Abstract
Fusarium head blight (FHB) is a major fungal disease that causes severe yield and quality loss in wheat. Biological control can be integrated with other management strategies to control FHB. For this purpose, Trichoderma gamsii strain T6085 is a potential biocontrol agent to limit the infection of F. graminearum and F. culmorum in wheat. However, the possible impacts of T. gamsii T6085 on the broader microbiome associated with the wheat plant are not currently understood. Therefore, we identified bacteria and fungi associated with different wheat tissues, including assessment of their relative abundances and dynamics in response to the application of T6085 and over time, using amplicon sequencing. Residues of the prior year’s wheat crop and the current year’s wheat spikes were collected at multiple time points, and kernel samples were collected at harvest. DNA was extracted from the collected wheat tissues, and amplicon sequencing was performed to profile microbiomes using 16S v4 rRNA amplicons for bacteria and ITS2 amplicons for fungi. Quantitative PCR was performed to evaluate the absolute abundances of F. graminearum and T. gamsii in different wheat tissues. Disease progression was tracked visually during the growing season, revealing that FHB severity and incidence were significantly reduced when T6085 was applied to wheat spikes at anthesis. However, treatment with T6085 did not lessen the F. graminearum abundance in wheat spikes or kernels. There were substantial changes in F. graminearum abundance over time; in crop residue, pathogen abundance was highest at the initial time point and declined over time, while in wheat spikes, pathogen abundance increased significantly over time. The predominant bacterial taxa in wheat spikes and kernels were Pseudomonas, Enterobacter, and Pantoea, while Alternaria and Fusarium were the dominant fungal groups. Although the microbiome structure changed substantially over time, there were no community-scale rearrangements due to the T6085 treatment. The work suggests several other taxa that could be explored as potential biocontrol agents to integrate with T6085 treatment. However, the timing and the type of T6085 application need to be improved to give more advantages for T6085 to colonize and reduce the F. graminearum inoculum in the field.
Introduction
Fusarium head blight (FHB) is a prominent disease of wheat worldwide. The spread of the FHB pathogens reduces wheat production and causes toxins to contaminate harvested wheat grains, decreasing both the quality and quantity of wheat products (Gilbert and Tekauz, 2000; McMullen et al., 2012). Fusarium is the genus of pathogens that cause FHB, with primary causal agents being F. graminearum and F. culmorum (Walter et al., 2010). Fusarium infection typically leads to the premature bleaching of wheat spikes, although the timing of infection can alter the symptoms. The wheat anthers provide the perfect place for the Fusarium spp. to infect. Therefore, infections occur predominantly during the flowering period of the wheat plant. Earlier infections, such as during early anthesis, can lead to a failure to produce kernels. However, later infection gives rise to diseased wheat kernels with a shriveled and wilted appearance. In addition, Fusarium infections can also arise after the development of wheat kernels and the Fusarium infected wheat kernels are contaminated with toxins released by the fungus (Strange et al., 1974; Kang and Buchenauer, 2000; Birr et al., 2020).
Fusarium graminearum overwinters within crop residue. In this phase of its lifecycle, the pathogen acquires nutrients from decaying plant materials and lives saprotrophically during the fall, winter and spring seasons (Sutton, 1982). Different Fusarium spp. may have different environmental optima. Therefore, the abundance of pathogenic species or strains varies with the climatic conditions in different locations. Although F. graminearum can survive and develop within a wide range of temperatures (10–30°C), warm and humid environmental conditions favor spore production and successful infection (Osborne and Stein, 2007). Further, F. graminearum produces two types of spores. Ascospores are produced within perithecia, the reproductive structures of the sexual stage of the fungus. Ascospores are forcibly ejected from perithecia, which assists in these spores reaching turbulent air and traveling long distances via the wind. In contrast, the asexual spores, known as macroconidia, are not actively discharged into the air and travel shorter distances through rain splash (Fernando et al., 1997; Gilbert and Fernando, 2004). Upon the infection of wheat kernels, Fusarium releases mycotoxins. Type B trichothecenes are the major mycotoxins associated with FHB in wheat. Deoxynivalenol (DON) and nivalenol (NIV) are the major Type B trichothecenes. These mycotoxins affect the baking quality of wheat-based food products. They cannot be easily eliminated during food processing due to the stability of the toxins under typical food processing conditions. Consumption of mycotoxin-contaminated foods can lead to health problems in humans (Gärtner et al., 2008; Pestka, 2010), and therefore maximum tolerable mycotoxin levels are regulated in foods.
Within the strategies available to manage FHB and reduce the risk of mycotoxins entering the food chain, biological control by beneficial microorganisms such as bacteria, yeasts, and filamentous fungi, is receiving increasing attention due to pressure to reduce the use of pesticides and as an alternative to transgenic plants in those countries where their use is not allowed (Collinge and Sarrocco, 2022). Furthermore, other currently available tools for FHB management, such as agronomic practices, fungicides, and resistant wheat varieties, do not assure complete control of the disease (Sarrocco and Vannacci, 2018).
Among beneficial filamentous fungi that can be used in FHB management, those belonging to Clonostachys and Trichoderma are the most promising (Galletti et al., 2020; Gimeno et al., 2021). Among the mechanisms by which these fungi can antagonize Fusarium spp., competition for crop residue and space at the infection court (spikes) seems particularly important (Sarrocco et al., 2019a). The FHB pathogens overwinter and produce infectious spores on crop residue and find in spikes at flowering, the most susceptible phenological stage for host infection (McMullen et al., 2012).
In this context, Trichoderma gamsii strain T6085 has already been demonstrated to be able to reduce the growth of FHB pathogens (Matarese et al., 2012), as well as to control FHB symptoms under both lab and field conditions (Sarrocco et al., 2013; Vicente et al., 2020). In addition, T6085 can compete for natural substrates against the most important Fusarium species involved in FHB (Sarrocco et al., 2019b; Lasinio et al., 2021). The ability of T6085 to endophytically colonize wheat roots (Sarrocco et al., 2021) and spike tissues (Sarrocco et al., 2013), combined with its ability to modulate the expression of defense-related genes in the host (Vicente et al., 2020), render this isolate an excellent candidate to be developed as the active ingredient of biopesticides to be used both in organic and integrated pest management strategies (Jensen et al., 2016).
However, pathogens such as Fusarium spp. are members of more complex microbiomes, including numerous other fungi and bacteria, that also live in association with different host tissues. These microbiomes are frequently divided among rhizosphere, phyllosphere, and endosphere communities (Turner et al., 2013), and extend to the residue that remains after a host plant has died (Bakker et al., 2017). Many beneficial functions are provided by the plant-associated microbiome, including facilitated nutrient uptake, promotion of plant growth and development, and induction of host plant resistance toward pathogens (Smith and Goodman, 1999). Therefore, as for other disease management strategies, it is relevant to assess how biological control approaches impact microbiomes to control pathogens. Current studies on the dynamics of microbiomes associated with various crops in response to the application of biocontrol agents have shown different impacts, but these are typically either transient (Chowdhury et al., 2013; Yin et al., 2013) or a minimal impact that did not create a measurable change in the microbial communities over time (Scherwinski et al., 2007; Perazzolli et al., 2014; Wei et al., 2016). This is the first study looking for a variation in wheat associated microbial community under the treatment of T. gamsii T6085 as a biocontrol agent to mitigate FHB in wheat. During this study we investigated the changes in microbial communities over time and incorporated different methods of introducing the biocontrol agent to compare and contrast their effectiveness.
This study aimed to evaluate the impact of T6085 on the natural fungal and bacterial communities that interact with various wheat tissues. It is crucial to determine the non-target effects on the microbiome due to the artificial inoculation of a biocontrol agent, as these could affect both plant and soil health. Therefore, the microbial communities associated with different wheat tissues were profiled, and their dynamics were observed at different time points in response to different T6085 treatments.
Materials and methods
Fungal material and inoculum preparation
Trichoderma gamsii strain T6085 has been studied for several years (Matarese et al., 2012). This strain was originally isolated from an uncultivated soil in Crimea (Ukraine), and its genome was sequenced and annotated (Baroncelli et al., 2016). Strain T6085 is deposited at the Fungal Collection of the Plant Pathology Lab (University of Pisa, Italy).
Inoculum of strain T6085 was prepared for application in the field trial using two different methods. For inoculation of wheat spikes, an aqueous suspension of spores (106 spores mL–1) was prepared on the day of application from cultures on potato dextrose agar (PDA) plates (24°C, 12 h light/12 h darkness for 2 weeks). For inoculation of crop residues from the prior cropping season, inoculum was prepared on whole millet grain. Briefly, 15 mL of aqueous spore suspension (105 spores mL–1, produced by cultivation on PDA as above) was used to inoculate 1 kg of millet seeds in sterile plastic bags (previously wet with 300 mL of distilled water and sterilized by autoclaving for 20 min at 121°C). The millet seeds were incubated at 24°C under 12 h light/12 h darkness for 10 days, then dried in a laminar flow hood and stored at 4°C until use.
Field experiment
Experimental design
An experiment was performed during the 2018–2019 cropping season to evaluate the effect of the application of T6085 on bacterial and fungal communities naturally associated with wheat residue, spikes and kernels. The experiment was established at the Center of Agro-environmental Research “Enrico Avanzi,” San Piero a Grado, Pisa (University of Pisa, Italy). Durum wheat (Triticum durum cv Tirex) was sown on 11 December 2018 in a no-tillage system, using a no-till seeder and applying about 400 seeds m–2 in rows 15 cm apart from each other. P and K mineral fertilizers were applied before sowing (30 and 107 kg ha–1, respectively); mineral nitrogen was applied splitting the total amount (170 kg ha–1) equally between the stages of beginning stem elongation and tillering. Weeds were controlled using a post-emergence herbicide application (Atlantis flex, a.i.: propoxycarbazone-sodium + mesosulfuron-methyl). The crop was harvested on the 24 July 2019.
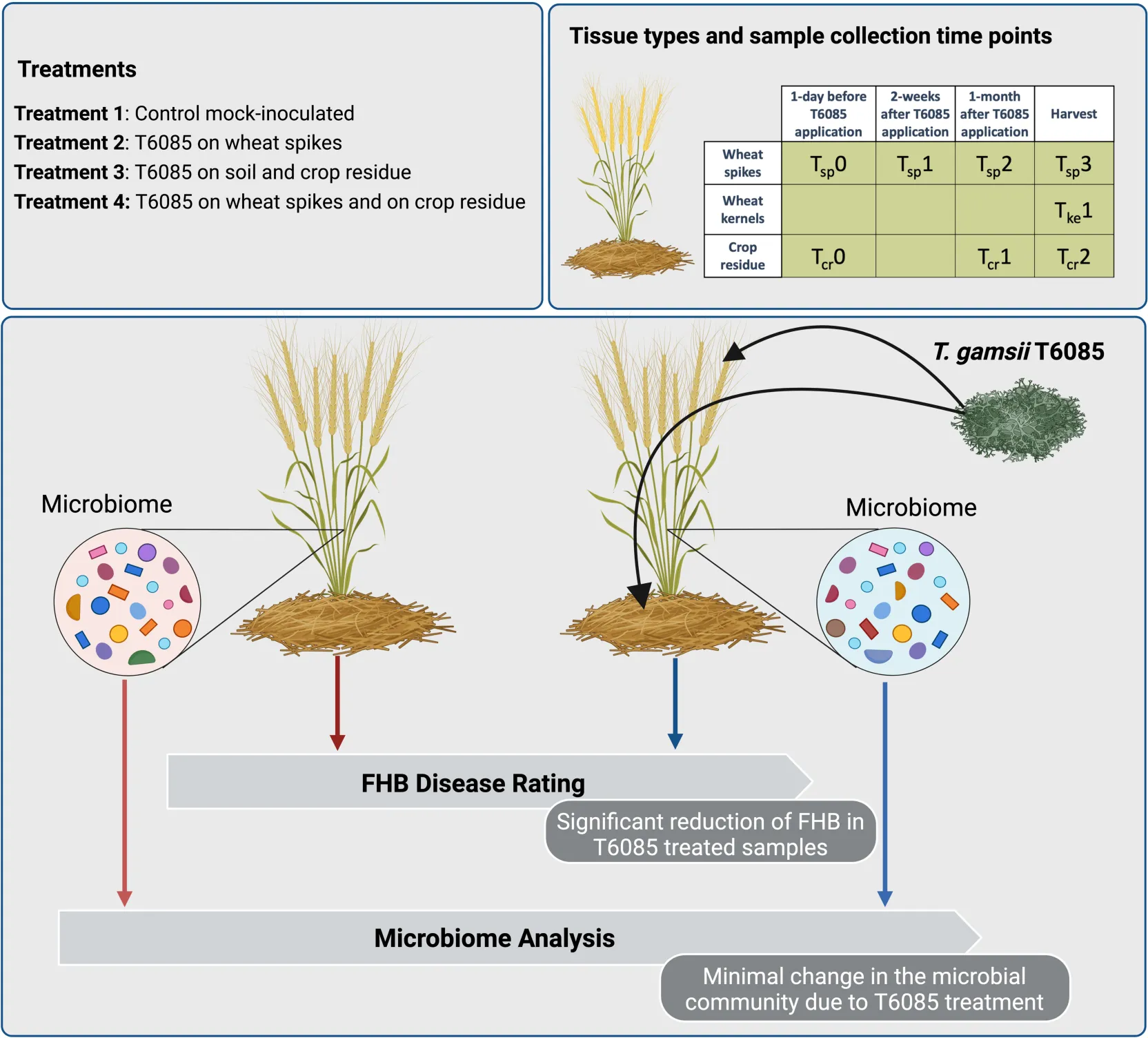
Plots were assigned using a randomized block design (randomization via Random.org) with five blocks and a plot size of 6 m wide × 9 m long (40 rows for plot). Four treatments were implemented: (1) Non-inoculated control; (2) wheat spikes at anthesis inoculated with T6085 (i.e., to place T6085 in competition with Fusarium at the site of infection); (3) crop residues from the previous cropping season (which was also of wheat) inoculated with T6085 (i.e., to create competition between T6085 and Fusarium at the site of spore production); (4) both spikes at anthesis and crop residues inoculated with T6085 (i.e., to combine biocontrol effects at both the site of inoculum production and at the site of infection). Natural Fusarium inoculum was relied upon for the development of FHB.
Inoculation and sampling of crop residues
At the beginning of stem extension (Feekes stage 6), T6085 was introduced to the soil surface of appropriate plots (i.e., Treatments 3 and 4) by distributing colonized millet seeds by hand (4 g m–2) while walking linearly into the field and spreading the inoculum all over the plot. For treatments with no inoculation of crop residues (i.e., Treatments 1 and 2), sterile millet seeds were distributed in the same manner and at the same rate. Samples of residue from the prior cropping season were collected 1 day before the introduction of T6085 (denoted as Tcr0, for “time 0, crop residue”), after 1 month (Tcr1), and just before the harvest (Tcr2). At each sample collection, a quadrat was marked by placing a 50 cm × 50 cm frame within the central region (2 × 3 m) of each plot and 50 pieces of wheat straw residue were collected by hand from within the quadrat (quadrat method). Residue pieces were cut longitudinally, washed three times for 1 min in sterile water to remove soil particles, dried in a sterile laminar flow hood, ground with a Cyclotech 1093 Sample mill and stored at –20°C until use for microbiome profiling.
Inoculation and sampling of wheat spikes
At anthesis, T6085 inoculum was applied on spikes (120 L ha–1) for treatments 2 and 4, while treatments 1 and 3 were given an equal volume of sterile water as a mock inoculum. Spikes were sampled using the quadrat method described above, taking a subsample of 10 spikes per sample. Spike sample collection took place 1 day before the introduction of T6085 to spikes (denoted as Tsp0, for “time 0, spike tissue”), 14 days post-inoculation (dpi) (Tsp1), 28 dpi (Tsp2), and at harvest (35 dpi; Tsp3). From each spike, 6 spikelets were subsampled (two from the third lower, two from the middle and two from the upper third of the spike) were collected and stored at –20°C until use for microbiome profiling.
Assessment of fusarium head blight severity and agronomic parameters
At time points Tsp1, Tsp2 and Tsp3, an additional 30 spikes from each plot were observed for FHB symptoms. Disease Incidence (DI) was defined as the % of spikes showing premature bleaching of some or all spikelets. Disease Severity (DS) was defined as the % of spikelets within a spike that were symptomatic. At the end of the cropping season, wheat was harvested, and several agronomical parameters were evaluated on sub-samples, such as spike number (count m–2), straw and spike fresh weight (g m–2) and total kernel weight (g m–2).
The DI and DS values, after angular transformation, and agronomical data were submitted to analysis of variance (ANOVA) and Tukey’s post hoc contrasts by using SygmaPlot 12.0 to evaluate the effect of T6085 application on disease development and crop productivity. The threshold for statistically significant effects was set at p ≤ 0.05.
Amplicon sequencing
Ground tissue samples were stored in DNA/RNA Shield (ZymoResearch) and shipped by express courier to Winnipeg, Canada. The DNA was extracted from a 0.1 g subsample using the GeneJet plant genomic DNA extraction kit (Thermo Fisher Scientific). The concentration of extracted DNA was diluted to a consistent 2 ng μL–1, based on spectrophotometric concentration measurement (NanoDrop One, Thermo Fisher Scientific). The ITS2 region of the fungi (employing ITS3_KYO2 forward primer and ITS4_KYO3 reverse primer) and the v4 region of 16S rRNA gene of bacteria (employing F515 forward primer and R806 reverse primer) were amplified (Caporaso et al., 2011; Toju et al., 2012). Primers were modified with 5′ overhangs for compatibility with later indexing steps and included variable length spacers to increase the signal diversity when sequencing through the primer region. The 20 μL PCR mixtures consisted of KAPA HiFi HotStart master mix (Roche), forward and reverse primers at 0.5 μM each, nuclease-free water, and template DNA at a final concentration of 0.8 ng μL–1. To inhibit the amplification of mitochondrial and chloroplast DNA from the host plant, mPNA and pPNA blockers (PNAbio) were included at 0.25 μM in the 16S rRNA amplification only (Table 1).
TABLE 1
| Reaction | Primers | PCR program |
| Amplicon PCR | ITS2 | Initial denaturation at 98°C for 3 min, 25 cycles of (denaturation at 98°C for 15 s, annealing 57°C for 10 s, extension 72°C for 1 min), final extension 72°C for 10 min |
| 16S rRNA | Initial denaturation at 98°C for 3 min, 25 cycles of (denaturation at 98°C for 15 s, annealing 57°C for 10 s, PNA clamping 75°C 10 s, extension 72°C for 1 min), final extension 72°C for 10 min | |
| Index PCR | Nextera XT index kit | Initial denaturation at 98°C for 3 min, 8 cycles of (denaturation at 98°C for 30 s, annealing 55°C for 30 s, extension 72°C for 30 s), final extension 72°C for 5 min |
| qPCR | Fus.GRA | Initial denaturation at 98°C for 3 min, 40 cycles of (denaturation at 98°C for 15 s, annealing/extension at 58°C for 30 s, plate read), melt curve analysis up to 65–95°C at 3 s/increment |
| Tgam | Initial denaturation at 98°C for 3 min, 40 cycles of (denaturation at 98°C for 15 s, annealing/extension at 56°C for 30 s, plate read), melt curve analysis up to 65–95°C at 3 s/increment |
The PCR conditions used for amplicon generation, indexing, and quantitative PCR.
Amplified PCR products were purified using KAPA HyperPure beads (Roche). Dual indices and adapters were attached via the Nextera XT Index Kit (Illumina), according to the manufacturer’s protocol. Indexed samples were pooled in equimolar quantities. Because some smaller than expected fragments (likely primer dimers) were present in the library, the pooled library was size-selected using a Flashgel recovery cassette (Lonza) and purified again with KAPA HyperPure beads. The amplicon size distribution of the pooled library was visualized using the TapeStation system (Agilent), and the concentration of MiSeq-compatible amplicons was determined using the KAPA Library Quantification Kit for Illumina Platforms (Roche). PhiX was spiked into each library at a rate of 7.5%. The samples were split across four sequencing runs. The MiSeq v2 reagent kit (2 × 250 bp) was used for v4 16s rRNA libraries, and the v3 reagent kit (2 × 300 bp) was used for ITS2 libraries. Negative controls (PCR reactions without template DNA) and mock communities were included in each sequencing run. For the bacterial libraries, the mock community was product MSA-3002 from the American Type Culture Collection, and for the fungal libraries, a mock community from Bakker (2018) was used. Unprocessed sequence data are available as accession PRJNA790613 at the NCBI SRA.
The sequence data were processed, and taxonomic identifications were assigned using CLC Genomics Workbench version 20.0.4 (Qiagen). The low-quality sequences (limit 0.05) and sequence ends containing ambiguous nucleotides (maximum 2) were trimmed off. Adapter sequences were trimmed and reads below a length of 15 were discarded. Reference-based operational taxonomic unit (OTU) clustering was performed at a 99% similarity percentage. To identify the likely taxonomic origin of fungal amplicons, the UNITE database v7.2 (99% with singletons, developer version) was used (Nilsson et al., 2018), while the SILVA 16S v132 99% database (Quast et al., 2013) was used for bacterial amplicons. Processed sequence variant lists, tables of abundances across samples, taxonomic affiliations, and metadata were stored as phyloseq objects in R (McMurdie and Holmes, 2013). Indices of diversity, including OTU richness, Shannon alpha diversity index, and Bray-Curtis pairwise dissimilarities, were calculated using the package vegan (Oksanen et al., 2022). Testing for community-scale differences among treatments via Adonis also used the package vegan. Box plots and ordinations were created using the package ggplot2 in R (Wickham, 2016). Differential expression analyses were performed using DEseq2 (Love et al., 2014). In testing for the differential relative abundance of OTUs, treatments were grouped according to whether or not T6085 was applied to the tissues in question; for example, with respect to kernel samples, the presence of T6085 was equivalent across Treatment 1 (non-inoculated control) and Treatment 3 (T6085 applied to crop residue only). OTU abundances were modeled on treatment, time point, the interaction between treatment and time point, and the blocking factor in the experimental design.
Quantitative PCR
Quantitative PCR (qPCR) was performed to calculate the densities of T. gamsii and F. graminearum within samples, using assays Fus.GRA and Tgam from Sarrocco et al. (2019b; Table 1). The qPCR mixture consisted of 10 μL of 2 × SYBR Green Master Mixture (Bio-Rad), forward and reverse primers at 0.5 μM each, nuclease-free water, and template DNA. Efficiencies when amplifying genomic DNA of the target organisms were 94.06% for the Fusa.GRA assay and 92.95% for the Tgam assay. The abundances of T. gamsii and F. graminearum were calculated using the standard curve method. Changes in F. graminearum densities over time and due to the presence of T. gamsii T6085 were analyzed by ANOVA, modeling log-transformed abundance values on treatment, time point, the interaction between treatment and time point, and the blocking factor in the experimental design. The Tukey HSD method was used to identify significant contrasts where the main effects were significant.
Results
Fusarium head blight development and severity in the field
At 14 dpi of T6085, there was no naturally occurring disease in any spikes collected. However, by 28 dpi of T6085, the typical symptoms of FHB blight, such as bleaching of some or all the spikelets within each spike, appeared. A reduction of DI was recorded in all the treatments in which T6085 was applied as spike and/or soil inoculum, with Treatment 2 (T6085 applied to flowering spikes) showing a significantly reduced percentage of affected spikes (Figure 1, p ≤ 0.05).
FIGURE 1

Fusarium head blight disease incidence, as assessed at 28 days post inoculation of T6085. Data are the mean (dark gray) + SD (soft gray) percentage of spikes showing visual symptoms of FHB (n = 5 replicates, with 30 spikes assessed per sample). Trt1 was the non-inoculated control. In Trt2, T. gamsii strain T6085 spore suspension was applied onto wheat spikes at anthesis. In Trt3, T6085 was applied to crop residue on the soil surface. In Trt4, T6085 was applied onto both spikes and crop residue at anthesis. Treatment means indicated by different letters are significantly different (ANOVA with Tukey HSD, p < 0.05).
The pattern of effects for DS was the same as was observed for DI; the percentage of affected spikelets within each spike was generally reduced in all the plots where T6085 was applied, but the decrease was only statistically significant in the contrast of Treatment 2 vs. the non-inoculated control (Figure 2, p ≤ 0.05).
FIGURE 2

Fusarium head blight disease severity as assessed at 28 days post inoculation of T6085. Data are the mean (dark gray) + SD (soft gray) percentage of spikelets within a spike showing visual symptoms of FHB (n = 5 replicates, at 30 spikes assessed per sample). Trt1 was the non-inoculated control. In Trt2, T. gamsii strain T6085 spore suspension was applied to wheat spikes at anthesis. In Trt3, T6085 was applied to crop residue at the soil surface. In Trt4, T6085 was applied to both spikes at anthesis and crop residue. Treatment means indicated by different letters are significantly different (ANOVA with Tukey HSD, p < 0.05).
By 35 dpi of T6085, differences among treatments in DI and severity were no longer apparent (data not shown), thus demonstrating that the beneficial effect of the antagonist did not last over a long time.
Agronomic parameters
At the end of the cropping season (on 24 July 2019), wheat was harvested from all plots, and agronomic factors were measured to evaluate the possible effects of the application of T6085 on wheat yield and yield components. There were no significant differences among treatments in spike density or the biomass of straw, spikes, or grain at harvest (Table 2; ANOVA with Tukey HSD, p > 0.05).
TABLE 2
| Treatment | Spike density (count m–2) | Straw fresh weight (g m–2) | Spike fresh weight (g m–2) | Grain yield (g m–2) |
| Treatment 1: Non-inoculated control | 285 a ± 53.4 | 249 a ± 45.6 | 342 a ± 74.9 | 225 a ± 82.3 |
| Treatment 2: T6085 to spikes | 302 a ± 49.6 | 259 a ± 33.7 | 361 a ± 73.5 | 248 a ± 60.5 |
| Treatment 3: T6085 to crop residue | 295 a ± 81.2 | 245 a ± 49.4 | 317 a ± 75.4 | 202 a ± 70.1 |
| Treatment 4: T6085 to spikes + crop residue | 309 a ± 45.3 | 250 a ± 31.7 | 341 a ± 57.1 | 231 a ± 42.2 |
Effect of treatments on some agronomic traits of wheat.
Values are means followed by a standard deviation of 5 plot-level replicates. Shared lowercase letters represent no statistically significant difference between treatments (ANOVA with Tukey HSD, p > 0.05).
The abundance of Fusarium graminearum in wheat tissues
The abundance of F. graminearum changed over time in crop residues and wheat spikes. In crop residues, F. graminearum abundance ranged from 4.03 × 10–5 to 8.57 × 10–5 ng μL–1 of DNA, while there was an extensive range in abundances in wheat spikes, from 3.11 × 10–5 to 2.44 × 10–3 ng μL–1 of DNA. In crop residues, the F. graminearum density was significantly higher at the initial time point (Tcr0), and it decreased over time (ANOVA with Tukey HSD, p ≤ 0.05). The abundance of F. graminearum in wheat spikes showed an opposite trend, as values were significantly higher at the last time point (Tsp3) compared to all earlier time points (Figure 3; ANOVA with Tukey HSD, p ≤ 0.05).
FIGURE 3

Abundance of F. graminearum over time in (A) crop residue and (B) wheat spikes. See the text for a complete description of the sampling time points (Tcr0, Tcr1, Tcr2; Tsp0, Tsp1, Tsp2, Tsp3). Individual data points are colored by treatment, where Trt1 (non-inoculated control), Trt2 (T6085 applied to spikes at anthesis), Trt3 (T6085 applied to crop residue) and Trt4 (T6085 applied to both spikes and to crop residue). Box plots indicate the 1st quartile, median and 3rd quartile. Different letters within each panel indicate significant differences between the time points (log-transformed F. graminearum abundances modeled on treatment, time point, the interaction between treatment and time point, and the blocking factor in the experimental design; Tukey HSD contrasts among time points, p ≤ 0.05).
Across experimental treatments, there were no significant changes in F. graminearum abundance due to the application of T6085 (Figure 4; ANOVA with Tukey HSD, p > 0.05).
FIGURE 4

The impact of T6085 application on F. graminearum abundance, as measured by quantitative PCR, in (A) crop residue; (B) wheat spikes; (C) wheat kernels. See text for a complete description of the sampling time points (Tcr0, Tcr1, Tcr2; Tsp0, Tsp1, Tsp2, Tsp3). The x-axis labels indicate the experimental treatments: Trt1 (non-inoculated control), Trt2 (T6085 applied to spikes at anthesis), Trt3 (T6085 applied to crop residue), Trt4 (T6085 applied to both spikes and to crop residue). Box plots indicate the 1st quartile, median and 3rd quartile. Within panels, there were no statistically significant differences among treatments (log-transformed F. graminearum abundances modeled on treatment, time point, the interaction between treatment and time point, and the blocking factor in the experimental design; p > 0.05 for treatment factor in all cases).
Variation in microbial diversity
The number of observed OTUs (i.e., richness) and Shannon diversity index were calculated to further understand the dynamics in the microbial community in different wheat tissues due to the T6085 treatment and over time. Crop residue had bacterial and fungal richness within the range between 1,343 and 2,889 OTUs and 74–178 OTUs, respectively. In both wheat spikes and kernels, fungal richness was less than 100 OTUs, while bacterial OTU richness was comparatively higher. The richness values did not change significantly in response to the application of T6085 (Table 3). However, there was a significant variation in the richness over time in bacterial and fungal communities residing in crop residue and wheat spikes (ANOVA with Tukey HSD, p ≤ 0.05). In crop residue, bacterial richness increased significantly from Tcr1 to Tcr2, and fungal richness increased significantly over time. In wheat spikes, bacterial richness was significantly lower at Tcr0 than at other time points, while the fungal richness significantly decreased from Tsp1 to Tsp2 (Table 4).
TABLE 3
| Type of tissue | Time point | Treatment* | Bacteria | Fungi | ||
| OTU richness | Shannon index | OTU richness | Shannon index | |||
| Residue | Tcr0 | Untreated | 1857.7 ± 222 | 5.8 ± 0 | 93.3 ± 12 | 2.5 ± 0 |
| Residue | Tcr1 | Treated | 2037.7 ± 245 a | 5.9 ± 0 a | 118.2 ± 18 a | 2.8 ± 0 a |
| Untreated | 1922.8 ± 332 a | 5.8 ± 0 a | 109.2 ± 25 a | 2.7 ± 0 a | ||
| Residue | Tcr2 | Treated | 2148.9 ± 360 a | 6.1 ± 0 a | 129.2 ± 19 a | 3.0 ± 0 a |
| Untreated | 1888.1 ± 209 a | 6.0 ± 0 a | 126.7 ± 28 a | 3.0 ± 0 a | ||
| Spikes | Tsp0 | Untreated | 14.4 ± 5 | 1.0 ± 0 | 5.6 ± 2 | 1.5 ± 0 |
| Spikes | Tsp1 | Treated | 35.6 ± 6 a | 2.0 ± 0 a | 6.6 ± 1 a | 1.7 ± 0 a |
| Untreated | 28.9 ± 4 a | 1.8 ± 0 a | 5.8 ± 1 a | 1.6 ± 0 a | ||
| Spikes | Tsp2 | Treated | 32.7 ± 8 a | 1.8 ± 0 a | 4.4 ± 2 a | 1.1 ± 1 a |
| Untreated | 32.5 ± 7 a | 1.8 ± 0 a | 4.5 ± 1 a | 1.2 ± 0 a | ||
| Spikes | Tsp3 | Treated | 32.6 ± 6 a | 2.1 ± 0 a | 5.1 ± 1 a | 1.5 ± 0 a |
| Untreated | 38.0 ± 6 a | 2.3 ± 0 a | 5.2 ± 1 a | 1.5 ± 0 a | ||
| Kernels | Tke1 | Treated | 206.4 ± 61 a | 1.7 ± 1 a | 55.7 ± 4 a | 2.0 ± 0 a |
| Untreated | 229.3 ± 70 a | 1.9 ± 1 a | 52.0 ± 6 a | 2.0 ± 0 a | ||
The microbial richness (number of OTUs) and Shannon index of diversity for bacteria and fungi in crop residue, wheat spikes and kernel tissues collected at different time points under different T6085 treatments.
Values shown are means ± standard deviation. There were no significant impacts of treatment with T6085 on these community characteristics (ANOVA with Tukey HSD, p > 0.05).
*Treatments were grouped according to whether or not T6085 was applied to the tissues in question; for example, with respect to kernel samples, the presence of T6085 was equivalent across Treatment 1 (non-inoculated control) and Treatment 3 (T6085 applied to crop residue only); therefore, these treatments were combined as “Untreated”.
TABLE 4
| Type of tissue | Time point | Bacteria | Fungi | ||
| OTU richness | Shannon index | OTU richness | Shannon index | ||
| Residue | Tcr0 | 1857.7 ± 222 b* | 5.8 ± 0 b | 93.3 ± 12 c | 2.5 ± 0 b |
| Tcr1 | 1999.4 ± 270 b | 5.9 ± 0 b | 115.2 ± 20 b | 2.8 ± 0 b | |
| Tcr2 | 2018.5 ± 316 a | 6.1 ± 0 a | 128.0 ± 23 a | 3.0 ± 0 a | |
| Spikes | Tsp0 | 14.4 ± 5 b | 1.1 ± 0 b | 5.6 ± 2 ab | 1.5 ± 1 ab |
| Tsp1 | 31.8 ± 6 a | 1.9 ± 0 a | 6.1 ± 1 a | 1.7 ± 0 a | |
| Tsp2 | 32.6 ± 7 a | 1.8 ± 0 a | 4.5 ± 2 b | 1.2 ± 1 b | |
| Tsp3 | 35.3 ± 7 a | 2.2 ± 0 a | 5.2 ± 1 ab | 1.5 ± 0 ab | |
| Kernels | Tke1 | 217.8 ± 65 | 1.8 ± 1 | 53.9 ± 6 | 2.0 ± 0 |
The microbial richness (number of OTUs) and Shannon index of diversity for bacteria and fungi in crop residue, wheat spikes and kernel tissues collected at different time points.
Values shown are means ± standard deviation.
*Among time points within tissue types, values followed by different lower-case letters were significantly different from each other (ANOVA with Tukey HSD, p < 0.05).
Similarly, the Shannon index of bacterial and fungal OTU diversity was not significantly changed by the application of T6085 (Table 3). However, there was a significant variation in the Shannon index over time in microbes associated with wheat spikes and crop residue (Table 4). The Shannon index of both bacteria and fungi increased over time, reaching a significantly higher value at Tcr2 compared to Tcr0 (ANOVA with Tukey HSD, p ≤ 0.05). Further, bacterial diversity was significantly higher at all later time points compared to Tsp0 in wheat spikes, while there was a significant increase in diversity in the fungal community associated with wheat spikes from Tsp1 to Tsp2 (ANOVA with Tukey HSD, p ≤ 0.05).
Variations in the microbial community due to treatment and over time
The changes in the microbial community due to the T6085 treatment were assessed using Bray-Curtis dissimilarities with principal coordinate analysis. Neither bacterial nor fungal community structures were appreciably changed due to the T6085 treatment (Adonis test, p > 0.05) (Figure 5).
FIGURE 5

Application of T6085 causes no broad changes in microbiomes of wheat. Plots are principal coordinates ordinations derived from pairwise community dissimilarity values (Bray-Curtis index). Panels represent bacterial communities from (A) crop residue; (B) wheat spikes; (C) wheat kernels; and fungal communities from (D) crop residue; (E) wheat spikes; (F) wheat kernels. Treatments have been grouped according to whether or not T. gamsii T6085 was applied to the tissue in question. Ellipses represent a 95% confidence level. The fungal and bacterial microbiomes did not show any significant community scale change due to T6085 treatment (Adonis test, p-values shown above each panel).
Even though there were no whole community rearrangements in response to the application of T6085, some individual taxa showed differential abundance due to the treatments in wheat spikes samples (DEseq, p ≤ 0.05) (Supplementary Table 1). These differentially abundant taxa included members of Massilla, Fusarium and Alternaria. Massilla showed a significantly higher abundance in T6085 treated samples collected at Tsp3, Alternaria showed a significantly lower abundance in T6085 treated wheat spikes samples collected at Tsp1, and a Fusarium sequence variant showed a reduced abundance in treated samples at Tsp2 while another Fusarium sequence variant showed a significantly higher abundance in T6085 treated samples collected at Tsp2.
We explored network analyses (i.e., patterns of association among sequence variants across samples) as an alternative approach to reveal whether broad community rearrangements could be observed. Interestingly, for microbiomes associated with wheat spikes, we observed only half as many significant associations among sequence variants for samples that had been treated with T6085, compared to samples that had not been treated with T6085 (Supplementary Figure 1). For example, edge density was 0.0193 for the network from untreated samples and was 0.00955 for the network from treated samples; similarly, the mean number of significant associations per sequence variant (i.e., mean degree per node) was double (3.06 vs. 1.51) for the network from untreated samples, compared to the network from treated samples (Supplementary Figure 1). However, at the scale of the network, we have only n = 1 observations, and so these comparisons must be taken with a grain of salt.
Despite the minimal variations in fungal and bacterial community structures due to the application of T6085, there were apparent changes in fungal and bacterial community composition over time in crop residue and wheat spikes (Figure 6). In crop residue samples, differentially abundant OTUs numbered 446 for the contrast between Tcr0 and Tcr1, 1 040 between Tcr0 and Tcr2, and 589 between Tcr1 and Tcr2. In wheat spike samples, differentially abundant OTUs numbered 76 between Tsp0 and Tsp1, 120 between Tsp0 and Tsp2, 111 between Tsp0 and Tsp3, 44 between Tsp1 and Tsp2, 68 between Tsp1 and Tsp3 and 69 between Tsp2 and Tsp3 (Supplementary Tables 2, 3). Thus, many microbial taxa associated with wheat demonstrated significant shifts in relative abundance over weeks.
FIGURE 6

Microbiomes associated with wheat tissue demonstrate progressive change over time. Plots are principal coordinates ordinations derived from pairwise community dissimilarity values (Bray-Curtis index). (A) Bacterial communities from crop residue; (B) fungal communities from crop residue; (C) bacterial communities from wheat spikes; (D) fungal communities from wheat spikes. See text for a complete description of the sampling time points (Tcr0, Tcr1, Tcr2; Tsp0, Tsp1, Tsp2, Tsp3). Ellipses represent a 95% confidence level.
The composition of microbiomes associated with wheat tissues
The ITS and 16S rRNA amplicon sequences that we observed were used to generate OTU tables, by which the relative abundances of different fungal and bacterial taxa could be evaluated. Crop residue showed a comparatively different microbial community composition than wheat spikes and kernels (Figure 7). Several bacterial genera were dominant in residue. These include Sphingomonas, Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, Methylobacterium, and Tychonema. The two most abundant fungi in the residue were unidentified taxa within the phylum Ascomycota. The highest abundance taxa belonged to the class Dothideomycetes and order Pleosporales. However, the second most abundant taxon was only identified up to the phylum level. In addition to these two taxa, Cyphellophora and Alternaria showed higher relative abundances in crop residue. In addition, Pseudomonas, Enterobacter, and Pantoea were the highly dominant bacterial genera in wheat spikes and kernels. These genera represented more than 60% of the bacterial 16S rRNA sequences observed. Alternaria and Fusarium were the predominant fungal genera in wheat spikes, and these two fungal genera accounted for more than 60% of the observed fungal ITS sequences from wheat spikes. Similarly, wheat kernels were highly dominated by Alternaria and Fusarium, which together accounted for more than 70% of the observed fungal sequences from wheat kernels (Figure 7).
FIGURE 7

The dominant taxa and average relative abundances of bacterial taxa in (A) crop residue; (B) wheat spikes; (C) wheat kernels; and fungal taxa in (D) crop residue; (E) wheat spikes; (F) wheat kernels. Within each panel, the legend lists the ten most abundant genera, differentiated by color. Treatments have been grouped according to whether or not T. gamsii T6085 was applied to the tissue in question, and mean values are shown.
Discussion
The field experiment discovered a significant reduction in FHB incidence and severity due to the T6085 application (Figures 1, 2). A reduction of FHB DS in response to T6085 application to wheat spikes has been observed previously (Sarrocco et al., 2013). Anthesis is the most favorable period for F. graminearum to infect the wheat spikes. In addition, T. gamsii can endophytically colonize the wheat spikes, and this characteristic of T6085 is important to its potential as a biological control agent. However, over time, the beneficial effect of T6085 in FHB reduction was diminished. Therefore, a reduction in disease was not observed at the Tsp3 time point. This fading of efficacy was expected both due to climatic conditions becoming particularly favorable to the development of the disease, and as T6085 was applied as an aqueous conidial suspension at a single time-point. The efficacy of biopesticides is strongly affected by conidial formulation and numbers, and application timing. Nevertheless, our observation of a significant reduction of symptoms on spikes at the Tsp2 time point confirms previous observations.
The abundance of F. graminearum in crop residue and wheat spikes changed significantly throughout the period during which data were collected (Figure 3). In residue, F. graminearum was most abundant at the on-set of the experiment, and its density decreased over time. Although Fusarium spp. grow well in nutrient-rich residue and use residues from wheat, maize or rape for survival and reproduction, the pathogen is a poor competitor among the crop residue associated microbiome. For example, according to Leplat et al. (2016), F. graminearum growth rate is higher within sterile soil than in non-sterilized soil. In the presence of an antagonist, the abundance of F. graminearum frequently decreases over time (Leplat et al., 2013). In addition, the weather can also influence Fusarium abundance in crop residues over time. For instance, the perithecial development rate is lower during dry weather conditions (Manstretta and Rossi, 2016). Therefore, the amount of F. graminearum present in soil can also vary with the humidity. However, the F. graminearum abundance in wheat spikes showed an opposite trend, where at the initial time points, the F. graminearum abundance was significantly lower in wheat spikes, and it rose significantly at times closer to harvest. In other words, F. graminearum growth and development were enhanced during the seed maturation. This could be a natural outcome of having more time for F. graminearum to proliferate during the maturation of wheat spikes, and suggests that a second application of a biocontrol agent after anthesis could help reduce the further spread of the pathogen on the spikelets.
During the field study, two methods were used to inoculate the T6085 into crop residue and wheat spikes to obtain efficient colonization of the fungi to sufficiently mitigate F. graminearum. The crop residue was inoculated using sterilized millet seeds inoculated with T6085 spores and these millet seeds were hand distributed throughout the wheat field. For wheat spikes inoculation, a T6085 spore suspension was used as it allowed for a uniform distribution of the fungal spores throughout the wheat head. However, the T6085 treatment had a minimal impact on the abundance of F. graminearum in crop residue, wheat spikes and kernels (Figure 4). The method used to inoculate T6085 in crop residue and wheat spikes may not have been sufficient to establish the fungus in wheat tissues. The amount of millet seeds spread within the field may not enough, or T. gamsii T6085 may have colonized different portions of wheat spikes compared to F. graminearum. Non-uniform colonization of T. gamsii T6085 in wheat spikes was observed in an earlier field experiment using the same biocontrol agent in the 2011 and 2012 growing seasons (Sarrocco et al., 2013). In that earlier experiment, T. gamsii T6085 colonized lower spikelets more efficiently than upper spikelets. Non-uniform colonization of wheat spikes by the fungus could be due to the downward movement of spores toward the bottom of the spike with gravity when applying it as a spore suspension. Modifications that could be incorporated to enhance the establishment of T. gamsii T6085 could include applying a higher amount of fungal inoculum onto crop residue, increasing the spore concentration within the inoculum, adding an adhesive compound to the spore suspension to promote retention on the plant tissue, and spraying the inoculum with greater uniformly throughout the spikes (Sarrocco et al., 2013).
Moreover, weather conditions also play an essential role. If the weather conditions are more favorable (heavy and frequent rainfalls during the flowering and grain filling period—in April and May in Italy) for the pathogen over the biocontrol agent, the biocontrol agent may not achieve the density required to inhibit the pathogen. In the present experiment, rainfall was 140 and 100 mm during April and May 2019, respectively, much higher than the respective 30 year average, i.e., 75 mm in April and 62 mm in May (Supplementary Figure 2). During a similar field experiment in the 2011 growing season (21 and 8 mm rainfall in April and May, respectively), a significant reduction in FHB DS (% of affected spikelets within a spike) was observed when T. gamsii T6085 was applied on wheat spikes at anthesis. However, when the same field experiment was performed in the following year (160 and 60 mm rainfall in April and May, respectively, comparable to rainfall occurred in 2019) a significant reduction in DS was not observed (Sarrocco et al., 2013). Therefore, the efficacy of biocontrol by T6085 under field conditions can depend on multiple variables, not least the weather conditions during the growing season.
The changes in the microbiome in response to the introduction of the biocontrol agent T. gamsii T6085 as a biocontrol agent was evaluated to understand off target effects of the T6085 treatment on the wheatfield. The number of observed taxa (OTU richness) and the Shannon diversity index are commonly used estimates to measure the biodiversity of microbial communities and it is crucial to calculate the diversity indices to see the alteration made by the T6085 (Spellerberg and Fedor, 2003; Brown et al., 2007). In this study, the impacts of T6085 on OTU richness and Shannon index of the microbial community associated with crop residue, wheat spikes, and kernels were minimal. Yet, OTU richness and Shannon index variation over time were evident (Table 4), which demonstrates a minimal impact on the microbial community scale differences by the T6085 treatment.
Observing patterns and relationships among samples based on multivariate characterization, such as a profile of taxon presence and abundance, frequently requires the use of techniques to reduce the dimensionality of the data. We combined pairwise dissimilarity scores with ordination techniques to aid in visualization and to compare the microbial community composition among treatments and time points. The results suggested that there were not any community-scale impacts when T6085 was applied as a biocontrol agent (Figure 5). Nevertheless, some fungal and bacterial taxa were differentially abundant in T6085 treated samples compared to control samples; these may represent taxa that interact most directly with T6085. Yet, the impact of plant tissue type and the plant development stage was more apparent than the impact of T6085 colonization on the variations in microbial community structure (Figure 6). Similar results were observed in several other studies when biocontrol agents were applied for the prevention of other pathogens, including R. solani (Grosch et al., 2006; Kröber et al., 2014), Phytophthora capsici (Cucu et al., 2020), and Verticillium dahliae (Scherwinski et al., 2007). Compared to the experimentally imposed treatments, these studies have also encountered a more significant effect on microbial communities in response to the plant growth stage or time progression. Environmental changes other than maturation or development of the host plant tissue are also likely to impact microbial community dynamics (Lauber et al., 2013; Massart et al., 2015; Araujo et al., 2019; Latz et al., 2021).
The fungi and bacteria associated with different plant tissues vary among the type of tissue. In addition, the identity of the dominant fungal and bacterial taxa can similarly differ among plant tissues. Therefore, it is interesting to see the changes in the microbiome according to the niche in which they reside. Our results showed that in the year following their growth, wheat residues remaining in the field were primarily dominated by two unidentified Ascomycota, Alternaria, Cyphellophora and several bacterial genera with higher diversity. Alternaria and Fusarium were the dominant fungal genera in spikes and kernels from the current year’s growth, while Pseudomonas, Enterobacter, and Pantoea were the predominant bacterial species (Figure 7). The dominancy of Fusarium and Alternaria in wheat phyllosphere has been previously observed in several studies (González et al., 2008; Hertz et al., 2016; Schiro et al., 2018). According to Hertz et al. (2016), the dominant pathogen in a given location may depend on the climatic conditions. In the same study, they made only sparse observations of F. graminearum in wheat due to the dry climatic conditions during anthesis. Both Fusarium and Alternaria are pathogenic to wheat plants and contaminate the seeds with toxic compounds harmful to consumers. The presence of the two primary pathogens may have reduced the colonization of other fungi in the wheat plant through competition (Müller et al., 2018; Hoffmann et al., 2021). However, the predominant bacterial species, including Pseudomonas, Enterobacter and Pantoea, have overcome the competition from the two fungal pathogens and have well colonized the wheat spikes and kernels.
Interestingly, all these predominant bacterial genera include plant-beneficial and potential biocontrol agents (Chen et al., 2017; Kumar et al., 2017; Müller et al., 2018). According to Müller et al. (2018), Pseudomonas species have shown potential biocontrol ability against F. graminearum. Yet, successful development of FHB even in the presence of naturally occurring Pseudomonas spp. suggests that at natural population densities, these bacteria may not effectively inhibit FHB. Artificial inoculation at higher densities or combining with another biocontrol agent like T. gamsii, may increase the likelihood that these organisms could mitigate F. graminearum infection in wheat. Bakker and McCormick (2019) also observed similar bacterial and fungal compositions in wheat kernels from Minnesota, United States. Their study showed more than 100 bacterial taxa and several dozen fungal taxa in an individual wheat kernel and more significant microbial composition changes over the years. They found a negative correlation between Fusarium load and the relative abundance of Sphingomonas in wheat kernels. Our results also showed a higher relative abundance of Sphingomonas in wheat kernels. Therefore, the genus Sphingomonas could be explored for potential biocontrol agents to fight against FHB in wheat (Bakker and McCormick, 2019).
One drawback we encountered in this study was that we did not identify Trichoderma sequences within the microbiome profiles by the workflow we used; given that T6085 was applied to the plots, we expected to observe this taxon in our microbiome profiles. The mock community control samples we used included Trichoderma reesei, but we found that the ITS2 sequences belonging to T. reesei could not be classified to narrower ranks than the family Hypocreaceae (data not shown). In observations from our biological samples, we observed OTUs that similarly could not be placed below Hypocreaceae; therefore, it is likely that T. gamsii was represented among these observations. Therefore, the method we used for fungal community profiling does not provide good discrimination among genera or species within this family. In addition, even though the ITS is considered a superior marker in species identification across the Kingdom fungi, the discriminatory power of ITS in Ascomycota is lower (Schoch et al., 2012).
Conclusion
Our results indicated the critical role of the plant development stage, tissue type, and environmental conditions in maintaining the microbial community structure. However, the application of T6085 has a limited impact on the microbial populations associated with wheat plants, which suggests that the off-target effects of the use of T6085 as a biocontrol agent are not a concern.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA790613.
Author contributions
SS, AB, GV, and MM designed and implemented the field experiment. AA performed the microbiome profiling and wrote the first draft of the manuscript. AB and AA worked in the portions of this work. All authors edited the manuscript and approved the final draft.
Funding
Funding was provided by the Canadian Agricultural Partnership and the Western Grains Research Foundation.
Acknowledgments
We thank Shaima Eissa for her assistance with sample processing, Wesely Johnson and Dr. Matthew McDougall for their assistance in operating the MiSeq instrument.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.972016/full#supplementary-material
Supplementary Figure 1Networks of co-associated sequence variants, for (A). wheat spikes that were only mock inoculated, and (B). wheat spikes that were inoculated with biocontrol agent Trichoderma gamsii strain T6085.
Supplementary Figure 2The weather data during 2018/2019 growing season. The bar graphs show the rainfall and the line graph shows the maximal (red) and the minimal (green) temperature from July 2018 to June 2019.
Supplementary Table 1List of sequence variants that were significantly differentially abundant (DEseq2, adjusted p < 0.05) between samples that were untreated vs. treated with T6085.
Supplementary Table 2Lists of bacterial sequence variants that were significantly differentially abundant (DEseq2, adjusted p < 0.05) between sampling time points within a given tissue type.
Supplementary Table 3Lists of fungal sequence variants that were significantly differentially abundant (DEseq2, adjusted p < 0.05) between sampling time points within a given tissue type.
References
1
AraujoR.DunlapC.BarnettS.FrancoC. M. M. (2019). Decoding wheat endosphere–rhizosphere microbiomes in Rhizoctonia solani–infested soils challenged by Streptomyces biocontrol agents.Front. Plant Sci.10:1038. 10.3389/fpls.2019.01038
2
BakkerM. G. (2018). A fungal mock community control for amplicon sequencing experiments.Mol. Ecol. Resour.18541–556. 10.1111/1755-0998.12760
3
BakkerM. G.McCormickS. P. (2019). Microbial correlates of Fusarium load and deoxynivalenol content in individual wheat kernels.Phytopathology109993–1002. 10.1094/PHYTO-08-18-0310-R
4
BakkerM. G.MoormanT. B.KasparT. C.ManterD. K. (2017). Isolation of cultivation-resistant oomycetes, first detected as amplicon sequences, from roots of herbicide-terminated winter rye.Phytobiomes J.124–35. 10.1094/PBIOMES-10-16-0011-R
5
BaroncelliR.ZapparataA.PiaggeschiG.SarroccoS.VannacciG. (2016). Draft whole-genome sequence of Trichoderma gamsii T6085, a promising biocontrol agent of Fusarium head blight on wheat.Genome Announc.4:e1747–e1715. 10.1128/genomeA.01747-15
6
BirrT.HaslerM.VerreetJ. A.KlinkH. (2020). Composition and predominance of Fusarium species causing Fusarium head blight in winter wheat grain depending on cultivar susceptibility and meteorological factors.Microorganisms8:617. 10.3390/microorganisms8040617
7
BrownR. L.ReillyL. A. J.PeetR. K. (2007). “Species Richness: Small Scale,”. Encyclopedia of Life SciencesNew York, NY: Wiley-Blackwell*. 10.1002/9780470015902.a0020488
8
CaporasoJ. G.LauberC. L.WaltersW. A.Berg-LyonsD.LozuponeC. A.TurnbaughP. J.et al (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample.Proc. Natl. Acad. Sci. U.S.A.1084516–4522. 10.1073/pnas.1000080107
9
ChenC.XinK.LiuH.ChengJ.ShenX.WangY.et al (2017). Pantoea alhagi, a novel endophytic bacterium with ability to improve growth and drought tolerance in wheat.Sci. Rep.7:41564. 10.1038/srep41564
10
ChowdhuryS. P.DietelK.RändlerM.SchmidM.JungeH.BorrissR.et al (2013). Effects of Bacillus amyloliquefaciens FZB42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community.PLoS One8:e68818. 10.1371/journal.pone.0068818
11
CollingeD. B.SarroccoS. (2022). Transgenic approaches for plant disease control: Status and prospects 2021.Plant Pathol.71207–225. 10.1111/ppa.13443
12
CucuM. A.GilardiG.PuglieseM.FerrocinoI.GullinoM. L. (2020). Effects of biocontrol agents and compost against the Phytophthora capsici of zucchini and their impact on the rhizosphere microbiota.Appl. Soil Ecol.154:103659. 10.1016/j.apsoil.2020.103659
13
FernandoW. G. D.PaulitzT. C.SeamanW. L.DutilleulP.MillerJ. D. (1997). Head blight gradients caused by Gibberella zeae from area sources of inoculum in wheat field plots.Phytopathology87414–421. 10.1094/PHYTO.1997.87.4.414
14
GallettiS.ParisR.CianchettaS. (2020). Selected isolates of Trichoderma gamsii induce different pathways of systemic resistance in maize upon Fusarium verticillioides challenge.Microbiol. Res.233:126406. 10.1016/J.MICRES.2019.126406
15
GärtnerB. H.MunichM.KleijerG.MascherF. (2008). Characterisation of kernel resistance against Fusarium infection in spring wheat by baking quality and mycotoxin assessments.Eur. J. Plant Pathol.12061–68. 10.1007/s10658-007-9198-5
16
GilbertJ.FernandoW. G. D. (2004). Epidemiology and biological control of Gibberella zeae / Fusarium graminearum.Can. J. Plant Pathol.26464–472. 10.1080/07060660409507166
17
GilbertJ.TekauzA. (2000). Review: Recent developments in research on fusarium head blight of wheat in Canada.Can. J. Plant Pathol.221–8. 10.1080/07060660009501155
18
GimenoA.LeimgruberM.KägiA.JennyE.VogelgsangS. (2021). UV protection and shelf life of the biological control agent Clonostachys rosea against Fusarium graminearum.Biol. Control158:104600. 10.1016/J.BIOCONTROL.2021.104600
19
GonzálezH. H. L.MoltóG. A.PacinA.ResnikS. L.ZelayaM. J.MasanaM.et al (2008). Trichothecenes and mycoflora in wheat harvested in nine locations in Buenos Aires Province, Argentina.Mycopathologia165105–114. 10.1007/s11046-007-9084-x
20
GroschR.ScherwinskiK.LottmannJ.BergG. (2006). Fungal antagonists of the plant pathogen Rhizoctonia solani: Selection, control efficacy and influence on the indigenous microbial community.Mycol. Res.1101464–1474. 10.1016/j.mycres.2006.09.014
21
HertzM.JensenI. R.JensenL. ØThomsenS. N.WindeJ.DueholmM. S.et al (2016). The fungal community changes over time in developing wheat heads.Int. J. Food Microbiol.22230–39. 10.1016/J.IJFOODMICRO.2016.01.018
22
HoffmannA.LischeidG.KochM.LentzschP.SommerfeldT.MüllerM. E. H. (2021). Co-cultivation of Fusarium, Alternaria, and Pseudomonas on wheat-ears affects microbial growth and mycotoxin production.Microorganisms9:443. 10.3390/microorganisms9020443
23
JensenD.KarlssonM.SarroccoS.VannacciG. (2016). “Biological control using microorganisms as an alternative to disease resistance,” in Plant Pathogen Resistance Biotechnology1st Edn, ed.CollingeD. B. (Hoboken, NJ: John Wiley & Sons).
24
KangZ.BuchenauerH. (2000). Cytology and ultrastructure of the infection of wheat spikes by Fusarium culmorum.Mycol. Res.1041083–1093. 10.1017/S0953756200002495
25
KröberM.WibbergD.GroschR.EikmeyerF.VerwaaijenB.ChowdhuryS. P.et al (2014). Effect of the strain Bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing.Front. Microbiol.5:252. 10.3389/fmicb.2014.00252
26
KumarA.MauryaB. R.RaghuwanshiR.MeenaV. S.Tofazzal IslamM. (2017). Co-inoculation with Enterobacter and rhizobacteria on yield and nutrient uptake by wheat (Triticum aestivum L.) in the alluvial soil under Indo-Gangetic Plain of India.J. Plant Growth Regul.36608–617. 10.1007/s00344-016-9663-5
27
LasinioG. J.PolliceA.PappalettereL.VannacciG.SarroccoS. (2021). A statistical protocol to describe differences among nutrient utilization patterns of Fusarium spp. and Trichoderma gamsii.Plant Pathol.701146–1157. 10.1111/PPA.13362
28
LatzM. A. C.KerrnM. H.SørensenH.CollingeD. B.JensenB.BrownJ. K. M.et al (2021). Succession of the fungal endophytic microbiome of wheat is dependent on tissue-specific interactions between host genotype and environment.Sci. Total Environ.759:143804. 10.1016/j.scitotenv.2020.143804
29
LauberC. L.RamirezK. S.AanderudZ.LennonJ.FiererN. (2013). Temporal variability in soil microbial communities across land-use types.ISME J.71641–1650. 10.1038/ismej.2013.50
30
LeplatJ.FribergH.AbidM.SteinbergC. (2013). Survival of Fusarium graminearum, the causal agent of Fusarium head blight. A review.Agron. Sustain. Dev.3397–111. 10.1007/s13593-012-0098-5
31
LeplatJ.HeraudC.GautheronE.ManginP.FalchettoL.SteinbergC. (2016). Colonization dynamic of various crop residues by Fusarium graminearum monitored through real-time PCR measurements.J. Appl. Microbiol.1211394–1405. 10.1111/JAM.13259
32
LoveM. I.HuberW.AndersS. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2.Genome Biol.15:550. 10.1186/S13059-014-0550-8/FIGURES/9
33
ManstrettaV.RossiV. (2016). Effects of temperature and moisture on development of Fusarium graminearum perithecia in maize stalk residues.Appl. Environ. Microbiol.82184–191. 10.1128/AEM.02436-15
34
MassartS.MargaritaM. M.HaissamJ. M. (2015). Biological control in the microbiome era: Challenges and opportunities.Biol. Control8998–108. 10.1016/j.biocontrol.2015.06.003
35
MatareseF.SarroccoS.GruberS.Seidl-SeibothV.VannacciG. (2012). Biocontrol of Fusarium head blight: Interactions between Trichoderma and mycotoxigenic Fusarium.Microbiology15898–106. 10.1099/mic.0.052639-0
36
McMullenM.BergstromG.de WolfE.Dill-MackyR.HershmanD.ShanerG.et al (2012). A unified effort to fight an enemy of wheat and barley: Fusarium head blight.Plant Dis.961712–1728. 10.1094/PDIS-03-12-0291-FE
37
McMurdieP. J.HolmesS. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data.PLoS One8:e61217. 10.1371/JOURNAL.PONE.0061217
38
MüllerT.RuppelS.BehrendtU.LentzschP.MüllerM. E. H. (2018). Antagonistic potential of fluorescent pseudomonads colonizing wheat heads against mycotoxin producing Alternaria and fusaria.Front. Microbiol.9:2124. 10.3389/fmicb.2018.02124
39
NilssonR. H.LarssonK. H.TaylorA. F. S.Bengtsson-PalmeJ.JeppesenT. S.SchigelD.et al (2018). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acid Res.47D259–D264.
40
OksanenJ.SimpsonG.BlanchetF.KindtR.LegendreP.MinchinP.et al (2022). Vegan: community ecology package. Package version 2.6-2. Available online at: https://CRAN.R-project.org/package=vegan
41
OsborneL. E.SteinJ. M. (2007). Epidemiology of Fusarium head blight on small-grain cereals.Int. J. Food Microbiol.119103–108. 10.1016/J.IJFOODMICRO.2007.07.032
42
PerazzolliM.AntonielliL.StorariM.PuopoloG.PancherM.GiovanniniO.et al (2014). Resilience of the natural phyllosphere microbiota of the grapevine to chemical and biological pesticides.Appl. Environ. Microbiol.803585–3596. 10.1128/AEM.00415-14
43
PestkaJ. J. (2010). Deoxynivalenol: Mechanisms of action, human exposure, and toxicological relevance.Arch. Toxicol.84663–679. 10.1007/s00204-010-0579-8
44
QuastC.PruesseE.YilmazP.GerkenJ.SchweerT.YarzaP.et al (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools.Nucleic Acids Res.41D590–D596. 10.1093/nar/gks1219
45
SarroccoS.EstebanP.VicenteI.BernardiR.PlainchampT.DomenichiniS.et al (2021). Straw competition and wheat root endophytism of Trichoderma gamsii T6085 as useful traits in the biological control of Fusarium head blight.Phytopathology1111129–1136. 10.1094/phyto-09-20-0441-r
46
SarroccoS.MatareseF.MonciniL.PachettiG.RitieniA.MorettiA.et al (2013). Biocontrol of Fusarium head blight by spike application of Trichoderma gamsii.J. Plant Pathol.95 (Supplement), S1.19–S1.27.
47
SarroccoS.MauroA.BattilaniP. (2019a). Use of competitive filamentous fungi as an alternative approach for mycotoxin risk reduction in staple cereals: State of art and future perspectives.Toxins11:701. 10.3390/toxins11120701
48
SarroccoS.ValentiF.ManfrediniS.EstebanP.BernardiR.PuntoniG.et al (2019b). Is exploitation competition involved in a multitrophic strategy for the biocontrol of Fusarium head blight?Phytopathology109560–570. 10.1094/PHYTO-04-18-0123-R
49
SarroccoS.VannacciG. (2018). Preharvest application of beneficial fungi as a strategy to prevent postharvest mycotoxin contamination: A review.Crop Prot.110160–170. 10.1016/J.CROPRO.2017.11.013
50
ScherwinskiK.WolfA.BergG. (2007). Assessing the risk of biological control agents on the indigenous microbial communities: Serratia plymuthica HRO-C48 and Streptomyces sp. HRO-71 as model bacteria.BioControl5287–112. 10.1007/s10526-006-9006-8
51
SchiroG.VerchG.GrimmV.MüllerM. E. H. (2018). Alternaria and Fusarium fungi: Differences in distribution and spore deposition in a topographically heterogeneous wheat field.J. Fungi4:63. 10.3390/jof4020063
52
SchochC. L.SeifertK. A.HuhndorfS.RobertV.SpougeJ. L.LevesqueC. A.et al (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi.Proc. Natl. Acad. Sci.1096241–6246. 10.1073/pnas.1117018109
53
SmithK. P.GoodmanR. M. (1999). Host variation for interactions with beneficial plant-associated microbes.Ann. Rev. Phytopathol.37473–491. 10.1146/annurev.phyto.37.1.473
54
SpellerbergI. F.FedorP. J. (2003). A tribute to Claude Shannon (1916-2001) and a plea for more rigorous use of species richness, species diversity and the “Shannon-Wiener” Index.Glob. Ecol. Biogeogr.12177–179. 10.1046/j.1466-822X.2003.00015.x
55
StrangeR. N.MajerJ. R.SmithH. (1974). The isolation and identification of choline and betaine as the two major components in anthers and wheat germ that stimulate Fusarium graminearum in vitro.Physiol. Plant Pathol.4277–290. 10.1016/0048-4059(74)90015-0
56
SuttonJ. C. (1982). Epidemiology of wheat head blight and maize ear rot caused by Fusarium graminearum.Can. J. Plant Pathol.4195–209. 10.1080/07060668209501326
57
TojuH.TanabeA. S.YamamotoS.SatoH. (2012). High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples.PLoS One7:e40863. 10.1371/journal.pone.0040863
58
TurnerT. R.JamesE. K.PooleP. S. (2013). The plant microbiome.Genome Biol.14:209. 10.1186/gb-2013-14-6-209
59
VicenteI.BaroncelliR.Moran-DiezM. E.BernardiR.PuntoniG.HermosaR.et al (2020). Combined comparative genomics and gene expression analyses provide insights into the terpene synthases inventory in Trichoderma.Microorganisms8:1603. 10.3390/microorganisms8101603
60
WalterS.NicholsonP.DoohanF. M. (2010). Action and reaction of host and pathogen during Fusarium head blight disease.New Phytol.18554–66. 10.1111/j.1469-8137.2009.03041.x
61
WeiF.HuX.XuX. (2016). Dispersal of Bacillus subtilis and its effect on strawberry phyllosphere microbiota under open field and protection conditions.Sci. Rep.6:22611. 10.1038/srep22611
62
WickhamH. (2016). ggpolt2 Elegant Graphics for Data Analysis. Use R! series.211. Available Online at: http://had.co.nz/ggplot2/book(accessed on Jul 11, 2022).
63
YinD.WangN.XiaF.LiQ.WangW. (2013). Impact of biocontrol agents Pseudomonas fluorescens 2P24 and CPF10 on the bacterial community in the cucumber rhizosphere.Eur. J. Soil Biol.5936–42. 10.1016/j.ejsobi.2013.09.001
Summary
Keywords
Fusarium graminearum, Triticum durum, 16S rRNA, ITS, microbiome, fusarium head blight
Citation
Alukumbura AS, Bigi A, Sarrocco S, Fernando WGD, Vannacci G, Mazzoncini M and Bakker MG (2022) Minimal impacts on the wheat microbiome when Trichoderma gamsii T6085 is applied as a biocontrol agent to manage fusarium head blight disease. Front. Microbiol. 13:972016. doi: 10.3389/fmicb.2022.972016
Received
17 June 2022
Accepted
18 August 2022
Published
23 September 2022
Volume
13 - 2022
Edited by
Martin Filion, Agriculture and Agri-Food Canada (AAFC), Canada
Reviewed by
Muzammil Hussain, State Key Laboratory of Mycology, Institute of Microbiology (CAS), China; Karolina Oszust, Institute of Agrophysics (PAN), Poland
Updates
Copyright
© 2022 Alukumbura, Bigi, Sarrocco, Fernando, Vannacci, Mazzoncini and Bakker.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sabrina Sarrocco, Sabrina.Sarrocco@unipi.itW. G. Dilantha Fernando, Dilantha.Fernando@umanitoba.caMatthew G. Bakker, Matthew.Bakker@umanitoba.ca
This article was submitted to Microbe and Virus Interactions with Plants, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.