- 1Centre for Applied Food Safety and Biotechnology, Department of Life Sciences, Central University of Technology, Bloemfontein, South Africa
- 2Unit for Environmental Sciences and Management, North-West University, Potchefstroom, South Africa
- 3Department of Life and Consumer Sciences, University of South Africa, Johannesburg, South Africa
- 4Thoracic Diseases Research Unit, Departments of Medicine and Biochemistry, Mayo Clinic College of Medicine, Rochester, MN, United States
Introduction: Carbapenem-resistant Pseudomonas aeruginosa (CRPA) represents a global threat, but the global distribution of carbapenem resistant bacteria remains a critical issue in public health.
Methods: We conducted a systematic review and meta-analysis on the global pooled prevalence estimate (PPE) of CRPA and their antibiotic resistance. The systematic review protocol was registered with PROSPERO (CRD42024579654). This study was carried out following the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines. Heterogeneity between studies was assessed using Cochrane Q test and I2 test statistics based on the random effects model. Comprehensive meta-analysis software v4.0 was used to analyze the pooled prevalence of CRPA.
Results: A total of 163 studies (both clinical and screening samples) containing a total of 58,344 cases from 39 countries were included in this study. The overall PPE of CRPA was 34.7% (95% CI: 0.316–0.37.8) for both clinical and screening samples. Meropenem had a PPE of 31.2% (95% CI: 0.272–0.352) and imipenem had the lowest PPE of 27.7% (95% CI: 0.238–0.319). Japan had the highest PPE at 98.2% (95% CI: 0.482–0.100) of CRPA, and the lowest was observed for Saudi Arabia at 13.9% (95% CI: 0. 064–0. 277). CRPA is widespread on five continents except Australia and Antarctica, while the highest PPE is in Europe at 47.6% (95% CI: 0.359–0.595) and the lowest in Asia at 32, 8% (95% CI: 0.293–0.364). The relatively higher PPE of CRPA was observed in Europe during the year interval 2014–2017 at 95.4% (95% CI: 0.388–0.999), followed by Africa from the year 2022–2024 with 38.5% (95% CI: 0.243–0.550). Ceftazidime was significantly higher in studies conducted before 2019 with a PPE of 44.7% (95% CI: 0.246% – 0.668), while CRPA after 2019 had a higher resistance to cefoperazone/sulbactam with a PPE of 17.3% (95% CI: 0.050–0.455).
Discussion: This review indicates that the prevalence of CRPA is generally high and varies significantly between countries. To prevent the emergence of CRPA and antibiotic resistance, future initiatives should prioritise strengthening laboratory capacity for early detection of antibiotic resistance.
1 Introduction
Pseudomonas aeruginosa is a Gram-negative, rod-shaped bacterium that belongs to the Pseudomonadaceae family (Marchaim et al., 2012; Abdalhamid et al., 2016). It is an opportunistic human pathogen that can cause a variety of infections in the blood, skin, respiratory tract, and urinary tract (Golle et al., 2017; Narimisa et al., 2024) and is common for healthcare-associated infections (HAIs) (Büchler et al., 2023). Antimicrobial resistance (AMR) is one of the greatest threats to public health worldwide, leading to significant increases in morbidity, mortality, and treatment failure for microbial infections, as well as economic losses to individuals and nations. The primary mechanisms by which resistance is mediated in Gram-negative bacteria are ampC-lactamases, carbapenemases, and extended spectrum lactamases (ESBL) (Schill et al., 2017; Marouf et al., 2023). Carbapenems are effective drugs against bacterial pathogens and resistance to them is considered a major threat to public health, especially among notorious nosocomial pathogens such as P. aeruginosa (Arowolo et al., 2023). Carbapenemases are categorized into three main classes: Class A (Klebsiella pneumoniae carbapenemase, KPC), Class B (metallo-β-lactamases, including NDM, VIM, and IMP), and Class D (oxacillinases, such as OXA-48-like carbapenemases) (Sheu et al., 2019).
The management of infections caused by this bacterium has become increasingly challenging due to the continued rise in antibiotic resistance and, more recently, the emergence of multidrug-resistant strains (Odoi et al., 2021; Deshwal et al., 2023). Carbapenems are used to treat infections caused by multidrug-resistant Gram-negative bacteria, but the selective pressure imposed by their use has contributed to the emergence of carbapenem-resistant strains (Rai et al., 2014; Sheu et al., 2019; Abubakar et al., 2022). Several studies reported antibiotic resistance of P. aeruginosa, including AMR against carbapenem (Çopur et al., 2021; Souza et al., 2021; Canton et al., 2022; Saha et al., 2022; Męcik et al., 2024; Pappa et al., 2024; Shahab et al., 2024; Yano et al., 2024). Treatment options can be severely restricted when bacteria develop resistance to carbapenem antibiotics, which are generally reserved for the treatment of multidrug-resistant bacterial infections (Deshwal et al., 2023).
Several systematic reviews of carbapenem-resistant CRPA worldwide have been published including study on carbapenem resistance in Acinetobacter baumannii and P. aeruginosa in sub-Saharan Africa, Carbapenemase-producing non-glucose-fermenting Gram-negative Bacilli in Africa, P. aeruginosa and Acinetobacter baumannii (Kindu et al., 2020), outbreak investigations after identifying CRPA (Büchler et al., 2023), and prevalence of meropenem-resistant P. aeruginosa in Ethiopia (Gobezie et al., 2024). Though there is limited information on comprehensive data available to estimate the global prevalence of CRPA and their antibiotic resistance. This systematic review and meta-analysis aimed to provide a comprehensive overview of the global prevalence of CRPA and their antibiotic resistance patterns, based on peer-reviewed published global data.
2 Materials and methods
2.1 Protocol registration
This study was registered in the International Prospective Register of Systematic Reviews (PROSPERO) with registration no: CRD42024579654.
2.2 Systematic review protocol
The Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines were used in the data extraction, screening, and analysis process, which have been confirmed on a checklist (Supplementary Table S1). This included searching database systems for potentially relevant articles, assessing their suitability for review and determining their relevance.
2.3 Search strategy
This study used several database systems, including Web of Science (https://lnkd.in/gAdNu8iz/ 03-04April/2024), Scopus (https://www.scopus.com/ 06/April/2024) Google Scholar (https://scholar.google.com/ 08-16/April/2024) ScienceDirect (https://lnkd.in/gBrgWFQ2/ 20-22/April/2024) PubMed (https://lnkd.in/gABzu4AD/ 22-25/April/2024) and EMBASE (https://www.embase.com/ 01-02/April/2024). The search strategy was not limited by language. Literature searches were conducted from 2014 to May, 2, 2024, using keywords comprising of “epidemiology,” “carbapenem,” “meropenem,” “imipenem,” “antibiotics resistant,” “Pseudomonas,” “multidrug resistance,” “VIM,” “IMP,” carbapenem-resistant Pseudomonas aeruginosa, “CRPA,” antibiotic resistance, “nosocomial,” “infections,” “health care related,” meropenem resistant, “antibiotics,” “antimicrobials,” “hospital setting.” Certain “MeSH” terms were used to find articles relevant to the study. The search strategy is detailed in Supplementary Table S2. The last search took place on 02 May 2024. No attempt was made to obtain further information or retrieve unpublished studies from the authors of the original manuscripts.
2.4 Study selection
The three authors TR, GK, and JN assessed the suitability of journal titles and abstracts for the inclusion and exclusion criteria. Two reviewers independently examined each study found through the search, focusing on the title, abstract, and selected full text. A third reviewer clarified any discrepancies. Chapter books, reviews, and conference abstracts were excluded. The full text of only English-language journal articles was incorporated. We screened titles and abstracts, then retrieved and downloaded relevant full-text articles via library resources and online databases. Full-text reviews of journal articles examining carbapenem-resistant P. aeruginosa and their resistance to antibiotics were selected.
2.5 Inclusion and exclusion criteria
Criteria for inclusion of studies were: (i) studies investigating the prevalence of carbapenem-resistant P. aeruginosa and their antibiotic resistance in humans. The following exclusion criteria were used, (ii) no total number of isolates, (iii) no abstracts, reviews, experiments, thesis, preprints and book chapters, and (iv) an unclear number of carbapenem-resistant P. aeruginosa and their antibiotic resistance.
2.6 Data extraction
To determine eligibility, full versions of potentially relevant articles were obtained by two authors independently (TR and GK) from the final selected studies. Data from each article were independently compiled and entered into a spreadsheet, including author names, publication year, location, total number of isolates, and total number of samples collected, which were entered into a Microsoft Excel for further analysis. Text, tables, and figures were used to extract data.
2.7 Quality appraisal
To confirm the methodological soundness of the research articles selected for quantitative synthesis, two authors independently used the Joanna Briggs Institute (JBI) Critical Appraisal Tools Checklist 2017 review guideline for prevalence studies (Ramatla et al., 2022). Studies that achieved a score of five or higher for the evaluation criteria were included.
2.8 Meta-analysis
Only journal articles that specifically addressed the antibiotic resistance of CRPA were included in the meta-analysis. We performed the meta-analysis using the Comprehensive Meta-Analysis (CMA) program Software v.4.0.1 The random effects model, equipped with 95% confidence intervals (CI), produced pooled estimates (PP) by using the reciprocal of the sample variance and a constant variable across the population effects to weigh each study. To assess Cochran heterogeneity (Q) within studies and percentage variation in prevalence, Higgins I2 (inverse variance) and the Cochran Q method were used. One can define low, moderate, or high heterogeneity as I2 values of less than 25, 50%, and more than 75%, respectively, while values close to 0% indicate no heterogeneity. A random effects model was used to create all pooled estimates. Statistical significance was determined by considering heterogeneity with a p-value less than 0.05 (p < 0.05). The p-values correspond to the heterogeneities between studies from a Chi-squared test of the null hypothesis that there is no heterogeneity. A subgroup analysis of study results was performed based on carbapenem, meropenem, imipenem, antibiotic resistance, countries, continents, published year, and carbapenem-resistant P. aeruginosa, and their antibiotic resistance. As part of the meta-analysis, subgroup analyzes that contained studies with fewer than three studies were excluded.
2.9 Publication bias
Publication bias was determined using an inverted funnel plot, Egger’s and Begg’s bias indicator tests, and the visual eye test. The Begg-Mazumdar bias indicator test was used to study the impact of publishing and selection bias.
3 Results
3.1 Search and screening results
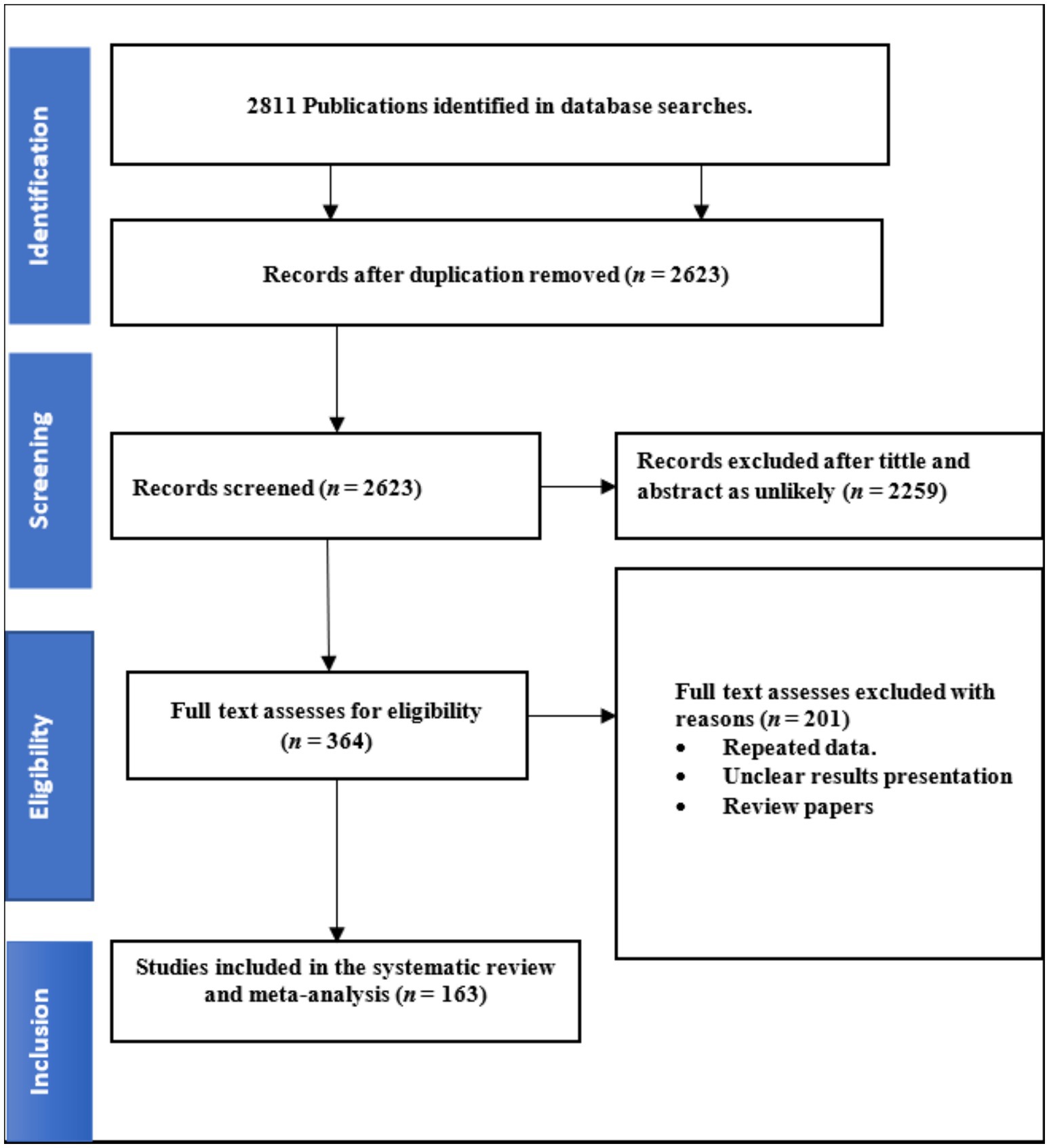
A total of 2,811 studies were retrieved after the initial search was conducted across four databases (Scopus, PubMed, ScienceDirect, EMBASE, Google Scholar and Web of Science), subsequently removing 188 duplicate articles. After removing duplicates and reviewing study titles and abstracts, 2,623 articles were excluded from further consideration. A total of 364 studies were initially considered eligible and were thus subjected to full text evaluation; thereafter, 163 studies were eligible for inclusion (Figure 1). The Joanna Briggs Institute (JBI) Critical Review Quality Assessment score ranges from 1 to 9. All 163 studies included in our analysis received a score of five or higher.
3.2 Characteristics of eligible studies
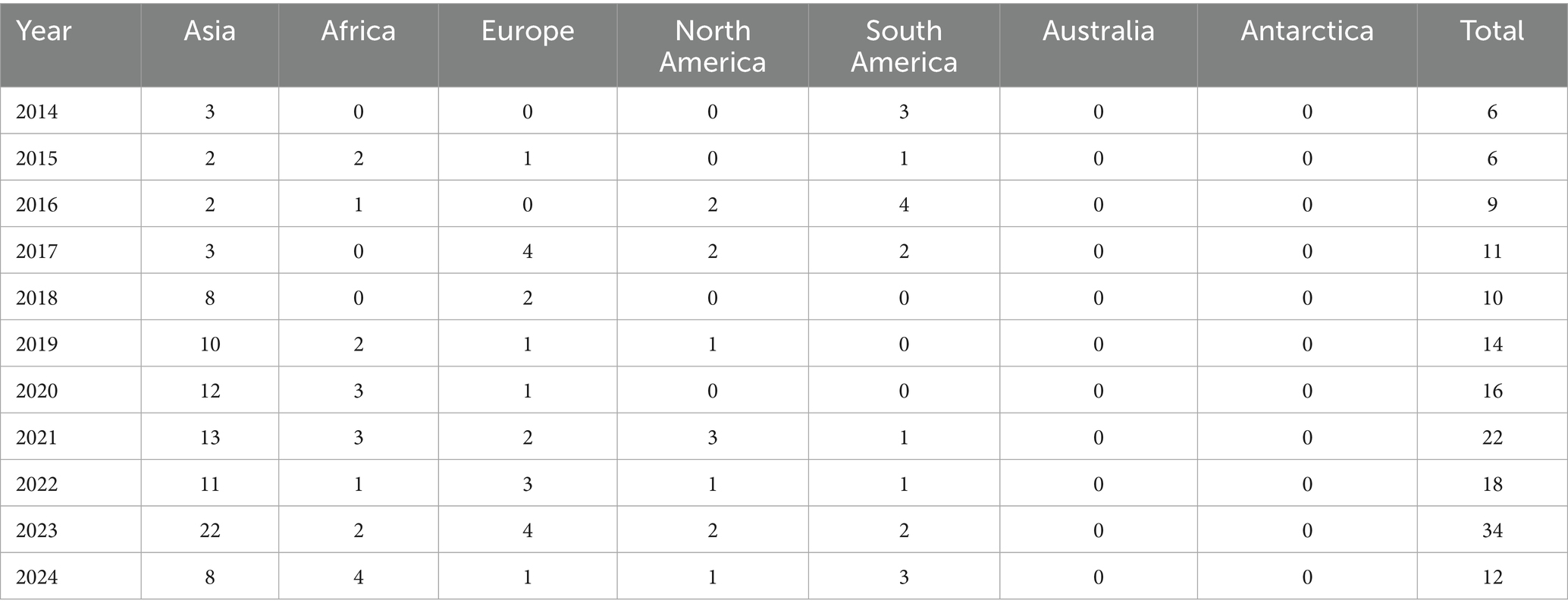
All included studies were published from 2014 to 2024, with most studies conducted between 2021 and 2024. Table 1 shows the annual distribution of published articles from all continents. Of the five continents, Asia had the highest number of published studies (n = 95), followed by Europe (n = 19), South America (n = 16), Africa (n = 15), and very few came from North America (n = 13). A total of 5 (3.06%) studies were conducted from more than one country/continent. The number of CRPA isolates confirmed worldwide ranged from 1 to 9,750 isolates. Numerous studies have examined the expression of antibiotic resistance genes for the carbapenem class, including blaIVIM 45 (25.6%), blaIIMP 22 (12.5%), blaINDM 18 (10.2%), blaIKPC 17 (9.7%), blaISPM 7 (9.7%), blaoxa48 7 (9.7%), blaISPM1 5 (2.8%), blaISIM 4 (2.3%), and blaIGIM 4(2.3%). Most CRPA isolates showed resistance to amikacin (n = 28; 15.9%), ceftazidime (n = 26; 14.8%), gentamicin (n = 25; 14.2%), cefepime (no = 25; 1.2%), ciprofloxacin (no = 24;1.2%) and Piperacillin-tazobactam (no = 20; 11.4%).
3.3 Meta-analysis results on overall prevalence
The global PPE of CRPA, as well as a summary of the subgroup analysis, are shown in Table 2. A total of 291,715 isolates were confirmed as CRPA, while 19,858 were confirmed to be CRPA multiple drug resistant (CRPA-MDR) isolates. Studies examining the prevalence of CRPA and their antibiotics in humans have found high heterogeneity based on methods, countries, continents, years and antibiotic resistance profiles (Tables 2, 3). The PPE of CRPA in the present study was 34.7% (0.347; 95% CI: 0.316–0.378, I2 = 99.2%) from 163 studies. Among studies reporting meropenem resistance, the PPE was 27.7% (0.277; 95% CI: 0.238–0.319, I2 = 98.7%), while imipenem had the least PPE of 31.2% (0.312; 95% CI: 0.272–0.352, I2 = 99.2%).
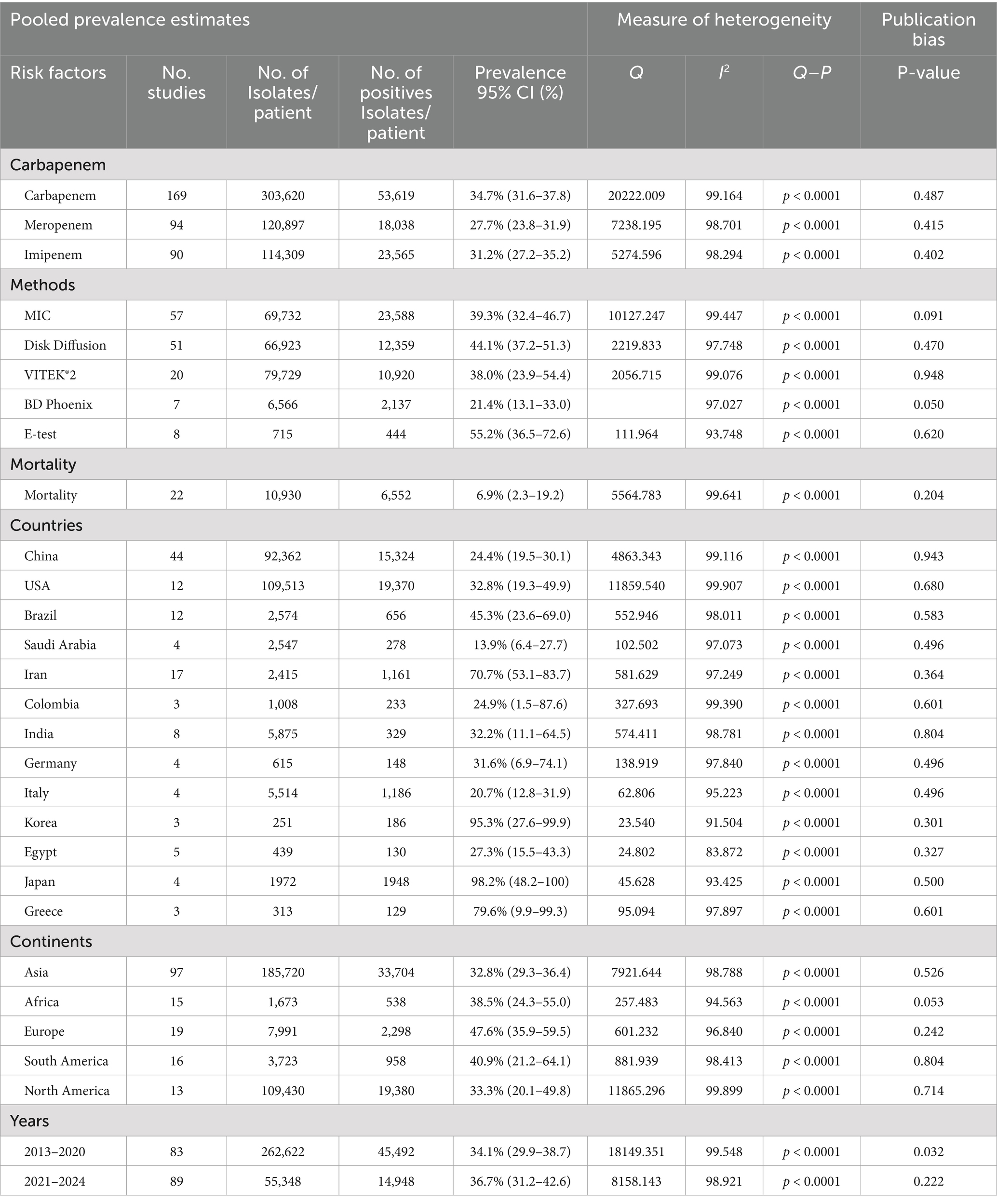
Table 2. Subgroup analysis of carbapenem-resistant Pseudomonas aeruginosa reported from 2014 to 2024.
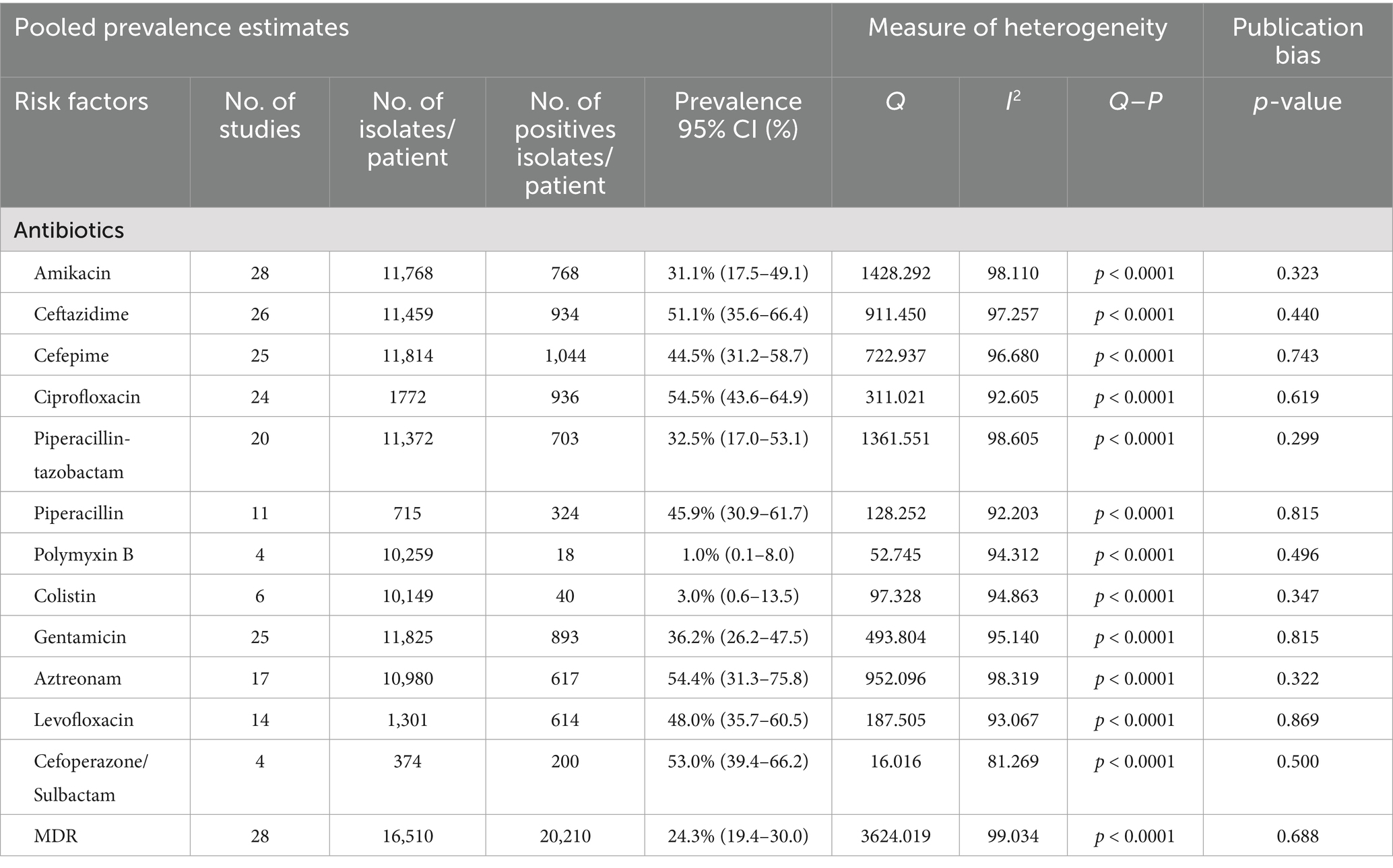
Table 3. Pooled prevalence estimates and 95% CI of antibiotic resistance profiles of carbapenem-resistant Pseudomonas aeruginosa.
3.3.1 Prevalence of carbapenem resistant Pseudomonas aeruginosa
The prevalence data in subgroups classified by the countries showed that studies from Japan registered the highest PPE of CRPA at 98.2% (0. 982; 95% CI: 0.482–0.100, Tau = 15.511, I2 = 93.4%), while Saudi Arabia had the lowest at 13.9% (0. 139; 95% CI: 0. 064–0. 277, Tau = 1.644, I2 = 91.1%) (Table 2). However, other countries were not included in the meta-analysis due to the small number of studies. Based on the continental distribution, which included only five of the seven continents, the highest prevalence of CRPA was reported in Europe with a PPE of 47.6% (0.476; 95% CI: 0.359–0.595, Tau = 0.779, I2 = 96.8%), followed by South America with a PPE of 40.9% (0.409; 95% CI: 0.212–0.641, Tau = 1.801, I2 = 98.4%) (Table 2). The CRPA PPE was below 40% on three continents: Africa 38.5% (0.385; 95% CI: 0.243–0.550, Tau = 0.656, I2 = 94.5%), North America 33.3% (0.333; 95% CI: 0.201–0.498, Tau = 0.777, I2 = 99.8%) and Asia 32.8% (0.328; 95% CI: 0.293–0.364, Tau = 1.016, I2 = 98.7%). Furthermore, a relatively higher PPE of CRPA was observed in Europe during the year interval 2014–2017 at 95.4% (0.954; 95% CI: 0.388–0.999, Tau = 14.677, I2 = 97.6%), followed by Africa from the year 2022–2024 with 87.5% (0.875; 95% CI: 0.449–0.636, Tau = 1.593, I2 = 89.6%) while Asia had a PPE of 54.4% (0.544; 95% CI: 0.449–0.636, Tau = 1.370, I2 = 97.6%) in 2018–2021(Table 2).
When it came to tests for antibiotic resistance, the E-test had the highest PPE at 55.2% (0.552; 95% CI: 0.65–0.726, Tau = 1.001, I2 = 93.7%), followed by MIC with a PPE of 39.3% (0.393; 95% CI: 0.324–0.46.7, Tau = 1.167, I2 = 99.4%), disk diffusion with a PPE of 44.1% (0.441; 95% CI: 0.372–0.513, Tau = 0.904, I2 = 97.7%), VITEK®2 with a PPE of 38.0% (0.380; 95% CI: 0. 239–0.544, Tau = 1.440, I2 = 99.1%), and lastly, PPE of 21.4% (0.214; 95% CI: 0.131–0.330, Tau = 0.487, I2 = 97.0%) for BD Phoenix.
3.3.2 Subgroup analysis by carbapenem-resistant Pseudomonas aeruginosa
We analyzed the antibiotic resistance of CRPA and 34 of the 163 studies reported resistance of carbapenem-resistant P. aeruginosa (Table 3). A subgroup study of antibiotic resistance found that the highest PPE of antibiotic resistance was against ciprofloxacin with 54.5% (0.545; 95% CI: 0.436–0.649, I2 = 92.6%) followed by aztreonam 54.4% (0.544; 95% CI: 0.313–758, I2 = 98.3%), cefoperazone/sulbactam 53.0% (0.530; 95% CI: 0.394–662, I2 = 81.3%), ceftazidime 51.1% (0.511; 95% CI: 0.356–0.664, I2 = 97.3%), levofloxacin 48.0% (0.480; 95% CI: 0.357–0.605, I2 = 93.1%), piperacillin 45.9% (0.459; 95% CI: 0.309–0.617, I2 = 92.2%), cefepime 44.5% (0.445; 95% CI: 0.312–0.587, I2 = 96.7%), gentamicin 36.2% (0.632; 95% CI; 0.262–0.475, I2 = 95.1%), piperacillin-tazobactam 32.5% (0.325; 95% CI; 0.170–0.531, I2 = 98.6%), amikacin 31.1% (0.311; 95% CI; 0.175–0.491, I2 = 98.1%), colistin 3.0% (0.003; 95% CI; 0.003–0.135, I2 = 94.9%), polymyxin B 1.0% (0.010; 95% CI; 0.0.001–0.080, I2 = 94.3%). The CRPA resistance rates in different continents are described in detail in Supplementary Table S3.
We conducted a meta-analysis of these antibiotics and compared antimicrobial resistance rates for CRPA before and after 2019. The rate of CRPA antimicrobial resistance to ceftazidime with PPE was 44.7% (0.447; 95% CI, 0.246–0.668%, I2 = 95.5) and was significantly higher in studies conducted before 2019, followed by ciprofloxacin with PPE at 41.2% (0.412; 95% CI, 0.256–0.588, I2 = 94.8). However, after 2019, CRPA showed significantly higher resistance to cefoperazone/sulbactam with a PPE of 17.3% (0.173; 95% CI, 0.050–0.455, I2 = 96.2) (Supplementary Table S4).
3.3.3 Subgroup analysis by carbapenem-resistant Pseudomonas aeruginosa-multidrug resistance
The study also analyzes the MDR of 28 of the 163 papers reporting on CRPA (Figure 2). Africa recorded the highest PPE at 63.2% (0.632; 95% CI; 0.211–0.917, Tau = 3.168, I2 = 96.9%). Followed by Asia at 28.4% (0.284; 95% CI; 0.151–0.469, Tau = 2.709, I2 = 98.9%) and finally North America with PPE of 17.7% (0.177; 95% CI; 0.106–0.280, Tau = 0.350, I2 = 99.7%). However, South America and Europe were not included in the analysis due to the small number of studies.
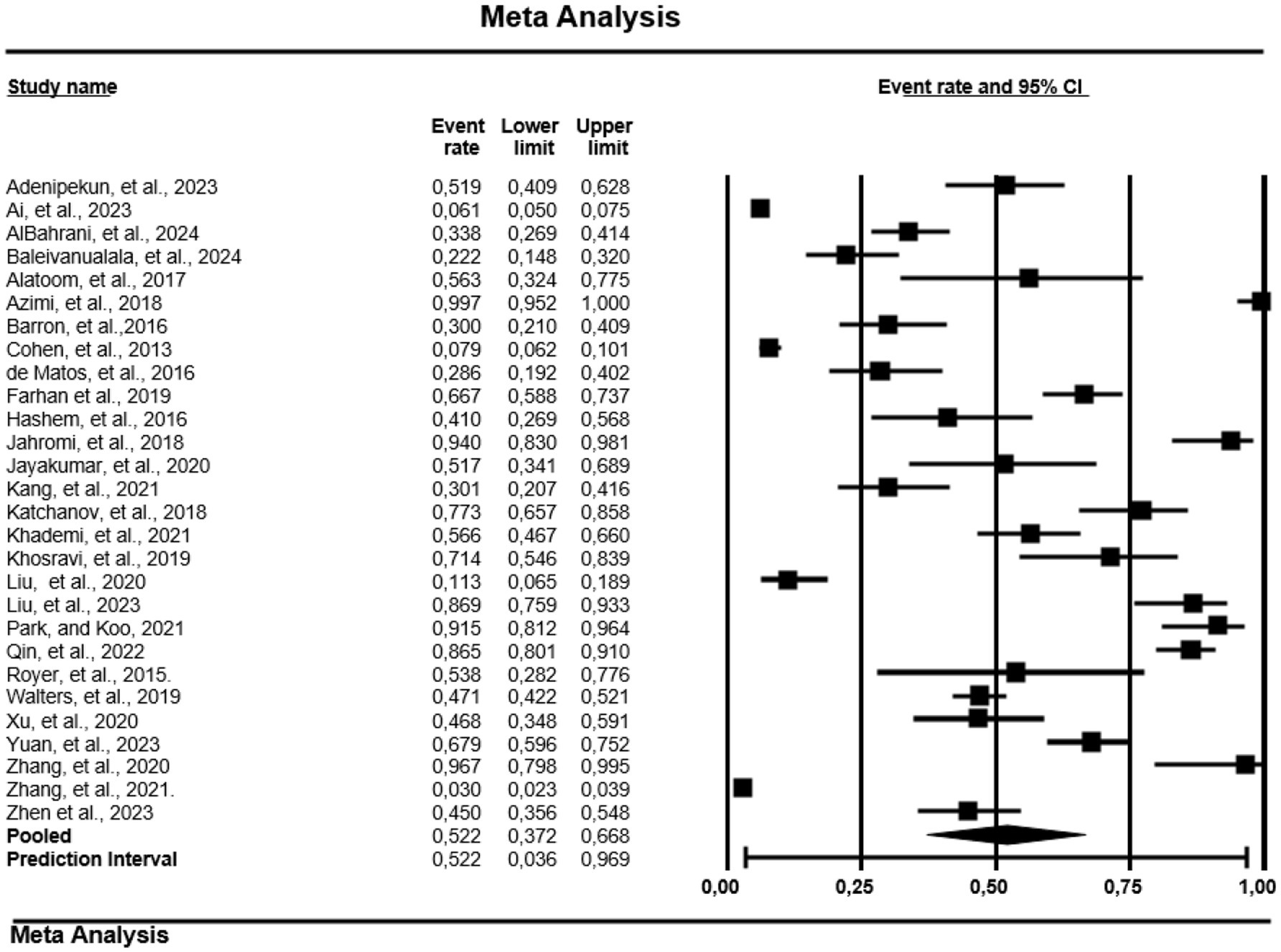
Figure 2. Forest plot showing the pooled estimates of multidrug resistance of carbapenem-resistant-P. aeruginosa globally. The diamond at the base indicates the pooled estimates from the overall studies.
3.3.4 Subgroup analysis by carbapenem-resistant Pseudomonas aeruginosa genes
The PPE of antibiotic resistance genes for CRPA (Table 4) considered in this meta-analysis were as follows: blaGIM with high PPE of 22.3% (0.223; 95% CI: 0.056–0.583, I2 = 94.2%), blaSPM 21.9% (0.219; 95% CI: 0.067–0.525, I2 = 94.3%), blaVIM 21.3% (0.213; 95% CI: 0.144–0.302, I2 = 96.6%), blaSPM1 19.9% (0.199; 95% CI: 0.088–0.390, I2 = 83.5%), blaSIM 15.9% (0.159; 95% CI: 0.041–0.455, I2 = 62.5%), blaIMP 14.2% (0.142; 95% CI: 0.078–0.243, I2 = 95.2%), blaNDM 8.4% (0.084; 95% CI: 0.038–0.177, I2 = 94.7%), and blaKPC 7.5% (0.075; 95% CI: 0.049–0.114, I2 = 88.5%). However, due to a low number of studies, genes for cefametazole, ceftriaxone, tobramycin, minocycline, ertapenem, carbenicillin, doripenem, tobramycin, and cefotaxime were not included in the analysis.
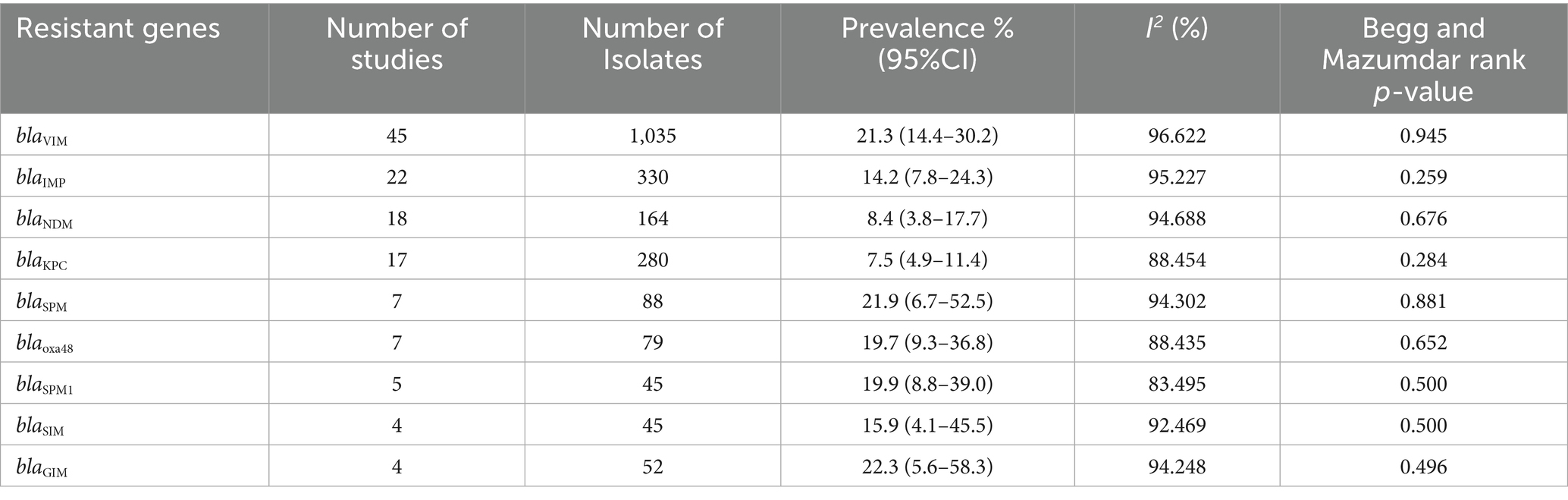
Table 4. Pooled prevalence rate and 95% CI of antibiotic resistance genes of carbapenem-resistant P. aeruginosa based on meta-analysis.
3.3.5 Prevalence of carbapenem-resistant Pseudomonas aeruginosa mortality
Regarding mortality, China recorded a PPE of 3.3% (0.033; 95% CI: 0.009–0.110, Tau = 3.400, I2 = 98.1%) in 153 patients with CRPA (Figure 3). However, countries like USA, Italy, Brazil, Omen, and Iran were not included in analysis due to low number of studies.
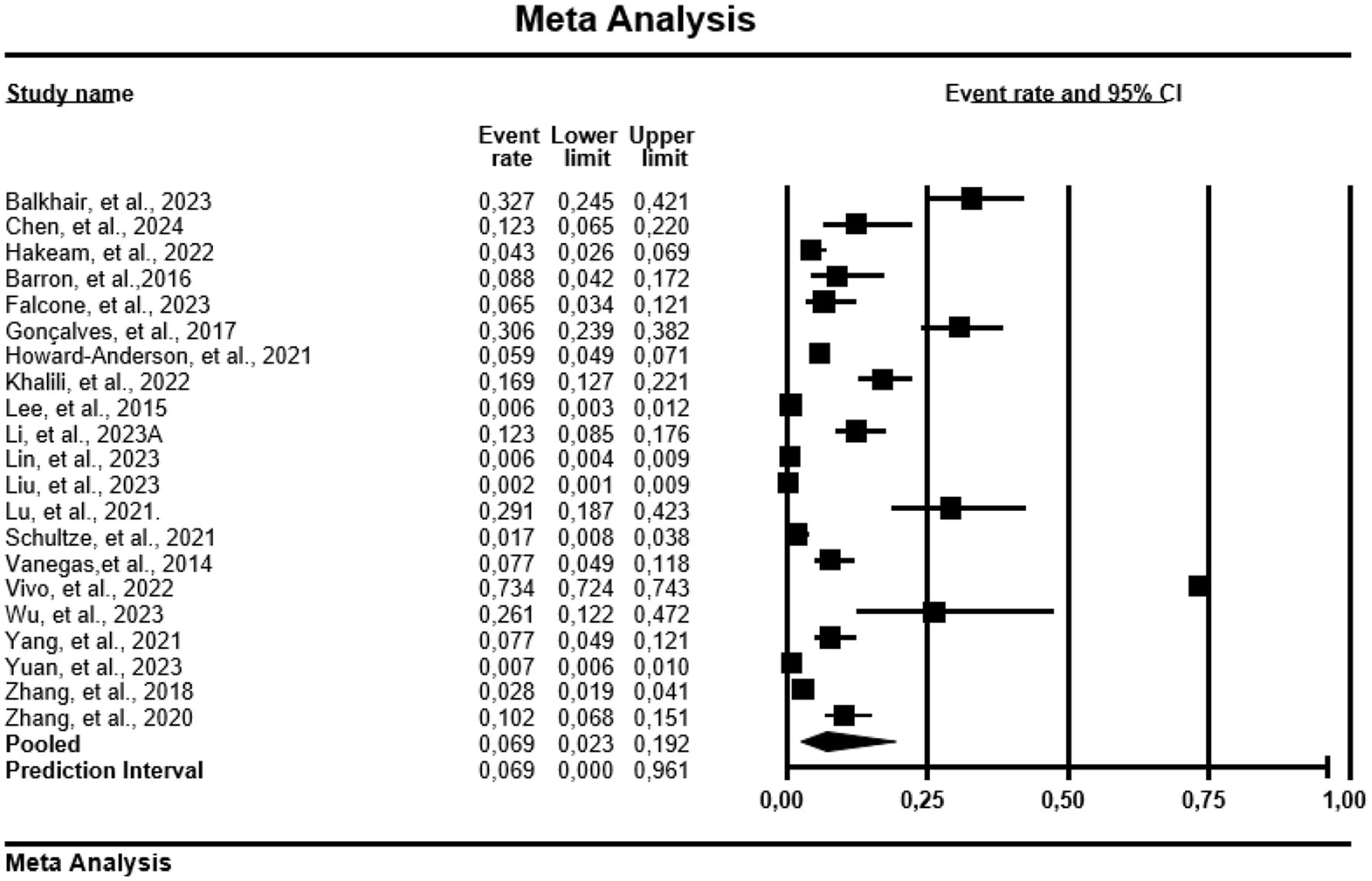
Figure 3. Forest plot showing mortality prevalence in carbapenem resistance-P. aeruginosa. The diamond at the base indicates the pooled estimates from the overall studies.
3.4 Risk of publication bias of included studies
The Begg and Mazumdar rank correlation test, Egger regression test, and funnel plots were additionally used to measure publication bias and check for symmetry. We examined publication bias using the funnel plot. The funnel plots of the estimates suggested publication bias (Supplementary Figure S1) for Asia, the year interval 2018–2021 (p = 0.025).
4 Discussion
In this systematic review and meta-analysis, we conducted an in-depth study to investigate the prevalence of carbapenem-resistant P. aeruginosa and its antibiotic resistance reported worldwide between 2014 and 2024, using over 163 peer reviewed published papers. Thus, we conducted a subgroup analysis of CRPA and their antibiotic resistance. Several articles that only reported on the prevalence of P. aeruginosa and not carbapenem-resistant strains were excluded from our systematic literature review. However, the aim of this study was to include only studies reporting on CRPA and their antibiotic resistance profiles.
Carbapenems are important broad-spectrum antimicrobials of last resort, therefore, resistance to them means increased infections, mortalities, length of hospital stays, and treatment costs (Arowolo et al., 2023). In this context, the results of our meta-analysis and systematic review of the prevalence of CRPA worldwide provide important new information about the terrain of antibiotic resistance. This study recorded an overall PPE of 34.7% for CRPA, with Asia, Africa, Europe, South America and North America having a PPE of 32.8, 38.5, 47.6, 40.9, 33.3 and 47.8%, respectively. This observation of prevalence is higher than that reported in sub-Saharan Africa in 2022 (8%), and the prevalence in the African continent of 21.36% (Kindu et al., 2020). Countries vary in the amount of antibiotics they consume, which may explain why different countries have different prevalence rates of resistant strains.
The current study revealed that the PPE for imipenem and meropenem is 31.2 and 27.7%, respectively. These results are comparable to the prevalence of 30% meropenem resistance reported in Turkey (Acar et al., 2019). Another study from Latin America shows a susceptibility rate of 57% to meropenem and 52% to imipenem among P. aeruginosa isolates (Labarca et al., 2016). Due to the increasing use of carbapenems to treat ESBL-producing infections, the number of carbapenem-resistant Enterobacteriaceae is increasing worldwide (Ramatla et al., 2023).
Studies used in this analysis show that the blaGIM, blaSPM, blaVIM, and blaGIM-1 genes are the most frequently occurring carbapenem-resistant genes in CRPA, with a PPE of 22.3, 21.9, 21.3, and 19.7%, respectively. The studies conducted in sub-Saharan Africa, China, and India reported the blaVIM and blaIMP genes as the most common carbapenemase genes detected in P. aeruginosa isolates (Verma et al., 2019; Wang et al., 2021; Arowolo et al., 2023). Carbapenem-resistant P. aeruginosa isolates have been associated with a variety of carbapenemases, including blaIMP, blaVIM, blaKPC, blaGES, blaNDM, blaGIM, and blaSPM (McCracken et al., 2019; Wang et al., 2021). Verona integron–encoded metallo-β-lactamase (blaVIM) is the most identified CRPA worldwide (Diene and Rolain, 2014). It is also the most common carbapenemase identified in P. aeruginosa in some countries like the United States (Kracalik et al., 2022). The metallo-β-lactamase blaGIM-1 (imipenemase) has so far only been found in clinical isolates of P. aeruginosa (Rieber et al., 2012). The blaKPC, blaNDM, and blaIMP exhibited a low prevalence. Class D oxacillinases (OXA)-type enzymes such as OXA- 48-like is one of the major classes of carbapenemases (Sheu et al., 2019). A number of OXA-carbapenemases, including the OXA-48-like carbapenemases, have proliferated within Enterobacterales and have emerged as a key mechanism for carbapenem resistance in these isolates across multiple countries (Poirel et al., 2012; Evans and Amyes, 2014; Ferous et al., 2024). Although data on the transfer dynamics of antimicrobial resistance genes (ARGs) between clinical environments are limited, there have been several instances where the same ARG was simultaneously identified on hospital surfaces and in patients (Andersson and Hughes, 2017; Lerminiaux and Cameron, 2019).
In this systematic review and meta-analysis, we also report the presence of CRPA and their antibiotic resistance. Using a random-effect model, the pooled prevalence of ciprofloxacin had the highest PPE of 54.5%. The literature reveals instances of ciprofloxacin-resistant P. aeruginosa (CRPAs) in chronic ear infections called suppurative otitis (Jang and Park, 2004; Jang et al., 2009), from patients diagnosed with P. aeruginosa nosocomial infection (Qin et al., 2022). There is a possibility that ciprofloxacin-resistant organisms first appeared in the human domain, since this antibiotic is frequently utilized in human healthcare (Ramatla et al., 2022). The AR against aztreonam had the second highest PPE of 54.4%. Aztreonam plays a crucial role in treating infections caused by metallo-β-lactamase (MBL)-producing carbapenem-resistant Enterobacterales and carbapenem-resistant CRPA (Ding et al., 2023). Therefore, the CRPA resistance to aztreonam is a concern for public health. The emergence of P. aeruginosa strains with multidrug resistance (MDR) is due to their possession of enormous genetic alterations that can develop a variety of factors associated with resistance to various antibiotics (Al-Abedi and Al-Mayahi, 2019). This study observed the proportion of public health impact of multidrug-resistant isolates (MDR) from 28 studies. We believe our results should generate hypotheses and stimulate additional research into the treatment of patients with CRPA.
Furthermore, the current study calculated the PPE for mortality, which is 8.6% from 14 studies. This prevalence is lower as compared to those reported before our study, ranging from 21.0 to 68.3% in Spain, Brazil, the USA, and China (Peña et al., 2012; Dantas et al., 2014; Buehrle et al., 2017; Shi et al., 2019; Vivo et al., 2022; Zhen et al., 2023). Therefore, our study supports the notion that patient mortality can be attributed to CRPA which is a very significant issue.
The subgroup analysis at the continental level showed that the PPE was higher in Europe (47.6%) and South America (40.9%). This is a higher prevalence than 15% reported in Ethiopia by Gobezie et al. (2024) for CRPA. We observed an increase in CRPA of 16.7 to 38.5% in Africa between 2018 and 2021 and 2022–2024, of 95.4 to 54.4% in Asia between 2014 and 2017 and 2018–2021 and around 19.6 to 50.9% in North America between 2018 and 2021 2021 and in between 2022 and 2024. These periodic increases suggest that there could be a global increase in CRPA infections, or the increased number of research funds could be a possible reason for this change.
Additionally, this study found that 56.4% of studies were conducted in Asia, compared to other continents such as Europe at 11.1%, South America (9.3%), Africa (8.7%) and North America (7.6%). No data were available from Australia/Oceania. Our findings were consistent with a previous study reporting that Asian countries have a high prevalence of P. aeruginosa due to chronic wound infections globally (Phan et al., 2023). The differences could be explained by the availability of research funding or the high number of P. aeruginosa infections in Asian countries compared to other continents. Furthermore, countries vary in the amount of antibiotics they consume, which may explain why different countries experience different prevalence rates of resistant strains.
5 Limitations
There are few limitations to our systematic review and meta-analysis: (a) The search strategy was limited to articles published in English, meaning that there may have been articles published in other languages that were missed. (b) The findings of some countries may not be accurate since the number of studies was limited. (c) Compared to other countries, some had more research reports than others. (d) There were no studies reported in Australia/Oceania continent. (e) No data were available in some countries. (f) When there is a high level of heterogeneity, it is difficult to evaluate the actual results of statistically significant publication bias tests. (g) The number of studies from some countries were unusually high, which may have influenced the overall estimate. (h) The pooled prevalence of some countries and continents was not calculated because there are few published studies. (i) Data from large scale surveillance systems was not included. (j) There may have been articles published in other languages that were missed since the search strategy was limited to articles published in English. (k) Nevertheless, meta-analyses that comprise fewer than ten studies or show high heterogeneity among studies might produce misleading outcomes from these evaluation tools. When heterogeneity is high, assessing the true results of statistically significant publication bias tests becomes very difficult. Given the significant variability among analyses, readers ought to be careful when analyzing pooled analyses and subgroups. (l) This study only examined mortality data from China, as other countries had insufficient studies to be included.
6 Conclusion
Our comprehensive meta-analysis provides important updated global prevalence of CRPA from 2014 to 2024, which appears to be increasing or spreading. There is an urgent demand for comprehensive surveillance studies, involving both hospitals and communities worldwide, to accurately measure the global prevalence of these multidrug-resistant pathogens. This study presents robust and valuable data that can serve as a useful reference for clinicians and researchers in informing them about the CRPA. Globally, alternative treatments for CRPA should be well researched, planned, and implemented.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
TR: Conceptualization, Investigation, Formal analysis, Writing – review & editing, Project administration, Writing – original draft, Methodology. JN: Conceptualization, Writing – review & editing, Supervision. KL: Writing – review & editing, Conceptualization, Data curation. OT: Writing – review & editing, Supervision. MM: Validation, Writing – review & editing. CA: Writing – review & editing, Methodology, Formal analysis. GK: Conceptualization, Writing – review & editing, Supervision, Methodology.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1599070/full#supplementary-material
Footnotes
References
Abdalhamid, B., Elhadi, N., Alabdulqader, N., Alsamman, K., and Aljindan, R. (2016). Rates of gastrointestinal tract colonization of carbapenem-resistant Enterobacteriaceae and Pseudomonas aeruginosa in hospitals in Saudi Arabia. New Microbes New Infect. 10, 77–83. doi: 10.1016/j.nmni.2016.01.014
Abubakar, U., Zulkarnain, A. I., Rodríguez-Baño, J., Kamarudin, N., Elrggal, M. E., Elnaem, M. H., et al. (2022). Treatments and predictors of mortality for carbapenem-resistant gram-negative bacilli infections in Malaysia: a retrospective cohort study. Trop. Med. Infect. Dis. 7:415. doi: 10.3390/tropicalmed7120415
Acar, A., Karaahmetoğlu, G., Akalın, H., and Altay, A. F. (2019). Pooled prevalence and trends of antimicrobial resistance in Pseudomonas aeruginosa clinical isolates over the past 10 years in Turkey: a meta-analysis. J. Glob. Antimicrob. Resist. 18, 64–70. doi: 10.1016/j.jgar.2019.01.032
Al-Abedi, K. J. H., and Al-Mayahi, F. A. (2019). Molecular detection of serine carbapenemase genes in carbapenem-resistant isolates of Pseudomonas aeruginosa recovered from patients in Al-Diwaniyah Province, Iraq. Iraq J. Pure Appl. Microbiol. 13, 1775–1782. doi: 10.22207/JPAM.13.3.53
Andersson, D. I., and Hughes, D. (2017). Selection and transmission of antibiotic-resistant bacteria. Microbiol. Spectr. 5:MTBP0013-2016. doi: 10.1128/microbiolspec.mtbp-0013-2016
Arowolo, M. T., Orababa, O. Q., Olaitan, M. O., Osibeluwo, B. V., Essiet, U. U., Batholomew, O. H., et al. (2023). Prevalence of carbapenem resistance in Acinetobacter baumannii and Pseudomonas aeruginosa in sub-Saharan Africa: a systematic review and meta-analysis. PLoS One 18:e0287762. doi: 10.1371/journal.pone.0287762x
Büchler, A. C., Shahab, S. N., Severin, J. A., Vos, M. C., and Voor in’t holt, A. F. (2023). Outbreak investigations after identifying carbapenem-resistant Pseudomonas aeruginosa: a systematic review. Antimicrob. Resist. Infect. Control 12:28. doi: 10.1186/s13756-023-01223-1
Buehrle, D. J., Shields, R. K., Clarke, L. G., Potoski, B. A., Clancy, C. J., and Nguyen, M. H. (2017). Carbapenem-resistant Pseudomonas aeruginosa bacteremia: risk factors for mortality and microbiologic treatment failure. Antimicrob. Agents Chemother. 61:e01243-16. doi: 10.1128/AAC.01243-16
Canton, R., Doi, Y., and Simner, P. J. (2022). Treatment of carbapenem-resistant Pseudomonas aeruginosa infections: a case for cefiderocol. Expert Rev. Anti-Infect. Ther. 20, 1077–1094. doi: 10.1080/14787210.2022.2071701
Çopur, Ç. A., Ertürk, A., Ejder, N., Rakici, E., Kostakoğlu, U., Esen Yıldız, İ. L. K. N. U. R., et al. (2021). Screening of antimicrobial resistance genes and epidemiological features in hospital and community-associated carbapenem-resistant pseudomonas aeruginosa infections. Infect. Drug Resist. 14, 1517–1526. doi: 10.2147/IDR.S299742
Dantas, R. C., Ferreira, M. L., Gontijo-Filho, P. P., and Ribas, R. M. (2014). Pseudomonas aeruginosa bacteraemia: independent risk factors for mortality and impact of resistance on outcome. J. Med. Microbiol. 63, 1679–1687. doi: 10.1099/jmm.0.073262-0
Deshwal, P. R., Aggarwal, M., Reddy, N. S., Fathima, R., and Tiwari, P. (2023). Association of β-lactam antimicrobial's exposure with carbapenem-resistant Pseudomonas aeruginosa infection: a cumulative meta-analysis. Glob. J. Health 7, 137–146. doi: 10.1016/j.glohj.2023.07.005
Diene, S. M., and Rolain, J. M. (2014). Carbapenemase genes and genetic platforms in gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 20, 831–838. doi: 10.1111/1469-0691.12655
Ding, L., Sun, Y., Zhang, Y., Shen, S., and Hu, F. (2023). In vivo development of aztreonam resistance in meropenem-resistant Pseudomonas aeruginosa owing to overexpression of the blaPDC-16. Microbiol. Spectr. 11:e0308022. doi: 10.1128/spectrum.03080-22
Evans, B. A., and Amyes, S. G. (2014). OXA β-lactamases. Clin. Microbiol. Rev. 27, 241–263. doi: 10.1128/CMR.00117-13
Ferous, S., Anastassopoulou, C., Pitiriga, V., Vrioni, G., and Tsakris, A. (2024). Antimicrobial and diagnostic stewardship of the novel β-lactam/β-lactamase inhibitors for infections due to Carbapenem-resistant Enterobacterales species and Pseudomonas aeruginosa. Antibiotics 13:285. doi: 10.3390/antibiotics13030285
Gobezie, M. Y., Hassen, M., Tesfaye, N. A., Solomon, T., Demessie, M. B., Kassa, T. D., et al. (2024). Prevalence of meropenem-resistant Pseudomonas Aeruginosa in Ethiopia: a systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 13:37. doi: 10.1186/s13756-024-01389-2
Golle, A., Janezic, S., and Rupnik, M. (2017). Low overlap between carbapenem resistant Pseudomonas aeruginosa genotypes isolated from hospitalized patients and wastewater treatment plants. PLoS One 12:e0186736. doi: 10.1371/journal.pone.0186736
Jang, C. H., and Park, S. Y. (2004). Emergence of ciprofloxacin-resistant pseudomonas in chronic suppurative otitis media. Clin. Otolaryngol. Allied Sci. 29, 321–323. doi: 10.1111/j.1365-2273.2004.00835.x
Jang, C. H., Park, H., Cho, Y. B., Choi, C. H., and Song, C. (2009). Antibacterial effect of octylcyanoacrylate against ciprofloxacin-resistant Pseudomonas aeruginosa isolates from patients with chronic suppurative otitis media. In Vivo 23, 183–185.
Kindu, M., Derseh, L., Gelaw, B., and Moges, F. (2020). Carbapenemase-producing non-glucose-fermenting gram-negative Bacilli in Africa, Pseudomonas aeruginosa and Acinetobacter baumannii: a systematic review and meta-analysis. Int. J. Microbiol. 2020, 1–18. doi: 10.1155/2020/9461901
Kracalik, I., Ham, D. C., McAllister, G., Smith, A. R., Vowles, M., Kauber, K., et al. (2022). Extensively drug-resistant carbapenemase-producing Pseudomonas aeruginosa and medical tourism from the United States to Mexico, 2018–2019. Emerg. Infect. Dis. 28, 51–61. doi: 10.3201/eid2801.211880
Labarca, J. A., Salles, M. J. C., Seas, C., and Guzmán-Blanco, M. (2016). Carbapenem resistance in Pseudomonas aeruginosa and Acinetobacter baumannii in the nosocomial setting in Latin America. Crit. Rev. Microbiol. 42, 276–292. doi: 10.3109/1040841X.2014.940494
Lerminiaux, N. A., and Cameron, A. D. (2019). Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65, 34–44. doi: 10.1139/cjm-2018-0275
Marchaim, D., Perez, F., Lee, J., Bheemreddy, S., Hujer, A. M., Rudin, S., et al. (2012). “Swimming in resistance”: co-colonization with carbapenem-resistant Enterobacteriaceae and Acinetobacter baumannii or Pseudomonas aeruginosa. Am. J. Infect. Control 40, 830–835. doi: 10.1016/j.ajic.2011.10.013
Marouf, S., Li, X., Salem, H. M., Ahmed, Z. S., Nader, S. M., Shaalan, M., et al. (2023). Molecular detection of multidrug-resistant Pseudomonas aeruginosa of different avian sources with pathogenicity testing and in vitro evaluation of antibacterial efficacy of silver nanoparticles against multidrug-resistant P. aeruginosa. Poult. Sci. 102:102995. doi: 10.1016/j.psj.2023.102995
McCracken, M. G., Adam, H. J., Blondeau, J. M., Walkty, A. J., Karlowsky, J. A., Hoban, D. J., et al. (2019). Characterization of carbapenem-resistant and XDR Pseudomonas aeruginosa in Canada: results of the CANWARD 2007–16 study. J. Antimicrob. Chemother. 74, iv32–iv38. doi: 10.1093/jac/dkz285
Męcik, M., Stefaniak, K., Harnisz, M., and Korzeniewska, E. (2024). Hospital and municipal wastewater as a source of carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa in the environment: a review. Environ. Sci. Pollut. Res. Int. 31, 48813–48838. doi: 10.1007/s11356-024-34436-x
Narimisa, N., Keshtkar, A., Dadgar-Zankbar, L., Bostanghadiri, N., Far, Y. R., Shahroodian, S., et al. (2024). Prevalence of colistin resistance in clinical isolates of Pseudomonas aeruginosa: a systematic review and meta-analysis. Front. Microbiol. 15:1477836. doi: 10.3389/fmicb.2024.1477836
Odoi, H., Boamah, V. E., Boakye, Y. D., and Agyare, C. (2021). Prevalence and phenotypic and genotypic resistance mechanisms of multidrug-resistant Pseudomonas aeruginosa strains isolated from clinical, environmental, and poultry litter samples from the Ashanti region of Ghana. J. Environ. Public Health 2021:9976064. doi: 10.1155/2021/9976064
Pappa, O., Louka, C., Karadimas, K., Maikousi, E., Tzoukmani, A., Polemis, M., et al. (2024). Emergence of NDM-1-producing Pseudomonas aeruginosa nosocomial isolates in Attica region of Greece. Microorganisms 12:1753. doi: 10.3390/microorganisms12091753
Peña, C., Suarez, C., Gozalo, M., Murillas, J., Almirante, B., Pomar, V., et al. (2012). Prospective multicenter study of the impact of carbapenem resistance on mortality in Pseudomonas aeruginosa bloodstream infections. Antimicrob. Agents Chemother. 56, 1265–1272. doi: 10.1128/AAC.05991-11
Phan, S., Feng, C. H., Huang, R., Lee, Z. X., Moua, Y., Phung, O. J., et al. (2023). Relative abundance and detection of Pseudomonas aeruginosa from chronic wound infections globally. Microorganisms 11:1210. doi: 10.3390/microorganisms11051210
Poirel, L., Potron, A., and Nordmann, P. (2012). OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67, 1597–1606. doi: 10.1093/jac/dks121
Qin, J., Zou, C., Tao, J., Wei, T., Yan, L., Zhang, Y., et al. (2022). Carbapenem resistant Pseudomonas aeruginosa infections in elderly patients: antimicrobial resistance profiles, risk factors and impact on clinical outcomes. Infect. Drug Resist. 15, 2301–2314. doi: 10.2147/IDR.S358778
Rai, S., Das, D., Niranjan, D. K., Singh, N. P., and Kaur, I. R. (2014). Carriage prevalence of carbapenem-resistant Enterobacteriaceae in stool samples: a surveillance study. Australas Med J 7, 64–67. doi: 10.4066/AMJ.2014.1926
Ramatla, T., Mafokwane, T., Lekota, K., Monyama, M., Khasapane, G., Serage, N., et al. (2023). “One health” perspective on prevalence of co-existing extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae: a comprehensive systematic review and meta-analysis. Ann. Clin. Microbiol. Antimicrob. 22:88. doi: 10.1186/s12941-023-00638-3
Ramatla, T., Tawana, M., Mphuthi, M. B., Onyiche, T. E., Lekota, K. E., Monyama, M. C., et al. (2022). Prevalence and antimicrobial resistance profiles of Campylobacter species in South Africa: a “one health” approach using systematic review and meta-analysis. Int. J. Infect. Dis. 125, 294–304. doi: 10.1016/j.ijid.2022.10.042
Rieber, H., Frontzek, A., and Pfeifer, Y. (2012). Emergence of metallo-β-lactamase GIM-1 in a clinical isolate of Serratia marcescens. Antimicrob. Agents Chemother. 56, 4945–4947. doi: 10.1128/AAC.00405-12
Saha, K., Kabir, N. D., Islam, M. R., Amin, M. B., Hoque, K. I., Halder, K., et al. (2022). Isolation and characterisation of carbapenem-resistant Pseudomonas aeruginosa from hospital environments in tertiary care hospitals in Dhaka, Bangladesh. J. Glob. Antimicrob. Resist. 30, 31–37. doi: 10.1016/j.jgar.2022.04.008
Schill, F., Abdulmawjood, A., Klein, G., and Reich, F. (2017). Prevalence and characterization of extended-spectrum β-lactamase (ESBL) and AmpC β-lactamase producing Enterobacteriaceae in fresh pork meat at processing level in Germany. Int. J. Food Microbiol. 257, 58–66. doi: 10.1016/j.ijfoodmicro.2017.06.010
Shahab, S. N., van Veen, A., Büchler, A. C., Saharman, Y. R., Karuniawati, A., Vos, M. C., et al. (2024). In search of the best method to detect carriage of carbapenem-resistant Pseudomonas aeruginosa in humans: a systematic review. Ann. Clin. Microbiol. Antimicrob. 23:50. doi: 10.1186/s12941-024-00707-1
Sheu, C. C., Chang, Y. T., Lin, S. Y., Chen, Y. H., and Hsueh, P. R. (2019). Infections caused by carbapenem-resistant Enterobacteriaceae: an update on therapeutic options. Front. Microbiol. 10:80. doi: 10.3389/fmicb.2019.00080
Shi, Q., Huang, C., Xiao, T., Wu, Z., and Xiao, Y. (2019). A retrospective analysis of Pseudomonas aeruginosa bloodstream infections: prevalence, risk factors, and outcome in carbapenem-susceptible and-non-susceptible infections. Antimicrob. Resist. Infect. Control 8:68. doi: 10.1186/s13756-019-0520-8
Souza, G. H. D. A. D., Rossato, L., Brito, G. T., Bet, G. M. D. S., and Simionatto, S. (2021). Carbapenem-resistant Pseudomonas aeruginosa strains: a worrying health problem in intensive care units. Rev. Inst. Med. Trop. Sao Paulo 63:e71. doi: 10.1590/S1678-9946202163071
Verma, N., Prahraj, A., Mishra, B., Behera, B., and Gupta, K. (2019). Detection of carbapenemase-producing Pseudomonas aeruginosa by phenotypic and genotypic methods in a tertiary care hospital of East India. J. Lab. Physicians 11, 287–291. doi: 10.4103/JLP.JLP_136_19
Vivo, A., Fitzpatrick, M. A., Suda, K. J., Jones, M. M., Perencevich, E. N., Rubin, M. A., et al. (2022). Epidemiology and outcomes associated with carbapenem-resistant Acinetobacter baumannii and carbapenem-resistant Pseudomonas aeruginosa: a retrospective cohort study. BMC Infect. Dis. 22:491. doi: 10.1186/s12879-022-07436-w
Wang, M. G., Liu, Z. Y., Liao, X. P., Sun, R. Y., Li, R. B., Liu, Y., et al. (2021). Retrospective data insight into the global distribution of carbapenemase-producing Pseudomonas aeruginosa. Antibiotics 10:548. doi: 10.3390/antibiotics10050548
Yano, H., Hayashi, W., Kawakami, S., Aoki, S., Anzai, E., Zuo, H., et al. (2024). Nationwide genome surveillance of carbapenem-resistant Pseudomonas aeruginosa in Japan. Antimicrob. Agents Chemother. 68:e0166923. doi: 10.1128/aac.01669-23
Zhen, S., Zhao, Y., Chen, Z., Zhang, T., Wang, J., Jiang, E., et al. (2023). Assessment of mortality-related risk factors and effective antimicrobial regimens for treatment of bloodstream infections caused by carbapenem-resistant Pseudomonas aeruginosa in patients with hematological diseases. Front. Cell. Infect. Microbiol. 13:1156651. doi: 10.3389/fcimb.2023.1156651
Keywords: pooled prevalence estimate, CRPA, antibiotic resistance, global, meta-analysis
Citation: Ramatla T, Nkhebenyane J, Lekota KE, Thekisoe O, Monyama M, Achilonu CC and Khasapane G (2025) Global prevalence and antibiotic resistance profiles of carbapenem-resistant Pseudomonas aeruginosa reported from 2014 to 2024: a systematic review and meta-analysis. Front. Microbiol. 16:1599070. doi: 10.3389/fmicb.2025.1599070
Edited by:
Taru Singh, Amity University, IndiaCopyright © 2025 Ramatla, Nkhebenyane, Lekota, Thekisoe, Monyama, Achilonu and Khasapane. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tsepo Ramatla, dHJhbWF0bGFAY3V0LmFjLnph
 Tsepo Ramatla
Tsepo Ramatla Jane Nkhebenyane1
Jane Nkhebenyane1