- 1Department of Surgical Diseases-2, West Kazakhstan Marat Ospanov Medical University, Aktobe, Kazakhstan
- 2Department of Oncology, West Kazakhstan Marat Ospanov Medical University, Aktobe, Kazakhstan
- 3Department of General Surgery, West Kazakhstan Marat Ospanov Medical University, Aktobe, Kazakhstan
- 4Scientific and Practical Center, West Kazakhstan Marat Ospanov Medical University, Aktobe, Kazakhstan
- 5Department of Natural Sciences, West Kazakhstan Marat Ospanov Medical University, Aktobe, Kazakhstan
Introduction: Human papillomavirus (HPV) has been increasingly implicated in the pathogenesis of breast cancer (BC), though its role remains controversial. Understanding HPV prevalence and genotype distribution across histological types and regions may clarify this potential association.
Methods: A comprehensive systematic review and meta-analysis was conducted using PubMed, Scopus, and Web of Science databases for studies published between January 1990 and April 2025. Eligible studies reported HPV prevalence in BC tissues stratified by histological classification. Non-English studies, reviews, and those lacking histological stratification were excluded. Data from 49 studies encompassing 4,173 BC cases were extracted. Pooled HPV prevalence and odds ratios (ORs) were calculated using random-effects models. Subgroup analyses were performed by histology, geographic region, and HPV genotype (16/18). Risk of bias was assessed using the Joanna Briggs Institute (JBI) checklist for cross-sectional studies and the Newcastle–Ottawa Scale for case-control designs.
Results: The pooled prevalence of HPV in BC tissues was 23% (95% CI: 18–28%), highest in invasive ductal carcinoma (24%). HPV-positive individuals exhibited a 3.6-fold higher risk of developing BC (OR = 3.63, 95% CI: 2.33–5.64), with the strongest association in invasive lobular carcinoma (OR = 4.41). HPV-18 showed a more consistent correlation with BC than HPV-16. Regional variation was observed, with Asian populations showing higher HPV prevalence and stronger associations.
Discussion: This meta-analysis suggests a significant association between HPV infection—particularly genotype 18—and breast cancer risk, especially in Asian regions and specific histological subtypes. These findings highlight the need for mechanistic studies and standardized molecular detection to elucidate the potential oncogenic role of HPV in breast tissue.
Systematic review registration: https://www.crd.york.ac.uk/PROSPERO/display_record.php?RecordID=1051960 identifier CRD420251051960
1 Introduction
Breast cancer (BC) is the most frequently diagnosed malignancy and the leading cause of cancer-related death among women worldwide (Smolarz et al., 2022). BC accounts for approximately 12% of all cancer cases globally, with over 2 million new cases reported in 2020 (Sedeta et al., 2023; Lukasiewicz et al., 2021). It is characterized by significant heterogeneity in its histopathological types, epidemiological factors, and clinical outcomes. Among these types, invasive lobular carcinoma (ILC), invasive ductal carcinoma (IDC), and ductal carcinoma in situ (DCIS) are the most prevalent. Understanding the potential infectious etiology of BC, including viral involvement, is crucial for advancing preventive and therapeutic strategies (Liu and Yu, 2023). The prevalence of BC is increasing, particularly in regions adopting Western lifestyle behaviors, such as South America, Africa, and Asia (Sedeta et al., 2023). Human papillomavirus (HPV), a DNA virus from the papillomaviridae family, predominantly targets epithelial tissues (Hareza et al., 2022). The virus is classified into non-oncogenic low-risk and oncogenic high-risk categories. Low-risk strains of HPV are typically responsible for the development of genital warts, whereas high-risk strains—particularly HPV types 16 and 18—are linked to the onset of cancers affecting the cervix, vulva, vagina, anus, penis, and the oropharyngeal region (Siddiqi and Ridker, 2019; Pesut et al., 2021). HPV’s oncogenic potential is primarily due to the E6 and E7 oncoproteins, which interfere with tumor suppressor proteins like p53 and pRB, promoting cell cycle disruption and carcinogenesis (Hareza et al., 2022).
HPV is primarily transmitted through sexual contact and, less commonly, via vertical transmission during childbirth (Malagon et al., 2021; Wierzbicka et al., 2023; Khayargoli et al., 2023; Freitas et al., 2013). Although these routes are well established, potential dissemination to mammary tissue remains hypothetical. Proposed mechanisms include hematogenous or lymphatic spread from other infected sites and retrograde ductal migration through the nipple–areolar complex (Lawson et al., 2015; Bodaghi et al., 2005; Akil et al., 2008; de Villiers et al., 2005; Purrahman et al., 2022; Blanco et al., 2021). While biologically plausible, direct evidence remains limited. Detections of HPV DNA and E6/E7 transcripts in nipple–areolar ducts and reports of circulating HPV DNA support biological plausibility for mammary tissue exposure.
The prospective correlation between HPV and breast carcinoma has emerged as a topic of significant scholarly discourse and investigation. While some studies suggest a possible link, others find no significant association (Purrahman et al., 2022). Previous studies investigating the occurrence of HPV DNA within BC tissues have reported widely varying rates, influenced by factors such as geographic region, population characteristics, histological subtype, and diagnostic methods. HPV types 16 and 18, which are recognized for their strong cancer-causing potential in cervical malignancies, have also been frequently identified in BC tissues. Nevertheless, their definitive role in the initiation or progression of BC is still uncertain.
To explore a possible causal relationship between HPV infection and BC development, we conducted an extensive systematic review and meta-analysis following the 2020 PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines. We aimed to evaluate the prevalence of HPV DNA in breast tumors quantitatively, explore associations with specific histological types (IDC, ILC, DCIS), and analyze trends in geographical and genotype-specific distribution. To our knowledge, this paper represents the first comprehensive meta-analysis that systematically categorizes findings by histopathological classification, enabling a more nuanced understanding of HPV’s potential oncogenic mechanisms across distinct BC phenotypes. Our work extends prior syntheses—including the updated meta-analysis by Awan et al. (2023)—by stratifying results a priori by histopathology (IDC, ILC, DCIS), geography, and genotype, and by formally exploring method-related heterogeneity. Compared with the recent comprehensive meta-analysis by Awan et al. (2023), the present review provides additional resolution by analyzing HPV prevalence and risk stratified by histological subtype (IDC, ILC, DCIS) and geographic region, incorporating studies published through 2025. The synthesized evidence may significantly contribute to the ongoing discourse regarding viral oncogenesis in BC and could inform the development of targeted screening protocols and therapeutic interventions.
2 Materials and methods
2.1 Search strategy
This thorough and systematic review of existing literature identified relevant studies that investigate the presence of HPV infection in patients diagnosed with BC. The study was registered in PROSPERO on May 19, 2025 (CRD420251051960). The protocol was registered retrospectively in PROSPERO after the database search was completed; no deviations from the initial search or analysis plan occurred. The search covered publications from January 1990 to April 2025 across three major databases: PubMed, Scopus, and Web of Science. A combination of controlled vocabulary (MeSH terms) and free-text keywords was used to maximize the sensitivity of the search strategy (Table 1). Search terms and selection criteria were harmonized with, but broadened beyond, prior frameworks (Awan et al., 2023), to capture histotype-specific reporting.
Table 1. Search strategy across databases for human papillomavirus (HPV) and breast cancer meta-analysis.
2.2 Selection of studies
Well-defined inclusion and exclusion criteria were delineated to guarantee the pertinence and integrity of the chosen studies. Inclusion criteria encompassed studies reporting HPV prevalence in BC tissues, stratified by histological types (IDC, ILC, and DCIS). Eligible studies included those with cross-sectional, case-based, or prevalence-focused designs. Research studies were excluded from consideration if they fulfilled any of the subsequent criteria: published in languages distinct from English; classified as letters, commentaries, reviews, case series, editorials, or commission reports; recognized as duplicate publications; did not report the raw data necessary to compute HPV prevalence or odds ratios (OR)—specifically, the number of BC and control samples and the counts of HPV-positive cases; or did not offer stratification based on histological tumor type. These stringent criteria were implemented to enhance methodological rigor, ensure data comparability across studies, and allow stratified analyses by histological types. Only English-language studies were included to ensure methodological consistency; we acknowledge this may introduce language bias, particularly for Asia and South America.
2.3 Data extraction
After the initial screening, three independent reviewers (AT, SB, and DK) evaluated the titles and abstracts of the studies that were identified, proceeding to full-text review when necessary. Throughout the search, 1,368 articles were screened, of which 49 (4,173 BC cases) met the eligibility criteria and were included in the final meta-analysis (Table 2—Summary table of studies reporting the presence of HPV in BC patients from 1992 to 2022) (Lawson et al., 2015; Akil et al., 2008; de Villiers et al., 2005; Hennig et al., 1999; Yu et al., 2000; Damin et al., 2004; Widschwendter et al., 2004; Kroupis et al., 2006; Choi et al., 2007; Duo et al., 2008; Khan et al., 2008; Heng et al., 2009; Mendizabal-Ruiz et al., 2009; Aguayo et al., 2011; Nascimento et al., 2024; Belachew et al., 2024; Mareti et al., 2023; Maldonado-Rodriguez et al., 2022; Alinezhadi et al., 2022; Gupta et al., 2021; Elagali et al., 2021; Tawfeik et al., 2020; Sher et al., 2020; Khodabandehlou et al., 2019; Salman et al., 2017; Balci et al., 2019; Antonsson et al., 2011; Herrera-Goepfert et al., 2011; Baltzell et al., 2012; Frega et al., 2012; Habyarimana et al., 2018; Ghaffari et al., 2018; Wang et al., 2017; Ngamkham et al., 2017; Naushad et al., 2017; Glenn et al., 2012; Islam et al., 2017; Doosti et al., 2016; Sigaroodi et al., 2012; Li et al., 2015; Gannon et al., 2015; Herrera-Goepfert et al., 2013; Liang et al., 2013; Pereira Suarez et al., 2013; Ahangar-Oskouee et al., 2014; Fu et al., 2015; Fernandes et al., 2015; Manzouri et al., 2014; Hong and Tang, 2014). Key variables identified comprised the initial author’s name, publication year, geographical study site, sample size, and HPV prevalence in BC cases, detected HPV genotypes, presence of coinfections, and the diagnostic methods used. Data extraction followed the PRISMA guidelines (Figure 1). The pooled HPV prevalence and odds ratios (ORs) were calculated using random-effects models. Subgroup analyses were performed by histology, region, and HPV genotype (HPV-16/18).
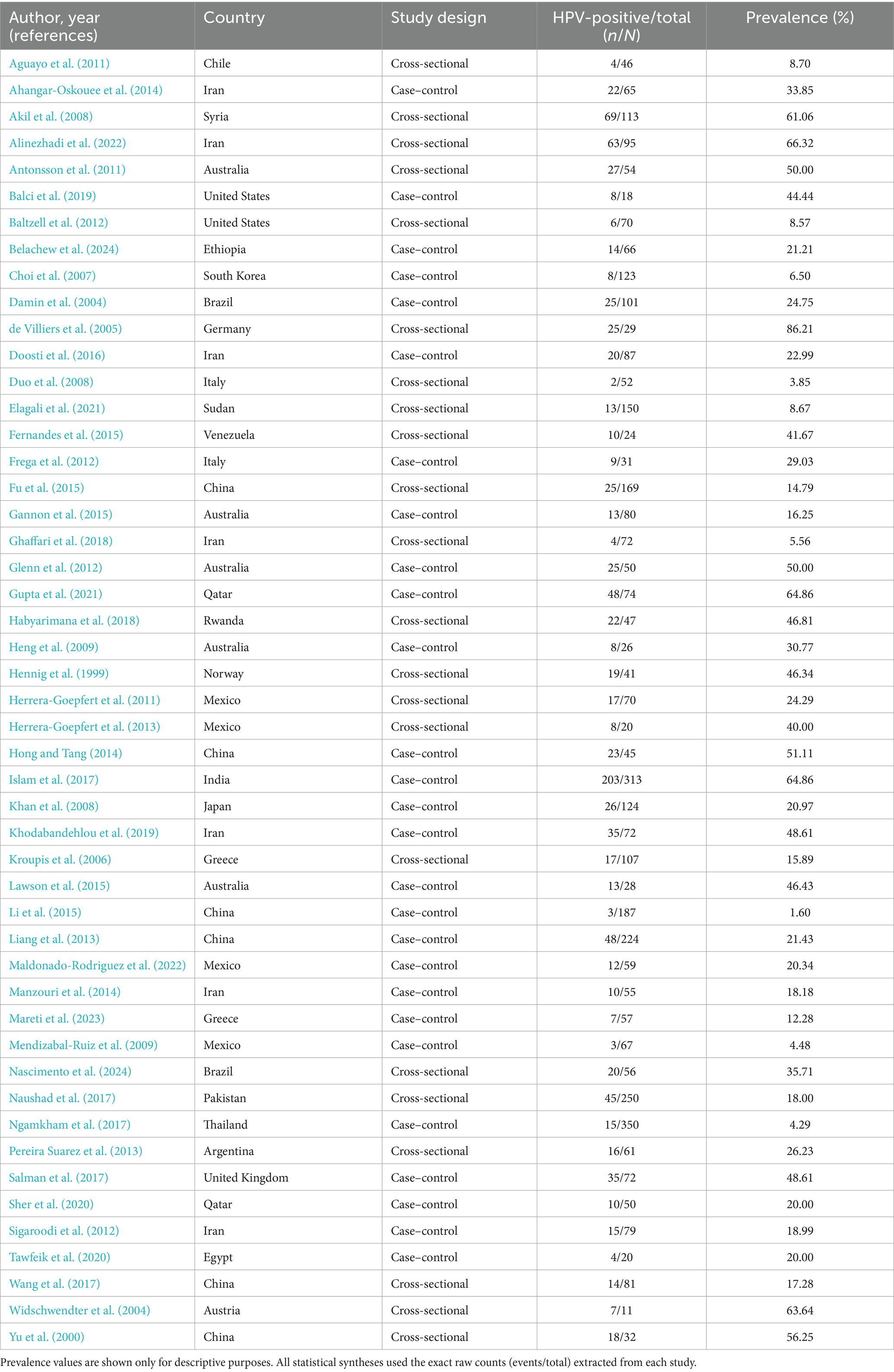
Table 2. Summary of the included studies for human papillomavirus (HPV) and breast cancer meta-analysis.
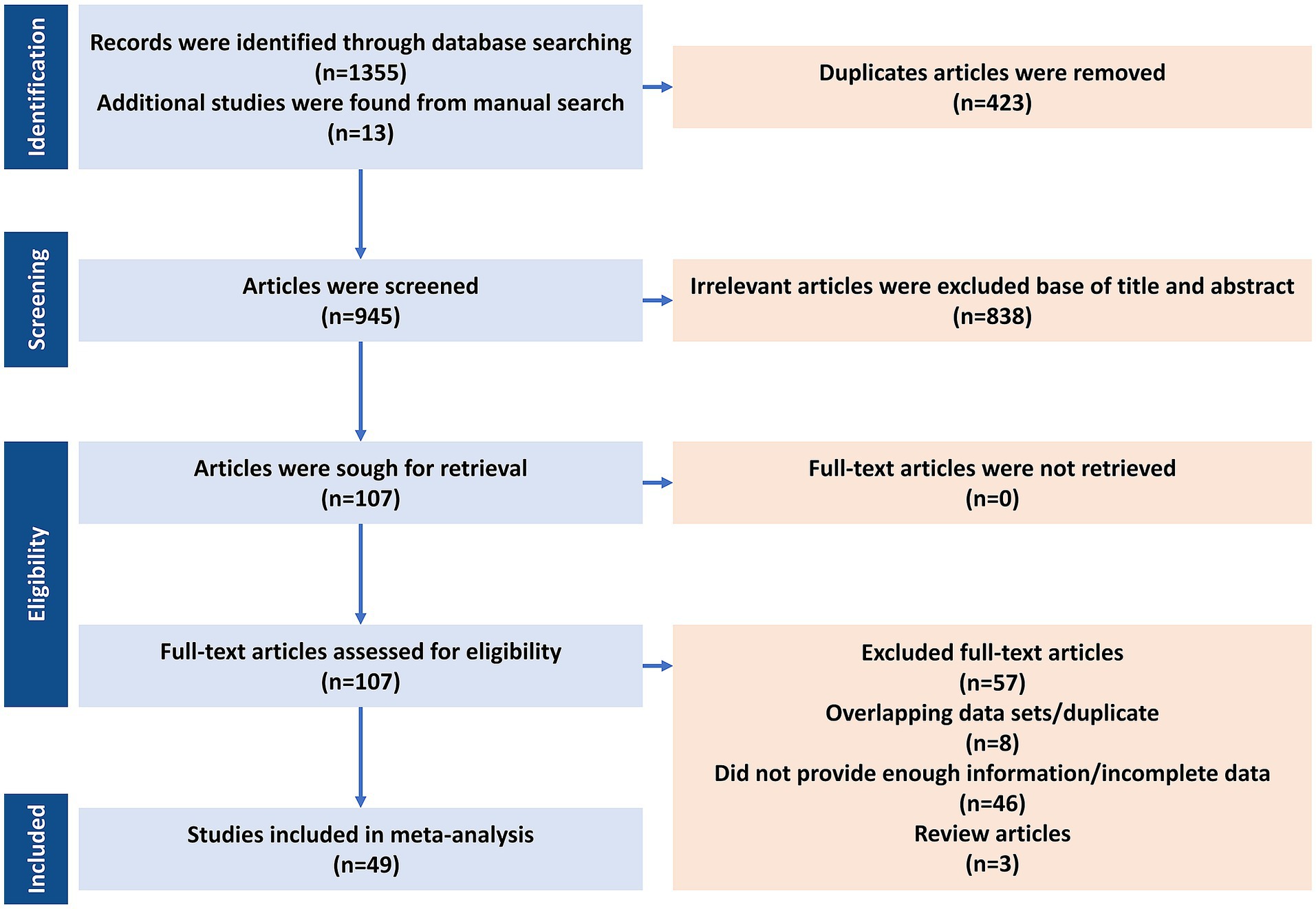
Figure 1. PRISMA flow diagram for the systematic search and article selection process (n = 49; 4,173 BC cases) for human papillomavirus (HPV) and breast cancer meta-analysis.
2.4 Quality assessment
Three reviewers (AT, SB, and DK) independently evaluated the methodological quality of each study included in the analysis. For cross-sectional studies, they applied the Joanna Briggs Institute (JBI) Critical Appraisal Checklist, while for case–control studies, the Newcastle–Ottawa Scale was used to assess the risk of bias. To ensure consistency and methodological integrity, any discrepancies in quality assessments were addressed through discussion until a consensus was reached among the reviewers.
2.5 Statistical analysis
Quantitative analyses were conducted after data extraction, using Microsoft Excel for preliminary organization and RevMan (version 5.4) together with R version (4.5.0) for advanced meta-analytic procedures. Odds ratios represented the odds of detecting HPV DNA in breast-cancer tissues compared with non-cancerous control breast tissues. Various R packages were utilized for the meta-analyses. The prevalence of HPV infection was estimated using the binomial distribution formula, with corresponding standard errors (SE). To accommodate the anticipated variability among studies, a random-effects model was applied. Heterogeneity was evaluated using Cochran’s Q test and the I2 statistic, with I2 values of 25, 50, and 75% indicating low, moderate, and high heterogeneity, respectively. Forest plots were generated to visually depict the effect sizes (ES) along with their 95% confidence intervals (95% CI). Subgroup analyses were also conducted to investigate possible sources of heterogeneity. The certainty of evidence was not assessed using the GRADE approach. We explored heterogeneity using subgroup analyses and meta-regression (metafor, REML) with moderators: (i) detection method (e.g., consensus PCR/nested PCR/hybrid capture/RT-PCR), (ii) specimen type (FFPE vs. fresh/frozen), and (iii) geographic region. For case–control studies, log-ORs were modeled against these moderators; for prevalence, logit-transformed proportions were used.
Prevalence (one-group) and odds-ratio (case–control) analyses were conducted separately. To address potential confounding by detection method or specimen handling, subgroup and meta-regression analyses were performed with moderators for assay type (consensus PCR, nested PCR, hybrid capture, RT-PCR) and tissue source (FFPE vs. fresh/frozen). Certainty of evidence (GRADE) was not performed due to observational designs and methodological heterogeneity. Studies with zero/near-zero cells were handled using continuity corrections per standard practice; resultant wide CIs indicate imprecision.
3 Results
3.1 Characteristics of included studies
A total of 1,381 records were identified, and 49 studies (4,173 BC cases) met the inclusion criteria (Figure 1). The final meta-analysis included 49 studies investigating the presence of HPV DNA in BC tissues, comprising a total of 4,173 BC cases. The analysis was conducted in two phases: (1) a one-group proportion meta-analysis to estimate the pooled prevalence of HPV among BC patients, and (2) a case–control meta-analysis comparing HPV prevalence between cancerous and non-cancerous breast tissues. The 86% prevalence in de Villiers et al. (2005) reflects small sample size and early detection platforms; exclusion in sensitivity analysis did not materially change pooled estimates.
Detailed per-study genotype data have been moved to Supplementary Table S1. Table 3 summarizes the 10 most frequent genotypes by continent. The analysis of HPV genotype distribution across the included studies revealed considerable variation in the prevalence of individual HPV types and co-infections (Supplementary Table S1). HPV-16 was the most frequently reported genotype, appearing in numerous studies with varying case numbers, followed by HPV-18, HPV-33, and HPV-31. Several studies also identified less common genotypes such as HPV-35, HPV-66, and HPV-58, while others reported the presence of low-risk types like HPV-6 and HPV-11. Notably, co-infections involving multiple HPV types were documented in many reports, suggesting a complex pattern of viral presence in affected individuals. The total number of cases per study ranged widely, with some studies focusing on a few specific genotypes and others providing broader screening results encompassing over 20 genotypes. This variation highlights both geographic and methodological differences in HPV detection and reporting, underlining the importance of comprehensive genotyping in understanding the epidemiology of HPV infections.
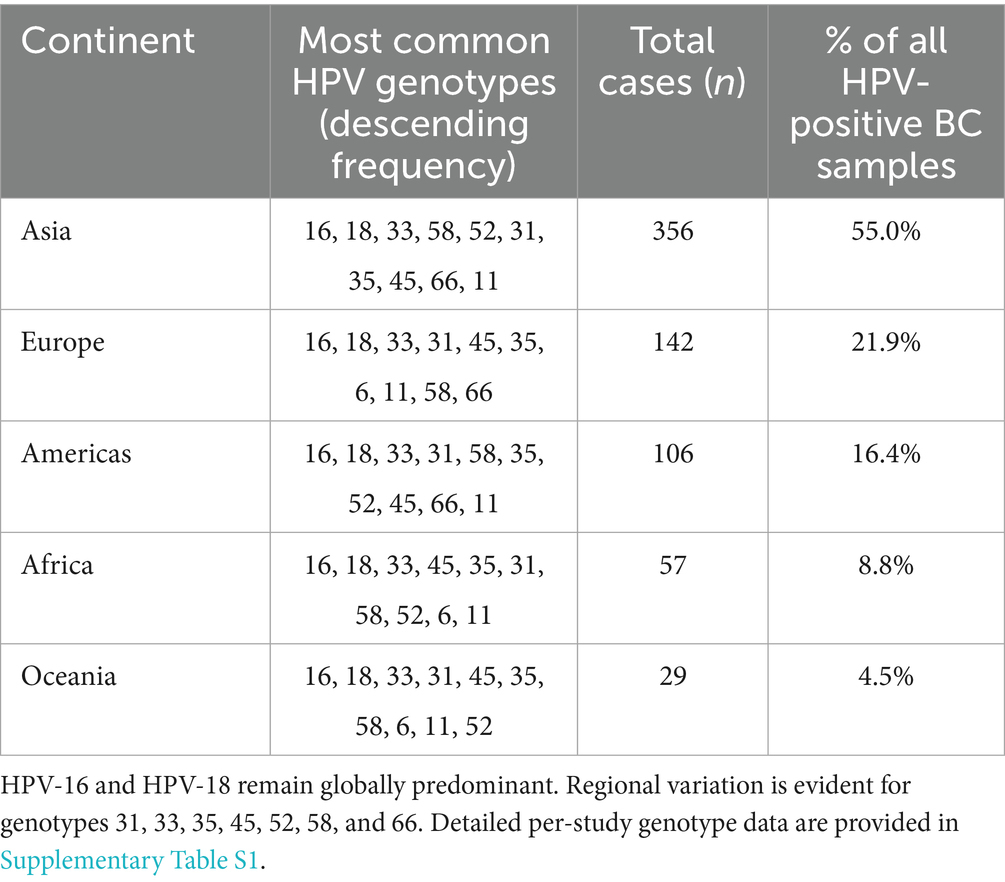
Table 3. Most frequent high-risk human papillomavirus (HPV) genotypes in breast cancer by continent (top 10 by frequency).
3.2 Methodological quality and bias risk
Quality assessment using JBI and Newcastle–Ottawa tools revealed that 85% of studies (42/49) demonstrated moderate quality (mean scores: 6/8 for cross-sectional, 6/9 for case–control) (Supplementary Tables S2, S3). Common limitations included incomplete adjustment for confounders (75% of studies) and variability in HPV detection protocols. Notably, heterogeneity was lowest for ILC (I2 = 11.6%) and DCIS (I2 = 25.9%), suggesting robust subtype-specific findings, while IDC exhibited substantial heterogeneity (I2 = 91.6%), likely reflecting methodological diversity across studies.
3.3 One-group proportion meta-analysis
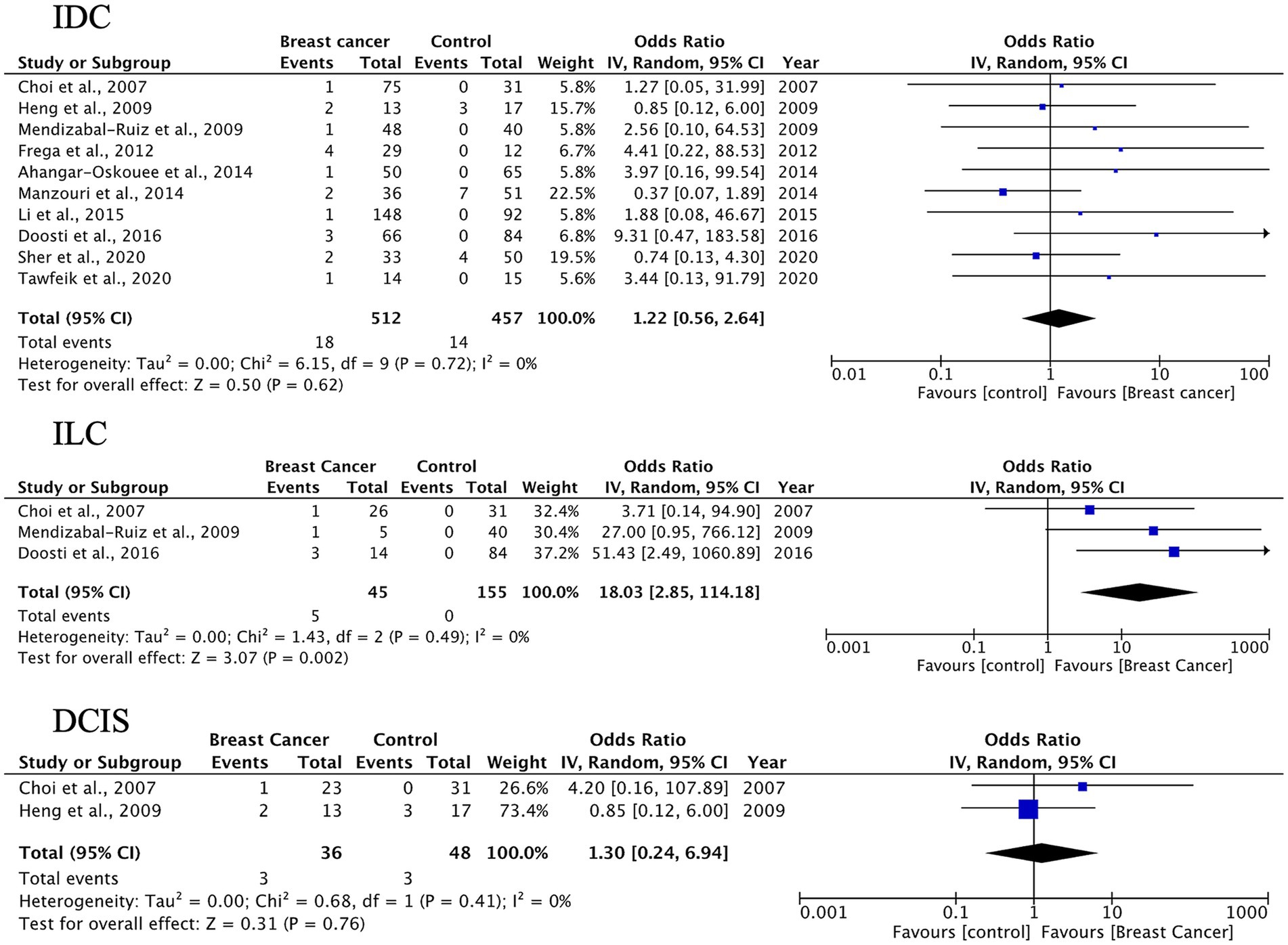
The pooled proportion of HPV-positive BC cases across all studies was 23% (95% CI: 18–28%). Subgroup analysis based on histological type yielded the following results (Figure 2): IDC: 24% (95% CI: 18–32%), I2 = 91.6%; ILC: 22% (95% CI: 13–35%), I2 = 11.6%; DCIS: 21% (95% CI: 13–32%), I2 = 25.9%; other histological types: 21% (95% CI: 12–33%), I2 = 50.5%.
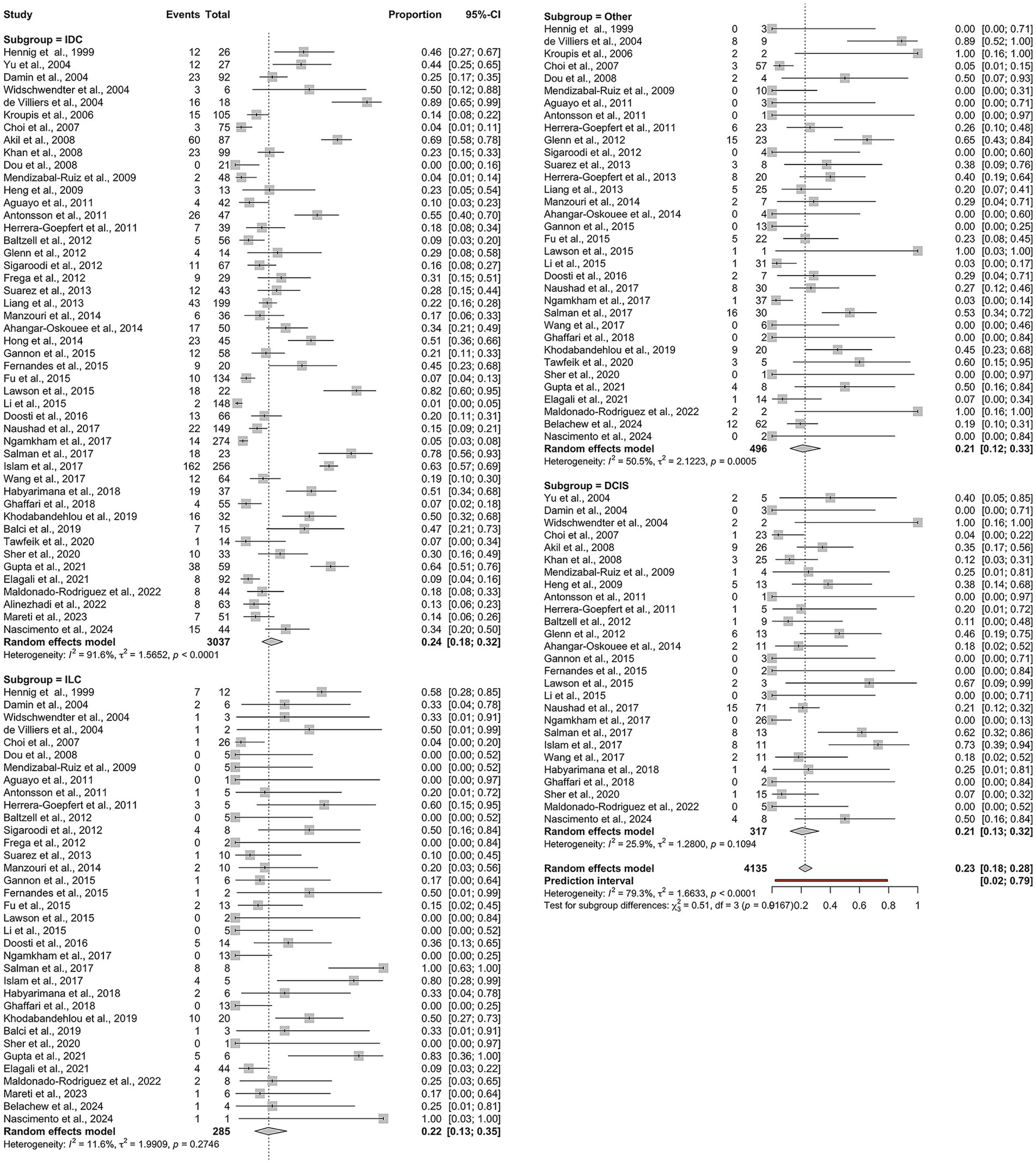
Figure 2. Overall pooled prevalence of human papillomavirus (HPV) across histological subtypes of breast cancer: IDC, ILC, DCIS, and other histological types.
While the overall heterogeneity was high (I2 = 79.3%, τ2 = 1.66, p < 0.0001), it varied significantly among the subgroups. Notably, ILC demonstrated minimal heterogeneity, indicating consistent findings across studies. In contrast, IDC showed substantial heterogeneity, suggesting considerable methodological or population-based variability between the studies. Given substantial heterogeneity, estimates for IDC should be interpreted as average associations across diverse settings rather than precise effects.
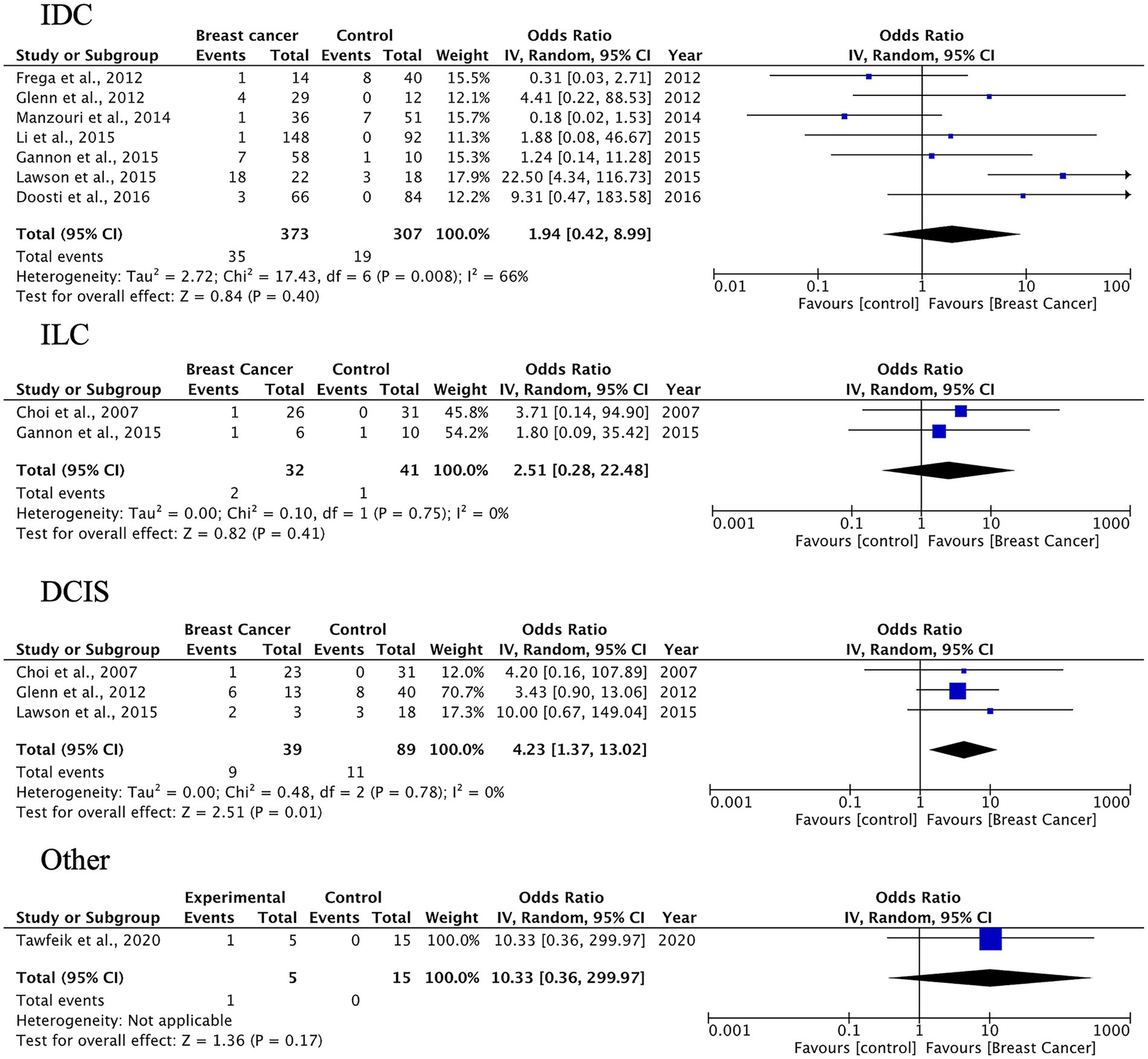
3.4 Case–control meta-analysis
A total of 27 case–control studies were included in the second analysis phase. Our analytical approach incorporated a three-tiered stratification of case–control studies, evaluating: (1) tumor histology, (2) geographic origin, and (3) HPV genotype (categorized as HPV-16, HPV-18). This multidimensional classification allowed for the detection of type-specific and region-dependent HPV carcinogenesis patterns. The pooled OR for HPV presence in BC tissues compared to control tissues was 3.63 (95% CI: 2.33–5.64, p < 0.00001), indicating a significantly higher prevalence of HPV in malignant samples (Figure 3). Subgroup-specific results were as follows: IDC: OR = 3.63 (95% CI: 2.33–5.66), I2 = 52%; ILC: OR = 4.41 (95% CI: 2.11–9.24), I2 = 35%; DCIS: OR = 3.10 (95% CI: 1.43–6.70), I2 = 38%; other types: OR = 3.28 (95% CI: 1.73–6.24), I2 = 49%. These results indicate a statistically significant association between HPV infection and BC across all histological subtypes. The strongest association was observed in ILC, while DCIS, which is a pre-invasive form, also showed a meaningful link, suggesting the potential role of HPV in early carcinogenic processes.
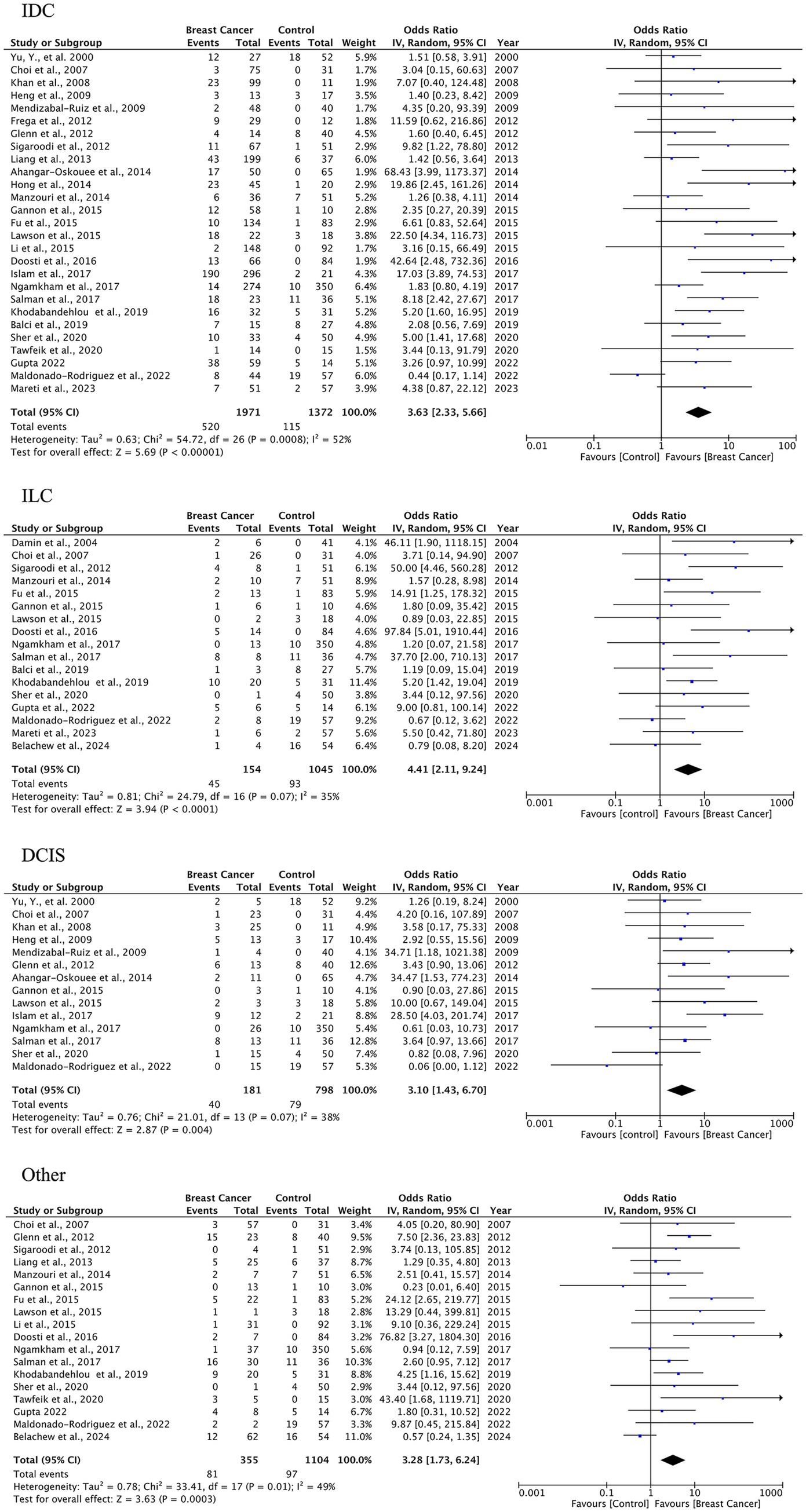
Figure 3. HPV prevalence in breast tissue specimens: case–control analysis by histological subtype (IDC, ILC, DCIS, others).
3.5 The incidence of HPV in BC varies by histological type and location
In analyzing IDC, the most common BC histotype, we observed an overall HPV prevalence of 24% (95% CI: 18–32%), with the highest detection rates in Asian populations (14 studies, n = 1,581 cases) and significant between-study heterogeneity (I2 = 91.6%). This association was less pronounced in other regions, likely reflecting variations in HPV genotype distribution and detection methodologies (Figure 4).
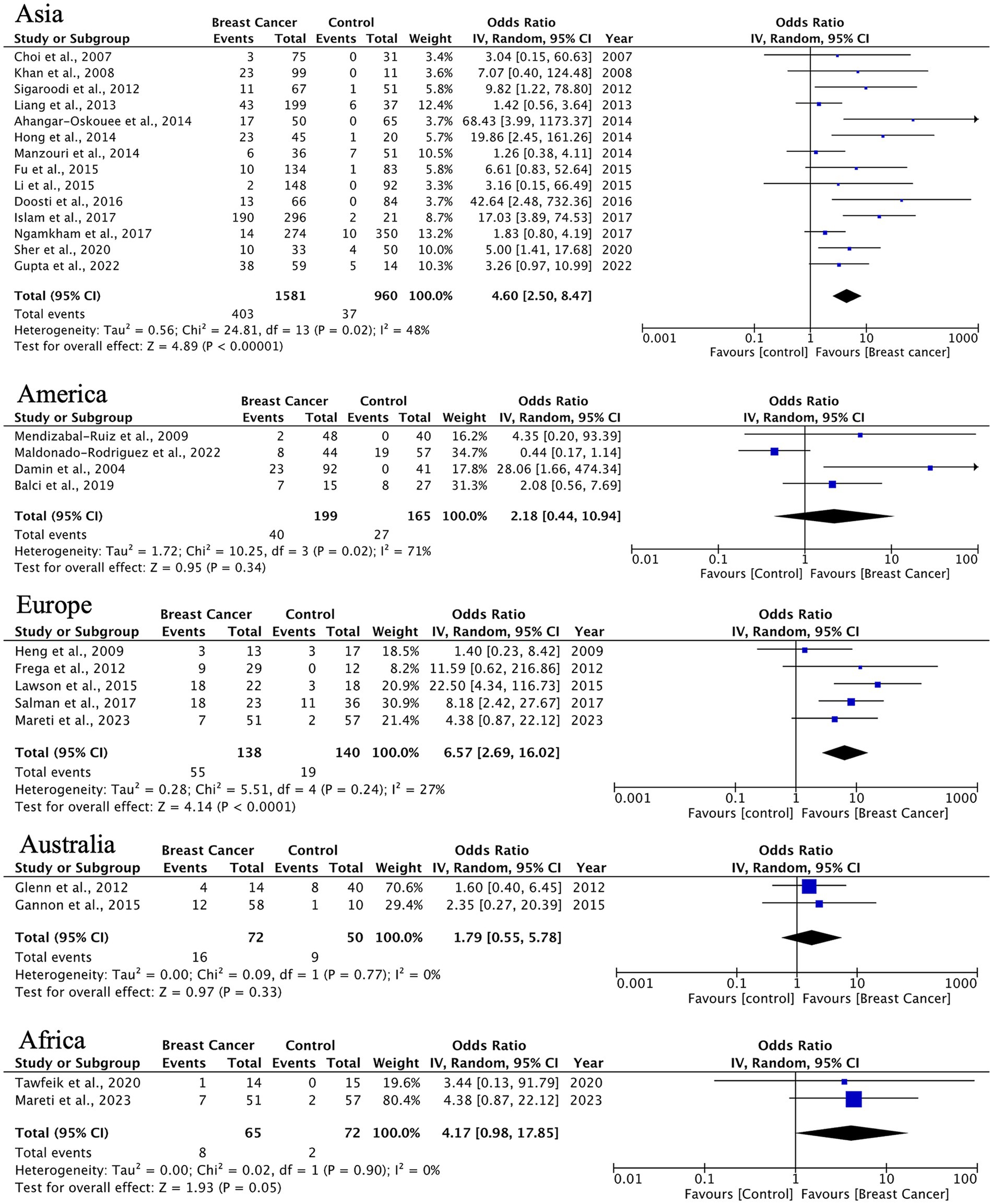
Figure 4. Geographic distribution of human papillomavirus (HPV) prevalence in IDC of the breast. Very wide CIs reflect low event counts and should be interpreted as imprecise estimates rather than robust effects.
HPV prevalence in ILC exhibits significant geographical variation. Asian studies (10 studies, n = 116 cases) demonstrate a strong HPV association (OR = 6.76, 95% CI: 2.76–16.57) with minimal heterogeneity (I2 = 25%), while other regions present non-significant associations (Americas: OR = 2.18, p = 0.33; Europe: OR = 3.43, p = 0.07) (Figure 5). The observed trend indicates possible differences in HPV oncogenic mechanisms between ductal and lobular carcinomas.
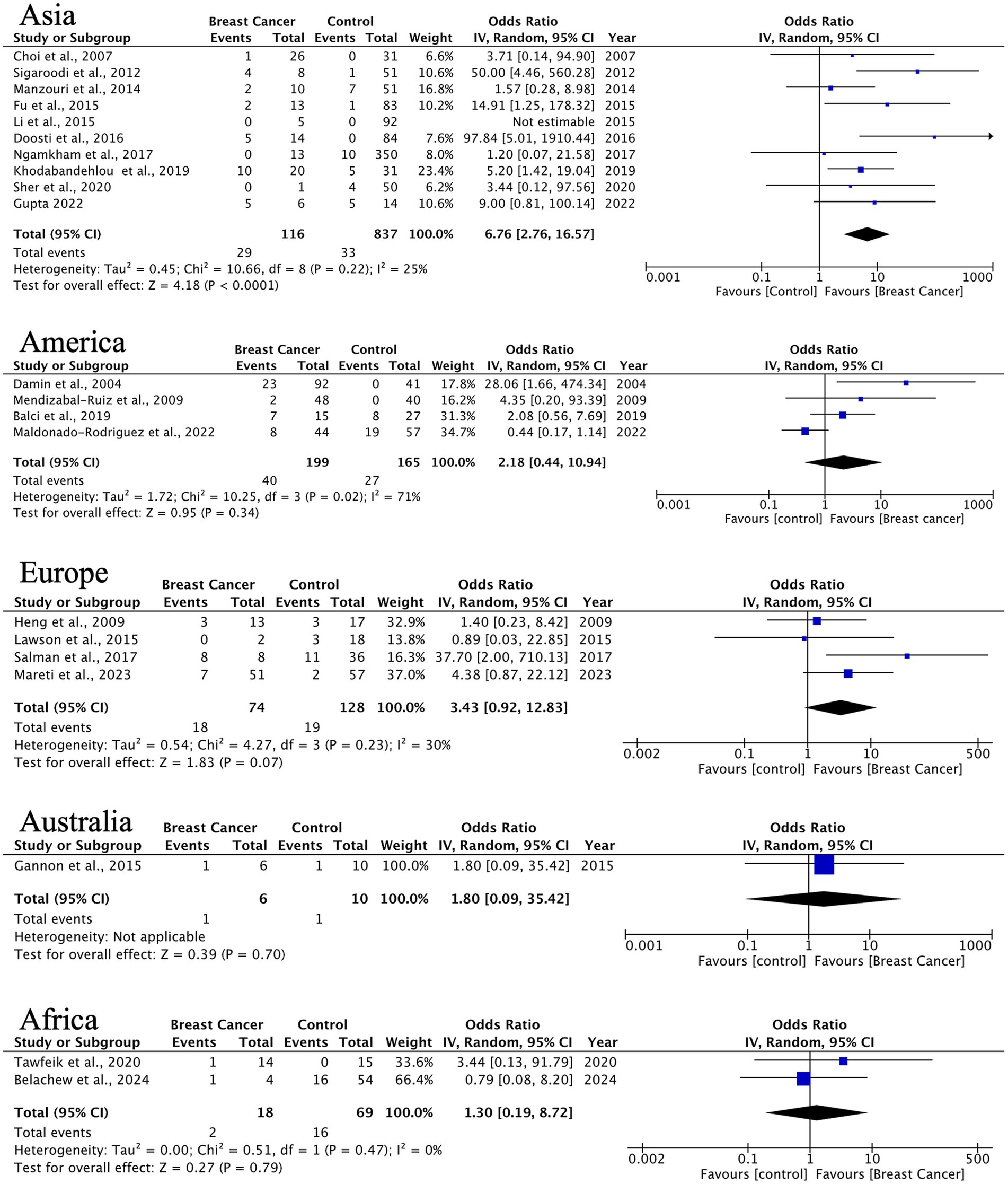
Figure 5. Geographic distribution of human papillomavirus (HPV) prevalence in invasive lobular breast cancer. Very wide CIs reflect low event counts and should be interpreted as imprecise estimates rather than robust effects.
For DCIS, we found statistically significant HPV associations in Europe (OR = 3.85, 95% CI: 1.46–10.14, p = 0.006) and borderline significance in Asia (OR = 3.29, p = 0.05). The European findings demonstrate consistent results across studies (I2 = 0%), which may support HPV’s potential role in early breast carcinogenesis (Figure 6).
![Four forest plots display the odds ratios for DCIS (Ductal Carcinoma In Situ) across different regions: Asia, America, Europe, and Australia. Each plot includes studies with events, total participants, weight, and confidence intervals. Asia shows a total odds ratio of 4.17 [1.02, 17.02], America shows 34.71 [1.18, 1021.38], Europe shows 3.85 [1.46, 10.14], and Australia shows 2.87 [0.83, 10.00]. Each plot includes measures of heterogeneity and tests for overall effects, denoted by diamond shapes. The x-axes range from 0.001 to 1000, indicating favors for control or breast cancer.](https://www.frontiersin.org/files/Articles/1712118/fmicb-16-1712118-HTML/image_m/fmicb-16-1712118-g006.jpg)
Figure 6. Geographic distribution of human papillomavirus (HPV) prevalence in ductal breast carcinoma in situ. Very wide CIs reflect low event counts and should be interpreted as imprecise estimates rather than robust effects.
Other histological types showed significant HPV associations in Asia (OR = 3.31, 95% CI: 1.70–6.45) and Europe (OR = 2.96, 95% CI: 1.13–7.79), although with limited sample sizes. Notably, results from Australia and Africa displayed extreme heterogeneity (I2 = 84–87%), indicating a need for standardized detection methods and larger studies in these regions (Figure 7).
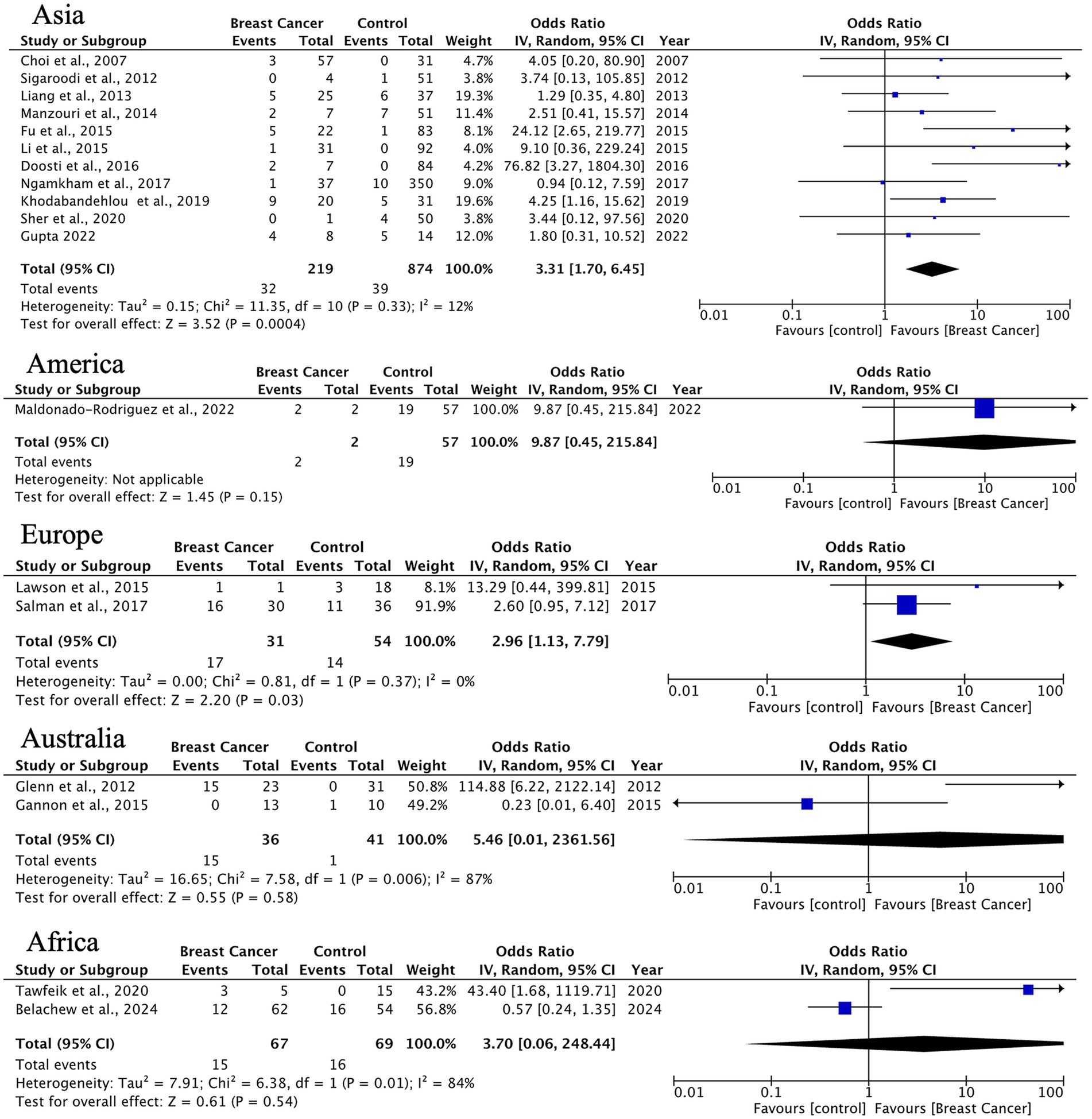
Figure 7. Geographic distribution of human papillomavirus (HPV) prevalence in uncommon breast carcinoma histotypes. Very wide CIs reflect low event counts and should be interpreted as imprecise estimates rather than robust effects.
3.6 The global prevalence of HPV 16/18 types in BC and the regional variations observed
The forest plots for HPV-16 and HPV-18 indicate a potential association with BC, though with varying levels of statistical significance and heterogeneity. The data for HPV-16 suggests a less consistent relationship, with the OR reflecting a non-significant positive association (Figure 8). Its clinical heterogeneity reflects diverse histological subtypes and molecular drivers that influence prognosis and therapeutic response.
Transcriptional activity subset. Five studies (~10%) assessed viral transcription (E6/E7 mRNA/protein); three reported positive signals. Where present, associations tended to be stronger, though sample sizes were limited.
HPV-18 appeared more frequently in breast-cancer samples than HPV-16 across several datasets; however, between-study variability precludes definitive conclusions regarding relative strength of association. Further research with larger sample sizes and more uniform study designs would help clarify these associations (Figure 9).
Figure 10 illustrate the global distribution of 21 HPV genotypes across five continents: Asia, Africa, the Americas, Europe, and Australia. HPV-16 emerged as the predominant type worldwide, accounting for 254 of 647 total cases (39%), with particularly high prevalence in Asia and Europe. HPV-18 was the second most common type, representing 115 cases (18%). The remaining 19 HPV types showed considerable geographic variation in their distribution patterns, with specific genotypes exhibiting regional predominance. For instance, HPV-31, 33, and 45 demonstrated 3- to 5-fold differences in prevalence between continents, while other oncogenic types (HPV-35, −52, −58) displayed even more pronounced regional clustering. These findings underscore both the universal dominance of HPV-16/18 in breast carcinoma across all studied populations and the distinct regional profiles for less common high-risk HPV types, which may reflect differences in viral evolution, population genetics, or environmental cofactors influencing genotype-specific oncogenesis.
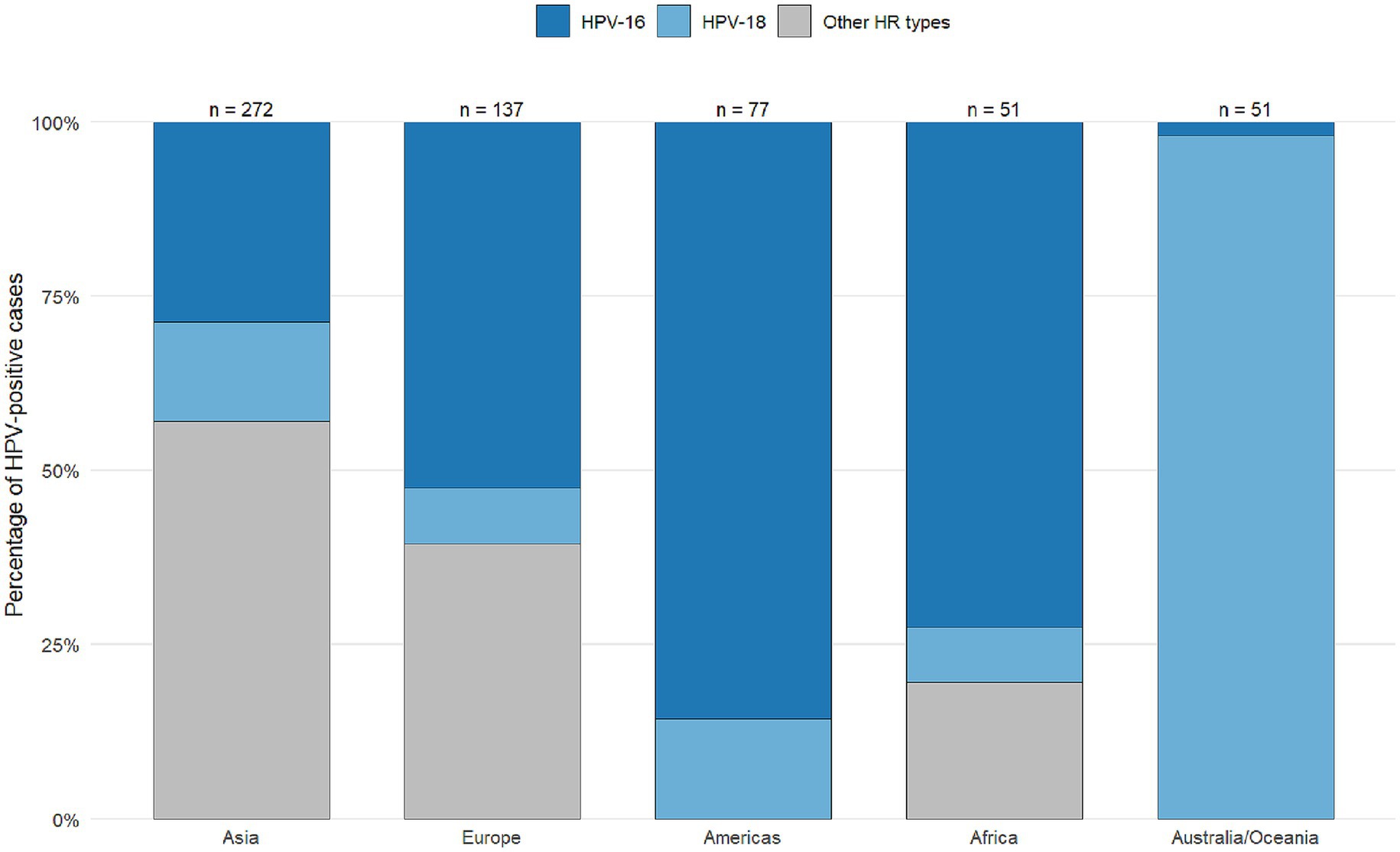
Figure 10. Distribution of human papillomavirus (HPV) types, including those other than 16/18, across various continents worldwide for studies reporting the presence of HPV in breast cancer patients from 1992 to 2022.
Heterogeneity exploration. In meta-regression of case–control log-ORs, detection method and specimen type were significant sources of variability (omnibus p < 0.05). Relative to consensus PCR, nested PCR tended to yield higher effect sizes, while hybrid capture/RT-PCR yielded lower estimates; fresh/frozen tissue showed higher detection than FFPE. Region remained a residual contributor after method/specimen adjustment, indicating both methodological and geographic components to heterogeneity. (Model details in Supplementary Table S4; influence diagnostics and residual plots in Supplementary Figure S1).
3.7 Publication bias and sensitivity analyses
Funnel plot asymmetry was detected for the IDC histotype (Egger’s test: p = 0.029; Begg’s test: p = 0.425) (Supplementary Table S5 and Figure 11). Trim-and-fill adjustment imputed 10 missing studies, reducing the IDC effect size to 34.9% (26.3–44.5%) while maintaining statistical significance (τ2 = 2.17, I2 = 93%). No adjustments were necessary for ILC or DCIS histotypes (Egger’s p > 0.3), underscoring the stability of these findings. Publication-bias diagnostics (Egger’s and Begg’s tests, trim-and-fill) were applied to the prevalence model to explore small-study effects; results should be interpreted qualitatively given high heterogeneity (I2 = 93%).
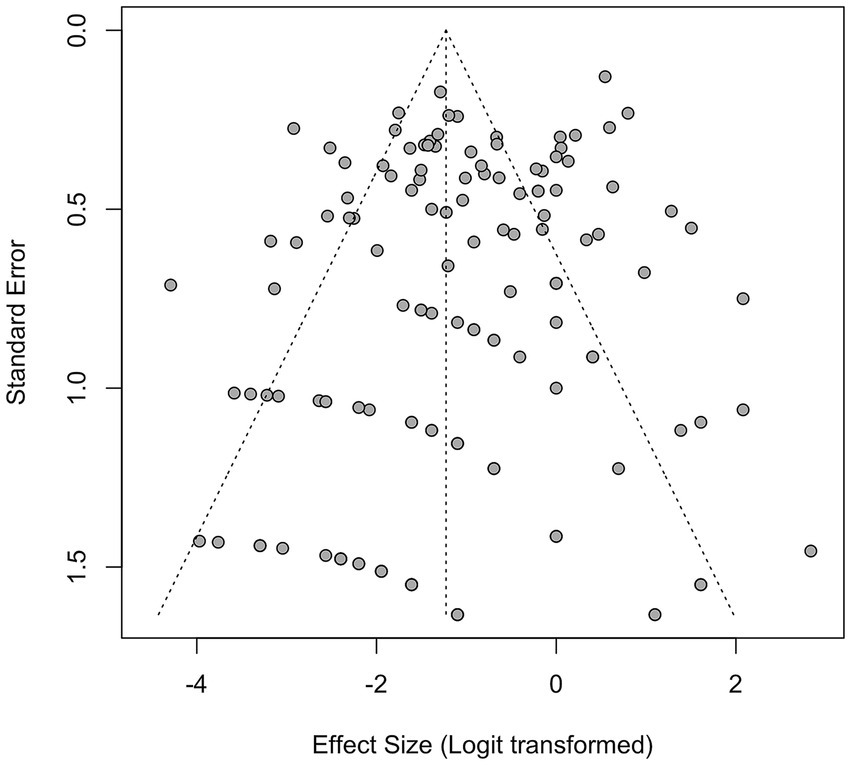
Figure 11. Funnel plot for all studies for studies reporting the presence of human papillomavirus (HPV) in breast cancer patients from 1992 to 2022.
4 Discussion
The pooled prevalence of HPV DNA detected in BC tissues was 23% (95% CI: 18–28%), with the highest rate observed in IDC at 24% (95% CI: 18–32%). HPV positivity was associated with higher odds of BC across histological subtypes, with the strongest signal in ILC. While heterogeneity was substantial for IDC, findings for ILC and DCIS were more consistent, suggesting a potential role for HPV in both invasive lobular disease and in situ lesions.
4.1 Comparison with prior evidence
Our pooled prevalence aligns with Awan et al. (2023), who reported elevated HPV detection in breast tumors across regions, though direct contrasts are limited by differences in inclusion windows and detection platforms. Importantly, our stratification by histopathology (IDC/ILC/DCIS) and genotype extends the literature by showing: (i) a more stable association in ILC (lower I2), and (ii) a relatively stronger and more consistent association for HPV-18 than for HPV-16. Findings from Gomes de Oliveira et al. (2022)—who focused on fresh tissues—support that detection can vary by specimen type; our sample-type analyses (see meta-regression below) similarly indicate that fresh/frozen vs. FFPE and detection method contribute materially to heterogeneity. Despite high I2 for IDC, random-effects pooling is appropriate to summarize between-study variability; consistent directional effects and sensitivity analyses support reporting a pooled estimate, while emphasizing caution.
While our findings corroborate those of Awan et al. (2023), the present review uniquely delineates histology-specific and genotype-specific associations, offering complementary insights into potential subtype-dependent viral oncogenesis. Even after accounting for assay and specimen moderators, the association’s direction remained consistent, suggesting that technical factors alone are unlikely to explain the signal.
4.2 Marked geographic variations in HPV prevalence and oncogenic influence
A significant finding of this study is the notable geographical variation in HPV prevalence and its relationship with BC. Studies conducted in Asia showed the highest prevalence and the strongest associations across all histological subtypes. For ILC, the OR reached 6.76 (95% CI: 2.76–16.57) with minimal heterogeneity, indicating a strong link between HPV infection and lobular carcinogenesis in Asian populations. In contrast, European studies primarily linked HPV to DCIS with an OR of 3.85 (95% CI: 1.46–10.14), suggesting a possible role of HPV in the initial phases of tumorigenesis in this group. These regional differences might reflect genuine biological variations in viral affinity, disparities in prevalent HPV genotypes, host genetic susceptibility, or differences in viral detection methodologies. Further exploration of these factors is necessary to clarify the underlying mechanisms driving these regional trends.
The meta-analysis showed distinct patterns regarding the associations of HPV-16 and HPV-18 with BC, emphasizing possible differences in their oncogenic mechanisms. Notably, HPV-18 exhibited a more consistent and stronger association with BC despite considerable variability among studies. This finding may relate to several biological and methodological considerations.
The observed geographic variations in HPV prevalence, particularly the stronger associations in Asian populations, may be influenced by environmental factors exacerbated by global warming, such as increased exposure to air pollutants or UV radiation, which can impair immune responses and potentially enhance HPV persistence (Shirkani and Shirkani, 2024). These environmental carcinogens could act synergistically with viral infections, warranting further investigation into their combined impact on breast carcinogenesis.
4.3 Roles of HPV-16 and HPV-18 in breast carcinogenesis
For HPV-18, the stronger association may reflect its unique oncogenic properties in breast tissue. The E6 and E7 oncoproteins of HPV-18 show a high affinity for degrading p53 and inactivating pRb, respectively, which could be particularly efficient in mammary epithelial cells. Additionally, differences in viral genome integration patterns between HPV-18 and other high-risk types may influence its carcinogenic potential. A plausible—but unproven—explanation is differential integration and oncoprotein expression (E6/E7). Only a minority of included studies evaluated transcriptional activity. HPV-18 is known to integrate into host DNA more frequently than HPV-16 in cervical cancer, resulting in sustained expression of E6/E7 and genomic instability (Lagstrom et al., 2021; Bodelon et al., 2016; Liu et al., 2016). A similar mechanism might function in breast tissue, where integrated HPV-18 DNA could drive malignant transformation more effectively. Furthermore, tissue-specific variations in viral entry receptors or host immune responses might favor HPV-18 persistence and oncogenesis in the breast microenvironment (Passmore and Williamson, 2016; Yo and Nuryanto, 2024).
The correlation of HPV-16 with BC was inconsistent and frequently non-significant. This discrepancy may arise from several confounding factors. First, detecting HPV-16 DNA in breast tissue could indicate contamination from adjacent skin or mucosal surfaces rather than involvement in viral carcinogenesis. Given the ubiquity of HPV-16 in anogenital and oropharyngeal cancers, we cannot rule out false-positive results due to sample handling or cross-contamination. Second, methodological differences across studies, such as variations in PCR primers, DNA extraction protocols, or the choice of paraffin-embedded versus fresh tissues, might impact HPV-16 detection rates. For instance, highly sensitive nested PCR studies may overestimate prevalence, whereas studies using sequencing methods could miss low viral loads. Third, biological differences in HPV-16 tropism for breast tissue might limit its oncogenic impact compared to HPV-18. If HPV-16 infects breast cells less efficiently or fails to integrate its genome stably, its contribution to malignant transformation would be weaker.
4.4 Clinical and public health implications
Although causality cannot be inferred from detection and case–control designs, HPV-18 appeared more frequently than HPV-16 in breast-cancer samples; however, between-study variability precludes firm conclusions regarding genotype-specific differences. Practically, these data motivate: (i) rigorous, contamination-resistant HPV testing in research biopsies; (ii) careful clinicopathologic correlation; and (iii) HPV vaccination might reduce HPV-related breast lesions if causal links are confirmed; current evidence is insufficient for policy recommendations.\. Until then, over-interpretation for screening or treatment is unwarranted.
4.5 Future research
Priorities include: (1) prospective, multi-region studies using standardized pre-analytic handling and orthogonal assays (DNA, E6/E7 mRNA, integration mapping, IHC/RNAscope) to verify active viral oncogenesis; (2) mechanistic models in mammary epithelium (E6/E7 expression, integration, APOBEC footprints); (3) robust case–control matching for sexual, reproductive, and environmental confounders; and (4) individual-participant-data (IPD) meta-analyses to harmonize histology, genotype, and method covariates.
4.6 Limitations
Study design & confounding: Predominantly observational designs with limited multivariable control. Differences in DNA extraction, assays (nested/consensus PCR, hybrid capture, RT-PCR), cut-offs, and specimen type (FFPE vs. fresh) influence detection. Our meta-regression indicates these factors materially contribute to heterogeneity. Funnel asymmetry for IDC suggests small-study effects; trim-and-fill attenuated—but did not eliminate—the signal. Over-representation of Asian cohorts may limit generalizability. Presence of viral DNA does not establish causation; markers of transcriptional activity (E6/E7 mRNA), integration, and on-pathway protein changes were inconsistently available. Language restriction to English may have excluded non-English studies (e.g., Chinese, Spanish, Persian) and could affect geographic comparisons. Absence of a formal GRADE assessment means findings are hypothesis-generating and not prescriptive for clinical policy. Early-generation assays may inflate detection in some reports.
5 Conclusion
This meta-analysis demonstrates a significant association between human papillomavirus (HPV) infection and breast cancer (BC), with a pooled prevalence of 23% in malignant tissues and a 3.6-fold higher odds of HPV detection compared with non-cancerous controls. These findings support a potential association of HPV in BC and underscore the need to strengthen the causality-focused evidence base through standardized detection protocols and mechanistic studies. Future research should prioritize large multi-regional cohorts and experimental models to determine whether HPV acts as a causal agent or co-factor in BC pathogenesis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
DK: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Formal analysis. SB: Methodology, Project administration, Writing – review & editing, Investigation, Supervision. BZ: Supervision, Validation, Writing – review & editing, Methodology, Visualization. AK: Validation, Visualization, Writing – review & editing, Formal analysis. NM: Validation, Visualization, Writing – review & editing, Methodology. SS: Formal analysis, Supervision, Writing – review & editing, Data curation. MA: Funding acquisition, Resources, Writing – review & editing, Data curation. ATu: Funding acquisition, Software, Writing – review & editing, Data curation, Formal analysis. ATa: Conceptualization, Methodology, Writing – original draft, Writing – review & editing, Formal analysis, Investigation.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by West Kazakhstan Marat Ospanov Medical University as part of the intra-university project “A comprehensive analysis of HPV-positive tumors in the mammary glands, lungs, and bowel: epidemiological, clinical, diagnostic, and genetic aspects.”
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1712118/full#supplementary-material
References
Aguayo, F., Khan, N., Koriyama, C., Gonzalez, C., Ampuero, S., Padilla, O., et al. (2011). Human papillomavirus and Epstein-Barr virus infections in breast cancer from Chile. Infect. Agent Cancer 6:7. doi: 10.1186/1750-9378-6-7
Ahangar-Oskouee, M., Shahmahmoodi, S., Jalilvand, S., Mahmoodi, M., Ziaee, A. A., Esmaeili, H. A., et al. (2014). No detection of 'high-risk' human papillomaviruses in a group of Iranian women with breast cancer. Asian Pac. J. Cancer Prev. 15, 4061–4065. doi: 10.7314/apjcp.2014.15.9.4061
Akil, N., Yasmeen, A., Kassab, A., Ghabreau, L., Darnel, A. D., and Al Moustafa, A. E. (2008). High-risk human papillomavirus infections in breast cancer in Syrian women and their association with Id-1 expression: a tissue microarray study. Br. J. Cancer 99, 404–407. doi: 10.1038/sj.bjc.6604503
Alinezhadi, M., Makvandi, M., Kaydani, G. A., Jazayeri, S. N., Charostad, J., Talaeizadeh, A. T., et al. (2022). Detection of high-risk human papillomavirus DNA in invasive ductal carcinoma specimens. Asian Pac. J. Cancer Prev. 23, 3201–3207. doi: 10.31557/APJCP.2022.23.9.3201
Antonsson, A., Spurr, T. P., Chen, A. C., Francis, G. D., McMillan, N. A., Saunders, N. A., et al. (2011). High prevalence of human papillomaviruses in fresh frozen breast cancer samples. J. Med. Virol. 83, 2157–2163. doi: 10.1002/jmv.22223
Awan, U. A., Khattak, A. A., Ahmed, N., Guo, X., Akhtar, S., Kamran, S., et al. (2023). An updated systemic review and meta-analysis on human papillomavirus in breast carcinogenesis. Front. Oncol. 13:1219161. doi: 10.3389/fonc.2023.1219161
Balci, F. L., Uras, C., and Feldman, S. M. (2019). Is human papillomavirus associated with breast cancer or papilloma presenting with pathologic nipple discharge? Cancer Treat. Res. Commun. 19:100122. doi: 10.1016/j.ctarc.2019.100122
Baltzell, K., Buehring, G. C., Krishnamurthy, S., Kuerer, H., Shen, H. M., and Sison, J. D. (2012). Limited evidence of human papillomavirus in [corrected] breast tissue using molecular in situ methods. Cancer 118, 1212–1220. doi: 10.1002/cncr.26389
Belachew, E. B., Desta, A. F., Mulu, A., Deneke, D. B., Tefera, D. A., Alemu, A., et al. (2024). High rate of high-risk human papillomavirus among benign and breast cancer patients in Ethiopia. PLoS One 19:e0298583. doi: 10.1371/journal.pone.0298583
Blanco, R., Carrillo-Beltran, D., Munoz, J. P., Corvalan, A. H., Calaf, G. M., and Aguayo, F. (2021). Human papillomavirus in breast carcinogenesis: a passenger, a cofactor, or a causal agent? Biology 10:804. doi: 10.3390/biology10080804
Bodaghi, S., Wood, L. V., Roby, G., Ryder, C., Steinberg, S. M., and Zheng, Z. M. (2005). Could human papillomaviruses be spread through blood? J. Clin. Microbiol. 43, 5428–5434. doi: 10.1128/JCM.43.11.5428-5434.2005
Bodelon, C., Untereiner, M. E., Machiela, M. J., Vinokurova, S., and Wentzensen, N. (2016). Genomic characterization of viral integration sites in HPV-related cancers. Int. J. Cancer 139, 2001–2011. doi: 10.1002/ijc.30243
Choi, Y. L., Cho, E. Y., Kim, J. H., Nam, S. J., Oh, Y. L., Song, S. Y., et al. (2007). Detection of human papillomavirus DNA by DNA chip in breast carcinomas of Korean women. Tumour Biol. 28, 327–332. doi: 10.1159/000124238
Damin, A. P., Karam, R., Zettler, C. G., Caleffi, M., and Alexandre, C. O. (2004). Evidence for an association of human papillomavirus and breast carcinomas. Breast Cancer Res. Treat. 84, 131–137. doi: 10.1023/B:BREA.0000018411.89667.0d
de Villiers, E. M., Sandstrom, R. E., zur Hausen, H., and Buck, C. E. (2005). Presence of papillomavirus sequences in condylomatous lesions of the mamillae and in invasive carcinoma of the breast. Breast Cancer Res. 7, R1–R11. doi: 10.1186/bcr940
Doosti, M., Bakhshesh, M., Zahir, S. T., Shayestehpour, M., and Karimi-Zarchi, M. (2016). Lack of evidence for a relationship between high risk human papillomaviruses and breast cancer in Iranian patients. Asian Pac. J. Cancer Prev. 17, 4357–4361.
Duo, D., Ghimenti, C., Migliora, P., Pavanelli, M. C., Mastracci, L., and Angeli, G. (2008). Identification and characterization of human papillomavirus DNA sequences in Italian breast cancer patients by PCR and line probe assay reverse hybridization. Mol. Med. Rep. 1, 673–677. doi: 10.3892/mmr_00000011
Elagali, A. M., Suliman, A. A., Altayeb, M., Dannoun, A. I., Parine, N. R., Sakr, H. I., et al. (2021). Human papillomavirus, gene mutation and estrogen and progesterone receptors in breast cancer: a cross-sectional study. Pan Afr. Med. J. 38:43. doi: 10.11604/pamj.2021.38.43.22013
Fernandes, A., Bianchi, G., Feltri, A. P., Perez, M., and Correnti, M. (2015). Presence of human papillomavirus in breast cancer and its association with prognostic factors. Ecancermedicalscience 9:548. doi: 10.3332/ecancer.2015.548
Frega, A., Lorenzon, L., Bononi, M., De Cesare, A., Ciardi, A., Lombardi, D., et al. (2012). Evaluation of E6 and E7 mRNA expression in HPV DNA positive breast cancer. Eur. J. Gynaecol. Oncol. 33, 164–167. doi: 10.12892/ejgo201202164
Freitas, A. C., Mariz, F. C., Silva, M. A., and Jesus, A. L. (2013). Human papillomavirus vertical transmission: review of current data. Clin. Infect. Dis. 56, 1451–1456. doi: 10.1093/cid/cit066
Fu, L., Wang, D., Shah, W., Wang, Y., Zhang, G., and He, J. (2015). Association of human papillomavirus type 58 with breast cancer in Shaanxi province of China. J. Med. Virol. 87, 1034–1040. doi: 10.1002/jmv.24142
Gannon, O. M., Antonsson, A., Milevskiy, M., Brown, M. A., Saunders, N. A., and Bennett, I. C. (2015). No association between HPV positive breast cancer and expression of human papilloma viral transcripts. Sci. Rep. 5:18081. doi: 10.1038/srep18081
Ghaffari, H., Nafissi, N., Hashemi-Bahremani, M., Alebouyeh, M. R., Tavakoli, A., Javanmard, D., et al. (2018). Molecular prevalence of human papillomavirus infection among Iranian women with breast cancer. Breast Dis. 37, 207–213. doi: 10.3233/BD-180333
Glenn, W. K., Heng, B., Delprado, W., Iacopetta, B., Whitaker, N. J., and Lawson, J. S. (2012). Epstein-Barr virus, human papillomavirus and mouse mammary tumour virus as multiple viruses in breast cancer. PLoS One 7:e48788. doi: 10.1371/journal.pone.0048788
Gomes de Oliveira, G., Gonçalves, A. K., Eleutério, J., and Pinheiro, L. G. P. (2022). Systematic review and meta-analysis of the papillomavirus prevalence in breast cancer fresh tissues. Breast Dis. 41, 123–132. doi: 10.3233/bd-201032
Gupta, I., Jabeen, A., Al-Sarraf, R., Farghaly, H., Vranic, S., Sultan, A. A., et al. (2021). The co-presence of high-risk human papillomaviruses and Epstein-Barr virus is linked with tumor grade and stage in Qatari women with breast cancer. Hum. Vaccin. Immunother. 17, 982–989. doi: 10.1080/21645515.2020.1802977
Habyarimana, T., Attaleb, M., Mazarati, J. B., Bakri, Y., and El Mzibri, M. (2018). Detection of human papillomavirus DNA in tumors from Rwandese breast cancer patients. Breast Cancer 25, 127–133. doi: 10.1007/s12282-018-0831-2
Hareza, D. A., Wilczynski, J. R., and Paradowska, E. (2022). Human papillomaviruses as infectious agents in gynecological cancers. Oncogenic properties of viral proteins. Int. J. Mol. Sci. 23:1818. doi: 10.3390/ijms23031818
Heng, B., Glenn, W. K., Ye, Y., Tran, B., Delprado, W., Lutze-Mann, L., et al. (2009). Human papilloma virus is associated with breast cancer. Br. J. Cancer 101, 1345–1350. doi: 10.1038/sj.bjc.6605282
Hennig, E. M., Suo, Z., Thoresen, S., Holm, R., Kvinnsland, S., and Nesland, J. M. (1999). Human papillomavirus 16 in breast cancer of women treated for high grade cervical intraepithelial neoplasia (CIN III). Breast Cancer Res. Treat. 53, 121–135. doi: 10.1023/a:1006162609420
Herrera-Goepfert, R., Khan, N. A., Koriyama, C., Akiba, S., and Perez-Sanchez, V. M. (2011). High-risk human papillomavirus in mammary gland carcinomas and non-neoplastic tissues of Mexican women: no evidence supporting a cause and effect relationship. Breast 20, 184–189. doi: 10.1016/j.breast.2010.11.006
Herrera-Goepfert, R., Vela-Chavez, T., Carrillo-Garcia, A., Lizano-Soberon, M., Amador-Molina, A., Onate-Ocana, L. F., et al. (2013). High-risk human papillomavirus (HPV) DNA sequences in metaplastic breast carcinomas of Mexican women. BMC Cancer 13:445. doi: 10.1186/1471-2407-13-445
Hong, L., and Tang, S. (2014). Does HPV 16/18 infection affect p53 expression in invasive ductal carcinoma? An experimental study. Pak. J. Med. Sci. 30, 789–792. doi: 10.12669/pjms.304.4534
Islam, S., Dasgupta, H., Roychowdhury, A., Bhattacharya, R., Mukherjee, N., Roy, A., et al. (2017). Study of association and molecular analysis of human papillomavirus in breast cancer of Indian patients: clinical and prognostic implication. PLoS One 12:e0172760. doi: 10.1371/journal.pone.0172760
Khan, N. A., Castillo, A., Koriyama, C., Kijima, Y., Umekita, Y., Ohi, Y., et al. (2008). Human papillomavirus detected in female breast carcinomas in Japan. Br. J. Cancer 99, 408–414. doi: 10.1038/sj.bjc.6604502
Khayargoli, P., Niyibizi, J., Mayrand, M. H., Audibert, F., Monnier, P., Brassard, P., et al. (2023). Human papillomavirus transmission and persistence in pregnant women and neonates. JAMA Pediatr. 177, 684–692. doi: 10.1001/jamapediatrics.2023.1283
Khodabandehlou, N., Mostafaei, S., Etemadi, A., Ghasemi, A., Payandeh, M., Hadifar, S., et al. (2019). Human papilloma virus and breast cancer: the role of inflammation and viral expressed proteins. BMC Cancer 19:61. doi: 10.1186/s12885-019-5286-0
Kroupis, C., Markou, A., Vourlidis, N., Dionyssiou-Asteriou, A., and Lianidou, E. S. (2006). Presence of high-risk human papillomavirus sequences in breast cancer tissues and association with histopathological characteristics. Clin. Biochem. 39, 727–731. doi: 10.1016/j.clinbiochem.2006.03.005
Lagstrom, S., Lovestad, A. H., Umu, S. U., Ambur, O. H., Nygard, M., Rounge, T. B., et al. (2021). HPV16 and HPV18 type-specific APOBEC3 and integration profiles in different diagnostic categories of cervical samples. Tumour Virus Res. 12:200221. doi: 10.1016/j.tvr.2021.200221
Lawson, J. S., Glenn, W. K., Salyakina, D., Clay, R., Delprado, W., Cheerala, B., et al. (2015). Human papilloma virus identification in breast cancer patients with previous cervical neoplasia. Front. Oncol. 5:298. doi: 10.3389/fonc.2015.00298
Li, J., Ding, J., and Zhai, K. (2015). Detection of human papillomavirus DNA in patients with breast tumor in China. PLoS One 10:e0136050. doi: 10.1371/journal.pone.0136050
Liang, W., Wang, J., Wang, C., Lv, Y., Gao, H., Zhang, K., et al. (2013). Detection of high-risk human papillomaviruses in fresh breast cancer samples using the hybrid capture 2 assay. J. Med. Virol. 85, 2087–2092. doi: 10.1002/jmv.23703
Liu, Y., Lu, Z., Xu, R., and Ke, Y. (2016). Comprehensive mapping of the human papillomavirus (HPV) DNA integration sites in cervical carcinomas by HPV capture technology. Oncotarget 7, 5852–5864. doi: 10.18632/oncotarget.6809
Liu, Y., and Yu, T. (2023). Clinicopathological characteristics and prognosis of triple-negative breast cancer invasive ductal carcinoma with ductal carcinoma in situ. J. Cancer Res. Clin. Oncol. 149, 11181–11191. doi: 10.1007/s00432-023-04895-9
Lukasiewicz, S., Czeczelewski, M., Forma, A., Baj, J., Sitarz, R., and Stanislawek, A. (2021). Breast cancer-epidemiology, risk factors, classification, prognostic markers, and current treatment strategies-an updated review. Cancer 13:4287. doi: 10.3390/cancers13174287
Malagon, T., MacCosham, A., Burchell, A. N., El-Zein, M., Tellier, P. P., Coutlee, F., et al. (2021). Sex- and type-specific genital human papillomavirus transmission rates between heterosexual partners: a Bayesian reanalysis of the HITCH cohort. Epidemiology 32, 368–377. doi: 10.1097/EDE.0000000000001324
Maldonado-Rodriguez, E., Hernandez-Barrales, M., Reyes-Lopez, A., Godina-Gonzalez, S., Gallegos-Flores, P. I., Esparza-Ibarra, E. L., et al. (2022). Presence of human papillomavirus DNA in malignant neoplasia and non-malignant breast disease. Curr. Issues Mol. Biol. 44, 3648–3665. doi: 10.3390/cimb44080250
Manzouri, L., Salehi, R., Shariatpanahi, S., and Rezaie, P. (2014). Prevalence of human papilloma virus among women with breast cancer since 2005–2009 in Isfahan. Adv. Biomed. Res. 3:75. doi: 10.4103/2277-9175.125873
Mareti, E., Vavoulidis, E., Papanastasiou, A., Maretis, T., Tsampazis, N., Margioula-Siarkou, C., et al. (2023). Evaluating the potential role of human papilloma virus infection in breast carcinogenesis via real-time polymerase chain reaction analyzes of breast fine needle aspiration samples from Greek patients. Diagn. Cytopathol. 51, 414–422. doi: 10.1002/dc.25130
Mendizabal-Ruiz, A. P., Morales, J. A., Ramirez-Jirano, L. J., Padilla-Rosas, M., Moran-Moguel, M. C., and Montoya-Fuentes, H. (2009). Low frequency of human papillomavirus DNA in breast cancer tissue. Breast Cancer Res. Treat. 114, 189–194. doi: 10.1007/s10549-008-9989-1
Nascimento, K. C. G., Sao Marcos, B. F., Fontes, P. H. B., Isidio, B. E. O., Leao, S. L., da Silva, G. R. P., et al. (2024). HPV detection in breast tumors and associated risk factors in Northeastern Brazil. Cells 13:1132. doi: 10.3390/cells13131132
Naushad, W., Surriya, O., and Sadia, H. (2017). Prevalence of EBV, HPV and MMTV in Pakistani breast cancer patients: a possible etiological role of viruses in breast cancer. Infect. Genet. Evol. 54, 230–237. doi: 10.1016/j.meegid.2017.07.010
Ngamkham, J., Karalak, A., Chaiwerawattana, A., Sornprom, A., Thanasutthichai, S., Sukarayodhin, S., et al. (2017). Prevalence of human papillomavirus infection in breast cancer cells from Thai women. Asian Pac. J. Cancer Prev. 18, 1839–1845. doi: 10.22034/APJCP.2017.18.7.1839
Passmore, J.-A. S., and Williamson, A.-L. (2016). Host immune responses associated with clearance or persistence of human papillomavirus infections. Curr. Obstet. Gynecol. Rep. 5, 177–188. doi: 10.1007/s13669-016-0163-1
Pereira Suarez, A. L., Lorenzetti, M. A., Gonzalez Lucano, R., Cohen, M., Gass, H., Martinez Vazquez, P., et al. (2013). Presence of human papilloma virus in a series of breast carcinoma from Argentina. PLoS One 8:e61613. doi: 10.1371/journal.pone.0061613
Pesut, E., Dukic, A., Lulic, L., Skelin, J., Simic, I., Milutin Gasperov, N., et al. (2021). Human papillomaviruses-associated cancers: an update of current knowledge. Viruses 13:2234. doi: 10.3390/v13112234
Purrahman, D., Avarvand, A. Y., Paradowska-Gorycka, A., Saki, N., Karimpourian, H., Jodat, H., et al. (2022). Association of human papillomavirus with breast cancer: a new perspective on an old debate. Future Oncol. 18, 2483–2494. doi: 10.2217/fon-2021-1158
Salman, N. A., Davies, G., Majidy, F., Shakir, F., Akinrinade, H., Perumal, D., et al. (2017). Association of high risk human papillomavirus and breast cancer: a UK based study. Sci. Rep. 7:43591. doi: 10.1038/srep43591
Sedeta, E. T., Jobre, B., and Avezbakiyev, B. (2023). Breast cancer: global patterns of incidence, mortality, and trends. J. Clin. Oncol. 41:10528. doi: 10.1200/JCO.2023.41.16_suppl.10528
Sher, G., Salman, N. A., Kulinski, M., Fadel, R. A., Gupta, V. K., Anand, A., et al. (2020). Prevalence and type distribution of high-risk human papillomavirus (HPV) in breast cancer: a Qatar based study. Cancer 12:1528. doi: 10.3390/cancers12061528
Shirkani, P., and Shirkani, A. (2024). Impact of global warming on cancer development: a review of environmental carcinogens and human immunogenetics. West Kazakh. Med. J. 66, 210–238. doi: 10.18502/wkmj.v66i3.15772
Siddiqi, H. K., and Ridker, P. M. (2019). Human papillomavirus infection. Circ. Res. 124, 677–678. doi: 10.1161/CIRCRESAHA.119.314719
Sigaroodi, A., Nadji, S. A., Naghshvar, F., Nategh, R., Emami, H., and Velayati, A. A. (2012). Human papillomavirus is associated with breast cancer in the north part of Iran. ScientificWorldJournal 2012:837191. doi: 10.1100/2012/837191
Smolarz, B., Nowak, A. Z., and Romanowicz, H. (2022). Breast cancer-epidemiology, classification, pathogenesis and treatment (review of literature). Cancer 14:2569. doi: 10.3390/cancers14102569
Tawfeik, A. M., Mora, A., Osman, A., Moneer, M. M., El-Sheikh, N., and Elrefaei, M. (2020). Frequency of CD4+ regulatory T cells, CD8+ T cells, and human papilloma virus infection in Egyptian Women with breast cancer. Int. J. Immunopathol. Pharmacol. 34:2058738420966822. doi: 10.1177/2058738420966822
Wang, Y. W., Zhang, K., Zhao, S., Lv, Y., Zhu, J., Liu, H., et al. (2017). HPV status and its correlation with BCL2, p21, p53, Rb, and survivin expression in breast cancer in a Chinese population. Biomed. Res. Int. 2017, 1–7. doi: 10.1155/2017/6315392
Widschwendter, A., Brunhuber, T., Wiedemair, A., Mueller-Holzner, E., and Marth, C. (2004). Detection of human papillomavirus DNA in breast cancer of patients with cervical cancer history. J. Clin. Virol. 31, 292–297. doi: 10.1016/j.jcv.2004.06.009
Wierzbicka, M., San Giorgi, M. R. M., and Dikkers, F. G. (2023). Transmission and clearance of human papillomavirus infection in the oral cavity and its role in oropharyngeal carcinoma—a review. Rev. Med. Virol. 33:e2337. doi: 10.1002/rmv.2337
Yo, E. C., and Nuryanto, K. H. (2024). Molecular and host lifestyle factors associated with persistent human papillomavirus infection and progression into cervical cancer: a literature review. Indones. J. Cancer 18, 226–232. doi: 10.33371/ijoc.v18i2.1068
Keywords: human papillomaviruses, breast neoplasms, meta-analysis, case–control studies, systematic review
Citation: Koishybayeva D, Balmagambetova S, Zhakiev B, Koishybayev A, Mussin NM, Sakhanova S, Aitmagambetova M, Tulyayeva A and Tamadon A (2025) The oncogenic role of human papillomavirus in breast cancer: a comprehensive systematic review and meta-analysis. Front. Microbiol. 16:1712118. doi: 10.3389/fmicb.2025.1712118
Edited by:
Mohammed Rohaim, Cairo University, EgyptReviewed by:
Abolfazl Jafari Sales, Islamic Azad University, Kazeroon, IranMeenu Jain, Gajara Raja Medical College, India
Copyright © 2025 Koishybayeva, Balmagambetova, Zhakiev, Koishybayev, Mussin, Sakhanova, Aitmagambetova, Tulyayeva and Tamadon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saule Balmagambetova, c2F1bGUuYmFsbWFnYW1iZXRvdmFAemttdS5reg==; Amin Tamadon; YW1pbnRhbWFkZG9uQHlhaG9vLmNvbQ==
†ORCID: Dana Koishybayeva, orcid.org/0009-0007-7942-0823
Saule Balmagambetova, orcid.org/0000-0003-4080-5383
Bazylbek Zhakiev, orcid.org/0000-0001-9128-6478
Arip Koishybayev, orcid.org/0000-0002-6164-8009
Nadiar M. Mussin, orcid.org/0000-0003-3600-8840
Svetlana Sakhanova, orcid.org/0000-0001-9786-6326
Marzhan Aitmagambetova, orcid.org/0000-0002-0346-5829
Anar B. Tulayeva, orcid.org/0000-0001-7149-0121
Amin Tamadon, orcid.org/0000-0002-0222-3035
 Dana Koishybayeva
Dana Koishybayeva Saule Balmagambetova2*†
Saule Balmagambetova2*† Nadiar M. Mussin
Nadiar M. Mussin Svetlana Sakhanova
Svetlana Sakhanova Anar Tulyayeva
Anar Tulyayeva Amin Tamadon
Amin Tamadon