Abstract
Introduction:
Understanding how microbial communities influence aroma profiles is critical to improving cigar quality. However, comparative studies examining the microbiome–aroma nexus across major cigar-producing regions remain limited.
Methods:
We integrated high-throughput metagenomic sequencing with volatile organic compound (VOC) profiling to investigate microbial community structure and aroma compounds in four Yunnan cigars and two Cuban cigars.
Results:
Bacillus spp. was consistently dominant across all samples, while Yunnan cigars exhibited higher microbial diversity. A total of 121 VOCs were detected, with nicotine, ylangene, δ-elemene, (R,S)-anatabine, and phenethyl alcohol identified as key aroma components. Nicotine, accounting for 29.7–55.0% of total VOC content, was positively correlated with Enterobacter and Escherichia, and negatively with Corynebacterium and Brachybacterium. Ylangene showed strong positive associations with Brachybacterium and Yaniella. After FDR correction, 26 differential VOCs were identified across cigar groups. KEGG pathway analysis revealed functional enrichment in carbohydrate, amino acid, and lipid metabolism. Principal component analysis indicated that the aroma complexity of certain Yunnan cigars, particularly YX4, approached that of Cuban cigars.
Discussion:
Our findings demonstrate that region-specific fermentation microbiota are intricately linked to the production of key VOCs. This work provides a scientific framework for optimizing cigar fermentation through microbial regulation and supports the potential for targeted microbial inoculation to enhance sensory quality and global competitiveness.
1 Introduction
Cigars are globally recognized as high-value cash crops, appreciated for their cultural heritage, complex flavor profiles, and high quality (Xing et al., 2021; Koszowski et al., 2018; Allem et al., 2019). Among the leading production regions, Cuba and Yunnan (China) have developed distinctive cigar products shaped by local environmental conditions and fermentation practices. Cuban cigars are renowned for their meticulous craftsmanship and controlled fermentations, often involving selected microbial inoculants to enhance flavor development (Viola et al., 2016). Yunnan cigars rely on spontaneous fermentation mediated by indigenous microbial communities and benefit from the region’s subtropical monsoon climate and traditional processing techniques (Jing et al., 2022; Chen et al., 2020).
As global demand for premium cigars continues to grow—projected to reach US $70.8 billion by 2032 with a CAGR of 4.6% —understanding the microbial and chemical determinants of cigar quality is increasingly important. As a prominent representative of traditional cigar markets, Cuba’s leading cigar manufacturer, Habanos S. A., achieved sales of USD 827 million in 2024, marking a 16% year-over-year increase (Allied Market Research, 2023). In China, the domestic cigar market is expanding even more rapidly, with a CAGR exceeding 7%. Yunnan cigars, primarily sold to cigarette manufacturers in provinces such as Shandong, Hubei, and Anhui, are in short supply, with 2024 sales at approximately 200 million yuan and an annual growth rate of 5–6% (RenrenDoc, 2025). These trends highlight Yunnan’s potential for both domestic dominance and global market expansion.
Fermentation is a key determinant of cigar quality, driving complex biochemical transformations through microbial metabolism, enzymatic activity, and interactions with environmental parameters such as temperature, humidity, and soil composition (Chattopadhyay et al., 2021; Zhang et al., 2024). These processes result in the generation of volatile organic compounds (VOCs), which contribute to characteristic aromas including nutty, floral, and coffee-like notes (Morris and Fiala, 2015; Krüsemann et al., 2018; Zheng et al., 2022). The metabolic activities of the microbial community encompass fatty acid and lipid biosynthesis, chlorogenic acid breakdown, carbohydrate degradation, protein catabolism, as well as amino acid and aromatic compound synthesis (Banožić et al., 2020; Su et al., 2024). Aldehydes and ketones, characterized by their carbonyl groups, contribute to tobacco’s distinctive aroma; esters add floral, fruity, and sweet notes; pyrazines impart nutty and roasted flavors; and furan compounds introduce a caramel-like scent (Chen et al., 2008; Villemur et al., 2009; Si et al., 2023).
Microbial communities associated with tobacco leaves—primarily bacteria and fungi—play pivotal roles in shaping these VOCs (Wu et al., 2023; Zhang et al., 2023). Bacterial genera frequently identified in tobacco include Bacillus, Sphingomonas, Stenotrophomonas, Erwinia, Pantoea, Lactococcus, Clostridium, Staphylococcus, Enterobacter, and Paenibacillus (Huang et al., 2010; Su et al., 2011). For instance, Pseudomonas efficiently degrades nicotine, while Bacillus breaks down lignin and carotene by secreting specific enzymes, yielding small aromatic compounds (Ye et al., 2017; Chopyk et al., 2017). Meanwhile, fungal such as Aspergillus, Penicillium, Alternaria, Chaetomium, Cladosporium, Rhizopus, Fusarium, Trichoderma, and Monascus are also common (Chen H. et al., 2018; Chen S. et al., 2018).
Environmental conditions, particularly temperature, humidity, and altitude, exert strong influences on microbial community composition and activity. For example, elevated temperatures in tropical and subtropical regions like Cuba and Yunnan promote the proliferation of thermophilic and mesophilic microorganisms that degrade carotenoids and terpenoids, leading to the formation of key aroma compounds such as ionone and dihydroactinidiolide (English et al., 1967; Maldonado-Robledo et al., 2003). High humidity favors the growth of fungi such as Aspergillus and Penicillium, enhancing floral and earthy VOCs (Liu et al., 2015; Wu et al., 2022). Additionally, altitude influences microbial diversity by altering soil pH and nutrient availability, while anthropogenic factors such as heavy metal pollution can suppress microbial richness and shift community structure (Ye et al., 2024; Liu et al., 2020).
Advances in omics technologies, including 16S rRNA gene sequencing, shotgun metagenomics, and metabolomics, have enabled comprehensive profiling of biological components such as microbial taxa, functional genes, protein expressions, and metabolic signatures within natural ecosystems (Zhang et al., 2021; Romdhane et al., 2022). These approaches have been increasingly applied to tobacco-related studies (Liu et al., 2021; Ye et al., 2021; Zhou et al., 2021), revealing the influence of geographic origin and leaf variety on VOCs (Tang et al., 2020; Yan et al., 2022; Zhengfeng et al., 2024). A thorough investigation of these aroma compounds not only elucidates how they differ across various cigar types but also provides a scientific framework for quality assessment (Wang et al., 2024).
However, systematic comparisons between major cigar-producing regions, such as Cuba and Yunnan, remain limited, particularly in elucidating how regional fermentation microbiota shape the aromatic profiles of cigars. We hypothesize that distinct microbial communities may be shaped by the unique environmental conditions of Yunnan and Cuba, drive differences in VOCs that define their respective cigar aromas, potentially enabling Yunnan cigars to achieve aromatic profiles comparable to those of Cuban cigars. To evaluate this hypothesis, we employed high-throughput metagenomic sequencing and GC–MS to analyze the microbial compositions and volatile profiles of cigars from both regions, aiming to elucidate the role of microbial communities in shaping region-specific aroma profiles. As the first integrated microbiome–metabolome comparison of Yunnan and Cuban cigars, our work provides mechanistic insights into aroma formation and offers a scientific basis for improving cigar quality through microbial modulation.
2 Materials and methods
2.1 Experimental materials
In this study, we collected four Yunnan cigars labeled YX1 (Huaning, 24°11′N, 102°55′E; altitude about 1,200 m), YX2 (Ganzhuang, 23°45′N, 102°2′E; 700–800 m), YX3 (Xinping, 24°40′N, 101°59′E; 500–550 m), and YX4 (Chengjiang, 24°40′N, 102°54′E; 1,650 m), along with two Cuban cigars labeled YX5 (Vuelta Abajo, 22°24′N, 83°42′W; 150 m) and YX6 (Vuelta Abajo, 22°24′N, 83°42′W; 100–200 m) for analysis. All the cigars were subjected to an identical fermentation process in 2020 and subsequently aged for 2 years before reaching their final form. The fermentation experiments were carried out with pest control, sorting, rehumidification, balancing, stacking, fermenting, repiling (five times), and unstacking. Repiling occurred immediately when the temperature reached 40–45°C. After that, the cigars were aged for 2 years under controlled conditions of 20°C and 70% humidity (Zhang et al., 2025a). The samples were selected to represent different varieties and geographical origins, as detailed in Table 1 Information of cigars. For each sample, we weighed out 6.0 g, divided it into six portions, and immediately froze the portions in sterile centrifuge tubes using liquid nitrogen. To maintain sample integrity, three portions from each group were stored at −20°C for GC–MS analysis while the remaining three were stored at −80°C for DNA extraction, in accordance with the requirements of each analytical procedure.
Table 1
| Place | Sample | Denomination of origin | Altitude |
|---|---|---|---|
| Yunnan | YX1 | Huaning (24°11′N, 102°55′E) | About 1,200 m |
| YX2 | Ganzhuang (23°45′N, 102°2′E) | 700–800 m | |
| YX3 | Xinping (24°40′N, 101°59′E) | 500–550 m | |
| YX4 | Chengjiang (24°40′N, 102°54′E) | About 1,650 m | |
| Cuba | YX5 | Vuelta Abajo (22°24′N, 83°42′W) Vuelta Abajo | About 150 m |
| YX6 | (22°24′N, 83°42′W) | 100–200 m |
Information of cigars.
2.2 Experimental methods
2.2.1 Metagenomic sequencing
DNA extraction, library construction, and sequencing were carried out at Beijing Novogene Co., Ltd., with each sample processed in triplicate. Approximately 1 μg of genomic DNA from each sample was randomly sheared into ~350 bp fragments using a Covaris ultrasonic disruptor (LE220R-plus, United States). Subsequent steps included end repair, addition of A-tails, adapter ligation, purification, and PCR amplification. The integrity and insert sizes of the resulting libraries were assessed by AATI analysis. Once the insert length met the requirements, the libraries were precisely quantified via Q-PCR to ensure an effective concentration above 3 nM. Libraries that passed quality control were then pooled based on effective concentration and target data yield, followed by PE150 sequencing (Illumina Novaseq 6,000, Illumina, San Diego, CA, United States).
2.2.2 Bioinformatic analysis
Raw Illumina data were preprocessed with Fastp,1 yielding clean reads for downstream analyses. We discarded paired reads if they contained adapter contamination, had more than 10% ambiguous nucleotides, or showed over 50% low-quality bases (Q < 5). The clean data were assembled using MEGAHIT with the meta-large parameter settings (−-end-to-end, −-sensitive, -I 200, −X 400). Scaffold sequences were broken at “N” junctions to generate contiguous sequences (scaftigs) without the presence of “N”.
Using default parameters, MetaGeneMark2 predicted open reading frames (ORFs) on scaftigs ≥ 500 bp for each sample (Karlsson et al., 2012; Oh et al., 2014; Qin et al., 2014). Any predicted segments under 100 nt were removed (Sunagawa et al., 2015). Redundant genes were clustered using CD-HIT3 with the parameters -c 0.95, −G 0, -aS 0.9, −g 1, and -d 0, producing a non-redundant initial gene catalog (Fu et al., 2012). Clean reads from each sample were aligned to this catalog via Bowtie2 (−-end-to-end, −-sensitive, -I 200, −X 400) to obtain gene read counts; genes with ≤ 2 reads in each sample were removed, resulting in the final gene set (Unigenes) for subsequent analyses (Li et al., 2014). Gene abundance was calculated based on the number of aligned reads (r) and the gene length (L) (Villar et al., 2015).
2.2.3 Volatile flavor compounds analysis
VOCs in cigars were analyzed via headspace solid-phase microextraction–gas chromatography–mass spectrometry (HS-SPME-GC–MS). Each 3.0 g sample was finely chopped, placed into three 20 mL headspace vials, ensuring biological replication. Each vail was spiked with 4 μL of a 5.0 μg/μL deuterated naphthalene (Naphthalene-d8) solution as an internal standard. The vials were sealed with silicone septa and incubated at 80°C for 20 min. An 85 μm CAR/PDMS fiber (MPS-Robotic, Gerstel, Germany) was then used to extract volatile compounds for 5 min at 80°C, followed by desorption at 250°C for 5 min.
Subsequent separation and identification of volatile flavor compounds were carried out on an Agilent 7890B gas chromatograph (Santa Clara, CA) equipped with a 5977B mass spectrometer and a DB-5MS capillary column (60 m × 0.25 mm × 0.25 μm film thickness). High-purity helium served as the carrier gas at a constant flow rate of 1 mL/min, with an injection port temperature of 250°C. The oven temperature was initially held at 40°C for 1 min, ramped at 5°C/min to 220°C, and maintained for 3 min. The transfer line and ion source were set to 270°C and 230°C, respectively, under electron impact (EI) ionization at 70 eV in full-scan mode (m/z 30–550). Compound identification was based on comparisons with authentic standards (when available) and the NIST 17 library.4 For semi-quantitative analysis, peak areas of individual compounds were normalized against the internal standard peak area (Naphthalene-d8) to determine relative concentrations (Zhang et al., 2024).
2.3 Data mining and statistical analysis
All experiments were conducted with at least three replicates per condition, and results are presented as mean values to ensure reliability and reproducibility. Heatmaps, principal component analysis (PCA), boxplots, and multiple comparisons were generated in Origin 2024 and GraphPad Prism 8.0.1. Additionally, we used Spearman’s correlation coefficients to examine the relationships between representative microbes and core VOCs, and performed network analyses in PyCharm Community Edition 2023.2.3. Kruskal-Wallis and Wilcoxon rank sum test are calculated in R (v4.2.2).
3 Results
3.1 Overview of the microbial community
After quality filtering, each of the 18 samples yielded between 67,247,870 and 99,945,732 high-quality sequences. Coverage analysis indicated that this sequencing depth was sufficient for saturation, ensuring robust capture of microbial diversity. As illustrated in Figure 1A, taxonomic analysis revealed that bacterial communities dominated the microbial composition in all samples, consistently accounting for close to or exceeding 50% of the total. This finding underscores the central role of bacteria in colonizing both Yunnan and Cuban cigars.
Figure 1
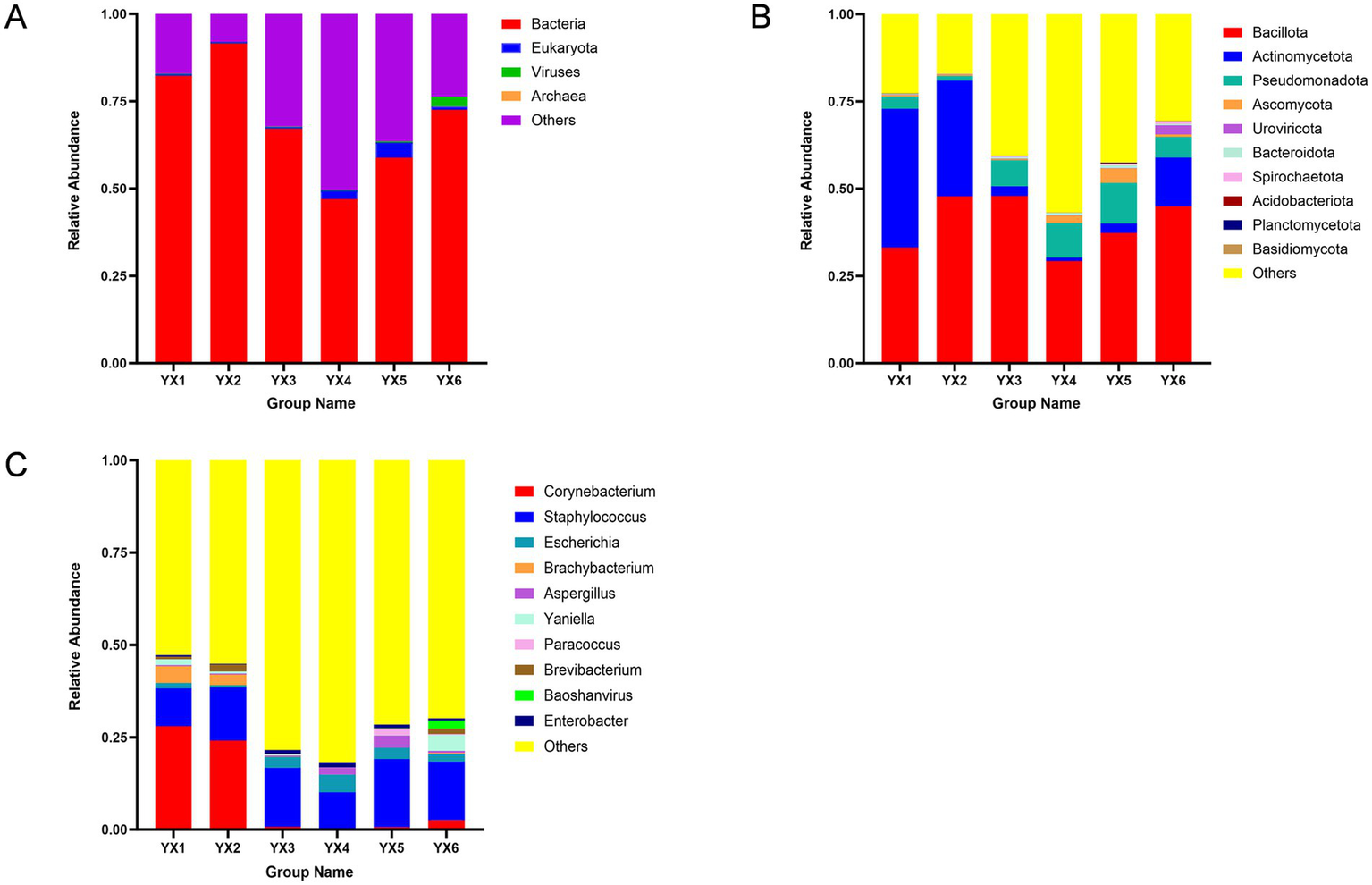
Microbial community composition of cigars at kindom (A), phylum (B) and genus (C) levels.
We classified microbial groups at two taxonomic levels, the top 10 phyla and genera by relative abundance were illustrated in the figures. At the phylum level, taxa with relative abundances above 0.5% were defined as dominant phyla, while at the genus level, those above 1% were considered dominant genera (Yao et al., 2024). At the phylum level (Figure 1B), Bacillota, Actinomycetota, and Pseudomonadota collectively accounted for 30.25–80.92% of total abundance. Bacteroidea also exhibited high abundance across all samples, suggesting a notable influence on the overall microbial composition. At the genus level (Figure 1C), Staphylococcus emerged as a dominant taxon, with high abundance in all six samples. Corynebacterium was particularly abundant in Yunnan cigars YX1 and YX2, making it a key genus in those two samples.
Despite these shared characteristics, microbial abundances varied substantially by geographic origin. For instance, Actinomycetota was significantly more abundant in YX1, YX2, and YX6 than in YX3, YX4, and YX5. Although Bacillota remained the most prevalent phylum overall, Pseudomonadota also had a noteworthy presence in YX3, YX4, and YX5. At the genus level, Paracoccus was found only in YX5, while Baoshanvirus appeared exclusively in YX6. In general, Yunnan cigars exhibit Bacillus dominance.
An alpha-diversity analysis was performed on the six cigar types. Microbial coverage for all samples approached 1 (Figure 2A), indicating high sequencing coverage and reliable sampling to reflect the cigar-associated microbial communities. The Chao1 index reflects community richness, whereas the Shannon and Simpson indices measure community diversity. The Chao1 index showed that Cuban cigars displayed higher bacterial richness than Yunnan cigars (Figure 2B), and YX5 also exhibited high species richness and evenness. Moreover, Yunnan cigars YX3 and YX4 are different from YX1 and YX2, demonstrated enhanced microbial richness and evenness (Figures 2C,D). Notably, YX4 exhibited clustering patterns more closely aligned with YX5.
Figure 2
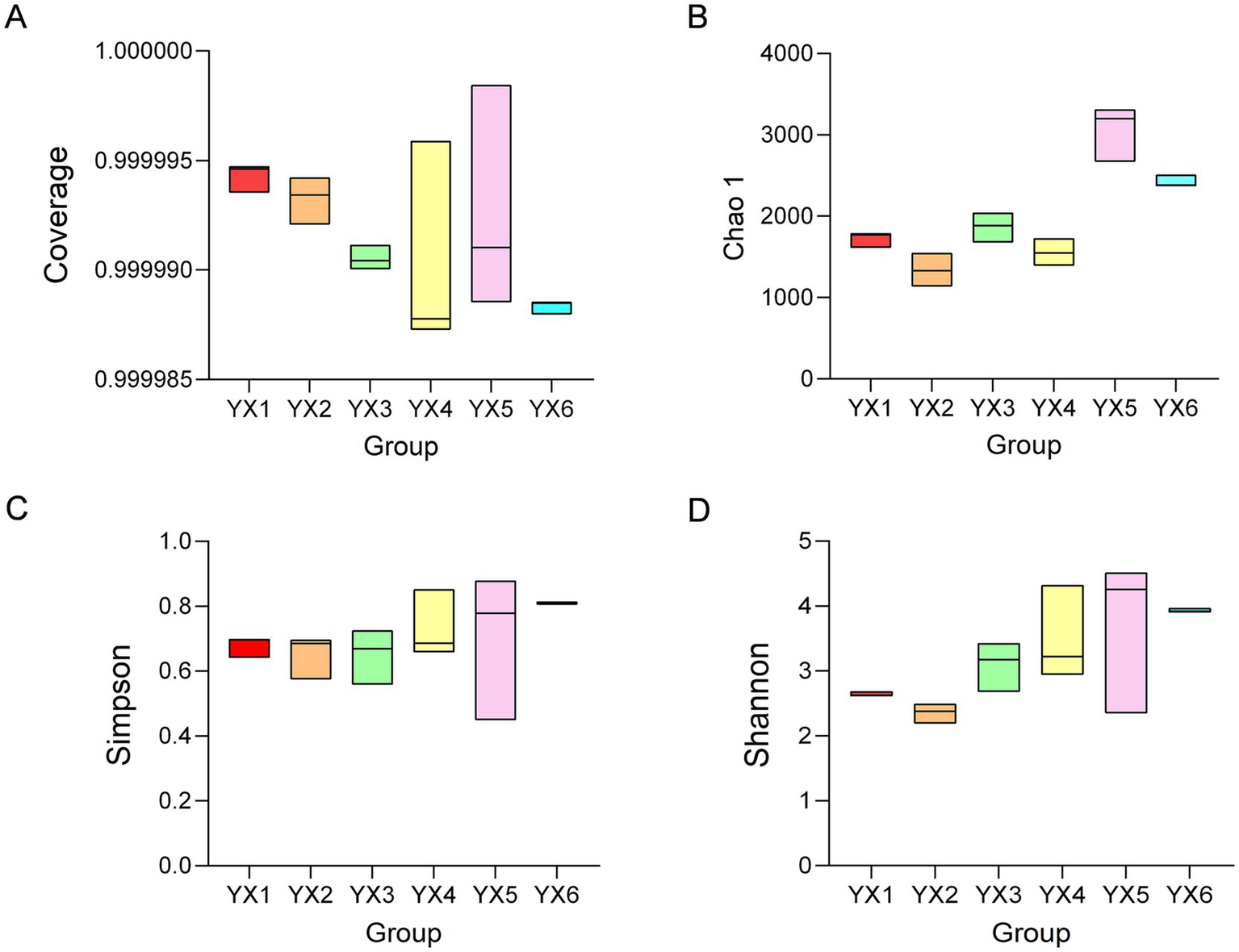
Alpha Indices. Coverage (A), Chao1 index (B), Shannon index (C), Simpson index (D).
Then, we use the principal coordinate analysis (PCoA) plot illustrates the variation in microbial community compositions across the six cigar samples (YX1–YX6) (Figure 3). The first two principal coordinates, PCoA 1 and PCoA 2, account for 69.54 and 18.04% of the total variance, respectively, capturing the majority of the dataset’s structural variability. Samples YX1 and YX2 are closely clustered in the left quadrant, signifying a high degree of similarity in their microbial profiles. Conversely, four samples clustered in the right quadrant, which means the composition of these microorganisms is significantly different from that of YX1 and YX2. Notably, YX5 exhibits greater dispersion, indicative of increased diversity in their microbial compositions. And the overlap observed between YX4 and YX6 suggests that the fermentation modifications applied to YX4 contribute to the convergence of its profile with that of premium Cuban cigars.
Figure 3
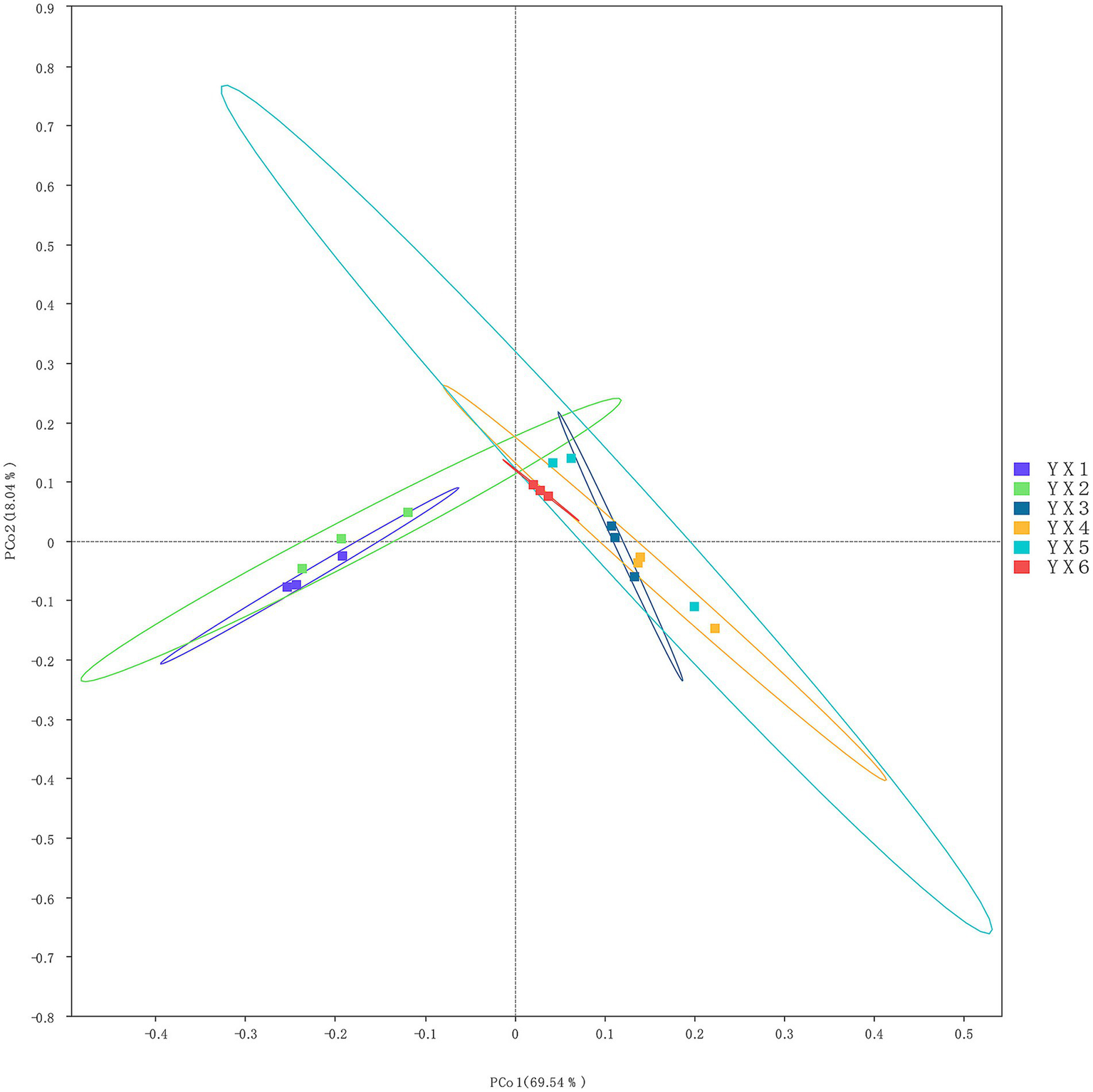
Beta diversity analysis of six cigar samples.
3.2 Predicted metabolic functions of microbial communities in cigars
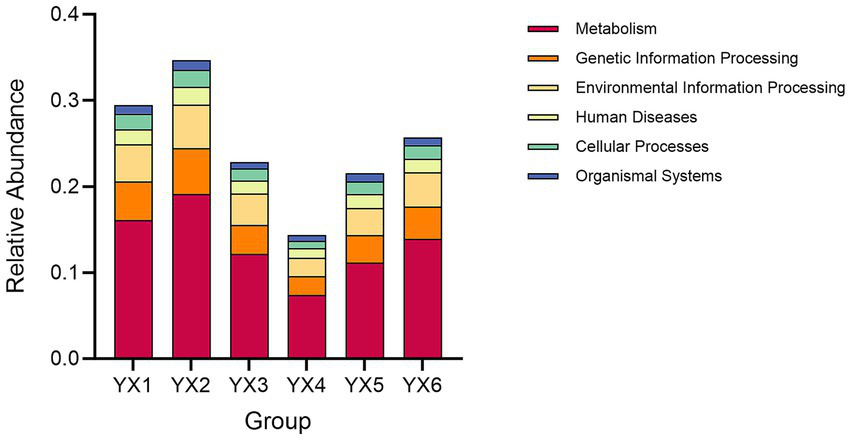
The KEGG database integrates gene and genome information with higher-level functional data, allowing for systematic analysis of gene function (Aoki-Kinoshita and Kanehisa, 2007). Cigar processing shapes microbial communities, impacting metabolic pathways and compound distributions. These metabolic dynamics are critical in shaping the chemical profiles that define a cigar’s flavor and quality. As shown in Figure 4, metabolism constituted a significant portion of functional activity in all groups, driving most of the between-group variability. Additionally, Figure 5 shows that the top five metabolic pathways in terms of relative abundance were carbohydrate metabolism, amino acid metabolism, energy metabolism, metabolism of cofactors and vitamins, and membrane transport. Furthermore, as shown in Supplementary Figure S1 “Unigenes KEGG” data, all the samples exhibited an enrichment of enzymes involved in the metabolism of terpenoids and polyketides, as well as related metabolic pathways. Consequently, differences in microbial composition across cigars intended for diverse purposes may lead to functional gene divergences, ultimately affecting flavor profiles.
Figure 4
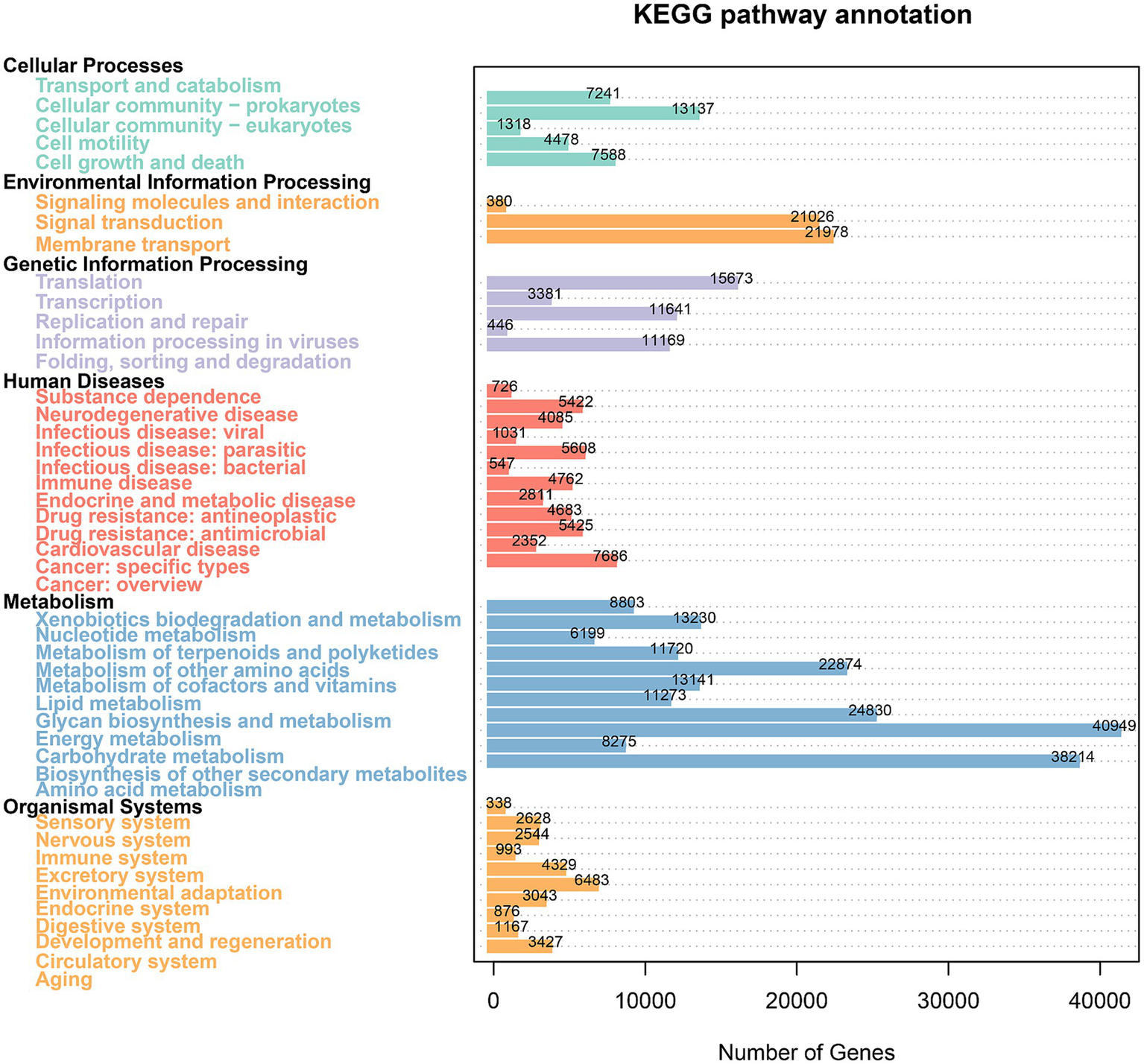
Analysis of groups contributions to KEGG.
Figure 5
The number of genes related to KEGG metabolic pathways.
LEfSe analysis was conducted to identify significant differences below the phylum level (Figure 6A). Circles from inner to outer represent microbial classifications from phylum to genus, and each color denotes taxa that differ significantly among groups. Figure 6B demonstrates the Linear Discriminant Analysis (LDA) of microbial taxa across experimental groups (YX1–YX6), where higher LDA scores indicate stronger discriminatory power and taxonomic significance of specific microbes within their respective groups. From the LEfSe, we identified 3 classes, 4 orders, and 5 families showing notable divergence. For example, Corynebacteriaceae was enriched in YX1, Lactobacillales in YX5, and Aspergillaceae in YX6. Compared with other groups, YX5 displayed more pronounced microbial differences, and YX1 also showed unique microbiotas. The low LDA scores of the characteristic bacteria in groups YX2, YX3, and YX4 indicate a high microbial diversity in these groups, but relatively fewer specific contributing bacteria. Although the main characteristic bacteria of these groups are not as significant as YX1, YX5, and YX6, they may represent the transitional stage of microbial communities during the fermentation process. Such significant variations may be tied to differences in fermentation processes, community structure, and species balance, all of which impact cigar quality.
Figure 6
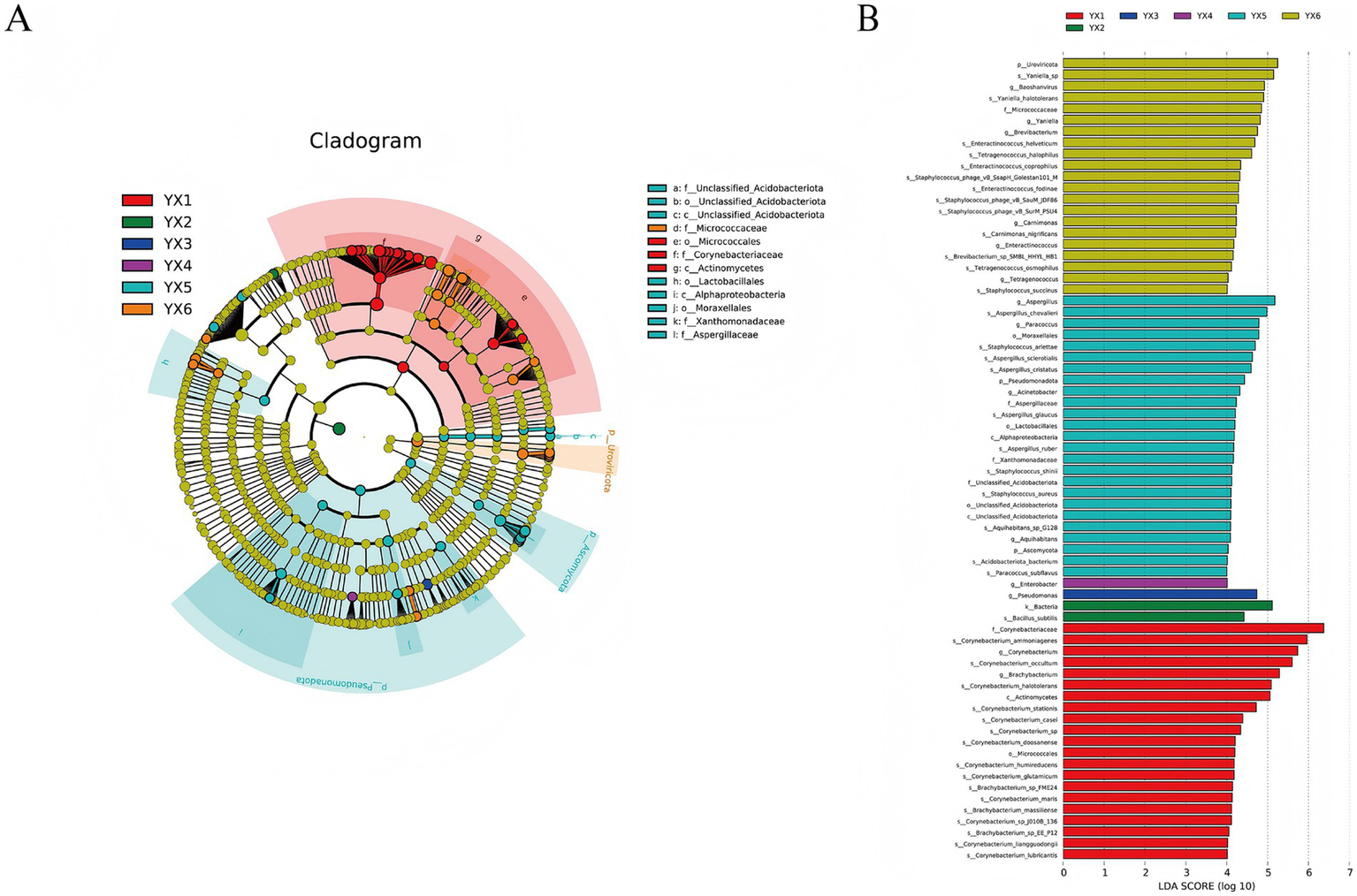
Differential microbes in different cigars. LEfSe (A) and LDA (B) analysis.
3.3 Profiles of volatile flavor compounds
We selected CAR/PDMS fiber for its suitability in capturing medium to low polarity compounds, optimizing target compound capture while minimizing interference. Mixed-coating fibers, like PDMS, enhance VOC coverage in multi-component analysis. The fiber’s excellent thermal stability ensures consistent GC–MS performance under high temperatures and supports repeated use. A DB-5 column, a non-polar option with a wide temperature range, was chosen to align with the fiber’s polarity, enabling efficient separation of medium to low polarity compounds with varying boiling points. This combination ensures reliable sensitivity and separation for the experiment.
To assess the differences in volatile flavor compounds among various cigars, HS-SPME-GC–MS analysis was conducted. Initially, 121 volatile organic compounds (VOCs) with identity scores exceeding 80 were preliminarily screened (Supplementary Table S2). These included 2 aldehydes, 8 ketones, 4 esters, 10 alkanes, 56 terpenes, 7 alkaloids, 9 alcohols, 1 phenol, and 24 other volatile components, with terpenes representing 46.28% of the VOCs.
Principal component analysis (PCA, Figure 7A) and heatmap (Supplementary Figure S1) were employed to examine distribution and diversity. PCA revealed close clustering of sample data points, suggesting high intra-group similarity. The heatmap showed that YX1, YX2, and YX6 exhibited more diverse VOC profiles: YX1 contained 68 distinct compounds, YX2 had 65, and YX6 had 66. In contrast, YX3 showed only 39 compounds, indicating a simpler volatile profile.
Figure 7
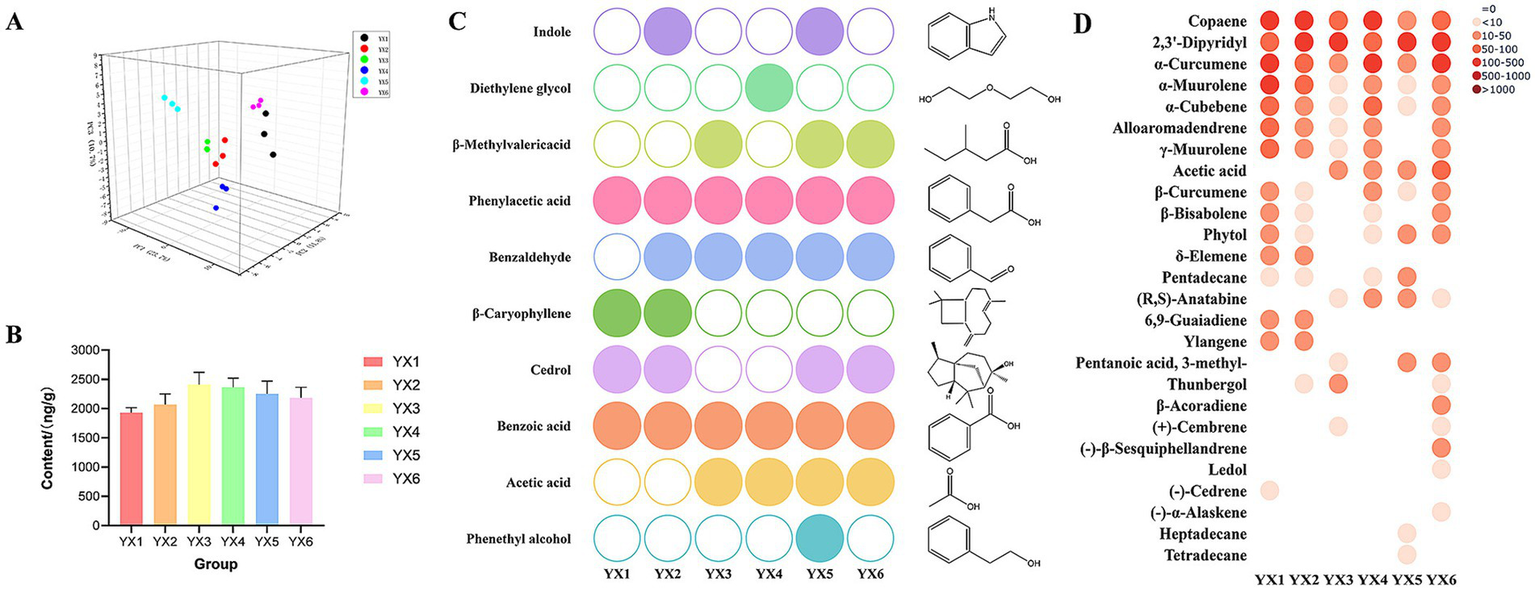
(A) PCA plot of six types of cigars based on aroma compounds; (B) content of nicotine in different group; (C) top 10 volatile compounds were identified using PCA; (D) FDR correction in VOC analysis.
Following the application of PCA for dimensionality reduction of the experimental data, the cumulative variance contribution rate of the first three principal components reached 57.8%. Nicotine, phenethyl alcohol, and acetic acid emerged as the primary characteristic variables, with corresponding variance explanation rates of 29.6, 14.8, and 13.4%, respectively. This analysis indicates that these three characteristic compounds make significant contributions to the explanation of variability within the dataset. Nicotine emerged as the most abundant compound, with average relative contents ranging from 1783.47 to 2246.41 ng/g (Figure 7B), accounting for 29.72–55.00% of the total volatile organic compounds (TVOCs). In comparison, YX3 has the highest nicotine content, while YX1 has the lowest. After that, we selected the top 10 compounds with ratings other than nicotine. These metabolites are phenethyl alcohol, acetic acid, benzoic acid, cedrol, β-caryophyllene, benzaldehyde, phenylacetic acid, β-methylvalericacid, diethylene glycol, indole. The identified compounds have been found to significantly contribute to the overall aroma profile of tobacco leaves. Figure 7C presents the distribution of these 10 metabolites across different groups of cigars. Within this figure, distinctively colored circular symbols represent the 10 types of differential metabolites, whereas unfilled circles indicate the absence of detectable volatile metabolites in the respective samples. It is readily apparent that Cuban cigars YX5 and YX6 exhibit a richer profile of volatile components, which significantly contribute to their distinct aroma and sensory attributes.
The Kruskal-Wallis non-parametric rank sum test was used for multiple group comparisons, and the Benjamini-Hochberg (FDR) method was applied to control the type I error rate. A significance threshold of α = 0.05 was set, and a corrected p-value less than 0.05 was considered statistically significant. Ultimately, 26 differentiating compounds were screened out from 121 compounds (Figure 7D). Notably, all smaples have copaene, 2,3′-dipyridyl, α-curcumene, α- murolene and α-cubebene. Subsequently, the Wilcoxon rank-sum test was conducted, and the Bonferroni method was used for multiple-comparison correction (Table 2 Wilcoxon rank sum test results). The results indicated that for the five pairs, namely YX1 vs. YX3, YX1 vs. YX5, YX2 vs. YX3, YX3 vs. YX6, and YX5 vs. YX6, the p-values were less than 0.05, suggesting significant differences among them. In contrast, for pairs such as YX1 vs. YX2, YX1 vs. YX4, YX1 vs. YX6, YX2 vs. YX4, YX2 vs. YX6, YX3 vs. YX4, YX3 vs. YX5, YX4 vs. YX5, and YX4 vs. YX6, the p-values were greater than 0.05, indicating no significant differences.
Table 2
| Group | YX1 | YX2 | YX3 | YX4 | YX5 |
|---|---|---|---|---|---|
| YX2 | 1.00000 | – | – | – | – |
| YX3 | 4.1e-05 | 0.00205 | – | – | – |
| YX4 | 0.17042 | 1.00000 | 0.50581 | – | – |
| YX5 | 0.00438 | 0.13002 | 1.00000 | 1.00000 | – |
| YX6 | 1.00000 | 1.00000 | 0.00016 | 1.00000 | 0.03150 |
Wilcoxon rank sum test results.
3.4 Relationship between microbial communities and VOCs
Spearman correlation heatmaps and clustering analyses were conducted to investigate the interplay between microbial taxa and volatile flavor compounds. We focused on the top 10 most abundant microbial genera in each sample (YX1–YX6), examining their correlations with identified compounds (R > 0.05, p < 0.05). The resulting heatmaps and network diagrams (Supplementary Figures S3–S8) indicate that YX3 (Yunnan) and YX5 (Cuban) displayed relatively sparse correlations, suggesting potentially fewer direct links between microbes and VOCs in these two samples. By contrast, YX1 (Yunnan) and YX6 (Cuban) exhibited dense correlation networks, pointing to more intimate relationships. Interestingly, certain microbes seem played contrasting roles in different systems. For instance, Baoshanvirus and Enterobacter were positively correlated with VOCs in YX3 but showed negative correlations in the other five cigar types. Aspergillus, the sole fungal genus among the top 10 microbial genera, exhibited close associations with compounds across all six samples. Notably, in sample YX3, Aspergillus demonstrated significant positive correlations with the majority of compounds, whereas in the other five samples, its associations with compounds were predominantly characterized by negative correlations.
Meanwhile, we selected the top 9 compounds based on the absolute correlation values between each bacterial genus and the associated compounds and a heatmap (Figure 8) was constructed. As is evident from the graph, Corynebacterium and Brachybacterium exhibited significant positive correlations to the nine specified organic compounds, whereas Staphylococcus demonstrated weak correlations across all tested metabolites. Besides, the compounds δ-elemene, β-calacorene, 6,9-guaiadene, and ylangene exhibited an influence on the microorganism that was inversely related to the effects observed for acetic acid, dihydroactinidiolide, nicotine, benzeneacetic acid, and (R, S)-anatabine. Specifically, δ-elemene, β-calacorene, 6,9-guaiadene, and ylangene exhibited positive correlations with the four microbial genera, namely Corynebacterium, Brachybacterium, Yaniella, and Brevibacterium. In contrast, negative correlations were observed between these compounds and Staphylococcus, Escherichia, Aspergillus, Baoshanvirus, and Enterobacter. Conversely, (R, S)-anatabine, benzeneacetic acid, nicotine, and dihydroactinidiolide showed negative correlations with Corynebacterium, Brachybacterium, Yaniella, and Brevibacterium, while positive correlations were detected with Staphylococcus, Escherichia, Aspergillus, Baoshanvirus, and Enterobacter. Furthermore, acetic acid, while exhibiting a weak positive correlation specifically with Yaniella, generally displayed a correlation pattern consistent with that observed for the latter group of compounds.
Figure 8
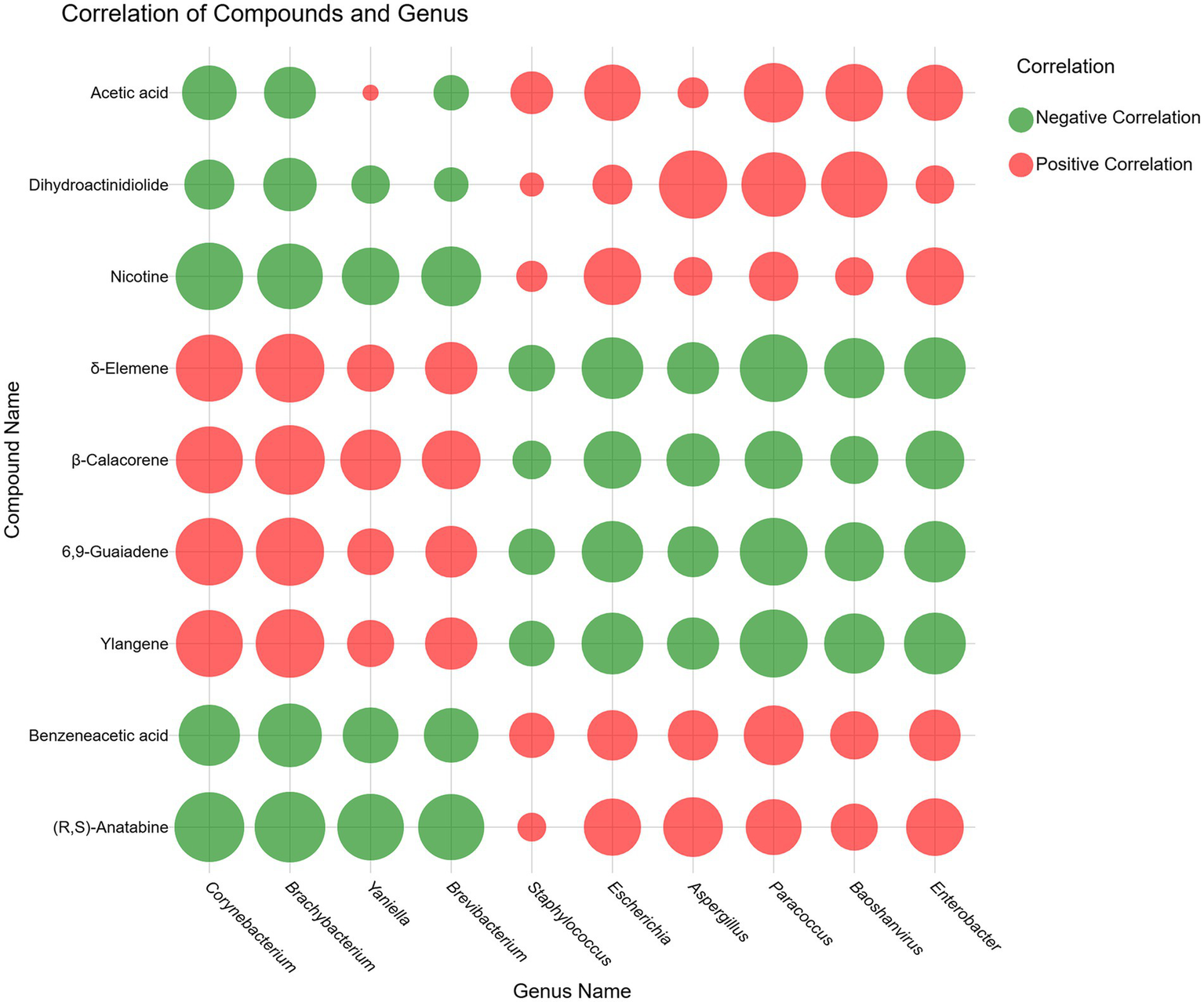
Microbial-VOCs correlation coefficient heatmap.
4 Discussion
In this study, we investigated six cigar samples—four from Yunnan and two from Cuba—to elucidate the interplay between microbial community structure and aroma composition. Our findings highlight the critical role of microbial composition and geographic factors in shaping the aroma profiles of cigars, providing new insights into the microbial and metabolic mechanisms underlying flavor development. The dominance of Bacillus spp. across all samples underscores its pivotal role in cigar fermentation, aligning with previous studies, which have demonstrated that Bacillus not only represents the dominant microbial genus in cigars but also possesses enzymatic capabilities to transform pungent precursors into desirable aromatic compounds (English et al., 1967; Xueru et al., 2022; Brandsch, 2006). Compared to other major cigar-producing regions, the microbial communities in cigars exhibit significant regional differences. For instance, in Yunnan cigars, Staphylococcus constitutes approximately 20% of the total microbial genera, whereas in Brazilian cigars, this proportion reaches as high as 79.66%. Additionally, the abundance of Aspergillus in Yunnan cigars is markedly lower than the 78.31% observed in Indonesian cigars. These disparities likely reflect the unique climatic conditions, fermentation processes, and tobacco raw materials characteristic of each region, which profoundly influence the composition and distribution of microbial communities (Zheng et al., 2022).
However, our analysis also revealed region-specific microbial signatures. Brachybacterium was identified as a characteristic microorganism in YX1 and YX2, and microbial taxa affiliated with the family Micrococcaceae predominated in Yunnan YX1, while the microbial communities in YX3 and YX4 were comparatively less diverse. Notably, YX4 exhibited a relatively high abundance of Aspergillus. Additionally, Paracoccus was identified as a characteristic microorganism in YX5, whereas Baoshanvirus was unique to YX6. Brachybacterium has ability to produce polyphenol oxidase, which facilitates the formation of pigments such as theaflavins and various aromatic compounds (Pan et al., 2023; Zuo et al., 2024). Micrococcaceae functionally linked to proteolytic and amino acid metabolic pathways, may underpin the formation of the initial flavor profile in cigar fermentation (Sims et al., 1986). Besides, the lower microbial abundance observed in YX3 and YX4 suggests a more advanced stage of fermentation, potentially reflecting their progression into a final equilibrium phase. The relatively high abundance of Aspergillus in YX4 may be attributed to the higher humidity levels in its fermentation environment. Moreover, the presence of Paracoccus is capable of effectively degrading phthalate compounds, playing a role in the removal of organic pollutants generated during the cigar production process (Xu et al., 2020). This suggests that the cultivation environment of YX5 may be contaminated to some extent. Currently, there are no reports demonstrating the function of Baoshanvirus in tobacco, it may represent a local microbial adaptation specific to the Vuelta Abajo region. These findings suggest that regional factors such as climate, soil, and fermentation practices significantly influence microbial diversity.
In terms of VOCs, nicotine plays a central role in cigar quality (Zhang et al., 2025b). Beyond its irritant moisturizing effect, is the most abundant compound across all samples, and it is also a key marker of product consistency (Yi et al., 2024). Nicotine biosynthesis involves the formation of a pyridine ring and a pyrrolidine ring. Key enzymes for the pyridine ring—aspartate oxidase (AO, EC 1.4.3.16), quinolinate synthase (QS, EC 2.5.1.72), and quinolinate phosphoribosyl transferase (QPT, EC 2.4.2.19)—were abundantly detected in sample YX5 and associated with Paracoccus, Staphylococcus, and Enterobacter (Brandsch, 2006). For the pyrrolidine ring, ornithine decarboxylase (ODC, EC 4.1.1.17) was found in Enterobacter, Aspergillus, Escherichia, and Paracoccus; PMT (EC 2.1.1.53) was absent in YX1 and YX2; and N-methylputrescine oxidase (MPO, EC 1.4.3.-) was abundant in YX5 and present at lower levels in YX6. These results suggest a potential positive correlation between nicotine levels and the presence of Paracoccus, Staphylococcus, Enterobacter, Aspergillus, and Escherichia. Additionally, in the nicotine degradation pathway, MPO1 (EC 1.4.3.21)—detected in all samples and linked to Brevibacterium—indicates that Brevibacterium may contributes to nicotine breakdown, aligning with its negative correlation with nicotine concentration.
Ylangene, a sesquiterpene with distinct floral and fruity notes, contributes significantly to cigar aroma complexity (Huayi et al., 2024). In this study, ylangene exhibited positive correlations with Brachybacterium and Corynebacterium, but negative correlations with Aspergillus, Enterobacter, Escherichia, Paracoccus, and Baoshanvirus. The key biosynthetic enzyme, sesquiterpene synthase (TPS2, EC: 4.2.3.-), was identified in YX3, YX4, YX5, and YX6. However, HS-SPME-GC–MS analysis revealed that ylangene was only detected in YX1 and YX2, not in the TPS2-positive samples. This discrepancy suggests that TPS2 expression alone may not determine ylangene presence, possibly due to regulatory or metabolic constraints. Although Aspergillus is a dominant fungus in tobacco fermentation, its weak correlation with ylangene implies a limited role in its biosynthesis. Nonetheless, Aspergillus contributes to overall aroma formation by producing enzymes such as amylase, protease, and cellulase, which degrade macromolecules in tobacco leaves into precursors of small aromatic compounds (Zhao et al., 2024).
(R, S)-Anatabine, a minor pyridine alkaloid in tobacco, is primarily synthesized via enzymatic pathways similar to those of nicotine. In this process, lysine is first converted to cadaverine by lysine decarboxylase (LDC), which is then cyclized by cyclase enzymes to form a piperidine ring structure (Leete, 1975). Degradation of (R, S)-anatabine has been demonstrated in Arthrobacter, which contains nicotine dehydrogenase, and in Pseudomonas, which harbors NicA2 (EC: 1.4.2.2) homologous enzymes. These enzymes have also been detected in YX2 and YX5 (Ruan et al., 2005; Sun et al., 2015). Experimental analyses reveal that the top 10 bacterial strains exhibit similar degradation patterns for (R, S)-anatabine and nicotine. Notably, Yaniella, Brevibacterium, Corynebacterium, Brachybacterium, and show negative correlations with both compounds, suggesting they likely possess NicA2 homologs and nicotine dehydrogenase, enabling the breakdown of tobacco alkaloids.
The KEGG pathway analysis further elucidated the metabolic basis of these microbial-chemical interactions. Pathways related to carbohydrate metabolism, amino acid biosynthesis, and lipid degradation were enriched in the microbial communities, producing precursors such as phenylalanine and tyrosine, which are critical for the synthesis of benzeneacetic acid and other aromatic compounds. Benzeneacetic acid is an intermediate in the degradation of phenylalanine, in which phenylalanine is converted to benzeneacetic acid through a series of reactions. In some bacteria, such as Escherichia coli, phenylacetate can be further metabolized into other compounds. The process involves the conversion of benzeneacetic acid to phenylacetyl-CoA (Ju et al., 2021). In addition, the phenylalanine metabolic pathway can also produce other aromatic compounds, such as benzoic acid and coumarin (Moore et al., 2002). Carbohydrate metabolism intermediates serve as substrates for various biosynthetic processes, including those producing aromatic amino acids (Pittard et al., 2005; Zhao et al., 2013).
Indole is a compound with a dual odor profile: it smells fecal at high concentrations but floral when highly diluted (Zeng et al., 2016). Its biosynthesis mainly derives from tryptophan metabolism. Typically, tryptophan is deaminated by tryptophanase to form indole-3-pyruvate, which is decarboxylated to indole-3-acetaldehyde and then oxidized to indole. Alternatively, tryptophan can be converted to indole-3-acrylic acid and subsequently decarboxylated to produce indole (Tang et al., 2023). This pathway varies across organisms: Escherichia coli synthesizes indole via tryptophanase (TnaA, EC: 4.1.99.1) (Watanabe and Snell, 1972); Bacillus subtilis uses tryptophan deaminase to generate indole-3-acrylic acid (Hoch and Crawford, 1973); Pseudomonas aeruginosa produces indole from tryptophan in soil and aquatic environments (Jayaraman et al., 2023); while Aspergillus niger converts anthranilic acid into indole through a distinct route (Arora et al., 2015). These findings illustrate the metabolic diversity underlying indole biosynthesis across taxa.
Acetic acid is a ubiquitous organic acid synthesized by various microorganisms through distinct enzymatic pathways. In aerobic conditions, Acetobacter and Gluconobacter oxidize ethanol to acetic acid via alcohol dehydrogenase (ADH, EC: 1.1.1.1) and aldehyde dehydrogenase (ALDH, EC: 1.2.1.3) (Mamlouk and Gullo, 2013; Mientus et al., 2017). Anaerobic microbes such as methanogens and sulfate-reducing bacteria also generate acetic acid through the acetyl-CoA pathway (Ragsdale, 2008). Metagenomic analysis revealed the presence of ADH and ALDH genes in Enterobacter, Paracoccus, Aspergillus, Staphylococcus, and Yaniella. Among them, Enterobacter was enriched in YX3 and YX5, Aspergillus was found in all samples, and Staphylococcus appeared only in YX5. In contrast, YX1 and YX2 showed lower abundance of these enzyme-producing genera. The acetate-degrading enzyme, acetyl-CoA synthetase (EC: 6.2.1.1), was annotated in Corynebacterium, Brachybacterium, and Brevibacterium, consistent with their negative correlation with acetate levels.
Globally, Cuban cigars are renowned for their craftsmanship, premium tobacco, and intricate fermentation process (Viola et al., 2016). In recent years, however, the global cigar landscape has diversified, with Nicaragua, the Dominican Republic, and Honduras gaining recognition through regional strengths and process innovation. Yunnan cigars from China have also emerged as strong competitors, driven by advances in fermentation science, improved tobacco cultivation, and policy support (Shuoli et al., 2023). Comparative analysis of four Yunnan and two Cuban cigars revealed that certain Yunnan samples exhibited microbial abundances comparable to or exceeding those of Cuban cigars, likely due to Yunnan’s distinctive climate, mineral-rich and soil (Wu et al., 2024). Moreover, some Yunnan cigars demonstrated notable aroma diversity and higher VOC concentrations, indicating potential quality attributes comparable to Cuban cigars (Yang et al., 2022). Overall, cigar flavor and quality result from a complex interplay of factors including terroir, tobacco variety, microbial ecology, and fermentation practices.
In summary, this study provides preliminary insights into the microbial communities and VOC profiles associated with Yunnan cigars. These findings may contribute to a deeper understanding of how microbial and chemical diversity might influence cigar quality. As future research continues to investigate the complex interactions between microbial metabolism and aroma formation, additional factors that shape cigar characteristics may be uncovered. Notably, the observed microbial diversity and VOC richness in Yunnan cigar samples appear to be comparable to those reported in Cuban cigars, which might suggest that regional cigar profiles are influenced by more than just geographic origin. These results may offer a new perspective for exploring the microbial basis of product quality and could inform future strategies in cigar fermentation and development.
Statements
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: accession number: PRJNA 1261466.
Author contributions
JP: Formal analysis, Writing – original draft, Data curation, Visualization, Investigation. M-DH: Writing – original draft, Investigation, Formal analysis. JW: Validation, Writing – review & editing, Supervision. J-XZ: Investigation, Conceptualization, Writing – original draft. BY: Project administration, Writing – original draft. H-HY: Software, Writing – original draft, Project administration. J-SH: Project administration, Writing – original draft. Y-LS: Project administration, Data curation, Writing – review & editing. X-RS: Writing – original draft, Resources, Conceptualization. W-GW: Supervision, Writing – review & editing, Formal analysis, Funding acquisition. L-DB: Funding acquisition, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by grants from the Yunnan Daguan Laboratory (Grant no. YNDG202402XJ02) and the Yunnan Tobacco Company, Lincang Branch (Grant no. 2025530900242002).
Conflict of interest
JW was employed by China Tobacco Yunnan Industrial Co., Ltd. J-XZ, BY, H-HY, and J-SH, were employed by Yunnan Tobacco Company, Lincang Branch. Y-LS, X-RS, and W-GW were employed by Yunnan Tobacco Company, Yuxi Branch.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that Gen AI was used in the creation of this manuscript. We use AI to simply polish the language of the article, which is not used for the analysis of pictures and data.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1597501/full#supplementary-material
Footnotes
1.^https://github.com/OpenGene/fastp
2.^http://topaz.gatech.edu/GeneMark/
References
1
AllemJ.-P.UppuS. P.Boley CruzT.UngerJ. B. (2019). Characterizing swisher little cigar–related posts on twitter in 2018: text analysis. J. Med. Internet Res.21:e14398. doi: 10.2196/14398
2
Allied Market Research. (2023). Cigar and cigarillos market. Available online at: https://www.alliedmarketre-search.com/cigar-cigarillos-market-A15361
3
Aoki-KinoshitaK. F.KanehisaM. (2007). Gene annotation and pathway mapping in KEGG. Methods Mol. Biol.396, 71–91. doi: 10.1007/978-1-59745-515-2_6
4
AroraP. K.SharmaA.BaeH. (2015). Microbial degradation of indole and its derivatives. J. Chem.:129159. doi: 10.1155/2015/129159
5
BanožićM.JokićS.AčkarĐ.BlažićM.ŠubarićD. (2020). Carbohydrates-key players in tobacco aroma formation and quality determination. Molecules25:1734. doi: 10.3390/molecules25071734
6
BrandschR. (2006). Microbiology and biochemistry of nicotine degradation. Appl. Microbiol. Biotechnol.69, 493–498. doi: 10.1007/s00253-005-0226-0
7
ChattopadhyayS.MalayilL.MongodinE. F.SapkotaA. R. (2021). A roadmap from unknowns to knowns: advancing our understanding of the microbiomes of commercially available tobacco products. Appl. Microbiol. Biotechnol.105, 2633–2645. doi: 10.1007/s00253-021-11183-4
8
ChenQ. L.CaiL.WangH. C.CaiL. T.GoodwinP.MaJ.et al. (2020). Fungal composition and diversity of the tobacco leaf phyllosphere during curing of leaves. Front. Microbiol.11:554051. doi: 10.3389/fmicb.2020.554051
9
ChenS.LiJ.LinJ.BaoK.FanJ.ZhangR.et al. (2018). High-throughput sequencing fungal community structures in aging tobacco strips from different growing areas and stalk positions. Tob. Sci. Technol.51, 12–19. doi: 10.16135/J.ISSN1002-0861.2017.0270
10
ChenC.LiX.YangJ.GongX.LiB.ZhangK.-Q. (2008). Isolation of nicotine-degrading bacterium Pseudomonas sp. Nic22, and its potential application in tobacco processing. Int. Microbiol.62, 226–231. doi: 10.1016/j.ibiod.2008.01.012
11
ChenH.WuH.YanB.ZhaoH.LiuF.ZhangH.et al. (2018). Core microbiome of medicinal plant salvia miltiorrhiza seed: a rich reservoir of beneficial microbes for secondary metabolism?Int. J. Mol. Sci.19:672. doi: 10.3390/ijms19030672
12
ChopykJ.ChattopadhyayS.KulkarniP.SmythE. M.HittleL. E.PaulsonJ. N.et al. (2017). Temporal variations in cigarette tobacco bacterial community composition and tobacco-specific nitrosamine content are influenced by brand and storage conditions. Front. Microbiol.8:358. doi: 10.3389/fmicb.2017.00358
13
EnglishC. F.BellE. J.BergerA. J. (1967). Isolation of thermophiles from broadleaf tobacco and effect of pure culture inoculation on cigar aroma and mildness. Appl. Microbiol.15, 117–119. doi: 10.1128/am.15.1.117-119.1967
14
FuL.NiuB.ZhuZ.WuS.LiW. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics28, 3150–3152. doi: 10.1093/bioinformatics/bts565
15
HochS. O. N.CrawfordI. P. (1973). Enzymes of the tryptophan pathway in three Bacillus species. J. Bacteriol.116, 685–693. doi: 10.1128/jb.116.2.685-693.1973
16
HuangJ.YangJ.DuanY.GuW.GongX.ZheW.et al. (2010). Bacterial diversities on unaged and aging flue-cured tobacco leaves estimated by 16S rRNA sequence analysis. Appl. Microbiol. Biotechnol.88, 553–562. doi: 10.1007/s00253-010-2763-4
17
HuayiX.WenZ.HuD.WeiZ.CaiwenT.YuJ.et al. (2024). The effect of anaerobic fermentation on the aroma components of cigar tobacco leaves. Hans J. Agric. Sci.14, 1339–1347. doi: 10.12677/hjas.2024.1412168
18
JayaramanS.Adhilaxmi KannanM.RajendhranN.JohnG. J.RamasamyT. (2023). Indole-3-acetic acid impacts biofilm formation and virulence production of Pseudomonas aeruginosa. Biofouling39, 800–815. doi: 10.1080/08927014.2023.2269537
19
JingL.WenjunZ.XiaoliB.LijunW.RuminD.WangL.et al. (2022). Comparative analysis of comprehensive quality of different cigars at home and abroad. Southwest China J. Agric. Sci.35, 81–89. doi: 10.16213/j.cnki.scjas.2022.1.011
20
JuZ.ShaoJ.ZhouM.JinJ.PanH.DingP.et al. (2021). Transcriptomic and metabolomic profiling reveal the p53-dependent benzeneacetic acid attenuation of silica-induced epithelial–mesenchymal transition in human bronchial epithelial cells. Cell Biosci.11, 30–18. doi: 10.1186/s13578-021-00545-0
21
KarlssonF. H.FåkF.NookaewI.TremaroliV.FagerbergB.PetranovicD.et al. (2012). Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun.3:1245. doi: 10.1038/ncomms2266
22
KoszowskiB.ThannerM. H.PickworthW. B.TaylorK. M.HullL. C.SchroederM. J. (2018). Nicotine content and physical properties of large cigars and cigarillos in the United States. Nicotine Tob. Res.20, 393–398. doi: 10.1093/ntr/ntx054
23
KrüsemannE. J.VisserW. F.CremersJ. W.PenningsJ.TalhoutR. (2018). Identification of flavour additives in tobacco products to develop a flavour library. Tob. Control.27, 105–111. doi: 10.1136/tobaccocontrol-2016-052961
24
LeeteE. (1975). Biosynthesis of anatabine and anabasine in Nicotiana glutinosa. J. Chem. Soc. Chem. Commun.1, 9–10. doi: 10.1039/c39750000009
25
LiJ.JiaH.CaiX.ZhongH.FengQ.SunagawaS.et al. (2014). An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol.32, 834–841. doi: 10.1038/nbt.2942
26
LiuL.HuangW.LyuX.HEX.CHENL.LIY. (2020). Effect of long-term biogas slurry application on fungal community in rhizosphere soil at areca taro fields. Fujian J. Agric. Sci.35, 1034–1042. doi: 10.19303/j.issn.1008-0384.2020.09.015
27
LiuJ.MaG.ChenT.HouY.YangS.ZhangK. Q.et al. (2015). Nicotine-degrading microorganisms and their potential applications. Appl. Microbiol. Biotechnol.99, 3775–3785. doi: 10.1007/s00253-015-6525-1
28
LiuF.WuZ.ZhangX.XiG.ZhaoZ.LaiM.et al. (2021). Microbial community and metabolic function analysis of cigar tobacco leaves during fermentation. Microbiolopen10:e1171. doi: 10.1002/mbo3.1171
29
Maldonado-RobledoG.Rodriguez-BustamanteE.Sanchez-ContrerasA.Rodriguez-SanojaR.SanchezS. (2003). Production of tobacco aroma from lutein. Specific role of the microorganisms involved in the process. Appl. Microbiol. Biotechnol.62, 484–488. doi: 10.1007/s00253-003-1315-6
30
MamloukD.GulloM. (2013). Acetic acid bacteria: physiology and carbon sources oxidation. Indian J. Microbiol.53, 377–384. doi: 10.1007/s12088-013-0414-z
31
MientusM.KostnerD.PetersB.LieblW.EhrenreichA. (2017). Characterization of membrane-bound dehydrogenases of Gluconobacter oxydans 621H using a new system for their functional expression. Appl. Microbiol. Biotechnol.101, 3189–3200. doi: 10.1007/s00253-016-8069-4
32
MooreB. S.HertweckC.HopkeJ. N.IzumikawaM.KalaitzisJ. A.NilsenG.et al. (2002). Plant-like biosynthetic pathways in bacteria: from benzoic acid to chalcone. J. Nat. Prod.65, 1956–1962. doi: 10.1021/np020230m
33
MorrisD. S.FialaS. C. (2015). Flavoured, non-cigarette tobacco for sale in the USA: an inventory analysis of internet retailers. Tob. Control.24, 101–102. doi: 10.1136/tobaccocontrol-2013-051059
34
OhJ.ByrdA. L.DemingC.ConlanS.BarnabasB.BlakesleyR.et al. (2014). Biogeography and individuality shape function in the human skin metagenome. Nature514, 59–64. doi: 10.1038/nature13786
35
PanY.WuQ. Y.LIL. L.ShiY. Z.TanZ. Y.ZhangJ.et al. (2023). Application of polyphenol oxidase-producing strains in improving the quality of cigar tobacco leaves. J. Henan Agric. Sci.52, 173–180. doi: 10.15933/j.cnki.1004?3268.2023.09.018
36
PittardJ.CamakarisH.YangJ. (2005). The TyrR regulon. Mol. Microbiol.55, 16–26. doi: 10.1111/j.1365-2958.2004.04385.x
37
QinN.YangF.LiA.PriftiE.ChenY.ShaoL.et al. (2014). Alterations of the human gut microbiome in liver cirrhosis. Nature513, 59–64. doi: 10.1038/nature13568
38
RagsdaleS. W. (2008). Enzymology of the wood–ljungdahl pathway of acetogenesis. Ann. N. Y. Acad. Sci.1125, 129–136. doi: 10.1196/annals.1419.015
39
RenrenDoc. (2025). Market status, supply and demand analysis, and investment evaluation of the Chinese cigar industry (2025–2030). Available online at: https://www.renrendoc.com/paper/402556410.html
40
RomdhaneS.SporA.AubertJ.BruD.BreuilM.-C.HallinS.et al. (2022). Unraveling negative biotic interactions determining soil microbial community assembly and functioning. ISME J.16, 296–306. doi: 10.1038/s41396-021-01076-9
41
RuanA.MinH.PengX.HuangZ. (2005). Isolation and characterization of Pseudomonas sp. strain HF-1, capable of degrading nicotine. Res. Microbiol.156, 700–706. doi: 10.1016/j.resmic.2005.02.010
42
ShuoliW.SongshuangD.RonghaoW.LinlinL.ChuangW.JianW.et al. (2023). Difference and correlation analysis of mineral element contents and sensory qualities between Yunnan province of China and Nicaragua cigar tobacco leaves. Crop Mag.3, 139–147. doi: 10.16035/j.issn.1001-7283.2023.03.019
43
SiH.ZhouK.ZhaoT.CuiB.LiuF.ZhaoM. (2023). The bacterial succession and its role in flavor compounds formation during the fermentation of cigar tobacco leaves. Bioresour. Bioprocess.10:74. doi: 10.1186/s40643-023-00694-9
44
SimsG. K.SommersL. E.KonopkaA. (1986). Degradation of pyridine by Micrococcus luteus isolated from soil. Appl. Environ. Microbiol.51, 963–968. doi: 10.1128/aem.51.5.963-968.1986
45
SuY.CuiY.FuK.BuL.SunY.ZhouQ.et al. (2024). Contribution of pectin-degrading bacteria to the quality of cigar fermentation: an analysis based on microbial communities and physicochemical components. Front. Microbiol.15:1481158. doi: 10.3389/fmicb.2024.1481158
46
SuC.GuW.ZheW.ZhangK. Q.DuanY.YangJ. (2011). Diversity and phylogeny of bacteria on Zimbabwe tobacco leaves estimated by 16S rRNA sequence analysis. Appl. Microbiol. Biotechnol.92, 1033–1044. doi: 10.1007/s00253-011-3367-3
47
SunW.LiY.McGuinnessL. R.LuoS.HuangW.KerkhofL. J.et al. (2015). Identification of anaerobic aniline-degrading bacteria at a contaminated industrial site. Environ. Sci. Technol.49, 11079–11088. doi: 10.1021/acs.est.5b02166
48
SunagawaS.CoelhoL. P.ChaffronS.KultimaJ. R.LabadieK.SalazarG.et al. (2015). Ocean plankton. Structure and function of the global ocean microbiome. Science348:1261359. doi: 10.1126/science.1261359
49
TangZ.ChenL.ChenZ.FuY.SunX.WangB.et al. (2020). Climatic factors determine the yield and quality of Honghe flue-cured tobacco. Sci. Rep.10:19868. doi: 10.1038/s41598-020-76919-0
50
TangJ.LiY.ZhangL.MuJ.JiangY.FuH.et al. (2023). Biosynthetic pathways and functions of indole-3-acetic acid in microorganisms. Microorganisms11:2077. doi: 10.3390/microorganisms11082077
51
VillarE.FarrantG. K.FollowsM.GarczarekL.SpeichS.AudicS.et al. (2015). Ocean plankton. Environmental characteristics of Agulhas rings affect interocean plankton transport. Science348:1261447. doi: 10.1126/science.1261447
52
VillemurR.LacasseM.MorinA. (2009). Monitoring the bacterial and fungal biota of eleven tobacco grades stored at three different locations. Contrib. Tob. Nicotine Res.23, 368–376. doi: 10.2478/cttr-2013-0871
53
ViolaA. S.GiovencoD. P.Miller LoE. J.DelnevoC. D. (2016). A cigar by any other name would taste as sweet. Tob. Control.25, 605–606. doi: 10.1136/tobaccocontrol-2015-052518
54
WangH.GuoD.ZhangM.WuG.ShiY.ZhouJ.et al. (2024). Correlation study on microbial communities and volatile flavor compounds in cigar tobacco leaves of diverse origins. Appl. Microbiol. Biotechnol.108:236. doi: 10.1007/s00253-024-13032-6
55
WatanabeT.SnellE. E. (1972). Reversibility of the tryptophanase reaction: synthesis of tryptophan from indole, pyruvate, and ammonia. Proc. Natl. Acad. Sci.69, 1086–1090. doi: 10.1073/pnas.69.5.1086
56
WuX.CaiW.ZhuP.PengZ.ZhengT.LiD.et al. (2022). Profiling the role of microorganisms in quality improvement of the aged flue-cured tobacco. BMC Microbiol.22:197. doi: 10.1186/s12866-022-02597-9
57
WuQ.PengZ.PanY.LiuL.LiL.ZhangJ.et al. (2023). Interaction analysis of tobacco leaf microbial community structure and volatiles flavor compounds during cigar stacking fermentation. Front. Microbiol.14:1168122. doi: 10.3389/fmicb.2023.1168122
58
WuY.ZhangH.WangW.KongG.LiZ.ZhangT.et al. (2024). Characterization of volatile organic compounds and aroma sensory properties in Yunnan cigar. J Anal Methods Chem.2024:9583022. doi: 10.1155/2024/9583022
59
XingL.YangJ.JiaY.HuX.LiuY.XuH.et al. (2021). Effects of ecological environment and host genotype on the phyllosphere bacterial communities of cigar tobacco (Nicotiana tabacum L.). Ecol. Evol.11, 10892–10903. doi: 10.1002/ece3.7861
60
XuY.MinhazulK. A.WangX.LiuX.LiX.MengQ.et al. (2020). Biodegradation of phthalate esters by Paracoccus kondratievae BJQ0001 isolated from Jiuqu (baijiu fermentation starter) and identification of the ester bond hydrolysis enzyme. Environ. Pollut.263:114506. doi: 10.1016/j.envpol.2020.114506
61
XueruS.LimengZ.LingduoB.ZhihuaH.XingQ.WenhuaC.et al. (2022). Study on the differences in microbial communities of aged cigar tobacco leaves from different regions in Yunnan province. Chin. Tob. Sci.43, 87–93. doi: 10.13496/j.issn.1007-5119.2022.05.014
62
YanT.ZhouP.LongF.LiuJ.WuF.ZhangM.et al. (2022). Unraveling the difference in the composition/content of the aroma compounds in different tobacco leaves: for better use. Chem. Med. Chem.2022, 1–10. doi: 10.1155/2022/3293899
63
YangX. Y.BarnettJ. R.YangT. (2022). Smoking environments in China: challenges for tobacco control. Berlin: Springer.
64
YaoL.ZhangT.ShanY.YangJ.YuJ.YangC.et al. (2024). The impact of fangxian huangjiu on the fermentation quality and microbial community dynamics of cigar wrapper leaves. Front. Bioeng. Biotechnol.12:1428750. doi: 10.3389/fbioe.2024.1428750
65
YeJ.DingY.QiX.XuJ.YangX.ZhangZ. (2021). Geographic and position-based variations in phyllospheric bacterial communities present on flue-cured tobacco. Appl. Microbiol. Biotechnol.105, 9297–9308. doi: 10.1007/s00253-021-11671-7
66
YeX. T.HuY.LiuQ.LuX. Y.LiuC. (2024). Characteristics of soil microbial communities in typical temperate glacial debris and retreat zones: a case study of the Azha and Midui glaciers. China Environ. Sci.44, 5108–5121. doi: 10.19674/j.cnki.issn1000-6923.2024.0175
67
YeJ.YanJ.ZhangZ.YangZ.LiuX.ZhouH.et al. (2017). The effects of threshing and redrying on bacterial communities that inhabit the surface of tobacco leaves. Appl. Microbiol. Biotechnol.101, 4279–4287. doi: 10.1007/s00253-017-8143-6
68
YiL.AnqiC.YingX.DanX.ChenW. (2024). Research progress on the prevalence and health hazards of cigar. Chin. J. Health Manag.18, 553–557. doi: 10.3760/cma.j.cn115624-20240314-00205
69
ZengL.ZhouY.GuiJ.FuX.MeiX.ZhenY.et al. (2016). Formation of volatile tea constituent indole during the oolong tea manufacturing process. J. Agric. Food Chem.64, 5011–5019. doi: 10.1021/acs.jafc.6b01742
70
ZhangQ.ChenX.DingY.KeZ.ZhouX.ZhangJ. (2021). Diversity and succession of the microbial community and its correlation with lipid oxidation in dry-cured black carp (Mylopharyngodon piceus) during storage. Food Microbiol.98:103686. doi: 10.1016/j.fm.2020.103686
71
ZhangM.GuoD.WangH.WuG.ShiY.ZhouJ.et al. (2024). Analyzing microbial community and volatile compound profiles in the fermentation of cigar tobacco leaves. Appl. Microbiol. Biotechnol.108:243. doi: 10.1007/s00253-024-13043-3
72
ZhangG.HeY.YangW.LiuZ.LinZ.ZhangT.et al. (2025a). Integrated microbiology and metabolomics analysis reveal the fermentation process and the flavor development in cigar tobacco leaf. Microbiol. Spectr.:e01029-24. doi: 10.1128/spectrum.01029-24
73
ZhangL.LiW.PengZ.ZhangJ. (2025b). Effect of microbial community on the formation of flavor components in cigar tobacco leaves during air-curing. BMC Microbiol.25:56. doi: 10.1186/s12866-025-03774-2
74
ZhangG.ZhaoL.LiW.YaoH.LuC.ZhaoG.et al. (2023). Changes in physicochemical properties and microbial community succession during leaf stacking fermentation. AMB Express13:132. doi: 10.1186/s13568-023-01642-8
75
ZhaoJ.LiQ.SunT.ZhuX.XuH.TangJ.et al. (2013). Engineering central metabolic modules of Escherichia coli for improving β-carotene production. Metab. Eng.17, 42–50. doi: 10.1016/j.ymben.2013.02.002
76
ZhaoJ.OuyangS.QiH.MaK.HuX.WangG.et al. (2024). Metabolomics and transcriptomics uncover the pectin hydrolysis during tobacco stem fermentation by Aspergillus niger. J. Clean. Prod.442:141005. doi: 10.1016/j.jclepro.2024.141005
77
ZhengT.ZhangQ.LiP.WuX.LiuY.YangZ.et al. (2022). Analysis of microbial community, volatile flavor compounds, and flavor of cigar tobacco leaves from different regions. Front. Microbiol.13:907270. doi: 10.3389/fmicb.2022.907270
78
ZhengfengL.TianQ.YuzhenX.LeZ.LuopingW.KaiD.et al. (2024). Similarity in the microbial community structure of tobacco from geographically similar regions. Sci. Rep.14:30933. doi: 10.1038/s41598-024-81565-x
79
ZhouJ.YuL.ZhangJ.LiuJ.ZouX. (2021). Dynamic characteristics and co-occurrence patterns of microbial community in tobacco leaves during the 24-month aging process. Ann. Microbiol.71:9. doi: 10.1186/s13213-021-01620-0
80
ZuoH.LvZ.GuoS.ZhouY.ShenC. (2024). Mixed processing improved the theaflavin, theanine and aroma components of congou black tea. Int. J. Food Sci. Technol.59, 8011–8026. doi: 10.1111/ijfs.17102
Summary
Keywords
cigar, metagenomic, volatile organic compounds, microbial community, microbial metabolism, microbiome
Citation
Pan J, Huang M-D, Wang J, Zhao J-X, Yang B, Yang H-H, Huang J-S, Su Y-L, Song X-R, Wang W-G and Bu L-D (2025) Unraveling the microbiome–aroma Nexus: a metagenomic and volatile compound analysis of Yunnan cigars. Front. Microbiol. 16:1597501. doi: 10.3389/fmicb.2025.1597501
Received
21 March 2025
Accepted
25 June 2025
Published
09 July 2025
Volume
16 - 2025
Edited by
Iftikhar Ahmed, National Agricultural Research Center, Pakistan
Reviewed by
Kiran Khandagale, Directorate of Onion and Garlic Research (ICAR), India
Ashok Gehlot, Institute of Himalayan Bioresource Technology (CSIR), India
Vikas Dadwal, Texas A and M University, United States
Updates
Copyright
© 2025 Pan, Huang, Wang, Zhao, Yang, Yang, Huang, Su, Song, Wang and Bu.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Wang, wangjin@iccas.ac.cn; Ling-Duo Bu, bulingduo@163.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.