- 1Department of Biochemistry, All India Institute of Medical Sciences, New Delhi, India
- 2Laboratory Oncology Unit, Dr. BRA-IRCH, All India Institute of Medical Sciences, New Delhi, India
Classical Non-homologous End Joining (NHEJ) pathway is the mainstay of cellular response to DNA double strand breaks. While aberrant expression of genes involved in this pathway has been linked with genomic instability and drug resistance in several cancers, limited information is available about its clinical significance in colon cancer. We performed a comprehensive analysis of seven essential genes, including XRCC5, XRCC6, PRKDC, LIG4, XRCC4, NHEJ1, and PAXX of this pathway, in colon cancer using multi-omics datasets, and studied their associations with molecular and clinicopathological features, including age, gender, stage, KRAS mutation, BRAF mutation, microsatellite instability status and promoter DNA methylation in TCGA colon cancer dataset. This analysis revealed upregulation of XRCC5, PRKDC, and PAXX in colon cancer compared to normal colon tissues, while LIG4 and NHEJ1 (XLF) displayed downregulation. The expression of these genes was independent of age and KRAS status, while XRCC5, PRKDC, and LIG4 exhibited reduced expression in BRAF mutant tumors. Interestingly, we observed a strong association between XRCC6, XRCC5, PRKDC and LIG4 overexpression and microsatellite instability status of the tumors. In multivariate analysis, high PAXX expression emerged as an independent prognostic marker for poor overall and disease specific survival. We also observed hypomethylation of PAXX promoter in tumors, which exhibited a strong correlation with its overexpression. Furthermore, PAXX overexpression was also associated with several oncogenic pathways as well as a reduction in numbers of tumor-infiltrating lymphocytes.
Introduction
Colorectal cancer (CRC) is the fourth most commonly diagnosed cancer and the third most common cause of cancer related deaths worldwide (Bray et al., 2018; Rawla et al., 2019). It represents a group of heterogeneous diseases that are characterized by a range of genomic and epigenomic alterations (The Cancer Genome Atlas Network, 2012). The knowledge regarding the molecular landscapes of CRCs is rapidly increasing, which has led to advancements in early detection methodologies and hence reduction of mortality rates (Arnold et al., 2017).
DNA double strand breaks (DSBs) are inherently induced during several physiological conditions, including stem cell differentiation, cell division, autophagy, and senescence. Homologous recombination repair (HRR), classical non-homologous end joining (c-NHEJ or NHEJ) and alternative end joining (Alt-EJ) are the three DNA damage repair systems, which efficiently repair DSBs, and thus maintain genomic stability during physiological stress (Chang et al., 2017). HRR functions by using a homologous DNA strand as a template to perform error free repair at DSB sites. Contrary to this, NHEJ is the primary DNA damage repair pathway and perform template independent repair of deleterious DSBs (Chang et al., 2017). Alt-EJ is a less characterized mechanism which works as a backup for both HRR and NHEJ in case of excessive DNA damage, and also utilizes micro-homologies between distant DNA sites for template dependent repair.
The core c-NHEJ system consists of Ku70/80 heterodimer (encoded by XRCC6 and XRCC5, respectively), XRCC4, DNA-dependent protein kinase catalytic subunit (DNA-PKcs, encoded by PRKDC), DNA Ligase 4 (encoded by LIG4), and XRCC4-like factor (XLF, encoded by NHEJ1). DNA damage sites are quickly recognized by Ku70/Ku80 heterodimer or Ku70 homodimer, which after binding to DNA free ends undergo allosteric change, thereby providing a scaffold for recruitment of DNA-PKcs. The protein kinase activity of Ku/DNA-PKcs complex recruits accessory factors to modify free DNA ends, which cannot be ligated directly (Davis et al., 2014). Then, XRCC4 and XLF also assemble at DSB sites, aligns the chromatin in the vicinity, and mediates recruitment of Ligase IV which carry out the final (ligation) step of the repair. These core components are sufficient to recognize DSBs, align the broken DNA fragments, and anneal them, fixing the DSBs (Chang et al., 2017).
PAXX (Paralog of XRCC4 and XLF; previously called C9orf142) is a recently characterized protein associated with the classical NHEJ pathway. It structurally resembles XRCC4 and XLF and facilitates the assembly of the core NHEJ complex at the DNA damage site (Ochi et al., 2015; Kumar et al., 2016). Although, PAXX and XLF perform overlapping functions and XLF can efficiently compensate for PAXX deficiency in colon cancer cells (Tadi et al., 2016), however, another study demonstrated that one protein between PAXX and XLF is essential for NHEJ repair, and PAXX also promotes Ku accumulation at DSBs (Liu et al., 2017). Interestingly, a recent study reported the synergistic role of PAXX, XRCC4, and XLF in the recruitment of DNA Pol λ as an accessory factor for DNA damage repair (Craxton et al., 2018). Although, these studies suggest that both PAXX and XLF perform overlapping but essential functions in NHEJ mediated DNA repair and influence drug resistance in solid tumors, the consequences and clinical implications of their altered expression in cancer patients have never been investigated. While XLF confers resistance to oxaliplatin and 5-fluorouracil in CRC cells (Liu et al., 2019), PAXX overexpression is associated with drug resistance in osteosarcoma cells (Ma et al., 2020).
Non-homologous end joining pathway genes in this CRC harbor both genetic and epigenetic alterations which promote cancer progression (Beggs et al., 2012; Mijnes et al., 2018). Variations at the 3′UTR of mRNA encoding DSB repair proteins have also been associated with a higher risk of CRC and poor outcome of the disease (Naccarati et al., 2015). Conventional cancer therapies including radiation and chemotherapy primarily exert their effect by inducing DSBs mediated cancer cell death. Therefore, the NHEJ pathway genes are considered as potential therapeutic targets to overcome drug resistance in CRC. Previous reports have analyzed the expression of NHEJ genes in different cancers, including some in colorectal cancer (Sishc and Davis, 2017). In the present study, we performed a comprehensive analysis of the core NHEJ pathway genes using well characterized multi-omics datasets to determine the deregulated expression pattern and clinical significance of NHEJ pathway genes in colon cancer.
Materials and Methods
Data Acquisition and Analysis
Oncomine1 a web online database was used to analyze the expression of mRNA encoded by NHEJ genes, in several colon cancer datasets. The parameters for comparing gene expression between normal and tumor tissues included mRNA data with a threshold of p < 0.01 with any fold change.
Gene expression and DNA methylation of colon cancer developed by The Cancer Genome Atlas (TCGA-COAD study) was extracted as fragment per kilobase million (FPKM) values from the UCSC Xena browser,2 and used for subsequent analysis. Similarly, information about clinical features and tumor mutation status of colon cancer patients of TCGA study was retrieved from cBioportal3 by selecting the TCGA PanCancer Atlas - Colorectal Adenocarcinoma study and selecting patients with colon adenocarcinoma in cancer type (Cerami et al., 2012; Gao et al., 2013).
High throughput total protein and phosphoprotein estimation data for 100 normal colon and 97 colon cancer tissues, generated by mass spectrometry (MS) in Clinical Proteomic Tumor Analysis Consortium (CPTAC) study was analyzed using UALCAN web server4 (Chen et al., 2019). The z-value, used to compare protein levels (depicted on the y-axis) represents the standard deviation from the median across samples. As described in UALCAN web server, log2 spectral count ratio values, downloaded from CPTAC colon cancer data were normalized within each sample profile and then normalized across samples to calculate z-values as relative protein levels. Available total protein and phosphoprotein levels of the NHEJ pathway were assessed using default parameters in the UALCAN web server.
MEXPRESS web server5 hosts the DNA methylation data from TCGA studies developed on “Illumina Human Methylation 450 Bead Chip” platform and provides access to methylation levels of designated CpG sites of the queried gene and its association with gene expression (Koch et al., 2015). For DNA methylation analysis, correlation of PAXX expression with the methylation status of its gene was determined using the MEXPRESS web server using default parameters.
Survival Analysis
Kaplan Meier plot was constructed along with log-rank test p-values using the “survminer” package in R statistical software (version 4.0.1). Briefly, patients were categorized into high and low expression groups based on median gene expression values in FPKM (extracted from UCSC Xena browser). Univariate analysis was performed for overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI) to establish the association of gene expression and clinicopathological parameters with patient outcome. Multivariate analysis was also performed for genes, which were significantly associated with prognosis in univariate analysis. Important clinical and molecular features, including age, gender, stage, histological subtype, KRAS status, and BRAF status were taken as covariates.
Pathway Analysis
Gene expression correlations of PAXX with whole gene expression profiles of colon cancer tissues from TCGA-COAD dataset were extracted from the cBioPortal web server (see text footnote 3). Briefly, PAXX expression was used as input in colon adenocarcinoma patient data from TCGA colon cancer (TCGA-COAD PanCancer study) dataset (Cerami et al., 2012; Gao et al., 2013) in cBioPortal. Then, by using the correlation module, the whole transcriptome correlations table of PAXX expression was retrieved. After filtering correlations with false detection rate normalized q-value < 0.05, genes were arranged by increasing value of Spearman’s correlation constant, thus creating a ranked gene file. The ranked gene file was further used as input for the pre-ranked GSEA module in the gene set enrichment analysis tool from Broad Institute6 with predefined molecular signature database hallmark gene set (version 7.1) as reference gene set for pathway enrichment (Liberzon et al., 2015). Genes enriched in the respective pathways were represented as direct image outputs along with calculated normalized enrichment score (NES), false discovery rate (FDR), and p-value.
Protein Interaction Analysis
Biophysical interactions of ORFeome-based complexes (BioPlex) network interactome tool,7 a large-scale interactome database based on affinity purification mass spectrometry (AP-MS) data of baits from the human ORFeome (Huttlin et al., 2017) was utilized to identify PAXX interacting proteins in colon cancer cells HCT-116. Then an interaction network of PAXX associated proteins in these cells was constructed using default parameters.
TISIDB Analysis
The tumor-immune interactions database (TISIDB)8 is an integral web portal for the interaction of tumor and immune system (Ru et al., 2019). This database enabled us to correlate PAXX gene expression and infiltration of different immune cells types including CD8 T cells (activated, central memory and effector memory), CD4 T cells (activated, central memory and effector memory), T helper cells (follicular, type 1 and 2), gamma delta T cells, B-cells (activated, immature and mature), dendritic cells (activated, plasmacytoid and immature), NK cells, macrophages, eosinophil, mast cell, neutrophils, and monocytes. Immune cell fractions were determined using the computational “deconvolution” approach, which is based on determining mRNA contribution from immune cells from the bulk tumor RNA-sequencing profile.
Statistical Analysis
Gene expression analyses were performed on Graphpad Prism (version 6). Mann-Whitney U-test was used for comparing gene expression between normal and colon cancer tissues. P-value < 0.05 was considered statistically significant. Wilcoxon paired t-test was applied for paired expression analysis between normal and colon cancer tissues. Level of significance denoted on the expression graphs were represented as ∗p-value < 0.05, ∗∗p-value < 0.01, ∗∗∗p-value < 0.001 and ****p-value < 0.0001. Patients were divided into two groups by median expression and a log-rank test was used to compare groups for Kaplan-Meier survival analysis. Univariate and multivariate survival data analysis were performed on Stata version 11.
Results
mRNA Expression Pattern of NHEJ Pathway Genes in Colon Cancer
To determine the expression pattern of core NHEJ genes in colon cancer, we performed Oncomine analysis for XRCC6 (Ku70), XRCC5 (Ku80), PRKDC (DNA-PKcs), XRCC4 (XRCC4), LIG4 (DNA ligase 4), NHEJ1 (XLF), and PAXX (PAXX/XLS). It provided the advantage of analyzing several datasets in parallel to assess the general expression pattern of these genes. This analysis revealed significant upregulation of five genes, (XRCC6, XRCC4, PRKDC, XRCC4, and PAXX) and downregulation of two (LIG4 and NHEJ1) NHEJ pathway genes, in tumor tissues compared to the normal tissues (Table 1).
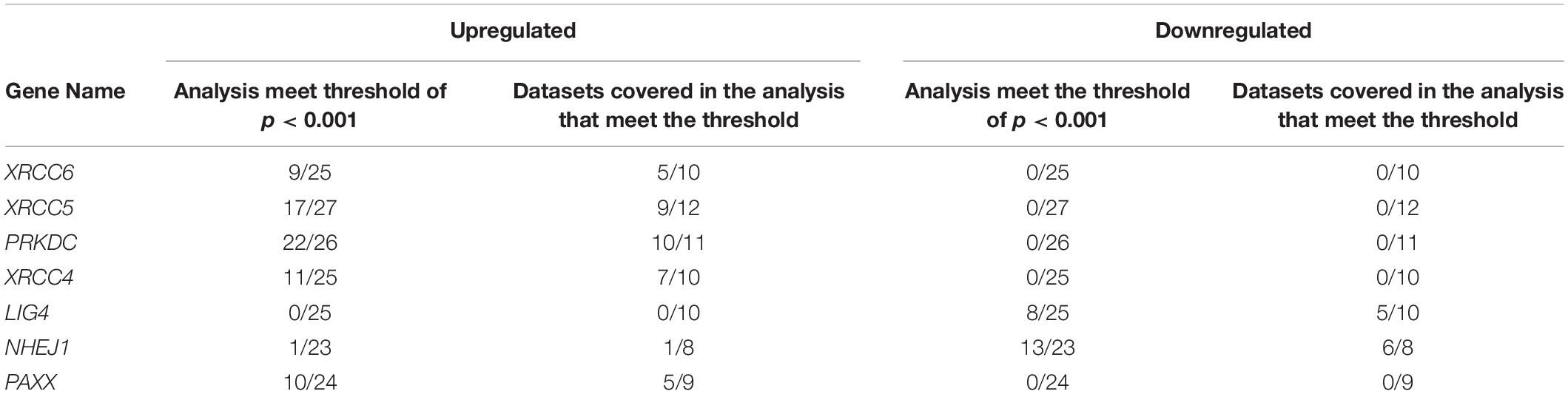
To corroborate our findings, we utilized a dataset of colon cancer from The Cancer Genome Atlas (TCGA) to compare the expression of NHEJ pathway genes between tumors and normal colon tissues. Consistent with the Oncomine analysis, comparison of all available normal (n = 41) and tumor tissues (n = 469) revealed overexpression of XRCC6, XRCC5, PRKDC, XRCC4, and PAXX in tumors compared to normal tissues, while LIG4 and NHEJ1 displayed lower expression in the tumor tissues (Figure 1A). However, analysis of 41 paired normal and tumor tissues revealed significant overexpression of only XRCC5, PRKDC, and PAXX genes in tumor tissues compared to the normal colon (Figures 1C,D,H, respectively), while LIG4 and NHEJ1 still displayed reduced expression (Figures 1F,G, respectively). Interestingly, in contrast to Oncomine analysis, XRCC6 and XRCC4 did not display differential expression between paired normal and tumor tissues (Figures 1B,E, respectively).
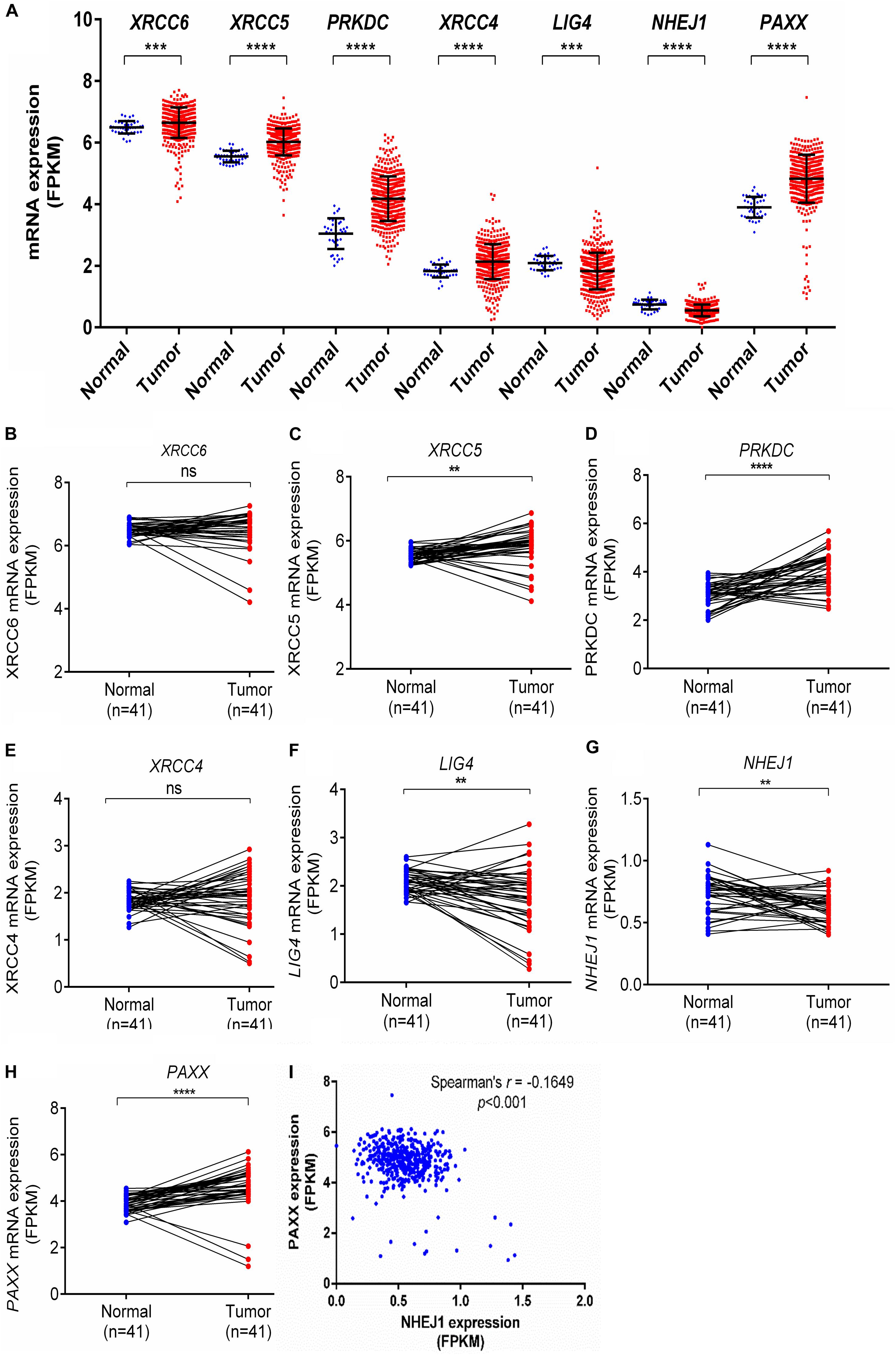
Figure 1. Comparison of mRNA expression of NHEJ pathway genes in normal colon and colon cancer tissues from TCGA-COAD dataset including total samples for seven NHEJ genes (A), and paired samples, including XRCC6 (B), XRCC5 (C), PRKDC (D), XRCC4 (E), LIG4 (F), NHEJ1 (G), and PAXX (H). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (I) Correlation between NHEJ1 and PAXX expression in the same dataset. FPKM, Fragments per kilo million bases.
Co-expression analysis among all NHEJ pathway genes in TCGA-COAD dataset revealed a negative correlation between PAXX and NHEJ1 expression (Figure 1I). While the expression of all other genes of this pathway exhibited positive correlations among them (Supplementary Table S1).
Expression of Proteins Encoded by NHEJ Pathway Genes in Colon Cancer
Further, the Clinical Proteomic Tumor Analysis Consortium (CPTAC) dataset, which consists of high throughput mass spectrometry based quantitative protein estimation data of colon cancer and respective normal colon tissues, was used to compare total and phosphorylated protein levels of NHEJ pathway in normal colon tissues (n = 100) and colon cancer (n = 97). Consistent with the Oncomine gene expression analysis, total protein levels of Ku70 (XRCC6), Ku80 (XRCC5), DNA-PKcs (PRKDC), XRCC4, and PAXX were found to be significantly higher in colon cancer tissue compared to normal colon tissues, while LIG4, which exhibited reduced mRNA expression in Oncomine analysis, also displayed higher total protein levels in tumors (p < 0.01 for all, Figures 2A–G). However, NHEJ1 protein levels in line with the Oncomine analysis were observed to be lower in tumor tissues compared to the controls (p < 0.001, Figure 2F).
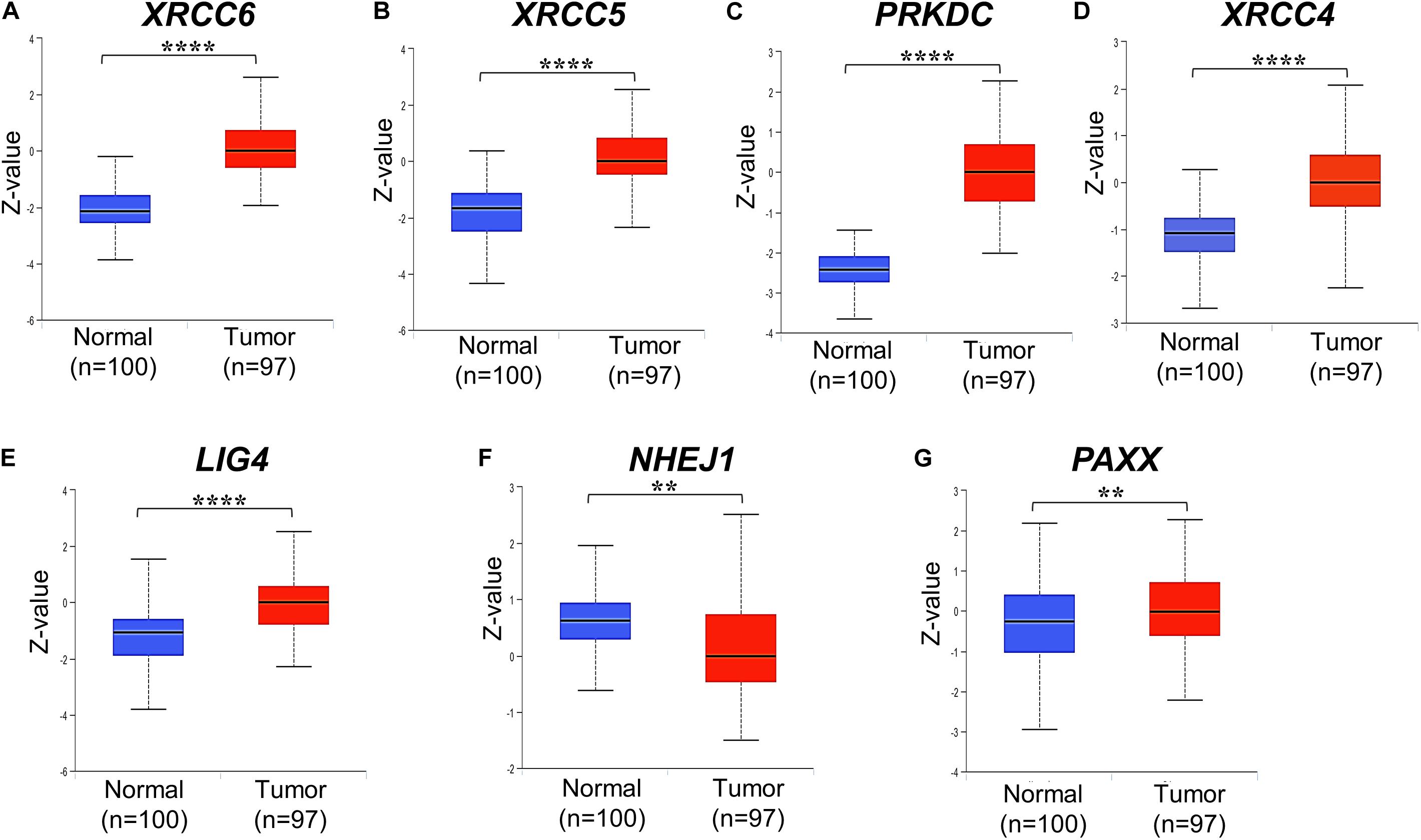
Figure 2. Total protein levels of NHEJ pathway genes in normal tissues and colon cancer tissues from CPTAC study, including (A) XRCC6 (B) XRCC5 (C) PRKDC (D) XRCC4 (E) LIG4 (F) NHEJ1 and (G) PAXX. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
DNA-PKcs has been shown to phosphorylate many of the core NHEJ factors in vitro, but most of these phosphorylations are non-essential for NHEJ function (Davis et al., 2014). We observed that some uncharacterized phosphorylated protein levels of Ku70 (XRCC6, position Ser520, and Thr455), DNA-PKcs (PRKDC, Ser893, Ser3995 and Ser3205), and PAXX (Ser148) were higher in colon cancer tissues compared to normal colon tissues (Supplementary Figures S1A–K), whereas phosphorylated XLF (NHEJ1, Ser287) was lesser in colon cancer tissues.
Interestingly, it has been previously demonstrated that PRKDC is phosphorylated at Ser3995 in response to IR radiation, by ATM serine/threonine kinase (ATM) protein, but this phosphorylation does not affect NHEJ repair (Neal et al., 2011). Further, Douglas et al. (2014) reported that DNA-PKcs is phosphorylated and dephosphorylated at Ser3205 by PLK1 (polo-like kinase 1) and PP6 (protein phosphatase 6), respectively during mitosis. Phospho-mimicry of PAXX phosphorylation at Ser134, Thr145, Ser148, and Ser152 has been reported to destabilize the PAXX-Ku-DNA ternary complex, but it does not affect the stimulation of LIG4/XRCC4 blunt-ended DNA-ligation activity by PAXX (Tadi et al., 2016). Therefore, the exact role of the modifications of DNA-PKcs and PAXX in NHEJ activity remains unclear and warrants further studies.
Associations of NHEJ Pathway Gene Expression With Clinicopathological Features in Colon Cancer
We analyzed associations of NHEJ pathway gene expression with other clinicopathological features, such as age, gender, histological type, stage, KRAS mutation status, BRAF mutation status, and microsatellite instability (MSI) status in colon cancer by performing a direct comparison between mRNA expressions of respective genes. None of the seven NHEJ pathway genes analyzed in the present study displayed any association with age (Supplementary Figures S2A–G). Only LIG4 was associated with gender and exhibited higher expression in males compared to females (Supplementary Figure S3E). Between two histological subtypes, no difference was observed in the expression of XRCC6, PRKDC, LIG4, and PAXX, while expression of XRCC5, XRCC4, and NHEJ1 was higher in adenocarcinoma compared to mucinous adenocarcinoma (Figure 3A). We further compared the mRNA expression of NHEJ genes between stage (I + II) group with stage (III + IV) group colon tumors. This analysis revealed reduced expression of XRCC6 in advanced stage group while LIG4 displayed elevated expression in the same group (Figure 3B). However, no difference in mRNA levels of XRCC5, PRKDC, XRCC4, NHEJ1, and PAXX between the two groups.
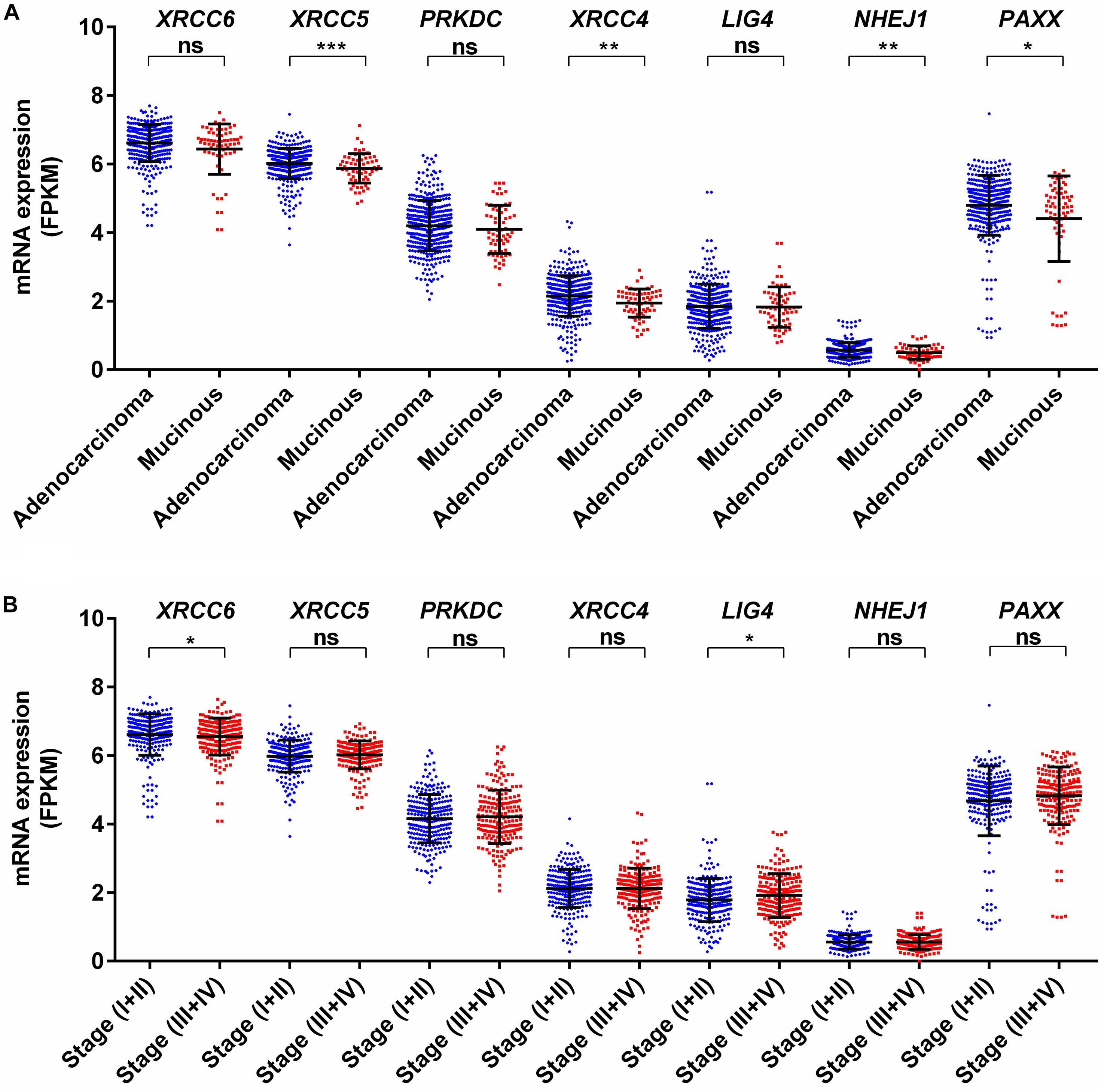
Figure 3. Association of mRNA expression of NHEJ pathway genes with (A) tumor histology, and (B) stage. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. FPKM, Fragments per kilo million bases.
KRAS mutations have been reported to enhance homologous recombination repair in preference to NHEJ in colorectal cancer cells (Kalimutho et al., 2017). In agreement with this report, we observed no difference for mRNA expression in all analyzed genes between KRAS wild type and mutant tumors (Figure 4A). Therefore, KRAS mediated oncogenic reprogramming does not seem to be involved in the altered NHEJ pathway in colon cancer. Concerning BRAF mutation in thyroid cancer, two reports have demonstrated that BRAF mutation promotes NHEJ activity through upregulation of NHEJ1 and it is also associated with radioresistance (Robb et al., 2018, 2019). In a melanoma cell line model, it has been shown that mutant BRAF inhibition may increase DNA damage by downregulation of NHEJ pathway genes, including XRCC6, XRCC5, and PRKDC (Fatkhutdinov et al., 2016). Our analysis revealed that BRAF mutant colon cancer did not harbor higher NHEJ1 expression compared to BRAF wild type tumors and three NHEJ pathway genes, XRCC5, PRKDC, and LIG4 are indeed lowly expressed in BRAF mutant tumors (Figure 4B). These results suggest that a detailed study of the NHEJ pathway concerning BRAF mutation in colon cancer is further warranted.
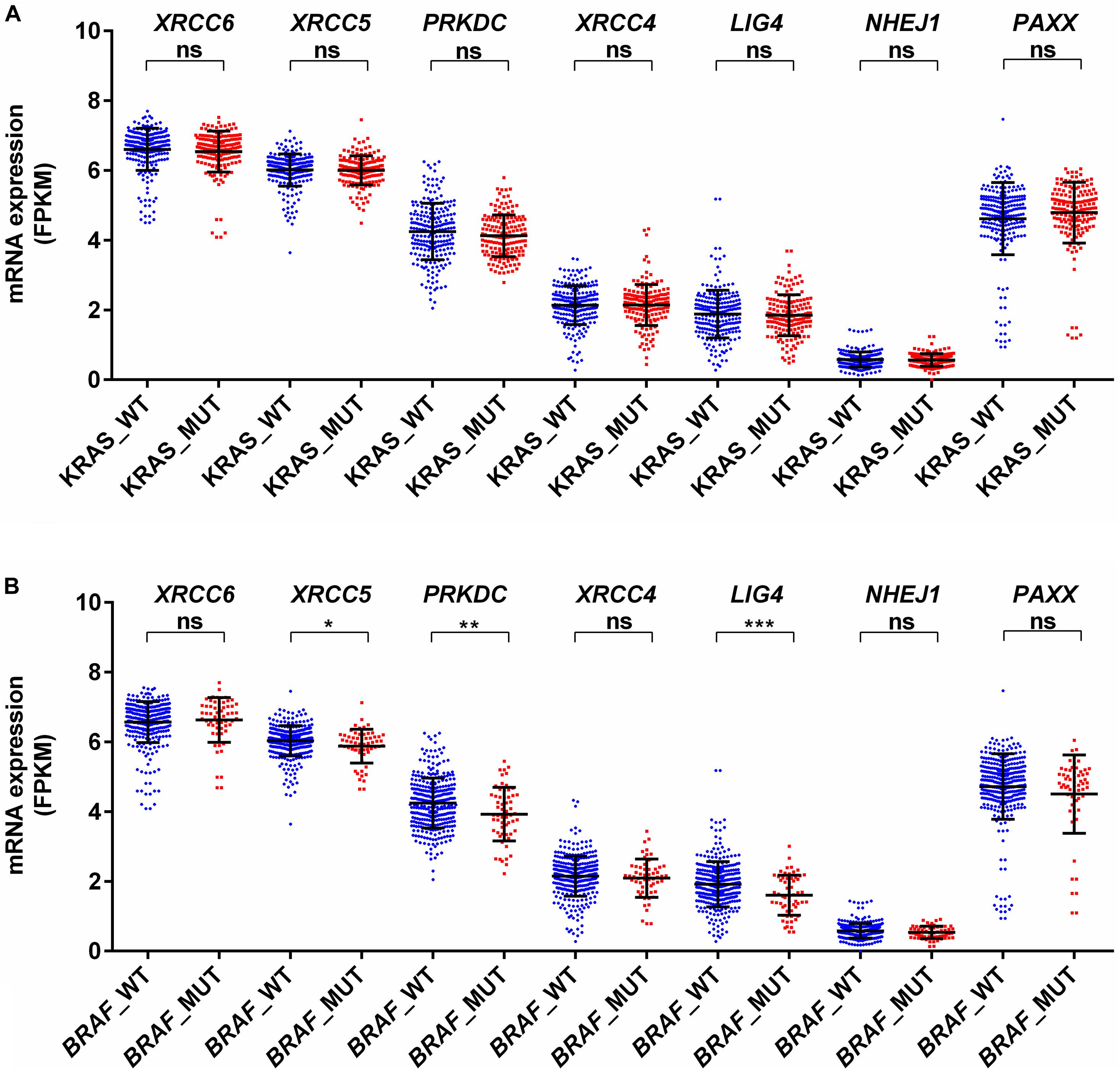
Figure 4. Association of mRNA expression of NHEJ pathway genes with (A) KRAS mutation status, and (B) BRAF mutation status. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. FPKM, Fragments per kilo million bases. WT, wild type; MUT, mutant.
Interestingly, a previous report suggests that the NHEJ pathway is impaired in several mismatch repair deficient colon cancer cell lines (Koh et al., 2005). We observed that expression of XRCC6 was higher in MSI-high tumors compared to MSI-low and microsatellite stable (MSS) tumors, while XRCC5, PRKDC, and LIG4 exhibited reduced expression in MSI-high tumors compared to both MSI-low and MSS tumors (Figures 5A–G).
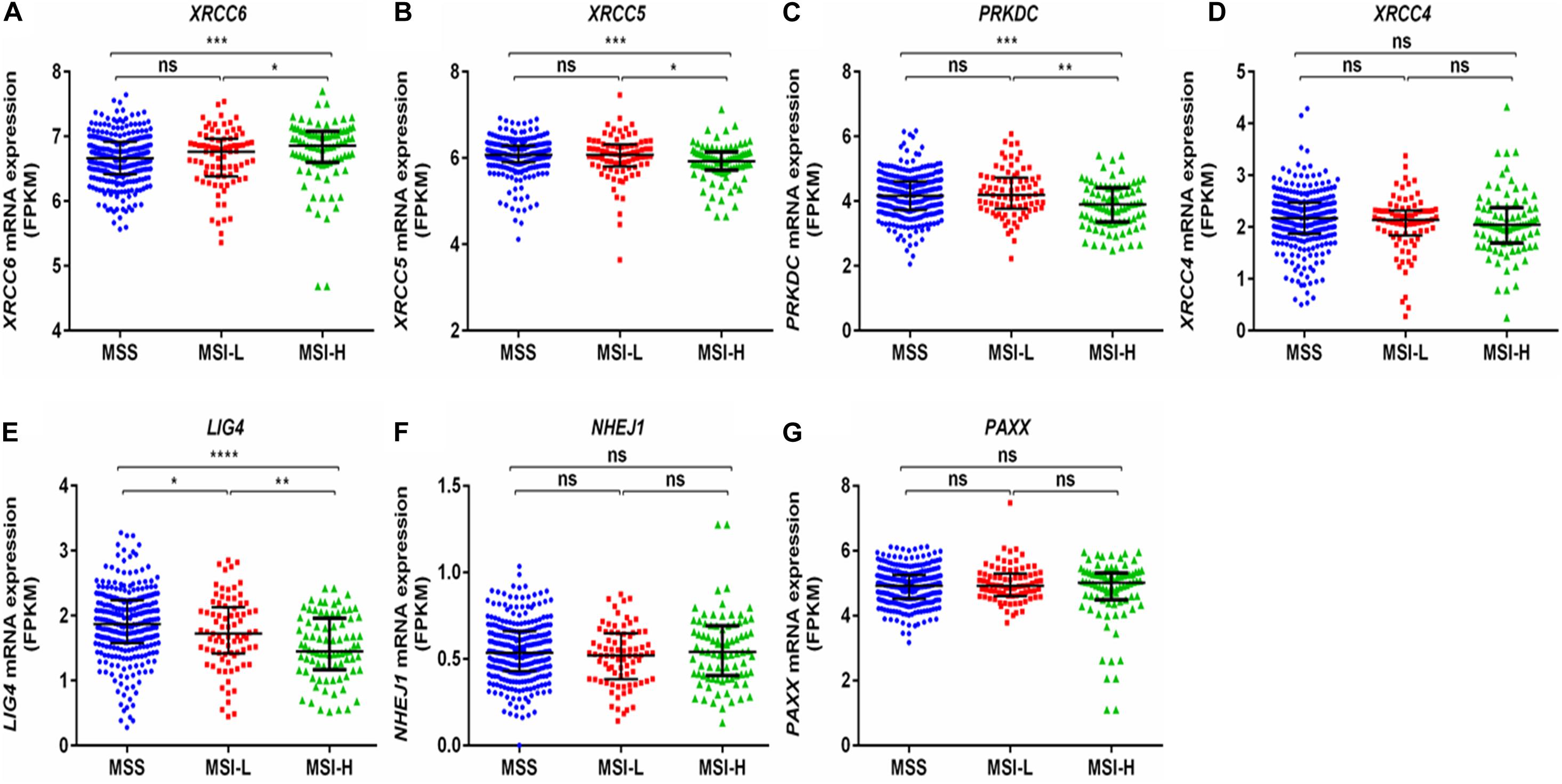
Figure 5. Comparison of mRNA expression of NHEJ pathway genes based on microsatellite instability status from TCGA-COAD dataset. (A) XRCC6 (B) XRCC5 (C) PRKDC (D) XRCC4 (E) LIG4 (F) NHEJ1, and (G) PAXX. FPKM, Fragments per kilo million bases. MSI-H, MSI-high; MSI-L, MSI-low; MSS, microsatellite stable. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Survival Analysis
To further determine the clinical significance of the expression of NHEJ pathway genes in colon cancer, we performed Kaplan-Meier survival analysis for overall survival (OS), disease-specific survival (DSS), progression-free interval (PFI), and disease-free interval (DFI) using TCGA colon cancer dataset. We observed that among all NHEJ pathway genes only elevated PAXX expression was associated with poor overall survival (p = 0.0011, Figure 6A), while other genes did not display significant association with OS (Supplementary Figure S4), DSS (Supplementary Figure S5), or PFI (Supplementary Figure S6). Interestingly, PAXX overexpression was also associated with poor DSS (p = 0.0011, Figure 6B), but not with PFI or DFI (Figures 6C,D, respectively). Furthermore, higher XRCC4 expression was associated with poor DFI (Supplementary Figure S7D).
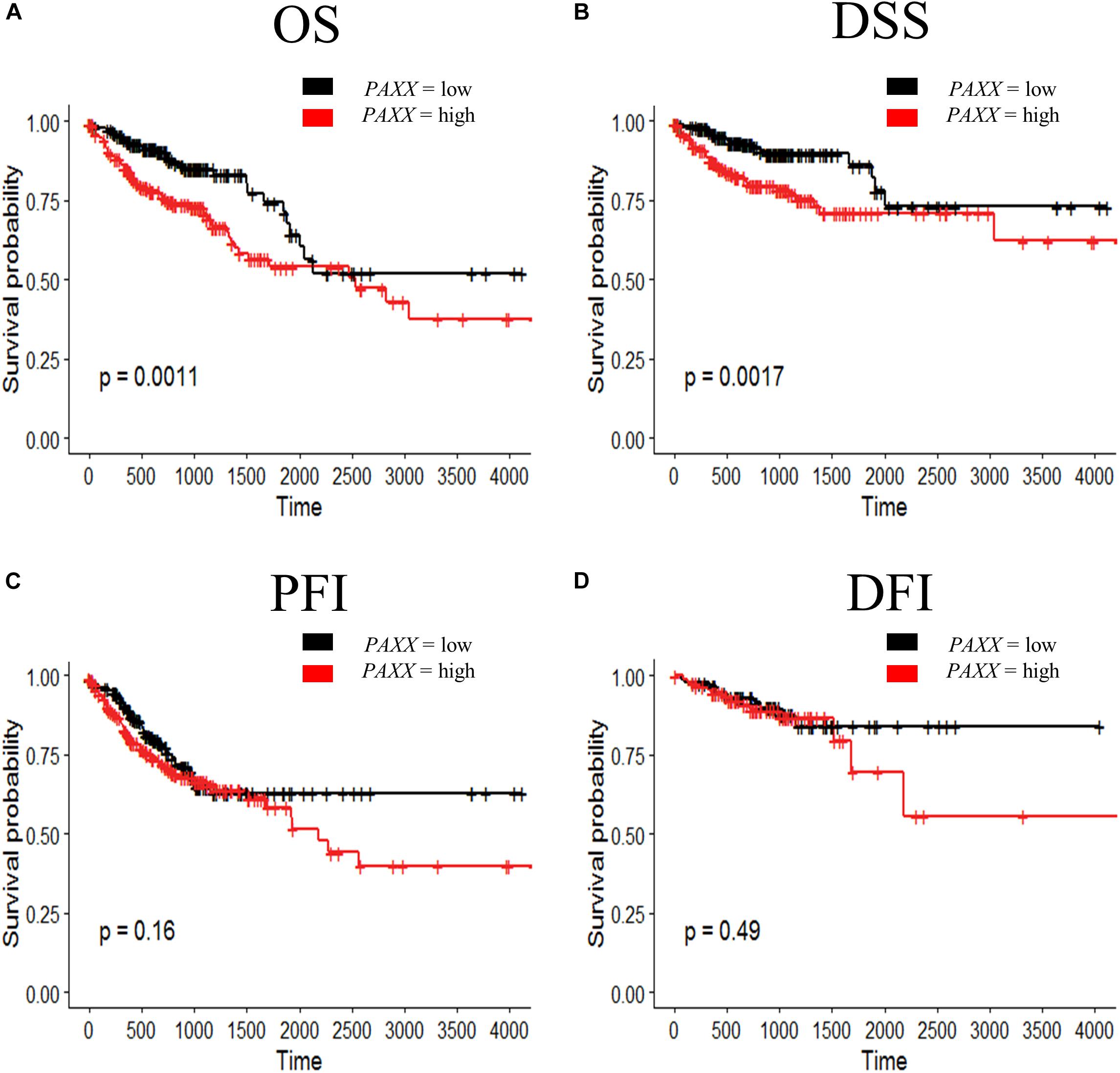
Figure 6. Kaplan-Meier survival curve for prognostic significance of PAXX gene expression in TCGA-COAD dataset, including (A) overall survival (OS), (B) disease-specific survival (DSS), (C) progression-free interval (PFI), and (D) disease-free interval (DFI). Survival probabilities are presented on the y-axis and time in days on the x-axis in all graphs. The log-rank test p-value has been depicted in respective graphs.
To assess the robustness of these gene products as prognostic biomarkers, we performed univariate analysis followed by a multivariate analysis using a Cox proportional hazards model. Gene expression was taken as a continuous variable while important clinical features including age, gender, stage, KRAS mutation, BRAF mutation, and MSI status were taken as covariates. The results of univariate analysis have been presented in Table 2. Interestingly, we observed that only PAXX overexpression was associated with poor OS and DSS, while the levels of other gene products were not associated with OS, DSS, PFI, or DFI. Therefore, the expression of PAXX was considered for multivariate analysis. Interestingly, in multivariate analysis, PAXX overexpression emerged as an independent marker for poor OS and DSS (Table 3).
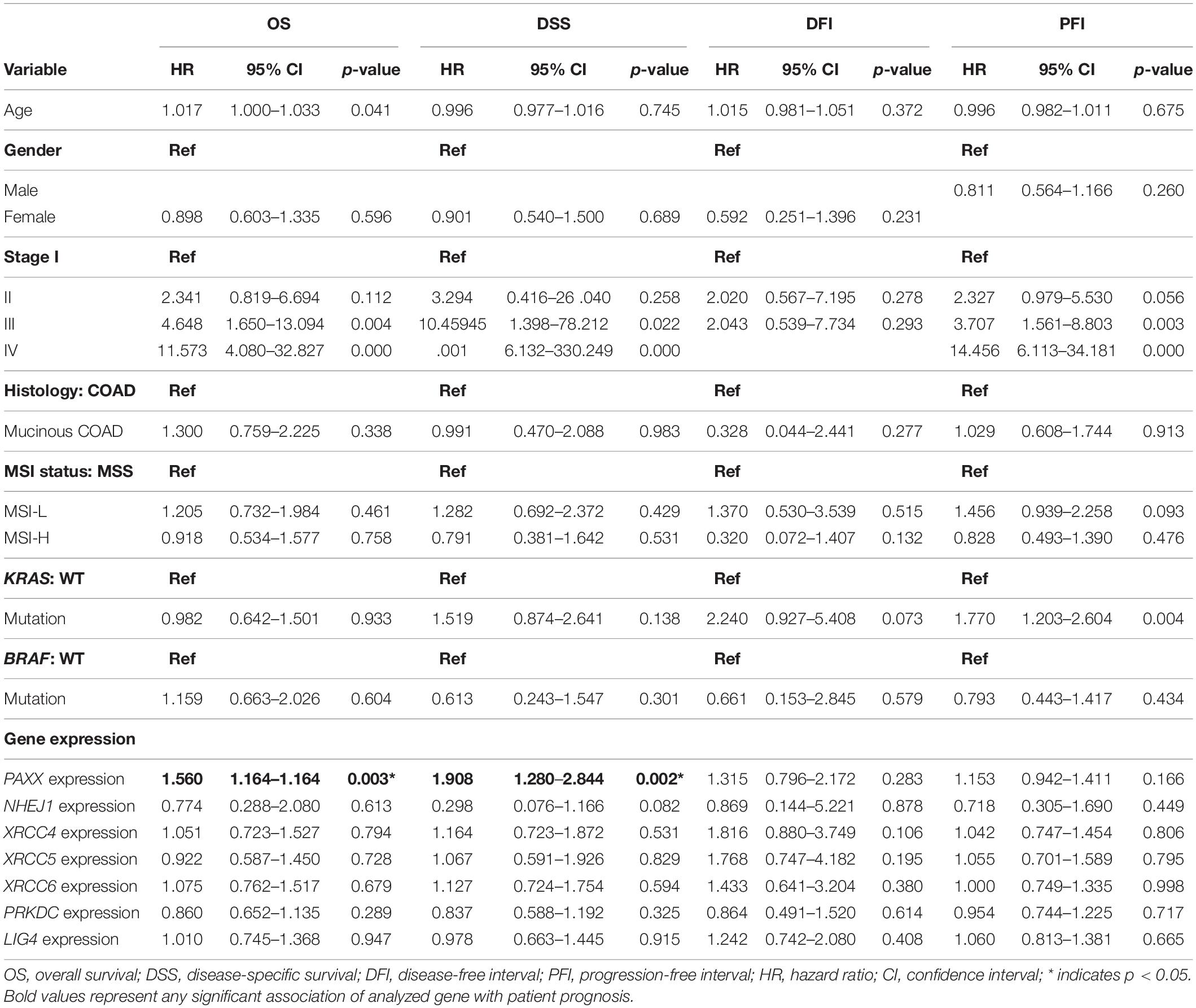
Table 2. Univariate analysis of the NHEJ pathway genes and clinicopathological parameters in TCGA-COAD dataset.
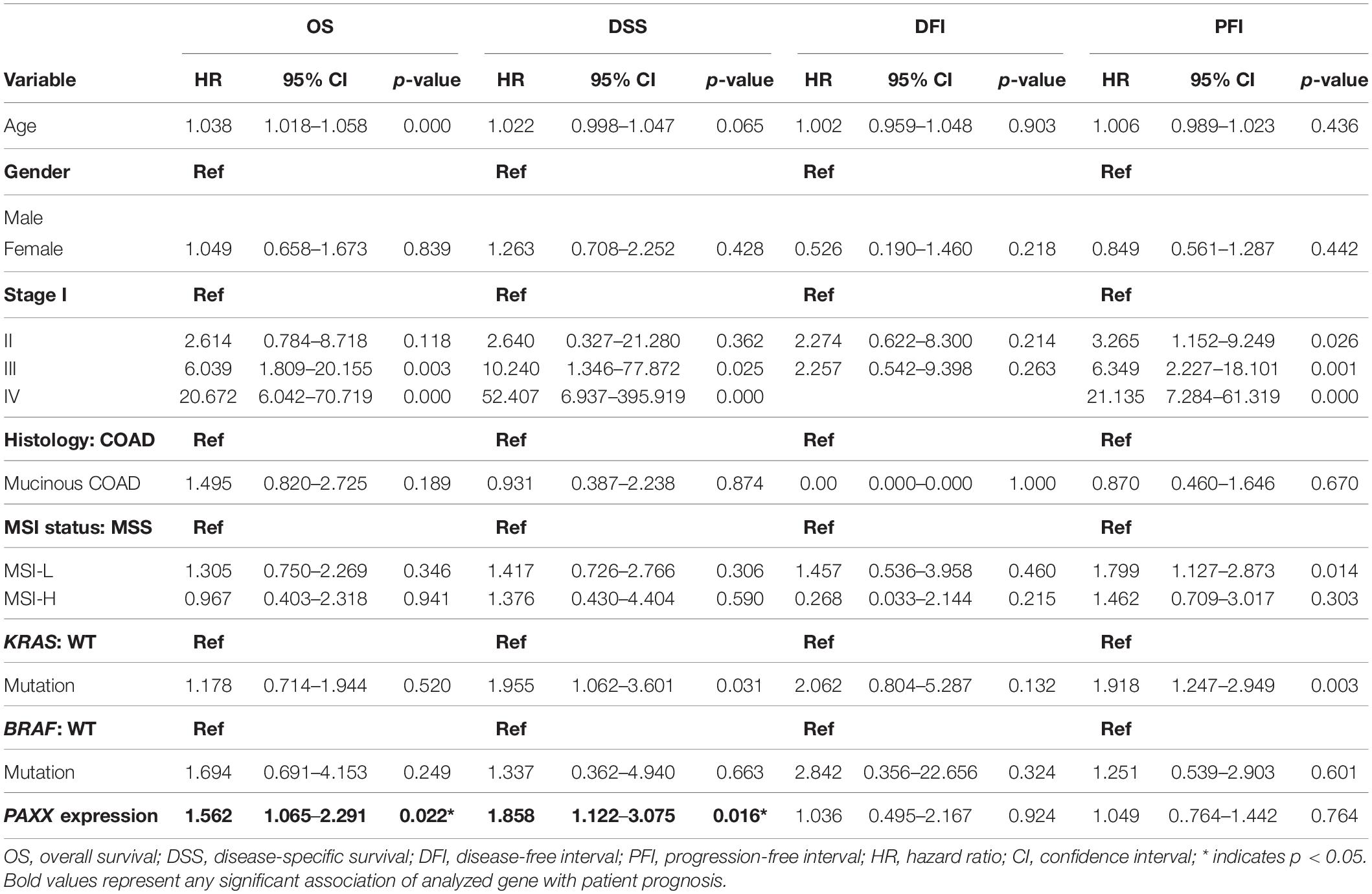
Table 3. Multivariate analysis of PAXX expression and clinicopathological parameters in TCGA-COAD dataset.
Methylation Analysis of the PAXX Gene
PAXX gene contains a CpG island spanning its transcription start site (TSS) and the first two exons (Figure 7A). Given this information, it was of interest to investigate the role of epigenetic modifications in the overexpression of PAXX in colon tumors. For this purpose, we assessed DNA methylation and paired RNA expression data of TCGA-COAD through the MEXPRESS web server. Pearson correlation analysis between methylation of five CpG sites of PAXX promoter and transcription of its gene revealed that DNA methylation of two distinct sites captured by probes, cg01126560 and cg25499748 exhibited significant negative correlation to PAXX gene expression in TCGA-COAD dataset (r = −0.232, p < 0.001 and r = −0.338, p < 0.001, respectively, Figure 7A). Further, the level of methylation of cg01126560 was lower in a group of all available colon cancer tissues compared to normal tissues (p < 0.0001, Figure 7B). Furthermore, a comparison of paired colon cancer tissues with respective normal tissues also revealed that colon cancer tissues exhibit lower methylation of cg01126560 (p < 0.0001, Figure 7C). These results suggested the involvement of methylation in transcriptional regulation of PAXX expression in colon carcinoma.
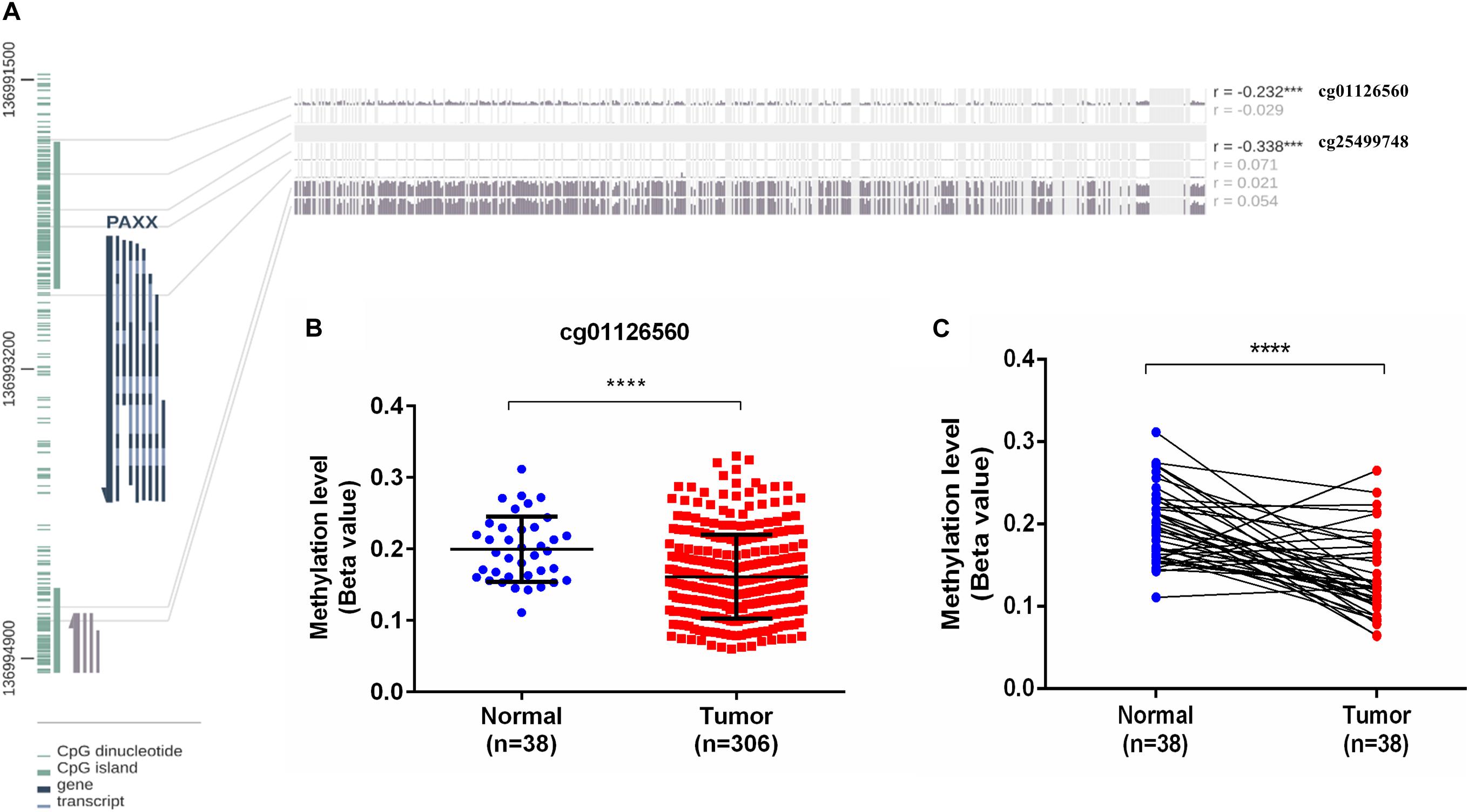
Figure 7. DNA methylation analysis of the PAXX gene from TCGA-COAD dataset. (A) Graphical representation of CpG Island and sites around PAXX genomic locus with Pearson’s correlation values of PAXX expression with methylation of different CpG sites. (B) Comparison of DNA methylation levels of cg01126560 between total normal and colon cancer tissues. (C) Comparison of DNA methylation levels of cg01126560 between paired normal colon and colon cancer tissues. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Cellular Pathways Associated With PAXX Expression in Colon Cancer
To assess the oncogenic pathways associated with PAXX expression in colon cancer, we performed gene set enrichment analysis (GSEA) for cancer hallmarks pathways using genes that exhibited significant correlations with PAXX. Among positively correlated pathways, PAXX expression exhibited the most significant correlation with oxidative phosphorylation (Figure 8A), besides other metabolic pathways including glycolysis (Figure 8F), fatty acid metabolism (Figure 8G), and adipogenesis (Figure 8H). We also observed a positive correlation of PAXX with DNA repair (Figure 8E), MYC targets (Figures 8B,C), E2F targets (Figure 8D), G2M checkpoint (Figure 8I), and reactive oxygen species (Figure 8J), pathways. Further, protein interaction data of PAXX protein in HCT-116 colon cancer cell line from “Bioplex 2.0” database also revealed interaction of PAXX with Werner syndrome ATP-dependent helicase (WRN), an established mediator of NHEJ pathway, supporting the involvement of PAXX in NHEJ pathway in colon cancer (Figure 8K). Interestingly, PAXX was also observed to interact with genes involved in glutathione metabolism, including glutathione peroxidase 1 and 7 (GPX1 and GPX7, respectively), which are primarily involved in protecting cells from oxidative stress, suggesting additional pro-tumor roles of PAXX in conferring therapeutic resistance to colon cancer cells.
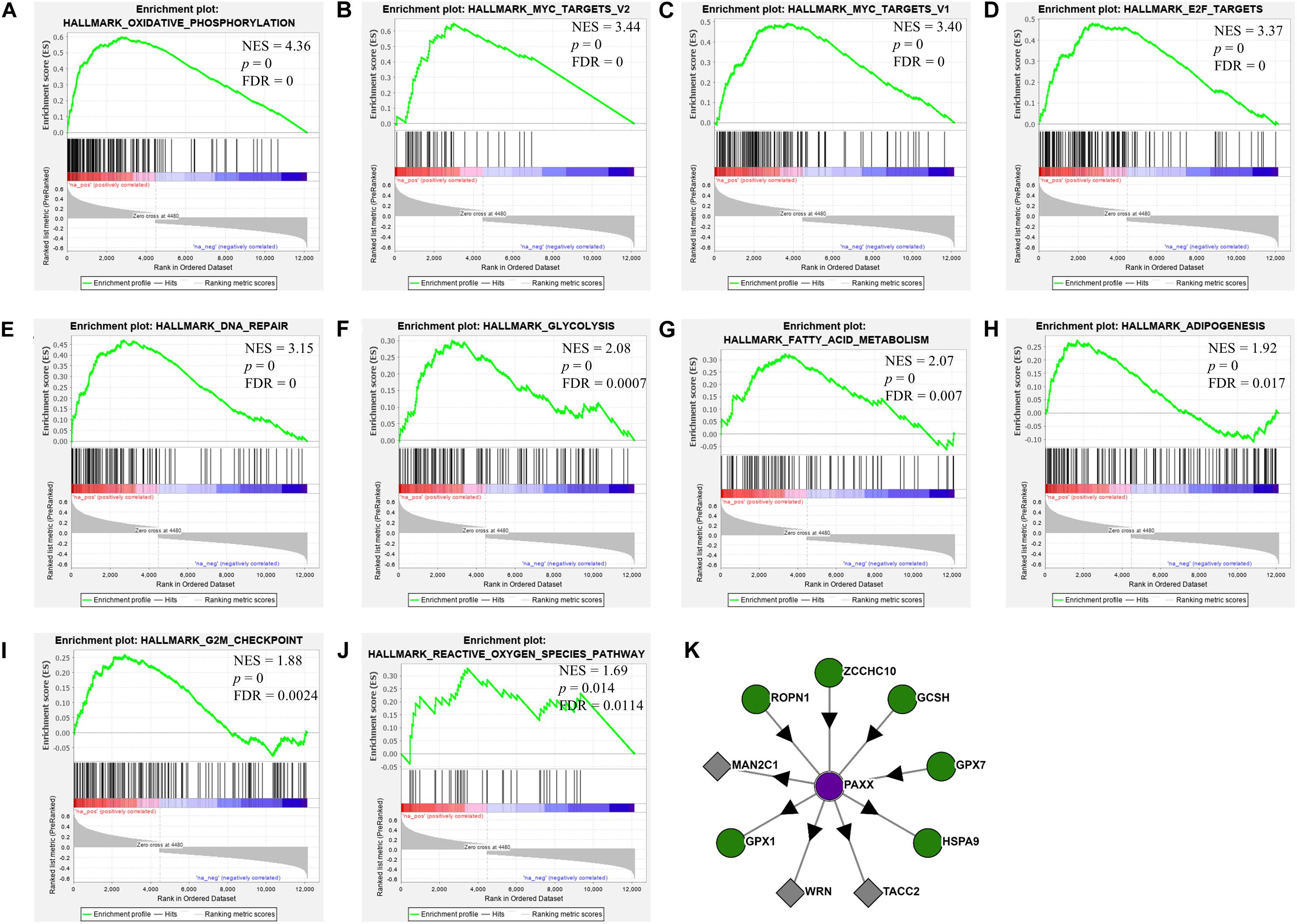
Figure 8. Gene set enrichment analysis of PAXX correlated genes in the TCGA-COAD dataset. Each plot (A–J) depicts positively enriched pathways of PAXX correlated genes with normalized enrichment score (NES), false discovery rate (FDR), and p-value depicted inside the respective pathway. (K) Depict results from the Bioplex 2.0 web server, showing protein-protein interactions of PAXX in HCT-116 colon cancer cell line.
We observed a significant negative correlation between PAXX expression and epithelial to mesenchymal transition pathway (Supplementary Figure S8A). Other pathways that exhibited a negative correlation with PAXX included downregulated genes in UV response, KRAS signaling, Hedgehog signaling, and angiogenesis (Supplementary Figures S8B–E). Interestingly, we also observed a negative correlation of PAXX with immunity associated pathways including inflammatory response, TGF beta signaling, and complement pathway (Supplementary Figures S8F–H). We further correlated PAXX expression with the computationally determined abundance of different tumor-infiltrating immune cells in TCGA-COAD dataset. PAXX was observed to be negatively correlated with twenty different immune cells, thereby suggesting the association of PAXX expression with overall reduced tumor immune infiltration in colon cancer (Supplementary Figure S9).
Discussion
Aberrations in the NHEJ pathway are common in cancers. Hosoi et al. reported elevated expression of Ku70 and Ku80 mRNA as well as proteins in colorectal carcinoma compared to the normal colon (Hosoi et al., 2004). In contrast, Beggs et al. reported reduced expression of Ku70 in colon cancer cells, which was associated with higher genomic instability (Beggs et al., 2012). In another study, it was observed that cytoplasmic Ku70 protein levels are higher in patients who do not respond to chemoradiotherapy, while Ku80 was lost in those patients (Pucci et al., 2017). Thus, previous studies have described both overexpression and downregulation of NHEJ pathway genes in colorectal cancer. Also, some of these studies have estimated mRNA levels while others have assessed protein expression. To resolve this paradox, we performed a comprehensive analysis of the core NHEJ pathway genes in colon cancer. Our analysis revealed elevated mRNA and protein expression of XRCC6 (Ku70) and XRCC5 (Ku80) in colon cancer compared to normal colon tissue. Furthermore, the overexpression pattern is more robust for XRCC5 as observed in paired normal and tumor tissue comparison, while XRCC6 did not exhibit significant difference. Indeed, we observed reduced XRCC6 expression in tumors at an advanced stage (stage III + IV) compared to the lower stage (stage I + II).
PRKDC exhibited overexpression in Oncomine analysis, TCGA dataset as well as CPTAC study suggesting consistent overexpression of this protein in colon cancer, both at the mRNA and protein levels. PRKDC expression was not associated with age, gender, stage, and histology. A previous study had also reported higher mRNA and protein levels of PRKDC in colorectal cancer tissues compared to normal tissues, which also exhibited a positive correlation with expression of XRCC6 and XRCC5 (Hosoi et al., 2004). In our analysis these three proteins exhibited a significant positive correlation with each other. Further, a recent report highlighted the dependency of colorectal cancer cells on PRKDC and also showed that PRKDC overexpression in colon cancer is associated with poor OS (Sun et al., 2016). While we observed a similar pattern of overexpression of PRKDC in colon cancer, its mRNA expression was not associated with any of the four types of survival parameters analyzed. Therefore, the collective data along with our results validate PRKDC overexpression as a potential therapeutic target in colon cancer.
Gene polymorphism in XRCC4 has been associated with CRC risk (Bau et al., 2010; Zhang and Hu, 2011). Our analysis revealed elevated levels of XRCC4 mRNA in Oncomine and protein data analysis, respectively, whereas the comparison of expression between paired normal and tumor tissues in TCGA dataset did not exhibit a significant difference in XRCC4 expression. A previous detailed report suggests that LIG4 protein levels are upregulated in colon cancer tissues and mediate Wnt/beta-catenin signaling induced radioresistance (Jun et al., 2016). In another study, quantitative RT–PCR in 61 paired normal colon and 393 CRCs demonstrated LIG4 downregulation in colon cancer tissues, which was further associated with its promoter hypermethylation (Kuhmann et al., 2014). While our results also suggest consistent downregulation of LIG4 mRNA expression in tumor cells, proteomic analysis displayed higher LIG4 levels in colon tumors. Furthermore, we observed higher expression of LIG4 in advanced stage tumors and male patients. Association of LIG4 mRNA expression with its protein levels and gender has not been reported and requires further exploration.
XLF (NHEJ1) was recently shown to enhance resistance to oxaliplatin and 5-fluorouracil in colorectal cancer cell lines (Liu et al., 2019). Association of higher XLF expression with drug resistance in hepatocellular carcinoma has also been reported (Yang and Wang, 2017). Contrary to these observations, we found consistent downregulation of XLF in colon cancer in our analysis, both at mRNA and the protein levels. Furthermore, its reduced mRNA expression was associated with the mucinous subtype, while no association was observed with the tumor stage. These results signify that although XLF is capable to induce drug resistance in CRC cells, its expression is nevertheless, downregulated in colon cancer. Intriguingly, we observed that NHEJ1 expression is negatively correlated with PAXX expression, and PAXX was observed to be consistently overexpressed in colon tumors compared to the normal tissues, both at the mRNA and protein levels. Interestingly, PAXX and XLF are functionally redundant (Kumar et al., 2016; Tadi et al., 2016), and also exhibit synthetic lethality (Liu et al., 2017). These results suggest that PAXX may preferentially function over XLF in DSB repair in colon cancer, which has been graphically represented in Figure 9.
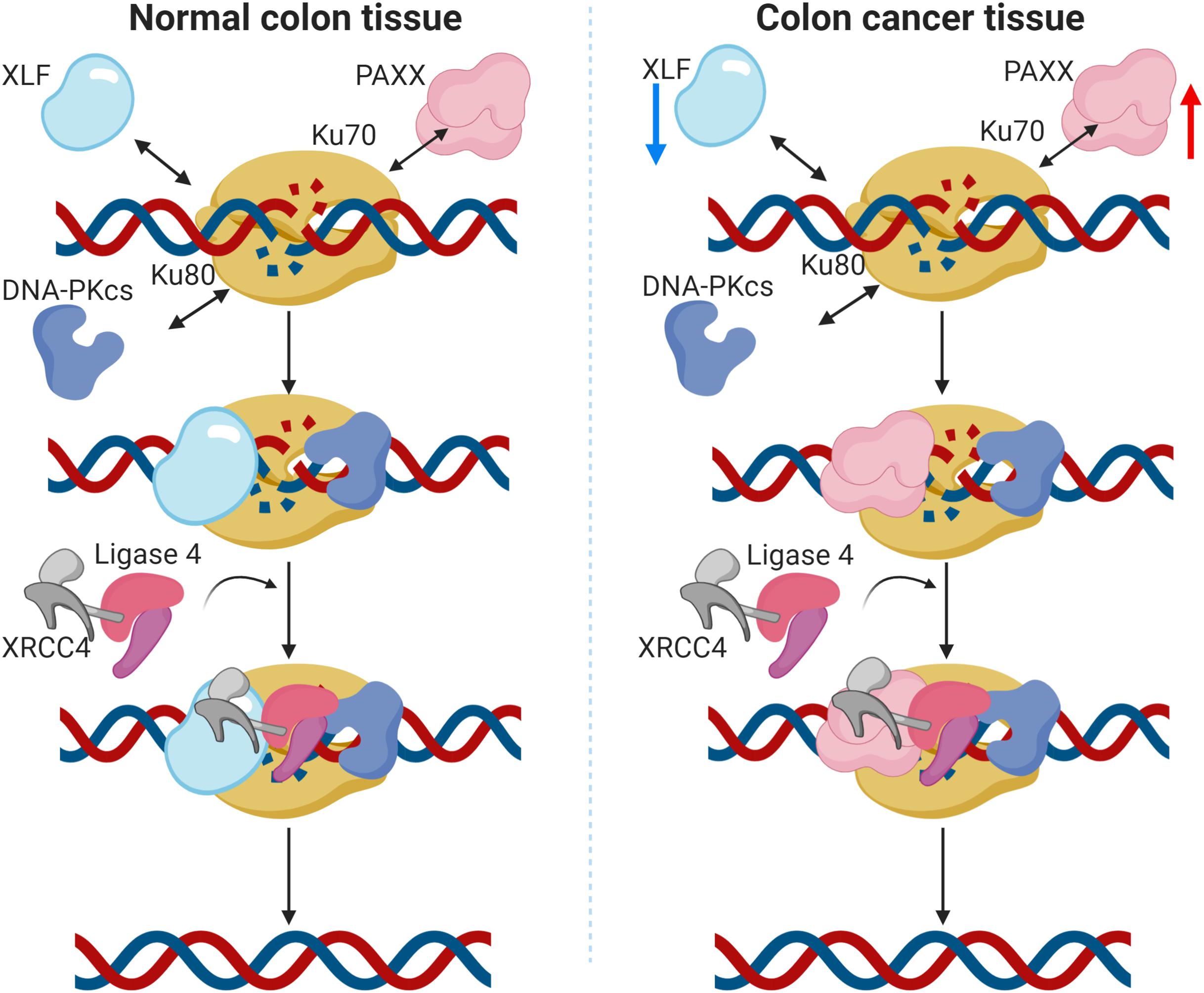
Figure 9. Graphical representation of the proposed function of PAXX in the NHEJ pathway in colon cancer. In normal colon tissues (left panel), DNA double strand breaks (DSBs) are actively identified by Ku70 and Ku80, followed by the recruitment of DNA-PKcs. The Ku/DNA-PKcs complex phosphorylates and recruits other accessory factors for DNA end processing. XRCC4 and XLF also bind to the DSB site and recruit DNA Ligase IV, which eventually seals the DSBs. PAXX has been demonstrated to work in the absence of XLF, as a backup in c-NHEJ repair (Tadi et al., 2016). In the case of colon cancer (right panel), protein levels of these proteins are altered. Notably, XLF is downregulated and PAXX is upregulated, suggesting PAXX may preferentially take over the XLF functions in colon cancer cells.
Our survival analysis revealed that among the NHEJ pathway genes analyzed in the current study, only PAXX emerged as an independent prognostic biomarker, while other NHEJ1 genes did not display any prognostic significance. In concordance to PAXX overexpression observed in colon tumors, higher expression of PAXX was associated with poor OS and DSS. Further, the expression and prognostic value of PAXX did not display any association with the stage and MSI status. DNA methylation analysis revealed a negative correlation of PAXX expression with its promoter methylation and the extent of methylation in this gene was found to be lower in tumors compared to the normal colon. We conclude from these results that PAXX expression in colon cancer is at least partly under epigenetic control.
As our results suggest the utility of PAXX as a potential therapeutic target in colon cancer, we performed gene set enrichment analysis to further determine the association of PAXX expression with underlying oncogenic pathways in colon cancer. In agreement with its established role in DNA repair, PAXX associated genes were highly enriched in DNA repair and cell cycle related processes. Recently, Yang et al., reported that PAXX also plays an important role in the base excision repair pathway and PAXX deficient cells display higher sensitivity to temozolomide in glioma cells (Yang et al., 2018). These results collectively suggest that PAXX may play important roles in different DNA repair pathways as well and PAXX may serve as a novel therapeutic target for DNA repair in cancer cells. Much before the detailed functions of PAXX were determined, Meyer et al. reported the association of PAXX overexpression with rapid leukemia establishment in a mouse model of human acute lymphocytic leukemia xenograft, and shorter time to relapse in the corresponding patients (Meyer et al., 2011). Other pathways associated with higher PAXX expression, were related to cell metabolism, including higher oxidative phosphorylation and glycolytic pathway while UV response, KRAS signaling, and angiogenesis pathways were associated with lower PAXX expression. While pathway analysis in the present study revealed close associations of PAXX expression with several other oncogenic pathways as well, it requires further exploration to provide causal relationships between PAXX expression and alterations of these pathways. Nevertheless, we observed that several immune system associated pathways including inflammatory response, TGF beta signaling, and complement pathway were negatively associated with PAXX expression. Furthermore, PAXX expression exhibited a negative association with the abundance of immune cells in the colon tumor microenvironment, which suggests its association with reduced overall infiltration of immune cells in colon cancer. Interestingly, reduced tumor inflammatory infiltrate is generally associated with poor prognosis in colorectal cancers (Mei et al., 2014). Thus our study provides novel insights into NHEJ pathway status in colon cancer and suggests the potential utility of PAXX as a novel prognostic marker and a therapeutic target in colon cancer.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Ethics Statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements.
Author Contributions
MA conceptualized the study. SC supervised the study and provided infrastructure to carry out this work. MA, SK, JS, and AC performed the data curation, interpretation, and statistical analysis. MA and SK wrote the original manuscript. AC and SC reviewed and edited the manuscript. All the authors approved the final manuscript.
Funding
This work is supported by the DBT/Wellcome Trust India Alliance Fellowship (grant number: IA/CPHI/17/1/503333) awarded to AC. SC acknowledges financial support from Indian Council of Medical Research (ICMR, India), (grant number: 2019-2914).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
MA acknowledges financial support as a fellowship from Council of Scientific and Industrial Research, Government of India. SK acknowledges financial support as fellowship from Department of Health Research, Government of India. JS acknowledges financial support as fellowship from Department of Biotechnology, Government of India.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2020.584053/full#supplementary-material
Footnotes
- ^ https://www.oncomine.org/resource/login.html
- ^ https://xena.ucsc.edu/
- ^ https://www.cbioportal.org/
- ^ http://ualcan.path.uab.edu/
- ^ https://mexpress.be/
- ^ https://www.gsea-msigdb.org/
- ^ https://bioplex.hms.harvard.edu
- ^ http://cis.hku.hk/software.html
References
Arnold, M., Sierra, M. S., Laversanne, M., Soerjomataram, I., Jemal, A., and Bray, F. (2017). Global patterns and trends in colorectal cancer incidence and mortality. Gut 66, 683–691. doi: 10.1136/gutjnl-2015-310912
Bau, D.-T., Yang, M.-D., Tsou, Y.-A., Lin, S.-S., Wu, C.-N., Hsieh, H.-H., et al. (2010). Colorectal cancer and genetic polymorphism of dna double-strand break repair gene XRCC4 in Taiwan. Anticancer. Res. 30, 2727–2730.
Beggs, A. D., Domingo, E., McGregor, M., Presz, M., Johnstone, E., Midgley, R., et al. (2012). Loss of expression of the double strand break repair protein ATM is associated with worse prognosis in colorectal cancer and loss of Ku70 expression is associated with CIN. Oncotarget 3, 1348–1355. doi: 10.18632/oncotarget.694
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. doi: 10.1158/2159-8290.CD-12-0095
Chang, H. H. Y., Pannunzio, N. R., Adachi, N., and Lieber, M. R. (2017). Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506. doi: 10.1038/nrm.2017.48
Chen, F., Chandrashekar, D. S., Varambally, S., and Creighton, C. J. (2019). Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 10, 1–15. doi: 10.1038/s41467-019-13528-0
Craxton, A., Munnur, D., Jukes-Jones, R., Skalka, G., Langlais, C., Cain, K., et al. (2018). PAXX and its paralogs synergistically direct DNA polymerase λ activity in DNA repair. Nat. Commun. 9:3877. doi: 10.1038/s41467-018-06127-y
Davis, A. J., Chen, B. P. C., and Chen, D. J. (2014). DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair. 17, 21–29. doi: 10.1016/j.dnarep.2014.02.020
Douglas, P., Ye, R., Trinkle-Mulcahy, L., Neal, J. A., De Wever, V., Morrice, N. A., et al. (2014). Polo-like kinase 1 (PLK1) and protein phosphatase 6 (PP6) regulate DNA-dependent protein kinase catalytic subunit (DNA-PKcs) phosphorylation in mitosis. Biosci. Rep. 34:e00113. doi: 10.1042/BSR20140051
Fatkhutdinov, N., Sproesser, K., Krepler, C., Liu, Q., Brafford, P. A., Herlyn, M., et al. (2016). Targeting RRM2 and Mutant BRAF is a novel combinatorial strategy for melanoma. Mol. Cancer Res. 14, 767–775. doi: 10.1158/1541-7786.MCR-16-0099
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:pl1. doi: 10.1126/scisignal.2004088
Hosoi, Y., Watanabe, T., Nakagawa, K., Matsumoto, Y., Enomoto, A., Morita, A., et al. (2004). Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int. J. Oncol. 25, 461–468.
Huttlin, E. L., Bruckner, R. J., Paulo, J. A., Cannon, J. R., Ting, L., Baltier, K., et al. (2017). Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509. doi: 10.1038/nature22366
Jun, S., Jung, Y.-S., Suh, H. N., Wang, W., Kim, M. J., Oh, Y. S., et al. (2016). LIG4 mediates Wnt signalling-induced radioresistance. Nat. Commun. 7, 1–13. doi: 10.1038/ncomms10994
Kalimutho, M., Bain, A. L., Mukherjee, B., Nag, P., Nanayakkara, D. M., Harten, S. K., et al. (2017). Enhanced dependency of KRAS-mutant colorectal cancer cells on RAD51-dependent homologous recombination repair identified from genetic interactions in Saccharomyces cerevisiae. Mol. Oncol. 11, 470–490. doi: 10.1002/1878-0261.12040
Koch, A., De Meyer, T., Jeschke, J., and Van Criekinge, W. (2015). MEXPRESS: visualizing expression, DNA methylation and clinical TCGA data. BMC Genomics 16:636. doi: 10.1186/s12864-015-1847-z
Koh, K. H., Kang, H. J., Li, L. S., Kim, N.-G., You, K. T., Yang, E., et al. (2005). Impaired nonhomologous end-joining in mismatch repair-deficient colon carcinomas. Lab. Invest. 85, 1130–1138. doi: 10.1038/labinvest.3700315
Kuhmann, C., Li, C., Kloor, M., Salou, M., Weigel, C., Schmidt, C. R., et al. (2014). Altered regulation of DNA ligase IV activity by aberrant promoter DNA methylation and gene amplification in colorectal cancer. Hum. Mol. Genet. 23, 2043–2054. doi: 10.1093/hmg/ddt599
Kumar, V., Alt, F. W., and Frock, R. L. (2016). PAXX and XLF DNA repair factors are functionally redundant in joining DNA breaks in a G1-arrested progenitor B-cell line. PNAS 113, 10619–10624. doi: 10.1073/pnas.1611882113
Liberzon, A., Birger, C., Thorvaldsdóttir, H., Ghandi, M., Mesirov, J. P., and Tamayo, P. (2015). The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425. doi: 10.1016/j.cels.2015.12.004
Liu, X., Shao, Z., Jiang, W., Lee, B. J., and Zha, S. (2017). PAXX promotes KU accumulation at DNA breaks and is essential for end-joining in XLF-deficient mice. Nat. Commun. 8:13816. doi: 10.1038/ncomms13816
Liu, Z., Yu, M., Fei, B., Sun, J., and Wang, D. (2019). Nonhomologous end joining key factor XLF enhances both 5-florouracil and oxaliplatin resistance in colorectal cancer. Onco Targets Ther. 12, 2095–2104. doi: 10.2147/OTT.S192923
Ma, W., Yang, L., Liu, H., Chen, P., Ren, H., and Ren, P. (2020). PAXX is a novel target to overcome resistance to doxorubicin and cisplatin in osteosarcoma. Biochem. Biophys. Res. Commun. 521, 204–211. doi: 10.1016/j.bbrc.2019.10.108
Mei, Z., Liu, Y., Liu, C., Cui, A., Liang, Z., Wang, G., et al. (2014). Tumour-infiltrating inflammation and prognosis in colorectal cancer: systematic review and meta-analysis. Br. J. Cancer 110, 1595–1605. doi: 10.1038/bjc.2014.46
Meyer, L. H., Eckhoff, S. M., Queudeville, M., Kraus, J. M., Giordan, M., Stursberg, J., et al. (2011). Early relapse in ALL is identified by time to leukemia in NOD/SCID mice and is characterized by a gene signature involving survival pathways. Cancer Cell 19, 206–217. doi: 10.1016/j.ccr.2010.11.014
Mijnes, J., Veeck, J., Gaisa, N. T., Burghardt, E., de Ruijter, T. C., Gostek, S., et al. (2018). Promoter methylation of DNA damage repair (DDR) genes in human tumor entities: RBBP8/CtIP is almost exclusively methylated in bladder cancer. Clin. Epigenet. 10:15. doi: 10.1186/s13148-018-0447-6
Naccarati, A., Rosa, F., Vymetalkova, V., Barone, E., Jiraskova, K., Di Gaetano, C., et al. (2015). Double-strand break repair and colorectal cancer: gene variants within 3′ UTRs and microRNAs binding as modulators of cancer risk and clinical outcome. Oncotarget 7, 23156–23169. doi: 10.18632/oncotarget.6804
Neal, J. A., Dang, V., Douglas, P., Wold, M. S., Lees-Miller, S. P., Meek, K., et al. (2011). Inhibition of homologous recombination by DNA-dependent protein kinase requires kinase activity, is titratable, and is modulated by autophosphorylation. Mol. Cell. Biol. 31, 1719–1733. doi: 10.1128/MCB.01298-10
Ochi, T., Blackford, A. N., Coates, J., Jhujh, S., Mehmood, S., Tamura, N., et al. (2015). PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 347, 185–188. doi: 10.1126/science.1261971
Pucci, S., Polidoro, C., Joubert, A., Mastrangeli, F., Tolu, B., Benassi, M., et al. (2017). Ku70, Ku80, and sClusterin: a cluster of predicting factors for response to neoadjuvant chemoradiation therapy in patients with locally advanced rectal cancer. Int. J. Radiat. Oncol. Biol. Phys. 97, 381–388. doi: 10.1016/j.ijrobp.2016.10.018
Rawla, P., Sunkara, T., and Barsouk, A. (2019). Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 14, 89–103. doi: 10.5114/pg.2018.81072
Robb, R., Yang, L., Shen, C., Saji, M., Ringel, M., and Williams, T. M. (2018). BRAF oncogenic activation induces radioresistance through non-homologous end-joining repair which is abrogated by targeted inhibition of braf mutational activity. Int. J. Radiat. Oncol. Biol. Phys. 102:S117. doi: 10.1016/j.ijrobp.2018.06.293
Robb, R., Yang, L., Shen, C., Wolfe, A., Webb, A., Zhang, X., et al. (2019). Inhibiting BRAF oncogene-mediated radioresistance effectively radiosensitizes BRAFV600E mutant thyroid cancer cells by constraining DNA double-strand break repair. Clin. Cancer Res. 25:clincanres.3625.2018. doi: 10.1158/1078-0432.CCR-18-3625
Ru, B., Wong, C. N., Tong, Y., Zhong, J. Y., Zhong, S. S. W., Wu, W. C., et al. (2019). TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 35, 4200–4202. doi: 10.1093/bioinformatics/btz210
Sishc, B. J., and Davis, A. J. (2017). The role of the core non-homologous end joining factors in carcinogenesis and cancer. Cancers 9:81. doi: 10.3390/cancers9070081
Sun, S., Cheng, S., Zhu, Y., Zhang, P., Liu, N., Xu, T., et al. (2016). Identification of PRKDC (Protein Kinase, DNA-Activated, Catalytic Polypeptide) as an essential gene for colorectal cancer (CRCs) cells. Gene 584, 90–96. doi: 10.1016/j.gene.2016.03.020
Tadi, S. K., Tellier-Lebègue, C., Nemoz, C., Drevet, P., Audebert, S., Roy, S., et al. (2016). PAXX Is an Accessory c-NHEJ Factor that Associates with Ku70 and Has Overlapping Functions with XLF. Cell Rep. 17, 541–555. doi: 10.1016/j.celrep.2016.09.026
The Cancer Genome Atlas Network (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. doi: 10.1038/nature11252
Yang, B., Fu, X., Hao, J., Sun, J., Li, Z., Li, H., et al. (2018). PAXX participates in base excision repair via interacting with Pol β and Contributes to TMZ resistance in glioma cells. J. Mol. Neurosci. 66, 214–221. doi: 10.1007/s12031-018-1157-4
Yang, S., and Wang, X. Q. (2017). XLF-mediated NHEJ activity in hepatocellular carcinoma therapy resistance. BMC Cancer 17:344. doi: 10.1186/s12885-017-3345-y
Keywords: PAXX, NHEJ1 gene, colon cancer, The Cancer Genome Atlas, DNA methylation, DNA Repair
Citation: Arora M, Kumari S, Singh J, Chopra A and Chauhan SS (2020) PAXX, Not NHEJ1 Is an Independent Prognosticator in Colon Cancer. Front. Mol. Biosci. 7:584053. doi: 10.3389/fmolb.2020.584053
Received: 16 July 2020; Accepted: 09 September 2020;
Published: 23 October 2020.
Edited by:
Amila Suraweera, Queensland University of Technology, AustraliaReviewed by:
Nikolay Mikhaylovich Borisov, Moscow Institute of Physics and Technology, RussiaChinnadurai Mani, Texas Tech University Health Sciences Center, United States
Copyright © 2020 Arora, Kumari, Singh, Chopra and Chauhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shyam S. Chauhan, c19zX2NoYXVoYW5AaG90bWFpbC5jb20=; Anita Chopra, Y2hvcHJhYW5pdGEyMDA1QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Mohit Arora
Mohit Arora Sarita Kumari
Sarita Kumari Jay Singh
Jay Singh Anita Chopra
Anita Chopra Shyam S. Chauhan
Shyam S. Chauhan