- 1Department of Laboratory Medicine, Nanjing Drum Tower Hospital Clinical College of Nanjing University of Chinese Medicine, Nanjing, Jiangsu, China
- 2Nanjing Municipal Centre for Disease Control and Prevention, Nanjing Municipal Key Laboratory of Public Health Testing, Nanjing, Jiangsu, China
- 3Jiangsu Key Laboratory of Laboratory Medicine, Department of Immunology, School of Medicine, School of Chemistry and Chemical Engineering, Jiangsu University, Zhenjiang, China
Background: The rise of carbapenem-resistant gram-negative bacteria (CRGNB) necessitates new therapeutic options such as cefiderocol.
Objective: To evaluate the in vitro efficacy of cefiderocol against clinical CRGNB and investigate associated resistance mechanisms.
Methods: A total of 370 CRGNB isolates were analyzed. Minimum inhibitory concentration (MIC) values were determined, and whole genome sequencing, efflux pump inhibition assays, and RT-qPCR were conducted to assess resistance-related mutations, gene loss, and expression changes.
Results: Cefiderocol demonstrated potent in vitro activity, with high susceptibility rates in C. freundii (100%), K. pneumoniae (93.3%), and E. hormaechei (92.2%), and notable activity against P. aeruginosa (80.0%) and Escherichia coli (76.8%). Efflux pump inhibition by Carbonyl Cyanide m-Chlorophenyl Hydrazone (CCCP) significantly reduced MICs in resistant strains. Key resistance mechanisms included β-lactamase gene variants (blaOXA-66, blaOXA-23, blaSHV-12), mutations in envZ, cirA, nuoC, ampC, and loss or altered expression of iron transporter genes (piuA, pirA, fepA).
Conclusion: Cefiderocol is highly effective against CRGNB; however, resistance may arise through diverse mechanisms, including efflux pump activity. Continued surveillance of emerging resistance is essential to guide its optimal clinical use.
1 Introduction
Carbapenem-resistant Gram-negative bacteria (CRGNB) have emerged as a significant global public health threat, posing substantial challenges in healthcare settings (Daoud and Dropa, 2023). These pathogens, including carbapenem-resistant Enterobacterales (CRE), carbapenem-resistant Pseudomonas aeruginosa (CRPA), and carbapenem-resistant Acinetobacter baumannii (CRAB), are associated with high morbidity and mortality due to limited therapeutic options and their ability to rapidly acquire and disseminate resistance genes, such as those encoding β-lactamases (Bassetti et al., 2021).
The global prevalence of CRGNB continues to rise, with notable regional variations (Jean et al., 2022). In Europe and North America, the incidence of CRE and CRAB has significantly increased, particularly in healthcare-associated infections (Müller et al., 2023; Duffy et al., 2023). In Asia, especially in countries like China and India, the spread of CRGNB has been exacerbated by high antibiotic consumption and inadequate infection control measures (Yin et al., 2023; Vijay et al., 2021). The Middle East and Latin America are also experiencing a growing burden of CRGNB infections, often linked to nosocomial outbreaks (Sha et al., 2017; Garcia-Betancur et al., 2021). These bacteria challenge healthcare systems worldwide, leading to prolonged hospital stays, increased healthcare costs, and limited therapeutic options (Li et al., 2024).
In response to the rising antimicrobial resistance, the development and evaluation of novel therapeutic agents have become urgent priorities. Cefiderocol, a novel siderophore cephalosporin antibiotic, employs a unique mechanism of action (as illustrated in Figure 1). By exploiting the bacterial iron transport system, cefiderocol can penetrate the outer membrane of Gram-negative bacteria (Ong'uti et al., 2022). This “Trojan horse” strategy allows cefiderocol to bypass common resistance mechanisms, such as alterations in porin channel and efflux pumps, which often limit the efficacy of other β-lactam antibiotics (Karakonstantis et al., 2022). Cefiderocol has been reported to be particularly effective against CRGNB, demonstrating stability against a wide array of β-lactamases, including metallo-β-lactamases and serine carbapenemases, thus enhancing its potential in treating severe infections caused by resistant pathogens (Wang et al., 2022; Sollima et al., 2024).
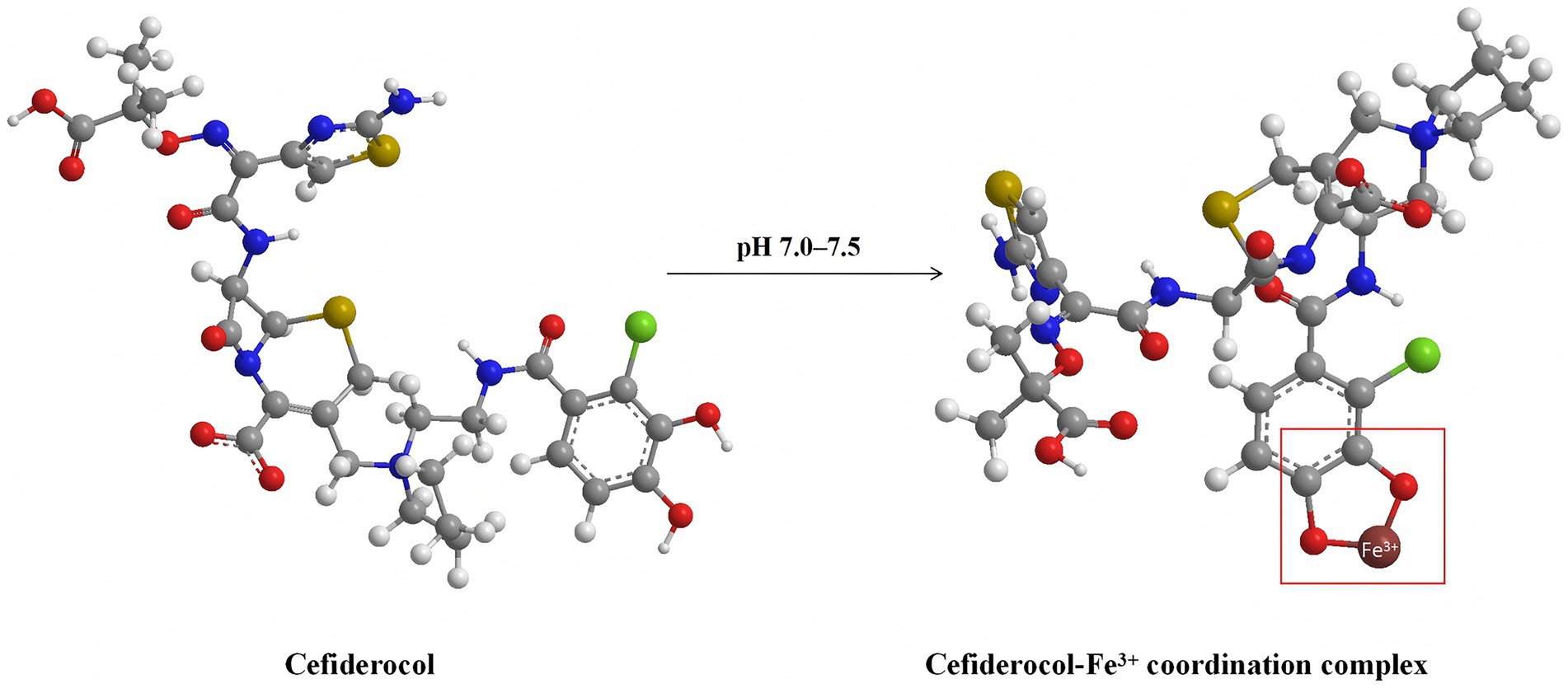
Figure 1. Molecular structures of cefiderocol (left) and its ferric iron (Fe3+) coordination complex (right) at physiological pH (7.0–7.5). This schematic depicts the chelation of cefiderocol with ferric iron, a critical step enabling its active uptake through bacterial iron transport systems. Atom colors are as follows: carbon (gray), oxygen (red), nitrogen (blue), sulfur (yellow), and iron (brown). The arrow indicates the pH-dependent formation of the cefiderocol–Fe3+ complex. The molecular model is the 3D structure described by MolView (https://molview.org).
Up to date, resistance mechanisms to cefiderocol include enzyme-mediated hydrolysis (e.g., metallo-β-lactamases and certain serine β-lactamases), mutations in porins leading to reduced outer membrane permeability, efflux pump overexpression, and modifications of target sites. Additionally, novel resistance determinants and adaptive mutations under antibiotic pressure have been reported. Notably, the amplification of specific β-lactamase genes, such as blaSHV-12, correlates with elevated resistance in Klebsiella pneumoniae (Moon and Huang, 2023). Altered membrane permeability exacerbates resistance through various mechanisms: mutations in iron transport-associated loci (envZ, tonB, cirA) reduce drug influx, while structural changes in the piuC gene (which encodes an outer membrane siderophore receptor) and regulatory mutations upstream of its operon impair cefiderophore uptake (Kriz et al., 2024). Porin mutations can increase cefiderocol Minimum inhibitory concentration (MIC) in isogenic K. pneumoniae (OmpK35, OmpK36) and P. aeruginosa (OprD) mutants (Ito et al., 2018). Enhanced efflux pump activity can expel cefiderocol, with target mutations in multidrug efflux regulators (such as baeS, czcS, nalC), antibiotic inactivation enzymes (ampR, dacB), and penicillin-binding protein genes (mrcB) playing key roles (Kriz et al., 2024).
Although cefiderocol has shown potent activity against CRGNB in previous studies (Yousefi et al., 2024; Piccica et al., 2023; de la Fuente et al., 2023), most investigations have been conducted outside of China, where resistance mechanisms and epidemiological patterns may differ. Regional data are therefore essential to assess whether these findings are generalizable to local clinical settings. In particular, little is known about the contribution of efflux pumps, resistance gene expression, and genomic variations to cefiderocol resistance among Chinese isolates.
To address this gap, the present study evaluated the in vitro antimicrobial activity of cefiderocol against a large collection of clinical CRGNB isolates, including CRE, CRPA, and CRAB. In addition, we investigated the role of efflux pumps through inhibition assays, quantified the expression of resistance-associated genes using RT-qPCR, and applied whole-genome sequencing (WGS) to identify gene mutations and loss events linked to cefiderocol resistance.
The objective of this study was to provide localized evidence on the efficacy and resistance mechanisms of cefiderocol in China, thereby contributing to both clinical decision-making and the global understanding of its role in managing CRGNB infections.
2 Materials and methods
2.1 Strain collection
A total of 370 CRGNB isolates were tested, including 58 K. pneumoniae, 99 Escherichia coli, 7 K. oxytoca, 51 E. hormaechei and 20 C. freundii, 82 A. baumannii, 53 P. aeruginosa. Due to the clonal dissemination of Carbapenem-resistant K. pneumoniae (CRKP) in our hospital, the major K. pneumoniae strains were collected from 15 hospitals in Nanjing city. The remaining strains were isolated from Nanjing Drum tower hospital. Among them, 99 E. coli, 7 K. oxytoca, 51 E. hormaechei, 20 C. freundii, and 53 P. aeruginosa were isolated between 2013 and 2021, and 82 A. baumannii strains were isolated between 2019 and 2022. All bacterial strains were obtained from various clinical specimens such as blood (n = 157), sputum (n = 75), urine (n = 63), secretion (n = 21), abdominal dropsy (n = 21), bile (n = 12), specimens with missing clinical source information (n = 8), catheter (n = 4), pleural effusion (n = 3), cerebrospinal fluid (n = 2), pus (n = 2), rectal swab (n = 1), and incision secretion (n = 1). The detailed distribution of isolation sources for each bacterial species is summarized in Supplementary Table S1.
2.2 Preparation of iron-depleted cation-adjusted Mueller–Hinton broth (ID-CAMHB)
Cefiderocol susceptibility testing was performed using iron-depleted cation-adjusted Mueller–Hinton broth (ID-CAMHB), prepared in accordance with Clinical and Laboratory Standards Institute (CLSI) 2024 guidelines (Clinical and Laboratory Standards Institute (CLSI), 2023). Briefly, 21.0 g of Mueller–Hinton (MH) medium was dissolved in 1 L of purified water, sterilized by autoclaving at 121 °C for 15 min, and cooled to room temperature. Chelex® 100 resin (100 g) was added, and the suspension was stirred with a magnetic stirrer for 6 h at room temperature. Following centrifugation, the supernatant was collected, and iron concentration was determined by inductively coupled plasma optical emission spectrometry (ICP-OES). When the iron concentration was confirmed to be < 0.03 μg/mL, the pH was adjusted to 7.2–7.4, and the medium was sequentially supplemented with ZnSO₄·7H₂O (final concentration 0.5–1.0 μg/mL), CaCl₂ (20.0–25.0 μg/mL), and MgCl₂·6H₂O (10.0–12.5 μg/mL). The final medium was sterilized by filtration through a 0.22 μm membrane, designated as ID-CAMHB, and stored at 4 °C until use.
2.3 MIC determination
Antimicrobial susceptibility testing was conducted using microbroth dilution for the following antibiotics: imipenem, ceftazidime, ceftriaxone, cefepime, amikacin, ciprofloxacin, levofloxacin, tobramycin, and gentamicin. Cefiderocol susceptibility was determined using iron-depleted cation-adjusted Mueller-Hinton broth, with a cefiderocol concentration range of 0.03125–32 μg/mL. E. coli ATCC25922 was used as a quality control. Results were interpreted according to CLSI2024 guideline (Clinical and Laboratory Standards Institute (CLSI), 2023).
2.4 Analysis of gene acquisition, mutations, and loss
WGS was performed on all 370 CRGNB isolates to investigate the genetic basis of cefiderocol resistance. Genomic DNA was extracted using standard protocols and sequenced using an Illumina platform. High-quality reads were assembled de novo, and annotation was conducted using Prokka. Comprehensive bioinformatics analyses were employed to identify gene acquisition events, point mutations, and gene loss associated with cefiderocol resistance. Specifically, resistance-related genes, including β-lactamases and iron transporter-associated genes, were examined. Comparative genomic analyses between cefiderocol-resistant and -susceptible isolates were conducted to detect significant mutations and gene deletions with A. baumannii ATCC 19606 (Accession Number: CP058289.1), E. coli K-12 MG1655 (Accession Number: NC_000913.3), K. pneumoniae ATCC 13883 (Accession Number: KN046818.1), and P. aeruginosa PAO1 (Accession Number: AE004091.2) being as reference genomes. Identified genetic alterations were further analyzed to elucidate their potential role in the emergence of cefiderocol resistance. The corresponding accession numbers for all sequenced isolates, including both cefiderocol-resistant and cefiderocol-susceptible strains, are now provided in Supplementary Table S2. WGS data of the cefiderocol-resistant isolates have been deposited in the NCBI GenBank database. The accession numbers are as follows: P. aeruginosa: 4087 (JBBXJG000000000), 19,686 (JBBXHS000000000), 12,218 (JBBXIN000000000), 17,774 (JBBXHW000000000), 3,077 (JBBXJJ000000000), 2,745 (JBBXJK000000000); A. baumannii: 10521 (JAVIKM000000000), 11,253 (JAVIKS000000000), 12,076 (JAVILL000000000), 12,150 (JAVILM000000000), 12,439 (JAVILT000000000), 14,118 (JAVIMK000000000), 14,179 (JAVIML000000000), 14,184 (JAVIMM000000000); E. coli: 3034 (JANWRX000000000), 14,449 (JANWPI000000000), 12,010 (JANWQC000000000), 14,109 (JANWPL000000000), 14,334 (JANWPJ000000000), 15,503 (JANWOY000000000), 16,769 (JANWOK000000000); E. hormaechei: 1707 (JANWOH000000000), 14,787 (JANWMY000000000); K. pneumoniae: njsetyy13 (VEON00000000), njxkyy19 (RZKE00000000).
2.5 Inhibition of efflux pump activity by carbonyl cyanide m-chlorophenyl hydrazone (CCCP) and reserpine
The role of efflux pumps in CRGNB resistance was evaluated using the CCCP and Reserpine inhibition tests. MIC changes were assessed in the presence and absence of CCCP (MedChemExpress, China) and Reserpine (MedChemExpress, China) at a final concentration of 50 μg/mL. Each isolate was inoculated into ID-CAMHB containing serial dilutions of cefiderocol. Overexpression of the efflux pump was considered as a positive phenotype when cefiderocol MIC was reduced by at least fourfold in the presence of CCCP and reserpine.
2.6 Gene expression was analyzed by RT-qPCR
Total RNA was extracted from bacteria using the RNAEX reagent (Accurate Biotechnology, Hunan, China) and the SteadyPure Universal RNA Extraction Kit (Accurate Biotechnology) according to the manufacturer’s instructions. The extracted RNA was reversely transcribed into cDNA using the Evo M-MLV RT Premix for qPCR (Accurate Biotechnology, Hunan, China). RT-qPCR was performed using a qPCR kit (Accurate Biotechnology, China). Relative RNA expression levels were analyzed using the 2−∆∆Ct method. All RT-qPCR analyses were conducted in triplicate biological replicates. The housekeeping gene used for P. aeruginosa was rpoD, while rpoB served as the housekeeping gene for both A. baumannii and K. pneumoniae. Primer sets are listed in Supplementary Table S3.
2.7 Construction of a phylogenetic tree
Assembled contigs in FASTA format from each sequencing run were submitted to the Centre for Genomic Epidemiology (CGE)1 (Kaas et al., 2014). Phylogenetic relationships were inferred using the web-based tool CSI Phylogeny 1.4 with stringent SNP filtering: minimum SNP quality of 30, mapping quality of 25, sequencing depth ≥10×, relative depth ≥10%, and Z-score ≥1.96. Heterozygous SNPs were optionally excluded, and a minimum distance of 10 bp between pruned SNPs was applied to reduce linkage bias. The tree was visualized using iTOL (Letunic and Bork, 2019).
2.8 Statistical analysis
Statistical analyses were performed using Microsoft Excel (Version 19.0, Microsoft Corporation, Redmond, WA, United States) and SPSS (Version 20.0). Descriptive statistics, including MIC50, MIC90, and MIC ranges, were calculated for all variables. Comparisons between groups with and without β-lactamase genes were performed using the non-parametric Mann–Whitney U test. Classification of cefiderocol-resistant and susceptible strains was analyzed using the Chi-squared test (with continuity correction) and Fisher’s exact test, while RT-qPCR data were evaluated using independent t-tests. Statistical significance was defined as p < 0.05. In figures, significance levels were indicated as follows: * p < 0.05, ** p < 0.01, *** p < 0.001, with non-significant differences marked as “ns.”
3 Result
3.1 Distribution and cumulative MIC of cefiderocol and other clinically used antibiotics against CRGNB isolates
Cefiderocol exhibited high antibacterial activity against the 370 CRGNB isolates. Notably, 16.2% of the isolates were susceptible to cefiderocol, showing a very low MIC of ≤0.03125 μg/mL, and 68.6% of isolates were inhibited at 2 μg/mL. At 4 μg/mL, the inhibition rate increased to 83.0%, with 100.0% inhibition achieved at 32 μg/mL. In comparison, imipenem exhibited lower initial activity, inhibiting only 1.1% of isolates at 0.25 μg/mL. Significant inhibition was observed starting at 16 μg/mL, with 98.5% of isolates inhibited, and 100.0% inhibition at ≥128 μg/mL. Other β-lactams, such as ceftazidime, ceftriaxone, and cefepime, showed variable effectiveness, with notable inhibition only at higher concentrations. Aminoglycosides and fluoroquinolones exhibited better activity, but their efficacy was generally lower than that of cefiderocol. The detailed susceptible data, including resistance percentages, are provided in Table 1 and Figure 2.
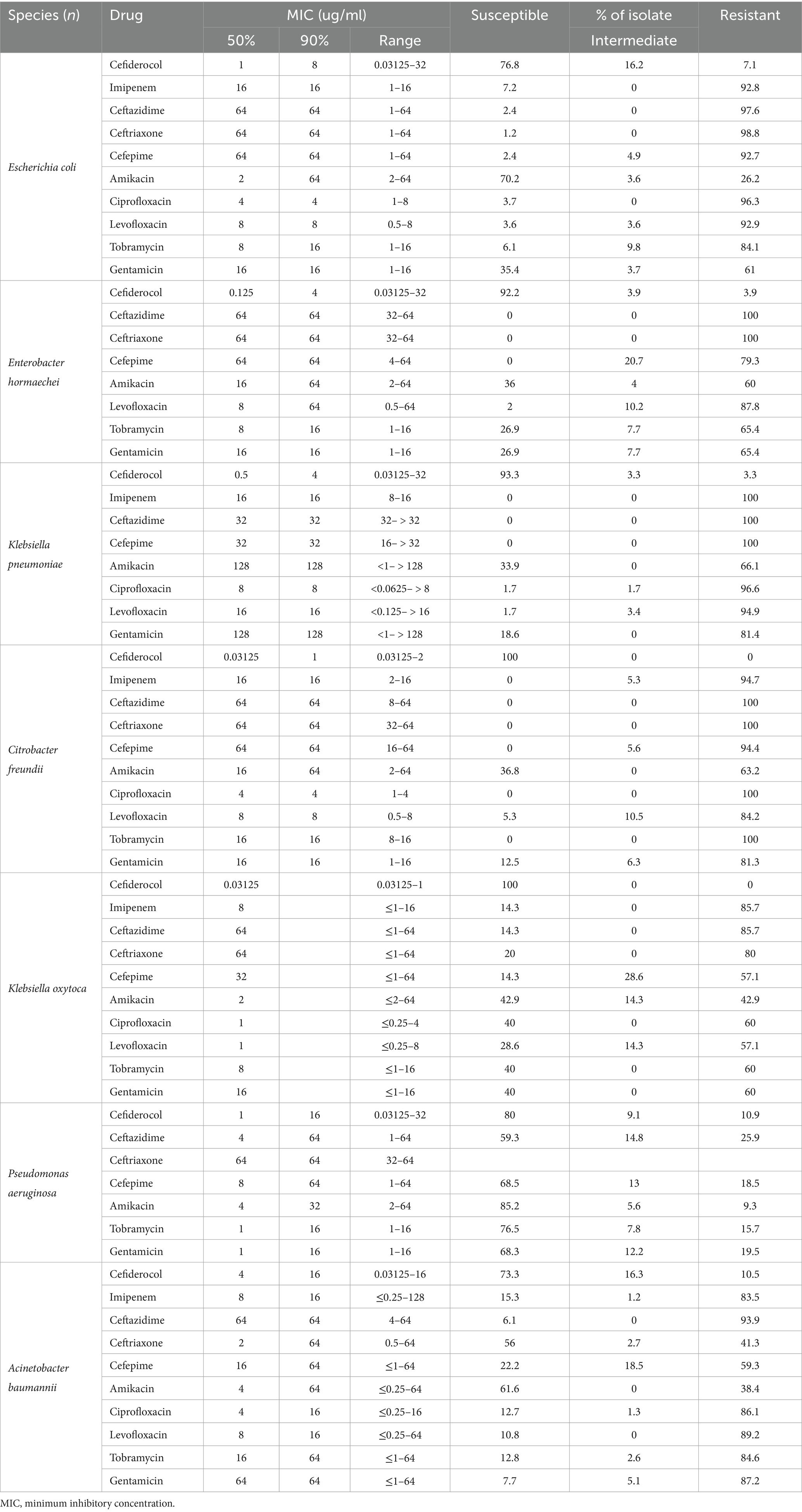
Table 1. Antimicrobial activities of cefiderocol and comparator agents against all carbapenem-resistant gram-negative bacteria isolates.
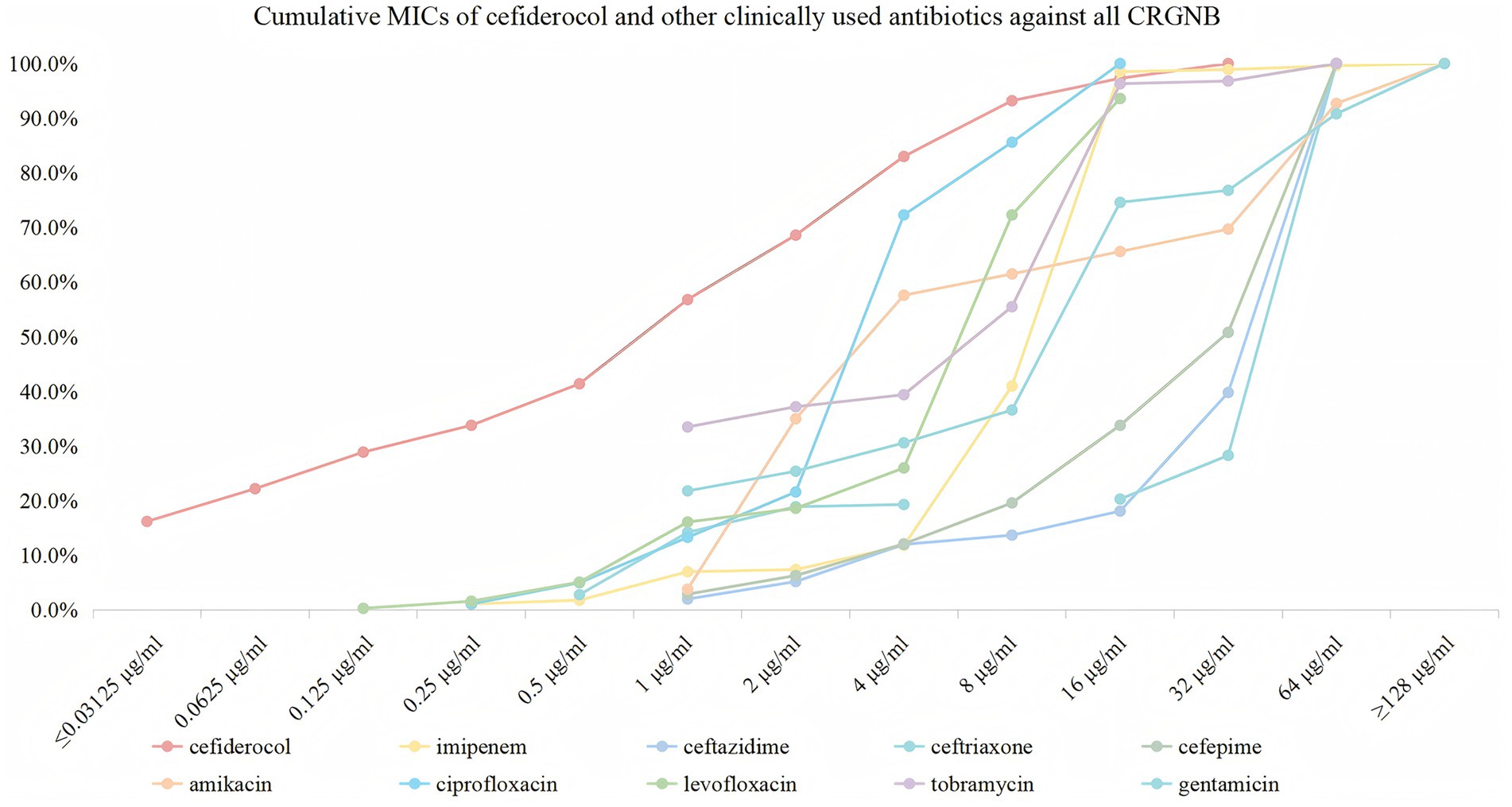
Figure 2. Cumulative MICs of cefiderocol and other clinically used antibiotics against all CRGNB. The figure illustrates the cumulative susceptibility of cefiderocol (red) and comparator antibiotics (imipenem, ceftazidime, ceftriaxone, cefepime, amikacin, ciprofloxacin, levofloxacin, tobramycin, and gentamicin) against CRGNB isolates. The x-axis represents MIC values (μg/ml) on a logarithmic scale (≤0.03125 to ≥128), and the y-axis shows the cumulative percentage of isolates inhibited at each concentration.
3.2 Antimicrobial efficacy of cefiderocol against carbapenem-resistant gram-negative bacterial isolates
Cefiderocol exhibits broad-spectrum antibacterial activity against CRGNB isolates. Specifically, C. freundii and K. oxytoca exhibited the highest susceptibility to cefiderocol (100.0%, resistance rates 0.0%), followed by K. pneumoniae (93.3% susceptible, 3.3% resistant), E. hormaechei (92.2% susceptible, 3.9% resistant), E. coli (76.8% susceptible, 7.1% resistant), and P. aeruginosa (80.0% susceptible, 10.9% resistant). A. baumannii demonstrated the lowest susceptibility, with 73.3% of isolates susceptible and 10.5% resistant. In stark contrast, other antibiotics, such as ceftazidime and ceftriaxone, exhibited high resistance levels, particularly with ceftazidime resistance in E. coli at 97.6% and ceftriaxone resistance in E. hormaechei at 100.0%.
Based on the MIC analysis of cefiderocol against various Gram-negative bacteria, the drug exhibited strong in vitro activity. For P. aeruginosa, cefiderocol shows effective inhibition, with a MIC50 of 1 μg/mL and MIC90 of 16 μg/mL. It is also effective against A. baumannii, with MIC50 at 4 μg/mL and MIC90 at 16 μg/mL, demonstrating good activity despite high resistance in this pathogen. E. coli showed high susceptibility with MIC50 of 1 μg/mL and MIC90 of 8 μg/mL. E. hormaechei and K. pneumoniae exhibited even higher sensitivity, with MIC50 values of 0.125 μg/mL and MIC90 of 0.5 μg/mL, respectively, highlighting excellent effectiveness. C. freundii and K. oxytoca were completely susceptible, with MIC50 values as low as 0.03125 μg/mL, emphasizing the potent activity of cefiderocol (Table 1; Figure 3).
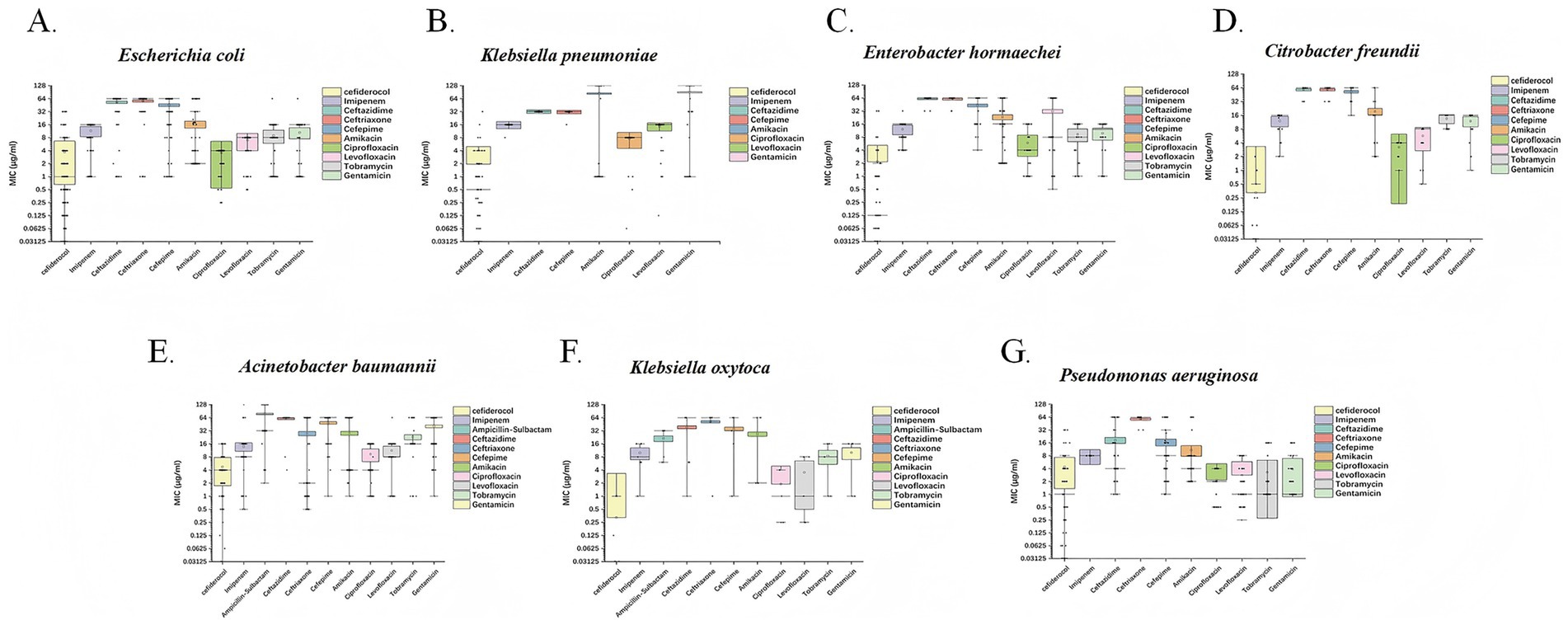
Figure 3. Box plots depicting the MIC (μg/mL) distributions of cefiderocol and other commonly used clinical antibiotics against seven CRGNB species. Each subplot corresponds to a distinct bacterial species: (A) E. coli, (B) K. pneumoniae, (C) E. hormaechei, (D) C. freundii, (E) A. baumannii, (F) K. oxytoca, and (G) P. aeruginosa. The x-axis represents different antibiotics, including cefiderocol, imipenem, ceftazidime, and others. The y-axis displays MIC values on a logarithmic scale, ranging from 0.03125 to 128 μg/mL.
3.3 Efflux pumps role in cefiderocol resistance using CCCP
Among the cefiderocol-resistant CRGNB strains, CCCP significantly reduced the MIC values of cefiderocol, with MICs for some strains dropping to below 0.03125 μg/mL, representing a reduction of over fourfold. In contrast, MIC values after inhibition with reserpine showed minimal changes (fold reduction near 1). However, in P. aeruginosa 4,087 and A. baumannii 10,521, the MIC values decreased by approximately twofold (Table 2).
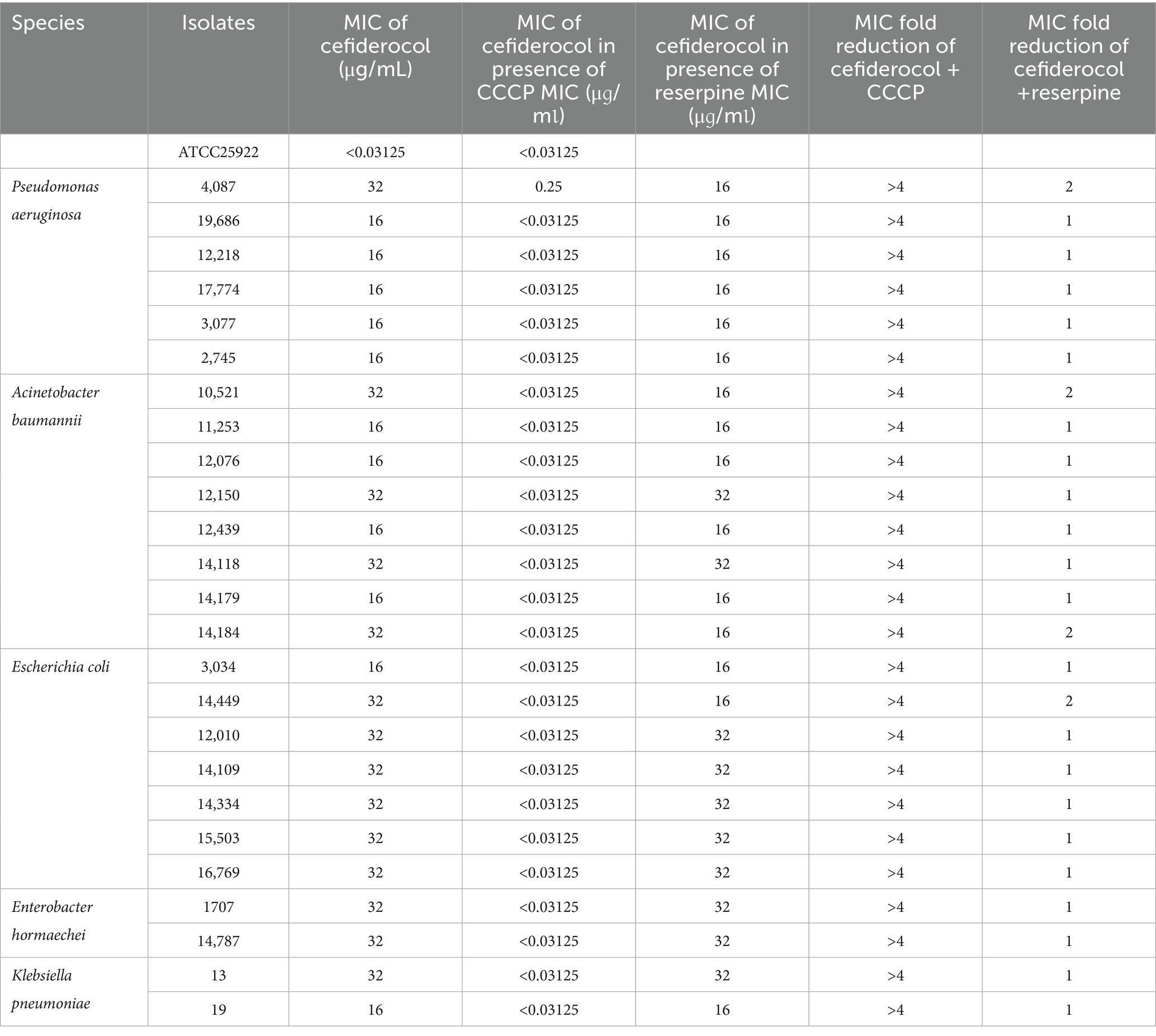
Table 2. Cefiderocol minimum inhibitory concentration values of resistant isolates in the presence or absence of 50 μg/mL CCCP or reserpine.
3.4 Correlation between β-lactamase gene and cefiderocol resistance
Among P. aeruginosa strains harboring blaPAO, 13.0% exhibited cefiderocol resistance. In contrast, resistance associated with other β-lactamase genes was confined to individual isolates. A. baumannii strains harboring blaOXA-23, blaOXA-66, blaTEM-1D, and blaADC-25 displayed MICs ranging from 0.03125 to 16 μg/mL, with most clustering at 4 μg/mL. K. pneumoniae strains carrying blaKPC-2 displayed a broader MIC range (0.03125–32 μg/mL), but most were concentrated around 0.03125 μg/mL. E. hormaechei exhibited MICs between 0.03125 and 32 μg/mL, with only one resistant strain and the rest remaining susceptible. Similarly, E. coli strains carrying blaTEM-1B and blaNDM-1 had MICs predominantly at 8 μg/mL, while those harboring other β-lactamase genes remained susceptible (Figure 4).
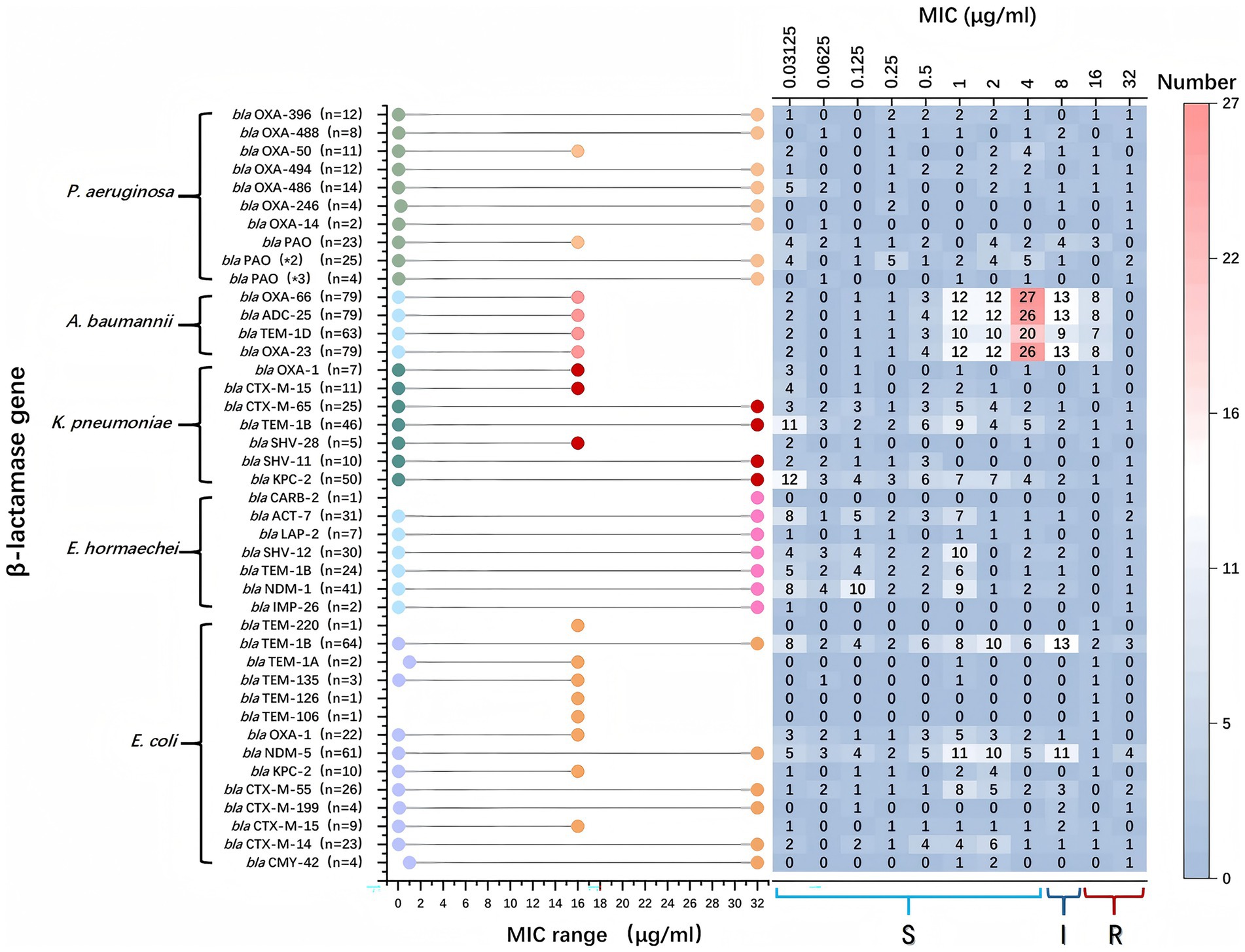
Figure 4. β-lactamase genes and their association with their minimum inhibitory concentration (MIC) ranges within various bacterial species. On the left, bacterial species and the specific β-lactamase genes they harbor are listed. The middle part of the chart uses colored dots to indicate the MIC ranges for bacteria with specific genes. The x-axis represents the MIC values in μg/ml, ranging from 0.03125 to 32. A higher MIC indicates stronger antibiotic resistance. The right section displays a heatmap, showing the distribution of bacterial strains at various MIC concentrations. Each cell contains the number of strains found at that MIC level, and the color gradient from light to dark indicates the strain count. Red represents the highest numbers, while blue represents lower numbers.
The predominant β-lactamase genes varied across species: In P. aeruginosa, blaPAO and blaOXA variants were most common; In A. baumannii, blaOXA-23, blaOXA-66, blaTEM-1D, and blaADC-25 predominated. In E. coli, blaNDM-5, blaTEM-1B, and blaCTX-M were the main genes, and in K. pneumoniae, blaKPC-2, blaSHV, blaTEM-1B, and various blaCTX-M variants were prevalent (Table 3).
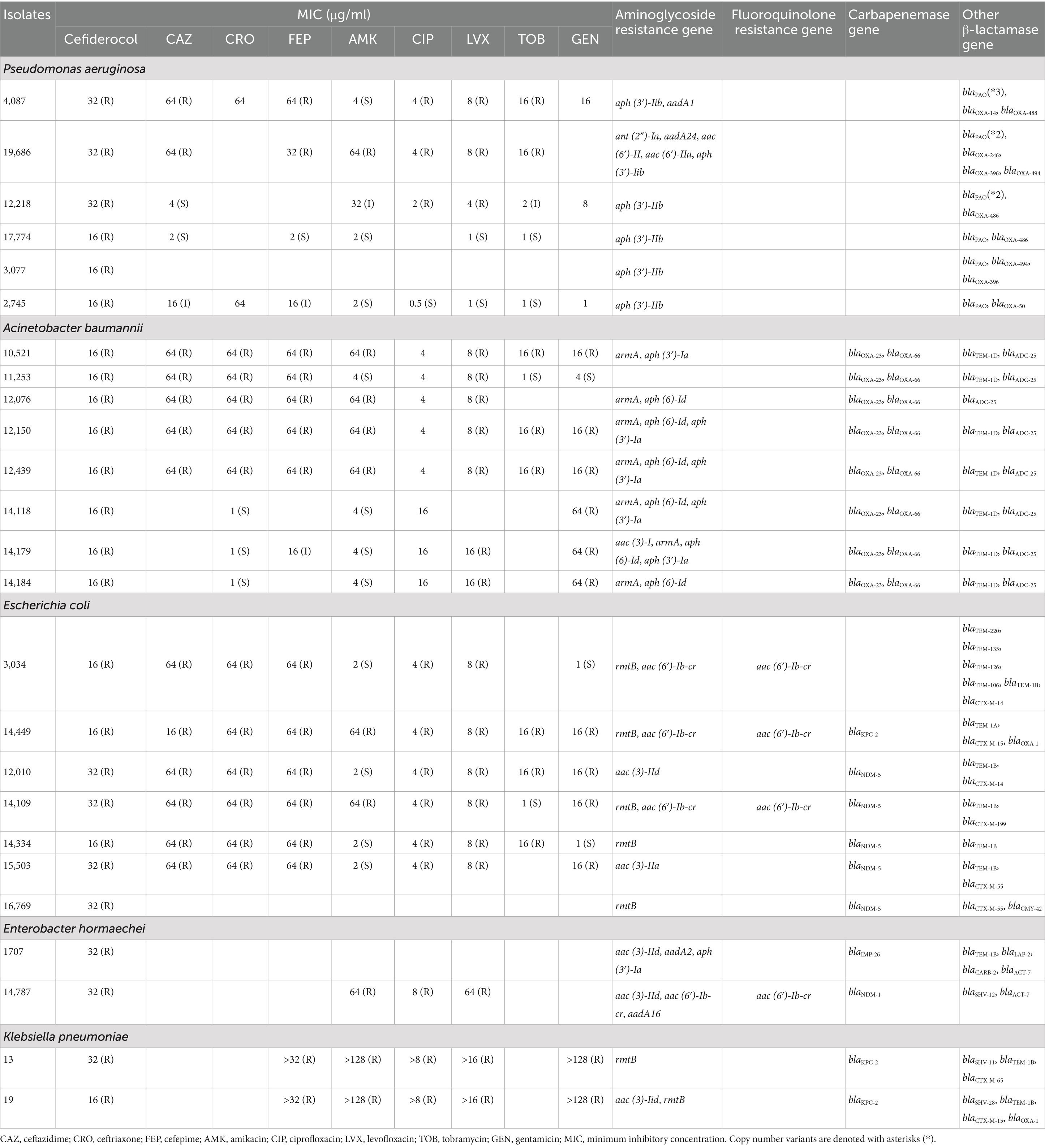
Table 3. Antimicrobial resistance profile and resistance genes of carbapenem-resistant gram-negative bacteria isolates resistant to cefiderocol.
The association between β-lactamase genes and cefiderocol resistance indicated a potential correlation between blaOXA-66 (p = 0.004) and blaOXA-23 (p = 0.050) in A. bumannii, and blaSHV-12 (p = 0.022) in E. hormaechei with cefiderocol resistance (Table 4). In contrast, other tested genes did not show a statistically significant correlation with resistance.
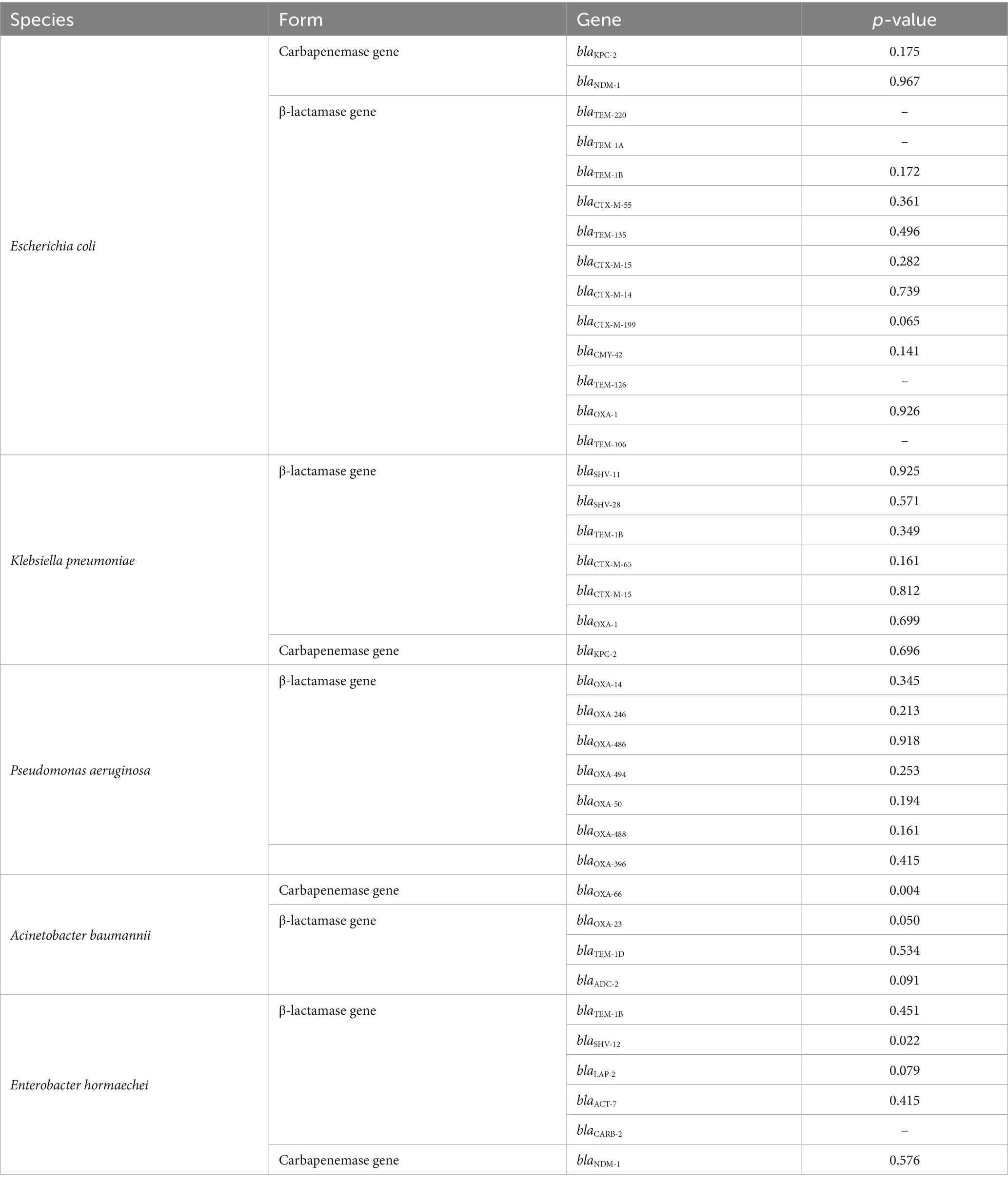
Table 4. Association between β-lactamase/carbapenemase genes and cefiderocol resistance analyzed by Mann–Whitney U rank-sum test.
3.5 Comparison of gene mutations in cefiderocol-resistant and cefiderocol-sensitive CRGNB
In K. pneumoniae, a missense mutation (c.442C > T, p.Pro148Ser) was detected in envZ, which encodes an osmoregulatory sensor protein (p = 0.036). In E. coli, multiple mutations were identified, particularly in cirA, encoding the iron-catecholate outer membrane transporter. Frameshift and missense mutations in cirA included c.1540_1545delC (p.Arg514fs, p = 0.006), c.1519_1535delG (p.Glu507fs, p = 0.006), and c.1395A > C (p.Glu465Asp., p = 0.004). Additionally, a frameshift mutation (c.268_269delAG, p.Ser90fs) was strongly associated with cefiderocol resistance (p < 0.001). Furthermore, a missense mutation in nuoC (c.1683_1685delC, p.Phe562Tyr) was identified (p = 0.006), suggesting a potential link between NADH-quinone oxidoreductase function and resistance. In E. hormaechei, mutations in ampC included c.953_956delTGGTinsCGGC (p.Val318Ala) and c.172C > T (p.Pro58Ser) (Table 5). These mutations were significantly more common in resistant strains (p < 0.05).
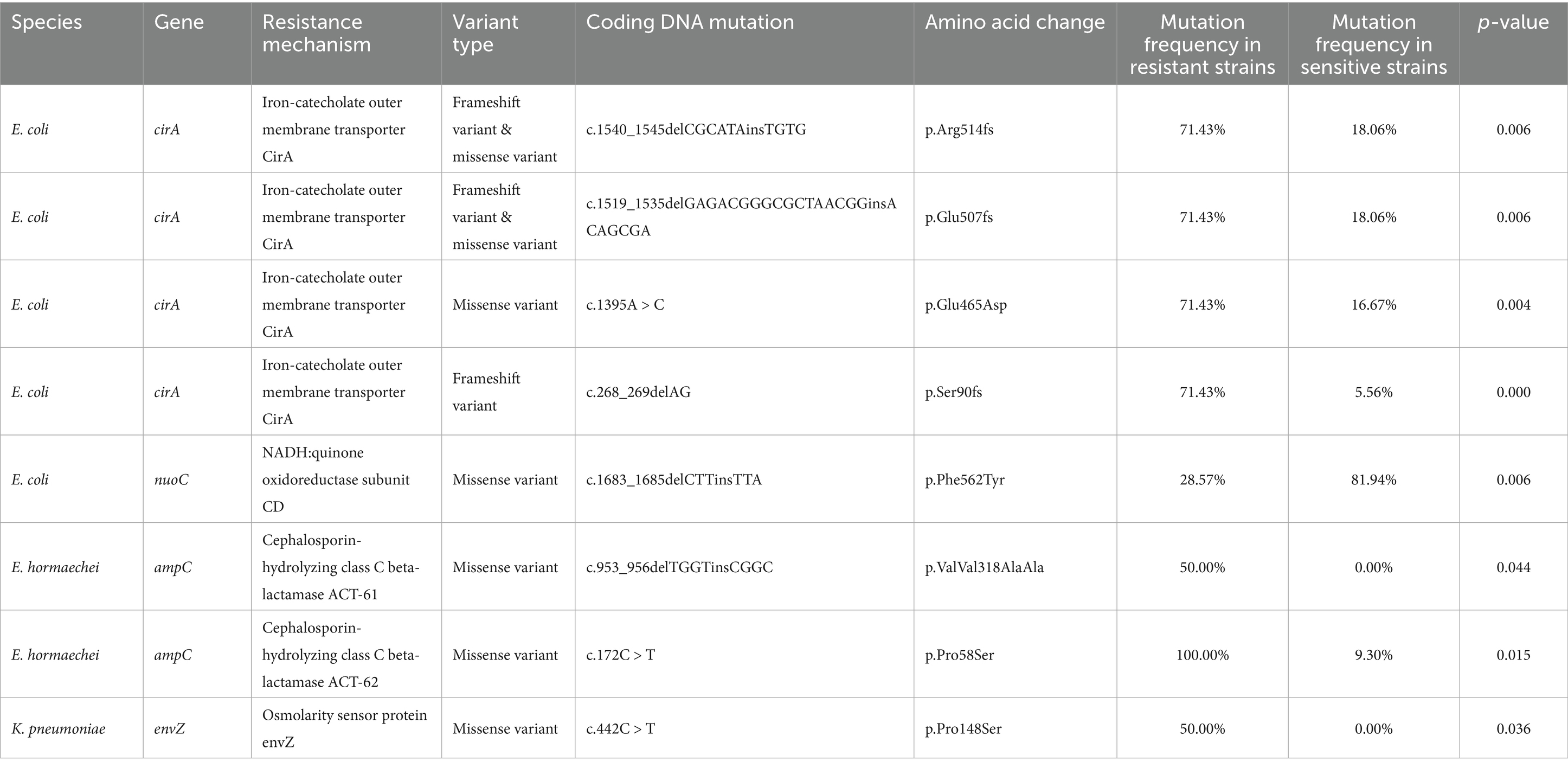
Table 5. Significant gene mutations in cefiderocol-resistant and -susceptible carbapenem-resistant gram-negative.
3.6 Comparison of gene loss in cefiderocol-resistant and cefiderocol-susceptible CRGNB
In P. aeruginosa, the iron-uptake receptor gene piuA showed a significantly higher genomic absence frequency in cefiderocol-resistant isolates (66.7%) compared with susceptible strains (43.9%), suggesting that loss of this gene may contribute to cefiderocol resistance. Other findings included rare ampD losses in P. aeruginosa, retention of ompK36 in K. pneumoniae, and minimal gene losses in A. baumannii and E. coli.
3.7 Differential expression of iron transporter-associated genes correlates with cefiderocol susceptibility across CRGNB
RT-qPCR analysis of resistance gene expression revealed differential expression across CRGNB strains (Figure 5). In P. aeruginosa, the siderophore synthase gene pirA exhibited marked downregulation in resistant isolates compared to susceptible counterparts [p < 0.05; Figure 5A (a)]. In contrast, the ferric iron receptor gene piuA did not show a statistically significant difference in expression between the two groups [p > 0.05; Figure 5A (b)].
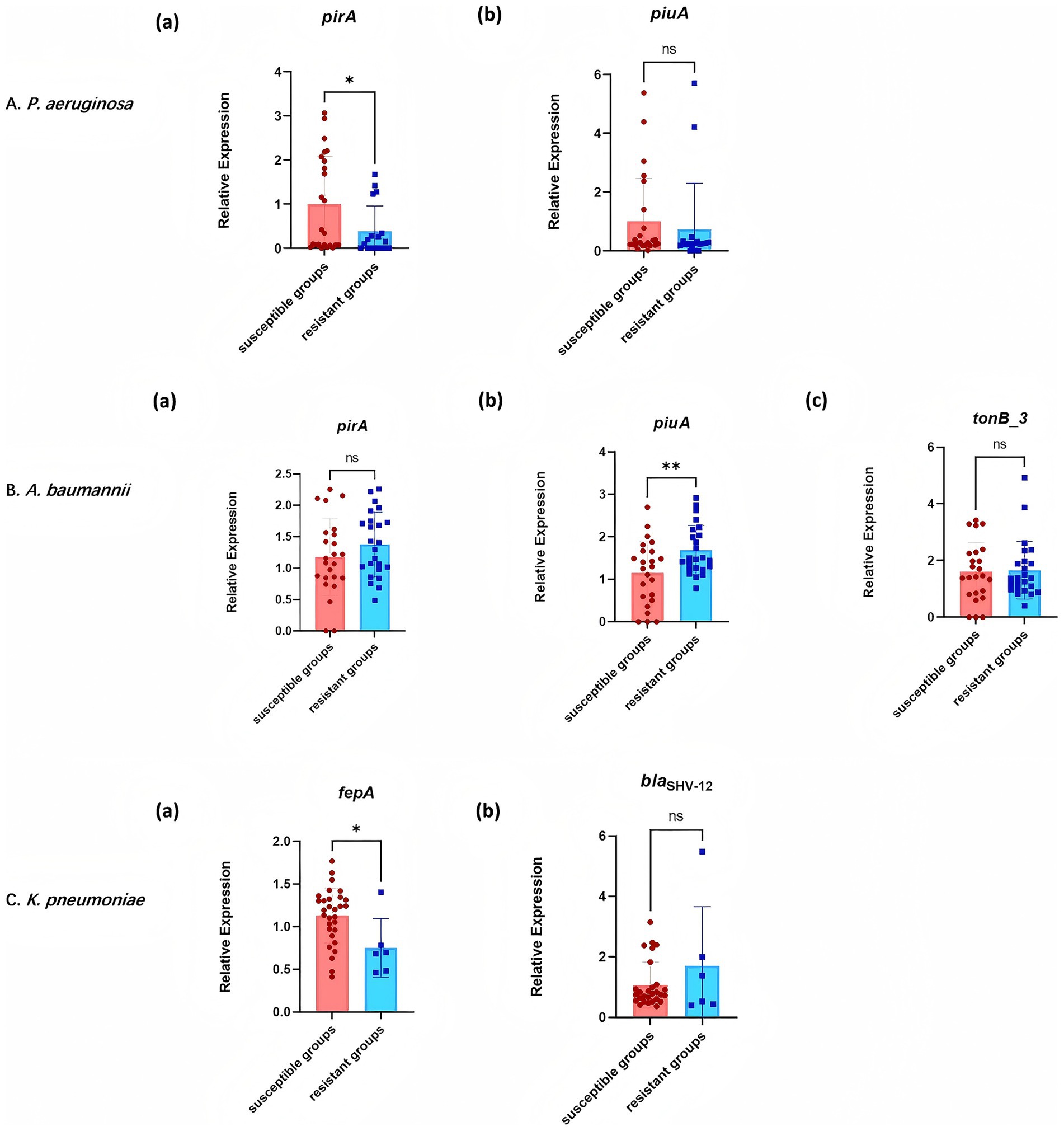
Figure 5. Expression of resistance genes in cefiderocol-resistant and cefiderocol-susceptible strains. (A) P. aeruginosa: Expression levels of pirA and piuA in resistant and susceptible groups. (B) A. baumannii: Expression levels of pirA, piuA, and tonB_3 in different groups. (C) K. pneumoniae: Expression levels of fepA and blaSHV-12 in resistant and susceptible strains. * p < 0.05, ** p < 0.01, *** p < 0.001.
In A. baumannii, an opposite trend was observed at the transcriptional level, with resistant strains exhibiting significant upregulation of piuA compared to susceptible strains [p < 0.01; Figure 5B (b)], while pirA expression remained statistically comparable across susceptibility groups [p > 0.05; Figure 5B (a)]. Moreover, the TonB-dependent energy transduction component tonB_3 maintained stable expression regardless of resistance status [p > 0.05; Figure 5B (c)].
For K. pneumoniae, the expression of the enterobactin transporter gene fepA was significantly downregulated in resistant isolates [p < 0.05; Figure 5C (a)], whereas the β-lactamase gene blaSHV-12 exhibited conserved transcriptional activity between resistant and susceptible groups [p > 0.05; Figure 5C (b)].
3.8 Phylogenetic analysis of CRGNB
Phylogenetic analysis indicated that the cefiderocol-resistant isolates of K. pneumoniae, P. aeruginosa, A. baumannii, and E. hormaechei did not cluster closely on the phylogenetic tree, indicating a lack of clonal relatedness among these strains analyzed (Figure 6). Thus, the same mutation within the specific genes may not be caused by clonal dissemination.
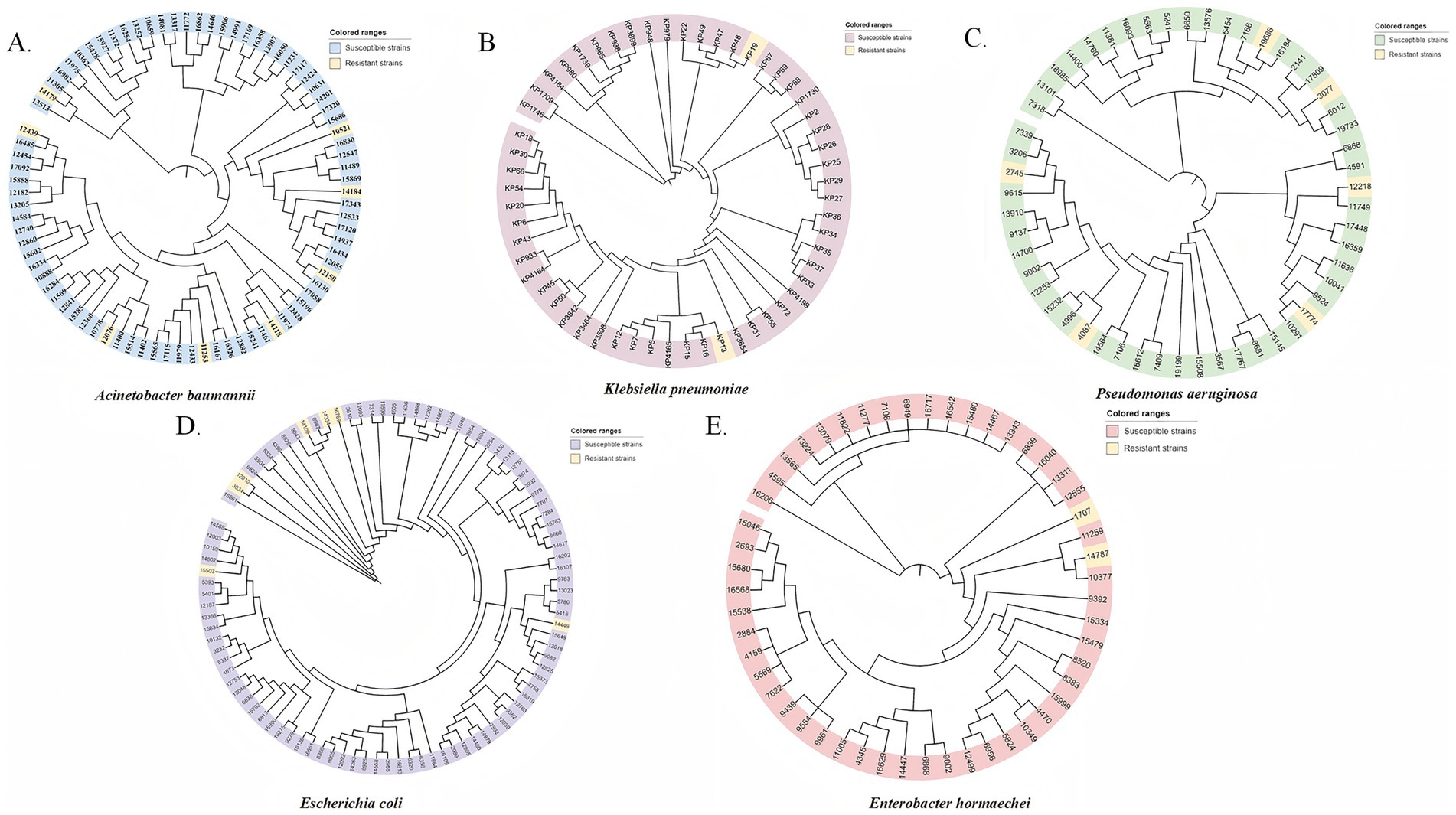
Figure 6. Phylogenetic trees of cefiderocol-resistant and susceptible CRGNB isolates based on SNP analysis. Phylogenetic trees were constructed for five major Gram-negative bacterial species, including (A) A. baumannii, (B) K. pneumoniae, (C) P. aeruginosa, (D) E. coli, and (E) E. hormaechei. Each isolate is represented at the tip of the tree and is color-coded based on its phenotypic susceptibility: resistant strains (yellow) and susceptible strains (gray or species-specific color tones). The clustering of resistant strains in certain branches indicates potential clonal dissemination. Scale bars represent the number of nucleotide substitutions per site.
4 Discussion
The emergence of CRGNB represents a critical challenge to global public health, as these pathogens display extensive drug resistance and readily acquire new mechanisms. Our study provides localized evidence from China, highlighting both the potent in vitro activity of cefiderocol and the diversity of resistance determinants that may undermine its efficacy.
Cefiderocol demonstrated broad-spectrum antibacterial activity, inhibiting 83.0% of isolates at 4 μg/mL and achieving complete inhibition at 32 μg/mL. Consistent with previous study, cefiderocol inhibited 92.1% of CRE, 86.5% of CRKP isolates, and 88.9% of CRAB isolates at similar MIC levels (Mushtaq et al., 2020). Such a high efficacy, particularly at lower concentrations, is largely attributed to its unique mechanism of action—exploiting iron–siderophore uptake systems to traverse the bacterial outer membrane—which enables it to overcome resistance mechanisms that limit traditional antibiotics such as imipenem and ceftazidime (Kaye et al., 2023).
Furthermore, our results are consistent with international studies reporting high cefiderocol efficacy against CRE and A. baumannii. Despite this potent activity, we observed slightly higher MIC₉₀ values for E. coli and K. pneumoniae compared with data from the U. S. and Italy (Albano et al., 2020; Padovani et al., 2023), and our study further revealed variability in MIC values that likely reflects regional differences in resistance patterns and testing methodologies. For example, K. pneumoniae in our study showed MIC₅₀ and MIC₉₀ values of 0.5 μg/mL and 4 μg/mL, which were slightly lower than those reported in another study from China (MIC₅₀ = 2 μg/mL, MIC₉₀ = 4 μg/mL) (Zhao et al., 2023). At the species level, C. freundii and K. pneumoniae exhibited the highest susceptibility (100.0 and 93.3%, respectively), whereas P. aeruginosa and A. baumannii showed comparatively lower rates (80.0 and 73.3%). These findings suggest species-specific differences in susceptibility that may reflect distinct resistance mechanisms, including efflux pump activity and outer membrane permeability changes. Importantly, efflux pump inhibition assays confirmed that proton-driven pumps play a major role in cefiderocol resistance, as CCCP restored susceptibility in resistant isolates by actively expelling a broad range of antibiotics and lower intracellular drug concentrations (Ren et al., 2024), emphasizing the central role of efflux activity (Brepoels et al., 2022). Furthermore, the distinct effects observed between reserpine (targeting ATP-dependent pumps) and CCCP (disrupting proton-driven pumps) suggest that proton-driven efflux systems are particularly critical in mediating cefiderocol resistance (Kabra et al., 2019).
Genomic and transcriptional analyses provided further resolution into resistance mechanisms. β-lactamase genes were widely distributed: blaPAO and various blaOXA variants in P. aeruginosa; blaOXA-23, blaOXA-66, and blaTEM-1D in A. baumannii; and blaNDM-5 and blaCTX-M in E. coli. Notably, cefiderocol resistance in E. hormaechei was associated with blaSHV-12, echoing prior evidence that amplification of this gene can drive resistance in K. pneumoniae (Liu et al., 2024). Moreover, mutations in ampC (p.Val318Ala and p.Pro58Ser) and envZ (regulator of outer membrane porin expression through the OmpR/EnvZ system) were significantly enriched in resistant isolates, reinforcing the role of regulatory and enzymatic pathways in shaping cefiderocol susceptibility (Shields et al., 2020; Yang et al., 2024). These findings are consistent with studies that reported ampC mutations mediating cross-resistance to ceftazidime/avibactam and cefiderocol, as well as envZ alterations modulating porin expression (Dahdouh et al., 2024).
Iron transport pathways also emerged as critical modulators of resistance. In P. aeruginosa, genomic loss of piuA was more frequent in resistant isolates (66.7%) than in susceptible strains (43.9%), suggesting that gene deletion disrupts siderophore-mediated uptake of cefiderocol (Moynie et al., 2017). Conversely, in A. baumannii, piuA was transcriptionally upregulated in resistant strains, consistent with reports that aberrant expression of siderophore receptors can alter drug transport dynamics (Nishimura et al., 2022). In K. pneumoniae, downregulation of fepA was correlated with resistance, emphasizing the essential role of iron-uptake porins in facilitating cefiderocol entry (Daoud et al., 2022). Together, these data highlight the multifaceted interplay of β-lactamase activity, efflux systems, and iron transport alterations in cefiderocol resistance.
From a clinical perspective, our results reinforce the potential of cefiderocol as a critical therapeutic option for CRGNB infections in China. However, the variable activity across species and the strong association with efflux- and iron-related mechanisms suggest that susceptibility testing should be integrated into routine clinical practice before cefiderocol use. Furthermore, the ability of efflux pump inhibitors to restore susceptibility raises the possibility of novel combination strategies, though translation into clinical application will require careful evaluation of toxicity and pharmacokinetic constraints. Molecular surveillance incorporating β-lactamase variants, efflux activity, and iron-uptake pathways may improve the precision of empirical therapy, enabling pathogen-specific treatment strategies and helping to preserve cefiderocol’s clinical utility.
This study has several limitations. First, as it was conducted at a single tertiary hospital, the findings may not be generalizable to other regions with different resistance profiles. Second, some isolates were collected in earlier years, which may not fully capture the most recent epidemiological trends. Third, the cross-sectional design provides only a snapshot of susceptibility, and longitudinal surveillance will be needed to monitor evolving resistance patterns over time. Finally, we did not examine the expression of porins and penicillin-binding proteins (PBPs), which have been reported in other studies to contribute to cefiderocol resistance. Future studies incorporating multiple centers, prospective longitudinal sampling, and additional resistance mechanisms will be valuable to provide a more comprehensive understanding of cefiderocol resistance.
5 Conclusion
In summary, our findings confirm that cefiderocol is highly active against diverse CRGNB isolates in China, but resistance driven by β-lactamases, efflux systems, and iron-uptake alterations poses a serious challenge. Continuous surveillance, incorporation of molecular resistance markers into routine diagnostics, and consideration of species-specific responses will be essential to optimize the clinical use of cefiderocol.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material. All the sequences were deposited into the NCBI Sequence Read Archive (SRA). The Whole Genome Shotgun BioProject for these isolates has been deposited at GenBank, the BioProject number was PRJNA882512.
Ethics statement
The studies involving humans were approved by the Ethics Committee of the Nanjing Drum Tower Hospital, Affiliated Hospital of Medical School, Nanjing University (The approval number is 2023-390). The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because due to anonymous and retrospective study design.
Author contributions
RY: Writing – original draft. JJ: Methodology, Visualization, Writing – original draft. LW: Formal analysis, Visualization, Writing – original draft. YZ: Validation, Visualization, Writing – original draft. HS: Project administration, Resources, Writing – original draft. JY: Investigation, Methodology, Writing – review & editing. XC: Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by the National Natural Science Foundation of China (Grant number 81902124) and Funding’s for Clinical Trials from the Affiliated Drum Tower Hospital, Medical School of Nanjing University (2021-LCYJ-PY-06).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1670179/full#supplementary-material
Footnotes
References
Albano, M., Karau, M. J., Schuetz, A. N., and Patel, R. (2020). Comparison of agar dilution to broth microdilution for testing in vitro activity of Cefiderocol against gram-negative Bacilli. J. Clin. Microbiol. 59:e00966-20. doi: 10.1128/JCM.00966-20
Bassetti, M., Russo, C., Vena, A., and Giacobbe, D. R. (2021). New antibiotics for the treatment of nonfermenting gram-negative bacteria. Curr. Opin. Infect. Dis. 34, 701–709. doi: 10.1097/QCO.0000000000000757
Brepoels, P., Appermans, K., Perez-Romero, C. A., Lories, B., Marchal, K., and Steenackers, H. P. (2022). Antibiotic cycling affects resistance evolution independently of collateral sensitivity. Mol. Biol. Evol. 39:msac257. doi: 10.1093/molbev/msac257
Clinical and Laboratory Standards Institute (CLSI) (2023). Performance standards for antimicrobial susceptibility testing. 33rd Edn. Berwyn, PA, USA: Clinical and Laboratory Standards Institute.
Dahdouh, E., Gomez-Marcos, L., Canada-Garcia, J. E., de Arellano, E. R., Sánchez-García, A., Sánchez-Romero, I., et al. (2024). Characterizing carbapenemase-producing Escherichia coli isolates from Spain: high genetic heterogeneity and wide geographical spread. Front. Cell. Infect. Microbiol. 14:1390966. doi: 10.3389/fcimb.2024.1390966
Daoud, L., Al-Marzooq, F., Moubareck, C. A., Ghazawi, A., and Collyns, T. (2022). Elucidating the effect of iron acquisition systems in Klebsiella pneumoniae on susceptibility to the novel siderophore-cephalosporin cefiderocol. PLoS One 17:e0277946. doi: 10.1371/journal.pone.0277946
Daoud, Z., and Dropa, M. (2023). Editorial: the global threat of carbapenem-resistant gram-negative bacteria, volume II. Front. Cell. Infect. Microbiol. 13:1196488. doi: 10.3389/fcimb.2023.1196488
de la Fuente, C., Rodríguez, M., Merino, N., Carmona, P., Machuca, I., Córdoba-Fernández, M., et al. (2023). Real-life use of cefiderocol for salvage therapy of severe infections due to carbapenem-resistant gram-negative bacteria. Int. J. Antimicrob. Agents 62:106818. doi: 10.1016/j.ijantimicag.2023.106818
Duffy, N., Li, R., Czaja, C. A., Johnston, H., Janelle, S. J., Jacob, J. T., et al. (2023). Trends in incidence of carbapenem-resistant Enterobacterales in 7 US sites, 2016 horizontal line 2020. Open Forum Infect. Dis. 10:ofad609. doi: 10.1093/ofid/ofad609
Garcia-Betancur, J. C., Appel, T. M., Esparza, G., Gales, A. C., Levy-Hara, G., Cornistein, W., et al. (2021). Update on the epidemiology of carbapenemases in Latin America and the Caribbean. Expert Rev. Anti-Infect. Ther. 19, 197–213. doi: 10.1080/14787210.2020.1813023
Ito, A., Sato, T., Ota, M., Takemura, M., Nishikawa, T., Toba, S., et al. (2018). In vitro antibacterial properties of Cefiderocol, a novel siderophore cephalosporin, against gram-negative Bacteria. Antimicrob. Agents Chemother. 62:e01454-17. doi: 10.1128/AAC.01454-17
Jean, S. S., Harnod, D., and Hsueh, P. R. (2022). Global threat of carbapenem-resistant gram-negative Bacteria. Front. Cell. Infect. Microbiol. 12:823684. doi: 10.3389/fcimb.2022.823684
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., and Lund, O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984
Kabra, R., Chauhan, N., Kumar, A., Ingale, P., and Singh, S. (2019). Efflux pumps and antimicrobial resistance: paradoxical components in systems genomics. Prog. Biophys. Mol. Biol. 141, 15–24. doi: 10.1016/j.pbiomolbio.2018.07.008
Karakonstantis, S., Rousaki, M., and Kritsotakis, E. I. (2022). Cefiderocol: systematic review of mechanisms of resistance, heteroresistance and in vivo emergence of resistance. Antibiotics (Basel) 11:723. doi: 10.3390/antibiotics11060723
Kaye, K. S., Naas, T., Pogue, J. M., and Rossolini, G. M. (2023). Cefiderocol, a siderophore cephalosporin, as a treatment option for infections caused by carbapenem-resistant Enterobacterales. Infect. Dis. Ther. 12, 777–806. doi: 10.1007/s40121-023-00773-6
Kriz, R., Spettel, K., Pichler, A., Schefberger, K., Sanz-Codina, M., Lötsch, F., et al. (2024). In vitro resistance development gives insights into molecular resistance mechanisms against cefiderocol. J. Antibiot. (Tokyo) 77, 757–767. doi: 10.1038/s41429-024-00762-y
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–w259. doi: 10.1093/nar/gkz239
Li, Q., Zhou, X., Yang, R., Shen, X., Li, G., Zhang, C., et al. (2024). Carbapenem-resistant gram-negative bacteria (CR-GNB) in ICUs: resistance genes, therapeutics, and prevention - a comprehensive review. Front. Public Health 12:1376513. doi: 10.3389/fpubh.2024.1376513
Liu, C., Yi, J., Lu, M., Yang, P., Du, C., Jiang, F., et al. (2024). Dynamic within-host cefiderocol heteroresistance caused by Bla(SHV-12) amplification in pandrug-resistant and hypervirulent Klebsiella pneumoniae sequence type 11. Drug Resist. Updat. 73:101038. doi: 10.1016/j.drup.2023.101038
Moon, S. H., and Huang, E. (2023). Cefiderocol resistance in Klebsiella pneumoniae is linked to SHV extended-spectrum beta-lactamase activities and functional loss of the outer membrane porin OmpK35. Microbiol. Spectr. 11:e0349622. doi: 10.1128/spectrum.03496-22
Moynie, L., Luscher, A., Rolo, D., Pletzer, D., Tortajada, A., Weingart, H., et al. (2017). Structure and function of the PiuA and PirA siderophore-drug receptors from Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob. Agents Chemother. 61:e02531-16. doi: 10.1128/AAC.02531-16
Müller, C., Reuter, S., Wille, J., Xanthopoulou, K., Stefanik, D., Grundmann, H., et al. (2023). A global view on carbapenem-resistant Acinetobacter baumannii. MBio 14:e0226023. doi: 10.1128/mbio.02260-23
Mushtaq, S., Sadouki, Z., Vickers, A., Livermore, D. M., and Woodford, N. (2020). In vitro activity of Cefiderocol, a siderophore cephalosporin, against multidrug-resistant gram-negative Bacteria. Antimicrob. Agents Chemother. 64:e01582-20. doi: 10.1128/AAC.01582-20
Nishimura, B., Escalante, J., Tuttobene, M. R., Subils, T., Mezcord, V., Pimentel, C., et al. (2022). Acinetobacter baumannii response to cefiderocol challenge in human urine. Scientific Reports 12:8763. doi: 10.1038/s41598-022-12829-7
Ong'uti, S., Czech, M., Robilotti, E., and Holubar, M. (2022). Cefiderocol: a new cephalosporin stratagem against multidrug-resistant gram-negative bacteria. Clin. Infect. Dis. 74, 1303–1312. doi: 10.1093/cid/ciab757
Padovani, M., Bertelli, A., Corbellini, S., Piccinelli, G., Gurrieri, F., and de Francesco, M. A. (2023). In vitro activity of Cefiderocol on multiresistant bacterial strains and genomic analysis of two Cefiderocol resistant strains. Antibiotics (Basel) 12:785. doi: 10.3390/antibiotics12040785
Piccica, M., Spinicci, M., Botta, A., Bianco, V., Lagi, F., Graziani, L., et al. (2023). Cefiderocol use for the treatment of infections by carbapenem-resistant gram-negative bacteria: an Italian multicentre real-life experience. J. Antimicrob. Chemother. 78, 2752–2761. doi: 10.1093/jac/dkad298
Ren, J., Wang, M., Zhou, W., and Liu, Z. (2024). Efflux pumps as potential targets for biofilm inhibition. Front. Microbiol. 15:1315238. doi: 10.3389/fmicb.2024.1315238
Sha, J., Li, Y., Chen, X., Hu, Y., Ren, Y., Geng, X., et al. (2017). Fatality risks for nosocomial outbreaks of Middle East respiratory syndrome coronavirus in the Middle East and South Korea. Arch. Virol. 162, 33–44. doi: 10.1007/s00705-016-3062-x
Shields, R. K., Iovleva, A., Kline, E. G., Kawai, A., McElheny, C. L., and Doi, Y. (2020). Clinical evolution of AmpC-mediated ceftazidime-avibactam and Cefiderocol resistance in Enterobacter cloacae Complex following exposure to cefepime. Clin. Infect. Dis. 71, 2713–2716. doi: 10.1093/cid/ciaa355
Sollima, A., Rossini, F., Lanza, P., Pallotto, C., Meschiari, M., Gentile, I., et al. (2024). Role of Cefiderocol in multidrug-resistant gram-negative central nervous system infections: real life experience and state-of-the-art. Antibiotics (Basel) 13:453. doi: 10.3390/antibiotics13050453
Vijay, D., Bedi, J. S., Dhaka, P., Singh, R., Singh, J., Arora, A. K., et al. (2021). Knowledge, attitude, and practices (KAP) survey among veterinarians, and risk factors relating to antimicrobial use and treatment failure in dairy herds of India. Antibiotics (Basel) 10:216. doi: 10.3390/antibiotics10020216
Wang, C., Yang, D., Wang, Y., and Ni, W. (2022). Cefiderocol for the treatment of multidrug-resistant gram-negative Bacteria: a systematic review of currently available evidence. Front. Pharmacol. 13:896971. doi: 10.3389/fphar.2022.896971
Yang, C., Wang, L., Lv, J., Wen, Y., Gao, Q., Qian, F., et al. (2024). Effects of different carbapenemase and siderophore production on cefiderocol susceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 68:e0101924. doi: 10.1128/aac.01019-24
Yin, L., Lu, L., He, L., Lu, G., Cao, Y., Wang, L., et al. (2023). Molecular characteristics of carbapenem-resistant gram-negative bacilli in pediatric patients in China. BMC Microbiol. 23:136. doi: 10.1186/s12866-023-02875-0
Yousefi, B., Kashanipoor, S., Mazaheri, P., Alibabaei, F., Babaeizad, A., Asli, S., et al. (2024). Cefiderocol in combating carbapenem-resistant Acinetobacter baumannii: action and resistance. Biomedicine 12:2532. doi: 10.3390/biomedicines12112532
Keywords: cefiderocol, carbapenem-resistant gram-negative bacteria, antimicrobial susceptibility, resistance mechanisms, efflux pump inhibitor, mutation
Citation: Yan R, Ji J, Wang L, Zou Y, Shen H, Yuan J and Cao X (2025) In vitro antimicrobial activity and resistance mechanisms of cefiderocol against clinical carbapenem-resistant gram-negative bacteria. Front. Microbiol. 16:1670179. doi: 10.3389/fmicb.2025.1670179
Edited by:
Spyros Pournaras, National and Kapodistrian University of Athens, GreeceReviewed by:
Subhasree Roy, National Institute of Cholera and Enteric Diseases (ICMR), IndiaSilpak Biswas, Calcutta School of Tropical Medicine, India
Daniela Talapan, The National Institute of Infectious Diseases Prof. Dr. Matei Bals, Romania
Cinara Feliciano, University of São Paulo, Brazil
Copyright © 2025 Yan, Ji, Wang, Zou, Shen, Yuan and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinhua Yuan, bmpjZGNraXR0eUAxNjMuY29t; Xiaoli Cao, Y2FvLXhpYW8tbGlAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Ruyu Yan
Ruyu Yan Jun Ji1†
Jun Ji1† Han Shen
Han Shen Xiaoli Cao
Xiaoli Cao