- 1College of Life Sciences, Inner Mongolia University, Hohhot, China
- 2Key Laboratory of Herbage and Endemic Crop Biology, Ministry of Education, Inner Mongolia University, Hohhot, China
- 3College of Forestry, Inner Mongolia Agricultural University, Hohhot, China
- 4Inner Mongolia Engineering Technology Research Center of Germplasm Resources Conservation and Utilization, Inner Mongolia University, Hohhot, China
- 5Vocational and Technical College, Inner Mongolia Agricultural University, Baotou, China
Leucocalocybe mongolica is an ecologically important grassland fungus that forms distinct fairy rings, which are hotspots of soil microbial activity. In this study, we investigated soil fungal communities across three spatial zones of L. mongolica fairy rings (inside, fruiting, and outside zones) in the Bayanbulak alpine grassland of Xinjiang, China. High-throughput sequencing, co-occurrence network analysis, and functional prediction tools (FUNGuild and PICRUSt2) were applied to assess community structure, interactions, and environmental drivers. The fruiting zone was dominated by L. mongolica, showing reduced fungal diversity, simplified microbial networks, and increased laccase activity, whereas the outside zone maintained higher diversity and more complex interactions. Redundancy analysis revealed strong correlations between fungal community composition and nitrogen availability, laccase activity, and pH. Functional predictions indicated spatially distinct trophic strategies, with saprotrophs enriched in the fruiting zone and ectomycorrhizal fungi dominant in other zones. These findings suggest that elevated laccase activity and nitrogen cycling may be key processes through which L. mongolica influences soil fungal zonation in grassland ecosystems.
1 Introduction
Leucocalocybe mongolica, an endemic and representative species in Chinese grasslands, is widely recognized as a valuable germplasm resource due to its distinctive aroma, large basidiocarps, and fleshy texture (Lu et al., 2017; Zaki et al., 2022). It is rich in bioactive compounds (e.g., peptides and polysaccharides) and essential nutrients, and studies have confirmed its diverse medicinal properties, including hepatoprotective, anticancer, antioxidant, and immunomodulatory effects (Duan et al., 2021; Wang et al., 2019; Ma et al., 2022; Zhao et al., 2020; You et al., 2014; Zaki et al., 2024). In natural grassland ecosystems, L. mongolica often forms type II fairy rings (Shantz and Piemeisel, 1917; Fidanza, 2007; Fidanza et al., 2016; Ito et al., 2017), which are characterized by concentric, dark green bands of grass with dense, belt-like distributions of fruiting bodies, clearly distinguishable from the surrounding vegetation. Unlike type I rings, which typically cause turf necrosis, or type III rings, which promote luxuriant plant growth without distinct fruiting belts, type II rings represent the most visually recognizable form in natural grasslands (Caesar-Tonthat et al., 2013). Fairy rings play a crucial ecological role by forming complex interactions among fungi, soil microbes, and plant roots, which may influence biodiversity at the ecosystem level. However, microbial diversity within the fruiting zone (ON) is often reduced due to the strong dominance of the ring-forming fungus (Bonanomi et al., 2012; Li et al., 2022). The fungi within these rings actively participate in nutrient cycling through organic matter decomposition, promoting nutrient availability and improving soil fertility (Lindahl and Tunlid, 2015). They also assist plants in water and nutrient absorption, enhancing plant resilience under environmental stress (Bonfante and Genre, 2010). In degraded or disturbed ecosystems, the fungal mycelial network can help stabilize soil structure, reduce erosion, and facilitate vegetation recovery, thus contributing to ecological restoration (Cairney, 2005; Van Der Heijden et al., 1998). As a dominant saprotroph, L. mongolica plays a pivotal role in regulating matter and energy flow in grassland ecosystems, reflecting the intricate relationships between fungi and their environment (Bonanomi et al., 2012).
In recent years, research on L. mongolica has been primarily concentrated in the Inner Mongolia region, focusing on its population genetic structure, soil microbial community composition, and metabolic functions (Lu et al., 2018; Duan and Bau, 2021a; Wang et al., 2022; Duan et al., 2022a,b). In addition, extensive studies in alpine grassland ecosystems of the Tibetan Plateau have investigated fairy rings formed by related fungi such as Floccularia luteovirens, systematically revealing their genetic variation, metabolomic characteristics, and microbial community structures (Xing et al., 2017; Xing et al., 2018; Tang et al., 2024). However, studies on L. mongolica populations in Xinjiang remain very limited. As a marginal population located at the western edge of the species’ distribution, the Xinjiang population may possess unique ecological adaptations and microbe-fungus interaction patterns. Given the distinct habitat conditions in Xinjiang, including differences in climate, soil types, and vegetation composition compared to Inner Mongolia and the Tibetan Plateau, this population is likely to exhibit specific characteristics in fungal community structure, functional traits, and responses to environmental factors.
This study focuses on L. mongolica populations in the Bayanbulak grassland in Hejing County, Xinjiang, utilizing high-throughput sequencing technology in combination with traditional ecological survey methods. The research primarily addresses the following scientific questions: (1) What are the composition and structural characteristics of soil fungal communities within the fairy rings of the Bayanbulak grassland population? (2) What are the interaction patterns within the soil microbial communities of the fairy rings? and (3) How does L. mongolica influence key soil factors and the functional traits of fungal communities? This study aims to fill the knowledge gap regarding L. mongolica in this region and provide scientific basis for formulating regional conservation strategies.
2 Materials and methods
2.1 Study site
The study area is located in the Bayanbulak Grassland (Hejing County), a temperate steppe region renowned for its abundant fairy rings (Supplementary Table S1). The Bayanbulak Grassland is located between 42°10′N–43°30′N and 82°32′E–86°15′E, at the junction of the eastern, southern, and western Tianshan Mountains in the Xinjiang Uygur Autonomous Region, China. This region has a typical temperate continental arid climate, with a mean annual temperature of −4.8 °C and an average annual precipitation of approximately 276 mm (Lü et al., 2023). The study area is characterized primarily by typical alpine swamp meadows, with expansive alpine Stipa purpurea grasslands on mountain slopes and foothills. Moist valley bottoms support the development of swamp meadows and marsh vegetation dominated by Carex, Kobresia, and Juncus species (http://lcj.xinjiang.gov.cn, accessed March 15, 2025). During the fruiting period of fungi in August to September 2024, four L. mongolica fairy rings were collected (Supplementary Table S1).
2.2 Soil sampling and measurements
In August 2024, field sampling of L. mongolica and its associated fairy ring was conducted in the Bayanbulak grassland of Hejing County. During the sampling process, surface litter such as dead branches and leaves was carefully removed, and detailed records were made of each sampling site’s geographic coordinates, sampling time, and other relevant information. Based on the ecological characteristics of the fairy rings, each sampling site was divided into three representative zones (Supplementary Figure S1): the outside zone (OUT), located 5 meters away from the fruiting area and unaffected by fungal mycelia or associated vegetation; the fruiting zone (ON), corresponding to the area where fruiting bodies are present; and the inside zone (IN), situated at the geometric center of the ring structure.
A standardized sampling procedure was adopted in this study. For each of the four fairy rings, soil samples were collected from the OUT and ON zones using a five-point sampling method, while three parallel samples were collected around the central point of the IN zone. In total, 52 original soil samples were obtained (4 fairy rings × 13 sampling points). After sieving through a 2 mm mesh, samples from the same zone within each ring were thoroughly homogenized to form composite samples, resulting in 12 composite samples (4 fairy rings × 3 zones). To ensure analytical reliability, three biological replicates were established for each composite sample, yielding a total of 36 samples (12 composite samples × 3 replicates) for subsequent analyses. All samples were immediately flash-frozen in liquid nitrogen after collection and stored at −80 °C for subsequent physicochemical measurements and high-throughput sequencing.
2.3 Soil chemical and physical characterization
Soil pH was measured using a digital pH meter in a soil–water suspension at a ratio of 1:2.5 (w/v) (Guo et al., 2020). Soil moisture content was determined using the gravimetric (drying) method. Ammonium nitrogen (NH₄⁺-N) and nitrate nitrogen (NO₃−-N) were quantified using a fully automated Kjeldahl nitrogen analyzer. Total nitrogen (TN), total phosphorus (TP), and total potassium (TK) in the soil were determined following the method described by Gao et al. (2019). Soil organic carbon (SOC) and available potassium (AK) were measured using commercial assay kits according to the manufacturer’s instructions (Suzhou Keming Biotechnology Co., Ltd., Suzhou, China). Soil enzyme activities were measured using commercial assay kits provided by Grace Biotechnology (China). The specific enzymes included: soil sucrase activity (S-SC, Cat. No.: G0302W96), soil cellulase activity (S-CL, Cat. No.: G0308W), soil laccase activity (SL, Cat. No.: G0325W), soil urease activity (S-UE, Cat. No.: G0301W96), soil FDA hydrolase activity (FDA, Cat. No.: G0322W), and soil chitinase activity (SJ, Cat. No.: G0335W). All measurements were performed strictly according to the manufacturer’s instructions.
2.4 DNA extraction and PCR amplification
Genomic DNA was extracted from soil samples using the CTAB method (Miao et al., 2014). The purity and concentration of the extracted DNA were assessed by 1% agarose gel electrophoresis. Appropriate amounts of DNA were then transferred into sterile centrifuge tubes and diluted to a final concentration of 1 ng/μL using sterile water. PCR amplification of the ITS1 region was performed using primers ITS1-1F-F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS1-1F-R (5′-GCTGCGTTCTTCATCGATGC-3′). All reactions were carried out in a 15 μL volume containing Phusion® High-Fidelity PCR Master Mix with GC Buffer, 2 μM of each forward and reverse primer, and approximately 10 ng of template DNA. The PCR amplification was performed under the following conditions: initial denaturation at 98 °C for 1 min; 30 cycles of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s, and extension at 72 °C for 30 s; followed by a final extension at 72 °C for 5 min. The PCR products were mixed with an equal volume of 1X TAE buffer and electrophoresed on a 2% agarose gel. Target DNA bands were excised and purified using the Universal DNA Extraction Kit (TianGen, China) according to the manufacturer’s instructions.
2.5 Libraries generated and Illumina sequencing
Library preparation was performed using the NEB Next® Ultra DNA Library Prep Kit. The constructed libraries were quality-checked with an Agilent 5,400 system and quantified by Q-PCR. Qualified libraries were sequenced on the Illumina platform. Raw sequences were processed through the DADA2 plugin in QIIME2 for quality filtered, denoised, merged, and non-chimeric to generate amplicon sequence variants (ASVs) (Callahan et al., 2016; Balvočiūtė and Huson, 2017; Caporaso et al., 2010). Representative ASV sequences were taxonomically annotated against the UNITE database version 8.2 (DeSantis et al., 2006).
2.6 Statistical analyses
All statistical analyses were performed using one-way ANOVA in IBM SPSS Statistics 27.0, with p < 0.05 considered statistically significant. Circos plots were generated using the R package “Circlize” to visualize the distribution of dominant fungal genera in the fairy rings of L. mongolica in Bayanbulak grassland. Potential microbial biomarkers were identified through linear discriminant analysis (LDA) effect size (LEfSe) method with an LDA score threshold >4.0, and the factorial Kruskal-Wallis test was set at α = 0.05 (Mandal et al., 2015; Segata et al., 2011). Alpha diversity metrics (Chao1, Shannon index, and Simpson’s reciprocal index) were calculated using the QIIME2 core-diversity plugin to assess within-sample diversity at the feature sequence level. Rarefaction curves of ASVs were generated in QIIME to evaluate sequencing depth adequacy. Inter-group comparisons of α-diversity indices were conducted using the Wilcoxon rank-sum test (IBM SPSS Statistics 27.0), with significance set at p < 0.05. Beta diversity was assessed using Bray-Curtis and weighted UniFrac distances to evaluate structural differences in microbial communities among samples, visualized through PCoA plots (Vázquez-Baeza et al., 2013). The statistical significance of beta diversity differences between groups was assessed using PERMANOVA (permutational multivariate analysis of variance). Co-occurrence networks were constructed and their topological properties analyzed using R packages “igraph,” “psych,” “WGCNA,” and “reshape2,” with network visualization performed in Gephi (Csardi, 2013; Revelle, 2023; Langfelder and Horvath, 2008; Wickham, 2020; Bastian et al., 2009). The Zi-Pi analysis of co-occurrence networks was conducted using R packages “ggplot2” and “igraph” and utilized Zi (range: 0–5) and Pi (range: 0–1) values to characterize node roles within the network (Han et al., 2022). Redundancy analysis (RDA) implemented in the “vegan” package was employed to explore potential relationships between microbial communities and environmental factors (Oksanen et al., 2013). PICRUSt2 and FUN-Guild were utilized to predict fungal community functions based on normalized ASV tables (Douglas et al., 2020; Nguyen et al., 2016). It should be noted that both methods rely on inference based on existing databases and algorithms, making them predictive analyses rather than experimentally verified results.
3 Result
3.1 Distinct diversity and assemblage structure of soil fungi across fairy ring zones of Leucocalocybe mongolica
High-throughput sequencing of soils from three fairy ring zones 2,640,608 high-quality reads, corresponding to 12,371 ASVs. Taxonomic annotation based on the UNITE database revealed a high annotation rate at the genus level, and the rarefaction curve reached saturation, indicating sufficient sequencing depth (Supplementary Figures S2b,c). Community composition analysis showed that Ascomycota, Basidiomycota, and Mortierellomycota dominated across all zones (Figure 1a). Notably, the relative abundance of Basidiomycota in the ON zone reached 52.06%, higher than that in the IN (36.66%) and OUT zones (35.53%), representing a 1.4-fold increase compared to both peripheral zones (Supplementary Figure S3). At the genus level, Leucocalocybe was the most characteristic taxon of the fruiting zone, while Inocybe, Geopora, and Cladosporium were also important contributors (Figure 1b). The ON and OUT zones shared 226 ASVs, and the IN zone also shared a considerable number of ASVs with the ON (171) and OUT (188) zones. In addition, all three zones shared a substantial core microbiome comprising 555 ASVs. Notably, each zone contained a considerable number of unique ASVs (IN: 1026; ON: 997; OUT: 1331), reflecting distinct fungal community structures in different zones (Figure 1c). Despite a shared core microbiome of 555 ASVs, each zone harbored many unique taxa, indicating strong spatial differentiation of fungal communities shaped by L. mongolica activity.
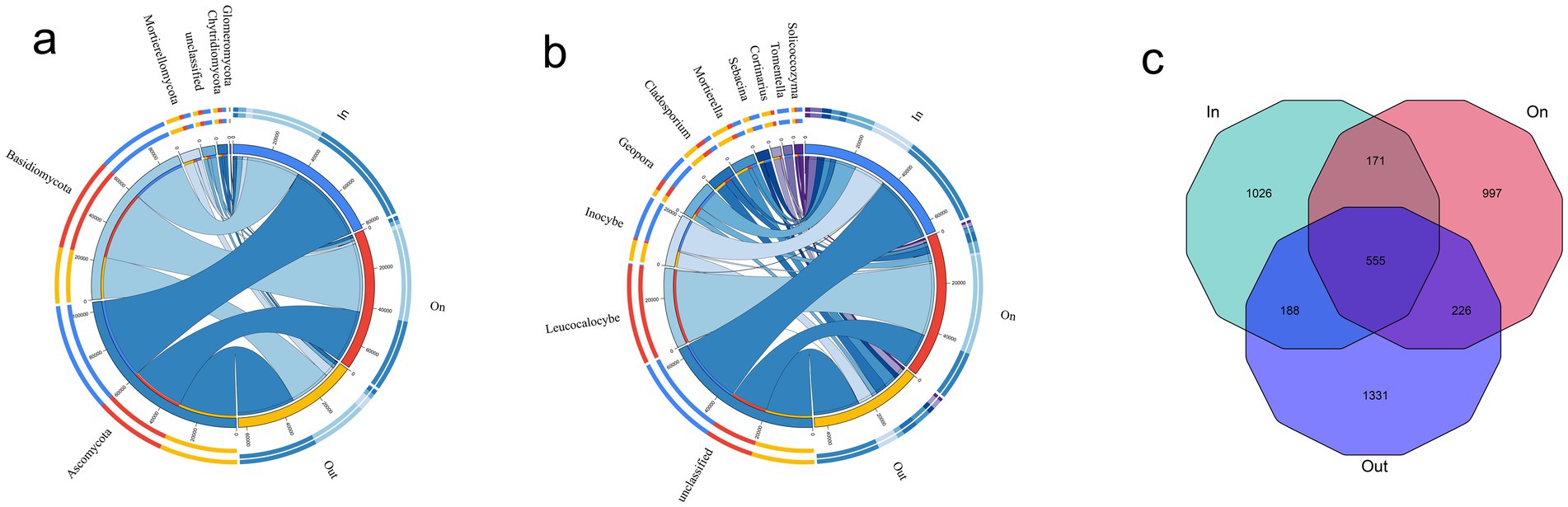
Figure 1. Fungal community composition analyses in different zones of L. mongolica fairy rings. (a) Phylum-level; (b) Genus-level; (c) Venn diagram of shared and unique ASVs across sample groups.
LEfSe analysis identified distinct fungal biomarkers across zones, reflecting strong habitat filtering within fairy rings (Figure 2). The IN zone was enriched with Sordariomycetes and several saprotrophic taxa, suggesting enhanced decomposition potential. The ON zone was dominated by Agaricomycetes, particularly Leucocalocybe, highlighting its competitive advantage in the fruiting zone. In contrast, the OUT zone supported diverse Ascomycota and Mortierellomycota lineages, including Mortierella and Hirsutella, indicating broader ecological strategies. Overall, these patterns demonstrate that L. mongolica activity drives sharp community differentiation through selective pressures in different microenvironments. Diversity analyses revealed clear spatial patterns: fungal richness and evenness were highest in the OUT zone, intermediate in the IN zone, and lowest in the ON zone (p < 0.05), indicating that L. mongolica activity suppresses community diversity within the fruiting zone (Figures 3a–c). Beta-diversity analysis further showed strong separation among zones (p < 0.01), with ON samples forming distinct clusters (Figures 3d,e). These patterns suggest that L. mongolica shapes fungal assemblages through microenvironmental modification and nutrient competition, reinforcing niche differentiation within fairy rings (Figure 3).
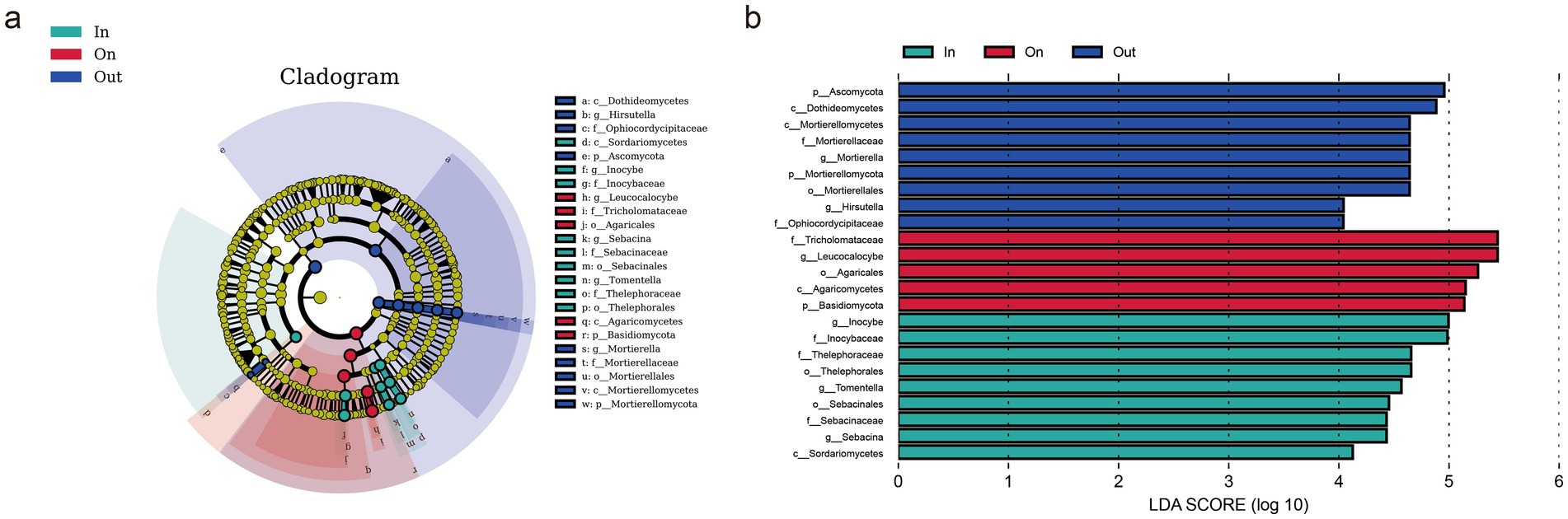
Figure 2. Comparison of differences in fairy ring soil fungi of L. mongolica sample plot (p: phylum; c: class; o: order; f: family; g: genus). (a) Cladogram is used to show taxonomic distribution of marker species in each group of samples; (b) Indicator microbial groups with LDA scores >4.0.
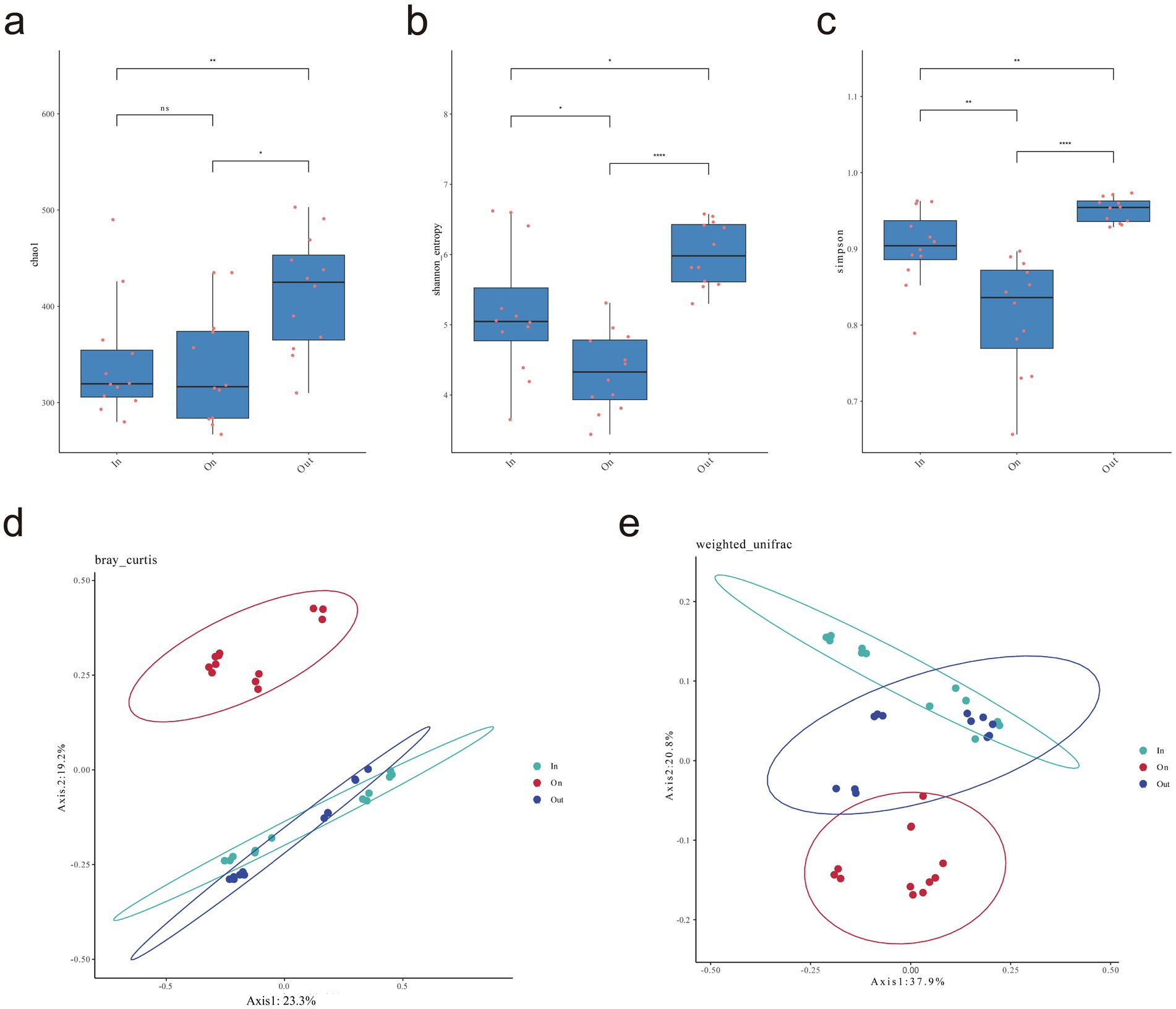
Figure 3. Analysis of soil fungal α and β diversity. (a) Chao1 index; (b) Shanon index; (c) Simpson index; (d) PCoA bray-curtis distance-based analysis (e) PCoA weighted UniFrac analysis. Using Wilcoxon rank sum test to analyze p-values. The “*” indicates a significant difference between the average values of two sets of samples, “*” represents p < 0.05, “**” p < 0.01, “***” p < 0.001. The statistical significance of beta diversity differences between groups was assessed using PERMANOVA.
3.2 Co-occurrence network of soil fungi
Based on co-occurrence network analysis, this study revealed distinct interaction patterns of soil fungal communities across different zones of L. mongolica fairy rings (Supplementary Table S2; Figure 4). The network in the IN zone comprised 196 nodes and 1,241 edges, exhibiting the highest connectivity density (average degree = 12.663) and clustering coefficient (0.533). This indicates a highly interconnected fungal community with relatively low modularity (0.268), suggesting a more complex network structure and a higher level of symbiotic relationships in this zone. In contrast, the ON zone network (201 nodes, 770 edges) exhibited distinct topological characteristics, with the lowest average degree (7.662) and clustering coefficient (0.433), but the highest modularity (0.416), indicating a more specialized and clearly partitioned interaction pattern dominated by L. mongolica. The OUT zone network (198 nodes, 945 edges) showed intermediate features, with an average degree (9.545) and modularity (0.378) falling between those of the IN and ON zones. In addition, the ASVs directly associated with L. mongolica included Thelebolus, Coprinellus, Penicillium, Claroideoglomus, and Chalara (|r| > 0.7, p < 0.05). Among these, only Thelebolus exhibited a positive interaction with L. mongolica, while the other four genera showed negative interactions. Notably, Coprinellus displayed strong positive associations with Penicillium, Claroideoglomus, and Chalara.
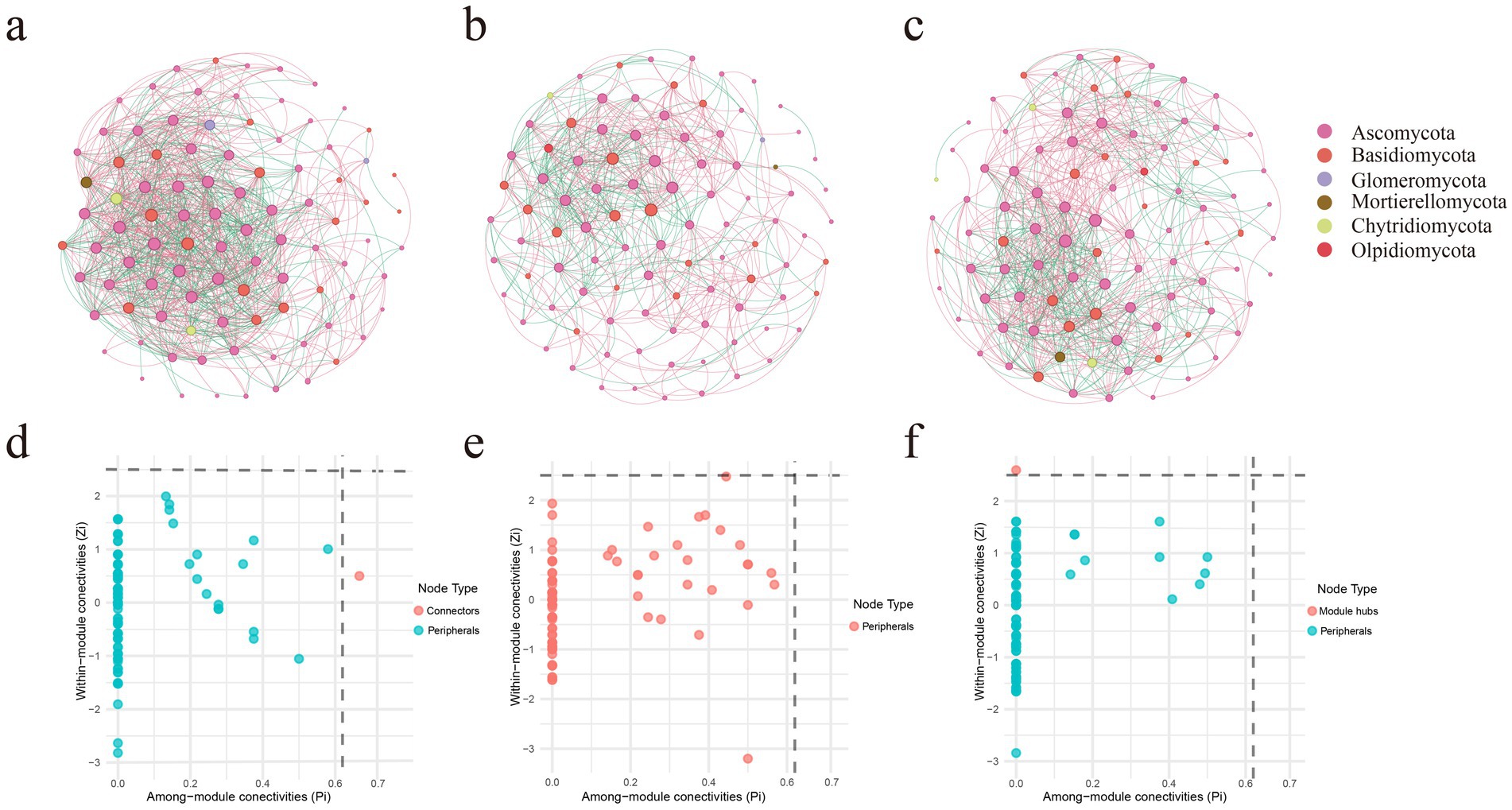
Figure 4. Co-occurrence network pattern of three different zones of L. mongolica fairy rings. (a–c) Represent the inside zone (IN), the fruiting zone (ON), and the outside zone (OUT), respectively. (d–f) Represent Zi-Pi graphs based on topological roles in networks of IN, ON, and OUT, respectively.
By calculating within-module connectivity (Zi) and among-module connectivity (Pi), we identified potential keystone species (Figures 4d–f). Ascomycota and Basidiomy-cota were the key fungal phyla across the three zones of the fairy ring. The ON zone exhibited a homogenized network structure, characterized by all ASVs being classified as peripheral nodes. In the IN zone, one ASV was identified as a connector, affiliated with the phylum Ascomycota and the genus Elasticomyces. In the OUT zone, one ASV was identified as a module hub, affiliated with the phylum Basidiomycota and the genus Hyphodontia. These keystone nodes in the IN and OUT zones may contribute to a higher level of ecological functional coordination.
3.3 Relationship between soil factors and fungal communities
The soil properties in different zones of L. mongolica fairy rings exhibit significant spatial heterogeneity (Supplementary Table S3). Notably, the ON zone displays unique physicochemical characteristics, with significantly lower soil pH compared to the In and OUT zones (p < 0.05), as well as elevated levels of ammonium and nitrate nitrogen. The OUT zone is characterized by optimal total nutrient levels, particularly with significantly higher total potassium and available potassium contents than the other zones (p < 0.05), while it has the lowest organic carbon content. In contrast, the IN zone exhibits the highest organic carbon content but relatively lower potassium levels. This heterogeneous nutrient distribution reflects the ecological functional differentiation among various zones of the fairy rings. Enzyme activity analysis revealed that the OUT zone exhibited significantly higher activities of soil cellulase, urease, FDA hydrolase, and chitinase compared to the ON zone (p < 0.05), where chitinase activity was the lowest among the three zones. These differences suggest that L. mongolica may selectively modulate soil enzyme activities by suppressing certain hydrolases while promoting cellulose degradation processes, thereby reshaping the soil functional profile within the ON zone. This finding provides important insight into the underlying mechanisms of fairy ring formation.
RDA further revealed functional differentiation among different zones of L. mongolica fairy rings in the Bayanbulak grassland. The first and second RDA axes explained 26.77 and 20.09% of the total variation, respectively (Figure 5), indicating that the model effectively captured the main environmental driving factors. The distinct separation of sample groups in the RDA ordination plot suggests significant differences in fungal community structure across the different zones of the L. mongolica fairy rings (permutation test, p < 0.01). Notably, the fungal community abundance in the fruiting zone was significantly positively correlated with ammonium nitrogen, nitrate nitrogen, and soil laccase activity, while showing a significant negative correlation with available potassium. This suggests potential nutrient competition between the fungal community and plants in nitrogen transformation and lignin degradation processes within this zone. In contrast, the fungal community in the out zone exhibited distinct enzymatic activity patterns, being positively correlated with urease and cellulase activities but negatively correlated with soil nitrogen content. This may reflect functional differentiation of microbial carbon and nitrogen metabolism in this zone. RDA of the top 10 dominant fungal genera further clarified that L. mongolica and Microdochium were strongly associated with soil ammonium nitrogen, nitrate nitrogen, and laccase activity, further highlighting the relationship between L. mongolica and ammonium-nitrate nitrogen and laccase activity.
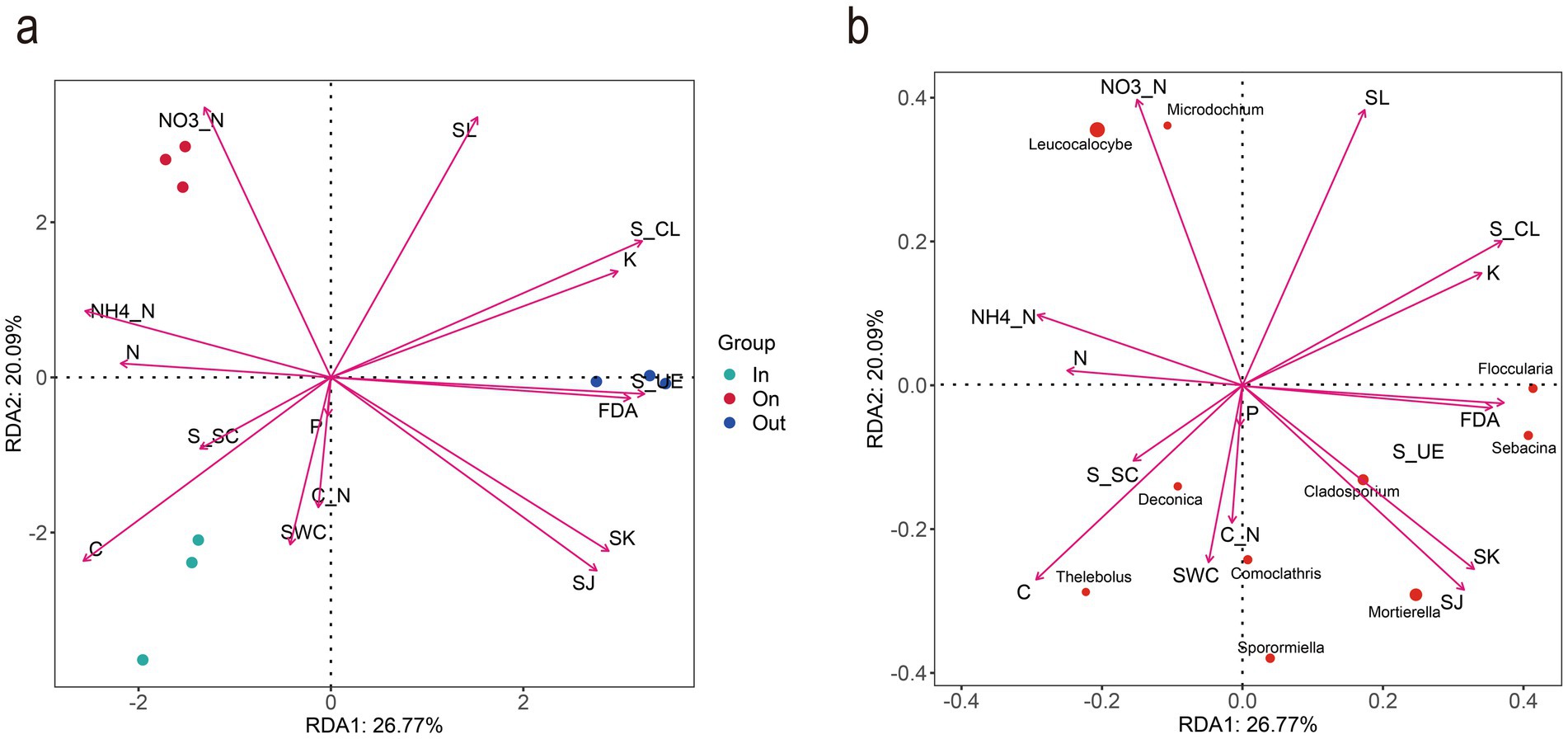
Figure 5. RDA ordination of fungal community-environment interactions. (a) Fungal genera distribution across zones (IN/ON/OUT) in relation to soil factors (RDA1: 26.77%; RDA2: 20.09%). (b) Top 10 genera in the ON zone and their correlations with soil properties.
3.4 Functional prediction of soil fungi
In this study, PICRUSt2 was employed to predict the functional profiles of MetaCyc metabolic pathways in soil fungal communities across three zones (Figures 6a,b). The results revealed that PWY-3781 (aromatic compound degradation pathway) and PWY-7279 (branched-chain amino acid biosynthesis pathway) were the common dominant metabolic pathways shared among the three zones, with their relative abundances consistently around 6% (Figure 6a). This indicates the conserved nature of these fundamental metabolic functions within soil fungal communities across different geographic locations. Multiple comparison analysis of the bar chart revealed that the ON zone exhibited significant site-specificity in certain metabolic pathways. Specifically, the abundances of PWY-5083 (polycyclic aromatic hydrocarbon degradation pathway) and PWY-6351 (chitin degradation pathway) were significantly higher than those in the other two zones (p < 0.05) (Figure 6b). In contrast, the abundances of PWY-5659 (carotenoid biosynthesis pathway) and PWY-66-422 (siderophore biosynthesis pathway) were significantly lower in the ON zone compared to other zones. These results suggest that fungi in the ON zone may employ distinct metabolic strategies to cope with oxidative stress and iron limitation.
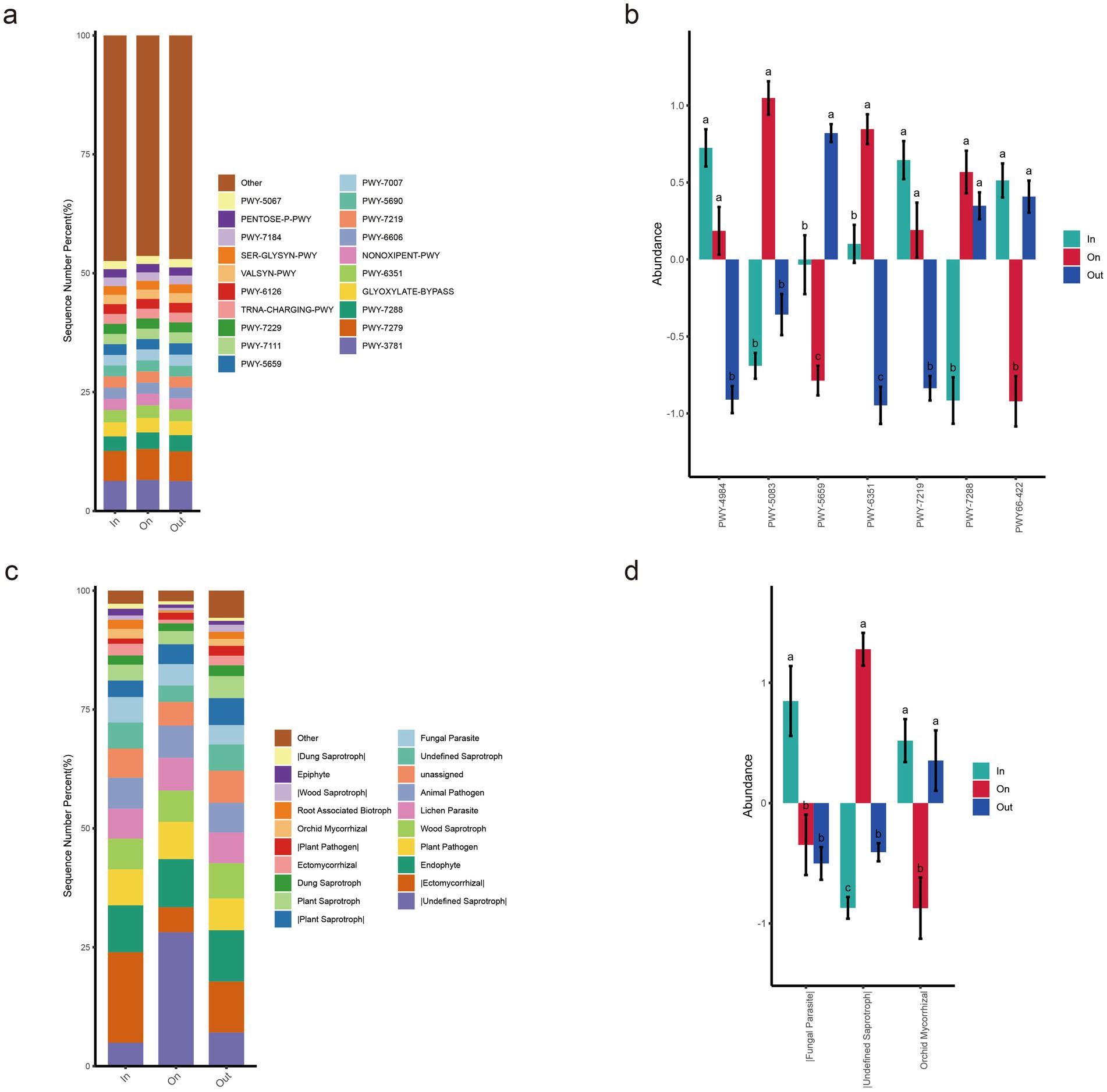
Figure 6. Functional prediction analysis of soil fungi based on PICRUSt2 (a,b) and FUNGuild (c,d). (a) Relative abundance distribution of fungal metabolic pathways predicted by PICRUSt2. (b) All significantly different MetaCyc pathways (p < 0.05) identified by ANOVA and Duncan’s test. (c) Relative abundance distribution of fungal metabolic pathways predicted by FUNGuild. (d) All significantly different functional guilds (p < 0.05) identified by ANOVA and Duncan’s test.
Functional prediction analysis based on FUNGuild (Figures 6c,d) revealed significant functional differentiation of soil fungal communities across different zones of the fairy ring. The ON zone exhibited distinct functional guild characteristics, with the relative abundance of Undefined Saprotrophs reaching 28.13%, which was significantly higher than those in the IN and OUT zones (p < 0.05) (Figure 6c). In contrast, the IN and OUT zones were dominated by the Ectomycorrhizal guild, with mean relative abundances of 18.9 and 10.7%, respectively. Notably, the abundances of four nutritional functional groups (Plant Pathogen, Wood Saprotroph, Lichen Parasite, and Animal Pathogen) remained consistently high across all zones. Specifically, the abundance of Orchid Mycorrhizal fungi in the ON zone was significantly lower than that in the IN and OUT zones (p < 0.05) (Figure 6d).
4 Discussion
4.1 Fungal community structure and ecological strategies in Leucocalocybe mongolica fairy rings
Distinct fungal community structures were observed among different zones of the fairy ring (Figure 1). The relative abundance of Basidiomycota in the ON zone reached 52.06%, which was significantly higher than that in the IN zone (36.66%) and the OUT zone (35.53%) (Supplementary Figure S3). This dominance was primarily driven by Leucocalocybe (family Tricholomataceae), whose LDA score exceeded 4.0, indicating that this species serves as a key biomarker for the ON zone. In addition, this study identified Cladosporium as another dominant fungal genus in the soil of L. mongolica fairy rings (Supplementary Figure S3b). Previous research has shown that secondary metabolites secreted by Cladosporium, including gibberellins, can promote plant growth (Răut et al., 2021). Therefore, we can speculate that Cladosporium plays a positive role in the formation of fairy rings and plant growth. The metabolites of Cladosporium interact beneficially with plants, while exhibiting antagonistic effects by inhibiting the germination and growth of several other fungal species (Moricca et al., 2005; Moricca et al., 2001).
F. luteovirens populations exhibit notable genetic variation and phylogenetic differentiation across altitudinal gradients (Xing et al., 2017). Moreover, microbial investigations revealed that its fairy rings harbor unique microbial communities and potential mycorrhiza-helper bacteria that facilitate plant growth (Xing et al., 2018). Recent metabolomic profiling further indicated geographical variation in secondary metabolites, which may contribute to the antioxidative capacity and ecological adaptation of F. luteovirens (Tang et al., 2024). In contrast, our study showed that the fruiting zone of L. mongolica fairy rings in the Bayanbulak grassland was characterized by sharply reduced fungal diversity, simplified microbial networks, and elevated laccase activity, highlighting a predominantly saprotrophic regulation strategy. This contrasts with the ectomycorrhizal symbiosis-based mechanisms reported for F. luteovirens. Such differences suggest that variation in ecological strategies between saprotrophic (L. mongolica) and ectomycorrhizal (F. luteovirens) fungi drives divergent pathways of nutrient competition, niche modification, and microbial interactions.
The Venn diagram analysis highlighted that although the three zones shared a substantial core microbiome (555 ASVs), each zone also harbored a large number of unique ASVs. This indicates that the fungal communities are strongly structured by spatial heterogeneity within the fairy ring ecosystem. The coexistence of a shared core microbiome and many zone-specific taxa suggests that while some fundamental ecological functions are maintained across zones (Duan and Bau, 2021a), distinct environmental conditions and fungal–plant interactions drive the differentiation of community assemblages in each zone. The high abundance of Leucocalocybe indicates its important role in maintaining the fundamental ecological functions of the fairy rings. Notably, the greater number of shared ASVs between the ON and OUT zones align with their spatial proximity observed in the PCoA analysis (Figures 2d,e), suggesting a certain degree of connection between the fungal communities in these two areas. Alpha diversity revealed the trend OUT > IN > ON (Figures 2a–c), consistent with previous reports (Wang et al., 2022; Duan et al., 2022a; Duan and Bau, 2021b; Kim et al., 2013; Ito et al., 2017), while β-diversity confirmed clear spatial separation among zones (Figures 2d,e), highlighting the strong influence of L. mongolica. Its vigorous mycelial growth preferentially exploits soil carbon and nitrogen sources, particularly by occupying the rhizosphere niche, effectively limiting the access of other fungi to nutrients and thereby suppressing their growth, resulting in a competitive exclusion effect (van Der Heijden et al., 2015; Liu et al., 2023; Liu et al., 2024). Furthermore, the mechanisms underlying these ecological dynamics primarily involve the secretion of inhibitory secondary metabolites (Keller, 2019; Bennett et al., 2017), and physical alteration of soil microhabitats (van Der Heijden et al., 2015). Together, these processes promote competitive exclusion and shape the unique fungal assemblage in the ON zone.
4.2 Topological features of microbial networks reveal the community restructuring mechanism of Leucocalocybe mongolica
The ON zone network showed higher modularity (0.416) but lower connectivity density (average degree 7.662) compared to the other zones, indicating reduced efficiency despite a compact core structure. These topological traits align with the low Shannon diversity (Figures 2a–c), β-diversity clustering (Figures 2d–f), and saprotroph dominance (Figure 6), suggesting that L. mongolica reshapes soil microecology by restructuring the fungal interaction network. In the co-occurrence network analysis, we further examined the ASVs directly associated with Leucocalocybe mongolica and their interaction patterns with other ASVs in the network. The results showed that Thelebolus exhibited a positive interaction with L. mongolica. Many species of Thelebolus are psychrophilic and display strong adaptability to cold and other extreme environments (Wicklow and Malloch, 1971; Mi et al., 2024). This characteristic suggests that Thelebolus may form a mutually beneficial relationship with L. mongolica within the fungal community, thereby enhancing their adaptive capacity and niche expansion potential in the soil environment. On the other hand, Coprinellus exhibited a negative interaction with L. mongolica, which may be attributed to the fact that both belong to Basidiomycota and are saprotrophic fungi (Hussain et al., 2018), thereby competing for similar soil nutrient resources. Claroideoglomus, as an arbuscular mycorrhizal fungus (Wu et al., 2024), may also compete with L. mongolica for root colonization sites or host-derived nutrients. Notably, Coprinellus showed strong positive associations with Penicillium, Claroideoglomus, and Chalara. This phenomenon may suggest that these taxa form functional cooperation or niche complementarity to cope with the environmental pressures imposed by L. mongolica. Such findings provide new insights into the assembly mechanisms of fungal communities under the influence of L. mongolica.
Combined with the network topology characteristics, it can be further inferred that L. mongolica may reshape the local microbial interaction network through three pathways. First, its intense resource competition (Morriën et al., 2017) likely eliminates numerous sensitive species, resulting in a reduction in network connectivity. Secondly, secondary metabolites secreted by fungi in the fairy ring may selectively promote the growth of specific tolerant species, forming more specialized interaction modules (Bennett et al., 2017). Third, L. mongolica plays a central role in fungal community assembly by establishing synergistic relationships with tolerant taxa (such as Thelebolus) to maintain community stability, while simultaneously leveraging competitive exclusion mechanisms to suppress the expansion of competing groups. Notably, no keystone taxa were detected in the ON-zone network, with all ASVs functioning as peripheral nodes. This phenomenon supports the “top-down” control hypothesis of L. mongolica as an absolute dominant species on community structure (Morriën et al., 2017), whereby it simplifies the network by suppressing interactions among other species and forming a self-centered network.
4.3 Nitrogen enrichment and laccase activity synergy in Leucocalocybe mongolica fairy rings
The fairy ring soil of L. mongolica exhibited elevated levels of ammonium and nitrate, consistent with previous studies (Duan et al., 2022a; Liu et al., 2024). Nitrogen is an essential nutrient for large fungi during mycelial growth and fruiting body development, as it is required for the biosynthesis of key cellular components such as proteins and nucleic acids (Kuypers et al., 2018; Carrasco and Preston, 2020). The lower pH observed in the fairy ring soil promotes the mineralization of organic nitrogen into NH₄⁺, while the increased NH₄⁺ concentration stimulates the proliferation of nitrifying microorganisms, thereby enhancing nitrogen utilization (Liu et al., 2024). In addition, unlike previous studies (Duan et al., 2022a), the cellulase activity in the soil of the fairy ring was significantly lower than that in the outer area, whereas laccase activity was significantly higher than in the other two areas. Laccase can catalyze the biodegradation of lignin and humic substances, thereby promoting the cycling of soil organic matter (Schuetz et al., 2014; Zavarzina et al., 2018). Aromatic compounds produced from lignin degradation may be utilized by certain nitrifying bacteria, thereby enhancing the nitrification process (Zheng et al., 2024). This could partly explain the higher nitrate nitrogen levels observed in the fairy ring zone. RDA indicated that the fungal community in the ring zone was significantly positively correlated with ammonium nitrogen, nitrate nitrogen, and laccase activity (Figure 5), further clarifying the relationship between environmental factors and fungal community composition. It should be noted that the physicochemical properties and enzyme activities in this study were measured in one representative fairy ring, whereas the fungal community sequencing was conducted across four independent rings. This design was adopted primarily due to resource constraints. Nevertheless, the observed patterns of nutrient availability and enzyme activities are consistent with previous reports on L. mongolica and other fairy ring fungi, and the inclusion of three biological replicates per zone ensures statistical reliability. We acknowledge that extending physicochemical and enzymatic analyses to multiple fairy rings would further enhance the generality of the results, and this represents an important direction for future research.
These fungus-driven nutrient changes have important ecological implications for grassland management and restoration. L. mongolica can enhance soil available nitrogen and serve as a natural biofertilizer for degraded grasslands; inoculation with such functional fungi can accelerate nutrient cycling, improve soil fertility, and promote vegetation recovery. Its elevated laccase activity enhances the decomposition of recalcitrant organic matter, facilitating organic matter turnover and nutrient release. This suggests that cultivating fungal communities with high lignin-degrading capacity could be an effective strategy to accelerate ecosystem restoration. Therefore, fairy ring–forming fungi not only provide ecological functions but also represent valuable biotic agents for sustainable grassland management, with microbial functions being key to restoring ecosystem productivity and stability.
4.4 Functional divergence and ecological adaptation of soil fungal communities in Leucocalocybe mongolica fairy rings
This study systematically elucidates the functional differentiation characteristics and ecological adaptation strategies of soil fungal communities in different zones of the L. mongolica fairy rings through MetaCyc metabolic pathway analysis and FUN-Guild functional prediction. The metabolic pathway analysis revealed that fundamental metabolic functions, such as PWY-3781 (aromatic compound degradation) and PWY-7279 (branched-chain amino acid biosynthesis), remained relatively stable across the three zones (6%), reflecting the conservatism of core metabolic functions in soil fungi. Notably, the enrichment of polycyclic aromatic hydrocarbon degradation (PWY-5083) and chitin degradation (PWY-6351) pathways observed in the ON zone (p < 0.05) corresponds with the high soil laccase activity detected in this area. Furthermore, a decrease in the siderophore biosynthesis pathway (PWY66-422) was found in the ON zone. Siderophores, produced by most fungal species, are iron-chelating agents involved in high-affinity iron uptake and intracellular processing (Happacher et al., 2023). The reduced siderophore biosynthesis pathway in this zone corroborates its lowest fungal diversity. Fungi in the ON zone may cope with oxidative stress and iron limitation through alter-native mechanisms, such as hyphal network transport, reflecting adaptive evolution to this unique microenvironment.
FUNGuild functional prediction further confirmed this functional differentiation. The high abundance of undefined saprotrophs (Figure 6c) in the ON zone indicates their significant role in organic matter decomposition and nutrient cycling (Liers et al., 2010; Allmér et al., 2009). This result is consistent with the enhanced organic matter degradation metabolic traits observed in the fairy ring zone, suggesting that L. mongolica may optimize carbon source utilization efficiency by reshaping saprotrophic functional groups. In contrast, the predominance of ectomycorrhizal fungi in the In and Out zones (Figure 6c) reflect a plant-fungal interaction network closer to the natural state in these areas (Tedersoo et al., 2020). Notably, the significantly reduced abundance of orchid mycorrhizal fungi in the ON zone (Figure 6d) may originate from allelopathic inhibition by L. mongolica, a phenomenon consistent with the allelopathic niche competition mechanism among fungi (Bennett et al., 2017). Such a synergistic “enzymatic degradation–allelopathy” interaction offers novel insights into the ecological competitive advantages of large fungi. It should be noted that functional annotations obtained from these two prediction approaches may involve certain biases, particularly given the incomplete coverage of fungal functional databases. Future studies should integrate metagenomics, transcriptomics, or enzymatic activity assays to more accurately elucidate the actual functions of fungal communities.
5 Conclusion
This study systematically elucidated the structure, diversity, and ecological functions of soil fungal communities across different zones of L. mongolica fairy rings in the Bayanbulak Grassland, Xinjiang. Our findings suggest that L. mongolica may establish a metabolic advantage in the fruiting zone by enhancing laccase activity and regulating nitrogen availability, which in turn could reshape microbial networks and suppress competing functional groups. Notably, the observed association between elevated laccase activity and nitrogen cycling in fairy ring soils provides preliminary evidence of a potential mechanism through which macro fungi influence belowground functional zonation.
These findings not only deepen ecological understanding but also hold direct practical significance for management. Protecting fungal diversity and the microenvironments of fairy rings—through measures such as limiting overgrazing and minimizing human disturbance—may help sustain ecosystem functions. Moreover, the functional roles of L. mongolica and its symbiotic fungi in nutrient cycling and plant stress tolerance suggest strong potential for ecological restoration of degraded grasslands, for instance by promoting natural expansion or assisted establishment of fairy rings. Considering the unique climatic and edaphic conditions of Xinjiang, region-specific monitoring and adaptive management will be essential to ensure effective conservation outcomes. Future work integrating metagenomics and metabolomics will be essential to unravel the molecular basis of these interactions and to translate them into practical restoration measures. Such efforts should also consider the ecological balance between L. mongolica and the associated microbial communities, to achieve synergistic protection of symbiotic microbes and plant functional groups (Nilsson et al., 2019).
Data availability statement
The datasets presented in this study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/, BioProject accession number PRJNA1335383, and SRA accession range SRR35646027 – SRR35646062.
Author contributions
SW: Writing – review & editing, Writing – original draft, Formal analysis, Methodology, Visualization, Conceptualization, Investigation. XW: Visualization, Writing – review & editing, Conceptualization, Investigation. JX: Formal analysis, Visualization, Investigation, Writing – review & editing. RN: Visualization, Writing – review & editing, Investigation. YW: Investigation, Writing – review & editing. JW: Writing – review & editing. FL: Funding acquisition, Writing – review & editing. QB: Supervision, Funding acquisition, Conceptualization, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32160335 and 32460793), the Opening/Autonomous Fund of Key Laboratory of Herbage and Endemic Crop Biology, Ministry of Education, Inner Mongolia University, Inner Mongolia Engineering Technology Research Center of Germplasm Resources Conservation and Utilization, Hohhot 010010, China, Project No. 21400-24151400, Inner Mongolia Autonomous Region Science and Technology Program (2025YFDZ0102), and the National Natural Science Foundation of Inner Mongolia (2025MS03144).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that Gen AI was used in the creation of this manuscript. During the preparation of this study, the authors utilized ChatGPT-4.0 to assist with English language polishing. The authors have reviewed and edited the output and take full responsibility for the content of this publication.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1667514/full#supplementary-material
References
Allmér, J., Stenlid, J., and Dahlberg, A. (2009). Logging-residue extraction does not reduce the diversity of litter-layer saprotrophic fungi in three Swedish coniferous stands after 25 years. Can. J. For. Res. 39, 1737–1748. doi: 10.1139/X09-096
Balvočiūtė, M., and Huson, D. H. (2017). SILVA, RDP, Greengenes, NCBI and OTT—how do these taxonomies compare? BMC Genomics 18, 1–8. doi: 10.1186/s12864-017-3501-4
Bastian, M., Heymann, S., and Jacomy, M. (2009). Gephi: an open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Web Soc. Media 3, 361–362. doi: 10.1609/icwsm.v3i1.13937
Bennett, J. A., Maherali, H., Reinhart, K. O., Lekberg, Y., Hart, M. M., and Klironomos, J. (2017). Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science 355, 181–184. doi: 10.1126/science.aai8212
Bonanomi, G., Mingo, A., Incerti, G., Mazzoleni, S., and Allegrezza, M. (2012). Fairy rings caused by a killer fungus foster plant diversity in species-rich grassland. J. Veg. Sci. 23, 236–248. doi: 10.1111/j.1654-1103.2011.01353.x
Bonfante, P., and Genre, A. (2010). Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis. Nat. Commun. 1:48. doi: 10.1038/ncomms1046
Caesar-Tonthat, T. C., Espeland, E., Caesar, A. J., Sainju, U. M., Lartey, R. T., and Gaskin, J. F. (2013). Effects of agaricus lilaceps fairy rings on soil aggregation and microbial community structure in relation to growth stimulation of western wheatgrass (Pascopyrum smithii) in eastern Montana rangeland. Microb. Ecol. 66, 120–131. doi: 10.1007/s00248-013-0194-3
Cairney, J. W. (2005). Basidiomycete mycelia in forest soils: dimensions, dynamics and roles in nutrient distribution. Mycol. Res. 109, 7–20. doi: 10.1017/S0953756204001753
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carrasco, J., and Preston, G. M. (2020). Growing edible mushrooms: a conversation between bacteria and fungi. Environ. Microbiol. 22, 858–872. doi: 10.1111/1462-2920.14765
Csardi, M. G. Package “igraph”. (2013). Available online at: https://igraph.org (Accessed May, 2025).
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Wicklow, D., and Malloch, D. (1971). Studies in the genus Thelebolus: temperature optima for growth and ascocarp development. Mycologia 63, 118–131. doi: 10.1080/00275514.1971.12019090
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Duan, M., Bao, H., and Bau, T. (2021). Analyses of transcriptomes and the first complete genome of L. mongolica provide new insights into phylogenetic relationships and conservation. Sci. Rep. 11:2930. doi: 10.1038/s41598-021-81784-6
Duan, M., and Bau, T. (2021a). Grassland fairy rings of L. mongolica represent the center of a rich soil microbial community. Braz. J. Microbiol. 52, 1357–1369. doi: 10.1007/s42770-021-00478-3
Duan, M., and Bau, T. (2021b). Initial sample processing can influence the soil microbial metabarcoding surveys, revealed by L. mongolica fairy ring ecosystem. Biotechnol. Biotechnol. Equip. 35, 1427–1438. doi: 10.1080/13102818.2021.1996272
Duan, M., Lu, M., Lu, J., Yang, W., Li, B., Ma, L., et al. (2022a). Soil chemical properties, metabolome, and metabarcoding give the new insights into the soil transforming process of fairy ring fungi L. mongolica. J. Fungi 8:680. doi: 10.3390/jof8070680
Duan, M., Lu, J., Yang, W., Lu, M., Wang, J., Li, S., et al. (2022b). Metabarcoding and metabolome analyses reveal mechanisms of Leymus chinensis growth promotion by fairy ring of Leucocalocybe mongolica. J. Fungi 8:944. doi: 10.3390/jof8090944
Fidanza, M. (2007). Characterization of soil properties associated with type-I fairy ring symptoms in turfgrass. Biologia 62, 533–536. doi: 10.2478/s11756-007-0105-5
Fidanza, M., Settle, D., and Wetzel, H. (2016). An index for evaluating fairy ring symptoms in turfgrass. HortScience 51, 1194–1196. doi: 10.21273/HORTSCI10915-16
Gao, X. S., Xiao, Y., Deng, L. J., Li, Q. Q., Wang, C. Q., Li, B., et al. (2019). Spatial variability of soil total nitrogen, phosphorus and potassium in Renshou County of Sichuan Basin, China. J. Integr. Agric. 18, 279–289. doi: 10.1016/S2095-3119(18)62069-6
Guo, Y., Chen, J., Tsolmon, B., He, A., Guo, J., Yang, J., et al. (2020). Effects of subsidence and transplanted trees on soil arbuscular mycorrhizal fungal diversity in a coal mining area of the loess plateau. Glob. Ecol. Conserv. 24:e01308. doi: 10.1016/j.gecco.2020.e01308
Han, Z., Xu, P., Li, Z., Lin, H., Zhu, C., Wang, J., et al. (2022). Microbial diversity and the abundance of keystone species drive the response of soil multifunctionality to organic substitution and biochar amendment in a tea plantation. GCB Bioenergy 14, 481–495. doi: 10.1111/gcbb.12926
Happacher, I., Aguiar, M., Yap, A., Decristoforo, C., and Haas, H. (2023). Fungal siderophore metabolism with a focus on Aspergillus fumigatus: impact on biotic interactions and potential translational applications. Essays Biochem. 67, 829–842. doi: 10.1042/EBC20220252
Hussain, S., Usman, M., Ahmad, H., Khan, J., and Khalid, A. N. (2018). The genus Coprinellus (Basidiomycota; Agaricales) in Pakistan with the description of four new species. MycoKeys 39, 41–61. doi: 10.3897/mycokeys.39.26743
Ito, A., Choi, J. H., Wu, J., Tanaka, H., Hirai, H., and Kawagishi, H. (2017). Plant growth inhibitors from the culture broth of fairy ring-forming fungus Lepista sordida. Mycoscience 58, 387–390. doi: 10.1016/j.myc.2017.05.001
Keller, N. P. (2019). Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 17, 167–180. doi: 10.1038/s41579-018-0121-1
Kim, M., Yoon, H., You, Y. H., Kim, Y. E., Woo, J. R., and Seo, Y. (2013). Metagenomic analysis of fungal communities inhabiting the fairy ring zone of Tricholoma matsutake. J. Microbiol. Biotechnol. 23, 1347–1356. doi: 10.4014/jmb.1306.06068
Kuypers, M. M., Marchant, H. K., and Kartal, B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276. doi: 10.1038/nrmicro.2018.9
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Li, Z., Liu, X., Zhang, M., and Xing, F. (2022). Plant diversity and fungal richness regulate the changes in soil multifunctionality in a semi-arid grassland. Biology 11:870. doi: 10.3390/biology11060870
Liers, C., Bobeth, C., Pecyna, M., Ullrich, R., and Hofrichter, M. (2010). DyP-like peroxidases of the jelly fungus Auricularia auricula-judae oxidize nonphenolic lignin model compounds and high-redox potential dyes. Appl. Microbiol. Biotechnol. 85, 1869–1879. doi: 10.1007/s00253-009-2173-7
Lindahl, B. D., and Tunlid, A. (2015). Ectomycorrhizal fungi–potential organic matter decomposers, yet not saprotrophs. New Phytol. 205, 1443–1447. doi: 10.1111/nph.13201
Liu, M., Wei, Y., Lian, L., Wei, B., Bi, Y., Liu, N., et al. (2023). Macrofungi promote SOC decomposition and weaken sequestration by modulating soil microbial function in temperate steppe. Sci. Total Environ. 899:165556. doi: 10.1016/j.scitotenv.2023.165556
Liu, M., Wei, Y., Lian, L., Zhang, J., Liu, N., Wilson, G. W., et al. (2024). Discovering the role of fairy ring fungi in accelerating nitrogen cycling to promote plant productivity in grasslands. Soil Biol. Biochem. 199:109595. doi: 10.1016/j.soilbio.2024.109595
Lu, T., Bao, H., and Bau, T. (2018). Genetic diversity and population structure of endemic mushroom Leucocalocybe mongolica in Mongolian plateau uncovered by EST-SSR markers. Biotechnol. Biotechnol. Equip. 32, 1195–1204. doi: 10.1080/13102818.2018.1510743
Lu, T., Bau, T., and Ohga, S. (2017). Physiological study of the wild edible mushroom L. mongolica. J. Fac. Agric. Kyushu Univ. 62, 1–8. doi: 10.5109/1798134
Lü, Z. L., Liu, B., Chang, F., Ma, Z. J., and Cao, Q. M. (2023). Relationship between plant functional diversity and ecosystem multifunctionality in Bayanbulak alpine meadow along an altitude gradient. Chin. J. Plant Ecol. 47, 822–832. doi: 10.17521/cjpe.2022.0195
Ma, L., Zhang, C., Xu, X., Wang, C., Liu, G., Liang, C., et al. (2022). Different facets of bacterial and fungal communities drive soil multifunctionality in grasslands spanning a 3500 km transect. Funct. Ecol. 36, 3120–3133. doi: 10.1111/1365-2435.14220
Mandal, S., Van Treuren, W., White, R. A., Eggesbø, M., Knight, R., and Peddada, S. D. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26:27663. doi: 10.3402/mehd.v26.27663
Mi, Z., Su, J., Yu, L., and Zhang, T. (2024). Comparative mitochondrial genomics of Thelebolaceae in Antarctica: insights into their extremophilic adaptations and evolutionary dynamics. IMA Fungus 15:33. doi: 10.1186/s43008-024-00164-7
Miao, T., Gao, S., Jiang, S., Kan, G., Liu, P., Wu, X., et al. (2014). A method suitable for DNA extraction from humus-rich soil. Biotechnol. Lett. 36, 2223–2228. doi: 10.1007/s10529-014-1591-5
Moricca, S., Ragazzi, A., and Assante, G. (2005). “Biological control of rust fungi by Cladosporium tenuissimum” in Rust diseases of willow and poplar. ed. A. R. Mccracken (Wallingford UK: CABI Publishing), 213–229.
Moricca, S., Ragazzi, A., Mitchelson, K. R., and Assante, G. (2001). Antagonism of the two-needle pine stem rust Fungi Cronartium flaccidum and Peridermium pini by Cladosporium tenuissimum in vitro and in planta. Phytopathology 91, 457–468. doi: 10.1094/PHYTO.2001.91.5.457
Morriën, E., Hannula, S. E., Snoek, L. B., Helmsing, N. R., Zweers, H., De Hollander, M., et al. (2017). Soil networks become more connected and take up more carbon as nature restoration progresses. Nat. Commun. 8:14349. doi: 10.1038/ncomms14349
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). Funguild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Nilsson, R. H., Anslan, S., Bahram, M., Wurzbacher, C., Baldrian, P., and Tedersoo, L. (2019). Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 17, 95–109. doi: 10.1038/s41579-018-0116-y
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’hara, R. B., et al. Package “vegan”. Community ecology package, version. (2013) 2, 1–295. Available online at: https://github.com/vegandevs/vegan/issues (Accessed March, 2025).
Răut, I., Călin, M., Capră, L., Gurban, A. M., Doni, M., Radu, N., et al. (2021). Cladosporium sp. isolate as fungal plant growth promoting agent. Agronomy 11:392. doi: 10.3390/agronomy11020392
Revelle, W. Psych: procedures for psychological, psychometric, and personality research. Northwestern University, Evanston, Illinois. R package version 2.3.6. (2023). Available online at: https://CRAN.R-project.org/package=psych (Accessed March, 2025).
Schuetz, M., Benske, A., Smith, R. A., Watanabe, Y., Tobimatsu, Y., Ralph, J., et al. (2014). Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 166, 798–807. doi: 10.1104/pp.114.245597
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60–R18. doi: 10.1186/gb-2011-12-6-r60
Shantz, H. L., and Piemeisel, R. L. (1917). Fungus fairy rings in eastern Colorado and their effect on vegetation. J. Agric. Res. 11, 191–246.
Tang, C., Fan, Y., Wang, T., Wang, J., Xiao, M., He, M., et al. (2024). Metabolomic profiling of Floccularia luteovirens from different geographical regions proposes a novel perspective on their antioxidative activities. Antioxidants 13:620. doi: 10.3390/antiox13050620
Tedersoo, L., Bahram, M., and Zobel, M. (2020). How mycorrhizal associations drive plant population and community biology. Science 367:eaba1223. doi: 10.1126/science.aba1223
Van Der Heijden, M. G., Klironomos, J. N., Ursic, M., Moutoglis, P., Streitwolf-Engel, R., Boller, T., et al. (1998). Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396, 69–72. doi: 10.1038/23932
van Der Heijden, M. G., Martin, F. M., Selosse, M. A., and Sanders, I. R. (2015). Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol. 205, 1406–1423. doi: 10.1111/nph.13288
Vázquez-Baeza, Y., Pirrung, M., Gonzalez, A., and Knight, R. (2013). EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2:16. doi: 10.1186/2047-217X-2-16
Wang, X., Bao, H., and Bau, T. (2019). Nutritional value and volatiles of the edible mushroom L. mongolica. Qual. Assur. Saf. Crops Foods 11, 679–685. doi: 10.3920/QAS2019.1585
Wang, Q., Wang, C., Wei, Y., Yao, W., Lei, Y., and Sun, Y. (2022). Soil microbes drive the flourishing growth of plants from L. mongolica fairy ring. Front. Microbiol. 13:893370. doi: 10.3389/fmicb.2022.893370
Wickham, H. reshape2: flexibly reshape data: a reboot of the reshape package. R package version 1.4.4. (2020). Available online at: https://CRAN.R-project.org/package=reshape2 (Accessed March, 2025).
Wu, X. H., Ma, C. Y., Jiang, H. J., Zhang, X. Y., Wang, H. M., Li, H. R., et al. (2024). Root endophyte-manipulated alteration in rhizodeposits stimulates Claroideoglomus in the rhizosphere to enhance drought resistance in peanut. J. Agric. Food Chem. 72, 20211–20223. doi: 10.1021/acs.jafc.4c05009
Xing, R., Gao, Q. B., Zhang, F. Q., Fu, P. C., Wang, J. L., Yan, H. Y., et al. (2017). Genetic variation and phylogenetic relationships of the ectomycorrhizal Floccularia luteovirens on the Qinghai-Tibet plateau. J. Microbiol. 55, 600–606. doi: 10.1007/s12275-017-7101-4
Xing, R., Yan, H. Y., Gao, Q. B., Zhang, F. Q., Wang, J. L., and Chen, S. L. (2018). Microbial communities inhabiting the fairy ring of Floccularia luteovirens and isolation of potential mycorrhiza helper bacteria. J. Basic Microbiol. 58, 554–563. doi: 10.1002/jobm.201700579
You, Q., Yin, X., Zhang, S., and Jiang, Z. (2014). Extraction, purification, and antioxidant activities of polysaccharides from Tricholoma mongolicum Imai. Carbohydr. Polym. 99, 1–10. doi: 10.1016/j.carbpol.2013.07.088
Zaki, A. H., Haiying, B., Mohany, M., Al-Rejaie, S. S., and Abugammie, B. (2024). The effect mechanism of ergosterol from the nutritional mushroom L. mongolica in breast cancer cells: protein expression modulation and metabolomic profiling using UHPLC-ESI-Q. Saudi Pharm. J. 32:102045. doi: 10.1016/j.jsps.2024.102045
Zaki, A. H., Zahid, M. T., and Haiying, B. (2022). Bioactive compounds of the culinary-medicinal mushroom L. mongolica (Agaricomycetes): pharmacological and therapeutic applications−a review. Int. J. Med. Mushrooms 24, 19–33. doi: 10.1615/IntJMedMushrooms.2022043854
Zavarzina, A. G., Semenova, T. A., Belova, O. V., Lisov, A. V., Leontievskii, A. A., and Ivanova, A. E. (2018). Laccase production and humic acids decomposition by microscopic soil fungi. Microbiology 87, 308–316. doi: 10.1134/S0026261718030153
Zhao, X., Bau, T., and Bao, H. (2020). Anti-tumor activity of polysaccharides obtained from L. mongolica using solid-state fermentation. Biotechnol. Biotechnol. Equip. 34, 841–849. doi: 10.1080/13102818.2020.1807406
Keywords: Leucocalocybe mongolica, fairy ring, soil fungal community, Bayanbulak grassland, microbial interaction network
Citation: Wang S, Wang X, Xu J, Na R, Wang Y, Wei J, Li F and Bao Q (2025) Soil fungal community structure and ecological functions in fairy rings of Leucocalocybe mongolica in Bayanbulak grassland. Front. Microbiol. 16:1667514. doi: 10.3389/fmicb.2025.1667514
Edited by:
Mukesh Kumar Malviya, Sage University, IndiaReviewed by:
Manoj Kumar Solanki, University of Silesia in Katowice, PolandRui Xing, Chinese Academy of Sciences (CAS), China
Copyright © 2025 Wang, Wang, Xu, Na, Wang, Wei, Li and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiuli Bao, YnFsMTk3ODc1QDEyNi5jb20=
 Senyuan Wang
Senyuan Wang Xingzhe Wang1,2
Xingzhe Wang1,2 Jiajia Xu
Jiajia Xu Jie Wei
Jie Wei