- 1The Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education), College of Life Sciences, Guizhou University, Guiyang, China
- 2Research Center of Karst Ecological Environment of Guizhou University, Guiyang, China
- 3Chishui Alsophila National Nature Reserve Administration, Chishui, China
- 4Guizhou Xishui National Nature Reserve Administration, Xishui, China
Introduction: The “rhizosphere effect” in plants occurs within approximately 5 mm from the root surface, where microbial communities exhibit distinct species composition and structural characteristics compared to non-rhizospheric soil. Root-associated fungi play crucial roles in nutrient acquisition enhancement, stress resistance improvement, organic matter decomposition, and carbon cycle promotion. Current research shows limited understanding of the rhizospheric fungal communities in Alsophila spinulosa, a rare and endangered plant species.
Methods: This study conducted a comparative analysis of fungal community composition and structural differences between A. spinulosa rhizospheric and non-rhizospheric soils. The functional roles of fungi in these distinct communities were systematically analyzed, with particular emphasis on identifying keystone fungal taxa within the rhizosphere, and explained the construction process of fungal communities.
Results: The results showed that there was no difference in α diversity between the rhizosphere and non-rhizosphere soil fungal communities of A. spinulosa, but the β diversity was significantly differentiated, indicating that the difference between rhizosphere and non-rhizosphere fungal communities was mainly reflected in species composition rather than species number. The two communities have common dominant phylum: Ascomycetes and Basidiomycetes, and common dominant genera: Mortierella and Saitozyma. The functional type was mainly saprotic. Linear discriminant analysis effect size (LEfSe) analysis revealed four biomarker genera (Arthopyrenia, Hypochnicium, Tremella, and Syncephalis) enriched in the A. spinulosa rhizospheric fungal community. Venn diagram analysis identified 169 core genera within this community, with Flavodon exclusively present in the rhizosphere. Mechanistic analysis of community assembly demonstrated that stochastic processes predominantly governed the structuring of rhizospheric fungal communities.
Discussion: In conclusion, this study elucidates the functional composition and assembly mechanisms of rhizospheric fungal communities in A. spinulosa, while identifying keystone fungal taxa potentially critical to its survival. Future investigations should: Quantify the specific contribution of Flavodon to A. spinulosa; Decipher the mechanistic linkages between these fungi and the plant’s stress resistance traits; Implement plant–soil-microbe synergistic restoration strategies to enhance natural regeneration capacity of A. spinulosa populations.
1 Introduction
Alsophila spinulosa is one of the few remaining rare woody fern species and represents an ancient relict plant that coexisted with dinosaurs (Zong et al., 2016). This species holds significant value not only for its medicinal properties but also for studies on speciation, phytogeography, paleobiology, and paleoenvironmental changes (Chen et al., 2008). However, A. spinulosa populations face severe threats due to short spore viability, difficult germination, prolonged life cycles, strict habitat requirements, and anthropogenic logging, necessitating urgent conservation measures (Cheng et al., 1990).
The rhizosphere is a micro-domain environment affected by plant root activity, ranging from 2 to 3 mm from the surface of fine roots. The interactions among plants, soil, and microorganisms in this area is very complicated (Zhou, 2011; Zhou et al., 2014). The roots of seed plants can act on the rhizosphere microbial flora by secreting sugars, amino acids, organic acids, and other substances, thus affecting the structure of the rhizosphere microbial community (Yuan et al., 2018; Huang et al., 2022). The composition and diversity of rhizosphere microbial species can also affect plant growth and development (Zhalnina et al., 2018). Of these species, rhizosphere soil fungi play an important role in regulating nutrient intake, promoting growth and development, improving the disease resistance of host plants, and maintaining the balance of rhizosphere microecosystems (Hannula et al., 2020; Powell and Rillig, 2018).
Compared with seed plants, the fibrous root structure of ferns is relatively simple, which is usually an adventitious root on the stem or bud, with no true apical meristem, underdeveloped vascular tissue, and low absorption and transport capacity (Dong et al., 2015; Sherwin Carlquist, 2001; Zhang, 2012). A. spinulosa, the only surviving woody fern today, has lignified main stems that can reach 6 m or more in height, but its absorption root anatomical characteristics are not significantly different from those of some small herbaceous ferns (Xiang et al., 2022). Research on ferns mainly focuses on vesicular-arbuscular (VA) mycorrhiza in the rhizosphere soil of ferns (Sha et al., 1998; Zhao, 1998). It has been suggested that VA mycorrhiza play a role in the nutrition and adaptation of ferns. This has been demonstrated in the fern Osmunda japonica (Ma and He, 2010), but not in A. spinulosa. Studies on the rhizosphere microorganisms of A. spinulosa have shown that bacterial community composition and diversity were greatly influenced by soil pH, while fungal community composition and diversity were greatly influenced by available phosphorus, organic carbon, sucrase, and urease (Che et al., 2024). Endophytic fungi in A. spinulosa exhibited higher diversity in leaf rachises compared to leaf blades (Zang et al., 2020). Nevertheless, the functional roles and assembly mechanisms of rhizospheric fungal communities in A. spinulosa remain insufficiently investigated, representing a critical research gap in understanding this relict species’ microbiome ecology. This study contributes to elucidating the unique rhizosphere microbiome assembly mechanisms in the endangered A. spinulosa, addressing a critical knowledge gap in microbial ecology of non-model plants. Simultaneously, it provides a microbiological foundation for developing targeted conservation strategies for this species.
2 Materials and methods
2.1 Study site
The Guizhou Chishui A. spinulosa National Nature Reserve is located in Chishui, Guizhou, China, with a latitudinal range of 28°20′19″–28°28′40″ N and a longitudinal range of 105°57′54″–106°7′7″ E″. It has obvious Danxia landform characteristics and a large number of A. spinulosa forest areas around the world (Liu, 2018). The reserve belongs to the middle subtropical humid monsoon climate zone, and the river valley has characteristics similar to the southern subtropical climate. There is no severe cold in winter, no scorching heat in summer, less sunshine, high temperature, high humidity, abundant precipitation, many cloudy and rainy days, and large vertical differences (Yang et al., 2011).
2.2 Sample collection
In the reserve, Jinfeng Mountain (JFS), Mentouxi Gou (MTXG), and Xujia Gou (XJG), which are densely distributed and less populated by human beings, were selected as sample plots. Three A. spinulosa individuals with good growth conditions and similar growth were randomly selected from different orientations in each plot, within a 0.5-meter radius centered on the main stem, surface soil was removed to expose A. spinulosa roots. Following the detachment of larger soil aggregates, the remaining soil adhering to the root system was collected as rhizosphere soil (R). Non-rhizosphere soil samples were collected from areas beyond the 0.5-meter radius (at depths of 0–20 cm) and at least 10 cm away from any visible roots (NR) (Riley and Barber, 1970). After removing impurities, such as stones, plant residues, and roots, the collected samples were quickly placed into sterile centrifuge tubes, numbered, and stored in liquid nitrogen. The samples collected in the same plot were divided into 2 groups according to rhizosphere or non-rhizosphere soil, and 18 soil samples from 6 groups were collected in 3 plots.
2.3 DNA extraction and sequencing
After genomic DNA was extracted from the sample, the quality of the extracted genomic DNA was detected using 1% agarose gel electrophoresis, and the DNA concentration and purity were determined using NanoDrop2000 (Thermo Scientific Company, Waltham, Massachusetts, United States). The ITS2 region was amplified using the primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′) using the extracted DNA as a template. The PCR products of the same sample were mixed and tested using 2% agarose gel electrophoresis. The PCR products were cut and recovered using an AchyPrep DNA Gel Recovery Kit (AXYGEN Company, San Carlos, California, United States), eluted by Tris_HCl, and tested by 2% agarose electrophoresis. Based on the preliminary quantitative results of electrophoresis, the PCR products were detected and quantified using the QuantiFluor™-ST Blue Fluorescence Quantification System (Promega Company, Madison, Wisconsin, United States). Purified amplicons were pooled in equimolar amounts and paired-end sequenced on the Illumina Nextseq2000 Platform (Illumina, San Diego, California, United States) according to the standard protocols of Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). The raw sequencing reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: PRJNA1033401).
2.4 Data analysis
Differences between sample groups were tested using the Kruskal–Wallis test. Principal coordinates analysis (PCoA) based on the Bray–Curtis distance was used to reveal the differences and similarities between the two groups of samples. Functional classification of fungi was carried out using the FUNGuild database. The linear discriminant analysis (LDA) effect size (LEfSe) was performed to identify the significantly abundant taxa (phylum to genera) of fungi among the different groups (LDA score > 3, p < 0.05). Venn diagrams were used to show the number and proportion of common and endemic fungi among groups. A phylogenetic tree was used to reveal the genetic relationships between each species in the sample during evolution. Employed a null model approach with 999 randomizations to calculate the beta nearest taxon index (βNTI), thereby evaluating the relative contributions of deterministic and stochastic processes in microbiome assembly. All statistical analyses were done with the free R software, version 2023.9.1.494.
3 Results
3.1 Fungal abundance and community functional composition
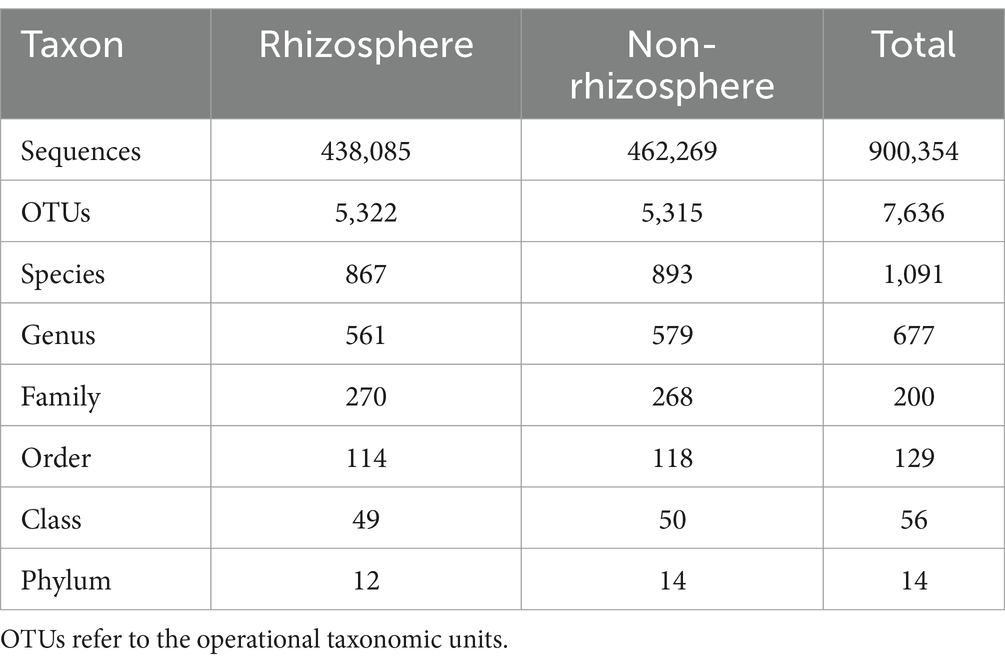
The sequencing data were spliced and underwent quality control and de-ligation to obtain an optimized sequence with an average length of 232 bp. The operational taxonomic unit (OTU) abundance table was obtained by clustering the optimized sequences at a similarity level of 97%. Sequences aligned to chloroplasts and mitochondria were removed, and the ITS database was used to annotate species information. At the phylum and genus levels, the abundance of fungal communities in the rhizosphere soil was lower than that in the non-rhizosphere soil, but there were no significant differences (Table 1).
Ascomycota and Basidiomycota were the most dominant phyla in both the rhizosphere and non-rhizosphere fungal communities (Figure 1a), and Mortierella and Saitozyma were dominant genera common between the two communities (Figure 1b).
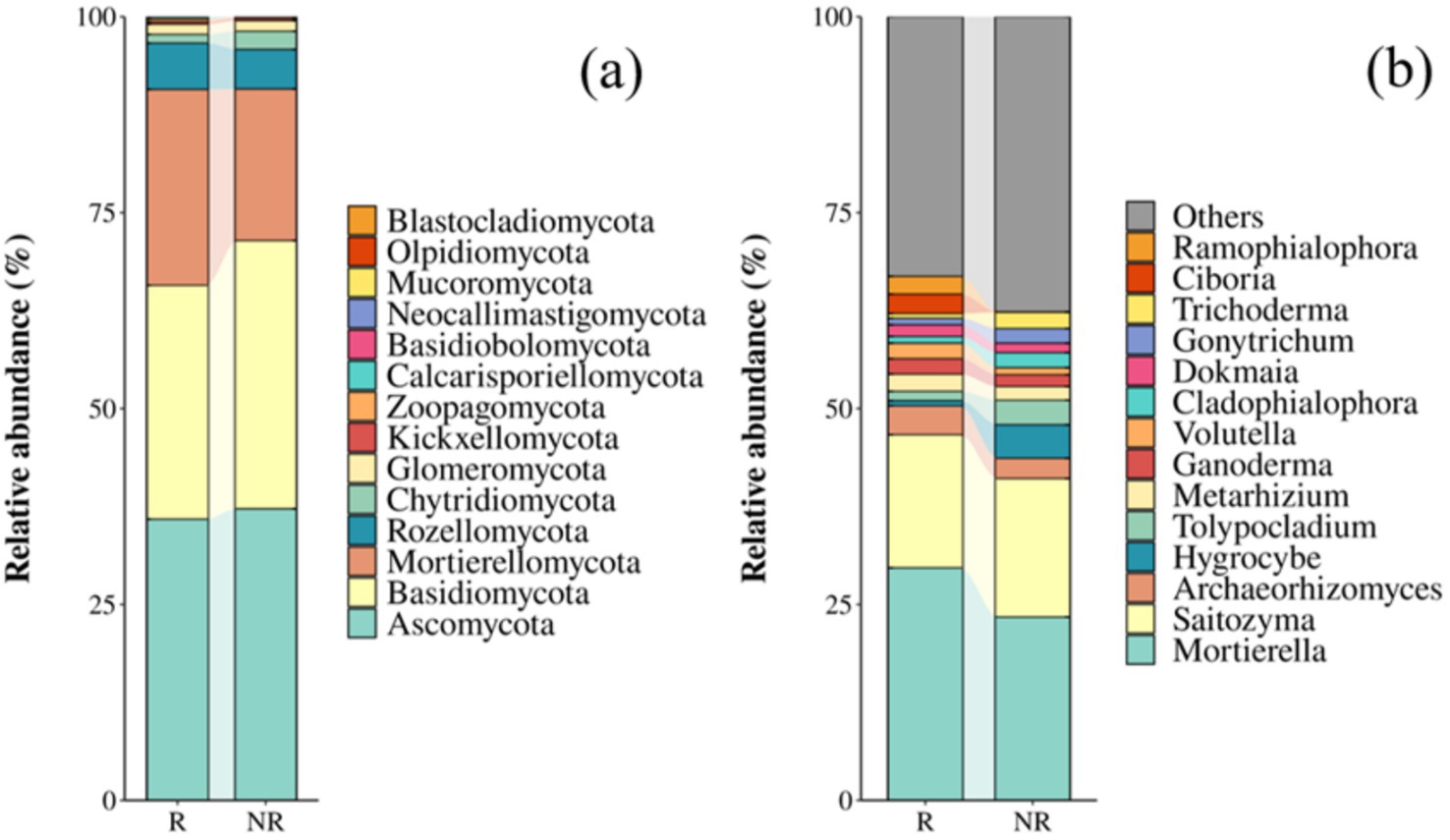
Figure 1. Relative abundance of fungal communities at the phylum (a) and genus (b) levels. R: rhizosphere fungal community; NR: non-rhizosphere fungal community.
Based on different nutritional methods, fungi were divided into 3 categories: pathotrophs, symbiotrophs, and saprotrophs. Based on these categories, fungi were divided into several guilds. Of the phyla and genera, 71.43% and 76.35%, respectively, were successfully annotated, and the credibility was assessed as “probable” and “highly probable.”
Based on the trophic mode (Figure 2a), the relative abundance of saprotrophs, pathotrophs, and pathotroph–saprotrophs were high in both the rhizosphere and non-rhizosphere fungal communities. For guilds (Figure 2b), both undefined saprotrophs and plant pathogens had a higher relative abundance, and the relative abundance of undefined saprotrophs in the rhizosphere fungal community was significantly lower than that in the non-rhizosphere fungal community (p < 0.05).
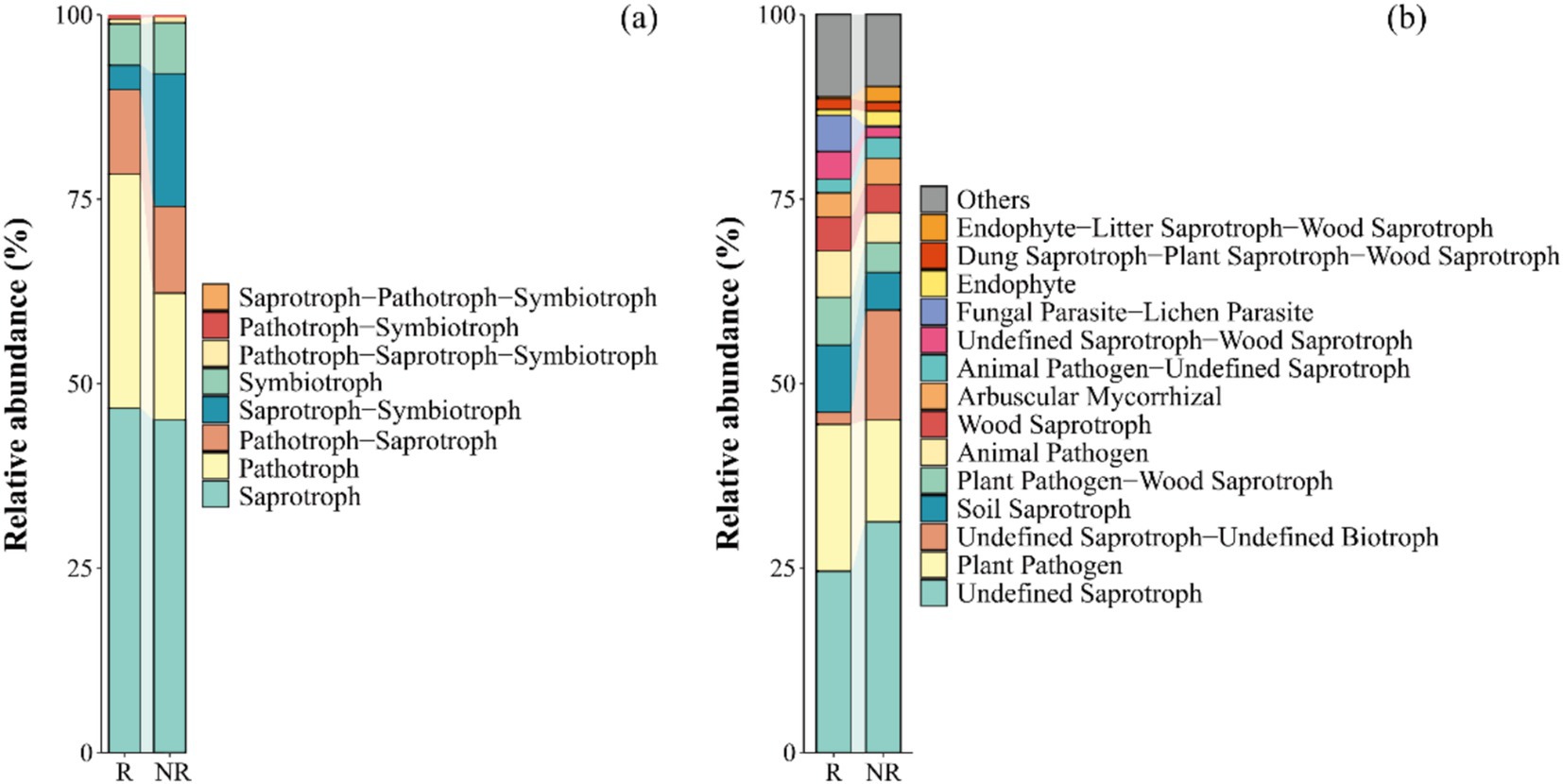
Figure 2. Relative abundance of different trophic modes (a) and guilds (b) in fungal communities. The trophic mode including 3 types: saprotroph, pathotroph, and symbiotroph. The guild has 12 types: animal pathogens, arbuscular mycorrhizal fungi, ectomycorrhizal fungi, ericoid mycorrhizal fungi, foliar endophytes, lichenicolous fungi, lichenized fungi, mycoparasites, plantpathogens, undefined root endophytes, undefined saprotrophs and wood saprotrophs. R: rhizosphere fungal community, NR: non-rhizosphere fungal community.
3.2 Diversity analysis of fungal communities
The richness and diversity of microbial communities were reflected by α-diversity analysis. The Ace, Chao, Shannon, Simpson, and Sob of fungal communities in the rhizosphere and non-rhizosphere soils of A. spinulosa did not show significant differences (p > 0.05). Therefore, the number and distribution of species were similar between the two communities (Figure 3).
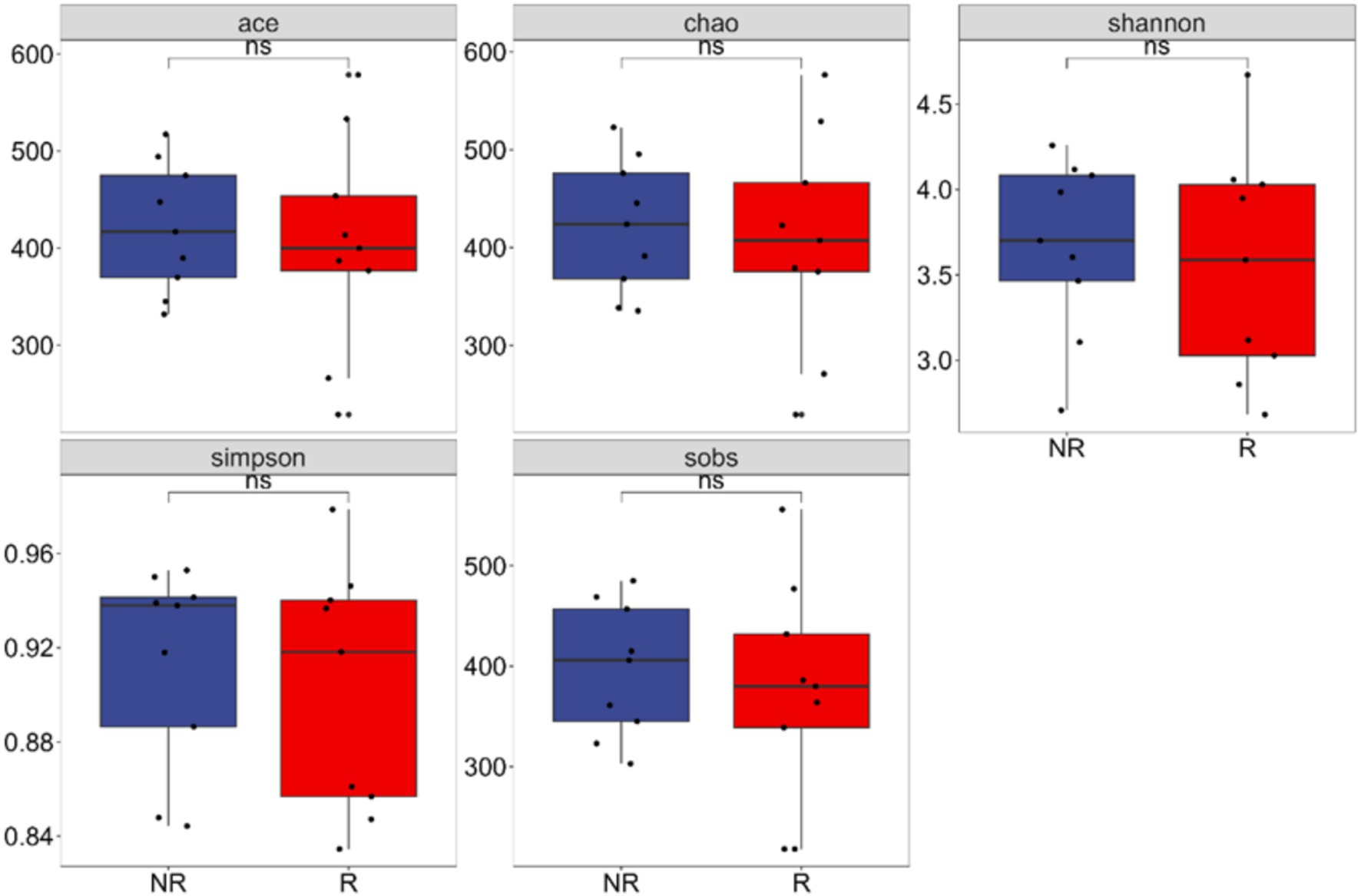
Figure 3. Fungal alpha diversity between the rhizosphere and non-rhizosphere of Alsophila spinulosa. Ace: abundance-based coverage estimator; sobs: species observed; R: rhizosphere fungal community, NR: non-rhizosphere fungal community; ns indicates that the difference is not significant (p > 0.05).
Based on the Bray–Curtis distance, PCoA was used to determine the degree of difference in rhizosphere and non-rhizosphere fungal communities of A. spinulosa with different habitats (Figure 4). Fungal communities from different habitats were significantly separated, but only the rhizosphere and non-rhizosphere fungal communities in XJG were separated in the same habitat.
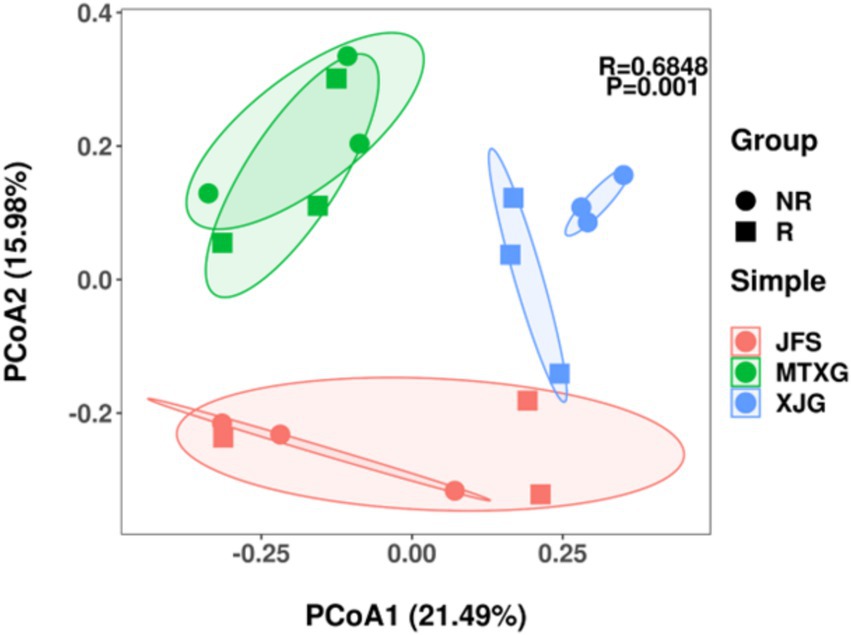
Figure 4. PCoA of fungal community composition based on the Bray–Curtis distance. R: rhizosphere fungal community, NR: non-rhizosphere fungal community; JFS: Jinfeng Mountain; MTXG: Mentouxi Gou; XJG: Xujia Gou.
3.3 Difference analysis of fungal composition
LEfSe identified biomarkers with significant differential impacts. Biomarkers in the rhizosphere and non-rhizosphere fungal communities appeared at both ends of the evolutionary tree, but biomarkers from the same group were close to each other on the evolutionary tree (Figure 5a). This suggests that the biomarkers in the same group had closer genetic relationships and more similar functions and that the biomarkers in the two groups that were more distant had different functions.
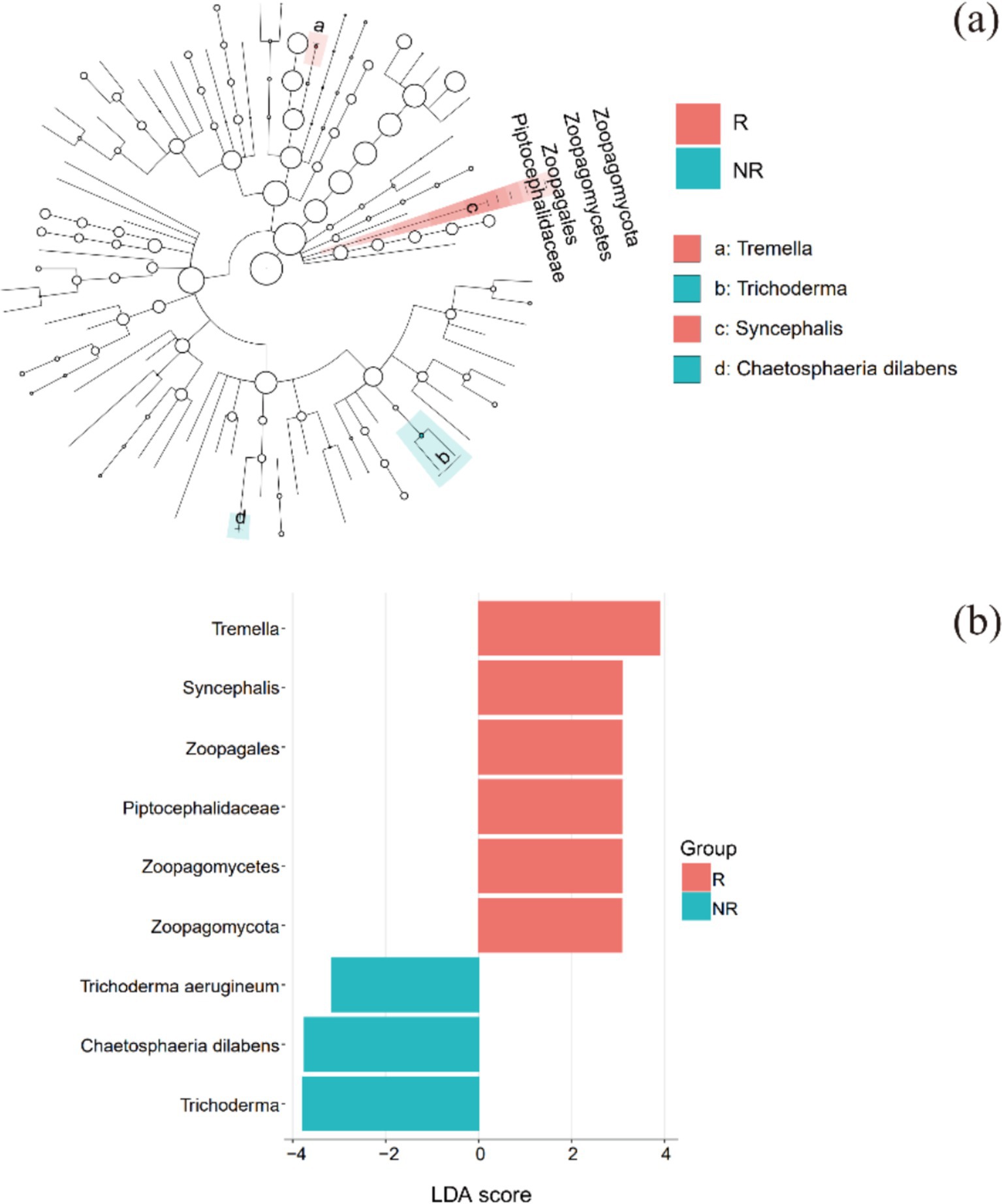
Figure 5. LEfSe analysis: (a) evolutionary branch diagram and (b) distribution histogram. R: rhizosphere fungal community, NR: non-rhizosphere fungal community.
At the phylum level, the biomarker in the rhizosphere was Zoopagomycota, a group of fungal parasites. At the genus level, biomarkers in the rhizosphere were Arthopyrenia, Hypochnicium, Tremella, and Syncephalis. Based on the FUNGuild results, Arthopyrenia was lichenized, meaning that it formed a symbiosis with algae and that they collectively formed a lichen. Hypochnicium was an undefined saprotroph, and Tremella was a fungal or lichen parasite. Syncephalis was a fungal parasite (Figure 5b).
There were only two biomarkers for non-rhizosphere fungal communities at the genus level: Chaetosphaeria and Trichoderma. Chaetosphaeria was an endophyte, litter saprotroph, or wood saprotroph, and Trichoderma was an undefined saprotroph.
The relative abundance of the above six biomarkers in rhizosphere (Arthopyrenia, Hypochnicium, Tremella, and Syncephalis) and non-rhizosphere (Chaetosphaeria and Trichoderma) fungal communities were compared. The relative abundance of these biomarkers was not high in the community. The highest in the rhizosphere community was Tremella, exceeding 1%, and the others were between 0.1 and 0.4%. The relative abundance of the 2 biomarkers in non-rhizosphere communities were both around 1% (Figure 6).
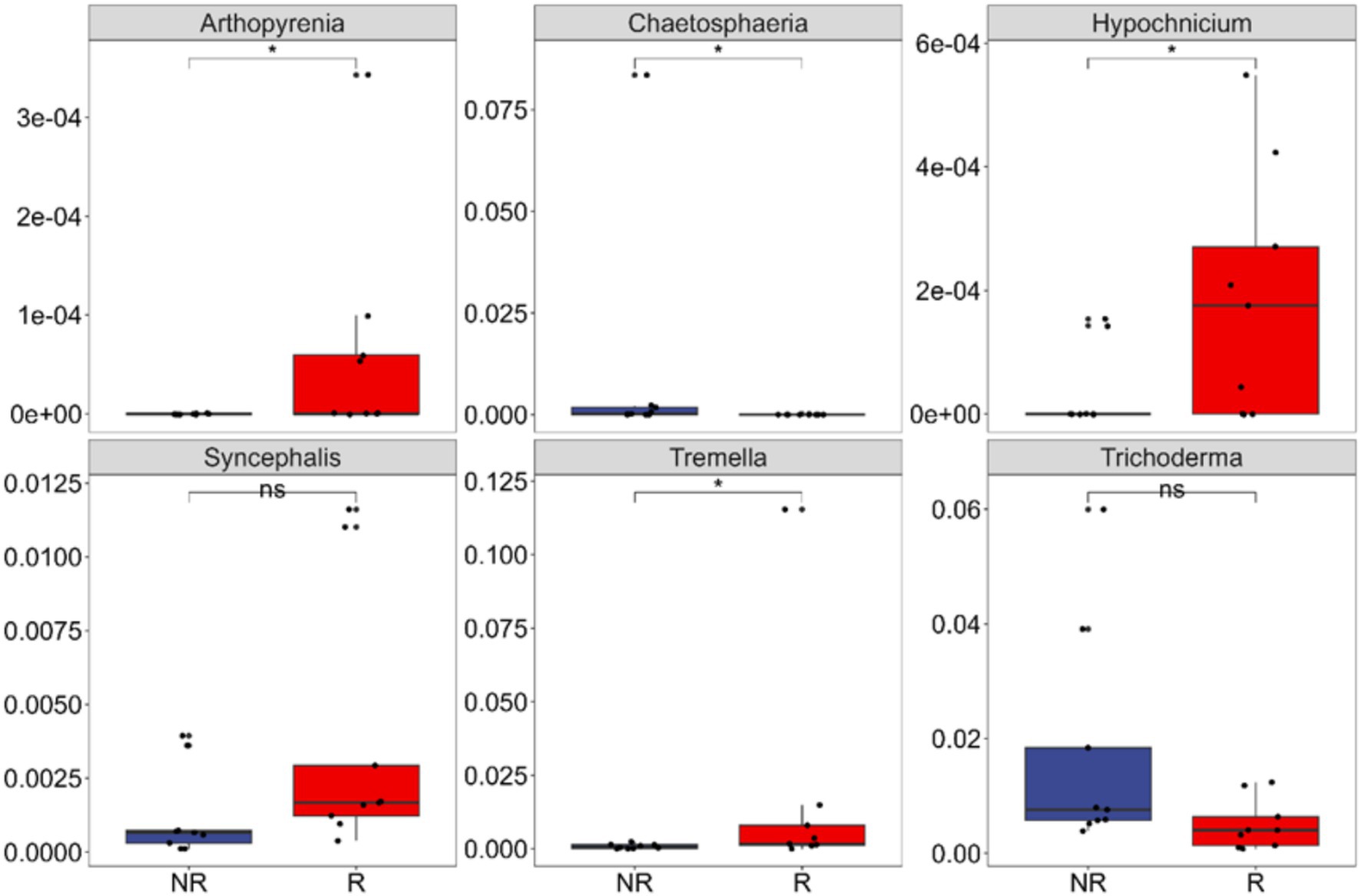
Figure 6. Differences in the relative abundance of each indicator species in rhizosphere (R) and non-rhizosphere (NR) soils at the genus level. Arthopyrenia, Hypochnicium, Tremella, and Syncephalis are biomarkers for rhizosphere fungal communities; Chaetosphaeria and Trichoderma are biomarkers for non-rhizosphere fungal communities. *0.01 < p ≤ 0.05; ns: p > 0.05.
3.4 Core fungi and endemic species
The core microbiome is considered a key component of the basic functions of holobionts (Dong et al., 2019). The genera that appeared in in all 3 habitats were the core fungi of the A. spinulosa rhizosphere soil fungal community; these 169 genera accounted for 30.12% of the genera in the rhizosphere community (Figure 7a). The most relatively abundant genera of the core fungi in the rhizosphere community were Mortierella and Saitozyma, which were in the wood saprotroph and fungal parasite/undefined saprotroph guilds, respectively (Figure 7b).
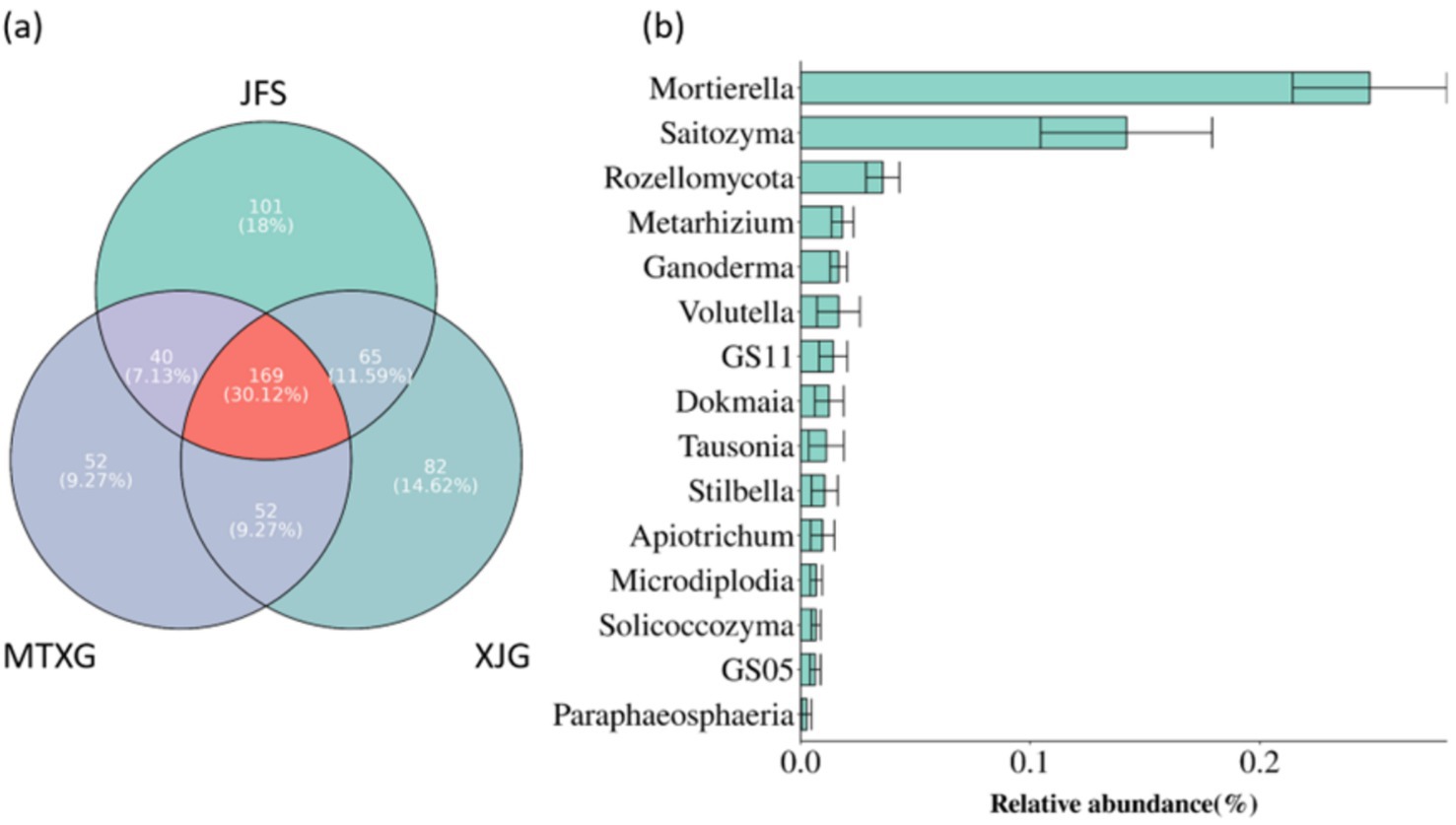
Figure 7. (a) Core fungi of rhizosphere fungal communities in three plots. (b) Top 15 most abundant genera among core fungi. JFS: Jinfeng Mountain; MTXG: Mentouxi Gou; XJG: Xujia Gou.
The core fungi, comprising 169 genera, in the rhizosphere fungal community of A. spinulosa were divided into 8 functional types: saprotroph, symbiotroph, pathotroph, saprotroph/symbiotroph, pathotroph/symbiotroph, pathotroph/saprotroph, pathogen/saprotroph/symbiotroph, and pathotroph/saprotroph/symbiotroph. In terms of abundance, there were more fungi with saprotrophic and symbiotic nutritional characteristics, but in terms of frequency of occurrence, saprotrophic fungi were the most common (Figure 8).

Figure 8. Relative abundance (a) and frequency (b) of different trophic types in rhizosphere core fungi.
Comparing all fungi present in the A. spinulosa rhizosphere those not in the rhizosphere revealed 98 unique genera in the rhizosphere community, accounting for 14.48% of the total (Figure 9a). Among these unique genera, those with a higher relative abundance were Mycenella, Myrothecium, and Henningsomyces. Mycenella was in the symbiotroph guild, and Myrothecium and Henningsomyces were in the saprotroph guild.
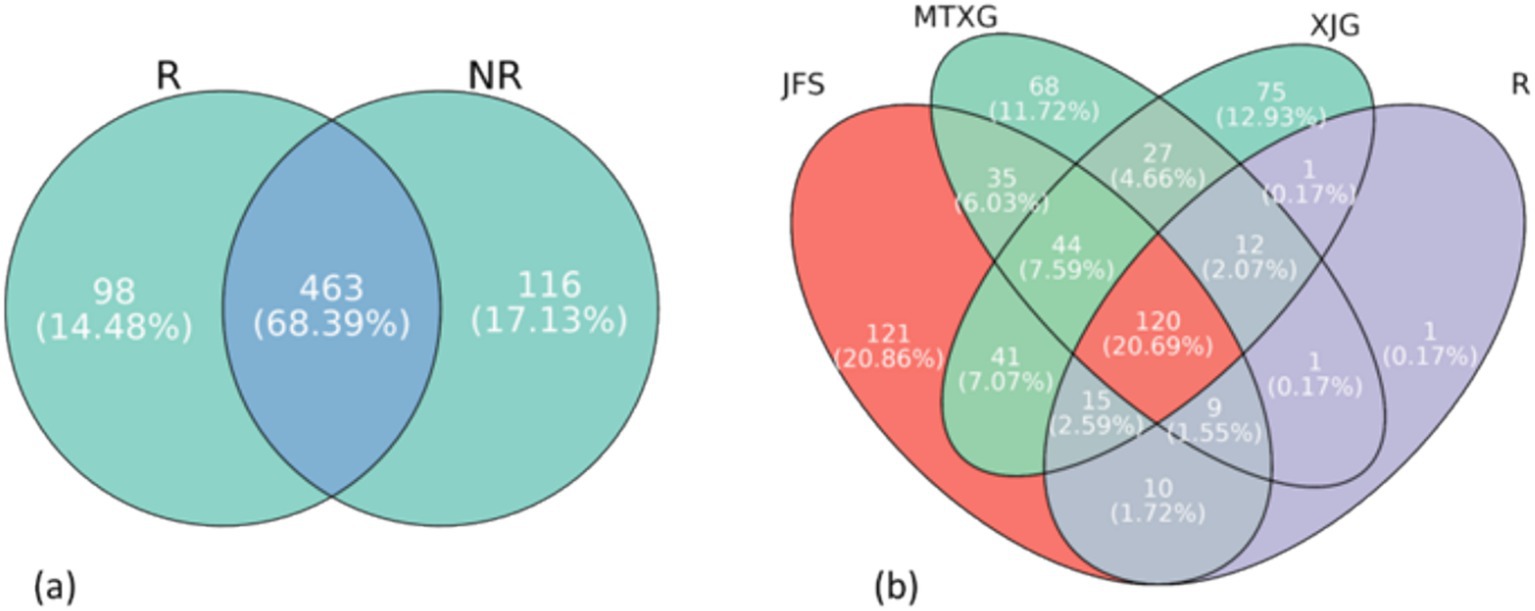
Figure 9. (a) Venn diagram of rhizosphere and non-rhizosphere soil fungi. R: rhizosphere fungal community, NR: non-rhizosphere fungal community. (b). Venn diagram of three non-rhizosphere habitats and rhizosphere soil fungi. JFS: Jinfeng Mountain; MTXG: Mentouxi Gou; XJG: Xujia Gou; R: the fungal genus within the rhizosphere fungal community.
The core fungi in the rhizosphere community were compared with those in non-rhizosphere communities. One genus, Flavodon, was found in the rhizosphere community but not in the non-rhizosphere community (Figure 9b), and its guild was undefined saprotroph.
3.5 Community assembly of rhizosphere and non-rhizosphere fungi
To evaluate the relative importance of certainty and randomness in the microbiome assembly, the β-closest taxon index (βNTI) and Raup–Crick index (RCBray) was calculated to infer the process of fungal community assembly changes in the rhizosphere and non-rhizosphere soil of A. spinulosa. The process of fungal community assembly in the rhizosphere and non-rhizosphere of A. spinulosa was significantly different (Figure 10a). Although both communities were dominated by random processes, there were more random components in the rhizosphere community (Figure 10b).
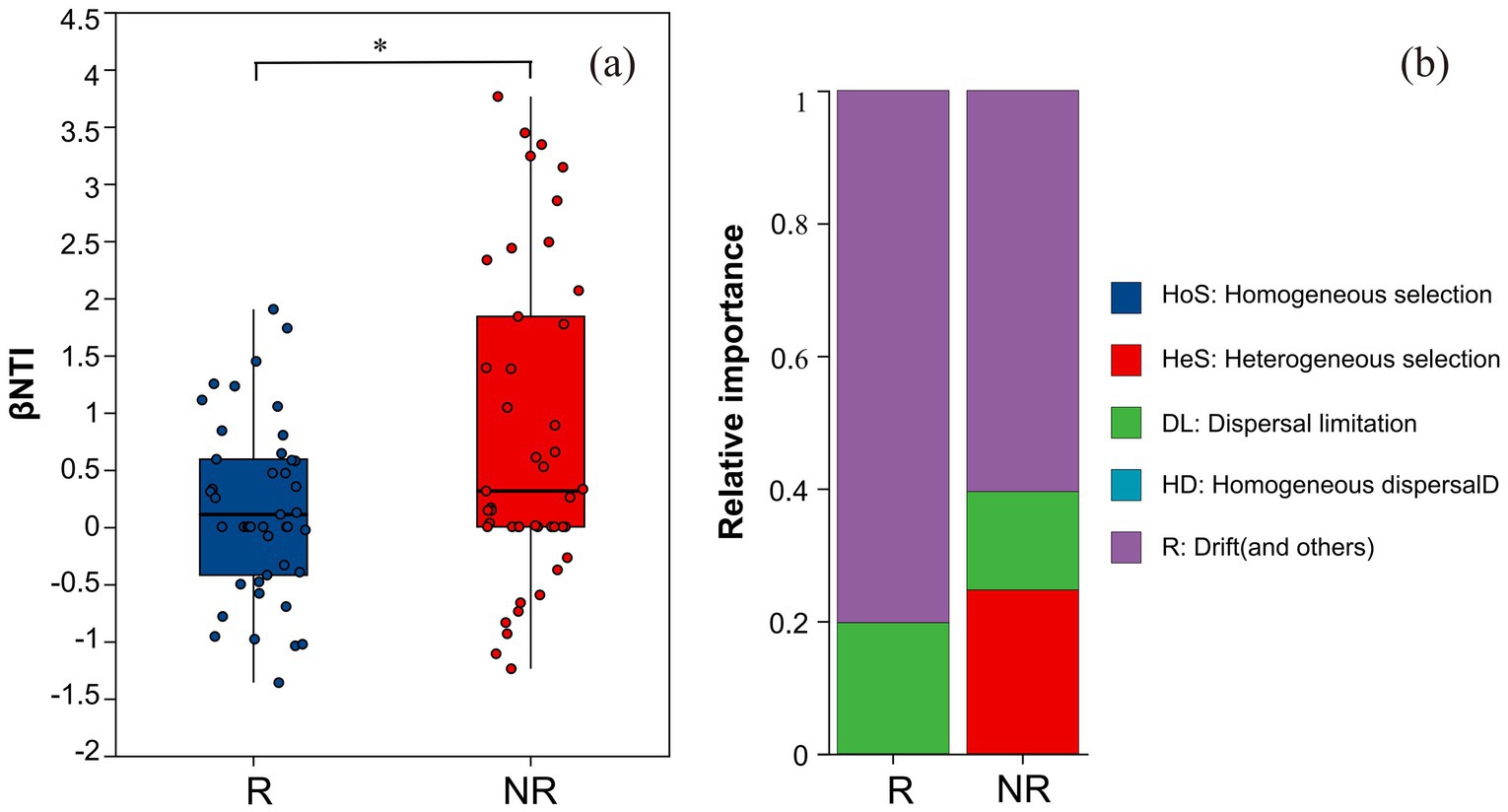
Figure 10. Analysis of community construction of rhizosphere and non-rhizosphere fungal communities in Alsophila spinulosa. (a) differences in βNTI between R and NR, (b) composition of ecological processes in R and NR. R: rhizosphere fungal community, NR: non-rhizosphere fungal community.
4 Discussion
4.1 Characteristics of rhizosphere and non-rhizosphere fungal communities
Ferns are the most primitive group of vascular plants. Although they have real roots, they are usually adventitious roots, and the root branch structure is simple with an underdeveloped vascular tissue (McAdam and Brodribb, 2012; Xiang et al., 2022). Mycorrhizal fungi often form symbiotic relationships with plants and have complex interactions with host plants and the environment, and this relationship is critical for plant growth and soil ecosystem health (Qu et al., 2023; Reese et al., 2018; Shao et al., 2021). In general, the richness and diversity of fungal communities in plant rhizosphere soil are higher than those in non-rhizosphere soil (Liu et al., 2021; Lu and Ang Zhang, 2006); however, this difference was not significant in the rhizosphere soil fungal community of A. spinulosa. After excluding sampling artifacts, these observed patterns likely reflect intrinsic biological characteristics of both the habitat and A. spinulosa itself. Both environmental stress and habitat homogenization may attenuate rhizosphere effects in plants—a hypothesis requiring further empirical validation through controlled studies. Furthermore, as an ancient relict species, A. spinulosa likely exhibits generalized symbiotic strategies with fungi rather than highly specific dependencies—a pattern consistent with early-diverging plant lineages’ microbial partnership characteristics.
4.2 Biomarkers in the rhizosphere community
In this study, four genera served as biomarkers for rhizosphere fungal communities. Hypochnicium is a genus of corticioid, wood-inhabiting fungi in Polyporales with a worldwide distribution. By decomposing wood, wood-inhabiting fungi release important elements, such as carbon, nitrogen, and phosphorus, into the soil and participate in nutrient cycling, promoting plant growth. Tremella and its accompanying fungi often decompose wood and promote nitrogen fixation. Syncephalis is a parasitic fungus that preys on protozoans, such as amoebas and nematodes, and inhabits them both internally and externally (Zhou, 2005). In summary, these fungi may play a role in providing nutrients and defending against diseases, but more research is needed to prove this. Both environmental stress and habitat homogenization may attenuate rhizosphere effects in plants—a hypothesis requiring further empirical validation through controlled studies.
4.3 Core and endemic fungi in the rhizosphere
Core microorganisms are closely related to the host and may have direct or indirect antagonism with other microorganisms, thus having positive or negative effects on plant growth (Busby et al., 2016; Matthew and Agler, 2016). The core fungi in the rhizosphere soil of A. spinulosa were dominated by saprophytes, and the most abundant genus, Mortierella, can be used to produce polyunsaturated fatty acids (Jin, 2016). At the same time, it also has an inhibitory effect on some plant pathogenic fungi (Yu et al., 2016). The second most abundant genus, Saitozyma, is a genus of the order Tremellales and is commonly found in soil and the plant rhizosphere (Leguina et al., 2019; Zhou et al., 2021; Zhou, 2020). These two genera can be regarded as beneficial fungi to a certain extent, but whether they have beneficial effects on A. spinulosa growth requires further research and verification.
Fungi endemic to the A. spinulosa rhizosphere (mentioned in Section 3.5) were mainly saprophytic fungi, and the most abundant were Mycenella, Myrothecium, and Henningsomyces. Myrothecium is widely found in plants and soils. Most species have strong cellulolytic abilities. Some species produce antibiotics, and some cause plant diseases (Wu, 1991). Henningsomyces is a wood-dwelling fungus that was first reported in China in 2007, and its specific functions have yet to be determined (Wei et al., 2007). Many dominant and unique saprophytic fungi in the A. spinulosa rhizosphere promote its growth to some extent, making them potential beneficial fungi.
The genus Flavodon was unique to the rhizosphere fungal community. Research has recently been conducted on Flavodon. Flavodon flavus is used in various industries for the degradation of lignin-related pollutants. Flavodon ambrosius can coexist with insects and grow on lignocellulose in dead trees. The cellulase activity of Flavodon sp. x10 is 1.388 U/mL, which is higher than that of the white rot fungi, Ganoderma applanatum, Trametes hirsuta, and Fomes fomentarius, giving it a significant advantage in the development and utilization of lignocellulose (Wang et al., 2023).
4.4 Community assembly of rhizosphere fungi
Based on βNTI and RCBray, the community construction process was divided into two deterministic processes, namely homogeneous selection and heterogeneous selection, and three stochastic processes, namely dispersal limitation, homogeneous dispersal, and drift and other ecological processes. The construction of the A. spinulosa rhizosphere fungal community was random and mainly affected by drift and other ecological processes. This shows that fungi are greatly affected by accidental events such as environmental changes, weather events, and habitat changes. One of the reasons why A. spinulosa is endangered is that it has strict habitat requirements. Environmental changes affect fungal communities, which may adversely impact the growth of A. spinulosa to some extent. In addition, the current insufficient or unstable environmental resources will decrease the fungal population size, thus increasing its sensitivity to drift and making some fungi dominate or disappear, which changes the functions of the community.
5 Conclusion
The disparities between rhizospheric and non-rhizospheric fungal communities in A. spinulosa soils primarily manifested in species composition rather than taxonomic abundance. The two most abundant genera in the rhizospheric fungal community, Mortierella and Saitozyma, play pivotal roles in soil material cycling and ecological system formation. The enriched biomarker genera and rhizosphere-unique genera within the fungal community may play distinct functional roles in A. spinulosa’s adaptation to specific habitats, such as lignin degradation and symbiotic interactions. Stochastic dispersal processes—such as spore propagation and ecological drift—dominated the assembly of rhizospheric communities, whereas deterministic processes (e.g., environmental filtering) exerted stronger influences on non-rhizospheric communities. This study delineates the assembly principles and ecological signatures of A. spinulosa-associated fungal communities, providing both a theoretical foundation for utilizing unique fungal functions and empirical support for implementing plant–soil-microbe synergistic restoration strategies in A. spinulosa conservation efforts.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA1033401.
Author contributions
ZH-M: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. YL-B: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing. YD-M: Investigation, Writing – review & editing. LF: Visualization, Writing – review & editing. CX-G: Investigation, Writing – review & editing. WD-Y: Investigation, Writing – review & editing. HQ-Q: Project administration, Resources, Writing – review & editing. MJ: Project administration, Resources, Writing – review & editing. YL-F: Methodology, Writing – review & editing. LY: Supervision, Writing – review & editing. CZ-F: Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Talent Introduction Scientific Research Project of Guizhou University (Gui-Da-Ren-Ji-He-Zi (2020) No. 27); Guizhou Provincial Basic Research Program (Natural Science) (No. Qian-Ke-He-Ji-Chu-ZK [2023] General-111); the Special program for key protected plants in national nature reserves (NO-20230101).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Busby, P. E., Peay, K. G., and Newcombe, G. (2016). Common foliar fungi of Populus trichocarpa modify Melampsora rust disease severity. New Phytol. 209, 1681–1692. doi: 10.1111/nph.13742
Che, B., Yang, W., He, Q., Jiang, Y., Zhang, B., and Chen, H. (2024). Soil microbial community composition and diversity in the rhizosphere of Alsophila spinulosa growing in different habitats within the Chishui Alsophila National Nature Reserve in Guizhou Province, China. Front. Microbiol. 15:1445255. doi: 10.3389/fmicb.2024.1445255
Chen, F. Z., Xiang, Q. X., and Li, S. H. (2008). Chemical constituents in the leaves of Alsophila spinulosa. Acta Botan. Boreali-Occiden. Sin. 28, 1246–1249.
Cheng, Z. Y., Tao, G. D., and Xu, Z. F. (1990). Discussion on the reasons why Alsophila spinulosa is endangered. Plant Diversity. 12, 186–190.
Dong, X., Wang, H., Gu, J., Wang, Y., and Wang, Z. (2015). Root morphology, histology and chemistry of nine fern species (pteridophyta) in a temperate forest. Plant Soil 393, 215–227. doi: 10.1007/s11104-015-2484-7
Dong, C. B., Zhang, Z. Y., Han, Y. F., and Liang, Z. Q. (2019). Research and application prospects of core microbiome. Mycosystema 38, 1–10. doi: 10.13346/j.mycosystema.180214
Hannula, S. E., Morriën, E., van der Putten, W. H., and de Boer, W. (2020). Rhizosphere fungi actively assimilating plant-derived carbon in a grassland soil. Fungal Ecol. 48:100988. doi: 10.1016/j.funeco.2020.100988
Huang, S. P., Jin, D. C., Li, Q. L., Wei, S. L., Tang, L. H., Huang, S. M., et al. (2022). Study on the characteristics and common rule of bacteria community in banana rhizosphere and non-rhizosphere soil. J. Southern Agricult. 53, 1253–1262. doi: 10.3969/j.issn.2095-1191.2022.05.007
Jin, T. (2016). A preliminary research on the quorum-sensing of filamentous Mortierella. Hubei: Huazhong University of Science and Technology.
Leguina, A. C. V., Mariano, J., Lacosegliaz, H. F. P. L., and Nieto-Peñalver, C. G. (2019). Copper alters the physiology of tomato rhizospheric isolates of Papiliotrema laurentii. Sci. Hortic. 243, 376–384. doi: 10.1016/j.scienta.2018.08.057
Liu, Q. (2018). The characteristics study of plant community in the Guizhou Chishui Alsophila spinulata National Nature Reserve. Chongqing: Southwest University.
Liu, M. X., Li, B. W., Sun, R. D., Zhang, Y. Y., Song, J. Y., Zhang, G. J., et al. (2021). Characteristics of culturable microbial communities in rhizosphere/non-rhizosphere soil of Ligularia virgaurea in alpine meadow elevation gradient. Acta Ecol. Sin. 41, 4853–4863. doi: 10.5846/stxb202004240987
Lu, Y. H., and Ang Zhang, F. S. (2006). The advances in rhizosphere microbiology. Soil. 38, 113–121. doi: 10.3321/j.issn:0253-9829.2006.02.001
Ma, Q., and He, Y. F. (2010). Study on VA mycorrhiza of Osmunda japonica. Hubei Agricult. Sci. 49, 901–903. doi: 10.14088/j.cnki.issn0439-8114.2010.04.049
Matthew, T., and Agler, J. S. C. S. (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14:e1002352. doi: 10.1371/journal.pbio.1002352
McAdam, S. A. M., and Brodribb, T. J. (2012). Stomatal innovation and the rise of seed plants. Ecol. Lett. 15, 1–8. doi: 10.1111/j.1461-0248.2011.01700.x
Powell, J. R., and Rillig, M. C. (2018). Biodiversity of arbuscular mycorrhizal fungi and ecosystem function. New Phytol. 220, 1059–1075. doi: 10.1111/nph.15119
Qu, Z., Li, Y., Xu, W., Chen, W., Hu, Y., and Wang, Z. (2023). Different genotypes regulate the microbial community structure in the soybean rhizosphere. J. Integr. Agric. 22, 585–597. doi: 10.1016/j.jia.2022.08.010
Reese, A. T., Lulow, K., David, L. A., and Wright, J. P. (2018). Plant community and soil conditions individually affect soil microbial community assembly in experimental mesocosms. Ecol. Evol. 8, 1196–1205. doi: 10.1002/ece3.3734
Riley, D., and Barber, S. A. (1970). Salt accumulation at the soybean (glycine max. (L.) Merr.) root-soil interface. Soil Sci. Soc. Am. J. 34, 154–155. doi: 10.2136/sssaj1970.03615995003400010042x
Sha, T., Zhao, Z. W., and Zhang, H. B. (1998). Fungi Flora in the rhizosphere soil of three species of pteridophytes. J. Yunnan Univ. (Nat. Sci. Ed.) 72, 68–69.
Shao, Q. Y., Dong, C. B., Han, Y. F., and Liang, Z. Q. (2021). Research progress in the rhizosphere microbiome of plants. J. Plant Nutr. Fertil. 27, 144–152. doi: 10.11674/zwyf.20203
Sherwin Carlquist, E. L. S. (2001). Vessels in ferns: structural, ecological, and evolutionary significance. Am. J. Bot. 88, 1–13. doi: 10.2307/2657121
Wang, B. T., Zhuo, Y., Jin, F. J., Yu, X. Y., Hu, S., Jin, L., et al. (2023). Enzyme activities of a high-efficiency cellulose degrading fungus Flavodon sp. x10. Mycosystema 42, 719–730. doi: 10.13346/j.mycosystema.220229
Wei, Y. L., Yuan, H. S., and Dai, Y. C. (2007). First report of Henningsomyces (Basidiomycetes) in China. J. Fungal Res. 4, 187–189. doi: 10.3969/j.issn.1672-3538.2007.04.001
Wu, W. P. (1991). Study on Hyphomycetes in Hebei ProvinceIII, four species of the Myrothecium Tode:Fr. J. Hebei Acad. Sci. 1, 69–74. doi: 10.16191/j.cnki.hbkx.1991.01.011
Xiang, W., Huang, D. L., and Zhu, S. D. (2022). Absorptive root anatomical traits of 26 tropical and subtropical fern species. Chinese J. Plant Ecol. 46, 593–601. doi: 10.17521/cjpe.2021.0328
Yang, G. B., Li, Y. Q., and Tu, Y. L. (2011). Investigation of eco-environment in Chishui Alsophila natural reserve. For. Grassl. Resour. Res., 94–100. doi: 10.3969/j.issn.1673-923X.2011.11.025
Yu, B. H., Xu, H., and Zhang, C. B. (2016). Identification of Fungi from Cordyceps sinensis and its antibacterial activity. Chin. J. Exp. Tradit. Med. Formulae 22, 36–40. doi: 10.13422/j.cnki.syfjx.2016080036
Yuan, J., Zhao, J., Wen, T., Zhao, M., Li, R., Goossens, P., et al. (2018). Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome 6:156. doi: 10.1186/s40168-018-0537-x
Zang, W., Krua, L. S. N., Shen, C., Du, Z. N., Sun, X., Sun, J. Q., et al. (2020). Species diversity and community composition of endophytic fungi from Alsophila spinulosa. Mycosystema 39, 731–742. doi: 10.13346/j.mycosystema.190438
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., Da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nature Microbiology. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhao, Z. W. (1998). VA mycorrhizal fungi in the rhizosphere soil of ferns in tropical and subtropical Yunnan. Acta Bot. Yunnanica. 20, 58–67.
Zhou, S. (2020). Studies on the diversity of endosphere and rhizosphere fungi on the Cypripedium japonicum Thunb. Shanghai: East China Normal University.
Zhou, X., Gao, D., Liu, J., Qiao, P. L., Zhou, X. G., Lu, H., et al. (2014). Changes in rhizosphere soil microbial communities in a continuously monocropped cucumber (Cucumis sativus L.) system. Eur. J. Soil Biol. 60, 1–8. doi: 10.1016/j.ejsobi.2013.10.005
Zhou, J., Xie, T. X., Liu, J. F., Lan, L. Y., Xu, Y. F., Liu, Z. Y., et al. (2021). Community structure and biological function of the root symbiotic fungi of wild Cymbidium ensifolium. Acta Microbiol Sin. 61, 2136–2153. doi: 10.13343/j.cnki.wsxb.20200692
Keywords: Alsophila spinulosa , fungal diversity, rhizosphere fungal, FUNGuild, community assembly
Citation: Hui-Min Z, Ling-Bin Y, Dong-Mei Y, Feng L, Xiu-Gang C, Dai-Yan W, Qin-Qin H, Jun M, Li-Fei Y, Yuan L and Zhi-Fei C (2025) Establishment and function of the rhizosphere fungal community in rare and endangered plant Alsophila spinulosa. Front. Microbiol. 16:1552748. doi: 10.3389/fmicb.2025.1552748
Edited by:
Mario Serrano, National Autonomous University of Mexico, MexicoReviewed by:
Modupe Stella Ayilara, North-West University, South AfricaXiawei Peng, Beijing Forestry University, China
Copyright © 2025 Hui-Min, Ling-Bin, Dong-Mei, Feng, Xiu-Gang, Dai-Yan, Qin-Qin, Jun, Li-Fei, Yuan and Zhi-Fei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Ling-Bin, bGJ5YW5AZ3p1LmVkdS5jbg==
 Zhang Hui-Min
Zhang Hui-Min Yan Ling-Bin
Yan Ling-Bin Yuan Dong-Mei1,2
Yuan Dong-Mei1,2 Liu Yuan
Liu Yuan Chen Zhi-Fei
Chen Zhi-Fei