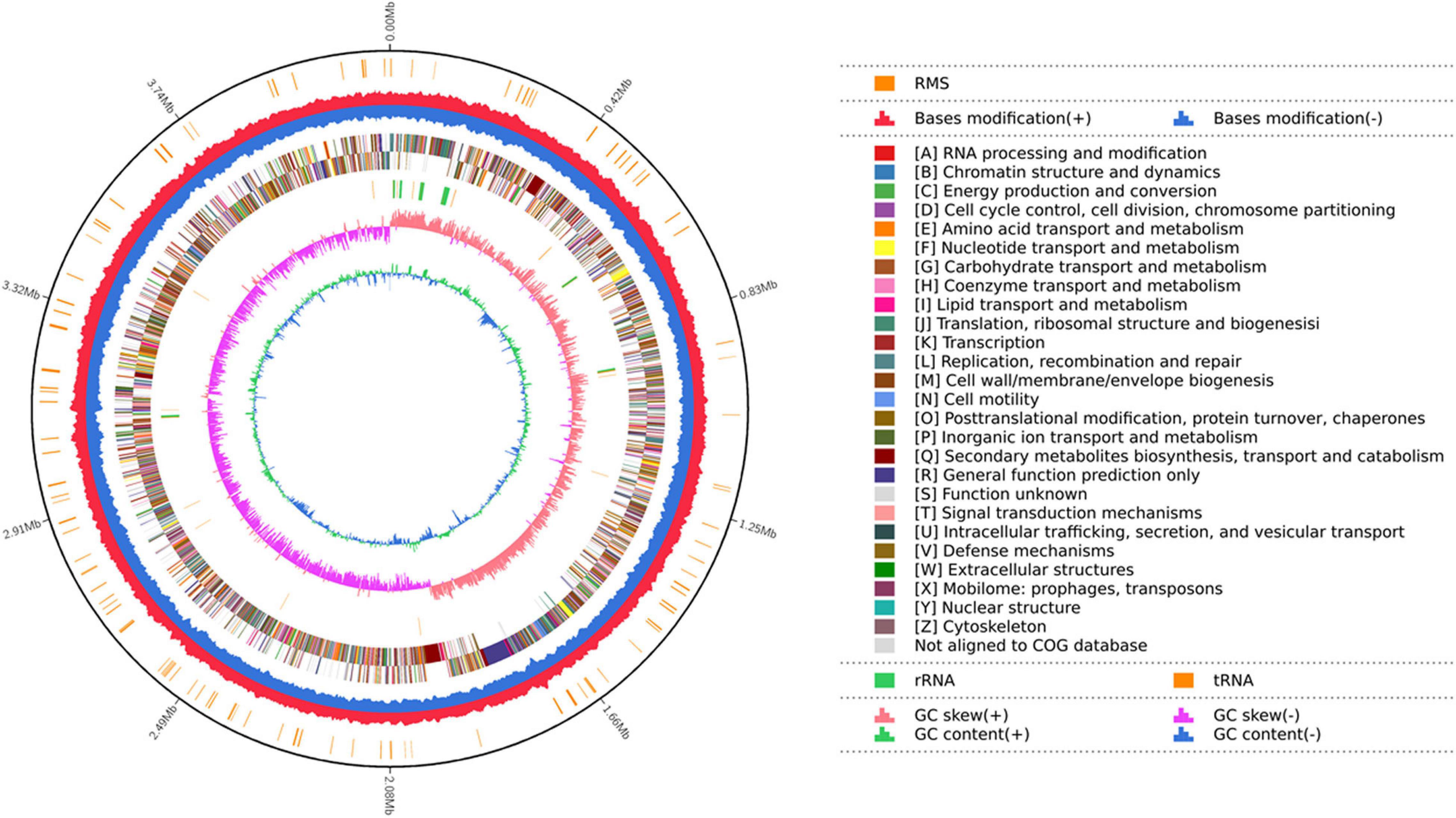
Figure 4. Map of the strain YB-04 genome. The distributions of circles from outwards to inwards are as follows: ring 1 for genome size (black line); ring 2 for the restriction modification system, for forward strand (red) and reverse strand (blue); ring 3 for COG classifications of protein-coding genes on the forward strand and reverse strand; ring 4 for the distribution of tRNAs (brown) and rRNAs (green); ring 5 for GC skew; ring 6 for GC content.
A phylogenetic tree based on 16S rRNA gene sequences of strain YB-04 and 10 other Bacillus isolates showed that strain YB-04, B. subtilis 168, and B. subtilis H1 clustered (Figure 5). Strain YB-04 and B. subtilis 168 had the maximum ANI value of 98.74%, followed by B. subtilis H1with 98.65%, which is higher than the cutoff of 95–96% for bacterial species identity. ANI values between strain YB-04 and the 8 other Bacillus species ranged from 71.05 to 77.22% (Figure 6). Therefore, strain YB-04 was identified as B. subtilis.
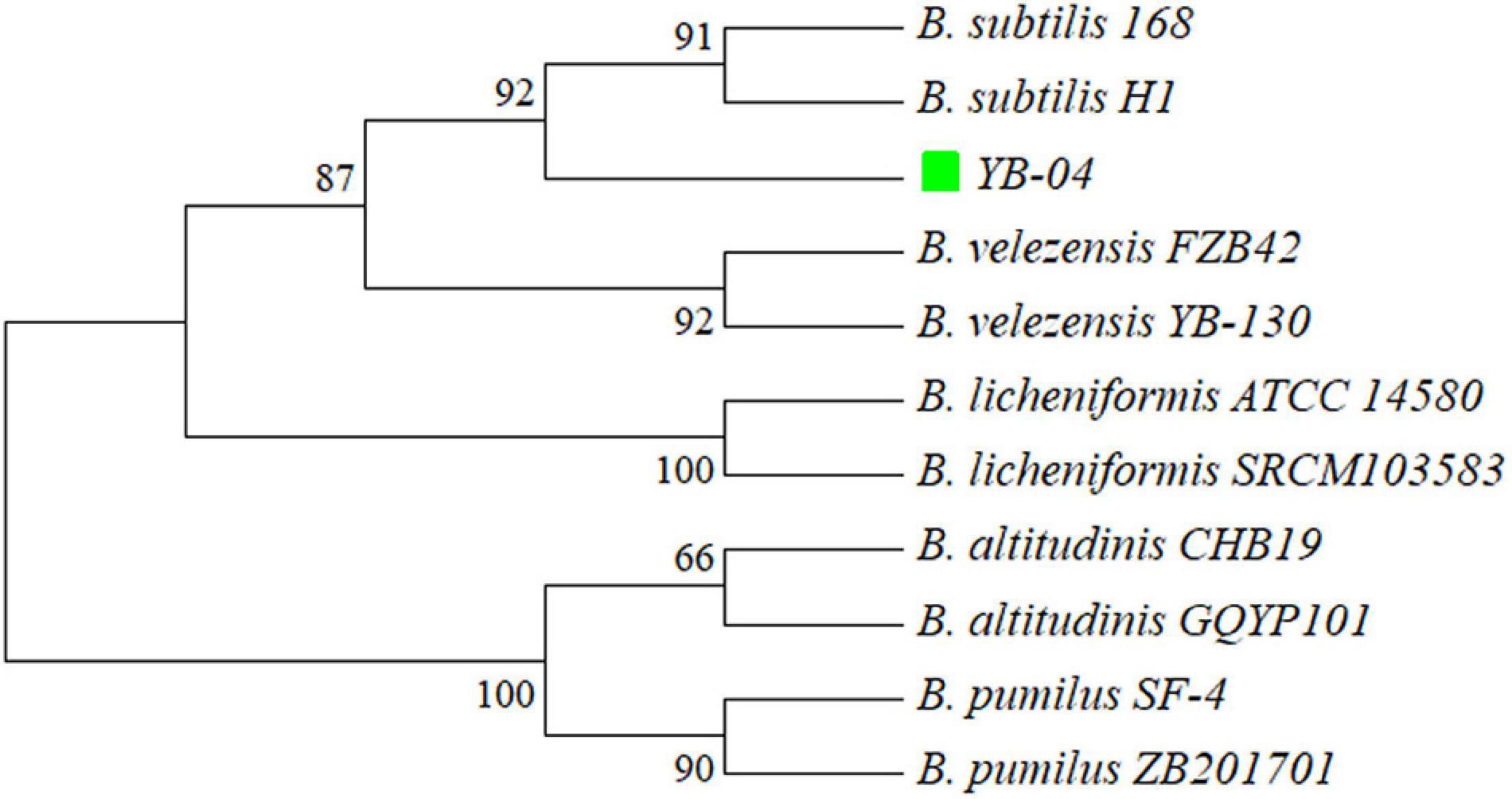
Figure 5. Phylogenetic tree of B. subtilis YB-04 and 10 other Bacillus species based on 16S rRNA sequences.
Genome Analysis of Selected Genes of Bacillus subtilis YB-04
The genome of B. subtilis YB-04 had 111 genes identified as putative CAZymes, namely, 2 auxiliary activities (AAs), 7 polysaccharide lyases (PLs), 15 carbohydrate-binding modules (CBMs), 19 carbohydrate esterases (CEs) 24 glycosyltransferases (GTs), and 51 glycoside hydrolases (GHs) (Figure 7 and Supplementary Table 5). Six of those were classified as both GHs and CBMs.
There were 13 gene clusters predicted to be responsible for the biosynthesis of secondary metabolites. At 100% similarity, there was each matching gene clusters for bacillaene, fengycin, bacillibactin, subtilin, subtilosin A, and bacilysin synthesis. There was also one gene cluster with 82% similarity to that for surfactin synthesis. There were 6 biosynthetic gene clusters with no similarity in the antiSMASH database that appeared to be novel biosynthetic gene clusters of secondary metabolites. Based on their matches to the antiSMASH database, these were one gene cluster each for types of lanthipeptide-class-i, Type III PKS, tRNA-dependent cyclodipeptide synthases, other unspecified ribosomally synthesized, post-translationally modified peptide products, and two gene clusters, each encoding for terpenes (Table 4).
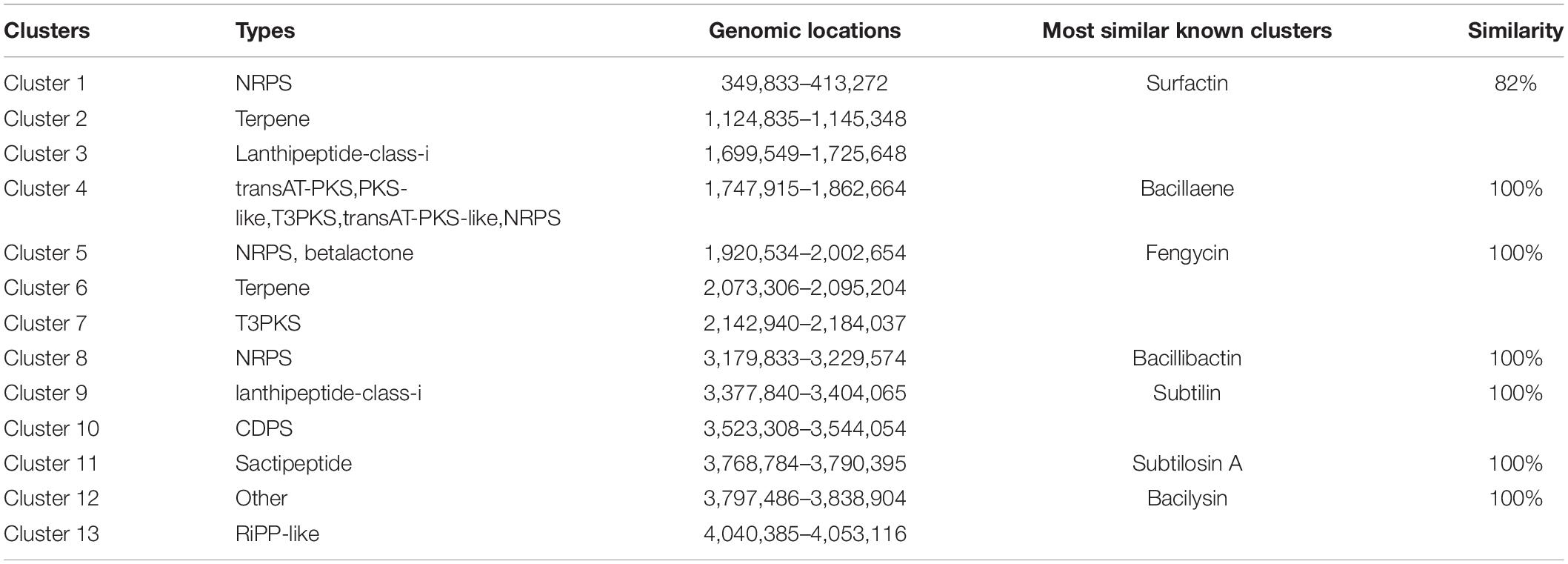
Table 4. List of the putative gene clusters encoding for secondary metabolites by antiSMASH in the B. subtilis YB-04 genome.
The genome of B. subtilis YB-04 contained predicted genes for an ATP-dependent phosphate uptake system PstABCS, phoPR operon for regulating Pho regulon in response to phosphate limitation, and alkaline phosphatase genes of phoA and phoD for phosphorus acquisition (Table 5). It also contained the nasABCDEF gene cluster for nitrite transport and reduction. Additionally, there were the potassium uptake system ktrABCD, a putative gamma-glutamylcyclotransferase YkqA for potassium assimilation, and a putative potassium efflux channel yugO.
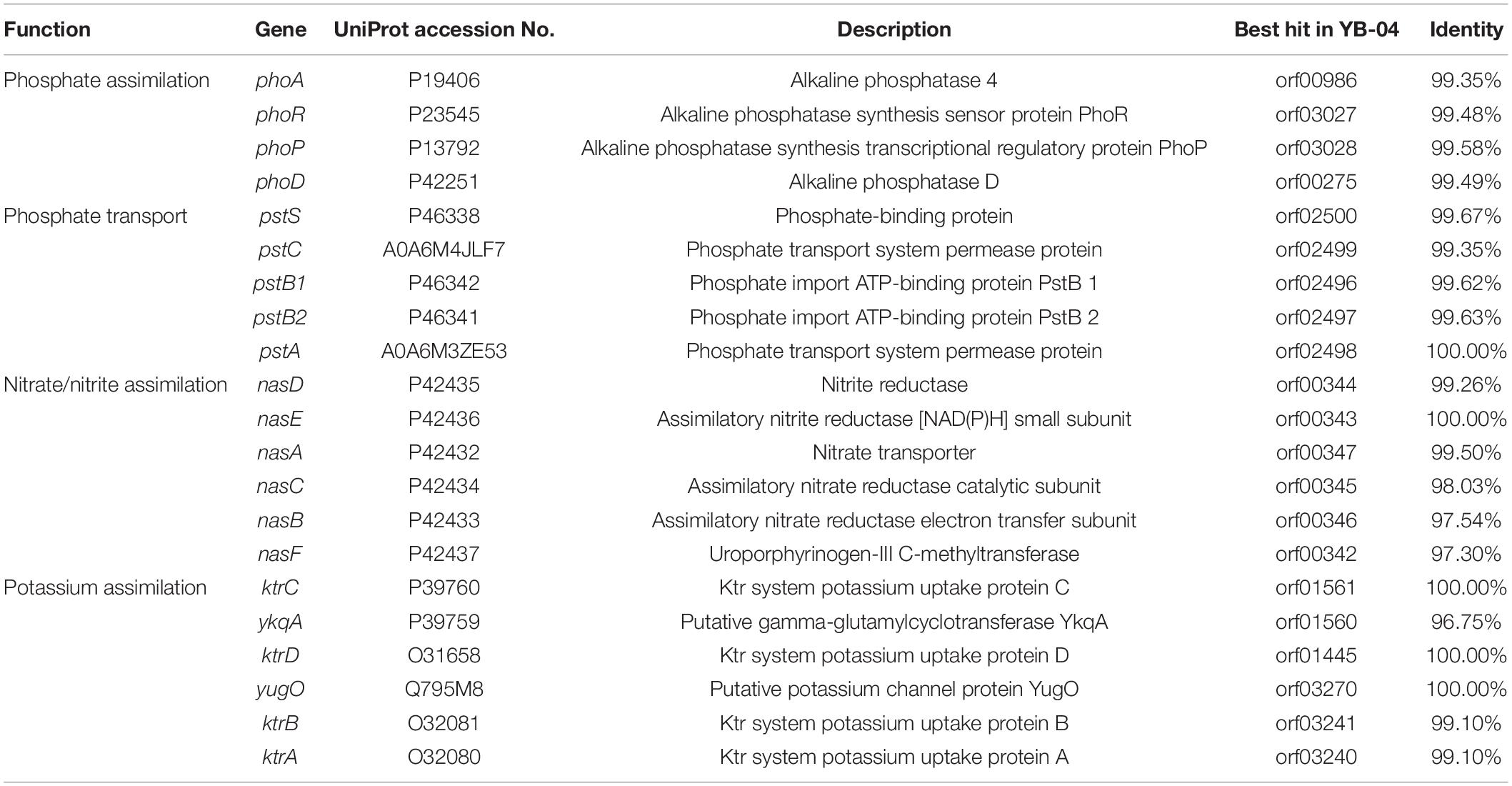
Table 5. Genes responsible for nitrogen, phosphorous, and potassium assimilation identified in the strain YB-04 genome.
Discussion
Fusarium wilt disease caused by Foc is one of the most devastating soil-borne diseases of cucumber, resulting in severe yield losses throughout the world (Zhou et al., 2018). While the use of BCAs to control plant diseases and PGPBs to improve plant growth is considered to be promising (Radhakrishnan et al., 2017; Koskey et al., 2021), new strains are needed to screen for greater efficiency, shelf life, and consistency. As part of that, an in-depth analysis of growth promotion and biocontrol traits will help in developing them into successful microbial products.
In this study, B. subtilis YB-04 was found to be a BCA with antagonistic activity against Foc in dual culture and significantly reduced Fusarium wilt caused by Foc at levels comparable to hymexazol, which is used to control the disease in China. It was also a PGPB with pronounced growth promotion on cucumber seedlings. Previously, B. subtilis B579, B. subtilis MBI600, and B. subtilis B068150 were also shown to significantly reduce cucumber Fusarium wilt and promote cucumber growth (Chen et al., 2010; Li et al., 2012; Samaras et al., 2020). Compared to those bacteria, the percentage reduction in the Fusarium wilt of cucumber by B. subtilis YB-04 was greater than those achieved by B. subtilis B068150, B. subtilis B579, or B. subtilis MBI600. The percentage of increased growth-promotion based on the shoot and root fresh weight and plant height by B. subtilis YB-04 was greater than those achieved by B. subtilis B579 or B. subtilis MBI600. Thus, B. subtilis YB-04 appears to be more effective as a BCA and PGPB than some of the previously described B. subtilis tested on cucumber.
To act as a BCA against plant pathogenic fungi, bacteria possess a number of mechanisms including synthesis of hydrolytic enzymes, production of antibiotics, and induction of systemic resistance (Morales-Cedeño et al., 2021; Saeed et al., 2021; Xu et al., 2021). In this study, B. subtilis YB-04 had all of those mechanisms. Hydrolytic enzyme activities including protease, amylase, cellulose, and β-1, 3-glucanase were present in pure cultures, and they can break down chitin, glucans, and other polymers in fungal cell walls, thus inhibiting the growth of fungal pathogens (Naglot et al., 2015; Li et al., 2019). Other B. subtilis BCAs with similar enzymes include B. subtilis BCC6327, B. subtilis ZIM3, and B. subtilis LR1 (Thakaew and Niamsup, 2013; Banerjee et al., 2017; Dai et al., 2020). Furthermore, a large number of CAZyme genes were found in the genome of B. subtilis YB-04, also suggesting that it has a strong capability to be antagonistic against fungal plant pathogens based on the potential degradation and use of fungal polymers as nutrient sources (Banani et al., 2015; Chen et al., 2018; Sui et al., 2020). In addition, gene clusters were found to be responsible for the biosynthesis of known secondary metabolites, including bacillaene, fengycin, bacillibactin, subtilin, subtilosin A, bacilysin, and surfactin, indicating antibiotic production by B. subtilis YB-04, which is common in Bacillus species (Xu et al., 2019; Su et al., 2020). Other B. subtilis BCAs found to produce antibiotics or have genes for antibiotic production included B. subtilis BSD-2 for lanthipeptide (Liu et al., 2016). Surfactin and fengycin can also be elicitors of induced systemic resistance in plants (Romero et al., 2007; Ongena et al., 2010). Many studies have reported that plant defense enzymes play important roles in disease resistance (Prasannath, 2017; Ji et al., 2020; Xu et al., 2021). Induction of the activities of defense-related enzymes in leaves following soil inoculation with B. subtilis YB-04 and Foc indicates a form of systemic resistance. Defense-related enzymes activities for PPO, SOD, CAT, PAL, and LOX could be induced by inoculation with B. subtilis YB-04 or Foc alone and the greatest increase was with the combination of B. subtilis YB-04 and Foc. Other B. subtilis BCAs causing host induction of defense-related enzyme activities include B. subtilis B579, B. subtilis SL-44, and B. subtilis CBR05 (Chen et al., 2010; Chandrasekaran and Chun, 2016; Wu et al., 2019).
All the growth parameters of cucumber seedlings measured in this study were increased with B. subtilis YB-04 treatment. Importantly, growth parameters were all increased much more in infected seedlings with Foc treated with B. subtilis YB-04 than those infected with Foc and treated with hymexazol. This indicates that B. subtilis YB-04 can improve plant growth while providing disease control, which would be an advantage over using hymexazol that did not promote growth. This was similar to Pseudomonas aeruginosa CQ-40 that controlled tomato gray mold caused by Botrytis cinerea and promoted the growth of tomato seedlings, whereas pyrimethanil only controlled the disease with a prevention effect of up to 64.71% (Wang et al., 2020).
To act as a PGPB, bacteria have a variety of mechanisms including the production of enzymes and siderophores for nutrient acquisition and phytohormones to promote growth, and enzymes to reduce the negative effects of various abiotic stresses (Glick, 2012; Saeed et al., 2021). B. subtilis YB-04 produced siderophores that can improve iron uptake and alleviate harmful effects of iron on plants that have been associated with enhanced plant growth (Haas, 2003; Dimkpa et al., 2009). The genome of B. subtilis YB-04 also contained genes responsible for nitrogen, phosphorous, and potassium assimilation. Plant growth and development depend on macronutrients, such as nitrogen, phosphorous, and potassium, that are mostly obtained from the soil and can be made more available to plants by soil microbes that have the ability to solubilize nutrients and transfer them to plants (Glick, 2012; Rana et al., 2020). Finally, B. subtilis YB-04 produced IAA, which may be taken up by the cucumber seedlings stimulating the transcriptional expression of IAA responsive genes and enhancing biomass (Spaepen et al., 2014; Jiang et al., 2020).
In summary, B. subtilis YB-04 appears to be an effective BCA against cucumber Fusarium wilt and an effective PGPB of cucumber seedlings. The BCA mechanisms could include induced systemic host resistance as indicated by greater host defense-related enzyme activities, and direct pathogen inhibition through secretion of extracellular enzymes and antibiotics. The PGPB mechanisms could include nutrient acquisition via siderophores and enzymes for fixing nitrogen and solubilizing potassium and phosphorus, and direct plant growth enhancement through increased amounts of indole acetic acid. Compared to other B. subtilis strains used as cucumber Fusarium wilt BCAs and PGPBs in cucumber, B. subtilis YB-04 is a more effective BCA than all those reported thus far and is a more effective PGPB than most reported so far. Thus, it appears to be a very promising novel beneficial B. subtilis strain for cucumber production.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
LY and WX conceived the research and designed the experiments. QY, XX, FY, XD, and WX performed the experiments and analyzed the data. WX prepared the manuscript draft. PG, BT, and LY critically revised the manuscript. All authors approved the final version of the manuscript.
Funding
This research was funded by the Central Government Guiding Local Projects (2020[44]), the Special Project for Science and Technology Innovation Team of Henan Academy of Agricultural Sciences (2022TD25), and the Outstanding Youth Science and Technology Fund of Henan Academy of Agricultural Sciences (2022YQ06).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.885430/full#supplementary-material
References
Ahn, P., Chung, H. S., and Lee, Y. H. (1998). Vegetative Compatibility Groups and Pathogenicity Among Isolates of Fusarium oxysporum f. sp. cucumerinum. Plant Dis. 82, 244–246. doi: 10.1094/PDIS.1998.82.2.244
Al-Naamani, L. S. H., Dobretsov, S., Al-Sabahi, J., and Soussi, B. (2015). Identification and characterization of two amylase producing bacteria Cellulosimicrobium sp. and Demequina sp. isolated from marine organisms. J. Agricult. Mar. Sci. 20, 8–15. doi: 10.24200/jams.vol20iss0pp8-15
Banani, H., Spadaro, D., Zhang, D., Matic, S., Garibaldi, A., and Gullino, M. L. (2015). Postharvest application of a novel chitinase cloned from Metschnikowia fructicola and overexpressed in Pichia pastoris to control brown rot of peaches. Int. J. Food Microbiol. 199, 54–61. doi: 10.1016/j.ijfoodmicro.2015.01.002
Banerjee, G., Nandi, A., and Ray, A. K. (2017). Assessment of hemolytic activity, enzyme production and bacteriocin characterization of Bacillus subtilis LR1 isolated from the gastrointestinal tract of fish. Archiv. Microbiol. 199, 115–124. doi: 10.1007/s00203-016-1283-8
Bauman, K. D., Butler, K. S., Moore, B. S., and Chekan, J. R. (2021). Genome mining methods to discover bioactive natural products. Nat. Prod. Rep. 38, 2100–2129. doi: 10.1039/d1np00032b
Blake, C., Christensen, M. N., and Kovacs, A. T. (2021). Molecular Aspects of Plant Growth Promotion and Protection by Bacillus subtilis. Mol. Plant Microbe Interact. 34, 15–25. doi: 10.1094/MPMI-08-20-0225-CR
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Chaisson, M. J., and Tesler, G. (2012). Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinform. 13:238. doi: 10.1186/1471-2105-13-238
Chandrasekaran, M., and Chun, S. C. (2016). Expression of PR-protein genes and induction of defense-related enzymes by Bacillus subtilis CBR05 in tomato (Solanum lycopersicum) plants challenged with Erwinia carotovora subsp. carotovora. Biosci. Biotechnol. Biochem. 80, 2277–2283. doi: 10.1080/09168451.2016.1206811
Chen, F., Wang, M., Zheng, Y., Luo, J., Yang, X., and Wang, X. (2010). Quantitative changes of plant defense enzymes and phytohormone in biocontrol of cucumber Fusarium wilt by Bacillus subtilis B579. World J. Microbiol. Biotechnol. 26, 675–684. doi: 10.1007/s11274-009-0222-0
Chen, P. H., Chen, R. Y., and Chou, J. Y. (2018). Screening and Evaluation of Yeast Antagonists for Biological Control of Botrytis cinerea on Strawberry Fruits. Mycobiology 46, 33–46. doi: 10.1080/12298093.2018.1454013
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Dai, J., Dong, A., Xiong, G., Liu, Y., Hossain, M. S., Liu, S., et al. (2020). Production of Highly Active Extracellular Amylase and Cellulase From Bacillus subtilis ZIM3 and a Recombinant Strain With a Potential Application in Tobacco Fermentation. Front. Microbiol. 11:1539. doi: 10.3389/fmicb.2020.01539
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Dimkpa, C. O., Merten, D., Svatoš, A., Büchel, G., and Kothe, E. (2009). Metal-induced oxidative stress impacting plant growth in contaminated soil is alleviated by microbial siderophores. Soil Biol. Biochem. 41, 154–162. doi: 10.1016/j.soilbio.2008.10.010
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Glick, B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012:963401. doi: 10.6064/2012/963401
Glickmann, E., and Dessaux, Y. (1995). A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Env. Microbiol. 61, 793–796. doi: 10.1128/aem.61.2.793-796
Gu, X., Zeng, Q., Wang, Y., Li, J., Zhao, Y., Li, Y., et al. (2021). Comprehensive genomic analysis of Bacillus subtilis 9407 reveals its biocontrol potential against bacterial fruit blotch. Phytopathol. Res. 3:4. doi: 10.1186/s42483-021-00081-2
Haas, H. (2003). Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Appl. Microbiol. Biotechnol. 62, 316–330. doi: 10.1007/s00253-003-1335-2
Ji, Z. L., Peng, S., Zhu, W., Dong, J. P., and Zhu, F. (2020). Induced resistance in nectarine fruit by Bacillus licheniformis W10 for the control of brown rot caused by Monilinia fructicola. Food. Microbiol. 92:103558. doi: 10.1016/j.fm.2020.103558
Jiang, Y., Wu, Y., Hu, N., Li, H., and Jiao, J. (2020). Interactions of bacterial-feeding nematodes and indole-3-acetic acid (IAA)-producing bacteria promotes growth of Arabidopsis thaliana by regulating soil auxin status. Appl. Soil Ecol. 147:103447. doi: 10.1016/j.apsoil.2019.103447
Kazanas, N. (1968). Proteolytic activity of microorganisms isolated from freshwater fish. Appl. Microb. 16, 128–132. doi: 10.1128/am.16.1.128-132
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Koskey, G., Mburu, S. W., Awino, R., Njeru, E. M., and Maingi, J. M. (2021). Potential Use of Beneficial Microorganisms for Soil Amelioration, Phytopathogen Biocontrol, and Sustainable Crop Production in Smallholder Agroecosystems. Front. Sustainab. Food Syst. 5:606308. doi: 10.3389/fsufs.2021.606308
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lagesen, K., Hallin, P., Rodland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Li, L., Ma, J., Li, Y., Wang, Z., Gao, T., and Wang, Q. (2012). Screening and partial characterization of Bacillus with potential applications in biocontrol of cucumber Fusarium wilt. Crop. Prot. 35, 29–35. doi: 10.1016/j.cropro.2011.12.004
Li, Z., Ye, X., Liu, M., Xia, C., Zhang, L., Luo, X., et al. (2019). A novel outer membrane β-1,6-glucanase is deployed in the predation of fungi by myxobacteria. ISME J. 13, 2223–2235. doi: 10.1038/s41396-019-0424-x
Liu, H., Yin, S., An, L., Zhang, G., Cheng, H., Xi, Y., et al. (2016). Complete genome sequence of Bacillus subtilis BSD-2, a microbial germicide isolated from cultivated cotton. J. Biotechnol. 230, 26–27. doi: 10.1016/j.jbiotec.2016.05.019
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Morales-Cedeño, L. R., Orozco-Mosqueda, M. D. C., Loeza-Lara, P. D., Parra-Cota, F. I, de los Santos-Villalobos, S., and Santoyo, G. (2021). Plant growth-promoting bacterial endophytes as biocontrol agents of pre- and post-harvest diseases:fundamentals, methods of application and future perspectives. Microbiolog. Res. 242:126612. doi: 10.1016/j.micres.2020.126612
Naglot, A., Goswami, S., Rahman, I., Shrimali, D. D., Yadav, K. K., Gupta, V. K., et al. (2015). Antagonistic Potential of Native Trichoderma viride Strain against Potent Tea Fungal Pathogens in North East India. Plant Pathol. J. 31, 278–289. doi: 10.5423/PPJ.OA.01.2015.0004
Nayak, S. K. (2021). Multifaceted applications of probiotic Bacillus species in aquaculture with special reference to Bacillus subtilis. Rev. Aquacult. 13, 862–906. doi: 10.1111/raq.12503
Ongena, M., Jourdan, E., Adam, A., Paquot, M., Brans, A., Joris, B., et al. (2010). Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microb. 9, 1084–1090. doi: 10.1111/j.1462-2920.2006.01202.x
Pan, H. Q., Li, Q. L., and Hu, J. C. (2017). The complete genome sequence of Bacillus velezensis 9912D reveals its biocontrol mechanism as a novel commercial biological fungicide agent. J. Biotechnol. 247, 25–28. doi: 10.1016/j.jbiotec.2017.02.022
Prasannath, K. (2017). Plant defense-related enzymes against pathogens: a review. AGRIEAST: J. Agricult. Sci. 11, 38–48. doi: 10.4038/agrieast.v11i1.33
Radhakrishnan, R., Hashem, A., and Abd Allah, E. F. (2017). Bacillus: a Biological Tool for Crop Improvement through Bio-Molecular Changes in Adverse Environments. Front. Physiol. 8:667–667. doi: 10.3389/fphys.2017.00667
Rana, K. L., Kour, D., Kaur, T., Devi, R., Yadav, A. N., Yadav, N., et al. (2020). Endophytic microbes: biodiversity, plant growth-promoting mechanisms and potential applications for agricultural sustainability. Antonie Van Leeuwenhoek 113, 1075–1107. doi: 10.1007/s10482-020-01429-y
Romero, D., de Vicente, A., Rakotoaly, R. H., Dufour, S. E., Veening, J. W., Arrebola, E., et al. (2007). The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol. Plant Microbe Interact. 20, 430–440. doi: 10.1094/MPMI-20-4-0430
Saeed, Q., Xiukang, W., Haider, F. U., Kucerik, J., Mumtaz, M. Z., Holatko, J., et al. (2021). Rhizosphere Bacteria in Plant Growth Promotion, Biocontrol, and Bioremediation of Contaminated Sites: A Comprehensive Review of Effects and Mechanisms. Int. J. Mol. Sci. 22:19. doi: 10.3390/ijms221910529
Samaras, A., Nikolaidis, M., Antequera-Gomez, M. L., Camara-Almiron, J., Romero, D., Moschakis, T., et al. (2020). Whole Genome Sequencing and Root Colonization Studies Reveal Novel Insights in the Biocontrol Potential and Growth Promotion by Bacillus subtilis MBI 600 on Cucumber. Front Microbiol 11:600393. doi: 10.3389/fmicb.2020.600393
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Analyt. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Spaepen, S., Bossuyt, S., Engelen, K., Marchal, K., and Vanderleyden, J. (2014). Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 201, 850–861. doi: 10.1111/nph.12590
Su, Z., Chen, X., Liu, X., Guo, Q., Li, S., Lu, X., et al. (2020). Genome mining and UHPLC-QTOF-MS/MS to identify the potential antimicrobial compounds and determine the specificity of biosynthetic gene clusters in Bacillus subtilis NCD-2. BMC Gen. 21:767. doi: 10.1186/s12864-020-07160-2
Sui, Y., Wisniewski, M., Droby, S., Piombo, E., Wu, X., and Yue, J. (2020). Genome Sequence, Assembly, and Characterization of the Antagonistic Yeast Candida oleophila Used as a Biocontrol Agent Against Post-harvest Diseases. Front Microbiol. 11:295. doi: 10.3389/fmicb.2020.00295
Teather, R. M., and Wood, P. J. (1982). Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Env. Microbiol. 43, 777–780. doi: 10.1128/aem.43.4.777-780
Thakaew, R., and Niamsup, H. (2013). Inhibitory Activity of Bacillus subtilis BCC 6327 Metabolites against Growth of Aflatoxigenic Fungi Isolated from Bird Chili Powder. Internat. J. Biosci. Biochem. Bioinform. 3, 27–32. doi: 10.7763/IJBBB.2013.V3.157
Wang, X., Zhou, X., Cai, Z., Guo, L., Chen, X., Chen, X., et al. (2020). A Biocontrol Strain of Pseudomonas aeruginosa CQ-40 Promote Growth and Control Botrytis cinerea in Tomato. Pathogens 10:1. doi: 10.3390/pathogens10010022
Wu, Z., Huang, Y., Li, Y., Dong, J., Liu, X., and Li, C. (2019). Biocontrol of Rhizoctonia solani via Induction of the Defense Mechanism and Antimicrobial Compounds Produced by Bacillus subtilis SL-44 on Pepper (Capsicum annuum L.). Front. Microbiol. 10:2676. doi: 10.3389/fmicb.2019.02676
Xu, W., Xu, L., Deng, X., Goodwin, P. H., Xia, M., Zhang, J., et al. (2021). Biological Control of Take-All and Growth Promotion in Wheat by Pseudomonas chlororaphis YB-10. Pathogens 10:7. doi: 10.3390/pathogens10070903
Xu, W., Zhang, L., Goodwin, P. H., Xia, M., Zhang, J., Wang, Q., et al. (2020). Isolation, Identification, and Complete Genome Assembly of an Endophytic Bacillus velezensis YB-130, Potential Biocontrol Agent Against Fusarium graminearum. Front. Microbiol. 11:598285. doi: 10.3389/fmicb.2020.598285
Xu, W.-F., Ren, H.-S., Ou, T., Lei, T., Wei, J.-H., Huang, C.-S., et al. (2019). Genomic and Functional Characterization of the Endophytic Bacillus subtilis 7PJ-16 Strain, a Potential Biocontrol Agent of Mulberry Fruit Sclerotiniose. Microb. Ecol. 77, 651–663. doi: 10.1007/s00248-018-1247-4
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Zhang, H., Yohe, T., Huang, L., Entwistle, S., Wu, P., Yang, Z., et al. (2018). dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101. doi: 10.1093/nar/gky418
Zhao, J., Mei, Z., Zhang, X., Xue, C., Zhang, C., Ma, T., et al. (2017). Suppression of Fusarium wilt of cucumber by ammonia gas fumigation via reduction of Fusarium population in the field. Sci. Rep. 7:43103. doi: 10.1038/srep43103
Zhou, J., Wang, M., Sun, Y., Gu, Z., Wang, R., Saydin, A., et al. (2017). Nitrate Increased Cucumber Tolerance to Fusarium Wilt by Regulating Fungal Toxin Production and Distribution. Toxins 9:3. doi: 10.3390/toxins9030100
Keywords: Fusarium oxysporum f.sp. cucumerinum, biocontrol agent, genome sequencing and assembly, Bacillus subitilis, growth promotion
Citation: Xu W, Yang Q, Yang F, Xie X, Goodwin PH, Deng X, Tian B and Yang L (2022) Evaluation and Genome Analysis of Bacillus subtilis YB-04 as a Potential Biocontrol Agent Against Fusarium Wilt and Growth Promotion Agent of Cucumber. Front. Microbiol. 13:885430. doi: 10.3389/fmicb.2022.885430
Received: 28 February 2022; Accepted: 06 May 2022;
Published: 09 June 2022.
Edited by:
Maurizio Ruzzi, University of Tuscia, ItalyReviewed by:
Orlando Borras-Hidalgo, Qilu University of Technology, ChinaDivjot Kour, Eternal University, India
Hesham Ali El Enshasy, University of Technology Malaysia, Malaysia
Copyright © 2022 Xu, Yang, Yang, Xie, Goodwin, Deng, Tian and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baoming Tian, MTUwNzEyNDY1NTZAMTYzLmNvbQ==; Lirong Yang, bHVja195bHJAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Wen Xu
Wen Xu Qian Yang
Qian Yang Fan Yang3†
Fan Yang3† Lirong Yang
Lirong Yang