- Huzhou Center for Disease Control and Prevention, Huzhou, China
Vibrio parahaemolyticus has emerged as a predominant cause of seafood-related infections globally. Despite this, a comprehensive analysis of its epidemiological traits and antimicrobial resistance patterns in Huzhou City remains lacking. Our study isolated 250 strains of V. parahaemolyticus from a total of 6,404 diarrhea patients sampled across six hospitals in Huzhou from 2021 to 2023. Epidemiological analysis revealed higher infection rates in warmer seasons, with the majority of cases occurring in individuals aged from 25 to 64. No significant gender-based difference was observed in the prevalence of V. parahaemolyticus. Serotype analysis identified O10:K4 as the predominant serotype. 93.20% (233/250) of clinical isolates harbored the tdh gene, while 4.0% (10/250) carrying the trh. Antimicrobial sensitivity testing indicated a strikingly high resistance rate of 95.56% (172/180) to cefazolin among clinical isolates. The cgMLST-based minimum spanning tree analysis revealed that the O4:KUT clinical isolates segregated into two distinct clusters, ST3 and ST2516, with considerable evolutionary divergence between them. In contrast, the O10:K4 and O3:K6 serotypes exhibited closer phylogenetic proximity. This study comprehensively characterizes V. parahaemolyticus in Huzhou, revealing critical insights into its epidemiology, virulence factors, and antibiotic resistance patterns, thereby enhancing our knowledge of its public health risks.
1 Introduction
Vibrio parahaemolyticus is a Gram-negative, spore-forming bacillus with halophilic properties (Preeprem et al., 2019), widely distributed in coastal areas, marine sediment, and seafood, and is prone to contaminate fish, shrimp, shellfish, and other aquatic products (Zhou et al., 2022). Consumption of undercooked or cross-contaminated food can lead to symptoms such as abdominal pain, diarrhea, acute gastroenteritis, and septicemia. V. parahaemolyticus is recognized as a major cause of foodborne disease outbreaks in many Asian countries (Letchumanan et al., 2014). In China, foodborne diseases are mainly caused by pathogenic microorganisms, with outbreaks primarily caused by V. parahaemolyticus, Salmonella and Staphylococcus aureus (Geng et al., 2019). V. parahaemolyticus is one of the most common causative agents of foodborne illness associated with ready-to-eat (RTE) foods (Xie et al., 2016). It is reported that in Zhejiang Province, pathogenic bacteria constitute the primary causative agents of foodborne disease outbreaks, with V. parahaemolyticus being the most prevalent species (Chen et al., 2023). Research indicates that V parahaemolyticus has emerged as the leading cause of foodborne diseases in Zhejiang Province, accounting for 58.4% of bacterial outbreaks (Chen et al., 2022). According to statistics, V. parahaemolyticus is also one of the main pathogens causing bacterial foodborne outbreaks and sporadic foodborne diarrhea cases in Huzhou City (Yan et al., 2024, 2020). This study selected 250 strains of V. parahaemolyticus isolated from sporadic diarrhea cases in at least, from six hospitals of Huzhou City from 2021 to 2023 and used real-time fluorescence PCR and microbroth dilution methods to detect virulence genes, serotypes, and drug sensitivity, in order to understand the epidemiological characteristics, presence of virulence genes, and the current status of antimicrobial resistance of V. parahaemolyticus in sporadic diarrhea cases in Huzhou City.
2 Materials and methods
2.1 Sample collection and V. parahaemolyticus serotyping
From January 2021 to December 2023, six hospitals including Wuxing District People's Hospital, Deqing County People's Hospital, Changxing County People's Hospital, Nanxun District People's Hospital, Anji County People's Hospital, and Huzhou First People's Hospital for foodborne disease surveillance in Huzhou City tested 6,404 diarrhea case specimens defined by having a daily bowel movement frequency of ≥3 times and abnormal stool characteristics (loose stools, watery stools, mucoid stools, or bloody stools, etc.) for V. parahaemolyticus (https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease), and a total of 250 V. parahaemolyticus strains were isolated. These included 88 isolations from 2021, 86 isolations from 2022, and 76 isolations from 2023. The number of isolates from each hospital per year was detailed in Table 1. The isolation, identification and serotyping of V. parahaemolyticus were performed as previously described (Zhang et al., 2024a). The serotyping of V. parahaemolyticus isolates was conducted using standardized slide agglutination assays with commercially available antisera (Denka Seiken Ltd., Tokyo, Japan). The complete serotyping scheme included 11 O (somatic) and 65 K (capsular) antigenic determinants. Isolates were classified into serotypes based on their unique O:K antigen combinations. Isolates that failed to demonstrate K-antigen agglutination were classified as K-untypeable (KUT) serotypes (Jones et al., 2012).
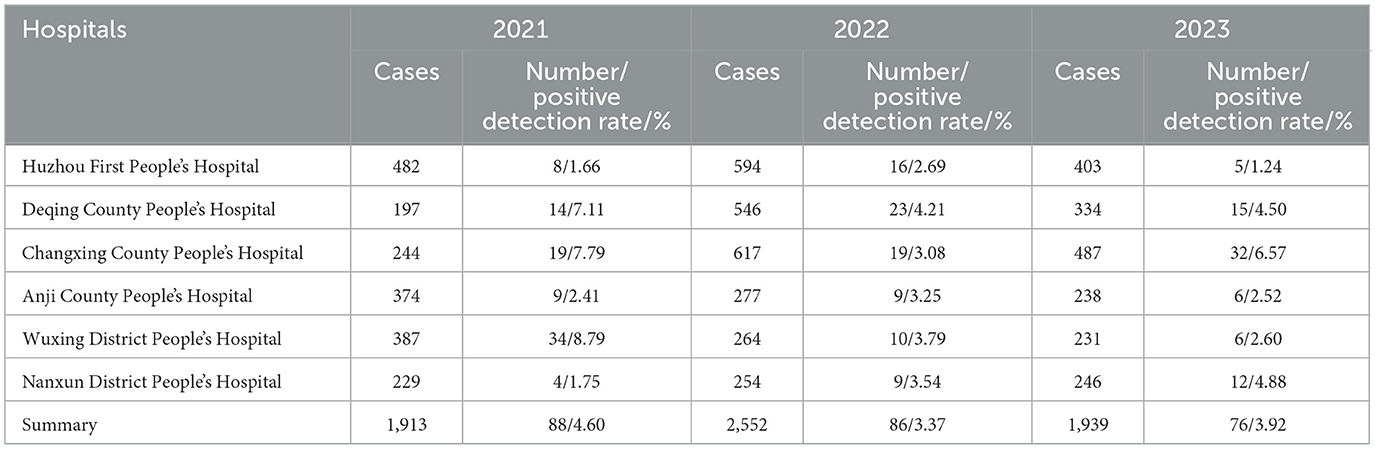
Table 1. Annual case numbers and positive detection rates from six hospitals in Huzhou City (2021–2023).
2.2 Statistical analysis of temporal distribution of cases
Strain distribution analysis was conducted in Excel according to specimen collection timelines, with data presentation optimized through bar chart visualization using KaleidaGraph 4.5. The tables were prepared using Microsoft Word.
2.3 Virulence gene detection
A fresh bacterial colony was suspended in 200 μL sterile water, heat-lysed at 100 °C for 10 min, and centrifuged at 10,000 × g for 5 min. The DNA-containing supernatant was collected and stored at −80 °C until further use (Yan et al., 2020). Pathogenic V. parahaemolyticus can produce either TDH, TRH, or both (Nishibuchi and Kaper, 1995). The gene tlh is widely considered to be a marker for V. parahaemolyticus (Bej et al., 1999; DePaola et al., 2003). The detection of tlh/tdh/trh virulence genes were performed according to the instructions of the V. parahaemolyticus (TLH, TDH, TRH) triplex real-time fluorescent PCR detection kit (Biogen Biological Co., Ltd., Shenzhen, China). The detailed method was performed as previously described (Zhang et al., 2024b). Quality control was performed using the manufacturer-provided negative and positive controls included in the kit to verify test accuracy.
2.4 Susceptibility test
In accordance with the Clinical and Laboratory Standard Institution guidelines (CLSI M100-S23) and ChinaPIN-2022-TYJS004, the microbroth dilution method was used to determine the minimal inhibitory concentration (MIC) of V. parahaemolyticus against antimicrobial agents. The 13 antimicrobial agents included penicillins: ampicillin (AMP) (64, 32, 16, 8, 4, 2, 1 μg/mL), ampicillin/sulbactam (AMS) (64/32, 32/16, 16/8, 8/4, 4/2, 2/1, 1/0.5 μg/mL); cephalosporins: cefotaxime (CTX) (16, 8, 4, 2, 1, 0.5, 0.25 μg/mL), ceftazidime (CAZ) (32, 16, 8, 4, 2, 1, 0.5 μg/mL), cefuroxime (CFX) (32, 16, 8, 4, 2, 1, 0.5 μg/mL), cefazolin (CFZ) (32, 16, 8, 4, 2, 1, 0.5, 0.25 μg/mL); carbapenems: imipenem (IPM) (8, 4, 2, 1, 0.5, 0.25 μg/mL); aminoglycosides: gentamicin (GEN) (32, 16, 8, 4, 2, 1 μg/mL); tetracyclines: tetracycline (TET) (32, 16, 8, 4, 2, 1 μg/mL); quinolones: nalidixic acid (NAL) (64, 32, 16, 8, 4, 2 μg/mL), ciprofloxacin (CIP) (32, 16, 8, 4, 2, 1, 0.5, 0.25, 0.12, 0.06, 0.03, 0.015 μg/mL); chloramphenicol: chloramphenicol (CHL) (64, 32, 16, 8, 4, 2 μg/mL); and sulfonamides: trimethoprim/sulfamethoxazole (SXT) (8/152, 4/76, 2/38, 1/19, 0.5/9.5, 0.25/4.75 μg/mL). The results were expressed as sensitive (S), intermediate (I), and resistant (R). Escherichia coli ATCC 25922 was included as a quality control strain.
2.5 Statistical analysis
Statistical processing was conducted using SPSS 23.0 software, and data analysis was performed with the chi-square test, with a difference considered statistically significant at p < 0.05.
2.6 Core genome multilocus sequence typing (cgMLST) profiling and minimum spanning tree-based phylogenetic analysis
We downloaded the 121 sequences of clinical isolates from 2021 to 2023, which were previously submitted to NCBI under BioProject accession numbers PRJNA1115946, PRJNA1072230, and PRJNA1071824 in our prior studies, for subsequent analysis. Detailed strain information was provided in Supplementary Table S1. For the characterization of V. parahaemolyticus, seven housekeeping genes (dnaE, gyrB, recA, dtdS, pntA, pyrC, and tnaA) were analyzed. A multilocus sequence typing (MLST) scheme was applied to the 121 genomes using mlst v2.23.0, generating an MLST profile for each isolate. The chewBBACA suite was employed for cgMLST analysis, beginning with schema construction using the CreateSchema module (Silva et al., 2018). Subsequently, allele identification was performed across 95% loci for each isolate through the AlleleCall module, producing a 95% allele profile matrix. To maintain analytical rigor, potential paralogous loci were systematically filtered using the RemoveParalogs module with default parameters, thereby eliminating sequences that could compromise cgMLST interpretation. The combination of alleles in each isolate formed an allelic profile that was used to generate minimum spanning trees (MSTs). The MSTs were constructed in BioNumerics using the cgMLST allele profiles as input data (Blanc et al., 2020). The unweighted pair group method with arithmetic mean (UPGMA) was used as the clustering algorithm. The pairwise distance was calculated by counting the number of pairwise allele differences.
3 Results
3.1 Basic case information
From 2021 to 2023, a total of 6,404 foodborne diarrhea case specimens were collected from six hospitals, yielding 250 isolates of V. parahaemolyticus, with an overall positive detection rate of 3.90%. Specifically, in 2021, 1,913 specimens were collected with a positive detection rate of 4.60%. The positive detection rates of Huzhou First People's Hospital, Deqing County People's Hospital, Changxing County People's Hospital, Anji County People's Hospital, Wuxing District People's Hospital, and Nanxun District People's Hospital were 1.66%, 7.11%, 7.79%, 2.41%, 8.79%, and 1.75%, respectively. In 2022, 2,552 specimens were collected with a rate of 3.37%. The positive detection rates for the six hospitals mentioned above were 2.69%, 4.21%, 3.08%, 3.25%, 3.79%, and 3.54%, respectively. In 2023, 1,939 specimens were collected with a rate of 3.92%. The six hospitals reported positive detection rates of 1.24%, 4.50%, 6.57%, 2.52%, 2.60%, and 4.88%, respectively (Table 1). The annual case numbers and positivity rates for the six specific hospitals from 2021 to 2023 are detailed in Table 1. The variation in the detection rate of V. parahaemolyticus across the different years was statistically significant (χ2= 4.413, p < 0.05).
3.2 Case population distribution
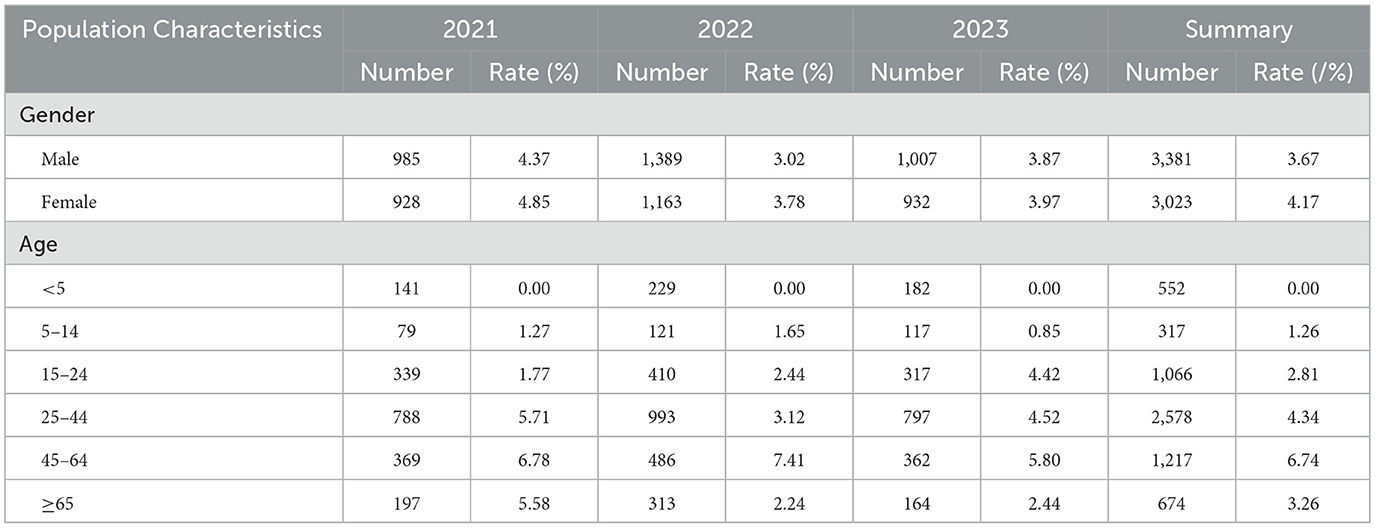
Among the 6,404 diarrhea infection cases, there were 3381 males and 3023 females.
Positive detection rates of V. parahaemolyticus in males and females were 3.67% and 4.17%, respectively, with no statistically significant difference between genders (χ2= 0.936, p > 0.05). The age distribution of infected cases ranged from 5 months to 97 years, with the highest infection rates in the 45–64 and 25–44 age groups at 6.74% and 4.34%, respectively, and no detection in the age group under 5 years (Table 2). The distribution of detection across different age groups was statistically significant (χ2= 59.822, p < 0.05).
3.3 Temporal distribution of cases
From 2021 to 2023, the detection rate of V. parahaemolyticus among foodborne diarrhea patients exhibited a clear seasonal trend. From March to May (spring), 28 positive cases of V. parahaemolyticus were identified from a total of 1,463 samples, resulting in a positive rate of 1.91%. From June to August (summer), 143 positive cases were identified from 2,136 samples, yielding a positive rate of 6.69%. From September to November (autumn), 78 out of 1,709 samples tested positive, with a positive rate of 4.56%. From December to February (winter), only 1 out of 1,096 samples tested positive, with the lowest positive rate of 0.091%. Positive cases were fewer during the winter and spring seasons, peaking in summer and autumn, especially from July to September (Figure 1).
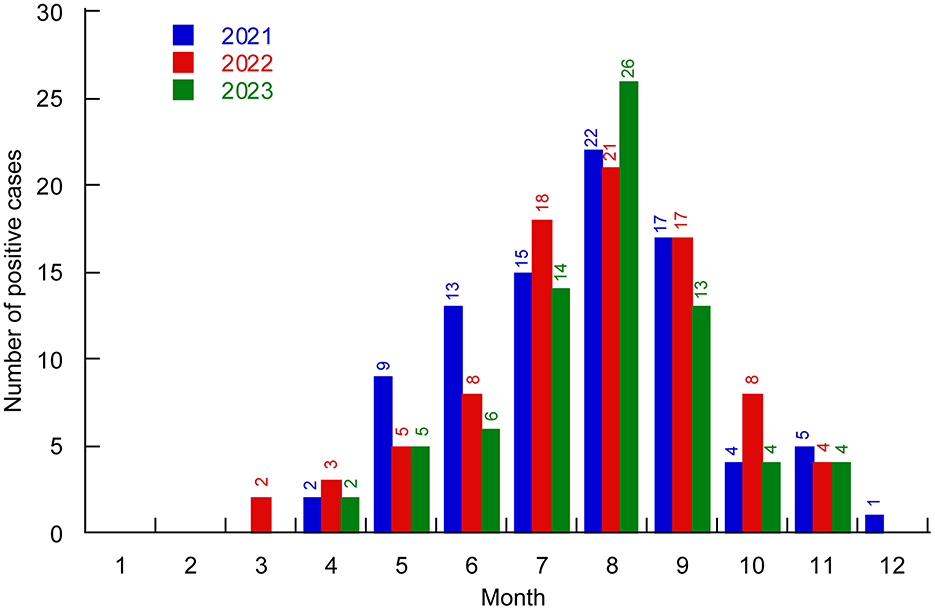
Figure 1. Distribution of V. parahaemolyticus positive cases in different months of Huzhou City from 2021 to 2023. Different colors represent different years.
3.4 Serovar distribution
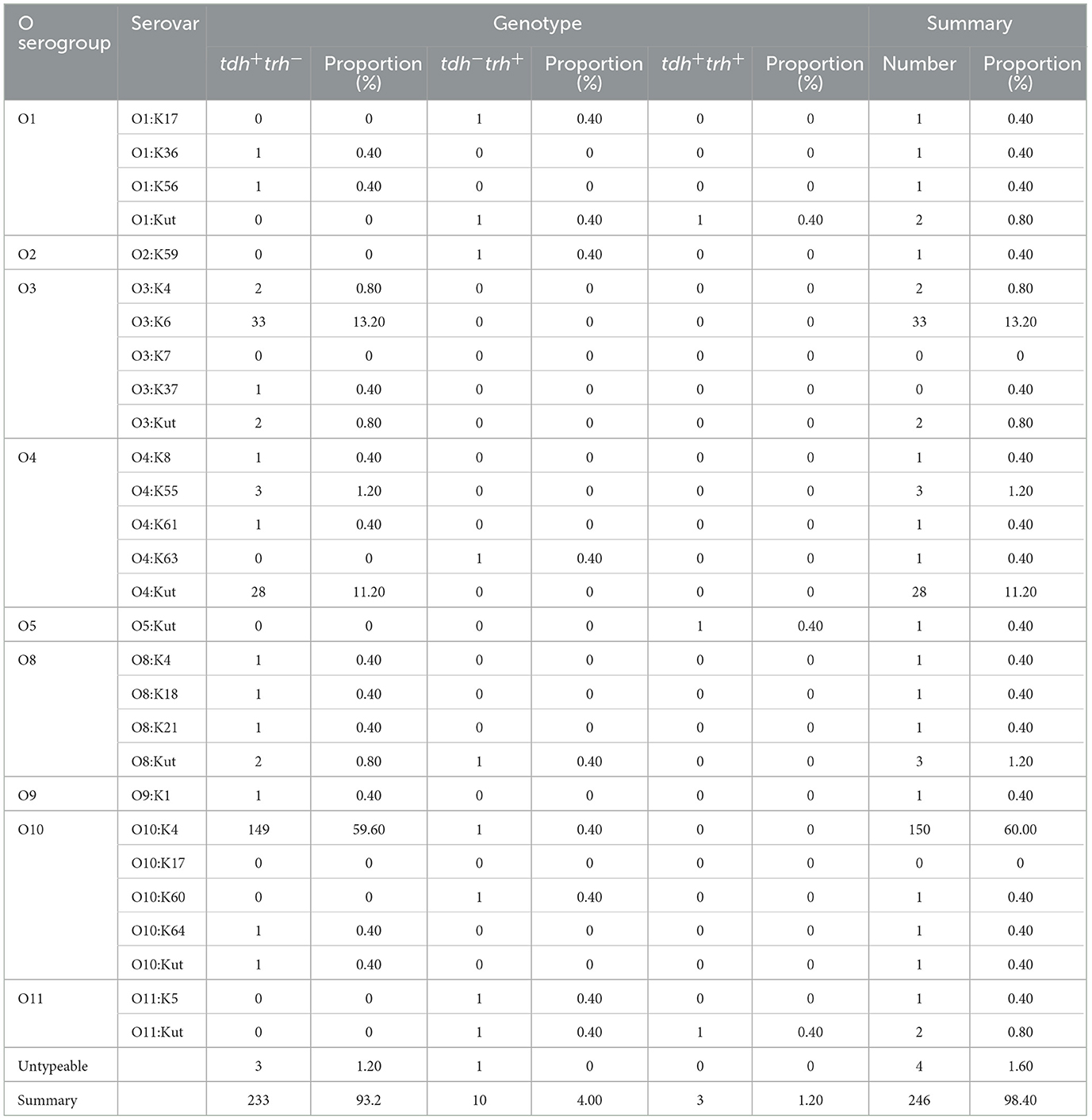
Among the 250 V. parahaemolyticus strains, 9 O serogroups and 28 serotypes were identified. The most prevalent serovars were O10:K4, constituting 60.00% of the strains (150 out of 250), followed by O3:K6 at 13.20% (33 out of 250), and O4:Kut at 11.20% (28 out of 250). A small fraction, 1.60% (4 out of 250), could not be typed.
In 2021, the strains were distributed across 6 O serogroups and 11 serotypes, with O10:K4 being the most dominant at 81.82% (72 out of 88), followed by O3:K6 at 5.69% (5 out of 88), and O4:Kut at 3.41% (3 out of 88). In 2022, the strains belonged to 5 O serogroups and 12 serotypes, with O10:K4 remaining the most prevalent at 40.70% (35 out of 86), O4:Kut at 27.91% (24 out of 86), and O3:K6 at 13.95% (12 out of 86). In 2023, the strains were categorized into 7 O serogroups and 15 serotypes, with O10:K4 continuing to dominate at 56.68% (43 out of 76), O3:K6 at 21.05% (16 out of 76), and O3:K4 at 3.95% (3 out of 76) (Table 3).
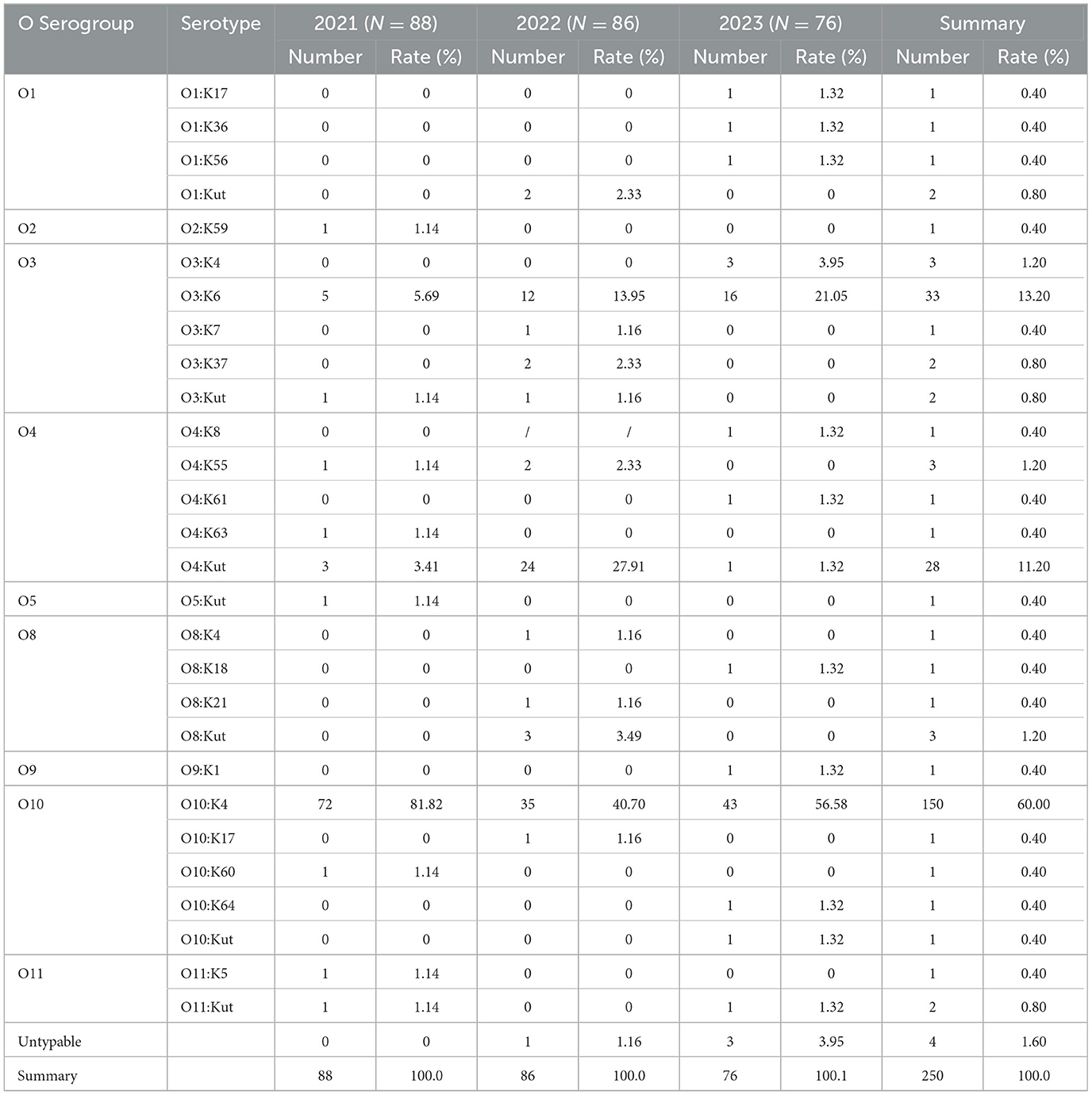
Table 3. Serovar distribution of V. parahaemolyticus in diarrhea cases in Huzhou city from 2021 to 2023.
3.5 Virulence gene detection results
All 250 strains of V. parahaemolyticus were identified as carrying the species-specific gene tlh. The most common virulence gene profile among these strains was tdh+, representing 93.20% of the total; trh+ was found in 4.00% of the strains; and the co-presence of both tdh+ and trh+ was observed in 1.20% (Table 4). We identified four isolates that carried only the tlh gene. tdh+ strains were predominantly found in serotypes O3:K6, O10:K4, and O4:KUT, whereas trh+ and tdh+trh+ strains were less prevalent among these three major serotypes. trh+ strains were distributed across multiple serotypes, including O1:K17, O1:KUT, O2:K59, O4:K63, O8:KUT, O10:K4, O10:K60, O11:K5, and O11:KUT. In contrast, tdh+trh+ strains were only detected in O1:KUT, O5:KUT, and O11:KUT.
3.6 Susceptibility test results
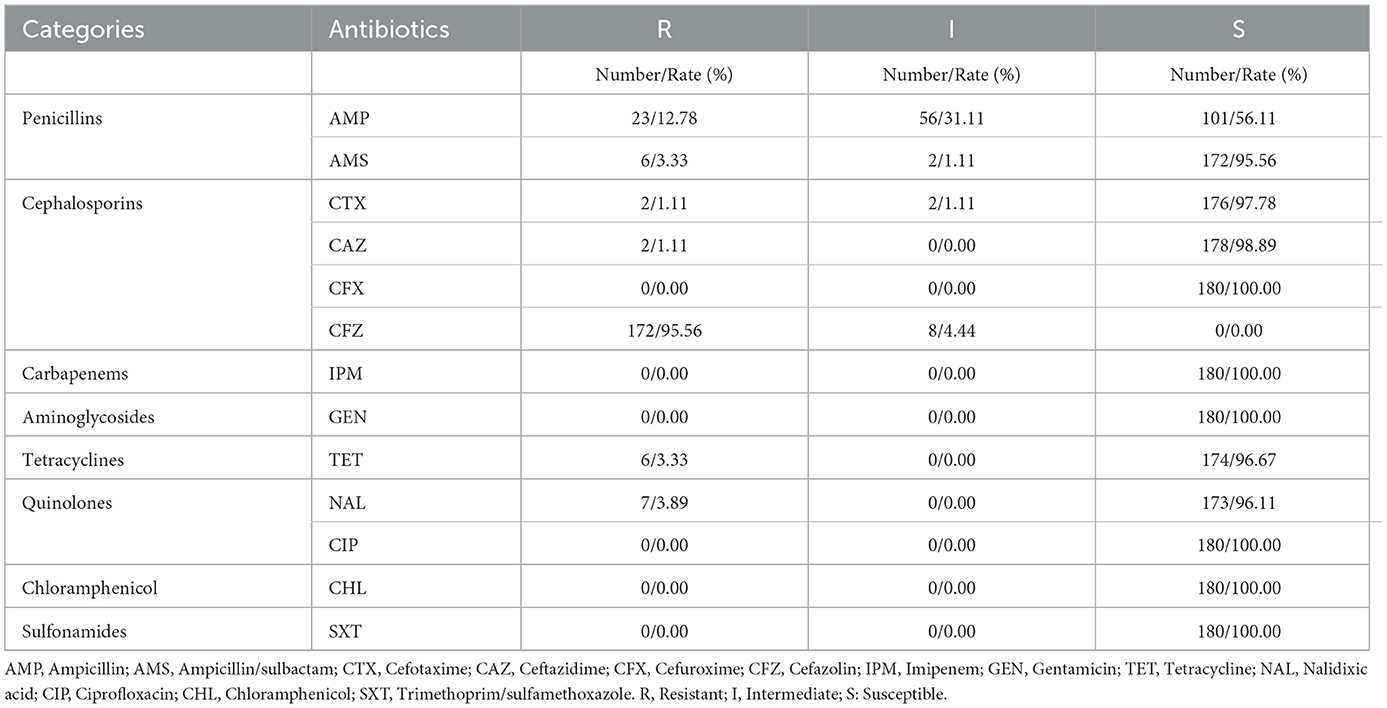
A random selection of 180 V. parahaemolyticus strains were subjected to susceptibility testing. The resistance rate to CFZ was as high as 95.56% (172 out of 180). The resistance rates for other antibiotics were: AMP at 12.78% (23 out of 180), NAL at 3.89% (7 out of 180), AMS and TET at 3.33% (6 out of 180), and CTX and CAZ at 1.11% (2 out of 180). In contrast, all strains were 100.0% sensitive to CFX, IPM, GEN, CHL, CIP, and SXT (Table 5).
3.7 Minimum spanning tree analysis
We performed cgMLST analysis on 121 clinical isolates, comprising 85 O10:K4, 15 O3:K6, and 21 O4:KUT strains, and constructed a minimum spanning tree. The results demonstrated that these isolates clustered into two major ST types (ST3 and ST2516) with significant genetic divergence. The O4:KUT serotype was distributed across both ST types. Interestingly, the O10:K4 serotype also separated into two distinct clusters, potentially evolving from O3:K6 strains (Figure 2).
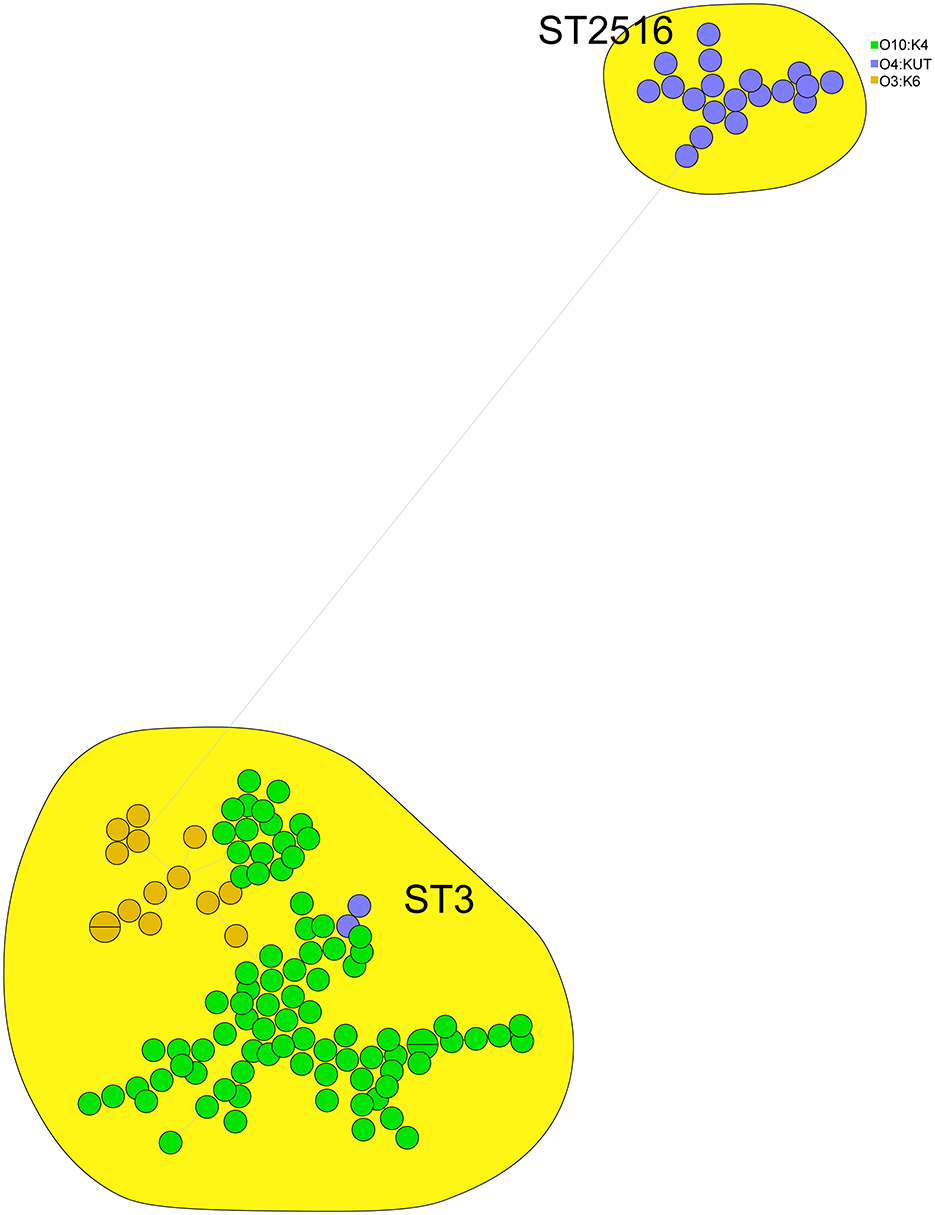
Figure 2. cgMLST based minimum spanning tree of 121 clinical V. parahaemolyticus isolates. Circle colors denote different serotypes (O3:K6, O4:KUT, O10:K4), while circle sizes reflect the number of isolates in each node.
4 Discussion
This study analyzed the infection status of V. parahaemolyticus in foodborne diarrhea cases in Huzhou City from January 2021 to December 2023, with a positive detection rate of 3.90% among 6,404 diarrhea cases. The detection rate is higher than the average found in Mainland China (2.08%) (Wang et al., 2021), likely attributed to Huzhou's geographic location and dietary customs. Huzhou City is rich in aquatic products, and its residents have a tradition of consuming raw or undercooked aquatic products. With the development of logistics, various live seafood products are increasingly sold in inland areas. According to the report, the contamination rate of V. parahaemolyticus in inland provinces has reached 23.14% (Pang et al., 2020). Therefore, there is a possibility of cross-contamination between seafood and freshwater products during sales and processing (Chen et al., 2021; Liao et al., 2015), leading to infections when people consume contaminated aquatic products.
The temporal distribution shows that V. parahaemolyticus infections can occur throughout the year except for January and February, with the majority of cases concentrated in July to September (Huang et al., 2023), which is consistent with the characteristic of high incidence in summer and autumn. According to Zhejiang Meteorological Bureau or Huzhou Meteorological Station reports, from 2021 to 2023, Huzhou experienced significant climatic variability driven by the alternating influences of El Niño-Southern Oscillation (ENSO) phenomena. The extended La Niña conditions (2020–2022) resulted in distinct seasonal patterns, characterized by colder-than-average winter temperatures and increased precipitation variability. Conversely, the emerging El Niño phase in 2023 contributed to intensified summer heatwaves and altered precipitation regimes, consistent with broader warming trends observed across eastern China (https://epmap.zjol.com.cn/jsb0523/202303/t20230310_25510472.shtml). High temperatures during these months facilitate bacterial growth and reproduction (Neil et al., 2023; Cantet et al., 2013; Fernandez-Velez et al., 2023). Our study has shown that sporadic diarrhea cases are mainly concentrated in middle-aged and young people (aged 25 to < 65 years), which was consistent with previous research (Wang et al., 2021), possibly because this age group has more opportunities to dine out compared to other age groups, and the abundance of night markets in summer and autumn increases the risk of infection when consuming undercooked or raw aquatic products (Li et al., 2020). Consistent with previous findings, the patients aged ≥65 years were the least likely to get infectious diarrhea (Gong et al., 2018). This diversity of age distribution might reflect a natural change in host immunity (Simon et al., 2015) and/or dietary habits that are related to age (Jiang et al., 2024). These findings of the effect of age on the pathogen detection might provide scientific evidence for finding the optimal timing to enhance prevention measures for V. parahaemolyticus.
V. parahaemolyticus produces thermolabile hemolysin (TLH), thermotolerant direct hemolysin (TDH), and thermotolerant direct hemolysin-related hemolysin (TRH), encoded by the tlh/tdh/trh genes are considered to be key indicators of V. parahaemolyticus pathogenicity (Lee et al., 2019; Broberg et al., 2011; Gutierrez West et al., 2013; Shirai et al., 1990). TDH and TRH are considered the most critical virulence factors of V. parahaemolyticus (Ceccarelli et al., 2013), with most clinical isolates producing one or both of these hemolysins (Letchumanan et al., 2014). The results of this study show that the positive rate of the tdh virulence gene is 93.20%, the trh virulence gene is 4.00% and the simultaneous positivity of tdh and trh virulence genes is 1.20% which was consistent with other study (Sun et al., 2022). Several studies have also reported that around 2.0% of the clinical strains do not contain tdh and/or trh (Li et al., 2014; Pazhani et al., 2014). Even in the absence of these hemolysins, V. parahaemolyticus remains pathogenic indicating that other virulence factors exist (Jones et al., 2012; Mahoney et al., 2010). It was reported that beyond hemolysins, the virulence mechanisms of V. parahaemolyticus critically depend on specialized secretion systems that mediate effector delivery into host cells (Orlova, 2015). Notably, the bacterium employs two functionally distinct type III secretion systems (T3SS): T3SS1 facilitates host cell death through the translocation of cytotoxic effectors (VopQ, VopS, VPA0450, and VopR/VP1638), while T3SS2 secretes a distinct repertoire of effectors (VopC, VopT, VopZ, VopA/P, VopV, VopL, and VPA1380) that collectively suppress host cell proliferation and induce cytotoxicity (Chatterjee et al., 2013). Additionally, the type VI secretion system (T6SS) contributes to pathogenicity through dual functionality—delivering virulence effectors not only into eukaryotic host cells but also competing bacterial cells (Sha et al., 2013; Li et al., 2019).
In 1996, the O3:K6 serotype of V. parahaemolyticus caused a large-scale food poisoning outbreak in India and subsequently became pandemic globally. O3:K6, along with 21 other serotypes, is known as the “pandemic clone” of V. parahaemolyticus and is the most frequently reported serotype worldwide (Okuda et al., 1997). Before 2021, the O3:K6 serotype was the main serotype in foodborne diarrhea cases in this region (Zhang et al., 2022). The results of this study show that from 2021 to 2023, the O10:K4 serotype accounted for 60.00% of foodborne diarrhea cases in Huzhou across all seasons, replacing O3:K6 as the dominant serotype in the region. In fact, in 2020, O10:K4 serotype was detected for the first time in Huzhou City. Afterwards, O10:K4 serotype surpassed O3:K6 as the new dominant serotype (Zhang et al., 2022). In recent years, the O10:K4 serotype has dominated in acute gastroenteritis outbreaks caused by V. parahaemolyticus in other provinces in China (Wu et al., 2024; Huang et al., 2022a,b). The emergence of serotype O10:K4 may be the response to host immunologic pressure (Huang et al., 2022a). A previous study indicates that O10:K4 strains and the genetic variant O3:K6 were placed in the same cluster, suggesting a possibility of transfer of the pathogenic genes (Huang et al., 2022b).
With the widespread and extensive use of antibiotics in clinical and breeding fields, the issue of bacterial resistance is becoming increasingly severe. The results of this study show that the resistance rate of V. parahaemolyticus to CFZ in Huzhou from 2021 to 2023 was as high as 95.56%; some strains exhibited resistance to AMS, CTX, CAZ, TET, and NAL. The resistance rate of local V. parahaemolyticus to CFZ was similar to that of clinical isolates from Nantong (99.1%) and from isolates from Nanjing (99.2%) (Zhou et al., 2022; Huang et al., 2023). However, it was significantly higher than the resistance rate reported for domestic clinical isolates a decade ago (50.4%) (Chen et al., 2016). A previous research has indicated that V. parahaemolyticus exhibits a notably high resistance to AMP in recent years (Dahanayake et al., 2020; Lopatek et al., 2018; Mok et al., 2021; Nishino et al., 2021; Vu et al., 2022). However, our study reveals that the organism demonstrates the highest resistance to CFZ, with resistance to AMP at a comparatively lower rate of 12.78%, aligning with previous findings (Zhang et al., 2024a), which may be related to the types of antibiotics used in aquaculture or clinical practice, leading to a decrease in ampicillin resistance. Currently, tetracycline, cephalothin, and quinolone drugs are considered first-line options for the treatment of V. parahaemolyticus infections (Elmahdi et al., 2016; Han et al., 2007; Tan et al., 2017). Earlier study reported 100% sensitivity of V. parahaemolyticus to the AMS (da Silva et al., 2021), our findings show a concerning development, with 3.33% of strains exhibiting resistance to this antibiotic regimen. The results of this study indicate that, apart from penicillins and cephalosporins, other antibiotics can be used as first-line treatments for V. parahaemolyticus. Therefore, continuous resistance monitoring of V. parahaemolyticus in foodborne diarrhea cases can timely grasp the trend of resistance changes, which is helpful for guiding rational clinical medication.
In this study, we identified two sequence types (ST3 and ST2516) among 121 clinical V. parahaemolyticus isolates (Figure 2). MLST analysis revealed that serotypes O10:K4, O3:K6 and O4:KUT belonged to these two STs, with O4:KUT strains distributed across both ST types and showing significant genetic divergence—a finding consistent with previous reports (Zhang et al., 2024b; Huang et al., 2022a). ST3 emerged as the predominant clinical type, aligning with prior studies that established its clinical significance as an epidemic clone circulating in Asia and America (Chen et al., 2016; Turner et al., 2013). These observations collectively demonstrate the crucial role of ST3 in human infections. Additionally, the O4:KUT strains of ST2516 warrant further investigation due to their potential clinical importance.
In summary, our analysis of V. parahaemolyticus infections across six hospitals in Huzhou (2021–2023) revealed that most cases occurred among middle-aged and young adults, with a distinct seasonal concentration during summer months. The predominant serotypes among clinical isolates were O10:K4, O3:K6, and O4:KUT. CgMLST analysis demonstrated significant phylogenetic divergence between some O4:KUT strains and the O10:K4/O3:K6 clusters.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
XW: Conceptualization, Investigation, Methodology, Software, Writing – original draft. LC: Data curation, Methodology, Writing – original draft. XZ: Methodology, Software, Writing – original draft. LJ: Investigation, Methodology, Software, Writing – review & editing. FD: Investigation, Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Medical Science and Technology Projects of Zhejiang Province, grant number 2024KY1658 and Natural Science Foundation of Zhejiang Province, grant number LTGY23C010001.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1551984/full#supplementary-material
References
Bej, A. K., Patterson, D. P., Brasher, C. W., Vickery, M. C., Jones, D. D., Kaysner, C. A., et al. (1999). Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods, 36, 215–225. doi: 10.1016/S0167-7012(99)00037-8
Blanc, D. S., Magalhaes, B., Koenig, I., Senn, L., and Grandbastien, B. (2020). Comparison of Whole Genome (wg-) and Core Genome (cg-) MLST (BioNumerics(TM)) Versus SNP Variant Calling for Epidemiological Investigation of Pseudomonas aeruginosa. Front. Microbiol. 11:1729. doi: 10.3389/fmicb.2020.01729
Broberg, C. A., Calder, T. J., and Orth, K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13, 992–1001. doi: 10.1016/j.micinf.2011.06.013
Cantet, F., Hervio-Heath, D., Caro, A., Le Mennec, C., Monteil, C., Quemere, C., et al. (2013). Quantification of Vibrio parahaemolyticus, Vibrio vulnificus and Vibrio cholerae in French Mediterranean coastal lagoons. Res. Microbiol. 164, 867–874. doi: 10.1016/j.resmic.2013.06.005
Ceccarelli, D., Hasan, N. A., Huq, A., and Colwell, R. R. (2013). Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell. Infect. Microbiol. 3:97. doi: 10.3389/fcimb.2013.00097
Chatterjee S. Chaudhury S. McShan A. C. Kaur K. De Guzman R. N. (2013) Structure biophysics of type III secretion in bacteria. Biochemistry. 52:2508–17. 10.1021/bi400160a
Chen, H., Dong, S., Yan, Y., Zhan, L., Zhang, J., Chen, J., et al. (2021). Prevalence and population analysis of Vibrio parahaemolyticus isolated from freshwater fish in Zhejiang Province, China. Foodborne Pathog. Dis. 18, 139–146. doi: 10.1089/fpd.2020.2798
Chen, L., Wang, J., Chen, J., Zhang, R., Zhang, H., Qi, X., et al. (2023). Epidemiological characteristics of Vibrio parahaemolyticus outbreaks, Zhejiang, China, 2010–2022. Front. Microbiol. 14:1171350. doi: 10.3389/fmicb.2023.1171350
Chen, L., Wang, J., Zhang, R., Zhang, H., Qi, X., He, Y., et al. (2022). An 11-year analysis of bacterial foodborne disease outbreaks in Zhejiang Province, China. Foods, 11:2382. doi: 10.3390/foods11162382
Chen, Y., Chen, X., Yu, F., Wu, M., Wang, R., Zheng, S., et al. (2016). Serology, virulence, antimicrobial susceptibility and molecular characteristics of clinical Vibrio parahaemolyticus strains circulating in southeastern China from 2009 to 2013. Clin. Microbiol. Infect. 22, 258e259-216. doi: 10.1016/j.cmi.2015.11.003
da Silva, L. V., Ossai, S., Chigbu, P., and Parveen, S. (2021). Antimicrobial and genetic profiles of Vibrio vulnificus and Vibrio parahaemolyticus isolated from the Maryland Coastal Bays, United States. Front. Microbiol. 12:676249. doi: 10.3389/fmicb.2021.676249
Dahanayake, P. S., Hossain, S., Wickramanayake, M., Wimalasena, S., and Heo, G. J. (2020). Manila clam (Ruditapes philippinarum) marketed in Korea as a source of vibrios harbouring virulence and beta-lactam resistance genes. Lett. Appl. Microbiol. 71, 46–53. doi: 10.1111/lam.13229
DePaola, A., Nordstrom, J. L., Bowers, J. C., Wells, J. G., and Cook, D. W. (2003). Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 69, 1521–1526. doi: 10.1128/AEM.69.3.1521-1526.2003
Elmahdi, S., DaSilva, L. V., and Parveen, S. (2016). Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: a review. Food Microbiol. 57, 128–134. doi: 10.1016/j.fm.2016.02.008
Fernandez-Velez, I., Bidegain, G., and Ben-Horin, T. (2023). Predicting the Growth of Vibrio parahaemolyticus in Oysters under varying ambient temperature. Microorganisms 11:1169. doi: 10.3390/microorganisms11051169
Geng, X., Li, W., Zhang, J., Fu, P., Han, H., Liu, J., et al. (2019). [Attribution analysis of food borne disease outbreaks in canteen from 2002 to 2016 in China]. Wei Sheng Yan Jiu, 48, 66–69.
Gong, X. H., Wu, H. Y., Li, J., Xiao, W. J., Zhang, X., Chen, M., et al. (2018). Epidemiology, aetiology and seasonality of infectious diarrhoea in adult outpatients through active surveillance in Shanghai, China, 2012–2016: a cross-sectional study. BMJ Open 8:e019699. doi: 10.1136/bmjopen-2017-019699
Gutierrez West, C. K., Klein, S. L., and Lovell, C. R. (2013). High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl. Environ. Microbiol. 79, 2247–2252. doi: 10.1128/AEM.03792-12
Han, F., Walker, R. D., Janes, M. E., Prinyawiwatkul, W., and Ge, B. (2007). Antimicrobial susceptibilities of Vibrio parahaemolyticus and Vibrio vulnificus isolates from Louisiana Gulf and retail raw oysters. Appl. Environ. Microbiol. 73, 7096–7098. doi: 10.1128/AEM.01116-07
Huang, A., Wang, Y., Xu, H., Jin, X., Yan, B., Zhang, W., et al. (2023). Antibiotic resistance and epidemiology of Vibrio parahaemolyticus from clinical samples in Nantong, China, 2018–2021. Infect. Drug Resist., 16, 7413–7425. doi: 10.2147/IDR.S432197
Huang, Y., Du, Y., Wang, H., Tan, D., Su, A., Li, X., et al. (2022a). New variant of Vibrio parahaemolyticus, sequence type 3, serotype O10:K4, China, 2020. Emerging Infect. Dis. 28, 1261–1264. doi: 10.3201/eid2806.211871
Huang, Y., Lyu, B., Zhang, X., Tian, Y., Lin, C., Shen, L., et al. (2022b). Vibrio parahaemolyticus O10:K4: an emergent serotype with pandemic virulence traits as predominant clone detected by whole-genome sequence analysis—Beijing Municipality, China, 2021. China CDC Wkly, 4, 471–477. doi: 10.46234/ccdcw2022.106
Jiang, D., Han, H., Guo, Y., Zhang, R., Zhan, L., Zhou, Y., et al. (2024). Epidemiological characteristics of sporadic foodborne diseases caused by Vibrio parahaemolyticus—China, 2013–2022. China CDC Wkly. 6, 1354–1359. doi: 10.46234/ccdcw2024.269
Jones, J. L., Ludeke, C. H., Bowers, J. C., Garrett, N., Fischer, M., Parsons, M. B., et al. (2012). Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J. Clin. Microbiol. 50, 2343–2352. doi: 10.1128/JCM.00196-12
Lee, Y., Choi, Y., Lee, S., Lee, H., Kim, S., Ha, J., et al. (2019). Occurrence of pathogenic Vibrio parahaemolyticus in seafood distribution channels and their antibiotic resistance profiles in S. Korea. Lett. Appl. Microbiol. 68, 128–133. doi: 10.1111/lam.13099
Letchumanan, V., Chan, K. G., and Lee, L. H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Li, L., Meng, H., Gu, D., Li, Y., and Jia, M. (2019). Molecular mechanisms of Vibrio parahaemolyticus pathogenesis. Microbiol. Res. 222, 43–51. doi: 10.1016/j.micres.2019.03.003
Li, Y., Xie, T., Pang, R., Wu, Q., Zhang, J., Lei, T., et al. (2020). Food-Borne Vibrio parahaemolyticus in China: prevalence, antibiotic susceptibility, and genetic characterization. Front. Microbiol. 11:1670. doi: 10.3389/fmicb.2020.01670
Li, Y., Xie, X., Shi, X., Lin, Y., Qiu, Y., Mou, J., et al. (2014). Vibrio parahaemolyticus, Southern Coastal Region of China, 2007–2012. Emerging Infect. Dis. 20, 685–688. doi: 10.3201/eid2004.130744
Liao, Y., Li, Y., Wu, S., Mou, J., Xu, Z., Cui, R., et al. (2015). Risk factors for Vibrio parahaemolyticus infection in a Southern Coastal Region of China. Foodborne Pathog. Dis. 12, 881–886. doi: 10.1089/fpd.2015.1988
Lopatek, M., Wieczorek, K., and Osek, J. (2018). Antimicrobial resistance, virulence factors, and genetic profiles of Vibrio parahaemolyticus from seafood. Appl. Environ. Microbiol. 84, e00537-18. doi: 10.1128/AEM.00537-18
Mahoney, J. C., Gerding, M. J., Jones, S. H., and Whistler, C. A. (2010). Comparison of the pathogenic potentials of environmental and clinical vibrio parahaemolyticus strains indicates a role for temperature regulation in virulence. Appl. Environ. Microbiol. 76, 7459–7465. doi: 10.1128/AEM.01450-10
Mok, J. S., Cho, S. R., Park, Y. J., Jo, M. R., Ha, K. S., Kim, P. H., et al. (2021). Distribution and antimicrobial resistance of Vibrio parahaemolyticus isolated from fish and shrimp aquaculture farms along the Korean coast. Mar. Pollut. Bull. 171:112785. doi: 10.1016/j.marpolbul.2021.112785
Neil, W. A., Hard, C., Bowers, J. C., and Jones, J. L. (2023). Levels of Vibrio parahaemolyticus in Pacific Oysters (Crassostrea gigas) from Washington State following ambient exposure and chilling. J. Food Prot. 86:100092. doi: 10.1016/j.jfp.2023.100092
Nishibuchi, M., and Kaper, J. B. (1995). Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63, 2093–2099. doi: 10.1128/iai.63.6.2093-2099.1995
Nishino, T., Suzuki, H., Mizumoto, S., Morinushi, H., Nagaoka, H., Goto, K., et al. (2021). Antimicrobial drug-resistance profile of Vibrio Parahaemolyticus isolated from Japanese Horse Mackerel (Trachurus Japonicus). Food Saf 9, 75–80. doi: 10.14252/foodsafetyfscj.D-21-00001
Okuda, J., Ishibashi, M., Hayakawa, E., Nishino, T., Takeda, Y., Mukhopadhyay, A. K., et al. (1997). Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35, 3150–3155. doi: 10.1128/jcm.35.12.3150-3155.1997
Orlova, E. V. (2015). A molecular syringe that kills cells. Nat. Struct. Mol. Biol. 22, 357–359. doi: 10.1038/nsmb.3021
Pang, R., Li, Y., Chen, M., Zeng, H., Lei, T., Zhang, J., et al. (2020). A database for risk assessment and comparative genomic analysis of foodborne Vibrio parahaemolyticus in China. Sci Data 7:321. doi: 10.1038/s41597-020-00671-3
Pazhani, G. P., Bhowmik, S. K., Ghosh, S., Guin, S., Dutta, S., Rajendran, K., et al. (2014). Trends in the epidemiology of pandemic and non-pandemic strains of Vibrio parahaemolyticus isolated from diarrheal patients in Kolkata, India. PLoS Negl. Trop. Dis. 8:e2815. doi: 10.1371/journal.pntd.0002815
Preeprem, S., Singkhamanan, K., Nishibuchi, M., Vuddhakul, V., and Mittraparp-Arthorn, P. (2019). Multiplex multilocus variable-number tandem-repeat analysis for typing of pandemic Vibrio parahaemolyticus O1:KUT isolates. Foodborne Pathog. Dis. 16, 104–113. doi: 10.1089/fpd.2018.2505
Sha, J., Rosenzweig, J. A., Kozlova, E. V., Wang, S., Erova, T. E., Kirtley, M. L., et al. (2013). Evaluation of the roles played by Hcp and VgrG type 6 secretion system effectors in Aeromonas hydrophila SSU pathogenesis. Microbiology 159, 1120–1135. doi: 10.1099/mic.0.063495-0
Shirai, H., Ito, H., Hirayama, T., Nakamoto, Y., Nakabayashi, N., Kumagai, K., et al. (1990). Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58, 3568–3573. doi: 10.1128/iai.58.11.3568-3573.1990
Silva, M., Machado, M. P., Silva, D. N., Rossi, M., Moran-Gilad, J., Santos, S., et al. (2018). chewBBACA: a complete suite for gene-by-gene schema creation and strain identification. Microb. Genom. 4:e000166. doi: 10.1099/mgen.0.000166
Simon, A. K., Hollander, G. A., and McMichael, A. (2015). Evolution of the immune system in humans from infancy to old age. Proc. Biol. Sci. 282:20143085. doi: 10.1098/rspb.2014.3085
Sun, J., Li, X., Hu, Z., Xue, X., Zhang, M., Wu, Q., et al. (2022). Characterization of Vibrio parahaemolyticus isolated from stool specimens of diarrhea patients in Nantong, Jiangsu, China during 2018–2020. PLoS ONE 17:e0273700. doi: 10.1371/journal.pone.0273700
Tan, C. W., Malcolm, T. T. H., Kuan, C. H., Thung, T. Y., Chang, W. S., Loo, Y. Y., et al. (2017). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from short mackerels (Rastrelliger brachysoma) in Malaysia. Front. Microbiol. 8:1087. doi: 10.3389/fmicb.2017.01087
Turner, J. W., Paranjpye, R. N., Landis, E. D., Biryukov, S. V., Gonzalez-Escalona, N., Nilsson, W. B., et al. (2013). Population structure of clinical and environmental Vibrio parahaemolyticus from the Pacific Northwest coast of the United States. PLoS ONE 8:e55726. doi: 10.1371/journal.pone.0055726
Vu, T. T. T., Hoang, T. T. H., Fleischmann, S., Pham, H. N., Lai, T. L. H., Cam, T. T. H., et al. (2022). Quantification and antimicrobial resistance of Vibrio parahaemolyticus in retail seafood in Hanoi, Vietnam. J. Food Prot. 85, 786–791. doi: 10.4315/JFP-21-444
Wang, L. P., Zhou, S. X., Wang, X., Lu, Q. B., Shi, L. S., Ren, X., et al. (2021). Etiological, epidemiological, and clinical features of acute diarrhea in China. Nat. Commun. 12:2464. doi: 10.1038/s41467-021-22551-z
Wu, X., Zhu, Y., Yan, W., Zhang, P., and Chen, L. (2024). Pathogenic characteristics of the Vibrio parahaemolyticus which caused a gastroenteritis outbreak event in Huzhou. FEMS Microbiol. Lett. 371:fnad130. doi: 10.1093/femsle/fnad130
Xie, T., Xu, X., Wu, Q., Zhang, J., and Cheng, J. (2016). Prevalence, molecular characterization, and antibiotic susceptibility of Vibrio parahaemolyticus from ready-to-eat foods in China. Front. Microbiol. 7:549. doi: 10.3389/fmicb.2016.00549
Yan, W., Ji, L., Dong, F., Chen, L., Yuan, R., Zhang, P., et al. (2024). Antimicrobial resistance and genomic analysis of Vibrio parahaemolyticus isolates from foodborne outbreaks, Huzhou, China, 2019–2023. Front. Microbiol. 15:1439522. doi: 10.3389/fmicb.2024.1439522
Yan, W., Ji, L., Xu, D., Chen, L., and Wu, X. (2020). Molecular characterization of clinical and environmental Vibrio parahaemolyticus isolates in Huzhou, China. PLoS ONE 15:e0240143. doi: 10.1371/journal.pone.0240143
Zhang, P., Ji, L., Yan, W., Chen, L., Zhu, X., Lu, Z., et al. (2024a). Whole-genome sequencing and transcriptome-characterized mechanism of streptomycin resistance in Vibrio parahaemolyticus O10: K4. Infect. Genet. Evol. 117:105540. doi: 10.1016/j.meegid.2023.105540
Zhang, P., Wu, X., Ji, L., Yan, W., Chen, L., Lu, Z., et al. (2024b). Prevalence and virulence of Vibrio parahaemolyticus isolated from clinical and environmental samples in Huzhou, China. BMC Genom. 25:1187. doi: 10.1186/s12864-024-11106-3
Zhang, P., Wu, X., Yuan, R., Yan, W., Xu, D., Ji, L., et al. (2022). Emergence and predominance of a new serotype of Vibrio parahaemolyticus in Huzhou, China. Int. J. Infect. Dis. 122, 93–98. doi: 10.1016/j.ijid.2022.05.023
Keywords: V. parahaemolyticus, virulence genes, antibiotic resistance, serovars, epidemiological features
Citation: Wu X, Chen L, Zhu X, Ji L and Dong F (2025) Epidemiological characteristics, virulence genes, and antimicrobial resistance analysis of Vibrio parahaemolyticus in diarrheal cases in Huzhou City from 2021 to 2023. Front. Microbiol. 16:1551984. doi: 10.3389/fmicb.2025.1551984
Received: 27 December 2024; Accepted: 14 August 2025;
Published: 29 August 2025.
Edited by:
Yolande Proroga, Experimental Zooprophylactic Institute of Southern Italy (IZSM), ItalyReviewed by:
Guodong Zhang, United States Food and Drug Administration, United StatesJorge Velazquez-Roman, Autonomous University of Sinaloa, Mexico
Copyright © 2025 Wu, Chen, Zhu, Ji and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fenfen Dong, MTUxNTY4NzQ4MjdAMTYzLmNvbQ==
 Xiaofang Wu
Xiaofang Wu Liping Chen
Liping Chen Fenfen Dong
Fenfen Dong