- 1School of Food Science and Engineering, Shaanxi University of Science and Technology, Xi’an, China
- 2Xi’an Key Laboratory of Antiviral and Antimicrobial-Resistant Bacteria Therapeutics Research, Xi’an, China
The anthocyanin compound cyanidin 3-O-glucoside (C3G) is a natural pigment widely used in food and nutraceutical industries. Its microbial synthesis by E. coli is a promising alternative to the traditional extraction methods. However, part of the synthesized C3G accumulates in the cytoplasm, thus potentially causing growth inhibition and product degradation. Therefore, it is necessary to enhance C3G secretion via exploration of native transporters facilitating C3G export. In this study, we report the screening and verification of native multidrug resistance transporters from 40 candidates in E. coli that can improve the extracellular C3G production when using catechin as the substrate. Overexpression of single transporter genes including fsr, yebQ, ynfM, mdlAB, and emrKY were found to increase C3G production by 0.5- to 4.8-fold. Genetic studies indicated that mdlAB and emrKY are vital transporters in the secretion of C3G. Our study reveals a set of new multidrug resistance transporters for the improvement of microbial biosynthesis of C3G and other anthocyanins.
Introduction
Anthocyanins are a group of colorful flavonoids with wide applications in the processing and production of food, cosmetics, and nutraceuticals (Blesso, 2019; Alappat and Alappat, 2020). The industrial production of anthocyanins mainly relies on plant extraction, which is constantly influenced by land supply and climate fluctuations (Rodríguez-Mena et al., 2023). A promising alternative is biosynthesis using engineered microbes as cell factories (Pandey et al., 2016; Yang et al., 2020; Courdavault et al., 2021; Yang et al., 2022).
Among all the known anthocyanin compounds, cyanidin 3-O-glucoside (C3G) with the highest natural abundance has been a research focus (Belwal et al., 2020). At present, C3G biosynthesis has been achieved in E. coli, Saccharomyces cerevisiae, Corynebacterium glutamicum, and Lactobacillus species (Yan et al., 2008; Levisson et al., 2018; Zha et al., 2018; Solopova et al., 2019), with the highest titer (439 mg/L) reported for E. coli using (+)-catechin as a precursor (Shrestha et al., 2019). This pathway contains two reactions sequentially catalyzed by anthocyanidin synthase (ANS) and 3-O-glycosyltransferase (3GT) using UDP-glucose as a sugar donor (Figure 1). The biosynthesis of C3G in E. coli has been engineered on the genetic, enzymatic, metabolic, cellular and fermentation levels for production enhancement (Yan et al., 2008; Lim et al., 2015; Zha and Koffas, 2017; Levisson et al., 2018; Solopova et al., 2019; Zha et al., 2020; Xu et al., 2022). However, a considerable amount of C3G accumulates inside cells possibly due to its high solubility in the aqueous phase and poor capability of diffusing across the cytoplasmic membrane and the outer membrane of E. coli (Lim et al., 2015; Zhao, 2015; Behrens et al., 2019). This negatively affects cell growth and metabolism, destabilizes C3G, and complicates the purification process, thereby decreasing the overall C3G titers.
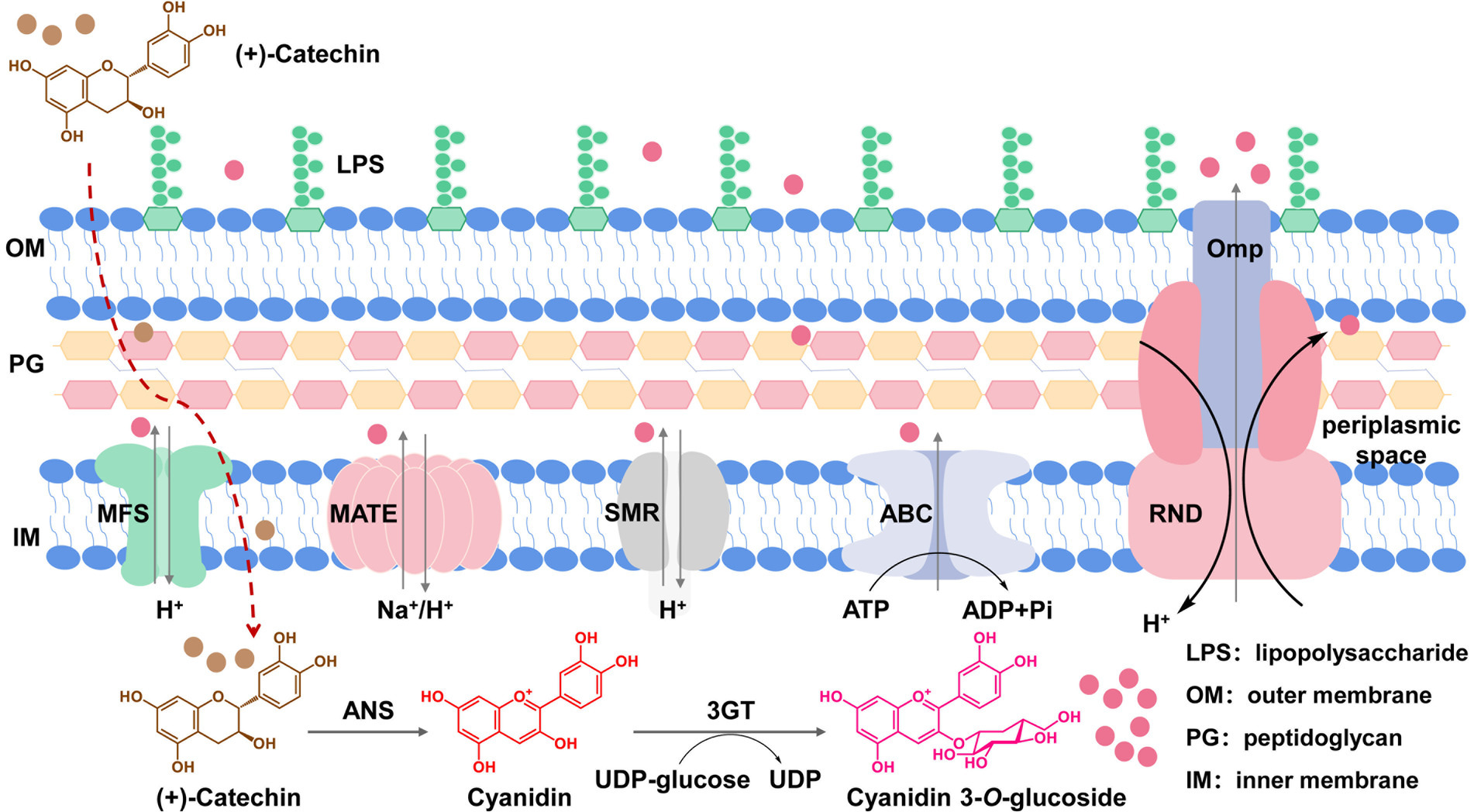
Figure 1. Efflux transporters in E. coli cells responsible for transportation of C3G out of cells. C3G is produced by ANS from Petunia hybrid and 3GT from Arabidopsis thaliana using catechin as the substrate. The produced C3G can be transported out of cells through native MDR transporters located in membranes.
A common approach of tackling the accumulation issue targets transporters to enhance product secretion (Wu et al., 2022). In the long history of evolution aiming at better adaptation to various harsh conditions, microorganisms have developed a set of membrane-bound multidrug resistance (MDR) transporters to pump out harmful substances from the cytoplasm and to reduce their damage to cell components (Jones et al., 2015; Lv et al., 2016). MDR transporters can be classified into five main categories, i.e., the ATP-binding cassette (ABC) superfamily transporters, the major facilitator superfamily (MFS) transporters, the small multidrug resistance (SMR) family transporters, the multi-drug and toxin compound extrusion (MATE) family transporters, and the resistance-nodulation-cell division (RND) superfamily transporters (Figure 1). MDR transporters can export antibiotics, toxic chemicals as well as some natural products (Du et al., 2018). Therefore, engineering of MDR transporters has been adopted in the microbial synthesis of natural products such as amorphadiene, resveratrol, caffeic acid, and reticuline (Zhang et al., 2016, 2017; Hu et al., 2018; Zhao et al., 2018; Yamada et al., 2022; Wang et al., 2023).
In E. coli, there is only one native transporter reported to date with C3G-exporting capability, i.e., the ABC transporter YadH, whose overexpression could increase C3G production by 15% (Lim et al., 2015). It is necessary to identify more effective transporters to further improve the bio-production. In our previous study, an MFS efflux transporter named MdtH was identified as a potent anthocyanin transporter, whose overexpression could increase the extracellular C3G level by 110% (Wu et al., 2024), demonstrating the potential of intrinsic E. coli transporters in the secretion of C3G. In this study, we constructed a library of all the MDR transporters of E. coli and tested their roles in the secretion and production of C3G. We identified a series of potential transporters responsible for C3G production via various mechanisms. Our study provides new targets for the engineering of C3G-producing microbes and will benefit the microbial biosynthesis of related anthocyanins.
Materials and methods
Bacterial strains and media
The strains used in this study are shown in Table 1. E. coli DH5α was used to construct plasmids and was grown in LB (lysogeny broth) medium supplemented with antibiotics when necessary. E. coli BL21 (DE3) was used for C3G production and transporter expression. The plasmid pETM6-At3GT-m-PhANS was transformed into E. coli BL21 (DE3) to construct the baseline C3G-producing strain (Cress et al., 2017). The plasmid pACYCDuet-1 was used to express transporter genes. C3G production was performed in M9 medium (pH 5.0) containing 4 g/L of glucose (Lim et al., 2015).
Construction of plasmids and recombinant strains
The plasmids and primers used in this study are shown in Table 1 and Supplementary Table S1, respectively. The transporter genes were amplified using the genomic DNA of E. coli BL21 (DE3) as the template, and subsequently cloned into the plasmid pACYCDuet-1 by Nde I and Xho I. For the co-overexpression of two transporter genes, the single overexpression plasmids were used and the second gene was cloned into the second ORF between BamH I and Not I.
The transporter knockout strains were constructed using the CRISPR/Cas9 tool developed in a previous study (Jiang et al., 2015). The knockout strains were formed by homologous recombination between the genome and the homologous arms, with ~200 bp gene sequences deleted in the ORF of the target transporter genes in the repair plasmids. The deletion was finally confirmed by PCR amplification of the extracted genomic DNA from the knockout strains and by gene sequencing of the amplified PCR products. The complementation strains were constructed by introducing the pACYCDuet-1 plasmids carrying the transporter genes into the knockout strains.
For all the cloning work, the enzymes were purchased from New England Biolabs, and kits for plasmid miniprep and gene cleanup were from TIANGEN Biotech. Primer synthesis and gene sequencing were performed by Sangon Biotech.
The plasmid pETM6-At3GT-m-PhANS and pACYCDuet-1 plasmids expressing MDR transporters were co-transformed into E. coli BL21 (DE3) using the traditional calcium chloride method.
Screening of MDR transporters to improve C3G production in 48-well plates
Cells were grown overnight at 37°C in 1 mL LB medium supplemented with 80 μg/mL ampicillin and 25 μg/mL chloramphenicol in 48-deep-well plates (VWR). Overnight cultures (20 μL) were sub-cultured into 1 mL of fresh LB medium containing the necessary antibiotics in 48-deep-well plates. Cells were grown at 37°C with shaking at 220 rpm, induced with 0.5 mM IPTG when OD600 (the optical density at 600 nm) reached 0.6, and further grown at 30°C with shaking for 3 h. Cells were then collected by centrifugation at 4,000 rpm and 4°C and resuspended in 1 mL of M9 medium (pH 5.0) with 500 mg/L (+)-catechin. The conversion process was then carried out at 30°C with shaking for 19 h. At the end of the fermentation, 200 μL of the culture supernatants were mixed with an equal volume of acidified methanol and centrifuged at 12,000 rpm for 10 min. The supernatants were used to quantify residual catechin and extracellular C3G.
C3G production in shake flasks
Glycerol stocks were inoculated into 3 mL of LB medium supplemented with 80 μg/mL ampicillin and 25 μg/mL chloramphenicol and grown overnight at 37°C. The overnight culture (1 mL) was sub-cultured into 50 mL of fresh LB medium containing the necessary antibiotics in 250 mL shake flasks. Cells were grown at 37°C with shaking at 200 rpm, induced with 0.5 mM IPTG when OD600 reached 0.6, and further grown for 3 h at 30°C and 200 rpm. Cells were then harvested by centrifugation at 4,000 rpm and 4°C and resuspended in 10 mL of M9 medium (pH 5.0) with 500 mg/L (+)-catechin in 100 mL shake flasks. Cells were incubated at 30°C and 200 rpm for 19 h, and then 200 μL of the culture supernatant were mixed thoroughly with an equal volume of acidified methanol and centrifuged at 12,000 rpm for 10 min. The supernatant was used for the quantification of extracellular C3G and residual catechin.
HPLC analysis of C3G and catechin
The fermentation products and residual catechin were analyzed by a Shimadzu Essentia LC-16 HPLC system with a diode array detector. A reversed phase C18 column (5 μm, 4.6 × 250 mm) was used to separate and quantify the compounds. The column temperature was maintained at 25°C. Water with 0.1% formic acid (solvent A) and acetonitrile with 0.1% formic acid (solvent B) were prepared as mobile phases at a flow rate of 1 mL/min. The elution program was as follows: 10–40% B at 0–10 min and 40–60% B at 10–15 min. C3G and catechin were detected and quantified by monitoring the absorbance at 520 nm and 280 nm, respectively. The C3G and (+)-catechin standards used in this study were purchased from Sigma-Aldrich.
Measurement of cell growth of the recombinant strains
Overnight cultures (1 mL) of diverse recombinant strains were sub-cultured into 50 mL of fresh LB medium containing the necessary antibiotics in 250 mL shake flasks, and were grown at 37°C and 200 rpm. Cell growth was monitored periodically by measuring OD600 in the absence of IPTG. To study cell growth under the induction condition, cells were grown to OD600 of ~0.6 and supplemented with 0.5 mM IPTG. The OD600 was then tracked periodically.
Results
Screening of potential MDR transporters related to C3G production in Escherichia coli
MDR transporters facilitate the outward flow of toxic compounds from the cytoplasm and enhance cell survival under harsh environmental conditions such as exposure to antibiotics and heavy metals (Du et al., 2018). In our previous study, we found that overexpression of an MFS transporter MdtH could increase the extracellular C3G production by around 110% due to better efflux (Wu et al., 2024). To identify other MDR transporters beneficial for extracellular C3G biosynthesis in E. coli, we constructed a transporter library containing the known or putative MDR transporter genes (Nishino and Yamaguchi, 2001) in E. coli as well as the genes encoding the TolC porin and TolC-involving tripartite efflux systems that are important for drug resistance (Table 2). Some of the MDR transporters that were already analyzed in our previous study were not included here. We overexpressed each of these genes individually under the C3G-producing background and analyzed the extent of extracellular specific C3G production in 48-deep-well plates. Genes that are present in the same operon were overexpressed together. Since the precursor catechin and the product C3G both present antimicrobial activities (Nakayama et al., 2013; Lim et al., 2015) and since the product C3G is unstable under neutral or basic conditions, we adopted a two-step biosynthetic procedure, in which cells were first grown in LB medium and induced for pathway enzyme expression for 3 h, and then shifted to M9 medium (pH 5) containing 500 mg/L of catechin for C3G synthesis.
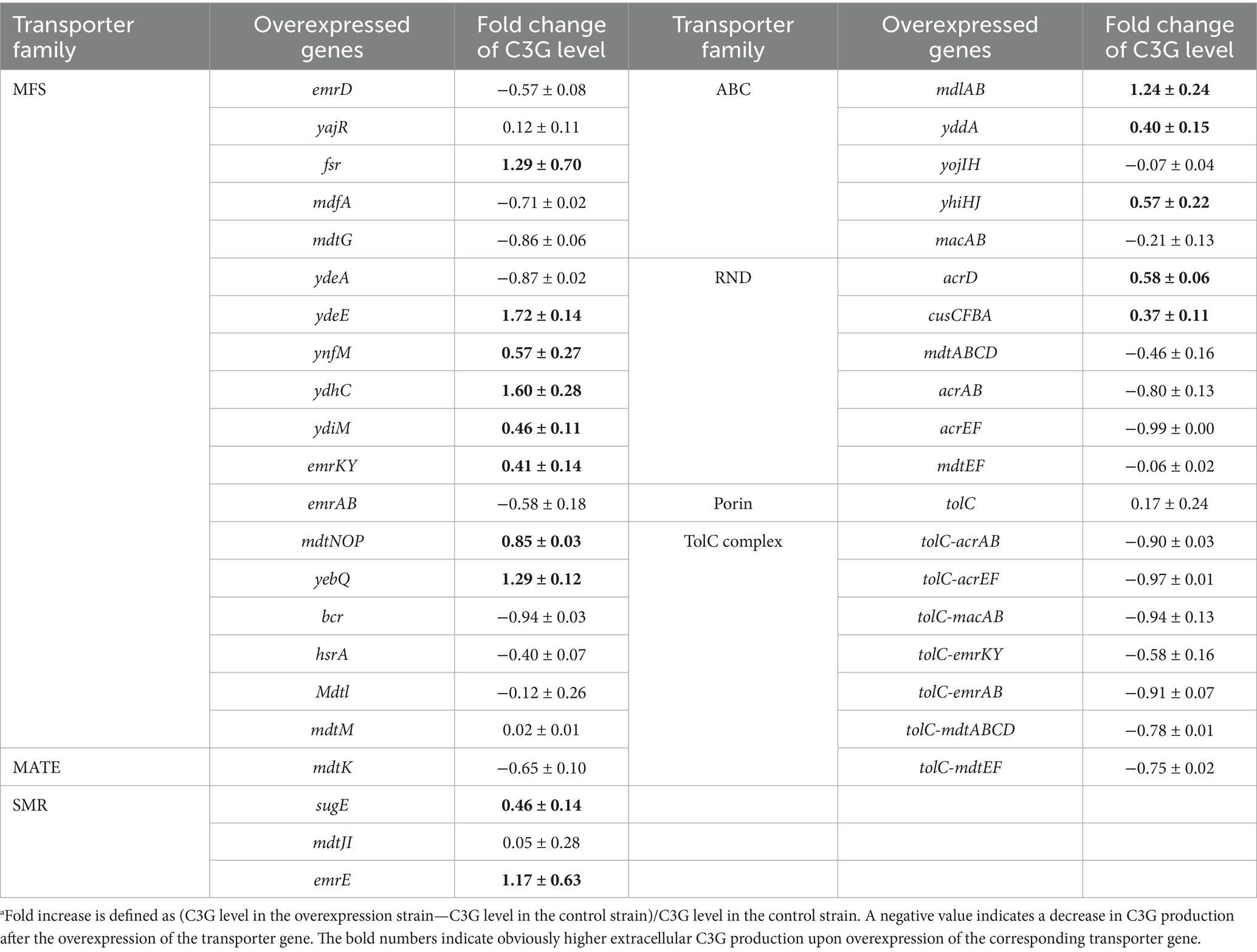
Table 2. Fold increasea in extracellular C3G production per cell OD600 in recombinant strains overexpressing diverse groups of MDR transporters in 48-deep-well plates as compared to the control strain.
As shown in Table 2, overexpression of fsr, ydeE, ynfM, ydhC, ydiM, emrKY, mdtNOP, yebQ, sugE, emrE, mdlAB, yddA, yhiHJ, acrD, or cusCFBA increased the extracellular level of C3G per OD600 by 0.37- to 1.72-fold, which was consistent with our hypothesis that the titer of C3G could be improved via efflux enhancement. Therefore, these transporter genes were selected for the subsequent analysis. It is interesting to note that overexpression of the transporter components that form tripartite efflux systems with TolC significantly impeded C3G biosynthesis, and the co-overexpression of tolC further enhanced such inhibitory effect, although a higher expression level of tolC alone had marginal effect on extracellular C3G concentration.
Overexpression of fsr, yebQ, ynfM, mdlAB, or emrKY could significantly increase extracellular C3G production
To verify the role of the selected MDR transporters in C3G biosynthesis, we conducted the bioconversion using the two-stage procedure in shake flasks. It is generally believed that the cultivation conditions in shake flasks can be controlled more stably than those in 48-well plates. Among the 15 selected transporter genes, overexpression of ynfM improved the extracellular synthesis per OD600 by 4.80-fold compared to the control strain that harbored the empty plasmid (Table 3), and upregulation of fsr, yebQ, emrKY or mdlAB increased specific C3G production by 0.54- to 4.12-fold. In comparison, the other transporter genes that were tested effective in 48-well-plates did not show positive correlations at a higher expression level with extracellular C3G production in shake flasks. Such a discrepancy may be attributed to the different extents of oxygen supply and shear stress.
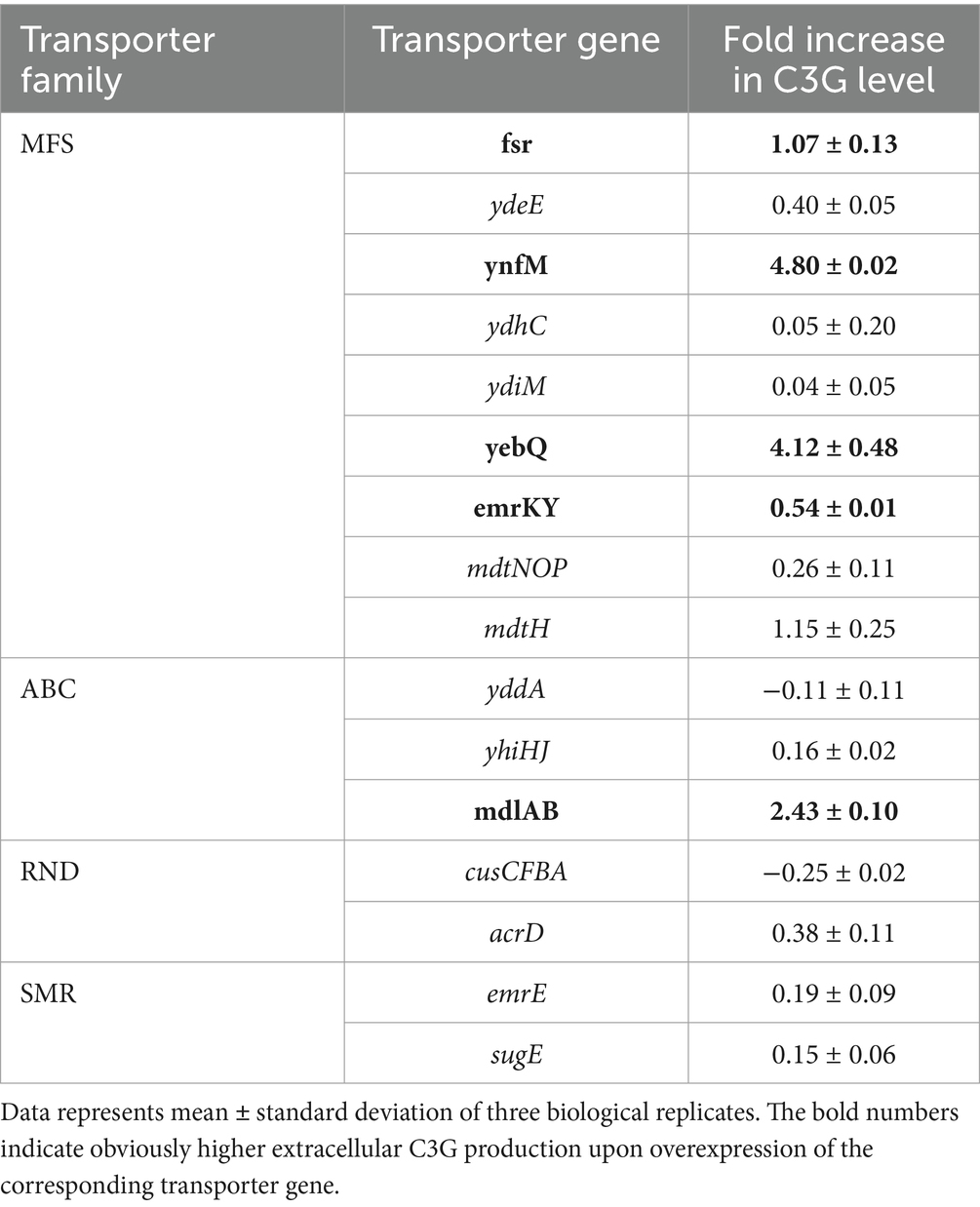
Table 3. The fold increase in extracellular C3G production per cell OD600 in recombinant strains individually overexpressing selected transporter genes in shake flasks relative to the control strains.
Genes yebQ, mdlAB, and emrKY were strongly correlated with C3G production
To investigate whether the phenotype of extracellular C3G production was indeed associated with the expression levels of the selected transporter genes, i.e., fsr, ynfM, yebQ, mdlAB, and emrKY, we constructed single-gene deletion and complementation strains, and analyzed C3G biosynthesis and catechin utilization by various recombinant strains. As shown in Figure 2A, overexpression of these genes greatly promoted extracellular C3G concentration compared with the control strain carrying the empty plasmid; deletion of yebQ, mdlAB, or emrKY decreased C3G production by 10–30% compared with the control strain, whereas deletion of fsr or ynfM had no impact on extracellular C3G concentration. Complementation of the corresponding genes in the knockout strains restored C3G production to levels similar to those in the overexpression strains. Moreover, despite large variations in C3G production by various overexpression strains, they consumed similar amounts of catechin except the mdlAB-related strains that utilized noticeably less catechin (Figure 2B). Taken into consideration both substrate usage and product formation, we found similar extracellular C3G yields obtained by the overexpression of ynfM, yebQ and mdlAB, which were 2.7- to 2.9-fold higher than that of the control strain. Interestingly, deletion of fsr or ynfM resulted in 0.61- or 0.82-fold higher yields, respectively (Figure 2C).
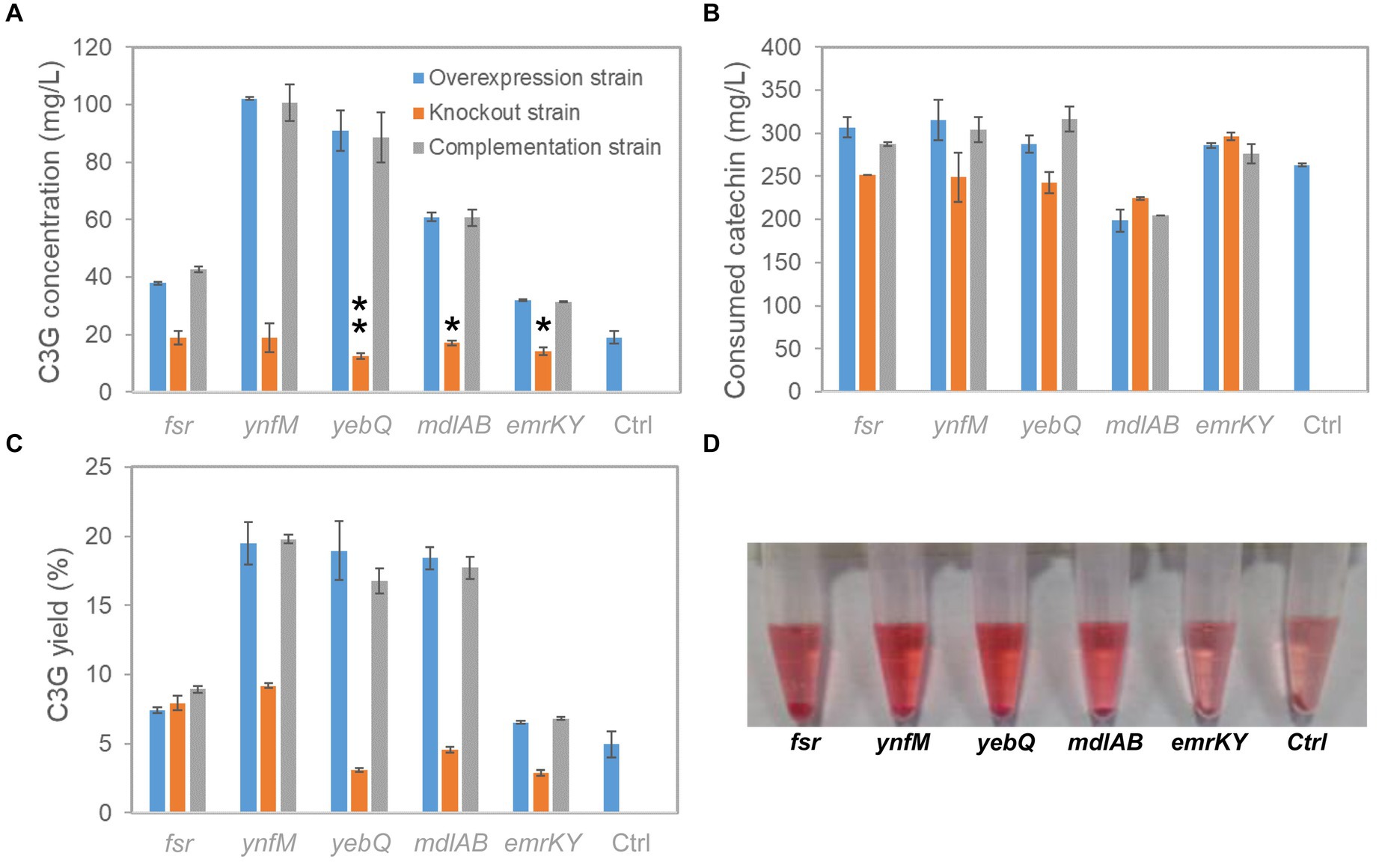
Figure 2. C3G production from catechin by recombinant E. coli strains expressing fsr, ynfM, yebQ, emrKY, and mdlAB. (A) The comparison of extracellular C3G titers by the overexpression strains, knockout strains and complementation strains related to transporter genes fsr, ynfM, yebQ, emrKY, and mdlAB. Statistical analysis was performed using two-tailed student t-test. *p < 0.5; **p < 0.01. (B) Utilization of catechin by different strains. (C) The extracellular yields of C3G from catechin by various strains. (D) The picture showing the supernatant of the final fermentation products by the recombinant strains overexpressing fsr, ynfM, yebQ, emrKY, and mdlAB. Data represents mean ± standard deviation of three biological replicates.
As C3G is naturally pink under acidic conditions, it is easy to judge the production using the colorimetric assay and by visual observation. Notably, the culture overexpressing ynfM or yebQ showed darker colors than the control strain and the other overexpression strains (Figure 2D). Additionally, the yebQ-overexpressing culture turned pink as early as 1 h after catechin supplementation (Supplementary Figure S1). In comparison, the control culture did not develop any pink color even after 4 h of biosynthesis.
Effect of transporter overexpression on cell growth
Generally, overexpression of transporters involves extensive folding and translocation of an array of proteins and their translocation to the limited membrane space, thereby causing excessive burden to the central cell metabolism (Jensen et al., 2017; Zhu et al., 2020). To understand how cell growth was affected by the expression levels of the five transporters identified in the present study, we monitored the growth curves of the overexpression, deletion, complementation, and the control strains. To rule out the possible effect of the vector pACYCDuet-1 that was adopted for episomal expression of the transporters, both the deletion and the control strains harbored the empty plasmid. As shown in Figure 3, without IPTG to induce the gene expression, the growth of all the recombinant strains was comparable to that of the control strain, suggesting that single deletion of the transporter genes had no apparent impact on cell growth. In contrast, IPTG induction of the transporter genes and the pathway genes reduced cell growth in the late-log phase and biomass accumulation in the stationary phase for the overexpression and the complementation strains when compared with the deletion and the control strains, although all the strains grew at similar rates at the mid-log phase (Figure 4). In addition, the control strain expressing ANS and 3GT showed greatly reduced biomass compared to the condition without IPTG addition.
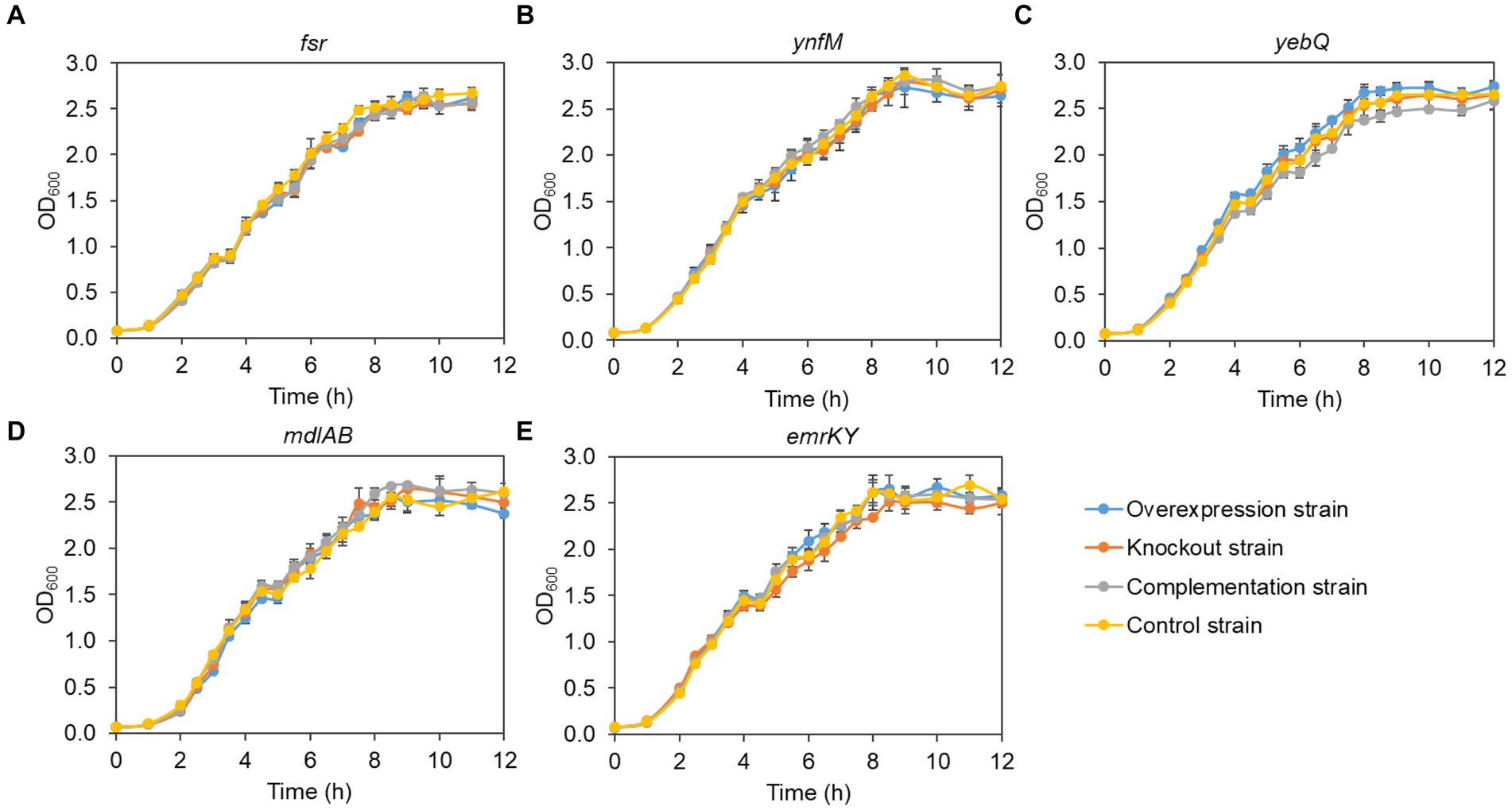
Figure 3. Growth curves of the overexpression strains, knockout strains and complementation strains regarding the transporter genes fsr (A), ynfM (B), yebQ (C), emrKY (D), and mdlAB (E) under the condition without IPTG induction. Data represents mean ± standard deviation of three biological replicates.
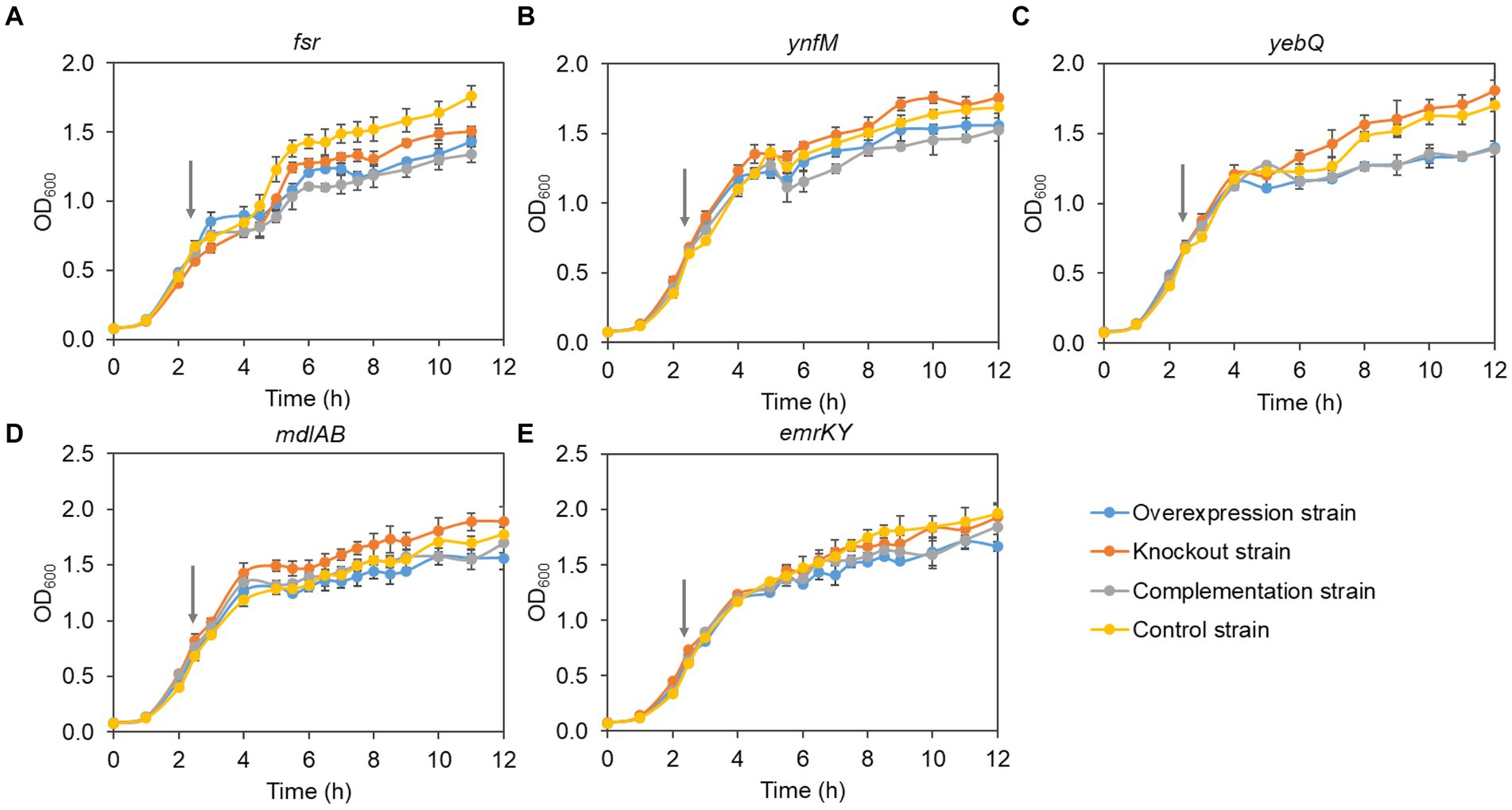
Figure 4. The cell growth of the overexpression strains, knockout strains and complementation strains of transporter genes fsr (A), ynfM (B), yebQ (C), emrKY (D), and mdlAB (E) with the induction of 0.5 mM IPTG. The red arrows indicate the time to add IPTG. Data represents mean ± standard deviation of three biological replicates.
Co-expression of transporter genes was not beneficial for C3G production
Given that the overexpression of ynfM, yebQ, mdlAB, and emrKY increased C3G production considerably, we explored whether the co-overexpression of these genes could further enhance the bio-production. The fsr gene was not included due to the fact that there was not much difference in the yield of extracellular C3G when this gene was overexpressed, deleted, or complemented. Compared with yebQ overexpression alone, the co-overexpression of yebQ together with ynfM, mdlAB, or emrKY reduced extracellular C3G production (Figure 5A). Similarly, overexpression of ynfM, which alone led to the highest extracellular C3G level, showed negative effects when co-expressed with mdlAB or emrKY. In contrast, the co-overexpression of mdlAB and emrKY resulted in a level of extracellular C3G production analogous to that when mdlAB alone was overexpressed. Co-expression of the transporter genes led to less utilization of catechin but did not cause much variation in the yield of extracellular C3G (Figures 5B,C).
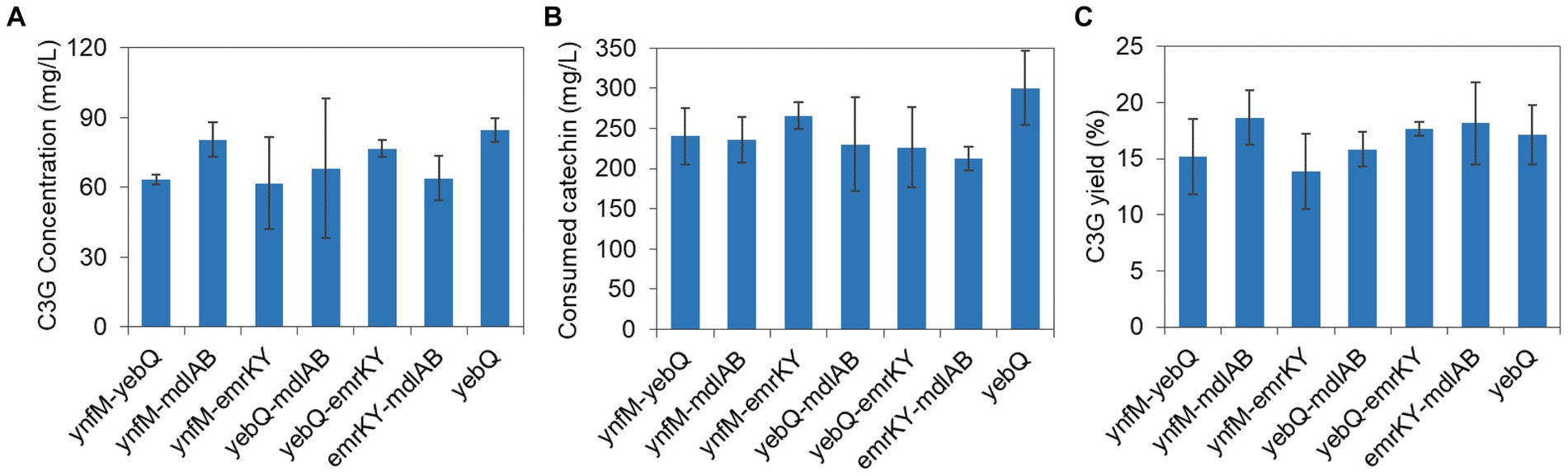
Figure 5. The extracellular C3G production (A), catechin utilization (B), and extracellular C3G yield (C) in recombinant strains co-overexpressing two transporter genes among ynfM, yebQ, mdlAB, and emrKY. The yebQ-overexpressing strain was used as a control. Data represents mean ± standard deviation of three biological replicates.
Double deletion of mdlAB and emrKY reduced C3G production significantly
Since single deletion of yebQ, mdlAB and emrKY reduced extracellular C3G production titer and the yields, and overexpression and complementation of each individual gene significantly improved both parameters, we wondered how critical these genes are for C3G production. To that end, we constructed double-knockout strains, and found that deletion of mdlAB/emrKY or mdlAB/yebQ combination reduced the extracellular C3G concentrations and the yields to levels lower than those in the single-knockout mutants, with the ΔmdlABΔemrKY double deletion leading to the largest extent of decline (Figure 6), while the substrate consumption of these two strains was similar to that of the control strain or the corresponding single deletion strains, suggesting that MdlAB and EmrKY are key to the secretion of C3G in E. coli. In contrast, ΔyebQΔemrKY mutation resulted in less extracellular C3G production than the Δyeb strain but more than the ΔemrKY strain. With slightly less catechin utilization, this double deletion strain generated C3G with an extracellular yield comparable to that in the ΔyebQ strain (Figure 6).
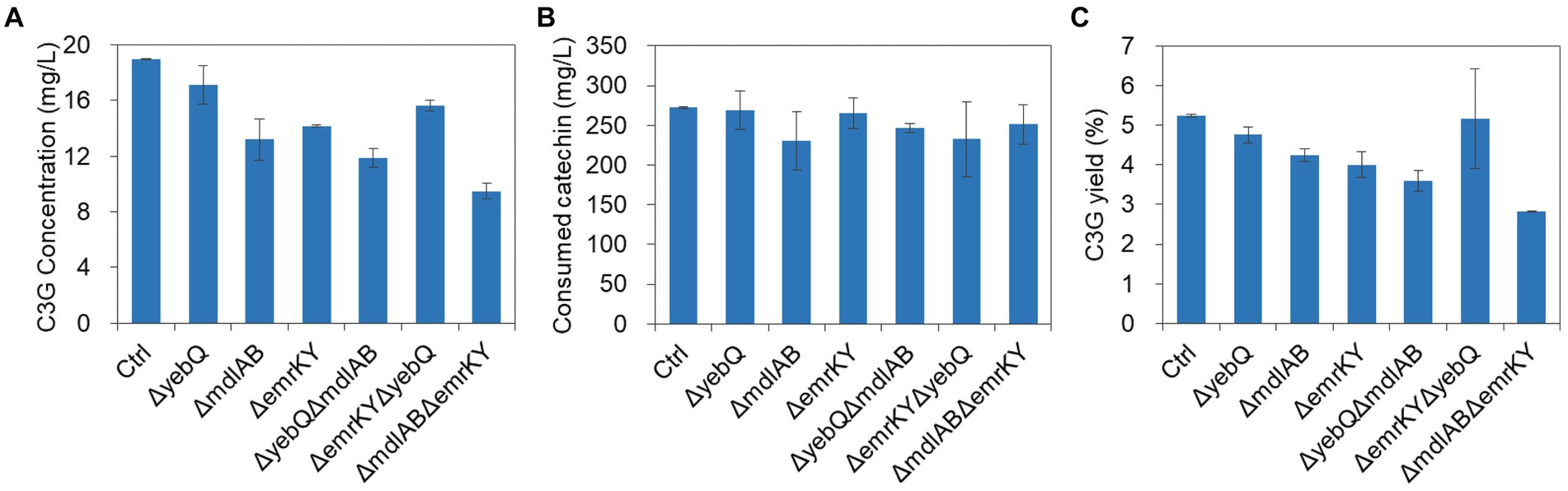
Figure 6. The extracellular C3G production (A), catechin utilization (B), and extracellular C3G yield (C) by double knockout strains of yebQ, emrKY, and mdlAB. Data represents mean ± standard deviation of three biological replicates.
Discussion
The bacterial cell envelope is armed with an array of transporters for material exchange, among which the efflux transporters provide protection against adverse environmental factors such as toxins and antimicrobials. Thereby, efflux transporters have been a research focus in the past few decades in the microbial synthesis of valuable compounds aiming at higher tolerance to or better secretion of the products. E. coli harbors a panel of MDR transporters, some of which are poorly characterized or even barely annotated. This makes it challenging to understand how anthocyanins, as well as other plant natural products that bear antimicrobial activities, migrate from the cytoplasm to the culture media and how their extracellular microbial synthesis can be improved. In the present study, we analyzed all the known MDR transporters in E. coli regarding their competence to enhance C3G efflux, and obtained five transporter genes, i.e., fsr, yebQ, ynfM, mdlAB, and emrKY, which could prominently strengthen C3G secretion and drive better extracellular synthesis.
Of the five positive transporters, MdlAB belongs to the ABC transporter superfamily and the other four are MFS transporters. Fsr may function as an efflux pump of toxic compounds, as the expression of fsr (fosmidomycin resistance) can confer resistance to fosmidomycin, trimethoprim, and carbonyl cyanide m-chlorophenylhydrazone (Fujisaki et al., 1996; Nishino and Yamaguchi, 2001). MdlA is a putative multidrug resistance-like ABC exporter. Its deletion has been reported to increase the total production of free fatty acids but not the extracellular level (Shin and Lee, 2017). MdlB is a putative ABC transporter that confers microbial tolerance to isopentenol (Foo et al., 2014). EmrKY is an efflux pump consisting of an inner membrane subunit and a periplasmic adaptor protein belonging to the major facilitator superfamily (Pasqua et al., 2019). Overexpression of emrKY can confer resistance to doxorubicin, rhodamine 6G, benzalkonium, erythromycin, and other toxic chemicals (Nishino and Yamaguchi, 2001). YebQ and YnfM have not been experimentally established as transporters. Overexpression of yebQ with its native regulatory gene is reported to confer hypersensitivity to trimethoprim (Nishino and Yamaguchi, 2001). In E. coli, YnfM has been predicted to function as an arabinose exporter and its overexpression together with its native regulatory element can sensitize cells to acriflavine (Nishino and Yamaguchi, 2001; Koita and Rao, 2012). The increased sensitivity may be related to the deleterious effect of these genes on cell growth when overexpressed. In accord with this, we observed less biomass accumulation at the stationary phase in cultures overexpressing yebQ or ynfM. Such damage may cause higher cell permeability, which in turn facilitates catechin uptake and C3G exportation and is unexpectedly beneficial for extracellular C3G production.
Overexpression of fsr, ynfM or yebQ increased C3G production as well as substrate consumption. It is very likely that the timely export of C3G lowers its intracellular concentration, thereby generating a driving force for the continuous uptake of catechin and conversion of catechin to C3G. Indeed, less catechin was utilized when these genes were deleted individually. The strain overexpressing mdlAB produced more extracellular C3G with less catechin compared with the control strain. Such a property led to a surge in the yield of extracellular C3G upon the overexpression or complementation of mdlAB, suggesting that this gene can be a potential starting point for strain optimization toward better C3G biosynthesis. It is worth mentioning that substrate utilization by mdlAB- and emrKY-deficient strains was very similar to that by the overexpression strains; nonetheless, the deletion strains produced much lower extracellular C3G than the overexpression strains. Given that catechin and C3G are structurally similar, a possible explanation is that MdlAB and EmrKY are involved in the export of both catechin and C3G, with the former to a lesser extent. With a high level of these transporters, cells on the one hand extrude more catechin to maintain a lower intracellular level for self-protection, and on the other hand export more C3G and correspondingly drives more influx of catechin. The combined effect is better C3G generation.
The co-overexpression of the positive transporter genes did not increase the production titer, suggesting that these genes do not have synergistic or additive roles in C3G biosynthesis. A higher level of efflux transporters can result in the loss of essential metabolites, which is detrimental to the efficiency and stability of the microbial cell factory. Moreover, despite considerable improvement in C3G biosynthesis with the overexpression of fsr, ynfM, yebQ, mdlAB, or emrKY, deletion of none of these genes completely blocked extracellular C3G accumulation. This clearly demonstrates that the extrusion of C3G is mediated by multiple transporters in a non-specific way, and that there is some extent of redundancy regarding the function of the MDR transporters in C3G efflux. The gene ynfM with the highest elevating effect on C3G production upon its overexpression appears not to be the most important transporter gene, as C3G level in its absence was comparable to that in the control strain. In comparison, double deletion of mdlAB/emrKY or mdlAB/yebQ further decreased both the extracellular product titer and the yield relative to the single deletion mutation, indicating that MdlAB is more important for C3G biosynthesis.
In summary, the present study identified a panel of MDR transporters closely associated with C3G biosynthesis, i.e., YnfM, YebQ, MdlAB, and EmrKY. These transporters considerably improve both the extracellular C3G concentration and the yield when overexpressed individually, with YnfM demonstrating the best effect. These transporters show redundant roles in C3G export while not displaying any synergistic or additive effect with one another, and it appears that MdlAB and EmrKY in combination are more critical than other transporters to the synthesis of C3G in E. coli. As transporters are vital for the intake of essential nutrients and extrusion of toxic metabolites, further production enhancement should focus on the fine regulation of their expression levels via strategies such as variations in the copy numbers and promoter strength, identification of transcription factors, optimization of the sequences in the ribosome binding sites, etc. These approaches will facilitate the establishment of a balance between normal cell growth, appropriate tolerance to the substrate and the product, and an acceptable level of product formation and export. This is the first systematic study investigating the role of all the intrinsic MDR transporters on the microbial production of anthocyanins. Our study potentiates the strategy of transporter identification for the biosynthesis of anthocyanins and related products in E. coli. Detailed investigations on the structural and biochemical properties of these transporters will further facilitate the construction of anthocyanin-producing microbes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
XW: Writing – original draft, Writing – review & editing, Conceptualization, Formal analysis, Funding acquisition, Project administration. RC: Writing – original draft, Formal analysis, Investigation. PL: Investigation, Writing – original draft, Methodology. JZ: Methodology, Writing – original draft, Conceptualization, Data curation, Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by The Key Research and Development Program of Shaanxi (2023-YBSF-260), Xi’an Science and Technology Plan Project (22GXFW0015), and National Natural Science Foundation of China (32170175), for research and publication.
Acknowledgments
We acknowledge the financial support from the Key Research and Development Program of Shaanxi (2023-YBSF-260), Xi’an Science and Technology Plan Project (22GXFW0015), and the National Natural Science Foundation of China (32170175).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1357794/full#supplementary-material
References
Alappat, B., and Alappat, J. (2020). Anthocyanin pigments: beyond aesthetics. Molecules 25:5500. doi: 10.3390/molecules25235500
Behrens, C. E., Smith, K. E., Iancu, C. V., Choe, J. Y., and Dean, J. V. (2019). Transport of anthocyanins and other flavonoids by the Arabidopsis ATP-binding cassette transporter AtABCC2. Sci. Rep. 9:437. doi: 10.1038/s41598-018-37504-8
Belwal, T., Singh, G., Jeandet, P., Pandey, A., Giri, L., Ramola, S., et al. (2020). Anthocyanins, multi-functional natural products of industrial relevance: recent biotechnological advances. Biotechnol. Adv. 43:107600. doi: 10.1016/j.biotechadv.2020.107600
Blesso, C. N. (2019). Dietary anthocyanins and human health. Nutrients 11:2107. doi: 10.3390/nu11092107
Courdavault, V., O'connor, S. E., Jensen, M. K., and Papon, N. (2021). Metabolic engineering for plant natural products biosynthesis: new procedures, concrete achievements and remaining limits. Nat. Prod. Rep. 38, 2145–2153. doi: 10.1039/D0NP00092B
Cress, B. F., Leitz, Q. D., Kim, D. C., Amore, T. D., Suzuki, J. Y., Linhardt, R. J., et al. (2017). CRISPRi-mediated metabolic engineering of E. coli for O-methylated anthocyanin production. Microb. Cell Factories 16:10. doi: 10.1186/s12934-016-0623-3
Du, D., Wang-Kan, X., Neuberger, A., Van Veen, H. W., Pos, K. M., Piddock, L. J. V., et al. (2018). Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol. 16, 523–539. doi: 10.1038/s41579-018-0048-6
Foo, J. L., Jensen, H. M., Dahl, R. H., George, K., Keasling, J. D., Lee, T. S., et al. (2014). Improving microbial biogasoline production in Escherichia coli using tolerance engineering. MBio 5:e01932. doi: 10.1128/mBio.01932-14
Fujisaki, S., Ohnuma, S., Horiuchi, T., Takahashi, I., Tsukui, S., Nishimura, Y., et al. (1996). Cloning of a gene from Escherichia coli that confers resistance to fosmidomycin as a consequence of amplification. Gene 175, 83–87. doi: 10.1016/0378-1119(96)00128-X
Hu, Y., Zhu, Z., Nielsen, J., and Siewers, V. (2018). Heterologous transporter expression for improved fatty alcohol secretion in yeast. Metab. Eng. 45, 51–58. doi: 10.1016/j.ymben.2017.11.008
Jensen, H. M., Eng, T., Chubukov, V., Herbert, R. A., and Mukhopadhyay, A. (2017). Improving membrane protein expression and function using genomic edits. Sci. Rep. 7:13030. doi: 10.1038/s41598-017-12901-7
Jiang, Y., Chen, B., Duan, C., Sun, B., Yang, J., and Yang, S. (2015). Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 81, 2506–2514. doi: 10.1128/AEM.04023-14
Jones, C. M., Hernández Lozada, N. J., and Pfleger, B. F. (2015). Efflux systems in bacteria and their metabolic engineering applications. Appl. Microbiol. Biotechnol. 99, 9381–9393. doi: 10.1007/s00253-015-6963-9
Koita, K., and Rao, C. V. (2012). Identification and analysis of the putative pentose sugar efflux transporters in Escherichia coli. PLoS One 7:e43700. doi: 10.1371/journal.pone.0043700
Levisson, M., Patinios, C., Hein, S., De Groot, P. A., Daran, J. M., Hall, R. D., et al. (2018). Engineering de novo anthocyanin production in Saccharomyces cerevisiae. Microb. Cell Factories 17:103. doi: 10.1186/s12934-018-0951-6
Lim, C. G., Wong, L., Bhan, N., Dvora, H., Xu, P., Venkiteswaran, S., et al. (2015). Development of a recombinant Escherichia coli strain for overproduction of the plant pigment anthocyanin. Appl. Environ. Microbiol. 81, 6276–6284. doi: 10.1128/AEM.01448-15
Lv, H., Li, J., Wu, Y., Garyali, S., and Wang, Y. (2016). Transporter and its engineering for secondary metabolites. Appl. Microbiol. Biotechnol. 100, 6119–6130. doi: 10.1007/s00253-016-7605-6
Nakayama, M., Shimatani, K., Ozawa, T., Shigemune, N., Tsugukuni, T., Tomiyama, D., et al. (2013). A study of the antibacterial mechanism of catechins: isolation and identification of Escherichia coli cell surface proteins that interact with epigallocatechin gallate. Food Control 33, 433–439. doi: 10.1016/j.foodcont.2013.03.016
Nishino, K., and Yamaguchi, A. (2001). Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183, 5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001
Pandey, R. P., Parajuli, P., Koffas, M. A. G., and Sohng, J. K. (2016). Microbial production of natural and non-natural flavonoids: pathway engineering, directed evolution and systems/synthetic biology. Biotechnol. Adv. 34, 634–662. doi: 10.1016/j.biotechadv.2016.02.012
Pasqua, M., Grossi, M., Scinicariello, S., Aussel, L., Barras, F., Colonna, B., et al. (2019). The MFS efflux pump EmrKY contributes to the survival of Shigella within macrophages. Sci. Rep. 9:2906. doi: 10.1038/s41598-019-39749-3
Rodríguez-Mena, A., Ochoa-Martínez, L. A., González-Herrera, S. M., Rutiaga-Quiñones, O. M., González-Laredo, R. F., and Olmedilla-Alonso, B. (2023). Natural pigments of plant origin: classification, extraction and application in foods. Food Chem. 398:133908. doi: 10.1016/j.foodchem.2022.133908
Shin, K. S., and Lee, S. K. (2017). Increasing extracellular free fatty acid production in Escherichia coli by disrupting membrane transport systems. J. Agric. Food Chem. 65, 11243–11250. doi: 10.1021/acs.jafc.7b04521
Shrestha, B., Pandey, R. P., Darsandhari, S., Parajuli, P., and Sohng, J. K. (2019). Combinatorial approach for improved cyanidin 3-O-glucoside production in Escherichia coli. Microb. Cell Factories 18:7. doi: 10.1186/s12934-019-1056-6
Solopova, A., Van Tilburg, A. Y., Foito, A., Allwood, W. J., Stewart, D., Kulakauskas, S., et al. (2019). Engineering Lactococcus lactis for the production of unusual anthocyanins using tea as substrate. Metab. Eng. 54, 160–169. doi: 10.1016/j.ymben.2019.04.002
Wang, L., Li, N., Yu, S., and Zhou, J. (2023). Enhancing caffeic acid production in Escherichia coli by engineering the biosynthesis pathway and transporter. Bioresour. Technol. 368:128320. doi: 10.1016/j.biortech.2022.128320
Wu, X., Liu, J., Liu, Z., Gong, G., and Zha, J. (2022). Microbial cell surface engineering for high-level synthesis of bio-products. Biotechnol. Adv. 55:107912. doi: 10.1016/j.biotechadv.2022.107912
Wu, X., Ren, J., Wang, J., Koffas, M. A. G., and Zha, J. (2024). A major facilitator superfamily transporter MdtH in Escherichia coli is involved in anthocyanin biosynthesis and secretion. Appl. Environ. Microbiol. 90:e0207923. doi: 10.1128/aem.02079-23
Xu, S., Li, G., Zhou, J., Chen, G., and Shao, J. (2022). Efficient production of anthocyanins in Saccharomyces cerevisiae by introducing anthocyanin transporter and knocking out endogenous degrading enzymes. Front. Bioeng. Biotechnol. 10:899182. doi: 10.3389/fbioe.2022.899182
Yamada, Y., Nakagawa, A., Sato, F., Minami, H., and Shitan, N. (2022). Transport engineering using tobacco transporter NtJAT1 enhances alkaloid production in Escherichia coli. Biosci. Biotechnol. Biochem. 86, 865–869. doi: 10.1093/bbb/zbac056
Yan, Y., Li, Z., and Koffas, M. A. (2008). High-yield anthocyanin biosynthesis in engineered Escherichia coli. Biotechnol. Bioeng. 100, 126–140. doi: 10.1002/bit.21721
Yang, D., Eun, H., Prabowo, C. P. S., Cho, S., and Lee, S. Y. (2022). Metabolic and cellular engineering for the production of natural products. Curr. Opin. Biotechnol. 77:102760. doi: 10.1016/j.copbio.2022.102760
Yang, D., Park, S. Y., Park, Y. S., Eun, H., and Lee, S. Y. (2020). Metabolic engineering of Escherichia coli for natural product biosynthesis. Trends Biotechnol. 38, 745–765. doi: 10.1016/j.tibtech.2019.11.007
Zha, J., and Koffas, M. A. G. (2017). Production of anthocyanins in metabolically engineered microorganisms: current status and perspectives. Synth. Syst. Biotechnol. 2, 259–266. doi: 10.1016/j.synbio.2017.10.005
Zha, J., Wu, X., and Koffas, M. A. G. (2020). Making brilliant colors by microorganisms. Curr. Opin. Biotechnol. 61, 135–141. doi: 10.1016/j.copbio.2019.12.020
Zha, J., Zang, Y., Mattozzi, M., Plassmeier, J., Gupta, M., Wu, X., et al. (2018). Metabolic engineering of Corynebacterium glutamicum for anthocyanin production. Microb. Cell Factories 17:143. doi: 10.1186/s12934-018-0990-z
Zhang, C., Chen, X., Stephanopoulos, G., and Too, H.-P. (2016). Efflux transporter engineering markedly improves amorphadiene production in Escherichia coli. Biotechnol. Bioeng. 113, 1755–1763. doi: 10.1002/bit.25943
Zhang, T., Song, R., Wang, M., Deng, L., Fan, L., and Wang, F. (2017). Regulating C4-dicarboxylate transporters for improving fumaric acid production. RSC Adv. 7, 2897–2904. doi: 10.1039/C6RA24727J
Zhao, J. (2015). Flavonoid transport mechanisms: how to go, and with whom. Trends Plant Sci. 20, 576–585. doi: 10.1016/j.tplants.2015.06.007
Zhao, Y., Wu, B. H., Liu, Z. N., Qiao, J., and Zhao, G. R. (2018). Combinatorial optimization of resveratrol production in engineered E. coli. J. Agric. Food Chem. 66, 13444–13453. doi: 10.1021/acs.jafc.8b05014
Keywords: cyanidin 3-O-glucoside, efflux transporter, anthocyanin, E. coli , multidrug resistance transporters
Citation: Wu X, Chen R, Liang P and Zha J (2024) Identification of Escherichia coli multidrug resistance transporters involved in anthocyanin biosynthesis. Front. Microbiol. 15:1357794. doi: 10.3389/fmicb.2024.1357794
Edited by:
Neha Arora, Skidmore College, United StatesReviewed by:
Guang-Rong Zhao, Tianjin University, ChinaPrem Pritam, University of Delaware, United States
Copyright © 2024 Wu, Chen, Liang and Zha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia Wu, d3V4aWFAc3VzdC5lZHUuY24=; Jian Zha, emhhamlhbjE5ODVAc3VzdC5lZHUuY24=
 Xia Wu1,2*
Xia Wu1,2* Rongxia Chen
Rongxia Chen Jian Zha
Jian Zha