- 1Key Laboratory of Endemic and Ethnic Diseases, Ministry of Education and School/Hospital of Stomatology Guizhou Medical University, Guiyang, China
- 2Department of Pathology, People’s Hospital of Qiandongnan, Qiandongnan Miao and Dong Autonomous Prefecture, Kaili, Guizhou, China
- 3Pediatric Intensive Care Unit, Guiyang Maternal and Child Health Care Hospital, Guiyang, China
- 4Key Laboratory of Microbiology and Parasitology of Education Department of Guizhou, Guizhou Medical University, Guiyang, China
- 5Collaborative Innovation Center for Prevention and Control of Endemic and Ethnic Regional Diseases Co-constructed by the Province and Ministry, Guiyang, China
Clostridioides difficile, a major cause of antibiotic-associated diarrhea and pseudomembranous colitis, is increasingly resistant to antibiotics and poses a significant threat due to its regulated virulence. The alternative sigma factor σ54 (rpoN) is known to regulate gene expression broadly, affecting microbial adaptation. Our study investigates how rpoN influences gene expression, physiology, and virulence in C. difficile. We used a modified CRISPR-Cpf1 system to create a rpoN deletion strain (∆rpoN) and a complemented strain (::rpoN) in the CD630 background, comparing their phenotypes and transcriptomes with the wild type. The ∆rpoN strain showed reduced motility and increased susceptibility to seven antibiotics, including β-lactams (amoxicillin, ampicillin, cefoxitin), nitroimidazoles (metronidazole), glycopeptides (vancomycin), fluoroquinolones (norfloxacin), and aminoglycosides (kanamycin). It also exhibited increased toxin gene expression, higher autolysis rates, and enhanced cytotoxicity and virulence in animal models. Additionally, rpoN deletion led to a decrease in glucose metabolic rate, which we attribute to the downregulation of glycolytic enzymes. Transcriptomic analysis indicated that reduced motility in ∆rpoN is due to downregulation of flagellar biosynthesis genes, while increased autolysis is linked to upregulation of autolysin genes like cwp19 and acd. The enhanced release of toxins due to higher autolysis rates contributes to the increased virulence of ∆rpoN. Our findings establish rpoN as a global regulator critical for antibiotic resistance, motility, metabolism, toxin production, and pathogenicity in C. difficile, suggesting its potential as a therapeutic target to mitigate virulence and resistance.
1 Introduction
Clostridioides difficile is a spore-forming, strictly anaerobic Gram-positive bacillus and a leading cause of antibiotic-associated diarrhea globally. Severe complications of C. difficile infection (CDI)—including pseudomembranous colitis, toxic megacolon, and sepsis—may arise from delayed or inaccurate diagnosis. CDI is further complicated by a 15%–35% recurrence rate, posing significant community and healthcare burdens (Finn et al., 2021). Due to its clinical urgency, the CDC classifies CDI as an immediate public health threat (CDC, 2024).
CDI initiates upon ingestion of environmental spores, which germinate into vegetative cells in the colon (Steindl et al., 2015). Antibiotic use disrupts the gut microbiota, creating a favorable niche for C. difficile spore germination into vegetative cells. Toxin A and Toxin B, the primary virulence factors, bind epithelial cell receptors and induce Rho GTPase glucosylation. This disrupts cytoskeletal integrity, triggering cell death and intestinal barrier breakdown. Subsequent influx of luminal contents into the lamina propria manifests as pseudomembranous colitis, characterized by inflammatory patches on the colon mucosa and symptoms such as severe diarrhea, abdominal pain, and fever (Fachi et al., 2024).
In prokaryotes, transcription initiation—a critical regulatory step for gene expression—involves complex mechanisms enabling adaptation to environmental changes (MacDonald et al., 2021). Since the discovery of the sigma (σ) factor in 1979, numerous studies have demonstrated its pivotal role in the function of RNA polymerase holoenzyme, influencing the transcription level of various genes (Borukhov and Nudler, 2003). Based on their functions, σ factors can be broadly classified into two categories: the σ70 family, which recognizes the −35/−10 region of promoters (Paget, 2015; Sun et al., 2021), and the Sigma-54 (σ54) family, which recognizes the −24/−12 region (Yang et al., 2015; Danson et al., 2019). The σ70 family, comprising four conserved regions, is the most abundant and primarily regulates gene expression during the exponential growth phase of bacteria (Mazumder and Kapanidis, 2019).
RpoN (σ54 family, also known as sigL) regulates the expression of numerous genes, thereby controlling various bacterial functions (Clark et al., 2022). In Clostridium acetobutylicum (Yang et al., 2020), rpoN mutants exhibit severely impaired butanol and ethanol production. Overexpression of rpoN significantly enhances the production of solvents such as acetone, butanol, and ethanol (Yang et al., 2020). Rukit et al. found that the rpoN gene in Listeria monocytogenes is involved in bacterial growth and host invasion, and its deletion enhances host invasion (Rukit et al., 2022). In Pseudomonas aeruginosa, rpoN has been demonstrated as a critical virulence regulator, and deleting the rpoN gene reduces its virulence (Lloyd et al., 2017). Additionally, the rpoN has been reported to participate in flagellum formation in Flavobacterium, thereby regulating the motility of the strain (Yu et al., 2021). Interestingly, studies have revealed that the rpoN regulates antibiotic resistance in clinical pathogens such as Pseudomonas aeruginosa. Inhibition of rpoN expression can increase the susceptibility of strains to antibiotics (Viducic et al., 2016; Lloyd et al., 2019). These findings indicate that rpoN is involved in multiple cellular processes and is crucial in regulating bacterial growth and metabolism.
Previous comparative genomic studies of 57 Clostridium species, by Nie et al., reconstructed the σ54 regulon in C. difficile and identified bEBPs and their regulatory modules (Nie et al., 2019). Soutourina et al. employed computational modeling, transcriptomics, and transcription start site mapping to investigate the SigL regulon in C. difficile strain 630Δerm, revealing its involvement in the Stickland reaction, a process that generates ATP and NAD+ and influences toxin production (Soutourina et al., 2020). Experimental validation confirmed the roles of CdsR and PrdR in cysteine and proline metabolism, respectively (Bouillaut et al., 2013; Gu et al., 2018). Andrew E. Clark’s work underscored the strain-specific nature of SigL’s effects on toxin production, sporulation, and cell surface properties (Clark et al., 2022). While prior studies identified σ54-associated regulons in C. difficile, strain-specific regulatory roles of RpoN remain unclear. To address this, we generated an rpoN-knockout in C. difficile CD630 (RT012) using CRISPR-Cpf1. Phenotypic and transcriptomic analyses revealed RpoN’s pleiotropic roles in toxin production, motility, biofilm formation, and virulence, expanding our understanding of its regulatory network.
2 Materials and methods
2.1 Bacterial strains and culture conditions
All Escherichia coli and Clostridioides difficile strains used in this study are shown in Supplementary Table S1. NEBExpress Competent Escherichia coli cells (C2523H, New England BioLabs) were used for gene cloning and plasmid construction. The CA434 was used as the donor strain for the conjugation transfer of shuttle vectors to C. difficile (Purdy et al., 2002). When necessary, chloramphenicol (6 μg/mL), kanamycin (50 μg/mL), and ampicillin (100 μg/mL) were added to the Luria-Bertani (LB) medium for transformants selection. C. difficile was grown anaerobically in the brain heart infusion (BHIS) medium supplemented with 5 g/L yeast and 1 g/L L-arginine at 37°C in an anaerobic chamber (AW500TG, Electrotek, United Kingdom). When necessary, streptomycin (15 μg/mL), cephalothin (8 μg/mL), D-cycloserine (250 μg/mL), and 5-fluoroorotic acid (2 mg/mL) were added to the BHIS medium for transformant selection.
2.2 Plasmids construction
All plasmids and primers used in this experiment are listed in Supplementary Table S1 (plasmids) and Supplementary Table S2 (primers). Restriction endonucleases were purchased from New England BioLabs (Beijing, China), and DNA polymerase and ClonExpress Multis One Step Cloning were purchased from Novozymes (C115, Nanjing, China).
To enhance the gene-targeting plasmids cure efficiency, we introduced the pyrF’ (orotidine-5′-phosphate decarboxylase) gene derived from Clostridium beijerinckii NCIMB 8052 in C. difficile 630 ∆pyrF mutant as a negative selection marker into the pWH34 plasmid backbone (Hong et al., 2018), and then inserted the sRNAP⸬crRNA (rpoN) gene-targeting fragment and rpoN homologous arms (rpoN_arms) to obtain the rpoN gene knockout plasmid pYJY3.
The detailed plasmid construction processes were as follows: Firstly, the pyrF’ gene was amplified with primer pair HW625/HW626 using the genomic DNA of Clostridium beijerinckii NCIMB 8052 as the template. The pyrF’ gene was ligated with XhoI-linearized pWH34 to obtain the plasmid pYJY3-S (Supplementary Figure S1B). Subsequently, the sRNAP⸬crRNA (rpoN) gene targeting fragment was amplified from pWH37 and simultaneously mutated using the primer pair HW520/HW521. Then, using the C. difficile 630 genome as a template, the upstream and downstream homologous arms of the rpoN gene were PCR-amplified using the primer pairs HW523/HW524 and HW525/HW526, respectively. Overlapping PCR connected the upstream and downstream homologous arms with the sRNAP⸬crRNA (rpoN) targeting fragment. The resulting amplicon was assembled to NotI-HF linearized pYJY3-S using the ClonExpress Multis One-Step Cloning kit. The resultant rpoN gene knockout plasmid was denoted pYJY3 (Supplementary Figure S1C).
To construct the rpoN and pyrF gene complementation vector, the pyrF and rpoN genes with their promoter were amplified from the C. difficile 630 genome using the primer pairs HW731/HW734 and HW684/HW685, respectively. The pyrF gene fragment was assembled with the HindIII-linearized pMTL82151 plasmid to produce pYJY2-S, and the rpoN gene fragment was assembled with the XhoI-linearized pYJY2-S plasmid to obtain rpoN and pyrF gene complementation vector pYJY4 (Supplementary Figure S3A).
2.3 Construction of ∆rpoN and ⸬rpoN mutants
Using the heat shock method, the rpoN gene knockout plasmid pYJY3 was introduced into the E. coli CA434 donor strain (Cañadas et al., 2019). The E. coli CA434 strain containing pYJY3 plasmid was used as a donor strain to conjugate the pYJY3 into C. difficile 630 ∆pyrF mutant (Hong et al., 2018). To screen ∆rpoN mutant: (1) The pYJY3-harboring thiamphenicol-resistant colonies (∆pyrF) were spread on BHIS solid medium containing 40 mmol/L lactose and 15 μg/mL Tm to screen ∆rpoN∆pyrF; (2) The deletion of the rpoN gene was verified using the primer pair HW557/HW558; (3) Obtained ΔrpoN∆pyrF mutants were spread on CDMM medium containing 2 mg/mL 5-fluoroorotic acid (5-FOA) to cure the pYJY3 plasmid; (4) The same plasmid conjugation method was used to introduce pYJY2-S and pYJY4 into the ΔrpoN∆pyrF mutant strain, resulting in complemented strains ΔrpoN∆pyrF⸬pyrF and ΔrpoN∆pyrF⸬pyrF⸬rpoN.
2.4 Evaluation of gene expression level using RT-qPCR
The WT, ΔrpoN, and ⸬rpoN C. difficile strains in the logarithmic growth phase were inoculated into fresh BHIS medium and incubated in the anaerobic chamber at 37°C to OD600 = 0.6. The cells were collected by centrifugation at 13,500×g for 3 min. Total RNA was extracted from WT, ΔrpoN, and ⸬rpoN using a bacterial total RNA extraction kit (DP430, TIANGEN, Beijing). The total RNA was then reverse-transcribed into cDNA using the FasKing gDNA Dispelling RT SuperMix (KR118, TIANGEN, Beijing). The expression levels of the rpoN gene in each strain were analyzed using the 16 s ribosomal RNA expression gene (rrs) as an internal reference. The amplification primers for rrs and rpoN genes were HW554/555 and HW693/694, respectively (Supplementary Table S2). The RT-qPCR system was prepared on ice with the 2 × HQ SYBR qPCR Mix (No Rox) kit as follow: 5 μL 2 × HO SYBR gPCR Mix, 0.2 μL Forward Primer (10 mM), 0.2 μL Reverse Primer (10 mM), 1 μL Template cDNA, and 3.6 μL ddH2O. The RT-qPCR reaction was performed on the CFX96 Real-Time PCR Detection System (CFX96 Conne, Bio-rad, United States). The data were statistically analyzed using Prism 10 software (Version10.0.3, GraphPad Software, Inc).
To determine the expression levels changes of toxin genes in the WT, ΔrpoN, and ⸬rpoN strains. These strains in the logarithmic growth phase were each transferred to fresh BHIS medium, with three replicates for each group. The strains were cultivated until they reached an OD600 of 0.8, after which the cells were collected and RNA was extracted. The 16S ribosomal RNA expression gene (rrs) was used as the reference gene. Primers HW554/555, HW887/888, and HW885/886 were used as amplification primers for the rrs, tcdA, and tcdB genes, respectively (Supplementary Table S2). The gene expression levels were detected by RT-qPCR.
2.5 Growth profile of strains
The WT, ΔrpoN, and ⸬rpoN strains were streaked onto solid BHIS agar plates, and single colonies were picked and transferred into BHIS liquid medium. The strains were cultured until they reached the logarithmic growth phase (OD600 = 0.5), then inoculated into fresh BHIS liquid medium at a 1% inoculation rate and cultured anaerobically at 37°C with three replicates for each group. The OD600 values were measured every 3 h using a cell density meter (Ultrospec 10, Amersham Biosciences, GE). Growth curves were plotted with time on the x-axis and log10(OD600) values on the y-axis.
2.6 Autolysis assay
Overnight cultures of C. difficile WT, ΔrpoN mutant, and ⸬rpoN strains were diluted to OD600 = 0.05 in BHI and incubated at 37°C until OD600 = 0.5. Bacterial cells were collected, washed twice, and resuspended in 50 mM potassium phosphate buffer (pH = 7.0), containing 0.01% Triton X-100, to OD600 = 0.5. The OD600 of the suspensions were then measured every 20 min at 37°C (OD600-M), the percent- age of unautolysed cells was calculated as (OD600-M/0.5)*100%. The untreated cells in the BHIS medium with the same inoculation ratio were set as the control group.
2.7 Motility assay
BHIS liquid medium was prepared by adding 0.5% agar to create a semi-solid BHIS medium. The WT, ΔrpoN, and ⸬rpoN strains in the logarithmic growth phase were streaked on plates and then incubated in an anaerobic chamber at 37°C for 48 h until colonies formed. Single colonies were inoculated into the semi-solid medium in a straight-neck glass tube (diameter 22 mm, specification 10 mL), with triplicates for each group. After cultivation under anaerobic conditions at 37°C for 12 h, the motility results of the strains were recorded.
2.8 Hydrogen sulfide (H2S) production
The production of H2S was determined by the formation of black bismuth sulfide (BS) precipitate through the reaction of bismuth chloride with hydrogen sulfide, with the optical density (OD405) serving as an indicator (Basic et al., 2015). The WT, ΔrpoN and ⸬rpoN strains were inoculated in BHIS medium. When the bacterial culture reached OD600 = 0.6, 100 μL of the bacterial suspension was reacted with an 100 μL of freshly prepared bismuth solution [0.4 M triethanolamine HCl, pH = 8.0; 10 mM bismuth chloride (III), 20 mM 5-phosphor-1-pyrroline, 20 mM EDTA, and 40 mM L-cysteine] were mixed in 96-well plates with 3 replicates per group and reacted in an anaerobic workstation at 37°C for 8 h. During the period, the OD405 optical density value is measured every 1 h, and the reaction curve is plotted with time as the x-axis and the OD405 values on the y-axis.
2.9 Scanning electron microscopy (SEM)
Single colonies of WT, ΔrpoN, and ⸬rpoN were picked and cultured in BHIS medium to OD600 = 0.6, then the bacterial cells were collected by centrifugation at 4,000×g for 3 min. The cell pellet was resuspended in a 2.5% glutaraldehyde and fixed overnight at 4°C. The bacterial strains were washed three times with phosphate-buffered saline (PBS) buffer (C10010500BT, Gibco, New York, United States), then dehydrated through graded ethanol of 50, 70, 90, and 100% (vol/vol) for 5 min each. The samples were then dried in a vacuum freeze dryer. After drying, the bacterial powder was carefully picked with a sterilized toothpick. It adhered to a carbon-conductive tape, followed by sputter coating with a gold film in a vacuum evaporator to enhance conductivity and image quality. Finally, a small amount of sample was attached to the tape on a glass slide, and the surface morphology of the strains was observed using a scanning electron microscope (S-3400, Hitachi, Japan).
2.10 Antibiotic susceptibility of ΔrpoN mutant
The antibiotics resistance of WT, ΔrpoN, and ⸬rpoN strains to metronidazole, vancomycin, amoxicillin, kanamycin, cefuroxime, norfloxacin, and ampicillin was determined using the series dilution method (Jorgensen and Ferraro, 2009). Each antibiotic was added to the first well of 96-well plate at 128 μg/mL and serially diluted by a factor of two times to create a gradient of antibiotic concentrations: 128, 64, 32, 16, 8, 4, 2, 1, 0.5, and 0.25 μg/mL. Then, 10 μL of WT, ΔrpoN, or ⸬rpoN strain with an OD600 of 0.6 was inoculated into each well, with triplicates for each strain. Finally, the cultures were incubated for 24 h at 37°C in an anaerobic chamber, and the OD600 values were read using a spectrophotometer (Varioskan LUX, Thermo Fisher Scientific, United States). Uninoculated blank controls were set up for all antibiotics. The OD600 values greater than or equal to 0.1 were considered the growth of C. difficile strain.
2.11 Single carbon source utilization assay
The WT, ΔrpoN, and ⸬rpoN strains were cultured to an OD600 of 0.6 and then inoculated into 5 mL of single carbon source medium (Ehsaan et al., 2016) at a 1% inoculation rate. The cultures were continuously incubated anaerobically at 37°C for 72 h. The OD600 values were recorded every 3 h during the logarithmic growth phase and every 12 h during the stationary phase, and growth curves were plotted accordingly.
2.12 Biofilm assays
The influence of rpoN on C. difficile biofilm formation was evaluated using a biofilm assay as previously reported (Pantaléon et al., 2015) with mirror modifications. Overnight C. difficile cultures were diluted 1: 200 in BHIS broth and distributed into 24-well plates, 2 mL per well. Plates were wrapped to prevent evaporation and incubated anaerobically at 37°C for 48 h. Following incubation, the culture supernatant was removed and then incubated at 37°C with 0.2% (w/v) crystal violet to fix and stain. The crystal violet was subsequently removed, and the biofilms were washed an additional two times with PBS and photographed. To measure biofilm formation, crystal violet retained by the biomass was released with anhydrous ethanol and quantitated by detecting the OD562 value in a technical quadruplicate for three individual wells.
2.13 Determine toxins level using western blot
In order to further elucidate variations in the protein levels of TcdA and TcdB in the different mutants, Western blot analysis was employed to detect the expression levels of TcdA and TcdB in WT, ΔrpoN, and ⸬rpoN strains. The supernatant was collected by centrifuging 5 mL of OD600 = 0.6 bacterial culture at 3,440×g, followed by sterile filtration through a 0.22 μm sterile filter membrane. The total protein content in the supernatant was determined using the BCA method (P0012S, Beyotime), and 6 μg of protein-containing supernatant was subjected to SDS-PAGE. Subsequently, the proteins in the gel were transferred to a PVDF membrane (IPVH00010, Merck) and subjected to immunoblotting using a 1:1,000 dilution of TcdA and TcdB antibodies (Clostridioides difficile, EPR23359-15/EPR23357-19, ab272720/ ab270452, Abcam). The blots were developed using a 1:10,000 dilution of horseradish peroxidase-conjugated rabbit anti-mouse IgG antibody (A0208, Beyotime), with triplicates for each sample.
2.14 Cell cytotoxicity assay
Vero cell line were selected for cytotoxicity assay with the WT, ΔrpoN and ⸬rpoN strains. Vero cells were cultured in DMEM (Dulbecco’s Modified Eagle’s Medium-high glucose) supplemented with 10% fetal bovine serum (Catalog Number, Gibco, United States) and 1% dual antibodies (10 mg/mL streptomycin, 10,000 U/mL penicillin). Vero cells were seeded onto 24-well plates and incubated at 37°C until the wells were confluent. The medium was then removed, and the cells were washed three times with sterile PBS buffer. Subsequently, 200 μL of DMEM (without antibiotics and serum) was added, and the cells were cultured for an additional 24 h. The WT, ΔrpoN, and ⸬rpoN strains were anaerobically incubated in an anerobic chamber until OD600 = 1.0, and 1 mL of the bacterial culture was centrifuged at 13,500×g for 5 min. The supernatant containing C. difficile toxins was filtered through a 0.22 μm sterile filter membrane (QFC06-SF022P25, Zeren Technology Co, Shenzhen, China). The toxin-containing supernatant was diluted from 0.5 × 101 to 0.5 × 1010, and 200 μL of each toxin dilution was used to infect the Vero cells. The cells were incubated overnight at 37°C. The cell morphology was observed and photographed under a light microscope at a magnification (CKX53SF, Olympus, Tokyo, Japan) of 200×, and the highest dilution factor that caused cell rounding in Vero cells was determined.
2.15 Construction of CDI animal model and the pathology of WT, ∆rpoN, and ⸬rpoN strains
Syrian golden hamsters (4–5 weeks old, ~100 g) were purchased from Liaoning Changsheng Biotechnology Co., Ltd. and housed 5 per cage with ad libitum access to food and water. Hamsters were acclimated for 1 week prior to the experiment. To disrupt the gut microbiota, hamsters in the experimental groups (WT, ΔrpoN, and ⸬rpoN) were administered a cocktail of antibiotics (kanamycin 0.8 mg/mL, gentamicin 0.07 mg/mL, polymyxin B 0.1135 mg/mL, metronidazole 0.43 mg/mL, and vancomycin 0.09 mg/mL) in drinking water for 7 days (Chen et al., 2008; Geeraerts et al., 2015; Mooyottu et al., 2017). Control animals received PBS. On day 9, all hamsters were subcutaneously injected with clindamycin (10 mg/kg) to further perturb the gut microbiota (Chen et al., 2008). On days 10, 12, and 14 post-clindamycin injection, hamsters were gavaged with different Clostridioides difficile strains or PBS (control). Each hamster received 100 μL of bacterial suspension (OD600 = 0.6, 4 × 105 CFU/mL). Body weight and fecal consistency were monitored daily.
2.16 Transcriptome data processing and analysis
The WT, ΔrpoN, and ⸬rpoN strains were inoculated into BHIS medium and cultured until the OD600 reached 0.6. The cultures were then centrifuged at 4,000×g for 10 min, the supernatant was removed, and the bacterial pellets of each strain were collected. The samples were transported with adequate dry ice in an foam box to Nanjing Personalomics Co., Ltd. for transcriptome sequencing. Initially, RNA extraction and quality assessment were performed. Once the samples were deemed satisfactory, the following steps were conducted for sequencing: (1) Removal of ribosomal RNA; (2) Enrichment and purification of mRNA; (3) Fragmentation of mRNA; (4) Construction of the sequencing library and quality assessment of the library; (5) Sequencing on the Illumina Novaseq 6,000 platform (Zhou et al., 2024).
Raw data organization, filtering, and quality assessment: after the samples are sequenced, sequence image files are obtained, and the sequencing platform software converts the data into raw data in FASTQ format with a size of 4 GB (Raw Data). The RNA-seq raw data (RNA-seq of Clostridioides difficile ΔrpoN mutant against wild-type control) was deposited in the ArrayExpress database under the accession number of E-MTAB-14512. The following treatments are applied to the raw data: Initially, data quality control is performed, which includes quality control of base quality distribution and Base Content of the sequenced data; Subsequently, to avoid interference with subsequent information analysis, further filtering is conducted on low-quality reads; Then, the obtained transcriptome data is aligned and analyzed against the reference genome GCF_000009205.2_ASM920v2_genomic.fna (NC_009089.1 and NC_008226.2) on NCBI; Finally, expression quantification analysis is performed on the measured genome. Since read counts are positively correlated with the actual expression levels of genes, gene length, and sequencing depth, to make the gene expression levels comparable between different genes and samples, the expression levels are normalized using FPKM (Fragments Per Kilobase of transcript per million mapped reads), and genes with an FPKM > 1 are considered to be expressed (Radakovits et al., 2012).
Cluster analysis of differentially expressed genes (Tatusov et al., 2003): Cluster analysis is used to determine the expression patterns of differentially expressed genes under different experimental conditions; genes with high expression correlation between samples are categorized into one group, which usually indicates that these genes have actual connections in certain biological processes or metabolic and signaling pathways. Therefore, we can discover unknown biological connections between genes through expression clustering. We use the R language Pheatmap package (version 1.0.12) for two-way hierarchical cluster analysis of the union of differential genes and samples in all comparison groups, clustering based on the expression levels of the same gene in different samples and the expression patterns of different genes in the same sample, using the Euclidean method to calculate distances, and the longest distance method (Complete Linkage) for clustering.
Functional enrichment analysis of differentially expressed genes (McClure et al., 2013): The DEseq software (version 1.44.0) is used to analyze the differentially expressed mRNA, with mRNA considered differentially expressed when the FoldChange (fold change) > 2 and the p-value < 0.05. A volcano plot of the differentially expressed mRNA is generated using the R language ggplot2 package (version 3.5.1). The differentially expressed mRNA target genes were subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. All target genes are mapped to each item in the GO database, the number of differentially expressed target genes in each item is calculated, and a hypergeometric distribution is used to calculate the significantly enriched target genes, with p ≤ 0.05 considered as significantly enriched pathways.
2.17 Statistical methods
Prism 10 (Version 10.0.3) was used for statistical analysis. Two-group comparisons were performed using students’ t-tests or non-parametric tests. Comparisons among multiple groups were performed using one-way ANOVA, while comparisons among multiple groups over time were conducted using two-way ANOVA, followed by Tukey’s multiple comparisons. The results were expressed as mean ± standard deviation, with a test level of α = 0.05, and P < 0.05 was statistically significant (n = 3). P > 0.1, n; *P < 0.05; **P < 0.01; ***P< 0.001; ****P < 0.0001.
3 Results
3.1 Phenotypic characterization of mutant strains
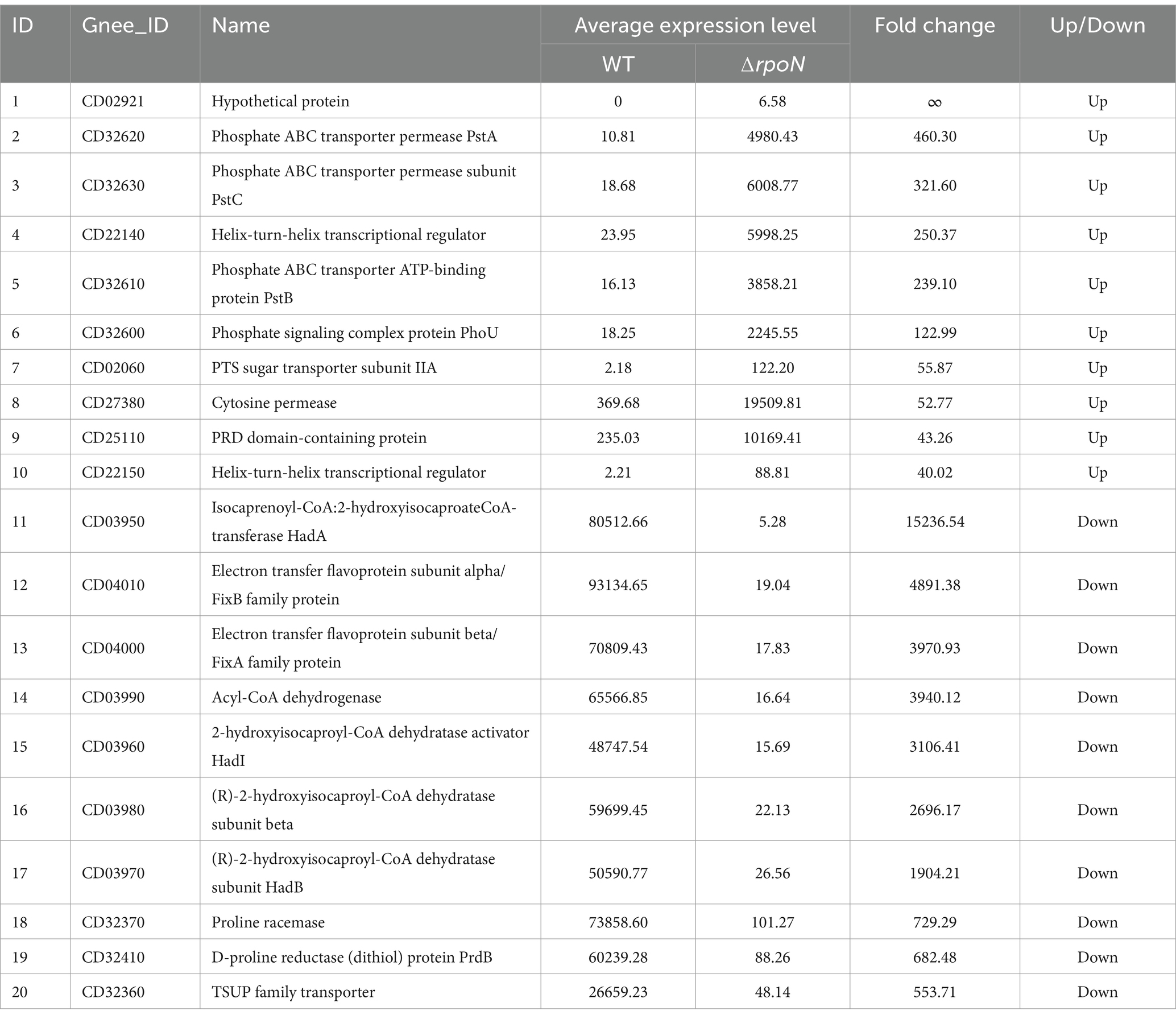
Initially, we verified the successful generation of the ΔrpoN [also known as sigL (Clark et al., 2022)] mutant strain by confirming its presence at both the genomic DNA and protein levels. RT-qPCR confirmed complete loss of rpoN expression in ΔrpoN, while complementation (⸬rpoN) restored expression to wild-type (WT) levels (Figure 1A). SDS-PAGE revealed the absence of the 52.1 kDa RpoN protein in ΔrpoN, which was rescued in ⸬rpoN (Figure 1B). The above results suggest that the construction of the C. difficile 630 strain ΔrpoN and ⸬rpoN strains was successful.
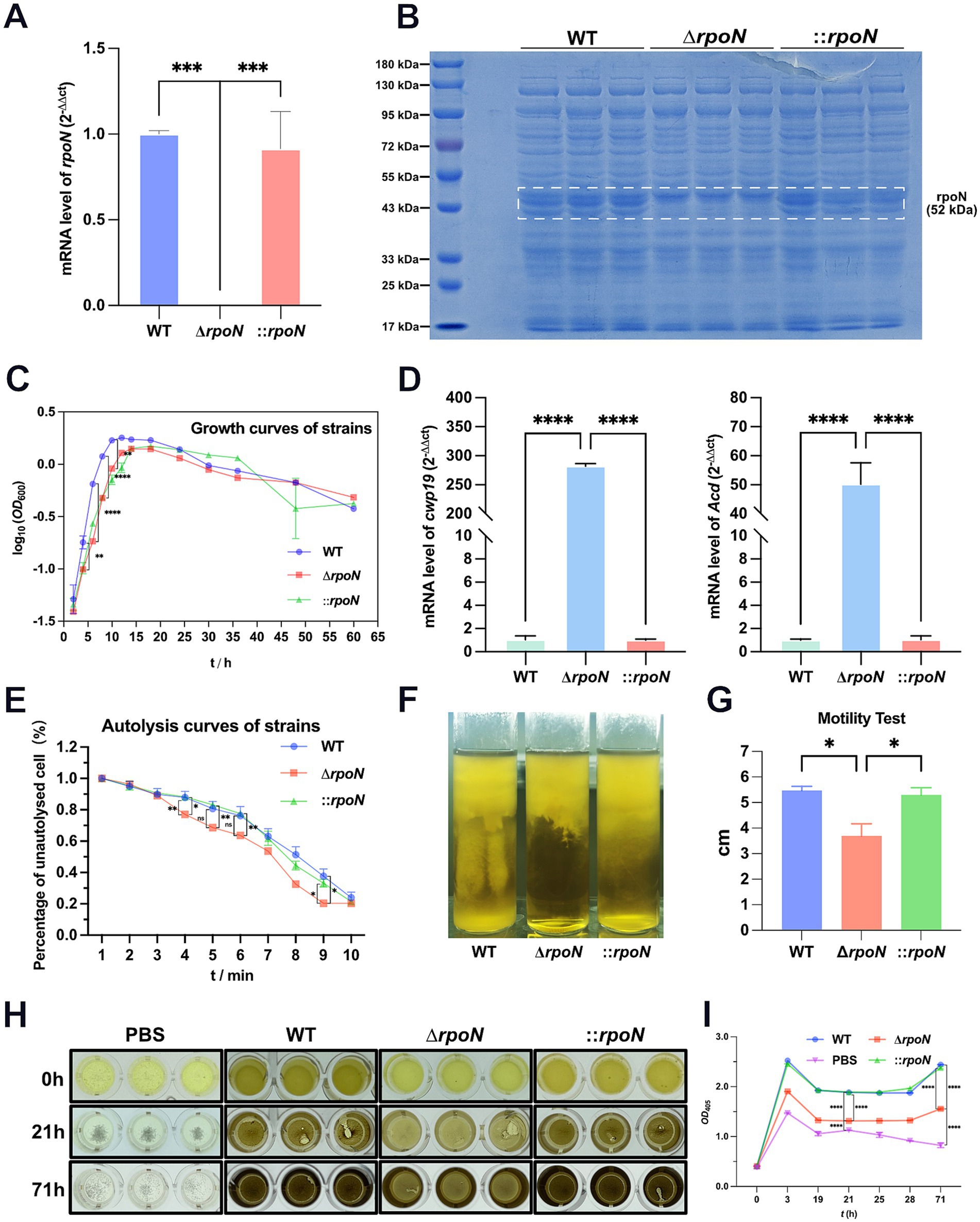
Figure 1. Phenotypic characterization of mutant strains. (A) RT-qPCR showed that in the expression of the rpoN gene no longer detectable in the ∆rpoN mutant, whereas it was restored in the ⸬rpoN complementary strain. Data are presented as mean ± SEM. Statistical significance was determined using one-way ANOVA, followed by Tukey’s post hoc test. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, **** p ≤ 0.0001; (B) Verification of ∆rpoN mutant by using SDS-PAGE. The results showed that the ∆rpoN mutant strain absented a 52 kDa protein band compared to the WT strain, and it was restored in the ⸬rpoN strain; lane M molecular weight marker (from top to bottom, 180 kDa, 130 kDa, 95 kDa, 72 kDa, 55 kDa, 43 kDa, 33 kDa, 25 kDa, and 17 kDa). (C) Growth curves of WT, ∆rpoN, and ⸬rpoN strains, the horizontal coordinate is incubation time (hours), and the vertical coordinate is cell turbidity at OD600; (D) The green, blue, and red bars indicate the expression levels of the cwp19 or Acd genes in the WT, ΔrpoN, and ⸬rpoN strains, respectively; (E) Cell autolysis rate of the WT, ∆rpoN, and ⸬rpoN strains, the horizontal coordinate is treatment duration of Triton X-100, the vertical coordinate is the percentage of unautolysed cells; (F,G) Comparison of motility of the WT, ∆rpoN, and ⸬rpoN strains, the motility of the ∆rpoN mutant strain was significantly decreased than that of the WT and the ⸬rpoN strains; (H,I) Comparison of hydrogen sulfide (H2S) production between the WT, ΔrpoN, and ⸬rpoN strains, the ΔrpoN mutant strain produced significantly less H2S than the WT, while the ⸬rpoN strain restored the ability to produce H2S. Data are presented as mean ± SEM. Statistical significance was determined using two-way ANOVA, followed by Tukey’s post hoc test in the growth curves, the autolysis curves and the H2S production curves. Motility test and the expression of autolysis-related genes were compared using one-way ANOVA. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
To assess the impact of rpoN on C. difficile 630 phenotype, we investigated changes in growth rate, motility, hydrogen sulfide (H2S) production, autolysis rate, and the expression of autolysis-related genes (Meouche and Peltier, 2018; Noori Goodarzi et al., 2022). During the logarithmic growth phase (0–12 h), the ΔrpoN strain exhibited a significantly reduced growth rate and maximum biomass compared to the WT strain. While all strains showed a short stationary phase, the ΔrpoN strain displayed a significantly higher autolysis rate than the WT. The ΔrpoN strain’s growth rate and maximum biomass were similar to those of the ⸬rpoN strain, but its autolysis rate was significantly lower than both the WT and ΔrpoN strains (Figure 1C).
To investigate the cause of the increased autolysis rate in the ΔrpoN strain, we quantified the expression of two known autolysis genes, cwp19 (Meouche and Peltier, 2018) and acd, using RT-qPCR (Noori Goodarzi et al., 2022). Results showed that both genes were significantly upregulated in the ΔrpoN strain (Figure 1D). Consistent with this, autolysis assays revealed a significantly higher autolysis rate for the ΔrpoN strain compared to the WT and ⸬rpoN strains (Figure 1E). Motility assays demonstrated that the ΔrpoN strain exhibited significantly reduced motility compared to both the WT and ⸬rpoN strains (p < 0.05) (Figures 1F,G). Furthermore, H2S production was significantly lower in the ΔrpoN strain compared to the WT and ⸬rpoN strains (Figures 1H,I). In summary, the ΔrpoN strain exhibited decreased growth rate, motility, and H2S production, coupled with an increased autolysis rate.
3.2 Changes in cell surface morphology
To investigate whether the increased autolysis rate of the ∆rpoN strain relates to changes in cell surface structure, we examined the surface morphology of WT, ∆rpoN, and ⸬rpoN strains using Scanning Electron Microscopy (SEM). Results showed abundant granules (Chu et al., 2016) attached to the surface of the WT strain (Figures 2A–D). In contrast, the ∆rpoN strain exhibited a significant reduction in granule attachment (Figures 2E–H). Importantly, the ⸬rpoN strain displayed granule attachment levels comparable to those of the WT strain (Figures 2I–L).
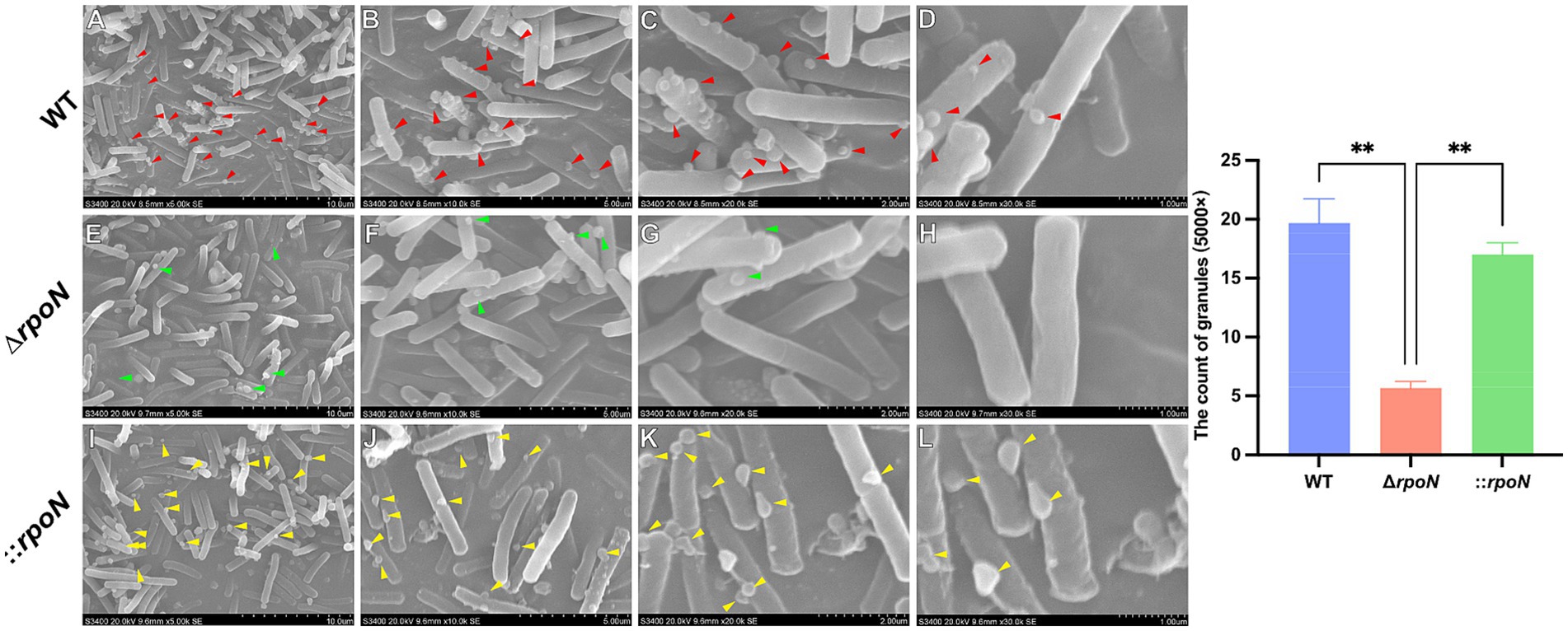
Figure 2. Surface morphology of WT, ∆rpoN and ⸬rpoN strains viewed by Scanning Electron Microscopy. (A–D) Cell surface morphology of the WT strain under ×5,000 (A), ×10,000 (B), ×20,000 (C), and ×30,000 (D) magnifications; (E–H) Cell surface morphology of ∆rpoN strain under ×5,000 (E), ×10,000 (F), ×20,000 (G), and ×30,000 (H) magnifications; (I–L) Cell surface morphology of ⸬rpoN strain under ×5,000 (I), ×10,000 (J), ×20,000 (K), and ×30,000 (L) magnifications. The arrows indicate cell surface deposits (granules). The blue, red, and green bars represent the number of granules in WT, ΔrpoN, and ⸬rpoN strains, respectively (Three fields of view at ×5,000 were selected, and each view was counted three times).
3.3 Deletion of rpoN significantly affects antibiotic resistance of Clostridioides difficile
RpoN is an alternative sigma factor involved in transcriptional regulation, microbial physiology, stress resistance, and pathogenicity. To assess the impact of rpoN on antibiotic resistance, we compared the resistance profiles of WT, ΔrpoN, and ⸬rpoN strains to amoxicillin (Figure 3A), ampicillin (Figure 3B), metronidazole (Figure 3C), vancomycin (Figure 3D), norfloxacin (Figure 3E), cefoxitin (Figure 3F), and kanamycin (Figure 3G). The ΔrpoN strain exhibited significantly reduced resistance to all tested antibiotics compared to the WT (Figures 3A–G). Resistance profiles of the ⸬rpoN strain partially restored to WT levels. These results indicate that the rpoN gene plays a significant role in modulating antibiotic resistance in C. difficile 630.
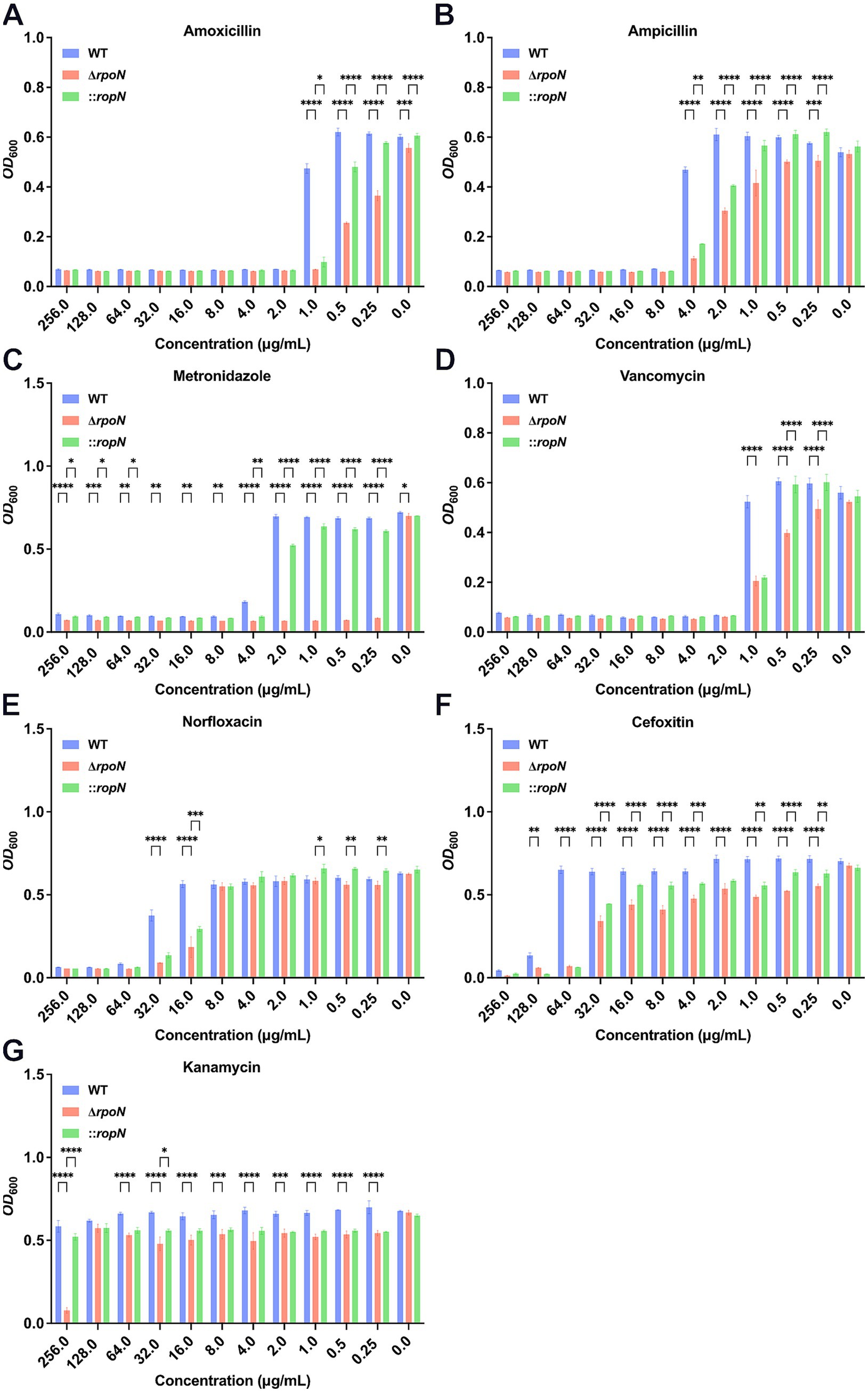
Figure 3. Antibiotic susceptibility of WT, ∆rpoN, and ⸬rpoN strains. The vertical axis represents the OD600 value, while the horizontal axis represents the concentration of different antibiotics (mg/mL). The blue, red, and green bars indicate the OD600 values of WT, ΔrpoN, and ⸬rpoN strains at different antibiotic concentrations. Compared to WT, the ΔrpoN strain exhibited significantly decreased susceptibility to amoxicillin (A), ampicillin (B), metronidazole (C), vancomycin (D), norfloxacin (E), cefotetan (F), and kanamycin (G), whereas the ⸬rpoN strain showed partial restoration of susceptibility to these antibiotics compared to WT.
3.4 Deletion of rpoN upregulates TcdA/TcdB expression and cytotoxicity in Clostridioides difficile
To further investigate the impact of rpoN on cytotoxicity, we assessed the virulence of WT, ΔrpoN, and ⸬rpoN strains using a Vero cell model. Vero cells were infected with graded dilutions (0.5 × 101 to 0.5 × 1010) of supernatant from each strain. Control cells (no supernatant) exhibited normal fusiform morphology with clear cell junctions (Figures 4A–C). Infection with undiluted supernatants from WT (Figure 4D), ΔrpoN (Figure 4E), and ⸬rpoN (Figure 4F) resulted in rounded, suspended cells with disrupted cell junctions. The highest supernatant dilutions that induced morphological changes in Vero cells were 0.5 × 107 (Figure 4G), 0.5 × 109 (Figure 4H), and 0.5 × 108 (Figure 4I) for WT, ΔrpoN, and ⸬rpoN, respectively. Consequently, the ΔrpoN strain exhibited a 100-fold increase in cytotoxicity compared to the WT, while the ⸬rpoN strain displayed intermediate cytotoxicity.
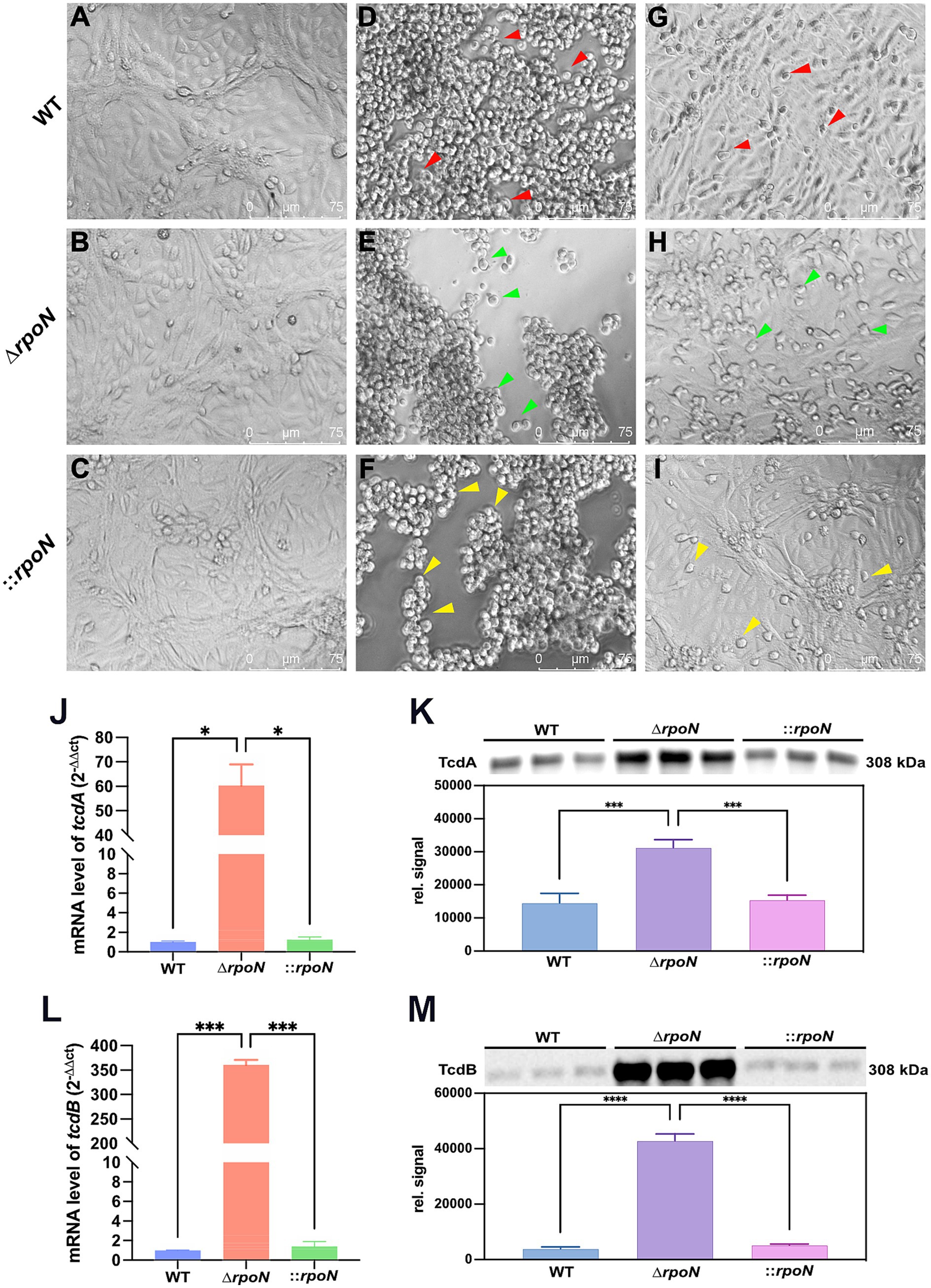
Figure 4. Cell toxicity of WT, ∆rpoN and ⸬rpoN strains. Supernatants of each strain were co-cultured with Vero cells, and morphological changes of Vero cells were observed under a light microscope. (A–C) Show the PBS control without toxins, where cells exhibit a healthy spindle shape. Vero cells cocultured with undiluted WT (D), ∆rpoN (E), and ⸬rpoN (F) supernatants became rounded and suspended (red, green, and yellow arrows); (G) Shows the results of Vero cells co-cultured with WT supernatant toxin diluted 0.5 × 107 times; (H) Shows the results of Vero cells co-cultured with ∆rpoN supernatant toxin diluted 0.5 × 109 times; (I) Shows the results of Vero cells co-cultured with ⸬rpoN supernatant toxin diluted 0.5 × 108 times; (J,L) Blue, red, and green bars represent the expression levels of tcdA or tcdB genes in WT, ∆rpoN, and ⸬rpoN strains, respectively; (K,M) Blue, purple, and pink bars represent the expression levels of TcdA or TcdB proteins in WT, ∆rpoN, and ⸬rpoN strains (detected by western blot), respectively.
To determine the mechanism underlying the increased cytotoxicity of the ΔrpoN strain, we examined the expression levels of tcdA and tcdB. RT-qPCR analysis revealed significantly increased mRNA expression of both toxin genes (Figures 4J,L). Western blot analysis confirmed a corresponding increase in TcdA and TcdB protein levels (Figures 4K,M). These findings indicate that deletion of rpoN leads to increased expression of TcdA and TcdB, resulting in enhanced cytotoxicity. This suggests that RpoN may act as a repressor of toxin gene expression in C. difficile.
3.5 The ΔrpoN mutant exhibits reduced biofilm formation
Biofilm formation contributes to C. difficile persistence in the gut and influences pathogenesis (Pantaléon et al., 2015; Chamarande et al., 2021). Screening for biofilm formation using a 24-well plate assay revealed that the ΔrpoN mutant formed less dense biofilms compared to the WT strain. Complementation of the mutant partially restored biofilm formation (Figures 5A,B).
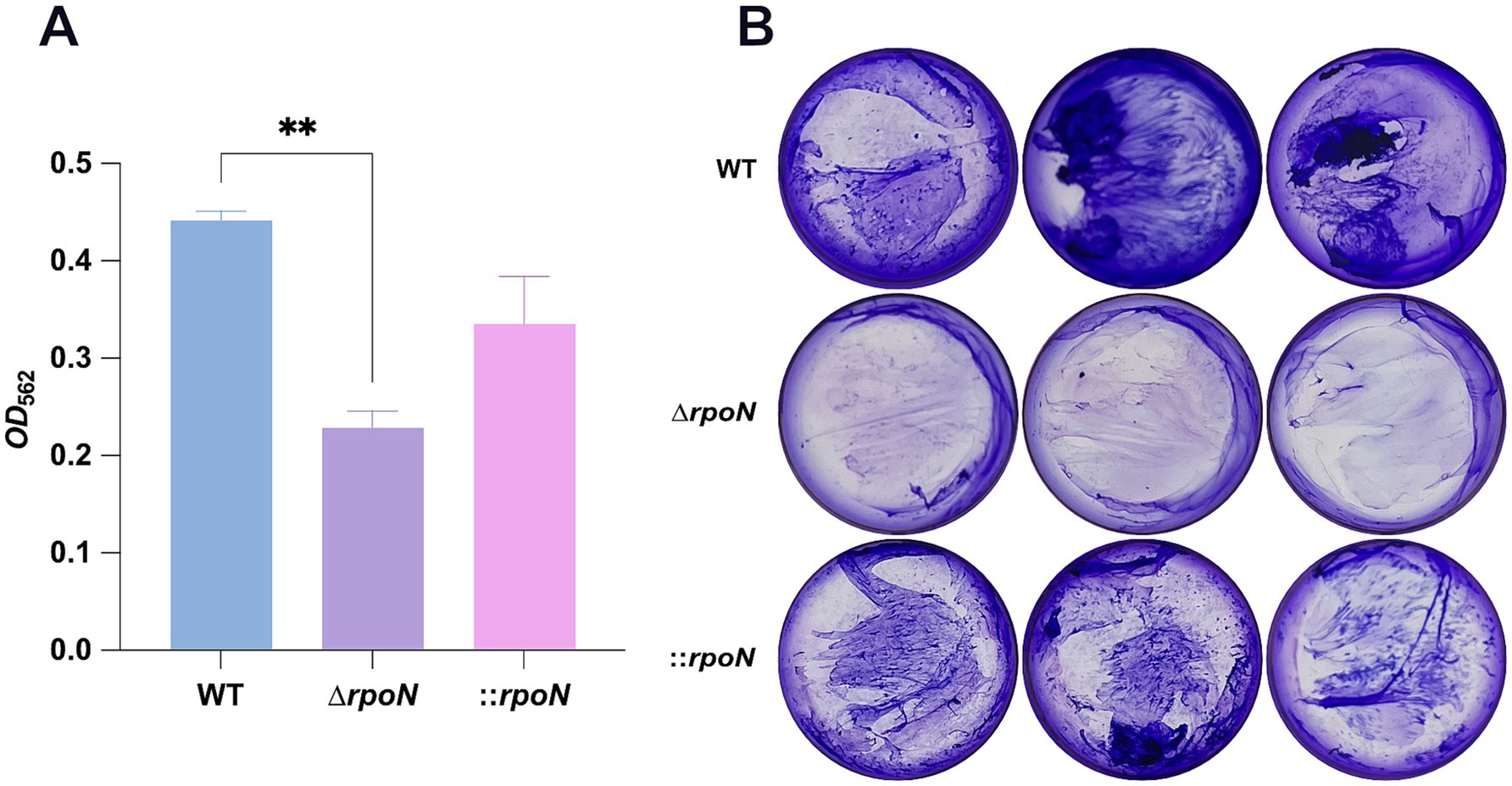
Figure 5. Biofilm assay for WT, ∆rpoN and ⸬rpoN strains. (A) Biofilm formation in WT, ∆rpoN and ⸬rpoN strains, which was assessed by measuring the retention of crystal violet by C. difficile biomass after 48 h; (B) Blue, purple, and pink bars represent the OD562 value of the WT, ∆rpoN and ⸬rpoN strains, which represent biofilm formation capability. The formed biofilm is reduced in ∆rpoN strain. Samples were grown in biological triplicate and compared using one-way ANOVA. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
3.6 Comparative transcriptomic analyses of the WT and ΔrpoN mutant gene expression profiles
To further investigate transcriptional changes in the ΔrpoN mutant compared to the WT strain, we performed comparative transcriptomic analyses during logarithmic growth. Overall, 600 genes were upregulated, 682 were downregulated, and 2,369 showed no significant change in expression (Figures 6A,B). As expected, expression of the rpoN gene was nearly undetectable.
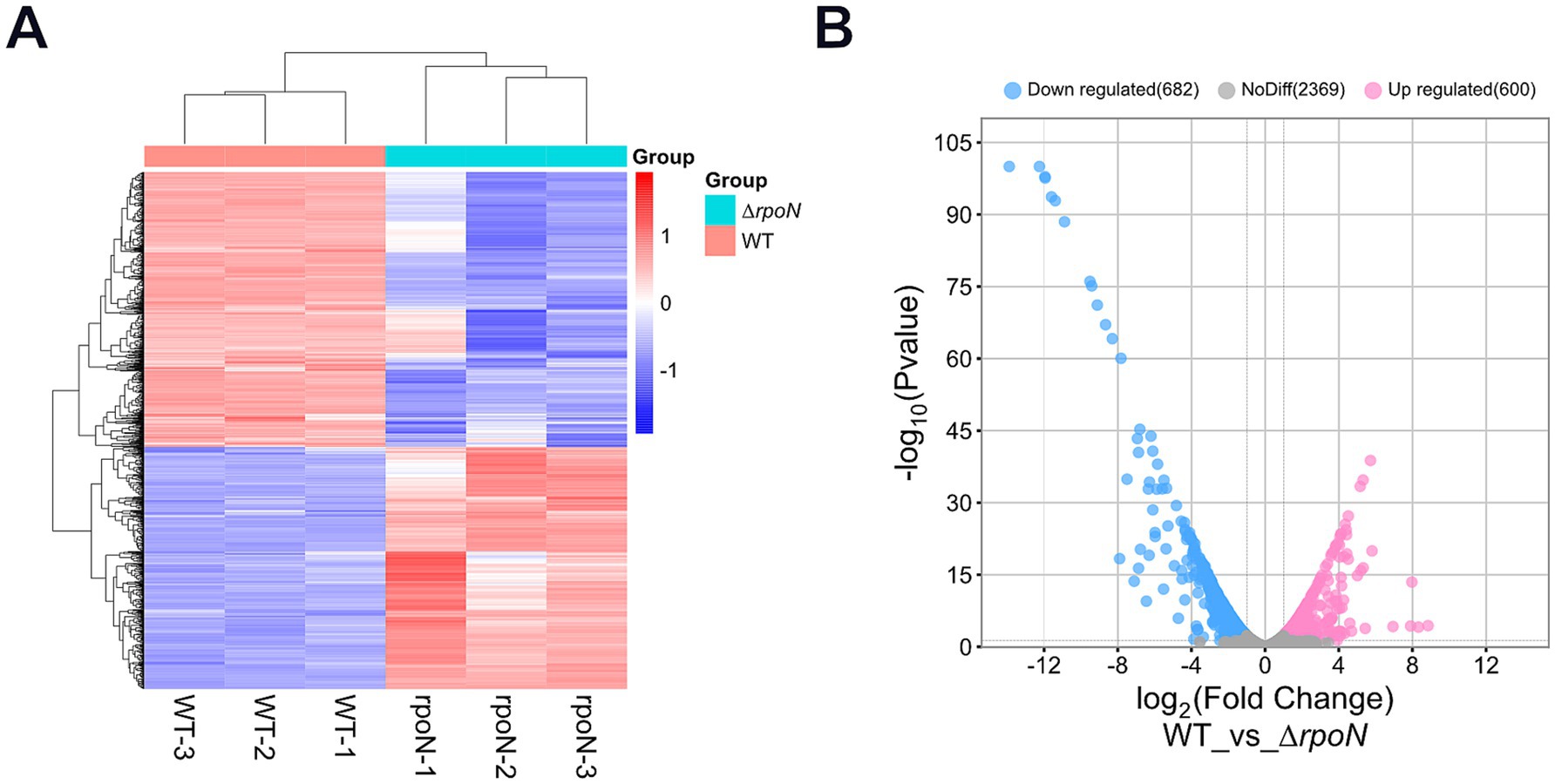
Figure 6. Transcriptome analysis. (A) Hierarchical clustering analysis of differentially expressed genes, with genes represented horizontally and samples vertically, each column representing a single sample; red indicates high-expression genes, and blue indicates low-expression genes; genes with high expression correlation are grouped together; (B) Volcano plot of differentially expressed genes, with the horizontal axis representing log2FoldChange and the vertical axis representing −log10(p-value). Blue dots on the left indicate downregulated genes in this group (682), pink dots on the right indicate upregulated genes (600), and grey dots represent genes with non-significant differential expression (2369).
The top 10 upregulated genes in the ΔrpoN mutant revealed significant enrichment in genes related to phosphate transport and transcriptional regulation. The phosphate transporter system (Pts), the transcription levels of phosphate ABC transporter permease (PstA), phosphate ABC transporter permease subunit (PtsC), phosphate ABC transporter ATP-binding protein (PstB), phosphate signaling complex protein (PhoU), and PTS sugar transporter subunit IIA were upregulated by 460, 322, 239, 123, and 56 times, respectively (Table 1). Factors related to transcriptional regulation, such as the helix-turn-helix transcriptional regulators (CD22140, CD22150) and the PRD domain-containing protein, were upregulated by 250, 40, and 43 times, respectively. Additionally, the hypothetical protein (CD02921) showed a transition from non-transcriptional to low-level transcription, and cytosine permease was upregulated by 53 times.
Conversely, the top 10 downregulated genes were largely associated with isovalerate metabolism and redox reactions. Genes encoding enzymes in the isovalerate pathway, such as isocaprenoyl-CoA-2-hydroxyisocaproate CoA-transferase (hadA), 2-hydroxyisocaproyl-CoA dehydratase activator (hadI), and subunits of (R)-2-hydroxyisocaproyl-CoA dehydratase (hadB), were downregulated by 15,237, 3,106, and 2,696 times, respectively. Electron transfer proteins, including electron transfer flavoprotein subunit alpha/FixB and beta/FixA family protein, were downregulated by 4,891 and 3,971 times, respectively. Acyl-CoA dehydrogenase was downregulated by 3,940 times. Proline racemase and D-proline reductase (dithiol) protein (PrdB) were downregulated by 729 and 682 times, respectively, and the TSUP family transporter by 554 times (Table 1).
3.7 Role of RpoN in virulence in a hamster model of CDI
To further evaluate the pathogenicity of the WT, ∆rpoN, and ⸬rpoN strains, we utilized a hamster infection model and assessed disease progression through phenotypic observations and colonic histopathology. Compared to the control (PBS) group, all experimental groups (WT, ∆rpoN, and ⸬rpoN) exhibited diarrhea, characterized by soft stool and wet tail. Concurrent with continuous antibiotic administration for 7 days (Chen et al., 2008; Geeraerts et al., 2015; Mooyottu et al., 2017), body weight gradually decreased in all infected groups. Following antibiotic discontinuation (days 8 to 10), body weight partially recovered in the experimental groups (Figures 7A,B).
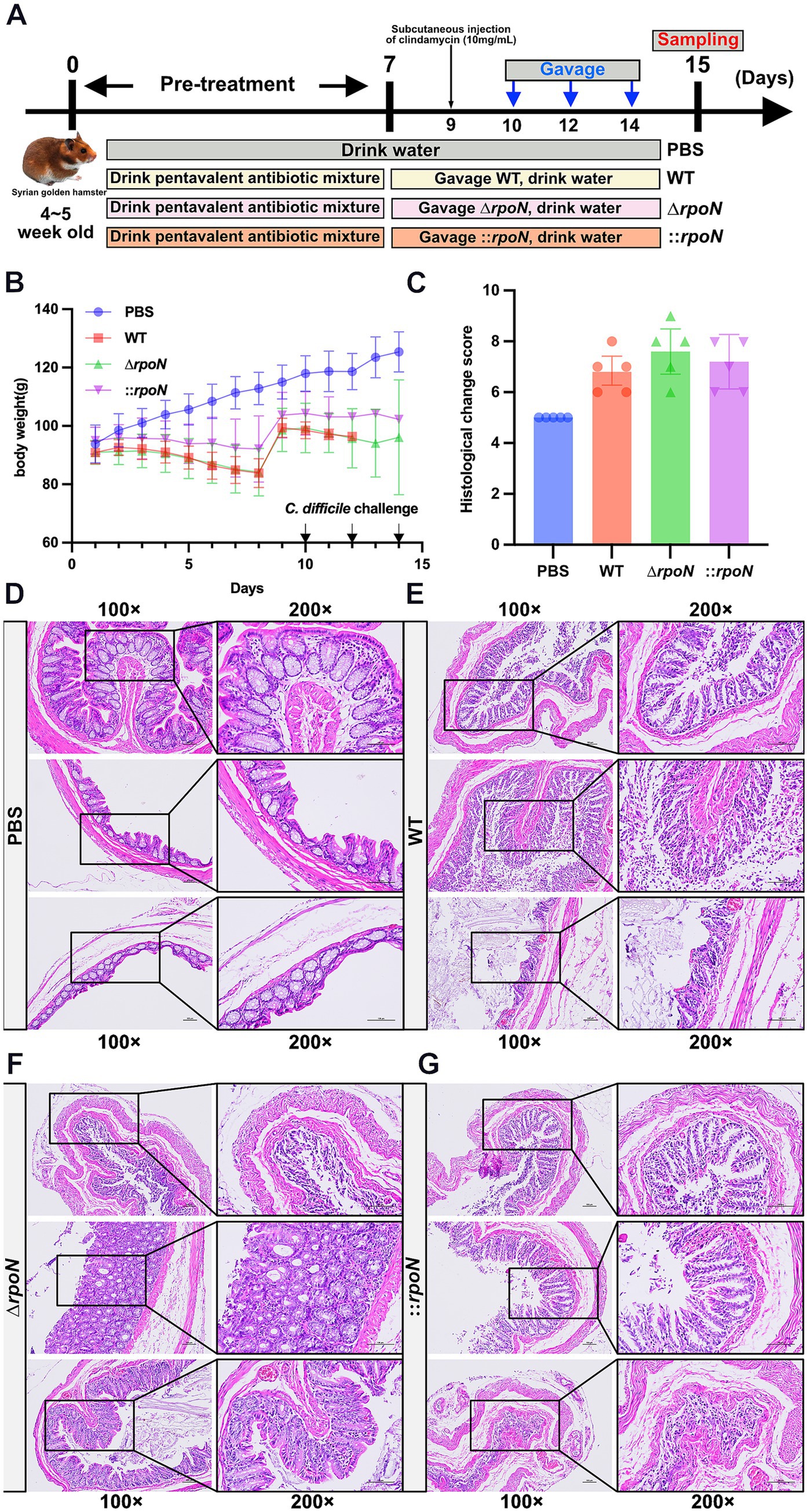
Figure 7. Construction of CDI animal model and the pathology of WT, ∆rpoN, and ⸬rpoN strains. (A) Schematic diagram of the construction process of CDI golden hamster model; (B) Daily weight change records of mice; (C) The H&E histopathological scores for the colon of the PBS, WT, ΔrpoN, and ⸬rpoN treated groups; (D–G) The H&E staining results for the PBS, WT, ΔrpoN, and ⸬rpoN groups. Data are expressed as mean ± SEM (n = 5 per group).
Histopathological analysis revealed differences in colonic tissue damage among the groups (Figure 7C). The control (PBS) group exhibited intact colonic morphology with clear layer structure, abundant glands, and regular arrangement of mucosal epithelial cells. Minimal damage was observed, with a few epithelial cells showing shrinkage and cytoplasmic staining, and a small number of inflammatory cells in the interstitium (Figure 7D). In the WT group, the colonic tissue retained clear layer structure, but mucosal epithelial cells were disorganized, with damage, detachment, and disintegration observed. Mild atrophy and loose arrangement of muscle layer cells were also noted, along with a few inflammatory cells (Figure 7E). The ∆rpoN group displayed the most severe colonic damage, characterized by loss of mucosal cells, disordered epithelial arrangement, extensive damage and disintegration of epithelial cells, sparse glands, incomplete or missing crypts, and obvious atrophy and sparse distribution of muscle layer cells. A large number of inflammatory cells were present in the interstitium (Figure 7F). Compared to the WT group, the ∆rpoN group showed increased crypt loss, epithelial surface damage and shedding, reduced mucus secretion, ulceration, and immune cell infiltration (Figures 7E,F). The ⸬rpoN group exhibited histopathology similar to the WT group, with clear layer structure, disorganized epithelial cells, some damage, detachment, and shrunken cell bodies, mild muscle layer atrophy, and a few inflammatory cells (Figure 7G). These findings suggest that the ∆rpoN strain exhibits increased pathogenicity in hamsters, correlating with the observed increase in toxin expression.
4 Discussion
The σ54 factor, essential for transcription in bacteria, forms a complex with RNA polymerase, enabling stable binding to promoter regions and initiating gene expression (Buck and Cannon, 1992). Consequently, σ54-dependent transcription factors exert broad regulatory effects on microbial physiology, influencing processes like transcription, carbon and nitrogen metabolism (Débarbouillé et al., 1991; Peng et al., 2015; Keffeler et al., 2021), toxin production, sporulation (Clark et al., 2022), motility (Douillard et al., 2009), biofilm formation (Da Silva Neto et al., 2008; Yang et al., 2020), secretion systems, and antibiotic synthesis (Liu et al., 2018). To investigate the role of σ54 in C. difficile, we generated an in-frame deletion mutant (∆rpoN) of the CD630 strain. Phenotypic analysis of this mutant revealed significant alterations, including reduced motility, autolysis rate, extracellular granule number, antibiotic resistance, H2S production, and glucose utilization. Notably, the ∆rpoN mutant exhibited significantly increased expression of the toxin genes tcdA and tcdB.
The increased expression of toxin genes in the ∆rpoN mutant aligns with previous observations in other C. difficile strains (Dubois et al., 2016), including BI-1 and CDC1 (Clark et al., 2022), and may result from competition between RpoN and the toxin-specific sigma factor TcdR for RNA polymerase, a mechanism consistent with the role of SigH (Saujet et al., 2011). Furthermore, upregulation of the AgrD1(CD27491)/AgrB1(CD27500) two-component system, approximately 2.78-fold and 2.29-fold respectively, likely contributes to the elevated expression of tcdA and tcdB (Ahmed et al., 2020). As expected, the increased production of TcdA and TcdB conferred enhanced virulence in a Vero cell model, and this increased pathogenicity was also confirmed in golden hamsters, suggesting that toxin gene expression is a primary driver of virulence. While other factors, such as adherence to intestinal epithelial cells and carbon source uptake, can also influence C. difficile pathogenicity (Janoir, 2016), our findings highlight the critical role of RpoN in regulating toxin production and subsequent virulence.
The ∆rpoN mutant of the CD630 strain also exhibited significantly increased autolysis rates, similar to observations in the CDC1 strain. However, unlike the BI-1 strain (Clark et al., 2022), it did not display cell aggregation. Interestingly, we found that the number of granules attached to the surface of the ΔrpoN strain was significantly reduced. Polyphosphate is widely distributed in both abiotic environments and living organisms. During microbial growth, phosphate is often actively acquired from external media and polymerized into longer-chain polymers via enzymatic reactions, serving as an intracellular storage depot. The formation of polyphosphate granules is intimately coupled to bacterial metabolic processes. Literature indicates that PG can bolster cell survival under stressful conditions, promote bacterial endospore formation (Lyratzakis et al., 2024), facilitate biofilm development (Zhang et al., 2005), and maintain outer membrane structure—for instance, a Pseudomonas aeruginosa PAO1 mutant deficient in ppk1 exhibits disrupted exopolymers and malformed outer membrane structure (Li et al., 2007). Therefore, the reduction in granule formation may lead to decreased stress resistance in C. difficile, which is consistent with our findings.
We observed significant upregulation of two autolysin genes in the ∆rpoN mutant: cell wall protein 19 (cwp19) and autolysin (acd). Cwp19 encodes a peptidoglycan-degrading enzyme with lytic transglycosylase activity (Wydau-Dematteis et al., 2018), while acd is an N-acetylglucosaminidase that hydrolyzes peptidoglycan bonds (Dhalluin et al., 2005). The increased expression of these autolysins likely contributes to the observed autolysis and may facilitate the release of toxins into the environment, potentially explaining the enhanced virulence observed in cell and animal models.
Consistent with previous reports demonstrating a role for rpoN in flagellum assembly regulation in other microorganisms (e.g., Liu et al., 2021, in Pseudomonas fluorescens), our KEGG analysis revealed a significant enrichment of flagellar assembly genes among downregulated cellular processes. Specifically, expression of genes encoding proteins involved in flagellum assembly, including fliL/M/N/Z/P/Q/R (CD02580/CD02700/CD02710/CD02590/CD02600/CD02610/CD02620), flhA/B (CD02630/CD02620), flgG (CD02680 and CD02690), fliC (CD02390), and fliA (CD02660), was significantly reduced. These genes encompass structural components, regulatory factors, and chemotaxis genes, suggesting that RpoN positively regulates flagellar assembly. Consequently, the reduced motility observed in the ∆rpoN mutant of C. difficile is directly linked to the downregulation of these flagellar genes.
Previous research has demonstrated that deletion of the rpoN gene can alter antibiotic susceptibility in various pathogenic bacteria. For instance, Pseudomonas fluorescens ∆rpoN mutants exhibit increased sensitivity to nine antibiotics (Liu et al., 2021), while a clinical Pseudomonas aeruginosa strain with a deleted rpoN gene shows increased susceptibility to several beta-lactams (Lloyd et al., 2019). Similarly, Borrelea burgdoeferi mutants with rpoN mutations display increased sensitivity to doxycycline (Sapi et al., 2016). However, the impact of rpoN deletion on antibiotic susceptibility is strain-dependent; in Pseudomonas aeruginosa, a ∆rpoN mutant in the logarithmic growth phase showed increased resistance to quinolones and carbapenems, linked to upregulation of pyoverdine expression (Viducic et al., 2007). In our study, the CD630 ∆rpoN mutant exhibited increased susceptibility to all seven antibiotics tested: β-lactams (amoxicillin, ampicillin, cefoxitin), nitroimidazoles (metronidazole), glycopeptides (vancomycin), fluoroquinolones (norfloxacin), and aminoglycosides (kanamycin). This increased susceptibility may be attributed to two primary mechanisms: (1) downregulation of antibiotic resistance genes, such as a 4.43-fold decrease in the expression of the vancomycin resistance gene VanT (CD16280, part of the VanC cluster, which influences vancomycin resistance), and (2) disruption of intracellular antioxidant mechanisms, evidenced by reduced synthesis of thiol compounds (CD00360, CD01180, CD07280, CD19170, CD20340, CD23800, CD23810, CD31740 oxidoreductase activity), potentially hindering the clearance of reactive oxygen species (ROS) generated under antibiotic (Albesa et al., 2004; Van Acker and Coenye, 2017). These findings suggest that RpoN represents a potential target for combination therapy in C. difficile infections.
Our transcriptomic analysis comparing WT and ∆rpoN mutants revealed significant downregulation of genes involved in glucose metabolism. Specifically, we observed a 6.69-fold and 7.68-fold decrease in expression of the sugar PTS system EIIA component (crr), and a 3.70-fold, 4.92-fold, 2.02-fold, 3.41-fold, 2.03-fold, 2.33-fold, 2.86-fold, 2.29-fold, and 2.01-fold reduction in ALDO (fructose-bisphosphate aldolase, class I), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), pgk (phosphoglycerate kinase), pyk (pyruvate kinase), aceE (pyruvate dehydrogenase E1 component), dlat (pyruvate dehydrogenase E2 component), dld (dihydrolipoyl dehydrogenase), and adh (alcohol dehydrogenase), respectively. This widespread downregulation likely contributes to the significantly reduced glucose utilization observed in the ∆rpoN mutant, suggesting that RpoN positively regulates glucose metabolism. Unlike observations in Gram-positive bacteria, regulation of nitrogen metabolism by RpoN was less prominent in C. difficile. We detected only modest upregulation of hcp (2.53-fold) and glnA (2.47-fold). Regarding cysteine metabolism, expression of cysK (cysteine synthase) and cysE (serine O-acetyltransferase) was dramatically reduced (68.68-fold and 57.33-fold, respectively) in the ∆rpoN mutant, leading to decreased L-cysteine synthesis. Given that L-cysteine can be converted to pyruvate and hydrogen sulfide (H2S), the observed reduction in RpoN-mediated cysteine synthesis may explain the decreased H2S production.
In conclusion, we constructed and characterized an in-frame deletion mutant of the rpoN gene in C. difficile through phenotypic analysis and RNA-Seq. Our results demonstrate that RpoN significantly impacts C. difficile motility, antibiotic susceptibility, toxin production, and virulence by regulating the expression of numerous genes. These findings establish RpoN as a global regulator in C. difficile and suggest it as a promising therapeutic target for reducing virulence and antibiotic resistance.
Data availability statement
The RNA-seq raw data (RNA-seq of Clostridioides difficile ΔrpoN mutant against wild-type control) was deposited in the ArrayExpress database (https://www.ebi.ac.uk/biostudies/arrayexpress) under the accession number of E-MTAB-14512.
Ethics statement
The animal study was approved by Ethics committee of Guizhou Medical University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YY: Investigation, Writing – original draft, Writing – review & editing. TH: Investigation, Writing – original draft, Writing – review & editing. JY: Investigation, Writing – original draft, Writing – review & editing. RS: Investigation, Writing – original draft, Writing – review & editing. LS: Investigation, Writing – original draft, Writing – review & editing. PL: Investigation, Writing – original draft, Writing – review & editing. YL: Investigation, Writing – original draft, Writing – review & editing. WM: Investigation, Writing – original draft, Writing – review & editing. JL: Supervision, Writing – original draft, Writing – review & editing. ZG: Supervision, Writing – original draft, Writing – review & editing. GC: Validation, Writing – original draft, Writing – review & editing. XQ: Supervision, Writing – original draft, Writing – review & editing. WH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Nos. 32170134, 32160015, 32460046); Excellent Young Talents Plan of Guizhou Medical University (2022)101.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1569627/full#supplementary-material
References
Ahmed, U. K. B., Shadid, T. M., Larabee, J. L., and Ballard, J. D. (2020). Combined and distinct roles of Agr proteins in Clostridioides difficile 630 sporulation, motility, and toxin production. mBio 11, e03190–e03120. doi: 10.1128/mBio.03190-20
Albesa, I., Becerra, M. C., Battán, P. C., and Páez, P. L. (2004). Oxidative stress involved in the antibacterial action of different antibiotics. Biochem. Biophys. Res. Commun. 317, 605–609. doi: 10.1016/j.bbrc.2004.03.085
Basic, A., Blomqvist, S., Carlén, A., and Dahlén, G. (2015). Estimation of bacterial hydrogen sulfide production in vitro. J. Oral Microbiol. 7:28166. doi: 10.3402/jom.v7.28166
Borukhov, S., and Nudler, E. (2003). RNA polymerase holoenzyme: structure, function and biological implications. Curr. Opin. Microbiol. 6, 93–100. doi: 10.1016/S1369-5274(03)00036-5
Bouillaut, L., Self, W. T., and Sonenshein, A. L. (2013). Proline-dependent regulation of Clostridium difficile Stickland metabolism. J. Bacteriol. 195, 844–854. doi: 10.1128/JB.01492-12
Buck, M., and Cannon, W. (1992). Specific binding of the transcription factor sigma-54 to promoter DNA. Nature 358:422. doi: 10.1038/358422a0
Cañadas, I. C., Groothuis, D., Zygouropoulou, M., Rodrigues, R., and Minton, N. P. (2019). RiboCas: a universal CRISPR-based editing tool for Clostridium. ACS Synth. Biol. 8, 1379–1390. doi: 10.1021/acssynbio.9b00075
CDC (2024). Antimicrobial resistance threats in the United States, 2021–2022. Antimicrobial resistance. Available online at: https://www.cdc.gov/antimicrobial-resistance/data-research/threats/update-2022.html (Accessed October 20, 2024).
Chamarande, J., Cunat, L., Caillet, C., Mathieu, L., Duval, J. F. L., Lozniewski, A., et al. (2021). Surface properties of Parabacteroides distasonis and impacts of stress-induced molecules on its surface adhesion and biofilm formation capacities. Microorganisms 9:1602. doi: 10.3390/microorganisms9081602
Chen, X., Katchar, K., Goldsmith, J. D., Nanthakumar, N., Cheknis, A., Gerding, D. N., et al. (2008). A mouse model of Clostridium difficile–associated disease. Gastroenterology 135, 1984–1992. doi: 10.1053/j.gastro.2008.09.002
Chu, M., Mallozzi, M. J. G., Roxas, B. P., Bertolo, L., Monteiro, M. A., Agellon, A., et al. (2016). A Clostridium difficile Cell Wall Glycopolymer locus influences bacterial shape, polysaccharide production and virulence. PLoS Pathog 12:e1005946. doi: 10.1371/journal.ppat.1005946
Clark, A. E., Adamson, C. C., Carothers, K. E., Roxas, B. A. P., Viswanathan, V. K., and Vedantam, G. (2022). The alternative sigma factor SigL influences Clostridioides difficile toxin production, sporulation, and cell surface properties. Front. Microbiol. 13:871152. doi: 10.3389/fmicb.2022.871152
Da Silva Neto, J. F., Koide, T., Abe, C. M., Gomes, S. L., and Marques, M. V. (2008). Role of σ54 in the regulation of genes involved in type I and type IV pili biogenesis in Xylella fastidiosa. Arch. Microbiol. 189, 249–261. doi: 10.1007/s00203-007-0314-x
Danson, A. E., Jovanovic, M., Buck, M., and Zhang, X. (2019). Mechanisms of σ54-dependent transcription initiation and regulation. J. Mol. Biol. 431, 3960–3974. doi: 10.1016/j.jmb.2019.04.022
Débarbouillé, M., Martin-Verstraete, I., Kunst, F., and Rapoport, G. (1991). The Bacilius subtilis sigL gene encodes an equivalent of c 54 from gram-negative bacteria. Proc. Natl. Acad. Sci. USA 88, 9092–9096. doi: 10.1073/pnas.88.20.9092
Dhalluin, A., Bourgeois, I., Pestel-Caron, M., Camiade, E., Raux, G., Courtin, P., et al. (2005). Acd, a peptidoglycan hydrolase of Clostridium difficile with N-acetylglucosaminidase activity. Microbiology 151, 2343–2351. doi: 10.1099/mic.0.27878-0
Douillard, F. P., Ryan, K. A., Hinds, J., and O’Toole, P. W. (2009). Effect of FliK mutation on the transcriptional activity of the σ 54 sigma factor RpoN in Helicobacter pylori. Microbiology 155, 1901–1911. doi: 10.1099/mic.0.026062-0
Dubois, T., Dancer-Thibonnier, M., Monot, M., Hamiot, A., Bouillaut, L., Soutourina, O., et al. (2016). Control of Clostridium difficile physiopathology in response to cysteine availability. Infect. Immun. 84, 2389–2405. doi: 10.1128/IAI.00121-16
Ehsaan, M., Kuehne, S. A., and Minton, N. P. (2016). “Clostridium difficile genome editing using pyrE alleles” in Clostridium difficile. eds. A. P. Roberts and P. Mullany (New York, NY: Springer New York), 35–52.
Fachi, J. L., Vinolo, M. A. R., and Colonna, M. (2024). Reviewing the Clostridioides difficile mouse model: insights into infection mechanisms. Microorganisms 12:273. doi: 10.3390/microorganisms12020273
Finn, E., Andersson, F. L., and Madin-Warburton, M. (2021). Burden of Clostridioides difficile infection (CDI) – a systematic review of the epidemiology of primary and recurrent CDI. BMC Infect. Dis. 21:456. doi: 10.1186/s12879-021-06147-y
Geeraerts, S., Ducatelle, R., Haesebrouck, F., and Van Immerseel, F. (2015). Bacillus amyloliquefaciens as prophylactic treatment for C lostridium difficile-associated disease in a mouse model. J. Gastroenterol.Hepatol. 30, 1275–1280. doi: 10.1111/jgh.12957
Gu, H., Yang, Y., Wang, M., Chen, S., Wang, H., Li, S., et al. (2018). Novel cysteine Desulfidase CdsB involved in releasing cysteine repression of toxin synthesis in Clostridium difficile. Front. Cell. Infect. Microbiol. 7:531. doi: 10.3389/fcimb.2017.00531
Hong, W., Zhang, J., Cui, G., Wang, L., and Wang, Y. (2018). Multiplexed CRISPR-Cpf1-mediated genome editing in Clostridium difficile toward the understanding of pathogenesis of C. difficile infection. ACS Synth. Biol. 7, 1588–1600. doi: 10.1021/acssynbio.8b00087
Janoir, C. (2016). Virulence factors of Clostridium difficile and their role during infection. Anaerobe 37, 13–24. doi: 10.1016/j.anaerobe.2015.10.009
Jorgensen, J. H., and Ferraro, M. J. (2009). Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis. 49, 1749–1755. doi: 10.1086/647952
Keffeler, E. C., Iyer, V. S., Parthasarathy, S., Ramsey, M. M., Gorman, M. J., Barke, T. L., et al. (2021). Influence of the alternative sigma factor RpoN on global gene expression and carbon catabolism in Enterococcus faecalis V583. mBio 12, e00380–e00321. doi: 10.1128/mBio.00380-21
Li, L., Rao, N. N., and Kornberg, A. (2007). Inorganic polyphosphate essential for lytic growth of phages P1 and fd. Proc. Natl. Acad. Sci. USA 104, 1794–1799. doi: 10.1073/pnas.0610763104
Liu, Y., Shi, H., Wang, Z., Huang, X., and Zhang, X. (2018). Pleiotropic control of antibiotic biosynthesis, flagellar operon expression, biofilm formation, and carbon source utilization by RpoN in Pseudomonas protegens H78. Appl. Microbiol. Biotechnol. 102, 9719–9730. doi: 10.1007/s00253-018-9282-0
Liu, X., Ye, Y., Zhu, Y., Wang, L., Yuan, L., Zhu, J., et al. (2021). Involvement of RpoN in regulating motility, biofilm, resistance, and spoilage potential of Pseudomonas fluorescens. Front. Microbiol. 12:641844. doi: 10.3389/fmicb.2021.641844
Lloyd, M. G., Lundgren, B. R., Hall, C. W., Gagnon, L. B.-P., Mah, T.-F., Moffat, J. F., et al. (2017). Targeting the alternative sigma factor RpoN to combat virulence in Pseudomonas aeruginosa. Sci. Rep. 7:12615. doi: 10.1038/s41598-017-12667-y
Lloyd, M. G., Vossler, J. L., Nomura, C. T., and Moffat, J. F. (2019). Blocking RpoN reduces virulence of Pseudomonas aeruginosa isolated from cystic fibrosis patients and increases antibiotic sensitivity in a laboratory strain. Sci. Rep. 9:6677. doi: 10.1038/s41598-019-43060-6
Lyratzakis, A., Kalogerakis, M., Polymerou, K., Spyros, A., and Tsiotis, G. (2024). Characterization of the intracellular polyphosphate granules of the phototrophic green sulfur bacterium Chlorobaculum tepidum. Biochim. Biophys. Acta Gen. Subj. 1868:130718. doi: 10.1016/j.bbagen.2024.130718
MacDonald, I. C., Seamons, T. R., Emmons, J. C., Javdan, S. B., and Deans, T. L. (2021). Enhanced regulation of prokaryotic gene expression by a eukaryotic transcriptional activator. Nat. Commun. 12:4109. doi: 10.1038/s41467-021-24434-9
Mazumder, A., and Kapanidis, A. N. (2019). Recent advances in understanding σ70-dependent transcription initiation mechanisms. J. Mol. Biol. 431, 3947–3959. doi: 10.1016/j.jmb.2019.04.046
McClure, R., Balasubramanian, D., Sun, Y., Bobrovskyy, M., Sumby, P., Genco, C. A., et al. (2013). Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 41:e140. doi: 10.1093/nar/gkt444
Meouche, I. E., and Peltier, J. (2018). Toxin release mediated by the novel autolysin Cwp19 in Clostridium difficile. Microb. Cell 5, 421–423. doi: 10.15698/mic2018.09.648
Mooyottu, S., Flock, G., Upadhyay, A., Upadhyaya, I., Maas, K., and Venkitanarayanan, K. (2017). Protective effect of carvacrol against gut dysbiosis and Clostridium difficile associated disease in a mouse model. Front. Microbiol. 8:625. doi: 10.3389/fmicb.2017.00625
Nie, X., Dong, W., and Yang, C. (2019). Genomic reconstruction of σ54 regulons in Clostridiales. BMC Genomics 20:565. doi: 10.1186/s12864-019-5918-4
Noori Goodarzi, N., Fereshteh, S., Azizi, O., Rahimi, H., Bolourchi, N., and Badmasti, F. (2022). Subtractive genomic approach toward introduction of novel immunogenic targets against Clostridioides difficile: thinking out of the box. Microb. Pathog. 162:105372. doi: 10.1016/j.micpath.2021.105372
Paget, M. (2015). Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomol. Ther. 5, 1245–1265. doi: 10.3390/biom5031245
Pantaléon, V., Soavelomandroso, A. P., Bouttier, S., Briandet, R., Roxas, B., Chu, M., et al. (2015). The Clostridium difficile protease Cwp84 modulates both biofilm formation and cell-surface properties. PLoS One 10:e0124971. doi: 10.1371/journal.pone.0124971
Peng, Q., Wang, G., Liu, G., Zhang, J., and Song, F. (2015). Identification of metabolism pathways directly regulated by sigma54 factor in Bacillus thuringiensis. Front. Microbiol. 6:407. doi: 10.3389/fmicb.2015.00407
Purdy, D., O’Keeffe, T. A. T., Elmore, M., Herbert, M., McLeod, A., Bokori-Brown, M., et al. (2002). Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 46, 439–452. doi: 10.1046/j.1365-2958.2002.03134.x
Radakovits, R., Jinkerson, R. E., Fuerstenberg, S. I., Tae, H., Settlage, R. E., Boore, J. L., et al. (2012). Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropsis gaditana. Nat. Commun. 3:686. doi: 10.1038/ncomms1688
Rukit, J., Boonmee, A., Kijpornyongpan, T., Tulsook, K., Baranyi, J., and Chaturongakul, S. (2022). Roles of alternative sigma factors in invasion and growth characteristics of listeria monocytogenes 10403S into human epithelial colorectal adenocarcinoma Caco-2 cell. Front. Microbiol. 13:901484. doi: 10.3389/fmicb.2022.901484
Sapi, E., Theophilus, P. A. S., Pham, T. V., Burugu, D., and Luecke, D. F. (2016). Effect of RpoN, RpoS and LuxS pathways on the biofilm formation and antibiotIc sensitivity of Borrelia burgdorferi. EuJMI 6, 272–286. doi: 10.1556/1886.2016.00026
Saujet, L., Monot, M., Dupuy, B., Soutourina, O., and Martin-Verstraete, I. (2011). The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J. Bacteriol. 193, 3186–3196. doi: 10.1128/JB.00272-11
Soutourina, O., Dubois, T., Monot, M., Shelyakin, P. V., Saujet, L., Boudry, P., et al. (2020). Genome-wide transcription start site mapping and promoter assignments to a sigma factor in the human Enteropathogen Clostridioides difficile. Front. Microbiol. 11:1939. doi: 10.3389/fmicb.2020.01939
Steindl, G., Fiedler, A., Huhulescu, S., Wewalka, G., and Allerberger, F. (2015). Effect of airborne hydrogen peroxide on spores of Clostridium difficile. Wien. Klin. Wochenschr. 127, 421–426. doi: 10.1007/s00508-014-0682-6
Sun, Z., Yakhnin, A. V., FitzGerald, P. C., Mclntosh, C. E., and Kashlev, M. (2021). Nascent RNA sequencing identifies a widespread sigma70-dependent pausing regulated by Gre factors in bacteria. Nat. Commun. 12:906. doi: 10.1038/s41467-021-21150-2
Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Kiryutin, B., Koonin, E. V., et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. doi: 10.1186/1471-2105-4-41
Van Acker, H., and Coenye, T. (2017). The role of reactive oxygen species in antibiotic-mediated killing of Bacteria. Trends Microbiol. 25, 456–466. doi: 10.1016/j.tim.2016.12.008
Viducic, D., Murakami, K., Amoh, T., Ono, T., and Miyake, Y. (2016). RpoN modulates Carbapenem tolerance in Pseudomonas aeruginosa through Pseudomonas quinolone signal and PqsE. Antimicrob. Agents Chemother. 60, 5752–5764. doi: 10.1128/AAC.00260-16
Viducic, D., Ono, T., Murakami, K., Katakami, M., Susilowati, H., and Miyake, Y. (2007). rpoN gene of Pseudomonas aeruginosa alters its susceptibility to quinolones and Carbapenems. Antimicrob. Agents Chemother. 51, 1455–1462. doi: 10.1128/AAC.00348-06
Wydau-Dematteis, S., El Meouche, I., Courtin, P., Hamiot, A., Lai-Kuen, R., Saubaméa, B., et al. (2018). Cwp19 is a novel lytic Transglycosylase involved in stationary-phase autolysis resulting in toxin release in Clostridium difficile. MBio 9, e00648–e00618. doi: 10.1128/mBio.00648-18
Yang, Y., Darbari, V. C., Zhang, N., Lu, D., Glyde, R., Wang, Y.-P., et al. (2015). Structures of the RNA polymerase-σ 54 reveal new and conserved regulatory strategies. Science 349, 882–885. doi: 10.1126/science.aab1478
Yang, B., Nie, X., Gu, Y., Jiang, W., and Yang, C. (2020). Control of solvent production by sigma-54 factor and the transcriptional activator AdhR in Clostridium beijerinckii. Microb. Biotechnol. 13, 328–338. doi: 10.1111/1751-7915.13505
Yu, C., Nguyen, D.-P., Yang, F., Shi, J., Wei, Y., Tian, F., et al. (2021). Transcriptome analysis revealed overlapping and special regulatory roles of RpoN1 and RpoN2 in motility, virulence, and growth of Xanthomonas oryzae pv. Oryzae. Front. Microbiol. 12:653354. doi: 10.3389/fmicb.2021.653354
Zhang, H., Gómez-García, M. R., Brown, M. R. W., and Kornberg, A. (2005). Inorganic polyphosphate in Dictyostelium discoideum: influence on development, sporulation, and predation. Proc. Natl. Acad. Sci. USA 102, 2731–2735. doi: 10.1073/pnas.0500023102
Zhou, H., He, J., Liu, R., Cheng, J., Yuan, Y., Mao, W., et al. (2024). Microenvironment-responsive metal-phenolic network release platform with ROS scavenging, anti-pyroptosis, and ECM regeneration for intervertebral disc degeneration. Bioactive Mater. 37, 51–71. doi: 10.1016/j.bioactmat.2024.02.036
Keywords: Clostridioides difficile, sigma-54, rpoN, antibiotic resistance, motility, toxin production, pathogenesis
Citation: Yang Y, Huang T, Yang J, Shao R, Shu L, Ling P, Lu Y, Ma W, Liao J, Guan Z, Cui G, Qi X and Hong W (2025) The sigma factor σ54 (rpoN) functions as a global regulator of antibiotic resistance, motility, metabolism, and virulence in Clostridioides difficile. Front. Microbiol. 16:1569627. doi: 10.3389/fmicb.2025.1569627
Edited by:
Muhammad Kamruzzaman, Westmead Institute for Medical Research, AustraliaReviewed by:
Duolong Zhu, Baylor College of Medicine, United StatesVineet Kumar, The University of Texas at Austin, United States
Copyright © 2025 Yang, Huang, Yang, Shao, Shu, Ling, Lu, Ma, Liao, Guan, Cui, Qi and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Hong, aG9uZ3dlaV8yMDE1QGhvdG1haWwuY29t; Xiaolan Qi, eGlhb2xhbjc2QDE2My5jb20=
†These authors have contributed equally to this work
 Ying Yang
Ying Yang Tingyu Huang1†
Tingyu Huang1† Guzhen Cui
Guzhen Cui Xiaolan Qi
Xiaolan Qi Wei Hong
Wei Hong