Abstract
Introduction:
Pseudomonas aeruginosa poses challenges in clinical and environmental contexts due to its capacity to colonize natural ecosystems and antibiotic resistance. This study characterized P. aeruginosa harboured by Diptera flies collected from illegal residential dumping sites and livestock (cattle, sheep, and goats) kraals in Potchefstroom, South Africa.
Methods:
The P. aeruginosa isolates were characterized using classical microbiological tests and species-specific gyrase B gene PCR assay. Antibiotic resistance (AR) was assessed on the isolates using disc diffusion assay (DDA). Additionally, PCR screened six virulence genes (exoS, plcN, plcH, toxA, lasB, and algD) among the isolates. Whole genome sequencing (WGS) was employed to confirm the identity and determine antibiotic resistance genes (ARGs) on selected isolates.
Results:
Culture-based and molecular assays showed that P. aeruginosa is prevalent in Diptera flies (Hemipyrellia spp., Synthesiomya spp., Chrysomya spp., Sarchophagidae spp., and Tabanus spp.) from livestock kraals (75%; n = 36/48) and dumping sites (48%; n = 23/48). The most detected virulent gene among the isolates was exoS (96.6%), followed by plcN and algD genes (83.1%), lasB (81.4%), toxA (76.3%), and plcH (47.5%). All P. aeruginosa isolates were resistant to metronidazole, sulphamethoxazole, cefazolin and amoxicillin based on DDA. The sulfonamide resistance sulI gene (88.1%) was the most detected ARG from the P. aeruginosa isolates, followed by acc(3)-IV (80.6%) coding for aminoglycoside. WGS revealed that P. aeruginosa isolates belong to the sequence type (ST3808), which is multidrug-resistant and contains ARGs for fosfomycin (fosA), ampicillin (blaOXA-50), chloramphenicol (catB7), beta-lactamase (blaPAO), and aminoglycoside (aph(3’)-IIb).
Discussion:
This study isolated ESBL-producing P. aeruginosa from various Diptera fly species collected from livestock kraals and residential dumping sites. This bacterium is important to “One Health” due to its multidrug resistance character and zoonotic nature. As a result, it requires consolidated control and management policies from the environmental, veterinary, and human health sectors.
1 Introduction
Diptera flies are among the most diverse groups of organisms that consist of 128 families and 124,000 species and serve as biological or mechanical vectors that carry and transmit multiple pathogenic bacterial species (Skevington and Dang, 2002; Taioe et al., 2017). Among the present bacterial species in Diptera flies is Pseudomonas aeruginosa, which can spread to other ecosystems, such as habitats for animals, humans, and the environment (Pang et al., 2019). It is a versatile, opportunistic bacterium that infrequently affects healthy individuals and is commonly associated with ventilator-associated pneumonia and nosocomial infections (Pang et al., 2019). Due to their mobility, eating, and excretory activities, Diptera flies can spread P. aeruginosa throughout settings high in organic waste (Boiocchi et al., 2019). This enhances the bacteria’s capacity to colonize various ecological niches, such as the surfaces of plants, water, and soil. The P. aeruginosa can be a major microbial participant in habitats where Diptera flies contribute to decomposition, helping to break down organic materials and affecting nutrient cycling processes (Pang et al., 2019).
It is well recognized that P. aeruginosa possesses a wide range of virulence factors, which enhance its pathogenicity and capacity to cause infections. Elastases (lasA and lasB), which break down elastin and other host proteins and cause tissue damage and invasion, are among the virulence factors (Faraji et al., 2016; Cathcart et al., 2011). Exotoxins also prevent protein synthesis in host cells by ADP-ribosylation of elongation factor-2, which results in cell death. In addition, other virulence factors include the Type VI Secretion System (T6SS), which influences bacterial competition and virulence, and the Type III Secretion System (T3SS), which tampers with cellular functions and immune responses. The coordination of various virulence factors’ expression, including biofilm formation and toxin production, is facilitated by the quorum sensing systems (Rhl and Las) (Deep et al., 2011).
Pseudomonas aeruginosa is known to pose a serious risk to both human and animal health due to its high level of intrinsic resistance to several different classes of antibiotics (Lister et al., 2009; Santajit and Indrawattana, 2016). Carbapenem-resistant P. aeruginosa produces extended-spectrum beta-lactamases (ESBLs) and metallo-beta-lactamases (MBLs) enzymes, causing resistance to various beta-lactam antibiotics (Paterson and Bonomo, 2005; Neyestanaki et al., 2014). Beta-lactams’ binding affinity may be decreased by mutations in Penicillin-Binding Proteins (PBPs) (Pandey and Cascella, 2021). Additionally, aminoglycosides are modified by enzymes that change them, making them ineffective. The resistance of P. aeruginosa to fluoroquinolones, tetracycline, and chloramphenicol, as well as the poor permeability of the outer membrane, are highly influenced by efflux pumps (Li et al., 1994).
Studies examining P. aeruginosa in Diptera flies are scarce, particularly in livestock and waste environmental settings. It is essential to comprehend these relationships and P. aeruginosa’s unique function in order to comprehend the traits of antibiotic resistance in this organism. Therefore, this study investigated the occurrence of P. aeruginosa and antimicrobial resistance patterns from Diptera flies collected in Ikageng township illegal residential dumping sites and livestock kraals in Matlwang village of Potchefstroom City in the North-West province of South Africa.
2 Materials and methods
2.1 Collection of Diptera flies
Diptera flies were collected from the illegal residential dumping site at Ikageng township (Coordinates: 26.7257° S; 27.0500° E) and from the Matlwang village livestock kraals (Coordinates: 26.4451° S; 26.5552° E) of Potchefstroom town in the North-West province of South Africa (Figure 1). Redtop disposable fly catcher traps were deployed for 14 days, and the flies were harvested every 2 days. Fly traps were set at two Ikageng illegal residential dumping sites which are 120 m from each other, while fly traps (~60 m from each other) were set in three livestock kraals in Matlwang village.
Figure 1
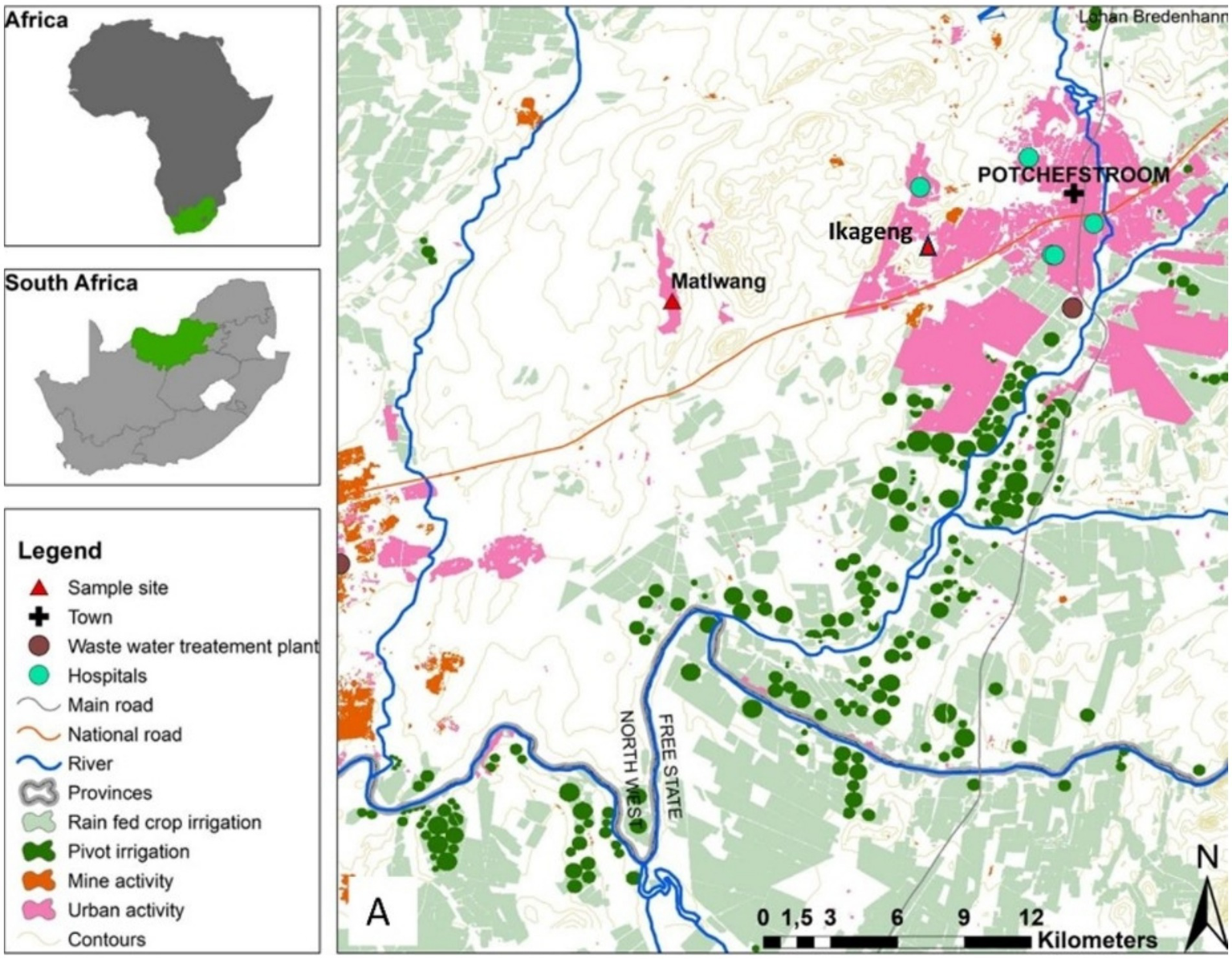
Map of the North West province in South Africa indicating Ikageng residential dumping site and Matlwang community area.
2.2 Identification of Diptera flies using microscopy
Diptera flies were separated according to morpho-species identification as described and then put in a 9:1 ratio of phosphate buffered saline (PBS) to glycerol. The Nikon SMZ1500 stereoscopic zoom microscope was utilized to identify the Diptera flies based on their size, shape, and color.
2.3 DNA extraction from Diptera flies
DNA was extracted from fly specimens using the DNeasy® Blood and Tissue kit according to the manufacturer’s instructions (QIAGEN, Germany). The quality and quantity of the extracted DNA was analyzed using spectrophotometry with a NanoDrop ND-100 system (NanoDrop Technologies, Wilmington, United States). The extracted DNA was stored at −20°C until used for PCR analysis.
2.4 Genetic identification of the flies
The Cytochrome Oxidase 1 (CO1) gene PCR assay was conducted for the identification of fly species using the following primers: LCO1: 5′-GGTCAACAAATCATAAAGATATTGG-3′ and HCO1: 5′-TAAACTTCAGGGTGACCAAAAATCA-3′. Each PCR reaction consisted of a total reaction of 25 μL consisting of 12.5 μL OneTaq® Quick-Load 2X Master Mix with Standard Buffer (0.4 mM dATP, 0.4 mM dCTP, 0.4 mM dGTP, 0.4 mMdTTP, 4 mM MgCl2, and loading buffer) (New England BioLabs, United States), 8.5 μL of nuclease-free water, 2 μL (5 ng/μl) of the template DNA, and 1 μL (10 μM) of each oligonucleotide primer. The PCR mixture was incubated on a ProFlex PCR system (Applied Biosystems, Singapore) with the following conditions: Initial denaturation at 94°C for 2 min, then 35 cycles of 94°C for 30 s, 47°C for 30 s and 72°C for 60 s, and final elongation at 72°C for 7 min (Monyama et al., 2023). Thereafter, PCR amplicons were sequenced with the BigDye Terminator sequencing kit v3.1 (Applied Biosystems) on the SeqStudio genetic analyzer at UESM sequencing facility of North-West University.
The sequenced gene were edited using FinchTV (Treves, 2010). The forward and reverse strands were linked together using BioEdit (Tippmann, 2004). The fly CO1 gene sequences were subjected to nucleotide Basic Local Alignment Search Tool (BLASTn) for confirmation of identical sequences available on the GenBank of the National Centre for Biotechnology Information (NCBI). Phylogenetic trees were created using maximum likelihood on Molecular Evolutionary Genetics Analysis (MEGAX) software (Kumar et al., 2018). A bootstrap value of 1,000 was employed to guarantee the tree’s confidence.
2.5 Culture isolation and biochemical characterization of Pseudomonas aeruginosa
A total of 96 pooled representative samples (5 flies per pool), comprising 45 from dumping sites and 51 from livestock kraals, were used in this study. These were randomly selected from the total collected fly population to ensure manageable yet statistically representative processing for downstream microbial analysis. Only flies morphologically identified as commonly present at both sites were included to ensure ecological comparability and minimize species-specific bias. Unknown or site-exclusive fly species were excluded to maintain consistency in species-related pathogen carriage assessment. To reduce surface contamination, the exterior surface of the flies was disinfected with 70% ethanol for 1 h. The flies were then crushed in phosphate buffer solution (PBS), followed by a 20 s vortexing. Thereafter, a volume of 1 mL of the PBS-fly homogenate was placed in 9 mL buffered peptone water (BPW) and incubated for 24 h with agitation at 37°C. A spread plate method was utilized where 100 μL of the BPW were transferred onto 90 mm agar plates of Cetrimide agar (Biolab) (Denissen et al., 2024). The spread plate method was carried out in duplicate to ensure that the results were harmonized. All the inoculated plates were incubated at 37°C for 24 h for culture isolation. The colonies grown were examined for morphology and pigment production and for gram-staining. Oxidation-Fermentation Test: This test was performed on presumptive Pseudomonas species to determine their oxidative reaction. Specifically, among the same samples of oxidase (+), catalase (+), and motility (+) activities (Nepali et al., 2018; Tohya et al., 2022), colonies were studied according to Abdalhamed et al. (2016) to confirm the Pseudomonas species. The nutrient agar (NA) (Biolab) plates were incubated for 24 h at 37°C to obtain pure cultures for molecular identification. Pseudomonas aeruginosa ATCC 27853 was used a positive control.
2.6 Genomic DNA extraction from Pseudomonas aeruginosa
DNA was extracted from overnight incubated cultures of P. aeruginosa using DNA Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. DNA quantification was conducted using the NanoDrop ND-100 system (NanoDrop Technologies, Wilmington, United States).
2.7 Identification of Pseudomonas aeruginosa using gyraseB gene PCR
Conventional PCR was conducted to identify Pseudomonas spp. isolated from Diptera flies targeting the P. aeruginosa gyraseB gene using the following primers: Forward: CCT GAC CAT CCG TCG CCA CAA, and Reverse: CGC AGC AGG ATG CCG ACG CC (Ahmadi et al., 2024). Each PCR reaction was conducted with a total mixture of 25 μL consisting of 12.5 μL OneTaq® Quick-Load 2X Master Mix with Standard Buffer (0.4 mM dATP, 0.4 mM dCTP, 0.4 mM dGTP, 0.4 mMdTTP, 4 mM MgCl2, and loading buffer) (New England BioLabs), 8.5 μL of nuclease-free water, 2 μL of the template DNA, and 1 μL of each oligonucleotide primer. PCR conditions were as follows: 96°C for 4 min, 35 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 1 min, and one step of final elongation at 72°C for 10 min with an infinite hold at 4°C. The amplified PCR products were electrophoresed on 1.5% agarose gel, stained with Ethidium Bromide (EtBr) and visualized under UV light.
2.8 The bacterial 16S rRNA gene PCR and sequencing
The 16S rRNA gene PCR assay was conducted using universal primer pair namely, 27F-GAGTTTGATCCTGGCTCAG and 1492R -GGTTACCTTGTTACGACTT (Ramatla et al., 2024a; Mlangeni et al., 2024). PCR reaction consisted of a total reaction of 25 μL consisting of 12.5 μL OneTaq® Quick-Load 2X Master Mix with Standard Buffer (0.4 mM dATP, 0.4 mM dCTP, 0.4 mM dGTP, 0.4 mMdTTP, 4 mM MgCl2, and loading buffer) (New England BioLabs), 8.5 μL of nuclease-free water, 2 μL of the template DNA, and 1 μL of each oligonucleotide primer. The following PCR conditions were used; 95°C for 5 min, 30 cycles of 95°C for 5 min, 55°C for 30 s, 72°C for 1 min and 72°C for 5 min followed by an infinite hold at 4°C. The amplified PCR products were electrophoresed as described above. PCR amplicons were sequenced with the BigDye Terminator sequencing kit v3.1 (Applied Biosystems) on the SeqStudio genetic analyzer at the UESM sequencing facility of North-West University.
2.9 Detection of virulence genes from Pseudomonas aeruginosa
Six P. aeruginosa virulence genes, namely, exoenzyme S (exoS), phospholipase N (plcN), phospholipase H (plcH), exotoxin A (toxA), elastas B (lasB), and alginate D (algD) were screened by PCR from P. aeruginosa DNA (Faraji et al., 2016; Ramatla et al., 2024a,b). PCR reaction consisted of a total reaction of 25 μL consisting of 12.5 μL OneTaq® Quick-Load 2X Master Mix with Standard Buffer (0.4 mM dATP, 0.4 mM dCTP, 0.4 mM dGTP, 0.4 mMdTTP, 4 mM MgCl2, and loading buffer) (New England BioLabs), 8.5 μL of nuclease-free water, 2 μL of the template DNA, and 1 μL of each oligonucleotide primer. PCR conditions are listed in Supplementary Table S1. The amplified PCR products were electrophoresed as described above.
2.10 Whole genome sequencing and bioinformatics analysis
The MGIEasy FS DNA Prep Kit (BGI, China) was used to create sequence libraries of the two P. aeruginosa strains by the manufacturer’s instructions. The strains, P311 and P37, were selected from Hemipyrellia spp., flies in residential dumping and livestock kraals sites, respectively. Selection was also based on the isolates’ proven resistance to more than two classes of antibiotics. Using a paired-end 150 nt approach, the produced libraries were sequenced using the BGI MGISEQ-2000 platform (BGI Shenzhen, China). FastQC version 0:10.1 (Andrews, 2017) was used to evaluate the quality of the sequenced reads. The MEGAHIT tool v.1.1.2 (Li et al., 2015) was used to de novo assemble the sequence paired-end trimmed reads. For the assembly, kmer sizes 21, 33, 55, 77, 99, and 127 were employed, and the minimum contig length was set at 500 bp. The KBase app (Arkin et al., 2018) was used with CheckM v1.1.6 (Parks et al., 2015) to evaluate individual assembled genomes’ quality and contamination percentage. Quast v 2.3 (Gurevich et al., 2013) assessed the draft genome assemblies. The Pub-MLST1 was used to identify the isolates (Jolley et al., 2018). We utilized kSNP within the IPGA tool, employing a k-mer length of 21, a minimum coverage of 10 reads, an error rate threshold of 0.01, genome coverage of at least 95%, and a SNP quality score threshold of 30 to ensure high-confidence SNP identification and accurate phylogenetic analysis as previously described (Magome et al., 2024). Moreover, only genomes with above 90% completeness and <5% contaminants were included in this analysis. We included 10 genomes of P. aeruginosa from South Africa isolated from the environment (n = 5), currently available, and humans (n = 5), including the reference strain PAO1. ABRicate pipeline was employed to screen for antibiotic resistance and virulence genes using the required databases previously described by Fono-Tamo et al. (2023). Briefly, the antibiotic resistance determinants were identified in each assembled genome using the ResFinder [−db ResFinder] (Feldgarden et al., 2019) with the minimum identity and coverage thresholds of 90 (−minid 90) and 70% (−mincov 70), respectively. Virulence factors in the sequenced genomes were mined using the Virulence Factor Database [−db vfdb] (Chen et al., 2005; Liu et al., 2019), using minimum identity and coverage thresholds of 90 (−minid 90) and 70% (−mincov 70), respectively.
2.11 Antimicrobial resistance test on Pseudomonas aeruginosa isolates
The disk diffusion assay (DDA), also known as the Kirby-Bauer method, was used to determine the antimicrobial profiles of P. aeruginosa isolates. Antimicrobial susceptibility was done for the following antibiotics: Aminoglycosides [Neomycin (N), Gentamicin (CN)], β-Lactam [Amoxicillin (AML)], nitroimidazole [Metronidazole (MTZ)], sulfonamide [Sulphamethoxazole (RL)] and cephalosporin [Cefazolin (KZ)]. A standardized inoculum (100 uL) was spread over the surface of Mueller-Hinton agar (MHA). The zone inhibition of the antibiotics was measured in millimeters (mm) according to the Clinical and Laboratory Standards Institute (CLSI), version 2023 (CLSI, 2023). It was classified as multidrug resistant (MDR) if resistant to at least three antibiotic classes (Ramatla et al., 2024b).
2.12 Determination of ESBL-producing isolates
ESBL-producing P. aeruginosa isolates were confirmed phenotypically on CHROMagar™ ESBL (CHROMagar, France), and plates were incubated aerobically at 37°C for 24 h. The agar differentiates between ESBL E. coli (red colony presentation) and ESBL P. aeruginosa isolates (blue-green) (Chenhaka et al., 2023). The isolates were chosen because they could produce the ESBL enzymes. The reference strains used in this study were non-pathogenic E. coli ATCC 10536 and ESBL-producing Pseudomonas aeruginosa ATCC 27853 (Microbiologics, United States).
2.13 Detection of antibiotic resistance genes in Pseudomonas aeruginosa isolates
The presence or absence of antibiotic-resistance genes was screened from P. aeruginosa isolates (Diab et al., 2018; Chow et al., 2001; Rahmani et al., 2013; Ramatla et al., 2024a). PCR reaction consisted of a total reaction of 25 μL consisting of 12.5 μL OneTaq® Quick-Load 2X Master Mix with Standard Buffer (0.4 mM dATP, 0.4 mM dCTP, 0.4 mM dGTP, 0.4 mMdTTP, 4 mM MgCl2, and loading buffer) (New England BioLabs), 8.5 μL of nuclease-free water, 2 μL of the template DNA, and 1 μL of each oligonucleotide primer. PCR conditions and primer sequences are shown in Supplementary Table S2. PCR amplicons were electrophoresed as described above.
3 Results
3.1 Morpho-species identification of the Diptera flies
A total of 1,422 and 2,793 flies were collected from the illegal residential dumping sites of Ikageng and the livestock kraals of Matlwang village, respectively. Captured Diptera flies, which were identified by microscopy, were categorized based on phenotypic traits such as body color, shape, and size. Hemipyrellia spp. (31%), Tabanus spp. (24%), and Chrysomya spp. (12%) were found in livestock kraals. The illegal residential dumping site revealed the presence of Hemipyrellia spp. (88%), Synthesiomyia spp. (2%), Sarcophagidae spp. (7%), and Chrysomya spp. (2%).
3.2 Molecular identification of the Diptera flies
An 1,888 bp region of the CO1 gene was used for genetic identification of the Diptera flies collected from Matlwang and Ikageng in Potchefstroom. BLAST nucleotide analysis identified the sequenced flies belonging to different genera species, supported by percentage identities between query cover and percentage identity with an E-value of 0.0. Hemipyrellia spp., Chrysomya spp., Synthesiomyia spp., and Sarcophagidae spp. are the four genera and species identified from the residential dumping site (Supplementary Figure S1). Two species belonging to the Hemipyrellia genus were found at the dumping site, identified as H. pulchra and H. fernandica. Chrysomya megacephala was also present at the dumping site, which clustered with C. megacephala isolates (MK075818.1 and MK075815.1), which were collected from Shandong in China. Given their close kinship, Chrysomya spp. and Hemipyrellia spp. belong in the same family as the Calliphoridae. Synthesiomyia nudiseta was one of the other taxa determined in this dumping site. The Sarcophagidae comprises S. tibalias and Sarcophagidae spp. in a different clade (Supplementary Figure S1). Three genera of species were identified in the livestock kraals of Matlwang as Hemipyrellia spp., Chrysomya spp., and Tabanus spp. (Supplementary Figure S2). The Hemipyrellia spp., specifically H. fernandica and H. ligurriens, were shown to belong to the same clade as the Chrysomya (Chrysomya marginals) in the Matlwang grouping. Tabanus spp. formed their own sub-clade, which was identified in this study from the livestock kraals. These were categorized as belonging to the relative minor sub-clades of the species, T. trivittatus voucher, T. birmanicus, and T. par.
3.3 Isolation and identification of Pseudomonas species using microbiological tests
In total, 59 Pseudomonas spp. were isolated and identified from 96 fly samples from residential dumping sites and livestock kraals. Pseudomonas spp. were isolated from Diptera flies, namely, Hemipyrellia spp., Synthesiomya spp., Chrysomya spp., and Sarchophagidae spp., collected from residential dumping sites. Furthermore, Pseudomonas spp. were isolated from Diptera flies, namely Hemipyrellia spp., Chrysomya spp., and Tabanus spp. and collected from the livestock kraals.
3.4 Molecular identification of Pseudomonas aeruginosa species
A total of 59 Pseudomonas spp. isolates from different fly species in the dumping site and livestock kraal were employed for P. aeruginosa identification. The species-specific gyraseB gene PCR identified 23 and 36 isolates as P. aeruginosa isolated from dumping and livestock kraals, respectively. The 16S rRNA gene PCR and sequencing confirmed these isolates as P. aeruginosa. The GenBank accession numbers were assigned to some of the represented strains (n = 5) as follows: sequence 1: OR122642; sequence 2: OR122643; sequence 3: OR122644; sequence 4: OR122645; sequence 5: OR122646.
3.5 Detection of virulent genes in Pseudomonas aeruginosa isolates by PCR
About 23 isolates from a residential dumping site from different fly genera, namely, Hemipyrellia spp. (n = 7), Synthesiomyia spp. (n = 7), Chrysomya spp. (n = 4) and Sarcophagidae spp. (n = 5), were tested for the presence and absence of these virulent genes. The exotoxin (toxA) and phospholipase (plcN) genes were detected in all (n = 23) P. aeruginosa isolates collected from residential dumping sites (Figure 2A). The elastase (lasB) gene was detected in all the P. aeruginosa isolates from Hemipyrellia spp., Sarcophagidae spp., and Synthesiomyia spp. The lasB gene was detected in (n = 3; 75%) of P. aeruginosa isolated from Chrysomya spp. The hemolytic phospholipase C (plcH) and the GDP-mannose 6-dehydrogenase (algD) gene were detected in all (100%) of the P. aeruginosa isolates from Chrysomya spp., Hemipyrellia spp. and Synthesiomyia spp. The plcH gene was present in (n = 4; 80%) of the isolates from Sarcophagidae spp. The exoenzyme S (exoS) gene was detected in all the isolates from fly-genera Synthesiomyia spp. and Chrysomya spp. (100%), Hemipyrellia spp. (n = 6; 86%), and present in (n = 4; 80%) of the Sarcophagidae spp.
Figure 2
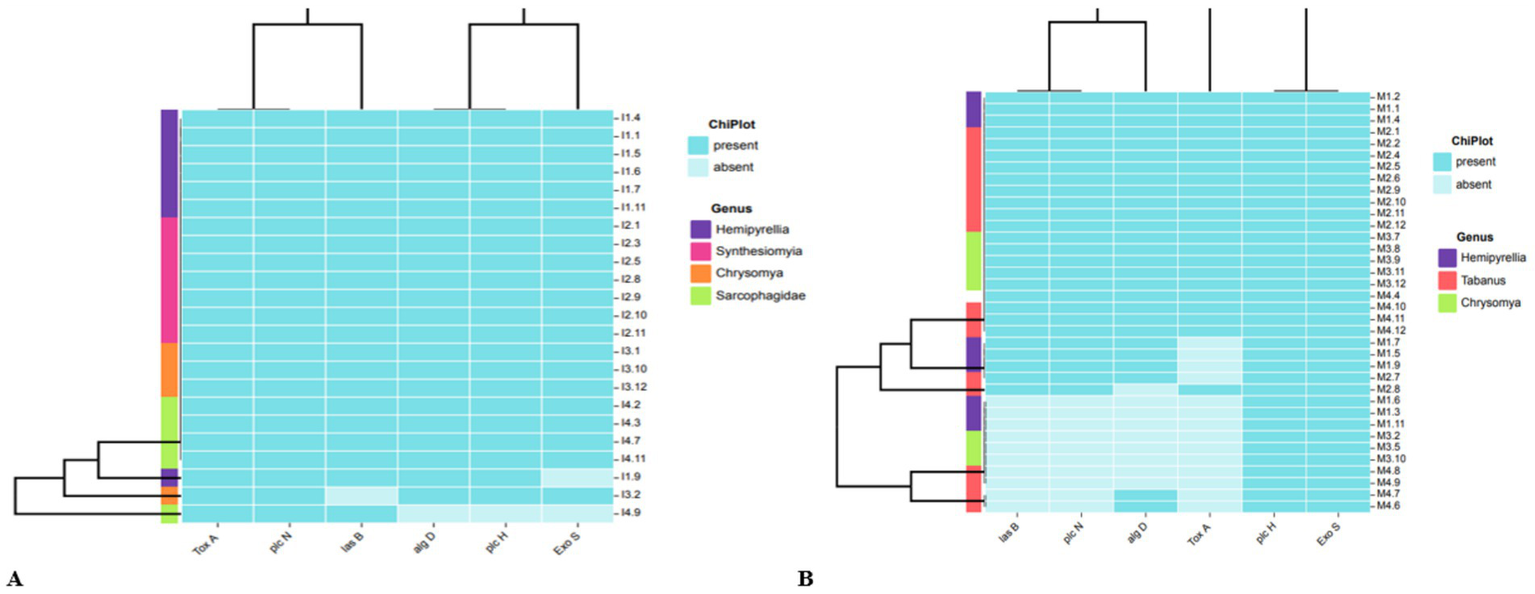
Phylogenetic clustering of the virulence genes that are present or absence in P. aeruginosa isolates in relation to fly-genera collected from (A) residential dumping site and (B) livestock kraals.
The livestock kraals identified 36 P. aeruginosa isolated from different fly genera, Hemipyrellia spp. (n = 9), Tabanus spp. (n = 17), and Chrysomya spp. (n = 8) and were profiled for the virulence genes (Figure 2B). The plcH and exoS genes were present in isolates investigated. The lasB gene was detected in Hemipyrellia spp. (n = 6; 66.7%), Tabanus spp. (n = 13; 76.47%), and Chrysomya spp. (n = 5; 62.5%), respectively. The algD gene was present in Hemipyrellia spp. (n = 6; 66.7%), Tabanus spp. (n = 14; 84.3%), and Chrysomya spp. (n = 5; 62.5%), respectively. The toxA gene was detected in 3 (33.33%) isolates from Hemipyrellia spp., (n = 12; 70.6%) in Tabanus spp., and (n = 5; 62.5%) in Chrysomya spp.
3.6 Antimicrobial susceptibility testing of Pseudomonas aeruginosa and determination of ESBL-producing isolates
An antimicrobial susceptibility test was performed to profile the 59 P. aeruginosa isolates isolated from residential dumping (Figure 3A) and livestock kraal sites (Figure 3B). All P. aeruginosa isolates investigated in this study were resistant to metronidazole, sulphamethoxazole, cefazolin, and amoxicillin. Thirteen percent of P. aeruginosa isolates from residential dumping sites were resistant to gentamicin, while 38.9% were resistant to neomycin. Meanwhile, 30.4 and 52.2% of isolates were classified as susceptible and intermediate to neomycin, respectively. Furthermore, 38.9% of P. aeruginosa isolated from livestock kraals were resistant to gentamicin. The extended-spectrum beta-lactamases (ESBL) were determined among all the confirmed P. aeruginosa isolates (n = 59). All the investigated P. aeruginosa isolates were considered to be ESBL-producing.
Figure 3
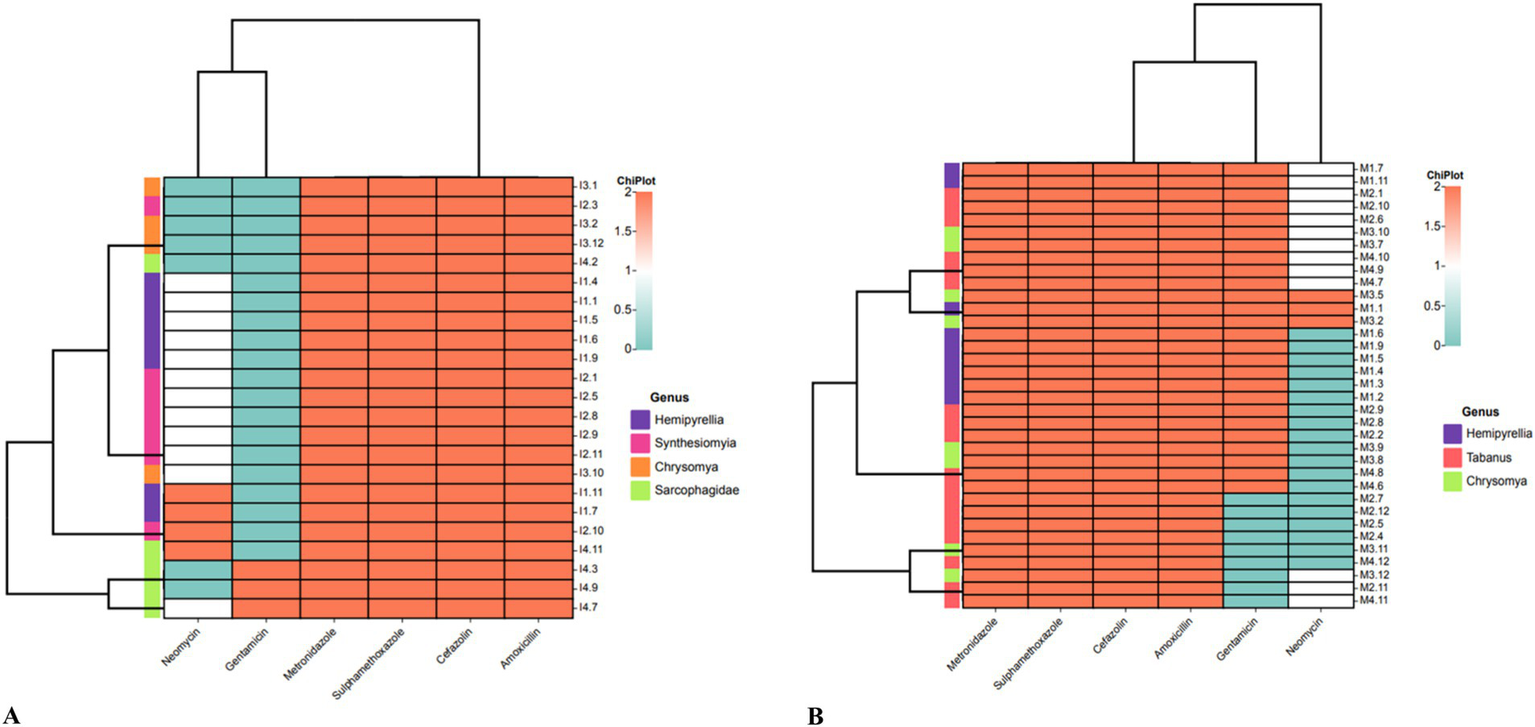
Heatmap analysis of the antibiotic susceptibility using the Kirby-Bauer method for P. aeruginosa isolates from flies collected at (A) residential dumping site and (B) livestock kraals.
3.7 Antibiotic resistance genes in Pseudomonas aeruginosa isolates detected by PCR
Four different antibiotic resistance genes (ARGs) were investigated in the P. aeruginosa isolates collected from residential dumping site (n = 23) and livestock kraal sites (n = 36). In this study, the detected ARGs included neomycin (aph (2″)-Ib), gentamicin (acc(3)-IV), metronidazole (rdxA), sulphamethoxazole (sulI) and amoxicillin (pbp1A). All P. aeruginosa isolates collected from residential dumping sites tested positive for the aminoglycoside acc(3)-IV gene that confers resistance to gentamicin. The PCR assay revealed the presence of the rdxA (43.5%) and sulI (86.9%) genes. The absence of the aph(2″)-Ib gene in P. aeruginosa isolates indicates their susceptibility to neomycin. The lack of the pbp1A gene is inconsistent with the DDA, whereby most of the isolates appeared to be resistant to amoxicillin.
Three ARGs were detected in P. aeruginosa isolates from livestock kraal, including acc(3)-IV (80.6%) and sulI (88.9%), genes that confer resistance to gentamicin and sulphamethoxazole, respectively. The pbp1A and aph (2″)-Ib genes that confer resistance to amoxicillin and neomycin, respectively, were absent in all the P. aeruginosa isolates. The absence of pbp1A gene is inconsistent with the disk diffusion assay, whereby all the isolates were resistant to amoxicillin. Four different β-lactam genes were examined among P. aeruginosa isolates from residential dumping (n = 23) and livestock kraal sites (n = 36) flies. The 4 β-lactam genes included blaOXA, blaSHV, blaTEM, and blaCTX-M. Only one P. aeruginosa isolate from the fly genus, Hemipyrellia spp. collected from livestock kraal consisted of the blaCTX-M gene.
3.8 Genome assembly of the Pseudomonas aeruginosa isolates
With an average of 150 bp paired-end reads, a total of 2,865,578 sequence reads for strain P311 and 2,996,628 for strain P7 were generated on the MGI sequencer. After trimming the sequence reads, 2,841,220 and 2,953,424 reads were determined from the strains P311 and P7, respectively. The genome features show that strain P37 is approximately 6.45 Mb, while strain P311 is ~6.37 Mb, which are both higher than the reference strain PAO1 which is 6.26 Mb. Strain P37 has a higher GC content, impacting the number of coding sequences determined. The G + C content of the sequenced strains and reference strain PAO1 is ~66%. Genome sequences of the two strains P37 and P311 isolated from Hemipyrellia spp. have been deposited in GenBank under the accession numbers JBDJPE000000000 P. aeruginosa P37 and JBDJPD000000000 P. aeruginosa P311. Genome assemblies are shown in Supplementary Table S3. Both the sequenced strains belong to the sequence type ST3808. The two strains group closely with P. aeruginosa strain WW isolated from South Africa, North West from water source.
3.9 Detection of virulence genes in Pseudomonas aeruginosa by whole genome sequencing
The use of whole genome sequencing on the selected P. aeruginosa strains isolated from Hemipyrellia spp. was confirmed as P. aeruginosa (Figure 4A). The strains clustered with P. aeruginosa strain ww isolated from water in the same Province. Whole genome sequencing identified 221 virulence genes in the sequenced P. aeruginosa P311 and P37 strains and the reference strain PAO1 (Figure 4B). Shared genes included flagellar motor proteins (MotC and MotD), flagellar biosynthetic proteins, type III secretion system proteins, phenazine biosynthesis proteins, paerucumarin biosynthesis proteins, pyoverdine biosynthesis proteins, general secretion pathway proteins, etc. In this study, only the type III secretion system effector ExoU phospholipase A2 activity was common in the sequenced strains, i.e., P311 and P37. About 19 virulence genes were unique to the reference strain PAO1. These include genes such as the (wzy) O-antigen chain length regulator, the type 4 fimbrial precursors PilA and PilB, the pyoverdine biosynthesis protein PvdJ- PvdD- PvdI complex, and the type VI secretion system substrate VgrG1b.
Figure 4
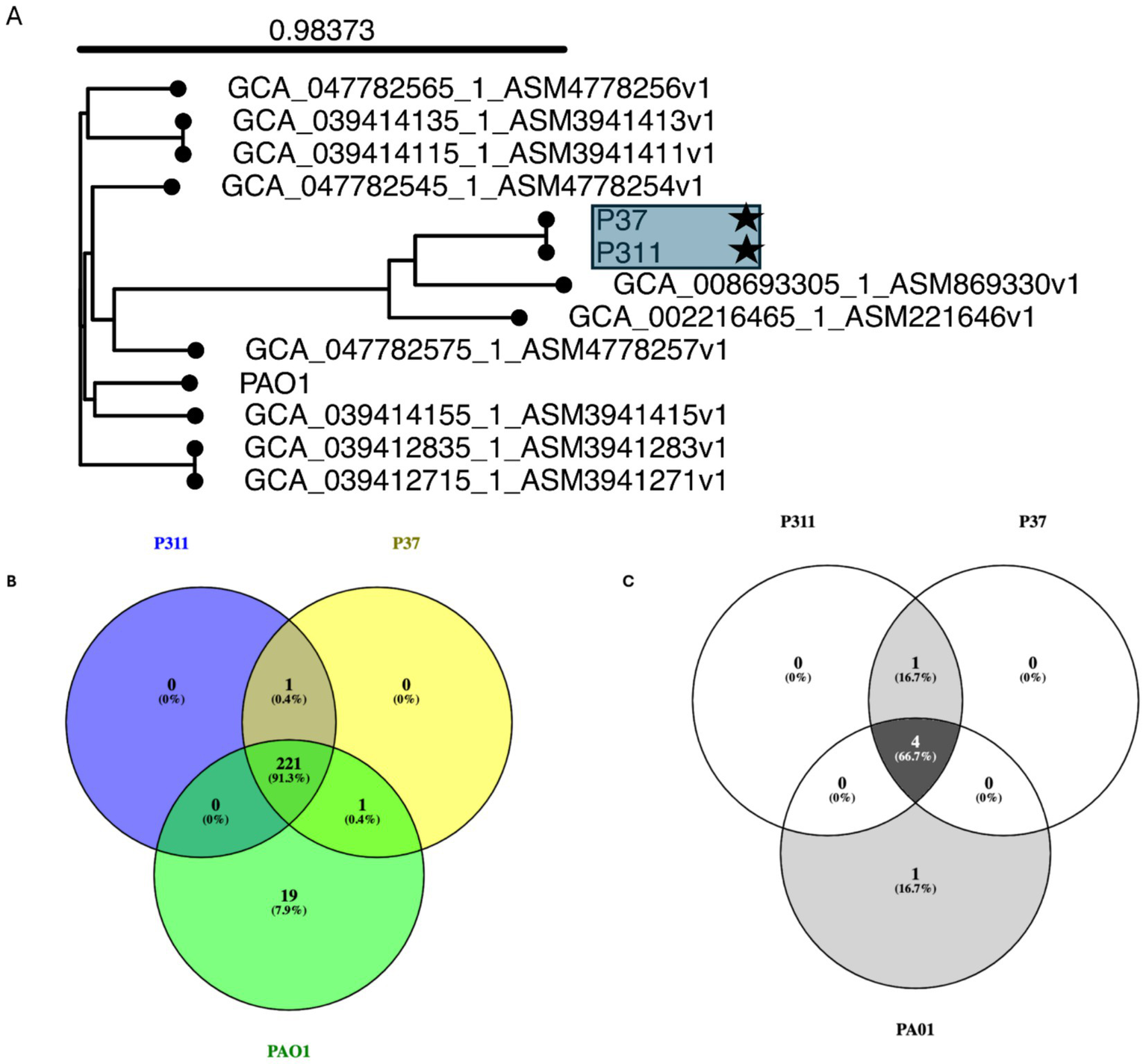
Whole genome-based phylogenetic tree using single nucleotide polymorphism inferring the evolutionary relationships among the sequenced Pseudomonas aeruginosa strains with antibiotic-resistant and virulence determinates. (A) Genome Blast Distance Phylogenies (GBDP) identified by TYGS between the sequenced P. aeruginosa strains (indicated by user strain) and related genomes. (B) Virulence genes and (C) antibiotic resistance genes that are shared among the sequenced strain and reference strain PAO1.
3.10 Antibiotic resistance genes detected by whole genome sequencing
The use of whole genome sequencing showed that the two sequenced P. aeruginosa strains, i.e., P37 and P311 are resistant to fosfomycin (fosA_4), chloramphenicol (catB_1), ampicillin (blaOXA-50_1), beta-lactamase (blaPAO), and aminoglycoside (aph(3′)-IIb_2) (Figure 4C; Supplementary Table S4). Moreover, the identified ARG profiles on the draft genomes of the sequenced P. aeruginosa strains were also found on the reference genome of P. aeruginosa strain PAO1 (AE004091.2). Differences were observed in the sequenced strains P311 and P37, consisting of the shared beta-lactamase blaPAO_4, while the reference PAO has the blaPAO_2 gene.
4 Discussion
This study investigated the antimicrobial resistance and virulence profiles of P. aeruginosa associated with the gut of Diptera flies collected from livestock kraals and residential dumping sites. Pseudomonas aeruginosa is an important opportunistic pathogen and a key factor in both nosocomial and community-acquired infections. Although it is frequently examined in clinical settings, its environmental reservoirs remain inadequately explored, especially in flies from anthropogenically impacted areas. This study addresses that gap, contributing to a One Health perspective on pathogen transmission dynamics at the intersection of humans, animals, and the environment. The study sheds light on how poor waste management in peri-urban settings contributes to the environmental persistence and amplification of P. aeruginosa, positioning these environments as critical, yet often neglected, hotspots in the antimicrobial resistance (AMR) cycle. This study shows that Diptera flies in residential and livestock-associated environments are carriers of multidrug-resistant Pseudomonas aeruginosa, harboring a range of antibiotic-resistance genes.
In this study, a significantly higher number of Diptera flies were associated with residential dumping sites as opposed to livestock kraals. This likely reflects the nutrient-rich and unhygienic conditions of dumping sites, which offer abundant resources for breeding (Goulson et al., 1999). In contrast, only a small number of Diptera species were linked to livestock, which suggests livestock kraals as a harsher habitat with fewer provisions and breeding space, where Diptera flies that are suited for such conditions might flourish, especially those that feed on feces and blood (Shety et al., 2022). The high density of flies in peri-urban dumping areas indicates a risk of pathogen transfer between human communities and nearby livestock operations, emphasizing the necessity of effective waste management for disease control.
Among the Diptera, Hemipyrellia spp. were found in substantial quantities in livestock kraal (31%) and the illegal residential dumping site (88%). They are known to proliferate in environments that include waste materials, dead animals, as well as animal and human excrement (Hore et al., 2017). Twenty-four percent of the Tabanus spp., also known as horseflies, were found exclusively in the livestock kraals. Female Tabanus spp. normally require a blood meal to reproduce, making them potential carriers of pathogenic bacteria (Snyman et al., 2020). In warm, humid regions of South Africa, such as the tropical climate of KwaZulu-Natal province, Tabanus spp. is highly prevalent (Esterhuizen, 2006). Seven percent of the Sarcophagidae spp. that belong to the family known as flesh flies were found in this study. Additionally, reports of Sarcophagidae spp. have been made in Cusco, Peru’s residential areas (Ly et al., 2018). Since Sarcophagidae spp. are ovoviviparous, they opportunistically deposit hatching or hatched maggots rather than eggs (Ren et al., 2018). The dumping site is an ideal environment for them to breed, given that there is an abundance of decaying and unsanitary materials to offer the maggots the highest chance of surviving (Ren et al., 2018). They typically deposit on carrion, decaying material, and feces. The detection of P. aeruginosa in environmentally abundant fly species such as Musca domestica and Hemipyrellia spp. suggests that these insects may play an active role in the environmental circulation of clinically significant resistance genes.
Pseudomonas aeruginosa is a known opportunistic pathogen with different virulence factors that enable it to reside in various host niches. These bacteria are a major global source of nosocomial and community-acquired illnesses. The virulence genes exotoxin A (toxA) and phospholipase N (plcN) were detected in all investigated isolates. The toxA gene is responsible for the regulation of exotoxin A synthesis and is known to cause tissue damage effectively and decrease the phagocytic activity of leukocytes in infected patients (Dong et al., 2015). Multidrug-resistant (MDR) P. aeruginosa strains isolated from patients have a substantially greater frequency of the toxA gene (76.6% of MDR strains) (Khosravi et al., 2016). However, there are no reports of the prevalence of toxA gene in Diptera flies. The plcN gene encodes for the phospholipase C enzyme, which disrupts host cell membranes and contributes to tissue damage (Terada et al., 1999). Another type of phospholipase is the plcH gene, which is responsible for hydrolysis phospholipids. This gene was detected in 99% of the isolates investigated in this study. The P. aeruginosa isolates in this study lacked the plcH gene and were augmented with other virulent genes that were absent, including alginate lyase (algD) and exoenzyme S (exoS). The P. aeruginosa is protected from the host defense mechanism by the algD gene during biofilm formation. This is achieved by the reduction of phagocytosis as well as the inhibition of the activation of complement proteins (Edward et al., 2023). While exoS impairs phagocytosis in the lungs, exacerbating infection (Faraji et al., 2016).
The lasB gene, encoding elastase that disrupts immune function, was found in 99% of P. aeruginosa isolated from Diptera flies collected from residential dumping sites. The virulence factor lasB in P. aeruginosa is responsible for proteolytic actions ranging from tissue damage to compromising the host immune system (Cathcart et al., 2011). However, this gene was absent in 16.6% of isolates from Hemipyrellia spp., Tabanus spp. (36%), and Chrysomya spp. (13.8%) in livestock kraals. The latter-mentioned strains, which lack other virulent genes such as toxA, lasB, and plcN, suggest that these P. aeruginosa can be classified as avirulent strains. The avirulent P. aeruginosa strains (70.3%) have been identified in clinical host material (Luzar and Montie, 1985). Nevertheless, these strains in the current study cannot be neglected as they showed multiple resistances to various antibiotics. This variation in virulence gene profiles suggests the circulation of both virulent and avirulent P. aeruginosa strains in environmental fly populations, mirroring clinical findings (Luzar and Montie, 1985) and underscoring the complex ecology of this pathogen.
Pseudomonas aeruginosa sensitivity to neomycin can vary based on a number of variables, such as the strain of the bacterium, its genetic composition, and any acquired resistance mechanisms it may have (Pang et al., 2019). Neomycin is effective against a wide range of bacterial strains, including certain isolates of P. aeruginosa (Uemura et al., 2017). This was evident as most isolates (88.14%) examined in this study were either intermediate or susceptible to neomycin. Furthermore, P. aeruginosa can withstand elevated neomycin concentrations and demonstrates enduring adaptive resistance, which results in cross-resistance to additional aminoglycoside antibiotics (Uemura et al., 2017). This should further be exploited using the minimum inhibition concentration assay, particularly as P. aeruginosa isolated from Diptera flies is not well-investigated globally. Based on DDA, all isolates were resistant to amoxicillin, supported by the blaOXA-50 gene, which was noticeable in the two genome-sequenced strains. The P. aeruginosa isolated from canine clinical cases found in South Africa also showed resistance against amoxycillin-clavulanic acid (99%) (Eliasi et al., 2020). Moreover, all isolates characterized in this study were considered ESBL-producers. However, these isolates did not detect the amoxicillin (Pbp1A) gene. In P. aeruginosa, the PbplA gene encodes a penicillin-binding protein. Enzymes called penicillin-binding proteins (PBPs) are part of the bacterial cell wall production process (Handfield et al., 1997). Beta-lactam antibiotics, such as cephalosporins and penicillins are directed toward them. One of the PBPs involved in cell wall formation in P. aeruginosa is PbplA, which is inhibited by beta-lactam antibiotics, preventing bacterial growth and causing cell death. A discrepancy between phenotypic and resistance to amoxicillin and the molecular detection of the Pbp1A gene by PCR. While PCR failed to amplify Pbp1A in several phenotypically resistant isolates, whole genome sequencing (WGS) of the two sequenced isolates confirmed the presence of Pbp1A and Pbp1B, as well as the associated lipoprotein activator LpoP, which is essential for the proper function of PBP1A. This inconsistency suggests that the PCR-based approach may have been limited by suboptimal primer design, possibly due to genetic variability in primer binding regions or sequence divergence in local P. aeruginosa populations. These findings highlight the importance of validating molecular detection methods against WGS and point to redesigning or optimizing primers to ensure accurate gene detection, mainly when correlating genotypic data with antibiotic resistance phenotypes. This discrepancy emphasizes the need to align molecular diagnostics with genomic evidence, particularly for environmental surveillance applications. The blaCTX-M gene was present among P. aeruginosa isolates (4.35%) harbored by flies from residential dumping sites and livestock kraals. The blaCTX-M is the least frequently isolated ESBL gene in P. aeruginosa strains among antibiotic-resistant strains (Shacheraghi et al., 2010). The lower prevalence of blaCTX-M gene (4.35%) in this study differs from that reported on isolates from patients in Mthatha City, Eastern Cape Province in South Africa, whereby 31.7% of the isolates tested positive (Hosu et al., 2021).
The acc(3)-IV gene is a type of aminoglycoside acetyltransferase encoded by the acc(3)-IV gene in bacteria, including P. aeruginosa. This enzyme acetylates aminoglycoside antibiotics, modifying them and decreasing their potency. In this study, about 88% of the isolates from the residential dumping site and the livestock kraal tested positive for this gene. Based on DDA, the isolated P. aeruginosa isolates (38.9%) from livestock kraals were also resistant to gentamicin, while 87% of the isolates from dumping sites were susceptible. Resistance to gentamicin has also been reported from P. aeruginosa isolated from dairy cattle, milk, the environment, and workers’ hands in Egypt (Badawy et al., 2023). In Iran, P. aeruginosa isolates from M. domestica flies showed a 49.1% resistance rate (Hemmatinezhad et al., 2015). Gentamicin is a routinely used antibiotic for treating P. aeruginosa infections; the existence of gentamicin-resistant P. aeruginosa strains is alarming, as resistance to this antibiotic can restrict treatment options and make managing the bacterium’s infections more difficult.
The sul1 gene, which is linked to sulfamethoxazole resistance, was detected in 87.72% of the P. aeruginosa isolates in the current study (Rahmani et al., 2013). Based on DDA, all isolates were confirmed to be resistant to sulphamethoxazole. The leading causes of P. aeruginosa resistance to sulfamethoxazole are mutations in topoisomerases, decreased expression of outer membrane proteins, and the synthesis of beta-lactamases and enzymes that alter aminoglycosides (Bonomo and Szabo, 2006). Nevertheless, none of the sul1 gene was detected in the whole genome sequenced strains of P. aeruginosa in this study, suggesting that isolates do not necessarily express gene mutations similarly or simultaneously. Whole genome sequencing of two sequenced P. aeruginosa isolates revealed that both belong to sequence type ST3808. This sequence type has not been widely reported in fly-associated P. aeruginosa and may represent an emerging lineage in non-clinical environments. Notably, phylogenetic analysis based on single nucleotide polymorphisms showed that the two ST3808 strains clustered closely with P. aeruginosa strain WW, a 2017 genome that was isolated from a water source in the North West province of South Africa.
This study significantly contributes to the One Health discourse by highlighting Diptera flies as often-overlooked vectors of AMR P. aeruginosa. Detecting clinically relevant resistance genes and virulence factors in flies from livestock and residential environments demonstrates the pathogen’s ability to cross ecological boundaries, reinforcing its zoonotic and environmental threat (Lister et al., 2009; Santajit and Indrawattana, 2016). Importantly, the study emphasizes several key implications for One Health. It establishes the zoonotic potential of Diptera flies, which can act as vectors for transmitting resistant P. aeruginosa among livestock, environmental sources, and human communities. Additionally, it points to the role of environmental reservoirs, particularly in peri-urban areas where inadequate waste management promotes the proliferation and persistence of pathogens.
The limitations of this study stem from excluding fly species that were not shared between the two sites in order to maintain comparability, we may have overlooked significant species-specific variations in pathogen carriage, thereby narrowing the ecological scope of our findings. Furthermore, excluding environmental and host samples, whether from livestock or humans, restricts our understanding of broader transmission dynamics and potential reservoirs within the One Health context, which merits further exploration. Moreover, the discrepancies noted between phenotypic antibiotic resistance profiles and genotypic findings, such as the resistance to amoxicillin in the absence of the pbp1A gene, underscore the complexity of antimicrobial resistance mechanisms that targeted PCR assays may not comprehensively capture. While whole genome sequencing was employed, only two isolates were analyzed, potentially limiting the representation of the genomic diversity of P. aeruginosa in these environments; subsequent work will involve sequencing additional isolates.
5 Conclusion
This study demonstrates that synanthropic Diptera fly species collected from illegal residential dumping sites and livestock kraals harbor multidrug-resistant P. aeruginosa, highlighting their potential role as vectors in disseminating clinically significant pathogens. Phenotypic analysis confirmed resistance to several antibiotics, notably amoxicillin, while molecular screening identified resistance genes such as bla_TEM, bla_SHV, bla_CTX-M, and efflux pump-related genes (mexA, oprM). This bacterium is well-known for being resilient and adaptable, especially in situations containing a significant number of organic materials. This bacterium is essential to “One Health” due to its multidrug resistance character and zoonotic nature. These findings suggest that flies could play a significant role in the environmental spread of antibiotic resistance, emphasizing the need for integrated “One Health” surveillance approaches that consider the interface of human, animal, and ecological health.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, JBDJPE000000000, JBDJPD000000000, OR122642–OR122646, MK075818.1h, and MK075815.1.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
LW: Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. IM: Data curation, Writing – review & editing. OT: Conceptualization, Data curation, Supervision, Writing – review & editing. KL: Conceptualization, Data curation, Funding acquisition, Supervision, Validation, Writing – review & editing. TR: Conceptualization, Formal analysis, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by NRF Incentive grant for rated researchers (GUN: CSUR23030681021) made available to KL.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1586811/full#supplementary-material
Footnotes
References
1
Abdalhamed A. Zeedan G. G. Hussein H. Abdeen E. (2016). Molecular and biochemical characterization of Pseudomonas species isolated from subclinical mastitis milk and ice cream and its susceptibility to allium sativum and commiphorra molmol plant extracts in Egypt. Int. J. Adv. Res.4, 1669–1679. doi: 10.21474/IJAR01/1378
2
Andrews S. (2017). FastQC: A quality control tool for high throughput sequence data, vol. 2010. Available at: https://cir.nii.ac.jp/crid/1370584340724053142
3
Ahmadi N. Salimizand H. Zomorodi A. R. Abbas J. E. Ramazanzadeh R. Haghi F. et al . (2024). Genomic diversity of β-lactamase producing Pseudomonas aeruginosa in Iran; the impact of global high-risk clones. Ann. Clin. Microbiol. Antimicrob.23:5. doi: 10.1186/s12941-024-00668-5
4
Arkin A. P. Cottingham R. W. Henry C. S. Harris N. L. Stevens R. L. Maslov S. et al . (2018). KBase: the United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol.36, 566–569. doi: 10.1038/nbt.4163
5
Badawy B. Moustafa S. Shata R. Sayed-Ahmed M. Z. Alqahtani S. S. Ali M. S. et al . (2023). Prevalence of multidrug-resistant Pseudomonas aeruginosa isolated from dairy cattle, Milk, environment, and workers’ hands. Microorganisms11:2775. doi: 10.3390/microorganisms11112775
6
Boiocchi F. Davies M. P. Hilton A. C. (2019). An examination of flying insects in seven hospitals in the United Kingdom and carriage of Bacteria by true flies (Diptera: Calliphoridae, Dolichopodidae, Fanniidae, Muscidae, Phoridae, Psychodidae, Sphaeroceridae). J. Med. Entomol.56, 1684–1697. doi: 10.1093/jme/tjz086
7
Bonomo R. A. Szabo D. (2006). Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis.43, S49–S56. doi: 10.1086/504477
8
Cathcart G. R. Quinn D. Greer B. Harriott P. Lynas J. F. Gilmore B. F. et al . (2011). Novel inhibitors of the Pseudomonas aeruginosa virulence factor LasB: a potential therapeutic approach for the attenuation of virulence mechanisms in pseudomonal infection. Antimicrob. Agents Chemother.55, 2670–2678. doi: 10.1128/AAC.00776-10
9
Chen L. Yang J. Yu J. Yao Z. Sun L. Shen Y. et al . (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res.33, D325–D328. doi: 10.1093/nar/gki008
10
Chenhaka L. H. Van Wyk D. A. Mienie C. Bezuidenhout C. C. Lekota K. E. (2023). The phylogenomic landscape of extended-spectrum β-lactamase producing Citrobacter species isolated from surface water. BMC Genomics24:755. doi: 10.1186/s12864-023-09867-4
11
Chow J. W. Kak V. You I. Kao S. J. Petrin J. Clewell D. B. et al . (2001). Aminoglycoside resistance genes aph(2″)-Ib and aac(6′)-Im detected together in strains of both Escherichia coli and Enterococcus faecium. Antimicrob. Agents Chemother.45, 2691–2694. doi: 10.1128/AAC.45.10.2691-2694.2001
12
CLSI (2023). Standards for antimicrobial susceptibility testing. 33rd Edn: Clinical and Laboratory Standards Institute. Available at: https://clsi.org/about/news/ast-news-update-june-2023-new-clsi-m100-ed33-updated-aminoglycoside-breakpoints-for-enterobacterales-and-pseudomonas-aeruginosa/
13
Deep A. Chaudhary U. Gupta V. (2011). Quorum sensing and bacterial pathogenicity: from molecules to disease. J. Lab. Phys.3, 4–11. doi: 10.4103/0974-2727.78553
14
Denissen J. Havenga B. Reyneke B. Khan S. Khan W. (2024). Comparing antibiotic resistance and virulence profiles of Enterococcus faecium, Klebsiella pneumoniae, and Pseudomonas aeruginosa from environmental and clinical settings. Heliyon10:e30215. doi: 10.1016/j.heliyon.2024.e30215
15
Diab M. El-Shenawy A. El-Ghannam M. Salem D. Abdelnasser M. Shaheen M. et al . (2018). Detection of antimicrobial resistance genes of Helicobacter pylori strains to clarithromycin, metronidazole, amoxicillin and tetracycline among Egyptian patients. Egypt. J. Med. Hum. Genet.19, 417–423. doi: 10.1016/j.ejmhg.2018.01.004
16
Dong D. Zou D. Liu H. Yang Z. Huang S. Liu N. et al . (2015). Rapid detection of pseudomonas aeruginosa targeting the toxA gene in intensive care unit patients from Beijing, China. Front. Microbiol.6:1100. doi: 10.3389/fmicb.2015.01100
17
Edward E. A. El Shehawy M. R. Abouelfetouh A. Aboulmagd E. (2023). Prevalence of different virulence factors and their association with antimicrobial resistance among Pseudomonas aeruginosa clinical isolates from Egypt. BMC Microbiol.23:161. doi: 10.1186/s12866-023-02897-8
18
Eliasi U. L. Sebola D. Oguttu J. W. Qekwana D. N. (2020). Antimicrobial resistance patterns of Pseudomonas aeruginosa isolated from canine clinical cases at a veterinary academic hospital in South Africa. J. S. Afr. Vet. Assoc.91:e1-e6. doi: 10.4102/jsava.v91i0.2052
19
Esterhuizen J. (2006). Seasonal abundance of horse flies (Diptera: Tabanidae) from two conservation areas in northeastern KwaZulu-Natal Province, South Africa. Afr. Entomol.14, 395–397. Available at: https://hdl.handle.net/10520/EJC32680
20
Faraji F. Mahzounieh M. Ebrahimi A. Fallah F. Teymournejad O. Lajevardi B. (2016). Molecular detection of virulence genes in Pseudomonas aeruginosa isolated from children with cystic fibrosis and burn wounds in Iran. Microb. Pathog.99, 1–4. doi: 10.1016/j.micpath.2016.07.013
21
Feldgarden M. Brover V. Haft D. H. Prasad A. B. Slotta D. J. Tolstoy I. et al . (2019). Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother.63, e00483–e00419. doi: 10.1128/AAC.00483-19
22
Fono-Tamo E. U. K. Kamika I. Dewar J. B. Lekota K. E. (2023). Comparative genomics revealed a potential threat of Aeromonas rivipollensis G87 strain and its antibiotic resistance. Antibiotics (Basel)12:131. doi: 10.3390/antibiotics12010131
23
Goulson D. Hughes W. O. Chapman J. W. (1999). Fly populations associated with landfill and composting sites used for household refuse disposal. Bull. Entomol. Res.89, 493–498. doi: 10.1017/S0007485399000644
24
Gurevich A. Saveliev V. Vyahhi N. Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics29, 1072–1075. doi: 10.1093/bioinformatics/btt086
25
Handfield J. Gagnon L. Dargis M. Huletsky A. (1997). Sequence of the ponA gene and characterization of the penicillin-binding protein 1A of Pseudomonas aeruginosa PAO1. Gene199, 49–56. doi: 10.1016/s0378-1119(97)00345-4
26
Hemmatinezhad B. Ommi D. Hafshejani T. T. Khamesipour F. (2015). Molecular detection and antimicrobial resistance of pseudomonas aeruginosa from houseflies (Musca domestica) in Iran. J. Venom. Anim. Toxins Incl. Trop. Dis.21:18. doi: 10.1186/s40409-015-0021-z
27
Hore G. Maity A. Naskar A. Ansar W. Ghosh S. Saha G. K. et al . (2017). Scanning electron microscopic studies on antenna of Hemipyrellia ligurriens (Wiedemann, 1830) (Diptera: Calliphoridae)-a blow fly species of forensic importance. Acta Trop.172, 20–28. doi: 10.1016/j.actatropica.2017.04.005
28
Hosu M. C. Vasaikar S. D. Okuthe G. E. Apalata T. (2021). Detection of extended spectrum beta-lactamase genes in pseudomonas aeruginosa isolated from patients in rural eastern cape province, South Africa. Sci. Rep.11:7110. doi: 10.1038/s41598-021-86570-y
29
Jolley K. A. Bray J. E. Maiden M. C. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.Org website and their applications. Wellcome Open Res.3:124. doi: 10.12688/wellcomeopenres
30
Khosravi A. D. Shafie F. Montazeri E. A. Rostami S. (2016). The frequency of genes encoding exotoxin a and exoenzyme S in Pseudomonas aeruginosa strains isolated from burn patients. Burns42, 1116–1120. doi: 10.1016/j.burns.2016.02.012
31
Kumar S. Stecher G. Li M. Knyaz C. Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.35, 1547–1549. doi: 10.1093/molbev/msy096
32
Li D. Liu C. M. Luo R. Sadakane K. Lam T. W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinctde bruijngraph. Bioinformatics31, 1674–1676. doi: 10.1093/bioinformatics/btv033
33
Li X. Z. Livermore D. M. Nikaido H. (1994). Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob. Agents Chemother.38, 1732–1741. doi: 10.1128/AAC.38.8.1732
34
Lister P. D. Wolter D. J. Hanson N. D. (2009). Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev.22, 582–610. doi: 10.1128/CMR.00040-09
35
Liu B. Zheng D. Jin Q. Chen L. Yang J. (2019). VFDB a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res.47, D687–D692. doi: 10.1093/nar/gky1080
36
Luzar M. A. Montie T. C. (1985). Avirulence and altered physiological properties of cystic fibrosis strains of Pseudomonas aeruginosa. Infect. Immun.50, 572–576. doi: 10.1128/iai.50.2.572-576.1985
37
Ly P. Aizenberg A. Martin T. Lopez M. Arturo Saldaña M. Hughes G. L. et al . (2018). Intestinal Myiasis caused by Sarcophaga spp. in Cusco, Peru: a case report and review of the literature. Case Rep. Infect. Dis.2018:3685439. doi: 10.1155/2018/3685439
38
Magome T. G. Ochai S. O. Hassim A. Bezuidenhout C. C. van Heerden H. Lekota K. E. (2024). A genome-based investigation of the Priestia species isolated from anthrax endemic regions in Kruger national park. Infect. Genet. Evol.123:105649. doi: 10.1016/j.meegid.2024.105649
39
Mlangeni L. N. Ramatla T. Lekota K. E. Price C. Thekisoe O. Weldon C. (2024). Occurrence, antimicrobial resistance, and virulence profiles of Salmonella Serovars isolated from wild reptiles in South Africa. Int. J. Microbiol.2024:5213895. doi: 10.1155/2024/5213895
40
Monyama M. C. Taioe O. M. Nkhebenyane J. S. van Wyk D. Ramatla T. Thekisoe O. M. (2023). Bacterial communities associated with houseflies (Musca domestica L.) inhabiting hospices in South Africa. Microorganisms11:1440. doi: 10.3390/microorganisms11061440
41
Nepali B. Bhattarai S. Shrestha J. (2018). Identification of Pseudomonas fluorescens using different biochemical tests. Int. J. Appl. Biol.2, 27–32. doi: 10.13140/RG.2.2.23860.40328
42
Neyestanaki D. K. Mirsalehian A. Rezagholizadeh F. Jabalameli F. Taherikalani M. Emaneini M. (2014). Determination of extended spectrum beta-lactamases, metallo-beta-lactamases and AmpC-beta-lactamases among carbapenem resistant Pseudomonas aeruginosa isolated from burn patients. Burns J. Int. Soc. Burn Inj.40, 1556–1561. doi: 10.1016/j.burns.2014.02.010
43
Pandey N. Cascella M. (2021). “Beta Lactam Antibiotics” in StatPearls, vol. 2021 (Treasure Island, FL: StatPearls Publishing).
44
Pang Z. Raudonis R. Glick B. R. Lin T. J. Cheng Z. (2019). Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv.37, 177–192. doi: 10.1016/j.biotechadv.2018.11.013
45
Parks D. H. Imelfort M. Skennerton C. T. Hugenholtz P. Tyson G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res.25, 1043–1055. doi: 10.1101/gr.186072.114
46
Paterson D. L. Bonomo R. A. (2005). Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev.18, 657–686. doi: 10.1128/CMR.18.4.657-686.2005
47
Rahmani M. Peighambari S. M. Svendsen C. A. Cavaco L. M. Agersø Y. Hendriksen R. S. (2013). Molecular clonality and antimicrobial resistance in Salmonella entericaserovars Enteritidis and Infantis from broilers in three northern regions of Iran. BMC Vet. Res.9:66. doi: 10.1186/1746-6148-9-66
48
Ramatla T. Ramaili T. Lekota K. Mileng K. Ndou R. Mphuthi M. et al . (2024a). Antibiotic resistance and virulence profiles of Proteus mirabilis isolated from broiler chickens at abattoir in South Africa. Vet. Med. Sci.10:e 1371. doi: 10.1002/vms3.1371
49
Ramatla T. Tutubala M. Motlhaping T. de Wet L. Mokgokong P. Thekisoe O. et al . (2024b). Molecular detection of Shiga toxin and extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolates from sheep and goats. Mol. Biol. Rep.51:57. doi: 10.1007/s11033-023-08987-0
50
Ren L. Shang Y. Chen W. Meng F. Cai J. Zhu G. et al . (2018). A brief review of forensically important flesh flies (Diptera: Sarcophagidae). For. Sci. Res.3, 16–26. doi: 10.1080/20961790.2018.1432099
51
Santajit S. Indrawattana N. (2016). Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int.2016, 1–8. doi: 10.1155/2016/2475067
52
Shacheraghi F. Shakibaie P. Noveiri H. (2010). Molecular identification of ESBL genesbla GES-1, blaVEB-1, blaCTX-M, blaOXA-1, blaOXA-4, blaOXA-10 and blaPER-1 in Pseudomonas aeruginosa strains isolated from burn patientsby PCR, RFLP and sequencing techniques. World Acad. Sci. Eng. Technol.4, 114–118.
53
Shety R. Dehuri M. Panda M. Mohanty B. (2022). Diversity and seasonal dynamics of dipteran flies infesting cattle and its habitation in Bhubaneswar, India. Int. J. Trop. Insect Sci.42, 983–988. doi: 10.1007/s42690-021-00612-6
54
Skevington J. H. Dang P. T. (2002). Exploring the diversity of flies (Diptera). Biodiversity3, 3–2. doi: 10.1080/14888386.2002.9712613
55
Snyman L. P. Neves L. Lempereur L. Bosman A. C. (2020). Overview of the horseflies (Diptera: Tabanidae) of South Africa: assessment of major collections for spatiotemporal analysis. Aust. J. Entomol.59, 549–560. doi: 10.1111/aen.12466
56
Taioe M. O. Motloang M. Y. Namangala B. Chota A. Molefe N. I. Musinguzi S. P. et al . (2017, 2017). Characterization of tabanid flies (diptera: tabanidae) in South Africa and Zambia and detection of protozoan parasites they are harbouring. Parasitology144, 1162–1178. doi: 10.1017/S0031182017000440
57
Terada L. S. Johansen K. A. Nowbar S. Vasil A. I. Vasil M. L. (1999). Pseudomonas aeruginosa hemolytic phospholipase C suppresses neutrophil respiratory burst activity. Infect. Immun.67, 2371–2376. doi: 10.1128/IAI.67.5.2371-2376.1999
58
Tippmann H. (2004). Analysis for free: comparing programs for sequence analysis. Brief. Bioinform.5, 82–87. doi: 10.1093/bib/5.1.82
59
Tohya M. Teramoto K. Watanabe S. Hishinuma T. Shimojima M. Ogawa M. et al . (2022). Whole-genome sequencing-based re-identification of Pseudomonas putida/fluorescens clinical isolates identified by biochemical bacterial identification systems. Microbiol. Spectr.10:e02491-21. doi: 10.1128/spectrum.02491-21
60
Treves D. S. (2010). Review of three DNA analysis applications for use in the microbiology or genetics classroom. J. Microbiol. Biol. Educ.11, 186–187. doi: 10.1128/jmbe.v11i2.205
61
Uemura S. Yokota S. I. Shiraishi T. Kitagawa M. Hirayama S. Kyan R. et al . (2017). Adaptive cross-resistance to aminoglycoside antibiotics in Pseudomonas aeruginosa induced by topical dosage of neomycin. Chemotherapy62, 121–127. doi: 10.1159/000449368
Summary
Keywords
Diptera flies, Pseudomonas aeruginosa , antibiotic resistance, virulence genes, WGS
Citation
de Wet L, Matle I, Thekisoe O, Lekota KE and Ramatla T (2025) Uncovering antibiotic resistance: extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa from dipteran flies in residential dumping and livestock environments. Front. Microbiol. 16:1586811. doi: 10.3389/fmicb.2025.1586811
Received
03 March 2025
Accepted
16 May 2025
Published
04 June 2025
Volume
16 - 2025
Edited by
Hazem Ramadan, Mansoura University, Egypt
Reviewed by
Ahmed Ali Al-Qahtani, King Faisal Specialist Hospital and Research Centre, Saudi Arabia
Rosario Morales-Espinosa, National Autonomous University of Mexico, Mexico
Updates
Copyright
© 2025 de Wet, Matle, Thekisoe, Lekota and Ramatla.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tsepo Ramatla, tramatla@cut.ac.za
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.