- 1School of Zhang Zhongjing Health Care and Food, Nanyang Institute of Technology, Nanyang, China
- 2Food and Pharmaceutical Engineering Institute, Guiyang University, Guiyang, China
- 3Inner Mongolia Meng Niu Dairy Industry (Group) Co. Ltd. R&D Center, Hohhot, China
- 4Heilongjiang Feihe Dairy Co., Ltd., Beijing, China
- 5Department of Immunology, College of Basic Medical Sciences, Shenyang Medical College, Shenyang, China
- 6Institute of Integrated Agricultural Development Research, Guizhou Academy of Agriculrural Sciences, Guiyang, China
- 7The Department of Food Science, Shenyang Medical College, Shenyang Medical College, Shenyang, China
The purpose of this study was to investigate the prevalence of Cronobacter spp. in commercial powdered infant formula (PIF) from nine provinces in China from March 2018 to September 2020, and to reveal the genotype, biofilm-forming ability, and antibiotic susceptibility of these isolates. A total of 27 Cronobacter strains, consisting of 22 Cronobacter sakazakii strains, 3 Cronobacter malonaticus strains, 1 Cronobacter turicensis strain, and 1 Cronobacter dublinensis strain, were isolated from 3,600 commercial PIF samples with a prevalence rate of 0.75%. Compared with the other 8 provinces, PIF from Shaanxi province had a higher prevalence rate (1.25%) of Cronobacter spp. These isolates were divided into 14 sequence types (STs), and 6 Cronobacter serotypes. The main Cronobacter STs were ST4, ST1, and ST64, and the dominant Cronobacter serotype was C. sakazakii serotype O2. Approximately 88.89% of Cronobacter isolates had a strong ability (OD595 > 1) to form biofilms on tinplate, among which the strains with ST4 were more dominant. All isolates were susceptible to ampicillin-sulbactam, ceftriaxone, cefotaxime, sulfadiazine, sulfadoxine, trimethoprim-sulfamethoxazole, gentamicin, tetracycline, ciprofloxacin, and colistin, while 55.56 and 96.30% isolates were resistant to cephalothin and vancomycin, respectively. Taken together, our findings highlighted the contamination status and characterization of Cronobacter spp. in commercial PIF from nine provinces of China, and provided guidance for the effective prevention and control of this pathogen in the production of PIF.
Introduction
Cronobacter spp. is an important opportunistic food-borne pathogen and can infect people of all ages (Sonbol et al., 2013; Kadlicekova et al., 2018). Due to lower immunity, newborns, especially low-birth-weight infants, are more likely to be infected by Cronobacter spp. compared to other groups of people, and suffer from bacteremia, necrotizing enterocolitis, meningitis, and even death (Caubilla-Barron et al., 2007; Fei et al., 2015). Currently, ingestion of powdered infant formula (PIF) contaminated with Cronobacter spp. is the leading cause of infection in newborns (Joseph and Forsythe, 2011; Yan et al., 2012). Therefore, the prevalence of Cronobacter spp. in commercial PIF has been a constant concern.
Genotyping of Cronobacter spp. isolated from PIF can reveal its molecular characterization, and contribute to the prevention and source tracking of this pathogen (Fei et al., 2018; Ling et al., 2021). Multilocus sequence typing (MLST) is considered a powerful tool for detection and genotyping of Cronobacter spp., because of discrimination at species level and information sharing platform (Joseph and Forsythe, 2012; Joseph et al., 2012; Li C. et al., 2020; Ling et al., 2021). To date, 3,506 Cronobacter spp. isolates have been classified into 794 sequence types (STs) using MLST method according to the records of Cronobacter PubMLST database (http://www.pubmlst.org/cronobacter). In addition, O-antigen serotyping based on lipopolysaccharide structure associated with the proinflammatory response of host to infection should also be implemented to increase the understanding of Cronobacter spp. for epidemiological purposes (Blazkova et al., 2015; Fei et al., 2017b).
Some studies have shown that secondary contamination during packaging and transportation is the main cause of Cronobacter spp. contamination in PIF, and the strong ability of Cronobacter spp. to form biofilms increases this possibility (Lee et al., 2012; Yang et al., 2013; Brandl et al., 2014). On the one hand, biofilm helps Cronobacter spp. firmly adhere to the surface of equipment and packaging materials, increasing the possibility of contamination of PIF with this pathogen (Aly et al., 2019). On the other hand, the existence of biofilm reduces the bactericidal effect of disinfectants and plays a protective role effect on Cronobacter spp. (Lehner et al., 2005). Therefore, the biofilm-forming ability of Cronobacter spp. isolated from PIF should be evaluated, which can contribute to the development of effective prevention and control measures of this pathogen.
In addition, the monitoring of antibiotic resistance of Cronobacter spp. is also necessary in order to update the effective antibiotic treatment regimen (Li Y. et al., 2020). For now, antibiotics are still the most effective therapeutic method for human Cronobacter infection (Fei et al., 2017a). However, long-term use of antibiotics or horizontal gene transfer are likely to result in increased antibiotic resistance in Cronobacter spp. (Holy et al., 2019). More importantly, the occurrence of multiple-drug-resistant Cronobacter strains can lead to the failure of antibiotics treatment, thereby increasing the epidemiological risk of this pathogen (Zeng et al., 2018; Li C. et al., 2020).
From the above considerations, we investigated the contamination by Cronobacter spp. in commercial PIF from supermarkets in nine provinces in China from March 2018 to September 2020. A total of 27 Cronobacter spp. isolates were detected, and genotyped using multilocus sequence typing (MLST) and O-antigen serotyping. Besides, the ability of biofilms formation on tinplate and antibiotic susceptibility of all isolates were tested.
Materials and Methods
Sampling
From March 2018 to September 2020, a total of 3,600 commercial PIF samples were randomly purchased from supermarkets in nine provinces of China, including Heilongjiang (n = 400), Xinjiang (n = 400), Jilin (n = 400), Hebei (n = 400), Henan (n = 400), Shaanxi (n = 400), Guizhou (n = 400), Yunnan (n = 400), and Fujian (n = 400). As shown in the Supplementary Figure 1, the nine provinces were distributed in the northeast, northwest, central, southwest, and southeast of China. The sample details are given in Supplementary Table 1, including collection region, collection time, manufacturers, place of origin, and raw milk types. Besides, all PIF samples are from local enterprises in China. After purchase, the packaging surfaces of all samples were sterilized with 75% alcohol, placed in sterile convenient insulated containers with ice packs, and quickly transferred to the laboratory.
Isolation and Identification of Cronobacter spp.
The suspected Cronobacter spp. were isolated from commercial PIF samples, and identified at the biochemical level according to the method as mentioned in the national food safety standard method for food microbiological examination as used in China GB4789.40-2010 (Ministry of Health of the People's Republic of China, 2010). Briefly, 100 g of PIF was fully dissolved in 900 ml sterile buffered peptone water followed by incubation at 36°C for 18 h. One milliliter culture was selectively enriched in 10 ml modified lauryl sulfate tryptose broth-vancomycin (mLST-Vm) medium with incubation at 44°C for 24 h. The above bacterial solution was spread on the Druggan-Forsythe-Iversen (DIF) medium, and the blue-green colonies were identified as the suspected colonies. All typical colonies were inoculated into tryptic soy broth, followed by cultivation at 37°C for 18 h. Then, genomic DNA of the culture was extracted using TIANamp Bacterial DNA Kit [Tiangen Biotech (Beijing) Co, Ltd, Beijing, China]. Finally, species-level identification of the isolates was performed using fusA gene sequencing, as previously reported (Fei et al., 2018).
MLST Analysis
Primer sequences and PCR amplification conditions for seven housekeeping genes (atpD, fusA, glnS, gltB, gyrB, infB, and ppsA) were provided by a previous report (Baldwin et al., 2009). The PCR amplified products were sequenced by Beijing Genomics Institute (BGI, Beijing China). Sequence type (ST) of isolate was determined by comparing it with known 7-loci profiles in the Cronobacter PubMLST database (http://www.pubmlst.org/cronobacter). A phylogenetic tree based on seven housekeeping genes was constructed using the Unweighted Pair Group Method with Arithmetic mean (UPGMA) algorithm in MEGA7.0, with 1,000 bootstrap replicates. The C. sakazakii ATCC 29544, C. sakazakii ATCC BAA-894, C. sakazakii ATCC 29004, C. sakazakii ATCC 12868, C. malonaticus CDC 105877, C. dublinensis LMG 23823, C. turicensis LMG 23827, C. universalis NCTC 9529, C. condimenti LMG 26250, and C. muytjensii ATCC 51329 were used as reference strains.
O-Antigen Serotype Analysis
The serotype-specific PCR assays was used to determine the O-antigen serotype (OT) of Cronobacter spp. isolates according to previous reports, as follows: C. sakazakii serotype (Cs) O1, CsO2, CsO3, CsO4, and CsO7 (Jarvis et al., 2011; Sun et al., 2012b), C. malonaticus serotype (Cm) O1 and CmO2 (Blazkova et al., 2015), C. turicensis serotype (Ct) O1 to CtO3 (Sun et al., 2012a; Jarvis et al., 2013), and C. dublinensis (Cd) O1 and CdO2 (Jarvis et al., 2013). The sequences of 12 pairs of primers used for O-antigen serotype analysis were shown in Supplementary Table 2.
Biofilm Formation by Cronobacter spp. on Tinplate
The biofilm biomass produced by Cronobacter spp. on tinplate was measured as described by Fei et al. (2021). In brief, 30 μl of Cronobacter pre-culture was inoculated into 24-well polystyrene plates containing 2 ml of TSB. Sterilized tinplate coupons (1 × 1 cm) were inserted vertically into the wells, followed by incubation at 37°C for 48 h to allow the isolates to form biofilms. After three gentle rinses with sterilized distilled water, the cultures on coupons were removed. The biofilms on coupons were stained with 0.1% crystal violet for 30 min, then rinsed to remove the crystal violet, followed by dissolution in 2 ml of 75% ethanol solution. The optical density (OD) value of the above-mentioned solution at wavelengths of 595 nm representing staining intensity was measured to assess the biofilm-forming ability of Cronobacter strains. The unincubated strain was used as a control to eliminate the background staining. The above experiments were repeated three times.
Antibiotic Susceptibility Testing
The Kirby–Bauer disk diffusion method was used to assess the antibiotic susceptibilities of Cronobacter isolates according to the guidelines of the Clinical Laboratory Standards Institute (CLSI, 2020). Nine categories of antibiotics were selected for the antibiotic susceptibility testing, β-Lactams: ampicillin (10 μg), amoxicillin (25 μg), cephalothin (30 μg), ampicillin-sulbactam (10:10 μg), cefotaxime (30 μg), and ceftriaxone (30 μg); macrolides: azithromycin (15 μg) and erythromycin (15 μg); sulfonamides: sulfadiazine (250 μg), sulfadoxine (250 μg), and trimethoprim-sulfamethoxazole (1.25/23.75 μg); aminoglycosides: streptomycin (10 μg) and gentamicin (10 μg); amphenicols: chloramphenicol (30 μg); tetracyclines: tetracycline (30 μg) and oxytetracycline (30 μg); glycopeptide: vancomycin (30 μg); fluoroquinolones: ciprofloxacin (5 μg) and pipemidic acid (30 μg); polypeptide: colistin (10 μg). Escherichia coli ATCC 25922 was used as the quality control strain. In accordance with the CLSI guidelines, if the diameter of inhibition zone (DIZ) against Cronobacter spp. isolates was greater than that of quality control strain, the antibiotic resistance was interpreted as sensitive (S); if it is less than, the antibiotic resistance was defined as resistant (R); if the DIZ was between R and S, the effect of antibiotics was expressed as intermediate (I).
Statistical Analysis
Biofilm formation test and antibiotic susceptibility test of isolates were carried out in triplicate. The data were presented in the form of mean ± standard deviation, and were statistically analyzed using ANOVA by the SPSS 20.0 software (SPSS Inc., Chicago, IL). P ≤ 0.05 was considered significant.
Results
Prevalence of Cronobacter spp. in Commercial PIF From Nine Provinces in China
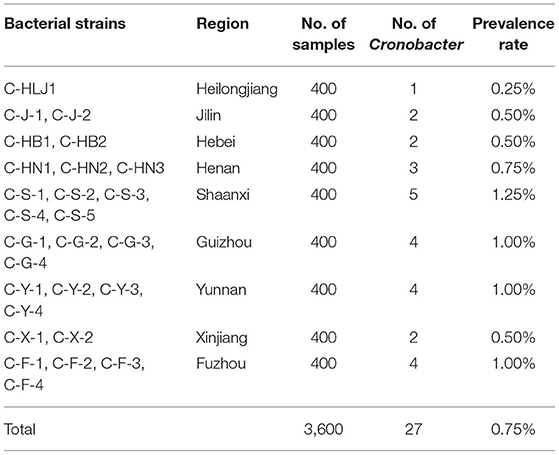
As shown in Table 1, the total prevalence rate of Cronobacter spp. in PIF from nine provinces in China was 0.75% (27/3600). The highest prevalence rate (1.25%, 5/400) of Cronobacter spp. was found in commercial PIF from Shaanxi province, followed by Guizhou (1.00%, 4/400), Yunnan (1.00%, 4/400), Fujian provinces (1.00%, 4/400), Henan (0.75%, 3/400), Jilin (0.50%, 2/400), Hebei (0.50%, 2/400), Xinjiang (0.50%, 2/400), and Heilongjiang (0.25%, 1/400).
MLST Analysis
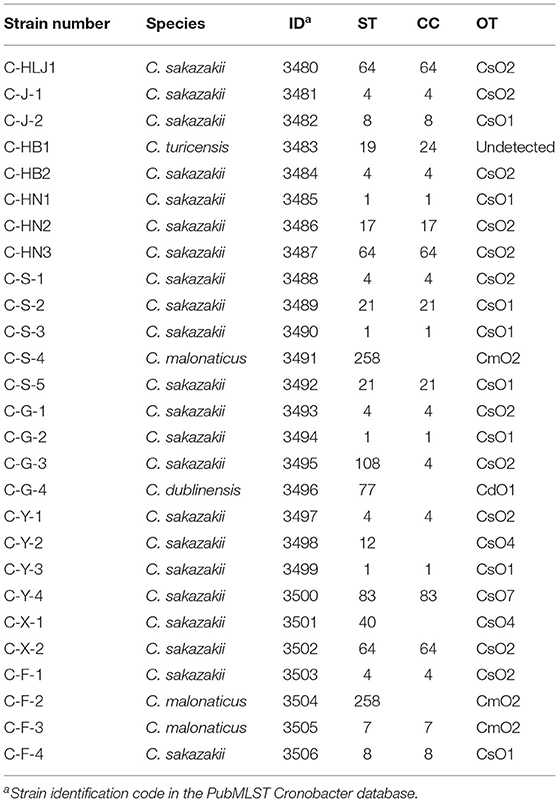
As shown in Table 2, 27 Cronobacter isolates included 22 C. sakazakii strains (81.48%), 3 C. malonaticus strains (11.11%), 1 C. turicensis strain (3.70%), and 1 C. dublinensis strain (3.70%), and were divided into 14 STs using MLST method. Among them, ST4 (6/27, 22.22%), ST1 (4/27, 14.81%), and ST64 (3/27, 11.11%) were the dominant Cronobacter STs, meanwhile, clonal complex (CC) 4 (7/27, 25.93%) was the main Cronobacter CC. In addition, Figure 1 showed a phylogenetic relationship of 27 isolates based on 3,036 bp-concatenated sequences of 7 housekeeping genes. These isolates were clustered into six groups using 98% similarity as the identification threshold. Group I (13/27, 48.15%) was the dominant group, and contains C. sakazakii ST4, ST8, ST64, ST83, and ST108. In addition, C. sakazakii ST4 was the most widely distributed, being found in all 6 provinces. However, a clear association between ST and collection region was not found.
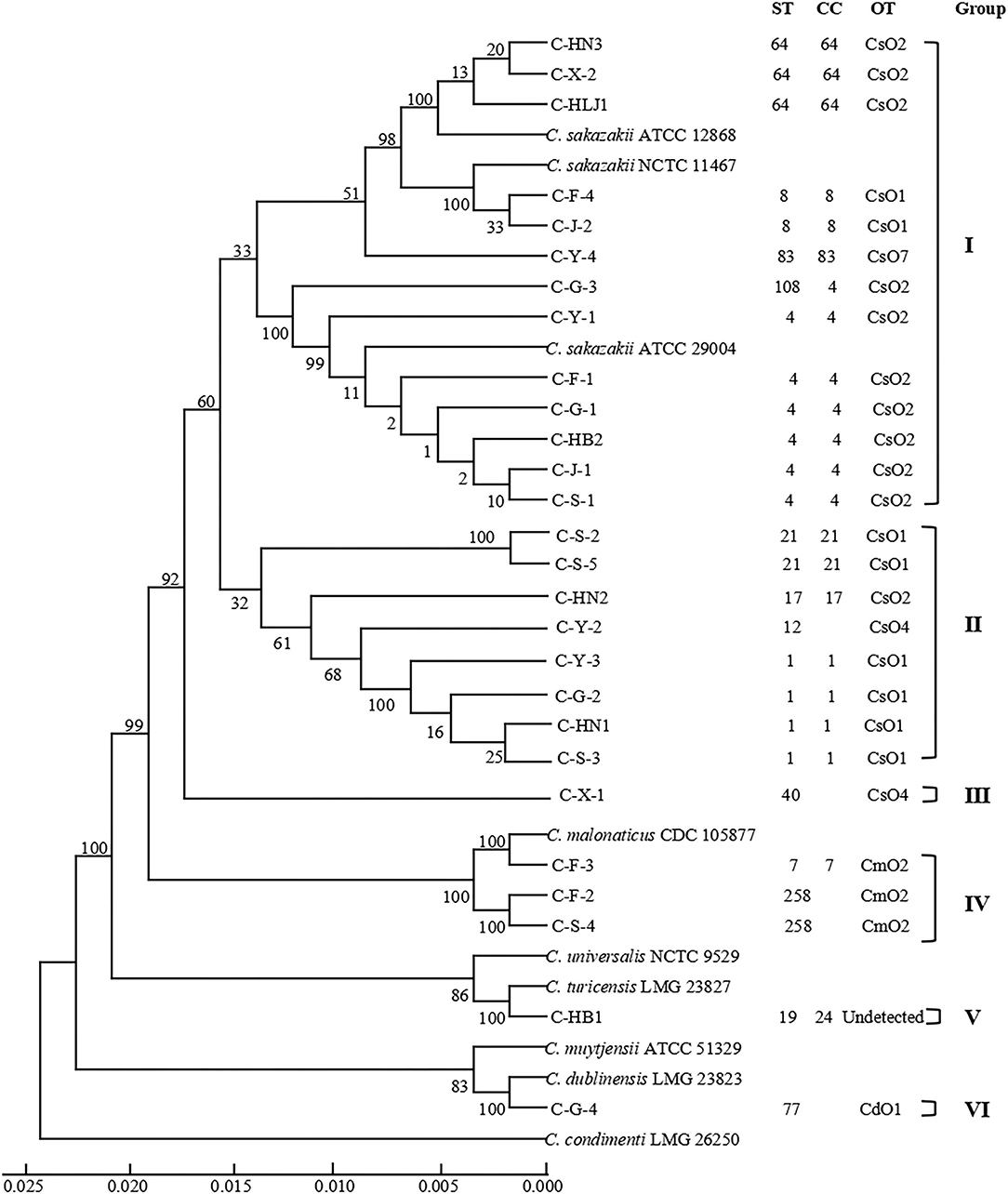
Figure 1. Phylogenetic relationship of 27 Cronobacter isolates containing 14 STs. The UPGMA tree based on 7 loci was constructed using MEGA7.0, with 1,000 bootstrap replicates. The Cronobacter sakazakii ATCC 29544, C. sakazakii ATCC BAA-894, C. sakazakii ATCC 29004, C. sakazakii ATCC 12868, Cronobacter malonaticus CDC 105877, Cronobacter dublinensis LMG 23823, Cronobacter turicensis LMG 23827, Cronobacter universalis NCTC 9529, Cronobacter condimenti LMG 26250, and Cronobacter muytjensii ATCC 51329 were used as the reference strains. Detailed information about each strain is added, including ST, CC, OT, and group.
O-Antigen Serotype Analysis
Table 2 showed that 27 Cronobacter strains were classified into 6 OTs. Twenty-two C. sakazakii isolates were divided into four C. sakazakii serotypes, among them, CsO2 (11/22, 50.00%) was the main OT of C. sakazakii, followed by CsO1 (8/22, 36.36%), CsO4 (2/22, 9.09%), and CsO7 (1/22, 4.55%). Three C. malonaticus isolates were identified as CmO2. Meanwhile, C. dublinensis C-G-4 (ST77) was detected as CdO1, while, the C. turicensis serotype of C. turicensis C-HB1 (ST19) was not found. Furthermore, the isolates with the same ST were classified into the same OT, however, the isolates with the same OT contained multiple STs, which suggested that the discrimination of O-antigen serotype analysis was lower than that of MLST.
Ability of Biofilm Formation of Cronobacter spp. on Tinplate
As shown in Figure 2, the biofilm-forming abilities of 27 Cronobacter isolates with 14STs on tinplate were tested. Among the 27 Cronobacter strains, the OD595 values of 24 isolates (88.89%), including 22 strains of C. sakazakii and 3 strains of C. malonaticus, were >1. Meanwhile, the OD595 values of three strains of C. sakazakii (C-F-1, C-G-1, and C-J-1) were >2, while the OD595 values of the three strains (C. sakazakii C-HN2, C. dublinensis C-HB1, and C. turicensis C-G-4) were <1. In addition, there was a correlation between the ST and biofilm-forming ability of Cronobacter strains. The ability to form biofilm of strains with ST4 (OD595 = 2.03 ± 0.14) was significantly stronger than that of strains with other STs (P < 0.05), followed by C. sakazakii ST12 (OD595 = 1.78 ± 0.13), C. sakazakii ST64 (OD595 = 1.66 ± 0.13), C. sakazakii ST1 (OD595 = 1.63 ± 0.33), and C. malonaticus ST258 (OD595 = 1.32 ± 0.16).
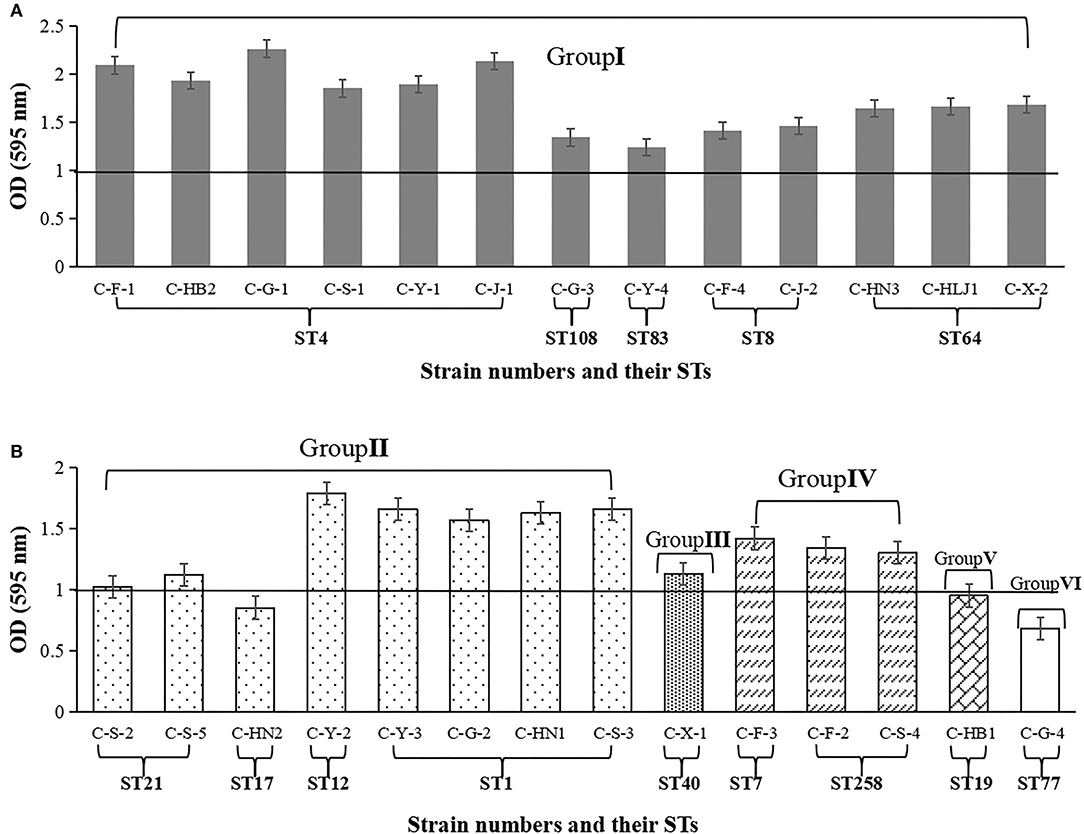
Figure 2. Biofilm biomass formed by 27 Cronobacter isolates on tinplate. The OD value at wavelength of 595 nm was used to indicate the biofilm biomass. (A) Cronobacter isolates belonging to group I. (B) Cronobacter isolates belonging to group II, group III group IV, group V, and group VI, respectively.
Antibiotic Susceptibility Analysis
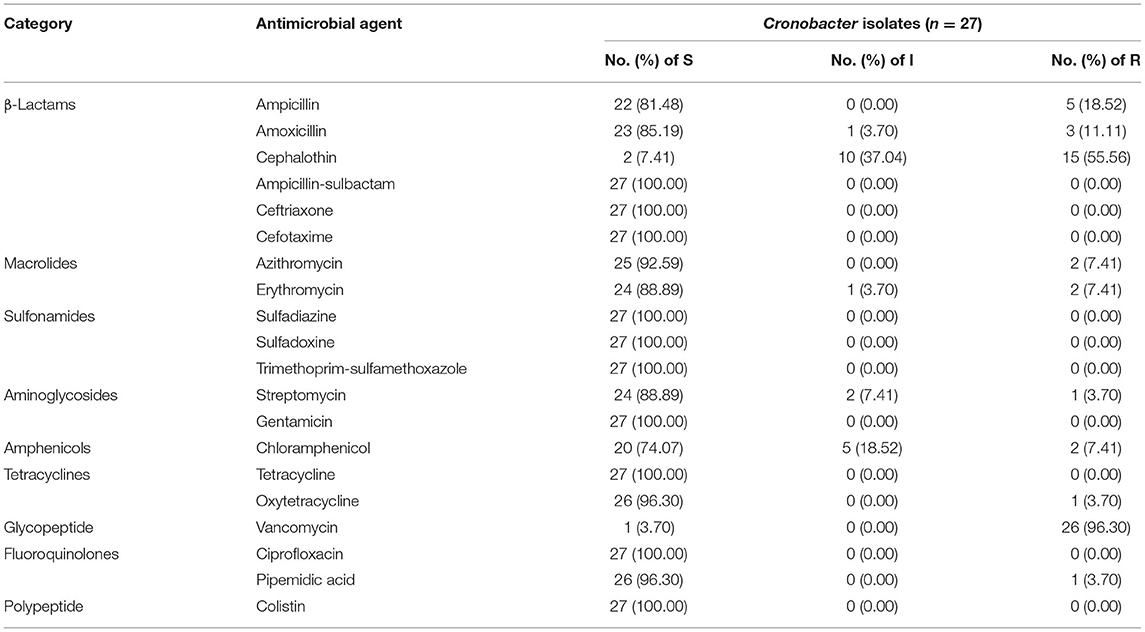
Table 3 showed the antibiotic susceptibilities of 27 Cronobacter isolates to 20 antibiotics (9 categories). All isolates were susceptible to ampicillin-sulbactam, ceftriaxone, cefotaxime, sulfadiazine, sulfadoxine, trimethoprim-sulfamethoxazole, gentamicin, tetracycline, ciprofloxacin, and colistin. The majority of isolates were susceptible to oxytetracycline (96.30%), pipemidic acid (96.30%), azithromycin (92.59%), erythromycin (88.89%), streptomycin (88.89%), amoxicillin (85.19%), and ampicillin (81.48%). While, 55.56 and 96.30% of strains were resistant to cephalothin and vancomycin, respectively. In addition, C. sakazakii C-Y-4 (ST83), C-S-2(ST21), and C-S-5(ST21) exhibited resistance to three antibiotics (ampicillin, cephalothin, and vancomycin).
Discussion
Over the years, many studies have been devoted to investigating the occurrence of Cronobacter spp. in PIF in the Chinese market, in order to provide an early warning of risk to consumers and public safety (Pan et al., 2014; Fei et al., 2017b; Zhang et al., 2017; Ling et al., 2021). Pan et al. (2014) detected 19 Cronobacter strains from 165 PIF samples, with a prevalence rate of 11.50%, from December 2011 to December 2012. Zhang et al. (2017) found that from 2011 to 2013, the prevalence rates of Cronobacter spp. in commercial PIF was 1.60% in Shandong, China. Further, Fei et al. (2017a,b) investigated the presence of Cronobacter spp. in PIF collected from Chinese retail markets from January 2015 to March 2017, and the prevalence rate was 2.80%. Ling et al. (2021) reviewed the incidences (Range from 0.90 to 23.10%) of Cronobacter spp. isolated from PIF in China from 2004 to 2018, which claimed to cover over 10,000 PIF samples in total. Compared with the above studies, in this study, the prevalence rate (0.75%) of Cronobacter spp. in commercial PIF was obviously reduced, indicating that these continuous studies have attracted the attention of government departments and manufacturers and contributed to the improvement of Cronobacter spp. control measures. It should be emphasized that almost all the methods for the isolation of Cronobacter spp. from China, including this study, were carried out according to the standard of China GB4789.40-2010 (Pan et al., 2014; Fei et al., 2017b; Zhang et al., 2017; Li et al., 2019; Li C. et al., 2020; Zeng et al., 2020). According to this national standard, the mLST-Vm medium was used for selective enrichment of bacteria solution, and its culture temperature was 44°C, which may lead to the failure of some Cronobacter strains to grow (International Standards Organization [ISO], 2017). On the other hand, the raw milk of PIF collected from Shaanxi province was goat milk, and the prevalence rate of this pathogen in these samples is the highest, whether there is a correlation between the two needs further study. In addition, we found that the prevalence rate of Cronobacter spp. was related to the distance between the place of production (Inner Mongolia) and the place of sale (except Shaanxi), and the farther the distance was, the higher the prevalence rate was (Supplementary Figure 1), which suggests that we need to strengthen the hygiene management in the PIF logistics process.
In this study, 27 Cronobacter strains isolated from commercial PIF contained four species, and were divided into 14 STs. Previous studies have shown that C. sakazakii, C. malonaticus, and C. turicensis are associated with the neonatal infections in seven species of Cronobacter spp. (Forsythe, 2005; Joseph et al., 2012), and the strains belonging to the above 3 species were detected in our study. Meanwhile, among the 14 STs, several important STs have been found. The details are as follows: C. sakazakii ST4 is the main ST leading to neonatal meningitis (Joseph et al., 2012), C. sakazakii ST1 is the dominant ST isolated from PIF (Sonbol et al., 2013), C. sakazakii ST12 is associated with infant necrotizing enterocolitis (Forsythe et al., 2014), and C. sakazakii ST8 is mainly isolated from clinical cases (Sonbol et al., 2013). In addition, C. sakazakii ST4, ST1, and ST64 were the dominant STs isolated from PIF in this study, which was consistent with previous studies (Fei et al., 2015, 2017b). But, ST64 was not the predominant ST of Cronobacter strains from PIF in other countries (Sonbol et al., 2013). Interestingly, C. sakazakii ST64 was also the dominant ST isolated from Chinese aquatic products, meat, and meat products (Li C. et al., 2020; Zeng et al., 2020). Therefore, C. sakazakii strains with ST64 should be heavily prevented and controlled for China's food industry.
As a supplement to MLST, the O-antigen serotype analysis was performed to reveal information related to the pathogenicity of Cronobacter isolates. The predominant serotype of Cronobacter isolates from commercial PIF in the current study was CsO2, which was closely correlated with the C. sakazakii ST4 causing neonatal meningitis. As comparisons, the main serotype of Cronobacter strains in aquatic products and edible mushrooms was CsO1 (Li et al., 2019; Li C. et al., 2020), as well as, CsO1 and CsO2 were the dominant serotypes of Cronobacter spp. in the spices, cereals, meat, and meat products (Li et al., 2017; Zeng et al., 2020). In addition, CsO3 was found in aquatic products, edible mushrooms, meat, and meat products (Li et al., 2019; Li C. et al., 2020; Zeng et al., 2020), but this serotype was not detected in our study.
Previous studies mainly focus on the biofilm formation of Cronobacter strains on stainless steel, however, most of this pathogen in PIF comes from re-contamination during packaging and logistics (Lee et al., 2012; Fei et al., 2015; Huang et al., 2020). Therefore, this study evaluated the biofilm-forming ability of Cronobacter isolates on tinplate that is the commonly used packaging material. In this study, we found that there was a certain relationship between the biofilm biomass and ST of Cronobacter spp., and the strains with ST4 had a higher ability to produce biofilm, which may be one reason why C. sakazakii ST4 is so prevalent in the environment. In addition, Gupta et al. (2018) revealed the ability of C. sakazakii to form biofilms on different materials, including stainless steel, polyurethane, polyvinyl chloride, and silicone, and found that polyvinyl chloride and silicone were the higher risk materials.
In order to better select effective antibiotics, 20 antibiotics belonging to 9 categories were used to test the antibiotic susceptibility of Cronobacter isolated from commercial PIF. In this study, all isolates were susceptible to all sulfonamides antibiotics, while most isolates were resistant to cephalothin and vancomycin, which showed similar resistance to most antibiotics as in previous studies (Fei et al., 2017b; Li et al., 2019; Li C. et al., 2020). Differently, compared with Cronobacter strains isolated from aquatic and meat products, the isolates in this study were more resistant to ampicillin and more susceptible to cephalothin and trimethoprim/sulfameth-oxazol (Li C. et al., 2020; Zeng et al., 2020). In addition, in this study, three isolates (11.11%) from PIF samples were found to be resistant to three antibiotics, which suggests that with the long-term use of antibiotics, multidrug-resistant strains are emerging, and that monitoring of antibiotic susceptibility of isolates should be continuous for early warning purposes.
Conclusions
In conclusion, we revealed the prevalence, genotype, biofilm formation, and antibiotic susceptibility of Cronobacter spp. in commercial PIF collected from nine provinces in China. Compared with before, a lower prevalence rate of Cronobacter spp. in PIF was found. C. sakazakii ST4, ST1, and ST64 were still the dominant STs in commercial PIF, and the isolates belonging to dominant STs had a stronger ability to produce biofilms. Sulfonamides were recommended for the treatment of Cronobacter infection. Our findings help to reveal the contamination status and characterization of Cronobacter spp. in PIF from Chinese markets, and provide the guidance for effectively preventing the contamination of this pathogen in PIF.
Data Availability Statement
The data presented in the study are deposited in the Cronobacter PubMLST database repository (http://www.pubmlst.org/cronobacter), accession number 3480-3506.
Author Contributions
PF, WY, YC, and ZM conceived and designed the experiments. PF, HJ, YM, GD, SL, XC, SJ, QX, and WY performed the experiments. PF, YC, and WY supervised the project. PF, XC, and YC analyzed the data. PF wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Natural Science Foundation of Henan Province (212300410137), Natural Science Foundation of Guizhou Province (Qiankehe Foundation-ZK [2022] General 008), Key Scientific Research Projects of Institutions of Higher Learning of Henan (21A550007), Liaoning Provincial Science and Technology Department issued the Key Research and Development Project of Tackling Key Problems and Industrialization (2017225065), and Innovation and Entrepreneurship Training Program of College Students of Henan Province (202110464027).
Conflict of Interest
ZM is employed by Inner Mongolia Meng Niu Dairy Industry Group Co., Ltd. SJ and QX are employed by Heilongjiang Feihe Dairy Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.900690/full#supplementary-material
Supplementary Figure 1. Sampling sites and milk source base of commercial PIF used in this study.
Supplementary Table 1. The specific information of all samples.
Supplementary Table 2. The sequences of 12 pairs of primers used for O-antigen serotype analysis.
References
Aly, M. A., Reimhult, E., Kneifel, W., and Domig, K. J. (2019). Characterization of biofilm formation by Cronobacter spp. isolates of different food origin under model conditions. J. Food Protect. 82, 65–77. doi: 10.4315/0362-028X.JFP-18-036
Baldwin, A., Loughlin, M., Caubilla-Barron, J., Kucerova, E., Manning, G., Dowson, C., et al. (2009). Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance which do not correlate with biotypes. BMC Microbiol. 9, 223. doi: 10.1186/1471-2180-9-223
Blazkova, M., Javurkova, B., Vlach, J., Goeselova, S., Karamonova, L., Ogrodzki, P., et al. (2015). Diversity of O antigens within the genus Cronobacter: from disorder to order. Appl. Environ. Microbiol. 81, 5574–5582. doi: 10.1128/AEM.00277-15
Brandl, H., Fricker-Feer, C., Ziegler, D., Mandal, J., Stephan, R., and Lehner, A. (2014). Distribution and identification of culturable airborne microorganisms in a Swiss milk processing facility. J. Dairy Sci. 97, 240–246. doi: 10.3168/jds.2013-7028
Caubilla-Barron, J., Hurrell, E., Townsend, S., Cheetham, P., Loc-Carrillo, C., Fayet, O., et al. (2007). Genotypic and phenotypic analysis of Enterobacter sakazakii strains from an outbreak resulting in fatalities in a neonatal intensive care unit in France. J. Clin. Microbiol. 45, 3979–3985. doi: 10.1128/JCM.01075-07
CLSI Clinical Laboratory Standards Institute (2020). Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standards Institute.
Fei, P., Jiang, Y., Feng, J., Forsythe, S. J., Li, R., Zhou, Y., et al. (2017a). Antibiotic and desiccation resistance of Cronobacter sakazakii and C. malonaticus isolates from powdered infant formula and processing environments. Front. Microbiol. 8,316. doi: 10.3389/fmicb.2017.00316
Fei, P., Jiang, Y., Gong, S., Li, R., Jiang, Y., Yuan, X., et al. (2018). Occurrence, genotyping, and antibiotic susceptibility of Cronobacter spp. in drinking water and food samples from Northeast China. J. Food Protect. 81, 456–460. doi: 10.4315/0362-028X.JFP-17-326
Fei, P., Jiang, Y., Jiang, Y., Yuan, X., Yang, T., Chen, J., et al. (2017b). Prevalence, molecular characterization, and antibiotic susceptibility of Cronobacter sakazakii isolates from powdered infant formula collected from Chinese retail markets. Front. Microbiol. 8,2026. doi: 10.3389/fmicb.2017.02026
Fei, P., Man, C., Lou, B., Forsythe, S. J., Chai, Y., Li, R., et al. (2015). Genotyping and source tracking of Cronobacter sakazakii and C. malonaticus isolates from powdered infant formula and an infant formula production factory in China. Appl. Environ. Microbiol. 81, 5430–5439. doi: 10.1128/AEM.01390-15
Fei, P., Xie, Q., Jiang, Y., Feng, H., Chang, Y., Kang, H., et al. (2021). Genotyping, antimicrobial susceptibility and biofilm formation of Bacillus cereus isolated from powdered food products in China. Foodborne Pathog. Dis. 18, 8–15. doi: 10.1089/fpd.2020.2802
Forsythe, S. J.. (2005). Enterobacter sakazakii and other bacteria in powdered infant milk formula. Matern. Child Nutr. 1, 44–50. doi: 10.1111/j.1740-8709.2004.00008.x
Forsythe, S. J., Dickins, B., and Jolley, K. A. (2014). Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genomics 15,1121. doi: 10.1186/1471-2164-15-1121
Gupta, T. B., Mowat, E., Brightwell, G., and Flint, S. H. (2018). Biofilm formation and genetic characterization of New Zealand Cronobacter isolates. J. Food Safety 38,e12430. doi: 10.1111/jfs.12430
Holy, O., Alsonosi, A., Hochel, I., Roderova, M., Zatloukalova, S., Mlynarcik, P., et al. (2019). Antibiotic susceptibility of Cronobacter spp. isolated from clinical samples. Pol. J. Microbiol. 68, 5–14. doi: 10.21307/pjm-2019-001
Huang, Y., Pei, Q., Deng, R., Zheng, X., Guo, J., Guo, D., et al. (2020). Inactivation efficacy of 405 nm LED against Cronobacter sakazakii biofilm. Front. Microbiol. 11,610077. doi: 10.3389/fmicb.2020.610077
International Standards Organization [ISO] (2017). Microbiology of the food chain: Horizontal method for the detection of Cronobacter spp. Geneva: International Standards Organization [ISO].
Jarvis, K. G., Grim, C. J., Franco, A. A., Gopinath, G., Sathyamoorthy, V., Hu, L., et al. (2011). Molecular characterization of Cronobacter lipopolysaccharide O-antigen gene clusters and development of serotype-specific PCR assays. Appl. Environ. Microbiol. 77, 4017–4026. doi: 10.1128/AEM.00162-11
Jarvis, K. G., Yan, Q. Q., Grim, C. J., Power, K. A., Franco, A. A., Hu, L., et al. (2013). Identification and characterization of five new molecular serogroups of Cronobacter spp. Foodborne Pathog. Dis. 10, 343–352. doi: 10.1089/fpd.2012.1344
Joseph, S., and Forsythe, S. J. (2011). Predominance of Cronobacter sakazakii sequence type 4 in neonatal infections. Emerg. Infect. Dis. 17, 1713–1715. doi: 10.3201/eid1709.110260
Joseph, S., and Forsythe, S. J. (2012). Insights into the emergent bacterial pathogen Cronobacter spp., generated by multilocus sequence typing and analysis. Front. Microbiol. 3,397. doi: 10.3389/fmicb.2012.00397
Joseph, S., Sonbol, H., Hariri, S., Desai, P., McClelland, M., and Forsythe, S. J. (2012). Diversity of the Cronobacter genus as revealed by multilocus sequence typing. J. Clin. Microbiol. 50, 3031–3039. doi: 10.1128/JCM.00905-12
Kadlicekova, V., Kajsik, M., Soltys, K., Szemes, T., Slobodnikova, L., Janosikova, L., et al. (2018). Characterisation of Cronobacter strains isolated from hospitalised adult patients. Anton. Leeuw. Int. J. G. 111, 1073–1085. doi: 10.1007/s10482-017-1008-2
Lee, Y. D., Park, J. H., and Chang, H. (2012). Detection, antibiotic susceptibility and biofilm formation of Cronobacter spp. from various foods in Korea. Food Control 24, 225–230. doi: 10.1016/j.foodcont.2011.09.023
Lehner, A., Riedel, K., Eberl, L., Breeuwer, P., Diep, B., and Stephan, R. (2005). Biofilm formation, extracellular polysaccharide production, and cell-to-cell signaling in various Enterobacter sakazakii strains: aspects promoting environmental persistence. J. Food Protect. 68, 2287–2294. doi: 10.4315/0362-028X-68.11.2287
Li, C., Zeng, H., Zhang, J., He, W., Ling, N., Chen, M., et al. (2019). Prevalence, antibiotic susceptibility, and molecular characterization of Cronobacter spp. isolated from edible mushrooms in China. Front. Microbiol. 10,283. doi: 10.3389/fmicb.2019.00283
Li, C., Zeng, H., Zhang, J., Luo, D., Chen, M., Lei, T., et al. (2020). Cronobacter spp. isolated from aquatic products in China: incidence, antibiotic resistance, molecular characteristic and CRISPR diversity. Int. J. Food Microbiol. 335,108857. doi: 10.1016/j.ijfoodmicro.2020.108857
Li, Y., Yu, H., Jiang, H., Jiao, Y., Zhang, Y., and Shao, J. (2017). Genetic diversity, antimicrobial susceptibility, and biofilm formation of Cronobacter spp. recovered from spices and cereals. Front. Microbiol. 8,2567. doi: 10.3389/fmicb.2017.02567
Li, Y., Zhang, Y., Zhang, L., Hu, Y., Hong, C., Xie, A., et al. (2020). Prevalence and genetic characteristics of Cronobacter spp. from food and human clinical stool samples in Wenzhou, China 2008-2018. Food Microbiol. 89, 103432. doi: 10.1016/j.fm.2020.103432
Ling, N., Jiang, Y., Zeng, H., Ding, Y., and Forsythe, S. (2021). Advances in our understanding and distribution of the Cronobacter genus in China. J. Food Sci. 86, 276–283. doi: 10.1111/1750-3841.15577
Ministry of Health of the People's Republic of China (2010). National Food Safety Standard: Food Microbiological Examnation: Enterobacter sakazakii. Beijing: China Standard Press.
Pan, Z., Cui, J., Lyu, G., Du, X., Qin, L., Guo, Y., et al. (2014). Isolation and molecular typing of Cronobacter spp. in commercial powdered infant formula and follow-up formula. Foodborne Pathog. Dis. 11, 456–461. doi: 10.1089/fpd.2013.1691
Sonbol, H., Joseph, S., McAuley, C. M., Craven, H. M., and Forsythe, S. J. (2013). Multilocus sequence typing of Cronobacter spp. from powdered infant formula and milk powder production factories. Int. Dairy J. 30, 1–7. doi: 10.1016/j.idairyj.2012.11.004
Sun, Y., Arbatsky, N. P., Wang, M., Shashkov, A. S., Liu, B., Wang, L., et al. (2012a). Structure and genetics of the O-antigen of Cronobacter turicensis G3882 from a new serotype, C. turicensis O2, and identification of a serotype-specific gene. Fems Immunol. Med. Mic. 66, 323–333. doi: 10.1111/j.1574-695X.2012.01013.x
Sun, Y., Wang, M., Wang, Q., Cao, B., He, X., Li, K., et al. (2012b). Genetic analysis of the Cronobacter sakazakii O4 to O7 O-antigen gene clusters and development of a PCR assay for identification of all C. sakazakii O serotypes. Appl. Environ. Microbiol. 78, 3966–3974. doi: 10.1128/AEM.07825-11
Yan, Q. Q., Condell, O., Power, K., Butler, F., Tall, B. D., and Fanning, S. (2012). Cronobacter species (formerly known as Enterobacter sakazakii) in powdered infant formula: a review of our current understanding of the biology of this bacterium. J. Appl. Microbiol. 113, 1–15. doi: 10.1111/j.1365-2672.2012.05281.x
Yang, S., Kim, S., Ryu, J.-H., and Kim, H. (2013). Inhibitory activity of Paenibacillus polymyxa on the biofilm formation of Cronobacter spp. on stainless steel surfaces. J. Food Sci. 78, M1036–M1040. doi: 10.1111/1750-3841.12168
Zeng, H., Lei, T., He, W., Zhang, J., Liang, B., Li, C., et al. (2018). Novel multidrug-resistant Cronobacter sakazakii causing meningitis in neonate, China, 2015. Emerg. Infect. Dis. 24, 2121–2123. doi: 10.3201/eid2411.180718
Zeng, H., Li, C., Ling, N., Zhang, J., Chen, M., Lei, T., et al. (2020). Prevalence, genetic analysis and CRISPR typing of Cronobacter spp. isolated from meat and meat products in China. Int. J. Food Microbiol. 321,108549. doi: 10.1016/j.ijfoodmicro.2020.108549
Keywords: Cronobacter spp., commercial powdered infant formula, genotype, biofilm formation, antibiotic susceptibility
Citation: Fei P, Jing H, Ma Y, Dong G, Chang Y, Meng Z, Jiang S, Xie Q, Li S, Chen X and Yang W (2022) Cronobacter spp. in Commercial Powdered Infant Formula Collected From Nine Provinces in China: Prevalence, Genotype, Biofilm Formation, and Antibiotic Susceptibility. Front. Microbiol. 13:900690. doi: 10.3389/fmicb.2022.900690
Received: 21 March 2022; Accepted: 19 April 2022;
Published: 27 May 2022.
Edited by:
Lin Lin, Jiangsu University, ChinaReviewed by:
Stephen Forsythe, Foodmicrobe.com, United KingdomDapeng Wang, Shanghai Jiao Tong University, China
Copyright © 2022 Fei, Jing, Ma, Dong, Chang, Meng, Jiang, Xie, Li, Chen and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiwei Yang, eXcxOTgyNjlAMTYzLmNvbQ==
 Peng Fei
Peng Fei He Jing1
He Jing1 Yunhe Chang
Yunhe Chang Shilong Jiang
Shilong Jiang