- 1School of Grassland, Xinjiang Agricultural University, Urumqi, China
- 2Key Laboratory of Grassland Resources and Ecology Autonomous Region, Urumqi, China
- 3Key Laboratory of Grassland Resources and Ecology, Ministry of Education, Urumqi, China
Introduction: As a pivotal restoration strategy for alleviating grassland degradation, long-term enclosure practices effectively eliminate livestock disturbances while facilitating ecosystem self-recovery. Understanding the dynamics of soil microbial respiration under enclosure management is crucial, as it provides a scientific foundation for optimizing grassland utilization and contributes to global research on the terrestrial carbon cycle.
Methods: We conducted a comparative study across three distinct enclosed grassland ecosystems in Xinjiang, China: temperate desert, temperate steppe, and mountain meadow. Through analyzing microbial community structure, diversity, assembly processes, and respiration rates between 9-year enclosed and grazed areas, we identified key ecological shifts.
Results: Three key advancements emerged: (1) Enclosure implementation led to a marked improvement in soil resource availability, triggering microbial community shifts from oligotrophic to eutrophic states with substantial biodiversity increases (bacterial diversity: 2.2–14%; fungal diversity: 12.4–27.2%); (2) Divergent assembly mechanisms were observed where surface soil bacterial communities (0–5 cm depth) transitioned from 55.6 to 100% deterministic processes, directly contrasting with fungal communities that shifted from 11.1 to 55.6% stochastic dominance; (3) Partial Least Squares Path Modeling (PLS-PM) revealed distinct ecosystem-specific regulatory mechanisms underpinning reduced microbial respiration.
Discussion: The PLS-PM analysis detailed these distinct mechanisms: soil property-induced microbial metabolic trade-offs enhanced carbon use efficiency in temperate desert (R2 = 0.951), plant-mediated microbial assembly processes promoted efficient carbon cycling in temperate steppe (R2 = 0.455), and plant-driven suppression of microbial biomass dominated respiratory reduction in mountain meadow (R2 = 0.883). The research establishes that enclosure achieves carbon sequestration through divergent pathways across ecosystems, providing critical insights for optimizing grassland management strategies and enhancing climate change mitigation efforts.
1 Introduction
As fundamental components of grassland ecosystems, soil microorganisms play a key role in decomposing plant biomass and organic residues, while simultaneously driving biogeochemical cycles and energy fluxes within these ecosystems. Their critical role in maintaining ecological equilibrium and serving as sensitive bioindicators of environmental change has been well documented (Huang et al., 2009). Contemporary microbial ecology research has undergone a paradigm shift toward investigating microbial community assembly mechanisms (Jiao et al., 2020; Stegen et al., 2012; Stegen et al., 2013; Nemergut et al., 2013; Dini-Andreote et al., 2015). Current scientific consensus holds that microbial assemblage dynamics are governed by an interplay of deterministic and stochastic processes (Stegen et al., 2012; Zhang et al., 2022; Wu et al., 2022; Dumbrell et al., 2010). Deterministic processes encompass both abiotic factors (environmental filtering and selection pressures) and biotic interactions (including microbial antagonism and mutualism), which collectively determine species survival, extinction probabilities, and relative abundance distributions within communities (Stegen et al., 2012; Vellend, 2010). Conversely, stochastic processes involve unpredictable disturbances, probabilistic dispersal limitations, and random birth-death events that occur independently of environmental gradients (Dini-Andreote et al., 2015; Chase and Myers, 2011). This theoretical framework has been extensively applied to elucidate microbial community formation (Fukami, 2015; Ernakovich et al., 2022) and provides novel insights into microbial responses to environmental perturbations (Nemergut et al., 2013; Dini-Andreote et al., 2015). Nevertheless, significant scientific debate persists regarding the relative contributions of deterministic versus stochastic processes in shaping microbial community structure and function (Nemergut et al., 2013; Ernakovich et al., 2022; Knelman and Nemergut, 2014; Tucker et al., 2016).
As the most biologically active components within soil ecosystems, soil microorganisms drive biogeochemical processes through their metabolic activities. Microbial respiration—the terminal process of organic carbon decomposition—serves as the primary conduit for soil-to-atmosphere CO₂ release, regulated by abiotic factors (temperature, moisture, soil properties) and ecosystem-specific variables (vegetation type, land management, anthropogenic disturbances), generating predictable and stochastic flux patterns (Raich and Schlesinger, 1992). Plant diversity enhances microbial biomass via substrate diversity and nutrient availability, establishing a direct link between floristic richness and elevated respiration rates (Lange et al., 2015; Eisenhauer et al., 2013; Khlifa et al., 2017; Ma and Chen, 2018; Tilman et al., 2001). These biome-specific regulatory mechanisms—though requiring empirical validation in carbon flux studies—provide a conceptual framework to interpret divergent respiration patterns across grasslands: deterministic assembly under resource enrichment (e.g., enclosure-induced nutrient availability) selects copiotrophic bacteria with high metabolic activity, accelerating labile carbon mineralization, while stochastic assembly in fungal communities promotes decomposition of complex substrates (e.g., lignin) and carbon stabilization through functional niche complementarity (Ernakovich et al., 2022; Knelman and Nemergut, 2014).
Grazing, as the most prevalent and cost-effective grassland management practice globally, often triggers vegetation degradation under unsustainable intensities, compromising multifunctionality through diminished ecological services (e.g., carbon sequestration, soil stabilization) and pastoral productivity (Dong, 2016). In response, grassland enclosures have been widely adopted to restore degraded ecosystems by eliminating herbivore pressure, thereby facilitating natural regeneration of vegetation and soil biota (Chen et al., 2018; Ju et al., 2024). Contemporary studies emphasize that enclosure-induced shifts in soil properties and plant communities restructure microbial assembly processes (deterministic vs. stochastic), which subsequently modulate microbial respiration and ecosystem carbon cycling (Hu et al., 2024). Nevertheless, critical gaps persist in understanding: (a) how soil–plant-microbe interactions collectively regulate respiration pathways; (b) the legacy effects of historical grazing on microbial functional redundancy; and (c) the metabolic constraints governing carbon flux trajectories during recovery (Wu et al., 2017).
Building upon this theoretical framework, our study conducted a comparative analysis of three grassland ecosystems subjected to 9-year grazing exclusion along the northern slope of the Tianshan Mountains: temperate desert, temperate steppe, and mountain meadow. Through comprehensive characterization of soil microbial community structure, diversity profiles, and respiratory activity across enclosed and grazed plots, we hypothesize that: (1) Bacteria-fungi assembly dichotomy: Functional divergence drives bacterial communities toward deterministic dominance via environmental filtering under resource enrichment (after enclosure), while fungal communities exhibit stochastic assembly through dispersal strategies and ecological drift in enriched environments (after enclosure); and (2) grazing exclusion induces distinct biome-specific pathways regulating soil microbial respiration: (i) in the fragile temperate desert, direct abiotic forcing via enclosure-triggered soil modifications will rapidly reconfigure nutrient dynamics to mediate respiration; (ii) in the semi-arid temperate steppe, vegetation recovery (e.g., increased dominance but declined diversity) will establish biotic control by restructuring microbial communities and assembly processes; and (iii) in the mesic mountain meadow, plant-driven suppression of microbial biomass will override other factors to ultimately limit respiratory carbon loss. Furthermore, we predict that these divergent pathways will hierarchically enhance ecosystem carbon retention under enclosure management. This investigation aims to elucidate how enclosure-induced changes in microbial assembly regulate soil microbial respiration, and thereby to establish predictive microbial assembly-ecosystem recovery linkages, identify biome-specific carbon cycling responses, and provide microbial-mediated management strategies for degraded grasslands.
2 Materials and methods
2.1 Study area
The research area encompasses Wenquan County of the Boltala Mongolian Autonomous Prefecture in the western region, as well as Fukang City and Qitai County, situated in the Changji Hui Autonomous Prefecture, to the east of the northern slope of the Tianshan Mountain. The fundamental details of the sample points are outlined in Figure 1 and Supplementary Table S1. Specifically, within the study area of Fukang City, the grassland type is classified as temperate desert, featuring sierozem soil and a sandy soil texture. The dominant flora comprises Haloxylon ammodendron and Seriphidium santolinum, with Kali collinum as an associated species. On the other hand, the grassland type in the Wenquan County research area is temperate steppe, characterized by chestnut soil and a loam soil texture. The primary flora includes Stipa capillata, Festuca ovina, and Artemisia frigida, with Aster altaicus as an associated species. Furthermore, the Qitai County research area is classified as mountain meadow, with chernozem soil and a loam soil texture. The dominant plant species is Alchemilla tianschanica, while the main associated species encompass Bromus inermis, Polygonum viviparum, Trifolium lupinaster, Carex liparocarpos, Astragalus membranaceus, and Geranium carolinianum, among others.
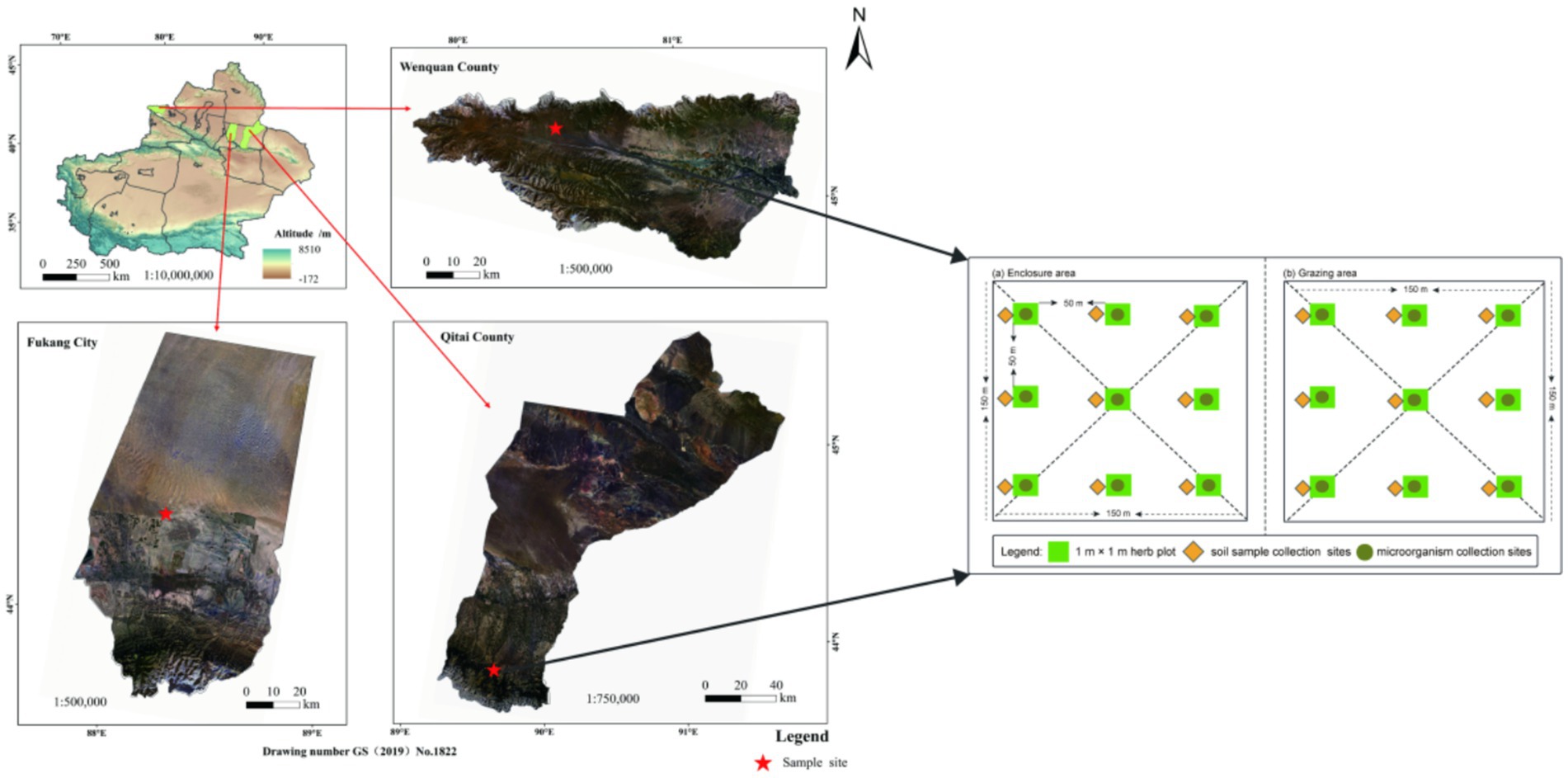
Figure 1. Digital elevation map showing the locations of the three sampled sites in the Tianshan Mountains.
2.2 Experiment design
The test sites were strategically positioned within the designated national fixed monitoring sites, each encompassing an area of approximately 3 × 104 m2. Two treatments are set in temperate desert (TD), temperate steppe (TS), and mountain meadow (MM), respectively: grazing exclusion plots (GE) and freely grazing plots (FG). Notably, all these exclusion plots have been enclosed since 2012 and remained enclosed for a duration of 9 years by the sampling year of 2021. The overall stocking rate for the three grassland sites ranged from 0.90 to 4.50 sheep units per hectare (hm2). The grazing intensities at these three sites fell within the category of moderate grazing in the local context, aligning with the carrying capacity of livestock in the area. Before enclosure, the enclosed areas exhibited comparable vegetation composition, community characteristics, and landforms to the control grazing areas.
2.3 Plant and soil sampling and determination
Vegetation surveys were conducted during peak biomass season (July 2021) across enclosure and grazed sites. Three 50-m spaced transects per treatment contained three 1 × 1 m quadrats (50-m intervals; 54 quadrats total). Plant species composition was recorded, with coverage (%) assessed by the point-intercept method, height (cm) by tape measure, density (plants·m−2) by direct counting, and above-ground biomass (AGB) by harvesting, oven-drying (105 °C/30 min → 80 °C/24 h), and weighing. Below-ground biomass (BGB) was determined from intact soil blocks (10 × 20 cm) after washing and drying [methods follow Liu et al. (2022)].
Soil sampling occurred concurrently within vegetation quadrats. Stratified samples (0–5 and 5–10 cm depths) were homogenized per layer, with subsamples stored at 4 °C (microbial assays) or air-dried (physicochemical analysis). Standard protocols quantified: (1) pH (soil:water = 5:1); (2) SOC (dichromate oxidation); (3) N/P fractions (Kjeldahl N; Mo-Sb colorimetric P); and (4) Microbial biomass (fumigation-extraction for MBC/MBN/MBP) (Liu et al., 2022) (Full analytical details in Appendix A). All datasets are original. Plant/soil data will be analyzed in a companion study currently in preparation, with no content overlap between manuscripts.
2.4 Soil microbial sampling and determination
In July 2021, the soil microorganism sample collection was carried out, and three typical samples were set up in each of the treatment area and the control area, with a sample line of 50 m, and the sample of three 1 m × 1 m on each sample line, and the sample space was about 50 m (Figure 1). First, the soil microorganism samples were collected from the soil 0–5 cm and the 5–10 cm layer, and each sample line was mixed evenly and then put in the car refrigerator (−20 °C) in a sealed bag and brought back to the laboratory. The study of soil microorganism samples by high-pass sequencing analysis of community structure and diversity, the research steps are: (1) to extract the total DNA in the sample using the Hongkey group DNA extraction kit. (2) the V3 ~ V4 variable zone sequence of the bacterium 16 s rRNA gene is the ITS1-1f zone of its rRNA gene, with the 338f-806r of the barcode sequence (the fungal derivative is the ITS1-1f-fdon ITS1-1f-r), and the PCR amplification is used to obtain the PCR product (Ju et al., 2024; Caporaso et al., 2011). (3) After the PCR product was built by quantitative and library, the Illumina MiSeq PE 300 platform was used to sequence the high pass, and to obtain the information of the base sequence of bacterial and fungal variable areas. (4) using the QIIME2.0 package, the OTU (taxonomic operational unit) cluster, the OTU represents the sequence and the SILVA database (Fungi for UNITE database) for the comparison analysis, and obtain the OTU corresponding classification unit and its corresponding quantity of abundance information (Jiang et al., 2022). (5) through the analysis of the microbial sequence, the abundance information of the microbiogenic cloud platform of the micro group, the discovery of α diversity, and so on. The analysis of the microbiological sequencing of the study was completed in the city of Shenzhen Microleague Technology Group Co., LTD (Jiang et al., 2022). The soil microbial respiration (SMR) was measured using an automatic soil aerobic culture system (He et al., 2013; Zhang et al., 2025).
The raw microbial sequence data from this study have been deposited in the Figshare repository and are publicly available under the DOI https://doi.org/10.6084/m9.figshare.30024817.v1 and at NCBI BioProject, accession PRJNA1338829 (Figshare permanent link: https://figshare.com/articles/figure/microbial_raw_sequence_data/30024817). These data are publicly available as of the date of publication.
2.5 Data statistics and analysis
Primary data preprocessing was performed using Microsoft Excel 2020 (Microsoft Corp.). Microbial α-diversity indices and soil microbial respiration metrics across grassland types were statistically analyzed through IBM SPSS Statistics 25.0 (IBM Corp.) employing a hierarchical analytical framework: (1) Before parametric tests, homogeneity of variances was assessed using Levene’s test (α = 0.05). For variables violating homogeneity (p < 0.05), Welch’s ANOVA or non-parametric Kruskal-Wallis tests were applied. (2) One-way ANOVA with post-hoc Tukey tests for inter-biome comparisons; (3) Multifactorial ANOVA to disentangle enclosure effects from ecosystem-type interactions; and (4) Independent-sample t-tests for paired enclosure vs. grazed plot analyzes. Microbial community assembly processes were quantified using the β-nearest taxon index (βNTI) and normalized stochasticity ratio (NST) computed through the “picante” (v1.8.2) and “NST” (v3.0.3) packages in R 4.2.1 (R Core Team). Deterministic versus stochastic dominance was assessed using established ecological thresholds (βNTI > 2 or βNTI < −2 indicates deterministic assembly; −2 < βNTI < 2 reflects stochastic dominance).
Partial Least Squares Path Modeling (PLS-PM) was performed using the plspm package (v0.4.9) in R 4.2.1 to elucidate the direct and indirect effects of environmental covariates on microbial respiration. This method was selected for its robustness with small sample sizes and its suitability for exploratory research and complex model structures. The analysis was designed to: (1) Decouple the direct and indirect effects along hypothesized pathways linking enclosure measures, soil properties (e.g., pH, SOC, TN), plant characteristics (e.g., above-ground biomass, below-ground biomass, richness), microbial properties (e.g., microbial biomass carbon, nitrogen), and microbial respiration; and (2) Test the proposed ecological mechanisms governing respiration dynamics in each grassland type. The model’s predictive accuracy and explanatory power were evaluated using the coefficient of determination (R2) for endogenous latent variables. The overall model quality was assessed by the Goodness-of-Fit (GoF) index. The significance of path coefficients was determined through a bootstrap validation procedure with 5,000 iterations to generate robust standard errors and confidence intervals.
All statistical graphics were programmatically generated using “ggplot2” (v3.4.2) in R 4.2.1, with subsequent vector refinement and compositional optimization performed in Adobe Illustrator 2021 (Adobe Inc.). Data are presented as mean ± standard error (SE).
3 Results
3.1 Effects of enclosure on microbial community structure of different grassland types
Bacterial phyla (Figure 2a): Actinobacteria, Proteobacteria, Acidobacteria, and Bacteroidetes dominated all grassland types and soil layers. After enclosure, Actinobacteria and Acidobacteria increased by 0.66–8.44% and 0.41–2.03% respectively, in the 0–5 cm layer of all grasslands, while Proteobacteria showed a 1.23% increase exclusively in a temperate steppe. Conversely, Bacteroidetes, Gemmatimonadetes, and Firmicutes decreased by 1.29–5.64%, 0.04–3.50%, and 0.05–0.53% in this layer. In the 5–10 cm layer, Actinobacteria increased by 2.60–9.41%, whereas Bacteroidetes, Verrucomicrobia, and Gemmatimonadetes decreased by 0.17–2.86%, 0.19–1.37%, and 0.28–4.09%, respectively.
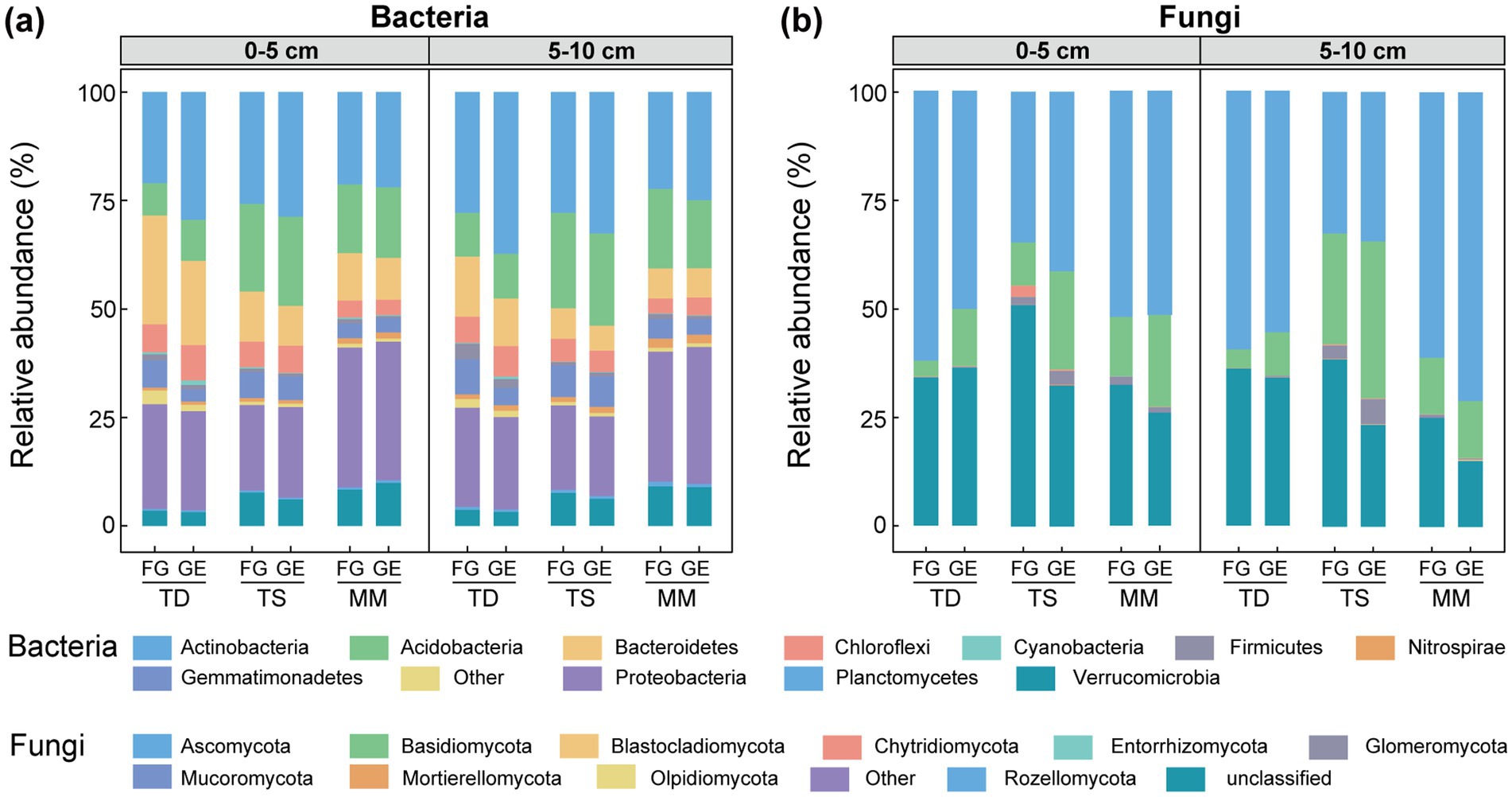
Figure 2. Effects of enclosure on microbial community structure at phylum level of different grassland types. (a) Bacteria relative abundance, (b) fungi relative abundance. TD (temperate desert), TS (temperate steppe), MM (mountain meadow), GE (grazing exclusion plots), FG (freely grazing plots).
Fungal phyla (Figure 2b): Ascomycota and Basidiomycota dominated all grasslands and soil layers, with other phyla (excluding Glomeromycota) accounting for <1% relative abundance. After enclosure, Basidiomycota increased by 7.54–12.64% in the 0–5 cm layer of all grasslands, while Ascomycota showed a 6.62% increase exclusively in a temperate steppe. In the 5–10 cm layer, Basidiomycota increased by 0.13–10.58% across all grasslands, while Ascomycota increased by 1.82–9.95% in temperate steppe and mountain meadow.
3.2 Effects of enclosure on microbial diversity in different grassland types
Enclosure-induced divergent bacterial alpha-diversity responses across grassland ecosystems and soil depths. After enclosure, in the 0–5 cm layer, the Chao1 index increased by 14.0% (p < 0.05) in the temperate desert but decreased by 10.0% (p < 0.05) in the mountain meadow, whereas no significant changes occurred in the temperate steppe [Figure 3A(a)]. At 5–10 cm depth, after enclosure, only the temperate desert exhibited a response, with a 2.2% increase in Shannon index [p < 0.05, Figure 3A(b)]. Beta diversity analysis (Bray-Curtis PCoA) demonstrated strong grassland-type-driven segregation (p < 0.001), with bacterial communities of temperate desert and steppe separated along PCoA axis 1 (32.49–32.51% variance explained), independent of soil depth [Figures 3A(c,d)].
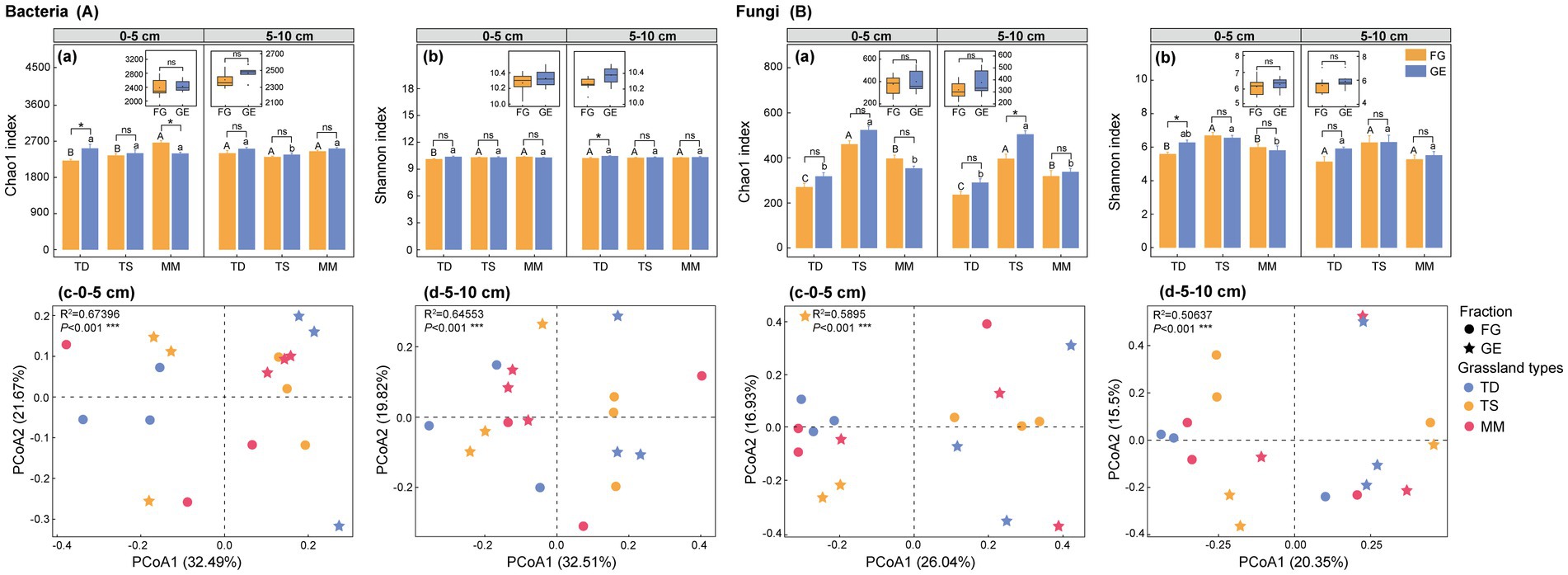
Figure 3. Effects of enclosure on microbial community diversity of different grassland types. (A) bacteria diversity index, (B) fungi diversity index, (a) bacterial and fungal Chao1 index, (b) bacterial and fungal Shannon index, (c) bacterial and fungal 0–5 cm soil layer microbial β diversity, (d) bacterial and fungal 5–10 cm soil layer microbial β diversity. All data points and error bars in histograms represent mean ± standard error. “*,” “**” and “***” respectively indicated significant (p < 0.05), extremely significant (p < 0.01) and (p < 0.001) differences among different treatments of the same grassland type as determined by independent-sample T tests, while “ns” indicated no significant (p > 0.05) differences; Capital letters: Significant differences between grazing areas of different grassland types (p < 0.05) as determined by one-way ANOVA followed by Duncan’s multiple range test; Lowercase letters: Differences between enclosure areas of different grassland types (p < 0.05) analyzed with the same method, where shared letters indicate no significant differences (p > 0.05); TD (temperate desert), TS (temperate steppe), MM (mountain meadow), GE (grazing exclusion plots), FG (freely grazing plots); Box plots: Combined analysis of enclosure vs. grazing effects across all three grassland types as determined by one-way ANOVA followed by Duncan’s multiple range test.
Fungal communities displayed contrasting layer-specific enclosure effects. After enclosure, in surface soil (0–5 cm), the Shannon index increased by 12.4% (p < 0.05) in the temperate desert, while subsurface soil (5–10 cm) showed a 27.2% Chao1 index rise (p < 0.05) in the temperate steppe [Figures 3B(a,b)]. Principal coordinates analysis (PCoA) demonstrated distinct stratification patterns: in 0–5 cm soil layer, temperate desert, and steppe ecosystems exhibited primary fungal community segregation along PCoA axis 1 [26.04% variance, Figure 3B(c)], and in 5–10 cm soil layer, temperate steppe communities showed predominant separation along PCoA axis 2 [15.5% variance, Figure 3B(d)].
3.3 Effects of enclosure on microbial assembly process of different grassland types
As shown in Figure 4, after grassland enclosure, the assembly of soil bacterial communities (βNTI) in both soil layers (0–5 cm and 5–10 cm) across all three grassland types shifted to complete dominance (100%) by deterministic processes [Figure 4A(a)]. Critically, in the 0–5 cm layer before enclosure, bacterial assembly exhibited a mixed process (55.6% deterministic + 44.4% stochastic), which transitioned entirely to deterministic processes (100%) following enclosure [Figure 4A(b)].
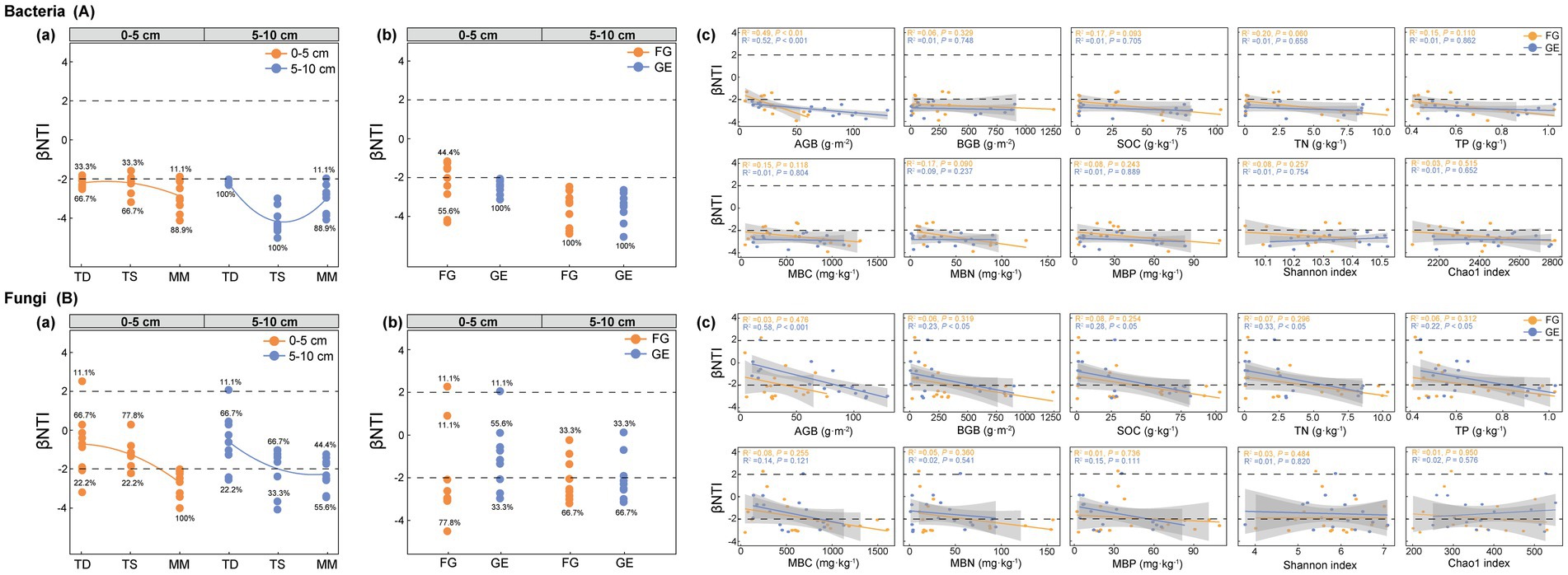
Figure 4. Effects of enclosure on microbial assembly in different grassland types. (A) Bacteria assembly process, (B) fungi assembly process, (a) microbial assembly process in different grassland types, (b) microbial assembly process in grazing and enclosure area. (c) Trends analyzed by linear regression; AGB (above-ground biomass), BGB (below-ground biomass), SOC (soil organic carbon), TN (total nitrogen), TP (total phosphorus), MBC (microbial biomass carbon), MBN (microbial biomass nitrogen), MBP (microbial biomass phosphorus), FG (freely grazing), GE (grazing exclusion).
For fungal communities under enclosure conditions, stochastic processes dominated assembly in both soil layers of the temperate desert and the temperate steppe, whereas deterministic processes prevailed in the mountain meadow [Figure 4B(a)]. In the 0–5 cm layer, pre-enclosure fungal assembly was predominantly deterministic (88.9%), but shifted to stochastic dominance (55.6%) post-enclosure [Figure 4B(b)]. This directional reversal in the topsoil, from mixed to deterministic for bacteria versus deterministic to stochastic for fungi, highlights an opposing response to enclosure management. These assembly shifts provide the mechanistic basis for the respiration patterns analyzed in Section 3.5.
By analyzing the linear relationships between plant properties, soil characteristics, microbial diversity, and soil microbial assembly processes, our results demonstrate that grassland enclosure management shifted microbial community assembly from predominantly stochastic toward deterministic processes; specifically, for bacterial βNTI, aboveground biomass (AGB) was the dominant driver [R2 = 0.52, p < 0.001; Figure 4A(c)], while for fungal βNTI, AGB showed the strongest correlation (R2 = 0.58, p < 0.001), with significant but weaker associations observed for belowground biomass (BGB: R2 = 0.23, p < 0.05), soil organic carbon (SOC: R2 = 0.28, p < 0.05), total nitrogen (TN: R2 = 0.33, p < 0.05), and total phosphorus (TP: R2 = 0.22, p < 0.05) [Figure 4B(c)].
3.4 Effects of enclosure on soil microbial respiration of different grassland types
Through the Figure 5 analysis, in the 0–5 cm layer, microbial respiration decreased by 71.3% (p < 0.01) in the temperate steppe and 31.8% (p < 0.05) in mountain meadow compared to pre-enclosure levels (Figure 5a). At 5–10 cm depth, only the temperate desert showed a marked decrease of 74.7% (p < 0.05) in microbial respiration after enclosure, while other grassland types remained statistically unchanged (Figure 5b).
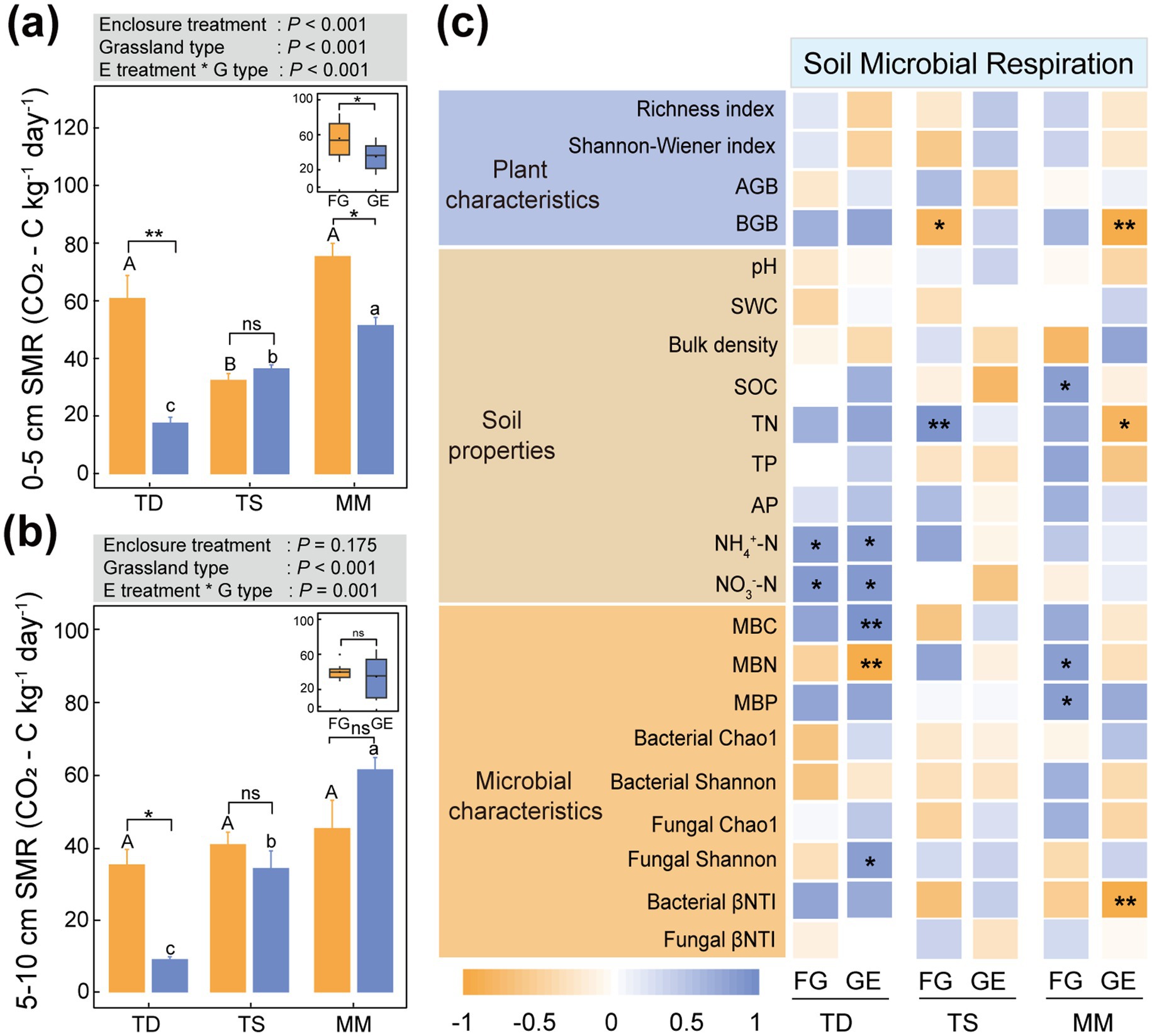
Figure 5. Effects of enclosure on soil microbial respiration of different grassland types. (a) 0–5 cm soil microbial respiration, (b) 5–10 cm soil microbial respiration. All data points and error bars in histograms represent mean ± standard error. “*,” “**” and “***” respectively indicated significant (p < 0.05), extremely significant (p < 0.01) and (p < 0.001) differences among different treatments of the same grassland type as determined by independent-sample T tests, while “ns” indicated no significant (p > 0.05) differences; Capital letters: Significant differences between grazing areas of different grassland types (p < 0.05) as determined by one-way ANOVA followed by Duncan’s multiple range test; Lowercase letters: Differences between enclosure areas of different grassland types (p < 0.05) analyzed with the same method, where shared letters indicate no significant differences (p > 0.05); TD (temperate desert), TS (temperate steppe), MM (mountain meadow), GE (grazing exclusion plots), FG (freely grazing plots); Gray box above histograms: Results of two-way ANOVA for the displayed metric; Box plots: Combined analysis of enclosure vs. grazing effects across all three grassland types as determined by one-way ANOVA followed by Duncan’s multiple range test. (c) Correlations between soil microbial respiration and other variables were quantified using Pearson’s method.
Pearson correlation analysis showed that soil microbial respiration was significantly correlated with NH4+-N, NO3−-N, MBC, MBN, and fungal Shannon index in the temperate desert after enclosure (p < 0.05 and p < 0.01). However, there were significant and extremely significant correlations with BGB, TN, and bacterial-βNTI in the mountain meadow (p < 0.05 and p < 0.01), but no significant correlations with each index in temperate steppe (Figure 5c).
3.5 Key drivers of soil microbial respiration under enclosure
The results of the structural equation modeling revealed that the factors influencing soil microbial respiration varied across grassland types (Figure 6). In the temperate desert, microbial respiration was primarily driven by the direct effects of soil properties and microbial biomass (p < 0.05, Figure 6a). Together with microbial diversity and assembly processes, these factors accounted for 95.1% of the variation in microbial respiration (Figure 6a). However, the total effects indicated that grazing exclusion had the strongest influence, suggesting that enclosure measures altered the habitat conditions of the temperate desert grassland, thereby indirectly affecting microbial respiration (Figure 6b). In the temperate steppe, microbial respiration was mainly influenced by plant traits (p < 0.05), followed by microbial assembly processes (Figures 6c,d). These factors, along with microbial diversity, collectively explained 45.5% of the variation in microbial respiration (Figure 6c). This pattern contrasted sharply with that observed in the temperate desert. In the mountain meadow, microbial respiration was directly affected by soil properties and microbial assembly processes (p < 0.01, Figure 6e), which, together with microbial biomass and diversity, explained 88.3% of the variation in microbial respiration (Figures 6e,f).
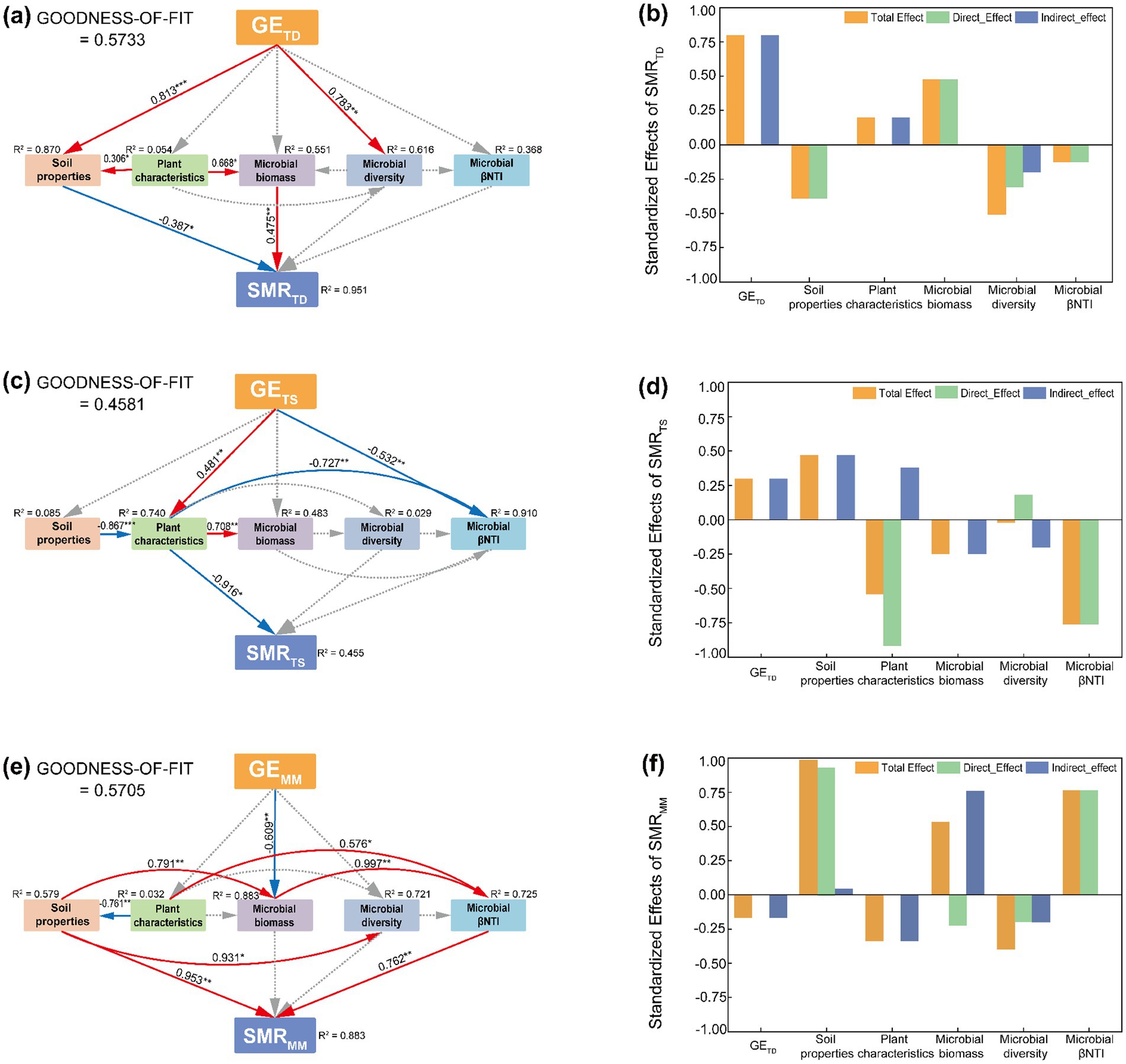
Figure 6. Analysis of soil microbial respiration path in different grassland types under the background of enclosure. (a) Temperate desert SEM, (b) temperate desert standardized effects of SMR, (c) temperate steppe SEM, (d) temperate steppe standardized effects of SMR, (e) mountain meadow SEM, (f) mountain meadow standardized effects of SMR; The numbers on the arrows represent the path coefficient, which are normalized by the mean of each parameter; The red, blue, and dashed gray arrows indicated significant positive correlations (p < 0.05), significantly negative correlation (p < 0.05) and no significant relationship (p > 0.05), respectively; The values near each variable represent the variance interpreted by model (R2); “*,” “**” and “***” respectively indicated significant (p < 0.05), extremely significant (p < 0.01) and (p < 0.001).
4 Discussion
4.1 Response of microbial community structure and diversity to enclosure
High-throughput sequencing revealed conserved phylum-level dominance patterns in grassland microbial communities, with bacterial assemblages consistently dominated by Actinobacteria, Proteobacteria, Acidobacteria, Bacteroidetes, Verrucomicrobia, and Chloroflexi, while fungal communities were primarily composed of Ascomycota and Basidiomycota (Figures 2a,b). These patterns align with established grassland microbial ecology frameworks, suggesting evolutionary conservation of core taxa with multifunctional adaptations to environmental fluctuations (Wei et al., 2020; Bachar et al., 2010; Chu et al., 2016; Zhang et al., 2013). Specifically, these conserved microbial guilds exhibit high soil niche plasticity, enabled by traits such as xerotolerance, hydric stress resilience, and metabolic versatility across moisture gradients. This adaptive capacity allows them to maintain functional dominance despite grassland-type variations (Li et al., 2019; Ren et al., 2018; Li et al., 2018). Following enclosure implementation, we observed systematic shifts in bacterial functional guilds: oligotrophic groups (Actinobacteria, Acidobacteria) increased in relative abundance, whereas copiotrophic phyla (Bacteroidetes, Firmicutes) declined. This successional transition from fast-growing to slow-growing microbial strategists likely reflects enhanced soil nutrient availability and vegetation diversity post-grazing exclusion (Zhou et al., 2017; Zhang et al., 2018), particularly through increased organic matter inputs that favored resource-competitive Actinobacteria and Acidobacteria. Concurrently, improved soil aeration under enclosure reduced the viability of anaerobic Firmicutes and Bacteroidetes, demonstrating how management-induced physicochemical changes cascade through microbial communities. Fungal communities demonstrated Basidiomycota enrichment across soil profiles, aligning with their lignin-degrading specialization (Lienhard et al., 2014; Ferreira de Araujo et al., 2017) and enclosure-induced accumulation of lignin-rich graminoid litter (He et al., 2019; Li et al., 2019).
Ecosystem-specific α-diversity trajectories emerged, with the xeric temperate desert exhibiting the strongest microbial re-assembly after enclosure, while mesic systems remained comparatively stable (Figure 3). New correlative evidence (Supplementary Figure S1) attributes these patterns to management-dependent plant–soil–microbe feedbacks: In the temperate desert, enclosure reversed bacterial linkages-from positive correlations with plant richness/SOC/TP/AP under grazing to negative associations—signaling a resource-driven regime shift where elevated plant inputs intensified competitive exclusion (Supplementary Figures S1a,b) (Cheng et al., 2016); simultaneously, fungi shifted from neutral to positive correlations with BGB/TN/AP, indicating nutrient-mediated functional specialization toward saprotrophic pathways under enrichment (Supplementary Figures S1a,b) (Zhou et al., 2024). In the temperate steppe, bacteria tightened phosphorus coupling (TP/AP, post-enclosure), revealing copiotrophic exploitation of newly available P; fungi decoupled from plant diversity (for Chao1), yet gained positive Shannon links with microbial biomass, demonstrating taxon-specific N-limitation persistence (Supplementary Figures S1c,d). In the mountain meadow, bacterial diversity strengthened ties to plant richness and MBC, confirming edaphic niche creation via litter-root synergy; fungal diversity maintained negative plant correlations, reflecting unwavering competitive exclusion by vegetation (Supplementary Figures S1e,f). Collectively, these data-anchored linkages establish that enclosure duration and ecosystem type jointly determine feedback loop directionality—from resource competition to niche optimization—refining grassland microbial models (Zhang et al., 2018; Zhang and Fu, 2022).
4.2 Taxon-specific assembly mechanisms and their environmental drivers under enclosure
Our null model analysis demonstrated that enclosure management induced divergent microbial assembly pathways: bacterial communities shifted toward deterministic selection (βNTI > − 2.0) in arid surface soils, while fungal assembly maintained stochastic dominance (+2.0 < βNTI < −2.0) (Figure 4). Crucially, these patterns were driven by distinct environmental factors: (1) Bacterial deterministic assembly was primarily governed by aboveground biomass (AGB) [R2 = 0.52, p < 0.001; Figure 4A(c)], where enhanced plant inputs intensified niche competition through litter-root synergy, optimizing selective pressures for competitive taxa (Wu et al., 2022; Fierer, 2008; Thuiller et al., 2005; Mason et al., 2011); (2) Fungal stochastic persistence occurred under multi-resource constraints involving belowground biomass (BGB: R2 = 0.23), soil organic carbon (SOC: R2 = 0.28), total nitrogen (TN: R2 = 0.33), and total phosphorus (TP: R2 = 0.22) [p < 0.05; Figure 4B(c)]. Consistent with Dini-Andreote et al. (2015), such heterogeneous resource availability amplifies stochasticity under moderate selection, enabling species-specific physiological adaptations that sustain biodiversity in resource-abundant environments (Chase, 2010).
These results refine classical niche theory (Bahram et al., 2018; Tripathi et al., 2012; Jiao et al., 2022): enclosure promotes deterministic bacterial assembly via plant-derived filters (AGB), aligning with observed competitive exclusion patterns (Section 4.1), while fungi respond to localized resource mosaics (BGB/SOC/TN/TP) through stochastic dynamics that facilitate functional diversification. This establishes taxon-specific environmental drivers as key mediators of microbial stabilization strategies under perturbation.
4.3 Response of microbial respiration to enclosure and its driving mechanisms
Our investigation demonstrates that grazing exclusion consistently suppressed soil microbial respiration across three distinct grassland ecosystems (Li et al., 2013), indicating its universal effectiveness in reducing carbon mineralization and potentially enhancing ecosystem carbon sequestration (Figures 5, 6). However, the underlying mechanisms driving this common outcome were strikingly divergent, highlighting the paramount importance of ecosystem-specific context in predicting the response to management interventions.
In the temperate desert grassland, the reduction in respiration was primarily governed by microbial physiological adaptation to nutrient limitation (Figures 6a,b) (Esch et al., 2017). Enclosure improved soil nutrient conditions and promoted microbial biomass accumulation, but it also induced strong stoichiometric imbalances (Supplementary Table S2). This triggered a fundamental metabolic trade-off, where microbes prioritized survival and biomass formation over respiratory activity. Consequently, the community shifted from a fast-growing, high-respiration state to a maintenance-oriented, energy-conserving state, a process consistent with the “microbial carbon pump” concept (Liang et al., 2017; Liang et al., 2019). Here, the direct negative effect of soil properties and microbial nutrient limitation overwhelmed the positive effect of increased biomass, leading to enhanced carbon retention efficiency.
Conversely, in the temperate steppe, the plant community emerged as the central mediator. Enclosure led to a decline in plant diversity, which itself exerted a strong negative effect on microbial respiration, likely through the production of poorer quality litter (Supplementary Table S3; Figures 6c,d) (Wang et al., 2014). Paradoxically, the net respiration decreased, a outcome resolved by a cascade of changes: enclosure fostered a more deterministic microbial assembly process, which selected for a community with a higher carbon use efficiency, thereby enhancing carbon retention—a finding that aligns with the newly identified primacy of microbial CUE in controlling soil carbon storage at a global scale (Figure 4) (Zhang et al., 2022; Tao et al., 2022). This efficiently assembled community effectively counteracted the upward pressure on respiration from both increased plant biomass and soil nutrients. Thus, the microbial response to environmental filtering, rather than plant traits directly, ultimately governed the carbon cycle.
In contrast to both, the mountain meadow exhibited a clear top-down control mechanism (Figures 6e,f) (Aqeel et al., 2023; Reynolds et al., 2003). Enclosure promoted plant growth, which directly and negatively impacted microbial biomass (Supplementary Tables S2, S3). This sharp decline in microbial abundance was the paramount factor suppressing respiration, as evidenced by a strong positive relationship between them (Total effect = 0.53). The reduction in biomass further weakened the microbial assembly process, which itself had a strong positive effect on respiration. This uncoupled the shift toward deterministic assembly from an increase in overall metabolic activity. Despite strong positive direct effects from soil properties, their influence was insufficient to offset the overwhelming suppressive effect of a diminished microbial pool.
Synthesis and Implications: Despite a common outcome, our study reveals three unique pathways through which enclosure moderates the soil carbon cycle: (1) Microbial Metabolic Trade-off in deserts, (2) Microbial Assembly-Mediated Efficiency in steppes, and (3) Plant-Induced Biomass Reduction in meadows. This spectrum of mechanisms—from primarily abiotic (soil nutrients) to biotic (plants and microbes)—underscores that effective ecosystem management must be context-dependent. Our findings move beyond a simple narrative of enclosure increasing carbon stocks and instead illuminate the complex biotic interactions that ultimately determine ecosystem carbon cycling efficiency. Future assessments of grazing exclusion policies should integrate these ecosystem-specific mechanisms to accurately predict their long-term impacts on grassland carbon sequestration.
5 Conclusion
Our study demonstrates that grazing enclosure consistently reduces microbial respiration across temperate desert, steppe, and mountain meadow ecosystems, indicating their universal potential to enhance carbon sequestration. However, this common outcome emerges through divergent ecosystem-specific mechanisms. In the temperate desert, reduced respiration was driven by microbial metabolic trade-offs in response to stoichiometric imbalances, enhancing carbon use efficiency. The temperate steppe response was mediated through biotic interactions, where enclosure filtered for a more deterministic microbial assembly process that suppressed respiration despite increased plant biomass. In contrast, the mountain meadow exhibited top-down control where plant-induced suppression of microbial biomass directly drove respiration declines.
While these findings reveal distinct pathways of carbon cycle regulation, the restricted spatial scale necessitates cautious extrapolation. Future research should prioritize temporal analyzes of microbial functional succession, age-dependent responses to enclosure duration, and long-term plant-microbe coevolution monitoring to refine predictive models and evidence-based strategies for ecosystem carbon management.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI BioProject, accession PRJNA1338829.
Author contributions
AJ: Methodology, Investigation, Data curation, Visualization, Writing – original draft, Software, Formal analysis, Conceptualization. DY: Visualization, Project administration, Conceptualization, Writing – review & editing, Resources, Funding acquisition, Supervision. ZS: Data curation, Investigation, Writing – original draft. NT: Data curation, Writing – original draft, Investigation. JA: Writing – original draft, Investigation, Data curation. AS: Investigation, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by the 2022 National Natural Science Foundation (No. 32260355). Sequencing and data analysis services were provided by Wekemo Tech Group Co., Ltd., Shenzhen, China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1594877/full#supplementary-material
References
Aqeel, M., Ran, J. Z., Hu, W. G., Irshad, M. K., Dong, L. W., Akram, M. A., et al. (2023). Plant-soil-microbe interactions in maintaining ecosystem stability and coordinated turnover under changing environmental conditions. Chemosphere 318:137924. doi: 10.1016/j.chemosphere.2023.137924
Bachar, A., Al-Ashhab, A., Soares, M. I. M., Sklarz, M. Y., Angel, R., Ungar, E. D., et al. (2010). Soil microbial abundance and diversity along a low precipitation gradient. Microb. Ecol. 60, 453–461. doi: 10.1007/s00248-010-9727-1
Bahram, M., Hildebrand, F., Forslund, S. K., Anderson, J. L., Soudzilovskaia, N. A., Bodegom, P. M., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560, 233–237. doi: 10.1038/s41586-018-0386-6
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chase, J. M. (2010). Stochastic community assembly causes higher biodiversity in more productive environments. Science 328, 1388–1391. doi: 10.1126/science.1187820
Chase, J. M., and Myers, J. A. (2011). Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 366, 2351–2363. doi: 10.1098/rstb.2011.0063
Chen, J., Luo, Y. Q., Xia, J. Y., Zhou, X. H., Niu, S. L., Shelton, S., et al. (2018). Divergent responses of ecosystem respiration components to livestock exclusion on the Qinghai Tibetan plateau. Land Degrad. Dev. 29, 1726–1737. doi: 10.1002/ldr.2981
Cheng, J. M., Jing, G. H., Wei, L., and Jing, Z. B. (2016). Long-term grazing exclusion effects on vegetation characteristics, soil properties and bacterial communities in the semi-arid grasslands of China. Ecol. Eng. 97, 170–178. doi: 10.1016/j.ecoleng.2016.09.003
Chu, H., Sun, H., Tripathi, B. M., Adams, J. M., Huang, R., Zhang, Y., et al. (2016). Bacterial community dissimilarity between the surface and subsurface soils equals horizontal differences over several kilometers in the western Tibetan plateau. Environ. Microbiol. 18, 1523–1533. doi: 10.1111/1462-2920.13236
Dini-Andreote, F., Stegen, J. C., Van Elsas, J. D., and Salles, J. F. (2015). Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. PNAS 112, E1326–E1332. doi: 10.1073/pnas.1414261112
Dong, Y. Q. (2016). The influence of the ban animal on the desert vegetation and the soil active organic carbon group of the moderate degraded ili. [dissertation thesis] Urumqi: Xinjiang Agricultural University.
Dumbrell, A. J., Nelson, M., Helgason, T., Dytham, C., and Fitter, A. H. (2010). Erratum: relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 4:1078. doi: 10.1038/ismej.2009.122
Eisenhauer, N., Dobies, T., Cesarz, S., Hobbie, S. E., Meyer, R. J., Worm, K., et al. (2013). Plant diversity effects on soil food webs are stronger than those of elevated CO2 and N deposition in a long-term grassland experiment. PNAS 110, 6889–6894. doi: 10.1073/pnas.1217382110
Ernakovich, J. G., Barbato, R. A., Rich, V. I., Schädel, C., Hewitt, R. E., Doherty, S. J., et al. (2022). Microbiome assembly in thawing permafrost and its feedbacks to climate. Glob. Change Biol. 28, 5007–5026. doi: 10.1111/gcb.16231
Esch, E. H., Lipson, D., and Cleland, E. E. (2017). Direct and indirect effects of shifting rainfall on soil microbial respiration and enzyme activity in a semi-arid system. Plant Soil 411, 333–346. doi: 10.1007/s11104-016-3027-6
Ferreira de Araujo, A. S., Bezerra, W. M., Dos Santos, V. M., Nunes, L. A. P. L., de Lyra, M. D. C. C. P., do Vale Barreto Figureueiredo, M., et al. (2017). Fungal diversity in soils across a gradient of preserved Brazilian Cerrado. J. Microbiol. 55, 273–279. doi: 10.1007/s12275-017-6350-6
Fierer, N. (2008). Microbial biogeography: patterns in microbial diversity across space and time. Accessing uncultivated microorganisms: from the environment to organisms and genomes and back, 95–115. Available online at: https://doi.org/10.1128/9781555815509.ch6.
Fukami, T. (2015). Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu. Rev. Ecol. Evol. Syst. 46, 1–23. doi: 10.1146/annurev-ecolsys-110411-160340
He, N. P., Wang, R. M., Gao, Y., Dai, J. Z., Wen, X. F., and Yu, G. R. (2013). Changes in thetemperature sensitivity of SOM decomposition with grassland succession: implications for soil C sequestration. Ecol. Evol. 3, 5045–5054. doi: 10.1002/ece3.881
He, M., Zhao, R., Tian, Q., Huang, L., Wang, X., and Liu, F. (2019). Predominant effects of litter chemistry on lignin degradation in the early stage of leaf litter decomposition. Plant Soil 442, 453–469. doi: 10.1007/s11104-019-04207-6
Hu, Y. G., Liu, W. J., Chang, J. C., Fan, Y. X., Hou, S. P., Zhang, Z. H., et al. (2024). Grazing exclusion-induced alterations of soil microbial biogeographic pattern and co-occurrence network across a Tibetan elevation gradient. Agric. Ecosyst. Environ. 376:109231. doi: 10.1016/j.agee.2024.109231
Huang, Y. M., An, S. S., and Xue, H. (2009). Responses of soil microorganisms C, N and respiration entropy to vegetation restoration in loess hilly region. Acta Ecol. Sin. 29, 2811–2818.
Jiang, A. J., Ding, Y. Q., Astaiken, J. L. H. T., Zhou, S. J., Nie, T. T., Li, C. Y., et al. (2022). The effect of the envelope on the characteristics of different grassland types of soil bacteria. Acta Agrestia Sinica 30, 2600–2608. doi: 10.11733/i.issn.1007-0435.2022.10.009
Jiao, S., Chu, H., Zhang, B., Wei, X., Chen, W., and Wei, G. (2022). Linking soil fungi to bacterial community assembly in arid ecosystems. iMeta 1:e2. doi: 10.1002/imt2.2
Jiao, S., Yang, Y., Xu, Y., Zhang, J., and Lu, Y. (2020). Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 14, 202–216. doi: 10.1038/s41396-019-0522-9
Ju, W. L., Fang, L. C., Shen, G. T., Delgado-Baquerizo, M., Chen, J., Zhou, G. Y., et al. (2024). New perspectives on microbiome and nutrient sequestration in soil aggregates during long-term grazing exclusion. Glob. Change Biol. 30:e17027. doi: 10.1111/gcb.17027
Khlifa, R., Paquette, A., Messier, C., Reich, P. B., and Munson, A. D. (2017). Do temperate tree species diversity and identity influence soil microbial community function and composition? Ecol. Evol. 7, 7965–7974. doi: 10.1002/ece3.3313
Knelman, J. E., and Nemergut, D. R. (2014). Changes in community assembly may shift the relationship between biodiversity and ecosystem function. Front. Microbiol. 5:424. doi: 10.3389/fmicb.2014.00424
Lange, M., Eisenhauer, N., Sierra, C. A., Bessler, H., Engels, C., Griffiths, R. I., et al. (2015). Plant diversity increases soil microbial activity and soil carbon storage. Nat. Commun. 6:6707. doi: 10.1038/ncomms7707
Li, H. Y., Yao, T., Gao, Y. M., Zhang, J. G., Ma, Y. C., Lu, X. W., et al. (2019). The relationship between the soil fungus community and the soil environmental factor was degraded. Acta Microbiol Sin. 59, 678–688. doi: 10.13343/j.cnki.wsxb.20180257
Li, H. Y., Yao, T., Zhang, J. G., Gao, Y. M., Ma, Y. C., Lu, X. W., et al. (2018). The relationship between the soil bacterial community and soil environmental factors of the east Qilian Mountain degradation. Chin. J. Appl. Ecol. 29, 3793–3801. doi: 10.13287/j.1001-9332.201811.037
Li, X., Zhang, C., Fu, H., Guo, D., Song, X., Wan, C., et al. (2013). Grazing exclusion alters soil microbial respiration, root respiration and the soil carbon balance in grasslands of the loess plateau, northern China. Soil Sci. Plant Nutr. 59, 877–887. doi: 10.1080/00380768.2013.862157
Liang, C., Amelung, W., Lehmann, J., and Kästner, M. (2019). Quantitative assessment of microbial necromass contribution to soil organic matter. Glob. Change Biol. 25, 3578–3590. doi: 10.1111/gcb.14781
Liang, C., Schimel, J. P., and Jastrow, J. D. (2017). The importance of anabolism in microbial control over soil carbon storage. Nat. Microbiol. 2, 17105–17106. doi: 10.1038/nmicrobiol.2017.105
Lienhard, P., Terrat, S., Prévost-Bouré, N. C., Nowak, V., Régnier, T., Sayphoummie, S., et al. (2014). Pyrosequencing evidences the impact of cropping on soil bacterial and fungal diversity in Laos tropical grassland. Agron. Sustain. Dev. 34, 525–533. doi: 10.1007/s13593-013-0162-9
Liu, H. X., Sun, Z. J., Dong, Y. Q., Yang, H. L., He, P. X., Yu, B. J., et al. (2022). Precipitation drives the accumulation of soil organic carbon in the sandy desert of the Junggar Basin, Northwest China. Ecol. Indic. 142:109224. doi: 10.1016/j.ecolind.2022.109224
Ma, Z., and Chen, H. Y. (2018). Positive species mixture effects on fine root turnover and mortality in natural boreal forests. Soil Biol. Boichem. 121, 130–137. doi: 10.1016/j.soilbio.2018.03.015
Mason, N. W., de Bello, F., Doležal, J., and Lepš, J. (2011). Niche overlap reveals the effects of competition, disturbance and contrasting assembly processes in experimental grassland communities. J. Ecol. 99, 788–796. doi: 10.1111/j.1365-2745.2011.01801.x
Nemergut, D. R., Schmidt, S. K., Fukami, T., O'Neill, S. P., Bilinski, T. M., Stanish, L. F., et al. (2013). Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356. doi: 10.1128/mmbr.00051-12
Raich, J. W., and Schlesinger, W. H. (1992). The global carbon dioxide flux in soil respiration and its relationship to vegetation and climate. Tellus B 44, 81–99. doi: 10.1034/j.1600-0889.1992.t01-1-00001.x
Ren, C., Zhang, W., Zhong, Z., Han, X., Yang, G., Feng, Y., et al. (2018). Differential responses of soil microbial biomass, diversity, and compositions to altitudinal gradients depend on plant and soil characteristics. Sci. Total Environ. 610-611, 750–758. doi: 10.1016/j.scitotenv.2017.08.110
Reynolds, H. L., Packer, A., Bever, J. D., and Clay, K. (2003). Grassroots ecology: plant-microbe-soil interactions as drivers of plant community structure and dynamics. Ecology 84, 2281–2291. doi: 10.1890/02-0298
Stegen, J. C., Lin, X., Fredrickson, J. K., Chen, X., Kennedy, D. W., Murray, C. J., et al. (2013). Quantifying community assembly processes and identifying features that impose them. ISME J. 7, 2069–2079. doi: 10.1038/ismej.2013.93
Stegen, J. C., Lin, X., Konopka, A. E., and Fredrickson, J. K. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6, 1653–1664. doi: 10.1038/ismej.2012.22
Tao, F., Huang, Y. Y., Hungate, B. A., Manzoni, S., Frey, S. D., Schmidt, M. W. I., et al. (2022). Microbial carbon use efficiency promotes global soil carbon storage. Nature 618, 981–985. doi: 10.1038/s41586-023-06042-3
Thuiller, W., Lavorel, S., and Araújo, M. B. (2005). Niche properties and geographical extent as predictors of species sensitivity to climate change. Glob. Ecol. Biogeogr. 14, 347–357. doi: 10.1111/j.1466-822X.2005.00162.x
Tilman, D., Reich, P. B., Knops, J., Wedin, D., Mielke, T., and Lehman, C. (2001). Diversity and productivity in a long-term grassland experiment. Science 294, 843–845. doi: 10.1126/science.1060391
Tripathi, B. M., Kim, M., Singh, D., Lee-Cruz, L., Lai-Hoe, A., Ainuddin, A. N., et al. (2012). Tropical soil bacterial communities in Malaysia: pH dominates in the equatorial tropics too. Microb. Ecol. 64, 474–484. doi: 10.1007/s00248-012-0028-8
Tucker, C. M., Shoemaker, L. G., Davies, K. F., Nemergut, D. R., and Melbourne, B. A. (2016). Differentiating between niche and neutral assembly in metacommunities using null models of β-diversity. Oikos 125, 778–789. doi: 10.1111/oik.02803
Vellend, M. (2010). Conceptual synthesis in community ecology. Q. Rev. Biol. 85:18306. doi: 10.1086/652373
Wang, Q., Wang, S., He, T., Liu, L., and Wu, J. (2014). Response of organic carbon mineralization and microbial community to leaf litter and nutrient additions in subtropical forest soils. Soil Biol. Biochem. 71, 13–20. doi: 10.1016/j.soilbio.2014.01.004
Wei, P., An, S. Z., Dong, Y. Q., Sun, Z. J., Bierdawulieti, X. H. Y., and Li, C. (2020). The diversity and community structure characteristics of desert soil bacteria in the jungjunga basin based on high hand sequencing. Acta Pratacul. Sin. 29, 182–190. doi: 10.11686/cyxb2019337
Wu, J., Feng, Y., Zhang, X., Wurst, S., Tietjen, B., Tarolli, P., et al. (2017). Grazing exclusion by fencing non-linearly restored the degraded alpine grasslands on the Tibetan plateau. Sci. Rep. 7:15202. doi: 10.1038/s41598-017-15530-2
Wu, M. H., Xue, K., Wei, P. J., Jia, Y. L., Zhang, Y., and Chen, S. Y. (2022). Soil microbial distribution and assembly are related to vegetation biomass in the alpine permafrost regions of the Qinghai-Tibet plateau. Sci. Total Environ. 834:155259. doi: 10.1016/j.scitotenv.2022.155259
Zhang, H. R., and Fu, G. (2022). The response of the development diversity of soil fungal community system in the northern alpine grassland. Acta Agrestia Sinica 30, 21–28. doi: 10.11733/i.issn.1007-0435.2022.01.003
Zhang, C., Liu, G., Song, Z., Wang, J., and Guo, L. (2018). Interactions of soil bacteria and fungi with plants during long-term grazing exclusion in semiarid grasslands. Soil Biol. Biochem. 124, 47–58. doi: 10.1016/j.soilbio.2018.05.026
Zhang, Y. D., Niu, D. C., Li, Q. W., Liu, H. Y., Wang, Y., Xu, J. R., et al. (2025). Nonlinear response of soil microbial network complexity to long-term nitrogen addition in a semiarid grassland: implications for soil carbon processes. Agric. Ecosyst. Environ. 380:109407. doi: 10.1016/j.agee.2024.109407
Zhang, Q., Wang, X., Zhang, Z., Liu, H., Liu, Y., Feng, Y., et al. (2022). Linking soil bacterial community assembly with the composition of organic carbon during forest succession. Soil Biol. Biochem. 173:108790. doi: 10.1016/j.soilbio.2022.108790
Zhang, X. F., Zhao, L., Xu, S. J. Jr., Liu, Y. Z., Liu, H. Y., and Cheng, G. D. (2013). Soil moisture effect on bacterial and fungal community in Beilu River (Tibetan plateau) permafrost soils with different vegetation types. J. Appl. Microbiol. 114, 1054–1065. doi: 10.1111/jam.12106
Zhou, S. J., Dong, Y. Q., Yang, H. L., Yang, S. W., Asitaiken, J. L. H. T., Liu, Z. Y., et al. (2024). Effects of grazing exclusion on soil properties, fungal community structure, and diversity in different grassland types. Ecol. Evol. 14:e11056. doi: 10.1002/ece3.11056
Keywords: enclosure, grassland types, microbial assembly process, microbial respiration, Partial Least Squares Path Modeling (PLS-PM)
Citation: Julihaiti A, Yiqiang D, Shijie Z, Tingting N, Anjing J and Shazhou A (2025) From arid deserts to mesic meadows: divergent pathways regulating microbial respiration under grassland enclosure. Front. Microbiol. 16:1594877. doi: 10.3389/fmicb.2025.1594877
Edited by:
Izzah Shahid, Public University of Navarre, SpainCopyright © 2025 Julihaiti, Yiqiang, Shijie, Tingting, Anjing and Shazhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong Yiqiang, eGpkeXExMjEwQDE2My5jb20=
 Asitaiken Julihaiti
Asitaiken Julihaiti Dong Yiqiang1,2,3*
Dong Yiqiang1,2,3* Jiang Anjing
Jiang Anjing