- 1College of Horticulture and Plant Protection, Inner Mongolia Agricultural University, Hohhot, China
- 2Sichuan Province Company of Tobacco Corporation in China, Chengdu, China
- 3Agriculture and Animal Husbandry Bureau of Bayannaoer City, Inner Mongolia (Rural Vitalization Bureau), Bayannaoer, China
- 4Key Laboratory of the Development and Resource Utilization of Biological Pesticide in Inner Mongolia, Hohhot, China
Superoxide dismutases (SODs) serve as the first line of defense against reactive oxygen species. Copper-zinc superoxide dismutase (Cu/Zn-SOD) is an enzyme whose activity depends on copper availability. Cu/Zn-SOD-1 induction is shown to be involved in the antioxidative and antiviral activity of acetylsalicylic acid in hepatitis C virus (HCV)-expressing cells. Here, RNA sequencing (RNA-seq) analysis identified SOD homologs (three NbCu/Zn-SOD, four NbFe-SOD, and two NbMn-SOD) that were differentially expressed in Nicotiana benthamiana during tobacco vein mottling virus (TVMV) infection. NbCu/Zn-SOD-1 was cloned from N. benthamiana and subsequently characterized. Encoding sequence and structural analyses of the NbCu/Zn-SOD-1 protein confirmed a conserved SOD enzyme domain, GFHLHEfGDtT, indicating that it is SOD-dependent and phylogenetically related to Cu/Zn-SOD in Nicotiana tabacum (XP 016486719.1). NbCu/Zn-SOD-1 was primarily localized in the cytoplasm. The transient expression of NbCu/Zn-SOD-1 led to a reduced accumulation of TVMV and PVY-Rosea1 (PVY-Ros1). In summary, our results suggest that NbCu/Zn-SOD-1 homologs participate in plant antiviral responses.
1 Introduction
Superoxide dismutase (SOD) was discovered in bovine erythrocytes and was officially named by McCord and Fridovich in 1969 (Mccord and Fridovich, 1969). SOD is a key enzyme in the intracellular antioxidant defense system. It catalyzes the dismutation reaction of superoxide anion (O₂−) to convert it into hydrogen peroxide (H₂O₂) and oxygen (O₂), which plays a vital role in plant stress resistance (Gupta et al., 2019). Therefore, the balance between intracellular SOD and H2O2 scavenging enzymes is crucial (Shah et al., 2001). The generated H₂O₂ can be further decomposed into water and oxygen under the influence of other antioxidant enzymes such as catalase (CAT), thereby reducing the intracellular reactive oxygen species (ROS) content, maintaining the intracellular redox balance, and protecting cells from oxidative damage (Yasmeen et al., 2013).
Depending on the metal prosthetic groups that bind to SODs, they are divided into three main types: copper/zinc superoxide dismutase (Cu/Zn-SOD), manganese superoxide dismutase (Mn-SOD), iron superoxide dismutase (Fe-SOD) (Zelko et al., 2002). In addition to these three SODs, Ni-SOD has been found in Streptomyces and Streptomyces coelicolor (Kim et al., 1996). Among them, Cu/Zn-SOD is the most abundant SOD in plants and typically plays a role in the cytoplasm, chloroplasts, and extracellular space of eukaryotes. Eukaryotic Cu/Zn-SOD is a 32 kDa protein that binds copper and zinc on each subunit. Non-covalent bonds form between subunits, and disulfide bonds and metal ions help stabilize the structure. The amino acid composition of Cu/Zn-SOD from different sources was not significantly different, showing clear evolutionary conservation and high sequence homology (Spagnolo et al., 2004). Among these, the sequence identity of SOD in human and bovine erythrocytes is as high as 80%. The highly conserved amino acid sequence of Cu/Zn-SOD suggests that it is crucial for maintaining the normal function of the enzyme.
Cu/Zn-SOD genes have been isolated from various plants, including longans, bananas, tomatoes, wheat, and Gossypium hirsutum (Lin and Lai, 2013; Feng et al., 2015; Feng et al., 2016; Tyagi et al., 2017; Wang et al., 2017). Several studies have demonstrated that Cu/Zn-SOD plays a crucial role in plant growth and development, fruit maturation, senescence, and responses to environmental stress, among other processes. In potato plants transfected with the Cu/Zn-SOD gene and exposed to salt stress, the activity of the SOD enzyme was significantly higher than in non-transgenic plants, indicating a strong antioxidant capacity and potential for salt tolerance.
In plant viruses, SODs are participated in the infection process. The early accumulation of ROS limits potato virus X (PVX) replication during symptomless extreme resistance (Hernández et al., 2016). A possible biochemical mechanism governing symptomless extreme virus resistance is the accumulation of ROS, which has a dual role in infected plants. Higher concentrations of ROS may promote programmed cell death of infected plant cells as well as the death or limitation of invading pathogens, such as viruses, because of their high toxicity (Halliwell and Gutteridge, 2015). By contrast, at low concentrations, ROS are crucial signaling compounds during the activation of host defense responses, including the induction of antioxidants, in healthy plant cells adjacent to infection sites (Levine et al., 1994; Dat et al., 2000; Thomas et al., 2002; Pogány et al., 2009). Accordingly, it has been known for decades that early (from 6 to 10 h after inoculation) nicotinamide adenine dinucleotide phosphate oxidase-dependent accumulation of superoxide and HO plays a definite role in hypersensitivity-associated resistance to plant viruses such as tobacco mosaic virus (TMV) (Doke and Ohashi, 1988; Rossetti and Bonatti, 2001).
This study revealed differential expression of nine SOD homologs in Nicotiana benthamiana during tobacco vein mottling virus (TVMV) infection, specifically identifying three NbCu/Zn-SODs, four NbFe-SODs, and two NbMn-SODs. NbCu/Zn-SOD-1 was cloned from N. benthamiana and further characterized. The effects of NbCu/Zn-SOD-1 on viral infection with TVMV and PVY-Ros1 were studied.
2 Materials and methods
2.1 Experimental materials
The vectors pDONR207, pEAQ-HT-DEST3, and Agrobacterium tumefaciens C58C1 were provided by Professor Juan Antonio García from the Centre National for Biotechnology (CNB), Spain. Escherichia coli DH5α competent cells were purchased from TaKaRa Bio (Osaka, Japan). The infectious clone of TVMV was described previously (Zhao et al., 2020).
2.2 Plant preparation
Nicotiana benthamiana seeds were sown in soil, and the plants were cultured in a greenhouse at approximately 22–26°C under a 16/8 h light/dark cycle. For infection or mechanical inoculation experiments, N. benthamiana plants with the same number of leaves and similar plant sizes (four-to-five leaves) were selected (Pasin et al., 2014).
2.3 Identification of SOD homologs from the transcriptome sequencing data
For transcriptome sequencing, N. benthamiana plants were infected with TVMV-infected clones, and healthy plants were used as controls (Zhao et al., 2020). Total RNA was extracted using the TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. Transcriptome sequencing and data analysis were performed. The transcriptome was generated from four complementary DNA (cDNA) libraries, comprising two biological replicates and two treatment sets. Manganese superoxide dismutases (NbSODs) were selected from the transcriptome sequencing data.
2.4 In silico sequence and structure analysis
A box plot was provided by LC Bio Technology Co., Ltd (Hangzhou, China). The heat map was drawn using TBtools software. Sequences were aligned using ESPript3.x and SnapGene software tools, and evolutionary history was inferred using Molecular Evolutionary Genetics Analysis Version 11 (MEGA 11) software (Kumar et al., 2016). The ExPaSy ProtParam tool was used to analyze the physicochemical properties of NbCu/Zn-SOD-1. The secondary structure of NbCu/Zn-SOD-1 was analyzed using NovoPro. The localization prediction of NbCu/Zn-SOD-1 using the PlantmPLoc tool and the ScanProsite tool was used to predict the conserved domain of NbCu/Zn-SOD-1, and Swiss-Model was used to predict the 3D structural models of remote sensing images (Waterhouse et al., 2018). NP 001234769.2 (Solanum lycopersicum Cu/Zn-SOD), XP 015058855.1 (Solanum pennellii Cu/Zn-SOD), XP 015164932.1 (Solanum tuberosum Cu/Zn-SOD), XP 016547975.1 (Capsicum annuum Cu/Zn-SOD), XP 060169665.1 (Lycium barbarum Cu/Zn-SOD), CAA32534.1 (Petunia Cu/Zn-SOD), NbCu/Zn-SOD-1 (N. benthamiana Cu/Zn-SOD), XP 016486719.1 (Nicotiana tabacum Cu/Zn-SOD), BAF 80585.1 (Populus alba Cu/Zn-SOD), and NP 001413319.1(Spinacia oleracea Cu/Zn-SOD) were used to obtain a structural similarity matrix and gene structure map between NbCu/Zn-SOD-1 and different species using TBtools software (Chen et al., 2020). A structural dendrogram of NbCu/Zn-SOD-1 among different species was obtained using MEGA 11 software.
2.5 Reverse transcription quantitative polymerase chain reaction (PCR) (RT-qPCR)
Transcript and gene expression levels were measured using fragments per kilobase of transcript per million mapped reads (FPKM). During differential gene expression analysis, the fold change denotes the expression ratio between two groups.
Total RNA was purified from virus-infected leaves and used in cDNA synthesis reactions. For reverse transcription, total RNA (approximately 1 μg) was converted to cDNA using SYBR Green Premix Pro Taq HS qPCR Kit (Rox Plus) (AG11718, Accurate Biology) according to the manufacturer’s instructions. Gene-specific primers (Supplementary Table S1) were designed using QuantStudio™ 3 and 5 Real-Time PCR Systems MAN0010407 (Thermo Fisher Scientific). The expression was normalized using Nbubiquitin as a reference, and the 2−ΔΔCT method was used to calculate the level of gene expression (Yue et al., 2023).
2.6 Construction of expression vector of NbCu/Zn-SOD-1 and its subcellular localization
Gene-specific primers for cloning were designed according to the NbCu/Zn-SOD-1 sequence (Supplementary Table S1). The NbCu/Zn-SOD-1 fragment was ligated into the PMD19-T vector and transformed into E. coli DH5α competent cells by heat shock, and then plated on Lysogeny Broth (LB) solid medium containing kanamycin and grown at 37°C overnight. Plasmids were extracted using the Tiagen Plasmid Extraction Kit, and restriction enzyme digestion was performed with Sap I and Xba I. The expression vector pEAQ-NbCu/Zn-SOD-1 was constructed using Gateway technology, as previously described (Sun et al., 2024). pDONR-NbCu/Zn-SOD-1 was digested with ApaL I. pEAQ-NbCu/Zn-SOD-1 was digested with ApaL I/SpeI (Supplementary Figure S1).
The NbCu/Zn-SOD-1 (663 bp) fragment was amplified from the T-NbCu/Zn-SOD-1 plasmid using the 1300GFP-NbCu/Zn-SOD-F/R primer. The empty vector, 1,300-GFP-EV, was digested using BamH I/Sal I to obtain a 10,497 bp fragment. NbCu/Zn-SOD-1 was ligated into the 1,300-GFP-EV vector to get the expression vector named 1,300-NbCu/Zn-SOD-1-GFP. The recombinant plasmids 1,300-NbCu/Zn-SOD-1-GFP. The homologous recombination method was used for connection and transformation into E. coli DH5a. Identification was performed using BamH I/Spe I digestion.
2.7 Inoculation of the virus
Agrobacterium C58C1 cells carrying pEAQ-NbCu/Zn-SOD-1, pEAQ-HT-DEST3, or pLX-TVMV were grown to OD600 = 1.0. Subsequently, a mixture of pEAQ-NbCu/Zn-SOD-1/TVMV was prepared at a V: V ratio of 1:1. As a control, co-infiltration of pEAQ-HT-DEST3 with each virus (V: V = 1:1) was used. Nine days post-inoculation (dpi), symptoms were observed and documented through photography. Three days after the infiltration of Agrobacterium carrying pEAQ-NbCu/Zn-SOD-1 into N. benthamiana, the PVY-Ros1 inoculum preparation in phosphate buffer, and mechanical inoculation were performed as previously described (Pasin et al., 2020).
2.8 Western blotting for virus detection
Approximately 0.2 g of infected leaf tissue was frozen in liquid nitrogen, ground into a powder, and added to a solution of 5% SDS, 2 times the volume. The mixture was then mixed thoroughly until homogenization. The mixture was then placed in a water bath at 95°C for 5 min, followed by centrifugation at 12,000 rpm and 4°C for 10 min. The addition of 2 times loading buffer (62.5 mM Tris–HCl, pH 6.8, 25% glycerol, 2% SDS, and 0.01% Bromophenol blue, and 250 mM dithiothreitol) solutions to the serum was performed in a 1:1 ratio. Subsequently, the mixture was reheated in a water bath at 95°C for 5 min and then placed on ice for 2 min. Afterward, it was centrifuged at 12,000 rpm and 4°C for 10 min. The resulting clear upper layer was collected for Western blotting as previously described (Yang et al., 2024).
Immunodetection was conducted using primary antibodies such as anti-TVMV coat protein (CP) for detecting TVMV and anti-PVY CP for detecting PVY-Ros1. Horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin (IgG; ab205718; Abcam) and mouse monoclonal antibodies were used as secondary antibodies. Protein signals were visualized by enhanced chemiluminescence.
2.9 Observation of subcellular localization
The infiltrated leaf tissue was cut, and temporary glass slides were made to observe fluorescent protein expression using a laser confocal microscope. The fluorescence of green fluorescent protein (GFP) was visualized with a Leica TCS SP52 confocal laser scanning microscope as described previously (Cheng et al., 2015; Wei and Wang, 2008; Cheng et al., 2015). GFP luminescence was observed at an excitation wavelength of 488 nm.
2.10 3,3′-Diaminobenzidine staining
Overexpression of NbCu/Zn-SOD-1 in N. benthamiana plants inoculated with TVMV or PVY-Ros1 was observed. After 9 days, leaves were soaked in 3,3′-diaminobenzidine (DAB) solution (1 mg/mL, pH 3.8), placed under light at 25°C for 12 h, and then transferred to boiling 95% ethanol to remove chlorophyll. After cooling, the leaves were placed in 95% ethanol for 3–5 h.
2.11 Determination of SOD enzyme activity
At 1, 3, 6, 9, and 12 days after virus infection, infected leaves were collected to measure the activity of SOD disease-resistance and defense enzymes. The kits (bixbio) were used to measure SOD activity.
3 Results
3.1 Expression of SOD homologs during TVMV viral infection
The TVMV infectious clone was infiltrated into the leaves of N. benthamiana plants with four to five leaves. Symptoms of infection were observed on day 9. Compared with the healthy control, the leaves of plants inoculated with TVMV showed apparent shrinkage and vein mottling (Figure 1A). Viral accumulation was detected by Western blot using a TVMV CP-specific antibody. A high accumulation of TVMV CP was detected in the tested samples, compared with the negative control of healthy plants (Figure 1B). This confirmed the successful infection of TVMV in N. benthamiana.
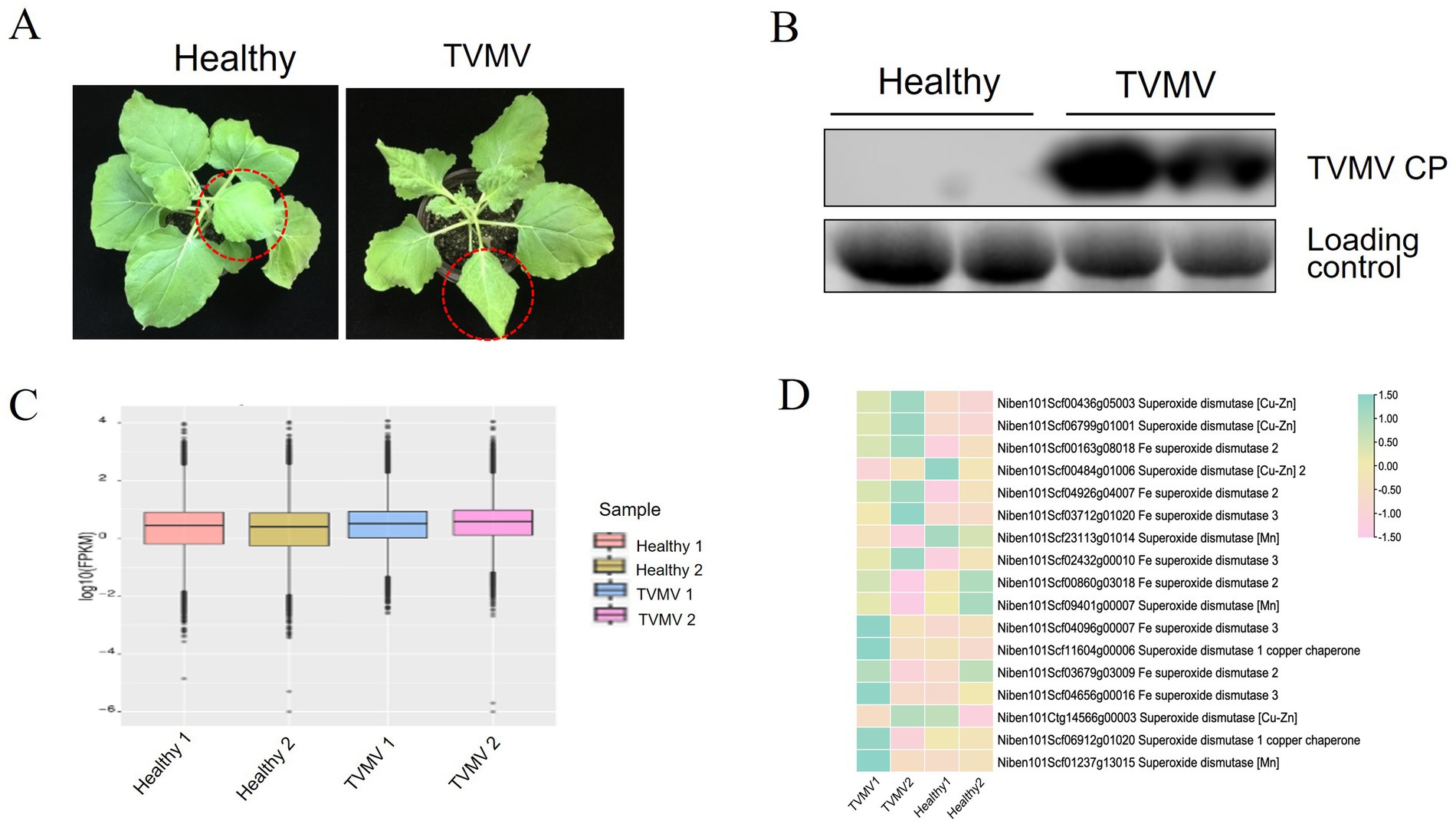
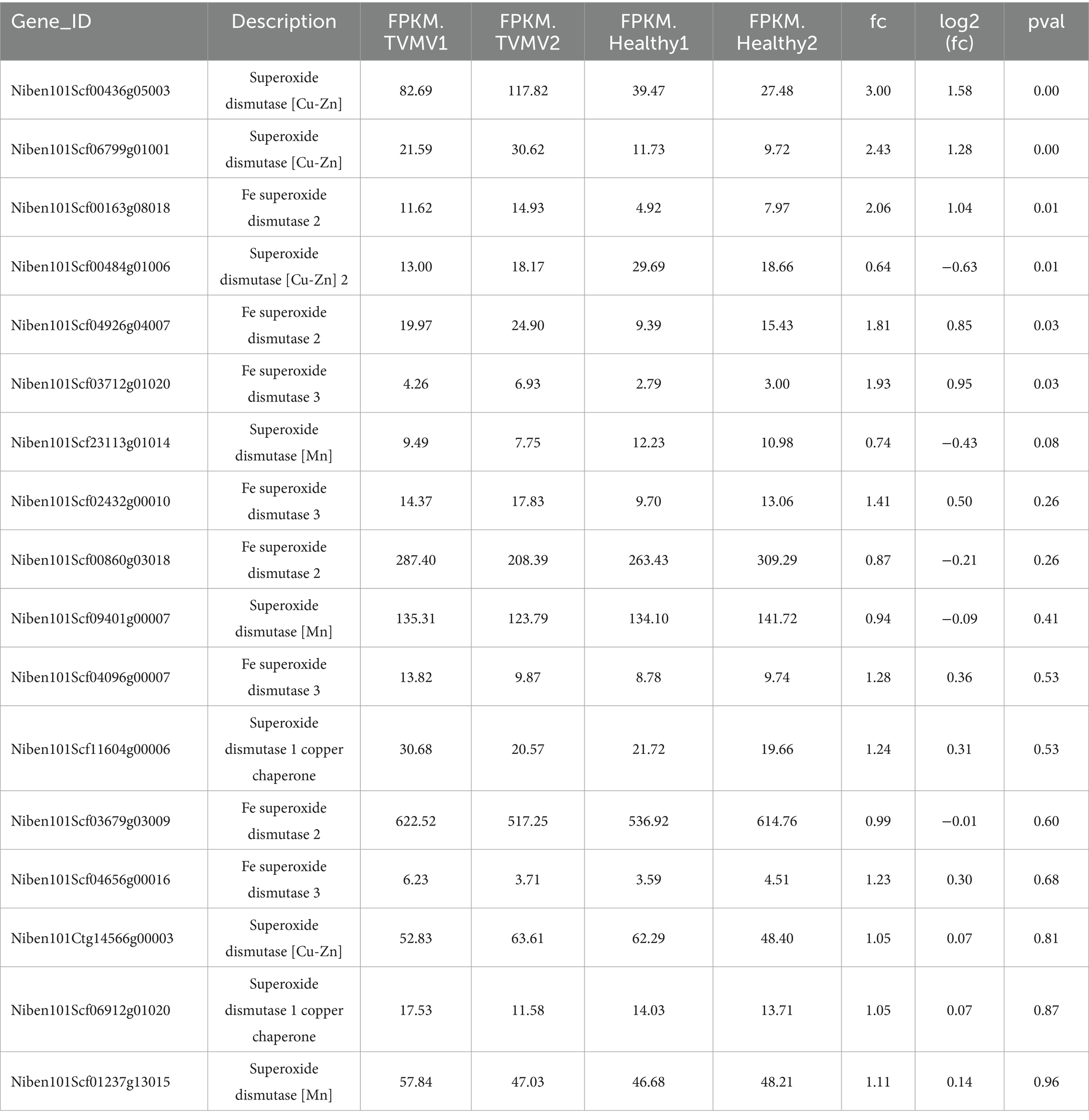
Figure 1. Expression of SOD homologs during TVMV viral infection. (A) Phenotypes of TVMV inoculated and control Nicotiana benthamiana plants. (B) Viral accumulation of TVMV was detected by Western blot with anti-TVMV CP. (C) Gene density of cDNA libraries. (D) Transcript expression of NbSOD homologs in Nicotiana benthamiana analyzed by RNA sequencing (RNA-seq).
To determine whether SOD homologs were regulated by TVMV infection, we performed a transcriptome sequencing assay using samples from N. benthamiana plants infected with TVMV at 9 dpi. The boxplot results showed that the TVMV treatment and healthy control groups had high repeatability (Figure 1C), suggesting the reliability of the data and enabling them to be used for further analysis. We found that 17 transcripts encoding SOD homologs were differentially expressed in TVMV-infected samples compared with healthy control plants (Table 1; Figure 1D).
3.2 Identification of NbCu/Zn-SOD-1 in Nicotiana benthamiana
Nine SOD homologs, including three NbCu/Zn-SOD, four NbFe-SOD2 and two NbMn-SOD homologs, were analyzed using RT-qPCR. The expression of three SODs (NbCu/Zn-SOD-1, NbCu/Zn-SOD-2, and NbCu/Zn-SOD-3) was upregulated and that of six SODs (NbFe-SOD2-1, NbFe-SOD2-2, NbFe-SOD2-3, NbFe-SOD2-4, NbMn-SOD-1, and NbMn-SOD-2) was downregulated after TVMV infection. The expression of NbCu/Zn-SOD-1 was significantly upregulated, whereas that of NbFe-SOD2-3 was significantly downregulated (Figure 2A). The amino acid composition of the SOD homologs ranged from 101 to 305 amino acids. The relative molecular weights of SOD homologs ranged from 10.3 34.9 kD. Among them, NbFe-SOD2-3, NbMn-SOD-1, and NbMn-SOD-2 were basic proteins with isoelectric points greater than 7, whereas the other six proteins were acidic proteins with isoelectric points below 7. Hydrophilicity analysis revealed that the Grand Average of Hydropathicity (GRAVY) values of the SOD proteins were negative, indicating they were classified as hydrophilic proteins. Prediction of subcellular localization showed that NbMn-SOD-1 and NbMn-SOD-2 were localized in the mitochondria, whereas other SOD homologs were localized in the chloroplasts (Table 2).
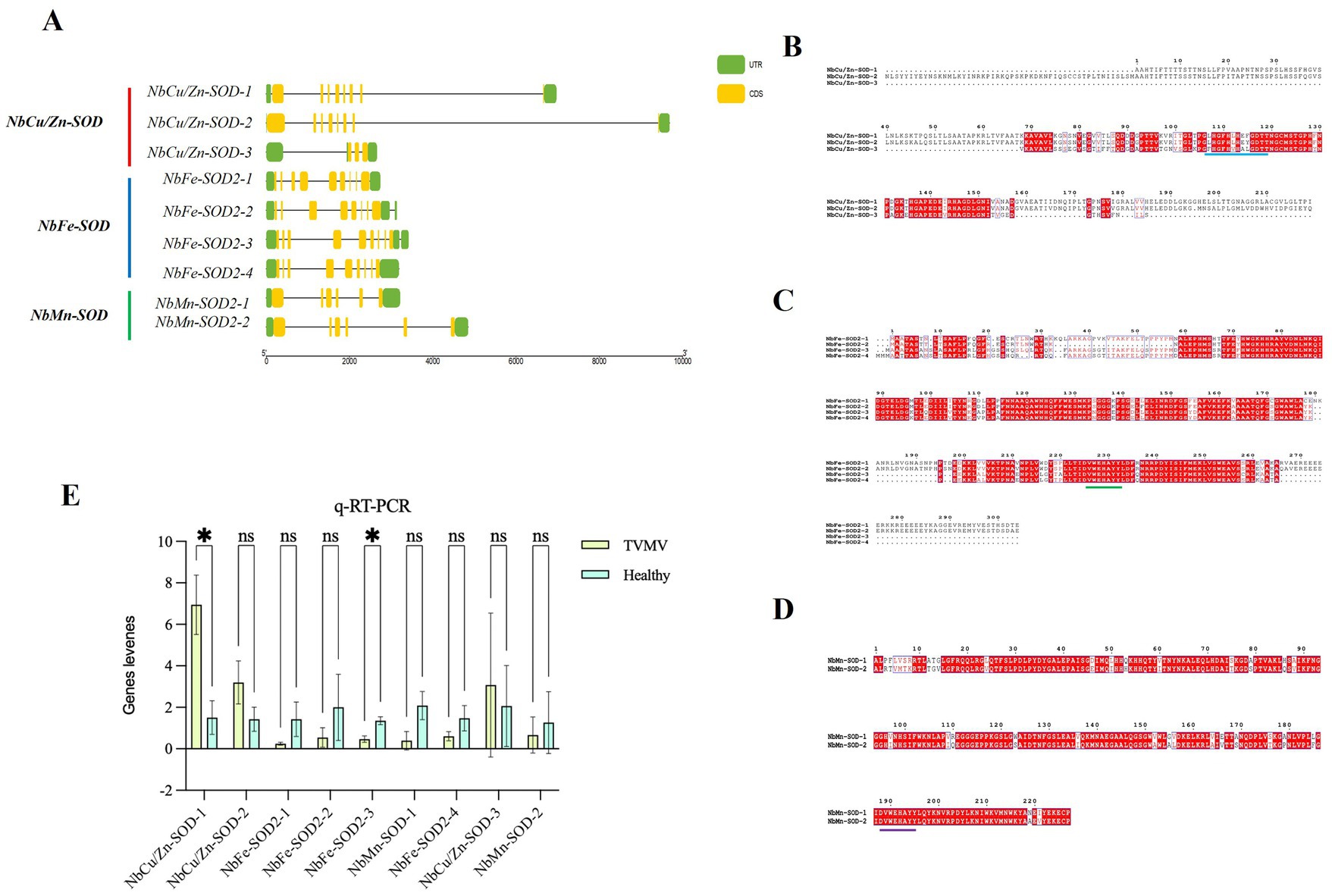
Figure 2. Identification of NbCu/Zn-SOD in Nicotiana benthamiana infected with TVMV. (A) The CDS-UTR structures and the multiple sequence alignment of SOD homologs in N. benthamiana. Yellow rectangles represent CDS, black lines represent exons, and green rectangles represent untranslated regions. (B) The multiple sequence alignment of Cu/Zn-SOD homologs of N. benthamiana; Dashes between amino acid alphabets indicate gaps that were introduced to optimize alignment; The background colors of alphabets represent the higher (white) to lower (red) variability of certain sites in sequences; The blue lines represent the shared conserved sequences. (C) The multiple sequence alignment of Fe-SOD homologs; The green lines represent the shared conserved sequences. (D) The multiple sequence alignment of Mn-SOD homologs; The purple line represents the shared conserved sequence. (E) Relative expression of SOD homologs after TVMV infection of N. benthamiana by RT-qPCR. *p < 0.05 and “ns” represents p > 0.5 by Student’s t-test.
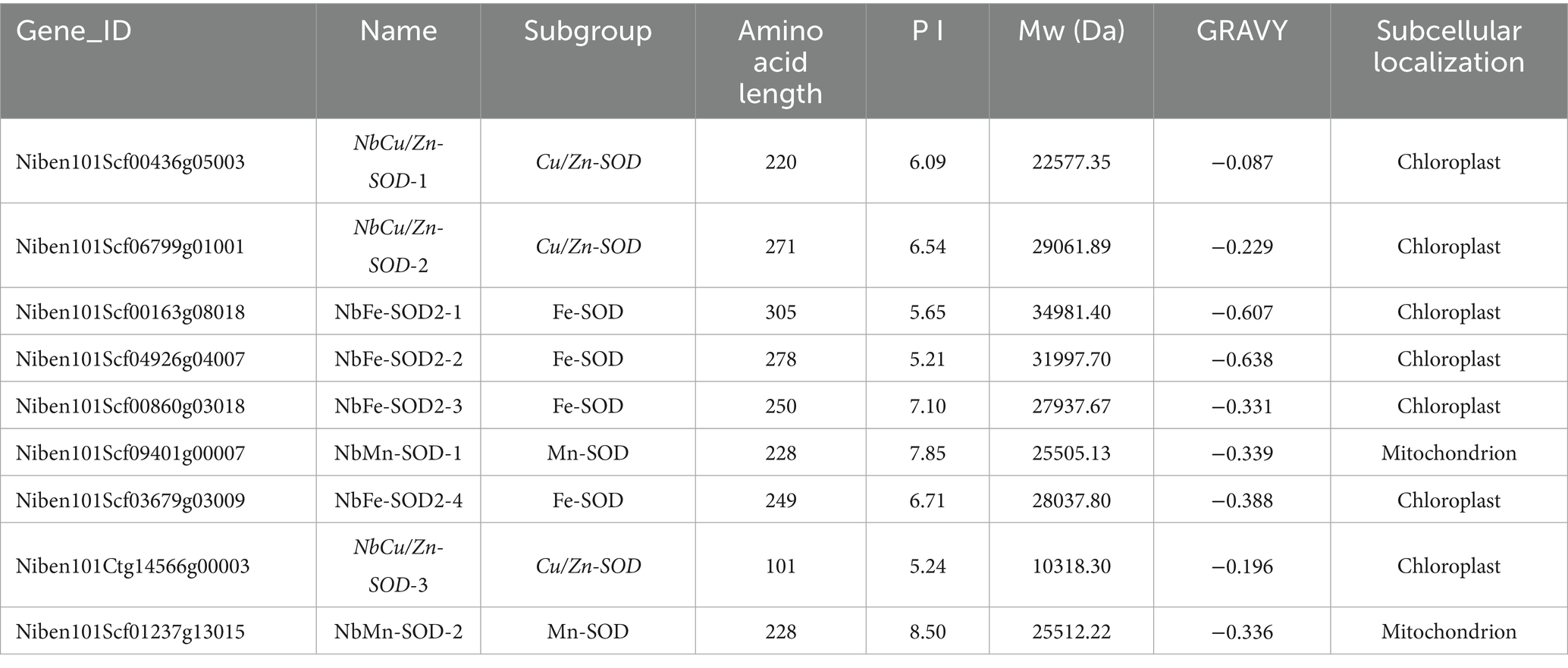
Table 2. The characteristics of SOD homologues in Nicotiana benthamiana and predicted subcellular localization.
The coding sequence (CDS)-untranslated region structure and multiple sequence alignments of SOD homologs in N. benthamiana were profiled. The length of the SOD locus in N. benthamiana was 2,658 to 9,691 bp. Three NbCu/Zn-SOD homologs contained three to nine CDS regions, and four NbFe-SOD2 homologs contained eight to nine CDS regions. NbMn-SOD homologs contained six CDS (Figure 2B). The amino acid sequences of SOD homologs of N. benthamiana were aligned using multiple sequence alignments. There was a conserved sequence “GFHLHEfGDtT” and “GFHVHAlGDtT” in the Cu/Zn-SOD amino acid sequence of N. benthamiana (Figure 2C). A conserved sequence “DvWEHAYY” was distributed on the amino acid sequence of Fe-SOD (Figure 2D). A conserved sequence “DvWEHAYY” was distributed in the amino acid sequence of Mn-SOD, which was the same as the conserved sequence distributed in the amino acid sequence of Fe-SOD (Figure 2E).
3.3 Characteristic of NbCu/Zn-SOD-1 in Nicotiana benthamiana
The secondary structure of NbCu/Zn-SOD-1 was analyzed using NovoPro. Sixteen α-helix and three β-folded structures were identified from the structure of NbCu/Zn-SOD-1(Figure 3A). The three-dimensional structure of NbCu/Zn-SOD-1 predicted by Swiss-Model showed a conserved core with high prediction confidence scores, which was flanked by less conserved termini with a variable number of disordered residues (Figure 3B). To confirm the structural correlation between the cloned NbCu/Zn-SOD-1 homologs and known Cu/Zn-SOD genes from other plant species, a reference structure obtained by X-ray crystallography or high-resolution modeling was used for structural comparison and phylogenetic tree alignment analysis. The structural similarity matrix and evolutionary tree showed that NbCu/Zn-SOD-1 was highly similar to the Cu/Zn-SOD of Nicotiana tabacum (Figures 3C,D).
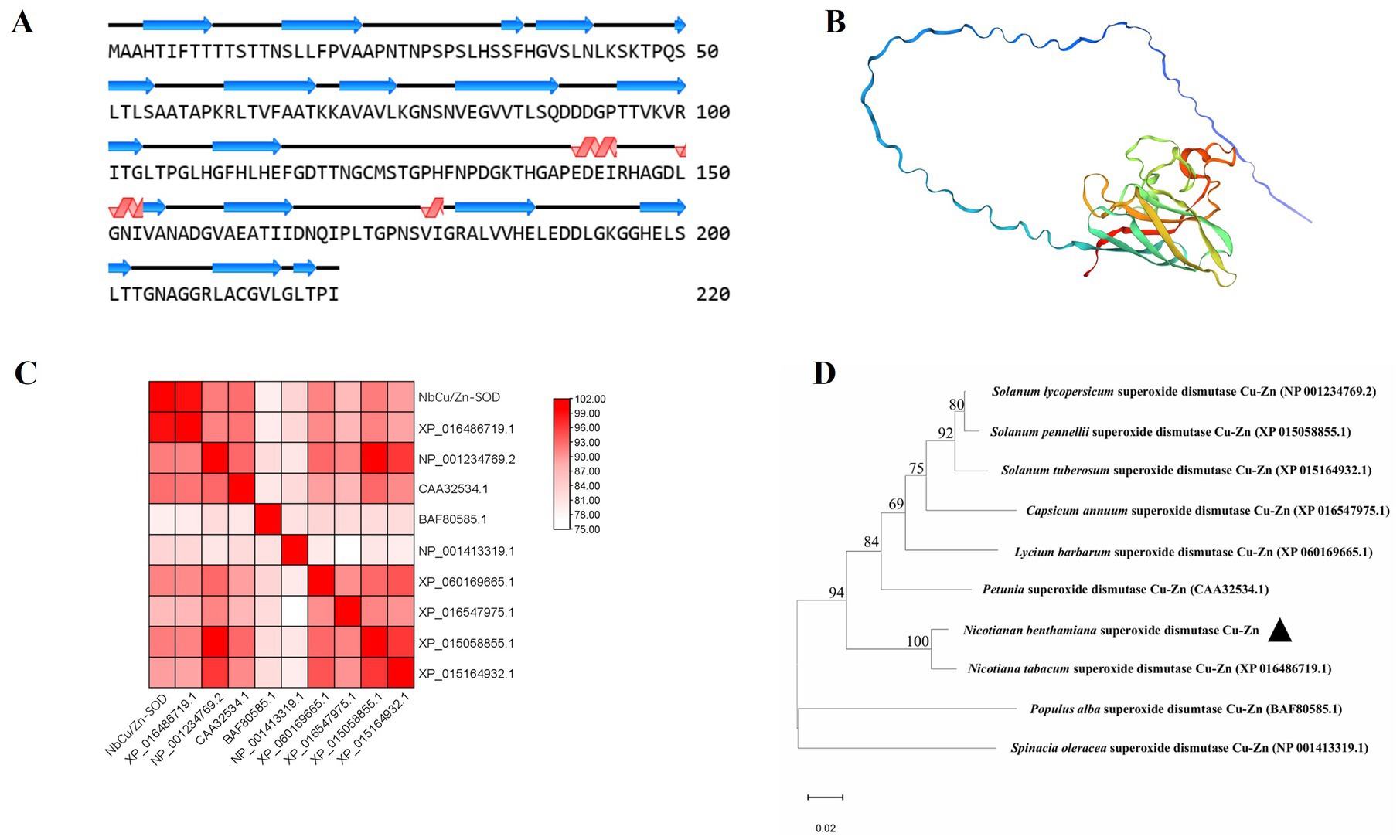
Figure 3. Structural modeling and analysis of NbCu/Zn-SOD-1. (A) Secondary structures derived from the predicted models of NbCu/Zn-SOD-1. (B) Tertiary structural models of NbCu/Zn-SOD-1; colors by prediction confidence scores. (C) The structural similarity matrix of NbCu/Zn-SOD-1; colors by Dali Z-scores. (D) The amino acid structural dendrogram of NbCu/Zn-SOD-1; branch support values are shown. Structural models of NbCu/Zn-SOD-1 were computed by MEGA 11.
3.4 Subcellular localization of NbCu/Zn-SOD-1 in Nicotiana benthamiana
To determine the subcellular localization of NbCu/Zn-SOD-1 in plant cells, we cloned the NbCu/Zn-SOD-1 sequence into the recombinant vector 1,300-GFP-EV. The NbCu/Zn-SOD-1 (663 bp) fragment was amplified from healthy N. benthamiana (Figure 4A). The vector 1,300-GFP-EV was digested using BamH and I/Sal I to obtain a 10,497 bp fragment (Figure 4B). NbCu/Zn-SOD-1 was ligated into the 1,300-GFP-EV vector to get the expression vector named 1,300-NbCu/Zn-SOD-1-GFP. The recombinant plasmid 1,300-NbCu/Zn-SOD-1-GFP was transformed into E. coli DH5a and verified by PCR (a 663 bp fragment) (Figure 4C) and restriction enzyme digestion with BamH I/Spe I (10,485 bp and 672 bp) (Figure 4D). The plasmid 1,300-NbCu /Zn-SOD-1-GFP was transformed into Agrobacterium C58C1 cells and infiltrated into N. benthamiana. Fluorescence was observed 3 dpi. GFP fluorescence was detected in the cytoplasm, indicating that NbCu/Zn-SOD-1 was localized in the cytoplasm (Figure 4E).
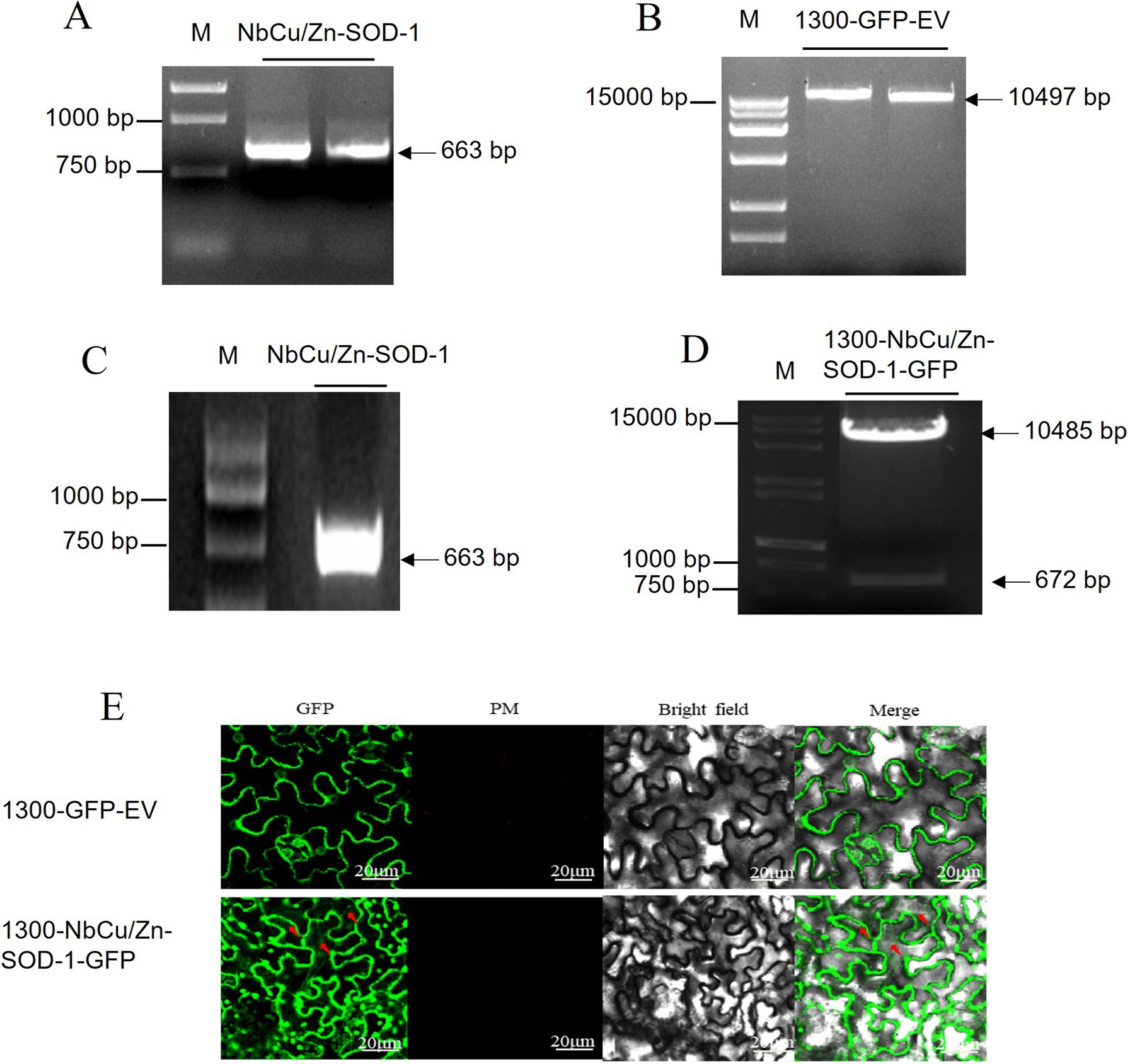
Figure 4. Construction of NbCu/Zn-SOD-1 subcellular vector and observation with subcellular localization. (A) NbCu/Zn-SOD-1 fragments obtained by PCR with primer 1,300-NbCu/Zn-SOD-F/R in agarose gel. (B) Digestion of 1,300-GFP-EV by BamH I/Sal I. (C) 1,300-NbCu/Zn-SOD-1-GFP fragments were amplified by PCR in Escherichia coli DH5a. (D) Digestion of 1,300-NbCu/Zn-SOD-1-GFP by BamH I/Spe I. Fragment sizes of the marker are shown on the right. (E) GFP luminescence was observed at an excitation wavelength of 488 nm. The 1,300-GFP-EV empty vector was used as a control.
3.5 Effect of NbCu/Zn-SOD-1 on plant viral infection
To determine the effect of NbCu/Zn-SOD-1 on plant viral infection, the NbCu/Zn-SOD-1 (663 bp) fragment from N. benthamiana was amplified and verified by sequencing (Supplementary Figures S1 and S2). The fragment of NbCu/Zn-SOD-1 was cloned into pEAQ-HT-D3 using a gateway recombination system to obtain pEAQ-NbCu/Zn-SOD-1. The plasmid pEAQ-NbCu/Zn-SOD-1 was transformed into Agrobacterium C58C1 and infiltrated into N. benthamiana leaves. The expression levels of NbCu/Zn-SOD-1 were detected using RT-qPCR. We found that the expression of NbCu/Zn-SOD-1 was significantly increased compared with that in healthy controls (Figure 5A). Thus, the transient expression of NbCu/Zn-SOD-1 was functional.
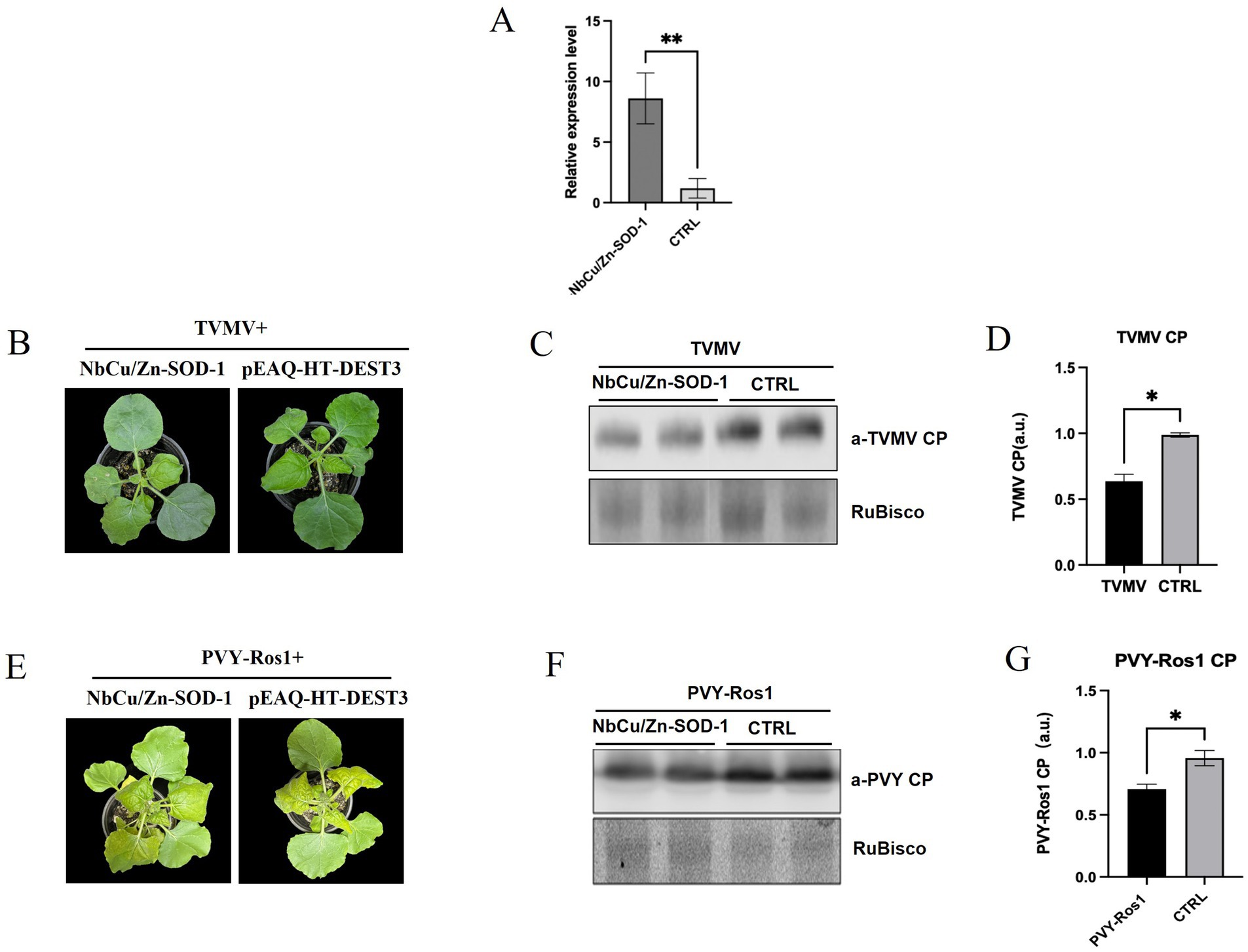
Figure 5. Effect of NbCu/Zn-SOD-1 on plant viral infection. (A) Plots show RT-qPCR transcript quantification values [mean ± standard deviation (SD)] in samples collected from leaves infiltrated with NbCu/Zn-SOD-1 overexpressing constructs. **p < 0.01 and *p < 0.05 by Student’s t-test. (B) Total plants and systemic leaves of overexpression NbCu/Zn-SOD-1 and empty vector (pEAQ-HT-DEST3) plants infected with TVMV as observed at 9 days post-inoculation (dpi). (C) Viral protein accumulation in upper uninoculated systemic leaves was measured by Western blot with an anti-TVMV coat protein (CP) serum; RuBisCO large subunit is shown as a loading control. (D,G) Plots show signal quantification values (mean ± SD). CTRL, control condition; p values by Student’s t-test are shown. (E) Total plants and systemic leaves of overexpression NbCu/Zn-SOD-1 and empty vector (pEAQ-HT-DEST3) plants infected with PVY-Ros1 as observed at 9 days post-inoculation (dpi). (F) Western blot with an anti-PVY coat protein (CP) serum.
pEAQ-NbCu/Zn-SOD-1 was then co-infected with the TVMV infectious clone. The upper leaves of TVMV-infected N. benthamiana displayed slight symptoms of shrinkage, while the control plants exhibited significantly stronger symptoms (Figure 5B). Virus accumulation in the upper leaves was analyzed by Western blotting 9 dpi. Western blot analysis showed that TVMV accumulation was significantly reduced in NbCu/Zn-SOD-1 overexpressing plants compared with the controls (Figures 5C,D). Three days after pEAQ-NbCu/Zn-SOD-1 overexpression, PVY-Ros1 was inoculated by mechanical inoculation, and symptoms of the plants were observed after 9 days. The number of plants overexpressing NbCu/Zn-SOD-1 was significantly reduced compared with the control (Figure 5E). Western blotting results were similar to those of TVMV, and the accumulation of PVY-Ros1 was reduced in plants overexpressing NbCu/Zn-SOD-1 compared with the control (Figures 5F,G). This suggests that NbCu/Zn-SOD-1 has a pronounced inhibitory effect on viral infection.
3.6 NbCu/Zn-SOD-1 enhances H2O2 quenching by increasing superoxide dismutase activity
After transient expression of NbCu/Zn-SOD-1 gene, SOD resistance defense enzyme activities in the leaves were detected after TVMV or PVY-Ros1 inoculation. The results showed that after co-injection of NbCu/Zn-SOD-1 gene and TVMV, SOD activity in plants increased from 1 to 9 days, reaching the highest value of 128.9 U/g on day 9, after which enzyme activity began to decline (Figure 6A). ROS accumulation in TVMV-infected plants was monitored using DAB staining, which showed lower levels of H2O2 in TVMV-infected plants than in control plants (Figure 6B). Three days after overexpressing NbCu/Zn-SOD-1 gene, N. benthamiana plants were mechanically inoculated with PVY-Ros1, and the upper leaves were used to detect SOD enzyme activity. SOD activity in plants reached the highest value of 318.2 U/mg on day 9, after which enzyme activity decreased slightly (Figure 6C). ROS accumulation in PVY-Ros1-infected plants was monitored using DAB staining, and the results showed no significant difference in H2O2 levels between PVY-Ros1-infected plants and control plants (Figure 6D). Transient expression of NbCu/Zn-SOD-1 gene increased SOD activity in N. benthamiana, thereby improving the defense of N. benthamiana against virus infection.
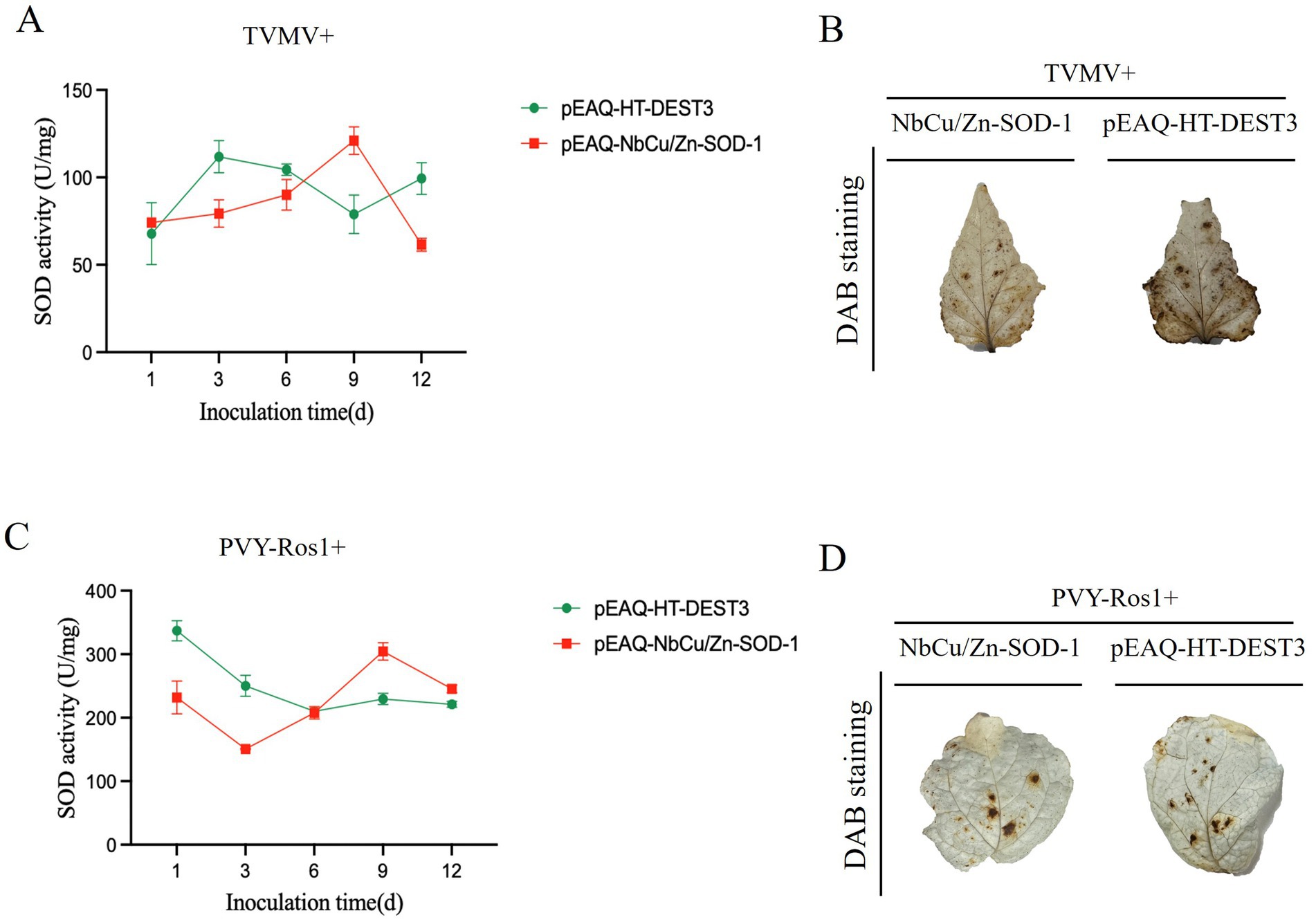
Figure 6. Effects of transient expression of NbCu/Zn-SOD-1 gene on SOD enzyme activity and ROS after TVMV and PVY-Ros1 infection. (A–C) Changes of SOD activity in Nicotiana benthamiana leaves with transient NbCu/Zn-SOD-1 gene after TVM or PVY-Ros1 inoculation. (B–D) After overexpression of NbCu/Zn-SOD-1, TVMV or PVY-Ros1 was inoculated, and the leaves of the system were taken to detect ROS in N. benthamiana leaves in situ. Leaves of a tobacco system inoculated with TVMV with an empty vector were used as controls.
4 Discussion
ROS are produced during the plant life processes. Superoxide anions form H2O2 under the catalysis of SOD, and H2O2 forms O2 through the catalysis of CAT (Fu et al., 2025). Under normal conditions, ROS in the plant are in a dynamic and stable state of production and clearance (Chen et al., 2022). When the plant is under stress, this balance is disrupted, and numerous ROS are produced, resulting in damage to the plant. Transgenic tobacco plants showed higher water content, less electron damage, lower malondialdehyde content, higher antioxidant enzyme activity, and higher survival rates than non-transgenic plants after prolonged drought and salt stress (Negi et al., 2015). When plants are infected with viruses, the redox reaction balance in the cells is disrupted, and many reactive oxygen species, such as O₂− and H₂O₂, are produced. These ROS attack lipids, proteins, and nucleic acids, leading to cell membrane damage, enzyme inactivation, and DNA mutations, which in turn promote virus propagation (Paolo et al., 2016).
ROS-scavenging systems play a crucial role in cellular functions because ROS are highly cytotoxic. The synchronous action of SOD, CAT, ascorbate peroxidase, monodehydroascorbate reductase, dehydroascorbate reductase, and glutathione reductase is part of the antioxidant system that protects plants from ROS damage (Kozi, 1999; Scandalios, 2005; Ashraf, 2009; Gill and Tuteja, 2010). Plants with high expression of these antioxidant enzymes are more tolerant to salt and oxidative stress. It has been shown that salt stress alters the levels of antioxidant enzymes to a large extent, providing tolerance to plants (Kim et al., 2005; Noreen and Ashraf, 2009; Wang et al., 2009; Tarchoune et al., 2010).
Cu/Zn-SOD plays a protective role in various types of tissue, protecting them from oxidative damage (Lewandowski et al., 2019). It is a critical enzyme in limiting reactive oxygen species in both the cytosol and the mitochondrial intermembrane space (Fetherolf et al., 2017). It plays a vital role in the balance of oxidation and antioxidants in the body, and is closely related to the occurrence and development of many diseases. Low concentrations of ROS play an essential role in plant disease resistance signaling. Cu/Zn-SOD may affect downstream signaling pathways by regulating ROS levels, such as activating mitogen-activated protein kinase cascade, or inducing the expression of genes related to disease resistance (Sumino et al., 2020). Cu/Zn-SOD may inhibit viral replication, assembly, or release by affecting the redox reaction state in cells. For example, certain viruses require a specific oxidative environment for replication, and Cu/ Zn-SOD mediated antioxidant effects may disrupt this environment, thereby limiting virus proliferation (Teoh et al., 2003).
In this study, we identified genes that were differentially expressed during TVMV infection, from which SOD homologs (three NbCu/Zn-SOD, four NbFe-SOD, and two NbMn-SOD) were selected. The qPCR results showed that the expression of NbCu/Zn-SOD-1 was significantly upregulated in tobacco plants infected with TVMV, with notable differences. This suggests that Cu/Zn-SOD-1 may prevent the harmful effects of ROS accumulation in plants and that SOD may play a key role in the antiviral process in plants.
In this study, the NbCu/Zn-SOD-1 gene was cloned, and its sequence was analyzed. The presence of the SOD enzyme activity domain was confirmed and was supported by secondary and tertiary structure prediction and modeling. The conserved domains are located in the central portion of these proteins. Phylogenetic analysis revealed that NbCu/Zn-SOD-1 was clustered with Cu/Zn-SOD (XP 016486719.1). This suggests that they may have engaged in similar activities. NbCu/Zn-SOD-1 subcellular localization results showed that NbCu/Zn-SOD-1 was localized in the cytoplasm.
We further investigated their roles in plant viral infections. The results showed that transient expression of NbCu/Zn-SOD-1 resulted in a decrease in the accumulation of TVMV and PVY-Ros1. This suggests that NbCu/Zn-SOD-1 homologs are involved in plant antiviral responses. Therefore, exploring the potential applications of NbCu/Zn-SOD-1 through bioengineering is an efficient way to improve anti-viral resistance in plants.
Data availability statement
The data supporting the findings of this study are available in the article and its Supplementary material or from the corresponding authors. The cloned gene sequences are available at NCBI (NbCu/Zn-SOD-1).
Author contributions
HW: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing, Formal analysis, Software, Validation, Visualization. JZ: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. ZM: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. ZS: Data curation, Formal analysis, Methodology, Writing – original draft, Software. DL: Data curation, Formal analysis, Methodology, Software, Writing – original draft, Resources. BL: Data curation, Methodology, Resources, Writing – original draft, Conceptualization. FY: Data curation, Methodology, Resources, Writing – original draft, Formal analysis, Software. CJ: Resources, Writing – original draft, Conceptualization, Investigation, Validation. HZ: Conceptualization, Investigation, Resources, Writing – original draft, Funding acquisition, Project administration, Supervision. MZ: Conceptualization, Investigation, Project administration, Resources, Supervision, Writing – original draft, Data curation, Methodology, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was “supported by grants for the major project of the National Science Foundation of China (grant·number: 31860489 to MZ) and the China Agriculture Research System of the Ministry of Finance (MOF) and the Ministry of Agriculture and Rural Affairs (MARA; grant number: CARS-07-C-3) to HZ.
Acknowledgments
The authors thank Zhiying Wang for technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1561731/full#supplementary-material
References
Ashraf, M. (2009). Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol. Adv. 27, 84–93. doi: 10.1016/j.biotechadv.2008.09.003
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H. Z., Lee, J., Lee, J. M., Han, M., Emonet, A., Lee, J., et al. (2022). MSD2, an apoplastic Mn-SOD, contributes to root skotomorphogenic growth by modulating ROS distribution in Arabidopsis. Plant Sci. 317:111192. doi: 10.1016/j.plantsci.2022.111192
Cheng, X., Deng, P., Cui, H., and Wang, A. (2015). Visualizing double-stranded RNA distribution and dynamics in living cells by dsRNA binding-dependent fluorescence complementation. Virology 485, 439–451. doi: 10.1016/j.virol.2015.08.023
Dat, J., Vandenabeele, S., Vranova, E., Montagu, M. V., Inze, D., and Breusegem, F. V. (2000). Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. 57, 779–795. doi: 10.1007/s000180050041
Doke, N., and Ohashi, Y. (1988). Involvement of an O2− generating system in the induction of necrotic lesions on tobacco leaves infected with tobacco mosaic virus. Physiol. Mol. Plant Pathol. 32, 163–175. doi: 10.1016/S0885-5765(88)80013-4
Feng, X., Lai, Z. X., Lin, Y. L., Lai, G. T., and Lian, C. L. (2015). Genome-wide identification and characterization of the superoxide dismutase gene family in Musa acuminata cv. Tianbaojiao (AAA group). BMC Genomics 16:823. doi: 10.1186/s12864-015-2046-7
Feng, K., Yu, J. H., Cheng, Y., Ruan, M. Y., Wang, R. Q., Ye, Q. J., et al. (2016). The SOD gene family in tomato: identification, phylogenetic relationships, and expression patterns. Front. Plant Sci. 7:1279. doi: 10.3389/fpls.2016.01279
Fetherolf, M. M., Boyd, S. D., Winkler, D. D., and Winge, D. R. (2017). Oxygen-dependent activation of cu,Zn-superoxide dismutase-1. Metallomics 9, 1047–1059. doi: 10.1039/C6MT00298F
Fu, X. Y., Chen, G., Ruan, X. Y., Kang, G. Z., Hou, D. Y., and Xu, H. W. (2025). Overexpression of OsPIN5b alters plant architecture and impairs cold tolerance in rice (Oryza sativa L.). Plan. Theory 14:1026. doi: 10.3390/plants14071026
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Gupta, A. S., Heinen, J. L., Holaday, A. S., Burke, J. J., and Allen, R. D. (2019). Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic cu/Zn superoxide dismutase. Proc. Natl. Acad. Sci. USA 90, 1629–1633. doi: 10.1073/pnas.90.4.1629
Halliwell, B., and Gutteridge, J. M. C. (2015). Free radicals in biology and medicine. 5th Edn. Oxford: Oxford University Press. doi: 10.1093/acprof:oso/9780198717478.001.0001
Hernández, J. A., Gullner, G., Clemente-Moreno, M. J., Künstler, A., Juhász, C., Díaz-Vivancos, P., et al. (2016). Oxidative stress and antioxidative responses in plant–virus interactions. Physiol. Mol. Plant Pathol. 94, 134–148. doi: 10.1016/j.pmpp.2015.09.001
Kim, E. J., Kim, H. P., Hah, Y. C., and Roe, J. H. (1996). Differential expression of superoxide Dismutases containing Ni and Fe/Zn in Streptomyces Coelicolor. Eur. J. Biochem. 241, 178–185. doi: 10.1111/j.1432-1033.1996.0178t.x
Kim, S. Y., Lim, J. H., Park, M. R., Kim, Y. J., Park, T. I., Seo, Y. W., et al. (2005). Enhanced antioxidant enzymes are associated with reduced hydrogen peroxide in barley roots under saline stress. J. Biochem. Mol. Biol. 38, 218–224. doi: 10.5483/BMBRep.2005.38.2.218
Kozi, A. (1999). The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 601–639.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Levine, A., Tenhaken, R., Dixon, R., and Lamb, C. (1994). H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79, 583–593. doi: 10.1016/0092-8674(94)90544-4
Lewandowski, U., Kepinska, M., and Milnerowicz, H. (2019). The copper-zinc superoxide dismutase activity in selected diseases. Eur. J. Clin. Investig. 49:10. doi: 10.1111/eci.13036
Lin, Y. L., and Lai, Z. X. (2013). Superoxide dismutase multigene family in longan somatic embryos: a comparison of CuZn-SOD, Fe-SOD, and Mn-SOD gene structure, splicing, phylogeny, and expression. Mol. Breed. 32, 595–615. doi: 10.1007/s11032-013-9892-2
Mccord, J. M., and Fridovich, I. (1969). Superoxide dismutase: an enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244, 6049–6055.
Negi, N. P., Shrivastava, D. C., Sharma, V., and Sarin, N. B. (2015). Overexpression of CuZnSOD from Arachis hypogaea alleviates salinity and drought stress in tobacco. Plant Cell Rep. 34, 1109–1126. doi: 10.1007/s00299-015-1770-4
Noreen, Z., and Ashraf, M. (2009). Assessment of variation in antioxidative defense system in salt-treated pea (Pisum sativum) cultivars and its putative use as salinity tolerance markers. J. Plant Physiol. 166, 1764–1774. doi: 10.1016/j.jplph.2009.05.005
Paolo, M., Simona, D., Anna, S., and Mariarosaria, S. (2016). The cu, Zn superoxide dismutase: not only a dismutase enzyme. Front. Physiol. 7:594. doi: 10.3389/fphys.2016.00594
Pasin, F., Shan, H. Y., García, B., Müller, M., León, D. S., Ludman, M., et al. (2020). Abscisic acid connects phytohormone signaling with RNA metabolic pathways and promotes an antiviral response that is evaded by a self-controlled RNA virus. Plant Commun. 1:100099. doi: 10.1016/j.xplc.2020.100099
Pasin, F., Simón-Mateo, C., and García, J. A. (2014). The hypervariable amino-terminus of P1 protease modulates Potyviral replication and host defense responses. PLoS Pathog. 10:e1003985. doi: 10.1371/journal.ppat.1003985
Pogány, M., Rad, U. V., Grün, S., Dongó, A., Pintye, A., Simoneau, P., et al. (2009). Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-Alternaria pathosystem. Plant Physiol. 151, 1459–1475. doi: 10.1104/pp.109.141994
Rossetti, S., and Bonatti, P. M. (2001). In situ histochemical monitoring of ozone- and TMV-induced reactive oxygen species in tobacco leaves. Plant Physiol. Biochem. 39, 433–442. doi: 10.1016/S0981-9428(01)01250-5
Scandalios, J. G. (2005). Oxidative stress: molecular perception and transduction of signals triggering antioxidant gene defenses. Braz. J. Med. Biol. Res. 38, 995–1014. doi: 10.1590/S0100-879X2005000700003
Shah, K., Kumar, R. G., Verma, S., and Dubey, R. S. (2001). Effect of cadmium on lipid peroxidation, superoxide anion generation and activities of antioxidant enzymes in growing rice seedlings. Plant Sci. 161, 1135–1144. doi: 10.1016/S0168-9452(01)00517-9
Spagnolo, L., Torö, I., D'orazio, M., O'neill, P., Pedersen, J. Z., Carugo, O., et al. (2004). Unique features of the sodC-encoded superoxide dismutase from Mycobacterium tuberculosis, a fully functional copper-containing enzyme lacking zinc in the active site - ScienceDirect. J. Biol. Chem. 279, 33447–33455. doi: 10.1074/jbc.M404699200
Sumino, Y., Kayo, Y., and Naoaki, I. (2020). Interaction between the ins/IGF-1 and p38 MAPK signaling pathways in molecular compensation of sod genes and modulation related to intracellular ROS levels in C. elegans - ScienceDirect. Biochemistry Biophysics Reports 23:100796. doi: 10.1016/j.bbrep.2020.100796
Sun, Z. Q., Liu, D. Y., Li, B., Yan, F. F., Wang, Y. H., Yang, T. Q., et al. (2024). 3'UTR of tobacco vein mottling virus regulates downstream GFP expression and changes in host gene expression. Front. Microbiol. 15:1477074. doi: 10.3389/fmicb.2024.1477074
Tarchoune, I., Sgherri, C., Izzo, R., Lachaal, M., Ouerghi, Z., and Navari-Izzo, F. (2010). Antioxidative responses of Ocimum basilicum to sodium chloride or sodium sulphate salinization. Plant Physiol. Biochem. 48, 772–777. doi: 10.1016/j.plaphy.2010.05.006
Teoh, M. L. T., Walasek, P. J., and Evans, D. H. (2003). Leporipoxvirus cu,Zn-superoxide dismutase (SOD) homologs are catalytically inert decoy proteins that bind copper chaperone for SOD. J. Biol. Chem. 278, 33175–33184. doi: 10.1074/jbc.M300644200
Thomas, S. K., Cutter, S. L., Hodgson, M., Gutekunst, M., and Jones, S. (2002). Use of spatial data and geographic technologies in response to the September 11 terrorist attack. Quick response research report #153. Boulder, CO: Natural Hazards Research and Applications Information Center, University of Colorado.
Tyagi, S., Sharma, S., Taneja, M., Shumayla,, Kumar, R., Sembi, J. K., et al. (2017). Superoxide dismutases in bread wheat (Triticum aestivum L.): comprehensive characterization and expression analysis during development and, biotic and abiotic stresses. Agri Gene 6, 1–13. doi: 10.1016/j.aggene.2017.08.003
Wang, W. B., Kim, Y. H., Lee, H. S., Kim, K. Y., Deng, X. P., and Kwak, S. S. (2009). Analysis of antioxidant enzyme activity during germination of alfalfa under salt and drought stresses. Plant Physiol. Biochem. 47, 570–577. doi: 10.1016/j.plaphy.2009.02.009
Wang, W., Zhang, X. P., Deng, F. N., Yuan, R., and Shen, F. F. (2017). Genome-wide characterization and expression analyses of superoxide dismutase (SOD) genes in Gossypium hirsutum. BMC Genomics 18:376. doi: 10.1186/s12864-017-3768-5
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Wei, T., and Wang, A. (2008). (2008). Biogenesis of cytoplasmic membranous vesicles for plant Potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI- and COPII-dependent manner. J. Virol. 82, 12252–12264. doi: 10.1128/JVI.01329-08
Yang, T. Q., Zhao, X. Y., Bai, J. J., Lv, W. X., Chen, Q., Hu, J., et al. (2024). Transcriptome analysis of genes involved in the pathogenesis mechanism of potato virus Y in potato cultivar YouJin. Front. Microbiol. 15:1353814. doi: 10.3389/fmicb.2024.1353814
Yasmeen, A., Basra, S. M. A., Farooq, M., Rehman, H. U., Hussain, N., and Athar, H. U. R. (2013). Exogenous application of moringa leaf extract modulates the antioxidant enzyme system to improve wheat performance under saline conditions. Plant Growth Regul. 69, 225–233. doi: 10.1007/s10725-012-9764-5
Yue, J. Y., Lu, Y., Sun, Z. Q., Guo, Y. Q., León, D. S., Pasin, F., et al. (2023). Methyltransferase-like (METTL) homologues participate in Nicotiana benthamiana antiviral responses. Plant Signal. Behav. 18:2214760. doi: 10.1080/15592324.2023.2214760
Zelko, I. N., Mariani, T. J., and Folz, R. J. (2002). Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 33, 337–349. doi: 10.1016/S0891-5849(02)00905-X
Keywords: superoxide dismutases, NbCu/Zn-SOD-1, Nicotiana benthamiana, tobacco vein mottling virus, antiviral resistance
Citation: Wang H, Zhang J, Meng Z, Sun Z, Liu D, Li B, Yan F, Jia C, Zhou H and Zhao M (2025) Cu/Zn superoxide dismutase homologs participate in Nicotiana benthamiana antiviral responses. Front. Microbiol. 16:1561731. doi: 10.3389/fmicb.2025.1561731
Edited by:
Jinliang Liu, Jilin University, ChinaReviewed by:
Susheel Kumar, National Botanical Research Institute (CSIR), IndiaZihao Xia, Shenyang Agricultural University, China
Copyright © 2025 Wang, Zhang, Meng, Sun, Liu, Li, Yan, Jia, Zhou and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingmin Zhao, TWluZ21pbnpoQDE2My5jb20=
 Haijuan Wang1
Haijuan Wang1 Zhenqi Sun
Zhenqi Sun Dongyang Liu
Dongyang Liu Mingmin Zhao
Mingmin Zhao