- 1School of Biological Engineering, Dalian Polytechnic University, Dalian, China
- 2National Engineering Research Center of Dairy Health for Maternal and Child, Beijing Sanyuan Foods Co. Ltd., Beijing, China
- 3Beijing Engineering Research Center of Dairy, Beijing Technical Innovation Center of Human Milk Research, Beijing Sanyuan Foods Co. Ltd., Beijing, China
- 4Key Laboratory of Dairy Science, Ministry of Education, Food Science College, Northeast Agricultural University, Harbin, China
- 5College of Life Sciences, Inner Mongolia University, Hohhot, China
Introduction: Hyperlipidemia, a prevalent metabolic disorder with rising global incidence, has become a major public health concern. Probiotics allow for a mild intervention strategy for hyperlipidemia management that has garnered increasing attention.
Methods: In this study, we investigated the therapeutic effects and underlying mechanisms of Lactobacillus paragasseri HM018 in hypercholesterolemic rats.
Results and discussion: We established three dosage groups (2.5 × 108, 5 × 108, and 1.5 × 109 CFU/rat), demonstrating that HM018 significantly reduced high-fat diet-induced serum total cholesterol, triglycerides, and low-density lipoprotein cholesterol levels, while ameliorating gut microbiota dysbiosis and decreasing the Firmicutes/Bacteroidetes ratio. Our transcriptomic analysis revealed that HM018 markedly upregulated Apoa1 expression both in the ileum and liver, while enhancing Abcg5/Abcg8 gene expression to promote β-sitosterol efflux. Concurrently, hepatic Cocs2/3 and Cish gene expression was downregulated, attenuating their inhibitory effects on hormonal and glucagon signaling, thereby improving glucose and lipid metabolism. Metabolomic profiling further indicated that HM018 significantly altered bile acid composition by modulating gut microbiota-mediated bile acid metabolism. In conclusion, Lactobacillus paracasei HM018 could ameliorate hyperlipidemia through multiple pathways, including gut microbiota modulation, hepatic lipid/glucose/bile acid metabolism improvement, and intestinal cholesterol efflux gene expression enhancement.
1 Introduction
Hyperlipidemia is a highly prevalent metabolic disorder worldwide (Su et al., 2022), characterized by elevated levels of total cholesterol (TC), triglycerides (TG), and low-density lipoprotein cholesterol (LDL-C), as well as reduced high-density lipoprotein cholesterol (HDL-C). The primary causes include high carbohydrate or fat intake, lack of exercise, and delayed intervention and treatment, leading to increased blood lipid levels (Khanali et al., 2023). Consequently, it triggers related diseases such as cardiovascular disease (CVD) and nonalcoholic fatty liver disease (NAFLD) (James et al., 2018; Roth et al., 2020). The prevalence and mortality rates of hyperlipidemia and its complications are increasing in various countries, making it a significant public health issue (James et al., 2018). In particular, CVDs induced by elevated blood lipid levels have become a major burden on healthcare services (Roth et al., 2020). Statins are commonly used drugs for treating hyperlipidemia (Lamb, 2020) and are particularly effective in reducing LDL-C levels. However, there is considerable interindividual variability in their lipid-lowering efficacy, which may be related to the composition of the gut microbiota (Jia et al., 2021; Hu et al., 2024).
The “gut microbiota” is a complex and dynamic community of microorganisms in the human intestine (Ross et al., 2024). The impact of a high-fat diet (HFD) on the structure of the gut microbiota has been well-documented. Dysbiosis of the gut microbiota leads to increased intestinal permeability, elevated levels of pro-inflammatory cytokines, insulin resistance, glucose intolerance, and a series of other issues (Howard et al., 2022). Gut microbes can influence blood lipid levels through metabolites such as short-chain fatty acids, lipopolysaccharide, and bile acids (Jia et al., 2021). Gut microbiota is an effective target for the prevention and treatment of metabolic syndromes. For example, Blautia producta, screened from human gut microbiota, inhibits lipid accumulation in cells and reduces lipid levels in the blood and liver of HFD-fed mice (Xu W. et al., 2023). Enterococcus faecium B6, isolated from obese children, promotes hepatic lipid accumulation, inflammation, and fibrosis through the bioactive metabolite tyramine, thereby contributing to NAFLD (Wei et al., 2024). Intervention with Lactococcus lactis subsp. Cremoris in Western diet-fed mice reduces hepatic steatosis and inflammation, as well as serum cholesterol and body mass index (Naudin et al., 2020).
Probiotics, particularly strains of the genus Lactobacillus, are reportedly significantly efficient in ameliorating dyslipidemia (Gao et al., 2021), although substantial functional heterogeneity exists among different strains (Patnode et al., 2021). In this study, we isolated a Lactobacillus strain, Lactobacillus paragasseri, from human milk. Based on the microbial taxonomic criteria (ANI < 95% and isDDH <70% for novel species designation), this strain represents a novel species reclassified from L. gasseri [exhibiting ANI values of 93.4–93.7% and isDDH values of 53.1–54.1% versus the reference strain ATCC 33323; (Tanizawa et al., 2018)], retaining an incompletely characterized mechanistic potential in improving hyperlipidemia. Through oral gavage administration, we systematically investigated the lipid-lowering mechanisms of human milk-derived L. paragasseri HM018 in high-fat diet-induced hyperlipidemic rats. Experimental results demonstrated that HM018 intervention significantly attenuated diet-induced serum triglyceride, total cholesterol, and low-density lipoprotein level increase. Integrated gut microbiota composition, fecal metabolomics, and ileal/hepatic transcriptomics analyses revealed the multifaceted regulatory effects of this probiotic on lipid metabolism.
2 Materials and methods
2.1 L. Paragasseri HM018 and culture medium
L. paragasseri HM018 is derived from healthy breast milk and is presently conserved at the China General Microbiological Culture Collection Center (CGMCC No. 19749). L. paragasseri HM018, isolated from breast milk, was preserved at the National Engineering Research Center for Dairy Health (Beijing, China). The strain was inoculated in De Man–Rogosa–Sharpe broth (Luqiao Co., Ltd., Beijing, China) at 37°C for 18 h and subcultured twice at a 1% inoculation rate to ensure bacterial activity. Cellular biomass was harvested via refrigerated centrifugation (4°C, 6,000 rpm, 15 min). The supernatant was removed, and the pellet was washed twice with sterile 0.01 M phosphate-buffered saline (PBS; 0.01 M concentration, sterile filtered, Solarbio). Finally, the bacterial suspension was adjusted to low-dose (2.5 × 108 colony-forming units [CFU] mL-1), medium-dose (5 × 108 CFU mL-1), and high-dose (1.5 × 109 CFU mL-1) concentrations for oral gavage.
2.2 Rat modeling and feeding
All animal procedures adhered to the Guidelines for the Care and Use of Laboratory Animals established by Beijing Union University andreceived approval from the Animal Ethics Committee of Beijing Union University (JCZX11-2306-1, Beijing, China).
Fifty adult male Sprague–Dawley rats (body weight 200 ± 20 g; Vital River Laboratories, Beijing, China), raised under specific pathogen-free (SPF) conditions, were acclimatized for one week under standard conditions with a 12-h light/dark cycle. Throughout the acclimatization period, food and water were freely available. Subsequently, the rats were randomly assigned to five experimental groups as follows: (1) a normal diet (ND) control group receiving standard chow and 2 mL of sterile PBS; (2) an HFD model group receiving an HFD and 2 mL of sterile PBS; and three treatment groups (3–5) receiving the HFD supplemented with varying concentrations of L. paragasseri HM018: (3) the low-dose group (LPL) received 2 mL of a 2.5 × 108 CFU/mL suspension; (4) the medium-dose group received 2 mL of a 5 × 108 CFU/mL suspension; and (5) the high-dose group (LPH) received 2 mL of a 1.5 × 109 CFU/mL suspension, all administered via gavage. All experimental diets were provided by Ke’ao Xieli Feed Co., Ltd. (Beijing, China).
After six weeks of intervention, following a 12-h fast after the final gavage, fecal samples were collected and incubated at 25 ± 1°C for 60 min. Subsequently, the rats were euthanized, and blood samples were collected, and Serum was obtained through centrifugation of blood samples at 3,000 × g for 15 min at 4°C. Tissue specimens (liver, epididymal adipose, ileal segments) were rapidly cryopreserved in liquid nitrogen vapor phase and maintained at −80°C.
2.3 Biochemical analysis
Serum biomarkers, including TG, TC, LDL-C, HDL-C, glucose, and glycated hemoglobin (GHb), were quantified using commercial assay kits (Nanjing Jiancheng Bioengineering Institute, China) following manufacturer protocols.
2.4 Hematoxylin and eosin (H&E) staining
Liver tissue samples from rats were fixed in 4% paraformaldehyde at room temperature for 24 h. Subsequently, the samples were dehydrated, embedded in paraffin, and sectioned into 5 μm slices. After dewaxing, the tissue sections were stained with hematoxylin and eosin (H&E) for histological observation.
2.5 Gene sequencing analysis of 16S rRNA
Using the OMEGA Mag-bind DNA Kit, genomic DNA was isolated from samples. The V3-V4 region of the 16S rRNA gene was then amplified with specific barcoded primers 341F (CCTAYGGGRBGCASCAG) and 806R (GGACTACNNGGGTATCTAAT). Polymerase chain reaction (PCR) products were analyzed using 2% agarose gel electrophoresis and subsequently purified using a Quant-iT PicoGreen dsDNA Assay Kit. Based on the preliminary quantification results from electrophoresis, the recovered PCR products were quantified using a fluorescence quantification system on a microplate reader (BioTek, FLx800). Library construction was performed using an Illumina TruSeq Nano DNA LT Library Prep Kit. The quality of the constructed libraries was assessed using an Agilent Bioanalyzer 2,100 and Promega QuantiFluor, followed by sequencing. Raw sequencing data were processed to remove primer sequences using Cutadapt, and quality control and annotation were performed using QIIME 2 with the SILVA database (version 138). Alpha and beta diversity indices were calculated using QIIME 2.
2.6 HPLC analysis
Fecal samples were collected from 10 rats per group (ND, HFD, and LPH groups; n = 30 total). Samples underwent liquid nitrogen flash-freezing and were maintained at −80°C Untargeted metabolomic analysis was performed at Shanghai Applied Protein Technology Co., Ltd. (Shanghai, China) using an ultra-high-performance liquid chromatography (UHPLC) system (1,290 Infinity II, Agilent Technologies) coupled with a quadrupole time-of-flight mass spectrometer (AB Sciex TripleTOF 6,600).
Fecal samples (50 mg) were homogenized with 400 μL methanol/acetonitrile (1:1, v/v) via vortex mixing, followed by sequential processing: 30-min sonication at 5°C (40 kHz), 30-min incubation at-20°C, and centrifugation (1,400 × g, 20 min, 4°C). The resulting supernatant was vacuum-dried and reconstituted in 120 μL acetonitrile/water (1:1, v/v). For quality assurance, 10 μL QC sample was introduced prior to final 5-min sonication/centrifugation, with processed supernatant ultimately transferred to LC–MS vials for analysis.
Chromatographic separation was performed on a Waters Acquity UPLC BEH Amide column (100 mm × 2.1 mm, 1.7 μm) employing hydrophilic interaction liquid chromatography (HILIC). The mobile phase consisted of (A) 25 mM ammonium acetate/ammonium hydroxide aqueous solution and (B) acetonitrile, with the following gradient profile: initial 95% B (0–0.5 min), linear gradient to 65% B (0.5–7.5 min), stepwise reduction to 40% B (7.5–7.6 min), isocratic hold (7.6–8.6 min), immediate return to 95% B (8.6–8.7 min), and 3-min column re-equilibration (8.7–11.7 min). The chromatographic system was maintained at 0.3 mL/min flow rate and 40°C.
Instrument parameters were configured as follows: ion source gases (60 psi), curtain gas (30 psi), 600°C source temperature, ±5,500 V ion spray voltage. The method employed 35 V (±15 eV) collision energy, ±60 V declustering potential, 4 Da isotope exclusion, and 10-cycle candidate ion-monitoring acquisition. Raw data were converted into mzXML format using ProteoWizard MSConvert. Peak detection, alignment, and annotation were performed using the XCMS workflow, with metabolites identified by matching accurate mass (m/z error <10 ppm) and MS/MS spectra against an in-house reference database.
2.7 Transcriptome analysis
Total RNA was extracted from the liver tissues of rats in the three groups (ND, HFD, and LPH) using the TRIzol reagent. RNA purity was evaluated by measuring the A260/A280 ratio using a Nanodrop ND-2000 spectrophotometer (Thermo Scientific, USA), and RNA integrity was assessed using an Agilent Bioanalyzer 4,150 system (Agilent Technologies, CA, USA). Subsequently, paired-end libraries were prepared using the ABclonal mRNA-seq Lib Prep Kit (ABclonal, China) following manufacturer protocols. Sequencing was conducted across dual platforms: Illumina Novaseq 6,000 and MGISEQ-T7 systems.
Sequencing reads were preprocessed using Trimmomatic (adapter removal/quality trimming). Genome alignment was performed using HISAT2. DESeq2 (v1.40.2) implemented in R was used to determine differentially expressed genes (criteria: |log2FC| > 1, FDR < 0.05). Enrichment analysis was performed using GO and KEGG pathways (clusterProfiler v4.10.0). Statistical significance was set at p < 0.05.
2.8 Gene expression profiling by real-time quantitative PCR
Rat liver and ileum tissue samples were collected. Total RNA was extracted using RNA-easy Isolation Reagent (R701-01, Vazyme, China), and RNA was reverse-transcribed into cDNA using the HiScript II Q RT SuperMix for qPCR (+gDNA wiper) (R223-01, Vazyme, China) kit. Subsequently, using ChamQ SYBR Master Mix (Q311-02, Vazyme, China) as the reaction system, real-time fluorescence quantitative PCR (RT-PCR) was carried out on an Applied Biosystems 7,500 Real-Time PCR System to detect the expression levels of the target genes (Abcg5, Abcg8, Socs2, and Cish). The primer sequences are as follows: for Abcg5, the forward primer is 5′-GTCCTTCAGCGTCAGCAACC-3′ and the reverse primer is 5′ -ATGGTCTGGCCACTCTCGAT-3′; for Abcg8, the forward primer is 5′-CACCCTAGACTCTAACTCCA-3′ and the reverse primer is 5′-GGAGCACTGGATAGTATTGG-3′ (Jang et al., 2022); for Socs2, the forward primer is 5′-GAACCACGCTGTCAAACT-3′ and the reverse primer is 5′-CTCCCACTCAGACTACCTATT-3′ (Dang et al., 2020); for Cish, the forward primer is 5′- TACCTCCGGGGATCTGGTTG −3′ and the reverse primer is 5′- CACGGGTGGTTTTGACTGAC -3′. Gadph (forward primer: 5′-GAAACCTGCCAAGTATGA-3′, reverse primer: 5′-GCTGTAGCCGTATTCATT-3′) was used as the internal reference gene for normalization analysis.
2.9 Statistical analysis
Graphical representations, including histograms, box plots, and heatmaps, were generated using Origin software (Version 2024, OriginLab, MA, USA). Statistical analyses were conducted using SPSS (Version 27.0.1, IBM, USA), employing one-way analysis of variance followed by Tukey’s post-hoc test for multiple comparisons. Data are presented as mean ± standard deviation (SD), with statistical significance set at p < 0.05. Additionally, bivariate correlations were evaluated using Spearman’s rank-order correlation method.
3 Results
3.1 HM018 ameliorates dyslipidemia and hepatic abnormalities in HFD-fed rats
After 6 weeks on a high-fat diet, serum TG and TC levels were notably increased in HFD group. In the probiotic intervention groups with varying doses, these parameters showed dose-dependent improvements. Additionally, the HFD group demonstrated significantly elevated LDL-cholesterol levels compared to the ND group. But was significantly reduced after probiotic intervention. However, HDL-C level, which is associated with lipid utilization, significantly decreased after HFD, and there was no change in its concentration after the intervention with HM018. Compared to the ND group, body weight, fasting glucose, and GHb levels showed no statistically significant variations across the experimental cohorts (Supplementary Figure S1). Histological analysis showed that HFD led to fat accumulation, and the ratio of the area of liver adipocytes in the HFD group was significantly higher than that in other groups. In summary, the abnormal elevation of key lipid markers (TG, TC, and LDL-C) confirmed the successful establishment of the HFD-induced rat model, while probiotic intervention significantly ameliorated dyslipidemia (Figure 1).
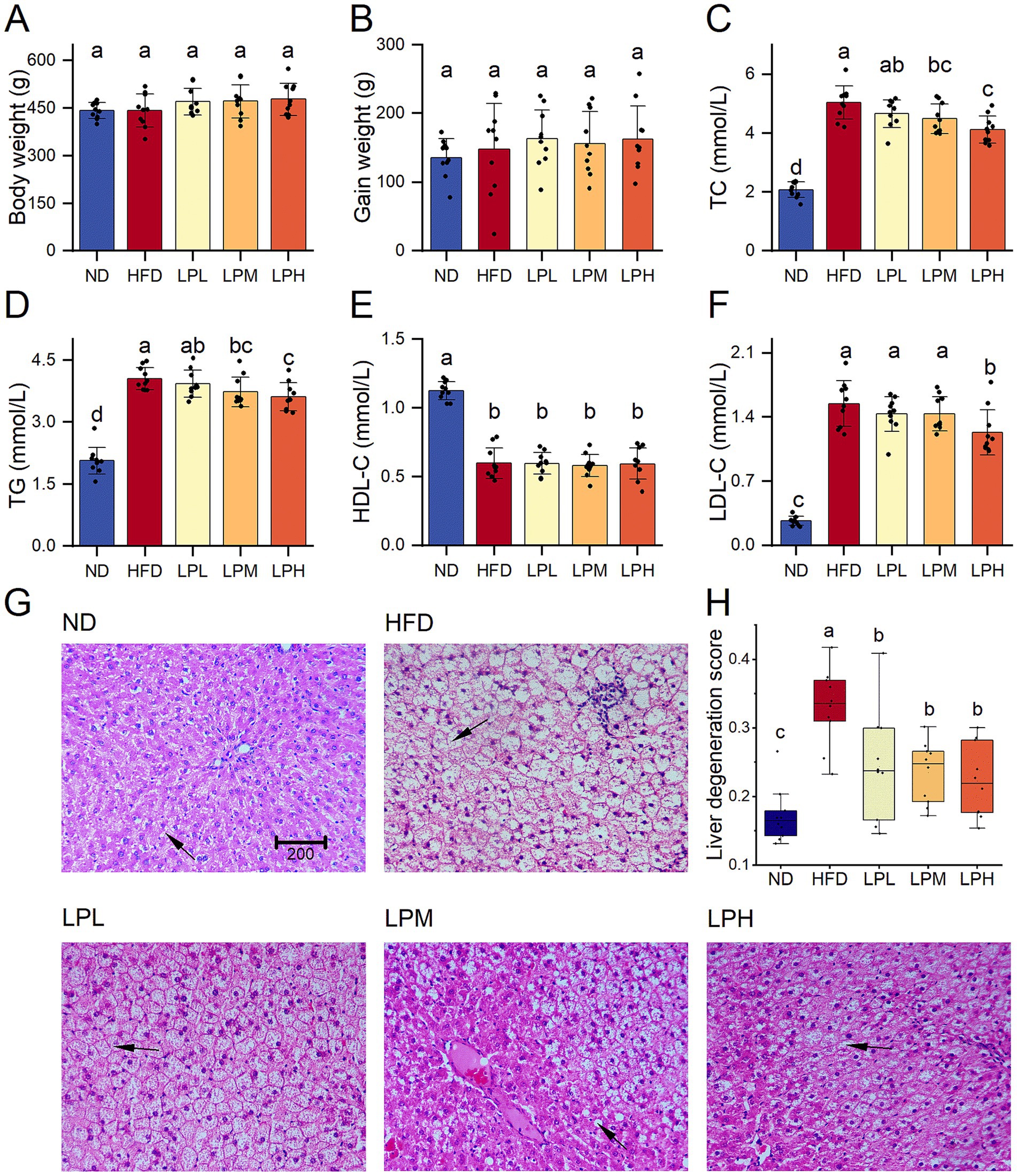
Figure 1. Effects of Lactobacillus paragasseri HM018 on dyslipidemia in rats. (A) Body weight. (B) Gain weight. (C) Serum total cholesterol (TC). (D) Serum triglycerides (TG). (E) High-density lipoprotein cholesterol (HDL-C). (F) Low-density lipoprotein cholesterol (LDL-C). (G) Hematoxylin and eosin staining of pathological sections of adipose tissue. Scale bar: 200 μm. (H) Quantitative analysis of the hepatic adipocyte area. ND: Normal diet; HFD: High-fat diet; LPL: Low-dose intervention group (2.5 × 108 CFU/rat); LPM: Medium-dose intervention group (5 × 108 CFU/rat); LPH: High-dose intervention group (1.5 × 109 CFU/rat); CFU: Colony-forming units. Significance groupings (p < 0.05) identified by Tukey’s test are represented through differential lowercase alphabetic annotations above corresponding boxplots within individual panels.
3.2 HM018 ameliorates the dysbiosis induced by an HFD
To elucidate the impact of HM018 supplementation on intestinal microbiome composition, 16S rRNA gene sequencing was conducted across thirty fecal specimens. A total of 3,914 amplicon sequence variants (ASVs) were obtained, with 1,189, 801, and 2,168 ASVs identified in the HFD model, experimental LPH, and control ND groups, respectively. The numbers of unique ASVs in the ND, HFD, and LPH groups were 1,423, 359, and 218, respectively (Supplementary Figure S2A).
Principal coordinates analysis revealed a significant separation in the gut microbiota structure between the HFD (green) and ND (red) groups after HFD feeding. Following HM018 intervention, the LPH group (blue) also exhibited a distinct microbiota composition compared to the HFD group. These results demonstrate significant HFD-induced perturbations in gut microbiota composition and that HM018 intervention modulated these changes (Figure 2A).
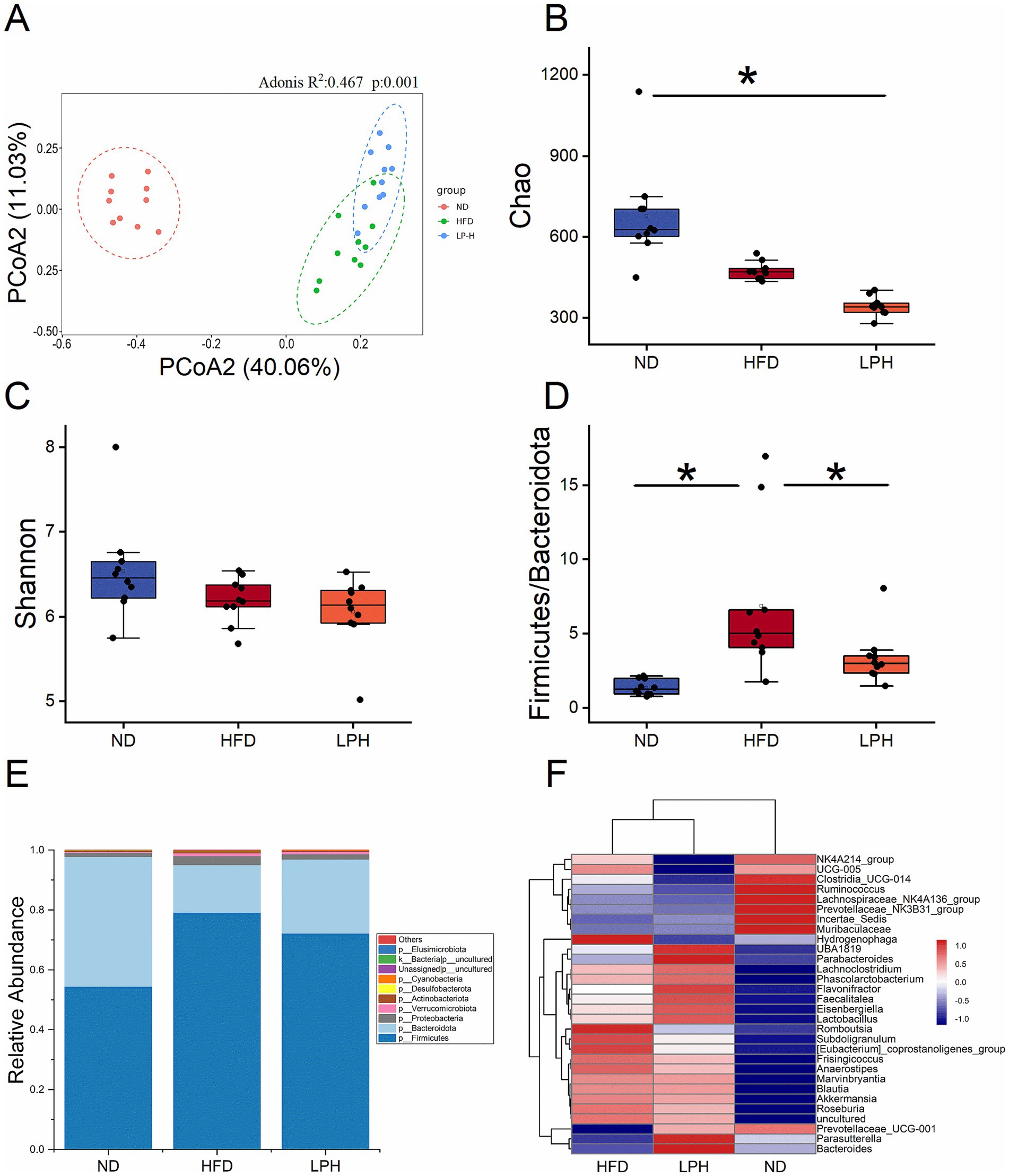
Figure 2. Effects of Lactobacillus paragasseri HM018 intervention on gut microbiota in high-fat diet-fed rats. (A) Principal coordinate analysis (PCoA) plot based on the Bray–Curtis distance matrix, illustrating structural differences in the gut microbiota among the three groups. (B) Chao1 and (C) Shannon indexes reflecting alpha diversity changes across groups. (D) Firmicutes-to-Bacteroidetes ratio in the gut microbiota. (E) Relative distribution of gut microbiota across groups. (F) Heatmap showing variations in the genus-level microbial composition. ND: Normal diet; HFD: High-fat diet; LPH: L. paragasseri HM018 intervention group. *p < 0.05, **p < 0.01 indicate statistical significance.
Alpha diversity analysis revealed that the HFD significantly reduced microbial richness (Chao1 and ACE indices) in the HFD group, and this reduction was further exacerbated after probiotic intervention (Figure 2B; Supplementary Figure S2B). While Shannon and Simpson diversity indices showed no statistically significant variations in the HFD cohort (p > 0.05), both metrics demonstrated consistent decreasing trends. In the dose-dependent analysis, the Chao1 and ACE indices gradually decreased with increasing probiotic dosage, whereas the Shannon and Simpson indices significantly increased (Figure 2C, Supplementary Figure S2C).
Phylum-level analysis identified five predominant bacterial taxa: Firmicutes, Bacteroidetes, Verrucomicrobia, Proteobacteria, and Actinobacteria. Compared with the ND and LPH groups, the relative abundance of Firmicutes was significantly increased in the HFD group, whereas that of Bacteroidetes was significantly decreased. The HFD significantly elevated the Firmicutes-to-Bacteroidetes ratio, which was significantly reduced after HM018 intervention (Figures 2D,E).
At the genus level, the heatmap analysis clustered the microbiota into three groups. Muribaculaceae, Clostridia_UCG-014, and Lachnospiraceae_NK4A136_group were the dominant genera in the ND group. Whereas, HFD significantly suppressed the abundance of Muribaculaceae and significantly increased the abundances of Blautia, Hydrogenophaga, Subdoligranulum, Romboutsia, Akkermansia, and [Eubacterium]_coprostanoligenes_group. In the LPH group, the abundances of Parasutterella, Bacteroides, Parabacteroides, and Prevotellaceae_UCG − 001 significantly increased after HM018 intervention (p < 0.05). In addition, UBA1819, Eisenbergiella, Lachnoclostridium, Roseburia, and Phascolarctobacterium abundances also increased (p > 0.05) (Figure 2F).
3.3 HM018 alters fecal bile acid composition
To delineate gut microbiota-metabolite interactions in hyperlipidemic models, we implemented LC–MS-based untargeted metabolomics profiling across three experimental cohorts (ND, HFD and LPH) Partial least squares-discriminant analysis (PLS-DA) revealed distinct clustering among the three groups, with significant separation between the LPH and HFD groups after HM018 intervention (Figure 3A). Comparative metabolomic analysis identified 1,134 differentially expressed metabolites (DEMs) in the HFD group relative to ND controls, including 288 downregulated and 847 upregulated metabolites. In contrast, the LPH group showed 166 upregulated and 90 downregulated metabolites compared to the HFD group (Figure 3B).
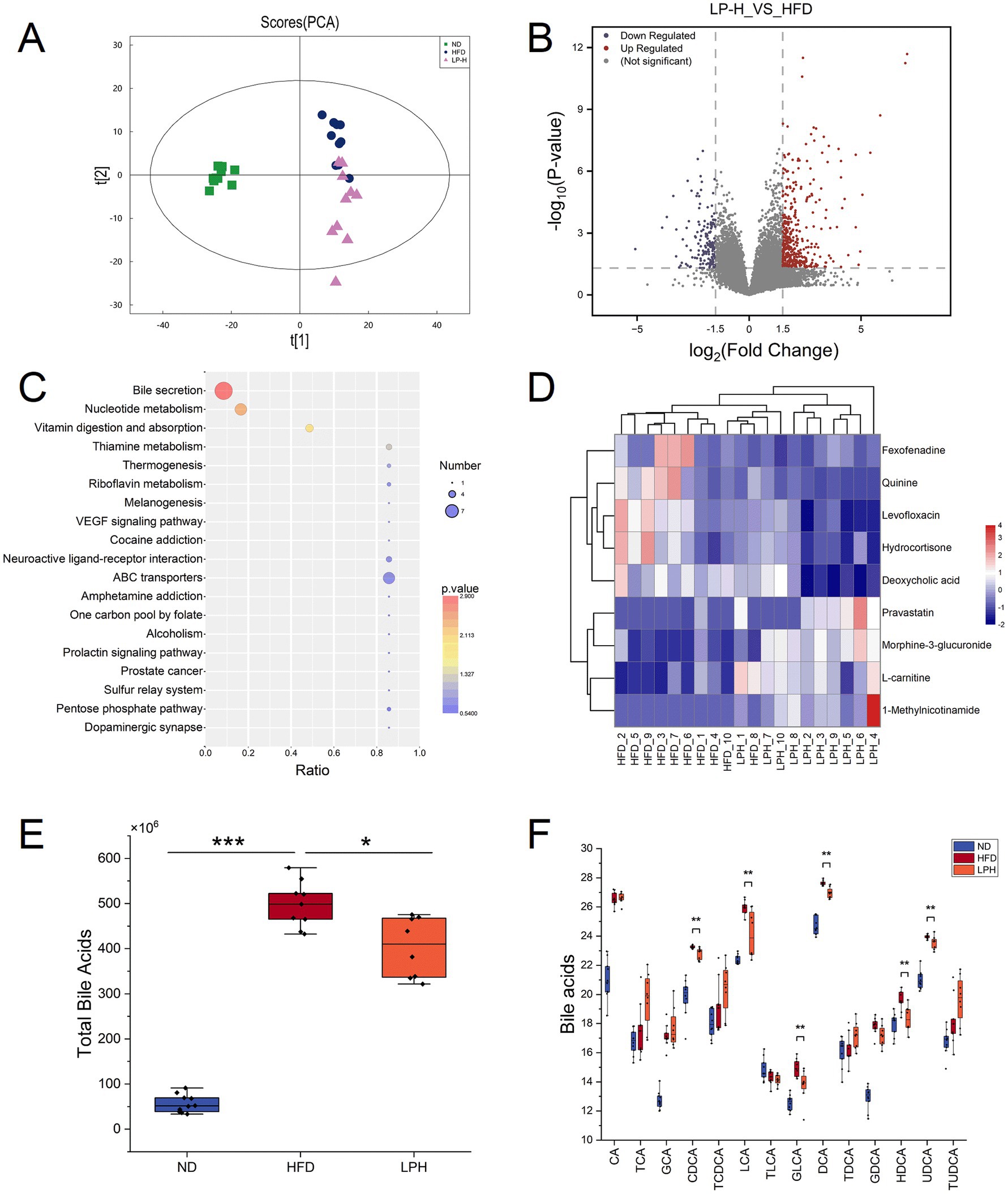
Figure 3. Effects of Lactobacillus paragasseri HM018 intervention on fecal metabolites in high-fat diet (HFD)-fed rats. (A) Principal component analysis (PCA) showing the overall differences in fecal metabolites between the intervention (LPH) and model (HFD) groups. (B) Volcano plot of differential metabolites in the LPH group compared to the HFD group, with red and blue dots representing significantly upregulated and downregulated metabolites, respectively. (C) Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of differential metabolites, with significantly enriched pathways displayed as a dot plot. (D) Heatmap of differential metabolites involved in bile acid metabolism, showing relative abundance changes of key bile acids in the LPH and HFD groups. (E) Differences in total bile acid content in feces before and after intervention. (F) Changes in key bile acids before and after L. paragasseri HM018 intervention, including cholic acid (CA), taurocholic acid (TCA), glycine-conjugated cholic acid (GCA), chenodeoxycholate (CDCA), taurochenodeoxycholic acid (TCDCA), lithocholic acid (LCA), taurolithocholic acid (TLCA), glycolithocholic acid (GLCA), deoxycholic acid (DCA), taurodeoxycholic acid (TDCA), glycine-conjugated deoxycholic acid (GDCA), hyodeoxycholic acid (HDCA), ursodeoxycholic acid (UDCA), and tauroursodeoxycholic acid (TUDCA). *p < 0.05, **p < 0.01 indicate statistical significance.
KEGG pathway enrichment analysis indicated that the differential metabolites were significantly enriched in pathways such as bile secretion, nucleotide metabolism, vitamin digestion and absorption, and thiamine metabolism (Figure 3C). Key metabolites involved in bile secretion included morphine-3-glucuronide, pravastatin, 1-methylnicotinamide, L-carnitine, fexofenadine, deoxycholic acid, quinine, hydrocortisone, and levofloxacin (Figure 3D). Additionally, bile acid homeostasis was significantly altered across the three groups: total bile acid levels were markedly elevated after HFD feeding but significantly reduced following HM018 intervention (Figure 3E). Compared to the HFD group, the LPH group exhibited significant downregulation of free bile acids, such as lithocholic acid (LCA), hyodeoxycholic acid (HDCA), chenodeoxycholate (CDCA), ursodeoxycholic acid (UDCA), and deoxycholic acid (DCA), and upregulation of conjugated bile acids such as taurocholic acid (TCA), glycine-conjugated cholic acid (GCA), taurochenodeoxycholic acid (TCDCA), taurodeoxycholic acid (TDCA), and tauroursodeoxycholic acid (TUDCA) (Figure 3F).
3.4 HM018 intervention improves lipid efflux and intestinal permeability
To elucidate host gene regulation by HM018-derived metabolites, we implemented transcriptomic sequencing of ileal specimens from three rat groups (15 samples total). Transcriptomic PCA clustering showed LPH group profiles were more similar to ND than HFD (Figure 4A). The LPH-HFD comparison yielded 213 significant DEGs, including 117 upregulated and 96 downregulated genes (Figure 4B).
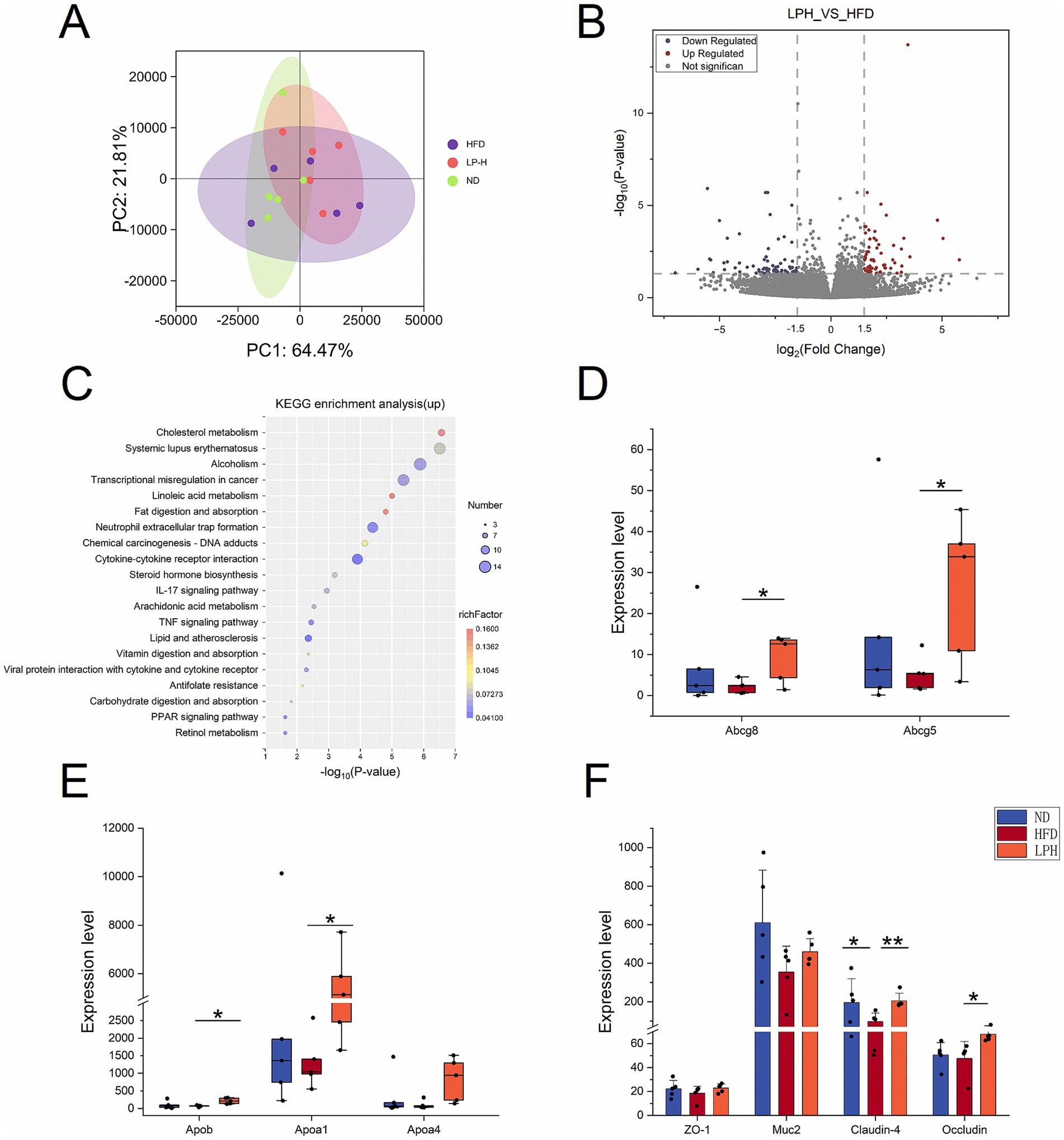
Figure 4. Effects of Lactobacillus paragasseri HM018 intervention on the ileal transcriptome in hyperlipidemic rats. (A) Principal component analysis (PCA) showing transcriptomic differences in ileal samples among the control (ND), model (HFD), and intervention (LPH) groups. (B) Volcano plot of differentially expressed genes in the ileum, with red and blue dots representing significantly upregulated and downregulated genes, respectively. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of upregulated genes, with enriched pathways displayed as a dot plot. (D) Expression levels of ATP-binding cassette transporters 5/8 (Abcg5/8) in ileal samples from the three groups. (E) Expression differences of apolipoprotein A-1 (Apoa1), apolipoprotein A-4 (Apoa4), and apolipoprotein B (Apob) among the three groups. (F) Transcriptional levels of intestinal tight junction components (ZO-1, Claudin-4, Occludin) and the goblet cell mucin 2(MUC2) in three treatment cohorts. *p < 0.05, **p < 0.01 indicate statistical significance.
KEGG pathway enrichment analysis demonstrated that the upregulated genes were significantly enriched in cholesterol metabolism, linoleic acid metabolism, fat digestion and absorption, vitamin digestion and absorption, arachidonic acid metabolism, steroid hormone biosynthesis, carbohydrate digestion and absorption, retinol metabolism, PPAR signaling pathway, and glutathione metabolism (Figure 4C). Notably, the cholesterol efflux-related genes Abcg5 and Abcg8 were markedly upregulated post-intervention, and this result was consistently verified by qPCR (Figure 4D; Supplementary Figure S3). Additionally, key regulators of lipid metabolism, including HDL-associated Apoa1 and intestinal lipid transporter Apoa4, demonstrated significant transcriptional upregulation (Figure 4E).
Downregulated genes were primarily enriched in pathways such as systemic lupus erythematosus, transcriptional dysregulation in cancer, alcoholism, and neutrophil extracellular trap formation (Supplementary Figure S3). To evaluate intestinal barrier integrity, we analyzed the expression of tight junction components (ZO-1, Claudin-4, Occludin) and the goblet cell marker Muc2. Both Occludin and Muc2 were significantly upregulated after HM018 intervention (Figure 4F).
3.5 HM018 intervention ameliorates hepatic glucose and lipid metabolism
To explore the effect of HM018 on rat livers, we performed RNA-seq analysis on liver samples from three groups: ND, LPH, and HFD. PCA revealed that the LPH and HFD groups exhibited more dispersed distributions than the ND group, indicating that both HFD and HM018 intervention significantly altered the hepatic transcriptomic profiles (Figure 5A). Further analysis identified 24 upregulated and 35 downregulated genes in the LPH group compared to those in the HFD group (Figure 5B). Additionally, Pathway enrichment profiling identified key lipid metabolism-related signaling cascades: JAK–STAT, TGF-β, FoxO transcriptional regulation, insulin signaling, adipocytokine communication, and diabetes pathways (Figure 5C; Supplementary Figure S4).
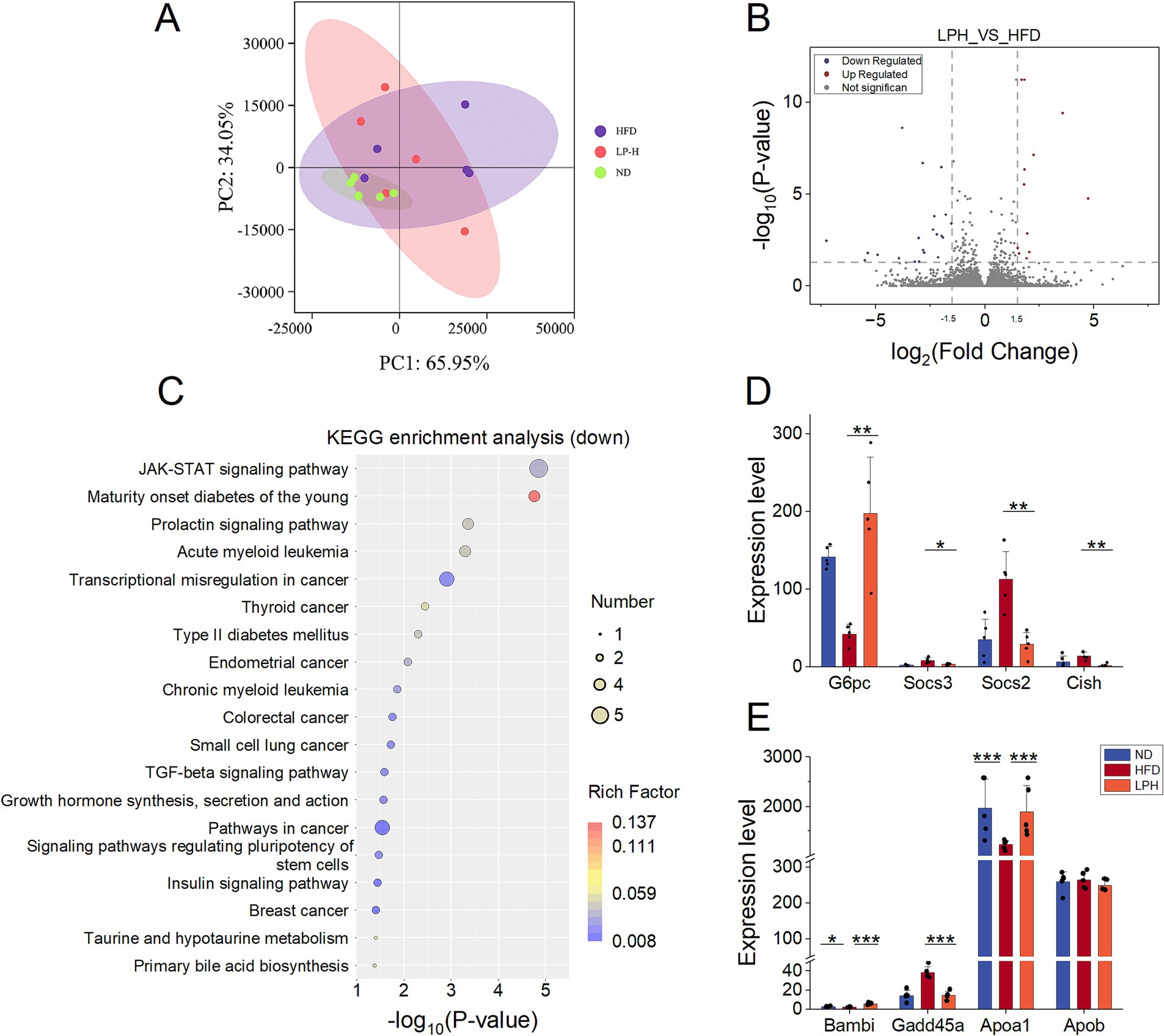
Figure 5. Effects of Lactobacillus paragasseri HM018 intervention on hepatic transcriptomics in hyperlipidemic rats. (A) Principal component analysis (PCA) revealed the differences in the transcriptomes of the ileum samples. (B) Volcano plot of differentially expressed genes in the liver. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of downregulated genes, with significantly enriched pathways displayed as a dot plot. (D) Expression levels of genes related to lipogenesis and metabolism in the liver, including glucose-6-phosphatase (G6PC), suppressor of cytokine signaling 2/3 (Socs2 and Socs3), and cytokine-inducible SH2-containing protein (Cish). (E) Expression differences of BMP and activin membrane-bound inhibitor (Bambi), growth arrest and DNA damage-inducible alpha (Gadd45a), apolipoprotein A-1 (Apoa1), and apolipoprotein B (Apob) among the three groups (*p < 0.05, **p < 0.01).
Among the genes related to lipogenesis, although not statistically significant, their transcriptional levels generally showed a downward trend after HM018 intervention (Supplementary Figure S4). Meanwhile, analysis of glucose metabolism-related genes revealed that HM018 intervention significantly upregulated glucose-6-phosphatase (G6pc) expression, while downregulating suppressor of cytokine signaling 2/3 (Socs2 and Socs3) and cytokine-inducible SH2-containing protein (Cish). Quantitative PCR (QPCR) results further confirmed that the expression of Socs2 and Cish in the LPH group was significantly reduced, validating this regulatory trend (Figure 5D; Supplementary Figure S4). Notably, HM018 intervention restored the expression of BMP and activin membrane-bound inhibitor (Bambi), growth arrest and DNA damage-inducible alpha (Gadd45a), and Apoa1 to levels comparable to those in the ND group. In contrast, Apob (low-density lipoprotein metabolism regulator) exhibited no significant expression differences across experimental groups (Figure 5E). These results suggest that HM018 may ameliorate hepatic metabolic disorders by multi-target regulation of key genes involved in glucose and lipid metabolism.
3.6 Integrated analysis of microbiome, metabolome, and transcriptome correlations
We first conducted a joint analysis of the four significantly altered bacterial phyla (Firmicutes, Bacteroidetes, Verrucomicrobia, and Proteobacteria) post-HM018 intervention using lipid profiles across the three experimental groups. The results showed that, except for Bacteroidetes, the relative abundances of Firmicutes, Verrucomicrobia, and Proteobacteria were inversely related to HDL levels, while demonstrating positive correlations with triglyceride, total cholesterol, and LDL levels (Supplementary Figure S5A). Genus-level analysis identified positive correlations between Blautia, Romboutsia, Subdoligranulum abundances and dyslipidemia severity scores, whereas Muribaculaceae, Incertae-Sedis, Lachnospiraceae_NK4A136_group, Ruminococcus, and Prevotellaceae_UCG-001 were significantly correlated with improved hyperlipidemia (Figure 6A).
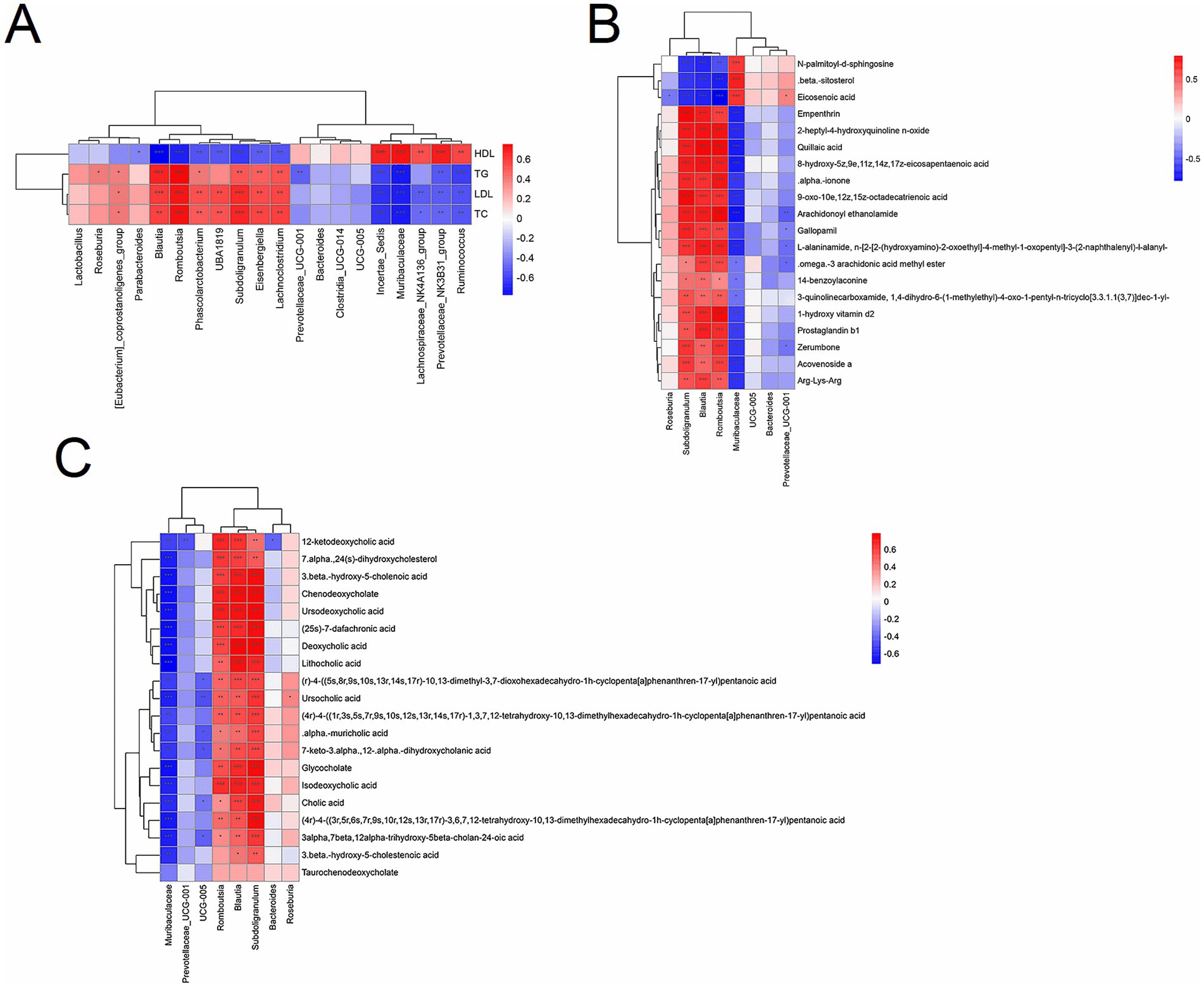
Figure 6. Correlation analysis between lipid profiles and the microbiome/metabolome. (A) Spearman’s correlation analysis between lipid profiles and gut microbiota at the genus level. (B) Spearman’s correlation analysis between gut microbiota and differential metabolites. (C) Spearman’s correlation analysis between gut microbiota and differential bile acids and their derivatives. The color gradient ranges from blue (negative correlation) to red (positive correlation). *p < 0.05, **p < 0.01, and ***p < 0.001 indicate significant correlations.
In the joint analysis of differential metabolites and lipid parameters, β-sitosterol, eicosenoic acid, and N-palmitoyl-D-sphingosine were strongly negatively correlated with TG, TC, and LDL (r < −0.7; p < 0.05), but strongly positively correlated with HDL (r > 0.7; p < 0.05). Increased concentrations of these metabolites were associated with better prognostic outcomes, whereas other metabolites were significantly enriched in hypercholesterolemic rats (|r| > 0.7) (Supplementary Figure S5B).
A combined analysis of differential metabolites and bacterial genera demonstrated that Muribaculaceae, a dominant genus in the ND group, showed strong positive correlations with eicosenoic acid, β-sitosterol, and N-palmitoyl-D-sphingosine, but negative correlations with other compounds (|r| > 0.7). In the intervention group, the increased abundance of Prevotellaceae_UCG-001 weakly correlated with eicosenoic acid and ω-3 arachidonic acid methyl ester (0.5 > |r| > 0.3). Notably, the dominant genera in the model group, Blautia and Subdoligranulum, displayed moderate negative correlations with β-sitosterol (−0.6 > r > −0.7) and positive correlations with compounds such as empenthrin (r > 0.3) (Figure 6B).
Furthermore, HM018 intervention significantly altered the composition of the bile acid pool in rats. Joint analysis of differential bile acids and their derivatives with bacterial genera revealed that Muribaculaceae was negatively correlated with most bile acids, whereas Blautia, Subdoligranulum, and Romboutsia showed significant positive correlations with bile acids (Figure 6C). These findings suggest that HM018 may ameliorate dyslipidemia by modulating the interaction between the gut microbiota and bile acid metabolism.
4 Discussion
This study demonstrates that L. paragasseri HM018 could significantly improve HFD-induced hyperlipidemia in rats, with marked serum TG, TC, and LDL level as well as adipocyte size reductions post-intervention (p < 0.05). With an increasing HM018 intervention dose, serum TC and TG levels displayed a gradient downward trend, and when the dose reached 109 CFU/rat, the TC, TG and LDL levels were significantly reduced (p < 0.05).
HFD alters gut microbiota composition, whereas probiotic intervention remodels the microbial structure and alleviates dysbiosis (Smirnova et al., 2022; Kappel et al., 2023). HFD increases the Firmicutes-to-Bacteroidetes ratio, which is associated with metabolic disorders such as dyslipidemia (Yasuzawa et al., 2022; Xu et al., 2023a), and elevates the abundance of Clostridiales within Firmicutes (Hewady et al., 2024), consistent with our findings. Obesity is closely linked to inflammation, as fat accumulation and localized inflammation reduce the expression of tight junction proteins (e.g., claudin-1, occludin, and ZO-1) (Chelakkot et al., 2018; Acciarino et al., 2024), thereby increasing intestinal permeability and triggering inflammation (Kim et al., 2024). HM018 intervention notably improved intestinal permeability.
Cholesterol, a key precursor of bile acid synthesis, is converted into primary bile acids in hepatocytes via the classical (mediated by CYP7A1) and alternative (mediated by CYP27A1) pathways. Subsequently, bile acid-CoA synthase and bile acid-CoA:amino acid N-acyltransferase catalyze the conjugation of primary bile acids with taurine or glycine to form bile salts stored in the gallbladder (Collins et al., 2023; Xu et al., 2023b). During the fed state, BAs are released into the gastrointestinal tract, where gut microbiota hydrolyze conjugated bile acids into free primary BAs via bile salt hydrolase (BSH), followed by 7α-dehydroxylase-mediated 7α-dehydroxylation to generate secondary BAs (DCA and LCA) (Long et al., 2017). High-cholesterol, low-fiber diets upregulate hepatic bile acid synthesis and elevate systemic bile acid levels (Chaudhari et al., 2021), further promoting lipid absorption and obesity. HM018 intervention significantly reduced total bile acid levels. HFD enriches gut microbiota with BSH and 7α-dehydroxylase activities, particularly Blautia, Eubacterium, and Clostridium species, leading to intestinal deoxycholic acid (DCA) accumulation and significant conjugated bile acid (T-α-MCA, T-β-MCA, TCA, TUDCA, and GCA) suppression, thereby establishing a pro-obesity bile acid metabolic disorder (Lin et al., 2019; Liu et al., 2021). External interventions could effectively modulate this dysbiosis by suppressing BSH-producing Clostridium while promoting Bacteroidetes and Akkermansia proliferation. This microbial remodeling reduces primary and increases conjugated bile acids, ultimately improving dyslipidemia and hepatic steatosis (Zhang et al., 2024). Recent studies have shown that a ketogenic diet inhibits gut microbial bile salt hydrolase, elevating circulating TDCA and TUDCA levels to reduce energy absorption, promote weight loss, and lower fasting glucose (Li et al., 2024). Elevated circulating TDCA, glycodeoxycholic acid, and glycoursodeoxycholic acid levels, along with reduced fecal DCA and UDCA levels, correlate with improved weight and glycemic control in patients with obesity (Chaudhari et al., 2021). Moreover, in the Lactobacillus plantarum HT121-associated hyperlipidemia improvement, serum TG, TC, and LDL levels significantly positively correlated with the relative abundance of the Blautia and Clostridium genera (Li et al., 2020). Our results demonstrate that HM018 intervention suppresses Blautia and Clostridium strains expressing BSH and 7α-dehydroxylase activities while promoting Bacteroidetes proliferation. This modulation significantly reduces free bile acid levels (including LCA, HDCA, CDCA, UDCA, and DCA; p < 0.05) and upregulates conjugated bile acids (TCA, GCA, TCDCA, TDCA, and TUDCA; 0.05 < p < 0.1), thereby effectively improving hyperlipidemia.
Reverse cholesterol transport (RCT), a critical process for eliminating excess cholesterol from the peripheral tissues to the liver, is driven by apolipoprotein A-I (ApoaI). Synthesized in the liver and intestine, ApoaI is a major component of HDL (Duong et al., 2008; Luo et al., 2020) and facilitates cholesterol efflux for biliary excretion (Liu et al., 2025). Mutations, enzymatic modifications, or metabolic alterations in ApoaI reduce HDL quality and contribute to dyslipidemia and CVDs (Bhale and Venkataraman, 2022). Transcriptomic analysis revealed that HM018 significantly upregulated ApoaI expression in the liver and ileum.
Under HFD conditions, the transintestinal cholesterol excretion (TICE) pathway, which is independent of biliary secretion and facilitated by Abcg5/8, becomes crucial in the elimination of cholesterol via feces (Luo et al., 2020; Xu et al., 2023b). Genetic variants of Abcg5/8 elevate plasma β-sitosterol and LDL cholesterol (Tada et al., 2022; Alenbawi et al., 2024). Our correlation analysis showed increased fecal β-sitosterol and cholesterol sulfate levels in the intervention group, suggesting that HM018 upregulates ileal Abcg5/8 transcription to enhance sterol excretion via TICE, thereby alleviating hyperlipidemia.
Several hepatic genes are associated with lipogenesis and metabolism. Among these, the upregulation of GADD45A promotes subcutaneous fat deposition and obesity (You et al., 2023b). Gadd45a knockout mice exhibited enhanced lipolysis, improved energy utilization, and increased resistance to HFD-induced obesity (You et al., 2023a). Bambi knockout exacerbates HFD-induced metabolic disorders, including hepatic steatosis, glucose intolerance, and insulin resistance (Chen X. et al., 2021), whereas Bambi overexpression suppresses obesity (Weber et al., 2023). Following HM018 intervention, the transcriptional level of Gadd45a was significantly downregulated, whereas that of Bambi was upregulated, indicating that HM018 improved insulin sensitivity and restored glucose metabolism. Additionally, HFD upregulated the transcriptional expression of Cish, Socs2, and Socs3. Previous studies have demonstrated that Cish-deficient mice fed an HFD exhibit reduced body weight and fat mass, along with improved insulin sensitivity (Chen G. et al., 2021; Lynch et al., 2024). However, glucose tolerance remains unchanged, accompanied by increased glucagon levels and sensitivity (Naser et al., 2022; Xiao et al., 2022). Socs3 is upregulated in obesity and inhibits leptin and insulin signaling (Palanivel et al., 2012), while its downregulation reduces hepatic lipid levels in obese Sprague–Dawley rats (Jiang et al., 2024). Socs2 upregulation promotes lipogenesis by negatively regulating growth hormone signaling and suppressing lipolysis (Yang et al., 2013). After HM018 intervention, hepatic expression of Cish, Socs2, and Socs3 was significantly downregulated, thereby attenuating their inhibitory effects on hormone and glucagon signaling (Gaudy et al., 2010; Palanivel et al., 2012; Zadjali et al., 2012). G6pc, a rate-limiting enzyme in glycogenolysis and gluconeogenesis, exhibited suppressed transcription in the HFD group. However, HM018 intervention restored G6pc expression to control levels. These findings suggest that HM018 may enhance glucagon sensitivity to promote glucose production, thereby modulating the utilization of glucose and lipids This study demonstrates that HM018 can significantly improve the lipid profile in high-cholesterol rats. However, it is important to note that there are species-specific differences in bile acid composition and binding between humans and rodents. Therefore, the lipid-lowering effects of HM018 in humans still need to be explored through further clinical studies (Wahlström et al., 2016).
In summary, L. paragasseri HM018 alleviated HFD-induced dyslipidemia by restoring hepatic and intestinal metabolic homeostasis. It modulated gut microbiota and bile acid metabolism to promote cholesterol excretion via the RCT and TICE pathways. Additionally, HM018 enhanced insulin sensitivity and glucose metabolism by modulating the expression of lipid-related genes.
5 Conclusion
This study demonstrates that L. paragasseri HM018 significantly ameliorates HFD-induced hyperlipidemia in rats. HM018 not only effectively regulates lipid profiles and remodels gut microbiota composition but also improves intestinal permeability. Furthermore, it modulates bile acid homeostasis, promotes RCT, and enhances cholesterol excretion. At the transcriptional level, HM018 significantly improves insulin sensitivity and alleviates glucose metabolism disorders. These findings provide a solid theoretical foundation for the potential application of HM018 as a therapeutic intervention in hyperlipidemia and related metabolic diseases.
Data availability statement
The data presented in the study are deposited in the Mendeley Data repository (https://data.mendeley.com), under the following DOI: 10.17632/r75rntjrnn.1.
Ethics statement
The animal study was approved by the Ethics Committee of Beijing Union University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
CY: Methodology, Conceptualization, Investigation, Writing – review & editing. XL: Writing – review & editing, Investigation, Supervision, Conceptualization, Methodology. MT: Writing – original draft, Data curation. LL: Writing – original draft, Data curation. XC: Writing – review & editing, Visualization. XY: Writing – original draft, Data curation. JH: Visualization, Writing – original draft. JZ: Supervision, Writing – review & editing. WQ: Writing – review & editing, Supervision. YZ: Writing – review & editing, Supervision, Methodology. LC: Writing – review & editing, Methodology, Supervision, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Special Funds for Guiding Local Scientific and Technological Development by the Central Government [Grant No. GuikeZY22096025], Beijing Science and Technology Plan [Grant No. Z221100006422012], Beijing Capital Agribusiness & Foods Group Science and Technology Project [SNSPKJ(2022)03].
Acknowledgments
We sincerely thank Shanghai Applied Protein Technology Co., Ltd. for their expert support and state-of-the-art resources, which were instrumental in generating the essential data for this study.
Conflict of interest
CY, XL, MT, LL, XC, XY, JH, JZ, WQ, and LC were employed by Beijing Sanyuan Foods Co. Ltd.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1599931/full#supplementary-material
References
Acciarino, A., Diwakarla, S., Handreck, J., Bergola, C., Sahakian, L., and McQuade, R. M. (2024). The role of the gastrointestinal barrier in obesity-associated systemic inflammation. Obes. Rev. 25:e13673. doi: 10.1111/obr.13673
Alenbawi, J., Al-Sarraj, Y. A., Umlai, U.-K. I., Kadhi, A., Hendi, N. N., Nemer, G., et al. (2024). Genome-wide association study and meta-analysis of phytosterols identifies a novel locus for serum levels of campesterol. Hum. Genomics 18:85. doi: 10.1186/s40246-024-00649-x
Bhale, A. S., and Venkataraman, K. (2022). Leveraging knowledge of HDLs major protein ApoA1: structure, function, mutations, and potential therapeutics. Biomed. Pharmacother. 154:113634. doi: 10.1016/j.biopha.2022.113634
Chaudhari, S. N., Harris, D. A., Aliakbarian, H., Luo, J. N., Henke, M. T., Subramaniam, R., et al. (2021). Bariatric surgery reveals a gut-restricted TGR5 agonist with anti-diabetic effects. Nat. Chem. Biol. 17, 20–29. doi: 10.1038/s41589-020-0604-z
Chelakkot, C., Ghim, J., and Ryu, S. H. (2018). Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 50, 1–9. doi: 10.1038/s12276-018-0126-x
Chen, G., Chen, J., Wu, J., Ren, X., Li, L., Lu, S., et al. (2021). Integrative analyses of mRNA expression profile reveal SOCS2 and CISH play important roles in GHR mutation-induced excessive abdominal fat deposition in the sex-linked dwarf chicken. Front. Genet. 11:610605. doi: 10.3389/fgene.2020.610605
Chen, X., Zhao, C., Xu, Y., Huang, K., Wang, Y., Wang, X., et al. (2021). Adipose-specific BMP and activin membrane-bound inhibitor (BAMBI) deletion promotes adipogenesis by accelerating ROS production. J. Biol. Chem. 296:100037. doi: 10.1074/jbc.RA120.014793
Collins, S. L., Stine, J. G., Bisanz, J. E., Okafor, C. D., and Patterson, A. D. (2023). Bile acids and the gut microbiota: metabolic interactions and impacts on disease. Nat. Rev. Microbiol. 21, 236–247. doi: 10.1038/s41579-022-00805-x
Dang, Y., Xu, J., Yang, Y., Li, C., Zhang, Q., Zhou, W., et al. (2020). Ling-gui-zhu-Gan decoction alleviates hepatic steatosis through SOCS2 modification by N6-methyladenosine. Biomed. Pharmacother. 127:109976. doi: 10.1016/j.biopha.2020.109976
Duong, P. T., Weibel, G. L., Lund-Katz, S., Rothblat, G. H., and Phillips, M. C. (2008). Characterization and properties of preβ-HDL particles formed by ABCA1-mediated cellular lipid efflux to apoA-I. J. Lipid Res. 49, 1006–1014. doi: 10.1194/jlr.M700506-JLR200
Gao, J., Wang, J., Zhao, L.-L., Yao, T.-T., Chen, Y., Ma, J., et al. (2021). Gut Lactobacillus level is a predictive marker for coronary atherosclerotic lesions Progress and prognosis in patients with acute coronary syndrome. Front. Cell. Infect. Microbiol. 11:687827. doi: 10.3389/fcimb.2021.687827
Gaudy, A. M., Clementi, A. H., Campbell, J. S., Smrcka, A. V., and Mooney, R. A. (2010). Suppressor of cytokine signaling-3 is a glucagon-inducible inhibitor of PKA activity and gluconeogenic gene expression in hepatocytes. J. Biol. Chem. 285, 41356–41365. doi: 10.1074/jbc.M110.159111
Hewady, S., Manuel, C. R., Pasquali, C., Koya, J., and Reznik, S. E. (2024). OM-85 attenuates high-fat diet-induced obesity, insulin resistance, gut dysbiosis and nonalcoholic steatohepatitis in a murine model. Biomed. Pharmacother. 181:117710. doi: 10.1016/j.biopha.2024.117710
Howard, E. J., Lam, T. K. T., and Duca, F. A. (2022). The gut microbiome: connecting diet, glucose homeostasis, and disease. Annu. Rev. Med. 73, 469–481. doi: 10.1146/annurev-med-042220-012821
Hu, L., Hu, B., Zhang, L., Hu, Y., Zhang, Y., Zhang, R., et al. (2024). Role of gut microbiota and metabolomics in the lipid-lowering efficacy of statins among Chinese patients with coronary heart disease and hypercholesterolemia. Front. Cell. Infect. Microbiol. 14:1408581. doi: 10.3389/fcimb.2024.1408581
James, S. L., Abate, D., Abate, K. H., Abay, S. M., Abbafati, C., Abbasi, N., et al. (2018). Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet 392, 1789–1858. doi: 10.1016/s0140-6736(18)32279-7
Jang, S., Lee, M.-S., Kang, S.-A., Kim, C.-T., and Kim, Y. (2022). Portulaca oleracea L. extract regulates hepatic cholesterol metabolism via the AMPK/MicroRNA-33/34a pathway in rats fed a high-cholesterol diet. Nutrients 14:3330. doi: 10.3390/nu14163330
Jia, X., Xu, W., Zhang, L., Li, X., Wang, R., and Wu, S. (2021). Impact of gut microbiota and microbiota-related metabolites on hyperlipidemia. Front. Cell. Infect. Microbiol. 11:634780. doi: 10.3389/fcimb.2021.634780
Jiang, W., Tan, J., Zhang, J., Deng, X., He, X., Zhang, J., et al. (2024). Polysaccharides from Dendrobium officinale improve obesity-induced insulin resistance through the gut microbiota and the SOCS3-mediated insulin receptor substrate-1 signaling pathway. J. Sci. Food Agric. 104, 3437–3447. doi: 10.1002/jsfa.13229
Kappel, B. A., De Angelis, L., Puetz, A., Ballanti, M., Menghini, R., Marx, N., et al. (2023). Antibiotic-induced gut microbiota depletion exacerbates host hypercholesterolemia. Pharmacol. Res. 187:106570. doi: 10.1016/j.phrs.2022.106570
Khanali, J., Ghasemi, E., Rashidi, M.-M., Ahmadi, N., Ghamari, S.-H., Azangou-Khyavy, M., et al. (2023). Prevalence of plasma lipid abnormalities and associated risk factors among Iranian adults based on the findings from STEPs survey 2021. Sci. Rep. 13:15499. doi: 10.1038/s41598-023-42341-5
Kim, S., Seo, S.-U., and Kweon, M.-N. (2024). Gut microbiota-derived metabolites tune host homeostasis fate. Semin. Immunopathol. 46:2. doi: 10.1007/s00281-024-01012-x
Lamb, Y. N. (2020). Rosuvastatin/ezetimibe: A review in hypercholesterolemia. Am. J. Cardiovasc. Drugs 20, 381–392. doi: 10.1007/s40256-020-00421-1
Li, X., Xiao, Y., Song, L., Huang, Y., Chu, Q., Zhu, S., et al. (2020). Effect of Lactobacillus plantarum HT121 on serum lipid profile, gut microbiota, and liver transcriptome and metabolomics in a high-cholesterol diet–induced hypercholesterolemia rat model. Nutrition 79-80:110966. doi: 10.1016/j.nut.2020.110966
Li, X., Yang, J., Zhou, X., Dai, C., Kong, M., Xie, L., et al. (2024). Ketogenic diet-induced bile acids protect against obesity through reduced calorie absorption. Nat. Metab. 6, 1397–1414. doi: 10.1038/s42255-024-01072-1
Lin, H., An, Y., Tang, H., and Wang, Y. (2019). Alterations of bile acids and gut microbiota in obesity induced by high fat diet in rat model. J. Agric. Food Chem. 67, 3624–3632. doi: 10.1021/acs.jafc.9b00249
Liu, X., Mao, B., Gu, J., Wu, J., Cui, S., Wang, G., et al. (2021). Blautia —a new functional genus with potential probiotic properties? Gut Microbes 13, 1–21. doi: 10.1080/19490976.2021.1875796
Liu, X., Zhang, Z., Aguirre, T., Shipton, M. L., Fu, L., Du, J., et al. (2025). Inhibiting IP6K1 confers atheroprotection by elevating circulating apolipoprotein A-I. Metabolism 163:156098. doi: 10.1016/j.metabol.2024.156098
Long, S. L., Gahan, C. G. M., and Joyce, S. A. (2017). Interactions between gut bacteria and bile in health and disease. Mol. Asp. Med. 56, 54–65. doi: 10.1016/j.mam.2017.06.002
Luo, J., Yang, H., and Song, B.-L. (2020). Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 21, 225–245. doi: 10.1038/s41580-019-0190-7
Lynch, D. M., Forrester, B., Webb, T., and Ciulli, A. (2024). Unravelling the druggability and immunological roles of the SOCS-family proteins. Front. Immunol. 15:1449397. doi: 10.3389/fimmu.2024.1449397
Naser, W., Maymand, S., Rivera, L. R., Connor, T., Liongue, C., Smith, C. M., et al. (2022). Cytokine-inducible SH2 domain containing protein contributes to regulation of adiposity, food intake, and glucose metabolism. FASEB J. 36:e22320. doi: 10.1096/fj.202101882R
Naudin, C. R., Maner-Smith, K., Owens, J. A., Wynn, G. M., Robinson, B. S., Matthews, J. D., et al. (2020). Lactococcus lactis subspecies cremoris elicits protection against metabolic changes induced by a Western-style diet. Gastroenterology 159, 639–651.e5. doi: 10.1053/j.gastro.2020.03.010
Palanivel, R., Fullerton, M. D., Galic, S., Honeyman, J., Hewitt, K. A., Jorgensen, S. B., et al. (2012). Reduced Socs3 expression in adipose tissue protects female mice against obesity-induced insulin resistance. Diabetologia 55, 3083–3093. doi: 10.1007/s00125-012-2665-3
Patnode, M. L., Guruge, J. L., Castillo, J. J., Couture, G. A., Lombard, V., Terrapon, N., et al. (2021). Strain-level functional variation in the human gut microbiota based on bacterial binding to artificial food particles. Cell Host Microbe 29, 664–673.e5. doi: 10.1016/j.chom.2021.01.007
Ross, F. C., Patangia, D., Grimaud, G., Lavelle, A., Dempsey, E. M., Ross, R. P., et al. (2024). The interplay between diet and the gut microbiome: implications for health and disease. Nat. Rev. Microbiol. 22, 671–686. doi: 10.1038/s41579-024-01068-4
Roth, G. A., Mensah, G. A., Johnson, C. O., Addolorato, G., Ammirati, E., Baddour, L. M., et al. (2020). Global burden of cardiovascular diseases and risk factors, 1990–2019. J. Am. Coll. Cardiol. 76, 2982–3021. doi: 10.1016/j.jacc.2020.11.010
Smirnova, E., Muthiah, M. D., Narayan, N., Siddiqui, M. S., Puri, P., Luketic, V. A., et al. (2022). Metabolic reprogramming of the intestinal microbiome with functional bile acid changes underlie the development of NAFLD. Hepatology 76, 1811–1824. doi: 10.1002/hep.32568
Su, X., Peng, H., Chen, X., Wu, X., and Wang, B. (2022). Hyperlipidemia and hypothyroidism. Clin. Chim. Acta 527, 61–70. doi: 10.1016/j.cca.2022.01.006
Tada, M. T., Rocha, V. Z., Lima, I. R., Oliveira, T. G. M., Chacra, A. P., Miname, M. H., et al. (2022). Screening of ABCG5 and ABCG8 genes for Sitosterolemia in a familial hypercholesterolemia Cascade screening program. Circ. Genom. Precis. Med. 15:e003390. doi: 10.1161/CIRCGEN.121.003390
Tanizawa, Y., Tada, I., Kobayashi, H., Endo, A., Maeno, S., Toyoda, A., et al. (2018). Lactobacillus paragasseri sp. nov., a sister taxon of Lactobacillus gasseri, based on whole-genome sequence analyses. Int. J. Syst. Evol. Microbiol. 68, 3512–3517. doi: 10.1099/ijsem.0.003020
Wahlström, A., Sayin, S. I., Marschall, H.-U., and Bäckhed, F. (2016). Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24, 41–50. doi: 10.1016/j.cmet.2016.05.005
Weber, F., Treeck, O., Mester, P., and Buechler, C. (2023). Expression and function of BMP and Activin membrane-bound inhibitor (BAMBI) in chronic liver diseases and hepatocellular carcinoma. Int. J. Mol. Sci. 24:3473. doi: 10.3390/ijms24043473
Wei, J., Luo, J., Yang, F., Feng, X., Zeng, M., Dai, W., et al. (2024). Cultivated Enterococcus faecium B6 from children with obesity promotes nonalcoholic fatty liver disease by the bioactive metabolite tyramine. Gut Microbes 16:2351620. doi: 10.1080/19490976.2024.2351620
Xiao, F., Deng, J., Jiao, F., Hu, X., Jiang, H., Yuan, F., et al. (2022). Hepatic cytokine-inducible SH2-containing protein (CISH) regulates gluconeogenesis via cAMP-responsive element binding protein (CREB). FASEB J. 36:e22541. doi: 10.1096/fj.202200870R
Xu, H., Fang, F., Wu, K., Song, J., Li, Y., Lu, X., et al. (2023a). Gut microbiota-bile acid crosstalk regulates murine lipid metabolism via the intestinal FXR-FGF19 axis in diet-induced humanized dyslipidemia. Microbiome 11:262. doi: 10.1186/s40168-023-01709-5
Xu, H., Xin, Y., Wang, J., Liu, Z., Cao, Y., Li, W., et al. (2023b). The TICE pathway: mechanisms and potential clinical applications. Curr. Atheroscler. Rep. 25, 653–662. doi: 10.1007/s11883-023-01147-6
Xu, W., Yu, J., Yang, Y., Li, Z., Zhang, Y., Zhang, F., et al. (2023). Strain-level screening of human gut microbes identifies Blautia producta as a new anti-hyperlipidemic probiotic. Gut Microbes 15:2228045. doi: 10.1080/19490976.2023.2228045
Yang, H. L., Feng, M., Tan, X., Yan, G. Y., and Sun, C. (2013). The role of SOCS2 in recombinant human growth hormone (rhGH) regulating lipid metabolism in high-fat-diet-induced obesity mice. Mol. Biol. Rep. 40, 2319–2326. doi: 10.1007/s11033-012-2313-5
Yasuzawa, T., Nishi, R., Ishitani, S., Matsuo, O., and Ueshima, S. (2022). Effects of Enzamin, a microbial product, on alterations of intestinal microbiota induced by a high-fat diet. Nutrients 14:4743. doi: 10.3390/nu14224743
You, W., Liu, S., Ji, J., Ling, D., Tu, Y., Zhou, Y., et al. (2023a). Growth arrest and DNA damage-inducible alpha regulates muscle repair and fat infiltration through ATP synthase F1 subunit alpha. J. Cachexia. Sarcopenia Muscle 14, 326–341. doi: 10.1002/jcsm.13134
You, W., Liu, S., Li, J., Tu, Y., and Shan, T. (2023b). GADD45A regulates subcutaneous fat deposition and lipid metabolism by interacting with Stat1. BMC Biol. 21:212. doi: 10.1186/s12915-023-01713-z
Zadjali, F., Santana-Farre, R., Vesterlund, M., Carow, B., Mirecki-Garrido, M., Hernandez-Hernandez, I., et al. (2012). SOCS2 deletion protects against hepatic steatosis but worsens insulin resistance in high-fat-diet-fed mice. FASEB J. 26, 3282–3291. doi: 10.1096/fj.12-205583
Zhang, K.-X., Zhu, Y., Song, S.-X., Bu, Q.-Y., You, X.-Y., Zou, H., et al. (2024). Ginsenoside Rb1, compound K and 20(S)-Protopanaxadiol attenuate high-fat diet-induced hyperlipidemia in rats via modulation of gut microbiota and bile acid metabolism. Molecules 29:1108. doi: 10.3390/molecules29051108
Keywords: hyperlipidemia, breast milk-derived probiotics, Lactobacillus paragasseri, gut microbiota, bile acid metabolism, cholesterol efflux
Citation: Yao C, Li X, Tang M, Liu L, Cai X, Yuan X, Hu J, Zhao J, Qiao W, Zhang Y and Chen L (2025) Lactobacillus paragasseri HM018 derived from breast milk ameliorates hyperlipidemia in high-cholesterol rats by modulating bile acid metabolism. Front. Microbiol. 16:1599931. doi: 10.3389/fmicb.2025.1599931
Edited by:
Laurent Dufossé, Université de la Réunion, FranceCopyright © 2025 Yao, Li, Tang, Liu, Cai, Yuan, Hu, Zhao, Qiao, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue Zhang, emhhbmd5dWUxMjRAMTI2LmNvbQ==; Lijun Chen, Y2hlbmxpanVuQHNhbnl1YW4uY29tLmNu
†These authors have contributed equally to this work
 Chunyu Yao1,2,3†
Chunyu Yao1,2,3† Mi Tang
Mi Tang Junying Zhao
Junying Zhao Lijun Chen
Lijun Chen