- 1Medical Laboratory Technology Department, Erbil Health and Medical Technical College, Erbil Polytechnic University, Erbil, Iraq
- 2Medical Laboratory Technology Department, Shaqlawa Technical College, Erbil Polytechnic University, Erbil, Iraq
Introduction: Pseudomonas aeruginosa is an opportunistic Gram-negative pathogen and a critical-priority organism according to the World Health Organization. Its increasing resistance to multiple antimicrobial classes, including carbapenems, poses a major challenge in treating infections among immunocompromised individuals, particularly burn and cancer patients.
Methods: This cross-sectional study investigated phenotypic resistance profiles, carbapenemase classifications using an advanced expert system, and the molecular prevalence of blaVIM and blaNDM genes in 50 clinical isolates from cancer, burn, and other immunocompromised patients in Erbil, Iraq.
Results: Multidrug resistance and carbapenem resistance were detected in 66.0 and 58.0% of isolates, respectively, with the highest burden among burn patients (93.8%). Carbapenem resistance was significantly associated with prior carbapenem exposure (p = 0.0044) and increased mortality (p = 0.0392). Carbapenemase-producing isolates classified by the advanced expert system exhibited universal multidrug resistance and more than 95% resistance to imipenem and meropenem. Molecular analysis identified blaVIM in 47.5%, blaNDM in 10.0%, and both genes in 30.0% of tested isolates, with blaNDM significantly associated with carbapenem resistance (p = 0.027). Resistance patterns varied by patient group and antibiotic class, with burn isolates demonstrating the highest rates.
Discussion/conclusion: These findings highlight the need for enhanced molecular surveillance, infection control, and antimicrobial stewardship in high-risk settings.
1 Introduction
Pseudomonas aeruginosa is a metabolically versatile, Gram-negative opportunistic pathogen that poses a major global healthcare threat due to its intrinsic resistance mechanisms and remarkable capacity to acquire additional resistance determinants. It thrives in nutrient-limited environments and hospital settings and is a leading cause of healthcare-associated infections (HAIs), particularly in immunocompromised individuals such as burn and cancer patients or those undergoing transplantation or intensive chemotherapy (Rossi et al., 2022; Wood et al., 2023). Clinical manifestations include ventilator-associated pneumonia, bloodstream infections, surgical site infections, and urinary tract infections, with mortality rates exceeding 50% in severe cases (Schwartz et al., 2024).
The emergence of multidrug-resistant (MDR) and carbapenem-resistant P. aeruginosa (CRPA) has prompted the World Health Organization to classify CRPA as a critical-priority pathogen for antimicrobial research and development (Tacconelli et al., 2018; Flores-Vega et al., 2025). Resistance arises from reduced outer membrane permeability, efflux pump upregulation, target site modification, and production of carbapenemases, particularly metallo-β-lactamases (MBLs) such as blaVIM and blaNDM, often encoded on mobile genetic elements (Karakonstantis et al., 2020; Brkic and Cirkovic, 2024).
Middle Eastern studies report a rising prevalence of these MBL genes, including blaNDM in 21% of CRPA isolates in Iraq (Alsaadi et al., 2020) and blaVIM in 19% of Iranian isolates (Vaez et al., 2018). In addition to resistance, P. aeruginosa expresses multiple virulence factors (e.g., toxA, lasB, exoS) linked to resistance mechanisms through shared regulatory networks (Moradali et al., 2017; Huang et al., 2025). Its large, adaptable genome (6.3–6.6 Mb) further complicates treatment (Grace et al., 2022; Hu et al., 2024).
Although strategies such as reverse vaccinology and immunoinformatics-based epitope prediction offer future promise, their clinical utility remains limited by genetic variability (Fereshteh et al., 2023; Zhu et al., 2025). Until then, the burden of CRPA continues to rise, particularly in vulnerable populations such as those with burns or cancer (Hasan et al., 2024a,b).
This study aimed to characterise the clinical and epidemiological features, resistance patterns, and molecular profiles of MDR P. aeruginosa isolates from burn, cancer, and other immunocompromised patients in Erbil, Iraq. Specifically, we assessed the prevalence and co-occurrence of blaVIM and blaNDM genes and their associations with phenotypic resistance patterns and AES-based carbapenemase classification to inform local antimicrobial stewardship and infection control efforts.
2 Materials and methods
2.1 Study design and population
A cross-sectional study was conducted between October 2024 and June 2025 across major public and private hospitals in Erbil, Iraq, including Nanakaly Hospital for Hematology and Oncology, Rizgary Teaching Hospital, the Burns and Plastic Surgery Hospital, Erbil Central Laboratory, Mihrabani Surgical Hospital, and additional private facilities. Hospitalized immunocompromised patients with culture-confirmed P. aeruginosa infections—attributed to malignancy, chemotherapy, burns, or chronic immunosuppressive conditions—were enrolled. Patients without confirmed infection or non-immunocompromised individuals were excluded. A total of 50 eligible patients were included and stratified into three clinical subgroups: cancer (n = 14), burn (n = 16), and other immunocompromised conditions (n = 20). Ethical approval was granted prior to data collection (see Section 2.7).
2.2 Specimen collection
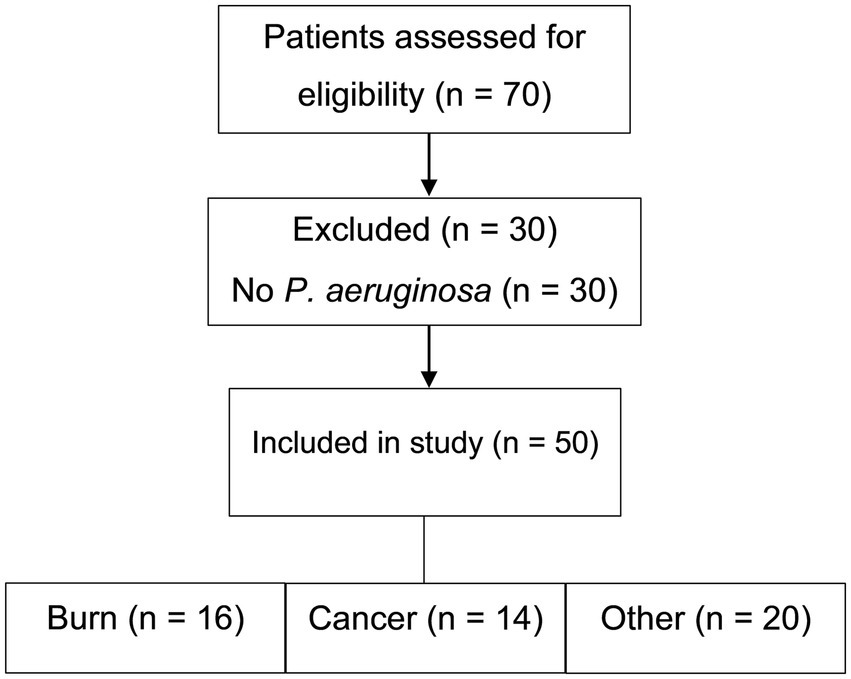
Seventy patients were initially screened for eligibility. Of these, 30 were excluded due to the absence of P. aeruginosa infection (Figure 1). Clinical specimens—including blood, urine, wound swabs, burn swabs, sputum, and bronchoalveolar lavage fluid—were collected using standard aseptic techniques and promptly transported to the microbiology laboratory. P. aeruginosa isolates were confirmed and preserved in brain heart infusion broth supplemented with 20% glycerol at −80 °C for molecular analysis. Only non-duplicate clinical isolates with confirmed identity and purity were included.
2.3 Microbiological identification and molecular confirmation
Bacterial isolation was performed using standard microbiological techniques. Clinical specimens were cultured on nutrient agar, MacConkey agar, and cetrimide agar and incubated at 37 °C for 24–48 h. Preliminary identification for P. aeruginosa was performed based on colony morphology, pigment production, oxidase test, catalase test, motility, and Gram staining. Confirmatory identification of P. aeruginosa was performed using the Vitek® 2 GN ID card system (bioMérieux, Marcy-l’Étoile, France). Molecular confirmation was performed by amplifying the conserved 16S rDNA gene via polymerase chain reaction (PCR) using previously validated primers (Jarjees et al., 2021). Quality control was ensured using the reference strain P. aeruginosa ATCC 27853.
2.4 Antimicrobial susceptibility testing (AST)
Isolates underwent susceptibility testing using the Vitek® 2 automated system with AST-N419, AST-N222, AST-XN20, or AST-417 cards (bioMérieux, Marcy-l’Étoile, France). Antibiotic classes tested included β-lactams, carbapenems, aminoglycosides, fluoroquinolones, and polymyxins. Where required, disk diffusion testing was performed in accordance with CLSI and EUCAST guidelines. Interpretations were based on minimum inhibitory concentrations (MICs) or inhibition zone diameters, and appropriate standard control strains (e.g., E. coli ATCC 25922, P. aeruginosa ATCC 27853) were included.
2.4.1 Definition of key variables
Multidrug-resistant (MDR) P. aeruginosa was defined as resistance to at least one agent in three or more antimicrobial categories, according to international consensus definitions. Carbapenem resistance was defined as resistance to at least one carbapenem (imipenem or meropenem), based on established clinical breakpoints. Prior carbapenem exposure referred to documented administration of any carbapenem within the 90 days preceding the date of culture collection. Mortality was defined as death occurring during the same hospitalization in which the P. aeruginosa infection was confirmed.
2.5 Molecular detection of blaVIM and blaNDM genes
2.5.1 DNA extraction
Bacterial DNA was extracted from isolates cultured in brain heart infusion broth using the Bacterial DNA Preparation Kit (Jena Bioscience, Germany). The concentration and purity of the extracted DNA were assessed using a NanoDrop™ spectrophotometer (Thermo Scientific, USA). DNA samples were stored at −20 °C until further use.
2.5.2 PCR amplification
PCR amplification of blaVIM (390 bp), blaNDM (621 bp), and 16S rDNA (956 bp) genes was performed in a final volume of 25 μL using a Techne thermal cycler (UK). Each reaction mixture contained 12.5 μL GoTaq® Green Master Mix (Promega, USA), 3 μL of genomic DNA template, 1.5 μL of each primer (10 μM; Macrogen, South Korea), and 6.5 μL of DNase/RNase-free water (Promega, USA). A positive control (P. aeruginosa ATCC 27853) and a negative control were included in all PCR runs. Primers were selected based on previously published sequences (Kazemian et al., 2019; Jarjees et al., 2021). Thermocycling parameters and primer sequences are detailed in Supplementary Table S1.
2.5.3 Agarose gel electrophoresis
PCR amplicons were separated on 2% agarose gels (Norgen Biotek, Canada) prepared in 1 × TBE buffer (Promega, USA). Gels were stained with Safe DNA Stain (SolarBio, China) and visualized using a UV transilluminator (Syngene, UK). A 1 kb DNA ladder (FroggaBio, Canada) was included in each run to estimate band sizes. The presence of target amplicons was confirmed by comparing observed bands with the expected product sizes. A complete list of laboratory instruments and Chemicals used is available in Supplementary Table S2.
2.6 Statistical analysis
All statistical analyses were performed using GraphPad Prism version 10.4.2 (GraphPad Software, San Diego, CA, USA). Categorical variables were analyzed using Chi-square or Fisher’s exact test, depending on expected frequencies. Associations between resistance gene carriage (blaVIM, blaNDM) and clinical variables were assessed using Fisher’s exact test.
One-way and two-way ANOVA, followed by Tukey’s post-hoc test, were employed to compare antibiotic resistance rates across patient groups and antibiotic classes. Assumptions of normality and homogeneity of variance were assessed using residual plots and appropriate statistical tests. A two-sided p-value < 0.05 was considered statistically significant. Missing data were handled using pairwise deletion. To reduce inter-site variability, standardized protocols were implemented across all participating laboratories. Effect sizes were not calculated due to the exploratory nature of the study and the relatively small sample size.
2.7 Ethical approval statement
Ethical approval for this study was obtained from the Medical Ethics Committee of Erbil Polytechnic University, Kurdistan Region, Iraq (Approval No. 25/0066 HRE; April 28, 2025). Written informed consent was obtained from all participants or their legal guardians prior to enrollment. The study was conducted in accordance with the principles of the Declaration of Helsinki and applicable institutional guidelines.
3 Results
3.1 Patient demographics and study population
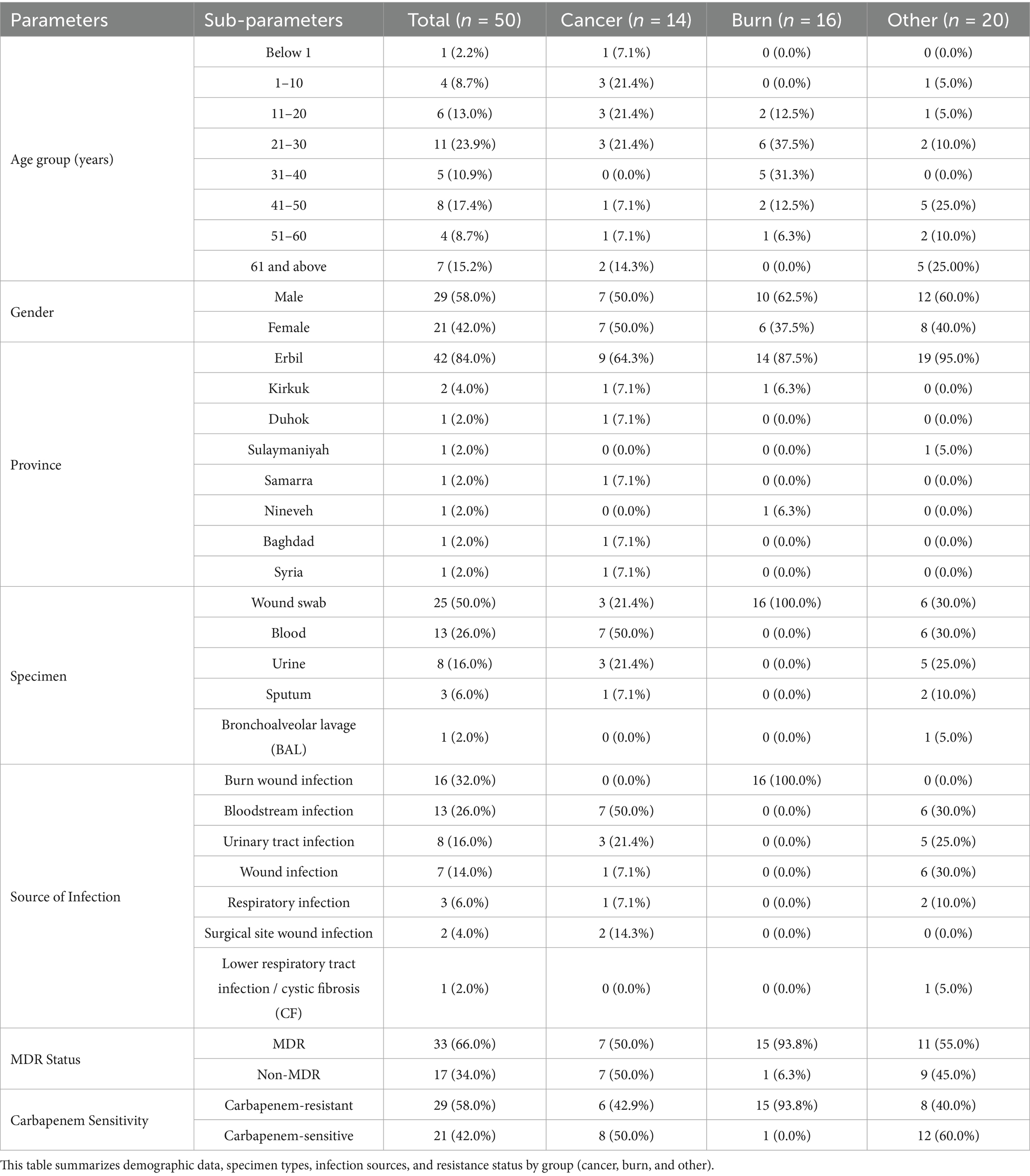
A total of 50 hospitalized immunocompromised patients with culture-confirmed P. aeruginosa infections were enrolled between October 2024 and June 2025. The cohort included 29 males (58.0%) and 21 females (42.0%), with the most common age range being 21–30 years (23.9%). Patients were stratified into three clinical groups: cancer (n = 14), burn (n = 16), and other immunocompromised conditions (n = 20). Figure 1 illustrates the patient screening and group allocation process. Detailed demographic and clinical characteristics are summarized in Table 1 and visualized in Supplementary Figures S1–S3.
Complete clinical and exposure data were available for the burn and cancer groups. However, for the “other” immunocompromised group, data on prior carbapenem exposure and mortality outcomes were not recorded. These missing values (n = 20 each) were excluded from the relevant analyses, as noted in the corresponding tables and figures.
Among the 50 isolates, 29 (58.0%) were classified as carbapenem-resistant P. aeruginosa (CRPA) and were selected for molecular analysis. A total of 40 isolates—including CRPA, non-CRPA, and one control strain—were subjected to PCR detection of blaVIM and blaNDM genes.
3.2 Clinical specimens and infection sources
Wound swabs accounted for the majority of specimens (50.0%), followed by blood (26.0%) and urine (16.0%). The most frequent infection source was burn wound infections (32.0%), followed by bloodstream (26.0%) and urinary tract infections (16.0%) (Table 1; Supplementary Figures S4, S5). Representative wound infections, including percutaneous endoscopic gastrostomy (PEG) site involvement, are shown in Supplementary Figure S6. Supplementary Figures S7 and S8 display P. aeruginosa cultures grown on Cetrimide Agar under UV illumination and the corresponding colony morphology, respectively.
Detailed characteristics of burn injuries—including mechanism of injury, burn degree, total body surface area (TBSA) affected, anatomical distribution, season, place of occurrence, and reason for admission—are summarized in Supplementary Table S3.
3.3 Antimicrobial resistance profiles
The highest resistance rates were observed for ceftolozane/tazobactam (84.2%), ceftazidime/avibactam (81.6%), and ceftazidime (73.5%). In contrast, colistin demonstrated the lowest resistance rate (5.0%). Among clinical subgroups, burn patients exhibited the highest resistance levels, followed by the cancer and other immunocompromised groups (Table 2; Figure 2). Comprehensive antimicrobial susceptibility data—including minimum inhibitory concentration (MIC) distributions and phenotypic resistance profiles—are provided in Supplementary Table S4.
3.4 Multidrug resistance and carbapenem resistance
Overall, 66.0% of P. aeruginosa isolates were classified as multidrug-resistant (MDR). MDR prevalence was highest among burn patients (93.8%), compared to cancer patients (50.0%) and the other immunocompromised group (55.0%) (Table 1; Figure 3). Carbapenem resistance followed a similar trend: burn (93.8%), cancer (42.9%), and other (40.0%).
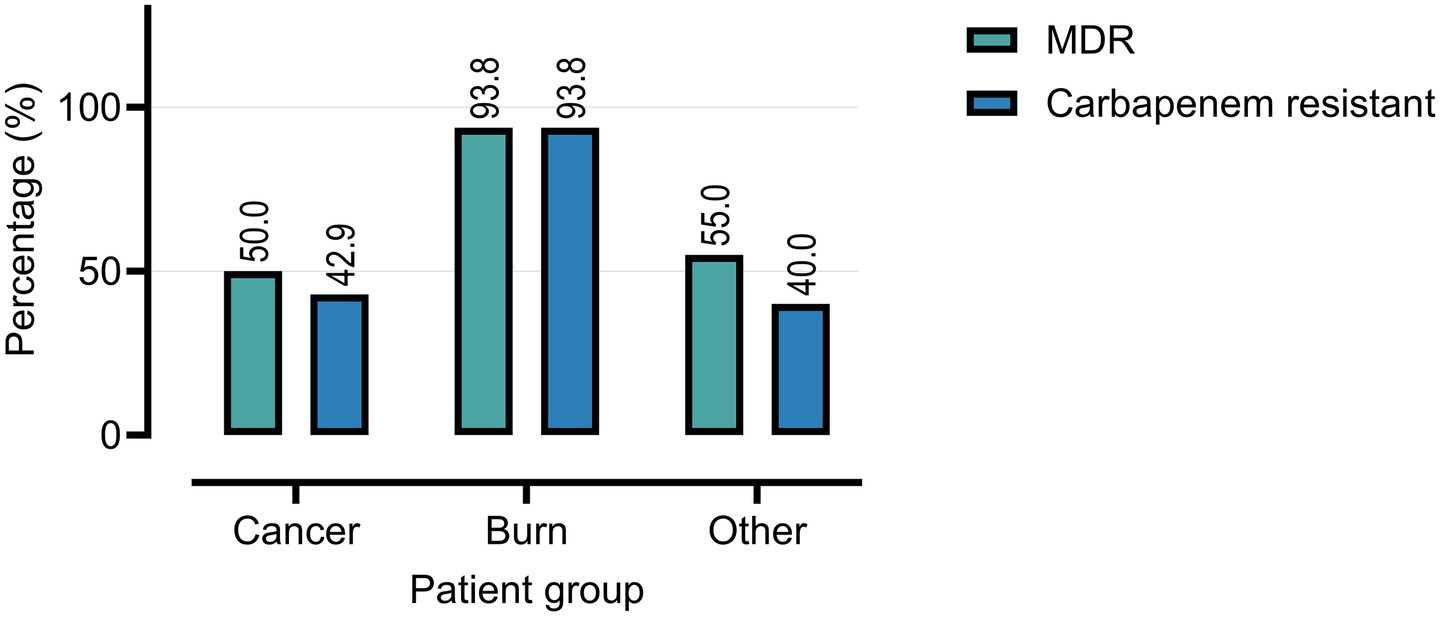
Figure 3. Prevalence of multidrug-resistant (MDR) and carbapenem-resistant P. aeruginosa isolates among clinical samples from cancer, burn, and other patient groups.
Prior carbapenem exposure was significantly associated with carbapenem resistance (76.2% vs. 57.1%, p = 0.0044), but showed no significant association with MDR status (p = 0.8011). Carbapenem-resistant isolates demonstrated significantly higher resistance across nearly all tested antibiotics compared to carbapenem-susceptible isolates (Table 3).

Table 3. Comparison of antibiotic resistance in carbapenem-resistant versus susceptible Pseudomonas aeruginosa (n = 50 each).
Mortality was also significantly higher in patients with prior carbapenem exposure (42.9%) compared to those without (28.6%, p = 0.0392) (Figures 4–6).

Figure 4. Carbapenem resistance is associated with prior use of carbapenems. Analysis includes burn and cancer groups only (n = 30); the other group lacked prior exposure data and was excluded.
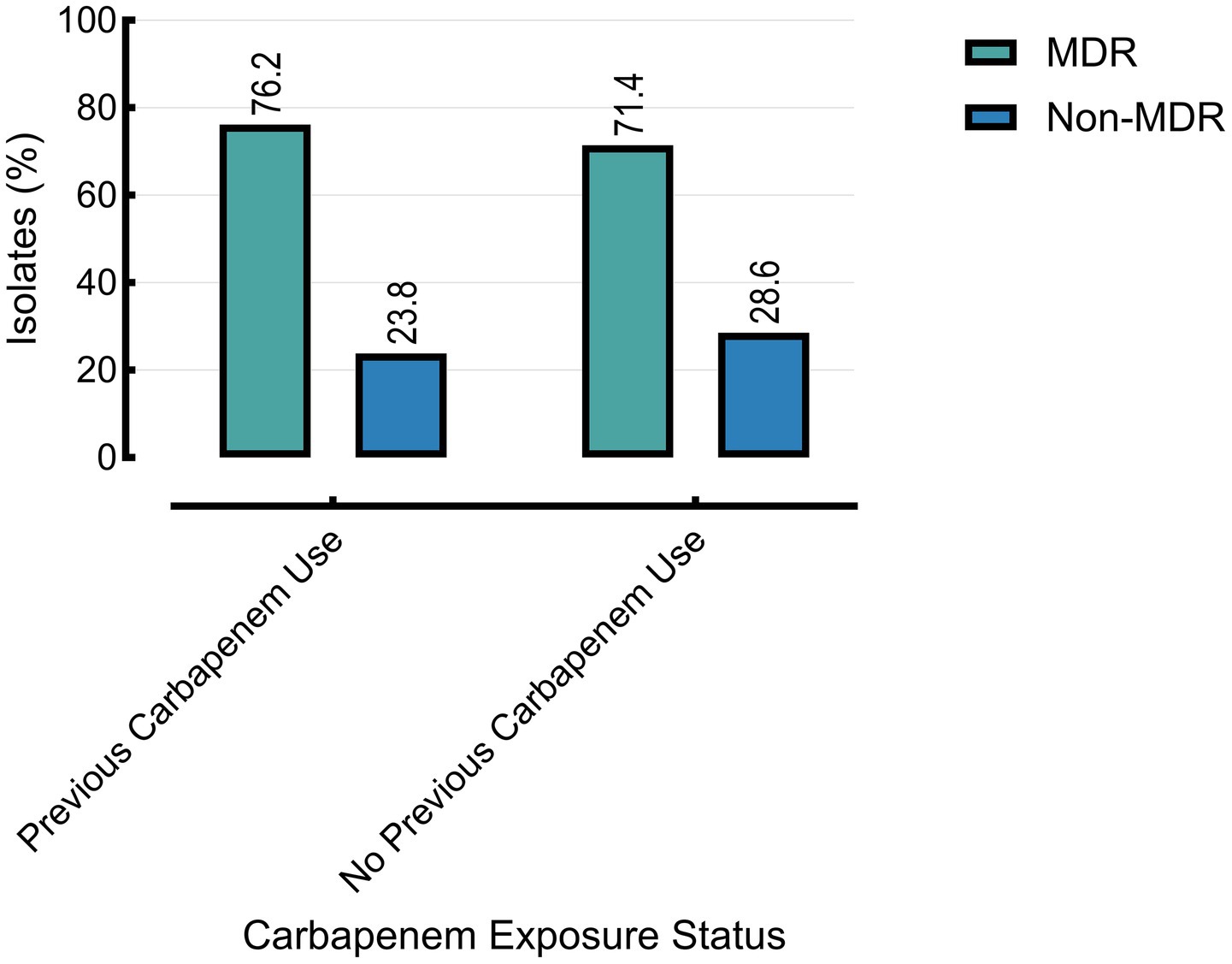
Figure 5. The multidrug-resistant (MDR) status is associated with prior carbapenem use. Other group excluded (n = 20) due to missing data on prior carbapenem exposure.

Figure 6. Mortality rate associated with prior carbapenem use. Analysis limited to burn and cancer groups (n = 30); mortality data were not available for the other group.
3.5 AES classification and resistance phenotypes
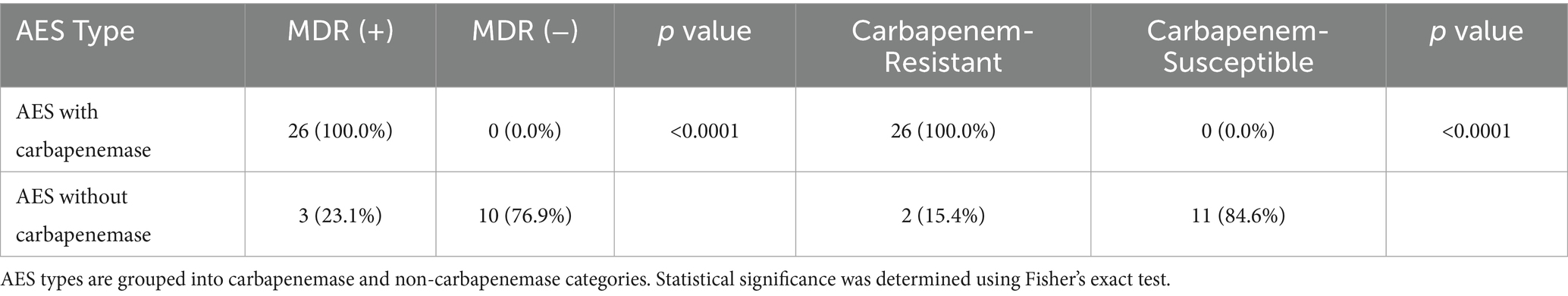
Carbapenemase production was identified in 40 of 50 P. aeruginosa isolates using the VITEK® 2 Advanced Expert System (AES). Among these, 26 isolates (65.0%) were classified as carbapenemase-producing subtypes. These isolates exhibited significantly higher rates of multidrug resistance (100%) and carbapenem resistance, including complete resistance to imipenem and 96.1% resistance to meropenem, compared to non-carbapenemase producers (MDR: 23.1%; carbapenem resistance: 15.4%; p < 0.0001).
Resistance profiles also varied significantly between AES subtypes. β-lactam resistance was higher among carbapenemase producers (p = 0.0014), as was quinolone resistance (p = 0.0016). Notably, colistin resistance was paradoxically higher among non-carbapenemase producers (77.6% vs. 5.0%; p < 0.0001) (Figure 7; Tables 4, 5).
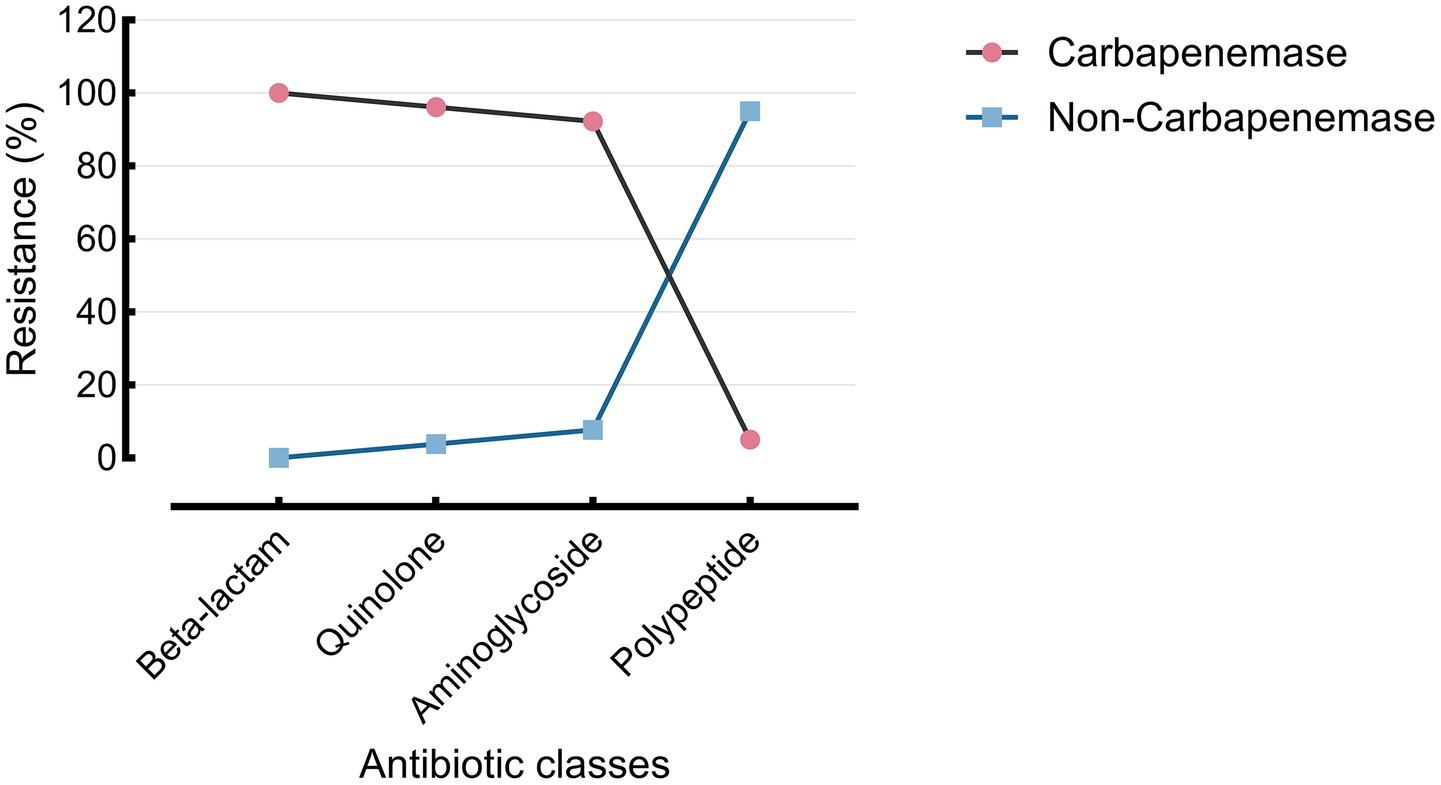
Figure 7. Resistance of the AES-predicted carbapenemase group across four antibiotic classes. Resistance rates across four major antibiotic classes are shown for AES-classified carbapenemase and non-carbapenemase P. aeruginosa isolates. Carbapenemase producers exhibited high resistance to β-lactams (100.0%), quinolones (96.2%), and aminoglycosides (92.3%), but low resistance to polypeptides (5.0%). Non-carbapenemase strains showed lower resistance across all classes, especially to polypeptides (5.0% vs. 77.6%).
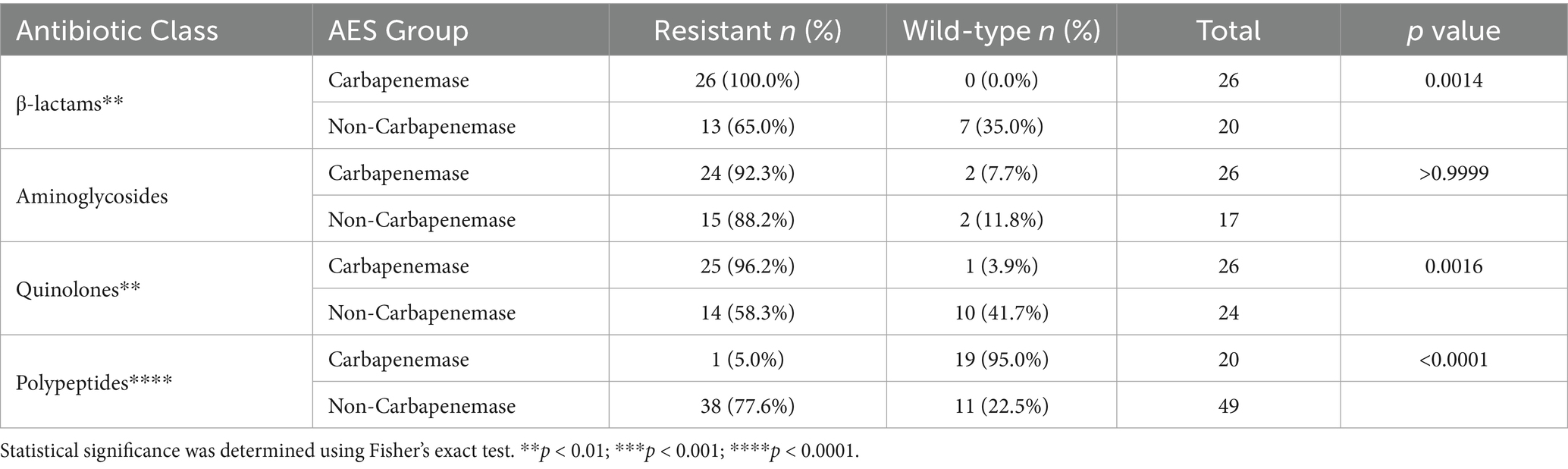
Table 5. Class-specific resistance patterns in AES carbapenemase vs. non-carbapenemase Pseudomonas aeruginosa.
3.6 Resistance patterns by patient group and antibiotic class (two-way ANOVA analysis)
Two-way ANOVA revealed that both antibiotic type (F(10, 20) = 6.979, p = 0.0001; accounting for 37.7% of the total variance) and patient group (F(2, 20) = 47.76, p < 0.0001; accounting for 51.5% of the variance) had statistically significant effects on resistance rates. Spearman’s rank correlation (Rs = −0.1825, p = 0.1547) indicated no evidence of heteroscedasticity; however, residuals failed to meet normality assumptions (p < 0.05 for all tests).
Tukey’s multiple comparison test revealed significantly higher resistance among isolates from burn patients compared to other groups. The most notable difference was observed between ceftazidime resistance in burn patients and colistin susceptibility in the other group (Δ = 119.1; 95% CI: 69.02–169.2; p < 0.0001). Supplementary Table S5 summarizes key pairwise comparisons; full results are available in Supplementary Table S6. An error-bar plot (mean difference ± 95% CI) visualizes selected comparisons such as CAZ: B vs. COL: O and FEP: B vs. COL: O (Figure 8). While some confidence intervals showed partial overlap, both main effects remained statistically robust (p < 0.0001).

Figure 8. Pairwise resistance differences (burn vs. other groups) across selected antibiotics. Mean resistance differences (±95% CI) between burn patients and other clinical groups for selected antibiotics, grouped by class. β-lactams showed consistently higher resistance in burn isolates. Imipenem (carbapenem), ciprofloxacin (quinolone), and gentamicin (aminoglycoside) also demonstrated significantly increased resistance in burn patients compared to colistin sensitivity in other groups (adjusted p < 0.0001).
To further explore trends within antibiotic classes, a stratified two-way ANOVA was performed. These sub-analyses demonstrated that the patient group was the primary source of variance across β-lactams, carbapenems, aminoglycosides, and fluoroquinolones, whereas differences between individual antibiotics within each class were generally not statistically significant. Detailed class-specific ANOVA results—including F-statistics, percentage variance explained, and residual diagnostics—are presented in Supplementary Figures S9–S14.
3.7 Prevalence and distribution of blaVIM and blaNDM genes
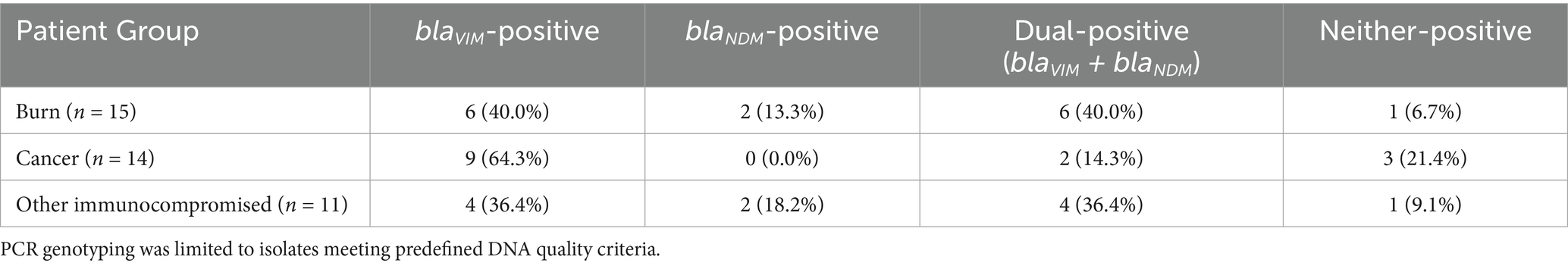
Among the 40 isolates tested by PCR, 19 (47.5%) were positive for blaVIM, 4 (10.0%) for blaNDM, and 12 (30.0%) co-harboured both genes. Five isolates (12.5%) were negative for both genes (Table 6). Gene distribution by patient group is summarised in Table 7. Burn patients showed the highest dual-carriage rate.
3.8 Association of resistance genes with AES and carbapenem phenotypes
No significant association was found between blaVIM presence and AES β-lactam resistance phenotype (p = 0.601) or carbapenem resistance (p = 0.686). However, blaNDM carriage was significantly associated with both AES β-lactam resistance phenotype (p = 0.0169) and carbapenem resistance (p = 0.027) (Supplementary Tables S7 and S8).
3.9 Co-occurrence of blaVIM and blaNDM
Twelve isolates (30.0%) co-harboured both genes. Fisher’s exact test showed no significant association between blaVIM and blaNDM co-occurrence (p > 0.9999) (Supplementary Table S9).
3.10 PCR validation and gel electrophoresis
All isolates were confirmed as P. aeruginosa via 16S rDNA PCR (956 bp). PCR yielded expected amplicons for blaVIM (390 bp) and blaNDM (621 bp). No non-specific bands were observed. Supplementary Figures S15–S20 show gel images for 16S rDNA; Supplementary Figures S21–S24 for blaVIM; and (Supplementary Figures S25, S26) for blaNDM.
4 Discussion
Pseudomonas aeruginosa is a major cause of healthcare-associated infections (HAIs) worldwide and is recognized by the WHO as a critical-priority pathogen due to its intrinsic resistance mechanisms and remarkable capacity to acquire additional resistance determinants (World Health Organization, 2017; Centers for Disease Control and Prevention (CDC), 2019; Flores-Vega et al., 2025). Globally, the prevalence of MDR P. aeruginosa ranges from 15 to 30% in some regions, while emerging evidence from Asia and Africa indicates pooled rates of around 46% (95% CI: 37.1–55.0) (Horcajada et al., 2019; Reyes et al., 2023).
In the Middle East, MDR rates vary considerably—from 75.6% in Egypt to 0% in Morocco, with intermediate rates of 7.3% in Saudi Arabia and 8.1% in Qatar. In the Levant, MDR prevalence ranges from 64.5% in Lebanon to 12.4% in Iraq, with carbapenem resistance increasingly driven by metallo-β-lactamase (MBL) production (Al-Orphaly et al., 2021).
The present study reports an MDR rate of 66.0% and carbapenem resistance of 58.0% among clinical isolates in Erbil, Iraq. Burn patients exhibited the highest burden (93.8% MDR), surpassing international reports, where MDR P. aeruginosa prevalence in burn units ranges from 15.2% in India, 19–23.1% in Pakistan and Brazil, to 64–72% in Algeria, Tanzania, and U.S. tertiary-care burn units (Elsheikh and Makram, 2024). Substantial rates were also observed in cancer patients (MDR: 50.0%, CRPA: 42.9%), consistent with previous reports in oncology populations (Othman et al., 2014; Abdulhak et al., 2025). While these values are lower than those observed in our burn unit, they remain higher than MDR rates reported in oncology settings in some high-income countries, which typically range between 25 and 40% (Horcajada et al., 2019). Elevated β-lactam resistance in cancer patients likely reflects frequent healthcare interactions, extended courses of broad-spectrum antibiotic therapy, neutropenia, chemotherapy-induced mucositis, immunosuppression, or institutional differences in infection prevention and control practices (Gottesdiener and Satlin, 2023).
Our burn unit data align with international reports that identify burn units as high-risk environments for MDR pathogen emergence due to frequent invasive procedures, immunosuppression, and heavy empirical antibiotic use (Hmissi et al., 2023; Ullah et al., 2023). Compared to the 64.5% MDR rate reported in Lebanon and 52.5% in Jordan (Al-Orphaly et al., 2021), our burn patient MDR prevalence is notably higher, underscoring the urgent need for strengthened infection prevention and control (IPC) in such units.
At the molecular level, the co-carriage of blaVIM and blaNDM genes was observed in 30% of PCR-tested isolates, with the highest prevalence among burn patients. Although blaVIM alone did not show a statistically significant association with phenotypic carbapenem resistance (p = 0.686), blaNDM carriage was significantly correlated with carbapenem resistance (p = 0.027). These findings align with prior studies identifying blaNDM as a key driver of carbapenem resistance (Alsaadi et al., 2020; Seyedi et al., 2022). The lack of association for blaVIM may reflect variability in expression, compensatory resistance mechanisms, or regional strain differences.
The VITEK® 2 Advanced Expert System (AES) provided additional insights into phenotypic resistance profiles. All AES-classified carbapenemase-producing isolates were also MDR and exhibited near-universal resistance to both imipenem (100%) and meropenem (96.1%), supporting its diagnostic utility in resource-limited settings where molecular testing may not be readily available (Chan and Leroi, 2021; Hackel and Bailey-Person, 2025; Laço et al., 2025).
The two-way ANOVA and Tukey’s post-hoc test revealed significant differences in resistance levels across patient groups and antibiotics. Burn patients exhibited significantly higher resistance to β-lactams and carbapenems, particularly ceftazidime, cefepime, and imipenem (p < 0.0001). While prior carbapenem exposure was significantly associated with carbapenem resistance (p = 0.0044), it was not predictive of MDR status (p = 0.8011). This suggests that intrinsic chromosomal mechanisms—such as AmpC β-lactamase overexpression, OprD porin loss, and efflux pump upregulation—may play a more dominant role in the development of MDR than antimicrobial exposure alone (Karakonstantis et al., 2020).
Moreover, the mortality rate was significantly higher among patients with prior carbapenem exposure (42.9%) compared to those without (28.6%, p = 0.0392), highlighting the clinical impact of antimicrobial resistance. While promising strategies such as reverse vaccinology and epitope-based vaccine design are under investigation (Fereshteh et al., 2023; Zhu et al., 2025), their translation into clinical practice remains distant. In the interim, robust surveillance, targeted antimicrobial stewardship, and enhanced infection control measures are critical to reducing CRPA transmission and associated morbidity and mortality.
5 Conclusion
Our study demonstrates the substantial burden of multidrug-resistant and carbapenem-resistant P. aeruginosa in immunocompromised patients, particularly those in burn units. The high frequency of resistance—especially among AES-classified carbapenemase producers and blaNDM-positive strains—highlights the need for urgent and targeted intervention. In resource-limited settings, the AES system offers a valuable surrogate for molecular testing, supporting rapid treatment decisions and infection control efforts. Integration of advanced diagnostics with routine susceptibility testing and molecular surveillance is essential to mitigate the growing threat of CRPA in healthcare settings.
Data availability statement
The raw data supporting the conclusions of this article, including MIC distributions, resistance profiles, pairwise comparisons, full statistical results, and gene -resistance associations (Tables S4-S9, including the.xlsx dataset), have been deposited in Figshare and are publicly available at: https://doi.org/10.6084/m9.figshare.29919473. Additional requests for data can be directed to the corresponding author at YmFocmEuaGFtYWRAZXB1LmVkdS5pcQ==.
Ethics statement
The studies involving humans were approved by Medical Ethics Committee of Erbil Polytechnic University, Erbil, Iraq (Approval No.: 25/0066 HRE; Date of Approval: 28 April 2025). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
BH: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. MM: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors gratefully acknowledge Dr. Dilshad Shexani, Specialist in General Surgery, GIT and Hepatobiliary Surgery at Rizgary Teaching Hospital and Zheen International Hospital, for his clinical collaboration and assistance in patient identification and access for sample collection. Appreciation is also extended to the physicians and clinical staff at participating hospitals in Erbil for their contributions to patient care and data provision. All individuals acknowledged have provided written permission to be named.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that Gen AI was used in the creation of this manuscript. Generative AI (ChatGPT, OpenAI) was used to assist in language editing, grammar improvement, and formatting suggestions during the preparation of this manuscript. The authors reviewed, verified, and take full responsibility for all content generated using AI tools.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1672531/full#supplementary-material. The complete dataset, including all supplementary figures and tables, has also been deposited in Figshare and is publicly available at: https://doi.org/10.6084/m9.figshare.29919473.
The Supplementary material includes 26 figures and 9 tables. Figures S1–S5 present demographic and clinical data; Figure S6 shows wound characteristics in cancer patients. Figures S7–S8 depict microbiological findings on cetrimide agar. Figures S9–S14 summarise statistical analyses of antibiotic resistance. Figures S15–S20 confirm 16S rDNA amplification by PCR. Figures S21–S24 and S25–S26 show amplification of blaVIM and blaNDM genes, respectively, with appropriate controls.
Table S1 provides thermocycling parameters and primer sequences. Table S2 lists laboratory instruments and chemicals. Burn injury characteristics are summarised in Table S3. Table S4 presents MIC distributions and resistance profiles. Table S5 outlines key pairwise comparisons; full results are in Table S6 (.xlsx). Tables S7 and S8 explore associations between blaVIM, blaNDM, AES phenotypes, and carbapenem resistance. Table S9 reports non-significant blaVIM, blaNDM co-occurrence.
References
Abdulhak, A., Zedan, H. H., El-Mahallawy, H. A., Sayed, A. A., Mohamed, H. O., and Zafer, M. M. (2025). Multidrug-resistant Pseudomonas aeruginosa in immunocompromised cancer patients: epidemiology, antimicrobial resistance, and virulence factors. BMC Infect. Dis. 25:804. doi: 10.1186/s12879-025-11182-0
Al-Orphaly, M., Hadi, H. A., Eltayeb, F. K., Al-Hail, H., Samuel, B. G., Sultan, A. A., et al. (2021). Epidemiology of multidrug-resistant Pseudomonas aeruginosa in the Middle East and North Africa region. mSphere 6:e00202-21. doi: 10.1128/mSphere.00202-21
Alsaadi, L. A., Al-Dulaimi, A., and Al-Taai, H. (2020). Prevalence of blaVIM, blaIMP and blaNDM genes in carbapenem-resistant Pseudomonas aeruginosa isolated from different clinical infections in Diyala, Iraq. Indian J. Public Health Res. Dev. 11:2264. doi: 10.37506/v11/i2/2020/ijphrd/195173
Brkic, S., and Cirkovic, I. (2024). Carbapenem-resistant Enterobacterales in the Western Balkans: Addressing gaps in European AMR surveillance map. Antibiotics (Basel) 13. doi: 10.3390/antibiotics13090895
Centers for Disease Control and Prevention (CDC) (2019). Antibiotic resistance threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services. Available online at: https://www.cdc.gov/antimicrobial-resistance/data-research/threats/index.html (Accessed 22 June 2025).
Chan, E., and Leroi, M. (2021). Evaluation of the VITEK 2 advanced expert system performance for predicting resistance mechanisms in Enterobacterales acquired from a hospital-based screening program. Pathology 53, 763–767. doi: 10.1016/j.pathol.2021.01.009
Elsheikh, R., and Makram, A. M. (2024). Multidrug-resistant organisms: the silent plight of burn patients. J. Burn Care Res. 45, 877–886. doi: 10.1093/jbcr/irae075
Fereshteh, S., Haririzadeh Jouriani, F., Noori Goodarzi, N., Torkamaneh, M., Khasheii, B., and Badmasti, F. (2023). Defeating a superbug: a breakthrough in vaccine design against multidrug-resistant Pseudomonas aeruginosa using reverse vaccinology. PLoS One 18:e0289609. doi: 10.1371/journal.pone.0289609
Flores-Vega, V. R., Partida-Sanchez, S., Ares, M. A., Ortiz-Navarrete, V., and Rosales-Reyes, R. (2025). High-risk Pseudomonas aeruginosa clones harbouring β-lactamases: 2024 update. Heliyon 11:e41540. doi: 10.1016/j.heliyon.2024.e41540
Gottesdiener, L. S., and Satlin, M. J. (2023). Global impact of antibacterial resistance in patients with hematologic malignancies and hematopoietic cell transplant recipients. Transpl. Infect. Dis. 25:e14169. doi: 10.1111/tid.14169
Grace, A., Sahu, R., Owen, D. R., and Dennis, V. A. (2022). Pseudomonas aeruginosa reference strains PAO1 and PA14: A genomic, phenotypic, and therapeutic review. Frontiers in Microbiology, 13. doi: 10.3389/fmicb.2022.1023523
Hackel, M., and Bailey-Person, M. (2025). Performance of the VITEK®2 advanced expert system (AES) as a rapid aid for reporting antimicrobial susceptibility testing in Pseudomonas aeruginosa and Acinetobacter spp. Open Forum Infect. Dis. 12:ofae631.2258. doi: 10.1093/ofid/ofae631.2258
Hasan, S. A., Raoof, W. M., and Ahmed, K. K. (2024a). Antibacterial activity of deer musk and Ziziphus spina-christi against carbapebem resistant gram negative bacteria isolated from patients with burns and wounds. Regul. Mech. Biosyst. 15, 267–278. doi: 10.15421/022439
Hasan, S. A., Raoof, W., and Ahmed, K. (2024b). First report of co-harboring bleomycin resistance gene (bleMBL) and carbapenemase resistance gene (blaNDM-1) Klebsiella pneumoniae in Iraq with comparison study among the sensitivity test, the BD Phoenix CPO detect test, and the RAPIDEC® CARBA NP test. Sib. J. Life Sci. Agric. 16, 208–237. doi: 10.12731/2658-6649-2024-16-4-1249
Hmissi, S., Raddaoui, A., Frigui, S., Abbassi, M. S., Achour, W., Chebbi, Y., et al. (2023). Detection of carbapenem-resistant Pseudomonas aeruginosa co-harbouring blaVIM-2 and blaGES-5 in burn patients. Acta Microbiol. Immunol. Hung. 70, 199–205. doi: 10.1556/030.2023.02089
Horcajada, J. P., Montero, M., Oliver, A., sorlí, L., Luque, S., Gómez-Zorrilla, S., et al. (2019). Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 32:e00031-19. doi: 10.1128/CMR.00031-19
Hu, Z., Zhou, L., Tao, X., Li, P., Zheng, X., Zhang, W., et al. (2024). Antimicrobial resistance survey and whole-genome analysis of nosocomial P. aeruginosa isolated from eastern province of China in 2016–2021. Ann. Clin. Microbiol. Antimicrob. 23:12. doi: 10.1186/s12941-023-00656-1
Huang, Q., Yan, K., and Li, G. (2025). Molecular characterization of virulent genes in Pseudomonas aeruginosa based on componential usage divergence. Sci. Rep. 15:11246. doi: 10.1038/s41598-025-95579-6
Jarjees, K., Jarjees, R., and Qader, G. (2021). Detection of blaCTX-M genes among extended spectrum beta lactamase producing Pseudomonas aeruginosa isolated from clinical specimens in Erbil. Indian J. Pharm. Sci. 83, 275–282. doi: 10.36468/pharmaceutical-sciences.spl.361
Karakonstantis, S., Kritsotakis, E. I., and Gikas, A. (2020). Treatment options for K. pneumoniae, P. Aeruginosa and A. baumannii co-resistant to carbapenems, aminoglycosides, polymyxins and tigecycline: an approach based on the mechanisms of resistance to carbapenems. Infection 48, 835–851. doi: 10.1007/s15010-020-01520-6
Kazemian, H., Heidari, H., Ghanavati, R., Ghaffourian, S., Yazdani, F., Sadeghifard, N., et al. (2019). Phenotypic and genotypic characterization of ESBL-, AmpC-, and carbapenemase-producing Klebsiella pneumoniae and Escherichia coli isolates. Med. Princ. Pract. 28, 547–551. doi: 10.1159/000500311
Laço, J., Martorell, S., Gallegos, M. D. C., and Gomila, M. (2025). Yearlong analysis of bacterial diversity in hospital sink drains: culturomics, antibiotic resistance and implications for infection control. Front. Microbiol. 15:1501170. doi: 10.3389/fmicb.2024.1501170
Moradali, M. F., Ghods, S., and Rehm, B. H. A. (2017). Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 7:39. doi: 10.3389/fcimb.2017.00039
Othman, N., Babakir-Mina, M., Noori, C. K., and Rashid, P. Y. (2014). Pseudomonas aeruginosa infection in burn patients in Sulaimaniyah, Iraq: risk factors and antibiotic resistance rates. J. Infect. Dev. Ctries. 8, 1498–1502. doi: 10.3855/jidc.4707
Reyes, J., Komarow, L., Chen, L., Ge, L., Hanson, B. M., Cober, E., et al. (2023). Global epidemiology and clinical outcomes of carbapenem-resistant Pseudomonas aeruginosa and associated carbapenemases (POP): a prospective cohort study. Lancet Microbe 4, e159–e170. doi: 10.1016/S2666-5247(22)00329-9
Rossi, E., Ghoul, M., and La Rosa, R. (2022). Editorial: Pseudomonas aeruginosa pathogenesis: virulence, antibiotic tolerance and resistance, stress responses and host-pathogen interactions. Front. Cell. Infect. Microbiol. 12:860314. doi: 10.3389/fcimb.2022.860314
Schwartz, B., Klamer, K., Zimmerman, J., Kale-Pradhan, P. B., and Bhargava, A. (2024). Multidrug resistant Pseudomonas aeruginosa in clinical settings: a review of resistance mechanisms and treatment strategies. Pathogens 13:975. doi: 10.3390/pathogens13110975
Seyedi, M., Yousefi, F., Naeimi, B., and Tajbakhsh, S. (2022). Phenotypic and genotypic investigation of metallo-β-lactamases in Pseudomonas aeruginosa clinical isolates in Bushehr, Iran. Iran. J. Basic Med. Sci. 25, 1196–1200. doi: 10.22038/IJBMS.2022.64359.14154
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Ullah, R., Amir, M., Anjum, S., Ur Rehman, M., Noorul Hasan, T., Sajjad Naqvi, S., et al. (2023). Presence of T3SS (exoS, exoT, exoU and exoY), susceptibility pattern and MIC of MDR-Pseudomonas aeruginosa from burn wounds. J. Infect. Dev. Ctries. 17, 1130–1137. doi: 10.3855/jidc.17580
Vaez, H., Khademi, F., Salehi-Abargouei, A., and Sahebkar, A. (2018). Metallo-β-lactamase-producing Pseudomonas aeruginosa in Iran: a systematic review and meta-analysis. Infez. Med. 26, 216–225.
Wood, S. J., Kuzel, T. M., and Shafikhani, S. H. (2023). Pseudomonas aeruginosa: infections, animal modeling, and therapeutics. Cells 12:199. doi: 10.3390/cells12010199
World Health Organization (2017). WHO publishes list of bacteria for which new antibiotics are urgently needed. Geneva: WHO. Available online at: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (Accessed 22 June 2025).
Keywords: Pseudomonas aeruginosa , multidrug resistance, carbapenem resistance, carbapenemase, blaVIM , blaNDM , cancer, burn
Citation: Hamad BK and Mahmud MA (2025) Molecular detection of blaVIM and blaNDM in multidrug-resistant Pseudomonas aeruginosa from cancer and burn patients in Erbil, Iraq. Front. Microbiol. 16:1672531. doi: 10.3389/fmicb.2025.1672531
Edited by:
Ziad Daoud, My Michigan Health System, United StatesReviewed by:
Sarah A. Hasan Babani, University of Kirkuk, IraqReem Polse, University of Zakho, Iraq
Copyright © 2025 Hamad and Mahmud. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bahra Kakamin Hamad, YmFocmEuaGFtYWRAZXB1LmVkdS5pcQ==
 Bahra Kakamin Hamad
Bahra Kakamin Hamad Muayad Ahmed Mahmud
Muayad Ahmed Mahmud