- 1Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
- 2Key Laboratory for Conservation and Utilization of Bio-Resource, Key Laboratory for Microbial Resources of the Ministry of Education, School of Life Sciences, Yunnan Institute of Microbiology, Yunnan University, Kunming, China
- 3College of Resources, Environmental Sciences and Chemistry, Chuxiong Normal University, Chuxiong, China
- 4Chuxiong Medical and Pharmaceutical College, Chuxiong, Yunnan, China
Desert ecosystems have increasingly piqued the interest of microbiologists seeking novel bioactive compounds, as they are viewed as a largely uncharted reservoir of extremophiles with remarkable resilience to severe conditions. The genus Kocuria, belonging to the phylum Actinomycetota, is particularly notable for its documented capacity to flourish in such extreme environmental conditions. In this study, a total of 21 Kocuria strains were isolated from various ecosystems. Using polyphasic taxonomy approaches, eight strains from desert soils (CPCC 205273T, CPCC 205300, CPCC 205290, CPCC 205236T, CPCC 205293, CPCC 205292T, CPCC 205315T, CPCC 205268T) were identified representing five new species of the genus Kocuria. Strains CPCC 205281 and CPCC 205293 were identified as siblings of strain CPCC 205236T, while strains CPCC 205290 and CPCC 205300 were identified as siblings of strain CPCC 205273T. Additionally, K. polaris and K. indcia were determined to be the later heterotypic synonym of K. rosea and K. marina, respectively. Genomic analysis and physiological assays demonstrated that these previously uncharacterized strains were tolerant to high-level salt concentration and UV radiation, key survival traits in desert environments. The fermentation analysis revealed that most strains produced high-level contents of indole-3-acetic acid (IAA), although the complete gene sets for IAA biosynthesis were found in only one strain. Comparative genome analysis further showed that genes related to carbohydrate metabolism and transports were significantly enriched in desert-derived Kocuria strains, indicating adaptation to desert habitats. Collectively, our findings enhance our understanding of Kocuria taxonomy and highlight their genetic adaptation strategies to extreme environments, with potential biotechnological applications.
1 Introduction
Extremophilic and extremotolerant Actinobacteria, including acid-tolerant, alkalitolerant, psychrotolerant, thermotolerant, halotolerant, haloalkalitolerant, and xerophiles, are a group of bacteria that have been less well-studied (Sayed et al., 2020). Particularly, as the largest continental ecosystem on earth with approximately 30% of the total land area, deserts (Neilson et al., 2012; Publications Office of the European Union, 2018), have increasingly attracted the interest of microbiologists in the quest for novel bioactive compounds. Actinobacteria found in desert environments can thrive in specific circumstances of pH or salinity and possess impressive gene clusters that generally enable them to produce secondary metabolites with distinct antibacterial properties., making it a valuable subject of study (Bull et al., 2016; Idris et al., 2017; Cordero et al., 2018).
The genus Kocuria, a large group of actinobacteria, stand out for its known ability to survive in extreme conditions. The genus Kocuria, belonging to the family Micrococcaceae, the order Micrococcales, the phylum Actinomycetota, was first proposed by Stackebrandt et al. (1995) with Kocuria rosea as the type species. To date, Kocuria encompasses 26 species with validly published and correct names (data from LPSN).1 Members of the genus Kocuria are Gram-stain-positive, non-endospore-forming, catalase-positive, coagulase-negative, non-hemolytic cocci, and typically colonize distinct types of ecosystems such as marine, desert, rhizoplane, and hyper-saline soil (Kovacs et al., 1999; Kim et al., 2004; Mayilraj et al., 2006; Wang et al., 2015). Accordingly, they have been documented to produce biotechnologically interesting metabolites (Haidar et al., 2018; Retamal-Morales et al., 2018), auxins, pigments (Samanta et al., 2016; Sahar et al., 2022), extracellular polysaccharides (Kumar and Sujitha, 2014), and antibiotics (Palomo et al., 2013), under different, occasionally harsh, environments.
Indole-3-acetic acid (IAA), a phytohormone, plays a significant role in bacterial-plant interactions and has recently been identified as a key factor in bacterial stress response mechanisms (Duca and Glick, 2020). For example, IAA-producing strains like Sinorhizobium meliloti demonstrate increased resistance to environmental stressors, such as desiccation and UV radiation, by enhancing the production of protective compounds such as lipopolysaccharides (LPS), biofilms, extracellular polysaccharides (EPS), and trehalose (Bianco et al., 2009; Imperlini et al., 2009). Additionally, recent studies suggest a complex interaction between IAA synthesis and bacterial susceptibility to antibiotics, offering new insights into the physiological importance of IAA beyond its role in plant-microbe symbiosis (Cerboneschi et al., 2016; Chowdhury et al., 2016). Notably, some rhizosphric Kocuria strains are recognized as plant-growth-promoting rhizobacteria (PGPR) due to their IAA production (Karnwal, 2019; Pantoja-Guerra et al., 2023). While metagenomics analysis revealed that Kocuria was also the key and even dominant community of the plant-free desert soil (Vikram et al., 2016; Sun et al., 2018). This paradox suggests that IAA’s role in Kocuria might extend beyond plant-microbe interactions, potentially serving as a key mediator of bacterial stress tolerance in extreme environments.
In this study, we isolated and characterized 21 Kocuria strains collected from various semi-arid and hyper-arid environments, with a focus on eight strains from desert soils. Through polyphasic taxonomy and comparative genomics, we sought to classify these strains, investigate their genomic adaptations to extreme environments, and explore their capacity for producing IAA—a key trait for stress resistance and plant-microbe interactions.
2 Materials and methods
2.1 Samples collection and Kocuria strains acquisition
Samples used for bacterial isolation were mainly collected from deserts, rhizosphere soil in forests, and other diverse ecosystems. The soil samples were transported and stored at 4°C in the dark until further processing. According to the procedure described in previous study (Deng et al., 2022), the samples were subjected to gradient dilution and then spread on one-tenth strength R2A medium, supplemented with aztreonam (to the final concentrate of 25 mg l−1) and potassium dichromate (to the final concentrate of 50 mg l−1) to prevent or stymie the growth of Gram-stain-negative bacteria and fungi that may be present. The plates were subsequently incubated at 37°C for 3–4 weeks till the separated colonies appeared. Then the well-grown colonies were picked and transferred onto the freshly prepared R2A plates, and then purified by repeat streaking. The purified strains were maintained on aqueous glycerol suspensions (20%, v/v) at −80°C for a long time preservation.
Strain CPCC 205236T and the other 11 strains were isolated from the desert soil samples collected from three different locations: Badain Jaran Desert (39°17′-39°31’N, 101°03′-103°00′E, 1,241–1,510 mH), Gurbantunggut Desert (44°48′-44°50’N, 88°16′-88°18′E, 600–620 mH), and Xinjiang Uygur Autonomous Region (43°13′-44°15’N, 84°45′-84°47′E, 2,620–2,637 mH). The remaining four soil-derived strains were obtained from the rhizosphere soil of an Oxytropis falcata located in Xinjiang province, China. In addition, four strains were recovered from the animal or human-associated ecosystems, including human skin and the feces of Crow. Only strain CPCC 204721 was isolated from the semi-arid air collected from Xinjiang province. Detailed information on the habitats of these newly isolated Kocuria strains was shown in Supplementary Table S1.
Strains K. rosea JCM 11614T and K. salina DSM 28714T, which were used as references for partial parallel experiments, were obtained from the RIKEN BioResource Research Center (JCM) and German Collection of Microorganisms and Cell Cultures GmbH (DSMZ), respectively.
2.2 Phylogenetic analysis based on 16S rRNA genes
The genomic DNA of the strains in this study was extracted and their 16S rRNA genes were amplified by PCR, according to the method previously described by Li et al. (2007). The sequences were quality-checked and assembled using Seqman program (Swindell and Plasterer, 1997). The assembled sequences were then compared with available 16S rRNA gene sequences of cultured species from the EzBioCloud2 platform (Yoon et al., 2017). Multiple sequence alignments and phylogenetic analysis were conducted on the molecular evolutionary genetics analysis (MEGA) software package (version 7.0) (Kumar et al., 2016) using neighbor-joining (NJ) (Saitou and Nei, 1987) and maximum-likelihood (ML) (Felsenstein, 1981) methods. Confidences of the topology of trees were evaluated with the 1,000 bootstrap resamplings (Felsenstein, 1985).
2.3 Genome sequencing, assembly, and annotation
Whole-genome sequencing of the new isolates was conducted on the Illumina HiSeq 4,000 platform (Illumina, SanDiego, CA, USA) at the Beijing Genomics Institute (Beijing, China). Genomic DNA was randomly sheared to construct three read libraries of length 300 bp using a Bioruptor ultrasonicator (Diagenode, Denville, NJ, USA) and physicochemical methods. Paired-end fragment libraries were then sequenced using manufacturer protocols. Low-quality reads (those with consecutive bases covered by fewer than five reads) were discarded using Fastq v1.4 (Chen et al., 2018), and the remaining reads were assembled with the SOAPdenovo v1.05 software program (Xie et al., 2014). The quality (index: completeness and contamination) of the draft genomes was accurately assessed by the CheckM v1.2.3 pipeline (Parks et al., 2015). Assembled genomes with completeness > 80% and contamination < 5% were annotated by using Prokka v1.14.5 (Seemann, 2014) (All assemblies passed the quality control threshold: completeness > 80% and contamination < 5%). Genes related to stress response and IAA production were identified by comparison to the Uniprot3 and Interpro4 databases. Furthermore, biosynthetic gene clusters (BGCs) were detected and characterized using antiSMASH version 6.0.5 All parameters for the prior bioinformatic software were default ones unless stated otherwise.
2.4 OGRIs and phylogenomics analysis
The overall genome relatedness indices (OGRIs) were used to determine the similarity between genomes useful for species delineation (Chun and Rainey, 2014). Average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values were calculated between the genomes of the newly isolated strains and their related species using the FastANI and the Genome-to-Genome Distance Calculator (GGDC, version 3.0)6 (Auch et al., 2010; Jain et al., 2018), respectively. To establish the phylogenomic relationships between the genus Kocuria, the single-copy orthologroups (SOG, one sequence per genome) were inferred by OrthoFinder v2.5.5 (Emms and Kelly, 2015, 2019) based on the annotated proteomes. For each SOG, a multiple sequence alignment was implemented using ClustalO v1.2.2 (Sievers et al., 2011), and the ambiguous sites were trimmed by trimAI v1.5.0 (Capella-Gutierrez et al., 2009) with the option “automated1.” A maximum-likelihood species tree was constructed based on a concatenation of these alignments using IQ-tree v2.3.6 (Nguyen et al., 2015; Minh et al., 2020) with 1,000 ultrafast bootstrap replicates, and the best model was selected by ModelFinder (Kalyaanamoorthy et al., 2017). In addition, MLSA (multilocus sequence analysis) was also implemented to evaluate the phylogenetic relationships. Forty conserved maker gene sequences were retrieved from genomes by using progenomes2 (Mende et al., 2019) with default options. Then, a phylogenetic tree was built using the same method as the phylogenomics based on the SOGs.
2.5 Growth conditions, morphological characteristics, and physiological tests
Physiological characteristics of strains CPCC 205293, CPCC 205236T, CPCC 205292T, CPCC 205300, CPCC 205315T, CPCC 205268T, CPCC 205273T, CPCC 205290, and two reference strains were determined following growth on various media, including on peptone yeast (PYG; 3% trypticase soy broth, 0.3% yeast extract, 1.5% agar; pH 7.0–7.2), tryptone soy agar (TSA), International Streptomyces Projects 2 medium (ISP 2), yeast extract sucrose (YM), Luria-Bertani, (R2A), International Streptomyces Projects 4 medium (ISP 4), and nutrient (Shirling and Gottlieb, 1966) media formulations, after incubation at 28°C for 1–2 weeks. The growth conditions at different temperatures were assayed in TSA medium at temperatures of 4, 10, 20, 26, 28, 30, 32, 37, 40, 45, 46, and 50°C for 14 d (Sun et al., 2017). The range of pH for growth (4.0–11.0 with 1 interval) was tested on TSA with a previously described buffer system (Xu et al., 2005). Salt (sodium chloride) tolerance was determined on TSA agar medium plates supplemented with NaCl concentrations of 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, and 20% (w/v). UV (Ultraviolet) radiation tolerance was tested according to the previously described procedures (Zhang et al., 2007). Growth after 10 days was identified as positive or negative relative to un-irradiated controls.
Gram staining reaction of the isolates was conducted as the method previously described (Magee et al., 1975). Colony appearance and pigment production were observed after incubation at 28°C on TSA medium. Cellular morphologies were observed by light microscopy (Zeiss Axio Scope. A1 Vario) and transmission electron microscopy (JEOL JEM-1010) after incubation for 5–7 days. The motility of cells was observed by light microscopy by first growing the strain on TSA semi-solid medium agar (0.3%, w/v). Oxidase activity was identified using an analytical profile index (API) oxidase reagent (bioMeriéux) according to the manufacturer’s instructions. Catalase activity was determined by bubble production in 3% (v/v) H2O2. Metabolic characteristics were determined using Biolog GEN III (MicroPlate), API 50CH, API 20NE, and API ZYM (bioMerieux) test kits following the manufacturer’s instructions. The tests for the formation of indole and H2S and the hydrolysis of gelatin, cellulose, and starch, were performed as previously described by Gonzalez et al. (1978).
2.6 Chemotaxonomic assays
Chemotaxonomic and molecular systematic studies of these eight isolates and reference strains were conducted with cells after cultivation in TSB (tryptone soy broth) medium at 28°C in shake flasks on a rotary shaker (150 r/min) until cells reached the logarithmic growth phase. Amino acids and peptides in whole-cell hydrolysates were analyzed by two-dimensional ascending thin-layer chromatography (TLC) on cellulose plates using solvent systems described by Schleifer and Kandler (1972). Sugar profiles were evaluated with TLC, as previously described by Komagata and Suzuki (1988). Analysis of respiratory quinones was carried out by the Identification Service of DSMZ (Braunschweig, Germany). Menaquinones were extracted according to Collins et al. (1977) and analyzed by High Performance Liquid Chromatography (HPLC) (Groth et al., 1997). Cellular fatty acids analysis was performed using the Microbial Identification System (Sherlock Version 6.0; MIDI database: ACTIN1) (Kroppenstedt, 1985; Sasser, 1990). Polar lipids were extracted and identified by two-dimensional TLC following the methods of (Minnikin et al., 1984).
2.7 Quantification of IAA synthesis
The bacterium culture was maintained at 30°C in LB medium supplemented with 120 mM methanol for 8 days at 200 rpm. Spectrophotometric quantification of indole-3-acetic acid (IAA) was evaluated by using Salkowski reagent (Ehmann, 1977). In brief, culture filtrate was collected by centrifugation at 7,000 rpm for 5 min. 0.5 mL aliquots of culture was reacted with 2 mL of Salkowski reagent [2% (w/v) FeCl₃·6H₂O in 35% (v/v) HClO₄], followed by the addition of 200 μL orthophosphoric acid (H3PO4, 85%) under vortex mixing. The reaction mixture was incubated in darkness for 30 min at ambient temperature (25 ± 1°C). Absorbance measurements were conducted at 530 nm using a UV–Vis spectrophotometer (Agilent Technologies, USA), with quantification achieved through a standard calibration curve generated from pure IAA (0–200 μg/mL; Fisher Scientific, Hampton, NH, USA). The experiment was repeated three times. The mean values of repetitions were calculated ± standard deviation (SD).
2.8 Comparison of functional genes
In this study, strains from desert-associated ecosystems were designated as DAK strains, whereas those from other ecosystems were designated as NDAK strains. To perform comparative analysis for desert-associated Kocuria strains (DAK) and other non-desert-associated Kocuria strains (NDAK), we constructed a genomic dataset consisting of 179 individuals affiliated to the genus Kocuria (21 strains were from this analysis and others were collected from the NCBI assembly database). Among which 25 strains with uncertain isolation sources were excluded. Genomes from the same strain with an ANI greater than 99.95% and alignment fraction exceeding 50% were marked as redundant genomes, and then they were dereplicated by filtering out one of the genomes at random. Finally, the remaining 143 genomes formed a representative Kocuria genome dataset (Supplementary Table S2). The comparison of functional genes between DAK and NDAK was performed according to the previous study (Levy et al., 2017). A random set with 15 genomes of NDAK was selected, and the mean and s.d of the phylogenetic distance in the set were calculated. This step was repeated 150 times, and then these 15 genomes of NDAK with minimum differences between their mean and s.d. of phylogenetic distances were selected for subsequent analysis. Open reading frames (ORFs) of these genomes were predicted using Prokka v1.14.5. The amino acid sequences of these putative ORFs were aligned against Cluster of Orthologous Groups (COG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases to obtain their corresponding annotations using eggnog Mapper v5.0 with default options (Huerta-Cepas et al., 2019). Phylogenetically informed principal component analysis (phylo-PCA) was used to visualize the differences in gene compositions between DAK and NDAK strains using the R package phytools v0.7–80 (Revell, 2024). The pan-genome-wide association studies (pan-GWAS) analysis was performed for identification of habitat-enriched KEGG orthogroups (KOs) using SCOARY v1.6.16 (Brynildsrud et al., 2016) depending on the KO presence/absence data set. Significant variants were determined using a cutoff of Bonferroni adjusted p < 0.05 with sensitivity and specificity greater than 80%.
3 Results and discussions
3.1 Identification of Kocuria spp.
Twenty-one strains were isolated from the desert soils, human skin, feces, and air samples from China (Supplementary Table S1). Comparative analysis of their 16S rRNA gene sequences revealed that these newly isolated strains belong to the family Micrococcaceae and were closely related to the genus Kocuria.
The comparative analysis of 16S rRNA gene sequences indicated that these newly isolated strains were members of the family Micrococcaceae with closely related to the genus Kocuria. Thirteen strains, including CPCC 205231, CPCC 205263, CPCC 205258, CPCC 205274, CPCC 205281, CPCC 205293, CPCC 205236T, CPCC 205273T, CPCC 205290, CPCC 205300, CPCC 205268T, CPCC 205315T, and CPCC 205292T, exhibited high 16S rRNA gene sequence similarities to K. polaris CMS 76orT (99.1–99.6%), K. rosea DSM 20447T (99.2–99.8%), K. himachalensis K07-05T (99.3–99.9%), and K. salina DSM 28714T (99.1–99.9%). Stains CPCC 204721, CPCC 205233, CPCC 205261, and CPCC 205297 shared the highest 16S rRNA gene similarity with K. indica NIO-1021T (99.86–99.93%). Strain CPCC 205235 showed the 99.7% similarity with K. carniphila CCM 132T; CPCC 104605 showed 100% identity with K. subflava YIM 13062T; while CPCC 205260 and CPCC 205295 shared 100% 16S rRNA gene sequence identity with the K. palustris DSM 11925T.
The 16S rRNA gene sequences of these 21 strains isolated from this study and 26 validated described type strains with correct names (Data from LPSN) were used to construct the phylogenetic trees. The phylogenetic tree based on the NJ method showed that these newly isolated strains fall into nine distinct phylogenetic groups denoted as Clade 1–9 (Figure 1A). However, Clades 1, 2, and 4 showed ambiguous phylogenetic placements between NJ and ML trees with low bootstrap values (Supplementary Figure S1). This ambiguity may be attributed to the high homogeneity of the 16S rRNA gene sequences within these clades, making it challenging to differentiate between strains using a single gene.
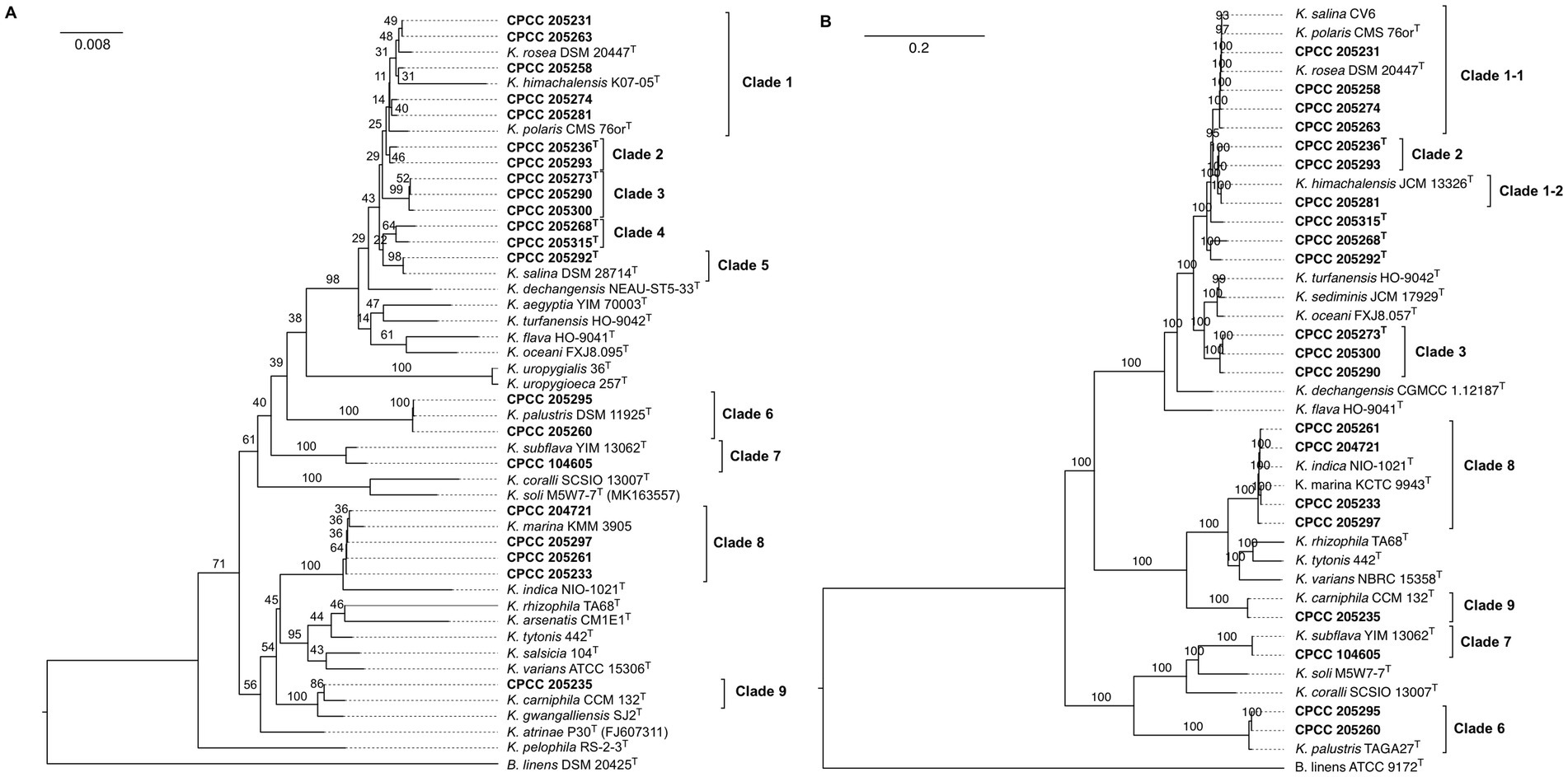
Figure 1. The phylogenetic analysis based on 16S rRNA gene (A) and core genome (B) of 21 newly isolated strains and 26 type strains of established species among the genus Kocuria. Strains isolated from this study are shown in bold. Bootstrap values that more than 70% are indicated for each node (1,000 replicates). Brevibacterium linens DSM 2042T was used as an outgroup.
3.2 Genome-based taxonomy
To further confirm the phylogenetic positions of these Kocuria strains, their genome sequences were determined and then subjected to phylogenomic tree reconstruction and MLSA with other reference genomes (Supplementary Table S1) (Brevibacterium linens DSM 2042T was used as the outgroup). Firstly, 40 universal markers were retrieved from these genomes, and aligned and concatenated to build an MLSA-based phylogeny. In addition, 662 single-copy orthogroups were inferred by OrthoFinder and then used to reconstruct a phylogenomic tree. Interestingly, phylogenomics and MLSA-based phylogeny provide a consistent and reliable phylogenetic topology (Supplementary Table S2; Figure 1B), especially on the internal ancestral nodes of the lower support phylogenetic groups recovered from the 16S rRNA-based phylogeny. Clade I with the lowest bootstrap values in 16S rRNA-based phylogeny segregated into two subclade (Clade 1–1 and Clade 1–2) with higher support inside the MLSA and phylogenomic trees. Strain CPCC 205281 and K. himachalensis JCM 13326T of Clade 1 formed a subclade (Clade 1–1), and it was distant from the other members of that clade (Figure 1B). The strain CPCC 205268T of Clade 4 were clustered with CPCC 205292T of Clade 5 with a bootstrap value of 100%, whereas another member of Clade 5 (K. salina CV6) was distributed in the Clade 1–1. In addition, CPCC 205215T was placed independently of the other Kocuria species and formed a monophyletic clade with a bootstrap value of 100%. The phylogenetic placements of the other six groups (Clade 2, 3, 6–9) were utterly consistent with that in the 16S rRNA gene tree.
The overall genome relatedness indices (OGRIs) values of 21 strains isolated from this study with their closely related strains were calculated by algorithms fastANI and GGDC. The strains within two subclades (Clade 1–1 and Clade 1–2) and six clades (Clade 2, 3, 6–9) exhibited dDDH and ANI values exceeding the suggested threshold for species delineation values of 70% for dDDH and 95–96% for ANI (Chun et al., 2018; Riesco and Trujillo, 2024), respectively, while the values between each subclade and/or clade were below these cut-off values (Supplementary Figures S3, S4). The ANI and dDDH values between strain CPCC 205268T and other Kocuria strains involved in this study were in a range of 78.5–91.3% and 20.1–41.7% (Supplementary Figures S3, S4), which were lower than the minimal threshold for species delineation. The same situation occurred between CPCC 205215T (ANI = 78.1–93.4%, dDDH = 20.0–51.3%) and CPCC 205292T (ANI = 87.1–92.4%, dDDH = 20.0–43.7%). The ANI and dDDH values for the studied strains were consistently below the 95–96 and 70% thresholds, respectively, confirming their distinction as new species.
Consequently, OGRIs combined with phylogenomics, the phylogeny of 16S rRNA gene indicated that 8 of 21 Kocuria strains (CPCC 205273T, CPCC 205290, CPCC 205300, CPCC 205268T, CPCC 205315T, CPCC 205292T, CPCC 205293, and CPCC 205236T) isolated from this study represent five new species, and others belong to the previous established species. In addition, genome-based taxonomic characterization demonstrated that K. polaris should be considered as a later heterotypic synonym of K. rosea and K. indica that as of K. marina, respectively.
3.3 Morphological, physiological, and biochemical features
The phenotypic properties of these eight newly isolated strains were compared with their closely related type strains to acquire a deeper understanding of their relationships. All strains were Gram-strain-positive, oxidase-negative, catalase-positive, non-motility, and aerobiotic coccoid. The key phenotypic characteristics of these eight isolates were compared with those of their closely related species in Table 1. Strain CPCC 205273T, CPCC 205290, and CPCC 205300 could be differentiated from K. rosea DSM 20447T by their positive alkaline phosphatase activity, while they did not grow at more than 50°C, and did not utilize D-galactose and D-melibiose. Strain CPCC 205236T and CPCC 205293 were positive for alkaline phosphatase but negative for cystine arylamidase; they could not grow at 50°C and utilize D-melibiose, and D-galactose, which distinguished them from K. rosea DSM 20447T. Differentials between CPCC 205315T, CPCC 205268T, and K. rosea DSM 20447T were hydrolysis of starch and assimilation of L-alanine. Moreover, CPCC 205292T was distinguished from K. salina DSM 28714T by its hydrolysis of cellulose and assimilation of L-alanine.
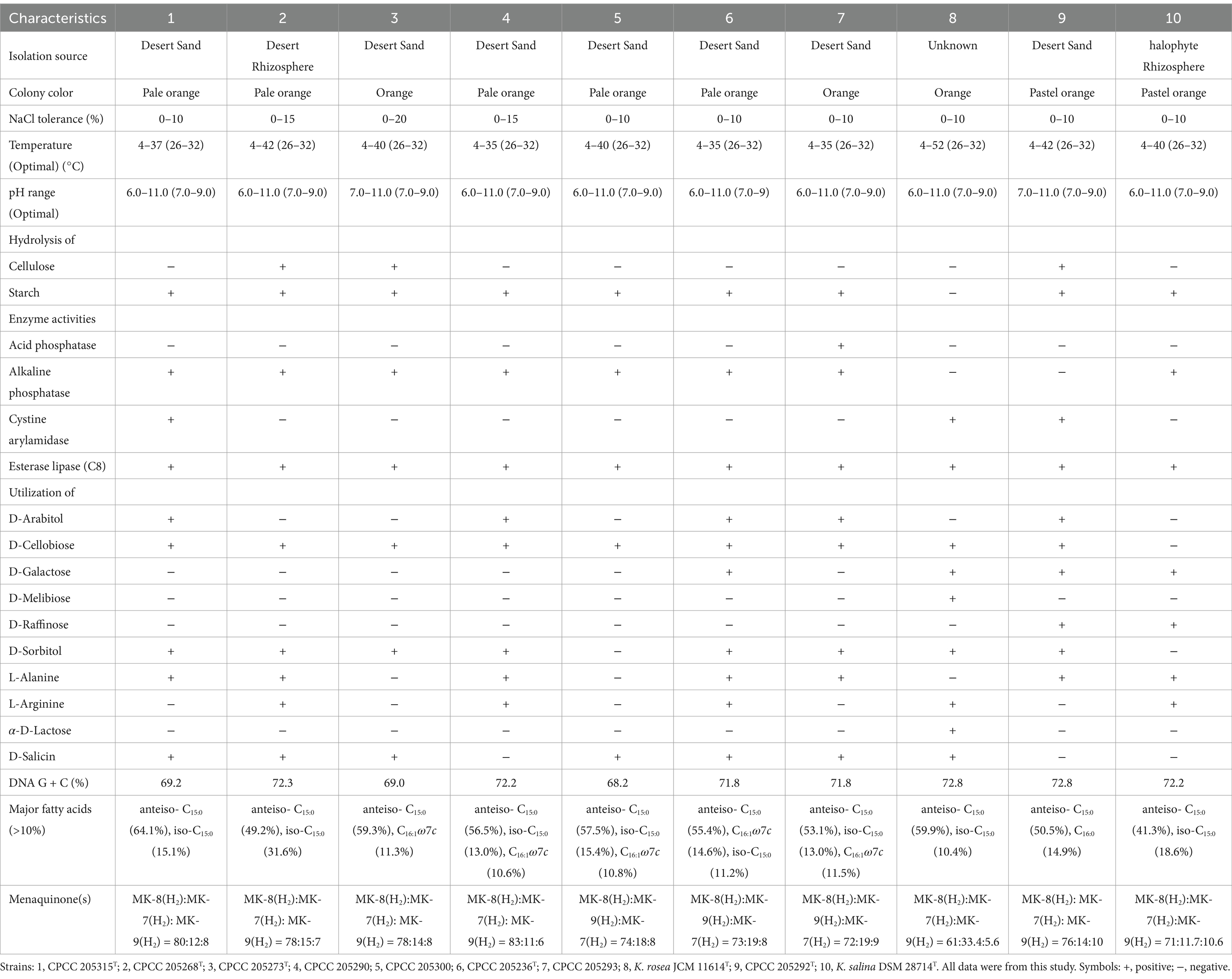
Table 1. Differentiating characteristics between the newly isolated strains and the related Kocuria reference strains.
The major polar lipids of these eight strains were diphosphatidylglycerol (DPG), and phosphatidylglycerol (PG), which is consistent with those of other Koruria species (Supplementary Figure S5). The anteiso-C15:0 was the predominant cellular fatty acid in all of these strains, in line with that of other members of the genus Kocuria. The other fatty acid compositions were detected in each strain but varied (Table 1; Supplementary Table S3). All strains contained MK-7(H2), MK-8(H2), and MK-9(H2) in their respiratory quinone systems. Quantitative differences in quinones between these previously uncharacterized strains and type strains of closely related species are given in Table 1.
On the basis of phylogenetic analysis, genomic relatedness, and phenotypic feature comparison, it was proposed that eight newly isolated strains should be classified as five new Kocuria species. The formal proposal of these new species is in the Species Descriptions section.
IAA (indole-3-acetic acid), a product of the amino acid L-tryptophan (L-Trp) (Yu et al., 2017), acts as a plant growth regulator. Several studies have revealed that a wide variety of bacteria have been able to produce the IAA such as Agrobacterium tumefaciens, Pseudomonas syringae, Streptomyces sp., Bacillus subtilis spp., Pseudomonas fluorescens, and Bacillus megaterium (Rushabh et al., 2020). IAA was detected and quantified in the fermentation broth of the eight newly isolated strains and two reference strains. Eight out of ten strains showed the ability to convert L-Trp into IAA. Quantification analysis revealed that the IAA content yield from the fermentation broth of CPCC 205236T, CPCC 205292T, CPCC 205293, CPCC 205290, CPCC 205315T, CPCC 205268T, K. rosea JCM 11614T, and K. salina DSM 28714T was 126.9 ± 5.7, 256.8 ± 8.9, 151.3 ± 4.3, 148.0 ± 7.9, 177.8 ± 0.35, 200.8 ± 3.4, 86.4 ± 5.2, and 257.6 ± 6.2 mg/L, respectively (Supplementary Figure S6).
3.4 Genome-wide screen for key genes associated with adaptation to extreme environments among Kocuria strains
Bacteria in the desert are subjected to a variety of physicochemical stresses, including low organic carbon and nitrogen availability, high UV irradiation, dryland salinity, and temperature extremes, microbes cope with these environmental stresses through unique physiological acclimation mechanisms (Georgiou et al., 2015; Allison, 2023). Therefore, the genes linked to stress response (Supplementary Table S4) were discovered in the genomes of the eight previously uncharacterized Kocuria strains isolated from the desert soil (Figure 2). Eight desert-derived Kocuria strains harbored high-abundance genes for the Opu transporter (opuABCD) and trehalose biosynthesis (otsAB and treS), both linked to osmoprotection under desiccation stress (Rath et al., 2020; Lee et al., 2023). Genes encoding heat (dnaJ, dnaK, and GRPE) and cold (cspA) shock proteins (Szabo et al., 1994) were also conserved across strains, supporting their adaption to extreme temperature fluctuations. Bacteria’s primary antioxidant defense systems, catalase (katE) enzymes (Lushchak, 2001), may not protect them from extreme and prolonged oxidative stress, leading them to activate SoxRS systems in response to redox-active compounds (Nunoshiba et al., 1993). Interestingly, all strains exhibited katE and the full set of genes associated with the SoxRS system (soxABDR). All strains contained uvrABCD and recADFNQG, which were separately involved in NER (nucleotide excision repair) mediated UV-radiation resistance and RecA-mediated repair of DNA double-strand breaks. Our observations suggested that these studied Kocuria strains isolated from the desert soil have a complete genetic basis for survival in extreme environments, like hyper-arid deserts.
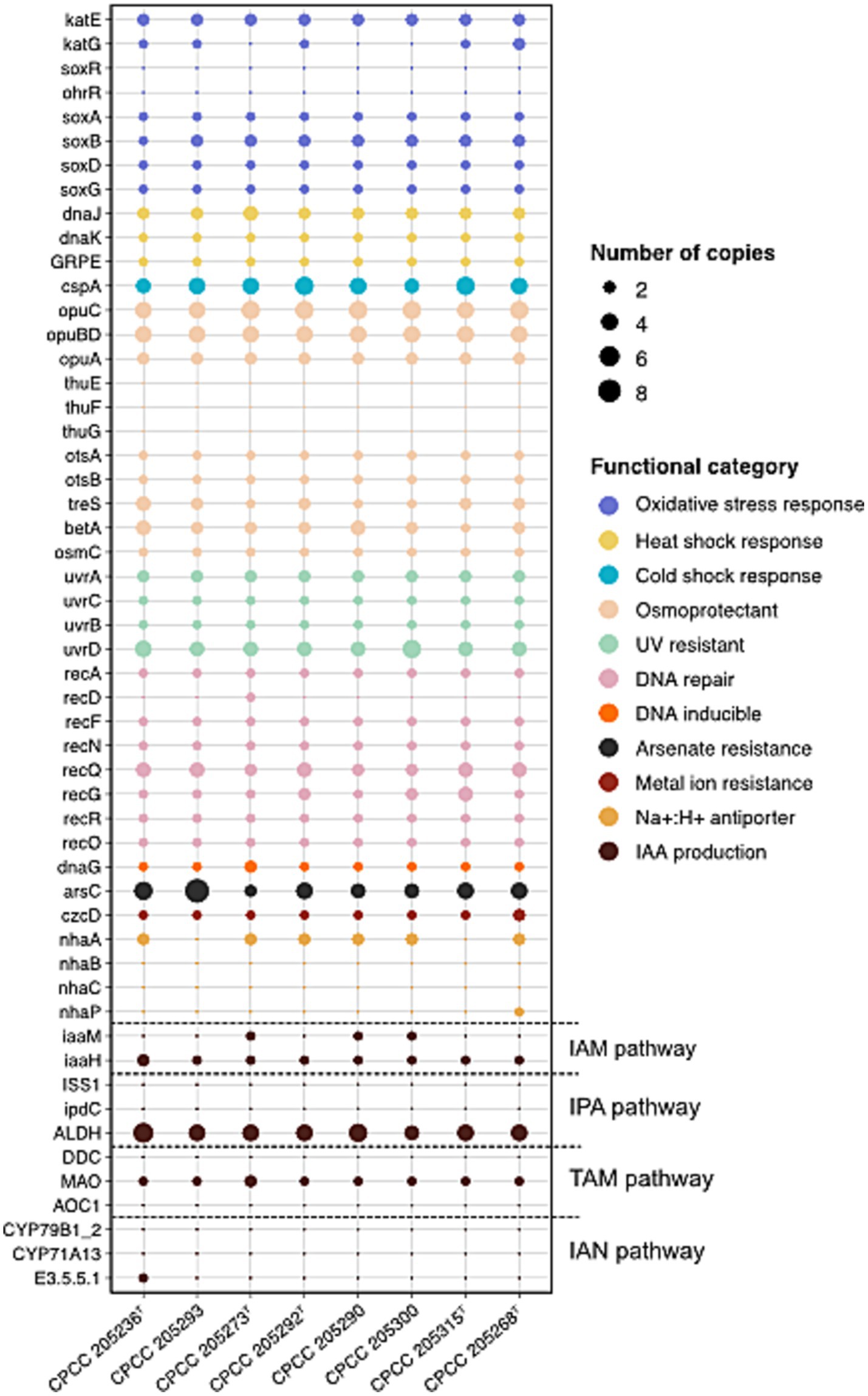
Figure 2. The distribution of genes involved in stress response and IAA production pathways in eight Kocuria strains isolated from desert soils.
In stress response mechanisms, IAA emerges as a key player in bacterial survival under extreme environmental conditions. Studies on Sinorhizobium meliloti have shown that the IAA-overproducing mutant exhibits enhanced resilience to UV irradiation, acidity, osmotic shock, and heat shock in comparison to the wild-type strain (Imperlini et al., 2009). In microorganisms, the L-Trp-dependent IAA biosynthesis is divided into five pathways according to the different intermediate metabolites: indole-3-acetamide (IAM), indole-3-pyruvic acid (IPA/IPyA), indole-3-acetonitrile (IAN), tryptamine (TAM), and TSO (L-Trp side-chain oxidase) pathways. Since the mechanism of the TSO pathway remains unknown, we excluded it and explored a pool of genes involved in the other four L-Trp-dependent pathways (Supplementary Table S4; Figure 2). Out of these four routes, the IAM pathway showed the most conserved genetic architecture in genomes of eight newly described Kocuria strains. While iaaH (encoding indoleacetamide hydrolase) was universally present, the key iaaM (encoding tryptophan-2-monooxygenase) was exclusively detected in genomes of strain CPCC 205273T, CPCC 205290, and CPCC 205300. Intriguingly, domain analysis (via InterProScan with default options) revealed uncharacterized sequences harboring a catalytic core domain of IaaM (Amino_oxidase) across all strains, suggesting the existence of IaaM isozymes with analogous functions. In addition, other actinobacteria phylogenetically related to Kocuria predominantly utilized the IAM pathway for IAA synthesis, supported by conserved iaaM/iaaH clusters (Lin and Xu, 2013; Myo et al., 2019). Therefore, these observations implied that these Kocuria strains employed the IAM pathway for IAA production, potentially relying on isozymes to compensate for incomplete iaaM in certain strains.
3.5 Secondary metabolite biosynthesis gene clusters
Actinobacteria, particularly those found in extreme habitats, possess a diverse range of unique bioactive chemicals through their secondary metabolism, which may have significant medical applications. Here, using antiSMASH, the secondary metabolite BGCs of these desert-associated previously uncharacterized strains were identified based on their genome sequence. Eight Kocuria strains exhibited six to nine BGCs with moderate similarities to previously described secondary metabolite biosynthetic gene clusters (Supplementary Table S5). These BGCs exhibited 5–100% similarities to documented secondary metabolite biosynthetic gene clusters including those for carotenoid, dactylocycline A, desferrioxamin B, microansamycin, stenothricin, glycopeptidolipid, ε-poly-L-lysine, oxalomycin B, salinipeptin A/B/C/D, FW0622 and branched-chain fatty acids. In addition, other unidentified secondary metabolite clusters were predicted that were attributable to those encoding NAPAA (Non-alpha poly-amino acids like e-Polylysin), RiPP-like (Post-translationally modified peptide like) compounds, indole, NRPS-like (Non-ribosomal peptide synthetase like) compounds, T3PKS (Type III polyketide synthases), linaridin, NI-siderophore, and terpene types (Supplementary Table S5).
3.6 Functional genes enrichment in desert-derived Kocuria strains
To decipher the underlying mechanism of desert adaptation among Kocuria, we conducted a comparative analysis for desert-associated Kocuria strains (DAK) and other non-desert-associated Kocuria strains (NDAK). The genome size of DAK was 4.24 Mb ± 0.51 on average, which was significantly larger than that of NDAK (3.09 Mb ± 0.34) (Mann–Whitney U test, p = 4e-8) (Figure 3A). However, the G + C content has no difference between DAK (70.4% ± 2.02%) and NDAK (69.7% ± 2.99%) (Mann–Whitney U test, p = 0.66) (Figure 3B). Based on 207 SOGs, we performed a phylogenomic analysis among the 143 Kocuria strains from different habitats to estimate their evolutionary relationships. A maximum-likelihood species tree showed that these Kocuria strains diverged into six clusters labeled as Cluster I-VI, respectively (Figure 3C). All of the DAK strains fell into Cluster IV except for CPCC 205295. Interestingly, by evaluating the size of their genomes, we found that the strains with relatively larger genomes were also mainly clustered into Cluster IV. It implied that the Kocuria strain might exhibit genomic expansion as a concomitant response to its adaptation to the desert environment.
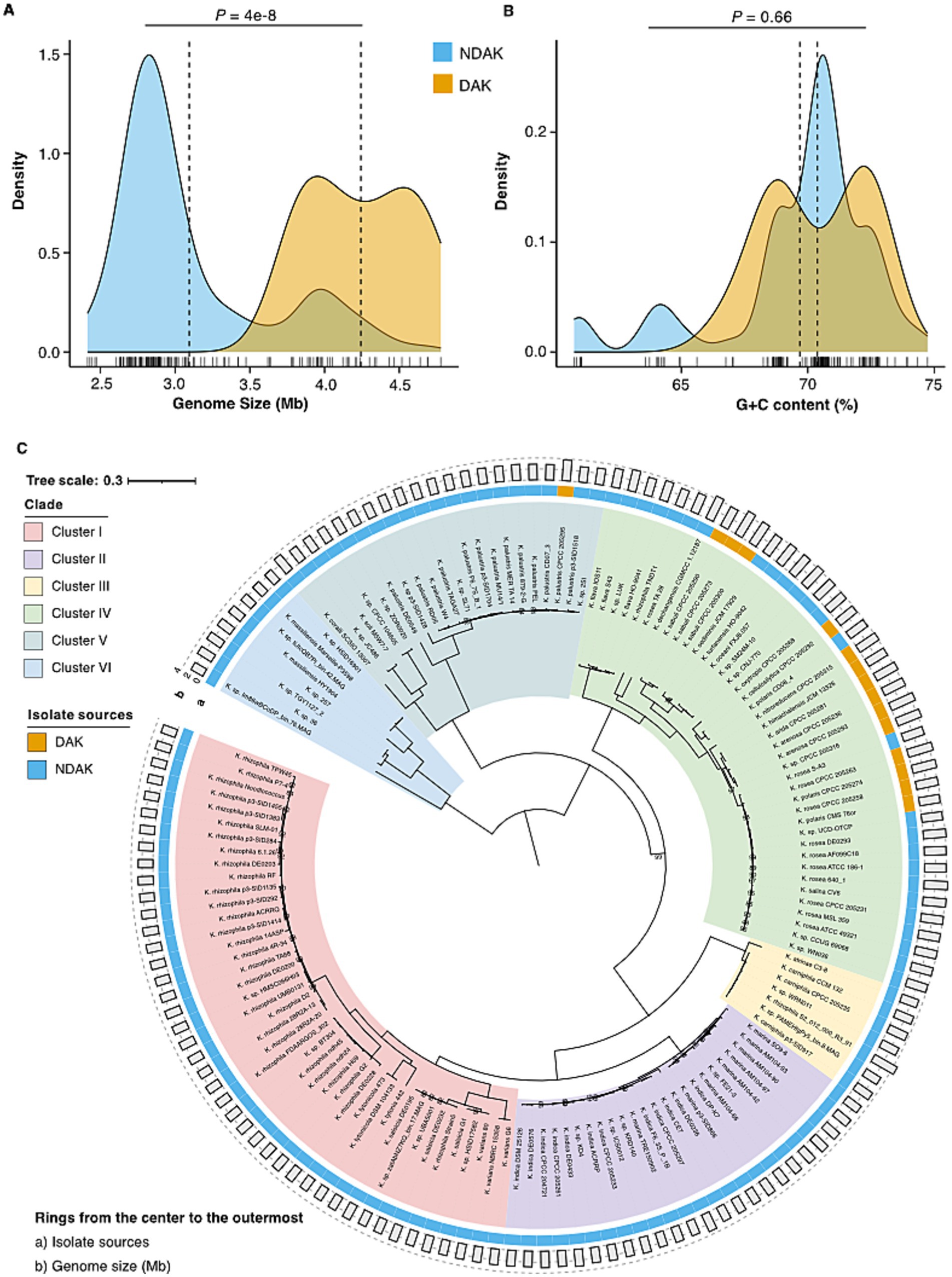
Figure 3. Genomic features comparison and phylogenomics of Kocuria strains. (A) The comparisons of genomic size and G + C content between DAK and NDAK strains. (B) The comparisons of G+C content between DAK and NDAK strains. (C) The phylogenetic tree based on the genome sequences of 143 Kocuria strains. Rings outside the tree are depicted by isolated sources (ring a) and genomic size (ring b). Bootstrap values that are equal to 100% are not shown. The scale bar denotes 0.3 substitutions per amino acid position.
The comparison of the core genome functions between DAK and NDAK strains was performed according to the methods of a previous study (See Methods). The results based on the COG database show that COG categories involved in the Intracellular trafficking, secretion, and vesicular transport (COG U), Cell wall/membrane/envelope biogenesis (COG M), carbohydrate transport and metabolism (COG G), and lipid transport and metabolism (COG I) were enriched in the DAK strains (Figure 4A top panel; Supplementary Table S6). As for the KEGG database, 21 pathways were significantly different between DAK and NDAK strains, and many of these metabolic pathways were related to environmental adaptation, including ABC transports, two-component system, and energy metabolism pathways (Figure 4A bottom panel; Supplementary Table S7) (Aujoulat et al., 2012; Ma et al., 2021). In particular, the relative abundance of starch and sucrose metabolism of DAK strains was two times the level of NDAK.
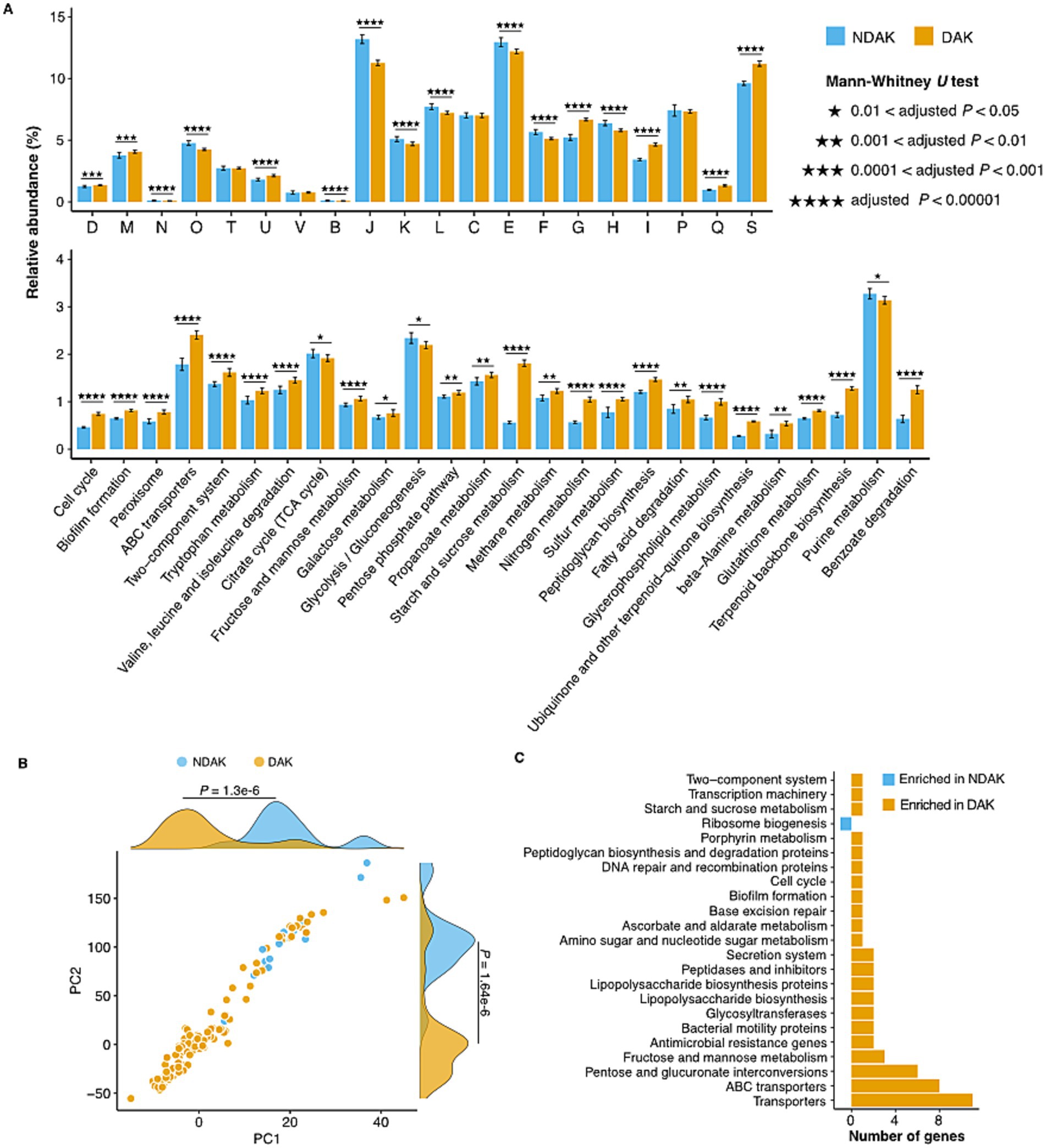
Figure 4. Enrichment analysis of functional categories in DAK and NDAK strains. (A) Pairwise comparison of core genomes among DAK and NDAK strains based on COG (top panel) and KEGG database (bottom panel). Bar charts represent the relative abundance of gene categories in statistical tests, and the error bars represent the standard deviations of relative abundance of categories in the core genomes of DAK or NDAK strains. (B) Phylogenetically informed principal-component analysis (phylo-PCA) of the KEGG pathways between DAK and NDAK strains. (C) Pan-GWAS analysis results showing typical KOs associated with desert environments by using SCOARY.
Genome-wide differences investigation of the KEGG pathways between DAK (15 strains) and NDAK (128 strains) strains was performed by phylogenetically informed principal component analysis (phylo-PCA) and pan-genome-wide association studies (Pan-GWAS) based on the presence/absence data of KOs. Firstly, phylo-PCA analysis shows that KEGG pathways in DAK and NDAK strains were divided into two clusters, which were significantly different (Mann–Whitney U test, p < 0.05) on both the PC1 and PC2 axes (Figure 4B), suggesting the distinct functional composition among DAK and NDAK strains. In addition, Pan-GWAS identified 30 KOs that were positively associated with desert habitat with an adjusted p value less than 0.05 (Figure 4C; Supplementary Table S8). The majority of these enriched KOs were assigned to the pathways of environmental signal response (transports and ABC transports) and carbohydrate metabolism (fructose and mannose metabolism and pentose and glucuronate interconversions), suggesting the major role in the desert adaptation of Kocuria strains.
4 Conclusion
In this study, we isolated and identified 21 Kocuria strains from diverse ecosystems. Based on the polyphasic taxonomic approaches, strains CPCC 205236T (together with CPCC 205293), CPCC 205273T, CPCC 205292T (together with CPCC 205290 and together with CPCC 205300), CPCC 205315T, and CPCC 205268T were verified to represent five new species of the genus Kocuria. Phenotypic and genomic analysis of these new strains demonstrated that the adaptation to desert niches may be achieved through various response strategies to UV radiation, carbon starvation, desiccation, and osmotic stress. Most of these newly isolated strains (6/8, 75%) could convert L-Trp to IAA by qualitative analysis of the fermentation, despite genome analysis revealing that only one of them processed the potential of IAA production. Moreover, the comparative genomics analysis revealed that the genes related to carbohydrate metabolism and transporters have been enriched in desert-associated Kocuria strains. Our findings underscored the critical role of Kocuria organisms in hyper-arid deserts and uncovered several genomic traits for adaption to the desert environment.
5 Taxonomic descriptions
5.1 Species descriptions
5.1.1 Kocuria arenosa sp. nov.
Kocuria arenosa (a.re.no’sa. L. fem. Adj. arenosa, sandy, dwelling in desert sand).
Cells are Gram-stain-positive, aerobic, coccoid (0.8–1.2 μm in diameter), non-motile, non-spore-forming. Colonies are 0.8–2.0 mm in diameter, convex with smooth, round, uniformly edged, opaque and pastel orange (sometimes pale orange or orange) in color on TSA medium. Growth temperature and pH range for growth are 4–42°C and pH 6.0–11.0, with optima growth at 26–32°C and pH 7.0–9.0. Can tolerate up to 10% (w/v) NaCl. Catalase-positive and oxidase-negative. Hydrolysis of starch are positive, but negetive for hydrolysis of gelatin and cellulose. Assimilation of D-arabitol, D-cellobios, D-, D-sorbitol, L-alanine, Inosine, gentiobiose, and salicin. Cells are positive for alkaline phosphatase, esterase lipase (C8), and β-glucosidase, but negetive for cystine arylamidase and β-galactosidase. IAA is detected from the fermentation broth. The polar lipids profile mainly contains diphosphatidylglycerol and phosphatidylglycerol. The major fatty acids (>10%) consist of anteiso-C15:0, C16:1 ω7c and iso-C15:0. The predominant quinone is MK-8(H2), with minor of MK-7(H2) and MK-9(H2).
The type strain, CPCC 205236T (= KCTC 59060T), was isolated from sand soil in Gurbantunggut Desert, China. The genome size is 4.2 Mbp, and the genomic DNA G + C content is 71.8%. The 16S rRNA gene and whole genome sequence of strain CPCC 205236T are publicly available with the accession numbers OR431689 and GCA_040981255.1, respectively.
5.1.2 Kocuria sabuli sp. nov.
Kocuria sabuli (sa’bu.li. L. gen. Neut. n. sabuli, of sand).
Cells are Gram-stain-positive, aerobic, coccoid (0.8–1.2 μm in diameter), non-motile by peritrichous flagella, non-spore-forming. Colonies are 1.0–2.0 mm in diameter, convex with smooth, round, uniformly edged, opaque and pale orange in color on TSA medium. Growth temperature and pH range for growth are 4–37°C and pH 6.0–11.0, with optima growth at 26–32°C and pH 7.0–9.0. Growth in the presence of up to 10 or 15% (w/v) NaCl. Catalase and oxidase negative. Hydrolysis of starch is positive and cellulose is negative. D-cellobiose, dextrin, gentiobiose, and L-pyroglutamic acid are assimilated. Cells are positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), and α-glucosidase and β-glucosidase and negative for acid phosphatase, lipase (C14), cystine arylamidase, α-galactosidase. The polar lipid profiles are diphosphatidylglycerol, phosphatidylglycerol. The major fatty acids (>10%) consist of anteiso-C15:0, and C16:1 ω7c. The predominant quinone is MK-8(H2), with minor amount of MK-7(H2) and MK-9(H2).
The type strain, CPCC 205273T (= KCTC 59062T), was isolated from soil from the Badain Jaran Desert, China. The genome size is 4.8 Mbp, and the genomic DNA G + C content is 69.0%. The 16S rRNA gene and whole genome sequence of strain CPCC 205273T are publicly available with the accession numbers OR431691 and GCA_040981185.1, respectively.
5.1.3 Kocuria cellulosilytica sp. nov.
Kocuria cellulosilytica (cel.lu.lo.si.ly‘ti.ca. N.L. neut. n. cellulosum, cellulose; N.L. adj. Lyticus, dissolving; N.L. fem. Adj. cellulosilytica, dissolving cellulose).
Cells are Gram-stain-positive, non-spore-forming, non-motile by flagella, and strictly aerobic. Single cell is coccoid (0.8–1.2 μm in diameter). Colonies are pastel orange. Growth temperature and pH range for growth are 4–42°C and pH 7.0–11.0, with optimum growth at 26–32°C and pH 7.0–9.0. Can tolerate up to 10% (w/v) NaCl. Positive for catalase, hydrolysis of cellulose, gelatin and starch, MR test, cystine arylamidase, esterase lipase (C8), valine arylamidase, and α-glucosidase, negative for acid phosphatase (C14), alkaline phosphatase, oxidase, lipase, trypsin, α-chymotrypsin, α-fucosidase, α-galactosidase, β-glucosidase, β-glucuronidase, β-galactosidase, α-mannosidase, H2S production, siderophore production, and VP test. D-arabitol, D-cellobiose, D-galactose, D-raffinose, D-sorbitol, and L-alanine can be assimilated but D-melibiose, D-salicin, L-arginine. α-D-lactose, and can not. The predominant quinones are MK-7(H2), MK-8(H2) and MK-9(H2). The fatty acid profile consists of the predominant components (> 10%) anteiso-C15:0 and C16:0.
The type strain, CPCC 205292T (=I19A-01430T = CGMCC 1.60067T), was isolated from a sand soil in the Badain Jaran Desert, China. The genome size is 3.9 Mbp, and the genomic DNA G + C content is 72.8%. The 16S rRNA gene and whole genome sequence of strain CPCC 205292T are publicly available with the accession numbers OR431698 and GCA_040981245.1, respectively.
5.1.4 Kocuria nitroreducens sp. nov.
Kocuria nitroreducens (ni.tro.re.du’cens. N.L. masc. n. nitras (gen. nitratis), nitrate; N.L. pref. Nitro-, pertaining to nitrate (in compound words); L. part. Pres. reducens, leading back, bringing back to a more reduced state; N.L. part. Adj. nitroreducens, reducing nitrate).
Cells are aerobic, Gram-stain-positive, coccoid (1.0–1.6 μm in diameter), non-motile and non-spore-forming. Colonies are 1.0–2.0 mm in diameter, convex with smooth, round, uniformly edged, opaque and pale orange in color on TSA medium. Growth temperature and pH range for growth are 4–37°C and pH 6.0–11.0, with optima growth at 26–32°C and pH 7.0–9.0. Cannot tolerate >10% (w/v) NaCl. Catalase-positive and oxidase-negative. Positive for nitrate reduction, gelatin hydrolyzation and starch hydrolysis; negative for cellulose hydrolyzation and H2S production. D-arabitol, D-cellobiose, D-salicin, D-sorbitol, L-alanine, gentiobiose, and inosine were assimilated. Activities of alkaline phosphatase, cystine arylamidase, esterase (C4), esterase lipase (C8), leucine arylamidase, naphthol-AS-B1-phosphohydrolase, valine arylamidase and β-glucosidase are detected; the following characteristics are absent: acid phosphatase, lipase (C14), N-acetyl-β-glucosaminidase, trypsin, α-chymotrypsin, α-fucosidase, α-galactosidase, α-glucosidase, β-glucuronidase, β-galactosidase, and α-mannosidase. Diphosphatidylglycerol, phosphatidylglycerol and are detected in the polar lipids extraction. The predominant quinone is MK-8(H2), with minor of MK-7(H2) and MK-9(H2). The fatty acid profile consists of the predominant components (> 10%) anteiso-C15:0 and iso-C15:0.
The type strain, CPCC 205315T (=KCTC 59068T), was isolated from soil in Gurbantunggut Desert, China. The genome size is 4.2 Mbp, and the genomic DNA G + C content is 69.2%. The 16S rRNA gene and whole genome sequence of strain CPCC 205315T are publicly available with the accession numbers OR431660 and GCA_042879195.1, respectively.
5.1.5 Kocuria oxytropis sp. nov.
Kocuria oxytropis (o.xy.tro’pis. N.L. gen. n. oxytropis, of Oxytropis, a plant genus, referring to the isolation of the type strain from Oxytropis falcata).
Cells are aerobic, Gram-stain-positive, non-motile, non-spore-forming, coccoid (0.5–0.8 μm in diameter). Colonies are 1.2–2.2 mm in diameter, convex with smooth, round, uniformly edged, opaque and pale orange in color on TSA medium. Growth temperature and pH range for growth are 4–42°C and pH 6.0–11.0, with optima growth at 26–32°C and pH 7.0–9.0. Can tolerate up to 15% (w/v) NaCl. Catalase-positive and oxidase-negative. Positive for cellulose hydrolyzation, gelatin hydrolyzation, nitrate reduction, starch hydrolysis; negative for H2S production. Assimilation of D-Cellobiose, Dextrin, D-Salicin, D-Serine, D-Sorbitol, inosine, L-Alanine, and L-Arginine. Activities of alkaline phosphatase, esterase lipase (C8), valine arylamidase, α-glucosidase, and β-glucosidase are detected. The following activities are not detected: acid phosphatase, α-galactosidase, β-galactosidase. Diphosphatidylglycerol and phosphatidylglycerol are detected in the polar lipid extraction. The predominant quinone is MK-8(H2), with minor of MK-7(H2) and MK-9(H2). The fatty acid profile (> 10%) consists of the anteiso-C15:0 and iso-C15:0.
The type strain, CPCC 205268T (= KCTC 59061T), was isolated from the rhizosphere soil of Oxytropis falcata. The genome size is 4.0 Mbp, and the genomic DNA G + C content is 72.3%. The 16S rRNA gene and whole genome sequence of strain CPCC 206268T are publicly available with the accession numbers OR431690 and GCF_040981195.1, respectively.
5.2 Emended species descriptions
5.2.1 Emended description of Kocuria rosea (Flügge 1886) (Stackebrandt et al., 1995)
Heterotypic synonym: Kocuria polaris (Reddy et al., 2003).
The description is based on the data reported in the (Stackebrandt et al., 1995; Reddy et al., 2003) and this study. Cells are coccoid or spherical (1·0–1·5 μm in diameter), occurring in pairs, tetrads or clusters. Non-motile, Gram-positive, and aerobic. Colonies appear smooth, round, uniformly edged, translucent, mucoid, circular, slightly convex, orange, pink, or red. Grows between 4 and 52°C, with optimum growth at 26–32°C. Grows occur at pH 7–12 and tolerate up to 10% NaCl. Catalase, cystine arylamidase, and lipase-positive. Negative for acid phosphatase, alkaline phosphatase, L-arginine decarboxylase, L-arginine dihydrolase, L-lysine decarboxylase, and β-galactosidase. Utilizes acetate, D-cellobiose, D-fructose, D-galactose, D-glucose, D-mannose, D-xylose, glycerol, L-glutamine, L-glycine, L-serine, L-threonine, rhamnose, and sorbitol as sole carbon sources, but can not utilize citrate, D-cysteine, D-ribose, D-sorbose, D-trehalose, dextran, dulcitol, L-histidine, L-isoleucine, L-lysine, L-methionine, L-proline, L-tryptophan, L-tyrosine, L-valine, lactic acid, melezitose, meso-erythritol, succinic acid, sucrose, thioglycollate, and β-hydroxybutyric acid as sole carbon sources. The major fatty acids are anteiso-C15:0, iso-C15:0. The predominant menaquinones are MK-7(H2) and MK-8(H2). The G + C content of the type strain genome is 72.75 L% and the approximate genome size is 3.88 Mbp. The accession numbers of the 16S rRNA gene and assembly genome sequence are X87756 and GCF_006717035.1, respectively.
The type strain is ATCC 186T (CCM 679T = CCUG 4312T = CIP 71.15T = DSM 20447T = IEGM 394T = IFO 3768T = JCM 11614T = LMG 14224T = NBRC 3768T = NCTC 7523T = NRRL B-2977T = VKM B-1823T = CMS 76orT = MTCC 3702T = DSM 14382T).
5.2.2 Emended description of Kocuria marina
Heterotypic synonym: Kocuria indica (Dastager et al., 2014).
The description is based on the data reported in the previous studies (Kim et al., 2004; Dastager et al., 2014). Cells are Gram-positive, aerobic, non-motile, and coccoid. Catalase-positive, oxidase-negative. Growth occurs in the presence of up to 15% NaCl, although its presence is not required for growth. The temperature range for growth is 4–45°C and optimum growth is observed at 28–30°C. Cellobiose, cupric acid, D-fructose, D-galactose, D-glucose, D-mannitol, D-mannose, D-ribose, inositol, L-rhamnose, lactose, melezitose, melibiose, phenylacetic acid, salicin, sucrose glycogen, trehalose, trisodium citrate, and turanose can be used as sole carbon sources for energy and growth, but not 2-ketogluconate, 5-ketogluconate, aesculin, amygdalin, arabitol, arbutin, D-adonitol, D-arabinose, D-lyxose, D-sorbitol, D-tagatose, dulcitol, erythritol, fucose, gentiobiose, glycerol, inulin, L-arabinose, L-sorbose, lactose, maltose, methyl α-D-gluocopyranoside, methyl α-D-mannopyranoside, methyl β-D-xylopyranoside, N-acetylglucosamine, potassium gluconate, raffinose, sucrose, xylitol, xylose. Positive for hydrolysis of starch. The major fatty acid (>10%) contains anteiso-C15:0, iso-C16:0 and anteiso-C17:0. The G + C content of the type strain genome is 68.82% and the approximate genome size is 2.79 Mbp. The accession numbers of the 16S rRNA gene and assembly genome sequence are AY211385 and GCF_014652975.1, respectively.
The type strain is CCUG 51442T (DSM 16420T = JCM 13363T = KCTC 9943T = KMM 3905T = NIO-1021T = NCIM 5455T = DSM 25126T = CCTCC AA 209050T).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
C-JL: Validation, Investigation, Software, Visualization, Writing – original draft. Z-MJ: Investigation, Validation, Writing – original draft, Resources. X-YZ: Validation, Writing – review & editing. H-HC: Validation, Writing – review & editing, Resources. L-YY: Resources, Validation, Writing – review & editing. G-FL: Validation, Writing – review & editing. Y-QZ: Resources, Validation, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the National Natural Science Foundation of China (32170021), CAMS Innovation Fund for Medical Sciences (CIFMS, 2021-I2M-1-055), Key project at the central government level the ability establishment of sustainable use for valuable Chinese medicine resources (2060302), Scientific Research Foundation of Yunnan Provincial Department of Education (2021J1121) and the National Infrastructure of Microbial Resources (NIMR-2024-3).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1547983/full#supplementary-material
Footnotes
1. ^https://lpsn.dsmz.de/genus/kocuria
2. ^https://www.ezbiocloud.net/
4. ^https://www.ebi.ac.uk/interpro/
References
Allison, S. D. (2023). Microbial drought resistance may destabilize soil carbon. Trends Microbiol. 31, 780–787. doi: 10.1016/j.tim.2023.03.002
Auch, A. F., Von Jan, M., Klenk, H.-P., and Göker, M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2, 117–134. doi: 10.4056/sigs.531120
Aujoulat, F., Roger, F., Bourdier, A., Lotthe, A., Lamy, B., Marchandin, H., et al. (2012). From environment to man: genome evolution and adaptation of human opportunistic bacterial pathogens. Genes 3, 191–232. doi: 10.3390/genes3020191
Bianco, C., Imperlini, E., and Defez, R. (2009). Legumes like more IAA. Plant Signal. Behav. 4, 763–765. doi: 10.4161/psb.4.8.9166
Brynildsrud, O., Bohlin, J., Scheffer, L., and Eldholm, V. (2016). Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 17:238. doi: 10.1186/s13059-016-1108-8
Bull, A. T., Asenjo, J. A., Goodfellow, M., and Gomez-Silva, B. (2016). The Atacama Desert: technical resources and the growing importance of novel microbial diversity. Ann. Rev. Microbiol. 70, 215–234. doi: 10.1146/annurev-micro-102215-095236
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cerboneschi, M., Decorosi, F., Biancalani, C., Ortenzi, M. V., Macconi, S., Giovannetti, L., et al. (2016). Indole-3-acetic acid in plant-pathogen interactions: a key molecule for in planta bacterial virulence and fitness. Res. Microbiol. 167, 774–787. doi: 10.1016/j.resmic.2016.09.002
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chowdhury, N., Kwan, B. W., and Wood, T. K. (2016). Persistence increases in the absence of the alarmone guanosine tetraphosphate by reducing cell growth. Sci. Rep. 6:20519. doi: 10.1038/srep20519
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., Da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Chun, J., and Rainey, F. A. (2014). Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 64, 316–324. doi: 10.1099/ijs.0.054171-0
Collins, M. D., Pirouz, T., Goodfellow, M., and Minnikin, D. E. (1977). Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230. doi: 10.1099/00221287-100-2-221
Cordero, R. R., Damiani, A., Jorquera, J., Sepulveda, E., Caballero, M., Fernandez, S., et al. (2018). Ultraviolet radiation in the Atacama Desert. Antonie Van Leeuwenhoek 111, 1301–1313. doi: 10.1007/s10482-018-1075-z
Dastager, S. G., Tang, S. K., Srinivasan, K., Lee, J. C., and Li, W. J. (2014). Kocuria indica sp. nov., isolated from a sediment sample. Int. J. Syst. Evol. Microbiol. 64, 869–874. doi: 10.1099/ijs.0.052548-0
Deng, Y., Han, X. F., Jiang, Z. M., Yu, L. Y., Li, Y., and Zhang, Y. Q. (2022). Characterization of three Stenotrophomonas strains isolated from different ecosystems and proposal of Stenotrophomonas mori sp. nov. and Stenotrophomonas lacuserhaii sp. nov. Front. Microbiol. 13:1056762. doi: 10.3389/fmicb.2022.1056762
Duca, D. R., and Glick, B. R. (2020). Indole-3-acetic acid biosynthesis and its regulation in plant-associated bacteria. Appl. Microbiol. Biotechnol. 104, 8607–8619. doi: 10.1007/s00253-020-10869-5
Ehmann, A. (1977). The van URK-Salkowski reagent — a sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives. J. Chromatogr. A 132, 267–276. doi: 10.1016/S0021-9673(00)89300-0
Emms, D. M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. doi: 10.1186/s13059-015-0721-2
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. doi: 10.1186/s13059-019-1832-y
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Georgiou, C. D., Sun, H. J., Mckay, C. P., Grintzalis, K., Papapostolou, I., Zisimopoulos, D., et al. (2015). Evidence for photochemical production of reactive oxygen species in desert soils. Nat. Commun. 6:7100. doi: 10.1038/ncomms8100
Gonzalez, C., Gutierrez, C., and Ramirez, C. (1978). Halobacterium vallismortis sp. nov. an amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can. J. Microbiol. 24, 710–715. doi: 10.1139/m78-119
Groth, I., Schumann, P., Rainey, F. A., Martin, K., Schuetze, B., and Augsten, K. (1997). Demetria terragena gen. Nov., sp. nov., a new genus of actinomycetes isolated from compost soil. Int. J. Syst. Bacteriol. 47, 1129–1133. doi: 10.1099/00207713-47-4-1129
Haidar, B., Ferdous, M., Fatema, B., Ferdous, A. S., Islam, M. R., and Khan, H. (2018). Population diversity of bacterial endophytes from jute (Corchorus olitorius) and evaluation of their potential role as bioinoculants. Microbiol. Res. 208, 43–53. doi: 10.1016/j.micres.2018.01.008
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernandez-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Idris, H., Goodfellow, M., Sanderson, R., Asenjo, J. A., and Bull, A. T. (2017). Actinobacterial rare biospheres and dark matter revealed in habitats of the Chilean Atacama Desert. Sci. Rep. 7:8373. doi: 10.1038/s41598-017-08937-4
Imperlini, E., Bianco, C., Lonardo, E., Camerini, S., Cermola, M., Moschetti, G., et al. (2009). Effects of indole-3-acetic acid on survival and on symbiotic nitrogen fixation and stem dry weight production. Appl. Microbiol. Biotechnol. 83, 727–738. doi: 10.1007/s00253-009-1974-z
Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Karnwal, A. (2019). Production of indole acetic acid by Kocuria rosea VB1 and Arthrobacter luteolus VB2 under the influence of L-tryptophan and maize root exudates. Biotechnologia 100, 29–35. doi: 10.5114/bta.2019.83209
Kim, S. B., Nedashkovskaya, O. I., Mikhailov, V. V., Han, S. K., Kim, K. O., Rhee, M. S., et al. (2004). Kocuria marina sp. nov., a novel actinobacterium isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 54, 1617–1620. doi: 10.1099/ijs.0.02742-0
Komagata, K., and Suzuki, K.-I. (1988). “4 lipid and Cell-Wall analysis in bacterial systematics” in Methods in microbiology. eds. R. R. Colwell and R. Grigorova (Cambridge, MA: Academic Press), 161–207.
Kovacs, G., Burghardt, J., Pradella, S., Schumann, P., Stackebrandt, E., and Marialigeti, K. (1999). Kocuria palustris sp. nov. and Kocuria rhizophila sp. nov., isolated from the rhizoplane of the narrow-leaved cattail (Typha angustifolia). Int. J. Syst. Bacteriol. 49, 167–173. doi: 10.1099/00207713-49-1-167
Kroppenstedt, R. M. (1985). “Fatty acid and menaquinone analysis of actinomycetes and related organisms” in Society of Applied Bacteriology. Technical series (London: Elsevier Science & Technology Books), 173–199.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kumar, C. G., and Sujitha, P. (2014). Kocuran, an exopolysaccharide isolated from Kocuria rosea strain BS-1 and evaluation of its in vitro immunosuppression activities. Enzym. Microb. Technol. 55, 113–120. doi: 10.1016/j.enzmictec.2013.10.007
Lee, J., Jeong, B., Bae, H. R., Jang, H. A., and Kim, J. K. (2023). Trehalose biosynthesis gene otsA protects against stress in the initial infection stage of Burkholderia-bean bug symbiosis. Microbiol. Spectr. 11:e0351022. doi: 10.1128/spectrum.03510-22
Levy, A., Salas Gonzalez, I., Mittelviefhaus, M., Clingenpeel, S., Herrera Paredes, S., Miao, J., et al. (2017). Genomic features of bacterial adaptation to plants. Nat. Genet. 50, 138–150. doi: 10.1038/s41588-017-0012-9
Li, W. J., Xu, P., Schumann, P., Zhang, Y. Q., Pukall, R., Xu, L. H., et al. (2007). Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int. J. Syst. Evol. Microbiol. 57, 1424–1428. doi: 10.1099/ijs.0.64749-0
Lin, L., and Xu, X. (2013). Indole-3-acetic acid production by endophytic Streptomyces sp. En-1 isolated from medicinal plants. Curr. Microbiol. 67, 209–217. doi: 10.1007/s00284-013-0348-z
Lushchak, V. I. (2001). Oxidative stress and mechanisms of protection against it in bacteria. Biochemistry 66, 476–489. doi: 10.1023/a:1010294415625
Ma, Y., Zhang, Y., Chen, K., Zhang, L., Zhang, Y., Wang, X., et al. (2021). The role of PhoP/PhoQ two component system in regulating stress adaptation in Cronobacter sakazakii. Food Microbiol. 100:103851. doi: 10.1016/j.fm.2021.103851
Magee, C. M., Rodeheaver, G., Edgerton, M. T., and Edlich, R. F. (1975). A more reliable gram staining technic for diagnosis of surgical infections. Am. J. Surg. 130, 341–346. doi: 10.1016/0002-9610(75)90398-0
Mayilraj, S., Kroppenstedt, R. M., Suresh, K., and Saini, H. S. (2006). Kocuria himachalensis sp. nov., an actinobacterium isolated from the Indian Himalayas. Int. J. Syst. Evol. Microbiol. 56, 1971–1975. doi: 10.1099/ijs.0.63915-0
Mende, D. R., Letunic, I., Maistrenko, O. M., Schmidt, T. S. B., Milanese, A., Paoli, L., et al. (2019). proGenomes2: an improved database for accurate and consistent habitat, taxonomic and functional annotations of prokaryotic genomes. Nucleic Acids Res. 48, D621–D625. doi: 10.1093/nar/gkz1002
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Minnikin, D. E., O'donnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241. doi: 10.1016/0167-7012(84)90018-6
Myo, E. M., Ge, B., Ma, J., Cui, H., Liu, B., Shi, L., et al. (2019). Indole-3-acetic acid production by Streptomyces fradiae NKZ-259 and its formulation to enhance plant growth. BMC Microbiol. 19:155. doi: 10.1186/s12866-019-1528-1
Neilson, J. W., Quade, J., Ortiz, M., Nelson, W. M., Legatzki, A., Tian, F., et al. (2012). Life at the hyperarid margin: novel bacterial diversity in arid soils of the Atacama Desert, Chile. Extremophiles 16, 553–566. doi: 10.1007/s00792-012-0454-z
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nunoshiba, T., Hidalgo, E., Li, Z., and Demple, B. (1993). Negative autoregulation by the Escherichia coli SoxS protein: a dampening mechanism for the soxRS redox stress response. J. Bacteriol. 175, 7492–7494. doi: 10.1128/jb.175.22.7492-7494.1993
Palomo, S., Gonzalez, I., De La Cruz, M., Martin, J., Tormo, J. R., Anderson, M., et al. (2013). Sponge-derived Kocuria and Micrococcus spp. as sources of the new thiazolyl peptide antibiotic kocurin. Mar. Drugs 11, 1071–1086. doi: 10.3390/md11041071
Pantoja-Guerra, M., Valero-Valero, N., and Ramírez, C. A. (2023). Total auxin level in the soil–plant system as a modulating factor for the effectiveness of PGPR inocula: a review. Chem. Biol. Technol. Agric. 10:6. doi: 10.1186/s40538-022-00370-8
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Publications Office of the European Union (2018). World atlas of desertification: Rethinking land degradation and sustainable land management. Luxembourg: Publications Office of the European Union.
Rath, H., Reder, A., Hoffmann, T., Hammer, E., Seubert, A., Bremer, E., et al. (2020). Management of osmoprotectant uptake hierarchy in Bacillus subtilis via a SigB-dependent antisense RNA. Front. Microbiol. 11:622. doi: 10.3389/fmicb.2020.00622
Reddy, G. S. N., Prakash, J. S. S., Prabahar, V., Matsumoto, G. I., Stackebrandt, E., and Shivaji, S. (2003). Kocuria polaris sp. nov., an orange-pigmented psychrophilic bacterium isolated from an Antarctic cyanobacterial mat sample. Int. J. Syst. Evol. Microbiol. 53, 183–187. doi: 10.1099/ijs.0.02336-0
Retamal-Morales, G., Mehnert, M., Schwabe, R., Tischler, D., Zapata, C., Chavez, R., et al. (2018). Detection of arsenic-binding siderophores in arsenic-tolerating Actinobacteria by a modified CAS assay. Ecotoxicol. Environ. Saf. 157, 176–181. doi: 10.1016/j.ecoenv.2018.03.087
Revell, L. J. (2024). Phytools 2.0: an updated R ecosystem for phylogenetic comparative methods (and other things). PeerJ 12:e16505. doi: 10.7717/peerj.16505
Riesco, R., and Trujillo, M. E. (2024). Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 74:6300. doi: 10.1099/ijsem.0.006300
Rushabh, S., Kajal, C., Prittesh, P., Amaresan, N., and Krishnamurthy, R. (2020). Isolation, characterization, and optimization of indole acetic acid–producing providencia species (7MM11) and their effect on tomato (Lycopersicon esculentum) seedlings. Biocatal. Agric. Biotechnol. 28:101732. doi: 10.1016/j.bcab.2020.101732
Sahar, A. M., Ghada, S. I., Maha, I. A. K., Ahmed, M. A.-H., Noor Mohammed, B., and And Mohamed, A.-Z. (2022). Production and partial characterization of yellow pigment produced by Kocuria flava isolate and testing its antioxidant and antimicrobial activity: life sciences-biochemistry for better diagnosis and therapy. Int. J. Life Sci. Pharma Res. 10, 58–66. doi: 10.22376/ijpbs/lpr.2020.10.2.L58-66
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Samanta, A. K., Chaudhuri, S., and Dutta, D. (2016). Antioxidant efficacy of carotenoid extract from bacterial strain Kocuria marina DAGII. Materials Today 3, 3427–3433. doi: 10.1016/j.matpr.2016.10.023
Sasser, M. (1990). Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl 20, 1–6.
Sayed, A. M., Hassan, M. H. A., Alhadrami, H. A., Hassan, H. M., Goodfellow, M., and Rateb, M. E. (2020). Extreme environments: microbiology leading to specialized metabolites. J. Appl. Microbiol. 128, 630–657. doi: 10.1111/jam.14386
Schleifer, K. H., and Kandler, O. (1972). Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477. doi: 10.1128/br.36.4.407-477.1972
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shirling, E. B., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species1. Int. J. Syst. Evol. Microbiol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Stackebrandt, E., Koch, C., Gvozdiak, O., and Schumann, P. (1995). Taxonomic dissection of the genus Micrococcus: Kocuria gen. Nov., Nesterenkonia gen. Nov., Kytococcus gen. Nov., Dermacoccus gen. Nov., and Micrococcus Cohn 1872 gen. Emend. Int. J. Syst. Bacteriol. 45, 682–692. doi: 10.1099/00207713-45-4-682
Sun, Y., Chen, H. H., Sun, H. M., Ai, M. J., Su, J., Yu, L. Y., et al. (2017). Naumannella huperziae sp. nov., an endophytic actinobacterium isolated from Huperzia serrata (Thunb.). Int. J. Syst. Evol. Microbiol. 67, 1867–1872. doi: 10.1099/ijsem.0.001882
Sun, Y., Shi, Y. L., Wang, H., Zhang, T., Yu, L. Y., Sun, H., et al. (2018). Diversity of bacteria and the characteristics of actinobacteria community structure in Badain Jaran Desert and Tengger Desert of China. Front. Microbiol. 9:1068. doi: 10.3389/fmicb.2018.01068
Szabo, A., Langer, T., Schroder, H., Flanagan, J., Bukau, B., and Hartl, F. U. (1994). The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc. Natl. Acad. Sci. USA 91, 10345–10349. doi: 10.1073/pnas.91.22.10345
Vikram, S., Guerrero, L. D., Makhalanyane, T. P., Le, P. T., Seely, M., and Cowan, D. A. (2016). Metagenomic analysis provides insights into functional capacity in a hyperarid desert soil niche community. Environ. Microbiol. 18, 1875–1888. doi: 10.1111/1462-2920.13088
Wang, K., Zhang, L., Liu, Y., Pan, Y., Meng, L., Xu, T., et al. (2015). Kocuria dechangensis sp. nov., an actinobacterium isolated from saline and alkaline soils. Int. J. Syst. Evol. Microbiol. 65, 3024–3030. doi: 10.1099/ijs.0.000372
Xie, Y., Wu, G., Tang, J., Luo, R., Patterson, J., Liu, S., et al. (2014). SOAPdenovo-trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30, 1660–1666. doi: 10.1093/bioinformatics/btu077
Xu, P., Li, W. J., Tang, S. K., Zhang, Y. Q., Chen, G. Z., Chen, H. H., et al. (2005). Naxibacter alkalitolerans gen. Nov., sp. nov., a novel member of the family 'Oxalobacteraceae' isolated from China. Int. J. Syst. Evol. Microbiol. 55, 1149–1153. doi: 10.1099/ijs.0.63407-0
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yu, X., Li, Y., Cui, Y., Liu, R., Li, Y., Chen, Q., et al. (2017). An indoleacetic acid-producing Ochrobactrum sp. MGJ11 counteracts cadmium effect on soybean by promoting plant growth. J. Appl. Microbiol. 122, 987–996. doi: 10.1111/jam.13379
Zhang, Y. Q., Sun, C. H., Li, W. J., Yu, L. Y., Zhou, J. Q., Zhang, Y. Q., et al. (2007). Deinococcus yunweiensis sp. nov., a gamma- and UV-radiation-resistant bacterium from China. Int. J. Syst. Evol. Microbiol. 57, 370–375. doi: 10.1099/ijs.0.64292-0
Glossary
IAA - Indole-3-acetic acid
MEGA - Molecular evolutionary genetics analysis
NJ - Neighbor-joining
ML - Maximum-likelihood
SOG - Single-copy orthologroups
MLSA - Multilocus sequences analysis
OGRIs - Overall genome relatedness indices
ANI - Average nucleotide identity
dDDH - Digital DNA–DNA hybridization
GGDC - Genome-to-Genome Distance Calculator
BGC - Biosynthetic gene cluster
PYG - Peptone yeast
TSA - Tryptone soya agar
ISP - International Streptomyces Project
YM - Yeast extract sucrose
TSB - Tryptone soya broth
UV - Ultraviolet
API - Analytical profile index
ORF - Open reading frame
COG - Cluster of Orthologous Group
KEGG - Kyoto Encyclopedia of Genes and Genome
phylo-PCA - Phylogenetically informed principal component analysis
TLC - Thin-layer chromatography
HPLC - High performance liquid chromatography
DPG - Diphosphatidylglycerol
PG - Phosphatidylglycerol
NER - Nucleotide excision repair
IAM - Indole-3-acetamide
IPA/IPyA - Indole-3-pyruvic acid
IAN - Indole-3-acetonitrile
TAM - Tryptamine
TSO - L-Trp side-chain oxidase
NAPAA - Non-alpha poly-amino acids like
RiPP-like - Post-translationally modified peptide like
NRPS-like - Non-ribosomal peptide synthetase like
T3PKS - Type III polyketide synthases
DAK - Desert-associated Kocuria strains
NDAK - Non-desert associated Kocuria strains
Pan-GWAS - Pan-genome-wide association studies
Keywords: extremophiles, Kocuria , polyphasic taxonomy, comparative genomics, IAA production
Citation: Li C-J, Jiang Z-M, Zhi X-Y, Chen H-H, Yu L-Y, Li G-F and Zhang Y-Q (2025) Genomic insights into Kocuria: taxonomic revision and identification of five IAA-producing extremophiles. Front. Microbiol. 16:1547983. doi: 10.3389/fmicb.2025.1547983
Edited by:
Edoardo Puglisi, Catholic University of the Sacred Heart, ItalyReviewed by:
Anthony Ayodeji Adegoke, University of Uyo, NigeriaAfef Najjari, Tunis El Manar University, Tunisia
Copyright © 2025 Li, Jiang, Zhi, Chen, Yu, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Qin Zhang, eXpoYW5nQGltYi5wdW1jLmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Cong-Jian Li1†
Cong-Jian Li1† Yu-Qin Zhang
Yu-Qin Zhang