- 1DNA Fingerprinting and Advanced Plant Virology Laboratory, AICRP-NSP, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Srinagar, India
- 2Dryland Agricultural Research Station (DARS), Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Budgam, India
- 3Division of Genetics and Plant Breeding, Faculty of Agriculture (FoA), Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Sopore, India
Bean common mosaic virus (BCMV) and bean common mosaic necrosis virus (BCMNV) are among the most challenging constraints for common bean production in Northern states of India due to their easy transmission through aphids and seeds. Highly valuable Indian common bean varieties and landraces are more susceptible to BCMV and BCMNV and very few varieties exhibit resistance to these viruses. Resistance towards these viruses is governed by a single dominant (I) gene and a few recessive genes (bc-1, bc-2, bc-3, bc-4, bc-ud, and bc-ur). This study aims to identify common bean genotypes bearing multiple resistant genes, each working with a different mode of action. A total of 123 genotypes of common beans were mechanically inoculated with BCMV and BCMNV isolates and molecular markers (SW13, ROC11, BCMV-CAPS, ENM-FWe/Rve) were used to identify the presence of two major resistant genes (I and bc-3). Out of these, 23 genotypes were found phenotypically resistant to both viruses. Furthermore, molecular screening was performed in which 13 hypersensitive resistant genotypes bearing a single dominant gene (I) were confirmed through SW13 and BCMV-CAPS markers. Additionally, ROC11/420, ENMF/R markers identified 4 genotypes bearing the recessive (bc-3) gene conferring complete resistance to the virus without executing hypersensitive response (HR). A valuable gene combination of both I, bc-3 (Ibc-3, Host group-12) genes in 3 genotypes was also established in the screened germplasm. However, in 3 phenotypically resistant genotypes, neither the I gene nor bc-3 gene was identified. The virus accumulation in the resistant genotypes was also understood properly through a time course experiment in a qPCR assay. This extensive identification of resistant common bean genotypes against BCMV and BCMNV can be readily included in the common bean breeding program of the Northern states of India for virus resistance.
1 Introduction
Bean common mosaic virus (BCMV) and bean common mosaic necrosis virus (BCMNV) are one of the most widespread viruses infecting common beans (Phaseolus vulgaris L.) and other cultivated legumes with worldwide distribution (Morales and Bos, 1988; Drijfhout and Morales, 2005; Morales, 2006). These viruses belong to the genus Potyvirus within the family Potyviridae. They possess a single-stranded (+) RNA genome of approximately 10 kilobases that encodes a large polyprotein precursor which is subsequently cleaved into 10 functional proteins (Adams et al., 2005; Olspert et al., 2015). Both viruses are non-persistently transmitted by probing aphids and are seed-borne, with a transmission efficiency of up to 80% (Wani et al., 2023; Morales, 2006). The major symptoms induced by BCMV on bean plants were mosaic, necrosis, chlorosis, etiolation and deformation, whereas BCMNV strains induce severe necrosis of bean plants (Drijfhout, 1978; Hamid et al., 2014; Rashid et al., 2022; Wani et al., 2023).
In India, especially in the Northern Himalayan region, valuable local bean varieties are becoming increasingly susceptible to diverse strains of BCMV and BCMNV, leading to heavy yield losses in hilly areas compared to the Northern plains. Also, BCMNV-induced whole plant necrosis was reported from Jammu and Kashmir (Hamid et al., 2014; Rashid et al., 2022) in which identifying resistant sources goes unrecognized. To overcome these viruses, we must identify and utilize resistant genotypes through germplasm screening and incorporate them into breeding programs. Resistance to BCMV and BCMNV is conferred by a single dominant (I) gene and six recessive genes namely bc-1, bc-2, bc-3, bc-4, bc-ud, and bc-ur (Ali, 1950; Soler-Garzón et al., 2024; Soler-Garzón et al., 2021a; Soler-Garzón et al., 2021b). The dominant (I) gene confers resistance against BCMV-BCMNV through a hypersensitive response (HR) on the primary leaves, whereas recessive genes confer absolute resistance toward specific viral strains (Drijfhout, 1978). Genotypes exhibiting HR present nervure localized necrosis on the primary inoculated leaves and demonstrate the presence of the dominant “I” gene for resistance (Drijfhout, 1978; Ogliari and Castano, 1992; Vallejos et al., 2006; Feng et al., 2015). At temperatures exceeding 30°C, some bean cultivars possessing the I gene experience strong whole plant necrosis (WPN), when infected with BCMNV and BCMV: NL-2 and NL-6 strains (Cadle-Davidson and Jahn, 2005). This interaction leads to a disease known as black root (Collmer et al., 2000; Feng et al., 2014). Hence, to prevent the WPN induced by BCMNV and BCMV (NL-2 and NL-6), genotypes bearing the “I” gene must be protected with extra “bc” recessive genes, especially in high-temperature areas (Collmer et al., 2000; Cadle-Davidson and Jahn, 2005; Feng et al., 2017). Although the non-necrotic strains cannot induce WPN in genotypes bearing the dominant I gene, the presence of an extra recessive gene will protect the bean plants from extreme HR when the temperature exceeds 30°C and provide additional protection (Ogliari and Castano, 1992).
Recessive resistance to BCMV-BCMNV is conferred by six recessive genes: four strain-specific bc-1, bc-2, bc-3, and bc-4 genes and two strain-unspecific bc-ur and bc-ud resistant genes (Soler-Garzón et al., 2021a; Soler-Garzón et al., 2021b; Soler-Garzón et al., 2024; Nisa et al., 2024). Strain-specific genes confer resistance to specific strains of BCMV-BCMNV. However, the bc-3 gene is exceptional in providing resistance to common beans against all strains of BCMV-BCMNV (Drijfhout, 1978; Miklas et al., 1998) except for a BCMV isolate 1755a (PG-VIII) that was able to overcome the bc-3 resistance (Feng et al., 2015). The strain-non-specific gene (bc-ud) is required for the complete action of the strain-specific genes (Drijfhout, 1978; Soler-Garzón et al., 2021a; Soler-Garzón et al., 2021b). A new gene “bc-ur” at the Bc-u locus on chromosome Pv05 was recently recognized as another strain-non-specific gene that interacts with bc-ud and also with the other recessive genes for conferring resistance against BCMV (Nisa et al., 2024; Soler-Garzón et al., 2024). To identify common bean varieties possessing these multiple resistance genes, reliable methods are necessary. Marker-assisted selection can be used as a consequence to efficiently select genotypes with desirable resistance genes. Molecular markers have been identified to be effective in identifying resistance genes in common beans against BCMV and BCMNV. Haley et al. (1994) identified a Randomly Amplified Polymorphic Marker (RAPD): OW13, linked to the I gene, which was later developed into a Sequence Characterized Amplified Region (SCAR) marker: SW13 (Melotto et al., 1996) that was more reliable and widely used. Molecular markers were also developed for the identification of recessive genes (Tang and Feng, 2022). Since most of these markers were dominant, progeny testing is necessary to distinguish between homozygous and heterozygous plants. Therefore, the use of co-dominant Cleaved Amplified Polymorphic Sequence (CAPS) marker was suggested for precise and efficient selection of BCMV-resistant genotypes compared to dominant markers (Bello et al., 2014).
As a proof of concept, the current study was conducted to identify common bean genotypes bearing the dominant I gene, recessive bc-3 gene and combination of both I and bc-3 genes (Ibc-3) for resistance with different modes of action based on phenotypic evaluation and molecular screening utilizing SCAR and CAPS markers. The pathogenicity and resistance mechanism during virus-plant interaction were studied. The resistant genes were later cloned, sequenced and aligned to identify the nucleotide differences and their implementation against virus resistance.
2 Materials and methods
2.1 Collection and maintenance of plant materials, viral isolates and inoculation method
123 genotypes of common beans, including 53 collections obtained from the National Bureau of Plant Genetic Resources (NBPGR)-New Delhi and 68 lines maintained by All India Co-ordinated Research Project (AICRP)- Seed Crops, AICRP-Pulses and Division of Genetics and Plant Breeding (Wadura, SKUAST-K) with truly unknown disease reaction to BCMV-BCMNV were evaluated in this study. All plants were grown under two treatments (T1: BCMV, T2: BCMNV) and three replicates in a growth chamber under ambient conditions of 26°C temperature and 70% relative humidity. For every genotype, non-inoculated control and mock-inoculated plants were maintained. The BCMV and BCMNV isolates (MW675689; OK094708), identified in our previous study (Rashid et al., 2022), were maintained through periodical propagation on Nicotiana benthamiana plants and were used to mechanically inoculate test plants at the primary leaf stage. The viral inoculum was prepared by homogenizing infected leaf tissue in 100 mM potassium phosphate buffer (pH 7.0) with 0.5% celite added directly to the viral inoculum just before inoculation (Wani et al., 2025). Ten days after planting, the first trifoliate leaves were mechanically inoculated with both BCMV and BCMNV isolates individually to identify the presence of the dominant hypersensitive I gene and recessive bc-3 gene. All the treatments were performed in triplicates with non-inoculated control and mock-inoculated plants maintained separately.
2.2 Screening of common bean germplasm against BCMV-BCMNV
2.2.1 Phenotypic evaluation
Disease reaction and symptoms of each virus were recorded every 2 days post inoculation (dpi) from 0 to 30 dpi and the genotype is classified as resistant/susceptible at 30th dpi. Disease severity was scored on a 0–3 scale according to Odu (Odu et al., 2004), where 0 = no disease symptoms/HR on plants, 1 = mild foliar disease symptoms, 2 = moderate foliar disease symptoms and 3 = severe distortion, malformation of leaves or stem and stunting. Resistant genotypes that exhibited no symptoms or HR were re-sown and re-inoculated to confirm resistance. Both symptomatic and asymptomatic plants were further tested using an RT-PCR assay to confirm the presence of the virus.
2.2.2 Genotypic evaluation
123 genotypes were screened for I and bc-3 gene resistance irrespective of the plant reaction to the virus (Table 2). Two SCAR markers (SW13 and ROC11) and two CAPS markers (BCMV-CAPS and ENM-FWe/Rve) were used for selection and their thermal conditions are given in Table 1. These markers were chosen due to their proven reliability and close linkage to the I and bc-3 genes. SCAR markers were initially used to identify the genotypes bearing resistant genes and CAPS markers were later used for revalidation. Pooled leaves of three replicates per genotype were collected at 30 dpi and stored in a deep freezer at −80°C. DNA was extracted from the young leaves of 10-day-old plants using a DNeasy plant mini kit (Qiagen). Total RNA was isolated from 100 mg of leaf tissue using TRIzol reagent as per the user guidelines (Invitrogen, Thermo Scientific). 1 μg of RNA was used as a template for cDNA synthesis using the Revert-Aid cDNA synthesis kit (Thermo Scientific) in a total volume of 20 μL. 2 μL of cDNA was used in the RT-PCR assay to test the presence of virus in both susceptible and resistant plants. PCR cycling conditions are listed in Table 1. PCR reactions were performed in 25 μL volumes, each containing 1 μL of 50 ng genomic DNA as a template, 12.5 μL of GoTaq green dye master mix (Promega, Madison, United States), 10 μM of each primer (20 pmol for SW13). For CAPS markers, 5 μL of PCR products were digested with 1 μL of 10X Reaction buffer (Thermo Scientific) and 1 μL of the restriction enzyme (Taq1 for I gene, Rsa1 for bc-3 gene) in a total volume of 15 μL. The digested products were then separated on a 2% agarose gel. The complete nucleotide sequence of both I and bc-3 genes corresponding to the BCMV-CAPS and ENM-FWe/Rve amplified product were purified and sanger sequenced at Medauxin genomics, Bangalore, http://www.medauxin.com/. The sequences were aligned and analyzed using the BioEdit 7.0 sequence alignment editor. BLASTn analysis of the ENM-FWe/Rve amplicon sequence was performed to identify similarities with known bc-3 gene sequences in reported cultivars.
2.3 qRT-PCR analysis
qPCR assay was carried out to study the best resistant genotypes inhibiting the replication and systemic movement of the virus in different resistant plants (I, bc-3, and Ibc-3). For assays, resistant genotypes possessing the I (WB-352), bc-3 (EC-127645), and Ibc-3 (EC-116117) genes for resistance were selected along with a highly susceptible plant (IC-437141). All plants were inoculated with BCMV-BCMNV isolates and RNA was extracted from these genotypes at two development stages (Day 4 and Day 8). For every genotype, uninoculated resistant plants were used as an untreated control. Reactions were performed in the Rotor-Gene Q Real-Time PCR system (QIAGEN) using a 20 μL reaction mixture, including 10 μL SYBR green master mix (Thermo Scientific), 1 μL each of forward and reverse primers (BCMV-CP and BCMNV, 10 μm, Table 1), 2 μL cDNA and 6 μL of Nuclease-free water. The cycling conditions were set at an initial hold of 95°C for 2 min, followed by cycling for 35 times at 95°C-30 s, 60°C-30 s and 72°C- 60 s. Following the final PCR cycle, melting curve analysis was performed on the samples by heating them from 70 to 95°C, with a rise of 0.3°C for each step for the detection of specific and non-specific PCR products. Each sample was assayed in three replicates, including the actin gene, used as an internal control. The comparative quantification report was used to construct the box plots using REST 2008 software (QIAGEN). For both qPCR and RT-PCR, BCMV and BCMNV specific primers (Rashid et al., 2022; Wani et al., 2025) were used to detect both viruses (Table 1).
3 Results
3.1 Plant reaction to BCMV/BCMNV
Based on the phenotypic evaluation from the screening experiment, 23 genotypes were found resistant to both BCMV and BCMNV. The resistant plants were categorized into two groups, hypersensitive and absolute, based on the presence or absence of necrosis symptoms on the inoculated leaves. The results were presented under three experimental groups based on the selection of resistant genotypes assisted by molecular markers: Group 1: characterized by the sole presence of the I gene, Group 2: characterized by the sole presence of the bc-3 gene, and Group 3: characterized by the presence of both I and bc-3 genes (Ibc-3).
3.2 Phenotypic and genotypic evaluation of plant materials bearing I gene
BCMV and BCMNV isolates induced typical mosaic, necrosis, mottling, leaf crinkling and deformations in susceptible cultivars and accurately distinguished the resistant genotypes carrying the dominant “II” gene from the susceptible “ii” genotypes. Of all the 123 plant materials screened for BCMNV resistance, 13 genotypes presented localized and systemic vein necrosis that suggested the presence of “I” gene conditioning resistance through “Temperature Insensitive Necrosis (TIN)” in bean plants (Figure 1b, Supplementary File S1). The tested BCMNV isolate induced initial necrotic lesions on resistant plant materials that appeared at 4 dpi and expanded quickly, reaching the entire leaf veins and a nervure necrotic vein reaction was noticed at 10–12 dpi (Figure 1b, Supplementary File S1). Similarly, from the tested 123 plant materials, 23 genotypes conferred resistance to BCMV. However, infection with BCMV didn’t induce TIN in those genotypes conditioning HR to BCMNV and conferring extreme immunity (Figure 1a). Also, few genotypes induced mild necrosis of primary leaves infected with BCMV, demonstrating the incomplete dominant nature of the I gene as described by Collmer et al. (2000), that I/i genotypes respond to BCMV infection also through an HR.

Figure 1. Resistant genotypes identified for the presence of the Dominant “I” gene. (a) All treatments inoculated with BCMV and a mock (SR-3, conditioning resistance to BCMV without HR). (b) All treatments inoculated with BCMNV and a mock (SR-3, conditioning resistance through an HR to BCMNV). (c) EC-325065 inoculated with BCMNV conditioning resistance through HR and without HR to BCMV; white arrows indicating the vein necrosis of primary leaves on inoculation with BCMNV at temperature < 30°C. Susceptible plants (IC-328558) do not possess I gene for resistance.
When a SCAR marker SW13 was utilized to screen all the genotypes for identifying the I gene, the marker consistently identified the plant materials presenting localized vein necrosis on the inoculated leaf. SW13 marker linked to the I gene amplified a 690 bp product from 16 genotypes (Table 2) that specified the presence of the dominant I gene for resistance (Figure 2a). However, three genotypes (EC-400444, EC-271540, and WB-6, Table 2), identified by the marker were susceptible to virus inoculation. They were later found to have mutations in the I gene at four nucleotide positions (Figure 3). To overcome these mutated forms, another co-dominant marker (BCMV-CAPS, Table 1) linked to the I gene was utilized. This marker is based on the presence of single nucleotide difference (A/G) between I gene-bearing resistant (II) and susceptible (ii) plants. Resistant plants have “A” allele while susceptible plants have “G” allele. PCR products (311 bp) of resistant genotypes on digestion with TaqI (Restriction site: TCGA) generated products of 201 and 110 bp, whereas susceptible plants remain un-cleaved (311 bp) by TaqI (Figure 2b) due to point mutations and absence of the restriction site (TCGA-TCGG). CAPS analysis based on these closely linked SNPs identified only true resistant plants bearing the I gene for resistance. PCR products from resistant and susceptible genotypes, containing the I gene corresponding to CAPS marker, were purified and sanger sequenced. Sequence alignment revealed point mutations between resistant and susceptible plants. One of these mutations created a Taq1 restriction site, as illustrated in Figure 3.
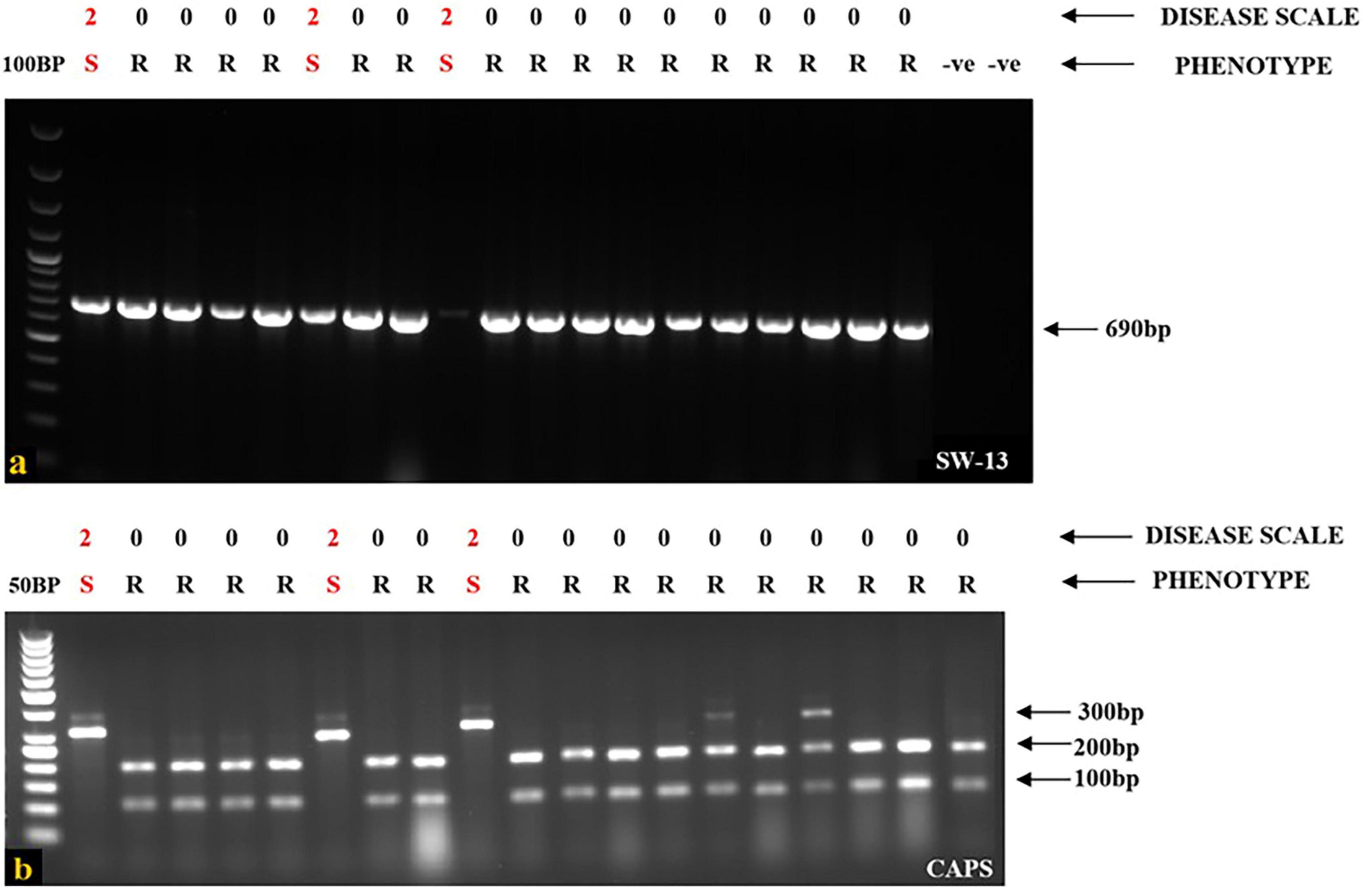
Figure 2. Identification of I gene for resistance in the screened genotypes. (a) SW13 marker identified hypersensitive resistant genotypes bearing I gene in which 3 cultivars are still susceptible with a disease scale of “2.” (b) CAPS marker identified and cleaved only resistant genotypes, whereas susceptible genotypes remain un-cleaved by Taq1. This marker yields two alleles. The resistant allele has two bands (200 and 100 bp), whereas the susceptible allele has a single band (311 bp). For both SW13 and CAPS markers, Lane 1–20, 1: Ladder (100 bp for SW13 and 50 bp for CAPS), 2 (EC-400444), 3 (EC-325065), 4(EC-400439), 5 (EC-13100), 6 (EC-127645), 7 (EC-271540), 8 (EC-405209), 9 (EC-116117), 10 (WB-6), 11 (WB-352), 12 (SR-3), 13 (WB-353 F), 14 (WB-N4), 15 (WB-N1), 16 (Local-2), 17 (Shalimar French bean-1), 18 (NSP-F1), 19 (NSP-F2), 20 (EC-325078), Lane 21, 22: Negative control for SW13 marker (DNA template from resistant plants).
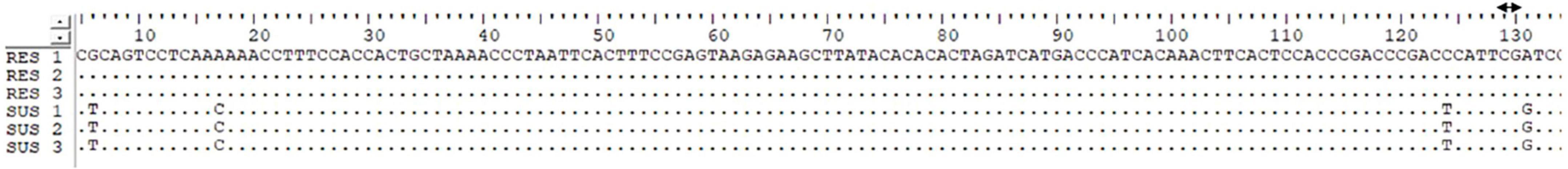
Figure 3. Illustrating sequence alignment of the I gene as identified by CAPS marker reveals four single-nucleotide polymorphisms (SNPs) that differentiate resistant and susceptible genotypes. A TaqI restriction site (TCGA) is present at nucleotide positions 126–130 in resistant genotypes, whereas a G-to-A substitution at position 128 results in the absence of this site in susceptible genotypes.
3.3 Phenotypic and genotypic evaluation of plant materials bearing the bc-3 gene
This resistant group included 4 genotypes that were resistant and symptomless after mechanical inoculation (BCMV and BCMNV) and were tested for the presence of the “bc-3” gene in PCR assays. Genotypes carrying this recessive resistance allele conferred complete immunity to both BCMV and BCMNV isolates (Figures 4a,b), without triggering any necrotic reaction on the inoculated leaves. Plant materials with the bc-3 gene never reacted with mosaic or necrosis and the only visible reaction was necrotic, in which pin-point necrotic spots developed on upper un-inoculated leaves in between 10 and 15 dpi infected with BCMNV (Figure 4) which were less prominent and did not extend into localized vein necrosis. No systemic necrosis can be exemplified as the plant materials are visibly healthy and recorded no symptoms throughout the 30-day phenotyping period.
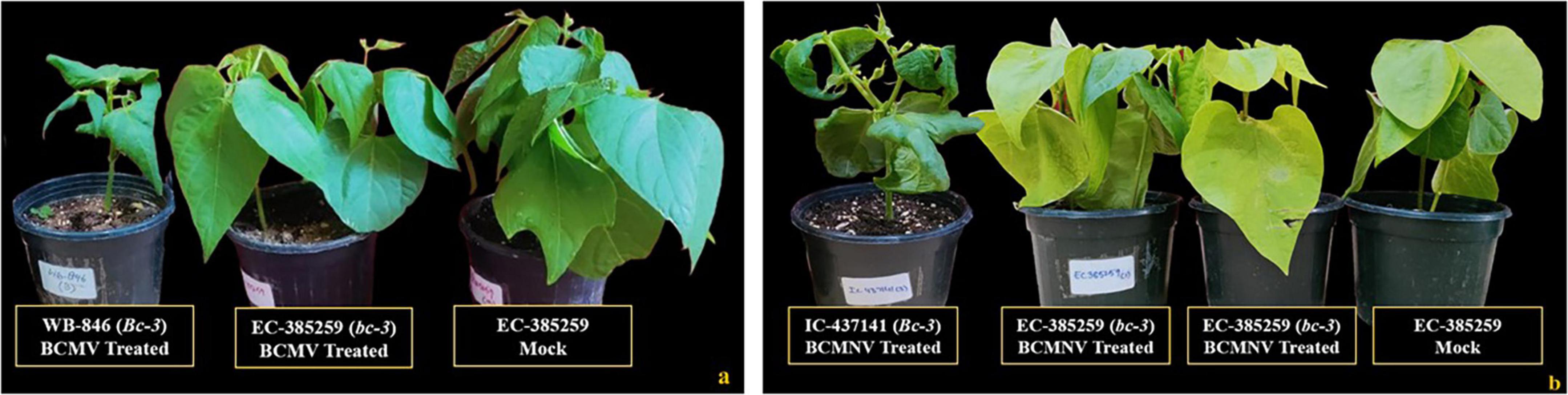
Figure 4. Resistant genotypes identified for the presence of the recessive “bc-3” gene. (a) All treatments inoculated with BCMV and a mock (EC-385259, conditioning resistance to BCMV). (b) All treatments inoculated with BCMNV and a mock (EC-385259, conditioning resistance to BCMNV). Susceptible plants (WB-846 and IC-437141) do not possess bc-3 gene for resistance.
The presence of the bc-3 gene in this group was confirmed by both ROC11 and ENM-FWe/Rve markers. The absence of SCAR marker ROC11/420 indicates the presence of bc-3 gene and its negative selection was initially used to eliminate the common bean genotypes lacking bc-3 disease resistance locus (Table 2). Subsequently, those genotypes bearing the bc-3 gene as identified by ROC11 marker were amplified using ENM-FWe/Rve marker. The genotypes which were not identified for the presence of bc-3 gene using ROC11 marker were not utilized to test with ENM-FWe/Rve marker. Digestion of PCR amplified products (541-bp fragment) with RsaI cleaved bc-3 carrying genotypes into 381- and 160-bp fragments, whereas the PCR products derived from the susceptible genotypes remain un-cleaved, due to the absence of mutations within the eIF4E gene. To confirm the presence of mutations in the eIF4E gene of resistant genotypes, the 541-bp PCR products were purified and sanger sequenced. Pairwise nucleotide sequence comparison was performed by using the Basic Local Alignment Search Tool (BLAST) and the partial coding sequence (541 bp) of genotypes carrying the bc-3 gene had a maximum identity of 100% to the published eIF4E gene sequence of the cultivar “IVT7214” (KT175572), reported to carry bc-3 gene.
3.4 Phenotypic and genotypic evaluation of plant materials bearing I and bc-3 gene
Three genotypes (EC-13100, EC-127645, EC-116117) bearing both I and bc-3 (Ibc-3) genes for resistance were identified from the screened germplasm. These genotypes presented no symptoms or any HR (Figures 5a,b) on both inoculated and upper un-inoculated leaves to both BCMV and BCMNV isolates. Hence, these genotypes were protected from BCMNV-induced necrosis and conditioned extreme resistance to the bean plants without subjecting them to HR. In this gene combinations (Ibc-3), the dominant I gene was protected from being necrotic by the presence of the bc-3 allele.
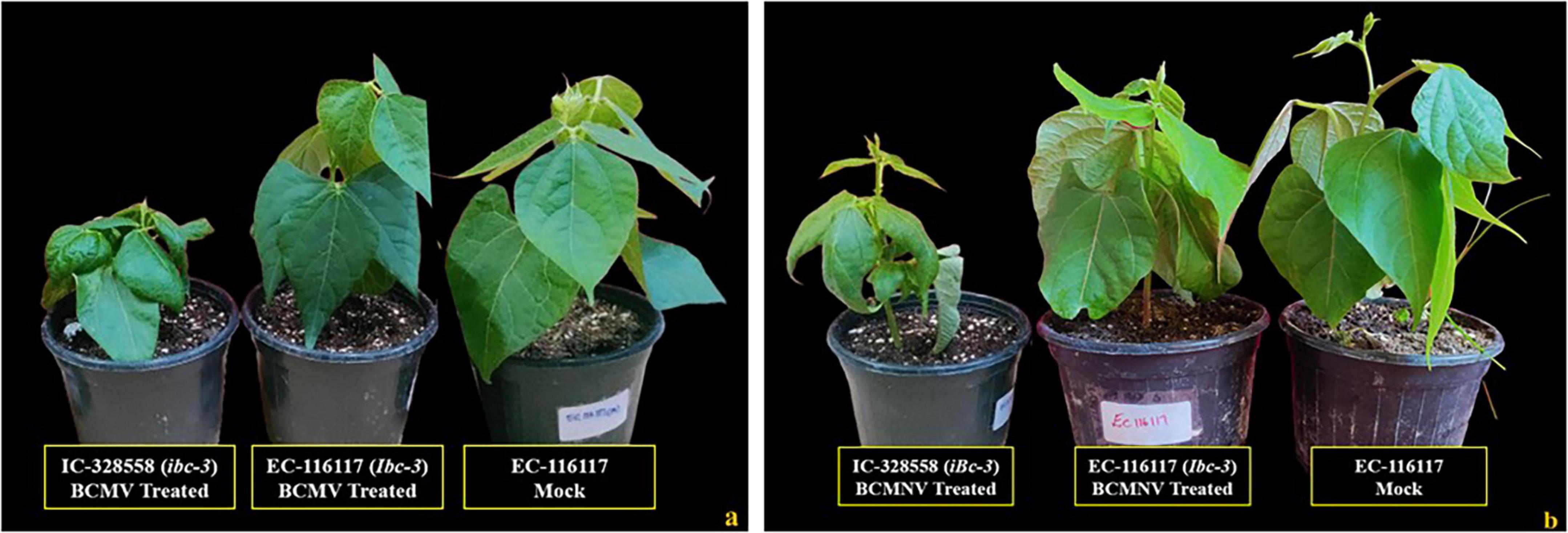
Figure 5. Resistant genotypes identified for the presence of both “I” and “bc-3” gene. (a) All treatments inoculated with BCMV and a mock (EC-116117, conditioning resistance to BCMV). (b) All treatments inoculated with BCMNV and a mock (EC-116117, conditioning resistance to BCMNV). Susceptible plants (IC-328558) do not possess both the I and bc-3 genes for resistance.
However, 3 resistant plant materials in which both I and bc-3 genes were not identified, the presence of other resistance genes or gene combination can be assumed, which protects the common beans against BCMV-BCMNV. Due to its minor influence on conferring resistance to the virus and also due to the limited availability of markers linked to these genes, its presence in the screened germplasm was not established. These genotypes conferred high resistance to the test isolates and the viral symptoms didn’t appear on both inoculated and upper uninoculated leaves till the entire phenotyping period of 30 dpi. In susceptible plants, the RT-PCR assay using BCMV and BCMNV primers yielded positive results, and did not detect the presence of the virus in resistant plants (Figure 6). Susceptible plants associated with the symptoms of mosaic, necrosis, leaf crinkling and puckering were amplified around 414 bp with BCMV CP primers and no BCMV presence was detected in resistant genotypes (Figures 6a,b). Whereas, BCMNV primers amplified the target virus with expected product size of 834 bp from susceptible genotypes (Figure 6c) and no BCMNV was detected from the RNA isolated from resistant genotypes.
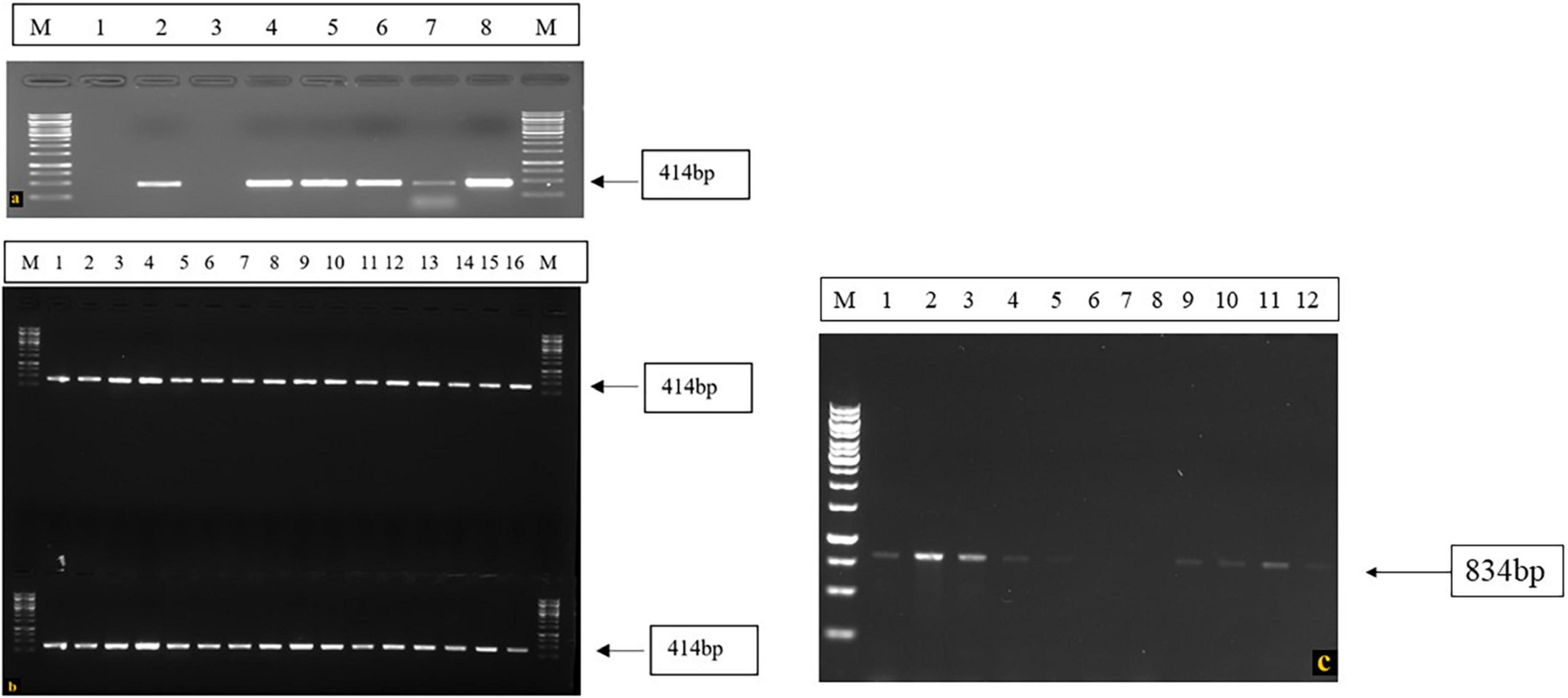
Figure 6. RT-PCR confirmation of presence of virus in the symptomatic and susceptible genotypes using BCMV-BCMNV CP primers, (a) M: 1 kb DNA ladder, 1 and 3: Negative Controls (RNA isolated from Resistant Plants), 2: BCMV Positive control, 4–8: RNA isolated from BCMV susceptible genotypes. (b) M: 1 kb DNA ladder, 1–16: BCMV Susceptible genotypes. (c) RT-PCR amplification of BCMNV in susceptible genotypes, Lane M: 1 kb DNA ladder, Lane 1: BCMNV positive control, Lane 2–5: Susceptible genotypes, Lane 6–8: Resistant genotypes (Negative control), Lane 9–12: Susceptible genotypes.
3.5 qPCR-based assessment of virus accumulation
Resistant phenotypes of bean cultivars differed based on the presence or absence of the I gene. The two test isolates induced either local or systemic necrosis in resistant cultivars (WB-352) carrying the I gene, but not in those genotypes carrying Ibc-3 (EC-116117) and bc-3 (EC-127645) resistant genes. qPCR analysis of the RNA isolated from the upper leaves of three different genotypes (I, bc-3, and Ibc-3) at the 4th and 8th day post-inoculation revealed varying levels of BCMV-BCMNV expression across the genotypes. Cultivars carrying the I gene alone exhibited low-level viral expression on Day 4, where initial necrotic lesions appeared on the inoculated leaves and an increased expression at day 8 (Figure 7a) that corresponds to extensive vein necrosis that usually appears at 10 dpi. In contrast, no viral spread was detected in Ibc-3 (Figure 7c) and bc-3 (Figure 7b) genotypes up to 8 dpi, and these plants remained visibly healthy throughout the 30-day observation period. This suggests that genotypes carrying the I gene alone would enable the virus to replicate and allow for systemic movement inside the host plants without the symptoms being expressed that were affected in the other genotypes (Ibc-3, bc-3), where its replication and movement were inhibited. The melting and amplification curves of the tested genotypes were provided in Supplementary File S2.
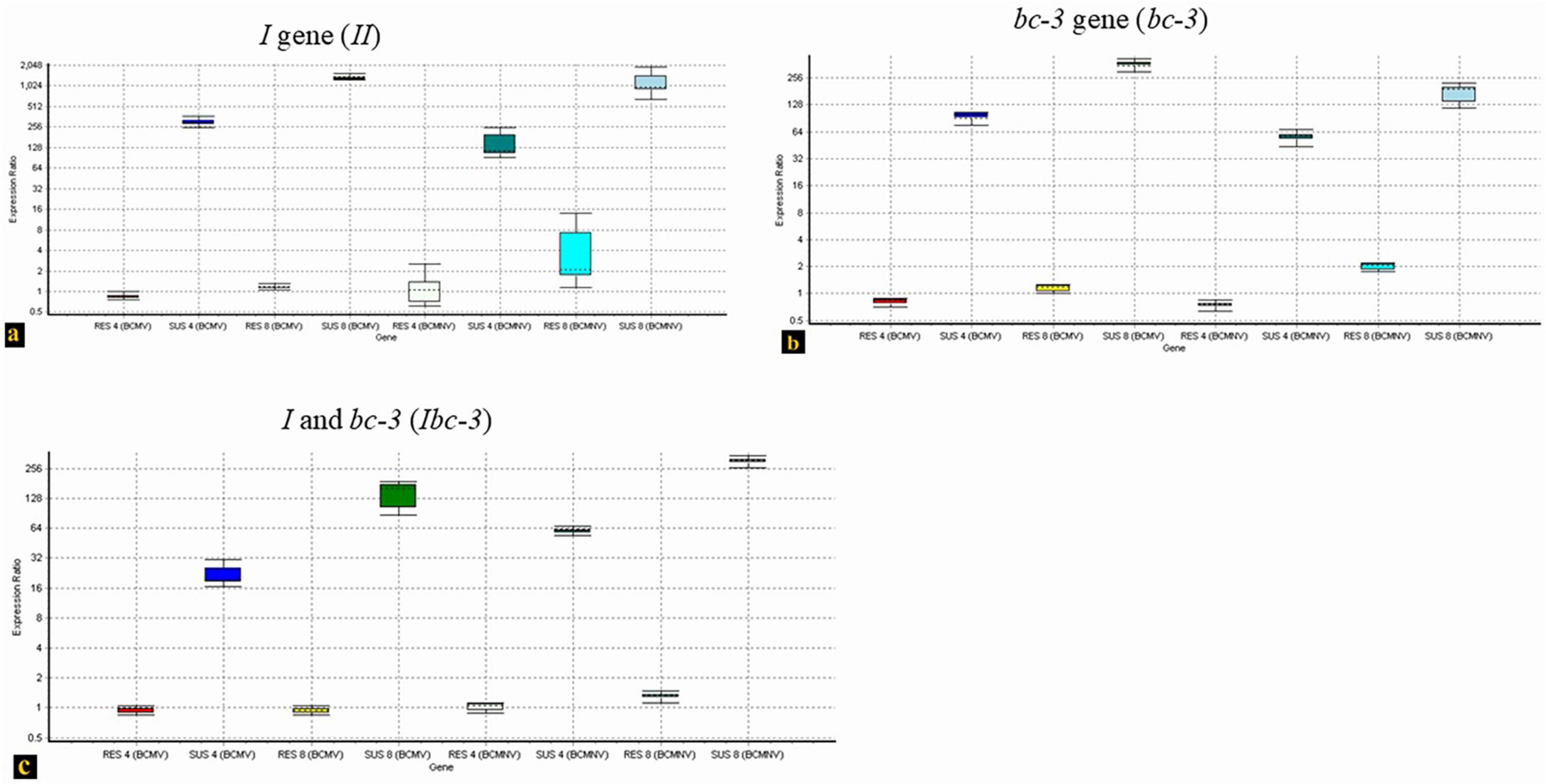
Figure 7. RT-qPCR based comparative quantification on the accumulation of BCMV-BCMNV in three resistant genotypes, (a) WB-352 (I). (b) EC-127645 (bc-3). (c) EC-116117 (Ibc-3) at two time points (Day 4 and Day 8). In three experiments (a–c), plants were inoculated with BCMV and BCMNV and its expression in the top leaves was analyzed in qPCR assay. The expression ratios of BCMV-BCMNV in both resistant and susceptible groups are represented by Whisker box plots and are calculated using the take-off and replication values of control plants (untreated) against treated group (resistant and susceptible plants) in REST software (QIAGEN).
4 Discussion
BCMV and BCMNV are two closely related viruses that pose a significant threat to global common bean production. These viruses can cause complete crop failure (up to 100% yield loss) and significantly reduce bean quality (Tang and Feng, 2022; Worrall et al., 2015). In response, identifying and developing common bean cultivars with broad genetic resistance to these viruses is crucial.
In this study, 123 common bean genotypes were inoculated to assess resistance and susceptibility to BCMV-BCMNV and to identify the plant materials bearing multiple resistant genes, each working with a different mode of action. Two forms of resistance were observed in the identified plant materials: some exhibited a hypersensitive reaction (HR) to the virus, while others remained resistant without exhibiting HR. Resistant plants exhibiting HR presented a vein necrosis of primary inoculated leaves between 7 and 10 dpi. Phenotypically, this necrotic vein reaction of inoculated primary leaves was the key feature in demonstrating the presence of the I gene (Feng et al., 2017; Collmer et al., 2000). This necrotic sign has been identified in most of the resistant cultivars (13 genotypes) in the screened germplasm and conferred resistance toward BCMV-BCMNV under controlled conditions (< 30°C). The evidence for the presence of the I gene was suggested by PCR using SW-13 and BCMV-CAPS markers. These results are in accordance with some previous studies that the I gene confers a high degree of resistance and complete immunity to BCMV strains below 30°C (Ali, 1950; Vallejos et al., 2006; Feng et al., 2017) and induces a necrotic reaction on primary inoculated leaves to BCMNV, regardless of the temperature. However, the wide use of common bean genotypes bearing the I gene has led to BCMNV-induced systemic necrosis and in response, its susceptibility can be prevented if the I gene is “protected” in the presence of one of the recessive genes (bc-1, bc-2, or bc-3) (Kelly et al., 1995; Singh and Schwartz, 2010; Pasev et al., 2014; Feng et al., 2018). We then focused on the presence of any recessive resistant gene in the remaining plant materials that were immune (without HR) to both BCMV and BCMNV. Interestingly, four resistant genotypes possessed the bc-3 gene, and three genotypes were in combination of both I and bc-3 genes (Ibc-3). However, some bean varieties with bc-3 resistance to BCMNV were also reported as susceptible to BCMV (Miklas et al., 1998). The recessive bc-3 gene encodes a mutated eukaryotic translation initiation factor (eIF4E) gene and was identified to be associated with resistance against BCMV-BCMNV in common beans. The VPg protein of the virus interacts with eIF4E proteins in plants and four-point mutations in the eIF4E gene impaired the interaction between eIF4E and VPg protein and also affected the replication of BCMV, thus conferring resistance in common beans (Naderpour et al., 2010). These amino acid polymorphisms in eIF4E were also reported previously to confer recessive resistance to Potyviruses in Pisum sativum against pea seed-borne mosaic virus, Capsicum annuum against chili veinal mottle virus and potato virus Y (PVY), Hordeum vulgare against barley yellow mosaic virus, Citrullus lunatus against zucchini yellow mosaic virus, Lycopersicum esculentum against potato virus Y (PVY) and tobacco etch virus (TEV) (Ashby et al., 2011; Hwang et al., 2009; Li et al., 2016; Ling et al., 2009; Piron et al., 2010; Ruffel et al., 2002; Ruffel et al., 2006).
A qPCR experiment was conducted to study the resistance conferred by the three resistant groups (I, bc-3, Ibc-3) that affect the systemic movement of the virus. This assay (Figure 7) also suggests that systemic spread of the virus was drastically affected in genotypes bearing Ibc-3 genes with both BCMV and BCMNV being unable to replicate independently and spread to the uninoculated upper leaves. However, the genotypes bearing the I gene demonstrate a difference in virus spread in uninoculated upper leaves when compared to those genotypes bearing bc-3 and Ibc-3 genes. The systemic movement of virus in the bean genotypes carrying I alleles here demonstrated that BCMV replication occurs inside the host plants. It was also proven in the screening experiment that, bean genotypes carrying the I gene were immune to BCMV and a few genotypes also conditioned resistance to BCMV through a mild necrosis of primary leaves. Previous experiments with bean lines carrying I alleles also demonstrated that BCMV replication depends on the I allele’s dosage (Cadle-Davidson and Jahn, 2005) and the virus could still replicate in genotypes carrying the I gene even at a low temperature (26°C) (Collmer et al., 2000). This suggests that BCMV/BCMNV strains are capable of replicating and undergoing intercellular movement within I-gene-containing common bean cultivars without expressing any viral symptoms and necrosis likely occurs when the virus spreads from the initially infected cell to a neighboring cell (Collmer et al., 2000; Cadle-Davidson and Jahn, 2005; Feng et al., 2017). So, the best combination of genes in breeding for BCMV-BCMNV resistance was likely the Ibc-3 combination as they confer a broad spectrum of genetic resistance (Miklas et al., 1998; Pasev et al., 2014; Singh and Schwartz, 2010; Worrall et al., 2015). This beneficial gene combination (Ibc-3) has been identified in some plant materials that provide promising resistance to BCMV-BCMNV also at temperatures above 30°C.
Hence, this study demonstrates the mechanism of resistance conferred by common bean cultivars bearing the dominant “I” and recessive “bc-3” genes and represents the influence of different genetic backgrounds on resistance to BCMV-BCMNV. Both SCAR (SW13 and ROC11) and CAPS (BCMV-CAPS and ENM-FWe/Rve) molecular markers used in this study were reliable for the identification of I and bc-3 genes. Importantly, there are no reported instances of recombinant viral strains overcoming resistance conferred by these genes in India. In this respect, this investigation was important to separate common bean cultivars with major genes for resistance. We can further introduce these genes directly into our elite common bean varieties that are susceptible to BCMV and can later be utilized in bean breeding program for virus resistance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
DM: Visualization, Writing – review & editing, Writing – original draft, Formal Analysis, Methodology, Validation, Software, Data curation, Conceptualization. SW: Validation, Writing – review & editing, Project administration, Formal Analysis, Supervision, Investigation, Software, Visualization, Methodology, Writing – original draft, Conceptualization. SB: Supervision, Data curation, Project administration, Conceptualization, Writing – review & editing, Resources, Funding acquisition. SR: Writing – review & editing, Investigation, Data curation, Validation, Formal Analysis, Visualization. AJ: Validation, Writing – review & editing, Methodology, Software. ZD: Resources, Funding acquisition, Visualization, Supervision, Project administration, Conceptualization, Writing – review & editing. SHW: Methodology, Conceptualization, Writing – review & editing, Project administration, Resources. PS: Data curation, Resources, Writing – review & editing, Supervision. GA: Supervision, Project administration, Writing – review & editing, Resources, Funding acquisition. AH: Conceptualization, Resources, Validation, Methodology, Writing – review & editing, Visualization, Investigation, Data curation, Formal Analysis, Supervision, Writing – original draft, Project administration, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by the SERB-DST (grant number: CRG/2022/003659), DST-PURSE (grant number: SR/PURSE/2022/124), and the ICAR- National Institute of Seed Science and Technology.
Acknowledgments
We acknowledge NBPGR-New Delhi, AICRP-NSP (Shalimar, SKUAST-K), and Division of Genetics and Plant Breeding (FOA, SKUAST-K) for providing seed material.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1614122/full#supplementary-material
References
Adams, M. J., Antoniw, J. F., and Beaudoin, F. (2005). Overview and analysis of the polyprotein cleavage sites in the family potyviridae. Mol. Plant Pathol. 6, 471–487. doi: 10.1111/j.1364-3703.2005.00296.x
Ali, M. A. (1950). Genetics of resistance to the common bean mosaic virus (bean virus 1) in the bean (Phaseolus vulgaris L.). Phytopathology 40, 69–79.
Ashby, J. A., Stevenson, C. E., Jarvis, G. E., Lawson, D. M., and Maule, A. J. (2011). Structure-based mutational analysis of eIF4E in relation to sbm1 resistance to pea seed-borne mosaic virus in pea. PLoS One 6:e15873. doi: 10.1371/journal.pone.0015873
Bello, M. H., Moghaddam, S. M., Massoudi, M., McClean, P. E., Cregan, P. B., and Miklas, P. N. (2014). Application of in silico bulked segregant analysis for rapid development of markers linked to Bean common mosaic virus resistance in common bean. BMC Genom. 15:903. doi: 10.1186/1471-2164-15-903
Cadle-Davidson, M. M., and Jahn, M. M. (2005). Resistance conferred against bean common mosaic virus by the incompletely dominant I locus of Phaseolus vulgaris is active at the single cell level. Arch. Virol. 150, 2601–2608. doi: 10.1007/s00705-005-0592-z
Collmer, C. W., Marston, M. F., Taylor, J. C., and Jahn, M. (2000). The I gene of bean: A dosage-dependent allele conferring extreme resistance, hypersensitive resistance, or spreading vascular necrosis in response to the potyvirus Bean common mosaic virus. Mol. Plant Microbe Interact. 13, 1266–1270. doi: 10.1094/MPMI.2000.13.11.1266
Drijfhout, E. (1978). Genetic interaction between Phaseolus vulgaris and bean common mosaic virus with implications for strain identification and breeding for resistance. Wageningen: Verslagen Van Landbouwkundige Onderzoekingen, Centre for Agricultural Publishing and Documentation.
Drijfhout, E., and Morales, F. (2005). “Bean common mosaic,” in Compendium of bean diseases, 2nd Edn, eds H. F. Schwartz, J. R. Steadman, R. Hall, and R. L. Forster (Saint Paul, MN: American Phytopathological Society), 60–62.
Feng, X., Guzmán, P., Myers, J. R., and Karasev, A. V. (2017). Resistance to Bean common mosaic necrosis virus conferred by the bc-1 gene affects systemic spread of the virus in common bean. Phytopathology 107, 893–900. doi: 10.1094/PHYTO-01-17-0013-R
Feng, X., Myers, J. R., and Karasev, A. V. (2015). Bean common mosaic virus isolate exhibits a novel pathogenicity profile in common bean, overcoming the bc-3 resistance allele coding for the mutated eIF4E translation initiation factor. Phytopathology 105, 1487–1495. doi: 10.1094/PHYTO-04-15-0108-R
Feng, X., Orellana, G. E., Myers, J. R., and Karasev, A. V. (2018). Recessive resistance to bean common mosaic virus conferred by the bc-1 and bc-2 genes in common bean (Phaseolus vulgaris) affects long-distance movement of the virus. Phytopathology 108, 1011–1018. doi: 10.1094/PHYTO-01-18-0021-R
Feng, X., Poplawsky, A. R., and Karasev, A. V. (2014). A recombinant of Bean common mosaic virus induces temperature-insensitive necrosis in an I gene-bearing line of common bean. Phytopathology 104, 1251–1257. doi: 10.1094/PHYTO-02-14-0048-R
Haley, S. D., Afanador, L., and Kelly, J. D. (1994). Identification and application of a random amplified polymorphic DNA marker for the I gene (Povyvirus resistance) in common bean. Phytopathology 84, 157–160. doi: 10.1094/Phyto-84-157
Hamid, A., Ahmad, M., Padder, B. A., Shah, M. D., Saleem, S., Sofi, T. A., et al. (2014). Pathogenic and coat protein characterization confirming the occurrence of Bean common mosaic virus on common bean (Phaseolus vulgaris) in Kashmir, India. Phytoparasitica 42, 317–322. doi: 10.1007/s12600-013-0362-5
Hwang, J., Li, J., Liu, W. Y., An, S. J., Cho, H., Her, N. H., et al. (2009). Double mutations in eIF4E and eIFiso4E confer recessive resistance to Chilli veinal mottle virus in pepper. Mol. Cells 27, 329–336. doi: 10.1007/s10059-009-0042-y
Johnson, W. C., Guzmán, P., Mandala, D., Mkandawire, A. B., Temple, S., Gilbertson, R., et al. (1997). Molecular tagging of the bc-3 gene for introgression into Andean common bean. Crop. Sci. 37, 248–254. doi: 10.2135/cropsci1997.0011183X003700010044x
Kelly, J. D., Afanador, L., and Haley, S. D. (1995). Pyramiding genes for resistance to Bean common mosaic virus. Euphytica 82, 207–212. doi: 10.1007/BF00029562
Li, H., Kondo, H., Kühne, T., and Shirako, Y. (2016). Barley yellow mosaic virus VPg is the determinant protein for breaking eIF4E-Mediated recessive resistance in barley plants. Front. Plant Sci. 7:1449. doi: 10.3389/fpls.2016.01449
Ling, K. S., Harris, K. R., Meyer, J. D., Levi, A., Guner, N., Wehner, T. C., et al. (2009). Non-synonymous single nucleotide polymorphisms in the watermelon eIF4E gene are closely associated with resistance to zucchini yellow mosaic virus. Theor. Appl. Genet. 120, 191–200. doi: 10.1007/s00122-009-1169-0
Melotto, M., Afanador, L., and Kelly, J. D. (1996). Development of a SCAR marker linked to the I gene in common bean. Genome 39, 1216–1219. doi: 10.1139/g96-155
Miklas, P., Lambert, S., Mink, G., and Silbernagel, M. (1998). Many beans with bc-3 resistance to BCMNV are susceptible to BCMV. Ann. Rept. Bean Improv. Coop. 41, 33–34.
Morales, F. J. (2006). “Common beans,” in Natural resistance mechanisms of plants to viruses, eds G. Loebenstein and J. P. Carr (The Netherlands: Springer), 367–382.
Morales, F. J., and Bos, L. (1988). Bean common mosaic virus. Descriptions of Plant Viruses, No. 337. Association of Applied Biologists. Available online at: http://www.dpvweb.net/dpv/showdpv.php?dpvno=337
Naderpour, M., Lund, O. S., Larsen, R., and Johansen, E. (2010). Potyviral resistance derived from cultivars of Phaseolus vulgaris carrying bc-3 is associated with the homozygotic presence of a mutated eIF4E allele. Mol. Plant Pathol. 11, 255–263. doi: 10.1111/j.1364-3703.2009.00602.x
Nisa, Q., Lateef, I., Nabi, A., Nabi, N., Fayaz, T., Bashir, A., et al. (2024). Northwestern Himalayan common beans: A treasure trove for breeding resistant bean cultivars for multiple foliar pathogens. Physiol. Mol. Plant Pathol. 134:102466. doi: 10.1016/j.pmpp.2024.102466
Odu, B. O., Asiedu, R., Hughes, J. A., Shoyinka, S., and Oladiran, A. O. (2004). Identification of resistance to Yam mosaic virus (YMV), genus Potyvirus in white Guinea yam (Dioscorea rotundata). Field Crops Res. 89, 97–105. doi: 10.1016/j.fcr.2004.01.009
Ogliari, J. B., and Castano, M. (1992). Identification of resistant germplasm to the bean common mosaic Virus-BCMV. Brasilia: Pesquisa Agropecuaria Brasileira.
Olspert, A., Chung, B. Y., Atkins, J. F., Carr, J. P., and Firth, A. E. (2015). Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep. 16, 995–1004. doi: 10.15252/embr.201540509
Pasev, G., Kostova, D., and Sofkova, S. (2014). Identification of genes for resistance to Bean common mosaic virus and Bean common mosaic necrosis virus in snap bean (Phaseolus vulgaris L.) breeding lines using conventional and molecular methods. J. Phytopathol. 162, 19–25. doi: 10.1111/jph.12149
Piron, F., Nicolaï, M., Minoïa, S., Piednoir, E., Moretti, A., Salgues, A., et al. (2010). An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS One 5:e11313. doi: 10.1371/journal.pone.0011313
Rashid, S., Wani, F., Ali, G., Sofi, T. A., Dar, Z. A., and Hamid, A. (2022). Viral metatranscriptomic approach to study the diversity of virus(es) associated with common bean (Phaseolus vulgaris L.) in the North-Western Himalayan region of India. Front. Microbiol. 13:943382. doi: 10.3389/fmicb.2022.943382
Ruffel, S., Dussault, M. H., Palloix, A., Moury, B., Bendahmane, A., Robaglia, C., et al. (2002). A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32, 1067–1075. doi: 10.1046/j.1365-313x.2002.01499.x
Ruffel, S., Gallois, J. L., Moury, B., Robaglia, C., Palloix, A., and Caranta, C. (2006). Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent pepper veinal mottle virus infection of pepper. J. Gen. Virol. 87(Pt 7), 2089–2098. doi: 10.1099/vir.0.81817-0
Singh, S., and Schwartz, H. F. (2010). Breeding common bean for resistance to diseases: A review. Crop. Sci. 50, 2199–2223. doi: 10.2135/cropsci2009.03.0163
Soler-Garzón, A., McClean, P. E., and Miklas, P. N. (2021a). Coding mutations in vacuolar protein-sorting 4 AAA+ ATPase endosomal sorting complexes required for transport protein homologs underlie bc-2 and new bc-4 gene conferring resistance to bean common mosaic virus in common bean. Front. Plant Sci. 12:769247. doi: 10.3389/fpls.2021.769247
Soler-Garzón, A., McClean, P. E., and Miklas, P. N. (2021b). Genome-Wide association mapping of bc-1 and bc-u reveals candidate genes and new adjustments to the host-pathogen interaction for resistance to bean common mosaic necrosis virus in common bean. Front. Plant Sci. 12:699569. doi: 10.3389/fpls.2021.699569
Soler-Garzón, A., McClean, P. E., and Miklas, P. N. (2024). The alleles bc-ud and bc-ur (previously bc-4 gene), representing coding mutations within Vps4 AAA+ ATPase ESCRT protein, interact with other genes to condition resistance to BCMV and BCMNV in common bean. Plant Genome 17:e20421. doi: 10.1002/tpg2.20421
Tang, M., and Feng, X. (2022). Bean common mosaic disease: Etiology, resistance resource, and future prospects. Agronomy 13:58. doi: 10.3390/agronomy13010058
Vallejos, C. E., Astua-Monge, G., Jones, V., Plyler, T. R., Sakiyama, N. S., and Mackenzie, S. A. (2006). Genetic and molecular characterization of the I locus of Phaseolus vulgaris. Genetics 172, 1229–1242. doi: 10.1534/genetics.105.050815
Wani, F., Rashid, S., Saleem, S., Ali, G., Mohiddin, F. A., and Hamid, A. (2025). Topical application of cocktail dsRNA induces plant resistance against bean common mosaic virus (BCMV). Appl. Biochem. Biotechnol. 197, 3431–3446. doi: 10.1007/s12010-025-05187-3
Wani, S., Nisa, Q., Fayaz, T., and Padder, B. A. (2023). “An overview of major bean diseases and current scenario of common bean resistance,” in Diseases in legume crops, eds U. C. Jha, H. Nayyar, K. D. Sharma, E. J. B. von Wettberg, P. Singh, and K. H. Siddique (Singapore: Springer), doi: 10.1007/978-981-99-3358-7_5
Keywords: BCMV, BCMNV, resistance, I-gene, bc-3 gene
Citation: Meghanath D, Wani S, Bashir S, Rashid S, Javaid A, Dar ZA, Wani SH, Sofi PA, Ali G and Hamid A (2025) Delineating the source of resistance to bean common mosaic virus (BCMV) and bean common mosaic necrosis virus (BCMNV) in common bean (Phaseolus vulgaris) cultivars of Jammu and Kashmir, a North-Western Himalayan region. Front. Microbiol. 16:1614122. doi: 10.3389/fmicb.2025.1614122
Received: 18 April 2025; Accepted: 30 May 2025;
Published: 24 June 2025.
Edited by:
Nazia Manzar, National Bureau of Agriculturally Important Microorganisms (ICAR), IndiaReviewed by:
Jose Trinidad Ascencio-Ibáñez, North Carolina State University, United StatesNagamani Sandra, Indian Agricultural Research Institute (ICAR), India
Copyright © 2025 Meghanath, Wani, Bashir, Rashid, Javaid, Dar, Wani, Sofi, Ali and Hamid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aflaq Hamid, ZmFsYWsxOUBnbWFpbC5jb20=
†These authors have contributed equally to this work and share first authorship
‡ORCID: Aflaq Hamid, orcid.org/0000-0001-6353-6070
 Dasari Meghanath
Dasari Meghanath Sumiah Wani1†
Sumiah Wani1† Shahjahan Rashid
Shahjahan Rashid Zahoor Ahmad Dar
Zahoor Ahmad Dar Shabir Hussain Wani
Shabir Hussain Wani Parvaze A. Sofi
Parvaze A. Sofi Aflaq Hamid
Aflaq Hamid