- Department of Biological Sciences, University of Limerick, Limerick, Ireland
Cultivation conditions, including plant species, variety, cultivation method, and seasonality, are all at least co-factors of epiphytic Listeria monocytogenes growth. Meanwhile, phyllosphere-associated bacteria were found to influence the colonization of invading pathogens. Thus, the main objective of this study was to determine whether cultivation conditions are factors in the development of the bacterial phyllosphere community on leafy vegetables, which consequently correlates positively or negatively with L. monocytogenes growth. Indeed, this study revealed that vegetable cultivation conditions are a more significant determinant of phyllosphere development than plant species. Of the identified phyllosphere-associated bacteria, the presence of Pseudomonadaceae had a positive correlation with L. monocytogenes populations on all tested produce. Hitherto, Pseudomonadaceae content appeared to be more critical for L. monocytogenes growth on spinach F1 Trumpet. For days 7–9 of storage, Pseudomonadaceae increased abundance on open field spinach F1 Trumpet were associated with L. monocytogenes’ most significant increase (0.94 log10 colony-forming unit (cfu) g−1). In contrast, Pseudomonadaceae content decreased for polytunnel spinach F1 Trumpet, and the corresponding L. monocytogenes populations remained unchanged. Carnobacteriaceae were present on spinach F1 Trumpet from the polytunnel but not on other spinach products, with higher associated L. monocytogenes growth. Pectobacteriaceae (genus Dickeya) increased for spinach F1 Trumpet polytunnel but decreased for other spinach produce with lower associated L. monocytogenes growth. Similarly, polytunnel rocket Esmee had an increasing relative abundance of Pectobacteriaceae, whereas it remained constant for polytunnel rocket Buzz. Compared to summer spinach F1 Trumpet produce, winter produce had significantly greater Streptococcaceae content and was correlated with a decrease in L. monocytogenes growth. Finally, higher phyllosphere alpha diversity putatively limited L. monocytogenes growth. Ultimately, this study revealed that cultivation conditions determine the bacterial phyllosphere community structure, which consequently influences L. monocytogenes growth.
1 Introduction
Leafy vegetables such as rocket and spinach are commonly consumed due to their vitamin, mineral, antioxidant, and phytochemical content (Colonna et al., 2016; Van der Avoort et al., 2018; Venu et al., 2019). To meet the demand for such leafy vegetables, global production of spinach has increased by 218% from 2001 to 2021 (FAO, 2021). Polytunnels enable all year-round production of such high-quality leafy vegetables in winter months or in countries where production may not be possible due to challenging weather conditions (Sagar, 2020). However, cultivation in polytunnels is also altering environmental conditions not only for plant growth but also for the growth of the plant microbiome.
While the increasing demand for vegetables has resulted in the adoption of cost-effective and fast production methods, less concern is given to the safety of their produce, that is, microbial contamination with foodborne pathogens such as Listeria monocytogenes (Balali et al., 2020). Potential sources of contamination include irrigation water and manures (re-harvest), as well as the handling of the produce (post-harvest) (Balali et al., 2020). In terms of L. monocytogenes growth on spinach and rocket produce, there have been conflicting results from studies with differing experimental and pre-harvest cultivation conditions (Sant'Ana et al., 2012; Lokerse et al., 2016; Söderqvist et al., 2017b; Ziegler et al., 2019; Culliney and Schmalenberger, 2020). However, Culliney and Schmalenberger (2022) revealed that cultivation conditions, that is, plant species and variety, cultivation method (polytunnel vs. open field), and seasonality of harvest, are at least partly responsible for differing levels of L. monocytogenes growth (Culliney and Schmalenberger, 2022).
Foodborne pathogens, such as L. monocytogenes do not grow in isolation but within a microbial community within the phyllosphere. The phyllosphere refers to the aerial parts of the plant, primarily the surface of the leaves, which harbor diverse and rich communities of bacteria, fungi, viruses, nematodes, and protozoans (Bashir et al., 2022). Plant species and genotype, as well as abiotic factors, such as geographical location, solar radiation, pollution, and nutrients, and biotic factors, including leaf age and presence of other microorganisms, are all drivers of the development of the phyllosphere (Xu et al., 2022). Although the phyllosphere harbors a highly diverse community, at the phylum level, the phyllospheres of different plant species, even from various geographical locations, exhibit high levels of similarity. They primarily consist of Pseudomonadota (Proteobacteria), Actinomycetota (Actinobacteria), Bacteroidota (Bacteroidetes), and Bacillota (Firmicutes) (Liu et al., 2020).
Phyllosphere-inhabiting microorganisms and their metabolites interact with their environment and may play protective roles against invading opportunistic foodborne pathogens (Saleem, 2021). A previous study revealed that bacterial isolates from ready-to-eat (RTE) lettuce influence the colonization of Listeria innocua in co-cultures (Francis and O’Beirne, 2002). However, a paucity of studies has investigated the in situ influence of the food microbiome or vegetable phyllosphere on the L. monocytogenes growth. A cultivation-based study did not identify any differences in resident bacteria present between cut leaves of broad-leaved endive associated with high and low levels of L. monocytogenes growth (Carlin et al., 1995). To date, there have been no attempts to correlate the phyllosphere bacteriome of rocket or kale with L. monocytogenes growth.
Lactic acid bacteria (LAB) are often naturally present as indigenous, spoilage bacteria and negatively impact L. monocytogenes due to their competitive growth capabilities (Østergaard et al., 2014). Additionally, LAB produce organic acids which reduce pH by lowering intracellular dissociation and intracellular leakage through porins or permeases to values beneath the pH at which L. monocytogenes performs optimally, that is, pH 7 (Webb et al., 2022). Moreover, LAB produce other metabolites or bio-preservative agents such as reuterin, bacteriocins, diacetyl, reutericyclin, organic acids, acetoin, and hydrogen peroxide (Ibrahim et al., 2021). Lactiplantibacillus plantarum is a LAB previously isolated from rocket produce, which harbors genes that encode for the production of Coagulin A and the active peptide Pediocin ACH. These can act as anti-listerial agents, thus displaying particular inhibition capacities of L. monocytogenes (Le Marrec et al., 2000; Espitia et al., 2016; Barbosa et al., 2021). Conversely, several members of the Pseudomonadaceae family cause hydrolysis of proteins, which could provide free amino acids likely to stimulate the L. monocytogenes growth (Marshall et al., 1992; Zilelidou and Skandamis, 2018). Pseudomonadaceae spp. can also increase nutrient availability, for example, carbon and nitrogen for pathogen colonization by altering ion transport across the plant cell plasma membranes (Hutchison, 1995). Additionally, P. putida has the ability to produce and release plant growth regulators, for example, indole-3-acetic acid, which promotes nutrient leakage and microbial fitness (Brandl and Lindow, 1998; Leveau and Lindow, 2005). Further research is needed to determine whether a higher diversity of the phyllosphere indigenous bacterial community is related to the reduction of the competitiveness of transient opportunistic pathogenic microorganisms (Darlison et al., 2019).
The objective of the present study was to utilize Illumina-based 16S amplicon sequencing to describe the bacterial composition of leafy vegetable phyllospheres. Different plant species (spinach, rocket, and kale), cultivars (F1 Trumpet vs. F1 Cello; and Buzz vs. Esmee), cultivation methods (polytunnel vs. open field), and seasonality (summer vs. winter spinach) were tested to identify the presence of certain bacteria of importance to L. monocytogenes growth. Changes in their relative abundance were correlated with shifts in the abundance of L. monocytogenes populations. This study hypothesized that differences in the relative abundance of certain phyllosphere-associated bacterial taxa attributed to differing cultivation conditions are essential co-factors responsible for divergent levels of L. monocytogenes growth. Consequently, the present study aimed to analyze the bacterial community structures of leafy vegetables cultivated differently, including spinach, rocket, and kale.
2 Materials and methods
2.1 Spinach, rocket, and kale produce
All spinach, rocket, and kale produce (Caryophyllales for spinach, Brassicales for rocket, and kale, referred here as species) used in this study were cultivated as described by Culliney and Schmalenberger (2022). A total of 160 samples from L. monocytogenes growth potential experiments were selected: open field and polytunnel spinach (F1 Trumpet; summer harvest), open field and polytunnel rocket (Buzz), polytunnel spinach (F1 Cello), polytunnel rocket (Esmee), open field spinach (F1 Trumpet; winter harvest). Samples were stored for days 0, 2, and 5 at 7°C and for days 7–9 at 12°C for days, where L. monocytogenes and total bacteria counts (TBCs) were enumerated on cultivation media (Culliney and Schmalenberger, 2022).
2.2 L. monocytogenes content of spinach, rocket, and kale produce
Growth experiments were executed as described in accordance with the European Union (EU) guidance document’s guidelines for conducting growth potential studies (European Union Reference Laboratory for Listeria monocytogenes (EURL Lm); EURL Lm, 2019). The rationale behind selecting these guidelines is to provide a robust representation of real-life scenarios involving low-level contaminations with the potential to grow under realistic storage conditions. Each sample consisted of 25 g of produce inoculated with 100 cfu g−1 of a three-strain mix of L. monocytogenes, that is, 959 (vegetable isolate), 1,382 (EURL Lm reference strain), and 6,179 (food processing plant isolate). The contents of each were transferred into separate stomacher bags and homogenized in 25 mL of phosphate-buffered saline (PBS) using a stomacher (Seward 400, AGB Scientific, Dublin, Ireland) for 120 s at a high speed (260 rpm). These homogenates were used for all types of microbial analysis.
Growth potentials (log10 cfu g−1) calculated from median values were open field spinach (F1 Trumpet; summer harvest) = 2.59, polytunnel spinach (F1 Trumpet) = 1.40, open field rocket (Buzz) = 1.28, polytunnel rocket (Buzz) = 1.45, polytunnel rocket (Esmee) = 1.23, polytunnel spinach (F1 Cello) = 1.84, polytunnel kale (Nero di Toscana) = 2.56, and open field spinach (F1 Trumpet; winter harvest) = 1.65 as described recently (Culliney and Schmalenberger, 2022).
The associated average L. monocytogenes counts (log10 cfu g−1) across the five time points (± the relative increase or decrease from the previous time point) are displayed in Table 1.
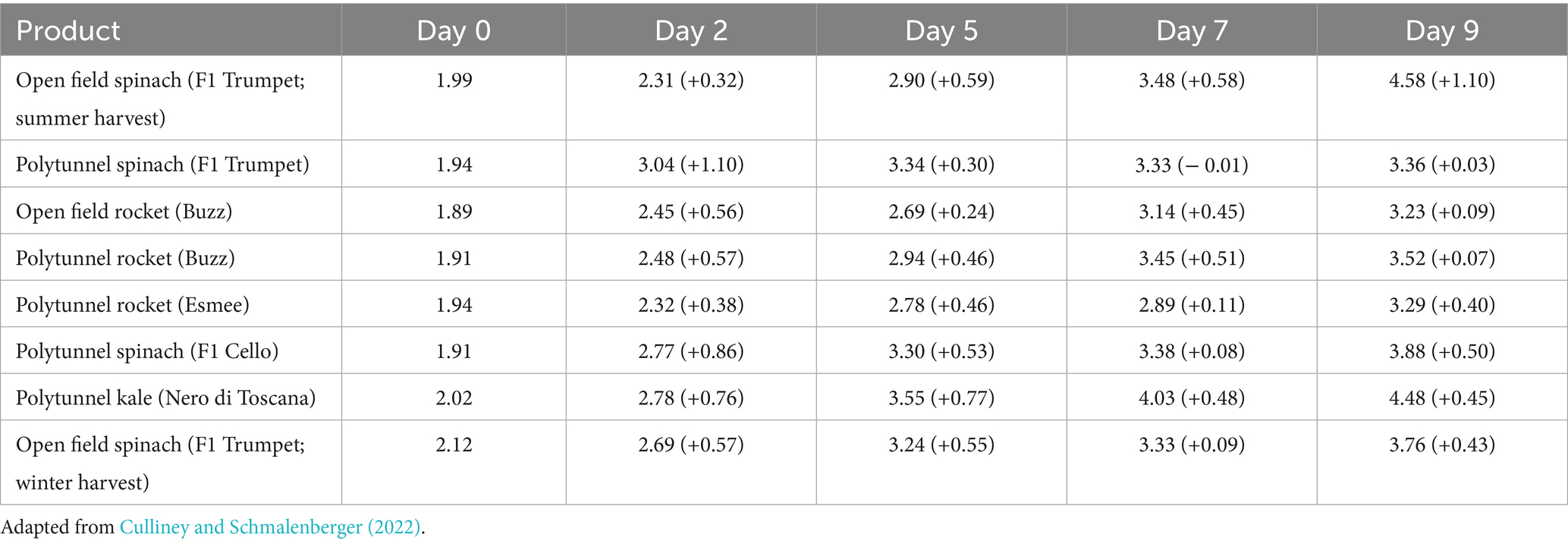
Table 1. Average Listeria monocytogenes counts (log10 cfu g−1 ± the relative increase or decrease from the previous time point) over time.
2.3 DNA extraction
The remaining homogenate suspensions obtained after microbial analysis were transferred into 50 mL conical tubes and centrifuged at 4,500g (15 min at 4°C). Supernatants were discarded, and the derived pellets were stored at −20°C. For DNA extraction, pellets were resuspended in 400 μL of PBS, and 100 μL was used for DNA extraction with the PowerFood DNA Isolation kit (MO BIO Laboratories, Carlsbad, CA, USA) according to the manufacturer’s instructions. The quantity and quality of the extracted DNA were determined with the Take3 plate in an Eon plate reader/incubator (BioTek, Winooski, VT, USA) (Culliney and Schmalenberger, 2024).
2.4 Next generation sequencing (NGS) analysis
All 160 samples were sent to the University of Minnesota Genomics Center (UMGC) for indexing and Illumina MiSeq (San Diego, CA) sequencing. Raw sequencing files were deposited in the Sequence Read Archive (National Center for Biotechnology Information [NCBI]) under the BioProject identification [ID] number: PRJNA117723.4. Bioinformatics analysis was performed using QIIME2 2021.11 (https://qiime2.org/) (Bolyen et al., 2019) as described recently by Culliney and Schmalenberger (2024). The paired-end sequences with quality of each group of 20 samples were demultiplexed and imported with metadata separately via a ManifestPhred33V2 file. This was followed by trimming and truncating (quality filtering at Q20) using the q2-dada2 plugin. Following this, the “qiime feature-table merge” and “qiime feature-table merge-seqs” plug-ins to merge feature tables and the representative amplicon sequence variants (ASV) were conducted, so the following group comparisons could be performed:
Comparison 1 (open field vs. polytunnel vs. plant species) consisted of open field spinach (F1 Trumpet), polytunnel spinach (F1 Trumpet), open field rocket (Buzz), and polytunnel rocket (Buzz).
Comparison 2 (variety vs. species) consisted of polytunnel spinach (F1 Trumpet), polytunnel rocket (Buzz), polytunnel rocket (Esmee), polytunnel spinach (F1 Cello), and polytunnel kale (Nero di Toscana).
Comparison 3 (seasonality) included open field spinach (F1 Trumpet; summer harvest) and open field spinach (F1 Trumpet; winter harvest).
Assigning taxonomic information to the ASV sequences was conducted using a pre-trained Naïve Bayes taxonomic classifier, which was trained on the Silva version 138.99% reference dataset where sequences were trimmed to represent only the region between the 515F/806R primers (V3–V4 region) as described previously (Culliney and Schmalenberger, 2025). Sequences not assigned to a phylum level, chloroplast, and mitochondrial sequences were removed using the filter-table method in the q2-taxa plugin. All subsequent analyses were conducted with both rarefied and unrarefied data. Even sampling depths for use in diversity metrics were for comparison 1: 11,519 → Retained 921,520 (29.48%) features in 80 (100.00%) samples at the specified sampling depth; for comparison 2: 3,117 → Retained 240,009 (9.99%) features in 77 (79.38%) samples at the specified sampling depth; and for comparison 3: 15,015 → Retained 600,600 (40.99%) features in 40 (100.00%) samples at the specified sampling depth. Alpha diversity metrics (observed ASVs, Shannon index, Pielou’s evenness, and Faith’s Phylogenetic Diversity) and beta diversity metrics (weighted unique fraction metric or UniFrac (Lozupone et al., 2007) and Bray–Curtis dissimilarity) using q2-diversity were estimated and viewed on Principal Coordinates Analysis (PCoA) Emperor plots. Analysis of Composition of Microbiomes (ANCOM) test in the q2-composition plugin was used to identify differentially abundant features. ANCOM identified individual taxa whose relative abundances are significantly different across groups. Relative abundance was calculated after conversion of the biome tables from QIIME2 to tsv files (phylum and family levels). Pearson’s correlation coefficient was determined to measure the strength and direction of the linear association between two variables (i.e., between L. monocytogenes populations and the corresponding relative abundance of each of the 20 most abundant families) for all groups over time (Sedgwick, 2012). Pearson’s correlation coefficient from <0.10 is a negligible correlation, 0.10–0.39 indicates weak correlations, 0.40–0.69 represents moderate correlations, while 0.70–0.89 indicates strong correlations, with >0.90 being very strong (Schober et al., 2018). Absolute abundances of bacterial taxa at the family and genus level were estimated by using the total heterotrophic counts published elsewhere (Culliney and Schmalenberger, 2022). Input, filtered, denoised, merged, non-chimeric reads, as well as chloroplast to total DNA content, are reported in Supplementary Tables S1–S3.
2.5 Statistical analysis
RStudio software (Posit, Boston, MA; version 4.1.1) was used for statistical analysis. In situations of normality (Shapiro–Wilk test) and homoscedasticity (Levene’s), a one-way analysis of variance (ANOVA) was conducted to compare input, filtered, denoised, merged, and non-chimeric reads between groups. The remainder of the statistical analysis for alpha and beta diversity metrics was conducted in QIIME2. For alpha diversity (observed ASVs, Shannon index, Pielou’s evenness, and Faith’s Phylogenetic Diversity (Faith, 1992)), comparisons among groups and pairwise comparisons were conducted through Kruskal–Wallis tests. Beta diversity was analyzed through the non-parametric permutation test, permutational multivariate analysis of variance (PERMANOVA) (999 permutations) (Anderson, 2017). Statistical significance was tested at p ≤ 0.05. In situations of normality (Shapiro–Wilk test) and homoscedasticity (Levene’s), a one-way ANOVA Tukey honestly significant difference (HSD) post hoc test applying Benjamini–Hochberg correction for multiple testing was conducted to compare relative abundances for all alpha diversity metrics and relative abundances across subgroups. In situations of non-normality, the Kruskal–Wallis rank sum test, using the kruskal.test function, and Dunn test post hoc analysis for multiple pairwise comparisons between groups were conducted, applying the Benjamini–Hochberg correction for multiple testing (false discovery rate was set at 10%). In situations of unequal variance, the oneway.test function was employed with var = F, and Games–Howell post hoc analysis.
3 Results
3.1 Comparison 1 (open field vs. polytunnel vs. plant species)
This section describes how alpha and beta diversities were shaped by the selection of different leafy vegetable plants (spinach and rocket) as well as other cultivation methods (open field and polytunnel) and how diversities evolved with storage. The primary assumption was that both plant species and environment affect alpha and beta diversities. In turn, differently developing phyllosphere communities were expected to affect L. monocytogenes growth over time, as well as the succession of the phyllosphere community over time.
3.1.1 Influence of cultivation method (polytunnel and open field) and plant species (spinach and rocket) on alpha diversity of a L. monocytogenes inoculated phyllosphere
On average, the richness and diversity for rocket (observed features, Shannon index and Faith’s Phylogenetic Diversity) were significantly greater for open field rocket (L. monocytogenes growth potential = 1.28 log10 cfu g−1) compared to polytunnel rocket produce (L. monocytogenes growth potential = 1.45 log10 cfu g−1). Pielou’s evenness was not significantly different between the open field rocket and polytunnel (p > 0.05). For spinach, Pielou’s evenness and diversity (Shannon index) were significantly higher for polytunnel spinach (L. monocytogenes growth potential = 1.40 log10 cfu g−1) compared to open field spinach (L. monocytogenes growth potential = 2.59 log10 cfu g−1). However, average observed features and Faith’s Phylogenetic Diversity values did not differ between spinach produce (p > 0.05). Except for Pielou’s evenness and Shannon index of polytunnel rocket, no other significant differences over time were observed for all alpha diversity metrics. Significant changes over time were only identified for polytunnel rocket (Shannon index and Pielou’s evenness) and open field rocket (Pielou’s evenness) (Supplementary Table S4), Rarefaction of sequencing reads did not influence alpha diversity metrics (Supplementary Table S5).
3.1.2 Influence of cultivation method (polytunnel and open field) and plant species (spinach and rocket) on beta diversity of a L. monocytogenes inoculated phyllosphere
All four groups, that is, open field rocket, polytunnel rocket, open field spinach, and polytunnel spinach produce, were all significantly different from each other (p = 0.001). When grouped by produce type, spinach and rocket produce were also significantly different (p = 0.001). Furthermore, when grouped by cultivation method, all polytunnel produce vs. all open field bacterial communities were significantly different (p = 0.001). While all four produce groups were significantly different, this was not always the case when compared at individual time points. The bacterial communities of all 5 time points of open field spinach were significantly different compared to polytunnel spinach produce (p = 0.026–0.038). The same was observed for polytunnel rocket compared to polytunnel spinach produce (p = 0.029–0.035). However, for open field vs. polytunnel rocket, significant differences between their bacterial communities were limited to days 0, 2, 5, and 9 (p = 0.019–0.037) and not day 7 (p = 0.057). Bacterial communities of open field rocket and spinach showed significant differences on day 0, 7, and 9 (p = 0.024–0.030) but not on day 5 or 7 (p = 0.069–0.084). Adjusting the p-value significance threshold with Benjamini–Hochberg correction did not change the statistical outcome of the tests. A visual representation of the bacterial beta diversity on a PCoA plot showed that communities of polytunnel rocket and spinach, as well as open field spinach and rocket, partially overlapped, while polytunnel rocket and open field rocket, as well as polytunnel spinach and open field spinach, were separated (Figure 1). Overall, beta diversity analyses highlighted the differences between the bacterial phyllosphere communities at the plant species and environment levels.
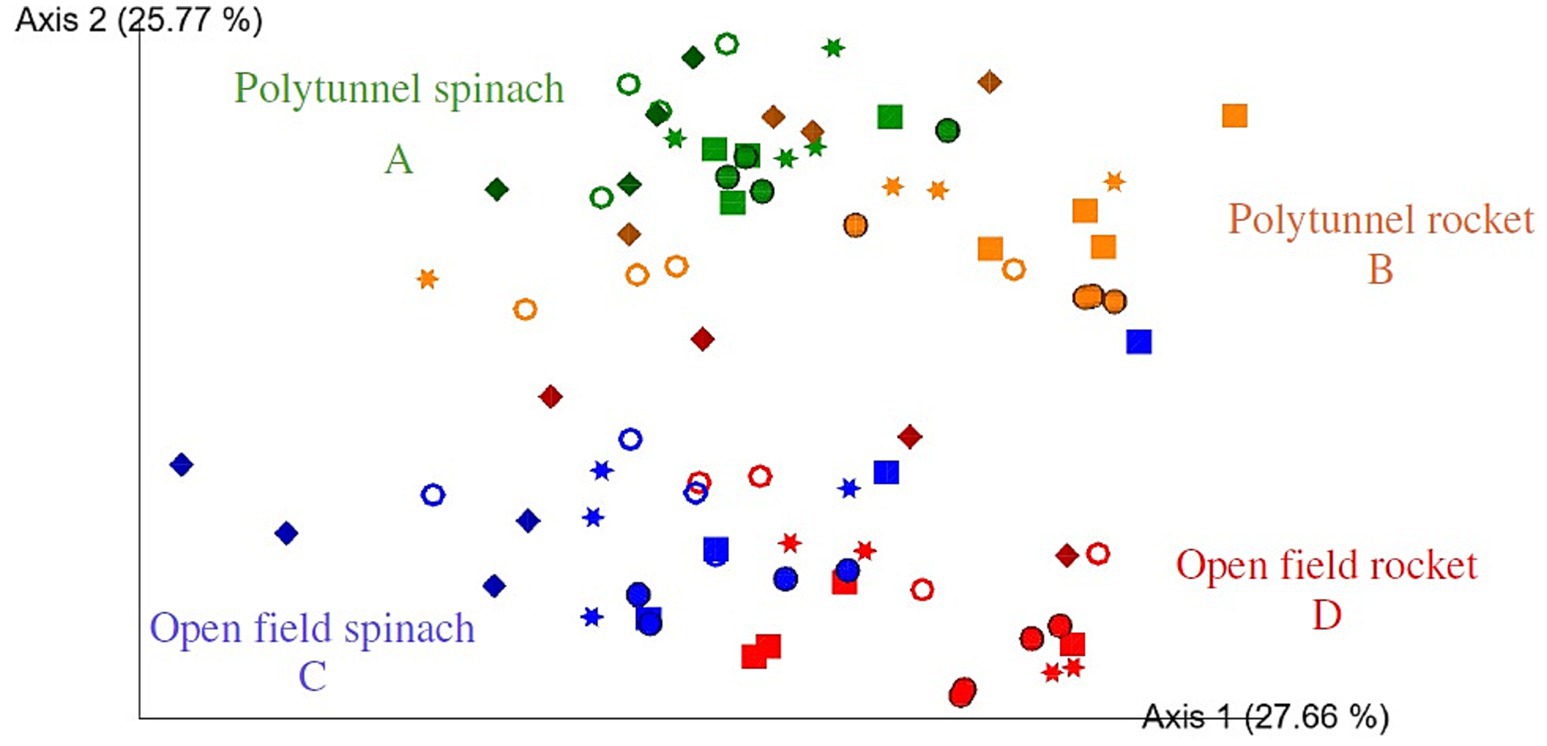
Figure 1. Two-dimensional Emperor (PCoA) plots showing beta diversity distances, that is, weighted UniFrac, among the different samples across open field rocket Buzz (red), open field spinach F1 Trumpet (blue), polytunnel rocket Buzz (orange), and polytunnel spinach F1 Trumpet (green) groups with rarefaction applied. Shapes revealed separations over time are day 0 = circle, day 2 = square, day 5 = star, day 7 = ring, and day 9 = diamond. Letters A-D indicate significant differences.
The phyllosphere of open field rocket produce changed significantly over time, that is, from days 0–9 and 2–9 (p = 0.028 and 0.030). For polytunnel rocket produce changes in phyllosphere structure occurred from days 0–9, 2–7, and 2–9 (p = 0.021–0.048). Open field spinach produce demonstrated significant changes in its phyllosphere from days 0–9, 2–9, and 5–9 (p = 0.014–0.030). Finally, polytunnel spinach exhibited the most significant changes in its phyllosphere community over time, that is, days 0–5, 0–7, 0–9, 2–7, 2–9, 5–7, and 5–9 (p = 0.019–0.041). Correcting the p-value significance threshold with Benjamini–Hochberg correction changed the outcome of only two statistical tests to non-significant, that is, polytunnel spinach produce from days 0–7 and polytunnel rocket produce from days 7–9. Overall, these findings demonstrate that bacterial phyllosphere communities change substantially during the storage period, even if they do not significantly change at each sampling time.
3.1.3 Influence of cultivation method (polytunnel and open field) and plant species (spinach and rocket) on phyla and family relative abundances of a L. monocytogenes inoculated phyllosphere
For all four groups, the three most abundant phyla were Pseudomonadota, Actinomycetota, and Bacteroidota, which comprised 89.64–94.82% of the phyllosphere bacterial communities (Figure 2). Over time, the total abundance of these three most abundant phyla ranged for (i) open field rocket from 88.96 to 94.44%, (ii) open field spinach from 92.28 to 96.15%, (iii) polytunnel rocket from 87.71 to 95.02%, and (iv) for polytunnel spinach from 88.21 to 91.26% of total phyla. At the phylum level, cultivation methods appeared to be a more influential determinant of relative bacterial community structure compared to plant species (Figure 2).
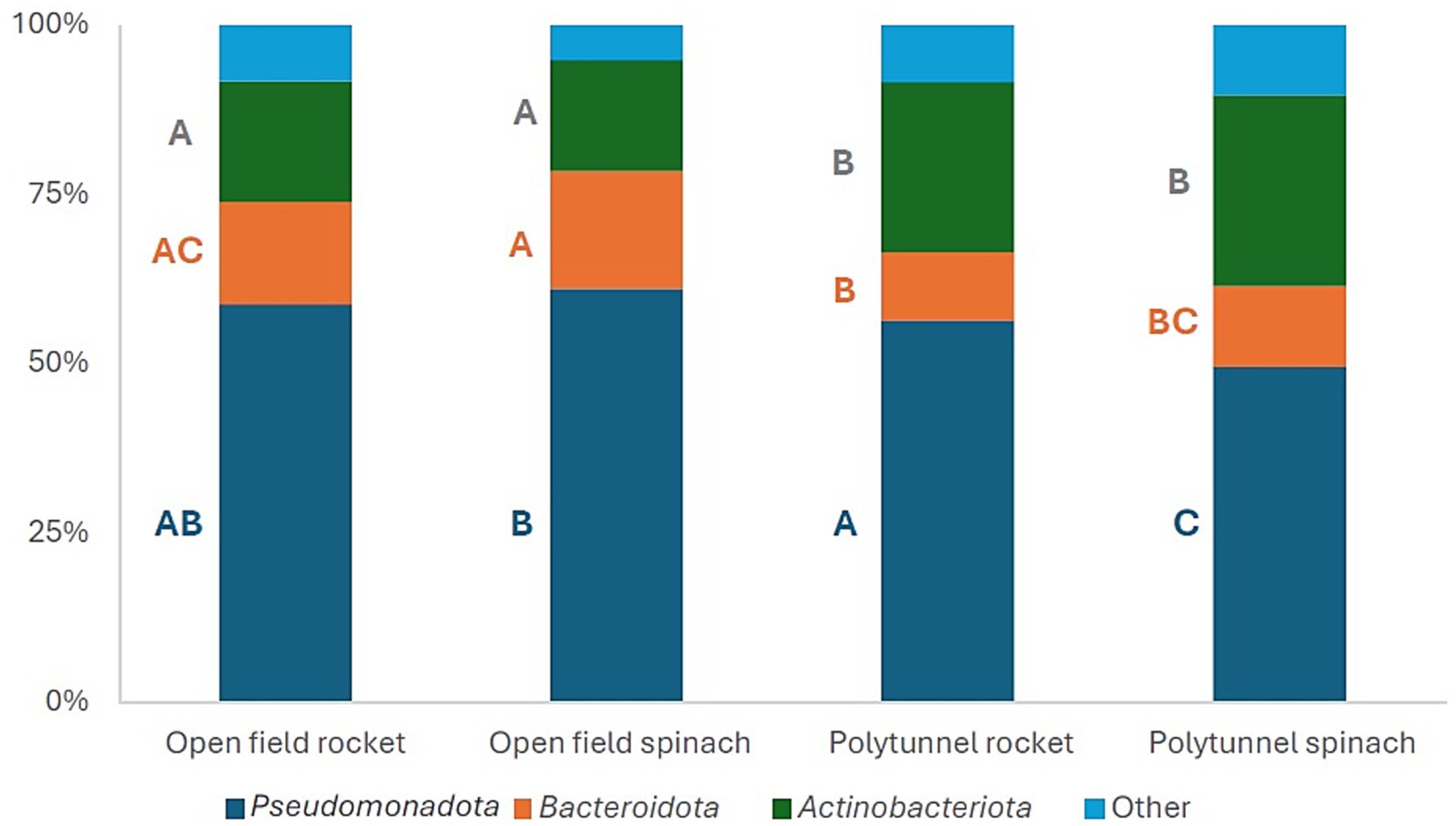
Figure 2. Mean relative abundances (%) of the three most abundant phyla of the 16S gene of the open field rocket Buzz, open field spinach F1 Trumpet, polytunnel rocket Buzz, and polytunnel spinach F1 Trumpet groups, with rarefaction applied. All remaining lower abundant phyla are combined in “Other.” A–C indicate significant differences between groups.
A total of 35 families common to all four groups were detected, albeit with some significant differences across groups and low relative abundances (Supplementary Table S6). Out of the 20 most abundant families of each group, 12 were shared by all four groups with substantially higher relative abundances. Open field rocket and polytunnel rocket produce shared 14 of their 20 most abundant families, four of which were significantly different in relative abundance. Open field rocket and open field spinach produce shared 16 families of their 20 most abundant, eight of which had significantly different relative abundances between the two groups. Open field spinach and polytunnel spinach produce had 16 families of their most abundant 20 in common, nine of which were significantly different. Polytunnel rocket and polytunnel spinach shared 16 out of 20 most abundant families, seven of which were significantly different (Table 2). Of the 15 families that showed differences in relative abundance, eight appeared to group by cultivation type (polytunnel and open field), while only four grouped by plant species. However, when total heterotrophic counts were used to estimate total abundances, the higher total abundance of bacteria in the spinach phyllospheres (open field and polytunnel) resulted in all but one family grouping according to plant species (Table 3 and Supplementary Table S7).
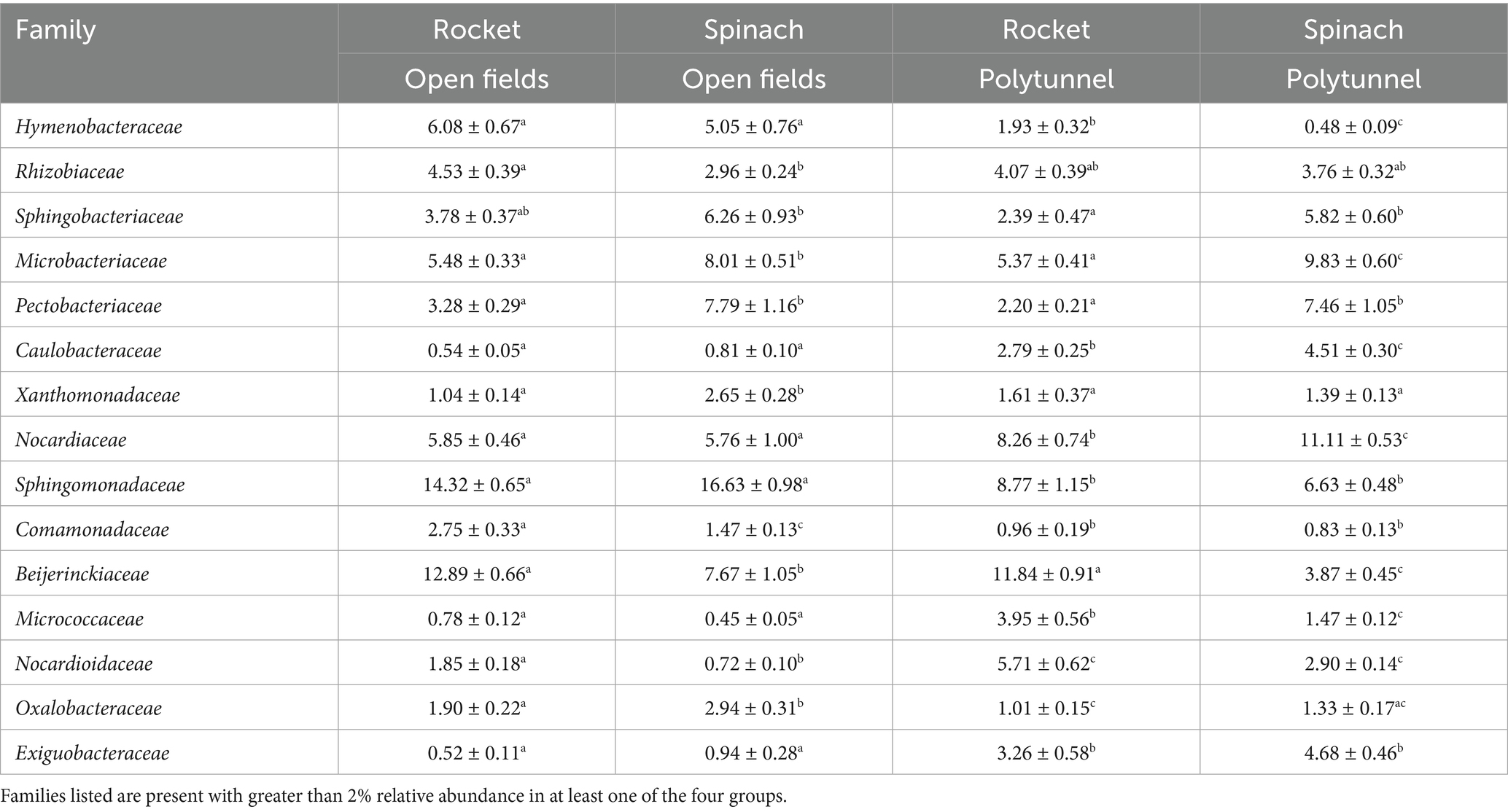
Table 2. Average relative abundance ± the standard error of families present different relative abundances in the phyllosphere of the open field vs. polytunnel and rocket vs. spinach with rarefaction applied. Letters a-c indicate significant differences.
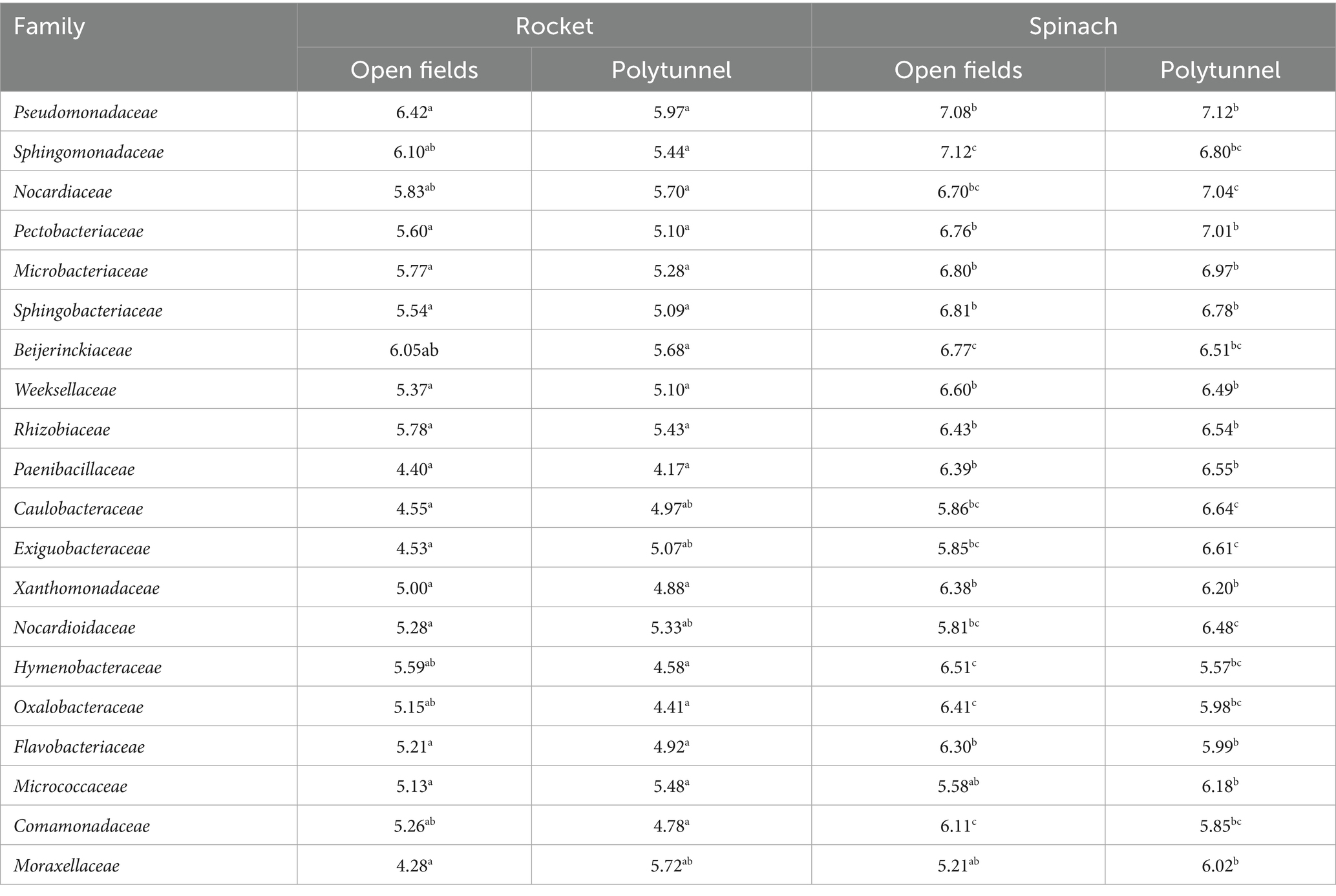
Table 3. Average absolute abundance (log 10, sequence data linked to total cfu counts) of the 20 most abundant families with significantly different abundances in the phyllosphere of the open field vs. polytunnel and rocket vs. spinach with rarefaction applied. Letters a-c indicate significant differences.
Overall, L. monocytogenes populations for all four groups showed common negative correlations with families Sphingomonadaceae and Beijerinckiaceae (Table 4 and Supplementary Tables S8–S11). Similarly, only two common positive correlations were identified, namely, Pseudomonadaceae and Xanthomonadaceae, between all four groups. L. monocytogenes populations of open field rocket had a strong positive correlation with three families, whereas a strong to very strong negative correlation was identified with seven families. L. monocytogenes populations of polytunnel rocket had a strong positive correlation with five families, and a strong to very strong negative correlation was revealed with seven families. For open field spinach L. monocytogenes populations showed a strong to very strong correlation with six families, while strong to very strong negative correlations were identified with only three. Finally, L. monocytogenes populations in polytunnel spinach showed a strong positive correlation with four families, and a strong to very strong negative correlation was identified among six families (Table 4).
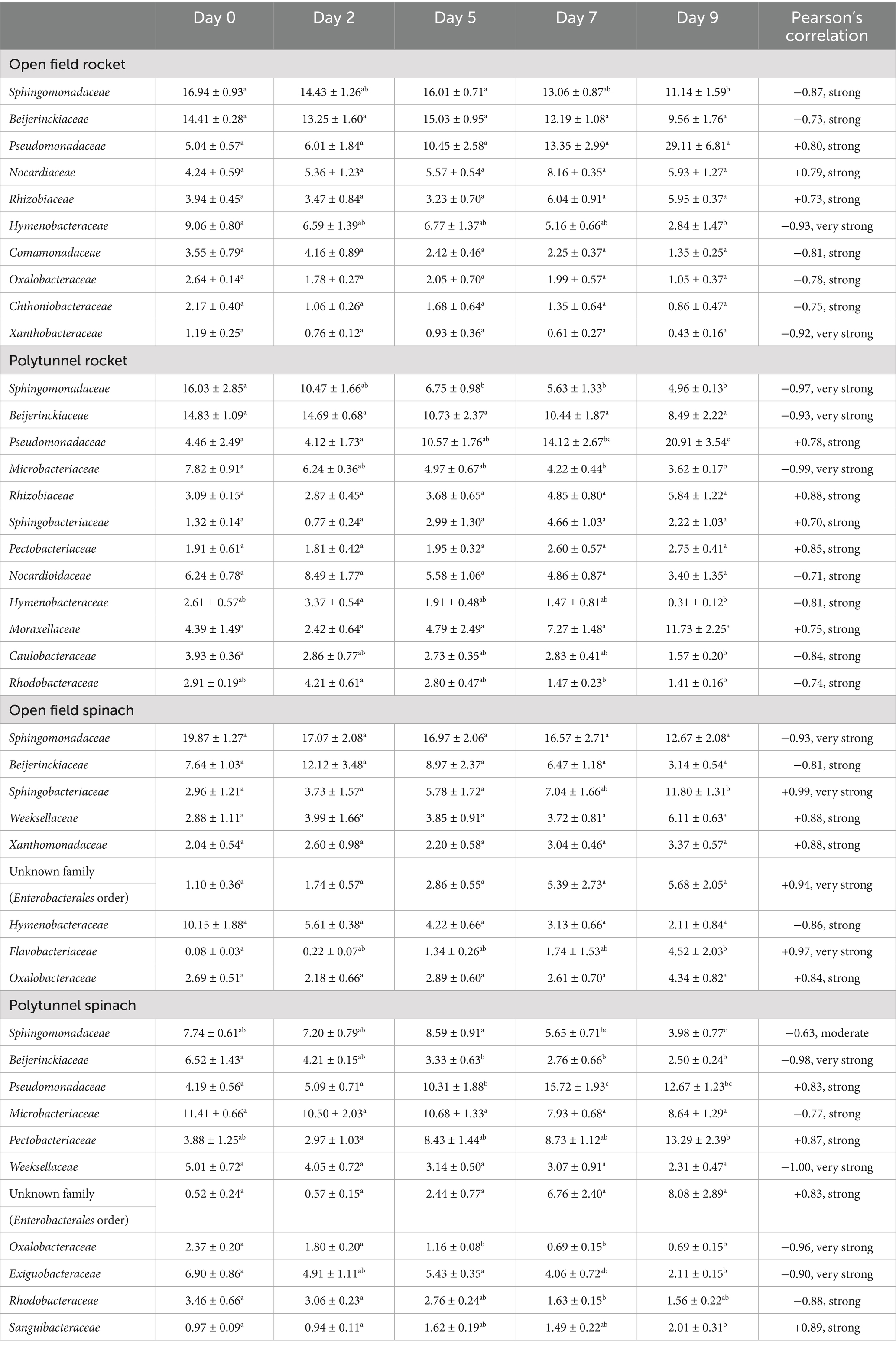
Table 4. Average relative abundance (% ± standard error) of families (16S ribosomal DNA [rDNA]) of open field, polytunnel, spinach and rocket across days 0, 2, 5, 7, and 9 rarefied with strong or very strong Pearson’s correlation coefficient (i.e., the strength and direction of the relationship between that specific family’s relative abundances and the corresponding Listeria monocytogenes populations over time). Letters a-c indicate significant differences between the groups.
Pseudomonadaceae content was not significantly different between all four groups (p = 0.277–0.849). On average, open field spinach displayed the highest average Pseudomonadaceae content, that is, 13.42%, followed by open field rocket 12.79%, polytunnel rocket 10.44%, and, finally, polytunnel spinach 9.60% (Supplementary Tables S8–S11). Therefore, open field spinach, which displayed the highest growth potential of 2.59 log10 cfu g−1 was associated with the highest average Pseudomonadaceae content, compared to spinach grown in a polytunnel setting, which displayed only 1.40 log10 cfu g−1. Relative abundance of Pseudomonadaceae content was compared for all four groups across the five different time points: At day 0, open fields spinach and open field rocket were significantly different (p < 0.001) and open field spinach and polytunnel spinach were significantly different (p < 0.001), remaining comparisons were not significantly different (p = 0.154–0.995). However, at days 2, 5, 7, and 9, no groups were significantly different from one another (p = 0.448–0.896, 0.161–0.984, 0.999, and 0.252–0.748). From days 7–9, Pseudomonadaceae content increased for open field rocket from 13.35 to 29.11% and polytunnel rocket from 14.12 to 20.91% (Supplementary Tables S8–S11). Open field spinach showed a moderate correlation (+0.66) between L. monocytogenes and Pseudomonadaceae compared to the strong positive correlation in polytunnel spinach. Similarly, open field and polytunnel rocket were strongly positively correlated between the two taxa (Table 4 and Supplementary Tables S8–S11).
Relative Pectobacteriaceae content (of which genus Dickeya was the sole genus) of polytunnel spinach produce displayed an increasing trend in relative abundance (3.9–13.3%) and strong positive correlation with L. monocytogenes populations from days 0–9 compared to open field spinach produce which displayed a decreasing trend (12.6–5.6%) and a moderate negative correlation with L. monocytogenes for the same period. The Pectobacteriaceae content remained consistently lower for rocket than for spinach. Moreover, the content of Pectobacteriaceae was positively correlated with L. monocytogenes in polytunnel rocket, which had a higher L. monocytogenes growth potential than open field rocket. Indeed, Pectobacteriaceae content of open field rocket correlated moderately negatively with decreasing L. monocytogenes populations (4.4–4.1%, Table 4 and Supplementary Tables S7–S10).
Polytunnel spinach retained the largest relative content of Lactobacillales (order level) (0.31%), followed by open field spinach (0.20%), open field rocket (0.16%), and, finally, polytunnel rocket (0.04%). Only the Lactobacillales content of open fields rocket vs. polytunnel rocket and polytunnel spinach vs. polytunnel rocket were significantly different (p = 0.019 and 0.027). Moreover, over time, significant differences were observed only at day 2, where the following comparisons were significantly different (p = 0.007, 0.019, and 0.019): open field rocket and polytunnel rocket; open field rocket and polytunnel spinach; and polytunnel rocket and open field spinach are significantly different. The relative abundance of Carnobacteriaceae (family of Lactobacillales) was significantly different between open field rocket and polytunnel spinach, as well as open field spinach and polytunnel spinach (p = 0.037 and 0.001), while all remaining group comparisons were not significantly different (p = 0.101–0.582). The Carnobacteriaceae content was on average 0.26% for polytunnel spinach, but was not at all present in open field spinach. On polytunnel and open field rocket, the relative abundance of Carnobacteriaceae was on average 0.01 and 0.02%, respectively.
Although detected and enumerated on Listeria selective agar, the Listeria genus, belonging to the Lactobacillales order, was not detected using NGS on open field spinach or open field rocket produce and was detected on only two of 20 samples belonging to polytunnel rocket produce, and in only one of 20 samples belonging to polytunnel spinach produce.
Overall, the majority of bacterial phyla and families were detected on spinach and rocket in both open fields and polytunnels. However, specific taxa and their change in abundance over time could be correlated with L. monocytogenes growth.
3.2 Comparison 2 (variety vs. species)
This section describes how their plant hosts shaped alpha and beta diversities at different taxonomic levels, that is, order (Caryophyllales for spinach, Brassicales for rocket and kale, referred here as species) vs. variety (Trumpet and Cello for spinach, Buzz and Esmee for rocket) and how diversity evolved during storage. The primary assumption was that both plant species and variety affect alpha and beta diversities. In turn, differently developing phyllosphere communities were expected to affect L. monocytogenes growth over time, as well as the succession of the phyllosphere community over time.
3.2.1 Influence of spinach and rocket cultivars as well as kale on alpha diversity of a L. monocytogenes inoculated phyllosphere
Rocket Buzz demonstrated the highest richness and diversity, followed by spinach F1 Trumpet, rocket Esmee, spinach F1 Cello, and, finally, kale Nero di Toscana (Supplementary Table S12). On average, observed features were all significantly different, with kale being the lowest, while rocket Buzz was the highest at day 5. For Faith’s Phylogenetic Diversity, kale Nero di Toscana and spinach F1 Cello were statistically similar; and spinach F1 Trumpet and rocket Esmee were statistically identical (p > 0.05). Only spinach F1 Trumpet and rocket Buzz had a significantly higher Shannon index than the remaining leafy vegetables. Spinach F1 Trumpet and rocket Buzz displayed the highest evenness (Pielou’s) that was substantially higher than for rocket Esmee, spinach F1 Cello, and kale Nero di Toscana.
Indeed, kale Nero di Toscana, with the lowest diversity, was associated with increased L. monocytogenes (growth potential = 2.56 log10 cfu g−1), and spinach F1 Cello, with the second-lowest diversity measurements, was associated with the second-highest growth potential, that is, 1.84 log10 cfu g−1. In contrast, the higher diversity groups spinach F1 Trumpet, rocket Esmee, and rocket Buzz were associated with lower growth potentials of L. monocytogenes (i.e., 1.23–1.45 log10 cfu g−1). Few significant differences were observed over time for alpha diversity metrics, which were limited to rocket Buzz for Shannon diversity (significantly highest on days 5 and 7) and Pielou’s evenness (significantly highest at day 5) (Supplementary Table S12).
The primary observation in this section is that differences in bacterial alpha diversity are related to plant taxonomic relatedness. These, in turn, may limit the growth and potential of L. monocytogenes when diversity is high.
3.2.2 Influence of spinach and rocket cultivars and kale on beta diversity of a L. monocytogenes inoculated phyllosphere
Based on the PCoA bi-plot, rocket Esmee was more separated on axis two from all other groups (Figure 3). Moreover, spinach F1 Cello and spinach F1 Trumpet partially overlapped, while spinach Trumpet also partially overlapped with rocket Buzz. However, significant differences were identified among all bacterial communities (p = 0.002–0.036), indicating that bacterial community structures are determined down to the plant variety level. When rocket and spinach varieties were grouped together, respectively, kale and rocket as well as kale and spinach were no longer significantly different (p = 0.140 and 0.059, respectively). However, spinach and rocket remained significantly different (p = 0.003). Adjusting the p-value significance threshold with Benjamini–Hochberg correction did not influence the outcome of the PERMANOVA tests. Comparisons within a vegetable variety over time were compromised for kale Nero di Toscana and rocket Esmee due to low sequence reads on days one to seven and day one, respectively.
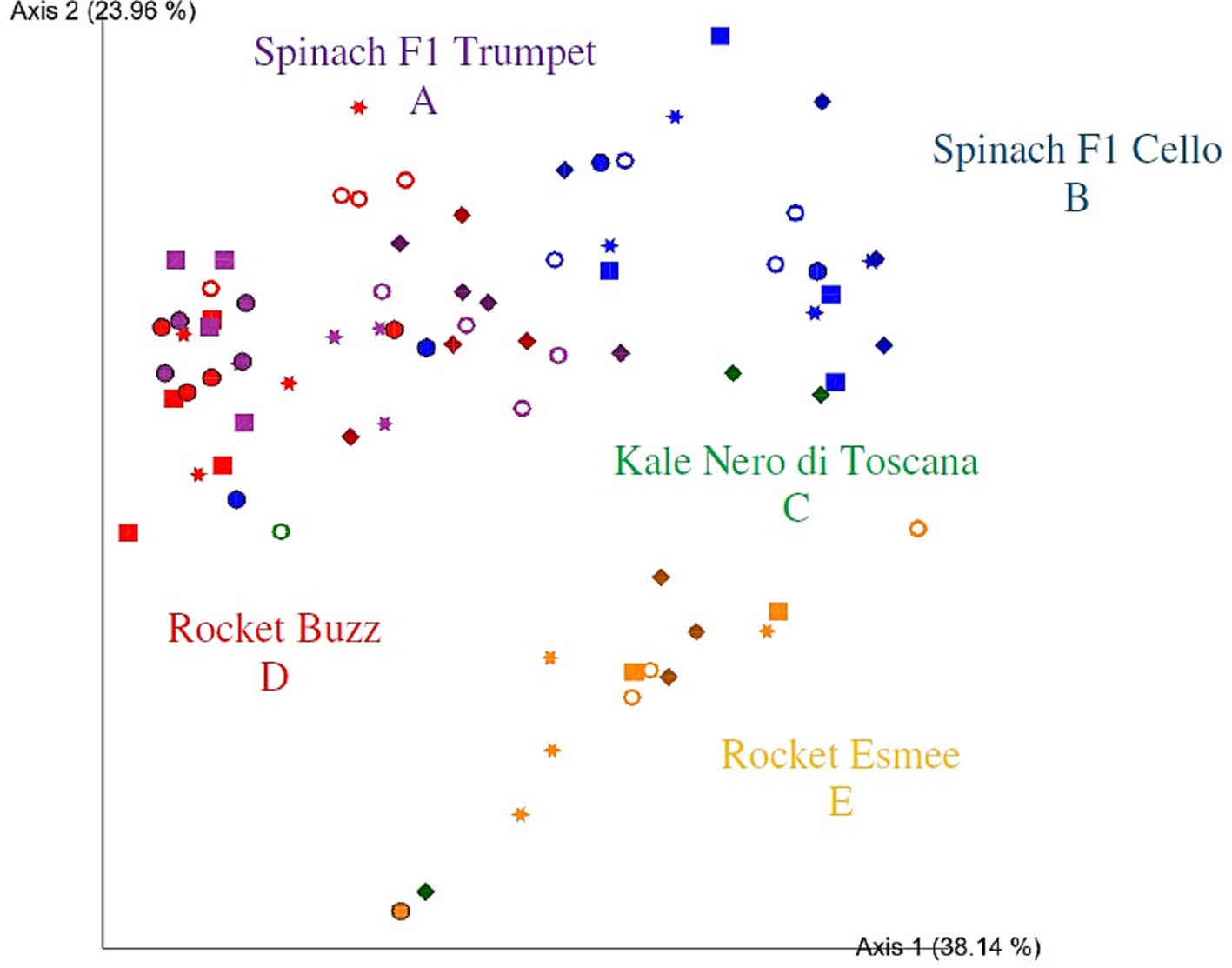
Figure 3. Two-dimensional Emperor (PCoA) plots showing beta diversity distances, that is, weighted UniFrac, among the different samples across polytunnel produce: rocket Esmee (orange), spinach F1 Cello (blue), kale Nero di Toscana (green), rocket Buzz (red) and spinach F1 Trumpet (purple) with rarefaction applied. Shapes revealed separations over time where day 0 = circle, day 2 = square, day 5 = star, day 7 = ring, and day 9 = diamond. 16 and 7 samples with low bacterial reads were removed for kale Nero di Toscana and rocket Esmee, respectively. Letters (A-E) indicate significant differences.
3.2.3 Influence of spinach and rocket cultivars and kale on phyla and family relative abundances of a L. monocytogenes inoculated phyllosphere
For all five groups, the four most abundant phyla were Pseudomonadota, Actinobacteriota, Bacteroidota, and Bacillota, which comprised 95.61–99.58% of the phyllosphere bacterial communities (Figure 4). Over time, the total abundance of these four most abundant phyla remained consistent across all five groups, ranging from 93.15 to 99.92%.
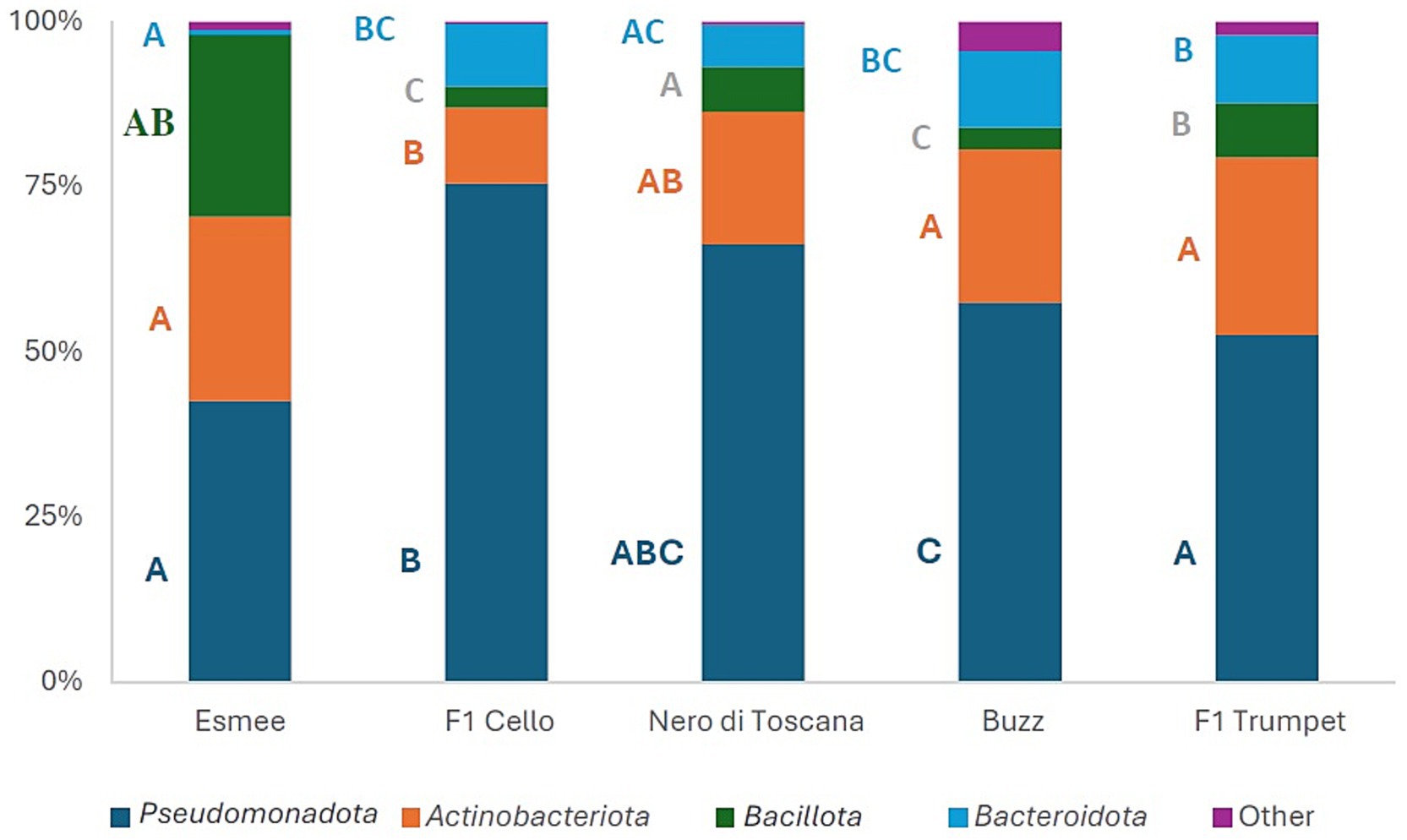
Figure 4. Mean relative abundances (%) of the four most abundant phyla of the 16S gene of the polytunnel produce: rocket Esmee, spinach F1 Cello, kale Nero di Toscana, rocket Buzz, and spinach F1 Trumpet, with rarefaction applied. All remaining lower abundant phyla are combined in “Other,” Letters A to C indicate significant differences between groups.
At the family level, 32 were common to all 5 groups, and their relative abundance was overall significantly affected by the leafy vegetable (Supplementary Table S13). Of the 20 most abundant families, 11, 3, 8, and 0 families showed significant changes in relative abundance over time for spinach F1 Trumpet, spinach F1 Cello, rocket Buzz, and rocket Esmee, respectively (Supplementary Tables S14–S17).
Spinach F1 Cello and Trumpet shared 17 out of the 20 most abundant families, whereas rocket varieties Esmee and Buzz only shared 14 families, of which the relative abundances of 10 were significantly different (p < 0.05). Since only four (days 7 and 9) kale samples were obtained with a sufficient number of reads for analysis, comparisons at the family level for kale were avoided.
Overall, L. monocytogenes populations for both spinach and rocket varieties exhibited only one common negative correlation with the Sphingomonadaceae and one common positive correlation with the Pseudomonadaceae (Table 4 and Supplementary Tables S14–S17). L. monocytogenes populations in spinach F1 Trumpet showed a strong positive correlation with Pseudomonadaceae, while a strong to very strong negative correlation was identified with six other families (Table 4). L. monocytogenes populations of spinach F1 Cello had a strong and very strong positive correlation with Flavobacteriaceae and Pseudomonadaceae, respectively, whereas a strong negative correlation was identified with four families. L. monocytogenes populations of rocket Buzz had a strong positive correlation with five families and a strong to very strong negative correlation with seven families. For rocket Esmee, a strong positive correlation with families Pseudomonadaceae and Xanthomonadaceae and a strong to very strong negative correlation was observed with eight families (Table 4 and Supplementary Tables S14–S17).
Spinach F1 Cello had an average higher, although not significant, Pseudomonadaceae content (19.0%) compared to spinach F1 Trumpet (9.6%). Rocket Esmee had a significantly (p < 0.05) higher average Pseudomonadaceae content (28.1%) compared to rocket Buzz (10.8%). However, at the genus level, absolute numbers (based on total heterotrophic counts) of Pseudomonas sp. are only clearly higher at days 2, 5, and 7 in variety Esmee when compared to Buzz (Supplementary Table S18). For both spinach varieties and rocket Buzz, Pseudomonadaceae content appeared to drastically and significantly increase (3.3–5.5-fold) over time. Pectobacteriaceae content (genus Dickeya) of polytunnel spinach F1 Trumpet produce displayed an increasing trend in relative abundance from days 0–9 (3.8–13.6%) compared to spinach F1 Cello produce, which displayed a decreasing trend (28.5–6.4%) for the same period. Moreover, the Pectobacteriaceae content (genus Dickeya) of polytunnel rocket Buzz from days 0–9 remained consistent (1.9–2.6%), whereas it increased substantially on rocket Esmee from 1.0 to 6.9%. Spinach F1 Cello had a Lactobacillales (order level) content of 0.03% compared to 0.35% for spinach F1 Trumpet. The Lactobacillales content of rocket Esmee and rocket Buzz was similar to F1 Cello (0.01 and 0.06%). The average Carnobacteriaceae content of spinach F1 Cello (L. monocytogenes growth potential = 1.84 log10 cfu g−1) was 0.03% and significantly different compared to 0.26% (0.69 and 0.33% at days 7 and 9, respectively) for spinach F1 Trumpet (p = 0.048). The Carnobacteriaceae content of the remaining groups ranged from 0.00 to 0.01%. Listeria (genus) content was only 0.01% for rocket Esmee and spinach F1 Cello, and not detected in rocket Buzz or spinach F1 Trumpet. When samples with a low number of reads were included in the analysis, Listeria was identified in 14 out of all 20 kale Nero di Toscana samples. In stark contrast, Listeria was detected in two of 20 samples in rocket Esmee (0.05 and 0.01%), spinach F1 Cello (0.04 and 0.07%), and rocket Buzz (both 0.01%), and in only one of 20 samples belonging to spinach F1 Trumpet (0.02%). Bacillaceae showed a strong negative correlation with L. monocytogenes growth in rocket Esmee, but not in Buzz or both spinach varieties. However, differences between Esmee and Buzz were also present at the genus level of Bacillaceae, with Bacillus sp. about four-fold higher in Esmee on days 2, 5, and 7 than in Buzz (Supplementary Table S18).
Similarly to findings at 3.1.3, the majority of bacterial phyla and families were detected on spinach, rocket, and kale. Similarly, specific taxa and their change in abundance over time appear to be correlated with L. monocytogenes growth.
3.3 Comparison 3 (seasonality)
This section describes how alpha and beta diversities were shaped by seasonality (winter and summer) in spinach Trumpet and how diversities evolved during storage. The primary assumption was that winter vs. summer production affects alpha and beta diversities. In turn, differently developing phyllosphere communities were expected to affect L. monocytogenes growth over time, as well as the succession of the phyllosphere community over time.
3.3.1 Influence of time of harvest on alpha diversity of a L. monocytogenes inoculated spinach phyllosphere
All alpha diversity metrics did not significantly change over time (p > 0.05). The number of observed features (ASVs) from summer produce (295–351) and winter produce (308–336) was statistically similar. The same findings were observed for Faith’s Phylogenetic Diversity (18.1–24.9). In contrast, the Shannon index was on average significantly greater for winter produce (6.2–6.8, L. monocytogenes growth potential = 1.65 log10 cfu g−1), compared to summer produce (5.8–6.4, L. monocytogenes growth potential = 2.59 log10 cfu g−1). However, these values did not change significantly over time (days 0–9) for either group (p > 0.05). Evenness was also considerably higher for winter produce (0.74–0.81) compared to summer produce (0.72–0.77). Compared to comparisons 1 and 3, here the changes of winter to summer produce had a less pronounced effect on alpha diversity.
3.3.2 Influence of growing season on beta diversity of a L. monocytogenes inoculated spinach phyllosphere
Based on the PCoA plot (Figure 5), separations were visually identified between winter and summer groups over time, evident between all data points, as confirmed by PERMAMOVA (p = 0.001). Adjusting the p-value significance threshold with Benjamini–Hochberg correction did not alter any significances. Separations for each day 0, 2, 5, 7, and 9 (summer vs. winter) were significant (p = 0.026–0.048). A visual separation according to time point within winter and summer produce was also clearly visible (Figure 5). For summer produce, statistically significant separations were observed over time for days 0–9, 2–9, and 5–9 (p = 0.022–0.032). For winter produce separations days (0–9, 2–9, 0–7, and 2–7) were significant (p = 0.026–0.034). However, after applying Benjamini–Hochberg correction, none of these results remained significantly different. Overall, when compared to alpha diversity, the beta diversity was affected by seasonality.
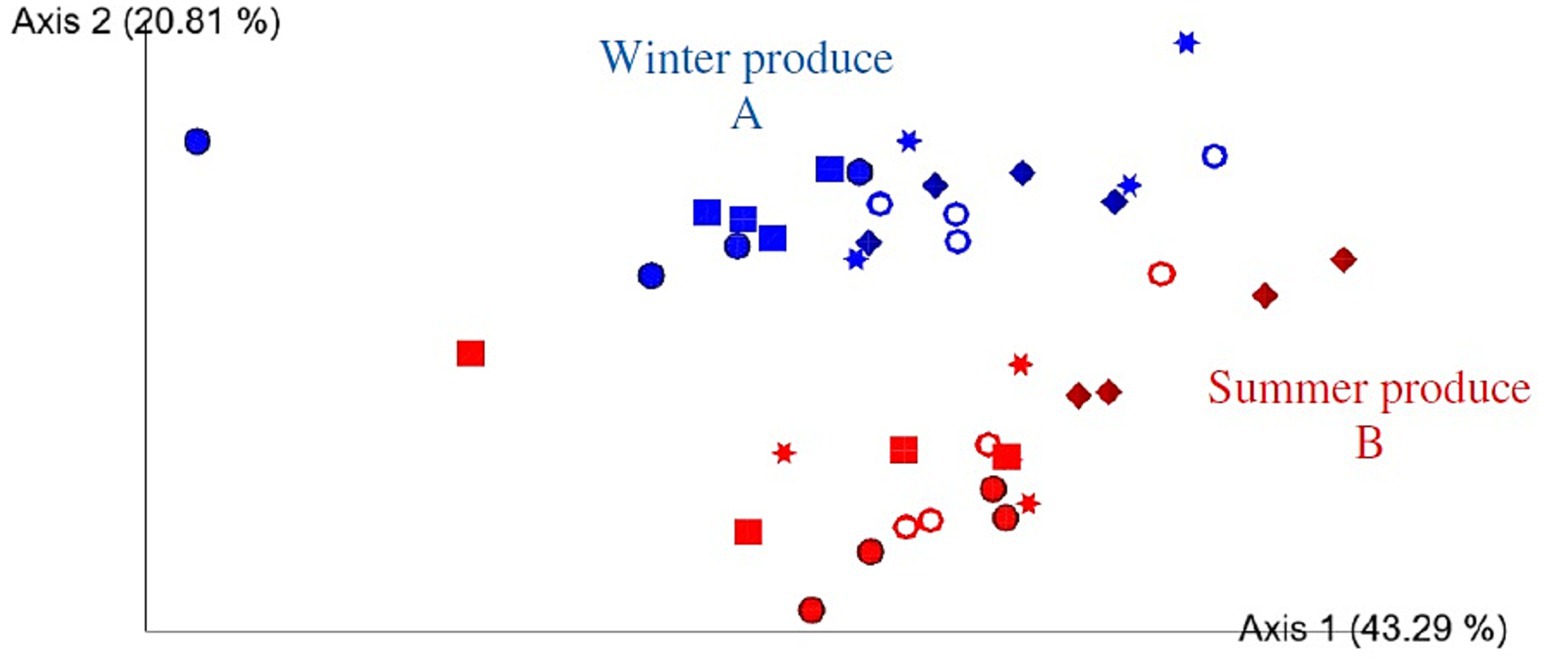
Figure 5. Two-dimensional Emperor (PCoA) plots showing beta diversity distances, that is, weighted UniFrac, among the different samples across open field spinach: winter (blue) and summer (red) produce, with rarefaction applied. Shapes revealed separations over time where day 0 = circle, day 2 = square, day 5 = star, day 7 = ring, and day 9 = diamond. A and B indicate significant differences.
3.3.3 Influence of time of harvest on phyla and family relative abundance of a L. monocytogenes inoculated spinach phyllosphere
For Winter and Summer produce, the most abundant four phyla were Pseudomonadota, Actinobacteriota, Bacteroidota, and Bacillota, which comprised 98.94 and 98.86% of the phyllosphere bacterial communities (Supplementary Figure S1). The only significant difference between summer and winter produce at the phylum level was that the Bacillota were significantly more abundant in the summer produce (p < 0.05). Over time (days 0–9), the total abundance of these four most abundant phyla remained consistent for both groups, ranging from 98.50 to 99.62%.
Winter and summer produce shared 31 families (Supplementary Table S19). However, 20 of those families had significantly different relative abundances between groups (p < 0.05). A total of 17 of the most abundant 20 families were shared between both groups, of which eight had significantly different relative abundances, that is, order Enterobacterales (family unknown), Sphingomonadaceae, Oxalobacteraceae, Rhizobiaceae, Caulobacteraceae, Nocardioidaceae, Rhodanobacteraceae, and Nocardiaceae.
ANCOM revealed 11 differentially abundant families, that is, Paenibacillaceae, order Saccharimonadales family Unknown, Myxococcaceae, Phormidiaceae, Deinococcaceae, Rhodobacteraceae, Spirosomaceae, Moraxellaceae, Rhodanobacteraceae, Hymenobacteraceae, and Nocardioidaceae, between winter and summer. Pseudomonadaceae content was not significantly different between the summer and winter produce (p = 0.905) or across all time points from days 0–9 (p = 0.075, 0.149, 0.255, 0.051, and 0.527). Similarly, Lactobacillales (order level) content was not significantly different between the summer and winter produce (p = 0.322) or across days 0–9 (p = 0.387, 0.638, 0.773, 0.767, and 0.314). Although Lactobacillales relative abundance was less than 1% for all produce, Lactobacillales content was on average higher for winter produce (0.34%), compared to summer produce (0.20%). The relative abundance of Lactobacillales, that is, Lactococcus genus, remained consistent throughout for winter produce, but for summer produce dropped from 0.52 to 0.22 to 0.03% from days 5–9, coinciding with increases in L. monocytogenes growth, such levels of L. monocytogenes growth which were not observed on winter produce. Moreover, in contrast to polytunnel spinach produce (Comparisons 1 and 2), Carnobacteriaceae was not present on open field spinach produce from summer or winter produce (Supplementary Table S19).
L. monocytogenes populations for the summer and winter groups showed five common negative correlations with the families Sphingomonadaceae, Microbacteriaceae, Beijerinckiaceae, Nocardiaceae, and Nocardioidaceae. Seven common positive correlations were identified with families Pseudomonadaceae, Sphingobacteriaceae, Weeksellaceae, an unknown family (Enterobacterales order), Rhizobiaceae, Oxalobacteraceae, and Xanthomonadaceae (Supplementary Tables S20, S21). L. monocytogenes populations of winter produce had a strong to very strong positive correlation with six families and a strong to very strong negative correlation with five families. L. monocytogenes populations in summer produce showed a strong to very strong positive correlation with six families and a strong to very strong negative correlation with four families (Table 5 and Supplementary Tables S20, S21). Similar to findings at 3.1.3 and 3.2.3, the majority of bacterial phyla and families were detected on spinach summer and winter produce. Again, specific taxa and their change in abundance over time appear to be correlated with L. monocytogenes growth. Although detected and enumerated on Listeria selective agar, the Listeria genus, belonging to the Lactobacillales order, was not detected using NGS on either winter or summer open field spinach produce (F1 Trumpet variety).
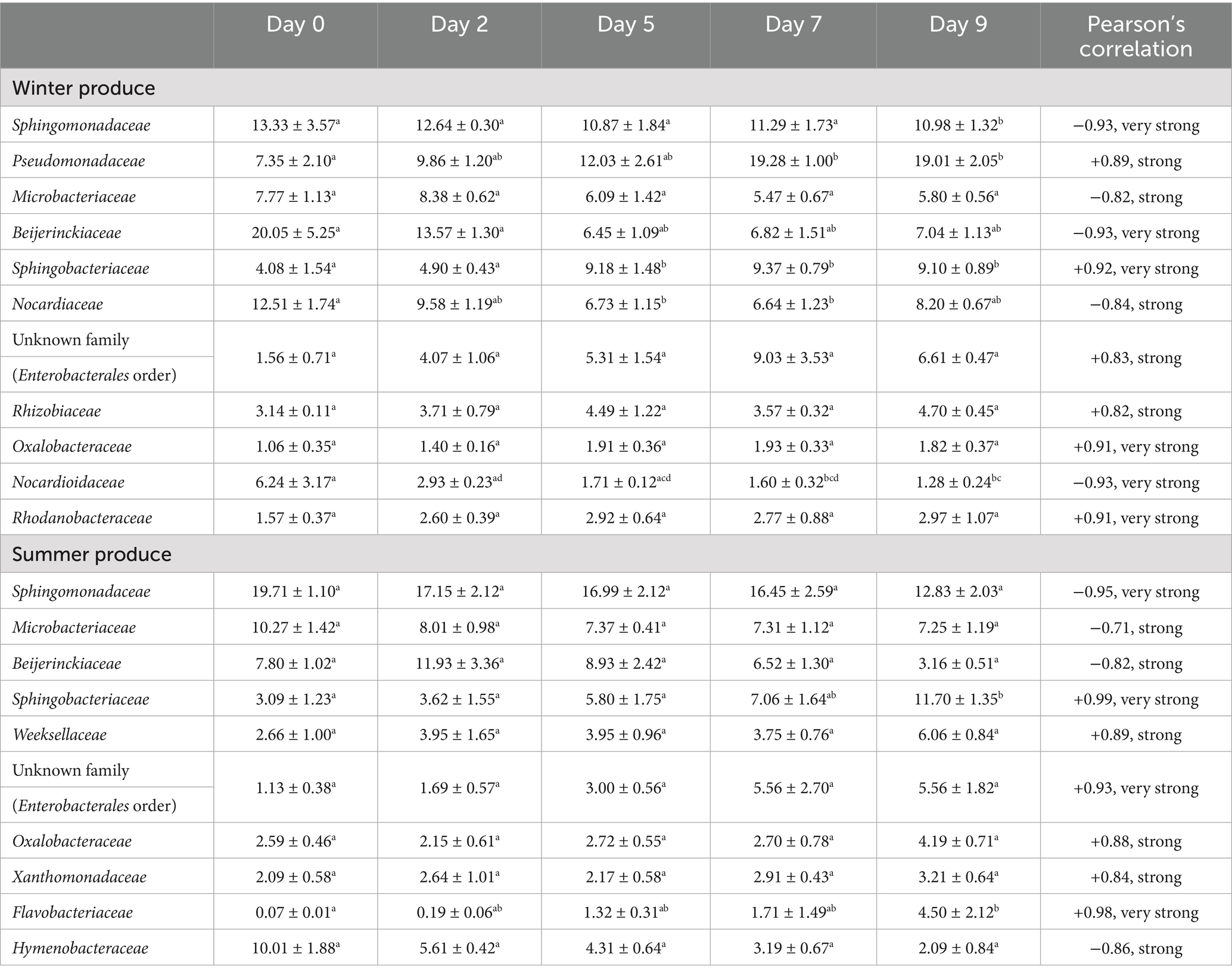
Table 5. Average relative abundance (% ± standard errors) of families (16S rDNA) of the summer open field spinach variety F1 Trumpet produce across days 0, 2, 5, 7, and 9 rarefied with strong to very strong Pearson’s correlation coefficient (i.e., the strength and direction of the relationship between that specific family’s relative abundances and the corresponding Listeria monocytogenes populations over time). Letters a–d indicate significant differences.
4 Discussion
The purpose of this study was to describe the influence of leafy vegetable cultivation conditions (cultivation method, plant species, cultivar, and season of harvest) on the development of the phyllosphere bacteriome and the effect on epiphytic L. monocytogenes growth.
4.1 Effects of cultivation conditions (open field vs. polytunnel), plant species (spinach and rocket), and cultivars (varieties)
Previous research assessing the effect of nitrogen fertilizer and leaf mineral content revealed that plant species alone, like spinach and rocket, influence the development of the phyllosphere (Darlison et al., 2019). However, the current study further revealed that the vegetable cultivation method had the strongest influence on the bacterial phyllosphere community structure. At the same time, plant species had a more pronounced effect on the overall abundance of phyllosphere bacteria. Here, polytunnel and open field cultivation of rocket and spinach displayed more similar phyllosphere bacterial communities compared to plant species alone. Additionally, the phyllosphere bacterial communities of various rocket and spinach cultivars were found to be significantly different in the present study. Previous research has identified the presence of microbe-plant variety interactions in field-grown lettuce. Dominated by Pseudomonadaceae and Enterobacteriaceae families, a clone library of three lettuce cultivars revealed significant differences between the relative abundances of genera belonging to the Enterobacteriaceae family, including Erwinia and Enterobacter (Hunter et al., 2010). While another study of the microbial diversity and structure of the phyllosphere of Alfalfa (Medicago sativa L.) identified significant effects of the season and the site where the plants were cultivated in open fields, no significant differences were detected between the two tested varieties (Zhang et al., 2022). While lettuce and alfalfa are bred for cultivation, and both plants are cultivated through a broad range of varieties, only lettuce is bred with the aim of human consumption of the leaves, as is the case for spinach and rocket. One may speculate that the breeding focus of lettuce, spinach, and rocket is primarily on the consumer experience of eating the leaves; thus, different varieties may differ more substantially in their leaf structure than this is the case for other plant varieties that are bred for livestock feeding.
4.2 Correlations between in situ phyllosphere taxa and inoculated L. monocytogenes growth
A novel aspect of the present study was the identification of the presence or absence of bacteria, and their shifts in relative abundance, which may be of potential importance to the L. monocytogenes growth. For example, Pseudomonadaceae, which are of high abundance and are associated with the hydrolysis of proteins into amino acids, can induce the stimulation of L. monocytogenes growth (Marshall et al., 1992; Zilelidou and Skandamis, 2018). Contrariwise, Lactobacillales that were present in low abundance are commonly associated with decreased L. monocytogenes survival due to their competitive growth abilities (Østergaard et al., 2014). Indeed, the L. monocytogenes growth-enhancing Pseudomonas species has previously been associated with spinach leaves of neutral pH (Babic et al., 1996). Additionally, as Pseudomonas species are pectolytic, their presence is positively correlated with the degradation and spoilage of such leafy vegetables, which increases during storage, as observed in the present study. Exposure to solar active radiation influenced the relative abundance of the Betaproteobacteria and Gammaproteobacteria, which is the class level of the Pseudomonadales order (Truchado et al., 2017). Relative abundances of Gammaproteobacteria were not significantly different with reductions in cumulative photosynthetically active radiation (PAR) from 4,889 to 3,602 μmol m−2 s−1, but were substantially higher when cumulative PAR was 3,115 μmol m−2 s−1. In the present study, the protection of spinach and rocket produce from PAR by cultivating in a polytunnel setting, compared to an open field, did not lead to significantly higher Pseudomonadaceae content.
In the present study, L. monocytogenes populations of all groups were positively correlated with Pseudomonadaceae content. In particular, Pseudomonadaceae content appeared to be most important for L. monocytogenes growth on spinach F1 Trumpet produce, especially from day 7 to 9. Relative increases from days 7–9 for open field spinach produce were associated with L. monocytogenes’ most significant increase during the same period. Conversely, when Pseudomonadaceae content decreased from days 7–9 for polytunnel spinach, the L. monocytogenes populations remained stationary. Indeed, amino acids hydrolyzed from proteins by Pseudomonadaceae are localized within the cellular tissue of leafy vegetables (Koseki and Isobe, 2005; Vacher et al., 2016). Open field spinach produce is likely exposed to more liquids on leaf surfaces due to wetter outdoor climatic conditions, potentially causing higher leaching of those nutrients for L. monocytogenes utilization compared to polytunnel produce (Tukey, 1970; Comte et al., 2012; Vacher et al., 2016; Kyere et al., 2019; Zhu et al., 2022). Overall, spinach contained higher total abundances of Pseudomonadaceae than rocket. The leaf physiology of rocket, that is, less surface area and fewer stomata (Maylani et al., 2020) might have prevented the release of some nutrients, that is, amino acids (hydrolyzed protein) for L. monocytogenes utilization (Culliney and Schmalenberger, 2022), thus limiting the growth of bacteria more than this is the case for spinach. However, at the genus level, higher numbers of Pseudomonas sp. on Esmee than on Buzz do not seem to influence the growth potentials of L. monocytogenes.
Moreover, a higher Lactobacillales content was associated with the lower L. monocytogenes growth potential compared to both rocket Buzz and spinach F1 Trumpet produce. A recent study of a mixed spinach salad containing chicken meat identified low levels of Lactobacillales content, consisting of only Carnobacteriaceae and Enterococcaceae, which increased from 0 to 1% at day 7 of storage at 15°C (Söderqvist et al., 2017a). There, the authors did not detect any Lactobacillales on plain baby spinach. In another study, storage of romaine lettuce over 14 days revealed a significant increase in Carnobacteriaceae’s relative abundance from 1.93 to 52.26% and a non-significant increase in Pseudomonadaceae content from 13.38 to 21.20% (Dharmarha et al., 2019). Both bacteriocin-producing, for example, Divercin AS7 and non-bacteriocin-producing species of Carnobacteria, C. divergens and C. maltaromaticum, have been demonstrated to be effective in vitro at minimizing epithelial cell invasion caused by L. monocytogenes Scott A (Pilchová et al., 2016) and Listeria spp. (Marković et al., 2022). Carnobacteria piscicola LK5 and 2762 strains suppressed the maximum population density reached by L. monocytogenes in brain heart infusion broth (Buchanan and Bagi, 1997). However, little of the L. monocytogenes maximum population density suppression was due to the strain’s bacteriocin production. Those authors suggested that the suppression potential of the strain C. piscicola 2762 was not caused by peroxide, pH depression, or oxygen depletion, but was caused by induced nutrient depletion. In the present study, Carnobacteriaceae were absent from open field spinach produce but present in significantly higher quantities on polytunnel spinach produce, particularly at days 7 and 9. This may have also inhibited the growth L. monocytogenes, leading to its lower growth potential. Moreover, spinach F1 Cello variety had no Carnobacteriaceae present, but significantly higher Pseudomonadaceae content (+9.43%) compared to spinach F1 Trumpet from the polytunnel setting. Thus, potentially explaining the higher growth potential of the spinach F1 Cello variety. However, albeit a higher Pseudomonadaceae content (+5.58%), polytunnel spinach F1 Cello may have caused less leaching of nutrients (hydrolyzed amino acids) due to being less exposed to rain and liquid on surface of the leaf, thus resulting in lower L. monocytogenes growth potential for spinach F1 Cello (1.84 log10 cfu g−1) compared to open field spinach F1 Trumpet (2.59 log10 cfu g−1).
4.3 Effect of cultivation conditions (open field, polytunnel, species, and variety) and bacterial taxa abundance on L. monocytogenes growth potential
In addition to Carnobacteriaceae, polytunnel spinach F1 Trumpet, which displayed a lower growth potential of L. monocytogenes, showed an increasing trend in Pectobacteriaceae content (genus Dickeya). In contrast, open field spinach, which was associated with a larger L. monocytogenes growth potential, exhibited a decreasing trend in Pectobacteriaceae content (genus Dickeya). Additionally, polytunnel spinach F1 Cello, which had a decreasing trend of Pectobacteriaceae content (genus Dickeya), was associated with a higher L. monocytogenes growth potential than polytunnel spinach F1 Trumpet. Similarly, rocket Esmee had an increasing trend in Pectobacteriaceae content (genus Dickeya), whereas rocket Buzz, with a consistently lower Pectobacteriaceae content (genus Dickeya), was associated with higher L. monocytogenes colonization. Pectobacteriaceae spp., in particular the genus Dickeya, is a necrotroph that is known to cause soft rot, where deterioration of vegetables occurs from the secretion of plant cell wall-degrading enzymes (Bellieny-Rabelo et al., 2019; Wasendorf et al., 2022). Additionally, Pectobacterium spp. are associated with a type VI secretion system, which also targets plant pathogens lacking cognate immunity proteins by secreting bactericidal effectors and further releasing low molecular weight bacteriocins, that is, carocin, pectocin, and carotovoricin (Shyntum et al., 2019). Moreover, Pectobacterium, Dickeya, and Serratia spp. produce the β-lactam antibiotic carbapenem (1-carbapen-2-em-3-carboxylic acid). However, leafy vegetable isolates of Pseudomonas sp., which putatively influenced L. monocytogenes growth on spinach in this study, have been found to possess antibiotic resistance genes toward β-lactam antibiotics such as meropenem and colistin (Yin et al., 2022).
4.4 Factors that affect the L. monocytogenes in situ growth
In 2016, Pectobacteriaceae was added to the Enterobacterales order. Prior to this, only a single Enterobacteraceae family existed for that order (Adeolu et al., 2016). In the present study, an unknown family from the Enterobacterales order was identified, with a relative abundance ranging from 0.00 to 33.46%. While the current study has no particular information on this new taxonomic bacterial group, the Enterobacteraceae of the same order possess the ability to produce colicins and microcins (Rebuffat, 2011). Microcins have proven ineffective against L. monocytogenes, but colicins produced with the help of the ColE1 gene are highly effective as an anti-listerial agent (Marković et al., 2022). Enterobacter spp., particularly Enterobacter cloacae, isolated from shredded iceberg lettuce, significantly reduced L. innocua colonization due to its nutritional competitiveness (Francis and O’Beirne, 2002).
Darlison et al. (2019) suggested that the influence of phyllosphere diversity on the proliferation of foodborne pathogens such as L. monocytogenes should be determined. Indeed, the significantly higher alpha diversity (Shannon index) of produce essentially appears to be correlated with lower L. monocytogenes growth potentials in the current study. However, the more diverse polytunnel rocket Buzz variety had more L. monocytogenes growth than the rocket Esmee variety. Indeed, higher and increasing Pectobacteriaceae content of Esmee, compared to the consistently low Pectobacteriaceae content, may be responsible for the 0.22 log10 cfu g−1 difference between those two growth potentials.
In the current study, seasonality was a significant driver of phyllosphere development in spinach. Bacterial diversity of the phyllosphere of Typha latifolia plants was not meaningfully influenced by short-term perturbations in weather conditions, such as rain events, but somewhat affected by seasonal climatic conditions and leaf-associated changes (Stone and Jackson, 2020). Darlison et al. (2019) suggested that annual variations resulting from varying weather conditions influenced phyllosphere communities of rocket and spinach. Although they could not rule out the effect of site-specific factors, as the produce was sampled in different parts of the same field over the 2 years. The present study accounted for site-specific factors by cultivating from the same location within both field and polytunnel settings, and also observed that weather parameters significantly influenced the spinach phyllosphere. Recently, the spinach phyllosphere has also been shown to be substantially influenced by seasonality (PERMANOVA, p < 0.003) (Ibekwe et al., 2021). An additional study revealed that the bacterial colonization of lettuce and rocket phyllosphere is also driven, at least in part, by seasonality (Dees et al., 2015).
4.5 Effects of total abundances of phyllosphere bacteria across plant species
To date, no previous studies have described the kale phyllosphere. Nevertheless, the kale endosphere has been recently studied (McNees et al., 2020). Across three different brands of store-purchased kale, Illumina sequencing of their endospheres revealed two common dominating Operational Taxonomic Units (OTUs) present were Pseudomonas and Enterobacteriaceae. In the present study, for kale, these, along with Micrococcaceae were also dominating families. Kale Nero di Toscana had the most similar content of Pseudomonadaceae as spinach F1 Cello. Although it demonstrated higher L. monocytogenes growth, due to the lower TBCs of kale (i.e., 2.80–4.74 log10 cfu g−1) and lower diversity, compared to rocket and spinach, less inhibition of the L. monocytogenes growth potentially occurs due to less competition for resources required for growth. Utilization of chloroplast-excluding protocols at the polymerase chain reaction (PCR) stage COMPETE (RInvT primer) (McManamon et al., 2019) or BLOCK (pPNA clamp) (Fitzpatrick et al., 2018; Culliney and Schmalenberger, 2024) as employed for open field spinach produce in a recent study, and would have been appropriate for rocket Esmee and kale Nero di Toscana. Their chloroplast-to-total DNA content was high, ranging from 58.00 to 97.30% (rocket Esmee) and 92.27 to 99.75% (kale Nero di Toscana), and thus could have prevented the exclusion of the 7 and 16 samples, respectively. Using the NGS approach, Listeria content was regularly detected on kale, but rarely occurred for rocket and spinach produce. The high TBCs of spinach and rocket may have contributed to this observation. Moreover, cultivation methods may not have detected cells that were at their viable but not-culturable (VBNC) stage (Müller and Ruppel, 2014). Thus, TBCs for all produce, including kale, may have been underestimated and, therefore, their total DNA content may have been associated with higher actual abundances. For example, a previous study used quantitative PCR (qPCR) and culturable techniques (TSA) to analyze lettuce samples from the same field and revealed that only 0.1–8.4% of TBCs were culturable bacteria (Rastogi et al., 2010). qPCR methods may be used in the future to enumerate TBC for this reason. However, qPCR-based quantifications may potentially overestimate bacterial population densities due to chloroplast co-amplification (Culliney and Schmalenberger, 2022) and multiple 16S ribosomal RNA (rRNA) gene copies per bacterial cell (Schmalenberger et al., 2001); hence, cultivation-dependent and independent approaches have biases. Furthermore, primer selection for 16S rRNA gene-based amplicon sequencing may also be responsible for an additional due to primer mismatch, which appears to be the case for L. monocytogenes 16S with the popular V3–V4 primers.
4.6 Pseudomonadaceae and Lactobacillales with putative contradicting effects on L. monocytogenes growth
Ibekwe et al. (2021) revealed that the four common dominating phyla Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria, which comprised 66.35% of their phyllosphere, were significantly different from the overall abundance, ranging from 88.21 to 99.92% of those four main phyla for spinach in the present study. Additionally, their Pseudomonadaceae content (0.49–11.5%) was, on average, lower than the Pseudomonadaceae content observed on spinach, rocket, and kale produce in this study. With an overall relative abundance of 35–53%, Pseudomonas has been referred to as the most commonly occurring genus in the spinach and rocket phyllospheres, even after being harvested in different seasons (spring and autumn) (Rosberg et al., 2021). Upon closer inspection of the Pseudomonadaceae family’s relative abundances, potential seasonal effects exist, especially for spinach. However, in the current study, the relative abundances of winter and summer open field spinach did not show significant differences. However, there was still a large difference of 0.94 log10 cfu g−1 between their L. monocytogenes growth potentials. LAB are more commonly detected on leafy produce cultivated in spring and summer compared to autumn and winter (Caponigro et al., 2010); however, the opposite was true in the current study. With a relative abundance of less than 1%, Lactobacillales may have been responsible for the significant growth potential difference. The Lactobacillales decreased from 0.52 to 0.22 to 0.03% from days 5–9 for summer produce, which was correlated with large increments in L. monocytogenes growth, which did not occur when Lactobacillales remained constant and on average in higher relative abundance for winter produce. More specifically, winter produce with lower L. monocytogenes growth had a significantly higher content of the Lactococcus genus (Streptococcaceae family; Lactobacillales order). Indeed, L. lactis subsp. lactis has been previously isolated from rocket leaves and is known as a bacteriocinogenic strain due to its ability to produce lantibiotic, which is an antimicrobial nisin variant that is highly effective as an anti-listerial agent on food products, including iceberg lettuce (Franz et al., 1997; Kruger et al., 2013; Ho et al., 2018; McManamon et al., 2019; Ho et al., 2021).
The remainder of phyllosphere-associated bacteria, which showed positive and negative correlations with L. monocytogenes populations identified in this study, did not appear to be potentially responsible for the conflicting epiphytic L. monocytogenes growth on spinach or rocket leaves. Correlations were determined using Pearson’s correlation, which is primarily used for linear relationships between two continuous variables, due to the normal distribution and increasing L. monocytogenes populations over time. However, a recent study revealed that Pearson’s can also be more efficient in testing a monotonic nonlinear relation compared to Spearman’s (van den Heuvel and Zhan, 2022). Future studies may use Spearman’s correlation as it evaluates the monotonic relationship between two continuous variables (Schober et al., 2018). Indeed, this approach is most often used for bacterial growth curves, which reach the stationary phase. In either case, such correlations must be interpreted with caution. For example, Zhao et al. (2021) revealed that association means that one variable provides information about another, whereas correlation means that two variables show an increasing or decreasing trend. Therefore, correlation implies an association, but not causation. Additionally, due to the absence of absolute numbers upon sequencing (Gloor et al., 2017), comparing relative abundances could lead to inaccurate conclusions when comparing phyllosphere microbiome over time or when comparing different phyllosphere communities, for example, kale or spinach, which have considerably different absolute cfu data, as relative data reflect a different amount of absolute numbers. Future studies should conduct correlations between absolute cfu data, that is, total bacterial populations and relative abundances from NGS datasets that are converted into absolute values via an additional qPCR step.
5 Conclusion
This study identified a link between leafy vegetable species, variety, and environmental growth conditions and the bacterial communities present on the leaf surface, that is, Pseudomonadaceae, Pectobacteriaceae, and Lactobacillales, such as Streptococcaceae and Carnobacteriaceae. Together, these factors are important in determining the growth potential of L. monocytogenes. However, the Pseudomonadaceae content appeared to be less critical for plant species with specific leaf surface characteristics, such as a narrow leaf surface area and a smaller number of stomata (e.g., rocket). Therefore, future studies should include leaf surface analyses in growth studies of L. monocytogenes on leafy vegetables. Due to the limitations of second-generation sequencing technologies in determining species-level identification of bacteria, a sequencing approach using third-generation amplicon sequencing techniques, as well as true metagenomics approaches, may reveal further insights into the functions of certain bacterial taxa in the phyllosphere and their abilities to aid or retard the L. monocytogenes growth.
Advancing aspects of microbial food safety for leafy vegetables may include future selection of varieties that are not only preferred due to their taste and sensory input during consumption but also due to their beneficial natural microbiome. Similarly, one could imagine a future where leafy vegetables are treated with probiotic foliar applications, where beneficial microbes are designed not only to be helpful for digestion but also helpful in suppressing foodborne pathogens.
EURL’s guidance document requires three batches for assessment of the growth potential of RTE products. These three batches are recommended to be from different production days. Although based on results from this study, this should be further updated to reflect produce with different seasonality. Moreover, as identified in the present study for spinach and rocket, the presence of certain phyllosphere or microbiome members could provide more in-depth information regarding L. monocytogenes growth potentials on RTE food products than TBC. Thus, the inclusion of NGS techniques could be considered an essential tool for assessing future challenges.
Microbiologists looking to describe the phyllosphere of kale or rocket (Esmee variety) should consider the use of chloroplast amplification blocking methods. This will reduce the number of samples discarded due to low bacterial reads, as occurred in the present study, thereby providing more detailed descriptions of phyllosphere-associated bacteria.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA1177234.
Author contributions
PC: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. AS: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The Department of Agriculture, Food and the Marine (DAFM) supported financially this research project (Listeria Challenge Studies, grant number: 17F/244).
Acknowledgments
We would like to thank the Department of Agriculture, Food and the Marine (DAFM) for funding this project (Listeria Challenge Studies, grant number: 17F/244) and our project partners for their valuable feedback.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1516740/full#supplementary-material
References
Adeolu, M., Alnajar, S., Naushad, S., and S. Gupta, R. (2016). Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. doi: 10.1099/ijsem.0.001485
Anderson, M. J. (2017). “Permutational multivariate analysis of variance (PERMANOVA)” in Wiley StatsRef: statistics reference online. John Wiley & Sons, Ltd, 1–15.
Babic, I., Roy, S., Watada, A. E., and Wergin, W. P. (1996). Changes in microbial populations on fresh cut spinach. Int. J. Food Microbiol. 31, 107–119. doi: 10.1016/0168-1605(96)00969-5
Balali, G. I., Yar, D. D., Afua Dela, V. G., and Adjei-Kusi, P. (2020). Microbial contamination, an increasing threat to the consumption of fresh fruits and vegetables in today’s world. Int. J. Microbiol. 2020, 1–13. doi: 10.1155/2020/3029295
Barbosa, J., Albano, H., Silva, B., Almeida, M. H., Nogueira, T., and Teixeira, P. (2021). Characterization of a Lactiplantibacillus plantarum R23 isolated from arugula by whole-genome sequencing and its bacteriocin production ability. Int. J. Environ. Res. Public Health 18:5515. doi: 10.3390/ijerph18115515
Bashir, I., War, A. F., Rafiq, I., Reshi, Z. A., Rashid, I., and Shouche, Y. S. (2022). Phyllosphere microbiome: diversity and functions. Microbiol. Res. 254:126888. doi: 10.1016/j.micres.2021.126888
Bellieny-Rabelo, D., Tanui, C. K., Miguel, N., Kwenda, S., Shyntum, D. Y., Moleleki, L. N., et al. (2019). Transcriptome and comparative genomics analyses reveal new functional insights on key determinants of pathogenesis and interbacterial competition in Pectobacterium and Dickeya spp. Appl. Environ. Microbiol. 85:e02050-18. doi: 10.1128/AEM.02050-18
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Author correction: reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37:1091. doi: 10.1038/s41587-019-0252-6
Brandl, M. T., and Lindow, S. E. (1998). Contribution of Indole-3-acetic acid production to the epiphytic fitness of Erwinia herbicola. Appl. Environ. Microbiol. 64, 3256–3263. doi: 10.1128/AEM.64.9.3256-3263.1998
Buchanan, R. L., and Bagi, L. K. (1997). Microbial competition: effect of culture conditions on the suppression of Listeria monocytogenes Scott a by Carnobacterium piscicola. J. Food Prot. 60, 254–261. doi: 10.4315/0362-028X-60.3.254
Caponigro, V., Ventura, M., Chiancone, I., Amato, L., Parente, E., and Piro, F. (2010). Variation of microbial load and visual quality of ready-to-eat salads by vegetable type, season, processor and retailer. Food Microbiol. 27, 1071–1077. doi: 10.1016/j.fm.2010.07.011
Carlin, F., Nguyen-The, C., and Da Silva, A. A. (1995). Factors affecting the growth of Listeria monocytogenes on minimally processed fresh endive. J. Appl. Bacteriol. 78, 636–646. doi: 10.1111/j.1365-2672.1995.tb03110.x
Colonna, E., Rouphael, Y., Barbieri, G., and De Pascale, S. (2016). Nutritional quality of ten leafy vegetables harvested at two light intensities. Food Chem. 199, 702–710. doi: 10.1016/j.foodchem.2015.12.068
Comte, I., Colin, F., Whalen, J. K., Grünberger, O., and Caliman, J.-P. (2012). “Agricultural practices in oil palm plantations and their impact on hydrological changes, nutrient fluxes and water quality in Indonesia” in Adv. Agron. ed. D. L. Sparks, (Academic Press), 116, 71–124.
Culliney, P., and Schmalenberger, A. (2020). Growth potential of Listeria monocytogenes on refrigerated spinach and rocket leaves in modified atmosphere packaging. Food Secur. 9:1211. doi: 10.3390/foods9091211
Culliney, P., and Schmalenberger, A. (2022). Cultivation conditions of spinach and rocket influence epiphytic growth of Listeria monocytogenes. Food Secur. 11:3056. doi: 10.3390/foods11193056
Culliney, P., and Schmalenberger, A. (2024). Bacterial community structure analysis on Listeria monocytogenes inoculated spinach leaves is affected by PCR based methods to exclude chloroplast co-amplification. bioRxiv. doi: 10.1101/2024.02.01.578417
Culliney, P., and Schmalenberger, A. (2025). Bacterial community structure analysis on Listeria monocytogenes inoculated spinach leaves is affected by PCR based methods to exclude chloroplast co-amplification. Microbe 6:100258. doi: 10.1016/j.microb.2025.100258
Darlison, J., Mogren, L., Rosberg, A. K., Grudén, M., Minet, A., Liné, C., et al. (2019). Leaf mineral content govern microbial community structure in the phyllosphere of spinach (Spinacia oleracea) and rocket (Diplotaxis tenuifolia). Sci. Total Environ. 675, 501–512. doi: 10.1016/j.scitotenv.2019.04.254
Dees, M. W., Lysøe, E., Nordskog, B., Brurberg, M. B., and Goodrich-Blair, H. (2015). Bacterial communities associated with surfaces of leafy greens: shift in composition and decrease in richness over time. Appl. Environ. Microbiol. 81, 1530–1539. doi: 10.1128/AEM.03470-14
Dharmarha, V., Guron, G., Boyer, R. R., Niemira, B. A., Pruden, A., Strawn, L. K., et al. (2019). Gamma irradiation influences the survival and regrowth of antibiotic-resistant bacteria and antibiotic-resistance genes on Romaine lettuce. Front. Microbiol. 10:710. doi: 10.3389/fmicb.2019.00710
Espitia, P. J. P., Otoni, C. G., and Soares, N. F. F. (2016). “Ediocin applications in antimicrobial food packaging systems” in Antimicrobial food packaging. ed. J. Barros-Velázquez, vol. 36 (Academic Press), 445–454.
EURL Lm (2019). Technical guidance document for conducting shelf-life studies on Listeria monocytogenes in ready-to-eat foods version 3-amended (21/02/2019). Maisons-Alfort, France: EURL Listeria monocytogenes, ANSES.
Faith, D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
FAO (2021). FAO (Food and Agriculture Organization of the United Nations), FAOSTAT, data, production, crops and livestock products. Available online at: https://www.fao.org/faostat/en/#data/TCL (Accessed February 15, 2023).
Fitzpatrick, C. R., Lu-Irving, P., Copeland, J., Guttman, D. S., Wang, P. W., Baltrus, D. A., et al. (2018). Chloroplast sequence variation and the efficacy of peptide nucleic acids for blocking host amplification in plant microbiome studies. Microbiome 6:144. doi: 10.1186/s40168-018-0534-0
Francis, G. A., and O’Beirne, D. (2002). Effects of the indigenous microflora of minimally processed lettuce on the survival and growth of Listeria innocua. Int. J. Food Sci. Tech. 33, 477–488. doi: 10.1046/j.1365-2621.1998.00199.x
Franz, C. M.a. P., Du Toit, M., Von Holy, A., Schillinger, U., and Holzapfel, W. H. (1997). Production of nisin-like bacteriocins by Lactococcus lactis strains isolated from vegetables. J. Basic Microbiol. 37, 187–196. doi: 10.1002/jobm.3620370307
Gloor, G. B., Macklaim, J. M., Pawlowsky-Glahn, V., and Egozcue, J. J. (2017). Microbiome datasets are compositional: and this is not optional. Front. Microbiol. 8:2224. doi: 10.3389/fmicb.2017.02224
Ho, V. T. T., Dong, A., Lo, R., and Turner, M. S. (2021). “Isolation and evaluation of anti-Listeria Lactococcus lactis from vegetal sources” in Listeria Monocytogenes. eds. E. M. Fox, H. Bierne, and B. Stessl, (Humana Press), 243–257.
Ho, V. T. T., Lo, R., Bansal, N., and Turner, M. S. (2018). Characterisation of Lactococcus lactis isolates from herbs, fruits and vegetables for use as biopreservatives against Listeria monocytogenes in cheese. Food Control 85, 472–483. doi: 10.1016/j.foodcont.2017.09.036
Hunter, P. J., Hand, P., Pink, D., Whipps, J. M., and Bending, G. D. (2010). Both leaf properties and microbe-microbe interactions influence within-species variation in bacterial population diversity and structure in the lettuce (Lactuca species) Phyllosphere. Appl. Environ. Microbiol. 76, 8117–8125. doi: 10.1128/AEM.01321-10
Hutchison, M. L. (1995). Role of biosurfactant and ion channel-forming activities of syringomycin tranransmembrane ion flux: a model for the mechanism of action in the plant-pathogen interaction. Mol. Plant-Microbe Interact. 8, 610–620. doi: 10.1094/MPMI-8-0610
Ibekwe, A. M., Ors, S., Ferreira, J. F. S., Liu, X., and Suarez, D. L. (2021). Influence of seasonal changes and salinity on spinach phyllosphere bacterial functional assemblage. PLoS One 16:e0252242. doi: 10.1371/journal.pone.0252242
Ibrahim, S. A., Ayivi, R. D., Zimmerman, T., Siddiqui, S. A., Altemimi, A. B., Fidan, H., et al. (2021). Lactic acid bacteria as antimicrobial agents: food safety and microbial food spoilage prevention. Food Secur. 10:3131. doi: 10.3390/foods10123131
Koseki, S., and Isobe, S. (2005). Growth of Listeria monocytogenes on iceberg lettuce and solid media. Int. J. Food Microbiol. 101, 217–225. doi: 10.1016/j.ijfoodmicro.2004.11.008
Kruger, M. F., Barbosa, M. D. S., Miranda, A., Landgraf, M., Destro, M. T., Todorov, S. D., et al. (2013). Isolation of bacteriocinogenic strain of Lactococcus lactis subsp. lactis from rocket salad (Eruca sativa mill.) and evidences of production of a variant of nisin with modification in the leader-peptide. Food Control 33, 467–476. doi: 10.1016/j.foodcont.2013.03.043
Kyere, E. O., Palmer, J., Wargent, J. J., Fletcher, G. C., and Flint, S. (2019). Colonisation of lettuce by Listeria Monocytogenes. Int. J. Food Sci. Technol. 54, 14–24. doi: 10.1111/ijfs.13905
Le Marrec, C., Hyronimus, B., Bressollier, P., Verneuil, B., and Urdaci, M. C. (2000). Biochemical and genetic characterization of coagulin, a new antilisterial bacteriocin in the pediocin family of bacteriocins, produced by Bacillus coagulans I4. Appl. Environ. Microbiol. 66, 5213–5220. doi: 10.1128/AEM.66.12.5213-5220.2000
Leveau, J. H. J., and Lindow, S. E. (2005). Utilization of the plant hormone Indole-3-acetic acid for growth by Pseudomonas putida strain 1290. Appl. Environ. Microbiol. 71, 2365–2371. doi: 10.1128/AEM.71.5.2365-2371.2005
Liu, H., Brettell, L. E., and Singh, B. (2020). Linking the phyllosphere microbiome to plant health. Trends Plant Sci. 25, 841–844. doi: 10.1016/j.tplants.2020.06.003
Lokerse, R. F. A., Maslowska-Corker, K. A., Van De Wardt, L. C., and Wijtzes, T. (2016). Growth capacity of Listeria monocytogenes in ingredients of ready-to-eat salads. Food Control 60, 338–345. doi: 10.1016/j.foodcont.2015.07.041
Lozupone, C. A., Hamady, M., Kelley, S. T., and Knight, R. (2007). Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. doi: 10.1128/AEM.01996-06
Marković, K. G., Grujović, M. Ž., Koraćević, M. G., Nikodijević, D. D., Milutinović, M. G., Semedo-Lemsaddek, T., et al. (2022). Colicins and microcins produced by Enterobacteriaceae: characterization, mode of action, and putative applications. Int. J. Environ. Res. Public Health 19:11825. doi: 10.3390/ijerph191811825
Marshall, D. L., Andrews, L. S., Wells, J. H., and Farr, A. J. (1992). Influence of modified atmosphere packaging on the competitive growth of Listeria monocytogenes and Pseudomonas fluorescens on precooked chicken. Food Microbiol. 9, 303–309. doi: 10.1016/0740-0020(92)80038-6
Maylani, E. D., Yuniati, R., and Wardhana, W. (2020). The effect of leaf surface character on the ability of water hyacinth, Eichhornia crassipes (Mart.) Solms. to transpire water. IOP Conf. Ser. Mater. Sci. Eng. 902:012070. doi: 10.1088/1757-899X/902/1/012070
Mcmanamon, O., Kaupper, T., Scollard, J., and Schmalenberger, A. (2019). Nisin application delays growth of Listeria monocytogenes on fresh-cut iceberg lettuce in modified atmosphere packaging, while the bacterial community structure changes within one week of storage. Postharvest Biol. Technol. 147, 185–195. doi: 10.1016/j.postharvbio.2018.10.002
Mcnees, C. R., Law, A. D., and Moe, L. A. (2020). Characterization of endophytic microbial communities in store-bought kale evaluated by different plant tissue homogenization methods. Phytobiomes J. 4, 211–216. doi: 10.1094/PBIOMES-08-19-0046-SC
Müller, T., and Ruppel, S. (2014). Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiol. Ecol. 87, 2–17. doi: 10.1111/1574-6941.12198
Østergaard, N. B., Eklöw, A., and Dalgaard, P. (2014). Modelling the effect of lactic acid bacteria from starter- and aroma culture on growth of Listeria monocytogenes in cottage cheese. Int. J. Food Microbiol. 188, 15–25. doi: 10.1016/j.ijfoodmicro.2014.07.012
Pilchová, T., Pilet, M.-F., Cappelier, J.-M., Pazlarová, J., and Tresse, O. (2016). Protective effect of Carnobacterium spp. against Listeria monocytogenes during host cell invasion using in vitro HT29 model. Front. Cell. Infect. Microbiol. 6:88. doi: 10.3389/fcimb.2016.00088
Rastogi, G., Tech, J. J., Coaker, G. L., and Leveau, J. H. J. (2010). A PCR-based toolbox for the culture-independent quantification of total bacterial abundances in plant environments. J. Microbiol. Methods 83, 127–132. doi: 10.1016/j.mimet.2010.08.006
Rebuffat, S. (2011). “Bacteriocins from gram-negative bacteria: a classification?” in Prokaryotic antimicrobial peptides. eds. D. Drider and S. Rebuffat (Springer Nature), New York: 55–72.
Rosberg, A. K., Darlison, J., Mogren, L., and Alsanius, B. W. (2021). Commercial wash of leafy vegetables do not significantly decrease bacterial load but leads to shifts in bacterial species composition. Food Microbiol. 94:103667. doi: 10.1016/j.fm.2020.103667
Sagar, L. (2020). “Exotic and uncommon vegetable production in poly tunnels” in Protected cultivation and smart agriculture. eds. S. Maitra, D. J. Gaikwad, and T. Shankar, (New Delhi: New Dehli Publishers), 15:146–160.
Saleem, B. (2021). “Phyllosphere microbiome: plant defense strategies” in Microbiomes and the global climate change. eds. S. A. Lone and A. Malik (Singapore: Springer Nature), 173–201.
Sant'ana, A. S., Barbosa, M. S., Destro, M. T., Landgraf, M., and Franco, B. D. G. M. (2012). Growth potential of Salmonella spp. and Listeria monocytogenes in nine types of ready-to-eat vegetables stored at variable temperature conditions during shelf-life. Int. J. Food Microbiol. 157, 52–58. doi: 10.1016/j.ijfoodmicro.2012.04.011
Schmalenberger, A., Schwieger, F., and Tebbe, C. C. (2001). Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67, 3557–3563. doi: 10.1128/AEM.67.8.3557-3563.2001
Schober, P., Boer, C., and Schwarte, L. A. (2018). Correlation coefficients. Anesth. Analg. 126, 1763–1768. doi: 10.1213/ANE.0000000000002864
Shyntum, D. Y., Nkomo, N. P., Shingange, N. L., Gricia, A. R., Bellieny-Rabelo, D., and Moleleki, L. N. (2019). The impact of type VI secretion system, bacteriocins and antibiotics on bacterial competition of Pectobacterium carotovorum subsp. brasiliense and the regulation of Carbapenem biosynthesis by Iron and the ferric-uptake regulator. Front. Microbiol. 10:02379. doi: 10.3389/fmicb.2019.02379
Söderqvist, K., Ahmed Osman, O., Wolff, C., Bertilsson, S., Vågsholm, I., and Boqvist, S. (2017a). Emerging microbiota during cold storage and temperature abuse of ready-to-eat salad. Infect. Ecol. Epidemiol. 7:1328963. doi: 10.1080/20008686.2017.1328963
Söderqvist, K., Lambertz, S. T., Vågsholm, I., Fernström, L.-L., Alsanius, B., Mogren, L., et al. (2017b). Fate of Listeria monocytogenes, pathogenic Yersinia enterocolitica, and Escherichia coli O157:H7 gfp+ in ready-to-eat salad during cold storage: what is the risk to consumers? J. Food Prot. 80, 204–212. doi: 10.4315/0362-028X.JFP-16-308
Stone, B. W. G., and Jackson, C. R. (2020). Seasonal patterns contribute more towards phyllosphere bacterial community structure than short-term perturbations. Microb. Ecol. 81, 146–156. doi: 10.1007/s00248-020-01564-z
Truchado, P., Gil, M. I., Reboleiro, P., Rodelas, B., and Allende, A. (2017). Impact of solar radiation exposure on phyllosphere bacterial community of red-pigmented baby leaf lettuce. Food Microbiol. 66, 77–85. doi: 10.1016/j.fm.2017.03.018
Tukey, H. B. (1970). The leaching of substances from plants. Annu. Rev. Plant Physiol. 21, 305–324. doi: 10.1146/annurev.pp.21.060170.001513
Vacher, C., Hampe, A., Porté, A. J., Sauer, U., Compant, S., and Morris, C. E. (2016). The phyllosphere: microbial jungle at the plant–climate interface. Annu. Rev. Ecol. Evol. Syst. 47, 1–24. doi: 10.1146/annurev-ecolsys-121415-032238
Van Den Heuvel, E., and Zhan, Z. (2022). Myths about linear and monotonic associations: Pearson’s r, spearman’s ρ, and Kendall’s τ. Am. Stat. 76, 44–52. doi: 10.1080/00031305.2021.2004922
Van Der Avoort, C. M. T., Van Loon, L. J. C., Hopman, M. T. E., and Verdijk, L. B. (2018). Increasing vegetable intake to obtain the health promoting and ergogenic effects of dietary nitrate. Eur. J. Clin. Nutr. 72, 1485–1489. doi: 10.1038/s41430-018-0140-z
Venu, S., Khushbu, S., Santhi, S., Rawson, A., Sunil, C. K., and Sureshkumar, K. (2019). “Phytochemical profile and therapeutic properties of leafy vegetables” in Plant and human health, volume 2. ed. K. R. H. Munir Ozturk, vol. 26 (Cham: Springer), 627–660.
Wasendorf, C., Schultz, D. L., Schmitz-Esser, S., Peters, N. T., and Dunning Hotopp, J. C. (2022). Genome sequences of soft rot-causing Pseudomonas isolates from spinach. Microbiol. Resour. Announc. 11, e00701–e00722. doi: 10.1128/mra.00701-22
Webb, L., Ma, L., and Lu, X. (2022). Impact of lactic acid bacteria on the control of Listeria monocytogenes in ready-to-eat foods. Food Qual. Saf. 6, 1–11. doi: 10.1093/fqsafe/fyac045
Xu, N., Zhao, Q., Zhang, Z., Zhang, Q., Wang, Y., Qin, G., et al. (2022). Phyllosphere microorganisms: sources, drivers, and their interactions with plant hosts. J. Agric. Food Chem. 70, 4860–4870. doi: 10.1021/acs.jafc.2c01113
Yin, Y., Zhu, D., Yang, G., Su, J., and Duan, G. (2022). Diverse antibiotic resistance genes and potential pathogens inhabit in the phyllosphere of fresh vegetables. Sci. Total Environ. 815:152851. doi: 10.1016/j.scitotenv.2021.152851
Zhang, M., Peng, C., Sun, W., Dong, R., and Hao, J. (2022). Effects of variety, plant location, and season on the phyllosphere bacterial community structure of alfalfa (Medicago sativa L.). Microorganisms 10:2023. doi: 10.3390/microorganisms10102023
Zhao, Z.-D., Zhao, N., and Ying, N. (2021). Association, correlation, and causation among transport variables of PM2.5. Front. Phys. 9:684104. doi: 10.3389/fphy.2021.684104
Zhu, Y. G., Xiong, C., Wei, Z., Chen, Q. L., Ma, B., Zhou, S. Y. D., et al. (2022). Impacts of global change on the phyllosphere microbiome. New Phytol. 234, 1977–1986. doi: 10.1111/nph.17928
Ziegler, M., Kent, D., Stephan, R., and Guldimann, C. (2019). Growth potential of Listeria monocytogenes in twelve different types of RTE salads: impact of food matrix, storage temperature and storage time. Int. J. Food Microbiol. 296, 83–92. doi: 10.1016/j.ijfoodmicro.2019.01.016
Keywords: variety, season, lactic acid bacteria, Pseudomonadaceae, Spinacia oleracea, Eruca sativa, Listeria monocytogenes
Citation: Culliney P and Schmalenberger A (2025) The cultivation conditions of leafy vegetables influence the structures of phyllosphere bacterial communities and ultimately impact the L. monocytogenes growth post-harvest. Front. Microbiol. 16:1516740. doi: 10.3389/fmicb.2025.1516740
Edited by:
Natalia Wiktorczyk-Kapischke, Nicolaus Copernicus University in Toruń, PolandReviewed by:
Aurel Maxim, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaAlex Fulano, Kenyatta University, Kenya
Copyright © 2025 Culliney and Schmalenberger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Achim Schmalenberger, YWNoaW0uc2NobWFsZW5iZXJnZXJAdWwuaWU=
 Paul Culliney
Paul Culliney Achim Schmalenberger
Achim Schmalenberger