- 1College of Plant Protection, Yunnan Agricultural University, Kunming, China
- 2Tea Research Institute, Hunan Academy of Agricultural Sciences, Changsha, China
As a biocontrol bacteria, Brevibacillus has been the subject of extensive research for agricultural applications. Antibacterial peptides (AMPs) are the main antibacterial products of Brevibacillus. This study isolated a strain of Br. brevis HNCS-1 from tea garden soil, and the strain has an antagonistic effect against five types of pathogens of tea diseases, namely Gloeosporium theae-sinensis, Elsinoe leucospira, Phyllosticta theaefolia, Fusarium sp., and Cercospora theae. To determine the genetic characteristics implicated in the biocontrol mechanism, the genome sequence of the HNCS-1 strain was obtained and analyzed further, and the data are deposited in the GenBank repository (No. CP128411). Comparative genomics analyses revealed that the HNCS-1 strain and 17 public Br. brevis share a core genome composed of 3,742 genes. Interestingly, only one non-ribosomal peptide synthetase (NRPS) gene cluster annotated as edeine is present in the core genome. And UHPLC-MS/MS detection results showd that edeine B and edeine A were the principal antibacterial peptides in the HNCS-1 strain. This study proves that edeine is the main antibacterial peptide of Br. brevis, and provides a new strategy for the identification of antibacterial products from other biocontrol bacteria.
Introduction
The tea plant (Camellia sinensis) is an important cash crop, and its beneficial metabolites are valuable for human health (Pan et al., 2022). Similar to other plants, tea plants are vulnerable to many diseases throughout their life cycle. Among them, fungal diseases are the most serious hazards to tea plants, resulting in a decline in tea production and quality. For example, tea white scab disease occurs year-round in the Chinese tea host production area, generally resulting in a reduction of approximately 10% in production and a reduction of over 50% in the sick tea garden (Zhou et al., 2019). Tea gray blight can reduce tea production by 10–20% (Sanjay et al., 2008). For a long time, chemical control has been the main means to address tea plant diseases. However, there are fewer pesticides registered for tea plant diseases, and long-term single and excessive use of chemical pesticides has caused problems such as pathogen resistance, pesticide residues, and environmental pollution. Therefore, tea plantations urgently need a sustainable, environmentally friendly, and anti-drug-resistance plant disease control technology to replace traditional chemical control. Microbial pesticides have become one of the ideal strategies for the comprehensive control of tea plant diseases due to their persistence and environmental friendliness.
Brevibacillus spp. is an essential source of biocontrol microorganisms and is widespread in nature, having been discovered in soil, flora, seawater, and the intestinal tracts of animals (Ruiu, 2013). There are 20 species in the genus, including 10 Bacillus species discovered previously (the Br. brevis cluster). Br. brevis is the parent species of the genus Brevibacillus (Shida et al., 1996). It has been reported in the disciplines of biological control of plant maladies, pollution degradation, and heavy metal remediation (Samrot et al., 2015; Che et al., 2016; Wang et al., 2016). Brevibacillus is one of the most studied bacterial groups, and it has been one of the most important producers of antimicrobial peptides (AMPs) (Yang and Yousef, 2018). AMPs play an important role in bacteriostasis (Xu et al., 2023). AMPs can be classified according to their biosynthetic pathways as ribosomally synthesized and post-translationally modified peptides (Ripps) and non-ribosomally synthesized peptides (NRPS). Currently, the preponderance of Brevibacillus AMPs are produced by non-ribosomal eptide synthetases. They consist of edeine (A, B, D, and F) (Czajgucki et al., 2006), gramicidin (A-C, S) (Govaerts et al., 2001; Kessler et al., 2004), gratisin (Tamaki et al., 1983), tyrocidine (A-D) (Mootz and Marahiel, 1997), BT1583 (Wu et al., 2005), tostadin (Song et al., 2012), tauramamide (Desjardine et al., 2007), brevistin (Shoji and Kato, 1976), spergualin (Takeuchi et al., 1981), loloatin (A-D) (Gerard et al., 1999), laterocidin (Xu et al., 2010), and tridecapeptide families (Yang et al., 2017). Brevibacillus spp. generate numerous Ripps, such as laterosporulin (Singh et al., 2012), laterosporulin10 (Baindara et al., 2016), and Bac-GM100 (Ghadbane et al., 2013). Most Brevibacillus AMPs exert their antimicrobial effect via cytoplasmic membrane damage, but edeine inhibits DNA synthesis and protein translation and synthesis at varying concentrations (Szer and Kurylo-Borowska, 1972; Dinos et al., 2004).
However, the diversity of types and structures of AMPs has also led to difficulties in the purification and identification of traditional chemical analysis techniques. Fortunately, the development of next-generation sequencing technology over the past two decades has consequently stimulated research on the comparative genomics of Br. brevis. More biocontrol strains of Br. brevis have had their entire genomes sequenced in order to investigate the relationships between antibacterial mechanisms and the underlying genetic diversity of Br. brevis genomes. With the advancement of bioinformatics prediction, AMP gene clusters have been continuously identified. The metabolic pathway database of AMPs, which can annotate the protein gene clusters of biocontrol bacteria at the gene level and predict the results of secondary metabolites, has been perpetually improved, making the rapid identification of AMPs feasible. In addition, with the continuous publication of multiple microbial genomes and the comparison of microbial genomes between different individuals of the same species, it is gradually recognized that a single reference genome cannot represent diversity within a species (Cuellar-Gaviria et al., 2023; Wan et al., 2023).
In this study, a strain of Br. brevis HNCS-1 with broad-spectrum antibacterial activity was isolated from the tea garden soil. To gain a comprehensive understanding of the biocontrol potential of strain HNCS-1, we sequenced the genome of Br. brevis HNCS-1. A comparative genomic analysis was conducted to characterize the pan-genome structure of this biocontrol bacterium, assembled with 17 public Br. brevis genomes. An NRPS gene cluster was found in the core genome, which may be related to the antibacterial ability of Br. brevis. This study provides a genetic context for future research on AMPs and provides a scientific basis for further optimizing the field applications of the microbial biopesticide derived from Br. brevis HNCS-1.
Materials and methods
Microbial culture and preservation
The cultivation and plant protection laboratory of the Tea Research Institute of the Hunan Academy of Agricultural Sciences isolated Br. brevis HNCS-1 from tea garden soil and deposited it in China Center for Type Culture Collection (No. M 2022713). The cultivation and plant protection laboratory isolated and preserved Gloeosporium theae-sinensis, Elsinoe leucospira, Phyllosticta theaefolia, Fusarium sp., and Cercospora theae.
The fungi were routinely cultivated at 26°C on a PDA medium (6 g potato extract, 20 g dextrose, 20 g agar, 1,000 ml H2O, pH 5.6 ± 0.2) and stored at 4°C on the same medium. Before using them in an experiment, they were activated on a PDA medium for 24 h at 26°C and then transferred by streaking. The HNCS-1 strain was stored in 20% glycerol at −80°C. It was activated by streaking at 37°C for 12 h on LB agar medium (10 g tryptone, 5 g yeast extract, 10 g NaCl, 15 g agar, 1,000 ml H2O, pH 7.0 ± 0.1).
Antibiotics activity of Brevibacillus brevis HNCS-1
Br. brevis HNCS-1 was inoculated into 500 ml Erlenmeyer flasks containing 200 ml of the LB liquid medium and cultivated at 37°C and 200 rpm for 12 h. Then, 10 ml of the seed cultures were transferred to 500 ml Erlenmeyer flasks containing the NB medium (10 g peptone, 3 g beef extract, 5g NaCl, 1,000 ml H2O, and pH 7.2 ± 0.2) and cultivated at 37°C and 180 rpm for 3 days. Br. brevis HNCS-1 fermentation broth was centrifuged at 4°C and 10,000 rpm for 10 min, and the supernatant was collected and filtered through a 0.22 μm filter.
On a PDA medium containing a 10% cell-free supernatant of Br. brevis HNCS-1, spore germination was used to determine the bacteriostatic activity against fungi. A fungus cake was made using a 5 mm diameter sterile cork borer and placed in the center of the plate. Plates of P. theaefolia, Fusarium sp., E. leucospira, and C. theae were incubated at 26°C for 12 days, 11 days, 22 days, 22 days, and 7 days, respectively. The experiment was performed thrice. The antagonistic activity of Br. brevis HNCS-1 was evaluated by inhibition rate (IR), which is calculated using the following formula:
where A is the diameter of tea fungus disks in the control treatment, B is the diameter of tea fungus disks with Br. brevis HNCS-1, and 5 is the diameter of the disks inoculated with tea fungus.
Genome sequencing, annotation, and alignment
The entire genome of Br. brevis HNCS-1 was sequenced using the Illumina and Nanopore platforms. Guangdong Magigene Technology Co., Ltd. performed this study in China. Following the standard protocol provided by Oxford Nanopore Technologies (ONT), the sequencing procedure includes sample quality detection, library construction, library quality detection, and library sequencing. SMRT Link v5.1.0 was chosen for the assembly of third-generation data alone, and Unicycle was chosen for the assembly of second-generation and third-generation data. tRNAscan-SE v1.3.1, rRNAmmer v1.2, and Rfam databases briefly predicted Glimmer, tRNAs, and rRNAs, respectively. PHAST identified prophage sequences within the genome assemblies. Island viewer predicted Genomic Islands (GIs).
The complete nucleotide sequence was also searched against Non-Redundant (NR) Protein Database, Swiss-Port, Cluster of Orthologous Groups (COGs), Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), Pfam, Carbohydrate-Active enZYmes (CAZy) database, Pathogen–Host Interactions (PHIs), Virulence Factor Database (VFDB), and Comprehensive Antibiotic Resistance Database (CARD) for functional annotation and further function assignment. Basic Local Alignment Search Tool (BLAST) and Diamond were used to compare the gene sequence with the reference database sequences. The finest matching results were selected based on identity, e-value, and score, which were the results of genome annotation. Circular maps of the genome were generated using Circos (Krzywinski et al., 2009).
Phylogenetic tree and pan-genome analysis
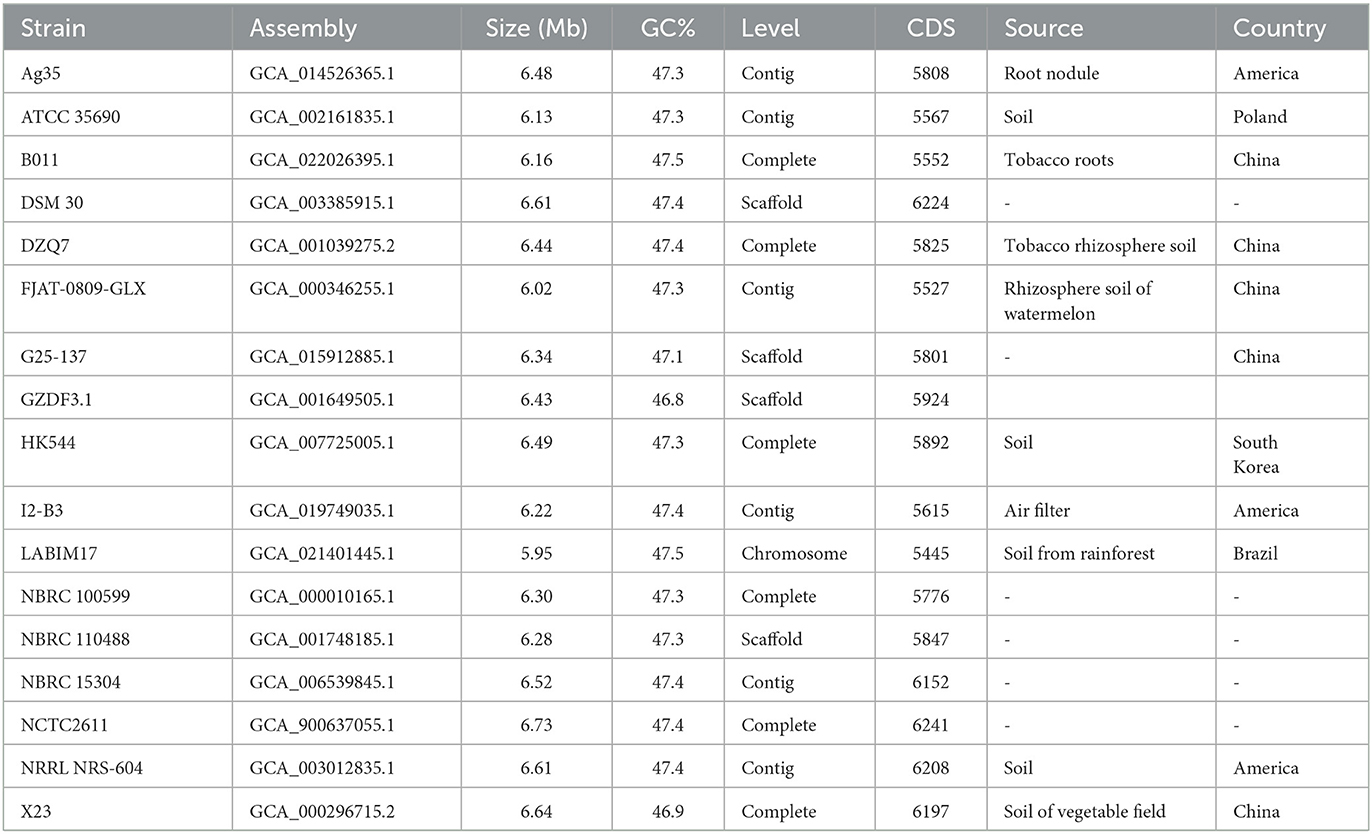
Br. brevis HNCS-1 and 17 other Br. brevis genomes were used to construct a phylogenetic tree, with B. subtilis ATCC 13952 serving as an out-group. Except for Br. brevis HNCS-1, all sequences were obtained from the National Center for Biotechnology Information (NCBI) (Table 1). OrthoFinder software (Emms and Kelly, 2019) was used to generate a maximum likelihood (ML) phylogenetic tree, which was visualized using Figtree v1.4.4. To evaluate the phylogeny of the Br. brevis strains, average nucleotide identity (ANI) was calculated using the FastANI software (Jain et al., 2018). The thermal map was then visualized using the TBtools software (Chen et al., 2020).
To identify the core and strain-specific genes, a pan-genome analysis was performed on 18 Br. brevis isolates using the BPGA v1.3 software, which implemented functional ortholog clustering using the amino acid sequences based on Gene Family (GF) approach (Chaudhari et al., 2016). The collection of genes shared by all 18 strains was referred to as the pan-genome, while the set of shared genes within the test genomes was referred to as their core genomes. For pan-genome genetic contexts, Heap's Law can be represented by the following formula:
where y represents the extent of the pan-genome, n represents the number of genomes, and k and γ are fitting parameters. According to this law, γ can be calculated as α = 1–γ, so when α < 1 (0 < γ < 1), the extent of the pan-genome increases unboundedly with the successive addition of new genomes and is considered open. In contrast, if α > 1 (γ < 0), the pan-genome trajectory approaches a plateau and can be considered closed as additional genomes are added (De Jesus et al., 2022).
Prediction of genes related to antibacterial activity
The NR database was chosen as the protein sequence database for our annotation system based on a balance of quality, comprehensiveness, and our practical needs. Using the complete nucleotide sequence of the Br. brevis HNCS-1 genome, the online tools antiSMASH 6.0 (Blin et al., 2021) were able to identify clusters of secondary metabolite genes.
Isolation and antibiotic activity of AMPs
One thousand milliliters of Br. brevis HNCS-1 supernatant was frozen at −80°C for 24 h and then concentrated to 20 ml by vacuum freeze drying. The concentrated sample (2 ml) was separated by column chromatography on Sephadex G-75 using an H2O-CH3OH gradient [H2O, H2O-CH3OH = 4:1 (v/v), H2O-CH3OH = 3:2 (v/v), H2O-CH3OH = 2:3 (v/v), H2O-CH3OH = 1:4 (v/v), CH3OH]. The eluent was collected using an EP tube, ranging from 0.7 ml to 1.0 ml per tube. The cell-free eluate's antibacterial activity against G. thea-sinensis was determined using agar diffusion on a PDA medium after it was filtered through a 0.22 μm filter membrane. In total, 1 ml of G. thea-sinensis spore suspension was distributed across the agar surface of the plates. Then, a sterilized cork borer was used to create 5 mm diameter wells with equal spacing in the agar. Overall, 3 days were spent incubating the dishes at 26°C. The experiment was performed thrice.
UHPLC-MS/MS method
For the separation, ultra-high-performance liquid chromatography (Exion, SCIEX) was utilized. The process utilized an Agilent ZORBAX SB-C18 column (2.1 mm × 150 mm, 2.7 μm) with acetonitrile (A) and 0.1% formic acid in water (B) as gradient elution mobile phases at a flow rate of 0.3 ml/min. The gradient elution protocol was as follows: 5% B from 0–5 min, 5–50% B from 5–20 min, 50–100% B from 20–30 min, and 100% B from 30–35 min. The temperature of the column was set to 35°C.
The X500R Q-TOF mass spectrometer (SCIEX, Framingham, MA, USA) was used to acquire untargeted mass spectral data. The LC effluent was injected into the mass spectrometer. ESI parameters were as follows: temperature: 500°C; ion source gas 1 and 2: 50 psi; curtain gas: 35 psi; CAD gas: 7 psi. Collision-induced dissociation at 35 ± 15 V in IDA mode was utilized to capture the MS and MS/MS spectra. Maximum candidate ions: 10; threshold for intensity: 600 cps; full scan mass range: 500–1,000 Da; ion discharge voltage: 5,500 V; collision energy: 35 V; collision energy spread: 15 V.
Results
Biocontrol activity of Brevibacillus brevis HNCS-1
Br. brevis HNCS-1 was evaluated for its antifungal activity against pathogenic fungi. The results demonstrated that HNCS-1 inhibited the mycelia proliferation of five tea fungal diseases significantly (Figure 1). Br. brevis HNCS-1 was isolated from tea garden soil and demonstrated broad-spectrum antagonistic activity. The mycelial growth of Phyllosticta theaefolia, Fusarium sp., Cercospora theae, and Gloeosporium theae-sinensis was completely inhibited by 10% Br. brevis HNCS-1 supernatant. Additionally, it significantly inhibits the mycelial proliferation of Elsinoe leucospira. Br. brevis HNCS-1 is a beneficial microbe with biotechnological application potential.
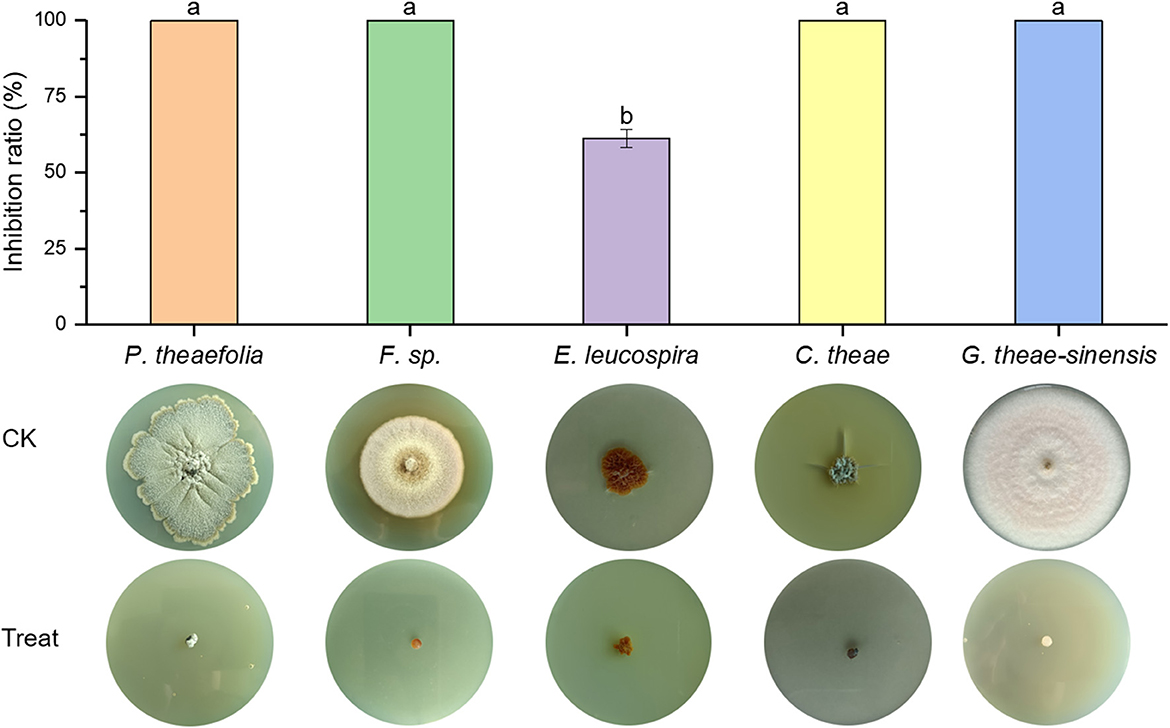
Figure 1. Antimicrobial activity of 10% Brevibacillus brevis HNCS-1 supernatant against five tea fungal diseases. The five tea fungal diseases from left to right are Phyllosticta theaefolia, Fusarium sp., Elsinoe leucospira, Cercospora theae, and Gloeosporium theae-sinensis. Different lowercase letters on the bar chart indicate significant differences in data at the 0.05 level.
General genome description of Brevibacillus brevis HNCS-1
The genome of the HNCS-1 strain consists of a single circular chromosome measuring 6,353,630 bp with a GC content of 47.15%; no plasmids were detected (GenBank CP128411). This chromosome genome consisted of 6,342 coding DNA sequences (CDS), which accounted for 87.50% of the genome. In addition, 15 sRNA, 44 rRNA, and 127 tRNA were predicted based on the chromosome sequence. In the genome, 31 Genomic Islands were identified, but no CRISPR repeat regions or prophages were found (Supplementary Figure 1).
In total, 5,796 and 4,080 identified genes were annotated as NR and Swiss-port, respectively, and 2,916 and 4,714 genes were classified into functional categories based on GO and COG designations, respectively (Supplementary Figures 2, 3). Overall, 5,810 KEGG pathways have been assigned (Supplementary Figure 4). Moreover, 4,808, 2,906, 1,190, 173, and 6 genes, respectively, were annotated in Pfm, PHI, VFDB, CAZyme, and CARD (Supplementary Figures 5, 6).
Whole-genome phylogenetic analysis of Brevibacillus brevis
The phylogenetic tree of 18 Br. brevis genomes and B. subtilis ATCC 13952 was constructed using the ML method and all orthogroups, with the ATCC 13952 strain serving as the root. It was found that HNCS-1, Br. brevis strains 17, and ATCC 13952 formed a large cluster, and different strains of Br. brevis revealed two groups (Figure 2A). In the meantime, strain HNCS-1 was a member of the same clade as strains G25-137, Ag35, ATCC 35690, GZDF3.1, NBRC100599, and LABIM17, but it was not a sister group to any other strains.
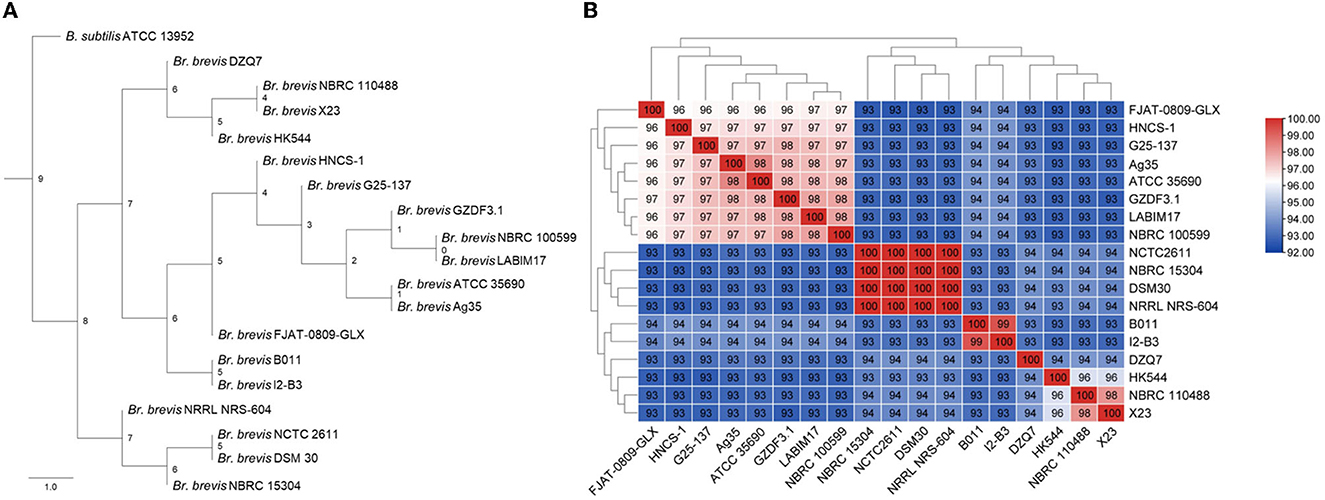
Figure 2. Phylogenetic tree of Brevibacillus brevis. (A) Maximum likelihood tree of different Br. brevis by 18 genomes and an out group (Bacillus subtilis ATCC 13952). (B) Heat map of average nucleotide identity values among different strains of Br. brevis revealing five groups.
To substantiate the results of the phylogenetic analysis, we also determined the ANI values of various strains. Strains with ANI values above 95% are regarded as belonging to the same species (Richter and Rossello-Mora, 2009). The results showed that the genetic relationship between ANI and phylogenetic trees was basically consistent, but ANI was more precise, dividing 18 strains of Br. brevis into 5 species (Figure 2B).
According to the results, they were not distinguished by the mixed trend of geographical origin and phylogenetic clustering of strains isolated from similar environments. The genome's phylogenetic clustering differed substantially from the strains' specific habitat classification.
The pan-genome features of Brevibacillus brevis
The BPGA software determined a pan-genome for the strain HNCS-1 and 17 sequenced Br. brevis strains by comparing pan-genome analyses of bacterial species by utilizing protein clustering data in the total pan-genome. There were 10,359 CDS in the total pan-genome of the 18 Br. brevis strains. Among the 10,359 protein-coding genes, 3,742 core genes represented 36.12% of the genes in the pan-genome of Br. brevis. The number of accessory gene families (3,961 genes) was greater than the number of core gene families (3,742 genes). Moreover, Br. brevis HNCS-1 contains 1,586 accessory genes. In addition, the strain HNCS-1 also contained the greatest number of distinct transcripts (560 genes) and has greater potential for gene exploration. Br. brevis strains NCTC2611 and DSM30 encoded the fewest number of specific strain genes with 4 and 6, respectively (Figure 3A).
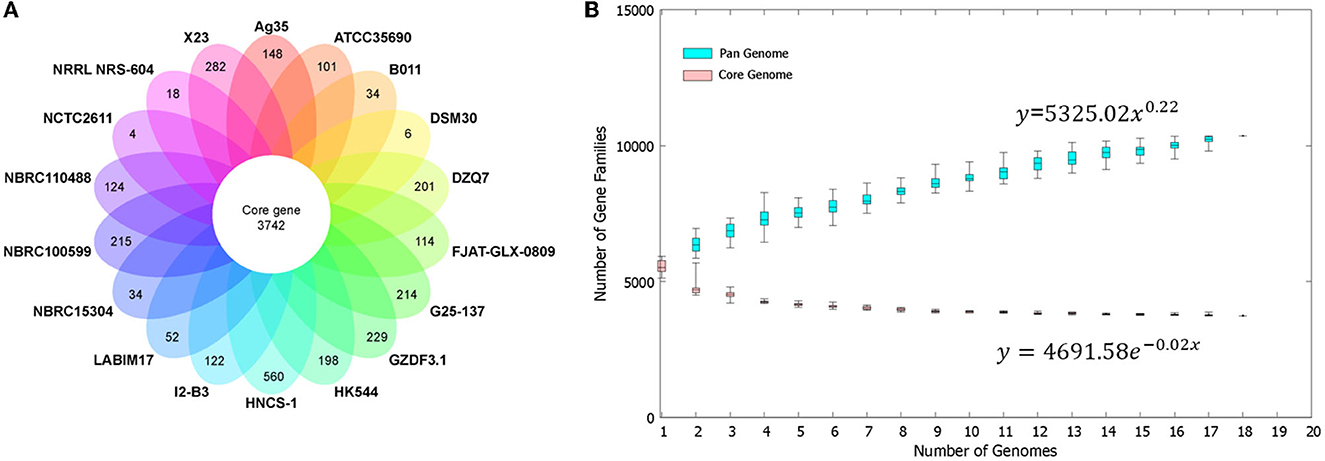
Figure 3. Pan-genome of 18 Br. brevis strains. (A) The number of unique CDS for each strain of the Br. brevis pan-genome. The inner circle shows the core genomes shared between all strains. The specific genes for each strain are indicated in each of the outer circles. (B) Curve development of pan (blue color) and core (pink color) genomes. The number of gene families is plotted in function of the genome number.
The number of Br. brevis genomes was plotted against the size of the pan-genome and the core genome. The pan-genome curve exhibited an asymptotic trend, indicating that 18 genomes were inadequate to characterize the complete gene repertoire of Br. brevis. According to the curve generated for these 18 genomes based on Heap's Law and least-square fit of exponential regression decay, the number of gene families in the pan-genome increases with the addition of each additional genome (γ = 0.22), indicating that the pan-genome of Br. brevis strains is currently open but may be closed soon (Figure 3B).
Analysis of genes encoding AMPs
It is noteworthy that the cell-free supernatant fermentation of Br. brevis inhibited numerous bacterial and fungal diseases. Additionally, the core genome sequence of Br. brevis was mined for AMP-encoding genes. Only one gene cluster implicated in edeine NRPS was identified in the core genome after NR annotation. The gene cluster likely contributes significantly to antimicrobial activity. The sequence similarity between the genes and the predicted ede BGC of Br. brevis Vm4 was high (Westman et al., 2013). The identified ede BGC in the Br. brevis HNCS-1 genome was structured identically to the Br. brevis Vm4 genome. In brief, the ede BGC (43.57 kb) in Br. brevis HNCS-1 contained 17 open reading frames, designated edeA through edeQ based on the homologous sequences in Br. brevis Vm4 (44.12 kb) (Figure 4A).
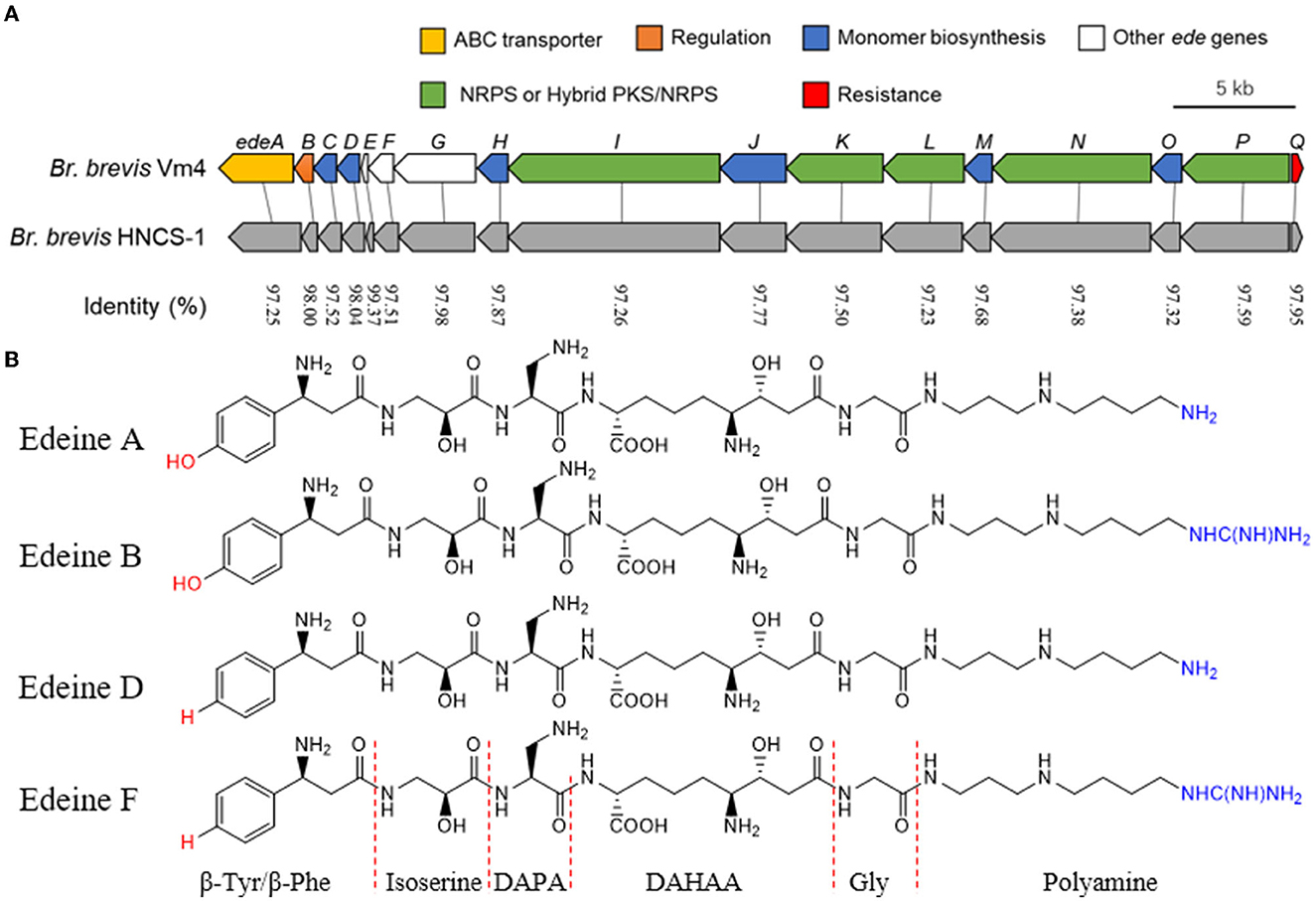
Figure 4. Characteristics of the ede BGC gene and structures of edeines. (A) Comparison of the ede BGC between Br. brevis Vm4 and Br. brevis HNCS-1. The sequences were compared by NCBI BLAST analysis. The percentage identity is displayed between homologous genes. (B) Chemical structure of edeine A, B, D, F. DAPA, 2,3-diaminopropionic acid; DAHAA, 2,6-diamino-7-hydroxyazaleic acid.
Additionally, the genome sequence of Br. brevis HNCS-1 was mined by antiSMASH software for the presence of gene-encoding AMPs. A total of 13 putative biosynthetic gene clusters were identified by the HNCS-1 strain genome (Supplementary Figure 7). Four putative gene clusters shared a high degree of similarity (>70% of genes shared similarity) with the petrobactin, tyrocidine, bacillopaline, and gramicidin gene clusters. Three putative gene clusters displayed modest similarity (<30% of genes displayed similarity) to the previously reported zwittermicin A, aurantinin B-D, and pacidamycin (1–7, D) gene clusters. Six putative gene clusters were not conserved in comparison with known clusters.
Purification and identification of AMPs
The antimicrobial peptide should be edeine, according to the results of genome annotation analysis. Edeines are a class of linear pentapeptides generated by Brevibacillus, a soil bacterium. Edeines A, B, D, and F exist as two isomers of α and β; however, only the α isomer possesses remarkable antibiotic properties (Figure 4B).
The antibacterial potential of eluent extracts containing antibacterial compounds was evaluated. Using an agar well diffusion assay, the antibacterial potential of eluent extracts from distinct collection tubes against G. theae-sinensis was determined. With an increase in eluent volume, the bacteriostatic activity initially increased steadily and then decreased dramatically (Figure 5A). The sample with the highest antibacterial activity was analyzed by UPLC-MS/MS for additional confirmation. According to the total ion chromatogram, the majority of the ultraviolet absorption is of short wavelength.
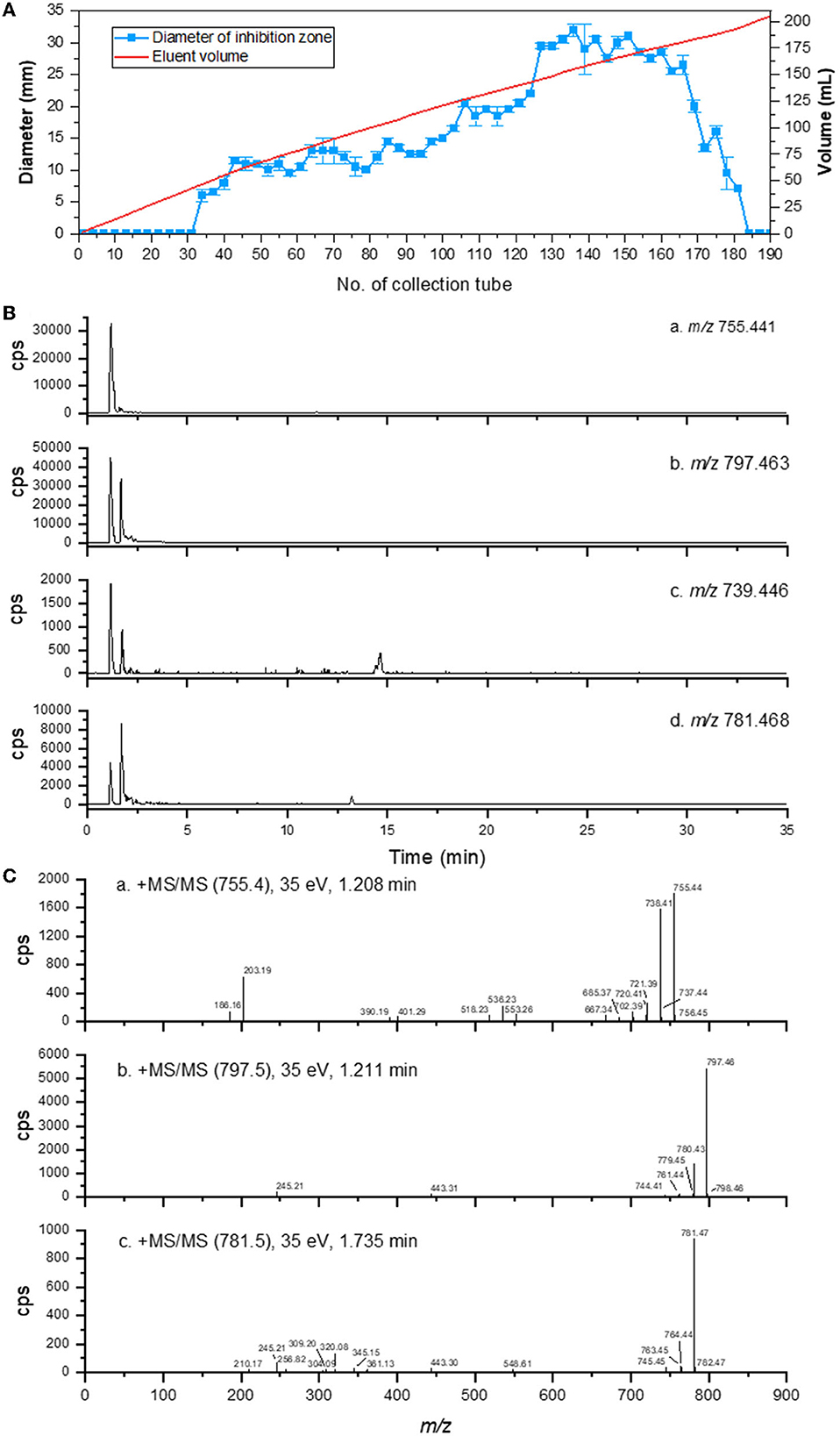
Figure 5. Purification and UHPLC-MS/MS analysis of AMPs from Brevibacillus brevis HNCS-1. (A) Antimicrobial activity of crude extract of HNCS-1 after elution with H2O/CH3OH. (B) The ion current of m/z 755, 797, 739, and 781. (C) MS/MS spectra of m/z 755, 797, and 781.
The theoretical molar masses of edeine A (C33H58N10O10), edeine B (C34H60N12O10), edeine D (C33H58N10O9), and edeine F (C34H60N12O9) are 754.4337, 796.4555, 738.4388, and 780.4606. Compound 1 (m/z 755), Compound 2 (m/z 797), Compound 3 (m/z 739), and Compound 4 (m/z 781) were extracted from the total ion current based on the ion mass spectral (m/z). Compound 1 (m/z 755) produced a [M+H]+ ion peak m/z 755.4132 (−1.7 ppm, C33H58N10O10) at a retention time of 1.182 min (Figure 5B-a). The electrospray ionization-mass spectrometry results indicated that its fragmentation pattern was identical to that of edeine A, including m/z 738, 737, 720, 667, 553, 536, 401, 390, and 203 (Figure 5C-a, Supplementary Figure 8). Compound 2 (m/z 797) produced a [M+H]+ ion peak m/z 797.4612 (−2.6 ppm, C34H60N12O10) at a retention time of 1.165 min (Figure 5B-b) with observable signals. The electrospray ionization-mass spectrometry results indicated that its fragmentation pattern was identical to that of edeine B, including m/z 780, 779, 761, 443, and 245 (Figure 5C-b, Supplementary Figure 9). Compound 3 (m/z 739) had faint signals at the retention time of 1.165 min (Figure 5B-c), but its peak value was too low to be analyzed further based on the available mass spectrum data. Compound 4 (m/z 781) produced a [M+H]+ ion peak m/z 781.4667 (−2.2 ppm, C34H60N12O9) at the retention time of 1.708 min (Figure 5B-d). The electrospray ionization-mass spectrometry results indicated that its fragmentation pattern was identical to that of edeine B, including m/z 780, 779, 761, 443, and 245 (Figure 5C-c, Supplementary Figure 10). To sum up, edeines were a class of highly polar AMPs with limited retention on reverse phase chromatography, which was consistent with the chromatographic retention property of the compounds discussed previously. Compounds 1, 2, and 4 were edeine A, B, and F, respectively. The chromatographic peak area suggested that edeine B had the maximum concentration, followed by edeine B and F had the lowest concentration.
In addition, to confirm the existence of secondary metabolites predicted by antiSMASH software, we extracted relevant data from the total ion current based on the theoretical molecular weight of petrobactin, tyrocidine, bacillopaline, and gramicidin, but they were not detected (Supplementary Figures 11–14).
Discussion
Allelopathy is a pervasive natural phenomenon in ecosystems, promoting and inhibiting the proliferation of diverse organisms. People have always been interested in the rational application of biocontrol's antagonistic effect on agricultural resistance and disease control. Tea plant diseases are difficult to control, resulting in substantial losses in yield and quality. Although chemical fungicides have some effect on the control of tea plant diseases, their misuse has resulted in severe environmental pollution issues. Utilizing biocontrol microorganisms to combat plant maladies is a popular topic of discussion. In certain instances, biocontrol microorganisms have been shown to be as effective as chemical fungicides in preventing plant diseases. Several bacterial species, such as Bacillus, Brevibacillus, and Pseudomonas, have been commercialized as biological control agents (Zhang et al., 2005; Paterson et al., 2017). Because of their extensive colonization capabilities, spore-forming Bacillus and Brevibacillus preparations are preferred for the development of commercial products. In this study, we isolated strain HNCS-1 of Br. brevis from tea garden soil that exhibits broad-spectrum resistance to five fungal diseases of tea plants. The HNCS-1 strain is a beneficial microorganism with prospective biotechnological application potential.
The HNCS-1 strain and 17 Br. brevis isolates from NCBI were subjected to a phylogenetic analysis using orthologous genes organized into four clusters. However, the strains of the same cluster originate from diverse habitats, indicating that Br. brevis is highly adaptable to its environment and can populate a variety of ecological niches. We examined the pan-genome of Br. brevis species. The results indicate that the pan-genome of Br. brevis is open and theoretically infinite, indicating that Br. brevis species tend to acquire new genes to improve adaptability. Bacteria could modify their genetic material to adapt to varying environmental conditions, resulting in increased niche diversity and larger pan-genomes (Konstantinidis and Tiedje, 2004). The HNCS-1 strain has more protein-coding genes (6,342) than other Br. brevis strains, making it more environmentally adaptable.
In addition, the traditional separation methods of antimicrobial substances have limitations. According to previous studies, this study tried to extract antimicrobial substances through the organic solvent extraction method, hydrochloric acid precipitation method, and ammonium sulfate precipitation method but never succeeded. Genome sequencing analysis and pan gene analysis open a new door for the identification of antimicrobial substances, which can quickly and accurately target antimicrobial substances and lay a theoretical foundation for the separation and application of antimicrobial substances.
A comparison of Br. brevis HNCS-1 proteins with those of other Br. brevis strains revealed 3,742 core genes. There is only one NRPS gene cluster annotated as edeine in the core genes. It is demonstrated that edeine is the antibacterial peptide at the center of Br. brevis. After a blast comparison, it was determined that the gene cluster of the HNCS-1 strain was extremely similar to the Br. brevis Vm4 gene cluster for edeine NRPS (identity >97%). To validate the accuracy of comprehensive genomic analysis, we used the UHPLC-MS/MS technique to identify edeine A, B, and F from the crude extract of the HNCS-1 strain, with edeine B and edeine A serving as the principal AMPs. The main species of edeine in the HNCS-1 strain differ from Br. brevis X23 (Du et al., 2022), which may be owing to the evolutionary tree's distant genetic relationship.
Edeine, as an AMP of the core genes of Br. brevis, should be abundant in the organism's secondary metabolites. Edeine was extracted from Br. brevis Vm4 and X23 by resin adsorption, respectively (Westman et al., 2013; Liu et al., 2022). However, ethylparaben from Br. brevis FJAT-0809-GLX was extracted by solvent extraction (Che et al., 2015), tostadin from Br. brevis XDH was extracted by ammonium sulfate precipitation (Song et al., 2012), siderophore from Br. brevis GZDF3 was extracted by ethanol precipitation (Sheng et al., 2020), and surfactin from Br. brevis KN8(2) was extracted by hydrochloric acid precipitation (Krishnan et al., 2019). The inability to extract edeine from the secondary metabolites of Br. brevis may have been caused by a mismatch in extraction techniques. Due to the ability of Br. brevis to acquire new genes from the environment, its antimicrobial peptides are diverse. The pan-genome analysis identified 560 HNCS-1 strain-specific genes. The unique genomes of the HNCS-1 strain require additional study. Additionally, genetic engineering is an excellent research direction. For instance, using in situ promoter engineering, the production of edeine in Br. brevis X23 was increased. Br. brevis HNCS-1 requires additional research, including the one strain many compounds approach (OSMAC) and genetic modification, to investigate novel compounds and yield enhancement.
Data availability statement
The data presented in the study are deposited in the GenBank repository, accession number CP128411.
Author contributions
CX, LZ, and XQ conceived and planned the experiments, funding acquisition, and revisions. WY and HY were involved in data and bioinformatics analyses, conducted experiments, and wrote the manuscript. XB and MH contributed to the interpretation of the results. QB and ZZ contributed to sample preparation. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (32072625 and 31660541), the Natural Science Foundation of Hunan Province (2021JJ30385), the Transformation and Industrialization Plan of Scientific and Technological Achievements of Hunan Province (2021NK1020), and the National Key Research and Development Plan (2022YFD1600800).
Acknowledgments
The authors would like to thank the Tea Research Institute, Hunan Academy of Agricultural Sciences for the assistance with experimental materials.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1198747/full#supplementary-material
References
Baindara, P., Singh, N., Ranjan, M., Nallabelli, N., Chaudhry, V., Pathania, G. L., et al. (2016). Laterosporulin10: a novel defensin like Class IId bacteriocin from Brevibacillus sp. strain SKDU10 with inhibitory activity against microbial pathogens. Microbiology 162, 1286–1299. doi: 10.1099/mic.0.000316
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Wezel, G. P., Medema, M. H., et al. (2021). AntiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6, 24373. doi: 10.1038/srep24373
Che, J. M., Liu, B., Chen, Z., Shi, H., Liu, G. H., Ge, C. B., et al. (2015). Identification of ethylparaben as the antimicrobial substance produced by Brevibacillus brevis FJAT-0809-GLX. Microbiol. Res. 172, 48–56. doi: 10.1016/j.micres.11007
Che, J. M., Ye, S. W., Liu, B., Deng, Y. Y., Chen, Q. Q., Ge, C. B., et al. (2016). Effects of Brevibacillus brevis FJAT-1501-BPA on growth performance, faecal microflora, faecal enzyme activities and blood parameters of weaned piglets. Antonie Van Leeuwenhoek Int. J. General Mol. Microbiol. 109, 1545–1553. doi: 10.1007/s10482-016-0756-8
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.06009
Cuellar-Gaviria, T. Z., García-Botero, C., Ju, K-. S., and Villegas-Escobar, V. (2023). The genome of Bacillus tequilensis EA-CB0015 sheds light into its epiphytic lifestyle and potential as a biocontrol agent. Front. Microbiol. 14, 5487. doi: 10.3389./fmicb.2023.1135487
Czajgucki, Z., Andruszkiewicz, R., and Kamysz, W. (2006). Structure activity relationship studies on the antimicrobial activity of novel edeine A and D analogues. J. Pept. Sci. 12, 653–662. doi: 10.1002/psc.775
De Jesus Aburjaile, L. C. L., Sousa, F. F., Felice, T. J., Soares, A. G., Alcantara, S. C., et al. (2022). Genomic characterization of lactobacillus delbrueckii strains with probiotics properties. Front. Bioinform 2, 912795. doi: 10.3389/fbinf.2022.912795
Desjardine, K., Pereira, A., Wright, H., Matainaho, T., Kelly, M., Andersen, R. J., et al. (2007). Tauramamide, a lipopeptide antibiotic produced in culture by Brevibacillus laterosporus isolated from a marine habitat: structure elucidation and synthesis. J. Nat. Prod. 70, 1850–1853. doi: 10.1021/np070209r
Dinos, G., Wilson, D. N., Teraoka, Y., Szaflarski, W., Fucini, P., Kalpaxis, D., et al. (2004). Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site RNA binding. Mol. Cell 13, 113–124. doi: 10.1016/s1097-2765(04)00002-4
Du, J., Zhang, C., Long, Q., Zhang, L., Chen, W., Liu, Q., et al. (2022). Characterization of a pathway-specific activator of edeine biosynthesis and improved edeine production by its overexpression in Brevibacillus brevis. Front. Plant Sci. 13, 1022476. doi: 10.3389/fpls.2022.1022476
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238. doi: 10.1186/s13059-019-1832-y
Gerard, J. M., Haden, P., Kelly, M. T., and Andersen, R. J. (1999). Loloatins A-D, cyclic decapeptide antibiotics produced in culture by a tropical marine bacterium. J. Nat. Prod. 62, 80–85. doi: 10.1021/np980219f
Ghadbane, M., Harzallah, D., Laribi, A. I., Jaouadi, B., and Belhadj, H. (2013). Purification and biochemical characterization of a highly thermostable bacteriocin isolated from Brevibacillus brevis strain GM100. Biosci. Biotechnol. Biochem. 77, 151–160. doi: 10.1271/bbb.120681
Govaerts, C., Orwa, J., Van Schepdael, A., Roets, E., and Hoogmartens, J. (2001). Structure elucidation of four related substances in gramicidin with liquid chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 15, 128–134. doi: 10.1002/1097-0231(20010130)15:2<128::AID-RCM205>3.0.CO;2-O
Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114. doi: 10.1038/s41467-018-07641-9
Kessler, N., Schuhmann, H., Morneweg, S., Linne, U., and Marahiel, M. A. (2004). The linear pentadecapeptide gramicidin is assembled by four multimodular nonribosomal peptide synthetases that comprise 16 modules with 56 catalytic domains. J. Biol. Chem. 279, 7413–7419. doi: 10.1074/jbc.M309658200
Konstantinidis, K. T., and Tiedje, J. M. (2004). Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA. 101, 3160–3165. doi: 10.1073/pnas.0308653100
Krishnan, N., Velramar, B., and Velu, R. K. (2019). Investigation of antifungal activity of surfactin against mycotoxigenic phytopathogenic fungus Fusarium moniliforme and its impact in seed germination and mycotoxicosis. Pestic. Biochem. Physiol. 155, 101–107. doi: 10.1016/j.pestbp.01010
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Liu, Q., Zhang, L., Wang, Y., Zhang, C., Liu, T., Duan, C., et al. (2022). Enhancement of edeine production in Brevibacillus brevis X23 via in situ promoter engineering. Microb. Biotechnol. 15, 577–589. doi: 10.1111/1751-7915.13825
Mootz, H. D., and Marahiel, M. A. (1997). The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J. Bacteriol. 179, 6843–6850. doi: 10.1128/jb.179.21.6843-6850.1997
Pan, S. Y., Nie, Q., Tai, H. C., Song, X. L., Tong, Y. F., Zhang, L. J., et al. (2022). Tea and tea drinking: China's outstanding contributions to the mankind. Chin. Med. 17, 27. doi: 10.1186/s13020-022-00571-1
Paterson, J., Jahanshah, G., Li, Y., Wang, Q., Mehnaz, S., Gross, H., et al. (2017). The contribution of genome mining strategies to the understanding of active principles of PGPR strains. FEMS Microbiol. Ecol. 93, 249. doi: 10.1093./femsec/fiw249
Richter, M., and Rossello-Mora, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Ruiu, L. (2013). Brevibacillus laterosporus, a pathogen of invertebrates and a broad-spectrum antimicrobial species. Insects 4, 476–492. doi: 10.3390/insects4030476
Samrot, A. V., Prasad, R. J., Rio, A. J., and Sneha, S. J. (2015). Bioprospecting of Brevibacillus brevis isolated from soil. Recent Pat. Biotechnol. 9, 42–49. doi: 10.2174/187220830901151111124333
Sanjay, R., Ponmurugan, P., and Baby, U. I. (2008). Evaluation of fungicides and biocontrol agents against grey blight disease of tea in the field. Crop Prot. 27, 689–694. doi: 10.1016/j.cropro.09014
Sheng, M. M., Jia, H. K., Zhang, G. Y., Zeng, L. N., Zhang, T. T., Long, Y. H., et al. (2020). Siderophore Production by rhizosphere biological control bacteria brevibacillus brevis GZDF3 of pinellia ternata and its antifungal effects on Candida albicans. J. Microbiol. Biotechnol. 30, 689–699. doi: 10.4014/jmb.1910.10066
Shida, O., Takagi, H., Kadowaki, K., and Komagata, K. (1996). Proposal for two new genera, Brevibacillus gen. nov. and Aneurinibacillus gen. nov. Int J Syst Bacteriol 46, 939–946. doi: 10.1099/00207713-46-4-939
Shoji, J., and Kato, T. (1976). The structure of brevistin. Studies on antibiotics from the genus Bacillus. X. J. Antibiot. 29, 380–389. doi: 10.7164/antibiotics.29.380
Singh, P. K., Chittpurna, Ashish, Sharma, V., Patil, P.B., and Korpole, S. (2012). Identification, purification and characterization of laterosporulin, a novel bacteriocin produced by Brevibacillus sp. strain GI-9. PLoS ONE 7, e31498. doi: 10.1371/journal.pone.0031498
Song, Z., Liu, Q. X., Guo, H., Ju, R. C., Zhao, Y. H., Li, J. Y., et al. (2012). Tostadin, a novel antibacterial peptide from an antagonistic microorganism Brevibacillus brevis XDH. Bioresour. Technol. 111, 504–506. doi: 10.1016/j.biortech.02051
Szer, W., and Kurylo-Borowska, Z. (1972). Interactions of edeine with bacterial ribosomal subunits. Selective inhibition of aminoacyl-tRNA binding sites. Biochim Biophys Acta 259, 357–368. doi: 10.1016/0005-2787(72)90310-3
Takeuchi, T., Iinuma, H., Kunimoto, S., Masuda, T., Ishizuka, M., Takeuchi, M., et al. (1981). A new antitumor antibiotic, spergualin: isolation and antitumor activity. J. Antibiot. 34, 1619–1621. doi: 10.7164/antibiotics.34.1619
Tamaki, M., Takimoto, M., Sofuku, S., and Muramatsu, I. (1983). Synthetic studies on gratisin. II. J. Antibiot. 36, 751–752. doi: 10.7164/antibiotics.36.751
Wan, X., Takala, T. M., Huynh, V. A., Ahonen, S. L., Paulin, L., Björkroth, J., et al. (2023). Comparative genomics of 40 Weissella paramesenteroides strains. Front. Microbiol. 14, 28. doi: 10.3389./fmicb.2023.1128028
Wang, L., Tang, L., Wang, R., Wang, X., Ye, J., Long, Y., et al. (2016). Biosorption and degradation of decabromodiphenyl ether by Brevibacillus brevis and the influence of decabromodiphenyl ether on cellular metabolic responses. Environ. Sci. Pollut. Res. Int. 23, 5166–5178. doi: 10.1007/s11356-015-5762-2
Westman, E. L., Yan, M., Waglechner, N., Koteva, K., and Wright, G. D. (2013). (2013). Self resistance to the atypical cationic antimicrobial peptide edeine of Brevibacillus brevis Vm4 by the N-acetyltransferase EdeQ. Chem. Biol. 20, 983–990. doi: 10.1016/j.chembiol.06010
Wu, X., Ballard, J., and Jiang, Y. W. (2005). Structure and biosynthesis of the BT peptide antibiotic from Brevibacillus texasporus. Appl. Environ. Microbiol. 71, 8519–8530. doi: 10.1128/AEM.71.12.8519-8530.2005
Xu, C., Qin, C., Zhang, R., Niu, W., and Shang, X. (2010). Solid-phase synthesis and antibiotic activities of cyclodecapeptides on the scaffold of naturally occurring Laterocidin. Bioorg. Med. Chem. Lett. 20, 164–167. doi: 10.1016/j.bmcl.11009
Xu, Y., Wang, Q., Dong, M., Song, H., Hang, B., Sun, Y., et al. (2023). Evaluation of the efficacy of the antimicrobial peptide HJH-3 in chickens infected with Salmonella Pullorum. Front. Microbiol. 14, 1102789. doi: 10.3389/fmicb.2023.1102789
Yang, X., Huang, E., and Yousef, A. E. (2017). Brevibacillin, a cationic lipopeptide that binds to lipoteichoic acid and subsequently disrupts cytoplasmic membrane of Staphylococcus aureus. Microbiol. Res. 195, 18–23. doi: 10.1016/j.micres.11002
Yang, X., and Yousef, A. E. (2018). Antimicrobial peptides produced by Brevibacillus spp.: structure, classification and bioactivity: a mini review. World J. Microbiology and Biotechnology 34, 4. doi: 10.1007./s11274-018-2437-4
Zhang, X., Zhang, B. X., Zhang, Z., Shen, W. F., Yang, C. H., Yu, J. Q., et al. (2005). Survival of the biocontrol agents Brevibacillus brevis ZJY-1 and Bacillus subtilis ZJY-116 on the spikes of barley in the field. J. Zhejiang Univ. Sci. B 6, 770–777. doi: 10.1631/jzus.2005.B0770
Keywords: Brevibacillus brevis, antagonistic activity, genome annotation, pan-genome analysis, antimicrobial peptides, edeine
Citation: Yang W, Yang H, Bao X, Hussain M, Bao Q, Zeng Z, Xiao C, Zhou L and Qin X (2023) Brevibacillus brevis HNCS-1: a biocontrol bacterium against tea plant diseases. Front. Microbiol. 14:1198747. doi: 10.3389/fmicb.2023.1198747
Received: 02 April 2023; Accepted: 15 August 2023;
Published: 13 September 2023.
Edited by:
Yasmeen Siddiqui, Universiti Putra Malaysia, MalaysiaReviewed by:
Bin Li, Zhejiang University, ChinaDixit Sharma, Central University of Himachal Pradesh, India
Copyright © 2023 Yang, Yang, Bao, Hussain, Bao, Zeng, Xiao, Zhou and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun Xiao, eC5jaHVuQHltYWlsLmNvbQ==; Lingyun Zhou, aG5jeXN6bHlAaHVuYWFzLmNu; Xiaoping Qin, cXhwOTlAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Wenbo Yang
Wenbo Yang Hui Yang2†
Hui Yang2† Mehboob Hussain
Mehboob Hussain Xiaoping Qin
Xiaoping Qin