- 1Department of Pharmacy, Nanchong Key Laboratory of Individualized Drug Therapy, Beijing Anzhen Nanchong Hospital of Capital Medical University & Nanchong Central Hospital, Nanchong, China
- 2Department of Laboratory, Beijing Anzhen Nanchong Hospital of Capital Medical University & Nanchong Central Hospital, Nanchong, China
Background: This study aims to investigate the distribution and drug resistance of Gram-negative bacteria in the intensive care unit (ICU) of a tertiary general hospital in Sichuan Province, with the goal of promoting rational antibiotic use and reducing multidrug resistance.
Methods: A retrospective analysis was conducted on the distribution and drug resistance of Gram-negative bacteria in ICU samples collected from January 2019 to December 2024.
Results: A total of 83,944 culture samples were analyzed, primarily blood (45.27%) and sputum (41.34%) specimens, with a steady increase in sample types annually. A total of 7,211 strains were isolated, 76.43% of which were from respiratory tract specimens. The predominant pathogens included Klebsiella pneumoniae (31.17%), Acinetobacter baumannii (30.11%), Escherichia coli (14.05%), and Pseudomonas aeruginosa (11.34%). The detection rates for carbapenem-resistant A. baumannii (CRAB) were 61.88%, carbapenem-resistant K. pneumoniae (CRKP) 29.28%, carbapenem-resistant P. aeruginosa (CRPA) 5.80%, and carbapenem-resistant E. coli (CREC) 3.04%. Susceptibility testing revealed fluctuating resistance rates for E. coli over the past 6 years. Notably, K. pneumoniae exhibited significant resistance to carbapenems (e.g., imipenem) and third-generation cephalosporins (e.g., ceftazidime).
Conclusion: From 2019 to 2024, the ICU experienced a severe problem with Gram-negative drug-resistant bacteria, particularly Enterobacteriaceae resistant to third-generation cephalosporins. A. baumannii isolates demonstrated resistance to most antibiotics, underscoring the need for continuous monitoring and the selection of effective antibiotics based on clinical practice.
1 Introduction
Antimicrobial resistance represents one of the most significant global public health challenges. In 2019, it was estimated that this issue contributed to approximately 4.95 million deaths worldwide, with a particularly pronounced impact in low- and middle-income countries (Murray, C. J. L. et al., 2022). In May 2024, the World Health Organization (WHO) revised its list of bacterial pathogens that pose the greatest threat to human health. Notably, antibiotic-resistant Gram-negative bacteria (GNB), including Acinetobacter baumannii and several pathogens within the Enterobacterales order, were highlighted as top priorities. These bacteria are especially concerning due to their ability to transfer resistance genes, their growing resistance to current treatments, and the severity of the infections they cause, which significantly contribute to the global disease burden. GNB are a leading etiology of nosocomial infections in intensive care units (ICUs), with their epidemiological distribution and antimicrobial resistance (AMR) profiles critically influencing the formulation of evidence-based anti-infective strategies. The escalating use of broad-spectrum antibiotics has precipitated a marked increase in multidrug-resistant (MDR) and extensively drug-resistant (XDR) GNB strains, particularly in ICUs, where these pathogens pose formidable challenges to infection prevention and patient outcomes. The evolution of AMR in GNB is multifactorial. It is driven by antibiotic selection pressure, suboptimal adherence to infection control protocols, and genetic mechanisms such as horizontal gene transfer and efflux pump overexpression (Dulanto Chiang and Dekker, 2019; ICPIC, 2019). ICU patients are particularly susceptible to drug-resistant infections due to immunosuppression, critical illness, and frequent exposure to invasive devices (e.g., mechanical ventilation, central venous catheters), which disrupt innate barriers and promote microbial colonization (Jiang et al., 2023). Furthermore, The heterogeneous and prolonged use of antibiotics in ICUs accelerates the clonal expansion and dissemination of resistant strains (Doron and Davidson, 2013). This study aimed to describe the epidemiological patterns and temporal AMR trends of GNB isolates in a tertiary hospital’s ICU. By analyzing retrospective microbiological surveillance data, we aimed to inform targeted antimicrobial stewardship interventions and optimize infection control policies. Understanding the dynamic interplay between pathogen distribution, resistance mechanisms, and clinical practices is crucial for mitigating the emergence of MDR/XDR pathogens and improving therapeutic efficacy.
2 Methodology
2.1 Sources of bacterial specimens
Bacterial specimens were obtained from patients admitted to the intensive care unit (ICU) of a large, tertiary-level, general teaching hospital in Sichuan Province between January 2019 and December 2024. Gram-negative bacilli strains isolated from sputum, urine, blood, and other clinical specimens were included in the study.
2.2 Isolation, culture, and identification of bacteria
All specimens were collected from ICU patients under sterile conditions. Blood samples were obtained via venipuncture into pre-labeled aerobic and anaerobic culture bottles. Sputum (both aspirated and expectorated), puncture fluid, and urine were promptly transported to the laboratory upon collection. Upon arrival, blood cultures were incubated using the BACTEC FX40 system, while sputum samples underwent Gram staining and quality assessment prior to inoculation onto agar plates. Bacterial isolation and culture were performed in strict adherence to the guidelines established by the Clinical and Laboratory Standards Institute (CLSI).
2.3 Antimicrobial susceptibility testing
The drug sensitivity testing was conducted in accordance with the guidelines recommended by the CLSI. Antimicrobial susceptibility was assessed using both automated instrumentation and the disk diffusion method. The minimum inhibitory concentrations (MICs) of antimicrobial agents were determined utilizing the Trek system. Susceptibility, intermediate, and resistance categories were assigned based on the breakpoints specified in the CLSI’s “Performance Standards for Antimicrobial Susceptibility Testing,” 31st edition, published in 2021. Definition of drug-resistant strains are listed in Supplementary material.
2.4 Statistical methods
Count data were presented as n (%) and trend p values were calculated using GraphPad Prism 10.0 (San Diego, United States), employing the chi-square test for trend. A p-value of less than 0.05 was considered indicative of statistical significance.
3 Results
3.1 Trend analysis of culture sample distribution and pathogen detection from 2019 to 2024
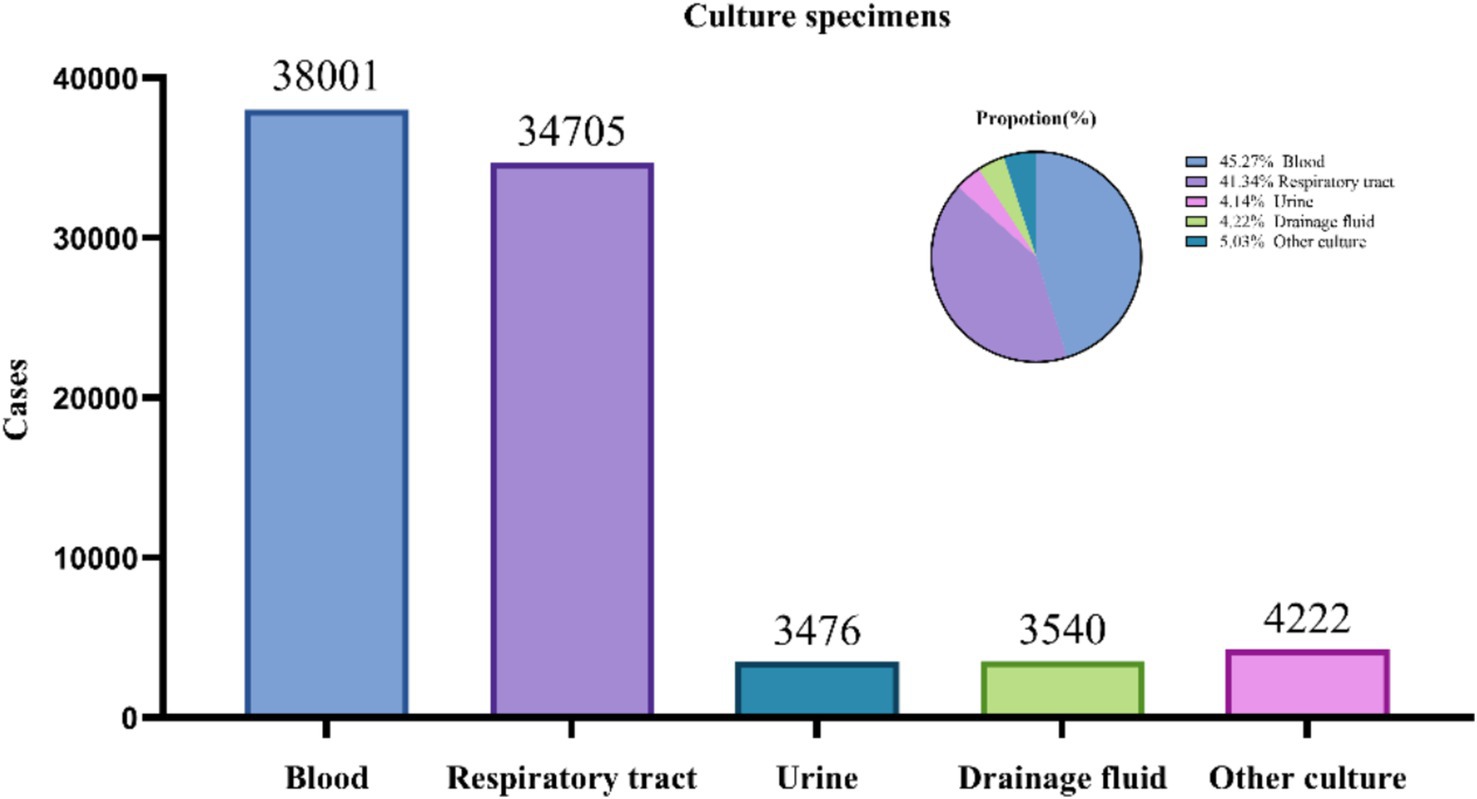
From 2019 to 2024, a total of 83,944 culture samples were collected. Blood samples (38,001; 45.27%) and respiratory tract samples (33,705; 41.34%) constituted the majority of the samples. The pathogens detected in drainage cultures (3,476; 4.14%), urine cultures (3,540; 4.22%), and other types of cultures (4,222; 5.03%) are summarized in Figure 1.
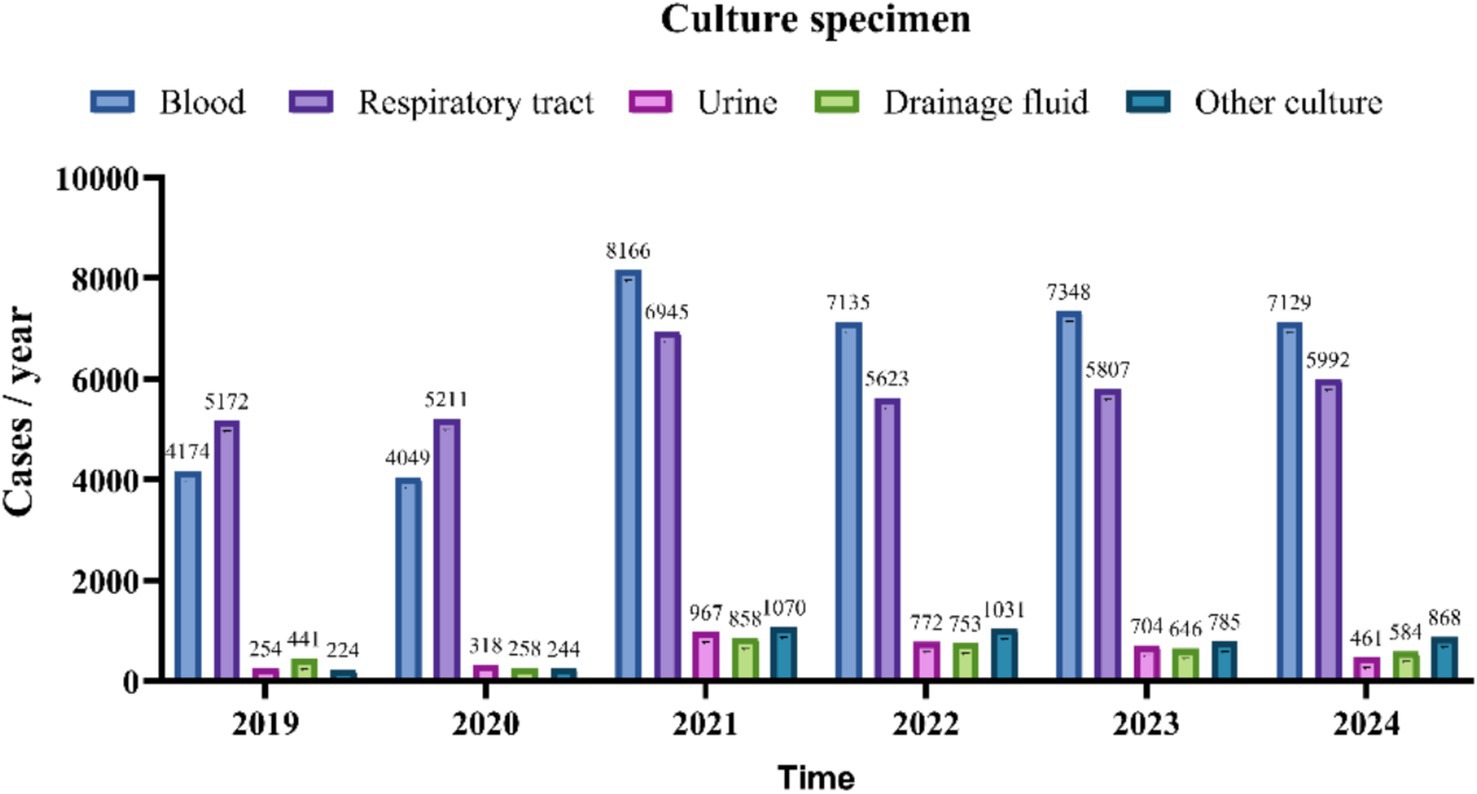
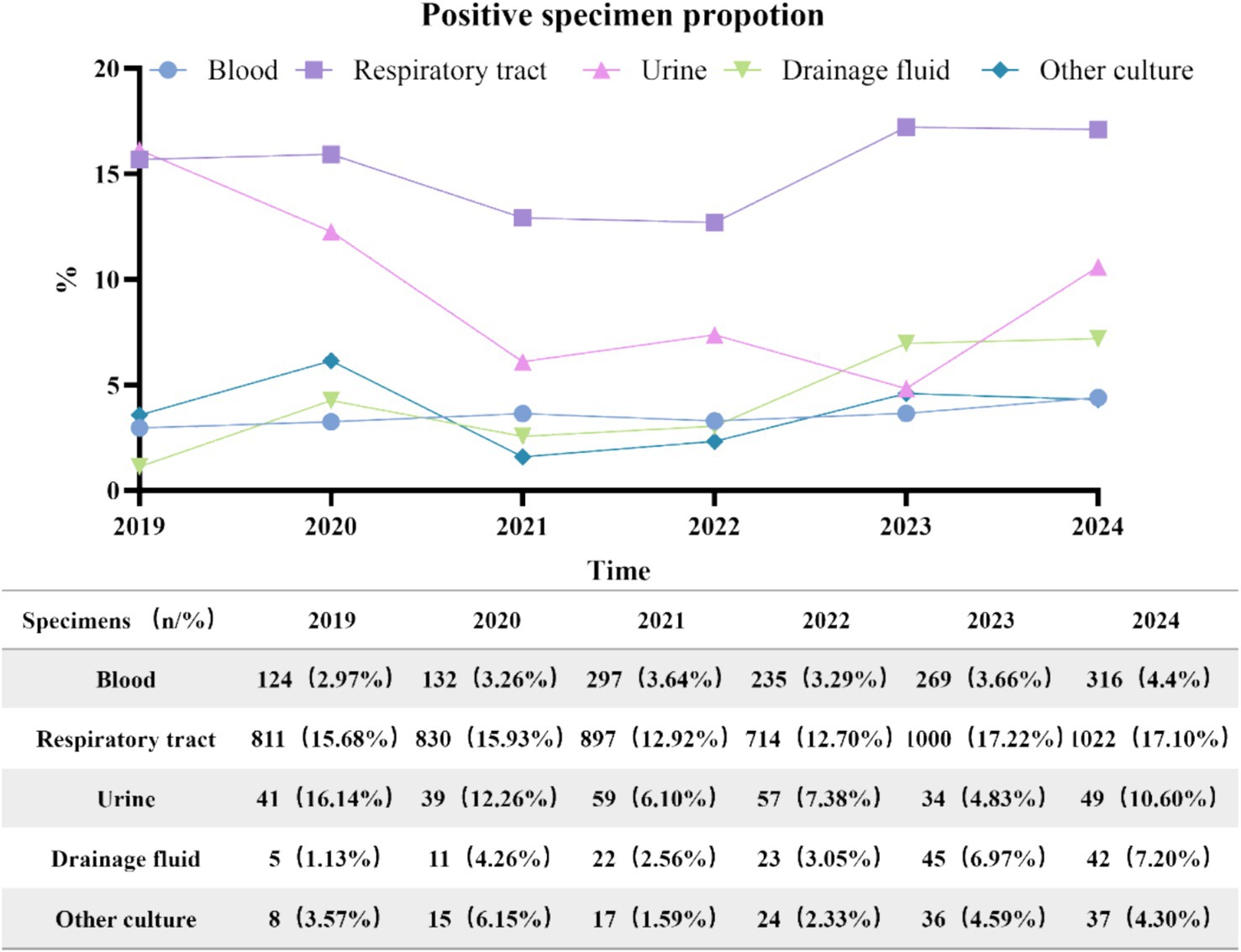
Between 2019 and 2024, the number of various types of culture specimens exhibited a consistent upward trend, as illustrated in Figure 2. A total of 7,211 positive specimens were cultured from 2019 to 2024. The annual distribution was as follows: 989 in 2019, 1,027 in 2020, 1,292 in 2021, 1,053 in 2022, 1,384 in 2023, and 1,466 in 2024. Specimen types included 5,274 respiratory specimens (73.1%), 1,373 blood specimens (19.0%), 148 drainage fluid specimens (2.1%), 279 urine specimens (3.9%), and 137 other types of specimens (1.9%). Respiratory and drainage fluid specimens demonstrated a progressive increase in positivity rates over the study period, while blood cultures maintained a stable proportion of positive results. In contrast, urine cultures exhibited an initial rise in positivity rates followed by a gradual decline after 2021, as illustrated in Figure 3.
3.2 Detection rate of Gram-negative bacteria in ICU during 2019–2024
This study examined 83,944 clinical isolates of Gram-negative bacteria collected from bloodstream, respiratory tract, urinary tract, and abdominal infections. Between 2019 and 2024, Klebsiella pneumoniae and the A. baumannii complex constituted 31.22 and 30.38% of the total cases, respectively. K. pneumoniae exhibited the highest overall burden, with a significant increase from 266 cases (27.01%) in 2019 to 470 cases (34.28%) in 2023, peaking that year. Although there was a slight decline to 400 cases (35.24%) in 2024, it remains a critical pathogen. Haemophilus influenzae demonstrated an alarming 11.6-fold increase from 5 cases to 56 cases, indicating emerging clinical significance. Serratia marcescens surged from 2 cases (0%) in 2019 to 29 cases (2.56%) in 2024, reflecting potential shifts in infection dynamics. Pseudomonas aeruginosa showed a sharp decline, dropping from 133 cases (13.50%) in 2019 to 61 cases (5.37%) in 2024. A. baumannii complex experienced significant fluctuations, peaking at 416 cases (42.23%) in 2019 before plummeting to 203 cases (19.41%) in 2022, followed by partial recovery to 314 cases (27.67%) in 2024. Stenotrophomonas maltophilia, Enterobacter cloacae complex, and Citrobacter freundii exhibited irregular patterns. Klebsiella oxytoca, Enterobacter aerogenes, and Burkholderia cepacia maintained consistently low counts. Refer to Table 1 for detailed data.
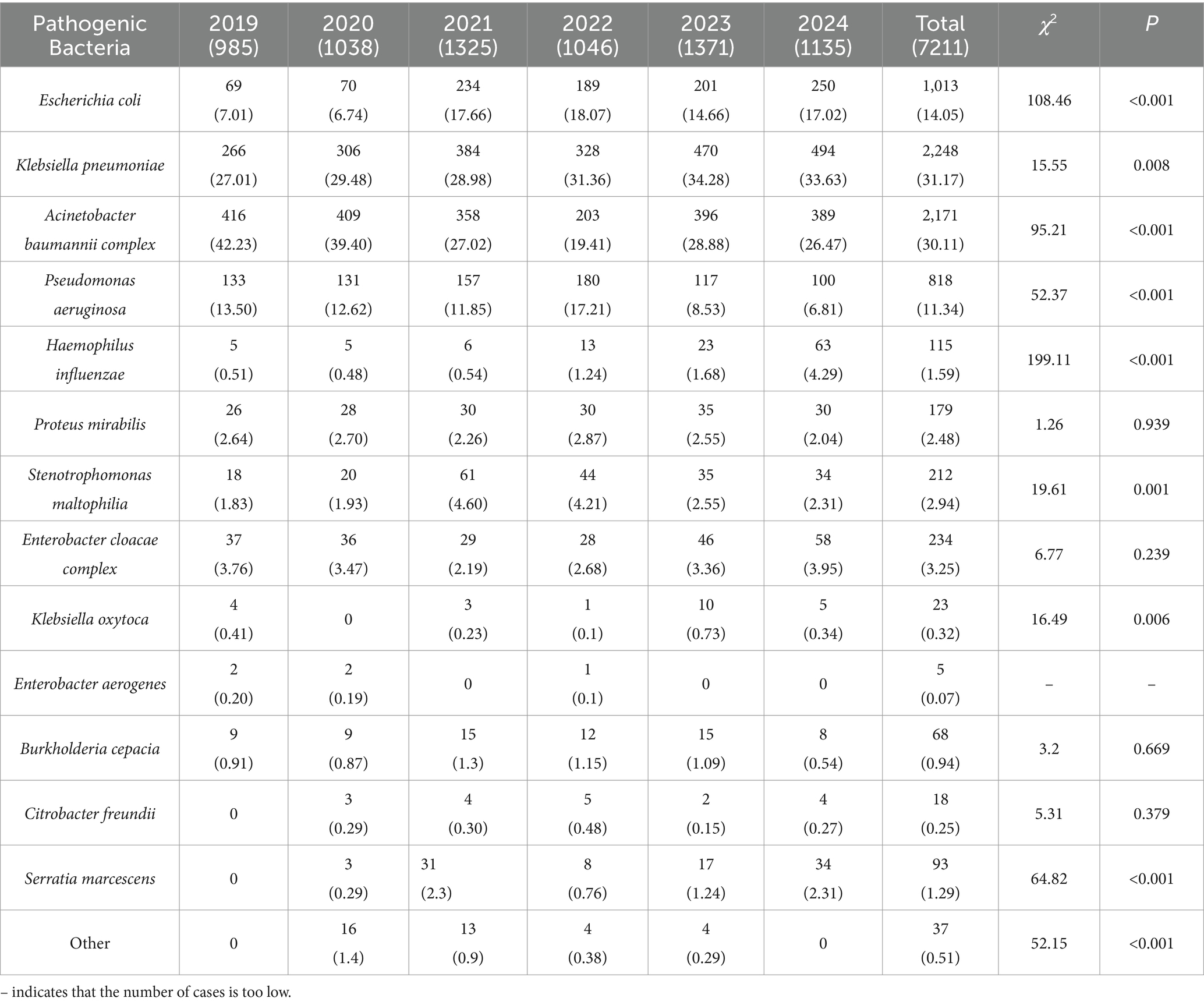
Table 1. Distribution and composition of pathogens in ICU: Gram-negative bacteria from 2019 to 2024 [n (%)].
3.2.1 Gram-negative bacteria across various infection sites
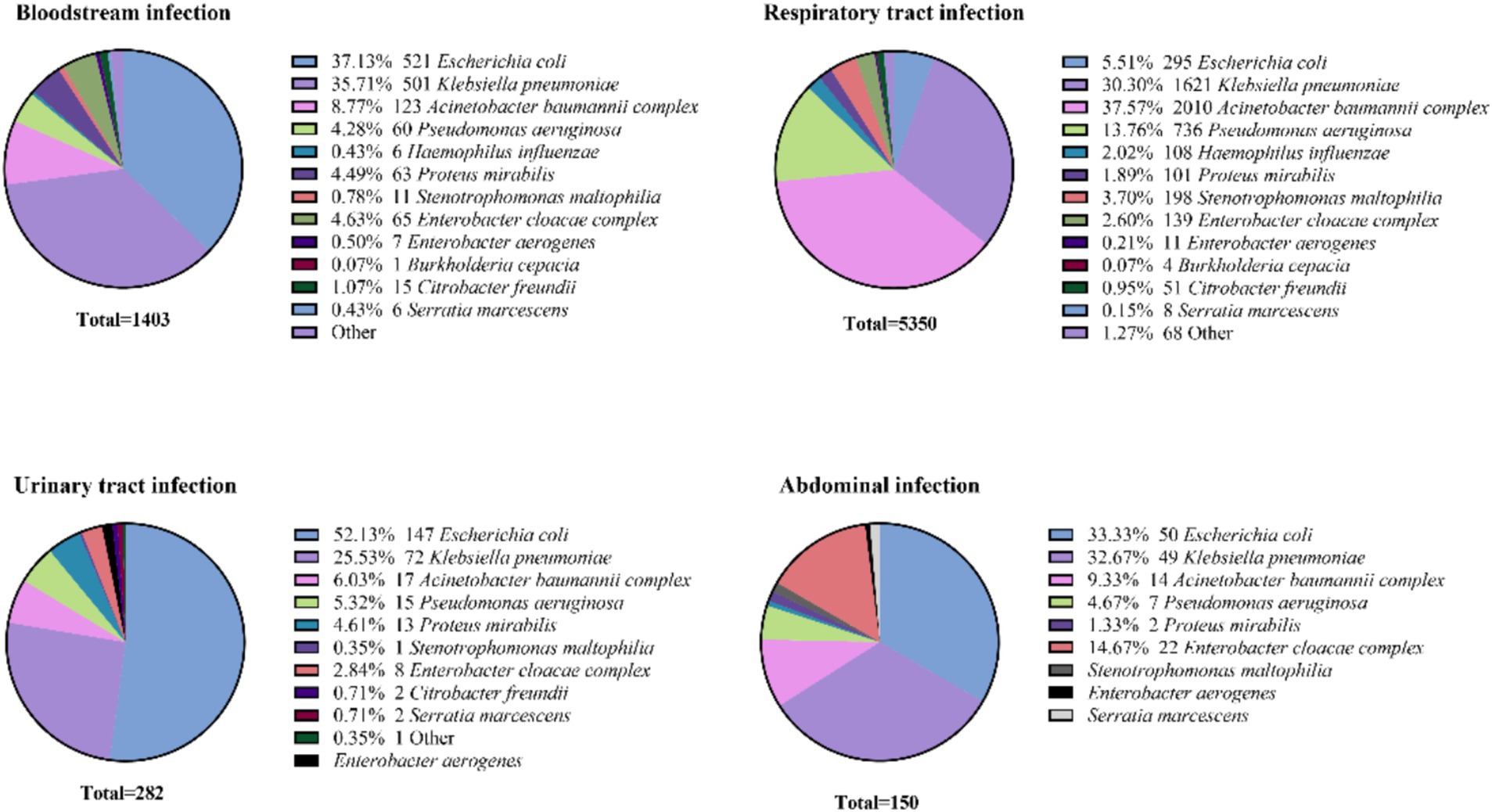
Escherichia coli was the predominant pathogen in urinary tract infections (52.13%, 147/295) and bloodstream infections (37.13%, 485/1403), followed by abdominal infections (33.33%, 50/150), while it was least isolation in respiratory tract infection (5.51%, 95/5350). In contrast, K. pneumoniae ranked second or third at all infection sites, with the highest detection rate observed in respiratory infections (30.30%, 1621/5350). P. aeruginosa and the A. baumannii complex were predominantly isolated from respiratory tract infections (13.76%, 736/5350 and 37.57%, 2010/5350, respectively), with lower frequencies in abdominal infections (4.67%, 6/150 and 9.33%, 11/150). B. cepacia showed moderate prevalence in bloodstream (0.07%, 15/1403) and respiratory infections (0.07%, 49/5133), as depicted in Figure 4.
3.2.2 Detection rates of multidrug-resistant Gram-negative bacteria in the ICU
We analyzed multidrug-resistant Gram-negative bacteria, including carbapenem-resistant E. coli and K. pneumoniae, as well as carbapenem-resistant P. aeruginosa and A. baumannii (Table 2). The average detection rates for carbapenem-resistant A. baumannii, E. coli, K. pneumoniae, and P. aeruginosa demonstrate a pattern of fluctuation.
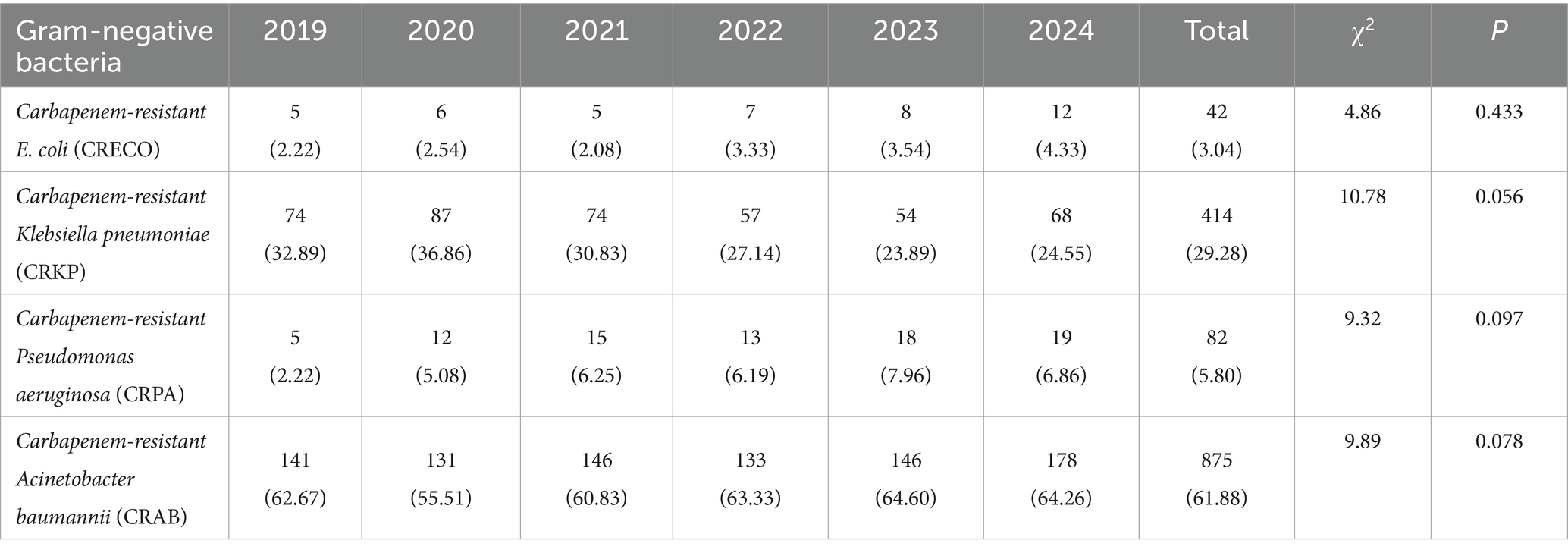
Table 2. Detection rates of multidrug-resistant Gram-negative bacteria in the ICU from 2019 to 2024 [n (%)].
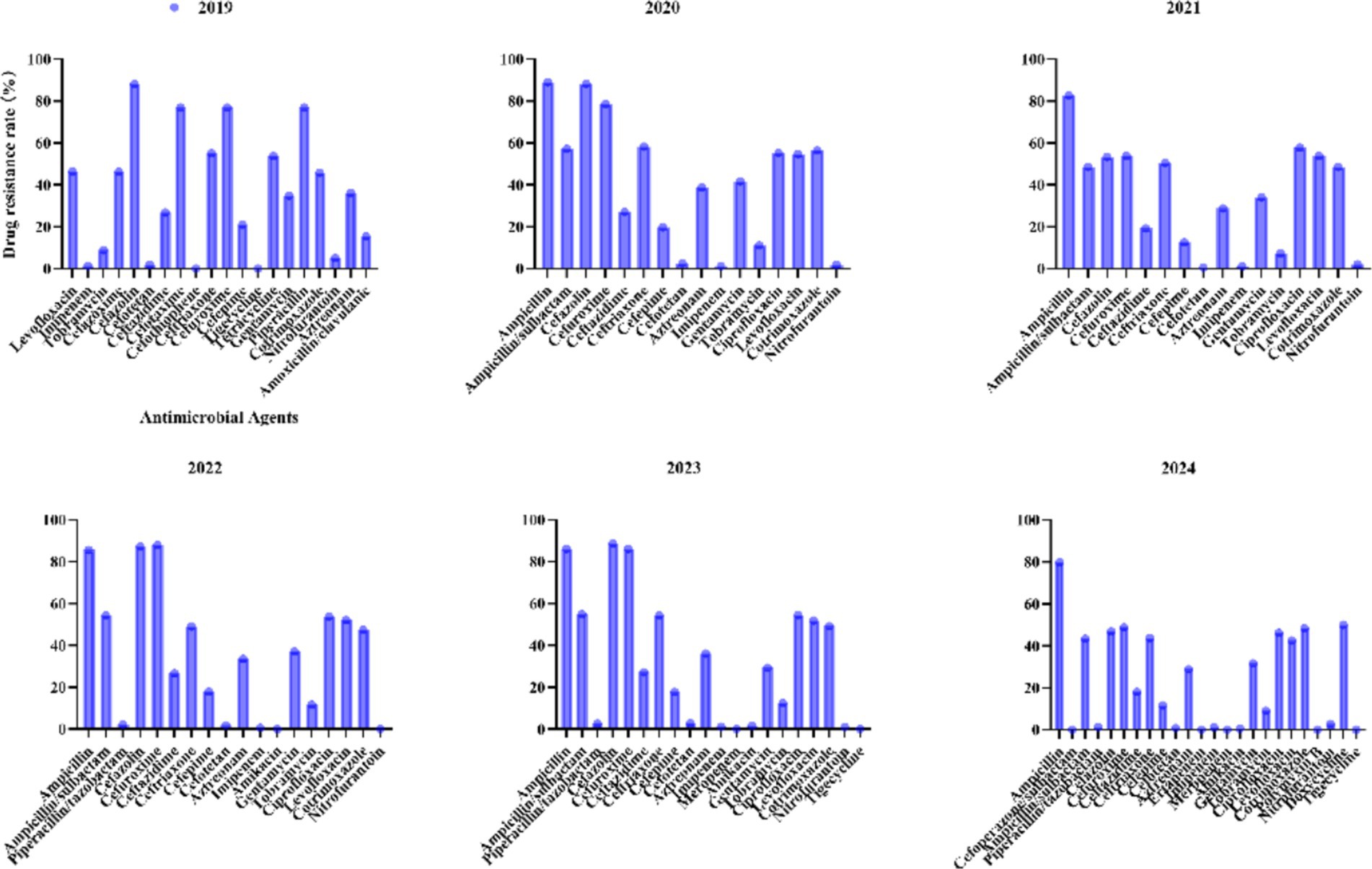
3.3 Antimicrobial resistance profiles of Escherichia coli to commonly used antibiotics from 2019 to 2024
For E. coli, recent years have witnessed significant fluctuations in antibiotic resistance patterns (Figure 5). Ampicillin has consistently exhibited high resistance levels (>79.8% annually), peaking at 89% in 2020 and marginally declining to 79.8% by 2024. Similarly, cephalosporins such as ceftriaxone showed an initial increase from 55.1% in 2019 to 54.3% in 2023, followed by a notable reduction to 43.7% in 2024, underscoring the need for continuous surveillance. Fluoroquinolones, including ciprofloxacin (from 55.2 to 46.1%) and levofloxacin (from 54.6 to 42.5%), demonstrated gradual declines between 2020 and 2024. Carbapenems (imipenem/meropenem) maintained exceptional efficacy, with resistance rates consistently ≤1.3%. Piperacillin/tazobactam exhibited minimal resistance (<2.8%), further decreasing to 1.3% by 2024. Nitrofurantoin continued to show negligible resistance (0–2.8%), highlighting its reliability for treating urinary tract infections.
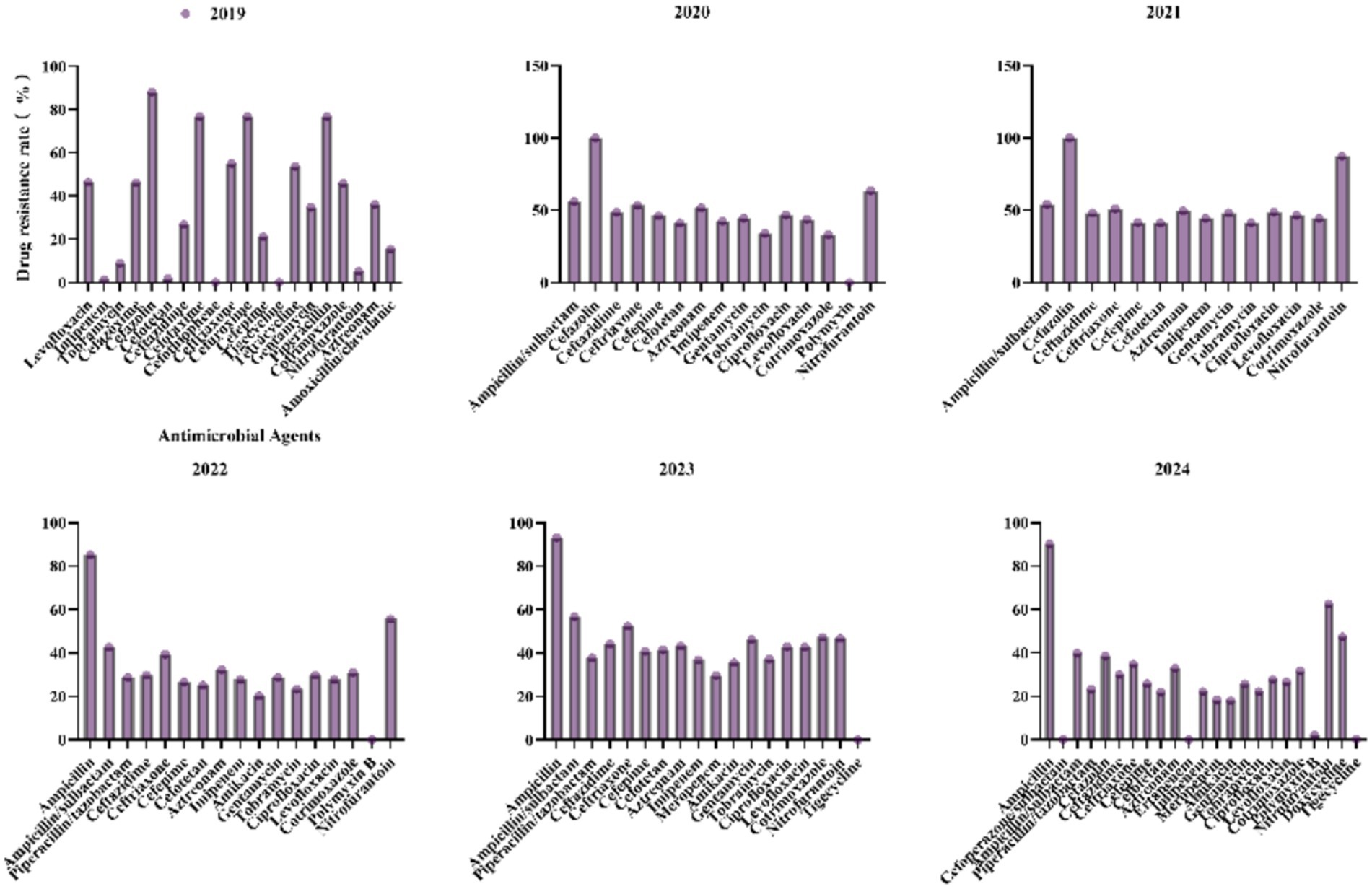
3.4 Antimicrobial resistance profiles of Klebsiella pneumoniae to commonly used antibiotics from 2019 to 2024
For K. pneumoniae, antibiotic resistance patterns have shown significant fluctuations in recent years (Figure 6). Ampicillin/Sulbactam resistance peaked at 56.9% in 2023 and then dropped to 39.8% in 2024. Ceftazidime resistance increased from 26.8% in 2019 to 48.6% in 2020, before decreasing to 29.9% in 2024. Piperacillin/Tazobactam resistance rose from 28.7% in 2022 to 37.7% in 2023, then fell to 23.1% in 2024, indicating intermittent selective pressures. Imipenem resistance sharply increased from 1.3% in 2019 to 36.8% in 2023, then declined to 22% in 2024, suggesting the emergence of carbapenemase-producing strains. Meropenem resistance was 29.7% in 2023 and decreased to 18.2% in 2024, highlighting a threat to last-line therapies. Resistance to Ciprofloxacin and Levofloxacin decreased from 46.9 and 43.8% in 2020 to 27.6 and 26.5% in 2024, likely due to reduced fluoroquinolone use. Polymyxin B and Tigecycline resistance remained low (0–2.1%), maintaining their efficacy against multidrug-resistant strains. The rise in carbapenem resistance underscores the need for robust antimicrobial stewardship programs and enhanced surveillance for carbapenemase genes.
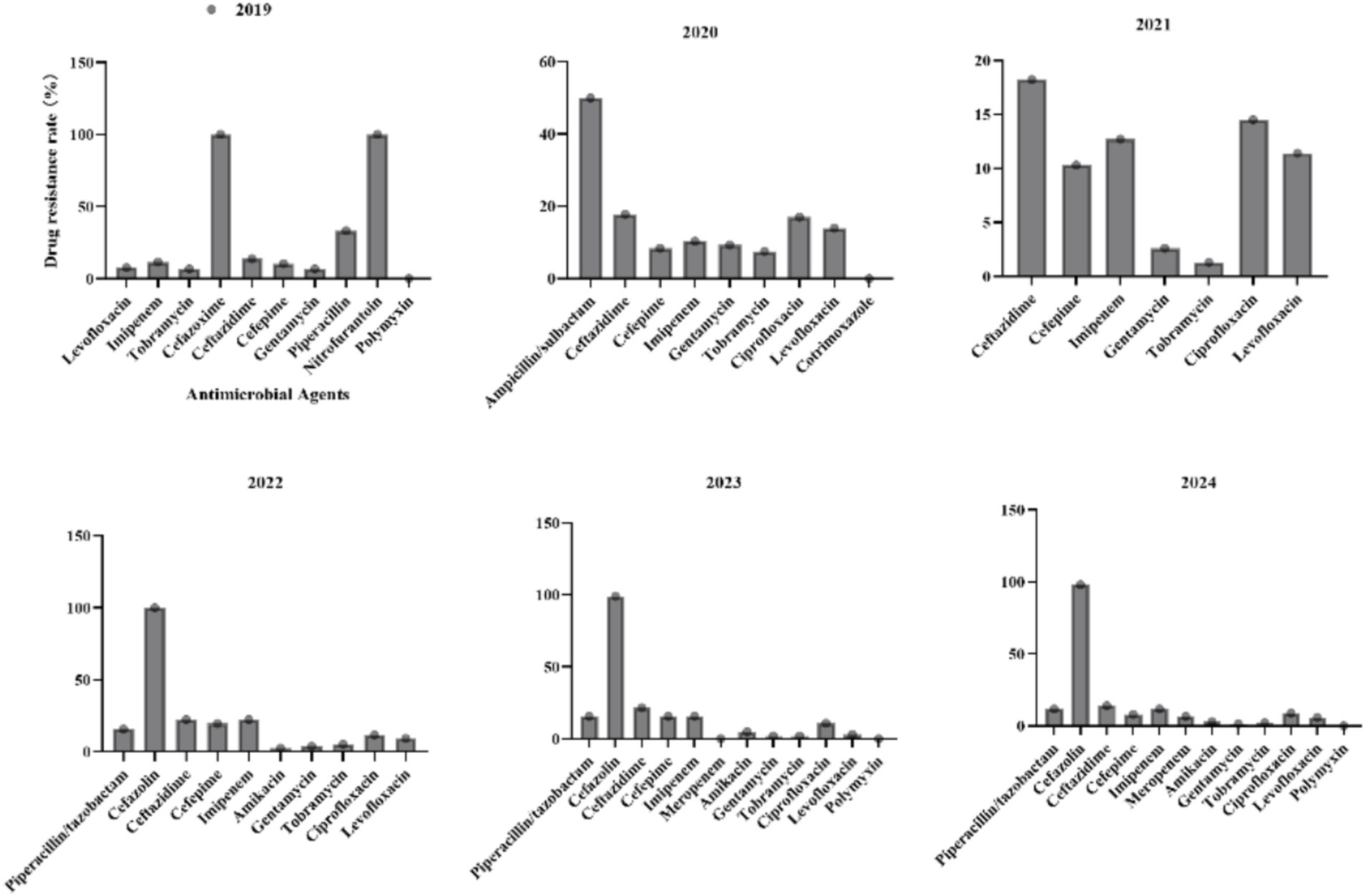
3.5 Antimicrobial resistance profiles of Pseudomonas aeruginosa to commonly used antibiotics from 2019 to 2024
For P. aeruginosa (Figure 7), the resistance to Ceftazidime initially increased from 13.8% in 2019 to 21.6% in 2023, followed by a significant reduction to 14% in 2024. Resistance to Imipenem exhibited cyclical fluctuations, peaking at 22.4% in 2022 before declining to 11.8% in 2024. Amikacin, Gentamicin, and Tobramycin maintained consistently low resistance rates (<10%) throughout the study period, highlighting their reliability for clinical use. Resistance to Ciprofloxacin and Levofloxacin fluctuated without a clear trend. Polymyxin demonstrated sustained effectiveness with no observed resistance (0%) across all reported years.
3.6 Antimicrobial resistance profiles of Acinetobacter baumannii complex to commonly used antibiotics from 2019 to 2024
For the A. baumannii complex (Figure 8), carbapenems such as imipenem and meropenem exhibited consistently high resistance rates (>88%) across all years, with meropenem reaching a peak of 91.7% in 2023. Tobramycin resistance increased steadily from 59.3% in 2019 to 79.9% in 2024. Although resistance to ampicillin/sulbactam has declined from 91.3% in 2020 to 66% in 2024, it remains critically elevated, thereby limiting its clinical utility. Resistance to levofloxacin showed significant fluctuations, decreasing sharply from 72.6% in 2019 to 39.8% in 2021, before increasing again to 69.9% in 2024. Polymyxin B and tigecycline maintained 0% resistance in 2024, underscoring their importance as viable options for treating multidrug-resistant infections.
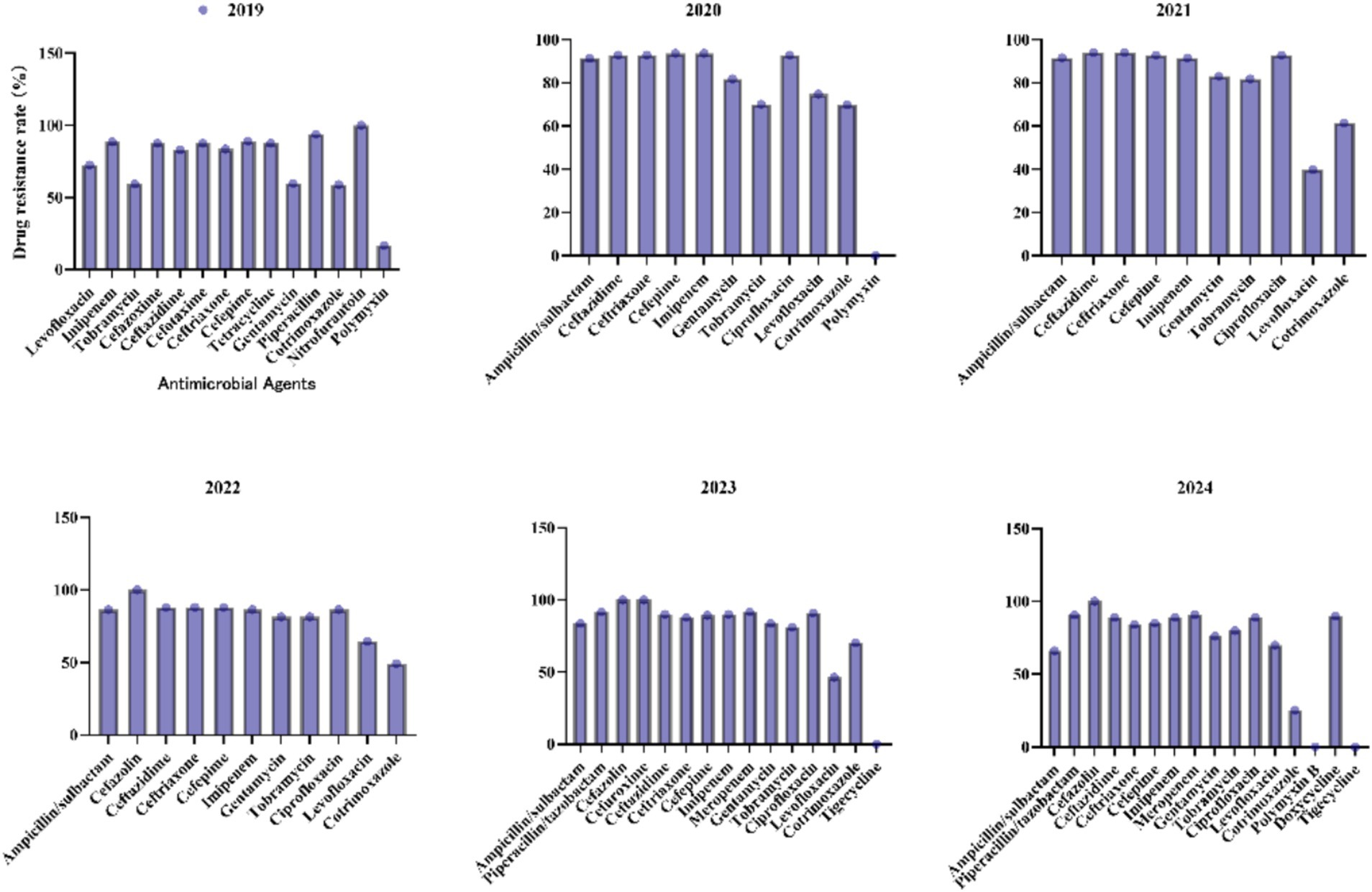
Figure 8. Antimicrobial resistance profiles of Acinetobacter baumannii complex to commonly used antibiotics.
4 Discussion
The intensive care unit (ICU) represents a critical battleground in the global AMR crisis, where the interplay of host vulnerability, invasive interventions, and microbial adaptability fosters the emergence and dissemination of MDR Gram-negative pathogens. This study delineates a concerning epidemiological landscape in western China, characterized by escalating carbapenem resistance in A. baumannii (CRAB: 62.14%) and K. pneumoniae (CRKP: 29.19%), mirroring global trends of ICU-acquired infections dominated by “ESKAPE” pathogens. The ICU environment—marked by immunosuppression, prolonged device use (e.g., ventilators, catheters), and frequent broad-spectrum antibiotic exposure—creates a permissive niche for MDR colonization and biofilm formation. Notably, bloodstream (45.27%) and respiratory (41.34%) infections predominate, reflecting the vulnerability of critically ill patients to invasive procedures and impaired mucosal barriers, which facilitate pathogen translocation and systemic dissemination (Vincent et al., 2020).
The dominance of K. pneumoniae and A. baumannii—accounting for over 60% of isolates, the fluctuating prevalence of A. baumannii complex 42.23% in 2019 to 26.47 in 2024. The marked decline in P. aeruginosa isolates (13.50 to 6.81%) contrasts with the rising prevalence of K. pneumoniae (27.01 to 33.63%), mirroring trends observed in Asia and Europe where K. pneumoniae has emerged as a dominant nosocomial pathogen due to its adaptability and carbapenemase-producing strains (Nordmann et al., 2012; Tacconelli et al., 2018). The 11.6-fold increase in H. influenzae and the emergence of S. marcescens (0 to 2.31%) suggest shifts in infection dynamics, possibly linked to changes in antibiotic prophylaxis or host immune status (Schelenz et al., 2016). However, its persistent dominance in respiratory infections (37.85%) highlights the urgent need for enhanced environmental decontamination and stewardship in ICUs. The staggering carbapenem resistance rates in A. baumannii (CRAB: 62.14%) and K. pneumoniae (CRKP: 29.19%) signal a regional epidemic of carbapenemase-producing strains, likely exacerbated by the overuse of empiric carbapenems in critically ill patients (Cortés et al., 2020). This aligns with national surveillance data from China, where CRAB prevalence in ICUs exceeds 50%, driven by clonal dissemination of blaOXA-23-harboring strains (Jiang et al., 2019; Liu and Liu, 2021). These findings align with global surveillance data, where carbapenemase genes (e.g., blaNDM, blaOXA-48) have disseminated rapidly in healthcare settings (Grundmann et al., 2017; Logan and Weinstein, 2017). The transient decline in CRKP resistance (22% in 2024) may reflect localized stewardship interventions, but the overall trajectory remains concerning. Similarly, the high resistance of A. baumannii to meropenem (91.7% in 2023) underscores the limited therapeutic options for these pathogens, necessitating strict adherence to carbapenem-sparing protocols (Tamma et al., 2022). To break the cycle of resistance, a dual focus on antimicrobial stewardship and environmental decontamination is essential. Machine learning models integrating real-time resistance data, patient comorbidities, and antibiotic consumption patterns could optimize empiric therapy (Feretzakis et al., 2020). Additionally, novel non-antibiotic adjuvants—such as phage therapy targeting CRAB biofilms or efflux pump inhibitors to restore carbapenem susceptibility—offer promise in preclinical studies (Strathdee et al., 2023). Locally, the high CRAB prevalence warrants investment in strict contact precautions and UV-C disinfection robots, which reduced CRAB transmission by 67% in a Korean ICU trial (Park et al., 2023).
The observed resistance patterns reveal pathogen-specific evolutionary strategies. A. baumannii’s pan-resistance to carbapenems and aminoglycosides correlates with its genomic plasticity, enabling rapid acquisition of resistance islands and efflux pump overexpression (Abd El-Rahman et al., 2023). In contrast, K. pneumoniae’s resistance to third-generation cephalosporins (e.g., ceftazidime) and carbapenems may stem from co-carriage of blaCTX-M-15 and blaNDM-5 on conjugative plasmids, facilitating horizontal gene transfer (Yang et al., 2023). The lower carbapenem resistance in E. coli (5.68%) compared to K. pneumoniae suggests ecological partitioning of resistance genes, possibly due to fitness costs associated with carbapenemase production in E. coli (Partridge et al., 2018). However, the fluctuating resistance of E. coli to cephalosporins highlights dynamic shifts in ESBL epidemiology, potentially linked to antibiotic cycling practices or community-acquired strain introductions (Tamma et al., 2021). The data necessitate a paradigm shift in ICU antimicrobial strategies. First, the high CRAB burden demands restrictive carbapenem use and adoption of rapid molecular diagnostics (e.g., PCR for blaOXA-51) to differentiate colonization from true infection, minimizing unnecessary treatment (van Belkum et al., 2019). Second, the rising CRKP prevalence calls for tailored empiric regimens: in regions with >20% CRKP, ceftazidime-avibactam or meropenem-vaborbactam should replace carbapenems as first-line therapy for sepsis, guided by local antibiograms (Tamma et al., 2021).
The E. coli maintained low carbapenem resistance (≤1.3%), consistent with its lesser propensity for carbapenemase production compared to Klebsiella and Acinetobacter species. However, the rising resistance to cephalosporins (e.g., ceftriaxone: 43.7%) signals the spread of extended-spectrum β-lactamases (ESBLs), urging the adoption of rapid diagnostics to guide targeted therapy (Paterson and Bonomo, 2005). The decline in fluoroquinolone resistance for E. coli (ciprofloxacin: 55.2 to 46.1%) and K. pneumoniae (levofloxacin: 43.8 to 26.5%) likely reflects reduced empirical use due to stewardship programs, as evidenced by similar trends in North America (Ramirez and Tolmasky, 2010). In contrast, the erratic resistance patterns of levofloxacin in A. baumannii (72.6–69.9%) and P. aeruginosa (7.7 to 5.9%) suggest pathogen-specific adaptive mechanisms, such as efflux pump overexpression or target mutations (van Belkum et al., 2019). Aminoglycosides retained moderate efficacy for P. aeruginosa (<10% resistance), but rising resistance in A. baumannii (tobramycin: 59.3–79.9%) and K. pneumoniae (gentamicin: 59.7–76%) highlights the gradual erosion of these agents’ utility. This trend parallels reports from high-resistance regions, where aminoglycoside-modifying enzymes have become widespread (Hooper and Jacoby, 2016).
While this single-center study provides granular insights into regional resistance dynamics, its retrospective design limits causal inference. The absence of genotypic data (e.g., whole-genome sequencing) precludes mapping resistance gene transmission networks. However, the large sample size (7,211 isolates) and longitudinal design (2019–2024) strengthen its validity as a sentinel report on MDR trends in western China.
5 Conclusion
The ICU continues to serve as a focal point for the evolution of multidrug-resistant (MDR) pathogens, driven by factors such as microbial resilience, therapeutic necessities, and environmental contamination. Addressing this challenge necessitates the integration of precision diagnostics, antibiotic stewardship programs, and advanced disinfection technologies, marking a paradigm shift from reactive treatment strategies to proactive ecological interventions aimed at disrupting resistance reservoirs.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Beijing Anzhen Nanchong Hospital of Capital Medical University & Nanchong Central Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by-product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
XC: Data curation, Writing – original draft. XL: Writing – review & editing. WR: Writing – review & editing. HL: Writing – review & editing, Data curation, Validation. SY: Project administration, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1587132/full#supplementary-material
References
Abd El-Rahman, O. A., Rasslan, F., Hassan, S. S., Ashour, H. M., and Wasfi, R. (2023). The RND efflux pump gene expression in the biofilm formation of Acinetobacter baumannii. Antibiotics 12:419. doi: 10.3390/antibiotics12020419
Cortés, A., Rooney, J., Bartley, D. J., Nisbet, A. J., and Cantacessi, C. (2020). Helminths, hosts, and their microbiota: new avenues for managing gastrointestinal helminthiases in ruminants. Expert Rev. Anti-Infect. Ther. 18, 977–985. doi: 10.1080/14787210.2020.1782188
Doron, S., and Davidson, L. E. (2013). Antimicrobial stewardship. Mayo Clin. Proc. 86, 1113–1123. doi: 10.4065/mcp.2011.0358
Dulanto Chiang, A., and Dekker, J. P. (2019). Efflux pump-mediated resistance to new beta lactam antibiotics in multidrug-resistant gram-negative bacteria. Commun. Med. 4, 1–9. doi: 10.1038/s43856-024-00591-y
Feretzakis, G., Loupelis, E., Sakagianni, A., Kalles, D., Lada, M., Christopoulos, C., et al. (2020). Using machine learning algorithms to predict antimicrobial resistance and assist empirical treatment. Stud. Health Technol. Inform. 272, 75–78. doi: 10.3233/SHTI200497
Grundmann, H., Glasner, C., Albiger, B., Aanensen, D. M., Tomlinson, C. T., Andrasević, A. T., et al. (2017). Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect. Dis. 17, 153–163. doi: 10.1016/S1473-3099(16)30257-2
Hooper, D. C., and Jacoby, G. A. (2016). Topoisomerase inhibitors: fluoroquinolone mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 6:5320. doi: 10.1101/cshperspect.a025320
ICPIC. (2019). Abstracts from the 5th International Conference on Prevention & Infection Control. Antimicrob Resist Infect Control 8 (Suppl 1), 148. doi: 10.1186/s13756-019-0567-6
Jiang, L., Liang, Y., Yao, W., Ai, J., Wang, X., and Zhao, Z. (2019). Molecular epidemiology and genetic characterisation of carbapenem-resistant Acinetobacter baumannii isolates from Guangdong Province, South China. J. Glob. Antimicrob. Resist. 17, 84–89. doi: 10.1016/j.jgar.2018.11.002
Jiang, H., Pu, H., and Huang, N. (2023). Risk predict model using multi-drug resistant organism infection from neuro-ICU patients: a retrospective cohort study. Sci. Rep. 13:15282. doi: 10.1038/s41598-023-42522-2
Liu, B., and Liu, L. (2021). Molecular epidemiology and mechanisms of Carbapenem-resistant Acinetobacter baumannii isolates from ICU and respiratory department patients of a Chinese university hospital. Infect Drug Resist 14, 743–755. doi: 10.2147/IDR.S299540
Logan, L. K., and Weinstein, R. A. (2017). The epidemiology of Carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215, S28–s36. doi: 10.1093/infdis/jiw282
Murray, C. J. L., Ikuta, K. S., Sharara, F., Swetschinski, L., Aguilar, G. R., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Nordmann, P., Gniadkowski, M., Giske, C. G., Poirel, L., Woodford, N., and Miriagou, V. (2012). Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Infect. 18, 432–438. doi: 10.1111/j.1469-0691.2012.03815.x
Park, S. M., Suh, J. W., Ju, Y. K., Kim, J. Y., Kim, S. B., Sohn, J. W., et al. (2023). Molecular and virulence characteristics of carbapenem-resistant Acinetobacter baumannii isolates: a prospective cohort study. Sci. Rep. 13:19536. doi: 10.1038/s41598-023-46985-1
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088–17. doi: 10.1128/CMR.00088-17
Paterson, D. L., and Bonomo, R. A. (2005). Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686. doi: 10.1128/CMR.18.4.657-686.2005
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Schelenz, S., Hagen, F., Rhodes, J. L., Abdolrasouli, A., Chowdhary, A., Hall, A., et al. (2016). First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob. Resist. Infect. Control 5:35. doi: 10.1186/s13756-016-0132-5
Strathdee, S. A., Hatfull, G. F., Mutalik, V. K., and Schooley, R. T. (2023). Phage therapy: from biological mechanisms to future directions. Cell 186, 17–31. doi: 10.1016/j.cell.2022.11.017
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Tamma, P. D., Aitken, S. L., Bonomo, R. A., Mathers, A. J., Van Duin, D., and Clancy, C. J. (2021). Infectious Diseases Society of America guidance on the treatment of extended-Spectrum β-lactamase producing Enterobacterales (ESBL-E), Carbapenem-resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 72, e169–e183. doi: 10.1093/cid/ciaa1478
Tamma, P. D., Aitken, S. L., Bonomo, R. A., Mathers, A. J., Van Duin, D., and Clancy, C. J. (2022). Infectious Diseases Society of America 2022 guidance on the treatment of extended-Spectrum β-lactamase producing Enterobacterales (ESBL-E), Carbapenem-resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 75, 187–212. doi: 10.1093/cid/ciac268
Van Belkum, A., Bachmann, T. T., Lüdke, G., Lisby, J. G., Kahlmeter, G., Mohess, A., et al. (2019). Developmental roadmap for antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 17, 51–62. doi: 10.1038/s41579-018-0098-9
Vincent, J. L., Sakr, Y., Singer, M., Martinloeches, I., Machado, F. R., Marshall, J. C., et al. (2020). Prevalence and outcomes of infection among patients in intensive care units in 2017. JAMA the. JAMA J. Am. Med. Assoc. 323:1478. doi: 10.1001/jama.2020.2717
Keywords: intensive care unit (ICU), Gram-negative bacteria, antimicrobial agents, antibiotic resistance, distribution patterns
Citation: Chen X, Liu X, Ren W, Li H and Yang S (2025) Distribution patterns and evolution of antimicrobial resistance in Gram-negative bacteria within the intensive care unit of a tertiary hospital from 2019 to 2024. Front. Microbiol. 16:1587132. doi: 10.3389/fmicb.2025.1587132
Edited by:
Maria Soledad Ramirez, California State University, Fullerton, United StatesReviewed by:
Okon Okwong Kenneth, Federal University, Wukari, NigeriaMaria Teresa Mascellino, Sapienza University of Rome, Italy
Copyright © 2025 Chen, Liu, Ren, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siyun Yang, NDEzMzA1MzQ0QHFxLmNvbQ==
 Xiaoying Chen
Xiaoying Chen Xin Liu1
Xin Liu1 Hongyan Li
Hongyan Li