- 1School of Environmental Science, University of Guelph, Guelph, ON, Canada
- 2Great Lakes Institute for Environmental Research, University of Windsor, Windsor, ON, Canada
- 3School of Earth and Space Exploration, Arizona State University, Tempe, AZ, United States
- 4Space Science & Astrobiology Division, NASA Ames Research Center, Moffett Field, CA, United States
Arthrobacter are commonly isolated from cold soil environments globally, including those that regularly reach sub-freezing temperatures, suggesting that Arthrobacter have significant potential for growth and activity under temperature and stress extremes. Arthrobacter agilis strain Ant-EH-1 was isolated from nutrient-poor, cold-arid mineral soils from Elephant Head, Antarctica and its growth and activity at sub-freezing temperatures were characterized in this study. We observed different optimal temperatures for cell division compared with aerobic heterotrophic respiration in A. agilis Ant-EH-1. Cell division was observed from at least −5 °C to 30 °C, with the optimal (fastest) growth rate occurring at 25 °C. Microbial respiration was measured from −5 °C to 30 °C with optimal (maximum CO2 produced) respiration occurring at 5 °C. Cold temperature optima of respiration compared with cell division could be indicative of adaptation to the cold and oligotrophic conditions of Elephant Head, where increased cell division under in situ conditions could lead to competition within the nutrient-poor soil matrix. The genome of A. agilis Ant-EH-1 was consistent with observations of cold-adapted activity and included genes related to cold stress, osmotic and oxidative stress, pigment biosynthesis, and potential scavenging of components from necromass. Microscopy revealed morphological differences in this isolate at sub-freezing temperatures, likely due to membrane or lipid modifications. Currently there are a limited number of organisms in culture that are capable of sub-zero growth, so characterisation of the growth and activity of subfreezing adapted microbiota is critical for understanding the ecology of Earth’s cryosphere, has broad biotechnological potential, and can also give insight into the limits for life on our planet or the potential for life on other cold planetary bodies.
Introduction
Arthrobacter species have been isolated from a broad variety of environments including soil (Lee et al., 2003), food (Irlinger et al., 2005), paintings (Heyrman et al., 2005), human clinical specimens (Mages et al., 2008; Funke et al., 1998; Hou et al., 1998), sea water (Chen et al., 2009), air (Li et al., 2004), ice (Liu et al., 2018; Kumar et al., 2015; Margesin et al., 2004), glacier cryoconites (Margesin et al., 2012), sub-glacial lakes (Singh et al., 2016), and Antarctic marine and lake sediment (Pindi et al., 2010; Chen et al., 2005; Reddy et al., 2000; Han et al., 2021). They are frequently isolated from extreme environments including the Antarctic and have a well-documented tolerance to cold temperatures globally (Pindi et al., 2010; Reddy et al., 2000; Dsouza et al., 2015; Gupta et al., 2004; Junge et al., 1998; Wang et al., 2009; Reddy, 2002; Cho et al., 2019; Vodickova et al., 2022; Mukhia et al., 2021). Elephant Head, located in Ellsworth Land, Antarctica, contains dry, ice-free soils and year-round sub-zero temperatures. Previous microbial activity (acetate mineralization) assays on soils from Elephant Head demonstrated that some, but not all, soils contained microbiota that could be active at the sub-zero conditions experienced in situ (Wood et al., 2024). Twenty-one bacterial isolates were previously cultivated from dry permafrost soils at Elephant Head as described in (Wood et al., 2024), with Arthrobacter the most prevalent genus, comprising seven of the 21 cultivated isolates. In this follow-up study, in order to determine whether these cultivated organisms are genetically adapted to the cold, and capable of activity in situ in the extreme Elephant Head environment, one Arthrobacter isolate capable of sub-zero growth (−5 °C) was chosen for further characterization of its cold adaptive capabilities and genomic traits.
Methods
Isolation and characterization
Arthrobacter agilis strain Ant-EH-1 was isolated from cold, dry surface soils collected from Elephant Head, Ellsworth Land, Antarctica (79°49.106’S 83°18.139 W). The average summer atmospheric temperature in Elephant Head is −10.3 °C, with a yearly average of −20.3 °C (McKay et al., 2019). Surface soils where A. agilis Ant-EH-1 was isolated from (“Site 1,” 0–10 cm depth), warm above 0 °C for only a few hundred hours during the year (an estimated ~500 h based on “Site 3” located 0.3 km away). Moisture content of the soils is less than 0.5%. Total organic carbon and nitrogen content is low (<0.07 and 0.007%, respectively; Wood et al., 2024).
Dry soil from Elephant Head was added to 1.5 mL of liquid media Reasoner’s 2A broth (R2B), incubated for 1 week at 15 °C, and then spread plated onto Reasoner’s 2A agar (R2A). The plate was incubated at 15 °C for an additional week. Pink-coloured colonies of A. agilis Ant-EH-1 were streaked for isolation and growth was characterized on R2A agar and in R2B liquid media at −10, −5, 0, 5, 15, 25, 30, and 37 °C. Growth in liquid media was measured via optical density (OD) at 600 nm using a spectrophotometer. Growth rate was calculated as the change in OD over time during the exponential phase of growth. Growth was also characterized on half strength and 1/10th strength R2A as well as on R2A agar plates amended with NaCl (5, 8, and 10%) at 15 °C to examine the salt tolerance of the isolate.
Acetate mineralization radiorespiration assay
Acetate mineralization by isolate A. agilis Ant-EH-1 was evaluated by a radiorespiration assay using radiolabeled acetate (1,2-14C) as a carbon substrate. Microcosms were set up in 20 mL serum vials in triplicate with triplicate autoclaved negative controls. Each 5 mL volume microcosm contained: 4860 uL R2B media; 20 uL 1,2-14C acetic acid (0.043 μCi (~95,000 disintegrations per minute, dpm)); 20 μL of unlabelled acetic acid (3.75 M); and 100 uL of 5.45×106 CFU/mL of A. agilis Ant-EH-1 liquid culture in R2B. Each microcosm also contained a vial of 0.5 mL 1 M potassium hydroxide (KOH) as a carbon dioxide trap. Sterile media (100 μL R2B) was added to negative controls to give the same final volume to all incubations. Microcosms were incubated at 30, 25, 15, 5, 0, and −5 °C. Measurements of KOH radioactivity (correlating with CO2 released) were taken periodically by liquid scintillation spectrometry on a Beckman Coulter (CA, USA) LS 6000SC and percent mineralization calculated as in Wood et al. (2024) and Goordial et al. (2016a).
DNA extraction and sequencing
Arthrobacter agilis Ant-EH-1 was grown on R2A at 15 °C for 1 week. Isolated colonies were suspended in 750 μL of Powerbead solution from the Qiagen DNeasy PowerLyzer PowerSoil kit and DNA extraction followed manufacturers protocol. DNA was eluted with 100 μL of DNAse-free water. Library preparation was completed using the Oxford Nanopore Rapid Sequencing Kit (SQK-RAD004) for use with the flongle flow cell following manufacturers protocol (Oxford Nanopore Technologies). A. agilis Ant-EH-1 DNA was loaded into three flongle flow cells and three replicate 24-h sequencing runs were carried out. High accuracy base calling was used for the first run and fast base calling was used for subsequent runs.
Genome analysis for adaptive traits
Sequence data from three sequencing reactions were concatenated together for assembly and analysis. Assembly was performed using Canu (v 2.2) (Koren et al., 2017). Genes were annotated using Prokka (v 1.14.6) (Seemann, 2014) and GhostKOALA (Kanehisa et al., 2016). Completeness of metabolic pathways was visualized with KEGG Decoder (Graham et al., 2018). Additional gene prediction and functional annotation was performed within the Integrated Microbial Genomes (IMG) platform developed by the Joint Genome Institute, Walnut Creek, CA, USA (Markowitz et al., 2009). The complete genome sequence of strain A. agilis Ant-EH-1 is available for public access on the Joint Genome Institute Integrated (JGI) Microbial Genomes & Microbiomes (IMG) under Gold Study ID: Gs0160646. The 16S rRNA gene was amplified using PCR (primers 27F – 5’-AGAGTTTGATCCTGGCTCAG-3′ and 1492R – 5’-TACGGYTACCTTGTTACGACTT – 3′) and Sanger sequenced at The Centre for Applied Genomics, Toronto, Canada. The Classifier tool of the Ribosomal Database Project (RDPII) v 2.13 (Wang et al., 2007) and the NCBI GenBank database were used to identify the isolate. The sequence can be found on NCBI GenBank under accession number OQ383637.
Scanning electron microscopy
Three A. agilis Ant-EH-1 liquid cultures grown in R2B were harvested for visualization with scanning electron microscopy (SEM): (1) growth at −5 °C for 16 months with visible flocculation (OD600 = 1.13); (2) growth at 5 °C for 16 months with no flocculation (OD600 = 1.57); (3) growth at 25 °C for 1 month with flocculation (OD600 = 1.4). Cultures were grown at −5 °C and 5 °C for extended periods to reflect the incubation period of the acetate mineralization assay and obtain sufficient biomass for SEM. Cultures were diluted in R2B to an OD600 of 1.0, and 1.5 mL of each diluted culture was centrifuged at 4,000 rpm for 4 min. Cultures without flocculation did not produce a visible pellet after initial centrifugation and were additionally centrifuged at 10,000 rpm for 4 min. Harvested cells were washed twice in phosphate buffer (35 mM K2HPO4 and NaH2PO4) with centrifugation for 4 min at 4,000 rpm for cultures with flocculation and 10,000 rpm for cultures without flocculation. Washed pellets were resuspended in 400 μL of phosphate buffer, of which 200 μL was placed on a carbon planchet and incubated for 30 min to allow cell adhesion. After adhesion, planchets were gently submerged in 0.075% ruthenium red and 2.5% glutaraldehyde in 100 mM HEPES pH 7.3 for 30 min to fix cells, then washed once in 100 mM HEPES pH 7.3 and twice in MilliQ water. Cells were then dehydrated by submerging the planchets sequentially in 50, 70, 80, 90, 100%, and a second round of 100% ethanol for 10 min each, followed by drying with a Denton DCP-1 Critical Point Dryer (NJ, USA). Planchets were then mounted on pin stubs with double-sided carbon tape and coated in gold with a Denton Desk V TSC sputter coater. Cells were imaged on a FEI Quanta FEG 250 SEM (OR, USA) at the University of Guelph Molecular and Cellular Imaging Facility (ON, CA).
Results and discussion
Growth and activity characteristics of Arthrobacter agilis Ant-EH-1
Arthrobacter agilis strain Ant-EH-1 grew on R2A media forming bright pink, circular colonies on plates, and pink colouration in liquid R2B media (Figure 1) and was capable of growth from −5 °C to 30 °C on solid and liquid media. Optimum (fastest) growth rate based on optical density occurred at 25 °C (Figure 1) in R2B media, a common nutrient media for growth of oligotrophic organisms. This optimum temperature is similar to other Arthrobacter isolates from polar environments (Supplementary file 1). A. agilis Ant-EH-1 was capable of growth at lower nutrient concentrations (two and ten-fold diluted R2A), and maintained its viability in culture after 1 year (e.g., could be re-streaked from R2A plates incubated at −5 °C for 1 year) indicating its ability to survive long periods of time at sub-zero temperatures without the input of new nutrients. A. agilis Ant-EH-1 was previously observed to be halotolerant, capable of growth in media supplemented with up to 8% NaCl (Wood et al., 2024), like many other cryophilic organisms capable of sub-zero growth (Goordial, 2021). Halotolerance is thought to be needed by microbiota for activity and survival in permafrost, as solutes are concentrated into brine veins at subfreezing temperatures.
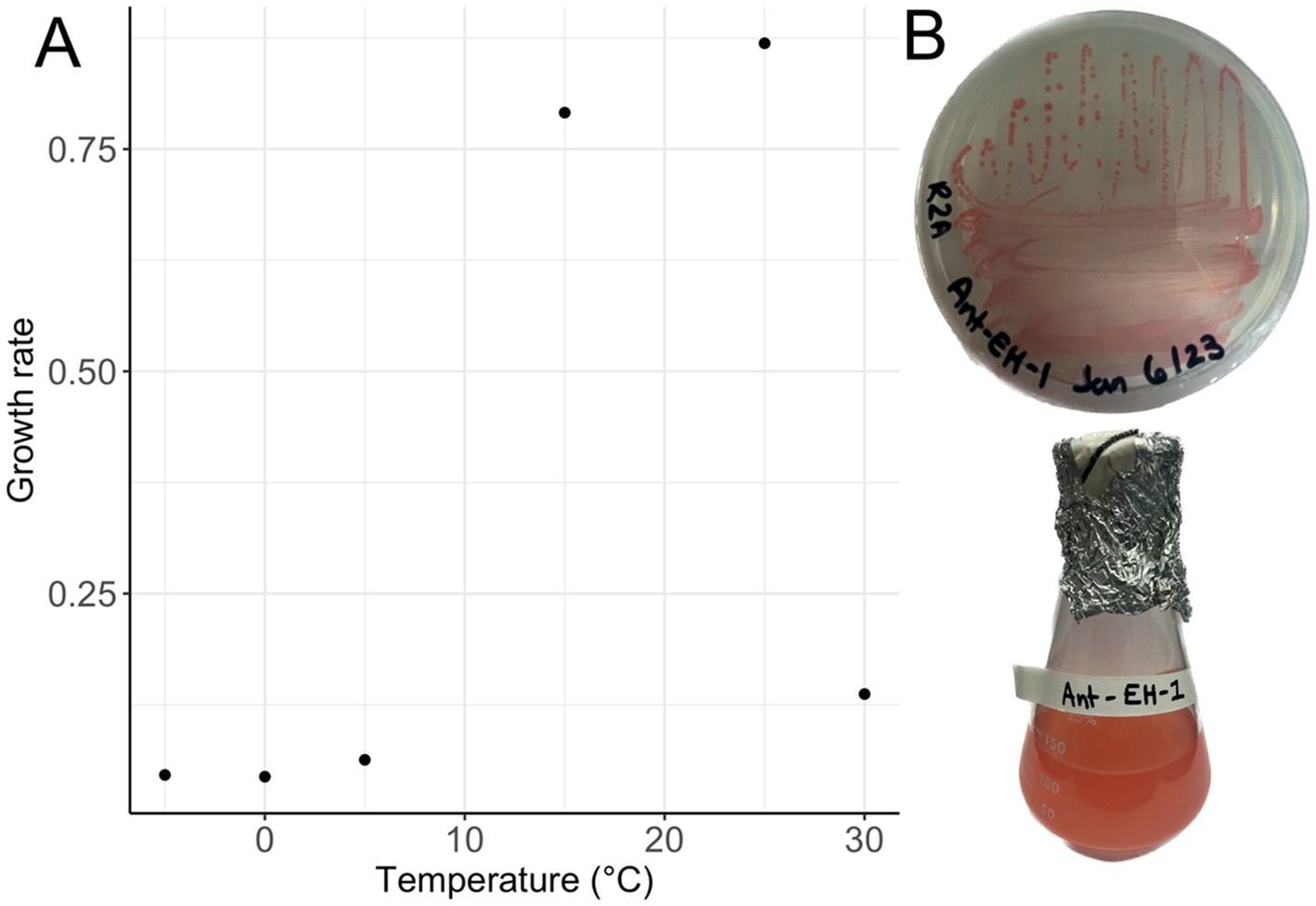
Figure 1. (A) Growth rate (h−1) of Arthrobacter agilis strain Ant-EH-1 in R2A from 30 °C to −5 °C based on optical density measurements (600 nm). (B) Arthrobacter agilis strain Ant-EH-1 colonies grown on R2A plate (top) and in liquid R2B (bottom).
Heterotrophic respiration of acetate was detected at all temperatures tested from −5 °C to 30 °C (Figure 2A). By day 100 of the incubation, 5 °C surpassed all warmer temperatures tested in total percent acetate mineralized. Temperatures below 5 °C demonstrated lag phases that were 50–80 days in length compared with warmer temperatures which had short to no lag phase observed. Total acetate respired in incubations at warmer temperatures (>5 °C) plateaued quickly (day 15–40) compared to 5 °C which did not plateau by the end of the experiment after 466 days (Figure 2A), inset. The highest cumulative percent mineralization of acetate occurred at 5 °C (45%), compared with all other temperatures tested (Figure 2B). The second highest acetate mineralization was observed at 0 °C with 5.6% mineralization.
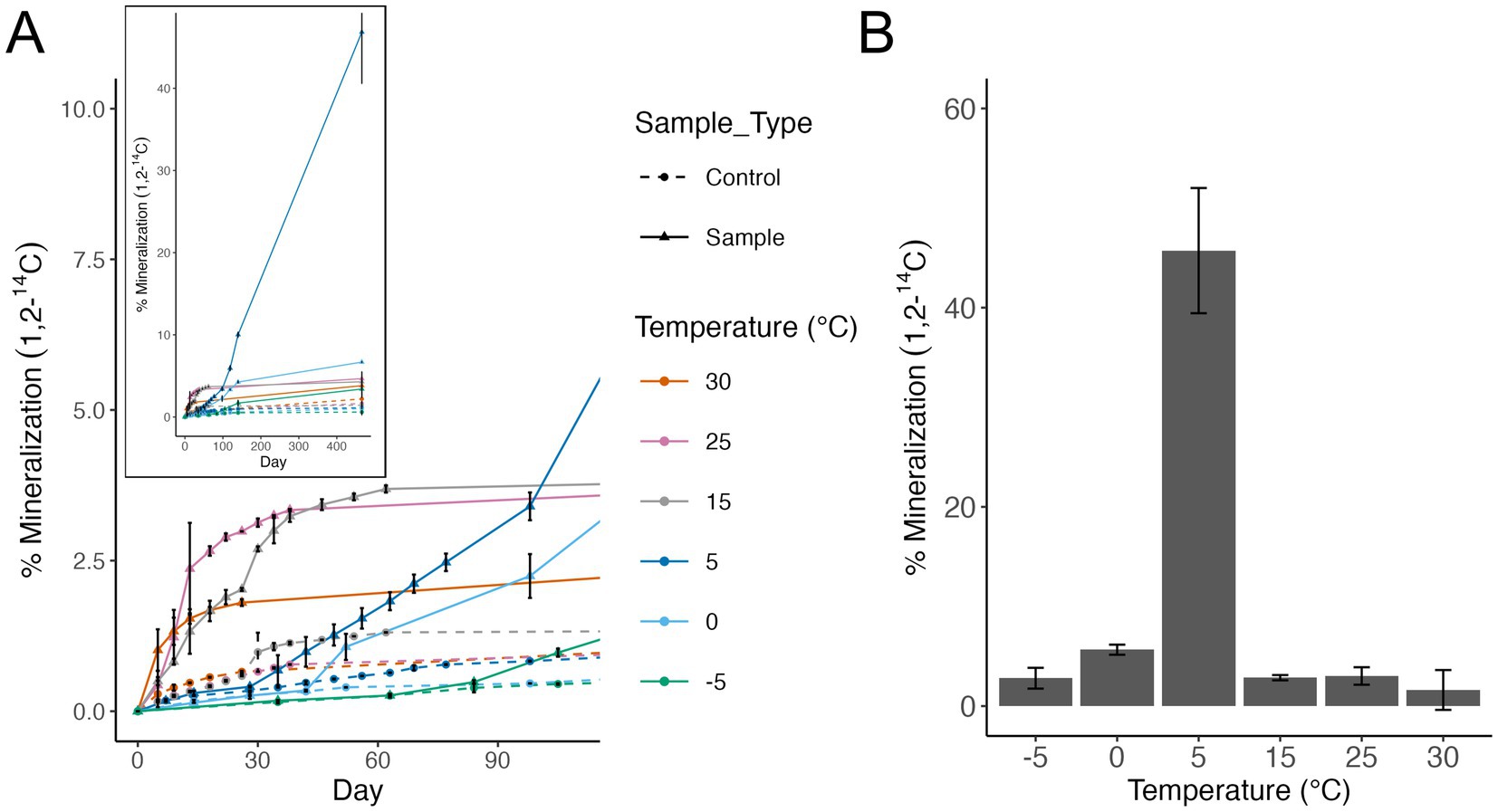
Figure 2. (A) Microbial activity assessed by the mineralization of radiolabeled acetate (1,2-14C) to carbon dioxide (CO2) by Arthrobacter agilis strain Ant-EH-1. Measurements are cumulative. Dashed lines show negative controls (sterile R2A media). Error bars show standard deviation of triplicate incubations. Inset shows activity to day 466. (B) Cumulative percent mineralization of radiolabeled acetate to carbon dioxide by A. agilis Ant-EH-1 after 466 days. Background levels in negative controls were subtracted from corresponding samples.
Arthrobacter genera have been cultivated from several Antarctic environments, and many are capable of growth at cold temperatures (0 to 5 °C) (Pindi et al., 2010; Chen et al., 2005; Reddy et al., 2000; Dsouza et al., 2015; Gupta et al., 2004; Wang et al., 2009; Reddy, 2002; Vodickova et al., 2022; Fong et al., 2001; Aislabie et al., 2013; Shen et al., 2021). Arthrobacter agilis Ant-EH-1 was closely related (similarity based on 16S rRNA gene) to other Arthrobacter from cold environments, including Antarctica (Supplementary Figure S2). To the best of our knowledge this is the first time that cell division has been documented via optical density for an Arthrobacter species at sub-zero temperatures as low as −5 °C. Prior studies stopped cultivation attempts at 4, 0 °C or −1 °C (Supplementary file 1), thus it is possible these isolates could be capable of cell division at lower temperatures. One prior study confirmed that an Arthrobacter species was capable of activity (measured via carbon dioxide production) down to −17 °C (Panikov and Sizova, 2006). A. agilis Ant-EH-1 had a maximum growth rate at 25 °C based on optical density measurements but showed significantly higher levels of activity at 5 °C based on radiorespiration assays. A. agilis Ant-EH-1 could be classified as a eurypsychrophile based on its ability to grow from −5 °C to 30 °C with a maximum growth rate above 20 °C (Raymond-Bouchard et al., 2018; Cavicchioli, 2016). Eurypsychrophiles can grow across a broad range of temperatures (temperature max > 30 °C, min below 0 °C) and have optimum growth rates around 20 °C (Raymond-Bouchard et al., 2018; Cavicchioli, 2016). However, fast growth rate at warmer temperatures does not necessarily indicate that these conditions are preferred by A. agilis Ant-EH-1. As suggested by Cavicchioli (2016) fast growth is not always better, especially in oligotrophic conditions such as those that exist in Elephant Head soils. Differences in activity and OD measurements suggests that A. agilis Ant-EH-1 employs different growth strategies at different temperatures. At warm temperatures it shows a rapid increase in activity and OD which plateaus quickly. At colder temperatures A. agilis Ant-EH-1 shows a slower initial increase in activity but then is able to sustain an active population for longer. Future transcriptomic analysis could help to determine if there is a different growth strategy employed at cold temperatures versus the slowed rates being a result of slowed kinetic reactions. This was demonstrated previously in transcriptomic studies of Psychrobacter sp. which found that it shifted from a fast-growing state at warmer temperatures (6–22 °C) to a resource efficiency state at cold temperatures (<4 °C) via downregulation of genes involved in energy metabolism (e.g., electron transport chain, TCA cycle) and biosynthesis (e.g., amino acid, nucleotide, ribosome, peptidoglycan synthesis) and upregulation of RNases and peptidases indicating a growth control response (Bergholz et al., 2009).
Cell envelope characteristics of A. agilis Ant-EH-1 across a temperature gradient
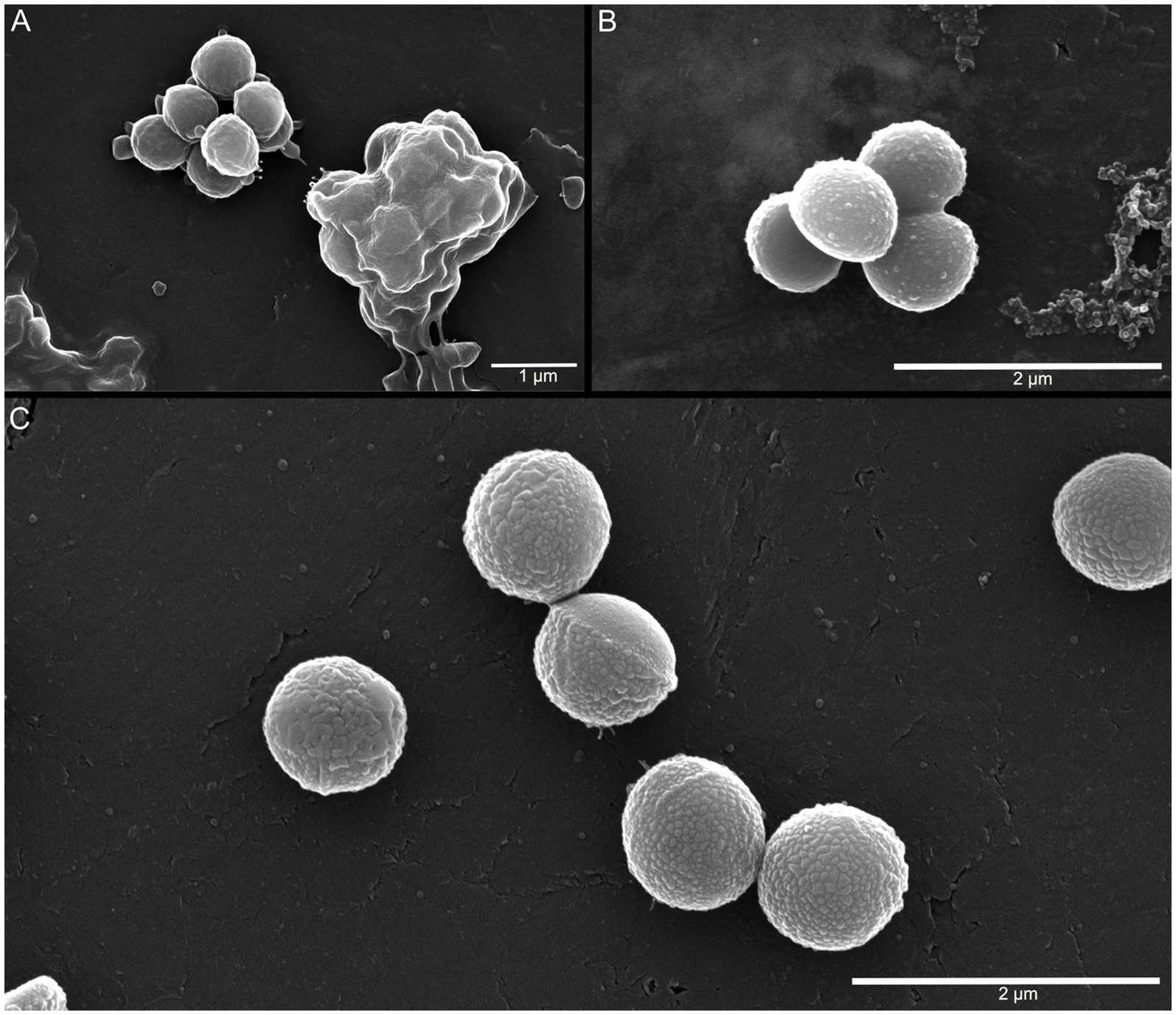
As the A. agilis Ant-EH-1 genome contained genes associated with cold-adaptive membrane and cell wall modifications, cultures grown at −5, 5, and 25 °C were visualized with SEM to identify potential temperature-dependent changes to the cell envelope. At all temperatures, cultures contained aggregated cells coated in extracellular polymeric substance (EPS)-like material (Figure 3). EPS have an array of functions in mediating cell--environment interactions, including formation of biofilms and aggregates and protection against environmental stressors (Costa et al., 2018). They are proposed to act as a cryoprotectant by limiting ice crystal formation and lowering the freezing point of water, among other potential mechanisms (De Maayer et al., 2014). Elevated EPS production at low and sub-zero temperatures has been observed in psychrophilic and psychrotolerant bacteria (Marx et al., 2009; Caruso et al., 2018), including A. agilis strain L77, a psychrotroph isolated from a subglacial lake (Singh et al., 2016).
Cells grown at 5 °C and −5 °C had nodule-like features partially (5 °C) or completely (−5 °C) covering the cell envelope (Figure 3), which may indicate cell wall modifications associated with cold adaptation. For example, dense nodular encrustations were observed in the psychrophile Planococcus halocryophilus Or1 grown at −15 °C as a result of peptidoglycan accumulation and calcium carbonate biomineralization (Mykytczuk et al., 2016). The genes associated with these accumulations in P. halocryophilus, peptidoglycan synthase (ftsI) and carbonic anhydrase (cab), were both identified in the A. agilis Ant-EH-1 genome. While the prominence of these nodules at −5 °C compared to 5 °C suggests a role in cold adaptation, it is unconfirmed whether these features were exclusive to A. agilis Ant-EH-1 grown at cold temperatures as thick EPS matrices at 25 °C may have obscured surface features. Additional transcriptomic and microscopic analyses are warranted to further investigate potential cold adaptive strategies.
General genome characteristics and cold adaptive and stress response genes in Arthrobacter agilis Ant-EH-1
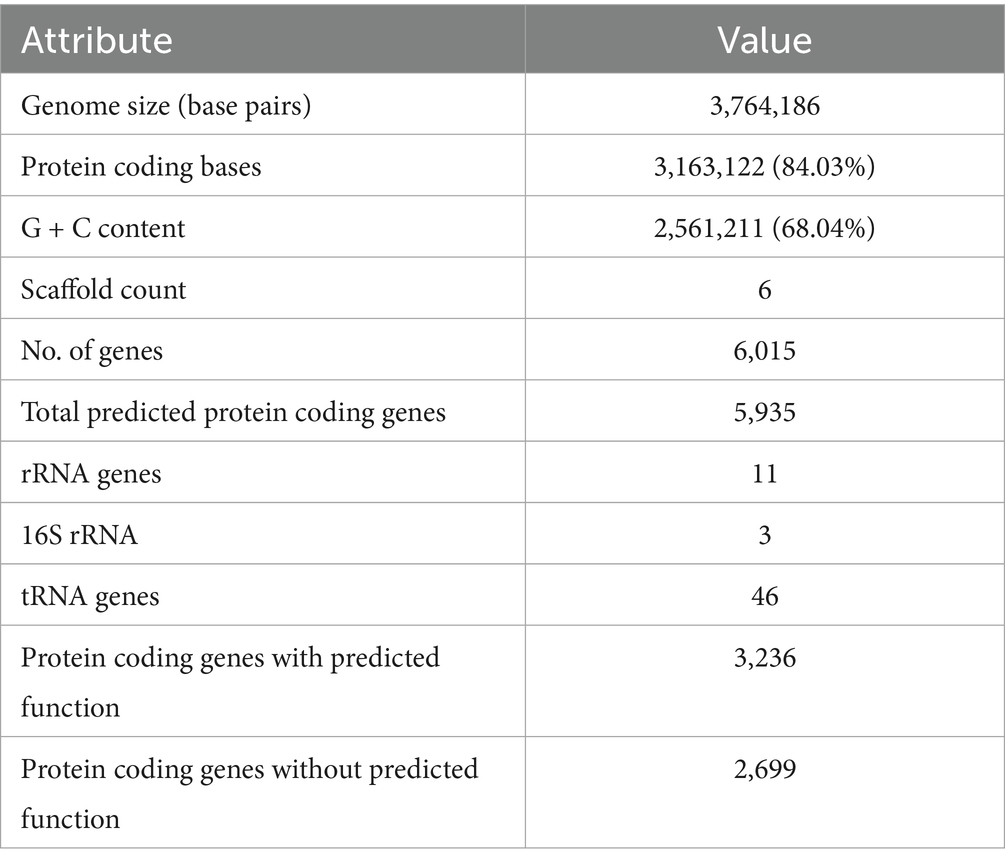
The Arthrobacter agilis Ant-EH-1 complete draft genome is 3,764,186 bp in length with 68.04% GC content. There were 5,935 protein encoding genes predicted, of which 3,236 were assigned a predicted function and the remaining annotated as hypothetical proteins (Table 1). Consistent with other psychrophiles and cryophiles the organism had genomic traits associated with cold adaptation, stress response, and DNA repair. While many of the genes and pathways are found in non-extremophilic organisms as well, these traits have been identified as being important in cryophiles and they are often present in multiple copies, or with redundant pathways as described below.
Cold and stress response
The Arthrobacter agilis Ant-EH-1 genome contains genes that facilitate general stress response (universal stress proteins, Kvint et al., 2003) as well as genes that facilitate cold adaptation via osmotic tolerance, oxidative stress, membrane and cell wall alterations, carotenoid biosynthesis, and DNA repair (Table 2; Supplementary materials). Coding regions for cold shock protein (cspA) were present in the genome. CspA can act as RNA chaperones and help with unfolding misfolded proteins and preventing misfolding during and after cold shock (Gottesman, 2018; Zhang and Gross, 2021). A. agilis Ant-EH-1 also has coding sequences for a variety of other chaperone proteins which can assist with protein folding under stressful conditions (clpB, dnaJ, dnaK) (Alam et al., 2021; Maillot et al., 2019).
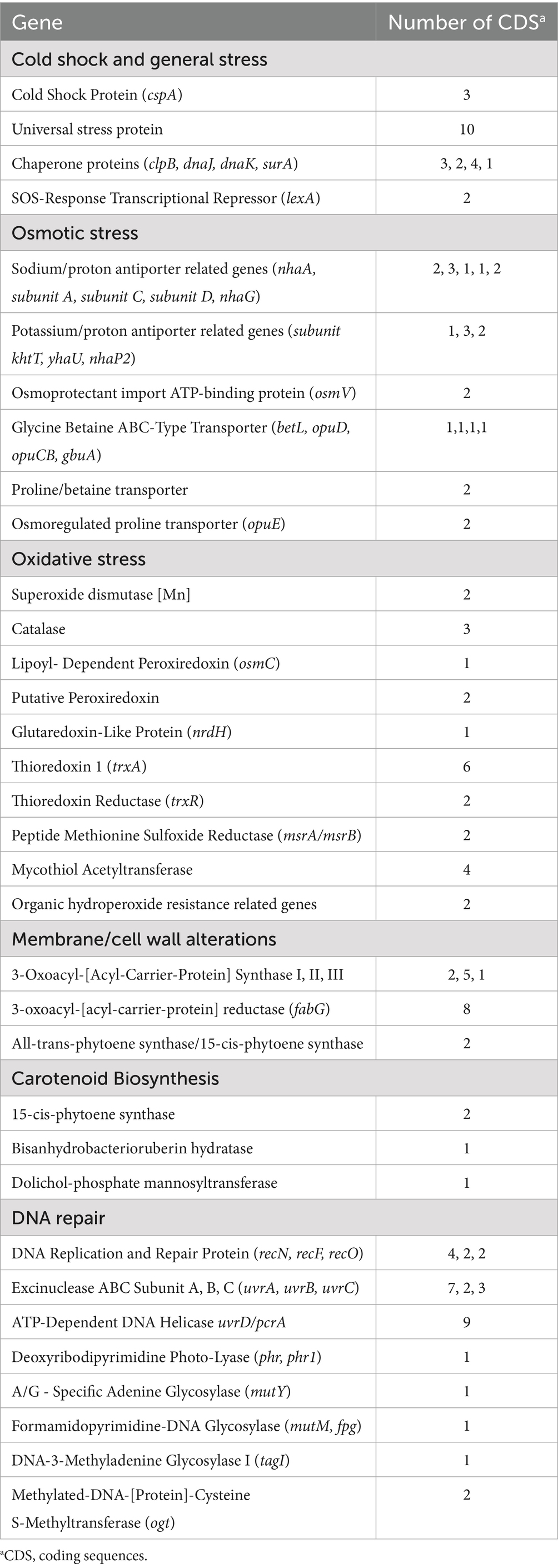
Table 2. Cold adaptation and stress response genes present in the Arthrobacter agilis strain Ant-EH-1 genome.
In dry permafrost environments, water activity is low because aridity and freezing temperatures maintain any water present as vapour or ice. Liquid water present under these conditions may be due to porosity and mineral substrate composition, as well as the presence of solutes. Solutes will become concentrated in liquid water films and pockets present at sub-zero temperatures in the soil matrix, resulting in high solute concentrations which bind to water molecules and further reduce water availability for use by microorganisms (Devoie et al., 2024). One of the most well understood adaptations to osmotic stress is the accumulation of compatible solutes, or small organic molecules within the cell which help to maintain turgor pressure, depress the freezing point of intracellular water to prevent ice crystal formation, and prevent protein aggregation (Chattopadhyay, 2002; Chin et al., 2010; Roberts, 2005). Genes for the import or synthesis of compatible solutes glycine betaine, proline, and trehalose were present (glycine betaine transport related genes [betL, opuD, opuCB, gbuA), proline/betaine transporter (opuAC), trehalose synthase (treS, treZ, treY)] (Table 2). Inorganic cations including sodium (Na+) and potassium (K+) ions can become toxic to the cell if accumulated to high concentrations. A. agilis Ant-EH-1 contains genes for Na+/K + antiporters (nhaA, nhaP, nhaD, mnhABCDEF) which transport Na + and K + out of the cell and uptake H+ in order to maintain intracellular pH and cell volume while avoiding cytotoxic Na+/K+ accumulation (Vimont and Berche, 2000; Bremer and Krämer, 2019; Padan and Schuldiner, 1994; Padan and Schuldiner, 1994).
Metabolism and adaptations for growth in oligotrophic conditions
Carbon utilization and storage
Arthrobacter agilis Ant-EH-1 carries out aerobic heterotrophic growth. Its genome encodes for complete glycolysis, pyruvate oxidation, tricarboxylic acid (TCA) cycle, and oxidative phosphorylation pathways, as well as a near-complete pentose phosphate cycle pathway (1 enzyme missing, transaldolase) (Table 3), though as this is a draft genome it is unclear whether this pathway is truly incomplete or instead reflects genome incompleteness. The A. agilis Ant-EH-1 genome also encodes for a complete glyoxylate shunt pathway (isocitrate lyase, AceA; and malate synthase, GlcB). The glyoxylate shunt bypasses the release of carbon dioxide (CO2) during the TCA cycle and may help conserve carbon in limiting environments. It was previously also identified in a bacterial isolate from nutrient-poor dry permafrost soils in University Valley, Antarctica where it was thought to help with carbon conservation in ~150,000-year-old permafrost (Goordial et al., 2016b).
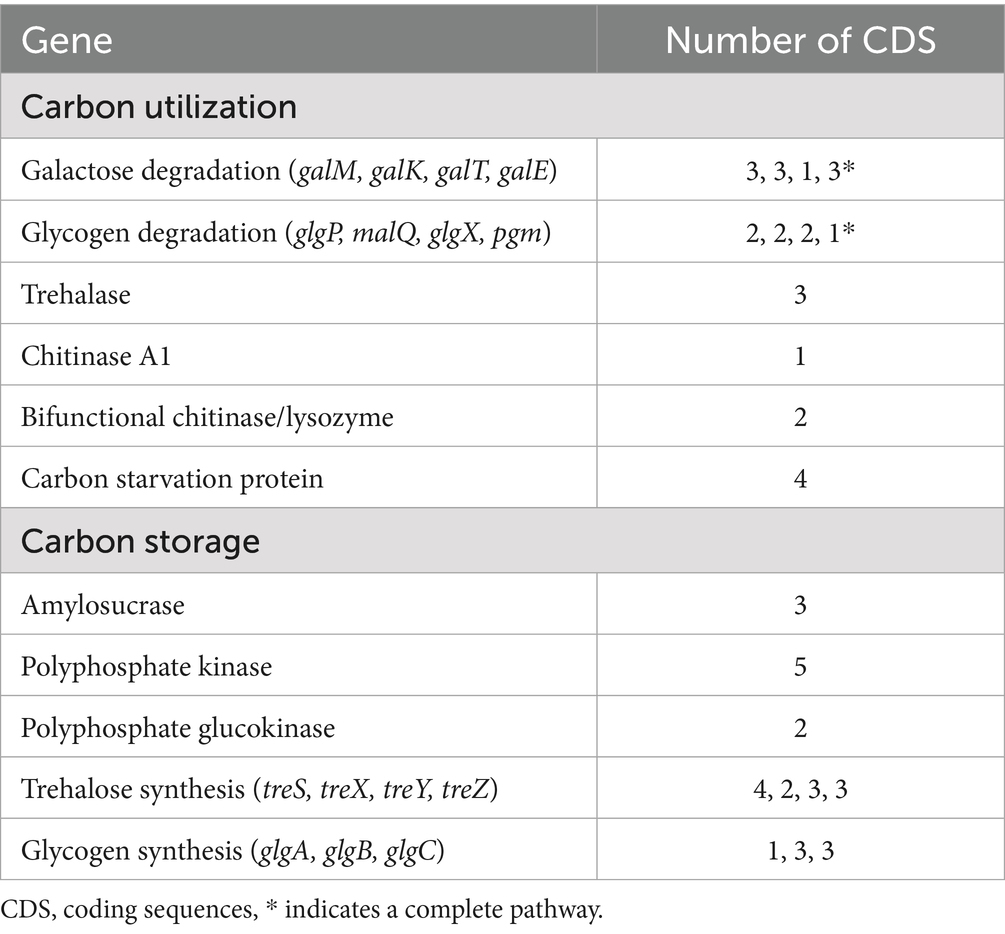
The A. agilis Ant-EH-1 genome contains coding sequences for the degradation of a variety of carbon compounds including a complete gluconeogenesis pathway, complete pathways for galactose and glycogen degradation, chitinases (chitinase A1, and bifunctional chitinase/lysozyme) for degradation of chitin and peptidoglycan, and trehalase for trehalose degradation (Table 3). Genetic traits associated with the ability to use carbon substrates derived from the degradation of cellular material (e.g., necromass components) were found to be a major component of the community metagenome in Elephant Head, Antarctica, soils from which A. agilis Ant-EH-1 was isolated (Wood et al., 2024). The isolate also has coding sequences for carbon starvation protein (cstA) which has been shown to regulate the cAMP-CRP-dependent carbon starvation response (Schultz and Matin, 1991). Genes for trace gas metabolisms of atmospheric hydrogen, carbon monoxide, and methane were not found within the genome (Leung and Greening, 2020; Bay et al., 2021).
A. agilis Ant-EH-I contains genes to synthesize compounds such as trehalose, glucan, glycogen, and polyphosphates (Table 3), which can be used for carbon and energy storage to enable survival in oligotrophic conditions. Other Antarctic Arthrobacter species are known to store carbon as glycogen (Dsouza et al., 2015). Trehalose may additionally serve as a protectant against numerous stressors including cold and desiccation, as well as a reserve of carbon (Dsouza et al., 2015; Elbein et al., 2003; Chen et al., 2011).
Conclusion
Arthrobacter agilis strain Ant-EH-1 is capable of cell division at −5 °C and has an optimum growth temperature of 25 °C. Though optimal cell division occurs at 25 °C, the highest amount of microbial respiration (activity) was measured at 5 °C. Its cold-active physiological traits were consistent with genome composition which encodes for functions related to cold adaptation including cold shock proteins, molecular chaperones, compatible solute transporters, oxidative stress response genes, and carotenoid biosynthesis genes. A. agilis Ant-EH-1 is also adapted for survival in oligotrophic conditions as demonstrated by genomic traits and its growth on low nutrient media (ten-fold diluted R2A). Its genome contains coding sequences for the catabolism of a variety of carbon compounds including components of cell walls from necromass. Its genome also contains genes for the synthesis of carbon and energy storage molecules such as trehalose, and polyphosphates. The ability of A. agilis strain Ant-EH-1 to divide and be active at −5 °C supports that the cold dry soils of Elephant Head contain viable microbial life which may be active in their environment, despite the difficulty in detecting such life in bulk soil analyses.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, OQ383637 https://gold.jgi.doe.gov/study?id=Gs0160646, Gs0160646.
Author contributions
CW: Methodology, Formal analysis, Validation, Data curation, Conceptualization, Writing – original draft, Visualization, Investigation, Writing – review & editing. EM: Methodology, Investigation, Writing – review & editing, Visualization. EH: Visualization, Writing – original draft, Formal analysis, Investigation. ET-R: Funding acquisition, Writing – review & editing, Visualization, Formal analysis. MW: Funding acquisition, Writing – review & editing. JG: Funding acquisition, Project administration, Formal analysis, Writing – review & editing, Supervision, Writing – original draft, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. We acknowledge funding from the NASA Exobiology program (80NSSC21K0480) to ET-R, JG, MW. The CIFAR Azrieli Global Scholar program, a CIFAR Fellowship and CIFAR Catalyst for funding to JG. Natural Sciences and Engineering Research Council of Canada (NSERC) for a Discovery grant RGPIN-2021-02585 to JG. CW received support from a NSERC CGS-M award, an Ontario Graduate Scholarship, a University of Guelph MacSon Entrance Scholarship and Polar Knowledge Canada Northern Scientific Training Program (NSTP) support. EH was supported by an NSERC Undergraduate student research assistantship (USRA) and EM an NSERC Postdoctoral Fellowship.
Acknowledgments
We also thank Chris McKay, Edward Balaban, and Barney and Robert Swan for their role in sampling the soil that Arthrobacter agilis Ant-EH-1 was isolated from.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1620620/full#supplementary-material
References
Aislabie, J. M., Lau, A., Dsouza, M., Shepherd, C., Rhodes, P., and Turner, S. J. (2013). Bacterial composition of soils of the Lake Wellman area, Darwin Mountains, Antarctica. Extremophiles 17, 775–786. doi: 10.1007/s00792-013-0560-6
Alam, A., Bröms, J. E., Kumar, R., and Sjöstedt, A. (2021). The role of ClpB in bacterial stress responses and virulence. Front. Mol. Biosci. 8:283. doi: 10.3389/fmolb.2021.668910
Bay, S. K., Dong, X., Bradley, J. A., Leung, P. M., Grinter, R., Jirapanjawat, T., et al. (2021). Trace gas oxidizers are widespread and active members of soil microbial communities. Nat. Microbiol. doi: 10.1038/s41564-020-00811-w
Bergholz, P. W., Bakermans, C., and Tiedje, J. M. (2009). Psychrobacter arcticus 273-4 uses resource efficiency and molecular motion adaptations for subzero temperature growth. J. Bacteriol. 191, 2340–2352. doi: 10.1128/JB.01377-08
Bremer, E., and Krämer, R. (2019). Responses of microorganisms to osmotic stress. Ann. Rev. Microbiol. 73, 313–334. doi: 10.1146/annurev-micro-020518-115504
Caruso, C., Rizzo, C., Mangano, S., Poli, A., Di Donato, P., Finore, I., et al. (2018). Production and biotechnological potential of extracellular polymeric substances from sponge-associated Antarctic bacteria. Appl. Environ. Microbiol. 84, e01624–e01617. doi: 10.1128/AEM.01624-17
Cavicchioli, R. (2016). On the concept of a psychrophile. ISME J. 10, 793–795. doi: 10.1038/ismej.2015.160
Chattopadhyay, M. K. (2002). The cryoprotective effects of glycine betaine on bacteria. Trends Microbiol. 10:311. doi: 10.1016/S0966-842X(02)02395-8
Chen, X. M., Jiang, Y., Li, Y. T., Zhang, H. H., Li, J., Chen, X., et al. (2011). Regulation of expression of trehalose-6-phosphate synthase during cold shock in Arthrobacter strain A3. Extremophiles 15, 499–508. doi: 10.1007/s00792-011-0380-5
Chen, Y. G., Tang, S. K., Zhang, Y. Q., Li, Z. Y., Yi, L. B., Wang, Y. X., et al. (2009). Arthrobacter halodurans sp. nov., a new halotolerant bacterium isolated from sea water. Antonie Van Leeuwenhoek 96, 63–70. doi: 10.1007/s10482-009-9336-5
Chen, M., Xiao, X., Wang, P., Zeng, X., and Wang, F. (2005). Arthrobacter ardleyensis sp. nov., isolated from Antarctic lake sediment and deep-sea sediment. Arch. Microbiol. 183, 301–305. doi: 10.1007/s00203-005-0772-y
Chin, J. P., Megaw, J., Magill, C. L., Nowotarski, K., Williams, J. P., Bhaganna, P., et al. (2010). Solutes determine the temperature windows for microbial survival and growth. PNAS 107, 7835–7840. doi: 10.1073/pnas.1000557107
Cho, Y. J., Cho, A., Hong, S. G., Choi, H. G., and Kim, O. S. (2019). Draft genome sequence of Arthrobacter oryzae TNBS02, a bacterium containing heavy metal resistance genes, isolated from soil of Antarctica. Microbiol Resour Announc 8, e01501–e01518. doi: 10.1128/MRA.01501-18
Costa, O. Y. A., Raaijmakers, J. M., and Kuramae, E. E. (2018). Microbial extracellular polymeric substances: ecological function and impact on soil aggregation. Front. Microbiol. 9:1636. doi: 10.3389/fmicb.2018.01636
De Maayer, P., Anderson, D., Cary, C., and Cowan, D. A. (2014). Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 15, 508–517. doi: 10.1002/embr.201338170
Devoie, É., Connon, R. F., Beddoe, R., Goordial, J., Quinton, W. L., and Craig, J. R. (2024). Disconnected active layers and unfrozen permafrost: a discussion of permafrost-related terms and definitions. Sci. Total Environ. 912:169017. doi: 10.1016/j.scitotenv.2023.169017
Dsouza, M., Taylor, M. W., Turner, S. J., and Aislabie, J. (2015). Genomic and phenotypic insights into the ecology of Arthrobacter from Antarctic soils. BMC Genomics 16:36. doi: 10.1186/s12864-015-1220-2
Elbein, A. D., Pan, Y. T., Pastuszak, I., and Carroll, D. (2003). New insights on trehalose: a multifunctional molecule. Glycobiology 13, 17R–27R. doi: 10.1093/glycob/cwg047
Fong, N., Burgess, M., Barrow, K., and Glenn, D. (2001). Carotenoid accumulation in the psychrotrophic bacterium Arthrobacter agilis in response to thermal and salt stress. Appl. Microbiol. Biotechnol. 56, 750–756. doi: 10.1007/s002530100739
Funke, G., Pagano-Niederer, M., Sjödén, B., and Falsen, E. (1998). Characteristics of Arthrobacter cumminsii, the most frequently encountered Arthrobacter species in human clinical specimens. J. Clin. Microbiol. 36, 1539–1543. doi: 10.1128/JCM.36.6.1539-1543.1998
Goordial, J. (2021). Cryomicrobial ecology: still much to learn about life left out in the cold. mSystems 6:10.1128/msystems.00852-21. doi: 10.1128/msystems.00852-21
Goordial, J., Davila, A., Lacelle, D., Pollard, W., Marinova, M. M., Greer, C. W., et al. (2016a). Nearing the cold-arid limits of microbial life in permafrost of an upper dry valley, Antarctica. ISME J. 10, 1613–1624. doi: 10.1038/ismej.2015.239
Goordial, J., Raymond-Bouchard, I., Zolotarov, Y., De Bethencourt, L., Ronholm, J., Shapiro, N., et al. (2016b). Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp. JG3 isolated from high elevation McMurdo Dry Valley permafrost, Antarctica. FEMS Microbiol. Ecol. 92, 1–11. doi: 10.1093/femsec/fiv154
Gottesman, S. (2018). Chilled in translation: adapting to bacterial climate change. Mol. Cell 70, 193–194. doi: 10.1016/j.molcel.2018.04.003
Graham, E. D., Heidelberg, J. F., and Tully, B. J. (2018). Potential for primary productivity in a globally-distributed bacterial phototroph. ISME J. 12, 1861–1866. doi: 10.1038/s41396-018-0091-3
Gupta, P., Reddy, G. S. N., Delille, D., and Shivaji, S. (2004). Arthrobacter gangotriensis sp. nov. and Arthrobacter kerguelensis sp. nov. from Antarctica. Int. J. Syst. Evol. Microbiol. 54, 2375–2378. doi: 10.1099/ijs.0.63110-0
Han, S. R., Kim, B., Jang, J. H., Park, H., and Oh, T. J. (2021). Complete genome sequence of Arthrobacter sp. PAMC25564 and its comparative genome analysis for elucidating the role of CAZymes in cold adaptation. BMC Genomics 22:403. doi: 10.1186/s12864-021-07734-8
Heyrman, J., Verbeeren, J., Schumann, P., Swings, J., and De Vos, P. (2005). Six novel Arthrobacter species isolated from deteriorated mural paintings. Int. J. Syst. Evol. Microbiol. 55, 1457–1464. doi: 10.1099/ijs.0.63358-0
Hou, X. G., Kawamura, Y., Sultana, F., Shu, S., Hirose, K., Goto, K., et al. (1998). Description of Arthrobacter creatinolyticus sp. nov., isolated from human urine. Int. J. Syst. Bacteriol. 48, 423–429. doi: 10.1099/00207713-48-2-423
Irlinger, F., Bimet, F., Delettre, J., Lefevre, M., and Grimont, P. A. (2005). Arthrobacter bergerei sp. nov. and Arthrobacter arilaitensis sp. nov., novel coryneform species isolated from the surfaces of cheeses. Int. J. Syst. Evol. Microbiol. 55, 457–462. doi: 10.1099/ijs.0.63125-0
Junge, K., Gosink, J. J., Hoppe, H. G., and Staley, J. T. (1998). Arthrobacter, Brachybacterium and Planococcus isolates identified from Antarctic Sea ice brine. Description of Planococcus mcmeekinii, sp. nov. Syst. Appl. Microbiol. 21, 306–314. doi: 10.1016/S0723-2020(98)80038-6
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Kumar, R., Singh, D., Swarnkar, M. K., Singh, A. K., and Kumar, S. (2015). Complete genome sequence of Arthrobacter sp. ERGS1:01, a putative novel bacterium with prospective cold active industrial enzymes, isolated from east Rathong glacier in India. J. Biotechnol. 214, 139–140. doi: 10.1016/j.jbiotec.2015.09.025
Kvint, K., Nachin, L., Diez, A., and Nyström, T. (2003). The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6, 140–145. doi: 10.1016/S1369-5274(03)00025-0
Lee, J. S., Lee, K. C., Pyun, Y. R., and Bae, K. S. (2003). Arthrobacter koreensis sp. nov., a novel alkalitolerant bacterium from soil. Int. J. Syst. Evol. Microbiol. 53, 1277–1280. doi: 10.1099/ijs.0.02492-0
Leung, P. M., and Greening, C. (2020). Greening lab metabolic marker gene databases. Melbourne, Australia: Monash University. Available online at: https://bridges.monash.edu/collections/Greening_lab_metabolic_marker_gene_databases/5230745
Li, Y., Kawamura, Y., Fujiwara, N., Naka, T., Liu, H., Huang, X., et al. (2004). Rothia aeria sp. nov., Rhodococcus baikonurensis sp. nov. and Arthrobacter russicus sp. nov., isolated from air in the Russian space laboratory Mir. Int. J. Syst. Evol. Microbiol. 54, 827–835. doi: 10.1099/ijs.0.02828-0
Liu, Q., Xin, Y. H., Chen, X. L., Liu, H. C., Zhou, Y. G., and Chen, W. X. (2018). Arthrobacter ruber sp. nov., isolated from glacier ice. Int. J. Syst. Evol. Microbiol. 68, 1616–1621. doi: 10.1099/ijsem.0.002719
Mages, I. S., Frodl, R., Bernard, K. A., and Funke, G. (2008). Identities of Arthrobacter spp. and Arthrobacter-like Bacteria encountered in human clinical specimens. J. Clin. Microbiol. 46, 2980–2986. doi: 10.1128/JCM.00658-08
Maillot, N. J., Honoré, F. A., Byrne, D., Méjean, V., and Genest, O. (2019). Cold adaptation in the environmental bacterium Shewanella oneidensis is controlled by a J-domain co-chaperone protein network. Commun. Biol. 2:323. doi: 10.1038/s42003-019-0567-3
Margesin, R., Schumann, P., Spröer, C., and Gounot, A. M. (2004). Arthrobacter psychrophenolicus sp. nov., isolated from an alpine ice cave. Int. J. Syst. Evol. Microbiol. 54, 2067–2072. doi: 10.1099/ijs.0.63124-0
Margesin, R., Schumann, P., Zhang, D. C., Redzic, M., Zhou, Y. G., Liu, H. C., et al. (2012). Arthrobacter cryoconiti sp. nov., a psychrophilic bacterium isolated from alpine glacier cryoconite. Int. J. Syst. Evol. Microbiol. 62, 397–402. doi: 10.1099/ijs.0.031138-0
Markowitz, V. M., Mavromatis, K., Ivanova, N. N., Chen, I. M. A., Chu, K., and Kyrpides, N. C. (2009). IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25, 2271–2278. doi: 10.1093/bioinformatics/btp393
Marx, J. G., Carpenter, S. D., and Deming, J. W. (2009). Production of cryoprotectant extracellular polysaccharide substances (EPS) by the marine psychrophilic bacterium Colwellia psychrerythraea strain 34H under extreme conditions. Can. J. Microbiol. 55, 63–72. doi: 10.1139/W08-130
McKay, C. P., Balaban, E., Abrahams, S., and Lewis, N. (2019). Dry permafrost over ice-cemented ground at elephant head, Ellsworth Land, Antarctica. Antarct. Sci. 31, 263–270. doi: 10.1017/S0954102019000269
Mukhia, S., Khatri, A., Acharya, V., and Kumar, R. (2021). Comparative genomics and molecular adaptational analysis of Arthrobacter from Sikkim Himalaya provided insights into its survivability under multiple high-altitude stress. Genomics 113, 151–158. doi: 10.1016/j.ygeno.2020.12.001
Mykytczuk, N. C. S., Lawrence, J. R., Omelon, C. R., Southam, G., and Whyte, L. G. (2016). Microscopic characterization of the bacterial cell envelope of Planococcus halocryophilus Or1 during subzero growth at −15 °C. Polar Biol. 39, 701–712. doi: 10.1007/s00300-015-1826-5
Padan, E., and Schuldiner, S. (1994). Molecular physiology of Na+/H+ antiporters, key transporters in circulation of Na+ and H+ in cells. Biochim. Biophys. Acta 1185, 129–151. doi: 10.1016/0005-2728(94)90204-6
Panikov, N. S., and Sizova, M. V. (2006). Growth kinetics of microorganisms isolated from Alaskan soil and permafrost in solid media frozen down to -35C. FEMS Microb. Ecol. 59, 500–512. doi: 10.1111/j.1574-6941.2006.00210.x
Pindi, P. K., Manorama, R., Begum, Z., and Shivaji, S. (2010). Arthrobacter antarcticus sp. nov., isolated from an Antarctic marine sediment. Int. J. Syst. Evol. Microbiol. 60, 2263–2266. doi: 10.1099/ijs.0.012989-0
Raymond-Bouchard, I., Tremblay, J., Altshuler, I., Greer, C. W., and Whyte, L. G. (2018). Comparative transcriptomics of cold growth and adaptive features of a eury- and steno-psychrophile. Front. Microbiol. 9:1565. doi: 10.3389/fmicb.2018.01565
Reddy, G. S. N. (2002). Arthrobacter roseus sp. nov., a psychrophilic bacterium isolated from an Antarctic cyanobacterial mat sample. Int. J. Syst. Evol. Microbiol. 52, 1017–1021. doi: 10.1099/00207713-52-3-1017
Reddy, G. S., Aggarwal, R. K., Matsumoto, G. I., and Shivaji, S. (2000). Arthrobacter flavus sp. nov., a psychrophilic bacterium isolated from a pond in McMurdo Dry Valley, Antarctica. Int. J. Syst. Evol. Microbiol. 50, 1553–1561. doi: 10.1099/00207713-50-4-1553
Roberts, M. F. (2005). Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 1:5. doi: 10.1186/1746-1448-1-5
Schultz, J. E., and Matin, A. (1991). Molecular and functional characterization of a carbon starvation gene of Escherichia coli. J. Mol. Biol. 218, 129–140. doi: 10.1016/0022-2836(91)90879-B
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shen, L., Liu, Y., Allen, M. A., Xu, B., Wang, N., Williams, T. J., et al. (2021). Linking genomic and physiological characteristics of psychrophilic Arthrobacter to metagenomic data to explain global environmental distribution. Microbiome 9:136. doi: 10.1186/s40168-021-01084-z
Singh, R. N., Gaba, S., Yadav, A. N., Gaur, P., Gulati, S., Kaushik, R., et al. (2016). First high quality draft genome sequence of a plant growth promoting and cold active enzyme producing psychrotrophic Arthrobacter agilis strain L77. Stand. Genomic Sci. 11, 1–9. doi: 10.1186/s40793-016-0176-4
Vimont, S., and Berche, P. (2000). NhaA, an Na(+)/H(+) antiporter involved in environmental survival of Vibrio cholerae. J. Bacteriol. 182, 2937–2944. doi: 10.1128/JB.182.10.2937-2944.2000
Vodickova, P., Suman, J., Benesova, E., Strejcek, M., Neumann-Schaal, M., Cajthaml, T., et al. (2022). Arthrobacter polaris sp. nov., a new cold-adapted member of the family Micrococcaceae isolated from Antarctic fellfield soil. Int. J. Syst. Evol. Microbiol. 72:5541. doi: 10.1099/ijsem.0.005541
Wang, F., Gai, Y., Chen, M., and Xiao, X. (2009). Arthrobacter psychrochitiniphilus sp. nov., a psychrotrophic bacterium isolated from Antarctica. Int. J. Syst. Evol. Microbiol. 59, 2759–2762. doi: 10.1099/ijs.0.008912-0
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wood, C., Bruinink, A., Trembath-Reichert, E., Wilhelm, M. B., Vidal, C., Balaban, E., et al. (2024). Active microbiota persist in dry permafrost and active layer from elephant head, Antarctica. ISME Commun. 4:ycad002. doi: 10.1093/ismeco/ycad002
Keywords: psychrophile, cryophile, extremophile, Arthrobacter , permafrost, cold-adaptation
Citation: Wood C, Magnuson E, Harrop E, Trembath-Reichert E, Wilhelm MB and Goordial J (2025) Subzero cell division, respiration, and genomic traits of cryophilic Arthrobacter agilis Ant-EH-1 isolated from cold-arid Antarctic mineral soils. Front. Microbiol. 16:1620620. doi: 10.3389/fmicb.2025.1620620
Edited by:
Tatiana A. Vishnivetskaya, The University of Tennessee, Knoxville, United StatesCopyright © 2025 Wood, Magnuson, Harrop, Trembath-Reichert, Wilhelm and Goordial. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacqueline Goordial, Z29vcmRpYWxAdW9ndWVscGguY2E=
 Claudia Wood1
Claudia Wood1 Ethan Harrop
Ethan Harrop Elizabeth Trembath-Reichert
Elizabeth Trembath-Reichert Jacqueline Goordial
Jacqueline Goordial