- 1School of Horticulture and Landscape, Yangzhou University, Yangzhou, Jiangsu, China
- 2Tibet Academy of Forest Trees, Lasa, China
Introduction: Pea–cucumber rotation combined with straw return as green manure is an environmentally friendly management strategy to suppress cucumber (Cucumis sativus L.) Fusarium wilt (FW) and alleviate continuous cropping obstacles.
Methods: We evaluated the variations in soil microbial compositions and nutrient levels between long-term cucumber monocropping and pea–cucumber rotation patterns via metagenomic sequencing and determination of soil properties.
Results: The study found that the bacterial communities exhibited marked diversity, whereas the α-diversity of fungal communities was significantly reduced. Based on the relative abundance of differential fungi and bacteria at the genus level, the genus Bacillus showed the highest abundance, with a two-fold increase, whereas Fusarium species exhibited a 4.9-fold reduction following the pea–cucumber rotation. Additionally, the contents of available nitrogen, potassium, and phosphorus in the soil increased by more than 1.3-fold after the rotation. Correlation analysis revealed that the genus Bacillus and available potassium were significantly and negatively correlated with Fusarium pathogens. Notably, the isolated B. pumilus and B. safensis strains significantly suppressed the growth of cucumber FW pathogens.
Discussion: These findings provide valuable insights for optimizing the combination of soil Bacillus populations and nutrient availability to maintain soil ecosystem health and improve cucumber growth and yield.
1 Introduction
Continuous cropping in agriculture often leads to soil sickness, which is primarily caused by the accumulation of harmful pathogens or toxic substances, a decline in key microbial taxa, and reduced fertility in the crop rhizosphere, ultimately resulting in decreased crop yield and quality (Banerjee et al., 2019; Ma H. et al., 2025; Ma Z. et al., 2025). In particular, the long-term practice of continuous monocropping for many crops has a negative impact on soil health (Sun et al., 2021). China is widely recognized as the largest producer and consumer of greenhouse vegetables, with cucumber (Cucumis sativus L.) being one of the most economically important varieties worldwide (Feng et al., 2020; Yuan and Zhang, 2021; Zhang et al., 2024; Xin et al., 2025; Nie et al., 2025). However, continuous cropping obstacles in cucumber cultivation are mainly associated with the accumulation of Fusarium oxysporum pathogens and root-knot nematodes, which greatly hinder sustainable cucumber production.
Various approaches have been employed to prevent soil sickness, including chemical inducers (Bai et al., 2020), biofertilizers (Aloo et al., 2022), microbial inoculants (Chang et al., 2022; Giovanardi et al., 2025), green manuring (Jin et al., 2019a), and soil fumigation (Tong et al., 2025). However, crop rotation remains an ancient and well-established agricultural practice that can reshape soil microbial communities, enhance associated beneficial microorganisms, and improve soil fertility to overcome soil sickness in continuously cropped fields, thereby promoting agroecosystem sustainability and productivity (Wu J. et al., 2025; Wu H. et al., 2025). Continuous cucumber cropping has been reported to decrease yield and quality (Sun et al., 2021; Chen et al., 2022), and soil under different cropping systems has been extensively investigated in intensive greenhouse regions. For example, green garlic–cucumber rotation can regulate soil physicochemical properties and microbial community dynamics, thereby improving the soil environment and cucumber productivity (Ding et al., 2020). Similarly, cress–cucumber rotation alters the diversity, structure, and composition of soil microbial communities, which exerts positive effects on cucumber growth and development (Gong et al., 2021). Pepper–cucumber rotation also restructures rhizobacterial communities, facilitating the colonization of the cucumber rhizosphere by two Pseudarthrobacter oxydans strains (RH60 and RH97) through the enrichment of palmitic acid in pepper root exudates, thereby alleviating cucumber root-knot nematode infection (Tian et al., 2025). Overall, the soil microbial environment exerts a crucial influence on below-ground processes and crop performance. Nevertheless, although crop rotation practices can foster beneficial microbial communities that decrease pest pressure, suppress pathogens, and improve plant health, further in-depth studies are still required to elucidate these mechanisms.
Accumulating evidence has shown that pea (Pisum sativum L.) rotation or direct application as green manure can enhance soil nutrient content and uptake, including nitrogen levels, water retention, and overall system productivity (Gan et al., 2015; Chaudhari et al., 2020; Mesfin et al., 2023). Notably, this practice also stimulates the enrichment of beneficial microbial communities containing plant growth-promoting microorganisms and biocontrol agents (Mazoyon et al., 2023), thereby affecting plant health and performance. For instance, canola–pea–barley rotation has been consistently reported to alter the composition of rhizosphere microbial communities, increase yield and oil content, and decrease disease pressure caused by Leptosphaeria and Alternaria (Town et al., 2023). Although pea has been widely used for crop rotation and as green manure in agricultural systems, the specific role of pea–cucumber rotation in alleviating the obstacles of continuous cucumber cropping and its effects on soil nutrients and microbial communities remain largely unexplored.
In this study, rhizosphere soils were collected from greenhouses subjected to long-term continuous cucumber cropping and from those subjected to pea–cucumber rotation with straw return as green manure, followed by high-throughput sequencing. The composition and diversity of soil bacterial and fungal communities showed significant changes. Among them, bacterial taxa belonging to the genus Bacillus exhibited the highest abundance, with a two-fold increase, whereas fungal taxa belonging to Fusarium species showed a 4.9-fold reduction after pea–cucumber rotation. In addition, the contents of available nitrogen (AN), available potassium (AK), and available phosphorus (AP) in the soil increased by 1.3-fold after rotation. Correlation analysis revealed significant relationships among soil bacteria, fungi, and nutrients. Specifically, the genus Bacillus and available potassium were significantly and negatively correlated with Fusarium pathogens, whereas available potassium was significantly and positively correlated with Bacillus. Furthermore, the isolated B. pumilus and B. safensis strains markedly suppressed the growth of cucumber Fusarium wilt (FW) pathogens. These findings underscore the importance of investigating the functional mechanisms of pea–cucumber rotation and green manure systems and identifying optimal combinations of beneficial soil bacteria and nutrients for sustainable cucumber production.
2 Materials and methods
2.1 Rhizosphere soil collection
Rhizosphere soil samples were collected from experimental greenhouse fields cultivated with cucumber or pea seedlings located at Yangzhou University (32°23′35.3″N 119°25′09.1″E, Yangzhou, China). Soil samples were obtained using a randomized complete block design from greenhouses with continuous cucumber cropping and pea–cucumber rotation patterns. Cucumber seeds were germinated overnight in the dark at 28 °C, and the resulting seedlings were grown in a controlled chamber under a 16 h/8 h light/dark cycle at 25 °C/18 °C (day/night), respectively. Cucumber seedlings at the five-true-leaf stage were transplanted into greenhouses under continuous cropping, where they were cultivated through multiple growing seasons until the final harvest. Pea (Pisum sativum L.) seeds were sown at a rate of 225 kg/ha with a row spacing of 20 cm. Narrow furrows of optimum depth were opened manually using a hand-operated furrow opener in greenhouses with long-term continuous cucumber cropping. When pea seedlings reached a height of 30–40 cm, they were incorporated into the soil as green manure via rotary tillage. Soil samples were then collected from the upper soil layer (10–20 cm) of cucumber monoculture plots (control, X_CK) and pea–cucumber rotation plots (treatment, X_YM) that had been established for more than 2 years, with five replicates per treatment.
2.2 DNA extraction, metagenomic sequencing, and analysis
Total genomic DNA was extracted from the different soil samples using a PowerSoil® DNA Isolation Kit (MO BIO Laboratories Inc., Carlsbad, CA, United States). DNA concentration and purity were determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), and DNA integrity was verified by 1% agarose gel electrophoresis. The extracted DNA was then used to analyze the composition and diversity of soil bacterial and fungal communities using high-throughput amplicon sequencing. Primer pairs 341F/806R and ITS1F/ITS2R were used to amplify the hypervariable V3–V4 region of the bacterial 16S rRNA gene and the ITS region of the fungal rRNA gene, respectively (Niem et al., 2020). The purified PCR products were sequenced on an Illumina MiSeq platform (Illumina, San Diego, USA) following the standard operating procedures. Sequence quality was assessed using FastQC to ensure high-quality reads for downstream analyses. The obtained sequences were processed and analyzed using the mothur and quantitative insights into microbial ecology (QIIME) (version 1.9.0) software packages to calculate α-diversity, β-diversity, and the relative abundance of operational taxonomic units (OTUs), as well as to perform taxonomic assignment and statistical analyses of bacterial and fungal communities (Yue et al., 2024). Finally, the microbial community data were correlated with the measured soil physicochemical properties to reveal potential relationships.
2.3 Determination of soil properties
The rhizosphere soil samples were divided into two groups—X_CK and X_YM—with five replicates per group. Soil physicochemical properties, including available nitrogen (AN), available phosphorus (AP), available potassium (AK), and organic matter (OM), were determined as previously described (Li et al., 2020; Yue et al., 2024).
2.4 Isolation of rhizosphere Bacillus strains
Bacillus strains were isolated from the rhizosphere soil samples using a combination of limiting dilution, antibiotic screening, PCR amplification, and sequencing (Das et al., 2025). Briefly, the soil samples were thoroughly homogenized and heat-treated at 65 °C for 2 h to enrich spore-forming bacteria. The treated samples were then suspended and serially diluted in a sterile 0.9% NaCl solution and spread onto nutrient agar plates, followed by incubation at 35 °C. PCR amplification of the bacterial 16S rRNA gene was performed using primer pairs 27F (5′-AGAGTTTGATC(AC)TGGCTCAG-3′) and 1492R (5′-ACGG(CT)TACCTTGTTACGACTT-3′) (Li et al., 2022). The resulting sequences were identified and analyzed using the nucleotide database of the National Center for Biotechnology Information (NCBI).
2.5 Effects of Bacillus strains on cucumber FW suppression
The isolated Bacillus strains, including B. cereus, B. pumilus, and B. safensis, were used to evaluate their antagonistic effects against Fusarium oxysporum f. sp. cucumerinum (FOC), the pathogen that causes cucumber FW. The antifungal activity of the three Bacillus strains was assessed using a colony diameter inhibition assay. Briefly, 1 mL of bacterial suspension (OD600 = 1.0) of each Bacillus strain was mixed with 18 mL of potato dextrose agar (PDA) medium, and a 6-mm agar plug of the FOC colony was placed in the center of each plate. The plates were prepared in triplicate. PDA plates containing an equal amount of sterile distilled water instead of bacterial suspension were used as controls. The diameter of the FOC colonies was measured to determine the inhibitory effect of each Bacillus strain.
2.6 Statistical analysis
The raw sequence data of soil bacterial and fungal communities were deposited in the NCBI database under Bioproject accession numbers PRJNA1312254 and PRJNA1312268, respectively. The experimental data were statistically analyzed using the general linear model procedure in SPSS (version 16.0; IBM Corp., NY, United States). Differences among the groups were assessed using analysis of variance (ANOVA) to determine significant effects.
3 Result
3.1 Effects of the pea–cucumber crop rotation model on the diversity of bacterial communities
High-throughput sequencing was used to analyze the composition of bacterial communities in soils from the pea–cucumber rotation (treatment, X_YM) and continuous cucumber cropping (control, X_CK) systems. Principal coordinate analysis (PCoA) revealed that the samples from the same treatment clustered closely together, and the five biological replicates from both control and treatment groups were clearly separated (Figure 1A). In addition, the weighted UniFrac distance differed significantly between the two groups (p = 0.0024) (Supplementary Figure 1A). The α-diversity indices (Richness, Chao1, and Shannon) showed a visible reduction under the rotation treatment, although the differences were not statistically significant (Supplementary Figure 1B).
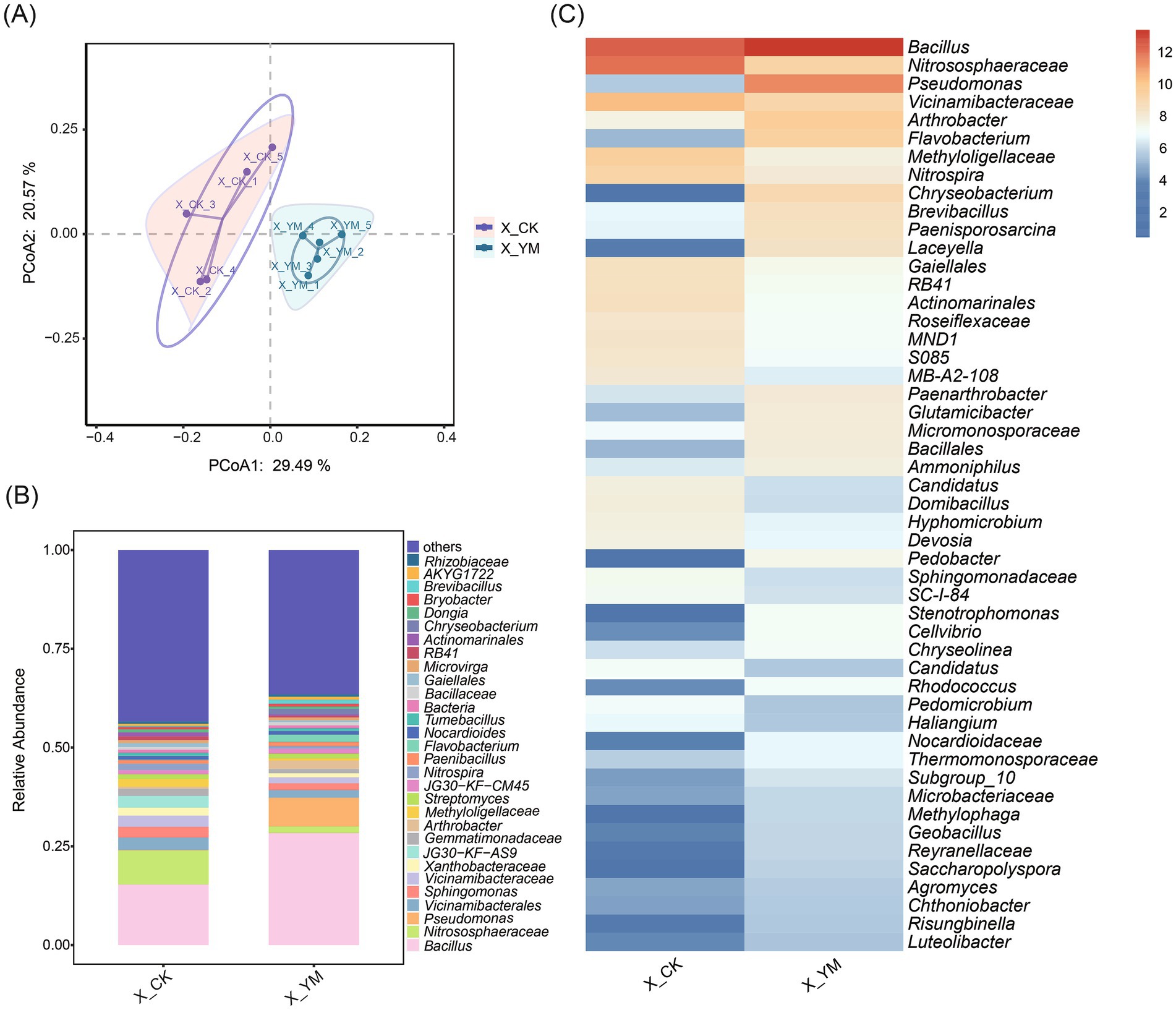
Figure 1. Changes in soil bacterial diversity after pea–cucumber rotation. (A) Principal coordinate analysis (PCoA) of bacterial communities under different cropping patterns. (B) Relative abundance of bacterial genera. (C) Heat map displaying the relative abundance of differential bacterial genera between treatments. X_YM and X_CK represent the samples collected from pea–cucumber rotation and continuous cucumber cropping soils, respectively.
The number of OTUs was used to characterize bacterial composition, and 409 core OTUs were identified (Supplementary Figure 1C). Taxonomic analysis indicated that the bacterial community exhibited marked diversity, with Bacillus species showing a significantly higher relative abundance under the pea–cucumber rotation treatment (Figure 1B). Based on abundance and |log2 fold change| > 1 criteria, the top 30 bacterial genera were selected to generate a heat map for differential comparison between the control and rotation treatments (Figure 1C; Supplementary Table 1). Among these, 14 genera showed significantly higher abundance, with the genus Bacillus exhibiting the greatest increase, two-fold, under the rotation treatment compared to the control (Figure 1C; Supplementary Figure 1D).
3.2 Dynamics of soil fungal communities after pea–cucumber crop rotation
To investigate the dynamics of soil fungal communities, α-diversity, β-diversity, and abundance differences of fungi were analyzed between the pea–cucumber rotation (treatment, X_YM) and continuous cucumber cropping (control, X_CK) soil samples. The α-diversity indices (Chao1, Shannon, and Pielou) showed a significant reduction in the pea–cucumber rotation soil sample (Figure 2A). PCoA demonstrated that the samples from the same treatment clustered closely together (Figure 2B). Taxonomic analysis revealed pronounced diversity in the distribution of fungal species (Supplementary Figure 2). Analysis of differential fungal abundance showed that 19 of the top 30 fungal species decreased significantly in the pea–cucumber rotation soil sample (Figure 2C; Supplementary Table 2), including pathogenic genera such as Cladosporium, Fusarium, and Alternaria, which cause cucumber black spot disease, cucumber Fusarium wilt, and cucumber Alternaria blight, respectively. In addition, reductions in the abundance of other soil-borne pathogens, including Didymella, Periconia, and Verticillium, were also observed; these pathogens are associated with soil-borne diseases and continuous cropping obstacles (Supplementary Figure 3). These results suggest that the reduction observed in fungal pathogens may be related to changes in bacterial community composition or abundance.
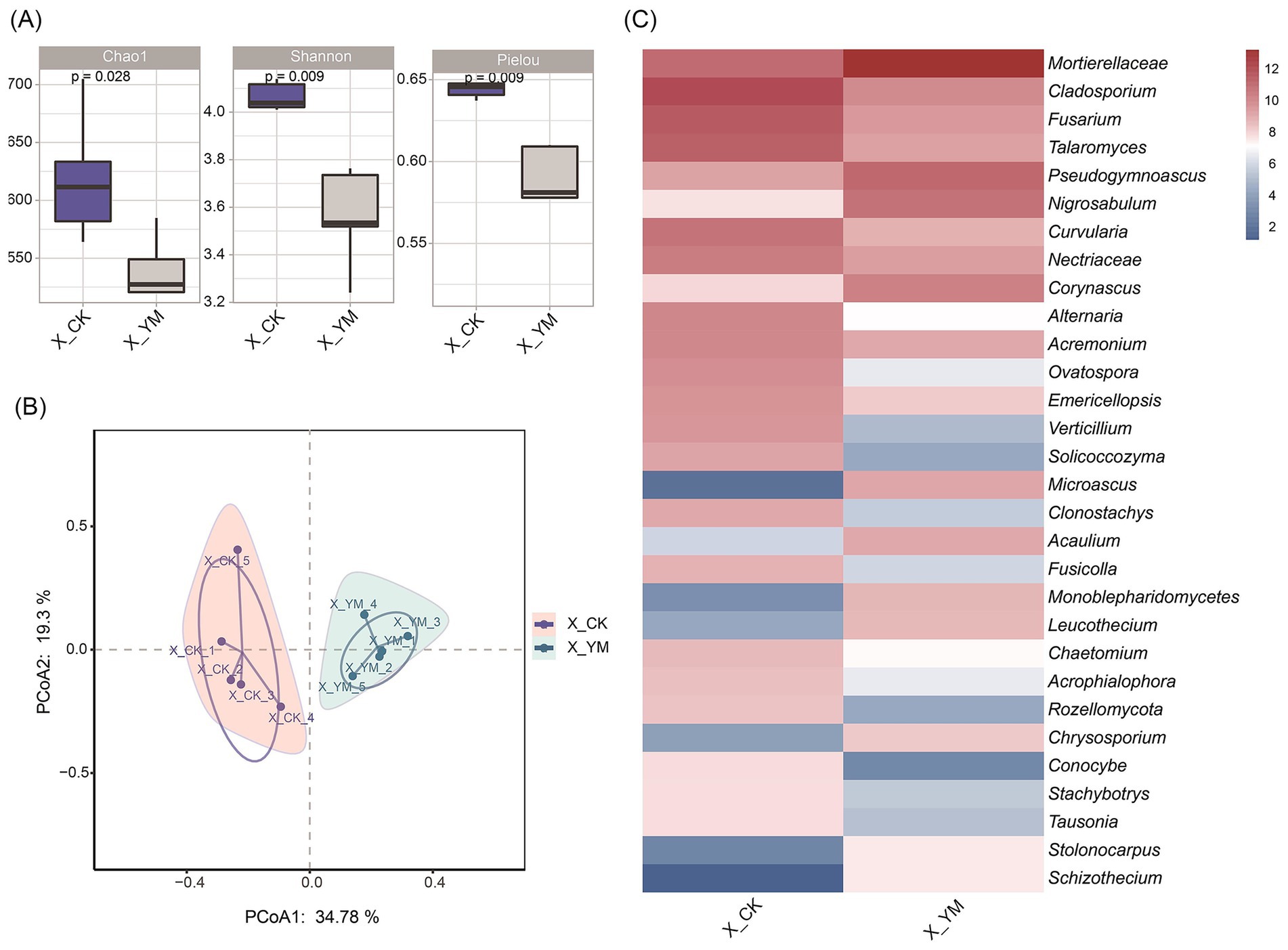
Figure 2. Composition and diversity of fungal communities after pea–cucumber rotation. (A) Alpha diversity indices (Chao1, Shannon, and Pielou) of fungal communities. (B) Principal coordinate analysis (PCoA) of fungal communities under different cropping patterns. (C) Heat map displaying the relative abundance of differential fungal genera between treatments. X_YM and X_CK represent the samples collected from pea–cucumber rotation and continuous cucumber cropping soils, respectively.
3.3 Impact of bacterial communities on fungal diversity and soil-borne pathogens
Many studies have confirmed that beneficial bacteria play a crucial role in the biological control of pathogens. Both bacterial and fungal communities were significantly altered by the pea–cucumber rotation treatment. To further examine the correlations between bacterial and fungal communities, the top 50 bacterial and fungal taxa with higher abundance and |log2 fold change| > 1 were selected for Procrustes analysis to quantify the similarity of spatial patterns between the two groups across samples. The results revealed a significant overall correlation between bacterial and fungal community structures (M2 ≈ 0.5161, p < 0.009; Figure 3A and Supplementary Table 3). The Mantel test was used to calculate the Mantel correlation coefficient (r) and its significance (p-value) between the α-diversity indices (Chao1 and Shannon) of bacterial communities and the abundance of soil-borne fungal pathogens. The results indicated that the genus Fusarium was significantly correlated with Verticillium and Periconia, although there was no significant correlation between overall bacterial diversity and soil-borne fungal pathogens (Figure 3B). In addition, Spearman correlation analysis was performed between the top 20 bacterial taxa and soil-borne pathogens, and the results were visualized as a heat map. The results showed that nine of the 20 bacterial taxa showed significant negative correlations with Fusarium and Verticillium pathogens (Figure 3C; Supplementary Table 4), with Bacillus species exhibiting the highest relative abundance. These findings suggest that Bacillus species may represent a group of beneficial bacteria enriched after pea–cucumber rotation treatment, contributing to the suppression of Fusarium pathogens in the soil.
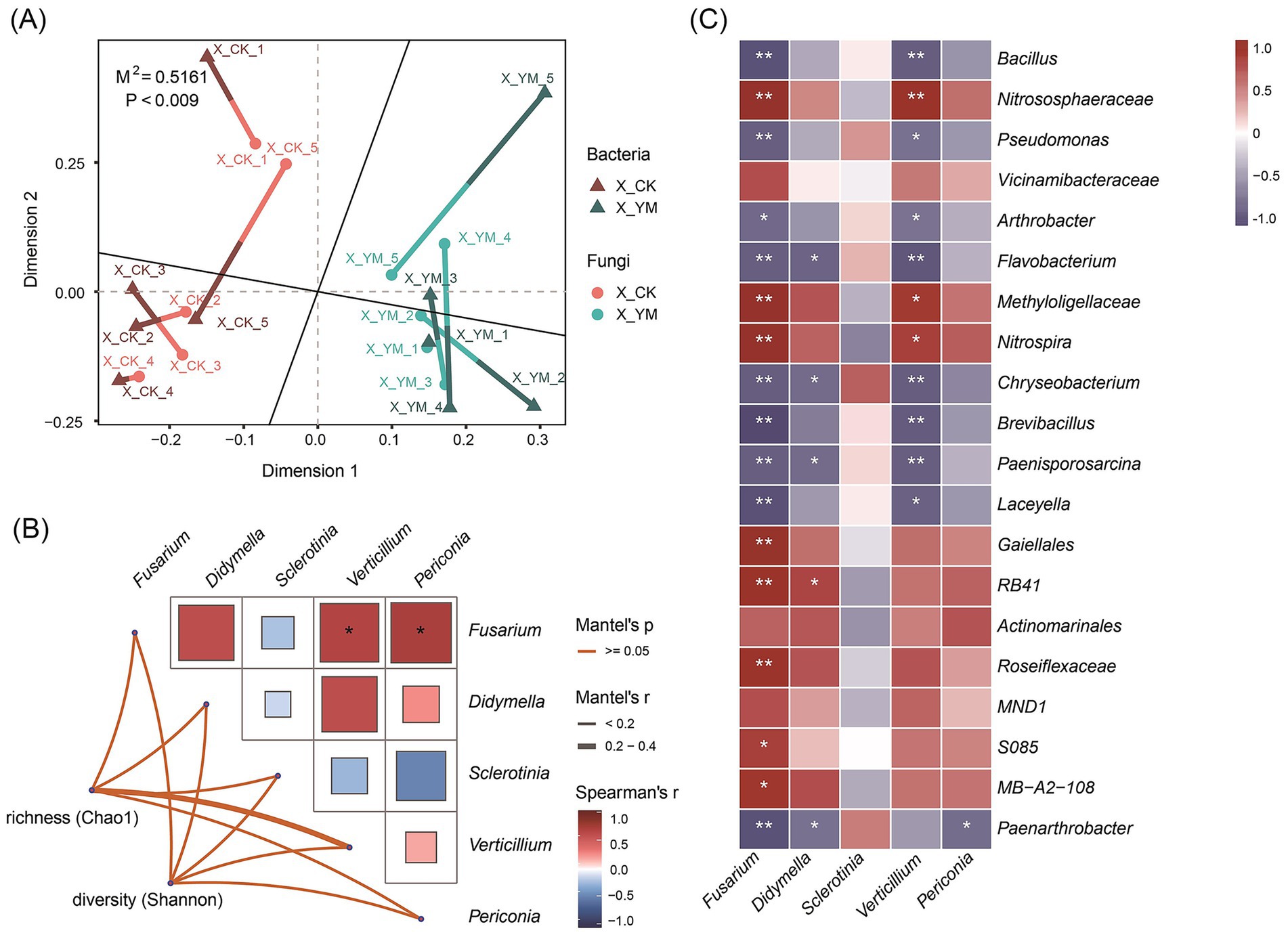
Figure 3. Correlation analysis between soil bacterial diversity and soil-borne fungal pathogens. (A) Procrustes analysis of the top 50 differential bacterial and fungal taxa. (B) The Mantel test showing correlations between bacterial diversity and soil-borne fungal pathogens. (C) Spearman correlation heat map displaying the relative abundance of differential bacterial taxa and soil-borne fungal pathogens. X_YM and X_CK represent the samples collected from pea–cucumber rotation and continuous cucumber cropping soils, respectively. The color scale indicates the correlation strength, with red indicating a positive correlation and blue indicating a negative correlation. “*” and “**” denote significance at p < 0.05 and p < 0.01, respectively.
3.4 Changes in soil nutrients after pea–cucumber crop rotation
The basic physicochemical properties of the soil, including AN, AK, AP, and OM, were measured and compared between the pea–cucumber rotation (X_YM) and continuous cucumber cropping (X_CK) treatments. The results showed that the contents of soil AN (135.17 mg/kg), AK (512.55 mg/kg), and AP (128.72 mg/kg) were significantly increased by more than 1.3-fold in the pea–cucumber rotation treatment compared to the continuous cucumber cropping control (Figure 4; Supplementary Table 5). In particular, the contents of soil AK and AP increased by 86.89 and 43.47%, respectively, after the pea–cucumber rotation treatment. However, the OM content did not differ significantly between the X_YM and X_CK groups. These findings indicate that implementing a pea–cucumber rotation system combined with straw return as green manure greatly alters soil nutrient composition, which may contribute to the regulation of soil microbial communities and the alleviation of continuous cropping obstacles.
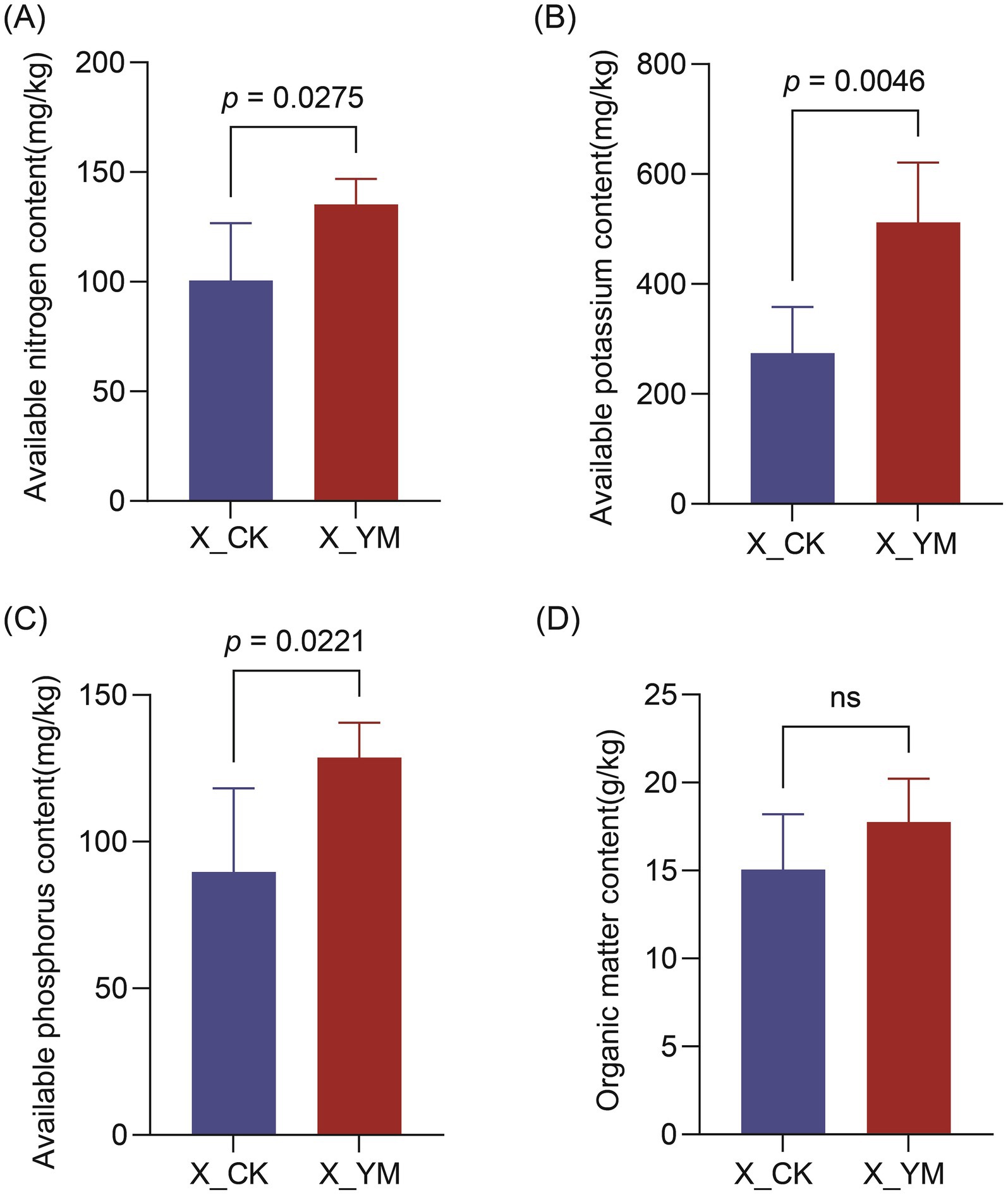
Figure 4. Contents of different soil nutrients: (A) available nitrogen, (B) available potassium, (C) available phosphorus, and (D) organic matter. X_YM and X_CK represent the samples collected from pea–cucumber rotation and continuous cucumber cropping soils, respectively. Values are presented as mean ± standard error (SE) of the five replicates. The p-values indicate significant differences between treatments.
3.5 Correlation analysis between soil nutrients and microbial communities
To explore the relationships between soil nutrients and microbial communities, the Mantel test was performed between the bacterial and fungal diversity indices (Chao1 and Shannon) and the matrix of soil nutrient variables by calculating the coefficient (r) and significance (p-value). The results showed that AP was significantly correlated with AN and AK (p < 0.01), and OM was significantly correlated with AN (p < 0.05). In addition, the Chao1 diversity index of both bacterial and fungal communities was significantly correlated with AP (p < 0.05) (Figure 5A). Moreover, the top 20 differential bacterial and fungal taxa were selected for correlation analysis with soil nutrient factors. A heat map was generated to visualize significant statistical correlations, providing a basis for interpreting the bacterial and fungal communities and their functional patterns. The results revealed that AK was significantly and positively correlated with nine bacterial taxa, including the genus Bacillus, and negatively correlated with seven bacterial taxa (Figure 5B; Supplementary Table 6). AK was also significantly and negatively correlated with seven fungal taxa, including the genera Fusarium and Verticillium (Figure 5C; Supplementary Table 7). These findings suggest that AK may be an important soil nutrient influencing bacterial and fungal community structures.
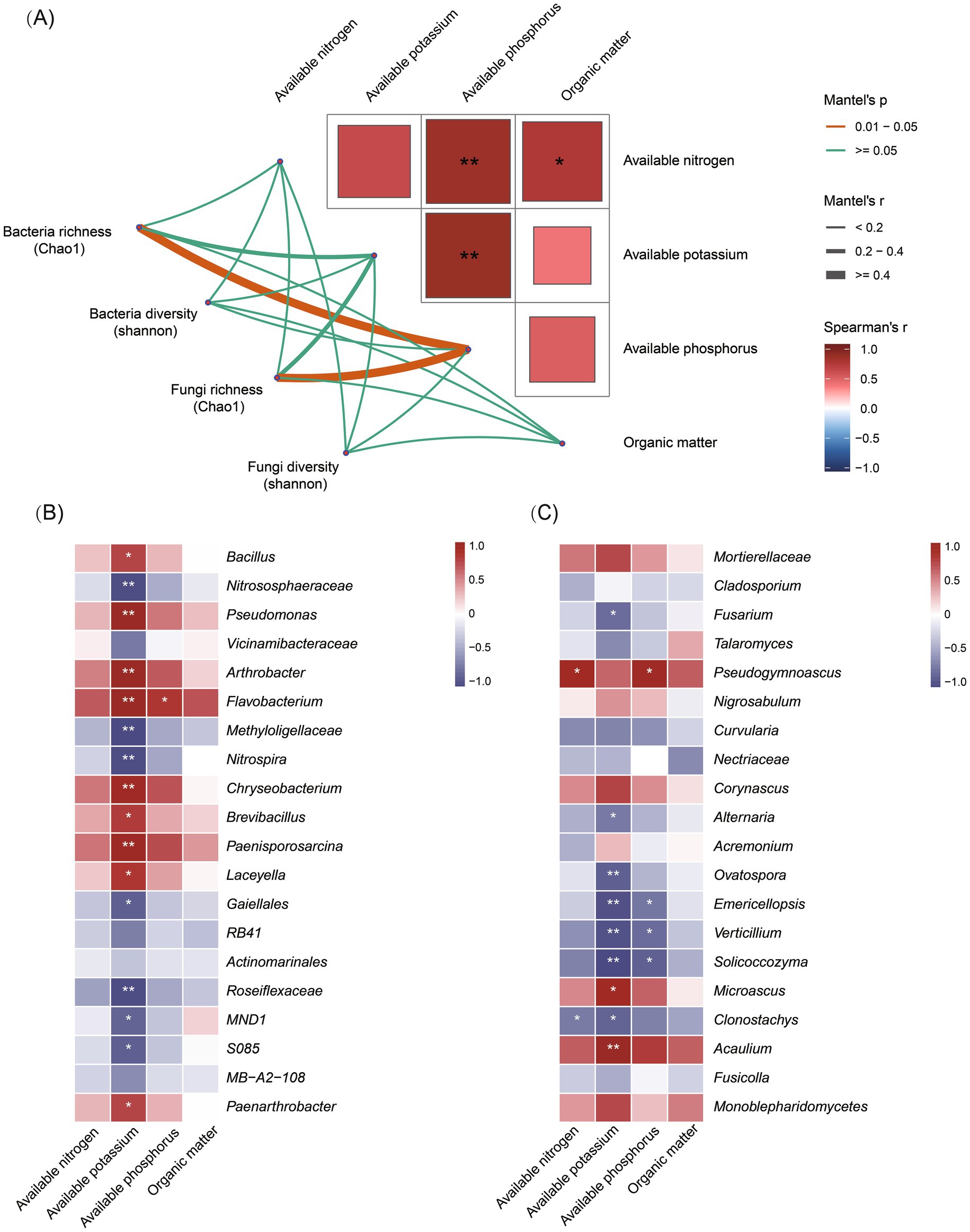
Figure 5. Correlation analysis between soil nutrients and microbial communities. (A) The Mantel test showing correlations between soil microbial diversity and nutrient variables. Spearman correlation heat map displaying the relative abundance of differential bacterial (B) and fungal taxa (C). The color scale indicates the correlation strength, with red indicating a positive correlation and blue indicating a negative correlation. “*” and “**” denote significance at p < 0.05 and p < 0.01, respectively.
In addition, AN was significantly and positively correlated with Pseudogymnoascus and negatively correlated with Clonostachys. AP was significantly and negatively correlated with Emericellopsis, Verticillium, and Solicoccozyma and positively correlated with Pseudogymnoascus (Figure 5C; Supplementary Table 7). Overall, these results indicate that soil nutrient variables strongly and significantly regulate the dynamics of bacterial and fungal communities.
3.6 Effects of Bacillus strains on cucumber FW suppression
Based on the above analysis, Bacillus species exhibited the highest abundance in soil after the pea–cucumber rotation treatment and showed significant negative correlations with Fusarium pathogens. Accordingly, Bacillus strains were isolated from the soil and evaluated for their ability to suppress FOC, the causal agent of cucumber Fusarium wilt. A total of three Bacillus strains (B. cereus, B. pumilus, and B. safensis) were isolated and identified using PCR amplification and sequencing (Supplementary Table 8). The antagonistic activity of these strains against FOC was assessed using colony diameter inhibition assays after incubation periods of 3, 5, 7, and 9 days. Compared to the control (FOC grown on medium without Bacillus strains), FOC colonies co-cultured with B. pumilus and B. safensis were significantly suppressed at multiple incubation time points (Figure 6; Supplementary Figure 4). Notably, B. safensis significantly suppressed FOC growth, reducing the colony size by 40.15 and 28.33% after 5 and 7 days of incubation, respectively (Figures 6A,B; Supplementary Figure 4B). These findings suggest that Bacillus strains exert positive effects on cucumber FW suppression. Therefore, it can be inferred that the combined enrichment of beneficial Bacillus species and soil nutrients may contribute to improved soil ecosystem health.
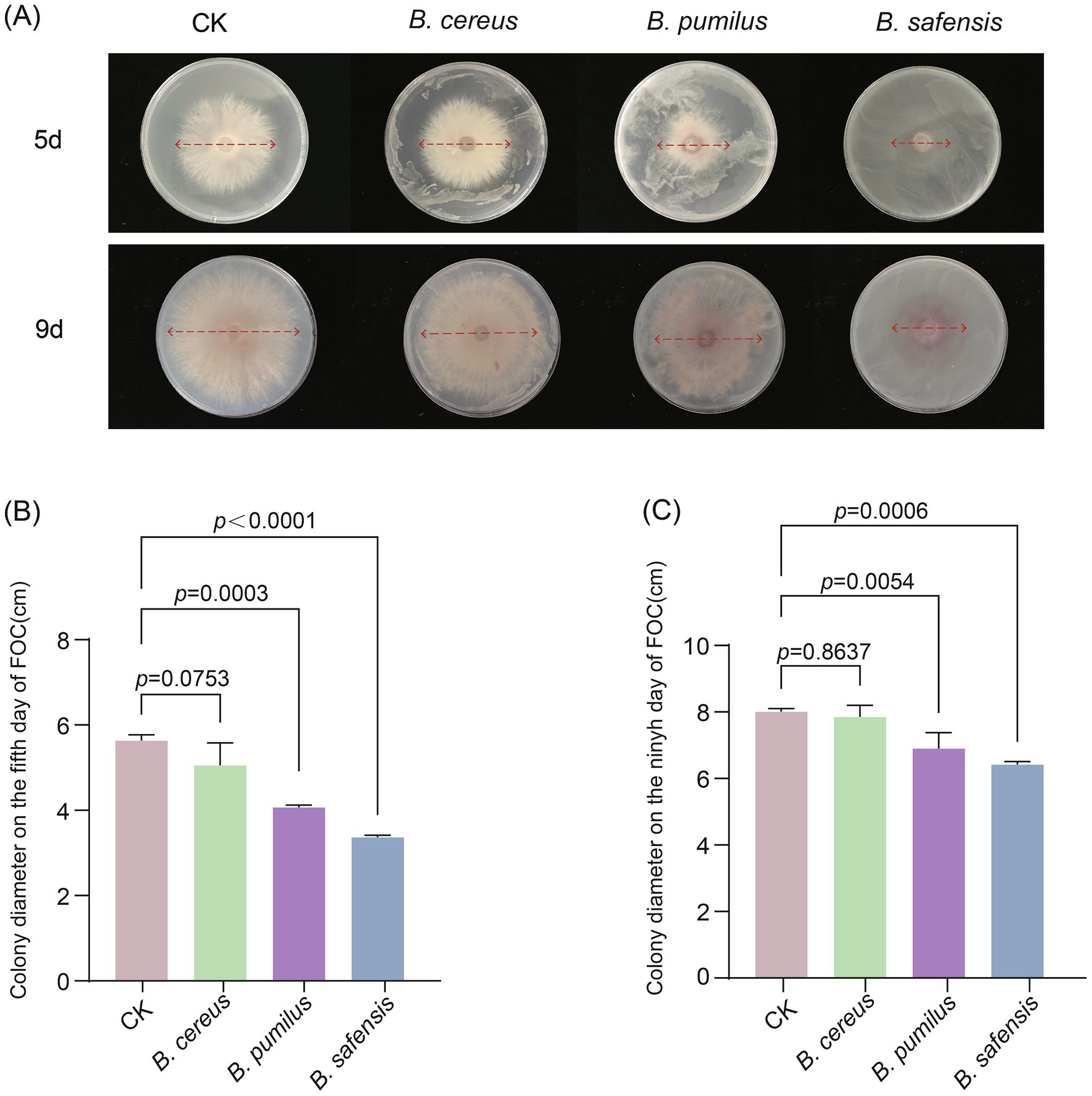
Figure 6. Effects of Bacillus strains on FOC suppression. (A) Differences in FOC growth on PDA media containing Bacillus strains after 5 and 9 days of incubation. Colony diameter of FOC after incubation with Bacillus strains for (B) 5 days and (C) 9 days. Values are presented as mean ± standard error (SE) of the five replicates. The p-values indicate significant differences between treatments.
4 Discussion
4.1 Negative effects of the continuous cucumber cropping system
Long-term continuous cropping often results in soil sickness, characterized by nutrient imbalance, accumulation of soil-borne pathogens, and subsequent declines in crop yield and quality (Banerjee et al., 2019; Wu et al., 2020; Chen et al., 2022). In this study, greenhouse soils under continuous cucumber cultivation (X_CK) exhibited high abundance of multiple harmful fungal pathogens, including the genera Fusarium, Cladosporium, Alternaria, Didymella, and Periconia, which are known to cause cucumber Fusarium wilt, black spot disease, Alternaria blight, gummy stem blight, and other diseases (Figure 2; Supplementary Figure 3). These findings highlight the challenges and negative effects associated with continuous cucumber cultivation. Previous studies have also demonstrated that prolonged cucumber monocropping in greenhouses adversely affects cucumber yield and quality, leading to decreased contents of vitamin C, soluble sugars, and proteins in cucumber (Sun et al., 2021). Moreover, with increasing years of continuous cucumber cropping (0, 4, 8, and 12 years), total soil salinity progressively increased, whereas soil pH significantly decreased (Huang et al., 2023). Similarly, an investigation of rhizosphere soils under 1, 3, 5, and 7 years of continuous cucumber cropping in solar greenhouses revealed that a longer cropping duration led to a gradual increase in the relative abundance of Proteobacteria, Chloroflexi, Gemmatimonadetes, Patescibacteria, and Firmicutes, accompanied by a significant decrease in Actinobacteria (Li et al., 2021).
4.2 Changes in microbial communities and soil nutrients regulated by the crop rotation model
In diverse agricultural systems, crop rotation can exert long-term effects by improving soil nutrient dynamics and enhancing the abundance of beneficial microorganisms (Wang et al., 2023; Wu J. et al., 2025). In this study, rotation with pea led to pronounced changes in rhizosphere bacterial and fungal communities and soil nutrient levels. In particular, potentially plant-beneficial bacteria (such as Bacillus spp.) showed a significant increase in abundance, whereas several pathogenic fungi, including Fusarium, Cladosporium, and Alternaria, were markedly reduced after pea–cucumber rotation (Figures 1, 2). In addition, the contents of soil AN, AK, and AP in the soil increased by more than 1.3-fold, with AK and AP increasing by 86.89 and 43.47%, respectively (Figure 4; Supplementary Table 5). Similar findings have been reported for other cucumber-based rotation patterns that reshape soil microbial communities and alter nutrient composition to suppress pathogen infections. For example, pepper–cucumber rotation promoted the enrichment of two Pseudarthrobacter oxydans strains, which markedly reduced root-knot nematode (Meloidogyne incognita) infection and diminished the accumulation of p-hydroxybenzoic acid in the soil (Tian et al., 2025). Similarly, cress–cucumber rotation greatly increased the relative abundance of potentially beneficial microorganisms, including Roseiflexus, Pseudallescheria spp., and Haliangium spp., thereby suppressing potential pathogenic fungi, such as Fusarium and Monographella spp., and improving cucumber yield (Gong et al., 2021). In addition, rotations with Indian mustard (Brassica juncea) and wild rocket (Diplotaxis tenuifolia (L.) DC.) were found to restructure rhizobacterial communities and enhance the populations of potential plant-beneficial microorganisms (such as Pseudomonas spp.), which significantly suppressed cucumber Fusarium wilt disease (Jin et al., 2019b). Collectively, these findings indicate that crop rotation models positively affect soil microbial ecology by enriching beneficial bacteria, reducing harmful pathogens, and improving nutrient availability, thereby promoting cucumber growth and yield. Nevertheless, the molecular mechanisms underlying these effects and the optimization and broader applicability of the pea–cucumber rotation model warrant further in-depth investigation.
4.3 Interactions between microbial communities and soil nutrients
Interactions between soil nutrients and microbial communities play critical roles in modulating soil fertility and health, thereby enhancing the natural resilience of soils against degradation and disease (Goodall et al., 2025). In this study, numerous bacterial taxa and soil physicochemical properties were found to be significantly correlated with pathogenic fungi. In particular, the abundance of Bacillus and AK content both increased significantly and were negatively correlated with the relative abundance of Fusarium pathogens. Moreover, AK was significantly and positively correlated with the relative abundance of Bacillus (Figures 3, 5). Accumulating evidence suggests that interactions between microbial communities and soil nutrients can positively affect plant resistance and productivity. For example, the application of a bio-organic fertilizer composed of organic matter (284 g/kg), total nitrogen (25.8 g/kg), total potassium (14.9 g/kg), total phosphorus (42 g/kg), and Bacillus amyloliquefaciens SQR9 (>107 CFU/g) in continuous cucumber cropping systems substantially altered soil microbial communities and physicochemical properties, leading to effective suppression of cucumber Fusarium wilt and increased yield (Huang et al., 2017). Furthermore, the diversity of soil microbial communities plays an important role in maintaining soil nutrient balance and ecosystem stability (Shu et al., 2024). Another study demonstrated that soil microbial communities could regulate organic matter dynamics, nutrient cycling—such as organic carbon, total phosphorus, and NH4+-N—and the degradation of soil pollutants (Han et al., 2024). Future studies should focus on elucidating the interactive relationships and regulatory networks between soil microbial communities (such as Bacillus spp.) and nutrients (such as AK) to improve soil ecosystem health and enhance the suppression of cucumber FW disease.
4.4 Functions of Bacillus strains in improving cucumber production and disease suppression
In recent years, various beneficial fungi and bacteria have been identified and applied to improve soil health, enhance plant resistance, and promote crop growth (Tzipilevich et al., 2021; Ma Y. et al., 2025; Moreira et al., 2025). For example, two Paenibacillus polymyxa strains, NSY50 and WLY78, have been reported to produce antifungal compounds that strongly suppress the growth of Fusarium oxysporum and regulate rhizospheric microbial communities, thereby promoting cucumber growth (Shi et al., 2017; Li and Chen, 2019; Du et al., 2022). Similarly, the application of Trichoderma harzianum SQR-T037 has been shown to rapidly degrade allelochemicals, significantly suppress Fusarium wilt, and alter microbial community composition in continuously cropped cucumber soils (Chen et al., 2011; Chen et al., 2012). In this study, two Bacillus strains, B. pumilus and B. safensis, were identified and exhibited strong suppression effects on FOC growth. In particular, B. safensis highly inhibited FOC growth, reducing the colony size by 40.15 and 28.33% after 5 and 7 days of incubation, respectively (Figure 6; Supplementary Figure 4). Previous studies have reported that numerous antagonistic bacteria belonging to the Bacillus genus, such as B. velezensis NH-1, B. siamensis NB92, B. amyloliquefaciens FH-1, B. ayatagriensis RMG6T, and B. amyloliquefaciens B2, are widely used to suppress cucumber FW pathogens and promote sustainable agricultural production (Luo et al., 2019; Wang J. et al., 2021; Wang H. et al., 2021; Das et al., 2025; Wang et al., 2025). Moreover, Bacillus species can activate beneficial microorganisms and secrete antifungal compounds that enhance disease resistance, thereby promoting the growth and health of cucumber (Wang and Sugiyama, 2024; Wang et al., 2025). Therefore, future studies should focus on exploring optimized combinations of Bacillus species with other beneficial microorganisms and identifying the specific antifungal metabolites secreted by Bacillus strains. These efforts will provide a more comprehensive understanding of the mechanisms underlying cucumber FW disease suppression and help alleviate the challenges of continuous cropping.
5 Conclusion
This study demonstrated that pea–cucumber rotation combined with straw return as green manure enhanced soil nutrient levels and reshaped the composition of complex microbial communities. The intricate correlations and interactions among soil nutrients, bacterial and fungal communities, and soil-borne pathogens were investigated, revealing their collective contribution to cucumber FW disease resistance. Two isolated Bacillus strains, B. pumilus and B. safensis, exhibited strong antagonistic activity against cucumber FW pathogens. Future studies should focus on elucidating the antifungal mechanisms of B. pumilus and B. safensis and exploring optimized combinations of Bacillus species, other beneficial microorganisms, and key soil nutrients to overcome continuous cropping obstacles and improve sustainable cucumber production.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
JX: Project administration, Validation, Data curation, Conceptualization, Methodology, Supervision, Writing – review & editing, Writing – original draft, Investigation, Software, Formal analysis. YY: Validation, Data curation, Methodology, Investigation, Writing – original draft, Software, Formal analysis. LP: Validation, Methodology, Data curation, Investigation, Writing – original draft, Formal analysis. NZ: Investigation, Project administration, Validation, Methodology, Conceptualization, Writing – review & editing, Data curation. DL: Validation, Writing – review & editing, Data curation, Methodology, Investigation. XC: Project administration, Writing – review & editing, Supervision, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the National Natural Science Foundation of China (grant nos. 32372690 and 31902015).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1697343/full#supplementary-material
Supplementary Figure 1 | Structure and diversity analyses of soil bacterial communities.
Supplementary Figure 2 | Structure and diversity analyses of soil fungal communities.
Supplementary Figure 3 | Relative abundances of soil-borne fungal pathogens.
Supplementary Figure 4 | Colony diameter of FOC after incubation with Bacillus strains for 3 days and 7 days.
Supplementary Table 1 | The abundance of soil bacterial communities, and the top 50 differential bacteria.
Supplementary Table 2 | The abundance of soil fungal communities, and the top 50 differential fungi.
Supplementary Table 3 | The Procrustes analysis of the top 50 differential bacteria and fungi.
Supplementary Table 4 | The significance (p value) and coefficient (r) value between the soil-borne fungal pathogens and top 20 differential bacteria.
Supplementary Table 5 | The content of different soil nutrients in the pea-cucumber rotation (X_YM) and cucumber continuous cropping soils (X_CK).
Supplementary Table 6 | The significance (p value) and coefficient (r) value between soil nutrients and top 20 differential bacteria.
Supplementary Table 7 | The significance (p value) and coefficient (r) value between soil nutrients and top 20 differential fungi.
Supplementary Table 8 | The identified information of rhizosphere Bacillus sp. strains.
References
Aloo, B. N., Tripathi, V., Makumba, B. A., and Mbega, E. R. (2022). Plant growth-promoting rhizobacterial biofertilizers for crop production: the past, present, and future. Front. Plant Sci. 13:1002448. doi: 10.3389/fpls.2022.1002448
Bai, Y., Chang, Y., Hussain, M., Lu, B., Zhang, J., Song, X., et al. (2020). Soil chemical and microbiological properties are changed by long-term chemical fertilizers that limit ecosystem functioning. Microorganisms 8:694. doi: 10.3390/microorganisms8050694
Banerjee, S., Walder, F., Buechi, L., Meyer, M., Held, A. Y., Gattinger, A., et al. (2019). Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 13, 1722–1736. doi: 10.1038/s41396-019-0383-2
Chang, F., Zhang, G., Zhang, L., Lyu, B., Liu, Z., Fan, Q., et al. (2022). Effects of biological fumigation combined with microbial agents on fungi community structure in continuous watermelon cropping soil. Chin. J. Eco-Agric. 30, 248–257. doi: 10.12357/cjea.20210473
Chaudhari, D., Rangappa, K., Das, A., Layek, J., Basavaraj, S., Kandpal, B. K., et al. (2020). Pea (Pisum sativum L.) plant shapes its rhizosphere microbiome for nutrient uptake and stress amelioration in acidic soils of the North-East Region of India. Front. Microbiol. 11:968. doi: 10.3389/fmicb.2020.00968
Chen, L. H., Huang, X. Q., Zhang, F. G., Zhao, D. K., Yang, X. M., and Shen, Q. R. (2012). Application of Trichoderma harzianum SQR-T037 bio-organic fertiliser significantly controls fusarium wilt and affects the microbial communities of continuously cropped soil of cucumber. J. Sci. Food Agric. 92, 2465–2470. doi: 10.1002/jsfa.5653
Chen, L., Yang, X., Raza, W., Li, J., Liu, Y., Qiu, M., et al. (2011). Trichoderma harzianum SQR-T037 rapidly degrades allelochemicals in rhizospheres of continuously cropped cucumbers. Appl. Microbiol. Biotechnol. 89, 1653–1663. doi: 10.1007/s00253-010-2948-x
Chen, X., Zhang, D., Li, Y., Li, H., Lou, J., Li, X., et al. (2022). Changes in rhizospheric microbiome structure and soil metabolic function in response to continuous cucumber cultivation. FEMS Microbiol. Ecol. 98:fiac129. doi: 10.1093/femsec/fiac129
Das, S., Mondal, R., Mandal, P., Kurt, H., Chakraborty, J., Islam, M. M., et al. (2025). Bacillus ayatagriensis sp. nov., a novel plant growth-promoting rhizobacteria strain isolated from mulberry rhizosphere. Sci. Rep. 15:26693. doi: 10.1038/s41598-025-05508-w
Ding, H., Ali, A., and Cheng, Z. (2020). Effect of green garlic/cucumber crop rotation for 3 years on the dynamics of soil properties and cucumber yield in Chinese anthrosol. J. Sci. Food Agric. 100, 362–370. doi: 10.1002/jsfa.10050
Du, N., Yang, Q., Guo, H., Xue, L., Fu, R., Dong, X., et al. (2022). Dissection of Paenibacillus polymyxa NSY50-induced Defense in cucumber roots against Fusarium oxysporum f. sp. cucumerinum by target metabolite profiling. Biology (Basel) 11:1028. doi: 10.3390/biology11071028
Feng, S., Zhang, J., Mu, Z., Wang, Y., Wen, C., Wu, T., et al. (2020). Recent progress on the molecular breeding of Cucumis sativus L. in China. Theor. Appl. Genet. 133, 1777–1790. doi: 10.1007/s00122-019-03484-0
Gan, Y., Hamel, C., O’Donovan, J. T., Cutforth, H., Zentner, R. P., Campbell, C. A., et al. (2015). Diversifying crop rotations with pulses enhances system productivity. Sci. Rep. 5:14625. doi: 10.1038/srep14625
Giovanardi, D., Biondi, E., Biondo, N., Quiroga, N., Modica, F., Puopolo, G., et al. (2025). Sustainable and innovative biological control strategies against Pseudomonas syringae pv. Tomato, Pseudomonas savastanoi pv. Phaseolicola and Xanthomonas spp. affecting vegetable crops: a review. Front. Plant Sci. 16:1536152. doi: 10.3389/fpls.2025.1536152
Gong, X., Shi, J., Zhou, X., Yuan, T., Gao, D., and Wu, F. (2021). Crop rotation with cress increases cucumber yields by regulating the composition of the rhizosphere soil microbial community. Front. Microbiol. 12:631882. doi: 10.3389/fmicb.2021.631882
Goodall, T., Busi, S. B., Griffiths, R. I., Jones, B., Pywell, R. F., Richards, A., et al. (2025). Soil properties in agricultural systems affect microbial genomic traits. FEMS Microbes 6:xtaf008. doi: 10.1093/femsmc/xtaf008
Han, C. L., Liang, D. F., Zhou, W. D., Xu, Q. Y., Xiang, M. X., Gu, Y. J., et al. (2024). Soil, plant, and microorganism interactions drive secondary succession in alpine grassland restoration. Plants 13:780. doi: 10.3390/plants13060780
Huang, N., Wang, W., Yao, Y., Zhu, F., Wang, W., and Chang, X. (2017). The influence of different concentrations of bio-organic fertilizer on cucumber Fusarium wilt and soil microflora alterations. PLoS One 12:e0171490. doi: 10.1371/journal.pone.0171490
Huang, S., Yu, J., Hou, D., Yue, H., Zhang, D., Li, Y., et al. (2023). Response of soil microbial community diversity to continuous cucumber cropping in facilities along the Yellow River irrigation area. PLoS One 18:e0289772. doi: 10.1371/journal.pone.0289772
Jin, X., Wang, J., Li, D., Wu, F., and Zhou, X. (2019b). Rotations with Indian mustard and wild rocket suppressed cucumber Fusarium wilt disease and changed rhizosphere bacterial communities. Microorganisms 7:57. doi: 10.3390/microorganisms7020057
Jin, X., Zhang, J. H., Shi, Y. J., Wu, F. Z., and Zhou, X. G. (2019a). Green manures of Indian mustard and wild rocket enhance cucumber resistance to Fusarium wilt through modulating rhizosphere bacterial community composition. Plant Soil 441, 283–300. doi: 10.1007/s11104-019-04118-6
Li, Y., and Chen, S. (2019). Fusaricidin produced by Paenibacillus polymyxa WLY78 induces systemic resistance against Fusarium wilt of cucumber. Int. J. Mol. Sci. 20:5240. doi: 10.3390/ijms20205240
Li, Y., Chi, J., Ao, J., Gao, X., Liu, X., Sun, Y., et al. (2021). Effects of different continuous cropping years on bacterial community and diversity of cucumber rhizosphere soil in solar-greenhouse. Curr. Microbiol. 78, 2380–2390. doi: 10.1007/s00284-021-02485-x
Li, J., Liu, P., Menguy, N., Benzerara, K., Bai, J., Zhao, X., et al. (2022). Identification of sulfate-reducing magnetotactic bacteria via a group-specific 16S rDNA primer and correlative fluorescence and electron microscopy: strategy for culture-independent study. Environ. Microbiol. 24, 5019–5038. doi: 10.1111/1462-2920.16109
Li, Y., Zeng, C., and Long, M. (2020). Variation of soil nutrients and bacterial community diversity of different land utilization types in Yangtze River basin, Chongqing municipality. PeerJ 8:e9386. doi: 10.7717/peerj.9386
Luo, W., Liu, L., Qi, G., Yang, F., Shi, X., and Zhao, X. (2019). Embedding Bacillus velezensis NH-1 in microcapsules for biocontrol of cucumber fusarium wilt. Appl. Environ. Microbiol. 85:e03128-18. doi: 10.1128/AEM.03128-18
Ma, Z., He, C., Tan, J., Jin, T., and Hua, S. (2025). Decreased nitrogen and carbohydrate metabolism activity leads to grain yield reduction in Qingke under continuous cropping. Plants (Basel) 14:2235. doi: 10.3390/plants14142235
Ma, H., Ren, Z., Luo, A., Fang, X., Liu, R., Wu, C., et al. (2025). Self-alleviation of continuous-cropping obstacles in potato via root-exudate-driven recruitment of growth-promoting bacteria. Plant Commun. 6:101372. doi: 10.1016/j.xplc.2025.101372
Ma, Y., Zuohereguli, K., Zhang, L., Kang, Y., Shi, L., Xu, H., et al. (2025). Soil microbial mechanisms to improve pear seedling growth by applying Bacillus and Trichoderma-amended biofertilizers. Plant Cell Environ. 48, 3968–3980. doi: 10.1111/pce.15395
Mazoyon, C., Hirel, B., Pecourt, A., Catterou, M., Gutierrez, L., Sarazin, V., et al. (2023). Sphingomonas sediminicola is an endosymbiotic bacterium able to induce the formation of root nodules in pea (Pisum sativum L.) and to enhance plant biomass production. Microorganisms 11:199. doi: 10.3390/microorganisms11010199
Mesfin, S., Gebresamuel, G., Haile, M., and Zenebe, A. (2023). Potentials of legumes rotation on yield and nitrogen uptake of subsequent wheat crop in northern Ethiopia. Heliyon 9:e16126. doi: 10.1016/j.heliyon.2023.e16126
Moreira, R. F., Pires, E. B. E., Sousa, O. F., Alves, G. B., Viteri Jumbo, L. O., Santos, G. R., et al. (2025). A novel neotropical Bacillus siamensis strain inhibits soil-borne plant pathogens and promotes soybean growth. Microorganisms 13:1366. doi: 10.3390/microorganisms13061366
Nie, J., Huang, H., Wu, S., Lin, T., Zhang, L., Lv, L., et al. (2025). Molecular regulation and domestication of parthenocarpy in cucumber. Nat Plants 11, 176–190. doi: 10.1038/s41477-024-01899-2
Niem, J. M., Billones-Baaijens, R., Stodart, B., and Savocchia, S. (2020). Diversity profiling of grapevine microbial Endosphere and antagonistic potential of endophytic Pseudomonas against grapevine trunk diseases. Front. Microbiol. 11:477. doi: 10.3389/fmicb.2020.00477
Shi, L., Du, N., Shu, S., Sun, J., Li, S., and Guo, S. (2017). Paenibacillus polymyxa NSY50 suppresses Fusarium wilt in cucumbers by regulating the rhizospheric microbial community. Sci. Rep. 7:41234. doi: 10.1038/srep41234
Shu, X. Y., Ye, Q. X., Huang, H., Xia, L. L., Tang, H., Liu, X. Y., et al. (2024). Effects of grazing exclusion on soil microbial diversity and its functionality in grasslands: a meta-analysis. Front. Plant Sci. 15:1366821. doi: 10.3389/fpls.2024.1366821
Sun, K., Fu, L., Song, Y., Yuan, L., Zhang, H., Wen, D., et al. (2021). Effects of continuous cucumber cropping on crop quality and soil fungal community. Environ. Monit. Assess. 193:436. doi: 10.1007/s10661-021-09136-5
Tian, T., Gheysen, G., Kyndt, T., Mo, C., Xiao, X., Lv, Y., et al. (2025). Pepper root exudate alleviates cucumber root-knot nematode infection by recruiting a rhizobacterium. Plant Commun. 6:101139. doi: 10.1016/j.xplc.2024.101139
Tong, Y., Du, H., Xiao, J., Zhou, B., Zheng, X., Deng, Y., et al. (2025). Watermelon wilt disease: causes, harms, and control measures. Front. Microbiol. 16:1601130. doi: 10.3389/fmicb.2025.1601130
Town, J. R., Dumonceaux, T., Tidemann, B., and Helgason, B. L. (2023). Crop rotation significantly influences the composition of soil, rhizosphere, and root microbiota in canola (Brassica napus L.). Environ. Microbiome 18:40. doi: 10.1186/s40793-023-00495-9
Tzipilevich, E., Russ, D., Dangl, J. L., and Benfey, P. N. (2021). Plant immune system activation is necessary for efficient root colonization by auxin-secreting beneficial bacteria. Cell Host Microbe 29, 1507–1520.e4. doi: 10.1016/j.chom.2021.09.005
Wang, H., Cai, X. Y., Xu, M., and Tian, F. (2021). Enhanced biocontrol of cucumber Fusarium wilt by combined application of new antagonistic Bacteria Bacillus amyloliquefaciens B2 and phenolic acid-degrading fungus Pleurotus ostreatus P5. Front. Microbiol. 12:700142. doi: 10.3389/fmicb.2021.700142
Wang, Y., Shi, M., Zhang, R., Zhang, W., Liu, Y., Sun, D., et al. (2023). Legume-potato rotation affects soil physicochemical properties, enzyme activity, and rhizosphere metabolism in continuous potato cropping. Chem. Biol. Technol. Agric. 10:132. doi: 10.1186/s40538-023-00508-2
Wang, C., Song, Z., Li, X., and Liu, Q. (2025). Bacterial community shifts in Fusarium-induced avocado root rot and the antagonistic potential of Bacillus siamensis NB92. Front. Microbiol. 16:1626537. doi: 10.3389/fmicb.2025.1626537
Wang, B., and Sugiyama, S. (2024). Microbial basis for suppression of soil-borne disease in crop rotation. Microorganisms 12:2290. doi: 10.3390/microorganisms12112290
Wang, J., Xu, S., Yang, R., Zhao, W., Zhu, D., Zhang, X., et al. (2021). Bacillus amyloliquefaciens FH-1 significantly affects cucumber seedlings and the rhizosphere bacterial community but not soil. Sci. Rep. 11:12055. doi: 10.1038/s41598-021-91399-6
Wu, J., Hu, S., Chen, J., Zhou, L., Yang, S., Zhou, N., et al. (2025). Soil microbial legacy mediated by buckwheat flavonoids enhances cabbage resistance to clubroot disease. Microbiome 13:176. doi: 10.1186/s40168-025-02166-y
Wu, H., Liu, E., Jin, T., Liu, B., Gopalakrishnan, S., Zhou, J., et al. (2025). Crop rotation increases Tibetan barley yield and soil quality on the Tibetan plateau. Nat. Food. 6, 151–160. doi: 10.1038/s43016-024-01094-8
Wu, X., Song, Y. Q., Yang, K. M., Gu, Y. A., Wei, Z., Kowalchuk, G. A., et al. (2020). Rhizosphere protists are key determinants of plant health. Microbiome 8:27. doi: 10.1186/s40168-020-00799-9
Xin, T., Zhang, Z., Zhang, Y., Li, X., Wang, S., Wang, G., et al. (2025). Recessive epistasis of a synonymous mutation confers cucumber domestication through epitranscriptomic regulation. Cell 188, 4517–4529.e15. doi: 10.1016/j.cell.2025.06.007
Yuan, Y., and Zhang, X. (2021). Comparison of agrochemicals allocation efficiency between greenhouse and open-field vegetables in China. Sci. Rep. 11:12807. doi: 10.1038/s41598-021-92316-7
Yue, Y., Hao, H., Wang, Q., Xiao, T., Zhang, Y., Chen, Q., et al. (2024). Dynamics of the soil microbial community associated with Morchella cultivation: diversity, assembly mechanism and yield prediction. Front. Microbiol. 15:1345231. doi: 10.3389/fmicb.2024.1345231
Keywords: cucumber Fusarium wilt, pea–cucumber crop rotation model, microbial diversity, soil nutrients, cucumber production
Citation: Xu J, Yao Y, Pan L, Zhang N, Li D and Chen X (2025) Pea–cucumber crop rotation suppresses Fusarium pathogens by reshaping soil microbial communities and enhancing nutrient availability. Front. Microbiol. 16:1697343. doi: 10.3389/fmicb.2025.1697343
Edited by:
Muhammad Zahid Mumtaz, Gansu Agricultural University, ChinaReviewed by:
Saowaluck Tibpromma, Qujing Normal University, ChinaAditi Bisht, Chandigarh University, India
Copyright © 2025 Xu, Yao, Pan, Zhang, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuehao Chen, eGhjaGVuQHl6dS5lZHUuY24=
 Jun Xu
Jun Xu Yuan Yao1
Yuan Yao1 Ningyuan Zhang
Ningyuan Zhang Xuehao Chen
Xuehao Chen