- 1College of Pharmacy, Guizhou University of Traditional Chinese Medicine, Guiyang, Guizhou, China
- 2School of Chinese Medicine, Hong Kong Baptist University, Hong Kong, Hong Kong SAR, China
- 3School of Food and Pharmaceutical Engineering, Guizhou Institute of Technology, Guiyang, Guizhou, China
During the survey on freshwater hyphomycetes in Guangxi, Guizhou and Hainan Provinces, China, five fresh collections were encountered. Based on their morphology, these five isolates were identified as belonging to Hermatomyces, Kirschsteiniothelia, Paramonodictys, Pleopunctum and Sparticola. Multi-gene phylogenetic analyses were performed for each genus, which resulted in the identification of five new species, namely Hermatomyces hainanensis, Kirschsteiniothelia ramus, Paramonodictys globosa, Pleopunctum guizhouense, and Sparticola irregularis. Detailed descriptions and illustrations of the morphological characteristics of these new taxa were provided. This research enriches the biodiversity of freshwater dematiaceous hyphomycetes.
1 Introduction
Freshwater fungi are a diverse and heterogeneous group that can be classified into different classes (Marvanová, 1980; Goh and Hyde, 1996; Shearer et al., 2009; Baschien et al., 2013; Luo et al., 2019; Dong et al., 2020; Yang et al., 2023). Calabon et al. (2022) listed 3,870 species occurring in freshwater habitats. They play essential roles, such as decomposers for submerged woody debris, in freshwater ecosystems (Wong et al., 1998; Grossart et al., 2019), and many of them possess unique biochemical properties that have great potential for various applications (Krauss et al., 2011; El-Elimat et al., 2021). Therefore, the study of fungal biodiversity in freshwater habitats is important, and five genera are involved in the present paper.
Hermatomyces was introduced by Spegazzini (1910) with the type species H. tucumanensis. This genus was previously placed in Lophiotremataceae (Tibpromma et al., 2016; Doilom et al., 2017). Hashimoto et al. (2017) excluded Hermatomyces from Lophiotremataceae and resurrected the family Hermatomycetaceae based on their phylogenetic analyses using SSU, ITS, LSU, tef1-α and rpb2 sequences. Asexual morph of Hermatomyces species is characterized by sporodochial conidiomata and dimorphic conidia, i.e., cylindrical conidia and lenticular conidia (Ellis, 1971; Chang, 1995; Tibpromma et al., 2016; Doilom et al., 2017; Koukol et al., 2018; Ren et al., 2021). Its sexual morph was recently reported by de Silva et al. (2022), which has dark brown to black ascomata with central ostiole, 8-spored, bitunicate asci, and broadly fusiform, hyaline, 1-septate ascospores.
Hawksworth (1985) proposed Kirschsteiniothelia as a sexual morphic genus, which was linked to the Dendryphiopsis asexual morph. Kirschsteiniotheliaceae was established by Boonmee et al. (2012) after observing the sexual-asexual morph connection based on a culture study. The order Kirschsteiniotheliales was subsequently proposed by Hernández-Restrepo et al. (2017). The dendryphiopsis-like asexual morph is characterized by macronematous, mononematous, apically branched conidiophores and cylindrical conidia with rounded ends (Ellis, 1971; Pratibha et al., 2010; Boonmee et al., 2012). Su et al. (2016) identified Kirschsteiniothelia submersa as a sporidesmium-like asexual morph, and then several novel Kirschsteiniothelia species with sporidesmium-like morphology were introduced, such as K. aquatica, K. cangshanensis, K. fluminicola and K. rostrata (Hyde et al., 2017; Bao et al., 2018). Sun et al. (2021) provided a detailed summary of Kirschsteiniothelia species.
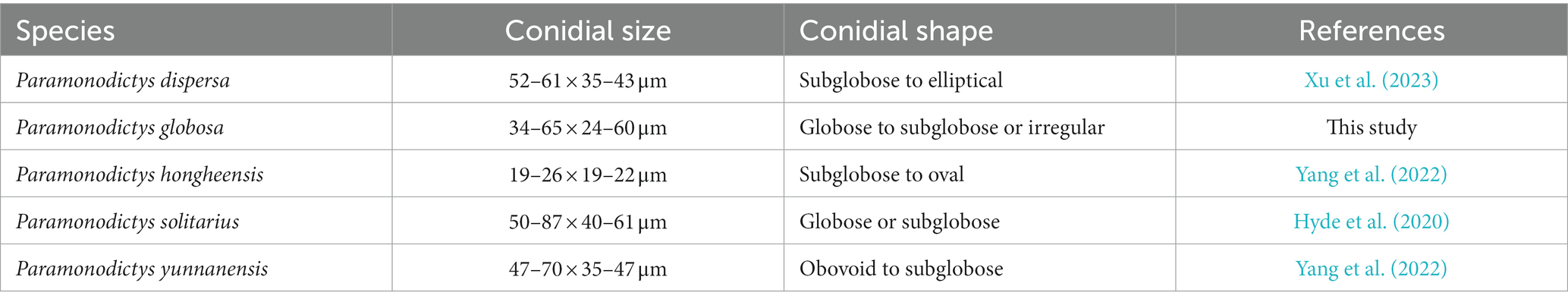
Hyde et al. (2020) introduced Paramonodictys for a monodictys-like species, which is characterized by present stroma and globose to subglobose, brown, muriform conidia. Paramonodictys is classified in Parabambusicolaceae (Pleosporales) (Hyde et al., 2020). To date, four species are accepted in this genus (Hyde et al., 2020; Yang et al., 2022; Xu et al., 2023), among which, P. dispersa, has been reported from a freshwater habitat (Xu et al., 2023).
Pleopunctum belonging to Phaeoseptaceae (Pleosporales) was introduced by Liu et al. (2019) based on the type species Pl. ellipsoideum, along with Pl. pseudoellipsoideum. Seven species were included in Pleopunctum, of which three species, viz. Pl. megalosporum, Pl. multicellularum and Pl. rotundatum were reported from freshwater habitats (Xu et al., 2023). The sexual morph of Pleopunctum has not been reported, and its asexual morph is characterized by sporodochial conidiomata, oval to ellipsoidal, brown, muriform conidia often with one or several hyaline, globose to ellipsoidal basal cells (Liu et al., 2019; Phukhamsakda et al., 2020; Boonmee et al., 2021; Senwanna et al., 2021; Wanasinghe et al., 2022; Xu et al., 2023). Hyaline phragmosporous or dictyosporous conidia have also been reported in this genus (Phukhamsakda et al., 2020; Senwanna et al., 2021; Wanasinghe et al., 2022).
Sparticola was introduced by Phukhamsakda et al. (2016) to accommodate S. forlicesenae, S. junci (type species) and S. triseptata. A fourth species, S. muriformis, was introduced by Karunarathna et al. (2017). All four of these terrestrial species have been found to exhibit sexual morphs in nature. Only S. junci produces a hyphomycetous asexual morph in culture, characterized by semi-macronematous to macronematous, pale brown to brown conidiophores, holoblastic conidiogenous cells, and irregular, brown to dark brown conidia (Phukhamsakda et al., 2016).
In this study, we introduce five new species collected from freshwater habitats in Guangxi, Guizhou and Hainan Provinces, China. Based on morphological characteristics and phylogenetic analyses, they are identified as Hermatomyces hainanensis sp. nov., Kirschsteiniothelia ramus sp. nov., Paramonodictys globosa sp. nov., Pleopunctum guizhouense sp. nov., and Sparticola irregularis sp. nov. Detailed descriptions and illustrations are provided for these five new taxa.
2 Materials and methods
2.1 Collections and examination of specimens
Fresh samples were collected from May 2021 to July 2022 in Guangxi, Guizhou and Hainan Provinces, China. The samples were incubated in moist plastic boxes at room temperature for 14 days. A Motic SMZ 168 Series dissecting microscope was used to check the specimen. Fruiting bodies of the new collections were examined and photographed with a Nikon ECLIPSE Ni compound microscope fitted with a Canon 90D digital camera. The software Tarosoft (R) Image Frame Work was used to take measurements of fungal structures, and Adobe Photoshop CC 2019 (Adobe Systems, USA) was used to prepare the photo-plates.
Single conidium isolations were carried out on potato dextrose agar (PDA) media (Senanayake et al., 2020). Germinated conidia were individually transferred to fresh PDA media plates and incubated in a constant temperature incubator at 25°C. Dried specimens were deposited in the Herbarium of Cryptogams, Kunming Institute of Botany Academia Sinica (HKAS), Kunming, China, and the herbarium of Guizhou Academic of Agriculture Sciences (GZAAS), Guiyang, China. Pure cultures were deposited in the Guizhou Culture Collection (GZCC), Guiyang, China. Fungal Names numbers were applied in Fungal Names (2023).1
2.2 DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from fresh mycelia growing on PDA medium for 1 month at 25°C using a DNA extraction kit (BioFlux, China). Four different gene regions, the nuclear large subunit rDNA (28S, LSU), the internal transcribed spacer (ITS), the translation elongation factor (tef1-α), and the RNA polymerase II subunit 2 (rpb2) were selected for study. Primer pairs LR0R/LR5 (Vilgalys and Hester, 1990), ITS5/ITS4 (White et al., 1990), EF1-983F/EF1-2218R (Rehner and Buckley, 2005) and fRPB2-5F/fRPB2-7cR (Liu et al., 1999) were used to amplify part of LSU, ITS, tef1-α and rpb2 loci, respectively. Polymerase chain reaction (PCR) was carried out in a 50 μL reaction volume containing 44 μL of 1.1 × T3 Supper PCR Mix (Qingke Biotech, China), 2 μL of forward and reverse primers, and 2 μL of DNA template. The PCR protocols were referred to Ma et al. (2023). And 1% agarose electrophoresis gels stained with ethidium bromide were used to examine the resulting PCR products. Successful PCR products were sequenced by Beijing Qingke Biotechnology Co., Ltd.
2.3 Phylogenetic analyses
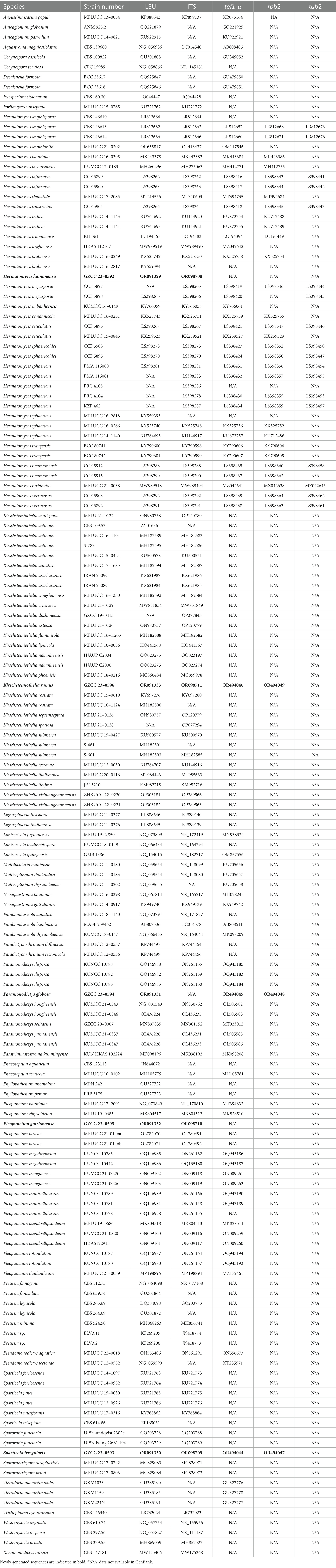
Sequences obtained from different primers were analyzed with related taxa determined by blastn search in NCBI. Alignments for different gene loci were automatically performed by online MAFFT version 7.2 Trimal v1.2 (Capella-Gutierrez et al., 2009) was used to remove ambiguously aligned regions and uninformative positions with gappyout option. Multi-gene alignments were combined using SequenceMatrix 1.7.8 (Vaidya et al., 2011). Alignments were checked visually using AliView (Larsson, 2014). Sequences derived in this study were deposited in GenBank (Table 1).
Maximum likelihood (ML) analyses were performed using IQ-TREE web server (Trifinopoulos et al., 2016). Substitution model was automatically tested. Ultrafast bootstrap (BS) analysis was implemented with 1,000 replicates. Maximum likelihood bootstrap values (ML-BS) equal or greater than 75% are marked near each node.
Bayesian inference (BI) analyses were carried out in MrBayes 3.2.6 (Ronquist et al., 2012) using a Markov Chain Monte Carlo (MCMC) algorithm. The best-fit substitution model GRT + I + G was decided for all four gene regions by MrModeltest 2.3 (Nylander, 2008) under the Akaike Information Criterion (AIC). Two parallel runs of four simultaneous Markov chains were performed for 1,000,000 generations. Trees were sampled every 1,000th generations. Burn-in phase was set at 25% and the remaining trees were used for calculating posterior probabilities (PP). PP values equal or greater than 0.95 are marked near each node.
Trees were visualized with FigTree v1.4.4,3 and the layouts were edited using Adobe Illustrator CS6 software (Adobe Systems, USA).
3 Taxonomy
Hermatomyces hainanensis J. Ma, Y.Z. Zhang & Y.Z. Lu, sp. nov., Figure 1.

Figure 1. Hermatomyces hainanensis (HKAS 129170, holotype). (A) Colonies on natural substrates. (B) Subicular hyphae. (C) Germinated conidium. (D–G) Cylindrical conidia. (H–J) Lenticular conidia. Scale bars: (B–J) = 20 μm.
Fungal Names number: FN571666.
Holotype: HKAS 129170.
Etymology: Referring to the location where the species was collected.
Saprobic on decaying wood in freshwater habitat. Sexual morph: Undetermined. Asexual morph: hyphomycetous. Colonies on natural substrate sporodochial, effuse, scattered, circular or subcircular, consisting of a brown sterile mycelial outer zone and an abundantly sporulating, dark brown to blackish brown center. Mycelium partly immersed, partly superficial, composed of pale to brown, branched, septate hyphae. Conidiophores 15–19 × 2–5 μm ( = 48 × 32 μm, n = 15), micronematous to semi-macronematous, unevenly cylindrical, geniculate, subhyaline to brown, septate, thick-walled. Conidiogenous cells 4.5–8 × 2.5–3 μm ( = 6.5 × 2.5 μm, n = 15), monoblastic, integrated, terminal, subcylindrical, subhyaline to pale brown. Conidia dimorphic, (1) cylindrical conidia 51–67 × 16–24 μm ( = 58.5 × 21 μm, n = 30), hyaline to subhyaline, often with a distinct dark brown pigmentation from the top downwards or at rim of the conidia, straight or broadly curved, phragmoseptate or muriform, sometimes with oblique septa, constricted at the septa, consisting of two columns from one or two basal cells, rounded at the apex; (2) lenticular conidia 44–52 × 29–39 μm ( = 48 × 32 μm, n = 30), ellipsoidal in front view, central cells dark brown to blackish brown, peripheral cells subhyaline to pale olivaceous brown, forming a distinct ring, muriform, constricted at the septa, smooth-walled, side views not observed.
Culture characteristics: Conidia were germinated on PDA medium and produced germ tubes within 18 h. Colonies grown on PDA are pale brown to brown, circular, surface flat, edge entire, reaching 36 mm diam. in 42 days at 25°C.
Material examined: China, Hainan Province, Qiongzhong Li and Miao Autonomous County, Baihualing Rainforest cultural tourism area, 18°98′ N, 109°82′ E, on rotting wood in a freshwater stream, 29 December 2021, Jian Ma, BH39 (HKAS 129170, holotype; GZAAS 23–0595, isotype), ex-type living culture GZCC 23-0592.
Notes: Hermatomyces hainanensis is similar to other Hermatomyces species with dimorphic conidia, such as H. bifurcatus, H. constrictus, H. iriomotensis, H. jinghaensis, H. krabiensis, H. megaspores, H. tucumanensis, and H. turbinatus (Chang, 1995; Tibpromma et al., 2016, 2017; Hashimoto et al., 2017; Koukol et al., 2018; Ren et al., 2021). Based on phylogenetic analyses, Hermatomyces hainanensis (GZCC 23-0592) is closely related to H. megasporus (CCF 5897 and CCF 5898) and H. reticulatus (CCF 5893 and MFLUCC 15-0843), although H. reticulatus only exhibits one type of conidia (Koukol et al., 2018). Despite some overlap in the sizes of the cylindrical (51–67 × 16–24 μm vs. 49.5–60.5 × 18–28 μm) and lenticular (44–52 × 29–39 μm vs. 49–56 × 37–46 μm) conidia of Hermatomyces hainanensis and H. megasporus (Koukol et al., 2018), the phylogenetic analyses suggest that they are distinct species (Figure 2). Comparisons of ITS sequences showed that there are 22 bp (in a total 886 bp, 2.5%) differences with 2 gaps between H. hainanensis (GZCC 23-0592) and H. megasporus (CCF 5898), and 10 bp (in a total 444 bp, 2.3%) differences with 1 gap between H. reticulatus (CCF 5893). Following the guidelines for species delineation (Jeewon and Hyde, 2016), we identify our collection as a new species.
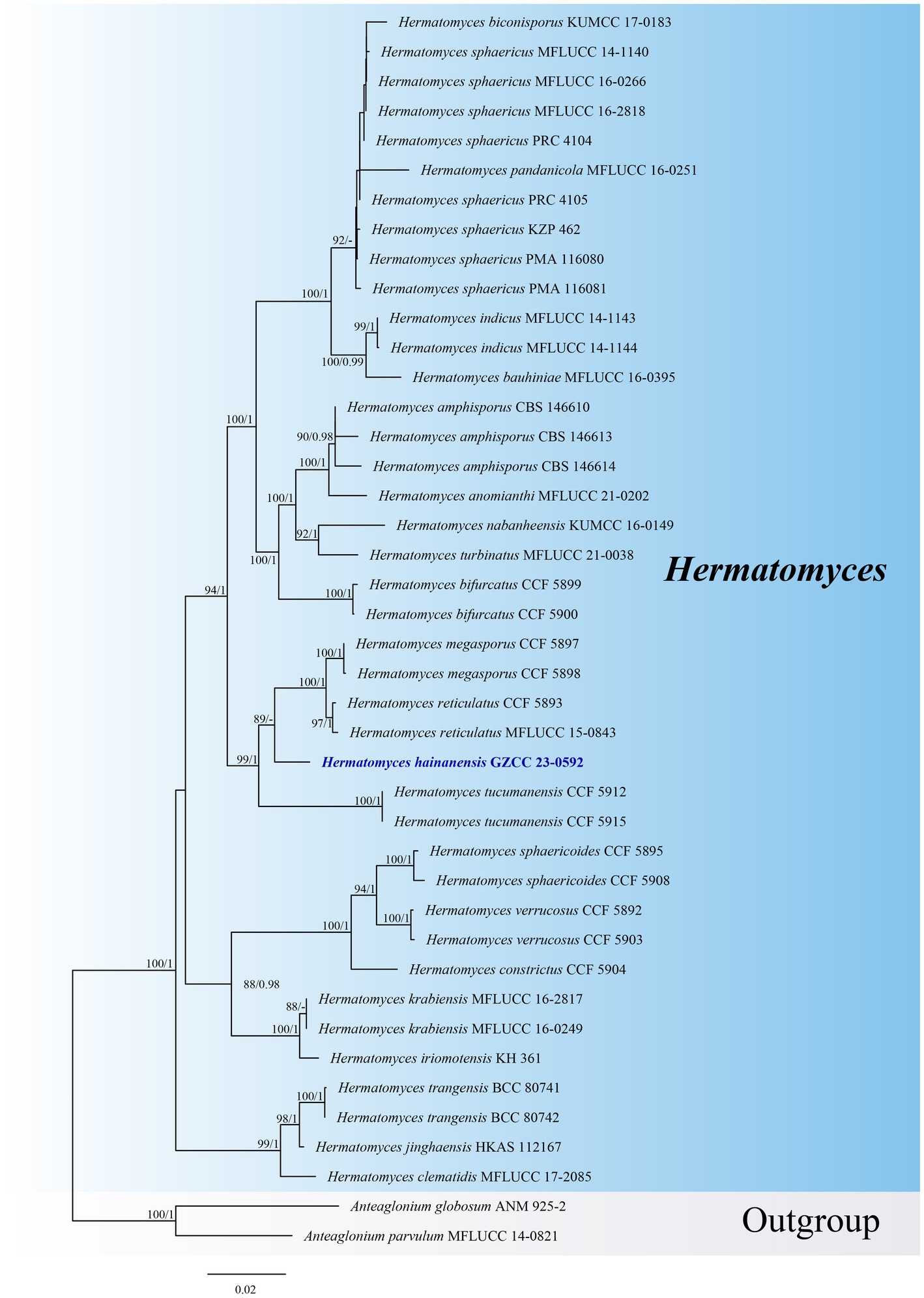
Figure 2. ML tree (−ln = 12797.020) based on the combined LSU-ITS-tef1-α-rpb2-tub rDNA sequences. The combined dataset comprises 41 strains, including the new collection. The alignment comprises 3,940 characters (LSU: 1–852, ITS: 853–1324, tef1-α: 1325–2260, rpb2: 2261–3295, tub: 3296–3940) including gaps. Among them, number of constant sites are 3,150, and number of parsimony informative sites are 597. Bootstrap support values for ML greater than 75% and PP greater than 0.95 are given near nodes as ML-BS/PP. The tree is rooted with Anteaglonium globosum (ANM 925–2) and Anteaglonium parvulum (MFLUCC 14–0821). The new taxon is indicated in bold and blue.
Kirschsteiniothelia ramus J. Ma, Y.Z. Zhang & Y.Z. Lu, sp. nov., Figure 3.

Figure 3. Kirschsteiniothelia ramus (HKAS 129167, holotype). (A,B) Colonies on natural substrates. (C–F) Conidiophores and conidia. (G–K) Conidia. Scale bars: (C–F) = 50 μm, (G–K) = 20 μm.
Fungal Names number: FN571667.
Holotype: HKAS 129167.
Etymology: Referring to the apically branched conidiophores.
Saprobic on decaying wood in freshwater habitat. Sexual morph: Undetermined. Asexual morph: hyphomycetous. Colonies on natural substrate effuse, dark brown, gregarious, velvety. Mycelium mostly immersed, composed of gray to brown, branched, septate hyphae. Conidiophores 102–248 × 5–11 μm ( = 174 × 8.5 μm, n = 15), macronematous, mononematous, erect, straight or flexuous, cylindrical, brown, slightly paler toward the apex, simple or mostly apically branched, septate, thick-walled. Conidiogenous cells 18–27 × 6.5–9 μm ( = 22.5 × 8 μm, n = 30), monotretic, integrated, terminal at the apex of stipe and fertile branches, pale brown to brown, doliiform or lageniform. Conidia 42–56 × 15–22 μm ( = 49.5 × 19.5 μm, n = 30), acrogenous, solitary, cylindrical, rounded at the apex, subtruncate at the base, pale olivaceous when young, brown when mature, 2–3-septate, with septa thickened and darkened, verruculose.
Culture characteristics: Conidia were germinated on PDA medium and produced germ tubes within 24 h. Colonies grown on PDA are gray to olivaceous, circular, surface flat, edge entire, reaching 20 mm diam. in 28 days at 25°C.
Material examined: China, Hainan Province, Yanoda Tropical rainforest scenic area, on submerged decaying wood in a freshwater stream, 23 October 2021, Jian Ma, Y13 (HKAS 129167, holotype; GZAAS 23-0599, isotype), ex-type living culture, GZCC 23-0596.
Notes: Kirschsteiniothelia ramus resembles other Kirschsteiniothelia species with dendryphiopsis-like asexual morphs (Sun et al., 2021). Phylogenetically (Figure 4), Kirschsteiniothelia ramus (GZCC 23-0596) is a sister taxon to K. lignicola (MFLUCC 10-0036). However, K. ramus has larger conidiophores (102–248 × 5–11 μm vs. 39–148 × 4–7 μm) and larger conidia (42–56 × 15–22 μm vs. 24.5–35 × 14–16 μm) than K. lignicola (Boonmee et al., 2012). Besides, conidia of K. ramus are 2–3-septate, while the latter is 1–2-septate (Boonmee et al., 2012). Furthermore, the ITS (471 bp) sequence variation between K. ramus (GZCC 23-0596) and K. lignicola (MFLUCC 10–0036) occurs in 31 positions, including 5 gaps. Therefore, we introduce Kirschsteiniothelia ramus as a new species.
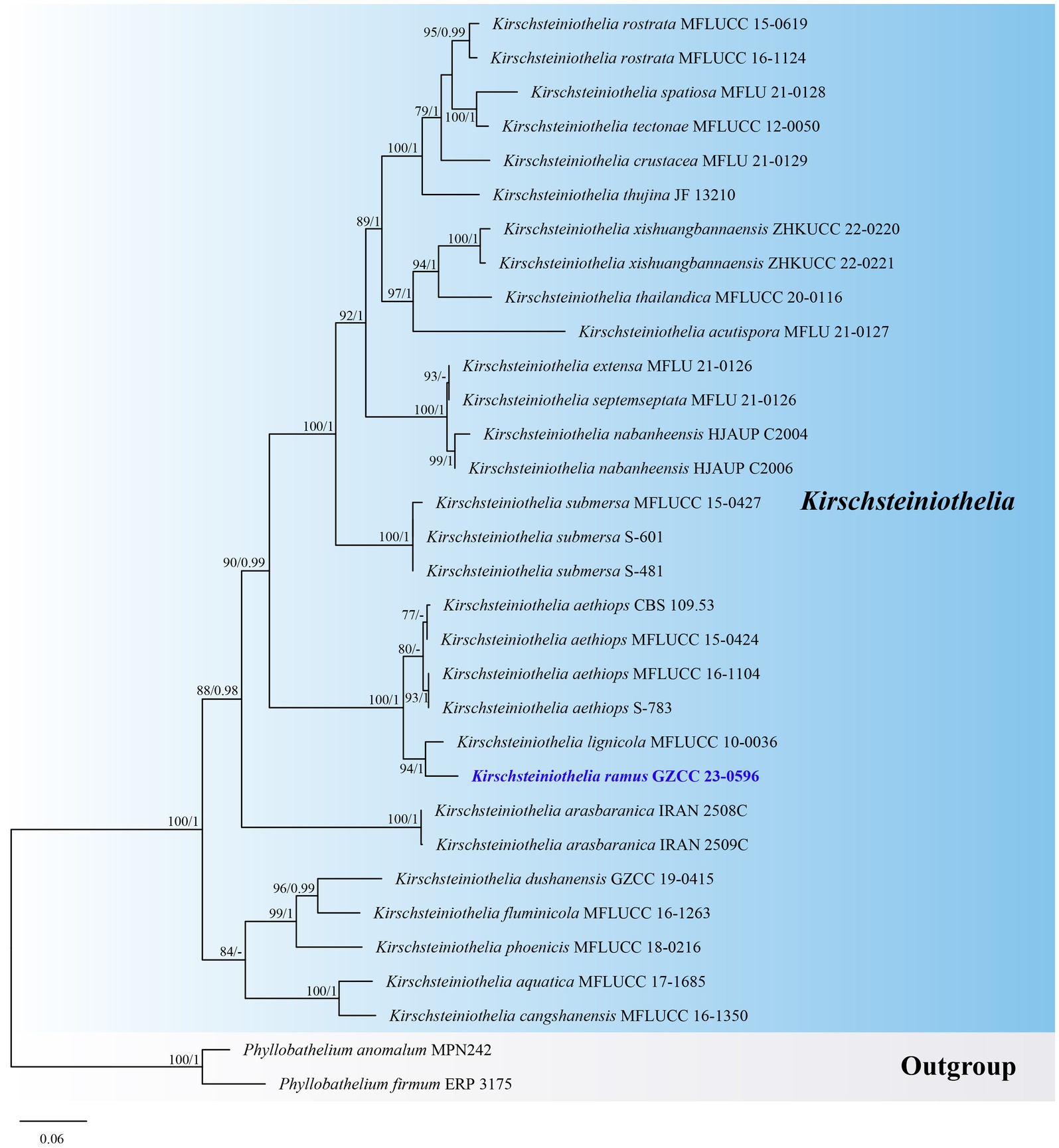
Figure 4. ML tree (−ln = 9010.980915) based on the combined LSU-ITS rDNA sequences. The combined dataset comprises 32 strains, including the new collection. The alignment comprises 1,461 characters (LSU: 1–872, ITS: 873–1461) including gaps. Among them, number of constant sites are 849, and number of parsimony informative sites are 474. Bootstrap support values for ML greater than 75% and PP greater than 0.95 are given near nodes as ML-BS/PP. The tree is rooted with Phyllobathelium anomalum (MPN242) and Phyllobathelium firmum (ERP 3175). The new taxon is indicated in bold and blue.
Paramonodictys globosa J. Ma, Y.Z. Zhang & Y.Z. Lu, sp. nov., Figure 5.
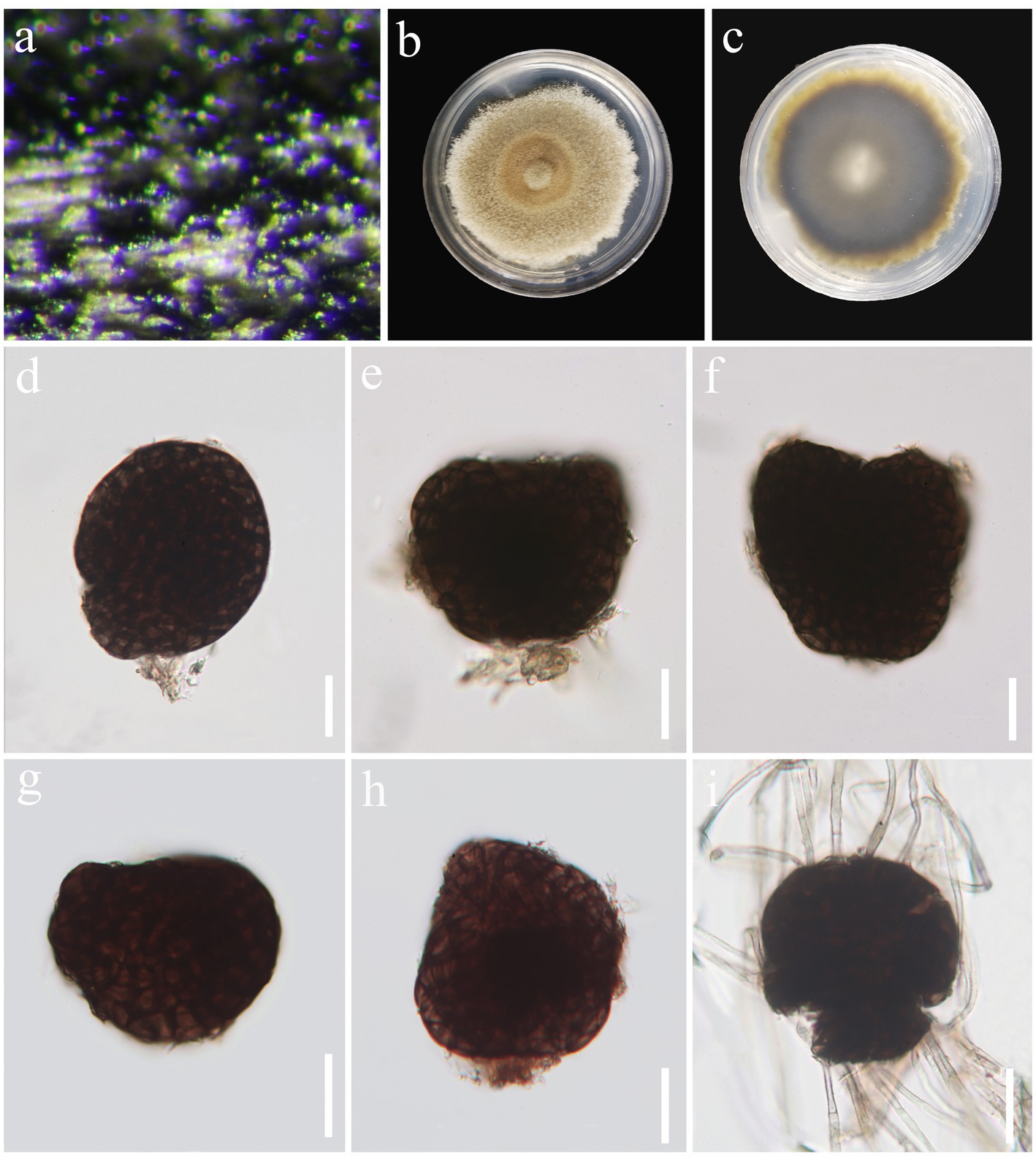
Figure 5. Paramonodictys globosa (HKAS 129169, holotype). (A) Colonies on natural substrates. (B,C) Colonies on PDA media. (D–H) Conidia. (I) Germinated conidium. Scale bars: (D–I) = 20 μm.
Fungal Names number: FN571668.
Holotype: HKAS 129169.
Etymology: Referring to the globose conidia.
Saprobic on decaying wood in freshwater habitat. Sexual morph: Undetermined. Asexual morph: hyphomycetous. Colonies on natural substrate superficial, effuse, scattered, black. Mycelium mostly immersed, composed of pale brown to brown, branched, septate hyphae. Stroma not observed. Conidiophores absent. Conidiogenous cells monoblastic. Conidia 34–65 × 24–60 μm ( = 54 × 46 μm, n = 30), solitary, globose to subglobose or irregular, usually broadly rounded at apex, subtruncate at base, olivaceous brown to dark brown, muriform, thickened and darkened at the septa, verrucous.
Culture characteristics: Conidia were germinated on PDA media and produced germ tubes within 24 h. Colonies grown on PDA are pale brown to brown, circular, surface umbonate, edge undulate, reaching 55 mm diam. in 42 days at 25°C.
Material examined: China, Guangxi Zhuang Autonomous Region, Hechi City, Nandan County, Pingzhou, on submerged decaying wood in a freshwater stream, 1 May 2021, Jian Ma, ND22 (HKAS 129169, holotype; GZAAS 23-0597, isotype), ex-type living culture, GZCC 23-0594.
Notes: A comparison of conidial sizes and shapes for the five accepted Paramonodictys species is provided in Table 2. The conidia of Paramonodictys globosa are larger than those of P. hongheensis. However, P. globosa has a similar conidial size to P. dispersa, P. solitarius and P. yunnanensis, meaning they cannot be distinguished based on morphology alone. In our phylogenetic analyses, P. globosa (GZCC 23-0594) formed a basal clade within the Paramonodictys group (Figure 6), indicating it is phylogenetically distinct. Therefore, we introduce Paramonodictys globosa as a new species based on both morphological and molecular evidence.
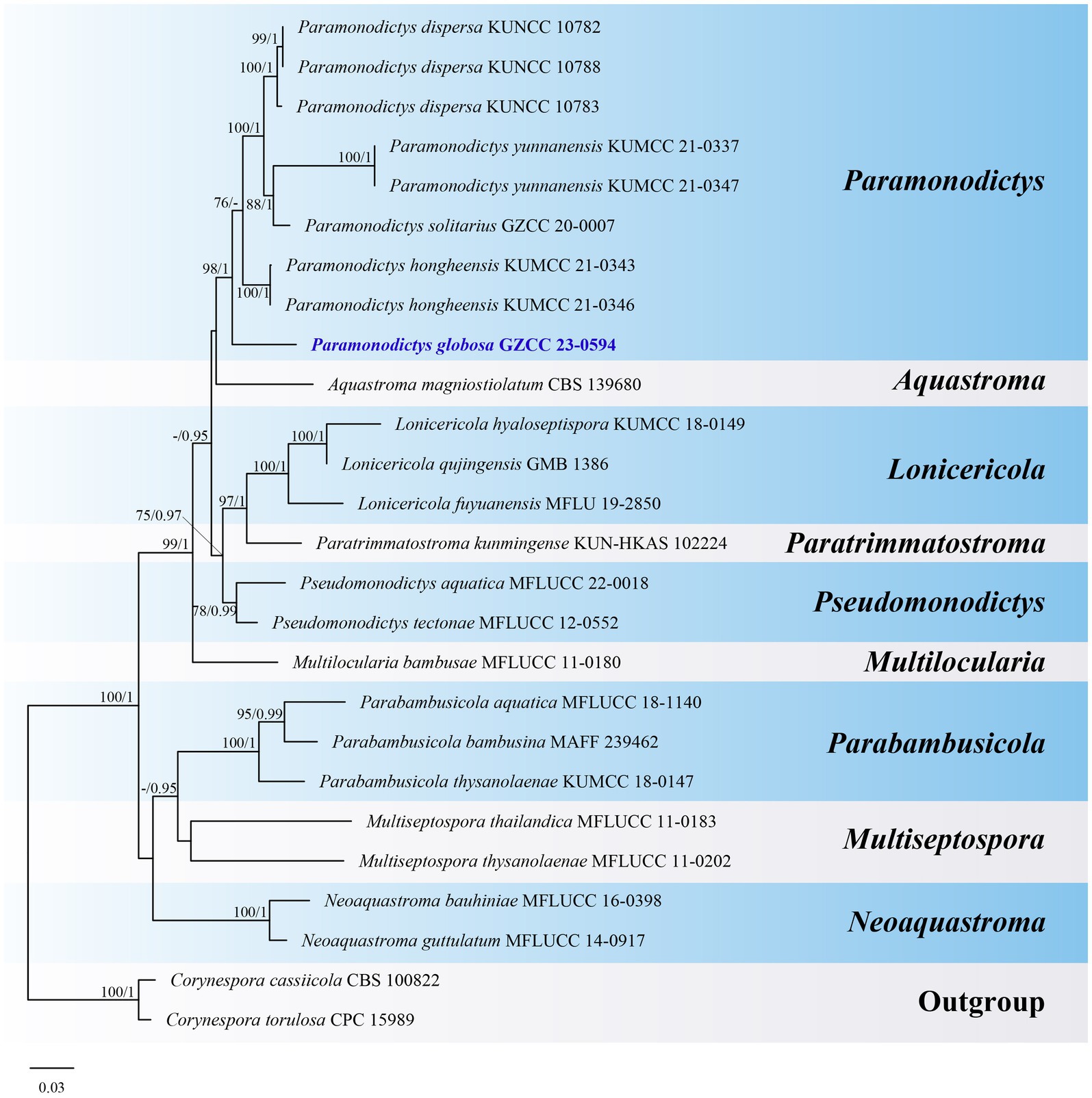
Figure 6. ML tree (−ln = 11817.931619) based on the combined LSU-ITS-tef1-α rDNA sequences. The combined dataset comprises 26 strains, including the new collection. The alignment comprises 2,548 characters (LSU: 1–847, ITS: 848–1366, tef1-α: 1367–2548) including gaps. Among them, number of constant sites are 1,802, and number of parsimony informative sites are 515. Bootstrap support values for ML greater than 75% and PP greater than 0.95 are given near nodes as ML-BS/PP. The tree is rooted with Corynespora cassiicola (CBS 100822) and Corynespora torulosa (CPC 15989). The new taxon is indicated in bold and blue.
Pleopunctum guizhouense J. Ma, N.G. Liu & Y.Z. Lu, sp. nov., Figure 7.

Figure 7. Pleopunctum guizhouense (HKAS 129171, holotype). (A) Colonies on natural substrates. (B,C) Colonies on PDA media. (D–J) Conidia. (K) Germinated conidium. Scale bars: (D–I) = 20 μm.
Fungal Names number: FN571669.
Holotype: HKAS 129171.
Etymology: Referring to the location where the species was collected.
Saprobic on decaying wood in freshwater habitat. Sexual morph: Undetermined. Asexual morph: hyphomycetous. Colonies on natural substrate superficial, brown, sporodochial, punctiform. Mycelium mostly immersed, composed of hyaline to pale brown, branched, septate hyphae. Conidiophores macronematous, mononematous, cylindrical, medium brown, simple or branched, septate, thick-walled. Conidiogenous cells monoblastic, integrated, terminal, medium brown. Conidia 45–64 × 26–29.5 μm ( = 57 × 28 μm, n = 30), acrogenous, solitary, oval to ellipsoidal, broadly obtuse at apex, truncate at base, median brown, darker at the apex, muriform, constricted at septa, smooth-walled, often with a hyaline, ellipsoidal to globose basal cell, 16–26 × 11–17 μm ( = 20 × 13.5 μm, n = 30).
Culture characteristics: Conidia were germinated on PDA media and produced germ tubes within 12 h. Colonies grown on PDA are white to pale brown in front view and brown in reverse view, circular, surface umbonate, edge undulate, reaching 35 mm diam. in 28 days at 25°C.
Material examined: China, Guizhou Province, Chishui City, Swan Castle Forest Park, on submerged decaying wood in a freshwater stream, 28 July 2022, Jian Ma, TEB11.1 (HKAS 129171, holotype; GZAAS23-0598, isotype), ex-type living culture, GZCC 23-0595.
Notes: Pleopunctum guizhouense (GZCC 23-0595) clusters together with Pl. menglaense (KUMCC 21-0025 and KUMCC 21-0026) with a weak support in the phylogenetic analyses (Figure 8). However, it can be differentiated from Pl. menglaense by its monomorphic conidia, which are brown and oval to ellipsoidal with a basal cell, while the latter has two types of conidia: spatulate to obovate, hyaline conidia, and brown, ellipsoidal to oblong conidia with 1–3 basal cells (Wanasinghe et al., 2022). The ITS (464 bp) sequence variation between Pl. guizhouense (GZCC 23-0595) and Pl. menglaense (KUMCC 21-0026) occurs in 16 positions, including 5 gaps. Considering these morphological differences and their distinct phylogenetic positions, we introduce our collection as a new species, named Pleopunctum guizhouense.
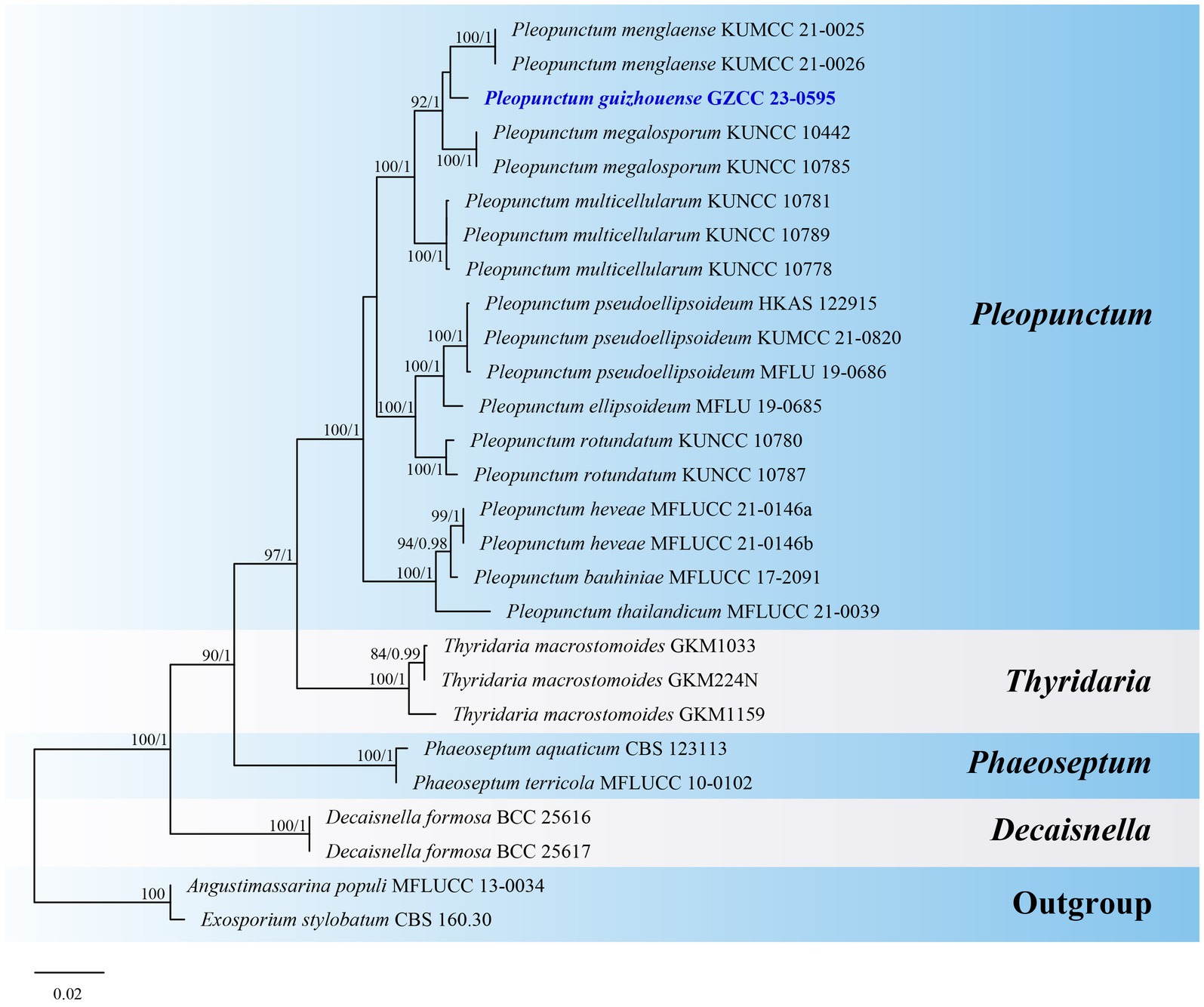
Figure 8. ML tree (−ln = 7049.253556) based on the combined LSU-ITS-tef1-α rDNA sequences. The combined dataset comprises 27 strains, including the new collection. The alignment comprises 2,305 characters (LSU: 1–851, ITS: 852–1385, tef1-α: 1386–2305) including gaps. Among them, number of constant sites are 1, 822, and number of parsimony informative sites are 411. Bootstrap support values for ML greater than 75% and PP greater than 0.95 are given near nodes as ML-BS/PP. The tree is rooted with Angustimassarina populi (MFLUCC 13-0034) and Exosporium stylobatum (CBS 160.30). The new taxon is indicated in bold and blue.
Sparticola irregularis J. Ma, N.G. Liu & Y.Z. Lu, sp. nov., Figure 9.

Figure 9. Sparticola irregularis (HKAS 129168, holotype). (A,B) Colonies on natural substrates. (C,D) Colonies on PDA media. (E–G) Conidiophores and conidia. (H–J) Conidia. (K) Germinated conidium. Scale bars: (E–G) = 50 μm, (H–K) = 20 μm.
Fungal Names number: FN571670.
Holotype: HKAS 129168.
Etymology: Referring to the irregular conidia.
Saprobic on decaying wood in freshwater habitat. Sexual morph: Undetermined. Asexual morph: hyphomycetous. Colonies on natural substrate effuse, black, velvety. Mycelium mostly immersed, composed of pale gray to pale brown, branched, septate hyphae. Conidiophores 97–162 × 9–11.5 μm ( = 127 × 10.5 μm, n = 15), macronematous, mononematous, erect, subcylindrical, sometimes with doliiform cells at upper part, dark brown and wider at base, paler and thinner toward to the apex, simple, occasionally branched, septate, thick-walled. Conidiogenous cells 11–15 × 5–6 μm ( = 13 × 5.5 μm, n = 15), monoblastic, integrated, terminal, subcylindrical or doliiform, brown. Conidia 33–53 × 33–51 μm ( = 45 × 40 μm, n = 30), acrogenous, solitary, irregular, brown to dark brown, muriform, with protuberant, truncate base.
Culture characteristics: Conidia were germinated on PDA media and produced germ tubes within 12 h. Colonies grown on PDA are gray to pale brown, irregular, surface flat, edge filiform, reaching 54 mm diam. in 42 days at 25°C.
Material examined: China, Hainan Province, Haikou City, Xiuying District, Ecological leisure trail, on decaying wood in a freshwater stream, 20°01′ N, 110°25′ E, 10 August 2021, Jian Ma, HK7 (HKAS 129168, holotype; GZAAS23-0596, isotype), ex-type living culture GZCC 23-0593.
Notes: Sparticola species are typically identified by their sexual morphology (Phukhamsakda et al., 2016; Karunarathna et al., 2017). Sparticola junci is the only species known to produce a hyphomycetous asexual morphology in culture, which is similar to that of our new collection in terms of conidial morphology (Phukhamsakda et al., 2016). Although our collection has larger conidiophores (97–162 × 9–11.5 μm vs. up to 35 × 4–7 μm) than those of S. junci, this difference might be attributed to variations in growth conditions (nature vs. culture). Unfortunately, we were unable to observe sporulation in our culture. Phylogenetically (Figure 10), Sparticola irregularis (GZCC 23-0593) forms a basal clade to the Sparticola group (ML-BS = 74%, PP = 0.93), and could represent a new genus because the LSU (823 bp) sequences comparison between S. irregularis (GZCC 23-0593) and S. junci (MFLUCC 15-0030) shows there are 28 position differences, including 2 gaps. However, considering that we only have one isolate, the evidence is insufficient to propose a new genus. Thus, we provisionally assign our new collection to Sparticola and introduce the new species Sparticola irregularis. Further fresh collections of Sparticola species or the discovery of the sexual morphology of S. irregularis may provide better resolution for its taxonomic identification.
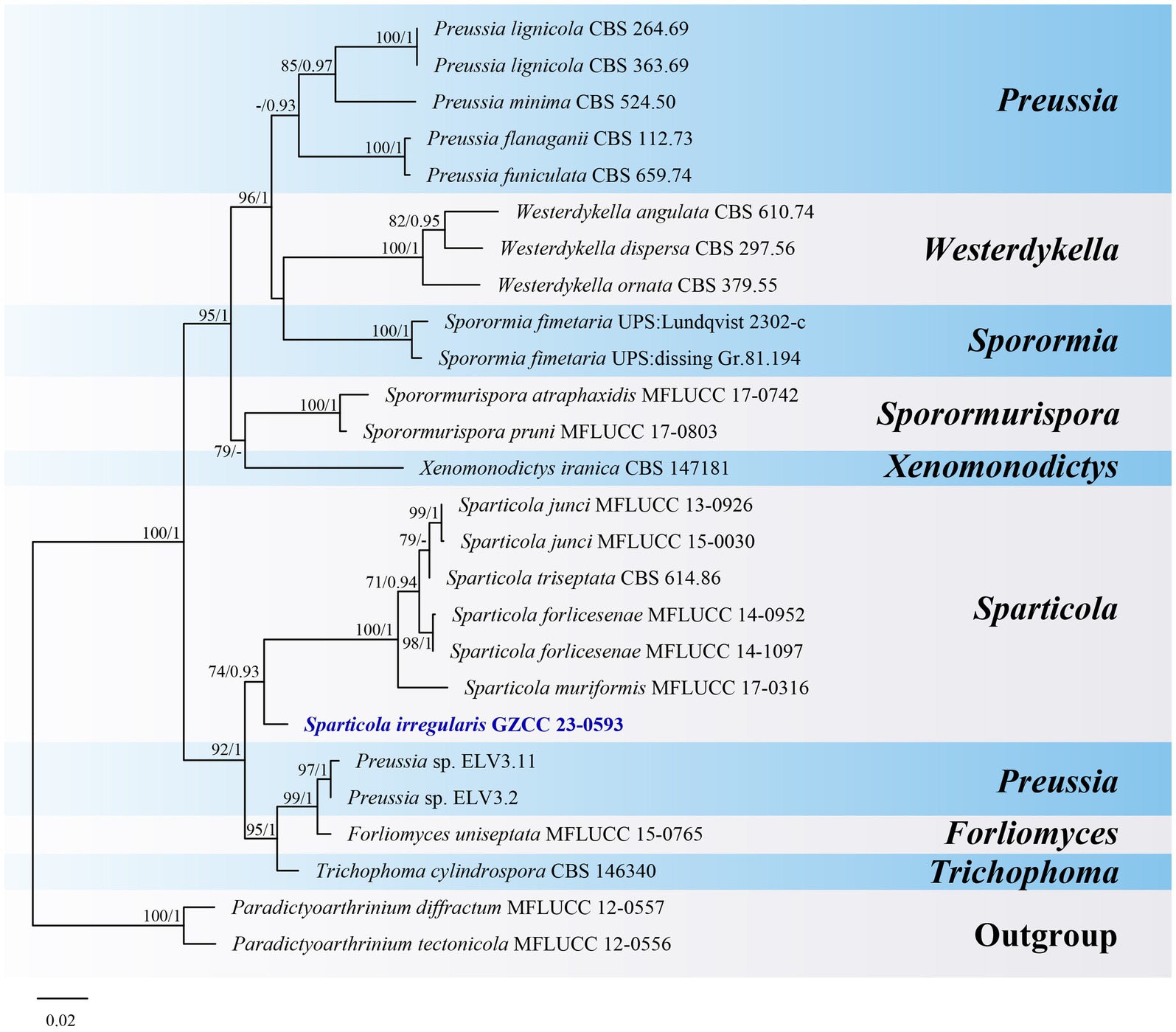
Figure 10. ML tree (−ln = 5776.431900) based on the combined LSU-ITS rDNA sequences. The combined dataset comprises 26 strains, including the new collection. The alignment comprises 1,364 characters (LSU: 1–847, ITS: 848–1364) including gaps. Among them, number of constant sites are 1,039, and number of parsimony informative sites are 265. Bootstrap support values for ML greater than 70% and PP greater than 0.90 are given near nodes as ML-BS/PP. The tree is rooted with Paradictyoarthrinium diffractum (MFLUCC 12–0557) and Paradictyoarthrinium tectonicola (MFLUCC 12-0556). The new taxon is indicated in bold and blue.
4 Discussion
In this study, we introduce five new species, namely Hermatomyces hainanensis, Kirschsteiniothelia ramus, Paramonodictys globosa, Pleopunctum guizhouense, and Sparticola irregularis. The discovery of these five new taxa enriches the freshwater fungi resources of China and further reveals the diverse morphology for this group of fungi. Hermatomyces, Kirschsteiniothelia, Paramonodictys and Pleopunctum have been reported from both terrestrial and freshwater habitats (Sun et al., 2021; Calabon et al., 2022; Xu et al., 2023). Among them, Hermatomyces and Kirschsteiniothelia have a worldwide distribution, while Pleopunctum are reported from China and Thailand (Liu et al., 2019; Phukhamsakda et al., 2020; Senwanna et al., 2021; Xu et al., 2023). To date, all published Paramonodictys species are described from China (Hyde et al., 2020; Yang et al., 2022; Xu et al., 2023). However, occurring at different altitudes suggests that Paramonodictys is highly adaptable to different environments and thus may also exist in other countries. Sparticola species mainly occur in Europe except for S. muriformis from China (Phukhamsakda et al., 2016; Karunarathna et al., 2017). In this study, we report Sparticola from freshwater habitat for the first time.
The classification of species in Hermatomyces, particularly H. sphaericus, has been widely debated. Koukol et al. (2018) considered H. chromolaenae, H. saikhuensis and H. tectonae as synonyms for H. sphaericus, a species that they regarded as monomorphic. Using the GCPSR method, Phukhamsakda et al. (2020) further supported this conclusion. Conversely, Tibpromma et al. (2018) identified H. biconisporus as a distinct species that produces two types of conidia, and clusters within H. sphaericus clade. They rejected Koukol’s treatment and suggested that H. sphaericus might be a species complex. The taxonomic statuses of two other species in the H. sphaericus clade, H. biconisporus and H. pandanicola, remain unresolved. However, Koukol and Delgado (2019) speculated that contamination during single spore isolation may have led to a mixture of conidia from H. sphaericus and H. biconisporus, while H. pandanicola might be a hybrid species, or the sequences in GenBank could have been provided erroneously (Koukol et al., 2018). Therefore, it is essential to collect fresh samples of H. biconisporus and H. pandanicola to resolve their taxonomic controversies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
JM provided the research materials. Y-ZZ and JM contributed to the methodology. N-GL performed the phylogenetic analyses. Y-ZZ, Q-LC, and N-GL wrote the original draft. Y-ZL, H-BC, and N-GL reviewed the draft. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was supported by the Guizhou Province, Science and Technology Department, Natural Science Fund, Qiankehe Grant No. [2018]1071 and the Guizhou University of Traditional Chinese Medicine, National Nature Supply Fund, Grant No. 2018YFC170810501, and the Innovation and Technology Fund in Hong Kong (MHP/023/20).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Bao, D. F., Luo, Z. L., Liu, J. K., Bhat, D. J., Sarunya, N., Li, W. L., et al. (2018). Lignicolous freshwater fungi in China III: three new species and a new record of Kirschsteiniothelia from northwestern Yunnan Province. Mycosphere 9, 755–768. doi: 10.5943/mycosphere/9/4/4
Baschien, C., Tsui, C. K.-M., Gulis, V., Szewzyk, U., and Marvanová, L. (2013). The molecular phylogeny of aquatic hyphomycetes with affinity to the Leotiomycetes. Fungal Biol. 117, 660–672. doi: 10.1016/j.funbio.2013.07.004
Boonmee, S., Ko, T. W. K., Chukeatirote, E., Hyde, K. D., Chen, H., Cai, L., et al. (2012). Two new Kirschsteiniothelia species with Dendryphiopsis anamorphs cluster in Kirschsteiniotheliaceae fam. nov. Mycologia 104, 698–714. doi: 10.3852/11-089
Boonmee, S., Wanasinghe, D. N., Calabon, M. S., Huanraluek, N., Chandrasiri, S. K. U., Jones, G. E. B., et al. (2021). Fungal diversity notes 1387–1511: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 111, 1–335. doi: 10.1007/s13225-021-00489-3
Calabon, M. S., Hyde, K. D., Jones, E. B. G., Luo, Z. L., Dong, W., Hurdeal, V. G., et al. (2022). Freshwater fungal numbers. Fungal Divers. 114, 3–235. doi: 10.1007/s13225-022-00503-2
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chang, H. S. (1995). Notes on Taiwan dematiaceous hyphomycetes, some species of the genera Exserticlava, Craspedodidymum and Hermatomyces. Bot. Bull. Acad. Sin. 36, 243–246.
de Silva, N. I., Hyde, K. D., Lumyong, S., Phillips, A. J. L., Bhat, D. J., Maharachchikumbura, S. S. N., et al. (2022). Morphology, phylogeny, host association and geography of fungi associated with plants of Annonaceae, Apocynaceae and Magnoliaceae. Mycosphere 13, 955–1076. doi: 10.5943/mycosphere/13/1/12
Doilom, M., Dissanayake, A. J., Wanasinghe, D. N., Boonmee, S., Liu, J.-K., Bhat, D. J., et al. (2017). Microfungi on Tectona grandis (teak) in northern Thailand. Fungal Divers. 82, 107–182. doi: 10.1007/s13225-016-0368-7
Dong, W., Wang, B., Hyde, K. D., McKenzie, E. H. C., Raja, H. A., Tanaka, K., et al. (2020). Freshwater Dothideomycetes. Fungal Divers. 105, 319–575. doi: 10.1007/s13225-020-00463-5
El-Elimat, T., Raja, H. A., Figueroa, M., Al Sharie, A. H., Bunch, R. L., and Oberlies, N. H. (2021). Freshwater fungi as a source of chemical diversity: a review. J. Nat. Prod. 84, 898–916. doi: 10.1021/acs.jnatprod.0c01340
Ellis, M.B. (1971). Dematiaceous hyphomycetes. Commonwealth Mycological Institute: Kew, Surrey, England.
Goh, T. K., and Hyde, K. D. (1996). Biodiversity of freshwater fungi. J. Ind. Microbiol. 17, 328–345. doi: 10.1007/BF01574764
Grossart, H.-P., Van den Wyngaert, S., Kagami, M., Wurzbacher, C., Cunliffe, M., and Rojas-Jimenez, K. (2019). Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 17, 339–354. doi: 10.1038/s41579-019-0175-8
Hashimoto, A., Matsumura, M., Hirayama, K., and Tanaka, K. (2017). Revision of Lophiotremataceae (Pleosporales, Dothideomycetes): Aquasubmersaceae, Cryptocoryneaceae, and Hermatomycetaceae fam. nov. Persoonia 39, 51–73. doi: 10.3767/persoonia.2017.39.03
Hawksworth, D. L. (1985). Kirschsteiniothelia, a new genus for the Microthelia incrustans-group (Dothideales). Bot. J. Linn. Soc. 91, 181–202. doi: 10.1111/j.1095-8339.1985.tb01144.x
Hernández-Restrepo, M., Gené, J., Castañeda-Ruiz, R. F., Mena-Portales, J., Crous, P. W., and Guarro, J. (2017). Phylogeny of saprobic microfungi from Southern Europe. Stud. Mycol. 86, 53–97. doi: 10.1016/j.simyco.2017.05.002
Hyde, K. D., Dong, Y., Phookamsak, R., Jeewon, R., Bhat, D. J., Jones, E. B. G., et al. (2020). Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 100, 5–277. doi: 10.1007/s13225-020-00439-5
Hyde, K. D., Norphanphoun, C., Abreu, V. P., Bazzicalupo, A., Thilini Chethana, K. W., Clericuzio, M., et al. (2017). Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Divers. 87, 1–235. doi: 10.1007/s13225-017-0391-3
Jeewon, R., and Hyde, K. D. (2016). Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7, 1669–1677. doi: 10.5943/mycosphere/7/11/4
Karunarathna, A., Phookamsak, R., Wanasinghe, D. N., Wijayawardene, N. N., Weerahewa, H. L. D., Khan, S., et al. (2017). Taxonomy and phylogeny of Sparticola muriformis sp. nov. on decaying grass. Mycosphere 8, 603–614. doi: 10.5943/mycosphere/8/4/9
Koukol, O., and Delgado, G. (2019). Do not forget Africa – revision of fungarium collections at Kew revealed a new species of Hermatomyces (Hermatomycetaceae, Pleosporales). Nova Hedwigia 109, 413–423. doi: 10.1127/nova_hedwigia/2019/0559
Koukol, O., Delgado, G., Hofmann, T. A., and Piepenbring, M. (2018). Panama, a hot spot for Hermatomyces (Hermatomycetaceae, Pleosporales) with five new species, and a critical synopsis of the genus. IMA Fungus 9, 107–141. doi: 10.5598/imafungus.2018.09.01.08
Krauss, G. J., Solé, M., Krauss, G., Schlosser, D., Wesenberg, D., and Bärlocher, F. (2011). Fungi in freshwaters: ecology, physiology and biochemical potential. FEMS Microbiol. Rev. 35, 620–651. doi: 10.1111/j.1574-6976.2011.00266.x
Larsson, A. (2014). AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30, 3276–3278. doi: 10.1093/bioinformatics/btu531
Liu, N. G., Hyde, K. D., Bhat, D. J., Jumpathong, J., and Liu, J. K. (2019). Morphological and phylogenetic studies of Pleopunctum gen. nov. (Phaeoseptaceae, Pleosporales) from China. Mycosphere 10, 757–775. doi: 10.5943/mycosphere/10/1/17
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Luo, Z. L., Hyde, K. D., Liu, J. K., Maharachchikumbura, S. S. N., Jeewon, R., Bao, D. F., et al. (2019). Freshwater Sordariomycetes. Fungal Divers. 99, 451–660. doi: 10.1007/s13225-019-00438-1
Ma, J., Xiao, X. J., Liu, N. G., Boonmee, S., Xiao, Y. P., and Lu, Y. Z. (2023). Morphological and multi-gene phylogenetic analyses reveal Pseudotubeufia gen. nov. and two new species in Tubeufiaceae from China. J. Fungi. 9:742. doi: 10.3390/jof9070742
Marvanová, L. (1980). New or noteworthy aquatic hyphomycetes. Clavatospora, Heliscella, Nawawia and Heliscina. Trans. Br. Mycol. Soc. 75, 221–231. doi: 10.1016/S0007-1536(80)80083-0
Nylander, J.A.A. (2008). MrModeltest 2.3. Uppsala: Department of Systematic Zoology, Uppsala University.
Phukhamsakda, C., Ariyawansa, H. A., Phillips, A. J. L., Wanasinghe, D. N., Bhat, D. J., McKenzie, E. H. C., et al. (2016). Additions to Sporormiaceae: introducing two novel genera, Sparticola and Forliomyces, from Spartium. Cryptogam. Mycol. 37, 75–97. doi: 10.7872/crym/v37.iss1.2016.75
Phukhamsakda, C., McKenzie, E. H. C., Phillips, A. J. L., Gareth Jones, E. B., Jayarama Bhat, D., Stadler, M., et al. (2020). Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 102, 1–203. doi: 10.1007/s13225-020-00448-4
Pratibha, J., Raghukumar, S., and Bhat, D. J. (2010). New species of Dendryphiopsis and Stauriella from Goa, India. Mycotaxon 113, 297–303. doi: 10.5248/113.297
Rehner, S. A., and Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.3852/mycologia.97.1.84
Ren, G. C., Wanasinghe, D. N., Monkai, J., Mortimer, P. E., Hyde, K. D., Xu, J. C., et al. (2021). Novel saprobic Hermatomyces species (Hermatomycetaceae, Pleosporales) from China (Yunnan Province) and Thailand. MycoKeys 82, 57–79. doi: 10.3897/mycokeys.82.67973
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Senanayake, I., Rathnayaka, A. R., Marasinghe, D. S., Calabon, M. S., Gentekaki, E., Lee, H. B., et al. (2020). Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11, 2678–2754. doi: 10.5943/mycosphere/11/1/20
Senwanna, C., Mapook, A., Samarakoon, M., Karunarathna, A., Wang, Y., Tang, A. M. C., et al. (2021). Ascomycetes on Para rubber (Hevea brasiliensis). Mycosphere 12, 1334–1512. doi: 10.5943/mycosphere/12/1/18
Shearer, C. A., Raja, H. A., Miller, A. N., Nelson, P., Tanaka, K., Hirayama, K., et al. (2009). The molecular phylogeny of freshwater Dothideomycetes. Stud. Mycol. 64, 145–153. doi: 10.3114/sim.2009.64.08
Spegazzini, C. (1910). Mycetes Argentinenses (Series V). Anales del Museo Nacional de Historia Natural Buenos Aires. Ser. 20, 329–467.
Su, H. Y., Hyde, K. D., Maharachchikumbura, S. S. N., Ariyawansa, H. A., Luo, Z. L., Promputtha, I., et al. (2016). The families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Divers. 80, 375–409. doi: 10.1007/s13225-016-0362-0
Sun, Y.-R., Jayawardena, R. S., Hyde, K. D., and Wang, Y. (2021). Kirschsteiniothelia thailandica sp. nov. (Kirschsteiniotheliaceae) from Thailand. Phytotaxa 490, 172–182. doi: 10.11646/phytotaxa.490.2.3
Tibpromma, S., Bhat, J., Doilom, M., Lumyong, S., Nontachaiyapoom, S., Yang, J. B., et al. (2016). Three new Hermatomyces species (Lophiotremataceae) on Pandanus odorifer from Southern Thailand. Phytotaxa 275, 127–139. doi: 10.11646/phytotaxa.275.2.4
Tibpromma, S., Hyde, K. D., Jeewon, R., Maharachchikumbura, S. S. N., Liu, J.-K., Bhat, D. J., et al. (2017). Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 83, 1–261. doi: 10.1007/s13225-017-0378-0
Tibpromma, S., Hyde, K. D., McKenzie, E. H. C., Bhat, D. J., Phillips, A. J. L., Wanasinghe, D. N., et al. (2018). Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Divers. 93, 1–160. doi: 10.1007/s13225-018-0408-6
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Vaidya, G., Lohman, D. J., and Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Wanasinghe, D. N., Ren, G. C., Xu, J. C., Cheewangkoon, R., and Mortimer, P. E. (2022). Insight into the taxonomic resolution of the pleosporalean species associated with dead woody litter in natural forests from Yunnan, China. J. Fungi 8:375. doi: 10.3390/jof8040375
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR protocols: a guide to methods and applications. eds. M. A. Innis, D. H. Gelfand, and J. J. Sninsky (New York: Academic Press), 282–287.
Wong, M. K. M., Goh, T. K., Hodgkiss, I. J., Hyde, K. D., Ranghoo, V. M., Tsui, C. K. M., et al. (1998). Role of fungi in freshwater ecosystems. Biodivers. Conserv. 7, 1187–1206. doi: 10.1023/A:1008883716975
Xu, R. J., Zhu, Y. A., Liu, N. G., Boonmee, S., Zhou, D. Q., and Zhao, Q. (2023). Taxonomy and phylogeny of hyphomycetous muriform conidial taxa from the Tibetan Plateau, China. J. Fungi 9:560. doi: 10.3390/jof9050560
Yang, J., Liu, L. L., Jones, E. B. G., Hyde, K. D., Liu, Z. Y., Bao, D. F., et al. (2023). Freshwater fungi from karst landscapes in China and Thailand. Fungal Divers. 119, 1–212. doi: 10.1007/s13225-023-00514-7
Keywords: 5 new taxa, asexual morph, biodiversity, morphology, phylogeny
Citation: Zhang Y-Z, Chen Q-L, Ma J, Lu Y-Z, Chen H-B and Liu N-G (2023) Morphological and multi-gene phylogenetic analyses reveal five new hyphomycetes from freshwater habitats. Front. Microbiol. 14:1253239. doi: 10.3389/fmicb.2023.1253239
Edited by:
Dian-Ming Hu, Jiangxi Agricultural University, ChinaReviewed by:
Saowaluck Tibpromma, Qujing Normal University, ChinaMingkwan Doilom, Zhongkai University of Agriculture and Engineering, China
Copyright © 2023 Zhang, Chen, Ma, Lu, Chen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ya-Zhou Zhang, eWF6aG91X3poYW5nMjAwOUAxNjMuY29t; Ning-Guo Liu, bGl1bmluZ2d1bzExQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Ya-Zhou Zhang1,2*†
Ya-Zhou Zhang1,2*† Qi-Lei Chen
Qi-Lei Chen Jian Ma
Jian Ma Yong-Zhong Lu
Yong-Zhong Lu Hu-Biao Chen
Hu-Biao Chen Ning-Guo Liu
Ning-Guo Liu