- 1Institute of Soil and Fertilizer, Academy of Agricultural Sciences/Fujian Key Laboratory of Plant Nutrition and Fertilizer, Fuzhou, China
- 2College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 3College of Resources and Environment, Institute of Environmental Microbiology, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China
Serratia marcescens is a gram-negative bacterium that is able to produce many secondary metabolites, such as the prominent red pigment prodigiosin (PG). In this work, a ptrA-disrupted mutant strain with reduced PG production was selected from Tn5 transposon mutants. RT–qPCR results indicated that ptrA promoted elevated transcription of the pig gene cluster in S. marcescens FZSF02. Furthermore, we found that ptrA also controls several other important biological functions of S. marcescens, including swimming and swarming motilities, biofilm formation, hemolytic activity, and stress tolerance. In conclusion, this study demonstrates that ptrA is a PG synthesis-promoting factor in S. marcescens and provides a brief understanding of the regulatory mechanism of ptrA in S. marcescens cell motility and hemolytic activity.
1. Introduction
S. marcescens is a widely distributed gram-negative, rod-shaped bacterium found in soil, water, plant surfaces, and insects (Grimont and Grimont, 1978) and is used for the production of many high-value products, including prodigiosin (Han et al., 2021), serratamolide (Clements et al., 2019), acetoin (Bai et al., 2015), althiomycin (Gerc et al., 2012), and 2,3-butanediol (Zhang et al., 2010). Therefore, S. marcescens has become an important microbial strain for industrial applications. In addition, S. marcescens is also a well-known opportunistic pathogen that infects plants, invertebrates, and vertebrates (Mahlen, 2011). S. marcescens has been shown to cause septicaemia (Wilfert et al., 1968), empyema (Nakashima et al., 1987), meningitis (Psautter et al., 1984) and other diseases.
Prodigiosin is a tripyrrole red secondary metabolite of microbial origin that has antibacterial (Lapenda et al., 2015), algicidal (Wei et al., 2020), antimalarial (Papireddy et al., 2011), immunomodulatory (Chang et al., 2011), and antitumor activities (Zhang et al., 2005; Pan et al., 2012). The synthesis of prodigiosin by S. marcescens is mainly controlled by 14 genes, including pigA-N, located in the same operon (Williamson et al., 2006). Environmental factors, such as nutrient deprivation, temperature, oxygen, pH, light, ionic strength, and phosphate availability, were also shown to affect prodigiosin generation (Solé et al., 1994; Bennett and Bentley, 2000; Slater et al., 2003; Giri et al., 2004; Williamson et al., 2006; Ryazantseva et al., 2012). Although the synthetic process of prodigiosin in S. marcescens has been elucidated in recent years, the understanding of the regulatory mechanisms responsible for prodigiosin biosynthesis in S. marcescens is still limited.
The protein encoded by ptrA is the metalloendopeptidase pitrilysin (Anastasi and Barrett, 1995). Pitrilysin has been generated by heterologous expression in Escherichia coli and was able to degrade small molecular polypeptides, such as insulin B chain, but could not degrade proteins, so it was also called an “oligopeptidase” type of endopeptidase (Anastasi et al., 1993).
In this study, the prodigiosin-producing wild-type strain S. marcescens FZSF02 was isolated from soil samples (Lin et al., 2019). In the work, a ptrA-disrupted mutant strain with reduced PG production was selected from Tn5 transposon mutants and ΔptrA was constructed in the wild-type strain FZSF02 with the homologous recombination method. The mechanism for positive regulation of prodigiosin production by PtrA protein was explored. Interestingly, PtrA also regulated swarming and swimming motilities, biofilm formation, hemolytic activity and stress resistance in S. marcescens. Our study data showed a novel regulator PtrA, which is important for various cellular processes in S. marcescens.
2. Materials and methods
2.1. Bacterial strains, plasmids, and culture conditions
S. marcescens FZSF02 is a prodigiosin-producing strain isolated from a soil sample (Lin et al., 2019). Mutant E35 displays reduced production of prodigiosin and was isolated from a Tn5 transposon insertion mutant library of strain FZSF02 (Jia et al., 2021). S. marcescens strains were incubated in Luria–Bertani (LB) medium (10 g tryptone, 5 g yeast extract and 5 g NaCl per liter) at 27°C or 37°C.
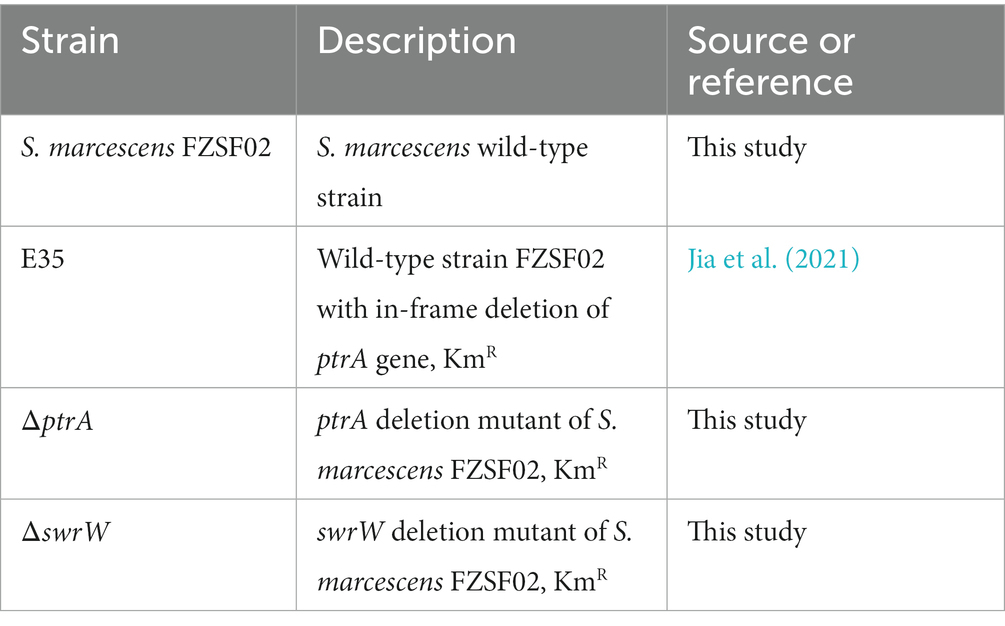
Antibiotics were added when required at the following final concentration: kanamycin (100 μg/mL). The bacterial strains and plasmids used in this study are listed in Table 1. The primers used in this study are listed in Table 2.
2.2. Construction of the ptrA gene deletion mutant
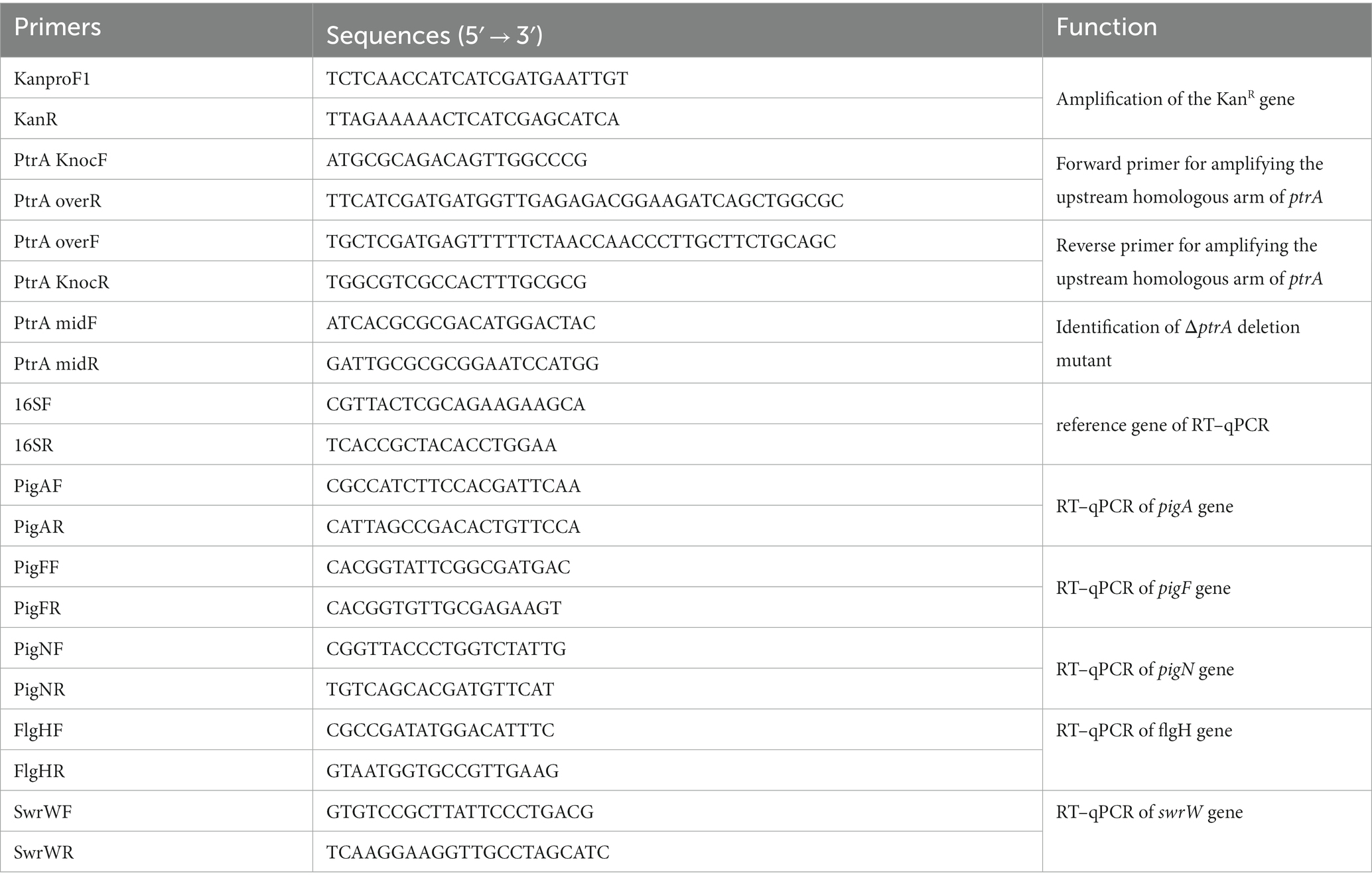
ptrA was screened and identified from previously constructed Tn5 transposon insertion mutant library of strain FZSF02 (Jia et al., 2021). The homologous recombination method was used to inactivate genes. Briefly, the forward homologous sequence of approximately 1,000 bp was amplified with primers PtrA knocF and PtrA overR, and the backward homologous sequence of approximately 1,000 bp was amplified with primers PtrA overF and PtrA knocR. The kanamycin resistance gene was amplified with primers KanproF and KanR; these three sequences were spliced by overlapping PCR. The overlapping PCR products were purified and then transformed into S. marcescens FZSF02 by using Gene Pulser Xcell (Bio-Rad) for electroporation for subsequent homologous recombination deletion of ptrA. Then, colonies containing deletions of the ptrA gene were selected on LB agar medium containing kanamycin at 27°C and identified by PCR, and ΔptrA was finally confirmed by sequence comparison.
2.3. Prodigiosin production assays
Determination of the prodigiosin yield of strains FZSF02 (WT) and ΔptrA was carried out with a modified method as reported previously (Kalivoda et al., 2010). Briefly, single colonies of WT and ΔptrA were selected from plates and inoculated into LB liquid medium overnight culture at 180 r/min at 37°C. One milliliter of the broth was transferred into 50 mL of fresh LB liquid medium and cultured with shaking at 27°C and 180 r/min for 24 h. The broth was properly diluted with acidified methanol (4 mL of 1 mol/L HCl and 96 mL of methanol) and shaken vigorously; after 10 min of standing, the mixture was centrifuged at 8,000 × g for 10 min, and the supernatant was removed for prodigiosin quantification. Prodigiosin production can be evaluated by the absorbance value at 535 nm (de Araújo et al., 2010). Optical densities of cultures were measured at 535 and 600 nm wavelength at time intervals of 0, 3, 6, 9, 12, 15, 18, 21, 24, 36 and 48 h, relative prodigiosin production of different strains was calculated by the value of A535/OD600, where the OD600 values of the fermentation broth represent the biomass of the strains. Experiments were independently replicated three times.
2.4. Growth curve measurements
To analyze the growth curve of strains FZSF02 and ΔptrA, the exponential-phase cells (OD600 of 1.0) of these two strains were inoculated in fresh LB medium at a 2% inoculation volume. Optical densities of cultures were measured at 600 nm wavelength at time intervals of 0, 3, 6, 9, 12, 15, 18, 21, 24, 36, and 48 h, and the growth curves were plotted as values at OD600 versus the incubation time. Experiments were independently replicated three times.
2.5. RNA extraction and quantitative real-time PCR
The samples were treated with the TransZol Up Plus RNA Kit (ER501; TransGen, Beijing, China) for total RNA extraction, and the total bacterial RNA was subjected to reverse transcription using FastKing gDNA Dispelling RT SuperMix (KR118; Tiangen, Beijing, China).
The RT-qPCRs were carried out in a final volume of 20 μL according to the manufacturer’s instructions, using 1 μL of the cDNA dilution as template, and were mixed with 0.4 μL of forward and reverse primers. The mixture was then exposed to RT–qPCR analysis using TransStart® Green qPCR SuperMix (AQ101-02; TransGen, Beijing, China) in the reaction in QuantStudio™ 6 Flex Real-Time PCR System Software (Applied Biosystems). Gene expression levels were measured using the 16S rRNA gene as a control using the 16SF and 16SR primers (Table 2). Gene expression levels of mutant strains were determined using the 2−ΔΔCT method with the relative fold-difference expression against the wild-type strain. The RT–qPCR from three biological replicates was analyzed, and three technical replicates were performed.
2.6. Motility assays
Swimming and swarming motility assays were performed by dropping 2 μL of the exponential phase cells (OD600 of 1.0) of both the FZSF02 and the ΔptrA strain onto the center of LB media plates with 0.3% (Swimming) or 0.7% (Swarming) BD agar (BD: Becton, Dickinson and Company), respectively (Soo et al., 2008, 2014). The swarming and swimming zones were observed, and the migration diameter was measured after incubation at 27°C for 12 h. Each group was independently repeated three times.
2.7. Biofilm analysis
The ability of the strains to form biofilms was assayed according to a previously reported method (Shanks et al., 2007). In detail, 1 mL of exponential-phase broth (OD600 of 1.0) was added to 50 mL of liquid LB medium, and then 200 μL of the mixed broth was added into the wells of 96-well microtiter plates (No. 655180. Greiner Bio-One). After 48 h of nonshaking incubation at 27°C, the optical density at OD600 was measured and recorded. After that, the biofilm was gently washed with distilled water five times and stained with 200 μL of crystal violet solution (0.1% w/v) for 15 min. Thereafter, the biofilm was gently washed with distilled water five times, and crystal violet was extracted with 200 μL of anhydrous ethanol. Finally, the optical density of the biofilm was measured at a wavelength of 595 nm using the SPECTRA MAX190 microplate reader (Molecular Devices). Biofilm yield ability was calculated by comparing the ratio of the value at OD595 to the value at OD600.
2.8. Stress tolerance assays
Fresh broth of FZSF02 and ΔptrA strains was adjusted to a density of OD600 1.0 with sterile water and used for the following tests. For H2O2, acid and osmotic tolerance assays, the broth was serially diluted (10-fold) with sterile water, and then 2 μL of the solutions were spotted onto LB agar medium containing 2 mM H2O2, 175 mM acetic acid and 3 M NaCl. For heat tolerance assays, the bacteria were treated at 50°C for 20 min, serially diluted (10-fold) with sterile water, and finally spotted onto LB agar medium. All plates were incubated for 24 h at 27°C.
Meanwhile, the CFU counting method was used to quantitatively evaluate the tolerance ability for H2O2, acid, osmotic and heat. For H2O2, the bacterial broth was treated with 2 mM H2O2 (prepared with 0.9% NaCl) for 5, 15, 30, and 45 min. For heat tolerance assays, the bacterial broth was treated at 50°C for 0, 5, 10, 15, and 20 min. For acid tolerance assays, the bacterial broth was treated with 175 mM acetic acid (prepared with 0.9% NaCl) for 0, 5, 10, 15 and 20 min. For osmotic tolerance assays, the bacterial broth was treated with 3 M NaCl for 0, 5, 10, 15 and 20 min. All treatments above were diluted (10-fold), spread on LB agar plates and incubated for 24 h at 27°C followed by CFU counting.
2.9. Catalase activity
Catalase activity was performed as previously described (de Ondarza, 2017). Briefly, the strains were cultured in liquid LB medium at 27°C and 180 r/min for 24 h. The cells in the broth were crushed by ultrasound and centrifuged at 4,000 r/min at 4°C for 10 min, and the subsequently obtained supernatant was a crude enzyme solution of catalase. To assay the catalase activity, 2.9 mL of 2 mM H2O2 (prepared by adding 0.15 mL of 30% H2O2 to 50 mL of PBS buffer) and 0.1 mL of diluted enzyme solution were mixed, and recording was immediately carried out at 240 nm every 20 s for a total time of 2 min. Catalase activity was calculated by the following formula: (ΔA240/min * dilution ratio)/(43.6/mol/min * sample volume * 0.001).
2.10. Hemolytic activity assay
Hemolytic activity was assayed as previously described (Di Venanzio et al., 2014; Ridder et al., 2021). Briefly, 2 μL of exponential-phase culture (OD600 = 1.0) was spotted on a blood agar plate (Hunan BKMAM Biotechnology Co., Ltd) and cultured at 27°C and 37°C for 5 days, and the diameters of the hemolytic transparent zones were measured. Experiments were independently repeated three times.
2.11. Statistical analysis
Student’s t tests or one-way ANOVA were used to compare significant differences between experimental groups, while multiple comparisons were conducted using one-way ANOVA with GraphPad Prism software. All experiments were independently replicated at least three times in this study.
3. Results and discussion
3.1. Identification of a prodigiosin synthesis activator PtrA
A mutant E35 with positively regulated synthesis of prodigiosin was isolated from a Tn5 transposon mutant library constructed with Serratia marcescens FZSF02. Transposon Tn5 was inserted in the coding region between 1,071 bp and 1,072 bp of the HMI62_20735 gene in E35, which encodes a protein with 100% identity to predicted PtrA (GenBank accession number QJU42755.1) (Figure 1A). PtrA of FZSF02 displayed 66.84, 66.84 and 66.53% similarities with E. coli CFT073 (Q8CVS2.1), Shigella flexneri (Q83QC3-1) and Salmonella enterica sp. LT2 (Q8ZMB5.1), respectively (Supplementary Figure S1).
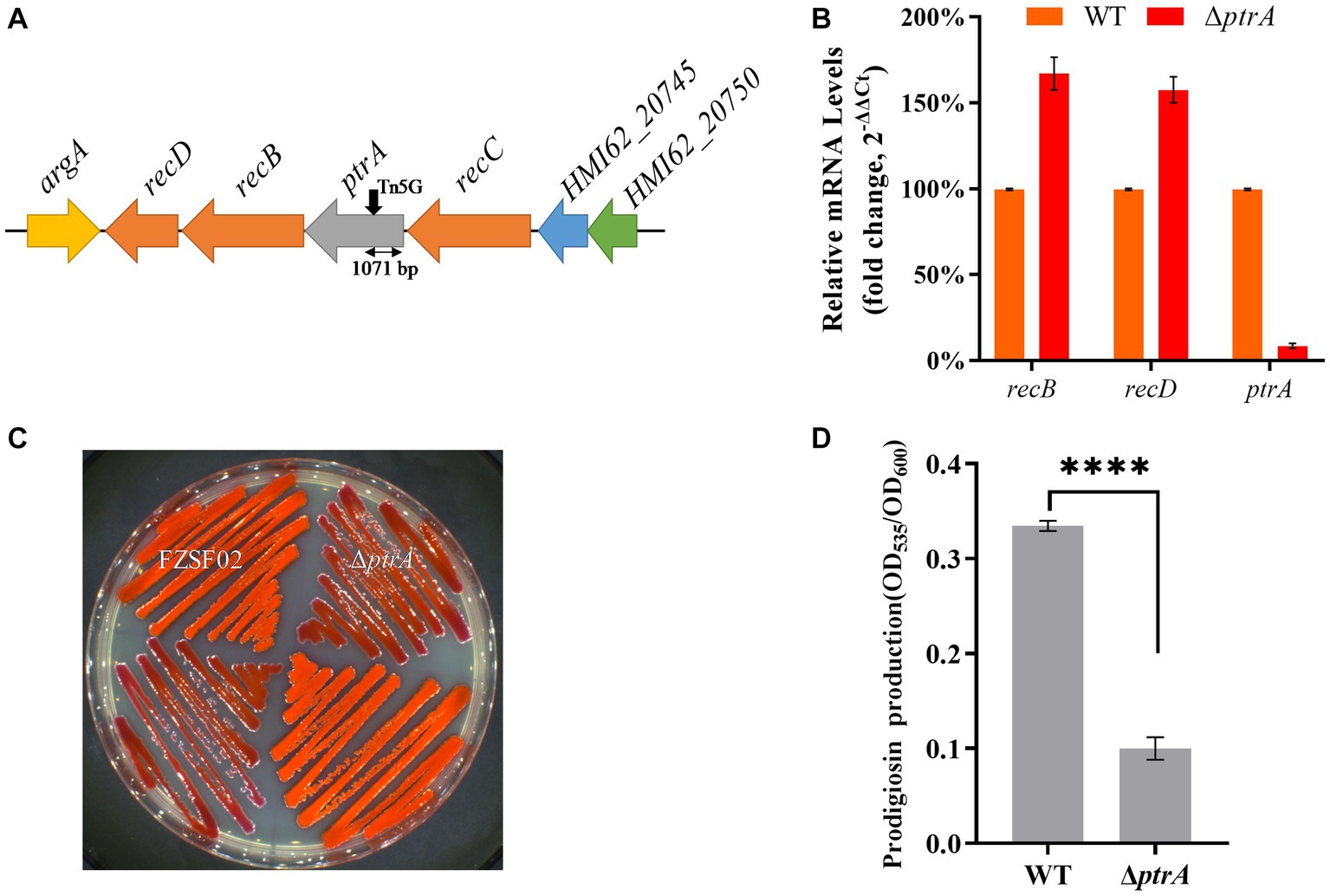
Figure 1. PtrA positively regulates the synthesis of prodigiosin in strain FZSF02. (A) The genetic locus identified in mutant E35. The panel shows the genetic map of the disrupted gene ptrA and its surroundings, and the black arrow indicates the position of the Tn5 insertion. (B) RT–qPCR analysis of the relative expression levels of recB and recD in WT and mutant ΔptrA. (C) Phenotype of the ptrA inactivated mutant strain ΔptrA. (D) Analysis of prodigiosin production in FZSF02 and ΔptrA. FZSF02 is wild-type S. marcescens, and ΔptrA is the ptrA-disrupted mutant. For (C), the experiments were independently replicated three times. Error bars represent standard deviations. Pairwise comparisons were performed using Student’s t test, while multiple comparisons were conducted using one-way ANOVA, ****p < 0.0001.
ptrA appeared to be the first gene in an operon, so polar effects are a concern. The effect of polar transcription on recB/D was verified by RT–qPCR, and the results showed that the relative expression levels of recB and recD in ΔptrA were upregulated by 1.67-fold and 1.58-fold, respectively, compared to the WT strain (Figure 1B). There was no significant difference, suggesting that the PtrA mutation was responsible for the phenotypes.
To further confirm the function of PtrA, a ptrA in-frame deletion strain ΔptrA was constructed (Figure 1C). The prodigiosin production of the mutant ptrA was only 0.3 times that of the wild-type strain FZSF02 (p < 0.0001, Figure 1D) when incubated for 48 h. The results suggested that ptrA encoded a protein, PtrA, that promoted prodigiosin synthesis in strain FZSF02.
3.2. PtrA affects the transcription level of the prodigiosin-associated pigA-pigN gene cluster
To study the effect of PtrA on bacterial growth ability, growth curves of WT and mutant ΔptrA were determined (Figure 2A). Compared to the wild-type strain FZSF02, the growth of ΔptrA was significantly slower from 0 to 36 h, which may be due to the synthesis of primary metabolites being decreased in ΔptrA. After 36 h, these two strains reached almost the same final biomass. At each sampling point, the OD535/OD600 value of ΔptrA was much lower than that of the wild-type strain (p < 0.0001, Figure 2B), which means that the ability of ΔptrA to synthesize prodigiosin was significantly reduced, and this decrease may be caused by the decreased growth ability and the low expression level of pig genes at 0–36 h. While after 36 h, this decrease was not due to a decrease in growth ability but probably due to lower expression levels of pig genes.
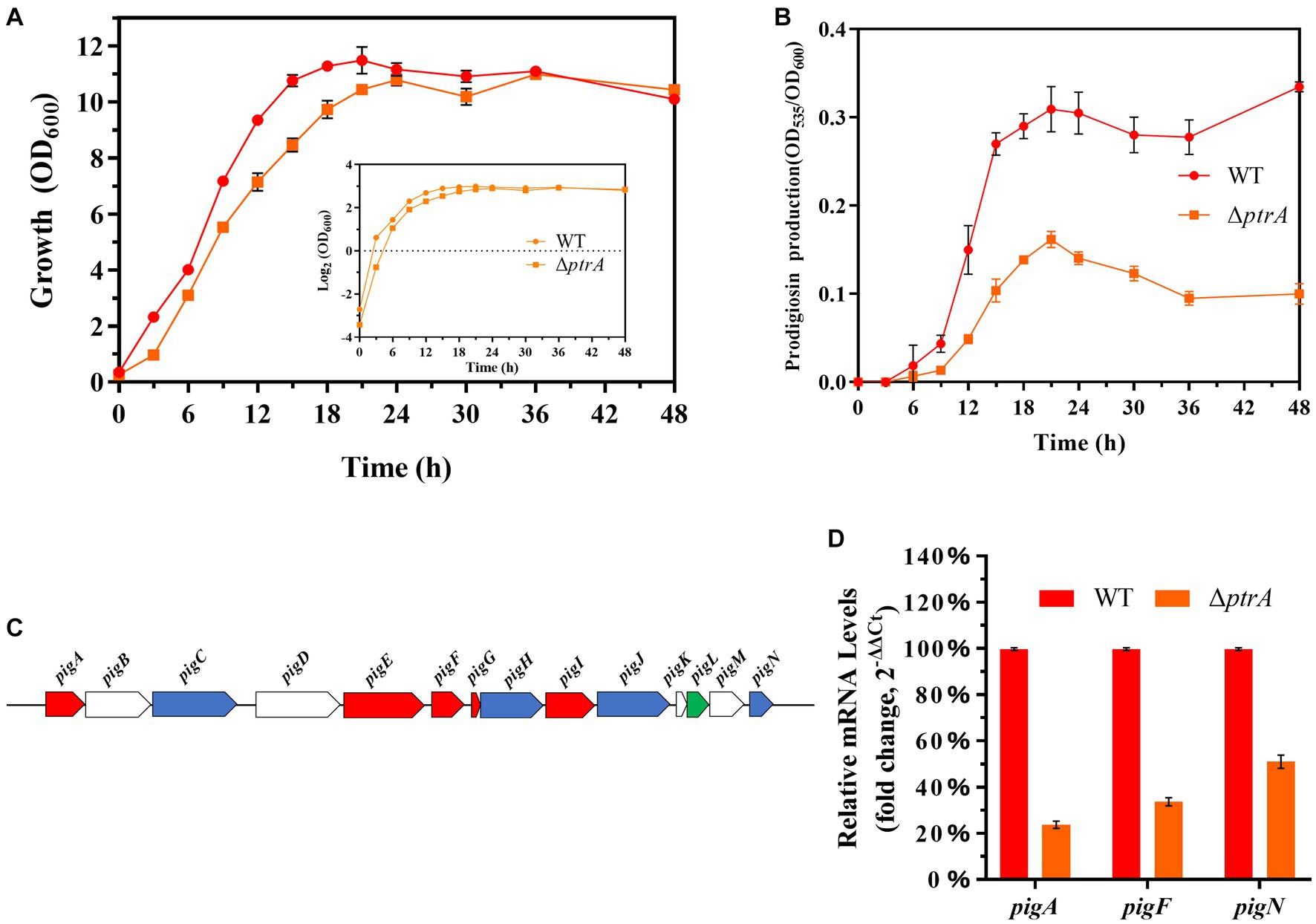
Figure 2. PtrA promotes the transcription of the pig gene cluster. (A) Growth curves of WT and ΔptrA strains by measuring OD600. (B) Unit cell production of prodigiosin of WT and ΔptrA strains at all time points. (C) The panel shows the genetic map of the pig gene cluster. (D) RT–qPCR analysis of the relative expression levels of the pigA, pigF, and pigN genes in the pig gene cluster in WT and mutant ΔptrA. For (A,B,D), the experiments were independently replicated three times. Error bars represent standard deviations.
The synthesis of prodigiosin by S. marcescens is mainly controlled by 14 genes, including pigA through pigN, which are located on the same operon (Figure 2C; Williamson et al., 2006). The results of real-time quantitative PCR (RT–qPCR) showed that the relative expression levels of pigA, pigF, and pigN in ΔptrA were reduced by 4.20-, 2.73-, and 1.96-fold, respectively (Figure 2D). These results suggest that PtrA promoted the synthesis of prodigiosin by positively regulating the expression of pig genes.
3.3. PtrA positively controls cell motility
Previous studies have shown that swimming and swarming are two modes of motility found in S. marcescens (Alberti and Harshey, 1990). In this study, the swimming distance of the mutant ΔptrA was significantly reduced compared to that of the wild-type strain FZSF02 when incubated on plates containing 0.3% (m/V) agar (Figure 3A, top; Figure 3B). For the swarming test, wild-type FZSF02 displayed an extended migration distance, but ΔptrA showed no migration distance (Figure 3A, bottom; Figure 3C), and the results indicated that PtrA positively affected the swimming and swarming functions of FZSF02.
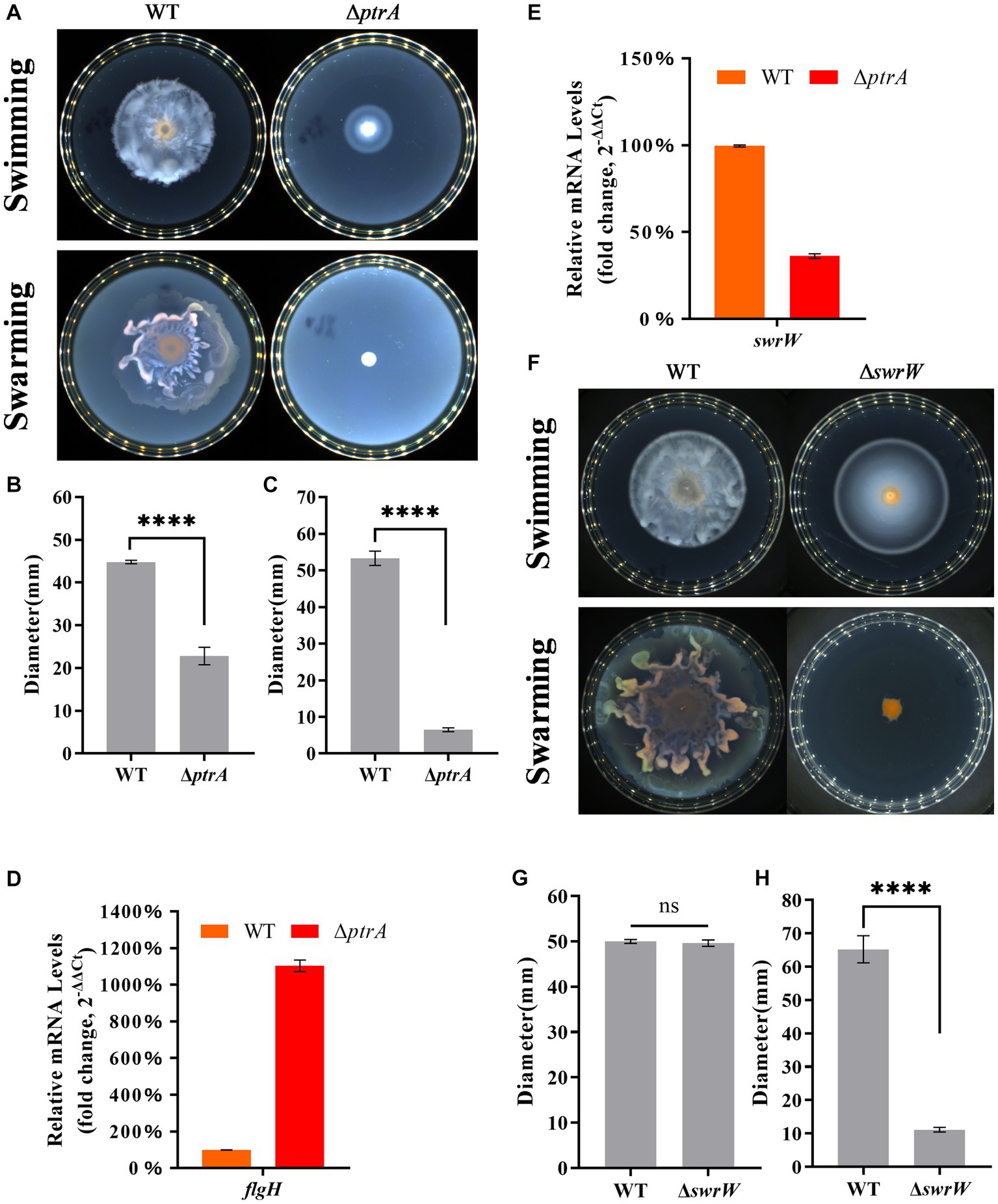
Figure 3. PtrA regulates cell motility in S. marcescens. (A) Swimming and swarming assay for WT and ΔptrA. (B) Colony diameter determination of WT and ΔptrA for the swimming assay. (C) Colony diameter determination of WT and ΔptrA for the swarming assay. (D) RT–qPCR analysis of the relative expression levels of flgH in flagellar synthesis in WT and mutant ΔptrA. (E) RT–qPCR analysis of the relative expression levels of swrW in WT and mutant ΔptrA. (F) Swimming and swarming assay for WT and ΔswrW. (G) Colony diameter determination of WT and ΔswrW for the swimming assay. (H) Colony diameter determination of WT and ΔswrW for the swarming assay. For (B–E,G,H), the experiments were independently replicated three times. Error bars represent standard deviations. Pairwise comparisons were performed using Student’s t test, while multiple comparisons were conducted using one-way ANOVA, ****p < 0.0001.
The motility of bacteria can be divided into swimming and swarming, and the flagellum has been shown to contribute to the motility of S. marcescens (Pan et al., 2020). To further analyze the relationship between PtrA and flagellum, the key gene flgH in the process of flagellar synthesis was selected for further analysis. Surprisingly, the RT–qPCR results showed that the relative expression level of flgH in ΔptrA was upregulated by 11.03-fold compared to the WT strain (Figure 3D). Surprisingly, ΔptrA displayed decreased motility but significantly upregulated flgH expression levels. However, it has been reported that serrawettin W1 (serratamolide) was distinguished from the flagellum in contributing to the swarming motility, while PigP mediated swarming motility of S. marcescens through control of serratamolide biosynthesis (Shanks et al., 2013). Serrawettin W1 was reported to act as a biosurfactant, reducing surface tension when S. marcescens swarms on a surface, and the gene swrW is required for biosynthesis of the biosurfactant serratamolide (Matsuyama et al., 1992; Li et al., 2005). To investigate whether swrW is downregulated in ΔptrA, the expression levels of swrW (HMI62_23130 gene) (GenBank accession number QJU42029.1) were assayed by RT–qPCR. The results showed that the relative expression levels of swrW in ΔptrA were downregulated by 2.76-fold (Figure 3E). At the same time, we constructed a swrW (HMI62_23130 gene) deletion mutant ΔswrW and performed swimming and swarming experiments. The results showed that the swimming movement migration distance of the mutant ΔswrW was no significant difference (Figure 3F, top; Figure 3G) and the swarming movement migration distance of the mutant ΔswrW was significantly reduced (Figure 3F, bottom; Figure 3H). These results demonstrated that ptrA positively regulated the swarming motility of S. marcescens FZSF02 by regulating the expression level of swrW but not flgH. The reason for flgH showing significantly upregulated expression levels in ΔptrA remains to be elucidated.
3.4. PtrA regulates biofilm formation and serrawettin W1 biosynthesis
S. marcescens is an opportunistic bacterium, and biofilm formation is a major requirement of pathogenesis. Biofilm formation in S. marcescens was linked to its ability to colonize, persist, and proliferate on either biological or inert surfaces (Choe et al., 2012). Therefore, we investigated whether PtrA was able to affect the ability to form biofilms in S. marcescens FZSF02. The results showed that biofilm production of mutant ΔptrA was significantly increased compared to FZSF02 (p < 0.0001, Figure 4A), indicating that PtrA negatively regulates S. marcescens biofilm synthesis.
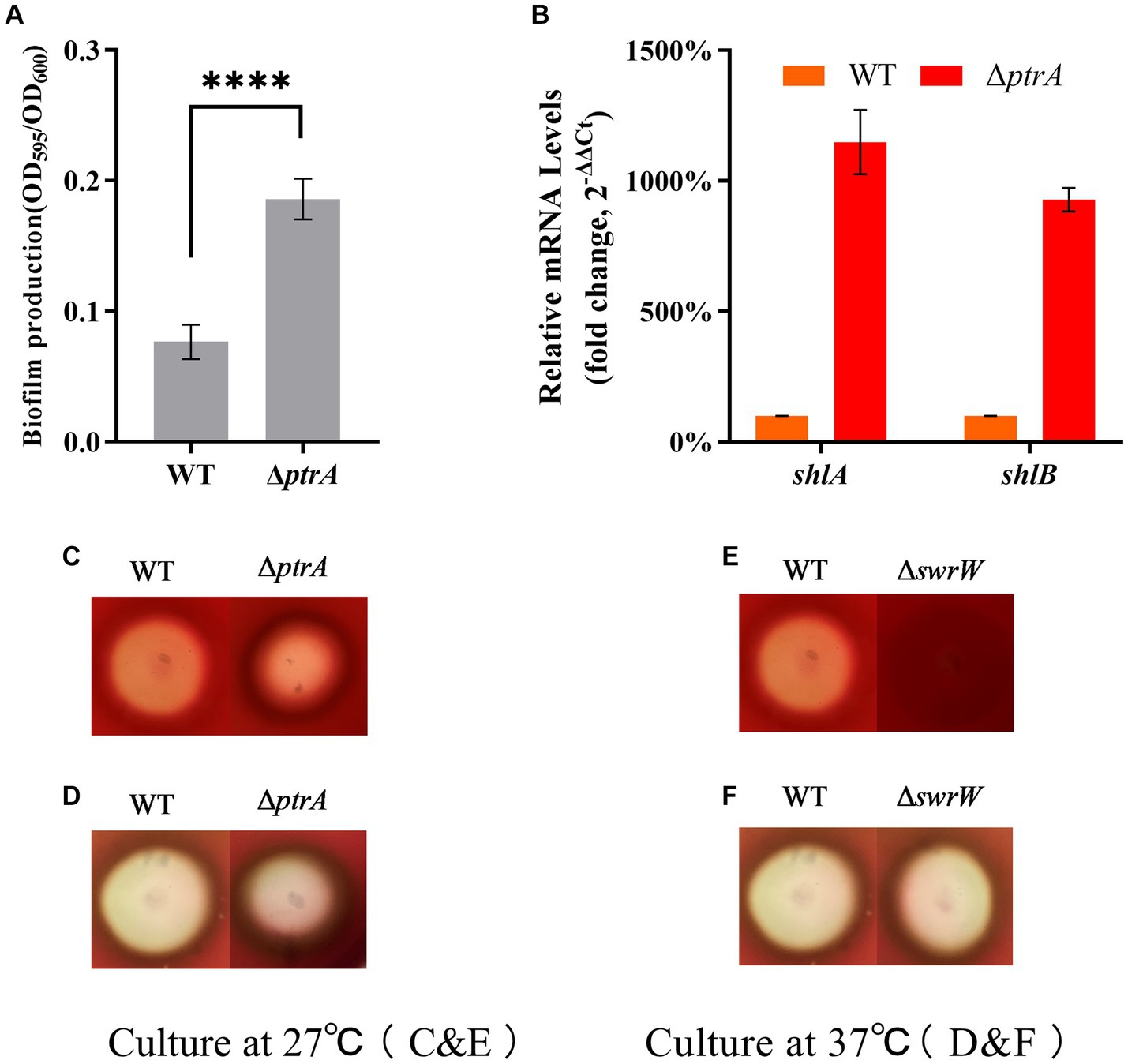
Figure 4. PtrA does not contribute to biofilm formation but contributes to serrawettin W1 biosynthesis in S. marcescens. (A) PtrA negatively regulates biofilm formation in WT. (B) RT–qPCR analysis of the relative expression levels of shlA and shlB in WT and mutant ΔptrA. (C) Hemolytic transparent circles of WT and ΔptrA at 27°C. (D) Hemolytic transparent circles of WT and ΔptrA at 37°C. (E) Hemolytic transparent circles of WT and ΔswrW at 27°C. (F) Hemolytic transparent circles of WT and ΔswrW at 37°C. For (A), the experiments were independently replicated three times. Error bars represent standard deviations. Pairwise comparisons were performed using Student’s t test, while multiple comparisons were conducted using one-way ANOVA, ****p < 0.0001.
Previous research has shown hemolytic activity in S. marcescens (Braun et al., 1985). Two genes, shlA and shlB, encode a hemolysin and an accessory protein necessary for activity, respectively, and both gene products are present in the outer membrane (Poole et al., 1988). The above results show that the biofilm production of ΔptrA is higher than that of WT, which means that the hemolysis of ΔptrA may be higher than that of WT. To test this idea, we performed hemolysis experiments on blood agar plates. The results showed that the hemolytic transparent zones of ΔptrA were significantly reduced compared to the WT at 27°C, indicating that the hemolytic activity was significantly reduced (Figure 4B); however, there was no significant difference in the hemolytic transparent zones between ΔptrA and WT at 37°C (Figure 4C).
To verify whether the hemolytic changes were caused by the changes in the expression levels of shlA and shlB, RT–qPCR analysis was performed on shlA and shlB. Surprisingly, the results showed that the relative expression levels of shlA and shlB in ΔptrA were upregulated by 11.49-fold and 9.28-fold, respectively, compared to the WT strain (Figure 4D). This indicates that the hemolysis of S. marcescens FZSF02 is not regulated by shlA and shlB. A literature review found that serrawettin W1 produced by S. marcescens is a surface-active exolipid with various functions supporting the behaviors of bacteria in surface environments (Matsuyama et al., 1989). Serrawettin W1 is a hemolytic factor produced by S. marcescens (Shanks et al., 2012), and PigP mediated hemolysis through control of serratamolide biosynthesis (Shanks et al., 2013). The hemolysis of WT and ΔswrW was determined, and the results showed that the hemolytic activity of ΔswrW also significantly decreased at 27°C (Figure 4E) and showed no significant change compared to that of the WT at 37°C (Figure 4F). A previous study showed that, similar to prodigiosin, the production of serrawettin W1 by S. marcescens is also temperature-regulated. That is, a positive product was produced at 27°C and inhibited at 37°C (Matsuyama et al., 1986). Overall, PtrA promoted serrawettin W1 biosynthesis and promoted hemolysis activity at 27°C.
3.5. PtrA plays an important role in stress tolerance in S. marcescens
Environmental stresses are usually active during the process of microbial fermentation and have a significant influence on microbial physiology (Guan et al., 2017). Hydrogen peroxide generated during oxidative stress is known to damage proteins, nucleic acids and cell membranes and has been implicated in cancer, aging, and several chronic neurodegenerative diseases and therefore presents a major challenge for aerobic organisms (Daroui et al., 2004). In this study, the H2O2 tolerance of ΔptrA was significantly decreased and only 2.49% of that of the WT survived (p < 0.0001, Figures 5A,F), indicating that PtrA was involved in cellular tolerance to H2O2. In bacteria, H2O2 is eliminated by catalase and peroxidase. To confirm whether the significantly reduced H2O2 tolerance of the mutant ΔptrA was due to the reduced production of catalase, catalase activities of WT and ΔptrA were assayed. The results showed that compared with WT, the catalase activity of ΔptrA was significantly decreased (p < 0.01, Figure 5B), indicating that one of the reasons for the significantly decreased H2O2 tolerance of ΔptrA was the decreased production of catalase.
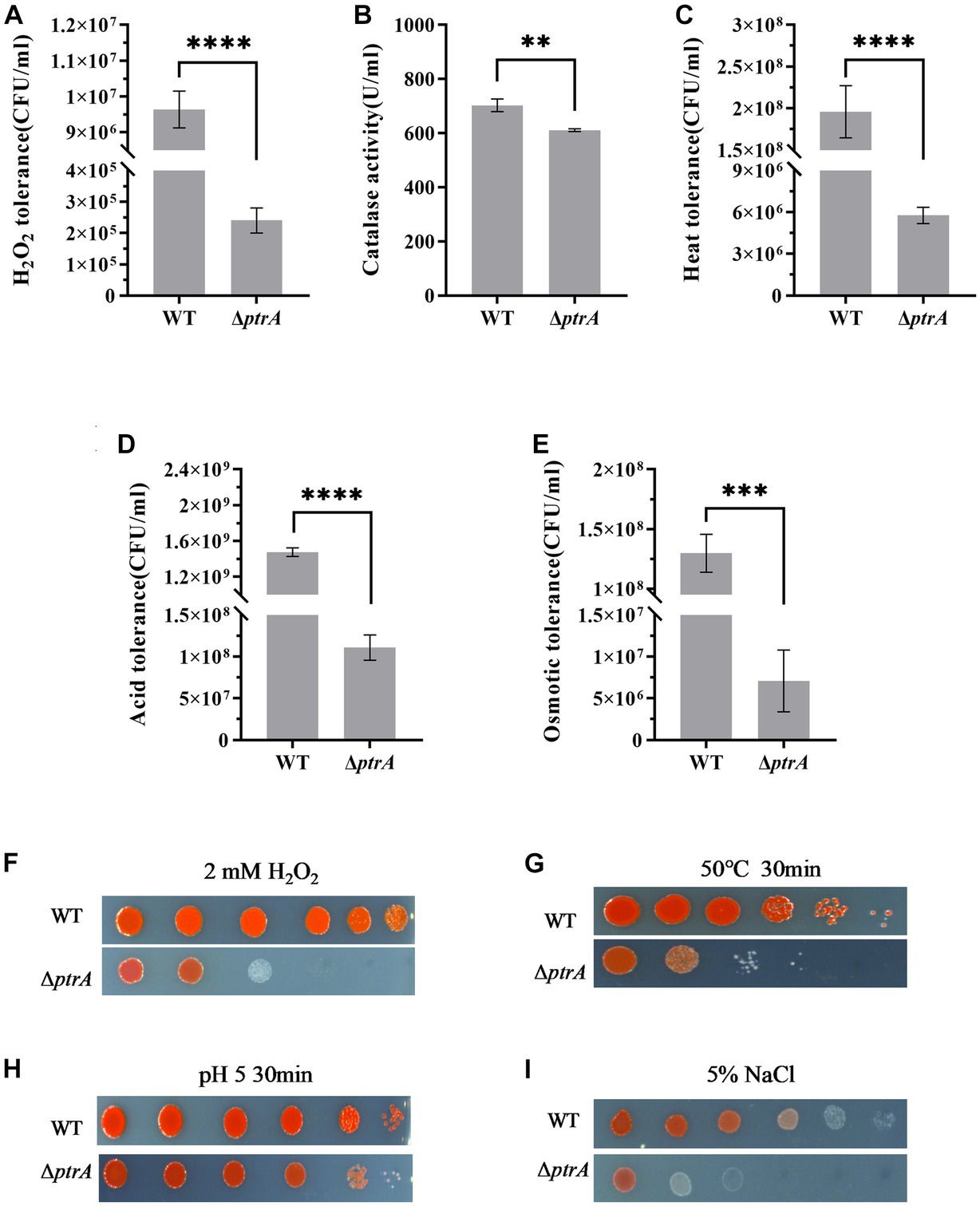
Figure 5. Influence of PtrA on H2O2 tolerance, heat tolerance, oxidative tolerance and acid tolerance. (A) H2O2 tolerance CFU counting of WT and ΔptrA. (B) PtrA positively regulates catalase activity in WT. (C) Heat tolerance CFU counting of WT and ΔptrA. (D) Acid tolerance CFU counting of WT and ΔptrA. (E) Osmotic tolerance CFU counting of WT and ΔptrA. (F) H2O2 tolerance spotting assay of WT and ΔptrA. (G) Heat tolerance spotting assay of WT and ΔptrA. (H) Acid tolerance spotting assay of WT and ΔptrA. (I) Osmotic tolerance spotting assay of WT and ΔptrA. For (A–D), the experiments were independently replicated three times. Error bars represent standard deviations. Pairwise comparisons were performed using Student’s t test, while multiple comparisons were conducted using one-way ANOVA, ****p < 0.0001.
Adaptation to changes in ambient temperature is a common feature of all living organisms (Richter et al., 2010). The heat tolerance of ΔptrA was significantly decreased and only 2.49% of that of the WT in survival (p < 0.0001, Figures 5C,G), indicating that PtrA is involved in the heat tolerance of S. marcescens.
To meet sudden and potentially lethal challenges, microorganisms must have effective mechanisms to withstand a variety of environmental stressors, acid resistance being one of the most common features (Waterman and Small, 2003). Therefore, the acid tolerance of ΔptrA was investigated, and the results showed that the acid tolerance of ΔptrA was significantly decreased and only 7.52% of that of the WT in survival (p < 0.0001, Figures 5D,H), strongly suggesting that PtrA plays an important role in the acid resistance of S. marcescens.
Bacteria inhabit natural and artificial environments with diverse and fluctuating osmolalities, one of the most ubiquitous stress factors encountered by microorganisms in their habitats (Wood, 2011). To investigate the effect of PtrA on the osmotic stress tolerance of S. marcescens, the osmotic tolerance of the FZSF02 and ΔptrA mutant strains was analyzed. The results showed that the osmotic tolerance of ΔptrA was significantly reduced, and only 5.48% of that was conferred by the WT (p < 0.001, Figures 5E,I), indicating that PtrA positively influences the osmotic tolerance of S. marcescens.
Although synthetic biology has experienced significant development, the efficiency of modifying and improving complex traits, such as stress tolerance and growth rates, is difficult to achieve because of the complex metabolic and regulatory networks involved. Therefore, adaptation is still a widely used method to improve microbial performance (Sandberg et al., 2019). The stress tole-rance in response to hydrogen peroxide, high temperature, acidic environment and high osmotic fluctuations was significantly reduced in ΔptrA mutants, displaying a significant reduction in the environmental adaptability of ΔptrA. The adaptability of microorganisms compensates for the lack of knowledge regarding the microbial ability to utilize nonpreferred substrates by rapidly improving the ability of microorganisms to utilize nonpreferred substrates (Tan et al., 2022). Therefore, a decrease in the adaptability of ΔptrA means a decrease in its viability. In other words, ptrA might be a potential gene for synthetic biology applications to improve the performance of various vital characteristics in bacterial strains.
The homolog of PtrA is Protease III in E. coli K-12, and it is devoid of activity toward aminopeptidase, carboxypeptidase, or esterase substrates but rapidly degrades small proteins (Cheng and Zipser, 1979). Studies on the biological functions of cell motility, hemolysis and stress adaptation have not been reported, to the best of our knowledge. Therefore, it could not be compared with other bacteria to determine whether PtrA in other bacteria also controlled the same cellular processes in Serratia marcescens.
4. Conclusion
In this paper, we studied the effect of PtrA on prodigiosin synthesis and other biological functions of S. marcescens FZSF02. Growth ability and prodigiosin significantly decreased in ΔptrA, and the reason for the decrease in prodigiosin production was not related to biomass; rather, it was associated with decreased expression of the pig gene cluster in ΔptrA. Mutation of ptrA negatively influenced the mobility and hemolytic activity of FZSF02 by lowering the expression level of swrW. The results also showed that a deletion of PtrA is not conducive to the survival of S. marcescens in environments with elevated levels of H2O2, heat, acid and osmotic stress. This might be the first time the function of PtrA was studied in detail. This study provided new insights into PtrA-dependent regulation of prodigiosin biosynthesis. Multiple effects on the growth, mobility, hemolytic activity, and stress adaptation of FZSF02 indicated that PtrA might be key for improving the performance of industrial microbial strains in the future.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
JL: investigation, formal analysis, methodology, and writing – original draft. YY: conceptualization, methodology, investigation, and writing – original draft. KZ: data curation, conceptualization, and validation. JZ: formal analysis and investigation. CR: investigation. JC: formal analysis and funding acquisition. XJ: conceptualization, methodology, writing – review and editing, funding acquisition, and project administration. All authors contributed to the article and approved the submittedversion.
Funding
This study was supported by the Exploration Program of Fujian Academy of Agricultural Sciences (ZYTS202217), Scientific Research in the Public Interest of Fujian Province (2020R1025003, 2021R1025002), Natural Science Foundation of Fujian Province of China (2021J01480), Chinese National Natural Science Foundation (31800068), Special Program for Extension Research of National Natural Science Foundation of Fujian Academy of Agricultural Sciences (AGY2018-1), and Science and Technology Innovation Team Program of Fujian Academy of Agricultural Sciences (CXTD2021002-3).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1240102/full#supplementary-material
References
Alberti, L., and Harshey, R. M. (1990). Differentiation of Serratia marcescens 274 into swimmer and swarmer cells. J. Bacteriol. 172, 4322–4328. doi: 10.1128/jb.172.8.4322-4328.1990
Anastasi, A., and Barrett, A. J. (1995). Pitrilysin. Methods Enzymol. 248, 684–692. doi: 10.1016/0076-6879(95)48045-5
Anastasi, A., Knight, C. G., and Barrett, A. J. (1993). Characterization of the bacterial metalloendopeptidase pitrilysin by use of a continuous fluorescence assay. Biochem. J. 290, 601–607. doi: 10.1042/bj2900601
Bai, F., Dai, L., Fan, J., Truong, N., Rao, B., Zhang, L., et al. (2015). Engineered Serratia marcescens for efficient (3R)-acetoin and (2R,3R)-2,3-butanediol production. J. Ind. Microbiol. Biotechnol. 42, 779–786. doi: 10.1007/s10295-015-1598-5
Bennett, J. W., and Bentley, R. (2000). Seeing red: the story of prodigiosin. Adv. Appl. Microbiol. 47, 1–32. doi: 10.1016/s0065-2164(00)47000-0
Braun, V., Günther, H., Neuss, B., and Tautz, C. (1985). Hemolytic activity of Serratia marcescens. Arch. Microbiol. 141, 371–376. doi: 10.1007/BF00428852
Chang, C. C., Wang, Y. H., Chern, C. M., Liou, K. T., Hou, Y. C., Peng, Y. T., et al. (2011). Prodigiosin inhibits gp91(phox) and iNOS expression to protect mice against the oxidative/nitrosative brain injury induced by hypoxia-ischemia. Toxicol. Appl. Pharmacol. 257, 137–147. doi: 10.1016/j.taap.2011.08.027
Cheng, Y. S., and Zipser, D. (1979). Purification and characterization of protease III from Escherichia coli. J. Biol. Chem. 254, 4698–4706. doi: 10.1016/S1773-2247(14)50126-8
Choe, H. S., Son, S. W., Choi, H. A., Kim, H. J., Ahn, S. G., Bang, J. H., et al. (2012). Analysis of the distribution of bacteria within urinary catheter biofilms using four different molecular techniques. Am. J. Infect. Control 40, e249–e254. doi: 10.1016/j.ajic.2012.05.010
Clements, T., Ndlovu, T., and Khan, W. (2019). Broad-spectrum antimicrobial activity of secondary metabolites produced by Serratia marcescens strains. Microbiol. Res. 229:126329. doi: 10.1016/j.micres.2019.126329
Daroui, P., Desai, S. D., Li, T. K., Liu, A. A., and Liu, L. F. (2004). Hydrogen peroxide induces topoisomerase I-mediated DNA damage and cell death. J. Biol. Chem. 279, 14587–14594. doi: 10.1074/jbc.M311370200
de Araújo, H. W., Fukushima, K., and Takaki, G. M. (2010). Prodigiosin production by Serratia marcescens UCP 1549 using renewable-resources as a low cost substrate. Molecules 15, 6931–6940. doi: 10.3390/molecules15106931
de Ondarza, J. (2017). Ozone sensitivity and catalase activity in pigmented and nonpigmented strains of Serratia marcescens. Open Microbiol. J. 11, 12–22. doi: 10.2174/1874285801711010012
Di Venanzio, G., Stepanenko, T. M., and García, V. E. (2014). Serratia marcescens ShlA pore-forming toxin is responsible for early induction of autophagy in host cells and is transcriptionally regulated by RcsB. Infect. Immun. 82, 3542–3554. doi: 10.1128/IAI.01682-14
Gerc, A. J., Song, L., Challis, G. L., Stanley-Wall, N. R., and Coulthurst, S. J. (2012). The insect pathogen Serratia marcescens Db10 uses a hybrid nonribosomal peptide synthetase-polyketide synthase to produce the antibiotic althiomycin. PLoS One 7:e44673. doi: 10.1371/journal.pone.0044673
Giri, A. V., Anandkumar, N., Muthukumaran, G., and Pennathur, G. (2004). A novel medium for the enhanced cell growth and production of prodigiosin from Serratia marcescens isolated from soil. BMC Microbiol. 4:11. doi: 10.1186/1471-2180-4-11
Grimont, P. A., and Grimont, F. (1978). The genus Serratia. Annu. Rev. Microbiol. 32, 221–248. doi: 10.1146/annurev.mi.32.100178.001253
Guan, N., Li, J., Shin, H. D., Du, G., Chen, J., and Liu, L. (2017). Microbial response to environmental stresses: from fundamental mechanisms to practical applications. Appl. Microbiol. Biotechnol. 101, 3991–4008. doi: 10.1007/s00253-017-8264-y
Han, R., Xiang, R., Li, J., Wang, F., and Wang, C. (2021). High-level production of microbial prodigiosin: a review. J. Basic Microbiol. 61, 506–523. doi: 10.1002/jobm.202100101
Jia, X., Liu, F., Zhao, K., Lin, J., Fang, Y., Cai, S., et al. (2021). Identification of essential genes associated with prodigiosin production in Serratia marcescens FZSF02. Front. Microbiol. 12:705853. doi: 10.3389/fmicb.2021.705853
Kalivoda, E. J., Stella, N. A., Aston, M. A., Fender, J. E., Thompson, P. P., Kowalski, R. P., et al. (2010). Cyclic AMP negatively regulates prodigiosin production by Serratia marcescens. Res. Microbiol. 161, 158–167. doi: 10.1016/j.resmic.2009.12.004
Lapenda, J. C., Silva, P. A., Vicalvi, M. C., Sena, K. X., and Nascimento, S. C. (2015). Antimicrobial activity of prodigiosin isolated from Serratia marcescens UFPEDA 398. World J. Microbiol. Biotechnol. 31, 399–406. doi: 10.1007/s11274-014-1793-y
Li, H., Tanikawa, T., Sato, Y., Nakagawa, Y., and Matsuyama, T. (2005). Serratia marcescens gene required for surfactant serrawettin W1 production encodes putative aminolipid synthetase belonging to nonribosomal peptide synthetase family. Microbiol. Immunol. 49, 303–310. doi: 10.1111/j.1348-0421.2005.tb03734.x
Lin, C., Jia, X., Fang, Y., Chen, L., Zhang, H., Lin, R., et al. (2019). Enhanced production of prodigiosin by Serratia marcescens FZSF02 in the form of pigment pellets. Electron. J. Biotechnol. 40, 58–64. doi: 10.1016/j.ejbt.2019.04.007
Mahlen, S. D. (2011). Serratia infections: from military experiments to current practice. Clin. Microbiol. Rev. 24, 755–791. doi: 10.1128/CMR.00017-11
Matsuyama, T., Kaneda, K., Nakagawa, Y., Isa, K., Hara-Hotta, H., and Yano, I. (1992). A novel extracellular cyclic lipopeptide which promotes flagellum-dependent and-independent spreading growth of Serratia marcescens. J. Bacteriol. 174, 1769–1776. doi: 10.1128/jb.174.6.1769-1776.1992
Matsuyama, T., Murakami, T., Fujita, M., Fujita, S., and Yano, I. (1986). Extracellular vesicle formation and biosurfactant production by Serratia marcescens. J. Gen. Microbiol. 132, 865–875. doi: 10.1099/00221287-132-4-865
Matsuyama, T., Sogawa, M., and Nakagawa, Y. (1989). Fractal spreading growth of Serratia marcescens which produces surface active exolipids. FEMS Microbiol. Lett. 61, 243–246. doi: 10.1016/0378-1097(89)90204-8
Nakashima, A. K., McCarthy, M. A., Martone, W. J., and Anderson, R. L. (1987). Epidemic septic arthritis caused by Serratia marcescens and associated with a benzalkonium chloride antiseptic. J. Clin. Microbiol. 25, 1014–1018. doi: 10.1128/jcm.25.6.1014-1018.1987
Pan, M. Y., Shen, Y. C., Lu, C. H., Yang, S. Y., Ho, T. F., Peng, Y. T., et al. (2012). Prodigiosin activates endoplasmic reticulum stress cell death pathway in human breast carcinoma cell lines. Toxicol. Appl. Pharmacol. 265, 325–334. doi: 10.1016/j.taap.2012.08.034
Pan, X., Sun, C., Tang, M., You, J., Osire, T., Zhao, Y., et al. (2020). LysR-type transcriptional regulator MetR controls prodigiosin production, methionine biosynthesis, cell motility, H2O2 tolerance, heat tolerance, and exopolysaccharide synthesis in Serratia marcescens. Appl. Environ. Microbiol. 86, e02241–e02219. doi: 10.1128/AEM.02241-19
Papireddy, K., Smilkstein, M., Kelly, J. X., Shweta, S. S. M., Alhamadsheh, M., Haynes, S. W., et al. (2011). Antimalarial activity of natural and synthetic prodiginines. J. Med. Chem. 54, 5296–5306. doi: 10.1021/jm200543y
Poole, K., Schiebel, E., and Braun, V. (1988). Molecular characterization of the hemolysin determinant of Serratia marcescens. J. Bacteriol. 170, 3177–3188. doi: 10.1128/jb.170.7.3177-3188.1988
Psautter, R. L., Mattman, L. H., and Legaspi, R. C. (1984). Serratia marcescens meningitis associated with a contaminated benzalkonium chloride solution. Infect. Control. 5, 223–225. doi: 10.1017/s019594170006015x
Richter, K., Haslbeck, M., and Buchner, J. (2010). The heat shock response: life on the verge of death. Mol. Cell 40, 253–266. doi: 10.1016/j.molcel.2010.10.006
Ridder, M. J., Daly, S. M., Hall, P. R., and Bose, J. L. (2021). Quantitative hemolysis assays. Methods Mol. Biol. 2341, 25–30. doi: 10.1007/978-1-0716-1550-8_4
Ryazantseva, I. N., Saakov, V. S., Andreyeva, I. N., Ogorodnikova, T. I., and Zuev, Y. F. (2012). Response of pigmented Serratia marcescens to the illumination. J. Photochem. Photobiol. B 106, 18–23. doi: 10.1016/j.jphotobiol.2011.08.006
Sandberg, T. E., Salazar, M. J., Weng, L. L., Palsson, B. O., and Feist, A. M. (2019). The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 56, 1–16. doi: 10.1016/j.ymben.2019.08.004
Shanks, R. M., Lahr, R. M., Stella, N. A., Arena, K. E., Brothers, K. M., Kwak, D. H., et al. (2013). A Serratia marcescens PigP homolog controls prodigiosin biosynthesis, swarming motility and hemolysis and is regulated by cAMP-CRP and HexS. PLoS One 8:e57634. doi: 10.1371/journal.pone.0057634
Shanks, R. M., Stella, N. A., Kalivoda, E. J., Doe, M. R., O’Dee, D. M., Lathrop, K. L., et al. (2007). A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J. Bacteriol. 189, 7262–7272. doi: 10.1128/JB.00859-07
Shanks, R. M., Stella, N. A., Lahr, R. M., Wang, S., Veverka, T. I., Kowalski, R. P., et al. (2012). Serratamolide is a hemolytic factor produced by Serratia marcescens. PLoS One 7:e36398. doi: 10.1371/journal.pone.0036398
Slater, H., Crow, M., Everson, L., and Salmond, G. P. (2003). Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and-independent pathways. Mol. Microbiol. 47, 303–320. doi: 10.1046/j.1365-2958.2003.03295.x
Solé, M., Rius, N., Francia, A., and Lorén, J. G. (1994). The effect of pH on prodigiosin production by non-proliferating cells of Serratia marcescens. Lett. Appl. Microbiol. 19, 341–344. doi: 10.1111/j.1472-765x.1994.tb00470.x
Soo, P. C., Horng, Y. T., Chang, Y. L., Tsai, W. W., Jeng, W. Y., Lu, C. C., et al. (2014). ManA is regulated by RssAB signaling and promotes motility in Serratia marcescens. Res. Microbiol. 165, 21–29. doi: 10.1016/j.resmic.2013.10.005
Soo, P. C., Horng, Y. T., Wei, J. R., Shu, J. C., Lu, C. C., and Lai, H. C. (2008). Regulation of swarming motility and flhDC(Sm) expression by RssAB signaling in Serratia marcescens. J. Bacteriol. 190, 2496–2504. doi: 10.1128/JB.01670-07
Tan, Y. S., Zhang, R. K., Liu, Z. H., Li, B. Z., and Yuan, Y. J. (2022). Microbial adaptation to enhance stress tolerance. Front. Microbiol. 13:888746. doi: 10.3389/fmicb.2022.888746
Waterman, S. R., and Small, P. L. (2003). The glutamate-dependent acid resistance system of Escherichia coli and Shigella flexneri is inhibited in vitro by L-trans-pyrrolidine-2,4-dicarboxylic acid. FEMS Microbiol. Lett. 224, 119–125. doi: 10.1016/S0378-1097(03)00427-0
Wei, J., Xie, X., Huang, F., Xiang, L., Wang, Y., Han, T., et al. (2020). Simultaneous microcystis algicidal and microcystin synthesis inhibition by a red pigment prodigiosin. Environ. Pollut. 256:113444. doi: 10.1016/j.envpol.2019.113444
Wilfert, J. N., Barrett, F. F., and Kass, E. H. (1968). Bacteremia due to Serratia marcescens. N. Engl. J. Med. 279, 286–289. doi: 10.1056/NEJM196808082790604
Williamson, N. R., Fineran, P. C., Leeper, F. J., and Salmond, G. P. (2006). The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 4, 887–899. doi: 10.1038/nrmicro1531
Wood, J. M. (2011). Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Annu. Rev. Microbiol. 65, 215–238. doi: 10.1146/annurev-micro090110-102815
Zhang, J., Shen, Y., Liu, J., and Wei, D. (2005). Antimetastatic effect of prodigiosin through inhibition of tumor invasion. Biochem. Pharmacol. 69, 407–414. doi: 10.1016/j.bcp.2004.08.037
Keywords: PtrA, stress tolerance, prodigiosin synthesis, cell motility, Serratia marcescens
Citation: Lin J, Yu Y, Zhao K, Zhao J, Rensing C, Chen J and Jia X (2023) PtrA regulates prodigiosin synthesis and biological functions in Serratia marcescens FZSF02. Front. Microbiol. 14:1240102. doi: 10.3389/fmicb.2023.1240102
Edited by:
Harold J. Schreier, University of Maryland, United StatesReviewed by:
Xuewei Pan, Jiangnan University, ChinaPryce Haddix, Auburn University at Montgomery, United States
Copyright © 2023 Lin, Yu, Zhao, Zhao, Rensing, Chen and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianbo Jia, eGJqMjAxMUAxNjMuY29t
 Junjie Lin
Junjie Lin Yanshuang Yu3
Yanshuang Yu3 Christopher Rensing
Christopher Rensing Jichen Chen
Jichen Chen Xianbo Jia
Xianbo Jia