- Henan Key Laboratory of Cereal and Oil Food Safety and Nutrition, College of Food Science and Engineering, Henan University of Technology, Zhengzhou, China
The phytopathogenic fungus Fusarium graminearum causes Fusarium head blight, which threatens agricultural yield and human health. We previously demonstrated that a plant-derived natural compound, 2-hydroxy-4-methoxybenzaldehyde (HMB), can inhibit F. graminearum. This study continued to investigate its antifungal mechanism. Compared to the control, HMB treatment at the minimum inhibitory concentration (MIC) significantly reduced ergosterol levels by 61.78%, indicating compromised fungal membrane integrity. Concurrently, intracellular reactive oxygen species (ROS) levels increased 22-fold, accompanied by a 146.03% increase in hydrogen peroxide (H₂O₂) content. Meanwhile, superoxide dismutase (SOD) activity increased, while catalase (CAT) activity declined, suggesting a marked change of redox metabolism upon HMB exposure. In addition, Quantitative Real-time PCR (qRT-PCR) analysis revealed that HMB treatment significantly regulated the expression of genes participating in ergosterol biosynthesis (Erg2, Erg5, Erg6, etc.), DON biosynthesis (up to 16 genes), the redox system (MnSOD, Cu/ZnSOD, GSS, and CAT), global regulators (LaeA, VeA, and VelB), and stress signaling pathways (Hog1, Ssk1, Ssk2, and Pbs2). These findings revealed the in-depth antifungal mechanism of HMB and proposed that HMB holds potential as an antifungal agent.
1 Introduction
Fusarium graminearum is a member of the phylum Ascomycota and is an important phytopathogenic fungus (Lee et al., 2009). It can infect various crops such as wheat, corn, barley, and rice, causing a spectrum of destructive diseases such as ear rot disease, gibberella stalk rot, and Fusarium head blight (FHB) (Ferrigo et al., 2016). Among the diseases, FHB poses significant threats to global food security (Ji et al., 2019). Severe FHB epidemics occur at least once every 4 or 5 years in countries such as China, the United States, and the European Union (Figueroa et al., 2017). From 2000 to 2018 in China, FHB affected over 4.5 million hectares of wheat fields each year, covering about 20% of the total planted area and causing annual yield losses exceeding 3.41 million metric tons (Chen et al., 2019). FHB outbreaks in 2015–2016 caused losses exceeding 1.4 billion dollars in the US (Wilson et al., 2018).
F. graminearum can also produce toxic secondary metabolites, including deoxynivalenol (DON), zearalenone (ZEN), and nivalenol (NIV) (Feizollahi and Roopesh, 2021; Häggblom and Nordkvist, 2015; Podgórska-Kryszczuk et al., 2022). Among these metabolites, DON (also known as vomitoxin) is the most prevalent toxin. When DON accumulates in contaminated crops, it can enter the food supply through unprocessed agricultural products during transportation and storage. Alarmingly, DON persists even after processing due to its heat-resistant nature, making it difficult to remove. As DON primarily affects the gut, immune system, and brain function, exposure to it could lead to intestinal irritation, vomiting, anorexia, weight loss, and immune suppression, posing significant risks to both human and animal health (Payros et al., 2016; Pestka, 2010). Therefore, controlling the growth of F. graminearum and the biosynthesis of DON is crucial for both crop production and food safety.
Currently, chemical fungicides (e.g., carbendazim, tebuconazole, and metconazole) are ubiquitously applied (Diao et al., 2018; Mendes et al., 2018). However, the emergence of resistant strains and the negative impacts on the environment and human health are serious problems thereof (Sun et al., 2018). Subsequently, recent studies have switched from chemical fungicides to plant-derived natural compounds, as they are rich in resources, safe for human and animals, environmentally friendly, and cost-effective, etc. Some natural compounds (e.g., glabridin, ferulic acid, myrcene, perillaldehyde, and myriocin) exhibit remarkable inhibitory effects against F. graminearum in recent published papers (Albayrak et al., 2023; Meng et al., 2024; Shao et al., 2021; Yan et al., 2023; Yang et al., 2021). Notably, cell membranes are common antifungal targets for the above-mentioned compounds.
2-Hydroxy-4-methoxybenzaldehyde (HMB) is an active compound in many herbals, such as Decalepis hamiltonii, Hemidesmus indicus, Mondia whitei, Periploca sepium, and Sclerocarya caffra (Subban and Mohan, 2003). It exhibits excellent antimicrobial, anti-inflammatory, hepatoprotective, and neuroprotective activities (Rathi et al., 2017). In particular, the antimicrobial spectrum of HMB is broad, including Escherichia coli, Staphylococcus epidermidis, Candida albicans, and Helicobacter pylori (Srikanta et al., 2011; Wang et al., 2010). Our research indicates that HMB is a more effective antifungal compound in comparison with its derivatives vanillin and o-vanillin against both Aspergillus flavus and F. graminearum (Li et al., 2024). Besides, compared to other compounds, the antifungal effect of HMB against F. graminearum is stronger than myrcene (MIC: 25 μg/μL) (Albayrak et al., 2023), perillaldehyde (MIC: 240 μg/L) (Meng et al., 2024), and similar to myriocin (MIC: 64 μg/mL) (Shao et al., 2021). Nevertheless, the mechanism of HMB against F. graminearum has not been thoroughly studied yet. In the present work, the ergosterol content was first determined. Then, we quantified the reactive oxygen species (ROS) production and the activities of related enzymes, as well as intermediate products within redox system. Moreover, in combination with transcriptome analysis and Quantitative Real-time PCR (qRT-PCR), relative expressions of important genes were quantified. This study provides in-depth insights in the antifungal and anti-mycotoxigenic mechanism of HMB, especially the relationship between redox system and DON biosynthesis. Meanwhile, the potential antifungal target(s) can also be identified and screened for effective controlling of F. graminearum in agricultural production and food preservation.
2 Materials and methods
2.1 Media and strains
Fusarium graminearum PH-1 was cultured on Potato Dextrose Agar (PDA), which was formulated with 20% (w/v) potato, 0.2% (w/v) dextrose, and 2% (w/v) agar at 28 ± 2°C. For the present study, mycelia were collected from Potato Dextrose Broth (PDB), a medium prepared with 2% (w/v) potato and 0.2% (w/v) dextrose. Five plugs of F. graminearum were cultured in 100 mL of carboxymethylcellulose sodium medium (CMC, 1.5% (w/v) carboxymethylcellulose sodium, 0.1% (w/v) yeast extract, 0.1% (w/v) NH₄NO₃, 0.1% (w/v) KH₂PO₄, and 0.05% (w/v) MgSO₄·7H₂O) for spore collection.
2.2 Ergosterol content determination
A suspension of 104 spores was added into 50 mL of PDB and incubated under shaking conditions (150 rpm) for 36 h. Subsequently, HMB (98%, CAS: 673–22-3) was supplemented into the cultures to achieve final concentrations of 0 (control), 1/4 MIC (50 μg/mL), 1/2 MIC (100 μg/mL), and MIC (200 μg/mL), as determined previously (Li et al., 2024). The treated cultures were further incubated for 36 h. Ergosterol extraction and quantification were performed according to the method described by Khan et al. (2010). Briefly, the harvested mycelia were ground by liquid nitrogen, 20 mg of mycelia were mixed with 5 mL of 25% (w/v) potassium hydroxide alcohol. The mixture was vortexed for 10 min and then heated at 85°C for 2 h. After cooling, N-heptane (3 mL) and sterile water (1 mL) were included. The biphasic system was vortexed for an additional 10 min and subsequently stored at −20°C overnight to facilitate phase separation. Absorbance of the upper phase were assayed at wavelengths of 230 nm and 282 nm with a UV-6100S double beam spectrophotometer (Shanghai Mapada Instruments Co., Ltd., China), and the ergosterol levels were then determined using the formula below:
where the E (%/cm) values of crystalline ergosterol and dehydroergosterol were 290 and 518, respectively.
2.3 ROS content determination
ROS content was evaluated using an assay kit (S0033S) from Beyotime Biotechnology (Shanghai, China). In short, the mycelia were treated with the reactive oxygen fluorescent probe (DCFH-DA) for 30 min, followed by observation with a fluorescence microscope (Leica Microsystems GmbH, Germany). Fluorescence intensity was subsequently assayed using ImageJ (v1.54, National Institutes of Health (NIH), Bethesda, MD, United States).
2.4 Determination of superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPx) activity
After grinding the mycelia in liquid nitrogen, 500 mg of the sample was suspended in phosphate buffered saline (PBS) (5 mL). The suspension was centrifuged at 12,000 rpm for 10 min at 4°C, and the supernatant was used as the crude enzyme extract for testing. SOD activity was assessed using a previously established method (Sahin et al., 2018) with minor modifications. The supernatant (50 μL) was thoroughly blended with PBS (4.5 mL), nitrotetrazolium blue chloride (0.3 mL), 220 mM methionine (0.3 mL), and 33 μM riboflavin (0.3 mL). After illumination at 4000 lux for 20 min at 25°C, the fluorescence intensity of the mixture was quantified at a wavelength of 560 nm. The formula below was utilized to determine SOD activity:
where A1 and A2 are the absorbance values at 560 nm for the control and sample groups, respectively. W (g) is the fresh weight of the mycelia, Vt (mL) represents the total volume of the enzyme extract, and Vs (mL) corresponds to the volume of enzyme solution used in the assay. The method for measuring CAT activity was as earlier established (Valenzuela- Cota et al., 2019). One hundred microliters of the crude enzyme extract were mixed with 1 mL of 0.3% (w/v) hydrogen peroxide (H₂O₂) and 2 mL of PBS. Subsequently, the absorbance was measured at a wavelength of 240 nm every minute. The method for calculating CAT activity was outlined as follows:
where Vs (mL) and Vt (mL) refer to the enzyme solution and crude enzyme extract volumes, respectively. ΔA240 indicates the variation in absorbance at 240 nm per minute, and the mycelial weight and reaction time are represented by W (g) and t (min), respectively. The kit from Nanjing Jiancheng Bioengineering Institute (NJBI) (H545-1-1) was used to assay the GPx activity.
2.5 H2O2 and glutathione (GSH) content determination
The H2O2 content was assessed by a commercial kit (BC3590) from Beijing Solarbio Science & Technology Co., Ltd. The GSH content was evaluated following a previously established method (Gill et al., 2012). One milligram of mycelia was mixed with 3 mL of 5% metaphosphoric acid, then centrifuged at 12,000 rpm for 10 min at 4°C. Subsequently, 1 mL of the supernatant was transferred to a solution containing 1 mL of PBS and 0.5 mL of 5,5’-Dithiobis-(2-nitrobenzoic acid) (DTNB), followed by incubation for 20 min at room temperature. The absorbance was measured at a wavelength of 412 nm. A standard curve was generated using GSH concentrations ranging from 0 to 50 μg/mL.
2.6 Transcriptome analysis
2.6.1 Library construction and sequencing
RNA was extracted from mycelia treated with 0 and 1/2 MIC (100 μg/mL) of HMB for 36 h using an RNA extraction kit (RC411-01, Vazyme Biotech, Inc., China). The RNA samples were then subjected to transcriptome sequencing by the Beijing Genome Institute (BGI). For mRNA enrichment from total RNA, poly-A mRNA was captured using Oligo (dT) magnetic beads. The mRNA was fragmented with a buffer, reverse-transcribed into cDNA using random N6 primers, and extended into double-stranded cDNA. The cDNA was end-repaired, 5′-phosphorylated, and a single ‘A’ overhang was added to the 3′ end. An adapter with a 3′ ‘T’ overhang was ligated. The product was PCR-amplified, denatured to single-stranded DNA, and circularized via a bridge primer, forming a single-stranded DNA library. The library was sequenced on an Illumina NovaSeq 6,000 platform to obtain raw sequencing data.
2.6.2 Bioinformatics analysis
The sequencing data were analyzed and filtered using SOAPnuke software (v1.5.6) (Chen et al., 2018). The cleaned data was conducted by the Dr. Tom multi-omics data mining system.1 The genome of F. graminearum PH-1 (NCBI accession number: GCF_000240135.3_ASM24013v3) was used as the reference, and comparative analysis was conducted with HISAT2 (Johns Hopkins University, USA) (Kim et al., 2015), which employs a hierarchical indexing approach for aligning spliced transcripts. Bowtie2 was adopted to map the clean data to comprehensive gene set, including both non-coding and known and novel coding transcripts (Langmead and Salzberg, 2012). To obtain a more comprehensive understanding of gene functions associated with phenotypic changes, gene expression levels were assessed using RSEM software (v1.3.1) (Li and Dewey, 2011), and hypergeometric tests were employed for GO and KEGG analysis.
2.7 qRT-PCR analysis
Reverse transcription and qRT-PCR were performed using the R433 and Q712 kits from Vazyme Biotech Co. Ltd. (Nanjing, China). Primers (sequences are in Supplementary Table S1) used in this experiment were designed and synthesized by Sangon Biotech Co. Ltd. (Shanghai, China). β-Tubulin was used as the control gene for data normalization, and the relative gene expression levels were calculated using the 2-ΔΔCT method.
2.8 Statistical analysis
The experiments were conducted at least three times in triplicate. The quantified results were presented as the mean ± SD (mean ± standard deviation) and the significance for ergosterol content, ROS fluorescence intensity, enzyme activities, as well as GSH and H₂O₂ levels was calculated by one-way ANOVA (p < 0.05) test. Statistical analysis of the qRT-PCR data was conducted using Student’s t-test with a significance threshold set at p < 0.05, and all analyses were performed via SPSS 27 (IBM, Armonk, NY, USA).
3 Results
3.1 HMB decreased ergosterol content
As the primary sterol molecule in fungal cell membranes, ergosterol not only contributes to the fluidity, permeability, and stability of cell membranes, but also plays important roles in various cellular processes, such as nutrient uptake, signal transduction, and ion transport (Jordá and Puig, 2020). As treatment with HMB disrupted the cell membrane integrity of F. graminearum (Li et al., 2024), we quantified ergosterol content. As illustrated in Figure 1, in comparison with the control group, treatment with different concentrations of HMB (1/4MIC, 1/2MIC, and MIC) reduced the content by 22.16, 36.13, and 61.78%, respectively.
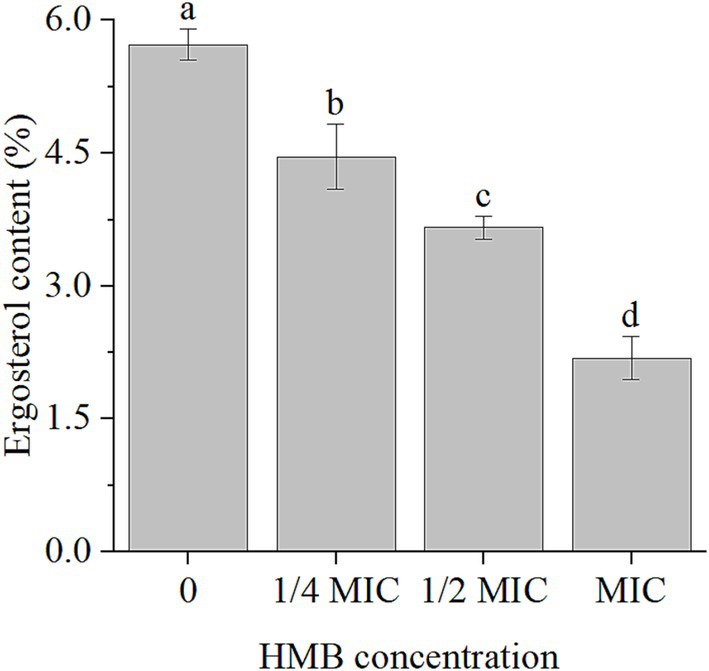
Figure 1. Content of ergosterol in F. graminearum treated with HMB at 0, 1/4, 1/2 and MIC. HMB: 2-Hydroxy-4-methoxybenzaldehyde; MIC: minimum inhibitory concentration. a-d significant difference (p < 0.05) according to Duncan’s multiple range test.
3.2 HMB disturbed redox metabolism of F. graminearum
As a typical product of lipid peroxidation, malondialdehyde (MDA) is commonly used as an indicator of oxidative damage to cell membranes. In our recent work, elevation of MDA content suggested that HMB disrupted lipid peroxidation in cell membranes (Li et al., 2024), leading to the assumption that the redox system was perturbed. To investigate this, we first examined whether HMB treatment leads to intracellular ROS accumulation. As shown in Figure 2, mycelial fluorescence intensity increased significantly with higher HMB concentrations (Figure 2A). Quantitative analysis showed that fluorescence intensities in the 1/2 MIC and MIC groups were approximately 9- and 22-fold higher, respectively, than those in the control (Figure 2B). These results demonstrate that HMB treatment triggers ROS accumulation in F. graminearum mycelia.
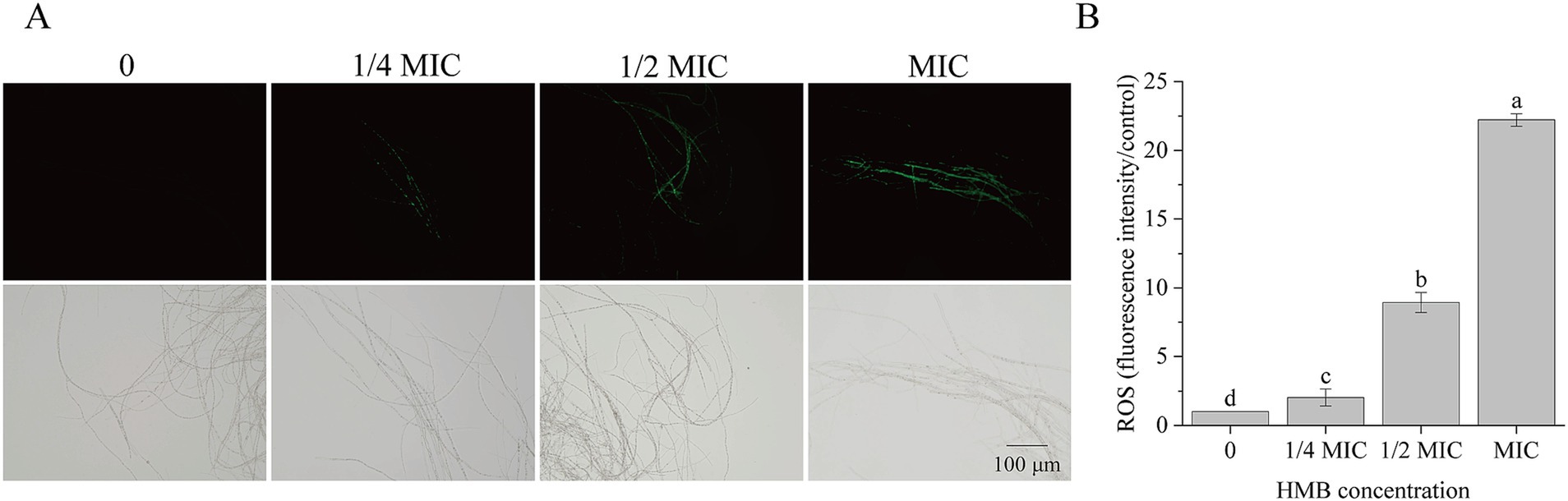
Figure 2. Influence of HMB (0, 1/4 MIC,1/2 MIC, and MIC) on ROS production. (A) Mycelia staining with CM-H2DCFDA (upper row: fluorescence images; lower row: bright field images); (B) Fluorescence intensity of ROS. a-c significant difference (p < 0.05) according to Duncan’s multiple range test.
Organisms have developed well-established mechanisms to counteract oxidative stress by eliminating elevated levels of cellular ROS. These mechanisms are generally classified into enzymatic and non-enzymatic systems (Koubaa and Brini, 2020). The enzymatic system contains CAT, SOD, peroxiredoxin, and GPx, and non-enzymatic system contains GSH, ascorbic acid, polyamines, flavonoids, alkaloids, and carotenoids (Heller and Tudzynski, 2011; Jamieson, 1998). As the first line of defense, SODs convert superoxide radicals (O2−) to H2O2, which is subsequently detoxified into water (H2O) by GPx and CAT (Fridovich, 1995; Heller and Tudzynski, 2011). To investigate the perturbation of the redox system in response to ROS accumulation, we measured the activities of SOD, CAT, and GPx, as well as the levels of GSH and H2O2. As illustrated in Figure 3A, SOD activity was elevated by 29.82, 118.82, and 172.52% in the 1/4 MIC, 1/2 MIC, and MIC groups, respectively. Meanwhile, the CAT activities were reduced by 13.64, 19.18, and 34.65%, respectively (Figure 3B). However, the GPx activity remained unchanged, even at the MIC group (Figure 3C). In combination with the catalyzing of GPx, GSH can be oxidized to oxidative glutathione (GSSG) during oxidative stress defense (Traynor et al., 2019). In this experiment, the GSH content did not show any significant variations (Figure 3D). However, in comparison with the control group, H₂O₂ content exhibited marked increases of 33.11, 96.36, and 146.03% for the 1/4 MIC, 1/2 MIC, and MIC groups, respectively (Figure 3E).
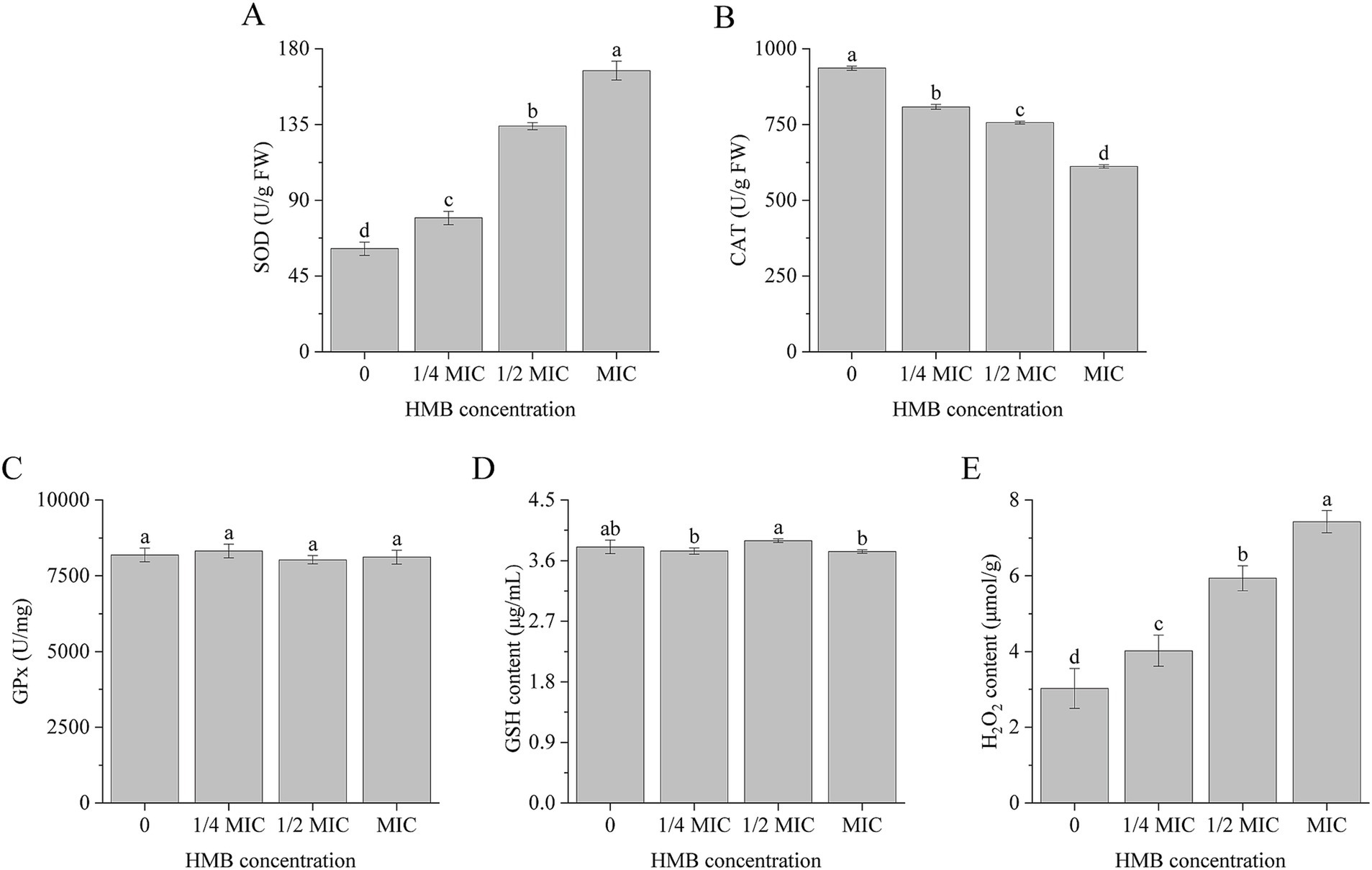
Figure 3. Activities of SOD (A), CAT (B), GPx (C), and content of GSH (D) and H2O2 (E). a-d significant difference (p < 0.05) according to Duncan’s multiple range test.
3.3 RNA-seq analysis
RNA-seq was conducted to examine the impact of HMB treatment on gene expressions in F. graminearum. The results shown in Figure 4A indicated that 1989 genes displayed differential expression, with 1,290 genes up-regulated and 699 genes down-regulated, while 11,327 genes showed no significant changes in expression. The GO enrichment analysis (Figure 4B) results categorized the differentially expressed genes (DEGs) into three groups: biological processes (BP), cellular components (CC), and molecular functions (MF). A total of 40 unique Gene Ontology (GO) categories (p < 0.05) were detected among the DEGs. In the context of Biological Processes (BP), the top three enriched categories were carbohydrate metabolic process, DNA-templated transcription, and rRNA processing. For Cellular Components (CC), the three most significantly represented categories were the integral component of the membrane, the cytoplasm, and the nucleus. For Molecular Functions (MF), the primary three categories were ATP binding, oxidoreductase activity, and transmembrane transporter activity. Among the top 40 annotated (KEGG) enrichment pathways, the first three were carbon metabolism, biosynthesis of amino acids and biosynthesis of cofactors (Figure 4C). As the content of ergosterol was reduced by HMB treatment, it is assumed that HMB treatment affects the expression of transcriptomic levels. Table 1 shows many genes participating in ergosterol biosynthesis, oxidative stress response, and DON biosynthesis were regulated. In detail, Erg2, Erg5, Erg6, Erg7, Erg9, Erg24, and Erg27 in ergosterol biosynthesis were down-regulated. Among them, two genes with an identity of approximately 74%, located on different chromosomes, both encode sterol 24-C-methyltransferase (Erg6). Meanwhile, the expressions of genes encoding CAT, MnSOD, and Cu/ZnSOD enzymes in the redox system were down-regulated, whereas the expressions of GSS and HYR1 encoding enzymes were up-regulated. In addition, 9 genes were downregulated and 11 genes were upregulated in the DON biosynthesis pathway.
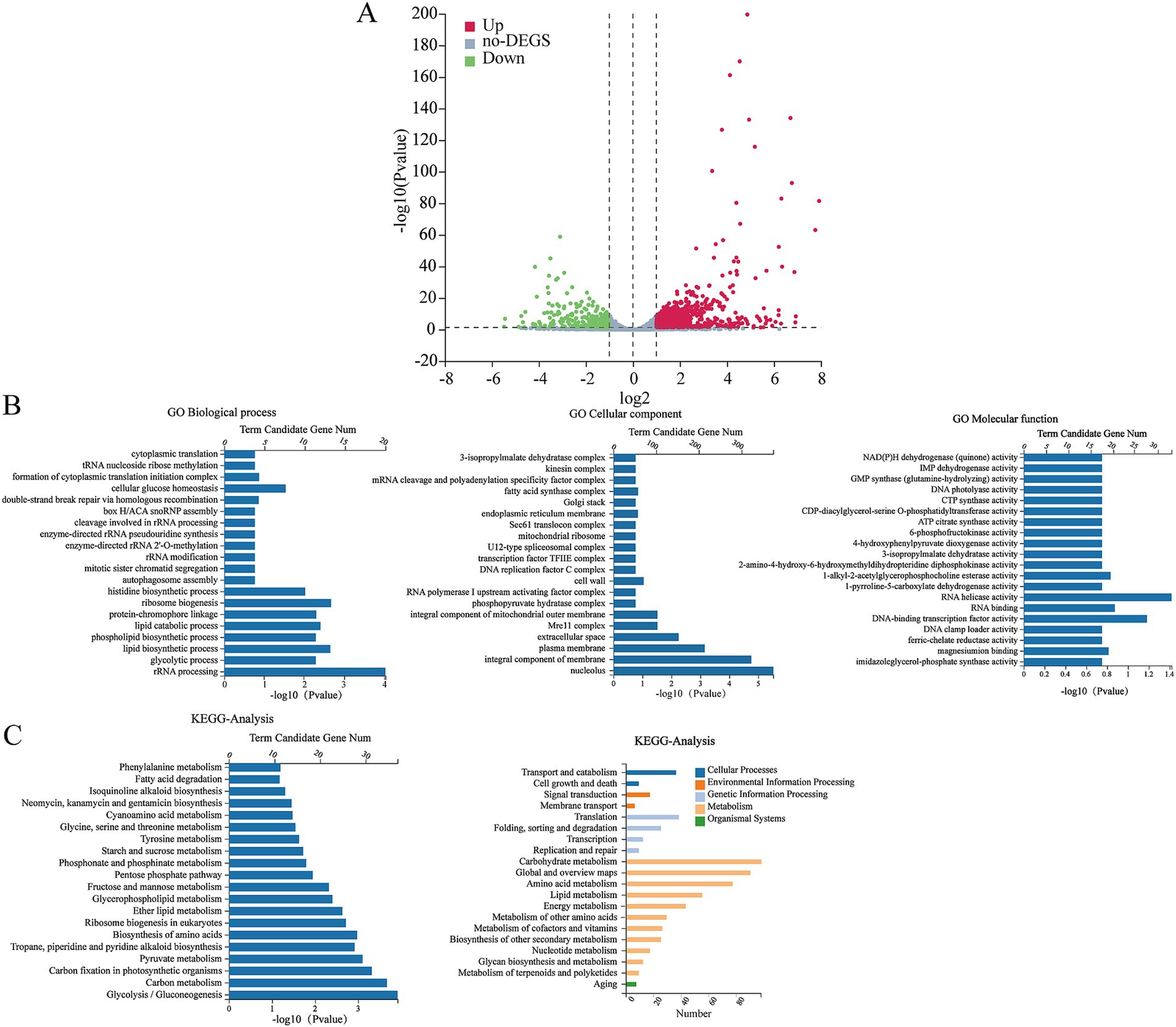
Figure 4. Volcano plots of DEGs after treatment of HMB at 1/2 MIC (A): The red dots and green dots represent up-regulated and down-regulated DEGs, respectively, and the grey dots represent genes with no significant changes in expression. Functional categorization of Biological Process (BP), Cell Components (CC), and Molecular Function (MF) of up- and down-regulated genes in GO (B). KEGG pathway classification of the top 20 differentially expressed genes (C).
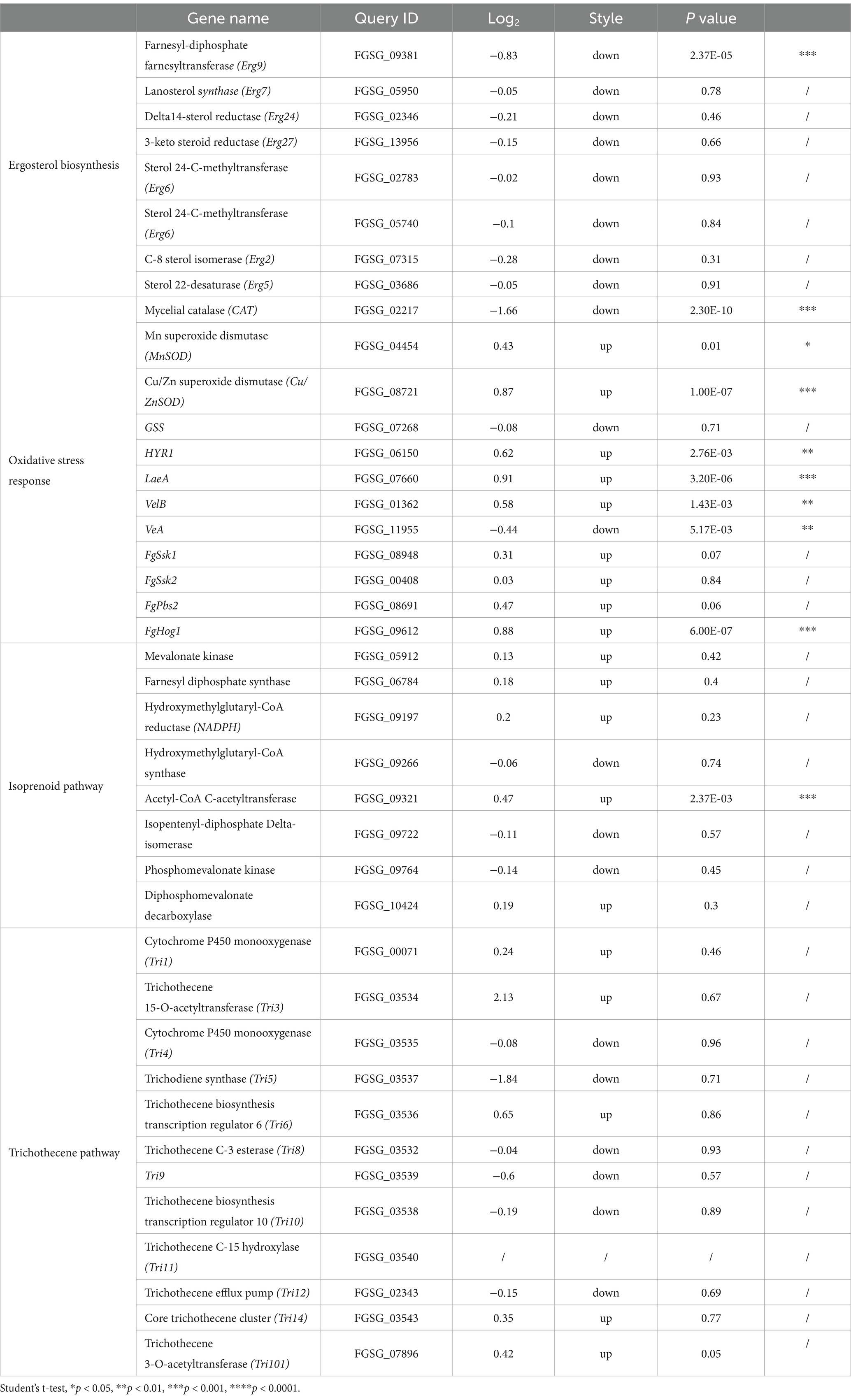
Table 1. Expression of genes participating in ergosterol biosynthesis, oxidative stress response, and DON biosynthesis of F. graminearum treated with HMB at 0 and 1/2 MIC.
3.4 HMB reduced the expression levels of genes in ergosterol biosynthesis
Ergosterol biosynthesis is an intricate process involving multiple enzymes (Supplementary Figure S1). The pathway begins with squalene synthase Erg9, which converts farnesyl pyrophosphate (FPP) into squalene, the key precursor for steroid synthesis. Squalene is then converted to lanosterol by squalene epoxidase Erg1 and lanosterol synthase Erg7 (Liu et al., 2019). Lanosterol undergoes a series of demethylation steps mediated by lanosterol C-14 demethylase Erg11 and the collaborative action of Erg24 to Erg27, producing zymosterol (Liu et al., 2011). Subsequently, zymosterol is converted into fecosterol through the catalyzation of C-24 methyltransferase Erg6. Fecosterol is then transformed into episterol by C-8 isomerase Erg2. In the final stages of the pathway, episterol is desaturated and reduced to ergosterol by Erg3, Erg5, and Erg4 (Han et al., 2023; Venegas et al., 2020). To confirm the expression changes of genes participating in ergosterol biosynthesis, qRT-PCR was conducted. As depicted in Figure 5, the expressions of Erg9 (FGSG_09381), Erg7 (FGSG_05950), Erg24 (FGSG_02346), Erg27 (FGSG_13956), Erg6 (FGSG_02783, FGSG_05740), Erg2 (FGSG_07315), and Erg5 (FGSG_03686) genes were downregulated significantly (p < 0.01), confirming that HMB treatment inhibited ergosterol biosynthesis.
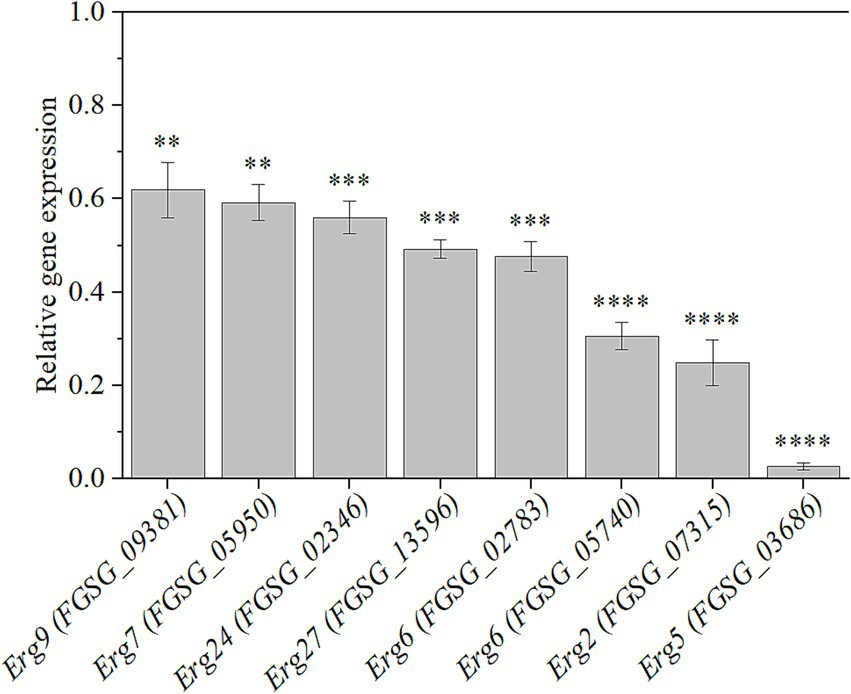
Figure 5. Relative expression levels of genes participating in ergosterol biosynthesis in response to HMB at 1/2 MIC. Black line represents normalized control expression at 1. Student’s t-test, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001.
3.5 HMB downregulated expressions of genes participating in DON biosynthesis and genes encoding redox enzymes and regulatory factors
As the biosynthesis of secondary metabolism was enriched in the RNA-seq analysis, we quantified the 20 DON biosynthetic genes expression after HMB treatment at 1/2 MIC by qRT-PCR. As illustrated in Figure 6, compared to the control group, Tri1 gene was upregulated about 2 folds (p < 0.05). However, 20 genes were down-regulated, and among which, up to 16 genes were significantly down-regulated (p < 0.05); for the remaining three genes (Tri5, Tri3, and Tri8), they were indistinctively down-regulated.
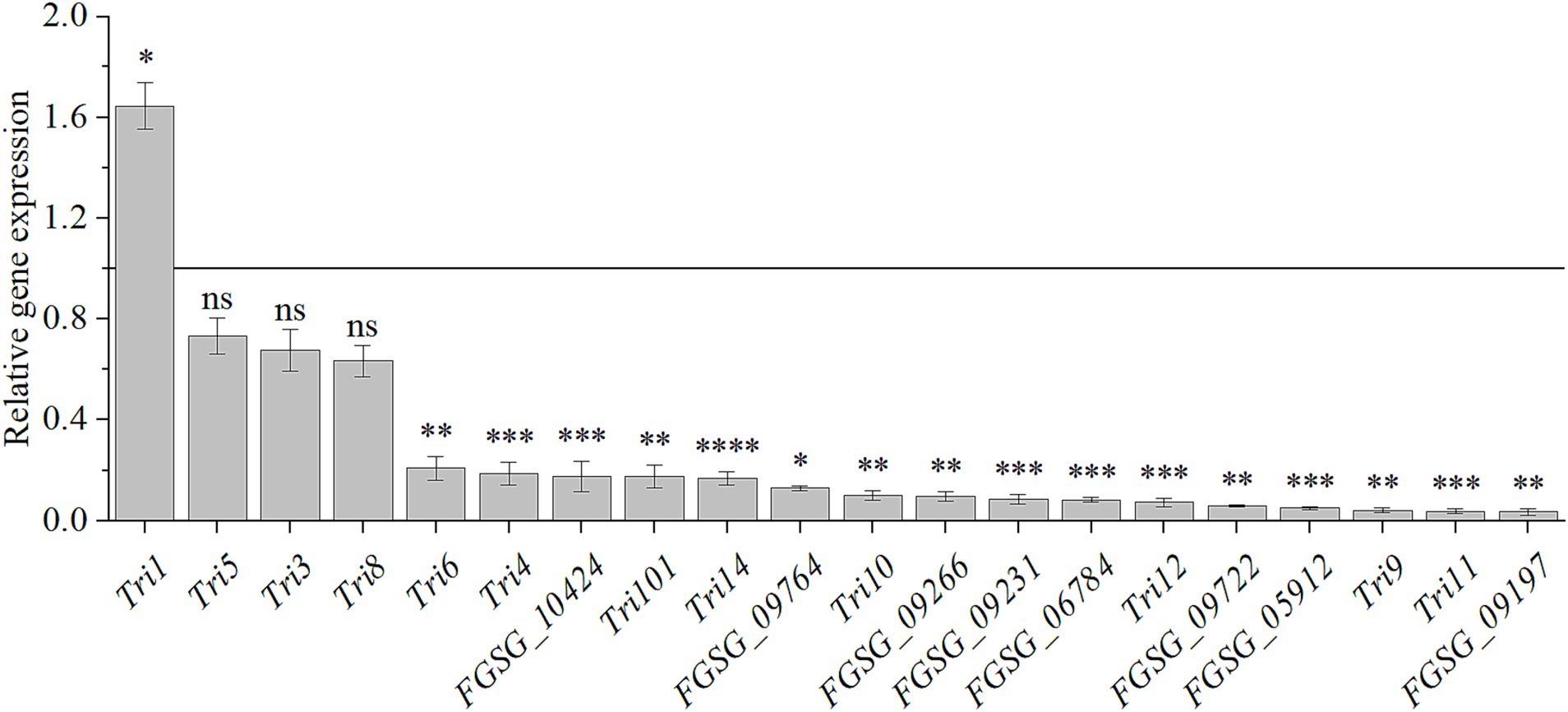
Figure 6. Relative expression levels of genes participating in DON biosynthesis in response to HMB at 1/2 MIC. Black line represents normalized control expression at 1. Student’s t-test, ns: not significant, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001.
Previous studies have found that HMB treatment significantly elevated MDA and glycerol levels in F. graminearum (Li et al., 2024), leading to lipid oxidation and osmotic stress responses that may interfere with redox homeostasis and the HOG-MAPK pathway, in order to test this conjecture the expression of genes related to oxidative, HOG-MAPK, and velvet complexes (LaeA, VelB, and VeA) was analyzed. As depicted in Figure 7, after HMB treatment, MnSOD showed a notable 2.92-fold upregulation (p < 0.001) and GSS and Cu/ZnSOD were indistinctively up-regulated. The expressions of CAT and HYR1 genes were decreased by 0.33-fold and 0.78-fold, respectively (p < 0.05). In addition, the expressions of global regulators (LaeA, VelB, and VeA) and 4 genes of the high osmolarity glycerol mitogen-activated protein kinase (HOG-MAPK) pathway (FgSsk1, FgSsk2, FgPbs2, and FgHog1) were markedly decreased (p < 0.05).
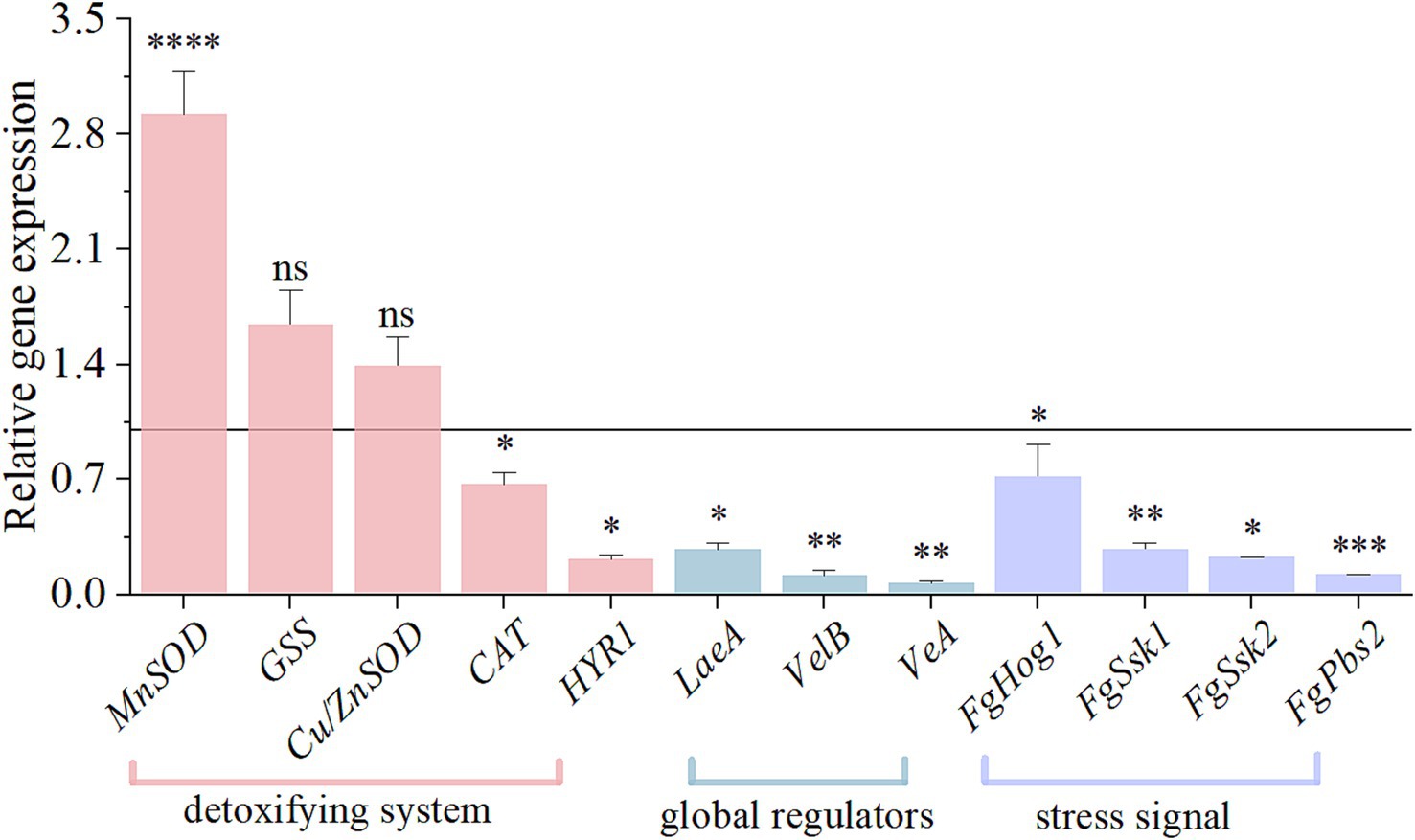
Figure 7. Effect of HMB on the relative expressions of genes involved in oxidative stress response in F. graminearum. Black line represents control expression level at 1. Student’s t-test, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001.
4 Discussion
Many plant-derived natural compounds are hydrophobic and prone to binding and disrupting cell membranes (Burt, 2004). The similar mode of action was also found in our publications (Li et al., 2021a; Li et al., 2021b; Li et al., 2021c; Li et al., 2020). Still, for HMB, the antifungal mechanism is still far from illustrated. Several commercial fungicides were developed as they could target ergosterol biosynthesis that is unique in fungi. For instance, azole fungicides (Ketoconazole, propiconazole, and tebuconazole) typically inhibits Erg11 activity (Qian et al., 2017). Allylamines and thiocarbamates target squalene epoxidase encoded by the Erg1 gene, while morpholines (fenpropimorph and amorolfine) downregulated the expression of Erg2 and Erg24 genes (Prasad et al., 2016). In natural product fungistatic assays, substances that disrupt ergosterol biosynthesis often act on multiple genes. For example, citral reduces the expression of the Erg7, Erg11, Erg6, Erg3, and Erg5 genes in Penicillium digitatum (OuYang et al., 2016), while cinnamaldehyde downregulates the gene expression of Erg11, Erg6, and Erg4 in Fusarium sambucinum (Wei et al., 2020). Thymol interferes with ergosterol biosynthesis in F. graminearum in a manner similar to azoles (Gao et al., 2016). In our present study, HMB treatment suppressed the expression of 8 genes in the ergosterol biosynthesis pathway, including Erg9 (FGSG_09381), Erg7 (FGSG_05950), Erg24 (FGSG_02346), Erg27 (FGSG_13956), Erg6 (FGSG_02783 and FGSG_05740), Erg2 (FGSG_07315), and Erg5 (FGSG_03686). Among them, the most strongly downregulated genes were located downstream, including Erg6 (FGSG_05740), Erg2, and Erg5. Hence, HMB inhibits the growth of F. graminearum by disrupting the cell membrane through downregulating the genes participating in ergosterol biosynthesis.
Under normal physiological conditions, ROS is generated through routine metabolic processes, with its production and scavenging maintained in a delicate balance (Dwyer et al., 2014). Excessive ROS accumulation could ultimately reduce cell viability and potentially cause cell death (Chen et al., 2023; Hwang et al., 2012). In this study, HMB treatment resulted in ROS accumulation, accompanied by increased SOD activity, decreased CAT activity, and unchanged GPx activity. This suggested that while the activity of SOD was boosted to counteract elevated ROS levels, the activities of other antioxidant enzymes (GPx and CAT) either remained unchanged or decreased, leading to sustained high intracellular ROS levels. In addition, as GSH levels remained relatively stable in response to HMB treatment, the non-enzymatic antioxidant is supposed to contribute little to ROS scavenging. Consistent with our hypotheses, H₂O₂ levels increased, likely caused by the differential regulations of these antioxidant enzymes, further contributing to ROS accumulation. Similar changes in enzyme activities and H2O2 levels were observed in A. flavus treated with paeonol (Li et al., 2022). However, other compounds exhibited varying effects on antioxidant enzymes besides causing ROS accumulation. For instance, methyl gallate reduced the activities of SOD, CAT, and POD (Liu et al., 2021), whereas α-thujone treatment increased CAT activity in F. graminearum (Teker et al., 2021). In addition, 4-propylphenol exposure significantly reduced GSH and GSSH levels in F. graminearum (Sun et al., 2024). In Candida glabrata, fluconazole treatment increased the GPx and SOD activity (Mahl et al., 2015). Therefore, redox systems in different fungi respond differently upon various compounds treatments.
DON biosynthesis occurs in two stages (Supplementary Figure S2): the first one is the isoprenoid pathway, which starts with acetyl-CoA and involves 7 reactions catalyzed by enzymes such as acetoacetyl-CoA synthase and HMG-CoA synthase. The final step in this pathway is the conversion of mevalonic acid to FPP. The second one is the trichothecene pathway, which involves 15 enzyme-mediated reactions converting FPP to DON. Initially, FPP is cyclized by Tri5 encoding trichodiene synthase to form trichodiene (TDN). After a series of catalyzation by the multifunctional cytochrome P450 monooxygenase (Tri4), isotrichotriol is formed (Oufensou et al., 2020). Then, Isotrichotriol undergoes non-enzymatic isomerization and cyclization to form isotrichodermol. Subsequently, Tri101, Tri11 and Tri3 then catalyze the formation of the first potentially toxic substance, calonectrin (CAL) (McCormick et al., 2011). In the last step, DON was produced by the catalyzation of Tri8 (McCormick et al., 2011). Tri6 and Tri10 are key regulatory genes in the production of DON (Seong et al., 2009). In detail, Tri6, a Cys2-His2-type transcription factor, regulates the Tri gene cluster consisting of Tri5, Tri12, and Tri4. As an upstream regulator, Tri10 indirectly controls DON biosynthesis by modulating the expression of downstream genes, such as Tri4 and Tri5, through the regulation of Tri6 (Ponts et al., 2007). Among the 16 downregulated DON biosynthesis genes, all 8 genes of the isoprenoid pathway were significantly down-regulated. Six genes (Tri4, Tri9, Tri11, Tri12, Tri14, and Tri101) located in the middle part of the biosynthetic pathway were significantly down-regulated except for Tri3, which was not significantly changed. Tri1 was significantly up-regulated, Tri5 and Tri8 genes, located at the beginning and end of the pathway, were not significantly regulated. For the two transcription factors, Tri6 and Tri10, they were also significantly down-regulated. Similarly, other compound treatments downregulated genes in the Tri gene cluster. For example, ferulic acid treatment downregulated the expressions of Tri4, Tri5, Tri6, Tri11, Tri12, Tri101, and Tri10 genes (Boutigny et al., 2009), while methyl gallate (MG) treatment downregulated the expressions of Tri3, Tri5, Tri8, Tri10, Tri11, and Tri12 genes (Liu et al., 2021). The downregulation of Tri5 gene was also found after myrcene treatment (Albayrak et al., 2023). However, the expression changes of other genes in the DON biosynthesis pathway were not mentioned in these papers.
Interestingly, our recent study showed that MDA content was significantly negatively correlated with DON content in F. graminearum after HMB treatment (Li et al., 2024). In addition, at the transcriptome level, mycotoxin production decreasing seems to be accompanied by the expression changes of redox enzyme genes. For instance, ethanol has been shown to inhibit the biosynthesis of aflatoxin B1 (AFB1), meanwhile, redox enzyme genes (Cat, Cat1, Cat2, CatA, and Cu, Zn superoxide dismutase gene SOD1) were upregulated and MnSOD gene was downregulated (Ren et al., 2020). Cinnamaldehyde treatment suppressed AFB1 production, and also upregulated the expressions of Cat1, CatA, SOD1, and MnSOD genes in A. flavus (Wang et al., 2019). Furthermore, other works have proved that several such genes are upstream regulators of DON biosynthesis. For example, knockout of the Cu/ZnSOD gene induced a significant decrease in DON production (Yao et al., 2016). Two mutants of SOD2 and SOD3 genes remarkably downregulated Tri5 and Tri6 genes expression and reduced 3-DON production (Furukawa et al., 2017). In the present work, HMB treatment up-regulated the expressions of MnSOD, GSS, and Cu/ZnSOD genes, while CAT and HYR1 genes were down-regulated. The gene expression results were consistent with the above enzyme activity results. Still, whether other redox enzymes (e.g., CAT, GSS, and HYR1) regulate DON biosynthesis requires further knockout and/or overexpression works.
Apart from redox enzymes, global regulators are known to regulate the DON biosynthetic gene cluster (Chen et al., 2019). Light is known to positively regulate DON biosynthesis in F. graminearum via specific regulation of the velvet complex (VelB/VeA/LaeA) (Chen et al., 2019), similar as those in A. flavus (Amare and Keller, 2014). In detail, the individual deletion of each of the three genes significantly downregulated the expressions of Tri5 and Tri6 genes (Jiang et al., 2012; Merhej et al., 2011; Yu et al., 2013). What’s more, according to the significantly elevated glycerol content in F. graminearum on exposure of HMB treatment, we supposed that the HOG-MAPK pathway was regulated, as this pathway responds to osmotic stress (Ochiai et al., 2007). Previous researches showed that several genes participating in HOG-MAPK pathway could regulate DON biosynthesis. In detail, three knockout mutants of FgSsk2, FgPbs2, and FgHog1 gene downregulated the transcript levels of Tri4 and Tri6 genes (Ochiai et al., 2007; Zheng et al., 2012). In this work, HMB treatment significantly downregulated the velvet complex genes (VelB, VeA, and LaeA) and HOG-MAPK pathway-related genes (FgSsk1, FgSsk2, FgPbs2, and FgHog1), indicating a collective contribution on the inhibition of DON biosynthesis.
5 Conclusion
As chemical fungicides are potential threats to environment (e.g., soil residues, resistance), human and animal health, green fungicide development is driven. In this study, the antifungal mechanism of HMB against F. graminearum was further elucidated. HMB inhibits mycelial growth by targeting the ergosterol biosynthesis, the anti-mycotoxigenic mechanism is probably involved with the participation of redox enzymes, velvet complex, and key factors in HOG-MAPK pathway. However, F. graminearum has diverse strains with varying physiological and pathogenic traits. Their sensitivity to compounds differs, so the inhibitory effects of HMB on other strains, apart from PH-1, need further study. Also, new transcriptional regulators, the relationship between redox system and the DON biosynthesis pathway still requires to be explored and elucidated. Based on these efforts and results, HMB holds a great promise in developing as an efficient fungicide in the preservation of agricultural products and food.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
HX: Data curation, Formal analysis, Investigation, Writing – original draft. YZ: Investigation, Writing – original draft. QL: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. We thank for the financial support of Cultivation Project of Tuoxin Team in Henan University of Technology (2024TXTD08), Henan Province Key Research and Development and promotion Project (Science and Technology Research) (252102110078), Cultivation Programme for Young Backbone Teachers in Henan University of Technology (0503/21421231), Key R&D projects in Henan Province (231111113300), and Double First-Class Discipline Construction Program of Henan University of Technology (0517-24410014).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1514170/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Ergosterol biosynthesis pathway.
SUPPLEMENTARY FIGURE S2 | DON biosynthesis pathway.
SUPPLEMENTARY TABLE S1 | Primer sequences used in qRT-PCR.
Footnotes
References
Albayrak, G., Yörük, E., Teker, T., and Sefer, Ö. (2023). Investigation of antifungal activities of myrcene on fusarium reference strains. Arch. Microbiol. 205:82. doi: 10.1007/s00203-023-03420-3
Amare, M. G., and Keller, N. P. (2014). Molecular mechanisms of aspergillus flavus secondary metabolism and development. Fungal Genet. Biol. 66, 11–18. doi: 10.1016/j.fgb.2014.02.008
Boutigny, A.-L., Barreau, C., Atanasova-Penichon, V., Verdal-Bonnin, M.-N., Pinson-Gadais, L., and Richard-Forget, F. (2009). Ferulic acid, an efficient inhibitor of type B trichothecene biosynthesis and tri gene expression in fusarium liquid cultures. Mycol. Res. 113, 746–753. doi: 10.1016/j.mycres.2009.02.010
Burt, S. (2004). Essential oils: their antibacterial properties and potential applications in foods-a review. Int. J. Food Microbiol. 94, 223–253. doi: 10.1016/j.ijfoodmicro.2004.03.022
Chen, Y., Chen, Y., Shi, C., Huang, Z., Zhang, Y., Li, S., et al. (2018). SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 7, 1–6. doi: 10.1093/gigascience/gix120
Chen, Y., Kistler, H. C., and Ma, Z. H. (2019). Fusarium graminearum trichothecene mycotoxins: biosynthesis, regulation, and management. Annu. Rev. Phytopathol. 57, 15–39. doi: 10.1146/annurev-phyto-082718-100318
Chen, L., Xu, X. X., Sun, Y. X., Xin, Q. H., Lv, Y. Y., Hu, Y. S., et al. (2023). Surfactin inhibits fusarium graminearum by accumulating intracellular ROS and inducing apoptosis mechanisms. World J. Microbiol. Biotechnol. 39:340. doi: 10.1007/s11274-023-03790-2
Diao, X., Hang, Y. Y., and Liu, C. L. (2018). The fungicidal activity of tebuconazole enantiomers against fusarium graminearum and its selective effect on DON production under different conditions. J. Agric. Food Chem. 66, 3637–3643. doi: 10.1021/acs.jafc.7b05483
Dwyer, D. J., Belenky, P. A., Yang, J. H., Macdonald, I. C., Martell, J. D., Takahashi, N., et al. (2014). Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. 111, E2100–E2109. doi: 10.1073/pnas.1401876111
Feizollahi, E., and Roopesh, M. R. (2021). Mechanisms of deoxynivalenol (DON) degradation during different treatments: a review. Crit. Rev. Food Sci. 62, 5903–5924. doi: 10.1080/10408398.2021.1895056
Ferrigo, D., Raiola, A., and Causin, R. (2016). Fusarium toxins in cereals: occurrence, legislation, factors promoting the appearance and their management. Molecules 21:627. doi: 10.3390/molecules21050627
Figueroa, M., Hammond-Kosack, K. E., and Solomon, P. S. (2017). A review of wheat disease-a field perspective. Mol. Plant Pathol. 19, 1523–1536. doi: 10.1111/mpp.12618
Fridovich, I. (1995). Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 64, 97–112. doi: 10.1146/annurev.bi.64.070195.000525
Furukawa, T., Yoshinari, T., and Sakuda, S. (2017). Intracellular superoxide level controlled by manganese superoxide dismutases affects trichothecene production in Fusarium graminearum. FEMS Microbiol. Lett. 364:fnx213. doi: 10.1093/femsle/fnx213
Gao, T., Zhou, H., Zhou, W., Hu, L. B., Chen, J., and Shi, Z. Q. (2016). The fungicidal activity of thymol against fusarium graminearum via inducing lipid peroxidation and disrupting ergosterol biosynthesis. Molecules 21:770. doi: 10.3390/molecules21060770
Gill, S. S., Khan, N. A., and Tuteja, N. (2012). Cadmium at high dose perturbs growth, photosynthesis and nitrogen metabolism while at low dose it up regulates sulfur assimilation and antioxidant machinery in garden cress (Lepidium sativum L.). Plant Sci. 182, 112–120. doi: 10.1016/j.plantsci.2011.04.018
Häggblom, P., and Nordkvist, E. (2015). Deoxynivalenol, zearalenone, and Fusarium graminearum contamination of cereal straw; field distribution; and sampling of big bales. Mycotoxin Res. 31, 101–107. doi: 10.1007/s12550-015-0220-z
Han, Z., Zong, Y., Zhang, X., Gong, D., Wang, B., Prusky, D., et al. (2023). Erg4 is involved in ergosterol biosynthesis, conidiation and stress response in Penicillium expansum. J. Fungi. 9:568. doi: 10.3390/jof9050568
Heller, J., and Tudzynski, P. (2011). Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu. Rev. Phytopathol. 49, 369–390. doi: 10.1146/annurev-phyto-072910-095355
Hwang, J. H., Hwang, I.-S., Liu, Q. H., Woo, E. R., and Lee, D. G. (2012). (+)-Medioresinol leads to intracellular ROS accumulation and mitochondria-mediated apoptotic cell death in Candida albicans. Biochimie 94, 1784–1793. doi: 10.1016/j.biochi.2012.04.010
Jamieson, D. J. (1998). Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14, 1511–1527. doi: 10.1002/(sici)1097-0061(199812)14:16<1511::Aid-yea356>3.0.Co;2-s
Ji, F., He, D., Olaniran, A. O., Mokoena, M. P., Xu, J., and Shi, J. (2019). Occurrence, toxicity, production and detection of fusarium mycotoxin: a review. FPPN 1, 1–14. doi: 10.1186/s43014-019-0007-2
Jiang, J., Yun, Y., Liu, Y., and Ma, Z. (2012). FgVELB is associated with vegetative differentiation, secondary metabolism and virulence in fusarium graminearum. Fungal Genet. Biol. 49, 653–662. doi: 10.1016/j.fgb.2012.06.005
Jordá, T., and Puig, S. (2020). Regulation of ergosterol biosynthesis in Saccharomyces cerevisiae. Genes 11:795. doi: 10.3390/genes11070795
Khan, A., Ahmad, A., Akhtar, F., Yousuf, S., Xess, I., Khan, L. A., et al. (2010). Ocimum sanctum essential oil and its active principles exert their antifungal activity by disrupting ergosterol biosynthesis and membrane integrity. Res. Microbiol. 161, 816–823. doi: 10.1016/j.resmic.2010.09.008
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Koubaa, S., and Brini, F. (2020). Functional analysis of a wheat group 3 late embryogenesis abundant protein (TdLEA3) in Arabidopsis thaliana under abiotic and biotic stresses. Plant Physiol. Bioch. 156, 396–406. doi: 10.1016/j.plaphy.2020.09.028
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lee, J., Chang, I.-Y., Kim, H., Yun, S.-H., Leslie, J. F., and Lee, Y.-W. (2009). Genetic diversity and fitness of Fusarium graminearum populations from rice in Korea. Appl. Environ. Microbiol. 75, 3289–3295. doi: 10.1128/aem.02287-08
Li, B., and Dewey, C. (2011). RSEM accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 1–16. doi: 10.1186/1471-2105-12-323
Li, Q., Wang, C., Xiao, H., Zhang, Y., and Xie, Y. (2024). 2-Hydroxy-4-methoxybenzaldehyde, a more effective antifungal aroma than vanillin and its derivatives against fusarium graminearum, destroys cell membranes, inhibits DON biosynthesis, and performs a promising antifungal effect on wheat grains. Front. Microbiol. 15:1359947. doi: 10.3389/fmicb.2024.1359947
Li, Q., Zhao, Y., Zhu, X., and Xie, Y. (2021a). Antifungal efficacy of paeonol on aspergillus flavus and its mode of action on cell walls and cell membranes. LWT 149:111985. doi: 10.1016/j.lwt.2021.111985
Li, Q., Zhao, Y., Zuo, X., Guo, F., Li, Y., and Xie, Y. (2022). Paeonol inhibits aspergillus flavus via disrupting ergosterol biosynthesis, redox metabolism, and aflatoxin biosynthesis on rice. LWT 163:113587. doi: 10.1016/j.lwt.2022.113587
Li, Q., Zhu, X., Xie, Y., and Liang, J. (2021b). Antifungal properties and mechanisms of three volatile aldehydes (octanal, nonanal and decanal) on aspergillus flavus. GOST 4, 131–140. doi: 10.1016/j.gaost.2021.07.002
Li, Q., Zhu, X. M., Xie, Y. L., and Ren, S. L. (2020). 2-hydroxy-4-methoxybenzaldehyde inhibits the growth of aspergillus flavus via damaging cell wall, cell membrane, manipulating respiration thus creating a promising antifungal effect on corn kernels. Int. J. Food Sci. Technol. 56, 178–184. doi: 10.1111/ijfs.14617
Li, Q., Zhu, X., Xie, Y., and Zhong, Y. (2021c). O-vanillin, a promising antifungal agent, inhibits aspergillus flavus by disrupting the integrity of cell walls and cell membranes. Appl. Microbiol. Biotechnol. 105, 5147–5158. doi: 10.1007/s00253-021-11371-2
Liu, X., Fang, X., Wang, S., Wu, D., Gao, T., Lee, Y.-W., et al. (2021). The antioxidant methyl gallate inhibits fungal growth and deoxynivalenol production in fusarium graminearum. FPPN 3, 1–9. doi: 10.1186/s43014-021-00070-0
Liu, X., Fu, J., Yun, Y. Z., Yin, Y. N., and Ma, Z. H. (2011). A sterol C-14 reductase encoded by FgERG24B is responsible for the intrinsic resistance of fusarium graminearum to amine fungicides. Microbiology 157, 1665–1675. doi: 10.1099/mic.0.045690-0
Liu, J. F., Xia, J. J., Nie, K. L., Wang, F., and Deng, L. (2019). Outline of the biosynthesis and regulation of ergosterol in yeast. World J. Microb. Biot. 35, 1–8. doi: 10.1007/s11274-019-2673-2
Mahl, C. D., Behling, C. S., Hackenhaar, F. S., De Carvalho E Silva, M. N., Putti, J., Salomon, T. B., et al. (2015). Induction of ROS generation by fluconazole in Candida glabrata: activation of antioxidant enzymes and oxidative DNA damage Diagn. Microbiol. Infect. Dis. 82, 203–208. doi: 10.1016/j.diagmicrobio.2015.03.019
Mccormick, S. P., Stanley, A. M., Stover, N. A., and Alexander, N. J. (2011). Trichothecenes: from simple to complex mycotoxins. Toxins 3, 802–814. doi: 10.3390/toxins3070802
Mendes, G. D. R. L., Ponte, E. M. D., Feltrin, A. C., Badiale-Furlong, E., and Oliveira, A. C. D. (2018). Common resistance to fusarium head blight in Brazilian wheat cultivars. Sci. Agric. 75, 426–431. doi: 10.1590/1678-992x-2016-0407
Meng, X., Wu, W., Niu, B., Liu, R., Chen, H., and Gao, H. (2024). Inhibitory effect and action mechanism of perillaldehyde on the fusarium graminearum in postharvest fresh ginger. Postharvest Biol. Tec. 209:112674. doi: 10.1016/j.postharvbio.2023.112674
Merhej, J., Urban, M., Dufresne, M., Hammond-Kosack, K. E., Richard-Forget, F., and Barreau, C. (2011). The velvet gene, FgVe1, affects fungal development and positively regulates trichothecene biosynthesis and pathogenicity in fusarium graminearum. Mol. Plant Pathol. 13, 363–374. doi: 10.1111/j.1364-3703.2011.00755.x
Ochiai, N., Tokai, T., Nishiuchi, T., Takahashi-Ando, N., Fujimura, M., and Kimura, M. (2007). Involvement of the osmosensor histidine kinase and osmotic stress-activated protein kinases in the regulation of secondary metabolism in fusarium graminearum. Biochem. Biophys. Res. Co. 363, 639–644. doi: 10.1016/j.bbrc.2007.09.027
Oufensou, S., Balmas, V., Azara, E., Fabbri, D., Dettori, M. A., Schüller, C., et al. (2020). Naturally occurring phenols modulate vegetative growth and deoxynivalenol biosynthesis in Fusarium graminearum. ACS Omega. 5, 29407–29415. doi: 10.1021/acsomega.0c04260
Ouyang, Q., Tao, N., and Jing, G. (2016). Transcriptional profiling analysis of Penicillium digitatum, the causal agent of citrus green mold, unravels an inhibited ergosterol biosynthesis pathway in response to citral. BMC Genomics 17:599. doi: 10.1186/s12864-016-2943-4
Payros, D., Alassane-Kpembi, I., Pierron, A., Loiseau, N., Pinton, P., and Oswald, I. P. (2016). Toxicology of deoxynivalenol and its acetylated and modified forms. Arch. Toxicol. 90, 2931–2957. doi: 10.1007/s00204-016-1826-4
Pestka, J. J. (2010). Deoxynivalenol: mechanisms of action, human exposure, and toxicological relevance. Arch. Toxicol. 84, 663–679. doi: 10.1007/s00204-010-0579-8
Podgórska-Kryszczuk, I., Solarska, E., and Kordowska-Wiater, M. (2022). Reduction of the fusarium mycotoxins: deoxynivalenol, nivalenol and zearalenone by selected non-conventional yeast strains in wheat grains and bread. Molecules 27:1578. doi: 10.3390/molecules27051578
Ponts, N., Pinson-Gadais, L., Barreau, C., Richard-Forget, F., and Ouellet, T. (2007). Exogenous H2O2 and catalase treatments interfere with tri genes expression in liquid cultures of fusarium graminearum. FEBS Lett. 581, 443–447. doi: 10.1016/j.febslet.2007.01.003
Prasad, R., Shah, A. H., and Rawal, M. K. (2016). Antifungals: mechanism of action and drug resistance. Yeast Membrane Transport. 892:304. doi: 10.1007/978-3-319-25304-6_14
Qian, H., Duan, M., Sun, X., Chi, M., Zhao, Y., Liang, W., et al. (2017). The binding mechanism between azoles and FgCYP51B, sterol 14α-demethylase of Fusarium graminearum. Pest Manag. Sci. 74, 126–134. doi: 10.1002/ps.4667
Rathi, N., Harwalkar, K., V, J., Sharma, A., and Rao, N. N. (2017). 2-hydroxy-4-methoxybenzaldehyde, an astounding food flavoring metabolite: a review. Asian J. Pharm. Clin. Res. 10, 105–110. doi: 10.22159/ajpcr.2017.v10i10.19729
Ren, Y., Jin, J., Zheng, M., Yang, Q., and Xing, F. (2020). Ethanol inhibits aflatoxin B1 biosynthesis in aspergillus flavus by up-regulating oxidative stress-related genes. Front. Microbiol. 10:2946. doi: 10.3389/fmicb.2019.02946
Sahin, U., Ekinci, M., Ors, S., Turan, M., Yildiz, S., and Yildirim, E. (2018). Effects of individual and combined effects of salinity and drought on physiological, nutritional and biochemical properties of cabbage (Brassica oleracea var. capitata). Sci. Hortic. 240, 196–204. doi: 10.1016/j.scienta.2018.06.016
Seong, K. Y., Pasquali, M., Zhou, X., Song, J., Hilburn, K., Mccormick, S., et al. (2009). Global gene regulation by fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol. Microbiol. 72, 354–367. doi: 10.1111/j.1365-2958.2009.06649.x
Shao, J., Pei, Z., Jing, H., Wang, L., Jiang, C., Du, X., et al. (2021). Antifungal activity of myriocin against Fusarium graminearum and its inhibitory effect on deoxynivalenol production in wheat grains. Physiol Mol Plant P. 114:101635. doi: 10.1016/j.pmpp.2021.101635
Srikanta, B. M., Harish Nayaka, M. A., and Dharmesh, S. M. (2011). Inhibition of Helicobacter pylori growth and its cytotoxicity by 2-hydroxy 4-methoxy benzaldehyde of Decalepis hamiltonii (Wight & Arn); a new functional attribute. Biochimie 93, 678–688. doi: 10.1016/j.biochi.2010.12.009
Subban, N., and Mohan, R. L. J. (2003). Determination of 2-hydroxy-4-methoxybenzaldehyde in roots of Decalepis hamiltonii (Wight & Arn.) and Hemidesmus indicus r.Br. J. AOAC Int. 86, 564–567. doi: 10.1093/jaoac/86.3.564
Sun, J., Li, W., Liu, Y. N., Lin, F. X., Huang, Z. H., Lu, F. X., et al. (2018). Growth inhibition of fusarium graminearum and reduction of deoxynivalenol production in wheat grain by bacillomycin D. J. Stored Prod. Res. 75, 21–28. doi: 10.1016/j.jspr.2017.11.002
Sun, S. F., Tang, N. W., Han, K., You, J. H., Liu, A. R., Wang, Q. Q., et al. (2024). Antifungal activity and mechanism of 4-propylphenol against fusarium graminearum, agent of wheat scab, and its potential application. J. Agric. Food Chem. 72, 5258–5268. doi: 10.1021/acs.jafc.3c09646
Teker, T., Sefer, Ö., Gazdağlı, A., Yörük, E., Varol, G. İ., and Albayrak, G. (2021). Α-thujone exhibits an antifungal activity against F. Graminearum by inducing oxidative stress, apoptosis, epigenetics alterations and reduced toxin synthesis. Eur. J. Plant Pathol. 160, 611–622. doi: 10.1007/s10658-021-02269-w
Traynor, A. M., Sheridan, K. J., Jones, G. W., Calera, J. A., and Doyle, S. (2019). Involvement of sulfur in the biosynthesis of essential metabolites in pathogenic fungi of animals, particularly aspergillus spp.: molecular and therapeutic implications. Front. Microbiol. 10, 564–567. doi: 10.3389/fmicb.2019.02859
Valenzuela- Cota, D. F., Buitimea-Cantúa, G. V., Plascencia-Jatomea, M., Cinco-Moroyoqui, F. J., Martínez-Higuera, A. A., and Rosas-Burgos, E. C. (2019). Inhibition of the antioxidant activity of catalase and superoxide dismutase from fusarium verticillioides exposed to a Jacquinia macrocarpa antifungal fraction. J. Environ. Sci. Health 54, 647–654. doi: 10.1080/03601234.2019.1622978
Venegas, M., Barahona, S., González, A. M., Sepúlveda, D., Zúñiga, G. E., Baeza, M., et al. (2020). Phenotypic analysis of mutants of ergosterol biosynthesis genes (ERG3 and ERG4) in the red yeast Xanthophyllomyces dendrorhous. Front. Microbiol. 11:1312. doi: 10.3389/fmicb.2020.01312
Wang, J. H., Liu, H., Zhao, J. L., Gao, H. F., Zhou, L. G., Liu, Z. L., et al. (2010). Antimicrobial and antioxidant activities of the root bark essential oil of Periploca sepium and its main component 2-hydroxy-4-methoxybenzaldehyde. Molecules 15, 5807–5817. doi: 10.3390/molecules15085807
Wang, P., Ma, L., Jin, J., Zheng, M., Pan, L., Zhao, Y., et al. (2019). The anti-aflatoxigenic mechanism of cinnamaldehyde in aspergillus flavus. Sci. Rep. 9:10499. doi: 10.1038/s41598-019-47003-z
Wei, J., Bi, Y., Xue, H., Wang, Y., Zong, Y., and Prusky, D. (2020). Antifungal activity of cinnamaldehyde against Fusarium sambucinum involves inhibition of ergosterol biosynthesis. J. Appl. Microbiol. 129, 256–265. doi: 10.1111/jam.14601
Wilson, W., Dahl, B., and Nganje, W. (2018). Economic costs of fusarium head blight, scab and deoxynivalenol. World Mycotoxin J. 11, 291–302. doi: 10.3920/wmj2017.2204
Yan, H., Meng, X. Y., Lin, X. F., Duan, N., Wang, Z. P., and Wu, S. J. (2023). Antifungal activity and inhibitory mechanisms of ferulic acid against the growth of fusarium graminearum. Food Biosci. 52:102414. doi: 10.1016/j.fbio.2023.102414
Yang, C., Xie, L., Ma, Y., Cai, X., Yue, G., Qin, G., et al. (2021). Study on the fungicidal mechanism of glabridin against fusarium graminearum. Pestic. Biochem. Physiol. 179:104963. doi: 10.1016/j.pestbp.2021.104963
Yao, S., Guo, Y., Wang, Y., Zhang, D., Xu, L., and Tang, W. (2016). A cytoplasmic cu-Zn superoxide dismutase SOD1 contributes to hyphal growth and virulence of fusarium graminearum. Fungal Genet. Biol. 91, 32–42. doi: 10.1016/j.fgb.2016.03.006
Yu, J.-H., Kim, H.-K., Lee, S., Jo, S.-M., Mccormick, S. P., Butchko, R. a. E., et al. (2013). Functional roles of FgLaeA in controlling secondary metabolism, sexual development, and virulence in fusarium graminearum. PLoS One 8:e68441. doi: 10.1371/journal.pone.0068441
Keywords: plant-derived natural compound, HMB, ergosterol biosynthesis, oxidative stress response, mycotoxin regulation, transcriptome analysis
Citation: Xiao H, Zhang Y and Li Q (2025) 2-Hydroxy-4-methoxybenzaldehyde (HMB) disrupts ergosterol biosynthesis, redox metabolism, and DON biosynthesis of Fusarium graminearum revealed by transcriptome analysis. Front. Microbiol. 16:1514170. doi: 10.3389/fmicb.2025.1514170
Edited by:
Alessandra Marcon Gasperini, University of Hertfordshire, United KingdomReviewed by:
Regina Sharmila Dass, Pondicherry University, IndiaJianhua Wang, Shanghai Academy of Agricultural Sciences, China
Copyright © 2025 Xiao, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Li, bHFAaGF1dC5lZHUuY24=
 Hongying Xiao
Hongying Xiao Yiming Zhang
Yiming Zhang Qian Li
Qian Li