- 1State Key Laboratory of Biobased Material and Green Papermaking (LBMP), School of Bioengineering, Qilu University of Technology (Shandong Academy of Sciences), Jinan, China
- 2Shandong Provincial Key Laboratory of Biophysics, Institute of Biophysics, Dezhou University, Dezhou, China
- 3Shandong Provincial Key Laboratory of Applied Microbiology, Ecology Institute, Qilu University of Technology (Shandong Academy of Sciences), Jinan, China
Phenazine-1-carboxylic acid (PCA) is a biologically active substance with the ability to prevent and control crop diseases. It was certified as a pesticide by the Ministry of Agriculture of China in 2011 and was named “Shenzimycin.” Lzh-T5 is a Pseudomonas chlororaphis strain found in the rhizosphere of tomatoes. This strain can produce only 230 mg/L of PCA. We used LDA-4, which produces the phenazine synthetic intermediate trans-2,3-dihydro-3-hydroxyanthranilic acid in high amounts, as the starting strain. By restoring phzF and knocking out phzO, we achieved PCA accumulation. Moreover, PCA production was enhanced after knocking out negative regulators, enhancing the shikimate pathway, and performing fed-batch fermentation, thus resulting in the production of 10,653 mg/L of PCA. It suggested that P. chlororaphis Lzh-T5 has the potential to become an efficiency cell factory of biologically active substances.
1. Introduction
Rice, one of the world’s major crops, occupies an important position in the global food supply chain (Molina et al., 2011; Zhao et al., 2022). Worldwide, rice is often affected by serious diseases and insect pests. Although chemical pesticides, such as tricyclazole and epoxiconazole, are often used to control these pests and diseases, they cause environmental and food pollution (Padovani et al., 2006; Yan et al., 2015; Garcia-Jaramillo et al., 2016; Meng et al., 2018; Medina et al., 2021; Sefiloglu et al., 2021; Li H. et al., 2022). Therefore, there are increasing restrictions on the use of chemical pesticides. This has resulted in biological pesticides becoming a good alternative (Sharma et al., 2020; Ahmad et al., 2021). In this context, phenazine-1-carboxylic acid (PCA), a type of biological pesticide, has been registered and certified as a Chinese pesticide and is named “Shenzimycin” owing to its good efficacy against diseases (Jin et al., 2015; Karmegham et al., 2020; Li et al., 2021; Sun et al., 2021).
Currently, PCA is primarily produced by the strain Pseudomonas aeruginosa, despite it being an opportunistic pathogen and a major harmful bacterium present in hospitals (Du et al., 2015; Walker and Moore, 2015; Zhou et al., 2016; Petitjean et al., 2021). Compared with P. aeruginosa, P. chlororaphis not only contains the phenazine-synthesizing gene cluster phzABCDEFG but it is also a safe strain and is more suitable as an engineered strain for the production of phenazine derivatives (Liu et al., 2016; Li et al., 2020).
Lzh-T5 is a P. chlororaphis strain normally found in the rhizosphere of tomatoes (Li et al., 2018). In addition to phzABCDEFG, it contains the phenazine modification gene phzO. PCA and 2-OH-PHZ can be detected in the fermentation product of the strain; therefore, it shows the potential to efficiently produce PCA (Li et al., 2018; Liu et al., 2021).
The shikimate pathway is a key metabolism pathway that is widely present in organisms, including P. chlororaphis (Liu et al., 2016; Li et al., 2020). A series of substances, such as coenzyme Q, aromatic amino acids, and phenazines, can be produced through the shikimate pathway (Talapatra and Talapatra, 2005; Liu et al., 2016). Trans-2,3-dihydro-3-hydroxyanthranilic acid (DHHA) is also an accumulation product of the shikimate pathway truncated in P. chlororaphis. Our previous study revealed that P. chlororaphis Lzh-T5 can produce DHHA (Liu et al., 2021).
In this study, we used a derivative strain of P. chlororaphis Lzh-T5, LDA-4, which possess high DHHA production capacity, as the starting strain and achieved PCA accumulation as a single product through genetic engineering. Gene expression and feed-bath strategies were used to increase the PCA production by the strain, resulting in an engineered strain that produced 10.65 g/L of PCA.
2. Materials and methods
2.1. Strains and bacterial culture
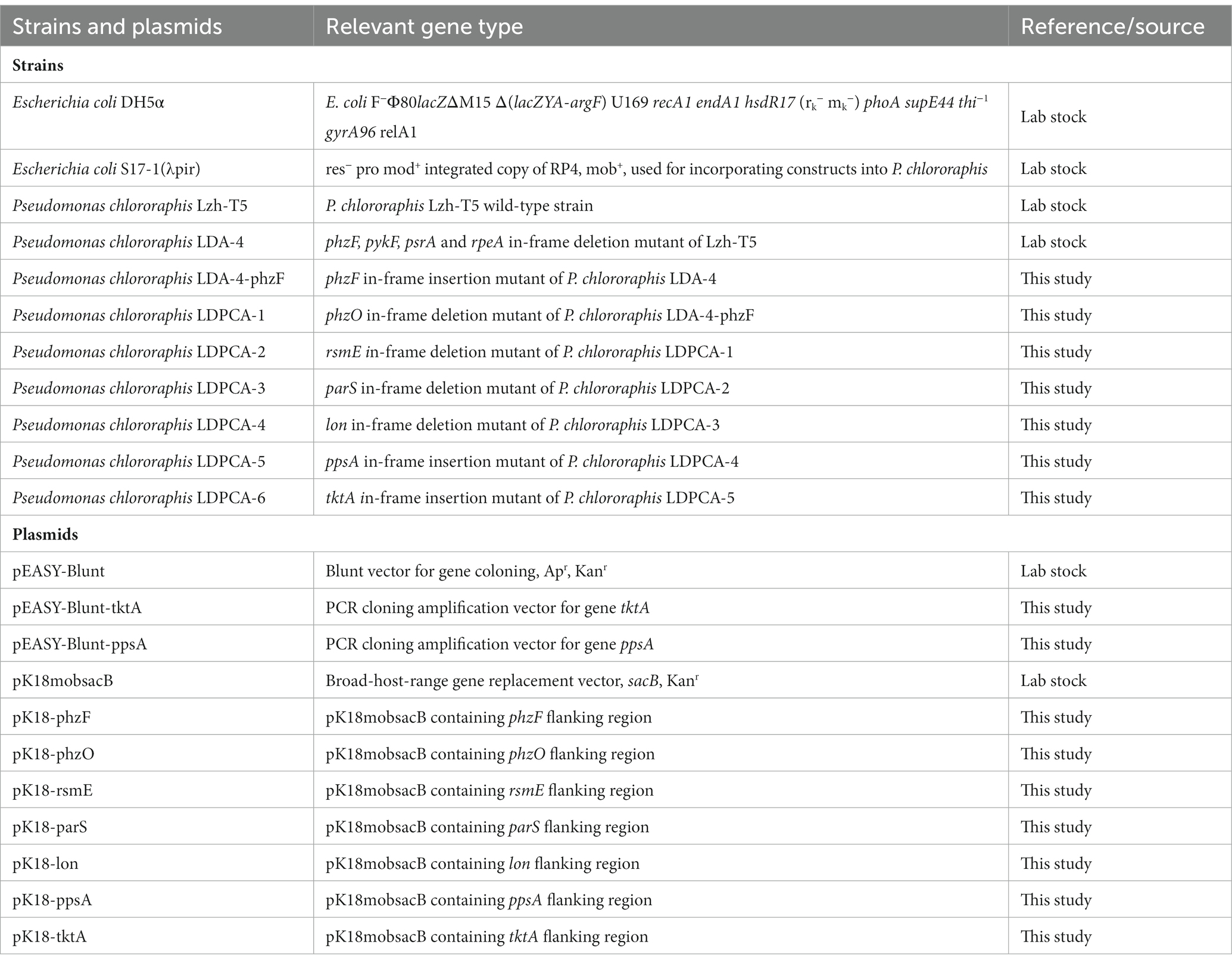
Different Escherichia coli and P. chlororaphis strains were used in this study (Table 1). E. coli is primarily used for gene knockout vector construction and biparental hybridization, whereas P. chlororaphis is primarily used for the construction of engineered strains producing phenazine derivatives. E. coli was cultured in LB medium at 37°C, and P. chlororaphis was cultured in King’s B medium at 28°C. The composition of these media has been described in our previous study (Li et al., 2020). Ampicillin and kanamycin sulfate were used as antibiotics at concentrations of 100 and 50 μg/ml, respectively.
2.2. Genetic engineering
The plasmids and primers used in this study are presented in Table 1 and Supplementary Table S1, respectively. phzF was restored into the genome of the LDA-4 strain through gene insertion. First, the primers for phzF insertion were designed according to the genome sequence of P. chlororaphis Lzh-T5 (Supplementary Table S1). Subsequently, phzF and its upstream and downstream fragment phzFUD was amplified through PCR using phzF-F1/phzF-R2 as primers and the Lzh-T5 genome as the template.
The restriction enzymes EcoRI and XbaI were used to digest the phzFUD fragment and the pK18mobSacB plasmid, respectively. After recovery through agarose gel electrophoresis, the recombinant plasmid pK18-phzFUD was obtained using T4 ligase. This plasmid was transformed into E. coli S17-1 (λ), resulting in the recombinant strains E. coli S17-1 (λ) and P. chlororaphis LDA-4; these strains were cultured respectively, and the pK18-phzFUD plasmid was introduced into pseudo-LDA-4 by biparental hybridization. The plasmid and the bacterial genome were double-exchanged on a King’s B medium plate containing 10% sucrose, and the phzF-restored strain was obtained through PCR.
Genes such as rsmE, parS, and lon have been knocked out from the genome of P. chlororaphis either individually or together, as described in our previous studies (Liu et al., 2016; Li et al., 2020).
2.3. Quantitative RT-PCR
Quantitative RT-PCR was performed to detect the transcriptional changes of related genes in different P. chlororaphis strains as described in our previous study (Liu et al., 2021). rpoD, a housekeeping gene of P. chlororaphis, was selected as the internal reference gene. The mRNA fold change was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
2.4. Fed-batch fermentation of strains
To maximize the efficacy of the engineered strains, we added the high-yielding strains LDPCA-6 to a 5-L fermenter to perform fed-batch fermentation with specific fermentation parameters according to our previous research (Li et al., 2020).
2.5. Separation, preparation, and detection of phenazine samples
PCA must be extracted from the fermentation broth and then detected and quantified by high-performance liquid chromatography (HPLC). We first extracted PCA from the fermentation broth using ethyl acetate and subjected it to reversed-phase C18 column (Agilent Technologies, 5 μm, 4.6*250 mm). Both 2-OH-PHZ and PCN are phenazine derivatives and exhibit similar material properties; the specific extraction and detection steps of these samples are described in our previous studies (Liu et al., 2016; Li et al., 2020).
3. Results
3.1. Restoration of phzF in LDA-4 resulted in the accumulation of phenazines
Lzh-T5 is a P. chlororaphis strain selected from the rhizosphere of tomato. It is known that its genome contains the key phzABCDEFG gene cluster for the synthesis of phenazine derivatives. The fermentation broth of this strain in KB would turn to orange (which is the color of 2-hydroxyphenazine), with the potential to produce phenazine derivatives. In our previous research, we blocked the biosynthesis of phenazine by knocking out phzF, which led to the accumulation of DHHA, an important chemical intermediate. We strengthened the synthetic pathway of DHHA via genetic engineering and obtained the engineered strain LDA-4. The yield of DHHA using LDA-4 reached 5.52 g/L in the shake flask (Liu et al., 2021). In the present study, we used LDA-4 to obtain high yield of PCA. DHHA accumulation occurred in LDA-4 owing to the knockout of phzF in the phenazine synthesis pathway (Figure 1). We restored the function of phzF in the genome of LDA-4 through genetic engineering to obtain the strain LDA-4-phzF. To confirm whether phzF could indeed play a role, we performed RT-PCR to verify its expression level in different strains. Our results revealed increased expression of phzF in LDA-4-phzF compared with that in LDA-4 (Figure 2A). At 28°C, the colony of the strain turned red (Figure 3). HPLC revealed three phenazine derivative peaks in LDA-4-phzF (Figure 4). Compared with the wild-type strain Lzh-T5, the yield of phenazines was increased to 3560.4 mg/L in LDA-4-phzF (Figures 5, 6).
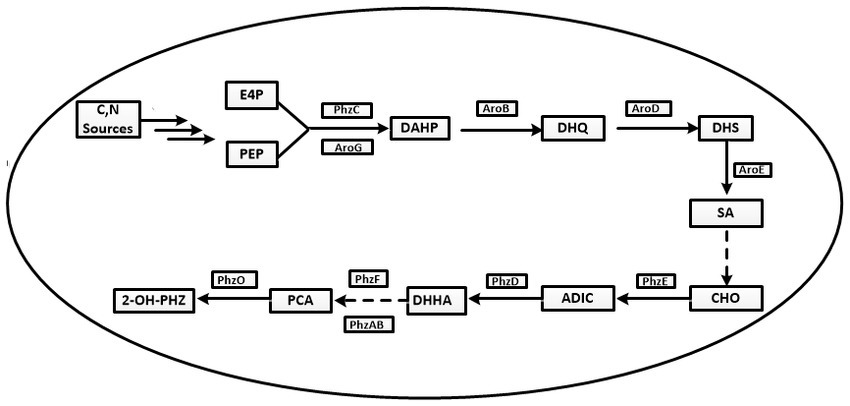
Figure 1. The synthetic pathway of phenazine-1-carboxylic acid and its derivatives in Pseudomonas chlororaphis. E4P, erythrose 4-phosphate; PEP, phosphoenolpyruvic acid; DAHP, 3-deoxy-darabinoheptulosonate-7-phosphate; DHQ, 3-dehydroquinic acid; DHS, 3-dehydroshikimic acid; SA, shikimic acid; CHO, chorismate; ADIC, 2-amino-4-deoxy branched acid; DHHA, Trans-2,3-dihydro-3-hydroxyanthranilic acid; PCA, Phenazine-1-carboxylic acid; 2-OH-PHZ, 2-hydroxyphenazine.
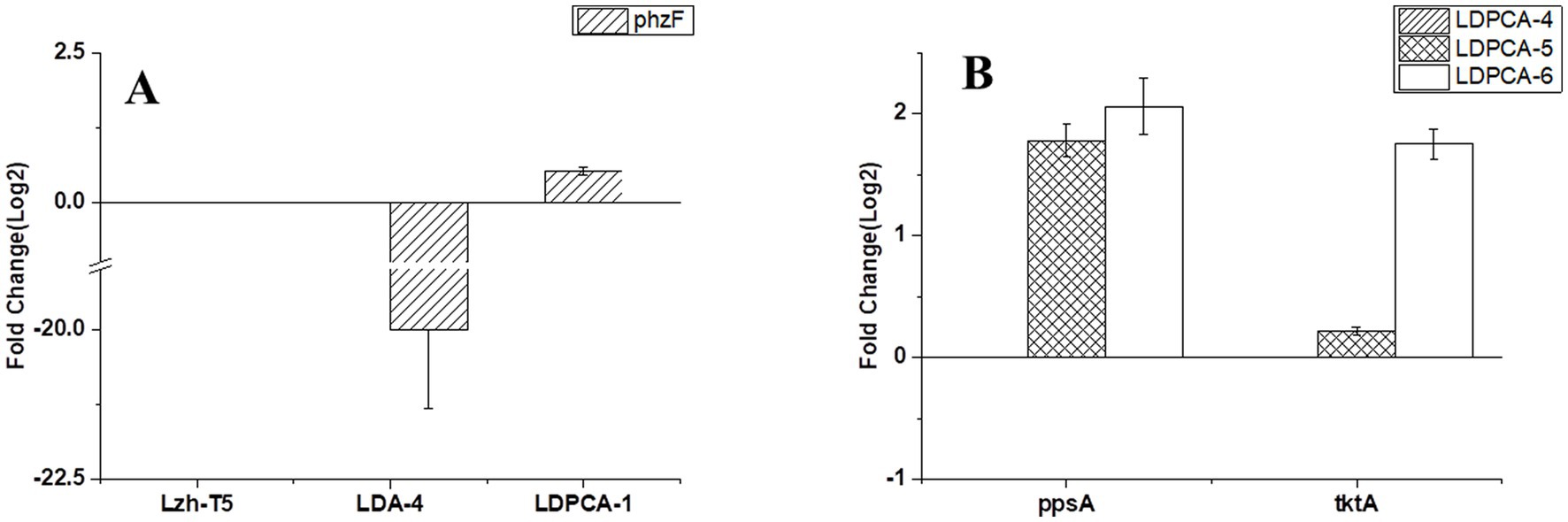
Figure 2. Transcriptional validation of different genes using Quantitative RT-PCR. (A) Transcriptional validation of phzF in different strains. (B) Transcriptional validation of tktA and ppsA in different strains. The data represent the means ± SD for three independent cultures.
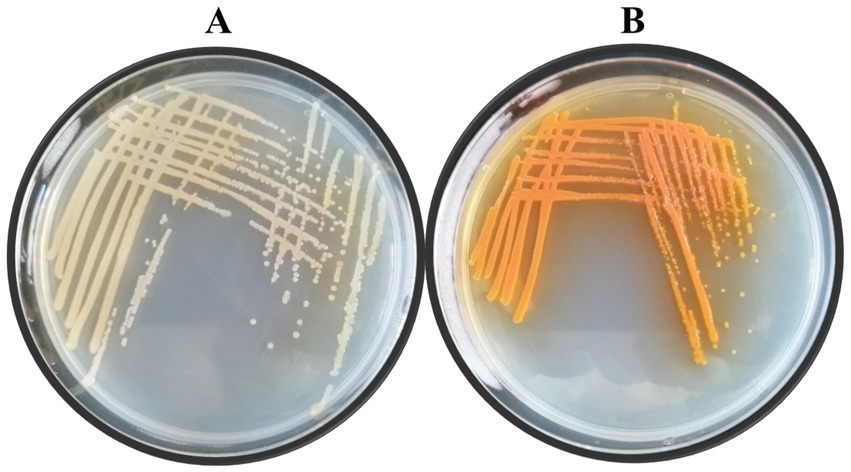
Figure 3. Colony morphology of different strains. (A) Pseudomonas chlororaphis LDA-4. (B) Pseudomonas chlororaphis LDA-4-phzF.
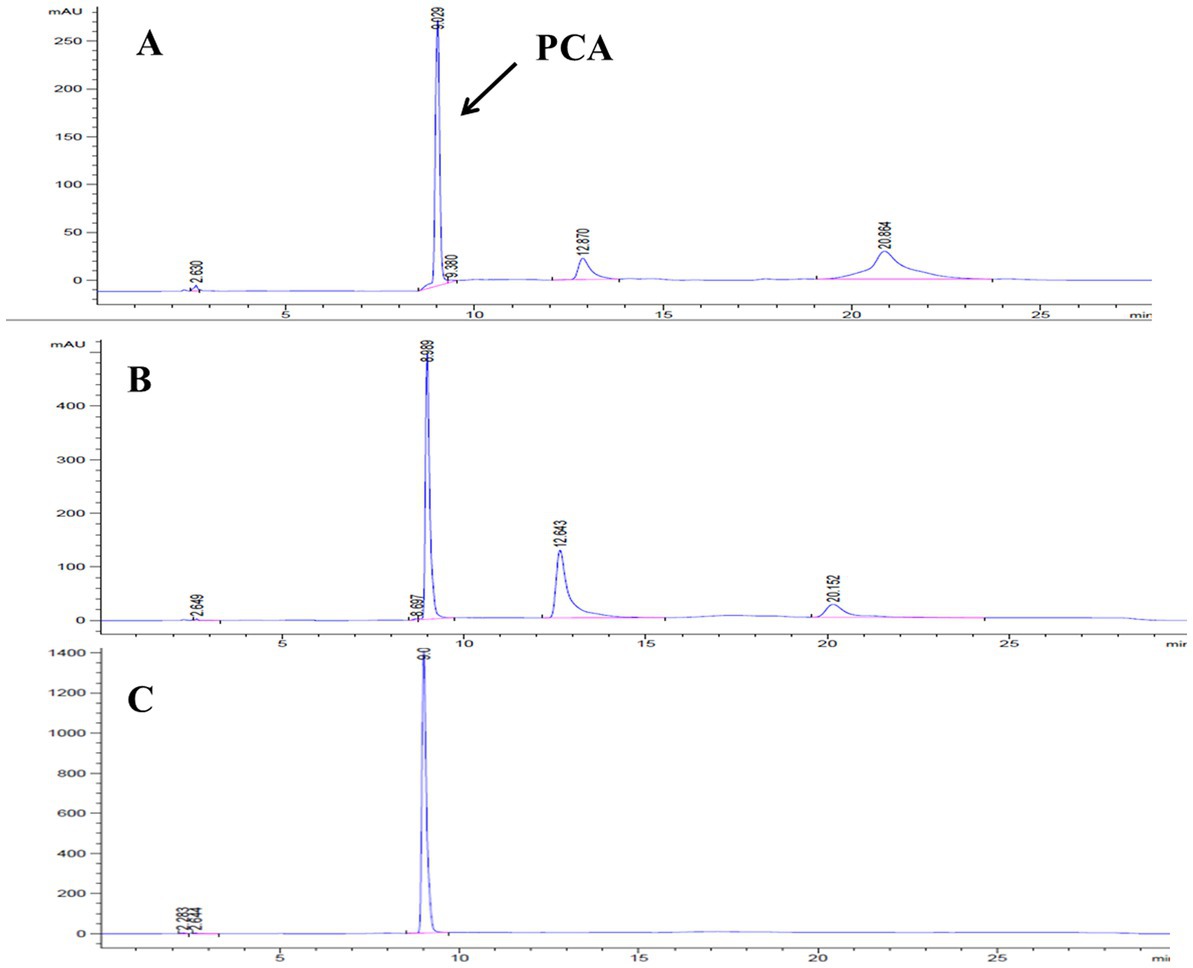
Figure 4. HPLC detection of fermentation broth of different strains. (A) Pseudomonas chlororaphis Lzh-T5. (B) Pseudomonas chlororaphis LDPCA-1. (C) Pseudomonas chlororaphis LDPCA-1.
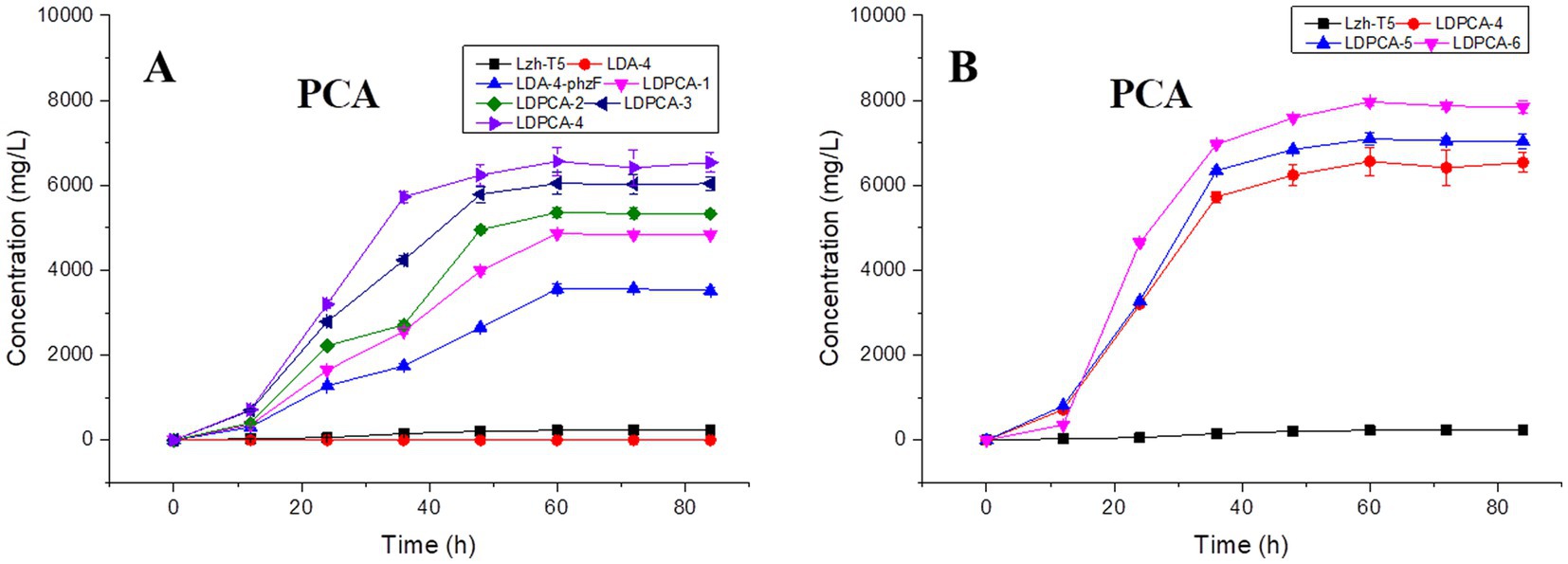
Figure 5. Effects of genetic modifications on PCA production. (A) PCA production of non-scar deletion strains. (B) PCA production of gene over-expression strains. The data represent the means ± SD for three independent cultures.
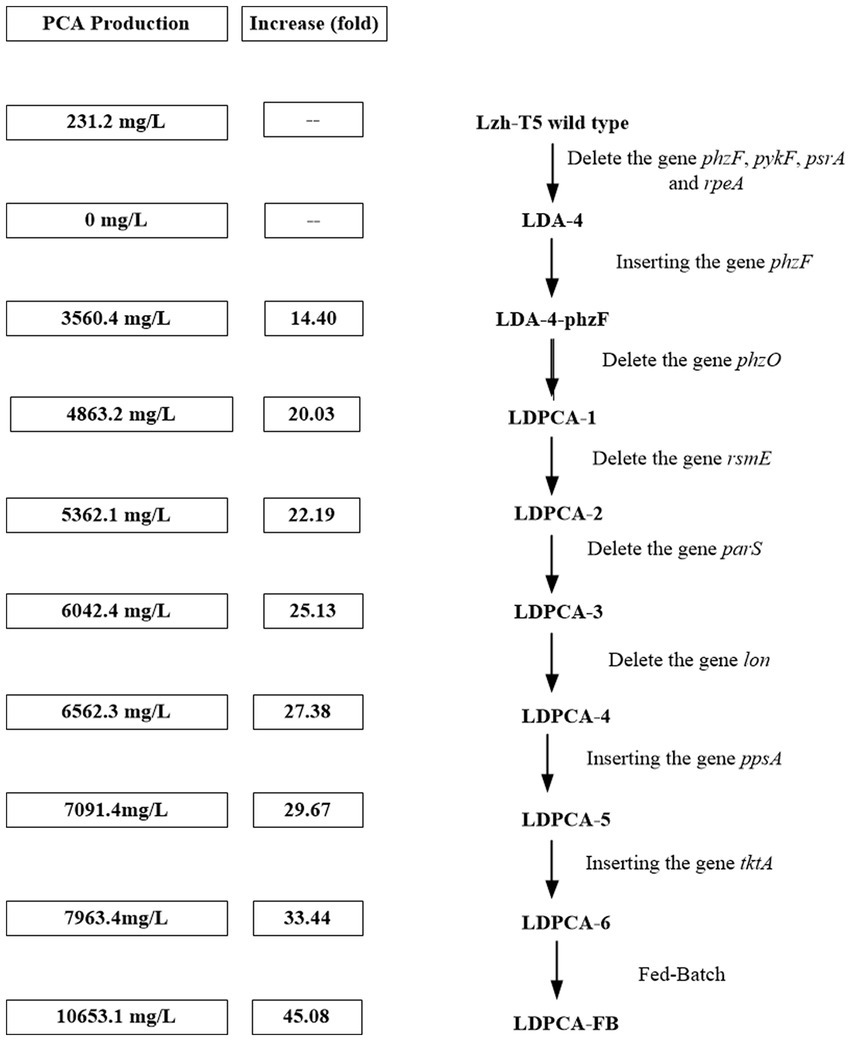
Figure 6. A summary of PCA production improve via genetic engineering operations in Pseudomonas chlororaphis.
3.2. Knockout of phzO resulted in PCA accumulation in the strain
After genome sequencing, the phenazine-related gene phzO was detected in P. chlororaphis Lzh-T5 (Li et al., 2018). The PhzO protein encoded by phzO is a phenazine-modified protein that catalyzes PCA conversion into 2-OH-PCA, which then spontaneously converts into 2-OH-PHZ (Chen et al., 2014). Therefore, both Lzh-T5 and its derivative strain LDA-4-phzF produce three phenazine derivatives, PCA, 2-OH-PCA, and 2-OH-PHZ (Liu et al., 2021). In this study, we knocked out phzO from the genome of the LDA-4-phzF strain using traceless knockout technology to obtain the LDPCA-1 strain. The metabolites of LDPCA-1 were extracted and detected using HPLC. Only PCA was accumulated in LDPCA-1 (Figure 4), with a yield of 4,863 mg/L, which was 20.9 times higher than that in the wild-type strain Lzh-T5 (Figures 5A, 6).
3.3. Enhancement of PCA production by knockout of negative regulatory factors
To enhance the production of PCA, several widely occurring negative regulator genes in P. chlororaphis were selected for knockout. We knocked out the negative regulatory factor rsmE from the genome of LDPCA-1 to obtain LDPCA-2 strain. HPLC revealed that PCA production by this strain increased to 5362.1 mg/L (Figures 5A, 6). Knockout of parS, another negative regulatory factor in the genome of LDPCA-2, resulted in the strain LDPCA-3, which presented with an increase in PCA yield to 6042.3 mg/L (Figures 5A, 6). The engineered strain LDPCA-4 was obtained by knocking out lon, and the PCA yield of this strain increased to 6562.3 mg/L (Figures 5A, 6).
3.4. Increasing PCA production by enhancing the shikimate pathway
Phenazine derivatives in Pseudomonas spp. are synthesized through the shikimate pathway. Enhancing this phenazine synthesis pathway can help promote PCA production. The shikimate pathway is an important metabolic pathway in Pseudomonas spp., which can not only synthesize phenazines but is also the lead pathway for coenzyme Q and aromatic amino acid synthesis (Talapatra and Talapatra, 2005; Liu et al., 2016). Phosphoenolpyruvate (PEP) and erythrose 4-phosphate (E4P) are the direct synthetic precursors of the shikimate pathway (Talapatra and Talapatra, 2005). Enhancing PEP and E4P supply can enhance the shikimate pathway (Rodriguez et al., 2014). Therefore, we selected ppsA and tktA, genes that can improve E4P and PEP production (Hu et al., 2017; Li et al., 2020). Accordingly, ppsA was integrated at the lon position of LDPCA-4 via homologous integration to obtain the recombinant strain LDPCA-5. Next, tktA was integrated at the parS position of the LDPCA-5 strain to obtain the recombinant strain LDPCA-6. RT-PCR revealed that the expression levels of tktA and ppsA in LDPCA-6 and LDPCA-5 were higher than those in LDPCA-4 (Figure 2B), suggesting that the genes integrated into the genome are transcribed and play a role in enhancing the shikimate pathway. HPLC detection revealed that the PCA production of LDPCA-5 and LDPCA-6 was increased, reaching 7091.4 and 7963.4 mg/L, respectively (Figures 5B, 6).
3.5. Enhancing PCA production by fed-batch fermentation
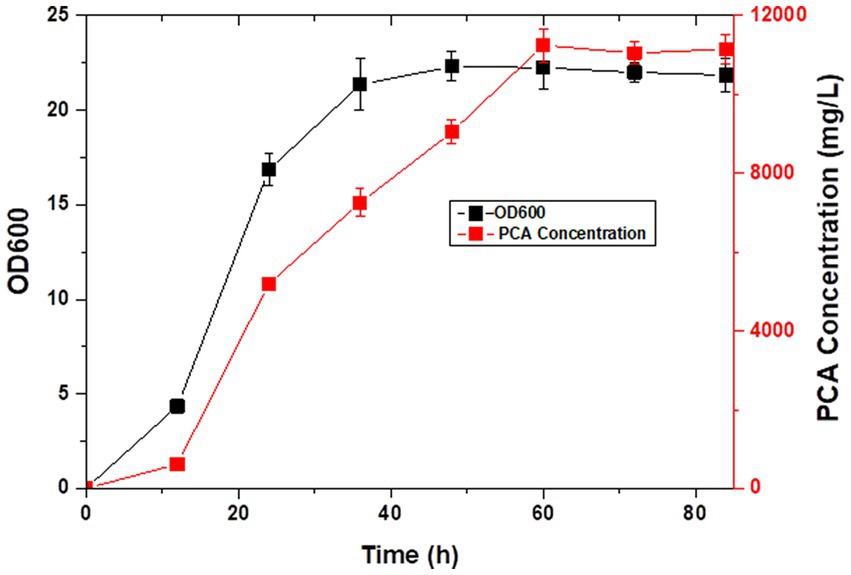
Pseudomonas spp. are aerobic bacteria; therefore, shake flask fermentation often limits the maximum efficiency of bacteria owing to insufficient oxygen supply. Moreover, after a period of growth, the efficiency of bacteria is limited due to lack of nutrients. In this study, we used the fed-batch method to ferment the engineered strain to improve PCA production by the bacterial strain. The fed-batch experiments were conducted in a 5-L fermenter after activating LDPCA-6 in a shake flask. PCA was extracted from the fermentation broth during the fermentation process. HPLC detection revealed that the bacterial growth reached a stable stage at 48 h after inoculation, and PCA production continued to increase for 60 h. The maximum yield was 10,653.1 mg/L, and the DCW yield was 2286.6 mg/g (Figures 6, 7).
4. Discussion
Phenazine derivatives are a class of biologically active substances that can be naturally produced by the Pseudomonas and Streptomyces spp. (Chin et al., 2000; Laursen and Nielsen, 2004; Mavrodi et al., 2006; Lugtenberg and Kamilova, 2009; Bilal et al., 2017). In this study, we focused on PCA production by Pseudomonas spp.
The synthesis steps of PCA by Pseudomonas are as follows. First PEP and E4P are converted to shikimate via the shikimate pathway. Then, shikimate is converted to PCA via enzymes encoded by the conserved phenazine synthesis gene cluster phzABCDEFG (Figure 1).
Different phenazine modification genes are present in Pseudomonas spp. that convert PCA into different phenazine derivatives, such as phzO, which converts PCA into 2-hydroxyphenazine; phzH, which converts PCA into phenazine-1-carboxamide; phzS, which converts PCA into 1-hydroxyphenazine; and phzS and phzM, which act simultaneously to convert PCA into pyocyanin (Chin et al., 2001; Mavrodi et al., 2001; Parsons et al., 2007; Greenhagen et al., 2008; Du et al., 2013).
Lzh-T5 is a strain of P. chlororaphis commonly found in the rhizosphere of tomatoes in Dezhou, China. Using genome sequencing, this strain was found to contain the typical phenazine synthesis gene cluster phzABCDEFG and a phenazine modification gene phzO. After fermentation, both PCA 2-OH-PCA and 2-OH-PHZ were present in the fermentation broth. We knocked out phzO using a traceless knockout method, which cut off the conversion of PCA into 2-OH-PHZ. After fermentation, HPLC detection revealed that the fermentation broth could accumulate PCA alone (Figure 4).
The wild strain Pseudomonas Lzh-T5 can produce PCA, although the yield is very low at only 220 mg/L. In our preliminary work, we used Lzh-T5 to produce DHHA through genetic engineering and obtained a yield of 11 g/L (Liu et al., 2021). DHHA is the synthetic precursor of phenazine derivatives in Pseudomonas and was synthesized by Pseudomonas after phzF knockout (Liu et al., 2021). Because of the metabolic modification of the DHHA-producing strain in this study, its metabolic flux in the shikimate pathway was stronger than that in the wild-type strain Lzh-T5. In this study, we selected the high DHHA-producing strain LDA-4 to produce PCA by restoring phzF.
Two-component signal transduction (TCST) systems are a class of regulatory systems that are widely present in bacteria and help bacteria adapt to changes in the surrounding environment. According to previous reports, Pseudomonas spp. contains a large number of TCST systems, such as the GacS/GacA and rpeA/rpeB systems (Bejerano-Sagie and Xavier, 2007; Peng et al., 2018). The GacS/GacA system is a TCST system that has been previously identified in Pseudomonas spp. Studies have reported that the GacS/GacA system affects the production of phenazines through intracellular small RNAs and negatively regulates the production of phenazines through rsmE (Liu et al., 2016; Li et al., 2020). We found rsmE to be present in the Lzh-T5 genome, and, after knocking out rsmE in the LDPCA-1 strain genome, PCA production increased from 4863.2 mg/L to 5362.1 mg/L. Compared with the GacS/GacA system, the parS/parR system has recently been discovered. Studies have also reported that the parS/parR system can regulate phenazine production in Pseudomonas (Liu et al., 2016; Peng et al., 2018). We obtained the strain LDPCA-3 by knocking out parS in the LDPCA-2 genome; moreover, HPLC revealed increased PCA production (Figures 5, 6). In addition to the TCST systems that can affect the production of phenazines, some enzymes can indirectly regulate phenazine synthesis through the TCST system. Reportedly, the protease LON has been reported to be a negative regulatory factor of phenazine derivatives (Laskowska et al., 1996; Whistler et al., 2000; Wang et al., 2013; Takeuchi et al., 2014; Liu et al., 2016). And LON protease affects the production of 2-hydroxyphenazine by affecting the stability of GacA protein (Liu et al., 2016; Li et al., 2020). After detection, LON was found in P. chlororaphis Lzh-T5. Our results indicate that LON negatively regulates PCA production (Figures 5, 6).
In Pseudomonas spp., the shikimate pathway is the leading pathway for phenazine synthesis. This pathway uses PEP and E4P as direct synthetic substrates (Talapatra and Talapatra, 2005). Previous research has shown that the expression of PEP synthase (encoded by ppsA) and transketolase (encoded by tktA) can increase the PEP and E4P pools in Pseudomonas cells, enhancing the shikimate pathway (Liu et al., 2016). In some previous studies, genes such as ppsA and tktA were overexpressed to promote the synthesis of phenazines, achieving good yields (Liu et al., 2016; Hu et al., 2017; Li et al., 2020). However, these studies generally used plasmids as vectors for overexpression. Plasmids are generally confirmed by antibiotics to ensure their stable existence in bacteria and are easily lost in the absence of antibiotics. Furthermore, when multiple genes are expressed simultaneously, the sequence order will affect the expression effect of different genes (Juminaga et al., 2012; Liu et al., 2016). These problems are not encountered when the overexpressed gene is introduced into the genome of the host bacterium. In this study, we used the principle of homologous recombination to introduce ppsA and tktA into the genome of Pseudomonas for over expression. The results of RT-PCR and PCA detection showed that the introduced ppsA and tktA promoted the shikimate pathway for PCA production (Figure 2).
PCA is a secondary metabolite of Pseudomonas spp. and needs to be produced during the growth of Pseudomonas spp. (Li S. et al., 2022; Olyaei and Sadeghpour, 2022). The growth and development of bacteria can be generally divided into the lag, logarithmic growth, and extinction phases. PCA production is more favorable with Pseudomonas in a stable state (Liu et al., 2016; Yue et al., 2018). Compared with batch fermentation, the fed-batch fermentation medium has rich ingredients, stable oxygen supply, and relatively constant pH value, which are more conducive to the rapid reproduction of microorganisms and the production of metabolites (Li et al., 2020). Therefore, the fed-batch process is the preferred fermentation mode. Moreover, PCA produced in this study is a secondary metabolite of Pseudomonas, and a large amount of accumulation will lead to feedback inhibition in the bacteria. The problems of substrate inhibition and catabolite inhibition can be overcome in the fed-batch process by the continuous addition of new medium. This significantly increases the yield of the target product. In this study, we added the PCA high-yielding strain LDPCA-6 to a 5-L fermenter for fed-batch fermentation. The HPLC results showed that the bacterial growth reached a stable stage at 60 h after inoculation, and the production of PCA continued to increase for 72 h. The maximum yield was 10,653.1 mg/L, which was 33.8% higher than that using batch culture.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
LL and KL conceived, designed the experiments, and drafted the manuscript. KL, ZL, XL, YX, and YC performed the experiments. KL, RW, LL, and PL analyzed the data. All authors read and approved the final manuscript.
Funding
This work was financially supported by the Qilu University of Technology of Cultivating Subject for Biology and Biochemistry (no. 202008), the International Technology Cooperation Project from Qilu University of Technology (Shandong Academy of Sciences) (no. 2022GH026), the Shandong Provincial Key Laboratory of Biophysics (no. FWL2021065), the Natural Science Foundation of Shandong Province (nos. ZR2019BC027 and ZR2020QC044), and the National Natural Science Foundation of China (no. 31800106). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1186052/full#supplementary-material
References
Ahmad, G., Khan, A., Khan, A. A., Ali, A., and Mohhamad, H. I. (2021). Biological control: a novel strategy for the control of the plant parasitic nematodes. Antonie Van Leeuwenhoek 114, 885–912. doi: 10.1007/s10482-021-01577-9
Bejerano-Sagie, M., and Xavier, K. B. (2007). The role of small RNAs in quorum sensing. Curr. Opin. Microbiol. 10, 189–198. doi: 10.1016/j.mib.2007.03.009
Bilal, M., Guo, S., Iqbal, H. M. N., Hu, H., Wang, W., and Zhang, X. (2017). Engineering Pseudomonas for phenazine biosynthesis, regulation, and biotechnological applications: a review. World J. Microbiol. Biotechnol. 33:191. doi: 10.1007/s11274-017-2356-9
Chen, M., Cao, H., Peng, H., Hu, H., Wang, W., and Zhang, X. (2014). Reaction kinetics for the biocatalytic conversion of phenazine-1-carboxylic acid to 2-hydroxyphenazine. PLoS One 9:e98537. doi: 10.1371/journal.pone.0098537
Chin, A. W. T. F., Bloemberg, G. V., Mulders, I. H., Dekkers, L. C., and Lugtenberg, B. J. (2000). Root colonization by phenazine-1-carboxamide-producing bacterium pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant-Microbe Interact. 13, 1340–1345. doi: 10.1094/MPMI.2000.13.12.1340
Chin, A. W. T. F., Thomas-Oates, J. E., Lugtenberg, B. J., and Bloemberg, G. V. (2001). Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol. Plant-Microbe Interact. 14, 1006–1015. doi: 10.1094/MPMI.2001.14.8.1006
Du, X., Li, Y., Zhou, Q., and Xu, Y. (2015). Regulation of gene expression in Pseudomonas aeruginosa M18 by phenazine-1-carboxylic acid. Appl. Microbiol. Biotechnol. 99, 813–825. doi: 10.1007/s00253-014-6101-0
Du, X., Li, Y., Zhou, W., Zhou, Q., Liu, H., and Xu, Y. (2013). Phenazine-1-carboxylic acid production in a chromosomally non-scar triple-deleted mutant Pseudomonas aeruginosa using statistical experimental designs to optimize yield. Appl. Microbiol. Biotechnol. 97, 7767–7778. doi: 10.1007/s00253-013-4921-y
Garcia-Jaramillo, M., Redondo-Gomez, S., Barcia-Piedras, J. M., Aguilar, M., Jurado, V., Hermosin, M. C., et al. (2016). Dissipation and effects of tricyclazole on soil microbial communities and rice growth as affected by amendment with alperujo compost. Sci. Total Environ. 550, 637–644. doi: 10.1016/j.scitotenv.2016.01.174
Greenhagen, B. T., Shi, K., Robinson, H., Gamage, S., Bera, A. K., Ladner, J. E., et al. (2008). Crystal structure of the pyocyanin biosynthetic protein PhzS. Biochemistry 47, 5281–5289. doi: 10.1021/bi702480t
Hu, H., Li, Y., Liu, K., Zhao, J., Wang, W., and Zhang, X. (2017). Production of trans-2,3-dihydro-3-hydroxyanthranilic acid by engineered Pseudomonas chlororaphis GP72. Appl. Microbiol. Biotechnol. 101, 6607–6613. doi: 10.1007/s00253-017-8408-0
Jin, K., Zhou, L., Jiang, H., Sun, S., Fang, Y., Liu, J., et al. (2015). Engineering the central biosynthetic and secondary metabolic pathways of Pseudomonas aeruginosa strain PA1201 to improve phenazine-1-carboxylic acid production. Metab. Eng. 32, 30–38. doi: 10.1016/j.ymben.2015.09.003
Juminaga, D., Baidoo, E. E., Redding-Johanson, A. M., Batth, T. S., Burd, H., Mukhopadhyay, A., et al. (2012). Modular engineering of L-tyrosine production in Escherichia coli. Appl. Environ. Microbiol. 78, 89–98. doi: 10.1128/AEM.06017-11
Karmegham, N., Vellasamy, S., Natesan, B., Sharma, M. P., Al Farraj, D. A., and Elshikh, M. S. (2020). Characterization of antifungal metabolite phenazine from rice rhizosphere Fluorescent pseudomonads (FPs) and their effect on sheath blight of rice. Saudi J. Biol. Sci. 27, 3313–3326. doi: 10.1016/j.sjbs.2020.10.007
Laskowska, E., Kuczynska-Wisnik, D., Skorko-Glonek, J., and Taylor, A. (1996). Degradation by proteases Lon, Clp and HtrA, of Escherichia coli proteins aggregated in vivo by heat shock; HtrA protease action in vivo and in vitro. Mol. Microbiol. 22, 555–571. doi: 10.1046/j.1365-2958.1996.1231493.x
Laursen, J. B., and Nielsen, J. (2004). Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chem. Rev. 104, 1663–1686. doi: 10.1021/cr020473j
Li, H., Li, Y., Wang, W., Wan, Q., Yu, X., and Sun, W. (2022). Uptake, translocation, and subcellular distribution of three triazole pesticides in rice. Environ. Sci. Pollut. Res. Int. 29, 25581–25590. doi: 10.1007/s11356-021-17467-6
Li, L., Li, Z., Yao, W., Zhang, X., Wang, R., Li, P., et al. (2020). Metabolic engineering of Pseudomonas chlororaphis Qlu-1 for the enhanced production of phenazine-1-carboxamide. J. Agric. Food Chem. 68, 14832–14840. doi: 10.1021/acs.jafc.0c05746
Li, Z., Li, X., Zeng, Q., Chen, M., Liu, D., Wang, J., et al. (2018). Genome sequence of Pseudomonas chlororaphis Lzh-T5, a plant growth-promoting rhizobacterium with antimicrobial activity. Genome Announc. 6:e00328-18. doi: 10.1128/genomeA.00328-18
Li, S., Yue, S. J., Huang, P., Feng, T. T., Zhang, H. Y., Yao, R. L. H., et al. (2022). Comparative metabolomics and transcriptomics analyses provide insights into the high-yield mechanism of phenazines biosynthesis in Pseudomonas chlororaphis GP72. J. Appl. Microbiol. 133, 2790–2801. doi: 10.1111/jam.15727
Li, X. J., Zhang, W., Zhao, C. N., Wu, Q. L., Li, J. K., and Xu, Z. H. (2021). Synthesis and fungicidal activity of phenazine-1-carboxylic triazole derivatives. J. Asian Nat. Prod. Res. 23, 452–465. doi: 10.1080/10286020.2020.1754400
Liu, K., Hu, H., Wang, W., and Zhang, X. (2016). Genetic engineering of Pseudomonas chlororaphis GP72 for the enhanced production of 2-hydroxyphenazine. Microb. Cell Fact. 15:131. doi: 10.1186/s12934-016-0529-0
Liu, K., Li, L., Yao, W., Wang, W., Huang, Y., Wang, R., et al. (2021). Genetic engineering of Pseudomonas chlororaphis Lzh-T5 to enhance production of trans-2,3-dihydro-3-hydroxyanthranilic acid. Sci. Rep. 11:16451. doi: 10.1038/s41598-021-94674-8
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Mavrodi, D. V., Blankenfeldt, W., and Thomashow, L. S. (2006). Phenazine compounds in Fluorescent pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 44, 417–445. doi: 10.1146/annurev.phyto.44.013106.145710
Mavrodi, D. V., Bonsall, R. F., Delaney, S. M., Soule, M. J., Phillips, G., and Thomashow, L. S. (2001). Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183, 6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001
Medina, M. B., Resnik, S. L., and Munitz, M. S. (2021). Optimization of a rice cooking method using response surface methodology with desirability function approach to minimize pesticide concentration. Food Chem. 352:129364. doi: 10.1016/j.foodchem.2021.129364
Meng, Z., Chen, X., Guan, L., Xu, Z., Zhang, Q., Song, Y., et al. (2018). Dissipation kinetics and risk assessments of tricyclazole during Oryza sativa L. growing, processing and storage. Environ. Sci. Pollut. Res. Int. 25, 35249–35256. doi: 10.1007/s11356-018-3445-5
Molina, J., Sikora, M., Garud, N., Flowers, J. M., Rubinstein, S., Reynolds, A., et al. (2011). Molecular evidence for a single evolutionary origin of domesticated rice. Proc. Natl. Acad. Sci. U. S. A. 108, 8351–8356. doi: 10.1073/pnas.1104686108
Olyaei, A., and Sadeghpour, M. (2022). A review on lawsone-based benzo[a]phenazin-5-ol: synthetic approaches and reactions. RSC Adv. 12, 13837–13895. doi: 10.1039/D2RA02139K
Padovani, L., Capri, E., Padovani, C., Puglisi, E., and Trevisan, M. (2006). Monitoring tricyclazole residues in rice paddy watersheds. Chemosphere 62, 303–314. doi: 10.1016/j.chemosphere.2005.05.025
Parsons, J. F., Greenhagen, B. T., Shi, K., Calabrese, K., Robinson, H., and Ladner, J. E. (2007). Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry 46, 1821–1828. doi: 10.1021/bi6024403
Peng, H., Zhang, P., Bilal, M., Wang, W., Hu, H., and Zhang, X. (2018). Enhanced biosynthesis of phenazine-1-carboxamide by engineered Pseudomonas chlororaphis HT66. Microb. Cell Fact. 17:117. doi: 10.1186/s12934-018-0962-3
Petitjean, M., Juarez, P., Meunier, A., Daguindau, E., Puja, H., Bertrand, X., et al. (2021). The rise and the fall of a Pseudomonas aeruginosa endemic lineage in a hospital. Microb. Genom. 7:000629. doi: 10.1099/mgen.0.000629
Rodriguez, A., Martinez, J. A., Flores, N., Escalante, A., Gosset, G., and Bolivar, F. (2014). Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds. Microb. Cell Fact. 13:126. doi: 10.1186/s12934-014-0126-z
Sefiloglu, F. O., Tezel, U., and Balcioglu, I. A. (2021). Validation of an analytical workflow for the analysis of pesticide and emerging organic contaminant residues in paddy soil and rice. J. Agric. Food Chem. 69, 3298–3306. doi: 10.1021/acs.jafc.0c06111
Sharma, A., Shukla, A., Attri, K., Kumar, M., Kumar, P., Suttee, A., et al. (2020). Global trends in pesticides: a looming threat and viable alternatives. Ecotoxicol. Environ. Saf. 201:110812. doi: 10.1016/j.ecoenv.2020.110812
Sun, X., Xu, Y., Chen, L., Jin, X., and Ni, H. (2021). The salt-tolerant phenazine-1-carboxamide-producing bacterium Pseudomonas aeruginosa NF011 isolated from wheat rhizosphere soil in dry farmland with antagonism against Fusarium graminearum. Microbiol. Res. 245:126673. doi: 10.1016/j.micres.2020.126673
Takeuchi, K., Tsuchiya, W., Noda, N., Suzuki, R., Yamazaki, T., and Haas, D. (2014). Lon protease negatively affects GacA protein stability and expression of the Gac/Rsm signal transduction pathway in Pseudomonas protegens. Environ. Microbiol. 16, 2538–2549. doi: 10.1111/1462-2920.12394
Walker, J., and Moore, G. (2015). Pseudomonas aeruginosa in hospital water systems: biofilms, guidelines, and practicalities. J. Hosp. Infect. 89, 324–327. doi: 10.1016/j.jhin.2014.11.019
Wang, D., Lee, S. H., Seeve, C., Yu, J. M., Pierson, L. S. 3rd, and Pierson, E. A. (2013). Roles of the Gac-Rsm pathway in the regulation of phenazine biosynthesis in Pseudomonas chlororaphis 30-84. Microbiology 2, 505–524. doi: 10.1002/mbo3.90
Whistler, C. A., Stockwell, V. O., and Loper, J. E. (2000). Lon protease influences antibiotic production and UV tolerance of Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 66, 2718–2725. doi: 10.1128/AEM.66.7.2718-2725.2000
Yan, B., Ye, F., and Gao, D. (2015). Residues of the fungicide epoxiconazole in rice and paddy in the Chinese field ecosystem. Pest Manag. Sci. 71, 65–71. doi: 10.1002/ps.3763
Yue, S., Bilal, M., Guo, S., Hu, H., Wang, W., and Zhang, X. (2018). Enhanced trans-2,3-dihydro-3-hydroxyanthranilic acid production by pH control and glycerol feeding strategies in engineered Pseudomonas chlororaphis GP72. J. Chem. Technol. Biotechnol. 93, 1618–1626. doi: 10.1002/jctb.5531
Zhao, W., Chou, J., Li, J., Xu, Y., Li, Y., and Hao, Y. (2022). Impacts of extreme climate events on future rice yields in global major rice-producing regions. Int. J. Environ. Res. Public Health 19:4437. doi: 10.3390/ijerph19084437
Keywords: phenazine-1-carboxylic acid, Pseudomonas chlororaphis , genetic engineering, non-scar deletion, fed-batch fermentation
Citation: Liu K, Li Z, Liang X, Xu Y, Cao Y, Wang R, Li P and Li L (2023) Biosynthesis and genetic engineering of phenazine-1-carboxylic acid in Pseudomonas chlororaphis Lzh-T5. Front. Microbiol. 14:1186052. doi: 10.3389/fmicb.2023.1186052
Edited by:
Samina Mehnaz, Forman Christian College, PakistanReviewed by:
Deeba Noreen Baig, Forman Christian College, PakistanMurat Ozdal, Atatürk University, Türkiye
Copyright © 2023 Liu, Li, Liang, Xu, Cao, Wang, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Li, bGlsaW5nMzM4MDI0MDBAcWx1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Kaiquan Liu
Kaiquan Liu Zhenghua Li
Zhenghua Li Xiaoli Liang1
Xiaoli Liang1 Piwu Li
Piwu Li