- 1Animal Husbandry and Veterinary Research Institute, Wuhan Academy of Agricultural Sciences, Wuhan, China
- 2College of Animal Science and Technology, Yangtze University, Jingzhou, China
To investigate the mechanisms by which Streptomyces thermovulgaris a2 (Sta2) enhances the reduction of antibiotic resistance genes (ARGs) in cattle manure composting, this study compared the effects of commercial microbial inoculant (CK) and its combination with Sta2 (ST). The results showed that the ST treatment extended the thermophilic phase (≥55 °C) to 18 days (compared to 11 days with CK) and increased the removal rates of tetG, sul1, ermQ, aac(6ʹ)-Ib-cr, and intI1/intI2 (by 4.8%–48.4%), simultaneously inhibiting the enrichment of sul2 and ermX. During the thermophilic phase, ST treatment slowed the decline in the abundances of key genera (e.g., Bacillus, Thermobacillus, Brachybacterium) and effectively promoted the growth of Actinomadura and Longispora within Actinobacteria. Redundancy analysis revealed that bacterial community succession (56.3%) and mobile genetic elements (MGEs, 30.7%) were key drivers of ARG dynamics, with intI1 and Firmicutes positively regulating most ARGs. Co-occurrence network analysis identified Lysinibacillus (harboring 9 ARG-MGE associations), Luteimonas (9), Brachybacterium (8), and the pathogen Corynebacterium (6) as multidrug resistant hosts. In summary, ST treatment enhanced the reduction of certain genes and multidrug-resistant host control by prolonging the thermophilic duration, reconstructing the microbial community composition, and effectively inhibiting intI1- and intI2-mediated horizontal gene transfer.
1 Introduction
The widespread use of antibiotics in animal husbandry has resulted in the continuous evolution and dissemination of drug-resistant bacteria and antibiotic resistance genes (ARGs), making livestock manure a significant reservoir of ARGs. For example, Qiu et al. (2022) detected 626 ARG subtypes across 20 classes in fecal samples, with five classes of resistance genes (β-lactams, multidrug, macrolides, aminoglycosides, and tetracyclines) accounting for 86.58% of the total. Once released into the environment through pathways such as rainwater runoff and manure application (Fang et al., 2018), ARGs can undergo horizontal transfer via mobile genetic elements (MGEs) like integrons and transposons or achieve vertical transmission through host proliferation (Duan et al., 2019; Zhu et al., 2023), posing threats to ecological environments and public health (Tong et al., 2022; Wang et al., 2022; Wang et al., 2023).
Aerobic composting is a common manure utilization technology. Most ARGs can be reduced through mechanisms such as high-temperature inactivation (Qian et al., 2016; Liu et al., 2023) and microbial competition (Liu et al., 2021), but the natural composting often leads to ARG residual or even enriched due to insufficient activity of indigenous microorganisms and unstable high-temperature phases (Zhu et al., 2023; Yang et al., 2024). For instance, tetG and sul2 abundance increases by 20–395 times after chicken manure composting (Wang et al., 2024); tetracycline resistance genes remain highly abundant in pig manure compost products (Zhu et al., 2023).
Globally, significant efforts have been directed toward improving ARG mitigation, with notable progress in process optimization (e.g., thermal pretreatment (Zhu et al., 2023) and semi-permeable membrane coverage (Sun et al., 2024)), the addition of amendments [e.g., biochar (Tong et al., 2022) and diatomite (Wei et al., 2022)], and microbial inoculation [e.g., composite microbial agents (Li et al., 2020)]. Among these, microbial inoculation is considered a leading strategy due to its environmental friendliness and cost-effectiveness.
The addition of microbial inoculants can improve ARG removal by reconstructing the microbial community and prolonging the thermophilic phase (Li et al., 2020; Cao et al., 2020). For instance, Bacillus megaterium can reduce the abundance of ARGs and sul1/sul2 enrichment by suppressing the activity of transposon Tn916/1545 (Guo et al., 2020); composite microbial inoculants enhance the removal of β-lactam resistance genes, tn916, and some tetracycline resistance genes by 3.7%–23.8% (Li et al., 2020). However, research has primarily focused on Bacillus-dominated inoculants (Duan et al., 2019; Li et al., 2020), while studies on thermotolerant actinomycetes (e.g., Streptomyces thermophilus) in reducing ARGs remain scarce.
Streptomyces thermophilus exhibits unique advantages due to its temperature adaptability and lignocellulose degradation capabilities. Inoculation with thermophilic Streptomyces can enhance composting efficiency. For example, S. thermoviolaceus LC-10 extends the thermophilic phase and accelerates lignocellulose degradation (Zhang et al., 2024), while Streptomyces thermovulgaris N3C07 shortens the composting cycle (Kocak et al., 2023). Bacillus-Streptomyces synergy enhances total nitrogen content (Zhou et al., 2024), though their collaborative regulation mechanism on ARGs remains unclear. The thermophilic Streptomyces strains autonomously screened during the high-temperature phase of composting (>50 °C) may exhibit stronger environmental adaptability and ARG reduction potential. However, their efficacy in eliminating ARGs, as well as their cooperative mechanisms with commercial agents, still requires in-depth investigation.
Therefore, the present study applied the thermophilic S. thermovulgaris strain a2 (Sta2), previously isolated from the thermophilic phase (>50 °C) of cattle manure composting and identified via 16S rRNA, into a composting system. The aim was to systematically investigate the reduction mechanisms of ARGs in cattle manure composting. By comparing the treatment effects of the commercial microbial agent alone and its combination with Sta2, the study aimed to explore (1) the dynamics of bacterial community succession, ARGs and MGEs during composting; and (2) the interactions among key physicochemical parameters, bacterial communities, and MGEs in driving ARGs, along with identifying dominant driving factors. This study is the first to apply isolated Sta2 in combination with commercial microbial agents for composting, focusing on elucidating its mechanistic role in the removal of ARGs. The findings will provide practical references for optimizing microbial agents to control ARG pollution in livestock and poultry manure.
2 Materials and methods
2.1 Experimental materials
The composting materials used in this study consisted of fresh cow manure, sawdust, and broiler farm bedding (grain hulls, chicken manure, etc.). The fresh manure and sawdust were obtained from a dairy farm (Wuhan, China); the air-dried broiler farm bedding was obtained from broiler farmers (Wuhan, China). The basic characteristics of the composting materials are presented in Supplementary Table S1. Commercial microbial agent (powder form, containing Bacillus, Coprococcus, Lactobacillus, and Saccharomyces among others, with a total viable count ≥1.0 × 1010 CFU/g) was provided by Wuhan Xurun Environmental Protection Technology Co. Strain Sta2 was cultivated under suitable conditions, and a bacterial solution with a total bacterial load of approximately 109 CFU/mL was prepared.
2.2 Experimental design and sample collection
The compost experiment was conducted from July to August 2022 in the rain shed of a dairy farm (Wuhan, China). An uncovered plastic box with an effective volume of 770 L was used as the compost container. The box was insulated with cotton and had evenly spaced ventilation holes (1 cm in diameter) on each side. The total weight of the raw materials used for composting was 400 kg (wet weight). A carbon to nitrogen ratio of 25:1 and a moisture content of approximately 58.0% were achieved by mixing fresh cow manure, broiler bedding, and sawdust in a ratio of 12:3:5 (wet weight). The well-mixed material was divided into two equal parts and loaded into two separate plastic boxes. Then the microbial agent was added, and the material was mixed thoroughly.
The experiment involved two groups, with one group assigned to each box. A commercial microbial agent (150 g) and pure water (600 mL) were added in the control group (labeled CK), whereas the commercial microbial agent (150 g) and Sta2 solution (600 mL) was added in the experimental treatment group (labeled ST). The compost was turned over manually on days 7, 14, 20, 25, and 30 to ensure adequate aeration. To achieve efficient composting, pure water was added on day 7 to adjust the water content to approximately 60% based on the results of water content measurements.
Representative samples were collected before pile turning on days 0 (initial feedstock, no bacterial agent), 3 (early thermophilic), 10 (mid-thermophilic), 20 (late thermophilic), and 35 (mature). A stratified, five-point sampling protocol was used: subsamples were taken from the surface of the pile at approximate depths of 10 cm (top), 30 cm (middle), and 50 cm (lower). For each layer, subsamples were combined, mixed, and homogenized via quartering. The three layer-specific samples were then pooled and underwent a final round of mixing and quartering to form a composite sample. This protocol was performed three times to obtain three technical replicates. Each sample weighed approximately 500 g and was divided into two equal portions, sealed in sterile bags, and stored at 4 °C (for physicochemical analysis) and −80 °C (for DNA analysis), respectively.
2.3 Measurement indicators and methods
The following determinations were repeated three times for all samples.
2.3.1 Temperature measurement
Temperature was measured using a wireless temperature monitor. A metal probe was inserted 30 cm into the center of the compost material. The ambient temperature was monitored in an area without direct sunlight. Temperatures were recorded at a fixed time each day.
2.3.2 pH and moisture content measurement
The pH and moisture content were determined in accordance with the methodology described by Li et al. (2020).
2.3.3 Nitrate-nitrogen (NO3-N) content analysis
The samples were extracted using 2 mol/L potassium chloride. The resulting extract was diluted according to a standard curve, and the NO3-N content was analyzed using a flow analyzer (AA3, SEAL Analytical, UK).
2.4 DNA extraction and quantitative polymerase chain reaction (qPCR)
Total DNA was extracted from samples using the FastDNA® Spin Kit for Soil (MP Biomedical, USA). The purity and concentration of DNA were determined using a Nanodrop ONEC spectrophotometer (Thermo Fisher Scientific, USA), and all extracts were of high quality (A260/A280 = 1.82 ± 0.02). The DNA was stored at −80 °C, and the relevant concentration data are presented in Supplementary Table S2.
The analysis targeted 22 genes, covering 7 tetracycline genes (tetB/P, tetC, tetG, tetQ, tetT, tetW, and tetX), 3 sulfonamide genes (sul1, sul2, and dfrA7), 2 macrolide genes (ermQ and ermX), 5 quinolone genes (gyrA, parC, qnrA, qnrC, and qnrS), 1 aminoglycoside gene (aac(6′)-Ib-cr), and 4 MGEs (intI1, intI2, Tn916/1545, and ISCRI) (Supplementary Table S4). This selection was based on their clinical relevance in dairy farming, documented environmental prevalence, and the role of MGEs in horizontal gene transfer (Oliver et al., 2020; Zhang et al., 2020; Qiu et al., 2022).
The target genes were screened by conventional PCR using gene-specific primers (Tsingke Biotechnology Co., Ltd., Beijing, China; sequences in Supplementary Table S3). Amplification products were verified by 1.5% agarose gel electrophoresis, and only samples with a single band of the correct size were analyzed by qPCR (Supplementary Table S4). Quantification of the target and 16S rRNA genes was performed in triplicate using a Bio-Rad CFX Connect™ real-time PCR system with DNA templates uniformly diluted to 10 ng/μL. Reaction specificity was verified by melting curve analysis. The standard curve showed a correlation coefficient (R2) ≥ 0.99, with the detection limit set at Ct = 31. The relative abundance of the target gene was calculated via the 2^(–ΔΔCt) method, using the 16S rRNA gene as the internal reference. Detailed reaction systems and thermal cycling conditions are provided in the Supplemental material (Liu et al., 2023; Li et al., 2020).
2.5 Amplicon sequencing
The V3-V4 region of the bacterial 16S rRNA gene was amplified using primers 341F/805R and sequenced on an Illumina MiSeq platform by Wuhan Benetech Co., Ltd. Raw sequencing reads were quality-filtered using Trimmomatic (v0.39). Subsequent processing within QIIME2 (v2022.3) involved denoising, paired-read merging, and chimera removal with the DADA2 plugin to generate amplicon sequence variants (ASVs). The ASV table was then filtered to remove rare ASVs and rarefied to the minimum sequencing depth for downstream statistical analysis.
2.6 Data processing and analysis
Data were analyzed using Microsoft Excel (2016), SPSS 25.0, and R (v4.2.2). Analyses included: t-tests for within-group differences of target genes; DESeq2 for differential microbial abundance; correlation analyses (Spearman/Pearson), and redundancy analysis (CANOCO 5.1). Principal coordinates analysis based on Bray–Curtis distance was conducted to examine community structure variation. Visualizations were created with GraphPad Prism 8.0.2 (trend lines), RAW Graphs 2.0 and Paysono platforms (heatmaps), and Gephi 0.9.2 (network diagrams).
3 Results
3.1 Changes in physicochemical parameters
The trends in characteristic physicochemical parameters during the composting process are illustrated in Figure 1. The temperatures of the two treatments rapidly increased to approximate 50 °C on day 1, remained above 50 °C on days 2–22, and then gradually decreased to ambient temperature (Figure 1A). The ST group sustained temperatures above 55 °C for 18 days, exceeding this duration for the CK group by 7 days and thereby meeting the sanitary requirements for fecal treatment. Both treatments exhibited a unimodal pH pattern (Figure 1B), peaking at approximately 8.8 on day 10 and declining gradually to ~7.0 by the end of compositing. Moisture content displayed an overall decreasing trend (Figure 1C), with a rapid decline from 0 to 5 days and a transient increase after moisture adjustment on day 7. The final moisture content decreased from 58.3% to 43.3% (CK) and 41.0% (ST), with the ST group achieving a 15.3% higher evaporation efficiency. The NO₃-N content (Figure 1D) exhibited stage-specific accumulation, initiated at 0.3 g/kg. It increased gradually during the first 20 days, followed by rapid accumulation from day 20–35, ultimately reaching 1.1 g/kg (CK) and 1.0 g/kg (ST).
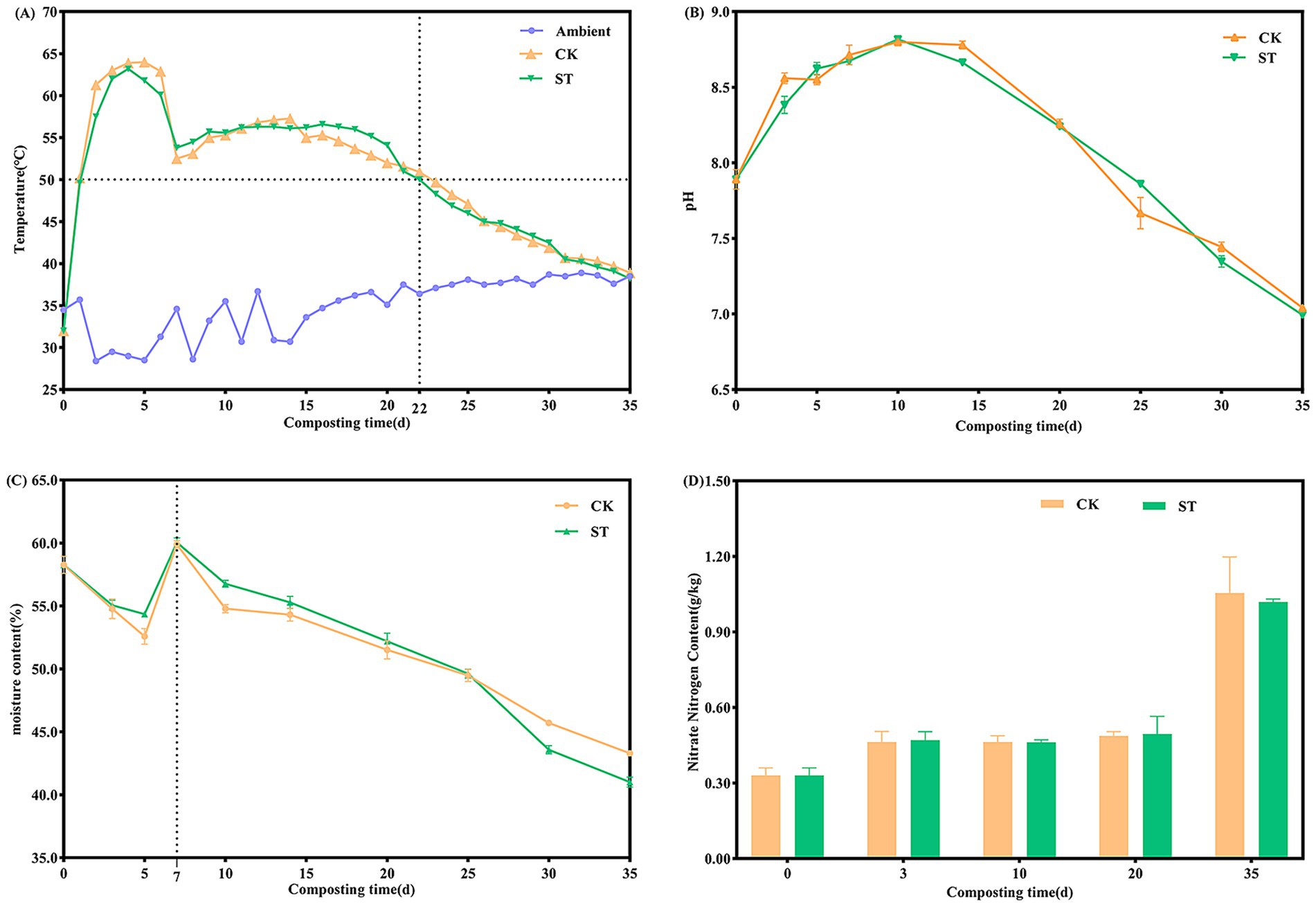
Figure 1. Changes in physicochemical parameters during composting: (A) temperature (on-line monitoring), (B) pH, (C) moisture content, and (D) NO₃-N content. Data points in B–D represent the mean of triplicate replicates, and error bars indicate the standard deviation.
3.2 Changes in ARGs and MGEs
A total of 12 target genes were detected in the composting feedstock, including tetracycline (tetG, tetW, and tetQ), sulfonamide (sul1 and sul2), macrolide (ermQ and ermX), quinolone (qnrC), aminoglycoside (aac(6′)-Ib-cr), as well as MGEs (intI1, intI2, Tn916/1545). These showed varying change trends during composting (Figure 2). Tetracycline ARGs (tetG, tetQ, tetW) generally decreased, with tetW showing a near-continuous decline (Figures 2A–C). Among the sulfonamide ARGs, sul1 decreased overall, whereas sul2 dropped during the high-temperature phase (days 3–10) before rebounding to exceed its initial level (Figures 2D,E). For other ARGs, ermQ exhibited a “decrease–increase–decrease” fluctuation pattern; qnrC and aac(6′)-Ib-cr underwent continuous attenuation; and ermX increased persistently (Figures 2F–I). Of the MGEs, intI1 and Tn916/1545 exhibited sustained overall declines, while intI2 decreased with fluctuations (Figures 2J–L).
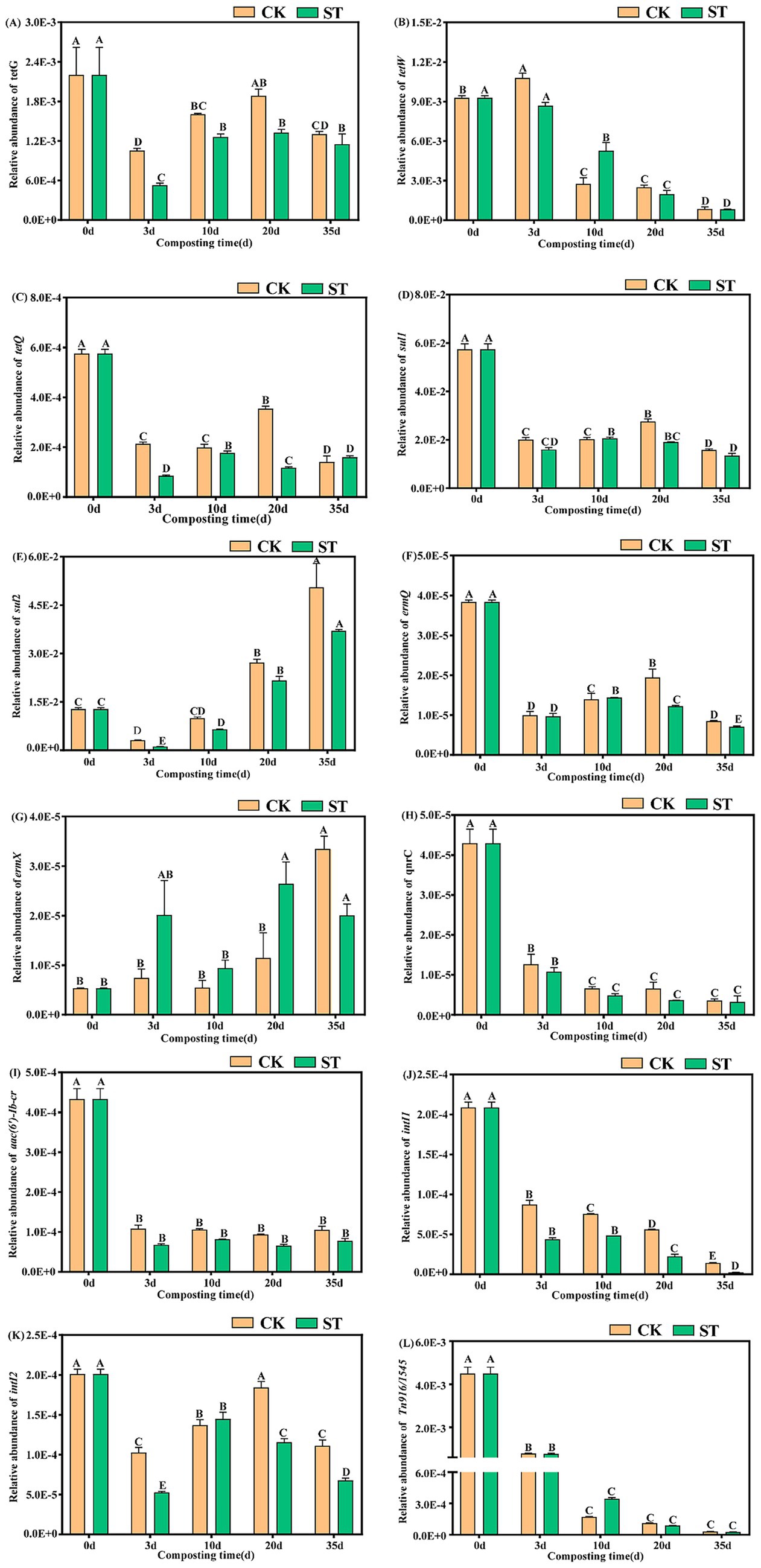
Figure 2. Changes in the relative abundances of ARGs and MGEs during composting. Different capital letters within the same group indicate significant differences (p < 0.01).
At the end of composting, the RAs of tetG, tetW, tetQ, sul1, ermQ, qnrC, and aac(6′)-Ib-cr decreased significantly by 40.7%–91.6% in the CK and 47.7%–92.5% in the ST compared to initial levels (p < 0.01). Similarly, intI1, intI2, and Tn916/1545 decreased by 44.6%–98.8% (CK) and 66.3%–99.4% (ST) (p < 0.01). In contrast, sul2 increased by 3.9-fold (CK) and 2.9-fold (ST) (p < 0.01), while ermX increased by 6.3-fold (CK) and 5.0-fold (ST) (p < 0.01). Compared with CK, the ST group exhibited 4.8%–17.2% greater removal of tetG, sul1, ermQ, and aac(6′)-Ib-cr, and 6.6% and 48.4% greater removal of intI1 and intI2, respectively. Concurrently, the enrichment of sul2 and ermX in ST were lower than those in CK (by 1.0-fold and 1.3-fold, respectively). In conclusion, the ST group enhanced the reduction of ARGs and MGEs, while effectively suppressing the enrichment of sul2 and ermX.
3.3 Changes in bacterial communities
Principal coordinates analysis (PCoA) based on Bray–Curtis distance revealed that PC1 and PC2 together explained 87.5% of the variation (Supplementary Figure S1). Samples from the same time points formed distinct clusters according to sampling time: 0-day samples formed a distinct cluster; 3-day and 10-day samples exhibited higher dispersion; and 20-day and 35-day samples showed the closest proximity. At the phylum level (top 10), the community exhibited a temperature-driven, staged succession (Figure 3A). At the initial stage (0 days), the community was dominated by Firmicutes (42.6%), Actinobacteria (17.2%), Bacteroidetes (15.0%), and Proteobacteria (12.9%), collectively accounting for 87.7% (Supplementary Table S5). In the early high-temperature phase (3 days), the RA of Firmicutes increased (CK 61.2%, ST 58.4%) (p < 0.05 vs. day 0), while those of the other three major phyla decreased. Subsequently, the Firmicutes abundance decreased progressively, while the Proteobacteria, Actinobacteria, and Bacteroidetes increased. Concurrently, Planctomycetes also exhibited substantial proliferation. By the late high-temperature phase (20 days), the microbial community structure had shifted to be dominated by these four phyla (70.8%–72.2%). Compared with the CK group, the ST group exhibited a smaller decrease in Firmicutes abundance (−50.8% vs. -54.8%) and a greater increase in Actinobacteria (+9.9% vs. +6.9%) during the thermophilic phase, while the increase in Planctomycetes was 6.7 percentage points lower (p > 0.05 for all comparisons). During the late composting stage (20–35 days), the ST group displayed a 5.0-percentage-point greater reduction in Actinobacteria than the CK group (p > 0.05). At the conclusion of composting, relative to initial levels, the abundance of Firmicutes (CK: −38.5%, ST: −38.9%; p < 0.05) and Actinobacteria (CK: −2.4%, ST: −1.5%; p < 0.05) declined, while those of Proteobacteria (CK: +15.0%, ST: +14.9%; p > 0.05), Bacteroidetes (CK: +3.5%, ST: +5.2%; p < 0.05), and Planctomycetes (CK: +11.3%, ST: +11.6%; p < 0.05) rose. The abundance of Chloroflexi increased to 6.7% (CK) and 4.1% (ST) (p < 0.05) (Supplementary Table S5).
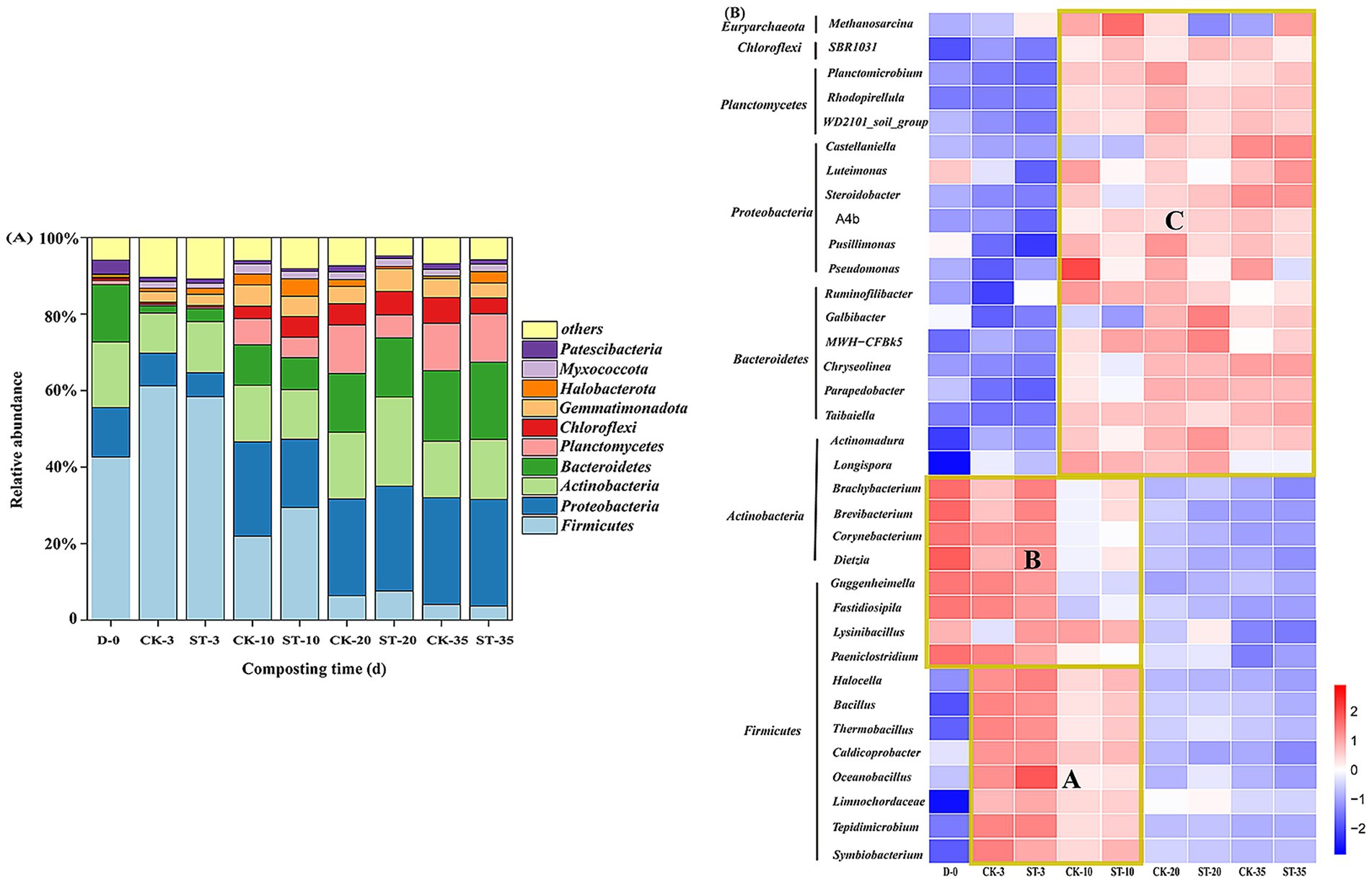
Figure 3. Changes in the relative abundances of the dominant phyla (A) and genera (B) during compositing (based on the mean values of three technical replicates).
At the genus level (top 35), the microbial communities clustered into three groups (A, B, and C) based on their abundance profiles across different composting phases (Figure 3B). Cluster A, comprising eight Firmicutes genera (e.g., Bacillus, Thermobacillus), increased sharply from 1.8% to 73.1%–78.5% (p < 0.001) by day 3, then declined to 5.4–6.9% (p < 0.001) by the end of composting (lower in the ST group, p > 0.05) (Supplementary Table S6). On day 10, the ST group abundance (39.1%) was higher than that of the CK group (28.9%) (p > 0.05). By day 20, four genera (e.g., Bacillus, Thermobacillus) within this cluster maintained higher proportions in the ST group than in the CK group. Cluster B, including four Firmicutes (e.g., Lysinibacillus) and four Actinobacteria genera (e.g., Corynebacterium), initially dominated (79.6%; 62.2% contributed by Actinobacteria) but declined to 1.1%–1.5% (p < 0.001) by the end (Supplementary Table S6), with Corynebacterium decreasing by >98.7% (p < 0.05). The ST group exhibited higher abundances of specific genera: on day 10, Actinobacteria genera and Fastidiosipila were 1.7- and 2.3-fold more abundant than in CK, respectively; by day 20, Lysinibacillus (1.4% vs. 0.6%, p < 0.05) and Brachybacterium (0.3% vs. 0.2%, p > 0.05) remained more abundant. Cluster C, comprising 19 genera from Bacteroidetes, Proteobacteria, Chloroflexi, and Euryarchaeota, increased from 18.5% to 91.6–93.5% (p < 0.001) by the end (Supplementary Table S6). Notably, key initial genera within this cluster, including Luteimonas (4.7%), Pusillimonas (2.6%), Pseudomonas (2.7%), and Methanosarcina (3.6%), exhibited divergent successional patterns: Luteimonas and Pusillimonas exhibited no substantial increase, whereas Pseudomonas abundance increased by 2.7-fold (CK, p < 0.05) and 0.41-fold (ST, p > 0.05) relative to initial levels. Methanosarcina peaked on day 10 (CK: 7.7%; ST: 11.4%, p > 0.05) then declining to 2.1% (CK) and 9.8% (ST) (p < 0.05). Actinomadura and Longispora increased to 12.6% (CK) and 19.5% (ST) (p < 0.001) on day 20 (Supplementary Table S7), and ultimately reached 26.1-fold (CK) and 54.5-fold (ST) (p < 0.001) of the initial abundance. Initially rare genera (abundance <0.2%), such as Steroidobacter (10.8%–11.4%), Castellaniella (7.4%–7.8%), and Chryseolinea (23.6%–25.1%) (p < 0.05), emerged as dominant taxa by the end (Supplementary Table S7). Additionally, the inoculated strain Sta2 was not detected in any samples. Overall, temperature dynamics drove significant microbial succession.
3.4 Correlation among microbiota, MGEs, physicochemical parameters, and ARGs
Redundancy analysis (RDA) was performed to analyze the effects of physicochemical parameters (pH, temperature, moisture, and NO3-N), the bacterial community (at the phylum level), and MGEs (intI1, intI2, and Tn916/1545) on the dynamics of ARGs during composting (Figure 4). The results showed that these variables collectively explained 93.7% of the total variation in ARGs (RDA1 58.8%, RDA2 34.9%). The bacterial community exhibited the highest contribution (56.3%), followed by MGEs (30.7%), while physicochemical parameters had the lowest contribution (6.7%). The bacterial community and MGEs collectively explained 87.0% of the variation in ARGs, with intI1 and Firmicutes responsible for 49.3% and 25.6% of the variation, respectively.
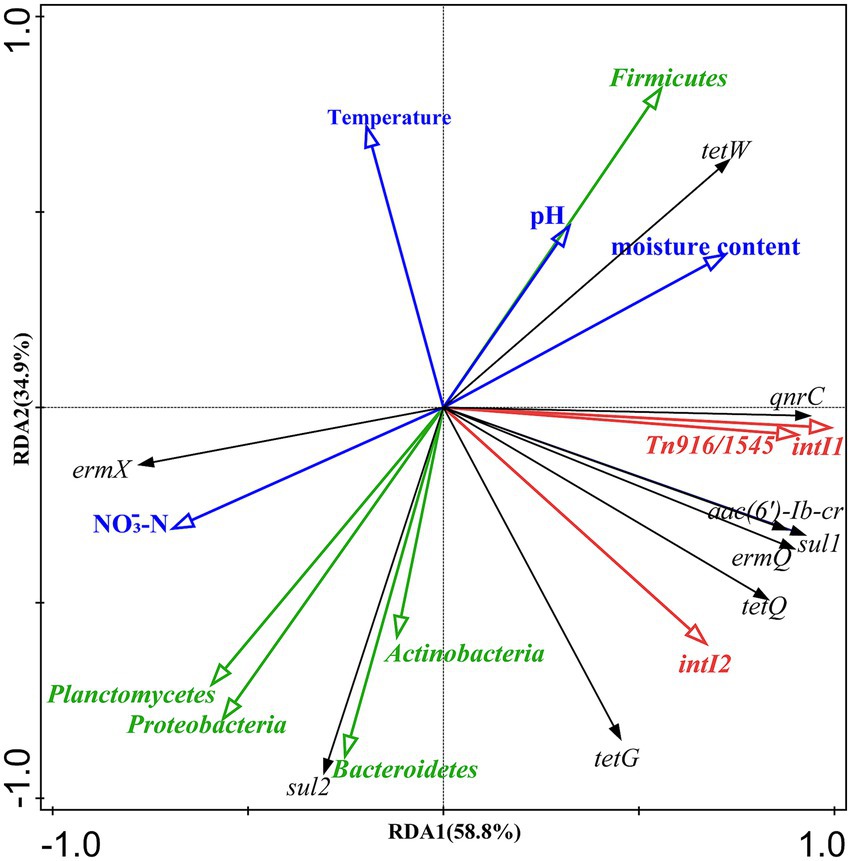
Figure 4. Redundancy analysis of bacterial phyla, physicochemical properties, ARGs and MGEs. The arrows represent different variable groups: bacterial phyla (green), physicochemical parameters (sky blue), ARGs (black), MGEs (red).
Spearman correlation analysis revealed distinct association patterns between bacterial phyla and ARGs/MGEs (Supplementary Figure S2). Firmicutes showed positive correlation with most targets, exhibiting significant positive correlations with tetW, qnrC, and MGEs (intI1, Tn916/1545) (p < 0.01). Actinobacteria was significantly positively correlated with tetG and sul2 (p < 0.01), and with intI2 (p < 0.05). Although generally negatively correlated with most genes, Proteobacteria showed significant positive correlations with sul2 and ermX (p < 0.01). Bacteroidetes was significantly positively correlated solely with sul2 (p < 0.01).
Pearson correlation analysis (Supplementary Figure S3) revealed specific associations between MGEs and ARGs. intI1 and Tn916/1545 showed significant positive correlations (p < 0.01) with five ARGs (e.g., tetQ, sul1); intI2 was significantly positively correlated (p < 0.01) with tetG, tetQ, and ermQ. MGEs were overall negatively correlated with sul2 and ermX, with a significant negative correlation between intI1 and ermX (p < 0.05). Moreover, sul2 and ermX showed weak, non-significant negative correlations (p > 0.05) with most other ARGs.
Regarding physicochemical parameters (Supplementary Figure S2), temperature was negatively correlated with most genes, showing highly significant correlations with tetG, sul2, aac(6′)-Ib-cr, and intI2 (p < 0.01). pH correlated negatively with sul2 (p < 0.01) and positively with tetW (p < 0.05). Moisture content showed significant positive correlations, whereas NO₃-N showed significant negative correlations, with the same five ARGs (e.g., tetW, sul1) and the three MGEs (p < 0.01).
3.5 Co-occurrence network of ARGs, MGEs, and microbial communities
Spearman correlation analysis (R > 0.8, p < 0.01) was used to construct a co-occurrence network among nine ARGs, three MGEs, and the top 35 most abundant bacterial genera (Figure 5). The results revealed significant positive correlations (p < 0.01) between all bacterial genera and the target genes. Based on co-occurrence patterns, Firmicutes (12 genera), Proteobacteria (6 genera), Actinobacteria (6 genera), and Bacteroidetes (5 genera) could represent the primary potential host phyla, collectively accounting for 82.9% of the associated genera. The number of genera associated with each gene varied widely: intI1 and tetG (24 each), sul2 and ermX (15 each), tetW (13), qnrC (4).
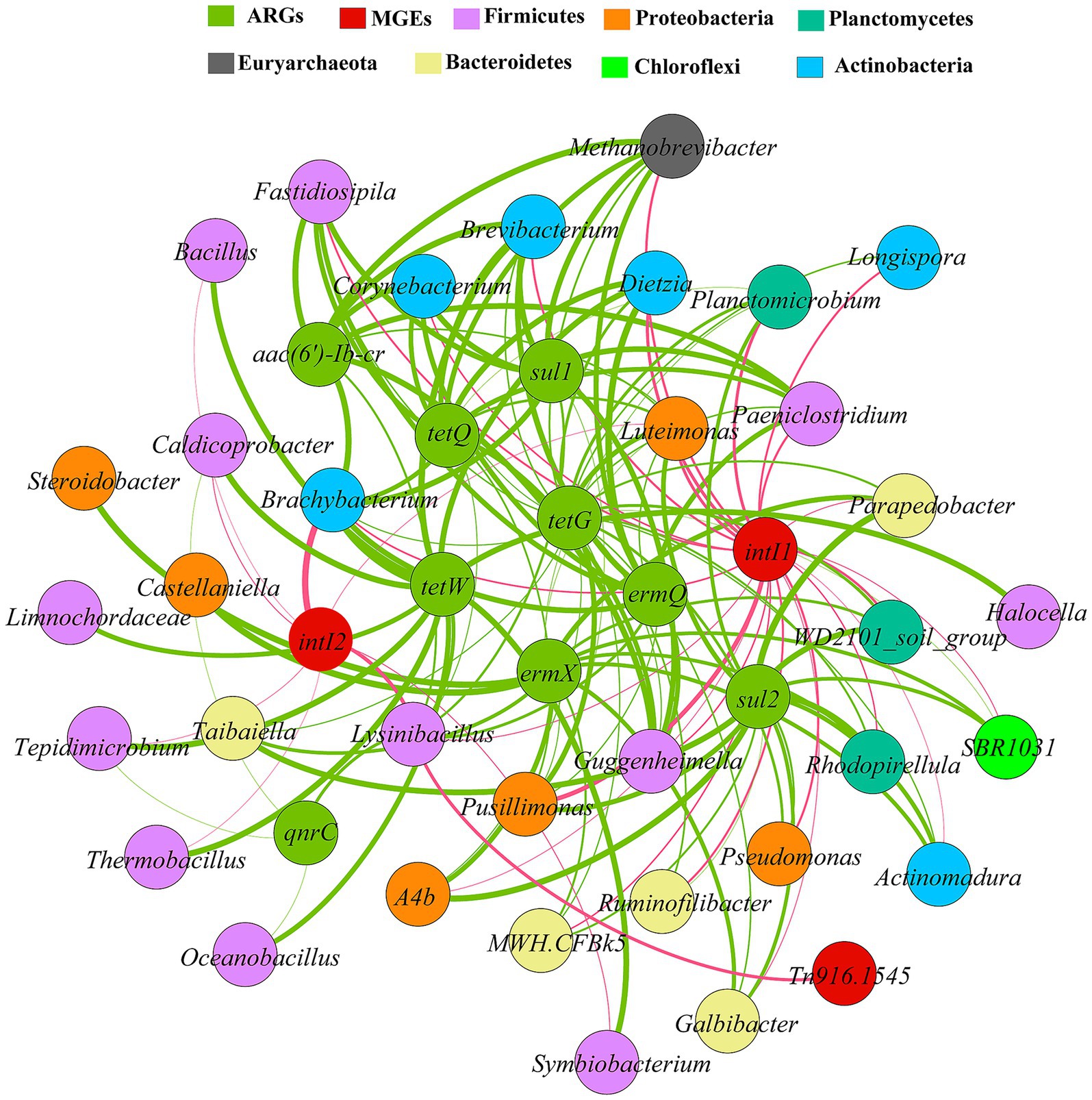
Figure 5. Network analysis of the co-occurrence of ARGs, MGEs and potential host bacteria. Connecting lines indicate significant positive correlations (p < 0.05) according to Spearman correlation coefficients (R > 0.8). The thickness of the line represents the magnitude of the correlation.
Multi-gene co-occurrence was prevalent. For example, Lysinibacillus was associated with nine genes: tetW, tetQ, sul1, qnrC, ermQ, aac(6′)-Ib-cr, intI1, intI2, and Tn916/1545. Similarly, Luteimonas was also associated with nine genes: tetG, tetQ, sul1, sul2, ermX, ermQ, aac(6′)-Ib-cr, intI1, and intI2, while Brachybacterium and Brevibacterium were linked to eight and seven genes, respectively. Furthermore, ermX and sul2 exhibited highly overlapping associated genera (sharing 13 out of 15), distributed across Bacteroidetes, Planctomycetes, and Proteobacteria. The integrons intI1 and intI2 were associated with 24 and 8 genera, respectively, and the transposon Tn916/1545 with one genus. Notably, Lysinibacillus was linked to three MGEs and Luteimonas to both integrons. Associations involving human pathogens were also identified: Corynebacterium with tetG, tetQ, sul1, ermQ, aac(6′)-Ib-cr, and intI1; Pseudomonas with tetG, sul2, and intI2; and Bacillus with tetW and intI2.
4 Discussion
In this study, co-inoculation extended the thermophilic phase (>55 °C) to 18 days, outperforming the 12-day duration achieved by single S. thermophilus inoculation (Zhang et al., 2024). This enhanced performance can be attributed to the synergistic interaction between the commercial inoculant and strain Sta2. Specifically, the unique role of Sta2 lies not in self-proliferation but in its synergistic stimulation with the commercial inoculant, which prompted targeted thermophile enrichment (Zhang et al., 2024). The resultant sustained microbial activity facilitated continuous organic matter decomposition and heat generation (Wei et al., 2019; Dong et al., 2024), thereby enhancing moisture evaporation in the ST group. During the cooling/maturation phase, the rapid accumulation of NO3-N aligned with the restored activity of nitrifying bacteria following temperature decrease (Yuan et al., 2018). The initial increase in pH, driven by ammonia released from organic matter decomposition, was followed by a gradual decline due to nitrification and organic acid accumulation (Liu et al., 2021). Collectively, these results indicate that the ST treatment enhanced the composting efficiency by extending the thermophilic phase, intensifying organic matter decomposition, and optimizing nitrogen transformation.
The findings demonstrate that addition of microbial agents during composting effectively remove most of the ARGs and MGEs (Li et al., 2020; Cao et al., 2020). Specifically, the reductions in tetracycline resistance genes align with literature (Duan et al., 2019). The lower removal of tetG (40.7%–47.7%) may be attributed to its efflux pump mechanism (Guo et al., 2019) and to the persistence of potential host bacteria (e.g., Actinomadura and Pseudomonas), which significantly correlated with tetG abundance (Figure 5). Sulfonamide resistance genes (sul1/sul2) declined primarily during the thermophilic phase, underscoring its importance for their removal (Qian et al., 2016). The persistent attenuation of sul1 may stem from thermal inactivation of host bacteria (Sun et al., 2024), while later-stage sul2 enrichment could be related to its broad host adaptability (Wei et al., 2022) or the presence of thermophilic hosts (Sun et al., 2024). The persistent declines in aac(6′)-Ib-cr, ermQ, and qnrC are also consistent with previous research (Wei et al., 2022; Li et al., 2022). In contrast, ermX enrichment may primarily stem from vertical transmission during host bacterial proliferation (Wang et al., 2021).
The significant reductions in three MGEs align with previous research findings (Liu et al., 2021; Cao et al., 2020). However, the removal efficiency of intI2 remained moderate (44.6%–66.3%), which may be associated with specific microbial community succession (Tong et al., 2022). This persistent intI2 may promote horizontal gene transfer, increasing the risk of ARG dissemination and potentially leading to the rebound of certain ARGs (Liu et al., 2023). Furthermore, while the set of target genes used was representative, it was not exhaustive. Future studies could employ expanded screening or metagenomics to achieve broader coverage of ARGs, including β-lactamases.
Collectively, microbial inoculants likely enhance ARG reduction by promoting the succession of the microbial community, accelerating organic matter degradation (Li et al., 2020; Cao et al., 2020), and thermally inactivating hosts or degrading ARGs (Chen et al., 2019). Notably, the ST group exhibited an enhanced effect, achieving greater removal efficiencies for several targets (particularly intI2) while also curbing the enrichment of sul2 and ermX. These results demonstrate that combining Sta2 with commercial microbial agents enhanced the reduction of resistance genes, as also reported for a composite microbial inoculant by Li et al. (2020). The primary mechanism may involve the introduction of Sta2 stimulating the proliferation of thermotolerant bacteria, which alters the community structure and prolongs the thermophilic phase via sustained organic matter degradation, leading to efficient killing or competitive inhibition of host bacteria (Wei et al., 2019; Duan et al., 2019; Wang et al., 2021). Concurrently, Sta2 inhibits the activity of intI1/intI2 to impede horizontal gene transfer, as previously observed (Duan et al., 2019; Liu et al., 2021). Thus, combining Sta2 with commercial inoculants is a viable strategy for enhancing the reduction of ARGs/MGEs while mitigating the enrichment of persistent ARGs.
The succession of microbial communities during composting displays temperature-driven phasic characteristics, supporting temperature as a key driver of microbial succession (Zhang et al., 2020; López et al., 2021). The greater dispersion during the thermophilic phase likely reflects the differential effects of the inoculants, whereas the convergence of samples during the cooling/maturation phase indicates the establishment of a stable microbial community structure.
At the phylum level, the initial dominance of Firmicutes, Actinobacteria, Bacteroidotes, and Proteobacteria agrees with typical cattle manure composition (Zhang et al., 2020). The thermophilic nature of Firmicutes accounts for its rapid proliferation during the early high-temperature phase (López et al., 2021), which may have competitively inhibited other phyla (Tong et al., 2022). Subsequent succession, characterized by a decreasing Firmicutes and increasing in Actinobacteria, Bacteroidetes, and Proteobacteria, followed the established trend reported in previous studies (Hu et al., 2019; Li et al., 2020). The late-stage increase of Chloroflexi is associated with its thermophilic nature (Wang et al., 2021). The functional importance of these phyla is well-established: Actinobacteria, Bacteroidetes, Proteobacteria, and Chloroflexi are recognized as key participants in lignocellulose degradation (Guo et al., 2021; Li et al., 2023), whereas Proteobacteria and Planctomycetes play crucial roles in nitrogen transformation (Tong et al., 2022; Zhou and Yao, 2020). Their collective rise in later stages suggests synergistic promotion of these processes. The introduction of Sta2 influenced community dynamics: during the thermophilic phase, it was associated with a smaller reduction in Firmicutes and a greater increase in Actinobacteria compared to CK group, indicating that Sta2 may promote actinobacterial proliferation (Liu et al., 2022). The concurrent smaller increase in Planctomycetes may result from competitive inhibition by Sta2 metabolites (Zhang et al., 2024). Furthermore, Actinobacteria underwent a greater reduction in the ST group during the cooling/maturation phase, likely because the introduction of Sta2 prolonged the high-temperature period, which accelerated organic matter decomposition and microbial succession (Dong et al., 2024). The final dominance of Proteobacteria, Bacteroidetes, and Planctomycetes indicating compost maturation (Li et al., 2020; Zhou and Yao, 2020).
Genus-level cluster analysis clarified the successional trajectory of functional microbial communities. The rapid proliferation of Cluster A genera (e.g., Bacillus, Thermobacillus) in the first 3 days was likely stimulated by exogenous microbial agents, enhancing microbial activity (Guo et al., 2020), promoting decomposition of organic matter and temperature rise (Zhou et al., 2019; Cao et al., 2020), and driving Firmicutes abundance dynamics. The sustainedly higher abundance of thermophilic genera (e.g., Bacillus, Thermobacillus) in the ST group during the high-temperature phase may be attributed to the extended high temperatures favoring their growth (Zhou et al., 2019), while its lower final abundance was likely due to the recolonization of mesophilic microorganisms and fungi during the cooling/maturation phase (Ezugworie et al., 2021).
The initial high abundance of Class B, particularly of Actinobacteria genera, is crucial for the transformation of organic compounds in the early stages, especially lignocellulose decomposition (Liu et al., 2022). Its significant decrease by the final phase, particularly Corynebacterium, was likely due to thermophilic-phase inhibition of mesophilic genera (Liu et al., 2020). The higher abundances of specific functional genera (e.g., Brachybacterium, Lysinibacillus) in the ST group during the thermophilic phase suggest that Sta2 may support the survival of these taxa by enhancing enzymatic activity and reshaping the microbial community (Zhao et al., 2017; Wei et al., 2019).
The dominance of Class C in the mid-to-late stages drove organic matter transformation and humification. Initially high-abundance genera (e.g., Luteimonas, Pseudomonas) likely originated primarily from feedstock, while their persistence may be linked to thermotolerance (Zhang et al., 2020; Li et al., 2023). Actinomadura and Longispora emerged as the primary Actinobacterial contributors in mid-late stages, effectively decomposing recalcitrant compounds (Wei et al., 2019). Their consistently higher abundance in the ST group suggests that Sta2 may have promoted their proliferation (Liu et al., 2022). Conversely, the smaller increase in Pseudomonas in the ST group may reflect inhibition from the prolonged thermophilic phase (Liu et al., 2020).
Moreover, several initially rare genera were enriched as dominant taxa by the conclusion of composting, such as Steroidobacter and Castellaniella for nitrogen transformation (Tong et al., 2022), and Chryseolinea for macromolecule degradation (Guo et al., 2021). This enrichment is imperative for compost maturation. Notably, the sustained higher abundance of Methanosarcina in the ST group may possibly be due to a positive effect of Sta2 introduction on its growth or local anaerobic microenvironments, though the responsible mechanisms require further study. In summary, Sta2 likely accelerates maturation by synergistically sustaining thermophilic-phase activity of heat-resistant bacteria (e.g., Bacillus, Thermobacillus, Brachybacterium), enhancing cellulose degraders (e.g., Actinomadura, Longispora), and driving functional microbiota succession (e.g., Castellaniella) in the maturation phase (Dong et al., 2024). The absence of the inoculated strain Sta2 in the sequencing data implies a lack of successful colonization, supporting its role as an “ecological initiator” that regulates indigenous microbes rather than proliferates itself (Zhang et al., 2024). Thus, this mode of action presents an inherently low risk for ARG dissemination. The genomic characterization of strain Sta2 remains a target for future research.
RDA revealed that the dynamic changes of ARGs during composting were primarily driven by microbial community succession, MGEs, and physicochemical parameters, which is consistent with previous studies (Wang et al., 2022). Among these factors, microbial communities exhibited the highest contribution rate, indicating their role as the primary driver of ARG evolution (Zhang et al., 2020; Zhao et al., 2021). Bacterial communities and MGEs collectively explained the vast majority of the variation in ARGs, directly implicating them as drivers (Liu et al., 2021). Notably, intI1 and the phylum Firmicutes exhibited the highest independent explanatory power for ARG variation, suggesting their crucial role in influencing ARG dynamics (Duan et al., 2019). The significant positive correlations of Firmicutes with tetW, qnrC, and key MGEs indicate that changes in this phylum may influence the dynamics of these genes and that specific taxa within it could be potential hosts (Qiu et al., 2022; Wang et al., 2024). The substantial decrease in Firmicutes abundance post-composting, especially in the ST group, was likely the primary factor causing the reductions in ARGs/MGEs and the enhanced removal efficiency of specific genes in the ST group (Duan et al., 2019). Similarly, Actinobacteria, significantly positively correlated with tetG, sul2, and intI2 might be associated with their persistence. Conversely, the increased abundance of Proteobacteria and Bacteroidetes during the late composting stage supported the maintenance of sul2 and ermX, aligning with their respective significant positive correlations.
The high contribution of MGEs to the variation of ARGs, particularly intI1, highlights their crucial role in ARG dynamics (Tong et al., 2022). The significant positive correlations of intI1 and Tn916/1545 with five ARGs, as well as of intI2 with tetG, tetQ, and ermQ, indicate that MGEs may play pivotal roles in the horizontal transfer of these ARGs (Tong et al., 2022; Li et al., 2022). The overall negative correlation between MGEs and sul2/ermX implies that horizontal transfer may not be the primary driver for these two genes (Wang et al., 2024). The significant post-composting reduction in MGE abundance confirms that inhibition of MGE-mediated horizontal transfer can effectively reduce ARGs (Wei et al., 2022; Zhu et al., 2023). The greater reductions of intI1 and intI2 in the ST group suggest that Sta2 may suppress MGE proliferation, consistent with the reported effect of microbial agents on intI1 (Duan et al., 2019). Furthermore, the weak, non-significant correlations of sul2 and ermX with most other ARGs indicate that their enrichment did not impede the removal of other genes, possibly due to differential ARG responses and host adaptation strategies during composting (Cao et al., 2020; Wang et al., 2024).
Moreover, physicochemical parameters can indirectly influence ARG dynamics by regulating microbial activity (Sun et al., 2024). The negative associations of temperature with most ARGs/MGEs suggest it may act as a primary driver of ARG variation (Ma et al., 2023; Sun et al., 2024). pH exhibited gene-specific effects, with its positive correlation with tetW and negative correlation with sul2, consistent with prior studies (Duan et al., 2019; Zhang et al., 2024). Moisture content positively correlated with multiple ARGs, aligning with the results of Zhang et al. (2020). Meanwhile, NO₃-N showed negative associations, which may be related to its indirect effects on potential host bacteria (Wang et al., 2020). These findings demonstrate that physicochemical parameters collectively shape ARG fate during composting by driving microbial succession (Liu et al., 2021). In summary, ARG reduction during composting is a synergistic process dominated by microbial community succession (Li et al., 2022), mediated by horizontal gene transfer via MGEs (Tong et al., 2022), and modulated by key physicochemical parameters (Wang et al., 2020). The introduction of Sta2 represents a promising strategy for reducing relevant ARGs through a greater reduction in Firmicutes abundance and suppressing MGE activity (Duan et al., 2019; Tong et al., 2022).
Co-occurrence network analysis revealed complex interactions among ARGs, MGEs, and microbial communities during composting. The network suggested that Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes could represent the primary potential host, which aligns with previous studies (Zhang et al., 2021; Qiu et al., 2022). A key finding was the heterogeneity in host range: intI1 and tetG were linked to a wide range of genera, pointing to a broad dissemination potential (Zhang et al., 2023); whereas the narrow associations of qnrC and Tn916/1545, may be attributed to competitive inhibition by compost inoculants (Wang et al., 2021). In this study, genera such as Lysinibacillus, Luteimonas, Brachybacterium, and Brevibacterium were significantly associated with multiple target genes. This finding supports the view that the widespread occurrence of multi-gene potential hosts can explain the simultaneous presence of multiple ARGs in the environment (Li et al., 2022). By the end of composting, the abundances of genera in Firmicutes and Actinobacteria had decreased, especially in the ST group, which correlates with the observed decline in most ARGs and MGEs, underscoring the link between host dynamics and gene fate (Qiu et al., 2022). Notably, ermX and sul2 shared a high degree of associated genera, suggesting potential co-transmission (Li et al., 2022). Their enrichment in the later stages of composting likely stems from the increased abundance of these shared host genera (Liu et al., 2023).
MGEs exhibited distinct association patterns. While intI1 was correlated with more genera than intI2, their overlapping genera (e.g., Luteimonas) and co-occurrence with transposons like Tn916/1545 suggests a possible synergistic role in ARG dissemination (Lima et al., 2020). Furthermore, pathogenic bacteria acting as key hosts pose potential risks (Wei et al., 2022). Several pathogenic bacteria were identified as multi-ARG-MGE potential hosts: Corynebacterium and Pseudomonas were associated with multiple ARGs and MGEs, such co-occurrence could potentially facilitate gene transfer (Zhou et al., 2021). While Corynebacterium was effectively removed by the end of composting, Pseudomonas and Bacillus (pathogenic genera, Wei et al., 2022) proliferated to varying degrees. This proliferation was significantly suppressed by the ST treatment, highlighting an advantage of the Sta2 inoculant.
5 Conclusion
Co-inoculation with Sta2 and commercial bacterial agents during composting effectively extended the high-temperature phase above 55 °C by 7 days, significantly reduced the relative abundances of most target genes, and enhanced the removal of specific ARGs (tetG, sul1, ermQ, and aac(6′)-Ib-cr) and MGEs (intI1 and intI2) by 4.8%–48.4%, while also inhibiting the enrichment of sul2 and ermX. Bacterial community succession and MGE-mediated horizontal gene transfer served as the core drivers for ARG reduction, with variations in intI1 and Firmicutes abundance exerting the most significant impact. Network analysis identified several genera, such as Lysinibacillus, Luteimonas, Brachybacterium, and the pathogen Corynebacterium, as potential hosts for multidrug-resistant genes. Furthermore, the strategy suppressed pathogens such as Corynebacterium and Pseudomonas, thereby enhancing compost biosafety. Overall, this approach enhanced ARG removal through sustained high-temperature sterilization, microbial community restructuring, and blocking ARG–MGE transmission, providing an effective solution for controlling ARG pollution and improving biosafety in livestock manure composting.
Data availability statement
The sequence data from this study have been submitted to the NCBI SRA under the accession number PRJNA1301025 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1301025).
Ethics statement
The article presents research on animals that do not require ethical approval for their study.
Author contributions
EJ: Conceptualization, Funding acquisition, Methodology, Project administration, Writing – original draft, Writing – review & editing. DG: Formal analysis, Investigation, Visualization, Writing – original draft. YZ: Data curation, Investigation, Writing – review & editing. PW: Resources, Writing – review & editing. JC: Writing – review & editing. PG: Formal analysis, Supervision, Writing – review & editing. PL: Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Tibet Autonomous Region Key R&D Project (XZ201801NB34) and the Wuhan Academy of Agricultural Sciences Scientific and Technological Innovation Project (XTCX2021002, XTCX2022002).
Acknowledgments
We thank Medjaden, Inc. for its assistance in the preparation of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1688304/full#supplementary-material
References
Cao, R., Ben, W., Qiang, Z., and Zhang, J. (2020). Removal of antibiotic resistance genes in pig manure composting influenced by inoculation of compound microbial agents. Bioresour. Technol. 317:123966. doi: 10.1016/j.biortech.2020.123966
Chen, C., Pankow, C. A., Oh, M., Heath, L. S., Zhang, L., Du, P., et al. (2019). Effect of antibiotic use and composting on antibiotic resistance gene abundance and resistome risks of soils receiving manure-derived amendments. Environ. Int. 128, 233–243. doi: 10.1016/j.envint.2019.04.043
Dong, W., Zhou, R., Li, X., Yan, H., Zheng, J., Peng, N., et al. (2024). Effect of simplified inoculum agent on performance and microbiome during cow manure-composting at industrial-scale. Bioresour. Technol. 393:130097. doi: 10.1016/j.biortech.2023.130097
Duan, M., Zhang, Y., Zhou, B., Wang, Q., Gu, J., Liu, G., et al. (2019). Changes in antibiotic resistance genes and mobile genetic elements during cattle manure composting after inoculation with Bacillus subtilis. Bioresour. Technol. 292:122011. doi: 10.1016/j.biortech.2019.122011
Ezugworie, F. N., Igbokwe, V. C., and Onwosi, C. O. (2021). Proliferation of antibiotic-resistant microorganisms and associated genes during composting: an overview of the potential impacts on public health, management and future. Sci. Total Environ. 784:147191. doi: 10.1016/j.scitotenv.2021.147191
Fang, H., Han, L., Zhang, H., Long, Z., Cai, L., and Yu, Y. (2018). Dissemination of antibiotic resistance genes and human pathogenic bacteria from a pig feedlot to the surrounding stream and agricultural soils. J. Hazard. Mater. 357, 53–62. doi: 10.1016/j.jhazmat.2018.05.066
Guo, Y., Chen, Q., Qin, Y., Yang, R., Yang, Q., Wang, Y., et al. (2021). Succession of the microbial communities and function prediction during short-term peach sawdust-based composting. Bioresour. Technol. 332:125079. doi: 10.1016/j.biortech.2021.125079
Guo, H., Gu, J., Wang, X., Nasir, M., Yu, J., Lei, L., et al. (2020). Elucidating the effect of microbial inoculum and ferric chloride as additives on the removal of antibiotic resistance genes from chicken manure during aerobic composting. Bioresour. Technol. 309:122802. doi: 10.1016/j.biortech.2020.122802
Guo, H., Gu, J., Wang, X., Yu, J., Nasir, M., Peng, H., et al. (2019). Responses of antibiotic and heavy metal resistance genes to bamboo charcoal and bamboo vinegar during aerobic composting. Environ. Pollut. 252, 1097–1105. doi: 10.1016/j.envpol.2019.05.014
Hu, T., Wang, X., Zhen, L., Gu, J., Zhang, K., Wang, Q., et al. (2019). Effects of inoculation with lignocellulose-degrading microorganisms on antibiotic resistance genes and the bacterial community during co-composting of swine manure with spent mushroom substrate. Environ. Pollut. 252, 110–118. doi: 10.1016/j.envpol.2019.05.078
Kocak, F., Tanir, S., Cetin, A., and Degirmenci, L. (2023). Simulatenous evaluation of composting experiments and metagenome analyses to illuminate the effect of Streptomyces spp. on organic matter degradation. World J. Microbiol. Biotechnol. 39:70. doi: 10.1007/s11274-023-03516-4
Li, K., Cao, R., Mo, S., Yao, R., Ren, Z., and Wu, J. (2020). Swine manure composting with compound microbial inoculants: removal of antibiotic resistance genes and their associations with microbial community. Front. Microbiol. 11:592592. doi: 10.3389/fmicb.2020.592592
Li, Y., Gu, J., Wang, X., Song, Z., Hu, T., Xie, J., et al. (2022). The fate of antibiotic resistance genes and their influential factors in swine manure composting with sepiolite as additive. Bioresour. Technol. 347:126727. doi: 10.1016/j.biortech.2022.126727
Li, X., Li, K., Wang, Y., Huang, Y., Yang, H., Zhu, P., et al. (2023). Diversity of lignocellulolytic functional genes and heterogeneity of thermophilic microbes during different wastes composting. Bioresour. Technol. 372:128697. doi: 10.1016/j.biortech.2023.128697
Lima, T., Domingues, S., and Silva, G. (2020). Manure as a potential hotspot for antibiotic resistance dissemination by horizontal gene transfer events. Vet. Sci. 7:E110. doi: 10.3390/vetsci7030110
Liu, Y., Ding, L., Wang, B., He, Q., and Wan, D. (2020). Using the modified pine wood as a novel recyclable bulking agent for sewage sludge composting: effect on nitrogen conversion and microbial community structures. Bioresour. Technol. 309:123357. doi: 10.1016/j.biortech.2020.123357
Liu, Z., Wei, Y., Li, J., and Ding, G. (2022). Integrating 16S rRNA amplicon metagenomics and selective culture for developing thermophilic bacterial inoculants to enhance manure composting. Waste Manag. 144, 357–365. doi: 10.1016/j.wasman.2022.04.013
Liu, Q., Wen, X., Li, X., Shan, Z., Cao, Z., Zhang, X., et al. (2023). Doxycycline induces the rebound of three tetracycline resistance genes during maturation of laying hen manure composting by increasing the abundance of potential host bacteria. J. Clean. Prod. 413:137516. doi: 10.1016/j.jclepro.2023.137516
Liu, B., Yu, K., Ahmed, I., Gin, K., Xi, B., Wei, Z., et al. (2021). Key factors driving the fate of antibiotic resistance genes and controlling strategies during aerobic composting of animal manure: a review. Sci. Total Environ. 791:148372. doi: 10.1016/j.scitotenv.2021.148372
Liu, Y., Zheng, L., Cai, Q., Xu, Y., Xie, Z., Liu, J., et al. (2021). Simultaneous reduction of antibiotics and antibiotic resistance genes in pig manure using a composting process with a novel microbial agent. Ecotoxicol. Environ. Saf. 208:111724. doi: 10.1016/j.ecoenv.2020.111724
López, M., Jurado, M., López-González, J., Estrella-González, M., Martínez-Gallardo, M., Toribio, A., et al. (2021). Characterization of thermophilic lignocellulolytic microorganisms in composting. Front. Microbiol. 12:697480. doi: 10.3389/fmicb.2021.697480
Ma, R., Wang, J., Liu, Y., Wang, G., Yang, Y., Liu, Y., et al. (2023). Dynamics of antibiotic resistance genes and bacterial community during pig manure, kitchen waste, and sewage sludge composting. J. Environ. Manag. 345:118651. doi: 10.1016/j.jenvman.2023.118651
Oliver, J., Gooch, C., Lansing, S., Schueler, J., Hurst, J., Sassoubre, L., et al. (2020). Invited review: fate of antibiotic residues, antibiotic-resistant bacteria, and antibiotic resistance genes in US dairy manure management systems. J. Dairy Sci. 103, 1051–1071. doi: 10.3168/jds.2019.16778
Qian, X., Sun, W., Gu, J., Wang, X., Sun, J., Yin, Y., et al. (2016). Variable effects of oxytetracycline on antibiotic resistance gene abundance and the bacterial community during aerobic composting of cow manure. J. Hazard. Mater. 315, 61–69. doi: 10.1016/j.jhazmat.2016.05.002
Qiu, T., Huo, L., Guo, Y., Gao, M., Wang, G., Hu, D., et al. (2022). Metagenomic assembly reveals hosts and mobility of common antibiotic resistome in animal manure and commercial compost. Environ. Microbiome. 17:42. doi: 10.1186/s40793-022-00437-x
Qiu, X., Zhou, G., and Wang, H. (2022). Nanoscale zero-valent iron inhibits the horizontal gene transfer of antibiotic resistance genes in chicken manure compost. J. Hazard. Mater. 422:126883. doi: 10.1016/j.jhazmat.2021.126883
Sun, B., Bai, Z., Li, R., Song, M., Zhang, J., Wang, J., et al. (2024). Efficient elimination of antibiotic resistome in livestock manure by semi-permeable membrane covered hyperthermophilic composting. Bioresour. Technol. 407:131134. doi: 10.1016/j.biortech.2024.131134
Tong, Z., Liu, F., Tian, Y., Zhang, J., Liu, H., Duan, J., et al. (2022). Effect of biochar on antibiotics and antibiotic resistance genes variations during co-composting of pig manure and corn straw. Front. Bioeng. Biotechnol. 10:960476. doi: 10.3389/fbioe.2022.960476
Wang, B., Chen, W., Sa, C., Gao, X., Chang, S., Wei, Y., et al. (2024). Dynamics of antibiotic resistance genes and the association with bacterial community during pig manure composting with chitin and glucosamine addition. Front. Microbiol. 15:1384577. doi: 10.3389/fmicb.2024.1384577
Wang, G., Gao, X., Cai, Y., Li, G., Ma, R., and Yuan, J. (2024). Dynamics of antibiotic resistance genes during manure composting: reduction in herbivores manure and accumulation in carnivores. Environ. Int. 190:108900. doi: 10.1016/j.envint.2024.108900
Wang, Q., Gu, J., Wang, X., Ma, J., Hu, T., Peng, H., et al. (2020). Effects of nano-zerovalent iron on antibiotic resistance genes and mobile genetic elements during swine manure composting. Environ. Pollut. 258:113654. doi: 10.1016/j.envpol.2019.113654
Wang, J., Gu, J., Wang, X., Song, Z., Dai, X., Guo, H., et al. (2021). Enhanced removal of antibiotic resistance genes and mobile genetic elements during swine manure composting inoculated with mature compost. J. Hazard. Mater. 411:125135. doi: 10.1016/j.jhazmat.2021.125135
Wang, G., Kong, Y., Yang, Y., Ma, R., Li, L., Li, G., et al. (2022). Composting temperature directly affects the removal of antibiotic resistance genes and mobile genetic elements in livestock manure. Environ. Pollut. 303:119174. doi: 10.1016/j.envpol.2022.119174
Wang, Z., Yun, H., Li, S., Ji, J., Khan, A., Fu, X., et al. (2023). Factors influencing the transfer and abundance of antibiotic resistance genes in livestock environments in China. Int. J. Environ. Sci. Technol. 20, 2197–2208. doi: 10.1007/s13762-022-04031-z
Wei, Y., Gu, J., Wang, X., Song, Z., Sun, W., Hu, T., et al. (2022). Elucidating the beneficial effects of diatomite for reducing abundances of antibiotic resistance genes during swine manure composting. Sci. Total Environ. 821:153199. doi: 10.1016/j.scitotenv.2022.153199
Wei, Y., Wu, D., Wei, D., Zhao, Y., Wu, J., Xie, X., et al. (2019). Improved lignocellulose-degrading performance during straw composting from diverse sources with actinomycetes inoculation by regulating the key enzyme activities. Bioresour. Technol. 271, 66–74. doi: 10.1016/j.biortech.2018.09.081
Yang, X., Sun, P., Liu, B., Ahmed, I., Xie, Z., and Zhang, B. (2024). Effect of extending high-temperature duration on ARG rebound in a co-composting process for organic wastes. Sustainability 16:5284. doi: 10.3390/su16135284
Yuan, J., Li, Y., Chen, S., Li, D., Tang, H., Chadwick, D., et al. (2018). Effects of phosphogypsum, superphosphate, and dicyandiamide on gaseous emission and compost quality during sewage sludge composting. Bioresour. Technol. 270, 368–376. doi: 10.1016/j.biortech.2018.09.023
Zhang, M., He, L., Liu, Y., Zhao, J., Zhang, J., Chen, J., et al. (2020). Variation of antibiotic resistome during commercial livestock manure composting. Environ. Int. 136:105458. doi: 10.1016/j.envint.2020.105458
Zhang, R., Liu, X., Wang, S., Fang, L., Sun, J., Liu, Y., et al. (2021). Distribution patterns of antibiotic resistance genes and their bacterial hosts in pig farm wastewater treatment systems and soil fertilized with pig manure. Sci. Total Environ. 758:143654. doi: 10.1016/j.scitotenv.2020.143654
Zhang, X., Ma, C., Zhang, W., Li, W., Yu, J., Xue, D., et al. (2020). Shifts in microbial community, pathogenicity-related genes and antibiotic resistance genes during dairy manure piled up. Microb. Biotechnol. 13, 1039–1053. doi: 10.1111/1751-7915.13551
Zhang, W., Yu, C., Yin, S., Chang, X., Chen, K., Xing, Y., et al. (2023). Transmission and retention of antibiotic resistance genes (ARGs) in chicken and sheep manure composting. Bioresour. Technol. 382:129190. doi: 10.1016/j.biortech.2023.129190
Zhang, J., Zou, Y., Wang, S., Zhang, W., Chen, Q., Wang, Q., et al. (2024). The inoculation of Bacillus paralicheniformis and Streptomyces thermoviolaceus enhances the lignocellulose degradation and microbial communities during spent mushroom substrate composting. Environ. Res. 263:120157. doi: 10.1016/j.envres.2024.120157
Zhao, W., Gu, J., Wang, X., Hu, T., Wang, J., Yu, J., et al. (2021). Effects of shrimp shell powder on antibiotic resistance genes and the bacterial community during swine manure composting. Sci. Total Environ. 752:142162. doi: 10.1016/j.scitotenv.2020.142162
Zhao, Y., Zhao, Y., Zhang, Z., Wei, Y., Wang, H., Lu, Q., et al. (2017). Effect of thermo-tolerant actinomycetes inoculation on cellulose degradation and the formation of humic substances during composting. Waste Manag. 68, 64–73. doi: 10.1016/j.wasman.2017.06.022
Zhou, G., Qiu, X., Wu, X., and Lu, S. (2021). Horizontal gene transfer is a key determinant of antibiotic resistance genes profiles during chicken manure composting with the addition of biochar and zeolite. J. Hazard. Mater. 408:124883. doi: 10.1016/j.jhazmat.2020.124883
Zhou, G., Qiu, X., Zhang, J., and Tao, C. (2019). Effects of seaweed fertilizer on enzyme activities, metabolic characteristics, and bacterial communities during maize straw composting. Bioresour. Technol. 286:121375. doi: 10.1016/j.biortech.2019.121375
Zhou, Z., Shi, X., Bhople, P., Jiang, J., Chater, C., Yang, S., et al. (2024). Enhancing C and N turnover, functional bacteria abundance, and the efficiency of biowaste conversion using Streptomyces-Bacillus inoculation. J. Environ. Manag. 358:120895. doi: 10.1016/j.jenvman.2024.120895
Zhou, Z., and Yao, H. (2020). Effects of composting different types of organic fertilizer on the microbial community structure and antibiotic resistance genes. Microorganisms 8:268. doi: 10.3390/microorganisms8020268
Keywords: cattle manure composting, Streptomyces thermovulgaris , microbe community, antibiotic resistance genes, mobile genetic elements, multidrug resistant host bacteria
Citation: Jin E, Gao D, Zhou Y, Wan P, Chen J, Gong P and Li P (2025) Co-inoculation with Streptomyces thermovulgaris and commercial microbial agents enhances the reduction of antibiotic resistance genes in cattle manure composting: driving mechanisms involving microbial communities and mobile genetic elements. Front. Microbiol. 16:1688304. doi: 10.3389/fmicb.2025.1688304
Edited by:
Tünde Pusztahelyi, University of Debrecen, HungaryCopyright © 2025 Jin, Gao, Zhou, Wan, Chen, Gong and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Gong, Z29uZ3BpbmcwOUBmb3htYWlsLmNvbQ==; Peng Li, bGlwZW5nQHlhbmd0emV1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Erguang Jin1†
Erguang Jin1† Ping Gong
Ping Gong Peng Li
Peng Li