Abstract
Ganoderma, a well-known genus in the Ganodermataceae family, has caused the extinction of several tree species due to its pathogenicity. This study explored the pathogenic effect of a newly identified Ganoderma species on trees and its competitive efficiency against Trichoderma species. Ganoderma camelum sp. nov. is characterized by small sessile basidiomata and a velvety, soft, camel-brown pileus. Phylogenetic analysis and ITS rDNA sequences indicated that the species were Trichoderma and Ganoderma camelum. Both fungal species competed antagonistically by secreting laccase. The laccase activity of G. camelum, with a value of 8.3 ± 4.0 U/mL, demonstrated the highest competitive activity against Trichoderma species. The laccase produced by T. atroviride (2.62 U/mL) was most effective in countering the pathogenic action of the novel G. camelum. The molecular weights of laccase were determined using SDS-PAGE (62.0 kDa for G. camelum and 57.0 kDa for T. atroviride). Due to the white rot induced by this Ganoderma species in the host tree, G. camelum showed the highest percentage inhibition of radial growth (76.3%) compared to T. atroviride (28.7%). This study aimed to evaluate the competitive antagonistic activity of Ganoderma and Trichoderma on malt extract agar media in the context of white rot disease in the host tree. This study concluded that the laccase from G. camelum caused weight loss in rubber wood blocks through laccase action, indicating tissue injury in the host species. Therefore, it was also concluded that G. camelum was more effective in pathogenic action of the host and resisted the biological action of T. atroviride. In principal components analysis (PCA), all the species associated with laccase exhibited a very strong influence on the variability of the system. The PIRG rate (percentage inhibition of radial growth) was strongly and positively correlated with laccase activity.
Introduction
Ganoderma is a genus widely distributed across desert, tropical, temperate, and agricultural ecosystems (Torres-Torres et al., 2015). Species within this genus exhibit diverse ecological relationships, including facultative, saprophytic, or parasitic associations with woody plants, particularly those in the Quercus-Pinus lineage (Torres-Torres et al., 2015). Ganoderma is a distinguished wood-decaying fungus, which affects both deceased and living trees (Pinar and Rodríguez-Couto, 2024).
These fungi also act as plant pathogens, causing basal stem rot in rubber, oil palm, and coconut trees (Kumari et al., 2024). As a basidiomycete, Ganoderma infects both young and mature trees, precipitating white rot through the decomposition of lignin, polysaccharides, and cellulose in hardwoods (Charpentier-Alfaro et al., 2023).
Various biotic factors, including basal rot caused by wound-colonizing fungi, contribute to the depletion of tree vegetation. In arid and semiarid regions, this disease targets the trunks, branches, and roots of woody plants (Al-Mosawi et al., 2024). Wood decay and discoloration are prevalent, and severe symptoms are observed in mature trees. This process unfolds over several years and remains imperceptible within the lifespan of the affected areas.
Ganoderma establishes colonization in a suitable host via the parasitic invasion of root masses and residual stumps, directly linking healthy roots with infected tissues within the soil (Shariffah-Muzaimah et al., 2015). Ganoderma camelum is also recognized as a wood rotter, which is believed to be an effective pathogen classified as a tree-dwelling (wood pathogenic) fungus.
The growth of Ganoderma is hindered by the presence of competitive antagonistic fungi, such as Trichoderma (Tong et al., 2020; Wang et al., 2022). Trichoderma, a soil-borne pathogen and an anamorphic form of Hypocrea (Ascomycota) (Kumar et al., 2023), induces biochemical responses in plants, thereby bolstering their defense against pathogens. Trichoderma Pers. is globally distributed and found in soil or decaying wood (Singh et al., 2020). A few Trichoderma species are renowned as industrial cellulose producers (Bischof et al., 2016) and are associated with diseases in mushrooms (Gangwar et al., 2018; An et al., 2022), including Ganoderma lucidum (Lu et al., 2016), Agaricus bisporus, Pleurotus ostreatus, and Lentinula edodes (Wang et al., 2016; Innocenti et al., 2019). These fungi are characterized by rapid growth and the production of a diverse array of degradative enzymes, e.g., laccase. Different Trichoderma species display antagonistic activity against other fungi through anastomosis around their hyphae, establishing a mycoparasitic relationship that curtails the activity and proliferation of pathogenic fungi affecting plants and trees (Chen et al., 2014; Yassin et al., 2021).
Investigations into the potential of laccase have encompassed various kingdoms (plants, bacteria, insects, and fungi), with fungal laccases demonstrating superior activity compared to other taxa. In higher fungi, laccase potential is particularly pronounced in its properties (Cázares-García et al., 2013; Loi et al., 2021). Fungal laccases are extensively involved in intracellular (extracellular) secretion processes, such as delignification, pathogenesis, and pigmentation (Tamano et al., 2022; Nazar et al., 2023; Rahman et al., 2024). Ganoderma, Pleurotus ostreatus, and Trametes versicolor are considered model organisms for laccase production (Yang et al., 2017), offering versatility and high potential for bioremediation strategies in various applications, including petrochemical, medical, textile, pesticide, and pharmaceutical waste treatment (Dong et al., 2023; Gutierrez-Rangel et al., 2024). Enzyme-assisted degradation of industrial and environmental effluents can be readily applied with multiple advantages (Al-Tohamy et al., 2023).
Despite the abundance of species, numerous regions remain unexplored, beyond the reach of researchers, and demand the undivided attention of mycologists. The species newly identified in this research was discovered in northern Pakistan, with its diminutive basidiome resembling those of the G. lucidum complex.
This study characterizes a novel Ganoderma species through morpho-anatomical, molecular, and phylogenetic analyses. The research also aimed to maximize laccase production by leveraging the competitive antagonistic interactions between Trichoderma and the newly identified Ganoderma species while also examining their wood pathogenic properties.
Materials and methods
Morpho-anatomical characterization, DNA extraction, PCR amplification, and phylogenetic analyses
Specimens of Ganoderma camelum were collected from Khanaspur Halipad, Abbottabad District, Khyber Pakhtunkhwa Province, Pakistan, between June 2018 and August 2019 and deposited under deposition numbers SCUF517 and SCUF518.
The morphological characterization of the fungus, including its color, shape, and other detailed features, were conducted according to the guidelines established by Corner (1983).
The thickness of the pileus was measured at the point where the width and length of the fruiting body intersect. The color of the pileus was determined using the color chart of Munsell (1975).
Microscopic analysis was executed by observing cross sections of the dried basidiomata, which were first soaked in a 5% potassium hydroxide (KOH) solution, then stained 1% Congo red, and subsequently visualized under a MX4300H compound light microscope (Meiji Techo Co., Ltd., Japan). At least 30 measurements were meticulously recorded at a magnification of 100X. For the spore measurements, 30 counts from two samples were presented as length × width (Nagy et al., 2010), with the apiculus excluded when not compressed. The microscopic characteristics were described in alignment with the methodology outlined by Cabarroi-Hernández et al. (2019).
Genomic DNA was isolated from the specimens using a modified CTAB technique, the ITS regions were analyzed and amplified using ITS1 and ITS2 primers (White et al., 1990). The PCR amplification process was performed within a 25 μL reaction volume, utilizing a master mix [DreamTaqGreen PCR Master Mix (2X), Fermentas].
The reaction mixture included 9.5 μL distilled water, 1 μL template DNA, 12.5 μL 2X PCR master mix, and 1 μL of each primer. The amplification protocol comprised 35 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 1 min, concluding with a final extension of 10 min at 72°C. The PCR products were purified and sequenced by TSINGKE Co., Ltd. (China).
The dataset included DNA sequences of the novel species, along with additional ITS sequences obtained from GenBankwww.ncbi.nlm.nih.gov/genbank/ and relevant literature. The sequences were automatically aligned using MAFFT and manually adjusted using CLUSTALW in BioEdit software. A phylogenetic tree was constructed using MEGA ver. 10 and RAxML (Hall, 1999; Katoh et al., 2019). The reliability of the tree was assessed through bootstrapping with 1,000 replicates.
Isolation and culturing
Ganoderma camelum was cultured by inoculating 1 cm sterile tissue segments onto Basidiomycete-selective malt extract agar (BSMEA). The BSMEA medium was prepared following the manufacturer’s guidelines (MEA) (Difco Laboratories, Franklin Lakes, NJ), augmented with streptomycin (100 mg/L) and benomyl 95% (4 mg/L) (Loyd et al., 2018b).
Terricolous fungi were isolated using the dilution plate technique with MEA, which was supplemented with Rose Bengal (1/15,000) and chloramphenicol (50 ppm) to inhibit bacterial growth (Smith and Dawson, 1944). Post-inoculation, the plates were incubated at 27°C for 10 days. Subsequently, the emerging colonies of Trichoderma were identified and enumerated. The samples of Ganoderma under study were subsequently preserved in the Suez Canal University Fungarium (SCUF) under the deposit numbers SCUF517 and SCUF518, respectively (https://ccinfo.wdcm.org/details?regnum=1180, accessed on 12 August 2024).
Qualitative assay of laccase
The production of laccase was assessed through the placement of 5 mm-diameter disks from 7-day-old colonies onto guaiacol-supplemented agar plates (Abdel-Azeem and Salem, 2012) and by direct inoculation into modified Czapek’s agar plates. These plates were then incubated in the dark at 28°C for 7 days. The development of a pronounced brown coloration beneath and around the fungal colony was interpreted as a positive response, indicative of guaiacol oxidation (Kalra et al., 2013).
Laccase production in submerged culture
Actively growing mycelia (five pieces of 5 mm diameter) were cultivated in 100 mL of basal nutritional medium (Umar et al., 2023) in an Erlenmeyer flask (250 mL) at 28°C and 150 rpm for 8 days from freshly prepared pure cultures of Ganoderma and Trichoderma (approximately 7 days incubation at 30°C). After incubation, the fungal broth culture containing mycelia was centrifuged at 10,000 rpm at 4°C for 20 min and then filtered using Whatman filter papers. The resulting extracellular fluid supernatant contained crude laccase, which was used for further research.
Quantitative assay of laccase
The supernatant, containing crude laccase, was employed to assess enzymatic activity by measuring the oxidation of the guaiacol substrate (Gao et al., 2011). For this quantification, a 50 mM sodium acetate buffer, adjusted to a pH of 4.5, was combined with 2 mM guaiacol. A solution comprising 1.5 mL of the crude enzyme supernatant, 1 mL of the sodium acetate buffer, and 1 mL of guaiacol was vigorously mixed for 30 s and then incubated at 30°C for 10 min (Chefetz et al., 1998). After incubation, absorbance was measured at 465 nm (465 = 12,100 M−1 cm−1). EA = (A* V)/(t * €* v), where E.A = enzyme activity (U/mL), A = absorbance at 465 nm, V = total volume of the reaction mixture (mL), v = enzyme volume (mL), t = incubation time (min), and € = extinction coefficient ( M−1 cm−1) (Gao et al., 2011).
Laccase purification and gel electrophoresis
The method described by Chefetz et al. (1998) was employed to purify the laccase. The filtrate was centrifuged at 13,000 rpm for 20 min at 10°C, after which the supernatant was precipitated with ammonium sulfate. The resulting precipitates were then dialyzed and loaded onto a DEAE-Cellulose anion-exchange column, which had been equilibrated with a 10 mM sodium acetate buffer (pH 5.5).
Subsequently, the laccase fraction was collected, concentrated, and dialyzed overnight, after which 3 mL of the DEAE-purified sample was applied to the column. Post-dialysis, the purity and molecular weights of the laccase were evaluated through SDS-PAGE analysis (Durán et al., 2002) and visualized using Coomassie Brilliant Blue R-250 staining. The relative molecular mass was estimated by comparison with standard molecular weight markers.
Ganoderma wood decay test
The Ganoderma wood degradation was evaluated using rubber wood blocks, each measuring 80 mm × 50 mm × 20 mm and weighing precisely (100 g). Initially, these blocks were immersed in distilled water overnight within plastic bags and subsequently subjected to autoclaving for 45 min at a temperature of 121°C. Subsequently, 100 mL of MEA broth was administered to each block, which was then autoclaved again under identical conditions. Following a 2 min cooling period within a laminar flow hood to facilitate the adequate absorption of the medium, Ganoderma sp. cultures derived from Petri plates were finely minced and introduced into sterile plastic bags. An additional 100 mL of MEA was added to the blocks, which were then incubated at ambient temperature for 120 days (Loyd et al., 2018b).
Control blocks without Ganoderma were included. The weight of both control wood blocks and the Ganoderma-infected wood blocks was meticulously recorded every 20 days to ascertain the degree of decay inflicted by the Ganoderma species.
Dual culture tests and percentage inhibition of radial growth
This experiment investigated the antagonistic relationship between Ganoderma and Trichoderma species. A 5 mm mycelium disk, each sourced from the periphery of an actively proliferating mycelium culture of both species, was excised and transferred to a distinct agar Petri plate. This disk was then permitted to propagate for 3 days at a controlled temperature of 25°C.
Control plates contained only Ganoderma cultures, and the study was replicated three times. The percentage inhibition of radial growth (PIRG) zones for both species was computed daily over 10 days, utilizing the formula delineated by Yazid et al. (2023).
In this formula, PIRG indicated “percentage inhibition of radial growth,” R1 indicated radial growth of the Ganoderma colony in the absence of Trichoderma, whereas R2 showed the radial growth of the Ganoderma colony in the presence of Trichoderma.
Slide culture method
A sterile, clean glass slide was inserted into 9-cm-diameter plates to facilitate the Ganoderma-Trichoderma interaction. An autoclaved, molten MEA layer was then applied onto the slide. The 5 mm disks, excised from 1-week-old colonies on the periphery of Ganoderma and Trichoderma isolates, were positioned 3 cm apart on the MEA surface, opposite each other across the slide. To mitigate drying, a small quantity of double-distilled water was added to the plate. Subsequently, the plate was incubated at 25°C for a week. Upon the completion of the incubation, the area where Ganoderma-Trichoderma hyphae interfaced was stained with lactophenol in cotton blue, and the slide was examined under a light microscope to assess any signs of mycelial penetration and cell wall degradation that occurred during the incubation period.
Statistical analysis
The mean values and standard deviations (±SD) from three biological replicates (n = 3) were presented in the data. These triplicate data sets transformed and were subsequently subjected to an ANOVA analysis using SPSS software. The mean differences were evaluated through the HSD (Tukey’s standardized range) test, with statistical significance set at Pd ≤ 0.05.
Principal component analysis (PCA) was employed to elucidate the relationships among the investigated cases and parameters. The statistical analyses were conducted using Statistica software (version 12.0, StatSoft Inc., Tulsa, OK, United States). Principal components analysis (PCA), ANOVA, and correlation determination were all performed at a significance level of a = 0.05. The data matrix utilized for the PCA statistical analysis of the chromatographic test results comprised three columns and 11 rows. The input matrix was automatically scaled.
Results
Molecular identification of Trichoderma and Ganoderma
ITS markers were employed to ascertain the Trichoderma species responsible for the highest laccase production. The potential species were identified by constructing a phylogenetic tree, which was generated using maximum likelihood analysis. The purified fungal mycelium was found to cluster into a distinct clade, closely related to other species, with high bootstrap values confirming the robustness of the tree’s topology (Table 1). Phylogenetic analysis facilitated the clear identification of the filamentous Trichoderma species (Figure 1). Multiple sequence alignments were executed with CLUSTALW in BioEdit software, followed by manual adjustment. Subsequently, phylogenetic trees were constructed from these alignments using MEGA Version 10.0 and RAxML, with the statistical significance of the tree being evaluated through bootstrapping with 1,000 replicates. The maximum likelihood tree topology for Ganoderma is depicted, exhibiting a 98% statistical bootstrap value, which supports the identification of a novel species (Figure 2; Table 2).
Table 1
| Taxon | GenBank No. | Country | Voucher/Strain/Isolate | References |
|---|---|---|---|---|
| Trichoderma harzianum | AF443928 | Mexico | G.J.S. 00-21 | Chaverri et al. (2003) |
| MW785562 | Pakistan | TH101 | Umar (2021) | |
| Trichoderma tomentosum | EU330958 | Canada | DAOM 178713A | Degenkolb et al. (2008) |
| DQ085432 | Canada | DAOM 178713A | Samuels (2004) | |
| Trichoderma velutinum | JX513903 | India | IIc2a | Prashantha et al. (2024) |
| JX513902 | India | IIA3b | Prashantha et al. (2024) | |
| Trichoderma viride | AY380909 | United States | ATCC 28038 | Holmes et al. (2004) |
| MW898148 | Pakistan | IAGST22 | This study | |
| Trichoderma aggressivum | FJ442607 | Ireland | CBS 100526 | Chaverri et al. (2003) |
| FJ442618 | Ecuador | DIS 252E | Chaverri et al. (2003) | |
| Trichoderma longipile | EU280074 | Canada | DAOM 1772271a | Hoyos-Carvajal et al. (2009) |
| KU516602 | Poland | 75Jb14 | Jankowiak et al. (2016) | |
| MW785564 | Pakistan | TL103 | This study | |
| Trichoderma virens | MW785563 | Pakistan | TV102 | Umar (2021) |
| KC479808 | Indonesia | GL2 | Shittu et al. (2017) | |
| Trichoderma cremeum | NR 134346 | United States | BPI 1112894 | Samuels (2004) |
| MW785565 | Pakistan | TC104 | This study | |
| Trichoderma beinartii | KX267803 | South Africa | PPRI 19281 | du Plessis et al. (2018) |
| MW785569 | Pakistan | TB109 | This study | |
| Trichoderma aureoviride | FJ998179 | China | FJD22 | Xia et al. (2011) |
| FJ998178 | China | FJD2 | Xia et al. (2011) | |
| Trichoderma pseudokoningii | MW785566 | Pakistan | TL105 | This study |
| NR 120296 | United States | NS19 | Kuhls et al. (1996) | |
| Trichoderma citrinoviride | MN187551 | Poland | Tc19-18Ig | Baturo-Cieśniewska et al. (2020) |
| MW785567 | Pakistan | TC106 | This study | |
| Hypocrea jecorina | AF362100 | Korea | KACC40517 | Park et al. (2005) |
| Trichoderma longibrachiatum | AY328041 | Hungary | UAMH 7956 | Szekeres et al. (2006) |
| AY328042 | Hungary | ATCC 208859 | Szekeres et al. (2006) | |
| AF362102 | Korea | T9 | Park et al. (2005) | |
| Trichoderma atroviride | AF456920 | United States | DAOM 222096 | Dodd et al. (2003) |
| AF011945 | Austria | UAMH 7956 | Kindermann et al. (1998) | |
| MW325977 | Pakistan | TAPU_07 | Umar (2021) | |
| Trichoderma erinaceum | DQ109534 | Peru | DIS 7 | Samuels et al. (2006) |
| Trichoderma strigosum | EU280114 | Guatemala | DAOM 234231 | Hoyos-Carvajal et al. (2009) |
| EU718074 | Germany | DMC 787b | Douanla-Meli et al. (2013) | |
| Trichoderma asperellum | JQ040317 | China | HNZZ1006 | Sun et al. (2012) |
| MW785568 | Pakistan | TAS107 | Umar (2021) | |
| EU280109 | Colombia | CIB T05 | Hoyos-Carvajal et al. (2009) | |
| Trichoderma pubescens | JQ272444 | United States | 16B5 | Baird et al. (2014) |
| DQ083016 | United States | DAOM 166162 | Samuels (2004) | |
| Trichoderma hamatum | FJ442658 | Ecuador | DIS 358H | Chaverri et al. (2003) |
| Protocrea farinosa | NR119700 | Austria | CBS 121551 | Jaklitsch et al. (2008) |
| Protocrea pallida | NR111329 | Austria | CBS 299.78 | Jaklitsch et al. (2008) |
Trichoderma species are used in phylogenetic analyses and are representative of each species used in this study.
Figure 1
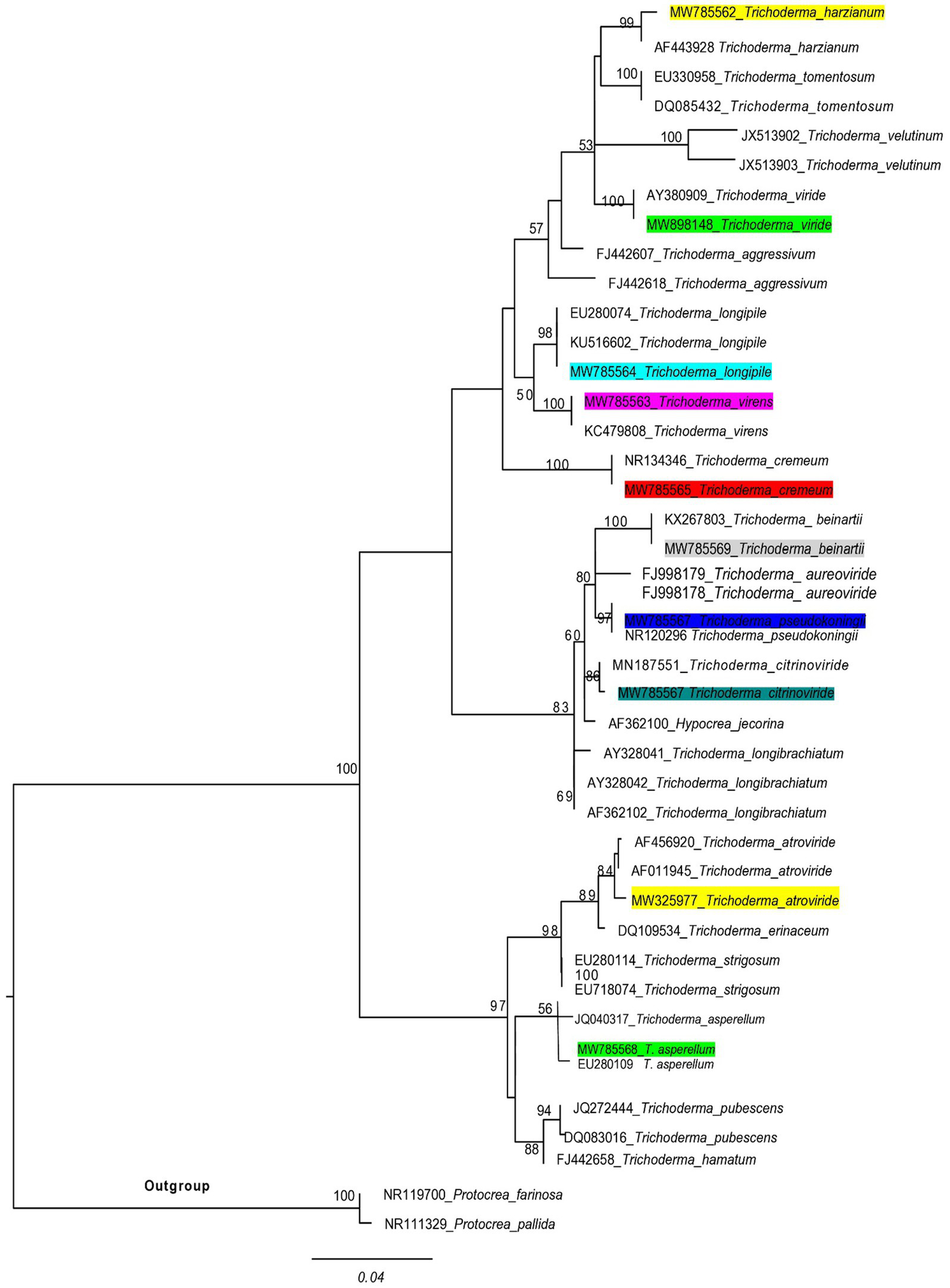
A phylogenetic tree of Trichoderma species was used in this experiment, and related taxa were based on ITS sequences generated by the maximum likelihood method. The tree was rooted using two species of Protocrea farinosa and Protocrea pallida. Bootstrap values (>50%) are shown at the branches.
Figure 2
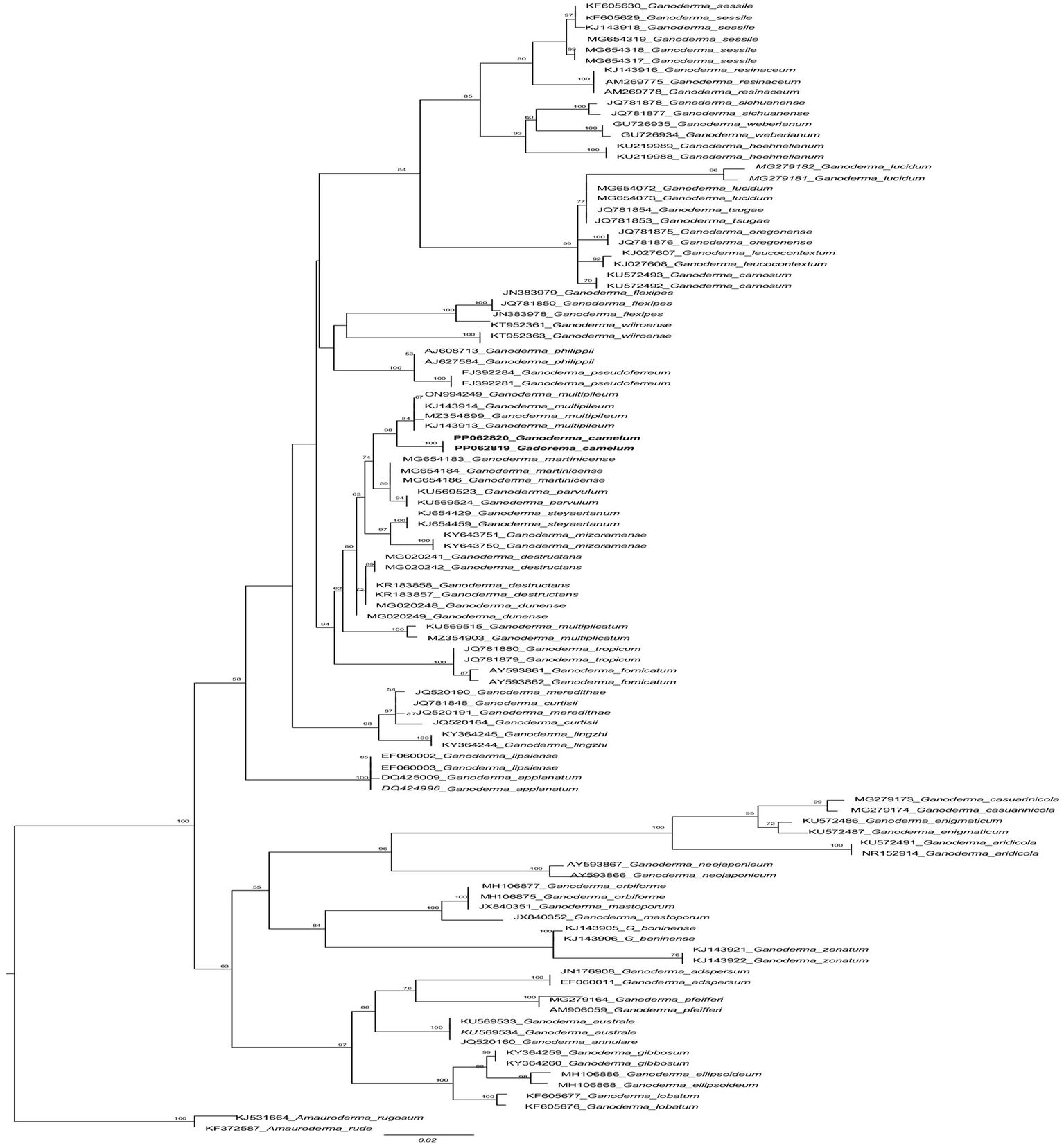
The phylogenetic tree of Ganoderma camelum used in this experiment and related taxa based on ITS sequences generated by the maximum likelihood method. The tree was rooted using two species from Amauroderma (Amauroderma rugosum and Amauroderma rude). Bootstrap values (>50%) are shown at the branches.
Table 2
| Taxon | Accession no. | Origin | Voucher/Strain/Isolate | References |
|---|---|---|---|---|
| Ganoderma sessile | KF605630 | United States | JV 1209/27 | Zhou et al. (2015) |
| KF605629 | United States | JV 1209/9 | Zhou et al. (2015) | |
| KJ143918 | United States | NY 00985711 | Zhou et al. (2015) | |
| MG654319 | United States | 228 DC | Loyd et al. (2018a) | |
| MG654318 | United States | 210FL | Loyd et al. (2018a) | |
| MG654317 | United States | 200MO | Loyd et al. (2018a) | |
| Ganoderma resinaceum | KJ143916 | Netherlands | CBS 194.76 | Loyd et al. (2018a) |
| AM269775 | Italy | DP1 | Guglielmo et al. (2007) | |
| AM269778 | Italy | G4/13 | Guglielmo et al. (2007) | |
| Ganoderma sichuanense | JQ781878 | China | Cui 7691 (BJFC) | Zhou et al. (2015) |
| JQ781877 | China | HMAS 42798 | Zhou et al. (2015) | |
| Ganoderma weberianum | GU726935 | India | GW-11 | Mohanty et al. (2011) |
| GU726934 | India | GW-10 | Mohanty et al. (2011) | |
| Ganoderma hoehnelianum | KU219989 | China | Dai 12096 | Song et al. (2016) |
| KU219988 | China | Dai11995 | Song et al. (2016) | |
| Ganoderma lucidum | MG279182 | China | Cui 14405 | Xing et al. (2018) |
| MG279181 | China | Cui 14404 | Xing et al. (2018) | |
| MG654072 | United States | UMNUT8 | Loyd et al. (2018a) | |
| MG654073 | United States | UMNUT9 | Loyd et al. (2018a) | |
| Ganoderma tsugae | JQ781854 | China | Yuan5649 | Loyd et al. (2018a) |
| JQ781853 | China | Dai3937 | Loyd et al. (2018a) | |
| Ganoderma oregonense | JQ781875 | United States | CBS 265.88 | Zhou et al. (2015) |
| JQ781876 | United States | CBS 266.88 | Zhou et al. (2015) | |
| Ganoderma leucocontextum | KJ027607 | China | GDGM443 03 | Li and Yuan (2015) |
| KJ027608 | China | GDGM44304 | Li and Yuan (2015) | |
| Ganoderma carnosum | KU572493 | Czech R | JV 8709/8 | Du et al. (2023) |
| KU572492 | Czech R | MJ 21/08 | Du et al. (2023) | |
| Ganoderma flexipes | JN383979 | China | Wei 5494 (IFP) | Zhou et al. (2015) |
| JN383978 | China | Wei5200 | Zhou et al. (2015) | |
| JQ781850 | China | Wei5491 | Cao et al. (2012) | |
| Ganoderma wiiroense | KT952361 | Ghana | UMN-20-GHA | Crous et al. (2015) |
| KT952363 | Ghana | UMN-21-GHA | Crous et al. (2015) | |
| Ganoderma philippii | AJ608713 | Malaysia | E7425 | Hapuarachchi et al. (2018) |
| AJ627584 | Malaysia | FRIM 589 | Hapuarachchi et al. (2018) | |
| Ganoderma pseudoferreum | FJ392284 | China | CATASGp008 | Costa-Rezende et al. (2017) |
| FJ392281 | China | CATASGp005 | Costa-Rezende et al. (2017) | |
| Ganoderma multipileum | ON994249 | China | HKAS 123775 | He et al. (2022) |
| KJ143914 | China | Dai 9447 | Zhou et al. (2015) | |
| MZ354899 | China | Cui 13597 | Sun et al. (2022) | |
| KJ143913 | China | CWN 04670 | Zhou et al. (2015) | |
| Ganoderma camelum | PP062819 | Pakistan | SCUF517 | This study |
| PP062820 | Pakistan | SCUF518 | This study | |
| Ganoderma martinicense | MG654184 | United States | 235TX | Loyd et al. (2018a) |
| MG654183 | United States | 232GA | Loyd et al. (2018a) | |
| MG654186 | United States | 248NC | Loyd et al. (2018a) | |
| Ganoderma parvulum | KU569523 | Colombia | CC16 | Bolaños et al. (2016) |
| KU569524 | Colombia | CC17 | Bolaños et al. (2016) | |
| Ganoderma steyaertanum | KJ654429 | Indonesia | IV-54-3 | Glen et al. (2014) |
| KJ654459 | Indonesia | 6-WN-15-M-A | Glen et al. (2014) | |
| Ganoderma mizoramense | KY643751 | India | UMN-MZ4 | Crous et al. (2017) |
| KY643750 | India | UMN-MZ5 | Crous et al. (2017) | |
| Ganoderma destructans | MG020241 | South Africa | CMW42140 | Tchotet Tchoumi et al. (2018) |
| MG020242 | South Africa | CMW42141 | Tchotet Tchoumi et al. (2018) | |
| KR183858 | South Africa | CMW43672 | Coetzee et al. (2015) | |
| KR183857 | South Africa | CMW4367 1 | Coetzee et al. (2015) | |
| Ganoderma dunense | MG020248 | South Africa | CMW42149 | Du et al. (2023) |
| MG020249 | South Africa | CMW 42150 | Du et al. (2023) | |
| Ganoderma multiplicatum | KU569515 | Colombia | CC8 | Bolaños et al. (2016) |
| MZ354903 | China | Dai 17395 | Sun et al. (2022) | |
| Ganoderma tropicum | JQ781880 | China | Yuan 3490 (IFP) | Zhou et al. (2015) |
| JQ781879 | China | Dai 9724 (IFP) | Zhou et al. (2015) | |
| Ganoderma fornicatum | AY593861 | China | AS5.539 | Wang and Yao (2005) |
| AY593862 | China | AS5.538 | Wang and Yao (2005) | |
| Ganoderma meredithae | JQ520190 | Korea | ATCC 64492 | Park et al. (2012) |
| JQ520191 | Korea | ASI 7140 | Park et al. (2012) | |
| Ganoderma curtisii | JQ781848 | Korea | CBS 100131 | Park et al. (2012) |
| JQ520164 | Korea | CBS 100132 | Park et al. (2012) | |
| Ganoderma lingzhi | KY364245 | South Korea | SFC20150624-06 | Jargalmaa et al. (2017) |
| KY364244 | South Korea | SFC20120721-08 | Jargalmaa et al. (2017) | |
| KY364245 | South Korea | SFC20150624-06 | Jargalmaa et al. (2017) | |
| Ganoderma lipsiense | EF060002 | Finland | NOR74/67/5 | Terho et al. (2007) |
| EF060003 | Finland | FIN R330 2la | Terho et al. (2007) | |
| Ganoderma applanatum | DQ425009 | China | GA165 | Costa-Rezende et al. (2017) |
| DQ424996 | China | GA117 | Costa-Rezende et al. (2017) | |
| Ganoderma casuarinicola | MG279173 | China | Dai 16336 | Du et al. (2023) |
| MG279174 | China | Dai 16337 | Du et al. (2023) | |
| Ganoderma enigmaticum | KU572486 | South Africa | Dai 15970 | Du et al. (2023) |
| KU572487 | South Africa | Dai 15971 | Du et al. (2023) | |
| Ganoderma aridicola | KU572491 | South Africa | Dai 12588 | Du et al. (2023) |
| NR152914 | South Africa | BJFC Dai 12588 | Xing et al. (2016) | |
| Ganoderma neojaponicum | AY593867 | China | AS5.542 | Wang and Yao (2005) |
| AY593866 | China | AS5.541 | Wang and Yao (2005) | |
| Ganoderma orbiforme | MH106877 | China | GACP1408 1329 | Hapuarachchi et al. (2018) |
| MH106875 | China | GACP1408 1235 | Hapuarachchi et al. (2018) | |
| Ganoderma mastoporum | JX840351 | China | TNMF0018835 | Wang et al. (2022) |
| JX840352 | China | TNM-F0018783 | Wang et al. (2022) | |
| Ganoderma boninense | KJ143905 | Japan | WD 2028 (FFPRI) | Zhou et al. (2015) |
| KJ143906 | Japan | WD 2085 | Zhou et al. (2015) | |
| Ganoderma zonatum | KJ143921 | United States | FL-02 (TNM) | Zhou et al. (2015) |
| KJ143922 | United States | FL-03 (TNM) | Zhou et al. (2015) | |
| Ganoderma adspersum | JN176908 | Italy | PF263 | Hapuarachchi et al. (2018) |
| EF060011 | Finland | ITA 39 | Terho et al. (2007) | |
| Ganoderma pfeifferi | MG279164 | China | Dai 12153 | Xing et al. (2018) |
| AM906059 | Italy | PLN 22 | Guglielmo et al. (2008) | |
| Ganoderma australe | KU569533 | Colombia | CTRA3 | Bolaños et al. (2016) |
| KU 569534 | Colombia | CTRA4 | Bolaños et al. (2016) | |
| Ganoderma annulare | JQ520160 | Korea | KCTC 16803 | Dai et al. (2011) |
| Ganoderma gibbosum | KY364259 | South Korea | SFC20130404-21 | Jargalmaa et al. (2017) |
| KY364260 | South Korea | SFC20140702-12 | Jargalmaa et al. (2017) | |
| Ganoderma ellipsoideum | MH106886 | China | GACP1408 1215 | Hapuarachchi et al. (2018) |
| MH106868 | China | GACP1408 0968 | Hapuarachchi et al. (2018) | |
| Ganoderma lobatum | KF605677 | United States | JV 0402/24 | Hapuarachchi et al. (2018) |
| KF605676 | United States | JV 1212/10J | Hapuarachchi et al. (2018) | |
| Amauroderma rugosum | KJ531664 | China | Cui 9011 | Li and Yuan (2015) |
| Amauroderma rude | KF372587 | China | GDGM25736 | Li et al. (2015) |
Species used for phylogenetic analyses of this study and their corresponding GenBank accession numbers.
Morpho-anatomical description of Trichoderma colonies and Ganoderma sp.
This study comprised 10 Trichoderma species and one new Ganoderma species evaluated for laccase activity. The colony characteristics of each species were determined (Table 3).
Table 3
| Sr No. | Species name | GenBank accessions | Conidia shape | Pigmentation | Colony appearance | PIRG rate (%) | Laccase activity (U/mL) |
|---|---|---|---|---|---|---|---|
| 1 | Ganoderma camelum | PP062819 | Absent | Green | White, branched, clamped | 76.3a | 8.3 ± 4.0 |
| 2 | Trichoderma harzianum | MW785562 | Globose to Subglobose | Green | Floccose/cottony white | 20cd | 1.12 ± 0.03 |
| 3 | T. viride | MW898148 | Globose | Gray to green | Highly intricate | 22.5abcd | 1.24 ± 0.05 |
| 4 | T. pseudokoningii | MW785566 | Oblong ellipsoidal | White | Ringed | 24abc | 1.16 ± 0.01 |
| 5 | T. cremeum | MW785565 | Oblong | Creamy | Circular | 25.6abc | 1.98 ± 0.09 |
| 6 | T. longipile | MW785564 | Oblong | Incarnate/green | Circular | 21cd | 1.84 ± 0.02 |
| 7 | T. atroviride | MW325977 | Globose to subglobose | Green | Highly intricate | 28.7ab | 2.62 ± 0.01 |
| 8 | T. citrinoviride | MW785567 | Ellipsoidal | Yellowish green | Rough and pigmented | 15.8de | 1.65 ± 0.01 |
| 9 | T. beinartii | MW785569 | Smooth oblong | Diffusible | Concentric zones | 20e | 0.42 ± 0.06 |
| 10 | T. asperellum | MW785568 | Subglobose/ovoidal | Green to dark green | Concentric rings | 18de | 1.25 ± 0.08 |
| 11 | T. virens | MW785563 | Ellipsoidal to obovoid | Light yellow/green | Floccose | 22abcd | 1.03 ± 0.07 |
List of laccase (U/mL) producing Ganoderma camelum and Trichoderma species with PIRG and colony characteristics.
Taxonomy
Ganoderma camelum A. Umar, sp. nov. (Figures 3A–D, 4A–E).
Figure 3

Ganoderma camelum (A,B). Basidiome upper surface (A: fresh, B: dried). (C1) Contextum and Tubes. (C2) Tubes. (C3) Contextum. (D) Lower pore surface. (Scale bars: A,B = 10 mm, C–E = 5 mm).
Figure 4
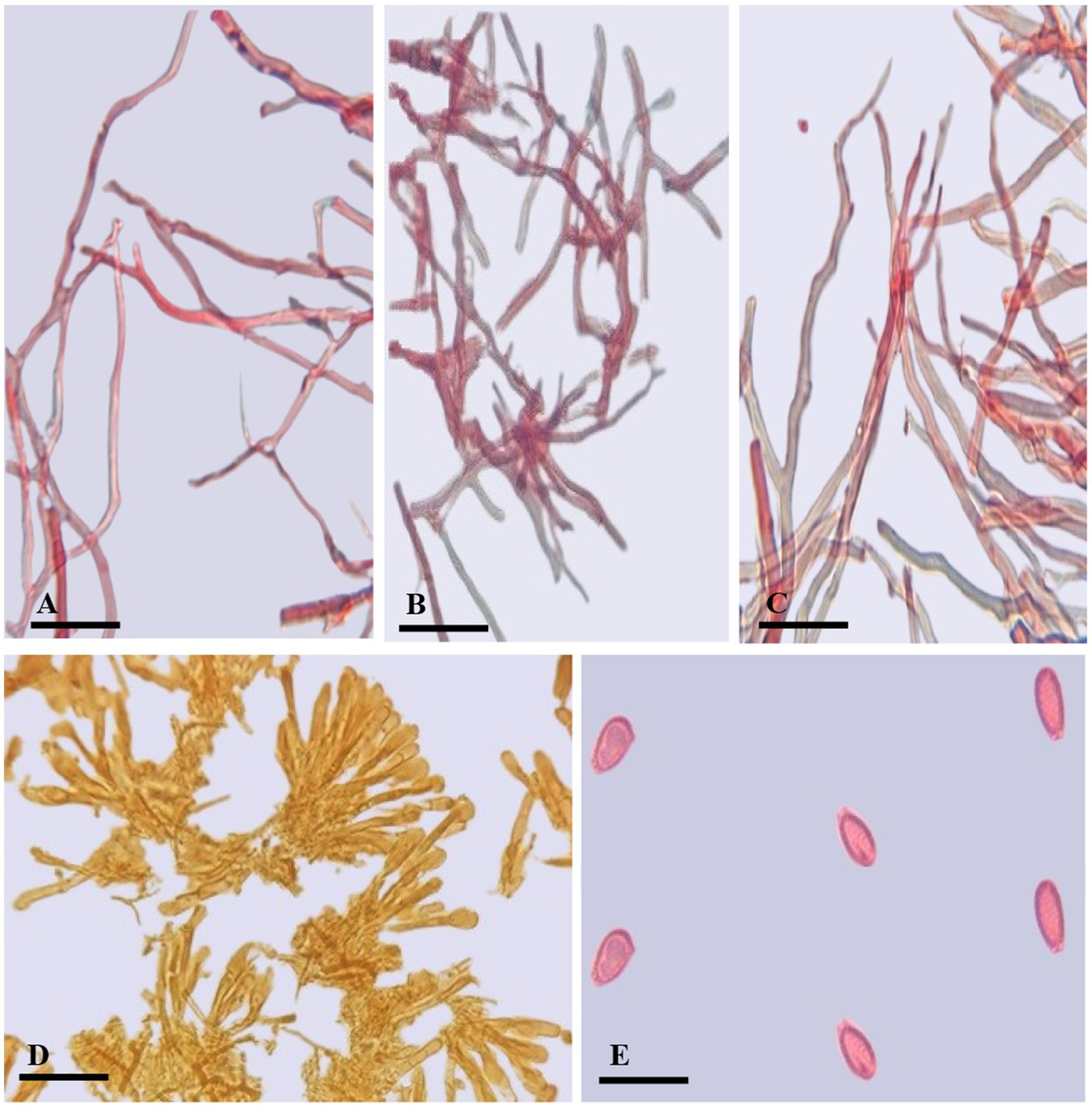
Ganoderma camelum (A) Generative hyphae. (B) Binding hyphae. (C) Skeletal hyphae. (D) Crustohymeniderm cells. (E) Basidiospores. (Scale bars: A–C = 5 μm, D–E = 10 μm).
MycoBank# 854703.
Diagnosis: In the phylogenetic tree (Figure 2), Ganoderma martinicense and G. multipileum are very closely matrixed to our new species of G. camelum. Morphologically, G. camelum is characterized by its sessile, delicate, and very soft velvety appearance of basidiomata; similarly, a sessile basidiome is found in G. martinicense, the closest species in the phylogenetic tree. The other closest species was G. multipileum, which rarely exhibits this characteristic and has stipitate basidiomata. Growth zones are found in both G. camelum and G. martinicense. The contextum is dark cinnamon brown in G. martinicense and light-brown to brown in G. multipileum, while G. camelum exhibited a soft light brown to camel brown contextum. The spores are larger in G. multipileum (7.3–) 8.0–11.5 (−12.2) × (5.3–) 5.5–7.8 (−8.3) μm and G. martinicense (8–) 8.8–10.5 (−11.3) × (5–) 5.5–7 (−7.2) μm, while smaller in G. camelum (4.7–5.2 × 2.3–3.6 μm).
Etymology: The species epithet “camelum” refers to camel brown color.
Holotype: PAKISTAN, Khyber Pakhtunkhwa, Abbottabad District, Khanspur Halipad (34° 1′ 16 N, 73° 25′ 40E, on the living stem of Pinus wallichiana, elevation 2,250 m above sea level, Aisha Umar, 6th June 2018, KPKHP31) (SCUF517: GenBank PP062819).
Description: Basidiomata sessile, convex, velvety, very soft, shallow waved five concentric zones, inner four zones tawny olive (10YR) to camel brown (5YR), outer zone pinkish buff (9/4R) to beige (9/6R); Margin 0.1–0.2 mm, very thin, white; Pileus 5.5–6.5 × 6.5–7.3 cm, glabrous, bumpy, verrucose, reniform, thick, margins thin, obtuse, slight radiating lines, soft flesh; Pores 110–153 × 125–130 μm, hard porous layer, yellow-brown (5YR) to tawny (10YR), subcircular or longitudinal; Tubes 0.2–0.3 mm long, min, non-stratified, ochraceous buff (6/8YR); Context 0.4–0.5 mm thick, light brown (6/6YR) soft velvety layer, beneath brown tawny (10YR) hard layer, milky cream, dry, fibrous and corky; Crustohymeniderm palissade club-shaped, clavate, pale yellow to yellow (5Y), 32.5–52.7 × 10.2–12.4 μm, smooth, double-walled, few two to three septate or few multi-septate cells; BASIDIOLES 6.5–13 × 4.5–7 μm, inverted pear-shaped to broadly clavate with big oil droplets; Basidiospores 4.7–5.2 × 2.3–3.6 μm AL = 4.6 μm, AW = 2.8 μm, Q = 1.63 (n = 30/1), ellipsoid with tapering ends, smooth, bitunicate, inter-walled pillars absent, laterally pointed; Hyphal System Trimitic (1) generative hyphae (septate, clamped, colorless, thin-walled, 2–3.4 μm), (2) skeletal hyphae (thick-walled, colorless, unbranched or few branches with distal end, 3.1–4.8 μm), and (3) binding hyphae (arboriform, colorless thick-walled, much-branched, 1.2–3.4 μm).
Additional specimen examined: PAKISTAN, Khyber Pakhtunkhwa, Abbottabad District, Khanspur Halipad (34° 1′ 16 N, 73° 25′ 40E), on the living stem of Pinus wallichiana, elevation 2,215 m above sea level, Aisha Umar, 25 August 2019, KPKHP32 (SCUF518:GenBank PP062820).
Results of the BLAST program
The consensus sequence of the ITS regions was subjected to a BLAST search utilizing the NCBI GenBank database, facilitating a comparison with a sequence database to ascertain species-level taxonomic data. An unknown or novel species is identified by situating it within an evolutionary context alongside other homologous sequences through phylogenetic analyses. The initial BLAST results for our sequences and consensus yielded 100 NCBI BLAST hits, of which over 58 were designated merely as “Ganoderma sp.” (merely the genus name was provided without specifying the species). These 58 sequences of Ganoderma species remained unnamed before our initial BLAST analysis. Consequently, this study has assigned species names to all previously unknown sequences associated with the genus Ganoderma. In the NCBI Query Cover, our novel species matched 100% with previously unidentified Ganoderma species. The initial BLAST revealed a 99.66% identification percentage, with an accuracy length of 619 (Accession PP062820.1) and 621 (Accession PP062819.1). The maximum score and total score for our new sequences were 1,081, suggesting that this represents a species not previously described.
Comparison of Ganoderma camelum with neighboring species of a phylogenetic tree
In the phylogenetic tree, several neighboring species are positioned in close proximity to our newly identified species. These include Ganoderma multipileum (Wang et al., 2009; Welti and Courtecuisse, 2010; Nguyen et al., 2023), G. martinicense (Loyd et al., 2018c), G. mizoramense (Crous et al., 2017), and G. parvulum (Torres-Torres and Guzmán-Dávalos, 2012) (Figure 2). Differentiation between G. camelum and G. martinicense from G. multipileum is evident through the presence of sessile basidiomata in the latter, as G. multipileum rarely exhibits this characteristic. The basidiomes of G. parvulum exhibit a range from sessile to stipitate, with a more frequent stipitate form akin to G. mizoramense. The basidiomata of G. mizoramense are pileate, stipitate, applanate, flabelliform, and devoid of any “growing zones,” contrasting with G. camelum. The pileal surface is smooth, laccate, radially rugose, slightly zonate with dark lines, and ranges from fully reddish brown to violet brown in G. parvulum. In contrast, it is small, soft, non-laccate and light camel brown in our new species. The upper pileus surface of G. mizoramense can be distinguished from G. camelum by its reddish brown (fresh) to liver-brown (dried) context, dark-brownish to dark reddish brown, with a white lower surface when fresh, as opposed to the light camel brown pileus and concolorous context observed in our new species.
Concentric growth zones are present in both G. camelum and G. martinicense. The contextum is dark cinnamon brown in G. martinicense and light brown to brown in G. multipileum, while our new species exhibits a soft velvety light brown or camel brown contextum. G. parvulum possesses a light pale ochraceous context (Torres-Torres and Guzmán-Dávalos, 2012) with dark horny (Murrill, 1902) or carob brown (Steyaert, 1980) resinaceous streaks, differing from our new species. The context in G. parvulum occasionally displays scattered yellow spots and a thin yellow line just below the crust, features absent in our new species. A uniform ochraceous or cinnamon context is characteristic of G. mizoramense.
In G. parvulum, the margins are slightly lobulated, ranging from white to pale yellow or grayish-yellow to yellowish orange, contrasting with the white and non-lobulated margins of G. camelum. Our species features a wide yellowish-brown porous surface, contrasting the white, yellowish white, or sun yellow surface in actively growing G. parvulum and the yellowish brown to brownish orange surface in dried pore surfaces.
Cuticular cells are cylindrical to slightly clavate, averaging 50 μm in length, while those in our new species are palissade club-shaped, clavate, pale yellow to yellow, smooth, double-walled, with few two-to three-septate or few multi-septate cells, and smaller in size (32.5–52.7 μm × 10.2–12.4 μm).
Ganoderma camelum has smaller basidiospores (4.7–5.2 μm × 2.3–3.6 μm), subglobose to ellipsoid, smooth, bitunicate, and lacks laterally pointed and inter-walled pillars. In contrast, G. parvulum features larger spores (8.1 μm × 5.9 μm), free to subfree very thin pillars. The basidiospores of G. mizoramense are brown, ellipsoid with a truncate base, verruculose, and larger (11.10 μm × 7.6 μm) than those of our new species. The spore size in G. multipileum and G. martinicense is larger (7.3–) 8.0–11.5 (−12.2) × (5.3–) 5.5–7.8 (−8.3) μm and (8–)8.8–10.5(−11.3) × (5–)5.5–7(−7.2) μm, respectively, while our new species exhibits smaller spores.
Screening of laccase-producing species
Four Trichoderma species, including T. cremeum, T. longipile, T. citrinoviride, and T. atroviride, were found to be capable of oxidizing guaiacol. Ganoderma exhibited the darkest maroon zone on the laccase detection plate, whereas Trichoderma demonstrated the highest laccase activity, as observed in Supplementary Figure S1A1,A2. Among the tested species, T. atroviride was identified as having the greatest laccase secretion potential, while T. citrinoviride presented the lowest. Consequently, T. atroviride secreted the highest amount of laccase, as depicted in Supplementary Figure S1B1,B2. On MEA media, T. atroviride produced pale green spores, with the prevalence of green indicating its dominant zones of operation. The growth rate within the Ganoderma zonal area was slower.
This study identified the most potent Trichoderma candidates under optimal conditions and compared their laccase activity to that of Ganoderma camelum, which had a laccase activity of 8.3 U/mL. Trichoderma atroviride was found to exhibit a laccase activity of 2.62 U/mL, establishing it as the most promising candidate for laccase production. The secondary productive species, T. cremeum, T. longipile, and T. citrinoviride, yielded laccase concentrations of 1.98 U/mL, 1.84 U/mL, and 1.65 U/mL, respectively (Table 3). The most robust candidate, T. atroviride, was selected for subsequent studies.
Partial purification of laccase
The purification of laccase from Trichoderma atroviride and Ganoderma camelum was carried out using 60% ammonium sulfate precipitation. After partial purification, the molecular weights of the laccase were determined using SDS-PAGE. Standard protein markers were employed to estimate the purified laccase, with the band positions post-staining aiding in this quantification. The molecular weights were approximately 57.0 kDa for T. atroviride and 62.0 kDa for G. camelum (Supplementary Figure S2).
Conflictual combat and potential of Ganoderma
The sequence of events commencing with reconnaissance culminates in the penetration of fungal pathogens, ultimately leading to host mortality. The secretion of laccase by G. camelum facilitated rapid mycelial advancement toward Trichoderma. In a competitive interaction, both species secreted laccase within their immediate environment, collaboratively executing the task. Ganoderma mycelium established physical contact with Trichoderma hyphae within the laccase oxidation zone, subsequently penetrating the lumen of the Trichoderma hyphae and assimilating its contents. Both Ganoderma and Trichoderma species released laccase enzymes or vied for space and nutrients during the antagonistic interaction.
Microscopic examination of the interaction between pathogenic G. camelum and T. atroviride revealed hyphal growth, followed by coiling, entanglement, and hooking around G. camelum. A change in medium color from white mycelium to purple indicated laccase secretion.
In vitro plate studies demonstrated that Ganoderma inhibited the growth of all Trichoderma species, which may be attributed to the higher molecular weight of laccase. An inhibition level exceeding 70%, as evaluated by the PIRG equation, was considered indicative of a species’ potential against another.
All Trichoderma species produced laccase at varying rates and inhibited G. camelum to different extents (Table 3). Trichoderma atroviride exhibited the highest PIRG score of 28.7%, which was statistically significant compared to other species. Regarding laccase production, five species were found to exhibit the highest protective effect against Ganoderma: T. atroviride, T. viride, T. virens, T. pseudokoningii, and T. cremeum, as determined by the PIRG evaluation.
Among these, T. atroviride secreted the highest amount of laccase, followed by T. cremeum, T. longipile, and T. citrinoviride. T. atroviride also effectively inhibited G. camelum, while T. cremeum, T. longipile, and T. citrinoviride also displayed rapid growth and purplish pigmentation after 5 days of incubation. Over a 10-day observation period, T. citrinoviride exhibited the smallest inhibitory zone against G. camelum development (76.3%) (Figures 5A–J).
Figure 5
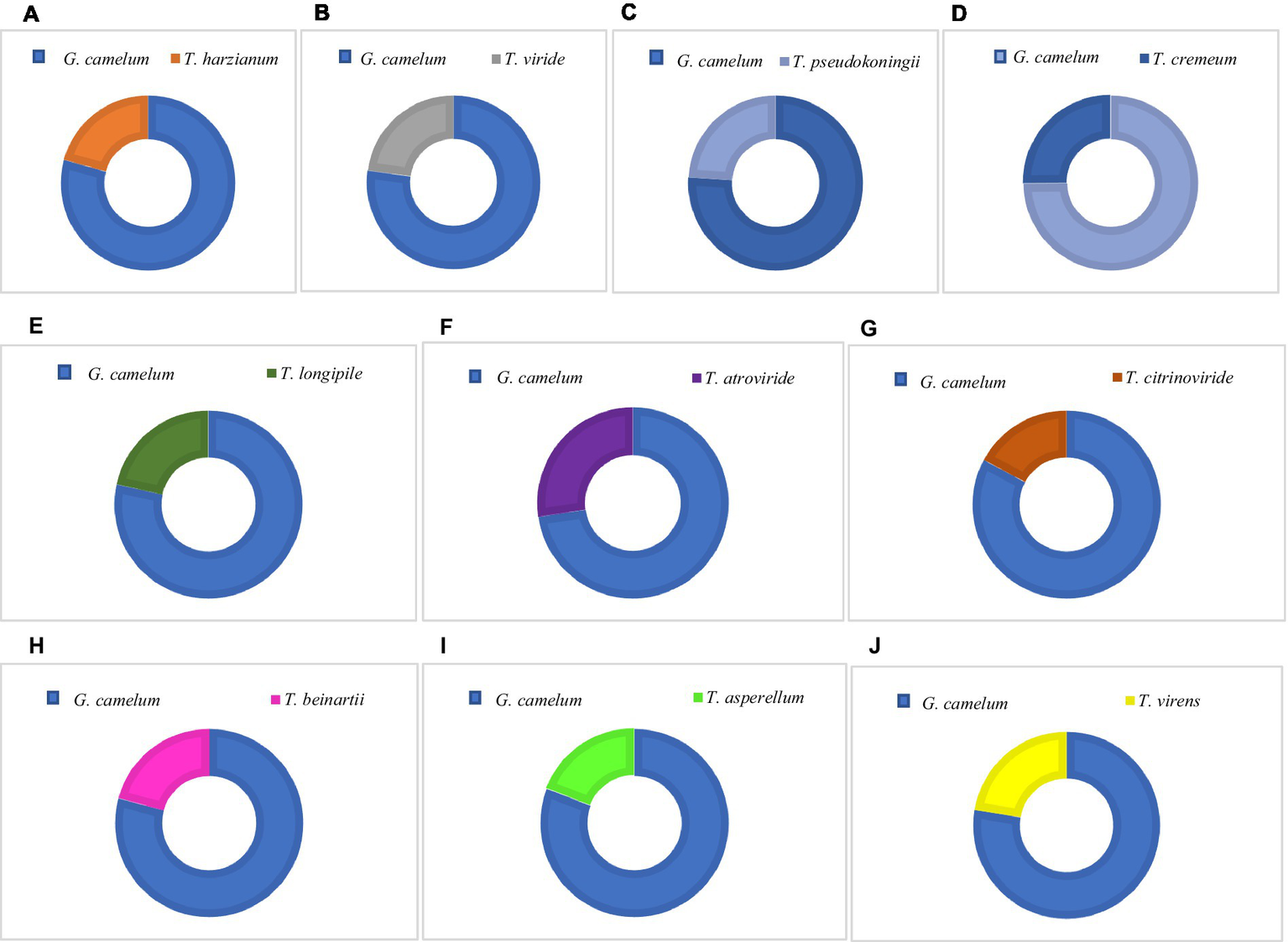
Percentage Inhibition of Radial Growth (PIRG) value of Trichoderma species and G. camelum.
The antagonistic interaction between Ganoderma and Trichoderma was confirmed through in vitro plate analysis, which revealed that Ganoderma suppressed the growth of all Trichoderma species. As assessed by the PIRG equation, an inhibition level above 70% was deemed indicative of a species’ antagonistic potential.
Microscopic inhibition of mycelium
Considerable alterations in hyphal morphology were detected in Ganoderma mycelia upon exposure to Trichoderma, in contrast to the control group.
Figure 4 reveals a robust and well-developed Ganoderma mycelium, characterized by its healthy, compact, and highly branched structure. In response, the mycelium of G. camelum adopted a highly branched structure to mitigate the growth of Trichoderma. This adaptation was facilitated by producing a copious amount of laccase, thereby maximizing its defensive potential.
The laccase zone expanded across the entire surface of the Petri plates; however, only a limited number of hyphal strands were colonized and obscured by Trichoderma spores.
The hyphal structure of T. atroviride showed signs of disruption, aggregation, shriveling, loss, flattening, and altered appearance. Mycelial damage was attributed to intense resource competition, ultimately inhibiting Trichoderma growth.
The attachment of Trichoderma spores to the fungal hyphae indicated a mycoparasitic interaction. Trichoderma sp. was observed to identify and encircle adjacent fungal hyphae, forming haustoria to penetrate cell walls as a defensive strategy (Supplementary Figure S4).
Wood decay and tree appearance
Airborne basidiospores of G. camelum are released through appropriate openings in the injured area of woody tissue near the ground. The basal region of the tree is prone to damage due to the moisture present in the soil.
Structural roots in the soil are damaged by basidiospores and gradually colonized, leading to tree death over several years. Structural roots anchor the tree in the soil, while fine feeder roots absorb moisture and nutrients daily.
Once structural roots are damaged, stability is at risk due to colonization by Ganoderma species. No effective prevention or control measures are available to overcome BSR disease until the tree’s demise. However, the Trichoderma species have also proved ineffective except for removing and replacing soil and trees. In this study, infected trees were compared with healthy ones as control. The vigor of Pinus wallichiana declined, resulting in structural weakness, slow growth, yellowing and shrinking leaves, susceptibility to wind damage, and, eventually, branch dieback.
Once colonized, the wood block becomes saturated with water, resulting in a fibrous, tender, porous, crumbly, and flaky texture. The wood becomes discolored, primarily in white hues with occasional yellowish tones. The weight of the block was measured every 20 days, decreasing from 98.2 g initially to 97.5 g after 40 days, 97.1 g after 60 days, and finally 95.3 g after 120 days (Figure 6). The infected areas develop a pale, patchy appearance due to the presence of mycelium.
Figure 6
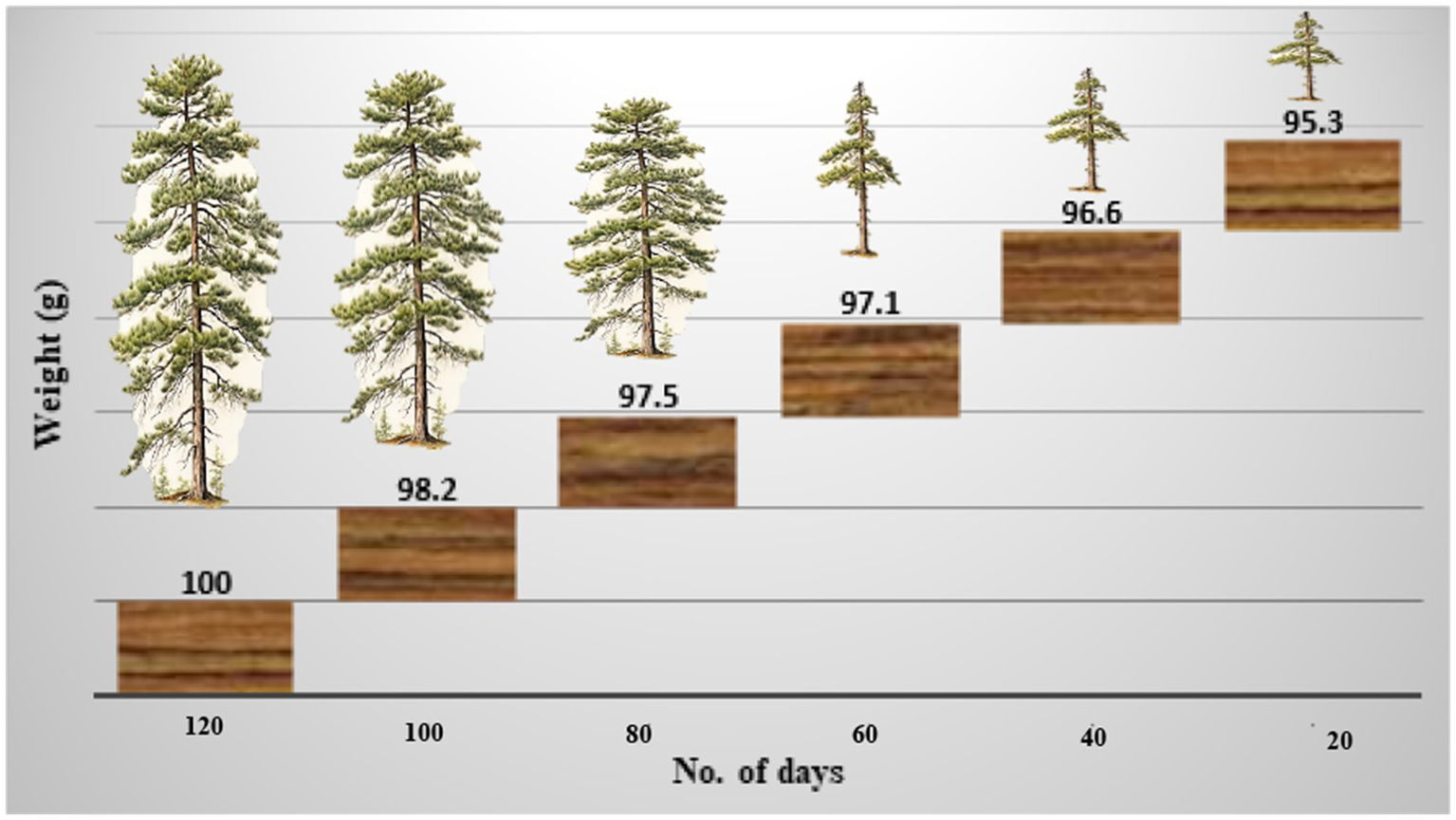
The weight of the wood block decreased over time by inoculation or infection of Ganoderma camelum sp. nov.
The woody stem becomes discolored, forming white patches due to laccase-induced degradation.
The mycelium growing within the softened wood secretes enzymes that degrade the cell wall components for energy and nourishment. Consequently, the wood undergoes whitening and bleaching due to oxidation by laccase, leading to lignin degradation.
Principal component analysis
Principal component analysis (PCA) was conducted, yielding two variables that accounted for 100% of the system’s variability. All parameters exerted a very strong influence on the variability of the system (Figure 7A). The PIRG rate and laccase activity parameters were strongly and positively correlated.
Figure 7
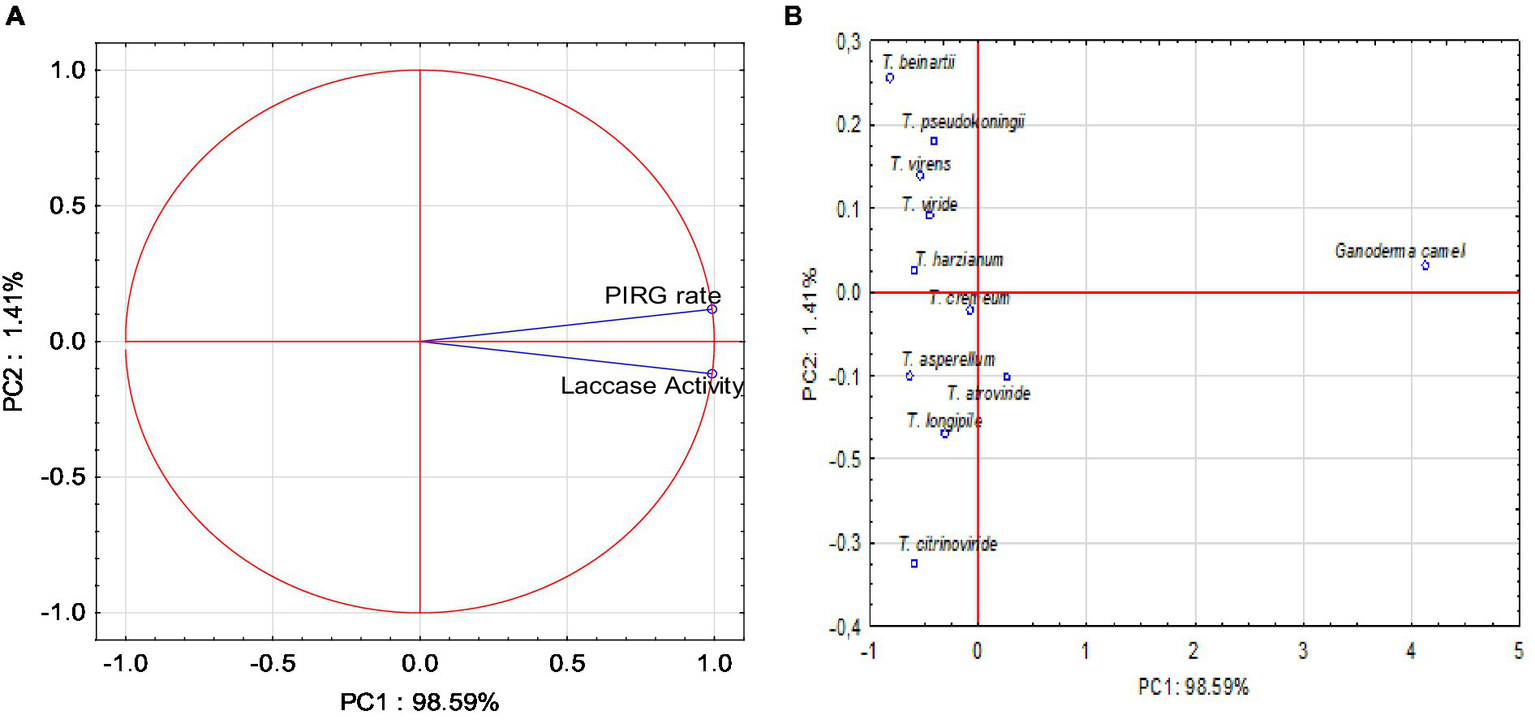
(A) Projection of variables: parameters on PC1 and PC2 loadings plot; (B) Projection of sample type on PC1 and PC2 scores plot.
The PCA analysis indicated that positive values of the first principal component (PC1) described the correlation and influence of the PIRG rate and laccase parameters, accounting for 98.59% of the variance. The conducted PCA analysis demonstrated that positive values of the first main component, PC1, described the types of fungi at 98.59%. Positive values of the first principal component, PC1, disclosed the Ganoderma type, and negative values depicted the Trichoderma type. The PCA analysis (Figure 7B) also revealed that the PIRG rate and laccase parameters were described by Ganoderma camelum.
Correlation matrix heat map
The heat map visually represents data, highlighting value variations through color contrasts. This graphical depiction serves as an effective means of illustrating the matrix’s values through a spectrum of colors. As detailed in Table 4, the correlation matrix shown the correlation coefficients for every pair of variables. In this study, the row and column labels consist of the “names of the variables” and the “numerical values of the calculated correlation coefficients,” which are explicitly listed within the table. The correlation coefficient, ranging from -1 to 1, indicated the strength of the linear relationship between the variables. The greater the absolute value, the more pronounced the relationship. Moreover, the sign of the correlation coefficient indicates whether the relationship between the studied variables is positive or negative.
Table 4
| r>= | −1 | −0.80 | −0.60 | −0.40 | −0.20 | 0 | 0.20 | 0.40 | 0.60 | 0.80 | 1 |
| PIRG rate | Laccase activity | ||||||||||
| PIRG rate | 1.000 | 0.972 | |||||||||
| Laccase activity | 0.972 | 1.000 | |||||||||
A heat map of the correlation matrix for the tested samples.
Discussion
Ganoderma camelum, a novel species discovered in Khyber Pakhtunkhwa, Abbottabad District, Khanspur Halipad, Pakistan, colonized on the stem of Pinus wallichiana. This species is characterized by a very soft, delicate, and velvety appearance of its basidiome, with a camel brown coloration and five distinct growth zones. The upper pileus layer is notably soft, contrasting with the slightly harder porous layer. The tubes are min, and the context is divided into two layers. The upper is soft and floccose, while the lower is flat, slightly hard, and composed. A unique feature of Ganoderma camelum is the presence of oval, bubble-like rounded bodies on the upper surface of its basidiome.
Phylogenetically, Ganoderma camelum forms a distinct clade with other Ganoderma species, supported by a robust statistical bootstrap value of 98%. This species is distinguished by its unique morphological characteristics and strong phylogenetic placement, thereby establishing it as a new species. Ganoderma camelum is differentiated from other Ganoderma species by its soft, delicate, and velvety nature, camel-brown coloration, and five growth zones. Its shelf-like shape further confirms its classification as a new species.
The potential for controlling BSR disease through biological means, where pathogenic Ganoderma species are present, has been observed. Basidiomycetes, specifically white rot fungi, degrade lignin and cellulose enzymatically, resulting in a light-colored, spongy, stringy mass that separates the firm heartwood and sapwood. White rot fungi commonly attack the hardwoods of deciduous trees, which are resistant to brown rot fungi.
Recently introduced control programs that utilize biological agents have yielded initial encouraging results in the fight against the diseases (Verma et al., 2022). The deployment of biological strategies presents a compelling alternative for managing stem rot diseases in trees, devoid of adverse environmental consequences. Unlike fungicides, biological agents colonize the rhizosphere without imparting toxic residues (Verma et al., 2023).
The integration of chemical fungicides into agricultural practices has become a fundamental aspect of crop management.
These substances, alongside pesticides, have been associated with various health issues, including cancer, respiratory ailments, and hormonal imbalances, depending on the level of exposure (Piel et al., 2019). As awareness of the detrimental impacts of excessive fungicide usage has grown, strategies for integrated pest management have been adopted to mitigate plant diseases. These approaches prioritize disease prevention and the comprehensive utilization of all available tools for plant disease management, factoring in their economic viability and toxicity. In this context, biological agents are increasingly proposed as an alternative to traditional fungicides (Ons et al., 2020). The concept of biocontrol, or biological control, often encounters confusion due to its varied interpretations in scholarly literature. Biocontrol is characterized by deploying a living organism to combat a specific plant pathogen through the secretion of diverse metabolites, antibiosis, parasitism, or competition for resources and space (Köhl et al., 2019). The use of living organisms to control plant diseases, not merely through direct antagonistic effects against plant pathogens but also via the induction of resistance, which activates the plant’s defense mechanisms, is also encompassed under the definition of a biocontrol agent (Raymaekers et al., 2020).
Ganoderma species are recognized for their role in wood decay, both in living trees and in decaying stumps or trunks, as documented across a global distribution spanning continents such as America, Asia, the Middle East, and Europe (He et al., 2022). These organisms are responsible for heart rot, a condition characterized by the growth within the central, non-living, woody tissues of standing trees. Notably, a single species of Ganoderma can target a multitude of host species, as observed in both temperate and tropical regions.
As biological control agents, pathogenic white fungi are frequently employed to manage plant and tree diseases. These fungi expedite wood degradation at a rate exceeding that of the pathogens. They vie for the same resources, compete for nutrients, synthesize inhibitory secondary metabolites, and possess the capacity to mycoparasitize the pathogens. Among other biological agents, Trichoderma competes with Ganoderma for wood resources by secreting laccase. Both species engage in a combative interaction, employing mycelial extension and laccase secretion to vie for space and nutrients (Figure 8).
Figure 8
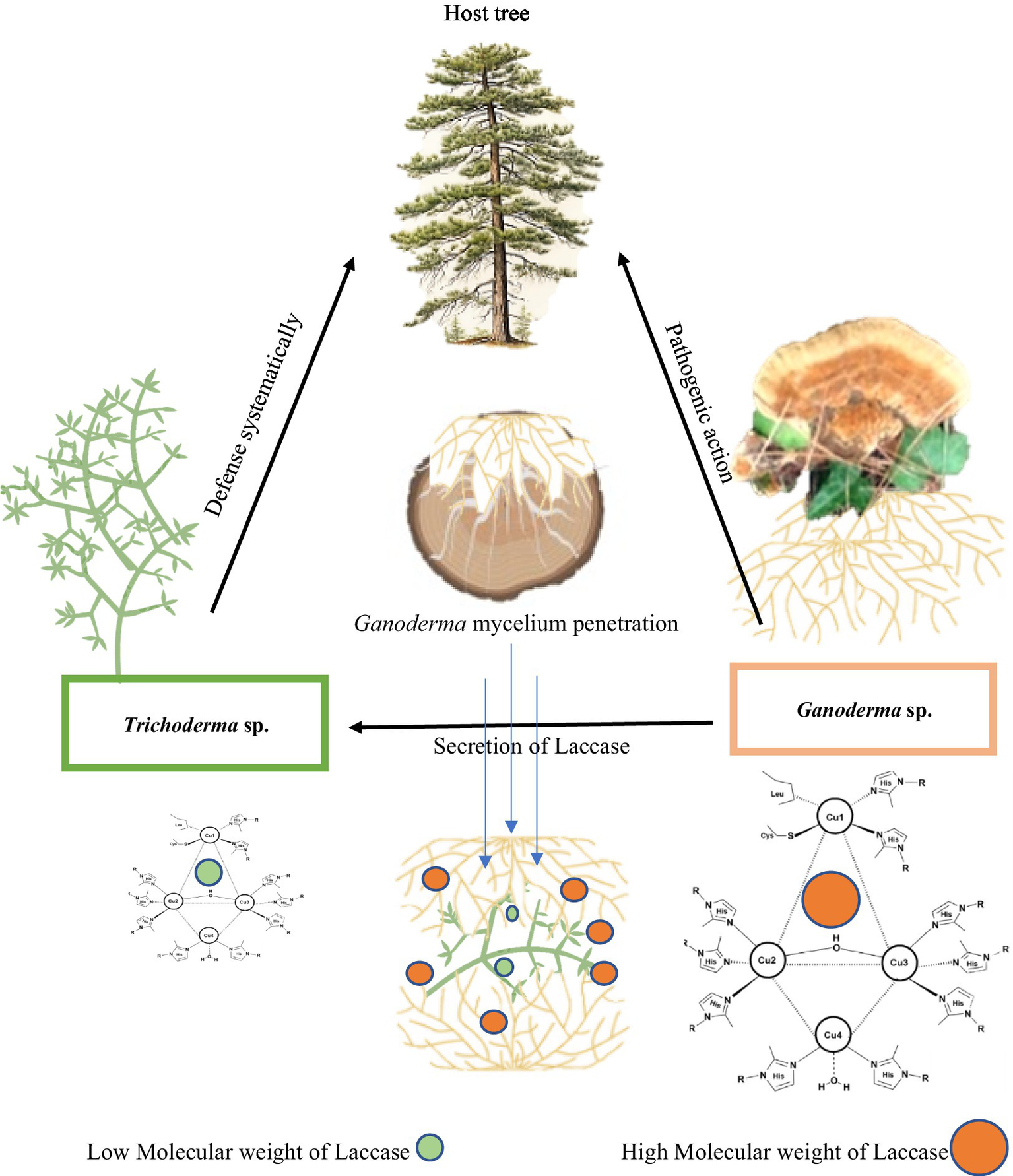
Mechanistic action of laccase of Ganoderma camelum against Trichoderma atroviride. Laccase of high molecular weight of Ganoderma antagonistically acts against the laccase of Trichoderma in competition of nutrients and space.
The occurrence of brown spots on Prosopis wood is attributed to the action of Ganoderma, which facilitates the removal of carbohydrates, ultimately resulting in a brownish, oxidized lignin residue. The absence of a fibrous texture indicates the swift degradation of cellulose (Yang et al., 2022). As the infection progresses, the wood exhibits shrinkage upon drying, with cross-linings becoming discernible. Notably, non-enzymatic processes characterize the initial stages of infection. The fungal organism secretes a suite of enzymes that traverse the cell wall, analogous to a pair of scissors severing chains of hemicellulose and cellulose into minute fragments (Shankar et al., 2024).
Spores of Ganoderma penetrate trees through natural incisions, stomatal openings, or during pollination via ovules. Given the global biodiversity, approximately 300,000 plant species coexist with a multitude of “endophytic microbes,” including fungal species that inhabit palms across tropical and temperate zones (Dhillon et al., 2023). Notably, Ganoderma boninense is the causative agent of the devastating BSR disease affecting oil palms in Malaysia and Southeast Asia. This pathogen specifically targets palms aged of 4–5 years in replanted areas or regions with recurrent coconut palm cultivation (Supriyanto et al., 2024). The economic impact of BSR is estimated to range from 70 to 470 million dollars. Oil palm plantations situated on peatlands are particularly susceptible to BSR, with current control measures being limited to environmentally benign biological interventions, such as the utilization of Trichoderma (Sajjan et al., 2024). The potential of Ganoderma-antagonistic fungi to serve as a biological control agent for BSR in oil palms underscores the importance of exploring sustainable management strategies for this disease.
Mushroom diseases, growth inhibitors, environmental microorganisms, and pathogens pose significant challenges to the cultivation of Ganoderma (Ke et al., 2019; An et al., 2022). This study elucidates the competitive dynamics between Trichoderma spp. and Ganoderma during growth. In the cultivation of Ganoderma, the degradation of its culture substrate releases abundant exogenous carbon and nitrogen resources into the casing soil, creating a distinctive ecosystem (Carrasco et al., 2019; Cai and Druzhinina, 2021). In such environments, numerous Trichoderma species have been identified near Ganoderma habitats (Wang et al., 2016; Oh et al., 2018; Allaga et al., 2021). Characterized by their broad-spectrum antagonistic activity against microorganisms (López-Bucio et al., 2015), Trichoderma spp. demonstrate a competitive growth tendency against Ganoderma species. Observations indicate that Trichoderma spp. exert various effects on G. lucidum, A. bisporus (Innocenti et al., 2019), and L. edodes (Wang et al., 2016).
Antagonistic activities, including the production of antifungal metabolites and/or enzymes, mycoparasitism, and ecological competition (Vinale et al., 2008; Anees et al., 2010; Mukherjee et al., 2012), may significantly contribute to the proliferation of these organisms and potentially compromise the growth of Ganoderma species. Conversely, the broad perspective of microbial competition and antagonistic action in Trichoderma makes it one of the front-line microorganism employed to control the different plant pathogens (Zin and Badaluddin, 2020).
Direct systems involve space or nutrient competition, enzyme production, pathogen enzyme inactivation, and parasitism (Saragih et al., 2022). Studies have predominantly employed a solitary control agent against a singular pathogen in the management of plant pathogens. Nevertheless, given the diverse environmental demands of various microorganisms, this strategy may not be universally effective across all the soil types. Moreover, naturally occurring biological control agents are more likely to function as mixed antagonistic communities than individual antagonist. These antagonistic blends exhibit greater stability and a broader spectrum of activity, thereby augmenting the dependability and effectiveness of biological control (Ongena and Jacques, 2008). Numerous antagonistic species, such as Trichoderma (Susanto et al., 2005), have effectively controlled Ganoderma colonization.
Symbiotic associations with host plants facilitate the mitigation of diverse stress forms, enhancement of plant metabolism, and increased biomass by various Trichoderma species (Susanto et al., 2005). A study by Go et al. (2019) elucidated that mycelial interaction is a primary method for evaluating the antagonistic efficacy of potent bioagents against fungal pathogens.
The antagonistic activity varied depending on the used medium (Saragih et al., 2022). Similar findings were observed by Mustafa et al. (2019), who indicated that PDA is the best medium for the growth of Trichoderma sp. and the performance of antagonistic activity. Naher et al. (2015) documented the antagonistic mechanism of Trichoderma sp. against the pathogen of G. boninense, wherein Trichoderma sp. entwined around the hyphae of G. boninense to counteract the pathogen.
Nascimento Brito et al. (2023) isolated three Trichoderma species from cocoa and rubber plants to counteract diseases caused by G. boninense. Culture experiments revealed that T. harzianum markedly inhibited the growth of G. boninense, with inhibition rates ranging from 47.86% (9 days) to 72.06% (14 days) (Rubio et al., 2017). The formation of an inhibition zone by Ganoderma species was visibly observed in this study.
The antagonistic impact of Phlebiopsis gigantea against Heterobasidion annosum, a root/butt rot pathogen affecting conifers, has been documented (Bruna et al., 2020). Similarly, Pycnoporus sanguineus, Grammothele fuligo, and Trametes lactinea, which naturally occur on oil palm trunks, have also been identified as possessing antagonistic activity against G. boninense (Naidu et al., 2015).
Endophytic pathogenic basidiomycetes have been investigated for their efficacy in combating fungal diseases in cacao (Rodríguez Velázquez et al., 2024). Schizophyllum commune and T. lactinea have demonstrated potential in controlling endophytic basidiomycetes (G. boninense) within oil palm plantations. Asymptomatic endophytic basidiomycetes can interfere with the activities of pathogens occupying the same ecological niche, as evidenced by the interaction between P. gigantea and H. annosum in conifers (Rodríguez Velázquez et al., 2024).
In conclusion, this study offers comprehensive insights into the identification of new Ganoderma species. The study highlights the potential challenges posed by laccases of Trichoderma species to Ganoderma species and also demonstrates the antagonistic dynamics between these fungi. To the best of our knowledge, this research presents novel data on the newly identified Ganoderma species and its pathogenic impact on host trees.
Continuous co-evolution within fungal pathogens results in the emergence of new races annually. Trichoderma species engage in complex interactions with other fungal species to address these evolving races. However, a profound understanding of the ecology of Ganoderma species is crucial for effectively managing the fungal diseases. Biotechnology holds promising potential for enhancing Trichoderma efficacy in controlling fungal pathogens and elucidating pathogenic mechanisms. The extensive use of fungicides has led to the proliferation of Trichoderma species to combat emerging pathogen races. Yet, the efficacy of individual applications is challenging to analyze. The application of any microbial agent necessitates careful consideration of technical performance and the safety of human health.
Moreover, Ganoderma species can be integrated with conventional chemicals for more rapid and effective disease management. The successful application of both species relies heavily on the contributions from research and developmental efforts across various industries, as well as support from governments, and non-governmental organizations.
Ultimately, while Trichoderma species have not completely eradicated the newly identified Ganoderma species, they have significantly reduced disease incidence with minimal crop damage. Trichoderma plays a pivotal role in managing plant-associated pathogens, predominantly soil-borne fungal species, but it remains ineffective against the pathogenicity of Ganoderma species. Despite of extensive research on Trichoderma, further investigation into its utilization and its interaction with the newly identified Ganoderma species is warranted.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
AU: Writing – original draft, Writing – review & editing. MSE: Writing – original draft. RMA: Writing – original draft. JMH: Writing – original draft. LD: Writing – review & editing. CW: Writing – original draft. JL: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was partly supported by the Chongqing Science Bureau (2024NSCQ-MSX3103).
Acknowledgments
The authors extend their appreciation to the Researchers Supporting Project (RSPD2024R418), King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1408521/full#supplementary-material
Abbreviations
ITS, Internal transcribed spacer; U/mL, Enzyme activity unit/mL; E.A, Enzyme activity; CTAB, Cetyl trimethylammonium bromide; MAFFT, Multiple Alignment using Fast Fourier Transform; MEGA, Molecular evolutionary genetics analysis; PIRG, Percentage inhibition of radial growth; SDS-PAGE, Sodium dodecyl sulfate-polyacrylamide gel electrophoresis; BSR, Basal stem rot; PCA, Principal components analysis.
References
1
Abdel-Azeem A. Salem F. (2012). Biodiversity of laccase producing fungi in Egypt. Mycosphere3, 900–920. doi: 10.5943/mycosphere/3/6/4
2
Allaga H. Zhumakayev A. Büchner R. Kocsubé S. Szűcs A. Vágvölgyi C. et al . (2021). Members of the Trichoderma harzianum species complex with mushroom pathogenic potential. Agronomy11:2434. doi: 10.3390/agronomy11122434
3
Al-Mosawi A. A. Abdulrazzaq A. K. Jamel D. S. Al-Rubaie W. L. Khedhair H. S. A. B. H. (2024). Evaluation of the inhibitory effect of Ganoderma extracts against the pathogenic fusarium wilt on potato plant. Dijlah J. Agric. Sci.2, 343–351.
4
Al-Tohamy R. Ali S. S. Xie R. Schagerl M. Khalil M. A. Sun J. (2023). Decolorization of reactive azo dye using novel halotolerant yeast consortium HYC and proposed degradation pathway. Ecotoxicol. Environ. Saf.263:115258. doi: 10.1016/j.ecoenv.2023.115258
5
An X.-Y. Cheng G.-H. Gao H.-X. Li X.-F. Yang Y. Li D. et al . (2022). Phylogenetic analysis of Trichoderma species associated with green mold disease on mushrooms and two new pathogens on Ganoderma sichuanense. J. Fungi8:704. doi: 10.3390/jof8070704
6
Anees M. Tronsmo A. Edel-Hermann V. Hjeljord L. G. Héraud C. Steinberg C. (2010). Characterization of field isolates of Trichoderma antagonistic against Rhizoctonia solani. Fungal Biol.114, 691–701. doi: 10.1016/j.funbio.2010.05.007
7
Baird R. Wood-Jones A. Varco J. Watson C. Starrett W. Taylor G. et al . (2014). Rhododendron decline in the great smoky mountains and surrounding areas: intensive site study of biotic and abiotic parameters associated with the decline. Sena13, 1–25. doi: 10.1656/058.013.0101
8
Baturo-Cieśniewska A. Pusz W. Patejuk K. (2020). Problems, limitations, and challenges in species identification of Ascomycota members on the basis of ITS regions. Acta Mycol55, 85–225. doi: 10.5586/am.5512
9
Bischof R. H. Ramoni J. Seiboth B. (2016). Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Factories15:106. doi: 10.1186/s12934-016-0507-6
10
Bolaños A. Bononi V. Gugliotta A. (2016). New records of Ganoderma multiplicatum (Mont.) pat. (Polyporales, Basidiomycota) from Colombia and its geographic distribution in South America. Check List12, 1–7. doi: 10.15560/12.4.1948
11
Bruna L. Klavina D. Zaluma A. Kenigsvalde K. Burneviča N. Nikolajeva V. et al . (2020). Efficacy of Phlebiopsis gigantea against Heterobasidion conidiospore and basidiospore infection in spruce wood. iForest Biogeosci Forest13, 369–375. doi: 10.3832/ifor3279-013
12
Cabarroi-Hernández M. Villalobos-Arámbula A. R. Torres-Torres M. G. Decock C. Guzmán-Dávalos L. (2019). The Ganoderma weberianum-resinaceum lineage: multilocus phylogenetic analysis and morphology confirm G. Mexicanum and G. parvulum in the Neotropics. MycoKeys59, 95–131. doi: 10.3897/mycokeys.59.33182
13
Cai F. Druzhinina I. S. (2021). In honor of John Bissett: authoritative guidelines on molecular identification of Trichoderma. Fungal Divers.107, 1–69. doi: 10.1007/s13225-020-00464-4
14
Cao Y. Wu S.-H. Dai Y.-C. (2012). Species clarification of the prize medicinal Ganoderma mushroom “Lingzhi.”. Fungal Divers.56, 49–62. doi: 10.1007/s13225-012-0178-5
15
Carrasco J. Tello M. L. de Toro M. Tkacz A. Poole P. Pérez-Clavijo M. et al . (2019). Casing microbiome dynamics during button mushroom cultivation: implications for dry and wet bubble diseases. Microbiology165, 611–624. doi: 10.1099/mic.0.000792
16
Cázares-García S. V. Vázquez-Garcidueñas S. Vázquez-Marrufo G. (2013). Structural and phylogenetic analysis of laccases from Trichoderma: a bioinformatic approach. PLoS One8:e55295. doi: 10.1371/journal.pone.0055295
17
Charpentier-Alfaro C. Benavides-Hernández J. Poggerini M. Crisci A. Mele G. Della Rocca G. et al . (2023). Wood-decaying Fungi: from timber degradation to sustainable insulating biomaterials production. Materials16:3547. doi: 10.3390/ma16093547
18
Chaverri P. Castlebury L. A. Samuels G. J. Geiser D. M. (2003). Multilocus phylogenetic structure within the Trichoderma harzianum/Hypocrea lixii complex. Mol. Phylogenet. Evol.27, 302–313. doi: 10.1016/s1055-7903(02)00400-1
19
Chefetz B. Chen Y. Hadar Y. (1998). Purification and characterization of laccase from Chaetomium thermophilium and its role in humification. Appl. Environ. Microbiol.64, 3175–3179. doi: 10.1128/AEM.64.9.3175-3179.1998
20
Chen J. Dou K. Gao Y. Li Y. (2014). Mechanism and application of Trichoderma spp. in biological control of corn diseases. Mycosystema33, 1154–1167. doi: 10.13346/j.mycosystema.140501
21
Coetzee M. P. A. Marincowitz S. Muthelo V. G. Wingfield M. J. (2015). Ganoderma species, including new taxa associated with root rot of the iconic Jacaranda mimosifolia in Pretoria, South Africa. IMA Fungus6, 249–256. doi: 10.5598/imafungus.2015.06.01.16
22
Corner E. J. H. (1983). The clavarioid Ramaria subgen. Echinoramaria. Pers. Mol. Phylog. Evol. Fungi12, 21–28.
23
Costa-Rezende D. H. Robledo G. L. Góes-Neto A. Reck M. A. Crespo E. Drechsler-Santos E. R. (2017). Morphological reassessment and molecular phylogenetic analyses of Amauroderma s.lat. Raised new perspectives in the generic classification of the Ganodermataceae family. Persoonia39, 254–269. doi: 10.3767/persoonia.2017.39.10
24
Crous P. W. Wingfield M. J. Burgess T. I. Hardy G. E. S. J. Barber P. A. Alvarado P. et al . (2017). Fungal planet description sheets: 558-624. Persoonia38, 240–384. doi: 10.3767/003158517X698941
25
Crous P. W. Wingfield M. J. Le Roux J. J. Richardson D. M. Strasberg D. Shivas R. G. et al . (2015). Fungal planet description sheets: 371-399. Persoonia35, 264–327. doi: 10.3767/003158515X690269
26
Dai Y.-C. Cui B.-K. Yuan H.-S. He S.-H. Wei Y.-L. Qin W.-M. et al . (2011). Wood-inhabiting Fungi in southern China. 4. Polypores from Hainan Province. Ann. Bot. Fenn.48, 219–231. doi: 10.5735/085.048.0302
27
Degenkolb T. Dieckmann R. Nielsen K. F. Gräfenhan T. Theis C. Zafari D. et al . (2008). The Trichoderma brevicompactum clade: a separate lineage with new species, new peptaibiotics, and mycotoxins. Mycol. Prog.7, 177–219. doi: 10.1007/s11557-008-0563-3
28
Dhillon B. Graham F. Laughinghouse H. D. Chakrabarti S. (2023). Assessment of endophytic fungal community in palm leaves and their biocontrol potential. PhytoFrontiers4, 296–304. doi: 10.1094/PHYTOFR-04-23-0053-R
29
Dodd S. L. Lieckfeldt E. Samuels G. J. (2003). Hypocrea atroviridis sp. nov., the teleomorph of Trichoderma atroviride. Mycologia95, 27–40. doi: 10.1080/15572536.2004.11833129
30
Dong C.-D. Tiwari A. Anisha G. S. Chen C.-W. Singh A. Haldar D. et al . (2023). Laccase: a potential biocatalyst for pollutant degradation. Environ. Pollut.319:120999. doi: 10.1016/j.envpol.2023.120999
31
Douanla-Meli C. Langer E. Talontsi Mouafo F. (2013). Fungal endophyte diversity and community patterns in healthy and yellowing leaves of Citrus limon. Fungal Ecol.6, 212–222. doi: 10.1016/j.funeco.2013.01.004
32
Du Z. Li Y. Wang X.-C. Wang K. Yao Y.-J. (2023). Re-examination of the holotype of Ganoderma sichuanense (Ganodermataceae, Polyporales) and a clarification of the identity of Chinese cultivated lingzhi. J. Fungi9:323. doi: 10.3390/jof9030323
33
du Plessis I. L. Druzhinina I. S. Atanasova L. Yarden O. Jacobs K. (2018). The diversity of Trichoderma species from soil in South Africa, with five new additions. Mycologia110, 559–583. doi: 10.1080/00275514.2018.1463059
34
Durán N. Rosa M. A. D’Annibale A. Gianfreda L. (2002). Applications of laccases and tyrosinases (phenoloxidases) immobilized on different supports: a review. Enzym. Microb. Technol.31, 907–931. doi: 10.1016/S0141-0229(02)00214-4
35
Gangwar O. Prakash Singh A. (2018). Trichoderma as an efficacious bioagent for combating biotic and abiotic stresses of wheat-a review. Agric. Rev.38, 48–54. doi: 10.18805/ag.R-1770
36
Gao H. Wang Y. Zhang W. Wang W. Mu Z. (2011). Isolation, identification and application in lignin degradation of an ascomycete GHJ-4. Afr. J. Biotechnol.10, 4166–4174. doi: 10.5897/AJB10.2250
37
Glen M. Yuskianti V. Puspitasari D. Francis A. Agustini L. Rimbawanto A. et al . (2014). Identification of basidiomycete fungi in I ndonesian hardwood plantations by DNA barcoding. Forest path.44, 496–508. doi: 10.1111/efp.12146
38
Go W. Z. H’ng P. S. Wong M. Y. Chin K. L. Ujang S. Noran A. S. (2019). Evaluation of Trichoderma asperellum as a potential biocontrol agent against Rigidoporus microporus Hevea brasiliensis. Arch. Phytopathol. Plant Protect.52, 639–666. doi: 10.1080/03235408.2019.1587821
39
Guglielmo F. Bergemann S. E. Gonthier P. Nicolotti G. Garbelotto M. (2007). A multiplex PCR-based method for the detection and early identification of wood rotting fungi in standing trees. J. Appl. Microbiol.103, 1490–1507. doi: 10.1111/j.1365-2672.2007.03378.x
40
Guglielmo F. Gonthier P. Garbelotto M. Nicolotti G. (2008). A PCR-based method for the identification of important wood rotting fungal taxa within Ganoderma, Inonotus s.l. and Phellinus s.l. FEMS Microbiol. Lett.282, 228–237. doi: 10.1111/j.1574-6968.2008.01132.x
41
Gutierrez-Rangel P. C. Mayolo-Deloisa K. Torres-Acosta M. A. (2024). Decisional tool development and application for techno-economic analysis of fungal laccase production. Bioresour. Technol.402:130781. doi: 10.1016/j.biortech.2024.130781
42
Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl. Acids Symp. Ser.734, 95–98. doi: 10.1021/bk-1999-0734.ch008
43
Hapuarachchi K. Karunarathna S. Raspé O. De Silva K. Thawthong A. Wu X. et al . (2018). High diversity of Ganoderma and Amauroderma (Ganodermataceae, Polyporales) in Hainan Island, China. Mycosphere9, 931–982. doi: 10.5943/mycosphere/9/5/1
44
He J. Han X. Luo Z.-L. Li E.-X. Tang S.-M. Luo H.-M. et al . (2022). Species diversity of Ganoderma (Ganodermataceae, Polyporales) with three new species and a key to Ganoderma in Yunnan Province, China. Front. Microbiol.13:1035434. doi: 10.3389/fmicb.2022.1035434
45
Holmes K. A. Schroers H.-J. Thomas S. E. Evans H. C. Samuels G. J. (2004). Taxonomy and biocontrol potential of a new species of Trichoderma from the Amazon basin of South America. Mycol. Prog.3, 199–210. doi: 10.1007/s11557-006-0090-z
46
Hoyos-Carvajal L. Orduz S. Bissett J. (2009). Genetic and metabolic biodiversity of Trichoderma from Colombia and adjacent neotropic regions. Fungal Genet. Biol.46, 615–631. doi: 10.1016/j.fgb.2009.04.006
47
Innocenti G. Montanari M. Righini H. Roberti R. (2019). Trichoderma species associated with green mould disease of Pleurotus ostreatus and their sensitivity to prochloraz. Plant Pathol.68, 392–398. doi: 10.1111/ppa.12953
48
Jaklitsch W. M. Põldmaa K. Samuels G. J. (2008). Reconsideration of Protocrea (Hypocreales, Hypocreaceae). Mycologia100, 962–984. doi: 10.3852/08-101
49
Jankowiak R. Bilański P. Paluch J. Kołodziej Z. (2016). Fungi associated with dieback of Abies alba seedlings in naturally regenerating forest ecosystems. Fungal Ecol.24, 61–69. doi: 10.1016/j.funeco.2016.08.013
50
Jargalmaa S. Eimes J. A. Park M. S. Park J. Y. Oh S.-Y. Lim Y. W. (2017). Taxonomic evaluation of selected Ganoderma species and database sequence validation. PeerJ5:e3596. doi: 10.7717/peerj.3596
51
Kalra K. Chauhan R. Shavez M. Sachdeva S. (2013). Isolation of laccase producing Trichoderma spp. and effect of PH and temperature on its activity. Int. J. Chem. Tech. Res.5, 2229–2235.
52
Katoh K. Rozewicki J. Yamada K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform.20, 1160–1166. doi: 10.1093/bib/bbx108
53
Ke L.-Q. Li P.-D. Xu J.-P. Wang Q.-S. Wang L.-L. Wen H.-P. (2019). Microbial communities and soil chemical features associated with commercial production of the medicinal mushroom Ganoderma lingzhi in soil. Sci. Rep.9:15839. doi: 10.1038/s41598-019-52368-2
54
Kindermann J. El-Ayouti Y. Samuels G. J. Kubicek C. P. (1998). Phylogeny of the genus Trichoderma based on sequence analysis of the internal transcribed spacer region 1 of the rDNA cluster. Fungal Genet. Biol.24, 298–309. doi: 10.1006/fgbi.1998.1049
55
Köhl J. Kolnaar R. Ravensberg W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci.10:845. doi: 10.3389/fpls.2019.00845
56
Kuhls K. Lieckfeldt E. Samuels G. J. Kovacs W. Meyer W. Petrini O. et al . (1996). Molecular evidence that the asexual industrial fungus Trichoderma reesei is a clonal derivative of the ascomycete Hypocrea jecorina. Proc. Natl. Acad. Sci. USA93, 7755–7760. doi: 10.1073/pnas.93.15.7755
57
Kumar R. Samanta P. Vijay Raj S. Bera P. Naimuddin M. (2023). Potential and prospects of Trichoderma in plant protection. Adv. Agric.2023, 1–11. doi: 10.1155/2023/5573662
58
Kumari A. Tapwal A. Thakur N. (2024). Ganoderma lucidum: insights on host range, diagnosis, and management strategies. J. Basic Microbiol.64:e2300769. doi: 10.1002/jobm.202300769
59
Li T.-H. Hu H.-P. Deng W.-Q. Wu S.-H. Wang D.-M. Tsering T. (2015). Ganoderma leucocontextum, a new member of the G. lucidum complex from southwestern China. Mycoscience56, 81–85. doi: 10.1016/j.myc.2014.03.005
60
Li M. Yuan H. (2015). Type studies on Amauroderma species described by J.D. Zhao et al. and the phylogeny of species in China. Mycotaxon130, 79–89. doi: 10.5248/130.79
61
Loi M. Glazunova O. Fedorova T. Logrieco A. F. Mulè G. (2021). Fungal laccases: the forefront of enzymes for sustainability. J. Fungi7:1048. doi: 10.3390/jof7121048
62
López-Bucio J. Pelagio-Flores R. Herrera-Estrella A. (2015). Trichoderma as biostimulant: exploiting the multilevel properties of a plant beneficial fungus. Sci. Hortic.196, 109–123. doi: 10.1016/j.scienta.2015.08.043
63
Loyd A. L. Barnes C. W. Held B. W. Schink M. J. Smith M. E. Smith J. A. et al . (2018a). Elucidating “lucidum”: distinguishing the diverse laccate Ganoderma species of the United States. PLoS One13:e0199738. doi: 10.1371/journal.pone.0199738
64
Loyd A. L. Held B. W. Linder E. R. Smith J. A. Blanchette R. A. (2018b). Elucidating wood decomposition by four species of Ganoderma from the United States. Fungal Biol.122, 254–263. doi: 10.1016/j.funbio.2018.01.006
65
Loyd A. L. Richter B. S. Jusino M. A. Truong C. Smith M. E. Blanchette R. A. et al . (2018c). Identifying the “mushroom of immortality”: assessing the Ganoderma species composition in commercial Reishi products. Front. Microbiol.9:1557. doi: 10.3389/fmicb.2018.01557
66
Lu B. Zuo B. Liu X. Feng J. Wang Z. Gao J. (2016). Trichoderma harzianum causing green mold disease on cultivated Ganoderma lucidum in Jilin Province, China. Plant Dis.100:2524. doi: 10.1094/PDIS-04-16-0422-PDN
67
Mohanty P. S. Harsh N. S. K. Pandey A. (2011). First report of Ganoderma resinaceum and G. weberianum from North India based on ITS sequence analysis and micromorphology. Mycosphere2, 469–474.
68
Mukherjee P. K. Horwitz B. A. Kenerley C. M. (2012). Secondary metabolism in Trichoderma—a genomic perspective. Microbiology158, 35–45. doi: 10.1099/mic.0.053629-0
69
Munsell (1975). Munsell Soil Color Charts. Baltimore: Munsell Color Co., Inc.
70
Murrill W. A. (1902). The Polyporaceae of North America. I. The genus Ganoderma. Bull. Torrey. Bot. Club29, 599–608. doi: 10.2307/2478682
71
Mustafa A. Naveed M. Abbas T. Saeed Q. Hussain A. Ashraf M. N. et al . (2019). Growth response of wheat and associated weeds to plant antagonistic rhizobacteria and fungi. Ital. J. Agron.14, 191–198. doi: 10.4081/ija.2019.1449
72
Nagy L. G. Vágvölgyi C. Papp T. (2010). Type studies and nomenclatural revisions in Parasola (Psathyrellaceae) and related taxa. Mycotaxon112, 103–141. doi: 10.5248/112.103
73
Naher L. Siddiquee S. Yusuf U. Mondal M. M. A. (2015). Issues of Ganoderma spp. and basal stem rot disease management in oil palm. Am. J. Agric. Sci.2, 103–107.
74
Naidu Y. Idris A. S. Nusaibah S. A. Norman K. Siddiqui Y. (2015). In vitro screening of biocontrol and biodegradation potential of selected hymenomycetes against Ganoderma boninense and infected oil palm waste. For. Pathol.45, 474–483. doi: 10.1111/efp.12197
75
Nascimento Brito V. Lana Alves J. Sírio Araújo K. de Souza Leite T. Borges de Queiroz C. Liparini Pereira O. et al . (2023). Endophytic Trichoderma species from rubber trees native to the Brazilian Amazon, including four new species. Front. Microbiol.14:1095199. doi: 10.3389/fmicb.2023.1095199
76
Nazar M. Xu Q. Zahoor Ullah M. W. Khan N. A. Iqbal B. et al . (2023). Integrated laccase delignification with improved lignocellulose recalcitrance for enhancing enzymatic saccharification of ensiled rice straw. Ind. Crop. Prod.202:116987. doi: 10.1016/j.indcrop.2023.116987
77
Nguyen T. T. T. Nguyen H. D. Bui A. T. Pham K. H. T. Van K. T. P. Tran L. T. et al . (2023). Phylogenetic analysis and morphology of Ganoderma multipileum, a Ganoderma species associated with dieback of the metropolitan woody plant Delonix regia (Boj. Ex hook.) Raf. in Vietnam. Sci. Prog.106:00368504231195503. doi: 10.1177/00368504231195503
78
Oh S.-Y. Park M. S. Cho H. J. Lim Y. W. (2018). Diversity and effect of Trichoderma isolated from the roots of Pinus densiflora within the fairy ring of pine mushroom (Tricholoma matsutake). PLoS One13:e0205900. doi: 10.1371/journal.pone.0205900
79
Ongena M. Jacques P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol.16, 115–125. doi: 10.1016/j.tim.2007.12.009
80
Ons L. Bylemans D. Thevissen K. Cammue B. P. A. (2020). Combining biocontrol agents with chemical fungicides for integrated plant fungal disease control. Microorganisms8:1930. doi: 10.3390/microorganisms8121930
81
Park Y.-J. Kwon O.-C. Son E.-S. Yoon D.-E. Han W. Nam J.-Y. et al . (2012). Genetic diversity analysis of Ganoderma species and development of a specific marker for identification of medicinal mushroom Ganoderma lucidum. AJMR6, 5417–5425. doi: 10.5897/AJMR12.846
82
Park M. S. Seo G. S. Bae K. S. Yu S. H. (2005). Characterization of Trichoderma spp. associated with green mold of oyster mushroom by PCR-RFLP and sequence analysis of ITS regions of rDNA. Plant Pathol. J.21, 229–236. doi: 10.5423/PPJ.2005.21.3.229
83
Piel C. Pouchieu C. Carles C. Béziat B. Boulanger M. Bureau M. et al . (2019). Agricultural exposures to carbamate herbicides and fungicides and central nervous system tumour incidence in the cohort AGRICAN. Environ. Int.130:104876. doi: 10.1016/j.envint.2019.05.070
84
Pinar O. Rodríguez-Couto S. (2024). Biologically active secondary metabolites from white-rot fungi. Front. Chem.12:1363354. doi: 10.3389/fchem.2024.1363354
85
Prashantha A. Suryanarayana V. Patil P. M. S. Krishnaraj P. U. Hegde R. V. (2024). Exploration of native Trichoderma spp. from different ecosystems of the Canara circle, Karnataka, India. Int. J. Environ. Clim. Change14, 239–249. doi: 10.9734/ijecc/2024/v14i34036
86
Rahman M. U. Ullah M. W. Shah J. A. Sethupathy S. Bilal H. Abdikakharovich S. A. et al . (2024). Harnessing the power of bacterial laccases for xenobiotic degradation in water: a 10-year overview. Sci. Total Environ.918:170498. doi: 10.1016/j.scitotenv.2024.170498
87
Raymaekers K. Ponet L. Holtappels D. Berckmans B. Cammue B. P. A. (2020). Screening for novel biocontrol agents applicable in plant disease management—a review. Biol. Control144:104240. doi: 10.1016/j.biocontrol.2020.104240
88
Rodríguez Velázquez N. D. Gómez De La Cruz I. Chávez Ramírez B. Estrada De Los Santos P. (2024). “Biological control of diseases in Theobroma cacao” in Biocontrol Agents for Improved Agriculture. eds. KumarA.SantoyoG.SinghJ. (Academic Press), 101–120. doi: 10.1016/B978-0-443-15199-6.00009-9
89
Rubio M. B. Hermosa R. Vicente R. Gómez-Acosta F. A. Morcuende R. Monte E. et al . (2017). The combination of Trichoderma harzianum and chemical fertilization leads to the deregulation of Phytohormone networking, preventing the adaptive responses of tomato plants to salt stress. Front. Plant Sci.8:294. doi: 10.3389/fpls.2017.00294
90
Sajjan U. Hubballi M. Pandey A. K. Devappa V. Maheswarappa H. P. (2024). Characterizations of Ganoderma species causing basal stem rot disease in coconut tree. 3 Biotech14:104. doi: 10.1007/s13205-023-03872-w
91
Samuels G. J. (2004). Changes in taxonomy, occurrence of the sexual stage and ecology of Trichoderma spp. Phytopathology94:S138.
92
Samuels G. J. Suarez C. Solis K. Holmes K. A. Thomas S. E. Ismaiel A. et al . (2006). Trichoderma theobromicola and T. Paucisporum: two new species isolated from cacao in South America. Mycol. Res.110, 381–392. doi: 10.1016/j.mycres.2006.01.009
93
Saragih R. Simanjorang M. Angrelina I. Irvanto D. Retnosari E. Murgianto F. et al . (2022). Exploration, identification, and in vitro antagonism test of Trichoderma spp. against Ganoderma spp. at PT Bumitama Gunajaya agro palm oil plantation, Central Kalimantan. IOP Conf. Ser. Earth Environ. Sci.976:012043. doi: 10.1088/1755-1315/976/1/012043
94
Shankar A. Saini S. Sharma K. K. (2024). Fungal-integrated second-generation lignocellulosic biorefinery: utilization of agricultural biomass for co-production of lignocellulolytic enzymes, mushroom, fungal polysaccharides, and bioethanol. Biomass Convers. Biorefinery14, 1117–1131. doi: 10.1007/s13399-022-02969-1
95
Shariffah-Muzaimah S. A. Idris A. S. Madihah A. Z. Dzolkhifli O. Kamaruzzaman S. Cheong P. C. H. (2015). Isolation of actinomycetes from rhizosphere of oil palm (Elaeis guineensis Jacq.) for antagonism against Ganoderma boninense. J. Oil Palm Res.27, 19–29.
96
Shittu O. G. Yemisi B. G. Damilola O. E. Dasola M. K. Augustine A. O. (2017). Isolation and identification of fungi associated with the rhizospheres of some economic trees. Ann. Food Sci. Technol.18, 696–699.
97
Singh S. K. Singh P. N. Maurya D. K. Rana S. (2020). “Advances in systematics, taxonomy, and conservation of Trichoderma species” in Trichoderma: Agricultural Applications and Beyond. eds. ManoharacharyC.SinghH. B.VarmaA. (Cham: Springer International Publishing), 1–20.
98
Smith N. R. Dawson V. T. (1944). The bacteriostatic action of rose bengal in media used for plate counts of soil fungi. Soil Sci.58, 467–472. doi: 10.1097/00010694-194412000-00006
99
Song J. Xing J.-H. Decock C. He X.-L. Cui B.-K. (2016). Molecular phylogeny and morphology reveal a new species of Amauroderma (Basidiomycota) from China. Phytotaxa260, 47–56. doi: 10.11646/phytotaxa.260.1.5
100
Steyaert R. L. (1980). Study of some Ganoderma species. Bull. Nat. Plantent. België50, 135–186. doi: 10.2307/3667780
101
Sun R. Liu Z. Fu K. Fan L. Chen J. (2012). Trichoderma biodiversity in China. J. Appl. Genet.53, 343–354. doi: 10.1007/s13353-012-0093-1
102
Sun Y.-F. Xing J.-H. He X.-L. Wu D.-M. Song C.-G. Liu S. et al . (2022). Species diversity, systematic revision and molecular phylogeny of Ganodermataceae (Polyporales, Basidiomycota) with an emphasis on Chinese collections. Stud. Mycol.101, 287–415. doi: 10.3114/sim.2022.101.05
103
Supriyanto S. Sulistyowati H. Arifin N. (2024). Status of basal stem rot disease on areca nut palm plantations in Kubu Raya District, West Kalimantan, Indonesia. Asian J. Agric.8, 41–49. doi: 10.13057/asianjagric/g080106
104
Susanto A. Sudharto P. S. Purba R. Y. (2005). Enhancing biological control of basal stem rot disease (Ganoderma boninense) in oil palm plantations. Mycopathologia159, 153–157. doi: 10.1007/s11046-004-4438-0
105
Szekeres A. Láday M. Kredics L. Varga J. Antal Z. Hatvani L. et al . (2006). Rapid identification of clinical Trichoderma longibrachiatum isolates by cellulose-acetate electrophoresis-mediated isoenzyme analysis. Clin. Microbiol. Infect.12, 369–375. doi: 10.1111/j.1469-0691.2005.01356.x
106
Tamano K. Takayama H. Yasokawa S. Sano M. Baker S. E. (2022). Major involvement of two laccase genes in conidial pigment biosynthesis in Aspergillus oryzae. Appl. Microbiol. Biotechnol.106, 287–300. doi: 10.1007/s00253-021-11669-1
107
Tchotet Tchoumi J. M. Coetzee M. P. A. Rajchenberg M. Wingfield M. J. Roux J. (2018). Three Ganoderma species, including Ganoderma dunense sp. nov., associated with dying Acacia cyclops trees in South Africa. Australas. Plant Pathol.47, 431–447. doi: 10.1007/s13313-018-0575-7
108
Terho M. Hantula J. Hallaksela A.-M. (2007). Occurrence and decay patterns of common wood-decay fungi in hazardous trees felled in the Helsinki City. For. Pathol.37, 420–432. doi: 10.1111/j.1439-0329.2007.00518.x
109
Tong X. Jiang H. Liang Y. Rao Y. Mei L. Wang Y. (2020). Waterlogging reduces soil colonization by antagonistic fungi and restores production in Ganoderma lucidum continuous cultivation. Crop Prot.137:105314. doi: 10.1016/j.cropro.2020.105314
110
Torres-Torres M. G. Guzmán-Dávalos L. (2012). The morphology of Ganoderma species with a laccate surface. Mycotaxon119, 201–216. doi: 10.5248/119.201
111
Torres-Torres M. G. Ryvarde L. Guzmán-Dávalos L. (2015). Ganoderma subgenus Ganoderma in Mexico. Rev. Mexic. Micolog.41, 27–45.
112
Umar A. (2021). Screening and evaluation of laccase produced by different Trichoderma species along with their phylogenetic relationship. Arch. Microbiol.203, 4319–4327. doi: 10.1007/s00203-021-02420-5
113
Umar A. Abid I. Antar M. Dufossé L. Hajji-Hedfi L. Elshikh M. S. et al . (2023). Electricity generation and oxidoreductase potential during dye discoloration by laccase-producing Ganoderma gibbosum in fungal fuel cell. Microb. Cell Factories22:258. doi: 10.1186/s12934-023-02258-0
114
Verma A. K. Bhojiya A. A. Upadhyay S. K. Yadav V. Singhal K. Abbas K. (2023). “Promising role of fungal symbiosis for eco-friendly green technology for environmental health” in Nano-Biofortification for Human and Environmental Health. eds. RajputV. D.El-RamadyH.UpadhyayS. K.MinkinaT.AhmedB.MandzhievaS. (Cham: Springer International Publishing), 237–266.
115
Verma A. Shameem N. Jatav H. S. Sathyanarayana E. Parray J. A. Poczai P. et al . (2022). Fungal endophytes to combat biotic and abiotic stresses for climate-smart and sustainable agriculture. Front. Plant Sci.13:953836. doi: 10.3389/fpls.2022.953836
116
Vinale F. Sivasithamparam K. Ghisalberti E. L. Marra R. Woo S. L. Lorito M. (2008). Trichoderma–plant–pathogen interactions. Soil Biol. Biochem.40, 1–10. doi: 10.1016/j.soilbio.2007.07.002
117
Wang G. Cao X. Ma X. Guo M. Liu C. Yan L. et al . (2016). Diversity and effect of Trichoderma spp. associated with green mold disease on Lentinula edodes in China. Microbiology5, 709–718. doi: 10.1002/mbo3.364
118
Wang D.-M. Wu S.-H. Su C.-H. Peng J.-T. Shih Y.-H. Chen L.-C. (2009). Ganoderma multipileum, the correct name for “G. lucidum” in tropical Asia. Bot. Stud.50, 451–458.
119
Wang D.-M. Yao Y.-J. (2005). Intrastrain internal transcribed spacer heterogeneity in Ganoderma species. Can. J. Microbiol.51, 113–121. doi: 10.1139/w04-118
120
Wang Y. Zeng L. Wu J. Jiang H. Mei L. (2022). Diversity and effects of competitive Trichoderma species in Ganoderma lucidum-cultivated soils. Front. Microbiol.13:1067822. doi: 10.3389/fmicb.2022.1067822
121
Welti S. Courtecuisse R. (2010). The Ganodermataceae in the French West Indies (Guadeloupe and Martinique). Fungal Divers.43, 103–126. doi: 10.1007/s13225-010-0036-2
122
White T. Bruns T. Lee S. Taylor J. Innis M. Gelfand D. et al . (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics in PCR Protocols: A Guide to Methods and Applications. eds. InnisM. A.GelfandD. H.SninskyJ. J.WhiteT. J., Academic Press. 315–322.
123
Xia X. Lie T. K. Qian X. Zheng Z. Huang Y. Shen Y. (2011). Species diversity, distribution, and genetic structure of endophytic and epiphytic Trichoderma associated with banana roots. Microb. Ecol.61, 619–625. doi: 10.1007/s00248-010-9770-y
124
Xing J.-H. Song J. Decock C. Cui B.-K. (2016). Morphological characters and phylogenetic analysis reveal a new species within the Ganoderma lucidum complex from South Africa. Phytotaxa266, 115–124. doi: 10.11646/phytotaxa.266.2.5
125
Xing J.-H. Sun Y.-F. Han Y.-L. Cui B.-K. Dai Y.-C. (2018). Morphological and molecular identification of two new Ganoderma species on Casuarina equisetifolia from China. MycoKeys34, 93–108. doi: 10.3897/mycokeys.34.22593
126
Yang J. Lin Y. Yang X. Ng T. B. Ye X. Lin J. (2017). Degradation of tetracycline by immobilized laccase and the proposed transformation pathway. J. Hazard. Mater.322, 525–531. doi: 10.1016/j.jhazmat.2016.10.019
127
Yang X. Sun S. Chen Q. Zhang Z. Wang J. Liu Y. et al . (2022). A polysaccharide of Ganoderma lucidum enhances antifungal activity of chemical fungicides against soil-borne diseases of wheat and maize by induced resistance. Agriculture12:55. doi: 10.3390/agriculture12010055
128
Yassin M. T. Mostafa A. A.-F. Al-Askar A. A. Sayed S. R. M. Rady A. M. (2021). Antagonistic activity of Trichoderma harzianum and Trichoderma viride strains against some fusarial pathogens causing stalk rot disease of maize, in vitro. J. King Saud Univ. Sci.33:101363. doi: 10.1016/j.jksus.2021.101363
129
Yazid S. N. E. Tajudin N. I. Razman N. A. A. Selamat J. Ismail S. I. Sanny M. et al . (2023). Mycotoxigenic fungal growth inhibition and multi-mycotoxin reduction of potential biological control agents indigenous to grain maize. Mycotoxin Res.39, 177–192. doi: 10.1007/s12550-023-00484-4
130
Zhou L.-W. Cao Y. Wu S.-H. Vlasák J. Li D.-W. Li M.-J. et al . (2015). Global diversity of the Ganoderma lucidum complex (Ganodermataceae, Polyporales) inferred from morphology and multilocus phylogeny. Phytochemistry114, 7–15. doi: 10.1016/j.phytochem.2014.09.023
131
Zin N. A. Badaluddin N. A. (2020). Biological functions of Trichoderma spp. for agriculture applications. Ann. Agric. Sci.65, 168–178. doi: 10.1016/j.aoas.2020.09.003
Summary
Keywords
growth rate, ITS, laccase, phylogeny, wood degradation
Citation
Umar A, Elshikh MS, Aljowaie RM, Hussein JM, Dufossé L, Wu C and Lu J (2024) Competitive antagonistic action of laccase between Trichoderma species and the newly identified wood pathogenic Ganoderma camelum. Front. Microbiol. 15:1408521. doi: 10.3389/fmicb.2024.1408521
Received
28 March 2024
Accepted
30 August 2024
Published
25 September 2024
Volume
15 - 2024
Edited by
Samantha Chandranath Karunarathna, Qujing Normal University, China
Reviewed by
Kalani Hapuarachchi, Southwest Forestry University, China
Muhammad Qasim, Shihezi University, China
Updates
Copyright
© 2024 Umar, Elshikh, Aljowaie, Hussein, Dufossé, Wu and Lu.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aisha Umar, ash.dr88@gmail.com; Junxing Lu, junxlu@163.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.