- Department of Clinical Research Center, The Children’s Hospital, Zhejiang University School of Medicine, National Clinical Research Center for Child Health, Hangzhou, China
Introduction: The discovery of novel antimicrobial mechanisms among existing clinical drugs is urgently needed. Disulfiram, an FDA-approved treatment for alcohol dependence, exhibits broad-spectrum antibacterial effects. However, its mechanism of action remains incompletely understood.
Methods: The antimicrobial activity of disulfiram was assessed using bacterial growth curves and colony-forming unit assays. Cytotoxicity was evaluated via propidium iodide staining and flow cytometry. Synergy with polymyxins or kanamycin was examined using checkerboard assays. RNA-seq was performed on disulfiram-treated E. coli, and differentially expressed genes were analyzed using the R package limma. Intracellular reactive oxygen species (ROS) levels were measured with fluorescent probes and flow cytometry.
Results: Disulfiram exhibited bacteriostatic, but not bactericidal, effects against E. coli and S. aureus. However, it significantly enhanced the bactericidal activity of colistin or kanamycin, both in vitro and in a murine E. coli infection model. Transcriptomic analysis revealed oxidative stress and zinc-related responses in disulfiram-treated E. coli. The bacteriostatic effects were reversed by the ROS scavenger N-acetyl-l-cysteine and zinc chelators, whereas zinc supplementation enhanced ROS production and growth inhibition.
Discussion: This study identifies a zinc-dependent ROS-mediated mechanism underlying the bacteriostatic activity of disulfiram. Although the in vivo concentrations of disulfiram during standard therapy are below its MIC, its synergistic effect with colistin suggests clinical relevance as an adjuvant. Disulfiram-induced redox stress and zinc modulation likely compromise bacterial antioxidant defenses and membrane integrity. These findings support further investigation of dithiocarbamate-based compounds as potential adjuvants or scaffolds for novel antimicrobial development.
1 Introduction
Bacterial diseases cause significant mortality globally and place a major burden on public health systems (GBD 2019 Antimicrobial Resistance Collaborators, 2022). Bacterial infections cause most cases of sepsis, an extreme host reaction to an infection. The number of sepsis-related deaths reached 11 million in 2017, resulting in great global health destruction (Darby et al., 2023; Bush and Bradford, 2020). The emergence of multidrug-resistant bacteria poses a further threat to existing antimicrobial treatment regimens. Currently, last-resort antibiotics, including colistin, are losing effectiveness due to the spread of resistance genes among bacteria (Darby et al., 2023; Bush and Bradford, 2020). Hence, the discovery of new antibiotics and the exploration of additional antimicrobial mechanisms of medicines currently used in the clinic are urgently needed.
Disulfiram has been the first FDA-approved drug for alcohol use disorder since 1951 (Suh et al., 2006). However, this chemical compound is so versatile that more than half a century later, studies continue to reveal additional medical uses. For example, disulfiram has antitumor effects on many types of human cancer (Lu et al., 2021); it inhibits neutrophil extracellular traps to limit SARS-CoV-2 lung injury (Adrover et al., 2022); and it normalizes the weight of obese mice (Bernier et al., 2020). Among these applications, one of the most attractive functions of disulfiram is to affect the growth of a broad spectrum of bacteria, against both Gram-negative pathogens (e.g., F. tularensis) and Gram-positive species (e.g., S. aureus) (Chen et al., 2023; Chen et al., 2020; Hamblin et al., 2019; Thakare et al., 2019) and even Mycoplasma (Meneguello et al., 2022). This function endows disulfiram with potential for the treatment of bacterial infections.
Nevertheless, the universal mechanisms underlying the antibacterial effects of disulfiram remain elusive. Studies have shown that disulfiram can inhibit aldehyde dehydrogenase-like proteins in F. tularensis or metallo-β-lactamases (MBLs) in Enterobacterales (Chen et al., 2020; Hamblin et al., 2019). However, some bacteria that disulfiram inhibits do not possess such proteins. Disulfiram forms a complex with copper and exerts synergistic killing effects on M. tuberculosis (Dalecki et al., 2015). However, this synergistic effect is observed primarily with mycobacteria. Also, the antibacterial activity of disulfiram may partly attribute to its electrophilic nature and ability to form disulfide bonds with thiol-containing bacterial proteins, thereby disrupting essential enzyme function. Nonetheless, the full mechanism and its therapeutic potential still warrant further investigation.
This study aimed to quantify the effect of disulfiram against bacteria and elucidate the possible mechanisms of disulfiram through transcriptomic and phenotypic assays. Through RNA sequencing and the use of reactive oxygen species (ROS) inhibitors, we exploited an additional mechanism by which disulfiram inhibits bacteria. We found that disulfiram halted bacterial growth by increasing zinc-dependent intracellular ROS levels, and ROS scavengers or zinc chelators could restore bacterial growth. These findings provide a potential therapeutic strategy for treating bacterial infectious diseases.
2 Materials and methods
2.1 Animals and ethics approval
C57BL/6 wild-type (WT) mice aged 6–8 weeks were purchased from Shanghai SLAC Laboratory Animal Cooperation. All the mice were housed in a specific pathogen-free and temperature-controlled standard environment in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals. The animal experimental protocols were approved by the Animal Review Committee of Zhejiang University School of Medicine and were in compliance with institutional guidelines.
2.2 Chemical compounds
The complete information for all antibiotics and compounds used in this study was listed as followed: disulfiram (MedChemExpress, HY-B0240, in DMSO), tetracyclin (Aladdin, T105493-10 g, in DMSO), colistin (Solarbio, C9810, in DMSO), meropenem (Aladdin, M427157-1 mL, in DMSO), ampicillin (Aladdin, A105483-5 g, in DMSO), sulbactam (Aladdin, E129319-1 g, in DMSO), N-Acetyl-L-cysteine (MedChemExpress, HY-B0215, in DMSO), ZnCl2 (MedChemExpress, HY-Y0420, in H2O), Tpen (Aladdin, N159625-250 mg, in DMSO), polymyxin B (Aladdin, P105490-1 g, in DMSO), kanamycin (Aladdin, K331597-200 mg, in DMSO), miconazole (Macklin, M909105-25 g, in DMSO), benzoquinone (Macklin, B802580-5 g, in DMSO), CuGlu (Aladdin, D133471-25 g, in H2O), TTM (Aladdin, A189030-200 mg, in DMSO), Ferric (III) Citrite (MedChemExpress, HY-B1645, in H2O), deferoxamine (MedChemExpress, HY-B0988, in DMSO), Dimethyl sulfoxide (MedChemExpress, HY-Y0320), coin oil (MedChemExpress, HY-Y1888).
2.3 Bacterial strains
The laboratory strains used in this study were obtained from the American Type Culture Collection (ATCC) and included Escherichia coli (ATCC 25922), Staphylococcus aureus (ATCC 25923), methicillin-resistant Staphylococcus aureus (MRSA; ATCC 43300), Klebsiella pneumoniae (ATCC 13883), Pseudomonas aeruginosa (ATCC 27853), and Acinetobacter baumannii (ATCC 17978). Clinical strains (E. coli and A. baumanii) were collected from Microbiology Laboratory of Children’s hospital, 2023–2024.
2.4 Determination of minimum inhibitory concentration (MIC50)
The MIC50 values of antibiotics were determined using the broth microdilution method according to CLSI guidelines. Bacterial suspensions (0.5 McFarland standard) were diluted 1:100 in cation-adjusted Mueller-Hinton broth and dispensed into 96-well plates containing two-fold serial dilutions of antibiotics (0.25–256 μg/mL). Plates were incubated at 37°C for 18–22 h. MIC50 was defined as the lowest concentration of antibiotic that inhibited ≥50% of bacterial growth, assessed by optical density at 600 nm (OD600). Experiments were performed in triplicate.
2.5 Bacterial killing analysis
Bacteria were cultured to log phase and diluted to an OD600 of 0.2–0.3. Disulfiram and colistin were added as indicated. The bacterial cells were subsequently washed 4 h later. The cell numbers were determined by dilution, incubation on Luria–Bertani agar plates at 37°C overnight, and calculation by multiplying the countable bacterial colony units by the number of dilutions.
2.6 Bacterial growth curve
Bacteria were cultured to log phase, diluted to 5 × 105 colony-forming units (CFUs)/ml in 200 μL of LB and placed in a 96-well microliter plate. The drugs and compounds were added as indicated. The growth curve of bacteria was monitored by measuring the absorbance at 600 nm using a microplate reader (SpectraMax190, Molecular Devices) over a 24 h period.
2.7 Checkerboard assay
Drug synergism was tested in checkerboard assays as described (Chen et al., 2023). Briefly, disulfiram and other antibiotics or compounds were twofold serially diluted in an 8 × 8 matrix within a 96-well microliter plate. Bacteria were grown to log phase and diluted in each well to 5 × 105 CFUs/ml in 200 μL of MHB. After 18–22 h of incubation at 37°C, the absorbance of each well at 600 nm was measured with a microplate reader (SpectraMax190, Molecular Devices). The fractional inhibitory concentration index (FICI) was calculated accordingly, and a value less than 0.5 demonstrated synergy.
2.8 Membrane permeability assay
The membrane permeability rate of the cells was determined by propidium iodide (PI, 2.5 μg/mL) staining and flow cytometry. A minimum of 20,000 events per sample were acquired using the PE fluorescence channel during flow cytometry analysis. The analysis was performed using a DxFLEX or a CytoFLEX LX (Beckman Coulter, United States).
2.9 In vivo model
We used an E. coli murine intra-abdominal infection model to analyze the synergistic effect of disulfiram and colistin in vivo. Briefly, E. coli bacteria were seeded on Luria–Bertani (LB) agar plates and cultured overnight. A single clone was selected for culture in liquid LB medium and incubated with shaking at 200 rpm at 37°C for another 12 h. The bacterial suspension was then prepared at a concentration of 7 × 106 colony-forming units (CFUs) per 100 μL of PBS. To induce intra-abdominal infection, the mice were intraperitoneally administered 100 μL of live E. coli suspension. Thirty minutes later, disulfiram (resolved in corn oil), colistin (resolved in corn oil) or control solvent (corn oil) was administered to the mice intraperitoneally as indicated. Mortality was assessed hourly for 72 h.
2.10 Microscopy
Bacteria were cultured to log phase and diluted to an OD600 of 0.2–0.3. Disulfiram and other antibiotics were added as indicated. Four hours later, the cells were inoculated on a slide and subjected to microscopic observation. Images were taken with an Olympus IX73 inverted microscope, and the cell lengths were measured using ImageJ software.
2.11 Motility assay
Motility assay was tested as described (Ejim et al., 2011). Briefly, swimming motility was observed on 0.3% (w/v) agar medium composed of 10 g/L trypticase peptone, 10 g/L NaCl and 5 g/L yeast extract. The bacterial cells were cultured to log phase, standardized to 2–3 × 106 CFUs/ml, point inoculated onto a swim plate and incubated for 20 h at 37°C. The swimming areas were calculated using ImageJ software.
2.12 RNA sequencing and differential expressed genes analysis
E. coli were grown to log phase and cultured with disulfiram or the solvent DMSO (n = 3 biologically independent samples) for 2 h. The cells were collected, total RNA was extracted using TRIzol® Reagent according to the manufacturer’s instructions (Invitrogen), and the genomic DNA was removed using DNase I (TaKara). The RNA was then quantified with an ND-2000 (NanoDrop Technologies) and subjected to library construction, sequencing and data analysis. The RNA-seq strand-specific libraries were prepared with a TruSeq RNA sample preparation kit from Illumina (San Diego, CA) using 5 μg of total RNA. In brief, rRNA was removed using a RiboZero rRNA removal kit (Epicenter) and fragmented in fragmentation buffer. cDNA synthesis, end repair, A-base addition and ligation of the Illumina-indexed adaptors were performed according to Illumina’s protocol. Paired-end libraries were sequenced on an Illumina NovaSeq 6000 (Shanghai BIOZERON Co., Ltd.). The raw paired-end reads were trimmed and quality controlled in Trimmomatic with the following parameters: SLIDINGWINDOW: 4:15 MINLEN:75 (version 0.36; http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic). Then, the clean reads were separately aligned to the reference genome in orientation mode using Rockhopper (http://cs.wellesley.edu/~btjaden/Rockhopper/) software. The differentially expressed mRNAs with a fold change > 2 or < 0.5 and p value < 0.05 were selected via the R package edgeR (https://bioconductor.org/packages/release/bioc/html/edgeR.html).
2.13 Clusters of transcriptomic changes
The transcriptomes from each chemical compound-treated E. coli experiment were downloaded from NCBI (colistin-treated E. coli: GSE220559; kanamycin-, H2O2-treated E. coli: GSE56133). Differentially expressed genes were analyzed using the bioconductor package limma (https://www.bioconductor.org/packages/release/bioc/html/limma.html). The clustering of transcriptomic changes was performed using the R package pheatmap (https://cran.r-project.org/web/packages/pheatmap/index.html) with the clustering method “ward. D.”
2.14 ROS measurement
Cellular ROS levels were measured using CellROX (Thermo Fisher Scientific, United States, C10444) probes. Briefly, bacteria were cultured to log phase, diluted to an OD600 of 0.2–0.3, cultured with the indicated compounds for 1 h, and then, the probes were loaded at 5 μM at 37°C for 30 min. The cells were washed twice and resuspended in PBS, and then flow cytometry was performed using a DxFLEX or a CytoFLEX LX (Beckman Coulter, United States). A minimum of 20,000 events per sample were acquired using the FITC fluorescence channel during flow cytometry analysis.
2.15 Data analysis
The data are shown as the means ± SDs. Differences were analyzed by Student’s t test or one-way ANOVA with Tukey’s post hoc analysis. Kaplan–Meier survival analysis was performed for survival experiments. A p value < 0.05 was considered statistically significant. Statistical analysis was carried out in GraphPad Prism 9.3 (GraphPad Software).
3 Results
3.1 Evaluation of disulfiram’s bacteriostatic activity
To understand the effects of disulfiram on bacteria, we first monitored the 50% minimal inhibitory concentration (MIC50) of disulfiram on the 6 common clinical bacteria including E. coli, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa and methicillin-resistant S. aureus (MRSA). The results revealed that disulfiram had relatively high MIC50 values against these bacteria, with values of 256 μg/mL against E. coli and P. aeruginosa and 32 μg/mL against S. aureus, A. baumannii and MRSA (Figure 1a). These concentrations are higher than typical clinical plasma levels, suggesting in vitro activity may not fully translate in vivo. Next, we tested whether disulfiram killed bacteria at high concentrations. We used tetracycline, a bacteriostatic drug (Maier et al., 2021), as a control. The results revealed that while tetracycline killed E. coli at 1 × MIC50 (0.5 μg/mL), disulfiram had nearly no killing effect on E. coli at 1 × MIC50 (256 μg/mL; n = 3, 95.53 ± 6.93% vs. 68.90 ± 10.18%, p = 0.0389), indicating that disulfiram had no bactericidal effect (Figure 1b). A microplate reader was then used to monitor the bacterial growth curve to further observe the effect of disulfiram on the growth of E. coli. Disulfiram delayed the exponential growth phase of E. coli in a concentration-dependent manner (Figure 1c). Moreover, disulfiram halted E. coli growth instantly at the exponential phase (Figure 1d), indicating that disulfiram has a fast bacteriostatic effect on E. coli. These results demonstrated that disulfiram is a fast bacteriostatic drug with no bactericidal effect.
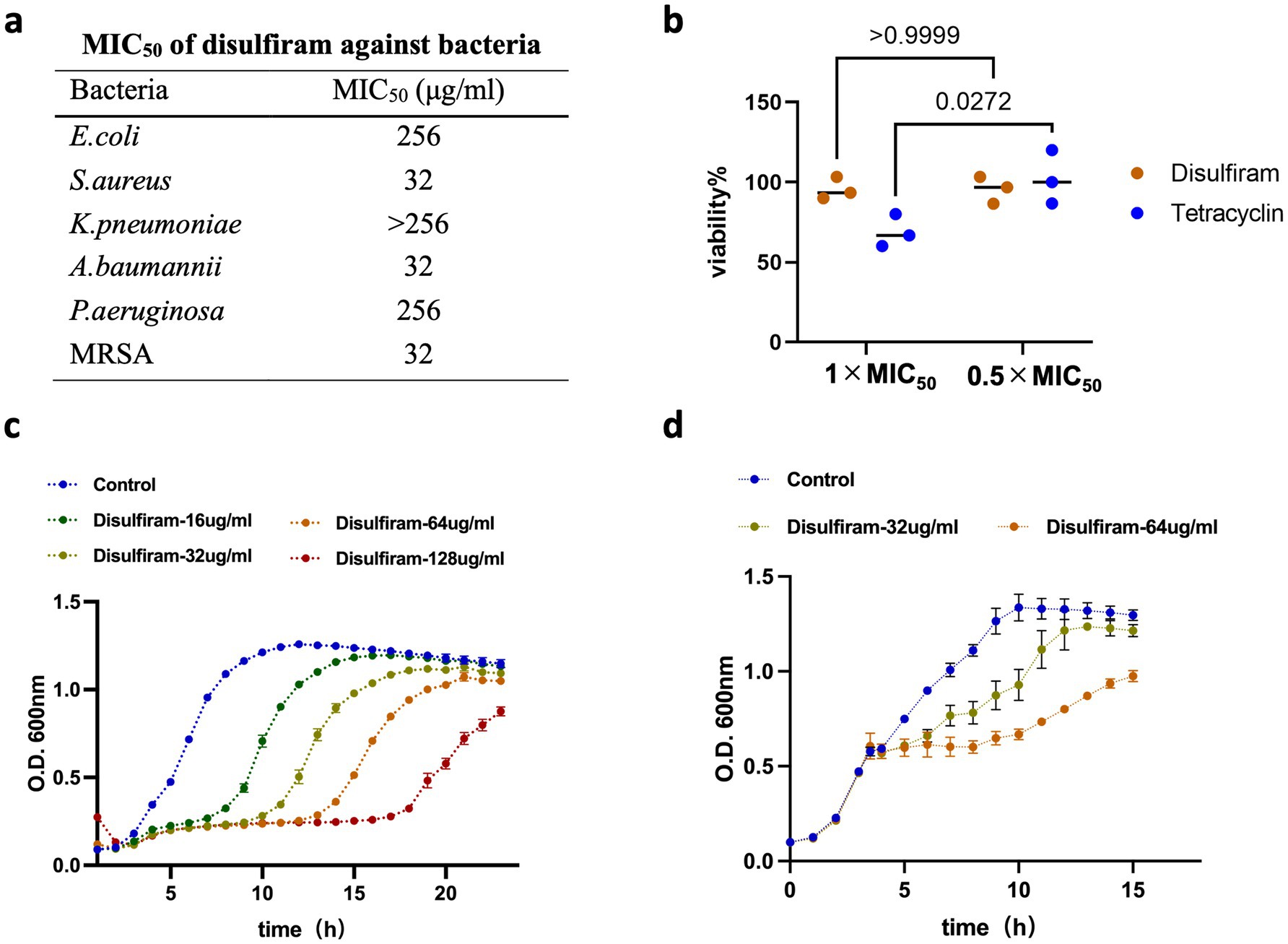
Figure 1. Evaluation of disulfiram’s bacteriostatic activity. (a) MIC50 values of disulfiram against multiple pathogens, including E. coli, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa and methicillin-resistant S. aureus (MRSA). (b) Viability of E. coli in the presence of tetracycline (MIC50: 0.5 μg/mL) or disulfiram (MIC50: 256 μg/mL). (c) Growth curve of E. coli treated with disulfiram at the indicated concentrations at the beginning of the experiment. (d) Growth curve of E. coli treated with disulfiram at the indicated concentration for 3.5 h. Data in c and d are the means ± SD of three biological replicates. p values were determined using unpaired, two-tailed Student’s t tests.
3.2 Combination effects of disulfiram and colistin on bacterial survival
Antibiotic sensitizers or adjuvants play important roles in preventing microbial infections through the collaboration of antibiotics (Wang et al., 2024). Fast bacteriostatic drugs are often used as adjuvants for stationary-phase bactericidal drugs. A recent study showed that disulfiram could bind to New Delhi metallo-β-lactamase 1 (NDM-1) and resensitize drug-resistant bacteria to colistin and meropenem (Chen et al., 2023). However, we found that 32 μg/mL disulfiram could reduce the MIC of colistin on E. coli by 4-fold, which did not involve antibiotic resistance genes (FICI = 0.375, Figure 2a). The combination of disulfiram and colistin amplified the bactericidal effect of colistin alone (Figure 2b; Supplementary Figure S2a). Similarly, disulfiram also synergized with polymyxin B (FICI = 0.25) or kanamycin (FICI = 0.375) against E. coli or with colistin (FICI = 0.375) or kanamycin (FICI = 0.5) against A. baumannii (Supplementary Figure S1). The clinical isolates that generally cause sepsis or Healthcare-Associated Infections (HAIs) may have different sensitivity patterns than lab strains. We have also included clinical isolated strains for assay. The result showed that disulfiram synergized with colistin against clinical isolated E. coli (FICI = 0.047) and A. baumannii (FICI = 0.125, Supplementary Figure S1). A cell permeability assay revealed that although disulfiram alone did not permeabilize the cell membrane, it augmented the cell membrane damage caused by colistin (n = 4, 19.48 ± 2.74% vs. 8.31 ± 1.13%, p = 0.0286, Figure 2c). Moreover, we applied disulfiram to an animal model of E. coli intraperitoneal infection and found that combination therapy with disulfiram and colistin resulted in better outcomes than treatment with colistin alone for the disease (n = 7–8, 70.83 ± 20.41 h vs. 51.02 ± 34.85 h, p = 0.013, Figure 2d; Supplementary Figure S2b). These results indicated that disulfiram could act as a potential antibiotic sensitizer or adjuvant with stationary-phase bactericidal antibiotics, including colistin, polymyxin B or kanamycin.
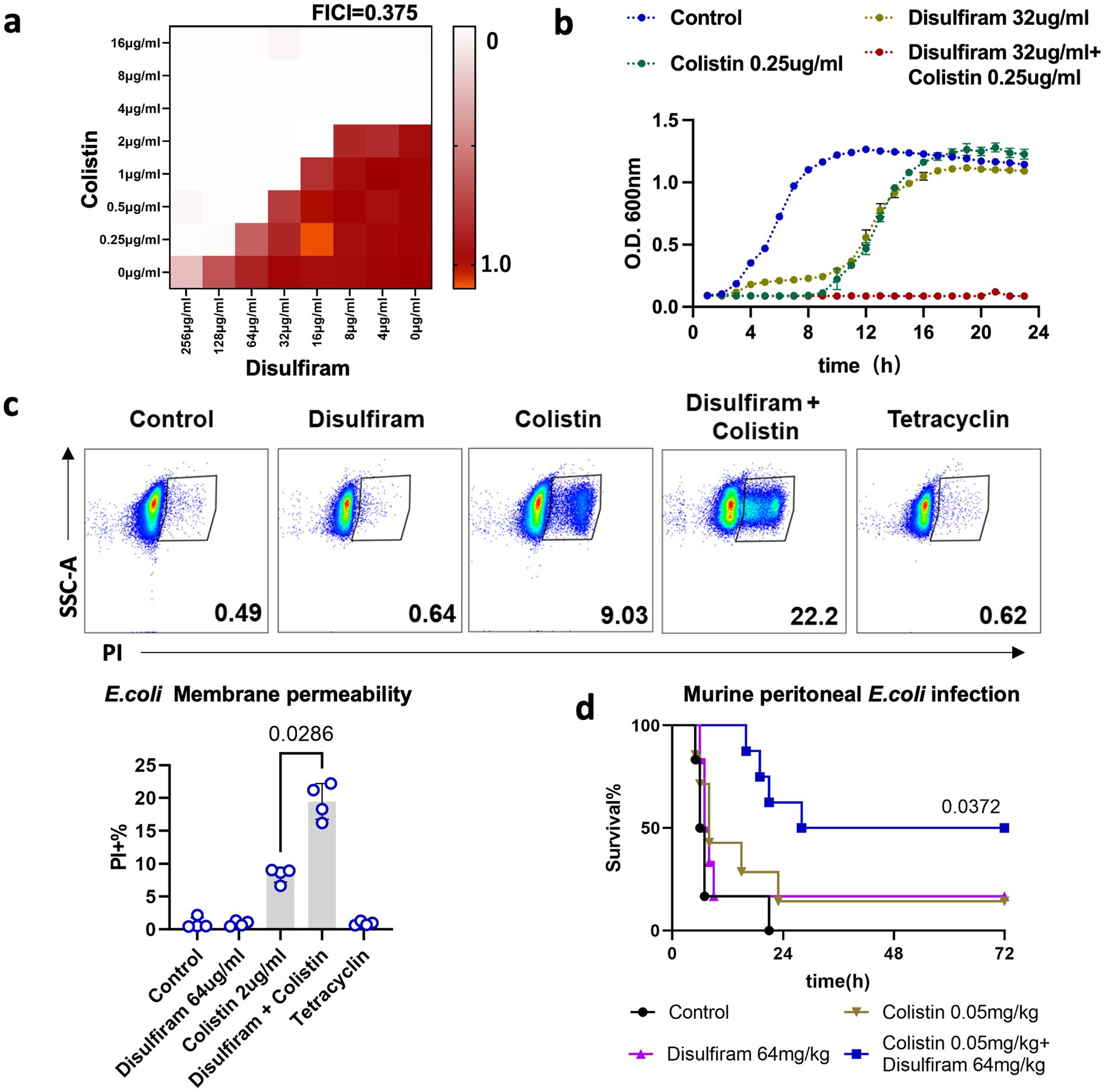
Figure 2. Combination effects of disulfiram and colistin on bacterial survival. (a) Checkerboard assay of disulfiram and colistin against E. coli. The optical density (OD) at 600 nm was measured after 20 h of incubation at 37°C. The data are expressed as the absorbance at 600 nm and represent biological replicates. (b) Growth curves of E. coli in the presence of disufiram and colistin for 24 h. The means ± SD of three biological replicates are shown. (c) Membrane permeability assay of E. coli treated with disulfiram (64 μg/mL), colistin (2 μg/mL) or tetracycline (0.5 μg/mL). p values were determined using the Mann–Whitney U test. (d) Survival rates of mice in the peritonitis–sepsis model (n = 6 ~ 8) over 72 h after infection with a lethal dose of E. coli (3.0 × 107 CFUs) treated with disulfiram or colistin alone or in combination. p values were determined using the Mantel–Cox test.
3.3 Analysis of bacterial motility and morphology under disulfiram treatment
Next, we sought to investigate the effect of disulfiram on E. coli. Swimming motility is vital to bacteria, as this process supports their movement toward resources. We found that under disulfiram treatment, the motility of E. coli was inhibited (At 64 μg/mL, n = 3, 100 ± 0% vs. 0 ± 0%, p = 0.0092, Figure 3a). This may result from decreased swimming ability or potential viability loss. Studies have shown that motility patterns are associated with bacterial cell shape and that the antibacterial cellular pathways of chemical compounds are associated with bacterial cytology profiles (Nonejuie et al., 2013; Wadhwa and Berg, 2022). Under microscopic observation, while sulbactam, ampicillin and meropenem, which are penicillin-binding protein (PBP)-targeting antibiotics, changed the length of E. coli, disulfiram did not have an effect (n = 30, 3.78 ± 1.04 μm vs. 3.77 ± 0.94 μm, p > 0.9999, Figure 3b), indicating that the target of disulfiram may not be bacterial PBPs (Penwell et al., 2015).
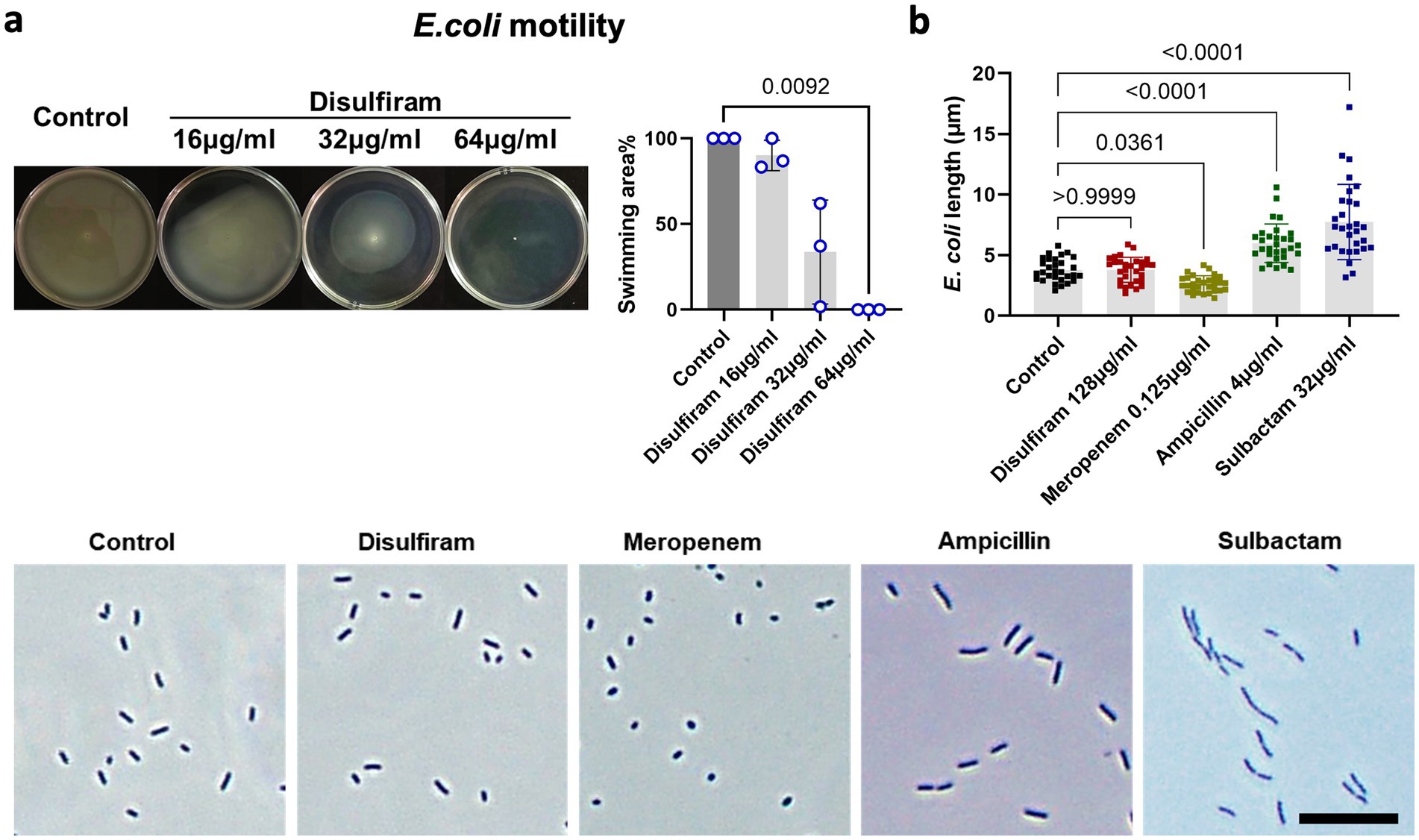
Figure 3. Analysis of bacterial motility and morphology under disulfiram treatment. (a) Swimming assay of E. coli treated with disulfiram at the indicated concentrations. Images represent biological replicates. (b) Microscopy images and lengths of E. coli treated with disulfiram, meropenem, ampicillin or sulbactam (0.5 × MIC50). Images represent biological replicates. p values were determined using one-way ANOVA.
3.4 Transcriptomic profiling of Escherichia coli in response to disulfiram exposure
To study the detailed mechanisms by which disulfiram inhibits bacterial growth in detail, we subjected disulfiram-treated E. coli to RNA sequencing (RNA-seq) to study the transcriptome changes. Compared with the control E. coli group, disulfiram-treated E. coli expressed higher levels of antioxidant-related genes, including hmpA, cysI, nrdH, and frmA; higher levels of the iron–sulfur cluster repair-related gene ytfE; lower levels of zinT and higher levels of zntA, two genes associated with zinc binding and zinc translocating; higher levels of the pyruvate uptake-related gene btsT; and lower levels of the nucleotide synthesis-related gene ygeW (Figure 4a). The top genes among the differentially expressed genes (DEGs) were hmpA, which encodes flavohemoglobin and is linked to the cell response to oxidative or nitrosative stress (Bonamore and Boffi, 2008). Azoles and quinones are reported to be inhibitors and substrates of flavohemoglobin (Moussaoui et al., 2018; Nobre et al., 2010). We first tested whether these compounds could augment the effect of disulfiram. The results of the checkerboard assay suggested that inhibiting or monitoring flavohemoglobin only had a very mild effect on disulfiram inhibition of E. coli (Supplementary Figures S2c,d), indicating that drugs or chemicals that target flavohemoglobin cannot efficiently enhance the effect of disulfiram. Pathway enrichment analysis further revealed that the differentially regulated genes were enriched in the iron-sulfer cluster binding, oxidoreductase activity and RNA binding pathways (Figure 4b). These results indicate that disulfiram may affect the growth of E. coli through oxidant activity, leading to iron-sulfer cluster damage and the suspension of genetic material synthesis. To provide a preliminary overview of potential similarities in gene expression profiles, we performed clustering analysis by comparing our transcriptomic data with publicly available datasets that examined RNA expression changes in E. coli in response to various bacteriostatic and bactericidal agents. The results revealed that the disulfiram-induced transcriptomic changes were closely associated with those caused by H2O2, which cause oxidative damage to pathogens (Schairer et al., 2012). In contrast, bactericidal drugs, including kanamycin or colistin, had relatively distinct transcriptomic changes (Figure 4c). Furthermore, we verified that the ROS inhibitor N-acetyl-L-cysteine (NAC) fully prevented the bacteriostatic effect of disulfiram on both E. coli and S. aureus (Figures 4d,e). These results confirmed that disulfiram inhibits bacterial growth by inducing ROS.
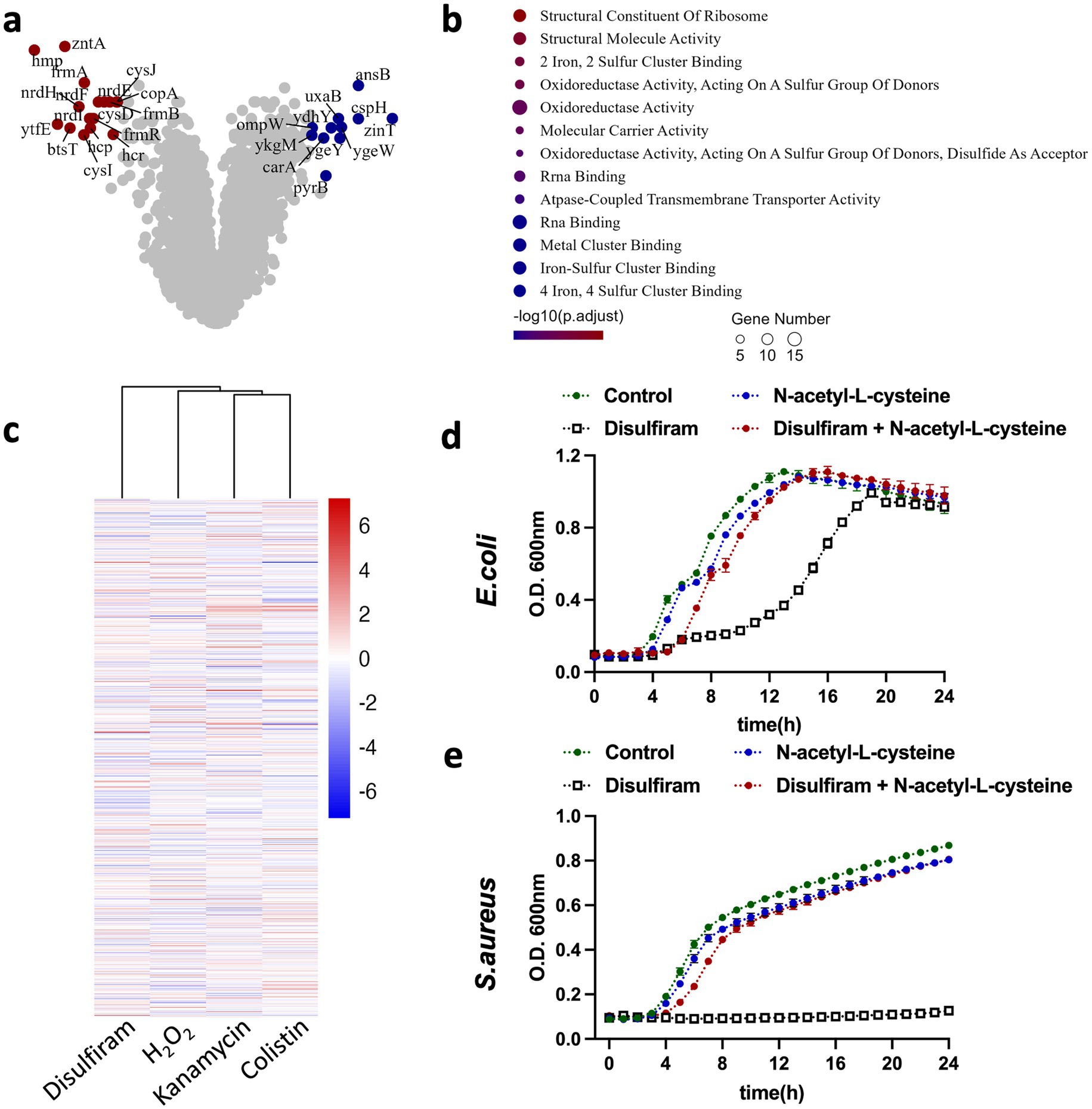
Figure 4. Transcriptomic profiling of E. coli in response to disulfiram exposure. (a) Transcriptome analysis of E. coli treated with disulfiram (64 μg/mL) for 4 h. Genes with increases or decreases of log [fold change] > 4 and p values < 0.01 are shown in the original list of 4,798 genes (n = 3 biological replicates). (b) Molecular function changes in differentially expressed genes in disulfiram (64 μg/mL, 4 h)-treated E. coli. Genes with increases or decreases of log [fold change] > 2 and p values < 0.05 were included and analyzed (n = 3 biological replicates). (c) Transcriptome profiles of E. coli treated with kanamycin, H2O2, disulfiram or colistin were clustered as described in the methods. A total of 3,218 genes were annotated universally across samples and analyzed. (d) Growth curves of E. coli in the presence of disufiram (32 μg/mL) and N-acetyl-L-cysteine (5 mM) for 24 h. (e) Growth curves of S. aureus in the presence of disufiram (32 μg/mL) and N-acetyl-L-cysteine (5 mM) for 24 h. Data in (d,e) are the means ± SD of three biological replicates.
3.5 Mechanistic investigation of disulfiram-mediated growth inhibition
The coeffect of metal ions is proposed as a biological mechanism of disulfiram (Skrott et al., 2017; Gao et al., 2022). Indeed, disulfiram and copper kill Mycobacterium tuberculosis in a synergetic manner (Dalecki et al., 2015). However, when copper gluconate was used as a Cu2+ supply in E. coli growth medium, we found the opposite effect that copper ions counteracted the inhibitory effect of disulfiram on E. coli growth (Supplementary Figure S3a). We proposed that copper ions and the metabolites of disulfiram (ditiocarb and diethyldithiocarbamate, which have bacteriostatic effects similar to those of disulfiram) could form chemical complexes (bis (diethyldithiocarbamate)–copper), which may cover the effective bacteriostatic group of disulfiram. Moreover, the copper chelator tetrathiomolybdate (TTM) had a very weak synergistic effect with disulfiram on the growth of E. coli (Supplementary Figure S3a). These results indicated that the disulfiram-mediated inhibition of bacterial growth was independent of copper ions.
As shown by the transcriptomic results, Fe-S cluster damage and repair participate in the mechanism of disulfiram. Ferric ions may induce ROS through the Fenton reaction. However, upon cotreatment with disulfiram, the ferric chelator DFO (deferoxamine) only weakly inhibited the growth of E. coli, whereas ferric citrate had no effect (Supplementary Figure S3b).
Zinc binding and zinc translocation may play a role in this process, as the zinT and zntA genes were upregulated in disulfiram-treated E. coli. Surprisingly, we found that while zinc ions augmented the inhibitory effect of disulfiram on both E. coli (Figure 5a) and S. aureus (Figure 5b), the zinc chelator TPEN (N, N, N′, N′-tetrakis- (2-pyridylmethyl) ethylenediamine) counteracted the inhibitory effect of disulfiram, fully restored the growth of E. coli (Figure 5c) and partly restored the growth of S. aureus (Figure 5d). These results suggested that zinc ions play important roles in the bacteriostatic mechanisms of disulfiram. Therefore, disulfiram has a ROS-dependent bacteriostatic effect. We next tested the relationship between ROS and zinc. We loaded CellROX, a fluorescent ROS probe, into bacteria as an indicator of ROS and found that while zinc ions augmented the disulfiram-induced increase in ROS level in E. coli (n = 3, 74.47 ± 4.98 vs. 65.80 ± 1.13, p = 0.0049) and S. aureus (n = 3, 129.0 ± 9.54% vs. 94.53 ± 8.63%, p = 0.0021), TPEN treatment fully suppressed this increase in both E. coli (n = 3, 64.13 ± 0.47% vs. 65.80 ± 1.13%, p = 0.9356) and S. aureus (n = 3, 47.97 ± 5.24% vs. 94.53 ± 8.63%, p = 0.0001, Figures 5d,e). These results suggested that the bacteriostatic effect of disulfiram is mediated by zinc ions, and that ROS levels depend on zinc.
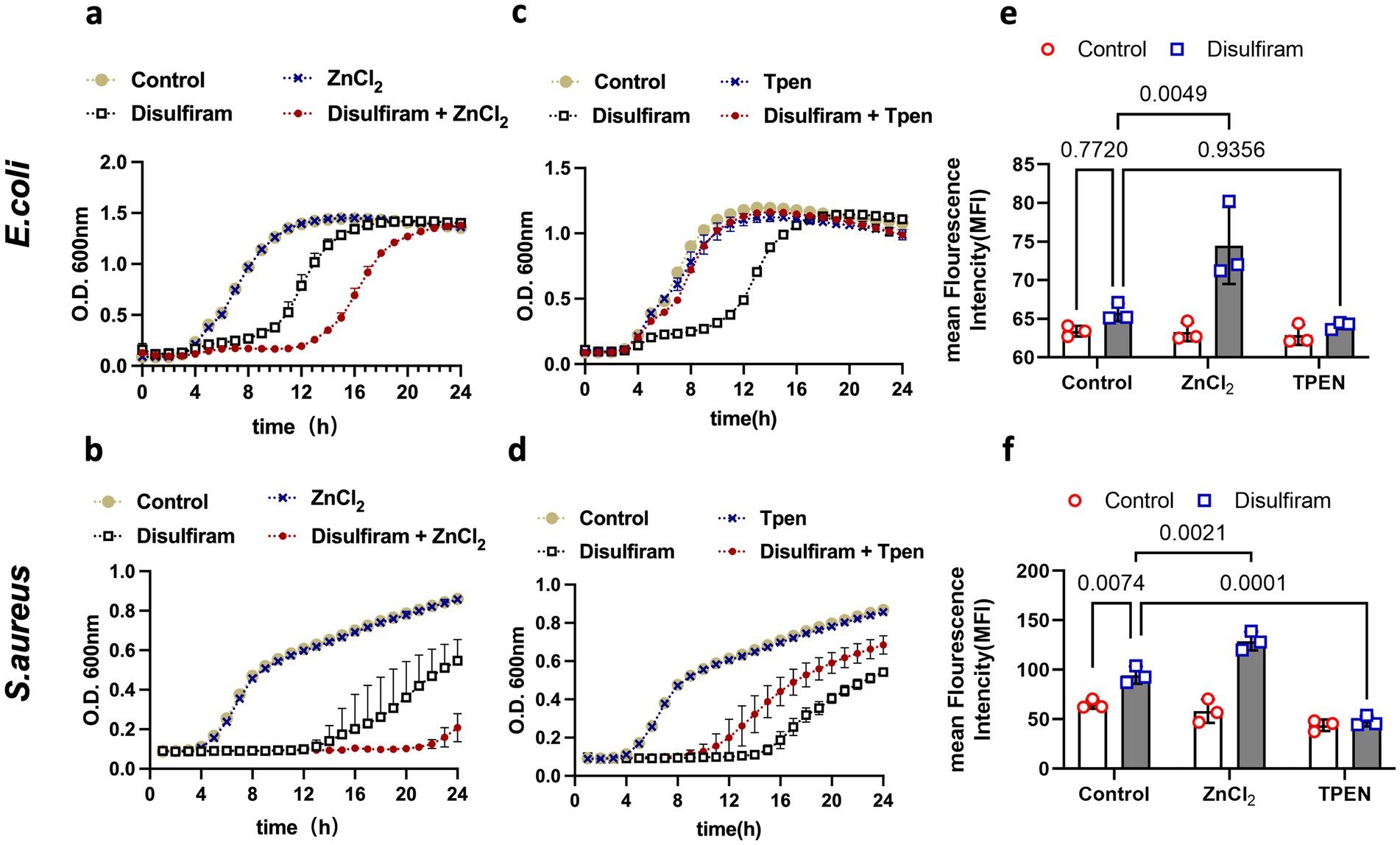
Figure 5. Mechanistic investigation of disulfiram-mediated growth inhibition. (a,c) Growth curves of E. coli treated with disulfiram (32 μg/mL), ZnCl2 (10 μM), or TPEN (10 μM) or their combination as indicated for 24 h. (b,d) Growth curves of S. aureus treated with disulfiram (32 μg/mL), ZnCl2 (10 μM), TPEN (10 μM) or their combination as indicated for 24 h. (e) ROS levels of E. coli treated with disulfiram (32 μg/mL), ZnCl2 (10 μM), TPEN (10 μM) or their combination as indicated. (f) ROS levels of S. aureus treated with disulfiram (32 μg/mL), ZnCl2 (10 μM), TPEN (10 μM) or their combination as indicated. The data in (a,d) are presented as the means ± SDs, and p values were determined using ordinary two-way ANOVA. The data in (a–d) are the means ± SD of three biological replicates.
4 Discussion
Our findings confirmed that disulfiram halted the growth of bacteria by inducing zinc-dependent ROS in bacteria, providing further mechanisms for the existing bacteriostatic drugs. While previous studies have reported the antibacterial effects of disulfiram and its role as a metallo-β-lactamase inhibitor or antibiotic adjuvant, our results are the first to identify zinc-dependent ROS generation as a key mechanism underlying its bacteriostatic activity.”
Even with maximal FDA-approved dosing (500 mg/day), disulfiram is only on the order of a few μg/mL in the plasma and this is lower than the reported antibacterial MIC₅₀ (~256 μg/mL). Thus, plasma disulfiram concentrations under typical therapy are too low to reach the concentrations required for direct antimicrobial activity. We speculated that an adjuvant effects of disulfiram at lower concentrations may be more clinically relevant than direct bacteriostatic activity. Therefore, future studies should evaluate whether the observed adjuvant effects at lower, clinically relevant concentrations may offer therapeutic benefit in combination with existing antibiotics. Disulfiram works in synergism with colistin. Disulfiram is reported to inhibit mRNA expression of mcr-1 and bind to MCR enzymes, thereby amplify membrane damage ability of colistin (Chen et al., 2023). However, in our study, the colistin resistance gene is not present or expressed in the E. coli 25,922 genome. This indicated that disulfiram may strengthen the ability of colistin by some other universal mechanisms. On one hand, disulfiram may weaken the outer membrane of bacteria via metal chelation, facilitating colistin’s penetration, on the other hand, transcriptomic analysis revealed disulfiram-induced oxidative stress pathways, which may amplify colistin’s bactericidal effect by overwhelming bacterial antioxidant defenses. This universal mechanism may potentially extend the usage of disulfiram in clinical circumstances. While disulfiram has demonstrated antibacterial activity in vitro, the in vivo mechanism remains to be fully elucidated. Our in vitro data of disulfiram suggested a substantial contribution of direct antibacterial action. However, additional immunological profiling will be needed to assess potential immune-mediated effects.
Bacteriostatic drugs are important in the treatment of multiple infectious diseases. For example, tigecycline, the next generation of tetracycline, is regarded as a last resort for multidrug-resistant (MDR) bacterial infection (Fang et al., 2020); linezolid is an important alternative in the treatment of vancomycin-resistant enterococci (VAE) or MRSA/MSSA (Bender et al., 2018; Kawasuji et al., 2023). However, these final-line antibiotics are now facing serious levels of resistance from bacteria. Currently available bacteriostatic drugs, including folate inhibitors (sulfonamides and trimethoprim), tetracyclines, and macrolides, either inhibit bacterial growth via folate synthesis inhibition or target ribosomal elements to affect protein synthesis (Sachin and Parag, 2021). Hydrogen peroxide, which generates ROS to kill bacteria, actually kills bacteria at a very high concentration (i.e., half of bacteria can be killed in 10 mM (0.03%) at 1 h) (Thomas et al., 1994). Studies have also reported that ROS do not participate in the bactericidal effects of multiple antibiotics (Keren et al., 2013). However, ROS can contribute to antibiotic lethality (Dwyer et al., 2014), which suggests that ROS inducers could be potentially important adjuvants for current bactericidal antibiotics. In our study, we found that disulfiram induced bacterial ROS, inhibited bacterial growth and augmented the effect of colistin in vitro and in vivo. These findings suggested that disulfiram or other dithiocarboxy acid-containing chemicals might be potential adjuvants with additional antimicrobial mechanisms.
H2O2, paraquat and NO are chemicals related to oxidative damage. H2O2 directly generates ROS; paraquat is a well-known ROS inducer; and NO causes oxidative stress through reactive nitrogen species (RNS) (Jomova et al., 2023). Disulfiram treatment of E. coli induced the expression of multiple redox detoxification genes, indicating that the redox cycle of bacteria is affected. However, how disulfiram induces ROS remains elusive, as dithiocarboxy acid tends to alter the redox cycle, and disulfide bonds alone do not affect bacterial growth. One possible explanation is that disulfiram induces a cellular stress response, during which bacteria produce reactive oxygen species as part of a nonspecific redox-balancing mechanism. However, this ROS production may ultimately contribute to self-damage and growth inhibition. It is also possible that this is not an active detoxification strategy but rather an unintended consequence of disrupted redox homeostasis.
Disulfiram-treated E. coli upregulated many Fe–S clusters that bind or repair associated transcripts. Fe–S clusters are required in critical biological processes, including gene expression, respiration and metabolism (Roche et al., 2013). This metallocofactor annotates more than 100 proteins in E. coli, representing approximately 3% of the proteome (Bak and Weerapana, 2023). Moreover, Fe is associated with oxidative reactions because redox enzymes typically select Fe-based centers due to their high redox and oxygen sensitivity. Thus, Fe–S clusters could be sensitive to oxidation and may degrade under certain conditions (Chen et al., 2023). Zinc is a relatively redox-inert metal and is the most frequent substitute for the Fe–S cofactor site to maintain protein functions (Pritts and Michel, 2022; Imlay et al., 2019). However, during disulfiram treatment, zinc deprivation maintains the growth of E. coli or S. aureus, and zinc supplementation facilitates growth inhibition and ROS production. These results contrast with the previous “helping” role of zinc, indicating that zinc may be detrimental to bacteria under such oxidative conditions.
Zinc is associated with ROS production in some unknown ways. Recently, reports have shown correlations between zinc and oxidative status in biological systems (Arriaza et al., 2022; Liao et al., 2023); however, the link between these factors remains elusive. As a bivalent cation, Zinc does not change its oxidation state in cells. Thus, it may participate in redox reactions in an indirect manner. Metallothionein, a thiol-containing protein, is one of the zinc-binding and zinc-transferring proteins that may alter redox status when interacts with metals. However, whether these proteins induce bacterial ROS under such conditions remains to be explored. Although our data strongly support a zinc-dependent ROS-mediated pathway, the precise molecular interactions between disulfiram, zinc homeostasis, and ROS generation require further study.
One of the major limitations of disulfiram is that it is not a strong antimicrobial drug. Nevertheless, the mechanism of disulfiram warrants further exploration, as understanding the modes of action of chemical groups such as dithiocarboxy acids could inform the development of novel antibiotics.
5 Conclusion
This study identifies a zinc-dependent induction of intracellular ROS as a key mechanism underlying the antibacterial activity of disulfiram. The growth-inhibitory effect was mitigated by ROS scavengers and zinc chelators, highlighting the role of zinc and oxidative stress. These findings offer mechanistic insights into disulfiram’s antibacterial action and support its potential for therapeutic repurposing.
Data availability statement
The raw data supporting the conclusions of this article is available in Supplementary material. RNA-seq data was available with GEO accession numbers of GSE299097 and SRA accession numbers of PRJNA1272480 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE299097).
Ethics statement
The animal study was approved by Animal Review Committee of Zhejiang University School of Medicine. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
QL: Writing – review & editing, Supervision, Software, Investigation, Methodology, Formal analysis, Writing – original draft, Resources, Validation, Funding acquisition, Data curation, Project administration, Visualization, Conceptualization. ZW: Writing – review & editing, Methodology. YP: Writing – review & editing, Methodology. YZ: Writing – review & editing, Methodology.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
We thank Qixing Chen from the Zhejiang University School of Medicine, for language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1619416/full#supplementary-material
References
Adrover, J. M., Carrau, L., Daßler-Plenker, J., Bram, Y., Chandar, V., Houghton, S., et al. (2022). Disulfiram inhibits neutrophil extracellular trap formation and protects rodents from acute lung injury and SARS-CoV-2 infection. JCI. Insight 7:e157342. doi: 10.1172/jci.insight.157342
Arriaza, K., Cuevas, C., Pena, E., Siques, P., and Brito, J. (2022). Impact of zinc on oxidative signaling pathways in the development of pulmonary vasoconstriction induced by hypobaric hypoxia. Int. J. Mol. Sci. 23:6974. doi: 10.3390/ijms23136974
Bak, D. W., and Weerapana, E. (2023). Monitoring Fe-S cluster occupancy across the E. coli proteome using chemoproteomics. Nat. Chem. Biol. 19, 356–366. doi: 10.1038/s41589-022-01227-9
Bender, J. K., Cattoir, V., Hegstad, K., Sadowy, E., Coque, T. M., Westh, H., et al. (2018). Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: towards a common nomenclature. Drug Resist. Updat. 40, 25–39. doi: 10.1016/j.drup.2018.10.002
Bernier, M., Mitchell, S. J., Wahl, D., Diaz, A., Singh, A., Seo, W., et al. (2020). Disulfiram treatment normalizes body weight in obese mice. Cell Metab. 32, 203–214.e4. doi: 10.1016/j.cmet.2020.04.019
Bonamore, A., and Boffi, A. (2008). Flavohemoglobin: structure and reactivity. IUBMB Life 60, 19–28. doi: 10.1002/iub.9
Bush, K., and Bradford, P. A. (2020). Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 33:e00047-19. doi: 10.1128/CMR.00047-19
Chen, C., Cai, J., Shi, J., Wang, Z., and Liu, Y. (2023). Resensitizing multidrug-resistant gram-negative bacteria to carbapenems and colistin using disulfiram. Commun. Biol. 6:810. doi: 10.1038/s42003-023-05173-7
Chen, J., Calderone, L. A., Pan, L., Quist, T., and Pandelia, M. E. (2023). The Fe and Zn cofactor dilemma. Biochim. Biophys. Acta Proteins Proteom. 1871:140931. doi: 10.1016/j.bbapap.2023.140931
Chen, C., Yang, K. W., Wu, L. Y., Li, J. Q., and Sun, L. Y. (2020). Disulfiram as a potent metallo-β-lactamase inhibitor with dual functional mechanisms. Chem. Commun. (Camb.) 56, 2755–2758. doi: 10.1039/C9CC09074F
Dalecki, A. G., Haeili, M., Shah, S., Speer, A., Niederweis, M., Kutsch, O., et al. (2015). Disulfiram and copper ions kill Mycobacterium tuberculosis in a synergistic manner. Antimicrob. Agents Chemother. 59, 4835–4844. doi: 10.1128/AAC.00692-15
Darby, E. M., Trampari, E., Siasat, P., Gaya, M. S., Alav, I., Webber, M. A., et al. (2023). Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 21, 280–295. doi: 10.1038/s41579-022-00820-y
Dwyer, D. J., Belenky, P. A., Yang, J. H., MacDonald, I., Martell, J. D., Takahashi, N., et al. (2014). Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. USA 111, E2100–E2109. doi: 10.1073/pnas.1401876111
Ejim, L., Farha, M. A., Falconer, S. B., Wildenhain, J., Coombes, B. K., Tyers, M., et al. (2011). Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 7, 348–350. doi: 10.1038/nchembio.559
Fang, L. X., Chen, C., Cui, C. Y., Li, X. P., Zhang, Y., Liao, X. P., et al. (2020). Emerging high-level Tigecycline resistance: novel tetracycline Destructases spread via the Mobile Tet (X). BioEssays 42:e2000014. doi: 10.1002/bies.202000014
Gao, X., Huang, H., Pan, C., Mei, Z., Yin, S., Zhou, L., et al. (2022). Disulfiram/copper induces immunogenic cell death and enhances CD47 blockade in hepatocellular carcinoma. Cancers (Basel) 14:4715. doi: 10.3390/cancers14194715
GBD 2019 Antimicrobial Resistance Collaborators (2022). Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the global burden of disease study 2019. Lancet 400, 2221–2248. doi: 10.1016/S0140-6736(22)02185-7
Hamblin, K. A., Flick-Smith, H., Barnes, K. B., Pereira-Leal, J. B., Surkont, J., Hampson, R., et al. (2019). Disulfiram, an alcohol dependence therapy, can inhibit the in vitro growth of Francisella tularensis. Int. J. Antimicrob. Agents 54, 85–88. doi: 10.1016/j.ijantimicag.2019.04.002
Imlay, J. A., Sethu, R., and Rohaun, S. K. (2019). Evolutionary adaptations that enable enzymes to tolerate oxidative stress. Free Radic. Biol. Med. 140, 4–13. doi: 10.1016/j.freeradbiomed.2019.01.048
Jomova, K., Raptova, R., Alomar, S. Y., Alwasel, S. H., Nepovimova, E., Kuca, K., et al. (2023). Reactive oxygen species, toxicity, oxidative stress, and antioxidants: chronic diseases and aging. Arch. Toxicol. 97, 2499–2574. doi: 10.1007/s00204-023-03562-9
Kawasuji, H., Nagaoka, K., Tsuji, Y., Kimoto, K., Takegoshi, Y., Kaneda, M., et al. (2023). Effectiveness and safety of linezolid versus vancomycin, Teicoplanin, or Daptomycin against methicillin-resistant Staphylococcus aureus bacteremia: a systematic review and meta-analysis. Antibiotics (Basel) 12:697. doi: 10.3390/antibiotics12040697
Keren, I., Wu, Y., Inocencio, J., Mulcahy, L. R., and Lewis, K. (2013). Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339, 1213–1216. doi: 10.1126/science.1232688
Liao, L. S., Chen, Y., Hou, C., Liu, Y. H., Su, G. F., Liang, H., et al. (2023). Potent zinc(II)-based immunogenic cell death inducer triggered by ROS-mediated ERS and mitochondrial ca(2+) overload. J. Med. Chem. 66, 10497–10509. doi: 10.1021/acs.jmedchem.3c00603
Lu, C., Li, X., Ren, Y., and Zhang, X. (2021). Disulfiram: a novel repurposed drug for cancer therapy. Cancer Chemother. Pharmacol. 87, 159–172. doi: 10.1007/s00280-020-04216-8
Maier, L., Goemans, C. V., Wirbel, J., Kuhn, M., Eberl, C., Pruteanu, M., et al. (2021). Unravelling the collateral damage of antibiotics on gut bacteria. Nature 599, 120–124. doi: 10.1038/s41586-021-03986-2
Meneguello, J. E., Murase, L. S., de Souza, J. V. P., de Oliveira, C. G., Ghiraldi-Lopes, L. D., Teixeira, J. J. V., et al. (2022). Systematic review of disulfiram as an antibacterial agent: what is the evidence? Int. J. Antimicrob. Agents 59:106578. doi: 10.1016/j.ijantimicag.2022.106578
Moussaoui, M., Misevičienė, L., Anusevičius, Ž., Marozienė, A., Lederer, F., Baciou, L., et al. (2018). Quinones and nitroaromatic compounds as subversive substrates of Staphylococcus aureus flavohemoglobin. Free Radic. Biol. Med. 123, 107–115. doi: 10.1016/j.freeradbiomed.2018.05.071
Nobre, L. S., Todorovic, S., Tavares, A. F. N., Oldfield, E., Hildebrandt, P., Teixeira, M., et al. (2010). Binding of azole antibiotics to Staphylococcus aureus flavohemoglobin increases intracellular oxidative stress. J. Bacteriol. 192, 1527–1533. doi: 10.1128/JB.01378-09
Nonejuie, P., Burkart, M., Pogliano, K., and Pogliano, J. (2013). Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci. USA 110, 16169–16174. doi: 10.1073/pnas.1311066110
Penwell, W. F., Shapiro, A. B., Giacobbe, R. A., Gu, R. F., Gao, N., Thresher, J., et al. (2015). Molecular mechanisms of sulbactam antibacterial activity and resistance determinants in Acinetobacter baumannii. Antimicrob. Agents Chemother. 59, 1680–1689. doi: 10.1128/AAC.04808-14
Pritts, J. D., and Michel, S. L. J. (2022). Fe-S clusters masquerading as zinc finger proteins. J. Inorg. Biochem. 230:111756. doi: 10.1016/j.jinorgbio.2022.111756
Roche, B., Aussel, L., Ezraty, B., Mandin, P., Py, B., and Barras, F. (2013). Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim. Biophys. Acta 1827, 455–469. doi: 10.1016/j.bbabio.2012.12.010
Sachin, M. P., and Parag, P. (2021). “Bactericidal and bacteriostatic antibiotics” in Infections and sepsis development. eds. N. Vincenzo, H. Lixing, and L. Jie (Rijeka: Intech Open). Ch. 1
Schairer, D. O., Chouake, J. S., Nosanchuk, J. D., and Friedman, A. J. (2012). The potential of nitric oxide releasing therapies as antimicrobial agents. Virulence 3, 271–279. doi: 10.4161/viru.20328
Skrott, Z., Mistrik, M., Andersen, K. K., Friis, S., Majera, D., Gursky, J., et al. (2017). Alcohol-abuse drug disulfiram targets cancer via p 97 segregase adaptor NPL4. Nature 552, 194–199. doi: 10.1038/nature25016
Suh, J. J., Pettinati, H. M., Kampman, K. M., and O'Brien, C. P. (2006). The status of disulfiram: a half of a century later. J. Clin. Psychopharmacol. 26, 290–302. doi: 10.1097/01.jcp.0000222512.25649.08
Thakare, R., Shukla, M., Kaul, G., Dasgupta, A., and Chopra, S. (2019). Repurposing disulfiram for treatment of Staphylococcus aureus infections. Int. J. Antimicrob. Agents 53, 709–715. doi: 10.1016/j.ijantimicag.2019.03.024
Thomas, E. L., Milligan, T. W., Joyner, R. E., and Jefferson, M. M. (1994). Antibacterial activity of hydrogen peroxide and the lactoperoxidase-hydrogen peroxide-thiocyanate system against oral streptococci. Infect. Immun. 62, 529–535. doi: 10.1128/iai.62.2.529-535.1994
Wadhwa, N., and Berg, H. C. (2022). Bacterial motility: machinery and mechanisms. Nat. Rev. Microbiol. 20, 161–173. doi: 10.1038/s41579-021-00626-4
Keywords: Escherichia coli, colistin, zinc, ROS, disulfiram
Citation: Luo Q, Wu Z, Pan Y and Zhang Y (2025) Disulfiram inhibits bacterial growth by inducing zinc-dependent reactive oxygen species. Front. Microbiol. 16:1619416. doi: 10.3389/fmicb.2025.1619416
Edited by:
John Osei Sekyere, University of Pretoria, South AfricaReviewed by:
Puneet Gandhi, Bhopal Memorial Hospital and Research Centre, IndiaRikhsan Kurniatuhadi, Tanjungpura University, Indonesia
Mohammed Mukhles, University of Anbar, Iraq
Copyright © 2025 Luo, Wu, Pan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinyu Luo, cXlsdW9Aemp1LmVkdS5jbg==
 Qinyu Luo
Qinyu Luo Zehua Wu
Zehua Wu