Abstract
Introduction:
Niclosamide (NIC) has significant potential as a clinical therapeutic agent for Clostridioides difficile infection (CDI); however, its strong hydrophobicity hampers its oral bioavailability, and its active effects against C. difficile remain unclear.
Methods:
Niclosamide-loaded controlled-release hyaluronic acid-modified poly (lactic-co-glycolic acid) naosphernes (NIC@PLGA-HAs) were synthesized using an oil-in-water emulsion technique and their effects on C. difficile cell growth, spore germination, biofilm formation, and NIC interaction sites with C. difficile toxin B (TcdB) were analyzed.
Results:
NIC@PLGA-HAs exhibited enhanced solubility and stability, with a water contact angle on a hydrophilic surface of 65.1° and a zeta potential of 31.57 ± 2.08 mV, and pH-responsive (pH 7.4) controlled-release characteristics compared to free NIC. The NIC@PLGA-HAs killed C. difficile vegetative cells at a minimum inhibitory concentration (MIC) of 4 μg/mL. When C. difficile cells were treated with NIC@PLGA-HAs at the 1/4 MIC, spore germination and biofilm formation were significantly inhibited compared to those in untreated cells (P < 0.01). NIC was found to interact with the receptor-binding domain of TcdB at 24 amino acid sites via an enthalpy-driven reaction (enthalpy change, 36.21 kJ/mol and entropy change, 212.9 J⋅mol/K). In vivo experimental findings in Mongolian gerbils indicated that NIC@PLGA-HAs outperformed free NIC in reducing pathological damage, diarrhea severity, weight loss, and TcdB production and enhanced the survival rate.
Conclusion:
These findings presented the therapeutic potential of NIC@PLGA-HAs with high solubility and stability, which simultaneously exerted multiple biological activities against C. difficile.
NIC@PLGA-Has present multiple effects against Clostridioides difficile. 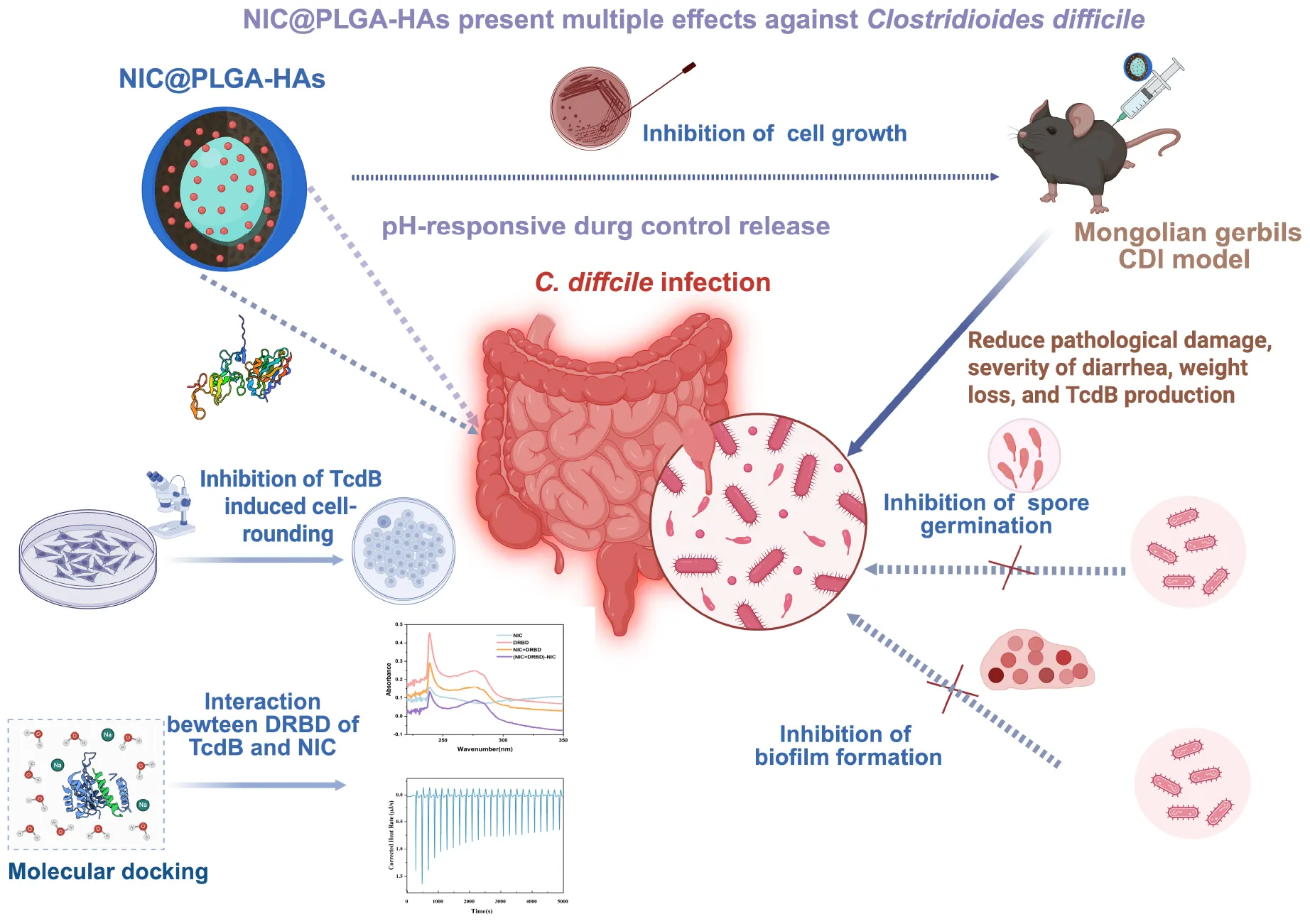
1 Introduction
Clostridioides difficile is a gram-positive spore-forming bacterium that produces toxin A, toxin B (TcdB), and binary toxin, leading to C. difficile infection (CDI) (Paparella et al., 2021). In patients who have used broad-spectrum antibiotics (Rupnik et al., 2009), immunosuppressants (Varma et al., 2022), or chemotherapy drugs (Crobach et al., 2023) for a long time, the destruction of the normal intestinal flora facilitates the invasion, colonization, and rapid multiplication of C. difficile. CDI symptoms include diarrhea, pseudomembranous colitis, intestinal perforation, toxic megacolon, and septic shock, which are life-threatening (Sandhu and McBride, 2018). In 2019, C. difficile was classified as a high-priority resistant organism by the United States Centers for Disease Control and Prevention (Smits et al., 2016; Centers for Disease Control and Prevention, 2019). The global burden of CDI underscores the urgent need for effective countermeasures (Smits et al., 2016; Centers for Disease Control and Prevention, 2019).
Metronidazole (Johnson et al., 2014) and vancomycin (Louie et al., 2011) are the primary antibiotics recommended for CDI treatment; however, the treatment failure and recurrence rates are high, partially because of gut microbiota disruption (Kelly and LaMont, 2008). Fidaxomicin, a new anti-clostridial agent, shows a similar therapeutic effect but has a lower recurrence rate and fewer treatment-related adverse events than vancomycin (Koon et al., 2018; Diaz-Pollan et al., 2023). However, the use of fidaxomicin also leads to gut microbiota disruption, making CDI treatment more challenging than ever (Tannock et al., 2010). Recently, therapeutic antibodies such as bezlotoxumab (Wilcox et al., 2017) and monoclonal (Lowy et al., 2010) or polyclonal (Steele et al., 2013) anti-C. difficile antibodies have been developed to neutralize C. difficile toxins. These antibodies play a crucial role in enhancing immune responses, reducing inflammation, restoring gut microbiota, and preventing CDI recurrence (Kelly et al., 2020), and act synergistically with antibiotics and fecal microbiota transplants (Drekonja et al., 2015; Hocquart et al., 2018). Despite their promising potential, several key challenges persist, such as low antibody specificity (Lawry et al., 2018), high production costs (Ooijevaar et al., 2018), potential immune responses (Madan and Petri, 2012), and uncertainties regarding optimal dosing and administration (DuPont, 2013).
Niclosamide (NIC), a United States Food and Drug Administration-approved anthelmintic drug for tapeworm infections (Xu et al., 2020), exhibited multifaceted mechanisms beyond its primary use (Chen et al., 2018). Although its mode of action remains incompletely characterized, known mechanisms included uncoupling of oxidative phosphorylation (Berube et al., 2018). Over recent years, it has been demonstrated that NIC was a multifunctional drug capable of inhibiting or regulating multiple signaling pathways and biological processes (Biersack, 2024). Its therapeutic potential extended beyond parasitic diseases to include (Wardle et al., 2024) cancers, bacterial and viral infections, and metabolic disorders (Berube et al., 2018; Chen et al., 2024). Previous studies indicated the ability of (Singh et al., 2022) NIC and related salicylanilide derivatives to inhibit Mycobacterium tuberculosis growth. NIC protected RAW264.7 macrophages and CHO cells from anthrax lethal toxin, as well as from Pseudomonas exotoxin and diphtheria toxin (Nestorovich and Bezrukov, 2014). Its mechanism may involve endosome (Wang et al., 2022) acidification, inhibition of the Gram-negative pathogen Pseudomonas aeruginosa, suppressing its quorum sensing response and production of the signaling molecule acyl homoserine lactone (Costabile et al., 2015). Transcriptomic analysis revealed NIC affected approximately 250 P. aeruginosa genes, with high specificity for quorum sensing-dependent targets (Francesco et al., 2013). It also suppressed surface motility, reduced secretion of virulence factors (elastase, pyocyanin, and rhamnolipids), and inhibited biofilm formation (Francesco et al., 2013). Oxyclozanide, a structural analog, demonstrated efficacy against the Gram-positive bacterium methicillin-resistant Staphylococcus aureus and exhibited bactericidal activity (Li et al., 2023). NIC inhibited severe acute respiratory syndrome coronavirus replication and protected Vero E6 cells from virus-induced cytopathic effects (Li et al., 2020). It has been also demonstrated that NIC was used as a potent, low-micromolar inhibitor of pH-dependent human rhinoviruses and influenza virus, Chikungunya virus, and Zika virus. In recent year, NIC has emerged as a potential candidate for repurposing owing to its safety profile and efficacy in treating CDI by protecting cells from the cytotoxic effects of all three C. difficile toxins (Zhang et al., 2022). NIC has been shown to improve symptoms related to both primary infection and recurrence in mouse models (Needham, 2024) and to preserve gut microbiota diversity and composition (Tam et al., 2018). However, the clinical application of NIC remains limited primarily due to two key factors: first, its inherent hydrophobicity substantially reduces bioavailability, especially following oral administration (Wahid et al., 2017; Meng et al., 2020; Liu et al., 2021); and second, the comprehensive biological activity of NIC against C. difficile has not yet been fully elucidated (Tam et al., 2018).
Recent advancements in the field of polymer-based oral drug delivery systems may hold the key to overcoming the low hydrophobicity of NIC (Fonte et al., 2015). Poly (lactic-co-glycolic acid) (PLGA) (Kumar et al., 2022), a prominent hydrophobic polymer, is widely used in drug delivery systems because of its biocompatibility (Chen et al., 2017) and biodegradability (Chatterjee and Chanda, 2022). Hyaluronic acid (HA) has emerged as a highly promising biopolymer for constructing drug delivery carriers owing to its inherent biocompatibility, non-immunogenicity, hydrophilicity, and bioresorbability (Samiraninezhad et al., 2023). Both PLGA and HA have been extensively studied (Martins et al., 2018). Our previous study demonstrated that hydrophilic HA-modified PLGA-HA microspheres, synthesized via microfluidic technology, exhibited sustained release behavior at pH 5.0 and 7.4, but not at pH 2.0 (Tai et al., 2023). The HA modification significantly enhanced the solubility of NIC and augmented its biological activity, thereby addressing the need for effective intestinal drug delivery carriers (Tai et al., 2023). The microfluidic synthesis of polymeric nano-delivery systems entails the optimization of reactant flow rates, selection of suitable polymers, and fine-tuning of synthesis conditions to achieve uniform particle size, stability, and efficient cargo encapsulation (Ran et al., 2017; Zhang et al., 2023).
In the present study, PLGA and HA were used to synthesize NIC-loaded nanospheres using the oil-in-water emulsion technique to assess the enhancement of the water solubility of NIC facilitated by the PLGA-HA nano-delivery system, and the biological activities of NIC@PLGA-HA nanoparticles (NIC@PLGA-HAs) against C. difficile were explored using in vivo and in vitro experiments.
2 Materials and methods
2.1 Reagents and cell lines
PLGA (50:50 mass ratio of lactic acid to glycolic acid, Mw 24,000–38,000, 76,000–115,000, acid-terminated) and NIC (mass fraction 98%) were purchased from Aladdin Reagent Co. Ltd. HA (amine-terminated Mw 80,000–150,000 Da) was purchased from Shanghai Macklin Biochemical Technology Co., Ltd. (Shanghai, China). Polyvinyl alcohol [type 105, alcoholysis degree, 98%–99% (mol/mol)] was purchased from Beijing WoKai Biological Technology Co. Ltd. All reagents are of biomedical grade and used as received. Caco-2 human epithelial colorectal adenocarcinoma cells (correctly identified by STR, freeze cells), fetal bovine serum (New Zealand Origin), streptomycin (United States Pharmacopeia Grade), and penicillin (United States Pharmacopeia Grade) were purchased from Shanghai Sangon Biotech Co. Ltd. Cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C with 5% CO2 and 95% humidity.
2.2 NIC@PLGA-HA synthesis
NIC@PLGA-HA was prepared using the oil-in-water emulsion technique (Tai et al., 2023). First, NIC (2.5, 3.0, and 3.5 mg) and PLGA (25.0 mg) were dissolved in ethyl acetate (5.0 mL), respectively. The solution was mixed with 10 mL of polyvinyl alcohol aqueous solution (1.2%, wt%) and 150 mg of HA until uniform consistency was achieved. Following sonication with the 100, 120, 150, 280, 350 w ultrasonic waves for 5, 10, and 15 min respectively, the mixture was stirred at room temperature until ethyl acetate was completely evaporated. The excess liquid was decanted by filtration, and the sediment was rinsed with deionized water, frozen at −80°C overnight, and lyophilized for 48 h.
2.3 NIC@PLGA-HA characterization
The chemical structures of the NIC@PLGA-HAs and their components were analyzed by Fourier Transform infrared spectroscopy (FT-IR, Nicolet iS 5; Thermo, Waltham, MA, United States) and X-ray diffraction (XRD, Bruker D8 Venture; Bruker Co., Germany). The light transmittance of the samples was measured using an Ultraviolet–Visible spectrophotometer (UV-vis, UV2600; Shimadzu, Japan). Their morphologies were observed by Transmission electron microscope (TEM, JEM 2100F; JEOL, Japan). The particle size and zeta potential distribution of the samples were determined using a particle size analyzer (Dynamic light scattering (DLS); Zetasizer Nano ZS90; Malvern Pananytical, Malvern, United Kingdom). The thermal stabilities of the NIC@PLGA-HAs were analyzed using a Diamond thermogravimetry analysis/differential thermal instrument (TG/DTA, PerkinElmer, United States). The hydrophilicity of the samples was determined by contact angle measurements (JY-82C; Chende, China). The detailed process has been previously described (Tai et al., 2023).
2.4 Determination of the drug-loading rate and drug release
To determine the drug-loading rate, 10 mg of freeze-dried NIC@PLGA-HAs was added to 5 mL of ethanol and placed in a shaker incubator (37°C, 100 × g) for 24 h until all non-encapsulated NIC was dissolved. After the solution was centrifuged at 14,000 × g at 4°C for 15 min, the free NIC absorbance was measured at 370 nm using an UV-vis spectrophotometer (UV-2600, Shimadzu). Finally, the concentration of free NIC in the solution was calculated using a calibration curve (A = 0.0594C + 0.0419, R2 = 0.9966), and the loading efficiency was calculated (Tai et al., 2023). To assess the drug release behavior, 10 mg of NIC and NIC@PLGA-HA were dispersed in 3 mL of release medium buffered solution. The solution was enclosed in dialysis bags (12,000 Da) and immersed in buffer solution (100 mL) in a shaking incubator (37°C, 100 × g). The cumulative release percentage was calculated as described in our previous report (Tai et al., 2023).
2.5 Measurement of the minimum inhibitory concentration (MIC) of NIC@PLGA-HAs
The MIC of NIC@PLGA-HAs were determined using an agar dilution assay according to the Clinical and Laboratory Standards Institute guidelines (Lewis, 2024). Three C. difficile standard strains [American Type Culture Collection (ATCC) BAA-1870, 700057, and 43255] and four clinical isolates were spotted three times on Brucella agar supplemented with hemin (5 μg/mL), vitamin K (1 μg/mL), and defibrillated sheep blood (5%, v/v) in the presence of NIC or NIC@PLGA-HA at concentrations ranging from 1 to 128 μg/mL using 2-fold serial dilutions. Vancomycin at concentrations ranging from 1 to 16 μg/mL using 2-fold serial dilutions was used as positive controls (Tam et al., 2018). The plates were incubated in GENbag Anaer (BioMérieux, Marcy l’Étoile, France) in an anaerobic chamber at 37°C for 48 h. MICs were determined in duplicate and were reported as the lowest concentration at which no colonies were observed.
2.6 Measurement of C. difficile biofilm formation
C. difficile biofilms were measured as reported previously (Dapa and Unnikrishnan, 2013), with minor modifications. An overnight culture of C. difficile cells (ATCC BAA-1870) was diluted to 1 × 106 colony-forming units (CFU)/mL in brain heart infusion-supplemented (BHIS) medium supplemented with 10 g/L glucose and 50 mM sodium phosphate buffer (pH 7.5). Then, 960 μL of the dilution was transferred into 24-well plates and incubated under anaerobic conditions at 37°C for 24 h. Then, 40 μL of BHIS with or without the 1/4 MIC of vancomycin, NIC, or NIC@PLGA-HA was added to the wells and incubated under anaerobic conditions at 37°C for 24 h. After that, the culture supernatant was removed, the wells were washed once with PBS and stained for 30 min with 0.1% (wt/vol) crystal violet in water. After the wells were washed twice with PBS, the bound crystal violet was solubilized by 95% ethanol. The absorbance at 570 nm was determined with a Synergy HT microplate reader (BioTek, CA, United States). Experiments were independently repeated three times.
2.7 Determination of C. difficile spore germination
C. difficile spores (ATCC BAA-1870) were prepared and purified as described previously (Sorg and Sonenshein, 2010). Briefly, 107 spores/mL suspended in BHIS medium supplemented with 0.1% taurocholate was mixed without and with the 1/4 MIC of NIC or NIC@PLGA-HAs and incubated in an anaerobic environment at 37°C for 48 h. At 2 days intervals from days 2 to 14, the culture was inoculated onto BHIS agar plates for further subculturing in an anaerobic environment at 37°C for 48 h, respectively. CFUs on each plate were counted. Experiments were independently repeated three times.
2.8 TcdB toxicity assay
C. difficile toxin B toxicity was performed as previously described (Tam et al., 2018). Caco-2 cells were seeded in a 24-well plate at 5 × 104 cells/well and cultured for 24 h. After mixing for 15 min, NIC@PLGA-HAs (10 ng/mL) and purified TcdB (1.6 ng/mL) prepared in our laboratory (Lin et al., 2024) were added to the wells. Purified TcdB alone at the same concentration was used as the positive control. The viability of Caco-2 cells was assessed at 12, 24, and 48 h using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega, Beijing, China). Morphologically, rounded cells were counted and calculated relative to the total number of cells.
2.9 Molecular modeling and kinetic simulation
Molecular docking simulation was conducted using AutoDock 4.2 by the Lamarckian genetic algorithm. The structure of the NIC was generated using KingDraw and optimized using the Gaussian 09 program. Geometry optimization was performed using the hybrid Becke, 3-parameter, Lee-Yang-Parr functional together with the transmembrane delivery and receptor-binding domain (DRBD) C, H, O, N, and F atoms. The crystal structure of DRBD (PDB ID: 4nc2) was obtained from Brook-Haven Protein Bank.1 Molecular dynamics simulation was employed to further refine the binding mode of the molecule-protein complex using the Desmond program. Both the protein and the molecule were parameterized using the OPLS4 force field and the SPC/E model was used as the water solvent. The molecule-protein complex was positioned within a cubic water box and solvated (Decherchi and Cavalli, 2020). Furthermore, for AlphaFold 3-specific docking, both the protein sequence and ligand structure were inputted into AlphaFold 3. The model predicted the bound complex by integrating ligand atomic coordinates and calculating binding affinity scores (Abramson et al., 2024; He et al., 2025).
2.10 Expression and purification of wide type and mutant of TcdB DRBD
Recombinant proteins were expressed and purified as previously reported (Chen et al., 2021). Gene sequences corresponding to TcdB DRBD with wide type (DRBDWT) and mutant (DRBDMT) were cloned into the pET-28a (+) vector (Sangon Biotech Co., Ltd.). DRBDMT was mutated in the amino acids as predicted above according to binding energies. Proteins expressed with a 6-His tag at the N-terminus were purified using Ni2+-NTA affinity chromatography (Sangon Biotech Co., Ltd.).
2.11 Ultraviolet-visible (UV-vis) absorption spectroscopy and isothermal titration calorimetry
Ultraviolet-visible spectra were recorded using an UV-vis spectrophotometer (UV2600, Shimadzu, Japan) to measure the interaction between DRBD and NIC in the wavelength range of 200–800 nm at room temperature (Wang et al., 2019). Isothermal titration calorimetry (ITC) was conducted on a NANOITC (TA Instruments, United States) at 25°C in a well containing 200 μL of reaction mixture (50 mM PBS, pH 7.4, 15% DMSO) and 12 μM TcdB-DRBD. The ligand was titrated into the protein solution over 25 injections of 2 μL for 6 s, with a 150 s equilibration period between injections. Data fit the model of a distinct binding site. The first injection for each sample was excluded from data fitting. A separate titration without TcdB-DRBD was performed for each condition to subtract the background heat of dilution (Gurung et al., 2017).
2.12 Animal model
The primary CDI model was established according to our previous report (Wan et al., 2024). Mongolian gerbils were purchased from the Animal Center of the Hangzhou Medical College. Mongolian gerbils were divided into five groups (N = 10 per group) and received mixed antibiotics in drinking water for 3 days. Animals were further treated with clindamycin (10 mg/kg) for 1 day. Mongolian gerbils in the four infected groups were orally administered 105 CFU of C. difficile spores (ATCC BAA-1870), and those in the non-infected control group were administered water with 5% DMSO. Animals in the three infection groups were treated with vancomycin, NIC, or NIC@PLGA-HA (20 mg/kg suspended in drinking water with 5% DMSO). The animals were monitored for signs of disease such as diarrhea, wet tails, and weight loss. After 4 days, the animals were euthanized and stool samples and intestinal tissues were collected. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of the Zhejiang Institute for Food and Drug Control, and were approved by the Animal Ethics Committee of the Zhejiang Institute for Food and Drug Control. The experimental animal license number is SCXK (Zhe) 2019-0002.
2.13 Statistical analysis
Data were statistically analyzed using GraphPad Prism version 10 (GraphPad Software, San Diego, CA, United States). Significant differences were determined using one-way analysis of variance (ANOVA). Statistical significance was set at P < 0.05.
3 Results and discussion
3.1 Characterization of NIC@PLGA-HAs
FT-IR, TEM, DLS, XRD, thermogravimetry (TG), and contact angle measurements were used to analyze the chemical composition, microscopic morphology, particle size, crystal structure, stability, and hydrophilicity of the NIC@PLGA-HAs (Figure 1). The chemical structures of NIC, PLGA, and HA are shown in Figure 1a. Compared with those of NIC, the FT-IR spectra of the NIC@PLGA-HAs exhibited two distinct absorption peaks for NIC at 1,328 and 1,653 cm–1, corresponding to the C-N and -C = O functional groups, respectively, indicating that NIC was successfully loaded into the PLGA-HAs (Figure 1b). Compared with those of PLGA, the FT-IR spectra of NIC@PLGA-HAs featured peaks at 1,748, 1,424, 1,172, and 1,611 cm–1, corresponding to the stretching vibrations of C = O, C-H, and C-O-C functional groups of PLGA and O-H, N-H, and C-O functional groups of HA, respectively, indicating that the PLGA-HA polymeric shell encapsulated the NIC.
FIGURE 1
Thermogravimetry and derivative thermogravimetry revealed sharp melting point peaks for unprocessed NIC, HA, and PLGA, which appeared at 307.7, 238.2, and 324.3°C respectively, and disappeared for NIC@PLGA-HAs, corroborating that NIC was loaded into the PLGA-HAs (Figure 1c and Supplementary Figure 1). For NIC@PLGA-HAs, a new peak appeared at 361.7°C, whereas that of NIC reduced in size. The peak had a large surface area and close contact between PLGA-HAs and NIC. These changes were considered to indicate a change in the NIC from the crystalline to the amorphous state and an increase in the thermal stability. XRD was used to confirm the crystal structures of NIC, HA, PLGA, and NIC@PLGA-HA (Figure 1d), confirming the amorphous nature of NIC@PLGA-HAs. The disappearance or reduced intensities of the XRD peaks are indicative of the amorphous nature of the nanoparticles compared with the unprocessed NIC.
TEM was used to investigate the micromorphology of NIC@PLGA-HAs with different preparation conditions (Figure 1e and Supplementary Figure 2). TEM images revealed that the NIC@PLGA-HAs maintained dispersibility and a uniform shape, suggesting that the particle size and shape of the NIC@PLGA-HAs were related to the concentration of NIC, ultrasonication time, and power. DLS and zeta potential measurements were straightforward table-top techniques that can be performed in standard laboratory settings to assess the hydrodynamic size and surface charge (Bhattacharjee, 2016). The diameters, mean diameter, size distribution, and polydispersity index of NIC@PLGA-HAs with different preparation conditions were verified using DLS (Figure 1f and Supplementary Figure 3), revealing the homogeneous particle hydrodynamic diameters of 2,533.7 nm for NIC particles and 259–530 nm for NIC@PLGA-HAs, depending on the NIC concentration, ultrasonication time, and power. The reduced particle size in NIC@PLGA-HAs was attributed to the hydrophilic surface of PLGA-HA counteracting NIC’s aggregation tendency caused by its low solubility and hydrophobicity. The polydispersity index value of NIC@PLGA-HAs ranged from 0.2 to 0.4, suggesting a moderate level of polydispersity. The zeta potentials of NIC@PLGA-HA synthesized under different ultrasonication time, the different NIC concentrations, and different powers were also measured (Figure 1f and Supplementary Figure 3). In principle, a system was considered electrostatically stable if the absolute zeta potential was above 20 mV (Chatterjee et al., 2019). The highest absolute zeta potential of 31.57 ± 2.08 mV was observed for NIC@PLGA-HAs synthesized by adding 3.5 mg NIC to the mixture using an ultrasound assay, as this lowered the probability of particle aggregation. NIC@PLGA-HAs were highly stable in solution, as evidenced by the particle size and zeta potential measurements, rendering them well suited for drug delivery applications. When PLGA and HA were physically mixed to form emulsified particles, the presence of HA may affect the final particle zeta potential (Hu et al., 2015). HA was a negatively charged polysaccharide, with the carboxyl groups on its molecular chains providing negative charges (Naik et al., 2017). When combined with PLGA (through physical adsorption or encapsulation within PLGA particles), the surface negative charges of HA increased the overall negative charge of the particles, thereby enhancing the zeta potential. Generally, the as-prepared NIC@PLGA-HAs exhibited high drug loading, a narrow size distribution, and both colloidal and thermal stability, demonstrating potential for manufacturability.
3.2 Hydrophilicity and NIC release behavior of NIC@PLGA-HAs
The water contact angle of NIC@PLGA-HAs was 65.1°, which was less than that of hydrophobic NIC (84.5°) (Figures 2a, b), demonstrating that PLGA-HA copolymers improved the hydrophilicity of NIC by connecting with other water-soluble substances, which was in agreement with previous findings (Naik et al., 2017; Wu H. et al., 2020). The critical micelle concentration (CMC) of NIC@PLGA-HAs was determined using a probe-free UV-vis spectroscopy (Ma et al., 2016). The CMCs of NIC and NIC@PLGA-HAs were shown in Figure 2. Through linear regression analysis, CMCs was determined to be 83.23 μg/mL for NIC and 300.86 μg/mL for NIC@PLGA-HAs. Upon exceeding the CMC, NIC@PLGA-HAs self-assembled into micelles in an aqueous medium. The hydrophobic nature of the NIC component within NIC@PLGA-HAs drove them to evade the aqueous environment, contributing to the formation of a hydrophobic micellar core. Therefore, an increase in the NIC concentration resulted in a corresponding increase in UV absorbance. Notably, the CMC of NIC@PLGA-HAs was higher than that of NIC, which could be attributed to the encapsulation by PLGA-HA. The increased CMC value indicated enhanced hydrophilicity (Ma et al., 2016), which aligned with the contact angle measurement results.
FIGURE 2
In vitro drug release from NIC@PLGA-HAs synthesized using the microfluidic method was pH-dependent, in line with our previous study (Tai et al., 2023). NIC releasing from PLGA-HAs was faster at pH 7.4 than at pH 5.0 and 2.0, demonstrating that NIC@PLGA-HAs had the pH-dependent nature of drug release and good solubility of PLGA-HA at the high pH, as reported previously (Martins et al., 2018). PLGA-HA had a primary carboxyl group and an ester group, which became deprotonated at pH 7.4, inducing the formation of soluble biopolymeric hydrogel networks (Wu H. et al., 2020). NIC@PLGA-HAs exhibited rapid burst release during the first 8 h, suggesting no chemical interactions between NIC and the PLGA-HA polymeric chains. After 8 h, the percentage of NIC released from PLGA-HAs was significantly higher than that of free NIC, and the release from NIC@PLGA-HAs was highly consistent with a quasi-first-order kinetic model (R2 = 0.97) (Figure 2c), which suggested that NIC adsorption occurred primarily via chemical or physical adsorption. The surface modification of PLGA nanoparticles with HA has numerous advantages, such as reducing the burst effect in drug release, facilitating zeta potential inversion for enhanced cellular adhesion and retention at the target site (Chen et al., 2017), and providing the opportunity to conjugate targeting ligands to surface amino groups (Chatterjee and Chanda, 2022). Therefore, this modification was beneficial to the binding between NIC and TcdB proteins, thereby likely improving the CDI treatment effect. Furthermore, the incorporation of HA enabled pH-dependent drug release, whereas PLGA-HA-modified nanoparticles offered dual protection against enzymatic and gastric degradation and reduced the gastrointestinal irritation caused by NIC via a controlled release mechanism (Chatterjee et al., 2019).
3.3 NIC@PLGA-HAs inhibit C. difficile vegetative cell growth, spore germination, and biofilm formation
The anti-clostridial activity of NIC@PLGA-HAs was evaluated using seven C. difficile strains (Table 1). NIC@PLGA-HAs inhibited the growth of all C. difficile strains tested, at concentrations ranging from 0.25 to 4 μg/mL. The half-maximal inhibitory concentration (IC50) was 1 μg/mL and the 90% inhibitory concentration (IC90) was 4 μg/mL. The minimal inhibitory concentration (MIC) of vancomycin ranged from 0.25 to 2 μg/mL (IC50 and IC90: 0.5 and 1 μg/mL). Therefore, the inhibitory capability of NIC@PLGA-HAs was comparable to that of vancomycin, the preferred therapeutic agent for primary and severe CDI (Hastey et al., 2017). However, the potential for prolonged NIC@PLGA-HAs exposure to induce resistance associated gene mutations remains experimentally unaddressed. The NIC@PLGA-HAs’ multi-component nature theoretically reduced selective pressure for drug resistance, but which should be further verified later (Su et al., 2021).
TABLE 1
| Strain | Agent (μg/mL) | ||
| Vancomycin | NIC | NIC@PLGA-HAs | |
| C. difficile (ATCC BAA-1870) | 2–4 | 4 | 2–4 |
| C. difficile (ATCC 700057) | 1 | 8 | 4 |
| C. difficile (ATCC 43255) | 1 | 16 | 4 |
| C. difficile (clinical isolate, ST35a) | 1 | 4 | 2 |
| C. difficile (clinical isolate, ST54) | 1 | 4-8 | 0.5 |
| C. difficile (clinical isolate, ST39) | 1 | 4 | 1–2 |
| C. difficile (clinical isolate, ST37) | 2 | 4 | 4 |
Minimum inhibitory concentratio (MIC) of three agents for inhibiting growth of C. difficile.
aST, sequence type.
C. difficile spore germination was a major factor in treatment failure of CDI and CDI recurrence (Zhu et al., 2018). Inhibition of C. difficile spore germination contributed to the effectiveness of CDI treatment and reduced CDI recurrence (Daou et al., 2019). In contrast to NIC, NIC@PLGA-HAs at the 1/4 MIC were able to inhibit C. difficile spore germination consistently 10 days after exposure (Figure 3a). However, C. difficile spore germination was distinctly decreased on the 12th day after exposure or not to NIC and NIC@PLGA-HAs at the 1/4 MIC, which phenomena should be further investigated. Overall, these results demonstrated that NIC@PLGA-HAs exhibited maximal inhibitory efficacy against C. difficile spore germination specifically during the initial 2–4 days (Semenyuk et al., 2014), suggesting heightened sensitivity to the outgrowth stage of spore germination (Horvat and Rupnik, 2018).
FIGURE 3
C. difficile biofilms were closely served as a niche for C. difficile spore formation (Barra-Carrasco and Paredes-Sabja, 2014). Additionally, C. difficile biofilm enhanced its persistence in the gut, increased its resistance to antibiotics, promoted CDI recurrence, and prolonged the duration of CDI (Stewart et al., 2020; Dicks, 2023). C. difficile treated with NIC or NIC@PLGA-HA at the 1/4 MIC produced significantly less biofilm than untreated C. difficile (P < 0.01), and no significant difference was found between NIC and NIC@PLGA-HA (Figure 3b).
3.4 NIC@PLGA-HAs bind to the delivery and receptor-binding domain (DRBD) domain of TcdB to suppress cytotoxicity
Effective treatment should target not only C. difficile cells and spores but also their toxins (Nooij et al., 2024; Tiseo et al., 2024). C. difficile toxins, particularly TcdB, damaged the colonic epithelium, leading to inflammation and severe diarrhea (Lyras et al., 2009). TcdB contained four structural domains, of which the DRBD recognized receptors on intestinal cells and assisted the glycosyltransferase domain to enter host cell cytosol (Kuehne et al., 2010). CDI has been effectively treated by blocking the interaction between DRBD of TcdB and host cell surface receptors (Dicks, 2023). Our previous study demonstrated that at a low concentration (10 ng/mL), NIC@PLGA-HA did not inhibit the growth of Caco-2 cells (Tai et al., 2023). The present study showed that TcdB (1.6 ng/mL)-induced cell rounding of Caco-2 cells was inhibited in the presence of NIC@PLGA-HA (10 ng/mL) (Figure 4a).
FIGURE 4
Based on the molecular docking between NIC and TcdB DRBDWT, DRBDMT was constructed by artificially mutating these amino acids (Supplementary Table 1). Nucleotide sequences with gene mutations were produced using the overlap PCR with the primers listed in (Supplementary Table 2). Fractions containing wild-type DRBDWT and mutant DRBDMT were verified by SDS-PAGE (Supplementary Figure 4). To gain a deeper insight into the binding mechanism between DRBD of TcdB and NIC, we simulated the molecular docking between NIC and TcdB DRBDWT and DRBDMT, respectively (Figure 4b). NIC entered the cavity of TcdB, and 24 amino acids (Table 2) spatially complemented the structure of NIC, thereby stabilizing the conformation of the resultant complex (Figure 4b-1) as described previously (Lawal et al., 2023). This association was characterized by the hydrogen bonding of hydrogen atoms within the molecule at distances of 2.9Å, 2.7Å, 2.4Å, 2.3Å, 1.8 Å, and etc. Hydrophobic and electrostatic interactions played a critical role in the binding between DRBD and NIC. The variance in residues from the DRBD that tethered to the NIC elucidated the observed diversity in binding outcomes. The results showed that NIC interacted unusually with the DRBDMT, forming an unstable complex compared to TcdB DRBDWT (Figure 4b-2). The AlphaFold3 was also used to further confirm the interaction binding sites. The results showed that NIC bound to significantly more sites on TcdB than those on mutated TcdB and exhibited higher binding energy (−6.30 Kcal/mol) (Supplementary Figure 5). The UV absorption spectra of NIC interacting with DRBD under simulated physiological conditions were shown in Figure 4c. Complex formation between DRBD and NIC induced clear peak changes at 275 and 230 nm compared to those for DRBD and NIC alone, confirming successful complexation. In the ITC analysis, the energy change curves indicated an endothermic process during DRBD titration with NIC. The endothermicity of the peaks decreased with continuous DRBD-NIC combination, displaying a gradual saturation process because of heat adsorption by the complex (Figure 4d). To eliminate interference from dilution heat in the curve-fitting process, an NIC DMSO solution was titrated into a PBS solution. The titration reactions indicated an endothermic dilution process. In contrast, the fitting results of the ITC curves obtained under identical experimental conditions for DRBDMT and NIC failed to demonstrate notable binding features, suggesting weak or absent binding interactions (Figure 4d). According to the Wiseman isotherm theory (Wang et al., 2019), an S-shaped fitted curve indicated strong interactions, whereas a parabolic curve suggested low affinity. Therefore, the interaction between the NIC and DRBDWT was stronger than between the NIC and DRBDMT.
TABLE 2
| Proteins | Binding energy (Kcal/mol) | Amino acid residues of binding sites | Length of hydrogen bonds (Å) |
| DRBD | −6.22 | ILE-4, ASN-2, ARG-31, ASP-72 | 2.8, 2.1, 3.5, 2.3 |
| ASP-112, ARG-115, SER-51, TYR-38 | 3.2, 2.7, 2.6, 2.5 | ||
| THR-101, ASP-152, GLU-781, SER-153 | 2.3, 1.8, 3.5, 3.2 | ||
| LYS-454, SER-470, ASN-472, ASP-431 | 1.7, 3.1, 2.1, 2.2 | ||
| LYS-231, ALA-226;ASN-2 | 2.7, 1.5; 3.0 | ||
| TYR-424, THR-449, ASN-448, ASN-276, and LYS-259 | 2.9, 3.3, 3.4, 1.8, and 1.9 | ||
| Mutated DRBD | −1.89 | ASN-276 and LYS-259 | 1.8 and 1.9 |
Interaction sites between Niclosamide (NIC) and DRBDWT/DRBDMT simulated by the molecular docking.
The thermodynamic parameters for the interaction between NIC and DRBD were listed in Table 2. The negative Gibbs energy change value (ΔG, −27.26 kJ/mol) indicated that the interaction between NIC and DRBD was a spontaneous process. NIC displayed the strongest binding affinity with DRBD, with a binding energy of −6.22 Kcal/mol. The low ΔG value of the NIC–DRBD complex indicated these interactions. The positive values of ΔH (36.21 kJ/mol) and ΔS (212.9J mol/K) supported that the binding between NIC and DRBD was an enthalpy-driven reaction (Figure 4d). Although this process required energy absorption, an increase in the entropy caused it to occur spontaneously. In biological processes, such phenomena were often involved in protein folding, molecular binding, or other intermolecular forces, and are typically accompanied by a transition from a structured to a less ordered state, naturally gravitating toward a state of equilibrium (Huang and Zhan, 2007).
Together, the above results indicated that the interaction between NIC and DRBD was multifaceted and involved hydrophobic forces, as well as hydrogen bonds and electrostatic interactions facilitated by ionic and polar residues (e.g., arginine and asparagine) in the adjacent ligand, contributing to complex stabilization. A previous study demonstrated that NIC inhibited TcdB toxicity by targeting pore formation (Tam et al., 2018). NIC has been documented to moderately elevate endosomal pH via a unique proton-shuttle mechanism, distinguishing it from other methods of endosomal deacidification, such as lysosomotropism (Jurgeit et al., 2012). Our findings suggested that NIC not only inhibited spore germination and biofilm formation, but also prevented TcdB-induced cell rounding by binding to the DRBD of TcdB.
3.5 In vivo treatment of an animal CDI model with NIC@PLGA-HAs
Mongolian gerbils challenged with C. difficile were treated with NIC, NIC@PLGA-HA, and vancomycin (the clinical CDI therapeutic agent used as positive control), respectively (Figure 5a). NIC and NIC@PLGA-HAs exhibited significant evidence of in vivo treatment efficacy in the CDI animal model. Typical CDI symptoms in Mongolian gerbils included severe weight loss on days 2 and 3 post-challenge, accompanied by diarrhea and a high mortality rate (Wan et al., 2024). NIC@PLGA-HAs remarkably protected Mongolian gerbils from death, as indicated by the higher survival rate than those in the vancomycin- and NIC-treated CDI groups (Figure 5b). All the treated animals showed less weight loss than the CDI control group (Figure 5c). NIC@PLGA-HAs inhibited toxin production more effectively than NIC, and was as effective as vancomycin (P < 0.001) (Figure 5d). Intestinal histopathological analysis indicated that animals in the CDI group exhibited infection characteristics, including increased inflammatory cell infiltration into intestinal tissues, enhanced mucus secretion, and significant epithelial cell shedding, which were significantly reduced in the NIC@PLGA-treated group (Figure 5e). NIC@PLGA-HAs and vancomycin protected Mongolian gerbils challenged with C. difficile at similar levels but via different mechanisms. Vancomycin directly killed C. difficile vegetative cells (Darkoh et al., 2022), and however NIC@PLGA-HAs not only inhibited the growth of C. difficile but also antagonize TcdB. Free NIC in its ethanolamine salt form (NEN) demonstrated inherent microbiota compatibility, causing no structural disruption and even increasing α-diversity in C. difficile-infected mice (Tam et al., 2018). While systemic PLGA delivery can reduce microbial diversity (Chaplin et al., 2020), the hyaluronic acid (HA) component counteracted this effect. Thus, the NIC@PLGA-HA nanocomposites theoretically combined NIC’s virulence-targeted action with HA’s microbiome-stabilizing properties while mitigating PLGA-associated risks, which was worthy of being studied in the later (Mao et al., 2021).
FIGURE 5
4 Conclusion
Our findings revealed that NIC@PLGA-HAs, with high solubility and stability and the pH-dependent nature of drug release, effectively killed C. difficile vegetative cells at a MIC of 4 μg/mL and simultaneously inhibited biofilm formation and spore germination. NIC interacted with TcdB at 24 amino acid sites within the DRBD, and however the key sites involved in NIC binding to the TcdB DRBD remained to be determined. Notably, NIC@PLGA-HAs showed superior treatment efficacy over free NIC in vivo, with increasing the survival rate and reducing CDI pathological injuries. In vitro and in vivo studies revealed that NIC@PLGA-HAs exhibited extended-release kinetics compared to free NIC, along with significantly improved aqueous solubility, enhanced chemical stability, and pH-responsive release behavior. These improvements might further improve oral bioavailability of NIC@PLGA-HAs compared to free NIC, and present superior inhibition of C. difficile spore germination and biofilm formation. We are also going to investigate synergistic interactions on treatment of CDI led by hypervirulent C. difficile strains by combining NIC@PLGA-HAs with vancomycin or fidaxomicin in the future studies. On the other hand, C. difficile drug resistance associated gene mutations induced by prolonged NIC@PLGA-HAs exposure should also be further investigated later.
Statements
Data availability statement
The original contributions presented in this study are included in this article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by the Animal Ethics Committee of Zhejiang Institute for Food and Drug Control (SCXK (Zhe) 2019-0002). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YT: Conceptualization, Writing – review and editing, Methodology, Writing – original draft, Funding acquisition, Visualization, Resources. MZ: Formal Analysis, Funding acquisition, Writing – original draft. YH: Formal Analysis, Methodology, Writing – original draft, Funding acquisition, Resources. HH: Data curation, Software, Writing – original draft, Investigation. FZ: Writing – original draft, Software, Data curation. MT: Software, Data curation, Writing – original draft. XS: Resources, Formal Analysis, Methodology, Investigation, Software, Writing – original draft. SW: Data curation, Conceptualization, Writing – original draft, Software. YC: Validation, Writing – review and editing, Investigation, Supervision. DJ: Funding acquisition, Validation, Resources, Writing – review and editing, Conceptualization. SL: Data curation, Formal Analysis, Methodology, Validation, Visualization, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This project was supported by the Zhejiang Provincial Natural Science Foundation key project (LXZ22H300001), Zhejiang Provincial Health and Construction Commission Foundation project (2024KY918, 2024ZL370), 2022 Institution-Specific Health Zhejiang Special Program project of Hangzhou Medical College (YS2022008), Medical Science and Technology Project of Zhejiang Province (2025KY1030), the National Natural Science Foundation of China (82372299), and the Basic Research Projects of Hangzhou Medical College (KYYB2023003).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1617631/full#supplementary-material
Footnotes
References
1
AbramsonJ.AdlerJ.DungerJ.EvansR.GreenT.PritzelA.et al (2024). Accurate structure prediction of biomolecular interactions with AlphaFold 3.Nature630493–500. 10.1038/s41586-024-07487-w
2
Barra-CarrascoJ.Paredes-SabjaD. (2014). Clostridium difficile spores: A major threat to the hospital environment.Fut. Microbiol.9475–486. 10.2217/fmb.14.2
3
BerubeB. J.CastroL.RussellD.OvechkinaY.ParishT. (2018). Novel screen to assess bactericidal activity of compounds against non-replicating Mycobacterium abscessus.Front. Microbiol.9:2417. 10.3389/fmicb.2018.02417
4
BhattacharjeeS. (2016). DLS and zeta potential – what they are and what they are not?J. Controll. Release235337–351. 10.1016/j.jconrel.2016.06.017
5
BiersackB. (2024). The antifungal potential of niclosamide and structurally related salicylanilides.Int. J. Mol. Sci.25:5977. 10.3390/ijms25115977
6
Centers for Disease Control and Prevention (2019). Antibiotic resistance threats in the United States.Washington, DC: US Department of Health and Human Services.
7
ChaplinA.GaoH.AsaseC.JosephsT.BoursierJ.ZwingermanN.et al (2020). Systemically-delivered biodegradable PLGA alters gut microbiota and induces transcriptomic reprogramming in the liver in an obesity mouse model.Sci. Rep.10:13786. 10.1038/s41598-020-69745-x
8
ChatterjeeM.ChandaN. (2022). Formulation of PLGA nano-carriers: Specialized modification for cancer therapeutic applications.Mater. Adv.3837–858. 10.1039/d1ma00600b
9
ChatterjeeS.HuiP. C. L.KanC. W.WangW. (2019). Dual-responsive (pH/temperature) pluronic F-127 hydrogel drug delivery system for textile-based transdermal therapy.Sci. Rep.9:11658. 10.1038/s41598-019-48254-6
10
ChenP.ZengJ.LiuZ.ThakerH.WangS.TianS.et al (2021). Structural basis for CSPG4 as a receptor for TcdB and a therapeutic target in Clostridioides difficile infection.Nat. Commun.12:3748. 10.1038/s41467-021-23878-3
11
ChenW.MookR. A.PremontR. T.WangJ. (2018). Niclosamide: Beyond an antihelminthic drug.Cell. Signal.4189–96. 10.1016/j.cellsig.2017.04.001
12
ChenX.ChenJ.LiB.YangX.ZengR.LiuY.et al (2017). PLGA-PEG-PLGA triblock copolymeric micelles as oral drug delivery system: In vitro drug release and in vivo pharmacokinetics assessment.J. Colloid Interface Sci.490542–552. 10.1016/j.jcis.2016.11.089
13
ChenX.YangS.ZhuB.XieS.LiX.LiuJ.et al (2024). Effects of environmentally relevant concentrations of niclosamide on lipid metabolism and steroid hormone synthesis in adult female zebrafish.Sci. Total Environ.910:168737. 10.1016/j.scitotenv.2023.168737
14
CostabileG.d’AngeloI.RampioniG.BondìR.PompiliB.AscenzioniF.et al (2015). Toward repositioning niclosamide for antivirulence therapy of Pseudomonas aeruginosa lung infections: Development of inhalable formulations through nanosuspension technology.Mol. Pharm.122604–2617. 10.1021/acs.molpharmaceut.5b00098
15
CrobachM. J. T.HornungB. V. H.VerduinC.VosM. C.HopmanJ.KumarN.et al (2023). Screening for Clostridioides difficile colonization at admission to the hospital: A multi-centre study.Clin. Microbiol. Infect.29891–896. 10.1016/j.cmi.2023.02.022
16
DaouN.WangY.LevdikovV. M.NandakumarM.LivnyJ.BouillautL.et al (2019). Impact of CodY protein on metabolism, sporulation and virulence in Clostridioides difficile ribotype 027.PLoS One14:e0206896. 10.1371/journal.pone.0206896
17
DapaT.UnnikrishnanM. (2013). Biofilm formation by Clostridium difficile.Gut Microbes4397–402. 10.4161/gmic.25862
18
DarkohC.KeitaK.OdoC.OyaroM.BrownE. L.AriasC. A.et al (2022). Emergence of clinical Clostridioides difficile isolates with decreased susceptibility to vancomycin.Clin. Infect. Dis.74120–126. 10.1093/cid/ciaa912
19
DecherchiS.CavalliA. (2020). Thermodynamics and kinetics of drug-target binding by molecular simulation.Chem. Rev.12012788–12833. 10.1021/acs.chemrev.0c00534
20
Diaz-PollanB.Carrasco MolinaS.MarceloC.De Gea GrelaA.Martínez-MartínP.Jiménez-GonzálezM.et al (2023). A comparison of the efficacy of treatment with fidaxomicin versus vancomycin in Clostridioides difficile infection.Cureus15:e48735. 10.7759/cureus.48735
21
DicksL. M. T. (2023). Biofilm formation of Clostridioides difficile, toxin production and alternatives to conventional antibiotics in the treatment of CDI.Microorganisms11:2161. 10.3390/microorganisms11092161
22
DrekonjaD.ReichJ.GezahegnS.GreerN.ShaukatA.MacDonaldR.et al (2015). Fecal microbiota transplantation for Clostridium difficile infection: A systematic review.Ann. Intern. Med.162630–638. 10.7326/M14-2693
23
DuPontH. L. (2013). Diagnosis and management of Clostridium difficile infection.Clin. Gastroenterol. Hepatol.111216–1223. 10.1016/j.cgh.2013.03.016
24
FonteP.AraújoF.SilvaC.PereiraC.ReisS.SantosH. A.et al (2015). Polymer-based nanoparticles for oral insulin delivery: Revisited approaches.Biotechnol. Adv.331342–1354. 10.1016/j.biotechadv.2015.02.010
25
FrancescoF. M.PillaiC. R.PichonC.AliS. O.BonfigliA.LacourS.et al (2013). New life for an old drug: The anthelmintic drug niclosamide inhibits Pseudomonas aeruginosa quorum sensing.Antimicrob. Agents Chemother.57996–1005. 10.1128/aac.01952-12
26
GurungA. B.BhattacharjeeA.Ajmal AliM.Al-HemaidF.LeeJ. (2017). Binding of small molecules at interface of protein-protein complex – a newer approach to rational drug design.Saudi J. Biol. Sci.24379–388. 10.1016/j.sjbs.2016.01.008
27
HasteyC. J.DaleS. E.NaryJ.CitronD.LawJ. H.Roe-CarpenterD. E.et al (2017). Comparison of Clostridium difficile minimum inhibitory concentrations obtained using agar dilution vs broth microdilution methods.Anaerobe4473–77. 10.1016/j.anaerobe.2017.02.006
28
HeX.LiJ.ShenS.XuH. E. (2025). AlphaFold3 versus experimental structures: Assessment of the accuracy in ligand-bound G protein-coupled receptors.Acta Pharmacol. Sin.461111–1122. 10.1038/s41401-024-01429-y
29
HocquartM.LagierJ. C.CassirN.SaidaniN.EldinC.KerbajJ.et al (2018). Early fecal microbiota transplantation improves survival in severe Clostridium difficile infections.Clin. Infect. Dis.66645–650. 10.1093/cid/cix762
30
HorvatS.RupnikM. (2018). Interactions between Clostridioides difficile and fecal microbiota in in vitro batch model: Growth, sporulation, and microbiota changes.Front. Microbiol.9:1633. 10.3389/fmicb.2018.01633
31
HuK.ZhouH.LiuY.LiuZ.LiuJ.TangJ.et al (2015). Hyaluronic acid functional amphipathic and redox-responsive polymer particles for the co-delivery of doxorubicin and cyclopamine to eradicate breast cancer cells and cancer stem cells.Nanoscale78607–8618. 10.1039/C5NR01084E
32
HuangX.ZhanC. G. (2007). How dopamine transporter interacts with dopamine: Insights from molecular modeling and simulation.Biophys. J.933627–3639. 10.1529/biophysj.107.110924
33
JohnsonS.LouieT. J.GerdingD. N.CornelyO. A.Chasan-TaberS.FittsD.et al (2014). Vancomycin, metronidazole, or tolevamer for Clostridium difficile infection: Results from two multinational, randomized, controlled trials.Clin. Infect. Dis.59345–354. 10.1093/cid/ciu313
34
JurgeitA.McDowellR.MoeseS.MeldrumE.SchwendenerR.GreberU. F. (2012). Niclosamide is a proton carrier and targets acidic endosomes with broad antiviral effects.PLoS Pathog.8:e1002976. 10.1371/journal.ppat.1002976
35
KellyC. P.LaMontJ. T. (2008). Clostridium difficile–more difficult than ever.N. Engl. J. Med.3591932–1940. 10.1056/NEJMra0707500
36
KellyC. P.PoxtonI. R.ShenJ.WilcoxM. H.GerdingD. N.ZhaoX.et al (2020). Effect of endogenous Clostridioides difficile toxin antibodies on recurrence of C. difficile infection.Clin. Infect. Dis.7181–86. 10.1093/cid/ciz809
37
KoonH. W.WangJ.MussattoC. C.OrtizC.LeeE. C.TranD. H. N.et al (2018). Fidaxomicin and OP-1118 inhibit Clostridium difficile toxin A- and B-mediated inflammatory responses via inhibition of NF-κB activity.Antimicrob. Agents Chemother.62:e01513-17. 10.1128/AAC.01513-17
38
KuehneS. A.CartmanS. T.HeapJ. T.KellyM. L.CockayneA.MintonN. P. (2010). The role of toxin a and toxin B in Clostridium difficile infection.Nature467711–713. 10.1038/nature09397
39
KumarA.SharipovM.TuraevA.AzizovS.AzizovI.MakhadoE.et al (2022). Polymer-based hybrid nanoarchitectures for cancer therapy applications.Polymers14:3027. 10.3390/polym14153027
40
LawalB.TsaiS. K.WuA. T. H.HuangH. S. (2023). In silico study of novel niclosamide derivatives, SARS-CoV-2 nonstructural proteins catalytic residue-targeting small molecules drug candidates.Arab. J. Chem.16:104654. 10.1016/j.arabjc.2023.104654
41
LawryB. M.JohnsonC. L.FlanaganK.SpoorsJ. A.McNeilC. J.WipatA.et al (2018). Species-specific detection of C. difficile using targeted antibody design.Anal. Chem.9013475–13482. 10.1021/acs.analchem.8b03349
42
LewisJ. S. (2024). Performance standards for antimicrobial susceptibility testing, 34th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
43
LiR.ZhangZ.HuangS.QiuL.LiX.ShanX.et al (2023). Synthesis, cytotoxicity, and pharmacokinetic evaluations of niclosamide analogs for anti-SARS-CoV-2.Eur. J. Med. Chem.253:115320. 10.1016/j.ejmech.2023.115320
44
LiZ.XuJ.LangY.FanS.KuoL.WeiD.et al (2020). JMX0207, a niclosamide derivative with improved pharmacokinetics, suppresses Zika virus infection both in vitro and in vivo.ACS Infect. Dis.62616–2628. 10.1021/acsinfecdis.0c00217
45
LinX.ZhangK.WangM.YangM.GuH.XueX.et al (2024). Expression and activity analysis of Clostridium difficile toxin B type 2.Chin. J. Zoonoses40498–503. 10.3969/j.issn.1002-2694.2024.00.076
46
LiuC.ArmstrongC. M.NingS.YangJ. C.LouW.LombardA. P.et al (2021). ARVib suppresses growth of advanced prostate cancer via inhibition of androgen receptor signaling.Oncogene405379–5392. 10.1038/s41388-021-01914-2
47
LouieT. J.MillerM. A.MullaneK. M.WeissK.LentnekA.GolanY.et al (2011). Fidaxomicin versus vancomycin for Clostridium difficile infection.N. Engl. J. Med.364422–431. 10.1056/NEJMoa0910812
48
LowyI.MolrineD. C.LeavB. A.BlairB. M.BaxterR.GerdingD. N.et al (2010). Treatment with monoclonal antibodies against Clostridium difficile toxins.N. Engl. J. Med.362197–205. 10.1056/NEJMoa0907635
49
LyrasD.O’ConnorJ. R.HowarthP. M.SambolS. P.CarterG. P.PhumoonnaT.et al (2009). Toxin B is essential for virulence of Clostridium difficile.Nature4581176–1179. 10.1038/nature07822
50
MaZ. G.MaR.XiaoX. L.ZhangY. H.ZhangX. Z.HuN.et al (2016). Azo polymeric micelles designed for colon-targeted dimethyl fumarate delivery for colon cancer therapy.Acta Biomater.44323–331. 10.1016/j.actbio.2016.08.021
51
MadanR.PetriW. A. (2012). Immune responses to Clostridium difficile infection.Trends Mol. Med.18658–666. 10.1016/j.molmed.2012.09.005
52
MaoT.SuC. W.JiQ.ChenC. Y.WangR.Vijaya KumarD.et al (2021). Hyaluronan-induced alterations of the gut microbiome protects mice against Citrobacter rodentium infection and intestinal inflammation.Gut Microbes13:1972757. 10.1080/19490976.2021.1972757
53
MartinsC.SousaF.AraújoF.SarmentoB. (2018). Functionalizing PLGA and PLGA derivatives for drug delivery and tissue regeneration applications.Adv. Healthcare Mater.7:1701035. 10.1002/adhm.201701035
54
MengF.HasanA.Mahdi Nejadi, BabadaeiM.Hashemi KaniP.Jouya TalaeiA.et al (2020). Polymeric-based microneedle arrays as potential platforms in the development of drugs delivery systems.J. Adv. Res.26137–147. 10.1016/j.jare.2020.07.017
55
NaikA.ShepherdD. V.ShepherdJ. H.BestS. M.CameronR. E. (2017). The effect of the type of HA on the degradation of PLGA/HA composites.Mater. Sci. Eng. C. Mater. Biol. Appl.70824–831. 10.1016/j.msec.2016.09.048
56
NeedhamD. (2024). Niclosamide: A career builder.J. Control. Release369786–856. 10.1016/j.jconrel.2023.07.016
57
NestorovichE. M.BezrukovS. M. (2014). Designing inhibitors of anthrax toxin.Exp. Opin. Drug Discov.9299–318. 10.1517/17460441.2014.877884
58
NooijS.VendrikK. E. W.ZwittinkR. D.DucarmonQ. R.KellerJ. J.KuijperE. J.et al (2024). Long-term beneficial effect of faecal microbiota transplantation on colonisation of multidrug-resistant bacteria and resistome abundance in patients with recurrent Clostridioides difficile infection.Genome Med.16:37. 10.1186/s13073-024-01306-7
59
OoijevaarR. E.van BeurdenY. H.TerveerE. M.GoorhuisA.BauerM. P.KellerJ. J.et al (2018). Update of treatment algorithms for Clostridium difficile infection.Clin. Microbiol. Infect.24452–462. 10.1016/j.cmi.2017.12.022
60
PaparellaA. S.AboulacheB. L.HarijanR. K.PottsK. S.TylerP. C.SchrammV. L. (2021). Inhibition of Clostridium difficile TcdA and TcdB toxins with transition state analogues.Nat. Commun.12:6285. 10.1038/s41467-021-26580-6
61
RanR.SunQ.BabyT.WibowoD.MiddelbergA. P. J.ZhaoC. X. (2017). Multiphase microfluidic synthesis of micro- and nanostructures for pharmaceutical applications.Chem. Eng. Sci.16978–96. 10.1016/j.ces.2017.01.008
62
RupnikM.WilcoxM. H.GerdingD. N. (2009). Clostridium difficile infection: New developments in epidemiology and pathogenesis.Nat. Rev. Microbiol.7526–536. 10.1038/nrmicro2164
63
SamiraninezhadN.AsadiK.RezazadehH.GholamiA. (2023). Using chitosan, hyaluronic acid, alginate, and gelatin-based smart biological hydrogels for drug delivery in oral mucosal lesions: A review.Int. J. Biol. Macromol.252:126573. 10.1016/j.ijbiomac.2023.126573
64
SandhuB. K.McBrideS. M. (2018). Clostridioides difficile.Trends Microbiol.261049–1050. 10.1016/j.tim.2018.09.004
65
SemenyukE. G.LaningM. L.FoleyJ.JohnstonR. F.KnightK. L.SavanovichD. H.et al (2014). Spore formation and toxin production in Clostridium difficile biofilms.PLoS One9:e87757. 10.1371/journal.pone.0087757
66
SinghS.WeissA.GoodmanJ.FiskM.KulkarniS.WhiteK.et al (2022). Niclosamide—a promising treatment for COVID- 19.Br. J. Pharmacol.1793250–3267. 10.1111/bph.15843
67
SmitsW. K.LyrasD.LacyD. B.WilcoxM. H.KuijperE. J. (2016). Clostridium difficile infection.Nat. Rev. Dis. Prim.2:16020. 10.1038/nrdp.2016.20
68
SorgJ. A.SonensheinA. L. (2010). Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid.J. Bacteriol.1924983–4990. 10.1128/JB.00610-10
69
SteeleJ.MukherjeeJ.ParryN.TziporiS. (2013). Antibody against TcdB, but not TcdA, prevents development of gastrointestinal and systemic Clostridium difficile disease.J. Infect. Dis.207323–330. 10.1093/infdis/jis669
70
StewartD.AnwarF.VedantamG. (2020). Anti-virulence strategies for Clostridioides difficile infection: Advances and roadblocks.Gut Microbes12:1802865. 10.1080/19490976.2020.1802865
71
SuY.ZhangB.SunR.LiuW.ZhuQ.ZhangX.et al (2021). PLGA-based biodegradable microspheres in drug delivery: Recent advances in research and application.Drug Deliv.281397–1418. 10.1080/10717544.2021.1938756
72
TaiY.TianM.ChenY.YouP.SongX.XuB.et al (2023). Preparation of PLGA microspheres loaded with niclosamide via microfluidic technology and their inhibition of caco-2 cell activity in vitro.Front. Chem.11:1249293. 10.3389/fchem.2023.1249293
73
TamJ.HamzaT.MaB.ChenK.BeilhartzG. L.RavelJ.et al (2018). Host-targeted niclosamide inhibits C. difficile virulence and prevents disease in mice without disrupting the gut microbiota.Nat. Commun.9:5233. 10.1038/s41467-018-07705-w
74
TannockG. W.MunroK.TaylorC.LawleyB.YoungW.ByrneB.et al (2010). A new macrocyclic antibiotic, fidaxomicin (OPT-80), causes less alteration to the bowel microbiota of Clostridium difficile-infected patients than does vancomycin.Microbiology1563354–3359. 10.1099/mic.0.042010-0
75
TiseoG.YahavD.AtamnaA.AvniT.CausseM.Pérez-NadalesE.et al (2024). Recurrent Clostridioides difficile infections in solid organ transplant recipients: The international CALIPSO study.J. Infect.89:106306. 10.1016/j.jinf.2024.106306
76
VarmaS.GreendykeW. G.LiJ.FreedbergD. E. (2022). Class-specific relationship between use of immunosuppressants and risk for community-acquired Clostridioides difficile infection.Clin. Infect. Dis.74793–801. 10.1093/cid/ciab567
77
WahidB.AliA.RafiqueS.IdreesM. (2017). Current status of therapeutic and vaccine approaches against zika virus.Eur. J. Intern. Med.4412–18. 10.1016/j.ejim.2017.08.001
78
WanS.YouP.ShiQ.HuH.ZhangL.ChenL.et al (2024). Gut microbiome changes in mouse, mongolian gerbil, and hamster models following Clostridioides difficile challenge.Front. Microbiol.15:1368194. 10.3389/fmicb.2024.1368194
79
WangC.LuH.LiX.YangK.TaoY.CaoQ.et al (2022). Identification of an anti-virulence drug that reverses antibiotic resistance in multidrug resistant bacteria.Biomed. Pharmacother.153:113334. 10.1016/j.biopha.2022.113334
80
WangN.HanX.LiJ.WangY.YuW.WangR.et al (2019). Comparative study of the bindings between 3-phenyl-1H-indazole and five proteins by isothermal titration calorimetry, spectroscopy and docking methods.J. Biomol. Struct. Dyn.374580–4589. 10.1080/07391102.2018.1554511
81
WardleM. T.AllenS. E.GamboaR.RodriguezS.GonzalezA. E.GarciaH. H.et al (2024). Mass chemotherapy with niclosamide for the control of Taenia solium: Population-based safety profile and treatment effectiveness.Lancet Reg. Health Am.38:100876. 10.1016/j.lana.2024.100876
82
WilcoxM. H.GerdingD. N.PoxtonI. R.KellyC.NathanR.BirchT.et al (2017). Bezlotoxumab for prevention of recurrent Clostridium difficile infection.N. Engl. J. Med.376305–317. 10.1056/NEJMoa1602615
83
WuH.LiuT.XuZ.QianJ.ShenX.LiY.et al (2020). Enhanced bacteriostatic activity, osteogenesis and osseointegration of silicon nitride/polyetherketoneketone composites with femtosecond laser induced micro/nano structural surface.Appl. Mater. Today18:100523. 10.1016/j.apmt.2019.100523
84
WuJ.ZhangZ.GuJ.ZhouW.LiangX.ZhouG.et al (2020). Mechanism of a long-term controlled drug release system based on simple blended electrospun fibers.J. Control. Release320337–346. 10.1016/j.jconrel.2020.01.020
85
XuJ.ShiP. Y.LiH.ZhouJ. (2020). Broad spectrum antiviral agent niclosamide and its therapeutic potential.ACS Infect. Dis.6909–915. 10.1021/acsinfecdis.0c00052
86
ZhangH.YangJ.SunR.HanS.YangZ.TengL. (2023). Microfluidics for nano-drug delivery systems: From fundamentals to industrialization.Acta Pharm. Sin. B133277–3299. 10.1016/j.apsb.2023.01.018
87
ZhangW.RanJ.ShangL.ZhangL.WangM.FeiC.et al (2022). Niclosamide as a repurposing drug against gram-positive bacterial infections.J. Antimicrob. Chemother.773312–3320. 10.1093/jac/dkac319
88
ZhuD.SorgJ. A.SunX. (2018). Clostridioides difficile biology: Sporulation, germination, and corresponding therapies for C. difficile infection.Front. Cell. Infect. Microbiol.8:29. 10.3389/fcimb.2018.00029
Summary
Keywords
loaded controlled-release nanospheres, Niclosamide, Clostridioides difficile, spore germination, biofilm formation, multiple effects
Citation
Tai Y, Zhang M, Han Y, Hu H, Lin S, Zhai F, Tian M, Song X, Wan S, Chen Y and Jin D (2025) Synthetic niclosamide-loaded controlled-release nanospheres with high solubility and stability exerting multiple effects against Clostridioides difficile. Front. Microbiol. 16:1617631. doi: 10.3389/fmicb.2025.1617631
Received
24 April 2025
Accepted
24 June 2025
Published
30 July 2025
Volume
16 - 2025
Edited by
Paolo Pellegrino, National Research Council (CNR), Italy
Reviewed by
M. Oves, King Abdulaziz University, Saudi Arabia
Jianhua Wang, Chinese Academy of Agricultural Sciences (CAAS), China
Laura Maria Slavu, University of Salento, Italy
Updates
Copyright
© 2025 Tai, Zhang, Han, Hu, Lin, Zhai, Tian, Song, Wan, Chen and Jin.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dazhi Jin, jind@hmc.edu.cnYu Chen, ychen@hmc.edu.cn
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.