- 1School of Biosciences, University of Nottingham, Sutton Bonington Campus, Sutton Bonington, United Kingdom
- 2Department of Biology, Ineos Oxford Institute for Antimicrobial Research (IOI), University of Oxford, Oxford, United Kingdom
- 3Department of Medical Microbiology, Division of Infection and Immunity, Cardiff University, Cardiff, United Kingdom
- 4School of Veterinary Science and Medicine, University of Nottingham, Sutton Bonington Campus, Sutton Bonington, United Kingdom
- 5Department of Mathematics and Applied Mathematics, University of Johannesburg, Johannesburg, South Africa
Third-generation cephalosporin-resistant Enterobacterales are ranked second on the World Health Organisation (WHO)’s Bacterial Priority Pathogens List. Amongst them, extended-spectrum β-lactamase-producing Escherichia coli (ESBL-Ec) are used by the WHO as sentinel organisms to monitor the spread of antibiotic resistance worldwide and are often associated with mobilisable multidrug resistance (MDR). However, we know less about how ESBL-producing genes spread in environmental E. coli. This study investigates how the blaCTX-M-15 gene from ESBL-Ec isolated on a UK dairy farm could transfer between strains. For this study, 39 E. coli were isolated from a single dairy farm over 4 months, using cefotaxime-supplemented selective media. All had similar antibiotic susceptibility test phenotypes, and PCR, whole genome sequencing (WGS), and resistance gene transmission experiments demonstrated they were all closely related. In silico multi-locus sequence typing and single-nucleotide polymorphism analysis showed that all 39 strains were Sequence Type 2325, but plasmid carriage differed. In total, 35 of the 39 ESBL-Ec strains were multidrug resistant, displaying blaCTX-M type cephalosporin resistance and resistance to fluoroquinolones and tetracyclines. WGS confirmed all 39 isolates carried the ISEcp1 mobile genetic element carrying the blaCTX-M-15 ESBL-producing gene, and the qnrS1 partial quinolone resistance gene in the chromosome. A total of 35 strains also carried tetAR within this ISEcp1 element. We found that sub-lethal levels of ampicillin, cloxacillin, and ceftazidime could enhance the transfer of ISEcp1 blaCTX-M-15 from the chromosome of these dairy farm strains into endogenous self-transmissible plasmids, which can themselves then transfer into and confer phenotypic antibiotic resistance in a recipient E. coli K-12 strain. In conclusion, we observed not only clonal dissemination of these environmentally occurring ESBL-producing strains within the farm environment but also showed experimentally that these strains had the ability to mobilise their ESBL producing genes, and that these and other resistance genes can be acquired or lost on transfer. This shows the importance of longitudinal monitoring of antibiotic resistance, especially in places with high prevalence or selective pressure for resistant bacteria.
1 Introduction
The World Health Organisation (WHO) has identified third-generation cephalosporin-resistant Enterobacterales as critical priority pathogens (WHO, 2024) and uses extended spectrum β-lactamase producing Escherichia coli (ESBL-Ec) as sentinel organisms in its Tricycle protocol for global AMR surveillance (WHO, 2021). ESBL-Ec is an important antimicrobial-resistant (AMR) organism due to its resistance to penicillin and cephalosporin antibiotics and because of the rapid worldwide spread of blaCTX-M type ESBLs (Cantón and Coque, 2006; Cantón et al., 2012), including in multi-drug-resistant clinical Enterobacterial strains (Ibrahim et al., 2016). The association of ESBL-producing blaCTX-M genes with mobile genetic elements such as plasmids and transposons has led to transmission and acquisition of resistance to human critical cephalosporin antibiotics in Enterobacterales in a wide range of human and animal environments, including in high-intensity livestock production such as dairy farms (Eckert et al., 2004; Liebana et al., 2013; Irrgang et al., 2017).
The insertion sequence (IS) element, ISEcp1, has played a key role in the acquisition and dissemination of blaCTX-M from the posited original source of this resistance, the soil-associated bacterium Kluyvera ascorbata (Humeniuk et al., 2002; Rossolini et al., 2008; Zong et al., 2010; Bevan et al., 2017; Afema et al., 2018). ISEcp1 assists DNA adjacent to where it has inserted in chromosomes or plasmids to move to other places in the genome, via a one-ended transposition mechanism that produces 5-bp target site duplications at the point the element inserts (Poirel et al., 2005; Kieffer et al., 2020). ISEcp1 is defined by and flanked by 14-bp Inverted Repeat left (IRL) and degenerate right IR sequences (IRR). Due to the recognition of an imperfect IRR sequence by ISEcp1, it can mobilise different-sized transposition units (Poirel et al., 2005) because downstream genes can be collected or lost as mobilisation takes place (Zong et al., 2010). ISEcp1 also provides the −35 and −10 promoter sequences for high-level expression of blaCTX-M (Poirel et al., 2005; Zong et al., 2010).
Previous studies have shown successful transposition of a cloned ISEcp1 in K. ascorbata from a chromosomal location into a plasmid that had been introduced into the strain, with ISEcp1 transposition enhanced in the presence of several β-lactam antibiotics (Lartigue et al., 2006; Nordmann et al., 2008). A further study by Hamamoto et al. (2020) demonstrated the transposition of ISEcp1 carrying blaCTX-M-14, from a plasmid construct location to a chromosomal location within an experimental E. coli strain. To the best of our knowledge, no published studies have explored transposition of a naturally occurring chromosomally encoded ISEcp1 or have addressed the question of whether endogenous plasmids in the host strain could then transfer mobilised ISEcp1 into a recipient strain and thus provide phenotypic cephalosporin resistance to it. Although previous studies have demonstrated enhanced transposition of ISEcp1 in the presence of ceftazidime, cefotaxime, and piperacillin (Lartigue et al., 2006; Nordmann et al., 2008), the presence of sub-lethal levels of other β-lactam antibiotics, such as those that might be encountered in therapeutically-treated dairy cattle, needs to be examined for their potential in promoting resistance dissemination via this mechanism.
The aim of this study was to investigate the carriage, dissemination, and transmission of blaCTX-M resistance in E. coli isolates from a UK dairy farm. Previously described Antimicrobial Susceptibility Testing (AST) data (Baker et al., 2022) of a collection of 811 confirmed E. coli dairy farm strains were used to select a subset of 39 ESBL isolates with a blaCTX type resistance phenotype. We used PCR to confirm the presence of ISEcp1 and blaCTX-M-15 in the 39 ESBL-Ec, which were then fully characterised by whole genome sequencing (WGS). We then interrogated whether the ISEcp1 element in these strains could transpose blaCTX-M-15 from the chromosome into their endogenous conjugative plasmids and then transfer cefotaxime resistance to a recipient E. coli strain. Next, we investigated whether the presence of sub-lethal levels of β-lactam antibiotics, such as those that could be encountered in therapeutically treated dairy cattle, might promote transfer of ISEcp1 blaCTX-M-15 resistance. In summary, we characterised a group of naturally occurring ESBL-Ec isolated over a 4-month period from a single dairy farm and then demonstrated that they can transfer antibiotic resistance into other E. coli strains.
2 Materials and methods
2.1 Bacterial strains and growth conditions
The 39 E. coli dairy farm isolates were chosen from a larger collection of 811 E. coli strains, taken as part of an integrated AMR study. Isolation and initial characterisation of strains are described in Baker et al. (2022) and Todman et al. (2024). Supplementary Table S1 shows the farm strains studied, date of isolation, sampling location, the selective media used for initial isolation, and the resistance profiles from the disc diffusion assay data supplied in Baker et al. (2022).
The 39 strains were selected on the basis of their ESBL type phenotype using the CLSI standard (CLSI, 2018) disc diffusion assays, which included resistance to the semi-synthetic penicillin, ampicillin (AMP), and to the cephalosporin, cefotaxime (CTX), but with susceptibility to the β-lactam/β-lactamase inhibitor combination amoxicillin/clavulanic acid (AMC) and to the cephamycin cefoxitin (FOX). A separate, but closely related ESBL isolate, EcoSL3110-774 (strain 774), with a similar antimicrobial susceptibility test (AST) profile, which was sequenced using PacBio as part of the dairy farm study (Baker et al., 2022), and it was also analysed in detail as part of this study. Strain 774 was found to contain an ISEcp1 blaCTX-M-15 element, and as this was complete and within one contig, this allowed for a detailed genetic environment to be constructed; however, no plasmids were found in strain 774.
The kanamycin (KAN) and rifampicin (RIF)-resistant E. coli K-12 strain CV601 encoding green fluorescent protein (GFP) (Smalla et al., 2000) was used as the recipient in all conjugation assays.
For all PCR and genetic work and reviving strains for AST assays, all strains were revived on or in either solid or liquid Lysogeny Broth (LB) Miller media (Sigma-Aldrich, USA) and at 37°C overnight for between 18 and 20 h. When revived in LB, cultures were grown overnight for 18–20 h at 37 °C with agitation at 180 RPM (Medline Scientific™ ISF-7100 Floor Standing Incubator Shaker, Fisher Scientific, UK).
2.2 Minimum inhibitory concentration (MIC) agar dilution assays
The 39 isolates were further characterised phenotypically using agar dilution MIC assays according to CLSI guideline methods (CLSI, 2018) but using EUCAST susceptibility breakpoints (EUCAST, 2022) (where available). A panel of 25 antibiotics was used in the MIC assays. These are listed in Supplementary Table S2 along with the concentration ranges tested for each antibiotic, the breakpoint utilised, and full details relating to the antibiotic discs used by Baker et al. (2022) in disc diffusion tests listed in Supplementary Table S3.
2.3 DNA extraction and purification
Total DNA for PCR was isolated from E. coli strains using the simple boiling method as previously described (Wei, 2013). PCR products requiring Sanger sequencing were purified using a NEB T1030 Monarch® PCR & DNA Cleanup kit according to the manufacturer’s instructions (NEB, USA). Genomic DNA (gDNA) extraction was conducted at the University of Cardiff, using a Qiagen QIAamp DNA Mini QIAcube kit using the QIAcube platform (QIAGEN, Germany) with an additional RNAase step. The gDNA was quantified using a Qubit v4.0 (Thermo Fisher Scientific, Loughborough, UK). The gDNA extracted was used to generate libraries for both the MiSeq (Illumina, USA) short read and MinION long read (Oxford Nanopore Technologies, UK) sequencing.
2.4 PCR
PCR experiments were used to determine the presence of ISEcp1 and blaCTX-M genes in isolates with a CTX-M type antibiotic resistance profile, and to confirm the presence of gfp in CV601 transconjugants. DreamTaq Green Mastermix (ThermoFisher Scientific, Loughborough, UK) was used according to the manufacturer’s instructions. Oligonucleotides were synthesised by Eurofins Genomics (Ebersberg, Germany) and are listed in Table 1. Oligonucleotides were used at a final working concentration of 10 pmol μl−1. All PCR conditions consisted of an initial denaturation at 95 °C for 5 min, followed by 30 cycles of 94 °C for 1 min, an annealing temperature (TA), annealing time, and extension at 72 °C specific to each gene (Table 1), and a final incubation at 72 °C for 10 min. PCR amplification of the blaCTX-M gene used the primers CTX-Fwd and CTX-Rvs (Dierikx et al., 2012), which amplified a 593 bp subfragment of the 876 bp blaCTX-M gene. An 846 bp subfragment of ISEcp1 was amplified using ISEcp1-Fwd and ISEcp1-Rvs (This study). PCR for gfp in the E. coli recipient strain CV601 used the primers Pgfp (up) and Pgfp (down), (Andersen et al., 1998), which amplified a 714 bp subfragment of the gfp gene. All PCR products were electrophoresed and visualised on a 1% TAE agarose gel run at 85 V for 1.5 h using a 100 bp Quick-Load® DNA Ladder (NEB, USA) as a marker and visualised using a Bio-Rad Universal Hood II-GelDoc System (Bio-Rad, USA).
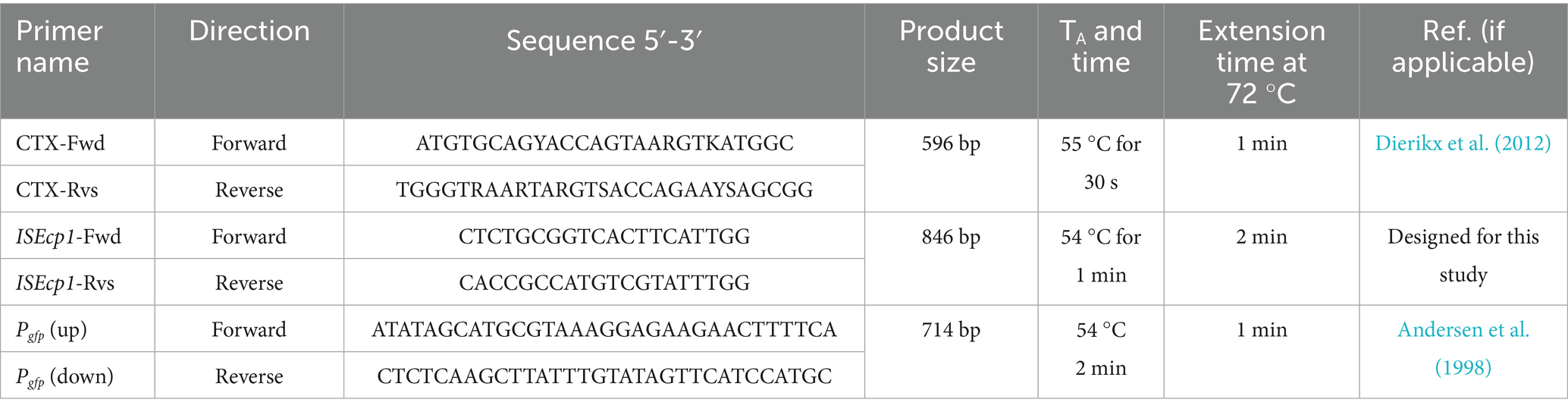
Table 1. Primer sets utilised within PCR analyses with gene specific TA and extension times at 72 °C given in seconds (secs) or minute(s) (min(s)).
2.5 Sanger sequencing, whole genome sequencing (WGS), assembly, and annotation
Sanger sequencing of PCR products was performed by Eurofins Genomics (Wolverhampton, UK). PacBio long-read WGS of strain 774 was conducted by the University of Liverpool, UK (Liverpool Genomics) using 10 kb libraries with 120 times coverage. Illumina short-read WGS and MinION long-read sequencing of ESBL-Ec isolates and transconjugants was carried out at the University of Cardiff, UK. Illumina sequencing library preparation was conducted using the Nextera XT v2 kit (Illumina, Cambridge, UK) with bead-based normalisation for library quantification measurements. Libraries were sequenced using the Illumina MiSeq using a v3 600-cycle kit (Illumina). The read length was 2×300 bp, before trimming. The gDNA for MinION sequencing was first subject to high-performance isolation and purification via Solid Phase Reversible Immobilisation (SPRI) bead clean up (Beckman-Coulter, High Wycombe, UK). Library preparation for MinION sequencing was conducted using the SQK-RBK110.96 rapid barcoding kit according to the manufacturer’s instructions (Oxford Nanopore Technologies (ONT), Oxford, UK). Sequencing was conducted on R9.4 flow cells (ONT, Oxford, UK). The rapid barcoding library kit generated read lengths of between 200 bp and 60 kb. Following quality trimming with Trimgalore (v0.6.4) and Filtlong (v0.2.1) for Illumina and ONT data, respectively, fastq raw sequences were hybrid assembled using Unicycler (v0.4.7) (Wick et al., 2017). Bioinformatic sequence analysis was completed using Geneious Prime (Dotmatics, Boston, USA), NCBI (National Institutes of Health, Maryland, USA), Snapgene (GSL Biotech LLC, Boston, USA) (all using standard parameters), and programmes available from the Centre for Genomic Epidemiology (CGE) set at standard parameters, including MLST 2.0 (Larsen et al., 2012), Res Finder 4.1 (Bortolaia et al., 2020), and Plasmid Finder 2.1 (Carattoli et al., 2014). The nucleotide sequence for ISEcp1 (accession number AJ242809) was downloaded from ISfinder, and a custom database was created within ABRicate (v1.0.0) to screen for the presence (and identify location) of the insertion sequence in all genomes. Whole genome sequences are deposited in NCBI under Bioproject number PRJNA1196928 with the following Accession numbers: SAMN45706859-SAMN45706910.
2.6 Core-genome phylogenetics and SNP distance comparison
A core genome phylogeny and SNP distance comparison were conducted on 37 of the 39 blaCTX isolates (with isolates EcoHS11212-878 (878) and EcoHS11212-880 (880) removed during quality filtering due to poor sequencing coverage and assembly) and 105 ST2325 genomes downloaded from Enterobase (downloaded April 2022 with available metadata on location and source recorded). Isolate EcoMHE1212-939 (939) with the best sequencing coverage and assembly was used as the representative reference genome for SNP-based data generation, with variant calling performed using Snippy (v4.6.0), using default parameters. Recombination sites were removed with Gubbins (v2.3.4), resulting in 13,143 SNPs in conserved genomic regions. The reference genome was annotated using Bakta (v1.9.3) and bedtools (v2.31.1) and was used to classify SNPs either within gene regions (n = 12,343 SNPs) or in intergenic regions (n = 826). Snp sites (v2.5.1) were used to extract SNP positions, and IQ-tree (v2.0) was used to generate a phylogeny. SNP pairwise distances were generated using snp-dists (v0.6). The phylogenetic tree was mid-rooted and annotated using iTOL v5.7 (Letunic and Bork, 2021).
2.7 Transposition experiments
Four transposition experiments were performed by adapting the methods of Lartigue et al. (2006) and Nordmann et al. (2008). To define the baseline frequency of transfer, transposition of ISEcp1 blaCTX-M-15 into endogenous plasmids and transfer of the plasmids into the E. coli K-12 CV601 recipient strain were performed in a non-selective (n/s) environment of LB broth with no added antibiotics. To determine if the presence of sub-lethal levels of the antibiotics could enhance the transfer frequency above the baseline transposition/transfer conditions, the experiments detailed above were performed in the presence of the third-generation cephalosporin ceftazidime (CAZ) and the penicillins ampicillin (AMP) and cloxacillin (CLOX), with the sub-lethal levels of antibiotic concentrations tested listed in Table 2. These antibiotics were chosen as they are commonly used in dairy farms globally, with CLOX often favoured for use in dry cow therapy (González Pereyra et al., 2015; Johnson et al., 2016; Breser et al., 2018; Liu et al., 2018; Whitfield and Laven, 2018; Rossi et al., 2019; McDougall et al., 2021). The third-generation cephalosporin (CAZ) was also chosen to act as a positive control for enhanced transposition, as previous studies by Lartigue et al. (2006) and Nordmann et al. (2008) demonstrated enhanced transposition of ISEcp1 in the presence of this antibiotic.
From an 18 to 20 h culture grown on non-selective LB agar at 37°C, a single colony suspension of the donors EcoSL1010-687 (687), EcoHS11212-876 (876), EcoMHE1801-956 (956), and EcoSS2501-961 (961) was made in 5 mL LB broth with and without added sub-inhibitory levels of antibiotics as listed in Table 2. A single colony suspension of the E. coli K-12 recipient CV601 (gfp, KanR, RifR) (Smalla et al., 2000) was also prepared in 5 mL LB broth containing 50 μg ml−1 of KAN and grown for 18 h at 37°C shaking at 180 RPM. Donor cultures were diluted 1 in 100 into 5 mL LB broth and grown for 3 h at 37°C, shaking at 120 RPM. The E. coli CV601 culture was centrifuged at 8,000 g for 5 min in 1 mL aliquots and the pellet washed twice in 1 mL of sterile Maximum Recovery Diluent (MRD; Sigma-Aldrich, USA), with a final resuspension in 1 mL MRD. Cultures were diluted using MRD to approximately 0.5–0.7 OD600 to achieve a culture containing ~1×108 CFU ml−1. Conjugation was performed using a 1:4 ratio of donor to recipient culture in a total volume of 1 mL. The conjugation mix was gently vortexed and then incubated at 37°C for 3 h without agitation. Mating was stopped by vigorous vortexing of the conjugation mix culture and placing the culture on ice. A 100–10−7 serial dilution in MRD was plated in duplicate onto LB agar containing 100 μg ml−1 AMP (according to the method of Nordmann et al., 2008; ISEcp1 selective marker) and 50 μg ml−1 KAN (CV601 selective marker) in 0.1 mL volumes. Plate counts for each donor and the recipient were also conducted by plating 100 μL in duplicate of the 100–10−7 serial dilutions in MRD, onto LB plus 100 μg ml−1 AMP for the donors and LB plus 50 μg ml−1 KAN for CV601. All plates were incubated for 18 h at 37 °C. Colonies were counted and CFU ml−1 calculations conducted on the following day.
Successful transconjugants were designated ‘transposition transconjugants’ (TT) as there had been transposition of ISEcp1 into an endogenous plasmid, followed by conjugation of the ISEcp1-containing plasmid to the recipient E. coli K-12 CV601 strain. Presumptive TT colonies from LB AMP100 KAN50 plates were selected for further confirmation by looking for expression of recipient GFP, detected if they fluoresced when exposed to UV illumination at a wavelength of 365 nm, using a UVGL58 UVP Dual Tube Handheld UV Lamp (ThermoFisher Scientific, Loughborough, UK). Colonies that fluoresced were then counted, and the CFU/ml−1 calculated. The transposition frequency was calculated as the ratio of CFU/ml of transconjugants to CFU/ml of donors. The plasmids contained within the donor strains were all cryptic, with no selectable genetic markers, so it was not possible to obtain a conjugation frequency for each plasmid alone. Therefore, the final transfer frequency was a combination of the transposition and conjugation frequencies. Transfer frequencies were calculated from counting fluorescent GFP-positive colonies. Colonies confirmed as a successful TT were restreaked onto double selective media purification plates, containing both AMP100 and KAN50 to create a pure culture, and these were banked as the confirmed TT strains for further study.
DNA was extracted using the simple boiling method (Wei, 2013) from single TT colonies from each of the purification plates. To ensure isolates were true transconjugants, rather than mutated donors, confirmation was achieved through PCR for blaCTX-M, ISEcp1, and gfp to show successful mobilisation of ISEcp1 in association with blaCTX-M and transfer into the recipient CV601 strain.
2.8 ISEcp1 plasmid typing and insert location
A total of 15 TTs were sequenced using Illumina short read and MinION long read WGS to further analyse the plasmid types that had transferred into the CV601 recipient. The genes located between the ISEcp1 IRs were also investigated to understand which genes had been transferred or lost during transposition. The sequences were first analysed through CGE online software1 using PlasmidFinder 2.1 (Joensen et al., 2014) to identify contigs containing plasmid replicons. Further sequence analysis was conducted in Geneious Prime, 2023 (version 2023.2.1),2 which included assessing the size of the new ISEcp1 genetic environments within the TT background, construction of the various ISEcp1 genetic environments and plasmid backbones, and searching the surrounding chromosome of the TTs.
ISEcp1 element sizes were assessed through the identification of first the IRL, and then the identification of the 5 bp repeats and the new IRR. The sequences of the TTs are available under Bioproject number PRJNA1196928; a full list of accession numbers is listed in Supplementary Table S4.
3 Results
3.1 MIC data for selected strains confirmed previously reported resistance
MIC analysis on the 39 strains for seven of the antibiotics confirmed previously reported phenotypic resistance data (Baker et al., 2022) for ampicillin (AMP), ceftazidime (CAZ), cefotaxime (CTX), cefpodoxime (CPD), cefquinome (CFQ), aztreonam (ATM), and tetracycline (TET). The full list of resistant MIC results is available in Supplementary Table S5.
All 39 strains displayed high-level resistance to AMP, CTX, CPD, and CFQ, consistent with blaCTX-M-15 carriage. Additionally, all strains were resistant to CAZ, but the MIC was at a much lower value of 16 μg ml−1 compared to the other third-generation cephalosporins, which had MICs of >512 μg ml−1 for CTX and 512 μg ml−1 for CPD. The MICs for CFQ and ATM were 128 μg ml−1 and 32 μg ml-1, respectively. EcoMHE1801-950 (950), EcoMHE1801-953 (953), EcoMHE1801-955 (955), and EcoMHE1801-956 (956) were the only isolates that were susceptible to TET, with an MIC of <2 μg ml−1. The remaining strains were TET resistant, which included seven isolates with an MIC of 128 μg ml−1 and the rest with an MIC of 64 μg ml−1. MIC results for all the strains for the remaining 19 antibiotics (Supplementary Table S6) tested indicated that they were sensitive.
3.2 All selected isolates contained blaCTX-M and ISEcp1
The 39 isolates selected for their ESBL phenotype were resistant to the β-lactam AMP, the third-generation cephalosporins CAZ, CTX, CPD, and the monobactam ATM (Baker et al., 2022). This confirmed AST testing data that suggested that the likely resistance mechanism was a blaCTX-M type ESBL. PacBio sequencing of ESBL isolate 774, (Baker et al., 2022) had already identified blaCTX-M-15 in the chromosome, in association with ISEcp1. PCR screening and Sanger sequencing of the remaining 38 strains (data not shown) confirmed they all carried both blaCTX-M and ISEcp1. CTX-M typing of the Sanger sequenced PCR products using NCBI BLAST searches, typed all 38 strains as blaCTX-M-15, which was confirmed as a single copy on the chromosome using WGS.
3.3 Sequence mapping demonstrates clonality of the ESBL-Ec isolates
Multi-locus sequence typing (MLST) of the WGS of the 39 blaCTX isolates determined that all were sequence type (ST) 2,325, indicating they could be closely related. ST2325 carrying ESBL has been found in livestock and animal produce; ducks (Yu et al., 2021), farmed rabbits (Silva et al., 2024), raw milk (Irrgang et al., 2017), pigs (Ding et al., 2021), and sheep (Atlaw et al., 2021), as well as in wildlife; gulls (Nesporova et al., 2024). ST2325 has also been found in humans and sewage (Zahra et al., 2018; Ding et al., 2021). To support the idea that ST2325 may form clonal clusters within other animal groups or in bovine-associated studies, a further 105 ST2325 isolates were downloaded from Enterobase (termed Enterobase isolates) for broader SNP distance comparison and phylogenetic analysis. The pairwise SNP distance comparison consisted of the 37 blaCTX isolates and the 1 reference genome: isolate 939 (the two isolates 878 and 880 were removed during quality filtering due to poor sequencing coverage and assembly, and isolate 939 was included as both an isolate and as a reference, so was counted twice, resulting in a total of 37 isolates plus the reference). This SNP distance comparison revealed that the 37 blaCTX isolates were within 0–6 pairwise SNPs, suggesting a clonal relationship (Figure 1). A few groups of ST2325 Enterobase isolates also formed genetic clusters on the phylogeny, suggesting the presence of several lineages (labelled Group 1–5, Figure 1). Only one ST2325 Enterobase isolate was close in similarity to this study’s isolate cohort, and this was a Spanish bovine isolate that was within <50 SNPs (coloured purple on the tree in Figure 1).
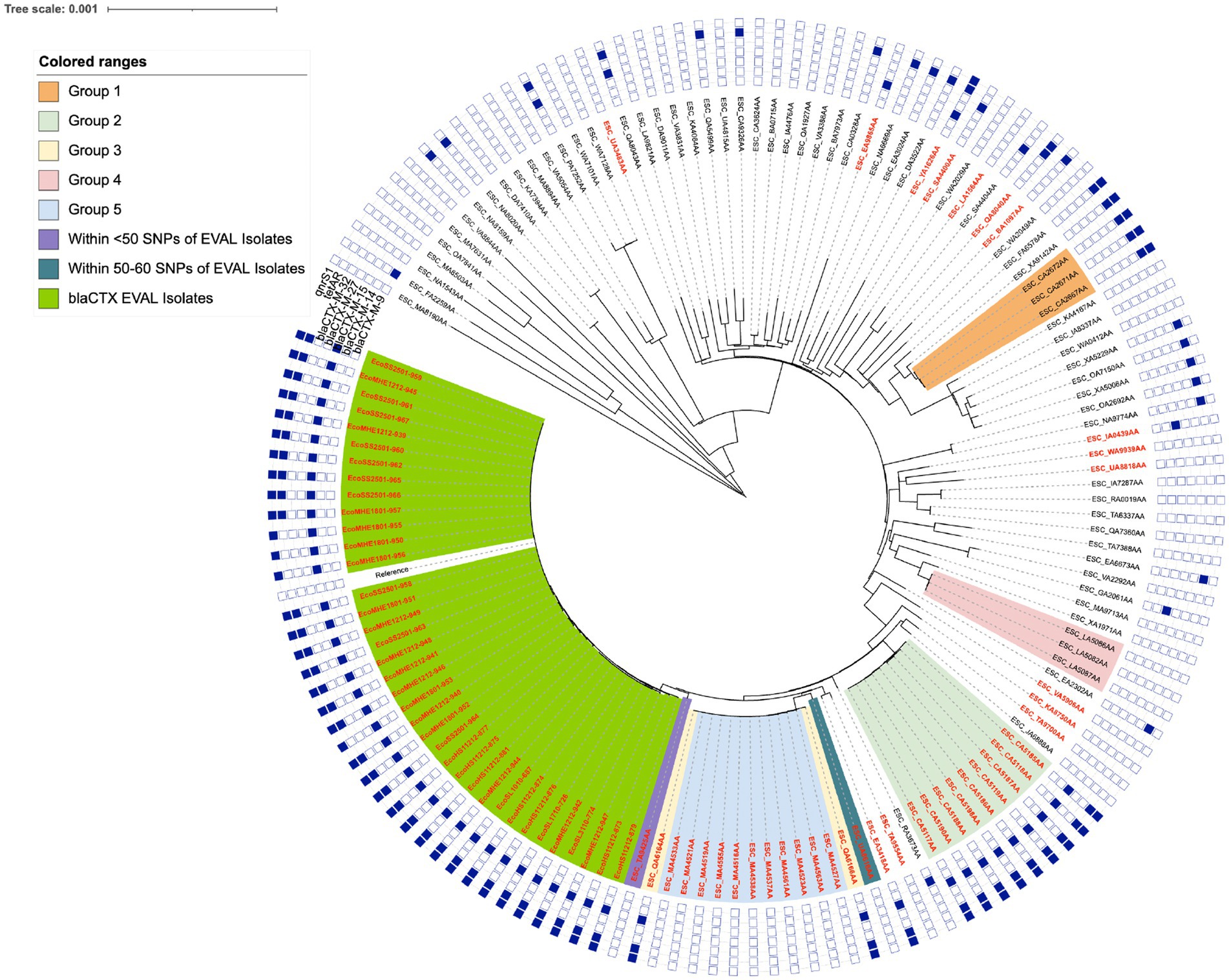
Figure 1. The whole genome phylogeny maximum likelihood tree generated using IQtree v2.0 with annotation achieved using the iTOL v.5.7, showing the 37 blaCTX isolates in combination with the 105 ST2325 genomes downloaded from Enterobase. The resistance gene carriage of each isolate is annotated around the outside of the tree, corresponding to the resistance genes qnrS1, blaCTX-M-32, blaCTX-M-27, blaCTX-M-15, blaCTX-M-14 and blaCTX-M-9, with positive carriage denoted as a filled blue square. Any isolates positive for ISEcp1 had the tree label shown in red and the 37 blaCTX isolates were highlighted in green. The colour range key and shaded clades on the tree, relates to groups of isolates that were identified as possible clonal groups from the SNP distance comparison.
Within the clonal groups from Enterobase, Group 1 was within 0–2 pairwise SNPs, Group 2 was within 0–6 pairwise SNPs, Group 3 was within 0–2 pairwise SNPs, Group 4 was within 0–2 pairwise SNPs, and Group 5 was within 0–4 pairwise SNPs. The blaCTX dairy farm isolates and each group alone appeared to be clonal but unrelated to any other groups or isolates. Only Groups 3 and 5 appeared to be clonally related to each other, as all isolates were within 1–4 SNPs of each other and they appeared on the same clade (Figure 1). No other groups appeared to be closely related to each other, the closest being the combined Group 3 and 5, which were within 58–72 SNPs of the blaCTX EVAL farms isolates. The two separate isolates ESC_TA9425AA (denoted as <50 SNPs of the dairy farm isolates on Figure 1) and ESC_UA8616AA (denoted as 50–60 SNPs of EVAL isolates on Figure 1) were within 36–45 and 49–58 SNPs of the blaCTX dairy farm isolates, respectively, and therefore were the most closely related of the Enterobase isolates to the blaCTX EVAL farms isolates. The SNP distance analysis clearly showed, however, that there were multiple sets of evidence for clustering of ST2325 isolates, but at the same time, there was some genomic diversity, which could suggest the international spread and adaptation of a successful clonal lineage. Full details of each group from the tree, the SNP information, full metadata for each isolate, including sampling geographical location and niche, and accession number are available in Supplementary Table S7. A complementary ANI approach provided similar genomic insights into the ST2325 EVAL and Enterobase collections, and details are available in Supplementary Note S2.
3.4 blaCTX resistance/mobile genetic determinants in the ST2325 collection
Of the 105 ST2325 Enterobase isolates, a total of 38 were found to be encoding ISEcp1, with 17 also encoding a blaCTX-M. All but 2 of the 17 were of blaCTX-M-15 type, with the remaining two of blaCTX-M-32 and blaCTX-M-27 type. The assemblies of the Enterobase genomes were largely derived from short-read-only sequencing data (which is indicative of limited quality for genomic context analyses), and therefore, it was difficult to be certain whether the ISEcp1 genetic environments were located chromosomally or in a plasmid. Manual inspection of the genome in all isolates positive for both ISEcp1 and blaCTX-M was conducted to confirm where ISEcp1 and blaCTX-M were in relation to one another. From this, ISEcp1 was in the same region as blaCTX-M in 16 of the 17 ISEcp1 and blaCTX-M positive Enterobase isolates, as the ISEcp1 transposase was located directly upstream of the blaCTX-M gene in the same contig. In the isolate encoding both ISEcp1 and blaCTX-M-32, the blaCTX-M-32 was found in the middle of a large contig surrounded by what appeared to be chromosomal DNA, which was different from the contig containing ISEcp1.
In addition, 13 of the 15 ISEcp1 and blaCTX-M-15 positive Enterobase isolates were found to be encoding both tetAR and qnrS1, with the remaining 2 of the 15 encoding either qnrS1 or tetAR alone. The ISEcp1 was located in the same region as tetAR in 2 isolates and in the same region as qnrS1 in 4 isolates. In only 1 isolate did both tetAR and qnrS1 appear to be located in the same region as ISEcp1. However, as stated above, due to the fragmented assemblies of the Enterobase genomes, it was unclear which contigs made up the entirety of the ISEcp1 genetic environment. Therefore, the relative locations of the tetAR, qnrS1, and ISEcp1 genes were merely an observation from the available data. The association of ISEcp1 with ST2325 showed that ISEcp1 was quite widespread throughout the 105 isolates, being found in 37.1%. In addition, the most commonly found blaCTX-M variant in association with ISEcp1 was blaCTX-M-15, which was the same as the blaCTX isolates in this study This could suggest that ST2325 may have an association with ISEcp1 and blaCTX-M-15 but considering the sample size available from Enterobase was small, it is difficult to be certain how widespread ISEcp1 and blaCTX-M-15 are throughout ST2325 isolates that are not represented in the database.
The highest represented niche was livestock, which made up 60% (64 isolates) of the Enterobase isolates examined. Within the livestock niche, the highest numbers were bovine samples, 31.4% (33 isolates), followed by poultry/avian 14.3% (15 isolates) and ovine/goat 9.5% (10 isolates). This could suggest that there is an association between ST2325 and bovine; however, bovine and poultry are more intensively farmed than sheep or goats (Pandey and Upadhyay, 2022) and therefore, the information available for those groups within the repositories may be limited compared to other higher-represented groups such as bovine and poultry. However, it is difficult to determine this, with the limited amount of data available to download for each animal group.
3.5 Whole genome sequencing of the 39 isolates identifies five plasmid replicon types and chromosomal carriage of all ARGs and ISEcp1 elements
Hybrid assembly of the WGS of the 39 isolates resulted in mostly complete chromosomes and plasmids, allowing high confidence in identifying where resistance genes were located in the genome and good accuracy for estimating plasmid sizes. Supplementary Table S8 shows the full assembly statistics and also includes the ORF number, contig number, contig numbers containing a plasmid, whether the plasmids were complete, overall genome size, overall %GC content, N50 number, and whether the chromosome was complete.
ResFinder results and manual searches of the WGS identified, in addition to blaCTX-M-15, the resistance genes: qnrS1, which provides low-level quinolone resistance (Allou et al., 2009) in all isolates, and tetAR, responsible for tetracycline resistance, in all but isolates 950, 953, 955, and 956. These latter isolates had shown susceptibility to tetracycline during AST disc and MIC assays. PlasmidFinder analysis showed that at least 5 plasmid replicon types were present in the isolates, including: IncFIC, IncFII, IncI1, IncI2, and IncX4. In addition to strain 774, two further isolates had no plasmids, and one isolate contained only a single plasmid. All of the plasmids lacked antibiotic resistance genes. Supplementary Table S8 gives full details of isolate plasmid carriage, replicon type, and the contig it was located in.
3.6 ISEcp1 carrying blaCTX-M-15 can transpose from environmental isolates into recipient strains
ISEcp1 blaCTX-M-15 transposition/conjugation experiments were undertaken using four representative isolates, 687, 876, 956, and 961, and the CV601 E. coli K-12 recipient strain. These donor strains carried endogenous plasmid replicon types IncFIC, IncFII, IncI1, IncI2, and IncX4 (Supplementary Table S8 details which plasmid replicon types were within each isolate). The method utilised the endogenous plasmids as vectors and included examples of all the plasmid replicon types observed in the 39 strains.
Of the 40 combinations of 4 donor strains and antibiotic-supplemented growth conditions used for transposition experiments (Table 3), under our experimental conditions, 23 resulted in the isolation of transconjugants. KAN- and AMP-resistant putative transposition transconjugant (TT) colonies that fluoresced under UV light were tested by PCR for the presence of blaCTX-M and ISEcp1 and for the E. coli K-12 recipient CV601 strain gfp gene. This confirmed that both blaCTX-M and ISEcp1 had successfully transferred into CV601 (data not shown).
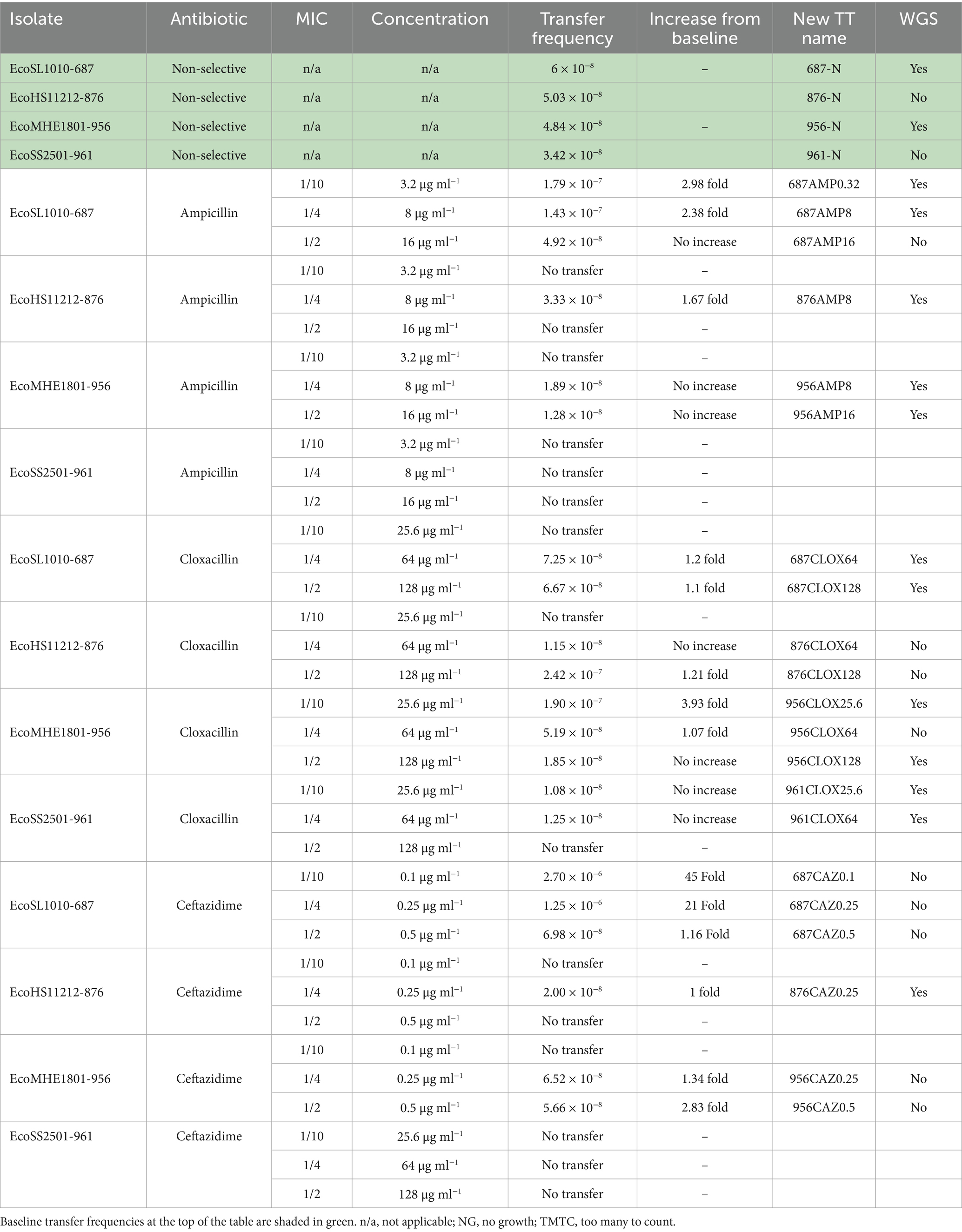
Table 3. Transfer frequencies of ISEcp1 element under selective and non-selective growth conditions.
3.7 Sublethal concentrations of antibiotics can lead to an enhanced transfer frequency of ISEcp1
The transfer frequency of the ISEcp1 element represents a combination of the initial transposition of ISEcp1 from the chromosome into a resident plasmid, followed by conjugation of the plasmid into the E. coli K-12 recipient strain CV601. Successful transposition/conjugation of the ISEcp1 element occurred in the absence as well as presence of AMP, CLOX, and CAZ in the LB media in the conjugation assays were carried out in, and at each of the concentrations of antibiotic that were used (Table 3). However, the frequency of transposition/conjugation varied with each donor strain. Furthermore, enhanced frequency of transposition and transfer into the CV601 host was successful with all the concentrations of CLOX and CAZ tested, but only with two concentrations of AMP (0.32 and 8 μg ml−1). The transfer frequency for each donor with each antibiotic and the increase from the baseline frequency of transfer and new TT names are also shown in Table 3.
The baseline transfer frequency of ISEcp1 from each donor to the recipient CV601, when isolates were grown in nonselective medium with a starting culture of 1 × 108 cells, was 6 × 10−8 with isolate 687 (around 1 in 16 million), 5.03 × 10−8 with isolate 876 (around 1 in 19 million), 4.84 × 10−8 with isolate 956 (around 1 in 20 million) and 3.42 × 10−8 with isolate 961 (around 1 in 29 million), which are lower than with artificially constructed donors (Lartigue et al., 2006; Nordmann et al., 2008). An enhanced frequency of transfer above the baseline frequency was only observed in 687, 876, and 956, and with only some of the antibiotics and concentrations tested.
The majority of enhanced transposition/ conjugation (transfer) frequencies were seen with 1/10 to 1/4 MIC concentrations of antibiotics, and this was evident for AMP in 687, which produced a slightly higher transfer frequency of 2.98-fold at 1/10 MIC than 2.38-fold at 1/4 MIC. In 687, CLOX had a slightly higher transfer frequency of 1.2 fold at 1/4 MIC than 1.1 fold at 1/2 MIC. In 956, CLOX showed a higher frequency of 3.93 fold at 1/10 MIC than 1.07 fold that was seen at 1/4 MIC. The biggest increase in enhanced transfer was seen with CAZ in 687, where there was a much higher frequency of transfer of 45-fold at 1/10 MIC than 21-fold at 1/4 MIC. In comparison, however, in 956, the enhanced frequency of transfer with CAZ was greater at 2.83 fold at 1/2 MIC than at 1.34 fold at 1/4 MIC. No enhanced transfer frequencies were observed with any antibiotics in 961, and there were no enhanced transfer frequencies with AMP in 876 and 956 or with CAZ in 876.
As shown in Table 3, of the 23 conjugations that show a successful transfer, 15 transconjugants (that represented a combination of each donor strain and each concentration of AMP and CLOX tested) were sequenced using Illumina short read and MinION long read with hybrid assembly.
The CGE MLST programme (Larsen et al., 2012) was used to confirm from the WGS data that all putative transconjugants except isolate 687AMP16 were of ST10, the same as the recipient CV601. Isolate 687AMP16 was found to be the same ST as the parent ST2325; this isolate was therefore discounted from any further analysis. The gfp gene was also located successfully within the genome sequence of all the TTs confirmed as being of ST10, further confirming that the TT strains were a result of plasmid transfers into CV601. The ISEcp1 blaCTX-M-15 elements were found to have inserted into all but the IncI2 plasmid replicon type plasmids during transfer to the recipient CV601 strain; partial plasmid maps showing the structure of the ISEcp1 elements are detailed in Supplementary Note S1 and full details of the insertion points with schematics of the IRR used by each TT and the TT genetic environments are detailed in Supplementary Tables S9, S10 and S11. These data show that in only two of the 13 (~15%) TT examined was the ISEcp1 element identical to that of the donor parent: three were larger, having gained additional chromosomal genes from the original host; the rest were smaller, having lost genes from the original ISEcp1 structure. Genes gained were from the Type 3 Secretion System (T3SS) located downstream of blaCTX-M-15 in the donor (Figure 2A). The most significant gene loss was that of the tetAR genes in nine TT, with six additionally losing the qnrS1 gene. These structural changes were all likely a result of the recognition of a new imperfect IRR used by ISEcp1 during transposition. From the data on the insertion sites in the plasmids, in some cases, a plasmid conjugation gene was interrupted, which could affect onward transmission of the ISEcp1elements. Thus, although many of the TT would be able to act as conjugation donors for the ISEcp1element with the blaCTX-M resistance genes, in only a minority would the tetAR and qnrS1genes also be transmitted, limiting their spread.
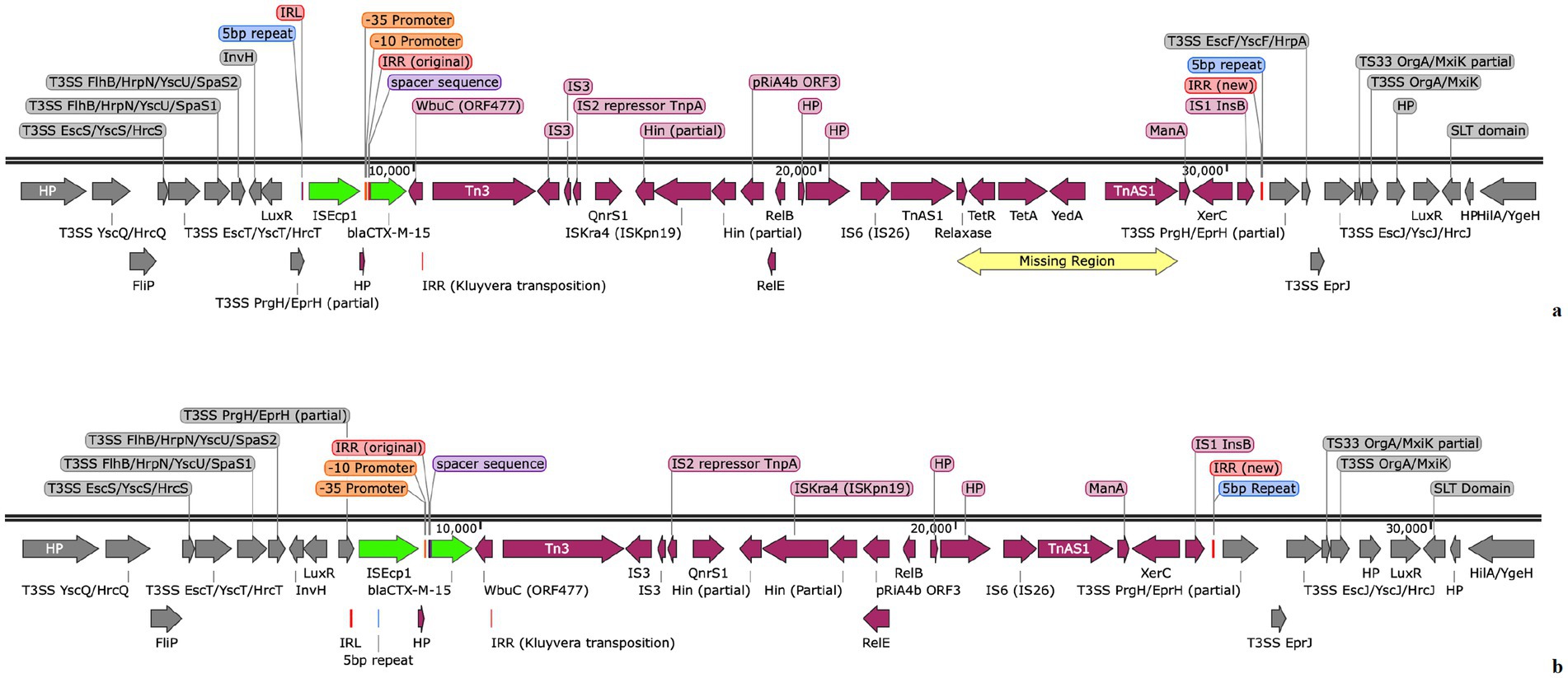
Figure 2. (A) ISEcp1 chromosomal environment of isolate EcoSL3110-774 from the PacBio sequence (ISEcp1 and blaCTX-M-15 are shown in green with the remaining genetic environment shown in maroon), showing the 5 bp repeats (blue), IRL and IRR (red) and the surrounding chromosome (grey) around the insertion of ISEcp1, with the area denoted as a yellow box and annotated as ‘missing region’, detailing the area missing from within the tetracycline susceptible isolates 950, 953, 955, 956 and 962 shown in (B) the smaller ISEcp1 element found in the chromosome of isolates 950, 953, 955, 956 and 962 which all were missing the region of TetAR, YedA and TnAS1. The ISEcp1 and blaCTX-M-15 are shown in green with the remaining genetic environment shown here in maroon, the 5 bp repeats are in blue, the IRL and IRR are in red and the surrounding chromosome around the insertion of ISEcp1 is in grey.
3.8 The ISEcp1 element provides the same level of phenotypic antibiotic resistance to the recipient strain as was in the donor strain
To determine the effects on phenotypic resistance from the transposition of ISEcp1 and transfer of resistance into the recipient CV601 strain, MICs were performed as described for the parent strains on the transposon transconjugants, to assess any changes in the level of resistance. A panel of 15 antibiotics was selected from the original MIC panel of 25 antibiotics (listed in Supplementary Table S2) that included AMP, CAZ, CTX, CPD, CFQ, ATM, AMC, FOX, ertapenem (ERT), neomycin (NEO), TET, nalidixic acid (NAL), ciprofloxacin (CIP), and enrofloxacin (ENR). Additionally, CLOX was included utilising a previously described breakpoint (Hertz et al., 2014).
The recipient strain CV601, which encodes a 3′-phosphotransferase aph3’, was only resistant to NEO in the panel of antibiotics, with an MIC of 64 μg ml-1, and was susceptible to all other antibiotics tested. However, following the transposition experiments, the recipient strain was resistant to AMP, CLOX, CAZ, CTX, CPD, CFQ, and ATM, with all TTs showing identical MICs of >512 μg ml−1, >512 μg ml−1, 16 μg ml−1, 512 μg ml−1, 128 μg ml-1, and 32 μg ml−1, respectively. These results were also identical to the MICs for the donor parent strains. The MICs for AMC, FOX, ERT, and NAL were also identical in all TTs and to the parent donor strains. Only 687CLOX128, 961CLOX64, and 687AMP0.32 were resistant to TET (with an MIC of 64 μg ml−1), which was identical to the parent donor strains. All other TTs were susceptible to TET, owing to the loss of the tetAR genes or their absence in the parent strains. The only other differences in MICs between the TTs and the parental strains were with CIP and ENR. In 8 of the TTs, which included 687-N, 687AMP0.32, 687CLOX64, 687CLOX128, 956AMP8, 956AMP16, 961CLOX25.6, and 961CLOX64, the MIC results for CIP and ENR were 0.25 μg ml−1 and 1 μg ml-1, respectively, which were the same as the parent MIC results. In the remaining TTs, which had lost qnrS1 during ISEcp1 transposition, the MIC for CIP and ENR gave the same MIC result as CV601 of ≤0.064 and ≤0.032, respectively. These MICs demonstrated that the level of phenotypic resistance was maintained following transposition of ISEcp1 from the parent donor strains into the recipient CV601, with high-level β-lactam resistance still present.
3.9 The ISEcp1 elements are fluid, losing or gaining resistance and virulence genes
The data from the isolate WGS and TTs together demonstrate that the ISEcp1 element is both capable of losing and gaining genes during transposition. Thirty-four of the isolates carried ISEcp1 elements, which were identical to each other and to that of the principal isolate 774 at 26,612 bp (Figure 2A). However, in isolates 950, 953, 955, 956, and 962, the ISEcp1 element was slightly smaller at 18,025 bp (Figure 2B). The smaller size of the ISEcp1 element in these five isolates was due to the absence of a 5,587 bp region encoding the tetracycline resistance and a relaxase, respectively tetAR, yedA, and TnAS1, (denoted as “Missing region” in Figure 2A) which supports the lack of phenotypic tetracycline resistance seen in the isolates 950, 953, 955, and 956. However, isolate 962 did show phenotypic resistance to tetracycline. In isolates 950, 953, 955, and 956, the tetAR region was completely absent from the genome; however, in isolate 962, a small, circularised region denoted as TnAS1 that encoded tetAR was found within a small contig separate from the ISEcp1 region, as shown in Figure 3. The 5,488 bp region of the contig in isolate 962 was identical to the TnAS1 region of all the other +tetAR ISEcp1 isolates. As already indicated (Section 3.7), many of the TT showed loss of genes during transposition, including resistance genes.
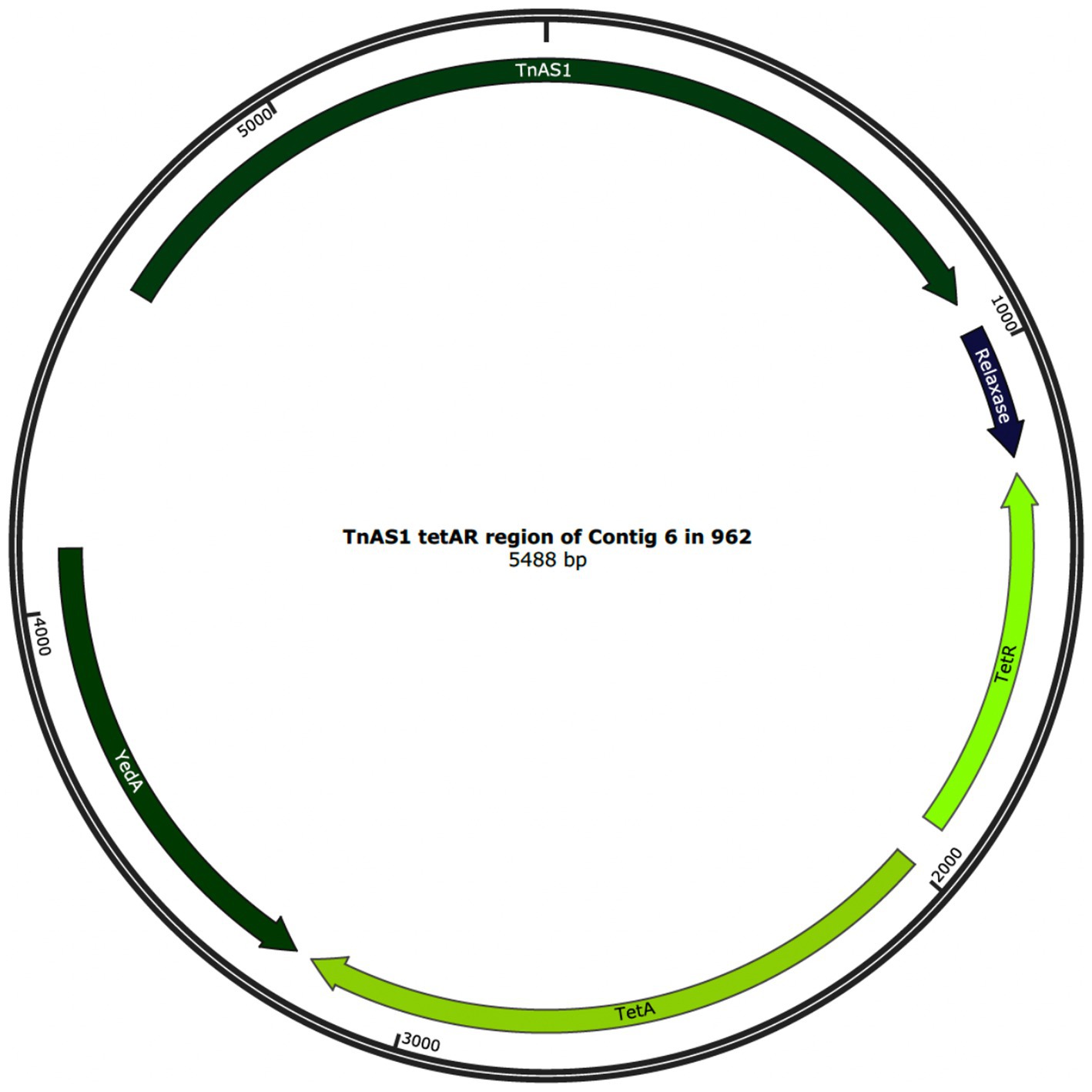
Figure 3. The annotated circularised genomic region of 5,488 bp in contig 6 from isolate 962, showing the complete TnAS1 transposase, relaxase, tetAR and yedA.
In addition, analysis of the TTs showed that three of the TTs—687AMP0.32, 687CLOX128, and 961CLOX64—had gained virulence genes from the T3SS by recognising a new imperfect IRR further along the genome during transfer (Section 3.7).
4 Discussion
We studied 39 ESBL-Ec isolated from a UK dairy farm to understand how related they were and whether the resistance genes they carried were mobile. We demonstrated that these strains were all ST2325 and within 0–6 SNPs of each other, isolated over 4 months, and from different parts of the dairy farm. WGS of the strains showed that the ESBL phenotype arises from chromosomal carriage of blaCTX-M-15, located on the mobile genetic element ISEcp1, which was identical in all but five isolates, where a truncated version of the element was present. We showed that the ISEcp1 element is fluid, able to gain or lose resistance or virulence genes following transposition; that this element and its resistance genes can be mobilised via plasmids and transfer to recipient strains; that this mobilisation can be enhanced by sublethal concentrations of antibiotics; and that mobilisation leads to acquisition of the resistance phenotype in the recipient strains, at a level that would be of clinical concern. To the best of our knowledge, this is the first time that environmentally occurring resistance genes from environmentally occurring E. coli have been shown to transfer using the strains’ own plasmids into a recipient E. coli strain, leading to phenotypic resistance. This is important evidence in favour of a One Health approach to AMR (Arnold et al., 2024). These results are also direct evidence for the relevance of the WHO Tricycle protocol for AMR (WHO, 2021), which specifically focusses on surveillance of ESBL-producing E. coli. Moreover, our results show that these resistances are dynamic and evolvable. They imply that surveillance must capture those AMR dynamics, whether through longitudinal sampling that can show whether these organisms are increasing or decreasing in prevalence or through sampling of selective agents, such as antibiotics or other chemicals that can select for transfer (Alav and Buckner, 2024), so that the risk of spread of resistance can be evaluated. Equally, a deep understanding of the dynamics of resistance evolution and spread also requires both genome sequencing and laboratory experimentation, which may be beyond the scope of the WHO protocol for some laboratories.
4.1 ISEcp1: a vehicle for blaCTX-M transmission
This study showed the ability of ISEcp1 to mobilise blaCTX-M-15 both in a nonselective and selective (sub-lethal levels of β-lactam antibiotics) laboratory conditions. Mobilisation of ISEcp1 in a non-selective environment suggested that the response to the experimental conditions, such as the ideal physiochemical conditions, high nutrient availability, and a stable temperature, was sufficient for mobilisation of ISEcp1 to occur. However, inclusion of sub-lethal levels of the antibiotics CAZ, AMP, and CLOX enhanced transposition of the ISEcp1 carrying blaCTX-M-15 and was likely the result of sub-lethal levels of antibiotics affecting the ISEcp1 element as opposed to blaCTX-M-15. This result was anticipated, as this effect has been previously described (Miller et al., 2004; Aminov, 2011; Beceiro et al., 2013). It is known that sub-lethal levels of β-lactam antibiotics can induce an SOS response, which may lead to increased mutagenic activity and genetic variability with resultant increased MGE mobilisation (Kuan et al., 1991; Capy et al., 2000; Foster, 2007). Other antibiotics, including ciprofloxacin, trimethoprim, and some other quinolones, may induce the SOS response, demonstrating that exposure to one antibiotic could result in the dissemination of resistance to an unrelated antibiotic (Hastings et al., 2004). Studies by Lartigue et al. (2006) and Nordmann et al. (2008) have shown that the presence of CAZ can enhance the transposition of an ISEcp1 element. Sub-lethal levels of CAZ enhanced transposition by up to 45-fold, and the other 2 antibiotics used in this study (AMP and CLOX) also enhanced transposition by up to 2.98 and 3.93-fold, respectively. Out of 36 TTs (not including the 4 baseline nonselective controls), 14 showed an enhanced frequency of transfer of ISEcp1 blaCTX-M-15 compared to the baseline frequency of transfer, which demonstrated that ISEcp1 transposition does not appear to be a rare event. Only a small number of strains were utilised in the enhanced transposition experiments, and many TTs were generated, with some transconjugant plates containing 1 colony from an initial culture plating of only 100 μL, which equates to ~ 1 × 108 CFU/mL. Although the assay we conducted arguably more closely approximates “real world” conditions for transposition into endogenous plasmids and transfer into a new host strain, the transfer rate was a combination of transposition and conjugation, so we cannot exclude other explanations such as enhanced conjugation effects occurring. Møller et al. (2017) demonstrated that high levels of cefotaxime (126 μg ml−1) were able to increase conjugation frequencies of an IncI1 plasmid. Following treatment with cefotaxime for 30 min, conjugation frequencies saw an 8.4-fold increase and after 60 min of treatment conjugation frequencies saw a 6.6-fold increase. Møller et al. (2017) also showed through qPCR that certain transfer genes were upregulated in response to cefotaxime treatment. Therefore, it cannot be ruled out that there was some influence on plasmid transfer from the antibiotics used in this study. However, transfer of the ISEcp1 elements was evident, as they had originally been chromosomally encoded in the parents and were of varying sizes in the resultant transconjugants, through loss or gain of genes.
4.2 ISEcp1 as a fluid genetic environment
ISEcp1 belongs to the IS1380 family of IS elements and uses a DDE transposase in a ‘copy-in’ mechanism (Poirel et al., 2005; Bouuaert and Chalmers, 2010). However, unlike other IS elements, it can mobilise downstream genes producing what have been termed ‘transposition units’. ISEcp1 mobilises via a one-ended transposition mechanism (Poirel et al., 2005; Kieffer et al., 2020) and is flanked by left and right Inverted Repeats (IRL and IRR). A common feature of ISEcp1 mobilisation is the recognition of an imperfect IRR (Poirel et al., 2003; Poirel et al., 2005; Lartigue et al., 2006; Dhanji et al., 2011; Zowawi et al., 2015; Sun et al., 2016; Hamamoto et al., 2020; Widyatama et al., 2021) and through this the collection of new genes may occur, into what have been termed transposition units (Zong et al., 2010; Widyatama et al., 2021; Yagi et al., 2021). Due to the recognition of an imperfect IRR, these transposition units can vary in size (Poirel et al., 2005). The result is that downstream genes may be collected or lost as mobilisation takes place (Zong et al., 2010), allowing for the capture of adjacent genes further downstream. ISEcp1 also provides the −35 and −10 promoter sequences for high level expression of blaCTX-M (Poirel et al., 2005; Zong et al., 2010).
The genetic environments of the ISEcp1 elements in the transconjugant plasmids varied significantly, and several of the ISEcp1 elements had lost both the tetAR region and qnrS1, whilst retaining blaCTX-M-15. Loss of these resistance genes showed that ISEcp1 transposition can also reduce the resistance gene content of the mobile element, through the recognition of an imperfect IRR within the ISEcp1 element, and consequent loss of resistance genes. Recognition of the new imperfect IRR is not random, as there is commonly some homology to the IRL, but it does appear to be somewhat random how far downstream from the IRL this recognition happens (Poirel et al., 2008). Therefore, gene gain or loss as a consequence of this degenerate sequence recognition phenomenon is equally possible, and the TTs analysed in this study appear to support this mechanism. Other mobile elements that mobilise by the recognition of variable different IRR sequences are Tn2 and the insertion sequence IS91. The right extremity of the mobile element is defined in both Tn2 and IS91 through this mechanism (Poirel et al., 2008).
The finding of a circularised TnAS1 (Figure 3) in isolate 962 shows the potential for tetAR to mobilise independently from the ISEcp1 element and suggests the TnAS1 may mobilise via either a copy out and paste in or a cut out and paste in mechanism (Bouuaert and Chalmers, 2010; Skipper et al., 2013).
Clearly, the mobility of the ISEcp1 element has implications in antibiotic resistance spread, pathogen evolution, and the silent colonisation of human and animal hosts by antibiotic-resistant E. coli. The plasticity of the ISEcp1 element extends beyond resistance genes and into the gain of virulence genes, as evidenced by the T3SS genes gained. The T3SS system within E. coli is an important factor, critical to virulence in pathogenic E. coli strains such as EPEC and EHEC. The T3SS delivers effector proteins to eukaryotic host cells, involved in the subversion of cellular processes, such as signalling pathways within the host, and results in attaching and effacing lesion creation (Ideses et al., 2005; Zhou et al., 2014). A good example of a foodborne pathogen that has a well-defined T3SS is EHEC O157, and healthy cattle are a known reservoir of EHEC O157 (Lim et al., 2010). This demonstrated that through ISEcp1 transposition, there is the potential for movement of important virulence genes, which might generate new variants of pathogens, which is a key fundamental biological process.
This acquisition and possible loss of downstream genes may have an impact on the evolution of gene content and, in particular, the “accessory genome” of a bacterium, by introducing or losing genes associated with resistance, virulence, or those genes involved in increased survival and colonisation of niche environments (Medini et al., 2005; Tettelin et al., 2008; Siguier et al., 2014).
4.3 Explaining the clonal expansion of the strains in the farm environment
The 39 ESBL E. coli isolates taken from the dairy farm over a 4-month period from the slurry tank, the muck heap, and heifer sheds were found to be clonal. This clone was resistant to cephalosporins that had been discontinued on the farm and to those still in use, such as the penicillins. Prior to this study, the last use of first-generation cephalosporins on this dairy farm was in April 2017, the last use of third-generation cephalosporins was in January 2016, and the last use of fourth-generation cephalosporins was in August 2015 (Baker et al., 2022; Todman et al., 2024). However, amoxicillin, benzylpenicillin, cloxacillin, penethamate, and oxytetracycline were used on the farm between 2015-18. The first blaCTX E. coli strain was isolated from the slurry tank on 10 October 2017 (strain 687), followed by strains isolated on 17 October (762), 31 October (774), 12 December (863–949), 18 January 2018 (950–957), and 25 January (958–96). There had therefore been a period of at least 20 and 24 months between the last use of third and fourth-generation cephalosporins, respectively, and at least 5 months between the last use of first-generation cephalosporins and the isolation of the first blaCTX E. coli strain, strain 687. This suggests that the use of first-, third-, or fourth-generation cephalosporins was not necessary to maintain this mobile genetic element in E. coli strains within the dairy farm, as the isolation of these strains occurred after those antibiotics had ceased to be used there. However, other β-lactam antibiotics were still in use, and because blaCTXM-15 confers resistance to them, their use may have been exerting a selective effect on carriage of this element.
There are five hypotheses behind the appearance and persistence of this clone: successful colonisation and growth of the ESBL-EC ST2325 strain within the herd, selective isolation media bias in strain recovery, selection by β-lactams, co-selection by tetracycline, or co-selection by copper and zinc ions.
This ESBL-Ec strain (the first isolate of which was strain 687) was first isolated at the end of a continuous period of sampling over nearly 2 years. This is consistent with the idea that the strain may have arisen either by a new strain arriving at the farm and becoming established, or an existing ST2325 strain acquiring the ISEcp1 element from another bacterium. It is clear from the sequence analysis that there had been multiple exchanges of plasmids within the different sequenced isolates. The global dissemination of ISEcp1 carrying blaCTX-M-15 has occurred both by clonal expansion of ST131 and by transfer of the element in Enterobacteriaceae (Awosile and Agbaje, 2021). That this has been clonal expansion and not repeated transfer of the ISEcp1 element is supported by the phylogenetics together with the identical nature of the ISEcp1 element in farm isolates, which, with the exception of the tetAR negative strains, did not demonstrate the high variability of this element evident in TTs.
Whilst we cannot exclude isolation bias, the fact that the ST2325 E. coli strain was established for at least 4 months on the farm and was isolated independently on each date amongst many other strains, mitigates against selective isolation bias.
We examined that it is unlikely that environmental β-lactam selection was selecting for ISEcp1, because β-lactams are known to be highly susceptible to hydrolysis. However, some studies appear to show that β-lactams may still be detected within environmental samples. It has been reported that levels of β-lactams are usually at the limit of detection (LOD) within wastewater/effluent or at very low levels, as they are often degraded quickly due to either β-lactamase activity or through their susceptibility to hydrolysis (Zhang and Li, 2011; Kulkarni et al., 2017; Rodriguez-Mozaz et al., 2020). Concentrations reported within effluent have been in the region of LOD – 99.4 ng L−1 for AMP (Lin et al., 2008; Rodriguez-Mozaz et al., 2020), 15 ng L−1 for CLOX (Watkinson et al., 2007) and LOD, 34 ng L−1 and <12 ng L−1 for CTX (Gulkowska et al., 2008). Despite hydrolysis, it is also possible that β-lactam breakdown products might still be able to select for resistance (Chio et al., 2023).
Co-selection by tetracycline may be more likely because the ISEcp1 blaCTX-M-15 element carried the tetAR tetracycline resistance genes in most isolates. Tetracycline was used extensively on the farm during 2015–2018, persists in the environment, and high levels of tetracycline resistance were observed (Baker et al., 2022). Finally, modelling of the farm environment predicted that cephalosporin resistance would be expected to be chromosomally encoded and could be driven by co-selection through high concentrations of copper and zinc in the dairy waste from the disposal of metal ion footbaths (Todman et al., 2024). The chromosomal carriage of the ISEcp1 blaCTX-M-15 element confirms the model prediction of co-selection and provides further evidence for the co-selection hypotheses for clonal expansion.
The information gained from the SNP distance comparison of the ST2325 Enterobase genomes together with the 37 blaCTX EVAL farms isolates showed that ST2325 appears to form small clonal groups. This was clear from the genomic data that came from studies reporting clear evidence of clonality in their isolates. Group 1 and 2 (Ludden et al., 2019), Group 3 (no reference available), Group 4 (Boehmer et al., 2018), Group 5 (Atlaw et al., 2021) and was also evident for the isolates of the blaCTX group analysed as part of this study. For the majority, the clonality was related to samples from the same studies and geographical area. The exceptions were Groups 3 and 5, which formed a clonal cluster on the tree, were within 1–4 SNPs of each other, and appeared to be from different studies. ST2325 is not a particularly widespread clone within the Enterobase genomes, and, in comparison to a significant dominant clone like ST131, which is represented with >14,000 genomes in Enterobase, the ST2325 numbers were relatively small. However, many of the Enterobase isolates were still only within 300–400 SNPs of each other. As the majority of the ST2325 isolates were identified from animals, with only a few from humans, this could possibly indicate a potential route ST2325 has taken from animals into the human population. However, as was previously stated, a greater number of samples would be required for this hypothesis to be looked at more rigorously.
5 Conclusion
Taken together, these results show the highly evolvable nature of ISEcp1 elements in environmentally occurring E. coli and their importance in the dissemination and transfer of antibiotic resistance. In environmental conditions, they and the blaCTX-M-15 gene they carry are capable of spreading through clonal expansion of the host strains. They are also capable of both acquiring and losing antibiotic resistance and virulence genes, and of horizontal transfer into recipient bacteria, especially under sublethal concentrations of antibiotics, and thus of mobilisation of clinically relevant resistance. Thus, it is essential to understand not just the prevalence of ESBL-producing E. coli as the WHO Tricycle One Health surveillance programme suggests, but also the conditions under which ISEcp1 elements can gain and spread resistance, and lose resistance genes, in order to effectively mitigate the global emergency of antimicrobial resistance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
CG-H: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. SH: Conceptualization, Data curation, Investigation, Methodology, Project administration, Writing – review & editing. KS: Data curation, Formal analysis, Methodology, Project administration, Software, Visualization, Writing – review & editing. TW: Data curation, Funding acquisition, Resources, Writing – review & editing. CO: Data curation, Methodology, Writing – review & editing. EP: Data curation, Methodology, Writing – review & editing. CH: Conceptualization, Investigation, Writing – review & editing. DS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Supervision, Writing – review & editing. CD: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing. JH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. CG-H was funded by the BBSRC Nottingham Doctoral Training Partnership (BB/M008770/1). This work was supported by the Antimicrobial Resistance Cross Council Initiative, supported by the seven United Kingdom research councils (NE/N019881/1).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1675089/full#supplementary-material
Footnotes
References
Afema, J. A., Ahmed, S., Besser, T. E., Jones, L. P., Sischo, W. M., and Davis, M. A. (2018). Molecular epidemiology of dairy cattle-associated Escherichia coli carrying blaCTX-M genes in Washington state. Appl. Environ. Microbiol. 84:e02430-17. doi: 10.1128/AEM.02430-17
Alav, I., and Buckner, M. M. C. (2024). Non-antibiotic compounds associated with humans and the environment can promote horizontal transfer of antimicrobial resistance genes. Crit. Rev. Microbiol. 50, 993–1010. doi: 10.1080/1040841X.2023.2233603
Allou, N., Cambau, E., Massias, L., Chau, F., and Fantin, B. (2009). Impact of low-level resistance to fluoroquinolones due to qnrA1 and qnrS1 genes or a gyrA mutation on ciprofloxacin bactericidal activity in a murine model of Escherichia coli urinary tract infection. Antimicrob. Agents Chemother. 53, 4292–4297. doi: 10.1128/AAC.01664-08
Aminov, R. I. (2011). Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158. doi: 10.3389/fmicb.2011.00158
Andersen, J. B., Sternberg, C., Poulsen, L. K., Bjorn, S. P., Givskov, M., and Molin, S. (1998). New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64, 2240–2246. doi: 10.1128/AEM.64.6.2240-2246.1998
Atlaw, N. A., Keelara, S., Correa, M., Foster, D., Gebreyes, W., Aidara-Kane, A., et al. (2021). Identification of CTX-M type ESBL E. coli from sheep and their abattoir environment using whole-genome sequencing. Pathogens 10:1480. doi: 10.3390/pathogens10111480
Arnold, K. E., Liang, G., McMahon, B. J., Fanning, S., Stekel, D. J., Pahl, O., et al. (2024). The need for One Health systems-thinking approaches to understand multiscale dissemination of antimicrobial resistance. The Lanc. Planet. Health, 8, e124–e133. doi: 10.1016/S2542-5196(23)00278-4
Awosile, B. B., and Agbaje, M. (2021). Genetic environments of plasmid-mediated blaCTXM-15 Beta-lactamase gene in Enterobacteriaceae from Africa. Microbiol. Res. 12, 383–394. doi: 10.3390/microbiolres12020026
Baker, M., Williams, A. D., Hooton, S. P. T., Helliwell, R., King, E., Dodsworth, T., et al. (2022). Antimicrobial resistance in dairy slurry tanks: a critical point for measurement and control. Environ. Int. 169:107516. doi: 10.1016/j.envint.2022.107516
Beceiro, A., Tomas, M., and Bou, G. (2013). Antimicrobial resistance and virulence: A successful or deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 26, 185–230. doi: 10.1128/CMR.00059-12
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Boehmer, T., Vogler, A. J., Thomas, A., Sauer, S., Hergenroether, M., Straubinger, R. K., et al. (2018). Phenotypic characterization and whole genome analysis of extended-spectrum beta-lactamase-producing bacteria isolated from dogs in Germany. PLoS One 13:e0206252. doi: 10.1371/journal.pone.0206252
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). Resfinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Bouuaert, C. C., and Chalmers, R. M. (2010). Gene therapy vectors: the prospects and potentials of the cut-and-paste transposons. Genetica 138, 473–484. doi: 10.1007/s10709-009-9391-x
Breser, M. L., Felipe, V., Bohl, L. P., Orellano, M. S., Isaac, P., Conesa, A., et al. (2018). Chitosan and cloxacillin combination improve antibiotic efficacy against different lifestyle of coagulase-negative staphylococcus isolates from chronic bovine mastitis. Sci. Rep. 8, 1–13. doi: 10.1038/s41598-018-23521-0
Cantón, R., and Coque, T. M. (2006). The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9, 466–475. doi: 10.1016/J.MIB.2006.08.011
Cantón, R., González-Alba, J. M., and Galán, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/FMICB.2012.00110/XML/NLM
Capy, P., Gasperi, G., Biémont, C., and Bazin, C. (2000). Stress and transposable elements: co-evolution or useful parasites? Heredity 85, 101–106. doi: 10.1046/j.1365-2540.2000.00751.x
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chio, H., Guest, E. E., Hobman, J. L., Dottorini, T., Hirst, J. D., and Stekel, D. J. (2023). Predicting bioactivity of antibiotic metabolites by molecular docking and dynamics. J. Mol. Graph. Model. 123:108508. doi: 10.1016/j.jmgm.2023.108508
CLSI (Ed.) (2018). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 11th Edn. Malvern, Pennsylvania, USA: CLSI Document M07-A11.
CLSI (Ed.) (2022). Performance standards for antimicrobial susceptibility testing. 32nd Edn. Malvern, Pennsylvania, USA: CLSI Document M100-S32.
Dhanji, H., Doumith, M., Hope, R., Livermore, D. M., and Woodford, N. (2011). ISEcp1-mediated transposition of linked blaCTX-M-3 and blaTEM-1b from the IncI1 plasmid pEK204 found in clinical isolates of Escherichia coli from Belfast, UK. J. Antimicrobial Chemother. 66, 2263–2265. doi: 10.1093/jac/dkr310
Dierikx, C. M., van Duijkeren, E., Schoormans, A. H., van Essen-Zandbergen, A., Veldman, K., Kant, A., et al. (2012). Occurrence and characteristics of extended-spectrum-β-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J. Antimicrob. Chemother. 67, 1368–1374. doi: 10.1093/jac/dks049
Ding, Y., Saw, W. Y., Tan, L. W. L., Moong, D. K. N., Nagarajan, N., Teo, Y. Y., et al. (2021). Extended-spectrum β-lactamase-producing and mcr-1-positive Escherichia coli from the gut microbiota of healthy Singaporeans. Appl. Environ. Microbiol. 87:e00488-21. doi: 10.1128/AEM.00488-21
Eckert, C., Gautier, V., Saladin-Allard, M., Hidri, N., Verdet, C., Ould-Hocine, Z., et al. (2004). Dissemination of CTX-M-type beta-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 48, 1249–1255. doi: 10.1128/AAC.48.4.1249-1255.2004
Foster, P. L. (2007). Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42, 373–397. doi: 10.1080/10409230701648494
González Pereyra, V., Pol, M., Pastorino, F., and Herrero, A. (2015). Quantification of antimicrobial usage in dairy cows and preweaned calves in Argentina. Prev. Vet. Med. 122, 273–279. doi: 10.1016/J.PREVETMED.2015.10.019
Gulkowska, A., Leung, H. W., So, M. K., Taniyasu, S., Yamashita, N., Yeung, L. W., et al. (2008). Removal of antibiotics from wastewater by sewage treatment facilities in Hong Kong and Shenzhen, China. Water Res. 42, 395–403. doi: 10.1016/j.watres.2007.07.031
Hamamoto, K., Tokunaga, T., Yagi, N., and Hirai, I. (2020). Characterization of blaCTX-M-14 transposition from plasmid to chromosome in Escherichia coli experimental strain. Int. J. Medical Microbiol. 310:151395. doi: 10.1016/j.ijmm.2020.151395
Hastings, P. J., Rosenberg, S. M., and Slack, A. (2004). Antibiotic-induced lateral transfer of antibiotic resistance. Trends Microbiol. 12, 401–404. doi: 10.1016/j.tim.2004.07.003
Hertz, F. B., Løbner-Olesen, A., and Frimodt-Møller, N. (2014). Antibiotic selection of Escherichia coli sequence type 131 in a mouse intestinal colonization model. Antimicrob. Agents Chemother. 58, 6139–6144. doi: 10.1128/AAC.03021-14
Humeniuk, C., Arlet, G., Gautier, V., Grimont, P., Labia, R., and Philippon, A. (2002). Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46, 3045–3049. doi: 10.1128/AAC.46.9.3045-3049.2002
Ibrahim, D. R., Dodd, C. E. R., Stekel, D. J., Ramsden, S. J., and Hobman, J. L. (2016). Multidrug resistant, extended spectrum β-lactamase (ESBL)-producing Escherichia coli isolated from a dairy farm. FEMS Microbiol. Ecol. 92:fiw013. doi: 10.1093/femsec/fiw013
Irrgang, A., Falgenhauer, L., Fischer, J., Ghosh, H., Guiral, E., Guerra, B., et al. (2017). CTX-M-15-producing E. coli isolates from food products in Germany are mainly associated with an IncF-type plasmid and belong to two predominant clonal E. coli lineages. Front. Microbio 8, 2318. doi: 10.3389/FMICB.2017.02318/BIBTEX
Ideses, D., Gophna, U., Paitan, Y., Chaudhuri, R. R., Pallen, M. J., and Ron, E. Z. (2005). A degenerate type III secretion system from septicemic Escherichia coli contributes to pathogenesis. J. Bacteriol. 187, 8164–8171. doi: 10.1128/JB.187.23.8164-8171.2005
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Johnson, A. P., Godden, S. M., Royster, E., Zuidhof, S., Miller, B., and Sorg, J. (2016). Randomized noninferiority study evaluating the efficacy of 2 commercial dry cow mastitis formulations. J. Dairy Sci. 99, 593–607. doi: 10.3168/JDS.2015-10190
Kieffer, N., Poirel, L., Mueller, L., Mancini, S., and Nordmann, P. (2020). ISEcp1-mediated transposition leads to fosfomycin and broad-spectrum cephalosporin resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 64:e00150-20. doi: 10.1128/AAC.00150-20
Kuan, C. T., Liu, S. K., and Tessman, I. (1991). Excision and transposition of Tn5 as an SOS activity in Escherichia coli. Genetics 128, 45–57. doi: 10.1093/genetics/128.1.45
Kulkarni, P., Olson, N. D., Raspanti, G. A., Goldstein, R. E. R., Gibbs, S. G., Sapkota, A., et al. (2017). Antibiotic concentrations decrease during wastewater treatment but persist at low levels in reclaimed water. Int. J. Environ. Res. Public Health 14:668. doi: 10.3390/ijerph14060668
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of Total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Lartigue, M.-F., Poirel, L., Aubert, D., and Nordmann, P. (2006). In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring beta-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob. Agents Chemother. 50, 1282–1286. doi: 10.1128/AAC.50.4.1282-1286.2006
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Liebana, E., Carattoli, A., Coque, T. M., Hasman, H., Magiorakos, A. P., Mevius, D., et al. (2013). Public health risks of enterobacterial isolates producing extended-spectrum β-lactamases or AmpC β-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin. Infect. Dis. 56, 1030–1037. doi: 10.1093/cid/cis1043
Lim, J. Y., Yoon, J. W., and Hovde, C. J. (2010). A brief overview of Escherichia coli O157:H7 and its plasmid O157. J. Microbiol. Biotechnol. 20, 5–14. doi: 10.4014/jmb.0908.08007
Lin, A. Y. C., Yu, T. H., and Lin, C. F. (2008). Pharmaceutical contamination in residential, industrial, and agricultural waste streams: risk to aqueous environments in Taiwan. Chemosphere 74, 131–141. doi: 10.1016/j.chemosphere.2008.08.027
Liu, G., Ding, L., Han, B., Piepers, S., Naqvi, S. A., Barkema, H. W., et al. (2018). Characteristics of Escherichia coli isolated from bovine mastitis exposed to subminimum inhibitory concentrations of Cefalotin or ceftazidime. Biomed. Res. Int. 2018, 1–10. doi: 10.1155/2018/4301628
Ludden, C., Raven, K. E., Jamrozy, D., Gouliouris, T., Blane, B., Coll, F., et al. (2019). One health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. mBio 10:e02693-18. doi: 10.1128/mBio.02693-18
McDougall, S., Penry, J., and Dymock, D. (2021). Antimicrobial susceptibilities in dairy herds that differ in dry cow therapy usage. J. Dairy Sci. 104, 9142–9163. doi: 10.3168/JDS.2020-19925
Medini, D., Donati, C., Tettelin, H., Masignani, V., and Rappuoli, R. (2005). The microbial pan-genome. Curr. Opin. Genet. Dev. 15, 589–594. doi: 10.1016/j.gde.2005.09.006
Miller, C., Thomsen, L. E., Gaggero, C., Mosseri, R., Ingmer, H., and Cohen, S. N. (2004). SOS response induction by β-lactams and bacterial defense against antibiotic lethality. Science 305, 1629–1631. doi: 10.1126/science.1101630
Møller, T. S. B., Liu, G., Boysen, A., Thomsen, L. E., Lüthje, F. L., Mortensen, S., et al. (2017). Treatment with cefotaxime affects expression of conjugation associated proteins and conjugation transfer frequency of an IncI1 plasmid in Escherichia coli. Front. Microbiol. 8:2365. doi: 10.3389/fmicb.2017.02365
Nesporova, K., Ruzickova, M., Tarabai, H., Krejci, S., Masarikova, M., Lausova, J., et al. (2024). Changing dynamics of antibiotic resistant Escherichia in Caspian gulls shows the importance of longitudinal environmental studies. Environ. Int. 186:108606. doi: 10.1016/j.envint.2024.108606
Nordmann, P., Lartigue, M.-F., and Poirel, L. (2008). β-Lactam induction of ISEcp1B-mediated mobilization of the naturally occurring blaCTX-M β-lactamase gene of Kluyvera ascorbata. FEMS Microbiol. Lett. 288, 247–249. doi: 10.1111/j.1574-6968.2008.01359.x
Pandey, H. O., and Upadhyay, D. (2022). “Global livestock production systems: classification, status, and future trends” in Emerging issues in climate smart livestock production. Cambridge, MA, USA: Academic Press, 47–70.
Poirel, L., Decousser, J. W., and Nordmann, P. (2003). Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47, 2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003
Poirel, L., Lartigue, M.-F., Decousser, J.-W., and Nordmann, P. (2005). ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49, 447–450. doi: 10.1128/AAC.49.1.447-450.2005
Poirel, L., Naas, T., and Nordmann, P. (2008). Genetic support of extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14, 75–81. doi: 10.1111/J.1469-0691.2007.01865.X
Rodriguez-Mozaz, S., Vaz-Moreira, I., Varela Della Giustina, S., Llorca, M., Barceló, D., Schubert, S., et al. (2020). Antibiotic residues in final effluents of European wastewater treatment plants and their impact on the aquatic environment. Environ. Int. 140:105733. doi: 10.1016/j.envint.2020.105733
Rossi, R. S., Amarante, A. F., Guerra, S. T., Latosinski, G. S., Rossi, B. F., Rall, V. L. M., et al. (2019). Efficacy of cefquinome and a combination of cloxacillin and ampicillin for treatment of dairy cows with Streptococcus agalactiae subclinical mastitis. PLoS One 14:e0216091. doi: 10.1371/journal.pone.0216091
Rossolini, G. M., D’Andrea, M. M., and Mugnaioli, C. (2008). The spread of CTX-M-type extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14, 33–41. doi: 10.1111/J.1469-0691.2007.01867.X
Siguier, P., Gourbeyre, E., and Chandler, M. (2014). Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol. Rev. 38, 865–891. doi: 10.1111/1574-6976.12067
Silva, A., Silva, V., Tavares, T., López, M., Rojo-Bezares, B., Pereira, J. E., et al. (2024). Rabbits as a reservoir of multidrug-resistant Escherichia coli: clonal lineages and public health impact. Antibiotics 13:376. doi: 10.3390/antibiotics13040376
Skipper, K. A., Andersen, P. R., Sharma, N., and Mikkelsen, J. G. (2013). DNA transposon-based gene vehicles – scenes from an evolutionary drive. J. Biomed. Sci. 20:92. doi: 10.1186/1423-0127-20-92
Smalla, K., Heuer, H., Gotz, A., Niemeyer, D., Krogerrecklenfort, E., and Tietze, E. (2000). Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl. Environ. Microbiol. 66, 4854–4862. doi: 10.1128/AEM.66.11.4854-4862.2000
Sun, J., Li, X. P., Yang, R. S., Fang, L. X., Huo, W., Li, S. M., et al. (2016). Complete nucleotide sequence of an IncI2 plasmid Coharboring blaCTX-M-55 and mcr-1. Antimicrob. Agents Chemother. 60, 5014–5017. doi: 10.1128/AAC.00774-16
Tettelin, H., Riley, D., Cattuto, C., and Medini, D. (2008). Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 11, 472–477. doi: 10.1016/j.mib.2008.09.006
Todman, H., Helliwell, R., King, L., Blanchard, A., Gray-Hammerton, C. J., Hooton, S. P., et al. (2024). Modelling the impact of wastewater flows and management practices on antimicrobial resistance in dairy farms. NPJ Antimicrobials Resistance 2:13. doi: 10.1038/s44259-024-00029-4
Watkinson, A. J., Murby, E. J., and Costanzo, S. D. (2007). Removal of antibiotics in conventional and advanced wastewater treatment: implications for environmental discharge and wastewater recycling. Water Res. 41, 4164–4176. doi: 10.1016/j.watres.2007.04.005
Wei, S.-H.. (2013). Escherichia coli contamination of pork carcasses in UK slaughterhouses. Available online at: http://eprints.nottingham.ac.uk/13477/1/Wei_thesis_2012_for_binding.pdf (Accessed August 6, 2019).
Whitfield, L. K., and Laven, R. A. (2018). A comparison of the effect of short-acting and long-acting cloxacillin-based dry-cow therapy on somatic cell counts after calving in cows also given internal teat sealants. N. Z. Vet. J. 66, 44–47. doi: 10.1080/00480169.2017.1386134
WHO. (2021). Integrated global surveillance on ESBL-producing E. coli using a “one health” approach: implementation and opportunities. Available online at: http://apps.who.int/bookorders. (Accessed June 4, 2025).
WHO. (2024). WHO bacterial priority pathogens list, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. Bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance, p. 72. Available online at: https://www.who.int/publications/i/item/9789240093461 (Accessed May 23, 2025).
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Widyatama, F. S., Yagi, N., Sarassari, R., Shirakawa, T., Le, D. T., Bui, M. H. T., et al. (2021). Analysis of the upstream genetic structures of the ISEcp1-blaCTX-M transposition units in Escherichia coli isolates carrying blaCTX-M obtained from the Indonesian and Vietnamese communities. Microbiol. Immunol. 65, 542–550. doi: 10.1111/1348-0421.12938
Yagi, N., Hamamoto, K., Thi Bui, K. N., Ueda, S., Tawata, S., Le, D. T., et al. (2021). A high-throughput sequencing determination method for upstream genetic structure (UGS) of ISEcp1-blaCTX-M transposition unit and application of the UGS to classification of bacterial isolates possessing blaCTX-M. J. Infect. Chemother. 27, 1288–1294. doi: 10.1016/J.JIAC.2021.04.001
Yu, Y., Cui, C. Y., Kuang, X., Chen, C., Wang, M. G., Liao, X. P., et al. (2021). Prevalence of tet(X4) in Escherichia coli from duck farms in Southeast China. Front. Microbiol. 12:716393. doi: 10.3389/fmicb.2021.716393
Zahra, R., Javeed, S., Malala, B., Babenko, D., and Toleman, M. A. (2018). Analysis of Escherichia coli STs and resistance mechanisms in sewage from Islamabad, Pakistan indicates a difference in E. coli carriage types between South Asia and Europe. J. Antimicrob. Chemother. 73, 1781–1785. doi: 10.1093/jac/dky109
Zhang, T., and Li, B. (2011). Occurrence, transformation, and fate of antibiotics in municipal wastewater treatment plants. Crit. Rev. Environ. Sci. Technol. 41, 951–998. doi: 10.1080/10643380903392692
Zhou, M., Guo, Z., Duan, Q., Hardwidge, P. R., and Zhu, G. (2014). Escherichia coli type III secretion system 2: A new kind of T3SS? Vet. Res. 45:32. doi: 10.1186/1297-9716-45-32
Zong, Z., Partridge, S. R., and Iredell, J. R. (2010). ISEcp1-mediated transposition and homologous recombination can explain the context of blaCTX-M-62 linked to qnrB2. Antimicrob. Agents Chemother. 54, 3039–3042. doi: 10.1128/AAC.00041-10
Keywords: AMR, ESBL, dairy, ISEcp1, transposition, ST2325
Citation: Gray-Hammerton CJ, Hooton SP, Sands K, Walsh TR, Orbegozo Rubio C, Portal EAR, Hudson C, Stekel DJ, Dodd CER and Hobman JL (2025) Sublethal concentrations of antibiotics enhance transmission of antibiotic resistance genes in environmental Escherichia coli. Front. Microbiol. 16:1675089. doi: 10.3389/fmicb.2025.1675089
Edited by:
Martin Gruhlke, Gesellschaft für Natur- und Wirkstoffforschung e.V., GermanyReviewed by:
Leonardo Gabriel Panunzi, CEA Saclay, FranceMichaela Projahn, Federal Institute for Risk Assessment (BfR), Germany
Copyright © 2025 Gray-Hammerton, Hooton, Sands, Walsh, Orbegozo Rubio, Portal, Hudson, Stekel, Dodd and Hobman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charlotte J. Gray-Hammerton, Y2hhcmxvdHRlLmdyYXktaGFtbWVydG9uQGJpb2xvZ3kub3guYWMudWs=
 Charlotte J. Gray-Hammerton
Charlotte J. Gray-Hammerton Steven P. Hooton1
Steven P. Hooton1 Claudia Orbegozo Rubio
Claudia Orbegozo Rubio Christopher Hudson
Christopher Hudson Dov J. Stekel
Dov J. Stekel Jon L. Hobman
Jon L. Hobman