- 1Bioscience Program, Biological and Environmental Science and Engineering Division, King Abdullah University of Science and Technology (KAUST), Thuwal, Saudi Arabia
- 2Environmental Science Program, Water Desalination and Reuse Center, King Abdullah University of Science and Technology (KAUST), Thuwal, Saudi Arabia
- 3Computational Biology Research Center, King Abdullah University of Science and Technology (KAUST), Thuwal, Saudi Arabia
- 4Services Hospital, Services Institute of Medical Sciences, Lahore, Pakistan
- 5Centre de Biologie Structurale (CBS), INSERM, CNRS, Université de Montpellier, Montpellier, France
Quorum quenching (QQ) is the enzymatic degradation of molecules used by bacteria for synchronizing their behavior within communities. QQ has attracted wide attention due to its potential to inhibit biofilm formation and suppress the production of virulence factors. Through its capacity to limit biofouling and infections, QQ has applications in water treatment, aquaculture, and healthcare. Several different QQ enzymes have been described; however, they often lack the high stability and catalytic efficiency required for industrial applications. Previously, we identified genes from genome sequences of Red Sea sediment bacteria encoding potential QQ enzymes. In this study, we report that one of them, named LrsL, is a metallo-β-lactamase superfamily QQ enzyme with outstanding catalytic features. X-ray crystallography shows that LrsL is a zinc-binding dimer. LrsL has an unusually hydrophobic substrate binding pocket that can accommodate a broad range of acyl-homoserine lactones (AHLs) with exceptionally high affinity. In vitro, LrsL achieves the highest catalytic efficiency reported thus far for any QQ enzyme with a Kcat/KM of 3 × 107. LrsL effectively inhibited Pseudomonas aeruginosa biofilm formation without affecting bacterial growth. Furthermore, LrsL suppressed the production of exopolysaccharides required for biofilm production. These features, and its capacity to regain its function after prolonged heat denaturation, identify LrsL as a robust and unusually efficient QQ enzyme for clinical and industrial applications.
Introduction
Quorum sensing (QS) is a chemical communication between bacteria that coordinates gene expression at the community level. QS is mediated by a diverse set of small molecules, among which acyl-homoserine lactones (AHLs) are the best-studied (Whiteley et al., 2017). AHLs play an important role in biofilm formation by various bacteria in many different environments (Eberl et al., 1996; Davies et al., 1998; Lee et al., 2018). Biofilms have detrimental consequences in health care and industries, such as water purification, shipping, and aquaculture (Salta et al., 2013; Rehman et al., 2019, 2020, 2021; Rumbaugh and Sauer, 2020). Once formed, biofilms are difficult to remove because of their increased resistance to antimicrobial and other physical and chemical cleaning procedures. Biofilms incur substantial economic costs, and the current biofilm prevention and treatment practices often negatively affect the environment. Therefore, sustainable and eco-friendly measures to mitigate biofilms are required.
The AHL molecules consist of a lactone ring linked to the acyl chain through an amide bond. The length of the acyl chain can vary from 4 to 18 carbon atoms and may carry an oxo or hydroxyl modification at the third carbon atom (Azimi et al., 2020). The QQ enzymes hydrolyzing the lactone ring of AHLs are called lactonases, whereas those cleaving the amide bond are amidases or acylases. Most QQ enzymes discovered so far are lactonases, whereas relatively few acylases have been identified (Dong et al., 2000; Wang et al., 2012; Tang et al., 2015). Lactonases belong to three protein superfamilies: (i) phosphotriesterase-like lactonases (PLL), (ii) paraoxonases, and (iii) metallo-β-lactamases (MBLs; Fetzner, 2015). AiiA was the first AHL lactonase discovered belonging to the MBL superfamily. It possesses a characteristic HXHXDH motif that binds two metal cations (Dong et al., 2000). Subsequently, AHL lactonases have been discovered in diverse bacteria exhibiting dramatic differences in kinetic parameters, stability, and substrate preference (Fetzner, 2015; Bergonzi et al., 2018).
Acyl-homoserine lactone-based QS plays a vital role in the pathogenicity of many gram-negative bacteria. For example, Pseudomonas aeruginosa, a human pathogen (Smith et al., 2002); Erwinia carotovora, a plant pathogen (Smadja et al., 2004); and Aeromonas hydrophila, a fish pathogen (Chu et al., 2013), use AHLs to regulate the production of virulence factors. The diseases caused by these pathogenic bacteria have a devastating influence on health, agriculture, and food security. Therefore, the inhibition of virulence factor production through the degradation of AHLs is an attractive strategy to reduce the pathogenicity of common bacteria. In contrast to bactericidal approaches, this so-called quorum quenching (QQ) was presented as a sustainable method that does not appear to induce resistance in target bacteria (Cegelski et al., 2008). However, these claims have been challenged, and it was suggested that certain QQ agents that enter the cells could exert selective pressure and impart resistance in bacteria (Defoirdt et al., 2010; Garcia-Contreras et al., 2013). It remains unclear whether QQ enzymes that do not enter the bacterial cell can give rise to resistance.
Although diverse chemical compounds exhibit a QS inhibitory effect, applying a QQ enzyme offers certain advantages. The key parameters for applying QQ enzymes as anti-infective agents are their substrate affinity, catalytic efficiency, and stability (Remy et al., 2020). Protein engineering may improve these factors (Billot et al., 2020). However, different applications require different QQ enzymes to suppress communication mediated through specific AHL molecules in specific environments. Given the widespread use of QS in nature and the diversity of potential QQ applications, there is a need to bio-prospect for novel QQ enzymes with a high affinity for specific AHL molecules, better stability, and improved catalytic activity.
Since their discovery, QQ enzymes have stirred much interest due to their potential use in several fields. Biotechnological applications of QQ enzymes can include those in human health, aquaculture, agriculture, biofouling, and biocorrosion (Billot et al., 2020). Successful application of QQ enzymes in industry requires a repertoire of QQ enzymes with varying substrate specificity, affinity, and stability under various conditions. With this purpose, we identified potential QQ enzymes from Red Sea sediment – a harsh and understudied environment (Renn et al., 2021). We gene-synthesized, expressed and purified the first QQ enzyme, LrsL, isolated from the genus Labrenzia. The 1.9 Å resolution crystal structure that we determined for LrsL reveals that it has an exceptionally hydrophobic substrate binding pocket, capable of accommodating a broad range of AHL molecules. Functional studies showed that LrsL exhibits the highest catalytic activity reported for any QQ enzyme thus far on a range of AHL molecules (C4-AHL, C6-AHL, and Oxo-C12-AHL). LrsL has the capability of regaining its functional three-dimensional fold after heat denaturation, which may be beneficial for biotech applications.
Materials and methods
Identification of LrsL
Previously, we identified seven QQ lactonase genes in Red Sea sediment bacteria genomes (Rehman and Leiknes, 2018). We selected all seven genes for expression in Escherichia coli (BL21 DE3) for the initial screening. The protein structures of QQ enzymes were predicted using SWISS-MODEL (Waterhouse et al., 2018). Unmodeled N- and C-terminal amino acids were assumed to be flexible and were removed before gene synthesis. The codons were optimized for expression in E. coli using an in-house Python script (Guo et al., 2021). The codon-optimized genes were gene-synthesized into expression plasmid pJEx411c (Guo et al., 2021), with the in-frame addition of a Twin-Strep-tag at the C-terminal, containing kanamycin resistance for selection. The seven plasmids were transformed into competent E. coli cells. Successful transformants were selected by plating on Luria–Bertani (LB) agar medium containing 50 μg/ml kanamycin (GoldBio, United States).
Bioinformatic analyses
Multiple sequence alignments were produced using the online version of MUSCLE (Edgar, 2004) and were edited using Jalview V-2 (Waterhouse et al., 2009). Phylogenetic analysis was performed in MEGA X using the maximum likelihood method with 500 bootstrap values (Kumar et al., 2018). AlphaFold (Jumper et al., 2021) models were calculated using the AlphaFold2_advanced Colab notebook.
Expression and purification of LrsL
Table 1 lists all strains and plasmids used in this study. An overnight culture of 10 ml of E. coli (BL21 DE3) harboring pJEx411c:LrsL plasmid was inoculated into fresh LB (1 l) with kanamycin (50 μg/ml; GoldBio, United States). The culture was incubated at 37°C with shaking at 200 rpm. At an optical density (OD)600 of 0.6, the expression of the QQ enzyme was induced using 250 μM isopropyl ß-D-1-thiogalactopyranoside (GoldBio, United States), and 500 μM ZnSO4 (Merck, Germany) was added, followed by incubation with shaking at 22°C for another 22 h. Cells were pelleted by centrifugation at 8000 g for 10 min at 4°C. The supernatant was discarded, and the cell pellet was stored at −80°C until further processing. The cell pellet was thawed in water at room temperature and resuspended in 5 ml of cold lysis buffer A (50 mM Tris, 200 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine (TCEP), and 250 μM ZnSO4, pH 7.8) for each 1 g of pellet and kept on ice. Benzonase (Millipore, United States) was added at 25 units/ml of buffer. Cells were lysed on a cell disruptor (Constant Systems Ltd.). Cell lysates were centrifuged in a prechilled centrifuge at 6,000 g for 10 min at 4°C. The supernatant was collected and recentrifuged at 35,000 g for 45 min at 4°C, and filtered using a miracloth (Millipore, United States). The LrsL protein was purified on an AKTA pure (GE Healthcare, United States) in the following sequence: Strep-tag affinity purification, ion-exchange, and gel-permeation chromatography. We used the StrepTrap HP (GE Healthcare) column and binding buffer B (50 mM Tris, 200 mM NaCl, and 1 mM TCEP, pH 8) for affinity purification. Elution was performed in a binding buffer containing 2.5 mM desthiobiotin. Fractions were pooled and concentrated to 5 ml using 10 K Amicon Ultra (Millipore) columns (centrifugation at 4000 g for 20 min). Concentrated proteins were dialyzed overnight at 4°C against buffer C (50 mM Tris, 1 mM TCEP, and 10 mM NaCl, pH 8) using 14–16 kDa membranes, subjected to the Mono-Q 10/100 Gl column (GE Healthcare) for ion-exchange purification, and recovered using a high salt buffer (50 mM tris, 1 M NaCl, and 1 mM TCEP, pH 8). Finally, the proteins were subjected to gel-filtration on a Superdex75 16/600 column (GE Healthcare) into buffer D (50 mM HEPES, 150 mM NaCl, and 250 μM ZnSO4, pH 7.8). After spin concentration, protein aliquots were flash-frozen in liquid nitrogen and stored at −80°C. The quality, purity and molecular mass of the recombinant proteins were measured using sodium dodecyl sulfate gel electrophoresis (Invitrogen). Protein concentrations were determined by absorbance on a NanoDrop spectrophotometer using sequence-specific extinction coefficients.1
Protein crystallization
Purified LrsL protein was used for crystallization and crystals were obtained using the sitting drop vapor diffusion method. LrsL crystals were obtained by equilibrating 300 nl of protein (10 mg/ml in 50 mM HEPES, 150 mM NaCl, and 250 μM ZnSO4, pH 7.8 buffer) with 300 nl of reservoir solution (1.6 M tri-sodium citrate tribasic) at 25°C using the Mosquito pipettor. Crystals were flash-cooled using liquid nitrogen without any cryo-protectant. Diffraction data were collected at Proxima 1 beamline (SOLEIL Synchrotron, France) at 100 K using a DECTRIS EIGER X 16 M detector (Proposal number 20210195). The data was processed in XDS (Kabsch, 2010).
Protein structure determination
Diffraction data was checked for possible contaminants using ContaMiner (Hungler et al., 2016). Initial phases were calculated using Balbes (Long et al., 2008). Automated rebuilding was performed by Arp wARP (Langer et al., 2008), followed by iterative manual rebuilding using COOT (Emsley et al., 2010) and automated refinement by Refmac (Murshudov et al., 1997). The protein model was evaluated using MolProbity (Davis et al., 2007).
Circular dichroism
Circular dichroism spectra measurements were performed in triplicates using JASCO spectropolarimeter, with a 1-mm path-length cell. LrsL was measured at 10 μM concentration in 2.5 mM bicine pH 8.3, 150 mM NaCl, and 0.25 mM ZnSO4 buffer. The measurement was performed in a temperature gradient starting at 25°C until 120°C. The sample was then cooled down to 25°C and re-measured. CD results were analyzed using CAPITO (Wiedemann et al., 2013).
In silico molecular docking analyses
The crystal structure of AidC, a Dizinc Quorum-Quenching Lactonase, in complex with N-hexnoyl-L-homoserine (PDB ID: 4zo3) and the crystal structure of LrsL from this study were used to perform docking analyses. The ligand structure of C6L homoserine was extracted from PDB ID: 4zo3 using PyMOL.2 Flexible docking was performed using AutoDock 4.2 (Goodsell et al., 1996) as described in (Vogeley et al., 2022), except that the size for the gridbox (x, y, z points) were set to 20 × 20 × 20 and centers for the grid were designated at X = 123.28, Y = 102.8, Z = 32.32 dimensions for PDB ID: 4zo3, whereas for LrsL these parameters were set to 20 × 20 × 20 and X = 123.16, Y = 102.74, Z = 32.85 dimensions, respectively. Final docking poses were analyzed using PyMOL.
Size exclusion chromatography
Size exclusion chromatography combined with multiangle light scattering was performed on a Superdex 200 10/300 column (Cytiva) on an Agilent HPLC setup. 5.5 mg/ml of LrsL were injected in the column and analyzed in 20 mM HEPES, pH 7.5, 200 mM NaCl, 1 mM TCEP, and 0.01% of NaN3. The molecular weight estimation and refractive index were calculated using Astra Multiangle Light Scattering software (Wyatt Technology).
Differential scanning fluorimetry
The experiment was performed as previously described (Momin et al., 2019). Briefly, thermal stability was assessed in 20 mM HEPES, pH 7.5, 200 mM NaCl, and 1 mM TCEP. Fluorescence was measured by heating the LrsL sample from 25°C to 99°C at 0.03°C/s. LrsL was used at a concentration of 10 μM, and fluorescent dye SYPRO Orange was used at a final concentration of 5X.
Characterization of QQ activity
The QQ activity of purified enzymes was investigated using Chromobacterium violaceum CV026 assay as described previously (Rehman and Leiknes, 2018). Briefly, Luria–Bertani (LB) agar plates were overlaid with 5 ml of 1/100th-dilution of an overnight culture of the biosensor strain CV026 mixed with LB soft agar (0.7%). After the soft agar was solidified, wells (6-mm) were created in the medium by using sterile pipette tips. These wells were filled with the LrsL and C6-AHLs mixture and incubated at 30°C for 24 h. C6-AHLs in buffer D and mixed with BSA were used as controls. The appearance of a purple halo around the well indicated intact C6-AHLs. On the other hand, LrsL degraded C6-AHLs, and therefore, the biosensor strain was not activated. Thus, halo formation was not observed. The minimum active concentration of LrsL was determined as the highest dilution that could degrade 10 μM C6-AHLs in 24 h, by preliminary studies and as determined by the C. violaceum assay (Mayer et al., 2018).
Kinetic characterization
Experiments were performed in a 200 ml reaction volume using a 96-well plate with a path length of 5.8 mm to calculate the catalytic parameters. For the AHL hydrolysis experiment, we used cresol purple (Alfa Aesar, United States) as a pH indicator, as previously described (Bergonzi et al., 2016, 2018). The hydrolysis of the lactone ring was monitored at 577 nm in the buffer (2.5 mM Bicine pH 8.3, 150 mM NaCl, 0.25 mM ZnSO4, 0.2 mM cresol purple, and 0.5% of DMSO, at 25°C) using different concentrations of enantiomerically pure AHLs ranging from 0 to 1 mM. A specific concentration of enzyme (1, 2, or 3 μM) was tested for each substrate. Each measurement was made in triplicate. The background rate was measured in the absence of the substrate and enzyme. The AHL degradation rates in the absence of the enzyme were measured as the control and found to be similar to the background rates. Catalytic parameters were calculated by fitting the data to the Michaelis–Menten equation using a rectangular hyperbola curve fitting model using SoftMax Pro 7.0.3 (Molecular Devices).
Biofilm assay
The overnight culture of P. aeruginosa was diluted to an OD of 0.05 and incubated with 1.14 μM of LrsL enzymes in round-bottom 96-well plates at 37°C. Biofilms were assayed 24 h after inoculation using the crystal violet staining method, as described previously (Coffey and Anderson, 2014). For the preformed biofilm assay, P. aeruginosa biofilm was grown for 24 h in 96-well plates. The culture medium was carefully removed and replenished with fresh LB containing 1.14 μM of LrsL. The plates were further incubated for 8 h at room temperature, and crystal violet staining was performed to quantify the attached biomass. We have used BSA as a negative control for the effects of adding protein to the experiment (Bergonzi et al., 2019). The concentrations of the enzymes were chosen based on our preliminary analysis and were consistent with previous studies (Zhang et al., 2017; Bergonzi et al., 2019).
Congo red binding assay
Production of exopolysaccharides was assessed using the previously described assay (Spiers et al., 2003). The overnight culture of P. aeruginosa PAO1 was inoculated with fresh LB at a 1:100 dilution. The LrsL and bovine serum albumin (BSA) were added to a final concentration of 1.14 μM. The cultures were incubated at 37°C for 72 h without shaking. The biomass was collected by centrifugation at 13,000 g for 5 min, and the supernatant was discarded. The pellet was resuspended in 1 ml of 20 μg/ml Congo red (CR) and incubated for 90 min with shaking at room temperature. The biomass and bound CR were sedimented by centrifugation at 16,000 g for 5 min. The supernatant was serially diluted, and optical density was measured at 490 nm. The LB media, PAO1 without LrsL, and PAO1 inoculated with BSA served as controls.
Statistical analyses
All statistical analyses were performed using Excel (v. 16.52 Microsoft) and data were plotted in GraphPad Prism 9.0.3
Results and discussion
Identification of LrsL as a promising QQ candidate
As the basis for this study, we used all the seven lactonase genes previously identified bioinformatically as QQ genes in genomes of Red Sea isolates (Rehman and Leiknes, 2018). Based on the predicted protein structures of these genes, we designed expression constructs for their catalytic domain (see Methods). In the initial screening of the recombinantly produced proteins, only the QQ gene from Labrenzia sp. VG12 (formerly QQ0007_VG12) encoding protein (ASP32504.1) exhibited good expression and degradation of AHLs (Supplementary Figure S1A). Therefore, we proceeded with the expression, purification, and further analyses of this protein, which we named LrsL (L for Labrenzia, rs for the Red Sea, and L for lactonase), representing the genus of the host bacteria, source of bacteria, and protein function. In the sequence used for gene synthesis, we removed the codons for the N-terminal 47 residues of LrsL, encompassing the secretion signal sequence and unstructured region. The resulting recombinant LrsL protein consists of 285 residues and has a calculated molecular mass of 30.8 kDa.
LrsL forms zinc-binding homodimers
The wild-type LrsL protein consists of 331 amino acid residues and carries an N-terminal twin-arginine translocation (TAT) signal sequence with a cleavage site between residue 38 and 39 as determined using SignalP analysis (Supplementary Figure S1B; Petersen et al., 2011). TAT signals generally serve to translocate folded proteins across membranes. To our knowledge, LrsL is the only QQ enzyme discovered to date that carries a TAT signal and is only the second enzyme that is potentially secreted extracellularly, the other one being MomL (Tang et al., 2015). These extracellular QQ enzymes were both discovered in a marine environment.
We established a multiple sequence alignment of LrsL and other well-studied lactonases (Figure 1A; Supplementary Figure S2). These include AhlS (BAK54003.1) from Solibacillus silvestris (Morohoshi et al., 2012), AidC (BAP32158.1) from Chryseobacterium sp. StRB126 (Wang et al., 2012), AhlD (AAP57766.1) from Arthrobacter sp. IBN110 (Park et al., 2003), AidP (WP_049694637.1) from Antarctic Planococcus versutus (See-Too et al., 2018), AhlK (VTR44369.1) from Klebsiella sp. (Chan, 2013), MomL (AIY30473.1) from Muricauda olearia (Tang et al., 2015), Aal (WP_021296945.1) from Alicyclobacillus acidoterrestris (Bergonzi et al., 2018), Gcl (WP017434252.1) from Parageobacillus caldoxylosilyticus (Bergonzi et al., 2016), AiiA (ACI96342.1) from Bacillus thuringiensis (Liu et al., 2005), and AiiB (WP_172691130.1) from Agrobacterium tumefaciens (Liu et al., 2007). The AiiA sequence in the NCBI database was truncated. We added 19 amino acids at the N-terminal to obtain the sequence given by (Liu et al., 2007). The HXHXDH~H (where X is any residue and ~ indicates a long separation) motif is considered the hallmark of MBL superfamily (Wang et al., 2012). All lactonases included shared the HXHXDH~H motif confirming they belong to the MBL superfamily (Figure 1A; Supplementary Figure S2; Dong et al., 2000; Wang et al., 2012). This motif is involved in metal ion coordination and is essential for the function of lactonases (Dong et al., 2002; Palzkill, 2013). AhlK carries an asparagine residue instead of a second aspartic acid at position 257 (Supplementary Figure S2, blue asterisk). The multiple sequence alignment demonstrated that His260 (numbering scheme corresponding to wild-type LrsL) was conserved in AidC, MomL, and LrsL. In contrast, AiiA, AiiB, Aal, Gcl, AhlK, AhlD, AhlS, and AidP all carry a conserved tyrosine in this position (Supplementary Figure S2, black asterisk). This tyrosine in AiiA and histidine in MomL are required for substrate binding and catalysis (Momb et al., 2008; Tang et al., 2015).
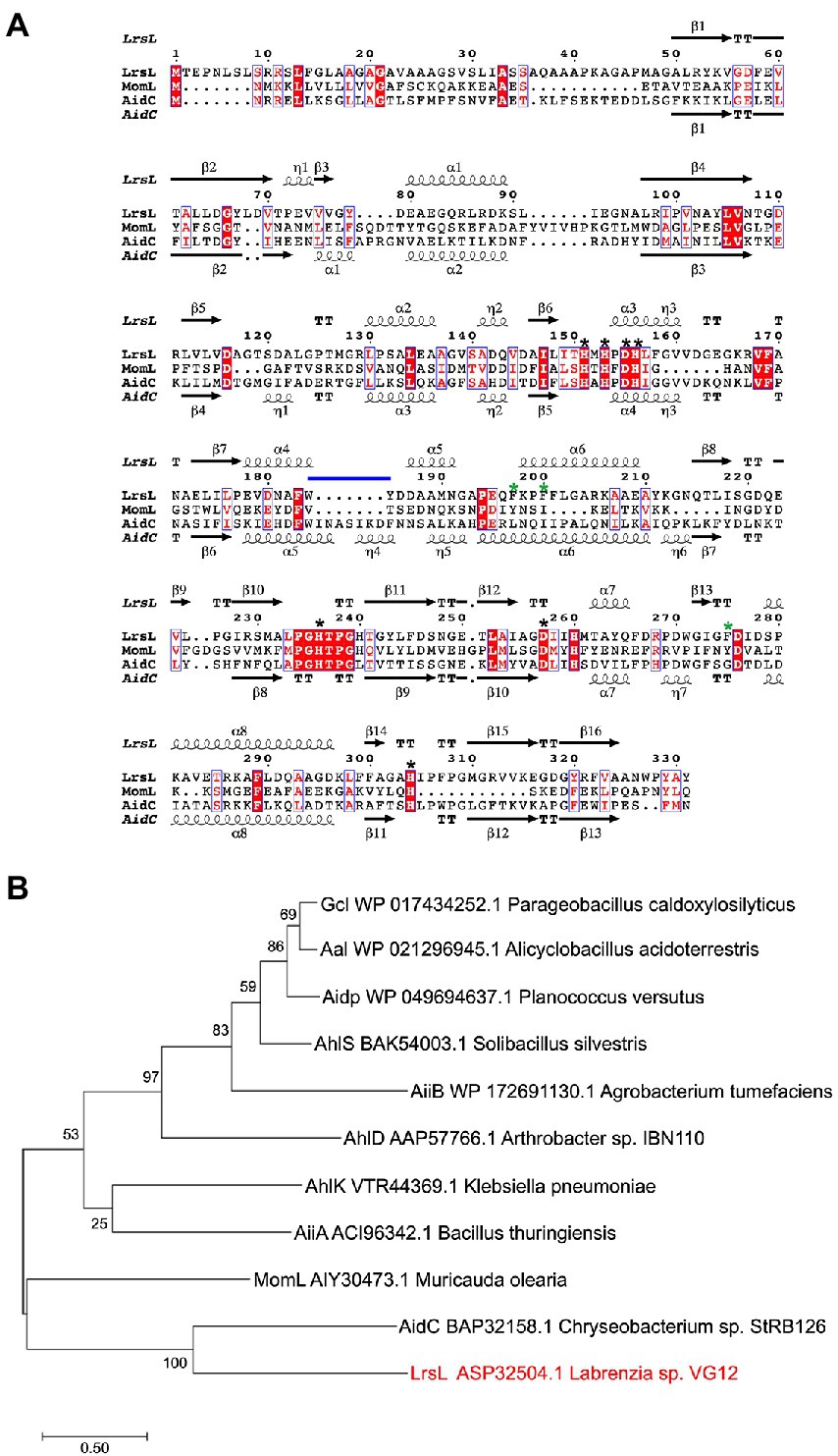
Figure 1. Phylogenetic analysis of QQ enzymes (A) annotated sequence alignment of LrsL, MomL and AidC prepared by Espript 3.0 (http://espript.ibcp.fr). Secondary structure of LrsL is shown above the sequence, and for AidC below. Conserved zinc coordination residues are marked with black asterisks. The three substrate-coordinating phenylalanines, found in LrsL, are marked with green asterisks. The atypical extension of the AidC loop between α5 and α6 is with a horizontal blue line. (B) Phylogenetic analysis of LrsL. A phylogenetic tree of LrsL and 10 other QQ enzymes was constructed using the Maximum Likelihood method. The end of each branch shows the name of the QQ enzyme, followed by the GenBank accession number and the name of the host bacteria. Branch node numbers represent bootstrap values and consensus inferred from 500 iterations.
The phylogenetic analysis revealed that the newly isolated LrsL was closely related to AidC, which forms stable zinc-binding homodimers (Figure 1B; Mascarenhas et al., 2015). The MBL superfamily contains monomeric, dimeric (Mascarenhas et al., 2015), and tetrameric enzymes (Ullah et al., 1998; Mercuri et al., 2001; Li de la Sierra-Gallay et al., 2005). To gain experimental insights into the mechanism of catalysis and self-assembly, we determined the three-dimensional structure of LrsL, showing residues 49–331 at a resolution of 1.9 Å (Figure 2A; Table 2). The crystals contain a symmetric LrsL dimer in the asymmetric unit, burying a total protein surface of 1,135 Å2 in the interface. Size exclusion chromatography–multiangle light scattering analysis confirmed that LrsL was eluted as a single peak with a molecular mass of 61.2 kDa corresponding to an LrsL dimer (Figure 2B). The LrsL dimeric arrangement resembles closely the one of AidC, with an r.m.s.d of 0.952 Å (Figure 2A). However, AidC has an extension of the loop between helices α5 and α6, which is absent in LrsL and most other lactonases (Figures 1A, 2A; Supplementary Figure S2). This loop extension covers one side of the substrate binding site and narrows substrate selectivity in AidC (Mascarenhas et al., 2015).
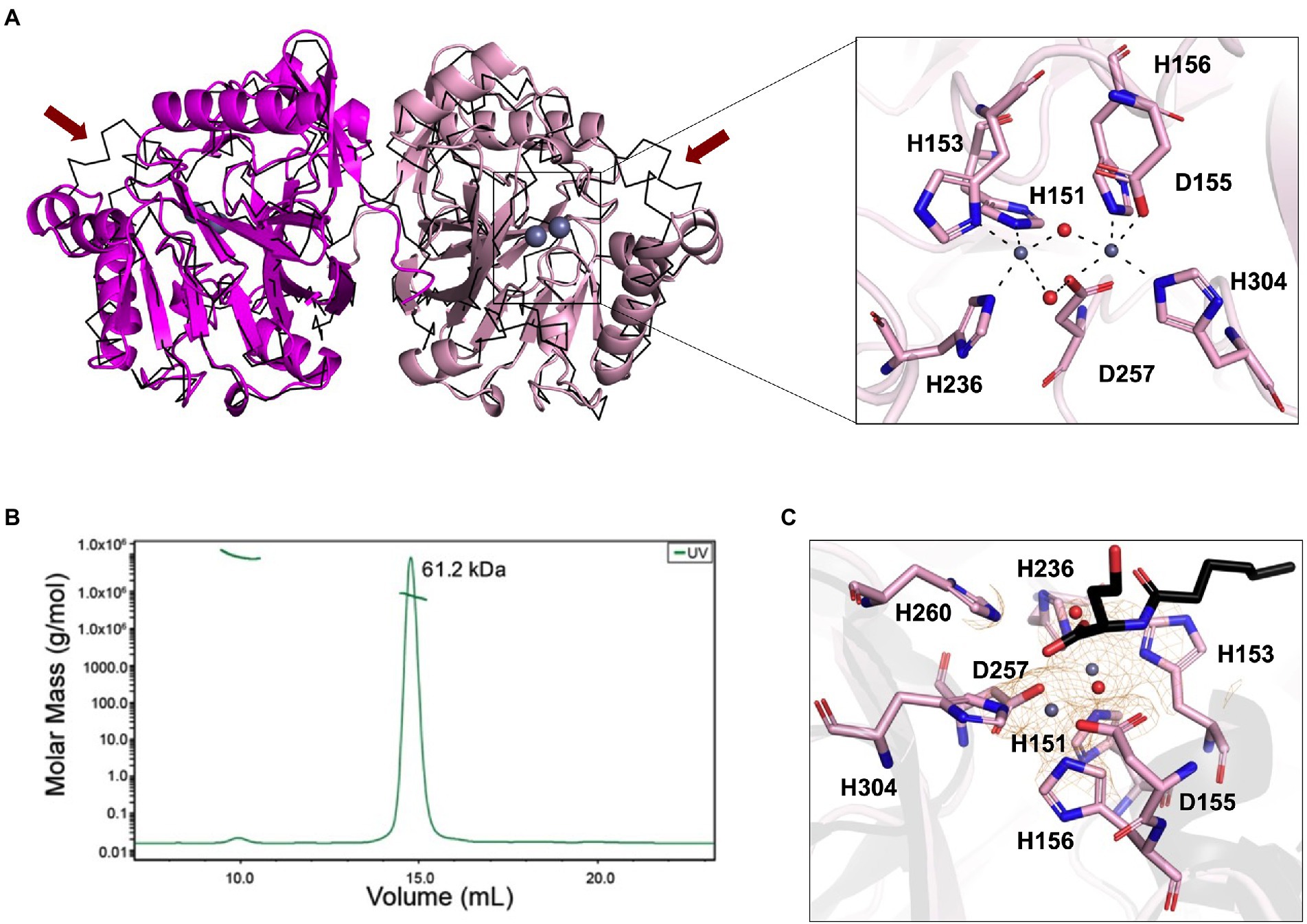
Figure 2. Structural analysis of LrsL. (A) Dimeric crystal structure of LrsL. (Left) The structure of LrsL (residue 49 to C-terminal) is shown as cartoon diagram, with one chain in magenta and the other chain in light pink. The C-alpha trace (black) of the experimental structure of the AidC dimer (PDB accession 4zo3) is superimposed. The two zinc ions included in each LrsL protein chain are shown in blue-gray. (Right) Zoom into the zinc-binding active site binding pocket of LrsL. The zinc-coordinating residues as highlighted in Figure 1A and Supplementary Figure S2 are marked as light pink sticks. Zinc ions are shown as blue-gray spheres. Zinc coordinating atoms are indicated by dashed lines from the atom to zinc. The red spheres indicate water molecules that contribute to zinc coordination in LrsL. (B) Size Exclusion Chromatography coupled with Multiangle Light Scattering (SEC-MALS) analysis showed that LrsL eluted as a single peak with a molecular weight of 60.2 kDa, corresponding to the molecular weight of an LrsL dimer. (C) 2Fo-Fc map (orange mesh) for the unliganded LrsL zinc ions and water molecules. The map is contoured at 2σ. The LrsL zinc-binding site is superimposed on the crystal structure of AidC:C6L complex (PDB 4zo3), showing electron density close to the C6L O1 atom in the AidC-C6L complex structure (C6L is shown as a stick model with carbon atoms in black). Residue coloring corresponds to the right panel in (A).
The crystal structure of LrsL exhibits a functional catalytic site where the conserved H151XH153XD155H156 ~ H236 ~ D257 ~ H304 (superscript show the position of amino acid in full-length protein) motif residues and one water molecule are coordinating two zinc ions as the metal cofactor, akin to AidC and other previously investigated lactonases (Dong et al., 2000; Figure 2A; Supplementary Figure S2). We crystallized LrsL without AHL but noticed an unoccupied electron density above the zinc molecules. This density, which we tentatively modeled as a water molecule, is in hydrogen-bonding distance to H260, and located in the same position as the AHL oxygen O1 of the AidC/N-hexanoyl-L-homoserine (C6L) complex (PDB id 4zo3; Figure 2C).
The crystal structure of LrsL was highly similar to the model predicted by AlphaFold2 (Jumper et al., 2021; r.m.s.d is 0.501 Å). Most residues, including those important for zinc binding, had been correctly placed by AlphaFold2 (Supplementary Figure S3). Minor differences were found in the side chain orientations of the phenylalanines F197, F200, and F275, and in the orientation of the helix carrying these residues. These regions have an elevated B-factor in the crystal structure, and hence the differences with the AlphaFold2 model may reflect this flexibility which could be related to catalysis/substrate binding.
LrsL robustly degrades C6-AHLs after heat treatment
Next, we investigated the ability of recombinant LrsL to degrade AHL molecules (Figure 3A). The incubation of affinity chromatography–purified LrsL with 10 μM C6-AHLs completely degraded these QS compounds, as determined using a C. violaceum CV026 biosensor assay on agar plates (Figure 3B). BSA was used as a control and did not degrade AHLs. The minimum active concentration of LrsL that could fully degrade 25 μM C6-AHLs in the biosensor assay was 0.23 μM (Figure 3A). This concentration is dependent on various factors, most importantly enzyme purity. The assay was performed using relatively low-purity LrsL (Figure 3A, second lane); hence, the obtained concentration should be considered an upper bound. Surprisingly, LrsL degraded C6-AHLs even after being heated in a dry bath at 110°C for 30 min, followed by cooling to room temperature and catalytic assaying. Highly thermostable lactonases, some showing a melting temperature above 100°C, have previously been reported for the PLL and MBL superfamilies, including the marine-derived MBL lactonase Aii20J (Merone et al., 2005; Hawwa et al., 2009; Cao et al., 2012; Hiblot et al., 2015; Mayer et al., 2015; Morohoshi et al., 2015; Bergonzi et al., 2018). However, Labrenzia was isolated from a mesophilic environment. Indeed, our thermal stability assay revealed that LrsL has a melting temperature Tm of only 58.3°C (Figure 3B). Circular dichroism spectrometry confirmed that LrsL was completely unfolded at 110°C but recovered its natural fold almost completely after cooling overnight. Hence, the capability of LrsL for maintaining its catalytic activity after heat treatment is explained by its robust refolding after heat denaturation (Figure 3C).
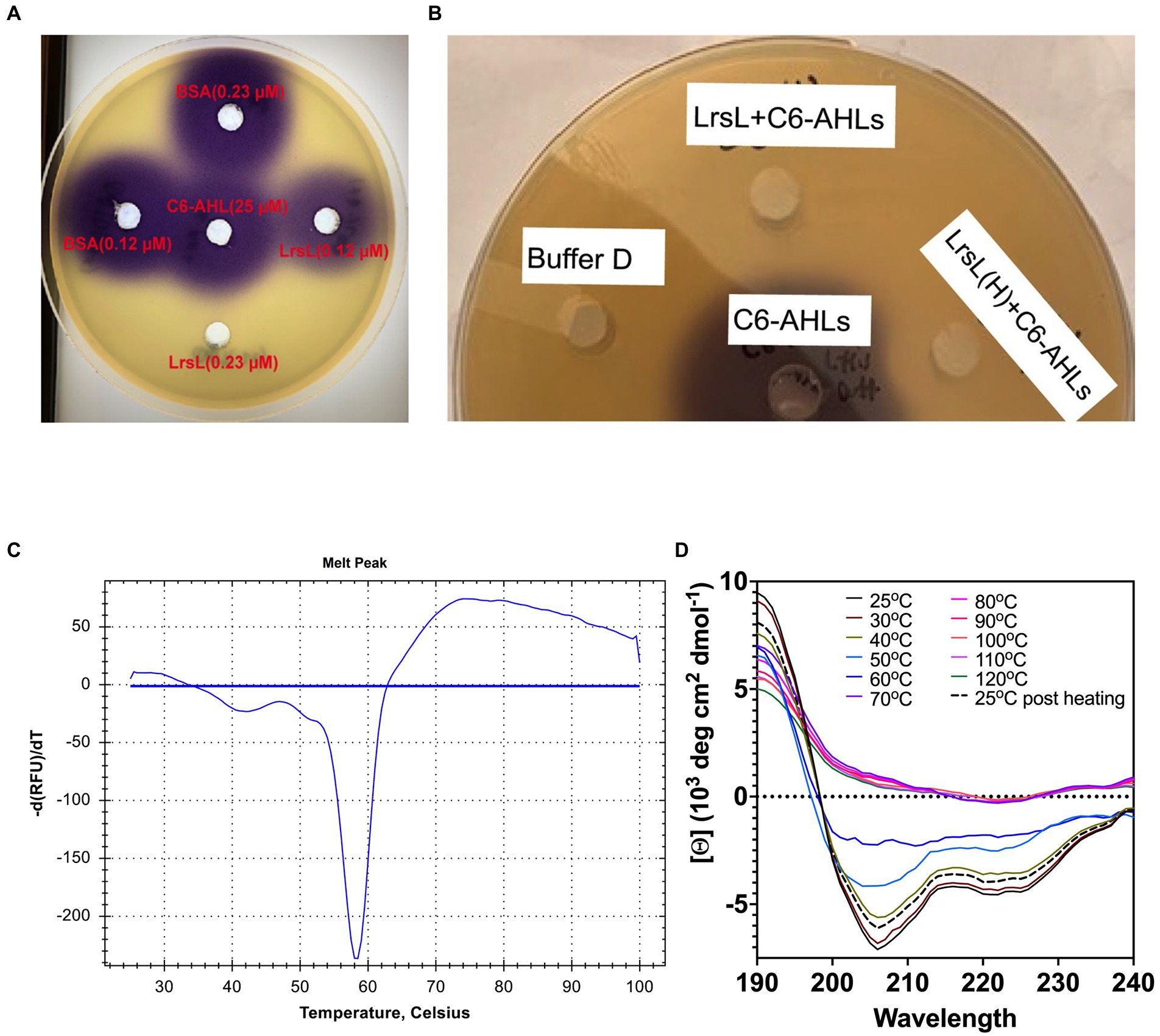
Figure 3. LrsL can degrade AHLs. (A) Two different concentrations (0.12 μM and 0.23 μM) of LrsL were incubated with 25 μM of C6-AHLs. The lower concentration (0.12 μM) of LrsL was not able to completely degrade AHLs; however, the higher concentration (0.23 μM) completely degraded AHLs. (B) LrsL degradation of AHL molecules. Affinity purified LrsL were incubated with 10 μM of C6-AHLs for 24 h, and the mixture was poured in wells of agar plate overlaid with Chromobacterium violaceum CV026. C6-AHLs in buffer D without LrsL was used as a positive control (middle well) and buffer D was used as a negative control. LrsL + C6AHLs indicates C6-AHLs treated with LrsL, while LrsL(H) + C6AHLs indicates C6-AHLs treated with heat-treated (110°C for 30 min) LrsL. The purple color development in the middle well (C6-AHLs) indicates intact AHLs, while no color indicates degradation or the absence of AHLs. (C) Differential Scanning Fluorimetry (DSF) thermal stability assay showing first derivative of melting curve. (D) Overlaid Circular Dichroism (CD) spectra of LrsL measured at temperatures from 25°C to 120°C (labeled appropriately by solid lines). The sample was cooled overnight and re-measured at 25°C (shown by a black dashed line).
Kinetic parameters of LrsL outperform those of other lactonases
Previous studies have demonstrated that AHL lactonases exhibit Km values in the range of 7.9 μM (Gcl) to 5.6 mM (AiiA) and catalytic turnover rates (Kcat) in the range of 8.9 s−1 (Gcl) to 91 s−1 (AiiA) measured against C6-AHLs (Momb et al., 2008; Bergonzi et al., 2016). Unusually low Michaelis constant Km values in the micromolar range were reported for AidC, suggesting it has a higher substrate affinity than other lactonases (Wang et al., 2012; Mascarenhas et al., 2015). Indeed, AidC ranks as one of the most efficient catalysts in terms of Kcat/Km values (Mascarenhas et al., 2015). The phylogenetic proximity to AidC suggested that LrsL is also an efficient catalyst for AHL degradation. However, there are also two important structural differences between AidC and LrsL, as revealed by the crystallographic LrsL structure, which may affect the catalytic efficiency and substrate selectivity. Firstly, the atypical α5 and α6 loop extension of AidC is in a position where it interferes with AHL chains beyond carbon C5, explaining the marked influence of AHL chain lengths on catalytic activity described for AidC (Mascarenhas et al., 2015). This AidC-specific loop extension is absent in LrsL. Therefore, we expected the catalytic activity of LrsL to be relatively independent of chain lengths. Secondly, LrsL shows a unique presence of three phenylalanines (F197, F200, and F275) in a position where they can coordinate the AHL tail carbons C1–C5 particularly well, suggesting a strong interaction with the substrate (Figures 1A, 4A; Supplementary Figure S2).
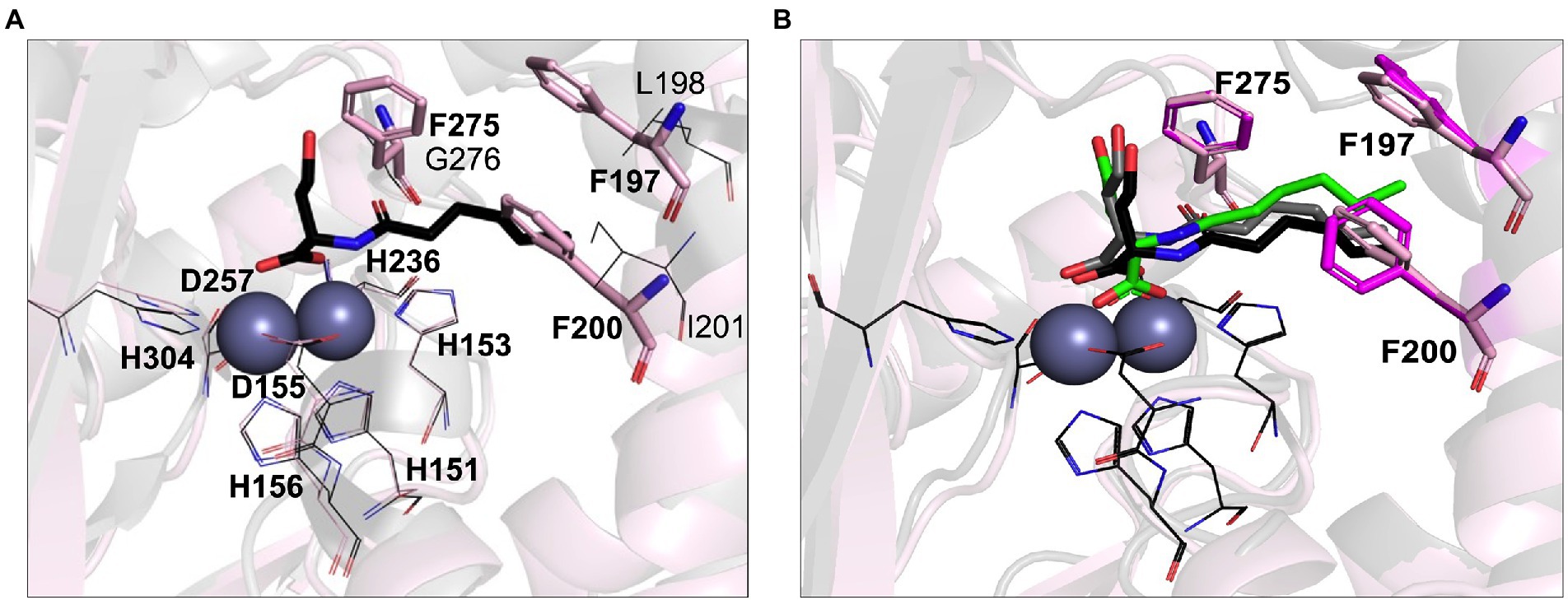
Figure 4. Computational AHL binding analysis. (A) The binding pocket of LrsL (light pink cartoon) is superimposed on AidC (PDB 4zo3; gray). The unique phenylalanine residues in the active site of LrsL are shown as light pink stick models. The corresponding AidC residues (leucine, isoleucine and glycine) are shown as dark gray lines. Conserved zinc coordination residues for LrsL (light pink lines) and AidC (black lines) are shown. Zinc ions are shown as blue-gray spheres. (B) Computational docking poses of C6L on AidC and LrsL. The crystallographic structure of the AidC:C6L complex is shown as a black cartoon, with C6L shown as black stick model. The redocked pose of C6L on AidC is represented as gray stick model and the docked pose of C6L on LrsL is shown as green stick model. The phenylalanine residues from the apo-crystal structure of LrsL are shown in light pink sticks, and from the C6L docked model of LrsL as magenta sticks. Conserved zinc coordination residue coloring and labeling corresponds to (A). Zinc ions are shown are blue-gray spheres.
To computationally explore the effect of the LrsL structural framework on substrate binding, we carried out in silico docking analyses. As a control and comparison, we used the crystallographic structure of AidC that had been determined with C6L (N-hexanoyl-L-homoserine) bound. Our docking procedure positioned C6L in the correct way onto AidC, validating our in silico approach (Figure 4B). C6L was computationally docked into the LrsL binding pocket in the same position, with the three phenylalanines engaging hydrophobic interactions with the carbon tail (Figure 4B). The calculated energy gain for C6L binding to LrsL was −9.1 kcal/mol, which was significantly larger than the −7.4 kcal/mol calculated for the (redocked) C6L binding to AidC. This analysis suggested that LrsL has an even higher affinity for the AHLs than AidC, and hence an even lower Km.
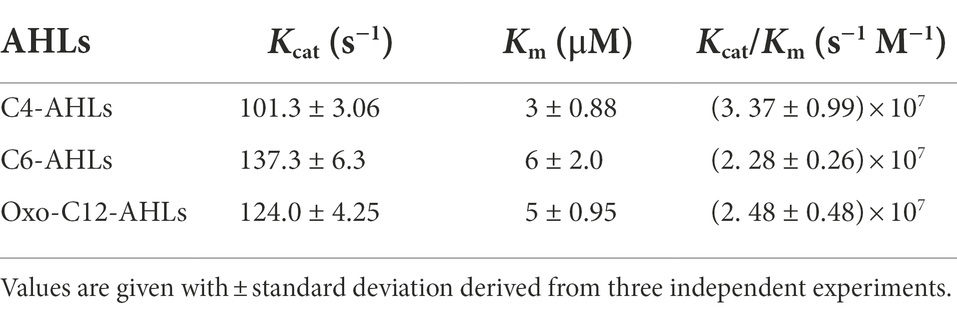
To experimentally assess the structure-based prediction that LrsL is an extremely efficient catalyst for AHLs with a range of tail lengths, we established the catalytic parameters of LrsL for three AHLs: C4-AHLs, C6-AHLs, and oxo-C12-AHLs (Table 3; Supplementary Figure S4). The C4-AHLs and oxo-C12-AHLs are used by the opportunistic pathogen P. aeruginosa to regulate virulence and biofilm formation. Indeed, LrsL exhibited a low Km of only 6 μM for C6-AHLs (Table 3). This Km value was markedly lower compared with other well-studied lactonases for C6-AHLs, such as AiiA and AiiB (Km = 1.6 mM and 5.6 mM, respectively; Liu et al., 2007; Momb et al., 2008), and with marine-derived MomL (Km = 790 μM; Tang et al., 2015). Further, LrsL also exhibited lower Km values than AidC (Km = 61 μM; Mascarenhas et al., 2015). Therefore, the substrate affinity of LrsL for C6-AHLs is higher than the substrate affinities of other previously reported AHL lactonases. As predicted, LrsL catalyzed C4-AHLs and oxo-C12-AHLs with Km values very similar to C6-AHLs (Table 3). The Km values are a limiting factor for lactonases, as the concentration of AHLs in natural and environmental settings usually ranges from pico to nanomoles per liter (Tan et al., 2014; Dal Bello et al., 2021). In biotech applications, lactonases with the lowest Km values are desirable, because they efficiently bind and degrade AHLs at lower concentrations, limiting the ability of bacteria to communicate.
Accordingly, the combination of a faster hydrolysis rate (Kcat) and higher substrate affinities (Km) in LrsL resulted in a catalytic efficiency (kcat/Km = 3 × 107 M−1) for LrsL that was substantially higher than for lactonases previously considered the most efficient catalysts (Table 3). For example, Gcl and AidC lactonases have catalytic efficiencies for C6-AHLs of 1.12 × 106 M−1 s−1 and 9.7 × 105 M−1 s−1, respectively (Mascarenhas et al., 2015; Bergonzi et al., 2016). Hence, based on the catalytic efficiency (kcat/Km), LrsL is currently the most efficient QQ enzyme available.
LrsL effectively inhibits biofilm formation
Next, we investigated the functional effects of the high in vitro catalytic rates of LrsL. We assessed the ability of LrsL to inhibit biofilm formation by P. aeruginosa PAO1, an important opportunistic pathogen commonly responsible for nosocomial infections. QS plays a vital role in biofilm formation by P. aeruginosa (Davies et al., 1998; Schuster et al., 2013), and an interference with its production of AHLs leads to the inhibition or formation of defective biofilms (Davies et al., 1998; Shih and Huang, 2002). Recently, it was confirmed that QQ enzymes reduce the biomass and density of biofilms formed by P. aeruginosa (Malesevic et al., 2020). We found that the application of 1.14 μM LrsL reduced the biofilm formation by more than 3-fold compared to untreated cultures and to cultures treated with BSA (Figure 5A). Similar to previous reports, we found no negative effect of LrsL on the growth of bacteria (Figure 5B; Bergonzi et al., 2018, 2019), further supporting that the use of QQ enzymes may not promote the development of resistance in bacteria.
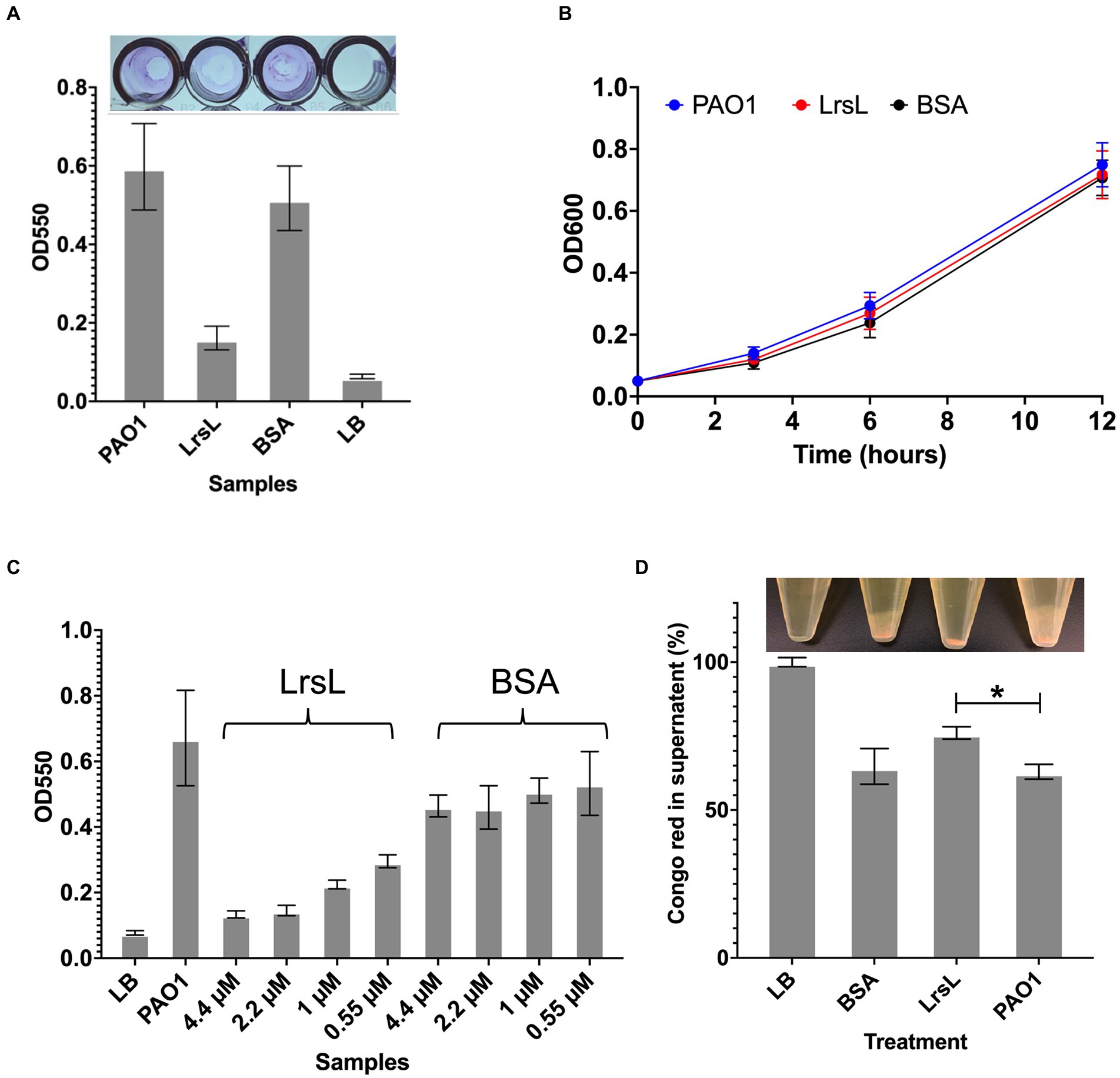
Figure 5. LrsL reduces biofilm formation. (A) The upper panel shows the photo of wells with biofilm formed by Pseudomonas aeruginosa PAO1 and various treatments. The lower panel shows the corresponding quantification of biofilms. The y-axis shows absorbance at 550 nm, and the x-axis shows biofilm formed by PAO1(with buffer D) and PAO1 with LrsL, and BSA. Unpaired student’s t-test was applied to measure the significance; value of p < 0.001 are indicated by asterisks. (B) The growth curves of P. aeruginosa (PAO1) and PAO1 treated with LrsL and BSA shows no significant difference. (C) LrsL reduces biofilm formation in a dose-dependent manner. Increasing the dose of LrsL increases the suppression of biofilm formation by P. aeruginosa. The y-axis shows absorbance at 550 nm, and the x-axis shows biofilm formed by PAO1 (with buffer D) and PAO1 treated with various concentrations of LrsL and BSA. The LrsL concentration was increased from 0.55 μM to 4.43 μM. As a control, PAO1 was also incubated with similar concentrations of BSA. For all the experiments, LB was used as a negative control. Error bars indicate the standard deviation of three biological replicates (eight wells each). (D) LrsL interferes with Pel production by P. aeruginosa. The upper panel show photographs of blank (LB) and P. aeruginosa cultures incubated with BSA, LrsL, and buffer D (PAO1). The lower panel shows the quantification of Pel using the Congo Red (CR) binding assay. For LB, all the CR remained in the supernatant, represented as 100%. In the presence of LrsL, Pel production was compromised, and more CR remained in supernatant compared to BSA and PAO1. The difference between CR in the supernatant of LrsL-treated and untreated PAO1 was significantly higher as measured by the student’s t-test, shown by an asterisk (value of p = 0.005). Error bars represent the standard deviation of three biological replicates.
The inhibitory effect of LrsL was dose-dependent. The application of 0.55 μM LrsL reduced the biofilm by 2.2-fold. Increasing the concentration of LrsL from 0.55 μM to 4.4 μM further reduced the biofilm by another 2.2-fold (Figure 5C). However, no dose-dependent response was observed for BSA, used as control. The treatment of an existing P. aeruginosa biofilm with LrsL for 6 h resulted in the decrease in the total biofilm thickness at the time of measurement by more than 50% compared with untreated or BSA-treated controls, without affecting bacterial growth (Supplementary Figure S5). It remains to be determined if the apparent biofilm decrease is solely the result of LrsL inhibiting the growth and/or renewal of the biofilm over the measurement period, or if LrsL can also disrupt preformed biofilms.
LrsL interferes with exopolysaccharide production
Normal P. aeruginosa biofilm production requires the exopolysaccharides Pel and Psl (Colvin et al., 2012). Therefore, we next asked whether LrsL interferes with biofilm production by causing changes in the production of Pel and/or Psl. Congo Red (CR) is a dye known to bind to Pel and Psl (Friedman and Kolter, 2004; Ma et al., 2006). When Pel and/or Psl are produced, they bind and precipitate CR, leaving a lower amount of CR in the supernatant. Consequently, bacterial strains deficient in Pel/Psl production reduce their binding to CR. (Friedman and Kolter, 2004; Ma et al., 2006). We, therefore, monitored supernatant CR to probe the effect of LrsL on Pel/Psl production.
Following P. aeruginosa PAO1 treatment with purified LrsL, we observed that most of the CR (76% ± 2.1) remained in the supernatant, suggesting suppressed production of Pel or Psl. In contrast, PAO1 and PAO1 treated with BSA exhibited a significantly lower amount of CR in the supernatant (63% ± 2.5 and 64% ± 6.03, respectively), inferring Pel production (Figure 5D). The effect of LrsL on Pel/Psl production was visible after the cultures were centrifuged. The LrsL-treated PAO1 lacked cloudy substances (likely comprising Pel/Psl and other extracellular polymeric substances) over the cell pellet, in contrast to PAO1 alone and PAO1 treated with BSA (Figure 5D, top). These results indicate that the application of LrsL disrupts the production of polysaccharides. Pel is upregulated by QS, and the addition of 3-oxo-C12-AHLs to QS mutants of P. aeruginosa restored Pel production (Sakuragi and Kolter, 2007). However, also Psl has been linked to QS (Attila et al., 2008). Hence, the relative contribution of each to biofilm inhibition by LrsL, as well as the molecular mechanisms linking QQ to decreased exopolysaccharide production remain to be investigated.
Conclusion
Biofilm formation or biofouling is a major problem in various fields because biofilms deteriorate materials and cause infections. Current antifouling and biofilm treatment procedures are toxic to the environment. The QQ enzymes may be a sustainable and eco-friendly class of agents that can be applied to counter biofilms and treat infections caused by pathogenic bacteria.
In this study, we characterized a novel QQ enzyme, LrsL, from the Red Sea sediment bacterium Labrenzia sp. VG12. Evolutionary analysis showed that LrsL belongs to the MBL superfamily whose members, such as GcL, MomL, and AiiA display broad substrate specificity (Wang et al., 2004; Mascarenhas et al., 2015; Tang et al., 2015; Bergonzi et al., 2019). Experimental and in silico analysis of LrsL indicated that LrsL can catalyze AHLs with varying tail lengths, and has evolved a unique hydrophobic mechanism to capture its substrates. The flexibility of the phenylalanine side chains and the absence of an AidC-like closure of the substrate binding pocket suggest that LrsL can also catalyze additional AHLs. We confirmed experimentally that LrsL is a broad spectrum QQ enzyme that degrades C4-AHLs, C6-AHLs, and 3-oxo-C12-AHLs with a catalytic efficiency of ~107 M−1 s−1. To date, this catalytic efficiency is the highest reported for any QQ lactonase. Furthermore, LrsL exhibited a very high affinity for different AHLs, making it an enzyme of choice in environments with low AHL concentrations. As an enzyme secreted in an ocean environment, this characteristic may be required to capture highly diluted substrates.
We found that purified LrsL suppresses the biofilm formation by P. aeruginosa, a ubiquitous bacterium involved in biofilm and nosocomial infections. Therefore, LrsL may help to prevent or limit biofilm formation on susceptible surfaces, such as medical devices and those submerged in water. Although LrsL will lose its structure and function at temperatures above 50°C, its capability to refold and regain function even after prolonged heat treatment at 120°C can facilitate its clinical and industrial applications.
Our experiments open the possibility that LrsL also breaks down existing P. aeruginosa biofilms. If confirmed through follow-up studies, this function would allow using LrsL and possibly other QQs to increase susceptibility of biofilm-forming bacteria to antimicrobials (Kiran et al., 2011). Thus, QQ enzymes would decrease the number of antimicrobials needed, which could limit the spread of antimicrobial resistance.
The LrsL-based disruption of exopolysaccharide production by P. aeruginosa may also have important implications for medical applications. Although the role of Pel and Psl in P. aeruginosa biofilm formation is strain-specific, both play a vital role in adherence to the surface and maintaining cell–cell interaction within a biofilm (Vasseur et al., 2005; Colvin et al., 2011, 2012). Furthermore, Pel protects biofilm bacteria against certain aminoglycoside antibiotics (Colvin et al., 2011). Therefore, interference with Pel production would likely compromise the ability of P. aeruginosa to form biofilms and render existing biofilms more susceptible to antimicrobial treatment (Kiran et al., 2011). This outcome is especially important, as P. aeruginosa is inherently resistant to antimicrobials, and biofilm formation increases the amount of antimicrobial that would be required. Therefore, even if QQ enzymes cannot completely remove biofilms, they may lower the amount of antimicrobial or cleaning chemicals required to clear biofilms.
Collectively, our work supports that QQ enzymes are promising agents to control biofouling and antimicrobial resistance in the clinic and in various industries.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material. Atomic coordinates for LrsL have been deposited in the Protein Data Bank (PDB) with accession code 7Y7U.
Author contributions
ZR, AM, RG, TI, and SA designed the experiments. ZR, AM, and AA performed the experiments. AM and SA carried out X-ray crystallography and computational analyses. ZR, AM, RG, and SA wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the King Abdullah University of Science and Technology (KAUST) through the baseline fund and Award No. URF/1/1976-36-01 from the Office of Sponsored Research.
Acknowledgments
We are thankful to Ana Otero from the University of Santiago de Compostela, Spain, for sharing the strain C. violaceum CV026. We would also like to thank A. S. Sandholu for his help with this work. We are grateful to P. Legrand for assistance and to the SOLEIL staff for smoothly running the facility.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.977673/full#supplementary-material
Footnotes
References
Attila, C., Ueda, A., and Wood, T. K. (2008). PA2663 (PpyR) increases biofilm formation in Pseudomonas aeruginosa PAO1 through the psl operon and stimulates virulence and quorum-sensing phenotypes. Appl. Microbiol. Biotechnol. 78, 293–307. doi: 10.1007/s00253-007-1308-y
Azimi, S., Klementiev, A. D., Whiteley, M., and Diggle, S. P. (2020). Bacterial quorum sensing during infection. Annu. Rev. Microbiol. 74, 201–219. doi: 10.1146/annurev-micro-032020-093845
Bergonzi, C., Schwab, M., and Elias, M. (2016). The quorum-quenching lactonase from Geobacillus caldoxylosilyticus: purification, characterization, crystallization and crystallographic analysis. Acta Crystallogr. F. Struct. Biol. Commun. 72, 681–686. doi: 10.1107/S2053230X16011821
Bergonzi, C., Schwab, M., Naik, T., Daude, D., Chabriere, E., and Elias, M. (2018). Structural and biochemical characterization of AaL, a quorum quenching Lactonase with unusual kinetic properties. Sci. Rep. 8:11262. doi: 10.1038/s41598-018-28988-5
Bergonzi, C., Schwab, M., Naik, T., and Elias, M. (2019). The structural determinants accounting for the broad substrate specificity of the quorum quenching Lactonase GcL. Chembiochem 20, 1848–1855. doi: 10.1002/cbic.201900024
Billot, R., Plener, L., Jacquet, P., Elias, M., Chabriere, E., and Daude, D. (2020). Engineering acyl-Homoserine lactone-interfering enzymes towards bacterial control. J. Biol. Chem. 295, 12993–13007. doi: 10.1074/jbc.REV120.013531
Cao, Y., He, S., Zhou, Z., Zhang, M., Mao, W., Zhang, H., et al. (2012). Orally administered thermostable N-acyl homoserine lactonase from bacillus sp. strain AI96 attenuates Aeromonas hydrophila infection in zebrafish. Appl. Environ. Microbiol. 78, 1899–1908. doi: 10.1128/AEM.06139-11
Cegelski, L., Marshall, G. R., Eldridge, G. R., and Hultgren, S. J. (2008). The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 6, 17–27. doi: 10.1038/nrmicro1818
Chan, K. G. (2013). Expression of Klebsiella sp. lactonase ahlK gene is growth-phase, cell-population density and N-acylhomoserine lactone independent. Front. Life Sci. 7, 132–139. doi: 10.1080/21553769.2013.833141
Chu, W. H., Liu, Y. W., Jiang, Y., Zhu, W., and Zhuang, X. Y. (2013). Production of N-acyl Homoserine lactones and virulence factors of waterborne Aeromonas hydrophila. Indian J. Microbiol. 53, 264–268. doi: 10.1007/s12088-013-0381-4
Coffey, B. M., and Anderson, G. G. (2014). Biofilm formation in the 96-well microtiter plate. Methods Mol. Biol. 1149, 631–641. doi: 10.1007/978-1-4939-0473-0_48
Colvin, K. M., Gordon, V. D., Murakami, K., Borlee, B. R., Wozniak, D. J., Wong, G. C. L., et al. (2011). The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7:e1001264. doi: 10.1371/journal.ppat.1001264
Colvin, K. M., Irie, Y., Tart, C. S., Urbano, R., Whitney, J. C., Ryder, C., et al. (2012). The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14, 1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x
Dal Bello, F., Zorzi, M., Aigotti, R., Medica, D., Fanelli, V., Cantaluppi, V., et al. (2021). Targeted and untargeted quantification of quorum sensing signalling molecules in bacterial cultures and biological samples via HPLC-TQ MS techniques. Anal. Bioanal. Chem. 413, 853–864. doi: 10.1007/s00216-020-03040-6
Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H., Costerton, J. W., and Greenberg, E. P. (1998). The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298. doi: 10.1126/science.280.5361.295
Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., et al. (2007). MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383. doi: 10.1093/nar/gkm216
Defoirdt, T., Boon, N., and Bossier, P. (2010). Can bacteria evolve resistance to quorum sensing disruption? PLoS Pathog. 6:e1000989. doi: 10.1371/journal.ppat.1000989
Dong, Y. H., Gusti, A. R., Zhang, Q., Xu, J. L., and Zhang, L. H. (2002). Identification of quorum-quenching N-acyl homoserine lactonases from bacillus species. Appl. Environ. Microbiol. 68, 1754–1759. doi: 10.1128/Aem.68.4.1754-1759.2002
Dong, Y. H., Xu, J. L., Li, X. Z., and Zhang, L. H. (2000). AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. U. S. A. 97, 3526–3531. doi: 10.1073/pnas.060023897
Eberl, L., Winson, M. K., Sternberg, C., Stewart, G. S., Christiansen, G., Chhabra, S. R., et al. (1996). Involvement of N-acyl-L-hormoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol. Microbiol. 20, 127–136. doi: 10.1111/j.1365-2958.1996.tb02495.x
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Emsley, P., Lohkamp, B., Scott, W. G., and Cowtan, K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501. doi: 10.1107/S0907444910007493
Fetzner, S. (2015). Quorum quenching enzymes. J. Biotechnol. 201, 2–14. doi: 10.1016/j.jbiotec.2014.09.001
Friedman, L., and Kolter, R. (2004). Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690. doi: 10.1046/j.1365-2958.2003.03877.x
Garcia-Contreras, R., Maeda, T., and Wood, T. K. (2013). Resistance to quorum-quenching compounds. Appl. Environ. Microbiol. 79, 6840–6846. doi: 10.1128/Aem.02378-13
Goodsell, D. S., Morris, G. M., and Olson, A. J. (1996). Automated docking of flexible ligands: applications of AutoDock. J. Mol. Recognit. 9, 1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6
Guo, K. Y., Wustoni, S., Koklu, A., Diaz-Galicia, E., Moser, M., Hama, A., et al. (2021). Rapid single-molecule detection of COVID-19 and MERS antigens via nanobody-functionalized organic electrochemical transistors. Nat. Biomed. Eng. 5, 666–677. doi: 10.1038/s41551-021-00734-9
Hawwa, R., Aikens, J., Turner, R. J., Santarsiero, B. D., and Mesecar, A. D. (2009). Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus. Arch. Biochem. Biophys. 488, 109–120. doi: 10.1016/j.abb.2009.06.005
Hiblot, J., Bzdrenga, J., Champion, C., Chabriere, E., and Elias, M. (2015). Crystal structure of VmoLac, a tentative quorum quenching lactonase from the extremophilic crenarchaeon Vulcanisaeta moutnovskia. Sci. Rep. 5, 8372. doi: 10.1038/srep08372
Holloway, B. W., Matsumoto, H., and Phibbs, P. V. Jr. (1986). The chromosome map of Pseudomonas aeruginosa PAO. Acta Microbiol. Pol. 35, 161–164.
Hungler, A., Momin, A., Diederichs, K., and Arold, S. T. (2016). ContaMiner and ContaBase: a webserver and database for early identification of unwantedly crystallized protein contaminants. J. Appl. Crystallogr. 49, 2252–2258. doi: 10.1107/S1600576716014965
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kabsch, W. (2010). XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132. doi: 10.1107/S0907444909047337
Kiran, S., Sharma, P., Harjai, K., and Capalash, N. (2011). Enzymatic quorum quenching increases antibiotic susceptibility of multidrug resistant Pseudomonas aeruginosa. Iran J Microbiol 3, 1–12.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Langer, G., Cohen, S. X., Lamzin, V. S., and Perrakis, A. (2008). Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179. doi: 10.1038/nprot.2008.91
Lee, K., Yu, H. R., Zhang, X. L., and Choo, K. H. (2018). Quorum sensing and quenching in membrane bioreactors: opportunities and challenges for biofouling control. Bioresour. Technol. 270, 656–668. doi: 10.1016/j.biortech.2018.09.019
Li de la Sierra-Gallay, I., Pellegrini, O., and Condon, C. (2005). Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature 433, 657–661. doi: 10.1038/nature03284
Liu, D. L., Lepore, B. W., Petsko, G. A., Thomas, P. W., Stone, E. M., Fast, W., et al. (2005). Three-dimensional structure of the quorum-quenching N-acyl homoserine lactone hydrolase from bacillus thuringiensis. Proc. Natl. Acad. Sci. U. S. A. 102, 11882–11887. doi: 10.1073/pnas.0505255102
Liu, D. L., Thomas, P. W., Momb, J., Hoang, Q. Y. Q., Petsko, G. A., Ringe, D., et al. (2007). Structure and specificity of a quorum-quenching lactonase (AiiB) from agrobacterium tumefaciens. Biochemistry 46, 11789–11799. doi: 10.1021/bi7012849
Long, F., Vagin, A. A., Young, P., and Murshudov, G. N. (2008). BALBES: a molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 64, 125–132. doi: 10.1107/S0907444907050172
Ma, L., Jackson, K. D., Landry, R. M., Parsek, M. R., and Wozniak, D. J. (2006). Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188, 8213–8221. doi: 10.1128/JB.01202-06
Malesevic, M., Stanisavljevic, N., Novovic, K., Polovic, N., Vasiljevic, Z., Kojic, M., et al. (2020). Burkholderia cepacia YtnP and Y2-aiiA lactonases inhibit virulence of Pseudomonas aeruginosa via quorum quenching activity. Microb. Pathog. 149:104561. doi: 10.1016/j.micpath.2020.104561
Mascarenhas, R., Thomas, P. W., Wu, C. X., Nocek, B. P., Hoang, Q. Q., Liu, D. L., et al. (2015). Structural and biochemical characterization of AidC, a quorum-quenching Lactonase with atypical selectivity. Biochemistry 54, 4342–4353. doi: 10.1021/acs.biochem.5b00499
Mayer, C., Muras, A., Romero, M., Lopez, M., Tomas, M., and Otero, A. (2018). Multiple quorum quenching enzymes are active in the nosocomial pathogen Acinetobacter baumannii ATCC17978. Front. Cell. Infect. Microbiol. 8:310. doi: 10.3389/fcimb.2018.00310
Mayer, C., Romero, M., Muras, A., and Otero, A. (2015). Aii20J, a wide-spectrum thermostable N-acylhomoserine lactonase from the marine bacterium Tenacibaculum sp. 20J, can quench AHL-mediated acid resistance in Escherichia coli. Appl. Microbiol. Biotechnol. 99, 9523–9539. doi: 10.1007/s00253-015-6741-8
McClean, K. H., Winson, M. K., Fish, L., Taylor, A., Chhabra, S. R., Camara, M., et al. (1997). Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology (Reading) 143, 3703–3711. doi: 10.1099/00221287-143-12-3703
Mercuri, P. S., Bouillenne, F., Boschi, L., Lamotte-Brasseur, J., Amicosante, G., Devreese, B., et al. (2001). Biochemical characterization of the FEZ-1 metallo-beta-lactamase of legionella gormanii ATCC 33297T produced in Escherichia coli. Antimicrob. Agents Chemother. 45, 1254–1262. doi: 10.1128/AAC.45.4.1254-1262.2001
Merone, L., Mandrich, L., Rossi, M., and Manco, G. (2005). A thermostable phosphotriesterase from the archaeon Sulfolobus solfataricus: cloning, overexpression and properties. Extremophiles 9, 297–305. doi: 10.1007/s00792-005-0445-4
Momb, J., Wang, C. H., Liu, D. L., Thomas, P. W., Petsko, G. A., Guo, H., et al. (2008). Mechanism of the quorum-quenching lactonase (AiiA) from bacillus thuringiensis. 2. Substrate modeling and active site mutations. Biochemistry 47, 7715–7725. doi: 10.1021/bi8003704
Momin, A. A., Hameed, U. F. S., and Arold, S. T. (2019). Passenger sequences can promote interlaced dimers in a common variant of the maltose-binding protein. Sci. Rep. 9:20396. doi: 10.1038/s41598-019-56718-y
Morohoshi, T., Tominaga, Y., Someya, N., and Ikeda, T. (2012). Complete genome sequence and characterization of the N-acylhomoserine lactone-degrading gene of the potato leaf-associated Solibacillus silvestris. J. Biosci. Bioeng. 113, 20–25. doi: 10.1016/j.jbiosc.2011.09.006
Morohoshi, T., Tominaga, Y., Someya, N., and Ikeda, T. (2015). Characterization of a novel thermostable N-acylhomoserine lactonase from the thermophilic bacterium Thermaerobacter marianensis. J. Biosci. Bioeng. 120, 1–5. doi: 10.1016/j.jbiosc.2014.11.014
Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255. doi: 10.1107/S0907444996012255
Palzkill, T. (2013). Metallo-beta-lactamase structure and function. Ann. N. Y. Acad. Sci. 1277, 91–104. doi: 10.1111/j.1749-6632.2012.06796.x
Park, S. Y., Lee, S. J., Oh, T. K., Oh, J. W., Koo, B. T., Yum, D. Y., et al. (2003). AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology-Sgm 149, 1541–1550. doi: 10.1099/mic.0.26269-0
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Rehman, Z. U., Ali, M., Iftikhar, H., and Leiknes, T. (2019). Genome-resolved metagenomic analysis reveals roles of microbial community members in full-scale seawater reverse osmosis plant. Water Res. 149, 263–271. doi: 10.1016/j.watres.2018.11.012
Rehman, Z. U., Fortunato, L., Cheng, T., and Leiknes, T. (2020). Metagenomic analysis of sludge and early-stage biofilm communities of a submerged membrane bioreactor. Sci. Total Environ. 701, 134682. doi: 10.1016/j.scitotenv.2019.134682
Rehman, Z. U., and Leiknes, T. (2018). Quorum-quenching bacteria isolated From Red Sea sediments reduce biofilm formation by Pseudomonas aeruginosa. Front. Microbiol. 9:1354. doi: 10.3389/fmicb.2018.01354
Rehman, Z. U., Vrouwenvelder, J. S., and Saikaly, P. E. (2021). Physicochemical properties of extracellular polymeric substances produced by three bacterial isolates From biofouled reverse osmosis membranes. Front. Microbiol. 12:668761. doi: 10.3389/fmicb.2021.668761
Remy, B., Plener, L., Decloquement, P., Armstrong, N., Elias, M., Daude, D., et al. (2020). Lactonase specificity is key to quorum quenching in Pseudomonas aeruginosa. Front. Microbiol. 11:762. doi: 10.3389/fmicb.2020.00762
Renn, D., Shepard, L., Vancea, A., Karan, R., Arold, S. T., and Rueping, M. (2021). Novel enzymes from the red sea brine pools: current state and potential. Front. Microbiol. 12:732856. doi: 10.3389/fmicb.2021.732856
Rumbaugh, K. P., and Sauer, K. (2020). Biofilm dispersion. Nat. Rev. Microbiol. 18, 571–586. doi: 10.1038/s41579-020-0385-0
Sakuragi, Y., and Kolter, R. (2007). Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 189, 5383–5386. doi: 10.1128/Jb.00137-07
Salta, M., Wharton, J. A., Blache, Y., Stokes, K. R., and Briand, J. F. (2013). Marine biofilms on artificial surfaces: structure and dynamics. Environ. Microbiol. 15, 2879–2893. doi: 10.1111/1462-2920.12186
Schuster, M., Sexton, D. J., Diggle, S. P., and Greenberg, E. P. (2013). Acyl-Homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 67, 43–63. doi: 10.1146/annurev-micro-092412-155635
See-Too, W. S., Convey, P., Pearce, D. A., and Chan, K. G. (2018). Characterization of a novel N-acylhomoserine lactonase, AidP, from Antarctic Planococcus sp. Microb. Cell Factories 17:179. doi: 10.1186/s12934-018-1024-6
Shih, P. C., and Huang, C. T. (2002). Effects of quorum-sensing deficiency on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. J. Antimicrob. Chemother. 49, 309–314. doi: 10.1093/jac/49.2.309
Smadja, B., Latour, X., Faure, D., Chevalier, S., Dessaux, Y., and Orange, N. (2004). Involvement of N-acylhomoserine lactones throughout plant infection by Erwinia carotovora subsp atroseptica (Pectobacterium atrosepticum). Mol. Plant-Microbe Interact. 17, 1269–1278. doi: 10.1094/Mpmi.2004.17.11.1269
Smith, R. S., Harris, S. G., Phipps, R., and Iglewski, B. (2002). The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J. Bacteriol. 184, 1132–1139. doi: 10.1128/jb.184.4.1132-1139.2002
Spiers, A. J., Bohannon, J., Gehrig, S. M., and Rainey, P. B. (2003). Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50, 15–27. doi: 10.1046/j.1365-2958.2003.03670.x
Tan, C. H., Koh, K. S., Xie, C., Tay, M., Zhou, Y., Williams, R., et al. (2014). The role of quorum sensing signalling in EPS production and the assembly of a sludge community into aerobic granules. ISME J. 8, 1186–1197. doi: 10.1038/ismej.2013.240
Tang, K., Su, Y., Brackman, G., Cui, F., Zhang, Y., Shi, X., et al. (2015). MomL, a novel marine-derived N-acyl homoserine lactonase from Muricauda olearia. Appl. Environ. Microbiol. 81, 774–782. doi: 10.1128/AEM.02805-14
Ullah, J. H., Walsh, T. R., Taylor, I. A., Emery, D. C., Verma, C. S., Gamblin, S. J., et al. (1998). The crystal structure of the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia at 1.7 A resolution. J. Mol. Biol. 284, 125–136. doi: 10.1006/jmbi.1998.2148
Vasseur, P., Vallet-Gely, I., Soscia, C., Genin, S., and Filloux, A. (2005). The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 151, 985–997. doi: 10.1099/mic.0.27410-0
Vogeley, C., Sondermann, N. C., Woeste, S., Momin, A. A., Gilardino, V., Hartung, F., et al. (2022). Unraveling the differential impact of PAHs and dioxin-like compounds on AKR1C3 reveals the EGFR extracellular domain as a critical determinant of the AHR response. Environ. Int. 158, 106989. doi: 10.1016/j.envint.2021.106989
Wang, W. Z., Morohoshi, T., Someya, N., and Ikeda, T. (2012). AidC, a novel N-Acylhomoserine Lactonase from the potato root-associated Cytophaga-Flavobacteria-Bacteroides (CFB) group bacterium Chryseobacterium sp strain StRB126. Appl. Environ. Microbiol. 78, 7985–7992. doi: 10.1128/Aem.02188-12
Wang, L. H., Weng, L. X., Dong, Y. H., and Zhang, L. H. (2004). Specificity and enzyme kinetics of the quorum-quenching N-acyl homoserine lactone lactonase (AHL-lactonase). J. Biol. Chem. 279, 13645–13651. doi: 10.1074/jbc.M311194200
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Whiteley, M., Diggle, S. P., and Greenberg, E. P. (2017). Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320. doi: 10.1038/nature24624
Wiedemann, C., Bellstedt, P., and Gorlach, M. (2013). CAPITO--a web server-based analysis and plotting tool for circular dichroism data. Bioinformatics 29, 1750–1757. doi: 10.1093/bioinformatics/btt278
Keywords: quorum quenching enzymes, biofilm, Pseudomonas aeruginosa, acyl-homoserine lactones, virulence factors
Citation: Rehman ZU, Momin AA, Aldehaiman A, Irum T, Grünberg R and Arold ST (2022) The exceptionally efficient quorum quenching enzyme LrsL suppresses Pseudomonas aeruginosa biofilm production. Front. Microbiol. 13:977673. doi: 10.3389/fmicb.2022.977673
Edited by:
Yang-Chun Yong, Jiangsu University, ChinaReviewed by:
Celia Mayer, University of Santiago de Compostela, SpainKarishma Bisht, Princeton University, United States
Zhen Fang, Jiangsu University, China
Copyright © 2022 Rehman, Momin, Aldehaiman, Irum, Grünberg and Arold. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zahid Ur Rehman, emFoaWQudXJyZWhtYW5Aa2F1c3QuZWR1LnNh; Stefan T. Arold, c3RlZmFuLmFyb2xkQGthdXN0LmVkdS5zYQ==
†These authors have contributed equally to this work
 Zahid Ur Rehman
Zahid Ur Rehman Afaque A. Momin
Afaque A. Momin Abdullah Aldehaiman
Abdullah Aldehaiman Tayyaba Irum4
Tayyaba Irum4 Raik Grünberg
Raik Grünberg Stefan T. Arold
Stefan T. Arold