- 1Department of Biological, Geological, and Environmental Sciences, University of Bologna, Bologna, Italy
- 2European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group for Legionella Infections (ESGLI), Basel, Switzerland
- 3Department of Civil, Chemical, Environmental, and Materials Engineering, University of Bologna, Bologna, Italy
- 4Department of Specialty, Diagnostic and Experimental Medicine, University of Bologna, Bologna, Italy
- 5Regional Reference Laboratory for Clinical Diagnosis of Legionellosis, Molecular Microbiology and Virology Unit, University Hospital-Policlinico Modena, Modena, Italy
- 6Laboratory of Microbial Ecology and Genomics of Microorganisms, Istituto Zooprofilattico Sperimentale delle Venezie, Legnaro, Italy
Legionella surveillance plays a significant role not only to prevent the risk of infection but also to study the ecology of isolates, their characteristics, and how their prevalence changes in the environment. The difficulty in Legionella isolation, identification, and typing results in a low notification rate; therefore, human infection is still underestimated. In addition, during Legionella surveillance, the special attention given to Legionella pneumophila leads to an underestimation of the prevalence and risk of infection for other species. This study describes the workflow performed during environmental Legionella surveillance that resulted in the isolation of two strains, named 8cVS16 and 9fVS26, associated with the genus Legionella. Traditional and novel approaches such as standard culture technique, MALDI-TOF MS, gene sequencing, and whole-genome sequencing (WGS) analysis were combined to demonstrate that isolates belong to a novel species. The strain characteristics, the differences between macrophage infectivity potential (mip), RNA polymerase β subunit (rpoB), and reference gene sequences, the average nucleotide identity (ANI) of 90.4%, and the DNA–DNA digital hybridization (dDDH) analysis of 43% demonstrate that these isolates belong to a new Legionella species. The finding suggests that, during the culture technique, special attention should be paid to the characteristics of the isolates that are less associated with the Legionella genus in order to investigate the differences found using more sensitive methods. The characterization of the two newly discovered isolates based on morphological, biochemical, and microscopic characteristics is currently underway and will be described in another future study.
Introduction
Legionella spp. are gram-negative intracellular pathogenic bacteria, are ubiquitous in water and soil, and are represented by more than 66 species, some of which are potentially capable to cause a severe form of pneumonia, called Legionnaires' disease (Jomehzadeh et al., 2019; Parte et al., 2020). Legionella pneumophila (Lp) is the most common infectious agent involved in Legionnaires' disease and consists of 15 different serogroups. Lp serogroup 1 (Lp1), according to epidemiological data, is mostly associated with human infections (European Centre for Disease Prevention Control, 2021; Rota et al., 2021). Nevertheless, other Legionella non-pneumophila species (non-Lp species) (i.e., L. anisa, L. rubrilucens, or L. longbeachae) are responsible for human infections that are typically acquired through inhalation of contamination aerosol (Muder and Yu, 2002; Matsui et al., 2010; Cunha et al., 2016; European Centre for Disease Prevention Control, 2021; Rota et al., 2021).
Artificial water distribution systems (WDSs) are considered the sites most associated with Legionella spp. proliferation and spread (Mercante and Winchell, 2015). The presence of biofilm, water stagnation due to low water flow, dead branches, or old pipelines are factors that make the WDS's facilities (e.g., large public or private buildings, companies, hospitals, or health facilities) a potential risk to the dissemination of Legionella spp. (Di Pippo et al., 2018; Nisar et al., 2020a,b; Totaro et al., 2020).
The main attention is given to the WDS of hospitals and healthcare facilities (HCF), where Legionella spp. contamination is considered a high-risk factor due to the presence of elderly or immunocompromised patients who are more susceptible to the infection (Spagnolo et al., 2013). Legionnaires' disease in Italy, in 2020, amounted to 34.3 cases per million inhabitants; therefore, Legionella colonization of hospitals or communities WDS represents one of the main public health concerns (Kyritsi et al., 2018; Rota et al., 2021; Brunello et al., 2022).
Starting from the epidemiological data, different countries have developed specific guidelines and promoted environmental monitoring programs in order to prevent and control Legionella infections, following the Water Safety Plan (WSP) approach, introduced by the World Health Organization (WHO) (World Health Organization, 2007). Through this approach, an ongoing Legionella surveillance program is suggested, also in the absence of cases, to undertake the most appropriate control and prevention measures.
The recent European Union (EU) Directive 2020/2184 suggests the prevention approach. This directive will be transposed in all the EU countries by January 2023 (European Parliament the Council of the European Union, 2020). In this directive, Legionella has been introduced as a new microbiological parameter for the evaluation of the drinking-water quality, highlighting the importance of the environmental monitoring of the WDS, starting from the water supply until the consumers' outlets.
Moreover, to achieve control of Legionella proliferation, the national and international directives point out several disinfection strategies, based on chemical disinfectants (i.e., chlorine dioxide, monochloramines, and hydrogen peroxide) and physical treatments [i.e., ultraviolet (UV) light and hot temperature treatments]. All of them have shown some advantages and disadvantages (Mcdonnell and Russell, 1999; Richardson et al., 2007; Lin et al., 2011; Mancini et al., 2015; Girolamini et al., 2019). However, some authors have shown how these treatments, especially the use of chemical disinfectants, could select resistant strains and introduce some changes in the bacterial genome. Therefore, the use of diagnostic methods able to follow the ecological and adaptive isolates changes should be the main objective to arise also during Legionella environmental surveillance (Jakubek et al., 2013; Girolamini et al., 2021b). Certainly, this aspect is strictly correlated with the sensitivity and specificity of detection and identification methods already available and in use. In recent times, the most common methods for Legionella isolation and identification in routinely environmental surveillance remain the culture technique and the latex agglutination test, despite the molecular approach as sequence-based typing (SBT) and subgrouping scheme based on monoclonal antibodies (MABs) for Lp (Helbig et al., 1997, 2002; Gaia et al., 2005; Ratzow et al., 2007) and macrophage infectivity potentiator (mip) for non-Lp species (Ratcliff et al., 1998; Ko et al., 2002; Pascale et al., 2021) are consolidated. Despite several approaches that have been developed through time, most of them are applied only when clusters or outbreaks occur and are carried out only by specialized or national reference laboratories.
The limits of all the methods, previously cited, are summarized as follows: (i) long incubation time of culture technique (at least 10 days); (ii) the agglutination test that is not able to recognize all Legionella species, showing also mis-identification or false-negative results; (iii) MABs, that can type only Lp and in particular for subtyping of Lp1, available to the national reference laboratories; (iv) SBT technique and gene sequencing that permits compare only some gene or short fragments of gene size (Helbig et al., 2002; Gaia et al., 2005; Orsini et al., 2011; Lück et al., 2013; Walker and McDermott, 2021).
Consequently, only in recent years with the data returned by whole-genome sequencing (WGS), it has been possible to obtain the most complete genome information, improving isolate typing as well as functional and drug susceptibility response. Moreover, the WGS analysis has opened new scenarios for the reconstruction of infection spread, establishing a correct relationship between environmental and clinical strains during the epidemiological investigation (Quainoo et al., 2017). However, even with its great usefulness, it is not routinely used to support environmental surveillance. Despite the high operational costs, the WGS pipelines could potentially reduce overall costs for the hospitals as well as in all facilities' practices through savings of indirect costs (Quainoo et al., 2017).
In the context of Legionella environmental monitoring, the primary goal is to improve WSPs, quantify the risk level, and identify isolates. Moreover, also during routine environmental surveillance, it is possible to isolate and characterize novel new Legionella species, suggesting how the water microflora is subject to continuous changes during the time and in response to the treatment (physical, chemical, and functional) undertaken (Li et al., 2017).
In this study, we described the workflow applied during Legionella environmental surveillance conducted for 10 years in an HCF that led to the isolation of a novel Legionella species. The knowledge acquired during the surveillance period regarding the WDS, the disinfection treatment, and the level of contamination were associated with traditional techniques (i.e., culture and latex agglutination test) as well as the innovative matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) technique, gene sequencing, and WGS analysis. All results obtained confirm the presence in HCF of two isolates not previously detected.
Materials and Methods
Characteristics of Healthcare Facility's Water Distribution System
The HCF involved in this study is a long-term care facility, built-in 2011, and made up of 78 dislocated inpatient rooms on three floors for a total of 120 beds.
The Legionella environmental surveillance program was started in 2012 with an elaboration of a Legionella risk assessment plan according to the Italian and Regional Guidelines (Italian National Institute of Health, 2015; Emilia-Romagna Region, 2017). Briefly, the Legionella monitoring was carried out two times a year, usually in the spring/summer and fall/winter. The Legionella concentration above the risk level as suggested by guidelines requires further sampling of positive samples (Italian National Institute of Health, 2015; Emilia-Romagna Region, 2017).
The WDS consists of cold water derived from municipal water, heated by two water heaters in parallel, one of them connected to the solar panel. The temperature of hot water at the supply outlets was about 50°C. The hot water system is treated with hydrogen peroxide and silver salt-based disinfectant (H2O2/Ag+). The continuous disinfection treatment provides a residual concentration to the distal outlets around 20 mg/L. The WDS schematic (Figure 1) was developed with Solid Edge 2022 V222.00.02.03 (Siemens Digital Industry Software Inc.).
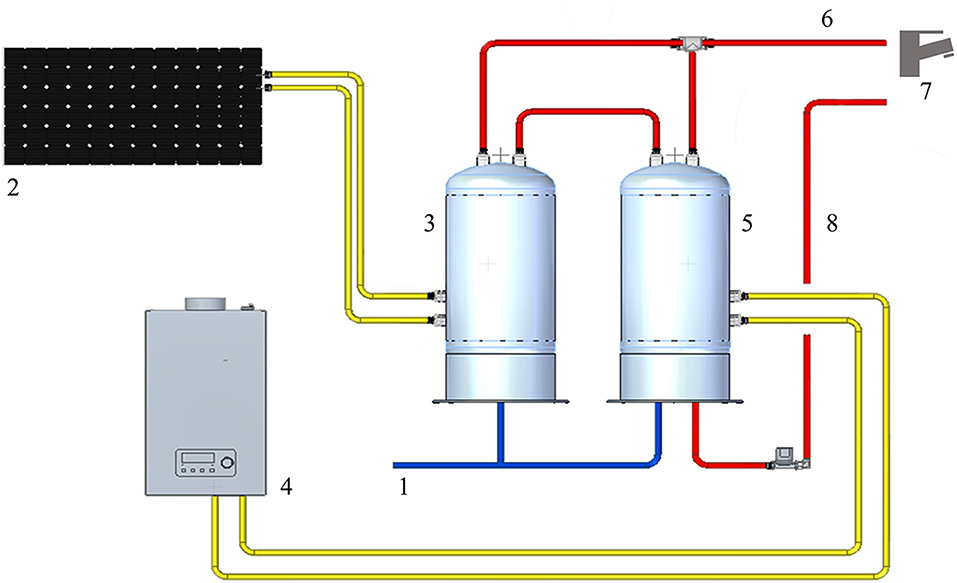
Figure 1. Representation of the HCF water distribution system: 1: cold-water supply; 2: solar panel; 3: solar heater water tank; 4: gas water heater; 5: heater water tank; 6: hot water output; 7: water distal outlets; 8: hot water return line.
Sample Collection
According to the Legionella risk assessment plan, hot- and cold-water samples were collected every 6 months. The sampling points were chosen in accordance with Italian Guidelines (Italian National Institute of Health, 2015), considering the building size, the number of inpatient rooms, the risk level of patients, and workers' exposure to bacteria, other than the facility's epidemiological data. All of these data are reported in the Legionella WSP developed since 2012.
Staring from the technical room, and following the hot and cold WDS, for each floor, four samples were chosen as follows: three hot water samples in the vicinity of, mid-way to, and away from the technical room, and one cold-water sample away from the water supply point.
A total of 30 sampling points were identified between the technical room (water supply point, hot water output, hot water return line, and hot water storage tanks) and HCF inpatient rooms (including showers, sinks, and toilet showerheads) and staff facilities. The criterion of rotation between inpatient rooms was applied.
Two liters of hot or cold water for each sample was collected in post-flushing modality (2 min), following the Italian National Unification and European Committee (UNI EN) International Standard Organization (ISO) 19458:2006 (EN ISO 19458:2006 Water quality - Sampling for microbiological analysis, 2006). During sampling, temperature and disinfectant residue values were measured and reported in distal outlets. The analysis was carried out on the same day of sampling.
Microbiological Analysis and Isolates Characterization
The Legionella isolation was performed using a standard culture technique in accordance with ISO 11731:2017 (International Organization for Standardization, 2017). Briefly, for the enumeration of Legionella, different aliquots of the sample (from 200 to 100 μl), which comes from filtered water (untreated), heat- and acid-treated, were seeded on glycine–polymyxin B–vancomycin–cycloheximide (GVPC) selective agar (Thermo Fisher Scientific, Diagnostic, Ltd., Basingstoke, UK) and incubated at 35 ± 2°C with 2.5% of CO2. The culture required a minimum of 10 for up to 15 days. Every 2 days, the plates were examined and the presumptive colonies were enumerated and sub-cultured on buffered charcoal yeast extract (BCYE) agar with and without L-cysteine (Cys+) and L-cysteine (Cys–) (Thermo Fisher Scientific, Diagnostic, Ltd., Basingstoke, UK). The Legionella colonies, growth only on BCYE Cys+, were identified using the Legionella latex agglutination test kit differentiating between Lp1, and Lp serogroups 2–14 (Lp2-14) and seven species of non-Lp species (Thermo Fisher Scientific, Ltd. Basingstoke, UK), based on manufacturing instructions.
MALDI-TOF MS Analysis
The isolates grown on BCYE Cys+, which returned positive or negative results for the Legionella agglutination test, were also analyzed by the MALDI Biotyper system (Bruker Daltonik GmbH, Bremen, Germany) as previously described (Pascale et al., 2020). Briefly, a fresh colony (24–48 h of incubation) was directly spotted in duplicate onto a MALDI Biotyper target plate, overlaid with 1 μl of the MALDI Biotyper matrix solution, and left to air dry before the next step. Spectra acquisition and processing were performed using the Microflex LT mass spectrometer (2,000–20,000 Da, linear positive mode) and the MALDI Biotyper Compass 4.1 software, whose library (version BDAL 7854) included the spectra of 39 Legionella strains.
The data were interpreted in accordance with the manufacturer's instructions. Briefly, when the instrument returned a log score ≥2.0 (“high confidence level”), the isolates were identified at the species level, while the genus was assigned for scores between 1.7 and 1.99 (“low confidence level”). In the presence of a score between 0.00 and 1.69, the isolates were considered as “not identified.”
A dendrogram based on hierarchical clustering analysis (HCA) of MALDI Biotyper spectra was developed using the MALDI Biotyper Compass Explorer software to generate a tree-like structure able to link the Legionella strains to each other using a linkage algorithm.
Sequencing of mip and rpoB Genes for Legionella Identification
InstaGene Matrix (Bio-Rad, Hercules, CA, USA) was used for DNA extraction that was quantified by Qubit fluorometer (Thermo Fisher Scientific, Paisley, UK).
mip and rpoB genes were used to perform the identification of the isolates according to Ratcliff et al. (1998), Ko et al. (2002), and Pascale et al. (2021). PCR products were visualized by electrophoresis on 2% agarose gel. BigDye kit was used for the sequencing reaction, and the sequences were analyzed on ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Raw sequencing data were assembled using CLC Main Workbench 7.6.4 software (QIAGEN, Redwood City, CA, USA). Furthermore, the mip and rpoB gene sequence comparison was performed using the Basic Local Alignment Search Tool (BLAST) search on the database of the National Center for Biotechnology Information (NCBI) and the database developed by the European Working Group for Legionella Infections [(EWGLI), renamed in ESCMID Study Group for Legionella Infections (ESGLI) from 2011]. The identification at the species level, considering the mip gene, was performed on the basis of an identity score between 98 and 100% compared to the sequences in the database (Fry et al., 2007) and considering the intervals variation of interspecies and intraspecies previously described by Ratcliff et al. (1998).
As reported in Ko et al. (2002), Adékambi et al. (2009), and Pascale et al. (2021) regarding the rpoB gene sequence, the threshold used for the identification was fixed at a 95% similarity percentage.
Phylogenetic and Allelic Diversity Analysis
To estimate the relationship between the isolates found and the strictly related Legionella species, a multiple sequence alignment (MSA) and a concatenated phylogenetic tree were developed on the mip and rpoB gene sequences. Manual editing was performed on the sequences, if required, trimming them to the same length as the reference sequence. The tree was built using software implemented in Geneious Prime's genome browser (Geneious Prime 2022.0.2; http://www.geneious.com) maintaining the default settings (Kearse et al., 2012). Through the MUSCLE algorithm, the nucleotide sequences were aligned (Edgar, 2004). FastTree (Price et al., 2010), a tool to deduce the approximate maximum likelihood of phylogenetic trees, was used to pass the resulting MSA. Jukes–Cantor was used, by FastTree, as a genetic distance model, and the Shimodaira–Hasegawa test was utilized to estimate the reliability of each split in the tree (default parameters) (Guindon et al., 2010). As in a cladogram, the lengths of the branches have been converted to be equal. Branch labels display the substitutions by the site.
Identification of Legionella by Whole-Genome Sequencing (WGS)
One hundred nanograms of DNA was used to prepare the library for the next-generation sequencing (NGS) carried out by Illumina Nextera XT DNA Library Preparation kit (Illumina, New England Biolabs, Ipswich, MA, USA). The Illumina NextSeq 500 platform (2 × 250 paired-end reads) was used to perform the sequencing.
Raw reads were assembled using TORMES v.1.2.0 (Quijada et al., 2019), an automated pipeline for whole bacterial genome analysis, using the default parameters. TORMES performed a sequence quality filtering (PRINSEQ v. 0.20.4) and a de novo genome assembly (SPAdes v. 13.4.1) (Bankevich et al., 2012). The generated contigs were passed to CSAR v1.1.1 (Chen et al., 2018), a scaffolding tool able to order and orient the contigs of the given draft genome based on one or more reference genomes of a related organism. Legionella sp. PC1000 (NZ_CP059400.1) was selected as a reference sequence, based on the output of KmerFinder (Hasman et al., 2014; Larsen et al., 2014; Clausen et al., 2018) that predicts prokaryotic species based on the number of overlapping k-mers, i.e., 16-mers, between the query genome and genomes in a reference database (NCBI). A further refinement was carried out by remapping the reads on the CSAR scaffolds, using the Geneious Prime 2022.0.2 software (http://www.geneious.com) (Kearse et al., 2012). Benchmarking Universal Single-Copy Orthologs (BUSCO) v.5.0.0 (Seppey and Manni, 2019) was performed to evaluate the completeness of the two genome assemblies. The final draft genomes were submitted to the GenBank requiring the annotation by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP v.4.3) (Tatusova et al., 2016).
The OrthoANI package (Yoon et al., 2017) was used to measure the intra- and inter-species genome similarities by average nucleotide identity (ANI) among the assembled draft genomes. Further, FastANI (Jain et al., 2018) through DFAST (Tanizawa et al., 2018) was performed against 13000 prokaryotic reference genomes from NCBI to assess the taxonomic identity. It was also measured based on BLAST+ (ANIb) and MUMmer (ANIm) using JSpeciesWS (Richter et al., 2016).
The relatedness of our strains to Legionella-known type strains was further analyzed by applying a digital DNA–DNA hybridization (dDDH) to the closest related strain based on the previous ANI outcome. The method was implemented via the Genome-to-Genome Distance Calculator 2.1 (GGDC) web service (http://ggdc.dsmz.de), retaining default parameters, using BLAST+ (Camacho et al., 2009) as a local alignment tool. The GGDC uses a Genome Blast Distance Phylogeny (GBDP) to infer genome-to-genome distances between pairs of entirely or partially sequenced genomes. Below the similarity threshold of the in silico DDH (70%) and the ANI analysis (95%), two strains are considered distinct species (Meier-Kolthoff et al., 2013; Kim et al., 2014).
Results
Legionella spp. Isolation and Features
Starting from 2012, the Legionella environmental monitoring returned mostly negative results (<50 colony formant unit (cfu)/L). Only two samples were positive for Legionella in March 2015 and 2016 with a concentration of 200 and 250 cfu/L, respectively. The samples were collected from the shower and toilet showerhead in two different inpatient rooms, both located on the ground floor. The water temperature measured was 45.8 and 44.0°C, with a disinfectant residue of 20 mg/L. Regarding the colonies' morphology, they are small (about 3 mm), gray-white, with a round shape, and well compact. Concerning the identification, both isolates grew well on BCYE Cys+ and showed a quick positive reaction to the latex agglutination test for non-Lp species.
In addition, when exposed under a Woods lamp (long-wavelength UV light at 365 nm), they showed blue-white autofluorescence (Figure 2).
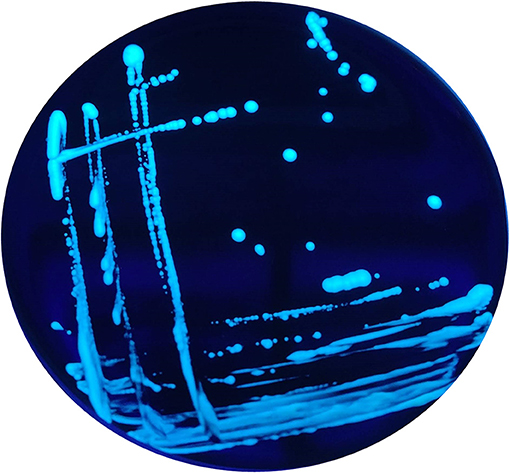
Figure 2. Blue-white autofluorescence of the non-Lp isolates grown on BCYE Cys+, found in 2015 and 2016, under Woods lamp (365 nm).
In particular, for further analysis, the strain isolated in 2015 was called 8cVS16, and the second one isolated in 2016 was named 9fVS26.
Considering that no other samples evaluated in the same year and during the subsequent monitoring yielded positive results for Legionella, the contamination found may be considered point-source contamination. Instead, the two strains were no longer detected in the water distribution system until December 2021, which was when the last sampling occurred.
MALDI-TOF MS Results
The MALDI Biotyper system identified both strains with a low confidence score (yellow color) as Legionella anisa. In particular, the score returned for 8cVS16 was 1.76 and for 9fVS26 was 1.78.
The dendrogram based on HCA, elaborated on the most closed Legionella species present in the instrument database, displays the 8cVS16 and 9fVS16 strains closed to each other and regrouped in one clade. This clade was well-separated by the two strictly related clades, represented by L. anisa and Legionella bozemanii reference strains (Figure 3). Therefore, the strains appeared very close to L. anisa, confirming the results returned by MALDI Biotyper software.
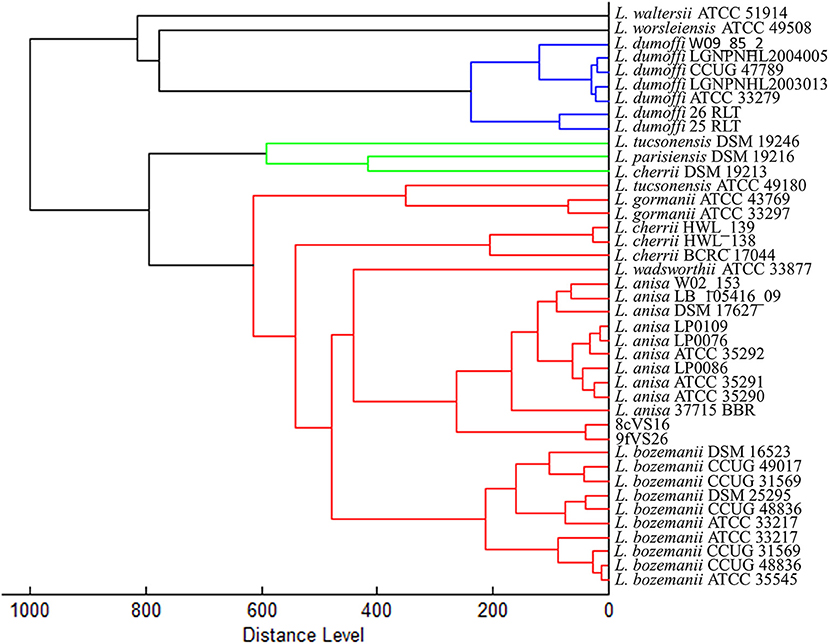
Figure 3. Dendrogram developed by HCA for 8cVS16 and 9fVS26 and related Legionella reference strains.
mip and rpoB Results
BLAST research on NCBI and the ESGLI database returned the best match for both strains L. anisa, reference strain ATCC 35292 (GenBank accession number GCA_900639785.1), with similarities of 96.7 and 92.4% for mip and rpoB, respectively.
Phylogenetic Analysis Results
The concatenated tree elaborated from mip and rpoB sequence genes (Figure 4) revealed the presence of two main clades, each containing four different subclades and each representing several Legionella species. Interestingly, the 8cVS16 and 9fVS26 strains were collocated into a monophyletic group comprising L. anisa, Legionella tucsonensis, and L. bozemanii strains. Moreover, it was particularly evident that they were strictly related to each other, forming a short branch representing a sister clade with the L. anisa ATCC 35292.
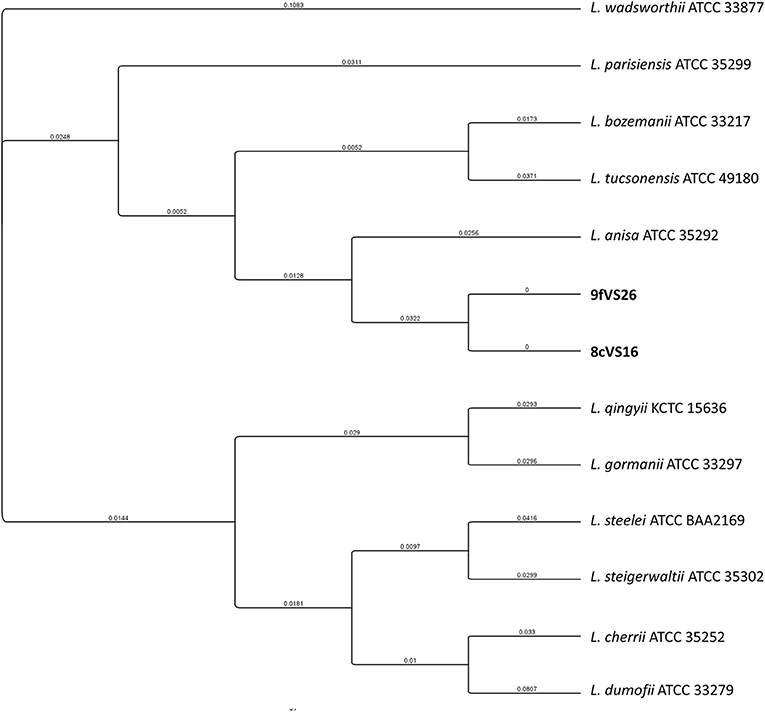
Figure 4. Phylogenetic tree of the two strains (8cVS16 and 9fVS26) and closely related Legionella species based on concatenation of two genes (mip and rpoB). Branch lengths are transformed to be equal like a cladogram. Branch labels display the substitutions per site.
WGS Analysis Results
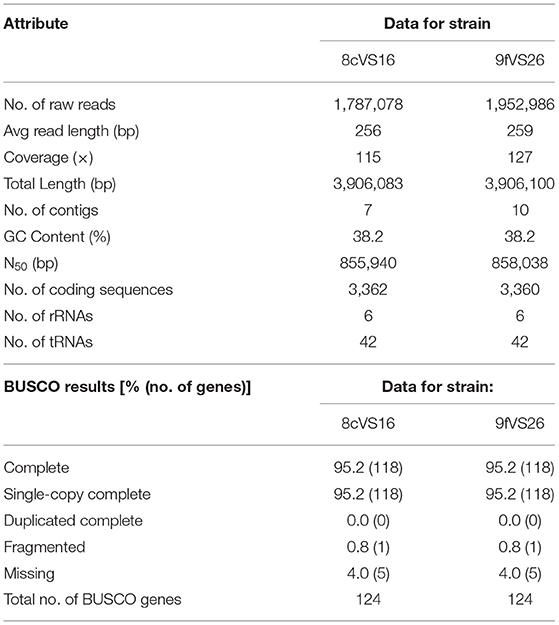
All the results obtained by the WGS analysis are summarized in Table 1. Briefly, the overall lengths of the genomes for 8cVS16 and 9fVS26, respectively, were 3,906,083 and 3,906,1003 bp, with a GC content of 38.2% for both.
In particular, BUSCO analysis, performed to evaluate the completeness of the two genome assemblies, indicates that the two genomes are near-complete, with a percentage of 95.2%.
Through the NCBI PGAP annotation required by GenBank to submit the draft genome, the following accession numbers were given: SRR17223244 and JAJTND000000000 for 8cVS16 and SRR17223245 and JAJSPM000000000 for 9fVS26.
Comparison, obtained from the OrthoANI package used to measure the intra- and inter-species genome similarities, returned the following values: 99.98% between 8cVS16 and 9fVS26, confirming that the two strains belong to the same species and are identical to each other.
The taxonomic identity, obtained by FastANI, returned an identity percentage of the closest strain for both 8cVS16 and 9fVS26. This strain was L. anisa strain WA-316-C3 (ATCC 35292) (GCA_900639785.1) with 90.74% ANI.
The analysis, performed by JSpeciesWS, confirmed the FastANI results with the following values: both 8cVS16 and 9fVS26 ANIb 90.08% compared with L. anisa WA-316-C3 (ATCC 35292), and ANIm 91.55% with L. anisa WA-316-C3 (ATCC 35292) for both 8cVS16 and 9fVS26 strains.
The analysis of our strains' relatedness to known types of strains using a digital DNA-DNA hybridization (dDDH) yielded the following results: a DDH value (generalized linear model (GLM)-based) of 43% for both 8cVS16 and 9fVS26 and a probability that DDH > 70% (i.e., same species) of 5.44% (via logistic regression).
Our results showed an ANI value of 90.74% and a dDDH value of 43%.
Discussion
The discovery and identification of novel bacterial species are events of great relevance, especially when they occur during routine environmental surveillance programs. The identification of novel species is much more frequent in the clinical setting, thanks to the most advanced phenotypic and genotypic technologies. In contrast, laboratories involved in environmental surveillance routinely use only the standard culture method, so detection of a potentially novel species relies heavily on the researcher's and technician's experience. The results obtained in this study prove that it is possible to detect isolates, which are difficult to identify or could be misclassified with standard methods, by applying more sensitive techniques such as genotyping and WGS, even during routine environmental monitoring, carried out in self-surveillance.
Commonly, also in the widely studied and known WDS, the selective stress induced by changes in water characteristics (e.g., pressure, temperature, inorganic, and organic compounds), as well as the continuous disinfection treatments, may occur during the selection of a novel species, which is never identified before (Mcdonnell and Russell, 1999; Girolamini et al., 2021a, 2022).
The HCF monitored, from 2012, did not show Legionella contamination, except for 2 years (2015 and 2016). During our surveillance, only one sample per year was found positive, with non-Lp species concentration over the level of risk fixed to <100 cfu/L. A continuous disinfection treatment (hydrogen peroxide and silver salt) was installed at the time of facility opening, considering the size of the facility and the characteristics of the patients (e.g., elderly, with chronic and immunocompromised diseases). According to other studies, the continuous dosage of this disinfectant can control Legionella proliferation, although, from HCS opening, Legionella was never detected (Lin et al., 2011). Therefore, we cannot know whether Legionella was always absent due to the presence of disinfectant, or whether the WDS characteristics and the fact that the structure was newly built, contributed to creating a “Legionella free” environment. The presence of punctual contamination in two outlets, during 10 years of monitoring, both on flexible shower tubes, which are poorly used, due to the presence of non-self-sufficient patients, suggests that probably these pipelines constituted a niche with a biofilm, were able to protect, and would promote Legionella survival (Storey et al., 2004; Kaplan, 2010; Mahapatra et al., 2015). Moreover, in these types of outlets, as in the showerhead tubes, it is difficult to undertake cleaning and maintenance practices due to the small size of pipelines. The substitution of the showerhead flexible tubes, performed after the communication of Legionella presence, could explain the absence of Legionella detection to date.
It is not a matter to be neglected, as already demonstrated, that the continuous supply of chemical substances in WDS could promote disinfection resistance of bacterial species and the selection of novel ones (Mcdonnell and Russell, 1999; Girolamini et al., 2021a, 2022). We have already demonstrated in other facilities how the disinfectant based on peroxide hydrogen and silver salt in the same environment produces a different action on Legionella, promoting L. anisa's resistance despite the high efficiency on Lp1 (Farhat et al., 2012; Girolamini et al., 2019, 2021b).
The culture technique, which is essential for the isolation, is not effective in correctly recognizing these novel isolates when combined with the common and rapid identification methods (e.g., agglutination test). Our results clearly showed that the two isolates presented features similar to well-known non-Lp species (e.g., L. anisa, L. bozemanii, and Legionella gormanii) and showed a positive agglutination reaction to multiple species latex reagents. Moreover, considering also the MALDI-TOF MS results, they could be misclassified as L. anisa.
In particular, the agglutination test has shown a positive result for non-Lp species antibodies and the phenotypic-proteomics analysis, provided by MALDI-TOF Biotyper, has identified the two strains as L. anisa with a low-confidential interval (yellow score), although the software can properly identify this species by assigning a green score, since the presence of its spectra in the instrument database. The results obtained were in accordance with the previous study (Moliner et al., 2010; Gaia et al., 2011; Svarrer and Uldum, 2012; Pascale et al., 2020), confirming the ability of MALDI-TOF MS to classify all the Legionella species contained in the database. The low confidence score returned for our isolates could be explained by the differences that 8cVS16 and 9fVS26 spectra showed with the L. anisa reference spectra contained in the instrument database. So, we can suppose that the differences at the genomic level were also reflected at the ribosomal protein level, which is the target of MALDI-TOF MS technique. These differences found at the proteomics level appear more evident in the dendrogram returned by HCA, where it is possible to observe how 8cVS16 and 9fVS26 form a separate clade with respect to the two main clades represented by L. anisa and L. bozemanii.
Starting from these considerations, the sequencing of Legionella characteristics genes is essential to obtain discrimination at interspecies and intraspecies levels.
Regarding the genomic analysis, mip and rpoB gene sequencing confirmed the previous results. The two isolates were associated with L. anisa, showing an identity percentage of 96.7 and 92.4%, for mip and rpoB, respectively, below the established threshold (Ratcliff et al., 1998; Ko et al., 2002; Adékambi et al., 2009; Pascale et al., 2021). The phylogenetic tree well-points out these differences, showing how the two isolates, despite their proximity to the L. anisa clade, represent a single subclade that needs to be further investigated.
The discrepancies found in sequences identity percentage have led us to investigate in more depth by applying the WGS technique. The genomes of 8cVS16 and 9fVS26 were then compared with the closest genome returned by FASTANI belonging to L. anisa (ATCC 35292). Although the whole-genome sequences for both isolates were almost completed, with a coverage of 95.2%, the ANI analysis returned a result of 90.74% and the dDDH analysis reported a value of 43% compared to the L. anisa ATCC 35292. Considering that the percentage obtained was under the established similarity thresholds (95 and 70%, respectively) (Meier-Kolthoff et al., 2013; Kim et al., 2014), we can consider the two isolates found belonging to a novel Legionella species.
More detailed studies on (i) morphology, including flagellar structures, (ii) Legionella growth tests at various temperatures and on culture media, (iii) biochemical, and (iv) antibiotic susceptibility tests will be the next steps in obtaining isolates characterization, resulting in the deposit of the type strain in culture collections and the most complete description of the novel species.
This study can be considered as a support tool for all laboratories that, during the phases of environmental surveillance, may find isolates with similar characteristics to the best-known species but with discrepancies in results. These laboratories are encouraged not to stop at the common identification tests but to continue the investigations in order to discover novel isolates and understand the dynamics that led to their development. The results of this study should not surprise us should we consider that the environment is undergoing profound changes (e.g., global warming), which can lead to an increase in temperature in both natural and artificial reservoirs. Moreover, the increasingly widespread use of disinfectants as a preventive strategy, other than changes in water characteristics and pipeline materials, leads to a strong impact on the selection of bacteria and the resistance development.
Therefore, acquiring extensive knowledge of these reservoirs and the events that can promote changes in the ecological niche of bacteria can help to prevent their spread in the man-made environment and contain the occurrence of cases, clusters, or outbreaks. In this context, environmental surveillance is critical, as is a proper risk assessment plan that takes into account changes in Legionella contamination over time. The latter will primarily focus on correct isolate identification and characterisation, as well as novel approaches able to recognise non-Lp species with unknown rates of infection and pathogenicity other than antibiotic resistance.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
LG, MRP, SSa, and SC conceived and designed the experiments and wrote the paper. LG, MRP, MM, FM, and SSp performed sample collection and culture experiments. MO performed the whole-genome sequencing. AG performed gene sequencing. LG, SSa, and MO performed the bioinformatics analysis. The manuscript has been read and approved by all named authors. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Dr. Miriam Cordovana for her support and suggestions for the MALDI-TOF MS analysis.
References
Adékambi, T., Drancourt, M., and Raoult, D. (2009). The rpoB gene as a tool for clinical microbiologists. Trends Microbiol. 17, 37–45. doi: 10.1016/j.tim.2008.09.008
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Brunello, A., Civilini, M., De Martin, S., Felice, A., Franchi, M., Iacumin, L., et al. (2022). Machine learning-assisted environmental surveillance of Legionella: a retrospective observational study in Friuli-Venezia Giulia region of Italy in the period 2002–2019. Inform. Med. Unlocked 28:100803. doi: 10.1016/j.imu.2021.100803
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chen, K. T., Liu, C. L., Huang, S. H., Shen, H. T., Shieh, Y. K., Chiu, H. T., et al. (2018). CSAR: A contig scaffolding tool using algebraic rearrangements. Bioinformatics 34, 109–111. doi: 10.1093/bioinformatics/btx543
Clausen, P. T. L. C., Aarestrup, F. M., and Lund, O. (2018). Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics 19:307. doi: 10.1186/s12859-018-2336-6
Cunha, B. A., Burillo, A., and Bouza, E. (2016). Legionnaires' disease. Lancet 387, 376–385. doi: 10.1016/S0140-6736(15)60078-2
Di Pippo, F., Di Gregorio, L., Congestri, R., Tandoi, V., and Rossetti, S. (2018). Biofilm growth and control in cooling water industrial systems. FEMS Microbiol. Ecol. 94, 1–13. doi: 10.1093/femsec/fiy044
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Emilia-Romagna Region (2017). Regional Guidelines for Surveillance and Control of Legionellosis. Delibera Della Giunta Regionale 12 Giugno 2017, N. 828. 2017. 2012. Available online at: https://bur.regione.emilia-romagna.it/bur/area-bollettini/bollettini-in-lavorazione/n-167-del-19-06-2017-parte-seconda.2017-06-19.6161668613/approvazione-delle-linee-guida-regionali-per-la-sorveglianza-e-il-controllo-della-legionellosi/allegato-1-linee-gu.
EN ISO 19458:2006 Water quality - Sampling for microbiological analysis (2006). Available online at: http://store.uni.com/catalogo/en-iso-19458-2006 (accessed February 2022)
European Centre for Disease Prevention and Control (2021). Legionnaires' Disease Annual Epidemiological Report for 2019. Stockholm Available online at: https://ecdc.europa.eu/sites/portal/files/documents/legionnaires-disease-annual-epidemiological-report.pdf.
European Parliament and the Council of the European Union (2020). Directive (EU) 2020/2184 of the European Parliament and of the Council of 16 December 2020 on the quality of water intended for human consumption (recast).
Farhat, M., Moletta-Denat, M., Frère, J., Onillon, S., Trouilhé, M. C., and Robine, E. (2012). Effects of disinfection on Legionella spp., eukarya, and biofilms in a hot water system. Appl. Environ. Microbiol. 78, 6850–6858. doi: 10.1128/AEM.00831-12
Fry, N. K., Afshar, B., Bellamy, W., Underwood, A. P., Ratcliff, R. M., Harrison, T. G., et al. (2007). Identification of Legionella spp. by 19 European reference laboratories: results of the European Working Group for Legionella Infections External Quality Assessment Scheme using DNA sequencing of the macrophage infectivity potentiator gene and dedicated online tools. Clin. Microbiol. Infect. 13, 1119–1124. doi: 10.1111/j.1469-0691.2007.01808.x
Gaia, V., Casati, S., and Tonolla, M. (2011). Rapid identification of Legionella spp. by MALDI-TOF MS based protein mass fingerprinting. Syst. Appl. Microbiol. 34, 40–44. doi: 10.1016/j.syapm.2010.11.007
Gaia, V., Fry, N. K., Afshar, B., Lück, P., Meugnier, H., Etienne, J., et al. (2005). Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J. Clin. Microbiol. 43, 2047–2052. doi: 10.1128/JCM.43.5.2047-2052.2005
Girolamini, L., Dormi, A., Pellati, T., Somaroli, P., Montanari, D., Costa, A., et al. (2019). Advances in Legionella control by a new formulation of hydrogen peroxide and silver salts in a hospital hot water network. Pathogens 8:209. doi: 10.3390/pathogens8040209
Girolamini, L., Salaris, S., Orsini, M., Pascale, M. R., Mazzotta, M., Grottola, A., et al. (2021a). Draft genome sequences of Legionella presumptive novel species isolated during environmental surveillance in artificial water systems. Microbiol. Resour. Announc. 10, e00307–21. doi: 10.1128/MRA.00307-21
Girolamini, L., Salaris, S., Orsini, M., Pascale, M. R., Mazzotta, M., Grottola, A., et al. (2022). Draft genome sequence of Legionella species isolated from drinking water in an Italian Industry. Microbiol. Resour. Announc. 11, 20–22. doi: 10.1128/mra.01152-21
Girolamini, L., Salaris, S., Pascale, M. R., Mazzotta, M., and Cristino, S. (2021b). Dynamics of Legionella community interactions in response to temperature and disinfection treatment: 7 years of investigation. Microb. Ecol. 83, 353–362. doi: 10.1007/s00248-021-01778-9
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hasman, H., Saputra, D., Sicheritz-Ponten, T., Lund, O., Svendsen, C. A., Frimodt-Moller, N., et al. (2014). Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol. 52, 139–146. doi: 10.1128/JCM.02452-13
Helbig, J. H., Bernander, S., Castellani Pastoris, M., Etienne, J., Gaia, V., Lauwers, S., et al. (2002). Pan-European study on culture-proven Legionnaires' disease: distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur. J. Clin. Microbiol. Infect. Dis. 21, 710–716. doi: 10.1007/s10096-002-0820-3
Helbig, J. H., Kurtz, J. B., Pastoris, M. C., Pelaz, C., and Lück, P. C. (1997). Antigenic lipopolysaccharide components of Legionella pneumophila recognized by monoclonal antibodies: possibilities and limitations for division of the species into serogroups. J. Clin. Microbiol. 35, 2841–2845. doi: 10.1128/jcm.35.11.2841-2845.1997
International Organization for Standardization (2017). ISO 11731:2017 Water quality — Enumeration of Legionella (accessed on February 2022).
Italian National Institute of Health (2015). Italian Guidelines for Prevention and Control of Legionellosis. Available online at: http://www.salute.gov.it/imgs/C_17_pubblicazioni_2362_allegato.pdf.
Jain, C., Rodriguez, -R., L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Jakubek, D., Le Brun, M., Leblon, G., Dubow, M., and Binet, M. (2013). The impact of monochloramine on the diversity and dynamics of Legionella pneumophila subpopulations in a nuclear power plant cooling circuit. FEMS Microbiol. Ecol. 85, 302–312. doi: 10.1111/1574-6941.12121
Jomehzadeh, N., Moosavian, M., Saki, M., and Rashno, M. (2019). Legionella and legionnaires' disease: an overview. J. Acute Dis. 8, 221–232. doi: 10.4103/2221-6189.272853
Kaplan, J. B. (2010). Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 89, 205–218. doi: 10.1177/0022034509359403
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, M., Oh, H. S., Park, S. C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Ko, K. S., Lee, H. K., Park, M. Y., Lee, K. H., Yun, Y. J., Woo, S. Y., et al. (2002). Application of RNA polymerase β-subunit gene (rpoB) sequences for the molecular differentiation of Legionella species. J. Clin. Microbiol. 40, 2653–2658. doi: 10.1128/JCM.40.7.2653-2658.2002
Kyritsi, M. A., Mouchtouri, V. A., Katsioulis, A., Kostara, E., Nakoulas, V., Hatzinikou, M., et al. (2018). Legionella colonization of hotel water systems in touristic places of Greece: association with system characteristics and physicochemical parameters. Int. J. Environ. Res. Public Health 15:2707. doi: 10.3390/ijerph15122707
Larsen, M. V., Cosentino, S., Lukjancenko, O., Saputra, D., Rasmussen, S., Hasman, H., et al. (2014). Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 52, 1529–1539. doi: 10.1128/JCM.02981-13
Li, Q., Yu, S., Li, L., Liu, G., Gu, Z., Liu, M., et al. (2017). Microbial communities shaped by treatment processes in a drinking water treatment plant and their contribution and threat to drinking water safety. Front. Microbiol. 8:2465. doi: 10.3389/fmicb.2017.02465
Lin, Y. E., Stout, J. E., and Yu, V. L. (2011). Controlling Legionella in hospital drinking water: an evidence-based review of disinfection methods. Infect. Control Hosp. Epidemiol. 32, 166–173. doi: 10.1086/657934
Lück, C., Fry, N. K., Helbig, J. H., Jarraud, S., and Harrison, T. G. (2013). “Typing Methods for Legionella,” in Legionella: Methods and Protocols, eds C. Buchrieser and H. Hilbi (Totowa, NJ: Humana Press), 119–148. doi: 10.1007/978-1-62703-161-5_6
Mahapatra, A., Padhi, N., Mahapatra, D., Bhatt, M., Sahoo, D., Jena, S., et al. (2015). Study of biofilm in bacteria from water pipelines. J. Clin. Diagnostic Res. 9, DC9–DC11. doi: 10.7860/JCDR/2015/12415.5715
Mancini, B., Scurti, M., Dormi, A., Grottola, A., Zanotti, A., and Cristino, S. (2015). Effect of monochloramine treatment on colonization of a hospital water distribution system by Legionella spp.: a 1 year experience study. Environ. Sci. Technol. 49, 4551–4558. doi: 10.1021/es506118e
Matsui, M., Fujii, S. I., Shiroiwa, R., Amemura-Maekawa, J., Chang, B., Kura, F., et al. (2010). Isolation of Legionella rubrilucens from a pneumonia patient co-infected with Legionella pneumophila. J. Med. Microbiol. 59, 1242–1246. doi: 10.1099/jmm.0.016089-0
Mcdonnell, G., and Russell, A. D. (1999). Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12, 147–179. doi: 10.1128/CMR.12.1.147
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Mercante, J. W., and Winchell, J. M. (2015). Current and emerging legionella diagnostics for laboratory and outbreak investigations. Clin. Microbiol. Rev. 28, 95–133. doi: 10.1128/CMR.00029-14
Moliner, C., Ginevra, C., Jarraud, S., Flaudrops, C., Bedotto, M., Couderc, C., et al. (2010). Rapid identification of Legionella species by mass spectrometry. J. Med. Microbiol. 59, 273–284. doi: 10.1099/jmm.0.014100-0
Muder, R. R., and Yu, V. L. (2002). Infection Due to Legionella species other than L. pneumophila. Clin. Infect. Dis. 35, 990–998. doi: 10.1086/342884
Nisar, M. A., Ross, K. E., Brown, M. H., Bentham, R., and Whiley, H. (2020a). Legionella pneumophila and protozoan hosts: implications for the control of hospital and potable water systems. Pathogens 9, 20–30. doi: 10.3390/pathogens9040286
Nisar, M. A., Ross, K. E., Brown, M. H., Bentham, R., and Whiley, H. (2020b). Water stagnation and flow obstruction reduces the quality of potable water and increases the risk of Legionelloses. Front. Environ. Sci. 8:611611. doi: 10.3389/fenvs.2020.611611
Orsini, M., Cristino, S., Grottola, A., and Romano-Spica, V. (2011). Bacteria misagglutination in Legionella surveillance programmes. J. Hosp. Infect. 79, 179–180. doi: 10.1016/j.jhin.2011.05.021
Parte, A. C., Carbasse, J. S., Meier-Kolthoff, J. P., Reimer, L. C., and Göker, M. (2020). List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Pascale, M. R., Mazzotta, M., Salaris, S., Girolamini, L., Grottola, A., Simone, M. L., et al. (2020). Evaluation of MALDI–TOF mass spectrometry in diagnostic and environmental surveillance of Legionella species: a comparison with culture and Mip-Gene sequencing technique. Front. Microbiol. 11:589369. doi: 10.3389/fmicb.2020.589369
Pascale, M. R., Salaris, S., Mazzotta, M., Girolamini, L., Fregni Serpini, G., Manni, L., et al. (2021). New insight regarding Legionella Non- Pneumophila species identification: comparison between the traditional mip gene classification scheme and a newly proposed scheme targeting the rpoB gene. Microbiol. Spectr. 9:e0116121. doi: 10.1128/Spectrum.01161-21
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490
Quainoo, S., Coolen, J. P. M., Sacha, A. F. T., van Hijum, C., Martijn, A., Huynen, c, W. J. G. M., et al. (2017). Whole-genome sequencing of bacterial pathogens : the future of nosocomial. Clin. Microbiol. Rev. 30, 1015–1064. doi: 10.1128/CMR.00016-17
Quijada, N. M., Rodríguez-Lázaro, D., Eiros, J. M., and Hernández, M. (2019). TORMES: An automated pipeline for whole bacterial genome analysis. Bioinformatics 35, 4207–4212. doi: 10.1093/bioinformatics/btz220
Ratcliff, R. M., Lanser, J. A., Manning, P. A., and Heuzenroeder, M. W. (1998). Sequence-based classification scheme for the genus Legionella targeting the mip gene. J. Clin. Microbiol. 36, 1560–1567. doi: 10.1128/JCM.36.6.1560-1567.1998
Ratzow, S., Gaia, V., Helbig, J. H., Fry, N. K., and Lück, P. C. (2007). Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J. Clin. Microbiol. 45, 1965–1968. doi: 10.1128/JCM.00261-07
Richardson, S. D., Plewa, M. J., Wagner, E. D., Schoeny, R., and DeMarini, D. M. (2007). Occurrence, genotoxicity, and carcinogenicity of regulated and emerging disinfection by-products in drinking water: a review and roadmap for research. Mutat. Res. Rev. Mutat. Res. 636, 178–242. doi: 10.1016/j.mrrev.2007.09.001
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Rota, M. C., Caporali, M. G., Bella, A., Scaturro, M., Giannitelli, S., and Ricci, M. L. (2021). Il sistema di sorveglianza della Legionellosi in Italia: i risultati del 2020. Boll. Epidemiol. Naz. 1, 32–38. Available at: https://www.epicentro.iss.it/ben/2020/4/sorveglianza-legionellosi-italia-2019.
Seppey, M., Manni, M., and Z., E. (2019). BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 1962, 227–245. doi: 10.1007/978-1-4939-9173-0_14
Spagnolo, A. M., Cristina, M. L., Casini, B., and Perdelli, F. (2013). Legionella pneumophila in healthcare facilities. Rev. Med. Microbiol. 24, 70–80. doi: 10.1097/MRM.0b013e328362fe66
Storey, M. V., Ashbolt, N. J., and Stenström, T. A. (2004). Biofilms, thermophilic amoebae and Legionella pneumophila – a quantitative risk assessment for distributed water. Water Sci. Technol. 50, 77–82. doi: 10.2166/wst.2004.0023
Svarrer, C. W., and Uldum, S. A. (2012). The occurrence of Legionella species other than Legionella pneumophila in clinical and environmental samples in Denmark identified by mip gene sequencing and matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 18, 1004–1009. doi: 10.1111/j.1469-0691.2011.03698.x
Tanizawa, Y., Fujisawa, T., and Nakamura, Y. (2018). DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34, 1037–1039. doi: 10.1093/bioinformatics/btx713
Tatusova, T., Dicuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Totaro, M., Mariotti, T., Bisordi, C., De Vita, E., Valentini, P., Costa, A. L., et al. (2020). Evaluation of legionella pneumophila decrease in hot water network of four hospital buildings after installation of electron time flow taps. Water 12:210. doi: 10.3390/w12010210
Walker, J. T., and McDermott, P. J. (2021). Confirming the presence of Legionella pneumophila in your water system: a review of current Legionella testing methods. J. AOAC Int. 104, 1135–1147. doi: 10.1093/jaoacint/qsab003
World Health Organization (2007). Legionella and the Prevention of Legionellosis. World Health Organization. Available online at: https://apps.who.int/iris/handle/10665/43233.
Keywords: mip sequencing, rpoB sequencing, MALDI-TOF MS, water distribution system, whole-genome sequencing (WGS)
Citation: Girolamini L, Pascale MR, Mazzotta M, Spiteri S, Marino F, Salaris S, Grottola A, Orsini M and Cristino S (2022) Combining Traditional and Molecular Techniques Supports the Discovery of a Novel Legionella Species During Environmental Surveillance in a Healthcare Facility. Front. Microbiol. 13:900936. doi: 10.3389/fmicb.2022.900936
Received: 21 March 2022; Accepted: 26 April 2022;
Published: 13 June 2022.
Edited by:
Ainsley Nicholson, Centers for Disease Control and Prevention (CDC), United StatesReviewed by:
Laura Gomez Valero, Institut Pasteur, FranceDanielle Wroblewski, New York State Department of Health, United States
Copyright © 2022 Girolamini, Pascale, Mazzotta, Spiteri, Marino, Salaris, Grottola, Orsini and Cristino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sandra Cristino, c2FuZHJhLmNyaXN0aW5vQHVuaWJvLml0
†These authors have contributed equally to this work
 Luna Girolamini
Luna Girolamini Maria Rosaria Pascale
Maria Rosaria Pascale Marta Mazzotta
Marta Mazzotta Simona Spiteri
Simona Spiteri Federica Marino
Federica Marino Silvano Salaris1
Silvano Salaris1 Antonella Grottola
Antonella Grottola Massimiliano Orsini
Massimiliano Orsini Sandra Cristino
Sandra Cristino